
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 23 September 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.555572
This article is part of the Research Topic Reproductive Barriers and Gene Introgression in Rice Species View all 16 articles
Interspecific and intraspecific hybrid sterility is a typical and common phenomenon of postzygotic reproductive barrier in rice. This is an indicator of speciation involved in the formation of new species or subspecies, and it significantly hampers the utilization of favorable genes from distant parents for rice improvement. The Oryza genus includes eight species with the same AA genome and is a model plant for studying the nature of hybrid sterility and its relationship with speciation. Hybrid sterility in rice is mostly controlled by nuclear genes, with more than 50 sterility loci genetically identified to date, of which 10 hybrid sterility loci or pairs were cloned and characterized at the molecular level. Comparing the mapping results for all sterility loci reported indicated that some of these loci from different species should be allelic to each other. Further research revealed that interactions between the multiple alleles at the hybrid sterility locus caused various genetic effect. One hypothesis for this important phenomenon is that the hybrid sterility loci are orthologous loci, which existed in ancient ancestors of rice. When one or more ancestors drifted to different continents, genetic divergence occurred because of adaptation, selection, and isolation among them such that various alleles from orthologous loci emerged over evolutionary time; hence, interspecific hybrid sterility would be mainly controlled by a few orthologous loci with different alleles. This hypothesis was tested and supported by the molecular characterization of hybrid sterility loci from S1, S5, Sa, qHMS7, and S27. From this, we may further deduce that both allelic and non-allelic interactions among different loci are the major genetic basis for the interspecific hybrid sterility between O. sativa and its AA genome relatives, and the same is true for intraspecific hybrid sterility in O. sativa. Therefore, it is necessary to raise the near-isogenic lines with various alleles/haplotypes and pyramided different alleles/haplotypes from sterile loci in the same genetic background aiming to study allelic and non-allelic interaction among different hybrid sterility loci in the AA genome species. Furthermore, the pyramiding lines ought to be used as bridge parents to overcome hybrid sterility for rice breeding purposes.
Reproductive barriers are very common and important phenomena in biology, being widely observed in animal and plant populations. Reproductive isolation was classified into pre-zygotic and post-zygotic isolation mechanisms (Smith, 1989). The former prevents the formation of hybrids, and the latter acts after the formation of a hybrid and includes hybrid necrosis, weakness, hybrid sterility, and lethality (Stebbins, 1950).
Hybrid sterility is generally thought to be the most pervasive post-zygotic isolating mechanism, which provides an initial driving force for genetic differentiation and thus plays a key role in speciation (Orr and Presgraves, 2000). One of the major challenges in biology is to understand the origin of species, so the relationship between the hybrid sterility and the formation of new species, or subspecies, is a subject of major interest in evolutionary biology (Darwin, 1859; Coyne and Orr, 2004). Yet, the introgression of favorable genes from distantly relatives through wide crossing and heterosis utilization between subspecies or species are hindered by the hybrid sterility.
Various causes have been ascribed to explain hybrid sterility, such as meiotic irregularities (Yao et al., 1958), chromosomal aberrations (Henderson, 1964), and cytoplasmic-nuclear interactions (Shinjyo, 1975). Recent reports have also speculated that structural variation at particular trait loci may contribute to the intersubspecific hybrid sterility between the O. sativa L. ssp. indica and O. sativa L. ssp. japonica accessions (Shen et al., 2017; Wang et al., 2018).
Nevertheless, most known cases of sterility arise from a disharmonious interaction between nuclear genes derived from their respective parents (Stebbins, 1958; Grant, 1981). Genes for hybrid sterility have been reported frequently in fungi, animals, and plants (Brideau et al., 2006; Lee et al., 2008; Bikard et al., 2009). This prominence indicates that gene interactions are critically involved in generating hybrid sterility.
Hybrid sterility occurs widely in hybrids of Asian cultivated rice species, especially between the Asian and African rice species, and between the cultivated rice species and their wild relatives. Because Oryza genus had broad genetic diversity and well-characterized genetic base, it is perhaps one of the best model plants to study the nature of hybrid sterility. Based upon published reports of more than 50 sterility loci genetically identified, and 10 hybrid sterility loci or pairs cloned and characterized at the molecular level, this review examines the links between the evolutionary relationship of the Oryza genus and hybrid sterility, the genetic models of hybrid sterility, the allelic variation of orthologous loci for hybrid sterility, the non-allelic interactions for hybrid sterility, and strategies for overcoming hybrid sterility in rice improvement.
The Oryza genus has eight AA genome species of diploid chromosome, of which six are wild and two are cultivated (Vaughan, 1989). One of the cultivated species, O. sativa L., has a global distribution and is now classified as two subspecies: O. sativa L. ssp. japonica and O. sativa L. ssp. indica. The japonica subspecies is further classified as three subpopulations, tropical, temperate, and aromatic, while the indica subspecies contains two subpopulations, indica and aus (Garris et al., 2005) or six subpopulations (Wang et al., 2018). Another cultivated species is O. glaberrima Steud., which is localized in West Africa and commonly referred to as African rice (Morishima et al., 1963). The six wild species are O. rufipogon Griff. and O. nivara Sharma et Shastry from Asia; O. longistaminata A. Chev. et Roehr. and O. barthii A. Chev. from sub-Saharan Africa; O. meridionalis Ng from Australia, and O. glumaepatula Steud. from South America (Vaughan et al., 2003). Hybrid sterility in rice was frequently observed between and within the AA genome species and was thought to serve as an important indicator for studying these species’ relationships, besides their morphological, physiological, ecological traits, and molecular markers (Kato et al., 1928; Chu et al., 1969; Morishima, 1969; Lu et al., 1998; Naredo et al., 1998; Lu et al., 2000; Vaughan et al., 2003).
The origin and evolutionary relationship of the AA genome nevertheless remain quite contentious, with different opinions and arguments being advanced. Most researchers now believe that the domestication of rice from wild to cultivated species occurred independently in Africa and Asia, respectively (Londo et al., 2006; Vaughan et al., 2008). Recent studies confirmed that O. glaberrima was domesticated directly from an O. barthii subgroup in a single domestication center along the Niger river (Wang et al., 2014).
One hypothesis is that japonica was derived from indica (Chang, 1976; Oka, 1988), while according to an alternative hypothesis both indica and japonica were originated independently from their wild ancestors (Second, 1982; Bautista et al., 2001; Cheng et al., 2003). Based on an analysis of their whole chloroplast genome sequences, the maternal genome of japonica may have been derived from O. rufipogon and that of indica may have originated from O. nivara, which would support the independent domestication theory (Wambugu et al., 2015). By employing the indica-japonica specific insertion/deletion markers to evaluate the genetic relationships within the genus Oryza, Chin et al. (2017) recently provided more evidence of indica and japonica evolving independently.
All the above studies concerned with rice’s origin and evolutionary history relied on evidence from either archaeological analyses or genetic markers. Given that the strength of reproductive isolation is significantly and positively correlated with parental divergence (Coyne and Orr, 2004), comparative analyses of the timing and evolutionary progression of genetic changes underlying reproductive isolation genes could us better understand the mechanisms of speciation (Moyle and Payseur, 2009). With the advent of molecular biology and in recent years genomic studies especially, the hybrid sterility genes, also called “speciation genes”, provide an unique angle of view for understanding the genetic, origin and evolution of the Oryza genus. For instance, the molecular evolution analysis of the S5 locus, one of the important female sterility loci in indica-japonica hybrids, revealed that indica and japonica subspecies of the Asian cultivated rice O. sativa were domesticated independently from wild species (Du et al., 2011). This result thus supported the independent origin theory of indica and japonica and was consistent with the finding reported by Wambugu et al. (2015). The cloning of S1 locus, which is the most important genetic factor causing the hybrid sterility between two cultivated species, provides experimental evidence for the independent origin of Asian cultivated rice and African cultivated rice (Xie et al., 2019b).
Two genetic models are generally accepted for explaining the genetic mechanism underpinning hybrid sterility in rice according to the Mendelian inheritance: allelic interaction (Kitamura, 1962) and epistatic interaction (Oka, 1957; Oka, 1974). The former proposes that a genetic interaction between two divergent alleles on a single locus causes the abortion of gametes carrying a specific allele, while the latter proposes that epistatic interactions between two loci cause hybrid sterility.
To date, more than 50 hybrid sterility loci in rice have been identified from gene mapping populations generated from various germplasm lines (Supplementary Table 1). Some of these loci cause pollen sterility, some cause female gamete abortion, and a few cause the abortion of both. Yet most of these loci seem to act independently on hybrid sterility, which could be explained by the one-locus interaction model; for those showing epistatic interactions, they fit the two-loci interaction model.
From the mapping loci, 10 hybrid sterility loci or pairs of epistatic interaction were cloned and characterized at the molecular level, for which the genetics of seven loci—S1, S5, S7, Sa, Sc, hsa1, and qHMS7—follow the one-locus interaction model (Chen et al., 2008; Long et al., 2008; Yang et al., 2012; Kubo et al., 2016; Yu et al., 2016; Shen et al., 2017; Xie et al., 2017; Koide et al., 2018; Yu et al., 2018; Xie et al., 2019b). Most loci contain two or more closely linked genes interacting to cause gamete sterility, which can take these recognized forms: a single Mendelian factor, such as two genes at Sa, qHMS7 and hsa1 locus (Long et al., 2008; Kubo et al., 2016; Yu et al., 2018); three genes at S1 and S5 loci (Yang et al., 2012; Xie et al., 2017; Koide et al., 2018; Xie et al., 2019b); and genomic structural variation at the Sc locus (Shen et al., 2017). All can be regarded as interaction between different alleles from a single Mendelian factor to control hybrid sterility.
The other three pairs (S27/28, DPL1/DPL2, and DGS1/DGS2) follow the two-loci interaction model (Mizuta et al., 2010; Yamagata et al., 2010; Nguyen et al., 2017). These gamete-essential genes were interchromosmal duplicated segment from their ancestral loci. The reciprocal loss of one of the duplicated genes caused the sterility in hybridization in the divergent species. Therefore, genetic segregation and recombination gave rise to gametes lacking any duplicated functional genes in the hybrids, thus leading to hybrid sterility (Mizuta et al., 2010; Yamagata et al., 2010; Nguyen et al., 2017).
However, the molecular mechanisms underlying each of the loci are in fact complex though the genetic basis of reproductive isolation seems simple according to classical genetic analysis. Not only are the genes involved in hybrid sterility characterized by very different functional categories, the way in which they interact to cause sterility is also distinct, and this topic has been reviewed before (Ouyang and Zhang, 2013; Xie et al., 2019a). However, it is necessary to recheck hybrid sterility from different perspectives.
In reviewing the progress made in the genetic mapping study of hybrid sterility loci/QTL between O. sativa and its AA genome relatives, six “hot spots” from different species were frequently identified (Figure 1). Comparing the mapping results for all these sterility loci suggested that some sterility loci arising from multiple species should be allelic to each other, since they were located to the same chromosome region and were identified as having similar genetic activity.
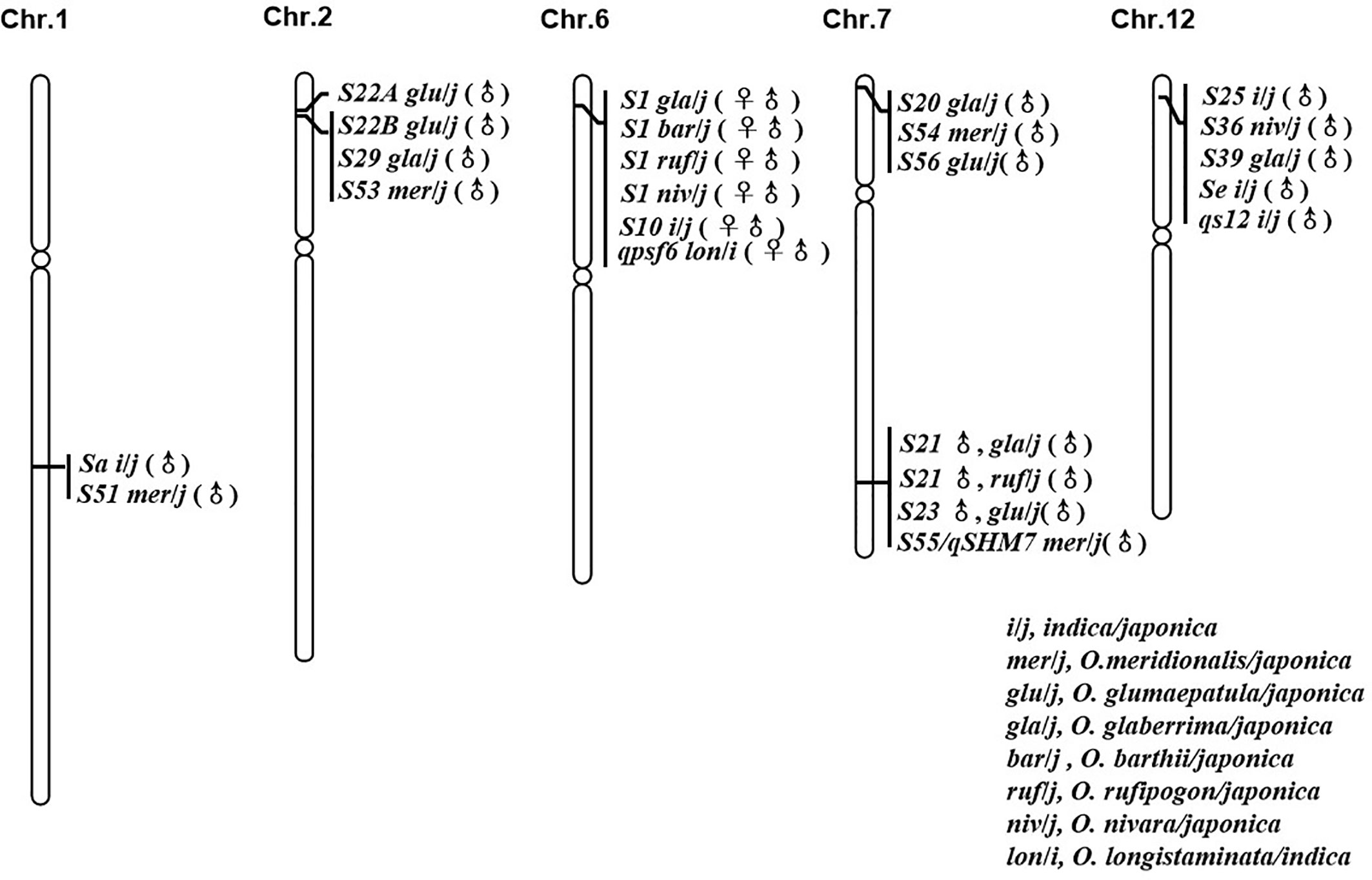
Figure 1 Loci with good co-linear relationship and similar genetic action for hybrid sterility in rice.
The interspecific hybrid sterility locus S1 on chromosome 6 was first identified in the cross between O. sativa (T65wx) and O. glaberrima (W025) (Sano et al., 1979). It is frequently reported by a large number of studies between the two cultivated rice species, O. sativa (indica/japonica) and O. glaberrima (Koide et al., 2008; Garavito et al., 2010; Zhou et al., 2010; Li J. et al., 2012). The S1 locus functions as a “gamete eliminator”: both male and female gametes carrying the allele of O. sativa are aborted in the heterozygote, for which the type of pollen abortion was of an empty abortion phenotype. The S10 locus, inducing both male and female gametes’ abortion in an intraspecific hybrid cross between T65wx (japonica type) and PTB 10 (indica type), was mapped onto the similar chromosome position as S1 on chromosome 6 (Sano et al., 1994), with some later studies indicating that S1 and S10 are allelic to each other (Heuer and Miézan, 2003; Zhu et al., 2005). Chen et al. (2009) detected one main-effect QTL, qpsf6, for pollen and spikelet fertility on the short arm of chromosome 6 close to the SSR marker RM587, from the cross between an indica rice cultivar RD23 of O. sativa and an accession of O. longistaminata. Comparing their position and effect indicated that this QTL coincides with the gamete eliminator S1 (Chen et al., 2009). Recently, a genetic study found that S1 also was a major sterility locus in the hybrid combinations obtained from O. sativa crossed with O. rufipogon, O. nivara, and O. barthii, respectively (Yang et al., 2016), which strongly suggests there was an orthologous hybrid sterility locus controlling hybrid sterility between O. sativa and its AA genome relatives in this area on rice chromosome 6.
The pollen killer locus S22, initially derived from the cross between O. glumaepatula and O. sativa, was subsequently identified as two closed but linked loci, S22A and S22B. Since the sterile alleles S22A and S22B are closely linked and contributed to hybrid sterility in the one-locus allelic interaction model, the S22A and S22B loci were regarded as a single Mendelian locus in their initial mapping experiments (Sobrizal et al., 2000a; Sakata et al., 2014). A pollen killer locus S29(t), which induced hybrid sterility in the cross between O. glaberrima and O. sativa, was mapped onto the similar position to S22 on chromosome 2, whose comparative mapping indicated that S29(t) corresponded to S22B (Hu et al., 2006; Sakata et al., 2014). S53(t), caused pollen sterility in an O. sativa–O. meridionalis hybrid, was identified on the same chromosome region harboring S22B (Li et al., 2018b). The male gametes carrying the O. sativa allele were viable whereas those carrying the O. glumaepatula/O. meridionalis allele were aborted in the S22B and S53(t) loci; hence, these loci from different species should be allelic to each other with multiple alleles. This implies there might be another orthologous hybrid sterility locus between the Asian cultivated rice O. sativa and the African rice O. glaberrima, and its wild relatives O. glumaepatula, O. meridionalis on rice chromosome 2.
The major locus Sa conferring the indica-japonica hybrid male sterility was identified on chromosome 1 with the genetic background of a japonica variety (Zhuang et al., 1999). Work by Li et al. (2018b) identified a pollen killer S51(t) on chromosome 1 from O. meridionalis. The sterile pollen in the heterozygotes of S51(t) and Sa showed a similarly empty abortion phenotype under I2-KI staining. Additionally, both S51(t)j and Saj, the japonica alleles, were aborted and were incapable of being transmitted to the progeny via the male gametes. According to a comparison of the location and the genetic mode of locus action, the S51(t) can be allelic to Sa (Li et al., 2018b).
The hybrid sterility locus S20 was identified on the distal end of the short arm of chromosome 7, from the cross between japonica rice (Taichung 65) and African rice Acc. IRGC104038 (Doi et al., 1999). An examination of map positions also revealed that S20 was highly likely to be allelic to S54(t) and S56(t), which were respectively identified in the hybridization crossed combinations between O. sativa (japonica) and O. meridionalis (Li et al., 2018b) and between O. sativa (japonica) and O. glumaepatula (Zhang et al., 2018). Just like S20, the japonica alleles of S54(t) and S56(t) were aborted and could not be inherited in progeny via male gametes, so plants with normal pollen fertility were homozygous for O. meridionalis and O. glumaepatula allele in the inbreeding population raised from the semisterile individuals (Li et al., 2018b; Zhang et al., 2018).
The hybrid sterility locus S21 was identified on the distal end of the long arm of chromosome 7, from the cross of japonica rice (Taichung 65) and African rice Acc. IRGC104038 (Doi et al., 1999). But a new allele of S21 was identified from O. rufipogon Acc. IRGC105715 (Miyazaki et al., 2007). Using the O. glumaepatula introgression lines with the background of O. sativa (Taichung 65), the pollen semi-sterile locus S23(t) was also located on the long arm of chromosome 7 and was thought to be allelic to S21 from O. glaberrima and O. rufipogon after comparing their map positions (Sobrizal et al., 2000b; Miyazaki et al., 2007). The pollen killer locus S55(t)/qHMS7, from O. meridionalis, has a map position similar to that of S21 identified from O. rufipogon, O. glaberrima, and S23(t) identified from O. glumaepatula (Doi et al., 1999; Sobrizal et al., 2000b; Miyazaki et al., 2007; Li et al., 2018b; Yu et al., 2018). This congruence pointed to a major pollen sterility locus on the long arm of chromosome 7 that is capable of inducing sterility in the hybridization combinations arising from O. sativa crossed with O. glaberrima, O. rufipogon, O. glumaepatula, and O. meridionalis, respectively, whose sequence analysis was based on the cloning of S23 and qHMS7 (Yu et al., 2018; Fang et al., 2019).
S36 and S25 are two F1 pollen sterility loci, both found on the distal end of the short arm of chromosome 12 from the interspecific cross of O. sativa (japonica) and O. nivara, and the intersubspecific cross of O. sativa, respectively (Kubo et al., 2001; Win et al., 2009). The pollen sterility locus Se and hybrid male sterility QTL qS12, both from the intersubspecific cross between japonica and indica, were mapped onto the short arm of chromosome 12 (Zhu et al., 2008; Zhang H. et al., 2011). Further, the pollen killer S39(t) identified from the interspecific cross between O. sativa ssp. japonica and O. glaberrima was likewise mapped onto the same chromosome region of S36 and S25 (Xu et al., 2014). Those pollen grains carrying the japonica allele were sterile in the heterozygous state. Having a similar map position and same genetic activity, S25, S36, S39, qS12, and Se might therefore be multiple alleles on the same locus responsible for hybrid sterility in rice.
The six hot spots mentioned above each includes at least two hybrid sterile alleles from different interspecific hybrid crosses, and researchers believe these loci should be allelic to each other, though their allelic relationship has yet to be tested. Nonetheless, the existence of these multiple alleles implies that the hybrid sterility among AA genome species of genus Oryza could be governed by a few orthologous loci. It is thus imperative to clarify the allelic relationships of these loci, although any robust confirmation of this allelism is difficult to obtain due to their diverse genetic backgrounds. Based upon this consideration, the near-isogenic lines (NILs) of 5 hybrid sterility loci (S1, S39, S44, S53, and S21/qHMS7) have been developed in the same genetic background, using 35 rice accessions from eight species of AA genome as donors and one japonica cultivar (Dianjingyou 1) as the recurrent parent. And the allelic relationship confirmation is ongoing.
One hypothesis for multiple alleles from different species is that these hybrid sterility loci are orthologous loci, which existed in ancient ancestors. When one or more ancestors drifted to different continents, genetic divergence occurred because of adaptation, selection, and isolation among them. With this accumulated genetic differentiation, the various alleles may divide the original function, lose it altogether, or introduce a new function (Lynch and Force, 2000). Hence, interspecific hybrid sterility would be mainly controlled by a few orthologous loci with different alleles/haplotypes. This hypothesis was proven by the molecular characterization of cloned hybrid sterility loci in rice.
The hybrid sterility locus qHMS7, which confers pollen semi-sterility in the hybrid between O. meridionalis and O. sativa ssp. japonica Dianjingyou 1 (DJY1), was cloned and molecularly characterized (Yu et al., 2018). The O. sativa allele of ORF2 (ORF2-D) and ORF3 (ORF3-D) respectively encoded the toxin and antidote, while ORF2-mer was non-functional and ORF3 was absent in O. meridionalis, which induced the pollens carrying the O. meridionalis allele to be aborted (Yu et al., 2018). The analyzed gene structure of the qHMS7 locus indicated that ORF2 was present in all sequenced accessions, with a total 27 haplotypes of ORF2 identified, yet only one haplotype of ORF3 (ORF3-D) was identified in parts of O. rufipogon and most of Asian-cultivated rice accessions, but none in O. meridionalis, O. longistaminata, O. barthii, and O. glaberrima (Yu et al., 2018). So, qHMS7 was not only detected in the cross of O. sativa–O. meridionalis (Yu et al., 2018; Li et al., 2018b) but also in the crosses of O. sativa–O. glaberrima and O. sativa–O. rufipogon as S21 (Doi et al., 1999; Miyazaki et al., 2007). The pollen semi-sterility locus S23 in O. sativa–O. glumaepatula has a similar map position to qHMS7 (Sobrizal et al., 2000b; Fang et al., 2019). Sequence analysis revealed that S23 was allelic to qHMS7, the ORF2 and ORF3 of S23 have the same “Toxin-Antidote” function when compared with qHMS7; however, the genetic effect of S23 differs from qHMS7 in two ways (Fang et al., 2019). On the one hand, F1 pollen fertility was semi-sterility (51.16 ± 1.29%) for qHMS7 and no homozygote for O. meridionalis was obtained, indicating the pollen grains carrying O. meridionalis allele had aborted completely; whereas, for S23, its F1 pollen fertility was higher (63.15 ± 13.49%) than that of qHMS7 and a few homozygotes of O. glumaepatula was found available, indicating that the pollen grains carrying the O. glumaepatula allele were not completely sterile (Yu et al., 2018; Fang et al., 2019). On the other hand, the pollen sterility of F1 hybrid could only be observed in a short photoperiods, but not in a long photoperiods at the S23 locus, yet no phenotypic differences were observed in different environments at the qHMS7 locus (Li et al., 2018b; Yu et al., 2018; Fang et al., 2019). The allele on O. sativa interacted with the respective alleles from O. meridionalis and O. glumaepatula, resulting in hybrid sterility with different genetic effects; accordingly, the allele on O. glumaepatula should differ from that on O. meridionalis. More research should reveal the nature of interactions occurring between the different haplotypes in rice.
The Sa locus was the first cloned and molecularly characterized hybrid sterility locus, from the indica-japonica cross, and it comprises two adjacent genes, SaF and SaM (Long et al., 2008). Most indica cultivars have the allele of SaM+SaF+, whereas all japonica cultivars contain SaM−SaF−. The alleles SaM+SaF+ and SaM+SaF− were variably present in Oryza species having the GG, FF, CC, EE, and AA genomes, including O. barthii, O. nivara, and O. sativa subsp. indica. Three alleles in particular SaM+SaF+, SaM+SaF−, and SaM−SaF− were found in the O. rufipogon populations. The mutation of SaM most likely arose in an O. rufipogon population with the allele SaM+SaF− and then generated the allele SaM−SaF−, whereas the variation in SaF occurred before the evolutionary split and gave rise to most of the current Oryza species (Long et al., 2008). We noticed that eight AA genome species possess the alleles of SaM+SaF+ and SaM+SaF−, but not O. sativa subsp. japonica, so it is readily inferred that the hybrid sterility locus S51(t) detected from O. meridionalis–japonica hybrid was one allele of Sa (Li et al., 2018b), with neutral allele Sa-n being identified from the indica rice and wild rice O. rufipogon (Long et al., 2008). It is reasonable to anticipate that, with further research, a tri-allelic system consisting of three types of Sa alleles (SaM+SaF+, SaM+SaF−, and SaM−SaF−) will emerge, which is able to control hybrid male sterility and fertility not only in O. sativa but also among AA genome species of Oryza.
The hybrid sterility locus S5, controlling female semi-sterility in hybrids between indica and japonica, consists of three closely linked genes ORF3, ORF4, and ORF5 that function together in a “Killer–Protector” system (Yang et al., 2012). The typical indica-like (ORF3+ORF4−ORF5+), japonica-like (ORF3−ORF4+ORF5−), and neutral (ORF3+ORF4+ORF5+ and ORF3−ORF4−ORF5−) alleles were found in wild rice accessions of O. rufipogon and O. nivara; this suggested the S5 locus already existed in wild relatives and the ancestors of indica and japonica rice, thus probably originating before domestication (Du et al., 2011; Yang et al., 2012). Presently, S5 has only been identified in the intersubspecific hybrid cross between indica and japonica, but according to the distribution of alleles in AA genome we think S5 also had effect for interspecific hybridization crosses.
Interspecific hybrid sterility locus S1 is also a tripartite “Killer-Protector” complex (Xie et al., 2019b). The African rice S1 allele is composed of three adjacent genes (S1A4, S1TPR, S1A6), while the Asian S1 allele includes only one gene, S1TP, which is a truncated form of S1TPR (Xie et al., 2017; Koide et al., 2018; Xie et al., 2019). Based on the analyzed gene structure of the S1 locus, the function structure A4-TPR-A6 exists in all examined accessions of O. glaberrima; and the nonfunction or dysfunction structureis present in all analyzed accessions of O. sativa, O. longistaminata, and O. rufipogon. The accessions of O. meridionalis and O. barthii possess both of the structures (Xie et al., 2019b).Total 7 haplotypes and 22 allelic variations of the S1 locus were detected in the Oryza species (Xie et al., 2019b). Further studying the relationship between hybrid sterility and the multiple allelic interactions is needed.
Taken together, the research findings above indicate the interaction between divergent alleles/haplotypes may lead to hybrid sterility, even their interaction mechanisms and genetic effects may be different. It can be further deduced that not only the allelic interactions among different alleles of orthologous loci, but also the non-allelic interactions among different loci, constitutes the major genetic basis for interspecific hybrid sterility between O. sativa and other AA genome species.
Benefit from the clone of S27 and S28, a pair of epistatic interaction loci caused F1 hybrid sterility in the hybrid pairs O. sativa–O. glumaepatula and O. sativa–O. nivara (Yamagata et al., 2010; Win et al., 2011), we can trace the interaction between different alleles that induce hybrid sterility. The S27 allele of O. sativa is composed of two tandem mitochondrial ribosomal protein L27 genes (mtRPL27a and mtRPL27b), and the S28 allele of O. sativa contains a loss-of-function allele for mtRPL27a. In brief, the S27 has normal functioning and S28 has no function in the O. sativa allele, whereas only the S28 has normal functioning and S27 is absent in the O. glumaepatula allele. The epistatic interaction induces the abortion of pollen grains carrying S27 and S28, both of which are inactive in F1 progeny of O. sativa and O. glumaepatula. Judging from a comparison of their genomic sequences, mtRPL27a at S28 on chromosome 4 is thought to be the most ancestral locus. With an interchromosomal duplication, a new copy of mtRPL27a at S27 on chromosome 8 was generated, and likewise mtRPL27b, also at S27 (Ueda et al., 2006). Among the eight AA genome species, the duplicated segment of S27 was widely observed, occurring in all except O. glumaepatula, and only part of the accessions of O. barthii and O. longistaminata lacked this segment (Yamagata et al., 2010). Further study showed that the interaction between S27 and S28 loci also causes the pollen sterility in the F1 hybrid of O. sativa and O. nivara, which inherited the duplicated segment of S27 (Win et al., 2011). Sequence analysis revealed that the structure of S27 allele in O. nivara is different from that of O. glumaepatula; in the former it is composed of two inactive genes (mtRPL27a, mtRPL27b) while in the latter species these two genes are absent. In short, S27-niv is a loss-of-function allele of S27. These results highlight that the mechanisms for hybrid sterility are likely different because of epistatic interactions between S28 loci and different alleles of S27 in divergent rice species.
The multiple alleles causing hybrid sterility in rice were analyzed at the molecular level for S5, Sa, qHMS7/S21/S23/S55, and S27. Meanwhile the cloning of hybrid sterility loci also proves the data support for the allelic relationship of these hybrid sterility loci mentioned above with same molecular location and similar genetic action. According to the latest research, at each hybrid sterility locus, the number of interacting alleles does not seem to be limited to three, of which one is neutral. It has been reported that four alleles characterize the S7 locus, S-7kn, S-7cp, S-7ai, and S-7n, which cause female sterility in the indica-japonica hybrid (Yanagihara et al., 1992). The genotype of S-7ai/S-7cp shows semi-sterility, in that the female gametes carrying S-7cp are aborted. But in the S-7ai/S-7kn genotype, only some of the female gametes possessing S-7ai are aborted. On the other hand, the S-7ai/S-7n, S-7n/S-7cp, andS-7n/S-7kn genotypes did not exhibit sterility as female gamete abortion (Yanagihara et al., 1992). The initial loci existing in the ancestral species likely differentiated into multiple alleles/haplotypes due to the emergence of genetic differentiation, which accumulated in each population; importantly, this variation was not harmful in the populations in which they arose. Of course, the variation that may affect fitness has been eliminated with the death of the individual. After hybridization between divergence populations, due to the incompatible interaction between two or more functionally diverged genes, the gametes carrying the weak alleles will be aborted, which may be nonfunctional, inactivated, or lack of corresponding protectors. This also explains why the cloned hybrid sterility loci involved in the one-locus systems are mostly composed of more than two linked genes. These closely linked genes, which are distinguished by minimal recombination, form a complex acted as a single Mendelian factor that can avoid inducing the suicidal killer/nonprotector allele in a hybrid that would result in a breakdown of the system (Xie et al., 2019a). The evolutionary history of cloned hybrid sterility genes has been described by many researchers (Long et al., 2008; Yamagata et al., 2010; Du et al., 2011; Kubo et al., 2016; Yu et al., 2018). Ouyang and Zhang (2013) proposed three evolutionary genetic models, those of parallel divergence, sequential divergence, and parallel-sequential divergence, to illustrate the process going from essential genes in the ancestral species to diverged genes based on the cloning data of hybrid sterility genes (Ouyang and Zhang, 2018). By disentangling the interaction between various alleles of these “speciation genes” which underpin the timing and evolutionary progression of genetic changes, we can understand the processes and patterns underlying the speciation and origin of AA genome species of rice further.
During the process by which cultivated rice was domesticated from wild ancestor thousands years ago, the cultivated rice’s genetic diversity has been gradually eroded. More importantly, the frequent use of a few elite parents in rice genetic improvement programs has exacerbated this decline in genetic diversity (Simmonds, 1976; Moncada et al., 2001). These challenges hinder our ability to attain further improvement in yield of new varieties while also making rice more susceptible to disease epidemics and pest outbreaks (Tanksley and McCouch, 1997). Thus, the relatives of O. sativa, which contain favorable genes (alleles), could be a valuable genetic resource for rice improvement via hybridization techniques. On the other hand, the utilization of heterosis in the grain yield of rice has boosted rice production in the past few decades. Current rice hybrids are mostly derived from the indica lines. Given their limited genetic diversity, intra-subspecific hybrid vigor now only provides rice breeders with limited yield increases, whereas the distant hybrids derived from intersubspecific even interspecific crosses performed stronger heterosis. However, severe reproductive isolation, such as that incurred by interspecific and intersubspecific hybrid sterility, limits the extensive utilization of these rice distant hybrids (Ikeda et al., 2009; Bolaji and Nwokeocha, 2014). Therefore, hybrid sterility studies should also aim to overcome the reproductive barrier, so as to enable breeders to utilize the valuable extant genetic resources and obtain the strong vigor of these hybrids.
Since hybrid sterility is partly caused by the negative interaction between divergent alleles in the background of the hybrid, replacing the divergent allele with the same or neutral alleles can eliminate the hybrid sterility on the corresponding locus. In this respect, the neutral alleles S5-n, Sa-n, Sb-n, Sd-n, and Se-n in the intersubspecific hybrid of indica-japonica (Ikehashi and Araki, 1986; Long et al., 2008; Li J. Q. et al., 2012), as well as S38-n, S39-n in the interspecific hybrid of O. sativa and O. glaberrima (Li et al., 2018a), have been identified. The use of these neutral alleles will give a strong promises on overcoming the interspecific and intersubspecific hybrid sterility. In most cases, as small number of sterility loci are involved in the hybrid cross, so it is necessary to pyramid several sterility loci alleles by using molecular marker assistant backcrossing methods. For example, Guo et al. (2016) polymerized four indica alleles of pollen sterility loci (Sb, Sc, Sd, Se) and the neutral allele of S5 locus, imparting the embryo sac sterility to the japonica variety, with eight indica-compatible japonica lines with different japonica backgrounds thus obtained. These indica-compatible japonica lines are highly compatible with the indica and effectively overcome the hybrid sterility in the intersubspecific hybrid. In the same vein, two neutral alleles of hybrid sterility loci were introgressed into indica rice HJX74 to develop HJX74 wide-compatible lines that were compatible with both japonica and indica testers (Guo et al., 2016).
Another promising strategy is to create hybrid-compatible lines with artificial neutral alleles generated by advanced molecular approaches, such as RNA interference technologies and gene-editing technology. For example, the neutral allele S1mut was created by mutagenesis and the heterozygous hybrids harboring S1mut/S1g and S1mut/S1s did not exhibit sterility (Koide et al., 2018). Artificial neutral S1 alleles could also be obtained by disrupting any one of the three genes in the S1-g (S1A4-S1TPR-S1A6), through a CRISPP/Cas9 knockout (Xie et al., 2019). The same is true for qHMS7 (Yu et al., 2018). The introduction of both Sa-n and Sc-n via CRISPR/Cas9 plant genome editing can assist in overcoming unwanted hybrid male sterility (Shen et al., 2017; Xie et al., 2017). A combination of these strategies may provide better assurance of hybrid fertility. With the further discovery and creation of neutral loci and the generation of intersubspecies and interspecies bridge parents, rice improvement will step into a new era.
The Bateson-Dobzhansky-Muller model posits that hybrid sterility arises from a disharmourose interaction between functionally diverged genes from the hybridizing distant parents (Bateson, 1909; Dobzhansky, 1937; Muller, 1942). This model was first proposed in tomato by Rick (1966) and supported by many other reports (Kitamura, 1962; Ikehashi and Araki, 1986; Sano, 1990). According to the model, independent mutations occur in diverging populations but without a reduction in fitness, becoming fixed; consequently, multiple alleles, including a neutral allele, present at the causal orthologous loci, which then interact negatively in the background of the hybrid (Nei et al., 1983). One locus allelic interaction model underpins the negative interaction occurring at a single locus as a consequence of the independent evolution of two alleles, thereby causing a significant reduction in the fitness of the heterozygote when compared with their homozygote parents. Such negative interaction can also occur between two independent loci affecting gamete development, the gametes carrying the recessive alleles at both loci aborted during gamete development, but gametes of other genotypes remained normal. In this review, we rechecked and updated current genetic models for hybrid sterility in rice. The multiple alleles should exist at each orthologous hybrid sterility locus including, but not limited to, the neutral allele. The major genetic basis for interspecific hybrid sterility between O. sativa and AA genome species in genus Oryza should be the interaction between various alleles at a single locus, and various alleles with other loci, and the same is true for intersubspecific hybrid sterility in O. sativa. Even some of the sterility loci identified in individual combinations/species existed like an orphan, but whether they harbor various alleles in other AA genome species or not requires more study.
Currently, our understanding of hybrid sterility remains limited to that within Asian cultivated rice and between Asian cultivated rice and other species of AA genome, along with a few reports on hybrid sterility among different ecotypes within O. sativa that are available. Meanwhile, the availability of information is very limited due to different backgrounds of the genetic materials used by various researchers. To better understand these processes, it is essential to identify, clone, and functionally characterize more hybrid sterility loci in the genus Oryza. Efforts should also extend to investigating both the genetic diversity and geographical distribution of the alleles of various loci at the species level, including extant wild relatives. Since allelic and non-allelic interactions among major sterility loci are the foundation for understanding the relationship between hybrid sterility and speciation, it is necessary to raise near-isogenic lines with various alleles/haplotypes and pyramided different loci, with the same genetic background, based on the cloning and molecule characteristics. Then, taking these data together, it would become possible to provide more evidence illuminating the origin of genes responsible for hybrid sterility and the evolutionary processes for the establishment of new species in Oryza genus and shed light on overcoming hybrid sterility in rice improvement.
DT proposed the insights. JL conceived and wrote the paper. JZ, YZ, YY, and QP reviewed and edited the manuscript.
This research was funded by grants from the National Natural Science Foundation of China (grant numbers 31991221, 31860372, and 31660380) and the Key Foundation Project of Yunnan Academy of Agricultural Sciences (grant numberYJZ201801).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Reviewer YK is currently organizing a Research Topic with one of the authors DT.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.555572/full#supplementary-material
Bateson, W. (1909). “Heredity and variation in modern light”. In: Darwin and modern Science. Ed. Seward, A. C. (Cambridge: Cambridge Univ. Press), 85–7101.
Bautista, N. S., Solis, R., Kamijima, O., Ishii, T. (2001). RAPD, RFLP and SSLP analyses of phylogenetic relationships between cultivated and wild species of rice. Genes Genet. Syst. 76, 71–79. doi: 10.1266/ggs.76.71
Bikard, D., Patel, D., Le Mette, C., Giorgi, V., Camilleri, C., Bennett, M. J., et al. (2009). Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323, 623e626. doi: 10.1126/science.1165917
Bolaji, A. O., Nwokeocha, C. C. (2014). Issues concerning reproductive isolation in a rice hybrid swarm involving Oryza sativa Linn., O.longistaminata A. Chev. et Roehr. and Oryza glaberrima Steud. located in Jebba Nigeria. Int. J. Biol. Chem. Sci. 7, 2040–2049. doi: 10.4314/ijbcs.v7i5.21
Brideau, N. J., Flores, H. A., Wang, J., Maheshwari, S., Wang, X., Barbashi, D. A. (2006). Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314, 1292–1295. doi: 10.1126/science.1133953
Chang, T. T. (1976). Origin, evolution, cultivation, dissemination, and Asian and African rice. Euphytica 25, 425–441. doi: 10.1007/BF00041576
Chen, J., Ding, J., Ouyang, Y., Du, H., Yang, J., Cheng, K., et al. (2008). A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. U. S. A. 105 (32), 11436–11441. doi: 10.1073/pnas.0804761105
Chen, Z., Hu, F., Xu, P., Li, J., Deng, X., Zhou, J., et al. (2009). QTL analysis for hybrid sterility and plant height in interspecific populations derived from a wild rice relative, Oryza longistaminata. Breed. Sci. 59, 441–445. doi: 10.1270/jsbbs.59.441
Cheng, C., Motohashi, R., Tsuchimoto, S., Fukuta, Y., Ohtsubo, H., Ohtsubo, E. (2003). Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 20, 67–75. doi: 10.1093/molbev/msg004
Chin, J. H., Lee, Y. J., Jiang, W., Koh, H. J., Thomson, M. J. (2017). Characterization of indica–japonica subspecies-specific InDel loci in wild relatives of rice (Oryza sativa L. subsp. indica kato and subsp. japonica kato). Genet. Resour. Crop Evol. 64 (2), 405–418. doi: 10.1007/s10722-016-0368-1
Chu, Y. E., Morishima, H., Oka, H., II. (1969). Reproductive barriers distributed in cultivated rice species and their wild relatives. Jpn. J. Genet. 44, 207–223. doi: 10.1266/jjg.44.207
Darwin, C. (1859). On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life, John Murray. London: Nabu Press.
Doi, K., Taguchi, K., Yoshimura, A. (1999). RFLP mapping of S20 and S21 for F1 pollen semi-sterility found in backcross progeny of Oryza sativa and O. glaberrima. Rice Genet. Newsl. 16, 65–68.
Du, H. Y., Ouyang, Y. D., Zhang, C. J., Zhang, Q. F. (2011). Complex evolution of S5, a major reproductive barrier regulator, in the cultivated rice Oryza sativa and its wild relatives. New Phytol. 191, 275–287. doi: 10.1111/j.1469-8137.2011.03691.x
Fang, C., Li, L., He, R., Wang, D., Wang, M., Hu, Q., et al. (2019). Identification of S23 causing both interspecific hybrid male sterility and environment-conditioned male sterility in rice. Rice 12, 10. doi: 10.1186/s12284-019-0271-4
Garavito, A., Guyot, R., Lozano, J., Gavory, F., Samain, S., Panaud, O., et al. (2010). A genetic model for the female sterility barrier between Asian and African cultivated rice species. Genetics 185 (4), 1425–1440. doi: 10.1534/genetics.110.116772
Garris, A. J., Tai, T. H., Coburn, J., Kresovich, S., Mccouch, S. (2005). Genetic structure and diversity in Oryza sativa L. Genetics 169, 1631–1638. doi: 10.1534/genetics.104.035642
Guo, J., Xu, X., Li, W., Zhu, W., Zhang, G. (2016). Overcoming inter-subspecific hybrid sterility in rice by developing indica-compatible japonica lines. Sci. Rep. 6, 26878. doi: 10.1038/srep26878
Henderson, M. T. (1964). Cytogenetic studies at the Louisianan agricultural experiment station on the nature of intervarietal hybrid sterility in Oryza sativa L. [M]// Rice genetics and Cytogenetics (Amsterdom: Elsevier Publishing Co), 147–153.
Heuer, S., Miézan, K. M. (2003). Assessing hybrid sterility in Oryza glaberrima andO sativa hybrid progenies by PCR marker analysis and crossing with wide compatibility varieties. Theor. Appl. Genet. 107, 902–909. doi: 10.1007/s00122-003-1325-x
Hu, F., Xu, P., Deng, X., Zhou, J., Li, J., Tao, D. (2006). Molecular mapping of a pollen killer gene S29(t) in Oryza glaberrima and colinear analysis with S22 in O.glumaepatula. Euphytica 151, 273– 278. doi: 10.1007/s10681-006-9146-z
Ikeda, R., Sokei, Y., Akintayo, I. (2009). Seed fertility of F1 hybrids between upland rice NERICA cultivars and Oryza sativa L. or O. glaberrima Steud. Breed. Sci. 59, 27–35. doi: 10.1270/jsbbs.59.27
Ikehashi, H., Araki, H. (1986). Genetics of F1 sterility in remote crosses of rice, pp. in Rice Genetics. Ed. Khush, G. S. (Manila, Philippines: International Rice Research Institute), 119–130.
Kato, S., Kosaka, H., Hara, S. (1928). On the affinity of rice varieties as shown by fertility of hybrid plants. Bull. Sci. Fac. Agric. Kyushu Univ. 3, 132–147.
Kitamura, E. (1962). Genetic studies on sterility observed in hybrids between distantly related varieties of rice, Oryza sativa L. Bull. Chugoku Agric. Exp. Station A8, 141–205.
Koide, Y., Onishi, K., Nishimoto, D., Baruah, A. R., Kanazawa, A., Sano, Y. (2008). Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179, 888–900. doi: 10.1111/j.1469-8137.2008.02490.x
Koide, Y., Ogino, A., Yoshikawa, T., Kitashima, Y., Saito, N., Kanaoka, Y., et al. (2018). Lineage-specific gene acquisition or loss is involved in interspecific hybrid sterility in rice. Proc. Natl. Acad. Sci. U. S. A. 115, E1955–E1962. doi: 10.1073/pnas.1711656115
Kubo, T., Eguchi, M., Yoshimura, A. (2001). A new gene for F1 pollen sterility located on chromosome 12 in japonica/indica cross of rice. Rice Genet. Newslett. 18, 54.
Kubo, T., Takashi, T., Ashikari, M., Yoshimura, A., Kurata, N. (2016). Two tightly linked genes at the hsa1 locus cause both F1 and F2 hybrid sterility in rice. Mol. Plant 9, 221–232. doi: 10.1016/j.molp.2015.09.014
Lee, H. Y., Chou, J. Y., Cheong, L., Chang, N. H., Yang, S. Y., Leu, J. Y. (2008). Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135 (6), 1065–1073. doi: 10.1016/j.cell.2008.10.047
Li, J. Q., Shahid, M. Q., Feng, J. H., Liu, X. D., Zhao, X. J., Lu, Y. G. (2012). Identification of neutral alleles at pollen sterility gene loci of cultivated rice (Oryza sativa L.) from wild rice (O. rufipogon Griff.). Plant Syst. Evol. 298, 33–42. doi: 10.1007/s00606-011-0520-5
Li, J., Xu, P., Zhou, J., Hu, F., Deng, X., Chen, Z., et al. (2012). Molecular Mapping of Sterility QTLs qSS-3, qSS-6a and qSS-7 as Single Mendelian Factors via NIL strategy. Rice Sci. 18 (2), 110–115. doi: 10.1016/S1672-6308(11)60016-4
Li, J., Zhou, J., Xu, P., Deng, X., Deng, W., He, M., et al. (2018a). Neutral alleles at hybrid sterility loci of Oryza glaberrima from AA genome relatives in genus Oryza. Breed. Sci. 68 (3), 343–351. doi: 10.1270/jsbbs.18006
Li, J., Zhou, J., Xu, P., Deng, X., Deng, W., Zhang, Y., et al. (2018b). Mapping five novel interspecific hybrid sterile loci between Oryza sativa and Oryza meridionalis. Breed. Sci. 68 (5), 516–523. doi: 10.1270/jsbbs.18001
Londo, J. P., Chiang, Y. C., Hung, K. H., Chiang, T. Y., Schaal, B. A. (2006). Phylogeography of Asian wild rice, Oryza rufifipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. U. S. A. 103, 9578–9583. doi: 10.1073/pnas.0603152103
Long, Y., Zhao, L., Niu, B., Su, J., Wu, H., Chen, Y., et al. (2008). Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. U. S. A. 105 (48), 18871–18876. doi: 10.1073/pnas.0810108105
Lu, B. R., Naredo, M. E. B., Juliano, A. B., Jackson, M. T. (1998). Taxonomic status of Oryza glumaepatula Steud. III. Assessment of genomic affinity among AA genome species from the New World, Asia, and Australia. Genet. Resour. Crop Evol. 45, 205–214. doi: 10.1023/A:1008686517357
Lu, B. R., Naedo, M. E. B., Juliano, A. B., Jackson, M. T. (2000). “Preliminary studies on taxonomy and biosystematics of the AA genome Oryza species (poaceae)” in Grasses, Systematics and Evolution Eds. Jacobs, S. W. L., Everett, J (CSIRO Publishing), 51–58.
Lynch, M., Force, A. G. (2000). The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156, 590–605. doi: 10.1086/316992
Miyazaki, Y., Doi, K., Yasui, H. (2007). Identification of a new allele of F1 pollen sterility gene, S21, detected from the hybrid between Oryza sativa and O. rufipogon. Rice Genet. Newslett. 23, 36–38.
Mizuta, Y., Harushima, Y., Kurata, N. (2010). Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. U. S. A. 107, 20417–20422. doi: 10.1073/pnas.1003124107
Moncada, P., Martinez, C., Borrero, J., Chatel, M., Gauch, H., Jr., Guimaraes, E., et al. (2001). Quantitative trait loci for yield and yield components in an Oryza sativa× Oryza rufipogon BC2F2 population evaluated in an upland environment. Theor. Appl. Genet. 102 (1), 41–52. doi: 10.1007/s001220051616
Morishima, H., Hinata, K., Oka, H., II. (1963). Comparison of modes of evolution of cultivated forms from two wild rice species, Oryza breviligulata ans O.perennis. Evolution 17, 170–181. doi: 10.1111/j.1558-5646.1963.tb03267.x
Morishima, H. (1969). Phenetic similarity and phylogenetic relationships among strains of Oryza perennis, estimated by methods of numerical taxonomy. Evolution 23, 429–443. doi: 10.2307/2406698
Moyle, L. C., Payseur, B. A. (2009). Reproductive isolation grows on trees. Trends Ecol. Evol. 24 (11), 591–598. doi: 10.1016/j.tree.2009.05.010
Naredo, M. E. B., Juliano, A. B., Lu, B. R., Jackson, M. T. (1998). Taxonomic status of Oryza glumaepatula steud. ii. hybridization between new world diploids and aa genome species from asia and australia. Genet. Resour. Crop Evol. 45 (3), 205–214. doi: 10.1023/A:1008634400519
Nei, M., Maruyama, T., Wu, C., II. (1983). Models of evolution of reproductiveisolation. Genetics 103, 557–579. doi: 10.1016/j.acra.2011.04.004
Nguyen, G., Yamagata, Y., Shigematsu, Y., Watanabe, M., Miyazaki, Y., Doi, K., et al. (2017). Duplication and loss of function of genes encoding RNA polymerase III subunit C4 causes hybrid incompatibility in rice. G3 Genes Genomes Genet. 7, 2565–2575. doi: 10.1534/g3.117.043943
Oka, H., II. (1957). Genic analysis for the sterility of hybrids between distantly related varieties of cultivated rice. J. Genet. 55 (3), 397–409. doi: 10.1007/BF02984059
Oka, H., II. (1974). Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics 77, 521–534.
Oka, H., II. (1988). Origin of cultivated rice: development in crop species (Tokyo: Jpn Sci Soc Press).
Orr, H. A., Presgraves, D. C. (2000). Speciation by postzygotic isolation: forces, genes and molecules. BioEssays 22, 1085–1094. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G
Ouyang, Y., Zhang, Q. (2013). Understanding reproductive isolation based on the rice model. Annu. Rev. Plant Biol. 64, 111–135. doi: 10.1146/annurev-arplant-050312-120205
Ouyang, Y., Zhang, Q. (2018). The molecular and evolutionary basis of reproductive isolation in plants. J. Genet. Genomics 45, 613–620. doi: 10.1016/j.jgg.2018.10.004
Rick, C. M. (1966). Abortion of male and female gametes in the tomato determined by allelic interaction. Genetics 53, 85–96. doi: 10.1016/j.plantsci.2015.09.010
Sakata, M., Yamagata, Y., Doi, K., Yoshimura, A. (2014). Two linked genes on rice chromosome 2 for F1 pollen sterility in a hybrid between Oryza sativa and O. glumaepatula. Breed. Sci. 64, 309–320. doi: 10.1270/jsbbs.64.309
Sano, Y., Chu, Y. E., Oka, H., II (1979). Genetic studies of speciation in cultivated rice, 1. Genic analysis for the F1 sterility between O. sativaL. and O. glaberrima Steud. Jpn. J. Genet. 54, 121–132. doi: 10.1266/jjg.54.121
Sano, Y., Sano, R., Eiguchi, M., Hirano, H.-Y. (1994). Gamete eliminator adjacent to the wx locus as revealed by pollen analysis in rice. J. Heredity 85 (4), 310–312. doi: 10.1093/oxfordjournals.jhered.a111465
Sano, Y. (1990). The genic nature of gamete eliminator in rice. Genetics 125, 183–191. doi: 10.1016/0735-0651(90)90042-E
Second, G. (1982). Origin of the genetic diversity of cultivated rice: study of the polymorphism scored at 40 isozyme loci. Jpn. J. Genet. 57, 25–57. doi: 10.1266/jjg.57.25
Shen, R., Wang, L., Liu, X., Wu, J., Jin, W., Zhao, X., et al. (2017). Genomic structural variation-mediated allelic suppression causes hybrid male sterility in rice. Nat. Commun. 8, 1310. doi: 10.1038/s41467-017-01400-y
Shinjyo, C. (1975). Genetical studies of cytoplasmic male sterility and fertility restoration in rice, Oryza sativa L. Sci. Bull. Coll. Agri. Univ. Ryukyus 22, 1–57.
Smith, J. M. (1989). “Macroevolution,” in Evolutionary Genetics (Oxford: Oxford University Press), 272–304.
Sobrizal, Y. M., Sanchez, P., Ikeda, K., Yoshimura, A. (2000a). Identification of a gene for male gamete abortion in backcross progeny of Oryza sativa and Oryza glumaepatula. Rice Genet. Newslett. 17, 59–61.
Sobrizal, Y. M., Sanchez, P., Ikeda, K., Yoshimura, A. (2000b). Mapping of F1 pollen semi-sterility gene found in backcross progeny of Oryza sativa L. and Oryza glumaepatula Steud. Rice Genet. Newsl. 17, 61–62.
Stebbins, G. L. (1958). The inviability, weakness, and sterility of interspecific hybrids. Adv. Genet. 9, 147–216. doi: 10.1016/S0065-2660(08)60162-5
Stebbins, G. (1950). “Isolation and the origin of species,” in Variation and Evolution in Plants (New York: Columbia University Press), 189–250.
Tanksley, S. D., McCouch, S. R. (1997). Seed banks and molecular maps: Unlocking genetic potential from the wild. Science 277, 1063–1066. doi: 10.1126/science.277.5329.1063
Ueda, M., Arimura, S., Yamamoto, M. P., Takaiwa, F., Tsutsumi, N., Kadowaki, K. (2006). Promoter shuffing at a nuclear gene for mitochondrial RPL27. Involvement of interchromosome and subsequent intrachromosome recombinations. Plant Physiol. 141, 702–710. doi: 10.1104/pp.105.075564
Vaughan, D. A., Morishima, H., Kadowaki, K. (2003). Diversity in the Oryza genus. Curr. Opin. Plant Biol. 6 (2), 139–146. doi: 10.1016/S1369-5266(03)00009-8
Vaughan, D. A., Ge, S., Kaga, A., Tomooka, N. (2008). “Phylogeny and Biogeography of the Genus Oryza,” in Rice Biology in the Genomics Era. Eds. Hirano, H.-Y., Hirai, A., Sano, Y., Sasaki, T. (Springer Berlin Heidelberg), 62, 219–234.
Vaughan, D. A. (1989). The genus Oryza L. current status of taxonomy. IRRI Research Paper Series No. 138 (Manila, Philippines: The International Rice Research Institute).
Wambugu, P. W., Brozynska, M., Furtado, A., Waters, D. L., Henry, R. J. (2015). Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci. Rep. 5, 13957. doi: 10.1038/srep13957
Wang, M., Yu, Y., Haberer, G., Marri, P. R., Fan, C., Goicoechea, J. L., et al. (2014). The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat. Genet. 46 (9), 982–988. doi: 10.1038/ng.3044
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Win, K. T., Kubo, T., Miyazaki, Y., Doi, K., Yamagata, Y., Yoshimura, A. (2009). Identifification of two loci causing F1 pollen sterility in inter- and intraspecifific crosses of rice. Breed. Sci. 59, 411–418. doi: 10.1270/jsbbs.59.411
Win, K. T., Yamagata, Y., Miyazaki, Y., Doi, K., Yasui, H., Yoshimura, A. (2011). Independent evolution of a new allele of F1 pollen sterility gene S27 encoding mitochondrial ribosomal protein L27 in Oryza nivara. Theor. Appl. Genet. 122, 385–394. doi: 10.1007/s00122-010-1454-y
Xie, Y., Xu, P., Huang, J., Ma, S., Xie, X., Tao, D., et al. (2017). Interspecific hybrid sterility in rice is mediated by OgTPR1 at the S1 locus encoding a peptidase-like protein. Mol. Plant 10, 1137– 1140. doi: 10.1016/j.molp.2017.05.005
Xie, Y., Shen, R., Chen, L., Liu, Y. G. (2019a). Molecular mechanisms of hybrid sterility in rice. Sci. China Life Sci. 62, 737–743. doi: 10.1007/s11427-019-9531-7
Xie, Y., Tang, J., Xie, X., Li, X., Huang, J., Fei, Y., et al. (2019b). An asymmetric allelic interaction drives allele transmission bias in interspecific rice hybrids. Nat. Commun. 10, 1–10. doi: 10.1038/S41467-019-10488-3
Xu, P., Zhou, J., Li, F., Hu, F., Deng, X., Feng, S., et al. (2014). Mapping three new interspecific hybrid sterile loci between Oryzasativa and O.glaberrima. Breed. Sci. 63, 476–482. doi: 10.1270/jsbbs.63.476
Yamagata, Y., Yamamoto, E., Aya, K., Win, K. T., Doi, K., Sobrizal, et al. (2010). Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 1494–1499. doi: 10.1073/pnas.0908283107
Yanagihara, S., Kato, H., Ikehashi, H. (1992). A new locus for multiple alleles causing hybrid sterility between an aus variety and javanica varieties in rice (Oryza sativa L.). Jpn. J. Breed. 42, 793–801. doi: 10.1270/jsbbs1951.42.793
Yang, J., Zhao, X., Cheng, K., Du, H., Ouyang, Y., Chen, J., et al. (2012). A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337, 1336e1340. doi: 10.1126/science.1223702
Yang, Y., Zhou, J., Li, J., Xu, P., Zhang, Y., Tao, D. (2016). Mapping QTLs for hybrid sterility in three AA genome wild species of Oryza. Breed. Sci. 66, 367–371. doi: 10.1270/jsbbs.15048
Yao, S. Y., Henderson, M., Jodon, N. E. (1958). Gryptic structural hybridity as a probable cause of sterility in intervarietal hybrids of cultivated rice, O. sativa. Cytologia 23, 46–55. doi: 10.1508/cytologia.23.46
Yu, Y., Zhao, Z., Shi, Y., Tian, H., Liu, L., Bian, X., et al. (2016). Hybrid sterility in rice (Oryza sativa L.) involves the tetratricopeptide repeat domain containing protein. Genetics 203, 1439e1451. doi: 10.1534/genetics.115.183848
Yu, X., Zhao, Z., Zheng, X., Zhou, J., Kong, W., Wang, P., et al. (2018). A selfifish genetic element confers non-Mendelian inheritance in rice. Science 360, 1130e1132. doi: 10.1126/science.aar4279
Zhang, Y., Zhao, Z., Zhou, J., Jiang, L., Bian, X., Wang, Y., et al. (2011). Fine mapping of a gene responsible for pollen semi-sterility in hybrids between Oryza sativa L. and O. glaberrima Steud. Mol. Breed. 28, 323–334. doi: 10.1007/s11032-010-9485-2
Zhang, Y., Zhou, J., Li, J., Yang, Y., Xu, P., Tao, D. (2018). Mapping of S56(t) responsible for interspecific hybrid sterility between Oryza sativa and Oryza glumaepatula. Breed. Sci. 68, 242–247. doi: 10.1270/jsbbs.17116
Zhou, J., Xu, P., Deng, X., Li, J., Hu, F., Ren, G., et al. (2010). Genetic dissection of a chromosomal region conferring hybrid sterility using multi-donors from Oryza glaberrima. Euphytica 175 (3), 395–407. doi: 10.1007/s10681-010-0188-x
Zhu, S., Jiang, L., Wang, C., Zhai, H., Li, D., Wan, J. (2005). The origin of a weedy rice ludao in China deduced by a genome wide analysis of its hybrid sterility genes. Breed. Sci. 55, 409–414. doi: 10.1270/jsbbs.55.409
Zhu, W., Li, W., Ding, X., Zhang, Z., Zeng, R., Zhu, H., et al. (2008). Preliminary identification of F1 pollen sterility gene S-e in Oryza sativa. J. South. China Agricult. Univ. 29, 1–5.
Keywords: allelic variation, hybrid sterility, interspecific hybrid, rice, Oryza sativa
Citation: Li J, Zhou J, Zhang Y, Yang Y, Pu Q and Tao D (2020) New Insights Into the Nature of Interspecific Hybrid Sterility in Rice. Front. Plant Sci. 11:555572. doi: 10.3389/fpls.2020.555572
Received: 25 April 2020; Accepted: 01 September 2020;
Published: 23 September 2020.
Edited by:
Ryo Fujimoto, Kobe University, JapanReviewed by:
Yohei Koide, Hokkaido University, JapanCopyright © 2020 Li, Zhou, Zhang, Yang, Pu and Tao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dayun Tao, dGFvZHkxMkBhbGl5dW4uY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.