- 1Department of Agricultural and Environmental Sciences, University of Milan, Milan, Italy
- 2Department of Agriculture and Life Sciences, Agricultural University of Georgia, Tbilisi, Georgia
- 3Faculty of Viticulture and Winemaking, Caucasus International University, Tbilisi, Georgia
- 4National Wine Agency of Georgia, Tbilisi, Georgia
Plasmopara viticola (Berk. et Curt.) Berl. and de Toni, the agent of downy mildew, is one of the most important pathogens of European grapevine (Vitis vinifera L.). Extensive evaluation of cultivated grapevine germplasm has highlighted the existence of resistant phenotypes in the Georgian (Southern Caucasus) germplasm. Resistance is shown as a reduction in disease severity. Unraveling the genetic architecture of grapevine response to P. viticola infection is crucial to develop resistant varieties and reduce the impact of disease management. The aim of this work was to apply a genome-wide association (GWA) approach to a panel of Georgian-derived accessions phenotyped for P. viticola susceptibility and genotyped with Vitis18kSNP chip array. GWA identified three highly significant novel loci on chromosomes 14 (Rpv29), 3 (Rpv30) and 16 (Rpv31) associated with a low level of pathogen sporulation. Rpv29, Rpv30, and Rpv31 loci appeared to be associated with plant defense genes against biotic stresses, such as genes involved in pathogen recognition and signal transduction. This study provides the first evidence of resistant loci against P. viticola in V. vinifera germplasm, and identifies potential target genes for breeding P. viticola resistant grapevine cultivars.
Introduction
Vitis vinifera L. is one of the most widely cultivated fruit tree species of agricultural interest and it is the only species of the Vitis genus extensively used in the global wine industry. According to the data collected in 2018, viticulture covers approximately 7.6 million hectares worldwide and produces more than 67 million tons of grapes1. Unfortunately, V. vinifera is also known as the most susceptible Vitis species to Plasmopara viticola (Berk. et Curt.) Berl. and de Toni, the oomycete causing grapevine downy mildew. P. viticola was introduced into France from North America during the XIX century together with American wild Vitis species and rapidly spread across Europe dividing into two genetically distinct groups (Fontaine et al., 2013; Maddalena et al., 2020). Structure analysis indicated that the European and Italian P. viticola populations is formed by two separate genetic clusters, distributed according to a geographical gradient (East-West) and climatic conditions (Fontaine et al., 2013; Maddalena et al., 2020). P. viticola is a polycyclic pathogen able to biotrophically grow on tissues (leaves, shoots, and clusters) of susceptible Vitis species and, particularly, V. vinifera. If adequate disease management strategies are not applied, the disease seriously affects yield in terms of on grape quality and quantity (Toffolatti et al., 2018b).
Resistant accessions within the North American non-vinifera species, such as Vitis riparia Michx., Vitis cinerea (Engelm. ex A.Gray) Engelm. ex Millard and Vitis labrusca L., and the Northeast Asian species (Vitis amurensis Rupr.), exhibit varying levels of resistance, ranging from moderate to high, due to co-evolution with the pathogen (Jürges et al., 2009). Several QTL (Quantitative Trait Loci), conferring downy mildew resistance, at different levels ranging from weak to total, were discovered in Vitis species background: Rpv1 and Rpv2 in Muscadinia rotundifolia Michaux (Merdinoglu et al., 2003; Wiedemann-Merdinoglu et al., 2006); Rpv3 and Rpv19 in Vitis rupestris Scheele (Welter et al., 2007; Bellin et al., 2009; Divilov et al., 2018; Vezzulli et al., 2019; Foria et al., 2020); Rpv4, Rpv7, Rpv11, Rpv17, Rpv18, Rpv20, and Rpv21, in unspecified American species (Fischer et al., 2004; Welter et al., 2007; Bellin et al., 2009; Divilov et al., 2018); Rpv5, Rpv6, Rpv9, and Rpv13 in V. riparia (Marguerit et al., 2009; Moreira et al., 2011); Rpv8, Rpv10, Rpv12, Rpv22, Rpv23, Rpv24, Rpv25, and Rpv26 in V. amurensis (Blasi et al., 2011; Schwander et al., 2012; Venuti et al., 2013; Song et al., 2018; Lin et al., 2019); Rpv14 in V. cinerea (Ochssner et al., 2016); Rpv15 and Rpv16 in Vitis piasezkii Maxim. (Pap et al., unpublished); Rpv27 in Vitis aestivalis Michx. (Sapkota et al., 2015, 2019); and Rpv28 (Bhattarai et al., in preparation; www.vivc.de).
The management of downy mildew on traditional V. vinifera varieties requires regular application of fungicides. It is estimated that in the European Union, viticulture accounts for approximately 70% of all agrochemicals used, most of which are applied to contain the agents of downy and powdery mildews. Nevertheless, the intensive use of chemicals is becoming more and more restricted because of their high costs, their risks to human health and their negative environmental impact due to the chemical residues detected in grapes, soil and aquifers. In addition, disease control could be difficult to attain in future because some P. viticola strains could develop site-specific fungicide resistances, leading to great difficulties in the management of disease, while the discovery of new modes of action is rare (Hollomon, 2015). The EU Directive 2009/128 for sustainable management of diseases caused by plant pathogens in Europe strongly recommends a reduction in the number of treatments in the field. Moreover, the application of Regulation 1107/2009, concerning the placement on the market of plant protection products, is causing a reduction in the active substances available. The exploitation of resistance sources is the best way to decrease the use of chemicals for disease management and to achieve an effective protection from P. viticola in an environmental friendly way. Breeders had already started crossing the susceptible V. vinifera varieties with American species in the XIX century, first in United States and then in Europe (Eibach and Töpfer, 2015; Migicovsky et al., 2016; Merdinoglu et al., 2018; Yobrégat, 2018). Nowadays, numerous varieties combining resistance traits from American and Asian species and the quality traits of V. vinifera are available (Reynolds, 2015). A comprehensive list of new resistant varieties can be accessed from the Vitis International Variety Catalog website (VIVC; www.vivc.de).
Finding new sources of resistance is of paramount importance in breeding for biotic stress resistance in a perennial crop, which has to be productive for years while maintaining its resistance characteristics at the same time: the main strategy for preventing the selection of pathogen strains able to overcome resistance is, in fact, pyramiding resistance genes in the crop variety (Eibach et al., 2007; Delmotte et al., 2016; Zini et al., 2019). Recently, unique resistance traits to the downy mildew agent have been reported in V. vinifera varieties (Bitsadze et al., 2015; Toffolatti et al., 2016) coming from the first domestication center of the species: Georgia, Southern Caucasus (Imazio et al., 2013). The resistance mechanism for one of these resistant cultivars, named Mgaloblishvili, has been studied in detail (Toffolatti et al., 2018a, 2020). After artificial inoculation, P. viticola growth and sporulation are significantly affected in Mgaloblishvili: the mycelium degenerates, sporangiophores show an altered morphology and lower numbers of sporangia are produced, without any evidences of the hypersensitive response that occurs in American species. From the transcriptomic point of view, its defense mechanism shows an overexpression of genes related to pathogen recognition through PAMP (pathogen-associated molecular patterns), DAMP (damage-associated molecular patterns), and effector receptors and ubiquitination, signaling pathway through ethylene, synthesis of antimicrobial compounds (such as monoterpenes and flavonoids) and fungal wall degrading enzymes, and the development of structural barriers (such as cell wall reinforcement). The discovery of resistance to P. viticola in V. vinifera promises fresh opportunities for grapevine breeding in terms of new resistant loci.
Breeding for disease resistance is a very time-consuming process (up to 25–30 years are required for a breeding program), because it needs the evaluation of resistance levels of the progeny and other important characteristics (yield and quality of vines), which are typically not achieved until the third year after planting. A way to considerably decrease the length of the breeding process (accelerating the process by up to 10 years) is the adoption of the marker-assisted selection (MAS) approach, which allows the targeted selection of progeny harboring the resistance loci (Eibach and Töpfer, 2015).
Identification of genomic loci associated with complex quantitative and qualitative traits was enabled by the development of QTL (quantitative trait locus) and GWA (genome wide association) mapping approaches, combining genetic and phenotypic data. QTL mapping is performed using segregating biparental populations, while GWA approach relies on historical recombination events which occurred in natural populations, germplasm collections and breeding materials (Korte and Farlow, 2013). Over the last 10 years, NGS (next-generation sequencing) technologies have made available numerous (from thousands to hundreds of thousands) SNP (single nucleotide polymorphism) markers to be used for GWA study (GWAS) in various plant and animal species (Bhat et al., 2016).
In grapevine, at least three high-density SNP arrays have been set up (Myles et al., 2010; Marrano et al., 2017; Laucou et al., 2018), and the most used SNP set is the Vitis18kSNP chip array, developed by the GrapeReSeq Consortium, re-sequencing the genome of 47 V. vinifera genotypes and 18 genotypes belonging to American Vitis species and holding 18,071 SNPs. This high-density SNP array has been demonstrated to be a valid method for mapping of both quantitative and qualitative traits (Laucou et al., 2018).
In the present work, the Vitis18kSNP chip array was used to genotype a panel of V. vinifera Georgian accessions to identify genomic regions and/or putative markers associated with P. viticola resistance in V. vinifera, through a GWA approach, to be used for MAS in further breeding programs.
Materials and Methods
Plant Materials
The panel of accessions analyzed in this study (Supplementary Table 1) accounted for 132 genotypes: 84 are seedlings of the Mgaloblishvili self-pollinated population, and 48 are genotypes belonging to the Georgian germplasm collection. The breeding-derived genotypes are maintained in the greenhouse of the Department of Agricultural and Environmental Sciences (DiSAA), located in Arcagna (Lodi, Italy) and the germplasm genotypes are planted in the DiSAA germplasm collection vineyard, located in Torrazza Coste (Pavia, Italy). Mgaloblishvili self-progeny was obtained in spring of 2012, by enclosing Mgaloblishvili inflorescences in paper bags before flowering. At harvesting, bunches were collected and the seeds were extracted from berries to be vernalized at 5°C for 2 months in humid sand. The vernalized seeds were placed in plates of polystyrene cups filled with rockwool and maintained at 20 to 25°C up to germination in a screenhouse. The seedlings were transplanted into 8-cm pots filled with a sand–peat mixture (7:3 in volume) and after 1 year were moved in 20-cm pots. The plants were regularly irrigated and maintained without mineral fertilization practice. In Figure 1, some stages of Mgaloblishvili self-pollination, seedling germination and plant maintenance in greenhouse are shown.

Figure 1. Some stages of Mgaloblishvili self-pollination (A,B), seedling germination (C) and plant maintenance in greenhouse (D).
Phenotyping
The degree of susceptibility to P. viticola was evaluated through experimental inoculation on leaf samples collected at the beginning of the 2015, 2016, and 2017 grapevine growing seasons, using the protocol described by Toffolatti et al. (2016). To maximize the genetic variability of the pathogen and allow the detection of accessions that were resistant to a wide range of pathogen strains, field populations of P. viticola were used for the experimental inoculations (Toffolatti et al., 2016). Recent studies demonstrated that the European and Italian P. viticola population is divided in two genetic clusters, separated over an east-west gradient (Fontaine et al., 2013; Maddalena et al., 2020). In this study, east and west populations of P. viticola coming from Italy, at S. Maria della Versa (Pavia; East population) and Casarsa della Delizia (Pordenone; West population), and Georgia (West), were mixed to perform experimental inoculations. Phenotypical evaluations were performed in triplicate. Briefly, three leaf discs (1.5 cm in diameter) were cut from three leaves collected from the 3rd–5th leaf starting from the shoot apex of the plants. The leaf disks were sprayed with 1 mL P. viticola sporangia suspension (5 × 104 sporangia⋅mL–1) and incubated in a humid chamber at 22°C for 10 days. Disease severity was estimated from the area covered by sporulation by calculating the Percentage Index of Infections (I%I) (Townsend and Heuberger, 1947; Toffolatti et al., 2016). The accessions with an average I%I lower than 25% along the three sampled years were considered resistant. The 25% threshold was chosen based on the I%I distribution. Box plot distribution of the three replicate values of the samples showed that only nine samples (ID 124, ID 122, ID LIB 56, ID 138, ID 109, ID L22A, ID M22F, ID M22A, ID M22E) showed I%I < 40%, while the others reached higher values (Supplementary Figure 1). The average I%I of these samples was 20 ± 5% (95% confidence interval). Therefore, 25% was the chosen threshold. The existence of differences between I%I recorded in different years was analyzed by Pearson’s correlation coefficient.
Resistance levels (RLs), expressed in percentage, were calculated for each accessions by using the following formula:
) where I%Ix is the average disease severity of the sample and I%IMAX is the maximum value of disease severity recorded (accession ID 157 M, I%I = 85.8%).
SNP Genotyping
The 132 genotypes were genotyped using the Vitis18kSNP array (Illumina Inc., San Diego, CA, United States), containing 18,071 SNPs. The genotyping of breeding-derived accessions (Mgaloblishvili seedlings) was performed in this work, while for germplasm genotypes the data were obtained by De Lorenzis et al. (2015). Genotyping was carried out on 200 ng of genomic DNA extracted from 100 mg of frozen young leaf tissue using NucleoSpin® Plant II (MACHEREY-NAGEL, Germany), according to the manufacturer’s protocol. DNA concentration and quality were checked by electrophoresis on agarose gel and by spectroscopy using a NanoDrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and Quant-iT dsDNA HS assay kit for Qubit 3.0 Fluorometer (Thermo Fisher Scientific). Genotyping analysis was performed by the laboratory of Fondazione Edmund Much (San Michele all’Adige, Trento, Italy).
Data Analysis
SNP data produced in this work (84 Mgaloblishvili seedlings) were filtered for samples showing a call quality value (p50GC) lower than 0.54 and loci with a GenTrain (GT) score value lower than 0.6 and a marker missing rate > 20% (De Lorenzis et al., 2015). The Mgaloblishvili self-pollinated population dataset and the SNP profiles of 48 varieties reported in De Lorenzis et al. (2015) were merged and filtered for minor allele frequency (MAF) > 5%.
MEGA 7.0 software (Kumar et al., 2016) was used to design a UPGMA (Unweighted Pair Group Method with Arithmetic Mean) phylogenetic tree, based on the Dice’s coefficient (Dice, 1945) distance matrix generated by PEAS 1.0 software (Xu et al., 2010). Principal Component Analysis (PCA) was carried out using adegenet package (Jombart, 2008) of R software (R Core Team), and the first two components values were plotted on a 2-D scatterplot. Structure analysis was carried out using LEA package (Frichot and François, 2015) of R software by varying the number of ancestral genetic groups (K) from 1 to 10 in ten repetition runs for each K value. The most likely K value was detected based on LEA cross-validation method.
The LD (linkage disequilibrium) estimation as Pearson’s squared correlation coefficient (r2) between each pair of molecular markers (Zhao et al., 2005) was evaluated using PLINK (Purcell et al., 2007) software. The pair-wise LD as r2 was calculated using the parameters –ld-window-r2 0,–ld-window 99999,–ld-window-kb 10000. The distances between loci were categorized into intervals of a fixed length (100 kb) and, for each interval, average r2 was calculated. The LD decay was visualized by plotting the average r2 per each interval from 0 up to 10 Mb by R software.
Association analysis was performed in R software using GAPIT package (Lipka et al., 2012). GLM (Generalized Linear Model), MLM (Mixed Linear Model), MLMM (Multiple Locus Mixed linear Model), FarmCPU (Fixed and random model Circulating Probability Unification) and SUPER (Settlement of MLM Under Progressively Exclusion Relationship) algorithms were tested. For fixed effect, Q-matrix (for K = 3), detected by LEA, was used as the covariate for association analysis accounting for population structure. The GWA algorithm performances were evaluated through quantile-quantile (QQ) plots. A conservative threshold for assessing SNP significance was calculated based on Bonferroni correction for a type I error rate of 0.05. The SNPs fitting a logistic regression, performed in PLINK software, were selected.
Candidate Gene Mining
Gene associated with SNP loci passing the Bonferroni-adjusted threshold were predicted based on the LD r2 threshold of 0.2 (Li et al., 2014), using the grapevine reference genome PN40024 (12X.v2 version) (Canaguier et al., 2017). The SNP loci mapping to reference genome was conducted using CLC Genomic Workbench software (v. 20.0) in advance sequence finder toolbox including negative strand. Nearby genes in linkage regions of stable SNP-trait associations with putative functions supposedly related to the P. viticola resistance trait were selected as candidates.
Results
Phenotypic and Genetic Diversity of Accession Panel
Phenotyping evaluations were performed for 3 years (2017–2019) and only genotypes scored with a I%I < 25% in the 3 years of evaluation were classified as resistant. Evaluation trials have shown an overall high susceptibility to P. viticola infection, with some accessions showing a large distribution of the data (Supplementary Figure 1). Nine out of 132 genotypes were resistant: five Mgaloblishvili seedlings (ID 124, 122, LIB 56, 138, 109), Mgaloblishvili and three varieties (Jani Bakhvis, Zerdagi, and Kamuri shavi) (Figure 2A and Supplementary Table 1). The samples showed a significant correlation among years (r > 0.991; N = 3; P < 0.043). RLs of the nine resistant genotypes ranged from 70 to 84% (Supplementary Table 1). None of the resistant genotypes showed HR in leaf tissues.
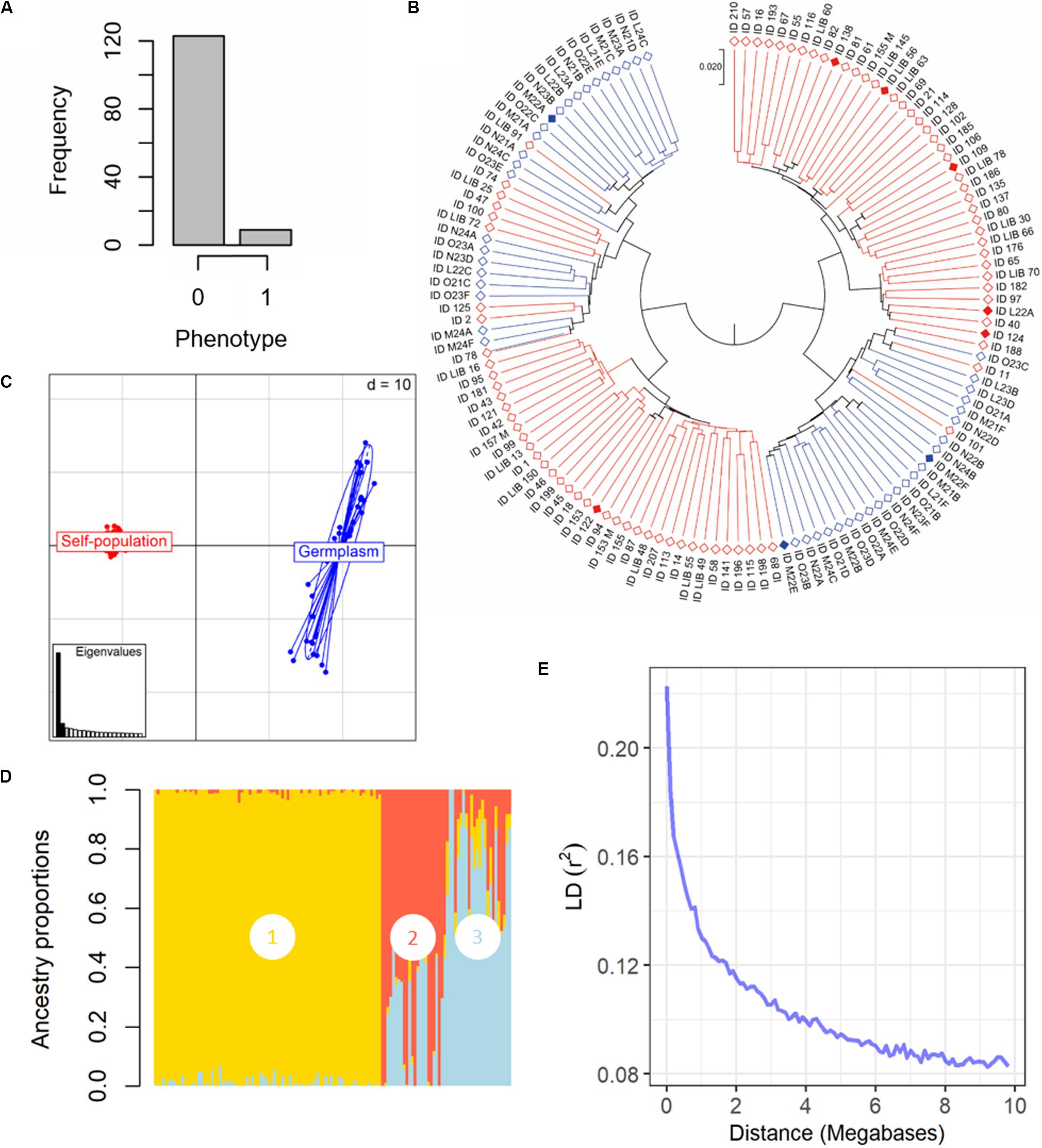
Figure 2. Phenotypical and genetic diversity in the panel of 132 grapevine accessions, belonging to the Mgaloblishvili self-pollinated population (84) and Georgian germplasm population (48), used for GWA analysis. The individuals were phenotyped for resistance to P. viticola infection and were genotyped using the Vitis18kSNP array. (A) Histogram summarizing the frequency of susceptible (0) vs. resistant (1) phenotypes. (B) UPGMA dendrogram showing relationships among individuals of Mgaloblishvili self- pollinated (red) and Georgian germplasm population (blue). Filled rhombus indicate resistant accessions (C) Scatterplot relationships among individuals of Mgaloblishvili self- pollinated (red) and Georgian germplasm population (blue), as represented by the first two principal components (PC1 along the horizontal axis, PC2 along the vertical axis) of PCA. (D) Admixture proportions as estimated by LEA package at K = 3, displayed in a barplot. Each sample is represented as a vertical bar, reflecting assignment probabilities to each of the three groups. Group 1: Mgaloblishvili self-pollinated individuals. Group 2 and 3: Georgian germplasm population individuals. (E) Decay of average linkage disequilibrium (LD r2) over distance (Mb).
The SNP genotyping data of the Mgaloblishvili self-pollinated population were merged with the ones of 48 Georgian cultivars (De Lorenzis et al., 2015). The final dataset accounted for 132 genotypes and 12,825 SNP loci (Supplementary Table 2). Clustering analysis discriminated the genotypes in two well distinct main groups (Figure 2B). In each main group, both breeding-derived genotypes and germplasm cultivars were included, though they were mainly clustered in well separated sub-groups. Resistant genotypes were distributed between the two main groups. The range of identity varied from 95 to 88%. PCA strongly differentiated Mgaloblishvili self-pollinated and germplasm individuals into two distinct groups (Figure 2C). The first two principal components (PCs) captured 33% of total explained variance (PC1 = 29% and PC2 = 4%). The two groups were separated along the PC1. As expected, the germplasm individuals showed a variability higher than the breeding-derived accessions. According to the cross-validation plot, structure analysis identified three ancestral populations (K = 3), one for Mgaloblishvili seedlings (group 1) and two for germplasm individuals (groups 2 and 3) (Figure 2D). The three resistant cultivars were assigned one to group 2 (Zerdagi) and two to group 3 (Jani Bakhvis and Kamuri shavi). The percentage of admixed genotypes (with a membership probability <80%) was 28%. All the admixed genotypes were detected among the cultivars (Supplementary Table 3). All the nine resistant genotypes showed a membership probability higher than 80%. LD decay was estimated for the entire dataset (Figure 2E). LD decreased with the increase in physical distance between marker loci. Average LD decay (r2 = 0.11) was observed after ∼2 Mb. The LD value dropped to 0.2 after ∼100 kb.
GWA Analysis
Different statistical models (GLM, MLM, MLMM, FarmCPU and SUPER) were tested for detecting associations for P. viticola resistance. Because structure analysis was able to capture the differences among the Georgian germplasm cultivars better than PCA, Q-matrix for K = 3 was used as covariate in the GWA analysis. The application of GLM, MLM and SUPER models allowed to account for stratification, although a relevant number of false positives was detected (Figures 3A,B,E). A significant SNP associated with P. viticola infection was identified in the three tested models: the SNP (chr14_21613512_C_T) located in the chromosome 14 at position 21,613,512 with a p-value of 4.01e-07, 5.09e-07 and 3.68e-10, respectively, for GLM, MLM and SUPER models. MLMM and FarmCPU models reduced false positive associations (Figures 3C,D). MLMM models detected one significant SNP associated with P. viticola infection, with a -log10 p-value above the Bonferroni-adjusted threshold, and two SNPs below the Bonferroni-adjusted threshold. The first SNP was the same detected by the GLM, MLM and SUPER models, with a p-value of 1.25e-08. The remaining two SNPs were li_T_C_chr16_21398409, located on chromosome 16 at position 21,398,409 and a p-value of 7.9e-06 and cn_C_T_chr3_16229046, located on chromosome 3 at position 16,229,046 and a p-value of 1.25e-05. FarmCPU model detected the same SNPs detected by MLMM model. chr14_21613512_C_T and cn_C_T_chr3_16229046 were above the Bonferroni-adjusted threshold, with p-values of 8.23e-08 and 8.18e-04, respectively, while li_T_C_chr16_21398409 was slightly below the threshold, with a p-value of 6.25e-03.
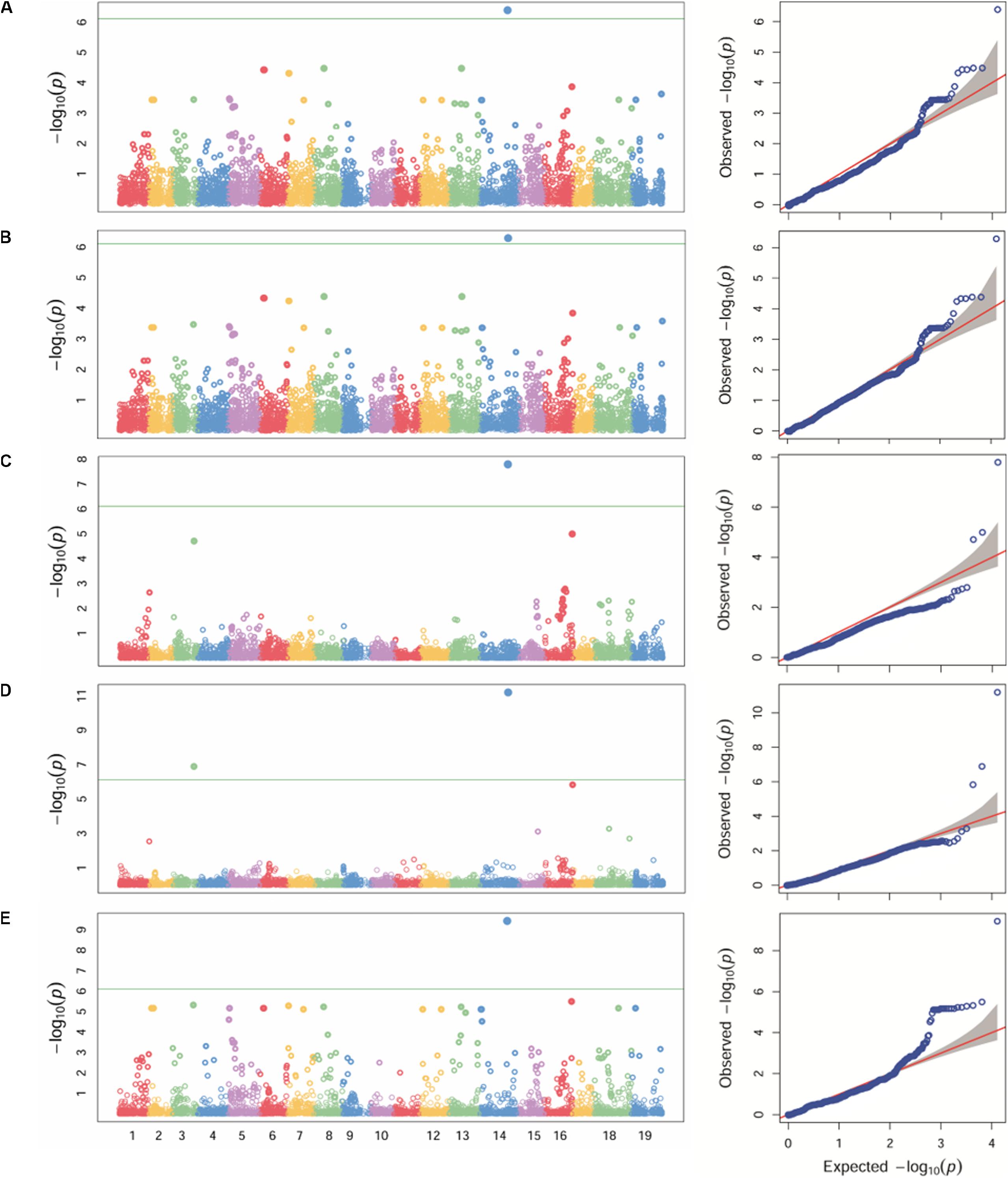
Figure 3. Manhattan plot (left) of -log10 p-values estimated for binary (resistant vs. susceptible) coded phenotypic response to P. viticola infection in the panel of 132 accessions genotyped by 18 k SNPs. Significant SNPs are circles above the Bonferroni-adjusted threshold (green horizontal line). Quantile-quantile plot (right) of expected vs. observed -log10 p-values. Association analysis results of GLM (A), MLM (B), MLMM (C), FarmCPU (D) and SUPER (E) algorithms.
For an approximate estimation of allelic effect, a logistic regression was fitted for the three significant SNPs. As observed by odds ratio, highly significant association was confirmed for chr14_21613512_C_T locus, followed by li_T_C_chr16_21398409 and cn_C_T_chr3_16229046 (Table 1).
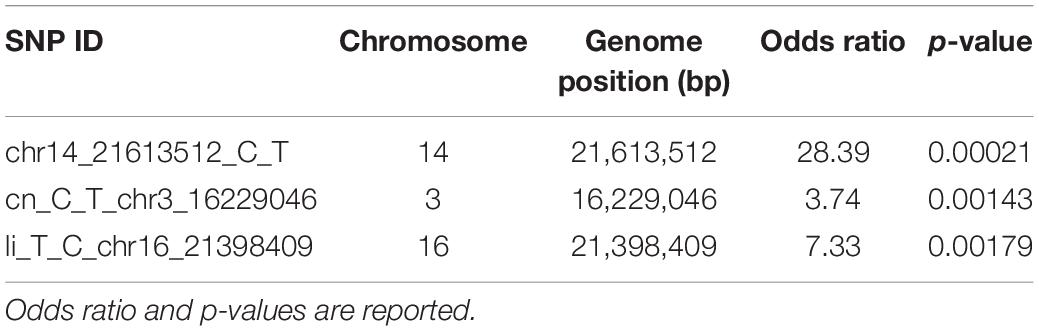
Table 1. Allelic effect estimation by logistic regression for SNP loci associated with P. viticola resistance traits.
Candidate Gene Prediction
The three SNP loci passing the Bonferroni-adjusted threshold were mapped to V. vinifera reference genome (PN40024 12X) to identify putative genes related to the P. viticola resistance trait. The LD value (r2) dropped to 0.2 after ∼100kb, for this reason a window of 100 kb upstream and downstream the most significant SNPs was chosen to search for candidate genes. Supplementary Table 4 reports the list of candidate genes in a window of 100 kb upstream and downstream the three SNPs associated to P. viticola resistance trait. Supplementary Table 5 reports the SNP allele information associated to these three regions.
The chr14_21613512_C_T locus mapped in the coding region of HEAT repeat-containing 5B protein (VIT_214s0006g03120) (Figure 4). The polymorphism (G → A) was non-synonymous giving rise to a change in the encoded amino acid, from aspartic acid (D) to asparagine (N). Upstream of this locus five genes were annotated: three of them encode for uncharacterized proteins (VIT_214s0006g03076, VIT_214s0006g03080, and VIT_214s0006g03100), and two for a probable cellulose synthase A catalytic subunit 8 [UDP-forming] (VIT_214s0006g03090) and an acyl-CoA-binding domain-containing protein 3-like (VIT_214s0006g03110). Downstream of this locus two genes were annotated, encoding for a probable carboxylesterase 17 and a plant cadmium resistance 4 protein (VIT_214s0006g03180 and VIT_214s0006g03190, respectively).
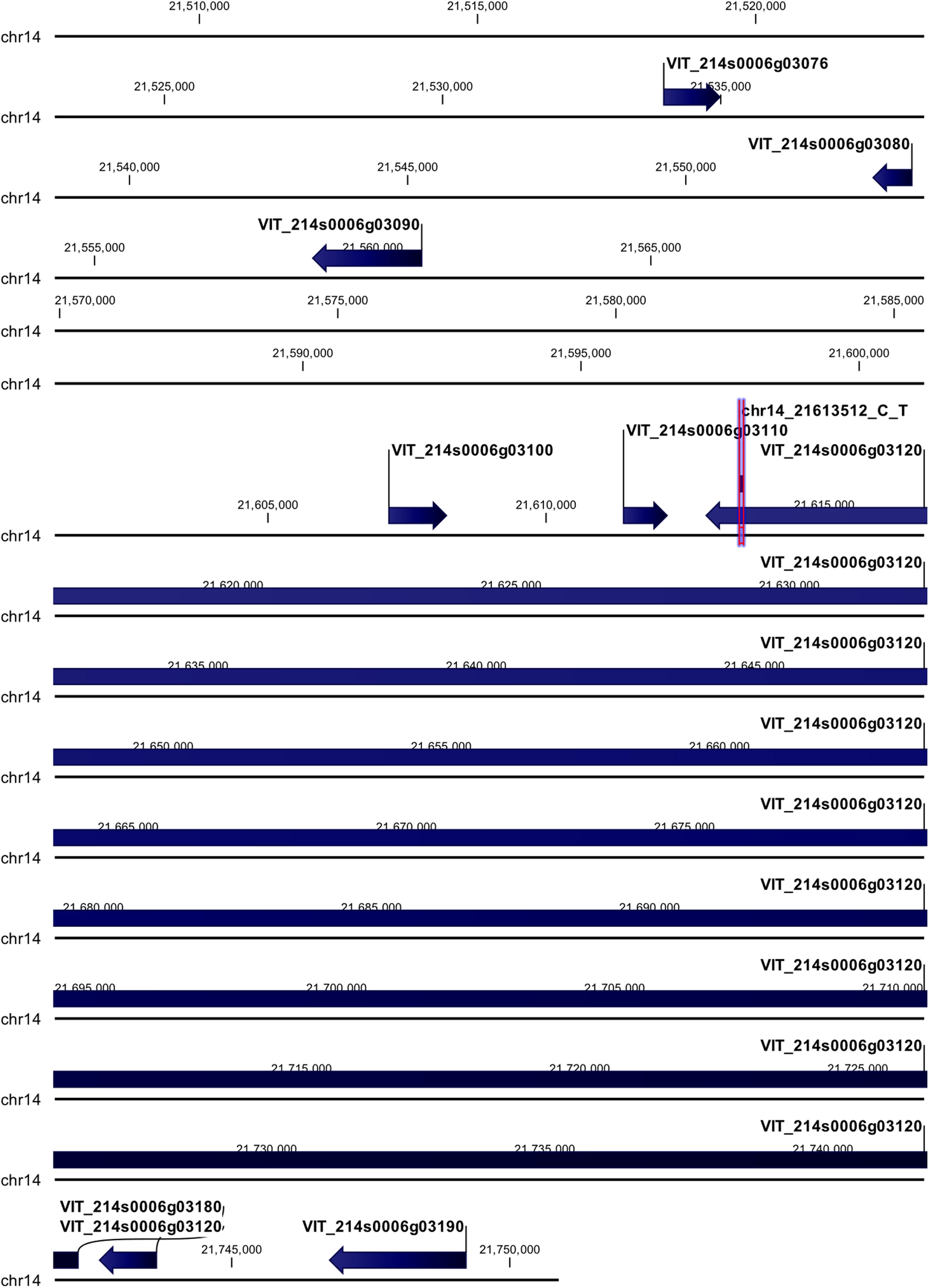
Figure 4. Annotation on grapevine reference genome PN40024 of chr14_21613512_C_T locus (Rpv29), on chromosome 14, associated with P. viticola resistance trait in V. vinifera. Locus is indicated with red arrows/violet bar, genes are indicated with blue arrows.
cn_C_T_chr3_16229046 and li_T_C_chr16_21398409 loci were mapped in intragenic regions (Figures 5, 6). The first locus was localized in a region including, upstream, an uncharacterized protein (VIT_203s0017g00420), a magnesium-dependent phosphatase 1 (VIT_203s0017g00410), an ubiquitin carboxyl-terminal hydrolase 21 (VIT_203s0017g00396), a MADS-box protein JOINTLESS-like (VIT_203s0017g00390), and a magnesium-dependent phosphatase 1-like (VIT_ 203s0017g00380), downstream, an uncharacterized protein (VIT_203s0017g00440), a MADS-box protein JOINTLESS-like (VIT_203s0017g00450) and an inositol transporter 1 (VIT_ 203s0017g00460). The second locus mapped in the genomic region including, upstream, two rust resistance kinase Lr10-like genes (VIT_216s0148g00020, VIT_216s0148g00010) and two genes encoding for uncharacterized proteins (VIT_216s0050g02810, VIT_216s0050g02800), and downstream, two rust resistance kinase Lr10-like genes (VIT_216s0148g00030 and VIT_216s0148g00040).
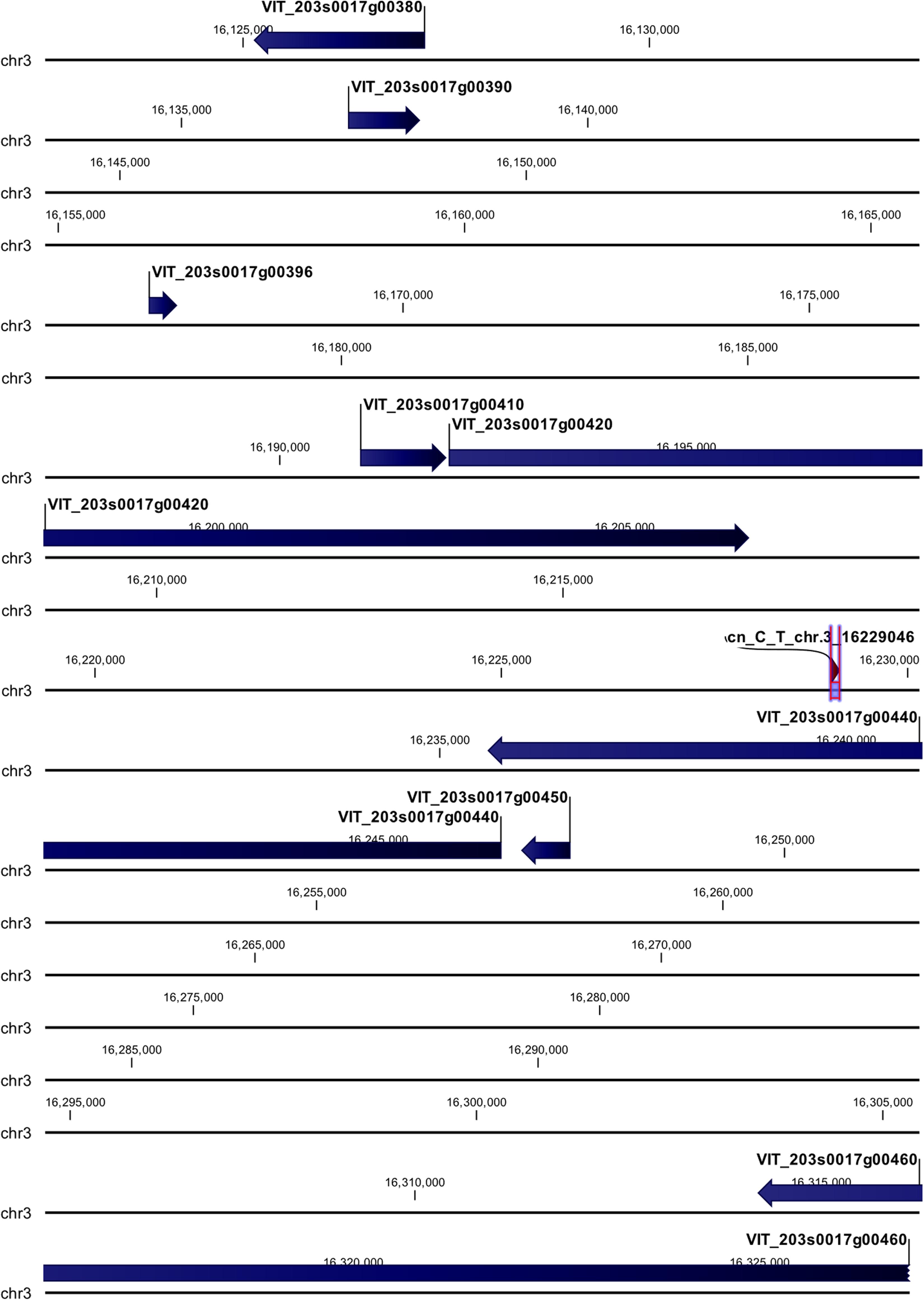
Figure 5. Annotation on grapevine reference genome PN40024 of cn_C_T_chr3_16229046 locus (Rpv30), on chromosome 3, associated with P. viticola resistance trait in V. vinifera. Locus is indicated with red arrows/violet bar, genes are indicated with blue arrows.
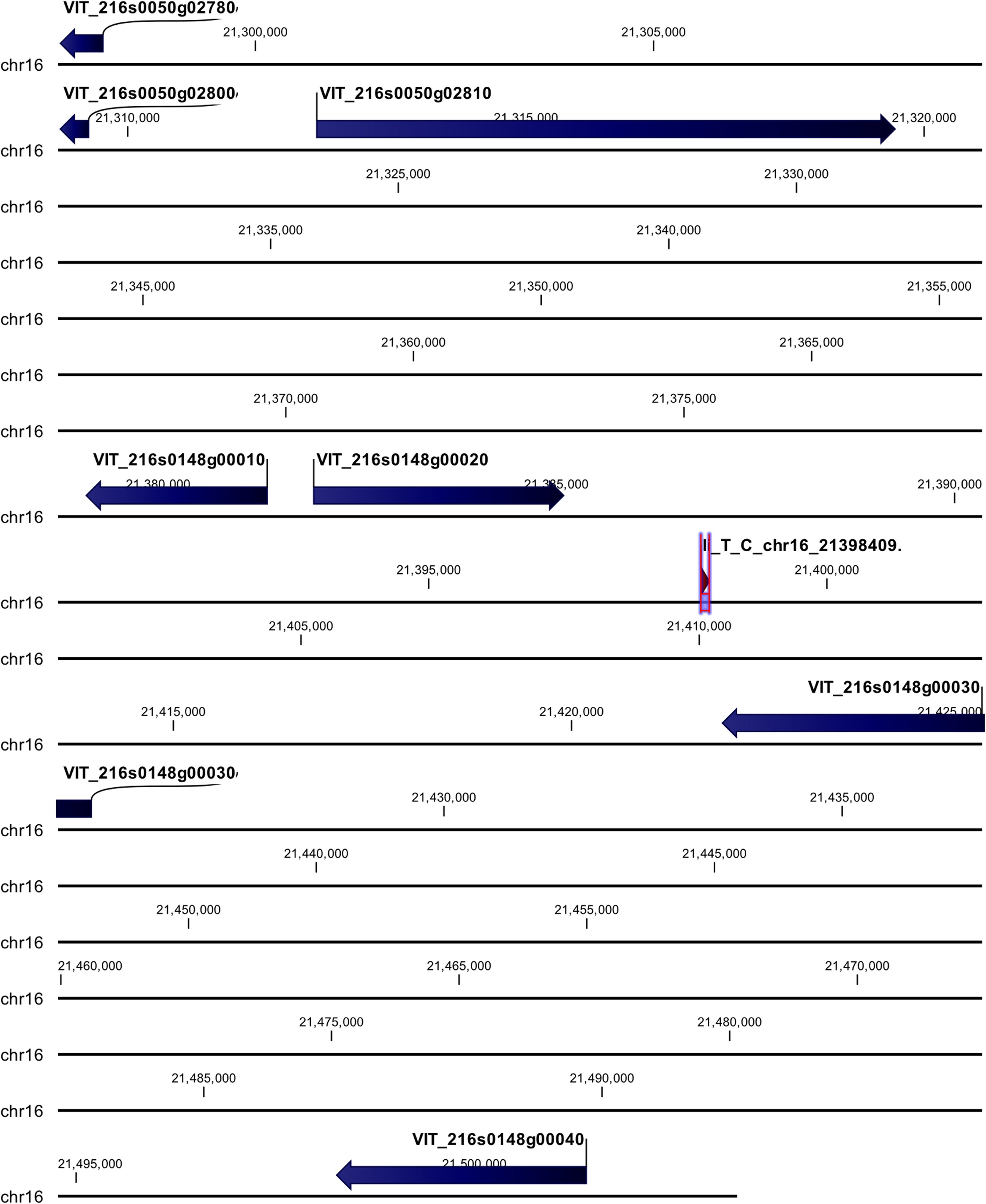
Figure 6. Annotation on grapevine reference genome PN40024 of li_T_C_chr16_21398409 locus (Rpv31), on chromosome 16, associated with P. viticola resistance trait in V. vinifera. Locus is indicated with red arrows/violet bar, genes are indicated with blue arrows.
Discussion
Downy mildew is one of the most important diseases affecting grapevines worldwide. So far, the sources of resistance were searched for in non-vinifera species, such as V. labrusca, V. aestivalis, V. riparia, V. rotundifolia and V. amurensis. The identification of resistant cultivars in the V. vinifera Georgian germplasm gave us the possibility to explore this promising material. In this work, a first insight was provided into quantitative resistance loci affecting downy mildew resistance traits in V. vinifera using an association mapping approach.
Grapevine Resistant Cultivars Belong to Different Georgian Regions
Experimental inoculations on 132 grapevine individuals belonging to the Mgaloblishvili seedling population and Georgian germplasm confirmed the high susceptibility of V. vinifera to P. viticola infection. Almost all breeding-derived and germplasm accessions were severely affected by the pathogen, developing medium to high I%I. Several accessions showed a large variability in the I%I distribution: this variability is frequently occurring in field assessment and in bioassays (Cadle-Davidson, 2008; Calonnec et al., 2013; Toffolatti et al., 2016) and could be related to several factors among which are the physiological state of the plant and the virulence of the pathogen. It is due to this variability that the experimental inoculations have been carried out in different years with mixed inocula: to identify those accessions that consistently showed a resistant behavior. A limited number of accessions (five breeding-derived and four germplasm accessions) clearly showed a reduced disease severity, which ranged from 5 to 25%. None of the accessions showed any necrotic spots, which are associated with HR, confirming that the defense mechanism is different from the one observed for North American and Asian Vitis species (Toffolatti et al., 2018a; Dry et al., 2019). The resistant cultivars showed different genetic origins. They were grouped in two different clusters and ancestral groups, characterized by cultivars having the same geographical provenance (Imazio et al., 2013; De Lorenzis et al., 2015). Zerdagi, a variety originated from Samegrelo province in the Western Georgia, was grouped with cultivars coming from Southern regions and Jani Bakhvis and Kamuri shavi with cultivars coming from the Western and Eastern regions.
Multi-Locus GWA Models Are the Best for Studying Complex Traits
The GWA approach was applied by genotyping 132 grapevine individuals with the 18k SNP genotyping array. A recent study has demonstrated the power of this array in detecting both known (such as berry color) and novel (such as acidity) loci related to phenotypic traits via GWA (Laucou et al., 2018). GWAS requires a genomic map with a marker density higher than the LD extent (Brachi et al., 2011). In our dataset, the average LD declined with the increase of the physical distance between markers, as already estimated in grapevine (Myles et al., 2010; Laucou et al., 2018; Figure 2E). The high LD levels observed in grapevine and the average inter-SNP spacing (about one SNP every ∼47 kbp, (Laucou et al., 2018), appear to be enough to tag associated loci.
Regarding the accuracy, the effectiveness of the GWA approach is strongly influenced by population stratification. Breeding-derived and germplasm accessions clearly showed genetic differentiation (Figures 2B–D). Since structure analysis was better able than PCA to capture the level of stratification, structure results were used as covariates for association analysis. Accounting for the complexity of phenotypic dataset and known population stratification, different algorithms, both single- (GLM, MLM, and SUPER) and multi-locus (MLMM and FarmCPU), for modeling marker-trait associations were tested. It is widely accepted that multi-locus GWAS models are superior to single-locus GWAS methods to identify association (Cui et al., 2018). In our study, multi-locus GWAS models detected the highest number of significant SNPs: FarmCPU = two (plus one just above the Bonferroni-adjusted threshold; MLMM = one (plus two Bonferroni-adjusted threshold); GLM, MLM and SUPER = one (Figure 3). Furthermore, our results confirm the usefulness of the Vitis SNP genotyping array in detecting loci associated with phenotypical traits (Laucou et al., 2018).
Three Novel SNP-Trait Associations to P. viticola Resistance Were Identified
To date, up to 28 QTL conferring resistance to downy mildew have been identified within wild Vitis species (Dry et al., 2019; www.vivc.de), but only two, Rpv1 and Rpv3 were characterized (Feechan et al., 2013; Eisenmann et al., 2019), mapping on chromosomes 12 and 18, respectively. Rpv1 is a NB-LRR (nucleotide-binding site leucine-rich repeat) receptor, while Rpv3 is associated with the biosynthesis of stilbenes. In our study, clear signals were identified on chromosomes 14, 3, and 16. The signal on chromosome 14, related to chr14_21613512_C_T locus, was recorded in all the five tested models, while the other two, related to cn_C_T_chr3_16229046 and li_T_C_chr16_21398409, were recorded only in MLMM and FarmCPU models, with some differences in the p-value. Among the 27 QTL already identified, three (Rpv8, 12, and 19) map on chromosome 14, while no QTL were found to map on chromosomes 3 and 16. Rpv8 and Rpv12 both mapped on the upper arm of chromosome 14 (Blasi et al., 2011; Venuti et al., 2013), while Rpv19 mapped on the lower arm, at around position 24 Mb (Divilov et al., 2018). Because the SNPs identified in this work do not physically co-locate to the QTL already identified, it is possible to conclude that the three loci are novel associations. We designated the locus on chromosome 14 (for chr14_21613512_C_T) Rpv29, the locus on chromosome 3 (cn_C_T_chr3_16229046) Rpv30 and the locus on chromosome 16 (li_T_C_chr16_21398409) Rpv31.
The logistic regression values (Table 1) indicated that the Rpv29 locus is the one having a major effect on the phenotype. Furthermore, the other two loci, Rpv30 and Rpv31, showed a statistically significant p-value as well, although the allelic effect estimation is lower. Nevertheless, since the resistance mechanism of accessions analyzed in this work did not show HR, it suggests that more than one locus are necessary to acquire the resistance.
Rpv29, Rpv30 and Rpv31 Are Markers Associated With Genes Related to P. viticola Resistance in V. vinifera
NB-LRR genes appeared to be associated with Rpv12 locus in the upper arm of chromosome 14 (Venuti et al., 2013). The SNP located on chromosome 14 (Rpv29) mapped in the coding region of HEAT repeat-containing 5B protein and the polymorphism leads to a non-synonymous amino acid substitution from aspartic acid to asparagine. Further studies are needed to better understand the effect at the protein level. HEAT motifs are tandemly repeated sequences of about 50 amino acid residues identified in a wide variety of eukaryotic proteins (Andrade et al., 2001). It was demonstrated that repeat proteins possess an intrinsic ability to bind peptides, acting as an integral component of protein complexes (Sharma and Pandey, 2016). HEAT repeat proteins, such as ILA, are required for plant immunity. In Arabidopsis thaliana, ILA is required for both non-host and basal resistance against Pseudomonas syringae, for resistance mediated by NB-LRR proteins and for systemic acquired resistance (SAR) (Monaghan and Li, 2010). NB-LRR proteins act as specific receptors of pathogen effectors, activating defense mechanisms leading to effector-triggered immunity (ETI) (Jones and Dangl, 2006). It is therefore tempting to speculate that the chr14_21613512_C_T locus could be involved in primary plant-pathogen interactions leading to both ETI and SAR. Nevertheless, further investigations are needed to confirm this result.
In a region spanning ∼100 kb upstream and downstream the Rpv29 locus, four genes, encoding for a probable cellulose synthase A catalytic subunit 8 [UDP-forming], an acyl-CoA-binding domain-containing protein 3-like, a probable carboxylesterase 17 and a plant cadmium resistance 4 protein, as well as three genes encoding for uncharacterized proteins, were mapped. All the candidate genes, except plant cadmium resistance 4 protein, appeared to be related to plant defense mechanism, based on the literature.
Cellulose synthases are involved in the secondary cell wall formation (Taylor et al., 2000). Structural modification, such as cell wall thickening, is one of the mechanisms adopted by plants to contrast the pathogen infection (Schulze-Lefert, 2004). A number of evidences proving the connection between cell wall structure and stress signaling, leading to enhanced production of hormones (such as jasmonate and ethylene) and to enhanced resistance to a broad range of pathogens were described (Ellis and Turner, 2001). Similarly to other organisms, Mgaloblishvili showed an up-regulation of genes, such as cellulose synthase-like protein G3 gene, that are involved in the transition from primary to secondary wall synthesis (Taylor et al., 1999).
Acyl-CoA binding proteins are thought to facilitate the transport of fatty acids/lipids among the cells (Kragelund et al., 1993). They are required for PAMP resistance to fungal pathogens, as described for A. thaliana against Botrytis cinerea and Colletotrichum higginsianum (Xia et al., 2012).
Carboxylesterases (CXEs) are a large family of enzymes, belonging to the α/β hydrolase fold superfamily, that hydrolyze ester, amide, and carbamate bonds (Putterill et al., 2003). They are involved in plant defense responses. Nicotiana tabacum, A. thaliana, and Capsicum annuum showed some CXEs involved in the plant-pathogen interaction, some of them related to hypersensitive response (Pontier et al., 1994; Kim et al., 2001; Putterill et al., 2003). In Vitis flexuosa, some CXEs were upregulated in response to Botrytis cinerea, Elsinoe ampelina, and Rhizobium vitis infection, indicating a putative role in defense mechanism during pathogen infection (Islam and Yun, 2016).
The cn_C_T_chr3_16229046 locus on chromosome 3 (Rpv30) was annotated close to predicted genes, such as MADS-box protein JOINTLESS-like, ubiquitin carboxyl-terminal hydrolase 21, magnesium-dependent phosphatase 1 and 1-like (MDP-1 and MDP-1-like) and inositol transporter 1 (INT1), and two genes encoding for uncharacterized proteins. All the candidate genes, except INT1, appeared to be related to plant defense mechanism, based on the literature.
MADS-domain transcription factors are proteins involved in multiple developmental pathways in plants, animals, and fungi (Castelán-Muñoz et al., 2019). JOINTLESS is a MADS-domain transcription factor, that together with MACROCALYX, induces the expression of AP2/ERF (ethylene response factor) 52 transcription factor in tomato during pre-abscission and abscission stages of pedicel (Nakano et al., 2014). Transcriptional data revealed that Mgaloblishvili defense mechanism is mediated mainly by ethylene (Toffolatti et al., 2018a). This MADS-domain transcription factor can be related to P. viticola resistance mechanism in V. vinifera.
Ubiquitin-protein hydrolases are involved in the processing of ubiquitinated proteins. Ubiquitination in plant cells modulates signaling mediated by PAMP receptors and leads to the accumulation of NB-LRR receptors (Furlan et al., 2012). In Mgaloblishvili, the ubiquitination process appeared to be activated, upregulating genes encoding for RING H2-type E3 ligases (Toffolatti et al., 2018a), activated in response to biotic and abiotic stresses and involved in ubiquitination (Mazzucotelli et al., 2006).
Protein phosphorylation, by a combined action of protein kinases and phosphatases, is a rapid post-translational control mechanism in the response to environmental stimuli, such pathogen elicitors, playing a major role in signal transduction pathways (Friso and van Wijk, 2015). Some DNA-binding proteins, with phosphatase activity, are able to bind defense-related genes and take part in their transcriptional regulation (i.e., DBP1 controlling transcription of the defense-related CEVI1 gene in A. thaliana during plant–virus interaction) (Carrasco et al., 2003). MDP-1 and MDP-1-like genes can be involved in the transcriptional regulation of some defense-related genes in the V. vinifera-P. viticola interactions.
The locus named li_T_C_chr16_21398409 (Rpv31) was annotated in linkage group including several rust resistance kinase Lr10-like genes. As already described above, Mgaloblishvili could recognize P. viticola through specific NB-LRR receptors, such as several Lr10 genes. It was demonstrated the Lr10 confers enhanced resistance to Puccinia triticina in Triticum aestivum (Feuillet et al., 2003). Frequently, NB-LRR genes occur in clusters. In Vitis, the Rpv12 locus accounts for 13 NB-LRR genes in a region of about 600 kb and it is part of a cluster of 46 NB-LRRs in the upper arm of chromosome 14 (Venuti et al., 2013). In our study, three Lr10-like genes (LOC100251517, LOC100256646, LOC100242248), spanning a region of about 47 kb on chromosome 16, appeared to be associated with the P. viticola resistance trait. Also, these three Lr10-like genes are part of a wider region, including a higher number of NB-LRR genes. In Toffolatti et al. (2018a), seven Lr10-like genes were differentially expressed (with a log2 fold-change value higher than 2) in Mgaloblishvili after P. viticola inoculation. Among them, four are located on chromosome 16, spanning a region of about 6 Mb. GWA results corroborate the involvement of these receptors in triggering the plant response. Indeed, during the infection process P. viticola has shown the expression of numerous different cytoplasmic and apoplastic effectors (Toffolatti et al., 2020) and their interaction with the NB-LRR receptors of the plant should be further investigated. Since no hypersensitive response (HR) was observed in the Georgian resistant accessions, due to absence of co-evolution with the pathogen, the involvement of the effector receptor Lr10 could be associated with an effector-triggered immunity not associated with HR. Indeed, HR is not always occurring in ETI (Jones and Dangl, 2006).
Conclusion
In this study, for the first time in V. vinifera, GWAS was used to identify loci associated with the resistance to P. viticola attack. The analysis provided evidence of three novel resistant loci (Rpv29, Rpv30, and Rpv31) in a panel of Georgian accessions, that they could be utilized for further genetic and breeding studies to select genotypes showing resistance to P. viticola infection. The three loci were found to co-locate in genomic regions enriched of genes associated with plant defense mechanism against biotic stress, suggesting both PAMP-triggered immunity and ETI-HR free response. Nevertheless, this hypothesis should be validated, by functionally characterize the candidate genes. Functional genomics approaches, such as CRISPR-based (Clustered Regularly Interspaced Short Palindromic Repeats) or RNA interference technologies, can help to functionally validate the candidate genes and, thus, to investigate which gene(s) is essential for resistance to P. viticola infection.
The great advantage provided by sources of resistance in V. vinifera germplasm, compared to the non-vinifera one, lies in the possibility to obtain crosses with cultivated varieties showing a good resistance level against a specific pathogen and, at the same time, able to provide a product free from the unpleasant characteristics usually imparted by the American vines, first of all the foxy flavor of the grapes. The discovery of resistant sources in the V. vinifera background is crucial to exploit favorable alleles already present in a germplasm, coupling at the same time good resistance to pathogen and good agronomic traits. Indeed, Caucasian accessions show very attractive characteristics for high-quality production also in the perspective of the climate change, such as late ripening, medium-size berries, avoidance of excessive sugar accumulation, smooth tannin and ability to maintain good level of acidity.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
GD, ST, OF, DM, and PB conceived the study. GM and NB performed phenotypical analysis. MS performed SNP profiling and analyzed the data. MS, GD, and ST wrote the manuscript. All authors approved the final version of the article.
Funding
The research was supported by University of Milan, DiSAA, Research Support Plan 2018, Linea 2 A, Project “Dal phenotyping al genome editing: strategie per limitare i danni da peronospora e legno nero in vite (ResVite),” National Wine Agency of Georgia within the “Research project for the Study of the Georgian Grapes and Wine Culture” and European Union within the project titled “FREECLIMB – Fruit Crops Adaptation To Climate Change In The Mediterranean Basin” in the frame of the Programme Partnership For Research And Innovation In The Mediterranean Area (PRIMA; call 2018).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The manuscript is dedicated to the memory of Prof. Annamaria Vercesi. The authors would like to thank Dr. Lesley Currah for supporting us on English Language Editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.562432/full#supplementary-material
Supplementary Figure S1 | Box-plot distribution of the I%I recorded by each grapevine accession, belonging to a Mgaloblishvili self-pollinated population (84) and Georgian germplasm population (48), following P. viticola inoculation. Resistant accessions are highlighted in red.
Supplementary Table S1 | List of grapevine accessions phenotyped for their resistance/susceptibility to P. viticola infection and genotyped by the Vitis18kSNP genotyping array. Phenotype column reports the resistance (1) or susceptibility to P. Viticola infection and resistance levels. The accessions showing a percentage of infection lower than 25%, in the three years of sampling (2017, 2018, and 2019), were considered resistant. Phenotypical evaluations were performed in triplicate.
Supplementary Table S2 | SNP profiles of 132 grapevine accessions, belonging to a Mgaloblishvili self-pollinated population (84) and Georgian germplasm population (48), genotyped at 18 k loci. Row data were filtered based on SNP call quality (p50GC) < 0.54, GenTrain score > 0.6, marker missing rate > 20% and minor allele frequency (MAF) > 5%, resulting in 12,825 SNP loci used for GWAS. “AA”: homozygous for dominant allele; “AB”: heterozygous for dominant allele; “BB”: homozygous for recessive allele; “NC”: missing data.
Supplementary Table S3 | Ancestry values at K = 3 detected for SNP profiles of 132 grapevine accessions, belonging to a Mgaloblishvili self-pollinated population (84) and Georgian germplasm population (48), genotyped at 18 k loci.
Supplementary Table S4 | List of candidate genes in a window of 100 kb upstream and downstream the three SNPs associated to resistance trait to P. viticola infection.
Supplementary Table S5 | Allele information on SNP loci detected in a region spanning 100 kb upstream and downstream the three loci associated (highlighted in gray) to the P. viticola resistance. In red: resistant genotypes.
Footnotes
References
Andrade, M. A., Petosa, C., Donoghue, S. I. O., Mu, C. W., and Delbru, M. (2001). Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18. doi: 10.1006/jmbi.2001.4624
Bellin, D., Peressotti, E., Merdinoglu, D., Wiedemann-Merdinoglu, S., Adam-Blondon, A. F., Cipriani, G., et al. (2009). Resistance to Plasmopara viticola in grapevine ‘Bianca’ is controlled by a major dominant gene causing localised necrosis at the infection site. Theor. Appl. Genet. 120, 163–176. doi: 10.1007/s00122-009-1167-2
Bhat, J. A., Ali, S., Salgotra, R. K., Mir, Z. A., Dutta, S., Jadon, V., et al. (2016). Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front. Genet. 7:221. doi: 10.3389/fgene.2016.00221
Bitsadze, N., Aznarashvili, M., Vercesi, A., Chipashvili, R., Failla, O., and Maghradze, D. (2015). Screening of Georgian grapevine germplasm for susceptibility to downy mildew (Plasmopara viticola). Vitis J. Grapevine Res. 54, 193–196. doi: 10.17660/ActaHortic.2014.1032.25
Blasi, P., Blanc, S., Prado, E., Rühl, E. H., Mestre, P., and Merdinoglu, D. (2011). Construction of a reference linkage map of Vitis amurensis and genetic mapping of Rpv8, a locus conferring resistance to grapevine downy mildew. Theor. Appl. Genet. 123, 43–53. doi: 10.1007/s00122-011-1565-0
Brachi, B., Morris, G. P., and Borevitz, J. O. (2011). Genome-wide association studies in plants: the missing heritability is in the field. Genome Biol. 12:232. doi: 10.1186/gb-2011-12-10-232
Cadle-Davidson, L. (2008). Variation within and between Vitis spp. for foliar resistance to the downy mildew pathogen Plasmopara viticola. Plant Dis. 92, 1577–1584. doi: 10.1094/PDIS-92-11-1577
Calonnec, A., Wiedemann-Merdinoglu, S., Delière, L., Cartolaro, P., Schneider, C., and Delmotte, F. (2013). The reliability of leaf bioassays for predicting disease resistance on fruit: a case study on grapevine resistance to downy and powdery mildew. Plant Pathol. 62, 533–544. doi: 10.1111/j.1365-3059.2012.02667.x
Canaguier, A., Grimplet, J., Di Gaspero, G., Scalabrin, S., Duchêne, E., Choisne, N., et al. (2017). A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genomics Data 14, 56–62. doi: 10.1016/j.gdata.2017.09.002
Carrasco, J. L., Ancillo, G., Mayda, E., and Vera, P. (2003). A novel transcription factor involved in plant defense endowed with protein phosphatase activity. EMBO J. 22, 3376–3384. doi: 10.1093/emboj/cdg323
Castelán-Muñoz, N., Herrera, J., Cajero-Sánchez, W., Arrizubieta, M., Trejo, C., García-Ponce, B., et al. (2019). MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 10:853. doi: 10.3389/fpls.2019.00853
Cui, Y., Zhang, F., and Zhou, Y. (2018). The application of multi-locus GWAS for the detection of salt-tolerance loci in rice. Front. Genet. 9:1464. doi: 10.3389/fpls.2018.01464
De Lorenzis, G., Chipashvili, R., Failla, O., and Maghradze, D. (2015). Study of genetic variability in Vitis vinifera L. germplasm by high-throughput Vitis18kSNP array: the case of Georgian genetic resources. BMC Plant Biol. 15:154. doi: 10.1186/s12870-015-0510-9
Delmotte, F., Bourguet, D., Franck, P., Guillemaud, T., Reboud, X., Vacher, C., et al. (2016). Combining selective pressures to enhance the durability of disease resistance genes. Front. Plant Sci. 7:1916. doi: 10.3389/fpls.2016.01916
Dice, L. R. (1945). Measures of the amount of ecologic association between species. Ecology 26, 297–302. doi: 10.2307/1932409
Divilov, K., Barba, P., Cadle, L., and Bruce, D. (2018). Single and multiple phenotype QTL analyses of downy mildew resistance in interspecific grapevines. Theor. Appl. Genet. 131, 1133–1143. doi: 10.1007/s00122-018-3065-y
Dry, I., Riaz, S., Fuchs, M., Sosnowski, M., and Thomas, M. (2019). “Scion breeding for resistance to biotic stresses,” in The Grape Genome, eds D. Cantu and A. M. Walker (Berlin: Springer), 319–347. doi: 10.1007/978-3-030-18601-2_15
Eibach, R., and Töpfer, R. (2015). “Traditional grapevine breeding techniques. Grapevine breeding programs for the wine industry,” in Grapevine Breeding Programs for the Wine Industry, ed. A. Reynolds (Amsterdam: Elsevier), 3–22. doi: 10.1016/b978-1-78242-075-0.00001-6
Eibach, R., Zyprian, E., Welter, L., and Töpfer, R. (2007). The use of molecular markers for pyramiding resistance genes in grapevine breeding. Vitis J. Grapevine Res. 46, 120–124. doi: 10.5073/vitis.2007.46.120-124
Eisenmann, B., Czemmel, S., Ziegler, T., Buchholz, G., Kortekamp, A., Trapp, O., et al. (2019). Rpv3 – 1 mediated resistance to grapevine downy mildew is associated with specific host transcriptional responses and the accumulation of stilbenes. BMC Plant Biol. 19:343. doi: 10.1186/s12870-019-1935-3
Ellis, C., and Turner, J. G. (2001). The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033. doi: 10.1105/tpc.13.5.1025
Feechan, A., Anderson, C., Torregrosa, L., Jermakow, A., Mestre, P., Wiedemann-Merdinoglu, S., et al. (2013). Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J. 76, 661–674. doi: 10.1111/tpj.12327
Feuillet, C., Travella, S., Stein, N., and Albar, L. (2003). Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc. Natl. Acad. Sci. U.S.A. 100, 15253–15258. doi: 10.1073/pnas.2435133100
Fischer, B., Salakhutdinov, I., Akkurt, M., Eibach, R., Edwards, K., Toepfer, R., et al. (2004). Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor. Appl. Genet. 108, 501–515. doi: 10.1007/s00122-003-1445-3
Fontaine, M. C., Austerlitz, F., Giraud, T., Labbé, F., Papura, D., Richard-Cervera, S., et al. (2013). Genetic signature of a range expansion and leap-frog event after the recent invasion of Europe by the grapevine downy mildew pathogen Plasmopara viticola. Mol. Ecol. 22, 2771–2786. doi: 10.1111/mec.12293
Foria, S., Copetti, D., Eisenmann, B., Magris, G., Vidotto, M., Scalabrin, S., et al. (2020). Gene duplication and transposition of mobile elements drive evolution of the Rpv3 resistance locus in grapevine. Plant J. 101, 529–542. doi: 10.1111/tpj.14551
Frichot, E., and François, O. (2015). LEA: an R package for landscape and ecological association studies. Methods Ecol. Evol. 6, 925–929. doi: 10.1111/2041-210X.12382
Friso, G., and van Wijk, K. J. (2015). Post-translational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487. doi: 10.1104/pp.15.01378
Furlan, G., Klinkenberg, J., and Trujillo, M. (2012). Regulation of plant immune receptors by ubiquitination. Front. Plant Sci. 3:238. doi: 10.3389/fpls.2012.00238
Hollomon, D. (2015). “Fungicide resistance: 40 years on and still a major problem,” in Fungicide Resistance in Plant Pathogens: Principles and a Guide to Practical Management, eds H. Ishii and D. Hollomon (Cham: Springer), 3–11. doi: 10.1007/978-4-431-55642-8_1
Imazio, S., Maghradze, D., De Lorenzis, G., Bacilieri, R., Laucou, V., This, P., et al. (2013). From the cradle of grapevine domestication: molecular overview and description of Georgian grapevine (Vitis vinifera L.) germplasm. Tree Genet. Genomes 9, 641–658. doi: 10.1007/s11295-013-0597-9
Islam, M. Z., and Yun, H. K. (2016). Identification and expression profiles of six transcripts encoding carboxylesterase protein in Vitis flexuosa infected with pathogens. Plant Pathol. J. 32, 347–356. doi: 10.5423/PPJ.OA.11.2015.0241
Jombart, T. (2008). ADEGENET: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. doi: 10.1093/bioinformatics/btn129
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jürges, G., Kassemeyer, H.-H., Dürrenberger, M., Düggelin, M., and Nick, P. (2009). The mode of interaction between Vitis and Plasmopara viticola Berk. & Curt. Ex de Bary depends on the host species. Plant Biol. 11, 886–898. doi: 10.1111/j.1438-8677.2008.00182.x
Kim, Y. S., Lee, H. H., Ko, M. K., Song, C. E., Bae, C.-Y., Lee, Y. H., et al. (2001). Inhibition of fungal appressorium formation by pepper (Capsicum annuum) Esterase. Mol. Plant Microbe Interact. 14, 80–85. doi: 10.1094/MPMI.2001.14.1.80
Korte, A., and Farlow, A. (2013). The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9, 1–9.
Kragelund, B. B., Andersen, K. V., Madsen, J. C., Knudsen, J., and Poulsen, F. M. (1993). Three-dimensional structure of the complex between acyl-coenzyme a binding protein and palmitoyl-coenzyme A. J. Mol. Biol. 230, 1260–1277. doi: 10.1006/jmbi.1993.1240
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Laucou, V., Launay, A., Bacilieri, R., Lacombe, T., Adam-Blondon, A.-F., Bérard, A., et al. (2018). Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS One 13:e0192540. doi: 10.1371/journal.pone.0192540
Li, X., Han, Y., Wei, Y., Acharya, A., Farmer, A. D., Ho, J., et al. (2014). Development of an alfalfa SNP array and its use to evaluate patterns of population structure and linkage disequilibrium. PLoS One 9:e84329. doi: 10.1371/journal.pone.0084329
Lin, H., Leng, H., Guo, Y., Kondo, S., Zhao, Y., Shi, G., et al. (2019). QTLs and candidate genes for downy mildew resistance conferred by interspecific grape (V. vinifera L. × V. amurensis Rupr.) crossing. Sci. Hortic. 244, 200–207. doi: 10.1016/j.scienta.2018.09.045
Lipka, A. E., Tian, F., Wang, Q., Peiffer, J., Li, M., Bradbury, P. J., et al. (2012). GAPIT: genome association and prediction integrated tool. Bioinformatics 28, 2397–2399. doi: 10.1093/bioinformatics/bts444
Maddalena, G., Delmotte, F., Bianco, P. A., De Lorenzis, G., and Toffolatti, S. L. (2020). Genetic structure of Italian population of the grapevine downy mildew agent, Plasmopara viticola. Ann. Appl. Biol. 176, 257–267. doi: 10.1111/aab.12567
Marguerit, E., Boury, C., Manicki, A., Donnart, M., Butterlin, G., Némorin, A., et al. (2009). Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 118, 1261–1278. doi: 10.1007/s00122-009-0979-4
Marrano, A., Birolo, G., Prazzoli, M. L., Lorenzi, S., Valle, G., and Grando, M. S. (2017). SNP-discovery by RAD-sequencing in a germplasm collection of wild and cultivated grapevines (V. vinifera L.). PLoS One 12:e0170655. doi: 10.1371/journal.pone.0170655
Mazzucotelli, E., Belloni, S., Marone, D., De Leonardis, A., Guerra, D., Di Fonzo, N., et al. (2006). The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr. Genomics 7, 509–522. doi: 10.2174/138920206779315728
Merdinoglu, D., Schneider, C., Prado, E., Wiedemann-Merdinoglu, S., and Mestre, P. (2018). Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 52, 203–209. doi: 10.20870/oeno-one.2018.52.3.2116
Merdinoglu, D., Wiedeman-Merdinoglu, S., Coste, P., Dumas, V., Haetty, S., Butterlin, G., et al. (2003). Genetic analysis of downy mildew resistance derived from Muscadinia rotundifolia. Acta Hortic. 603, 451–456. doi: 10.17660/ActaHortic.2003.603.57
Migicovsky, Z., Sawler, J., Money, D., Eibach, R., Miller, A. J., Luby, J. J., et al. (2016). Genomic ancestry estimation quantifies use of wild species in grape breeding. BMC Genomics 17:478. doi: 10.1186/s12864-016-2834-8
Monaghan, J., and Li, X. (2010). The heat repeat protein ILITYHIA is required for plant immunity. Plant Cell Physiol. 51, 742–753. doi: 10.1093/pcp/pcq038
Moreira, F. M., Madini, A., Marino, R., Zulini, L., Stefanini, M., Velasco, R., et al. (2011). Genetic linkage maps of two interspecific grape crosses (Vitis spp.) used to localize quantitative trait loci for downy mildew resistance. Tree Genet. Genomes 7, 153–167. doi: 10.1007/s11295-010-0322-x
Myles, S., Chia, J.-M., Hurwitz, B., Simon, C., Zhong, G. Y., Buckler, E., et al. (2010). Rapid genomic characterization of the genus Vitis. PLoS One 5:e8219. doi: 10.1371/journal.pone.0008219
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Ochssner, I., Hausmann, L., and Töpfer, R. (2016). Rpv14, a new genetic source for Plasmopara viticola resistance conferred by Vitis cinerea. Vitis 55, 79–81. doi: 10.5073/vitis.2016.55.79-81
Pontier, D., Godiard, L., Marco, Y., and Roby, D. (1994). hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant/pathogen interactions. Plant J. 5, 507–521. doi: 10.1046/j.1365-313X.1994.05040507.x
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
Putterill, J. J., Plummer, K. M., Newcomb, R. D., and Marshall, S. D. G. (2003). The carboxylesterase gene family from Arabidopsis thaliana. J. Mol. Evol. 57, 487–500. doi: 10.1007/s00239-003-2492-8
Sapkota, S., Chen, L.-L., Schreiner, K., Ge, H., and Hwang, C.-F. (2015). A phenotypic study of Botrytis bunch rot resistance in Vitis aestivalis-derived ‘Norton’ grape. Trop. Plant Pathol. 40, 279–282. doi: 10.1007/s40858-015-0028-6
Sapkota, S., Chen, L.-L., Yang, S., Hyma, K. E., Cadle-Davidson, L., and Hwang, C.-F. (2019). Construction of a high-density linkage map and QTL detection of downy mildew resistance in Vitis aestivalis-derived ‘Norton.’. Theor. Appl. Genet. 132, 137–147. doi: 10.1007/s00122-018-3203-6
Schulze-Lefert, P. (2004). Knocking on the heaven’s wall: pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 7, 377–383. doi: 10.1016/j.pbi.2004.05.004
Schwander, F., Eibach, R., Fechter, I., Hausmann, L., Zyprian, E., and Töpfer, R. (2012). Rpv10: a new locus from the Asian Vitis gene pool for pyramiding downy mildew resistance loci in grapevine. Theor. Appl. Genet. 124, 163–176. doi: 10.1007/s00122-011-1695-4
Sharma, M., and Pandey, G. K. (2016). Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 6:1218. doi: 10.3389/fpls.2015.01218
Song, S., Fu, P., and Lu, J. (2018). “Downy mildew resistant QTLs in Vitis amurensis ‘Shuang Hong’ grapevine,” in Proceedings of the XIIth International Grapevine Breeding and Genetics Conference, Bordeaux.
Taylor, N. G., Laurie, S., and Turner, S. R. (2000). Multiple cellulose synthase catalytic subunits are required for cellulose synthesis in Arabidopsis. Plant Cell 12:2529. doi: 10.2307/3871246
Taylor, N. G., Scheible, W.-R., Cutler, S., Somerville, C. R., and Turner, S. R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11, 769–779. doi: 10.1105/tpc.11.5.769
Toffolatti, S. L., De Lorenzis, G., Brilli, M., Moser, M., Shariati, V., Tavakol, E., et al. (2020). Novel aspects on the interaction between grapevine and Plasmopara viticola: dual-RNA-Seq analysis highlights gene expression dynamics in the pathogen and the plant during the battle for infection. Genes 11:261. doi: 10.3390/genes11030261
Toffolatti, S. L., De Lorenzis, G., Costa, A., Maddalena, G., Passera, A., Bonza, M. C., et al. (2018a). Unique resistance traits against downy mildew from the center of origin of grapevine (Vitis vinifera). Sci. Rep. 8:12523. doi: 10.1038/s41598-018-30413-w
Toffolatti, S. L., Maddalena, G., Salomoni, D., Maghradze, D., Bianco, P. A., and Failla, O. (2016). Evidence of resistance to the downy mildew agent Plasmopara viticola in the Georgian Vitis vinifera germplasm. Vitis J. Grapevine Res. 55, 121–128. doi: 10.5073/vitis.2016.55.121-128
Toffolatti, S. L., Russo, G., Campia, P., Bianco, P. A., Borsa, P., Coatti, M., et al. (2018b). A time-course investigation of resistance to the carboxylic acid amide mandipropamid in field populations of Plasmopara viticola treated with anti-resistance strategies. Pest Manag. Sci. 74, 2822–2834. doi: 10.1002/ps.5072
Townsend, G., and Heuberger, J. (1947). Methods for estimating losses caused by disease in fungicide experiments. Plant Dis. Rep. 27, 340–343.
Venuti, S., Copetti, D., Foria, S., Falginella, L., Hoffmann, S., Bellin, D., et al. (2013). Historical introgression of the downy mildew resistance gene Rpv12 from the asian species Vitis amurensis into grapevine varieties. PLoS One 8:e0061228. doi: 10.1371/journal.pone.0061228
Vezzulli, S., Malacarne, G., Masuero, D., Vecchione, A., Dolzani, C., Goremykin, V., et al. (2019). The Rpv3-3 haplotype and stilbenoid induction mediate downy mildew resistance in a grapevine interspecific population. Front. Plant Sci. 10:234. doi: 10.3389/fpls.2019.00234
Welter, L. J., Göktürk-Baydar, N., Akkurt, M., Maul, E., Eibach, R., Töpfer, R., et al. (2007). Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breed. 20, 359–374. doi: 10.1007/s11032-007-9097-7
Wiedemann-Merdinoglu, S., Prado, E., Coste, P., Dumas, V., Butterlin, G., Bouquet, A., et al. (2006). “Genetic analysis of resistance to downy mildew derived from Muscadinia rotundifolia,” in Proceedings of the Ninth International Conference on Grape Genetics and Breeding, Udine, Italy 2-6 July, Udine.
Xia, Y., Yu, K., Gao, Q., Wilson, E. V., Navarre, D., Kachroo, P., et al. (2012). Acyl CoA binding proteins are required for cuticle formation and plant responses to microbes. Front. Plant Sci. 3:224. doi: 10.3389/fpls.2012.00224
Xu, S., Gupta, S., and Jin, L. I. (2010). PEAS V1.0: a package for elementary analysis of SNP data. Mol. Ecol. Resour. 10, 1085–1088. doi: 10.1111/j.1755-0998.2010.02862.x
Yobrégat, O. (2018). Introduction to resistant vine types: a brief history and overview of the situation. OENO One 52, 241–246. doi: 10.20870/oeno-one.2018.52.3.2220
Zhao, H., Nettleton, D., Soller, M., and Dekkers, J. C. M. (2005). Evaluation of linkage disequilibrium measures between multi-allelic markers as predictors of linkage disequilibrium between markers and QTL. Genet. Res. 86, 77–87. doi: 10.1017/S001667230500769X
Keywords: Georgia, GWAS, NB-LRR, SNP, Southern Caucasus, biotrophic plant pathogen, marker-assisted breeding
Citation: Sargolzaei M, Maddalena G, Bitsadze N, Maghradze D, Bianco PA, Failla O, Toffolatti SL and De Lorenzis G (2020) Rpv29, Rpv30 and Rpv31: Three Novel Genomic Loci Associated With Resistance to Plasmopara viticola in Vitis vinifera. Front. Plant Sci. 11:562432. doi: 10.3389/fpls.2020.562432
Received: 15 May 2020; Accepted: 17 September 2020;
Published: 08 October 2020.
Edited by:
Soren K. Rasmussen, University of Copenhagen, DenmarkReviewed by:
Sabine W. Merdinoglu, INRA Centre Colmar, FranceWalter Chitarra, Council for Agricultural and Economics Research (CREA), Italy
Copyright © 2020 Sargolzaei, Maddalena, Bitsadze, Maghradze, Bianco, Failla, Toffolatti and De Lorenzis. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella De Lorenzis, Z2FicmllbGxhLmRlbG9yZW56aXNAdW5pbWkuaXQ=; Silvia Laura Toffolatti, c2lsdmlhLnRvZmZvbGF0dGlAdW5pbWkuaXQ=
 Maryam Sargolzaei
Maryam Sargolzaei Giuliana Maddalena
Giuliana Maddalena Nana Bitsadze
Nana Bitsadze David Maghradze
David Maghradze Piero Attilio Bianco
Piero Attilio Bianco Osvaldo Failla
Osvaldo Failla Silvia Laura Toffolatti
Silvia Laura Toffolatti Gabriella De Lorenzis
Gabriella De Lorenzis