
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 July 2020
Sec. Virology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01098
This article is part of the Research Topic Next-Generation Sequencing and CRISPR-Cas Editing in Plant Virology View all 17 articles
 Yoo-Joung Yoon1†
Yoo-Joung Yoon1† Jelli Venkatesh1†
Jelli Venkatesh1† Joung-Ho Lee1
Joung-Ho Lee1 Jinhee Kim2
Jinhee Kim2 Hye-Eun Lee2
Hye-Eun Lee2 Do-Sun Kim2
Do-Sun Kim2 Byoung-Cheorl Kang1*
Byoung-Cheorl Kang1*Many of the recessive virus-resistance genes in plants encode eukaryotic translation initiation factors (eIFs), including eIF4E, eIF4G, and related proteins. Notably, eIF4E and its isoform eIF(iso)4E are pivotal for viral infection and act as recessive resistance genes against various potyviruses in a wide range of plants. In this study, we used Clustered Regularly Interspaced Palindromic Repeats/CRISPR-associated protein 9 (CRISPR/Cas9)-mediated targeted mutagenesis to test whether novel sequence-specific mutations at eIF4E1 in Solanum lycopersicum (tomato) cv. Micro-Tom could confer enhanced resistance to potyviruses. This approach produced heritable homozygous mutations in the transgene-free E1 generation. Sequence analysis of eIF4E1 from E0 transgenic plants expressing Cas9 and eIF4E-sgRNA transcripts identified chimeric deletions ranging from 11 to 43 bp. Genotype analysis of the eIF4E1-edited lines in E0, E1, and E2 transgenic tomato plants showed that the mutations were transmitted to subsequent generations. When homozygous mutant lines were tested for resistance to potyviruses, they exhibited no resistance to tobacco etch virus (TEV). Notably, however, several mutant lines showed no accumulation of viral particles upon infection with pepper mottle virus (PepMoV). These results indicate that site-specific mutation of tomato eIF4E1 successfully conferred enhanced resistance to PepMoV. Thus, this study demonstrates the feasibility of the use of CRISPR/Cas9 approach to accelerate breeding for trait improvement in tomato plants.
Potyviridae is the largest family of plant RNA viruses, which account for about 30% of known plant viruses, cause considerable damage to crop plants (Ward and Shukla, 1991; Cui and Wang, 2019). The potyviruses tobacco etch virus (TEV), potato virus Y (PVY), chilli veinal mottle virus (ChiVMV), pepper mottle virus (PepMoV), and pepper veinal mottle virus (PVMV) infect numerous solanaceous plants, including tomato, potato, and pepper (Kothari et al., 2010; Zhao et al., 2014; Hančinský et al., 2020). Potyviruses, such as PepMoV and TEV are potential threat to tomato crop (Wintermantel, 2011; Melzer et al., 2012).
Eukaryotic translation initiation factor (eIF) genes, such as eukaryotic translation initiation factor 4E (eIF4E), eukaryotic translation initiation factor (Iso) 4E (eIF(iso)4E), and eukaryotic translation initiation factor 4G (eIF4G), are required for RNA viruses to maintain their lifecycle (Sanfacon, 2015; Revers and García, 2015). The number of eIF4E family members is species-dependent. In tomato, the eIF4E family consists of two eIF4E homologs (eIF4E1 and eIF4E2), one eIF(iso)4E homolog (Lebaron et al., 2016). Notably, several eIF genes confer recessive resistance to one or more potyviruses in Solanaceae crops (Kang et al., 2005a; Ruffel et al., 2006; Kang et al., 2007; Lee et al., 2013). A single recessive resistance locus pot-1 encoding eIF4E1 protein was identified from the tomato wild relative, Solanum habrochaites accession PI247087. eIF4E1 confers resistance to several potyviruses, including PVY, TEV, and PepMoV (Ruffel et al., 2005; Kang et al., 2005a; Wang, 2015). Knockout mutants of tomato eIF4E2 and eIF(iso)4E were reported to be fully susceptible to potyviruses, thus suggesting prominent role of eIF4E1 in potyviral resistance in Solanaceous crops (Robaglia and Caranta, 2006; Charron et al., 2008).
The eIF protein eIF4E is a component of a multiprotein complex that aids the initiation of protein translation by enabling recognition and interaction with the mRNA cap structure and recruitment of ribosomes. The viral genome-linked protein (VPg) of potyviruses is covalently connected to the 5′ end of viral RNA and acts as an analog of the eukaryotic mRNA cap structure during protein translation (Wang, 2015). The physical interaction between the host factor, eIF4E (or its homolog eIF(iso)4E), and VPg is crucial for potyvirus infectivity (Miyoshi et al., 2006; Robaglia and Caranta, 2006; Hwang et al., 2009; Kim et al., 2013; Kim et al., 2014; Tavert-Roudet et al., 2017; de Oliveira et al., 2019). Moreover, mutations in these host factors can inhibit the interaction between the host factor and the VPg, and consequently inhibit viral proliferation and host infection (Duprat et al., 2002; Lellis et al., 2002; Sato et al., 2005; Rodríguez-Hernández et al., 2012).
The majority of naturally occurring plant recessive resistance genes have been mapped to mutations in genes encoding the isoforms of the translation initiation factors eIF4E and eIF4G that hinder their interactions with viral RNAs or proteins (Kang et al., 2005a; Ruffel et al., 2006; Le Gall et al., 2011; Wang and Krishnaswamy, 2012; Lee et al., 2013). Natural recessive resistance to potyviruses has been exploited in numerous breeding programs (Kang et al., 2005b; Ruffel et al., 2006; Lee et al., 2013). However, conventional breeding requires massive backcrossing to introgress the trait of interest into an elite background; furthermore, the availability of favorable alleles in natural populations is limited. New alleles can be created by random mutagenesis (Nicaise, 2014), but this requires labor-intensive, time-consuming screening of large populations to select mutants with desirable properties.
Advances in genome-editing tools have accelerated site-directed mutagenesis in crops. Clustered regularly interspersed palindromic repeats (CRISPR)/CRISPR-associated protein 9 (CRISPR/Cas9) is a targeted genome-editing technique derived from the adaptive immune mechanism of Staphylococcus pyogenes against bacteriophages (Hsu et al., 2014). Since its first report in 2012, CRISPR/Cas9 has become the technology of choice for genome editing due to its ease, low cost, and significantly shorter timeframe for construct preparation compared to those of other genome-editing tools, such as zinc finger nucleases (ZFN) and transcription activator-like effector nucleases (TALENs) (Cong and Zhang, 2015). CRISPR/Cas9 has been utilized for site-directed mutagenesis in microbes, animals, human cells, and plants (Jiang et al., 2013; Wu et al., 2013; Zhang et al., 2017; Shapiro et al., 2018). Precise genome editing of host factors can be deployed for development of recessive genetic resistance against viral diseases in plants (Wang, 2015). However, the application of CRISPR/Cas9 tools to improve plant resistance to pathogens has not been widely explored, with only a few reports to date (Chandrasekaran et al., 2016; Pyott et al., 2016; Wang et al., 2016; Peng et al., 2017; Gomez et al., 2019). Recently, CRISPR/Cas9 genome editing of plants for potyvirus resistance has been reported. For instance, knockout of the gene eIF(iso)4E in the model crop plant Arabidopsis thaliana was shown to confer resistance to turnip mosaic virus (TuMV) (Pyott et al., 2016). In another study, CRISPR/Cas9-mediated knockout of eIF4E in cucumber resulted in broad-spectrum viral resistance to several plant viruses, including cucumber vein yellowing virus (CVYV), zucchini yellow mosaic virus (ZYMV), and papaya ringspot mosaic virus-w (PRSV-W) (Chandrasekaran et al., 2016). Similarly novel allelic variants of rice eIF4G generated through CRISPR/Cas9 mediated genome editing conferred resistance to rice tungro spherical virus (Macovei et al., 2018). Thus, targeted genome editing can be expected to accelerate plant breeding for disease-resistant crop plants by facilitating the introduction of precise and predictable genetic changes directly into an elite strain background.
In this study, to introduce allelic variations in the eIF4E gene, we mutated eIF4E1 in the tomato cultivar Micro-Tom using CRISPR/Cas9 technology and Agrobacterium-mediated transformation, and bred the E0 transgenic plants carrying eIF4E1 mutations to produce E1 and E2 progeny. We then tested the resistance of the homozygous mutant lines to the potyviruses TEV and PepMoV. The homozygous Micro-Tom mutant lines carrying genome-edited eIF4E1 were resistant to PepMoV, although not to TEV, and showed normal plant growth and development after PepMoV challenge. Furthermore, by segregation in the E1 generation, we were able to select virus-resistant plants that carried an edited eIF4E1 gene or genes but not the introduced transgene. Altogether, this study thus provides important information for understanding and analyzing tomato CRISPR/Cas9 mutants, and accelerating breeding for trait improvement.
Seeds of Solanum lycopersicum cv. Micro-Tom were surface sterilized in 70% ethanol for 1 min and in 2% NaOCl with one drop of Tween 20 for 15 min, and then rinsed four or five times with sterilized water. Seeds were germinated on 1/2 MS medium (Murashige and Skoog, 1962) containing 20 g/L sucrose and 8 g/L agar agar. All cultures were grown at 24°C with a 16 h light/8 h dark cycle under cool fluorescent light. Cotyledons of 7–8-day-old seedlings, before the appearance of the first true leaves, were used as explants for tissue culture.
eIF4E is a plant cellular translation initiation factor essential for potyvirus infection, and mutations in the eIF4E gene can confer resistance to potyviruses. In this study, we aimed to use CRISPR/Cas9-mediated targeted genome editing of eIF4E1 (GenBank: AY723733) to develop potyvirus-resistant tomato plants. The gene phytoene desaturase (PDS) (GenBank: EF650011), encoding the key enzyme in carotenoid biosynthesis, was used as a control gene to test genome-editing efficiency due to the easily detectable photobleached phenotype of PDS mutants. To design sgRNAs, we identified appropriate target sgRNA sequences using the CCTop - CRISPR/Cas9 target online predictor (https://crispr.cos.uni-heidelberg.de/index.html). sgRNAs targeting the first exon with high prediction scores were used for genome editing (Figure 1), since mutations in the 5′ region or first exon would increase the chance of creating nonfunctional proteins by causing frameshifts or early stop codons. These sgRNAs were cloned under the control of the AtU6-26 promoter into a binary vector (pHSE401) carrying a maize codon-optimized Cas9 gene driven by the CaMV 35S promoter. The 23-bp sequences of the corresponding primers, PDSgRNA_F and PDSgRNA_R (or eIF4E1gRNA_F and eIF4E1gRNA_R), flanked by a BsaI recognition site, were annealed and cloned into a BsaI site in the vector pHSE401 by Golden Gate cloning according to a previously reported method (Xing et al., 2014). CRISPR/Cas9 vectors were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation.
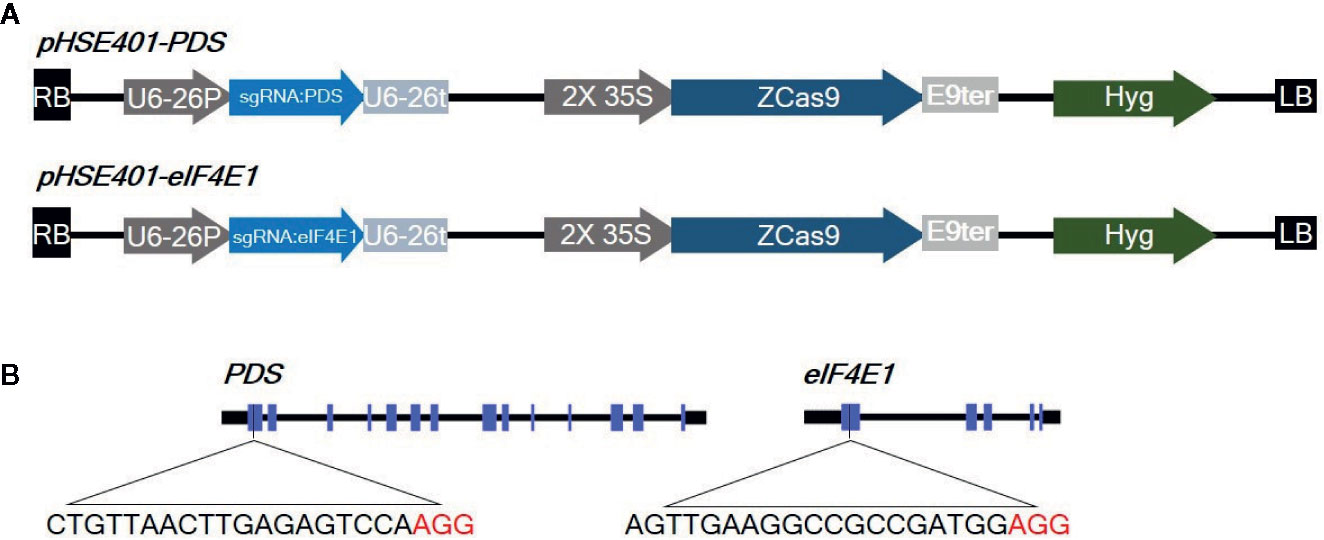
Figure 1 Diagrams of the CRISPR/Cas9 vector constructs and sgRNA target sites (A) Diagram of the cassettes (pHSE401-PDS and pHSE401-eIF4E1) expressing the maize codon-optimized Cas9 gene under the control of the 35S promoter, and the PDS- and eIF4E-sgRNA sequence driven by the Arabidopsis U6-26 polymerase III promoter. (B) Diagram of the CRISPR/Cas9 target sites within PDS and eIF4E1. The sgRNA target sequences are shown in black letters, followed by protospacer-adjacent motif (PAM) sequences in red.
A single Agrobacterium colony was inoculated into 15 mL of liquid LB medium containing 50 μg/mL kanamycin and 50 μg/mL rifampicin and incubated in a shaking incubator at 28°C for 12–16 h until it reached an OD600 of 0.6. The Agrobacterium suspension was then centrifuged at 8,000 rpm for 10 min at 20°C to collect the pellet. The pellet was completely resuspended in liquid 1/2 MS medium containing 3% sucrose and 200 μM acetosyringone. Cotyledons from 7–8-day-old seedlings were excised under sterile conditions, and then the tip of each cotyledon was removed, sectioned transversely into two fragments, and incubated adaxial side down in a preculture medium consisting of MS with 30 g/L sucrose, 1 mg/L 1-naphthaleneacetic acid (NAA), 1 mg/L benzylaminopurine (BAP) for 2 days, and 8 g/L agar agar. Explants were co-cultured in the Agrobacterium suspension for 20 min, transferred to sterile filter paper to briefly drain excess suspension, and then placed on the same medium used for preculture and incubated for 2 days. Then, explants were transferred to shoot induction medium consisting of MS with 30 g/L sucrose, 2 mg/L trans-zeatin riboside, 0.1 mg/L indole-3-acetic acid (IAA), 20 mg/L hygromycin, 250 mg/L carbenicillin, and 8 g/L agar agar for 4–6 weeks. Explants with shoot buds were moved to shoot elongation medium consisting of MS with 30 g/L sucrose, 1 mg/L trans-zeatin riboside, 0.1 mg/L IAA, 20 mg/L hygromycin, 250 mg/L carbenicillin, and 8 g/L agar agar. Tomato shoots of about 2 cm height were cut and transferred to rooting medium consisting of MS with 30 g/L sucrose, 1 mg/L IAA, 10 mg/L hygromycin, 250 mg/L carbenicillin, and 8 g/L agar agar. Rooted plants were transferred into plastic pots containing potting mixture (Hanarum, Minong Fertilizer, Korea) and kept in a growth room maintained at 24°C and a 16 h light/8 h dark cycle.
Genomic DNA (gDNA) was extracted from leaf samples of putative transgenic plants by the cetyltrimethylammonium bromide (CTAB) method (Porebski et al., 1997). gDNA was quantified by NanoDrop spectrophotometer (Nanodrop Technology, Inc., Wilmington, DE, USA) and diluted to 100 ng/μL. The status of putative transgenic plants was confirmed by PCR using Hpt (Hygromycin phosphotransferase) and Cas9 gene-specific primers (Table 1). PCR conditions were as follows: 94°C for 5 min, followed by 34 cycles of denaturing at 94°C for 30 s, annealing from 55°C for 30 s, 72°C extension for 30 s, and then final extension at 72°C 5 min. Total RNA was extracted from young leaf tissues using an MG RNAzol kit according to the manufacturer’s instructions (MGmed, Seoul, Korea). The integrity and concentration of the total RNA were analyzed on a 1% agarose gel and a NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, USA), respectively. One microgram of total RNA was used to synthesize complementary DNA (cDNA) using an EasyScript Reverse Transcriptase kit (TransGen, Beijing, China) with oligo(dT) primers. The resulting cDNAs were used for further expression analysis. RT-PCR was performed as described for gene confirmation using 1 µL cDNA as a template.
The transgenic plants were genotyped for mutation detection using primers flanking sgRNA target regions. PCR products were purified using a LaboPass PCR clean-up kit (Cosmo Genetech, Seoul, Korea). The purified amplicons were also cloned into the TA cloning vector, pMD20-T (Mighty TA-cloning kit, TAKARA, Shiga, Japan), according to the manufacturer’s instructions, and positive colonies were selected by blue/white colony selection. Plasmids were extracted from least five positive clones and sequenced using M13F and M13R primers at Bionics (Seoul, Korea). To identify CRISPR/Cas9 induced mutations, DNA sequence alignments were performed using Lasergene’s SeqMan program (DNASTAR, Madison, WI, USA). Mutations in E1 progeny was detected by directly sequencing the PCR amplicons of the target region as well as by sequencing positive TA clones.
For virus inoculum preparation, frozen stocks of TEV-HAT and PepMoV-Vb1, which were stored at –80°C, were used to inoculate 3-week-old Nicotiana benthamiana plants. Frozen inocula were ground in 0.1 M potassium phosphate buffer (pH 7.0), mixed with 400-grit carborundum, and rubbed on the lower leaves of N. benthamiana. After 10–20 min of inoculation, leaves were washed with distilled water (Hull, 2009). To inoculate the tomato plants, infected N. benthamiana leaves were collected and inocula were prepared as mentioned above. Tomato plants with two fully expanded leaves were used for viral inoculation. Two pairs of cotyledons were inoculated; inoculated and non-inoculated control plants (mock) were grown in a growth chamber (16 h light and 8 h night under white fluorescent light). To ensure viral infection, plants were reinoculated 7 days after the first inoculation.
After viral inoculation, plants were monitored regularly for the appearance of symptoms. Leaf tissue was tested for the presence of virus using double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA), according to the manufacturer’s instructions (Agdia, Inc. Elkhart, IN, USA). Virus accumulation was tested at 7/20 days post inoculation (DPI). DAS-ELISA was performed to detect the accumulation of the coat protein (CP) of TEV or PepMoV. Three replicates of inoculated and upper non-inoculated leaves of E1 lines were used for ELISA analysis. Absorbance of samples at 405 nm was measured using a microplate reader (Biotek, VT, USA). The statistical significance of the data was performed with Student’s t-test using Microsoft Excel 2016 (Microsoft, Redmond, WA, USA).
To develop tomato plants with edited PDS and eIF4E1 genes, we delivered the pHSE401-PDS and pHSE401-eIF4E1 CRISPR/Cas9 constructs harboring the respective sgRNAs into S. lycopersicum cv. Micro-Tom via Agrobacterium transformation. We transferred putatively genome-edited shoots regenerated from the callus to shoot elongation medium and allowed them to elongate, and then cut elongated shoots and transferred them to rooting medium to encourage root formation. We observed the photobleached phenotype in four out of 113 PDS-edited explants transformed with the pHSE401-PDS construct (Figure 2A), demonstrating about 3.5% gene-editing efficiency of the PDS gene with both copies expected to be edited. However, the PDS-edited, photobleached shoots failed to develop into rooted shoots (Figure 2A). After sampling the photobleached shoots of the PDS-edited explants and putative eIF4E1-edited plants, we confirmed the integration of the transfer DNA (T-DNA) by genomic DNA PCR of the samples using Cas9- and Hpt-specific primers (Supplementary Figure S1A). We obtained PCR products with the expected amplicon sizes from the photobleached shoots, demonstrating the successful integration of Cas9 and Hpt (Supplementary Figure S1A).
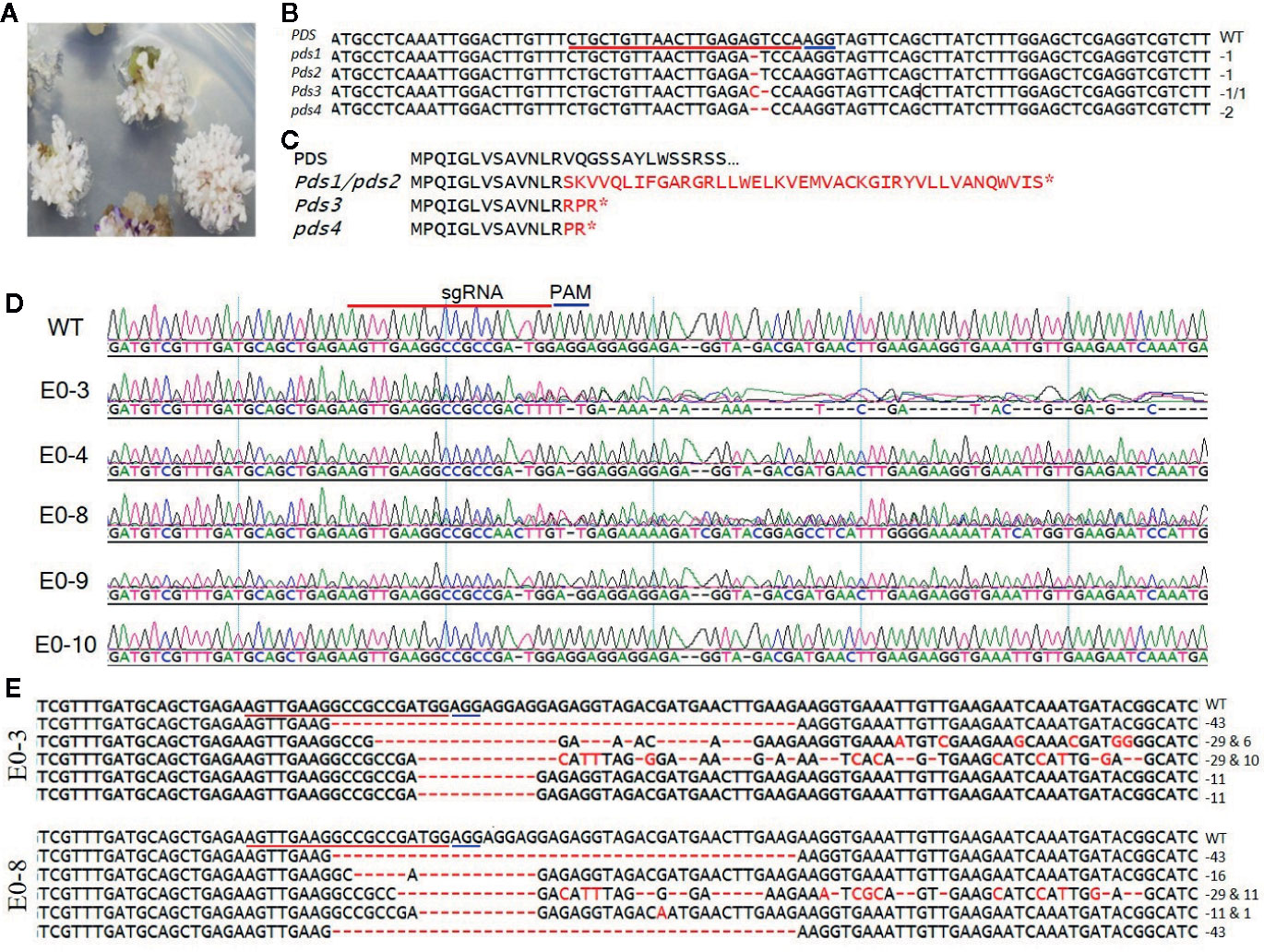
Figure 2 Sequence analysis of the PDS- and eIF4E1-targeted plants. (A) PDS-targeted explants showing the photobleached phenotype. (B) Mutation detection in PDS-targeted shoots by direct sequencing of PCR product in target region. (C) Predicted amino acid sequence alignment of PDS genes from genome-edited plants. Mutations causing early stop codons or changes in protein sequence are indicated with star symbol. (D) Representative chromatograms produced by direct sequencing of PCR products, displayed for sequence alignment of the mutated eIF4E1 genes from the E0-3, E0-4, E0-8, E0-9, and E0-10 transgenic line. All transgenic lines with exception of the E0-10 showed mixed peaks in the target region due to their varied indels as compared with the wild-type sequence (WT) and are regarded as CRISPR/Cas9-induced mutant lines. (E) DNA sequences of mutated eIF4E1 in the E0-3 and E0-8 line. The number of mutations of each type revealed by random sequencing of TA clones of PCR products. DNA deletions are denoted by red dashes, and deletion sizes (in nucleotides) are marked on the right side of the sequence. The sgRNA target sequence is underlined in red and the PAM motif in blue.
We also regenerated 22 putative eIF4E1-edited transgenic plants, which we tested for the presence of the transgenes, the Cas9 and Hpt genes by PCR using transgene-specific primers (Supplementary Figure S1B). Sixteen of the 22 plants contained both Cas9 and Hpt, with a transformation efficiency of 72%. Furthermore, RT-PCR results confirmed the expression of the Cas9 gene in all the PCR positive plants (Supplementary Figure S1C). No morphological changes were observed in eIF4E1-edited plants.
To confirm that the CRISPR/Cas9 editing had introduced mutations in the PDS gene, we performed PCR amplification of the target region from the albino tomato mutants using primers (Table 1) that flanked the sgRNA target, and then sequenced the PCR products by Sanger sequencing. Sequence analysis showed two types of sequence variations: one- and two-nucleotide deletions with breakpoints 3 bp upstream of the protospacer-adjacent motif (PAM) sequence (Figure 2B). These deletions resulted in frameshift mutations causing early stop codons, preventing the expression of functional PDS protein (Figure 2C). Similarly, we identified CRISPR/Cas9-induced mutations in eIF4E1 by sequencing the PCR amplicons of the sgRNA target-flanking region from the genomic DNA of 16 Cas9-positive tomato lines. Sequencing of the sgRNA target region revealed no mutant homozygous lines among the E0 plants; 15 of the 16 putative transgenic plants showed mixed sequence peaks at the sgRNA target site compared to the wild-type target sequence (Figure 2D), indicating that a mixture of mutant (reflective of CRISPR-Cas9-induced mutation) and wild-type alleles of eIF4E1 were present. A single plant, E0-10 (Figure 2D) showed sequence peaks similar to that of wild-type target sequence, indicating no editing the in target region despite showing expression of the Cas9 gene. To further characterize the mutations in the E0 plants, we randomly selected the E0-3 and E0-8 transgenic lines for further investigation by TA cloning and sequencing of the target region. The sequencing results for the TA clones from E0-3 and E0-8 revealed four different alleles of eIF4E1 (Figure 2E), with indels and/or substitutions at various positions both in proximity to the sgRNA target and outside the immediate sgRNA target region. The presence of a mixture of different mutant alleles in each E0 line suggested that active somatic mutation was occurring in the edited plants. Therefore, we advanced selected E0 plants to the E1 and E2 generations to create homozygous mutant lines.
Among the eIF4E1 mutants, we selected the E0-3 and E0-8 plants from which to generate E1 lines. First, we allowed the E0-3 and E0-8 plants to self-pollinate to produce E1 lines. We then extracted gDNA from leaf samples of the E1 progeny and tested these for the presence of transgene by PCR with transgene-specific primers. Among 19 plants derived from E0-3, 13 carried transgenes (1, 2, 3, 4, 9, 10, 12, 13, 15, 16, 17, 18, and 19) and 6 did not (5, 6, 7, 8, 11, and 14), and among seven plants from the E0-8 plant, five carried transgenes (3, 4, 5, 6, and 7) and two did not (1 and 2) (Figure 3), indicative of the transgene-heterozygous status of the E0 plants.
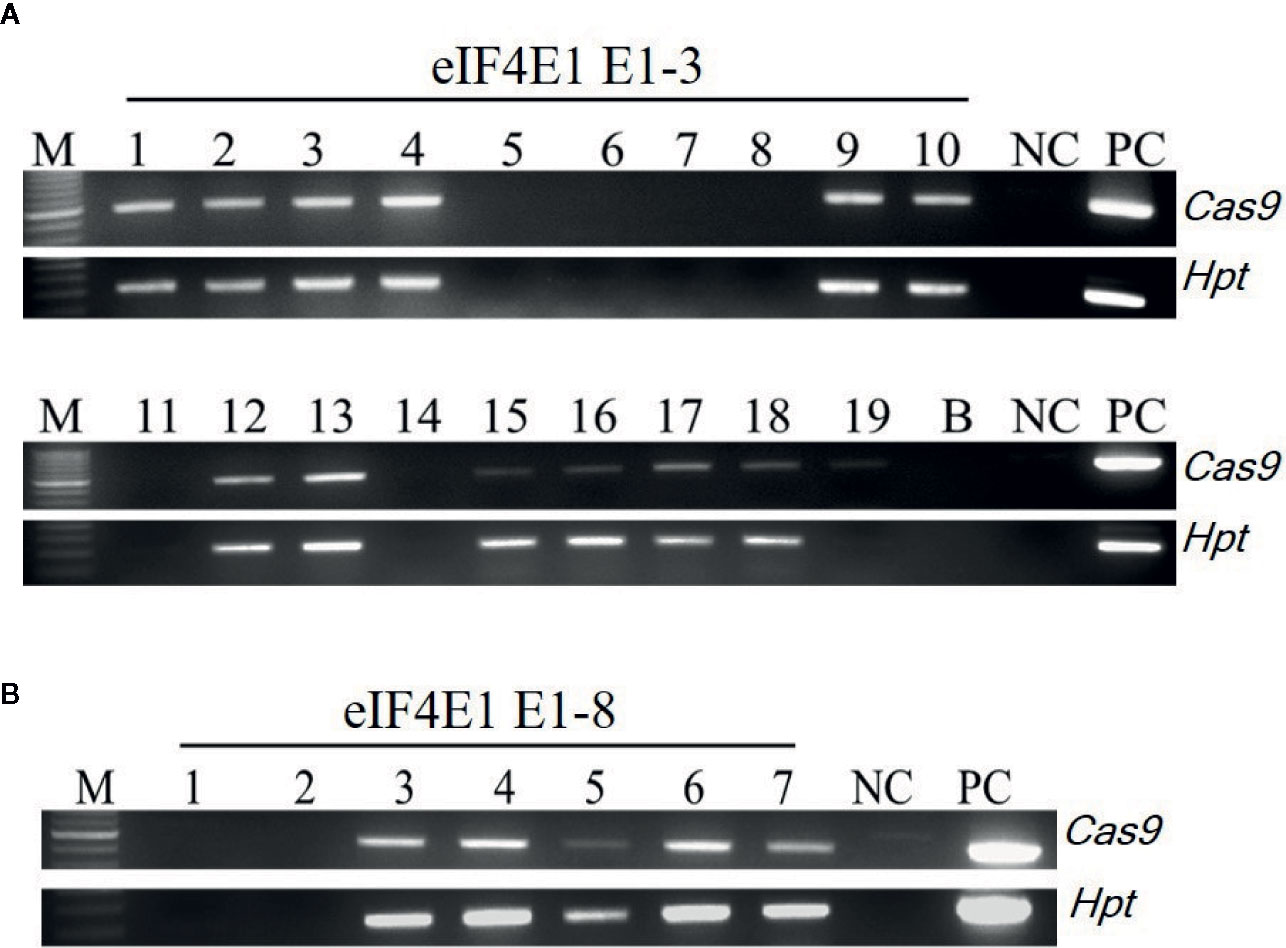
Figure 3 Transgene screening of eIF4E1-edited E1 progeny. (A, B) Transgene screening of eIF4E1-targeted E1 progeny of E0-3 (A; 19 progeny) and E0-8 (B; 7 progeny) by genomic DNA PCR. NC, wild type (negative control); PC, Cas9 plasmid (positive control), Hpt, Hygromycin phosphotransferase marker gene.
To reveal sequence variations of E1 plants, we performed TA cloning and sequencing of amplicons of the target region in E1 plants derived from lines E0-3 and E0-8. New mutation patterns were evident in the E1 lines as compared to the E0 plant, and two lines (E1 3-8 and E1 3-17) derived from E0-3 were homozygous for the expected 43-bp deletion (Figure 4A). The E1 3-11 line was biallelic, carrying two deletions of 43 and 11 bp; two other lines (E1 3-9 and 3-15) showed a 29-bp deletion and a tandem insertion of 38 bp of adjacent repeat sequence, respectively (Figure 4C); and the E1 3-19 line carried mixed mutations, with deletions of the unexpected sizes of 12, 13, and 15 bp (Figure 4A). The three lines (E1 8-3, 8-5, 8-7) derived from the E0-8 plant were homozygous for the expected 11-bp, 43-bp, and 29-bp deletions and 38-bp insertion of a repeat sequence (Figures 4A, C). The E1 8-1 line was biallelic, with a 43-bp deletion and a 29-bp deletion and 38-bp repeat-sequence insertion, whereas the E1 8-4 line had mixed mutations along with a wild-type allele (Figure 4B). The presence of new mutation patterns in E1 lines that differed from those in the E0 plants suggested that active somatic mutation was occurring in the E0 plants (Figures 2E and 4). Notably, one line, E1 3-8, had a homozygous edited eIF4E1 gene but no T-DNA. Overall, several homozygous and biallelic lines were recovered in the E1 generation (Table 2), along with a smaller proportion of mosaic mutants carrying different allelic variants (Table 2).
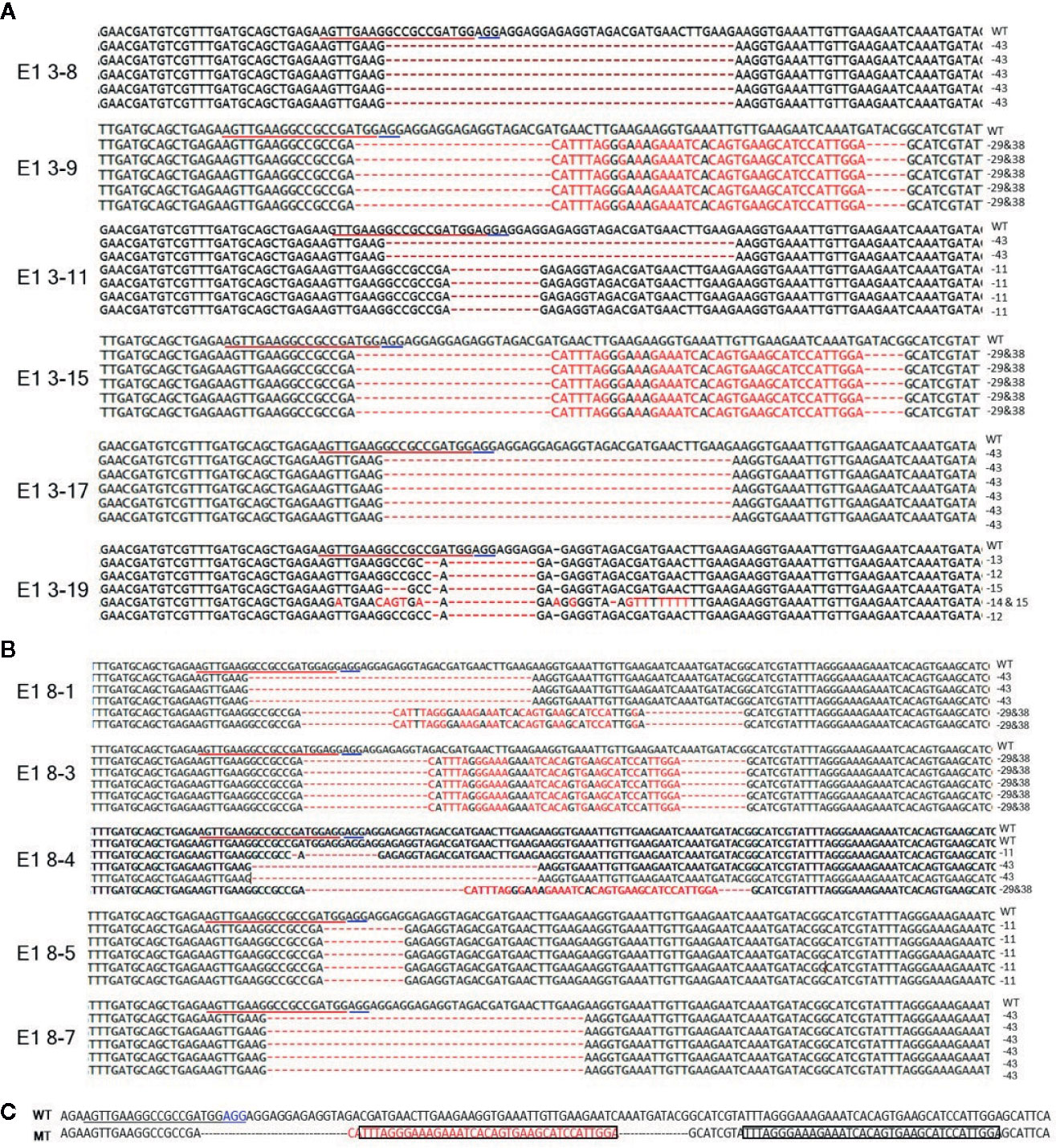
Figure 4 Genome-edited sequences of eIF4E1 from E1 transgenic lines. (A, B) DNA sequences of the eIF4E1 mutant lines derived from E0-3 (A) and E0-8 (B). (C) Insertion of a 38-bp repeat sequence (boxed) from sgRNA adjacent region. The sgRNA target sequence is underlined in red and the PAM motif in blue. WT, wild type; MT, mutant lines (E1 3-9, E1 3-15, E1 8-1, and E1 8-3).
To investigate whether these CRISPR/Cas9-derived eIF4E1 mutants conferred resistance to TEV, we inoculated E1 3-8 (homozygous line) and E1 3-11 (biallelic line) plants with TEV (Figure 5). TEV symptoms appeared as early as 7 DPI; the wild-type plant showed typical TEV symptoms, including vein clearing and several small chlorotic spots in the leaves. Both eIF4E1 mutant lines showed similar symptoms (Figure 5A). To confirm virus infection, we performed DAS-ELISA analysis using inoculated lower and uninoculated upper leaves. We detected similar high amounts of virus coat protein accumulation in the E1 3-8 and E1 3-11 lines, indicating that the eIF4E1-edited lines were susceptible to TEV-HAT (Figure 5B). We next assessed whether CRISPR/Cas9 eIF4E1 mutants could confer resistance to another potyvirus, PepMoV, by challenging three E1 mutant lines (E1 3-8, E1 3-11, and E1 3-15) with PepMoV. PepMoV coat protein accumulated to a high level in both the inoculated and uninoculated upper leaves of the susceptible wild-type control (Figure 5), whereas no coat protein accumulated in any of the three mutant lines in either the inoculated lower or uninoculated systemic leaves, confirming that the eIF4E1-edited lines were resistant to PepMoV (Figure 5C).
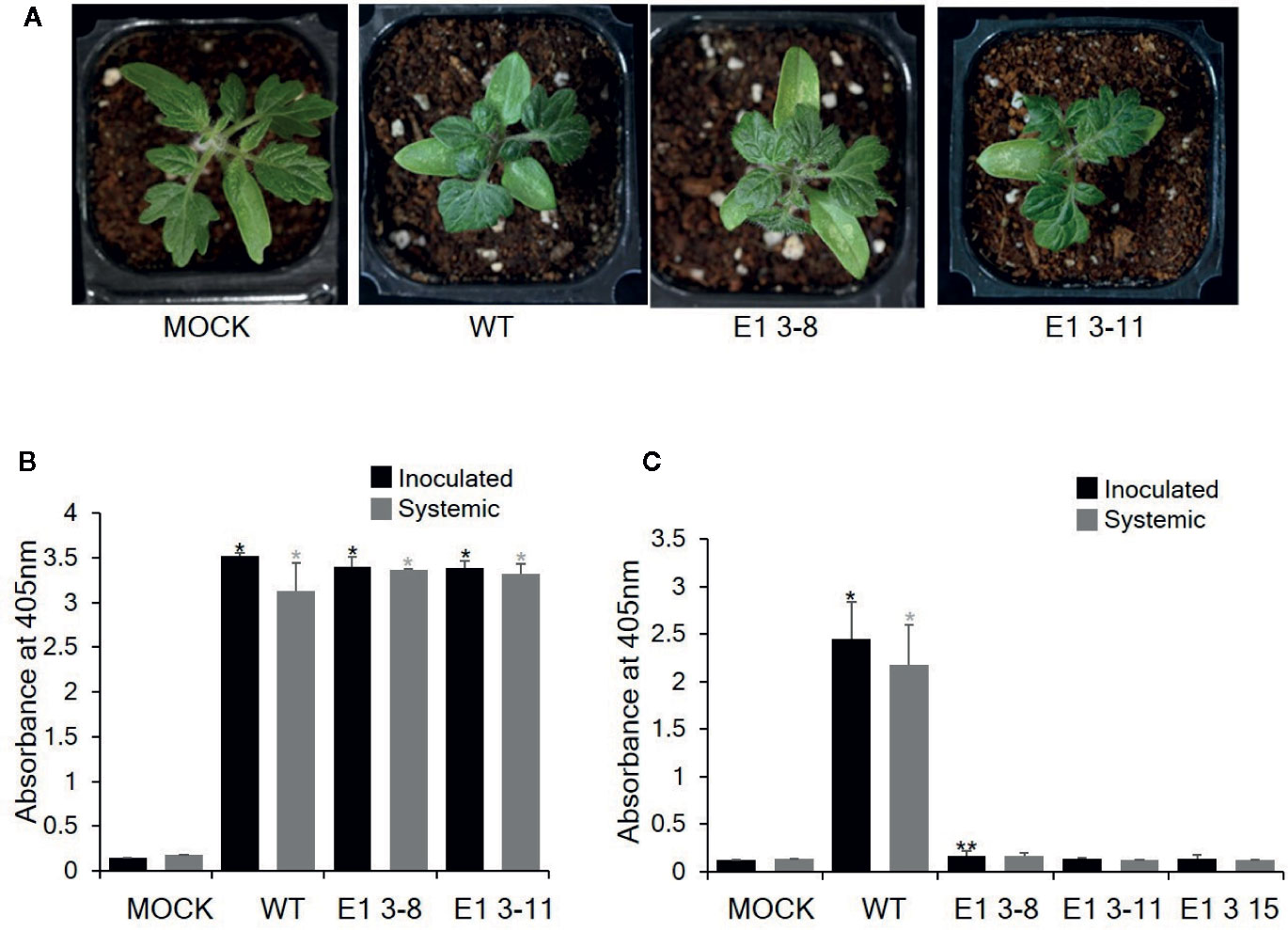
Figure 5 Resistance analysis of eIF4E1 mutant lines challenged with potyviruses. (A) Phenotypes of eIF4E1 mutant lines infected with TEV at 7 DPI. (B, C) Analysis of the accumulation of TEV-HAT (B) and PepMoV (C) in inoculated and systemic leaves by DAS-ELISA. Error bars indicate mean values of replicates ± SD. Asterisks indicate significant differences (*P < 0.01; **P < 0.05) between ‘mock′ and ‘treatments′ according to Student’s t‐test.
Here we subjected the eIF4E1 gene to CRISPR/Cas9 gene editing with the aim of creating potyvirus resistance in S. lycopersicum Micro-Tom, as recessive resistance genes are considered to be confer more durable resistance than dominant resistance (R) genes (Kang et al., 2005a; Borrelli et al., 2018). In this study, we developed CRISPR/Cas9-edited tomato PDS and eIF4E1 mutants through Agrobacterium-mediated delivery of CRISPR/Cas9 and sgRNA reagents. Sequencing of the PDS gene target region revealed that DNA double-strand edits occurred 3 bp upstream of the PAM site, resulting in indels of one or two nucleotides in PDS, as reported previously (Garneau et al., 2010; Jinek et al., 2012). All the indel mutations led to early stop codons, causing loss of PDS gene function. In the eIF4E-edited lines, indels occurred at various positions near the sgRNA target and in some cases extending beyond the sgRNA target and PAM region, creating premature stop codons and truncated eIF4E1 proteins (Supplementary Data File 1). Notably, we observed a 38-bp insertion of a repeat sequence as well as a 29-bp deletion in several of the gene-edited lines, E1 3-9, E1 3-15, E1 8-1, and E1 8-3; the mechanism leading to the insertion of this adjacent repeat sequence will require further investigation.
Unlike PDS-targeted photobleached plants, the eIF4E1-targeted mutant plants in the E0 generation were chimeric, with mixtures of different allelic variations, and the E1 generation comprised a mixture of homozygous, biallelic, and mosaic mutants. Increasing evidence suggests that transgenic plants can be chimeric in the E0 generation. For example, chimeric lines have also been reported in T0 (E0) genome-edited Arabidopsis, tomato, rice, and soybean (Brooks et al., 2014; Feng et al., 2014; Ma et al., 2015; Pan et al., 2016; Pyott et al., 2016; Liu et al., 2020). One study identified only three non-mosaic mutants among nearly 300 lines of Arabidopsis created by editing of the gene RRP42 (Yan et al., 2017). The mosaicism of the E1 mutants could then be due to the fact that in some edited lines, one or more wild-type alleles might escape detection in the E0 generation but then be detected in later generations (Brooks et al., 2014; Zhang et al., 2014; Zhang et al., 2019; Liu et al., 2020). Furthermore, other new alleles created from late-arising chimeric tissues might also be overlooked in E0 plants, resulting in different flowers carrying different alleles (Brooks et al., 2014). In agreement with this contention, in the present study, some of the E1 plants produced by gene editing of eIF4E1 were chimeric, and furthermore mutations different from those identified in E0 plants were observed in E1 plants.
Notwithstanding these complications, we advanced the E0 plants carrying diverse alleles of the target gene to the next generation, and selected E1 lines carrying homozygous alleles from segregating populations, as reported in previous studies (Xu et al., 2015; Chandrasekaran et al., 2016; Pyott et al., 2016). Accordingly, sequence analysis of the E1 lines identified both homozygous and biallelic eIF4E1 mutants, both of which would be expected to stably pass on their mutant status to their descendants (Chandrasekaran et al., 2016), consistent with this, homozygous mutant alleles were stably inherited in the E2 progeny.
Previous studies in pepper suggest that eIF4E is a key factor in the interaction between viruses and their hosts. Mutations in eIF4E cause conformational shifts in the encoded proteins, interrupting the interaction between VPg and eIF4E and conferring plant resistance to the virus at the cellular level (Kang et al., 2005a). This implied that site-directed mutagenesis could be used to create dominant negative mutations or targeted silencing of host genes (Azevedo et al., 2002; Kang et al., 2005b; Kang et al., 2007). We therefore assessed our eIF4E-edited E1 lines, with or without the Cas9 transgene, for resistance to potyviruses. Edited plants carrying mutations in eIF4E1 showed resistant to PepMoV. Consistent with this, we did not detect accumulation of coat protein in either inoculated or uninoculated leaves. However, E1 plants carrying mutations in eIF4E1 showed typical TEV symptoms, and ELISA analysis confirmed the presence of systemic infection. Viral coat protein accumulated to high levels in both inoculated and uninoculated upper leaves of the homozygous E1 3-8 and biallelic E1 3-11 lines. These results contrast with the findings of an earlier study indicating that transgenic tomato plants overexpressing a recessive resistance allele of eIF4E (pvr1) from Capsicum chinense showed dominant resistance to TEV (Kang et al., 2007). The transgenic expression of the Capsicum eIF4E mutant allele was suggested to perturb the interactions required for viral susceptibility in a heterologous host system (Kang et al., 2007). However, the susceptibility of the edited plants to TEV in tomato that we observed here could be due to the redundant activities of eIF4E homologs (eIF4E1 and eIF4E2) coupled with the absence of a dominant negative allele whose protein product could interfere with the endogenous wild‐type proteins and inhibit viral infection (Kang et al., 2007). Furthermore, an eIF4E1-knockout tomato plant selected from a TILLING population was reported to be susceptible to potyviruses because TEV could utilize either eIF4E1 or eIF4E2 for its replication in tomato (Gauffier et al., 2016). Thus, development of a TEV virus-resistant tomato line is hindered by the gene redundancy of eIF4E homologs. eIF4E1 and eIF4E2 double-knockout plants would be expected to show a broad-spectrum resistance to a wide range of potyviruses (Gauffier et al., 2016); however, care should be taken to ensure that mutations in both candidate genes do not impair the plant growth and development, as often occurs with double mutants (Mazier et al., 2011; Gauffier et al., 2016).
We used CRISPR/Cas9 gene editing to induce mutations in the tomato eIF4E1 gene by A. tumefaciens-mediated transformation. Evaluation of the mutation and inheritance patterns of this gene in the E0 and later generations by sequencing revealed high proportions of chimeric mutant lines in the E0 generation and of homozygous and biallelic mutants of eIF4E1 in the E1 generation. The CRISPR/Cas9-mediated gene mutations were stably transmitted to the E1 and E2 descendants irrespective of the presence or absence of T-DNA. Although the presence of redundant eIF4E homologs apparently precluded TEV resistance in the eIF4E1 genome-edited Micro-Tom plants, an edited eIF4E1 with an early stop codon conferred resistance to PepMoV. Thus, our system enabled successful CRISPR/Cas9 editing of tomato eIF4E1, resulting in resistance to a common viral pathogen of Solanaceae; further research is planned to obtain homozygous E0 plants and to extend this work to pepper (Capsicum species) to produce strains with broad-spectrum virus resistance.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Y-JY and JV designed the constructs and performed the transformation. Y-JY, D-SK, JK, and H-EL characterized the transgenics and identified the mutants. Y-JY, J-HL and JV performed viral inoculations and ELISA. JV wrote the manuscript. B-CK, D-SK, JK, H-EL, and JV revised the manuscript. B-CK conceived and designed the research. All authors contributed to the article and approved the submitted version.
This work was carried out with the support of the Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01327801, PJ01327803), Rural Development Administration, Republic of Korea.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01098/full#supplementary-material
Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. doi: 10.1126/science.1067554
Borrelli, V. M., Brambilla, V., Rogowsky, P., Marocco, A., Lanubile, A. (2018). The enhancement of plant disease resistance using CRISPR/Cas9 technology. Front. Plant Sci. 9:1245.S:1245. doi: 10.3389/fpls.2018.01245.S
Brooks, C., Nekrasov, V., Lippman, Z. B., Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Charron, C., Nicolai, M., Gallois, J. L., Robaglia, C., Moury, B., Palloix, A., et al. (2008). Natural variation and functional analyses provide evidence for co-evolution between plant eIF4E and potyviral VPg. Plant J. 54, 56–68. doi: 10.1111/j.1365-313X.2008.03407.x
Cong, L., Zhang, F. (2015). Genome engineering using CRISPR-Cas9 system. Methods Mol. Biol. 1239, 197–217. doi: 10.1007/978-1-4939-1862-1_10
Cui, H., Wang, A. (2019). The biological impact of the hypervariable N-terminal region of potyviral genomes. Annu. Rev. Virol. 6, 255–274. doi: 10.1146/annurev-virology-092818-015843
de Oliveira, L. C., Volpon, L., Rahardjo, A. K., Osborne, M. J., Culjkovic-Kraljacic, B., Trahan, C., et al. (2019). Structural studies of the eIF4E–VPg complex reveal a direct competition for capped RNA: Implications for translation. Proc. Natl. Acad. Sci. U. S. A. 116, 24056–24065. doi: 10.1073/pnas.1904752116
Duprat, A., Caranta, C., Revers, F., Menand, B., Browning, K. S., Robaglia, C. (2002). The Arabidopsis eukaryotic initiation factor (iso) 4E is dispensable for plant growth but required for susceptibility to potyviruses. Plant J. 32, 927–934. doi: 10.1046/j.1365-313x.2002.01481.x
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Garneau, J. E., Dupuis, M.È., Villion, M., Romero, D. A., Barrangou, R., Boyaval, P., et al. (2010). The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67. doi: 10.1038/nature09523
Gauffier, C., Lebaron, C., Moretti, A., Constant, C., Moquet, F., Bonnet, G., et al. (2016). A TILLING approach to generate broad-spectrum resistance to potyviruses in tomato is hampered by eIF4E gene redundancy. Plant J. 85, 717–729. doi: 10.1111/tpj.13136
Gomez, M. A., Lin, Z. D., Moll, T., Chauhan, R. D., Hayden, L., Renninger, K., et al. (2019). Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 17, 421–434. doi: 10.1111/pbi.12987
Hančinský, R., Mihálik, D., Mrkvová, M., Candresse, T., Glasa, M. (2020). Plant viruses infecting Solanaceae family members in the cultivated and wild environments: A Review. Plants 9:667. doi: 10.3390/plants9050667
Hsu, P. D., Lander, E. S., Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Hull, R. (2009). Mechanical inoculation of plant viruses. Curr. Protoc. Microbiol. 13, 16B–166. doi: 10.1002/9780471729259.mc16b06s13
Hwang, J., Li, J., Liu, W. Y., An, S. J., Cho, H., Her, N. H., et al. (2009). Double mutations in eIF4E and eIFiso4E confer recessive resistance to Chilli veinal mottle virus in pepper. Mol. Cells 27, 329–336. doi: 10.1007/s10059-009-0042-y
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188–e188. doi: 10.1093/nar/gkt780
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Kang, B. C., Yeam, I., Jahn, M. M. (2005a). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. doi: 10.1146/annurev.phyto.43.011205.141140
Kang, B. C., Yeam, I., Frantz, J. D., Murphy, J. F., Jahn, M. M. (2005b). The pvr1 locus in Capsicum encodes a translation initiation factor eIF4E that interacts with Tobacco etch virusTobacco etch virus VPg. Plant J. 42, 392–405. doi: 10.1111/j.1365-313X.2005.02381.x
Kang, B. C., Yeam, I., Li, H., Perez, K. W., Jahn, M. M. (2007). Ectopic expression of a recessive resistance gene generates dominant potyvirus resistance in plants. Plant Biotechnol. J. 5, 526–536. doi: 10.1111/j.1467-7652.2007.00262.x
Kim, J., Kang, W. H., Yang, H. B., Park, S., Jang, C. S., Yu, H. J., et al. (2013). Identification of a broad-spectrum recessive gene in Brassica rapa and molecular analysis of the eIF4E gene family to develop molecular markers. Mol. Breed. 32, 385–398. doi: 10.1111/mpp.12120
Kim, J., Kang, W. H., Hwang, J., Yang, H. B., Dosun, K., Oh, C. S., et al. (2014). Transgenic Brassica rapa plants over-expressing eIF(iso)4E variants show broad-spectrum Turnip mosaic virus (TuMV) resistance. Mol. Plant Pathol. 15, 615–626. doi: 10.1111/mpp.12120
Kothari, S. L., Joshi, A., Kachhwaha, S., OchoaAlejo, N. (2010). Chilli peppers-a review on tissue culture and transgenesis. Biotechnol. Adv. 28, 35–48. doi: 10.1016/j.biotechadv.2009.08.005
Le Gall, O., Aranda, M. A., Caranta, C. (2011). “Plant resistance to viruses mediated by translation initiation factors,” in The Recent Advances in Plant Virology. Eds. Caranta, C., Aranda, M. A., Tepfer, M., López-Moya, J. (Wymondham, Norfolk: Caister Academic Press), 177–194.
Lebaron, C., Rosado, A., Sauvage, C., Gauffier, C., German-Retana, S., Moury, B., et al. (2016). A new eIF4E1 allele characterized by RNAseq data mining is associated with resistance to PVY in tomato albeit with a low durability. J. Gen. Virol. 97, 3063–3072. doi: 10.1099/jgv.0.000609
Lee, H. R., An, H. J., You, Y. G., Lee, J., Kim, H. J., Kang, B. C., et al. (2013). Development of a novel codominant molecular marker for Chili veinal mottle virus resistance in Capsicum annuum L. Euphytica 193, 197–205. doi: 10.1007/s10681-013-0897-z
Lellis, A. D., Kasschau, K. D., Whitham, S. A., Carrington, J. C. (2002). Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF (iso) 4E during potyvirus infection. Curr. Biol. 12, 1046–1051. doi: 10.1016/s0960-9822(02)00898-9
Liu, H., Wang, K., Jia, Z., Gong, Q., Lin, Z., Du, L., et al. (2020). Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J. Exp. Bot. 71, 1337–1349. doi: 10.1093/jxb/erz529
Ma, X., Zhang, Q., Zhu, Q., Liu, W., Chen, Y., Qiu, R., et al. (2015). A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 8, 1274–1284. doi: 10.1016/j.molp.2015.04.007
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Cermak, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Mazier, M., Flamain, F., Nicolaï, M., Sarnette, V., Caranta, C. (2011). ). Knock-down of both eIF4E1 and eIF4E2 genes confers broad-spectrum resistance against potyviruses in tomato. PLoS One 6, e29595. doi: 10.1371/journal.pone.0029595
Melzer, M. J., Sugano, J. S., Cabanas, D., Dey, K. K., Kandouh, B., Mauro, D., et al. (2012). First report of Pepper mottle virus infecting tomato in Hawaii. Plant Dis. 96, 917. doi: 10.1094/PDIS-02-12-0147-PDN
Miyoshi, H., Suehiro, N., Tomoo, K., Muto, S., Takahashi, T., Tsukamoto, T., et al. (2006). Binding analyses for the interaction between plant virus genome-linked protein (VPg) and plant translational initiation factors. Biochimie 88, 329–340. doi: 10.1016/j.biochi.2005.09.002
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant 5, 473–479. . doi: 10.1111/j.1399-3054.1962.tb08052.x
Nicaise, V. (2014). Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5:660:660. doi: 10.3389/fpls.2014.00660
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6:24765. doi: 10.1038/srep24765
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Porebski, S., Bailey, L. G., Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15, 8–15. doi: 10.1007/BF02772108
Pyott, D. E., Sheehan, E., Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 17, 1276–1288. doi: 10.1111/mpp.12417
Revers, F., García, J. A. (2015). Molecular biology of potyviruses. Adv. Virus Res. 92, 101–199. doi: 10.1016/bs.aivir.2014.11.006
Robaglia, C., Caranta, C. (2006). Translation initiation factors: a weak link in plant RNA virus infection. Trends Plant Sci. 11, 40–45. doi: 10.1016/j.tplants.2005.11.004
Rodríguez-Hernández, A. M., Gosalvez, B., Sempere, R. N., Burgos, L., Aranda, M. A., Truniger, V. (2012). Melon RNA interference (RNAi) lines silenced for Cm-eIF4E show broad virus resistance. Mol. Plant Pathol. 13, 755–763. doi: 10.1111/j.1364-3703.2012.00785.x
Ruffel, S., Gallois, J. L., Lesage, M. L., Caranta, C. (2005). The recessive potyvirus resistance gene pot-1 is the tomato orthologue of the pepper pvr2-eIF4E gene. Mol. Genet. Genomics 274, 346–353. doi: 10.1007/s00438-005-0003-x
Ruffel, S., Gallois, J. L., Moury, B., Robaglia, C., Palloix, A., Caranta, C. (2006). Simultaneous mutations in translation initiation factors eIF4E and eIF(iso)4E are required to prevent pepper veinal mottle virus infection of pepper. J. Gen. Virol. 87, 2089–2098. doi: 10.1099/vir.0.81817-0
Sanfacon, H. (2015). Plant translation factors and virus resistance. Viruses 7, 3392–3419. doi: 10.3390/v7072778
Sato, M., Nakahara, K., Yoshii, M., Ishikawa, M., Uyeda, I. (2005). Selective involvement of members of the eukaryotic initiation factor 4E family in the infection of Arabidopsis thaliana by potyviruses. FEBS Lett. 579, 1167–1171. doi: 10.1016/j.febslet.2004.12.086
Shapiro, R. S., Chavez, A., Collins, J. J. (2018). CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat. Rev. Microbiol. 16, 333–339. doi: 10.1038/s41579-018-0002-7
Tavert-Roudet, G., Anne, A., Barra, A., Chovin, A., Demaille, C., Michon, T. (2017). The Potyvirus particle recruits the plant translation initiation factor eIF4E by means of the VPg covalently linked to the viral RNA. Mol. Plant Microbe Interact. 30, 754–762. doi: 10.1094/MPMI-04-17-0091-R
Wang, A., Krishnaswamy, S. (2012). Eukaryotic translation initiation factor 4E-mediated recessive resistance to plant viruses and its utility in crop improvement. Mol. Plant Pathol. 13, 795–803. doi: 10.1111/j.1364-3703.2012.00791.x
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS One 11, e0154027. doi: 10.1371/journal.pone.0154027
Wang, A. (2015). Dissecting the molecular network of virus-plant interactions: the complex roles of host factors. Annu. Rev. Phytopathol. 253, 45–66. doi: 10.1146/annurev-phyto-080614-120001
Ward, C. W., Shukla, D. D. (1991). Taxonomy of potyviruses: current problems and some solutions. Intervirology 32, 269–296. doi: 10.1159/000150211
Wintermantel, W. M. (2011). A comparison of disinfectants to prevent spread of potyviruses in greenhouse tomato production. Plant Health Prog. 12, 19. doi: 10.1094/PHP-2011-0221-01-RS
Wu, Y., Liang, D., Wang, Y., Bai, M., Tan, W., Bao, S., et al. (2013). Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell 13, 659–662. doi: 10.1016/j.stem.2013.10.016
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14:327. doi: 10.1186/s12870-014-0327-y
Xu, R. F., Li, H., Qin, R. Y., Li, J., Qiu, C. H., Yang, Y. C., et al. (2015). Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 5:11491. doi: 10.1038/srep11491
Yan, X., Yan, Z., Han, Y. (2017). RRP42, a subunit of exosome, plays an important role in female gametophytes development and mesophyll cell morphogenesis in Arabidopsis. Front. Plant Sci. 8:981:981. doi: 10.3389/fpls.2017.00981
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhang, Z., Zhang, Y., Gao, F., Han, S., Cheah, K. S., Tse, H. F., et al. (2017). CRISPR/Cas9 genome-editing system in human stem cells: current status and future prospects. Mol. Ther. Nucleic Acids 9, 230–241. doi: 10.1016/j.omtn.2017.09.009
Zhang, S., Zhang, R., Gao, J., Gu, T., Song, G., Li, W., et al. (2019). Highly efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumefaciens-mediated CRISPR/Cas9 system. Int. J. Mol. Sci. 20, 4257. doi: 10.3390/ijms20174257
Keywords: CRISPR/Cas9, eukaryotic translation initiation factor 4E (eIF4E), genome editing, potyvirus, phytoene desaturase (PDS), Solanum lycopersicum cv. Micro-Tom
Citation: Yoon Y-J, Venkatesh J, Lee J-H, Kim J, Lee H-E, Kim D-S and Kang B-C (2020) Genome Editing of eIF4E1 in Tomato Confers Resistance to Pepper Mottle Virus. Front. Plant Sci. 11:1098. doi: 10.3389/fpls.2020.01098
Received: 21 May 2020; Accepted: 03 July 2020;
Published: 24 July 2020.
Edited by:
Henryk Hanokh Czosnek, Hebrew University of Jerusalem, IsraelReviewed by:
Aiming Wang, Agriculture and Agri-Food Canada (AAFC), CanadaCopyright © 2020 Yoon, Venkatesh, Lee, Kim, Lee, Kim and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byoung-Cheorl Kang, Yms1NEBzbnUuYWMua3I=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.