- 1Department of Botany, Indira Gandhi National Tribal University (IGNTU), Amarkantak, India
- 2Department of Life Science, Central University of Karnataka, Kalaburagi, India
- 3Department of Plant Sciences, School of Life Sciences, University of Hyderabad (UoH), Hyderabad, India
- 4Department of Biotechnology, ICAR-Indian Institute of Rice Research (IIRR), Hyderabad, India
- 5ICAR-Crop Improvement Division, Indian Institute of Pulses Research (IIPR), Kanpur, India
Rice is the most important food crop worldwide and sustainable rice production is important for ensuring global food security. Biotic stresses limit rice production significantly and among them, bacterial blight (BB) disease caused by Xanthomonas oryzae pv. oryzae (Xoo) is very important. BB reduces rice yields severely in the highly productive irrigated and rainfed lowland ecosystems and in recent years; the disease is spreading fast to other rice growing ecosystems as well. Being a vascular pathogen, Xoo interferes with a range of physiological and biochemical exchange processes in rice. The response of rice to Xoo involves specific interactions between resistance (R) genes of rice and avirulence (Avr) genes of Xoo, covering most of the resistance genes except the recessive ones. The genetic basis of resistance to BB in rice has been studied intensively, and at least 44 genes conferring resistance to BB have been identified, and many resistant rice cultivars and hybrids have been developed and released worldwide. However, the existence and emergence of new virulent isolates of Xoo in the realm of a rapidly changing climate necessitates identification of novel broad-spectrum resistance genes and intensification of gene-deployment strategies. This review discusses about the origin and occurrence of BB in rice, interactions between Xoo and rice, the important roles of resistance genes in plant’s defense response, the contribution of rice resistance genes toward development of disease resistance varieties, identification and characterization of novel, and broad-spectrum BB resistance genes from wild species of Oryza and also presents a perspective on potential strategies to achieve the goal of sustainable disease management.
Introduction
Rice (Oryza sativa L.) is an important staple food crop for more than 3.5 billion people across the world (Khush, 2005), provides 27 percent of the calories and 20 percent of protein required for the global population, and remains a major source of nutrition in developing and underdeveloped countries (FAO, 2004). Notwithstanding the progress witnessed in rice improvement over the last seven decades, the present rate of increase in rice yields are not adequate to keep pace with a rapidly growing population. The global demand of rice is estimated to rise by 26% in next 25 years, demanding an increase in its production from 676 million tones (mt) to 852 mt over the same period across the globe (Khush, 2013). Exacerbating the scenario, this production goal has to be achieved in the face of shrinking agricultural lands, dwindling water resources, declining soil productivity, and, most importantly, increasing cost of labor and other inputs. In parallel, we also need to improve rice production incrementally to combat and overcome the constantly evolving pathogen and pest populations and develop resilience in rice in the realm of rapidly changing climatic conditions.
Biotic stresses such as insect pests (brown plant hoppers, stem borers, etc.) and diseases such as bacterial blight caused by Xanthomonas oryzae pv. oryzae, rice blast caused by Magnaporthe oryzae and sheath blight caused by Rhizoctonia solani substantially reduce rice yields globally (Savary et al., 1998). The reduction in rice yield by bacterial blight (BB) is reported to be 50% (Khush et al., 1989), and during severe infection, it can reduce yield up to 81% (Srinivasan and Gnanamanickam, 2005), making it one of the most devastating diseases of rice (Ou, 1985). The conventional remedies recommended for managing BB disease, such as use of chemicals and antibiotics, biological control agents, and cultural practices, have limited utility and remain ineffective, especially when the disease occurs in epidemic proportion (Sundaram et al., 2008; Gnanamanickam, 2009).
Improving host-plant immunity has been considered as one of the best choices available for achieving economical and sustainable management of BB disease in a durable manner (Mundt, 2014; Pradhan et al., 2015). Understanding host resistance mechanisms and immunity against disease-causing pathogens like Xoo and their mutual interactions has been a topic of intense research in the past two decades. The evolving tools and techniques of plant molecular biology have been instrumental in getting vital insights into host-pathogen interactions and developing strategies for broad-spectrum, durable resistance. Chen and Ronald (2011) highlighted the nuances associated with innate immunity of rice against diverse pathogen elicitors. Typically, on encountering a biotic stress, the host plant reduces or enhances its susceptibility to a pathogen due to the existence of molecular cross talks between the pathogens themselves and between the pathogens and their host plant (Atkinson and Urwin, 2012). Interactions among pathways associated with response and tolerance/resistance to abiotic and biotic stresses have been established, and new insights have been gained on hormonal signaling pathways associated with antagonistic or synergistic interactions between biotic and abiotic stresses (Denancé et al., 2013; Jain et al., 2017). Therefore, understanding host-pathogen communication has been a longtime pursuit of plant biologists and plant pathologists.
Modern omics approaches like genomics, transcriptomics, proteomics, metabolomics, interactomics, etc. can be helpful in identification of the genes and their products, which are involved in pathogen perception by the host and also the response manifested by the host against the pathogen attack. Several resistance genes from different plant species have been identified, molecular mapped, cloned, and characterized (Sanseverino et al., 2010; Gururani et al., 2012). These genes have been assembled into five classes based on predicted protein domains (Gururani et al., 2012). Hm1 gene of maize, which encodes a reductase, represents the first class. The HC toxin of Cochliobolous carbonum race 1 is inactivated by Hm1 gene (Johal and Briggs, 1992). Pto gene belong to the second class, which encode membrane-associated serine-threonine kinase. It provides resistance to Pseudomonas syringae pv. tomato. The cytoplasmic receptor kinase protein represents the third class. It includes Rps2 and Rpm1, N, L6, Prf, and Xa1 gene of Arabidopsis, tobacco, flax, tomato, and rice, respectively (Salmeron et al., 1996; Yoshimura et al., 1998). Tomato Cf gene represents the fourth class wherein Cf gene encodes LRR motifs in extracellular domain and a short C-terminal tail in the intracellular domain (Dixon et al., 1996). The rice Xa21 represent the fifth class, which encodes a receptor kinase like protein, and it confers broad spectrum resistance to Xoo (Song et al., 1995).
In May 2014, the genome sequences of 3,000 strains of rice have been published by a joint effort of Chinese Academy of Agricultural Science (CAAS) and International Rice Research Institute, Philippines (Li et al., 2014). This project undoubtedly will provide the clear insights on utilizing host resistance genes belonging to one or more above mentioned classes of resistance genes for obtaining durable resistance against disease like BB (Li et al., 2014). In this article, we have reviewed the molecular mechanisms that are associated with interaction between rice and Xoo, pathways of resistance and susceptibility in rice, and the application of modern biotechnology approaches for breeding durable, broad-spectrum BB resistance rice varieties.
Molecular Events Associated With Infection By Xoo And Resistance Against The Pathogen
Plant-Pathogen Communication and Symptomatology
Xanthomonas oryzae pv. oryzae (Xoo) is a gram negative, non-spore forming, rod shaped bacterium, which is motile with a single polar flagellum. Individual cells show a range from around 0.7 to 2.0 µm in length and from 0.4 to 0.7 µm in width and require an optimal temperature between 25 and 30°C for their growth (Bradbury, 1984). Unlike mammals, plants have a complex cell wall and bacteria need to get through this barrier to gain access to nutrients. This is achieved by the bacteria through destruction of the cell wall barrier by means of secreting cell wall degrading enzymes (CDEs) such as lipase/esterase (LipA), cellulase (ClsA), cellobiosidase (CbsA), xylanase (XynB), etc., which is one of the most effective virulence strategy adapted by bacterial pathogens (Agrios, 1997; Rajeshwari et al., 2005; Jha et al., 2007; Malukani et al., 2020). However, receptors like WAKL21.2 predict the damage caused by Xoo CDEs and recruit the components of immunity (Malukani et al., 2020). Xoo gets into rice leaf tissues generally via wounds or natural openings such as hydathodes (Ou, 1985). Subsequently, it multiplies and flourishes in the intercellular spaces (apoplast) beneath the epithelial cells. Thereafter, it disseminates to other parts of plants through the xylem vessels (Noda and Kaku, 1999). After few days, xylem vessels are filled by bacteria and its exo-polysacchride (EPS). The bacterial with its exudates can be observed on the leaf surface (Figures 1 and 2), as they come out through hydathodes. This is considered as a clear-cut symptom of the disease and most importantly it serves as a source of secondary inoculum (Mew et al., 1993).
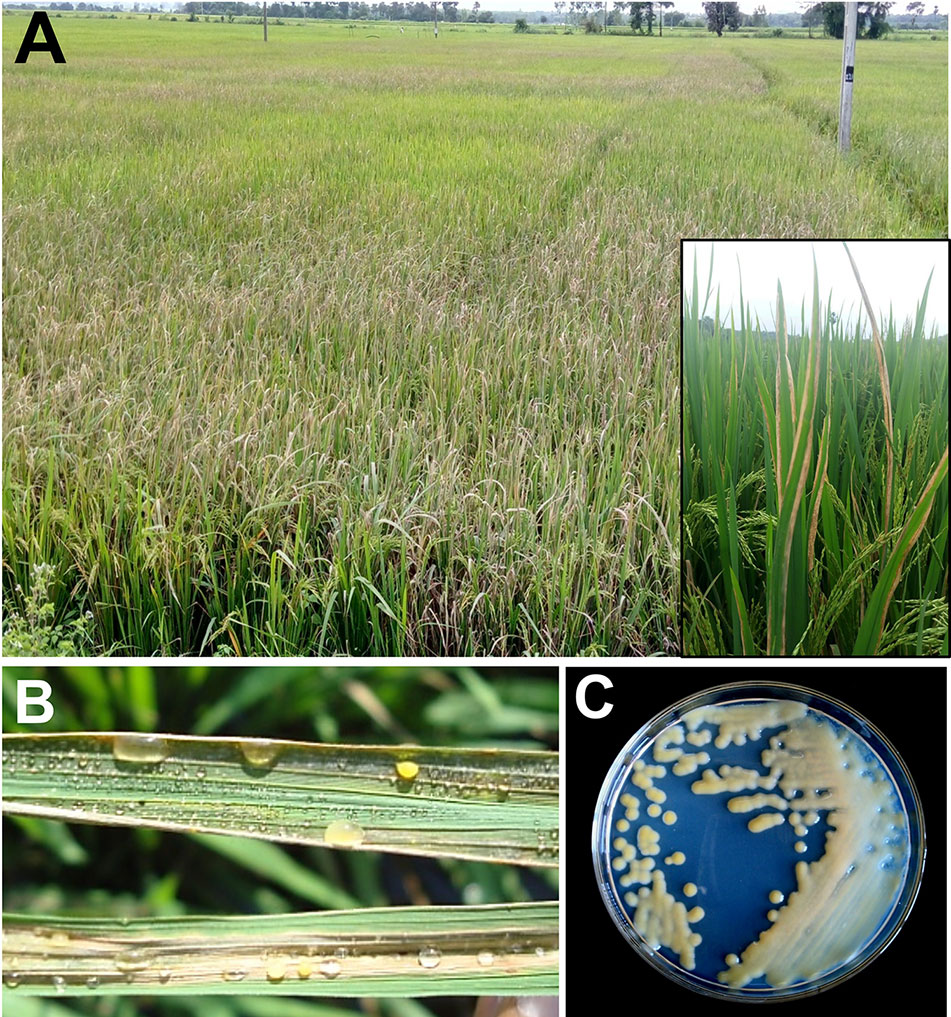
Figure 1 Bacterial blight of rice. (A) Closer view of infected plants; (B) bacterial ooze on infected leaf; (C) Xanthomonas oryzae pv. oryzae (Xoo) colonies on culture plate.
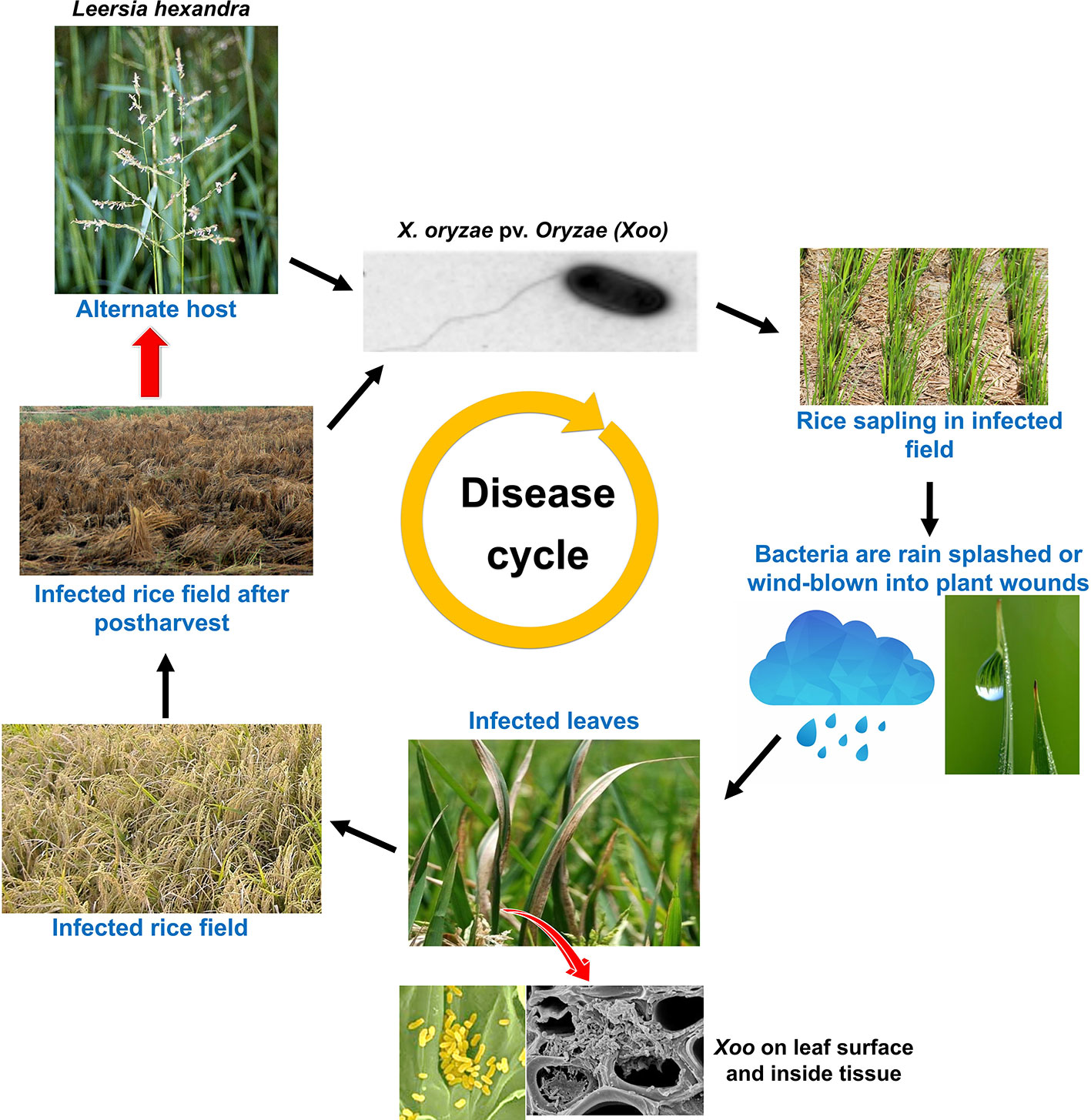
Figure 2 Disease life cycle of the rice bacterial blight caused by bacteria- Xoo, including the influence of disease secondary host plant on disease severity.
Many plant pathogenic bacteria including Xoo use type III secretion system to transport virulence proteins and enzymes to disrupt host signaling and hijacks host metabolism for their growth and development. The proteins secreted by type III secretion system are called effector protein, which includes transcription activator like (TAL) effector and non-TAL effector proteins (White et al., 2009; Scholze and Boch, 2011). TAL effector protein supports the proliferation of Xoo and establishment of infection in host plant by altering host transcription machinery through upregulation of selected host genes required for multiplication of the pathogen, whereas non-TAL effector protein promotes virulence through suppression of host innate immunity. Few TAL effectors and their cognate effector (E) genes, viz., AvrXa10/Xa10, AvrXa23/Xa23, and AvrXa27/Xa27, have been cloned from rice (Hopkins et al., 1992; Gu et al., 2005; Tian et al., 2014), and virulence function of others TAL effectors, TalC, pthXo1, pthXo2, pthXo3, pthXo6, and pthXo7, have been studied. The PthXo1 TAL effector persuades the expression of host susceptibility gene Os8N3 (nodulin 3 gene family; renamed as OsSWEET11), which encodes a membrane protein associated with sugar transport (Yang B. et al., 2006). In the cultivar Nipponbare, it activates virulence by inducing Os8N3/SWEET11 gene (Figure 3A) (Yang S. et al., 2006). It has been reported that the recessive xa13 resistance allele arose due to mutation in the promoter region of Os8N3/SWEET11 (Chu et al., 2006). Another host susceptibility gene, SWEET14 is targeted by many TALEs (viz., AvrXa7, PthXo3, TalC, and TalF; Oliva et al., 2019) to trigger the release of sugar molecules in the apoplast required by the pathogen as nutrient source (Streubel et al., 2013). Mutation within effector binding element (EBE) of AvrXa7 in the Os11N3/SWEET14 promoter resulted in disease resistance against Xoo (Li T. et al., 2012). Similarly, deletion in the EBE of Xa7 in wild rice confers broad spectrum resistance to BB (Hutin et al., 2015). Two other TAL effectors PthXo6 and PthXo7 promote the transcription of host genes OsTFX1 and OsTFIIAγ1, respectively (Sugio et al., 2007). One more TAL effector gene pthXo8 (homolog of pthXo6) has been found to be involved in manipulation of small RNA pathway of the host (Yang and White, 2004).
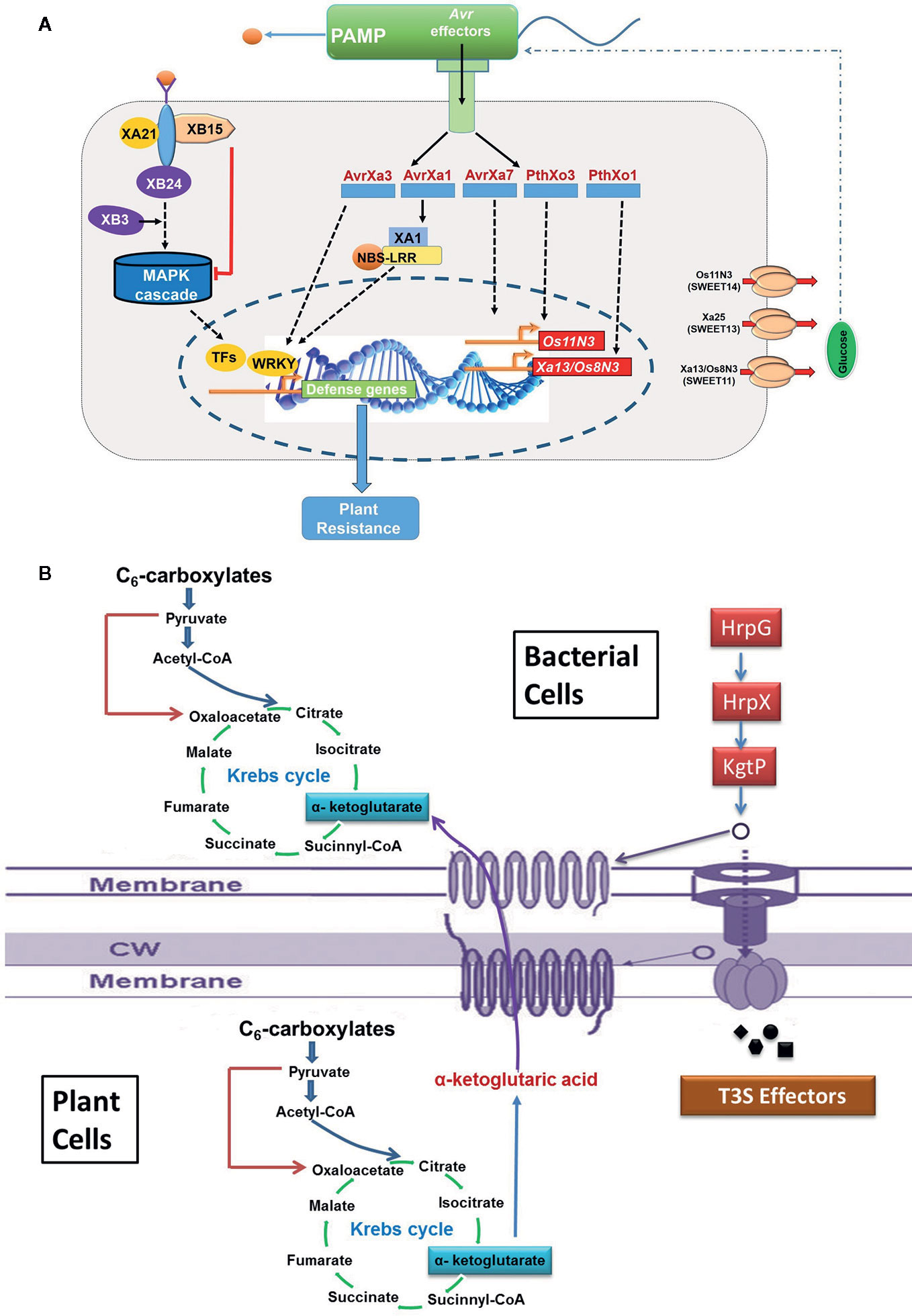
Figure 3 The schematic representation for the molecular signaling involved during host and pathogen interaction. (A) Hijacking host key genes (SWEET11/13/14) by pathogen; (B) utilization of host metabolic resources like Kreb’s intermediate by pathogen.
Xoo genome also encodes a type II secretion system. Proteins secreted by type II secretion system possess secretion signal at N terminal and are transported to periplasmic space (Voulhoux et al., 2001; Jha et al., 2005). Type II secreted (TIIS) proteins are mostly toxins and enzymes that targets diverse components of the host defense system. In addition, type II secretion system secretes variety of carbohydrate degrading enzymes like cellulases, pectate lyases, xylanases, and polygalacturonases (Cianciotto and White, 2017), thus weakening the cell wall. It has been noted that rice plants perceive type II protein and in response, hypersensitive reaction is inducted (Jha et al., 2005). Mutation in genes encoding type II secretion system diminishes the virulence of Xoo in the host, thus clearly demonstrating the importance of TIIS in plant pathogenesis (Cianciotto and White, 2017). Xoo targets and alters different host gene products and TAL effectors to amend the host physiology to have favorable effects on host susceptibility. The ability of plants to detect the adverse effects and speed of response against the pathogen determines the host fate. It has also been found that plants down-regulate the level of auxin in response to pathogen attack for enhancing disease resistance (Navarro et al., 2006). In addition, ABA suppresses the basal defense mechanism of rice against virulent Xoo strains and likely to function as virulence factor. An enhanced level of ABA is known to increase susceptibility of rice to Xoo by mediating the SA defense mechanism (Xu et al., 2013). Xoo also produce autoinducers (hormone like molecule) to detect the local population density (quorum sensing; QS) (Karatan and Watnick, 2009; Pradhan and Chatterjee, 2014). Based on the quorum sensing, bacteria regulate their gene expression pattern to effectively parasitize the plant cells (Karatan and Watnick, 2009). Various signaling molecules engage in QS including N-acylhomoserine lactones (AHLs), autoinducers-2 (AI-2), diffusible signal factors (DSFs), and oligopeptides (Deng et al., 2010). Further, the Xoo secretes large amount of extracellular polysaccharide (EPS; extra cellular polysaccharide high molecular-weight sugar molecules), which choke the xylem and cause typical wilting symptoms. EPS has an important role to play as it enhances pathogenicity by protecting the bacteria from antimicrobial compounds of the host plants (Leigh and Coplin, 1992; Dharmapuri and Sonti, 1999). All of the above mechanisms contribute together to promote pathogenesis (Leigh and Coplin, 1992). A comprehensive representation of the mode of Xoo infection in rice is illustrated in Figures 1 and 2.
Host-Mediated Disease Resistance
The pathogen infects by evading or compromising the host defense responses. In doing so, the pathogen escapes the recognition by host receptors, mitigates or inhibits downstream signaling in the host, or takes over the host signaling mechanism to favor establishment of disease. To counter this, plants have also developed several receptors and sensors that interact with microbial components and nullify their effect. A unique strategy has been adopted to improve the immunity in crops by enhancing the recognition spectrum of the host plant’s own immune system (Lacombe et al., 2010). It involves the transfer of pathogen-associated molecular pattern (PAMP) like perception system across plant families and provides broad-spectrum disease resistance. Plant immunity has been categorized into two levels based on microbial component recognition. The first and second level of immunity is known as basal immunity [PAMPs-triggered immunity (PTI)] and gene-for-gene resistance [effector triggered immunity (ETI)], respectively (Jones and Dangl, 2006; Monaghan and Zipfel, 2012). Both PTI and ETI are mediated by receptor kinase proteins localized in plasma membrane and nucleotide binding (NB) leucine-rich repeat (LRR) proteins and other factors localized in cytoplasm respectively (Jones and Dangl, 2006; Macho and Zipfel, 2014). PTI provides quantitative resistance, and ETI provides qualitative resistance in plant pathogen interaction (Zhang and Wang, 2013). Rice-Xoo interaction is an exclusive example of qualitative resistance, i.e., major gene conferred resistance (Zhang and Wang, 2013). The major disease resistance genes of rice, which provide resistance to Xoo, fall under either ETI or PTI or may fall under an additional mechanism, different from ETI or PTI (Hu et al., 2017).
One important and unique feature that is typical of qualitative resistance of rice against Xoo is that one third of the major disease resistance genes are recessive genetically (Zhang and Wang, 2013; Hu et al., 2017). Out of the 44 known R-genes, at least 11 have been cloned and characterized (Xa1, Xa3/Xa26, Xa4, xa5, Xa10, xa13, Xa21, Xa23, xa25, Xa27, and xa41) (Table 1) (Tian et al., 2014; Wang et al., 2014a; Wang et al., 2014b; Cao et al., 2018). Of the remianing R genes, at least nine have been fine-mapped on different chromosome so far, viz., Xa2, Xa4, Xa7, Xa22, Xa30, Xa33, Xa38, Xa39, and Xa40 (http://www.mshigen.nig.ac.jp/rice/oryzabase/gene/list). Interestingly, mutation in rice lines have also produced important R genes/alleles such as Xa1, xa5, xa13, Xa23, xa25, Xa26/Xa3, Xa27, and xa41 (Nakai et al., 1988; Song et al., 1995; Yoshimura et al., 1998; Gao et al., 2002; Lee et al., 2003; Iyer et al., 2004; Sun et al., 2004; Gu et al., 2005; Chu et al., 2006; Liu et al., 2011; Wang et al., 2015; Hutin et al., 2015) (Table 1). Different R-genes encodes different types of proteins wherein Xa1 encodes NB-LRR type protein (Yoshimura et al., 1998) and confers resistance to Xoo isolates by recognizing TALEs (Ji et al., 2016). Xa21 and Xa3/Xa26 encode plasma membrane localized LRR receptor like kinase proteins and confer race specific resistance to Xoo (Song et al., 1995; Li et al., 2012). Xa4 encode cell wall associated protein kinase and boosts resistance to Xoo by strengthening the cell wall (Hu et al., 2017). The recessive gene xa5 encodes gamma subunit of the basal transcription factor IIA 5 (TFIIAγ5) and is a substitution variant of a single amino acid V39E (Iyer and McCouch, 2004; Yuan et al., 2016). The non-variant version of the basal transcription factor is required for survival of Xoo in rice. The genes, xa13, xa25, and xa41, encode transmembrane proteins (Chu et al., 2006; Liu et al., 2011; Hutin et al., 2015; Cheng et al., 2017), which are basically sugar transporters, and the dominant alleles of these genes are specifically induced by TALEs produced by the pathogen for establishing infection. Xa10 encodes an ER membrane protein, which elicits Ca2+ depletion in ER membrane inducing host cell death (Tian et al., 2014). Xa23 is known to be an executor R gene that encodes a protein with 113 amino acid residues. The transcription of Xa23 is triggered by AvrXa23, a TALE from Xoo (Wang et al., 2015). Xa27 encodes apoplast protein, which triggers thickening of the secondary cell wall of the vascular bundle elements (Gu et al., 2004). Both dominant and recessive like Xa1, Xa4, Xa21, xa5, and xa13 confer race specific resistance to Xoo, respectively, whereas the recessive alleles of genes such as xa1, xa4, and xa21 and dominant alleles of Xa5 and Xa13 are susceptible to Xoo (Zhang and Wang, 2013). The cloned R genes, its cognate Avr genes, and the nature of the resistance of R genes have been summarized in Table 2.
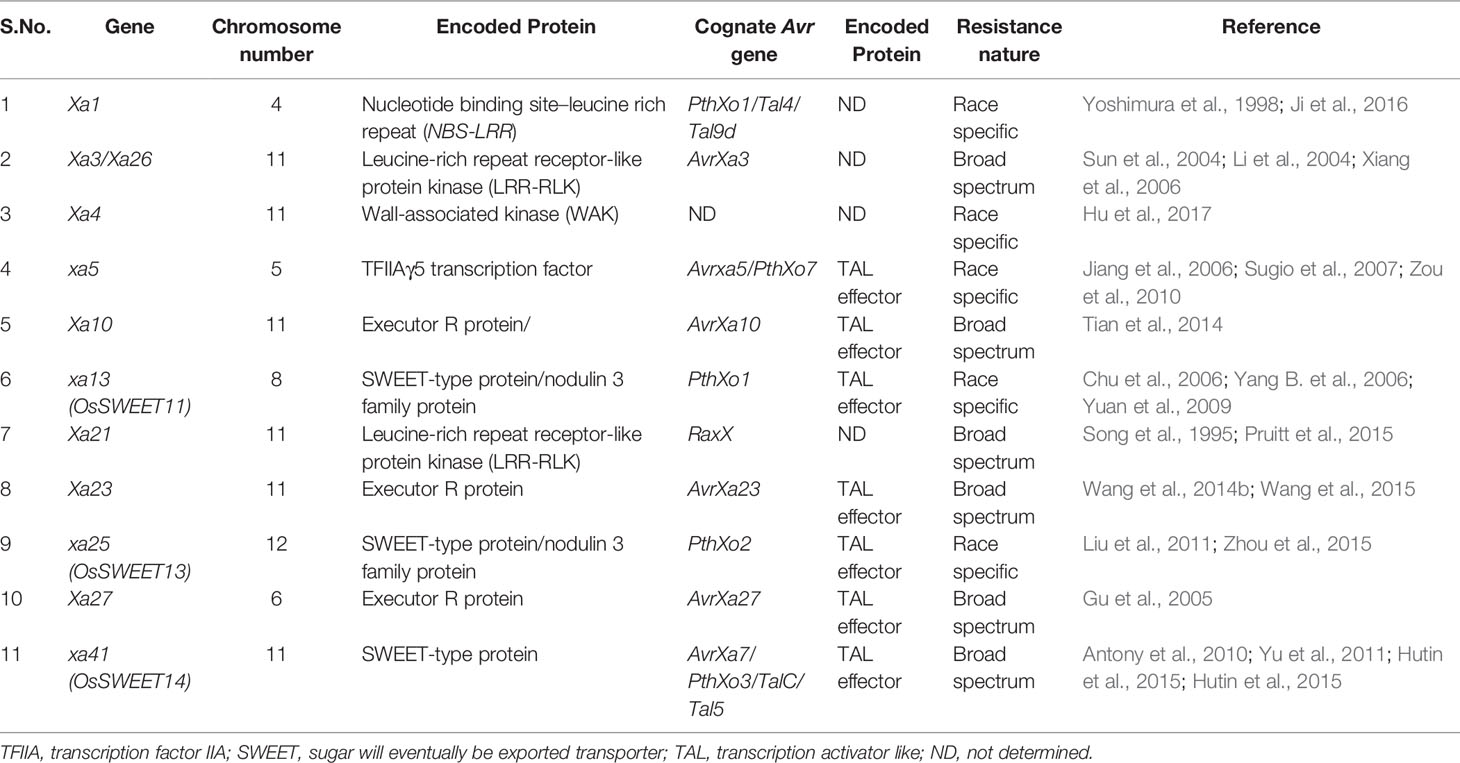
Table 2 List of cloned rice R genes, cognate Xanthomonas oryzae Avr genes, and nature of resistance of R genes (adapted from Jiang et al., 2020).
Introgression of Novel BB Resistance Genes From Wild Relatives of Rice
The genus Oryza includes two cultivated species of rice, i.e., O. sativa and O. glaberrima (2n = 24, genome type AA) and 22 wild species (2n = 24, 48) containing an array of genome types, including those belonging to AA, BB, CC, BBCC, CCDD, EE, FF, GG, KKLL, and HHJJ (Goicoechea et al., 2010). The Oryza, belonging to wild species are considered as a repository of genetic diversity that can be an asset for crop improvement. Although wild relatives of rice are atrocious to the cultivated varieties in terms of many agronomic traits, they are potential genetic resource with tremendous genetic diversity (Ali et al., 2010). Presence of adaptive traits that are often lacking in cultivars renders them vulnerable to several biotic and abiotic stresses, while a majority of the wild rice can withstand harsh biotic and abiotic environmental conditions. Cultivated rice has been the source of many BB resistance genes, and introgression of these genes into elite varieties/hybrids has been done through conventional breeding or also through marker-assisted breeding (MAB). However, transfer of genes from wild species to cultivated types brings with it a set of challenges such as hybrid sterility, linkage drag, and incompatibility barriers. So far, a handful of BB resistance genes have been identified and introgressed from related wild species of Oryza into cultivars (Nino-Liu et al., 2006; Sanchez et al., 2013). With the advent of molecular and genomic tools such as trait-associated DNA markers, high-throughput marker-assisted genotyping rapid identification of BB resistant sources and the process of their introgression into elite cultivars can be accelerated tremendously.
The first BB resistance gene to be cloned and characterized in the rice was Xa21. It was originally identified and introgressed from an accession of O. longistaminata (AA genome), and the gene encodes a receptor kinase like protein and provides broad-spectrum resistance against BB (Song et al., 1995). Xa21 has been transferred to several rice cultivars and hybrids through marker-assisted breeding (MAB) (Williams et al., 1996; Singh et al., 2001; Perez et al., 2008; Sundaram et al., 2008; Balachiranjeevi et al., 2018). Additional broad-spectrum BB R-gene Xa23 (encoding executor R protein) was transferred into Asian cultivated rice from O. rufipogon (AA) (Zhang et al., 2001). Various wild species of Oryza pertaining to secondary gene pool have also assisted as a source of BB R-genes such as Xa27 from O. minuta (BBCC) and Xa29(t) from O. officinalis (CC) (Amante-Bordeos et al., 1992; Tan et al., 2004; Gu et al., 2005). Other BB resistance genes isolated from wild relatives and characterized with the help of molecular markers and genomic tools include Xa10 (Gu et al., 2008; Tian et al., 2014), Xa30 (O. rufipogon) (Jin et al., 2007), Xa32 (O. australiensis) (Zheng et al., 2009), and xa32 (O. meyeriana) (Ruan et al., 2008), Xa32t (O. australiensis), Xa33 (O. nivara) (Natrajkumar et al., 2012), Xa35t (O. minuta), and Xa38 (O. nivara) (Cheema et al., 2008). A novel locus on chromosome 12 of O. latifolia (wild allotetraploid rice species) was identified recently, and it confer race specific resistance of Xoo strain PXO339 (Angeles-Shim et al., 2020). Based on these developments, it can be inferred that wild relatives of Oryza are expected to contribute significantly in developing durable BB resistant rice varieties. Whole genome sequencing of wild rice will expedite identification of resistance genes from the wild relatives of rice and may offer insights about the the pathways associated with the evolution of different resistance genes.
In additional to wild rice resources, it is generally accepted that durable and broad-spectrum resistance against plant dieseases can be enhanced by deployment of quantitative trait loci (QTLs) along with major genes so that both vertical and horizontal resistance can be achieved. In the recent past, a few studies highlight the role of QTLs with respect to resistance/tolerance against BB of rice. Even though few QTLs associated with tolerance/resistance to BB have been reported earlier, most of these QTLs mapped closely to already identify major resistance genes (Li et al., 1999). Five major QTLs were maped on various chromosomes for African resistant Xoo strains Xoo. Various loci on different chromosomes such as 1, 7, 9, 10, and 11 explained as much as 13%, 37%, 13%, 11% and 15% of phenotypic variation in terms of resistance, respectively (Djedatin et al., 2016). A major qBBS11 was identified by composite interval mapping of MAGIC population derived from Japonica, and it explained 31.25% of the phenotypic variation (Kim and Reinke, 2019), and this QTL was later renamed as Xa43(t). On closer examination, xa34(t) has been identified to co-localize along with qABB-1 on rice chromosome 1, which is a resistance QTL induced by the African Xoo strain (Chen et al., 2011).
Analyzing Resistance Gene Analogues (RGAs) As A Novel Tool To Identify Blight Resistance Gene
Resistance genes analogs (RGAs) are a large class of disease resistance associated genes and they can be categorized into two major groups, namely NBS-LRR and transmembrane LRR (TM-LRR) (Hammond-Kosack and Jones, 1997; Sekhwal et al., 2015). Others include pentatricopeptide repeats (PPRs) and apoplastic peroxidases. NBS-LRR class of RGAs targets effector protein of pathogen, thus mediate effector triggered immunity (ETI) in host cell, whereas TM-LRR class of RGAs mediates PTI (Chisholm et al., 2006). NBS-LRR represents the most abundant and best-known family of RGAs contributing to disease resistance in plants (Porter et al., 2009). Analysis of whole genome sequences of japonica Nipponbare and indica 93-11 suggested presence of RGAs in pseudogenes, with 347 RGAs in Nipponbare and 345 in 93-11 as pseudogenes (Luo et al., 2012). Interestingly, most of the identified pseudogenes have strong identity with one or the other NBS protein (Liu et al., 2011). Further, many studies have shown that the RGAs are randomly distributed on chromosome either in large or small clusters (Ghazi et al., 2009), for example, 50% NBS and 74.3% NBS-LRR class of RGAs were found to be clustered in rice (Yang S. et al., 2006). The distribution of RGAs in clustered manner potentially functions like reservoir of genetic variation, which may be responsible for bringing the evolution of new R genes (Michelmore and Meyers, 1998; Young, 2000; Zhou et al., 2007). On the long are of chromosome 11, cluster of six Xa21 like RGAs was reported. The NBS-LRR containing genes cluster was also predicted at 0.6 Mb away from Xa21, which indicates the existance of extra NBS-LRR–type genes for activation and expression of the Xa21 gene (Ghazi et al., 2009). Therefore, from the application point of view, RGAs provide enormous opportunities as they can be used as candidates’ genes for R-gene mapping and cloning, co-localization of QTLs, SNP marker development, and for resistance breeding (Liu et al., 2007; Huang et al., 2012).
Modern Approaches for Development of BB Resistant Rice
Molecular Breeding for BB Resistance in Rice
Efforts have been made to improve resistance of rice against BB through conventional and modern breeding techniques. This is achieved through standard crossing and/or backcrossing an elite rice variety/hybrid with the genotype carrying the resistance gene to BB. The practice not only reduces the use of chemical pesticides but also offers a sustainable way for management of this disease. Presence of several virulent bacterial strains in the rice growing areas throughout the world necesssitates cultivation of such rice varieties, which are endowed with multiple resistance genes as gene-pyramids. To date, identification of 44 R genes conferring resistance to diverse Xoo races has been completed (Busungu et al., 2016; Neelam et al., 2019); majority of these identified genes come from O. sativa ssp. indica or japonica. As mentioned earlier, a set of resistance genes have been also identified from wild species of rice such as O. longistaminata, O. rufipogon, O. minuta, and O. officinalis (Bhasin et al., 2012; Kumar et al., 2012; Wang et al., 2014a; Zhang et al., 2014; Kim et al., 2015) (Table 1).
It is pertinent to note that 14 R-genes (xa5, xa8, xa13, xa15, xa19, xa20, xa24, xa25, xa26b, xa28, xa31, xa32, xa33, and xa34) out of 44 known R-genes are recessive, while the others are dominant in their inheritance, and Xa27 has shown both dominant and semi-dominant inheritance in different genetic backgrounds (Gu et al., 2004; Chen et al., 2011; Kim et al., 2015; Cao et al., 2018) (Table 1). The incorporation of several resistance (R) genes has been facilitated through marker-assisted backcrossing (MABC) or conventional backcross breeding, and resistance breeding has played a significant role in defending rice from the attack by the pathogen (Sundaram et al., 2008; Sundaram et al., 2009; Perumalsamy et al., 2010; Hari et al., 2011; Hari et al., 2013; Pandey et al., 2013; Kim et al., 2015; Balachiranjeevi et al., 2015; Abhilash et al., 2016; Abhilash et al., 2017). A rice derived BB resistance gene Xa38 was incorporated into a BB susceptible rice variety PB1121 and APMS 6B (a rice maintainer line), either singly or in combination with other BB resistance genes, using a modified MABC approach, and improved lines showed broad spectrum of resistance against different Xoo races (Ellur et al., 2016; Yugander et al., 2019). Rice varieties carrying single resistance genes (e.g., Xa4) are not recommended for long term cultivation, as acute selection pressure on the pathogen results in rapid evolution of compatibility between Xoo and rice (Mew et al., 1992). Harnessing broad-spectrum resistance through pyramiding multiple resistance genes can be helpful to avoid such breakdowns. The probability of breakdown in cases of resistance conferred by two or more genes in a single genotype is much lower than that of a single gene controlling resistance (Mundt, 1990). For instance, R-genes in combination (Xa4/xa5 and xa5/Xa21) offer higher level of resistance compared to both parental level and single gene (Sattari et al., 2014; Pradhan et al., 2016). Huang et al. (1997) developed four-gene pyramid lines comprising Xa4, xa5, xa13, and Xa21 genes in IR24 cultivar genetic background through MABC, and the gene-pyramid lines showed broad spectrum disease resistance. Several research groups have performed BB gene pyramiding in rice using MABC techniques, e.g., marker-assisted introgression of xa5, xa13, and Xa21 in the genetic background of PR106, Samba Mahsuri, Triguna, and Jalmagna (Singh et al., 2001; Sundaram et al., 2008; Sundaram et al., 2009; Pradhan et al., 2015). Similarly, xa13 and Xa21genes pyramiding was carried out in the genetic background of Pusa Basmati 1 (Joseph et al., 2004). Table 3 offers a comprehensive list of BB resistance genes that have been deployed or in the process of deployment in rice through MAB.
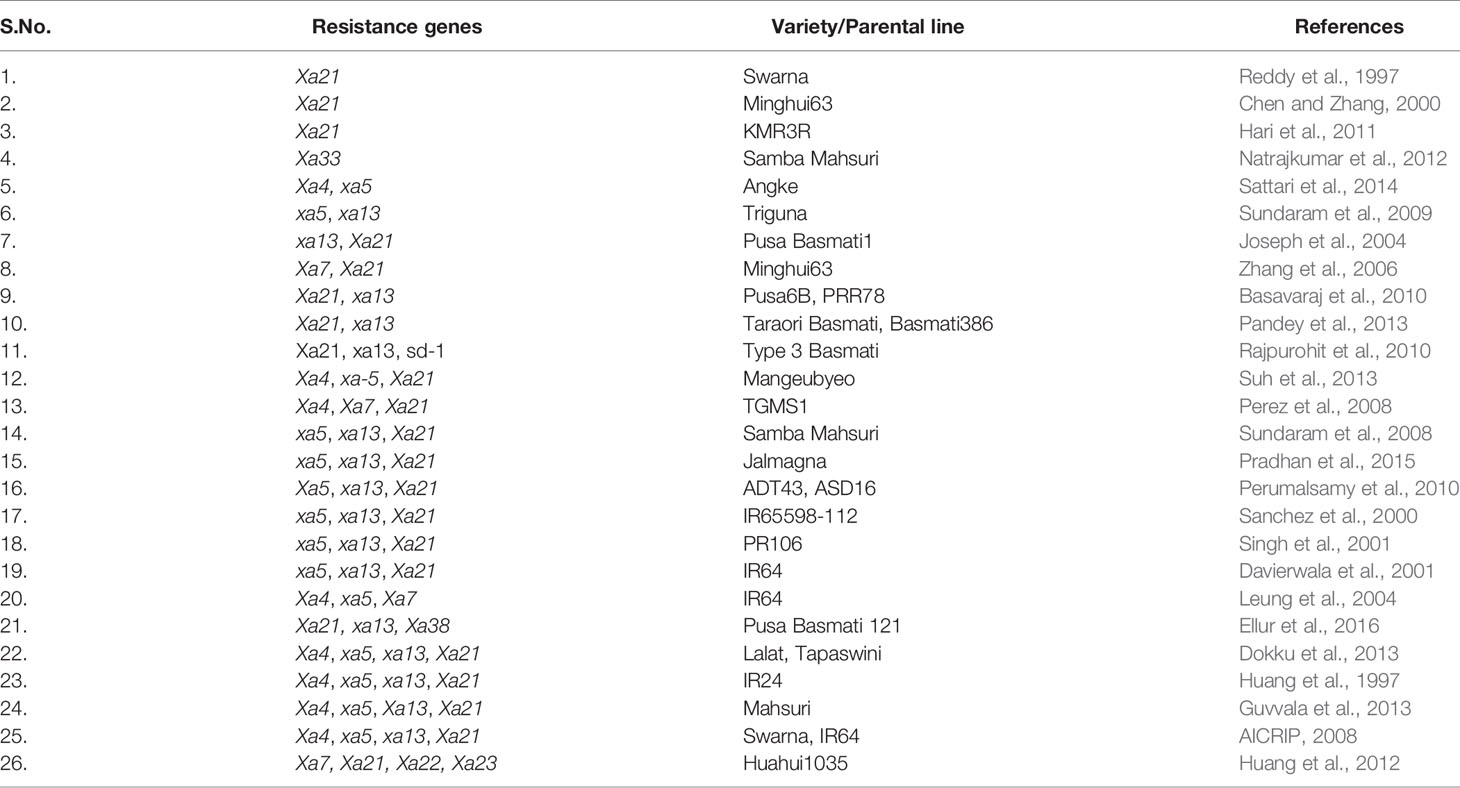
Table 3 Cultivars improved for bacterial blight resistance through breeding/marker-assisted breeding.
Development of Transgenic Rice Resistant to BB
A potential strategy to control BB disease is the genetic transformation of elite cultivars using cloned resistance genes. Compared to conventional breeding, it is less time-consuming and avoids the problem of linkage drag. The first transgenic line containing Xa21, T-309, was developed in japonica rice by Wang et al. (1996). Later, Xa21 was introduced into several varieties such as IR 72, MH 63, and IR 51500 (Datta et al., 2002; Tu et al., 1998; Tu et al., 2000). Similarly, an elite restorer line genetically transformed with Xa21 has shown marked level of resistance to BB while retaining its original traits (Zhai et al., 2004). Field trials of Xa21 transgenic rice conducted in India, Philippines, and China led to the identification of BB-resistant lines in transgenic IR 72 (Datta, 2004). Transgenic lines carrying one or the other Xa genes were developed for functional characterization of the gene; however, none of these could be commercialized due to regulatory and policy bottlenecks. Hence, conventional breeding combined with marker-assisted breeding or genomics assisted breeding is the preferred strategy for developing resistant lines/varieties to control BB disease.
Mining of Novel Alleles of BB Resistance Genes
Mining superior alleles from different gene pools of any crop provides opportunity to access novel and effective alleles for biotic and abiotic stresses, which can be deployed in plant breeding for cultivar development (Ramkumar et al., 2010). To identify novel or superior allele of the known gene among the population, PCR-based approach is widely used. In this method, PCR amplification of homologs from different wild and cultivated germplasm is performed and analyze the PCR amplicon. This method is known as allele mining, and it has been widely studied for BB resistance genes such as Xa7 (Utami et al., 2013), Xa27 (Bimolata et al., 2013), Xa26, Xa21, and xa5 (Bimolata et al., 2015), etc. This method will also reveal the degree of conservation among genes and other regulatory regions across the species. As mentioned earlier, fast adaptation of the pathogen races causes the failure of disease resistance in varieties containing single resistance genes and even two genes. Therefore, discovery of novel sources of resistance becomes crucial to match the plasticity of rapidly evolving pathogenic Xoo strains (Barry et al., 2007). In this context, surveying the genic/allelic variation available in landraces (Borba et al., 2009), traditional varieties, and wild relatives (Barbier, 1989) of rice will contribute to obtain effective and durable BB resistant varieties. One such example is Xa7, which has been used to develop BB resistance rice varieties (Perez et al., 2008; Utami et al., 2010). Allele mining approach was applied in the local rice accessions of Indonesia (Parekaligolara) to isolate resistant alleles of Xa7 (Utami et al., 2013). Subsequently, these accessions containing variant alleles with respect to Xa7 have been used as for developing new BB resistant rice lines (Utami et al., 2013). Analysis of variation in amino acid residues between the resistant and susceptible lines has revealed co-linear non-synonymous substitutions of lysine-cysteine-valine to serine-serine-threonine, respectively. Another BB resistance gene, Xa27 provides resistance to rice against only those strains of Xoo harboring the avirulence gene avrXa27. Both dominant and recessive alleles of Xa27 gene code for same protein without any changes in the protein sequence, but polymorphism exists in their promoter sequences. Deletion of three nucleotides AGA at 51st position in the promoter of recessive allele (i.e., non-functional allele) of the gene has been reported (Gu et al., 2005). Twenty-seven alleles of Xa27 gene have been identified in O. nivara and O. sativa at both promoter as wel as 5´UTR region. Such nucleotide diversity analysis will certainly help in diversification of gene function and enhancing the intensity of BB resistance (Bimolata et al., 2013). The nucleotide diversity analysis of naturally occurring BB resistance genes (Xa21, Xa26, and xa5) alleles was carried out in diverse cultivars of Oryza species and their wild relatives (Bimolata et al., 2013). The highest singleton variable sites (SVS) and nucleotide diversity were reported in Xa26, whereas maximum frequency of single nucleotide polymorphisms (SNPs) was observed in Xa21. Many substitutions and InDels resulted in nucleotide and amino acid polymorphism at Xa21 and Xa26 loci, which also have pathogen recognition LRR domain, and finally resulted in non-functional gene. Transition bias was reported in all the three alleles of Xa21, Xa26, and xa5, where G to A transition was favored more (Bimolata et al., 2015). Functional characterization of the new alleles will help in deciphering their actual roles in resistance against the pathogen. Alternative promising approach is RNA interference (RNAi) technique, which is used to silence molecules involved in regulating resistace genes negatively (Gust et al., 2010; Li C. et al., 2012). The final outcome results as a stronger and durable defense response, which leads to reduce disease manifestation and progression.
Mutagenesis and TILLING
Targeting induced local lesions in genomes (TILLING) is a reverse genetic non-transgenic technique that is exploited to detect induced mutations in the target genes for the improvement of both plant and animal species (Barkley and Wang, 2008). Recently, with the aim to develop disease resistance varieties and creating useful genetic variation for multiple traits, TILLING population have been generated in many economically important plants like wheat (Fitzgerald et al., 2010), barley (Talamè et al., 2008), tomato (Piron et al., 2010), sunflower (Sabetta et al., 2011), and melon (González et al., 2011) by employing either chemical or physical mutagens. In rice, Till and colleagues (2007) could successfully generate a high-density TILLING population (1 mutation/250–300 kb) in Nipponbare variety of rice. Wu et al. (2005) demonstrated the efficacy of TILLING approach for identifying mutant rice lines with enhanced resistance against BB, rice blast and tungro virus, with a frequency ranging from 0.01 to 0.1%. They generated 60,000 IR64 TILLING mutant lines by using chemical and physical mutagens, out of which 38,000 unique lines were advanced to M4 generation for forward and reverse TILLING. In rice, the technique has been employed to isolate various rice mutants by targeting important agro-economical genes such as OsBZIP for rice blast resistance (Till et al., 2007); OsTPS1c OsDREB, OsSNAC1, OsAKT1, OsHKT6, OsNSCC2, OsHAK11, OsSOS1, OsAHP1, and OsPLA1 for abiotic resistance (Till et al., 2007; Suzuki et al., 2008; Casella et al., 2013; Hwang et al., 2016); OsSD1 for regulating plant height (Casella et al., 2013); OsHd1 and OsSAD flowering (Suzuki et al., 2008; Casella et al., 2013); OsACOS12 for fertility (Li et al., 2016); and OsBADH2 for aroma (Casella et al., 2013).
TILLING population can also be used for forward genetic studies. Screening of 60,000 IR64 (rice indica cultivar) mutants led to identification of several loss or gain of resistance mutants of showing resistance or enhanced susceptibility to BB, blast, and tungro diseases (Wu et al., 2005). EMS induced mutant in the genetic background of Nagina 22 rice variety has been developed and utilized for various genetic studies in India (Sevanthi et al., 2018). It may be worthwhile to screen the rice lines for resistance against BB and other biotic stress and identify novel genetic variations. In addition to the above, we have generated a large sized EMS mutagenized population of Samba Mahsuri and screened them for resistance against bacterial blight. Preliminary results show that few of the mutant lines of Samba Mahsuri display enhanced resistance (Ershad Gopi et al., 2017). These mutagenized populations can be shared with researchers or breeders for rapid screening for a range of phenotypes, and the TILLING populations can serve as a public resource for the research community. For instance, IRRI, Philippines has distributed around 15,000 mutant lines of IR64 to researchers worldwide for detection of novel phenotypes, including sensitivity to plant hormones, phytic acid abundance, response to salinity and drought, and non-host resistance (Wu et al., 2005). A public TILLING support tool (http://tilling.ucdavis.edu/index.php/RiceTilling) and rice mutant database (IRIS; http://www.iris.irri.org) have been established in rice.
Sequencing Based High-Throughput Mutation Detection Systems
In rice, NGS based TILLING protocols are well documented (Burkart-Waco et al., 2016) and it is a matter of time before the strategy is widely adopted to identify genes associated with resistance/susceptibility against bacterial blight pathogen in rice. Unlike NGS, where one can obtain the exact information of nucleotide base change and its position caused due to mutagen, high-resolution melting (HRM) identifies the mutation based on the differences in the melting curve of fragments of mutant and wild type. HRM technique is particularly important for analyzing the target genes that consist of multiple exons of smaller lengths. Though NGS and HRM techniques are still costlier, their capacity to generate results within a very short span of time is indeed encouraging. NGS- and HRM-based TILLING or Eco-TILLING strategy could uncover a new set of genes controlling BB resistance, and hence expedite the progress of developing new rice cultivars with greater level of resilience. In this context, it is worthwhile to note that International Rice Research Institute (IRRI), Philippiens along with other collaborators have sequenced ~ 3000 rice accessions and the rice lines can be mined for novel alleles of major, cloned and well characterized genes conferring BB resistance.
Genome Editing
Breeding traits of agronomic significance largely relies upon existing allelic variation and involves repeated cycles of crossing and selection to obtain a crop genotype with desired level of improvements, which may consume considerable time and efforts (Sundaram et al., 2014). These limitations can be overcome by using emerging genome editing technologies. Genome editing based on artificial nucleases is a transformative technology that has the ability to modify plant genomes in an accurate and expectable manner. So far, four sequence-specific nucleases, i.e., transcription activator-like effector nucleases (TALENs) (Christian et al., 2010), zinc finger nucleases (ZFNs) (Kim et al., 1996), meganucleases (Smith et al., 2006) and the CRISPR/Cas (CRISPR-associated) nucleases, have been successfully used in genome editing in many crop plants (Bortesi and Fischer, 2015). In principle, all these technologies can be used for modifying plant traits such as disease resistance. For example, TALEN technology was applied successfully to mutate a BB susceptible gene, Os11N3/OsSWEET14 promoter in rice. The inability of the effector to bind to the promoter of OsSWEET14, ultimately resulted in BB resistance (Li et al., 2012). The Os11N3 is a sucrose-efflux transporter family gene (i.e., SWEET gene) whose expression gets activated by Xoo effectors for the pathogen’s nutritional needs. By editing the effector-binding element (EBE) of Os11N3, the virulence function of effectors produced by Xoo was abolished, leading to improved BB resistance (Li et al., 2013). CRISPR/Cas9 mutagenesis of another susceptibility gene encoding sucrose transporter OsSWEET13 was performed to achieve BB resistance. Xoo effector/TALE PthXo2 induces the expression of OsSWEET13 in host, which subsequently resulted in establishment of Xoo and host susceptibility (Zhou et al., 2015; Borrelli et al., 2018). The expression of OsSWEET13 gene has been evidenced to be activated by binding of the TALE, PthXo2 to EBE of its promoter sequence (Xu et al., 2019). Genome edited lines showed significantly higher level of resistance to pathogen strains possessing the TALE. This study exemplified the fact that the technology can be applied to elite rice varieties to edit multiple genes simultaneously or sequentially to provide stronger and durable resistance against majority of BB strains. Furthermore, better understanding of SWEET genes and CRISPR/Cas9-mediated genome editing tool has helped in producing broad spectrum resistance in Kitaake, IR64 and Ciherang-Sub1 rice varieties through genome editing (Oliva et al., 2019; Xu et al., 2019). Recently, CRISPR/Cas9-mediated genome editing in the promoter region EBEs of OsSWEET14 gene has been demonstrated to confer resistance in Super Basmati rice lines against Xoo strain carrying AvrXa7 TALE (Zafar et al., 2020).
Among the currently available nuclease-based genome editing tools, CRISPR/Cas system is the latest and more popular technology, which relies on RNA-guided engineered nucleases (Jinek et al., 2012). CRISPR/Cas method employed for genome editing consists of a Cas9 endonuclease targeting a specific sequence of the genome defined by a single guide RNA. The CRISPR/Cas technology is simple and efficient, more importantly, has the ability to cleave even methylated DNA (Hsu et al., 2013), and has less or no off-target mutations (Shen et al., 2014). Therefore, CRISPR/Cas system is more versatile for editing plant genomes with highly methylated CpG sites (Miao et al., 2013) and will the most desirable system for editing sequences of rice susceptibility genes like OsSWEET11, OsSWEET13, OsSWEET14, etc. Broad spectrum resistance against Xoo was reported in the transgenic rice lines, in which promoter region EBEs sequence of OsSWEET11, OsSWEET13, and OsSWEET14 were edited through CRISPR/Cas9 technology (Xu et al., 2019). Transgenic wheat plants with mutated mildew resistance locus (MLO) obtained by CRISPR/Cas9, and TALEN technologies showed improved resistance to powdery mildew (Wang et al., 2014). With constant refinements in the technical aspects with respect to specific targeting of the desired gene sequence with precision, genome editing will certainly be a method of choice for developing disease resistant varieties in rice.
Genomic Selection (GS)
Current crop breeding approach largely depends on the robust phenotyping and deployment of genetic markers. Major limitation of marker-assisted selection in plant breeding is use of biparental mapping population for QTL prediction and its applicability with traits associated with major effect genes but may work for polygenic traits that are controlled by many genes of small-effect, and in general, such traits are crucial for the improvement of new crop varieties (Heffner et al., 2009; Crossa et al., 2017). To improve the crop selection procedure, breeders have now adopting a newer model—a black box approach ‘Genomic Selection Model’ that does not solely depend on the prior knowledge about the effect or function of individual genetic markers. In fact, GS involves huge set of phenotyping surveillance along with all molecular marker information which avoid biased marker effect estimates; thereby, it can capture more of the variations that appears due to small-effect QTLs (Heffner et al., 2009). Further, GS is an improved form of MAS which concurrently estimates a genomic estimated breeding value for all locus, haplotype, or marker across the entire genome of each genotype. Therefore, genomic selection offers opportunity to increase grain yield and quality by rapid selection of superior genotypes and accelerating breeding cycle in less time. In recent years, GS and genomic-enabled prediction (GP) have been studied in rice for enhancing grain yield by analyzing the genetics and the statistical complexity, which includes environment interaction with genotype that control trait phenotype (Spindel et al., 2015; Xu et al., 2018). The genomic prediction model upon cross validating 363 elite breeding lines from IRRI predicted with an accuracy that ranged from 0.31 to 0.34 for grain yield and 0.63 for flowering time (Spindel et al., 2015). Similarly, Xu et al. (2018) used 575 rice hybrids as a training population, and 362,760 potential hybrids were used to predict agronomic traits such as branch number (primary and secondary) and per panicle grain number and primary branch with accuracy of 36.12, 61.80, and 75.83%, respectively. It can be expected that, in coming years, rice breeding will deploy GS model and GP prediction to rapidly identify novel loci associated with BB resistance and other productivity related traits and quickly use them in breeding programmes.
Molecular Omics Approaches for Studying Rice-Xoo Interaction
Transcriptome Analysis
In the last 15 years, more than 70 key genes providing resistance to different plant pathogen have been identified, cloned and characterized from different plant species (Sharma et al., 2009; Sanseverino et al., 2010). Pathogen incursion can alter the transcript levels of various host plant genes (Iqbal et al., 2005), and several techniques have been developed in recent years to study the differential expression pattern of host genes associated with response to pathogen attack and resistance against it. Some of these expression profiling techniques are cDNA microarray (Schena et al., 1995), cDNA- amplified fragment length polymorphisms (AFLP) (Vuylsteke et al., 2007), suppression subtractive hybridization (Diatchenko et al., 1996), serial analysis of gene expression (SAGE) (Velculescu et al., 1995; Mardis, 2008), digital gene expression (DGE) (Audic and Claverie, 1997), qPCR (Mortazavi et al., 2008), etc. Transcriptome analysis helps elucidate key genes and pathways participating in defense signaling during plant pathogen interaction (Glazebrook, 2001).
cDNA microarray study of a transgenic rice (TP309-Xa21)-Xoo (P6 and K1avirulent and virulent strain respectively) interaction observed 454 and 498 DEGs in the incompatible and compatible interactions, respectively, of which co-regulated genes were 237 (Li Q. et al., 2006). Ethylene receptor-like protein, ethylene-insensitive protein, protein phosphatase, and ADH were upregulated only in rice-Xoo incompatible interaction (Li Q. et al., 2006). A genome-wide identification of defense response genes was performed in xa13 gene mediated resistance plants wherein 702 unique expressed sequences triggered by xa13 were identified (Chu et al., 2004). Sequence analysis showed induction of homologs to putative R-genes encoding NBS-LRR and XA21 like protein. There was also induction of gene homologs related to host-pathogen interaction reported in other plant species, such as PR proteins, peroxidases, WRKY transcription factors, GST, RNA helicases, ubiquitins, catalase, ankyrin-like protein, and cytochrome P450 (Chu et al., 2004). Microarray analysis after infection by BXO43 strain of Xoo in the resistant variety of rice, Ajaya (IET 8585), and susceptible variety, IR24, has revealed the differential expression of 274 genes between and susceptible genotypes (Grewal et al., 2012). Out of 274 genes, 152 and 122 were reported to be up- and down-regulated, respectively, in IET8585 compared to IR24. Some of the major up-regulated transcripts include chitinase precursor, WRKY69, Hin 1, DREB1B, NB-ARC domain containing protein, glutathione S-transferase (GST), cytochrome P450, harpin-induced protein, lipoxygenase, and flavanoid 3-monooxygenase (Grewal et al., 2012). It was interesting to note that several defense related signaling molecules such as MAPKKK17 (Menges et al., 2008), MAPKKK3 (Zipfel et al., 2004), and PP2C were found to be up-regulated (Grewal et al., 2012). Wen et al. (2002) also conducted an expression profile of 12 defense response genes where they observed constitutive expression of these genes but significantly induced under the influence of Xoo and the fungal pathogen causing blast disease, Pyricularia grisea. The RNA-Seq analysis of resistant (CBB23; a rice line carrying Xa23 gene) and a susceptible (JG30) rice gentotypes led to identify several DEGs post infection by Xoo strain PXO99A. Moreover, several of the DEGs were up-regulated in CBB23 were related to immune responses like peroxidase, phytosulfokinase, RLKs, serine/threonine kinase, TFs (WRKY, NAC, MYB, bZIP, AP2/ERF etc.) and phytohormones (SA and ET) (Tariq et al., 2018). In another RNA-Seq analysis, where CBB23 was challenged with PXO99A and its mutant P99M2 exploited 1235 DEGs at defferent time point (Wang et al., 2019). Publicly available Xoo infected rice microarray data analysis revealed the importance of mitochondria and chloroplast as an arena for up-regulated and down-regulated genes in response to Xoo infection (Kong et al., 2020). Therefore, the genes up-regulated in resistance varieties can be targeted for improvement through molecular breeding and transgenic approaches, and the rice genes that are up-regulated in susceptibility reaction against Xoo could be potential targets for silencing or genome editing.
Proteomics Analysis of Products Encoded by Resistance Genes
Completion of sequencing of rice and Xoo genomes are significant accomplishments in host-pathogen interaction study. Even though genomes can evolve rapidly because of either transposable elements movement or from epigenetic changes, they are in general contemplated as highly static compared to their extremely dynamic proteomes. Therefore, there has been increased focus on elucidating functional aspect of proteins involved in rice-Xoo interaction. Comparative proteomics is emerging as a promising approach to develop a global understanding of protein expression under various conditions including attack by pathogen on the plant host (Agrawal and Rakwal, 2011; Ding et al., 2012). Using proteomic approaches, different biological processes including protein–protein interaction, post-translational modification, protein expression, etc. could be successfully analyzed during plant development, particularly during stress conditions (Hashiguchi et al., 2010). The induction of PR5 protein post nitrogen application was reported in case of M. grisea infection (Konishi et al., 2001). Stress related proteins such as superoxide dismutase (SOD), heat shock proteins (HSP), and dehydrins are induced in plant post rice yellow mottle virus (RYMV) inoculation (Ventelon-Debout et al., 2004). Proteomic study of sub-cellular organelles has been performed such as plasma membrane, vacuolar membrane, mitochondria, and chloroplast. Cytosolic and membrane protein study revealed the activation of proteins related to defense such as thaumatin-like protein (TLP) (PR5), probenazole-inducible protein 1 (PBZ10), (SOD), and peroxiredoxin (Mahmood et al., 2006). Transgenic rice lines overexpressing a TLP showed moderate level of resistance. After proteomic analysis, 440 protein spots were detected, where 10 proteins, including TLPs, were differentially expressed (five up-regulated and five down-regulated). TLP, ATP synthase B chain, glycine cleavage H protein, and 2-Cys peroxiredoxin were significantly up-regulated, whereas glycerol aldehyde dehydrogenase, salt induced protein, transketolase, and oxygen evolving enhancer protein 2 were down-regulated. Out of 10 differentially regulated protein spots, two did not show a substantial match with any known proteins (Mahmood et al., 2006). A study on plasma membrane (PM) proteome in rice revealed the involvement of at least eight PM-associated proteins in BB defense out of 20 protein spots induced after Xoo infection (Chen et al., 2007). These proteins were H+-ATPase, prohibitin (OsPHB2), hypersensitive-induced response protein (OsHIR1), quinone reductase, zinc finger, universal stress protein (USP), C2 domain protein, HSP, and protein phosphatase. A stable somatic hybrid line SH76 was developed using wild rice O. meyeriana and japonica rice cultivar (8411), which proteomic analysis revealed differential expression of 77 proteins including stress related proteins such as putative glutathione S- transferase, ascorbate peroxidase, and mitochondrial chaperonin-60 (Yu et al., 2008). Interestingly, differential induction stress associated proteins have been reported upon Xoo challenge, suggesting the likelihood of participation of mutual stress pathways. Some of the important candidate proteins activated in O. longistaminata post BB infection include cyclin-dependent kinase C, germin-like protein, putative r40c1, glutathione-dependent dehydroascorbate reductase 1 (GSH-DHAR1), and Ent-isokaur-15-ene synthase (Kumar et al., 2015).
Metabolomics Analysis of Compounds From Resistant Plants
Cell signaling is the first molecular event that occurs during pathogen infection. Plant produces volatile metabolites/hormones such as ethylene, methyl jasmonate, methyl salicylate and nitric oxide as key mediators of host response to pathogen/pest infection and for systemic acquired resistance (Heuberger et al., 2014). These hormones conjugates with other metabolites like jasmonate and isoleucine to provide immunity (Staswick and Tiryaki, 2004). During plant-pathogen interaction, many small size molecules of different class such as homoserine, asparagine, and sphingolipids mediate signaling (Heuberger et al., 2014) and regulated through cross-talk between hormones like ethylene-jasmonate, nitric oxide, and jasmonate (Lorenzo et al., 2003; Jian and Wu, 2005).
Metabolism impacts cellular physiology and plays an essential role in biology. Recent advances in metabolomic analytical tool provides opportunity to dissect the layers of plant metabolic regulation, thus allowing bridging the gap between genome and the phenome (Metallo and Vander-Heiden, 2013). It also aids in identifying signature metabolites linked to agronomic traits, thereby plays dynamic role in crop improvement (Kumar et al., 2017; Sharma et al., 2018). Like other omics approach, metabolomics has the ability to examine the global expression of small metabolites that are involved in signaling, and morphological, physio-chemical responses produced during plant-pathogen interaction. In the past few years, researchers have put an effort to address the diverse mechanisms that rice plants use to adapt its metabolism during infection by Xoo and also discuss how metabolic flux alteration can be used to identify central regulatory nodes under pathological cell physiology. A resistant rice genotype responds to BB pathogenic invasion by increasing carotenoids, total phenolic, and flavonoid contents (Kumar et al., 2013). Boosted levels of flavonoids (cyanidin 3-galactoside, cyanidin 3-glucoside, delphinidin 3-arabinoside, and delphinidin 3-galactoside) in black scented rice protects it from diseases and these compounds are also known to have health promoting effects on human (Asem et al., 2015).
A high resolution metabolite QTL (mQTL) analysis of rice recombinant inbred line (RIL) population revealed ~ 2,800 mQTL associated with 900 metabolites (Gong et al., 2013). A major mQTL for aromatically acylated flavonoids co-located within a 0.5-Mb region on chromosome 10, which consists of one acyltransferase gene OsAT1 conferring BB resistance in rice by regulating lysophosphatidylcholines, has been identified (Mori et al., 2007; Gong et al., 2013). Leaf samples are excellent source to study the resistance mechanism in rice. Hence, evaluation of susceptible versus resistant mutants, varieties, or genotypes is an ultimate tool to interpret resistance mechanisms and identify defense related metabolites. The leaf metabolome was examined to identify the metabolites that might be responsible for differences between BB susceptible, wild type cultivar, and resistant transgenic rice plant (http://www.agilent.com/cs/library/applications/5989-6234EN.pdf). Studies suggest distinct subsets of metabolites at pre and post-invasion might coordinate during BB infection. For example, 42 metabolites could be predicted to be associated with BB resistance, 22 metabolites were connected to infection response, 25 metabolites could be formed by bacteria or in response to it, and a total of 170 metabolites were identified, which differentially expressed between the two-contrast line (Fisher and Sana, 2007). Recently, seed metabolome of a BB resistant line, C418/Xa23, which was generated through marker assisted breeding, and a transgenic variety C418-Xa21 were compared with the wild type susceptible progenitor (C418) (Wu et al., 2012). The study revealed distinct metabolite pattern in the seed of resistant line with significant decline in few common metabolites: amino acids (alanine, glycine, and tyrosine), organic acids (ferulic acid, succinic acid, and malic acid), and glycerol. Additionally, linoleic acid emerged as specific signature metabolite in the seed of resistant breeding line. Possibly, these metabolites regulate the Kreb’s cycle and amine biosynthesis, which drive the metabolic state and cell physiology (Figure 3B). There is a possibility to use these metabolites as novel discriminatory metabolites to identify BB resistant rice genotypes.
System Biology Approach to Understand Rice-Xoo Interaction For Developing Strategies For Durable Resistance
Consolidating findings emanating from multiple omics platforms contributes to an improved understanding of metabolic pathways, genes, and gene-interaction networks responsible for the phenotypic changes that accompany plant-microbes interactions. A growing body of literature that integrates metabolomics, transcriptomics, and proteomics analyses suggests that significant metabolic alterations happen during plant-pathogen interactions (Wan et al., 2002; Oksman-Caldentey and Saito, 2005; Hollywood et al., 2006; Figueiredo et al., 2008; Tan et al., 2009; Choi et al., 2010; Sana et al., 2010; Paternain and Campion, 2013). However, similar approach for studying BB resistance in rice is presently lacking. To the best of our knowledge, only one study is conducted in rice to establish correlations between the metabolome and transcriptome (Sana et al., 2010). Examination of the metabolic profile of a resistant rice variety infected with Xoo suggested significant up-regulation of compounds such as pigments, rutin, fatty acids, and lipids in the resistant plant. The study compared the differential expression of genes in relation to these metabolite products and the corresponding enzymes, and their regulatory pathways. For instance, the transcriptomics and metabolomics data revealed strong correlation between decreased in glutamate levels with increased expression of glutamate decarboxylase, which encodes for an enzyme that catalyzes decarboxylation of glutamate to GABA in the Xoo challenged plants. Similarly, the increased expression of Phenylalanine ammonia lyase (gene product regulates phenyl propanoid pathway) was in coordination with the elevated level of phenylalanine in the Xoo challenged resistant plant. However, the expression of isocitrate lyase, b-1,3-glucanase and chitinase showed negative correlation with the metabolite data in the resistant plant as these genes are involved in degradation of fungal cell wall and provide resistance against fungal pathogen (O’Toole et al., 2001). We anticipate a remarkable increase in the studies that combine different omics platforms to provide better insights into BB resistance in rice.
Epigenetics: A New Way to Improve Trait Understanding and Manipulation
To evaluate the epigenetic control responsible for resistance against Xoo, methylated regions of rice genome were using methylation sensitive amplified polymorphism (MSAP) technique, and cytosine methylation was screened by the bisulfite mapping technique (Akimoto et al., 2007). The rice seed were treated with 5-azadeoxycytidine (inhibitor of DNA methylation), and the progeny were grown in field. Among 1000 seeds treated with 5-azadeoxycytidine, only 35 seedlings survived and, out of that, two showed dwarf phenotypes. In contrast to susceptible wild type, line-2 showed constitutive expression of Xa21G and resistance against BB (Akimoto et al., 2007). Normally, the promoter remained at hyper-methylated condition, which silences Xa21G gene to cause susceptibility to Xoo. Besides DNA methylation, epigenetic regulatory pathways also regulate the initial step of plant-bacteria interaction through small RNAs (sRNAs), such as small interfering RNAs (siRNAs) and micro RNAs (miRNAs) (Carvalho et al., 2016). In Arabidopsis and legumes, miR393 is induced during pathogenic infection and confers resistance to associative and endophytic diazotrophic bacteria (AEDB) by attenuation of auxin signaling pathways (Navarro et al., 2006; Subramanian et al., 2008). Similarly, enhanced expression of miR160 in Arabidopsis during bacterial infection indicates its possible involvement in defense response (Fahlgren et al., 2007). In contrast, the down-regulation of miR160 and miR393 in maize caused suppression of defense response (Thiebaut et al., 2014). Genome editing tools such as ZFNs and TALEs were used for targeted epigenetic modifications in plants (Gaj et al., 2013). Presently, the CRISPR/Cas9 technology has become the widely accepted genome editing tool for plant modification including epigenetic modification (Malzahn et al., 2017; Van de Wiel et al., 2017). In mammalian system, targeted enhanced CpG methylation has been successfully demonstrated by using CRISPR/Cas9-DNMT3A as epigenetic modifier (Vojta et al., 2016). In future, such epigenetic modifications are expected to be routinely deployed for targeted methylation for disease resistance.
Impact of Climate On BB Resistance Genes
The classical concept of Plant Pathology describes a plant disease as the result of interaction of host, pathogen and environment, commonly referred to as ‘disease triangle’. Thus, variations in any parameters of environment can remarkably affect the disease consequence. Several studies have shown that the effectiveness of ‘R’-gene mediated defense response can be substantially influenced by temperature variation (de Jong et al., 2000; Uauy et al., 2005; Webb et al., 2010; Zhu et al., 2010; Zhao et al., 2016). Webb et al. (2010) reported that near isogenic line IRBB4 owning the BB resistance gene Xa4 exhibited much longer lesion at higher temperature regime (35:27°C, day:night) than in lower temperature regimes (29:21°C, day:night). Similarly, Dossa et al. (2017) observed that drought stress significantly reduced Xa4 mediated BB resistance in rice. On the contrary, efficacy of BB resistance gene, Xa7 was significantly improved at higher temperature (35:31°C, day:night) compared to lower temperature (29:21°C, day:night) in limiting BB severity and Xoo population (Webb et al., 2010). In a detailed study, Cohen et al. (2017) found that the level of Xa7-mediated BB resistance was comparatively stronger and faster at higher temperature as compared to lower temperature regime. These contrasting scenarios raise concerns about durability of single R gene and different gene combinations in the scenario of a rapidly changing climate. Recently, Dossa et al. (2020) demonstrated that near isogenic line IRBB 67 (owning BB resistance genes Xa4 and Xa7) did not show any difference in lesion length at both lower and higher temperature regimes, indicating that the reduced effectiveness of Xa4 at higher temperature did not affect the resistance level of IRBB 67.
Perspective and Conclusions
The research achievements in the recent past have contributed to improved understanding of resistance mechanisms in rice against Xoo infection and also the pathways associated with susceptibility. Deploying multiple sets of carefully selected R-genes as gene-pyramids holds promise for developing improved rice cultivars/hybrids with durable and broad-spectrum BB resistance. Identification and utilization of new resistant genes/alleles, tapping the genetic variability in diverse germplasm, and generation novel variation in existing genes through gene-editing will be crucial to achieve sustained production of rice for ever-increasing population. Mapping and characterization of different BB R-genes have made marker-assisted selection a valuable tool to develop durable BB disease resistance in rice. Moreover, strategies for gene discovery based on genomics and proteomics together with transgene validation through genetic transformation are increasingly helping us understand the functional profiles of candidate genes. Research on this aspect has been immensely benefited from the increasing information on structure and function of major BB resistance genes. Although a smaller number of major R-genes have been cloned and characterized as of now, RGAs and DNA markers linked to resistance trait have been routinely deployed in BB resistant genotypes breeding. The possibility to surveil genomic variations and the evolution of virulence-avirulence factors have expanded further with the availability of complete genome sequences of rice and Xoo. Domestication of rice has indeed narrowed the genetic diversity particularly with respect to diverse allelic forms of resistance genes that may exist in wild relatives of Oryza. Hence, it becomes imperative to conduct large-scale survey of wild rice species to characterize novel BB resistance genes and also novel alleles of known resistance genes so that they can be gainfully used for rice improvements. In our opinion, the need of the hour is to make best possible use of information and resources available not only in rice but also in other related crop species so as to achive durable resistance against BB disease. For example, investigations on how wheat crop avoids infection by Xoo or how rice avoids infection by the wheat rust pathogen, Puccinia gramins fsp. Tritici may offer clues for developing broad spectrum resistance in rice against multiple pathogens including Xoo. Interestingly, the focus of the research is now making a paradigm shift from individual genes to the whole plant systems. We anticipate that a better-coordinated inter-disciplinary research may reduce redundancy and competition in specific area along with dedicating greater attention to previously unexplored research areas.
Author Contributions
AK, RK, DS, SD, and AB designed the article. AK, RK, DS, SD, PS, and HS wrote the article. MP, NS, IG, GL, and RS corrected and improved the article. All authors contributed to the article and approved the submitted version.
Funding
AK sincerely thanks University Grants Commission (UGC) start-up grant (no. F.30-392/2017 (BSR) and Madhya Pradesh Council of Science and Technology (Endt. no. 3879/CST/R&D/BioSci/2018) for the funding to the laboratory. RK thanks Science and Engineering Research Board (SERB), Department of Science and Technology (DST) for extending financial support (project #PDF/2016/002758). RS, GL, and PS thank the Indian Council of Agricultural Research, Department of Biotechnology, Department of Science and Technology and Council for Scientific and Industrial Research, Government of India for the generous funding and infrastructural support. SD sincerely thanks University Grants Commission (UGC) for start-up grant (no. F.30-420/2018 (BSR).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abhilash, V. A., Balachiranjeevi, C. H., Bhaskar, Naik, S., Rambabu, R., Rekha, G., Hajira, S. K., et al. (2016). Development of gene-pyramid lines of the elite restorer line, RPHR-1005 possessing durable bacterial blight and blast resistance. Front. Plant Sci. 7:1195. doi: 10.3389/fpls.2016.01195
Abhilash, V. A., Balachiranjeevi, C. H., Naik, S. B., Rekha, G., Rambabu, R., Harika, G., et al. (2017). Marker-assisted pyramiding of bacterial blight and gall midge resistance genes into RPHR-1005, the restorer line of the popular rice hybrid DRRH-3. Mol. Breed. 37, 86. doi: 10.1007/s11032-017-0687-8
Agrawal, G. K., Rakwal, R. (2011). Rice proteomics: A move to-ward expanded proteome coverage to comparative and functional proteomics uncovers the mysteries of rice and plant biology. Proteomics 11, 1630–1649. doi: 10.1002/pmic.201000696
AICRIP. (2008). Varietal Improvement. 4th ed. (Hyderabad, India: All India Coordinated Rice Improvement Project Directorate of Rice Research Rajendranagar). Vol. 1,
Akimoto, K., Katakami, H., Kim, H. J., Ogawa, E., Sano, C. M., Wada, Y., et al. (2007). Epigenetic inheritance in rice plants. Ann. Bot. 100, 205–217. doi: 10.1093/aob/mcm110
Ali, M. L., Sanchez, P. L., Yu, S. B., Lorieux, M., Eizenga, G. C. (2010). Chromosome segment substitution lines: a powerful tool for the introgression of valuable genes from Oryza wild species into cultivated rice (O. sativa). Rice 3, 218–234. doi: 10.1007/s12284-010-9058-3
Amante-Bordeos, A., Sitch, L. A., Nelson, R., Dalmacio, R. D., Oliva, N. P., Aswidinnoor, H., et al. (1992). Transfer of bacterial blight and blast resistance from the tetraploid wild rice Oryza minuta to cultivated rice, Oryza sativa. Theor. Appl. Genet. 84, 345–354. doi: 10.1007/BF00229493
Angeles-Shim, R. B., Shim, J., Vinarao, R. B., Lapis, R. S., Singleton, J. J. (2020). A novel locus from the wild allotetraploid rice species Oryza latifolia Desv. confers bacterial blight (Xanthomonas oryzae pv. oryzae) resistance in rice (O. sativa). PLoS One 15 (2), e0229155. doi: 10.1371/journal.pone.0229155
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell. 22, 3864–3876. doi: 10.1105/tpc.110.078964
Asem, I. D., Imotomba, R. K., Mazumder, P. B., Laishram, J. M. (2015). Anthocyanin content in the black scented rice (Chakhao): its impact on human health and plant defense. Symbiosis 66, 47–54. doi: 10.1007/s13199-015-0329-z
Atkinson, N. J., Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Audic, S., Claverie, J. M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. doi: 10.1101/gr.7.10.986
Balachiranjeevi, C. H., Bhaskar, N. S., Abhilash, V., Akanksha, S., Viraktamath, B. C., Madhav, M. S., et al. (2015). Marker-assisted introgression of bacterial blight and blast resistance into DRR17B, an elite, fine-grain type maintainer line of rice. Mol. Breed. 35, 151. doi: 10.1007/s11032-015-0348-8
Balachiranjeevi, C. H., Naik, B., Kumar, A., Harika, G., Mahadev Swamy, H. K., Hajira, Sk., et al. (2018). Marker-assisted pyramiding of two major, broad-spectrum bacterial blight resistance genes, Xa21 and Xa33 into an elite maintainer line of rice, DRR17B. PLoS One 13, e0201271. doi: 10.1101/368712
Barbier, P. (1989). Genetic variation and ecotypic differentiation in wild rice Oryza rufipogon: Population differentiation in life-history traits and isozymes. Jpn. J. Genet. 64, 259–271. doi: 10.1266/jjg.64.259
Barkley, N. A., Wang, M. L. (2008). Application of TILLING and EcoTILLING as reverse genetic approaches to elucidate the function of genes in plants and animals. Curr. Genom. 9, 212–226. doi: 10.2174/138920208784533656
Barry, M. B., Pham, J. L., Noyer, J. L., Billot, C., Courtois, B., Ahmadi, N. (2007). Genetic diversity of the two cultivated rice species (O. sativa and O. glaberrima) in maritime Guinea. Evidence for interspecifc recombination. Euphytica 154, 127–137. doi: 10.1007/s10681-006-9278-1
Basavaraj, S. H., Singh, V. K., Singh, A., Singh, A. (2010). Singh A. Marker–assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol. Breed. 26, 293–305. doi: 10.1007/s11032-010-9407-3
Bhasin, H., Bhatia, D., Raghuvanshi, S., Lore, J. S., Sahi, G. K., Kaur, B. (2012). New PCR-based sequence-tagged site marker for bacterial blight resistance gene Xa38 of rice. Mol. Breed. 30, 607–611. doi: 10.1007/s11032-011-9646-y
Bimolata, W., Kumar, A., Sundaram, R. M., Laha, G. S., Qureshi, I. A., Reddy, G. A. (2013). Analysis of nucleotide diversity among alleles of the major bacterial blight resistance gene Xa27 in cultivars of rice (Oryza sativa) and its wild relatives. Planta 238, 239–305. doi: 10.1007/s00425-013-1891-3
Bimolata, W., Kumar, A., Reddy, M. S. K., Sundaram, R. M., Laha, G. S., Qureshi, I. A., et al. (2015). Nucleotide diversity analysis of three major bacterial blight resistance genes in rice. PLoS One 10, e0120186. doi: 10.1371/journal.pone.0120186
Borba, T. C., Mendes, C. D., Guimarães, É.P., Brunes, T. O., Fonseca, J. R., Brondani, R. V., et al. (2009). Genetic variability of Brazilian rice landraces determined by SSR markers. Pesq. Agropec. Bras. Brasília. 44, 706–712. doi: 10.1590/S0100-204X2009000700009
Borrelli, V. M., Brambilla, V., Rogowsky, P., Marocco, A., Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9:1245. doi: 10.3389/fpls.2018.01245
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Bradbury, J. F. (1984). “Genus II. XanthomonasDowson,” in Bergey"s Manual of Systematic Bacteriology. Eds. Krieg, N. R., Holt, J. G. (Baltimore: Williams & Wilkins), 199–210.
Burkart-Waco, D., Tsai, H., Ngo, K., Henry, I. M., Comai, L., Tai, T. H. (2016). “Biotechnologies for Plant Mutation Breeding: Protocols,” in Next-Generation Sequencing for Targeted Discovery of Rare Mutations in Rice Eds. Jankowicz-Cieslak, J., Tai, T. H., Kumlehn, J., Till, B. J. (Cham, Switzerland: Springer International Publishing), pp, 323–340. doi: 10.1007/10.1007
Busungu, C., Taura, S., Sakagami, J. I., Ichitani, K. (2016). Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24. Breed. Sci. 66, 636–645. doi: 10.1270/jsbbs.16062
Cao, J., Zhang, M., Xiao, J., Li, X., Yuan, M., Wang, S. (2018). Dominant and Recessive Major R Genes Lead to Different Types of Host Cell Death During Resistance to Xanthomonas oryzae in Rice. Front. Plant Sci. 9:1711. doi: 10.3389/fpls.2018.01711
Carvalho, T. L., Ballesteros, H. G., Thiebaut, F., Ferreira, P. C., Hemerly, A. S. (2016). Nice to meet you: genetic, epigenetic and metabolic controls of plant perception of beneficial associative and endophytic diazotrophic bacteria in non-leguminous plants. Plant Mol. Biol. 90, 561–574. doi: 10.1007/s11103-016-0435-1
Casella, L., Greco, R., Bruschi, G., Wozniak, B., Dreni, L., Kater, M., et al. (2013). TILLING in European rice: Hunting mutations for crop improvement. Crop Sci. 53, 2550–2562. doi: 10.2135/cropsci2012.12.0693
Cheema, K. K., Grewal, N. K., Vikal, Y., Sharma, R., Lore, J. S., Das, A., et al. (2008). A novel bacterial blight resistance gene from Oryza nivara mapped to 38 Kb region on chromosome 4L and transferred to Oryza sativa L. Genet. Res. 90, 397–407. doi: 10.1017/S0016672308009786
Chen, H., Wang, S., Zhang, Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92 (7), 750–754. doi: 10.1094/PHYTO.2002.92.7.750
Chen, X., Ronald, P. C. (2011). Innate immunity in rice. Trends Plant Sci. 16, 451–459. doi: 10.1016/j.tplants.2011.04.003
Chen, S., Zhang, Q. F. (2000). Improvement of bacterial blight resistance of hybrid rice by molecular marker-assisted selection. J. Huazhong Agric. Univ. 19, 183–189.
Chen, F., Yuan, Y., Li, Q., He, Z. (2007). Proteomic analysis of rice plasma membrane reveals proteins involved in early defense response to bacterial blight. Proteomics 7, 1529–1539. doi: 10.1002/pmic.200500765
Chen, S., Liu, X., Zeng, L., Ouyang, D., Yang, J., Zhu, X. (2011). Genetic analysis and molecular mapping of a novel recessive gene xa34(t) for resistance against Xanthomonas oryzae pv. oryzae. Theor. Appl. Genet. 122, 1331–1338. doi: 10.1007/s00122-011-1534-7
Cheng, Q., Mao, W., Xie, W., Liu, Q., Cao, J., Yuan, M., et al. (2017). Characterization of a disease susceptibility locus for exploring an efficient way to improve rice resistance against bacterial blight. Sci. China Life Sci. 60, 298–306. doi: 10.1007/s11427-016-0299-x
Chisholm, S. T., Coaker, G., Day, B., Staskawicz, B. J. (2006). Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124, 803–814. doi: 10.1016/j.cell.2006.02.008
Choi, Y. J., Yun, H. K., Park, K. S., Noh, J. H., Heo, Y. Y., Kim, S. H., et al. (2010). Transcriptional profiling of ESTs responsive to Rhizobium vitis from “Tamnara” grapevines (Vitis sp.). J. Plant Physiol. 167, 1084–1092. doi: 10.1016/j.jplph.2010.02.005
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi: 10.1534/genetics.110.120717
Chu, Z., Ouyang, Y., Zhang, J., Yang, H., Wang, S. (2004). Genome wide analysis of defense responsive genes in bacterial blight resistance of rice mediated by the recessive R gene xa13. Mol. Genet. Genomics 271, 111–120. doi: 10.1007/s00438-003-0964-6
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., et al. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Gene Dev. 20, 1250–1255. doi: 10.1101/gad.1416306
Cianciotto, N. P., White, R. C. (2017). Expanding role of type II secretion in bacterial pathogenesis and beyond. Infect. Immun. 85, e00014–e00017. doi: 10.1128/IAI.00014-17
Cohen, S. P., Liu, H., Argueso, C. T., Pereira, A., Vera Cruz, C., Verdier, V., et al. (2017). RNA-Seq analysis reveals insight into enhanced rice Xa7-mediated bacterial blight resistance at high temperature. PLoS One 12 (11), e0187625. . doi: 10.1371/journal.pone.0187625
Crossa, J., Pérez-Rodríguez, P., Cuevas, J., Montesinos-López, O., Jarquín, D., de los Campos, G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011
Datta, K., Baisakh, N., Thet, K. M., Tu, J., Datta, S. (2002). Pyramiding transgenes for multiple resistance in rice against bacterial blight, yellow stem borer and sheath blight. Theor. Appl. Genet. 106, 1–8. doi: 10.1007/s00122-002-1014-1
Davierwala, A. P., Reddy, A. P., Lagu, M. D., Ranjekar, P. K., Gupta, V. S. (2001). Marker assisted selection of bacterial blight resistance genes in rice. Biochem. Genet. 39, 170–183. doi: 10.1023/A:1010282732444
de Jong, C. F., Takken, F. L., Cai, X., de Wit, P. J., Joosten, M. H. (2000). Attenuation of Cf-mediated defense responses at atelevated temperatures correlates with a decrease in elicitor-binding sites. Mol. Plant-Microbe Interact. 15 (10), 1040–1049. doi: 10.1094/MPMI.2002.15.10.1040
Denancé, N., Sánchez-Vallet, A., Goffner, D., Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Deng, Y., Wu, J. E., Tao, F., Zhang, L. H. (2010). Listening to a new language: DSF-based quorum sensing in Gram-negative bacteria. Chem. Rev. 111, 160–173. doi: 10.1021/cr100354f
Dharmapuri, S., Sonti, R. V. (1999). A transposon insertion in the gumG homologue of Xanthomonas oryzae pv.oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol. Lett. 179, 53–59. doi: 10.1111/j.1574-6968.1999.tb08707.x
Diatchenko, L., Lau, Y. F., Campbell, A. P., Chenchik, A., Moqadam, F., Huang, B., et al. (1996). Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. 93, 6025–6030. doi: 10.1073/pnas.93.12.6025
Ding, C., You, J., Wang, S., Liu, Z., Li, G., Wang, Q., et al. (2012). A proteomic approach to analyze nitrogen and cytokinin-responsive proteins in rice roots. Mol. Biol. Rep. 39, 1617–1626. doi: 10.1007/s11033-011-0901-4
Dixon, M. S., Jones, D. A., Keddie, J. S., Thomas, C. M., Harrison, K., Jones, J. D. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. doi: 10.1016/S0092-8674(00)81290-8
Djedatin, G., Ndjiondjop, M. N., Sanni, A., Lorieux, M., Verdier, V., Ghesquiere, A. (2016). Identification of novel major and minor QTLsassociated with Xanthomonas oryzae pv. Oryzae (African strains) resistance in rice (Oryza sativaL). Rice 9, 18. doi: 10.1186/s12284-016-0090-9
Dokku, P., Das, K. M., Rao, G. J. N. (2013). Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker assisted selection. Euphytica 192, 87–96. doi: 10.1007/s10681-013-0878-2
Dossa, G. S., Torres, R., Henry, A., Oliva, R., Maiss, E., Vera Cruz, C., et al. (2017). Rice response to simultaneous bacterial blight and drought stress during compatible and incompatible interactions. Eur. J. Plant Pathol. 147, 115–127. doi: 10.1007/s10658-016-0985-8
Dossa, G. S., Quibod, I., Atienz ana-Grande, G., Olivca, R., Maiss, E., Vera Cruz, C., et al. (2020). Rice pyramided line IRBB67 (Xa4/Xa7)homeostasis under combined stress of high temperature and bacterial blight.Sci. Rep. 10, 683. doi: 10.1038/s41598-020-57499-5
Ellur, R. K., Khanna, A., Bhowmick, P. K., Vinod, K. K., Nagarajan, M., Mondal, K. K., et al. (2016). Marker-aided incorporation of Xa38, a novel bacterial blight resistance gene, in PB1121 and comparison of its resistance spectrum with xa13+Xa21. Sci. Rep. 6, 29188. doi: 10.1038/srep29188
Ershad, M., Gopi, P., Suneel, B., Laha, G. S., Sundaram, R. M., Padma kumari, A. P., et al. (2017). Identification of Bacterial Blight Resistant Mutant Lines in the Mutagenized Samba Mashuri (BPT 5204) Population. ICAR-IIRR News Lett. 15, 234–235.
Fahlgren, N., Howell, M. D., Kasschau, K. D., Chapman, E. J., Sullivan, C. M., Cumbie, J. S. (2007). High-throughput sequencing of Arabidopsis microRNAs: Evidence for frequent birth and death of miRNA genes. PLoS One 2, e219. . doi: 10.1371/journal.pone.0000219
FAO (Food and Agriculture Organization) (2004). Rice is life (Italy: FAO International Year of Rice, Rice and Human Nutrition).
Figueiredo, A., Fortes, A. M., Ferreira, S., Sebastiana, M., Choi, Y. H., Sousa, L., et al. (2008). Transcriptional and metabolic profiling of grape (Vitis vinifera L.) leaves unravel possible innate resistance against pathogenic fungi. J. Exp. Bot. 59, 3371–3381. doi: 10.1093/jxb/ern187
Fisher, S., Sana, T. (2007). “,” in Metabolomic profiling of bacterial leaf blight in rice, vol. 2. (Santa Clara, CA: Agilent Technologies), 16.
Fitzgerald, T. L., Kazan, K., Li, Z., Morell, M. K., Manners, J. M. (2010). A high-throughput method for the detection of homologous gene deletions in hexaploid wheat. BMC Plant Biol. 10, 264. doi: 10.1186/1471-2229-10-264
Gaj, T., Gersbach, C. A., Barbas, C. F. (2013). ZFN, TALEN and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004
Gao, D. Y., Xu, Z. G., Chen, Z. Y., Sun, L. H., Sun, Q. M., Lu, F. (2002). Identification of a resistance gene to bacterial blight (Xanthomonas oryzae pv. oryzae) in a somaclonal mutant HX-3 of indica rice. Yi Chuan Xue Bao 29, 138–143.
Ghazi, I. A., Srivastava, P. S., Dalal, V., Gaikwad, K., Singh, A. K., Sharma, T. R. (2009). Physical mapping, expression analysis and polymorphism survey of resistance gene analogues on chromosome 11 of rice. J. Biosci. 34, 251–261. doi: 10.1007/s12038-009-0029-z
Glazebrook, J. (2001). Genes controlling expression of defense responses in Arabidopsis-status. Curr. Opin. Plant Biol. 4, 301–308. doi: 10.1016/S1369-5266(00)00177-1
Gnanamanickam, S. S., Priyadarisini, V. B., Narayanan, N. N., Vasudevan, P., Kavitha, S. (1999). An overview of bacterial blight disease of rice and strategies for its management. Curr. Sci. 77, 1435–1443.
Gnanamanickam, S. S. (2009). “Biological control of bacterial blight of rice,” in Biological control of rice diseases, vol. 8 . Ed. Gnanamanickam, S. S. (New York: Springer), 67–78.
Goicoechea, J. L., Ammiraju, J. S., Marri, P. R., Chen, M., Jackson, S., Yu, Y. (2010). The future of rice genomics: Sequencing the collective Oryza genome. Rice 3, 89–97. doi: 10.1007/s12284-010-9052-9
Gong, L., Chen, W., Gao, Y., Liu, X., Zhang, H., Xu, C. (2013). Genetic analysis of the metabolome exemplified using a rice population. Proc. Natl. Acad. Sci. 10, 20320–20325. doi: 10.1073/pnas.1319681110
González, M., Xu, M., Esteras, C., Roig, C., Monforte, A. J., Troadec, C., et al. (2011). Towards a TILLING platform for functional genomics in Piel de Sapo melons. BMC Res. Notes 4, 289. doi: 10.1186/1756-0500-4-289
Grewal, R., Gupta, S., Das, S. (2012). Xanthomonas oryzae pv oryzae triggers immediate transcriptomic modulations in rice. BMC Genomics 13, 49. doi: 10.1186/1471-2164-13-49
Gu, K., Tian, D., Yang, F. W., Wu, L., Sreekala, C., Wang, D., et al. (2004). High-resolution genetic mapping of Xa27 (t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 108, 800–807. doi: 10.1007/s00122-003-1491-x
Gu, K., Yang, B., Tian, D., Wu, L., Wang, D., Sreekala, C. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Gu, K., Sangha, J. S., Li, Y., Yin, Z. (2008). High-resolution genetic mapping of bacterial blight resistance gene Xa10. Theor. Appl. Genet. 116, 155–163. doi: 10.1007/s00122-007-0655-5
Guo, S. B., Zhang, D. P., Lin, X. H. (2010). Identification and mapping of a novel bacterial blight resistance gene Xa35(t)originated from Oryza minuta. Sci. Agric. Sin. 43, 2611–2618.
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., Park, S. W. (2012). Plant disease resistance genes: current status and future directions. Physiol. Mol. Plant Pathol. 78, 51– 65. doi: 10.1016/j.pmpp.2012.01.002
Gust, A. A., Brunner, F., Nürnberger, T. (2010). Biotechnological concepts for improving plant innate immunity. Curr. Opin. Biotechnol. 21, 204–210. doi: 10.1016/j.copbio.2010.02.004
Guvvala, L. D., Koradi, P., Shenoy, V., Marella, L. S. (2013). Making an Indian traditional rice variety Mahsuri, bacterial blight resistant using marker-assisted selection. J. Crop Sci. Biotech. 16, 111–121. doi: 10.1007/s12892-013-0009-6
Hammond-Kosack, K. E., Jones, J. D. (1997). Plant disease resistance genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 575–607. doi: 10.1146/annurev.arplant.48.1.575
Hari, Y., Srinivasarao, K., Viraktamath, B. C., Hari Prasad, A. S., Laha, G. S., Ahmed, M. I., et al. (2011). Marker-assisted improvement of a stable restorer line, KMR-3R and its derived hybrid KRH2 for bacterial blight resistance and grain quality. Plant Breed. 130, 608–616. doi: 10.1111/j.1439-0523.2011.01881.x
Hari, Y., Srinivasarao, K., Viraktamath, B. C., Hari Prasad, A. S., Laha, G. S., Ahmed, M. I., et al. (2013). Marker-assisted introgression of bacterial blight and blast resistance into IR 58025B, an elite maintainer line of rice. Plant Breed. 132, 586–594. doi: 10.1111/pbr.12056
Hashiguchi, A., Ahsan, N., Komatsu, S. (2010). Proteomics application of crops in the context of climatic changes. Food Res. Int. 43, 1803–1813. doi: 10.1016/j.foodres.2009.07.033
He, Q., Li, D., Zhu, Y., Tan, M., Zhang, D., Lin, X. (2006). Fine mapping of Xa2, a bacterial blight resistance gene in rice. Mol. Breed. 17, 1–6. doi: 10.1007/s11032-005-8698-2
Heffner, L., Sorrells, M. E., Jannink, J. L. (2009). Genomic selection for crop improvement. Crop Sci. 49, 1–12. doi: 10.2135/cropsci2008.08.0512
Heuberger, A. L., Robison, F. M., Lyons, S. M., Broeckling, C. D., Prenni, J. E. (2014). Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 5:291. doi: 10.3389/fpls.2014.00291
Hollywood, K., Brison, D. R., Goodacre, R. (2006). Metabolomics: Current technologies and future trends. Proteomics 6, 4716–4723. doi: 10.1002/pmic.200600106
Hopkins, C. M., White, F. F., Choi, S. H., Guo, A., Leach, J. E. (1992). Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 5, 451–459. doi: 10.1094/MPMI-5-451
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Hu, K., Cao, J., Zhang, J., Xia, F., Ke, Y., Zhang, H., et al. (2017). Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement. Nat. Plants 3, 17009. doi: 10.1038/nplants.2017.9
Huang, N., Angeles, E. R., Domingo, J., Magpantay, G., Singh, S., Zhang, G., et al. (1997). Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor. Appl. Genet. 95, 313–320. doi: 10.1007/s001220050565
Huang, B., Xu, J. Y., Hou, M. S., Ali, J., Mou, T. M. (2012). Introgression of bacterial blight resistance genes Xa7, Xa21, Xa22 and Xa23 into hybrid rice restorer lines by molecular marker-assisted selection. Euphytica 187, 449–459. doi: 10.1007/s10681-012-0758-1
Hutin, M., Sabot, F., Ghesquière, A., Koebnik, R., Szurek, B. (2015). A knowledge-based molecular screen uncovers a broad-spectrum Os SWEET 14 resistance allele to bacterial blight from wild rice. Plant J. 84, 694–703. doi: 10.1111/tpj.13042
Hwang, J. E., Jang, D. S., Lee, K. J., Ahn, J. W., Kim, S. H., Kang, S. Y., et al. (2016). Identification of gamma ray irradiation-induced mutations in membrane transport genes in a rice population by TILLING. Genes Genet. Syst. 91, 245–256. doi: 10.1266/ggs.15-00052
Iqbal, M. J., Yaegashi, S., Ahsan, R., Shopinski, K. L., Lightfoot, D. A. (2005). Root response to Fusarium solani f. sp. glycines: temporal accumulation of transcripts in partially resistant and susceptible soybean. Theor. Appl. Genet. 110, 1429–1438. doi: 10.1007/s00122-005-1969-9
Iyer, A. S., McCouch, S. R. (2004). The rice bacterial blight resistance gene xa5 encodes a novel form of disease resistance. Mol. Plant Microbe Interact. 17, 1348–1354. doi: 10.1094/MPMI.2004.17.12.1348
Iyer, A. S., Jiang, H., Huang, L., McCouch, S. R. (2004). Genetic and functional characterization of the rice bacterial blight disease resistance gene xa5. Phytopathol 98, 289–295. doi: 10.1094/PHYTO-98-3-0289
Jain, P., Singh, P. K., Kapoor, R., Khanna, A., Solanke, A. U., Krishnan, S. G., et al. (2017). Understanding host-pathogen interactions with expression profiling of NILs carrying rice-blast resistance Pi9 gene. Front. Plant Sci. 8:93. doi: 10.3389/fpls.2017.00093
Jha, G., Rajeshwari, R., Sonti, R. V. (2005). Bacterial type two secretion system secreted proteins: double-edged swords for plant pathogens. Mol. Plant-Microbe Interact. 18, 891–898. doi: 10.1094/MPMI-18-0891
Jha, G., Rajeshwari, R., Sonti, R. V. (2007). Functional interplay between two Xanthomonas oryzae pv. oryzae secretion systems in modulating virulence on rice. Mol. Plant Microbe Interact. 20, 31–40. doi: 10.1094/MPMI-20-0031
Ji, Z., Ji, C., Liu, B., Zou, L., Chen, G., Yang, B. (2016). Interfering TAL effectors of Xanthomonas oryzae neutralize R-gene-mediated plant disease resistance. Nat. Commun. 7, 1–9. doi: 10.1038/ncomms13435
Jian, W. W., Wu, J. Y. (2005). Nitric oxide is involved in methyl jasmonate-induced defense responses and secondary metabolism activities of Taxus cells. Plant Cell Physiol. 46, 923–930. doi: 10.1093/pcp/pci098
Jiang, G. H., Xia, Z. H., Zhou, Y. L., Wan, J., Li, D. Y., Chen, R. S. (2006). Testifying the rice bacterial blight resistance gene xa5 by genetic complementation and further analyzing xa5 (Xa5) in comparison with its homolog TFIIAgamma1. Mol. Genet. Genomics 275, 354–366. doi: 10.1007/s00438-005-0091-7
Jiang, N., Yan, J., Liang, Y., Shi, Y., He, Z., Wu, Y., et al. (2020). Resistance Genes and their Interactions with Bacterial Blight/Leaf Streak Pathogens (Xanthomonas oryzae) in Rice (Oryza sativa L.)-an Updated Review. Rice 13, 1–12. doi: 10.1186/s12284-019-0358-y
Jin, X. W., Wang, C. L., Yang, Q., Jiang, Q. X., Fan, Y. L., Liu, G. C., et al (2007). Breeding of near-isogenic line CBB30 and molecular mapping of Xa30(t), a new resistance gene to bacterial blight in rice. Sci. Agric. Sin. 40 (6), 1094–1100.
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johal, G. S., Briggs, S. P. (1992). Reductase activity encoded by the HM1 disease resistance gene in maize. Science 258, 985–987. doi: 10.1126/science.1359642
Jones, J. D., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Joseph, M., Gopalakrishnan, S., Sharma, R. K., Singh, V. P., Singh, A. K., Singh, N. K., et al. (2004). Combining bacterial blight resistance and basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breed. 13, 377–387. doi: 10.1023/B:MOLB.0000034093.63593.4c
Kaku, H., Ogawa, T. (2001). Genetic Analysis of the relationship between the browning reaction and bacterial blight resistance gene Xa3 in rice. J. Gen. Plant Pathol. 67, 228–230. doi: 10.1007/PL00013017
Karatan, E., Watnick, P. (2009). Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol. Mol. Biol. Rev. 73, 310–347. doi: 10.1128/MMBR.00041-08
Kaur, R., Grewal, N., Das, A., Vikal, Y., Singh, J., Bharaj, T. S., et al. (2005). Inheritance of bacterial blight resistance in two accessions of wild rice, Oryza nivara. Rice Genet. Newsl. 22, 21.
Khush, G. S., Angeles, E. R. (1999). A new gene for resistance to race 6 of bacterial blight in rice, Oryza sativa. Rice Genet. Newsl. 16, 92– 93.
Khush, G. S., Mackill, D. J., Sidhu, G. S. (1989). “Breeding rice for resistance to bacterial blight. Bacterial Blight of Rice,” in Proceedings of the International Workshop on Bacterial Blight Rice (Manila, Philippines: IRRI), 207–217.
Khush, G. S. (2005). What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 59, 1–6. doi: 10.1007/s11103-005-2159-5
Khush, G. S. (2013). Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant Breed. 132, 433–436. doi: 10.1111/pbr.1991
Kim, S. (2018). Identification of novel recessive gene xa44(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip. Theor. Appl. Genet. 131, 2733–2743. doi: 10.1007/s00122-018-3187-2
Kim, S.-M., Reinke, R. F. (2019). A novel resistance gene for bacterial blight in rice Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 14 (2), e0211775. doi: 10.1371/journal.pone.0211775
Kim, Y. G., Cha, J., Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Kim, S. M., Suh, J. P., Qin, Y., Noh, T. H., Reinke, R. F., Jena, K. K. (2015). Identification and fine-mapping of a new resistance gene, Xa40, conferring resistance to bacterial blight races in rice (Oryza sativa L.). Theor. Appl. Genet. 128, 1933–1943. doi: 10.1007/s00122-015-2557-2
Kong, W., Ding, L., Xia, X. (2020). Identification and characterization of genes frequently responsive to Xanthomonas oryzae pv. oryzae and Magnaporthe oryzae infections in rice. BMC Genomics 21, 21. doi: 10.1186/s12864-019-6438-y
Konishi, H., Ishiguro, K., Komatsu, S. A. (2001). Proteomics approach towards understanding blast fungus infection of rice grown under different levels of nitrogen fertilization. Proteomics 1, 1162–1171. doi: 10.1002/1615-9861(200109)1:9<1162::AID-PROT1162>3.0.CO;2-S
Korinsak, S., Sriprakhon, S., Sirithanya, P., Jairin, J., Vanavichit, A., Toojinda, T. (2009). Identification of microsatellite markers (SSR) linked to a new bacterial blight resistance gene xa33 (t) in rice cultivar ‘Ba7’. Maejo. Int. J. Sci. Tech. 3, 235–247.
Kumar, P. N., Sujatha, K., Laha, G. S., Rao, K. S., Mishra, B., Viraktamath, B. C., et al. (2012). Identification and fine-mapping of Xa33 a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathol 102, 222–228. doi: 10.1094/PHYTO-03-11-0075
Kumar, A., Gul, M. Z., Zeeshan, A., Bimolata, W., Qureshi, I. A., Ghazi, I. A. (2013). Differential antioxidative responses of three different rice genotypes during bacterial blight infection. Aust. J. Crop Sci. 7, 1893–1900.
Kumar, A., Bimolata, W., Kannan, M., Kirti, P. B., Qureshi, I. A., Ghazi, I. A. (2015). Comparative proteomics reveals differential induction of both biotic and abiotic stress response associated proteins in rice during Xanthomonas oryzae pv. oryzae infection. Funct. Integr. Genomic 15, 425–437. doi: 10.1007/s10142-014-0431-y
Kumar, R., Bohra, A., Pandey, A. K., Pandey, M. K., Kumar, A. (2017). Metabolomics for plant improvement: status and prospects. Front. Plant Sci. 8:1302. doi: 10.3389/fpls.2017.01302
Kurata, N., Yamazaki, Y. (2006). Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 140, 12–17. doi: 10.1104/pp.105.063008
Lacombe, S., Rougon-Cardoso, A., Sherwood, E., Peeters, N., Dahlbeck, D., Van Esse, H. P., et al. (2010). Interfamily transfer of a plant pattern-recognition receptor confers broad-spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. doi: 10.1038/nbt.1613
Lee, K. S., Khush, G. S. (2000). Genetic analysis of resistance to bacterial blight, Xanthomonasoryzae pv. oryzae rice. Rice Genet. Newsl. 17, 72.
Lee, K. S., Rasabandith, S., Angeles, E. R., Khush, G. S. (2003). Inheritance of resistance to bacterial blight in 21 cultivars of rice. Phytopathol 93, 147–152. doi: 10.1094/PHYTO.2003.93.2.147
Leigh, J. A., Coplin, D. L. (1992). Exopolysaccharides in plant–bacterial interactions. Annu. Rev. Microbiol. 46, 307–346. doi: 10.1146/annurev.mi.46.100192.001515
Leung, H., Wu, J., Liu, B., Bustaman, R., Sridhar, K., Singh, E., et al. (2004). Sustainable disease resistance in rice: currentand future strategies. Proceedings of the 4th international crop science congress, Brisbane, Australia, 26 September - 1 October.
Li, Z. K., Luo, L. J., Mei, H. W., Paterson, A. H., Zhao, X. Z., Zhong, D. B., et al. (1999). A “defeated” rice resistance gene acts as a QTL against a virulent strain of Xanthomonas oryzae pv. oryzae. Mol. Gen. Genet. 261, 58–63. doi: 10.1007/s004380050941
Li, P., Long, J. Y., Huang, Y. C., Zhang, Y., Wang, J. S. (2004). AvrXa3: a novel member of avrBs3 gene family from Xanthomonas oryzae pv. oryzae has a dual function. Prog. Nat. Sci. 14, 767–773. doi: 10.1080/10020070412331344311
Li, Z. K., Arif, M., Zhong, D. B., Fu, B. Y., Xu, J. L., Domingo-Rey, J., et al. (2006). Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc. Natl. Acad. Sci. 103, 7994–7999. doi: 10.1073/pnas.0507492103
Li, Q., Chen, F., Sun, L., Zhang, Z., Yang, Y., He, Z. (2006). Expression profiling of rice genes in early defense responses to blast and bacterial blight pathogens using cDNA microarray. Physiol. Mol. Plant Pathol. 68, 51–60. doi: 10.1016/j.pmpp.2006.06.002
Li, C., Wei, J., Lin, Y., Chen, H. (2012). Gene silencing using the recessive rice bacterial blight resistance gene xa13 as a new paradigm in plant breeding. Plant Cell Rep. 31, 851–862. doi: 10.1007/s00299-011-1206-8
Li, H. J., Li, X. H., Xiao, J. H., Wing, R. A., Wang, S. P. (2012). Ortholog alleles at Xa3/Xa26 locus confer conserved race-specific resistance against Xanthomonas oryzae in rice. Mol. Plant 5, 281–290. doi: 10.1093/mp/ssr079
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, T., Huang, S., Zhou, J., Yang, B. (2013). Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 6, 781–789. doi: 10.1093/mp/sst034
Li, J. Y., Wang, J., Zeigler, R. S. (2014). The 3,000 rice genomes project: new opportunities and challenges for future rice research. GigaScience 3, 1–3. doi: 10.1186/2047-217X-3-8
Li, Y., Li, D., Guo, Z., Shi, Q., Xiong, S., Zhang, C., et al. (2016). OsACOS12, an orthologue of Arabidopsisacyl-CoA synthetase5, plays an important role in pollen exine formation and anther development in rice. BMC Plant Biol. 16, 256. doi: 10.1186/s12870-016-0943-9
Liu, H. X., Liu, F. Q., Hu, B. S., Yang, W. F., Chen, Z. Y., Xu, Z. G. (2004). Virulence of Xanthomonas oryzae pv. oryzae on rice near-isogenic lines with single resistance gene and pyramiding lines in China. Agric. Sci. China 3, 764–769.
Liu, J., Liu, X., Dai, L., Wang, G. (2007). Recent progress in elucidating the structure, function and evolution of disease resistance genes in plants. J. Genet. Genom. 34, 765–776. doi: 10.1016/S1673-8527(07)60087-3
Liu, Q., Yuan, M., Zhou, Y. A., Li, X., Xiao, J., Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ. 34, 1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Luo, S., Zhang, Y., Hu, Q., Chen, J., Li, K., Lu, C., et al. (2012). Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family. Plant Physiol. 159, 197–210. doi: 10.1104/pp.111.192062
Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54, 263–272. doi: 10.1016/j.molcel.2014.03.028
Mahmood, T., Jan, A., Kakishima, M., Komatsu, S. (2006). Proteomic analysis of bacterial blight defense-responsive proteins in rice leaf blades. Proteomics 6, 6053–6065. doi: 10.1002/pmic.200600470
Malukani, K. K., Ranjan, A., Hota, S. J., Patel, H. K., Sonti, R. V. (2020). Dual Activities of Receptor-Like KinaseOsWAKL21.2 Induce Immune Responses. Plant Physiol. 183 (3), 1345–1363. doi: 10.1104/pp.19.01579
Malzahn, A., Lowder, L., Qi, Y. (2017). Plant genome editing with TALEN and CRISPR. Cell Biosci. 24, 7–21. doi: 10.1186/s13578-017-0148-4
Mardis, E. R. (2008). Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402. doi: 10.1146/annurev.genom.9.081307.164359
Menges, M., Dóczi, R., Ökrész, L., Morandini, P., Mizzi, L., Soloviev, M., et al. (2008). Comprehensive gene expression atlas for the Arabidopsis MAP kinase signalling pathways. New Phytol. 179, 643–662. doi: 10.1111/j.1469-8137.2008.02552.x
Metallo, C. M., Vander-Heiden, M. G. (2013). Understanding metabolic regulation and its influence on cell physiology. Mol. Cell 49, 388–398. doi: 10.1016/j.molcel.2013.01.018
Mew, T. W., Vera Cruz, C. M., Medalla, E. S. (1992). Changes in race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis. 76, 1029–1032. doi: 10.1094/PD-76-1029
Mew, T. W., Alvarez, A. M., Leach, J. E., Swings, J. (1993). Focus on bacterial blight of rice. Plant Dis. 77, 5–12. doi: 10.1094/PD-77-0005
Mew, T. W. (1987). Current status and future prospects of research on bacterial blight of rice. Ann. Rev. Phytopathol. 25, 359–382. doi: 10.1146/annurev.py.25.090187.002043
Miao, L., Wang, C., Zheng, C., Che, J., Gao, Y., Wen, Y., et al. (2010). Molecular mapping of a new gene for resistance to rice bacterial blight. Sci. Agric. Sin. 43, 3051–3058.
Miao, J., Guo, D., Zhang, J., Huang, Q., Qin, G., Zhang, X., et al. (2013). Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 23, 1233–1236. doi: 10.1038/cr.2013.123
Michelmore, R. W., Meyers, B. C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. doi: 10.1101/gr.8.11.1113
Monaghan, J., Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Mori, M., Tomita, C., Sugimoto, K., Hasegawa, M., Hayashi, N., Dubouzet, J. G., et al. (2007). Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol. Biol. 63, 847–860. doi: 10.1007/s11103-006-9130-y
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., Wold, B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628. doi: 10.1038/nmeth.1226
Mundt, C. C. (1990). Probability of mutation to multiple virulence and durability of resistance gene pyramids. Phytopathol 80, 221–223. doi: 10.1094/Phyto-80-221
Mundt, C. C. (2014). Durable resistance: a key to sustainable management of pathogens and pests. Infect. Genet. Evol. 27, 446–455. doi: 10.1016/j.meegid.2014.01.011
Nakai, H., Nakamura, S., Kuwahara, S., Saito, M. (1988). Genetic studies of an induced rice mutant resistant to multiple races of bacterial leaf blight. Rice Genet. Newsl. 5, 101.
Natrajkumar, P., Sujatha, K., Laha, G. S., Srinivasa Rao, K., Mishra, B., Viraktamath, B. C., et al (2012). Identification and fine-mapping of Xa33, a novel gene for resistance to Xanthomonas oryzae pv. oryzae. Phytopathology 102, 222. doi: 10.1094/PHYTO-03-11-0075
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Neelam, K., Mahajan, R., Gupta, V., Bhatia, D., Gill, B. K., Komal, R., et al. (2019). High-resolution genetic mapping of a novelbacterial blight resistance gene xa-45 (t) identified from Oryza glaberrima and transferred to Oryzasativa. Theor. Appl. Genet. 6, 1–7. doi: 10.1007/s00122-019-03501-2
Nino-Liu, D. O., Ronald, P. C., Bogdanove, A. J. (2006). Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol. Plant Pathol. 7, 303–324. doi: 10.1111/j.1364-3703.2006.00344.x
Noda, T., Kaku, H. (1999). Growth of Xanthomonas oryzae pv. oryzae in planta and in guttation fluid of rice. Ann. Phytopathol. Soc Japan 65, 9–14. doi: 10.3186/jjphytopath.65.9
Ogawa, T. (1988). Near-isogenic lines as international differentials for resistance to bacterial blight of rice. Rice Genet. Newsl. 5, 106–109.
Oksman-Caldentey, K. M., Saito, K. (2005). Integrating genomics and metabolomics for engineering plant metabolic pathways. Curr. Opin. Biotechnol. 16, 174–179. doi: 10.1016/j.copbio.2005.02.007
Oliva, R., Ji, C., Atienza-Grande, G., Huguet-Tapia, J. C., Perez-Quintero, A., Li, T., et al. (2019). Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 37, 1344–1350. doi: 10.1038/s41587-019-0267-z
O’Toole, J. C., Toenniessen, G. H., Murashige, T., Harris, R. R., Herdt, R. W. (2001). “The Rockefeller Foundation’s international program on rice biotechnology,” in Rice Genetics IV. Eds. Khush, G. S., (New Delhi: Science Publishers and Los Baños, Philippines: International Rice Research Institute). 39–62.
Oryzabase (2006). Integrated Rice Science Database, Japan: National Institute of Genetics and Kyushu University. http://www.shigen.nig.ac.jp/rice/oryzabase/.
Pandey, M. K., Rani, N. S., Sundaram, R. M., Laha, G. S., Madhav, M. S., Rao, K. S., et al. (2013). Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection. Mol. Breed. 31, 239–246. doi: 10.1007/s11032-012-9779-7
Paternain, L., Campion, J. (2013). Metabolomics and transcriptomics of metabolic disorders. Curr. Nutr. Rep. 2, 199–206. doi: 10.1007/s13668-013-0062-2
Perez, L. M., Redoña, E. D., Mendioro, M. S., Cruz, C. M., Leung, H. (2008). Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermos sensitive genetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica 164, 627–636. doi: 10.1007/s10681-008-9653-1
Perumalsamy, S., Bharani, M., Sudha, M., Nagarajan, P., Arul, L., Saraswathi, R., et al. (2010). Functional marker-assisted selection forbacterial leaf blight resistance genes in rice (Oryza sativa L.). Plant Breed. 129, 400–406. doi: 10.1111/j.1439-0523.2009.01705.x
Piron, F., Nicolai, M., Minoia, S., Piednoir, E., Moretti, A., Salgues, A., et al. (2010). An induced mutation in tomato eIF4E leads to immunity to two potyviruses. PLoS One 5, e11313. doi: 10.1371/journal.pone.0011313
Porter, B. W., Chittoor, J. M., Yano, M., Sasaki, T., White, F. F. (2003). Development and mapping of markers linked to the rice bacterial blight resistance gene Xa7. Crop Sci. 43, 1484–1492. doi: 10.2135/cropsci2003.1484
Porter, B. W., Paidi, M., Ming, R., Alam, M., Nishijima, W. T., Zhu, Y. J. (2009). Genome-wide analysis of Carica papaya reveals a small NBS resistance gene family. Mol. Genet. Genom. 281, 609–626. doi: 10.1007/s00438-009-0434-x
Pradhan, B. B., Chatterjee, S. (2014). Reversible non-genetic phenotypic heterogeneity in bacterial quorum sensing. Mol. Microbiol. 92, 557–569. doi: 10.1111/mmi.12575
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., et al. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deep water rice variety, Jalmagna. Rice 8, 51. doi: 10.1186/s12284-015-0051-8
Pradhan, S. K., Nayak, D. K., Pandit, E., Behera, L., Anandan, A., Mukherjee, A. K., et al. (2016). Incorporation of bacterial blight resistance genes into lowland rice cultivars through marker assisted backcross breeding. Phytopathol 106, 710–718. doi: 10.1094/PHYTO-09-15-0226-R
Pruitt, R. N., Schwessinger, B., Joe, A., Thomas, N., Liu, F., Albert, M., et al. (2015). The rice immune receptor XA21 recognizes atyrosine-sulfated protein from a gram-negative bacterium. Sci. Adv. 1, e1500245. doi: 10.1126/sciadv.1500245
Rajeshwari, R., Jha, G., Sonti, R. V. (2005). Role of an in planta expressed xylanase of Xanthomonas oryzae pv. oryzae in promoting virulence on rice. Mol. Plant-Microbe Interact. 18, 830–837. doi: 10.1094/MPMI-18-0830
Rajpurohit, D., Kumar, R., Kumar, M., Paul, P., Awasthi, A., Basha, P. O., et al. (2010). Pyramiding of two bacterial blight resistance and a semidwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 178, 111–126. doi: 10.1007/s10681-010-0279-8
Ram, T., Laha, G. S., Gautam, S. K., Deen, R., Madhav, M. S., Brar, D. S., et al. (2010). Identification of new gene introgressed from Oryza brachyantha with broad-spectrum resistance to bacterial blight of rice in India. Rice Genet. Newsl. 25, 57.
Ramkumar, G., Sakthivel, K., Sundaram, R. M., Neeraja, C. N., Balachandran, S. M., Rani, N. S., et al (2010). Allele mining in crops: prospects and potentials. Biotechnol. Adv. 28, 451–461. doi: 10.1016/j.biotechadv.2010.02.007
Reddy, J. N., Baraoidan, M. R., Bernardo, M. A., George, M. L. C., Sridhar, R. (1997). Application of marker-assisted selection in rice for bacterial blight resistance gene, Xa21. Curr. Sci. 73, 873–875.
Ruan, H. H., Yan, C. Q., An, D. R., Liu, R. H., Chen, J. P. (2008). Identifying and mapping new gene xa32 (t) for resistance to bacterial blight (Xanthomonas oryzae pv. oryzae, Xoo) from Oryzae meyeriana L. Acta Agric. Bor. Sin. 17, 170–174.
Sabetta, W., Alba, V., Blanco, A. (2011). Montemurro C. sunTILL: a TILLING resource forgene function analysis in sunflower. Plant Methods 7, 20. doi: 10.1186/1746-4811-7-20
Salmeron, J. M., Oldroyd, G. E., Rommens, C. M., Scofield, S. R., Kim, H. S., Lavelle, D. T., et al. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86, 123–133. doi: 10.1016/S0092-8674(00)80083-5
Sana, T. R., Fischer, S., Wohlgemuth, G., Katrekar, A., Jung, K. H., Ronald, P. C., et al. (2010). Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. Oryzae Metabolomics 6, 451–465. doi: 10.1007/s11306-010-0218-7
Sanchez, A. C., Brar, D. S., Huang, N., Khush, G. S. (2000). Sequence tagged site markers-assisted selection for three bacterial blight resistance genes in rice. Crop Sci. 40, 792–797. doi: 10.2135/cropsci2000.403792x
Sanchez, P. L., Wing, R. A., Brar, D. S. (2013). “The wild relative of rice: genomes and genomics,” in Genetics and genomics of rice, plant gene. Eds. Zhang, Q., Wing, R. A. (New York: Springer), pp 616–626.
Sanseverino, W., Roma, G., De Simone, M., Faino, L., Melito, S., Stupka, E., et al. (2010). PRGdb: a bioinformatics platform for plant resistance gene analysis. Nucleic Acid Res. 38, D814–D821. doi: 10.1093/nar/gkp978
Sattari, A., Fakheri, B., Noroozi, M., Moazami, K. (2014). Leaf blight resistance in rice: a review of breeding and biotechnology. Int. J. Farm Allied Sci. 3, 895–902.
Savary, S., Elazegui, F. A., Teng, P. S. (1998). Assessing the representativeness of data on yield losses due to rice disease in tropical. Asia Plant Dis. 84, 357–369. doi: 10.1094/PDIS.2000.84.3.357
Schena, M., Shalon, D., Davis, R. W., Brown, P. O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. doi: 10.1126/science.270.5235.467
Scholze, H., Boch, J. (2011). TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47–53. doi: 10.1016/j.mib.2010.12.001
Sekhwal, M. K., Li, P., Lam, I., Wang, X., Cloutier, S., You, F. M. (2015). Disease resistance gene analogs (RGAs) in plants. Int. J. Mol. Sci. 16, 19248–19290. doi: 10.3390/ijms160819248
Sevanthi, A. M. V., Kandwal, P., Kale, P. B., Prakash, C., Ramkumar, M. K., Yadav, N., et al. (2018). Whole genome characterization of a few EMS-induced mutants of upland rice variety Nagina 22 reveals a staggeringly high frequency of SNPs which show high phenotypic plasticity towards the wild-type. Front. Plant Sci. 9:1179 . doi: 10.3389/fpls.2018.01179
Sharma, T. R., Das, A., Kumar, S. P., Lodha, M. L. (2009). Resistance gene analogues as a tool for rapid identification and cloning of disease resistance genes in plants: A review. J. Plant Biochem. Biotechnol. 18, 01–11. doi: 10.1007/BF03263289
Sharma, K., Sarma, S., Bohra, A., Mitra, A., Sharma, N. K., Kumar, A. (2018). “Plant metabolomics: An emerging technology for crop improvement,” in New Visions in Plant Science, 1st ed. Ed. Çelik, O. (London, UK: IntechOpen), 65–79.
Shen, B., Zhang, W., Zhang, J., Zhou, J., Wang, J., Chen, L., et al. (2014). Efficient genome modification by CRISPR-Cas9 nickase with minimal off-target effects. Nat. Methods 11, 399–402. doi: 10.1038/nmeth.2857
Sidhu, G. S., Khush, G. S., Mew, T. W. (1978). Genetic analysis of bacterial blight resistance in seventy our cultivars of rice, Oryza sativa L. Theor. Appl. Genet. 53, 105 –1111. doi: 10.1007/BF00272687
Singh, R. J., Khush, G. S., Mew, T. W. (1983). A new gene for resistance to bacterial blight in rice. Crop Sci. 23, 558–560. doi: 10.2135/cropsci1983.0011183X002300030026x
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., et al. (2001). Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 102, 1011–1015. doi: 10.1007/s001220000495
Singh, K., Vikal, Y., Singh, S., Leung, H., Dhaliwal, H. S., Khush, G. S. (2002). Mapping of bacterial blight resistance gene xa8 using microsatellite markers. Rice Genet. Newsl. 19, 94– 97.
Smith, J., Grizot, S., Arnould, S., Duclert, A., Epinat, J. C., Chames, P., et al. (2006). A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 34, e149. doi: 10.1093/nar/gkl720
Song, W. Y., Wang, G. L., Chen, L. L., Kim, H. S., Pi, L. Y., Holsten, T., et al. (1995). A receptor kinase like protein encoded by the rice disease resistance gene Xa21. Science 270, 1804–1806. doi: 10.1126/science.270.5243.1804
Spindel, J., Begum, H., Akdemir, D., Virk, P., Collard, B., Redona, E., et al. (2015). Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 11, e1004982. doi: 10.1371/journal.pgen.1004982
Srinivasan, B., Gnanamanickam, S. (2005). Identification of a new source of resistance in wild rice, Oryza rufipogon to bacterial blight of rice caused by Indian strains of Xanthomonas oryzae pv. oryzae. Curr. Sci. 88, 1229–1231.
Staswick, P. E., Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200, 808–819. doi: 10.1111/nph.12411
Subramanian, S., Fu, Y., Sunkar, R., Barbazuk, W. B., Zhu, J. K., Yu, O. (2008). Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9, 160. doi: 10.1186/1471-2164-9-160
Sugio, A., Yang, B., Zhu, T., White, F. F. (2007). Two type III effector genes of Xanthomonas oryzae pv. oryzae control the induction of the host genes OsTFIIAg1 and OsTFX1 during bacterial blight of rice. Proc. Nat. Acad. Sci. 104, 10720–10725. doi: 10.1073/pnas.0701742104
Suh, J. P., Jeung, J. U., Noh, T. H., Cho, Y. C., Park, S. H., Park, H. S., et al. (2013). Development of breeding lines with three pyramided resistance genes that confer broad-spectrum bacterial blight resistance and their molecular analysis in rice. Rice 6, 5–12. doi: 10.1186/1939-8433-6-5
Sun, X., Cao, Y., Yang, Z., Xu, C., Li, X., Wang, S., et al. (2004). Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J. 37, 517–527. doi: 10.1046/j.1365-313X.2003.01976.x
Sundaram, R. M., Vishnupriya, M. R., Biradar, S. K., Laha, G. S., Reddy, G. A., Rani, N. S., et al. (2008). Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422. doi: 10.1007/s10681-007-9564-6
Sundaram, R. M., Vishnupriya, M. R., Laha, G. S., Rani, N. S., Rao, P. S., Balachandran, S. M. (2009). Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety. Biotechnol. J. 4, 400–407. doi: 10.1002/biot.200800310
Sundaram, R. M., Chatterjee, S., Oliva, R., Laha, G. S., Cruz, C. V., Leach, J. E., et al. (2014). Update on bacterial blight of rice: Fourth international conference on bacterial blight. Rice 7, 12. doi: 10.1186/s12284-014-0012-7
Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H., Moriguchi, K., et al. (2008). MNU-induced mutant pools and high-performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279, 213–223. doi: 10.1007/s00438-007-0293-2
Talamè, V., Bovina, R., Sanguineti, M. C., Tuberosa, R., Lundqvist, U., Salvi, S. (2008). TILLMore, a resource for the discovery of chemically induced mutants in barley. Plant Biotechnol. J. 6, 477–485. doi: 10.1111/j.1467-7652.2008.00341.x
Tan, G. X., Ren, X., Weng, Q. M., Shi, Z. Y., Zhu, L. L., He, G. C. (2004). Mapping of a new resistance gene to bacterial blight in rice line introgressed from Oryza officinalis. Acta Genet. Sin. 31, 724–729.
Tan, K. C., Ipcho, S. V., Trengove, R. D., Oliver, R. P., Solomon, P. S. (2009). Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol. Plant Pathol. 10, 703–715. doi: 10.1111/j.1364-3703.2009.00565.x
Tariq, R., Wang, C. L., Qin, T. F., Xu, F., Tang, Y., Gao, Y., et al. (2018). Comparative transcriptome profiling of rice near-isogenic line carrying Xa23 under infection of Xanthomonas oryzae pv. Oryzae. Int. J. Mol. Sci. 19, 717. doi: 10.3390/ijms19030717
Taura, S., Ogawa, T., Yoshimura, A., Ikeda, R., Iwata, N. (1992). Identification of a recessive resistance gene to rice bacterial blight of mutant line XM6, Oryzae sativa L. Japan. J. Breed. 42, 7–13.
Thiebaut, F., Rojas, C. A., Grativol, C., Motta, M. R., Vieira, T., Regulski, M., et al. (2014). Genome-wide identification of microRNA and siRNA responsive to endophytic beneficial diazotrophic bacteria in maize. BMC Genomics 15, 766. . doi: 10.1186/1471-2164-15-766
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effector-dependent resistance protein Xa10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26, 497–515. doi: 10.1105/tpc.113.119255
Till, B. J., Cooper, J., Tai, T. H., Colowit, P., Greene, E. A., Henikoff, S., et al. (2007). Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7, 19. doi: 10.1186/1471-2229-7-19
Tu, J., Ona, I., Zhang, Q., Mew, T. W., Khush, G. S., Datta, S. K. (1998). Transgenic rice variety IR72 with Xa21 is resistant to bacterial blight. Theor. Appl. Genet. 97, 31–36. doi: 10.1007/s001220050863
Tu, J., Datta, K., Khush, G. S., Zhang, Q., Datta, S. K. (2000). Field performance of Xa21 transgenic indica rice (Oryza sativa L.), IR72. Theor. Appl. Genet. 101, 15–20. doi: 10.1007/s001220051443
Uauy, C., Brevis, J. C., Chen, X., Khan, I., Jackson, L., Chicaiza, O., et al. (2005). High temperature adult-plant (HTAP) stripe rust resistance gene Yr36 from Triticum turgidum ssp. Dicoccoides is closely linked to the grain protein content locus Gpc-B1. Theor. Appl. Genet. 112, 97–105. doi: 10.1007/s00122-005-0109-x
Utami, D. W., Septiningsih, E. M., Triny, T. S., Fatimah, Y. (2010). Allele mining of bacterial blight resistance gene, Xa7 on Indonesian local rice germplasm. J. AgroBiogen. 6, 1–9. doi: 10.21082/jbio.v6n1.2010.p1-9
Utami, D. W., Lestari, P., Koerniati, S. (2013). A Relative expression of Xa7 gene controlling bacterial leaf blight resistance in Indonesian local rice population (Oryza sativa L.). J. Crop Sci. Biotech. 16, 1–7. doi: 10.1007/s12892-012-0091-1
Van de Wiel, C. C., Schaart, J. G., Lotz, L. A., Smulders, M. J. (2017). New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol. Rep. 11, 1–8. doi: 10.1007/s11816-017-0425-z
Velculescu, V. E., Zhang, L., Vogelstein, B., Kinzler, K. W. (1995). Serial analysis of gene expression. Science 270, 484–487. doi: 10.1126/science.270.5235.484
Ventelon-Debout, M., Delalande, F., Brizard, J. P., Diemer, H., Van Dorsselaer, A., Brugidou, C. (2004). Proteome analysis of cultivar-specific deregulations of Oryza sativaindica and O. sativa japonica cellular suspensions undergoing rice yellow mottle virus infection. Proteomics 4, 216–225. doi: 10.1002/pmic.200300502
Vojta, A., Dobrinić, P., Tadić, V., Bočkor, L., Korać, P., Julg, B., et al. (2016). Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 44, 5615–5628. doi: 10.1093/nar/gkw159
Voulhoux, R., Ball, G., Ize, B., Vasil, M. L., Lazdunski, A., Wu, L. F., et al. (2001). Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J. 20, 6735–6741. doi: 10.1093/emboj/20.23.6735
Vuylsteke, M., Peleman, J. D., van-Eijk, M. J. T. (2007). AFLP technology for DNA fingerprinting. Nat. Protoc. 2, 1387–1398. doi: 10.1038/nprot.2007.175
Wan, J., Dunning, F. M., Bent, A. F. (2002). Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct. Integr. Genomics 2, 259–273. doi: 10.1007/s10142-002-0080-4
Wang, G. L., Song, W. Y., Ruan, D. L., Sideris, S., Ronald, P. C. (1996). The cloned gene, Xa21, confers resistance to multiple X. oryzae pv. oryzae isolates in transgenic plants. Mol. Plant Microbe Interct. 9, 850–855. doi: 10.1094/MPMI-9-0850
Wang, W., Zhai, W., Luo, M., Jiang, G., Chen, X., Li, X., et al. (2001). Chromosome landing at the bacterial blight resistance gene Xa4 locus using a deep coverage rice BAC library. Mol. Gen. Genomics 265, 118–125. doi: 10.1007/s004380000382
Wang, C., Tan, M., Xu, X., Wen, G., Zhang, D., Lin, X. (2003). Localizing the bacterial blight resistance gene, Xa22(t), to a 100-Kilobase bacterial artificial chromosome. Phytopathol 93, 1258–1262. doi: 10.1094/PHYTO.2003.93.10.1258
Wang, C., Wen, G., Lin, X., Liu, X., Zhang, D. (2009). Identification and fine mapping of the new bacterial blight resistance gene, Xa31(t), in rice. Eur. J. Plant Pathol. 123, 235–240. doi: 10.1007/s10658-008-9356-4
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wang, C., Fan, Y., Zheng, C., Qin, T., Zhang, X., Zhao, K. (2014a). High-resolution genetic mapping of rice bacterial blight resistance gene Xa23. Mol. Genet. Genomics 289, 745–753. doi: 10.1007/s00438-014-0848-y
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2014b). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant Pathol. 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wang, C., Tariq, R., Ji, Z., Wei, Z., Zheng, K., Mishra, R., et al. (2019). Transcriptome analysis of a rice cultivar reveals the differentially expressed genes in response to wild and mutant strains of Xanthomonas oryzae pv. oryzae. Sci. Rep. 9, 3757. doi: 10.1038/s41598-019-39928-2
Webb, K. M., Ona, I., Bai, J., Garrett, K., Mew, T., Cruz, V., et al. (2010). A benefit of high temperature: increased effectiveness of a rice bacterial blight disease resistance gene. New Phytol. 185 (2), 568–576. doi: 10.1111/j.1469-8137.2009.03076.x
Wen, J. Q., Oono, K., Imai, R. (2002). Two novel mitogen-activated protein signaling components, OsMEK1 and OsMAP1, are involved in a moderate low-temperature signaling pathway in rice. Plant Physiol. 129, 1880–1891. doi: 10.1104/pp.006072
White, F. F., Potnis, N., Jones, J. B., Koebnik, R. (2009). The type III effectors of Xanthomonas. Mol. Plant Pathol. 10, 749–766. doi: 10.1111/j.1364-3703.2009.00590.x
Williams, C. E., Wang, B., Holsten, T. E., Scambray, J., da Silva, F. D. (1996). Ronald PC. Markers for selection of rice Xa 21 disease resistance gene. Theor. Appl. Genet. 93, 1119–1122. doi: 10.1007/BF00230134
Wu, J. L., Wu, C., Lei, C., Baraoidan, M., Bordeos, A., Madamba, M. R., et al. (2005). Chemical- and irradiation-induced mutants of indica rice IR64 for forward and reverse genetics. Plant Mol. Biol. 59, 85–97. doi: 10.1007/s11103-004-5112-0
Wu, J., Yu, H., Dai, H., Mei, W., Huang, X., Zhu, S., et al. (2012). Metabolite profiles of rice cultivars containing bacterial blight-resistant genes are distinctive from susceptible rice. Acta Biochim. Biophy. Sin. (Shanghai) 44, 650–659. doi: 10.1093/abbs/gms043
Xiang, Y., Cao, Y. L., Xu, C. Q., Li, X., Wang, S. (2006). Xa3, conferring resistance for rice bacterial blight and encoding a receptor kinase-like protein, is the same as Xa26. Theor. Appl. Genet. 113, 1347–1355. doi: 10.1007/s00122-006-0388-x
Xu, J., Audenaert, K., Hofte, M., De Vleesschauwer, D. (2013). Abscisic acid promotes susceptibility to therice leaf blight pathogen Xanthomonas oryzae pv. oryzae by suppressing salicylic acid-mediateddefenses. PLoS One 8, e67413. doi: 10.1371/journal.pone.0067413
Xu, Y., Wang, X., Ding, X., Zheng, X., Yang, Z., Xu, C., et al. (2018). Genomic selection of agronomic traits in hybrid rice using an NCII population. Rice 11, 32. doi: 10.1186/s12284-018-0223-4
Xu, Z., Xu, X., Gong, Q., Li, Z., Li, Y., Wang, S., et al. (2019). Engineering broad-spectrum bacterial blight resistance by simultaneously disrupting variable TALE-binding elements of multiple susceptibility genes in rice. Mol. Plant 12 (11), 1434–1446. doi: 10.1016/j.molp.2019.08.006
Yang, B., White, F. F. (2004). Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant-Microbe Interact. 17, 1192–1200. doi: 10.1094/MPMI.2004.17.11.1192
Yang, Z., Sun, X., Wang, S., Zhang, Q. (2003). Genetic and physical mapping of a new gene for bacterial blight resistance in rice. Theor. Appl. Genet. 106, 1467–1472. doi: 10.1007/s00122-003-1205-4
Yang, B., Sugio, A., White, F. F. (2006). Os8N3 is a host disease susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yang, S., Feng, Z., Zhang, X., Jiang, K., Jin, X., Hang, Y., et al. (2006). Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol. Biol. 62, 181–193. doi: 10.1007/s11103-006-9012-3
Yoshimura, A., Mew, T. W., Khush, G. S., Omura, T. (1983). Inheritance of resistance to bacterial blight in rice cultivar Cas 209. Phytopathology 73, 1409–1412. doi: 10.1094/Phyto-73-1409
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. 95, 1663–1668. doi: 10.1073/pnas.95.4.1663
Young, N. D. (2000). The genetic architecture of resistance. Curr. Opin. Plant Biol. 3, 285–290. doi: 10.1016/S1369-5266(00)00081-9
Yu, C. L., Yan, S. P., Wang, C. C., Hu, H. T., Sun, W. N., Yan, C. Q., et al. (2008). Pathogenesis-related proteins in somatic hybrid rice induced by bacterial blight. Phytochemistry 69 (10), 1989–1996. doi: 10.1016/j.phytochem.2008.04.006
Yu, Y., Streubel, J., Balzergue, S., Champion, A., Boch, J., Koebnik, R. (2011). Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol. Plant-Microbe Interact. 24, 1102–1113. doi: 10.1094/MPMI-11-10-0254
Yuan, M., Chu, Z., Li, X., Xu, C., Wang, S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol. 50, 947–955. doi: 10.1093/pcp/pcp046
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., et al. (2016). A host basal transcription factor is a keycomponent for infection of rice by TALE-carrying bacteria. eLife 5, e19605. doi: 10.7554/eLife.19605
Yugander, A., Sundaram, R. M., Singh, K., Prasad, M. S., Prasad, A. H., Madhav, M. S., et al. (2019). Marker assisted introgression of a major bacterial blight resistance gene, Xa38 into a rice maintainer line, APMS 6B. Indian Phytopathol. 72, 35–41. doi: 10.1007/s42360-018-00111-8
Zafar, K., Khan, M. Z., Amin, I., Mukhtar, Z., Yasmin, S., Arif, M., et al. (2020). Precise CRISPR-Cas9 Mediated Genome Editing in Super Basmati Rice for Resistance Against Bacterial Blight by Targeting the Major Susceptibility Gene. Front. Plant Sci. 11:575:575. doi: 10.3389/fpls.2020.00575
Zhai, W., Chen, C., Zhu, X., Chen, X., Zhang, D., Li, X., et al. (2004). Analysis of T-DNA-Xa21 loci and bacterial blight resistance effects of the transgene Xa21 in transgenic rice. Theor. Appl. Genet. 109, 534–542. doi: 10.1007/s00122-004-1670-4
Zhang, Q., Lin, S. C., Zhao, B. Y., Wang, C. L., Yang, W. C., Zhou, Y. L., et al. (1998). Identification and tagging of a new gene for resistance to bacterial blight (Xanthomonas oryzae pv oryzae) from O. rufipogon. Rice Genet. Newsl. 15, 138–142.
Zhang, H., Wang, S. (2013). Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195. doi: 10.1016/j.pbi.2013.02.008
Zhang, Q., Wang, C. L., Zhao, K. J., Zhao, Y. L., Caslana, V. C., Zhu, X. D., et al. (2001). The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet. Newsl. 18, 71–72.
Zhang, J., Li, X., Jiang, G., Xu, Y., He, Y. (2006). Pyramiding of Xa7 and Xa21 for the improvement of disease resistance to bacterial blight in hybrid rice. Plant Breed. 125, 600–605. doi: 10.1111/j.1439-0523.2006.01281.x
Zhang, F., Zhuo, D. L., Zhang, F., Huang, L. Y., Wang, W. S., Xu, J. L., et al. (2014). Xa39, a novel dominant gene conferring broad-spectrum resistance to Xanthomonas oryzae pv. oryzae in rice. Plant Pathol. 64, 568–575. doi: 10.1111/ppa.12283
Zhao, F., Li, Y., Chen, L., Zhu, L., Ren, H., Lin, H., et al. (2016). Temperature dependent defence of Nicotiana tabacum against Cucumber mosaic virus and recovery occurs with the formation of dark green islands. J. Plant Biol. 59 (3), 293–301. doi: 10.1007/s12374-016-0035-2
Zheng, C. K., Wang, C. L., Yu, Y. J., Liang, Y. T., Zhao, K. J. (2009). Identification and molecular mapping of Xa32(t),a novel resistance ene for bacterial blight (Xanthomonas oryzae pv. oryzae) in rice.Acta Agron. Sin. 35, 1173–1180. doi: 10.1016/S1875-2780(08)60089-9
Zhou, B., Dolan, M., Sakai, H., Wang, G. L. (2007). The genomic dynamics and evolutionary mechanism of the Pi2/9 locus in rice. Mol. Plant-Microbe Interact. 20, 63–71. doi: 10.1094/MPMI-20-0063
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhu, Y., Qian, W., Hua, J. (2010). Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 6 (4), e1000844. doi: 10.1371/journal.ppat.1000844
Zipfel, C., Robatzek, S., Navarro, L., Oakeley, E. J., Jones, J. D., Felix, G., et al. (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428, 764–767. doi: 10.1038/nature02485
Keywords: rice, bacterial blight, resistance breeding, marker-assisted selection, genome editing, CRISPR/Cas, plant defense, transcriptome
Citation: Kumar A, Kumar R, Sengupta D, Das SN, Pandey MK, Bohra A, Sharma NK, Sinha P, Sk H, Ghazi IA, Laha GS and Sundaram RM (2020) Deployment of Genetic and Genomic Tools Toward Gaining a Better Understanding of Rice-Xanthomonasoryzae pv. oryzae Interactions for Development of Durable Bacterial Blight Resistant Rice. Front. Plant Sci. 11:1152. doi: 10.3389/fpls.2020.01152
Received: 26 May 2020; Accepted: 15 July 2020;
Published: 04 August 2020.
Edited by:
Elizabeth R. Waters, San Diego State University, United StatesReviewed by:
Gerbert Sylvestre Dossa, Food and Agriculture Organization of the United Nations, ItalyMeng Yuan, Huazhong Agricultural University, China
Copyright © 2020 Kumar, Kumar, Sengupta, Das, Pandey, Bohra, Sharma, Sinha, Sk, Ghazi, Laha and Sundaram. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raman Meenakshi Sundaram, rms_28@rediffmail.com; Anirudh Kumar, anirudh.kumar@igntu.ac.in
†Present address: Manish K. Pandey, Research Program-Genetic Gains, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Hyderabad, India
 Anirudh Kumar
Anirudh Kumar