
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 August 2020
Sec. Aquatic Photosynthetic Organisms
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01259
This article is part of the Research TopicIce and Snow AlgaeView all 10 articles
Glycerol, a compatible solute, has previously been found to act as an osmoprotectant in some marine Chlamydomonas species and several species of Dunaliella from hypersaline ponds. Recently, Chlamydomonas reinhardtii and Dunaliella salina were shown to make glycerol with an unusual bidomain enzyme, which appears to be unique to algae, that contains a phosphoserine phosphatase and glycerol-3-phosphate dehydrogenase. Here we report that two psychrophilic species of Chlamydomonas (C. spp. UWO241 and ICE-MDV) from Lake Bonney, Antarctica also produce high levels of glycerol to survive in the lake’s saline waters. Glycerol concentration increased linearly with salinity and at 1.3 M NaCl, exceeded 400 mM in C. sp. UWO241, the more salt-tolerant strain. We also show that both species expressed several isoforms of the bidomain enzyme. An analysis of one of the isoforms of C. sp. UWO241 showed that it was strongly upregulated by NaCl and is thus the likely source of glycerol. These results reveal another adaptation of the Lake Bonney Chlamydomonas species that allow them to survive in an extreme polar environment.
Glycerol is well-known for its ability to mitigate environmental stresses such as freezing and hyperosmotic conditions in a wide variety of organisms, including yeast (Wang et al., 2001), insects (Bentz and Mullins, 1999; Bennett et al., 2005), and vertebrates (Raymond, 1992; Layne, 1999). It is also well-known as an osmoprotectant in some algae, including salt pond isolates of Dunaliella (Ben-Amotz and Avron, 1973; Sussman and Avron, 1981) and several marine isolates of Chlamydomonas (Ahmad and Hellebust, 1986; Miyasaka and Ikeda, 1997; Miyasaka et al., 1998). As a compatible solute, glycerol prevents osmotic water loss and thus a build-up of intracellular salt concentrations. Two psychrophilic Chlamydomonas species, Chlamydomonas sp. UWO241 (Neale and Priscu, 1995) (hereafter Chlamy-UWO) and Chlamydomonas sp. ICE-MDV (Li et al., 2016) (hereafter Chlamy-ICE) were isolated from Lake Bonney, a permanently ice-covered lake in Antarctica. The lake has a steep halocline, in which the salinity reaches about 150 PSU (equivalent to about 2.1 M NaCl, or 4.3 x the salinity of seawater) at a depth of 40 m (Spigel et al., 2018). Chlamy-UWO’s ability to survive in this environment has been intensively studied for over two decades. Among its adaptation are a photochemical apparatus well-suited for a cold, saline and low light environment (Morgan et al., 1998), more fluid membranes (Morgan-Kiss et al., 2002) and ice-binding proteins (IBPs) to prevent freeze-thaw injury (Raymond and Morgan-Kiss, 2013). Less is known about Chlamy-ICE which was isolated more recently: it also possesses IBPs (Raymond and Morgan-Kiss, 2017) but appears to have differences in acclimatory ability compared to Chlamy-UWO (Cook et al., 2019). Much more is known about the cold adaptations of a closely-related Antarctic pyschrophile Chlamydomonas sp. ICE-L (Cvetkovska et al., 2017). Because both Chlamy-UWO and Chlamy-ICE are adapted to a saline environment, we wished to know whether they also produce glycerol as an osmoprotectant or cryoprotectant.
Until recently, the pathways used by algae to produce glycerol have been a matter of speculation. Glycogen, or one of its products, dihydroxyacetone phosphate (DHAP), has been identified as the source of glycerol in a number of organisms, including yeast, (Modig et al., 2007), Arabidopsis (Caparros-Martin et al., 2007), a bacterium (Larrouy-Maumus et al., 2013), mountain pine beetle (Fraser et al., 2017), and rainbow smelt (Raymond, 1995). In the DHAP pathway, DHAP is converted to glycerol-3-phosphatate (G3P) by a nicotinamide-adenine dinucleotide (NAD+)-dependent glycerol-3 phosphate dehydrogenase (GPDH), which is then converted to glycerol by a phosphoserine phosphatase (PSP). The problem is that algae typically have several GPDHs and phosphatases, and attempts to identify the enzymes involved have been until recently unsuccessful.
Dunaliella salina has a bidomain enzyme that includes an N-terminal PSP and a C-terminal NAD+-dependent GPDH (He et al., 2007). It was proposed as a candidate for glycerol production directly from DHAP (He et al., 2007), but initial attempts to confirm its activity were unsuccessful (He et al., 2009). However, this enzyme still seemed like the most likely source of glycerol. During our study of IBPs in Chlamy-UWO (Raymond and Morgan-Kiss, 2013), we noticed that it produced glycerol as an osmoprotectant, i.e., glycerol production increased with increasing salinity. We searched the Chlamy-UWO genome for a Dunaliella-like bidomain enzyme and found at least two homologs. We then examined the expression of one of the homologs and found that it strongly increased with increasing salinity, as reported in a master’s thesis (Stahl, 2014). This suggested that this enzyme alone was capable of producing glycerol from DHAP
Recently, Morales-Sanchez et al. (2017) showed that a similar bidomain enzyme in Chlamydomonas reinhardtii was sufficient to produce glycerol directly from DHAP, and He Q. H. et al. (2020) finally confirmed that the Dunaliella salina bidomain protein was sufficient to produce glycerol. Here, we report our test of the hypothesis that Chlamy-UWO uses a similar bidomain enzyme to produce glycerol. We also show that Chlamy-ICE is capable of producing high levels of glycerol under saline conditions and has similar bidomain enzymes.
Chlamydomonas sp. UWO241 [Chlamy-UWO, also referred to as C. raudensis in some earlier studies, e.g., (Dolhi et al., 2013)] and Chlamydomonas sp. ICE-MDV (Chlamy-ICE) were previously isolated from the east lobe of Lake Bonney, Antarctica. Chlamy-UWO was isolated from below the permanent chemocline in the deep photic zone (17 m) where the salinity is similar to the salinity of seawater but rapidly increases with increasing depth (Spigel et al., 2018). Its natural depth range is not known. Chlamy-ICE was recovered from a depth of 13 m, where the salinity is markedly lower (Li et al., 2016). Both strains were maintained in Bold’s basal medium (BBM) supplemented with the indicated amounts of NaCl in a temperature/light regime of 8°C/50 μmol · m−2 · s−1 at Miami University. Chlamydomonas sp. UWO241 is deposited in the Bigelow Algal collection (CCMP1619). Chlamydomonas sp. ICE-MDV is available upon request to RMK.
Fresh cells were shipped overnight to the University of Nevada Las Vegas (UNLV) with ice packs. The temperature varied by less than 2°C during shipment. For each sample, 1.50 ml of cell culture was centrifuged in pre-weighed tubes. The supernatant was removed and the remaining medium was removed with a fine, drawn-out pipet. The tubes were reweighed to obtain the pellet weights (about 3 mg), sealed, and stored at -25°C. For glycerol measurement, the tubes were thawed, and the bottoms of the tubes were subjected to two freeze-thaw cycles in liquid nitrogen to break the cells. The pellets were suspended in 1.00 ml DI water, vortexed to release glycerol and centrifuged at 16,000 x g for 2 min at 4°C to yield a clear supernatant. Subsequent testing of the pellet showed that virtually all of the glycerol was released from the cells. Glycerol was quantified enzymatically with Free glycerol reagent (Sigma no. F6428), which develops a 540 nm (purple) color in the presence of glycerol. Intracellular glycerol concentration (mM) in a cell pellet was calculated as Mg x 1,000/(MW x Mp x f), where Mg is the mass of glycerol released from the pellet, MW is the molecular weight of glycerol (92 g mol-1), Mp is the mass of the pellet, and f is fraction of the cell mass that is water. The latter was estimated as 0.7 based on measurements of Chlamydomonas pulsatilla (Ahmad and Hellebust, 1986). This calculation assumed that the extracellular water content in the pellets was zero. If extracelular water were present in the pellets, the actual glycerol concentrations would be higher than those reported.
Sequences of the enzymes described in this study were assembled from transcriptome and genome data obtained from Chlamy-UWO (Raymond and Morgan-Kiss, 2013) and Chlamy-ICE (Raymond and Morgan-Kiss, 2017). The transcriptomes were obtained with 454 and Illumina sequencing, respectively. Additional DNA and mRNA reads needed to complete assembly of Chlamy-UWO isoform 3 were kindly provided by David Smith (University of Western Ontario). Gene expression levels were expressed as FPKM (fragments per kilobase per million reads) values: FPKM = r/R/L, where r is the number of unique reads for a given isoform with e-values less than 1e-20 (each read was assigned only to the isoform that it most closely matched), R is the number of millions of reads in the transcriptome, and L is the length of the gene in kb. The transcriptomes were searched for genes similar to the bidomain enzyme in Dunaliella salina (AAX56341). Several isoforms of the gene were found and assembled. Introns were identified by comparing these sequences with the genomic data. Sequences for the qPCR reference genes were obtained from the Chlamy-UWO transcriptome.
Chloroplast signals were predicted with ChloroP 1.1 (http://www.cbs.dtu.dk/services/ChloroP/) (Emanuelsson et al., 1999). PSP and GPDH domains were identified with NCBI’s conserved domain database https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi. A Neighbor-Joining phylogenetic tree was constructed with Mega X (Tamura et al., 2011), using two non-chlorophyte bidomain proteins to root the tree. We thank Armin Hallmann (University of Bielefeld, Germany) for correcting the bidomain sequence of Volvox carteri used in the tree.
Cultures of Chlamy-UWO were grown at 8°C and 50 μmol m-2 s-1 in BBM supplemented with a range of salt concentrations (10, 300, 700, and 1,300 mM NaCl) until samples reached the mid exponential phase. Total RNA was extracted with a Qiagen RNeasy mini kit (No. 74104) as per manufacturer’s instructions. Residual genomic DNA was removed by Ambion DNase (Thermo Fisher Scientific, Waltham, MA). RNA was reverse transcribed with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) as specified by the manufacturer. Expression of the Chlamy-UWO PSP-GPDH isoform 2 was quantified by real-time quantitative PCR using the ΔΔCq method (Liu et al., 2012) and a Bio-Rad CFX Connect Real-time thermal cycler. Histone H2B (histh2b) and 40S ribosomal protein S10 (rps10) (GenBank accessions MT362546 and MT362547, respectively) were used as reference genes, as their expressions were fairly stable over the conditions used. Primers are shown in Table S1.
A 3D model of Chlamy-UWO isoform 1 was predicted with Swiss Model (Waterhouse et al., 2018) (https://swissmodel.expasy.org/) using the structure of the Dunaliella salina bidomain protein (6iuy.1.A) (He Q. H. et al., 2020) as template. In the 580-a.a. region of overlap, the two proteins had an identity of 54%. The free energy of the model was then minimized (from -22.8 to -30.9 MJ/mol) with the Yasara energy minimization server (Krieger et al., 2009) (http://www.yasara.org/minimizationserver.htm) and displayed with the Yasara viewer (Waterhouse et al., 2018) (http://www.yasara.org/). Stereo views were obtained by rotating the molecules 3° around the vertical axis.
Intracellular glycerol levels in Chlamy-UWO and Chlamy-ICE increased with increasing NaCl concentration. Both species maintained glycerol levels of about 150 mM at 700 mM NaCl (Figure 1A). At 1,300 mM NaCl, Chlamy-UWO cells reached over 400 mM glycerol while Chlamy-ICE cells were unable to grow. In contrast, glycerol levels in the supernatant of centrifuged dense cultures of Chlamy-UWO were very low (0.4 mM at 700 m NaCl and 1.1 mM at 1,300 mM NaCl), indicating that the cells were maintaining a strong gradient between the intracellular and extracellular environments.
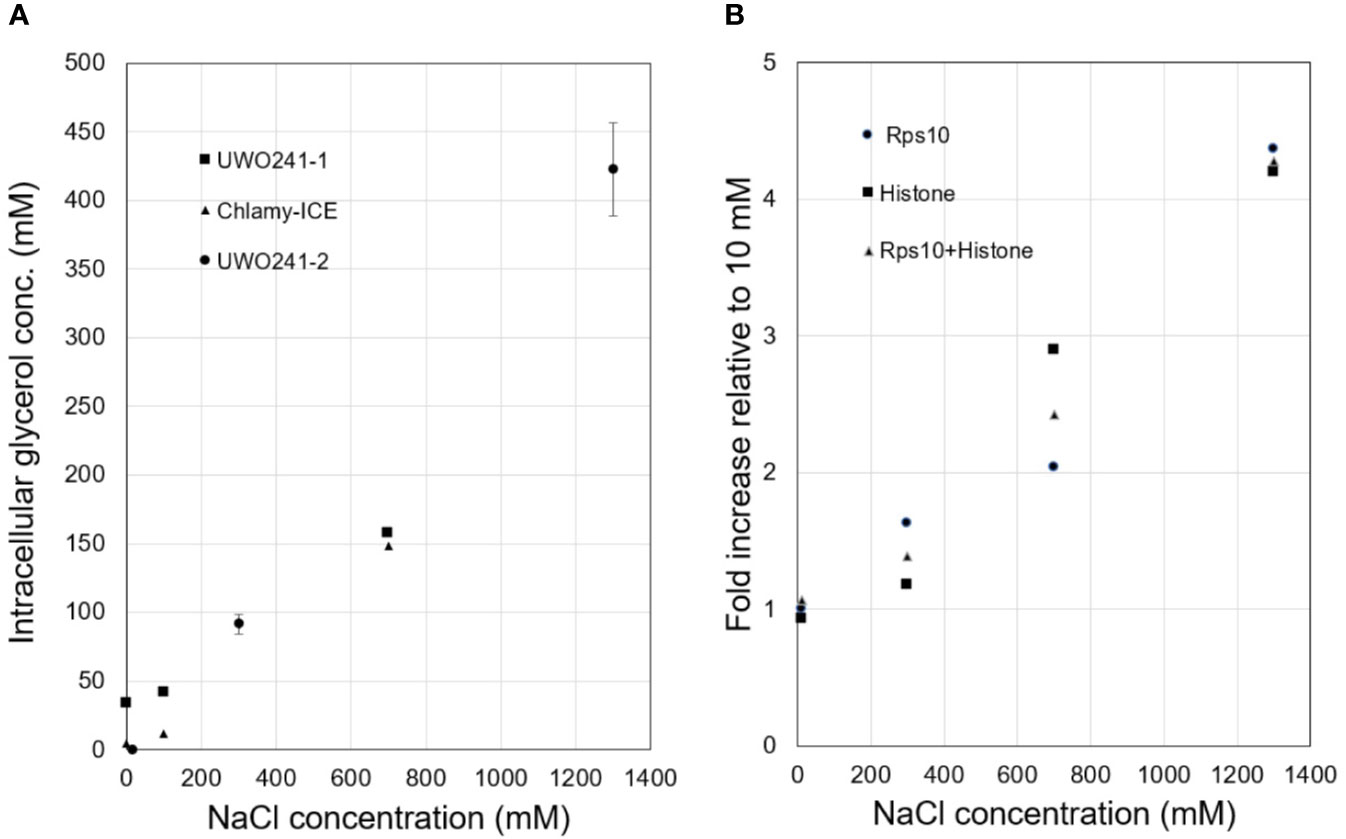
Figure 1 Responses of Lake Bonney Chlamydomonas species to increasing salinity. (A), Glycerol production in Chlamy-UWO and Chlamy-ICE. Two independent measurements were made for Chlamy-UWO. For UWO-2, three measurements were made for each sample. (B), Expression of PSP/GPDH isoform 2 of Chlamy-UWO using different reference genes.
We thus searched the Chlamy-UWO transcriptome for enzymes similar to the proposed PSP/GPDH that had been found in Dunaliella (He et al., 2007). Three complete isoforms could be assembled. We selected one of the isoforms to see if it could account for the increased glycerol production at increased salinity. The mRNA expression of Chlamy-UWO isoform 2 increased about four-fold as the NaCl concentration increased to 1,300 mM, regardless of the reference gene (Figure 1B). For the reference gene combination Rps10+Histone, whose points were more linear than those of the single reference genes, the increase was significant at the p <0.01 level (linear regression). This increase closely paralleled the increase in glycerol concentration. This, together with the recent findings in C. reinhardtii and D. salina, strongly supports the idea that the bidomain enzymes are a major source (and possibly the main source) of glycerol in Chlamy-UWO.
When the Chlamy-ICE transcriptome became available in 2016, it was also examined for homologs of the bidomain proteins. It appeared to have seven such homologs, but only five of them could be completely assembled (Table 1). For each of the Lake Bonney species, the expression levels of the different isoforms varied considerably (Table 1). In the approximately 600-a.a. conserved region that includes both enzyme domains, the identities of the Chlamy-UWO and Chlamy-ICE isoforms to C. reinhardtii GPD3 are about 60% (Table 1). To gain insights into the relationships of the enzymes in the three Chlamydomonas species, their exon/intron structures were compared. The numbers of introns (and their locations) differed considerably among the three Chlamydomonas species (Table 1), suggesting that their divergence occurred long ago.
The eight complete isoforms of Chlamy-UWO and Chlamy-ICE consist of an N-terminal chloroplast-targeting signal, a PSP domain and a C-terminal NAD+-dependent GDPH domain (Figure 2A), similar to the structure in Dunaliella (He Q. H. et al., 2020). A 3D model of isoform 1 of the Chlamy-UWO enzyme predicted from the structure of the D. salina enzyme is shown in Figure 2B. The region that could be modeled ranged from Thr60 at the start of the PSP domain (lower part of molecule in Figure 2B) to Phe639 near the end of the GPDH domain (upper part). The model is similar to the structure of the Dunaliella protein, with two nearly independent domains connected by a short link. The key residues forming the binding sites of DHAP, NAD+, and glycerol-3-P in Dunaliella (He Q. H. et al., 2020) are conserved in the Chlamy-UWO isoform (Figure S1). As a test of the accuracy of the model, the predicted binding sites of DHAP and NAD+ on the GPDH domain of the Chlamydomonas protein were compared to those in Dunaliella. The residues that form the binding sites in the two structures as well as their locations are nearly the same (Figure S2), supporting the accuracy of the model.
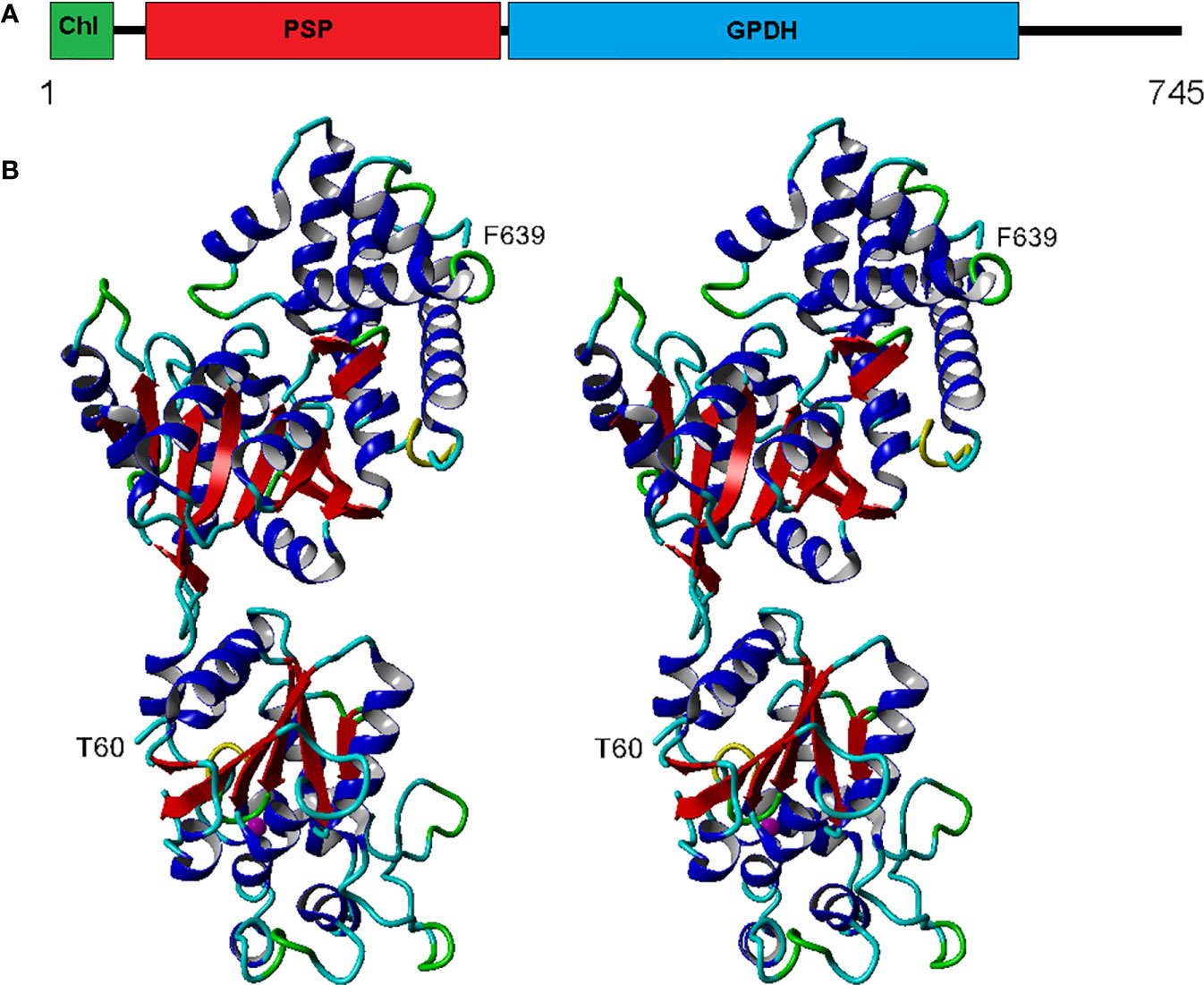
Figure 2 Structure of Chlamy-UWO PSP/GPDH isoform 1. (A), Domain structure, consisting of an N-terminal chloroplast-targeting signal, a phosphoserine phosphatase domain and a glycerol-3-phosphate dehydrogenase domain. (B), Stereoview of the molecule generated by Swiss-Model using the PSP/GPDH of D. salina as template. The lower portion, beginning at Thr60, shows the PSP domain with an embedded magnesium ion (olive). The upper portion, ending at Phe639, shows the GPDH domain. Color codes: red, beta sheet; blue, alpha helix; cyan, coil; green, turn; yellow, 310 helix.
To better understand the evolution of the Chlamydomonas bidomain proteins, a phylogenetic tree of the conserved PSP/GPDH domains was constructed by the neighbor-joining method (Figure 3). The tree was rooted on homologous proteins from two non-chlorophytes, a rhodophyte (Porphyridium purpureum) and a primitive relative of the fungi (Sphaeroforma arctica) that were recently submitted to GenBank. Within each Chlamydomonas species, the isoforms clustered with high bootstrap values, suggesting that within each species, the isoforms diverged from a single gene. C. reinhardtii, Chlamy-UWO and Chlamy-ICE formed a group that clusters separately from Dunaliella, but otherwise the three Chlamydomonas clusters appear weakly related, in agreement with the considerable differences in exon structures of the three species. These findings, together with the finding of the P. purpureum protein, raise the possibility that the chlorophyte bidomain proteins have an ancient origin, possibly dating back to a common ancestor of the chlorophytes and rhodophytes.
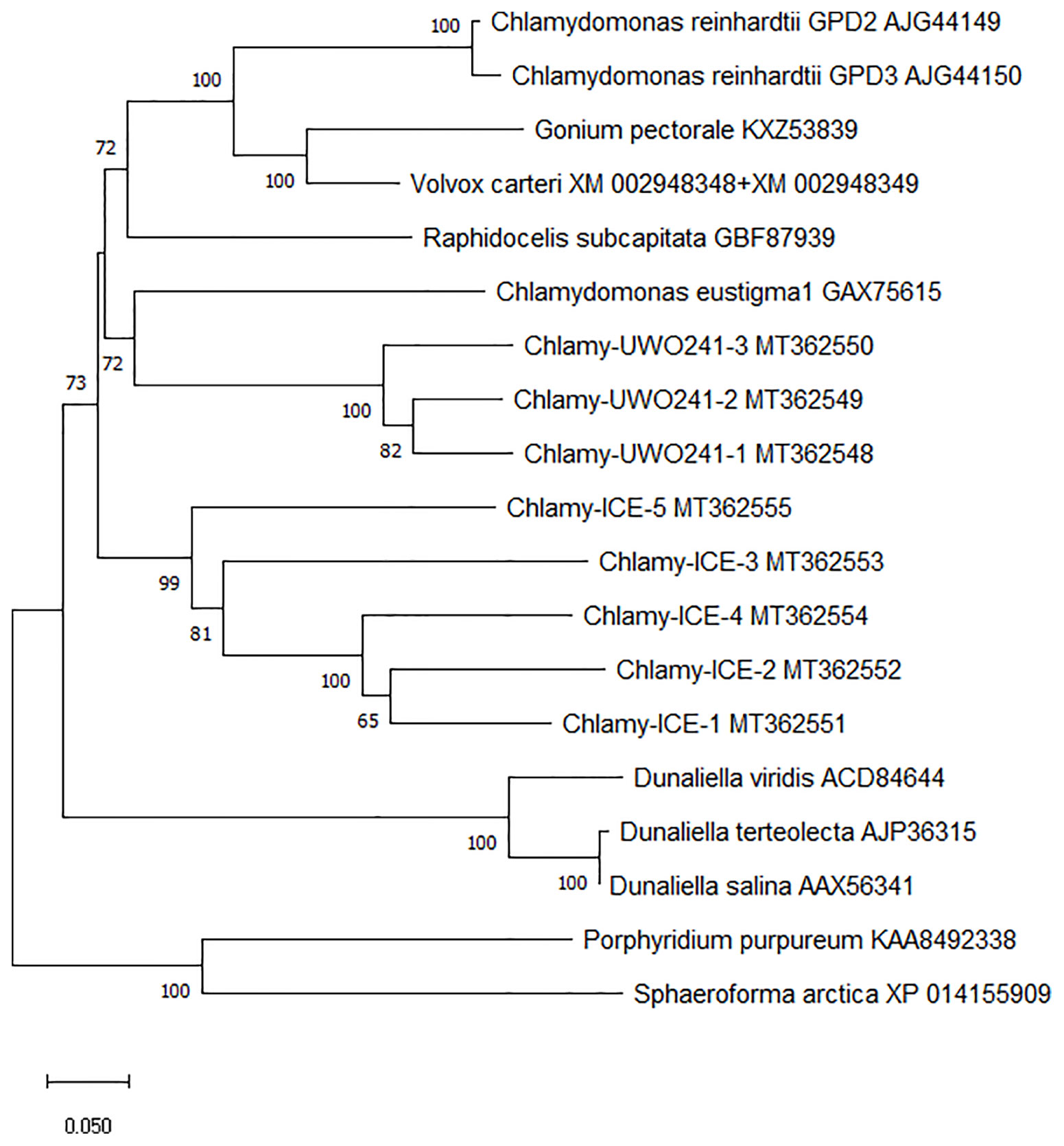
Figure 3 Neighbor-joining phylogenetic tree of the ~600 a.a. region containing the PSP and GPDH domains of PSP/GPDH bidomain enzymes of 10 chlorophytes. Bootstrap values less than 65% are not shown.
In view of the importance of glycerol as a compatible solute in animal and plant physiology, it is surprising that so little was known about how it was synthesized until recently. Previous studies referred to the enzymes involved only generically without identifying specific genes, and thus not really “nailing down” the pathway. Recently, the specific enzymes involved in glycerol synthesis have been identified in a bacterium (Larrouy-Maumus et al., 2013), a fish (Raymond, 2015), and, as highlighted here, two species of algae (Morales-Sanchez et al., 2017; He Q. H. et al., 2020). The algal enzymes are especially interesting because of their novel form in which the two enzymes needed to convert DHAP into glycerol are fused into a single enzyme. Although these double enzymes are largely limited to chlorophytes, the recent finding of homologous genes in two unicellular organisms (P. purpureum and S. arctica) that have links to some of the earliest eukaryotes (Mendoza et al., 2002; Bhattacharya et al., 2013) suggests that the fused gene may have evolved early in the history of eukaryotes.
The present results show that multiple copies of similar enzymes are found in two glycerol-producing Chlamydomonas species from Antarctica. Furthermore, the expression of a representative isoform from Chlamy-UWO was strongly upregulated by high salinity and the upregulation was associated with an increase in glycerol production. This result was supported by a recent proteomic study of Chlamy-UWO in which enzymes classified as NAD+-dependent GPDHs showed a six-fold increase when NaCl concentration was increased from near zero to 700 mM (Kalra et al., 2020). These results indicate that the bidomain enzyme is a major source of glycerol in Chlamy-UWO, and most likely in Chlamy-ICE as well. Using glycerol as an osmoprotectant, Chlamy-UWO and Chlamy-ICE can survive in salinities greater than the salinity of seawater (at least 700 and 1300 mM NaCl for Chlamy-ICE and Chlamy-UWO, respectively; equivalent to about 1.4x and 2.5x the salinity of seawater, respectively). The highest glycerol levels observed in Chlamy-ICE and Chlamy-UWO were 150 and 420 mM, respectively (Figure 1A). (Chlamy-ICE may not need higher levels of glycerol since it appears to live in shallower and thus less saline waters than Chlamy-UWO). For comparison, C. reinhardtii, a non-halotolerant species, can tolerate a maximum salinity of 200 mM NaCl (León and Galván, 1994) (but no more than 100 mM NaCl in our laboratory) and accumulates only about 26 mM glycerol in medium containing 100 mM KCl. (Husic and Tolbert, 1986). On the other hand, the marine coastal C. pulsatilla can produce much higher levels of glycerol, 1,450 mM when grown in double-strength seawater (Ahmad and Hellebust, 1986).
It should be noted that the glycerol levels in the Lake Bonney species are not enough to balance the osmolarity of the external medium. For example, at 1,300 mM NaCl (~2.6 Osm), the glycerol level in Chlamy-UWO was only about 0.4 Osm. Thus, the cells appear to be increasing the concentrations of additional osmolytes to maintain osmotic equilibrium. In addition to glycerol, proline and sucrose were found to accumulate at high levels in high-salt-grown Chlamy-UWO cultures (Kalra et al., 2020). In C. pulsatilla, in which glycerol contributed only about 57% to the intracellular osmolality, large increases in sodium and chloride ions made important contributions to maintaining osmotic equilibrium, but sugars and amino acids did not appear to have a significant role (Ahmad and Hellebust, 1986).
C. reinhardtii has four other GPDHs designated GPD1, GPD4, GPD5, and the mitochondrial mtGPD (Morales-Sanchez et al., 2017). tBlastn analyses of contigs assembled from the Chlamy-UWO and Chlamy-ICE transcriptomes indicate that both species have close homologs of each of these proteins (data not shown). We cannot rule out the possibility that these GPDHs also have a role in glycerol production, although GPD4 in C. reinhardtii was not upregulated by 125 mM NaCl (Morales-Sanchez et al., 2017).
That all the Lake Bonney PSP/GPDH isoforms have a chloroplast targeting signal suggests that they function in the chloroplast, as was concluded for the bidomain enzymes in C. reinhardtii (Morales-Sanchez et al., 2017) and D. salina (He Q. H. et al., 2020). This seems reasonable in view of the fact that the enzymes could act directly on DHAP produced by the Calvin cycle in the chloroplast. However, several chlorophyte bidomain proteins in the databank do not appear to have chloroplast signals as judged by ChloroP, which suggests they function in the cytoplasm.
Because Chlamy-UWO and Chlamy-ICE are living in an environment that is constantly exposed to freezing, as demonstrated by their expression of numerous ice-binding proteins (Raymond and Morgan-Kiss, 2013; Raymond and Morgan-Kiss, 2017), glycerol, by lowering the freezing point of the intracellular medium, could also act as a cryoprotectant. These might be the only organisms to use glycerol as both an osmoprotectant and a cryoprotectant.
Most of the known bidomain glycerol enzymes belong to chlorophytes, which in addition to Chlamydomonas and Dunaliella, include Gonium, Raphidocelis, Micractinium, Chlorella, Chloropicon, Volvox, and Tetrabaena. Many of these species are freshwater species and so it remains to be seen whether these species also produce glycerol and for what purpose. It also remains to be seen whether other marine Chlamydomonas species that make glycerol (Ahmad and Hellebust, 1986; Miyasaka et al., 1998) also have these enzymes. Finally, little is known about how Chlamydomonas spp. sense changes in salinity. Recent studies on osmosensing in microalgae (Suescún-Bolívar and Thomé, 2015; Charneco et al., 2018; He Q. et al., 2020) have implicated the possible involvement of mitogen-activated protein (MAP) kinases. The behavior of these genes in response to increases in salinity should thus be interesting.
In summary, we show that two Antarctic extremophiles, in addition to having several adaptations to low light and low temperature, also have adapted to high salinity by producing glycerol as an osmoprotectant (and possibly a cryoprotectant), and at least one of them (Chlamy-UWO) achieves this by using an unusual bidomain enzyme that can make glycerol directly from DHAP.
Publicly available datasets were analyzed in this study. The Chlamydomonas sp. UWO241 transcriptome is available under GenBank accession PRJNA575885.
All authors contributed equally to this study.
This study was partially funded by NSF grant 1637708 and DOE Grant DE-SC0019138 to RM-K.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank David Smith, Marina Cvetkovska, Xi Zhang, and Norm Hüner (University of Western Ontario) for providing additional sequence data for Chlamy-ICE. JR thanks the School of Life Sciences, UNLV for providing laboratory facilities for this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01259/full#supplementary-material
Table S1 | qPCR primers used in this study.
Figure S1 | Alignment of PSP/GPDH domains of Chlamy-ICE and Dunaliella salina showing conservation of ligand binding sites. The binding sites in Dunaliella are from He Q. H. et al. (2020). Red underline, PSP domain; blue underline, GPDH domain. Blue asterisks, glycerol-3 phosphate; green asterisks, NAD; red asterisks, DHAP. Black background, identical a.a.s; gray background, similar a.a.s. Numbering refers to position in the Chlamy-ICE protein. The region shown (from T60 to F639) is the same region shown in Figure 2B.
Figure S2 | Stereo views comparing the ligand binding sites in the GPDH domain of the PSP/GPDH protein in Dunaliella salina (top) and Chlamy-UWO isoform 2 (bottom). The Dunaliella structure was determined by X-ray crystallography. The Chlamy-UWO structure is predicted by SwissModel based on the Dunaliella structure. The binding sites for Dunaliella were obtained from He Q. H. et al. (2020). The amino acid residues forming the binding sites are shown. Red, DHAP binding site; green, NAD binding site. These amino acid residues correspond to the red and green asterisks, respectively, in Figure S1.
Ahmad, I., Hellebust, J. A. (1986). The role of glycerol and inorganic-ions in osmoregulatory responses of the euryhaline flagellate Chlamydomonas-pulsatilla wollenweber. Plant Physiol. 82, 406–410. doi: 10.1104/pp.82.2.406
Bhattacharya, D., Price, D. C., Chan, C. X., Qiu, H., Rose, N., Ball, S., et al. (2013). Genome of the red alga Porphyridium purpureum. Nature Commun. 4, 1941.
Ben-Amotz, A., Avron, M. (1973). The role of glycerol in the osmotic regulation of the halophilic alga Dunaliella parva. Plant Physiol. 51, 875–878. doi: 10.1104/pp.51.5.875
Bennett, V. A., Sformo, T., Walters, K., Toien, O., Jeannet, K., Hochstrasser, R., et al. (2005). Comparative overwintering physiology of Alaska and Indiana populations of the beetle Cucujus clavipes (Fabricius): roles of antifreeze proteins, polyols, dehydration and diapause. J. Exp. Biol. 208, 4467–4477. doi: 10.1242/jeb.01892
Bentz, B. J., Mullins, D. E. (1999). Ecology of mountain pine beetle (Coleoptera: Scolytidae) cold hardening in the intermountain west. Environ. Entomol. 28, 577–587. doi: 10.1093/ee/28.4.577
Caparros-Martin, J. A., Reiland, S., Kochert, K., Cutanda, M. C., Culianez-Macia, F. A. (2007). Arabidopsis thaliana AtGpp1 and AtGpp2: two novel low molecular weight phosphatases involved in plant glycerol metabolism. Plant Mol. Biol. 63, 505–517. doi: 10.1007/s11103-006-9104-0
Charneco, G. O., Parages, M. L., Camarena-Gómez, M. T., Jiménez, C. (2018). Phosphorylation of MAP Kinases crucially controls the response to environmental stress in Dunaliella viridis. Environ. Exp. Bot. 156, 203–213. doi: 10.1016/j.envexpbot.2018.08.030
Cook, G., Teufel, A., Kalra, I., Li, W., Wang, X., Priscu, J., et al. (2019). The Antarctic psychrophiles Chlamydomonas spp. UWO241 and ICE-MDV exhibit differential restructuring of photosystem I in response to iron. Photosynthesis Res. 141, 209–228. doi: 10.1007/s11120-019-00621-0
Cvetkovska, M., Hüner, N. P. A., Smith, D. R. (2017). Chilling out: the evolution and diversification of psychrophilic algae with a focus on Chlamydomonadales. Polar Biol. 40, 1169–1184. doi: 10.1007/s00300-016-2045-4
Dolhi, J. M., Maxwell, D. P., Morgan-Kiss, R. M. (2013). Review: the Antarctic Chlamydomonas raudensis: an emerging model for cold adaptation of photosynthesis. Extremophiles 17, 711–722. doi: 10.1007/s00792-013-0571-3
Emanuelsson, O., Nielsen, H., Von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. doi: 10.1110/ps.8.5.978
Fraser, J. D., Bonnett, T. R., Keeling, C. I., Huber, D. P. W. (2017). Seasonal shifts in accumulation of glycerol biosynthetic gene transcripts in mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Curculionidae), larvae. Peerj 5, e3284. doi: 10.7717/peerj.3284
He, Q., Qiao, D., Bai, L., Zhang, Q., Yang, W., Li, Q., et al. (2007). Cloning and characterization of a plastidic glycerol 3-phosphate dehydrogenase cDNA from Dunaliella salina. J. Plant Physiol. 164, 214–220. doi: 10.1016/j.jplph.2006.04.004
He, Y., Meng, X., Fan, Q., Sun, X., Xu, Z., Song, R. (2009). Cloning and characterization of two novel chloroplastic glycerol-3-phosphate dehydrogenases from Dunaliella viridis. Plant Mol. Biol. 71, 193–205. doi: 10.1007/s11103-009-9517-7
He, Q. H., Toh, J. D., Ero, R., Qiao, Z., Kumar, V., Serra, A., et al. (2020). The unusual di-domain structure of Dunaliella salina glycerol-3-phosphate dehydrogenase enables direct conversion of dihydroxyacetone phosphate to glycerol. Plant J. 102, 153–164. doi: 10.1111/tpj.14619
He, Q., Lin, Y., Tan, H., Zhou, Y., Wen, Y., Gan, J., et al. (2020). Transcriptomic profiles of Dunaliella salina in response to hypersaline stress. BMC Genomics 21, 115. doi: 10.1186/s12864-020-6507-2
Husic, H. D., Tolbert, N. E. (1986). Effect of Osmotic Stress on Carbon Metabolism in Chlamydomonas reinhardtii. Accumulation Glycerol. as an Osmoregul. Solute 82, 594–596. doi: 10.1104/pp.82.2.594
Kalra, I., Wang, X., Cvetkovska, M., Jeong, J., Mchargue, W., Zhang, R., et al. (2020). Chlamydomonas sp. UWO 241 exhibits high cyclic electron flow and rewired metabolism under high salinity. Plant Physiol. 183, 588–601. doi: 10.1104/pp.19.01280
Krieger, E., Joo, K., Lee, J., Lee, J., Raman, S., Thompson, J., et al. (2009). Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins-Structure Funct. Bioinf. 77, 114–122. doi: 10.1002/prot.22570
Larrouy-Maumus, G., Biswas, T., Hunt, D. M., Kelly, G., Tsodikov, O. V., De Carvalho, L. P. S. (2013). Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 110, 11320–11325. doi: 10.1073/pnas.1221597110
Layne, J. R. (1999). Freeze tolerance and cryoprotectant mobilization in the gray treefrog (Hyla versicolor). J. Exp. Zool. 283, 221–225. doi: 10.1002/(SICI)1097-010X(19990215)283:3<221::AID-JEZ1>3.0.CO;2-Q
León, R., Galván, F. (1994). Halotolerance studies on Chlamydomonas reinhardtii: glycerol excretion by free and immobilized cells. J. Appl. Phycol. 6, 13–20. doi: 10.1007/BF02185898
Li, W., Podar, M., Morgan-Kiss, R. M. (2016). Ultrastructural and single-cell-level characterization reveals metabolic versatility in a microbial eukaryote community from an ice-covered Antarctic lake. Appl. Environ. Microbiol. 82, 3659–3670. doi: 10.1128/AEM.00478-16
Liu, C. L., Wu, G. T., Huang, X. H., Liu, S. H., Cong, B. L. (2012). Validation of housekeeping genes for gene expression studies in an ice alga Chlamydomonas during freezing acclimation. Extremophiles 16, 419–425. doi: 10.1007/s00792-012-0441-4
Mendoza, L., Taylor, J. W., Ajello, L. (2002). The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Ann. Rev. Microbiol. 56, 315–344.
Miyasaka, H., Ikeda, K. (1997). Osmoregulating mechanism of the halotolerant green alga Chlamydomonas, strain HS-5. Plant Sci. 127, 91–96. doi: 10.1016/S0168-9452(97)00125-8
Miyasaka, H., Ohnishi, Y., Akano, T., Fukatsu, K., Mizoguchi, T., Yagi, K., et al. (1998). Excretion of glycerol by the marine Chlamydomonas sp. strain W-80 in high CO2 cultures. J. Ferment. Bioeng. 85, 122–124. doi: 10.1016/S0922-338X(97)80367-4
Modig, T., Granath, K., Adler, L., Liden, G. (2007). Anaerobic glycerol production by Saccharomyces cerevisiae strains under hyperosmotic stress. Appl. Microbiol. Biotechnol. 75, 289–296. doi: 10.1007/s00253-006-0821-8
Morales-Sanchez, D., Kim, Y., Terng, E. L., Peterson, L., Cerutti, H. (2017). A multidomain enzyme, with glycerol-3-phosphate dehydrogenase and phosphatase activities, is involved in a chloroplastic pathway for glycerol synthesis in Chlamydomonas reinhardtii. Plant J. 90, 1079–1092. doi: 10.1111/tpj.13530
Morgan, R. M., Ivanov, A. G., Priscu, J. C., Maxwell, D. P., Huner, N. P. A. (1998). Structure and composition of the photochemical apparatus of the antarctic green alga, Chlamydomonas subcaudata. Photosynthesis Res. 56, 303–314. doi: 10.1023/A:1006048519302
Morgan-Kiss, R., Ivanov, A. G., Williams, J., Mobashsher, K., Huner, N. P. A. (2002). Differential thermal effects on the energy distribution between photosystem II and photosystem I in thylakoid membranes of a psychrophilic and a mesophilic alga. Biochim. Biophys. Acta 1561, 251–265. doi: 10.1016/S0005-2736(02)00352-8
Neale, P. J., Priscu, J. C. (1995). The photosynthetic apparatus of phytoplankton from a perennially ice-covered Antarctic lake: acclimation to an extreme shade environment. Plant Cell Physiol. 36, 253–263. doi: 10.1093/oxfordjournals.pcp.a078757
Raymond, J. A., Morgan-Kiss, R. (2013). Separate Origins of Ice-Binding Proteins in Antarctic Chlamydomonas Species. PloS One 8 (3), e59186. doi: 10.1371/journal.pone.0059186
Raymond, J. A., Morgan-Kiss, R. (2017). Multiple ice-binding proteins of probable prokaryotic origin in an antarctic lake alga, Chlamydomonas sp. ICE-MDV (Chlorophyceae). J. Phycol. 53, 848–854. doi: 10.1111/jpy.12550
Raymond, J. A. (1992). Glycerol is a colligative antifreeze in some northern fishes. J. Exp. Zool. 262, 347–352. doi: 10.1002/jez.1402620316
Raymond, J. (1995). Glycerol synthesis in the rainbow smelt Osmerus mordax. J. Exp. Biol. 198, 2569–2573.
Raymond, J. A. (2015). Two potential fish glycerol-3-phosphate phosphatases. Fish Physiol. Biochem. 41, 811–818. doi: 10.1007/s10695-015-0048-7
Spigel, R. H., Priscu, J. C., Obryk, M. K., Stone, W., Doran, P. T. (2018). The physical limnology of a permanently ice-covered and chemically stratified Antarctic lake using high resolution spatial data from an autonomous underwater vehicle. Limnol. Oceanogr. 63, 1234–1252. doi: 10.1002/lno.10768
Stahl, S. E. (2014). Photooxidative stress response in mesophilic and psychrophilic strains of Chlamydomonas raudensis: a comparative study. Thesis (Miami, OH: Miami University).
Suescún-Bolívar, L. P., Thomé, P. E. (2015). Osmosensing and osmoregulation in unicellular eukaryotes. World J. Microbiol. Biotechnol. 31, 435–443. doi: 10.1007/s11274-015-1811-8
Sussman, I., Avron, M. (1981). Characterization and partial-purification of DL-glycerol-1-phosphatase from Dunaliella salina. Biochim. Biophys. Acta 661, 199–204. doi: 10.1016/0005-2744(81)90004-8
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Wang, Z. X., Zhuge, J., Fang, H. Y., Prior, B. A. (2001). Glycerol production by microbial fermentation: A review. Biotechnol. Adv. 19, 201–223. doi: 10.1016/S0734-9750(01)00060-X
Keywords: Chlamydomonas, glycerol synthesis, phosphoserine phosphatase, glycerol-3-phosphate dehydrogenase, Antarctica, Lake Bonney
Citation: Raymond JA, Morgan-Kiss R and Stahl-Rommel S (2020) Glycerol Is an Osmoprotectant in Two Antarctic Chlamydomonas Species From an Ice-Covered Saline Lake and Is Synthesized by an Unusual Bidomain Enzyme. Front. Plant Sci. 11:1259. doi: 10.3389/fpls.2020.01259
Received: 18 June 2020; Accepted: 30 July 2020;
Published: 20 August 2020.
Edited by:
Linda Nedbalová, Charles University, CzechiaReviewed by:
Aharon Oren, Hebrew University of Jerusalem, IsraelCopyright © 2020 Raymond, Morgan-Kiss and Stahl-Rommel. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James A. Raymond, cmF5bW9uZEB1bmx2Lm5ldmFkYS5lZHU=
†Present address: Sarah Stahl-Rommel, JES Tech, Houston, TX, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.