
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 03 September 2020
Sec. Plant Biotechnology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.532771
Pre-harvest sprouting (PHS) often results in reduced grain yield and quality and is a major problem in cereal production. Improved seed dormancy would inhibit PHS. Here we show that seed-specific overexpression of two SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) genes SPL12 and IPA1 enhances seed dormancy and inhibits PHS without noticeable effects on shoot architecture in rice. In addition, seed-specific overexpression of IPA1 also increases grain size and thus improves grain productivity. Furthermore, our results suggest that SPL12 enhances the seed dormancy through directly regulating many genes in the gibberellin (GA) pathway. This research provides an efficient method to suppress PHS and will facilitate breeding elite crop varieties.
In agriculture, pre-harvest sprouting (PHS) often results in severe grain yield loss, as well as reduced grain quality and germinability (Li et al., 2004; Chono et al., 2006), and thus imposes a major threat to agriculture and food security. In crops, seed dormancy is regarded as an important agronomic trait due to its key roles in controlling PHS and seed longevity (Gubler et al., 2005; Kottearachchi et al., 2006; Wu et al., 2016). Improved seed dormancy would inhibit PHS and increase seed longevity. Therefore, controlling seed dormancy is a very important goal in agriculture worldwide.
Seed dormancy is first acquired during seed mature stage, and the phytohormone abscisic acid (ABA) plays a fundamental role in establishing this primary seed dormancy. ABA positively regulates seed dormancy, and as seeds mature, ABA level gradually increases to establish seed dormancy (Kang et al., 2015). Besides ABA, another key factor determining seed dormancy is the phytohormone gibberellins (GAs) (Shu et al., 2016; Tuan et al., 2018). GAs promote the transition from dormancy to germination, and antagonize the effects of ABA on seed dormancy. Therefore, the balance between the effects of ABA and GAs is crucial in determining the status of seed dormancy. Many other factors, such as the phytohormone auxins, temperature, light, and air humidity, also have effects on seed dormancy, and most of these factors affect seed dormancy through the ABA and/or GA pathways (Finch-Savage and Footitt, 2017). Although major progress in studying seed dormancy have been made, the detailed molecular mechanisms underlying seed dormancy are largely unknown, and crop breeders often lack effective methods to control seed dormancy.
The conserved microRNA miR156, together with its targets SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factor genes, plays important roles in many aspects of plant development (Chen et al., 2010; Preston and Hileman, 2013). In crops, the miR156/SPL module regulates grain yield through modulating plant shoot architecture and grain size (Wang, 2014; Wang and Wang, 2015). In rice, the miR156 target gene Ideal Plant Architecture 1 (IPA1) encodes SPL14 and was recently considered as a “green revolution” gene for improving grain yield (Lu et al., 2013; Wang and Wang, 2017). The IPA1 quantitative trait locus (QTL) allele ipa1-1d relieves its suppression by miR156 and thus confers an ideal plant architecture with fewer tillers, stronger culms, and more grains per panicle (Jiao et al., 2010). Recently, miR156 was reported to negatively regulate seed dormancy, and IPA1 mediated part of the effects of miR156 on seed dormancy by directly regulating many genes in the GA pathway (Miao et al., 2019). However, the roles of other SPL genes such as SPL12 in seed dormancy are still unknown.
Here, to explore the roles of SPL genes in seed development, we examined the effects of SPL12 and IPA1 overexpression (POle18:SPL12 and POle18:IPA1) on seed dormancy and grain size. Our results showed that overexpression of IPA1 and SPL12 driven by the seed-specific Ole18 promoter (Wu et al., 1998) enhanced seed dormancy and inhibited PHS without obvious effects on plant shoot architecture. In addition, IPA1 seed-specific overexpression also increased grain size and improved grain productivity. Furthermore, our results showed impaired GA pathway in SPL12 overexpression lines and revealed in vivo associations of SPL12 with the promoters of many GA biosynthetic, deactivating and signaling genes, suggesting that SPL12 positively regulates seed dormancy through directly regulating the genes in the GA pathway. Our results provide an effective method to inhibit PHS and will facilitate breeding elite crop varieties.
To construct POle18:SPL12 and POle18:IPA1 vectors, the Ole18 promoter (a 1249-bp fragment upstream the start codon in the Ole18 gene) (Wu et al., 1998) was first fused respectively with SPL12 and IPA1-encoding sequences through fusion PCR technology, and then the POle18:SPL12 and POle18:IPA1 fragments were introduced into the intermediate vector pDONR207 using Gateway™ BP Clonase™ II Enzyme Mix (Invitrogen, Cat. no. 11789-020). After sequencing characterization, the POle18:SPL12 and POle18:IPA1 fragments were subcloned from the right pDONR207 (POle18:SPL12) and pDONR207 (POle18:IPA1) clones, respectively, into the pGWB4 vector using Gateway™ LR Clonase™ II Enzyme Mix ((Invitrogen, Cat. no. 11791-020). The resulting overexpression vectors were transformed into the Japonica rice variety Zhonghua 11 (ZH11) through Agrobacterium-mediated method.
To construct P35S:SPL12-FLAG overexpression vector, the SPL12-encoding sequence was cloned into intermediate vector pDONR207 using Gateway™ BP Clonase™ II Enzyme Mix (Invitrogen, Cat. no. 11789-020), and then the SPL12-encoding sequence was subcloned from the pDONR207 (P35S:SPL12-FLAG) clone into the pGWB11 vector using Gateway™ LR Clonase™ II Enzyme Mix ((Invitrogen, Cat. no. 11791-020). The resulting P35S:SPL12-FLAG overexpression vector was transformed into the Japonica rice variety Xiushui 134 (XS134) through Agrobacterium-mediated method.
The transgenic plants were grown in the paddy field. The seeds from the transgenic plants were sowed in Hangzhou (China) in early June.
Total RNA was extracted using RNAprep Pure Plant Kit (TIANGEN, Cat. No. DP432, China). Reverse transcription was performed using PrimeScriptTM RT reagent Kit (Takara, Cat. No. RR047A, Japan). Real-time PCR assays were performed using SYBR® Premix Ex Taq™ II (Takara, Cat. No. RR820A, Japan), and the Bio-Rad CFX96 real-time PCR instrument was used in our study. The sequences of gene-specific primers for the target genes were listed in Table S3, and rice Ubiquitin gene (LOC_Os05g06770) was used as an internal reference gene, and qPCR reactions were performed in three biological replicates.
ChIP assays were performed using ab117137-ChIP Kit-Plants (abcam, Lot. GR323561-19, England). Firstly, we harvested 1 g seedling shoots from P35S:SPL12-FLAG transgenic plants when the seedlings were grown to about 15 cm in the paddy field. Then the chromatin complexes were isolated, sonicated and incubated with anti-FLAG antibody named M185-3LL (Japan). Precipitated chromatin DNA was analyzed by quantitative PCR using SYBR® Premix Ex Taq™ II (Takara, Cat. No. RR820A, Japan). Primer sequences for the real-time PCR were listed in Table S4, and qPCR reactions were performed in three biological replicates.
Undehusked fresh seeds without drying treatment were first soaked in water for two days (16 h light/8 h dark cycles at 30°C), and then continued to germinate at 30°C. The germination rate was counted every two days.
PHS was investigated in the paddy field of Hangzhou at the normal harvest time. In every line, all of the seeds from a plant were investigated in the assays, and the sprouting data of every line were obtained from at least three plants. The PHS investigations were finished in 1 day.
Seedlings of the wild type, POle18:SPL12 and POle18:IPA1 were grown in Hangzhou paddy field under natural conditions. The area per plot was 90 cm×60 cm, and 24 plants were cultivated in each plot with planting density of 15 cm×15 cm.
Plant materials were ground into powder in liquid nitrogen, and extracted with methanol/water (8/2) at 4°C. The extract was centrifuged at 12,000 g under 4°C for 15 min. The supernatant was collected and evaporated to dryness under nitrogen gas stream, and then reconstituted in methanol/water (3/7). The solution was centrifuged and the supernatant was collected for LC-MS analysis. The LC-MS analysis was conducted with the API6500 Q TRAP LC/MS/MS system, equipped with an ESI Turbo Ion-Spray interface, operating in a positive ion mode and controlled by Analyst 1.6 software (AB Sciex).
Disrupting MIR156 genes was recently reported to enhance seed dormancy in rice (Miao et al., 2019), and transcriptomic data published previously showed that among the miR156 target genes, IPA1 and SPL12 were expressed most highly in the embryos of fresh wet seeds (freshly harvested seeds without drying treatment, and termed fresh seeds hereinafter), suggesting that both IPA1 and SPL12 were involved in seed dormancy controlling. Thus, we overexpressed IPA1 and SPL12 using the rice seed-specific Ole18 promoter (Wu et al., 1998), and at least twenty-five independent lines of each vector were obtained. Germination analyses using fresh seeds revealed that seed-specific overexpression lines of SPL12 and IPA1 showed obviously slower germination than the wild type (Figures 1A–F). These results indicate that overexpression of SPL12 and IPA1 enhances seed dormancy. In addition, we also investigated the PHS rates of POle18:SPL12 and POle18:IPA1 overexpression lines and the wild type in the paddy field of Hangzhou, and found that overexpression of SPL12 and IPA1 significantly suppressed PHS (Figures 1G, H).
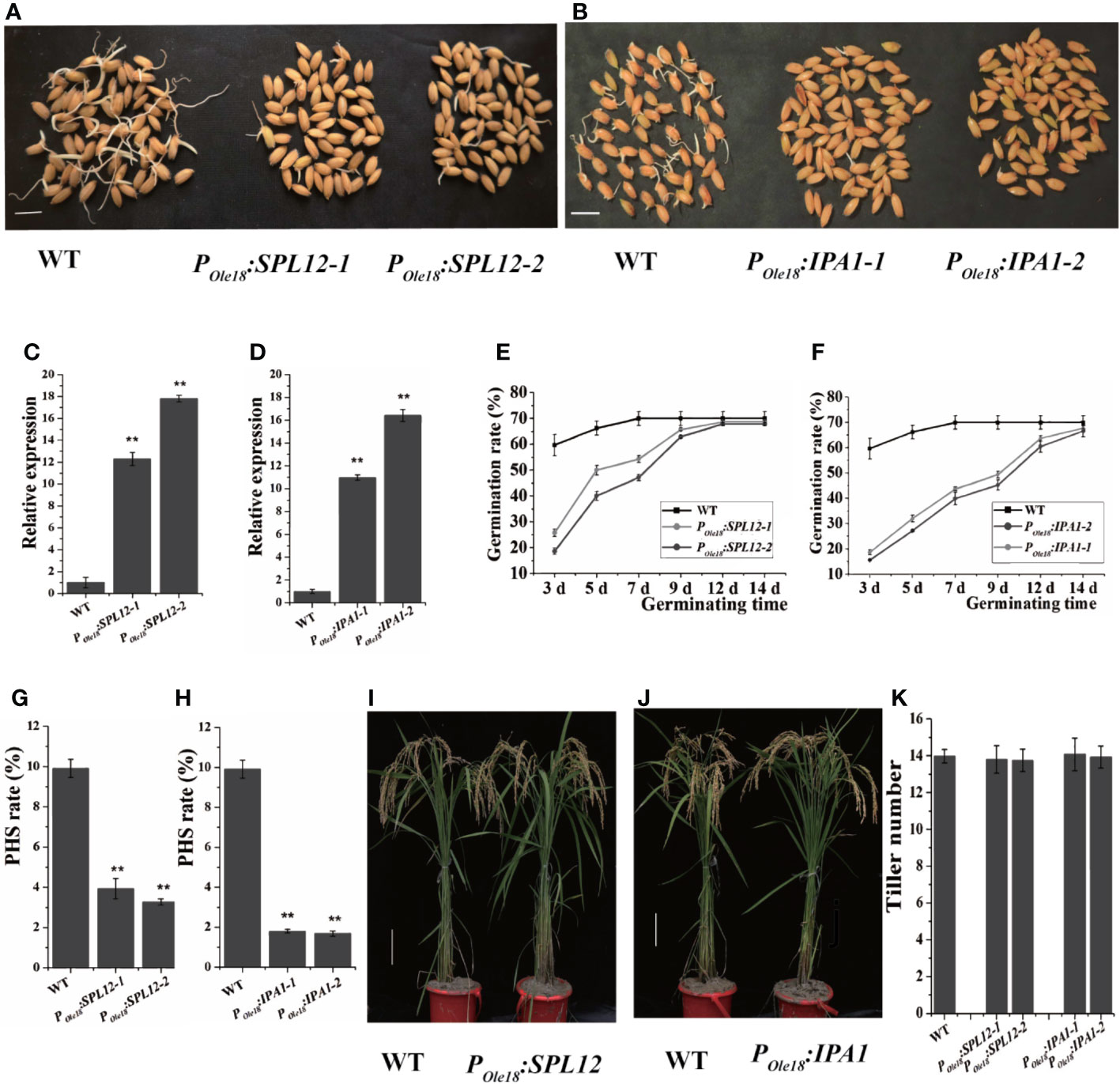
Figure 1 Seed-specific overexpression of SPL12 and IPA1 enhanced seed dormancy. (A) Germination of fresh seeds of POle18:SPL12 and wild-type lines. Scale bar, 1 cm. (B) Germination of fresh seeds of POle18:IPA1 and wild-type lines. Scale bar, 1 cm. (C) Relative expression levels of SPL12 in the fresh seed embryos of wild-type and POle18:SPL12 lines. (D) Relative expression levels of IPA1 in the fresh seed embryos of wild-type and POle18:IPA1 lines. (E) Germination rates of fresh seeds of wild-type and POle18:SPL12 lines. (F) Germination rates of fresh seeds of wild-type and POle18:IPA1 lines. (G) PHS rates of wild-type and POle18:SPL12 lines. (H) PHS rates of wild-type and POle18:IPA1 lines. (I) Wild-type and POle18:SPL12 plants at the mature stage. Scale bar, 10 cm. (J) Wild-type and POle18:IPA1 plants at the mature stage. Scale bar, 10 cm. (K) Tiller numbers of wild-type, POle18:SPL12 and POle18:IPA1 plants. The double asterisks (**) represent significant difference calculated by the Student’s t-test at P < 0.01. Values are means ± SE (n = 3).
In crops, the miR156/SPL module plays key roles in modulating plant shoot architecture. Thus, we examined the plant shoot architectures of the SPL12 and IPA1 overexpression lines, and observed no obvious differences between the overexpression lines and the wild type (Figures 1I–K). In fact, besides seed dormancy and grain size, we observed no obvious differences between the overexpression lines and the wild type.
In crops, the miR156/SPL module has important roles in modulating the grain size and shape. Thus, we compared the grain sizes and shapes of the overexpression lines to the wild type. We did not observe obvious differences in grain size and shape between the SPL12 overexpression lines and the wild type (Figures 2A–D), suggesting that SPL12 plays negligible roles in modulating grain size and shape. Unlike the situation in SPL12 overexpression lines, IPA1 overexpression significantly increased grain lengths and 1000-grain weights (Figures 2A–D). Consistently, plot grain yield test revealed that IPA1 overexpression slightly increased the grain yield (Figure 2E).
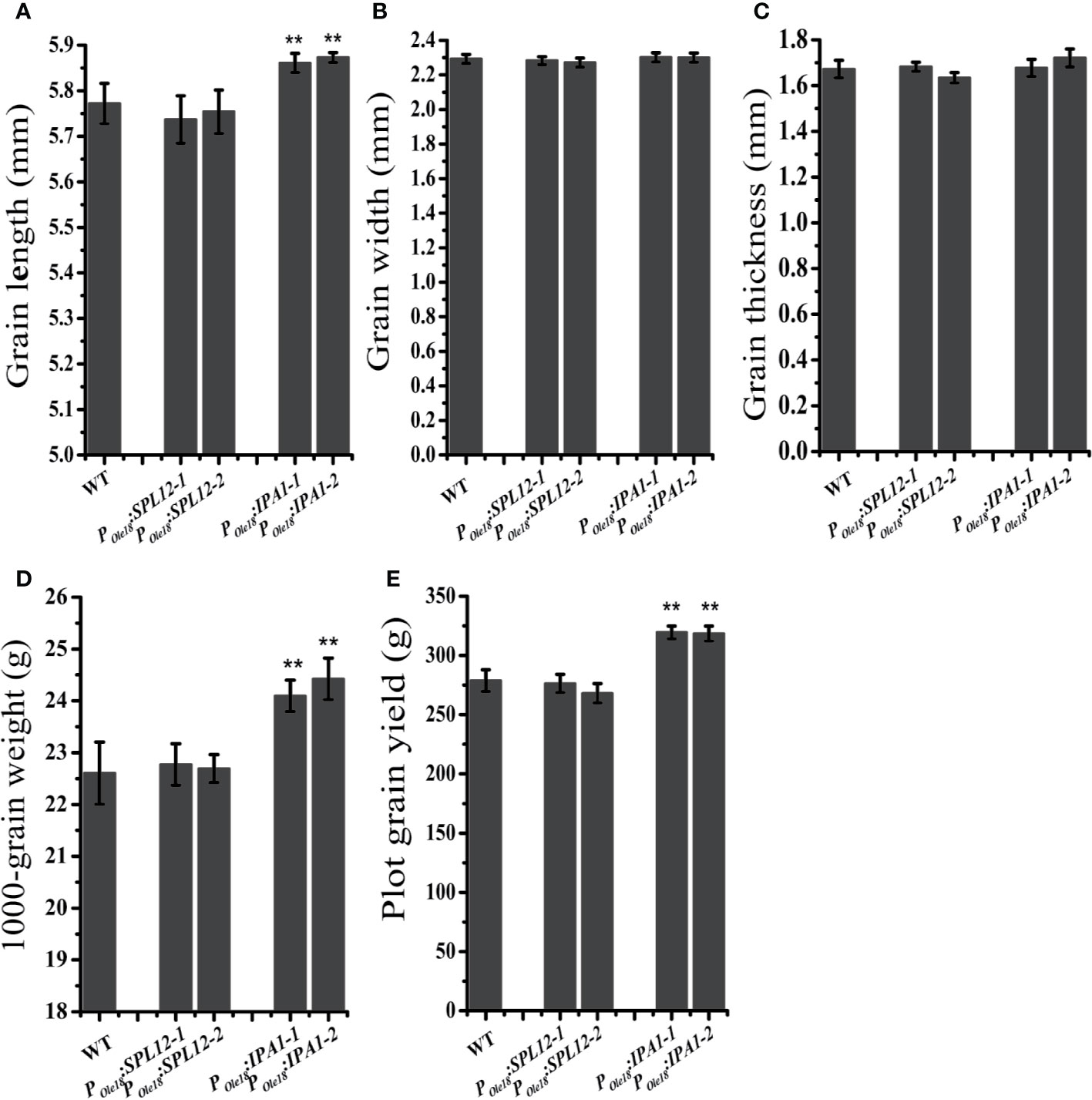
Figure 2 Overexpression of IPA1 but not SPL12 increased grain size. (A) Grain lengths of wild-type, POle18:SPL12 and POle18:IPA1 plants. (B) Grain width of wild-type, POle18:SPL12 and POle18:IPA1 plants. (C) Grain thickness of wild-type, POle18:SPL12 and POle18:IPA1 plants. (D) 1,000-grain weights of wild-type, POle18:SPL12 and POle18:IPA1 plants. (E) Plot grain yield of wild-type, POle18:SPL12 and POle18:IPA1 plants. The double asterisks (**) represent significant difference calculated by the Student’s t-test at P < 0.01. Values are means ± SE (n = 3).
To explore how overexpression of SPL12 enhanced seed dormancy, we analyzed the transcriptomes of the fresh seed embryos from a SPL12 overexpression line (designated SPL12-2) and the wild type, and the result did not suggest enhanced ABA pathway in the SPL12 overexpression line compared to the wild type. Phytohormone measuring revealed that compared to the wild type, the fresh seed embryos of SPL12 and IPA1 overexpression lines contained decreased ABA levels (Figure 3A). These results suggest that overexpression of SPL12 and IPA1 does not enhance seed dormancy through the ABA pathway.
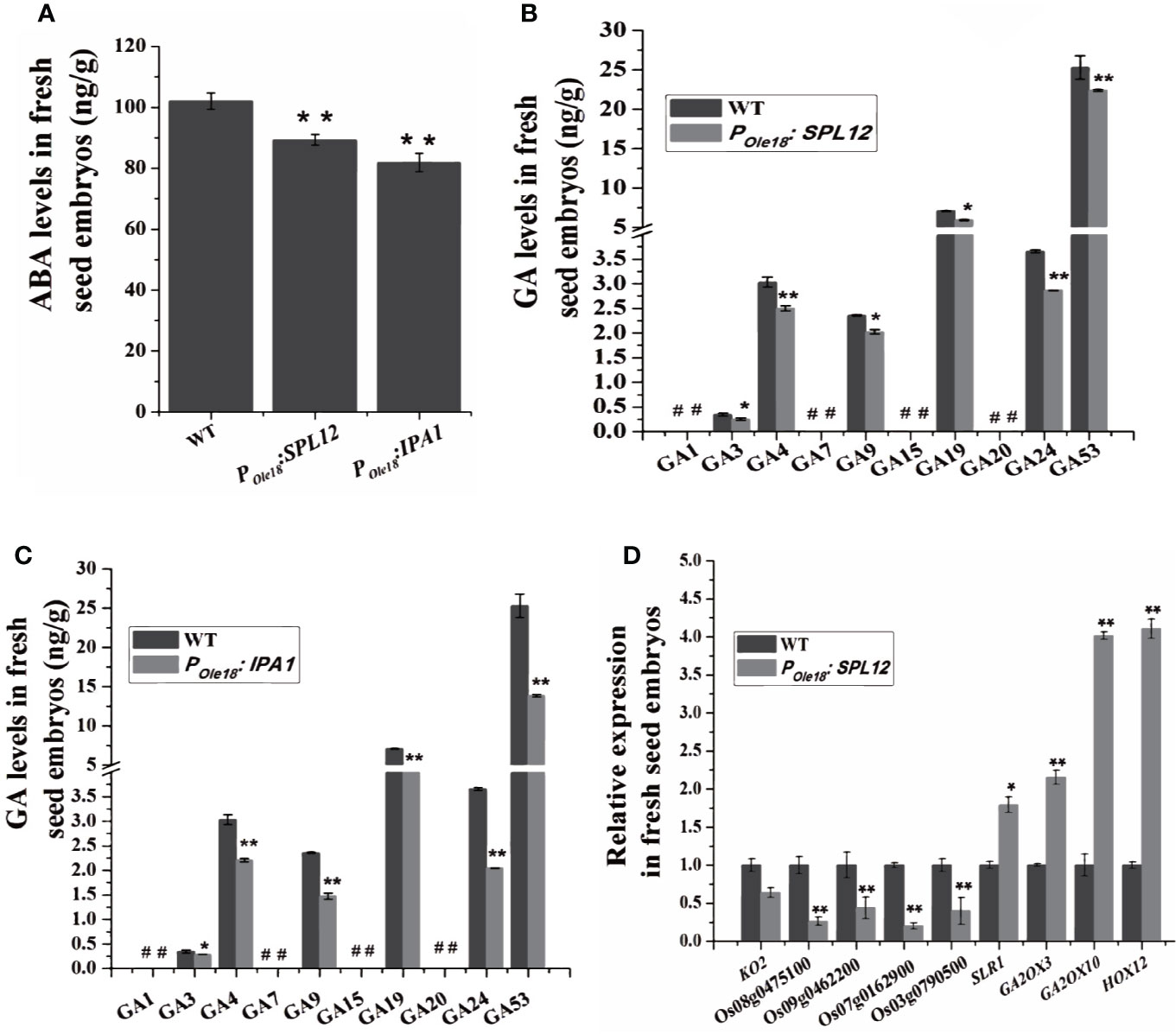
Figure 3 Overexpression of SPL12 enhanced seed dormancy through GA pathway. (A) ABA levels in wild-type, POle18:SPL12, and POle18:IPA1 fresh seed embryos. (B) GA levels in wild-type and POle18:SPL12 fresh seed embryos. #, undetectable GAs. (C) GA levels in wild-type and POle18:IPA1 fresh seed embryos. #, undetectable GAs. (D) Relative expression levels of GA biosynthetic, signaling, and deactivating DEGs in fresh seed embryos. Three biological replications were performed in each assay. The double asterisks (**) represent significant difference calculated by the Student’s t-test at P < 0.01, and single asterisk (*) means P < 0.05. Values are means ± SE (n = 3).
Then, we measured GA levels in the fresh seed embryos. The results showed that the levels of two bioactive GAs, GA3, and GA4, were lower in SPL12 and IPAI overexpression lines than in the wild type (Figures 3B, C). We could not find other bioactive GAs (GA1 and GA7) in the fresh seed embryos of both overexpression and wild-type lines.
Consistent with the recent report that the GA pathway was impaired by the MIR156 gene disruption, our transcriptome analyses showed that SPL12 overexpression down-regulated some key GA biosynthetic and putative GA receptor genes but up-regulated some GA deactivating genes (Table S1 and Figure S1), suggesting impaired GA pathway in the fresh seed embryos of SPL12 overexpression lines. In the SPL12-2 overexpression line compared to the wild type, a total of 3632 differentially expressed genes (DEGs), including 2239 up-regulated and 1393 down-regulated genes, were identified (ratio ≥ 2 or ≤ 0.5, and false discovery rate (FDR) < 0.05) (Table S2). Among the DEGs identified in the SPL12-2 overexpression line compared to the wild type, three important GA deactivating genes (GA2ox3, GA2ox10, and HOX12) were up-regulated, whereas four putative GA receptor genes (Os07g0162900, Os08g0475100, Os09g0462200, and Os03g0790500) were obviously down-regulated by the overexpression (Table S1). When lowering the threshold for DEGs to ratio ≥ 1.5 or ≤ 0.75 (FDR < 0.05), we found that a key GA biosynthetic gene KO2 was also down-regulated, whereas SLR1, one negative regulator of GA signaling, was up-regulated (Table S1). These results were validated by RT-qPCR (Figure 3D and Table S3). These results suggest that SPL12 enhances seed dormancy through the GA pathway.
Since SPL12 is a transcription factor, it is possible that SPL12 directly regulates GA-related genes. SPL family proteins were reported to directly bind GTAC motifs to regulate gene expression (Birkenbihl et al., 2005; Lu et al., 2013). Through sequence analyses of the 1000-bp promoter regions, we found that most of the GA-related DEGs identified above contained multiple GTAC motifs. Thus, we conducted ChIP-qPCR assays to explore the possible interactions between SPL12 and the promoters of GA-related DEGs. The results revealed direct interactions between SPL12 and 9 GA-related genes (Figure 4). The 9 genes included one GA biosynthetic gene KO2, four putative GA receptor genes (Os07g0162900, Os03g0790500, Os09g0462200, and Os08g0475100), three GA deactivating genes (GA2ox3, GA2ox10, and HOX12), and SLR1. These results suggest that SPL12 controls the GA pathway through directly regulating the GA biosynthetic, signaling, and deactivating genes.
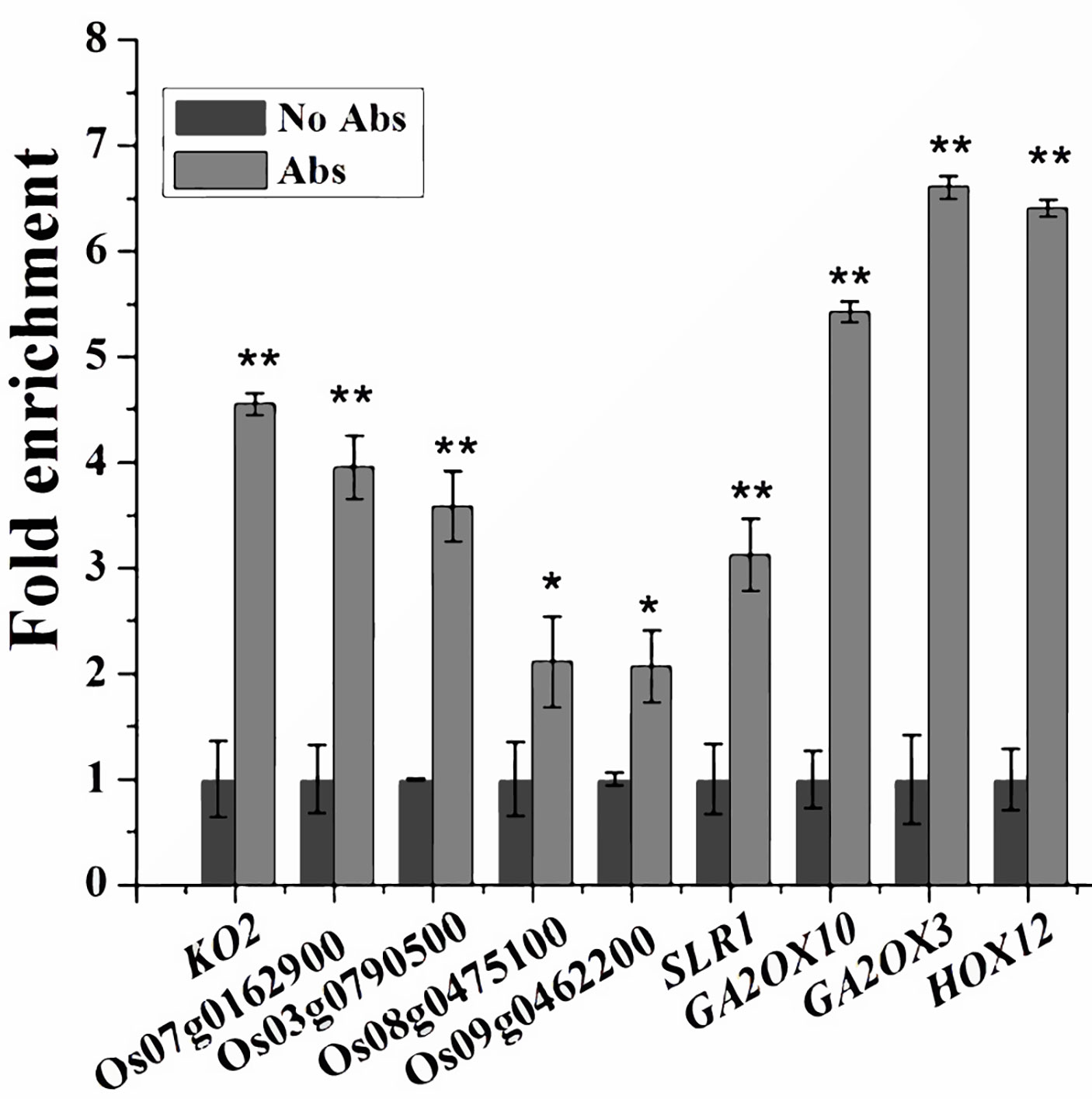
Figure 4 Associations between SPL12 and the promoters of GA biosynthetic, signaling, and deactivating genes. Primers for the ChIP-qPCR were listed in Values are means ± SE (n = 3). The double asterisks (**) represent significant difference calculated by the Student’s t-test at P < 0.01, and single asterisk (*) means P < 0.05. The fold enrichment was normalized against the promoter of rice Ubiquitin.
In crops, PHS often results in severe grain yield loss and reduced grain quality. Improved seed dormancy would inhibit PHS and increase seed longevity (Gubler et al., 2005; Kottearachchi et al., 2006; Wu et al., 2016). The balance between the effects of ABA and GAs is crucial in determining the status of seed dormancy (Shu et al., 2016; Tuan et al., 2018). In rice, miR156 was reported to negatively regulate seed dormancy, and IPA1 mediated part of the effects of miR156 on seed dormancy (Miao et al., 2019). Among the SPL family genes, SPL13, IPA1, and SPL16 were reported to positively regulate seed size (Wang et al., 2012; Wang et al., 2015; Si et al., 2016). However, the roles of SPL12 in seed dormancy and seed size are still unknown.
In this study, we found that seed-specific overexpression of SPL12 and IPA1 improved seed dormancy with negligible effects in rice plant architecture (Figures 1I–K). SPL12-2 overexpression line enhanced seed dormancy more intensely than SPL12-1 overexpression line, which was consistent with higher SPL12 expression level in SPL12-2 overexpression line than that in SPL12-1 overexpression line. Seed-specific overexpression of IPA1 significantly increased seed lengths, 1,000-grain weights and the grain yield, but seed-specific overexpression SPL12 had negligible effects in seed size. IPA1 but not SPL12 increased grain size, and we inferred that it could be due to the different downstream genes regulated by SPL12 and IPA1.
Phytohormone measuring revealed that compared to the wild type, the fresh seed embryos of POle18:SPL12-2 and POle18:IPA1-2 overexpression lines contained slightly decreased ABA levels, and consistently, the transcriptome analyses did not reveal enhanced ABA pathway in POle18:SPL12 fresh seed embryos compared to the wild type, suggesting that overexpression of SPL12 and IPA1 did not enhance seed dormancy through the ABA pathway. The phytohormone measuring also revealed decreased bioactive GA (GA3 and GA4) levels, suggesting that SPL12 may enhance seed dormancy through the GA pathway. Previous study showed that IPA1 mediated the effects of miR156 on seed dormancy through regulating multiple genes in the GA pathway (Miao et al., 2019). Considering the similarity between IPA1 and SPL12, we inferred that SPL12 also enhanced seed dormancy through the GA pathway.
Our transcriptomic analyses supported this speculation. The transcriptomic analyses in wild-type and POle : SPL12 fresh seed embryos showed that a key GA biosynthetic gene KO2 and four putative GA receptor genes (Os07g0162900, Os08g0475100, Os09g0462200, and Os03g0790500) were obviously down-regulated by SPL12 overexpression, whereas a negative regulator of GA signaling SLR1 and three important GA deactivating genes (GA2ox3, GA2ox10, and HOX12) were up-regulated by SPL12 overexpression, suggesting impaired GA pathway in the fresh seed embryos of POle18:SPL12 overexpression lines. In addition, ChIP-qPCR assays demonstrated association of SPL12 with the promoters of a GA biosynthetic genes, four putative GA receptor genes, three deactivating genes and one negative regulator of GA signaling gene SLR1, suggesting that SPL12 enhanced seed dormancy through directly regulating multiple genes in the GA pathway.
In general, our study supports that seed-specific overexpression of SPL12 and IPA1 enhances seed dormancy through directly regulating multiple genes in the GA pathway. IPA1 seed specific overexpression also increases grain size and improves grain productivity. This research provides an efficient method to suppress PHS and will facilitate breeding elite crop varieties.
All datasets generated for this study are included in the article/Supplementary Material.
CM and MQ designed the study and constructed all vectors. MQ and YZ performed the transgenic experiments, analyzed data and wrote the main manuscript. YY and MQ analyzed data. CM and SL oversaw the entire study.
This work was supported by the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (Grant No. 2016C02056-1), the National Natural Science Foundation of China for Young Scientists (Grant. no. 31800241) and the Program for Changjiang Scholars and Innovative Research Team in University of the Ministry of Education of China (No. IRT17R99).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.532771/full#supplementary-material
Birkenbihl, R. P., Jach, G., Saedler, H., Huijser, P. (2005). Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J. Mol. Biol. 352, 590–596.10. doi: 10.1016/j.jmb.2005.07.013
Chen, X., Zhang, Z., Liu, D., Zhang, K., Li, A., Mao, L. (2010). SQUAMOSA promoter-binding protein-like transcription factors: star players for plant growth and development. J. Integr. Plant Biol. 52, 946–951. doi: 10.1111/j.1744-7909.2010.00987.x
Chono, M., Honda, I., Shinoda, S., Kushiro, T., Kamiya, Y., Nambara, E., et al. (2006). Field studies on the regulation of abscisic acid content and germinability during grain development of barley: molecular and chemical analysis of pre-harvest sprouting. J. Exp. Bot. 57, 2421–2434. doi: 10.1093/jxb/erj215
Finch-Savage, W. E., Footitt, S. (2017). Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 68, 843–856. doi: 10.1093/jxb/erw477
Gubler, F., Millar, A. A., Jacobsen, J. V. (2005). Dormancy release, ABA and pre-harvest sprouting. Curr. Opin. Plant Bio. 8, 183–187.16. doi: 10.1016/j.pbi.2005.01.011
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kang, J., Yim, S., Choi, H., Kim, A., Lee, K. P., Lopez-Molina, L., et al. (2015). Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6, 1–10. doi: 10.1038/ncomms9113
Kottearachchi, N. S., Uchino, N., Kato, K., Miura, H. (2006). Increased grain dormancy in white-grained wheat by introgression of preharvest sprouting tolerance QTLs. Euphytica 152, 421–428. doi: 10.1007/s10681-006-9231-3
Li, C., Ni, P., Francki, M., Hunter, A., Zhang, Y., Schibeci, D., et al. (2004). Genes controlling seed dormancy and pre-harvest sprouting in a rice-wheat-barley comparison. Funct. Integr. Genom. 4, 84–93. doi: 10.1007/s10142-004-0104-3
Lu, Z., Yu, H., Xiong, G., Wang, J., Jiao, Y., Liu, G., et al. (2013). Genome-Wide binding analysis of the transcription activator IDEAL PLANT ARCHITECTURE1 reveals a complex network regulating rice plant architecture. Plant Cell. 25, 3743–3759. doi: 10.1105/tpc.113.113639
Miao, C., Wang, Z., Zhang, L., Yao, J., Hua, K., Liu, X., et al. (2019). The grain yield modulator miR156 regulates seed dormancy through the gibberellin pathway in rice. Nat. Commun. 10, 1–12. doi: 10.1038/s41467-019-11830-5
Preston, J. C., Hileman, L. C. (2013). Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 4, 1–13. doi: 10.3389/fpls.2013.00080
Shu, K., Liu, X., Xie, Q., He, Z. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Tuan, P. A., Kumar, R., Rehal, P. K., Toora, P. K., Ayele, B. T. (2018). Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci. 9, 668. doi: 10.3389/fpls.2018.00668
Wang, H., Wang, H. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8, 677–688. doi: 10.1016/j.molp.2015.01.008
Wang, B., Wang, H. (2017). IPA1: A new “green revolution” Gene? Mol. Plant 10, 779–781. doi: 10.1016/j.molp.2017.04.011
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, J. (2014). Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 65, 4723–4730. doi: 10.1093/jxb/eru246
Wu, L., Wang, L., Chen, P., Chen, L., Tzen, J. (1998). Genomic cloning of 18 kDa oleosin and detection of triacylglycerols and oleosin isoforms in maturing rice and postgerminative seedlings. J. Biochem. 123, 386–391. doi: 10.1093/oxfordjournals.jbchem.a021949
Keywords: seed dormancy, seed germination, SPL12, IPA1, gibberellin pathway
Citation: Qin M, Zhang Y, Yang Y, Miao C and Liu S (2020) Seed-Specific Overexpression of SPL12 and IPA1 Improves Seed Dormancy and Grain Size in Rice. Front. Plant Sci. 11:532771. doi: 10.3389/fpls.2020.532771
Received: 05 February 2020; Accepted: 13 August 2020;
Published: 03 September 2020.
Edited by:
Paulo Arruda, Campinas State University, BrazilReviewed by:
Jirui Wang, Sichuan Agricultural University, ChinaCopyright © 2020 Qin, Zhang, Yang, Miao and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunbo Miao, Y2h1bmJvbWlhb0AxNjMuY29t; Shenkui Liu, c2hlbmt1aWxpdUBuZWZ1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.