- 1Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 2Center for Bioenergy Innovation, Oak Ridge National Laboratory, Oak Ridge, TN, United States
- 3State Key Laboratory of Subtropical Silviculture, School of Forestry and Biotechnology, Zhejiang A&F University, Hangzhou, China
- 4Biology Department, Brookhaven National Laboratory, Upton, NY, United States
Within the context of global warming, long-living plants such as perennial woody species endure adverse conditions. Among all of the abiotic stresses, drought stress is one of the most detrimental stresses that inhibit plant growth and productivity. Plants have evolved multiple mechanisms to respond to drought stress, among which transcriptional regulation is one of the key mechanisms. In this review, we summarize recent progress on the regulation of drought response by transcription factor (TF) families, which include abscisic acid (ABA)-dependent ABA-responsive element/ABRE-binding factors (ABRE/ABF), WRKY, and Nuclear Factor Y families, as well as ABA-independent AP2/ERF and NAC families, in the model plant Arabidopsis. We also review what is known in woody species, particularly Populus, due to its importance and relevance in economic and ecological processes. We discuss opportunities for a deeper understanding of drought response in woody plants with the development of high-throughput omics analyses and advanced genome editing techniques.
Introduction
Plant growth and development are affected by environmental stimuli including biotic and abiotic stresses. Drought stress is one of the most deleterious abiotic stresses to plants caused by limited rainfall, rising temperatures, and insufficient water availability and has become an increasing concern because of global climate change (Yamaguchi-Shinozaki and Shinozaki, 2006; Nakashima et al., 2014; Gupta et al., 2020a). Drought stress causes a series of injuries in terms of plant physiological, biochemical, and metabolic impacts, which result in plant growth retardation, cell damage, and leads to loss of crop yield and quality (Joshi et al., 2016). The sessile nature of plants requires them to develop effective responsive mechanisms, including escape (i.e., accelerating flowering), avoidance (i.e., maintaining high internal water content), and tolerance (i.e., maintaining growth under low internal water content) to adapt to drought stress (Salehi-Lisar and Bakhshayeshan-Agdam, 2016; Gupta et al., 2020a).
In response to drought stress, stomatal closure is an early and rapid response to avoid water loss, although it also can lead to negative effects on photosynthesis (Raghavendra et al., 2010). Morphological changes, including wax biosynthesis on leaf surface and enhanced root growth are other strategies adopted by plants to reduce respiration and improve water uptake, respectively, (Aharoni et al., 2004; Meng et al., 2019). Secondly, plants produce functional proteins, such as dehydrins and late embryogenesis-abundant proteins (LEA), as well as wide spectrum of metabolites to relieve potential osmotic damage from drought stress. Thirdly, plants produce antioxidant enzymes to scavenge reactive oxygen species (ROS) and protect cells from oxidative damage induced by drought stress (Claeys and Inze, 2013). Noticeably, stomatal closure is through a complex membrane transporter system to quickly respond to drought stress (Raghavendra et al., 2010; Gong et al., 2020). Most of the other responses are typically under the control of transcriptional regulation in which transcription factors (TFs) play the pivotal roles (Takahashi et al., 2018).
Much of the progress of drought response studies has been made in the model plants Arabidopsis thaliana and rice (Oryza sativa) over the past decades (Shinozaki and Yamaguchi-Shinozaki, 2006; Joshi et al., 2016). As a stress hormone, abscisic acid (ABA) is abruptly synthesized in response to different stresses, including drought, enhancing plant drought tolerance through closing stomata and restraining plant growth (Zeevaart, 1980; Zhu, 2016). ABA is perceived by the PYR/PYL/RCAR receptors, which activate downstream TFs, such as ABA-responsive element (ABRE)-binding proteins (AREBs)/ABRE-binding factors (ABFs) by cascade kinase reactions. Activated ABA signaling enhances drought tolerance by inducing stress-responsive genes. However, expression of some drought-responsive genes is independent of ABA. For example, RD29A and ERD1 can be induced by drought in an ABA-independent pathway. Their expression is controlled by other TFs families, such as APETALA2/ethylene-responsive factor (AP2/ERF), and NAM, ATAF1/2, and CUC2 (NAC) (Takahashi et al., 2018).
Similar to Arabidopsis and rice, woody plant species are also negatively affected by drought, including reduced plant growth, inhibited wood formation, and increased susceptibility to pathogens (Fernàndez-Martínez et al., 2013; Yin et al., 2014; Polle et al., 2019). With the approaches of genetics and omics, studies on response of woody plants have identified many important TFs, and the transcriptional regulation of drought response is emerging (Estravis-Barcala et al., 2019). In this review, we will provide an update on the recent progress of the role of TFs in drought response in the model plant, Arabidopsis, and highlight findings in the woody genus, Populus.
ABA-Dependent Drought Response Pathway
The ABA signaling pathway consists of receptor RCAR/PYR/PYLs, protein phosphatase PP2C, kinase SnRK2s (SnRK2.2, SnRK2.3, and SnRK2.6), and the targeting substrates (Umezawa et al., 2010; Guo et al., 2011). Once bound and activated by ABA, PYR/PYL/RCARs form a trimeric complex with PP2Cs, which inhibits the phosphatase activity of PP2Cs. SnRK2s are then released from the association with PP2Cs. Released SnRK2s can be activated by autophosphorylation, and, in turn, phosphorylate the downstream TFs and ion channel proteins (Fujita et al., 2009; Ma et al., 2009; Park et al., 2009; Umezawa et al., 2009). Among SnRK2 targets, AREBs/ABFs are the downstream TFs in the ABA signaling pathway. Some other TF families, such as WRKY, MYB, and NF-Ys, are also involved in drought response and adaption (Singh and Laxmi, 2015).
AREBs/ABFs
Gene expression microarray and RNA-seq approaches have been used extensively to identify drought-responsive genes. Through the analysis of cis-acting promoter elements, 8-bp-long ABRE, PyACGTGGC, was identified in the promoter in 82% of dehydration-responsive genes in Arabidopsis (Maruyama et al., 2012). ABRE are bound by AREBs/ABFs, which belong to the bZIP TF family. Four members of AREBs/ABFs (AREB1, AREB2, ABF3, and ABF1), whose expression are induced by both dehydration and ABA treatment, were reported to regulate drought response through the ABA-dependent pathway. The quadruple mutant of areb1 areb2 abf3 abf1 showed a drought-sensitive phenotype and reduced sensitivity to ABA (Yoshida et al., 2010, 2015). ENHANCED EM LEVEL (EFL), belonging to bZIP TF family, formed a protein complex with GIGANTEA (GI) to regulate diurnal ABA biosynthesis contributing to drought tolerance (Baek et al., 2020a). Noticeably, an ABRE is required to co-locate with other copies of ABRE or coupling elements (CEs) to activate ABA-responsive genes (Figure 1). Since ABA regulates most of its target genes through AREBs/ABFs, the ABRE element is recognized as a key signature for drought-responsive genes regulated by the ABA-dependent pathway.
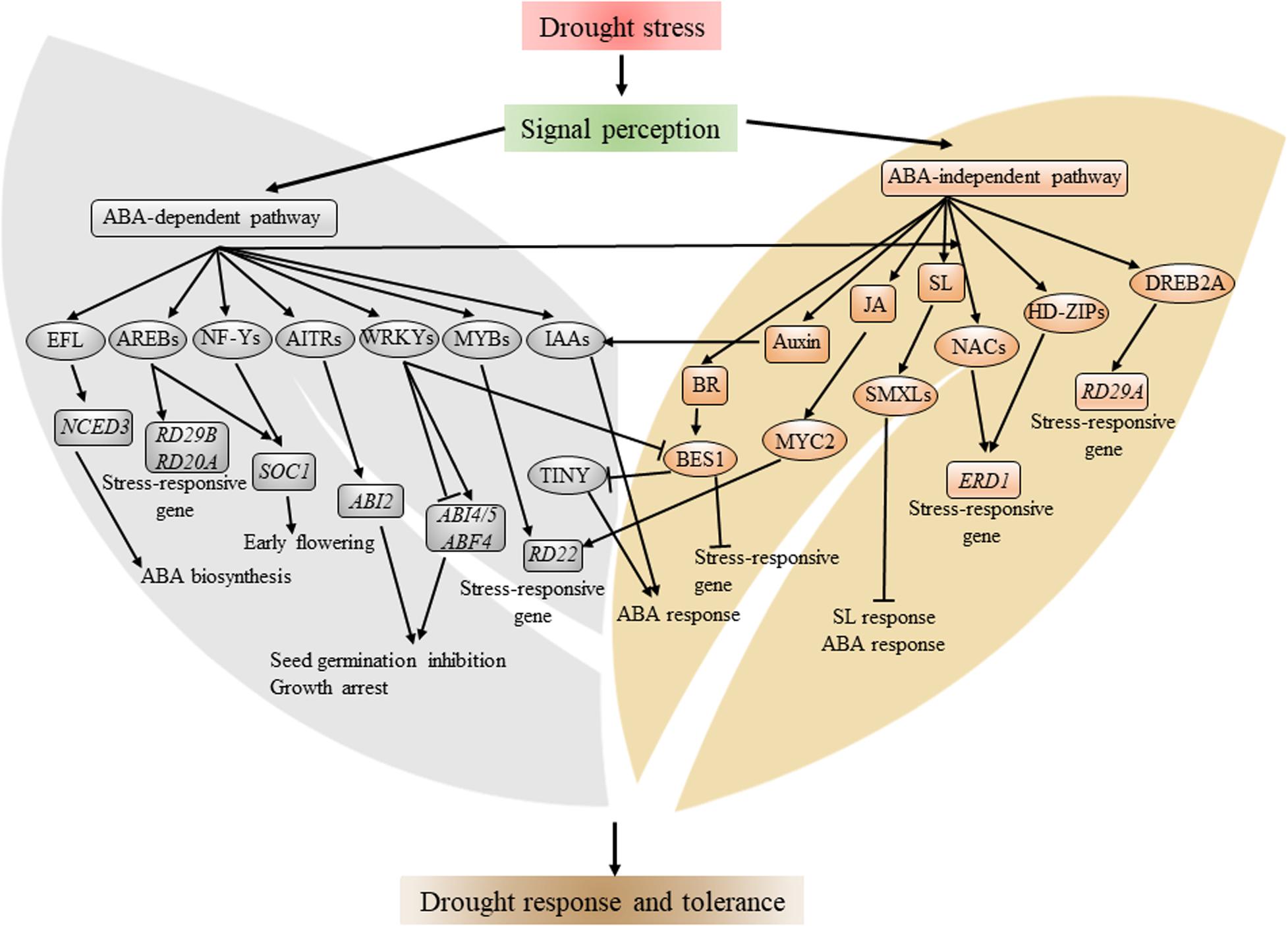
Figure 1. Schematic representation of major transcription factors and transcriptional network involved in drought stress response. Drought stress is perceived by plant in an unclear mechanism, which induces the biosynthesis of ABA. ABA signaling, in turn, activates several TF families, including AREB, NF-Y, WRKY, and MYB. There are ABA-independent TFs involved in drought stress, such as DREB2A, HD-ZIP, MYC2, and several NACs. These TFs, individually or cooperatively, regulate plant growth and development, metabolism, and expression of stress-inducible genes, such as RD22 and ERD1. Arrows and bars indicate activation and inhibition of the expression of downstream genes, respectively. Components in the gray background denote the ABA-dependent pathway, and components in brown background denote the ABA-independent pathway.
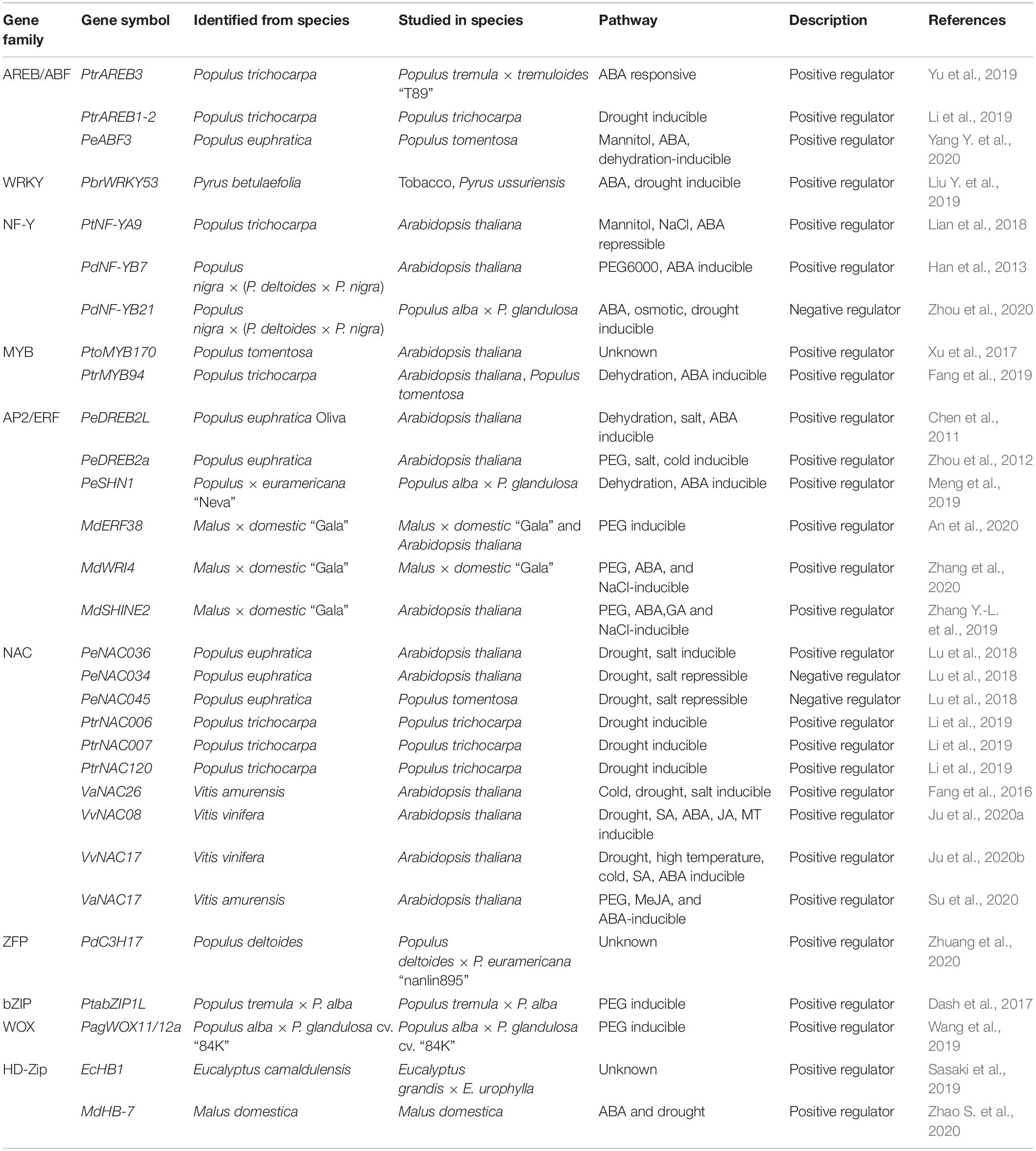
Recently, many ABA signaling components have been reported to be involved in drought response in woody plants. Overexpression of the ABA receptor, PtPYRL1 or PtPYRL5, increased drought tolerance (Yu et al., 2017). Accordingly, Arabidopsis transgenic plants overexpressing Populus PP2C genes negatively regulated drought tolerance and showed enhanced water loss (Arshad and Mattsson, 2014; Chen et al., 2015). There are 14 putative AREB/ABF members encoded in the Populus genome. Eight of them were upregulated upon exogenous ABA treatment, whereas the other six members were downregulated (Ji et al., 2013). Transgenic plants overexpressing PtrAREB3 showed a strong drought tolerance phenotype under drought conditions with compromised biomass production (Yu et al., 2019). PeABF3, isolated from Populus euphratica, was induced by dehydration and ABA treatment. Overexpression of PeABF3 enhance drought tolerance by promoting ABA-induced stomatal closure (Yang Y. et al., 2020). Li et al. (2019) identified four ABRE1 homologs in P. trichocarpa, three of which (PtrAREB1-2, PtrAREB1-3, and PtrAREB1-4) were induced by drought treatment. Knocking down of PtrAREB1 showed reduced drought tolerance in transgenic poplar (Table 1). PtrAREB1-2 was reported to recruit HAT complex proteins, ADA2b and GCN5, to increase H3K9 acetylation and activate the expression of downstream genes, such as PtrNACs (Li et al., 2019). Besides AREB/ABFs, another bZIP TF PtabZIP1L contributed to drought tolerance by increasing lateral root formation and modulating the biosynthesis of the drought tolerance-related metabolites (Dash et al., 2017). In conclusion, ABA biosynthesis and the ABA signaling pathway play pivotal roles in drought stress response in woody plants, such as poplar.
WRKYs
Some other TF families do not recognize the signature ABRE motif but play an important role in ABA-dependent stress response (Singh and Laxmi, 2015). As one of the largest plant TF families, the WRKY TF family features the conserved WRKY domain which shows potential DNA-binding activity. WRKYs recognize the W-box (TTGACC/T) that has been found in the promoters of many biotic and abiotic stress-responsive genes (Chen et al., 2019). WRKY18, WRKY40, and WRKY60 negatively regulate ABA-responsive genes (i.e., ABI4, ABI5, and ABF4) by directly binding to their promoters under normal conditions, and the wrky40 mutant showed ABA-hypersensitivity phenotypes. Expression of WRKY18, WRKY40, and WRKY60 are induced by water deficiency as well as ABA, and ABA also promotes the ABAR-WRKY40 interaction to relieve the negative effect of WRKY40 on ABI5 expression (Chen et al., 2010; Shang et al., 2010). WRKY63/ABO3 is induced by ABA treatment, and the abo3 mutant is more sensitive to drought stress (Ren et al., 2010). WRKY46, WRKY54, and WRKY70 have also been identified to be involved in BR-regulated drought response, and the wrky46 wrky54 wrky70 triple mutant shows more tolerance to drought stress (Chen et al., 2017). In addition to negatively regulating drought stress, WRKY40 and WRKY70 are known repressors in plant immunity, and it will be interesting to investigate their roles in the crosstalk of pathogen and drought stress, given that it has been reported that drought stress-induced ABA biosynthesis inhibits salicylic acid (SA)-mediated plant immunity (Chen et al., 2019; Gupta et al., 2020b). Other WRKY TFs, such as WRKY28 and WRKY21, were reported to positively and negatively regulate plant response to drought stress, respectively, (Babitha et al., 2013; Zhao K. X. et al., 2020; Figure 1).
Genome-wide identification of the WRKY TF family have been carried out in several woody plants, such as poplar and grapevine (He et al., 2012; Jiang et al., 2014; Wang et al., 2014). Over 100 putative PtrWRKY genes have been identified in Populus, and several WRKY members in group III showed enhanced expression in response to drought treatment. It would be interesting to further investigate the function of these drought-responsive PtrWRKYs, such as PtrWRKY89, whose expression is strongly induced by ABA and PEG treatment (He et al., 2012; Wang et al., 2015). Recently, a drought and ABA-induced PbrWRKY53 was isolated from Pyrus betulaefolia. Overexpression of PbrWRKY53 increased drought tolerance in tobacco and Pyrus ussuriensis, whereas the PbrWRKY53 knock-down lines showed compromised drought tolerance in P. ussuriensis with reduced PbrNCED1 and PbrNCED3 expression levels (Liu Y. et al., 2019; Table 1). The function of WRKY TFs in biotic stress response has been well-studied in woody plants (Duan et al., 2015; Jiang et al., 2017; Liu G. et al., 2019). However, their function in abiotic stress response, especially in drought response, needs to be deeply investigated.
NF-Ys
NF-Ys consist of three subunits (NF-YA, NF-YB, and NF-YC) and recognize the CCAAT box in the promoters of target genes. Arabidopsis AtNF-YA5 is induced by drought stress in an ABA-dependent manner, and the mutant nf-ya5 shows hypersensitivity to drought stress, whereas overexpression of NF-YA5 enhances drought tolerance (Li et al., 2008). Other members in NF-YA family, such as AtNF-YA3, AtNF-YA7, and AtNF-YA10 are positive regulators of drought response (Leyva-Gonzalez et al., 2012). AtNF-YB2, induced by dehydration stress in both ABA-dependent and ABA-independent manners, also positively regulates drought tolerance (Sato et al., 2019). It was also reported that AtNF-YC3/4/9 enhances drought-escape-responsive flowering by interacting with ABF3/4 and activates SOC1 expression under drought stress (Hwang et al., 2019; Figure 1).
There are 52 NF-Y genes, including 13 NF-YAs, 20 NF-YBs, and 19 NF-YCs identified in the P. trichocarpa genome. Among 13 PtNF-YA genes, 11 of them were found to contain the ABRE element in their promoter regions, which correlates with their expressions being induced by abiotic stresses. Noticeably, PtNF-YA2 and PtNF-YA4 were induced by polyethylene glycol-simulated drought treatment (Liu et al., 2020). Although the expression of PtNF-YA9 was not induced by dehydration, Arabidopsis plants overexpressing PtNF-YA9 showed high tolerance to drought stress by promoting ABA-dependent stomatal closure in leaves (Lian et al., 2018). Similarly, PdNF-YB7 identified from Populus nigra × (P. deltoides × P. nigra) was also reported to increase drought tolerance in Arabidopsis by regulating ABA-dependent dehydration response (Han et al., 2013). Overexpression of PdNF-YB21, another PdNF-YB, enhanced root growth and drought tolerance, while the knockout mutant showed the opposite phenotypes in poplar [Populus nigra × (P. deltoides × P. nigra)] (Zhou et al., 2020). Further investigation showed that PdNF-YB21 interacts with a B3 domain TF PdFUSCA3 and activates the expression of ABA biosynthesis genes (Suzuki and McCarty, 2008). Accumulation of ABA in the roots of PdNF-YB21 overexpression plants increased root growth and enhanced drought tolerance by promoting IAA transport (Zhou et al., 2020; Table 1). Collectively, these results indicate that NF-Y TFs have important functions in drought response in the ABA-dependent pathway in woody plants.
MYBs and Other TF Families
For some drought-responsive genes, such as RD22 and AtADH1, their expression is dependent on ABA. However, the ABRE cis-elements are not found in their promoters. Further studies identified two cis-elements, specifically MYBRS (C/TAACNA/G) and MYCRS (CANNTG), in their promoters. The MYB TF family member AtMYB2 and the bHLH TF AtMYC2 have been identified to recognize these elements, respectively, (Abe et al., 1997, 2003). AtAMYB96 is another MYB TF involved in drought response, and the myb96-1 mutant showed susceptibility to drought, whereas transgenic plants overexpressing AtMYB96 showed enhanced drought tolerance. In addition to the activation of target genes in response to ABA, AtMYB96 was reported to negatively regulate ABA-repressible genes RHO GTPASE OF PLANTS (ROPs) by recruiting HDA15 (Seo et al., 2009; Lee and Seo, 2019; Figure 1).
There are other TF families that regulate drought response via the ABA-dependent pathway. ABA-induced transcriptional repressors (AITRs) belong to a novel TF family, and the double mutant of aitr2 aitr5 and triple mutant of aitr2 aitr5 aitr6 show strong drought tolerance. Further analysis shows that these AITRs are able to target and repress key regulators in ABA signaling, such as ABI2 (Tian et al., 2017). Auxin-sensitive Aux/IAA transcriptional repressors are also reported to be involved in drought response. The triple mutant of iaa5 iaa6 iaa19 shows decreased drought tolerance. Further analysis indicated that IAA5/6/19, induced by drought and ABA, regulate stomatal closure by adjusting glucosinolate levels (Salehin et al., 2019).
As one of the largest TF families, there are over 190 MYB TFs identified in poplar (Wilkins et al., 2009). PtoMYB170 was identified from Populus tomentosa (Chinese white poplar), and overexpression of PtoMYB170 in Arabidopsis increased drought tolerance by promoting dark-induced stomatal closure (Xu et al., 2017). Overexpression of PtrMYB94 increased drought tolerance in both poplar and Arabidopsis. Further analysis revealed that PtrMYB94 enhanced drought tolerance in an ABA-dependent manner by activating ABA-responsive genes and increasing ABA content (Fang et al., 2019; Table 1). Compared with Arabidopsis, studies of MYB TFs in drought response in poplar are still limited.
ABA-Independent Drought Response Pathway
Although many TFs are induced by ABA and function in the ABA-dependent signaling pathway, there are some TFs whose expression is highly induced by water deficiency, but not primarily mediated by ABA biosynthesis or signaling pathway (Yoshida et al., 2014). DREB2s and some NACs belong to this category.
AP2/ERFs
Dehydration-responsive element DRE (A/GCCGAC) was identified in many drought-responsive genes, which is recognized by DREB2 proteins. DREB2A and DREB2B belong to the AP2/ERF (Apetala2 and ethylene-responsive factors) TF family (Maruyama et al., 2012). The expression of DREB2A is slightly induced by ABA, but highly induced by dehydration, supporting the ABA-independent manner in response to drought stress (Kim et al., 2011). Overexpression of the constitutively active DREB2A (DREB2A-CA) increases drought tolerance. The active form of DREB2A-CA lacks the PEST sequence that is a negative regulatory domain for DREB2A, and makes this protein unstable under non-stressful conditions (Sakuma et al., 2006). Further analysis revealed that protein stability of DREB2A is controlled by DRIP1 and DRIP2 through the ubiquitin/26S proteasome system under normal conditions (Qin et al., 2008). TINY belongs to AP2/ERF family in Arabidopsis. tiny tiny2 tiny3 triple mutant showed hypersensitive phenotypes to drought stress whereas transgenic plants overexpressing TINY enhanced drought tolerance. TINY positively regulates drought-responsive genes and promotes ABA-mediated stomatal closure by interacting with BES1 in BR signaling (Xie et al., 2019). Other ERF/AP2 TF family members such as HARDY (HRD), TG/RAP2.4A, and AtERF74 were also reported to positively regulate drought tolerance, while AtRAP2.1 negatively regulates drought response (Karaba et al., 2007; Dong and Liu, 2010; Zhu et al., 2014; Yao et al., 2017; Figure 1).
In woody species, there are 202 and 149 AP2/ERFs (consisting of ERF, RAV, APW, and Soloist subfamilies) in Populus and Vitis vinifera, respectively, (Zhuang et al., 2008; Licausi et al., 2010). PeSHN1, an AP2/ERF member isolated from Populus × euramericana “Neva,” was reported to promote wax biosynthesis by targeting PeLACS2 and other related genes. It was concluded that poplar plants overexpressing PeSHN1 enhance drought tolerance by increasing wax biosynthesis and reducing transpiration (Meng et al., 2019). Orthologs of Arabidopsis DREB2A, PeDREB2a and PeDREB2L from P. euphratica were shown to be involved in drought response. Overexpression of PeDREB2a or PeDREB2L in Arabidopsis enhances drought tolerance although the potential PEST sequences were identified within the protein sequence. These results suggest that PeDREB2s are more stable than DREB2A and may function as the constitutively active form (Chen et al., 2011; Zhou et al., 2012). PagERF35, whose encoding gene is induced by drought treatment, can directly bind to the DRE motifs in the promoter of PagWOX11/12a. PagWOX11/12a, belonging to the WUSCHEL-related homebox (WOX) TF familiy, positively regulates drought tolerance by increasing root growth. In addition, co-expression analysis and transcriptional activation assay suggest that ERF and WOX may form a regulatory module responding to drought stress (Wang et al., 2019). MdERF38 was shown to interact with MdMYB1 to promote anthocyanin biosynthesis in response to drought stress in apple (An et al., 2020). Another two AP2/ERF TFs, MdSHINE2 and MdWRI4, were also positively involved in drought tolerance by regulating wax biosynthesis in Arabidopsis (Zhang Y.-L. et al., 2019; Zhang et al., 2020; Table 1).
NACs and Other TF Families
NAM, ATAF, and CUC (NAC) TFs are encoded by the largest plant-specific NAC gene family. Many NACs induced by drought stress or ABA were reported to bind NACRS(CGTG/A) sequence in the promoters of drought stress-responsive genes (Puranik et al., 2012). Some NACs regulate drought response through the ABA-dependent pathway, while the other NACs do so through the ABA-independent pathway. ERD1 is a dehydration-responsive gene whose expression is induced by drought stress, but not by ABA (Nakashima et al., 1997). Further analysis revealed that the ERD1 promoter contains both NACRS (CATGTG) and zinc finger homeodomain recognition sequences (ZFHDR, CACTAAATTGTCAC). ANAC019, ANAC055, ANAC072/RD26, and ZFHD1 were reported to bind to the promoter of ERD1. Overexpression of ZFHD1 or co-overexpression of ZFHD1 and ANACs show induced ERD1 expression and drought tolerance (Tran et al., 2007; Singh and Laxmi, 2015). Interestingly, ANAC072/RD26 regulates other drought-responsive genes and enhances drought tolerance in an ABA-dependent manner (Fujita et al., 2004). Recently, phosphorylation of RD26 by BIN2 was reported to be required for the activation of RD26 in drought-responsive genes (Jiang et al., 2019). JUB1 is a drought-induced NAC TF. Overexpression of JUB1, driven by either CaMV35S promoter or the RD29A promoter, enhances drought response, and JUB1 was positively regulated by HD-Zip class I TF AtHB13 (Ebrahimian-Motlagh et al., 2017). SUPPRESSOR OF MORE AXILLARY GROWTH2 (MAX2)-LIKE6 (SMXL6), SMXL7, and SMXL8 belonging to SAMX1-LIKE (SMXL) family, in addition to acting as transcriptional repressors in strigolactone signaling, negatively regulate drought response. Transcriptomic and physiological evidence suggested that these three SMXL proteins regulate drought response in both the ABA-dependent and ABA-independent pathways (Wang et al., 2020; Yang T. et al., 2020; Figure 1).
There are 163 NAC genes identified in P. trichocarpa (Hu et al., 2010). Three drought-responsive NAC genes were identified from P. euphratica, including PeNAC034, PeNAC036, and PeNAC045. PeNAC036 is induced, while PeNAC034 and PeNAC045 are repressed by drought stress. Consequently, overexpression of PeNAC036 in Arabidopsis enhances drought tolerance, while overexpression of PeNAC034 in Arabidopsis reduces drought tolerance. Poplar plants overexpressing PeNAC045 also showed a drought-sensitive phenotype (Lu et al., 2018). It was proposed that P. euphratica may adapt to a drought environment by activating and repressing different sets of NAC genes (Lu et al., 2018). PtrNAC006, PtrNAC007, and PtrNAC120, regulated by PtrAREB1, were shown to be positive regulators in drought response (Li et al., 2019). In grapevine, several NAC TFs, including VvNAC26, VvNAC08, VvNAC17, and VaNAC17, were reported to enhance drought tolerance in Arabidopsis (Fang et al., 2016; Su et al., 2020; Ju et al., 2020a, b; Table 1). A novel CCCH zinc finger TF, PdC3H17, was found to positively regulate drought response in poplar. Overexpression of PdC3H17 confers drought tolerance by enhancing the ROS-scavenging abilities, as well as maintaining water potential in stem (Zhuang et al., 2020). A homeodomain leucine zipper (HD-Zip) TF, EcHB1, identified from Eucalyptus camaldulensis, was shown to enhance drought tolerance by increasing photosynthetic efficiency, while reducing leaf area in Eucalyptus (Sasaki et al., 2019). MdHB-7, another HD-Zip TF from Malus domestica, promoted drought tolerance by enhancing ABA content, stomatal closure, and ROS detoxification (Zhao S. et al., 2020).
Epigenetic Regulation of Drought Response
Epigenetic regulations, including DNA methylation, histone modifications, chromatin remodeling, and small RNA, contribute to abiotic stress responses (Chang et al., 2019). Increasing evidence has demonstrated the role of histone modification and DNA methylation in response to drought stress. A histone demethylase, JMJ17, demethylates H3K4 under dehydration conditions (Huang et al., 2019). The jmj17 mutant shows high H3K4me3 levels in drought-responsive genes, such as OST1 and ABF3, and enhances target gene expression and drought tolerance (Huang et al., 2019). LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) belongs to the PRC1 complex, which mediates transcriptional repression of drought-related TF genes, such as ANAC019 and ANAC055. The lhp1 mutant enhances drought tolerance by promoting the expression of drought-responsive genes (Ramirez-Prado et al., 2019). HISTONE DEACETYLASE 9 (HDA9) positively regulates drought response by interacting with ABI4 and maintaining ABA homeostasis in response to drought stress (Baek et al., 2020b).
Populus methylome studies revealed that drought stress changed the DNA methylation level of TF coding genes and further altered their expression pattern (Liang et al., 2014). An RNA m6A methyltransferase coding gene from poplar, MTA, was shown to enhance drought tolerance by promoting trichome and root development (Lu et al., 2020). As mentioned above, several histone modification enzymes, such as AtHDA15 and PtrADA2b-PtrGCN5, were found to interact with key TFs, such as AtMYB96 and PtrAREB1-2, to regulate drought response in Arabidopsis and poplar (Lee and Seo, 2019; Li et al., 2019). Epigenetic regulation is also involved in stress memory by regulating gene expression, in which TFs may also participate. Using periodically combined drought and heat treatments, several stress-related memory genes were identified through tissue-specific transcriptomic profiling studies in poplar. Among them, the homologs of Arabidopsis HOMEOBOX7 (HB7) were proposed as the most prominent candidates enhancing plant photosynthesis during stress recovery (Georgii et al., 2019). In conclusion, epigenetic regulation and stress memory play important roles in drought response and tolerance, but their regulatory roles have not been well defined and deserve further investigations.
Crosstalk Between ABA-Dependent and ABA-Independent Drought Response Pathways
Plants respond to drought stress through complex regulatory networks. TFs play key roles by regulating large numbers of downstream genes, as well as interacting with other TFs. Transcriptional regulatory networks in drought response can be predicted by analyzing transcriptome data (Zhang C. et al., 2018). Crosstalks between the ABA pathway and other pathways have been revealed to regulate drought response. For example, SMXLs from SL signaling are negatively involved in drought response by regulating both SL-responsive genes and ABA-responsive genes (Yang T. et al., 2020). Accumulated evidence supports the physical interactions among TFs from ABA-dependent and ABA-independent pathways. ABA-dependent AREBs/ABFs were found to interact with DREB2A, leading to synergistic activation of the drought-responsive genes such as RD29A (Lee et al., 2010). AREB1 and AREB2 were also found to interact with ABA-independent ANAC096, and their interactions enhance transcriptional activities of the AREBs (Xu et al., 2013). JA-regulated MYC2 was found to interact with the ABA-responsive ABF3, supporting the crosstalk between JA signaling and ABA signaling in response to drought stress (Liu et al., 2018). BES1 from BR signaling regulates drought response by interacting with WRKY, AP2/ERF, and NAC TFs (Nolan et al., 2020). Collectively, drought response is regulated by the interplay between ABA-dependent and ABA-independent pathways (Yoshida et al., 2014).
Complexities and Research Strategies of Drought Response in Woody Plants
Long-lived trees experience drought stress together with other biotic and abiotic stresses. High temperatures are the most common stress occurring simultaneously with drought in the field, and trees can respond differently to a single stress versus combined stresses (Zandalinas et al., 2017). Jia et al. (2017) reported that heat and drought stresses shared responsive genes in Populus simonii. The hierarchical genetic regulatory networks involving several TF, such as ERF1 and RD26, were formed during these combined stress treatments. It was proposed that the co-expression networks contributed to single and/or combined stress responses by regulating hormone biosynthesis and ROS production (Jia et al., 2017). Ozone (O3) is another phytotoxic air pollutant. Combined effects of high ozone and drought stresses were investigated in poplar through transcriptome analysis. Twelve core TFs were predicted to be master regulators in response to the combination of high O3 and drought stresses. Further analysis on differentially-expressed genes (DEGs) indicated that Populus plants respond to O3 and/or drought by regulating isoprene biosynthesis and the ABA-dependent pathways (Zhang J. et al., 2019). The molecular response of combined drought and low nitrogen stress has also been studied in Populus. RNA-seq analyses of Populus simonii roots in response to drought and low nitrogen stresses revealed that drought positively regulates ammonium uptake and amino acid metabolism, which, in turn, promote drought tolerance (Zhang C. et al., 2018).
The roles of different microbes and endophytes in drought tolerance have also been revealed in poplar recently. Khan et al. (2016) reported that combined application of endophytes greatly enhanced plant biomass in Populus deltoides × P. nigra clone OP-367 in response to drought. Further analyses revealed that phytohormone production of these endophytes and ROS detoxification of inoculated plants contributed to drought tolerance (Khan et al., 2016). Using an RNA-seq approach, it was found that cyclic drought treatment increased the phytobiome in taxa which could benefit the host P. deltoides plants in terms of disease response, ROS metabolism, and photosynthesis (Garcia et al., 2018).
Transcriptome studies, such as RNA-seq, have been used to explore the complexities of drought stress combined with other environment factors in woody plants (Estravis-Barcala et al., 2019). The advantage of transcriptome study is that it not only unravels the transcriptional regulatory networks under any stress combination but also provides detailed transcriptional events in genotype-, tissue-, and developmental stage-specific bases (Cohen et al., 2010). Transcriptomic analyses can become more effective when combined with other omics approaches, such as metabolomics, since metabolite profiling can provide additional insights by integrating diverse transcriptomic responses (Hamanishi et al., 2015). The large number of key TFs and transcriptional regulatory networks identified from RNA-seq studies can deepen our understanding of drought response and tolerance in woody plants.
Future Perspectives
The transcriptional regulatory network of drought response was built extensively in the model plant Arabidopsis during the last few decades. In comparison with model plants, knowledge of drought response in woody species is still limited. Drought response varies with species and genotypes. Recently developed high-throughput omics strategies shed light on the poplar response to drought and the transcriptional regulation underlying drought tolerance. For example, genome-wide association mapping studies (GWAS) and Expression Quantitative Trait Locus (eQTL) mapping have become effective tools to identify the genetic loci underlying variation in economically important phenotypic traits and transcriptional regulation (Muchero et al., 2018; Zhang J. et al., 2018). Systems biology approaches including genomics, transcriptomics, proteomics, metabolomics, and phenomics may help facilitate the identification and functional characterization of TFs regulating drought response in woody plants (De Ollas et al., 2019). The adoption of CRISPR/Cas9 technique in trees provides the power of precisely editing the genomic loci to uncover genes in the drought response pathway (Bewg et al., 2018).
Most studies have been conducted with single-stress studies conducted in laboratory conditions. Forest trees often endure multiple stresses, such as heat and drought in field. It is imperative to verify the function of drought-related TFs and observe their impact on phenotypic expression under field conditions. The models generated from multistress studies will deepen our understanding on drought stress response and provide genetic engineering targets to create drought-tolerant and high-yield woody plants.
Notice
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the United States Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Author Contributions
WM and J-GC conceived the study. TY drafted the manuscript. JZ, MX, GY, TT, WM, and J-GC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Center for Bioenergy Innovation, the Plant-Microbe Interfaces Scientific Focus Area and the Genomics-Enabled Plant Biology for Determination of Gene Function program by the Office of Biological and Environmental Research in the United States Department of Energy Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the United States Department of Energy under contract DE-AC05-00OR22725. JZ is funded by the Zhejiang A&F University Research and Development Fund Talent Startup Project.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., and Pereira, A. (2004). The SHINE Clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
An, J.-P., Zhang, X.-W., Bi, S.-Q., You, C.-X., Wang, X.-F., and Hao, Y.-J. (2020) The ERF transcription factor MdERF38 promotes drought stress-induced anthocyanin biosynthesis in apple. Plant J. 101, 573–589. doi: 10.1111/tpj.14555
Arshad, M., and Mattsson, J. (2014). A putative poplar PP2C-encoding gene negatively regulates drought and abscisic acid responses in transgenic Arabidopsis thaliana. Trees 28, 531–543. doi: 10.1007/s00468-013-0969-967
Babitha, K. C., Ramu, S. V., Pruthvi, V., Mahesh, P., Nataraja, K. N., and Udayakumar, M. (2013). Co-expression of AtbHLH17 and AtWRKY28 confers resistance to abiotic stress in Arabidopsis. Transgenic Res. 22, 327–341. doi: 10.1007/s11248-012-9645-9648
Baek, D., Kim, W.-Y., Cha, J.-Y., Park, H. J., Shin, G., Park, J., et al. (2020a). The gigantea-enhanced em level complex enhances drought tolerance via regulation of abscisic acid synthesis. Plant Physiol. 184, 443–458. doi: 10.1104/pp.20.00779
Baek, D., Shin, G., Kim, M. C., Shen, M., Lee, S. Y., and Yun, D.-J. (2020b). Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Front. Plant Sci. 11:143. doi: 10.3389/fpls.2020.00143
Bewg, W. P., Ci, D., and Tsai, C.-J. (2018). Genome editing in trees: from multiple repair pathways to long-term stability. Front. Plant Sci. 9:1732. doi: 10.3389/fpls.2018.01732
Chang, Y. N., Zhu, C., Jiang, J., Zhang, H., Zhu, J. K., and Duan, C. G. (2019). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi: 10.1111/jipb.12901
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., and Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40 and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10:281. doi: 10.1186/1471-2229-10-281
Chen, J., Nolan, T. M., Ye, H., Zhang, M., Tong, H., Xin, P., et al. (2017). Arabidopsis WRKY46, WRKY54, and WRKY70 transcription factors are involved in brassinosteroid-regulated plant growth and drought responses. Plant Cell 29, 1425–1439. doi: 10.1105/tpc.17.00364
Chen, J., Xia, X., and Yin, W. (2011). A poplar DRE-binding protein gene, PeDREB2L, is involved in regulation of defense response against abiotic stress. Gene 483, 36–42. doi: 10.1016/j.gene.2011.05.010
Chen, J., Zhang, D., Zhang, C., Xia, X., Yin, W., and Tian, Q. (2015). A putative PP2C-encoding gene negatively regulates ABA signaling in Populus euphratica. PLoS One 10:e0139466. doi: 10.1371/journal.pone.0139466
Chen, X., Li, C., Wang, H., and Guo, Z. (2019). WRKY transcription factors: evolution, binding, and action. Phytopathol. Res. 1:13. doi: 10.1186/s42483-019-0022-x
Claeys, H., and Inze, D. (2013). The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 162, 1768–1779. doi: 10.1104/pp.113.220921
Cohen, D., Bogeat-Triboulot, M.-B., Tisserant, E., Balzergue, S., Martin-Magniette, M.-L., Lelandais, G., et al. (2010). Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11:630. doi: 10.1186/1471-2164-11-630
Dash, M., Yordanov, Y. S., Georgieva, T., Tschaplinski, T. J., Yordanova, E., and Busov, V. (2017). Poplar PtabZIP1-like enhances lateral root formation and biomass growth under drought stress. Plant J. 89, 692–705. doi: 10.1111/tpj.13413
De Ollas, C., Morillón, R., Fotopoulos, V., Puértolas, J., Ollitrault, P., Gómez-Cadenas, A., et al. (2019). Facing climate change: biotechnology of iconic mediterranean woody crops. Front. Plant Sci. 10:427. doi: 10.3389/fpls.2019.00427
Dong, C.-J., and Liu, J.-Y. (2010). The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 10:47. doi: 10.1186/1471-2229-10-47
Duan, Y., Jiang, Y., Ye, S., Karim, A., Ling, Z., He, Y., et al. (2015). PtrWRKY73, a salicylic acid-inducible poplar WRKY transcription factor, is involved in disease resistance in Arabidopsis thaliana. Plant Cell Rep. 34, 831–841. doi: 10.1007/s00299-015-1745-1745
Ebrahimian-Motlagh, S., Ribone, P. A., Thirumalaikumar, V. P., Allu, A. D., Chan, R. L., Mueller-Roeber, B., et al. (2017). Jungbrunnen1 confers drought tolerance downstream of the HD-zip I transcription factor AtHB13. Front. Plant Sci. 8:2118. doi: 10.3389/fpls.2017.02118
Estravis-Barcala, M., Mattera, M. G., Soliani, C., Bellora, N., Opgenoorth, L., Heer, K., et al. (2019). Molecular bases of responses to abiotic stress in trees. J. Exp. Bot. 71, 3765–3779. doi: 10.1093/jxb/erz532
Fang, L., Su, L., Sun, X., Li, X., Sun, M., Karungo, S. K., et al. (2016). Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 67, 2829–2845. doi: 10.1093/jxb/erw122
Fang, Q., Wang, X., Wang, H., Tang, X., Liu, C., Yin, H., et al. (2019). The poplar R2R3 MYB transcription factor PtrMYB94 coordinates with abscisic acid signaling to improve drought tolerance in plants. Tree Physiol. 40, 46–59. doi: 10.1093/treephys/tpz113
Fernàndez-Martínez, J., Zacchini, M., Elena, G., Fernández-Marín, B., and Fleck, I. (2013). Effect of environmental stress factors on ecophysiological traits and susceptibility to pathogens of five Populus clones throughout the growing season. Tree Physiol. 33, 618–627. doi: 10.1093/treephys/tpt039
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004) A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313X.2004.02171.x
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
Garcia, B. J., Labbé, J. L., Jones, P., Abraham, P. E., Hodge, I., Climer, S., et al. (2018). Phytobiome and transcriptional adaptation of Populus deltoides to acute progressive drought and cyclic drought. Phytobiomes J. 2, 249–260. doi: 10.1094/pbiomes-04-18-0021-r
Georgii, E., Kugler, K., Pfeifer, M., Vanzo, E., Block, K., Domagalska, M. A., et al. (2019). The Systems architecture of molecular memory in poplar after abiotic stress. Plant Cell 31, 346–367. doi: 10.1105/tpc.18.00431
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Guo, J., Yang, X., Weston, D. J., and Chen, J.-G. (2011). Abscisic acid receptors: past, present and futureF. J. Integr. Plant Biol. 53, 469–479. doi: 10.1111/j.1744-7909.2011.01044.x
Gupta, A., Rico-Medina, A., and Caño-Delgado, A. I. (2020a). The physiology of plant responses to drought. Science 368, 266–269. doi: 10.1126/science.aaz7614
Gupta, A., Sinha, R., Fernandes, J. L., Abdelrahman, M., Burritt, D. J., and Tran, L. P. (2020b). Phytohormones regulate convergent and divergent responses between individual and combined drought and pathogen infection. Crit. Rev. Biotechnol. 40, 320–340. doi: 10.1080/07388551.2019.1710459
Hamanishi, E. T., Barchet, G. L. H., Dauwe, R., Mansfield, S. D., and Campbell, M. M. (2015). Poplar trees reconfigure the transcriptome and metabolome in response to drought in a genotype- and time-of-day-dependent manner. BMC Genomics 16:329. doi: 10.1186/s12864-015-1535-z
Han, X., Tang, S., An, Y., Zheng, D. C., Xia, X. L., and Yin, W. L. (2013). Overexpression of the poplar NF-YB7 transcription factor confers drought tolerance and improves water-use efficiency in Arabidopsis. J. Exp. Bot. 64, 4589–4601. doi: 10.1093/jxb/ert262
He, H., Dong, Q., Shao, Y., Jiang, H., Zhu, S., Cheng, B., et al. (2012). Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 31, 1199–1217. doi: 10.1007/s00299-012-1241-1240
Hu, R., Qi, G., Kong, Y., Kong, D., Gao, Q., and Zhou, G. (2010). Comprehensive analysis of NAC domain transcription factor gene family in Populus trichocarpa. BMC Plant Biol. 10:145. doi: 10.1186/1471-2229-10-145
Huang, S., Zhang, A., Jin, J. B., Zhao, B., Wang, T.-J., Wu, Y., et al. (2019). Arabidopsis histone H3K4 demethylase JMJ17 functions in dehydration stress response. New Phytol. 223, 1372–1387. doi: 10.1111/nph.15874
Hwang, K., Susila, H., Nasim, Z., Jung, J. Y., and Ahn, J. H. (2019). Arabidopsis ABF3 and ABF4 transcription factors act with the NF-YC complex to regulate SOC1 expression and mediate drought-accelerated flowering. Mol. Plant 12, 489–505. doi: 10.1016/j.molp.2019.01.002
Ji, L., Wang, J., Ye, M., Li, Y., Guo, B., Chen, Z., et al. (2013). Identification and characterization of the populus AREB/ABF subfamily. J. Integr. Plant Biol. 55, 177–186. doi: 10.1111/j.1744-7909.2012.01183.x
Jia, J., Zhou, J., Shi, W., Cao, X., Luo, J., Polle, A., et al. (2017). Comparative transcriptomic analysis reveals the roles of overlapping heat-/drought-responsive genes in poplars exposed to high temperature and drought. Sci. Rep. 7:43215. doi: 10.1038/srep43215
Jiang, H., Tang, B., Xie, Z., Nolan, T., Ye, H., Song, G.-Y., et al. (2019). GSK3-like kinase BIN2 phosphorylates RD26 to potentiate drought signaling in Arabidopsis. Plant J. 100, 923–937. doi: 10.1111/tpj.14484
Jiang, Y., Duan, Y., Yin, J., Ye, S., Zhu, J., Zhang, F., et al. (2014). Genome-wide identification and characterization of the Populus WRKY transcription factor family and analysis of their expression in response to biotic and abiotic stresses. J. Exp. Bot. 65, 6629–6644. doi: 10.1093/jxb/eru381
Jiang, Y., Guo, L., Ma, X., Zhao, X., Jiao, B., Li, C., et al. (2017). The WRKY transcription factors PtrWRKY18 and PtrWRKY35 promote Melampsora resistance in Populus. Tree Physiol. 37, 665–675. doi: 10.1093/treephys/tpx008
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7:1029. doi: 10.3389/fpls.2016.01029
Ju, Y.-L., Min, Z., Yue, X.-F., Zhang, Y.-L., Zhang, J.-X., Zhang, Z.-Q., et al. (2020a). Overexpression of grapevine VvNAC08 enhances drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 151, 214–222. doi: 10.1016/j.plaphy.2020.03.028
Ju, Y.-L., Yue, X.-F., Min, Z., Wang, X.-H., Fang, Y.-L., and Zhang, J.-X. (2020b). VvNAC17, a novel stress-responsive grapevine (Vitis vinifera L.) NAC transcription factor, increases sensitivity to abscisic acid and enhances salinity, freezing, and drought tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 146, 98–111. doi: 10.1016/j.plaphy.2019.11.002
Karaba, A., Dixit, S., Greco, R., Aharoni, A., Trijatmiko, K. R., Marsch-Martinez, N., et al. (2007). Improvement of water use efficiency in rice by expression of HARDY an Arabidopsis drought and salt tolerance gene. Proc. Natl. Acad. Sci. U. S. A. 104, 15270–15275. doi: 10.1073/pnas.0707294104
Khan, Z., Rho, H., Firrincieli, A., Hung, S. H., Luna, V., Masciarelli, O., et al. (2016). Growth enhancement and drought tolerance of hybrid poplar upon inoculation with endophyte consortia. Curr. Plant Biol. 6, 38–47. doi: 10.1016/j.cpb.2016.08.001
Kim, J. S., Mizoi, J., Yoshida, T., Fujita, Y., Nakajima, J., Ohori, T., et al. (2011). An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol. 52, 2136–2146. doi: 10.1093/pcp/pcr143
Lee, H. G., and Seo, P. J. (2019). MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis. Nat. Commun. 10:1713. doi: 10.1038/s41467-019-09417-9411
Lee, S.-J., Kang, J.-Y., Park, H.-J., Kim, M. D., Bae, M. S., Choi, H.-I., et al. (2010). DREB2C interacts with ABF2, a bZIP protein regulating abscisic acid-responsive gene expression, and its overexpression affects abscisic acid sensitivity. Plant Physiol. 153, 716–727. doi: 10.1104/pp.110.154617
Leyva-Gonzalez, M. A., Ibarra-Laclette, E., Cruz-Ramirez, A., and Herrera-Estrella, L. (2012). Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7:e48138. doi: 10.1371/journal.pone.0048138
Li, S., Lin, Y.-C. J., Wang, P., Zhang, B., Li, M., Chen, S., et al. (2019). The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 31, 663–686. doi: 10.1105/tpc.18.00437
Li, W. X., Oono, Y., Zhu, J., He, X. J., Wu, J. M., Iida, K., et al. (2008). The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20, 2238–2251. doi: 10.1105/tpc.108.059444
Lian, C., Li, Q., Yao, K., Zhang, Y., Meng, S., Yin, W., et al. (2018). Populus trichocarpa PtNF-YA9, a multifunctional transcription factor, regulates seed germination, abiotic stress, plant growth and development in Arabidopsis. Front. Plant Sci. 9:954. doi: 10.3389/fpls.2018.00954
Liang, D., Zhang, Z., Wu, H., Huang, C., Shuai, P., Ye, C.-Y., et al. (2014). Single-base-resolution methylomes of populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet. 15:S9. doi: 10.1186/1471-2156-15-S1-S9
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., and Perata, P. (2010). Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11:719. doi: 10.1186/1471-2164-11-719
Liu, G., Zeng, H., Li, X., Wei, Y., and Shi, H. (2019). Functional analysis of MaWRKY24 in transcriptional activation of autophagy-related gene 8f/g and plant disease susceptibility to soil-borne fusarium oxysporum f. sp. cubense. Pathogens 8:264. doi: 10.3390/pathogens8040264
Liu, R., Wu, M., Liu, H. L., Gao, Y. M., Chen, J., Yan, H. W., et al. (2020). Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Physiol. Plant doi: 10.1111/ppl.13084 (Online ahead of print)
Liu, S., Lv, Z., Liu, Y., Li, L., and Zhang, L. (2018). Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 41, 624–637. doi: 10.1590/1678-4685-gmb-2017-0229
Liu, Y., Yang, T., Lin, Z., Gu, B., Xing, C., Zhao, L., et al. (2019). A WRKY transcription factor PbrWRKY53 from Pyrus betulaefolia is involved in drought tolerance and AsA accumulation. Plant Biotechnol. J. 17, 1770–1787. doi: 10.1111/pbi.13099
Lu, L., Zhang, Y., He, Q., Qi, Z., Zhang, G., Xu, W., et al. (2020). MTA, an RNA m(6)A Methyltransferase, enhances drought tolerance by regulating the development of trichomes and roots in poplar. Intl. J. Mol. Sci. 21:2462. doi: 10.3390/ijms21072462
Lu, X., Zhang, X., Duan, H., Lian, C., Liu, C., Yin, W., et al. (2018). Three stress-responsive NAC transcription factors from Populus euphratica differentially regulate salt and drought tolerance in transgenic plants. Physiol. Plant. 162, 73–97. doi: 10.1111/ppl.12613
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Meng, S., Cao, Y., Li, H., Bian, Z., Wang, D., Lian, C., et al. (2019). PeSHN1 regulates water-use efficiency and drought tolerance by modulating wax biosynthesis in poplar. Tree Physiol. 39, 1371–1386. doi: 10.1093/treephys/tpz033
Muchero, W., Sondreli, K. L., Chen, J.-G., Urbanowicz, B. R., Zhang, J., Singan, V., et al. (2018). Association mapping, transcriptomics, and transient expression identify candidate genes mediating plant–pathogen interactions in a tree. Proc. Natl. Acad. Sci. U. S. A. 115, 11573–11578. doi: 10.1073/pnas.1804428115
Nakashima, K., Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). A nuclear gene, erd1, encoding a chloroplast-targeted Clp protease regulatory subunit homolog is not only induced by water stress but also developmentally up-regulated during senescence in Arabidopsis thaliana. Plant J. 12, 851–861. doi: 10.1046/j.1365-313X.1997.12040851.x
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Nolan, T. M., Vukašinović, N., Liu, D., Russinova, E., and Yin, Y. (2020). Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell 32, 295–318. doi: 10.1105/tpc.19.00335
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Polle, A., Chen, S. L., Eckert, C., and Harfouche, A. (2019). Engineering drought resistance in forest trees. Front. Plant Sci. 9:1875. doi: 10.3389/fpls.2018.01875
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Ramirez-Prado, J. S., Latrasse, D., Rodriguez-Granados, N. Y., Huang, Y., Manza-Mianza, D., Brik-Chaouche, R., et al. (2019). The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 100, 1118–1131. doi: 10.1111/tpj.14502
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Salehi-Lisar, S. Y., and Bakhshayeshan-Agdam, H. (2016). “Drought stress in plants: causes, consequences, and tolerance,” in Drought Stress Tolerance in Plants, Vol 1: Physiology and Biochemistry, eds M. A. Hossain, S. H. Wani, S. Bhattacharjee, D. J. Burritt, and L.-S. P. Tran (Cham: Springer International Publishing), 1–16. doi: 10.1007/978-3-319-28899-4_1
Salehin, M., Li, B., Tang, M., Katz, E., Song, L., Ecker, J. R., et al. (2019). Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 10:4021. doi: 10.1038/s41467-019-12002-12001
Sasaki, K., Ida, Y., Kitajima, S., Kawazu, T., Hibino, T., and Hanba, Y. T. (2019). Overexpressing the HD-Zip class II transcription factor EcHB1 from Eucalyptus camaldulensis increased the leaf photosynthesis and drought tolerance of Eucalyptus. Sci. Rep. 9:14121. doi: 10.1038/s41598-019-50610-50615
Sato, H., Suzuki, T., Takahashi, F., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2019). NF-YB2 and NF-YB3 have functionally diverged and differentially induce drought and heat stress-specific genes. Plant Physiol. 180, 1677–1690. doi: 10.1104/pp.19.00391
Seo, P. J., Xiang, F., Qiao, M., Park, J. Y., Lee, Y. N., Kim, S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Singh, D., and Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895. doi: 10.3389/fpls.2015.00895
Su, L., Fang, L., Zhu, Z., Zhang, L., Sun, X., Wang, Y., et al. (2020). The transcription factor VaNAC17 from grapevine (Vitis amurensis) enhances drought tolerance by modulating jasmonic acid biosynthesis in transgenic Arabidopsis. Plant Cell Rep. 39, 621–634. doi: 10.1007/s00299-020-02519-x
Suzuki, M., and McCarty, D. R. (2008). Functional symmetry of the B3 network controlling seed development. Curr. Opin. Plant Biol. 11, 548–553. doi: 10.1016/j.pbi.2008.06.015
Takahashi, F., Kuromori, T., Sato, H., and Shinozaki, K. (2018). “Regulatory gene networks in drought stress responses and resistance in plants,” in Survival Strategies in Extreme Cold and Desiccation: Adaptation Mechanisms and Their Applications, eds M. Iwaya-Inoue, M. Sakurai, and M. Uemura (Singapore: Springer), 189–214. doi: 10.1007/978-981-13-1244-1_11
Tian, H., Chen, S., Yang, W., Wang, T., Zheng, K., Wang, Y., et al. (2017). A novel family of transcription factors conserved in angiosperms is required for ABA signalling. Plant Cell Environ. 40, 2958–2971. doi: 10.1111/pce.13058
Tran, L. S., Nakashima, K., Sakuma, Y., Osakabe, Y., Qin, F., Simpson, S. D., et al. (2007). Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49, 46–63. doi: 10.1111/j.1365-313X.2006.02932.x
Umezawa, T., Nakashima, K., Miyakawa, T., Kuromori, T., Tanokura, M., Shinozaki, K., et al. (2010). Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. doi: 10.1093/pcp/pcq156
Umezawa, T., Sugiyama, N., Mizoguchi, M., Hayashi, S., Myouga, F., Yamaguchi-Shinozaki, K., et al. (2009). Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 17588–17593. doi: 10.1073/pnas.0907095106
Wang, L., Wang, B., Yu, H., Guo, H., Lin, T., Kou, L., et al. (2020). Transcriptional regulation of strigolactone signalling in Arabidopsis. Nature 583, 277–281. doi: 10.1038/s41586-020-2382-x
Wang, L.-Q., Li, Z., Wen, S.-S., Wang, J.-N., Zhao, S.-T., and Lu, M.-Z. (2019). WUSCHEL-related homeobox gene PagWOX11/12a responds to drought stress by enhancing root elongation and biomass growth in poplar. J. Exp. Bot. 71, 1503–1513. doi: 10.1093/jxb/erz490
Wang, M., Vannozzi, A., Wang, G., Liang, Y. H., Tornielli, G. B., Zenoni, S., et al. (2014). Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Hortic Res. 1:14016. doi: 10.1038/hortres.2014.16
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., and Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Direct 10:48. doi: 10.1186/s13062-015-0076-73
Wilkins, O., Nahal, H., Foong, J., Provart, N. J., and Campbell, M. M. (2009). Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol. 149, 981–993. doi: 10.1104/pp.108.132795
Xie, Z., Nolan, T., Jiang, H., Tang, B., Zhang, M., Li, Z., et al. (2019). The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 31, 1788–1806. doi: 10.1105/tpc.18.00918
Xu, C., Fu, X., Liu, R., Guo, L., Ran, L., Li, C., et al. (2017). PtoMYB170 positively regulates lignin deposition during wood formation in poplar and confers drought tolerance in transgenic Arabidopsis. Tree Physiol. 37, 1713–1726. doi: 10.1093/treephys/tpx093
Xu, Z. Y., Kim, S. Y., Hyeon do, Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, T., Lian, Y., Kang, J., Bian, Z., Xuan, L., Gao, Z., et al. (2020). The Suppressor of MAX2 1 (SMAX1)-Like SMXL6, SMXL7 and SMXL8 act as negative regulators in response to drought stress in Arabidopsis. Plant Cell Physiol. 61, 1477–1492. doi: 10.1093/pcp/pcaa066
Yang, Y., Li, H.-G., Wang, J., Wang, H.-L., He, F., Su, Y., et al. (2020). ABF3 enhances drought tolerance via promoting ABA-induced stomatal closure by directly regulating ADF5 in Populus euphratica. J. Exper. Bot. eraa383. doi: 10.1093/jxb/eraa383
Yao, Y., He, R. J., Xie, Q. L., Zhao, X. H., Deng, X. M., He, J. B., et al. (2017). ETHYLENE RESPONSE FACTOR 74 (ERF74) plays an essential role in controlling a respiratory burst oxidase homolog D (RbohD)-dependent mechanism in response to different stresses in Arabidopsis. New Phytol. 213, 1667–1681. doi: 10.1111/nph.14278
Yin, H., Chen, C. J., Yang, J., Weston, D. J., Chen, J.-G., Muchero, W., et al. (2014). Functional genomics of drought tolerance in bioenergy crops. Crit. Rev. Plant Sci. 33, 205–224. doi: 10.1080/07352689.2014.870417
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
Yu, D., Wildhagen, H., Tylewicz, S., Miskolczi, P. C., Bhalerao, R. P., and Polle, A. (2019). Abscisic acid signalling mediates biomass trade-off and allocation in poplar. New Phytol. 223, 1192–1203. doi: 10.1111/nph.15878
Yu, J., Ge, H., Wang, X., Tang, R., Wang, Y., Zhao, F., et al. (2017). Overexpression of pyrabactin resistance-like abscisic acid receptors enhances drought, osmotic, and cold tolerance in transgenic poplars. Front. Plant Sci. 8:1752. doi: 10.3389/fpls.2017.01752
Zandalinas, S. I., Balfagón, D., Arbona, V., and Gómez-Cadenas, A. (2017). Modulation of antioxidant defense system is associated with combined drought and heat stress tolerance in citrus. Front. Plant Sci. 8:953. doi: 10.3389/fpls.2017.00953
Zeevaart, J. A. (1980). Changes in the levels of abscisic acid and its metabolites in excised leaf blades of xanthium strumarium during and after water stress. Plant Physiol. 66, 672–678. doi: 10.1104/pp.66.4.672
Zhang, C., Meng, S., Li, M., and Zhao, Z. (2018). Transcriptomic insight into nitrogen uptake and metabolism of Populus simonii in response to drought and low nitrogen stresses. Tree Physiol. 38, 1672–1684. doi: 10.1093/treephys/tpy085
Zhang, C.-L., Wang, Y.-X., Hu, X., Zhang, Y.-L., Wang, G.-L., You, C.-X., et al. (2020). An apple AP2/EREBP-type transcription factor, MdWRI4, enhances plant resistance to abiotic stress by increasing cuticular wax load. Environ. Exp. Bot. 180:104206. doi: 10.1016/j.envexpbot.2020.104206
Zhang, J., Gao, F., Jia, H., Hu, J., and Feng, Z. (2019). Molecular response of poplar to single and combined ozone and drought. Sci. Total Environ. 655, 1364–1375. doi: 10.1016/j.scitotenv.2018.11.195
Zhang, J., Yang, Y., Zheng, K., Xie, M., Feng, K., Jawdy, S. S., et al. (2018). Genome-wide association studies and expression-based quantitative trait loci analyses reveal roles of HCT2 in caffeoylquinic acid biosynthesis and its regulation by defense-responsive transcription factors in Populus. New Phytol. 220, 502–516. doi: 10.1111/nph.15297
Zhang, Y.-L., Zhang, C.-L., Wang, G.-L., Wang, Y.-X., Qi, C.-H., You, C.-X., et al. (2019). Apple AP2/EREBP transcription factor MdSHINE2 confers drought resistance by regulating wax biosynthesis. Planta 249, 1627–1643. doi: 10.1007/s00425-019-03115-3114
Zhao, K. X., Chu, S. S., Zhang, X. D., Wang, L. P., Rono, J. K., and Yang, Z. M. (2020). AtWRKY21 negatively regulates tolerance to osmotic stress in Arabidopsis. Environ. Exp. Bot. 169:103920. doi: 10.1016/j.envexpbot.2019.103920
Zhao, S., Gao, H., Jia, X., Wang, H., Ke, M., and Ma, F. (2020). The HD-Zip i transcription factor MdHB-7 regulates drought tolerance in transgenic apple (Malus domestica). Environ. Exp. Bot. 180:104246. doi: 10.1016/j.envexpbot.2020.104246
Zhou, M.-L., Ma, J.-T., Zhao, Y.-M., Wei, Y.-H., Tang, Y.-X., and Wu, Y.-M. (2012). Improvement of drought and salt tolerance in Arabidopsis and Lotus corniculatus by overexpression of a novel DREB transcription factor from Populus euphratica. Gene 506, 10–17. doi: 10.1016/j.gene.2012.06.089
Zhou, Y., Zhang, Y., Wang, X., Han, X., An, Y., Lin, S., et al. (2020). The root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by ABA-mediated IAA transport in Populus. New Phytol. 227, 407–426. doi: 10.1111/nph.16524
Zhu, D., Wu, Z., Cao, G., Li, J., Wei, J., Tsuge, T., et al. (2014). TRANSLUCENT GREEN, an ERF family transcription factor, controls water balance in arabidopsis by activating the expression of Aquaporin genes. Mol. Plant 7, 601–615. doi: 10.1093/mp/sst152
Zhu, J.-K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029
Zhuang, J., Cai, B., Peng, R. H., Zhu, B., Jin, X. F., Xue, Y., et al. (2008). Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 371, 468–474. doi: 10.1016/j.bbrc.2008.04.087
Keywords: Populus, Arabidopsis, transcription factor, transcriptional regulation, abscisic acid, drought response
Citation: Yao T, Zhang J, Xie M, Yuan G, Tschaplinski TJ, Muchero W and Chen J-G (2021) Transcriptional Regulation of Drought Response in Arabidopsis and Woody Plants. Front. Plant Sci. 11:572137. doi: 10.3389/fpls.2020.572137
Received: 12 June 2020; Accepted: 25 November 2020;
Published: 08 January 2021.
Edited by:
Vicent Arbona, University of Jaume I, SpainReviewed by:
Parvathi Madathil Sreekumar, Kerala Agricultural University, IndiaQibin Ma, South China Agricultural University, China
Copyright © 2021 This work is authored by Yao, Zhang, Xie, Yuan, Tschaplinski, Muchero and Chen on behalf of the U.S. Government and, as regards Yao, Zhang, Xie, Yuan, Tschaplinski, Muchero and Chen and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wellington Muchero, bXVjaGVyb3dAb3JubC5nb3Y=; Jin-Gui Chen, Y2hlbmpAb3JubC5nb3Y=
 Tao Yao
Tao Yao Jin Zhang
Jin Zhang Meng Xie
Meng Xie Guoliang Yuan1,2
Guoliang Yuan1,2 Timothy J. Tschaplinski
Timothy J. Tschaplinski Wellington Muchero
Wellington Muchero Jin-Gui Chen
Jin-Gui Chen