
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 09 July 2020
Sec. Plant Nutrition
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.01016
This article is part of the Research Topic Heavy Metal Toxicity in Plants: Recent Insights on Physiological and Molecular Aspects View all 14 articles
 Hanmei Du1†
Hanmei Du1† Ying Huang1†
Ying Huang1† Min Qu1
Min Qu1 Yihong Li1
Yihong Li1 Xiaoqi Hu1
Xiaoqi Hu1 Wei Yang1
Wei Yang1 Hongjie Li1
Hongjie Li1 Wenzhu He2
Wenzhu He2 Jianzhou Ding1
Jianzhou Ding1 Chan Liu1
Chan Liu1 Shibin Gao1
Shibin Gao1 Moju Cao1
Moju Cao1 Yanli Lu1
Yanli Lu1 Suzhi Zhang1*
Suzhi Zhang1*Aluminum (Al) toxicity is the primary limiting factor that affects crop yields in acid soil. However, the genes that contribute to the Al tolerance process in maize are still poorly understood. Previous studies have predicted that ZmAT6 is a novel protein which could be upregulated under Al stress condition. Here, we found that ZmAT6 is expressed in many tissues and organs and can be dramatically induced by Al in both the roots and shoots but particularly in the shoots. The overexpression of ZmAT6 in maize and Arabidopsis plants increased their root growth and reduced the accumulation of Al, suggesting the contribution of ZmAT6 to Al tolerance. Moreover, the ZmAT6 transgenic maize plants had lower contents of malondialdehyde and reactive oxygen species (ROS), but much higher proline content and even lower Evans blue absorption in the roots compared with the wild type. Furthermore, the activity of several enzymes of the antioxidant system, such as peroxidase (POD), superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX), increased in ZmAT6 transgenic maize plants, particularly SOD. Consistently, the expression of ZmSOD in transgenic maize was predominant upregulated by Al stress. Taken together, these findings revealed that ZmAT6 could at least partially confer enhanced tolerance to Al toxicity by scavenging ROS in maize.
Acidic soils refer to soil with pH ≤ 5.5, occupying almost 30% of the arable soil and 50% of the potential cultivated land (von Uexküll and Mutert, 1995). Aluminum (Al), the third abundant element in the earth’s crust, can be converted into soluble and toxic Al3+ which is the major limiting factor for plants’ growth in acidic soil (Kochian et al., 2004). Aluminum toxicity could rapidly inhibit the elongation of the root system even in a micromolar concentration, then affect the absorption of water and nutrients, and eventually resulting in the decline of crop yield (Kinraide, 1990; Ma and Furukawa, 2003; Kochian et al., 2004). Improving the physiological and genetic tolerance to Al in crops has long been a challenging problem for researchers.
To cope with Al stress, plants have evolved a series of aluminum resistant mechanisms. The primary Al-tolerance mechanisms in plants refer to the exclusion tolerance and internal tolerance (Kochian, 1995; Ma et al., 1997; Ma, 2000; Daspute et al., 2017). The common feature of the Al exclusion mechanisms is to prevent Al from entering the root apex by the excretion of detoxified organic acids (OAs) and Pi ligands to the apoplast or rhizosphere. The internal tolerance mechanism refers to the sequestration of Al into vacuoles and its detoxification by chelation (Ma, 2000; Zheng and Yang, 2005). Both mechanisms are controlled by the expression of a series of genes. In recent decades, many genes related to Al tolerance have been identified, such as organic acid transporter genes (ALMTs and MATEs) (Hoekenga et al., 2006; Furukawa et al., 2007; Liu et al., 2013), antioxidative stress-related genes (Wu et al., 2017), also including those encoding aluminum transporter (Nramps and ABC transporter, aquaporins) (Huang et al., 2012; Li et al., 2014; Kochian et al., 2015; Lu et al., 2017; Wang et al., 2017), enzymes related to cell wall polysaccharide metabolism (XTHs) (Zhu et al., 2012; Zhu et al., 2014), and transcription factor (STOP, WRKY, ASR, NAC) (Iuchi et al., 2007; Ding et al., 2013; Arenhart et al., 2016; Li et al., 2018; Lou et al., 2019).
Previous studies have reported that one of an important component of the plant’s reaction to toxic levels of Al is oxidative stress, because Al3+ can induce the increase of active reactive oxygen species (ROS) and lipid peroxidase related enzyme activities in plants such as soybean, Arabidopsis, wheat, and maize (Cakmak and Horst, 1991; Keith et al., 1998; Giannakoula et al., 2010; Xu et al., 2012; Sun et al., 2017). In other words, the exposure of plants to an excessive amount of Al usually leads to the overproduction of ROS (Yamamoto et al., 2001; Exley, 2004) and lipid peroxidation, resulting in dysfunctional organelles, serious cell damage, and even cell death (Yamamoto et al., 2003; Šimonovičová et al., 2004; Kochian et al., 2005). The major source of ROS in Al stressed plants is the activated plasma membrane NAPDH oxidase which can lead to the production of O2·− and H2O2 (Sagi and Fluhr, 2001). Al3+ can quickly cross the plasma membrane and activate the Fenton reaction in the cytoplasm which increases the content of ROS in cells (Taylor et al., 2000).
To protect the plants from Al-triggered oxidative stress, they have evolved two defense ways, including enzyme-catalyzed antioxidant system and non-enzymatic system, to decrease ROS production, detoxify ROS, and stimulate the recovery from ROS-induced damages (Ahmad et al., 2010; Chowra et al., 2017; Daspute et al., 2017). The enzyme-catalyzed antioxidant system mainly improve the activity of antioxidant enzymes which include peroxidase (POD), superoxide dismutase (SOD), catalase (CAT), and ascorbate peroxidase (APX) (Ezaki et al., 2013), or increase the expression level of antioxidant enzyme genes (Irany et al., 2018). While the non-enzymatic antioxidants are ascorbate (AsA) and glutathione (GSH) (Xu et al., 2012). Ectopic overexpression of wheat WMnSOD1 and alternative oxidase gene improved Al tolerance in transgenic Brassica napus plants and tobacco cells, respectively (Basu et al., 2001; Panda et al., 2013). Transgenic Arabidopsis plants, which overexpressed three glutathione S-transferase genes and two peroxidase genes of tobacco, were endowed with strong aluminum tolerance (Ezaki et al., 2000). Besides, it was also reported some upstream gene, such as OsPIN2, and PEPC and PPDK, which are in positive control of the expression of antioxidant enzyme gene, could also enhance the tolerance of transgenic rice plants to Al toxicity via reducing the production of ROS, improving the activity ROS-scavenging enzyme, and weakening lipid peroxidation (Wu et al., 2014; Zhang et al., 2018). Additionally, it was also believed that the synthesis of cysteine-rich proteins can reduce the production of ROS. Fortunately, one of this protein had been identified as an Al tolerance gene that regulated the transcription by STOP1-like protein (ART1) in rice (Xia et al., 2013).
ZmAT6 (aluminum tolerance 6) gene, which encodes an unknown protein and is upregulated by Al stress, was identified from gene chip data in our previous study (Xu et al., 2017). In this study, we aimed to investigate the role that ZmAT6 played during Al stress. The Al tolerance-related phenotype of ZmAT6 was assessed in transgenic maize plants and Arabidopsis, as well as various indices of Al tolerance. The mechanism underlining the involvement of ROS scavenging in the ZmAT6-mediated antagonization of Al toxicity was explored.
The Al-tolerant maize inbred line 178, which has a high value of relative root growth (RRG = 45%) in our previous study (Xu et al., 2017), was used in this study. The seeds of 178 were first sterilized with 75% (w/v) alcohol for 2 min, then with 2% (w/v) NaOCl for 8 min, and finally germinated in quartz sand for 7 d under 28°C, 60% relative humidity and a photoperiod of 16 h/8 h (light/dark) cycle. After germinating, the seedlings were transferred to Hoagland’s solution (Hoagland and Arnon, 1950) and grown for 5 days to the three-leaf stage for further treatment. The nutrient solution was adjusted to pH 5.8 with HCl and renewed every two days. The greenhouse conditions were now set as 14 h/28°C and 10 h/22°C day–night cycle, 70% relative humidity and 300 μmol m−2s−1 intense luminosity.
Multiple sequence alignment of ZmAT6 (GRMZM5G886177) and its homologs was performed using DNAMAN (LynonBiosoft). Promoter analysis was performed by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), and the cis-elements are listed in Table S2.
Analysis of the Expression of ZmAT6 by Semi-Quantitative RT-PCR and Real-Time PCR
The tissues and organs 10 days after pollination, including the roots, stems, leaves, ears, tassels, and kernels, were collected from 178; the seedlings were treated with 0.5 mM CaCl2 solution (pH 4.2) at 28°C overnight before treatment with Al and then transferred into the same solution containing additively 60 µM AlCl3 (pH 4.2) and treated for 0, 6, 12, 24, 48, and 96 h. All the tissues were immediately frozen in liquid nitrogen and stored at −80°C. Semi-quantitative RT-PCR and real-time PCR were carried out as previously described (Lin et al., 2014). Three biological replicates were performed for each experiment. The primers used for ZmAT6 were listed in Table S3.
The subcellular localization of the ZmAT6 protein was predicted by the WoLF PSORT program (http://wolfpsort.org) (Horton et al., 2007). Moreover, the full length CDS of theZmAT6 gene was ligated to pCAMBIA2300 to establish the ZmAT6:GFP vector.
The full length coding sequence of the ZmAT6 gene was amplified and ligated to the CPB vector behind the cauliflower mosaic virus 35S (CaMV35S) promoter to construct the p35S:ZmAT6 vector. The p35S:ZmAT6 vector was transformed into immature maize callus of 18-599 by an Agrobacterium-mediated method (Huang et al., 2018) and into Arabidopsis by the flowering dip method (Clough and Bent, 1998). The transgenic plants of maize and Arabidopsis were confirmed by PCR amplification and harvested individually. The homozygous seeds of T4 generation were used for future experiments.
The seedlings of the transgenic maize and wild type (WT) 18-599 were transferred to Hoagland’s solution with or without 200 μM AlCl3 (pH 4.2). After two weeks of culture, the fresh weights of the underground part and the upper part of the ground were measured separately.
The root lengths of the transgenic maize plants and wild type were measured and treated as the initial length after pretreatment with 0.5 mM CaCl2 for 12 h. The maize plants were exposed to 60 μM AlCl3 for 3 days, the final length of the root was recorded, and the RRG was calculated (Sasaki et al., 2004; Sasaki et al., 2006). The root tips (approximately 1 cm) were collected and measured as previously described to determine the Al content (Osawa and Matsumoto, 2001).
After treatment with 60 μM AlCl3 for 24 h, the roots of transgenic maize plants and WT were immediately collected and dyed as described by Baker and Mock (Baker and Mock, 1994). Lipid peroxidation was determined by measuring the MDA content using the thiobarbituric acid method (Cakmak and Horst, 1991). Free proline determination was assessed as described by Bates (Bates et al., 1973).
After treatment with 60 μM AlCl3 for 24 h, the active oxygen content was measured in the seedlings of transgenic maize and WT (Jana and Choudhuri, 1982). The super oxygen free radical (O2·−) was measured by the hydroxylamine method (Elstner and Heupel, 1976).
After Al treatment, roots were collected immediately, and 0.5 g of the root sample was homogenized with 50 mM sodium phosphate buffer (pH 7.0) containing 3 ml 1 mM ethylenediaminetetraacetic acid (EDTA). Homogenates were then centrifuged at 4°C for 20 min at 15,365 g, and the supernatants were used to determine the enzyme activity. The whole extraction procedure was carried out at 4°C.
The activities of SOD, POD, CAT, and APX were determined as previously described (Aebi, 1974; Giannopolitis and Ries, 1977; Nakano and Asada, 1981; Zheng and Huystee, 1992).
The relative expression of antioxidant enzyme genes in WT and one of the maize transgenic line L5 (Figure S2) was detected by RT-PCR, and the primers were listed in Table S3.
All experiments were repeated at least three times, and data were represented as the mean ± SD of the replicates. Student’s t tests at p < 0.05 and 0.01 were performed to identify significant differences between observation values using the SPSS21 software. The figures were drawn using the Origin 8.0 software (Origin Lab Corporation, Northampton, MA, USA).
Using gene specific primers, the complete CDS sequence of ZmAT6 was isolated from the aluminum tolerance maize line 178. The 771 bp open reading frame (ORF) of ZmAT6 has three exons and encodes a 27.79 kDa non-transmembrane protein with a predicted pI of 5.80 (Figure S1, Table S1). Multiple sequence alignment showed that ZmAT6 shared a higher similarity, up to 75.19%, with the rice homolog (Os01g0731600), higher than its Arabidopsis counterpart (At1g78780) (51.56%) (Figure 1). In addition, a promoter analysis of ZmAT6 revealed that it included but was not limited to the binding sites GGN(T/g/a/C)V(C/A/g)S(C/G) of ART1 (Table S2), the major transcriptional factor that regulates the series of Al response genes in rice. However, protein prediction did not reveal any particular structure of ZmAT6.
To better understand the functions of ZmAT6, its expression patterns in several organs, including roots, stems, leaves, ears, tassels and kernels, at different growth stages were monitored using semi-quantitative reverse transcription PCR (RT-PCR). ZmAT6 was moderately expressed in all the organs with the exception of its weak expression in ears (Figure 2A). Moreover, to investigate the pattern of expression of ZmAT6 on Al exposure, the mRNA abundance of ZmAT6 was monitored further in roots and shoots under a time-course Al stress treatment from 0 to 96 h. As shown in Figure 2B, the transcription of ZmAT6 was dramatically upregulated after Al treatment in the roots or the shoots. The Al-inducted expression level of ZmAT6 was quite stable in the roots during the whole process. Alternatively, ZmAT6 in the shoots exhibited a much higher level of expression than in the roots. The mRNA abundance of ZmAT6 peaked at time point six and then gradually decreased during prolonged Al stress. These results indicated that ZmAT6 was sensitive to the fluctuating environmental Al3+ status in seedlings and was involved in the tolerance to aluminum toxicity in maize.
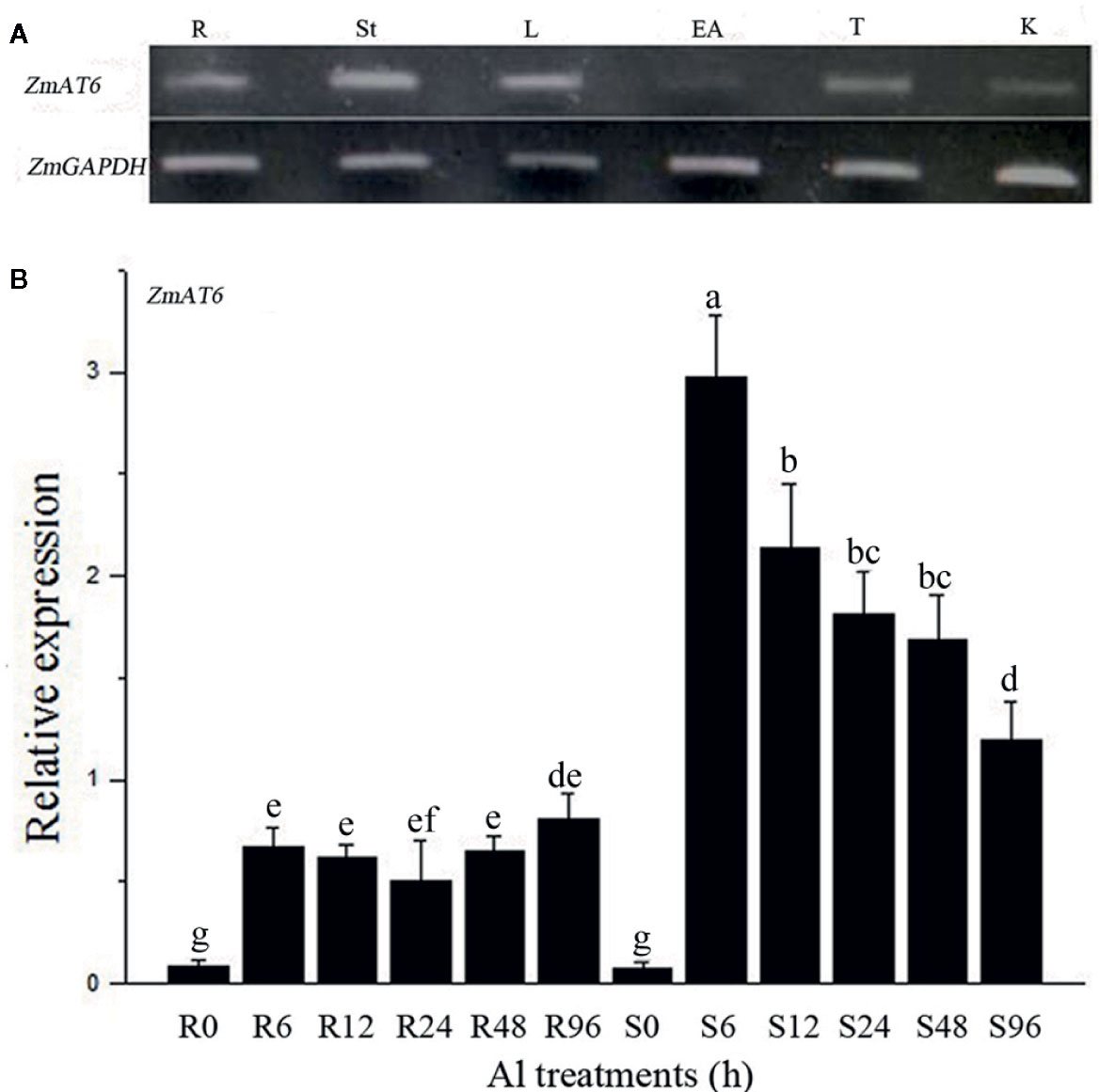
Figure 2 The expression pattern of the ZmAT6 gene. (A) Tissue and organ expression pattern of ZmAT6 in adult maize plants. The letter above the columns of expression data refer to: R, root; St, stem; L, leaf; EA, ear; T, tassel; and K, kernel. (B) Transcription of ZmAT6 in the roots (R) and shoot (S) was quantified at 0, 6, 12, 24, 48, and 96 h after Al treatment (60 μM AlCl3, pH = 4.2). ZmGAPDH was used as internal reference gene. The values were presented as mean ± SD (n = 3) and marked with different letters to indicate statistic significant difference at P < 0.05 (student’s t test).
Using the WoLF PSORT program, ZmAT6 protein was predicted to be localized in the chloroplast or cytoplasm. To detect the subcellular localization of ZmAT6 protein, the coding region of ZmAT6 was fused with the 3′ end of the GFP gene and driven by the 35S promoter. The GFP gene alone under the control of the 35S promoter served as the control. The subcellular localization of ZmAT6 was determined in a transient expression system in Nicotiana benthamiana leaves. The result indicated that the 35S:ZmAT6:GFP fusion protein appeared to be localized in the chloroplast but not the cytoplasm (Figure 3).
To further investigate the function of ZmAT6 under Al stress, the entire ORF of ZmAT6 was inserted into the binary vector CPB (Figures S2A, B), and the recombinant expression vector p35S:ZmAT6 was transformed into maize inbred line 18-599. Finally, 10 independent transgenic plants of T0 generation were identified and confirmed by PCR detection (Figure S2C). Among them, three homozygous lines (L3, L5, and L10) of T3 generation with higher ZmAT6 expression (Figure S2D) were propagated and selected for analysis.
To determine whether the overexpression of ZmAT6 enhanced the tolerance of the transgenic plants to Al, we assessed the plant growth of the three overexpressed transgenic lines of ZmAT6 (OE-ZmAT6) and the wild type (18-599, WT) under normal and Al stress conditions. At the beginning, the transgenic plants grew normally like the WT in a hydroponic culture (pH 4.2). Nevertheless, when cultivated in the same solution containing 200 μM AlCl3, all of the OE-ZmAT6 plants uniformly showed a high tolerance to Al. The mean value of RRG of OE-ZmAT6 plants was 90%, much higher than that of the WT (70%) (Figures 4A, D). As for the root fresh weight, the mean value of ZmAT6 transgenic lines was about 87% heavier than that of WT (Figure 4B), indicating a less inhibition of OE-ZmAT6 plants when subjected to Al stress. Moreover, the vigorous leaf growth of the OE-ZmAT6 transgenic plants could also be verified by the significant increase in the shoot fresh weights (Figures 4A, C). Concerning the Al content in root tips, the mean value of 24.63 μg/g fresh weight (FW) was lower in the OE-ZmAT6 plants than the 37.23μg/g FW in the WT (Figure 4E).
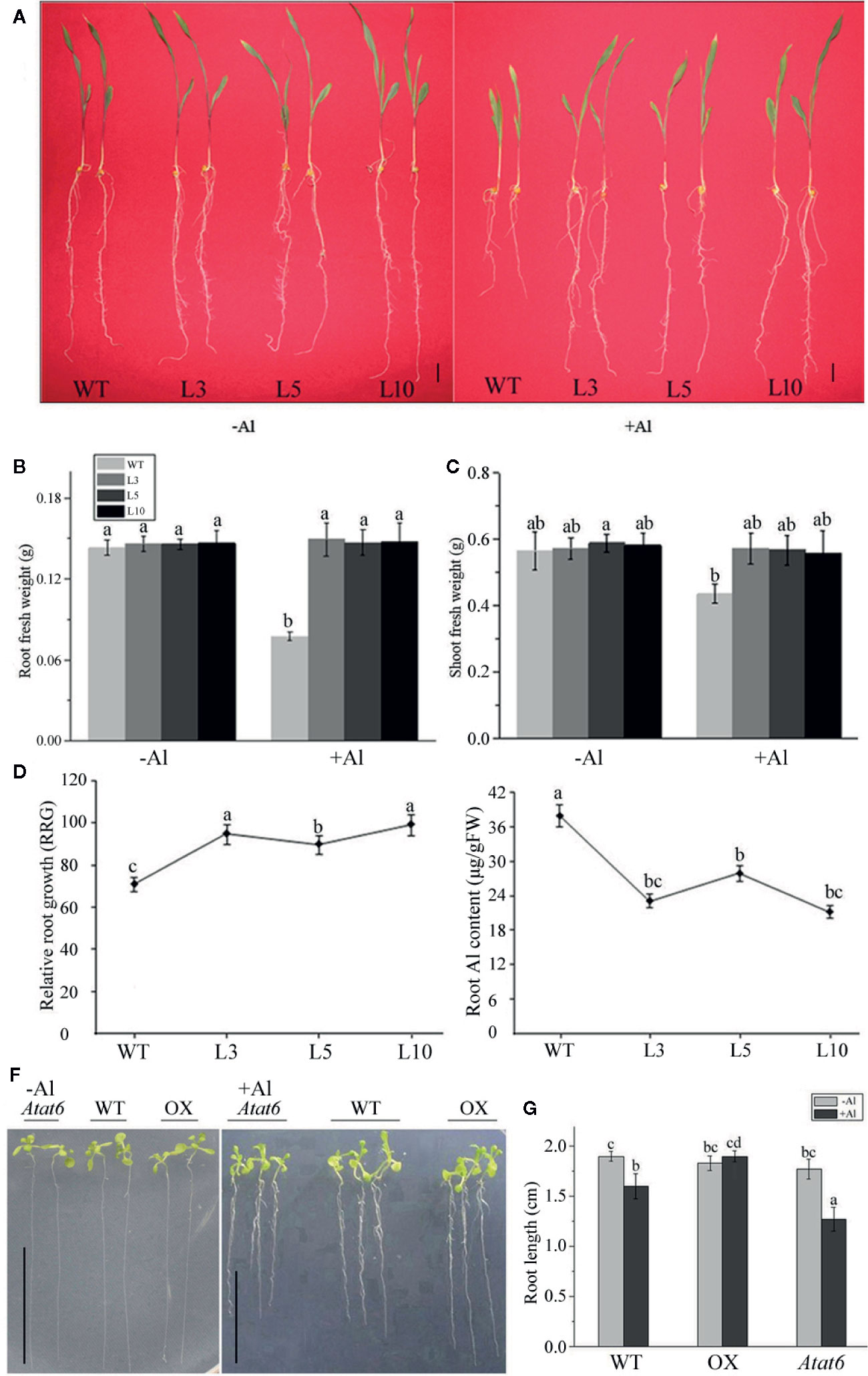
Figure 4 ZmAT6 overexpression enhanced aluminum tolerance in both transgenic maize and Arabidopsis. (A) Wild type (WT) and OE-ZmAT6 transgenic plants were grown on hydroponic culture with or without Al treatment (60 μM AlCl3, pH 4.2). The corresponding (B) root fresh weight, (C) shoot fresh weight, (D) relative root growth (RRG), and (E) aluminum (Al) content. Values represent mean ± SD (n = 3–5). Scale bar: 2.0 cm. Different letters indicate significant differences (P < 0.01) (multiple comparison). (F) Wild type Col-0 and mutants Atat6 and ZmAT6 overexpressed (OX) transgenic Arabidopsis plants grown with or without aluminum treatment (pH 4.2) and the corresponding (G) root length. Scale bar: 1.0 cm. Values represent mean ± SD (n ≥ 3). Different letters indicate significant differences (P < 0.01) (multiple comparison).
In addition, evidence from its Arabidopsis homolog AtAT6 reinforced the fact that AtAT6 could enhance or decrease aluminum tolerance via overexpression or mutation (Figure S3, Figures 4F, G). In comparison with the WT Col-0, the root length of the Atat6 mutant (SALK_082224) was much shorter, while those of the AtAT6-overexpressed (OX) Arabidopsis plants were much longer on Al exposure (Figure 4G).
Furthermore, physiological indices related to Al-stress tolerance, including Evans blue staining, malondialdehyde (MDA) content, and proline (Pro) content, were also measured in three OE-ZmAT6 transgenic lines and WT maize plants on Al exposure. As shown in Figure 5 and Figure S4, the results indicated that the mean values of the Evans blue uptake (1.80 OD 600/g) and MDA content (3.14 μM/μg FW) in the OE-ZmAT6 lines were significantly lower than those of the WT under Al treatment. In contrast, the Pro content (15.80 μM/g FW) was clearly higher than that in the WT (11.77 μM/g FW) when the plants suffered from Al toxicity (Figure 5C). These results verified the fact that overexpression of ZmAT6 conferred tolerance to Al toxicity in OE-ZmAT6 plants and reduced the oxidative damage of maize under Al stress.
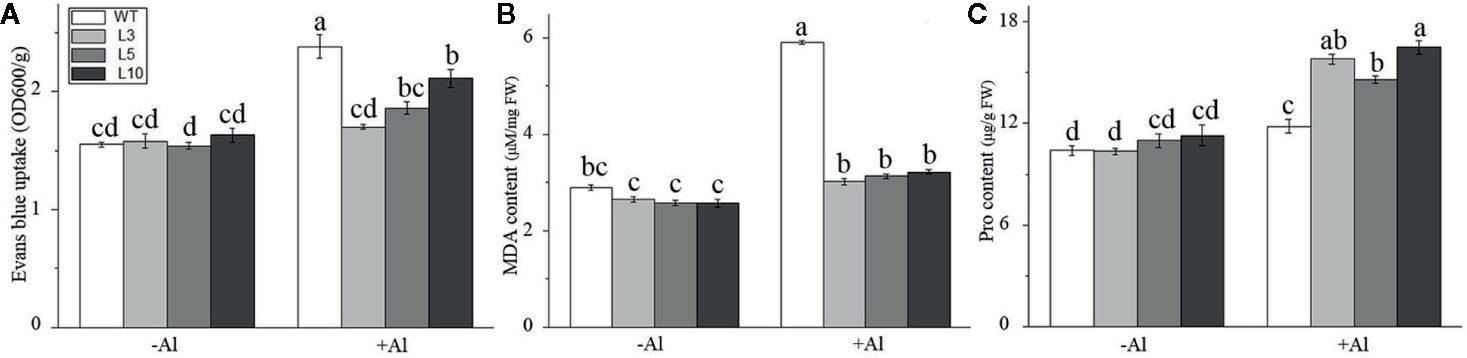
Figure 5 Determination of physiological indexes related to aluminum tolerance. (A) Evans blue, (B) Malondialdehyde (MDA) and (C) Proline (Pro) in the wild type (WT) and three OE-ZmAT6 transgenic maize lines (L3, l5, and L10). Values represent mean ± SD (n = 10). Different letters indicate significant differences (P < 0.01) (multiple comparison).
To investigate whether ZmAT6 is related to the scavenging of ROS, we measured the content of hydrogen peroxide (H2O2) and the rate of production of the superoxide anion in the three OE-ZmAT6 lines and the WT. Under normal conditions, no distinct difference could be detected between the OE-ZmAT6 lines and WT regarding these two indices. Nevertheless, both H2O2 and the superoxide anion had increased in all the plants tested during Al stress, and the amplitude was even higher in the WT than in OE-ZmAT6 plants (Figures 6A, B).The increase of H2O2 content and productive rate of superoxide anion in WT against OE-ZmAT6 lines under Al stress were 0.38:0.09 (μM/L FW) and 0.24:0.11 (μM/mg) min, respectively. These results indicated that ZmAT6 played an important role in the scavenging of the excessive ROS caused by Al stress, which endowed transgenic maize plants with the capacity to tolerate aluminum toxicity by ROS cleaning.
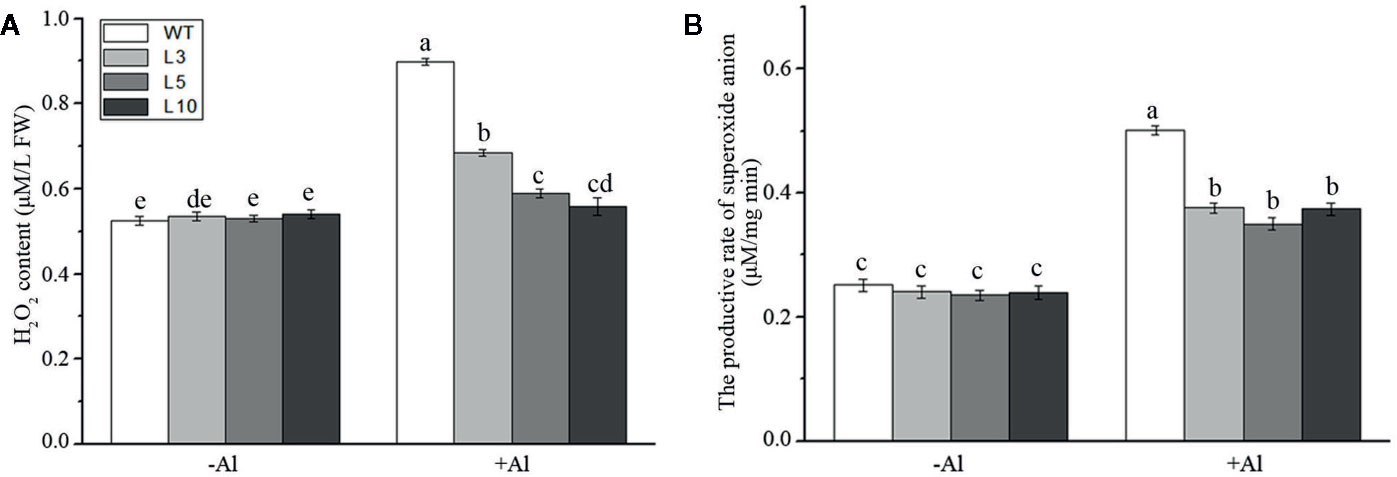
Figure 6 Determination of ROS under aluminum stress. (A) Hydrogen peroxide content and (B) the productive rate of superoxide anion in wild type (WT) and three OE-ZmAT6 transgenic maize lines (L3, L5, and L10). Values represent mean ± SD (n = 3). Different letters indicate significant differences (P < 0.01) (multiple comparison).
To investigate the factors affecting the ZmAT6-mediated scavenging of ROS, we examined the activity of several enzymes usually involved in the antioxidant system. It was notable that the SOD activity of three OE-ZmAT6 lines was significantly higher than that of the WT despite Al treatment. After treatment with 60 μM AlCl3 (pH 4.2), the mean value of SOD activity of the OE-ZmAT6 plants was dramatically increased, up to 244.4 U/g (Figure 7A). The activities of POD, CAT, and APX remained almost the same in the WT plants as in the OE-ZmAT6 plants but decreased separately to 28.07, 22.92, and 14.77%, respectively, when subjected to Al stress (Figures 7B–D).
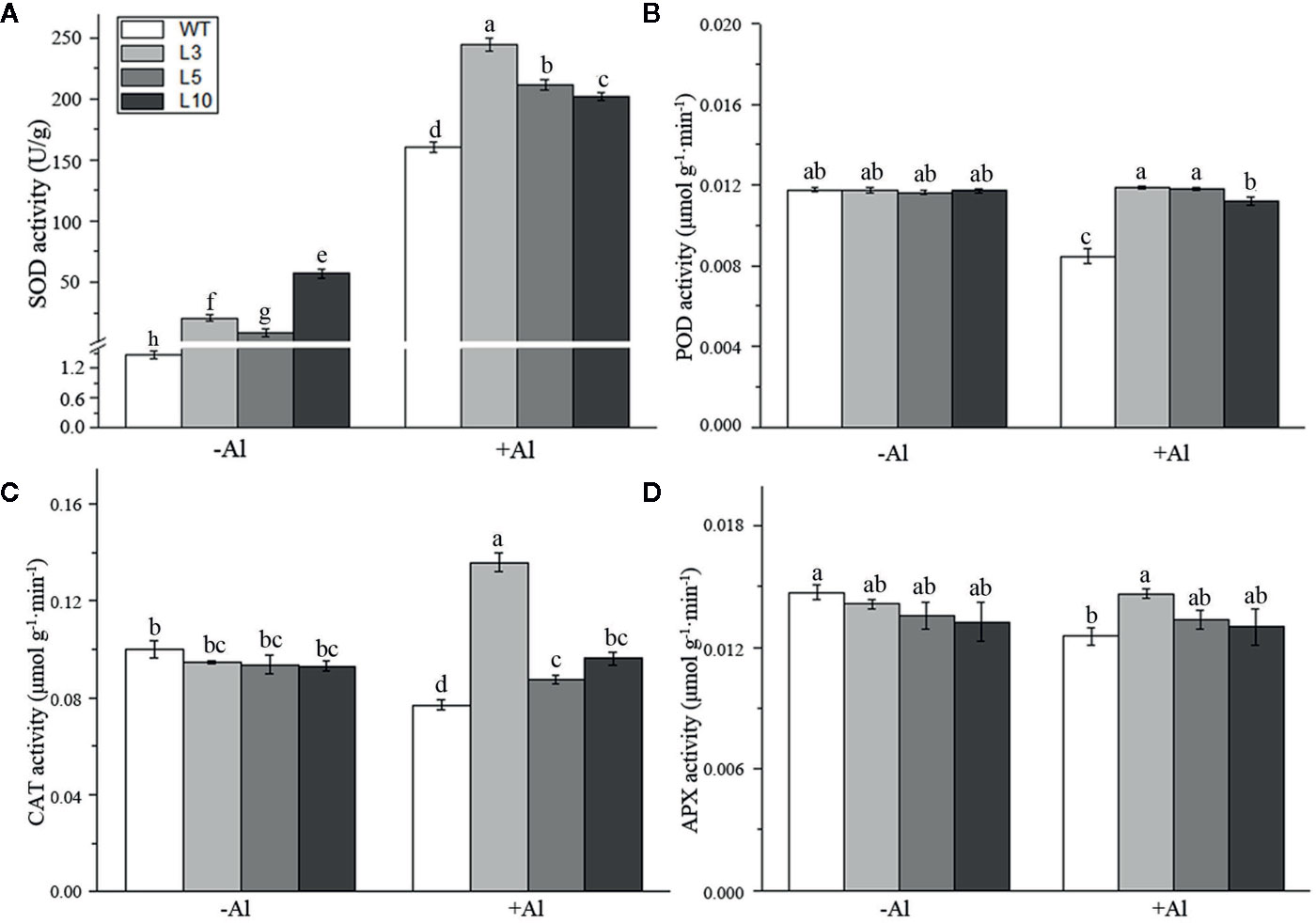
Figure 7 Determination of the activity of several antioxidant enzymes. The activities of antioxidant-related enzymes (A) SOD, (B) POD, (C) CAT, and (D) APX were detected. Values represent mean ± SD (n = 4). Different letters indicate significant differences (P < 0.05) (multiple comparison).
In addition, we also monitored the expression patterns of three antioxidant-enzyme genes on Al exposure. As shown in Figures 8A, B, the expression of ZmSOD and ZmPOD could be upregulated by Al stimulus in all tested plants with the exception of ZmPOD in WT, consistent with the enhanced activities of antioxidant enzymes (Figure 7). As for ZmSOD and ZmPOD, both of them exhibited a much higher level of expression either in the root or leaf of OE-ZmAT6 plant than in WT. In particular, the expression of ZmCAT in the root of WT was higher than in OE-ZmAT6 plant (Figure 8C). These results suggested that both the regulation on gene expression and activity of antioxidant enzymes contributed to the enhanced Al tolerance of OE-ZmAT6 plant in maize.
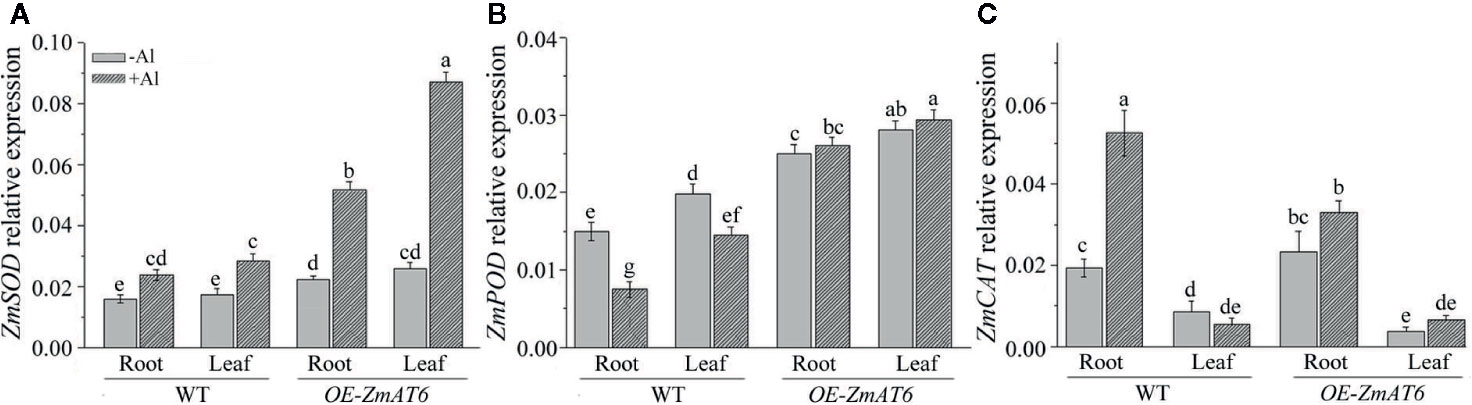
Figure 8 The relative expression of antioxidant enzyme genes. The relative expression levels of (A) ZmSOD, (B) ZmPOD, and (C) ZmCAT were determined with or without Al treatment in WT and OE-ZmAT6 transgenic maize (L5). ZmGAPDH was used as internal reference gene. Values represent mean ± SD (n = 3). Different letters indicate significant differences (P < 0.05) (multiple comparison).
Some plants evolved many mechanisms to tolerate Al toxicity. ZmAT6 was first identified as an Al-induced gene with an unpredicted function from our previous microarray data (Xu et al., 2017). The unique indication was that its rice or tholog was regulated by OsART1, a master transcriptional factor that controls more than 30 Al response genes by binding to the GGTCC (GGN(T/g/a/C)V(C/A/g)S(C/G)) site of its promoter (Yamaji et al., 2009; Tsutsui et al., 2011). Similarly, we also found a GGNVS site in the promoter of ZmAT6 (Table S2). In addition, ZmAT6 expresses in many tissues and organs, and it could be rapidly induced by Al stress in both the roots and shoots of maize (Figure 2). These results suggested that ZmAT6 may be a downstream gene in maize directly targeted by a transcription factor such as rice OsART1 when the plants are under Al stress. An exploration of the OsART1 equivalent in maize by strategies such as homolog query and gel shift assay could confirm this hypothesis.
The inhibition of the root length was an initial detection of Al toxicity in plants (Matsumoto and Motoda, 2012). Plants with strong Al tolerance usually exhibited high relative root growth (Ma et al., 2018; Badia et al., 2020). In this study, the RRG of OE-ZmAT6 transgenic maize was more than 20% higher than those in the WT. Meanwhile, the mean value of the total fresh weight and the fresh weight of roots were more than 30% and even over 50% weightier than that of WT after exposure to Al, respectively (Figures 4A–D). Attributed to the ectopic overexpression of ZmAT6, the clear improvement of the Al tolerance was exhibited not only in transgenic maize plants but also in transgenic wild type Arabidopsis and at6 mutant (Figures 4F, G). Moreover, the content of aluminum in the root of OE-ZmAT6 plant is rather lower than that in the WT (Figure 4E), underlying an antagonism to Al toxicity.
In addition, a semi reduction of the superoxide radical ion caused by aluminum toxicity may lead to oxidative stress and serious cell damage. The enzyme-catalyzed antioxidant system and the non-enzymatic antioxidant system of plants could scavenge excessive ROS and reduce the cell damage caused by aluminum toxicity (Achary et al., 2008; Irany et al., 2018). The intensity of Evans blue staining and the content of MDA in the cells of target tissue could reflect the lipid peroxidation of the cell membrane. In this study, the significantly lower Evans blue uptake (Figure 5A, Figure S4) and MDA content (Figure 5B) of the OE-ZmAT6 plants suggested that aluminum stress caused less damage to the cell membrane. This was consistent with the results of Sun (Sun et al., 2017) and Yu (Yu et al., 2018), who reported that the MDA content in the root tips of wheat increased significantly under aluminum treatment, particularly in sensitive genotype plants, and the higher Evans Blue uptake in the root apexes was due to the Al-induced oxidative stress. Moreover, proline, a type of antioxidant that favors ROS removal (Rodriguez and Redman, 2005), had a significantly higher content in tolerant plants than in sensitive ones (Ashraf and Foolad, 2007). The obviously increased content of proline in the OE-ZmAT6 plants under Al stress in this study was highly consistent with the previous study of Giannakoula (Giannakoula et al., 2008), who found that the proline content in the Al-tolerant genotype maize increased with the concentration of Al.
Previous studies have documented that the excessive accumulation of ROS in plants under aluminum stress is the primary reason for oxidative stress and the decisive factor that inhibited root elongation (Tamás et al., 2006; Giannakoula et al., 2010). Giannakoula (Giannakoula et al., 2010) and Sun (Sun et al., 2017) had found that O2·− and H2O2 could predominantly accumulate in the roots of Al-sensitive maize and wheat, respectively, after exposure to Al. Compared with the wild type, overexpression of AtPrx64 in transgenic tobacco also showed less root growth inhibition, lower H2O2 content, and less MDA accumulation following Al exposure (Wu et al., 2017). In our study, the content of H2O2 and the productive rate of the superoxide anion in the ROS of OE-ZmAT6 maize plants were significantly lower than those in the WT under Al stress (Figure 6), indicating the involvement of AT6 in ROS scavenging. Indeed, the activities of the antioxidant enzymes POD, CAT, and APX of the OE-ZmAT6 plants had no obvious change before and after Al treatment even though there was a significant difference between the transgenic lines and WT (Figures 7B–D). The only exception was SOD, which had significantly higher activities in the OE-ZmAT6 plants than that in the WT before or after Al stress (Figure 7A). Consistently, the expression level of ZmSOD was also predominately upregulated by Al stimulus (Figure 8A), suggesting that SOD played a crucial role in effectively removing excess ROS from OE-ZmAT6 maize cells and maintaining the balance of ROS.
Coincidentally, a number of studies also found that the enzyme-catalyzed antioxidant system is involved in Al stress antagonism even though the major scavenger enzyme was not the same. Darkó et al. (Darkó et al., 2004) found that the activity of the antioxidant enzymes in the wheat root tips changed noticeably under Al stress, and the activity of APX in Al-sensitive genotypes was significantly higher, while those of SOD and CAT were lower than those of the Al-tolerant genotype. However, the activities of SOD and POD were significantly improved in Al-sensitive maize after Al treatment, while the activities of SOD, POD and CAT exhibited no distinction in the Al-tolerant genotypes (Boscolo et al., 2003). Therefore, the ROS scavenging responses of antioxidant enzymes under Al stress conditions varied depending on the species and genotypes. Alternatively, the overexpression of genes encoding ROS-scavenging enzymes in several plant species had documented that their activities enhanced the tolerance of Al (Panda et al., 2013; Sun et al., 2017; Irany et al., 2018). In this study, ZmAT6 enhanced the aluminum tolerance of maize by increasing the expression level of ZmSOD gene and improving the activity of the antioxidant enzymes SOD in the antioxidant enzymatic system.
In conclusion, we demonstrated that the chloroplast-located gene ZmAT6 could be quickly induced by Al stress and could enhance the tolerance to Al toxicity when overexpressed in transgenic maize and Arabidopsis. ZmAT6 could antagonize the Al toxicity via at least two ROS removal approaches: increasing the activity of antioxidant enzymes SOD and the content of antioxidant proline.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
HD and YH carried out the experiments, analyzed the data, and drafted the manuscript. JD, HL, XH, MQ, YLi, and WY contributed to sample collection and data analysis. WH, MC, SG, and YLu contributed with consultation. SZ designed the experiment and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China [No. 30800687, 31071434], and the Major Project of Education Department in Sichuan [No. 15ZA0022].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.01016/full#supplementary-material
Achary, V. M., Jena, S., Panda, K. K., Panda, B. B. (2008). Aluminium induced oxidative stress and DNA damage in root cells of Allium cepa L. Ecotoxicol. Environ. Saf. 70, 300–310. doi: 10.1016/j.ecoenv.2007.10.022
Aebi, H. (1974). “Catalase, method of enzymatic analysis,” in Method of enzymatic analysis 3. Ed. Bergmeyer, H. U. (New York: Academic), 673–684.
Ahmad, P., Jaleel, C. A., Salem, M. A., Nabi, G., Sharma, S. (2010). Roles of enzymatic and nonenzymatic antioxidants in plants during abiotic stress. Crit. Rev. Biotechnol. 30, 161–175. doi: 10.3109/07388550903524243
Arenhart, R. A., Schunemann, M., Bucker Neto, L., Margis, R., Wang, Z. Y., Margis-Pinheiro, M. (2016). Rice ASR1 and ASR5 are complementary transcription factors regulating aluminium responsive genes. Plant Cell Environ. 39, 645–651. doi: 10.1111/pce.12655
Ashraf, M., Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59, 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Badia, M. B., Maurino, V. G., Pavlovic, T., Arias, C. L., Pagani, M. A., Andreo, C. S., et al. (2020). Loss of function of Arabidopsis NADP-malic enzyme 1 results in enhanced tolerance to aluminum stress. Plant J. 101, 653–665. doi: 10.1111/tpj.14571
Baker, C. J., Mock, N. M. (1994). An improved methods for monitoring cell death in cell suspension and leaf disc assays using evans blue. Plant Cell Tiss. Org. Cult. 39, 7–12. doi: 10.1007/BF00037585
Basu, U., Good, A. G., Taylor, G. J. (2001). Transgenic Brassica napus plants overexpressing aluminum-induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminum. Plant Cell Environ. 24, 1278–1269. doi: 10.1046/j.0016-8025.2001.00783.x
Bates, L. S., Waldren, R. P., Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Boscolo, P. R. S., Menossi, M., Jorge, R. A. (2003). Aluminum-induced oxidative stress in maize. Phytochemistry 62, 181–189. doi: 10.1016/S0031-9422(02)00491-0
Cakmak, I., Horst, W. J. (1991). Effect of aluminium on lipid peroxidation, superoxide dismutase, catalase, and peroxidase activities in root tips of soybean (Glycine max). Physiol. Plant. 83, 463–468. doi: 10.1111/j.1399-3054.1991.tb00121.x
Chowra, U., Yanase, E., Koyama, H., Panda, S. K. (2017). Aluminium-induced excessive ROS causes cellular damage and metabolic shifts in black gram Vigna mungo (L.) Hepper. Protoplasma 254, 293–302. doi: 10.1007/s00709-016-0943-5
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplifed method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. Cell Mol. Biol. 16, 735. doi: 10.1046/j.1365-313x.1998.00343.x
Darkó, É, Ambrus, H., Stefanovits-Bányai, É, Fodor, J., Bakos, F., Barnabás, B. (2004). Aluminium toxicity, Al tolerance and oxidative stress in an Al-sensitive wheat genotype and in Al-tolerant lines developed by in vitro microspore selection. Plant Sci. 166, 583–591. doi: 10.1016/j.plantsci.2003.10.023
Daspute, A. A., Sadhukhan, A., Tokizawa, M., Kobayashi, Y., Panda, S. K., Koyama, H. (2017). Transcriptional regulation of aluminum-tolerance genes in higher plants: clarifying the underlying molecular mechanisms. Front. Plant Sci. 8, 1358. doi: 10.3389/fpls.2017.01358
Ding, Z. J., Yan, J. Y., Xu, X. Y., Li, G. X., Zheng, S. J. (2013). WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 76, 825–835. doi: 10.1111/tpj.12337
Elstner, E. F., Heupel, A. (1976). Inhibition of nitrite formation from hydroxylammoniumchloride: a simple assay for superoxide dismutase. Anal. Biochem. 70, 616–620. doi: 10.1016/0003-2697(76)90488-7
Exley, C. (2004). The pro-oxidant activity of aluminum. Free Radic. Biol. Med. 36, 380–387. doi: 10.1016/j.freeradbiomed.2003.11.017
Ezaki, B., Gardner, R. C., Ezaki, Y., Matsumoto, H. (2000). Expression of aluminum-induced genes in transgenic Arabidopsis plants can ameliorate aluminum stress and/or oxidative stress. Plant Physiol. 122, 657–665. doi: 10.1104/pp.122.3.657
Ezaki, B., Jayaram, K., Higashi, A., Takahashi, K. (2013). A combination of five mechanisms confers a high tolerance for aluminum to a wild species of Poaceae, Andropogon virginicus L. Environ. Exp. Bot. 93, 35–44. doi: 10.1016/j.envexpbot.2013.05.002
Furukawa, J., Yamaji, N., Wang, H., Mitani, N., Murata, Y., Sato, K., et al. (2007). An aluminum-activated citrate transporter in barley. Plant Cell Physiol. 48, 1081–1091. doi: 10.1093/pcp/pcm091
Giannakoula, A., Moustakas, M., Mylona, P., Papadakis, I., Yupsanis, T. (2008). Aluminum tolerance in maize is correlated with increased levels of mineral nutrients, carbohydrates and proline, and decreased levels of lipid peroxidation and Al accumulation. J. Plant Physiol. 165, 385–396. doi: 10.1016/j.jplph.2007.01.014
Giannakoula, A., Moustakas, M., Syros, T., Yupsanis, T. (2010). Aluminum stress induces up-regulation of an efficient antioxidant system in the Al-tolerant maize line but not in the Al-sensitive line. Environ. Exp. Bot. 67, 487–494. doi: 10.1016/j.envexpbot.2009.07.010
Giannopolitis, C. N., Ries, S. K. (1977). Superoxide dismutases I. Occurrence in higher plants. Plant Physiol. 59, 309–314. doi: 10.1104/pp.59.2.309
Hoagland, D. R., Arnon, D., II (1950). The water-culture method for growing plants without soil. Univ. Galif. Agric. Exp. Sta. Circ. 347, 357–359. doi: 10.1016/S0140-6736(00)73482-9
Hoekenga, O. A., Maron, L. G., Piñeros, M. A., Cancado, G. M., Shaff, J., Kobayashi, Y., et al. (2006). AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. PNAS 103, 9738–9743. doi: 10.1073/pnas.0602868103
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, 585–587. doi: 10.1093/nar/gkm259
Huang, C. F., Yamaji, N., Chen, Z., Ma, J. F. (2012). A tonoplast-localized half-size ABC transporter is required for internal detoxification of aluminum in rice. Plant J. 69, 857–867. doi: 10.1111/j.1365-313X.2011.04837.x
Huang, S., Gao, J., You, J., Liang, Y., Guan, K., Yan, S., et al. (2018). Identification of STOP1-like proteins associated with aluminum tolerance in sweet sorghum (Sorghum bicolor L.). Front. Plant Sci. 9, 258. doi: 10.3389/fpls.2018.00258
Irany, R. P., Anny , C. D. L., Tatiane , L. P., Raquel , A. A., Maria , D. C. P. B. (2018). Gene expression and antioxidant enzymatic activity in passion fruit exposed to aluminum. Afr. J. Agric. Res. 13, 115–120. doi: 10.5897/ajar2017.12834
Iuchi, S., Koyama, H., Iuchi, A., Kobayashi, Y., Kitabayashi, S., Ikka, T., et al. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. PNAS 104, 9900–9905. doi: 10.1073/pnas.0700117104
Jana, S., Choudhuri, M. A. (1982). Glycolate metabolism of three submersed aquatic angiosperms during ageing. Aquat Bot. 12, 345–354. doi: 10.1016/0304-3770(82)90026-2
Keith, D., Richards, E. J. S., Yogesh, K. S., Keith, R. D., Richard, C. G. (1998). Aluminum induces oxidative stress genes in Arabdopsis thaliana. Plant Physiol. 116, 409–418. doi: 10.1104/pp.116.1.409
Kinraide, T. B. (1990). Assessing the rhizotoxicity of the aluminate ion, Al(OH)4–. Plant Physiol. 93, 1620–1625. doi: 10.1104/pp.93.4.1620
Kochian, L. V., Hoekenga, O. A., Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kochian, L. V., Piñeros, M. A., Hoekenga, O. A. (2005). The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 274, 175–195. doi: 10.1007/s11104-004-1158-7
Kochian, L. V., Pineros, M. A., Liu, J., Magalhaes, J. V. (2015). Plant adaptation to acid soils: The molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66, 571–598. doi: 10.1146/annurev-arplant-043014-114822
Kochian, L. V. (1995). Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Biol. 46, 237–260. doi: 10.1146/annurev.pp.46.060195.001321
Li, J. Y., Liu, J., Dong, D., Jia, X., McCouch, S. R., Kochian, L. V. (2014). Natural variation underlies alterations in Nramp aluminum transporter (NRAT1) expression and function that play a key role in rice aluminum tolerance. Proc. Natl. Acad. Sci. U. S. A. 111, 6503–6508. doi: 10.1073/pnas.1318975111
Li, G. Z., Wang, Z. Q., Yokosho, K., Ding, B., Fan, W., Gong, Q. Q., et al. (2018). Transcription factor WRKY22 promotes aluminum tolerance via activation of OsFRDL4 expression and enhancement of citrate secretion in rice (Oryza sativa). New Phytol. 219, 149–162. doi: 10.1111/nph.15143
Lin, Y. A., Zhang, C. L., Lan, H., Gao, S. B., Liu, H. L., Liu, J., et al. (2014). Validation of potential reference genes for qPCR in maize across abiotic stresses, hormone treatments, and tissue types. PloS One 9, e95445. doi: 10.1371/journal.pone.0095445
Liu, M. Y., Chen, W. W., Xu, J. M., Fan, W., Yang, J. L., Zheng, S. J. (2013). The role of VuMATE1 expression in aluminium-inducible citrate secretion in rice bean (Vigna umbellata) roots. J. Exp. Bot. 64, 1795–1804. doi: 10.1093/jxb/ert039
Lou, H. Q., Fan, W., Jin, J. F., Xu, J. M., Chen, W. W., Yang, J. L., et al. (2019). A NAC-type transcription factor confers aluminium resistance by regulating cell wall-associated receptor kinase 1 and cell wall pectin. Plant Cell Environ. 43, 463–478. doi: 10.1111/pce.13676
Lu, M., Wang, Z., Fu, S., Yang, G., Shi, M., Lu, Y., et al. (2017). Functional characterization of the SbNrat1 gene in sorghum. Plant Sci. 262, 18–23. doi: 10.1016/j.plantsci.2017.05.010
Ma, J. F., Furukawa, J. (2003). Recent progress in the research of external Al detoxification in higher plants: a minireview. J. Inorg. Biochem. 97, 46–51. doi: 10.1016/s0162-0134(03)00245-9
Ma, J. F., Hiradate, S., Nomoto, K., Iwashlta, T., Matsumoto, H. (1997). Internal detoxification mechanism of Al in hydrangea (identification of Al form in the leaves). Plant Physiol. 113, 1033–1039. doi: 10.1104/pp.113.4.1033
Ma, Q., Yi, R., Li, L., Liang, Z., Zeng, T., Zhang, Y., et al. (2018). GsMATE encoding a multidrug and toxic compound extrusion transporter enhances aluminum tolerance in. Arabidopsis Thaliana BMC Plant Biol. 18, 212. doi: 10.1186/s12870-018-1397-z
Ma, J. F. (2000). Role of organic acids in detoxification of aluminum in higher plants. Plant Cell Physiol. 41, 383–390. doi: 10.1093/pcp/41.4.383
Matsumoto, H., Motoda, H. (2012). Aluminum toxicity recovery processes in root apices. Possible association with oxidative stress. Plant Sci. 185–186, 1–8. doi: 10.1016/j.plantsci.2011.07.019
Nakano, Y., Asada, K. (1981). Hydrogen peroxide is pcavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol. 22, 867–880. doi: 10.1093/oxfordjournals.pcp.a076232
Osawa, H., Matsumoto, H. (2001). Possible involvement of protein phosphorylation in aluminum-responsive malate efflux from wheat root apex. Plant Physiol. 126, 411–420. doi: 10.1104/pp.126.1.411
Panda, S. K., Sahoo, L., Katsuhara, M., Matsumoto, H. (2013). Overexpression of alternative oxidase gene confers aluminum tolerance by altering the respiratory capacity and the response to oxidative stress in tobacco cells. Mol. Biotechnol. 54, 551–563. doi: 10.1007/s12033-012-9595-7
Rodriguez, R., Redman, R. (2005). Balancing the generation and elimination of reactive oxygen species. PNAS 102, 3175–3176. doi: 10.1073/pnas.0500367102
Sagi, M., Fluhr, R. (2001). Superoxide production by plant homologues of the gp91(phox) NADPH oxidase. Modulation of activity by calcium and by tobacco mosaic virus infection. Plant Physiol. 126, 1281–1290. doi: 10.1104/pp.126.3.1281
Sasaki, T., Yamamoto, Y., Ezaki, B., Katsuhara, M., Ahn, S. J., Ryan, P. R., et al. (2004). A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37, 645–653. doi: 10.1111/j.1365-313X.2003.01991.x
Sasaki, T., Ryan, P. R., Delhaize, E., Hebb, D. M., Ogihara, Y., Kawaura, K., et al. (2006). Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol. 47, 1343–1354. doi: 10.1093/pcp/pcl002
Šimonovičová, M., Huttová, J., Mistrík, I., Široká, B., Tamás, L. (2004). Root growth inhibition by aluminum is probably caused by cell death due to peroxidase-mediated hydrogen peroxide production. Protoplasma 224, 91–98. doi: 10.1007/s00709-004-0054-6
Sun, C., Liu, L., Zhou, W., Lu, L., Jin, C., Lin, X. (2017). Aluminum Induces Distinct Changes in the Metabolism of Reactive Oxygen and Nitrogen Species in the Roots of Two Wheat Genotypes with Different Aluminum Resistance. J. Age Food Chem. 65, 9419–9427. doi: 10.1021/acs.jafc.7b03386
Tamás, L., Huttova, J., Mistrik, I., Simonovicova, M., Siroka, B. (2006). Aluminium-induced drought and oxidative stress in barley roots. J. Plant Physiol. 163, 781–784. doi: 10.1016/j.jplph.2005.08.012
Taylor, G. J., McDonald-Stephens, J. L., Hunter, D. B., Bertsch, P. M., Elmore, D., Rengel, Z., et al. (2000). Direct Measurement of Aluminum Uptake and Distribution in Single Cells of. Chara Corallina Plant Physiol. 123, 987–996. doi: 10.1104/pp.123.3.987
Tsutsui, T., Yamaji, N., Ma, J. F. (2011). Identification of a cis-acting element of ART1, a C2H2-type zinc-finger transcription factor for aluminum tolerance in rice. Plant Physiol. 156, 925–931. doi: 10.1104/pp.111.175802
von Uexküll, H. R., Mutert, E. (1995). Global extent, development and economic-impact of acid soils. Plant Soil 171, 1–15. doi: 10.1007/bf00009558
Wang, Y., Li, R., Li, D., Jia, X., Zhou, D., Li, J., et al. (2017). NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, 5047–5052. doi: 10.1073/pnas.1618557114
Wu, D., Shen, H., Yokawa, K., Baluska, F. (2014). Alleviation of aluminium-induced cell rigidity by overexpression of OsPIN2 in rice roots. J. Exp. Bot. 65, 5305–5315. doi: 10.1093/jxb/eru292
Wu, Y. S., Yang, Z. L., How, J. Y., Xu, H. N., Chen, L. M., Li, K. Z. (2017). Overexpression of a peroxidase gene (AtPrx64) of Arabidopsis thaliana in tobacco improves plant’s tolerance to aluminum stress. Plant Mol. Biol. 95, 157–168. doi: 10.1007/s11103-017-0644-2
Xia, J., Yamaji, N., Ma, J. F. (2013). A plasma membrane-localized small peptide is involved in rice aluminum tolerance. Plant J. 76, 345–355. doi: 10.1111/tpj.12296
Xu, F. J., Li, G., Jin, C. W., Liu, W. J., Zhang, S. S., Zhang, Y. S., et al. (2012). Aluminum-induced changes in reactive oxygen species accumulation, lipid peroxidation and antioxidant capacity in wheat root tips. Biol. Plant 51, 89–96. doi: 10.1007/s10535-012-0021-6
Xu, L. M., Liu, W., Cui, B. M., Wang, N., Ding, J. Z., Liu, C., et al. (2017). Aluminium Tolerance Assessment of 141 Maize Germplasms in a Solution Culture. Universal J. Agric. Res. 5, 1–9. doi: 10.13189/ujar.2017.050101
Yamaji, N., Huang, C. F., Nagao, S., Yano, M., Sato, Y., Nagamura, Y., et al. (2009). A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell. 21, 3339–3349. doi: 10.1105/tpc.109.070771
Yamamoto, Y., Kobayashi, Y., Matsumoto, H. (2001). Lipid peroxidation is an early symptom triggered by aluminum, but not the primary cause of elongation inhibition in pea roots. Plant Physiol. 125, 199–208. doi: 10.1104/pp.125.1.199
Yamamoto, Y., Kobayashi, Y., Devi, S. R., Rikiishi, S., Matsumoto, H. (2003). Oxidative stress triggered by aluminum in plant roots. Plant Soil 255, 239–243. doi: 10.1023/A:1026127803156
Yu, Y., Zhou, W., Zhou, K., Liu, W., Liang, X., Chen, Y., et al. (2018). Polyamines modulate aluminum-induced oxidative stress differently by inducing or reducing H2O2 production in wheat. Chemosphere 212, 645–653. doi: 10.1016/j.chemosphere.2018.08.133
Zhang, Y. H., Wang, E. M., Zhao, T. F., Wang, Q. Q., Chen, L. J. (2018). Characteristics of Chlorophyll Fluorescence and Antioxidant-Oxidant Balance in PEPC and PPDK Transgenic Rice under Aluminum Stress. Russian J. Plant Physiol. 65, 49–56. doi: 10.1134/s1021443718010211
Zheng, X., Huystee, R. B. V. (1992). Peroxidase regulated elongation of segments from peanut hypocotyls. Plant Sci. 81, 47–56. doi: 10.1016/0168-9452(92)90023-F
Zheng, S. J., Yang, J. L. (2005). Target sites of aluminum phytotoxicity. Biol. Plant. 49, 321–331. doi: 10.1007/s10535-005-0001-1
Zhu, X. F., Shi, Y. Z., Lei, G. J., Fry, S. C., Zhang, B. C., Zhou, Y. H., et al. (2012). XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in. Arabidopsis Plant Cell. 24, 4731–4747. doi: 10.1105/tpc.112.106039
Zhu, X. F., Wan, J. X., Sun, Y., Shi, Y. Z., Braam, J., Li, G. X., et al. (2014). Xyloglucan Endotransglucosylase-Hydrolase17 Interacts with Xyloglucan Endotransglucosylase-Hydrolase31 to Confer Xyloglucan Endotransglucosylase Action and Affect Aluminum Sensitivity in Arabidopsis. Plant Physiol. 165, 1566–1574. doi: 10.1104/pp.114.243790
Keywords: aluminum toxicity, ZmAT6, maize, reactive oxygen species, antioxidant
Citation: Du H, Huang Y, Qu M, Li Y, Hu X, Yang W, Li H, He W, Ding J, Liu C, Gao S, Cao M, Lu Y and Zhang S (2020) A Maize ZmAT6 Gene Confers Aluminum Tolerance via Reactive Oxygen Species Scavenging. Front. Plant Sci. 11:1016. doi: 10.3389/fpls.2020.01016
Received: 24 April 2020; Accepted: 22 June 2020;
Published: 09 July 2020.
Edited by:
Rafaqat Ali Gill, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Hong Zhai, China Agricultural University, ChinaCopyright © 2020 Du, Huang, Qu, Li, Hu, Yang, Li, He, Ding, Liu, Gao, Cao, Lu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzhi Zhang, c3V6aGkxMDI2QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.