
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 October 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.572135
This article is part of the Research TopicVernalization and Flowering Time: Celebrating 20 Years of FLCView all 15 articles
Narrow-leafed lupin (Lupinus angustifolius L.) is a moderate-yielding legume crop known for its high grain protein content and contribution to soil improvement. It is cultivated under photoperiods ranging from 9 to 17 h, as a spring-sown (in colder locations) or as an autumn-sown crop (in warmer regions). Wild populations require a prolonged cold period, called vernalization, to induce flowering. The key achievement of L. angustifolius domestication was the discovery of two natural mutations (named Ku and Jul) conferring vernalization independence. These mutations are overlapping deletion variants in the promoter of LanFTc1, a homolog of the Arabidopsis thaliana FLOWERING LOCUS T (FT) gene. The third deletion, named here as Pal, was recently found in primitive germplasm. In this study, we genotyped L. angustifolius germplasm that differs in domestication status and geographical origin for LanFTc1 alleles, which we then phenotyped to establish flowering time and vernalization responsiveness. The Ku and Jul lines were vernalization-independent and early flowering, wild (ku) lines were vernalization-dependent and late flowering, whereas the Pal line conferred intermediate phenotype. Three lines representing ku, Pal, and Ku alleles were subjected to gene expression surveys under 8- and 16-h photoperiods. FT homologs (LanFTa1, LanFTa2, LanFTc1, and LanFTc2) and some genes selected by recent expression quantitative trait loci mapping were analyzed. Expression profiles of LanFTc1 and LanAGL8 (AGAMOUS-like 8) matched observed differences in flowering time between genotypes, highlighted by high induction after vernalization in the Ku line. Moreover, these genes revealed altered circadian clock control in Pal line under short days. LanFD (FD) and LanCRLK1 (CALCIUM/CALMODULIN-REGULATED RECEPTOR-LIKE KINASE 1) were negatively responsive to vernalization in Ku and Pal lines but positively responsive or variable in ku, whereas LanUGT85A2 (UDP-GLUCOSYL TRANSFERASE 85A2) was significantly suppressed by vernalization in all lines. Such a pattern suggests the opposite regulation of these gene pairs in the vernalization pathway. LanCRLK1 and LanUGT85A2 are homologs of A. thaliana genes involved in the FLOWERING LOCUS C (FLC) vernalization pathway. Lupins, like many other legumes, do not have any FLC homologs. Therefore, candidate genes surveyed in this study, namely LanFTc1, LanAGL8, LanCRLK1, and LanUGT85A2, may constitute anchors for further elucidation of molecular components contributing to vernalization response in legumes.
Narrow-leafed lupin (Lupinus angustifolius L.) is a legume plant that is cultivated as “green” manure or as a grain crop for animal feed or human consumption (Kurlovich, 2002). The exploitation of L. angustifolius as a crop is advantageous in many respects. First, L. angustifolius cultivation has a positive influence on soil fertility due to the mobilization of soil-bound phosphorus and diazotrophic nitrogen fixation (Evans et al., 1987; Lambers et al., 2013). Moreover, L. angustifolius grains are characterized by high protein content and the use of them in livestock farming systems has many benefits in terms of the economic and environmental impact (Abraham et al., 2019). As seed alkaloid content was reduced by breeding below 0.01% of seed dry weight, a bitter taste, a typical feature of lupins, was fully eliminated from modern cultivars (Kamel et al., 2016). Additionally, the postharvest stubble can be now safety grazed by livestock, because the risk of lupinosis disease was diminished by the introduction of Phomospis stem blight resistance genes (Mulholland et al., 1976; Jago et al., 1982; Cowling et al., 1987; Williamson et al., 1994; Shankar et al., 2002; Ksia̧żkiewicz et al., 2020). Furthermore, L. angustifolius is currently being promoted in human food markets as a result of its nutritional, metabolomic, and other health benefits (Foyer et al., 2016; Kouris-Blazos and Belski, 2016).
Cultivated L. angustifolius germplasm primarily originated from the western Mediterranean basin (Mousavi-Derazmahalleh et al., 2018b). Natural adaptation of wild L. angustifolius populations to the Mediterranean climate is the requirement of the prolonged cold period (i.e., vernalization) during germination or juvenile phase to induce flowering (Gladstones and Hill, 1969; Rahman and Gladstones, 1972; Landers, 1995; Adhikari et al., 2012). The selection of early phenology was based on the removal of the vernalization requirement, a major achievement in the domestication of L. angustifolius, enabling temperature-independent sowing (French and Buirchell, 2005). L. angustifolius is grown in various environments ranging from the subtropics (Australia) and Mediterranean regions (including Morocco, Spain, southern France, and Italy), through temperate oceanic climates (such as the United Kingdom, northern France, and Benelux), humid continental climates (for example in Germany, the Baltic countries, and Ukraine), though to the subarctic zone, localized as far north as 60° latitude (Russia). In warmer regions it is usually autumn-sown, whereas in colder regions it is exclusively spring-sown (Annicchiarico and Carroni, 2009).
Lupin crops are cultivated in various photoperiod conditions, ranging from about 9 h (autumn sowing in Australia) to 16–17 h (spring sowing in Baltic countries and Russia) (Ksia̧żkiewicz et al., 2017). The vernalization requirements of wild populations are so demanding that they can only be completely fulfilled by spring sowing in northern locations (Adhikari et al., 2012). Worldwide, L. angustifolius cultivation is based on vernalization independence selected from only two natural mutations (named Ku and Jul) that were discovered in domesticated germplasm just a little over half a century ago (Mikołajczyk, 1966; Gladstones and Hill, 1969). The use of only two donors of early flowering in L. angustifolius breeding, followed by a strong selection of key agronomic traits, have resulted in a lack of phenological diversity in domesticated germplasm (Stefanova and Buirchell, 2010; Berger et al., 2012; Cowling, 2020). Therefore, further adaptation of this crop to new agronomic conditions resulting from a rapidly changing climate, may only be possible with the incorporation of novel genetic resources of intermediate phenology.
During recent years L. angustifolius was supplemented with numerous molecular resources, including bacterial artificial chromosome libraries carrying nuclear DNA inserts, consecutively improved linkage maps with sequence-defined markers, transcriptome assemblies, and a progressively updated genome sequence of the reference cultivar Tanjil (Kasprzak et al., 2006; Nelson et al., 2006; Gao et al., 2011; Yang et al., 2013; Kamphuis et al., 2015; Wyrwa et al., 2016; Hane et al., 2017; Zhou et al., 2018; Kozak et al., 2019). These resources were harnessed to reveal the genetic identity of Ku, which was found to be a homolog of a FLOWERING LOCUS T (FT) gene, named LanFTc1 (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017). FT gene is a well-recognized floral integrator gene, promoting flowering in response to environmental conditions signaled by photoperiod, vernalization, and circadian clock pathways (Turck et al., 2008). The functional mutation underlying the domesticated Ku allele in L. angustifolius was assigned to the 1423 bp deletion in the promoter region of LanFTc1, carrying potential binding sites for several transcription factors acting as FT gene repressors in Arabidopsis thaliana (Nelson et al., 2017). Interestingly, the second L. angustifolius early phenology mutation, Jul, was recently revealed to be the third LanFTc1 allele, in the form of a 5162 bp deletion in the promoter region, fully encompassing that 1423 bp Ku deletion (Taylor et al., 2019). Screening of germplasm resources with the LanFTc1 markers resulted in the identification of a fourth LanFTc1 allele in wild population line originating from Palestine (a country annotated as Israel in Australian collection or Egypt in Polish gene bank), carrying 1208 bp deletion partially overlapping with domesticated Ku deletion (Rychel, 2018; Taylor et al., 2019). This Palestinian allele was named here as Pal.
Some genes involved in flowering time regulation are under strict circadian clock control and their expression levels fluctuate during a day (Shim et al., 2017). One such example in Arabidopsis is the FT gene, which expression is correlated with the stability of CONSTANS (CO) protein and shows two peaks, the first in the morning (lower) and the second in the evening (higher) (Turck et al., 2008). This CO-dependent regulation is facilitated by enhancer protein binding to specific sites in the promoter sequence, located approximately 5 kbp upstream of the first codon, which results in forming a DNA loop bringing all the components together (Cao et al., 2014). Interestingly, L. angustifolius allelic variants of the LanFTc1 promoter have the distal binding sequences (CCAAT-boxes) preserved (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017; Taylor et al., 2019). This study sought to explore whether the altered length of the LanFTc1 promoter is associated with the altered circadian clock control of LanFTc1 and other related genes.
Taking into consideration the (i) role of FT genes in integrating key environmentally responsive pathways, (ii) demonstrated allelic variability of LanFTc1 gene associated with flowering time, and (iii) wide range of environmental variables occurring at lupin cultivation sites, we decided to explore the photoperiod and vernalization responsiveness in L. angustifolius. Here, the L. angustifolius germplasm differing in domestication status and geographical origin was genotyped for LanFTc1 alleles and phenotyped for flowering time and vernalization responsiveness. Then, the transcriptomic response of candidate genes from the vernalization pathway in early (ku), intermediate (Pal), and late flowering (Ku) L. angustifolius germplasm was assayed under contrasting photoperiod and vernalization conditions, accounting the influence of circadian rhythm control. This study discusses the hypothetical function of these genes in terms of flowering time regulation and response to vernalization.
Ninety-two L. angustifolius lines were subjected to genotyping and phenotyping. Lines were derived from the European Lupin Gene Resources Database maintained by the Poznań Plant Breeding Ltd. station located in Wiatrowo. The lines originated from 13 countries, including Spain (32 lines), Poland (15), Australia (11), Russia (11), Germany (7), Italy (4), Belarus (3), Israel (2), Algeria, Morocco, Palestine, Portugal and Republic of South Africa (1). Accession numbers and information on the domestication status and country of origin are provided in Supplementary Table 1.
Young leaf tissue from three biological replicates per plant was sampled. Frozen (−80°C) plant tissue (50 mg) was homogenized using TissueLyser II (Qiagen, Hilden, Germany) and two stainless steel beads (ø 5 mm) in 2 ml tubes (Eppendorf, Hamburg, Germany). DNA was isolated using DNeasy Plant Mini Kit (Qiagen). PCR was performed using GoTaq Long PCR Master Mix (Promega, Mannheim, Germany) and published LanFTc1_INDEL2 primers (Taylor et al., 2019), provided here for reference in Supplementary Table 2. PCR conditions were as follows: initial denaturation (94°C for 2 min), then 35 cycles composed of denaturation (94°C for 30 s), annealing (62°C for 30 s), and elongation (72°C for 5 min), followed by the final extension (72°C for 10 min). Products were resolved by agarose gel electrophoresis and SYBR Safe DNA staining (Invitrogen, Carlsbad, CA, United States) and visualized on UV-transilluminator (Uvitec, Thermo Fisher Scientific, Waltham, MA, United States). Wild P27255 allele (ku, without deletion) was encoded as “A,” Palestinian allele (Pal, 1208 bp deletion) as “B,” 83A:476 allele (Ku, 1423 bp deletion) as “C,” and Krasnolistny allele (Jul, 5162 bp deletion) as “D.”
Vernalization was carried out by placing imbibed seeds for 21 days at 5°C in darkness on moist filter paper in Petri dishes. Non-vernalized control plants were sown five days before the end of the vernalization procedure and kept at ∼21°C to maintain a similar thermal time (Huyghe, 1991). Plants were cultivated in a greenhouse maintained by the Institute of Plant Genetics, the Polish Academy of Sciences, Poznań, Poland (52°26′N 16°54′E) during growing seasons of 2014 (sowing of vernalized plants on 14.05) and 2015 (sowing of vernalized plants on 25.03) under ambient long day photoperiod (12–17 h). The greenhouse was equipped with automatic heating to keep the minimum air temperature above 18°C. Passive cooling was maintained by a temperature-dependent ventilation system (activated at 22°C). Air temperature (daily mean and maximum) and daily sunshine hours recorded by the nearby localized meteorological station (Poznań-Ławica, 5.1 km) as well as the theoretical photoperiod hours calculated for this latitude (covering 120 days from sowing date for both years) were provided for reference in Supplementary Tables 3, 4. Flowering time was recorded as the number of days from sowing date of vernalized plants until the first fully colored petal was observed. The average number of plants sampled in 2014 was 5.6 for the non-vernalized variant (min. 3, max. 6) and 6.9 for the vernalized variant (min. 4, max. 7), whereas in 2015 it was 4.7 for the non-vernalized variant (min. 4, max. 5) and 5.0 for the vernalized variant.
Based on the results of LanFTc1 allele genotyping and vernalization responsiveness phenotyping, three accessions were selected for gene expression profiling: P27255 from Morocco (96,234, carrying wild allele ku), 83A:476 from Australia (96,233, carrying domesticated allele Ku), and Palestyna from Palestine (95,799, carrying intermediate allele Pal). The vernalization of lines was performed as described above. Non-vernalized control plants were sown five days before the end of the vernalization procedure. Plants from both variants were cultivated in climatic chambers with controlled humidity (40–50% day, 70–80% night) and temperature (22°C day, 18°C night). Two types of photoperiods were studied, short day (SD, 8 h, from 8 AM to 4 PM) and long day (LD, 16 h, from 4 AM to 8 PM). Young leaves from five biological replicates were sampled every week at two times of day to follow a circadian rhythm, at 9 AM, and 3 PM under SD or at 7 AM and 6 PM under LD. Plant material was immediately frozen in liquid nitrogen and stored at −80°C. Three replicates with similar plant phenology (growth rate and time to flowering) were subjected to gene expression profiling. Taking into consideration observed flowering time, from 2 to 4 terms were selected for gene expression, representing the period from about 2 weeks before flowering to the flowering date and/or few days after flowering (Supplementary Table 5).
Frozen young leaf tissue (50 mg) was homogenized using TissueLyser II (Qiagen) and two stainless steel beads (ø 5 mm) in 2 ml tubes (Eppendorf). RNA isolation was performed using the SV Total RNA Isolation System (Promega) without any alterations to the protocol. RNA concentration and purity were measured using NanoDrop 2000 (Thermo Fisher Scientific) and A260/A280 ratio. RNA integrity was visualized by 1% agarose gel electrophoresis of denatured samples in 1 × TAE buffer. RNA concentration was equalized to 1000 ng/μl in nuclease-free water. First-strand cDNA synthesis was performed using GoScript (TM) Reverse Transcription System (Promega) and 5 μg of total RNA per sample.
The set of genes selected for quantitative PCR included, among others, four L. angustifolius representatives of FT clade, namely LanFTa1 (Lup021189, XM_019571501.1), LanFTa2 (XM_019596455.1), LanFTc1 (Lup015264, XM_019601808.1), and LanFTc2 (Lup005674, XM_019565316.1) (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017). First names in parentheses correspond to gene names provided in the L. angustifolius pseudochromosome assembly paper (Hane et al., 2017), whereas the second names address L. angustifolius NCBI Reference Sequences LupAngTanjil_v1.0. Moreover, our assay endeavored also some candidate genes revealed by recent L. angustifolius (eQTL mapping) study (Plewiński et al., 2019) to be associated with the vernalization response, as follows: LanUGT85A2 (Lup002110, XM_019574900.1), LanCRLK1 (Lup011808, XM_019603391.1), LanAGL8 (Lup018485, XM_019583439.1), and LanFD (Lup018024, XM_019567853.1). Alignment of coding sequences to the L. angustifolius genome assembly provided evidence that LanAGL8 and LanFD are present in single copies.
Moreover, some major A. thaliana components of the flowering induction pathway were considered, as follows: LEAFY (LFY, AT5G61850), APETALA1 (AP1, AT1G69120), VERNALIZATION INSENSITIVE 3 (VIN3, AT5G57380), and VERNALIZATION 5 (VRN5, AT3G24440). Multiple sequence alignment revealed that all these genes have putatively three copies in the L. angustifolius genome, namely Lup006312 (XM_019602464.1), Lup012189 (XM_019558971.1) and Lup027481 (XM_019607325.1) for LFY; Lup021855 (XM_019605469.1), Lup024348 (XM_019588203.1) and Lup006876 (XM_019572960.1) for AP1; Lup009440 (XM_019586787.1), Lup013437 (XM_019598860.1) and Lup026125 (XM_019608083.1) for VIN3; and Lup009144 (XM_019591910.1), Lup018692 (XM_019567058.1) and Lup032778 (XM_019590742.1) for VRN5. Analysis of leaf transcriptome data obtained for the L. angustifolius linkage mapping population, covering developmental phases from juvenile to generative growth after partial vernalization (Plewiński et al., 2019), revealed negligible expression of all LFY and AP1 homologs (∼0.07 reads per kilobase million, RPKM), very low expression of VRN5 (∼0.56 RPKM) and high expression of VIN3 copies (∼25.07 RPKM) (Supplementary Table 6). Therefore, VRN5 and VIN3 homologs were selected for the expression assay, whereas LFY and AP1 homologs were discarded. Reference genes validated in previous L. angustifolius quantitative gene expression studies were selected for this assay, namely LanDExH7 (Lup023733, XM_019579367.1), and LanTUB6 (Lup032899, XM_019581544.1) (Taylor et al., 2016, 2019; Nelson et al., 2017). Primers were designed in Geneious Prime (Auckland, New Zealand) using Primer3 (Kearse et al., 2012; Untergasser et al., 2012). Due to the high similarity between particular copies, all three VRN5 homologs were analyzed together using one primer pair. The remaining genes were profiled on one by one basis. Designed primers and expected product sizes are provided in Supplementary Table 2.
In prior experiments, a CFX Connect Real-Time PCR Detection System (Bio-Rad Polska, Warsaw, Poland) was calibrated using Melt Calibration Kit (Bio-Rad Polska) according to the manufacturer’s protocol. A standard curve was developed to assess the performance of the quantitative PCR assay and its dynamic range following recent recommendations (Svec et al., 2015). Analyzed genes were amplified using GoTaq G2 Flexi DNA Polymerase (Promega) and subjected to 1% agarose gel electrophoresis. Amplicons were excised from a gel, extracted with the aid of QIAquick Gel Extraction Kit (Qiagen), quantified using NanoDrop 2000 (Thermo Fisher Scientific), and outsourced (Genomed Ltd., Warsaw, Poland) for direct Sanger sequencing on ABI PRISM 3130 Genetic Analyzer XL (Applied Biosystems, Hitachi). A series of dilutions in concentrations ranging from 1 to 10–10 of the original templates were prepared for every gene using an initial volume of 20 μl to reduce the sampling error. 6 replicates per each concentration were performed using the iTaq Universal SYBR Green Supermix (Bio-Rad Polska). A two-step PCR protocol was exploited according to the protocol. A calculation of R2 and PCR efficiency values was done in Bio-Rad CFX Manager 3.1. The values obtained are provided in Supplementary Table 7.
The quantitative PCR analysis of gene expression was performed using 96-well PCR plates and two reference genes (LanDExH7 and LanTUB6). Inter-run calibration sample (LanTUB6) and no template control were used on all plates. Three biological replicates were analyzed for each time point, and all samples were run in 3 technical repeats. High-resolution PCR product melting in the range of temperature from 65 to 85°C was performed after PCR to control the specificity of amplification. Melt profiles were inspected for the amplification of unspecific products, highlighted by the presence of melting peaks at different temperatures than those obtained during standard curve preparation. Calculations of ΔΔCq were performed in Bio-Rad CFX Manager 3.1 taking into consideration PCR efficiency values and results obtained for both reference genes. The final computations (mean value and standard deviation) and visualization (graphs) were performed in Microsoft Excel 2010.
Calculations were performed to check the influence of circadian clock (expression in the evening divided by expression in the morning), growth phase (maximum expression at analyzed date divided by maximum expression at the first date), vernalization (fold change of expression after vernalization), and genotype (comparison of expression levels observed in studied lines, including Ku/Pal, Ku/ku, and Pal/Ku, performed for all data points). The values obtained are provided in Supplementary Data Sheet 1. The statistical significance of these quotients was tested using a t test for the mean ratio as proposed by Hauschke et al. (1999), Tamhane and Logan (2004)). Calculations were made in R (R Core Team, 2013) with custom script using “t.test.ratio” function from the ratios package. In the first step, the equal variance was tested. If this condition was satisficed classical t-test formula was used, otherwise, Welch’s t-test formula was used (Welch, 1947). P-values were rounded up to four decimal places and are provided in the Supplementary Data Sheet 1.
Indel variation in the promoter region of the major flowering time gene LanFTc1 (Figure 1) was recently revealed to be associated with flowering time and vernalization responsiveness in L. angustifolius (Nelson et al., 2017; Taylor et al., 2019). To evaluate allelic composition in germplasm exploited by European lupin breeders, ninety-two L. angustifolius accessions, encompassing 43 primitive populations or landraces, 23 cross derivatives or breeding lines, 25 cultivars, and one mutant, derived from the European Lupin Gene Resources Database, were screened with primers flanking polymorphic LanFTc1 promoter region (Table 1). As expected, wild LanFTc1 allele (ku) was found mainly in primitive accessions collected in the Mediterranean Basin as well as in a few old domesticated materials originating from Russia, Poland, Germany, and the Republic of South Africa. One wild population line, Palestyna originating from Palestine, was found to carry the shortest variant of deletion (1208 bp), named here as Pal. Alleles Ku (1423 bp deletion) and Jul (5162 bp deletion) were found only in domesticated germplasm – the first one was present mostly in released cultivars, whereas the second one was typically in breeding materials at a different stage of improvement. Accessions carrying Ku originated primarily from Australia, Germany, and Poland, whereas those carrying Jul from Poland and Russia. Results of marker screening are provided in Supplementary Table 8.
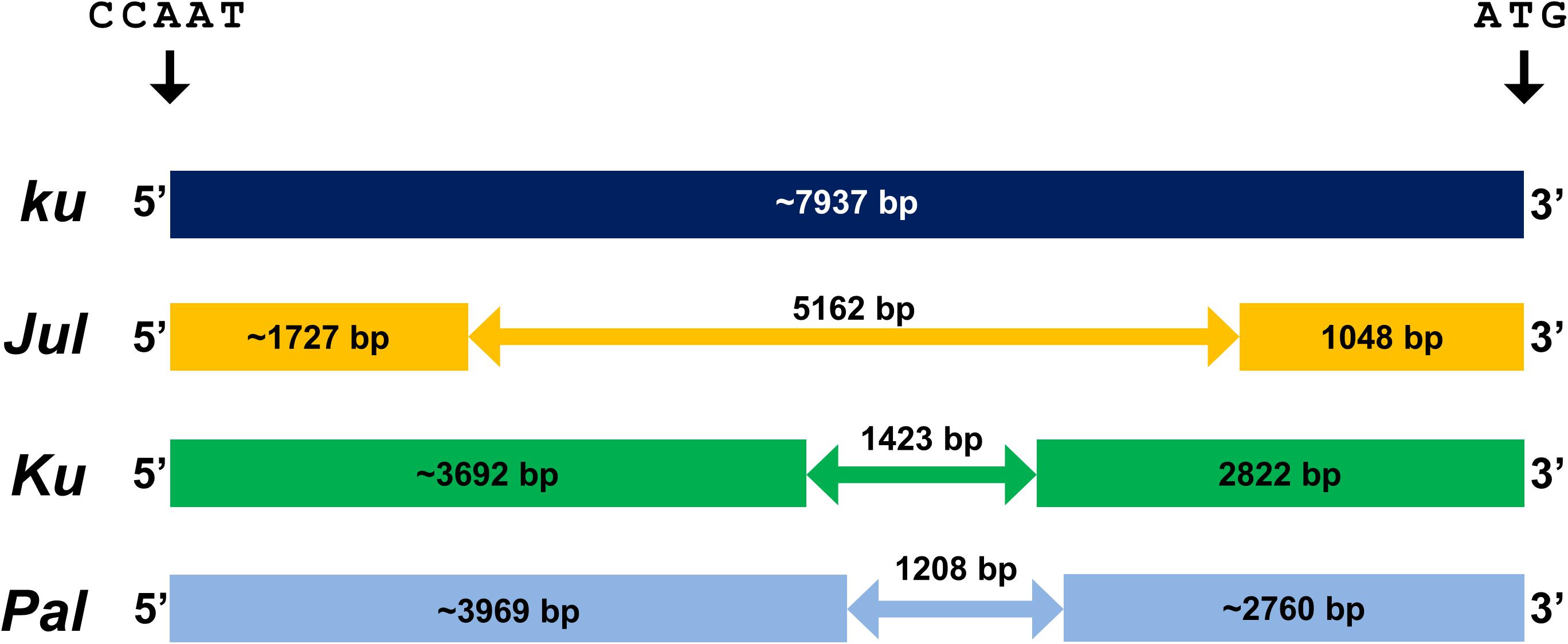
Figure 1. Indel variation in the promoter sequence of LanFTc1 gene controlling flowering induction in L. angustifolius. Allele ku is typical for wild populations, alleles Jul and Ku are present only in domesticated germplasm, whereas allele Pal was found only in wild germplasm from Palestine. The position is given in relation to the first nucleotide of CCAAT-box (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017; Taylor et al., 2019).
Ninety-two L. angustifolius accessions with known LanFTc1 promoter allele composition were evaluated for phenotypic response to vernalization in two consecutive years, 2014 and 2015 (Table 2). Lines carrying wild ku allele revealed the longest vegetative phase, ranging from 61.2 ± 0.9 to 116.2 ± 3.0 days in ‘2014 and from 79.0 ± 5.8 to 101.5 ± 3.9 days in ‘2015. These lines demonstrated also high vernalization responsiveness, highlighted by the acceleration of flowering after vernalization by 13.9 to 66.4 days in 2014, and by 11.6 to 35.6 days in 2015. The line carrying Pal allele showed intermediate phenology and flowered about 20-35 days quicker than the average ku genotype, namely after ∼53 days from sowing in ‘2014 and ∼68 days in ‘2015. The vernalization procedure advanced flowering induction in this line by about two weeks. Accessions carrying domesticated Ku allele were very early and started flowering after 37.2 ± 0.4 to 46.2 ± 8.9 days in 2014, and after 54.0 ± 0.0 to 59.6 ± 2.6 days in 2015. This set contained germplasm low-responsive to vernalization, which accelerated flowering time by up to ∼8.6 days, as well as some truly thermoneutral accessions, which flowered at the same time regardless of the vernalization procedure. Lines carrying domesticated European Jul allele also revealed very early phenology, manifested by the onset of flowering after 37.0 ± 1.3 to 41.0 ± 5.9 days in 2014, and after 52.2 ± 1.5 to 60.8 ± 1.8 days in 2015. Most Jul lines were fully thermoneutral. On average, Jul lines flowered earlier than Ku lines in both years, however taking into consideration standard errors resulting from variability between biological replicates, differences between mean values were not statistically significant. Results of time to flowering and vernalization responsiveness in relation to LanFTc1_INDEL2 marker polymorphism are provided in Supplementary Table 8 (2014) and Supplementary Table 9 (2015).
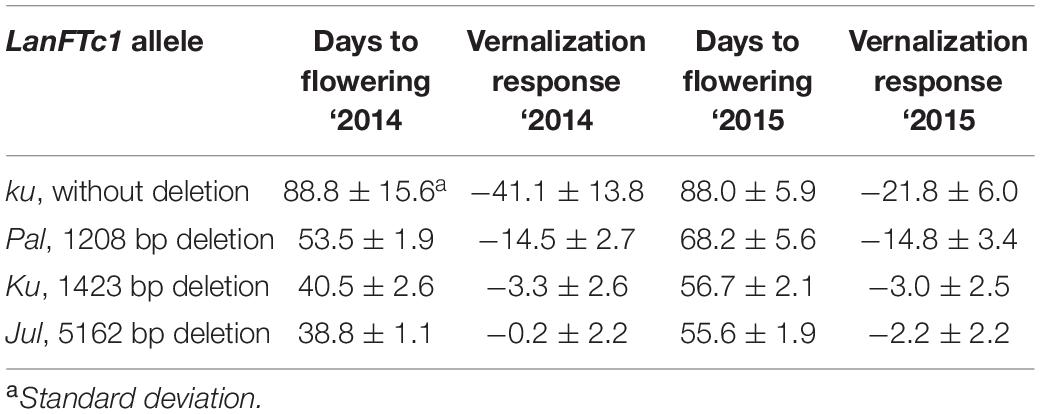
Table 2. Comparison of flowering time and vernalization responsiveness of L. angustifolius germplasm carrying Jul, Ku, Pal, and ku LanFTc1 alleles, cultivated in a greenhouse under natural long days (LD).
Based on the results of LanFTc1_INDEL2 marker screening and vernalization responsiveness, three lines were selected for vernalization response phenotyping under 8-h (SD) and 16-h (LD) photoperiods: 84A:476 carrying domesticated Ku allele (early flowering, thermoneutral), Palestyna carrying wild Pal allele (moderately flowering and responsive to vernalization), and P27255 carrying wild ku allele (late flowering and highly responsive to vernalization). 83A:476 was revealed to be the earliest line in both photoperiods, followed by Palestyna (Table 3). Under SD, 83A:476 accelerated transition between phases in response to vernalization by about 5 days, whereas under LD by about 3 days. These responses were higher in Palestyna, amounting to about 12-19 days and 3-5 days, respectively. P27255 did not flower during the experiment (90 days) in all variants except vernalized plants under LD.
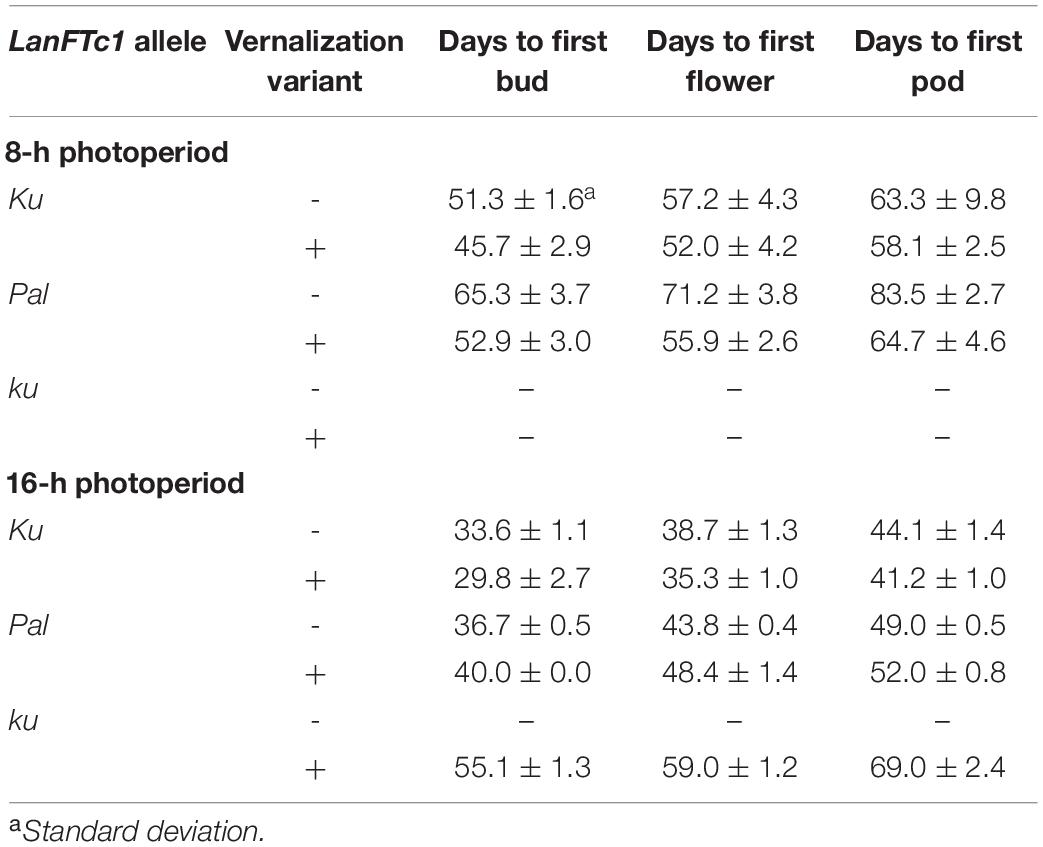
Table 3. Number of days to first bud, flower, and pod in L. angustifolius germplasm carrying Ku, Pal, and ku LanFTc1 alleles, cultivated under 8- and 16-h photoperiods.
83A:476 (Ku), Palestyna (Pal), and P27255 (ku) grown under SD and LD conditions were used for gene expression profiling (Supplementary Data Sheet 1). The vernalization responsiveness of analyzed genes was calculated as a mean fold change of expression in vernalized plants compared to non-vernalized control (averaged across all day terms and dates measured for a particular line), as summarized in Table 4. The circadian clock responsiveness were calculated as a mean fold change of expression occurring between the morning and the evening terms (averaged across all dates measured for a particular line) and are provided in Table 5. The trend in expression level during plant growth was calculated as a fold change of expression occurring between the first and the last term (based on maximum daily values) and is provided in Table 6.
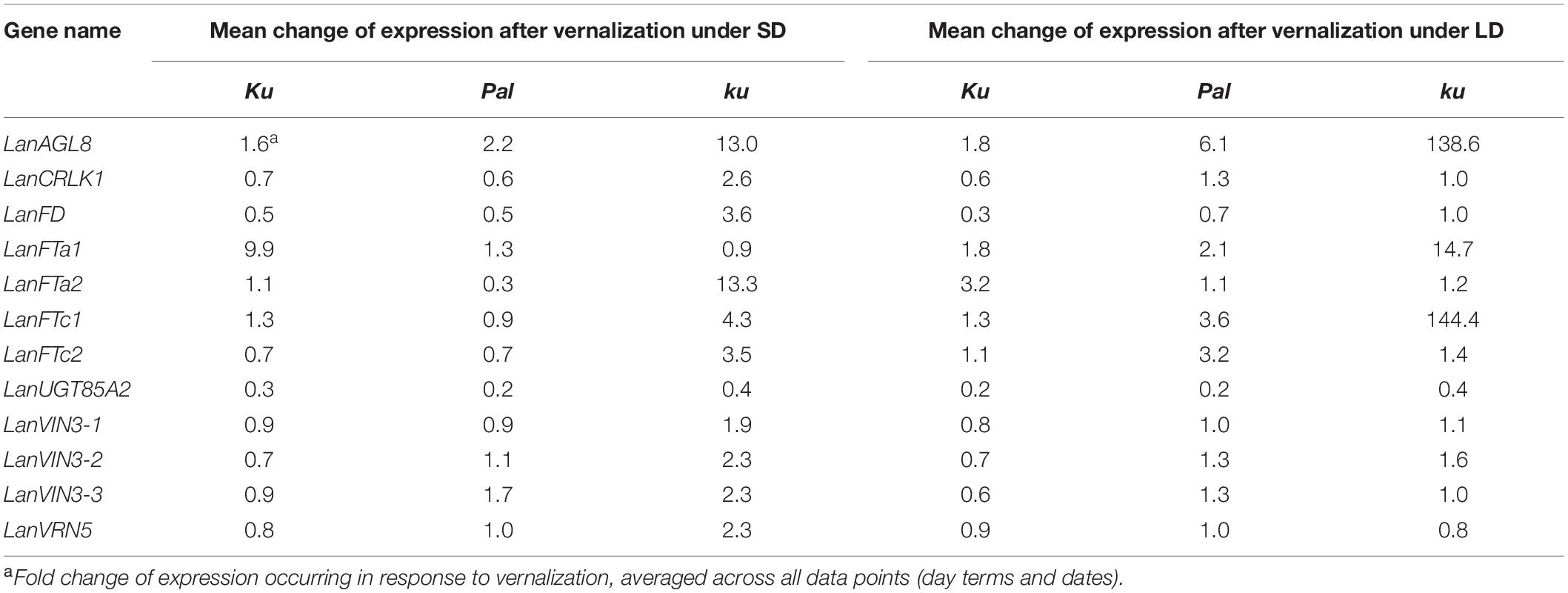
Table 4. Vernalization responsiveness of analyzed genes in L. angustifolius germplasm carrying Ku, Pal, and ku alleles, cultivated under 8-h (SD) and 16-h (LD) photoperiods.
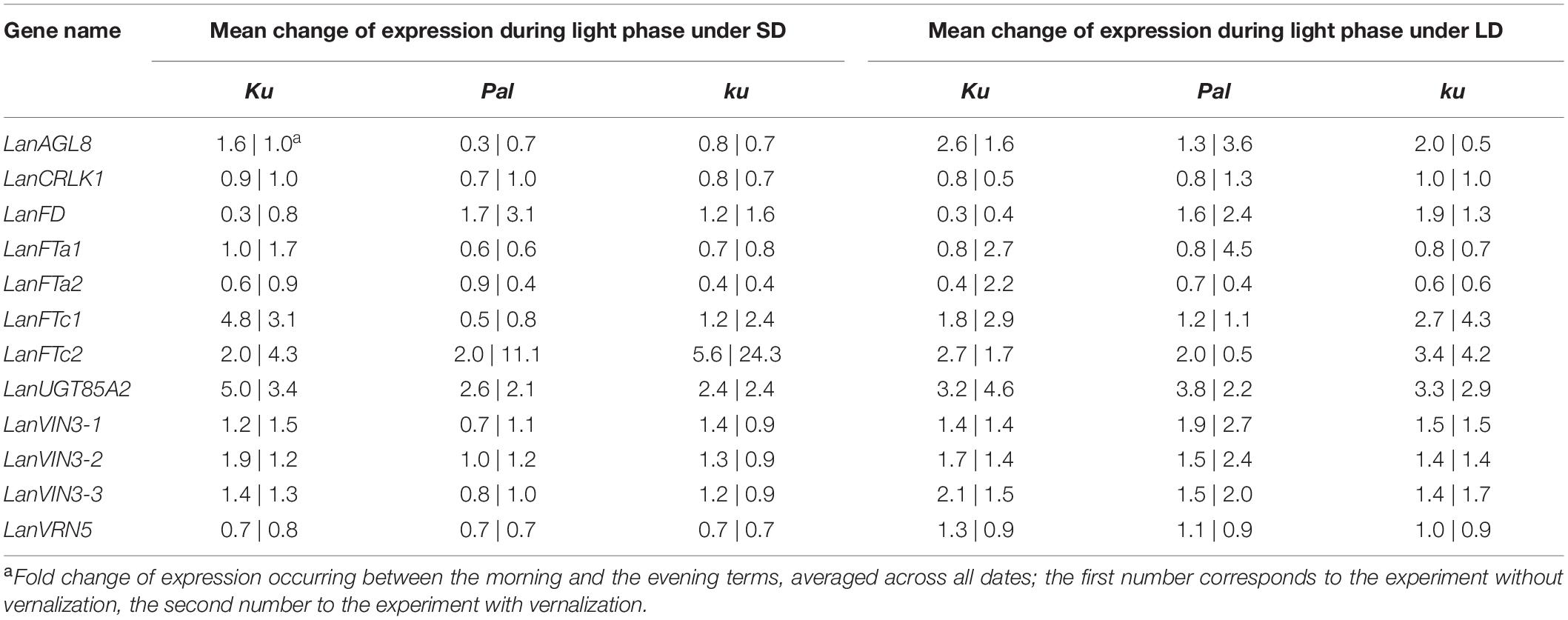
Table 5. Circadian clock responsiveness of analyzed genes in L. angustifolius germplasm carrying Ku, Pal, and ku alleles, cultivated under 8-h (SD) and 16-h (LD) photoperiods without vernalization and with vernalization.
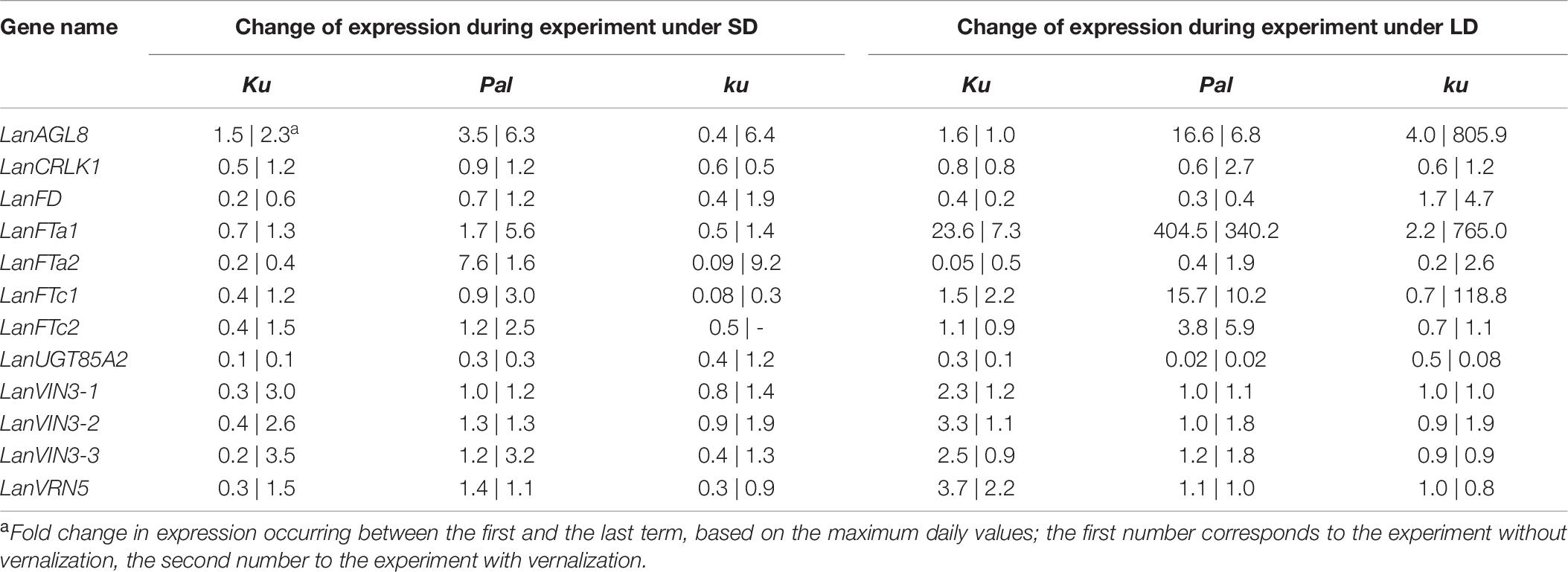
Table 6. Change of expression level of analyzed genes in L. angustifolius germplasm carrying Ku, Pal, and ku alleles, during plant growth under 8-h (SD) and 16-h (LD) photoperiods.
First, we analyzed the expression of the LanFTc1 gene, which is considered as the major controller of vernalization responsiveness and early flowering in L. angustifolius (Nelson et al., 2017; Taylor et al., 2019). The studied genotypes showed different patterns of LanFTc1 expression in response to photoperiod, vernalization, and circadian rhythm.
Under SD, LanFTc1 expression in non-vernalized plants was the highest in 83A:476 and the lowest in P27255 (Figure 2A). Indeed, LanFTc1 expression in 83A:476 was up to 2.4 times higher than in Palestyna (P = 0.0016) and 460-2213 times higher than in P27255 (P = 0.0000). After vernalization, LanFTc1 expression in 83A:476 was up to 4.5 times higher than in Palestyna (P = 0.0064) and up to 686 times higher than in P27255 (P = 0.0029). The difference of LanFTc1 expression between 83A:476 and P27255 increased during plant development in both vernalization variants, whereas for the pair 83A:476 and Palestyna it was decreasing. Under LD, LanFTc1 expression was the highest in Palestyna and the lowest in P27255 (Figure 2B). Namely, in non-vernalized plants, expression in Palestyna was 1.8-5.4 times higher than in 83A:476 (P = 0.0125) and 121-2642 times higher than in P27255 (P = 0.0085), whereas in vernalized plants it was 1.5-13.8 times higher in Palestyna than in 83A:476 (not significant, NS) and 24-276 times higher than in P27255 (P = 0.0094). The difference to Palestyna was increasing during plant growth for 83A:476 in both vernalization variants and P27255 in the absence of vernalization.
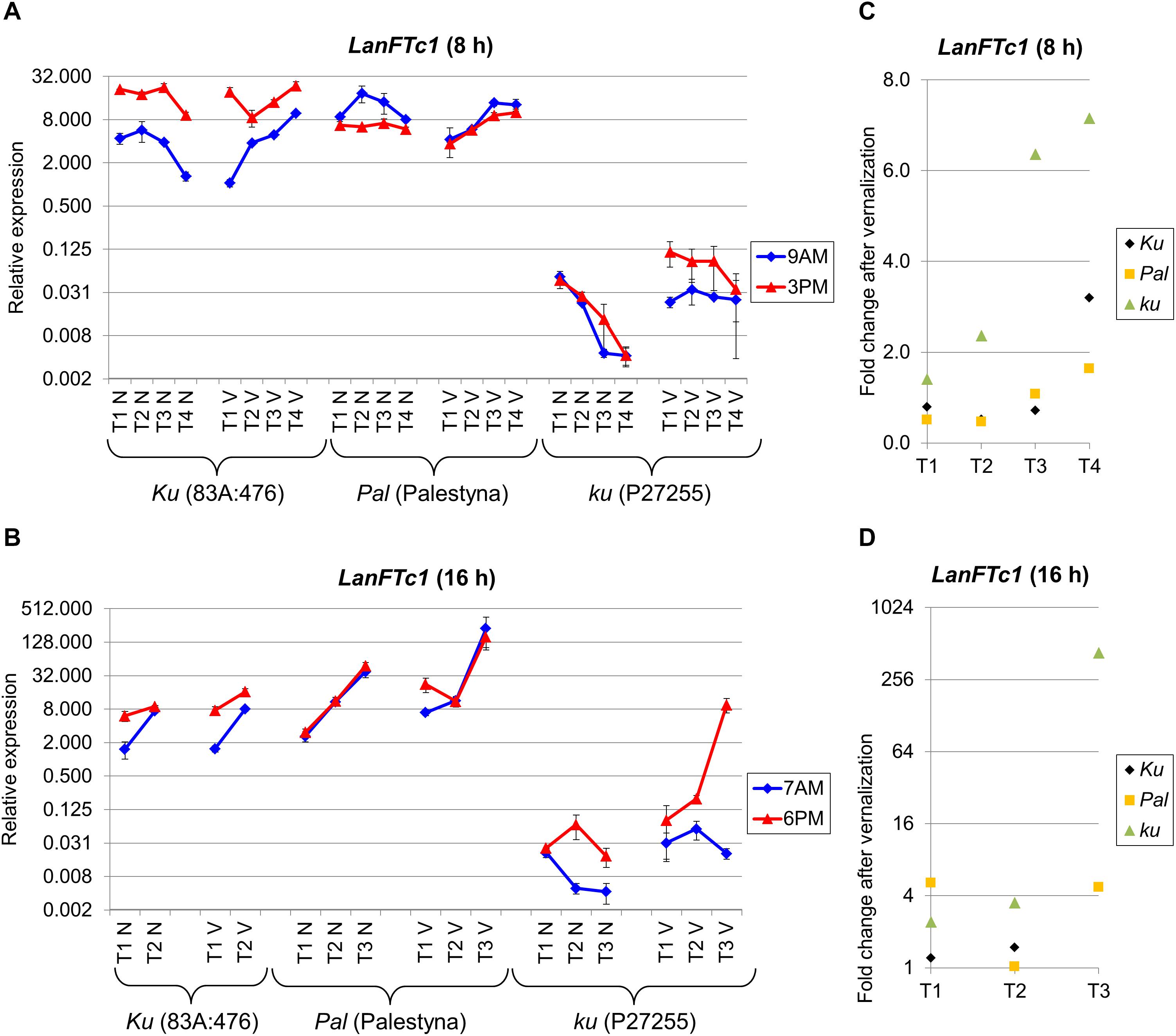
Figure 2. Gene expression profile of the LanFTc1 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under an 8-h photoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates. A logarithmic scale was used to accommodate observed large differences in gene expression values.
Vernalization influence on LanFTc1 expression in P27255 was manifested by up to a 7.2-fold increase under SD (NS) (Figure 2C) and up to 427-fold increase under LD (P = 0.0125) (Figure 2D). The vernalization effect under SD was changing in 83A:476 from a 0.5-fold decrease (P = 0.0115) to a 3.2-fold increase (P = 0.0094) and similarly in Palestyna from a 0.5-fold decrease (P = 0.0291) to 1.6-fold increase (NS). This effect under LD was neutral in 83A:476 and positive in Palestyna, up to 5.1-fold increase (P = 0.0351).
The circadian clock regulation differed between genotypes and partially between environments. In 83A:476, LanFTc1 expression was generally higher in the evening than in the morning in all combinations of the photoperiod and vernalization (up to 18.4-fold increase, P = 0.0008). In Palestyna, LanFTc1 expression under SD was usually higher in the morning (up to 2.9-fold increase, P = 0.0172), whereas under LD this effect was variable. In P27255, LanFTc1 expression was higher in the evening, especially under LD after vernalization (P = 0.0046).
Besides LanFTc1, three other FT genes, namely LanFTa1, LanFTa2, and LanFTc2, were analyzed to complement our perspective on FT clade transcriptional activity in L. angustifolius response to major environmental cues. LanFTa1 gene expression under SD did not reveal any significant trend during plant growth and differences between genotypes were also usually not significant (Figure 3A). The relative level of expression was rather low (mean 0.38). A different pattern was observed under LD, when the expression was highly induced before flowering compared to the first term, namely 24-fold (P = 0.045) in non-vernalized 83A:476 (7-fold in vernalized, NS), 405-fold (P = 0.0051) in non-vernalized Palestyna (340-fold in vernalized, P = 0.0015), and 765-fold in vernalized P27255 (Figure 3B). In non-vernalized P27255 LanFTa1 expression remained at a low level but this genotype did not flower in such conditions.
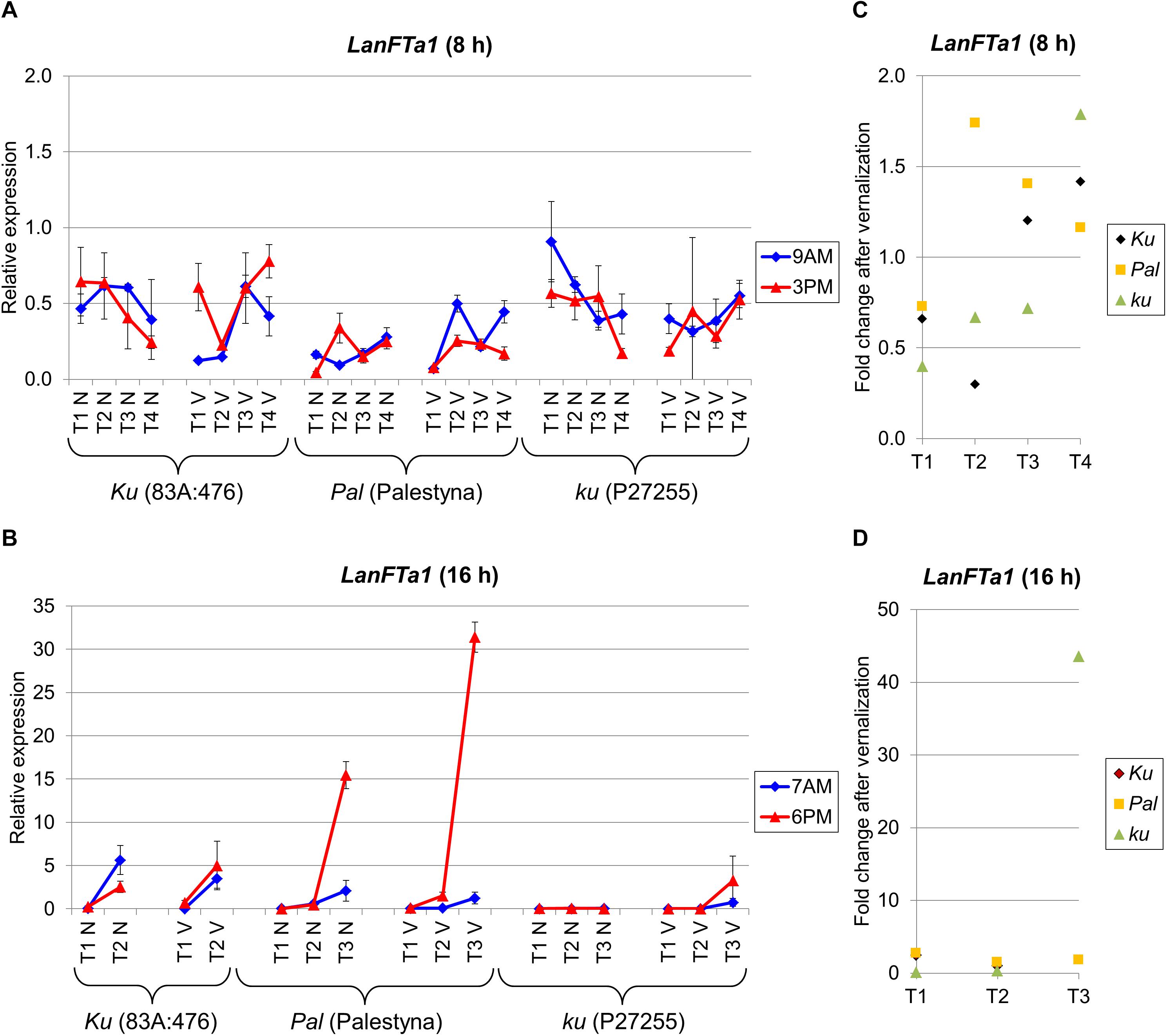
Figure 3. Gene expression profile of the LanFTa1 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under 8-h photoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates.
The vernalization effect on LanFTa1 expression under LD in 83A:476 and Palestyna was slightly positive, up to 2.5-fold (NS) and 2.8-fold (P = 0.0011) increase, respectively (Figures 3C,D). In P27255, the vernalization influence was initially moderately negative (0.14-fold and 0.35-fold decrease, P = 0.0002) but became highly positive just before flowering (43.6-fold increase). The circadian regulation was not stable across terms and genotypes, however, when LanFTa1 was induced before flowering in Palestyna under LD, its expression was significantly higher in the evening than in the morning (up to 25.1-fold increase, P = 0.0004).
LanFTa2 gene expression was very low in both photoperiods, amounting to mean values of 0.095 under SD and 0.004 under LD (Supplementary Figures 1A,B). There was remarkable induction of expression in P27255 after vernalization under SD (up to 38-fold, P = 0.0002), however, the relative expression values achieved were much lower than those observed for LanFTc1 and LanFTa1 (Supplementary Figures 1C,D). This pattern was not recreated under LD.
LanFTc2 expression was on a low level in all lines under both photoperiods (Supplementary Figures 2A,B). A moderate induction by vernalization in P27255 under SD (up to 7.3-fold increase, P = 0.0187) and in Palestyna under LD (up to 4.7-fold increase, P = 0.0023) was observed but obtained levels were less than half of the mean expression obtained for control genes (Supplementary Figures 2C,D).
Besides homologs constituting the L. angustifolius FT clade (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017), four novel candidate genes (LanAGL8, LanFD, LanUGT85A2, and LanCRLK1), recently considered to be putatively involved in Ku-based response (Plewiński et al., 2019), were profiled in this study. LanAGL8 is an L. angustifolius homolog of the A. thaliana FRUITFULL gene participating in flowering time control, meristem identity, and fruit development (Mandel and Yanofsky, 1995; Gu et al., 1998). LanAGL8 expression was consecutively increasing during plant growth for all studied combinations of lines, photoperiod, and vernalization variants except non-vernalized P27255 under SD. However, there were considerable differences in the observed expression levels between genotypes. Under SD without vernalization, expression of the LanAGL8 gene in 83A:476 (Ku) was approximately 7-17 times higher (P = 0.0028) than in Palestyna (Pal) and 1420-6090 times higher (P = 0.0035) than in P27255 (ku) (Figure 4A). The influence of vernalization resulted in a reduction of differences in LanAGL8 expression between 83A:476 and Palestyna by 11–23%, and between 83A:476 and P27255 by 30–92%. Under LD without vernalization, LanAGL8 expression in 83A:476 was higher up to 12-fold (P = 0.0086) than in Palestyna and up to 1805-fold (P = 0.0001) than in P27255 (Figure 4B). The application of vernalization reduced these differences considerably. To summarize, Palestyna revealed an intermediate LanAGL8 expression profile, however, it was much more like 83A:476 than P27255.
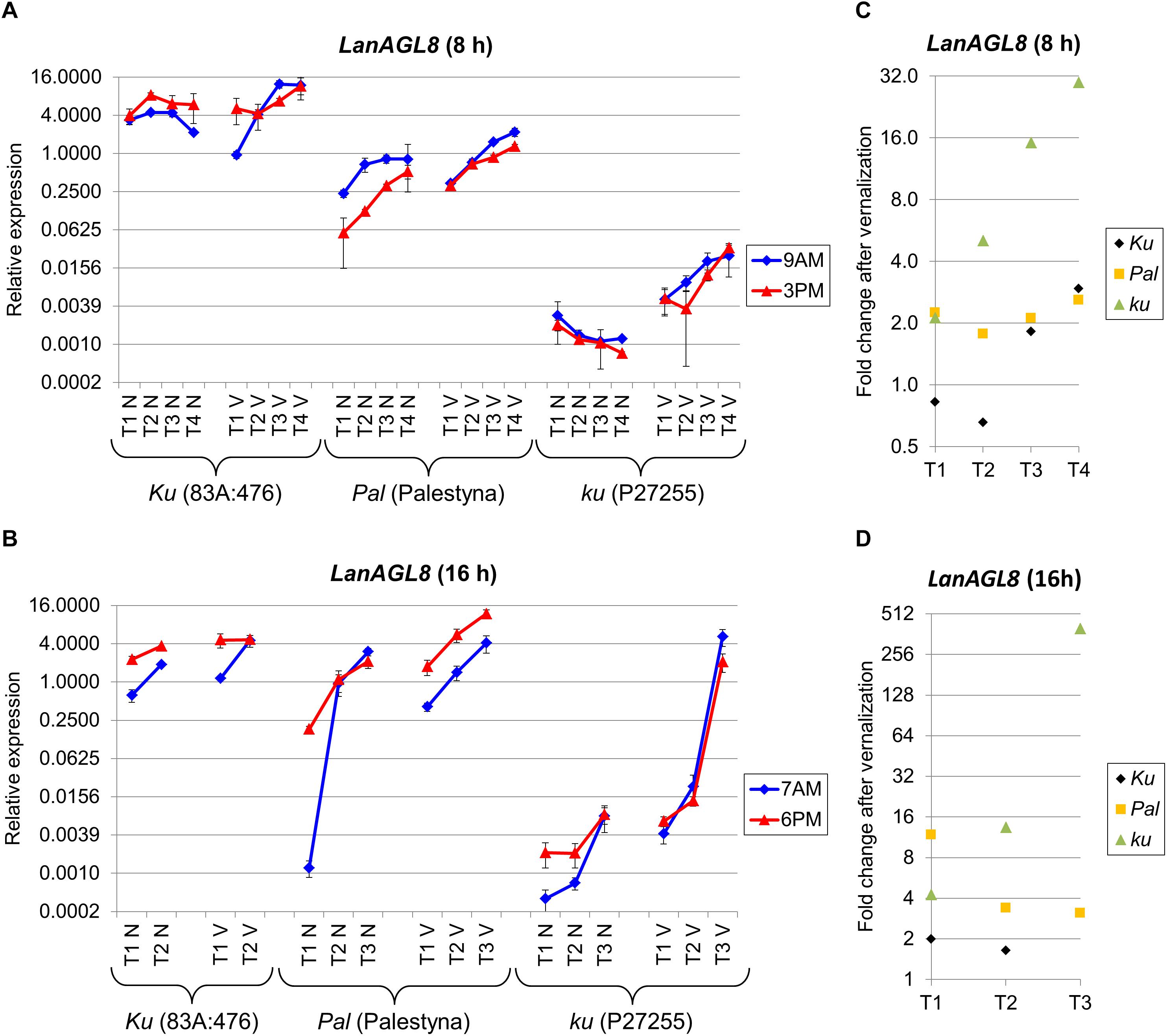
Figure 4. Gene expression profile of the LanAGL8 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under 8-h photoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates. A logarithmic scale was used to accommodate observed large differences in gene expression values.
The LanAGL8 gene revealed high positive responsiveness to vernalization in P27255 in both photoperiods, and this effect was consecutively increasing during plant growth until flowering. Namely, the change of LanAGL8 expression in P27255 after vernalization was raised from 2.1-fold (NS) to 29.8-fold (P = 0.0175) under SD (Figure 4C), and from 4.3-fold (P = 0.0121) to 398.2-fold (P = 0.0004) under LD (Figure 4D). Vernalization was also inductive in Palestyna, providing an up to 2.6-fold increase (P = 0.0329) of LanAGL8 expression under SD (effect stable across sampling terms) and up to 11.8-fold increase (P = 0.0297) under LD (effect consecutively decreasing). In the 83A:476 line, the influence of vernalization was the lowest, yielding up to a 2.9-fold increase of LanAGL8 expression under SD (P = 0.0066) and up to a 2.0-fold increase under LD (P = 0.0315).
The LanAGL8 gene revealed diversified circadian clock regulation between genotypes and environments. Under SD, its expression was higher in the morning than in the evening in Palestyna and P27255, whereas in 83A:476 this relation was the opposite. Moreover, vernalization partially diminished these differences. Under LD, LanAGL8 expression was usually higher in the evening terms for all lines, however, during the growth of vernalized P27255 this trend reversed.
LanCRLK1 is an L. angustifolius homolog of the A. thaliana CRLK1 gene which participates in response to low temperature in a calcium-dependent manner (Yang et al., 2010). Changes in LanCRLK1 expression level were usually not related to the progress of plant growth, except P27255 cultivated under SD, which revealed a stable decrease of LanCRLK1 both in non-vernalized and vernalized variants. The expression of LanCRLK1 was higher under SD than under LD. Moreover, under SD without vernalization, LanCRLK1 expression was the highest in Palestyna (up to 2.7-fold increase compared to 83A:476, P = 0.0002) and the lowest in P27255 (about 0.4-fold decrease, P = 0.0099). However, after vernalization, expression was highest in P27255 (up to a 3.5-fold increase compared to 83A:476, P = 0.0005) and lowest in 83A:476 (Figure 5A). The differences in the LanCRLK1 expression between genotypes under LD were generally not significant (Figure 5B).
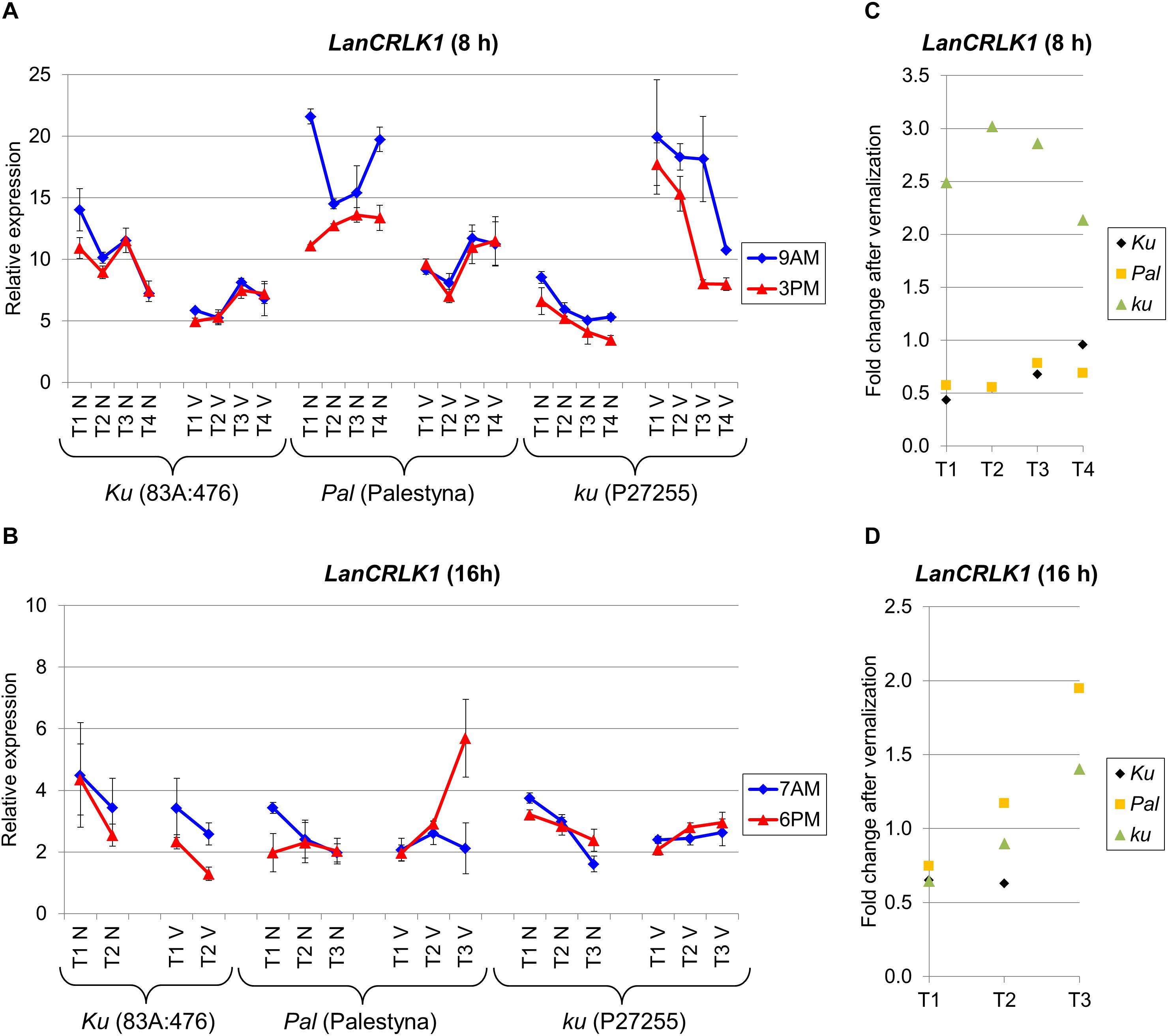
Figure 5. Gene expression profile of the LanCRLK1 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under 8-h photoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates.
The effect of vernalization on LanCRLK1 expression under SD was negative (from 0.4-fold to 0.7-fold decrease, P = 0.0009) or neutral in 83A:476, negative in Palestyna (from 0.6-fold to 0.8-fold decrease, P = 0.0000), and positive in P27255 (up to 3.0 fold increase, P = 0.0000) (Figure 5C). Under LD, the vernalization effect was also negative in 83A:476 (0.6-fold decrease, NS), however, in the remaining two lines, the effect changed during plant growth from negative to positive: from a 0.7-fold decrease to 1.9-fold increase in Palestyna (all NS) and from 0.6-fold decrease (P = 0.0001) to 1.4-fold increase in P27255 (P = 0.0107) (Figure 5D). The influence of the circadian clock on LanCRLK1 expression was variable in both photoperiods and usually not significant.
LanFD is an L. angustifolius homolog of the A. thaliana FD gene which triggers flowering based on FT-mediated signaling (Abe et al., 2005). LanFD expression in non-vernalized plants was usually the highest at the first sampling date and decreased during plant growth in both photoperiods, except in P27255 cultivated under LD. However, in vernalized plants, this decreasing trend was diminished or even reversed, as observed for P27255. Under SD, LanFD expression was the highest in Palestyna (1.1–5.6 times higher than in 83A:476, P = 0.0024) and the lowest in P27255 (0.3–0.9 times lower than in 83A:476, P = 0.0123) (Figure 6A). Interestingly, these relations were changed after vernalization as follows: LanFD expression in P27255 compared to 83A:476 was 3.2–12.2 times higher (P = 0.0011), whereas in Palestyna 2.1–7.4 times higher than in 83A:476 (P = 0.0001). Under LD without vernalization, differences in LanFD expression between genotypes were rather low, accounting for up to a 2.3-fold increase in Palestyna (P = 0.0205) and up to a 5.0-fold increase in P27255 (P = 0.0000) compared to 83A:476. However, vernalization exaggerated these contrasts and observed LanFD expression in P27255 was up to 25.7 times higher than in 83A:476 (P = 0.0078) and up to 5.5 times higher than in Palestyna (P = 0.0032) (Figure 6B).
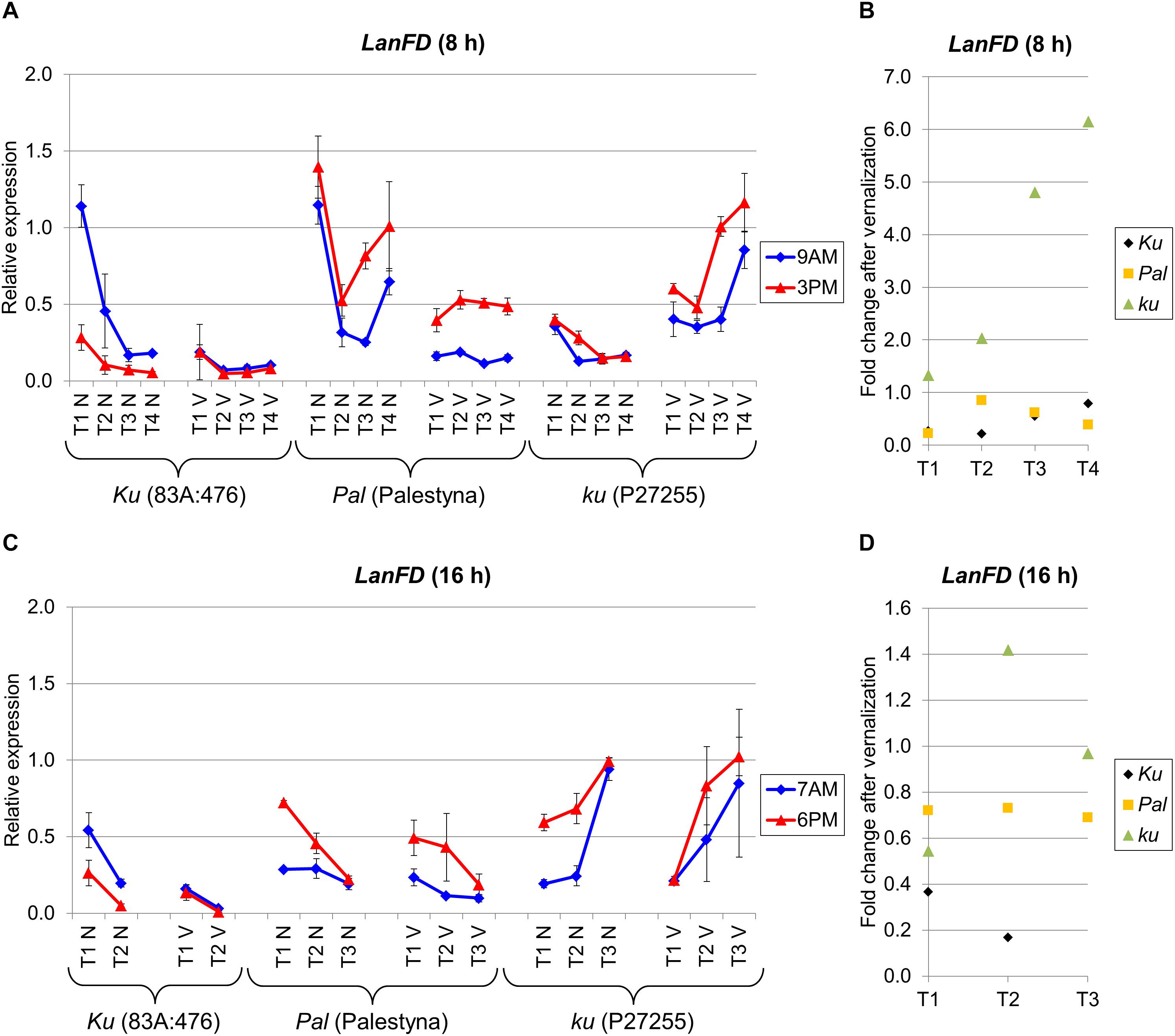
Figure 6. Gene expression profile of the LanFD gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under 8-h photoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates. A logarithmic scale was used to accommodate observed large differences in gene expression values.
The effect of vernalization on LanFD expression was negative in 83A:476 and Palestyna under both photoperiods (0.17-fold to 0.85-fold decrease), positive in P27255 under SD (up to 6.2-fold increase), and unstable in P27255 under LD (from 0.5-fold decrease to 1.4-fold increase) (Figures 6C,D). Genotypes differed in the circadian clock control of LanFD expression. Generally, in 83A:476 the levels were higher in the morning (up to 4.4 fold-increase), whereas in Palestine and P27255 in the evening (up to 4.5-fold and 3.1-fold increase, respectively).
LanUGT85A2 is considered as an L. angustifolius homolog of the A. thaliana UDP-glycosyltransferase 85A2 gene. The expression of this gene was the highest at the first sampling date in all lines and consecutively decreased during the experiment. Palestyna was revealed to have the biggest decrease of LanUGT85A2 during the transition from juvenile to generative phase, whereas P27255 had the lowest. Indeed, this decrease of LanUGT85A2 expression during plant growth under SD reached approximately 0.1-fold for 83A:476 (P = 0.0114), 0.3-fold for Palestyna (P = 0.0033) and 0.4-fold for P27255 (P = 0.0003), whereas under LD about 0.3-fold for 83A:476 (P = 0.0501), 0.02-fold for Palestyna (P = 0.0182) and 0.5-fold for P27255 (P = 0.0196).
Under SD, the highest first-term LanUGT85A2 expression was observed in 83A:476, about 1.7-fold higher than in Palestyna (P = 0.0448) and P27255 (NS) (Figure 7A). These differences were doubled by vernalization. Contrary, under LD, the highest first-term expression was in P27255, 1.1-fold higher than in Palestyna (NS) and 3.7-fold higher than in 83A:476 (P = 0.0006), and these differences were tripled by vernalization (Figure 7B).
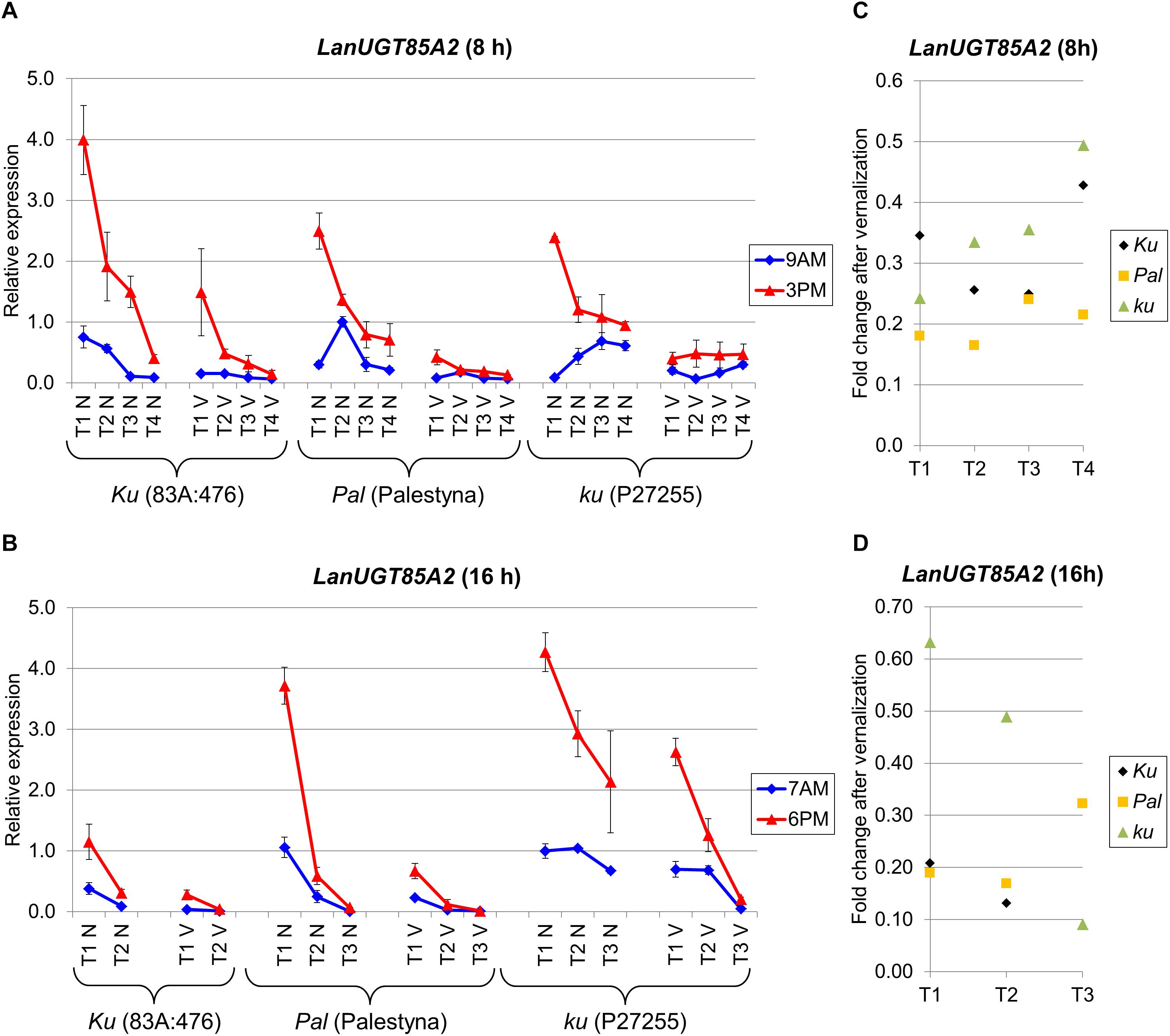
Figure 7. Gene expression profile of the LanUGT85A2 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku). (A) expression under 8-hphotoperiod, (B) expression under 16-h photoperiod, (C) vernalization response under 8-h photoperiod, (D) vernalization response under 16-h photoperiod. T1-T4 stands for sampling terms (Supplementary Table 5), V for vernalized plants, and N for non-vernalized plants. Timespan of photoperiods: 8-h from 4 AM to 8 PM, 16-h from 4 AM to 8 PM. Two references were used for normalization (LanDExH7 and LanTUB6) and one sample (LanTUB6) for inter-run calibration. Error bars indicate a standard deviation of 3 biological replicates, each representing a mean of 3 technical replicates.
Vernalization was observed to have a stable negative effect on LanUGT85A2 gene expression, highlighted by an average 0.29-fold decrease under SD (Figure 7C) and a 0.28-fold decrease under LD (Figure 7D). Response to vernalization was the weakest in P27255 (0.38-fold) and the strongest in Palestyna (0.21-fold). Such a disproportion in vernalization response, combined with the observed difference in decreasing trend during plant growth, resulted in the highest LanUGT85A2 expression in the P27255 line at the end of the experiment under both photoperiods (up to 31-fold increase compared to 83A:476 under LD, P = 0.0233).
The direction of circadian clock control was coherent across all genotypes and environments. LanUGT85A2 expression was higher in the evening than in the morning for all terms, both with and without vernalization. Under SD, it increased during the day up to 13.9-fold in 83A:476 (P = 0.0165), 8.3-fold in Palestyna (P = 0.0075) and 28.2-fold in P27255 (P = 0.0000). Under LD, it increased up to 7.4-fold in 83A:476 (P = 0.0326), 12.8-fold in Palestyna (P = 0.0005) and 4.4-fold in P27255 (P = 0.002).
The L. angustifolius genome contains three homologs (named here as LanVIN3-1, LanVIN3-2, and LanVIN3-3) a VIN3 gene that is involved in the vernalization response in A. thaliana (Sung and Amasino, 2004). Therefore, the transcriptional activity of these genes was also profiled. In general, LanVIN3-1 revealed the highest expression, whereas LanVIN3-3 the lowest (see Supplementary Figures 3–5).
All three copies revealed differences in expression levels between genotypes, usually in descending order, 83A:476 – Palestyna – P27255. Maximum differences between 83A:476 and P27255 expression under LD reached 5.9-fold (LanVIN3-1, P = 0.0016), 25.6-fold (LanVIN3-2, P = 0.0061) and 8.4-fold (LanVIN3-3, P = 0.0059), whereas under SD these values were as follows: 4.8-fold (LanVIN3-1, P = 0.0135), 10.8-fold (LanVIN3-2, P = 0.0036) and 5.5-fold (LanVIN3-3, P = 0.0072). The expression levels of all three genes under LD were usually significantly higher in the evening than in the morning in all lines, whereas under SD, diurnal variations were frequently not significant or variable. Photoperiod conditions had a significant influence on expression changes during plant growth, especially for 83A:476. The expression levels of these genes in non-vernalized 83A:476 under LD increased by 2.3-fold (LanVIN3-1, P = 0.0008), 3.3-fold (LanVIN3-2, P = 0.0048) and 2.5-fold (LanVIN3-3, P = 0.0036) whereas under SD they decreased by up to 0.32-fold (LanVIN3-1, P = 0.0000), 0.37-fold (LanVIN3-2, P = 0.0110), and 0.24-fold (LanVIN3-3, P = 0.0009). The vernalization effect differed between genotypes and photoperiods. Under LD it was significant for 83A:476 (repression of all three genes, up to 0.43-fold) and Palestyna (induction of LanVIN3-2 and LanVIN3-3, up to 2.0-fold), whereas under SD it was significant for 83A:476 (changing from repression, up to 0.18-fold, to induction, up to 2.3-fold) and P27255 (induction, up to 3.6-fold). To summarize, all LanVIN3 homologs revealed different transcriptomic responses to vernalization than the LanFTc1 and LanAGL8 genes.
Besides VIN3, VRN5 protein is also considered to participate in the vernalization-induced epigenetic silencing in A. thaliana (Greb et al., 2007). Therefore, the expression of the latter gene was also profiled to supplement the analysis. All three L. angustifolius copies, LanVRN5-1, LanVRN5-2, and LanVRN5-3, were analyzed using one universal primer pair (Supplementary Figure 6). LanVRN5 clade revealed an expression profile relatively similar to LanVIN3, especially to LanVIN3-2. Like LanVIN3 genes, LanVRN5 in 83A:476 revealed an increase of expression during plant growth under LD and decrease under SD. All lines showed a significant decrease of expression during the light phase under SD and usually not a significant diurnal trend under LD. In both photoperiods without pre-sowing vernalization, 83A:476 revealed a significantly higher expression of LanVRN5 than Palestyna and P27255. However, the vernalization responsiveness of LanVRN5 considerably differed between photoperiods and genotypes. Under LD, only P27255 was responsive (showing repression), whereas under SD all lines responded but in different ways and with changing response during plant growth: namely from repression to induction in 83A:476, from induction to repression in Palestyna, and by continuously increasing induction in P27255. In vernalized plants under SD, LanVRN5 expression was the highest in P27255.
FT is a major flowering time integrator gene, gathering signals from several pathways that detect environmental conditions (Turck et al., 2008). First studies on the conservation of A. thaliana genes in legumes have revealed the presence of the well-preserved homologs of many flowering time regulatory genes, including some representatives of FT clade found in Pisum sativum L., Medicago truncatula L., and Glicyne max (L.), Merrill (Hecht et al., 2005). When the first gene-based linkage map of L. angustifolius was published, it highlighted a conserved collinear block shared between the fragment of M. truncatula chromosome 7 and the so-called linkage group LG01 (currently NLL-10), carrying major early flowering locus Ku (Nelson et al., 2006). It was later revealed that the syntenic region in M. truncatula contained several FT homologs (Young et al., 2011). The development of a bacterial artificial chromosome library for the L. angustifolius nuclear genome opened a possibility of gene cloning by DNA hybridization and the sequencing of selected clones (Kasprzak et al., 2006). Such an approach, combined with novel high-throughput sequencing techniques and gene expression profiling, resulted in the identification of a candidate gene for Ku and enabled the formulation of the hypothesis that a 1423 bp deletion in the promoter region of this gene is a causal mutation conferring early flowering phenotype (Nelson et al., 2017). Then, two other overlapping deletion variants, covering 1208 bp (Pal) and 5162 bp (Jul) were identified (Taylor et al., 2019). In the A. thaliana, a variation in promoter length was found to modulate the photoperiodic response of FT, and some important regulatory blocks contributing to this response were identified (Adrian et al., 2010; Liu et al., 2014). However, comparative mapping revealed a low sequence similarity between LanFTc1 and A. thaliana FT promoters, except RE-alpha and CCAAT boxes which were found at expected positions and indicated that FT promoter length in L. angustifolius may be at least as big as in A. thaliana (up to 7 kb) (Ksia̧żkiewicz et al., 2016). All recognized LanFTc1 deletions conferring Ku, Pal, and Jul alleles are located downstream of the pair of CCAAT boxes marking a putative beginning of the functional promoter (Ksia̧żkiewicz et al., 2016; Nelson et al., 2017; Taylor et al., 2019). Thus, L. angustifolius Ku, Pal, and Jul alleles have both distal and proximal regions preserved (Taylor et al., 2019). However, deletion sequences encompassed candidate binding sites for many transcription factors, including those already evidenced to be involved in FT regulation in model plants (Nelson et al., 2017; Taylor et al., 2019). In A. thaliana, similarly to L. angustifolius, the functional FT promoter indels also retained distal and proximal regions (Adrian et al., 2010; Liu et al., 2014). On the contrary, the study involving the FT promoter from cotton provided evidence that the proximal region might play an important role in this species (Sang et al., 2019). To our knowledge, a model revealed for L. angustifolius is the only known legume example of FT promoter indel variation. However, in many legumes, FT genes were found to be associated with flowering traits. In the economically most important legume crop worldwide, a soybean, Glycine max, mutations in FT genes are responsible for at least three loci conferring early/late flowering, namely E9 (GmFT2a), E10 (GmFT4), and qDTF-J1 (GmFT5a) (Takeshima et al., 2016; Zhao et al., 2016; Samanfar et al., 2017). Moreover, natural variations of the GmFT2b sequence are associated with soybean adaption to high−latitude regions (Chen et al., 2020). In M. truncatula, vernalization responsiveness and early flowering are conferred by the FTa1 gene, whereas photoperiod response by the FTb gene (Laurie et al., 2011; Putterill et al., 2013). In pea (Pisum sativum), the FTa1 gene corresponds to the pea GIGAS locus, which is essential for flowering under LD and promoting flowering under SD (Hecht et al., 2011). In chickpea, a major quantitative trait locus (QTL) for the flowering time under SD conditions was mapped in the region containing a cluster of three FT genes (FTa1-FTa2-FTc), which collectively showed upregulated expression in domesticated germplasm (Ortega et al., 2019). In the sister lupin crop species, white lupin (L. albus L.), one of the four major QTLs conferring early flowering and partial vernalization independence was found associated with the FTa1 gene (Rychel et al., 2019). L. angustifolius genome contains two FTa and two FTc genes, which putatively arose from single copies by lineage-specific duplication, whereas the whole FTb subclade is absent (Ksia̧żkiewicz et al., 2016). Indeed, L. angustifolius was recently used as a reference species in several phylogenetic studies addressing the influence of whole-genome and local duplications on the evolutionary fate of selected legume-specific and plant-wide gene clades (Przysiecka et al., 2015; Narożna et al., 2017; Szczepaniak et al., 2018; Czyż et al., 2020). The differences in the expression profiles for FTa and FTc genes, as established in the present study, provided novel evidence supporting the hypothesis on a functional divergence of particular duplicates.
In the present study, based on experiments performed in a greenhouse under natural LD (12–17 h photoperiod), Ku and Jul alleles were found to be associated with early flowering and vernalization independence, Pal with slightly delayed flowering and partial vernalization responsiveness, and ku with late flowering and high vernalization dependence. The same phenomenon was reported for the recent study performed in phytotron under natural 10–12 h photoperiod (intermediate between SD and LD) in Australia (Taylor et al., 2019). However, we observed relatively high differences in flowering time for early lines (Ku, Pal, and Jul) in a greenhouse between 2014 and 2015 repeats, accounting for about two weeks on average (earlier in 2014). These differences can be attributed to the variation in temperature which occurred during these experiments. During the first 70 days of the 2014 experiment, we observed 31 days with maximum temperature ≥25°C and 9 days with ≥30°C, whereas in ‘2015 these numbers were much lower, 3 and 0 days, respectively. The higher temperature could advance flowering because it is shown to strongly accelerate flowering in model plants as well as in many other plant species (Parmesan and Yohe, 2003; Thines et al., 2014). Moreover, the average photoperiod in 2014 was about 2 h longer than in 2015 due to differences in sowing terms. Indeed, our subsequent controlled-environment study revealed that even early L. angustifolius germplasm is responsive to LD conditions and accelerated transition between particular developmental phases by about 18–25 days compared to SD. The observed phenology of the Pal allele can be very beneficial for L. angustifolius cultivation in the era of changing climate, especially in Europe and Australia where the majority of worldwide lupin production occurs. Thus, the European land climate experienced rapid warming in recent decades, resulting in the mean year temperature surge to approximately 2°C above the 1910–1960 average (NOAA, 2020). Climate warming raised multiple challenging issues for grain legume breeders, including higher water deficits and severe drought periods, propagation of pests and diseases as well as de-regulation of temperature-based control of growth and development processes (Vadez et al., 2012; Scheelbeek et al., 2018; Lippmann et al., 2019). Affected regulatory pathways include, among others, the flowering time control (Nelson et al., 2010). The rapid flowering of domesticated germplasm may favor drought escape and adaptation for spring sowing in higher latitudes (Annicchiarico et al., 2010, 2018; Berger et al., 2017). However, the observed extension of the vegetation period has raised the demand for germplasm with intermediate phenology and cross-environment adaptation. Such research was recently initiated in L. albus in three European locations contrasting sowing time (autumn or spring) and climate type (Annicchiarico et al., 2019). Climatic variables were also addressed in an L. angustifolius genome-wide association study, providing some candidate polymorphisms that await further exploitation (Mousavi-Derazmahalleh et al., 2018a). L. angustifolius Pal allele confers flowering time and vernalization responsiveness phenotype intermediating between domesticated and wild lines. As this phenotype is consistent within the large range of photoperiod conditions (8, 10–12, and 16–17 h), it may be found applicable for all regions where lupins are currently cultivated.
Previous studies have highlighted the negative association of LanFTc1 and LanAGL8 gene expression with the number of days to flowering in L. angustifolius, mapping population and the positive direction of such association for LanFD, LanCRLK1, and LanUGT85A2 genes (Nelson et al., 2017; Plewiński et al., 2019). The present study revealed that these genes differ in vernalization responsiveness between genotypes and photoperiods. LanFTc1 and LanAGL8 genes were found to be highly induced by vernalization in wild germplasm, whereas LanUGT85A2 was found to be significantly suppressed (Table 4). LanAGL8 protein sequence revealed the highest similarity to A. thaliana FRUITFULL (FUL, AGAMOUS-LIKE 8, AT5G60910) and APETALA1 (AP1, AGAMOUS-LIKE 7, AT1G69120) genes. Both AP1 and FUL play a role in floral meristem identity but have different functions. AP1 controls the formation of sepals and petals whereas FUL is involved in cauline leaf and fruit development (Irish and Sussex, 1990; Gu et al., 1998). These genes revealed tissue-specific expression during generative organ development (Irish and Sussex, 1990; Mandel and Yanofsky, 1995; Klepikova et al., 2016). In the present study, high levels of LanAGL8 expression were revealed in leaf tissue. Moreover, the expression profiles and vernalization responsiveness of LanAGL8 and LanFTc1 were very similar. Both genes revealed comparable circadian clock control, i.e., morning induction in Pal line under short days. Confronting these observations with the information on a 100% association between the LanFTc1 genotype and flowering time phenotype in a large germplasm collection (Nelson et al., 2017; Taylor et al., 2019), the conclusion can be raised that LanAGL8 acts putatively downstream of LanFTc1 in L. angustifolius, in which LanAGL8 may perform a similar function, like its homolog in cereals, an AP1-like gene called VRN1, which regulates the transition from vegetative to generative phase in response to vernalization and is expressed in many organs, including leaves (Trevaskis et al., 2003; Yan et al., 2003). Indeed, the wheat homolog of FT, (VRN3), activates expression of VRN1 in leaves and shoot apical meristem, promoting flowering under inductive long days (Li et al., 2015). As a MADS box transcription factor, VRN1 binds to many targets in the genome and may regulate many genes, linking vernalization and photoperiod pathways (Deng et al., 2015). Moreover, the allelic diversity of VRN1 copies provides wide plasticity of temperature-based responses in winter wheat (Dixon et al., 2019).
In this study, we also revealed differences in the vernalization responsiveness of a LanFD gene between early and late flowering germplasm. In wheat, FD−like, VRN3, and 14−3−3 proteins form together a florigen activation complex which can bind the VRN1 promoter, therefore a variation in FD expression may modulate the effect of mobile florigen signal (Li et al., 2015). Results obtained in this study, are supported by a significant correlation between the LanFD gene expression profile and vernalization responsiveness in the L. angustifolius mapping population (Plewiński et al., 2019), which indicates that LanFD may contribute to FT regulatory function, especially in the wild, vernalization-responsive germplasm.
Our recent expression quantitative trait loci (eQTL) mapping study provided transcriptomic evidence for the contribution of several genes acting in C-repeat binding factor (CBF) cold responsiveness (LanCRLK1), and in UDP-glycosyltransferases (LanUGT85A2) pathways in the vernalization response via LanFTc1 in L. angustifolius (Plewiński et al., 2019). LanCRLK1 is a homolog of A. thaliana CALCIUM/CALMODULIN-REGULATED RECEPTOR-LIKE KINASE 1, which is the first component in the cold responsiveness pathway (Yang et al., 2010). Downstream genes in this pathway, the C-repeat binding factors (CBF) and INDUCER OF CBF EXPRESSION 1 (ICE1), provide regulatory links to FLOWERING LOCUS C (FLC) (Kim et al., 2004; Lee et al., 2015). The present study has highlighted the positive vernalization responsiveness of the LanCRLK1 gene but only in wild germplasm under SD. In other genotype x environment combinations, the response was quasi thermoneutral. This finding is coherent with the general observation that CBF cold responsiveness pathway is downregulated and less effective under LD conditions than under SD (Lee and Thomashow, 2012). The expression profile of LanCRLK1 did not provide convincing evidence on the contribution of this gene in the vernalization responsiveness of L. angustifolius. Nevertheless, the reduction of LanCRLK1 expression in the evening, combined with the decreasing trend during development that was revealed in this study for vernalized ku line under LD, may explain the direction of the association between the LanCRLK1 expression pattern and the vernalization responsiveness observed in the L. angustifolius mapping population e-QTL study, which was also performed under LD with partial vernalization (Plewiński et al., 2019). The question arises as to whether these differences in the LanCRLK1 gene expression profile between the Ku/Pal and the ku lines, may have consequences in terms of cold acclimation and freezing tolerance of early flowering lines. A negative correlation between early phenology and cold acclimation could be a very undesirable trait hampering the autumn sowing of L. angustifolius in many regions of Southern Europe.
This study evidenced the negative response of LanUGT85A2 to vernalization in all genotypes under both photoperiods. The genotypes explored in this study revealed different responses to photoperiod, and under SD, the LanUGT85A2 expression was highest in the early flowering line, whereas under LD, it was in late flowering. Indeed, during the L. angustifolius mapping population e-QTL assay, which was performed under LD with mild vernalization, LanUGT85A2 expression revealed a significant positive correlation with the late-flowering phenotype (Plewiński et al., 2019). LanUGT85A2 is a representative of the UDP-glycosyltransferases protein family. In A. thaliana, a relatively close homolog of this gene, UGT87A2, promotes flowering in the vernalization and gibberellin pathways by repression of FLC (Wang B. et al., 2012). Similarly, ectopic over-expression in tobacco of a putative glycosyltransferase gene 1, PtGT1, derived from poplar (Populus tomentosa Carr.), resulted in an early flowering phenotype (Wang Y.-W. et al., 2012). Contrary, another A. thaliana homolog, UGT84A2, delays flowering by activation of the indole-3-butyric acid (IBA) pathway, leading to down-regulation of AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8 genes, and, consequently FT (Zhang et al., 2017). Taking into consideration the direction of the association between LanUGT85A2 expression and time to flowering, the latter mechanism seems to be more probable in L. angustifolius than those of UGT87A2 and PtGT1.
This research highlighted the hypothetical involvement of FLC-related genes in L. angustifolius vernalization-dependent flowering time regulation. However, legume genomes, except for soybean, generally do not have FLC homologs (Hecht et al., 2005; Lyu et al., 2020). Nevertheless, other genes from the vernalization pathway, including some potential activators and repressors of FLC, are present (Hane et al., 2017). Interestingly, soybean has retained one FLC copy within its genome and does not require vernalization to initiate flowering. FLC in this species is involved in long-term low temperature-triggered late flowering by inhibiting FT gene expression (Lyu et al., 2020).
In Arabidopsis, vernalization-dependent silencing of the FLC gene is performed by VIN3 and VRN5 proteins which contribute to H3K27me3 and H3K9me2 enrichments during cold periods (Sung and Amasino, 2004; Greb et al., 2007; Kim and Sung, 2013). Despite their similar function in A. thaliana, VIN3 and VRN5 genes differ in expression profiles and vernalization responsiveness. Namely, VIN3 expression in A. thaliana is very low and highly induced during vernalization, but soon after the end of the cold period, it decreases again to pre-vernalization level; whereas the VRN5 gene is constitutively expressed at much higher levels than VIN3 with additional high upregulation after the end of vernalization (Kim and Sung, 2013). In our study, LanVIN3 genes revealed significantly higher relative expression than LanVRN5. Apart from this difference, expression profiles of LanVRN5 and LanVIN3 homologs were similar to each other. Moreover, this research provided lines of evidence for the high expression of three LanVIN3 homologs in early flowering 83A:476 lines without vernalization, highlighting the opposite effects of vernalization for particular genotypes. The observed differences in vernalization responses and expression profiles of LanVIN3 and LanVRN5 genes did not match observed differences in phenotypes (time to flowering). This may indicate that LanVIN3 and LanVRN5 genes are not involved in the vernalization response in L. angustifolius.
All datasets presented in this study are included in the article/Supplementary Material.
SR-B designed experiments and performed plant phenotyping for time to flowering and vernalization responsiveness, plant genotyping with molecular markers, and gene expression profiling by quantitative PCR. PP performed RNA isolation and quantification. RG participated in the concept of the study and the interpretation of the results. BK contributed to data analysis and statistics. MK analyzed the data, interpreted the results, prepared the figures, and drafted the manuscript. All authors approved the final manuscript.
This study was supported by the Polish National Science Centre Project No. 2014/15/N/NZ9/03919.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank Magdalena Tomaszewska from the Department of Genomics of the Institute of Plant Genetics, Polish Academy of Sciences for help in performing plant experiments. The authors also thank Bogdan Wolko from the same department for mentoring during the project implementation.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.572135/full#supplementary-material
Supplementary Table 1 | Accession numbers and information on domestication status and country of origin of L. angustifolius lines used in the study.
Supplementary Table 2 | Primers used in the study.
Supplementary Table 3 | Air temperature (daily mean and maximum) and daily sunshine hours recorded by the local/nearest meteorological station (Poznań-Ławica, 5.1 km) and theoretical photoperiod hours calculated for this location during the 2014 experiment.
Supplementary Table 4 | Air temperature (daily mean and maximum) and daily sunshine hours recorded by the local/nearest meteorological station (Poznań-Ławica, 5.1 km) and theoretical photoperiod hours calculated for this location during the 2015 experiment.
Supplementary Table 5 | Sampling terms selected for gene expression profiling.
Supplementary Table 6 | Leaf expression of selected flowering-related genes (reads per kilobase million, RPKM) derived from the transcriptome sequencing of L. angustifolius mapping population and correlation with vernalization responsiveness (ku).
Supplementary Table 7 | R2 and PCR efficiency values obtained for standard curves.
Supplementary Table 8 | Comparison of LanFTc1_INDEL2 marker scores and time to flowering in response to vernalization observed during ‘2014 experiment.
Supplementary Table 9 | Comparison of LanFTc1_INDEL2 marker scores and time to flowering in response to vernalization observed during ‘2015 experiment.
Supplementary Figure 1 | Gene expression profile of the LanFTa2 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Figure 2 | Gene expression profile of the LanFTc2 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Figure 3 | Gene expression profile of the LanVIN3-1 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Figure 4 | Gene expression profile of the LanVIN3-2 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Figure 5 | Gene expression profile of the LanVIN3-3 gene in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Figure 6 | Gene expression profile of LanVRN5 genes in response to photoperiod and vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Supplementary Data Sheet 1 | Gene expression data and calculations for LanAGL8, LanCRLK1, LanFD, LanFTa1, LanFTa2, LanFTc1, LanFTc2, and LanUGT85A2 genes, obtained for two photoperiod variants (8- and 16-h) in response to vernalization in three lines (83A:476, Palestyna, and P27255) carrying different LanFTc1 alleles (Ku, Pal, and ku).
Abe, M., Kobayashi, Y., Yamamoto, S., Daimon, Y., Yamaguchi, A., Ikeda, Y., et al. (2005). FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1056. doi: 10.1126/science.1115983
Abraham, E. M., Ganopoulos, I., Madesis, P., Mavromatis, A., Mylona, P., Nianiou-Obeidat, I., et al. (2019). The use of lupin as a source of protein in animal feeding: genomic tools and breeding approaches. Int. J. Mol. Sci. 20:851. doi: 10.3390/ijms20040851
Adhikari, K. N., Buirchell, B. J., and Sweetingham, M. W. (2012). Length of vernalization period affects flowering time in three lupin species. Plant Breed. 131, 631–636. doi: 10.1111/j.1439-0523.2012.01996.x
Adrian, J., Farrona, S., Reimer, J. J., Albani, M. C., Coupland, G., and Turck, F. (2010). cis-Regulatory elements and chromatin state coordinately control temporal and spatial expression of FLOWERING LOCUS T in Arabidopsis. Plant Cell 22, 1425–1440. doi: 10.1105/tpc.110.074682
Annicchiarico, P., and Carroni, A. M. (2009). Diversity of white and narrow-leafed lupin genotype adaptive response across climatically-contrasting Italian environments and implications for selection. Euphytica 166, 71–81. doi: 10.1007/s10681-008-9836-9
Annicchiarico, P., Harzic, N., and Carroni, A. M. (2010). Adaptation, diversity, and exploitation of global white lupin (Lupinus albus L.) landrace genetic resources. Field Crops Res. 119, 114–124. doi: 10.1016/j.fcr.2010.06.022
Annicchiarico, P., Nazzicari, N., Ferrari, B., Harzic, N., Carroni, A. M., Romani, M., et al. (2019). Genomic prediction of grain yield in contrasting environments for white lupin genetic resources. Mol. Breed. 39:142.
Annicchiarico, P., Romani, M., and Pecetti, L. (2018). White lupin (Lupinus albus) variation for adaptation to severe drought stress. Plant Breed. 137, 782–789. doi: 10.1111/pbr.12642
Berger, J. D., Buirchell, B. J., Luckett, D. J., and Nelson, M. N. (2012). Domestication bottlenecks limit genetic diversity and constrain adaptation in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 124, 637–652. doi: 10.1007/s00122-011-1736-z
Berger, J. D., Shrestha, D., and Ludwig, C. (2017). Reproductive strategies in mediterranean legumes: trade-offs between phenology, seed size and vigor within and between wild and domesticated Lupinus species collected along aridity gradients. Front. Plant Sci. 8:548. doi: 10.3389/fpls.2017.00548
Cao, S., Kumimoto, R. W., Gnesutta, N., Calogero, A. M., Mantovani, R., and Holt, B. F. III (2014). A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 26, 1009–1017. doi: 10.1105/tpc.113.120352
Chen, L., Cai, Y., Qu, M., Wang, L., Sun, H., Jiang, B., et al. (2020). Soybean adaption to high-latitude regions is associated with natural variations of GmFT2b, an ortholog of FLOWERING LOCUS T. Plant Cell Environ. 43, 934–944. doi: 10.1111/pce.13695
Cowling, W. A. (2020). “Genetic diversity in narrow-leafed lupin breeding after the domestication bottleneck,” in The Lupin Genome, eds K. B. Singh, L. G. Kamphuis, and M. N. Nelson, (Cham: Springer International Publishing), 1–17. doi: 10.1007/978-3-030-21270-4_1
Cowling, W. A., Hamblin, J., Wood, P. M., and Gladstones, J. S. (1987). Resistance to Phomopsis stem blight in Lupinus angustifolius L. Crop Sci. 27, 648–652. doi: 10.2135/cropsci1987.0011183x002700040007x
Czyż, K. B., Ksia̧żkiewicz, M., Koczyk, G., Szczepaniak, A., Podkowiński, J., and Naganowska, B. (2020). A tale of two families: whole genome and segmental duplications underlie glutamine synthetase and phosphoenolpyruvate carboxylase diversity in narrow-leafed lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 21:2580. doi: 10.3390/ijms21072580
Deng, W., Casao, M. C., Wang, P., Sato, K., Hayes, P. M., Finnegan, E. J., et al. (2015). Direct links between the vernalization response and other key traits of cereal crops. Nat. Commun. 6:5882.
Dixon, L. E., Karsai, I., Kiss, T., Adamski, N. M., Liu, Z., Ding, Y., et al. (2019). VERNALIZATION1 controls developmental responses of winter wheat under high ambient temperatures. Development 146:dev172684. doi: 10.1242/dev.172684
Evans, J., Turner, G. L., O’connor, G. E., and Bergersen, F. J. (1987). Nitrogen fixation and accretion of soil nitrogen by field-grown lupins (Lupinus angustifolius). Field Crops Res. 16, 309–322. doi: 10.1016/0378-4290(87)90069-4
Foyer, C. H., Lam, H. M., Nguyen, H. T., Siddique, K. H., Varshney, R. K., Colmer, T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2:16112.
French, R. J., and Buirchell, B. (2005). Lupin: the largest grain legume crop in Western Australia, its adaptation and improvement through plant breeding. Austr. J. Agric. Res. 56, 1169–1180. doi: 10.1071/ar05088
Gao, L.-L., Hane, J. K., Kamphuis, L. G., Foley, R., Shi, B.-J., Atkins, C. A., et al. (2011). Development of genomic resources for the narrow-leafed lupin (Lupinus angustifolius): construction of a bacterial artificial chromosome (BAC) library and BAC-end sequencing. BMC Genomics 12:521. doi: 10.1186/1471-2164-12-521
Gladstones, J., and Hill, G. (1969). Selection for economic characters in Lupinus angustifolius and L. digitatus. 2. Time of flowering. Austr. J. Exp. Agric. 9, 213–220. doi: 10.1071/ea9690213
Greb, T., Mylne, J. S., Crevillen, P., Geraldo, N., An, H., Gendall, A. R., et al. (2007). The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr. Biol. 17, 73–78. doi: 10.1016/j.cub.2006.11.052
Gu, Q., Ferrandiz, C., Yanofsky, M. F., and Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517.
Hane, J. K., Ming, Y., Kamphuis, L. G., Nelson, M. N., Garg, G., Atkins, C. A., et al. (2017). A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: insights into plant-microbe interactions and legume evolution. Plant Biotechnol. J. 15, 318–330. doi: 10.1111/pbi.12615
Hauschke, D., Kieser, M., and Hothorn, L. A. (1999). Proof of safety in toxicology based on the ratio of two means for normally distributed data. Biometric. J. J. Math. Methods Biosci. 41, 295–304. doi: 10.1002/(sici)1521-4036(199906)41:3<295::aid-bimj295>3.0.co;2-2
Hecht, V., Foucher, F., Ferrandiz, C., Macknight, R., Navarro, C., Morin, J., et al. (2005). Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol. 137, 1420–1434. doi: 10.1104/pp.104.057018
Hecht, V., Laurie, R. E., Vander Schoor, J. K., Ridge, S., Knowles, C. L., Liew, L. C., et al. (2011). The pea GIGAS gene is a FLOWERING LOCUS T homolog necessary for graft-transmissible specification of flowering but not for responsiveness to photoperiod. Plant Cell 23, 147–161. doi: 10.1105/tpc.110.081042
Huyghe, C. (1991). Winter growth of autumn-sown white lupin (Lupinus albus L.) main apex growth model. Ann. Bot. 67, 429–434. doi: 10.1093/oxfordjournals.aob.a088160
Irish, V. F., and Sussex, I. M. (1990). Function of the apetala-1 gene during Arabidopsis floral development. Plant Cell 2, 741–753. doi: 10.2307/3869173
Jago, M. V., Peterson, J. E., Payne, A. L., and Campbell, D. G. (1982). Lupinosis: response of sheep to different doses of phomopsin. Austr. J. Exp. Biol. Med. Sci. 60, 239–251. doi: 10.1038/icb.1982.29
Kamel, K. A., Święcicki, W., Kaczmarek, Z., and Barzyk, P. (2016). Quantitative and qualitative content of alkaloids in seeds of a narrow-leafed lupin (Lupinus angustifolius L.) collection. Genet. Resourc. Crop Evol. 63, 711–719. doi: 10.1007/s10722-015-0278-7
Kamphuis, L. G., Hane, J. K., Nelson, M. N., Gao, L., Atkins, C. A., and Singh, K. B. (2015). Transcriptome sequencing of different narrow-leafed lupin tissue types provides a comprehensive uni-gene assembly and extensive gene-based molecular markers. Plant Biotechnol. J. 13, 14–25. doi: 10.1111/pbi.12229
Kasprzak, A., Safár, J., Janda, J., Dolezel, J., Wolko, B., and Naganowska, B. (2006). The bacterial artificial chromosome (BAC) library of the narrow-leafed lupin (Lupinus angustifolius L.). Cell. Mol. Biol. Lett. 11, 396–407.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kim, D.-H., and Sung, S. (2013). Coordination of the vernalization response through a VIN3 and FLC gene family regulatory network in Arabidopsis. Plant Cell 25, 454–469. doi: 10.1105/tpc.112.104760
Kim, H.-J., Hyun, Y., Park, J.-Y., Park, M.-J., Park, M.-K., Kim, M. D., et al. (2004). A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat. Genet. 36, 167–171. doi: 10.1038/ng1298
Klepikova, A. V., Kasianov, A. S., Gerasimov, E. S., Logacheva, M. D., and Penin, A. A. (2016). A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 88, 1058–1070. doi: 10.1111/tpj.13312
Kouris-Blazos, A., and Belski, R. (2016). Health benefits of legumes and pulses with a focus on Australian sweet lupins. Asia Pac. J. Clin. Nutr. 25, 1–17. doi: 10.1039/9781788015721-00001
Kozak, B., Galek, R., Zalewski, D., and Sawicka-Sienkiewicz, E. (2019). Preliminary genetic map of a new recombinant inbred line population for narrow-leafed lupin (Lupinus angustifolius L.). Agronomy 9:653. doi: 10.3390/agronomy9100653
Ksia̧żkiewicz, M., Nazzicari, N., Yang, H., Nelson, M. N., Renshaw, D., Rychel, S., et al. (2017). A high-density consensus linkage map of white lupin highlights synteny with narrow-leafed lupin and provides markers tagging key agronomic traits. Sci. Rep. 7:15335.
Ksia̧żkiewicz, M., Rychel, S., Nelson, M. N., Wyrwa, K., Naganowska, B., and Wolko, B. (2016). Expansion of the phosphatidylethanolamine binding protein family in legumes: a case study of Lupinus angustifolius L. FLOWERING LOCUS T homologs, LanFTc1 and LanFTc2. BMC Genomics 17:820. doi: 10.1186/s12864-016-3150-z
Ksia̧żkiewicz, M., Wójcik, K., Irzykowski, W., Bielski, W., Rychel, S., Kaczmarek, J., et al. (2020). Validation of Diaporthe toxica resistance markers in European Lupinus angustifolius germplasm and identification of novel resistance donors for marker-assisted selection. J. Appl. Genet. 61, 1–12. doi: 10.1007/s13353-019-00521-y
Kurlovich, B. S. (2002). Lupins: Geography, Classification, Genetic Resources and Breeding. St. Petersburg: Bogouslav Kourlovitch.
Lambers, H., Clements, J. C., and Nelson, M. N. (2013). How a phosphorus-acquisition strategy based on carboxylate exudation powers the success and agronomic potential of lupines (Lupinus, Fabaceae). Am. J. Bot. 100, 263–288. doi: 10.3732/ajb.1200474
Landers, K. F. (1995). Vernalization responses in narrow-leafed lupin (Lupinus angustifolius) genotypes. Austr. J. Agric. Res. 46, 1011–1025. doi: 10.1071/ar9951011
Laurie, R. E., Diwadkar, P., Jaudal, M., Zhang, L., Hecht, V., Wen, J., et al. (2011). The Medicago FLOWERING LOCUS T homolog, MtFTa1, is a key regulator of flowering time. Plant Physiol. 156, 2207–2224. doi: 10.1104/pp.111.180182
Lee, C.-M., and Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 109, 15054–15059. doi: 10.1073/pnas.1211295109
Lee, J.-H., Jung, J.-H., and Park, C.-M. (2015). INDUCER OF CBF EXPRESSION 1 integrates cold signals into FLOWERING LOCUS C-mediated flowering pathways in Arabidopsis. Plant J. 84, 29–40. doi: 10.1111/tpj.12956
Li, C., Lin, H., and Dubcovsky, J. (2015). Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 84, 70–82. doi: 10.1111/tpj.12960
Lippmann, R., Babben, S., Menger, A., Delker, C., and Quint, M. (2019). Development of wild and cultivated plants under global warming conditions. Curr. Biol. 29, R1326–R1338.
Liu, L., Adrian, J., Pankin, A., Hu, J., Dong, X., Von Korff, M., et al. (2014). Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat. Commun. 5:4558.
Lyu, J., Cai, Z., Li, Y., Suo, H., Yi, R., Zhang, S., et al. (2020). The floral repressor GmFLC-like is involved in regulating flowering time mediated by low temperature in soybean. Int. J. Mol. Sci. 21:1322. doi: 10.3390/ijms21041322
Mandel, M. A., and Yanofsky, M. F. (1995). The Arabidopsis AGL8 MADS box gene is expressed in inflorescence meristems and is negatively regulated by APETALA1. Plant Cell 7, 1763–1771. doi: 10.1105/tpc.7.11.1763
Mikołajczyk, J. (1966). Genetic studies in Lupinus angustifolius. Part. III. Inheritance of the alkaloid content, seed hardness and length of the growing season in blue lupin. Genet. Polon. 7, 181–196.
Mousavi-Derazmahalleh, M., Bayer, P. E., Nevado, B., Hurgobin, B., Filatov, D., Kilian, A., et al. (2018a). Exploring the genetic and adaptive diversity of a pan-mediterranean crop wild relative: narrow-leafed lupin. Theor. Appl. Genet. 131, 887–901. doi: 10.1007/s00122-017-3045-7
Mousavi-Derazmahalleh, M., Nevado, B., Bayer, P. E., Filatov, D. A., Hane, J. K., Edwards, D., et al. (2018b). The western mediterranean region provided the founder population of domesticated narrow-leafed lupin. Theor. Appl. Genet. 131, 2543–2554. doi: 10.1007/s00122-018-3171-x
Mulholland, J., Coombe, J., and Dann, P. (1976). Use of oat, lupin and field pea stubbles by grazing sheep. Austr. J. Exp. Agric. 16, 467–471. doi: 10.1071/ea9760467
Narożna, D., Ksia̧żkiewicz, M., Przysiecka, Ł, Króliczak, J., Wolko, B., Naganowska, B., et al. (2017). Legume isoflavone synthase genes have evolved by whole-genome and local duplications yielding transcriptionally active paralogs. Plant Sci. 264, 149–167. doi: 10.1016/j.plantsci.2017.09.007
Nelson, M. N., Berger, J. D., and Erskine, W. (2010). Flowering time control in annual legumes: prospects in a changing global climate. CAB Rev. 5, 1–14.
Nelson, M. N., Ksia̧żkiewicz, M., Rychel, S., Besharat, N., Taylor, C. M., Wyrwa, K., et al. (2017). The loss of vernalization requirement in narrow-leafed lupin is associated with a deletion in the promoter and de-repressed expression of a Flowering Locus T (FT) homologue. New Phytol. 213, 220–232. doi: 10.1111/nph.14094
Nelson, M. N., Phan, H. T. T., Ellwood, S. R., Moolhuijzen, P. M., Hane, J., Williams, A., et al. (2006). The first gene-based map of Lupinus angustifolius L.-location of domestication genes and conserved synteny with Medicago truncatula. Theor. Appl. Genet. 113, 225–238. doi: 10.1007/s00122-006-0288-0
NOAA (2020). National Centers for Environmental Information, Climate at a Glance: Global Time Series. Washington, DC: NOAA.
Ortega, R., Hecht, V. F. G., Freeman, J. S., Rubio, J., Carrasquilla-Garcia, N., Mir, R. R., et al. (2019). Altered expression of an FT cluster underlies a major locus controlling domestication-related changes to chickpea phenology and growth habit. Front. Plant Sci. 10:824. doi: 10.3389/fpls.2019.00824
Parmesan, C., and Yohe, G. (2003). A globally coherent fingerprint of climate change impacts across natural systems. Nature 421, 37–42. doi: 10.1038/nature01286
Plewiński, P., Ksia̧żkiewicz, M., Rychel-Bielska, S., Rudy, E., and Wolko, B. (2019). Candidate domestication-related genes revealed by expression quantitative trait loci mapping of narrow-leafed lupin (Lupinus angustifolius L.). Int. J. Mol. Sci. 20:5670. doi: 10.3390/ijms20225670
Przysiecka, Ł, Ksia̧żkiewicz, M., Wolko, B., and Naganowska, B. (2015). Structure, expression profile and phylogenetic inference of chalcone isomerase-like genes from the narrow-leafed lupin (Lupinus angustifolius L.) genome. Front. Plant Sci. 6:268. doi: 10.3389/fpls.2015.00268
Putterill, J., Zhang, L., Yeoh, C. C., Balcerowicz, M., Jaudal, M., and Gasic, E. V. (2013). FT genes and regulation of flowering in the legume Medicago truncatula. Funct. Plant Biol. 40, 1199–1207. doi: 10.1071/fp13087
R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Rahman, M., and Gladstones, J. (1972). Control of lupin flower initiation by vernalization, photoperiod and temperature under controlled environment. Austr. J. Exp. Agric. 12, 638–645. doi: 10.1071/ea9720638
Rychel, S. (2018). Charakterystyka Genów Uczestnicza̧cych w Procesie Indukcji Kwitnienia u Łubinu Wa̧skolistnego (Lupinus angustifolius L.) i Łubinu Białego (L. albus L.). PhD Thesis. Warsaw: Polish Academy of Sciences.
Rychel, S., Ksia̧żkiewicz, M., Tomaszewska, M., Bielski, W., and Wolko, B. (2019). FLOWERING LOCUS T, GIGANTEA, SEPALLATA and FRIGIDA homologs are candidate genes involved in white lupin (Lupinus albus L.) early flowering. Mol. Breed. 39:43.
Samanfar, B., Molnar, S. J., Charette, M., Schoenrock, A., Dehne, F., Golshani, A., et al. (2017). Mapping and identification of a potential candidate gene for a novel maturity locus, E10, in soybean. Theor. Appl. Genet. 130, 377–390. doi: 10.1007/s00122-016-2819-7
Sang, N., Cai, D., Li, C., Sun, Y., and Huang, X. (2019). Characterization and activity analyses of the FLOWERING LOCUS T promoter in Gossypium Hirsutum. Int. J. Mol. Sci. 20, 4769. doi: 10.3390/ijms20194769
Scheelbeek, P. F. D., Bird, F. A., Tuomisto, H. L., Green, R., Harris, F. B., Joy, E. J. M., et al. (2018). Effect of environmental changes on vegetable and legume yields and nutritional quality. Proc. Natl. Acad. Sci. U.S.A. 115, 6804–6809. doi: 10.1073/pnas.1800442115
Shankar, M., Sweetingham, M. W., and Cowling, W. A. (2002). Identification of alleles at two loci controlling resistance to Phomopsis stem blight in narrow-leafed lupin (Lupinus angustifolius L.). Euphytica 125, 35–44.
Shim, J. S., Kubota, A., and Imaizumi, T. (2017). Circadian clock and photoperiodic flowering in Arabidopsis: CONSTANS is a hub for signal integration. Plant Physiol. 173, 5–15. doi: 10.1104/pp.16.01327
Stefanova, K. T., and Buirchell, B. (2010). Multiplicative mixed models for genetic gain assessment in lupin breeding. Crop Sci. 50, 880–891. doi: 10.2135/cropsci2009.07.0402
Sung, S., and Amasino, R. M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. doi: 10.1038/nature02195
Svec, D., Tichopad, A., Novosadova, V., Pfaffl, M. W., and Kubista, M. (2015). How good is a PCR efficiency estimate: recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantific. 3, 9–16. doi: 10.1016/j.bdq.2015.01.005
Szczepaniak, A., Ksia̧żkiewicz, M., Podkowiński, J., Czyż, K. B., Figlerowicz, M., and Naganowska, B. (2018). Legume cytosolic and plastid acetyl-coenzyme-A carboxylase genes differ by evolutionary patterns and selection pressure schemes acting before and after whole-genome duplications. Genes 9:563. doi: 10.3390/genes9110563
Takeshima, R., Hayashi, T., Zhu, J., Zhao, C., Xu, M., Yamaguchi, N., et al. (2016). A soybean quantitative trait locus that promotes flowering under long days is identified as FT5a, a FLOWERING LOCUS T ortholog. J. Exp. Bot. 67, 5247–5258. doi: 10.1093/jxb/erw283
Tamhane, A. C., and Logan, B. R. (2004). Finding the maximum safe dose level for heteroscedastic data. J. Biopharmac. Stat. 14, 843–856. doi: 10.1081/bip-200035413
Taylor, C. M., Jost, R., Erskine, W., and Nelson, M. N. (2016). Identifying stable reference genes for qRT-PCR normalisation in gene expression studies of narrow-leafed lupin (Lupinus angustifolius L.). PLoS One 11:e0148300. doi: 10.1371/journal.pone.0148300
Taylor, C. M., Kamphuis, L. G., Zhang, W., Garg, G., Berger, J. D., Mousavi-Derazmahalleh, M., et al. (2019). INDEL variation in the regulatory region of the major flowering time gene LanFTc1 is associated with vernalization response and flowering time in narrow-leafed lupin (Lupinus angustifolius L.). Plant, Cell Environ. 42, 174–187. doi: 10.1111/pce.13320
Thines, B. C., Youn, Y., Duarte, M. I., and Harmon, F. G. (2014). The time of day effects of warm temperature on flowering time involve PIF4 and PIF5. J. Exp. Bot. 65, 1141–1151. doi: 10.1093/jxb/ert487
Trevaskis, B., Bagnall, D. J., Ellis, M. H., Peacock, W. J., and Dennis, E. S. (2003). MADS box genes control vernalization-induced flowering in cereals. Proc. Natl. Acad. Sci. U.S.A. 100, 13099–13104. doi: 10.1073/pnas.1635053100
Turck, F., Fornara, F., and Coupland, G. (2008). Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu. Rev. Plant Biol. 59, 573–594. doi: 10.1146/annurev.arplant.59.032607.092755
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Vadez, V., Berger, J. D., Warkentin, T., Asseng, S., Ratnakumar, P., Rao, K. P. C., et al. (2012). Adaptation of grain legumes to climate change: a review. Agron. Sustain. Dev. 32, 31–44.
Wang, B., Jin, S.-H., Hu, H.-Q., Sun, Y.-G., Wang, Y.-W., Han, P., et al. (2012). UGT87A2, an Arabidopsis glycosyltransferase, regulates flowering time via FLOWERING LOCUS C. New Phytol. 194, 666–675. doi: 10.1111/j.1469-8137.2012.04107.x
Wang, Y.-W., Wang, W.-C., Jin, S.-H., Wang, J., Wang, B., and Hou, B.-K. (2012). Over-expression of a putative poplar glycosyltransferase gene, PtGT1, in tobacco increases lignin content and causes early flowering. J. Exp. Bot. 63, 2799–2808. doi: 10.1093/jxb/ers001
Welch, B. L. (1947). The generalization ofstudent’s’ problem when several different population variances are involved. Biometrika 34, 28–35. doi: 10.2307/2332510
Williamson, P. M., Highet, A. S., Gams, W., Sivasithamparam, K., and Cowling, W. A. (1994). Diaporthe toxica sp. nov., the cause of lupinosis in sheep. Mycol. Res. 98, 1364–1368. doi: 10.1016/s0953-7562(09)81064-2
Wyrwa, K., Ksia̧żkiewicz, M., Szczepaniak, A., Susek, K., Podkowiński, J., and Naganowska, B. (2016). Integration of Lupinus angustifolius L. (narrow-leafed lupin) genome maps and comparative mapping within legumes. Chromosome Res. 24, 355–378. doi: 10.1007/s10577-016-9526-8
Yan, L., Loukoianov, A., Tranquilli, G., Helguera, M., Fahima, T., and Dubcovsky, J. (2003). Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. U.S.A. 100, 6263–6268. doi: 10.1073/pnas.0937399100
Yang, H., Tao, Y., Zheng, Z., Zhang, Q., Zhou, G., Sweetingham, M. W., et al. (2013). Draft genome sequence, and a sequence-defined genetic linkage map of the legume crop species Lupinus angustifolius L. PLoS One 8:e64799. doi: 10.1371/journal.pone.0064799.t002
Yang, T., Chaudhuri, S., Yang, L., Du, L., and Poovaiah, B. W. (2010). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. J. Biol. Chem. 285, 7119–7126. doi: 10.1074/jbc.m109.035659
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524.
Zhang, G.-Z., Jin, S.-H., Li, P., Jiang, X.-Y., Li, Y.-J., and Hou, B.-K. (2017). Ectopic expression of UGT84A2 delayed flowering by indole-3-butyric acid-mediated transcriptional repression of ARF6 and ARF8 genes in Arabidopsis. Plant Cell Rep. 36, 1995–2006. doi: 10.1007/s00299-017-2225-x
Zhao, C., Takeshima, R., Zhu, J., Xu, M., Sato, M., Watanabe, S., et al. (2016). A recessive allele for delayed flowering at the soybean maturity locus E9 is a leaky allele of FT2a, a FLOWERING LOCUS T ortholog. BMC Plant Biol. 16:20.
Keywords: vernalization, photoperiod, flowering, expression, FLOWERING LOCUS T, duplication, deletion
Citation: Rychel-Bielska S, Plewiński P, Kozak B, Galek R and Ksia̧żkiewicz M (2020) Photoperiod and Vernalization Control of Flowering-Related Genes: A Case Study of the Narrow-Leafed Lupin (Lupinus angustifolius L.). Front. Plant Sci. 11:572135. doi: 10.3389/fpls.2020.572135
Received: 12 June 2020; Accepted: 24 September 2020;
Published: 26 October 2020.
Edited by:
Christian Jung, University of Kiel, GermanyReviewed by:
Dong-Hwan Kim, Chung-Ang University, South KoreaCopyright © 2020 Rychel-Bielska, Plewiński, Kozak, Galek and Ksia̧żkiewicz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michał Ksia̧żkiewicz, bWtzaUBpZ3IucG96bmFuLnBs
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.