- Zhengzhou Fruit Research Institute, Chinese Academy of Agricultural Sciences, Zhengzhou, China
Kiwifruit bacterial canker, caused by the bacterial pathogen Pseudomonas syringae pv. actinidiae (Psa), is a destructive disease in the kiwifruit industry globally. Consequently, understanding the mechanism of defense against pathogens in kiwifruit could facilitate the development of effective novel protection strategies. The Non-expressor of Pathogenesis-Related genes 1 (NPR1) is a critical component of the salicylic acid (SA)-dependent signaling pathway. Here, a novel kiwifruit NPR1-like gene, designated AeNPR1a, was isolated by using PCR and rapid amplification of cDNA ends techniques. The full-length cDNA consisted of 1952 base pairs with a 1,746-bp open-reading frame encoding a 582 amino acid protein. Homology analysis showed that the AeNPR1a protein is significantly similar to the VvNPR1 of grape. A 2.0 Kb 5′-flanking region of AeNPR1a was isolated, and sequence identification revealed the presence of several putative cis-regulatory elements, including basic elements, defense and stress response elements, and binding sites for WRKY transcription factors. Real-time quantitative PCR results demonstrated that AeNPR1a had different expression patterns in various tissues, and its transcription could be induced by phytohormone treatment and Psa inoculation. The yeast two-hybrid assay revealed that AeNPR1a interacts with AeTGA2. Constitutive expression of AeNPR1a induced the expression of pathogenesis-related gene in transgenic tobacco plants and enhanced tolerance to bacterial pathogens. In addition, AeNPR1a expression could restore basal resistance to Pseudomonas syringae pv. tomato DC3000 (Pst) in Arabidopsis npr1-1 mutant. Our data suggest that AeNPR1a gene is likely to play a pivotal role in defense responses in kiwifruit.
Introduction
Kiwifruit (Actinidia spp) is a perennial deciduous vine originating from China, and it is an emerging economic fruit tree. It has long been called “the king of VC” because of its unique flavor and high nutrient composition, being rich in vitamin C (Huang et al., 2013; Huang, 2016). Over the past few decades, kiwifruit has become an important horticultural crop, and numerous varieties have been commercialized. However, the majority of the currently cultivated Actinidia chinensis (i.e., Hort16A, Hongyang) and Actinidia deliciosa (i.e., Hayward) cultivars and their pollinators are highly susceptible to the infection of kiwifruit bacterial canker, a major kiwifruit disease caused by Pseudomonas syringae pv. actinidiae (Psa). The pathogen was first recognized and recorded in Japan in 1984 (Serizawa et al., 1989) and was subsequently described in Korea, Italy, and other kiwifruit production areas (Koh et al., 1994; Everett et al., 2011). Psa is the primary factor limiting kiwifruit production, and it is responsible for considerable economic losses in the kiwifruit industry globally. Therefore, a comprehensive understanding of kiwifruit disease defense mechanisms would facilitate the management and decrease of the effects of pathogen infection and the development of effective strategies for controlling Psa. In recent years, the genomes of several kiwifruit cultivars (i.e., A. chinensis Hongyang, Red5, and A. eriantha White) have been sequenced, which is convenient for the exploration of genes involved in defense responses and the study of regulatory mechanisms (Huang et al., 2013; Pilkington et al., 2018; Tang et al., 2019).
Plants have evolved complex regulatory networks of defenses to recognize and combat invading microbes and herbivorous insects, such as hypersensitive response (HR), systemic acquired resistance (SAR), and induced systemic resistance (ISR) (Kinkema et al., 2000). Such defense systems are tightly coordinated through the activities of several phytohormones, especially salicylic acid (SA). SA is a defense signal molecule that participates in resistance against biotrophic and hemibiotrophic pathogens by inducing SAR (Pieterse et al., 2009; An and Mou, 2011). SAR is one of the most commonly induced defense mechanisms and is typically triggered when plants are invaded locally by pathogens to provide long-lasting and broad-spectrum protection for the whole plant (Gao et al., 2015). The establishment of SAR involves the generation and transport of signals to uninfected distal tissues, which is highly dependent on the higher levels of in vivo SA, since blocking of SA accumulation inhibits SAR induction (Shine et al., 2019; Zhang and Li, 2019). When plants are attacked by a pathogen, SA concentrations increase from a basal level to a high level both locally and systemically. In addition, in many plants, exogenous application of SA or its functional analogs, such as 2, 6-dichloroisonicotinic acid (INA) and benzo (1,2,3) thiadiazole-7-carbothioic acid S-methylester (BTH), can also induce SAR and enhance plant resistance to disease (Görlach et al., 1996; Wu et al., 2012).
Despite the underlying mechanisms of the relationship between SA and SAR have not yet been elucidated comprehensively, Extensive studies in model plant Arabidopsis have shown that the Non-expressor of Pathogenesis-Related genes 1 (NPR1), also known as Non-inducible immunity 1 (NIM1) or Salicylic acid insensitive 1 (SAI1), played a crucial role in salicylic acid-mediated SAR (Loake and Grant, 2007; Chen et al., 2019). NPR1 belongs to a small NPR1-like family that contains four NPR1homologs (AtNPR1-AtNPR4) and two BOP homologs (AtBOP1 and AtBOP2) in Arabidopsis. Phylogenetic analysis of this family revealed three functionally distinct clades (Zhang et al., 2006; Kuai and Després, 2016). The most widely studied member of the NPR1-like family is the NPR1 gene. NPR1 was originally identified in a screen of Arabidopsis mutants that were unable to express the PR genes during pathogen invasion (Cao et al., 1994). Mutant npr1 plants not only compromised SAR response in the presence of the SA or SA analogs, exhibiting little expression of PR genes, but also increased susceptibility to pathogens (Cao et al., 1997). The Arabidopsis NPR1 (AtNPR1) gene encodes a protein with a BTB/POZ (Broad-complex, Tramtrack and Bric-a-brac/Pox virus, and Zinc finger) domain and an ankyrin repeats domain, both of which mediate protein-protein interactions (Cao et al., 1997; Sandhu et al., 2009; Backer et al., 2019). It also possesses several functional amino acid residues, such as Cys82, Cys150, Cys155, Cys160, and Cys216, which are crucial for oligomer-monomer transition. Cys156 facilitates NPR1 oligomerization in vivo via S-nitrosylation, while Arg432 in the C-terminus serve for SA binding (Mou et al., 2003; Tada et al., 2008; Ding et al., 2018; Wang et al., 2020). In Arabidopsis, the NPR1 is expressed constitutively at low levels and encodes an oligomerized, cytosolic protein. During pathogenic infection or SA treatment, the NPR1 oligomer in the cytoplasm disintegrates into its monomeric form, which translocates into the nucleus to interact with several TGA family members, and subsequently induces the transcription of PR genes (Kinkema et al., 2000; Fan and Dong, 2002). The redox modification and subsequent translocation to the nucleus of NPR1 are both necessary for SAR (Mou et al., 2003).
It has been confirmed that NPR1 functions as the central regulator in plant defense networks, including the cross-talk between SA-JA defense pathways and the jasmonic acid (JA)/ethylene (ET)-dependent induced responses (Spoel et al., 2003; Leon-Reyes et al., 2009; Pieterse et al., 2009). The over-expression of AtNPR1 transcript in Arabidopsis plants led to enhanced resistance to both bacterial and fungal pathogens, such as Pseudomonas syringae, Peronospora parasitica, and Hyaloperonospora arabidopsidis, in a dosage-dependent manner (Cao et al., 1998; Friedrich et al., 2001). Similarly, ectopic expression of AtNPR1 in other crops including rice, wheat, tomato, soybean, apple, strawberry, citrus, and olive also resulted in resistance to a wide range of pathogens (Chern et al., 2001; Lin et al., 2004; Makandar et al., 2006; Malnoy et al., 2007; Matthews et al., 2014; Dutt et al., 2015; Silva et al., 2015; Robertson et al., 2018; Narváez et al., 2020). In transgenic cotton and tobacco plants, over-expression of AtNPR1 conferred resistance to Rotylenchulus reniformis and Spodoptera litura, respectively (Meur et al., 2008; Parkhi et al., 2010). Moreover, homologs of AtNPR1 have been isolated and characterized in several economically important crops. For example, constitutive expression of the apple homolog of NPR1, MhNPR1, resulted in the activation of PR genes and enhanced resistance against Podosphaera leucotricha in transgenic apple (Chen et al., 2012). In addition, OsNPR1/NH1 overexpression in rice conferred resistance to bacterial blight, while silencing of the gene exhibited enhanced susceptibility to the pathogen (Yuan et al., 2007; Bai et al., 2011). Most recently, mulberry MuNPR1 overexpression in transgenic Arabidopsis enhanced resistance against Pst and inhibited callose deposition (Xu et al., 2019). In periwinkle, silencing of CrNPR1 significantly repressed CrPR1a induction and accelerated periwinkle leaf yellowing symptom development following Tobacco rattle virus (TRV) infection (Sung et al., 2019). The results of the studies above suggest that similar defense mechanisms exist across many crop species, and manipulating the expression of NPR1 or its ortholog could be an effective strategy of improving crop disease resistance (Malnoy et al., 2007; Yuan et al., 2007; Silva et al., 2018).
In the present study, we isolated and characterized a novel full-length NPR1 homolog, designated AeNPR1a, from a kiwifruit (A. eriantha) with high resistance to Psa. The sub-cellular localization of AeNPR1a based on the expression of GFP fusion proteins in Arabidopsis protoplasts indicated its presence in nuclei. Analysis of the 5′-upstream region identified potential cis-elements responsible for the regulation of the expression of AeNPR1a. Subsequently, we investigated the tissue expression levels of the gene and its expression profiles following treatment with SA, methyl jasmonate (MeJA), and Psa using RT-qPCR. Interaction between AeNPR1a and AeTGA2 was assayed using the yeast two-hybrid system. The effect of AeNPR1a over-expression on the growth of pathogenic bacteria in transgenic plants was also explored.
Materials and Methods
Plant Materials
The plant material used in the present study is A. eriantha, and plants were grown at the Kiwifruit Germplasm Repository of the National Horticulture Germplasm Resources Center in Zhengzhou, Henan province, China (N 34°42′, E 113°41′, 106 m). Several 4-year-old A. eriantha plants were selected for the collection of samples for tissue-specific expression analysis. All tissues were immediately frozen with liquid nitrogen and stored at −80°C until their subsequent use. Each biological sample was collected from three individual plants in the present study.
Cloning of Full-Length AeNPR1a cDNA and Genomic DNA
Total RNA for cDNA synthesis was extracted from kiwifruit leaves using a modified CTAB method (Japelaghi et al., 2011). The RNA samples were treated with 2 U DNase I (Takara, Dalian, China) for 30 min at 37°C and then purified further. The first-strand cDNA was synthesized using approximately 1 μg of total RNA and the Oligo d(T)18 adaptor primer by RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) according to the manufacturer’s instructions.
To clone the full-length cDNA of the NPR1-like gene from A. eriantha, two pairs of primers (Supplementary Table S1) were designed based on the partial coding sequence of AeNPR1a, which has been annotated in our previous RNA-seq data. The 5′ and 3′ rapid amplification of cDNA ends (RACE) strategies were performed using the SMART™ RACE cDNA Amplification Kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instruction. Thus, a pair of gene-specific primers, NPR1-01 and NPR1-02 (Supplementary Table S1), was designed based on the 5′-UTR and 3′-UTR for full-length cDNA. The PCR products were purified using a DNA Gel Purification Kit (GENEray, Shanghai, China), subcloned into the pMD18-T vector (Takara, Dalian, China), and at least three clones were selected for sequencing. The sequence was uploaded to the GenBank database of National Center of Biotechnology Information (NCBI; accession no. MN544777). Furthermore, the DNA from young leaves was extracted using a genomic DNA purification kit (Promega, Madison, WI, USA) and treated with RNase I (Takara, Dalian, China). AeNPR1a gene cloning from genomic DNA was also performed based on the full-length cDNA, and then, the target sequence was purified and sequenced as above.
Sequence Alignment and Phylogenetic Analysis
The amino acid sequence of AeNPR1a was predicted using NCBI Open Reading Frame (ORF) finder, and was then submitted for analysis by SMART (Letunic and Bork, 2018). The BLAST search program was used for gene sequence similarity searches in NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The exon/intron organization of NPR1 genes was generated using the Gene Structure Display Server 2.0 (http://gsds.cbi.pku.edu.cn/). For protein sequence analysis, the coding sequence of AcNPR1a was also isolated from Actinidia chinensis based on the AeNPR1a and genomic information. Sequences with high homology to AeNPR1a and AcNPR1a were aligned using Clustal W (https://www.ebi.ac.uk/Tools). To identify NPR1-like proteins in kiwifruit, sequence of the known Arabidopsis thaliana NPR1-like proteins were queried against A. chinensis and A. eriantha genomic databases (Pilkington et al., 2018; Tang et al., 2019) using BLAST. Subsequently, phylogenetic analysis and phylogenetic tree construction were performed using the Maximum Likelihood method in MEGA 5 (Tamura et al., 2011), with bootstrap confidence values from 1,000 replicates.
Isolation and Bioinformatics Analysis of AeNPR1a Promoter
To obtain the 5′-flanking region of AeNPR1a, genomic DNA was digested with EcoRV, DraI, StuI, and PvuII (TaKaRa, Dalian, China). Then, the digested DNA were purified and ligated separately with Genome Walker adaptors using the BD Universal Genome Walker™ kit (Clontech, Mountain View, CA, USA) according to the manufacturer′s instructions. Nested PCR was carried out using the prepared DNA as a template with the adaptor primers and gene specific primers (Supplementary Table S1). The final purified PCR products were cloned into pMD18-T vector (TaKaRa, Dalian, China) and sequenced. To identify the putative regulatory motifs of AeNPR1a promoter, bioinformatic analyses were carried out using PLACE (Higo et al., 1999) and PlantCARE (Lescot et al., 2002) databases.
Subcellular Localization Assay
The full-length open reading frame (ORF) of AeNPR1a without the termination codon was cloned into the pBI121-EGFP vector to generate a fusion construct under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The empty vector was used as a control. After sequence confirmation, these recombinant plasmids were transformed into Arabidopsis mesophyll protoplasts and onion epidermal cells as previously described (Yoo et al., 2007). Nuclei were stained with 50 μg·ml−1 4′,6-diamidino-2-phenylindole (DAPI; Sigma, St. Louis, MO, USA) after a 24-h incubation, and then, the GFP fluorescence signal was observed and imaged with a laser confocal microscope (Zeiss, Oberkochen, Germany). The excitation lines were configured at 405 and 488 nm.
Treatment of A. eriantha With SA, MeJA, and Psa Pathogen
To further investigate the induction mechanism of AeNPR1a, the spring shoots of 2-year-old plants were sprayed with SA (5 mM) (Sigma, St. Louis, MO, USA) and MeJA (0.1 mM) (Sigma, St. Louis, MO, USA), separately. Control plants for each treatment were treated with sterile distilled water containing equal amounts of the solvent (0.1% ethanol v/v) used for hormone preparation. Leaf samples were harvested from control and hormone-treated plants after 0, 6, 12, 24, 48, 72, and 96 h for RNA isolation. For bacterial infection, Psa-2 (Biovar 3) was grown on King’s B agar medium at 25°C for 2 days. Subsequently, a single colony was inoculated in liquid King’s B medium and incubated on a shaker for 16 to 20 h at 25°C until the OD600 reached 0.6. The bacterial cells were collected by centrifugation at 4000×g for 10 min and diluted to the desired concentration (106 colony forming units (CFU)·ml−1) with 10 mM MgCl2 for plant inoculation. The bacterial suspension was infiltrated on the abaxial surface of 2-year-old plant leaves with sterile syringes, and then, the inoculated plants were kept in a growth chamber at 80% relative humidity and 25°C. Control plants were mock-inoculated with 10 mM MgCl2 and incubated separately to prevent cross-contamination. Leaf samples from the control and bacteria inoculated plants were harvested at different time points and immediately frozen in liquid nitrogen and stored at −80°C. All the treatments had three replicates.
Gene Expression Analysis by Real-Time Quantitative RT-PCR
The spatio-temporal expression patterns of AeNPR1a in different tissues and its expression profiles in treated and control leaves were tested by quantitative real-time RT-PCR. RNA extraction and cDNA synthesis were carried out as described above.
For quantitative RT-PCR analysis, cDNA was diluted with RNase-free water before use. Real-time PCR of target genes was performed with specific primers using a LightCycler™ 480 Real-time PCR system with 2×SYBR Green Master Mix (Roche Applied Science, Mannheim, Germany). Kiwifruit β-actin was used as the internal reference gene. Primer sequences are listed in Supplementary Table S1. Real-time PCR products were amplified with 1-μl template of the RT reaction mixture, 10-μl 2×SYBR Green Master Mix, 0.5 μl each forward and reverse primer (10 μmol·μl−1), and water, with a final volume of 20 μl. The qRT-PCR amplification conditions were 95°C for 5 min, and then, 40 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 15 s. Data were evaluated by calibrator-normalized relative quantification with efficiency correction using the RelQuant software version 1.01 or the LightCycler™ 480 software version 1.5 (Roche Applied Science, Mannheim, Germany) according to the 2—△△CT method. Three biological replicates and four technical replicates were assayed.
Yeast Two-Hybrid Assays
The experimental procedures of yeast two-hybrid assays were performed using the BD Matchmaker system (Clontech, Mountain View, CA, USA). The entire ORF and different regions from AeNPR1a were digested using Nde I (Takara, Dalian, China) and BamH I (Takara, Dalian, China) and subcloned into pGBKT7 vector, and the ORF of AeTGA2 was cloned into the pGADT7 vector. All constructs were transformed into AH109 competent yeast cells according to the manufacturer′s instruction. The co-transformed yeast clones were also streaked on selective medium without tryptophan, histidine, leucine, and adenine, but supplemented with 20 μg·ml−1 5-bromo-4-chloro-3-indoxyl-α-D-galactopyranoside (X-α-Gal) (Sigma, St. Louis, MO, USA) to further confirm positive interactions.
Construction of Expression Vector and Genetic Transformation
To produce a vector for the constitutive expression of AeNPR1a, Bgl II (Takara, Dalian, China) and BstE II (Takara, Dalian, China) sites were introduced at the end of the full-length cDNA sequence of AeNPR1a by PCR amplification, and then, the fragment was digested and subcloned into the pCAMBIA 1301 vector (CAMBIA, Canberra, Australia) driven by CaMV35S promoter. The resulting recombinant plasmid was sequenced to verify the absence of PCR errors.
For plant transformation, the 35S::AeNPR1a vector was transferred into Agrobacterium tumefaciens strain EHA105 using the freeze-thawing method (Holsters et al., 1978). Transformation of tobacco (Nicotiana tabacum cv. NC89) was performed using the leaf disc co-cultivation method as described by Krügel et al. (Krügel et al., 2002). The transgenic seedlings were regenerated under 10 mg·L-1 hygromycin B selection, transferred to soil, and grown in a growth chamber at 23 ± 1°C under 16h/8h light/dark conditions. To investigate whether the AeNPR1a gene was homologous to Arabidopsis NPR1, the 35S::AeNPR1a vector was also transformed into the Arabidopsis npr1-1 mutant via the floral dip method (Clough and Bent, 1998). Transgenic plants of the T2 or T3 generation and single-copy lines were used in further analyses.
Pathogen Challenge of Transgenic Plants
AeNPR1a transgenic lines were evaluated for disease resistance to both Pst and Psa pathogens as previously described with a few modifications (Liu et al., 2005). Briefly, the strains were re-suspended to 106 CFU· ml−1 in 10 mM MgCl2 (Sigma, St. Louis, MO, USA) solution. The leaves of 4-week-old transgenic and non-transgenic tobacco were inoculated by infiltration with 50-μl bacterial suspensions for Pst and Psa resistance evaluation, and infection of Arabidopsis was performed on 4-week-old npr1, wild-type, and transgenic plants by spraying for Pst resistance evaluation. The inoculated plants were placed in a growth chamber under 16-h/8-h light/dark conditions with 80% relative humidity at a constant temperature of 25°C. Three days after inoculation, leaf punches were collected from treated leaves, surface sterilized, and ground in 1 ml sterile 10 mM MgCl2. The suspensions were serially diluted and plated on King’s B agar medium. After 2 days of incubation at 25°C, |the average number of CFU per leaf disk was calculated and statistically analyzed. For phenotypic observation, the leaves of the control mock plants (inoculated with 10 mM MgCl2) and pathogen-inoculated plants were photographed 7 days after infection. The inoculation of each strain was repeated three times, and mean values and standard deviations were obtained with three leaves from three independent plants.
Statistical Analysis
Statistical analysis was carried out using the SPSS statistics program (SPSS Inc., Chicago, USA). Data were analyzed using Student’s t-test for pairwise comparisons and Duncan’s multiple range test for multiple comparisons. Significant differences were evaluated at P < 0.05.
Results
Amino Acid Sequence Alignment and Phylogenetic Analysis of AeNPR1a
The full length sequence of AeNPR1a (GenBank accession no. MN544777) was isolated from A. eriantha by the RACE method. The sequence was identical to the corresponding gene in the published sequenced genome. The open reading frame of AeNPR1a encoded a deduced 582 amino acid protein of 64.26 kDa, and the predicted isoelectric point (pI) was 5.82. Alignment of the deduced amino acid sequence revealed that AeNPR1a is most closely related to AcNPR1a (98.11% identity), which originates from the same genus, with only 10 residues varying, followed by VvNPR1 (74.06% identity), StNPR1 (73.21% identity), NtNPR1 (71.94% identity), and AtNPR1 (52.86% identity) (Figure 1). The genomic structure of AeNPR1a was similar to that of the other NPR1 orthologs, sharing four exons and three introns (Figure 2A). Protein sequence comparison showed that AeNPR1a contained two conserved domains, a BTB/POZ domain and an ankyrin repeat domain (Figures 1, 2B). Eight highly conserved cysteine residues were identified in AeNPR1a, which contained the five Cys residues that are required for oligomer-monomer transition of AtNPR1. However, functionally relevant residues Cys156, described for oligomerization, are not present in AeNPR1a (Figure 1). Nonetheless, a putative nuclear localization signal (NLS) and a penta-amino acid motif (LENRV) were present in the C-terminal of AeNPR1a (Figure 1).
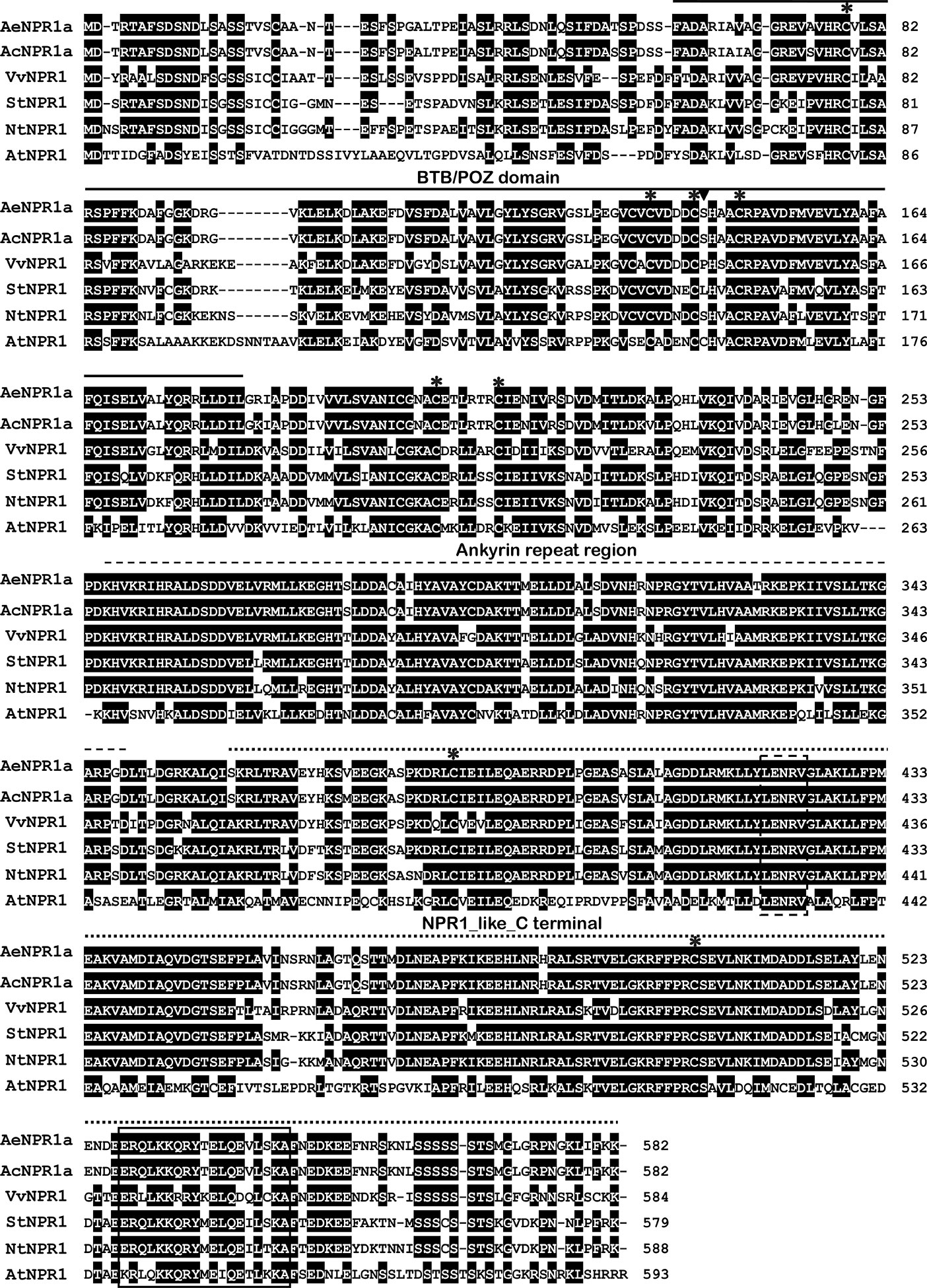
Figure 1 Multiple alignment of AeNPR1a protein with other NPR1 homologs. The BTB domain, Ankyrin repeat region and NPR1_like_C terminal are indicated with solid lines, broken lines and dotted lines, respectively. The conserved cysteine residues among the proteins are labeled by asterisks. The Cys156 relative to Arabidopsis is marked by inverted triangle. The putative nuclear localization signal (NLS) and LENRV are highlighted by solid line box and dashed line box, respectively. AcNPR1a (PSS20797), VvNPR1 (XP_002281475), StNPR1 (XP_006357709), NtNPR1 (AF480488), and AtNPR1 (AT1G64280).
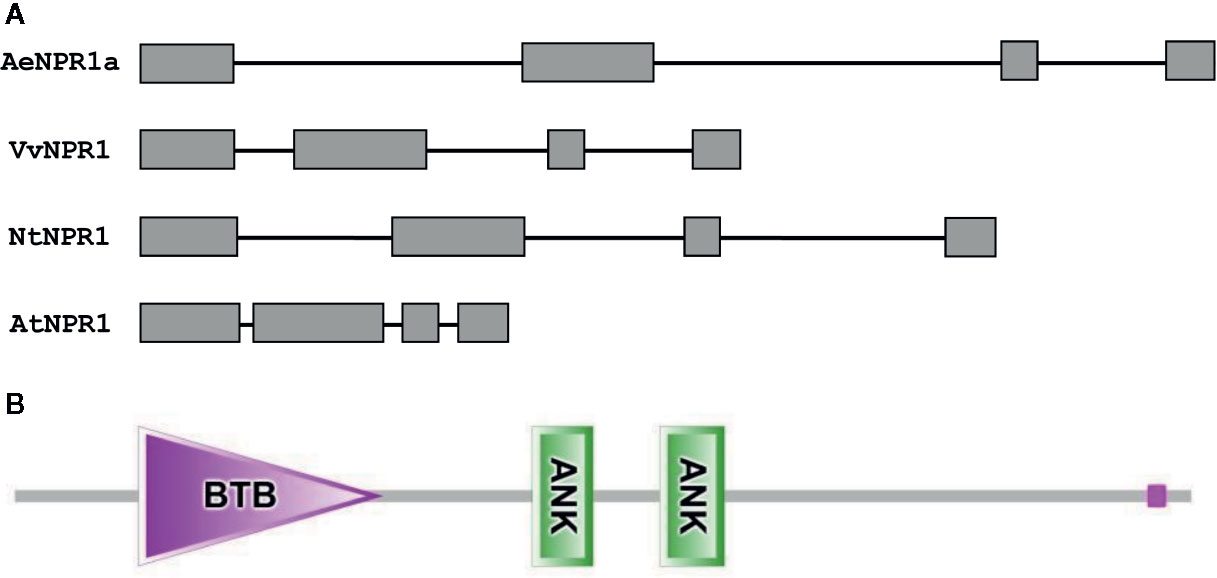
Figure 2 Analysis of AeNPR1a gene and protein structure. (A) Comparison of the exon-intron structures of AeNPR1a gene homologs. Exons and introns are represented by boxes and black lines, respectively. (B) The conserved domains of the AeNPR1a protein after analyzed by SMART.
To reveal the evolutionary relationship between AeNPR1a and other NPR1-like proteins from various plant species, the AeNPR1a and AeNPR1a-like protein sequences obtained in the present study were subjected to phylogenetic analysis with other NPR1 homologs. As shown in Figure 3, the tree was grouped into two major clades, AeNPR1a was the most closely related to AcNPR1a, AcNPR1b, AeNPR1b, as well as VvNPR1, clustering with the clade containing AtNPR1 and AtNPR2. The other five members from A. Chinensis and A. eriantha were grouped into clade II along with AtNPR3 and AtNPR4. The data suggest that AeNPR1a may be a homolog of NPR1 in kiwifruit.
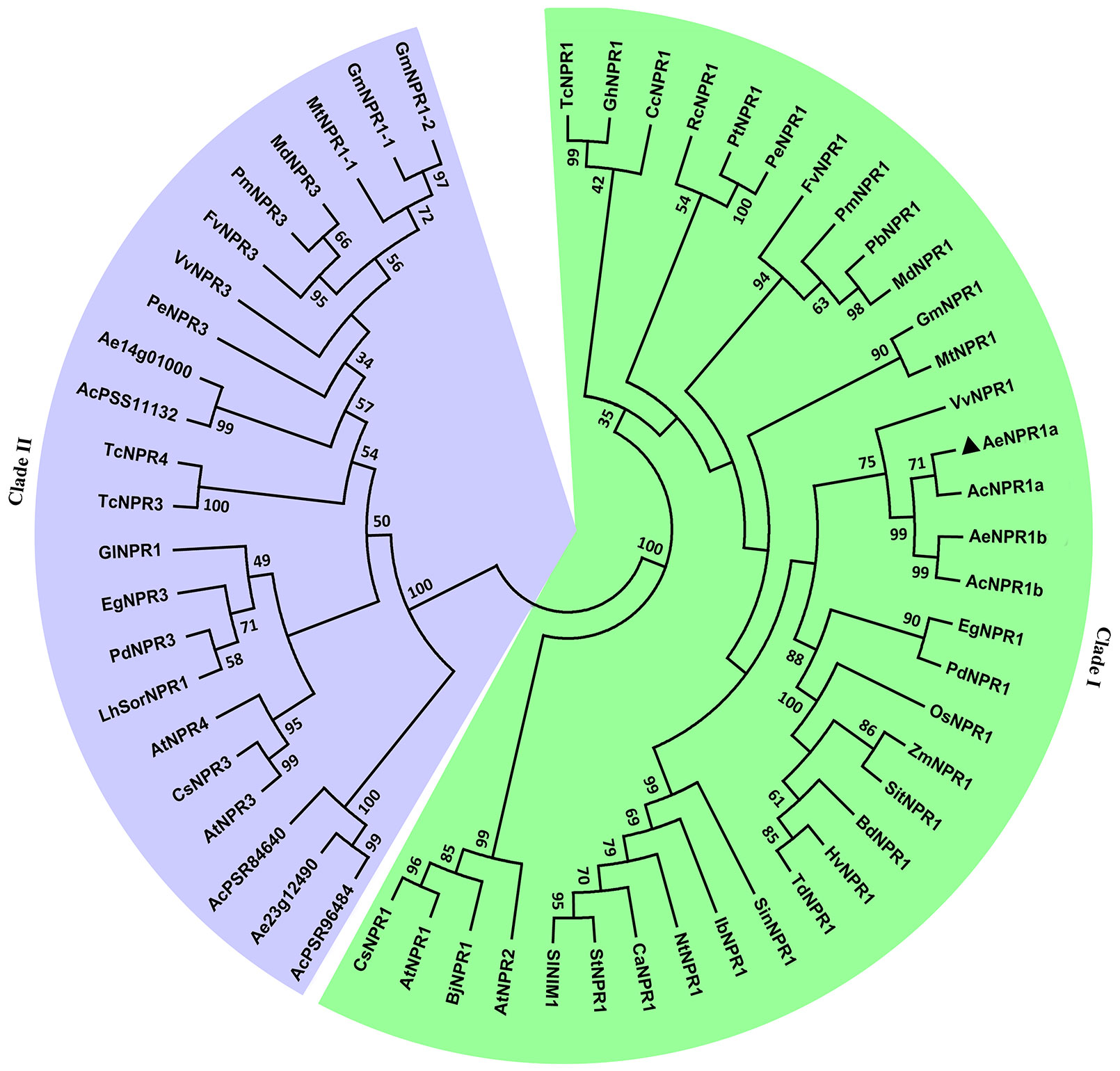
Figure 3 Phylogenetic relationship of AeNPR1a and other NPR1 homologs from different plant species. The tree was generated using the Maximum Likelihood method with 1,000 bootstrap replicates. AeNPR1a is indicated by a triangle. The accession numbers and species names for the NPR1 homolog sequences used in the analysis were summarized in Supplementary Table S2.
Subcellular Localization of AeNPR1a
To examine the subcellular localization of AeNPR1a, the coding region of AeNPR1a without the stop codon was fused to the 5′ end of the enhanced green fluorescent protein (EGFP) reporter gene under the control of CaMV35S promoter. Subsequently, the fusion construct 35S::AeNPR1a-EGFP was introduced into Arabidopsis mesophyll protoplasts and onion epidermal cells. GFP fluorescence was observed under a confocal fluorescent microscope. The results indicated that the fluorescence of AeNPR1a-EGFP was predominantly detected in the nucleus (Figure 4 and Supplementary Figure S1). In contrast, the control vector expressing free EGFP yielded fluorescence in both the nucleus and cytoplasm (Figure 4 and Supplementary Figure S1). Therefore, AeNPR1a appeared to be primarily a nuclear protein in these cell types.
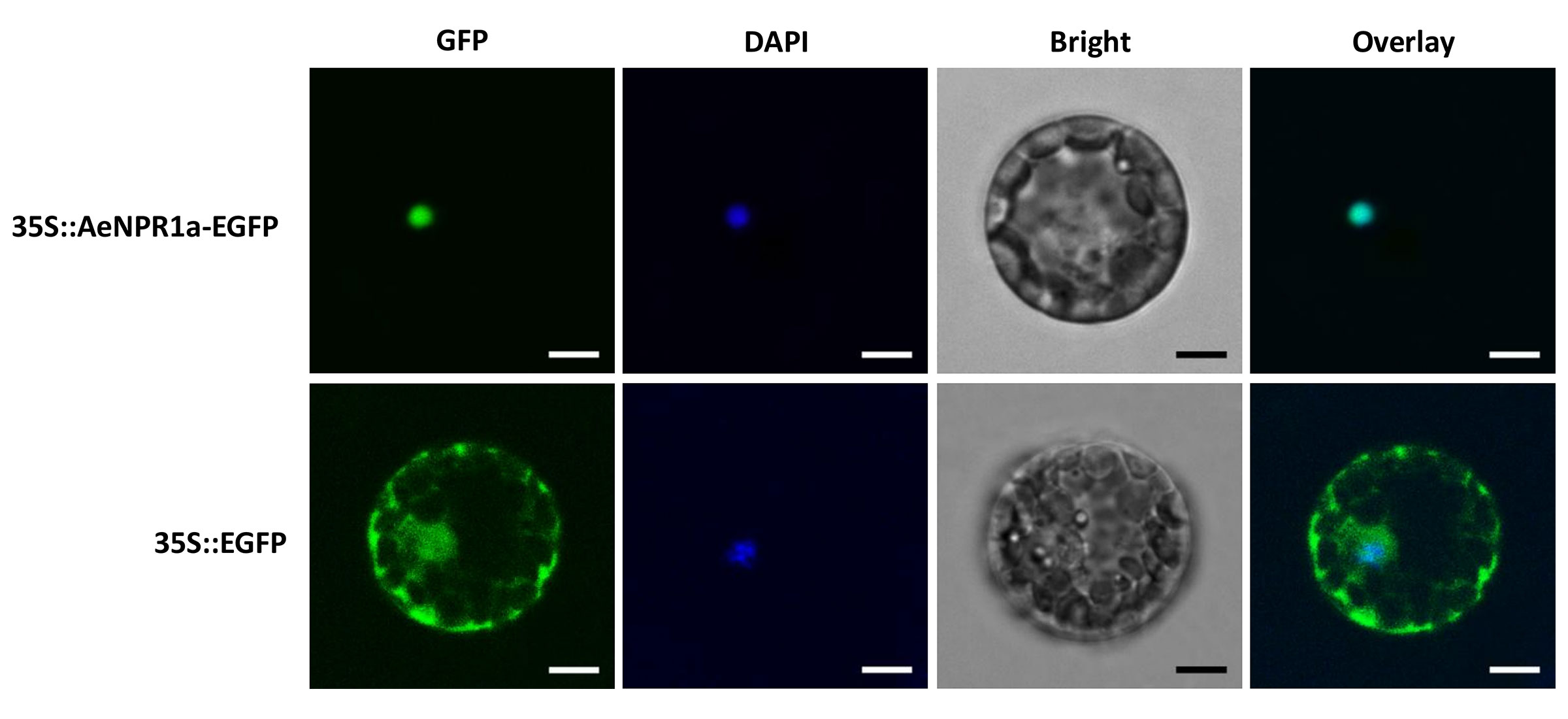
Figure 4 Subcellular localization of AeNPR1a protein. The constructs of 35S::AeNPR1a-EGFP and 35S::EGFP were transformed into Arabidopsis mesophyll protoplast cells by PEG-mediated transformation. Fluorescence signals were examined by a laser confocal scanning microscope. Nuclei of the cells were visualized DAPI staining. Overlay: merged GFP and DAPI images. Scale bars = 5 μm.
Characterization of Potential Cis-Acting Elements in the AeNPR1a Promoter
The 2000bp 5′-flanking region of the AeNPR1a gene was isolated by chromosome walking. Meanwhile, the sequence was compared with the promoter of AcNPR1a in the A. chinensis genome database, which showed that there were many differences between the two sequences. Sequence alignment of the promoter and the AeNPR1a 5’-UTR region showed that the transcription start site (TSS) was located at −239 bp upstream of the ATG codon (Figure 5). Bioinformatic analysis revealed a putative TATA box and two CAAT box were located at the −335 and −415/−435 regions, respectively. Four W-box, which is recognized specifically by SA-induced WRKY DNA binding proteins, were identified at −126, −762, −1,459, and −1664 positions. Two potential RAV1AAT elements and a TC-rich repeats that participate in defense and stress responses were located at positions −499, −1,412, and −687. Two 5′ UTR pyrimidine-rich stretches existed in the −149 to −95 regions, which conferred high transcription. Furthermore, we also noted the presence of several cis-acting elements associated with phytohormone and abiotic stress regulation such as TCA-element, TGACG-motif, AuxRR-core, GARE-motif, MYC, and LTR (Figure 5), which are known involved in SA, MeJA, auxin, gibberellin, dehydration, and low temperature-associated responses, respectively.
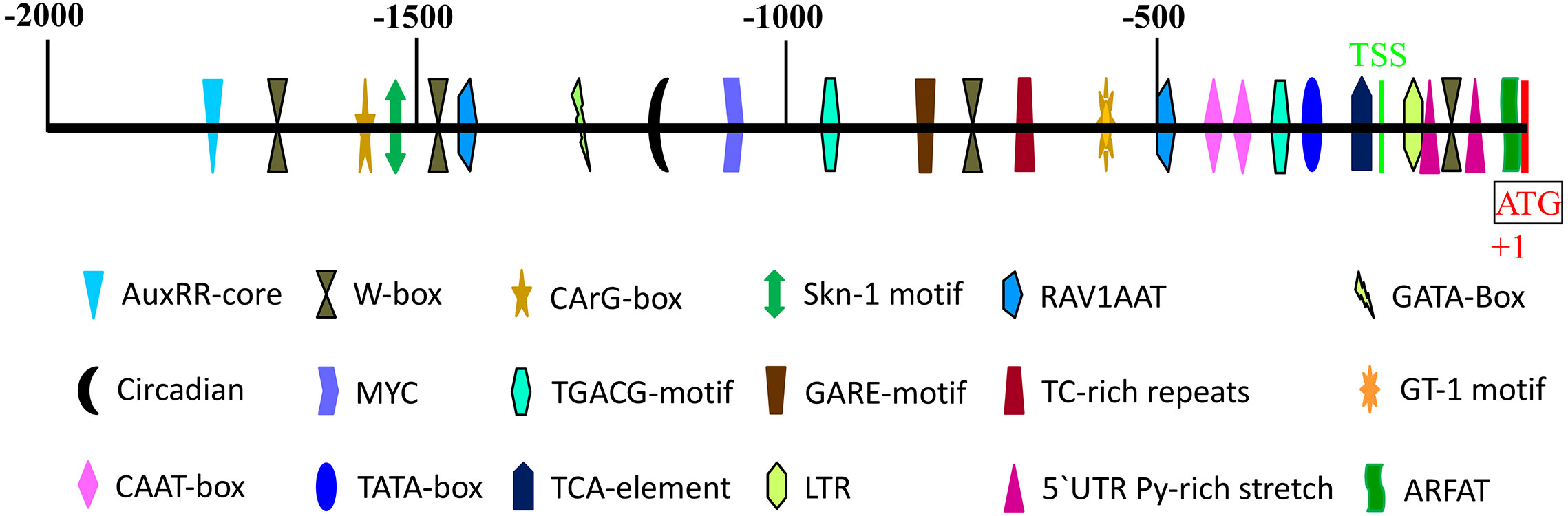
Figure 5 Schematic representation of predicted cis-acting regulatory elements in AeNPR1a promoter region. The colored shapes represent different cis-elements as indicated. The scale on top of the bar represents the distance relative to the translational start condon (+1). TSS, transcription start site.
Expression Analysis of AeNPR1a in Different Tissues and in Response to Hormone Treatments and Psa Infection
Quantitative real-time RT-PCR analyses were performed to examine the expression levels of AeNPR1a in various tissues and under different treatment conditions. As shown in Figure 6A, the transcripts of AeNPR1a were detected in almost all the tissues investigated, while the levels of expression varied considerably among different tissues. The highest levels of AeNPR1a expression were observed in the mature leaf (8.9-fold, compared the levels in the young stem), followed by young fruit and mature stems (2.5-fold), and the lowest expression levels were detected in the alabastrum (Figure 6A).
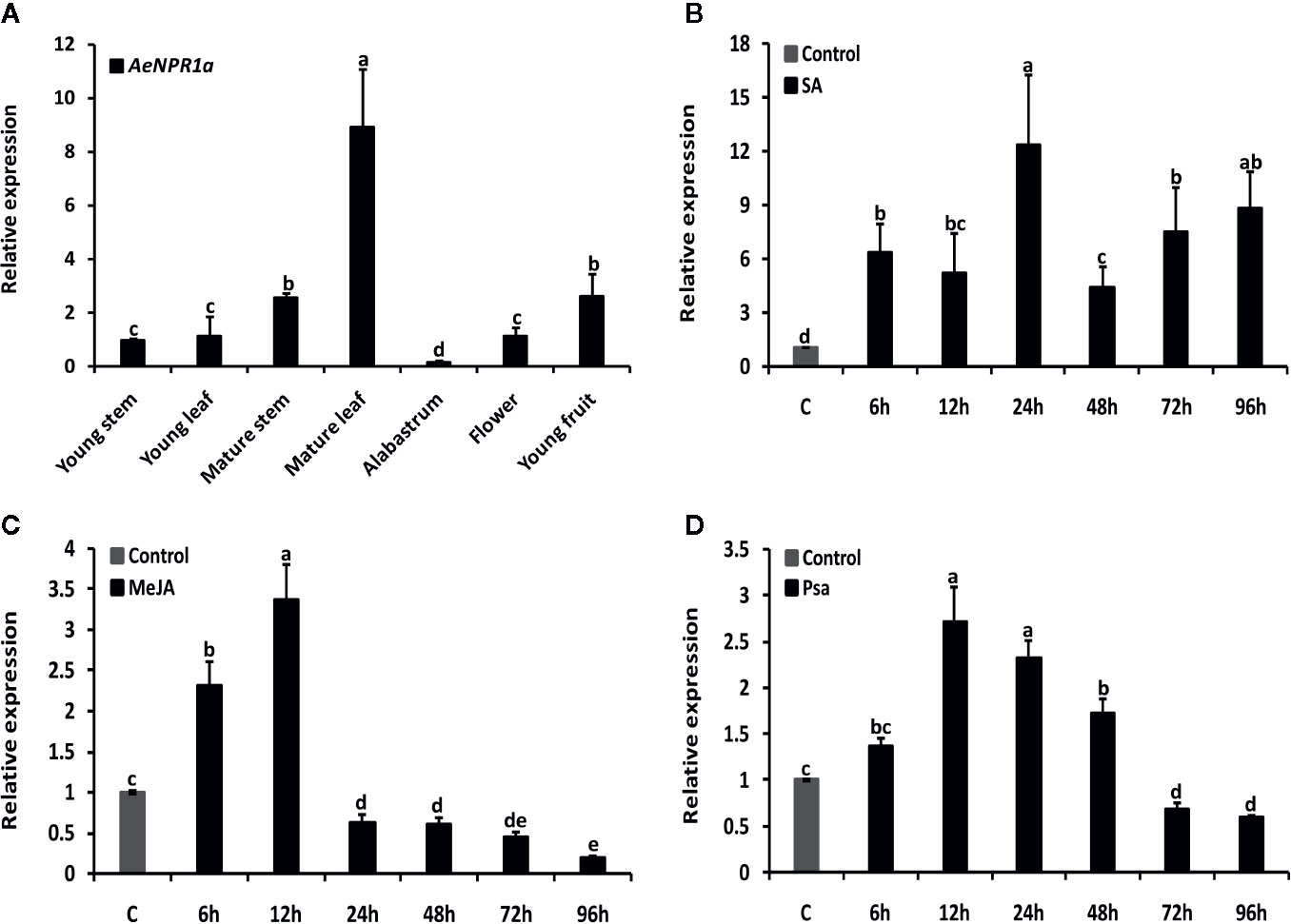
Figure 6 Expression patterns of AeNPR1a in A. eriantha. (A) Expression profiles of AeNPR1a in different tissues. The expression in different tissues was calibrated using expression in the young stem. (B–D) Expression analyses of AeNPR1a in leaves at various time points after SA, MeJA treatment, and Psa infection, respectively. β-actin was used to normalize the samples. The gene expression level at each time point is shown as relative to the mock, which was set to 1. Data represent the mean ± standard deviation (SD) of three biological replicates. Bars with different letters are significantly different (P < 0.05) according to Duncan’s multiple tests. The experiments were repeated three times with similar results.
It has been previously reported that exogenous plant defense molecules, such as SA, JA, and so on, can induce expression of NPR1 or its homologous genes, to activate plant disease resistance responses (Kunkel and Brooks, 2002; Ali et al., 2017; Neeley et al., 2019). Here, the temporal expression profiles of AeNPR1a in leaves following treatment with SA, MeJA as well as infection with Psa, were analyzed by real-time qRT-PCR, respectively. As shown in Figure 6, when plants were treated with SA, AeNPR1a expression was induced significantly and had increased up to the peak level 24 h after treatment (Figure 6B). Upon MeJA treatment, AeNPR1a transcription was up-regulated after 6 h of treatment application, and the maximum expression levels were observed at 12 h, with rapid decline at later time points (Figure 6C). To further explore the defensive role of AeNPR1a, plants were inoculated with Psa pathogen and we monitored changes in AeNPR1a expression. After inoculation, AeNPR1a transcription increased slightly at 6 h, with a single peak 12 h post inoculation (Figure 6D).
AeNPR1a Could Interact With AeTGA2
Previous reports suggest that NPR1 interacts with TGA family members in Arabidopsis (Zhang et al., 1999). Here, the AeTGA2 full-length cDNA sequence was obtained, phylogenetic analyses of protein sequences suggested that AeTGA2 was grouped together with other TGA2 homologs (Figure 7A). To further explore the potential interaction between AeNPR1a and AeTGA2 in kiwifruit, we performed yeast two-hybrid assays. The different regions of AeNPR1a were cloned in a pGBKT7 vector containing a DNA-binding domain to construct BD bait, and the ORF of AeTGA2 was cloned into the pGADT7 vector with a DNA activation domain to construct AD prey (Figure 7B). The bait and prey plasmids were co-transformed into yeast cells and selected on QOD mediums. As shown in Figure 7B, both the complete AeNPR1a(1-582) and the AeNPR1a(1-360) truncation that included the BTB and ANK domains showed interaction with AeTGA2. However, neither the N-terminal AeNPR1a(1-182) nor C-terminal AeNPR1a(361-582) alone can interact with AeTGA2 (Figure 7B). The results indicated the interaction between AeNPR1a and AeTGA2.
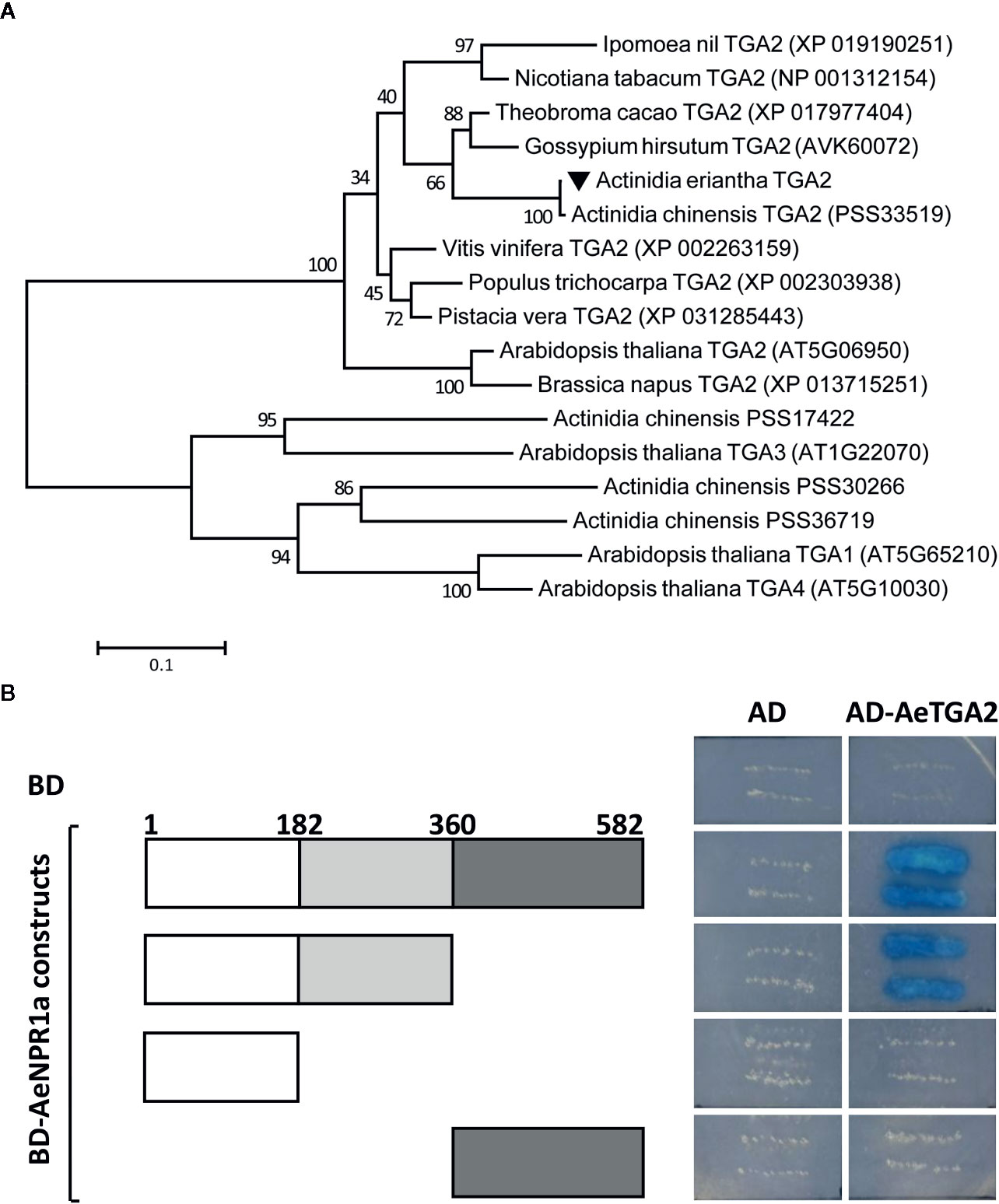
Figure 7 AeNPR1a interacts with AeTGA2. (A) Phylogenetic analysis of AeTGA2 with other TGA proteins from different plant species. The tree was generated using Maximum-Likelihood method with 1,000 bootstrap replicates. GenBank IDs of each protein sequence are attached in the brackets behind the species names. (B) Yeast two-hybrid assay of protein interaction between AeNPR1a and AeTGA2. Transformed yeast cells were streaked on QOD (SD/-Trp/-Leu/-His/-Ade) medium supplemented with 20 μg/ml X-α-Gal. Yeast cells co-transformed with pGBKT7 and pGADT7, pGBKT7-AeNPR1a constructs and pGADT7 were used as negative control.
Ectopic Expression of AeNPR1a Enhanced Disease Resistance in Transgenic Tobacco
In order to assess the function of the AeNPR1a, we constitutively expressed AeNPR1a in tobacco. A total of 14 independent transgenic lines were generated by Agrobacterium-mediated transformation, and transgene expression was confirmed by semi-quantitative RT-PCR analysis (Figure 8A). These transgenic lines appeared normal in their growth and development compared to wild-type plants (Supplementary Table S3 and Supplementary Figure S2). Three transgenic lines were randomly selected to generate T2 generation plants for further investigation.
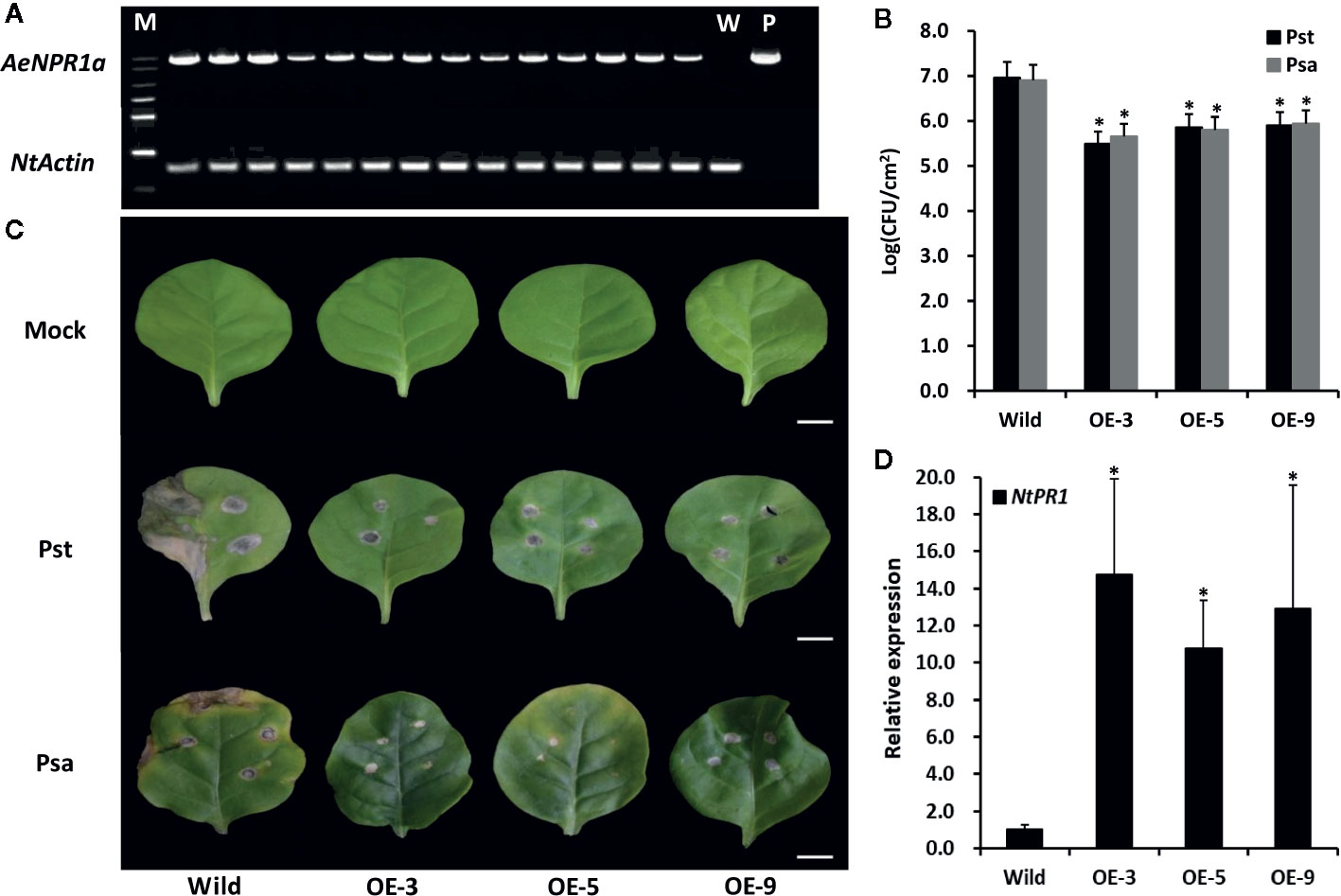
Figure 8 Overexpression of AeNPR1a in transgenic tobacco display enhanced resistance to bacterial pathogen. (A) RT-PCR analysis of AeNPR1a expression in T2 transgenic tobacco plants. M, DNA marker; W, the wild-type tobacco; P, the positive control (pCAMBIA plasmid). NtActin was used as internal control. (B) Growth of Pseudomonas syringae pv. tomato DC3000 (Pst) and Pseudomonas syringae pv. Actinidiae (Psa) in wild-type and AeNPR1a transgenic plants (OE-3, OE-5, OE-9). The leaves of 4-week-old wild-type and transgenic plants were inoculated with Pst and Psa, respectively. Bacterial population was monitored 3 days after inoculation using serial dilution. Each data point represents the mean and standard errors of three samples from three individual plants. The asterisks represent statistically significant differences between the wild-type and AeNPR1a transgenic plants (P < 0.05). CFU: Colony forming unit. (C) Disease symptoms from representative leaves of wild-type and transgenic plants at 7 days after inoculation with Pst, Psa, or 10 mM MgCl2 (Mock). Bars = 1 cm. (D) Transcript analysis of NtPR1 gene in transgenic plants by quantitative PCR. NtActin was used to normalize the samples. Asterisks indicate a significant difference (Student’s t-test, P < 0.05). The results are presented from three independent experiments.
To determine whether AeNPR1a over-expression confers resistance to pathogenic bacteria, the transgenic and wild-type plants were inoculated with Pst and Psa, separately. As shown in Figure 8B, the bacterial populations of both Pst and Psa in the wild-type leaf discs increased significantly when compared to the levels in the AeNPR1a transgenic lines 3 days after inoculation. Seven days after infection, wild-type leaves displayed severe necrotic lesions (Figure 8C). Although the AeNPR1a transgenic lines also exhibited disease symptoms, the lesions degree in necrotic phenotype was comparatively lower than wild-type (Figure 8C). Generally, the expression of PR genes has been considered a marker for plant disease resistance. To understand whether pathogen resistance in AeNPR1a transgenic lines was related to PR induction, the expression of NtPR1 in the transgenic tobacco lines were analyzed. As shown in Figure 8D, the transcript levels of NtPR1 in transgenic plants increased significantly relative to the levels in the wild-type. These results demonstrated that AeNPR1a overexpression in tobacco confers enhanced resistance to bacterial pathogens, and the resistance could be associated with increased expression of PR gene.
AeNPR1a Complements Resistance to Pseudomonas syringae pv. Tomato DC3000 in Arabidopsis npr1-1 Mutant
To further investigate if AeNPR1a has functions similar to Arabidopsis NPR1, the 35S::AeNPR1a construct was transformed into the Arabidopsis npr1-1 mutant for stable transformants by Agrobacterium-mediated transformation (Figure 9A). Three independent T3 transgenic lines expressing AeNPR1a and npr1-1 mutant, along with wild-type plants, were infected with bacterial Pst suspended in 10 mM MgCl2, or 10 mM MgCl2 alone as a mock. Three days after inoculation, the population of Pst on infected leaves was measured to quantify the disease symptoms. As shown in Figure 9B, the levels of bacterial growth in infected npr1-1 mutants were more than 3 times the levels in the wild-type. Three transgenic lines expressing AeNPR1a had significant decreases in bacterial growth when compared to the levels in npr1-1 mutant (Figure 9B). After 7 days of inoculation, the leaves of npr1-1 mutants showed severe necrotic lesions, while such symptoms in transgenic plants were restricted (Figure 9C). In addition, the over-expression of AeNPR1a in the npr1-1 mutant significantly elevated the AtPR1 expression (Figure 9D). Overall, the observations above indicated that AeNPR1a can functionally complement the basal resistance to Pst in the npr1-1 mutant plants.
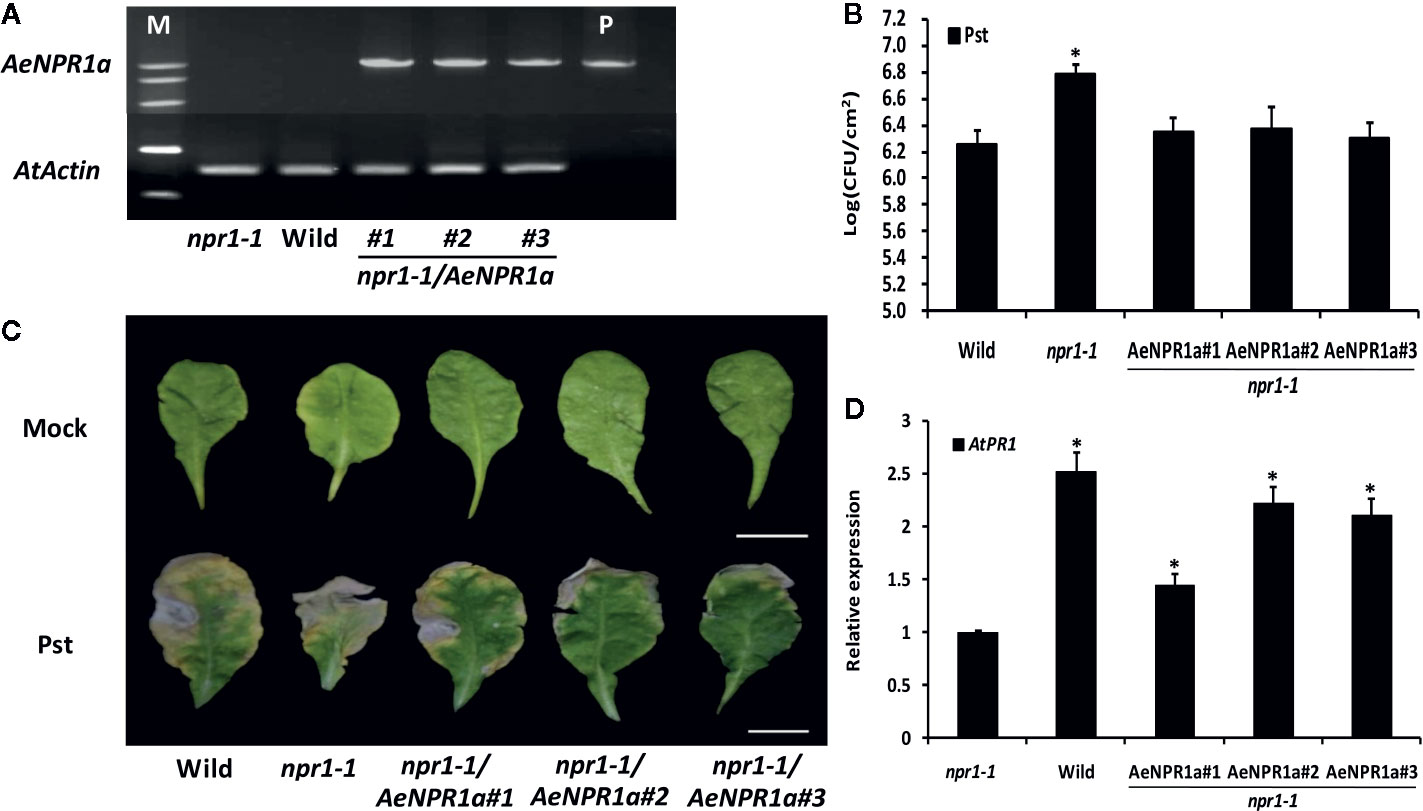
Figure 9 Complementation of the Arabidopsis npr1-1 mutants by AeNPR1a. (A) RT-PCR analysis of AeNPR1a expression was performed with cDNA prepared from leaves of npr1-1, wild type and transgenic npr1-1 mutant lines. M, DNA marker; P, the positive control (pCAMBIA plasmid). AtActin was used as internal control. (B) Bacterial growth of Pseudomonas syringae pv. tomato DC3000 (Pst) on leaves of 4-week-old wild-type, npr1-1, and three individual npr1-1/AeNPR1a transgenic plants. Samples were taken from infected leaves 3 days after inoculation to quantify colony-forming units (CFU). Data represent the means ± standard errors of three biological replicates, and each containing three leaf disks from three individual plants. The asterisk indicates statistically significant differences from the wild-type (P < 0.05). (C) Symptoms on leaves infected with Pst or mock (10 mM MgCl2) at 7 days post-inoculation. Bars = 1 cm. (D) Analysis of AtPR1 gene expression in npr1-1, wild type and transgenic plants. Values are shown as mean ± standard deviation (SD) for three replicates, and representative results are presented from three independent experiments. Asterisks indicate a significant difference (Student’s t-test, P < 0.05%).
Discussion
Plants employ diverse signal transduction pathways to defend themselves against infection by pathogens. SAR is a one of the major defense pathways that can confer long-lasting resistance in plants, and SAR establishment is mediated predominantly by NPR1 in Arabidopsis (Gao et al., 2015; Zhang et al., 2019). It has been revealed that NPR1 is involved in various biotic defense or abiotic stress signaling pathways (Cao et al., 1998; Wu et al., 2012; Withers and Dong, 2016). Although homologs of the NPR1 gene have been isolated and characterized in several plant species, our understanding of the function of NPR1 homologs in kiwifruit remains poor. In the present study, a novel NPR1-like gene (AeNPR1a) from kiwifruit was characterized. The intron/exon organization of AeNPR1a exhibited structures similar to NPR1 in Arabidopsis, as well as in other species. The derived amino acid sequence of AeNPR1a shows 52.86% identity to Arabidopsis NPR1, and they shared typical features, such as a BTB/POZ domain and an ankyrin repeat, which are highly conserved among all NPR1-type proteins of monocotyledonous and dicotyledonous plants (Cao et al., 1997; Silva et al., 2018; Backer et al., 2019). These domains are responsible for the co-activation of TGA transcription factors and protein-protein interactions (Rochon et al., 2006; Ding et al., 2018). The cysteine residues, Cys82, Cys150, Cys155, Cys160, and Cys216 are highly conserved among all the sequences, and they could be involved in the oligomer-monomer transition of NPR1 or NPR1-like proteins (Mou et al., 2003). It has been demonstrated that the S-nitrosylation of NPR1 at Cys156 could facilitate its oligomerization in Arabidopsis (Tada et al., 2008). Cys156 in AeNPR1a was altered to a Serine residue. Previous studies have reported that although Cys156 in the GmNPR1-1 protein of soybean is substituted, GmNPR1-1 still complements NPR1 function in the npr1-1 mutant, which might involve other residues, to maintain GmNPR1-1 protein homeostasis (Sandhu et al., 2009). This means that some other conserved residues of AeNPR1a might also play critical roles in defense regulation in kiwifruit. In addition, the AeNPR1a protein also has a potential nuclear localization motif in the C-terminus (Figure 1). The data suggest that the AeNPR1a protein shared some common characteristics with NPR1 from other plants.
Phylogenetic analysis revealed that NPR1-like proteins in kiwifruit were clustered in different clades; the AeNPR1a was closer to VvNPR1 of Vitis vinifera and grouped in the same clade as AtNPR1, which is essential for SAR establishment (Cao et al., 1997; Dong, 2004). It have been reported that AtNPR2, as a paralog of AtNPR1, can complement npr1 mutants and play a role in SA perception (Castelló et al., 2018). The phylogenic relation of AeNPR1a and AeNPR1b implied that AeNPR1b might also act as component of the SA signal pathway in kiwifruit. According to previous studies, nuclear localization of NPR1 is essential for its function, which is directed by the NLS in the C-terminus (Kinkema et al., 2000). In Arabidopsis, NPR1 is mainly localized in the cytoplasm in the form of oligomers in conditions with no biotic stress. Upon SA induction, monomeric NPR1 is released from the oligomers and transported into the nucleus (Mou et al., 2003; Maier et al., 2011). However, in the present study, unlike AtNPR1, which also showed a small amount of fluorescence in the plasma membrane of onion epidermal cells, the transiently expressed AeNPR1a-EGFP fusion protein was predominantly located in the nucleus of Arabidopsis protoplast cells and onion epidermal cells (Figure 4 and Supplementary Figure S1). Differences in species or key amino acid residues could have promoted the translocation of AeNPR1a into the nucleus. Consistently, constitutive nuclear localization has also been reported by transient expression of StoNPR1 and VvNPR1, even in the absence of SA induction (Le Henanff et al., 2009; Jue et al., 2014). The results suggest that AeNPR1a is primarily localized in the nucleus under non-inductive condition.
The specific gene expression is mainly driven by the promoter. To reveal the regulatory mechanism of AeNPR1a expression, a 2.0 Kb AeNPR1a promoter was isolated, and several cis-acting regulatory elements, including phytohormone and defense responsive elements, were identified, which suggested that AeNPR1a could be regulated by plant hormones and stress factors. The expression patterns of AeNPR1a were analyzed by qRT-PCR, and it was expressed constitutively in different tissues, particularly in mature leaves. The observed expression patterns differed from the high levels of expression of NPR1 homologs in roots and stems of avocado and soybean, respectively (Sandhu et al., 2009; Backer et al., 2015). We also present evidence that AeNPR1a transcript is expressed in reproductive tissues. Previous studies have demonstrated that NPR1 expression can be induced by exogenous signaling molecules, such as SA, JA, and ethylene, to activate plant defense responses and confer resistance to pathogens (Kunkel and Brooks, 2002; Leon-Reyes et al., 2009; Wang et al., 2017). AeNPR1a transcription was induced not only by treatment with SA or MeJA but also by pathogen infection, which may be associated with the presence of phytohormone and defense responsive elements in the promoter. In contrast, the expression of BjNPR1 in mustard was not altered after MeJA treatment (Ali et al., 2017). The discrepancy could be attributed to variation of the mechanism of signal molecules in different species (Kunkel and Brooks, 2002; Caarls et al., 2015). In addition, AeNPR1a expression was induced rapidly by each treatment in 24 h, suggesting that the gene might participate in the early stages of defense responses to treatment with exogenous signaling molecules and pathogen infection. Similar results were also observed in rice, lily, and Mimulus lewisii (Yuan et al., 2007; Wang et al., 2017; He and Shi, 2018). In Arabidopsis, the interaction between NPR1 and TGA2 family proteins is known to be important for the activation of defense genes, and conferring resistance to secondary infection (Fan and Dong, 2002; Silva et al., 2018). Therefore, the potential interaction between AeNPR1a and AeTGA2 was assessed using the yeast two-hybrid system. Our results not only support the existence of an interaction between the two proteins but also highlighted some regions of AeNPR1a protein that are critical for the interaction. However, whether AeNPR1a-AeTGA2 interaction regulates PR genes transcription to trigger SAR responses in kiwifruit still requires verification.
Numerous molecular studies have shown that NPR1 or NPR1-like genes confer resistance to bacterial and fungal pathogens in various transgenic plants, and the resistance was associated with PR expression (Cao et al., 1998; Le Henanff et al., 2011; Zhang et al., 2012; Ali et al., 2017). Transgenic citrus plants expressing AtNPR1 exhibited significant up-regulation of PR1 expression and enhanced resistance to citrus canker and Huanglongbing (Zhang et al., 2010; Robertson et al., 2018). Ectopic expression of Malus hupehensis MhNPR1 in tobacco induced the transcription of PR genes and facilitated salt and osmotic stress tolerance, in addition to enhanced resistance to Botrytis cinerea (Zhang et al., 2012; Zhang et al., 2014). Similarly, in the present study, the over-expression of AeNPR1a in transgenic tobacco plants triggered disease resistance against both Pst and Psa bacterial pathogens, as transgenic lines had reduced pathogen proliferation and limited disease spread to non-infected leaf parts. It has been reported that over-expression AtNPR1 in transgenic strawberry plants improved resistance to pathogens, but the plants displayed numerous undesirable traits with regard to growth and development, such as decreased plant height, canopy density, and defective fruits (Silva et al., 2015). The transgenic Arabidopsis plants that express mulberry MuNPR1 produced an early flowering phenotype (Xu et al., 2019). However, AeNPR1a transgenic tobacco exhibited normal phenotypes and did not exhibit any detrimental morphological traits under our experimental conditions, and similar results have been reported in cotton, citrus, and mustard (Parkhi et al., 2010; Dutt et al., 2015; Ali et al., 2017). Notably, the transcript levels of the SAR marker NtPR1 were increased significantly in transgenic tobacco lines without an exogenous inducer. A possible explanation for the phenomenon is AeNPR1a transcription in transgenic lines achieved a certain threshold that is required for NtPR1 activation, or biotic stress from T-DNA insertions might persist in transgenic tobacco plants.
The npr1 mutant Arabidopsis plants fail to induce SAR in response to various inducing agents and display increased susceptibility to pathogen infections (Cao et al., 1994). In the present study, the effects of complementation of the Arabidopsis npr1-1 mutant with AeNPR1a were analyzed. Over-expression of AeNPR1a in the npr1-1 mutant increased resistance to Pst infection, which is consistent with previous observations in other plant species, including rice and Gladiolus (Yuan et al., 2007; Zhong et al., 2015). Consequently, we conclude that AeNPR1a is orthologous to Arabidopsis NPR1 and might participate in mechanisms of defense against pathogens in kiwifruit.
In summary, the results of the presented study demonstrate that AeNPR1a can be induced by phytohormones and Psa infection. We provide evidence that AeNPR1a can interact with AeTGA2. In addition, ectopic expression of AeNPR1a in transgenic tobacco and Arabidopsis npr1-1 mutant confers resistance against bacterial pathogens, based on the observed differences in the degrees of severity of disease symptoms between transformed and untransformed lines. Our results could facilitate efforts to enhance disease resistance in kiwifruit through biotechnological approaches that target endogenous defense mechanisms. In future research, over-expression or knockout of AeNPR1a could be carried out in kiwifruit, and transgenic plants would subsequently be challenged under biotic and abiotic stresses. In addition, the promoter function of the gene will be characterized to further elucidate the defense pathways and networks mediated by AeNPR1a. The finding of such studies would facilitate the genetic improvement of kiwifruit and lead to the development of more resistant resources.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
L-MS and J-BF participated in research design. L-MS carried out most of the experiments and wrote the manuscript. L-MS and J-BF were responsible for generating the data and for interpretation of the results. M-ML performed the real-time PCR experiments. X-JQ and J-YC contributed in the statistical analyses. MZ carried out genetic transformation. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Key Research and Development Program of China (2019YFD1000200), the National Natural Science Foundation of China (grant no. 31701778), the Agricultural Science and Technology Innovation Program (ASTIP) (CAAS-ASTIP-2020-ZFRI-04), and Modern agricultural industry technology system of Henan province (S2014-11).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
An earlier version of this paper was presented as an abstract at the IX International Conference on kiwifruit.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.551201/full#supplementary-material
References
Ali, S., Mir, Z. A., Tyagi, A., Mehari, H., Meena, R. P., Bhat, J. A., et al. (2017). Overexpression of NPR1 in Brassica juncea Confers Broad Spectrum Resistance to Fungal Pathogens. Front. Plant Sci. 8, 1693. doi: 10.3389/fpls.2017.01693
An, C., Mou, Z. (2011). Salicylic Acid and its Function in Plant Immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x
Backer, R., Mahomed, W., Reeksting, B. J., Engelbrecht, J., Ibarra-Laclette, E., van den Berg, N. (2015). Phylogenetic and expression analysis of the NPR1-like gene family from Persea americana (Mill.). Front. Plant Sci. 6, 300. doi: 10.3389/fpls.2015.00300
Backer, R., Naidoo, S., van den Berg, N. (2019). The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and Related Family: Mechanistic Insights in Plant Disease Resistance. Front. Plant Sci. 10, 102. doi: 10.3389/fpls.2019.00102
Bai, W., Chern, M., Ruan, D., Canlas, P. E., Sze-to, W. H., Ronald, P. C. (2011). Enhanced disease resistance and hypersensitivity to BTH by introduction of an NH1/OsNPR1 paralog. Plant Biotechnol. J. 9, 205–215. doi: 10.1111/j.1467-7652.2010.00544.x
Caarls, L., Pieterse, C. M. J., Van Wees, S. C. M. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. doi: 10.3389/fpls.2015.00170
Cao, H., Bowling, S. A., Gordon, A. S., Dong, X. (1994). Characterization of an Arabidopsis Mutant That Is Nonresponsive to Inducers of Systemic Acquired Resistance. Plant Cell. 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., Dong, X. (1997). The Arabidopsis NPR1 Gene That Controls Systemic Acquired Resistance Encodes a Novel Protein Containing Ankyrin Repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Cao, H., Li, X., Dong, X. (1998). Generation of broad-spectrum disease resistance by overexpression of an essential regulatory gene in systemic acquired resistance. Proc. Natl. Acad. Sci. 95, 6531–6536. doi: 10.1073/pnas.95.11.6531
Castelló, M. J., Medina-Puche, L., Lamilla, J., Tornero, P. (2018). NPR1 paralogs of Arabidopsis and their role in salicylic acid perception. PloS One 13, e0209835. doi: 10.1371/journal.pone.0209835
Chen, X.-K., Zhang, J.-Y., Zhang, Z., Du, X.-L., Du, B.-B., Qu, S.-C. (2012). Overexpressing MhNPR1 in transgenic Fuji apples enhances resistance to apple powdery mildew. Mol. Biol. Rep. 39, 8083–8089. doi: 10.1007/s11033-012-1655-3
Chen, J., Mohan, R., Zhang, Y., Li, M., Chen, H., Palmer, I. A., et al. (2019). NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8. Plant Physiol. 181, 289–304. doi: 10.1104/pp.19.00124
Chern, M.-S., Fitzgerald, H. A., Yadav, R. C., Canlas, P. E., Dong, X., Ronald, P. C. (2001). Evidence for a disease-resistance pathway in rice similar to the NPR1-mediated signaling pathway in Arabidopsis. Plant J. 27, 101–113. doi: 10.1046/j.1365-313x.2001.01070.x
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ding, Y., Sun, T., Ao, K., Peng, Y., Zhang, Y., Li, X., et al. (2018). Opposite Roles of Salicylic Acid Receptors NPR1 and NPR3/NPR4 in Transcriptional Regulation of Plant Immunity. Cell 173, 1454–1467. doi: 10.1016/j.cell.2018.03.044
Dong, X. (2004). NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. doi: 10.1016/j.pbi.2004.07.005
Dutt, M., Barthe, G., Irey, M., Grosser, J. (2015). Transgenic Citrus Expressing an Arabidopsis NPR1 Gene Exhibit Enhanced Resistance against Huanglongbing (HLB; Citrus Greening). PloS One 10, e0137134. doi: 10.1371/journal.pone.0137134
Everett, K. R., Taylor, R. K., Romberg, M. K., Rees-George, J., Fullerton, R. A., Vanneste, J. L., et al. (2011). First report of Pseudomonas syringae pv. actinidiae causing kiwifruit bacterial canker in New Zealand. Australas. Plant Dis. Notes 6, 67–71. doi: 10.1007/s13314-011-0023-9
Fan, W., Dong, X. (2002). In Vivo Interaction between NPR1 and Transcription Factor TGA2 Leads to Salicylic Acid–Mediated Gene Activation in Arabidopsis. Plant Cell. 14, 1377–1389. doi: 10.1105/tpc.001628
Friedrich, L., Lawton, K., Dietrich, R., Willits, M., Cade, R., Ryals, J. (2001). NIM1 Overexpression in Arabidopsis Potentiates Plant Disease Resistance and Results in Enhanced Effectiveness of Fungicides. Mol. Plant-Microbe Interact. 14, 1114–1124. doi: 10.1094/mpmi.2001.14.9.1114
Gao, Q.-M., Zhu, S., Kachroo, P., Kachroo, A. (2015). Signal regulators of systemic acquired resistance. Front. Plant Sci. 6, 228. doi: 10.3389/fpls.2015.00228
Görlach, J., Volrath, S., Knauf-Beiter, G., Hengy, G., Beckhove, U., Kogel, K. H., et al. (1996). Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell. 8, 629–643. doi: 10.1105/tpc.8.4.629
He, X., Shi, Y. (2018). Cloning and characterization of a Mimulus lewisii NPR1 gene involved in regulating plant resistance to Rhizoctonia solani. Plant Biotechnol. 35, 349–356. doi: 10.5511/plantbiotechnology.18.0820a
Higo, K., Ugawa, Y., Iwamoto, M., Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Holsters, M., de Waele, D., Depicker, A., Messens, E., van Montagu, M., Schell, J. (1978). Transfection and transformation of Agrobacterium tumefaciens. Mol. Gen. Genet. MGG 163, 181–187. doi: 10.1007/bf00267408
Huang, S., Ding, J., Deng, D., Tang, W., Sun, H., Liu, D., et al. (2013). Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 4, 2640. doi: 10.1038/ncomms3640
Huang, H. (2016). “Introduction,” in Kiwifruit: the genus Actinidia. Ed. Huang, H. (San Diego: Academic Press), 1–7.
Japelaghi, R. H., Haddad, R., Garoosi, G.-A. (2011). Rapid and Efficient Isolation of High Quality Nucleic Acids from Plant Tissues Rich in Polyphenols and Polysaccharides. Mol. Biotechnol. 49, 129–137. doi: 10.1007/s12033-011-9384-8
Jue, D.-w., Yang, L., Shi, C., Chen, M., Yang, Q. (2014). Cloning and characterization of a Solanum torvum NPR1 gene involved in regulating plant resistance to Verticillium dahliae. Acta Physiol, Planta. 36, 2999–3011. doi: 10.1007/s11738-014-1671-0
Kinkema, M., Fan, W., Dong, X. (2000). Nuclear Localization of NPR1 Is Required for Activation of PR Gene Expression. Plant Cell. 12, 2339–2350. doi: 10.1105/tpc.12.12.2339
Koh, Y., Chung, H., Cha, B., Lee, D. (1994). Outbreak and spread of bacterial canker in kiwifruit. Korean J. Plant Pathol. 10, 68–72.
Krügel, T., Lim, M., Gase, K., Halitschke, R., Baldwin, I. T. (2002). Agrobacterium-mediated transformation of Nicotiana attenuata, a model ecological expression system. Chemoecology 12, 177–183. doi: 10.1007/pl00012666
Kuai, X., Després, C. (2016). Defining Arabidopsis NPR1 orthologues in crops for translational plant immunity. Can. J. Plant Pathol. 38, 25–30. doi: 10.1080/07060661.2016.1149106
Kunkel, B. N., Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Le Henanff, G., Heitz, T., Mestre, P., Mutterer, J., Walter, B., Chong, J. (2009). Characterization of Vitis vinifera NPR1 homologs involved in the regulation of Pathogenesis-Related gene expression. BMC Plant Biol. 9:54. doi: 10.1186/1471-2229-9-54
Le Henanff, G., Farine, S., Kieffer-Mazet, F., Miclot, A.-S., Heitz, T., Mestre, P., et al. (2011). Vitis vinifera VvNPR1.1 is the functional ortholog of AtNPR1 and its overexpression in grapevine triggers constitutive activation of PR genes and enhanced resistance to powdery mildew. Planta 234, 405–417. doi: 10.1007/s00425-011-1412-1
Leon-Reyes, A., Spoel, S. H., De Lange, E. S., Abe, H., Kobayashi, M., Tsuda, S., et al. (2009). Ethylene Modulates the Role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in Cross Talk between Salicylate and Jasmonate Signaling. Plant Physiol. 149, 1797–1809. doi: 10.1104/pp.108.133926
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, D493–D496. doi: 10.1093/nar/gkx922
Lin, W.-C., Lu, C.-F., Wu, J.-W., Cheng, M.-L., Lin, Y.-M., Yang, N.-S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 13, 567–581. doi: 10.1007/s11248-004-2375-9
Liu, G., Holub, E. B., Alonso, J. M., Ecker, J. R., Fobert, P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. doi: 10.1111/j.1365-313X.2004.02296.x
Loake, G., Grant, M. (2007). Salicylic acid in plant defence-the players and protagonists. Curr. Opin. Plant Biol. 10, 466–472. doi: 10.1016/j.pbi.2007.08.008
Maier, F., Zwicker, S., Huckelhoven, A., Meissner, M., Funk, J., Pfitzner, A. J. P., et al. (2011). NONEXPRESSOR OF PATHOGENESIS-RELATED PROTEINS1 (NPR1) and some NPR1-related proteins are sensitive to salicylic acid. Mol. Plant Pathol. 12, 73–91. doi: 10.1111/j.1364-3703.2010.00653.x
Makandar, R., Essig, J. S., Schapaugh, M. A., Trick, H. N., Shah, J. (2006). Genetically Engineered Resistance to Fusarium Head Blight in Wheat by Expression of Arabidopsis NPR1. Mol. Plant-Microbe Interact. 19, 123–129. doi: 10.1094/mpmi-19-0123
Malnoy, M., Jin, Q., Borejsza-Wysocka, E. E., He, S. Y., Aldwinckle, H. S. (2007). Overexpression of the apple MpNPR1 gene confers increased disease resistance in malus x domestica. Mol. Plant-Microbe Interact. 20, 1568–1580. doi: 10.1094/mpmi-20-12-1568
Matthews, B. F., Beard, H., Brewer, E., Kabir, S., MacDonald, M. H., Youssef, R. M. (2014). Arabidopsis genes, AtNPR1, AtTGA2 and AtPR-5, confer partial resistance to soybean cyst nematode (Heterodera glycines) when overexpressed in transgenic soybean roots. BMC Plant Biol. 14:96. doi: 10.1186/1471-2229-14-96
Meur, G., Budatha, M., Srinivasan, T., Rajesh Kumar, K. R., Dutta Gupta, A., Kirti, P. B. (2008). Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Physiol. Planta. 133, 765–775. doi: 10.1111/j.1399-3054.2008.01098.x
Mou, Z., Fan, W., Dong, X. (2003). Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Narváez, I., Pliego Prieto, C., Palomo-Ríos, E., Fresta, L., Jiménez-Díaz, R. M., Trapero-Casas, J. L., et al. (2020). Heterologous Expression of the AtNPR1 Gene in Olive and Its Effects on Fungal Tolerance. Front. Plant Sci. 11, 308. doi: 10.3389/fpls.2020.00308
Neeley, D., Konopka, E., Straub, A., Maier, F., Pfitzner, A. J. P., Pfitzner, U. (2019). Salicylic acid-driven association of LENRV and NIMIN1/NIMIN2 binding domain regions in the C-terminus of tobacco NPR1 transduces SAR signal. bioRxiv 543645. doi: 10.1101/543645
Parkhi, V., Kumar, V., Campbell, L. M., Bell, A. A., Shah, J., Rathore, K. S. (2010). Resistance against various fungal pathogens and reniform nematode in transgenic cotton plants expressing Arabidopsis NPR1. Transgenic Res. 19, 959–975. doi: 10.1007/s11248-010-9374-9
Pieterse, C. M. J., Leon-Reyes, A., Van der Ent, S., Van Wees, S. C. M. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Pilkington, S. M., Crowhurst, R., Hilario, E., Nardozza, S., Fraser, L., Peng, Y., et al. (2018). A manually annotated Actinidia chinensis var. chinensis (kiwifruit) genome highlights the challenges associated with draft genomes and gene prediction in plants. BMC Genomics 19, 257. doi: 10.1186/s12864-018-4656-3
Robertson, C. J., Zhang, X., Gowda, S., Orbović, V., Dawson, W. O., Z., M. (2018). Overexpression of the Arabidopsis NPR1 protein in citrus confers tolerance to Huanglongbing. J. Citrus Pathol. 5, 38911
Rochon, A., Boyle, P., Wignes, T., Fobert, P. R., Després, C. (2006). The Coactivator Function of Arabidopsis NPR1 Requires the Core of Its BTB/POZ Domain and the Oxidation of C-Terminal Cysteines. Plant Cell. 18, 3670–3685. doi: 10.1105/tpc.106.046953
Sandhu, D., Tasma, I. M., Frasch, R., Bhattacharyya, M. K. (2009). Systemic acquired resistance in soybean is regulated by two proteins, orthologous to Arabidopsis NPR1. BMC Plant Biol. 9:105. doi: 10.1186/1471-2229-9-105
Serizawa, S., Ichikawa, T., Takikawa, Y., Tsuyumu, S., Goto, M. (1989). Occurrence of Bacterial Canker of Kiwifruit in Japan. Japanese J. Phytopathol. 55, 427–436. doi: 10.3186/jjphytopath.55.427
Shine, M. B., Xiao, X., Kachroo, P., Kachroo, A. (2019). Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 279, 81–86. doi: 10.1016/j.plantsci.2018.01.001
Silva, K. J. P., Brunings, A., Peres, N. A., Mou, Z., Folta, K. M. (2015). The Arabidopsis NPR1 gene confers broad-spectrum disease resistance in strawberry. Transgenic Res. 24, 693–704. doi: 10.1007/s11248-015-9869-5
Silva, K. J. P., Mahna, N., Mou, Z., Folta, K. M. (2018). NPR1 as a transgenic crop protection strategy in horticultural species. Hortic. Res. 5, 15. doi: 10.1038/s41438-018-0026-1
Spoel, S. H., Koornneef, A., Claessens, S. M. C., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell. 15, 760–770. doi: 10.1105/tpc.009159
Sung, Y.-C., Lin, C.-P., Hsu, H.-J., Chen, Y.-L., Chen, J.-C. (2019). Silencing of CrNPR1 and CrNPR3 Alters Plant Susceptibility to Periwinkle Leaf Yellowing Phytoplasma. Front. Plant Sci. 10, 1183. doi: 10.3389/fpls.2019.01183
Tada, Y., Spoel, S. H., Pajerowska-Mukhtar, K., Mou, Z., Song, J., Wang, C., et al. (2008). Plant Immunity Requires Conformational Charges of NPR1 via S-Nitrosylation and Thioredoxins. Science 321, 952. doi: 10.1126/science.1156970
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tang, W., Sun, X., Yue, J., Tang, X., Jiao, C., Yang, Y., et al. (2019). Chromosome-scale genome assembly of kiwifruit Actinidia eriantha with single-molecule sequencing and chromatin interaction mapping. Giga Sci. 8, giz027. doi: 10.1093/gigascience/giz027
Wang, L., Guo, Z., Zhang, Y., Wang, Y., Yang, G., Yang, L., et al. (2017). Overexpression of LhSorNPR1, a NPR1-like gene from the oriental hybrid lily ‘Sorbonne’, conferred enhanced resistance to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Physiol. Mol. Biol. Plants. 23, 793–808. doi: 10.1007/s12298-017-0466-3
Wang, W., Withers, J., Li, H., Zwack, P. J., Rusnac, D.-V., Shi, H., et al. (2020). Structural basis of salicylic acid perception by Arabidopsis NPR proteins. Nature. doi: 10.1038/s41586-020-2596-y
Withers, J., Dong, X. (2016). Posttranslational Modifications of NPR1: A Single Protein Playing Multiple Roles in Plant Immunity and Physiology. PloS Pathogens. 12, e1005707. doi: 10.1371/journal.ppat.1005707
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 Protein Is a Receptor for the Plant Defense Hormone Salicylic Acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xu, Y.-Q., Wang, H., Qin, R.-L., Fang, L.-J., Liu, Z., Yuan, S.-S., et al. (2019). Characterization of NPR1 and NPR4 genes from mulberry (Morus multicaulis) and their roles in development and stress resistance. Physiol. Planta. 167, 302–316. doi: 10.1111/ppl.12889
Yoo, S. D., Cho, Y. H., Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1038/nprot.2007.199
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhang, Y., Li, X. (2019). Salicylic acid: biosynthesis, perception, and contributions to plant immunity. Curr. Opin. Plant Biol. 50, 29–36. doi: 10.1016/j.pbi.2019.02.004
Zhang, Y., Fan, W., Kinkema, M., Li, X., Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U. States A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Y., Cheng, Y. T., Qu, N., Zhao, Q., Bi, D., Li, X. (2006). Negative regulation of defense responses in Arabidopsis by two NPR1 paralogs. Plant J. 48, 647–656. doi: 10.1111/j.1365-313X.2006.02903.x
Zhang, X., Francis, M. I., Dawson, W. O., Graham, J. H., Orbović, V., Triplett, E. W., et al. (2010). Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 128, 91–100. doi: 10.1007/s10658-010-9633-x
Zhang, J.-Y., Qiao, Y.-S., Lv, D., Gao, Z.-H., Qu, S.-C., Zhang, Z. (2012). Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biol. 14, 46–56. doi: 10.1111/j.1438-8677.2011.00483.x
Zhang, J.-Y., Qu, S.-C., Qiao, Y.-S., Zhang, Z., Guo, Z.-R. (2014). Overexpression of the Malus hupehensis MhNPR1 gene increased tolerance to salt and osmotic stress in transgenic tobacco. Mol. Biol. Rep. 41, 1553–1561. doi: 10.1007/s11033-013-3001-9
Zhang, J., Ren, Z., Zhou, Y., Ma, Z., Ma, Y., Hou, D., et al. (2019). NPR1 and Redox Rhythmx: Connections, between Circadian Clock and Plant Immunity. Int. J. Mol. Sci. 20:1211. doi: 10.3390/ijms20051211
Keywords: kiwifruit, NPR1, disease resistance, Pseudomonas syringae pv. actinidiae, systemic acquired resistance, salicylic acid
Citation: Sun L-M, Fang J-B, Zhang M, Qi X-J, Lin M-M and Chen J-Y (2020) Molecular Cloning and Functional Analysis of the NPR1 Homolog in Kiwifruit (Actinidia eriantha). Front. Plant Sci. 11:551201. doi: 10.3389/fpls.2020.551201
Received: 12 April 2020; Accepted: 01 September 2020;
Published: 16 September 2020.
Edited by:
Andrea Chini, National Center for Biotechnology (CNB), SpainReviewed by:
Hirofumi Nakagami, Max Planck Institute for Plant Breeding Research, GermanyYuelin Zhang, University of British Columbia, Canada
Copyright © 2020 Sun, Fang, Zhang, Qi, Lin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Bao Fang, ZmFuZ2ppbmJhb0BjYWFzLmNu
 Lei-Ming Sun
Lei-Ming Sun Jin-Bao Fang*
Jin-Bao Fang* Min Zhang
Min Zhang Xiu-Juan Qi
Xiu-Juan Qi Miao-Miao Lin
Miao-Miao Lin Jin-Yong Chen
Jin-Yong Chen