- Department of Plant Agriculture, University of Guelph, Guelph, ON, Canada
There is a need to increase wheat productivity to meet the food demands of the ever-growing human population. However, accelerated development of high yielding varieties is hindered by drought, which is worsening due to climate change. In this context, germplasm diversity is central to the development of drought-tolerant wheat. Extensive collections of these genetic resources are conserved in national and international genebanks. In addition to phenotypic assessments, the use of advanced molecular techniques (e.g., genotype by sequencing) to identify quantitative trait loci (QTLs) for drought tolerance related traits is useful for genome- and marker-assisted selection based approaches. Therefore, to assist wheat breeders at a critical time, we searched the recent peer-reviewed literature (2011-current), first, to identify wheat germplasm observed to be useful genetic sources for drought tolerance, and second, to report QTLs associated with drought tolerance. Though many breeders limit the parents used in breeding programs to a familiar core collection, the results of this review show that larger germplasm collections have been sources of useful genes for drought tolerance in wheat. The review also demonstrates that QTLs for drought tolerance in wheat are associated with diverse physio-morphological traits, at different growth stages. Here, we also briefly discuss the potential of genome engineering/editing to improve drought tolerance in wheat. The use of CRISPR-Cas9 and other gene-editing technologies can be used to fine-tune the expression of genes controlling drought adaptive traits, while high throughput phenotyping (HTP) techniques can potentially accelerate the selection process. These efforts are empowered by wheat researcher consortia.
Introduction
Bread wheat (Triticum aestivum L.) is one of the world’s major cereal crops with global production of 756.7 million tons in 2017 (FAO, 2018). The world’s population is expected to exceed 9 billion by 2050, requiring at least a 60% increase in wheat yield (United Nations, 2019). An increase in wheat yield from the current level of 1% per year to at least 1.6% is deemed necessary to address this challenge (GCARD, 2012). This is further challenged by projected unpredictable rainfall patterns associated with climate change which are expected to lead to more drought events (IPCC, 2013). However, the degree of impact on final yield depends on the growth stage and the intensity and duration of stress events (Daryanto et al., 2016; Sarto et al., 2017).
Like many other crops, drought affects wheat at all growth stages (HongBo et al., 2005; Saeidi et al., 2015; Saeidi and Abdoli, 2015; Wang et al., 2015; Sarto et al., 2017; Ding et al., 2018). Some of the growth stage-specific physio-morphological traits associated with drought tolerance in wheat include: early vigor (Rebetzke et al., 1999), coleoptile length (Rebetzke et al., 2007), leaf chlorophyll content (Khayatnezhad et al., 2011; Kira et al., 2015; Ramya et al., 2016), glaucousness (waxiness) for photo-protection (Merah et al., 2000; Bi et al., 2017), leaf rolling (Kadioglu and Terzi, 2007), carbon isotope discrimination (Kumar and Singh, 2009), flag leaf senescence (Verma et al., 2004; Hafsi et al., 2013) and plant height (Su et al., 2019). Also, root system architecture (RSA) traits are fundamental targets for breeding drought-tolerant wheat varieties (Lopes and Reynolds, 2010). A breeding program that selects for these physio-morphological traits has the potential to contribute to drought stress tolerance in wheat, as recently reviewed (Khadka et al., 2020).
In general, breeding can be accelerated by exploiting the diversity of genetic resources as sources of alleles that enhance desirable traits. More than 800,000 wheat accessions, including local landraces and synthetics, are conserved in genebanks globally (FAO, 1998). A large proportion of these accessions are accessible to wheat breeders. Recent advancements in genomics (e.g., genotype by sequencing, see below) have enabled exploration of this genetic diversity, leading to the discovery of markers and associated quantitative trait loci (QTLs) that can be utilized in marker assisted selection and genomic selection to accelerate variety development (Huang and Han, 2014). Wheat has a large and complex genome (Marcussen et al., 2014; Shi and Ling, 2017; Uauy, 2017; Abrouk et al., 2018). Very recently, however, an annotated reference whole genome sequence for bread wheat was released, which described 107,891 high-confidence level genes (IWGSC, 2018). This release has provided a significant opportunity to use genetic resources for exploring the wheat genome, and selecting alleles that encode desirable physio-morphological traits associated with drought tolerance.
There have been excellent recent reviews on the progress in breeding wheat and other cereals for tolerance to abiotic stress including drought (Mohammadi, 2018; Choudhary et al., 2019; Gupta et al., 2020). However, a unique feature of this current paper is that it reviews the different genetic resources that have been or can be exploited to accelerate the breeding of wheat for drought tolerance. The paper comprehensively updates recent progress (2011–2020) on the discovery of QTLs that promote drought tolerance in wheat, expands the list of associated physio-morphological traits and provides helpful details to breeders (e.g., QTL source populations). The QTL section is now the most up-to-date review on the topic. Furthermore, this paper also presents very recent advances in genetic engineering including the CRISPR/Cas9 genome editing system that has been used to target genes conferring drought tolerance.
Utilization of Genetic Variation From Diverse Sources
Use of diverse germplasm is key to the development of drought-tolerant wheat varieties. As noted in Table 1, T. aestivum landraces are one of the major groups of genetic resources valuable for breeding drought-tolerant wheat (Mwadzingeni et al., 2017). They have complex morphological diversity and are mostly grown in low input environments (Padulosi et al., 2012) which make them more adapted to stress (Padulosi et al., 2012; Lopes et al., 2015a). For example, a group of Creole wheat landraces (the landraces introduced to Mexico from Europe) showed better adaptation to different abiotic stresses, including drought, due to the presence of rare but beneficial alleles (Vikram et al., 2016). Similarly, the Japanese landrace “Aka Komugi” is one of the sources of the dwarfing Rht8c allele (Lopes et al., 2015a; Grover et al., 2018) that contributes to breeding drought tolerant wheat. This is because Rht8c promotes a higher-yielding semi-dwarf phenotype, but does not reduce the length of the coleoptile and thus permits deep sowing (Lopes et al., 2015a). Furthermore, in a study that evaluated 21 genotypes, nine wheat landraces exhibited drought tolerance based on a stress susceptibility index (SSI) (Sareen et al., 2014).
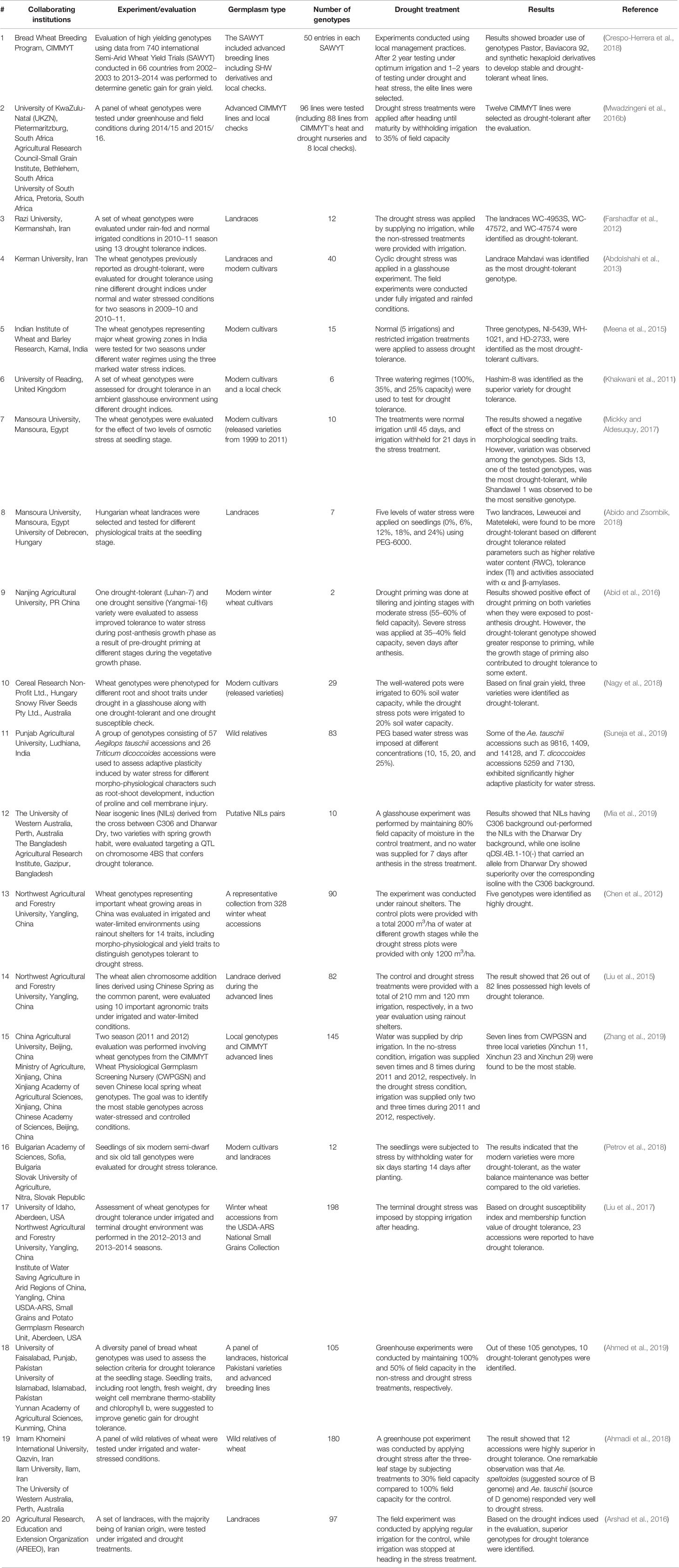
Table 1 Recent peer reviewed reports (2011-current) of germplasm evaluation for assessing drought tolerance in wheat.
In addition to landraces of T. aestivum, other domesticated wheat species such as T. compactum Host, T. sphaerococcum Perc., T. durum Desf, T. turgidum T. turanicum Jakubz (“Kumut”) and T. polonicum L., are also sources of valuable alleles to enable breeding of wheat for drought tolerance (Janni et al., 2018; Szabo-Hever et al., 2018; Guellim et al., 2019; Nemtsev et al., 2019). Wild relatives of wheat and other related genera such as Aegilops are additional valued sources of germplasm for breeding drought-tolerant varieties (Table 1).
The rye (Secale cereale L.) derived wheat genotypes, also known as the 1B/1R chromosome translocation lines, have been utilized by different wheat breeding programs (Kumar et al., 2003; Hoffmann, 2008; Ren et al., 2017). These 1B/1R chromosome translocation genotypes consist of the long arm of wheat chromosome 1B including its centromere, and the short arm or a portion of rye chromosome 1R (Cai and Liu, 1989; Heslop-Harrison et al., 1990). These genotypes are also important sources of abiotic stress tolerance including drought-tolerance (Kumar et al., 2003; Rajaram, 2005). Hoffmann (2008) demonstrated that the 1B/1R translocation wheat genotypes possess greater drought tolerance. However, this may further depend on the genetic background of the recipient wheat genotype (Monneveux et al., 2003; Singh et al., 1998) as shown by Tahmasebi et al. (2015) where 1B/1R translocation genotypes were not suitable sources of diversity for improving drought tolerance.
The challenges in utilizing these landraces and wild relatives are often associated with epistatic and pleiotropic effects of some genes, leading to linkage drag (Sehgal et al., 2015). However, these unfavorable genetic effects can be minimized through recurrent backcrossing with elite genetic materials and selection. In general, the development of modern varieties has reduced the genetic diversity of bread wheat, and this reduction in diversity shows both spatial and temporal trends (Rauf et al., 2010). Studies have shown that there is extensive reduction of nucleotide diversity in the A and B genome in modern wheat, including elite lines, compared to their progenitors (Haudry et al., 2007; Dreisigacker et al., 2008). Nonetheless, elite germplasm is a convenient source of genetic variation and has been considerably utilized in breeding for drought-tolerant wheat (Table 1). Most of the breeding programs around the world work on elite materials to develop drought-tolerant cultivars despite the advantages that landraces and wild relatives potentially carry. The major reason behind this is that in elite materials, there is less linkage drag associated with co-inheritance of undesirable and defective genes and rare alleles. As a result, elite materials have fewer challenges related to pleiotropic and epistatic gene effects in breeding populations (Sehgal et al., 2015; Mwadzingeni et al., 2017). The advantages offered by modern elite genetic materials compared to landraces and wild relatives make enhancement of genetic gain and selection response more rapid.
As a way to move forward, synthetic hexaploid wheats (SHWs) have been developed to widen the genetic variation in modern wheat. SHWs have deployed ancestral genomes using interspecific hybridization techniques (Reynolds et al., 2007; Khan et al., 2016). The SHW lines (AABBD′D′) have been developed by artificial hybridization of tetraploid durum wheat (T. turgidum, AABB) and diploid wild goat grass (Aegilops tauschiii, D′D′) (Reynolds et al., 2007; Jafarzadeh et al., 2016; Khan et al., 2016; Li A. et al., 2018; Rosyara et al., 2019). A study involving CIMMYT SHWs has demonstrated the importance of the D′ genome of Ae. tauschii in maintaining genetic diversity as well as improving genetic gain (Rosyara et al., 2019). The diversity in the D′ genome may be contributing to increased stress tolerance in SHW lines (Khan et al., 2016). In one study, six SHW lines were compared to four winter wheat cultivars grown in the U.S. Great Plains under drought stress (Becker et al., 2016). One of the SHW lines was shown to be superior under drought conditions for root morphological traits including deep root biomass and length of the longest root. In another study, an evaluation of 33 Ae. tauschii accessions and corresponding SHW lines revealed a wide range of variability for drought tolerance (Sohail et al., 2011). CIMMYT’s efforts to develop SHW lines based on 600 genebank Ae. tauschii accessions has been successful, as indicated by the encouraging adoption of SHW cultivars in India and South-West China; of these, at least 30% have been reported to be drought-tolerant (Aberkane et al., 2019). Similarly, Song et al. (2017) found that six out of 34 SHW lines were drought-tolerant, and these SHW lines showed high antioxidant activities (superoxidase and peroxidase activities) that minimize drought-associated oxidative cellular damage, and thus improve drought tolerance (Zhang and Kirkham, 1994; Laxa et al., 2019). In this study, the SHWs also demonstrated superior drought tolerance over the parental lines (Song et al., 2017). Similarly, Lopes and Reynolds (2011) observed that under drought conditions, four synthetic hexaploid derived lines out-yielded the parental lines by an average of 26%. Combined, these studies suggest that there are opportunities to exploit broad genetic variation in SHWs to improve different agronomic traits including drought tolerance in bread wheat.
Furthermore, mutation breeding, including using elite germplasm, can be another avenue to create variants suitable for drought adaptation. Among the cereals, wheat is one of the major crops in which mutation breeding has been employed for cultivar development. For example, “Sharbati Sonora,” an early maturing wheat cultivar developed by gamma radiation of a Mexican cultivar, made a major contribution to wheat production in India (Raina et al., 2016). Using gamma ray radiation, 11 drought-tolerant wheat mutant lines were identified in a recent study (Sen et al., 2017). Although mutation breeding is less common compared to other breeding methods, it has potential to generate novel stress tolerance alleles.
The major sources of the above mentioned genetic resources are national and international genebanks that maintain extensive collections of wheat landraces, wild relatives, breeding populations, obsolete varieties, and modern elite varieties. The diversity of wheat germplasm deposited in these genebanks appears to be under-utilized in breeding wheat for drought tolerance, though there are growing initiatives to evaluate and identify accessions that are suitable for wheat breeding programs. These initiatives include: The Wheat Pre-breeding Project (https://www.cwrdiversity.org/partnership/wheat-pre-breeding-project/), The Wheat Improvement Strategic Programme (WISP) Consortium (http://www.wheatisp.org/Consortium/WISP.php) and Seeds of Discovery (SeeD) (https://seedsofdiscovery.org/) (Singh S. et al., 2018). Genesys (https://www.genesys-pgr.org/welcome) is an online portal that hosts information about plant genetic resources deposited in genebanks. Out of almost 320,000 wheat accessions documented in Genesys, 23% are landraces, and 7% are wild relatives. The International Maize and Wheat Improvement Centre (CIMMYT) (http://www.cimmyt.org/seed-request/) hosts the most extensive collection of wheat germplasm (102,375 accessions). USDA-ARS genebanks hold 67,615 wheat accessions: most of them are maintained at the National Small Grains Collection (https://www.ars.usda.gov/pacific-west-area/aberdeen-id/small-grains-and-potato-germplasm-research/docs/national-small-grains-collection/) in Aberdeen, ID. Wild species are held at the Wheat Genetic Resource Center (WGRC) (https://www.k-state.edu/wgrc/), and additional materials at the Germplasm Resources Information Network (GRIN) (https://www.ars-grin.gov/). The Australian Grains Genebank (http://www.seedpartnership.org.au/associates/agg) has 42,624 accessions, and the International Center for Agriculture Research in the Dry Areas (ICARDA) (http://www.icarda.org/) has 41,471 accessions. ICARDA and CIMMYT together are managing 1,570 accessions of Ae. tauschii (Aberkane et al., 2019). Other organizations maintaining wheat germplasm are: Navadanya in India (http://www.navdanya.org/), the Svalbard Global Seed Vault in Norway (https://www.croptrust.org/our-work/svalbard-global-seed-vault/) and the National Bureau of Plant Genetic Resources (NBPGR) in India (http://www.nbpgr.ernet.in/). However, a limitation associated with the Svalbard, Navadanya, and NBPGR collections is their restricted access.
Once genetic diversity has been identified for crossing, a double haploid (DH) strategy can be employed. The DH lines possess identical copies of chromosomes in the genome which are complete homozygotes: this allows quick fixation of desired alleles. Wheat DH populations (Fleury et al., 2010; Yan et al., 2017) show potential in improving drought tolerance, as they enhance selection efficiency by improved additive genetic variance for complex quantitative traits (Dashti et al., 2007; Mwadzingeni et al., 2017). The DH lines are considered important for QTL x environment interactions, as trait means are estimated more efficiently, resulting in precise selection due to complete homozygosity (Shamasbi et al., 2017). DH breeding, augmented by advanced molecular tools, has potential to contribute to breeding drought-tolerant wheat. Different studies in wheat have shown the potential of DH lines as sources of stress tolerance. For example, Fatima et al. (2018) identified five drought-tolerant DH lines out of 84, which were tested under control and drought stress conditions. However, DH lines are associated with negative effects from colchicine treatment, and also gametoclonal and somaclonal variation that affect plant performance. Furthermore, a high level of homozygosity may make DH lines inferior to conventionally developed inbred lines (Niemirowicz-Szczytt, 1997). Other major challenges associated with DH in breeding for drought tolerance are financial expense and unique technical expertise involving plant cell and tissue culture techniques.
Molecular Breeding of Wheat for Drought Tolerance
Identification of QTLs for Drought Tolerance Related Traits
By employing the above genetic diversity, conventional linkage mapping, using biparental populations and double haploid lines, has commonly been used to locate QTLs/genes associated with target traits, including those associated with drought (Sallam et al., 2016; Zhao et al, 2018; Li L. et al., 2019). The objective of these studies is to facilitate marker/genome assisted selection. As the mapping populations used in traditional linkage mapping are derived by hybridization between two parents (most cases), and have limited genetic variation, only low natural allelic diversity can be captured which results in low resolution QTLs (Sallam et al., 2016; Zhao et al., 2018). Furthermore, the large genome size of wheat, which has different epistatic interactions among the QTLs, in addition to a large number of genes influencing a trait, have reduced the identification of useful QTLs (Ashraf, 2010). However, more recently, genome-wide association studies (GWAS) studies have been used to identify marker-trait associations for the trait(s) of interest (Corvin et al., 2014; Scherer and Christensen, 2016). GWAS represents a powerful alternate to linkage mapping. GWAS facilitates the exploration of the genetic variation of complex quantitative traits such as drought, controlled by several genes and their interactions (Kooke et al., 2016). GWAS exploits linkage disequilibrium (LD) resulting from variants at a locus caused by different factors such as historical mutations, natural and artificial selection, and other forces (Wang et al., 2012; Huang and Han, 2014; Visscher et al., 2017). The method accommodates natural populations with diverse genetic backgrounds (Huang and Han, 2014) and hence takes advantage of the diversity available in the genebanks described above. The efficiency of GWAS depends upon individual factors such as the number of loci for a trait that segregates in the population, genetic architecture, and size of the study population (Visscher et al., 2017). For more precise detection of QTLs/genes, some studies have combined linkage mapping and GWAS (Sallam et al., 2016; Shi W. et al., 2017; Liu et al., 2018; Zhao et al., 2018, Li G. et al., 2019).
GWAS and linkage mapping of drought tolerance QTLs have been enabled by the availability of a high-quality reference genome for common wheat along with next-generation sequencing (NGS) technologies (Kilian and Graner, 2012; Ray and Satya, 2014; Ramirez-Gonzalez et al., 2015; IWGSC, 2018). Genotype by sequencing (GBS), a DNA sequencing method that follows the NGS protocol, does not require prior genome sequence information and has enormous potential to genotype complex genomes such as in wheat (Poland et al., 2012; Mwadzingeni et al., 2016a; Chung et al., 2017).
Using these methodologies, important QTLs for wheat physio-morphological traits associated with drought tolerance have been reported in various studies. Many earlier studies have identified genomic regions associated with different physiological traits such as carbon isotope discrimination (ΔC) on chromosomes 1BL, 2BS, 3BS, 4AS, 4BS, 5AS, 7AS, and 7BS (Rebetzke et al., 2008); flag leaf senescence on chromosomes 2B and 2D (Verma et al., 2004); coleoptile length on chromosomes 4B and 6A (Rebetzke et al., 2001); seedling vigor on chromosome 6A (Spielmeyer et al., 2007); and canopy temperature on 1B, 2B, 3B, 4A, and 5A (Pinto et al., 2010). Building upon these earlier discoveries, the last few years have shown significant advances. For example, Tura et al. (2020) mapped yield QTLs under drought in a double haploid population on chromosomes 4A, 5B, and 7A. In a comprehensive GWAS study involving a panel of 210 European elite wheat lines, Touzy et al. (2019) discovered 24, 31, and 28 QTLs associated with drought tolerance, respectively, under low, medium and high water stress conditions. Bhatta et al. (2018) identified 90 marker-trait associations (MTAs) related to yield and associated traits under limited water conditions in a GWAS study with GBS markers using 123 synthetic hexaploid wheat lines. Similarly, QTLs for drought-related traits such as the drought susceptibility index (DSI), normalized difference vegetative index (NDVI), and leaf traits (including green leaf area, leaf senescence, and flag leaf phenotypes) were detected on chromosomes 1B, 4A, 6B, 5B, 7A, and 7B (Edae et al., 2014) in a GWAS involving a spring wheat panel. Table 2 summarizes QTLs reported for drought tolerance related traits on different chromosomes of the wheat genome, published in the peer reviewed literature from 2011 onwards.
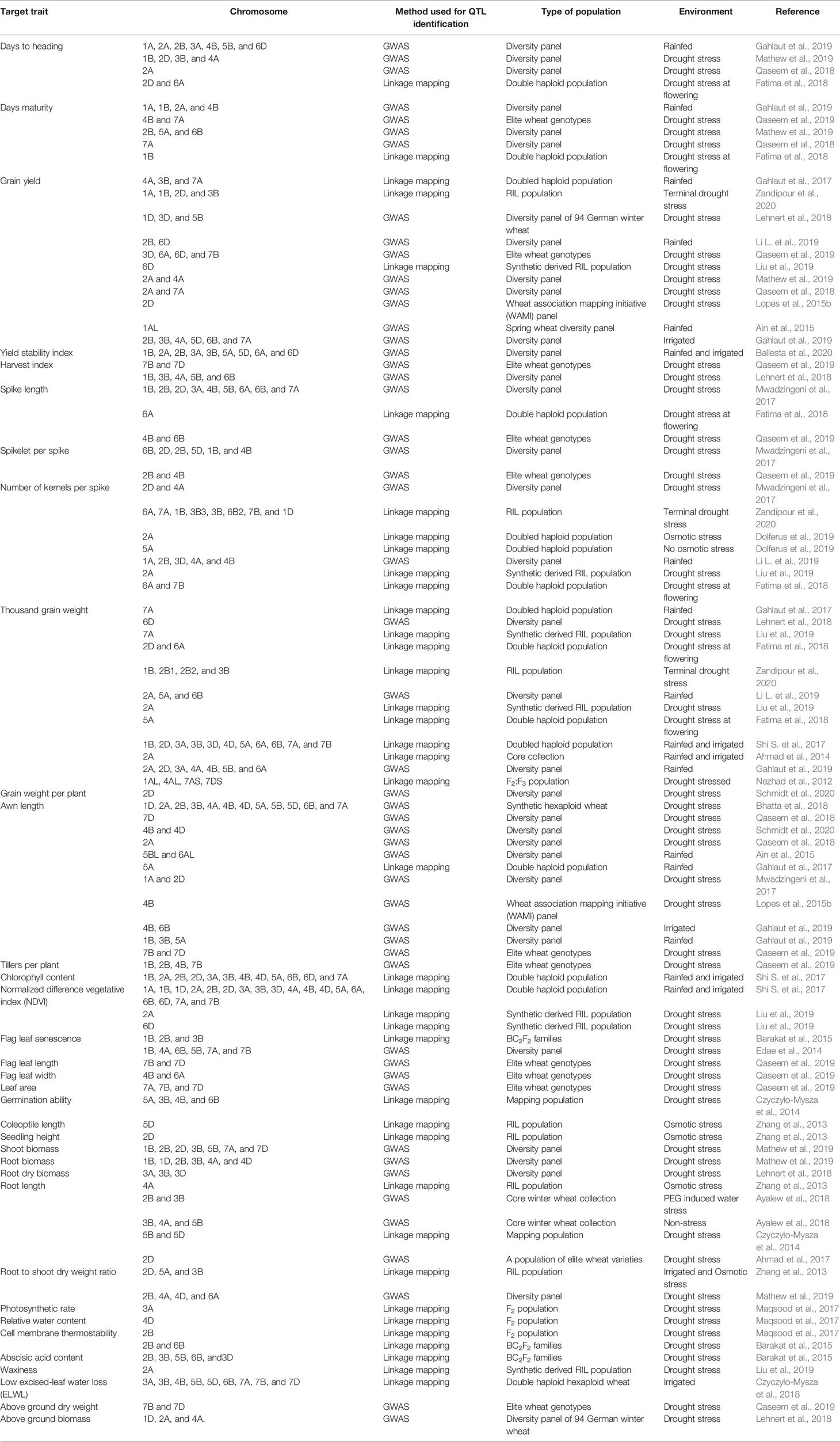
Table 2 Recent peer reviewed reports (2011-current) of the important physio-morphological traits associated with drought tolerance and locations of QTLs associated with these traits in wheat.
A number of the QTLs reported in Table 2 that are associated with different physio-morphological traits that contribute to wheat grain yield under drought, appear to be restricted to specific genetic backgrounds and environments (Griffiths et al., 2012; Acuna-Galindo et al., 2015). To determine which QTLs are relevant across genetic backgrounds and environments, some studies have used a meta-analysis approach to identify common QTLs discovered in different studies. For example, in a QTL meta-analysis recently conducted by Soriano and Alvaro (2019), 634 QTLs retrieved from earlier studies were projected on a consensus map having 7352 markers, which resulted in 94 consensus QTL regions, of which 35 were associated with root architecture and response to moisture stress. Similarly, in another QTL meta-analysis, Acuna-Galindo et al. (2015) reported 43 meta-QTLs related to drought and heat stress of which 20 were specific to drought stress tolerance. These results suggest that QTL meta-analysis, along with better estimations of QTL effects, may maximize the benefits of past drought QTL studies (Acuna-Galindo et al., 2015; Gupta et al., 2017; Soriano and Alvaro (2019).
Once candidate genes or loci have been identified, another approach is to use allele-specific markers to identify favorable alleles (haplotypes) that promote drought tolerance in a defined population. For example, Khalid et al. (2019) in a study comprising 213 advanced lines derived from synthetic hexaploid wheat and elite cultivars (CIMMYT and Pakistan), identified favorable alleles at five candidate genes associated with drought adaptation, including those that encode a cell wall invertase gene (TaCwi-A1) that converts sucrose into glucose and fructose, a dehydration responsive element binding protein (Dreb1) and COMT-3B that promotes lignin under water stress. The cell wall invertase gene discovery was consistent with an earlier study involving 348 modern Chinese cultivars which identified a favorable allele (haplotype) at another invertase gene (TaCWI-5D) on chromosome 5D in terms of promoting drought tolerance in wheat (Jiang et al., 2015).
Combined, these advances in genome sequencing, QTL and favorable allele identification, have provided wheat breeders with numerous targets for introgression and selection of drought tolerance promoting alleles that are potentially stable across environments and genetic backgrounds.
Genome Engineering Techniques
Genome engineering techniques, including gene pyramiding, gene stacking, and transgenics, broaden the methods available to breed wheat for tolerance to abiotic stress including drought (Budak et al., 2015; Mwadzingeni et al., 2016a). Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) is a bacterium-derived genome editing system; in this system, a DNA fragment encoding a non-coding RNA sequence is designed to target and then cleave a chromosomal DNA target of interest, within the Cas9 protein complex (Garneau et al., 2010; Budak et al., 2015; Zhang et al., 2018; Kumar et al., 2019; Okada et al., 2019; El-Mounadi et al., 2020; Shinwari et al., 2020). Recently, Wang et al. (2018) used CRISPR/Cas9 to increase grain size in wheat. Kim et al. (2018) also successfully established a CRISPR/Cas9 genome editing system in wheat protoplasts to edit the stress-responsive factor genes TaDREB2 and TaERF3. Since some essential regulatory genes that control the biosynthesis of metabolites associated with drought tolerance have been identified (Yang et al., 2010), the CRISPR/Cas9 gene editing technology could potentially be used to target such genes and traits in the future (Singh B. et al., 2018).
In addition, CRISPR/Cas9 mediated base editing (Zong et al., 2017; Li C. et al., 2018) and prime editing (Anzalone et al., 2019) techniques could be potentially utilized in breeding for drought tolerant wheat in the future as alternatives to the standard CRISPR/Cas9 genome editing system. Gene editing using CRISPR technologies, which depends on double strand breaks induced by the Cas9 protein (Garneau et al., 2010), has limitations associated with the delivery of donor DNA to the targeted cell types due to the low frequency of homologous recombination in plants (Molla and Yang, 2019; Hassan et al., 2020). The advantage of base editing technology over the standard CRISPR technology is: it does not require double stranded breaks and can simply perform substitution mutations allowing four kinds of modifications (C/G-to-T/A and A/T-to-G/C) (Komor et al., 2016; Gaudelli et al., 2017; Li C. et al., 2018; Molla and Yang, 2019). However, as base editing does not create indels (Komor et al., 2016; Gaudelli et al., 2017), an alternative prime editing technology which also does not require double stranded DNA cleavage, has been engineered to introduce indels; it also allows all 12 base to base conversions (Anzalone et al., 2019). As the prime editing method overcomes the above-stated limitations and enables different mutations (point mutations and indels) in wheat protoplasts (Lin et al., 2020), it may be a game changer technology in the future in terms of engineering drought tolerant wheat (Hassan et al., 2020; Marzec and Hensel, 2020).
In addition to gene editing, there is evidence suggesting that wheat genetic improvement is also possible through genetic engineering. For instance, transfer into wheat of the AISAP gene encoding a stress-associated protein from the halophyte grass/Mediterranean saltgrass (Aeluropus littoralis) has been shown to enhance the germination rate, biomass, and grain yield of wheat under osmotic- and salinity stress (Ben-Saad et al., 2012). A recent study (Ayadi et al., 2019) demonstrated that overexpression of the wheat aquaporin gene TdPIP2 in transgenic wheat improved drought tolerance. Similarly, the overexpression of the Escherichia coli CspA gene (modified as SeCspA) in transgenic wheat lowered the rate of water loss and maintained higher chlorophyll and proline under drought compared to non-transgenic plants (Yu et al., 2017). Additionally, improved drought tolerance (enhanced growth, delayed senescence, higher relative water content, higher photosynthetic rate, and higher total chlorophyll content) was observed in transgenic wheat in which an Arabidopsis thaliana cysteine protease (OVERLY TOLERANT TO SALT-1, OTS1) was over-expressed (using a ubiquitin promoter) compared to non-transformed wheat plants (le Roux et al., 2019). It has also been reported that overexpression of a fructan exohydrolase encoding gene (1-FEH w3) promotes higher fructan remobilization under drought which contributes positively to grain yield (Zhang et al., 2015; Hou et al., 2018). Several other studies have shown that overexpression of different drought responsive genes confers greater drought tolerance in transgenic wheat including the genes AtHDG11 (Li et al., 2016), TaCIPK23 (Cui et al., 2018), TaBZR2 (Cui et al., 2019), TaWRKY2 (Gao et al., 2018), TaWRKY1, and TaWRKY33 (He et al., 2016), and TaPYL4 (Mega et al., 2019). Interestingly, TaSnRK2.9, which is a sucrose non-fermenting 1-related protein kinase gene cloned from bread wheat, elevated ABA content in tobacco (Nicotiana tabacum) resulting in greater drought tolerance (Feng et al., 2019).
Therefore, both gene editing and genetic engineering show promise in terms of achieving drought tolerance in wheat. However, time, effort, skill, financial resources, regulatory issues, and public acceptance are major constraints to wider adoption of these techniques in wheat research programs.
Future Perspectives
The world is currently facing the prospect of food insecurity due to increasing drought. An assessment of genetic gain over fifty years of wheat breeding at CIMMYT showed an increase in grain yield by ~18 kg/ha per year under drought (Mondal et al., 2020) but this rate will be insufficient to meet demand. As this review has shown, promising approaches to improve drought tolerance in wheat include exploration of wheat diversity in genebanks, identification of genome-wide QTLs, validation of putative QTLs, and introgression of markers, genomic segments or candidate genes. However, undertaking this pipeline de novo is a major challenge in terms of time, skill, and resources. Therefore, the QTL meta-analysis approach (Acuna-Galindo et al., 2015; Gupta et al., 2017; Soriano and Alvaro, 2019) could be an immediate option that utilizes the results of already-existing QTLs associated with drought tolerance in wheat. Another promising near-term technique is genome editing using CRISPR/Cas9, given the potential it has shown to improve traits such as male sterility, grain weight, protein content, and powdery mildew resistance (Zhang Y. et al., 2017; Wang et al., 2018; Zhang et al., 2018; Okada et al., 2019). Furthermore, the pioneering CRISPR-based technologies including base editing and prime editing may have potential future. Though transgenic wheat offers potential, public acceptance may limit its adoption. By contrast, synthetic hexaploid wheats (SHWs) have shown promise in terms of drought tolerance and simplify the use of existing wild genetic resources (Lopes and Reynolds, 2011; Sohail et al., 2011; Becker et al., 2016; Aberkane et al., 2019; Laxa et al., 2019).
Apart from the use of wider germplasm, the growth environment in which selection and breeding is undertaken is important as well as the phenotyping strategies employed. The tradition of breeding for drought tolerance in wheat has been to compare grain yield in a water deficit environment compared to an optimal environment (Khakwani et al., 2011). However, the complex nature of drought (e.g., drought on sandy soil is more severe than on clay soil) complicates these efforts in terms of their actual impact on yield stability (Hoover et al., 2018). Furthermore, the large and complex nature of the wheat genome makes breeding for drought-tolerant wheat challenging. Therefore, adoption of a comprehensive strategy to develop drought-tolerant wheat varieties is needed. Such a strategy requires precise phenotyping in a water deficit environment (Fleury et al., 2010) as the efficiency of genomics also rests on advances in phenomics (Araus and Cairns, 2014; Fahlgren et al., 2015; Bai et al., 2016; Fernandez et al., 2017; Zhang X. et al., 2017). Many ground-based and aerial phenotyping platforms have been developed recently, with advances made in remote sensing, aeronautics, and computing (White et al., 2012; Araus and Cairns, 2014). In addition, new bioinformatics platforms have shown potential to overcome challenges associated with the management of the high volume of data generated by precision phenotyping tools (Shi et al., 2016; Singh et al., 2016; Coppens et al., 2017; Araus et al., 2018; Yang et al., 2020). These include Minimum Information About a Plant Phenotyping Experiment (MIAPPE), a platform that enables harmonization of phenotyping experiments (Bolger et al., 2019); and Crop Ontology, a platform that promotes proper use of genotypic and phenotypic data through data annotation (Shrestha et al., 2012).
Global agricultural research institutions such as CIMMYT and ICARDA have already initiated precision phenotyping coupled with NGS protocols to improve the utilization of wheat genebank materials and their development into pre-breeding materials for stress tolerance breeding programs (Crossa et al., 2016; Singh S. et al., 2018; Singh et al., 2019). However, other national and international institutions holding wheat genetic materials may need to prioritize similar efforts in order to deploy potential germplasm into wheat breeding programs. Traditional efforts may be revived to generate new allelic diversity for drought tolerance traits through mutagenesis involving chemical mutagens, ultraviolet light, and high energy radiation (Chen et al., 2014). Similarly, to discover the genetic basis of complex traits, transcriptomics, proteomics, and metabolomics offer promise (Reddy et al., 2014). For example, the information generated concerning different metabolic pathways have potential to add value in terms of developing targeted strategies that modulate the expression of genes associated with stress tolerance (Yang et al., 2010).
Further enhancement and strengthening of current global research collaborations is another way forward. CIMMYT is using the concept of 12 mega environments (ME) (http://wheatatlas.org/megaenvironments), defined as broad geographic regions with similar biotic and abiotic stresses, as well as agronomic practices and consumer preferences, to develop and promote wheat cultivars for wider adoption. One immediate activity of this collaboration could be to support the initiation of genomic characterization of genebank materials in developing countries. In the same vein, a very promising global initiative is the “The Heat and Drought Wheat Improvement Consortium (HeDWIC)” (https://www.hedwic.org), which is a worldwide platform of wheat scientists from more than 90 countries working on drought and heat tolerance. Similarly, “The International Wheat Yield Partnership (IWYP)” (https://iwyp.org) is a recent global consortium of partners including public and private research organizations, that aims to improve the genetic potential of wheat grain yield by 50% throught collaborative efforts in two decades. It is hoped that such global scientific cooperation in wheat breeding has the potential to generate new drought-tolerant varieties to combat climate change.
Author Contributions
KK and AN conceptualized the manuscript, while KK undertook the literature review, analysis, and wrote the manuscript, and AN and MR edited the manuscript.
Funding
The manuscript is a part of PhD thesis (KK) supported by a grant to MR from the Canadian International Food Security Research Fund (CIFSRF), jointly funded by Global Affairs Canada and the International Development Research Centre (IDRC, Ottawa). The PhD program is also partially supported by grants to AN by the Agricultural Adaptation Council of Canada; Grain Farmers of Ontario, Canada; and SeCan, Canada.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to dedicate this article to AN, the lead author’s PhD supervisor who passed away during the preparation of this manuscript due to pancreatic cancer. The authors thank Prof. P. Stephen Baenziger (University of Nebraska) for helpful comments on an earlier thesis version of this manuscript. The authors are grateful to Global Affairs Canada, IDRC Canada, Agricultural Adaptation Council, Canada; Grain Farmers of Ontario, Canada; and SeCan, Canada, for financial support. The authors also acknowledge the intellectual contributions of the Nepal Agriculture Research Council (NARC) and the International Maize and Wheat Improvement Centre (CIMMYT).
References
Abdolshahi, R., Safarian, A., Nazari, M., Pourseyedi, S., Mohamadi-Nejad, G. (2013). Screening drought-tolerant genotypes in bread wheat (Triticum aestivum L.) using different multivariate methods. Arch. Agron. Soil Sci. 59, 685–704. doi: 10.1080/03650340.2012.667080
Aberkane, H., Payne, T., Kishi, M., Amri, A., Jamora, N. (2019). Reaching into the past to tackle new challenges: Improving wheat by conserving wild " goat grass ". Genebank Impacts Brief No. 2. CGIAR Genebank Platform (Bonn (Germany): Crop Trust). Available at: https://repository.cimmyt.org/bitstream/handle/10883/20626/61255.pdf?sequence=1&isAllowed=y.
Abid, M., Tian, Z., Ata-ul-karim, S. T., Liu, Y., Cui, Y., Zahoor, R., et al. (2016). Improved tolerance to post-anthesis drought stress by pre-drought priming at vegetative stages in drought-tolerant and -sensitive wheat cultivars. Plant Physiol. Biochem. 106, 218–227. doi: 10.1016/j.plaphy.2016.05.003
Abido, W. A. E., Zsombik, L. (2018). Effect of water stress on germination of some Hungarian wheat landraces varieties. Acta Ecol. Sin. 38, 422–428. doi: 10.1016/j.chnaes.2018.03.004
Abrouk, M., Stritt, C., Muller, T., Keller, B., Roulin, A. C., Krattinger, S. G. (2018). High-throughput genotyping of the spelt gene pool reveals patterns of agricultural history in Europe. bioRxiv 481424. doi: 10.1101/481424
Acuna-Galindo, M. A., Mason, R. E., Subramanian, N. K., Hays, D. B. (2015). Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop Sci. 55, 477–492. doi: 10.2135/cropsci2013.11.0793
Ahmad, M. Q., Khan, S. H., Khan, A. S., Kazi, A. M., Basra, S. M. A. (2014). Identification of QTLs for drought tolerance traits on wheat chromosome 2A using association mapping. Int. J. Agric. Biol. 16, 862–870.
Ahmad, I., Ali, N., Ahmad, H., Innamullah (2017). “Association mapping of root traits for drought tolerance in bread wheat,” in Wheat Improvement, Management and Utilization. Eds. Wanyera, R., Owuoche, J. (London: INTECH), 39–57. doi: 10.5772/67242
Ahmadi, J., Pour-Aboughadareh, A., Fabriki-Ourang, S., Mehrabi, A. A., Siddique, K. H. M. (2018). Screening wheat germplasm for seedling root architectural traits under contrasting water regimes: potential sources of variability for drought adaptation. Arch. Agron. Soil Sci. 64, 1351–1365. doi: 10.1080/03650340.2018.1432855
Ahmed, H. G. M. D., Sajjad, M., Li, M., Azmat, M. A., Rizwan, M., Maqsood, R. H., et al. (2019). Selection criteria for drought-tolerant bread wheat genotypes at seedling stage. Sustain 11, 1–17. doi: 10.3390/su11092584
Ain, Q., Rasheed, A., Anwar, A., Mahmood, T., Imtiaz, M., Mahmood, T., et al. (2015). Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Front. Plant Sci. 6, 743. doi: 10.3389/fpls.2015.00743
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi: 10.1038/s41586-019-1711-4
Araus, J. L., Cairns, J. E. (2014). Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 19, 52–61. doi: 10.1016/j.tplants.2013.09.008
Araus, J. L., Kefauver, S. C., Zaman-Allah, M., Olsen, M. S., Cairns, J. E. (2018). Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 23, 451–466. doi: 10.1016/j.tplants.2018.02.001
Arshad, Y., Zahravi, M., Hassanpour, J. (2016). Identification of sources for drought tolerance in local bread wheat landraces. Iran. J. Genet. Plant Breed. 5, 22–31.
Ashraf, M. (2010). Inducing drought tolerance in plants: Recent advances. Biotechnol. Adv. 28, 169–183. doi: 10.1016/j.biotechadv.2009.11.005
Ayadi, M., Brini, F., Masmoudi, K. (2019). Overexpression of a wheat aquaporin gene, tdpip2;1, enhances salt and drought tolerance in transgenic durum wheat cv. Maali. Int. J. Mol. Sci. 20:2389. doi: 10.3390/ijms20102389
Ayalew, H., Liu, H., Börner, A., Kobiljski, B., Liu, C., Yan, G. (2018). Genome-wide association mapping of major root length QTLs under PEG induced water stress in wheat. Front. Plant Sci. 871, 1759. doi: 10.3389/fpls.2018.01759
Bai, G., Ge, Y., Hussain, W., Baenziger, P. S., Graef, G. (2016). A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput. Electron. Agric. 128, 181–192. doi: 10.1016/j.compag.2016.08.021
Ballesta, P., Mora, F., Del Pozo, A. (2020). Association mapping of drought tolerance indices in wheat: QTL-rich regions on chromosome 4A. Sci. Agric. 77, e20180153. doi: 10.1590/1678-992X-2018-0153
Barakat, M. N., Moustafa, K. A., Elshafei, A. A., Salem, A. K. (2015). Identification of QTLs for four physiological traits in an advanced backcross population of wheat under drought stress. Plant Omics J. 8, 122–129.
Becker, S. R., Byrne, P. F., Reid, S. D., Bauerle, W. L., McKay, J. K., Haley, S. D. (2016). Root traits contributing to drought tolerance of synthetic hexaploid wheat in a greenhouse study. Euphytica 207, 213–224. doi: 10.1007/s10681-015-1574-1
Ben-Saad, R., Ben-Ramdhan, W., Zouari, N., Azaza, J., Mieulet, D., Guiderdoni, E., et al. (2012). Marker-free transgenic durum wheat cv. Karim expressing the AlSAP gene exhibits a high level of tolerance to salinity and dehydration stresses. Mol. Breed. 30, 521–533. doi: 10.1007/s11032-011-9641-3
Bhatta, M., Morgounov, A., Belamkar, V., Baenziger, P. S. (2018). Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int. J. Mol. Sci. 19, 3011. doi: 10.3390/ijms19103011
Bi, H., Kovalchuk, N., Langridge, P., Tricker, P. J., Lopato, S., Borisjuk, N. (2017). The impact of drought on wheat leaf cuticle properties. BMC Plant Biol. 17, 85. doi: 10.1186/s12870-017-1033-3
Bolger, A. M., Poorter, H., Dumschott, K., Bolger, M. E., Arend, D., Osorio, S., et al. (2019). Computational aspects underlying genome to phenome analysis in plants. Plant J. 97, 182–198. doi: 10.1111/tpj.14179
Budak, H., Hussain, B., Khan, Z., Ozturk, N. Z., Ullah, N. (2015). From genetics to functional genomics: Improvement in drought signaling and tolerance in wheat. Front. Plant Sci. 6:1012:1012. doi: 10.3389/fpls.2015.01012
Cai, X., Liu, D. (1989). Identification of a 1B/1R wheat-rye chromosome translocation. Theor. Appl. Genet. 77, 81–83. doi: 10.1007/BF00292320
Chen, X., Min, D., Ahmad, T., Hu, Y. (2012). Evaluation of 14 morphological, yield-related and physiological traits as indicators of drought tolerance in Chinese winter bread wheat revealed by analysis of the membership function value of drought tolerance (MFVD). Field Crop Res. 137, 195–201. doi: 10.1016/j.fcr.2012.09.008
Chen, L., Hao, L., Parry, M. A. J., Phillips, A. L., Hu, Y. G. (2014). Progress in TILLING as a tool for functional genomics and improvement of crops. J. Integr. Plant Biol. 56, 425–443. doi: 10.1111/jipb.12192
Choudhary, M., Wani, S. H., Kumar, P., Bagaria, P. K., Rakshit, S., Roorkiwal, M., et al. (2019). QTLian breeding for climate resilience in cereals: progress and prospects. Funct. Integr. Genomics 19, 685–701. doi: 10.1007/s10142-019-00684-1
Chung, Y. S., Choi, S. C., Jun, T. H., Kim, C. (2017). Genotyping-by-sequencing: A promising tool for plant genetics research and breeding. Hortic. Environ. Biotechnol. 58, 425–431. doi: 10.1007/s13580-017-0297-8
Coppens, F., Wuyts, N., Inzé, D., Dhondt, S. (2017). Unlocking the potential of plant phenotyping data through integration and data-driven approaches. Curr. Opin. Syst. Biol. 4, 58–63. doi: 10.1016/j.coisb.2017.07.002
Corvin, A., Craddock, N., Sullivan, P. F. (2014). Genome-wide association studies: A primer. Psychol. Med. 40, 1063–1077. doi: 10.1017/S0033291709991723
Crespo-Herrera, L. A., Crossa, J., Huerta-Espino, J., Vargas, M., Mondal, S., Velu, G., et al. (2018). Genetic gains for grain yield in CIMMYT’s semi-arid wheat yield trials grown in suboptimal environments. Crop Sci. 58, 1890–1898. doi: 10.2135/cropsci2018.01.0017
Crossa, J., Jarquín, D., Franco, J., Pérez-Rodríguez, P., Burgueño, J., Saint-Pierre, C., et al. (2016). Genomic prediction of gene bank wheat landraces. G3 Genes Genom. Genet. 6, 1819–1834. doi: 10.1534/g3.116.029637
Cui, X., Du, Y., Fu, J., Yu, T., Wang, C., Chen, M., et al. (2018). Wheat CBL-interacting protein kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 18, 93. doi: 10.1186/s12870-018-1306-5RESEARCH
Cui, X.-Y., Gao, Y., Guo, J., Yu, T.-F., Zheng, W.-J., Liu, Y.-W., et al. (2019). BES / BZR transcription factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 180, 605–620. doi: 10.1104/pp.19.00100
Czyczyło-Mysza, I., Marcińska, I., Skrzypek, E., Cyganek, K., Juzoń, K., Karbarz, M. (2014). QTL mapping for germination of seeds obtained from previous wheat generation under drought. Cent. Eur. J. Biol. 9, 374–382. doi: 10.2478/s11535-013-0273-y
Czyczyło-Mysza, I. M., Marcińska, I., Skrzypek, E., Bocianowski, J., Dziurka, K., Rančić, D., et al. (2018). Genetic analysis of water loss of excised leaves associated with drought tolerance in wheat. PeerJ 6, e5063. doi: 10.7717/peerj.5063
Daryanto, S., Wang, L., Jacinthe, P. A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS One 11, e0156362. doi: 10.1371/journal.pone.0156362
Dashti, H., Yazdi-Samadi, B., Ghannadha, M., Naghavi, M., Quarri, S. (2007). QTL analysis for drought resistance in wheat using doubled haploid lines. Int. J. Agric. Biol. 9, 98–102.
Ding, J., Huang, Z., Zhu, M., Li, C., Zhu, X., Guo, W. (2018). Does cyclic water stress damage wheat yield more than a single stress? PLoS One 13, e0195535. doi: 10.1371/journal.pone.0195535
Dolferus, R., Thavamanikumar, S., Sangma, H., Kleven, S., Wallace, X., Forrest, K., et al. (2019). Determining the genetic architecture of reproductive stage drought tolerance in wheat using a correlated trait and correlated marker effect model. G3 Genes Genom. Genet. 9, 473–489. doi: 10.1534/g3.118.200835
Dreisigacker, S., Kishii, M., Lage, J., Warburton, M. (2008). Use of synthetic hexaploid wheat to increase diversity for CIMMYT bread wheat improvement. Aust. J. Agric. Res. 59, 413–420. doi: 10.1071/AR072250004-9409/08/050413
Edae, E., Byrne, P. F., Haley, S. D., Lopes, M. S., Reynolds, M. P. (2014). Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 127, 791–807. doi: 10.1007/s00122-013-2257-8
El-Mounadi, K., Morales-Floriano, M. L., Garcia-Ruiz, H. (2020). Principles, applications, and biosafety of plant genome editing using CRISPR-Cas9. Front. Plant Sci. 11, 56. doi: 10.3389/fpls.2020.00056
Fahlgren, N., Gehan, M. A., Baxter, I. (2015). Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 24, 93–99. doi: 10.1016/j.pbi.2015.02.006
FAO (1998). The state of the world"s plant genetic resources for food and agriculture (Rome, Italy: Food and Agriculture Organization of the United Nations). Available at: http://www.fao.org/3/a-w7324e.pd.
FAO (2018). Crop prospects and food situation. Quarterly global report (Rome, Italy: Food and Agriculture Organization of the United Nations). Available at: http://www.fao.org/3/i8764en/I8764EN.pdf.
Farshadfar, E., Saeidi, M., Honarmand, S. J. (2012). Evaluation of drought tolerance screening techniques among some landraces of bread wheat genotypes. Eur. J. Exp. Biol. 2, 1585–1592.
Fatima, S., Chaudhari, S. K., Akhtar, S., Amjad, M. S., Akbar, M., Iqbal, M. S., et al. (2018). Mapping QTLs for yield and yield components under drought stress in bread wheat (Triticum aestivum L.). Appl. Ecol. Environ. Res. 16, 4431–4453. doi: 10.15666/aeer/1604_44314453
Feng, F., Wang, L., Wu, Y., Luo, Q., Zhang, Y., Qui, D., et al. (2019). TaSnRK2.9, a sucrose non-fermenting 1-related protein kinase gene, positively regulates plant response to drought and salt stress in transgenic tobacco. Front. Plant Sci. 9, 2003. doi: 10.3389/fpls.2018.02003
Fernandez, M. G. S., Bao, Y., Tang, L., Schnable, P. S. (2017). A high-throughput, field-Based phenotyping technology for tall biomass crops. Plant Physiol. 174, 2008–2022. doi: 10.1104/pp.17.00707
Fleury, D., Jefferies, S., Kuchel, H., Langridge, P. (2010). Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 61, 3211–3222. doi: 10.1093/jxb/erq152
Gahlaut, V., Jaiswal, V., Tyagi, B. S., Singh, G., Sareen, S., Balyan, H. S., et al. (2017). QTL mapping for nine drought-responsive agronomic traits in bread wheat under irrigated and rain-fed environments. PLoS One 12, e0182857. doi: 10.1371/journal.pone.0182857
Gahlaut, V., Jaiswal, V., Singh, S., Balyan, H. S., Gupta, P. K. (2019). Multi-locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci. Rep. 9, 19486. doi: 10.1038/s41598-019-55520-0
Gao, H., Wang, Y., Xu, P., Zhang, Z. (2018). Overexpression of a WRKY transcription factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 9, 997. doi: 10.3389/fpls.2018.00997
Garneau, J. E., Dupuis, M.-E., Villion, M., Romero, D., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–72. doi: 10.1038/nature09523
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D., II, et al. (2017). Programmable base editing of A.T to G.C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi: 10.1038/nature24644
GCARD (2012). Breakout session P1.1 National Food Security – The Wheat Initiative – an International Research Initiative for Wheat Improvement. Second Global Conference on Agricultural Research for Development, Punta del Este, Uruguay (29 Oct-1 Nov, 2012). Available at: http://www.fao.org/docs/eims/upload/306175/Briefing Paper (3)-Wheat Initative - Hélène Lucas.pdf.
Griffiths, S., Simmonds, J., Leverington, M., Wang, Y., Fish, L., Sayers, L., et al. (2012). Meta-QTL analysis of the genetic control of crop height in elite European winter wheat germplasm. Mol. Breed. 29, 159–171. doi: 10.1007/s11032-010-9534-x
Grover, G., Sharma, A., Gill, H. S., Srivastava, P., Bains, N. S. (2018). Rht8 gene as an alternate dwarfing gene in elite Indian spring wheat cultivars. PLoS One 13, e0199330. doi: 10.1371/journal.pone.0199330
Guellim, A., Catterou, M., Chabrerie, O., Tetu, T., Hirel, B., Dubois, F., et al. (2019). Identification of phenotypic and physiological markers of salt stress tolerance in durum wheat (Triticum durum desf.) through integrated analyses. Agronomy 9, 844. doi: 10.3390/agronomy9120844
Gupta, P., Balyan, H., Gahlaut, V. (2017). QTL analysis for drought tolerance in wheat: Present status and future possibilities. Agronomy 7, 5. doi: 10.3390/agronomy7010005
Gupta, P. K., Balyan, H. S., Sharma, S., Kumar, R. (2020). Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor. Appl. Genet. 133, 1569–1602. doi: 10.1007/s00122-020-03583-3
Hafsi, M., Hadji, A., Guendouz, A., Maamari, K. (2013) Relationship between flag leaf senescence and grain yield in durum wheat grown under drought conditions. J. Agron. 12, 69–77.
Hassan, M., Yuan, G., Chen, J., Tuskan, G. A., Yang, X. (2020). Review article prime editing technology and its prospects for future applications in plant biology research. BioDesign Res. 2020, 9350905. doi: 10.34133/2020/9350905
Haudry, A., Cenci, A., Ravel, C., Bataillon, T., Brunel, D., Poncet, C., et al. (2007). Grinding up wheat: A massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 24, 1506–1517. doi: 10.1093/molbev/msm077
He, G. H., Xu, J. Y., Wang, X. Y., Liu, J. M., Li, P. S., Chen, M., et al. (2016). Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 16, 116. doi: 10.1186/s12870-016-0806-4
Heslop-Harrison, J. S., Leitch, A. R., Schwarzacher, T., Anamthawat-Jonsson, K. (1990). Detection and characterization of 1B/1R translocations in hexaploid wheat. Heredity 65, 385–392. doi: 10.1038/hdy.1990.108
Hoffmann, B. (2008). Alteration of drought tolerance of winter wheat caused by translocation of rye chromosome segment 1RS. Cereal Res. Commun. 36, 269–278. doi: 10.1556/CRC.36.2008.2.7
HongBo, S., ZongSuo, L., MingAn, S., ShiMeng, S., ZanMin, H. (2005). Investigation on dynamic changes of photosynthetic characteristics of 10 wheat (Triticum aestivum L.) genotypes during two vegetative-growth stages at water deficits. Colloids Surf. B. Biointerf. 43, 221–227. doi: 10.1016/j.colsurfb.2005.05.005
Hoover, D., Wilcox, K. R., Young, K. (2018). Experimental droughts with rainout shelters: A methodological review. Ecosphere 9, e02088. doi: 10.1002/ecs2.2088
Hou, J., Huang, X., Sun, W., Du, C., Wang, C., Xie, Y., et al. (2018). Accumulation of water-soluble carbohydrates and gene expression infile:///C:/Users/Kamal/Desktop/Additional references-1B/fpls-09-02003.pdf1fehw3, TaSnRK.pdf wheat stems correlates with drought resistance. J. Plant Physiol. 231, 182–191. doi: 10.1016/j.jplph.2018.09.017
Huang, X., Han, B. (2014). Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 65, 531–551. doi: 10.1146/annurev-arplant-050213-035715
IPCC (2013). “Summary for policymakers”, in Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the International Panel on Climate Change. Eds. Stocker, T., Qin, D., Plattner, G., Tignor, M., Allen, S., Boschung, J. (Cambridge, United Kingdom and New York, NY, USA: Cambridge University Press). Available at: https://www.ipcc.ch/report/ar5/wg1/.
IWGSC (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361, eaar7191. doi: 10.1126/science.aar7191
Jafarzadeh, J., Bonnett, D., Jannink, J. L., Akdemir, D., Dreisigacker, S., Sorrells, M. E. (2016). Breeding value of primary synthetic wheat genotypes for grain yield. PLoS One 11, e0162860. doi: 10.1371/journal.pone.0162860
Janni, M., Bozzini, T., Di Giovanni, M., Moscetti, I., Lupi, R., Gennaro, A., et al. (2018). First production of wild emmer (Triticum turgidum ssp. dicoccoides) transgenic plants. Plant Cell. Tissue Organ Cult. 132, 461–467. doi: 10.1007/s11240-017-1342-0
Jiang, Y., Jiang, Q., Hao, C., Hou, J., Wang, L., Zhang, H., et al. (2015). A yield associated gene TaCWI, in wheat: its function, selection and evolution in global breeding revealed by haplotype analysis. Theor. Appl. Genet. 128, 131–143. doi: 10.1007/s00122-014-2417-5
Kadioglu, A., Terzi, R. (2007). A dehydration avoidance mechanism: Leaf rolling. Bot. Rev. 73, 290–302. doi: 10.1663/0006-8101(2007)73[290:ADAMLR]2.0.CO;2
Khadka, K., Earl, H., Raizada, M. N., Navabi, A. (2020). A physio-morphological trait-based approach for breeding drought-tolerant wheat. Front. Plant Sci. 11, 715. doi: 10.3389/fpls.2020.00715
Khakwani, A. A., Dennett, M. D., Munir, M. (2011). Drought tolerance screening of wheat varieties by inducing water stress conditions. Songklanakarin J. Sci. Technol. 33, 135–142.
Khalid, M., Afzal, F., Gul, A., Amir, R., Subhani, A., Ahmed, Z., et al (2019) Molecular characterization of 87 functional genes in wheat diversity panel and their association with phenotypes under well-watered and water-limited conditions. Front. Plant Sci. 10, 717. doi: 10.3389/fpls.2019.00717
Khan, Z., Qazi, J., Rasheed, A., Mujeeb-Kazi, A. (2016). Diversity in D-genome synthetic hexaploid wheat association panel for seedling emergence traits under salinity stress. Plant Genet. Resour. 15, 488–495. doi: 10.1017/S1479262116000198
Khayatnezhad, M., Zaeifizadeh, M., Gholamin, R. (2011). Effect of end-season drought stress on chlorophyll fluorescence and content of antioxidant enzyme superoxide dismutase enzyme (SOD) in susceptible and tolerant genotypes of durum wheat. Afr. J. Agric. Res. 6, 6397–6406. doi: 10.5897/AJAR11.250
Kilian, B., Graner, A. (2012). NGS technologies for analyzing germplasm diversity in genebanks. Brief. Funct. Genomics 11, 38–50. doi: 10.1093/bfgp/elr046
Kim, D., Alptekin, B., Budak, H. (2018). CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genomics 18, 31–41. doi: 10.1007/s10142-017-0572-x
Kira, O., Linker, R., Gitelson, A. (2015). Non-destructive estimation of foliar chlorophyll and carotenoid contents: Focus on informative spectral bands. Int. J. Appl. Earth Obs. Geoinf. 38, 251–260. doi: 10.1016/j.jag.2015.01.003
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi: 10.1038/nature17946
Kooke, R., Kruijer, W., Bours, R., Becker, F., Kuhn, A., Geest, H., et al. (2016). Genome-wide association mapping and genomic prediction elucidate the genetic architecture of morphological traits in arabidopsis. Plant Physiol. 170, 2187–2203. doi: 10.1104/pp.15.00997
Kumar, S., Singh, B. (2009). Effect of water stress on carbon isotope discrimination and Rubisco activity in bread and durum wheat genotypes. Physiol. Mol. Biol. Plants 15, 281–286. doi: 10.1007/s12298-009-0032-8
Kumar, S., Kumar, N., Balyan, H. S., Gupta, P. K. (2003). 1BL.1RS translocation in some Indian bread wheat genotypes and strategies for its use in future wheat breeding. Caryologia 56, 23–30. doi: 10.1080/00087114.2003.10589303
Kumar, R., Kaur, A., Pandey, A., Mamrutha, H. M., Singh, G. P. (2019). CRISPR-based genome editing in wheat: a comprehensive review and future prospects. Mol. Biol. Rep. 46, 3557–3569. doi: 10.1007/s11033-019-04761-3
Laxa, M., Liebthal, M., Telman, W., Chibani, K., Dietz, K. J. (2019). The role of the plant antioxidant system in drought tolerance. Antioxidants 8:94. doi: 10.3390/antiox8040094
le Roux, M. L., Kunert, K. J., van der Vyver, C., Cullis, C. A., Botha, A. M. (2019). Expression of a small ubiquitin-like modifier protease increases drought tolerance in wheat (Triticum aestivum L.). Front. Plant Sci. 10, 266. doi: 10.3389/fpls.2019.00266
Lehnert, H., Serfling, A., Friedt, W., Ordon, F. (2018). Genome-wide association studies reveal genomic regions associated with the response of wheat (Triticum aestivum L.) to mycorrhizae under drought stress conditions. Front. Plant Sci. 9, 1728. doi: 10.3389/fpls.2018.01728
Li, L., Zheng, M., Deng, G., Liang, J., Zhang, H., Pan, Z., et al. (2016). Overexpression of AtHDG11 enhanced drought tolerance in wheat (Triticum aestivum L.). Mol. Breed. 36, 23. doi: 10.1007/s11032-016-0447-1
Li, C., Zong, Y., Wang, Y., Jin, S., Zhang, D., Song, Q., et al. (2018). Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 19, 59. doi: 10.1186/s13059-018-1443-z
Li, A., Liu, D., Yang, W., Kishii, M., Mao, L. (2018). Synthetic hexaploid wheat: Yesterday, today and tomorrow. Engineering 4, 552–558. doi: 10.1016/j.eng.2018.07.001
Li, G., Xu, X., Tan, C., Carver, B. F., Bai, G., Wang, X., et al. (2019). Identification of powdery mildew resistance loci in wheat by integrating genome-wide association study (GWAS)and linkage mapping. Crop J. 7, 294–306. doi: 10.1016/j.cj.2019.01.005
Li, L., Mao, X., Wang, J., Chang, X., Reynolds, M., Jing, R. (2019). Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell Environ. 42, 2540–2553. doi: 10.1111/pce.13577
Lin, Q., Zong, Y., Xue, C., Wang, S., Jin, S., Zhu, Z., et al. (2020). Prime genome editing in rice and wheat. Nat. Biotechnol. 38, 582–585. doi: 10.1038/s41587-020-0455-x
Liu, C., Yang, Z., Hu, Y. (2015). Drought resistance of wheat alien chromosome addition lines evaluated by membership function value based on multiple traits and drought resistance index of grain yield. Field Crop Res. 179, 103–112. doi: 10.1016/j.fcr.2015.04.016
Liu, Y., Bowman, B., Hu, Y. G., Liang, X., Zhao, W., Wheeler, J., et al. (2017). Evaluation of agronomic traits and drought tolerance of winter wheat accessions from the USDA-ARS National Small Grains Collection. Agronomy 7, 51. doi: 10.3390/agronomy7030051
Liu, K., Sun, X., Ning, T., Duan, X., Wang, Q., Liu, T., et al. (2018). Genetic dissection of wheat panicle traits using linkage analysis and a genome-wide association study. Theor. Appl. Genet. 131, 1073–1090. doi: 10.1007/s00122-018-3059-9
Liu, C., Sukumaran, S., Claverie, E., Sansaloni, C., Dreisigacker, S., Reynolds, M. (2019). Genetic dissection of heat and drought stress QTLs in phenology-controlled synthetic-derived recombinant inbred lines in spring wheat. Mol. Breed. 39, 34. doi: 10.1007/s11032-019-0938-y
Lopes, M. S., Reynolds, M. P. (2010). Partitioning of assimilates to deeper roots is associated with cooler canopies and increased yield under drought in wheat. Funct. Plant Biol. 37, 147–156. doi: 10.1071/FP09121
Lopes, M., Reynolds, M. P. (2011). Drought adaptive traits and wide adaptation in elite lines derived from resynthesized hexaploid wheat. Crop Sci. 51, 1617–1621. doi: 10.2135/cropsci2010.07.0445
Lopes, M. S., El-basyoni, I., Baenziger, P. S., Singh, S., Royo, C., Ozbek, K., et al. (2015a). Exploiting genetic diversity from landraces in wheat breeding for adaptation to climate change. J. Exp. Bot. 66, 3477–3486. doi: 10.1093/jxb/erv122
Lopes, M. S., Dreisigacker, S., Peña, R. J., Sukumaran, S., Reynolds, M. P. (2015b). Genetic characterization of the wheat association mapping initiative (WAMI) panel for dissection of complex traits in spring wheat. Theor. Appl. Genet. 128, 453–464. doi: 10.1007/s00122-014-2444-2
Maqsood, R. H., Amjid, M. W., Saleem, M. A., Shabbir, G., Khaliq, I. (2017). Identification of genomic regions conferring drought tolerance in bread wheat using ISSR markers. Pakistan J. Bot. 49, 1821–1827.
Marcussen, T., Sandve, S. R., Heier, L., Spannagl, M., Pfeifer, M., et al. (2014). Ancient hybridizations among the ancestral genomes of bread wheat. Science 345, 1250092. doi: 10.1126/science.1250092
Marzec, M., Hensel, G. (2020). Prime editing: Game changer for modifying plant genomes trends in plant science. Trends Plant Sci. xx, 1–3. doi: 10.1016/j.tplants.2020.05.008
Mathew, I., Shimelis, H., Shayanowako, A., II, Laing, M., Chaplot, V. (2019). Genome-wide association study of drought tolerance and biomass allocation in wheat. PLoS One 14, e0225383. doi: 10.1371/journal.pone.0225383
Meena, R. P., Tripathi, S. C., Chander, S., Chookar, R. S., Verma, M. A., Sharma, R. K. (2015). Identifying drought-tolerant wheat varieties using different indices. SAARC J. Agric. 13, 148–161. doi: 10.3329/sja.v13i1.24188
Mega, R., Abe, F., Kim, J. S., Tsuboi, Y., Tanaka, K., Kobayashi, H., et al. (2019) Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 5, 153–159. doi: 10.1038/s41477-019-0361-8
Merah, O., Deléens, E., Souyris, I., Monneveux, P. (2000). Effect of glaucousness on carbon isotope discrimination and grain yield in durum wheat. J. Agron. Crop Sci. 185, 259–265. doi: 10.1046/j.1439-037X.2000.00434.x
Mia, S., Liu, H., Wang, X., Yan, G. (2019). Multiple near-isogenic lines targeting a QTL hotspot of drought tolerance showed contrasting performance under post-anthesis water stress. Front. Plant Sci. 10, 271. doi: 10.3389/fpls.2019.00271
Mickky, B. M., Aldesuquy, H. S. (2017). Impact of osmotic stress on seedling growth observations, membrane characteristics and antioxidant defense system of different wheat genotypes. Egypt. J. Basic Appl. Sci. 4, 47–54. doi: 10.1016/j.ejbas.2016.10.001
Mohammadi, R. (2018). Breeding for increased drought tolerance in wheat: A review. Crop Pasture Sci. 69, 223–241. doi: 10.1071/CP17387
Molla, K. A., Yang, Y. (2019). CRISPR/Cas-mediated base editing: Technical considerations and practical applications. Trends Biotechnol. 37, 1121–1142. doi: 10.1016/j.tibtech.2019.03.008
Mondal, S., Dutta, S., Crespo-Herrera, L., Huerta-Espino, J., Braun, H. J., Singh, R. P. (2020). Fifty years of semi-dwarf spring wheat breeding at CIMMYT: Grain yield progress in optimum, drought and heat stress environments. F. Crop Res. 250, 107757. doi: 10.1016/j.fcr.2020.107757
Monneveux, P., Reynolds, M. P., Zaharieva, M., Mujeeb-Kazi, A. (2003). Effect of T1BL.1RS chromosome translocation on bread wheat grain yield and physiological related traits in a warm environment. Cereal Res. Commun. 31, 371–378. doi: 10.1007/bf03543367
Mwadzingeni, L., Shimelis, H., Dube, E., Laing, M. D., Tsilo, T. J. (2016a). Breeding wheat for drought tolerance: Progress and technologies. J. Integr. Agric. 15, 935–943. doi: 10.1016/S2095-3119(15)61102-9
Mwadzingeni, L., Shimelis, H., Tesfay, S., Tsilo, T. J. (2016b). Screening of bread wheat genotypes for drought tolerance using phenotypic and proline analyses. Front. Plant Sci. 7, 1276. doi: 10.3389/fpls.2016.01276
Mwadzingeni, L., Shimelis, H., Rees, D. J. G., Tsilo, T. J. (2017). Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS One 12, e0171692. doi: 10.1371/journal.pone.0171692
Nagy, É., Lehoczki-Krsjak, S., Lantos, C., Pauk, J. (2018). Phenotyping for testing drought tolerance on wheat varieties of different origins. South Afr. J. Bot. 116, 216–221. doi: 10.1016/j.sajb.2018.03.009
Nemtsev, B. F., Nemtsev, A. B., Goncharov, N. P., Kurkova, S. V. (2019). Spring common wheat breeding lines produced on the basis of distant hybridization: ecological strain testing in Bagan. Curr. Challenges Plant Genet. Genom. Bioinform. Biotechnol. 2019, 30–33. doi: 10.18699/icg-plantgen2019-07
Nezhad, K. Z., Weber, W. E., Röder, M. S., Sharma, S., Lohwasser, U., Meyer, R. C., et al. (2012). QTL analysis for thousand-grain weight under terminal drought stress in bread wheat (Triticum aestivum L.). Euphytica 186, 127–138. doi: 10.1007/s10681-011-0559-y
Niemirowicz-Szczytt, K. (1997). Excessive homozygosity in doubled haploids - Advantages and disadvantages for plant breeding and fundamental research. Acta Physiol. Plant 19, 155–167. doi: 10.1007/s11738-997-0032-7
Okada, A., Arndell, T., Borisjuk, N., Sharma, N., Watson-haigh, N. S., Tucker, E. J., et al. (2019). CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 17, 1905–1913. doi: 10.1111/pbi.13106
Padulosi, S., Bergamini, N., Lawrence, T. (2012). “On-farm conservation of neglected and underutilized species: Status, trends, and novel approaches to deal with climate change,” Proceedings of the International Conference Friedrichsdorf, Frankfurt, 14–16 June, 2011 (Bioversity International, Rome), 307.
Petrov, P., Petrova, A., Tashev, I. D. T., Brestic, K. O. M., Misheva, S. (2018). Relationships between leaf morpho-anatomy, water status and cell membrane stability in leaves of wheat seedlings subjected to severe soil drought. J. Agron. Crop Sci. 204, 219–227. doi: 10.1111/jac.12255
Pinto, R. S., Reynolds, M. P., Mathews, K. L., McIntyre, C. L., Olivares-Villegas, J. J., Chapman, S. C. (2010). Heat and drought adaptive QTL in a wheat population designed to minimize confounding agronomic effects. Theor. Appl. Genet. 121, 1001–1021. doi: 10.1007/s00122-010-1351-4
Poland, J., Endelman, J., Dawson, J., Rutkoski, J., Wu, S. Y., Manes, Y., et al. (2012). Genomic selection in wheat breeding using genotyping-by-sequencing. Plant Genome 5, 103–113. doi: 10.3835/Plantgenome2012.06.0006
Qaseem, M. F., Qureshi, R., Muqaddasi, Q. H.. (2018) Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. PLoS One 13, e0199121. doi: 10.1371/journal.pone.0199121
Qaseem, M. F., Qureshi, R., Shaheen, H., Shafqat, N. (2019). Genome-wide association analyses for yield and yield-related traits in bread wheat (Triticum aestivum L.) under pre-anthesis combined heat and drought stress in field conditions. PLoS One 14, e0213407. doi: 10.1371/journal.pone.0213407
Raina, A., Laskar, R., Khursheed, S., Amin, R., Tantray, Y., Parveen, K., et al. (2016). Role of mutation breeding in crop improvement- past, present and future. Asian Res. J. Agric. 2, 1–13. doi: 10.9734/arja/2016/29334
Rajaram, S. (2005). Role of conventional plant breeding and biotechnology in future wheat production. Turkish J. Agric. For. 29, 105–111. doi: 10.3906/tar-0404-1
Ramirez-Gonzalez, R. H., Segovia, V., Bird, N., Caccamo, M., Uauy, C. (2015). “Next generation sequencing enabled genetics in hexaploid wheat,” in Advances in Wheat Genetics: From Genome to Field. Proceedings of 12th International Wheat Genetics Symposium. Eds. Ogihara, Y., Takumi, S., Handa, H. (Yokohama, Japan: Springer Open), 201–209. Available at: https://www.springer.com/gp/book/9784431556749.
Ramya, P., Singh, G. P., Jain, N., Singh, P. K., Pandey, M. K., Sharma, K., et al. (2016). Effect of recurrent selection on drought tolerance and related morpho-physiological traits in bread wheat. PLoS One 11, e0156869. doi: 10.1371/journal.pone.0156869
Rauf, S., Teixeira, J. A., Ali, A., Abdul, K. (2010). Consequences of plant breeding on genetic diversity. Int. J. Plant Breed. 4, 1–21.
Ray, S., Satya, P. (2014). Next generation sequencing technologies for next generation plant breeding. Front. Plant Sci. 5, 367. doi: 10.3389/fpls.2014.00367
Rebetzke, G. J., Richards, R. A., Fischer, V. M., Mickelson, B. J. (1999). Breeding long coleoptile, reduced height wheats. Euphytica 106, 159–168. doi: 10.1023/A:1003518920119
Rebetzke, G. J., Appels, R., Morrison, A. D., Richards, R. A., McDonald, G., Ellis, M. H., et al. (2001). Quantitative trait loci on chromosome 4B for coleoptile length and early vigour in wheat (Triticum aestivum L.). Aust. J. Agric. Res. 52, 1221–1234. doi: 10.1071/ar01042
Rebetzke, G. J., Ellis, M. H., Bonnett, D. G., Richards, R. A. (2007). Molecular mapping of genes for Coleoptile growth in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 114, 1173–1183. doi: 10.1007/s00122-007-0509-1
Rebetzke, G. J., Condon, A. G., Farquhar, G. D., Appels, R., Richards, R. A. (2008). Quantitative trait loci for carbon isotope discrimination are repeatable across environments and wheat mapping populations. Theor. Appl. Genet. 118, 123–137. doi: 10.1007/s00122-008-0882-4
Reddy, S. K., Liu, S., Rudd, J. C., Xue, Q., Payton, P., Finlayson, S. A., et al. (2014). Physiology and transcriptomics of water-deficit stress responses in wheat cultivars TAM 111 and TAM 112. J. Plant Physiol. 171, 1289–1298. doi: 10.1016/j.jplph.2014.05.005
Ren, T., Tang, Z., Fu, S., Yan, B., Tan, F., Ren, Z., et al. (2017). Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 8, 799. doi: 10.3389/fpls.2017.00799
Reynolds, M., Dreccer, F., Trethowan, R. (2007). Drought-adaptive traits derived from wheat wild relatives and landraces. J. Exp. Bot. 58, 177–186. doi: 10.1093/jxb/erl250
Rosyara, U., Kishii, M., Payne, T., Sansaloni, C. P., Singh, R. P., Braun, H. J., et al. (2019). Genetic contribution of synthetic hexaploid wheat to CIMMYT’s spring bread wheat breeding germplasm. Sci. Rep. 9, 12355. doi: 10.1038/s41598-019-47936-5
Saeidi, M., Abdoli, M. (2015). Effect of drought stress during grain filling on yield and its components, gas exchange variables, and some physiological traits of wheat cultivars. J. Agric. Sci. Technol. 17, 885–898.
Saeidi, M., Ardalani, S., Jalali-Honarmand, S., Ghobadi, M. E., Abdoli, M. (2015). Evaluation of drought stress at vegetative growth stage on the grain yield formation and some physiological traits as well as fluorescence parameters of different bread wheat cultivars. Acta Biol. Szeged 59, 35–44.
Sallam, A., Arbaoui, M., El-Esawi, M., Abshire, N., Martsch, R. (2016). Identification and verification of QTL associated with frost tolerance using linkage mapping and GWAS in winter Faba bean. Front. Plant Sci. 7, 1098. doi: 10.3389/fpls.2016.01098
Sareen, S., Sharma, P., Tiwari, V., Sharma, I. (2014). Identifying wheat landraces as genetic resources for drought and heat tolerance. Res. Crop 15, 846–851. doi: 10.5958/2348-7542.2014.01421.1
Sarto, M. V. M., Sarto, J. R. W., Rampim, L., Bassegio, D., da Costa, P. F., Inagaki, A. M. (2017). Wheat phenology and yield under drought: A review. Aust. J. Crop Sci. 11, 941–946. doi: 10.21475/ajcs17.11.08.pne351
Scherer, A., Christensen, G. B. (2016). Concepts and relevance of genome-wide association studies. Sci. Prog. 99, 59–67. doi: 10.3184/003685016X14558068452913
Schmidt, J., Tricker, P. J., Eckermann, P., Kalambettu, P., Garcia, M., Fleury, D. (2020). Novel alleles for combined drought and heat stress tolerance in wheat. Front. Plant Sci. 10, 1800. doi: 10.3389/fpls.2019.01800
Sehgal, D., Vikram, P., Sansaloni, C. P., Ortiz, C., Pierre, C., Saint, et al. (2015). Exploring and mobilizing the gene bank biodiversity for wheat improvement. PLoS One 10, e0132112. doi: 10.1371/journal.pone.0132112
Sen, A., Ozturk, I., Yaycili, O., Alikamanoglu, S. (2017). Drought tolerance in irradiated wheat mutants studied by genetic and biochemical markers. J. Plant Growth Regul. 36, 669–679. doi: 10.1007/s00344-017-9668-8
Shamasbi, F. V., Jamali, S. H., Sadeghzadeh, B., Mandoulakani, B. A. (2017). Genetic mapping of quantitative trait loci for yield-affecting traits in a barley doubled haploid population derived from clipper × sahara 3771. Front. Plant Sci. 8, 688. doi: 10.3389/fpls.2017.00688
Shi, X., Ling, H. (2017). Current advances in genome sequencing of common wheat and its ancestral species. Crop J. 6, 15–21. doi: 10.1016/j.cj.2017.11.001
Shi, Y., Thomasson, J. A., Murray, S. C., Pugh, N. A., Rooney, W. L., Shafian, S., et al. (2016). Unmanned aerial vehicles for high- throughput phenotyping and agronomic research. PLoS One 11, e0159781. doi: 10.5061/dryad.65m87
Shi, W., Hao, C., Zhang, Y., Cheng, J., Zhang, Z., Liu, J., et al. (2017). A combined association mapping and linkage analysis of kernel number per spike in common wheat (Triticum aestivum L.). Front. Plant Sci. 8, 1412. doi: 10.3389/fpls.2017.01412
Shi, S., Azam, F., II, Li, H., Chang, X., Li, B., Jing, R. (2017). Mapping QTL for stay-green and agronomic traits in wheat under diverse water regimes. Euphytica 213, 246. doi: 10.1007/s10681-017-2002-5
Shinwari, Z. K., Jan, S. A., Nakashima, K., Yamaguchi-Shinozaki, K. (2020). Genetic engineering approaches to understanding drought tolerance in plants. Plant Biotechnol. Rep. 14, 151–162. doi: 10.1007/s11816-020-00598-6
Shrestha, R., Matteis, L., Skofic, M., Portugal, A., McLaren, G., Hyman, G., et al. (2012). Bridging the phenotypic and genetic data useful for integrated breeding through a data annotation using the Crop Ontology developed by the crop communities of practice. Front. Physiol. 3, 326. doi: 10.3389/fphys.2012.00326
Singh, R. P., Huerta-Espino, J., Rajaram, S., Crossa, J. (1998). Agronomic effects from chromosome translocations 7DL.7AG and 1BL.1RS in spring wheat. Crop Sci. 38, 27–33. doi: 10.2135/cropsci1998.0011183X003800010005x
Singh, A., Ganapathysubramanian, B., Singh, A. K., Sarkar, S. (2016). Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 21, 110–124. doi: 10.1016/j.tplants.2015.10.015
Singh, B., Kukreja, S., Goutam, U. (2018). Milestones achieved in response to drought stress through reverse genetic approaches. F1000Research 7, 1311. doi: 10.12688/f1000research.15606.1
Singh, S., Vikram, P., Sehgal, D., Burgueño, J., Sharma, A., Singh, S. K., et al. (2018). Harnessing genetic potential of wheat germplasm banks through impact-oriented-prebreeding for future food and nutritional security. Sci. Rep. 8, 12527. doi: 10.1038/s41598-018-30667-4
Singh, N., Wu, S., Raupp, W. J., Sehgal, S., Arora, S., Tiwari, V., et al. (2019). Efficient curation of genebanks using next generation sequencing reveals substantial duplication of germplasm accessions. Sci. Rep. 9, 650. doi: 10.1038/s41598-018-37269-0
Sohail, Q., Inoue, T., Tanaka, H., Eltayeb, A. E., Matsuoka, Y., Tsujimoto, H. (2011). Applicability of Aegilops tauschii drought tolerance traits to breeding of hexaploid wheat. Breed. Sci. 61, 347–357. doi: 10.1270/jsbbs.61.347
Song, Q., Liu, C., Goudia, D., Chen, L., Hu, Y. (2017). Drought resistance of new synthetic hexaploid wheat accessions evaluated by multiple traits and antioxidant enzyme activity. F. Crop Res. 210, 91–103. doi: 10.1016/j.fcr.2017.05.028
Soriano, J. M., Alvaro, F. (2019). Discovering consensus genomic regions in wheat for root-related traits by QTL meta-analysis. Sci. Rep. 9, 10537. doi: 10.1038/s41598-019-47038-2
Spielmeyer, W., Hyles, J., Joaquim, P., Azanza, F., Bonnett, D., Ellis, M. E., et al. (2007). A QTL on chromosome 6A in bread wheat (Triticum aestivum) is associated with longer coleoptiles, greater seedling vigour and final plant height. Theor. Appl. Genet. 115, 59–66. doi: 10.1007/s00122-007-0540-2
Su, Y., Wu, F., Ao, Z., Jin, S., Qin, F., Liu, B., et al. (2019). Evaluating maize phenotype dynamics under drought stress using terrestrial lidar. Plant Methods 15, 11. doi: 10.1186/s13007-019-0396-x
Suneja, Y., Gupta, A. K., Bains, N. S. (2019). Stress adaptive plasticity: Aegilops tauschii and Triticum dicoccoides as potential donors of drought associated morpho-physiological traits in wheat. Front. Plant Sci. 10, 211. doi: 10.3389/fpls.2019.00211
Szabo-Hever, A., Zhang, Q., Friesen, T. L., Zhong, S., Elias, E. M., Cai, X., et al. (2018). Genetic diversity and resistance to Fusarium head blight in synthetic hexaploid wheat derived from Aegilops tauschii and diverse Triticum turgidum subspecies. Front. Plant Sci. 871, 1829. doi: 10.3389/fpls.2018.01829
Tahmasebi, S., Heidari, B., Pakniyat, H., Dadkhodaie, A. (2015). Consequences of 1BL/1RS translocation on agronomic and physiological traits in wheat. Cereal Res. Commun. 43, 554–566. doi: 10.1556/0806.43.2015.016
Touzy, G., Rincent, R., Bogard, M., Lafarge, S., Dubreuil, P., Mini, A., et al. (2019). Using environmental clustering to identify specific drought tolerance QTLs in bread wheat (T. aestivum L.). Theor. Appl. Genet. 132, 2859–2880. doi: 10.1007/s00122-019-03393-2
Tura, H., Edwards, J., Gahlaut, V., Garcia, M., Sznajder, B., Baumann, U., et al. (2020). QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor. Appl. Genet. 133, 239–257. doi: 10.1007/s00122-019-03454-6
Uauy, C. (2017). Wheat genomics comes of age. Curr. Opin. Plant Biol. 36, 142–148. doi: 10.1016/j.pbi.2017.01.007
United Nations (2019). World Population Prospects 2019: Highlights (ST/ESA/SER.A/423) (United Nations: Department of Economic and Social Affairs, Population Division). Available at: http://www.ncbi.nlm.nih.gov/pubmed/12283219.
Verma, V., Foulkes, M. J., Sylvester-Bradley, R., Caligari, P. D. S., Snape, J. W. (2004). Mapping quantitative trait loci for flag leave senescence as a yield determinant in winter wheat under optimal and drought stressed environments. Euphytica 135, 255–263.
Vikram, P., Franco, J., Burgueño-Ferreira, J., Li, H., Sehgal, D., Saint Pierre, C., et al. (2016). Unlocking the genetic diversity of Creole wheats. Sci. Rep. 6, 23092. doi: 10.1038/srep23092
Visscher, P. M., Wray, N. R., Zhang, Q., Sklar, P., Mccarthy, M., II, Brown, M. A., et al. (2017). 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22. doi: 10.1016/j.ajhg.2017.06.005
Wang, M., Jiang, N., Jia, T., Leach, L., Cockram, J., Waugh, R., et al. (2012). Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. Theor. Appl. Genet. 124, 233–246. doi: 10.1007/s00122-011-1697-2
Wang, X., Vignjevic, M., Liu, F., Jacobsen, S., Jiang, D., Wollenweber, B. (2015). Drought priming at vegetative growth stages improves tolerance to drought and heat stresses occurring during grain filling in spring wheat. Plant Growth Regul. 75, 677–687. doi: 10.1007/s10725-014-9969-x
Wang, M., Wang, S., Liang, Z., Shi, W., Gao, C., Xia, G. (2018). From genetic stock to genome editing: gene exploitation in wheat. Trends Biotechnol. 36, 160–172. doi: 10.1016/j.tibtech.2017.10.002
White, J. W., Andrade-Sanchez, P., Gore, M. A., Bronson, K. F., Coffelt, T. A., Conley, M. M., et al. (2012). Field-based phenomics for plant genetics research. Field Crop Res. 133, 101–112. doi: 10.1016/j.fcr.2012.04.003
Yan, G., Liu, H., Wang, H., Lu, Z., Wang, Y., Mullan, D., et al. (2017). Accelerated generation of selfed pure line plants for gene identification and crop breeding. Front. Plant Sci. 8, 1786. doi: 10.3389/fpls.2017.01786
Yang, S., Vanderbeld, B., Wan, J., Huang, Y. (2010). Narrowing down the targets: Towards successful genetic engineering of drought-tolerant crops. Mol. Plant 3, 469–490. doi: 10.1093/mp/ssq016
Yang, W., Feng, H., Zhang, X., Zhang, J., Doonan, J. H., Batchelor, W. D., et al. (2020). Crop phenomics and high-throughput phenotyping: Past decades, current challenges, and future perspectives. Mol. Plant 13, 187–214. doi: 10.1016/j.molp.2020.01.008
Yu, T. F., Xu, Z. S., Guo, J. K., Wang, Y. X., Abernathy, B., Fu, J. D., et al. (2017). Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 7, 44050. doi: 10.1038/srep44050
Zandipour, M., Majidi Hervan, E., Azadi, A., Khosroshahli, M., Etminan, A. (2020). A QTL hot spot region on chromosome 1B for nine important traits under terminal drought stress conditions in wheat. Cereal Res. Commun. 48, 17–24. doi: 10.1007/s42976-020-00017-0
Zhang, J., Kirkham, M. B. (1994). Drought-stress-induced changes in activities of superoxide dismutase, catalase, and peroxidase in wheat species. Plant Cell Physiol. 35, 785–791. doi: 10.1093/oxfordjournals.pcp.a078658
Zhang, H., Cui, F., Wang, L., Li, J., Ding, A., Zhao, C., et al. (2013). Conditional and unconditional QTL mapping of drought-tolerance-related traits of wheat seedling using two related RIL populations. J. Genet. 92, 213–231. doi: 10.1007/s12041-013-0253-z
Zhang, J., Xu, Y., Chen, W., Dell, B., Vergauwen, R., Biddulph, B., et al. (2015). A wheat 1-FEH w3 variant underlies enzyme activity for stem WSC remobilization to grain under drought. New Phytol. 205, 293–305. doi: 10.1111/nph.13030
Zhang, X., Huang, C., Wu, D., Qiao, F., Li, W., Duan, L., et al. (2017). High-throughput phenotyping and QTL mapping reveals the genetic architecture of maize. Plant Physiol. 173, 1554–1564. doi: 10.1104/pp.16.01516
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Zhang, Y., Li, D., Zhang, D., Zhao, X., Cao, X., Dong, L., et al. (2018). Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 94, 857–866. doi: 10.1111/tpj.13903
Zhang, Y., Wang, Z., Fan, Z., Li, J., Gao, X., Zhang, H., et al. (2019). Phenotyping and evaluation of CIMMYT WPHYSGP nursery lines and local wheat varieties under two irrigation regimes. Breed. Sci. 67, 55–67. doi: 10.1270/jsbbs.18104
Zhao, X., Luo, L., Cao, Y., Liu, Y., Li, Y., Wu, W., et al. (2018). Genome-wide association analysis and QTL mapping reveal the genetic control of cadmium accumulation in maize leaf. BMC Genomics 19, 91. doi: 10.1186/s12864-017-4395-x
Keywords: drought tolerance, genetic resources, landraces, quantitative trait loci mapping, wheat, climate change
Citation: Khadka K, Raizada MN and Navabi A (2020) Recent Progress in Germplasm Evaluation and Gene Mapping to Enable Breeding of Drought-Tolerant Wheat. Front. Plant Sci. 11:1149. doi: 10.3389/fpls.2020.01149
Received: 11 November 2019; Accepted: 15 July 2020;
Published: 04 August 2020.
Edited by:
Thomas Miedaner, University of Hohenheim, GermanyReviewed by:
Deepmala Sehgal, International Maize and Wheat Improvement Center, MexicoAwais Rasheed, Quaid-i-Azam University, Pakistan
Karl Kunert, University of Pretoria, South Africa
Copyright © 2020 Khadka, Raizada and Navabi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kamal Khadka, a2FtYWwua2hhZGthMDExQGdtYWlsLmNvbQ==
†Deceased
 Kamal Khadka
Kamal Khadka Manish N. Raizada
Manish N. Raizada Alireza Navabi†
Alireza Navabi†