- 1Department of Biotechnology, Vignan’s Foundation for Science, Technology and Research (Deemed to be University), Guntur, India
- 2Department of Chemistry, Manipal University Jaipur, Jaipur, India
- 3Department of Genetics, Osmania University, Hyderabad, India
- 4Biochemistry Division, National Institute of Nutrition-ICMR, Hyderabad, India
- 5Department of Botany, Osmania University, Hyderabad, India
- 6Department of Biotechnology and Bioinformatics, Birla Institute of Scientific Research, Jaipur, India
- 7Genomix CARL, Pulivendula, India
Lysine (Lys) is indispensable nutritionally, and its levels in plants are modulated by both transcriptional and post-transcriptional control during plant ontogeny. Animal glutamate receptor homologs have been detected in plants, which may participate in several plant processes through the Lys catabolic products. Interestingly, a connection between Lys and serotonin metabolism has been established recently in rice. 2-Aminoadipate, a catabolic product of Lys appears to play a critical role between serotonin accumulation and the color of rice endosperm/grain. It has also been shown that expression of some lysine-methylated proteins and genes encoding lysine-methyltransferases (KMTs) are regulated by cadmium even as it is known that Lys biosynthesis and its degradation are modulated by novel mechanisms. Three complex pathways co-exist in plants for serine (Ser) biosynthesis, and the relative preponderance of each pathway in relation to plant development or abiotic stress tolerance are being unfolded slowly. But the phosphorylated pathway of L-Ser biosynthesis (PPSB) appears to play critical roles and is essential in plant metabolism and development. Ser, which participates indirectly in purine and pyrimidine biosynthesis and plays a pivotal role in plant metabolism and signaling. Also, L-Ser has been implicated in plant responses to both biotic and abiotic stresses. A large body of information implicates Lys-rich and serine/arginine-rich (SR) proteins in a very wide array of abiotic stresses. Interestingly, a link exists between Lys-rich K-segment and stress tolerance levels. It is of interest to note that abiotic stresses largely influence the expression patterns of SR proteins and also the alternative splicing (AS) patterns. We have checked if any lncRNAs form a cohort of differentially expressed genes from the publicly available PPSB, sequence read archives of NCBI GenBank. Finally, we discuss the link between Lys and Ser synthesis, catabolism, Lys-proteins, and SR proteins during plant development and their myriad roles in response to abiotic stresses.
Introduction
Amino acids are organic compounds, which contain amine (-NH2) and carboxyl C(=O)OH) functional groups along with a side chain (R group). As amino acids are building blocks of proteins, they participate in the synthesis of hormones in animals and in peptide hormone synthesis in plants (Hirakawa et al., 2017). In the case of plants, amino acids also take part in the synthesis of several secondary plant products. Amino acids that cannot be synthesized by some mammals and humans are generally known as “essential,” and lysine (Lys) is one among them (Bright and Shewry, 1983). The pathway of Lys is always a target for the development of herbicides in addition to increasing nutritional value in cereals. Lys is entangled in histone modifications and, therefore, is associated with epigenome and stress biology in plants (Yuan et al., 2013). Lys is the most limiting in all major cereal grains and is therefore considered as a nutritionally significant amino acid (Wenefrida et al., 2009; Galili and Amir, 2013). This is precisely the reason why Lys is a target for crop improvement. Poor people suffer from deficiencies in the essential amino acids like Lys and methionine. If crop plants contain less Lys, the nutritional value of such crops is also reduced by more than 50% and leads to imbalances in the amino acids. But breeding methods have led to the accumulation of Lys in vegetative tissues, which is in fact deleterious to the growth of plants (Ghislain et al., 1995). Therefore, pathways for Lys synthesis in plants have been identified and also the corresponding genes that encode them, keeping in view of their genetic transformation as an alternative approach. But such an effort requires identification of seed specific promoters, which help in the accumulation of more Lys in seeds/grains (Amir and Tabe, 2006). Most importantly, Lys acts as a precursor for the metabolic pathway implicated in plant stress response and also its development as revealed from the studies of Arruda et al. (2000) and Galili et al. (2001). Negrutiu et al. (1984) isolated for the first time, a Lys over-producing mutant from the protoplast cultures of Nicotiana sylvestris. This mutant was a result of altered expression of one of the biosynthetic pathway genes encoding a dihydrodipicolinate synthase (DHDPS). Reduced sensitivity to feedback inhibition in transgenic plants with overexpressed bacterial DHDPS (Perl et al., 1992; Shaul and Galili, 1992, 1993) helped to discover the Lys biosynthetic pathway and its modulation. Further, Karchi et al. (1995) dissected out that Lys biosynthesis and catabolism are coordinately regulated during the development of a seed in tobacco. Thus, for the first time the amino acid Lys has been correlated with plant development. Galili et al. (2001) later reviewed the catabolic events of Lys and implicated them not only to development but also to abiotic stress tolerance. A large body of information suggests that the regulatory networks associated with Lys biosynthesis and catabolism are intertwined largely with plant tissues and organ specificity and interactions between diverse metabolic fluxes.
Another important amino acid in the body of plants is serine (Ser), which helps to form the phospholipids necessary for signal transduction. L-Ser is not only a proteinogenic amino acid, but also it participates in catalytic functions of diverse enzymatic reactions in plants. Importantly, Ser takes an active part in the biosynthesis of many biomolecules such as amino acids, phospholipids, and sphingolipids that are obligatory for cell proliferation (Kalhan and Hanson, 2012). Ser gets phosphorylated by kinases and participates in signaling mechanisms (Chaneton et al., 2012). In plants, multiple biosynthetic pathways exist for Ser. The photorespiratory glycolate pathway appears as the major one, but non-photorespiratory phosphorylated pathway also exists.
This review discusses dehydration proteins known as dehydrins (DHNs), which are members of the LEA protein family group 2 or D-11 (Close, 1997). Since they do not have proper tertiary structures, they are described as “protein clouds” or “cooked spaghetti” (Uversky, 2013, 2016). DHNs are hydrophobic, with more than 50% of the residues being charged or polar in nature and 25% being either alanine or glycine. DHNs contain Lys- and Ser-rich residue and are associated with stress tolerance (Malik et al., 2017). Both Lys and Ser are essential, the former for humans, and the later for plants. Interestingly, both of them are important in plants as a link between metabolism and development. This review focuses on the climacteric links that have been established in recent times about Lys and Ser biosynthesis and catabolism, their association with plant growth and development, abiotic stress tolerance, and also Lys- and Ser-rich proteins, functional significance, and their remarkable ability to bestow stress tolerance in plants.
Two Lys Biosynthetic Pathways Exist in Plants but Differ From That of Prokaryotes
In nature, two pathways namely diaminopimelate (DAP) pathway (Figure 1A) and α-aminoadipate (AAA) have been identified for biosynthesis of Lys (Figure 1B). The first pathway belongs to the aspartate derived biosynthetic family, involved also in the synthesis of threonine, methionine, and isoleucine (Galili, 1995; Velasco et al., 2002; Hudson et al., 2005). The second pathway is part of the glutamate biosynthetic family, involving α-ketoglutarate and acetyl coenzyme A (as seen in Thermus thermophilus; Miyazaki et al., 2004; Xu et al., 2006). While the DAP pathway operates in prokaryotes and plants, the AAA pathway in yeast, protists, and higher fungi. In the DAP pathway, aspartate and pyruvate act as precursors for the biosynthesis of Lys via the intermediate DAP. This pathway adds carbon groups to aspartate to yield Lys (Figure 1A), but exhibits feedback regulation either by Lys or by threonine (Velasco et al., 2002; Hudson et al., 2005). Both pathways are not known to operate in any single organism/plant till date. In higher plants, Lys is synthesized in plastids and no evidence exists for its cytosolic synthesis (Hudson et al., 2006). Glutamate first gets converted to aspartate by aspartate aminotransferase enzyme (AAT). Aspartate kinase (AK), the first enzyme of the pathway catalyzes aspartate to 3-aspartic semialdehyde (3-ASA). While light and photosynthetic activity in the daytime positively affect 3-ASA biosynthesis especially in young leaves (Figure 1A), darkness stimulates the degradation of aspartate to asparagine. Isoenzymes of AK exist as monofunctional polypeptides, which have the Lys-sensitive kinase activity. Interestingly, the bifunctional enzymes AK/homoserine dehydrogenase (HSD) contain threonine-sensitive AK as well as HSD isozymes linked on a single polypeptide. Threonine partially feedback regulates HSD enzyme and, therefore, Lys production is affected (Galili, 2002; Figure 1A). Some of the factors affecting Lys biosynthesis in plants are shown in Table 1. Vauterin et al. (1999) have shown that activity of DHDPS is high in meristems, in the vasculature of leaves, roots, stems, carpels, styles, stigma, pollen, and young embryos. In line with this, they demonstrated that DHDPS promoter exhibits cell type-specific expressions, indicating its multiple functions in diverse tissues. Synthesis of Lys by transcriptional activation in a tissue specific manner clearly implicates regulation of the enzyme at the RNA level. The enzyme L,L-diaminopimelate aminotransferase (LL-DAP-AT) is a novel variant that catalyzes tetrahydrodipicolinate to L,L-DAP and then helps in the synthesis of Lys in lower plants (Physcomitrella patens) as well as higher plants like A. thaliana, soybean, and spinach (Hudson et al., 2006). This indicates that the enzyme is highly conserved among diverse plant species. Further, in A. thaliana, this enzyme catalyzes a reversible reaction. DHDPS and dihydrodipicolinate reductase (DHDPR) enzymes catalyze the two vital steps in Lys biosynthesis (Figure 1A). It is arguable, however, if it has a tetrameric or dimeric arrangement since it is not yet distinctly known. Tetrahydrodipicolinate (THDPA) is converted to meso-2,6-diaminopimelate (m-DAP) catalyzed by different enzymes. Hudson et al. (2006) reported a novel and specific LL-DAP-AT that directly converts THDPA to LL-DAP in A. thaliana (regarded as DAP-AT pathway). Interestingly, LL-DAP-AT operates efficiently in the forward/biosynthetic direction in plants (Hudson et al., 2006). Diaminopimelate epimerase is the last enzyme in the pathway, which converts m-DAP to Lys (Figure 1A).
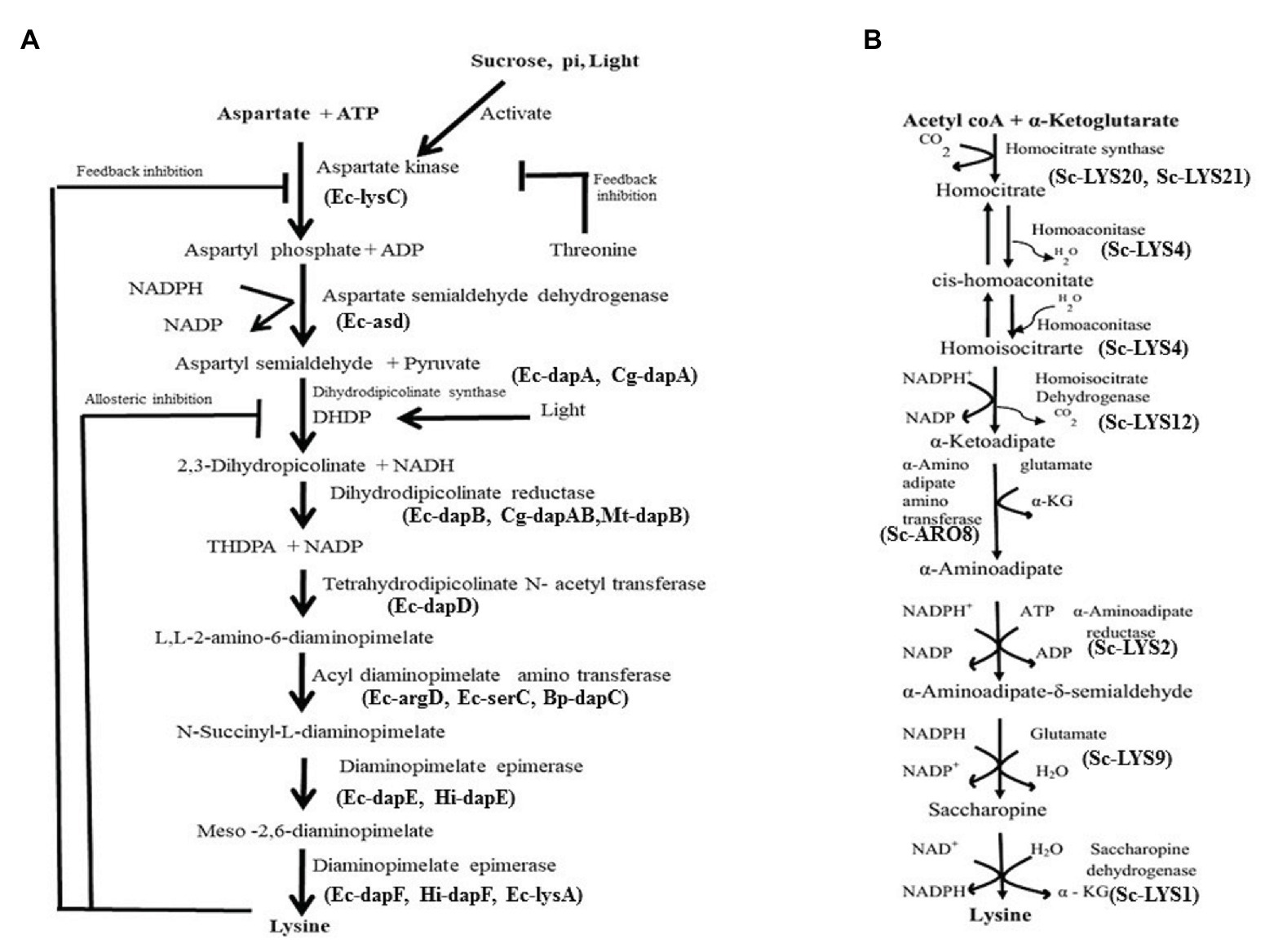
Figure 1. (A) Lysine biosynthesis pathway and its feedback regulation in plants. Ec, Escherichia coli; Sc, S. cerevisiae; Hi, Haemophilus influenzae. (B) Biosynthesis pathway of lysine via aminoadipate. The pathway operates mostly in fungi.
Transgenic Plants Accumulate High Lys Content
Transgenic lines were generated by over expressing bacterial Lys biosynthetic pathway genes in several plants. While seeds were wrinkled in high-Lys producing transgenic Glycine max (Falco et al., 1995), seed germination retarded in A. thaliana (Zhu and Galili, 2003, 2004), and the endosperm hardened in the QPM mutant of maize (Gibbon et al., 2003). Long et al. (2013), and Yang et al. (2016) generated transgenic rice by overexpressing a combination of bacterial genes that encode AK and DHDPS and also by inhibiting Lys ketoglutarate (α-KG) reductase/saccharopine dehydrogenase (LKR/SDH). These transgenic lines of rice displayed 60-fold increase in Lys and thus biofortified, but endosperm color was dark-brown. Surprisingly, the seed germination and subsequent performance of these plants when evaluated in the field conditions did not show any change in their nutrition. Overexpression of tryptophan decarboxylase 3, an enzyme involved in serotonin (sometimes called the happy chemical, seen in human brain mostly) biosynthesis proved that tryptophan (a remote precursor of serotonin) and serotonin are responsible for dark-brown phenotype of endosperm in high-Lys producing lines (Kanjanaphachoat et al., 2012; Yang et al., 2016, 2018). They also proposed that 2-aminoadipate (an intermediate in Lys catabolism) might play a role between jasmonate signaling, elevated serotonin levels, and the endosperm color. Using a seed-specific promoter, Zhu and Galili (2003, 2004) overexpressed a bacterial DHDPS gene in a knockout A. thaliana mutant lacking the bifunctional enzyme α-aminoadipic semialdehyde synthase (AASS) that contains both LKR/SDH activities (in bacteria and fungi, the two enzymes are encoded by separate genes; Zhu et al., 2002). Transgenic A. thaliana seeds displayed 64-fold increase in free Lys levels. Houmard et al. (2007) achieved significant increase in the amount of Lys in transgenic maize kernels by RNA interference technique through endosperm specific suppression of LKR/SDH gene. Possibilities exist therefore for attaining high Lys containing crop plants as nutrient/food supplements to both animals and humans using its degradative pathway genes. In the light of the above facts, a deeper understanding of the metabolism and its networks are needed before we create Lys/nutrient-rich crop plants acceptable to the consumers.
Lys Catabolism Produces Glutamate and Stress-Related Metabolites, Which Help to Overcome Abiotic Stress
If amino acids (cysteine or Lys for example) are accumulated in high concentrations, they may be toxic to the plants (Zhu and Galili, 2004), hence, they must be degraded. The saccharopine pathway (Figure 2) is the primary pathway for Lys degradation, which occurs in the liver (in animals) specifically within mitochondria (Galili et al., 2001). This is the reverse of the AAA pathway (Figure 1B), and in plants and animals, the first two steps in the catabolism are catalyzed by LKR/SDH activities. Lys combines with α-KG to form saccharopine by the enzyme LKR (Figure 2), which ultimately generates glutamate and other stress-related metabolites. Also, Lys has been found vital during seed germination, where it serves as a source of carbon and energy by feeding the intermediates (like α-KG) of TCA cycle to the growing seedlings (Afendi et al., 2012). Thus, the mechanistic insights into Lys degradation and its intermediates serving as the precursors for generation of energy (NADH) via the electron transport chain have been furnished (Araujo et al., 2010).
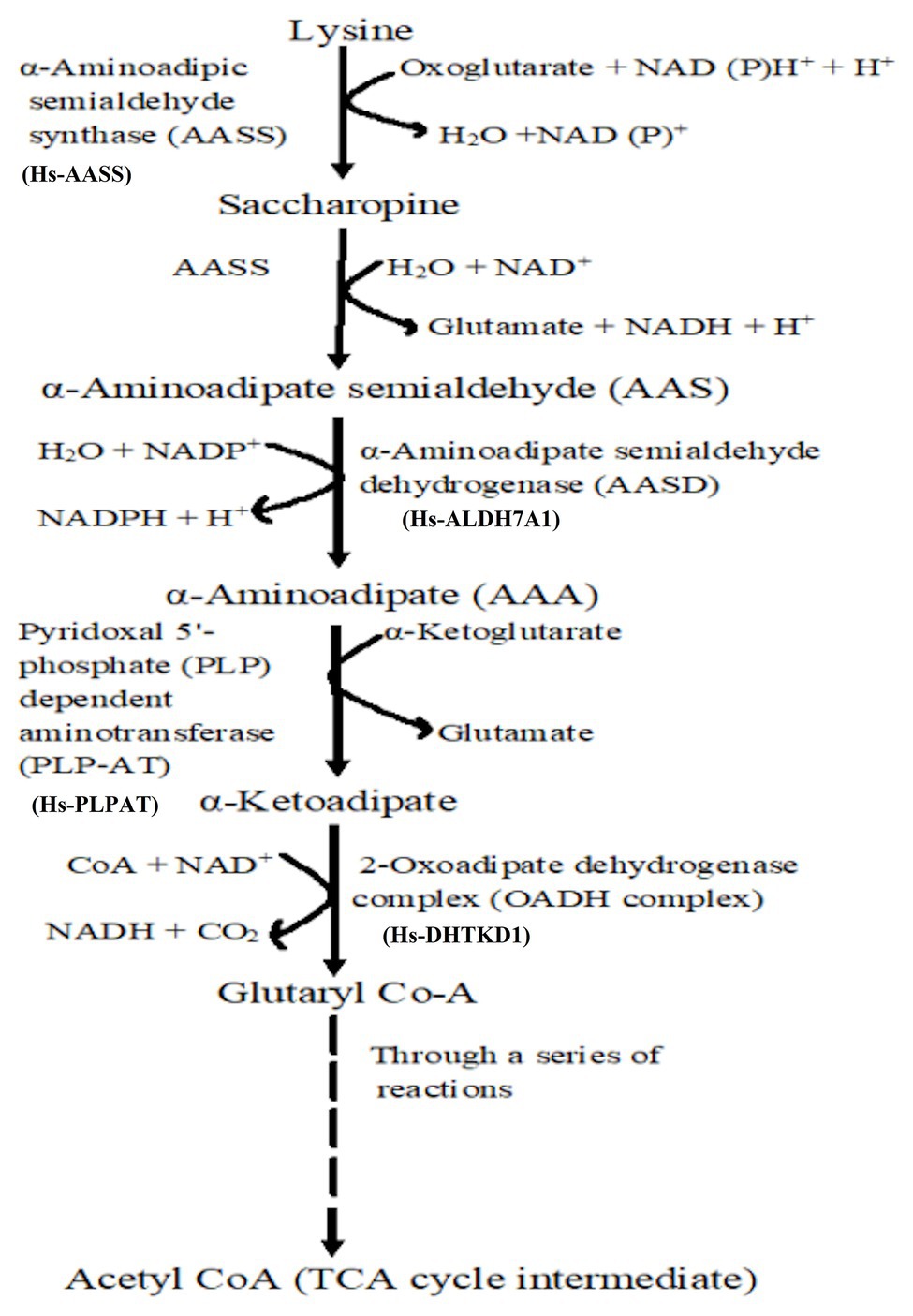
Figure 2. Lysine catabolism via saccharopine is the prominent pathway. Broken line indicates that through several reactions, glutaryl Co-A is converted to TCA cycle intermediate acetyl coenzyme A for energy generation.
Lys serves as an alternative respiratory substrate under a variety of stresses, including drought, and senescence under extended darkness (Urano et al., 2009; Joshi et al., 2010). Flexibility/remobilization of molecules are necessary since it helps the plants to maintain metabolic homeostasis and supply energy, the primary necessity to survive. Consistent with this, under salinity and drought stress conditions, contents of Lys and other amino acids increased in wheat, potato, and safflower, which displayed tolerance to stress (Saeedipour and Moradi, 2012; Muttucumaru et al., 2015; Zafari and Ebadi, 2016) and high expression of LKR/SDH and aminoadipic semialdehyde dehydrogenase (AASADH) genes involved in saccharopine pathway (Brocker et al., 2010; Less et al., 2011). Further, ectopic expression of AASADH resulted in the generation of stress-tolerant plants (Rodrigues et al., 2006). In radish, drought stress was relieved with Lys application in chelation with Zn as seed priming (Noman et al., 2018). Supporting such studies, Wang et al. (2013) observed higher Lys and protein contents in transgenic maize lines in comparison with wild type plants. Such transgenics exhibited not only substantial growth but also salt stress tolerance. Upregulation of saccharopine pathway genes directs Lys to degrade to α-aminoadipate semialdehyde (AAS) and glutamate, which in turn regulates homologs of animal glutamate receptors involved in plant development (Galili et al., 2001). Arruda et al. (2000) pointed out that the saccharopine pathway described above might be accountable for the biosynthesis of several regulatory molecules implicated in root growth and senescence under both biotic and abiotic stresses. Further, Rizwan et al. (2017) demonstrated the alleviation of metal stress in wheat upon foliar spray of Lys. It has been found that variability exists among different populations of A. halleri for Cd accumulation (Stein et al., 2017; Corso et al., 2018). Such an intraspecific variability has prompted Serre et al. (2020) to investigate the molecular mechanisms associated with metal tolerance, especially the Lys-methylated proteins in plants. The effect of Cd on Lys-methylated proteins and KMTs were studied in A. thaliana, A. lyrata, and A. halleri, which have differential tolerance to Cd. Gene expression, protein mass spectrometric, and immunoblotting techniques revealed significant expressions in Lys-methylated proteins and regulation of genes coding KMTs by Cd (Serre et al., 2020). Interruption of KMT gene in A. thaliana displayed a significant increase in Cd tolerance in comparison with wild type plants. Further, a knock-out mutant of the calmodulin Lys methyltransferase gene exhibited enhanced Cd tolerance. These results suggest that the regulation of nonhistone proteins by Lys methylation play pivotal roles in A. thaliana plants to Cd stress. The results are novel and unexpected, hinting us to explore more about lysine-methylated proteins and KMTs in future to generate plants for phytoremediation. But in the developing maize endosperm, saccharopine pathway is induced by exogenous supply of lysine and repressed by salt stress. In young maize coleoptiles, LKR/SDH and AASADH were induced at the transcript level by abiotic stress. Only the AASADH protein accumulates in the stressed tissues, but not the LKR/SDH. These results indicate that in the developing seeds, the saccharopine pathway is used for pipecolic acid synthesis, although proline plays a role in abiotic stress response. Thus, saccharopine pathways in maize seed development and stress response differ from dicots (Kiyota et al., 2015). Batista-Silva et al. (2019) demonstrated rapid detoxification of Lys in the leaves of A. thaliana during initial stages of stress recovery. Their results indicate that amino acid-derived secondary metabolites increase when the stress is being relieved and further showed an increase in the levels of amino acids that act as precursors for secondary plant product biosynthesis.
Multiple Pathways Co-Exist for Ser Biosynthesis in Plants
Ser participates in the biosynthetic pathways of several key biomolecules essential for synthesis of other nitrogen bases (purines, pyrimidines, and thymidine), amino acids (cysteine, glycine, and tryptophan), phospholipids, sphingolipids, and cell proliferation (Ros et al., 2014; Mattaini et al., 2016). Cysteine in turn is needed for the biosynthesis of both methionine and homocysteine. Ser biosynthesis in plants proceeds via different pathways: glycolate pathway, which is associated with photorespiration and two non-photorespiratory pathways and the phosphorylated and the glycerate pathways. Existence or coexistence of three distinct pathways in plants for Ser biosynthesis reveals the complex nature of its regulatory processes. Out of the three, glycolate pathway (Figure 3A) has been considered as the prime pathway (Tolbert, 1980; Douce et al., 2001). Photorespiration has been considered as the main source of Ser in plants since this pathway is associated with it; therefore, it has been assumed that, at least, in photosynthetic tissues the quantitative contribution of this pathway to Ser pool would be higher. For this reason, the non-photorespiratory pathways have been considered of minor importance to date. Ser is synthesized through glycolate, glycerate, and PPSB pathways, which are described below.
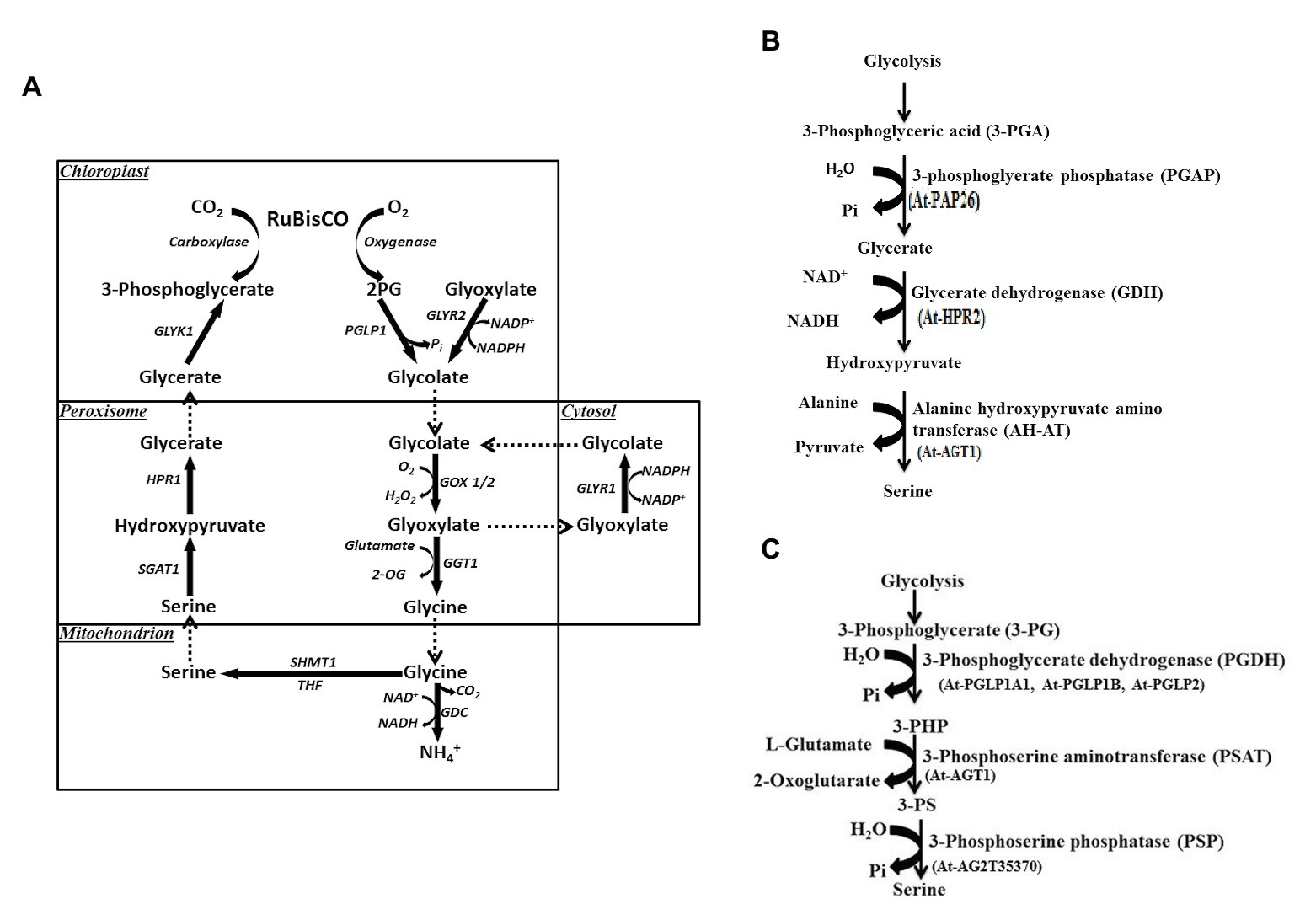
Figure 3. (A) Biosynthesis of serine via glycolate pathway during photorespiration. 2PG, 2-phosphoglycolate; PGLP1, 2PG phosphatase 1; GLYR1/2, glyoxylate reductase 1/2, glutamate glyoxylate aminotransferase 1; GDC, glycine decarboxylase complex; THF, tetrahydrofolate; SHMT1, serine hydroxymethyl transferase 1; SGAT1, serine glyoxylate aminotransferase 1; HPR, hydroxypyruvate reductase 1; GLYK1, glycerate kinase 1. (B) Serine biosynthesis via glycerate pathway. At, Arabidopsis thaliana. (C) Phosphorylated pathway of L-serine biosynthesis (PPSB). 3-PG, 3-phosphoglycerate; 3-PHP, 3-phosphohydroxypyruvate; 3-PS, 3-phosphoserine; and At, Arabidopsis thaliana.
Biosynthesis of Ser in Plants
Through the glycolate pathway, L-Ser is synthesized during plant photorespiration within the mitochondrial matrix. In this pathway, one molecule of glycine is decarboxylated and deaminated by a fascinating complex enzyme called glycine decarboxylase (GDC). Both carbon dioxide (CO2) and ammonia (NH3) are formed with the release of NADH from NAD+ (Douce et al., 2001). In a complex reaction, the methylene carbon of glycine molecule is transferred to tetrahydrofolate (THF) to form methylene tetrahydrofolate (5,10-CH2-THF). Methylene THF now combines with the second molecule of glycine to form L-Ser catalyzed by a Ser hydroxymethyltransferase (SHMT) enzyme (Figure 3A). This is a pyridoxal 5'-phosphate-dependent enzyme, which catalyzes the reversible reaction of Ser to glycine in either a tetrahydrofolate-dependent or tetrahydrofolate-independent manner. One-carbon units that result from the activity of SHMT, have been found vital in proliferating cells (Wu et al., 2017). The glycolate pathway has also biological significance like the other two. For instance, Arabidopsis SHMT mutant (shm1-1) was isolated by Somerville and Ogren (1981), which shows a strong photorespiratory phenotype that is rescued under high CO2 (Voll et al., 2006). One of the SHMT1-deficient mutants defective in GLU1 gene has been reported in A. thaliana (Jamai et al., 2009), which encodes ferredoxin-dependent glutamate synthase located in chloroplasts (Fd-GOGAT). This enzyme is associated with the photorespiratory reassimilation of NH3 and primary nitrogen assimilation. An important observation made by Jamai et al. (2009) shows that Fd-GOGAT is targeted to the mitochondria as well as chloroplasts. Interaction of Fd-GOGAT has been found essential for photorespiratory SHMT activity (Table 1). Thus, a complex regulation occurs in this high flux pathway. Both SHM1 and SHM2 are targeted to the mitochondria and operate redundantly in one carbon metabolism of non-photorespiring especially during lignification of vascular cells. Interestingly, double mutant lacking both mitochondrial SHMTs shows a lethal phenotype even under non-photorespiratory conditions, which point out the non-redundant role(s) of both isoforms and the importance of C1 metabolism for plant development (Engel et al., 2011). Five more SHM isoforms were predicted and targeted to different subcellular compartments like cytosol, plastids, and nucleus. Functional information is available for the plastidic isoform SHM3 (Zhang et al., 2010), which is believed to be involved in general one-carbon metabolism along with cytosolic isoforms SHM4 and SHM5 (Engel et al., 2011).
Ser is also synthesized in leaves through glycerate pathway. It is well-known that 3-phosphoglycerate (3-PG) is synthesized during glycolytic pathway (Figure 3B). L-Ser is formed from 3-PG by a dephosphorylation reaction (Kleczkowski and Givan, 1988) and includes part of the reverse sequence of the photorespiratory cycle. 3-PG forms glycerate and the reaction is catalyzed by 3-phosphoglycerate phosphatase (PGAP). Glycerate is then converted to hydroxypyruvate by glycerate dehydrogenase (GDH) enzyme (Figure 3B). In the last reaction, alanine-hydroxypyruvate aminotransferase (AH-AT) enzyme and glycine hydroxypyruvate aminotransferase (GH-AT) convert hydroxypyruvate to L-Ser. While one reaction takes place in the cytosol (PGAP), the remaining three (GDH, AH-AT, and GH-AT) take place in peroxisomes through enzymatic pathways (Kleczkowski and Givan, 1988; Greenler et al., 1989). To date, several genes involved in the glycerate pathway have been identified. However, no specific genes have been characterized and/or cloned to account specifically for glycerate pathway, so its functional significance remains little unclear (Ros et al., 2013; Igamberdiev and Kleczkowski, 2018).
One of the Ser biosynthetic pathways, PPSB, is conserved across bacteria, plants, and animals (Fell and Snell, 1988; Ho and Saito, 2001) and operates in plastids in plants. 3-PG generated via plastidial glycolysis and Calvin cycles acts as a precursor for the synthesis of Ser (Figure 3C). In the three sequential reactions, 3-PG is first converted to 3-phosphohydroxypyruvate (3-PHP) by the enzyme D-3-phosphoglycerate dehydrogenase 1 (PGDH1), EC 1.1.1.95; Slaughter and Davies, 1968). In this conversion, the cofactor NAD+ is reduced to NADH. 3-PHP is then converted to 3-phosphoserine (3-PS) by a transamination reaction catalyzed by 3-phosphoserine aminotransferase (PSAT, EC2.6.1.52; Hanford and Davies, 1958). Wulfert and Krueger (2018) for the first time provided the genetic evidences for the PSAT genes. PSAT1-silenced lines displayed a strong inhibition of shoot and root growth and hypersensitivity to the inhibition of the photorespiratory Ser biosynthesis at higher CO2 levels (Table 1). Such lines also showed accumulation of certain amino acids, due to increased assimilation of NH3. Knockdown of PSAT1 alters the amino acid metabolism in plants (Wulfert and Krueger, 2018). PSAT catalyzes the transfer of the amino group of glutamate to 3-hydroxypyruvate, resulting in 2-oxoglutarate and 3-phosphoserine. It is interesting to note that PSAT1 is essential for light and sugar-dependent growth promotion in A. thaliana (Wulfert and Krueger, 2018). During this biosynthetic pathway, glutamate is converted to 2-oxoglutarate, an intermediate in the TCA cycle that generates energy. 3-PS is finally catalyzed by 3-phosphoserine phosphatase (PSP, EC 3.1.3.3) to form Ser (Figure 3C). The enzymatic and genetic evidence for these enzymes were achieved by Benstein et al. (2013), Cascales-Miñana et al. (2013), and Toujani et al. (2013). While three genes for PGDHs were described, two genes encoding for PSAT noticed in the TAIR,1 cloned and observed to have activity in vitro (Ho et al., 1998, 1999a,b; Ho and Saito, 2001). Coexistence of multiple genes for PPSB and the presence of corresponding enzymes in different tissues indicate that they may perform diverse functions. This fact needs to be further investigated, but there is evidence supporting it (Ho and Saito, 2001; Waditee et al., 2007; Benstein et al., 2013). The genes associated with PPSB are expressed in several tissues (Ros et al., 2014) based on which it is argued that this pathway acts as a link between metabolism and development. It has been demonstrated that PGDH1 expression has been modulated by high CO2 levels (Benstein et al., 2013). They demonstrate that the content of Ser is reduced in PGDH1-silenced plants exposed to high CO2. They are of the opinion that expression of PGDH2 is not modulated by high CO2. Furthermore, PGDH1 and PGDH3 genes have been found regulated in A. thaliana by light-dark regimes in photosynthetic tissues (Toujani et al., 2013; Table 1). On the other hand, in non-photosynthetic tissues, PGDH family of genes are equally expressed under light-dark regimes indicating that PPSB pathway (Figure 3C) may be critical for both photosynthetic and non-photosynthetic tissues (Table 1). Unlike PGDH family of genes, where their expression pattern is tightly regulated at organ level, PSP1 gene is expressed in all organs (Cascales-Miñana et al., 2013; Toujani et al., 2013). This could be due to the presence of a unique PSP1 gene. Thus, several pieces of evidence point out that the PPSB pathway is highly relevant in photosynthetic tissues during dark conditions, when photorespiration does not operate.
Ser Metabolism is Essential for Plant Development and Abiotic Stress Tolerance
Though biological significance of PPSB has been demonstrated and found essential for the development of male gametophyte, pollen, embryo, and postembryonic root growth (Cascales-Miñana et al., 2013; Toujani et al., 2013; Ros et al., 2014, and the references therein), its metabolic implications have not been completely understood. In PPSB-deficient mutants, adenosine 5'-phosphosulfate reductase genes, sulfate transporters were upregulated (Anoman et al., 2019) with enhanced flux of 35S into thiol biosynthesis, mostly in root tissues, and also their transport and allocation. Thus, deficiency of PPSB activity perturbs sulfur homeostasis between photosynthetic and non-photosynthetic tissues (Anoman et al., 2019). Besides supplying carbon to the one-carbon pool (necessary for the biosynthesis of thymidylate and methionine), Ser is implicated in signaling mechanisms (Ser is phosphorylated by kinases; Antonov et al., 2014; Ros et al., 2014; Mattaini et al., 2016). Double mutants of gapcp1gapcp2 arrested not only primary root growth, but also showed defects in pollen development. Supplementation of L-Ser to the gapcp1gapcp2 mutant roots resulted in the recovery of root growth (Muñoz-Bertomeu et al., 2009). Further, it has been noticed that embryo and male gametophyte become lethal in mutant plants lacking Ser palmitoyltransferase, an enzyme associated with sphingolipid biosynthesis (Chen et al., 2006; Dietrich et al., 2008). Associated with the above, in psp1 and pgdh1 homozygous mutant, development of embryo in Arabidopsis is totally arrested inferring the importance of Ser in all the above cases (Benstein et al., 2013; Cascales-Miñana et al., 2013; Toujani et al., 2013). Similarly, in PSP1- and PGDH1-deficient Arabidopsis mutants, root development was inhibited, indicating poor auxin biosynthesis in such mutants. In plants, tryptophan (produced by condensation of Ser and indole) acts as a precursor for auxin biosynthesis (Tzin and Galili, 2010; Dunn, 2012), so lack of Ser in the cells leads to root developmental defects. Benstein et al. (2013) further noticed inhibition of leaf initiation when plants containing reduced levels of PGDH activity were grown under elevated levels of CO2. Ser is also implicated in folate metabolism. Mutants deficient in tetrahydrofolate metabolism show defects both in root and embryo development. It is however opined that Ser is not solely responsible for root development, and multiple factors/processes might be involved along with Ser. Interestingly, D-Ser (a derivative of L-Ser) has been found to act as a signaling molecule between male gametophyte and pistil communications (Michard et al., 2011). Thus, these observations infer that Ser metabolic pathways are critical for plant ontogeny. Alterations in PPSB gene expressions perturb not only the TCA cycle but also amino acid biosynthesis (Cascales-Miñana et al., 2013; Toujani et al., 2013).
SHMT1 functions in the photorespiratory pathway and play a critical role in controlling cell damage provoked by high light (photooxidative stress) and salt stresses and in restricting pathogen-induced cell death (Moreno et al., 2005). Further, SHMT1, located in mitochondria, converts glycine to Ser, and Liu et al. (2019) reported complementation of A. thaliana shm1-1 (photorespiratory growth phenotype) by SHMT1 wild type plants. They found that reduced SHMT activity led to a decreased stomatal closure in response to ABA and salt stress. Transgenic lines, which showed reduced SHMT activity, were not only more sensitive to salt stress but also to drought stress recovery. In line with this, Batool et al. (2018) propose that cysteine is the limiting factor for ABA biosynthesis in the early stages of drought conditions in guard cells and potentially in other cell types of the leaf. Thus, its explicit glycolate pathway has a role to play in plant abiotic stress. Glycerate pathway has been linked to γ-aminobutyric acid (GABA) shunt, which affects plant growth as well as development throughout life cycle and accumulates rapidly and contributes in response to biotic and abiotic stresses (Ramos-Ruiz et al., 2019). Ser formed in glycerate and PPSB pathways acts as a precursor of glycine, formate, and glycolate, which accumulate under stress conditions (Igamberdiev and Kleczkowski, 2018). A hypothesis has been presented by Igamberdiev and Kleczkowski (2018) on the regulation of redox balance in stressed plant cells via participation of the reactions connected with glycerate and PPSB pathways. Thus, a link between carbon and nitrogen metabolism exists, which is regarded as vital in maintaining cellular redox and energy levels under stress conditions.
Accumulation of Ser was noticed in plants exposed to low and high temperatures, flooding, and high salt stress levels (Stewart and Larher, 1980; Kaplan et al., 2004; Bocian et al., 2015; Li et al., 2017) as well as combined stresses of drought and heat (Hossain et al., 2017). Overexpression of PGDH gene isolated from Aphanothece halophytica (a cyanobacterial species) lead to elevated salt and cold stress tolerance in A. thaliana (Waditee et al., 2007), indicating the involvement of PGDH enzyme in abiotic stresses. In line with this, studies conducted by Rosa-Téllez et al. (2020) have demonstrated that the response observed to salt stress depends on the isoform studied. Thus, while PGDH1 activity could be relevant for plant tolerance to salinity, the function of PGDH3 seems detrimental under such environmental conditions. Lines overexpressing PGDH1 (OexPGDH1) accumulate less proline and raffinose (stress markers) in roots under salt stress than lines overexpressing PGDH3 (OexPGDH3). Moreover, the levels of oxidized glutathione (GSSG; derived from Cys) increased under salt stress in OexPGDH3 as compared to both OexPGDH1 and wild type plants. Ser is utilized in many biosynthetic pathways and thereby contributes to nucleotide synthesis, methylation reactions, and production of the reducing power. Both glutathione and NADPH help in antioxidant defense in plants (Yang and Vousden, 2016). Glutathione acts as a precursor of phytochelatins and helps in chelating toxic metals, and needed for the detoxification of methylglyoxal, a cytotoxic and an emerging signaling molecule in plant abiotic stress responses and tolerance. Further, glutathione has been found to impact translation and subsequent changes. Interestingly, PPSB could affect the ABA signal transduction and thus trigger the downstream genes under cold and salt stress conditions. In support of this statement, double mutants like gapcp1gapcp2, which have an impaired phosphorylation pathway are insensitive to ABA (Muñoz-Bertomeu et al., 2011a,b), suggesting that Ser pathway somehow triggers ABA signals for environmental stress tolerance. Ser acts as a precursor for the synthesis of glycine betaine, an important osmotic agent, and a scavenger of ROS. Some species like rice and Arabidopsis do not produce glycine betaine, but in such a case, glutathione (a derivative of cysteine) scavenges ROS generated during abiotic stress (Rosa-Téllez et al., 2020). The relevance and importance of catabolism of amino acids as an alternative respiratory substrate has been demonstrated during drought or short light periods (Araujo et al., 2010; Engqvist et al., 2011; Krüßel et al., 2014). It appears therefore that a tight relationship exists between Lys and Ser metabolism during abiotic stress recovery.
DHNs Contain Lysine-Rich Residues Involved in Abiotic Stress Tolerance
Late embryogenesis abundant proteins (LEAs) are hydrophilic and thermostable in nature. This property helps them to interact not only with biomolecules, but also metal ions (Liu et al., 2013; Cuevas-Velazquez et al., 2017). DHNs are group II LEA proteins and are widely distributed in bryophytes, gymnosperms, and angiosperms (Yu et al., 2018), accumulate during embryogenesis and induced in vegetative tissues following exposure to diverse environmental stresses (Battaglia et al., 2008). They are low molecular weight (ranging 9–200 kD), modular, intrinsically disordered proteins (IDPs), and lack well-defined three-dimensional structures (Riley et al., 2019). But SbDHN5 (from Sorghum bicolor) has been discovered as an ordered protein with phosphorylation sites (Nagaraju et al., 2018). Hence, it would be of interest to find out the overexpression of SbDHN5 in crop plants and their resistance to abiotic stresses. Proximate in vitro evidences exist that DHNs protect plants in myriad ways, such as buffering of ion sequestration (Alsheikh et al., 2003), hydrate water (Bokor et al., 2005), chaperone activity (Abedini et al., 2017), membrane binding and stabilization (Xing et al., 2011), enzyme cryoprotection (Hughes and Graether, 2011; Drira et al., 2013), and in scavenging ROS (Hara et al., 2013). DHNs contain Lys-rich K-segment, which is prevalent in all DHNs with EKKGIMDKIKEKLPG (15 amino acids) and located near the C-terminus (Close, 1996; Malik et al., 2017). K-segments participate in forming class A2 amphipathic α-helix that protect both enzymes and membranes (Baker et al., 1988). More than one K-segment is also noticed in few DHN proteins, but with several conserved residues (Koag et al., 2009; Graether and Boddington, 2014). DHNs have four different types of conserved sequence motifs namely K-, S-, Y-, ([V/T]D] [E/Q]YGNP), and ф segments. Quite intriguing is that no position in the K-segment is conserved totally (Graether and Boddington, 2014). Without any exception, all DHNs contain a minimum of one copy of Lys-rich K-segment located near the C-terminal. In some DHNs, more than one K-segment, but with distinct amino acid sequences may be present. The conserved residues among the K-segment include Lys-Ile-Lys-Glu in the core, Lys-Leu-Pro-Gly in the C-terminal, and Glu-Lys-Lys in the N-terminal regions (Graether and Boddington, 2014). However, the amino acid residues in K-segment among lower and higher plants may differ. In gymnosperms, K-segment shares a variable sequence like (Q/E)K(P/A)G(M/L)LDKIK(A/Q)(K/M)(I/L)PG, while in higher plants, it has EKKGIMDKIKEKLPG (Close, 1996; Jarvis et al., 1996). While K-segment is associated with plant development besides stress tolerance, the conserved sequence of Y is homologous to that of chaperone molecules (Martin et al., 1993). On the other hand, S-segment contains Ser cascade sequence SSSSSSSD, and this segment is observed mostly as a single copy in DHNs. S-segment is a phosphorylatable patch of 4–10 Ser residues and can transfer DHNs from cytoplasm to the nucleus (Goday et al., 1994). Stretches of sequence with a variable length are usually noticed and S-segment also participates in plant development alongside abiotic stress tolerance. It is also of interest to note that in YnSKn-type DHNs, the K- and S-segments are linked by a fixed motif GXGGRRKK (where X can be any amino acid), indicating a functional linkage between K- and S-segments. While the motif GXGG is highly pliable and interacts with negatively charged phosphoserines with K-segment, motif RRKK appears to be a nuclear localization signal (Jensen et al., 1998; Malik et al., 2017). In between the conserved motifs, there are ф segments with small, polar, and charged amino acids. Ф-segments are stretches of sequences with variable lengths of amino acids. Based on the sequence and the number of K, S, and Y segments, DHNs have been further classified in five subfamilies/subgroups such as KnS, YnSKn, Kn, YnKn, and SKn (Mundy and Chua, 1988). While YnSKn-type DHNs are expressed mostly under desiccation and salt stresses, Kn, SKn, and KnS are upregulated by cold, desiccation, and salt stresses (Graether and Boddington, 2014). The presence of S (Ser) motif preceding the K (Lys) motif (YnSKn, SKn, and SnKS) and also frequent occurrence of one of such subclasses YnSKn in monocots has been noticed by Abedini et al. (2017). Interestingly, out of 13 HvDHNs, 10 YnSKn members were noticed in a drought-tolerant barley (Kosová et al., 2011), and three in Sorghum bicolor out of six detected (Nagaraju et al., 2018). DHNs have been observed in vegetative tissues grown under control conditions, inferring that they play a key role in plant growth. The Y2K4-type DHN MtCAS31 (from Medicago truncatula) by interacting with AtICE1 (induces CBF expression 1) has been shown to associate with stomatal development, increasing the drought tolerance by decreasing the stomatal density of transgenic A. thaliana (Xie et al., 2012). DHN5 when overexpressed in A. thaliana showed different responses to biotic (as an antibacterial and antifungal factor) and abiotic stresses (Drira et al., 2015, 2016) besides protecting lactate dehydrogenase, β-glucosidase, and glucose oxidase from cold and heat stresses (Brini et al., 2010; Drira et al., 2013).
The DHNs are localized mostly in the cytoplasm, nucleus, plasma membrane, and mitochondria (Hara et al., 2013). It is interesting to note that many S-segment-containing DHNs are localized to the nucleus inferring that the S-segment moves to nucleus besides Kn and YnKn types (Wisniewski et al., 1999; Lin et al., 2012). Graether and Boddington (2014) discovered that several of Kn, SKn, KnS, YnSKn, and YnKn DHNs were upregulated during low temperature (cold), desiccation, and salt stresses. The His-flanking K-segments (the major functional component) have been found to bind to membranes and play a major role during stress response (Eriksson et al., 2011). Further, a correlation exists between the number of K-segments and abiotic stress tolerance. In line with this statement, K-segment of wheat DHN WZY2 has been found to protect plants from temperature stress (Yang et al., 2015). In this regard, it is interesting to note that the derivative containing two K-segments (WZY2) displays remarkable cold and heat stress tolerance than the truncated derivative without K-segments. This implies that K-segment is the major functional component of WZY2 (Yang et al., 2015). Interestingly, among the spliced DHN1a_s (YSK2) and unspliced DHN1a_u (YS), only the spliced variant exhibited resistance to cold and drought stresses and to Botrytis cinerea (Rosales et al., 2014).
Ser-Rich Proteins are Implicated in Abiotic Stress as Well as in Plant Development
Lazar et al. (1995) first identified serine/arginine-rich (SR) proteins using monoclonal antibodies raised against a Ser phospho epitope in the arginine/serine-rich (RS) domain. SR proteins now appear as a highly conserved family of RNA-binding protein members in eukaryotes and regarded as crucial alternative splicing (AS) regulators of pre-mRNAs (Palusa et al., 2010), thereby increasing the transcript complexity. SR proteins range in size from 21 to 41 kDa in A. thaliana, and have two RNA recognition motifs at the N- and C-terminal RS domains, rich in SR dipeptides. AS has been recognized as a means of plant adaptation to a changing environment and is controlled in a tissue- and development-specific manner especially under abiotic stress conditions. These proteins play crucial roles in maintaining genome stability (Xiao et al., 2007), promoting transcriptional elongation (Lin et al., 2008), and cell cycle progression (Zhong et al., 2009). Many environmental stresses modulate AS patterns of SR proteins, phosphorylation status, and subcellular distribution in plants. In the promoter regions, Chen et al. (2019) found 92 development-, stress-, and hormone-related cis-elements. This implies that SR proteins play an important role during plant development and in response to environmental stresses. SR proteins were predicted to interact with other SR and non-SR proteins, inferring their association in other functions. Developmental defects were noticed with the overexpression of Arabidopsis SRp30, RSZ33, or mutations in SC35 and SCL genes (Lopato et al., 1999; Maria et al., 2003; Yan et al., 2017), implying the involvement of SR proteins in plant development. The number of SR proteins may vary in different taxa (16 in Physcomitrella patens, 18 in Brachypodium, 22 in rice, 25 in Brassica rapa, and 40 in wheat; Melo et al., 2020), which are divided into many subfamilies. Expression levels of the SCL30a, SCL28, and SCL33 genes were altered upon treatment with ABA (Cruz et al., 2014), indicating that SR proteins respond to ABA. Zhang et al. (2014) noticed that splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Besides its response to high light intensities (Tanabe et al., 2008), Arabidopsis SR45 participates as a suppressor to innate immunity (Zhang et al., 2017). Likewise, SR protein RSZ21 obtained from Arachis has been found to play a role in plant defense and HR-like cell death (Kumar and Kirti, 2012). Yoon et al. (2018) discovered altered expression levels of 78.6% genes (22 out of 28) and 60.7% of AS patterns in Brassica rapa in response to abiotic stresses. The highest expressions were detected when plants were exposed to oxidative, cold, and heat treatments. Interestingly, cold and heat stresses caused the most AS events. These studies point out that type of abiotic stress largely influences the expression patterns of SR proteins and also the AS patterns. Using CRISPR/Cas9-mediated plant genome engineering, Butt et al. (2019) targeted each rice SR locus and produced single knockouts. Such a study is extremely vital and forms a useful resource material to understand the role of SR proteins in plant development as well as abiotic stresses.
Do ncRNAs Have a Role in the Regulation of Lys and Ser Pathways and its Interactions?
Major part of the genome is non-coding and is transcribed into non-coding RNAs (ncRNAs) only, which is known to play a regulatory role. Among these ncRNAs, long non-coding RNAs (lncRNAs) are known to be involved in regulation of gene expression. However, very little is known about the role of ncRNAs in providing tolerance against biotic stress including plant diseases. The interactions of lncRNAs with proteins in plants using a system genomic approach is promising and is relevant to ascertain characteristic trait biology relationships. In the recent past, several databases for genes encoding lncRNAs in plants have become available, which include greeNC (Gallart et al., 2016), CantataDB (Szczesniak et al., 2016), PLncDB (Jin et al., 2013) etc. However, these databases lack bona fide entries and have poor annotations. In plants, lncRNAs are known to perform multiple biological functions, which include the following: phosphate homeostasis (Bazin and Bailey-Serres, 2015), flowering (Swiezewski et al., 2009), photomorphogenesis (Wang et al., 2014), stress response (Yuan et al., 2018), fertility (Liu et al., 2017), etc. The lncRNAs are also known to function as target mimics of miRNAs (Aung et al., 2006; Pant et al., 2008) and regulate post-translational processes via protein modifications and protein-protein interactions (PPIs; Liu et al., 2015), etc. Song and Zhang (2017) have extensively reviewed the characteristics, identification, and functions of lncRNAs in response to various stresses. The authors delve into a greater understanding and need of nutrient deficiency and the factors associated with abiotic stress. We also argue that the identification of regulatory elements associated with a disease and validating them through NGS has been a routine task. In this process, distinct signatures in the form of lncRNAs in plants would be ideal candidates for studying agronomically important traits. It is a challenge to identify ncRNAs that were not characterized earlier and to find if they are specific to any trait/genotype. For example, the miRNA target analysis also divulged that DHNs are targeted by 51 miRNAs responsive to abiotic stress. The gene expressions are common and well-coordinated under diverse abiotic stress conditions and DHNs are no exception. The role of DHNs under different abiotic stress conditions, the regulatory networks of DHN genes, and their physiological functions have been discussed (Yu et al., 2018). In S. bicolor, transcript expressions were higher in roots, stems, and leaves in comparison with inflorescences (Nagaraju et al., 2018). While all DHN genes exhibited high expressions in stems under cold, heat, salt, and drought stresses, SbDHN2 displayed the highest expression under multiple stresses in all the tissues of S. bicolor (Nagaraju et al., 2018). These results infer that the involvement of SbDHN2 of YnS group in a wide array of stresses. In the recent past, studies on proteome diversity revealed that lncRNAs play a very important role in serine/arginine (SR) regulations (Fesenko et al., 2017). Furthermore, the role of lncRNAs in nonsense-mediated mRNA decay was identified under differential alternative splicing in relation to altitude, not the abiotic stress, which is beyond the scope of this review. Furthermore, Ser and Lys metabolism together are not explicitly known.
Conclusions and Outlook
Both Lys and Ser are key amino acids, involved in plant ontogeny and also connected with abiotic stress tolerance. The biosynthetic pathways are multiple and complex. Among the pathways that exist, the PPSB appears to play a critical role during growth and development. Diverse mechanisms associated with biosynthesis and catabolism of these two important amino acids however, need to be elucidated and also their interaction with other pathways. Attempts were made to enrich the seeds of rice and others with Lys, but still problems exist with regard to seed morphology. These shortcomings need to be overcome in future in order to make high Lys-containing lines available to the public. Further, it is concluded that Lys is catabolized to serve as an alternative respiratory substrate since photosynthetic activity is impaired under abiotic stress conditions. Thus, plants get detoxified by supplementing the much-needed energy during the time of abiotic stress recovery. This vital function of Lys is in addition to its regular participation in protein synthesis and other metabolic activities. Such functions to the amino acids are novel and were not predicted earlier. Many functions of Lys and Ser during growth and development have come into light recently, though the mechanistic explanations are not clear. Further, Lys- and Ser-rich proteins and their induction during abiotic stress reveals the crucial role they play in a wide spectrum of abiotic stresses. Identification of the cis- and trans-acting elements associated with the activation/suppression of the biosynthetic pathway genes and subsequent events that take place in the downstream remains elucidated. Finally, lncRNAs appear to play a role in these processes by interacting with serine kinase and several novel proteins. Mapping those miRNAs and lncRNAs, their interaction and the key roles they play during the regulation of these processes is certainly the biggest challenge for future research.
Author Contributions
PBK wrote the first draft with sections on Serine and split by all other authors. RS developed the figures. PS wrote sections on lncRNAs. PBK, PS, and RP proofread the manuscript before all the agreeing to submission. All authors contributed to the revisions and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
References
Abedini, R., Golmohammadi, F. G., PishkamRad, R., Pourabedi, E., Jafarnezhadi, A., Shobbar, Z. S., et al. (2017). Plant dehydrins: shedding light on structure and expression patterns of dehydrin gene family in barley. J. Plant Res. 130, 747–763. doi: 10.1007/s10265-017-0941-5
Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Ikeda, S., et al. (2012). KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53:e1. doi: 10.1093/pcp/pcr165
Alsheikh, M. K., Heyen, B. J., and Randall, S. K. (2003). Ion binding properties of the dehydrin ERD14 are dependent upon phosphorylation. J. Biol. Chem. 278, 40882–40889. doi: 10.1074/jbc.M307151200
Amir, R., and Tabe, L. (2006). “Molecular approaches to improving plant methionine content” in Plant genetic engineering. Metabolic engineering and molecular farming II. Vol. 8. eds. P. K. Jaiwal and R. P. Singh (Huston, Texas, USA: Studium Press LLC), 1–26.
Anoman, A. D., Flores-Tornero, M., Benstein, R. M., Blau, S., Rosa-Téllez, S., Bräutigam, A., et al. (2019). Deficiency in the phosphorylated pathway of serine biosynthesis perturbs sulfur assimilation. Plant Physiol. 180, 153–170. doi: 10.1104/pp.18.01549
Antonov, A., Agostini, M., Morello, M., Minieri, M., Melino, G., and Amelio, I. (2014). Bioinformatics analysis of the serine and glycine pathway in cancer cells. Oncotarget 5, 11004–11013. doi: 10.18632/oncotarget.2668
Araujo, W. L., Ishizaki, K., Nunes-Nesi, A., Larson, T. R., Tohge, T., Krahnert, I., et al. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22, 1549–1563. doi: 10.1105/tpc110075630
Arruda, P., Kemper, E. L., Papes, F., and Leite, A. (2000). Regulation of lysine catabolism in higher plants. Trends Plant Sci. 5, 324–330. doi: 10.1016/S1360-1385(00)01688-5
Aung, K., Lin, S., Wu, C. C., Huang, Y. T., Su, C. L., and Chiou, T. J. (2006). pho2, a phosphate over accumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141, 1000–1011. doi: 10.1104/pp.106.078063
Baker, J., Van Dennsteele, C., and Dure, L. (1988). Sequence and characterization of 6 lea proteins and their genes from cotton. Plant Mol. Biol. 11, 277–291. doi: 10.1007/BF00027385
Batista-Silva, W., Rugen, B., Nunes-Nesi, A., Arruda, W. L., Braun, H., and Hildebrandt, T. M. (2019). The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 42, 1630–1644. doi: 10.1111/pce.13518
Batool, S., Uslu, V. V., Rajab, H., Ahmad, N., Waadt, R., Geiger, D., et al. (2018). Sulfate is incorporated into cysteine to trigger ABA production and stomatal closure. Plant Cell 30, 2973–2987. doi: 10.1105/tpc.18.00612
Battaglia, M., Olvera-Carrillo, Y., Garciarrubio, A., Campos, F., and Covarrubias, A. A. (2008). The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. doi: 10.1104/pp108120725
Bazin, J., and Bailey-Serres, J. (2015). Emerging roles of long non-coding RNA in root developmental plasticity and regulation of phosphate homeostasis. Front. Plant Sci. 6:400. doi: 10.3389/fpls.2015.00400
Benstein, R. M., Ludewig, K., Wulfert, S., Wittek, S., Gigolashvili, T., Frerigmann, H., et al. (2013). Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25, 5011–5029. doi: 10.1105/tpc.113.118992
Bocian, A., Zwierzykowski, Z., Rapacz, M., Koczyk, G., Ciesiolka, D., and Kosmala, A. (2015). Metabolite profiling during cold acclimation of Lolium perenne genotypes distinct in the level of frost tolerance. J. Appl. Genet. 56, 439–449. doi: 10.1007/s13353-015-0293-6
Bokor, M., Csizmók, V., Kovács, D., Bánki, P., Friedrich, P., Tompa, P., et al. (2005). NMR relaxation studies on the hydrate layer of intrinsically unstructured proteins. Biophys. J. 88, 2030–2037. doi: 10.1529/biophysj.104.051912
Bright, S. W. J., and Shewry, P. R. (1983). Improvement of protein quality in cereals. CRC Crit. Rev. Plant Sci. 1, 49–93. doi: 10.1080/07352688309382171
Brini, F., Saibi, W., Amara, I., Gargouri, A., Masmoudi, K., and Hanin, M. (2010). Wheat dehydrin DHN-5 exerts a heat-protective effect on β-glucosidase and glucose oxidase activities. Biosci. Biotechnol. Biochem. 74, 1050–1054. doi: 10.1271/bbb.90949
Brocker, C., Lassen, N., Estey, T., Pappa, A., Cantore, M., Orlova, V. V., et al. (2010). Aldehyde dehydrogenase 7A1 (ALDH7A1) is a novel enzyme involved in cellular defense against hyperosmotic stress. J. Biol. Chem. 285, 18452–18463. doi: 10.1074/jbc.M109.077925
Butt, H., Piatek, A., Li, L., Reddy, A. S. N., and Mahfouz, M. M. (2019). Multiplex CRISPR mutagenesis of the serine/arginine-rich (SR) gene family in rice. Genes 10:596. doi: 10.3390/genes10080596
Cascales-Miñana, B., Muñoz-Bertomeu, J., Flores-Tornero, M., Anoman, A. D., Pertusa, J., Alaiz, M., et al. (2013). The phosphorylated pathway of serine biosynthesis is essential both for male gametophyte and embryo development and for root growth in Arabidopsis. Plant Cell 25, 2084–2101. doi: 10.1105/tpc.113.112359
Chaneton, B., Hillmann, P., Zheng, L., Martin, A. C., Maddocks, O. D., Chokkathukalam, A., et al. (2012). Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462. doi: 10.1038/nature11540
Chen, M., Han, G., Dietrich, C. R., Dunn, T. M., and Cahoon, E. B. (2006). The essential nature of sphingolipids in plants as revealed by the functional identification and characterization of the Arabidopsis LCB1 subunit of serine palmitoyltransferase. Plant Cell 18, 3576–3593. doi: 10.1105/tpc.105.040774
Chen, S., Li, J., Liu, Y., and Li, H. (2019). Genome-wide analysis of serine/arginine-rich protein family in wheat and Brachypodium distachyon. Plan. Theory 8:188. doi: 10.3390/plants8070188
Close, T. J. (1996). Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol. Plant. 97, 795–803. doi: 10.1111/j.1399-3054.1996.tb00546.x
Close, T. J. (1997). Dehydrins: a commonalty in the response of plants to dehydration and low temperature. Physiol. Plant. 100, 291–296. doi: 10.1111/j1399-30541997tb04785x
Corso, M., Schvartzman, M. S., Guzzo, F., Souard, F., Malkowski, E., Hanikenne, M., et al. (2018). Contrasting cadmium resistance strategies in two metallicolous populations of Arabidopsis halleri. New Phytol. 218, 283–297. doi: 10.1111/nph.14948
Cruz, T. M. D., Carvalho, R. F., Richardson, D. N., and Duque, P. (2014). Abscisic acid (ABA) regulation of Arabidopsis SR protein gene expression. Int. J. Mol. Sci. 15, 17541–17564. doi: 10.3390/ijms151017541
Cuevas-Velazquez, C. L., Reyes, J. L., and Covarrubias, A. A. (2017). Group 4 late embryogenesis abundant proteins as a model to study intrinsically disordered proteins in plants. Plant Signal. Behav. 12, 10893–10903. doi: 10.1080/15592324.2017.1343777
Dellero, Y., Jossier, M., Glab, N., Oury, C., Tcherkez, G., and Hodges, M. (2016). An Arabidopsis thaliana photorespiratory glycolate oxidase mutant exhibits altered carbon allocation and leaf senescence after a transfer from high CO2 to air. J. Exp. Bot. 67, 3149–3163. doi: 10.1093/jxb/erw054
Dietrich, C. R., Han, G., Chen, M., Berg, R. H., Dunn, T. M., and Cahoon, E. B. (2008). Loss-of-function mutations and inducible RNAi suppression of Arabidopsis LCB2 genes reveal the critical role of sphingolipids in gametophytic and sporophytic cell viability. Plant J. 54, 284–298. doi: 10.1111/j.1365-313X.2008.03420.x
Douce, R., Bourguignon, J., Neuburger, M., and Rébeillé, F. (2001). The glycine decarboxylase system: a fascinating complex. Trends Plant Sci. 6, 167–176. doi: 10.1016/S1360-1385(01)01892-1
Drira, M., Hanin, M., Masmoudi, K., and Brini, F. (2016). Comparison of full-length and conserved segments of wheat dehydrin DHN-5 overexpressed in Arabidopsis thaliana showed different responses to abiotic and biotic stress. Funct. Plant Biol. 43, 1048–1060. doi: 10.1071/FP16134
Drira, M., Saibi, W., Amara, I., Masmoudi, K., Hanin, M., and Brini, F. (2015). Wheat dehydrin K-segments ensure bacterial stress tolerance, antiaggregation and antimicrobial effects. Appl. Biochem. Biotechnol. 175, 1–12. doi: 10.1007/s12010-015-1502-9
Drira, M., Saibi, W., Brini, F., Gargouri, A., Masmoudi, K., and Hanin, M. (2013). The K-segments of the wheat dehydrin DHN-5 are essential for the protection of lactate dehydrogenase and β-glucosidase activities in vitro. Mol. Biotechnol. 54, 643–650. doi: 10.1007/s12033-012-9606-8
Dunn, M. F. (2012). Allosteric regulation of substrate channeling and catalysis in the tryptophan synthase bienzyme complex. Arch. Biochem. Biophys. 519, 154–166. doi: 10.1016/j.abb.2012.01.016
Engel, N., Ewald, R., Gupta, K. J., Zrenner, R., Hagemann, M., and Bauwe, H. (2011). The presequence of Arabidopsis serine hydroxymethyltransferase SHM2 selectively prevents import into mesophyll mitochondria. Plant Physiol. 157, 1711–1720. doi: 10.1104/pp.111.184564
Engqvist, M. K., Kuhn, A., Wienstroer, J., Weber, K., Jansen, E. E. W., Jakobs, C., et al. (2011). Plant D-2-hydroxyglutarate dehydrogenase participates in the catabolism of lysine especially during senescence. J. Biol. Chem. 286, 11382–11390. doi: 10.1074/jbc.M110.194175
Eriksson, S. K., Kutzer, M., Procek, J., Grobner, G., and Harryson, P. (2011). Tunable membrane binding of the intrinsically disordered dehydrin Lti30, a cold-induced plant stress protein. Plant Cell 23, 2391–2404. doi: 10.1105/tpc.111.085183
Falco, S. C., Guida, T., Locke, M., Mauvais, J., Sanders, C., Ward, R. T., et al. (1995). Transgenic canola and soybean seeds with increased lysine. Biotechnology 13, 577–582. doi: 10.1038/nbt0695-577
Fell, D. A., and Snell, K. (1988). Control analysis of mammalian serine biosynthesis. Feedback inhibition on the final step. Biochem. J. 256, 97–101. doi: 10.1042/bj2560097
Fesenko, I., Khazigaleeva, R., Kirov, I., Kniazev, A., Glushenko, O., Babalyan, K., et al. (2017). Alternative splicing shapes transcriptome but not proteome diversity in Physcomitrella patens. Sci. Rep. 7:2698. doi: 10.1038/s41598-017-02970-z
Galili, G. (1995). Regulation of lysine and threonine synthesis. Plant Cell 7, 899–906. doi: 10.2307/3870045
Galili, G. (2002). New insights into the regulation and functional significance of lysine metabolism in plants. Annu. Rev. Plant Biol. 53, 27–43. doi: 10.1146/annurevarplant53091401110929
Galili, G., and Amir, R. (2013). Fortifying plants with the essential amino acids lysine and methionine to improve nutritional quality. Plant Biotechnol. J. 11, 211–222. doi: 10.1111/pbi.12025
Galili, G., Tang, G., Zhu, X., and Gakiere, B. (2001). Lysine catabolism: a stress and development super-regulated metabolic pathway. Curr. Opin. Plant Biol. 4, 261–266. doi: 10.1016/S1369-5266(00)00170-9
Gallart, P. A., Pulido, H., de Lagran, I. A. M., Sanseverino, W., and Cigliano, A. (2016). GREENC: a Wiki-based database of plant lncRNAs. Nucleic Acids Res. 44, D1161–D1166. doi: 10.1093/nar/gkv1215
Ghislain, M., Frankard, V., and Jacobs, M. (1995). A dinucleotide mutation in dihydrodipicolinate synthase of Nicotiana sylvestris leads to lysine overproduction. Plant J. 8, 733–743. doi: 10.1046/j.1365-313X.1995.08050733.x
Gibbon, B. C., Wang, X., and Larkins, B. A. (2003). Altered starch structure is associated with endosperm modification in quality protein maize. Proc. Natl. Acad. Sci. U. S. A. 100, 15329–15334. doi: 10.1073/pnas.2136854100
Goday, A., Jensen, B., Culiañez-Macià, F., Mar Albà, M., Figueras, M., Serratosa, J., et al. (1994). The maize abscisic acid-responsive protein Rab17 is located in the nucleus and interacts with nuclear localization signals. Plant Cell 6, 351–360. doi: 10.1105/tpc.6.3.351
Graether, S. P., and Boddington, K. F. (2014). Disorder and function: a review of the dehydrin protein family. Front. Plant Sci. 5:576. doi: 10.3389/fpls.2014.00576
Greenler, J. M., Sloan, J. S., Schwartz, B. W., and Becker, W. M. (1989). Isolation, characterization and sequence analysis of a full-length cDNA clone encoding NADH-dependent hydroxypyruvate reductase from cucumber. Plant Mol. Biol. 13, 139–150. doi: 10.1007/BF00016133
Hanford, J., and Davies, D. D. (1958). Formation of phosphoserine from 3-phosphoglycerate in higher plants. Nature 182, 532–533. doi: 10.1038/182532a0
Hara, M., Kondo, M., and Kato, T. A. (2013). KS-type dehydrin and its related domains reduce cu-promoted radical generation and the histidine residues contribute to the radical-reducing activities. J. Exp. Bot. 64, 1615–1624. doi: 10.1093/jxb/ert016
Hirakawa, Y., Torii, K. U., and Uchida, N. (2017). Mechanisms and strategies shaping plant peptide hormones. Plant Cell Physiol. 58, 1313–1318. doi: 10.1093/pcp/pcx069
Ho, C. L., Noji, M., and Saito, K. (1999a). Plastidic pathway of serine biosynthesis molecular cloning and expression of 3-phosphoserine phosphatase from Arabidopsis thaliana. J. Biol. Chem. 274, 11007–11012. doi: 10.1074/jbc.274.16.11007
Ho, C. L., Noji, M., Saito, M., and Saito, K. (1999b). Regulation of serine biosynthesis in Arabidopsis crucial role of plastidic 3-phosphoglycerate dehydrogenase in non-photosynthetic tissues. J. Biol. Chem. 274, 397–402. doi: 10.1074/jbc.274.1.397
Ho, C. L., Noji, M., Saito, M., Yamazaki, M., and Saito, K. (1998). Molecular characterization of plastidic phosphoserine aminotransferase in serine biosynthesis from Arabidopsis. Plant J. 16, 443–452. doi: 10.1046/j.1365-313x.1998.00313.x
Ho, C. L., and Saito, K. (2001). Molecular biology of the plastidic phosphorylated serine biosynthetic pathway in Arabidopsis thaliana. Amino Acids 20, 243–259. doi: 10.1007/s007260170042
Hossain, M. S., Persicke, M., ElSayed, A. I., Kalinowski, J., and Dietz, K. J. (2017). Metabolite profiling at the cellular and subcellular level reveals metabolites associated with salinity tolerance in sugar beet. J. Exp. Bot. 68, 5961–5976. doi: 10.1093/jxb/erx388
Houmard, N. M., Mainville, J. L., Bonin, C. P., Huang, S., Luethy, M. H., and Malvar, T. M. (2007). High-lysine corn generated by endosperm-specific suppression of lysine catabolism using RNAi. Plant Biotechnol. J. 5, 605–614. doi: 10.1111/j.1467-7652.2007.00265.x
Hudson, A. O., Bless, C., Macedo, P., Chatterjee, S. P., Singh, B. K., Gilvarg, C., et al. (2005). Biosynthesis of lysine in plants: evidence for a variant of the known bacterial pathways. Biochim. Biophys. Acta 1721, 27–36. doi: 10.1016/j.bbagen.2004.09.008
Hudson, A. O., Singh, B. K., Leustek, T., and Gilvarg, C. (2006). An LL-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol. 140, 292–301. doi: 10.1104/pp.105.072629
Hughes, S., and Graether, S. P. (2011). Cryoprotective mechanism of a small intrinsically disordered dehydrin protein. Protein Sci. 20, 42–50. doi: 10.1002/pro.534
Igamberdiev, A. U., and Kleczkowski, L. A. (2018). The glycerate and phosphorylated pathways of serine synthesis in plants: the branches of plant glycolysis linking carbon and nitrogen metabolism. Front. Plant Sci. 9:318. doi: 10.3389/fpls.2018.00318
Jamai, A., Salome, P. A., Schilling, S. H., Weber, A. P. M., and McClung, C. R. (2009). Arabidopsis photorespiratory serine hydroxymethyltransferase activity requires the mitochondrial accumulation of ferredoxin dependent glutamate synthase. Plant Cell 21, 595–606. doi: 10.1105/tpc.108.063289
Jarvis, S. B., Taylor, M. A., MacLeod, M. R., and Davies, H. V. (1996). Cloning and characterisation of the cDNA clones of three genes that are differentially expressed during dormancy-breakage in the seeds of douglas fir (Pseudotsuga menziesii). J. Plant Physiol. 147, 559–566. doi: 10.1016/S0176-1617(96)80046-0
Jensen, A. B., Goday, A., Figueras, M., Jessop, A. C., and Pages, M. (1998). Phosphorylation mediates the nuclear targeting of the maize Rab17 protein. Plant J. 13, 691–697. doi: 10.1046/j.1365-313X.1998.00069.x
Jin, J., Liu, J., Wang, H., Wong, L., and Chua, N. M. (2013). PLncDB: plant long non-coding RNA database. Bioinformatics 29, 1068–1071. doi: 10.1093/bioinformatics/btt107
Joshi, V., Joung, J. G., Fei, Z., and Jander, G. (2010). Interdependence of threonine, methionine and isoleucine metabolism in plants: accumulation and transcriptional regulation under abiotic stress. Amino Acids 39, 933–947. doi: 10.1007/s00726-010-0505-7
Kalhan, S. C., and Hanson, R. W. (2012). Resurgence of serine: an often neglected but indispensable amino acid. J. Biol. Chem. 287, 19786–19791. doi: 10.1074/jbc.R112.357194
Kanjanaphachoat, P., Wei, B. Y., Lo, S. F., Wang, I. W., Wang, C. S., Yu, S. M., et al. (2012). Serotonin accumulation in transgenic rice by over-expressing tryptophan decarboxlyase results in a dark brown phenotype and stunted growth. Plant Mol. Biol. 78, 525–543. doi: 10.1007/s11103-012-9882-5
Kaplan, F., Kopka, J., Haskell, D. W., Zhao, W., Schiller, K. C., Gatzke, N., et al. (2004). Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 136, 4159–4168. doi: 10.1104/pp.104.052142
Karchi, H., Miron, D., Ben-Yaacov, S., and Galili, G. (1995). The lysine-dependent stimulation of lysine catabolism in tobacco seed requires calcium and protein phosphorylation. Plant Cell 7, 1963–1970. doi: 10.1105/tpc.7.11.1963
Kiyota, E., Pena, I. A., and Arruda, P. (2015). The saccharopine pathway in seed development and stress response of maize. Plant Cell Environ. 38, 2450–2461. doi: 10.1111/pce.12563
Kleczkowski, L. A., and Givan, C. V. (1988). Serine formation in leaves by mechanisms other than the glycolate pathway. J. Plant Physiol. 132, 641–652. doi: 10.1016/S0176-1617(88)80223-2
Koag, M. C., Wilkens, S., Fenton, R. D., Resnik, J., Vo, E., and Close, T. J. (2009). The K-segment of maize DHN1 mediates binding to anionic phospholipid vesicles and concomitant structural changes. Plant Physiol. 150, 1503–1514. doi: 10.1104/pp109136697
Kosová, K., Vítámvás, P., Prášil, I. T., and Renaut, J. (2011). Plant proteome changes under abiotic stress-contribution of proteomics studies to understanding plant stress response. J. Proteomics 74, 1301–1322. doi: 10.1016/j.jprot.2011.02.006
Krüßel, L., Junemann, J., Wirtz, M., Birke, H., Thornton, J. D., Browning, L. W., et al. (2014). The mitochondrial sulfur dioxygenase ETHYLMALONIC ENCEPHALOPATHY PROTEIN1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol. 165, 92–104. doi: 10.1104/pp.114.239764
Kumar, K. R. R., and Kirti, P. B. (2012). Novel role for a serine/arginine-rich splicing factor, AdRSZ21 in plant defense and HR-like cell death. Plant Mol. Biol. 80, 461–476. doi: 10.1007/s11103-012-9960-8
Lazar, G., Schaal, T., Maniatis, T., and Goodman, H. M. (1995). Identification of a plant serine-arginine-rich protein similar to the mammalian splicing factor SF2/ASF. Proc. Natl. Acad. Sci. U. S. A. 92, 7672–7676. doi: 10.1073/pnas.92.17.7672
Less, H., Angelovici, R., Tzin, V., and Galili, G. (2011). Coordinated gene networks regulating Arabidopsis plant metabolism in response to various stresses and nutritional cues. Plant Cell 23, 1264–1271. doi: 10.1105/tpc.110.082867
Li, M., Guo, R., Jiao, Y., Jin, X., Zhang, H., and Shi, L. (2017). Comparison of salt tolerance in soja based on metabolomics of seedling roots. Front. Plant Sci. 8:1101. doi: 10.3389/fpls.2017.01101
Lin, S., Coutinho-Mansfield, G., Wang, D., Pandit, S., and Fu, X. D. (2008). The splicing factor SC35 has an active role in transcriptional elongation. Nat. Struct. Mol. Biol. 15, 819–826. doi: 10.1038/nsmb.1461
Lin, C. H., Peng, P. H., Ko, C. Y., Markhart, A. H., and Lin, T. Y. (2012). Characterization of a novel Y2K-type dehydrin VrDhn1 from Vigna radiata. Plant Cell Physiol. 53, 930–942. doi: 10.1093/pcp/pcs040
Liu, Y., Mauve, C., Lamothe-Sibold, M., Guerard, F., and Glab, N. (2019). Photorespiratory serine hydroxymethyltransferase 1 activity impacts abiotic stress tolerance and stomatal closure. Plant Cell Environ. 42, 2567–2583. doi: 10.1111/pce.13595
Liu, Y., Wang, L., Xing, X., Sun, L., Pan, J., Kong, X., et al. (2013). ZmLEA3, a multifunctional group 3 LEA protein from maize (Zea mays L.) is involved in biotic and abiotic stresses. Plant Cell Physiol. 54, 944–959. doi: 10.1093/pcp/pct047
Liu, Y., Wang, L., Zhang, T., Yang, X., and Li, D. (2017). Functional characterization of KS-type dehydrin ZmDHN13 and its related conserved domains under oxidative stress. Sci. Rep. 7:7361. doi: 10.1038/s41598-017-07852-y
Liu, H., Yu, C., Li, H., Ouyang, B., Wang, T., Zhang, J., et al. (2015). Overexpression of ShDHN, a dehydrin gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses in tomato. Plant Sci. 231, 198–211. doi: 10.1016/j.plantsci.2014.12.006
Long, X., Liu, Q., Chan, M., Wang, Q., and Sun, S. S. (2013). Metabolic engineering and profiling of rice with increased lysine. Plant Biotechnol. J. 11, 490–501. doi: 10.1111/pbi.12037
Lopato, S., Kalyna, M., Dorner, S., Kobayashi, R., Krainer, A. R., and Barta, A. (1999). atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 13, 987–1001. doi: 10.1101/gad.13.8.987
Malik, A. A., Veltri, M., Boddington, K. F., Singh, K. K., and Graether, S. P. (2017). Genome analysis of conserved dehydrin motifs in vascular plants. Front. Plant Sci. 8:709. doi: 10.3389/fpls.2017.00709
Maria, K., Sergiy, L., and Andrea, B. (2003). Ectopic expression of atRSZ33 reveals its function in splicing and causes pleiotropic changes in development. Mol. Biol. Cell 14, 3565–3577. doi: 10.1091/mbc.e03-02-0109
Martin, J., Geromanos, S., Tempest, P., and Hartl, F. U. (1993). Identification of nucleotide-binding regions in the chaperonin proteins GroEL and GroES. Nature 366, 279–282. doi: 10.1038/366279a0
Mattaini, K. R., Sullivan, M. R., and Heiden, M. G. V. (2016). The importance of serine metabolism in cancer. J. Cell Biol. 214, 249–257. doi: 10.1083/jcb.201604085
Melo, J. P., Kalyna, M., and Duque, P. (2020). Current challenges in studying alternative splicing in plants: the case of Physcomitrella patens SR proteins. Front. Plant Sci. 11:286. doi: 10.3389/fpls.2020.00286
Michard, E., Lima, P. T., Borges, F., Silva, A. C., Portes, M. T., Carvalho, J. E., et al. (2011). Glutamate receptor-like genes form Ca2+ channels in pollen tubes and are regulated by pistil D-serine. Science 332, 434–437. doi: 10.1126/science.1201101
Miyazaki, T., Miyazaki, J., Yamane, H., and Nishiyama, M. (2004). α-Aminoadipate aminotransferase from an extremely thermophilic bacterium, Thermus thermophilus. Microbiology 150, 2327–2334. doi: 10.1099/mic.0.27037-0
Moreno, J. I., Martin, R., and Castresana, C. (2005). Arabidopsis SHMT1, a serine hydroxymethyltransferase that functions in the photorespiratory pathway influences resistance to biotic and abiotic stress. Plant J. 41, 451–463. doi: 10.1111/j.1365-313X.2004.02311.x
Mundy, J., and Chua, N. H. (1988). Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 7, 2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x
Muñoz-Bertomeu, J., Anoman, A. D., Toujani, W., Cascales-Miñana, B., Flores-Tornero, M., and Ros, R. (2011a). Interactions between abscisic acid and plastidial glycolysis in Arabidopsis. Plant Signal. Behav. 6, 157–159. doi: 10.4161/psb.6.1.14312
Muñoz-Bertomeu, J., Bermúdez, M. A., Segura, J., and Ros, R. (2011b). Arabidopsis plants deficient in plastidial glyceraldehyde-3-phosphate dehydrogenase show alterations in abscisic acid (ABA) signal transduction: interaction between ABA and primary metabolism. J. Exp. Bot. 62, 1229–1239. doi: 10.1093/jxb/erq353
Muñoz-Bertomeu, J., Cascales-Miñana, B., Mulet, J. M., Baroja-Fernández, E., Pozueta-Romero, J., Kuhn, J. M., et al. (2009). Plastidial glyceraldehyde-3-phosphate dehydrogenase deficiency leads to altered root development and affects the sugar and amino acid balance in Arabidopsis. Plant Physiol. 151, 541–558. doi: 10.1104/pp.109.143701
Muttucumaru, N., Powers, S. J., Elmore, J. S., Mottram, D. S., and Halford, N. G. (2015). Effects of water availability on free amino acids, sugars, and acrylamide-forming potential in potato. J. Agric. Food Chem. 63, 2566–2575. doi: 10.1021/jf506031w
Nagaraju, M., Reddy, P. S., Kumar, S. A., Kumar, A., Suravajhala, P., Ali, A., et al. (2018). Genome-wide in silico analysis of dehydrins in Sorghum bicolor, Setaria italica and Zea mays and quantitative analysis of dehydrin gene expressions under abiotic stresses in Sorghum bicolor. Plant Gene 13, 64–75. doi: 10.1016/j.plgene.2018.01.004
Negrutiu, I., Cattoir-Reynearts, A., Verbruggen, I., and Jacobs, M. (1984). Lysine overproducer mutants with an altered dihydrodipicolinate synthase from protoplast culture of Nicotiana sylvestris (Spegazzini and Comes). Theor. Appl. Genet. 68, 11–20. doi: 10.1007/BF00252303
Noman, A., Ali, Q., Maqsood, J., Iqbal, N., Javed, M. T., Rasool, N., et al. (2018). Deciphering physio-biochemical, yield, and nutritional quality attributes of water-stressed radish (Raphanus sativus L.) plants grown from Zn-Lys primed seeds. Chemosphere 195, 175–189. doi: 10.1016/j.chemosphere.2017.12.059
Okamura, E., and Hirai, M. Y. (2017). Novel regulatory mechanism of serine biosynthesis associated with 3-phosphoglycerate dehydrogenase in Arabidopsis thaliana. Sci. Rep. 7:3533. doi: 10.1038/s41598-017-03807-5
Palusa, S. G., Ali, G. S., and Reddy, A. S. (2010). Alternative splicing of pre-mRNAs of Arabidopsis serine/arginine-rich proteins: regulation by hormones and stresses. Plant J. 49, 1091–1107. doi: 10.1111/j.1365-313X.2006.03020.x
Pant, B. D., Buhtz, A., Kehr, J., and Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Perl, A., Shaul, O., and Galili, G. (1992). Regulation of lysine synthesis in transgenic potato plants expressing a bacterial dihydrodipicolinate synthase in their chloroplasts. Plant Mol. Biol. 19, 815–823. doi: 10.1007/BF00027077
Ramos-Ruiz, R., Martinez, F., and Knauf-Beiter, G. (2019). The effects of GABA in plants. Cogent Food Agric. 5:1670553. doi: 10.1080/23311932.2019.1670553
Riley, A. C., Ashlock, D. A., and Graether, S. P. (2019). Evolution of the modular, disordered stress proteins known as dehydrins. PLoS One 14:e0211813. doi: 10.1371/journal.pone.0211813
Rizwan, M., Ali, S., Hussain, A., Ali, Q., Shakoor, M. B., Zia-ur-Rehman, M., et al. (2017). Effect of zinc-lysine on growth, yield and cadmium uptake in wheat (Triticum aestivum L.) and health risk assessment. Chemosphere 187, 35–42. doi: 10.1016/j.chemosphere.2017.08.071
Rodrigues, S. M., Andrade, M. O., Gomes, A. P. S., DaMatta, F. M., Baracat-Pereira, M. C., and Fontes, E. P. B. (2006). Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. J. Exp. Bot. 57, 1909–1918. doi: 10.1093/jxb/erj132
Ros, R., Cascales-Miñana, B., Segura, J., Anoman, A. D., Toujani, W., Flores-Tornero, M., et al. (2013). Serine biosynthesis by photorespiratory and non-photorespiratory pathways: an interesting interplay with unknown regulatory networks. Plant Biol. 15, 707–712. doi: 10.1111/j.1438-8677.2012.00682.x
Ros, R., Muñoz-Bertomeu, J., and Krueger, S. (2014). Serine in plants: biosynthesis, metabolism, and functions. Trends Plant Sci. 19, 564–569. doi: 10.1016/j.tplants.2014.06.003
Rosales, R., Romero, I., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2014). The crucial role of ф and K-segments in the in vitro functionality of Vitis vinifera dehydrin DHN1a. Phytochemistry 108, 17–25. doi: 10.1016/j.phytochem.2014.10.006
Rosa-Téllez, S., Anoman, A. D., Alcantara-Enguidanos, A., Garza-Aguirre, R. A., Alseekh, S., and Ros, R. (2020). PGDH family genes differentially affect Arabidopsis tolerance to salt stress. Plant Sci. 290:110284. doi: 10.1016/j.plantsci.2019.110284
Saeedipour, S., and Moradi, F. (2012). Stress-induced changes in the free amino acid composition of two wheat cultivars with difference in drought resistance. Afr. J. Biotechnol. 11, 9559–9565. doi: 10.5897/AJB11.3870
Serre, N. B. C., Sarthou, M., Gigarel, O., Figuet, S., Corso, M., Choulet, J., et al. (2020). Protein lysine methylation contributes to modulating the response of sensitive and tolerant Arabidopsis species to cadmium stress. Plant Cell Environ. 43, 760–774. doi: 10.1111/pce.13692
Shaul, O., and Galili, G. (1992). Increased lysine synthesis in tobacco plants that express high levels of bacterial dihydrodipicolinate synthase in their chloroplasts. Plant J. 2, 203–209. doi: 10.1111/j.1365-313X.1992.00203.x
Shaul, O., and Galili, G. (1993). Concerted regulation of lysine and threonine synthesis in tobacco plants expressing bacterial feedback-insensitive aspartate kinase and dihydrodipicolinate synthase. Plant Mol. Biol. 23, 759–768. doi: 10.1007/BF00021531
Slaughter, J. C., and Davies, D. D. (1968). The isolation and characterization of 3-phosphoglycerate dehydrogenase from peas. Biochem. J. 109, 743–748. doi: 10.1042/bj1090743
Somerville, C. R., and Ogren, W. L. (1981). Photorespiration-deficient mutants of Arabidopsis thaliana lacking mitochondrial serine transhydroxymethylase activity. Plant Physiol. 67, 666–671. doi: 10.1104/pp.67.4.666
Song, Y., and Zhang, D. (2017). The role of long noncoding RNAs in plant stress tolerance. Methods Mol. Biol. 1631, 41–68. doi: 10.1007/978-1-4939-7136-7_3
Stein, R. J., Höreth, S., de Melo, J. R., Syllwasschy, L., Lee, G., Garbin, M. L., et al. (2017). Relationships between soil and leaf mineral composition are element-specific, environment-dependent and geographically structured in the emerging model Arabidopsis halleri. New Phytol. 213, 1274–1286. doi: 10.1111/nph.14219
Stewart, G. R., and Larher, F. (1980). “Accumulation of amino acids and related compounds in relation to environmental stress” in Amino acids and derivatives. Springer Berlin Heidelberg: Academic Press, 609–635.
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Szczesniak, M. W., Rosikiewicz, W., and Makałowska, I. (2016). CANTATAdb: a collection of plant long non-coding RNAs. Plant Cell Physiol. 57:e8. doi: 10.1093/pcp/pcv201
Tanabe, N., Kimura, A., Yoshimura, K., and Shigeoka, S. (2008). “Identification of interacting factors with a high-light responsible SR protein, atSR45a, involved in the regulation of alternative splicing in Arabidopsis” in Photosynthesis. Energy from the Sun. Dordrecht, The Netherlands: Springer, 1347–1350.
Tolbert, N. E. (1980). “Photorespiration” in The biochemistry of plants. Vol. 2. ed. D. D. Davis (New York: Academic Press), 487–523.
Toujani, W., Muñoz-Bertomeu, J., Flores-Tornero, M., Rosa-Téllez, S., Anoman, A. D., Alseekh, S., et al. (2013). Functional characterization of the plastidial 3-phosphoglycerate dehydrogenase family in Arabidopsis. Plant Physiol. 163, 1164–1178. doi: 10.1104/pp.113.226720
Tzin, V., and Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3, 956–972. doi: 10.1093/mp/ssq048
Urano, K., Maruyama, K., Ogata, Y., Morishita, Y., Takeda, M., Sakurai, N., et al. (2009). Characterization of the ABA-regulated global responses to dehydration in Arabidopsis by metabolomics. Plant J. 57, 1065–1078. doi: 10.1111/j.1365-313X.2008.03748.x
Uversky, V. N. (2013). A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 22, 693–724. doi: 10.1002/pro.2261
Uversky, V. N. (2016). Dancing protein clouds: the strange biology and chaotic physics of intrinsically disordered proteins. J. Biol. Chem. 291, 6681–6688. doi: 10.1074/jbc.R115.685859
Vauterin, M., Frankard, V., and Jacobs, M. (1999). The Arabidopsis thaliana dhdps gene encoding dihydrodipicolinate synthase, key enzyme of lysine biosynthesis, is expressed in a cell-specific manner. Plant Mol. Biol. 39, 695–708. doi: 10.1023/A:1006132428623
Velasco, A. M., Leguina, J. I., and Lazcano, A. (2002). Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 55, 445–449. doi: 10.1007/s00239-002-2340-2
Voll, L. M., Jamai, A., Renne, P., Voll, H., McClung, C. R., and Weber, A. P. (2006). The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol. 140, 59–66. doi: 10.1104/pp.105.071399
Waditee, R., Bhuiyan, N. H., Hirata, E., Hibino, T., Tanaka, Y., Shikata, M., et al. (2007). Metabolic engineering for betaine accumulation in microbes and plants. J. Biol. Chem. 282, 34185–34193. doi: 10.1074/jbc.M704939200
Wang, Y., Fan, X., Lin, F., He, G., Terzaghi, W., Zhu, D., et al. (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U. S. A. 111, 10359–10364. doi: 10.1073/pnas.1409457111
Wang, M., Liu, C., Li, S., Zhu, D., Zhao, Q., and Yu, J. (2013). Improved nutritive quality and salt resistance in transgenic maize by simultaneously overexpression of a natural lysine-rich protein gene, SBgLR, and an ERF transcription factor gene, TSRF1. Int. J. Mol. Sci. 14, 9459–9474. doi: 10.3390/ijms14059459
Wenefrida, I., Utomo, H. S., Blanche, S. B., and Linscombe, S. D. (2009). Enhancing essential amino acids and health benefit components in grain crops for improved nutritional values. Recent Pat. DNA Gene Seq. 3, 219–225. doi: 10.2174/187221509789318405
Wisniewski, M., Webb, R., Balsamo, R., Close, T. J., Yu, X. M., and Griffith, M. (1999). Purification, immunolocalization, cryoprotective, and antifreeze activity of PCA60: A dehydrin from peach (Prunus persica). Physiol. Plant. 105, 600–608. doi: 10.1034/j.1399-3054.1999.105402.x
Wu, M., Wanggou, S., Li, X., Liu, Q., and Xie, Y. (2017). Overexpression of mitochondrial serine hydroxyl-methyltransferase 2 is associated with poor prognosis and promotes cell proliferation and invasion in gliomas. Onco Targets Ther. 10, 3781–3788. doi: 10.2147/OTT.S130409
Wulfert, S., and Krueger, S. (2018). Phosphoserine aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front. Plant Sci. 9:1712. doi: 10.3389/fpls.2018.01712
Xiao, R., Sun, Y., Ding, J. H., Lin, S., Rose, D. W., Rosenfeld, M. G., et al. (2007). Splicing regulator SC35 is essential for genomic stability and cell proliferation during mammalian organogenesis. Mol. Cell. Biol. 27, 5393–5402. doi: 10.1128/MCB.00288-07
Xie, C., Zhang, R. X., Qu, Y. T., Miao, Z. Y., Zhang, Y. Q., Shen, X. Y., et al. (2012). Overexpression of MtCAS31 enhances drought tolerance in transgenic Arabidopsis by reducing stomatal density. New Phytol. 195, 124–135. doi: 10.1111/j.1469-8137.2012.04136.x
Xing, X., Liu, Y., Kong, X., Liu, Y., and Li, D. (2011). Overexpression of a maize dehydrin gene, ZmDHN2b, in tobacco enhances tolerance to low temperature. Plant Growth Regul. 65, 109–118. doi: 10.1007/s10725-011-9580-3
Xu, H., Andi, B., Qian, J., West, A. H., and Cook, P. F. (2006). The α-aminoadipate pathway for lysine biosynthesis in fungi. Cell Biochem. Biophys. 46, 43–64. doi: 10.1385/CBB:46:1:43
Yan, Q., Xia, X., Sun, Z., and Fang, Y. (2017). Depletion of Arabidopsis SC35 and SC35-like serine/arginine-rich proteins affects the transcription and splicing of a subset of genes. PLoS Genet. 13:e1006663. doi: 10.1371/journal.pgen.1006663
Yang, M., and Vousden, K. H. (2016). Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662. doi: 10.1038/nrc.2016.81
Yang, Q. Q., Zhang, C. Q., Chan, M. L., Zhao, D. S., Chen, J. Z., Wang, Q., et al. (2016). Biofortification of rice with the essential amino acid lysine: molecular characterization, nutritional evaluation, and field performance. J. Exp. Bot. 67, 4285–4296. doi: 10.1093/jxb/erw209
Yang, W., Zhang, L., Lv, H., Li, H., Zhang, Y., Xu, Y., et al. (2015). The K-segments of wheat dehydrin WZY2 are essential for its protective functions under temperature stress. Front. Plant Sci. 6:406. doi: 10.3389/fpls.2015.00406
Yang, Q. Q., Zhao, D. S., Zhang, C. Q., Wu, H. Y., Li, Q. F., Gu, M. H., et al. (2018). A connection between lysine and serotonin metabolism in rice endosperm. Plant Physiol. 176, 1965–1980. doi: 10.1104/pp.17.01283
Yoon, E. K., Krishnamurthy, P., Kim, J. A., Jeong, M. J., and Lee, S. I. (2018). Genome-wide characterization of Brassica rapa genes encoding serine/arginine-rich proteins: expression and alternative splicing events by abiotic stresses. J. Plant Biol. 61, 198–209. doi: 10.1007/s12374-017-0391-6
Yu, Z., Wang, X., and Zhang, L. (2018). Structural and functional dynamics of dehydrins: a plant protector protein under abiotic stress. Int. J. Mol. Sci. 19:3420. doi: 10.3390/ijms19113420
Yuan, J., Li, J., Yang, Y., Tan, C., Zhu, Y., Hu, L., et al. (2018). Stress-responsive regulation of long non-coding RNA polyadenylation in Oryza sativa. Plant J. 93, 814–827. doi: 10.1111/tpj.13804
Yuan, L., Liu, X., Luo, M., Yang, S., and Wu, K. (2013). Involvement of histone modifications in plant abiotic stress responses. J. Integr. Plant Biol. 55, 892–901. doi: 10.1111/jipb.12060
Zafari, M., and Ebadi, A. (2016). Effects of water strass and brassinosteroid (24-epibrassinolide) on changes of some amino acids and pigments in safflower (Cartamus tinctorius L.). J. Curr. Res. Sci. 1:711.
Zhang, W., Du, B., Liu, D., and Qi, X. (2014). Splicing factor SR34b mutation reduces cadmium tolerance in Arabidopsis by regulating iron-regulated transporter 1 gene. Biochem. Biophys. Res. Commun. 455, 312–317. doi: 10.1016/j.bbrc.2014.11.017
Zhang, X. N., Shi, Y., Powers, J. J., Gowda, N. B., Zhang, C., Ibrahim, H. M. M., et al. (2017). Transcriptome analyses reveal SR45 to be a neutral splicing regulator and a suppressor of innate immunity in Arabidopsis thaliana. BMC Genomics 18:772. doi: 10.1186/s12864-017-4183-7
Zhang, Y., Sun, K., Sandoval, F. J., Santiago, K., and Roje, S. (2010). One-carbon metabolism in plants: characterization of a plastid serine hydroxymethyltransferase. Biochem. J. 430, 97–105. doi: 10.1042/BJ20100566
Zhong, X. Y., Wang, P., Han, J., Rosenfeld, M. G., and Fu, X. D. (2009). SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol. Cell 35, 1–10. doi: 10.1016/j.molcel.2009.06.016
Zhu, X., and Galili, G. (2003). Increased lysine synthesis coupled with a knockout of its catabolism synergistically boosts lysine content and also transregulates the metabolism of other amino acids in Arabidopsis seeds. Plant Cell 15, 845–853. doi: 10.1105/tpc.009647
Zhu, X., and Galili, G. (2004). Lysine metabolism is concurrently regulated by synthesis and catabolism in both reproductive and vegetative tissues. Plant Physiol. 135, 129–136. doi: 10.1104/pp.103.037168
Zhu, X., Tang, G., and Galili, G. (2002). The activity of the Arabidopsis bifunctional lysine-ketoglutarate reductase/saccharopine dehydrogenase enzyme of lysine catabolism is regulated by functional interaction between its two enzyme domains. J. Biol. Chem. 277, 49655–49661. doi: 10.1074/jbc.M205466200
Keywords: lysine metabolism, serine metabolism, abiotic stress, plant ontology, stress tolerance
Citation: Kishor PBK, Suravajhala R, Rajasheker G, Marka N, Shridhar KK, Dhulala D, Scinthia KP, Divya K, Doma M, Edupuganti S, Suravajhala P and Polavarapu R (2020) Lysine, Lysine-Rich, Serine, and Serine-Rich Proteins: Link Between Metabolism, Development, and Abiotic Stress Tolerance and the Role of ncRNAs in Their Regulation. Front. Plant Sci. 11:546213. doi: 10.3389/fpls.2020.546213
Edited by:
Giuseppe Forlani, University of Ferrara, ItalyReviewed by:
Sara Rosa-Téllez, University of Valencia, SpainBalasubramani Subramani Paranthaman, Florida Agricultural and Mechanical University, United States
Copyright © 2020 Kishor, Suravajhala, Rajasheker, Marka, Shridhar, Dhulala, Scinthia, Divya, Doma, Edupuganti, Suravajhala and Polavarapu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: P. B. Kavi Kishor, cGJrYXZpQHlhaG9vLmNvbQ==
 P. B. Kavi Kishor1*
P. B. Kavi Kishor1*