- Key Laboratory for Tree Breeding and Germplasm Improvement, Southern Modern Forestry Collaborative Innovation Center, College of Forestry, Nanjing Forestry University, Nanjing, China
Although the CRISPR/Cas9 system has been widely used for crop breeding, its application for the genetic improvement of trees has been limited, partly because of the outcrossing nature and substantial genomic heterozygosity of trees. Shanxin yang (Populus davidiana × P. bolleana), is a commercially important poplar clone that is widely grown in northern China. An established transformation protocol for this interspecific hybrid enables researchers to simultaneously investigate the efficiency and specificity of the CRISPR/Cas9-mediated manipulation of a highly heterozygous genome. Using the phytoene desaturase gene (PDS) as an example, we revealed that the CRISPR/Cas9 system could efficiently edit the Shanxin yang genome. Two sgRNAs were designed and incorporated into a single binary vector containing the Cas9 expression cassette. Among 62 independent transgenic lines, 85.5% exhibited an exclusively albino phenotype, revealing the total loss of PDS function. The Illumina sequencing results confirmed the targeted mutation of PdbPDS homologs induced by CRISPR/Cas9, and small insertions/deletions were the most common mutations. Biallelic and homozygous knockout mutations were detected at both target sites of the T0 transformants. Off-target activity was detected for sgRNA2 with a frequency of 3.2%. Additionally, the SNP interference of targeting specificity was assessed based on the sequence variation among PdbPDS homologs. A single mismatch at 19- or 10-bp from the PAM was tolerated by the CRISPR/Cas9 system. Therefore, multiple homologous genes were simultaneously edited despite the presence of a mismatch between the sgRNA and the target site. The establishment of a viable CRISPR/Cas9-based strategy for editing the Shanxin yang genome will not only accelerate the breeding process, but may also be relevant for other economically or scientifically important non-model plants species.
Introduction
The CRISPR/Cas9 system is a revolutionary technology for plant breeding because of its simplicity, efficiency, and versatility (Bortesi and Fischer, 2015; Gao, 2018). This system consists of two essential components, the Cas9 nuclease and a single guide RNA (sgRNA), which can induce double-strand breaks at specific genomic sites (Jinek et al., 2012). The cellular repair of CRISPR/Cas9-mediated double-strand breaks through non-homologous end joining may result in gene modifications (Bortesi and Fischer, 2015). The ability to reprogram CRISPR/Cas9 to target any gene of interest using an engineered sgRNA has enabled plant breeders to develop new varieties with desired characteristics, including higher yields, improved quality, and increased resistance to disease and climate change. For example, CRISPR/Cas9-mediated inactivation of OsGW5 in rice significantly increased the grain width and weight (Liu et al., 2017); low-gluten wheat was created by modifying the α-gliadin gene array (Sanchez-Leon et al., 2018); and the CRISPR-mediated genomic deletion of the SlMlo1 locus generated tomato plants resistant to powdery mildew (Nekrasov et al., 2017).
Between 2013 and 2019, CRISPR/Cas9 technology has been successfully used to modify plant species from 45 genera across 24 families (Shan et al., 2020). However, the application of the CRISPR/Cas9 system to genetically alter woody perennials has been limited to only the following 10 species: apple (Nishitani et al., 2016), cacao (Fister et al., 2018), cassava (Odipio et al., 2017), citrus (Jia et al., 2017), coffee (Breitler et al., 2018), grape (Nakajima et al., 2017), kiwifruit (Wang et al., 2018), Parasponia andersonii (Van Zeijl et al., 2018), pomegranate (Chang et al., 2019), and poplar (Fan et al., 2015; Zhou et al., 2015). Recalcitrance to transformation and/or difficulties in regeneration are two predominant issues preventing the more widespread application of CRISPR in woody plants (Bewg et al., 2018). Moreover, their outcrossing nature and highly heterozygous genomes represent additional challenges to the genetic editing of tree species. The considerable abundance of single nucleotide polymorphisms (SNPs) in outcrossing species may restrict or even abolish CRISPR/Cas9-mediated genome editing (Jinek et al., 2012). In Populus species, one SNP altering the protospacer adjacent motif (PAM) site from NGG to NGA insulated the genome completely from editing (Zhou et al., 2015).
Shanxin yang (Populus davidiana × P. bolleana) is an elite aspen hybrid poplar bred for its cold and drought tolerance (Li et al., 2005). Because of its rapid growth, narrow crown, and stress resistance, Shanxin yang is an economically important cultivar that is widely grown in the northern provinces of China. Furthermore, an efficient transformation and regeneration system has been established for this hybrid poplar (Wang et al., 2011). Thus, many economically valuable traits can be improved via genetic modification. In this study, using the phytoene desaturase gene (PDS) as an example, we demonstrate the highly efficient editing of the Shanxin yang genome via a CRISPR/Cas9 system. Additionally, with the similar but not identical sequences of PdbPDS gene fragments, the effects of SNPs on genome editing efficiency were investigated. The establishment of a CRISPR/Cas9-based method for genetically manipulating in Shanxin yang may greatly facilitate the breeding of improved poplar cultivars. Moreover, the practical approach presented herein may be applicable for other valuable woody perennials that have highly heterozygous genomes but limited genomic resources.
Materials and Methods
Cloning of PdbPDS Fragments and Selection of CRISPR/Cas9 Target Sites
Genomic DNA was extracted from young leaves according to a modified CTAB (cetyltrimethyl ammonium bromide) method (Porebski et al., 1997). On the basis of the conserved sequence in two Populus trichocarpa PDS genes, primers F1 (5’-GTTGAATTTGGTTTTGGAGAA-3’) and R1 (5’- CATCTCTTGCTTCAAGCAATA-3’) were designed to amplify the 5’ end fragments of PDS genes in Shanxin yang. The PCR amplification was completed with KOD DNA polymerase (Toyobo, Japan) in a total volume of 25 μL, with the following reaction conditions: 94°C for 3 min; 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 50 s; and 72°C for 10 min. The PCR products were purified with the AxyPrep™ DNA Gel Extraction Kit (Axygen Scientific, USA), cloned into the pEASY-Blunt vector (Transgen Biotech, China), and then analyzed by Sanger sequencing (Sangon Biotech, China). All possible Cas9 target sites within the obtained sequences were identified with the online software CRISPR-GE (http://skl.scau.edu.cn/, Xie et al., 2017). Two target sites were selected based on their genomic locations, GC contents, and potential off-target scores.
Vector Construction and Plant Transformation
A gene-editing vector for PdbPDS expressing two sgRNAs was constructed as described by Xing et al. (2014). Briefly, the two target sites were incorporated into the following four primers: DT1-BsF: 5′-ATATATGGTCTCGATTGTGAGTGCATTCAACTTGAGCGTT-3′, 5′-DT1-F0: TG TGAGTGCATTCAACTTGAGCGTTTTAGAGCTAGAAATAGC-3′, DT2-R0: 5′-AAC AGACCGGACCTTGATAACACAATCTCTTAGTCGACTCTAC-3′, and DT2-BsR: 5′-ATTATTGGTCTCGAAACAGACCGGACCTTGATAACACAA-3′. The italicized letters represent the target sequence. With these four primers, a PCR fragment (626 bp) was amplified from pCBC-DT1T2 and then incorporated between the BsaI sites of pHSE401 by Golden Gate cloning. The resulting pHSE401-2gR-PdbPDS recombinant plasmid was inserted into Agrobacterium tumefaciens (strain EHA105) cells according to a freeze–thaw method (Chen et al., 1994). The transformed clones with the correct sequence were identified by colony PCR and sequencing.
Shanxin yang seedlings were transformed with a slightly modified version of the method described by Wang et al. (2011). Specifically, leaf disks excised from aseptic seedlings were inoculated by swirling in the A. tumefaciens cultures for approximately 20 min. After the co-cultivation, the infected explants were washed several times with sterilized distilled water, and then pre-cultivated for one week on medium containing 200 mg/L cefotaxime and 150 mg/L timentin. The explants were then transferred to the shoot induction medium supplemented with 200 mg/L cefotaxime, 150 mg/L timentin, and 10mg/L hygromycin (Hyg). After the Hyg-resistant shoots were approximately 1.5 cm long, they were transferred to the rooting medium.
Detection of Mutations
Genomic DNA was extracted from the leaves of each transgenic line and used as the template to amplify a Hyg gene (1,150 bp) with primers HygF (5′- CTACAAATCTATCTCTCTCG-3′) and HygR (5′- TATCTGGGAACTACTCACAC-3′) and the following PCR conditions: 94°C for 3 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 70 s; and 72°C for 5 min. The PCR products were visualized on 1% agarose gels stained with GelRed (Generay, China). All of the Hyg-positive DNA samples as well as two wild-type (WT) DNA samples were used as templates to amplify the fragment spanning two target sites. The first round of PCR amplification was completed with primers F2 (5′-TTGGTTTTGGAGAAATGAGT-3′) and R2 (5′-GAAGAACGAAAGGATGAAGA-3′), and the following PCR conditions: 94°C for 3 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s; and 72°C for 5 min. The amplification was verified by agarose gel electrophoresis. During the second round of PCR amplification, 12 barcoded forward primers and eight barcoded reverse primers were used to add a unique barcode to each sample. The PCR conditions were as follows: 98°C for 2 min; 30 cycles of 98°C for 10 s, 60°C for 15 s, and 72°C for 5 s; and 72°C for 5 min. Three indexed PCR-free libraries (each containing 32 samples) were then prepared with the TruSeq DNA Sample Preparation Kit (Illumina, USA). The libraries were sequenced with the Illumina HiSeq 2000 system.
Based on the barcode sequence, reads of each transgenic line were separated using Perl script. Raw reads were trimmed for quality using prinseq v0.20.4 (Schmieder and Edwards, 2011), and paired-reads were assembled using PANDAseq (Masella et al., 2012). After barcode trimming, representative sequences that have a frequency higher than 5% within a given sample were retained and aligned using DNAMAN v.8 (Lynnon Corporation).
Results
Detection of PDS Genes in Shanxin Yang and Selection of sgRNA Targets
The AtPDS3 protein sequence was used to identify two homologous genes (Potri.014G148700 and Potri.002G235200) in the P. trichocarpa genome (version 3.1, https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Ptrichocarpa_er). These genes shared a sequence identity of 92.01% at the amino acid level. The intra-genome synteny data in the Plant Genome Duplication Database (Lee et al., 2012) indicate these two PtPDS genes are paralogs that arose from segmental or whole-genome duplication events. To detect the PDS genes in Shanxin yang, a primer pair (F1 and R1) targeting a conserved sequence between the two PtPDS genes was designed to flank the first three exons (Figure 1A). Unexpectedly, two PCR products were amplified, one with the expected size of 730 bp, and another approximately 100 bp smaller (Figure 1B). Both amplified products were purified from agarose gels, cloned, and sequenced. A sequence alignment revealed that the longer product included two SNP-containing allelic fragments (PdbPDS1-1 and PdbPDS1-2), which showed a higher similarity with Potri.014G148700 (Figure 1C). The shorter product comprised only one amplicon (PdbPDS2), which displayed a higher identity with Potri.002G235200. A comparison with Potri.002G235200 uncovered a 114 bp deletion in the first putative exon of PdbPDS2 (Figure 1D). Because Shanxin yang is a diploid species, the unique PdbPDS2 sequence suggests that the two alleles of this gene might be homozygous within the amplified fragment (PdbPDS2-1 and PdbPDS2-2).
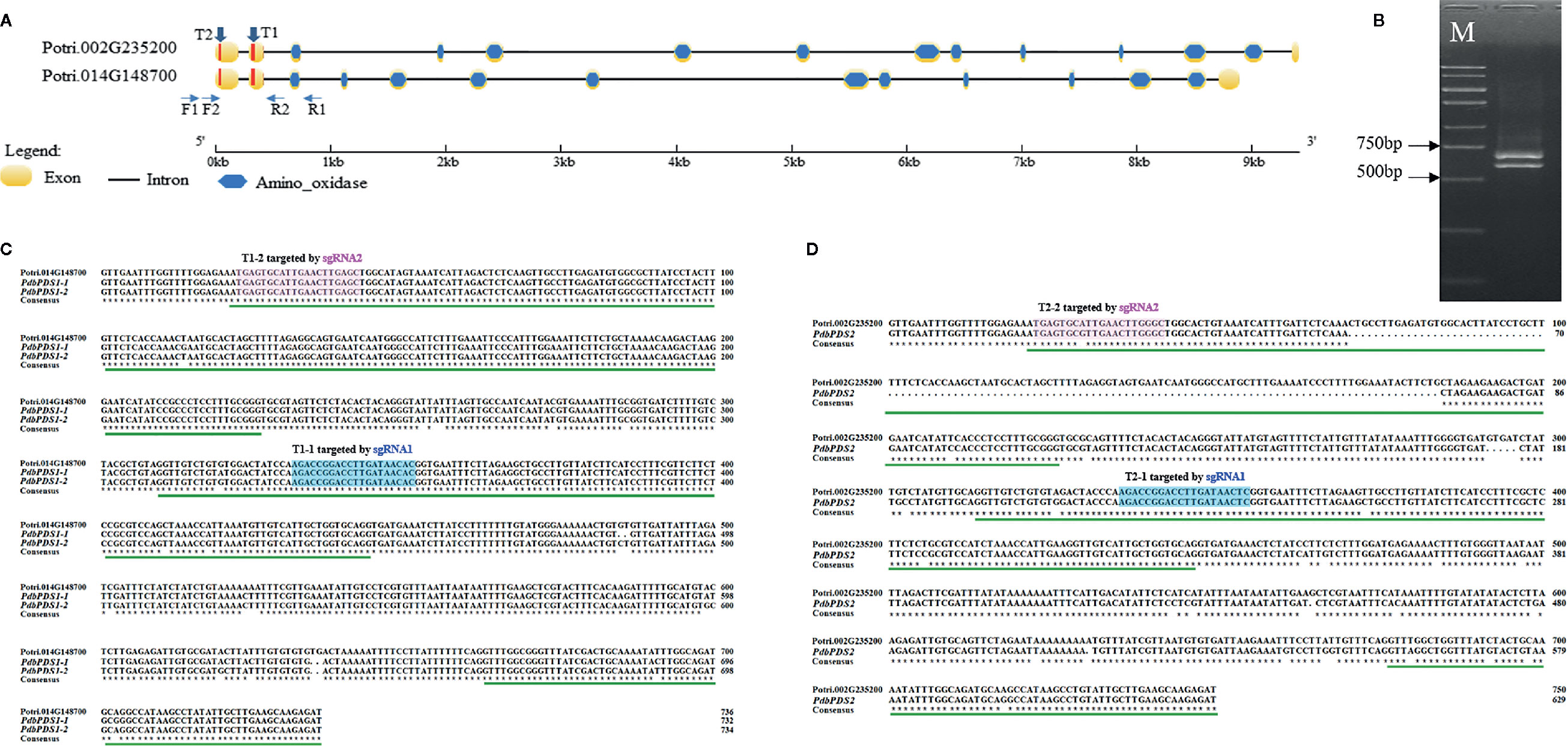
Figure 1 Targets locations and sequence alignments. (A) Structures of PtPDS genes in P. trichocarpa. T1 and T2 indicate the targets locations. F1 and R1 were used to amplify Shanxin yang PDS genes fragments. F2 and R2 were used to amplify the target region. (B) PCR amplification of PdbPDS genes fragments. (C) Alignment of two PdbPDS1 allele fragments and Potri.014G148700. (D) Alignment of one PdbPDS2 allele fragment and Potri.002G235200. In (C, D), the green lines below aligned sequences indicate predicted exons within the amplified fragments; and the blue and pink boxes represent potential target sites that may be bind by sgRNA1 and sgRNA2, respectively. The first number in each target name represents the gene number, while the second number represents the number of sgRNA, e.g., T1-1 means the target site in PdbPDS1 that can be bounded by sgRNA1.
On the basis of the allelic variation of PdbPDS1, two sgRNAs (sgRNA1 and sgRNA2) were designed to target two conserved sites located (Figure 1A). Candidate targets were also identified in PdbPDS2. The sgRNA1 might bind to T1-1 and T2-1, which were located in the second exons of two alleles of two genes, respectively (blue boxes in Figures 1C, D); and the sgRNA2 might bind to T1-2 and T2-2, which were located in the first exons of two alleles of two genes (pink boxes in Figures 1C, D). The sgRNA1 sequence perfectly matched the T1-1 (Figure 2A), while the T2-1 harbored a SNP 19 bp away from the PAM (Figure 2B). Therefore, the sgRNA1 had two perfectly matched sites (T1-1) and two single-mismatched sites (T2-1). To assess the SNP interference of sgRNA specificity at different positions, a mismatch was introduced into the sgRNA2 sequence (G replaced with C) 9 bp from the PAM (Figure 2C). Additionally, two naturally existing SNPs were detected in the corresponding site of PdbPDS2 (T2-2, Figure 2D). Thus, the sgRNA2 had two imperfect target sites (T1-2, 1 bp mismatch) and two potential off-target sites (T2-2, 3 bp mismatch). A single binary vector containing two sgRNA expression cassettes was constructed, in which sgRNA1 and 2 were expressed under the control of U6-29P and U6-26P, respectively (Figure 2E).
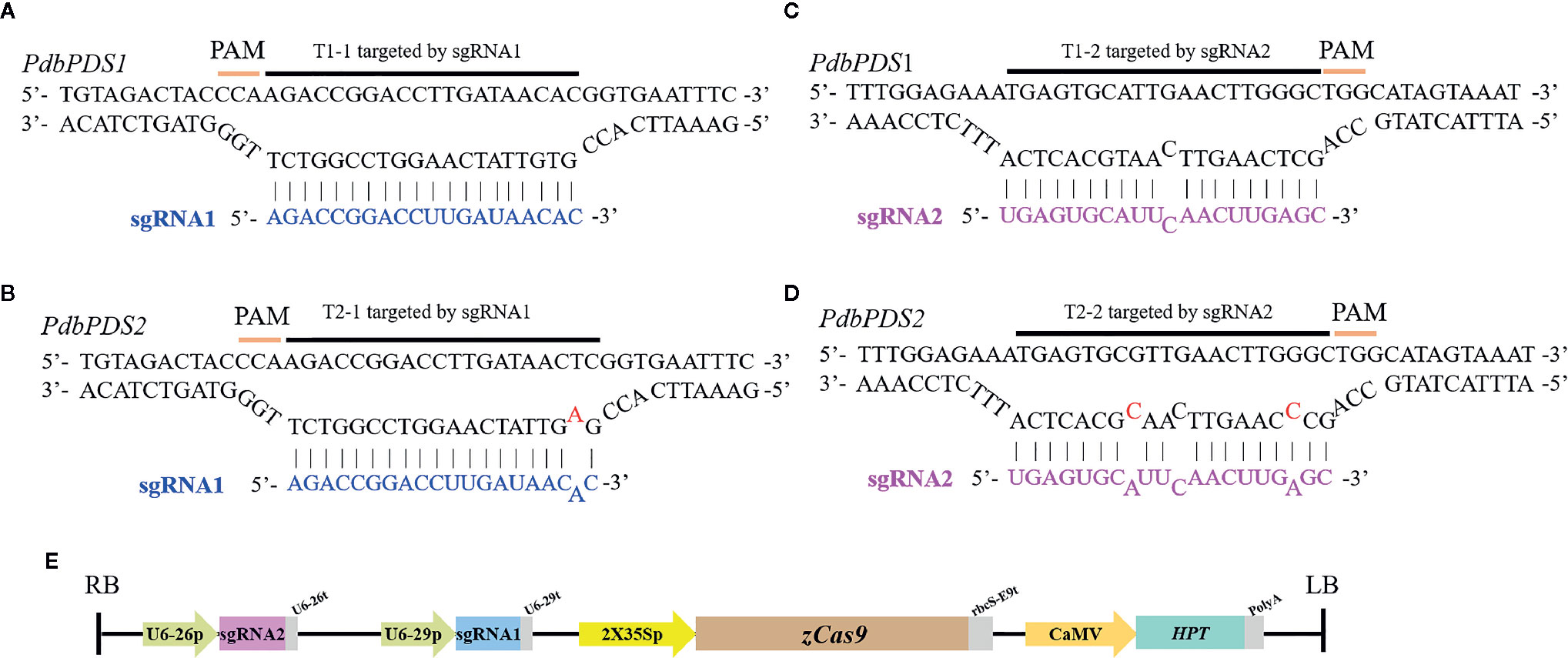
Figure 2 Schematic diagram of the complementarity between the sgRNAs and target sites, as well as the vector construction. (A) Perfect complementarity between sgRNA1 and T1-1. (B) Incomplete base-pairing between sgRNA1 and T2-1. (C) Incomplete base-pairing between sgRNA2 and T1-2. (D) Three mismatches in the putative off-target site T2-2. (E) T-DNA region of the constructed gene editing vector. The abbreviations are referred to Xing et al. (2014). U6-26p and U6-29p, two Arabidopsis U6 gene promoters; U6-26t and U6-29t, two U6-26 terminators with downstream sequence; 2x35Sp, 2x35S promoter; zCas9, Zea mays codon-optimized Cas9; rbcS-E9t, rbcS E9 terminator; CaMV, CaMV 35S promoter; Hpt, hygromycin phosphotransferase; PolyA, CaMV polyA signal. The sequence identity between 2x35Sp and CaMV is 94.55%.
Phenotypes of CRISPR/Cas9 Transgenic Poplar
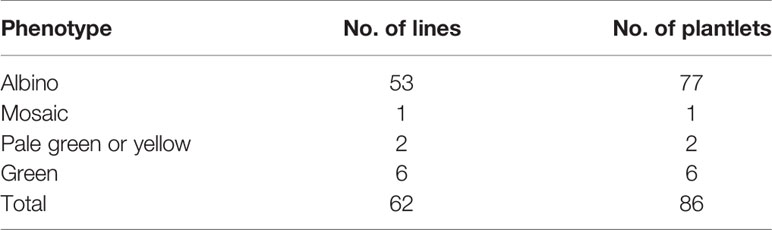
The pHSE401-2gR-PdbPDS recombinant plasmid containing the Cas9 expression cassette, two sgRNA cassettes, and the hygromycin phosphotransferase (HPT) gene (selection marker) was transformed into Shanxin yang leaf disks via Agrobacterium-mediated transformation. Approximately two months later, Hyg-resistant shoot buds were regenerated with the following four phenotypes: total albino (Figure 3A), variegated with both albino and green leaves (Figure 3B), pale green (Figure 3C), or normal green (i.e., same as the WT) (Figure 3D). The regenerated buds were transferred to the selective rooting medium. Finally, we obtained 84 putative transgenic plantlets from 62 independent lines (Table 1). A PCR analysis revealed that the HPT gene was present in all of the T0 seedlings (Figure S1).

Figure 3 Representative Shanxin yang phenotypes resulting from PDS gene mutations. Total albino (A), variegated (B), and pale green (C) phenotypes of the regenerated shoots. (D) Wild-type (WT) Shanxin yang shoots. (E) Comparison between the 1-month-old albino and WT seedlings.
Among the 62 transgenic lines, 53 (85.5%) exhibited an exclusively albino phenotype, reflecting the total loss of PDS function. Additionally, one line exhibited a mosaic phenotype, whereas two lines were pale green or yellow (Table 1). The remaining six lines were phenotypically the same as the WT control. The albino plantlets were weak and small (Figure 3E), and died during acclimatization.
Characterization of the Targeted Editing of the PdbPDS Genes
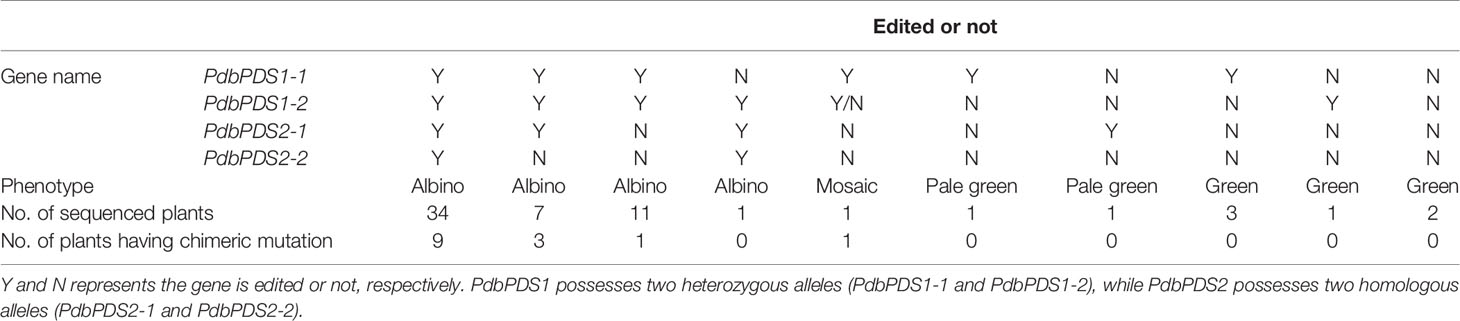
With the F2/R2 primer pair, DNA segments spanning the two targets were amplified for 62 transgenic plants representing the 62 different lines (Table 1). Each PCR product comprised three different fragments, which were derived from PdbPDS1-1, PdbPDS1-2, and PdbPDS2. Amplicons of each sample were uniquely barcoded and subjected to Illumina sequencing. Mutations to at least one target site were detected in 60 of the 62 analyzed plants (i.e., 96.8%) (Table S1). The transgenic plants exhibited an albino phenotype when more than two alleles of the two genes was edited, whereas they were pale green or normal green when only one gene was mutated (Table 2). The mosaic plant contained chimeric mutations, in which five distinct PdbPDS1 alleles were detected (Table S1). Moreover, chimeric mutations were also identified in 13 albino plants, each of which had at least three different PdbPDS1 alleles (Table 2 and Table S1). Therefore, chimerism occurred in the transformants at a rate of about 23.3%.
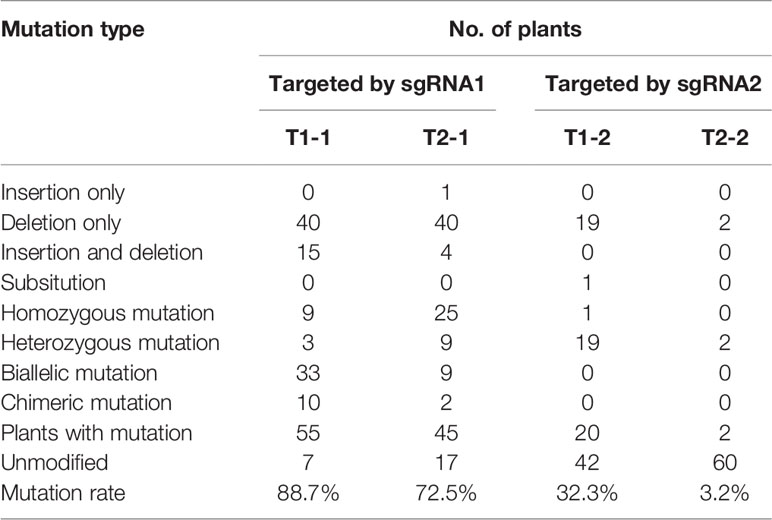
A total of 119 gene-editing events were identified in 60 plants with PDS mutations (Table S1). The mutation type and mutagenesis rate were analyzed in detail for every target site. Small insertions/deletions (indels) were prevalent among the editing events (Table 3), with eight, five, and two different types of indels in T1-1, T2-1, and T1-2, respectively (Figure 4). The analyses of T1-1 and T1-2 in PdbPDS1 revealed mutations in at least one allele in 55 and 20 plants, respectively (Table 3). Of the 55 plants, 42 had mutations in both alleles (76.4%), including nine homozygous mutations and 33 biallelic mutations (Table 3), whereas among the 20 plants, only one had mutations at T1-2 in both alleles (Table 3). Notably, 12 plants with modifications to both T1-1 and T1-2 in PdbPDS1-1 and four plants with four altered targets in two PdbPDS1 alleles were identified (Table S1), but no large deletion or insertion was detected, suggesting that Cas9 cleaved the two targets in one gene sequentially rather than simultaneously. Regarding the analyses of T2-1 in PdbPDS2, mutations in at least one allele were detected in 45 plants, of which 34 had mutations in both alleles (75.6%), including 25 homozygous mutations and nine biallelic mutations (Table 3).

Figure 4 Targeted mutations of PdbPDS genes induced by CRISPR/Cas9. The top row of each panel is the WT sequence of the given sgRNA, and the protospacer adjacent motif (PAM) is underlined. Deletions are indicated by red dashes. Insertions and substitutions are indicated by red letters. The number in brackets represents the number of plants with the specific mutant allele.
The sgRNA1 was intended to two alleles of two PdbPDS genes (T1-1 and T2-1). Among the 46 plants containing mutations introduced by sgRNA1, 42 (i.e., nearly 90%) had mutations in more than one gene and 27 (i.e., 43.5%) had mutations in all four genes (Table S2). Moreover, the editing rate of sgRNA1 at T1-1 was 88.7%, but at T2-1, the rate decreased to 72.5% (Table 3) because of the presence of a SNP in the T2-1 sequence (Figure 2B). Although the single-base mismatch was distal to the PAM site, it significantly decreased the editing efficiency of sgRNA1 (P < 0.05, χ2 test). Compared with the high efficiency of sgRNA1, the editing rate of sgRNA2 at T1-2 was only 32.2%, which can be partly explained by the mismatch in the middle of the target site (Figure 2C) as well as the relatively low efficiency of sgRNA2 itself. Additionally, T2-2 was predicted to be a potential off-target site, harboring three mismatches (Figure 2D). Among the 62 sequenced plants, two had mutations at this site (i.e., 3.2%) (Figure 4).
Discussion
Shanxin yang is a typical interspecific outcrossing woody perennial species with a highly heterozygous genome. Using a pair of conserved primers, we amplified three different PDS genes sequences, including two haplotypes of PdbPDS1 and one haplotype of PdbPDS2. PdbPDS1 is a heterozygous locus, while PdbPDS2 can be either hemizygous or homozygous. If PdbPDS2 only presents on one of the chromatids (hemizygous), the wild-type allele cannot be detected in the PdbPDS2 edited plants unless the edited line is a chimeric plant. We obtained nine lines showing heterozygous mutations in PdbPDS2, i.e., both the edited and unedited PdbPDS2 were identified in these lines (Table 3). In the nine PdbPDS2 edited lines, PdbPDS1 was edited simultaneously. Based on the editing results of PdbPDS1, seven of the nine edited plants are not chimeras. Therefore, PdbPDS2 should possess two homologous alleles in Shanxin yang. According to the gene editing results (Table 2), PdbPDS1 and PdbPDS2 may be both functional, and they regulate the chlorophyll biosynthesis in a dose-dependent manner. If more than two alleles of the two genes were edited, the transgenic plants appeared albino.
Two sgRNAs were designed to evaluate the efficiency and specificity of the CRISPR/Cas9-mediated genomic mutations in Shanxin yang. Our data revealed that sgRNA1 was able to simultaneously edit multiple genes, and 43.5% of the transgenic plants had mutations at all four target sites (Table S2). Multigene editing via CRISPR/Cas9 guided by a single sgRNA has been reported for many plant species (Lawrenson et al., 2015; Braatz et al., 2017; Gao et al., 2017; Yu et al., 2018), but the efficiencies of the multigene mutations were not always provided. In a previous study of tetraploid potato (Solanum tuberosum), only 2% of the regenerated lines had mutations in all four alleles (Andersson et al., 2017). Additionally, Zhang et al. (2016) reported that 10% of 80 T0 mutants had all six alleles simultaneously knocked out in hexaploid wheat (Triticum aestivum L.). Compared with strategies involving the expression of multiple sgRNAs for multiplex genome editing (Minkenberg et al., 2017), the use of a single sgRNA has the advantage of minimizing the risk of off-target mutations (Yu et al., 2018).
Notably, sgRNA1 is identical to the target region in PdbPDS1, whereas it differs from the target in PdbPDS2 because of a SNP 19bp from the PAM. However, homozygous and biallelic mutations in PdbPDS2 were still frequently detected (Table 3). This observation is consistent with the results of earlier studies in which genes with highly similar but not identical sequences were edited simultaneously by a single minimally stringent sgRNA (Lawrenson et al., 2015; Endo et al., 2016; Andersson et al., 2017; Braatz et al., 2017; Gao et al., 2017; Kanazashi et al., 2018). For example, in rice (Oryza sativa), three CDK genes can be mutated by one sgRNA even with an imperfect target match (Endo et al., 2016). In allotetraploid cotton (Gossypium hirsutum L.), an sgRNA with a mismatch 12 bp upstream of the PAM reportedly can mutate in all four GhCLA1 homoeoalleles (Gao et al., 2017). Moreover, the mutation of three or four closely related genes with one incompletely matched sgRNA has been reported for Brassica oleracea and soybean (Glycine max) (Lawrenson et al., 2015; Kanazashi et al., 2018). These results suggest that although site-specific mutagenesis is highly desirable and necessary for characterizing gene functions, the tolerance of mismatches (as few as one or two nucleotide differences) in the target region can be exploited for multi-homologous or multi-paralogous gene editing, especially for polyploid plants.
The 8–12 nucleotides sequence upstream of the PAM is the seed region critical for target affinity and specificity (Semenova et al., 2011; Wiedenheft et al., 2011; Belhaj et al., 2015). Previous studies proved that a single-base mismatch within the 12 bp adjacent to the PAM can prevent the genomic cleavage by Cas9 (Sapranauskas et al., 2011; Jinek et al., 2012; Cong et al., 2013). In the current study, the mismatch in sgRNA2 was 10 bp from the PAM, but 32.2% of the transgenic plants had mutations at the target site (T1-2). This result is in accordance with the findings of Hinz et al. (2016), who demonstrated that a mismatch at a position +10 nucleotides from the PAM minimally affects Cas9 activity in vitro. Similarly, in another study, the sgRNA targeting four BnALC alleles in oilseed rape (Brassica napus) contained a SNP 10 nucleotides upstream of the PAM (Braatz et al., 2017). In contrast, the inhibitory effect of a single SNP on gene editing efficiency was observed in an earlier investigation of poplar [P. tremula × alba (717-1B4)], in which one SNP near or within the PAM prevented the Cas9-mediated cleavage (Zhou et al., 2015). These results support the previous finding that a SNP located distally from the PAM often has a limited effect on Cas9 activity (Jinek et al., 2012; Pattanayak et al., 2013). However, these studies collectively suggest that the length of the seed region varies among different systems, and more work is needed to elucidate the effects of SNPs on CRISPR/Cas9-mediated genome editing.
Off-target activity is another major concern of the CRISPR/Cas9 system. In this study, T2-2 was a putative off-target site of sgRNA2 in PbdPDS2. Although T2-2 has three mismatches with the sgRNAs, of the 62 transgenic plants, two (3.2%) had off-target mutations at this site. A high-frequency of off-target mutations has been reported for human cells and mouse embryos (Fu et al., 2013; Aryal et al., 2018). In plants, off-target mutagenesis was rare in some studies (Bilichak and Eudes, 2016; Wolt et al., 2016; Li et al., 2019), but was detected at frequencies of 1.6–13% in other studies (Upadhyay et al., 2013; Zhang et al., 2014; Jacobs et al., 2015; Sauer et al., 2016). The potential high frequency of off-target mutations in Arabidopsis thaliana was recently revealed by Zhang et al. (2018). Moreover, the frequency of off-target mutations apparently increases in the T2 progeny (Zhang et al., 2018). Therefore, special attention should be paid to limit the risk of off-target mutation, especially for perennial plants, such as trees, since the CRISPR/Cas9 system may function for many months or even years in these plant species.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
JW performed the experiments and prepared the manuscript. HW helped in analyzing the sequencing data. YC and TY designed the study and helped to draft the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Jianhua Wei (Forestry Research Institute of the Chinese Academy of Forestry) for providing the Shanxin yang clone, and Prof. Qijun Chen (China Agricultural University) for providing the CRISPR/Cas9 binary vector pHSE401 and the plasmid pCBC-DT1T2. This study was funded by the National Key Research and Development Plan of China (2016YFD0600101), the National Natural Science Foundation of China (31800562). It was also supported by the Qing Lan Talent Support Program at Jiangsu Province and the Youth Elite Science Sponsorship Program by CAST (YESS20160121).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00996/full#supplementary-material
References
Andersson, M., Turesson, H., Nicolia, A., Fält, A. S., Samuelsson, M., Hofvander, P. (2017). Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 36 (1), 117–128. doi: 10.1007/s00299-016-2062-3
Aryal, N. K., Wasylishen, A. R., Lozano, G. (2018). CRISPR/Cas9 can mediate high-efficiency off-target mutations in mice in vivo. Cell Death Dis. 9 (11), 1–3. doi: 10.1038/s41419-018-1146-0
Belhaj, K., Chaparro-Garcia, A., Kamoun, S., Patron, N. J., Nekrasov, V. (2015). Editing plant genomes with CRISPR/Cas9. Current opinion in biotechnology. 32, 76–84. doi: 10.1016/j.copbio.2014.11.007
Bewg, W. P., Ci, D., Tsai, C. J. (2018). Genome editing in trees: from multiple repair pathways to long-term stability. Frontiers in plant science. 9, 1732. doi: 10.3389/fpls.2018.01732
Bilichak, A., Eudes, F. (2016). “Genetic engineering of plants using Zn fingers, TALENs, and CRISPRs,” in Genome stability. Eds. Kovalchuk, I., Kovalchuk, O. (New York, NY: Cambridge, Academic Press), 187–201.
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 (1), 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Braatz, J., Harloff, H. J., Mascher, M., Stein, N., Himmelbach, A., Jung, C. (2017). CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174 (2), 935–942. doi: 10.1104/pp.17.00426
Breitler, J. C., Dechamp, E., Campa, C., Rodrigues, L. A. Z., Guyot, R., Marraccini, P., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis has the potential to accelerate the domestication of Coffea canephora. Plant Cell. Tissue Organ Cult. (PCTOC) 134 (3), 383–394. doi: 10.1007/s11240-018-1429-2
Chang, L., Wu, S., Tian, L. (2019). Effective genome editing and identification of a regiospecific gallic acid 4-O-glycosyltransferase in pomegranate (Punica granatum L.). Horticult. Res. 6 (1), 1–15. doi: 10.1038/s41438-019-0206-7
Chen, H., Nelson, R. S., Sherwood, J. L. (1994). Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 16 (4), 664–668.
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339 (6121), 819–823. doi: 10.1126/science.1231143
Endo, A., Masafumi, M., Kaya, H., Toki, S. (2016). Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci. Rep. 6, 38169. doi: 10.1038/srep38169
Fan, D., Liu, T., Li, C., Jiao, B., Li, S., Hou, Y., et al. (2015). Efficient CRISPR/Cas9-mediated targeted mutagenesis in Populus in the first generation. Sci. Rep. 5, 12217. doi: 10.1038/srep12217
Fister, A. S., Landherr, L., Maximova, S. N., Guiltinan, M. J. (2018). Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 9, 268. doi: 10.3389/fpls.2018.00268
Fu, Y., Foden, J. A., Khayter, C., Maeder, M. L., Reyon, D., Joung, J. K., et al. (2013). High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 31 (9), 822–826. doi: 10.1038/nbt.2623
Gao, W., Long, L., Tian, X., Xu, F., Liu, J., Singh, P. K., et al. (2017). Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 8, 1364. doi: 10.3389/fpls.2017.01364
Gao, C. (2018). The future of CRISPR technologies in agriculture. Nat. Rev. Mol. Cell Biol. 19 (5), 275–276. doi: 10.1038/nrm.2018.2
Hinz, J. M., Laughery, M. F., Wyrick, J. J. (2016). Nucleosomes selectively inhibit Cas9 off-target activity at a site located at the nucleosome edge. J. Biol. Chem. 291 (48), 24851–24856. doi: 10.1074/jbc.c116.758706
Jacobs, T. B., LaFayette, P. R., Schmitz, R. J., Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15 (1), 16. doi: 10.1186/s12896-015-0131-2
Jia, H., Xu, J., Orbović, V., Zhang, Y., Wang, N. (2017). Editing citrus genome via SaCas9/sgRNA system. Front. Plant Sci. 8, 2135. doi: 10.3389/fpls.2017.02135
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., Charpentier, E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337 (6096), 816–821. doi: 10.1126/science.1225829
Kanazashi, Y., Hirose, A., Takahashi, I., Mikami, M., Endo, M., Hirose, S., et al. (2018). Simultaneous site-directed mutagenesis of duplicated loci in soybean using a single guide RNA. Plant Cell Rep. 37 (3), 553–563. doi: 10.1007/s00299-018-2251-3
Lawrenson, T., Shorinola, O., Stacey, N., Li, C., Østergaard, L., Patron, N., et al. (2015). Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 16 (1), 258. doi: 10.1186/s13059-015-0826-7
Lee, T. H., Tang, H., Wang, X., Paterson, A. H. (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41 (D1), D1152–D1158. doi: 10.1093/nar/gks1104
Li, Z., Dong, Y., Zhao, L. (2005). Cuttage seedling-raising technique of Populus davidiana× bolleana. J. Northeast For. Univ. 33 (4), 100–101. doi: 10.13759/j.cnki.dlxb.2005.04.036
Li, J., Manghwar, H., Sun, L., Wang, P., Wang, G., Sheng, H., et al. (2019). Whole genome sequencing reveals rare off-target mutations and considerable inherent genetic or/and somaclonal variations in CRISPR/Cas9-edited cotton plants. Plant Biotechnol. J. 17 (5), 858–868. doi: 10.1111/pbi.13020
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3 (5), 1–7. doi: 10.1038/nplants.2017.43
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., Neufeld, J. D. (2012). PANDAseq: paired-end assembler for illumina sequences. BMC Bioinf. 13 (1), 31. doi: 10.1186/1471-2105-13-31
Minkenberg, B., Wheatley, M., Yang, Y. (2017). “CRISPR/Cas9-enabled multiplex genome editing and its application,” in Progress in molecular biology and translational science, vol. 149. (New York, NY: Cambridge, Academic Press), 111–132. doi: 10.1016/bs.pmbts.2017.05.003
Nakajima, I., Ban, Y., Azuma, A., Onoue, N., Moriguchi, T., Yamamoto, T., et al. (2017). CRISPR/Cas9-mediated targeted mutagenesis in grape. PloS One 12 (5), e0177966. doi: 10.1371/journal.pone.0177966
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7 (1), 1–6. doi: 10.1038/s41598-017-00578-x
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6 (1), 1–8. doi: 10.1038/srep31481
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 8, 1780. doi: 10.3389/fpls.2017.01780
Pattanayak, V., Lin, S., Guilinger, J. P., Ma, E., Doudna, J. A., Liu, D. R. (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 31 (9), 839. doi: 10.1038/nbt.2673
Porebski, S., Bailey, L. G., Baum, B. R. (1997). Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Rep. 15 (1), 8–15. doi: 10.1007/bf02772108
Sanchez-Leon, S., Gil-Humanes, J., Ozuna, C. V., Gimenez, M. J., Sousa, C., Voytas, D. F., et al. (2018). Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16 (4), 902–910. doi: 10.1111/pbi.12837
Sapranauskas, R., Gasiunas, G., Fremaux, C., Barrangou, R., Horvath, P., Siksnys, V. (2011). The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 39 (21), 9275–9282. doi: 10.1093/nar/gkr606
Sauer, N. J., Narvaez-Vasquez, J., Mozoruk, J., Miller, R. B., Warburg, Z. J., Woodward, M. J., et al. (2016). Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 170 (4), 1917–1928. doi: 10.1104/pp.15.01696
Schmieder, R., Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27 (6), 863–864. doi: 10.1093/bioinformatics/btr026
Semenova, E., Jore, M. M., Datsenko, K. A., Semenova, A., Westra, E. R., Wanner, B., et al. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. 108 (25), 10098–10103. doi: 10.1073/pnas.1104144108
Shan, S., Soltis, P. S., Soltis, D. E., Yang, B. (2020). Considerations in adapting CRISPR/Cas9 in nongenetic model plant systems. Appl. Plant Sci., 8 (1), e11314. doi: 10.1002/aps3.11314
Upadhyay, S. K., Kumar, J., Alok, A., Tuli, R. (2013). RNA-guided genome editing for target gene mutations in wheat. G3: Genes Genom. Genet. 3 (12), 2233–2238. doi: 10.1534/g3.113.008847
Van Zeijl, A., Wardhani, T. A., Seifi Kalhor, M., Rutten, L., Bu, F., Hartog, M., et al. (2018). CRISPR/Cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Front. Plant Sci. 9, 284. doi: 10.3389/fpls.2018.00284
Wang, H., Wang, C., Liu, H., Tang, R., Zhang, H. (2011). An efficient Agrobacterium-mediated transformation and regeneration system for leaf explants of two elite aspen hybrid clones Populus alba× P. berolinensis and Populus davidiana× P. bolleana. Plant Cell Rep. 30 (11), 2037–2044. doi: 10.1007/s00299-011-1111-1
Wang, X., Tu, M., Wang, D., Liu, J., Li, Y., Li, Z., et al. (2018). CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 16 (4), 844–855. doi: 10.1111/pbi.12832
Wiedenheft, B., van Duijn, E., Bultema, J. B., Waghmare, S. P., Zhou, K., Barendregt, A., et al. (2011). RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. 108 (25), 10092–10097. doi: 10.1073/pnas.1102716108
Wolt, J. D., Wang, K., Sashital, D., Lawrence-Dill, C. J. (2016). Achieving plant CRISPR targeting that limits off-target effects. Plant Genome 9 (3), 1–8. doi: 10.3835/plantgenome2016.05.0047
Xie, X., Ma, X., Zhu, Q., Zeng, D., Li, G., Liu, Y. G. (2017). CRISPR-GE: a convenient software toolkit for CRISPR-based genome editing. Mol. Plant 10 (9), 1246–1249. doi: 10.1016/j.molp.2017.06.004
Xing, H. L., Dong, L., Wang, Z. P., Zhang, H. Y., Han, C. Y., Liu, B., et al. (2014). A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14 (1), 327. doi: 10.1186/s12870-014-0327-y
Yu, Z., Chen, Q., Chen, W., Zhang, X., Mei, F., Zhang, P., et al. (2018). Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in Arabidopsis. J. Integr. Plant Biol. 60 (5), 376–381. doi: 10.1111/jipb.12622
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12 (6), 797–807. doi: 10.1111/pbi.12200
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7 (1), 1–8. doi: 10.1038/ncomms12617
Zhang, Q., Xing, H. L., Wang, Z. P., Zhang, H. Y., Yang, F., Wang, X. C., et al. (2018). Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol. Biol. 96 (4-5), 445–456. doi: 10.1007/s11103-018-0709-x
Keywords: CRISPR/Cas9, Populus davidiana × P. bolleana, PDS gene, gene editing, single nucleotide polymorphism effect, off-target
Citation: Wang J, Wu H, Chen Y and Yin T (2020) Efficient CRISPR/Cas9-Mediated Gene Editing in an Interspecific Hybrid Poplar With a Highly Heterozygous Genome. Front. Plant Sci. 11:996. doi: 10.3389/fpls.2020.00996
Received: 15 March 2020; Accepted: 17 June 2020;
Published: 03 July 2020.
Edited by:
Chandrashekhar Pralhad Joshi, Michigan Technological University, United StatesReviewed by:
Keming Luo, Southwest University, ChinaWilliam Patrick Bewg, University of Georgia, United States
Copyright © 2020 Wang, Wu, Chen and Yin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingnan Chen, Y2hlbnlpbmduYW5AbmpmdS5lZHUuY24=
 Jie Wang
Jie Wang Huaitong Wu
Huaitong Wu Yingnan Chen
Yingnan Chen Tongming Yin
Tongming Yin