
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 12 June 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00872
This article is part of the Research TopicNew Insights Into Seed Metabolites: From Research to ApplicationView all 8 articles
Pulses are one of the most important categories of food plants, and Pea (Pisum sativum L.) as a member of pulses is considered a key crop for food and feed and sustainable agriculture. Integrative multi-omics and microsymbiont impact studies on the plant's immune system are important steps toward more productive and tolerant food plants and thus will help to find solutions against food poverty. Didymella pinodes is a main fungal pathogen of pea plants. Arbuscular mycorrhizal fungi (AMF) promote plant growth and alleviate various stresses. However, it remained unclear as to how the AMF effect on seed metabolism and how this influences resistance against the pathogen. This study assesses the AMF impacts on yield components and seed quality upon D. pinodes infection on two different P. sativum cultivars, susceptible versus tolerant, grown in pots through phenotypic and seed molecular analyses. We found that AMF symbiosis affects the majority of all tested yield components as well as a reduction of disease severity in both cultivars. Seeds of mycorrhizal pea plants showed strong responses of secondary metabolites with nutritional, medicinal, and pharmaceutical attributes, also involved in pathogen response. This is further supported by proteomic data, functionally determining those primary and secondary metabolic pathways, involved in pathogen response and induced upon AMF-colonization. The data also revealed cultivar specific effects of AMF symbiosis that increase understanding of genotype related differences. Additionally, a suite of proteins and secondary metabolites are presented, induced in seeds of P. sativum upon AMF-colonization and pathogen attack, and possibly involved in induced systemic resistance against D. pinodes, useful for modern breeding strategies implementing microsymbionts toward increased pathogen resistance.
A sustainable supply of crops to feed human societies could be the most important humanitarian action and its impact on declining of current and future food restrictions on this planet should be more sensed especially in regions with the highest food insecurity. Hence, the stable food supply is completely dependent on the sustainable production of crops by increasing yield and quality improvement through the development of natural potentials locally. To supply the food sustainably, large-scale production of pulses or legumes is inevitable (Fernandez-Aparicio et al., 2010). Pea (Pisum sativum L.) is one of the major legumes in the world (FAOSTAT1, updated 2019). Moreover, the use of microsymboints is in line with sustainable agriculture (Sayeed Akhtar et al., 2011).
Considering the importance of pulses as a source of vegetable protein in food basket and global need to improve their quantity and quality sustainably should be the most urgent need particularly in regions where people are suffering from the highest poverty rate. Thus, the study of microsymbionts which affect yield components of legumes especially on seed and upon stress conditions not only due to better use of natural growth promoters in agricultural systems but also most probably reduces the losses caused by abiotic and biotic stresses. Arbuscular mycorrhizal fungi (AMF) have interactions with a broad range of plants in nature (Smith and Read, 2008, as cited in Wehner et al., 2010). AMF inoculation of crops due to an increase in yield and growth (Hamel and Plenchette, 2007, as cited in Horii and Ishii, 2014). Promoting tolerance against water deficit and growth has been observed in papaya plants associated with AMF (Cruz et al., 2000, as cited in Horii and Ishii, 2014).
Chemical properties, physiology, and seed development of soybean could be influenced by AMF root colonization (Bethlenfalvay et al., 1997). Bona et al. (2016) noted to positive effects of AMF root colonization in prior studies on quality and yield in strawberry (Castellanos-Morales et al., 2010; Castellanos-Morales et al., 2012; Lingua et al., 2013; Bona et al., 2015) and saffron (Aimo et al., 2010). Also, the inoculation of Allium sativum with AMF has enhanced the yield and growth (Borde et al., 2009, as cited in Bona et al., 2016).
The formed symbiosis between AMF and maize (Zea mays) affected its seed proteome through modulation of enzymes related to functional categories such as stress, nucleotide metabolism, energy, storage, and development (Bona et al., 2016). Colonization of Amorpha fruticosa roots by Glomus mosseae has been modified the root proteome (Song et al., 2015). The G. mosseae by promoting cell integrity and osmotic stress depletion in wheat protected the root system against drought stress (Bernardo et al., 2017). The protein synthesis, metal handling, RNA metabolism, and reactive oxygen species (ROS) are influenced in leaf proteome of pea plants inoculated with AMF (Desalegn et al., 2016). In leaves of AMF colonized Medicago truncatula plants, not only genes related to synthesis of jasmonic acid (JA), flavonoid, abscisic acid (ABA), and terpenoids were up-regulated but also flavonoids and anthocyanins were intensified (Adolfsson et al., 2017).
Biotic stresses such as plant diseases are serious threats for food plants and consequently for food sources (Strange and Scott, 2005). One of the main diseases known in P. sativum is ascochyta blight caused by D. pinodes (Moussart et al., 1998). D. pinodes as the most abundant pathogen among seeds of P. sativum (Deneufbourg et al., 1994, as cited in Moussart et al., 1998) causes the poor quality and yield reduction (Khan et al., 2013). Under the field conditions, disease management of this pathogen is difficult in comparison with the control of other diseases upon P. sativum (Khan et al., 2013).
Chemical treatment of crop seeds is considered one of the major strategies in integrated control of ascochyta blight (Davidson and Kimber, 2007). On the other hand, using chemical compounds is not only a sustainable solution for the control of plant diseases but also acts as a destroyer of the environment or against sustainable agriculture systems. Therefore, symbiosis formation between food plants and microsymbionts with bio-control potential could be used as stable and safe disease management. Promoting of resistance against biotic stress in plants by AMF is proved (Azcón-Aguilar and Barea, 1996, as cited in Rebollo Couto et al., 2013).
Dehne (1982) reviewed the former studies on the role of mycorrhizae for effective protection of varied plant species against fungal pathogens such as Pythium ultimum (Stewart and Pfleger, 1977), Phytophthora cinnamomi (Bäertschi et al., 1982), P. megasperma (Chou and Schmitthenner, 1974), P. parasitica (Schenck et al., 1977; Davis and Menge, 1980), F. oxysporum cucumerinum (Dehne, 1977), F. oxysporum lycopersici (Dehne and Schoenbeck, 1979), Rhizoctonia solani (Stewart and Pfleger, 1977), Cylindrocladium scoparium (Barnard, 1977), Phoma terrestris (Becker, 1976), Pyrenochaeta terrestris (Safir, 1968), Olpidium brassicae (Schoenbeck and Dehne, 1979; Schoenbeck and Dehne, 1981), and Thielaviopsis basicola (Baltruschat and Schonbeck, 1972; Schoenbeck and Dehne, 1977).
Replacing common chemical fertilizers and toxicant with AMF has been focused in line with sustainable agriculture (Gange et al., 1999; Harrier and Watson, 2004; Wang et al., 2008; Lecomte et al., 2011, as cited in Rebollo Couto et al., 2013). Additionally, AMF could be applied as a bio-control agent and growth promoter (Sayeed Akhtar et al., 2011). It was found that AMF, directly and indirectly, inhibit the prevalence of fungal root pathogens (Wehner et al., 2010). The role of AMF in strengthening the host plant upon pathogen infection by up-regulation of defensive pathways and signals has been described (Haneef Khan et al., 2010).
Some studies have investigated the AMF impacts on the plant's growth at the maturity stage (Bethlenfalvay et al., 1994; Bethlenfalvay et al., 1997; Al-Karaki and Clark, 1999). Prior researches have mainly investigated the role of AMF on soil-borne pathogens in non-legumes and few investigations have attended to AMF impacts as a bio-control agent on aboveground fungal pathogens including ascochyta blight of P. sativum caused by D. pinodes. Further, most studies have analyzed the growth parameters of host plants upon mycorrhizal colonization rather than yield components. For instance, recent studies that have investigated the D. pinodes infection of P. sativum upon different treatments including mycorrhizal plants, were focusing on the leaves where no clear disease reduction in mycorrhizal treatments compared with non-mycorrhizal was reported (Desalegn et al., 2016; Turetschek et al., 2017). In general, only a few studies exist, using multilevel-omics approaches by integrating plant morphological and molecular phenotyping (Zivy et al., 2015).
Here, we assess the integrative effects of AMF on several yield components, growth parameters, seed secondary metabolome, and -proteome upon tolerant and susceptible pea cultivars against D. pinodes as an aboveground pathogen in pots. This study answers the following research questions: I) Does AMF affect above-belowground growth and yield parameters in pea plants? II) How does AMF promote the productivity and seed quality of P. sativum from the phenotypic-omics perspective? III) Has AMF-symbiosis a protecting effect on seed quality of tolerant and susceptible pea cultivars upon pathogen attack? IV) Do AMF influence the seed secondary metabolome and proteome? and V) How do AMF affect the response of the seed metabolism upon pathogen infection?
In line with processes carried out before (Ranjbar Sistani et al., 2017) and according to Begum et al. (2001), the disinfected pea seeds (ethanol 95%) were washed by ultrapure water and then, were kept (20 min) in bleach 5% and rinsed by ultrapure water several times. Finally, the soaked seeds (4 h) were considered for planting (Begum et al., 2001). The pre-germinated pea seeds during 3 days on the sterile perlite-vermiculate substrate were planted in pots containing sterilized soil with the following chemical properties: K2PO 120 mg/L, N 7 mg/L, pH 5.6 and P 13 mg/L (Desalegn et al., 2016). The pots were kept under the following conditions: 600 µmol m−2 s−1 lighting (14-h day/10-h night), humidity 60%–70%, and 22°C day/16°C night (Ludidi et al., 2007; Larrainzar et al., 2014). Treatments consisting of tolerant [cultivar Protecta (cv. Pr)] and susceptible [cultivar Messire (cv. Me)] genotypes, symbiont (M = mycorrhizal, NM = non˗mycorrhizal) and pathogen (infected/diseased = I and uninfected/healthy = U) were arranged in pots. Four plants per biological replicate (three biol. replicates) and two technical replicates per each treatment were considered. For metabolomics, proteomics, and phenotypic evaluations of seed, a hundred seeds per each biological replicate were considered. A modified recipe of B&D (Broughton and Dilworth, 1970) nutrient solution (KNO3 1011.03 ppm, MgSO4·7H2O 61.65 ppm, ZnSO4·7H2O 0.14 ppm, Fe-citrate 2.63 ppm, MnSO4·H2O 0.17 ppm, CaCl2 147 ppm, CoSO4·7H2O 0.028 ppm, K2SO4 43.5 ppm, Na2MoO4·2H2O 0.024 ppm, CuSO4·5H2O 0.05 ppm and H3BO3 0.12 ppm) was used for mycorrhizal treatments and B&D solution containing KH2PO4 (68 ppm) was applied for non-AMF treatments (Desalegn et al., 2016). An overview of the experimental setup can be found in Figure 1.
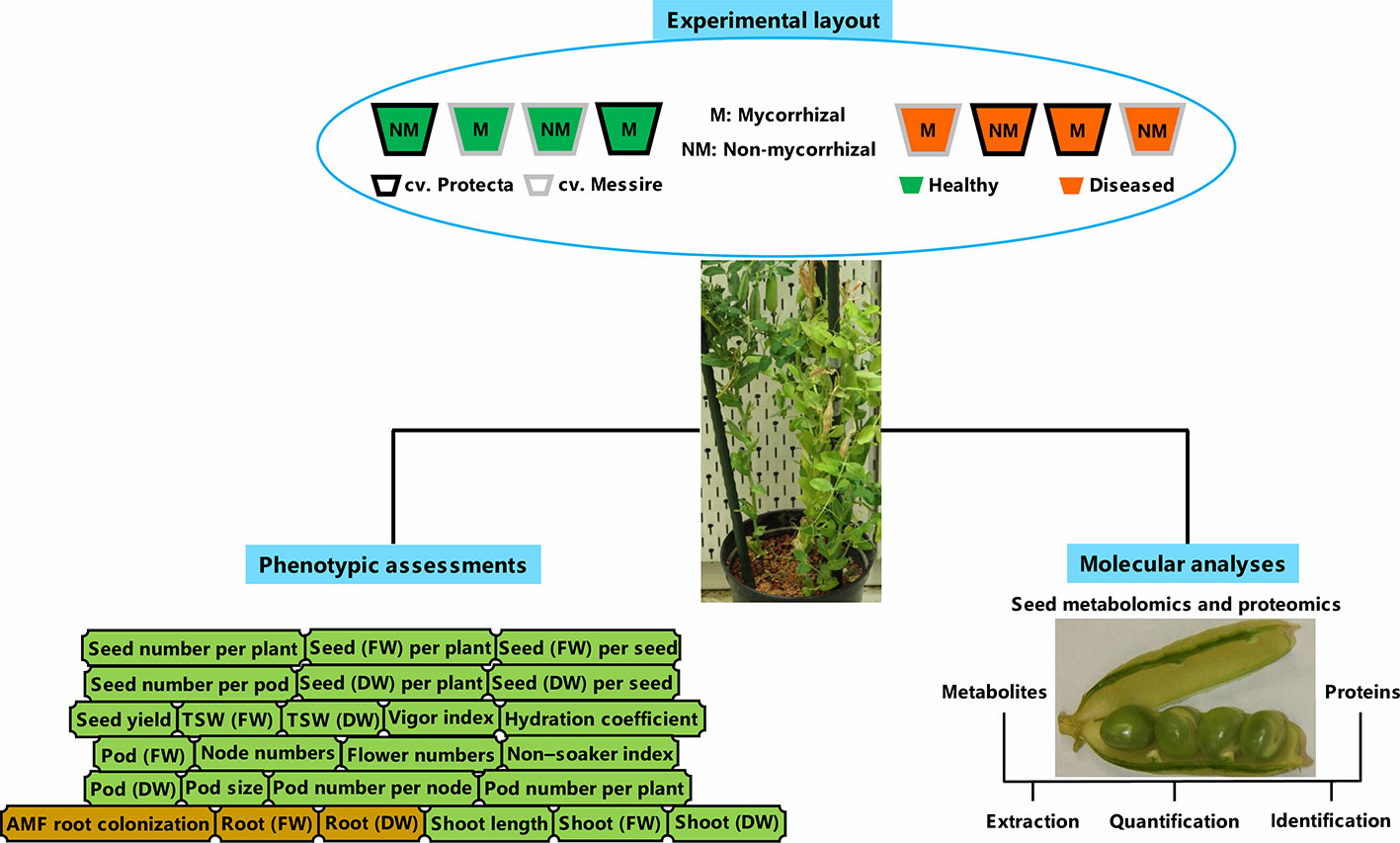
Figure 1 Schematic summary of experimental layout, molecular, and phenotyping analyses. FW, fresh weight; DW, dry weight.
Commercial mycorrhizal inoculum SYMBIVIT® (Symbiom Ltd., Czech Republic, www.symbiom.cz) containing five naturally occurring Glomerales species: Claroideoglomus etunicatum, C. claroideum, Rhizophagus irregularis, Funneliformis geosporus, and F. mosseae, was used. For mycorrhizal symbiosis establishment, 5 g of mycorrhizal inoculant (Horii and Ishii, 2014) according to company recipe and a modified method of Al-Karaki and Clark (1999) was added into each planting hole. Seeds were placed on the top of the inoculant (Al-Karaki and Clark, 1999).
According to our previous study (Ranjbar Sistani et al., 2017) and in detail: the prepared pathogen (D. pinodes) by Rubiales Lab in Spain was multiplied on Potato Dextrose Agar (PDA) and incubated upon 12-h photoperiod and 22°C (Davidson et al., 2012). Incubated colonies of pathogen after a week was used to make fungal suspension by adding Milli-Q (ultrapure) water and scratching of colonies upon sterile conditions (Zimmer and Sabourin, 1986; Carrillo et al., 2013). The autoclaved cheesecloth was applied to filter the prepared suspension and the filtered pathogen suspension with concentration 3 × 105 spores/ml was selected for inoculation (Zimmer and Sabourin, 1986; Carrillo et al., 2013). The fungal suspension plus TWEEN 20 (120 μl/100 ml) was handled to inoculate the pea seedlings after 20 days of planting separately from healthy (uninfected) treatments that were sprayed by a mixture of water and TWEEN 20 (Carrillo et al., 2013). Then, inoculated seedlings were kept under transparent covers during 48 h (Carrillo et al., 2013; Okorska et al., 2014) and after this period, infected seedlings were set apart for one week (12-h photoperiod, 21 ± 2°C) (Garry et al., 1998). Afterward, all seedlings were arranged as described in Performing Experiments and Planting, the aerial parts of infected pea plants by D. pinodes were regularly and periodic investigated by using a USB digital microscope (25X-400X, BMSCI, Japan) to image and computing the lesions (Ranjbar Sistani et al., 2017). The captured dimensions of lesions were applied to calculate the area of lesions and finally disease severity assay (Hwang et al., 2006; Carrillo et al., 2013). To complete the disease severity assessments, the infection rate of seeds in diseased pea plants was specified through the paper towel and PDA methods (Xue et al., 1996; Gaurilˇcikien˙e et al., 2012; Mahmoud et al., 2013) besides the area of lesions.
The pea plants were harvested when pods and seeds were ripe (BBCH 81-88). The root samples were prepared for investigation of mycorrhizal root colonization when the seeds were matured (Al-Karaki and Clark, 1999; Horii and Ishii, 2014). According to the modified method of Jin et al., 2013, soil particles were removed from the root samples using tap water, washed again with Milli-Q (ultrapure) water, and then dried with a paper towel. For each pot, subsamples of about 0.1–0.2 g were separated from the root system, freshly weighted (FW1) and immediately the fresh weight of remaining root was recorded (FW2) besides determining of the dry weight of that larger sample (DW2) [(Giovannetti and Mosse, 1980; Plant & Mycorrhiza Root Lengths2)]. The small subsamples of pea roots were boiled (10% KOH, 15 min) and the boiled root samples were washed by using water (Vierheilig et al., 1998). For staining, the cleaned root samples were put in boiling ink-acetic acid (5%, 3 min), and then, the stained root samples were washed (20 min) with acidified water by acetic acid (Vierheilig et al., 1998).
The mycorrhizal colonization of P. sativum roots was studied based on the gridline intersect method (Giovannetti and Mosse, 1980). On average, 20 stained root samples per pot were investigated. The count of intersections between lines, NM roots (R1), and M roots (R2) were recorded (INVAM2). The percentage of root colonization by AMF was calculated per each root sample by the following formula (Giovannetti and Mosse, 1980; Jin et al., 2013; INVAM2):
Moreover, the dry weight of the small subsample (DW1) was calculated by the following equation: DW1 = FW1 × DW2/FW2 (INVAM2). For the larger subsample that was not stained, the mycorrhizal root length (R3) was estimated [R3 = R2 × DW2 / DW1] (INVAM2). Total mycorrhizal root length (TMRL) and total plant root length (TPRL) were determined by the following formulas: TMRL = R2 + R3; TPRL = R1 + R3 (INVAM2).
In compliance with prior work (Ranjbar Sistani et al., 2017), the total weight of pea seeds per each pot was recorded and the seed yield (kg/ha) formula that was mentioned in Sajid et al. (2012) work was adapted to determine the seed yield per treatment (kg pot−1). The physical quality of pea seeds was evaluated through quantification of absorption and hardness rates of pea seeds per treatment that were kept in water (16 h) and then were assessed by hydration coefficient (%) and non-soakers (%) formulas (Abdelgani et al., 1999). The vigor index of pea seeds was quantified based on seed germination rate and seedling length measurements by using rolled paper towels (Abdul Baki and Anderson, 1973; Farrag and Moharam, 2012).
Mature seeds of P. sativum were freeze-dried by liquid nitrogen and then were coldly milled by Retsch mixer mill MM 400 (Retsch, Germany) (Ranjbar Sistani et al., 2017).
The seed proteins were extracted according to an adapted protocol of Wienkoop et al. (2008): 1.5 ml of extraction buffer [PMSF (1 mM), sucrose (0.7 M), EDTA (5 mM), Milli-Q water, PVPP (1% w/v), DTT (5 mM), and 50 mM Tris-HCl (pH 7.5)] was added to 50 mg of resulting powder from seed milling and then homogenization was completed by adding 1.5 ml Roti®-Phenol (Carl Roth GmbH, Karlsruhe, Germany). After that, the shaken mixtures (30 min, 4°C) were centrifuged (4°C, 4,000 × g, 30 min) and the resulting supernatants were precipitated with acetone at −20°C overnight. Then, the supernatant was centrifuged (4°C, 4,000 × g, 15 min), and then air-dried resulting pellet was dissolved with urea buffer [urea (8 M), HEPES (50 mM, pH 7.8)]. After protein concentration determination (Bradford, 1976), the protein pre-digestion (150 μg), was done by adding endoproteinase Lys-C sequencing grade (30°C, 5 h; Roche, Germany) followed by trypsin digestion, adding Poroszyme immobilized trypsin beads (5 μl; Applied Biosystems, Germany) and overnighting incubation in a hybridization oven (37°C). Afterward, digested proteins were desalted by using C18-SPEC 96-well plates (Agilent technologies) followed by an arrangement of desalting output in a vacuum centrifuge concentrator and eventually, storage at −80°C. For mass spectrometry of P. sativum seed, dried samples (1 μg per sample) were dissolved in a mixture containing formic acid (0.1%) and acetonitrile (2%) and were analyzed by Orbitrap Elite Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Bremen, Germany) that was connected to a 1D nano LC (UltiMate 3000, Thermo Fisher Scientific) through loading into a column (EASY-Spray, PepMap C18, >2 µm particles, 100 Å pore size, 15 cm × 50 µm ID) (PepMap RSLC, Thermo scientific). MS settings were: top 20 MS2 scans (CID activation), charge state screening enabled with the rejection of unassigned and +1 charge states, scan range (350–1,800 m/z), repeat (count and duration: 1 and 30 s), required signal threshold (Min. 1,000) and exclusion (list size and duration: 500 and 60 s).
The MaxQuant (version 1.6.0.16, Cox and Mann, 2008) was used with the following parameters: label-free quantification (MS/MS, large and min ratio: 2), missed cleavages (max: 2), min peptide length (unspecific search): 6, main and first search peptide tolerances: 4.5, 20 ppm respectively, FDR and PSM: 0.01, match tolerances (centroid: 7.5 and isotope: 2 ppm), variable modifications (max: 5) and activated decoy mode revert. For a sequence database in the Andromeda search engine (Cox et al., 2011), a constructed FASTA protein database (Desalegn et al., 2016; Turetschek et al., 2016) was used. Fasta, mercator, and MS data files with the identifier PXD006617 were stored in the PRIDE database of ProteomeXchange Consortium (Vizcaíno et al., 2016).
A modified protocol of De Vos et al. (2007) was applied for seed metabolomics as described before (Ranjbar Sistani et al., 2017): the mixed ground pea seed (100 mg) with 1 ml methanol (80%) were arranged in a filled ultrasonic bath with ice and water (10 min) followed by centrifugation (10 min, 21,000 × g). The resulting supernatant was dried in a vacuum centrifuge concentrator and then each dried sample was mixed with 50 μl [methanol (50%) + FA (0.1%)] and was centrifuged. After that, the reserpine (3μl, Sigma-Aldrich) was added to each sample which was diluted (1:10) by methanol (5%) in FA (0.1%) and were centrifuged again. For mass spectrometry, 20 μl of each supernatant was used. The NanoESI LC-MS/MS was carried out by using LTQ-Orbitrap XL Hybrid Ion Trap-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, Germany) with HPLC column and same parameters setting to proteomics analysis except for required signal (min 50,000) and scan range (130–1,800 m/z).
Annotation, quantification, and identification of the extracted metabolites were carried out as previously described (Ranjbar Sistani et al., 2017), with some changes: for identification and quantification of metabolites, Xcalibur (version 2.3.26, Thermo Fisher Scientific Inc.) was used for conversion of mass spectrometry output (RAW files) to CDF files. The resulting CDF files were used as input of MET-COFEA (Zhang et al., 2014). Then, the output files of MET-COFEA were loaded into the MET-XAlign (Zhang et al., 2015) tool. In parallel, the created mzXML files from the conversion of RAW files by MassMatrix (version 3.9, Case Western Reserve University3) were processed by using ProtMAX 2012_rev.2.14 (Egelhofer et al., 2013) as well as a spectral survey (Wang et al., 2016) and peak integration (Xcalibur 2.2, Thermo Scientific). The resulting data from MET-XAlign and ProtMAX outputs were used to complete the metabolite identification by using HMDB4 (Wishart et al., 2018), METLIN5 (Smith et al., 2005; Guijas et al., 2018), KNApSAcK family6 (Afendi et al., 2012). The metabolite candidates were finalized through literature reviews.
The normality of data was checked by using the Shapiro-Wilk test and to find the significant differences in treatments, ANOVA with Tukey HSD (P-value < 0.05) as a post hoc test, and the Kruskal-Wallis test were applied by using STATGRAPHICS Centurion 187. To study the major impacts of factors (microsymbiont, cultivar, and pathogen) and their interactions for phenotypic data, multifactor (three-way) ANOVA plus Tukey HSD procedure (P-value < 0.05) was performed in STATGRAPHICS Centurion 187. To analyze the correlations among all phenotypic variables, Pearson correlation coefficients through STATGRAPHICS Centurion 187 were calculated. Only proteins and metabolites that were reproducibly detected in all three biol. replicates (averages of two technical replicates per biological replicate) of at last one treatment were considered for quantification (Ranjbar Sistani et al., 2017). Filling of missed values base on a prior distribution followed by z-transformation was performed for independent component analysis (ICA) and cluster analysis by using COVAIN (Sun and Weckwerth, 2012) (Updated 2019; MATLAB R2017a, The MathWorks, Inc., Natick, Massachusetts, United States.). The q-values were calculated using an online Shiny application8 of q-value R package9 (Storey, 2002; Storey, 2003; Storey and Tibshirani, 2003; Storey et al., 2004; R Core Team, 2013).
The mean mycorrhizal colonization in roots of inoculated pea plants was ranged between 64.78% and 80.13% (Table S1). Although the mean AMF colonization between two cultivars was not significantly different, cultivar Protecta (cv. Pr) indicated higher colonization (11%) compared to cultivar Messire (cv. Me) (Table S1). Similarly, mycorrhizal colonization was not significantly affected by pathogen infection (Table S1), however, uninfected pea plants were more colonized (12%) by AMF than infected plants (Table S1). Although, TMRL and TPRL were not significantly different between the two cultivars, cv. Pr had higher root lengths (mycorrhizal-colonized and not) in comparison with cv. Me (Table S1). Root (FW and DW) were strongly and positively correlated with TMRL and TPRL (Table S2). Also the symbiont factor showed a significant impact on root fresh and dry weights (Table 1).
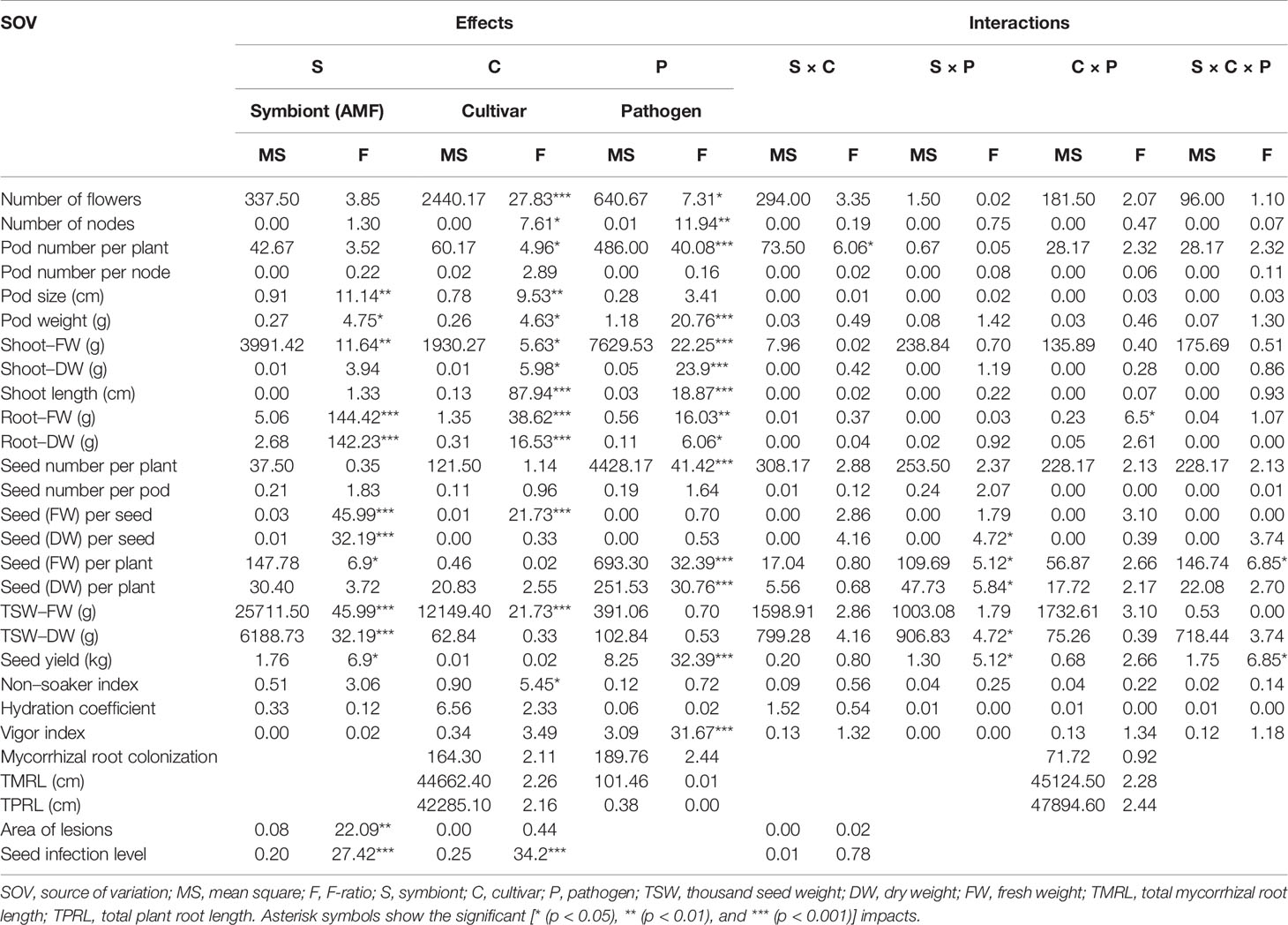
Table 1 Three-way ANOVA of main effects of AMF, pathogen, cultivar type, and their interactions impact on above–belowground yield and growth components of P. sativum.
The following results are according to multifactor ANOVA to determine the variability of phenotypic parameters under the influence of three factors (symbiont, cultivar, and pathogen) besides comparison of treatments (M vs. NM) per each phenotypical variable. Although symbiont (AMF) factor had a significant effect on pod size, pod weight, seed (FW and DW) per seed, seed (FW) per plant, thousand seed weight TSW (FW and DW), seed yield, root (FW and DW), and shoot (FW) (Table 1), the yield components showed no significant changes between M and NM treatments (Tables 2 and 3). Cultivar and pathogen factors separately indicated significant effects on pod number per plant, pod weight, shoot length, number of flowers, number of nodes, shoot (FW and DW), and root (FW and DW) (Table 1). Cultivar and symbiont factors individually showed a significant influence on pod size, seed (FW) per seed, and TSW (FW) (Table 1). Also, the pathogen factor had a significant impact on seed (FW) per plant, seed yield, seed (DW) per plant, and seed number per plant (Table 1). Among seed physical properties, cultivar and pathogen factors separately demonstrated a significant effect on non-soaker and vigor indexes respectively (Table 1). Among yield components, symbiont, cultivar, and pathogen factors had significant effects separately on pod weight while the triple interaction effect of these factors on seed (FW) per plant and seed yield were significant (Table 1). Additionally, a significant and positive correlation was observed between seed number per plant and mycorrhizal colonization percentage while the none-soaker index was significantly and negatively correlated with AMF colonization, TMRL, and TPRL (Table S2). Although the majority of yield components showed no significant changes between M and NM treatments (Tables 2 and 3) but the seed (FW and DW) per seed and TSW (FW and DW) in cv. Pr were significantly enhanced in M plants vs. NM under healthy and diseased conditions (Table 2). Furthermore, root (FW and DW) were significantly enhanced in mycorrhizal (M) vs. non˗mycorrhizal (NM) plants upon healthy and diseased across both cultivars (Table 3).
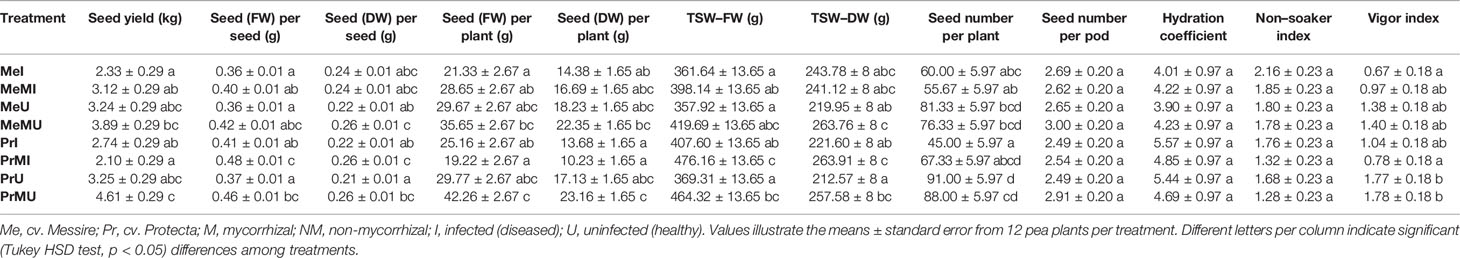
Table 2 The yield components and seed physical properties of pea plants upon different treatments and conditions.
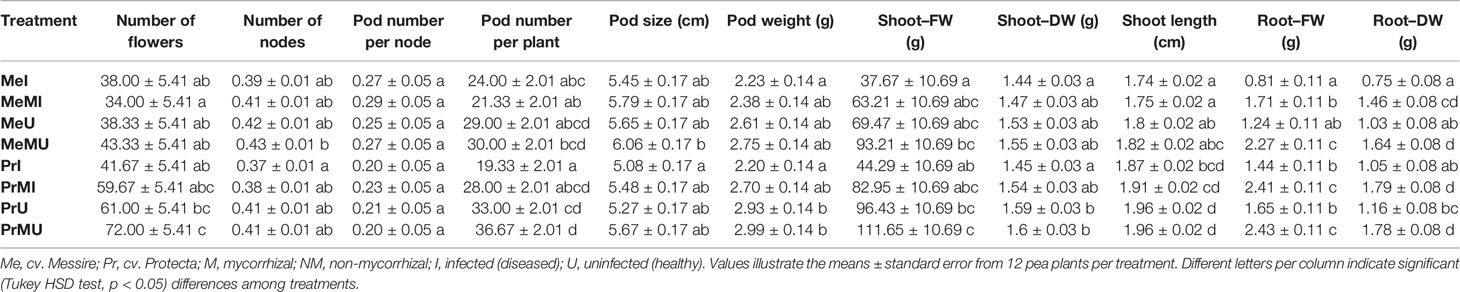
Table 3 The above–belowground growth parameters in different treatments of P. sativum under varied conditions.
The disease severity of P. sativum plants infected by D. pinodes was determined by the evaluation of the seed infection level and area of lesions. Although the mean area of lesions between the two cultivars was not significantly different, cv. Pr had lower lesions area (Table 4) and significantly lower levels of seed infection (Tukey HSD test, p < 0.05, nearly 2 folds) compared with cv. Me (Table 4). We found that AMF treatment of cv. Me resulted in a significantly lower level (Tukey HSD test, p < 0.05, nearly 2 folds) of seed infection (Table 4). Also, the inoculation of AMF significantly decreased the mean area of lesions in both infected cultivars (Table 4).
In total, 47 metabolites from 12 chemical classes and families were identified. The identified metabolites mostly belonged to glycerophospholipids, flavonoids, isoflavonoids, prenol lipids, carboxylic acids and derivatives, and organooxygen compounds (Table S3A). Glycerophosphoinositol phosphates (PIPs) as well as 2',4',5-Trihydroxy-7,8-[2-(1-methylethenyl) furo] isoflavone and lupinisoflavone A could not be assigned separately (due to identical chemical formula, retention time and m/z) and were therefore assigned as compound groups of PIPs and putative metabolites (*2',4',5-Tri_Lup_Isoflavon), respectively (Table S3A). Compared to our previous study, we increased our metabolite identification due to the additional use of the KNApSAcK family (http://kanaya.naist.jp/KNApSAcK_Family/) database. Eighty-seven percent of identified metabolites were significantly changed (ANOVA, Kruskal-Wallis, Tukey HSD tests, P-value < 0.05, and ≥ two-fold change) among treatments (M vs. NM of diseased plants across both cultivars) (Table S3B). Most of the metabolites that significantly increased in M vs. NM treatments were induced upon pathogen infection and mainly in cv. Pr (Table S3B). For example, (Z)-3-Oxo-2-(2-pentenyl)-1-cyclopenteneacetic acid, cyanidin 3-sophoroside 5-glucoside, luteone 7-glucoside, and stachyose were significantly enhanced in the seed metabolome of M vs. NM of cv. Pr under pathogen infection (Table S3B).
In contrast, L-2-Amino-3-(oxalylamino) propanoic acid (or L-3-Amino-2-(oxalylamino) propanoic acid), N-(Carbethoxyacetyl)-4-chloro-L-tryptophan, and vignatic acid A were only significantly accumulated in M vs. NM treatments of cv. Me against pathogen infection (Table S3B).
Most of the increased seed metabolites of M plants upon infection were carboxylic acids and derivatives (Tables S3A, B). Furthermore, some metabolites including 2,3-Dihydroxy-2,4-cyclopentadien-1-one, L-DOPA 3'-glucoside, soyasapogenol C and syoyualdehyde were significantly enhanced in M vs. NM of diseased treatments (Table S3B).
Independent of M or NM treatments, pathogen-infected versus healthy plants (I vs. U) showed a significant increase of malvidin 3-rutinoside-5-glucoside, peonidin 3-rhamnoside 5-glucoside, PI (20:5(5Z,8Z,11Z,14Z,17Z)/0:0), wistin, *2',4',5-Tri_Lup_Isoflavon, sativan, 6''-O-Malonylwistin, abscisic acid and kudzusaponin SA1 (Table S3B). In contrast, soyasapogenol C, syoyualdehyde, (Z)-3-Oxo-2-(2-pentenyl)-1-cyclopenteneacetic acid, 8-Galactopyranosyl-6-glucopyranosyl-4',5-dihydroxy-7-methoxyflavone, luteone 7-glucoside, N-Jasmonoylisoleucine and stachyose levels were significantly reduced in I vs. U treatments (Table S3B).
Among metabolites that showed a significant difference between two cultivars, 8-Galactopyranosyl-6-glucopyranosyl-4',5-dihydroxy-7-methoxyflavone, delphinidin 3-lathyroside 5-glucoside, (Z)-3-Oxo-2-(2-pentenyl)-1-cyclopenteneacetic acid, 6-O-b-D-Fructofuranosyl-2-deoxy-D-glucose, indole-3-acetamide, were remarkably enhanced in cv. Pr (Table S3B).
Only proteins without missing values (LFQ intensities) across replicates of the same treatment were analyzed statistically. The resulting 1,325 proteins were functionally categorized by applying the Mercator pipeline (Lohse et al., 2014) and MapMan tool (Thimm et al., 2004) (Table S4). Table S4 illustrates the comprehensive seed proteomics data including categorized proteins, protein descriptions, mapped functional classes, statistical analyses, and fold change (FC) ratio for all treatments. Overall, remarkable changes (ANOVA, Kruskal-Wallis, Tukey HSD test, and P-value < 0.05 and ≥ two-fold change) were found in 25% of mapped proteins by comparing M vs. NM under diseased and healthy conditions in both cultivars (Table S4).
Among mapped functional categories that were significantly changed (P-value < 0.05 and ≥ two-fold change) in M vs. NM, the category of protein degradation had the highest number of proteins (Table S4). Most of the identified proteins were involved in protein degradation and thus several of those were found with significantly increased levels in M vs. NM treatments (Table S4). Among M vs. NM treatments under healthy conditions, protein degradation, protein synthesis, and stress categories in cv. Pr and plastid (PS) in cv. Me had the largest number of proteins changed upon M (Table S4). Protein functional categories such as amino acid metabolism (Figure 2; frv2_55445, frv2_85107, frv2_60349, frv2_112647, frv2_48897 and frv2_83949), miscellaneous (misc) (Figure 2; frv2_61709, frv2_111740, Q9FN08, frv2_46536, frv2_82443, P93479, frv2_103111, frv2_103598, frv2_78262, and frv2_83016) and redox (Table S4; frv2_125364, frv2_76239, frv2_47501, frv2_76239, frv2_77791, frv2_81620, and Q9FF55) showed a significant increase in M vs. NM upon disease and healthy conditions in both cultivars.
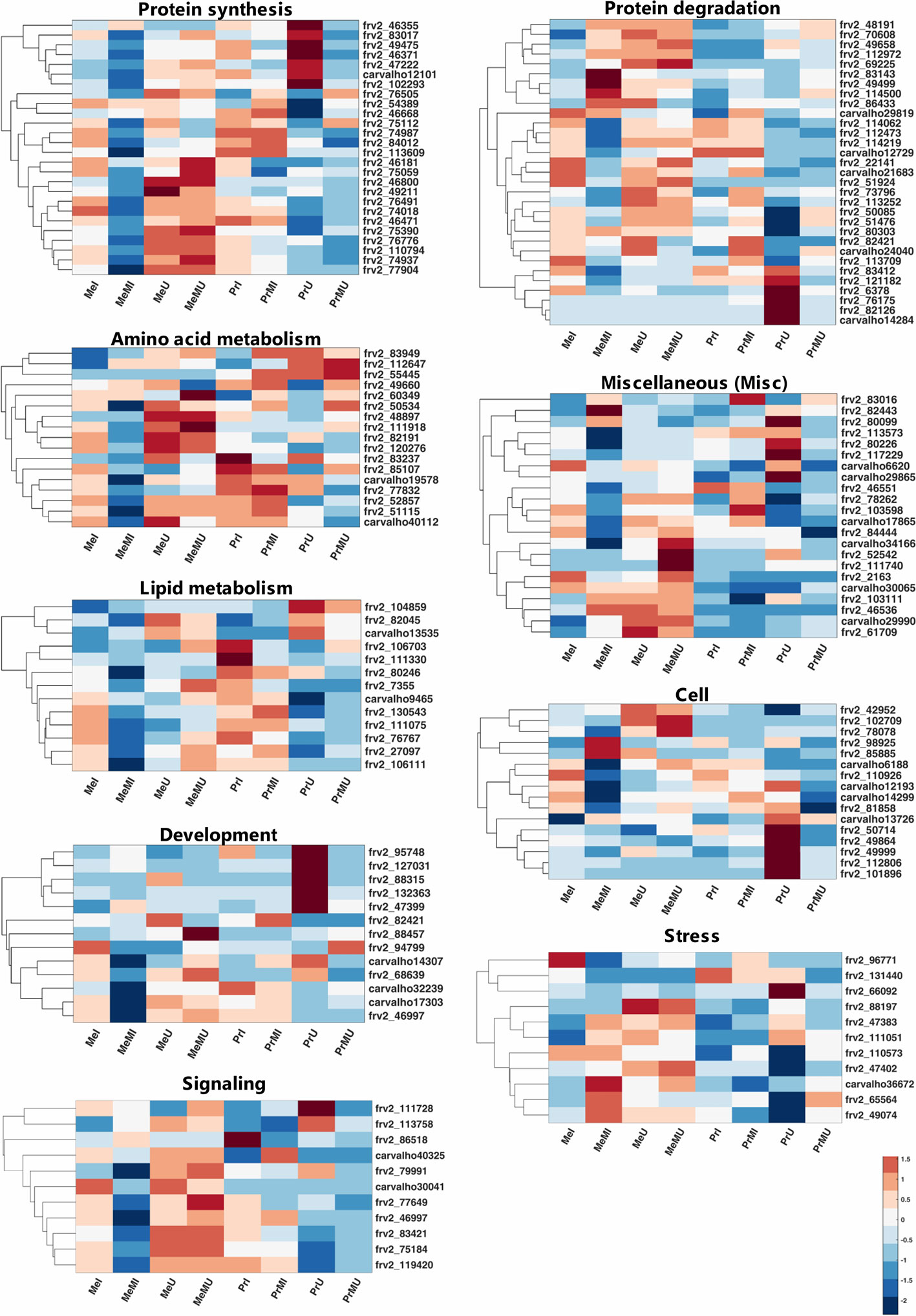
Figure 2 Clustered heatmaps of mapped functional categories which including ≥ 10 proteins of seed proteins significantly changed (Kruskal-Wallis; ANOVA, Tukey HSD test; p < 0.05 and ≥ two-fold change) in M compared with NM treatments upon I and U conditions across both cultivars. The heatmap cells are z-transformed of mean LFQ intensities per treatment. Me, cv. Messire; Pr, cv. Protecta; M, mycorrhizal; NM, non-mycorrhizal; I, infected (diseased); U, uninfected (healthy).
Under pathogen infection, several categories including DNA (Table S4; frv2_98925 and frv2_50256), mitochondrial electron transport (Table S4; frv2_77583, frv2_103383, frv2_81222, and frv2_53151), nucleotide metabolism (Table S4; frv2_87000, frv2_81447, frv2_112313, and frv2_48788), oxidative pentose phosphate (OPP) (Table S4; Q43848 and Q9SZE1), protein amino acid (aa) activation (Table S4; Q9ZPI1 and frv2_121810) and signaling (Figure 2; frv2_113758 and Q96453) were remarkably enhanced in M vs. NM of both cultivars. In contrast, the protein synthesis category (Figure 2; frv2_75059, frv2_46668, frv2_75112, frv2_54389, and frv2_76505) indicated a notable accumulation in M vs. NM exclusively under healthy conditions. In M vs. NM, the categories development (Figure 2; frv2_47399, frv2_88457, and frv2_94799) and PS (Table S4; Q43848, frv2_80442, frv2_77556, frv2_84713, and Q9SHE8) categories only in cv. Me besides C1-metabolism (Table S4; frv2_90362 and frv2_86521) and glycolysis (Table S4; frv2_114679, frv2_112199, frv2_47599, and Q88C93) uniquely in cv. Pr were significantly pronounced upon disease and health conditions.
Comparing of I vs. U upon M and NM treatments in both cultivars indicated that levels of several proteins engaged in amino acid metabolism, cell, DNA, lipid metabolism, metal handling, misc, mitochondrial electron transport, nucleotide metabolism, photosynthesis (PS), redox and signaling were significantly reduced while abundances of some proteins involved in proteins synthesis were remarkably enhanced (Table S4). Interestingly, some levels of proteins involved in protein degradation and stress categories were either notably accumulated or reduced in I vs. U (Table S4). Upon pathogen infection, the largest functional categories were protein synthesis (increased levels) and protein degradation (decreased levels) (Table S4). Some proteins assigned to fermentation, major CHO metabolism, and protein targeting were remarkably accumulated only in the seed proteome of NM treatments of both cultivars against pathogen infection. In contrast, some proteins of C1-metabolism, development, protein post-translational modification, TCA cycle, biodegradation of xenobiotics, and transport categories showed a significant reduction (Table S4).
Comparison of cultivars across all treatments including M and NM under disease and healthy conditions demonstrated that several proteins of N-metabolism, secondary metabolism, and signaling in cv. Me besides some proteins of amino acid metabolism and misc in cv. Pr were significantly (p < 0.05) and distinctively enhanced (≥2-fold) (Table S4). Moreover, several levels of proteins involved in cofactor and vitamin metabolism were significantly intensified in both cultivars of NM treatments against pathogen infection (Table S4). Numbers of identified proteins from OPP and protein post-translational modification classes in cv. Me in addition to some TCA cycle-related proteins in cv. Pr showed a significant enhancement in NM treatments upon pathogen infection (Table S4).
The proteins from main functional classes (including >2 proteins) with significantly increased fold change ratio (p < 0.05, FC ≥ 2) in M vs. NM under healthy and disease conditions of both cultivars are presented in Figure S1. Amino acid metabolism, misc and redox categories were only the main protein functional groups with enhanced FC significantly upon all paired comparisons of M vs. NM (Figure S1A). The most increased FC in M vs. NM belonged to protein synthesis in cv. Pr under healthy conditions (Figure S1A). Hormone metabolism and OPP had the highest increased FC in M treatments of cv. Me and cv. Pr respectively against pathogen infection (Figure S1A). Under pathogen infection, proteins associated with mitochondrial electron transport, nucleotide metabolism, OPP, protein amino acid (aa) activation and DNA were exclusively and largely accumulated in M vs. NM of both cultivars while, protein synthesis was uniquely promoted in M treatments of cultivars under healthy conditions (Figure S1A). When comparing M vs. NM, a significant increase of C1-metabolism, glycolysis, major CHO metabolism, and minor CHO metabolism were observed only in cv. Pr besides the notable accumulation of proteins related to development, PS, and TCA cycle individually in cv. Me were observed (Figure S1A). Protein degradation, protein synthesis, and stress functional classes were strongly increased in seed proteome of all I vs. U treatments in both cultivars, and on average among these categories, protein synthesis had the highest summed FC ratio (Figure S1B). Under disease conditions, engaged proteins in cell and biodegradation of xenobiotics illustrated noteworthy enhancement exclusively in cv. Pr while development, protein post-translational modification, and redox were highly intensified only in cv. Me (Figure S1B). The proteins included in signaling, stress, and TCA cycle were significantly promoted in cv. Pr compared with cv. Me upon M and NM treatments of both healthy and diseased (Figure S1C). Among these protein functional categories, stress-related proteins were highly increased in Pr vs. Me than proteins mapped in signaling and TCA cycle (Figure S1C).
To determine the metabolites and proteins with the highest impact on discrimination of treatments under stress conditions caused by pathogen, an ICA was performed (Figure 3). Only intensities of identified seed metabolites and proteins with significant alteration (ANOVA, Kruskal-Wallis, Tukey HSD tests, P-value < 0.05, and ≥ two-fold change) in M vs. NM upon diseased conditions of both cultivars were analyzed. The integrated seed metabolomics and proteomics data displayed distinctive separations of cultivars (Me vs. Pr) on IC2 and mycorrhizal symbiont (M vs. NM) on IC3 (Figure 3). Figure 3 shows the ten highest positive and negative loadings responsible for these discriminations are depicted in separate bar plots.
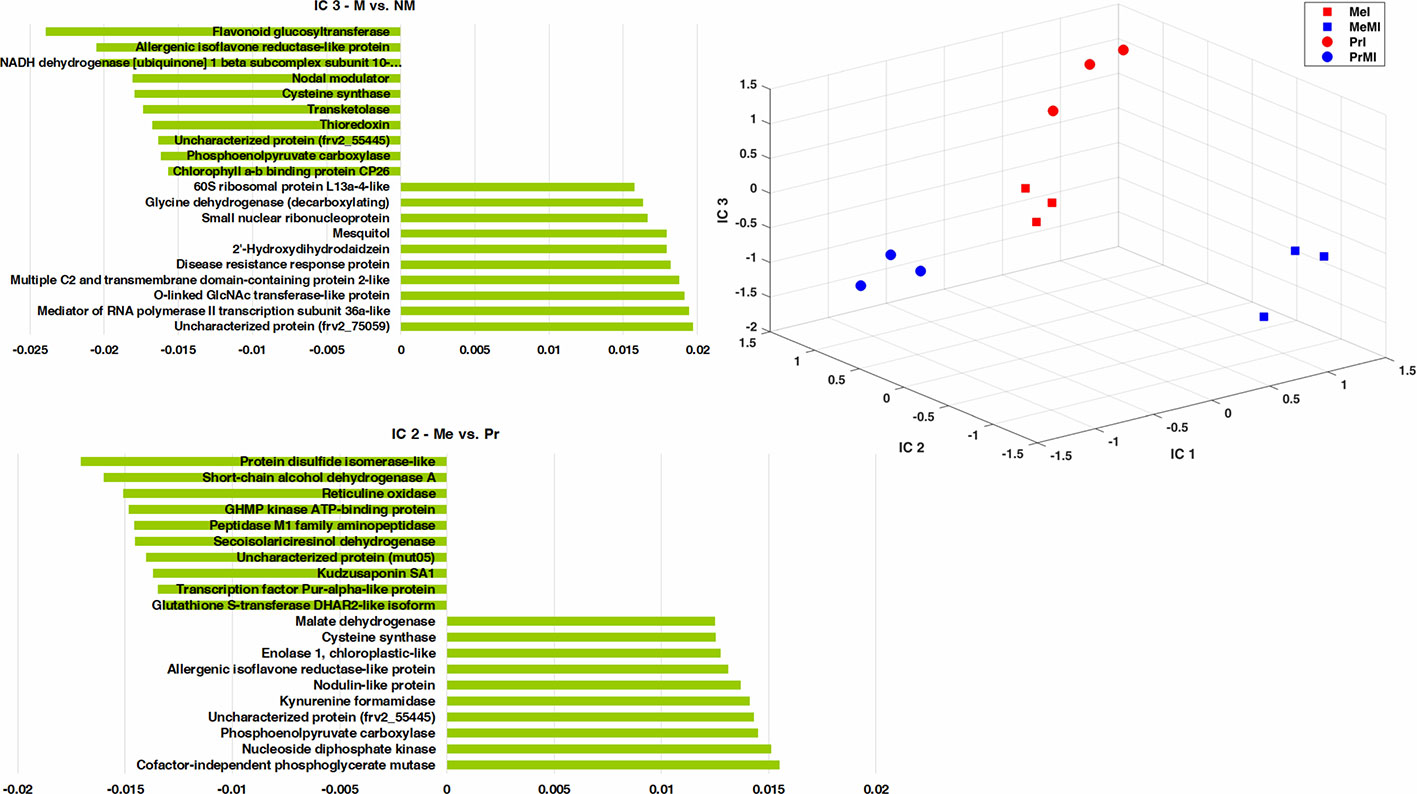
Figure 3 ICA of integrated seed metabolomics-proteomics data including remarkably changed (Kruskal-Wallis; ANOVA, Tukey HSD test; p < 0.05 and ≥ two-fold change) seed metabolites and proteins in M vs. NM upon diseased conditions of both cultivars. Numeric data were z-transformed. The loadings graphs illustrate the components with the top ten of the highest loadings (> 0.01 and < −0.01) on IC2 and IC3. Me, cv. Messire; Pr, cv. Protecta; M, mycorrhizal; I, infected (diseased).
Kudzusaponin SA1 and a Nodulin-like protein (frv2_125506) had considerable effects on the separation of Me vs. Pr in disease treatments (Figure 3). Some proteins like phosphoenolpyruvate carboxylase, allergenic isoflavone reductase-like protein, and cysteine synthase were highly involved in discrimination of M vs. NM besides Me vs. Pr upon diseased treatments (Figure 3 and Table S5). An uncharacterized protein (frv2_55445) belonged to amino acid metabolism (degradation, arginine) had a high impact on the separation between both cultivars (Me vs. Pr) and M vs. NM in diseased treatments (Figure 3 and Table S5). Two proteins (small nuclear ribonucleoprotein and mediator of RNA polymerase II transcription subunit 36a-like) mapped in RNA category were recognized with high influences on the separation of M vs. NM among diseased treatments (Figure 3 and Table S5).
Cofactor-independent phosphoglycerate mutase (glycolysis) and protein disulfide isomerase-like (redox) had the highest impact on the separation of Me vs. Pr under diseased conditions (Figure 3 and Table S5). Furthermore, an uncharacterized protein (frv2_75059) from protein synthesis class as well as flavonoid glucosyltransferase (secondary metabolism) indicated a strong effect on discrimination of M vs. NM upon diseased treatments (Figure 3 and Table S5). Among disease treatments, two metabolites including 2'-hydroxydihydrodaidzein (isoflavonoid) and mesquitol (flavonoid) showed a remarkable impact on separating M vs. NM (Figure 3 and Table S5).
The data were then further reduced to focus on the most relevant protein groups potentially involved in Induced Systemic Resistance (ISR). For this, proteins that showed increased levels in M vs. NM of healthy plants overlapping with significantly increased levels upon pathogen infection of NM plants were selected for MapMan visualization (Figure 4). Names of the protein groups which were visualized, can be drawn from supplemental Table S6 as they were in the same order. Figure 4A schematically shows the priming effect where protein levels of AMF symbiotic and healthy plants were induced compared to NM plants. During pathogen attack these protein levels also were increased in NM plants while they were not changed in infected M plants as their levels were already induced (primed) (Figure 4). Altogether, cv. Protecta shows a strong overlap of induced proteins (Figure 4B and Table S6) between healthy M treated and stressed non-symbiotic (PrI) plants while only one protein was also slightly further increased upon infection in M plants (MI). In contrast, only a few proteins (four proteins) were induced in healthy M plants of cv. Messire and overlapping with a stress response of non-symbiotic plants (Figure 4C and Table S6). The strongest responses of the seed metabolism of cv. Protecta (in terms of numbers) were proteins related to primary metabolism such as gluconeogenesis and amino acid synthesis and secondary metabolism-related proteins, including lipid and hormone (jasmonate) regulating proteins. In terms of fold changes, two proteins of yet unknown function revealed the highest induction (>100 fold) upon AMF symbiosis of healthy and after infection of NM Protecta plants (Table S6). The four proteins possibly involved in a priming effect in cv. Messire were a major intrinsic protein, a Polyvinylalcohol dehydrogenase-like protein (signaling), a Cytochrome b5-like protein, and a storage protein (legumin) (Table S6).
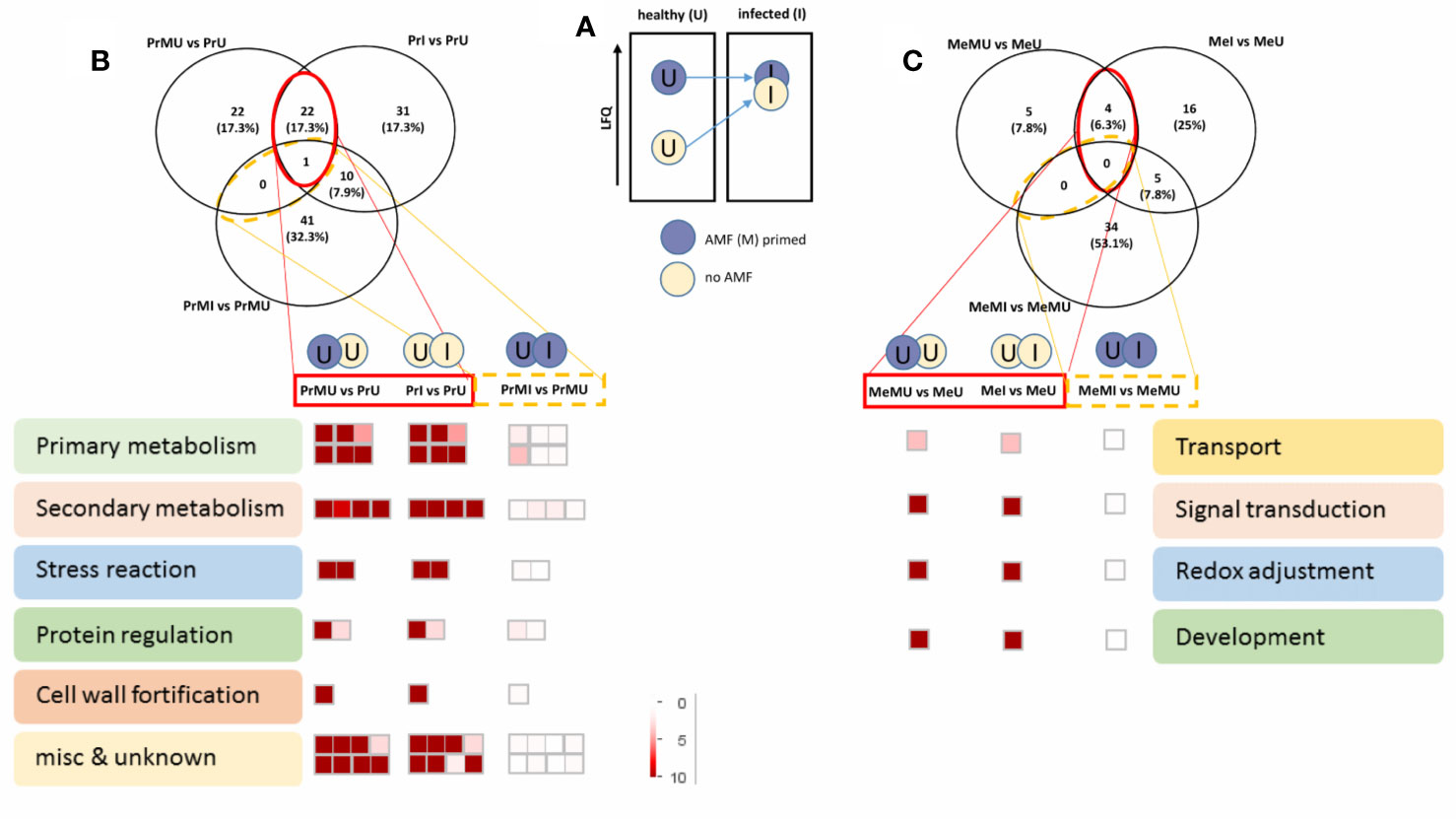
Figure 4 Overview of protein regulated pathways, involved in AMF priming. (A) Schematic concept of the AMF induced priming effect. Venn diagram and Mapman overview of selected overlapping proteins, significantly induced upon AMF treatment of healthy and infected non-symbiotic plants of cv. Protecta (B) and cv. Messire (C) (see also Supplemental Table S6). LFQ, Label-Free Quantification.
Thus far, major studies of mycorrhizal impacts on plants have been focused on some phenotypical components and/or improvement of nutritional properties in plants under optimal environmental growth conditions (Bethlenfalvay et al., 1994; Abdel-Fattah, 1997; Bethlenfalvay et al., 1997; Al-Karaki and Clark, 1999; Jin et al., 2013; Horii and Ishii, 2014; Kavitha and Nelson, 2014; Young et al., 2015; Abdel-Fattah et al., 2016). Even though there are increasing numbers of studies using omics technologies to investigate mycorrhizal symbiosis (Fontana et al., 2009; Pedone-Bonfim et al., 2013; Rebollo Couto et al., 2013; Laparre et al., 2014; Rivero et al., 2015; Saia et al., 2015; Bona et al., 2016; Larsen et al., 2016; Adolfsson et al., 2017; Li et al., 2017; Hill et al., 2018), the integration of several omics levels including metabolomics and proteomics with phenomics analyses is still a research gap. Also, most of this type of studies have concentrated on vegetative parts including shoot (Scheublin and van der Heijden, 2006), root (Schliemann et al., 2008; Song et al., 2015), leaf (Pedone-Bonfim et al., 2013; Schweiger et al., 2014; Desalegn et al., 2016; Adolfsson et al., 2017; Turetschek et al., 2017), and tuber (Lu et al., 2015). The current study investigates the mycorrhizal (M) impacts on above- and belowground yield and growth parameters integrating metabolomics (secondary metabolites) and proteomics analyses of seeds in two P. sativum cultivars in response to pathogen stress.
The successful colonization of plants by AMF is influenced by temperature, nutritional properties of soil, type of mycorrhizal symbiont, plant species, and light conditions (Smith and Smith, 1996; Johnson et al., 1997, as cited in Jansa et al., 2008). In this study, the high average AMF colonization (70%) of pea roots indicated a stable symbiosis between AMF species and the root system of P. sativum cultivars similar to our previous observations (Desalegn et al., 2016). Moreover, the considerable enhancement of root biomass in mycorrhiza symbiotic (M) plants compared with non-mycorrhizal (NM) plants in both cultivars and under disease stress prove the high impact of AMF on the root system as a plant growth-promoting symbiont in line with reported observations (Kavitha and Nelson, 2014; Oruru et al., 2017; Wang et al., 2018). Here, the remarkable increase of root biomass in M vs. NM plants besides significant positive correlation among root biomass (FW and DW) as well as between total mycorrhizal and plant root length (TMRL and TPRL) confirm the mutual connection between mycorrhizal root colonization and architecture of root system that was found before by Sinclair et al. (2014) in colonized roots of strawberry. However, to reach optimal AMF promoted crop productivity, the choice of AMF species that match with the various crops is determinative (Van Geel et al., 2016). Chen et al. (2017) have concluded that the application of AMF in a combined form of several mycorrhizal species is more effective compared with single inoculation on cucumber. This is important, as we can say that the AMF mixture, we used in this study, is effective even if we cannot distinguish between the putative differences in efficiency of the different species. Hence, although our approach is more close to nature, a differential analysis of the various AMF species remains a future task. Nevertheless, the mutual impact of AMF on root growth and vice versa, as indicated previously (Atkinson et al., 1994, as cited in Sinclair et al., 2014), corroborates our results for both pea cultivars. Further studies on AMF species and inoculum composition may further increase growth-promoting efficiency. Interestingly, the lower TMRL and TPRL in cv. Messire than cv. Protecta related not only to different root structures but also indicate the dissimilar response of the two cultivars to mycorrhizal root colonization by the same AMF species. Similarly, we previously observed an individual cultivar root-microsymbiont colonization pattern between cv. Messire and Protecta when inoculated with Rhizobium (Turetschek et al., 2017). Different AMF root colonization and root architecture were also observed in olive cultivars (Chatzistathis et al., 2013). Oruru et al., 2017 not only found the dissimilarities in AMF root colonization between modern and wild cultivars of cowpea, they also reviewed the differences among cultivars of tomato (Steinkellner et al., 2012), wheat (Tarawaya, 2003), maize (Njeru et al., 2013), and barley (Zhu et al., 2003). However, not much is known about the reason for and consequences of these root architectural differences in relation to symbiotic efficiency and hence, further root phenotyping (incl. molecular) studies will be needed. Finally, it is noteworthy that no significant negative effect of the pathogen on AMF colonization compared to healthy plants could be found regardless of cultivars. In contrast, our prior study (Ranjbar Sistani et al., 2017) showed significant reduction of Rhizobium density isolated from nodulated roots caused by D. pinodes. These results indicate that AMF root colonization is more stable under pathogen attack compared to Rhizobium symbiosis.
In general, our results show that AMF as belowground symbionts also promote the growth of the above-ground parts of pea plants. Like in our previous study (Ranjbar Sistani et al., 2017), which focused on effects of bacterial (Rhizobium) symbiosis on pea seeds, here, we found that AMF had a significant impact on seed yield, seed (FW and DW) per seed, TSW (FW and DW), seed (FW) per plant, pod weight, and pod size. In earlier studies, the notable influence of AMF root colonization on seed yield enhancement (Cely et al., 2016; Adeyemi et al., 2017), seed weight per plant (Abdel-Fattah et al., 2016), pod weight (Abdel-Fattah et al., 2016; Adeyemi et al., 2017), and hundred-seeds weight (Kavitha and Nelson, 2014; Abdel-Fattah et al., 2016) has been observed. Specific proteins such as glutamine synthetase (N-metabolism), enolase (glycolysis), the protein disulfide isomerase (redox), malate dehydrogenase (TCA cycle), peptidyl-prolyl cis-trans isomerase (cell) were remarkably enhanced in seed proteome of M vs. NM pea plants. These proteins have been recognized previously as essential proteins during Brassica campestris seed development (Li et al., 2012). The data indicate that AMF influence seed growth and development of P. sativum cultivars.
Interestingly, Indole-3-acetamide (IAM), crucial for the synthesis of the auxin indole-3-acetic acid (IAA) (Mano et al., 2010), was reduced in our M treated plants. IAA is included in AMF symbiosis relation (Ludwig-Müller and Güther, 2007) and plant growth-regulation (Kögl and Kostermans, 1934; Went and Thimann, 1937). The role of AMF in promoting the synthesis of auxin and consequently enhancement of root-hair growth in Poncirus trifoliate was reported (Liu et al., 2018). The depletion of IAM upon AMF treatment might indicate that it was required for the formation of IAA, not detected in our study. Nevertheless, enhanced IAA production of seeds is not only of advantage for seed germination and development but also for microsymbiont attraction and interaction (Fu et al., 2015).
Strong genotype related impacts of AMF on pod weight in okra (Nwangburuka et al., 2012) have been reported earlier. Proteome analysis also revealed that secondary metabolism, general stress, and redox response and also proteins of the primary metabolism of amino acid synthesis and glycolysis are more enhanced in cv. Protecta compared to cv. Messire. Interestingly, most of the AMF responsive proteins of those metabolic pathways are also involved in pathogen defense.
The majority of studies associated with biocontrol and impact of mycorrhiza against plant pathogens are restricted to soil-borne and root fungal phytopathogens (Vierheilig et al., 2008). Here, the pathogen factor had significantly negative effects on aboveground parameters. The intensity of several proteins mapped for many functional classes including C1-metabolism, cell, development, DNA, metal handling, nucleotide metabolism, protein post-translational modification, redox, biodegradation of xenobiotics, signaling, TCA cycle, and transport were considerably reduced under stress caused by the pathogen. These negative effects were much more pronounced in cv. Messire compared to Protecta. These findings show the broad metabolic dampening effects of D. pinodes on P. sativum seeds, particularly for cv. Messire especially in the absence of AMF. Protecta, in contrast, shows a stronger increase in several proteins of the signaling, stress response, development, amino acid, glycolysis, and of the secondary metabolism, which might be related to the enhanced protection of cv. Protecta against the pathogen. Here, several of our previously identified metabolites and proteins involved in general pathogen resistance can be confirmed (Ranjbar Sistani et al., 2017).
For instance, we previously found proteins of the Late Embryo Abundant (LEA)-related family among the most significantly involved in pathogen resistance response (Ranjbar Sistani et al., 2017). Several groups of those (frv2_129189; frv2_25524, frv2_11113; frv2_54342, frv2_79176, P28639, frv2_128555) again showed a significant increase upon pathogen attack, however, independent of M treatment. LEA proteins are mainly known to be induced during seed development and in resistance to dehydration (Battaglia and Covarrubias, 2013). Our data confirm the findings of our previous work that LEA proteins are important for enhanced seed pathogen resistance and are not only involved in drought stress tolerance (Ranjbar Sistani et al., 2017).
Likewise, vicilin storage proteins (frv2_74601 and frv2_80935) were again found with a stronger increase under infection in cv. Protecta, however, slightly but not significantly stronger in M vs. NM treated plants. Nevertheless, this effect was more pronounced in rhizobia symbiotic plants (Ranjbar Sistani et al., 2017).
Also, the phytohormone abscisic acid (ABA) is a known component in plant environmental stress reactions (Luo et al., 2009; Song et al., 2014; as cited in Porcel et al., 2012; Adolfsson et al., 2017). A higher ABA content has been detected in leaves of Medicago truncatula colonized by AMF (Adolfsson et al., 2017). In Arabidopsis seeds, it has been reported that salicylic acid application induced ABA signaling leading to increased synthesis of ABA-regulated proteins, such as LEA proteins, dehydrins, and heat shock proteins, (Rajjou et al., 2006). Although here, ABA was notably enhanced in seeds upon pathogen infection, this accumulation was not enhanced in AMF symbiotic plants. Thus, our data support a role of ABA during pathogen defense and enhanced LEA protein levels independent on AMF symbiosis.
Interestingly, we found several plastidic proteins in pea seeds strongly depleted in both cultivars upon pathogen defense, independent of AMF symbiosis. Plastids are important components for embryos development in P. sativum (Smith et al., 1990). The role of chloroplast in the seeds biotic stress response, however, has not been described before. We suggest that the huge breakdown of plastidic proteins, we observed upon infection, might serve as a source for the enhanced secondary metabolite synthesis. The findings are in line with Fondevilla et al. (2011) who found that genes assigned to the primary metabolism were mostly down-regulated in accession P665, a resistant genotype, compared to Messire as susceptible pea cultivar against D. pinodes infection. Altogether, a common pathogen response of both cultivars is evident through enhanced levels of proteins involved in (iso) flavonoid production, of which the most significant and common compound identified was Sativan. Unfortunately, not much is known about the function of this isoflavone. Our data, however, support that it is involved in pathogen defense, especially of NM treated plants.
We found a significant reduction in seed infection levels, as well as lower levels of leaf lesion areas, when comparing M vs. NM treated plants of both cultivars. Remarkably, with an almost 2-fold reduction of seed infection levels the bio-control potential of mycorrhizal symbiosis against D. pinodes was very similar to the potential of rhizobial symbiosis demonstrated in our previous study (Ranjbar Sistani et al., 2017). This is particularly interesting because our earlier studies on AMF symbiosis showed no reduction in leaflets disease severity upon D. pinodes attack (Desalegn et al., 2016; Turetschek et al., 2017). Nevertheless, it has been reported that AMF protect plants against pathogens through the dampening of phytopathogenic effects (Borowicz, 2001). Also, AMF inhibiting impact on disease caused by Xanthomonas translucens on leaves of wheat was observed (Fiorilli et al., 2018). Altogether, the significant decrease of seed infection besides smaller lesion areas in AMF treated plants highlights the bio-control potential of AMF as a belowground microsymbiont reducing disease severity caused by an aboveground pathogen, in line with the previously reviewed findings by Jung et al. (2012), reporting the role of AMF in the reduction of infections caused by Botrytis cinerea (Møller et al., 2009; Pozo et al., 2010), Alternaria solani (Fritz et al., 2006; de la Noval et al., 2007), and Magnaporthe grisea (Campos-Soriano et al., 2012). Additionally, the high potential of AMF to control B. cinerea in tomato has been identified (Fiorilli et al., 2011). Nevertheless, compared to Rhizobium inoculation (Desalegn et al., 2016), AMF seems to have less significant effects on pea cultivar growth promotion and leaf protection but a similar impact on seed pathogen resistance. This study together with our earlier work (Ranjbar Sistani et al., 2017) revealed that both mycorrhizal and rhizobial below-ground symbionts are dominant against above-ground pathogen invasion. Besides, the below-ground symbiotic activity by AMF or Rhizobium reduced the systematic infection caused by the above-ground pathogen. Both studies indicate that the efficiency of these fungal and bacterial microsymbionts could be influenced by cultivar type and pathogen infection, however, their general bio-control function and infection inhibition is cultivar independent.
Levels of Vignatic acid A were increased in seeds of M treatments of cv. Messire upon pathogen infection. Vignatic acid A is known as a preventing compound with a promoting impact on resistance against bruchid beetle pest (Sugawara et al., 1996).
This cultivar specific effect becomes even more complex when comparing metabolic pathways that are induced by AMF symbiosis of healthy plants and are potentially involved in pathogen response. This specific mechanism (priming) of the plant metabolism by microsymbionts such as AMF, before infection by biotrophic and hemibiotrophic pathogens, is known as ISR (Cameron et al., 2013). Primed plants have already developed a certain metabolic defense apparatus, which reduces or even prevents pathogen attack. For instance, phytohormone JA plays a role in inducing systemic resistance against stresses (Luo et al., 2009; Song et al., 2014 as cited in Staudinger et al., 2016; Adolfsson et al., 2017; Bernardo et al., 2017) and also during plant development (Bernardo et al., 2017). Also, JA has a high impact on mycorrhizal colonization especially in higher plants (Gutjahr et al., 2015, as cited in Bernardo et al., 2017). According to our results, a protein related to the synthesis of JA (lipoxygenase frv2_95615 and frv2_84258) was highly increased in M plants (Table S4). Furthermore, data indicate that the JA-responsive protein (frv2_84258) is involved in ISR (Figure 4 and Table S6). Additionally, we identified N-jasmonoyl isoleucine, and it was significantly enhanced in cv. Protecta, similar to the levels of JA in our previous study (Ranjbar Sistani et al., 2017), but this time strongly induced by AMF instead of Rhizobium symbiosis. The JA has a role in regulating of pea reaction against D. pinodes (Fondevilla et al., 2011). These findings support that cv. Protecta is not only a more resistant genotype but is also more receptive to ISR mechanisms by AMF. Hence, our data support that the effect of AMF treatment on the seed metabolism and in line with our previous findings of Rhizobium treatments is not due to a difference in P-regime but rather depending on the cultivar and due to a global change in metabolic homeostasis of the whole plant induced by microsymbiosis. Although pathogenesis-related (PR) proteins are known to be commonly involved in pathogen defense (van Loon et al., 2006), we found only a few of them involved in pea leaf and seed protection (Desalegn et al., 2016; Ranjbar Sistani et al., 2017). Here again, we found possible isoforms (frv2_110573; frv2_74661; frv2_41448; frv2_88778) induced upon pathogen infection in cv. Protecta seeds only. Interestingly, they were already induced by AMF symbiosis and further accumulated upon stress. Besides PR proteins, the detection of proteins involved in cell wall fortification, such as proteins of the monolignol pathway, seemed limited. Nevertheless, we found a UDP-glucosyltransferase as well as UDP-glucose 4-epimerase and UDP-glucose 6-dehydrogenase possibly involved in cell wall synthesis. These three enzymes were induced upon pathogen attack of cv. Protecta, but only UDP-glucosyltransferase (frv2_103598) was also enhanced in seeds of AMF symbiotic plants (cv. Protecta). Furthermore, Cyanidin 3-sophoroside 5-glucoside seemed involved in better seed pathogen resistance upon AMF symbiosis of cv. Protecta, being only induced upon infection of M treated plants. These findings suggest that AMF does not only prime.
The 2,3-Dihydroxy-2,4-cyclopentadien-1-one was also accumulated upon pathogen infection in both cultivars with a more significant effect on cv. Protecta. Very striking were also the high levels of several proteins of amino acid metabolism and glycolysis in cv. Protecta. Protein levels showed a strong increase under pathogen infection of non-mycorrhizal cv. Protecta, but some were also induced in healthy M treated plants and did not much further increase in Protecta seeds of M plants after pathogen attack, indicative for a primed primary metabolism as previously described by Schwachtje et al. (2018). By the production of pyruvate, the entry metabolite of the TCA cycle, glycolysis plays a role in energy supply and thus the production of defense metabolites (Rojas et al., 2014). Hence, the data suggest that the cultivar related difference in the primary metabolism seems another reason for the better performance of cv. Protecta.
Nevertheless, some defense mechanisms were also found to be more pronounced in cv. Messire. For instance, flavonoid Quercetin 3-(6''-acetylgalactoside)-7-rhamnoside strongly accumulated upon pathogen infection, especially in cv. Messire and most strongly in seeds of M treated plants. Similarly, Pisatin as specific phytoalexin of P. sativum has been induced against pathogen attack (De Wit-Elshove, 1969). Its synthesis pathway has previously been observed to be induced in the leaf proteome of Rhizobium inoculated pea plants under infection caused by D. pinodes (Desalegn et al., 2016). In contrast, pisatin involvement in rhizobial induced seed protection could not be detected in our previous study (Ranjbar Sistani et al., 2017). Interestingly, in the present study, a considerable AMF induced enhancement of proteins and metabolites of the secondary metabolism such as proteins of the flavonoid biosynthesis, especially the accumulation of isoflavone reductase and chalcone isomerase, along with increased levels of flavonoids and isoflavonoids, mainly in seeds of cv. Messire under disease stress was found. This finding supports that pisatin biosynthesis is induced in seeds of M plants upon pathogen infection.
Although, AMF showed cultivar specific effects, particularly for cv. Protecta, our results also indicated a general enhancement of pathogen resistance through AMF symbiosis (Figure 5). For example, G-proteins, known as signal transduction regulators (Smith et al., 1998; van Heusden, 2009; Smith et al., 2011; Parua and Young, 2014, as cited in Sun et al., 2018) have a role in plant responses to various stress conditions (Roberts et al., 2002; Lozano-Duran and Robatzek, 2015; Li et al., 2016, as cited in Sun et al., 2018). Mycorrhizal induced accumulation of G-proteins in wheat roots against drought stress was also reported (Bernardo et al., 2017). In our study, the group of proteins (frv2_86036; frv2_118099; Q9C5W6; frv2_94136) was particularly increased in seeds of AMF-associated plants under pathogen disease, indicating a possible role in ISR signaling.
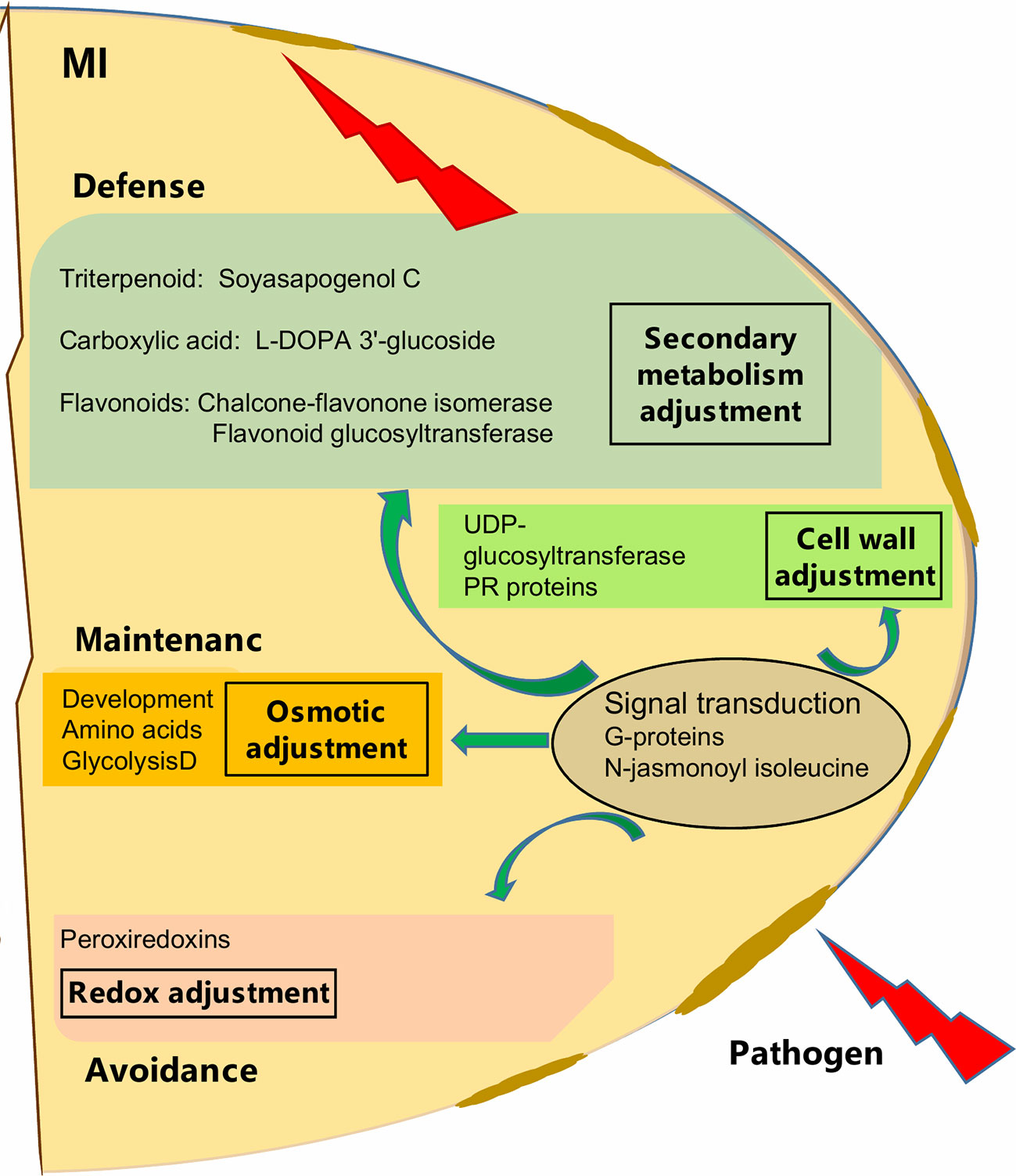
Figure 5 Schematic overview of mycorrizal induced response of pea seed (grown in pots) against D. pinodes infection. Seed metabolites and proteins significantly (Tukey HSD, p < 0.05; n = 3; Supplementary Table S3B and S4) enhanced with the highest ICA loadings (> 0.01 and < −0.01; Supplementary Table S5) are shown. MI, Mycorrhizae inoculated, and pathogen-infected.
In general, we found a similar molecular defense pattern induced by either AMF like Rhizobium against D. pinodes, described in our prior study (Ranjbar Sistani et al., 2017). This is not surprising, as the host builds fortification and munitions. Hence, Flavonoids and Soysapogenol C were important components in secondary metabolism adjustment in both Rhizobium- and AMF-induced responses. Interestingly, however, L-DOPA 3'-glucoside, discussed above, was significantly induced by AMF and in response to stress and not found responsive to Rhizobium-symbiosis. Also, the main proteins involved in signal transduction, mentioned above, were differentially induced. While, AMF enhanced levels of G-proteins and phosphatase 2C seems Rhizobium specific (Ranjbar Sistani et al., 2017). The analysis of secondary metabolites is still in its infancy and identification remains a bottleneck. Thus, major symbiont and pathogen involved differences and further key secondary metabolites regulation pathogen defense, remain to be identified in the future. The difference might also lie in response rate and/or induction level that might be different not only between cultivars but also depending on symbiosis, terms that need further evaluation.
Taken together, among common stress responses, several proteins and metabolites have been found AMF and cultivar specifically regulated, demonstrating different defense strategies. While genotype-specific resistance pattern is a common base in plant breeding, not much is known about genotype-specific microsymbiont induced priming effects.
In this study, we demonstrated that AMF is not only promoting growth and yield of pea plants but also protects seed yield upon pathogen attack. The seed metabolic response of two cultivars with varying susceptibility to the pathogen D. pinodes was analyzed. Besides, with the growth-promoting effect of AMF microsymbiont, we demonstrate its positive impacts on the resistance of the plants against the pathogen. We found that the genotypic effects are strong for both the general and the AMF induced pathogen response.
Altogether, cv. Protecta, the less susceptible genotype, accumulates more proteins of the secondary metabolism involved in lipid, flavonoid, and phytohormone production as well as of the primary amino acid and glycolysis metabolism. Besides, cv. Protecta showed a stronger induction (priming) of seed metabolic pathways upon AMF symbiosis, which led to a dampened and less severe but more effective response to pathogen attack. Nevertheless, pathogen infection was also reduced in cv. Messire when interacting with AMF. However, infection levels were similar compared to cv. Protecta without AMF symbiosis and thus AMF impact less effective. Thus, the data demonstrate AMF and genotype-specific pathogen defense strategies. This study demonstrates that AMF as below-ground microsymbiont not only promote the growth above-below ground parts of pea plants but enhance the resistance of P. sativum plants and hence protects seed quantity. Also, AMF have a notable influence on the seed metabolome and proteome, influencing seed metabolism and thus nutritional and medicinal pea seeds quality.
Although, mycorrhizal and rhizobial symbionts in singular applications could be prescribed for improvement of pea seed protection, little is known about impacts of co-inoculation, often occurring in nature, and therefore investigations are demanded.
We believe that sustainable development of food resources for food poverty elimination can be strongly supported through the enhancement of productivity and quality of crops by using microbial symbiotic potentials. Focusing on plant species supplying food and microsymbiont interaction studies on improving production- and seed quality-related strategies under stress conditions besides promoting current ecological properties should be a priority in plant and agricultural studies. We recommend more studies related to efficiency assessment of soil microorganisms on food plants productivity upon abiotic and biotic stresses under field conditions with considering the environmental viabilities.
The datasets generated for this study can be found in the PRIDE PXD006617.
Experiments, including phenotyping, proteomics, and secondary metabolites analyses were carried out by NRS. Data mining was performed by NRS and SW. NRS and SW wrote the manuscript. SW, GD, and H-PK conceived the work. The manuscript was revised by GD and H-PK.
We appreciated the support from COST Action FA1306.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the gardeners of the division of Molecular Systems Biology (Mosys, Faculty of Life Sciences, University of Vienna), Thomas Joch, and Andreas Schröfl. Thanks also to Sonja Tischler for running and maintaining the mass spectrometry facility of Mosys division. We thank Core Facility Cell Imaging and Ultrastructure Research (CIUS, Faculty of Life Sciences, University of Vienna) for supplying microscopy equipment, Dr. Irene Lichtscheidl-Schultz, and Dr. Ingeborg Lang for their instructions about microscopy equipment. Also, we warmly thank Dr. Wenchao Zhang for his supportive and effectual instructions about MET-COFEA and MET-XAlign tools.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00872/full#supplementary-material
Figure S1 | Comparison of functional categories (including > 2 proteins) of seed proteome significantly (Kruskal-Wallis; ANOVA, Tukey HSD test; p < 0.05) increased (summed increased fold change ≥ 2) in M vs. NM, I vs. U and Me vs. Pr. Me: cv. Messire, Pr: cv. Protecta, M: mycorrhizal, NM: non-mycorrhizal, I: infected (diseased), U: uninfected (healthy).
Abdel-Fattah, G. M., Shukry, W. M., Shokr, M. M. B., Ahmed, M. A. (2016). Application of mycorrhizal technology for improving yield production of common bean plants. Int. J. Appl. Sci. Biotechnol. 4, 191–197. doi: 10.3126/ijasbt.v4i2.15103
Abdel-Fattah, G. M. (1997). Functional activity of va-mycorrhiza (Glomus mosseae) in the growth and productivity of soybean plants grown in sterilized soil. Folia Microbiol. 42, 495–502. doi: 10.1007/BF02826560
Abdelgani, M. E., Elsheikh, E. A. E., Mukhtarb, N. O. (1999). The effect of Rhizobium inoculation and chemical fertilization on seed quality of fenugreek. Food Chem. 64, 289–293. doi: 10.1016/S0308-8146(98)00098-3
Abdul Baki, A. A., Anderson, J. D. (1973). Vigor determination in soybean seed by multiple criteria. Crop Sci. 13, 630–633. doi: 10.2135/cropsci1973.0011183X001300060013x
Adeyemi, N., Sakariyawo, O., Atayese, M. (2017). Yield and yield attributes responses of soybean (Glycine max L. Merrill) to elevated CO2 and arbuscular mycorrhizal fungi inoculation in the humid transitory rainforest. Not. Sci. Biol. 9, 233–241. doi: 10.15835/nsb9210002
Adolfsson, L., Nziengui, H., Abreu, I. N., Šimura, J., Beebo, A., Herdean, A., et al. (2017). Enhanced Secondary- and Hormone Metabolism in Leaves of Arbuscular Mycorrhizal Medicago truncatula. Plant Physiol. 175, 392–411. doi: 10.1104/pp.16.01509
Afendi, F. M., Okada, T., Yamazaki, M., Hirai-Morita, A., Nakamura, Y., Nakamura, K., et al. (2012). KNApSAcK family databases: integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 53, e1(1–e112. doi: 10.1093/pcp/pcr165
Aimo, S., Gosetti, F., D'Agostino, G., Gamalero, E., Gianotti, V., Bottaro, M., et al. (2010). Use of arbuscular mycorrhizal fungi and beneficial soil bacteria to improve yield and quality of saffron (Crocus sativus L.). ISHS Acta Hortic. 850, 159–162. doi: 10.17660/ActaHortic.2010.850.25
Al-Karaki, G. N., Clark, R. B. (1999). Mycorrhizal influence on protein and lipid of durum wheat grown at different soil phosphorus levels. Mycorrhiza 9, 97–101. doi: 10.1007/s005720050006
Atkinson, D., Berta, G., Hooker, J. E. (1994). “Impact of mycorrhizal colonisation on root architecture, root longevity and the formation of growth regulators,” in Impact of Arbuscular Mycorrhizas on Sustainable Agriculture and Natural Ecosystems. Eds. Gianinazzi, S., Schüepp, H. (Birkhäuser, Basel: ALS Advances in Life Sciences). doi: 10.1007/978-3-0348-8504-1_8
Azcón-Aguilar, C., Barea, J. M. (1996). Arbuscular mycorrhizas and biological control of soil-borne plant pathogens–an overview of the mechanisms involved. Mycorrhiza 6, 457–464. doi: 10.1007/s005720050147
Bäertschi, H., Gianinazzi-Pearson, V., Vegh, I. (1982). Vesiculararbuscular mycorrhiza and root rot disease (Phytophthora cinnamomi Rands) development in Chamaecyparis lawsoniana. Pythopathol 102, 213–218. doi: 10.1111/j.1439-0434.1981.tb03382.x
Baltruschat, H., Schonbeck, F. (1972). The influence of endotrophic mycorrhiza on the infestation of tobacco by Thielaviopsis basicola. Phytopathol 84, 172–188. doi: 10.1111/j.1439-0434.1975.tb03558.x
Barnard, E. L. (1977). The mycorrhizal biology of Liriodendron ruliptfera L. and its relationship to Cylindrocladium root rot. [dissertation] ([Durham]: Duke Univ., NC), 147. pp.
Battaglia, M., Covarrubias, A. A. (2013). Late embryogenesis abundant (LEA) proteins in legumes. Front. Plant Sci. 25, 4:190. doi: 10.3389/fpls.2013.00190
Becker, W. N. (1976). Quantification of onion vesicular-arbuscular mycorrhizae and their resistance to Pyrenochaeta terrestris. [dissertation] ([Urbana]: Univ. Illinois.), 72 pp.
Begum, A., Leibovitch, S., Migner, P., Zhang, F. (2001). Inoculation of pea (Pisum sativum L.) by Rhizobium leguminosarum bv. viceae preincubated with naringenin and hesperetin or application of naringenin and hesperetin directly into soil increased pea nodulation under short season conditions. Plant Soil. 237, 71–80. doi: 10.1023/A:1013328906027
Bernardo, L., Morcia, C., Carletti, P., Ghizzoni, R., Badeck, F. W., Rizza, F., et al. (2017). Proteomic insight into the mitigation of wheat root drought stress by arbuscular mycorrhizae. J. Proteomics S1874-3919 (17), 30108–30102. doi: 10.1016/j.jprot.2017.03.024
Bethlenfalvay, G. J., Mihara, K. L., Schreiner, R. P. (1994). Mycorrhizae alter protein and lipid contents and yield of pea seeds. Crop Sci. 34, 998–1003. doi: 10.2135/cropsci1994.0011183X003400040031x
Bethlenfalvay, G. J., Paul Schreiner, R., Mihara, K. L. (1997). Mycorrhizal fungi effects on nutrient composition and yield of soybean seeds. J. Plant Nutr. 20, 581–591. doi: 10.1080/01904169709365276
Bona, E., Lingua, G., Manassero, P., Cantamessa, S., Marsano, F., Todeschini, V., et al. (2015). AM fungi and PGP pseudomonads increase flowering, fruit production, and vitamin content in strawberry grown at low nitrogen and phosphorus levels. Mycorrhiza 25, 181–193. doi: 10.1007/s00572-014-0599-y
Bona, E., Scarafoni, A., Marsano, F., Boatt, L., Copetta, A., Massa, N., et al. (2016). Arbuscular mycorrhizal symbiosis affects the grain proteome of Zea mays: a field study. Sci. Rep. 6, 26439. doi: 10.1038/srep26439
Borde, M., Dudhane, M., Jite, P. K. (2009). Role of bioinoculant (AM Fungi) increasing in growth, flavor content and yield in Allium sativum L. under field condition. Not. Bot. Horti. Agrobot. Cluj-Napoca 37, 124–128. doi: 10.15835/nbha3723289
Borowicz, V. A. (2001). Do arbuscular mycorrhizal fungi alter plant-pathogen relations? Ecology 82, 3057–3068. doi: 10.2307/2679834
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Broughton, W., Dilworth, M. (1970). “Plant nutrient solutions,” in Handbook for Rhizobia; Methods in Legume–Rhizobium Technology. Eds. Somasegaran, P., Hoben, H. J. (Honolulu, HI: University of Hawaii).
Cameron, D. D., Neal, A. L., van Wees, S. C., Ton, J. (2013). Mycorrhiza-induced resistance: more than the sum of its parts? Trends Plant Sci. 18, 539–545. doi: 10.1016/j.tplants.2013.06.004
Campos-Soriano, L., García-Martínez, J., San Segundo, B. (2012). The arbuscular mycorrhizal symbiosis promotes the systemic induction of regulatory defense-related genes in rice leaves and confers resistance to pathogen infection. Mol. Plant Pathol 13, 579–592. doi: 10.1111/j.1364-3703.2011.00773.x
Carrillo, E., Rubiales, D., Pérez-de-Luque, A., Fondevilla, S. (2013). Characterization of mechanisms of resistance against Didymella pinodes in Pisum spp. Eur. J. Plant Pathol. 135, 761–769. doi: 10.1007/s10658-012-0116-0
Castellanos-Morales, V., Villegas, J., Wendelin, S., Vierheilig, H., Eder, R., Cárdenas-Navarro, R. (2010). Root colonisation by the arbuscular mycorrhizal fungus Glomus intraradices alters the quality of strawberry fruits (Fragaria × ananassa Duch.) at different nitrogen levels. J. Sci. Food Agric. 90, 1774–1782. doi: 10.1002/jsfa.3998
Castellanos-Morales, V., Villegas-Moreno, J., Vierheilig, H., Cárdenas-Navarro, R. (2012). Nitrogen availability drives the effect of Glomus intraradices on the growth of strawberry (Fragaria x ananassa Duch.) plants. J. Sci. Food Agric. 92, 2260–2264. doi: 10.1002/jsfa.5618
Cely, M. V. T., de Oliveira, A. G., de Freitas, V. F., de Luca, M. B., Barazetti, A. R., dos Santos, I. M. O., et al. (2016). Inoculant of arbuscular mycorrhizal fungi (Rhizophagus clarus) increase yield of soybean and cotton under field conditions. Front. Microbiol. 7, 720. doi: 10.3389/fmicb.2016.00720
Chatzistathis, T., Orfanoudakis, M., Alifragis, D., Therios, L. (2013). Colonization of Greek olive cultivars' root system by arbuscular mycorrhiza fungus: root morphology, growth, and mineral nutrition of olive plants. Sci. Agric. 70, 185–194. doi: 10.1590/S0103-90162013000300007
Chen, S., Zhao, H., Zou, C., Li, Y., Chen, Y., Wang, Z., et al. (2017). Combined inoculation with multiple arbuscular mycorrhizal fungi improves growth, nutrient uptake and photosynthesis in cucumber seedlings. Front. Microbiol. 8, 2516. doi: 10.3389/fmicb.2017.02516
Chou, L. G., Schmitthenner, A. F. (1974). Effect of Rhizobium japonicum and Endogone mosseae on soybean root rot caused by Pythium ultimum and Phytophthora megasperma var. sojae. Plant Dis. Rep. 58, 221–225.
Cox, J., Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., Mann, M. (2011). Andromeda: A peptide search engine Integrated into the maxquant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Cruz, A. F., Ishii, T., Kadoya, K. (2000). Effects of arbuscular mycorrhizal fungi on tree growth, leaf water potential, and levels of 1-aminocyclopropane-1-carboxylic acid and ethylene in the roots of papaya under water-stress conditions. Mycorrhiza 10, 121–123. doi: 10.1007/s005720000067
Davidson, J. A., Kimber, R. B. E. (2007). Integrated disease management of ascochyta blight in pulse crops. Eur. J. Plant Pathol. 119, 99–110. doi: 10.1007/s10658-007-9132-x
Davidson, J., Krysinska-Kaczmarek, M., McKay, H. (2012). Comparison of cultural growth and in planta quantification of Didymella pinodes, Phoma koolunga and Phoma medicaginis var. pinodella, causal agents of ascochyta. Mycologia 104, 93–101. doi: 10.3852/11-118
Davis, R. M., Menge, J. A. (1980). Influence of Glomus fasciculatus and soil phosphorus on Phytophthora root rot of citrus. Phytopathology 70, 447–452. doi: 10.1094/Phyto-70-447
de la Noval, B., Pérez, E., Martínez, B., León, O., Martínez-Gallardo, N., Délano-Frier, J. (2007). Exogenous systemin has a contrasting effect on disease resistance in mycorrhizal tomato (Solanum lycopersicum) plants infected with necrotrophic or hemibiotrophic pathogens. Mycorrhiza 17, 449–460. doi: 10.1007/s00572-007-0122-9
De Vos, R., Moco, S., Lommen, A., Keurentjes, J. J., Bino, R. J., Hall, R. D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2, 778–791. doi: 10.1038/nprot.2007.95
De Wit-Elshove, A. (1969). The role of pisatin in the resistance of pea plants-some further experiments on the breakdown of pisatin. Netherlands J. Plant Pathol. 75, 164–168. doi: 10.1007/BF02137212
Dehne, H. W., Schoenbeck, F. (1979). The influence of endotrophic mycorrhiza on plant diseases. 1. Colonization of tomato plants by Fusarium oxysporum f. sp. lycopersici. Phytopathol 95, 105–110. doi: 10.1111/j.1439-0434.1979.tb01584.x
Dehne, H. W. (1977). Untersuchungen Ober den Einfluss der endotrophen Mycorrhiza auf die Fusarium-Welke an Tomate und Gurke. [Dissertation] ([Bonn]: Univ. Bonn., W. Germany), 150.
Dehne, H. W. (1982). Interaction between vesicular-arbuscular mycorrhizal fungi and plant pathogens. Phytopathology 72, 1114–1119.
Deneufbourg, F., Seguin, B., Molinero, V. (1994). Les traitements de semences contre l'anthracnose. Bull. Des. Semences 129, 39–41.
Desalegn, G., Turetschek, R., Kaul, H. P., Wienkoop, S. (2016). Microbial symbionts affect Pisum sativum proteome and metabolome under Didymella pinodes infection. J. Proteomics 143, 173–187. doi: 10.1016/j.jprot.2016.03.018
Egelhofer, V., Hoehenwarter, W., Lyon, D., Weckwerth, W., Wienkoop, S. (2013). Using ProtMAX to create high-mass-accuracy precursor alignments from label-free quantitative mass spectrometry data generated in shotgun proteomics experiments. Nat. Protoc. 8, 595–601. doi: 10.1038/nprot.2013.013
Farrag, E., Moharam, M. (2012). Pathogenic fungi transmitted through cucumber seeds and safely elimination by application of peppermint extract and oil. Not. Sci. Biol. 4, 83–91. doi: 10.15835/nsb437969
Fernandez-Aparicio, M., Garcia-Garrido, J. M., Ocampo, J. A., Rubiales, D. (2010). Colonisation of field pea roots by arbuscular mycorrhizal fungi reduces Orobanche and Phelipanche species seed germination. Weed Res. 50, 262–268. doi: 10.1111/j.1365-3180.2010.00771.x
Fiorilli, V., Catoni, M., Francia, D., Cardinale, F., Lanfranco, L. (2011). The arbuscular mycorrhizal symbiosis reduces disease severity in tomato plants infected by Botrytis cinerea. J. Plant Pathol. 93, 237–242. doi: 10.1400/169610
Fiorilli, V., Vannini, C., Ortolani, F., Garcia-Seco, D., Chiapello, M., Novero, M., et al. (2018). Omics approaches revealed how arbuscular mycorrhizal symbiosis enhances yield and resistance to leaf pathogen in wheat. Sci. Rep. 258 (1), 9625. doi: 10.1038/s41598-018-27622-8
Fondevilla, S., Küster, H., Krajinski, F., Cubero, J. I., Rubiales, D. (2011). Identification of genes differentially expressed in a resistant reaction to Mycosphaerella pinodes in pea using microarray technology. BMC Genomics 12, 28. doi: 10.1186/1471-2164-12-28
Fontana, A., Reichelt, M., Hempel, S., Gershenzon, J., Unsicker, S. B. (2009). The effects of arbuscular mycorrhizal fungi on direct and indirect defense metabolites of Plantago lanceolata L. J. Chem. Ecol. 35, 833–843. doi: 10.1007/s10886-009-9654-0
Fritz, M., Jakobsen, I., Lyngkjær, M. F., Thordal-Christensen, H., Pons-Kühnemann, J. (2006). Arbuscular mycorrhiza reduces susceptibility of tomato to Alternaria solani. Mycorrhiza 16, 413–419. doi: 10.1007/s00572-006-0051-z
Fu, S. F., Wei, J. Y., Chen, H. W., Liu, Y. Y., Lu, H. Y., Chou, J. Y. (2015). Indole-3-acetic acid: A widespread physiological code in interactions of fungi with other organisms. Plant Signal Behav. 10 (8), e1048052. doi: 10.1080/15592324.2015.1048052
Gange, A. C., Lindsay, D. E., Ellis, L. S. (1999). Can arbuscular mycorrhizal fungi be used to control the undesirable grass Poa annua on golf courses? J. Appl. Ecol. 36, 909–919. doi: 10.1046/j.1365-2664.1999.00456.x
Garry, G., Jeuffroy, M. H., Ney, B., Tivoli, B. (1998). Effects of ascochyta blight (Mycosphaerella pinodes) on the photosynthesizing leaf area and the photosynthetic efficiency of the green leaf area of dried-pea (Pisum sativum). Plant Pathol. 47, 473–479. doi: 10.1046/j.1365-3059.1998.00259.x
Gaurilˇcikien˙e, I., ˇCesnuleviˇcien˙e, R., Janusauškait˙e, D., Ronis, A. (2012). Management of seed borne root and foot rot diseases of pea (Pisum sativum L.) with a fungicide seed treatment. Žemdirbystė (Agriculture). 99, 77–84.
Giovannetti, M., Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Guijas, C., Montenegro-Burke, J. R., Domingo-Almenara, X., Palermo, A., Warth, B., Hermann, G., et al. (2018). METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 90, 3156–3164. doi: 10.1021/acs.analchem.7b04424
Gutjahr, C., Siegler, H., Haga, K., Iino, M., Paszkowski, U. (2015). Full establishment of arbuscular mycorrhizal symbiosis in rice occurs independently of enzymatic jasmonate biosynthesis. PloS One 10 (4), e0123422. doi: 10.1371/journal.pone.0123422
Hamel, C., Plenchette, C. (2007). Mycorrhizae in crop production (Binghampton: Haworth food and agricultural products press). doi: 10.1094/Phyto-68-1377
Haneef Khan, Md., Meghvansi, M. K., Panwar, V., Gogoi, H. K., Singh, L. (2010). Arbuscular mycorrhizal fungi-induced signaling in plant defence against phytopathogens. J. Phytol. 2, 53–69.
Harrier, L. A., Watson, C. A. (2004). The potential role of arbuscular mycorrhizal (AM) fungi in the bioprotection of plants against soil-borne pathogens in organic and/ or other sustainable farming systems. Pest Manag. Sci. 60, 149–157. doi: 10.1002/ps.820
Hill, E. M., Robinson, L. A., Abdul-Sada, A., Vanbergen, A. J., Hodge, A., Hartley, S. E. (2018). Arbuscular mycorrhizal fungi and plant chemical defence: effects of colonisation on aboveground and belowground metabolomes. J. Chem. Ecol. 44, 198–208. doi: 10.1007/s10886-017-0921-1
Horii, S., Ishii, T. (2014). Effect of arbuscular mycorrhizal fungi and their partner bacteria on the growth of sesame plants and the concentration of sesamin in the seeds. Am. J. Plant Sci. 5, 3066–3072. doi: 10.4236/ajps.2014.520323
Hwang, S. F., Zhang, R., Chang, K. F., Gossen, B. D., Turnbull, G. D., Bing, D. J. (2006). Comparison of three methods for assessing disease reaction to Mycosphaerella pinodes in field pea (Pisum sativum). J. Plant Dis. Prot. 113, 20–23.
Jansa, J., Smith, F. A., Smith, S. E. (2008). Are there benefits of simultaneous root colonization by different arbuscular mycorrhizal fungi? New Phytol. 177, 779–789. doi: 10.1111/j.1469-8137.2007.02294.x
Jin, H., Germida, J. J., Walley, F. L. (2013). Impact of arbuscular mycorrhizal fungal inoculants on subsequent arbuscular mycorrhizal fungi colonization in pot-cultured field pea (Pisum sativum L.). Mycorrhiza 23, 45–59. doi: 10.1007/s00572-012-0448-9
Johnson, N. C., Graham, J. H., Smith, F. A. (1997). Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol. 135, 575–585. doi: 10.1046/j.1469-8137.1997.00729.x
Jung, S. C., Martinez-Medina, A., Lopez-Raez, J. A., Pozo, M. J. (2012). Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 38, 651–664. doi: 10.1007/s10886-012-0134-6
Kögl, F., Kostermans, D. G. F. R. (1934). Hetero-auxin als Stoffwechselprodukt niederer pflanzlicher Organismen. Isolierung aus Hefe. 13. Mitteilung über pflanzliche Wachstumsstoffe. Hoppe-Seyler´s Z. für pPysiol. Chem. 228, 113–121. doi: 10.1515/bchm2.1934.228.3-6.113
Kavitha, T., Nelson, R. (2014). Effect of arbuscular mycorrhizal fungi (AMF) on growth and yield of sunflower (Helianthus annuus L.). J. Exp. Biol. Agric. Sci. 2 (2S), 226–232.
Khan, T. N., Timmerman-Vaughanb, G. M., Rubialesc, D., Warkentind, T. D., Siddiquee, K. H. M., Erskinea, W., et al. (2013). Didymella pinodes and its management in field pea: Challenges and opportunities. Field Crops Res. 148, 61–77. doi: 10.1016/j.fcr.2013.04.003
Laparre, J., Malbreil, M., Letisse, F., Portais, J. C., Roux, C., Bécard, G., et al. (2014). Combining metabolomics and gene expression analysis reveals that propionyl- and butyryl-carnitines are involved in late stages of arbuscular mycorrhizal symbiosis. Mol. Plant 7, 554–566. doi: 10.1093/mp/sst136
Larrainzar, E., Gil-Quintana, E., Seminario, A., Arrese-Igor, C., González, E. M. (2014). Nodule carbohydrate catabolism is enhanced in the Medicagotruncatula A17-Sinorhizobium medicae WSM419 symbiosis. Front. Microbial. 5, 447. doi: 10.3389/fmicb.2014.00447
Larsen, P. E., Sreedasyam, A., Trivedi, G., Desai, S., Dai, Y., Cseke, L. J., et al. (2016). Multi-omics approach identifies molecular mechanisms of plant-fungus mycorrhizal interaction. Front. Plant Sci. 6, 1061. doi: 10.3389/fpls.2015.01061
Lecomte, J., St-Arnaud, M., Hijri, M. (2011). Isolation and identification of soil bacteria growing at the expense of arbuscular mycorrhizal fungi. FEMS Microbiol. Lett. 317, 43–51. doi: 10.1111/j.1574-6968.2011.02209.x
Li, W., Gao, Y., Xu, H., Zhang, Y., Wang, J. (2012). A proteomic analysis of seed development in Brassica campestri L. PloS One 7 (11), e50290. doi: 10.1371/journal.pone.0050290
Li, T., Sun, Y., Ruan, Y., Xu, L., Hu, Y., Hao, Z., et al. (2016). Potential role of D-myo-inositol-3-phosphate synthase and 14-3-3 genes in the crosstalk between Zea mays and Rhizophagus intraradices under drought stress. Mycorrhiza 26, 1–15. doi: 10.1007/s00572-016-0723-2
Li, Z., Yao, Q., Dearth, S. P., Entler, M. R., Castro Gonzalez, H. F., Uehling, J. K., et al. (2017). Integrated proteomics and metabolomics suggests symbiotic metabolism and multimodal regulation in a fungal-endobacterial system. Environ. Microbiol. 19, 1041–1053. doi: 10.1111/1462-2920.13605
Lingua, G., Bona, E., Manassero, P., Marsano, F., Todeschini, V., Cantamessa, S., et al. (2013). Arbuscular mycorrhizal fungi and plant growth-promoting pseudomonads increases anthocyanin concentration in strawberry fruits (Fragaria x ananassa var. selva) in conditions of reduced fertilization. Int. J. Mol. Sci. 14, 16207–16225. doi: 10.3390/ijms140816207
Liu, C. Y., Zhang, F., Zhang, D. J., Srivastava, A. K., Wu, Q. S., Zou, Y. N. (2018). Mycorrhiza stimulates root-hair growth and IAA synthesis and transport in trifoliate orange under drought stress. Sci. Rep. 8 (1), 1978. doi: 10.1038/s41598-018-20456-4
Lohse, M., Nagel, A., Herter, T., May, P., Schroda, M., Zrenner, R., et al. (2014). Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37, 1250–1258. doi: 10.1111/pce.12231
Lozano-Duran, R., Robatzek, S. (2015). 14-3-3 proteins in plant-pathogen interactions. Mol. Plant Microbe Interact. 28, 511–518. doi: 10.1094/MPMI-10-14-0322-CR
Lu, F. C., Lee, C. Y., Wang, C. L. (2015). The influence of arbuscular mycorrhizal fungi inoculation on yam (Dioscorea spp.) tuber weights and secondary metabolite content. PeerJ 24, 3:e1266. doi: 10.7717/peerj.1266
Ludidi, N. N., Pellny, T. K., Kiddle, G., Dutilleul, C., Groten, K., Van Heerden, P. D. R., et al. (2007). Genetic variation in pea (Pisum sativum L.) demonstrates the importance of root but not shoot C/N ratios in the control of plant morphology and reveals a unique relationship between shoot length and nodulation intensity. Plant Cell Environ. 30, 1256–1268. doi: 10.1111/j.1365-3040.2007.01699.x
Ludwig-Müller, J., Güther, M. (2007). Auxins as Signals in Arbuscular Mycorrhiza Formation. Plant Signal Behav. 2 (3), 194–196. doi: 10.1111/j.1399-3054.2006.00812.x
Luo, Z. B., Janz, D., Jiang, X., Göbel, C., Wildhagen, H., Tan, Y., et al. (2009). Upgrading root physiology for stress tolerance by ectomycorrhizas: insights from metabolite and transcriptional profiling into reprogramming for stress anticipation. Plant Physiol. 151, 1902–1917. doi: 10.1104/pp.109.143735
Møller, K., Kristensen, K., Yohalem, D., Larsen, J. (2009). Biological management of gray mold in pot roses by coinoculation of the biocontrol agent Ulocladium atrum and the mycorrhizal fungus Glomus mosseae. Biol. Control. 49, 120–125. doi: 10.1016/j.biocontrol.2009.01.015
Mahmoud, S. Y. M., Hosseny, M. H., EL-Shaikh, K. A. A., Obiadalla, A. H. A., Mohamed, Y. A. (2013). Seed borne fungal pathogens associated with common bean (Phaseolus vulgaris L.) seeds and their impact on germination. J. Environ. Stud. 11, 19–26.
Mano, Y., Nemoto, K., Suzuki, M., Seki, H., Fujii, I., Muranaka, T. (2010). The AMI1 gene family: indole-3-acetamide hydrolase functions in auxin biosynthesis in plants. J. Exp. Bot. 61 (1), 25–32. doi: 10.1093/jxb/erp292
Moussart, A., Tivoli, B., Lemarchand, E., Deneufbourg, F., Roi, S., Sicard, G. (1998). Role of seed infection by the Ascochyta blight pathogen of dried pea (Mycosphaerella pinodes) in seedling emergence, early disease development and transmission of the disease to aerial plant parts. Eur. J. Plant Pathol. 104, 93–102. doi: 10.1023/A:1008673914537
Njeru, E. M., Avio, L., Sbrana, C., Turrini, A., Bocci, G., Bàrberi, P., et al. (2013). First evidence for a major cover crop effect on arbuscular mycorrhizal fungi and organic maize growth. Agron. Sustain. Dev. 34, 841–848. doi: 10.1007/s13593-013-0197-y
Nwangburuka, C. C., Olawuyi, O. J., Oyekale, K., Ogunwwenmo, K. O., Denton, O., Daramola, D. S., et al. (2012). Effect of arburscular mycorrhizea (AM), poultry manure (PM), NPK fertilizer and the combination of AM-PM on the growth and yield of okra (Abelmoschus esculentus). Nat. Sci. .10 (9), 35–41.
Okorska, S., Michalczyk, D., Okorski, A., Piotrowicz-Cieslak, A., Pupel, P., Glowacka, K., et al. (2014). Variability of PSPAL1 (Phenylalanine Ammonia-lyase Gene-1) proximal promoter sequence and expression in pea challenged with Mycosphaerella pinodes. Czech J. Genet. Plant Breed. 50, 163–170. doi: 10.17221/149/2013-CJGPB
Oruru, M. B., Njeru, E. M., Pasquet, R., Runo, S. (2017). Response of a wild-type and modern cowpea cultivars to arbuscular mycorrhizal inoculation in sterilized and non-sterilized soil. J. Plant Nutr. 41, 90–101. doi: 10.1080/01904167.2017.1381728
Parua, P. K., Young, E. T. (2014). Binding and transcriptional regulation by 14-3-3 (Bmh) proteins requires residues outside of the canonical motif. Eukaryot. Cell. 13, 21–30. doi: 10.1128/EC.00240-13
Pedone-Bonfim, M. V., Lins, M. A., Coelho, I. R., Santana, A. S., Silva, F. S., Maia, L. C. (2013). Mycorrhizal technology and phosphorus in the production of primary and secondary metabolites in cebil (Anadenanthera colubrina (Vell.) Brenan) seedlings. J. Sci. Food Agric. 93, 1479–1484. doi: 10.1002/jsfa.5919
Plant & Mycorrhiza Root Lengths. International culture collection of (vesicular) arbuscular mycorrhizal fungus (INVAM) (West Virginia University). Retrieved from: https://invam.wvu.edu/methods/mycorrhizae/mycorrhizalength Last updated: 10.05.2019.
Porcel, R., Aroca, R., Ruiz-Lozano, J. M. (2012). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron. Sustain. Dev. 32, 181–200. doi: 10.1007/s13593-011-0029-x
Pozo, M. J., Jung, S. C., López-Ráez, J. A., Azcón-Aguilar, C. (2010). “Impact of arbuscular mycorrhizal symbiosis on plant response to biotic stress: The role of plant defence mechanisms,” in Arbuscular mycorrhizas: physiology and function. Eds. Koltai, H., Kapulnik, Y. (Dordrecht: Springer). doi: 10.1007/978-90-481-9489-6_9
R Core Team (2013). R: A language and environment for statistical computing (Vienna, Austria: R Foundation for Statistical Computing). URL http://www.R-project.org/.
Rajjou, L., Belghazi, M., Huguet, R., Robin, C., Moreau, A., Job, C., et al. (2006). Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Phys. 141, 910–923. doi: 10.1104/pp.106.082057
Ranjbar Sistani, N., Kaul, H.-P., Desalegn, G., Wienkoop, S. (2017). Rhizobium impacts on seed productivity, quality, and protection of Pisum sativum upon disease stress caused by Didymella pinodes: phenotypic, proteomic, and metabolomic traits. Front. Plant Sci. 8, 1961. doi: 10.3389/fpls.2017.01961
Rebollo Couto, M. S., Lovato, P. E., Wipf, D., Dumas-Gaudot, E. (2013). Proteomic studies of arbuscular mycorrhizal associations. Adv. Biol. Chem. 3, 48–58. doi: 10.4236/abc.2013.31007
Rivero, J., Gamir, J., Aroca, R., Pozo, M. J., Flors, V. (2015). Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 6, 598. doi: 10.3389/fmicb.2015.00598
Roberts, M. R., Salinas, J., Collinge, D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. doi: 10.1023/A:1021261614491
Rojas, C. M., Senthil-Kumar, M., Tzin, V., Mysore, K. S. (2014). Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 5, 17. doi: 10.3389/fpls.2014.00017
Safir, G. (1968). The influence of vesicular-arbuscular mycorrhiza on the resistance of onion to Pyrenochaeta terrestris. [Master"s thesis] ([Urbana]: Univ. Illinois.), 36 pp.
Saia, S., Ruisi, P., Fileccia, V., Di Miceli, G., Amato, G., Martinelli, F. (2015). Metabolomics suggests that soil inoculation with arbuscular mycorrhizal fungi decreased free amino acid content in roots of durum wheat grown under N-limited, P-rich field conditions. PloS One 10 (6), e0129591. doi: 10.1371/journal.pone.0129591
Sajid, M., Khan, M. A., Rab, A., Shah, S. N. M., Arif, M., Jan, I., et al. (2012). Impact of nitrogen and phosphorus on seed yield and yield components of Okra cultivars. J. Anim. Plant Sci. 22, 704–707.
Sayeed Akhtar, M., Siddiqui, Z. A., Wiemken, A. (2011). “Arbuscular mycorrhizal fungi and Rhizobium to control plant fungal diseases,” in alternative farming systems, biotechnology, drought Stress and ecological fertilisation. Sustainable Agriculture Reviews, 6 . Ed. lichtfouse, E. (Springer, Dordrecht: Alternative Farming Systems, Biotechnology, Drought Stress and Ecological Fertilisation). doi: 10.1007/978-94-007-0186-1_9
Schenck, N. C., Ridings, W. H., Cornell, J. A. (1977). Interaction of two vesicular-arbuscular mycorrhizal fungi and Phytophthora parasitica on two citrus root stocks. (Abstr.), in: Proc. Third North Am. Conf. Mycorrhizae, Atlanta: University of Georgia. p. 9.
Scheublin, T. R., van der Heijden, M. G. (2006). Arbuscular mycorrhizal fungi colonize nonfixing root nodules of several legume species. New Phytol. 172, 732–738. doi: 10.1111/j.1469-8137.2006.01858.x
Schliemann, W., Ammer, C., Strack, D. (2008). Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry 69, 112–146. doi: 10.1016/j.phytochem.2007.06.032
Schoenbeck, F., Dehne, H. W. (1977). Damage to mycorrhizal cotton seedlings by Thielaviopsis basicola. Plant Dis. Rep. 61, 266–268.
Schoenbeck, F., Dehne, H. W. (1979). Untersuchungen zum Einfluss der endotrophen Mycorrhiza auf Planzenkrankheiten, 4. Pilzliche Sprossparasiten, Olpidium brassicae, TMV. Z. Pflanzenkrankh. Pflanzenschutz 86, 103–112.
Schwachtje, J., Fischer, A., Erban, A., Kopka, J. (2018). Primed primary metabolism in systemic leaves: a functional systems analysis. Sci. Rep. 8, 216. doi: 10.1038/s41598-017-18397-5
Schweiger, R., Baier, M. C., Persicke, M., Müller, C. (2014). High specificity in plant leaf metabolic responses to arbuscular mycorrhiza. Nat. Commun. 22, 5:3886. doi: 10.1038/ncomms4886
Sinclair, G., Charest, C., Dalpé, Y., Khanizadeh, S. (2014). Influence of colonization by arbuscular mycorrhizal fungi on three strawberry cultivars under salty conditions. Agric. Food Sci. 23, 146–158. doi: 10.23986/afsci.9552
Smith, F. A., Smith, S. E. (1996). Mutualism and parasitism: diversity in function and structure in the ‘arbuscular' (VA) mycorrhizal symbiosis. Adv. Bot. Res. 22, 1–43. doi: 10.1016/S0065-2296(08)60055-5
Smith, A. M., Quinton-Tulloch, J., Denyer, K. (1990). Characteristics of plastids responsible for starch synthesis in developing pea embryos. Planta 80, 517–523. doi: 10.1007/BF02411449
Smith, A., Ward, M. P., Garrett, S. (1998). Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17, 3556–3564. doi: 10.1093/emboj/17.13.3556
Smith, C. A., O'Maille, G., Want, E. J., Qin, C., Trauger, S. A., Brandon, T. R., et al. (2005). METLIN: a metabolite mass spectral database. Ther. Drug Monit. . 27, 747–751. doi: 10.1097/01.ftd.0000179845.53213.39
Smith, A. J., Daut, J., Schwappach, B. (2011). Membrane proteins as 14-3- 3 clients in functional regulation and intracellular transport. Physiology 26, 181–191. doi: 10.1152/physiol.00042.2010
Song, Y. Y., Ye, M., Li, C., He, X., Zhu-Salzman, K., Wang, R. L., et al. (2014). Hijacking common mycorrhizal networks for herbivore induced defence signal transfer between tomato plants. Sci. Rep. 4, 3915. doi: 10.1038/srep03915
Song, F., Qi, D., Liu, X., Kong, X., Gao, Y., Zhou, Z., et al. (2015). Proteomic analysis of symbiotic proteins of Glomus mosseae and Amorpha fruticosa. Sci. Rep. 5, 18031. doi: 10.1038/srep18031
Staudinger, C., Mehmeti-Tershani, V., Gil-Quintana, E., Gonzalez, E. M., Hofhansl, F., Bachmann, G., et al. (2016). Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteomics 136, 202–213. doi: 10.1016/j.jprot.2016.01.006
Steinkellner, S., Hage-Ahmed, K., García-Garrido, J. M., Illana, A., Ocampo, J. A., Vierheilig, H. (2012). A comparison of wild-type, old and modern tomato cultivars in the interaction with the arbuscular mycorrhizal fungus Glomus mosseae and the tomato pathogen fusarium oxysporum f. sp. Lycopersici. Mycorrhiza. 22, 189–194. doi: 10.1007/s00572-011-0393-z
Stewart, E. L., Pfleger, F. L. (1977). Development of poinsettia as influenced by endomycorrhizae, fertilizer and root rot pathogens Pythium ultimum and Rhizoctonia solani. Florist's Rev. 159, 79–80.
Storey, J. D., Tibshirani, R. (2003). Statistical significance for genome-wide studies. Proc. Natl. Acad. Sci. U. S. A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Storey, J. D., Taylor, J. E., Siegmund, D. (2004). Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J. R. Statist. Soc. Ser. B. 66, 187–205. doi: 10.1111/j.1467-9868.2004.00439.x
Storey, D. (2002). A direct approach to false discovery rates. J. R. Statist. Soc Ser. B. 64, 479–498. doi: 10.1111/1467-9868.00346
Storey, J. D. (2003). The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31, 2013–2035. doi: 10.1214/aos/1074290335
Strange, R. N., Scott, P. R. (2005). Plant disease: a threat to global food security. Annu. Rev. Phytopathol. 43, 83–116. doi: 10.1146/annurev.phyto.43.113004.133839
Sugawara, F., Ishimoto, M., Le-Van, N., Koshino, H., Uzawa, J., Yoshida, S., et al. (1996). Insecticidal peptide from mungbean: a resistant factor against infestation with azuki bean weevil. J. Agric. Food Chem. 44, 3360–3364. doi: 10.1021/jf960166c
Sun, X., Weckwerth, W. (2012). COVAIN: a toolbox for uni- and multivariate statistics, timeseries and correlation network analysis and inverse estimation of the differential Jacobian from metabolomics covariance data. Metabolomics 8, 81–93. doi: 10.1007/s11306-012-0399-3
Sun, Z., Song, J., Xin, X., Xie, X., Zhao, B. (2018). Arbuscular mycorrhizal fungal 14-3-3 proteins are involved in arbuscule formation and responses to abiotic stresses during AM symbiosis. Front. Microbiol. 9, 91. doi: 10.3389/fmicb.2018.00091
Tarawaya, K. (2003). Arbuscular mycorrhizal dependency of different plant species and cultivars. Soil Sci. Plant Nutr. 49, 655–668. doi: 10.1080/00380768.2003.10410323
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). Mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Turetschek, R., Lyon, D., Desalegn, G., Kaul, H. P., Wienkoop, S. (2016). “A proteomic workflow using high-throughput de novo sequencing towards complementation of genome information for improved comparative crop science,” in Proteomics in Systems Biology. Methods in Molecular Biology, vol. 1394 . Ed. Reinders, J. (New York, NY: Humana Press).
Turetschek, R., Desalegn, G., Epple, T., Kaul, H.-P., Wienkoop, S. (2017). Key metabolic traits of Pisum sativum maintain cell vitality during Didymella pinodes infection: cultivar resistance and the microsymbionts' influence. J. Proteomics 169, 189–201. doi: 10.1016/j.jprot.2017.03.001
Van Geel, M., De Beenhouwer, M., Lievens, B., Honnay, O. (2016). Crop-specific and single-species mycorrhizal inoculation is the best approach to improve crop growth in controlled environments. Agron. Sustain. Dev. 36 (2), 37. doi: 10.1007/s13593-016-0373-y
van Heusden, G. P. (2009). 14-3-3 Proteins: insights from genome-wide studies in yeast. Genomics 94, 287–293. doi: 10.1016/j.ygeno.2009.07.004
van Loon, L. C., Rep, M., Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Vierheilig, H., Coughlan, A. P., Wyss, U., Piche, Y. (1998). Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl. Environ. Microbiol. 64, 5004–5007. doi: 10.1128/AEM.64.12.5004-5007.1998
Vierheilig, H., Steinkellner, S., Khaosaad, T., Garcia-Garrido, J. M., Varma, A. (2008). “The biocontrol effect of mycorrhization on soilborne fungal pathogens and the autoregulation of the AM symbiosis: One mechanism, two effects?,” in Mycorrhiza (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-540-78826-3_15
Vizcaíno, J., Csordas, A., Del-Toro, N., Dianes, J., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, 447–456. doi: 10.1093/nar/gkv1145
Wang, Y. Y., Vestberg, M., Walker, C., Hurme, T., Zhang, X., Lindström, K. (2008). Diversity and infectivity of arbuscular mycorrhizal fungi in agricultural soils of the Sichuan Province of mainland China. Mycorrhiza 18, 59–68. doi: 10.1007/s00572-008-0161-x
Wang, L., Nägele, T., Doerfler, H., Fragner, L., Chaturvedi, P., Nukarinen, E., et al. (2016). System level analysis of cacao seed ripening reveals a sequential interplay of primary and secondary metabolism leading to polyphenol accumulation and preparation of stress resistance. Plant J. 87, 318–332. doi: 10.1111/tpj.13201
Wang, Y., Wang, M., Li, Y., Wu, A., Huang, J. (2018). Effects of arbuscular mycorrhizal fungi on growth and nitrogen uptake of Chrysanthemum morifolium under salt stress. PloS One 13 (4), e0196408. doi: 10.1371/journal.pone.0196408
Wehner, J., Antunes, P. M., Powell, J. R., Mazukatow, J., Rillig, M. C. (2010). Plant pathogen protection by arbuscular mycorrhizas: A role for fungal diversity? Pedobiologia 53, 197–201. doi: 10.1016/j.pedobi.2009.10.002
Wienkoop, S., Morgenthal, K., Wolschin, F., Scholz, M., Selbig, J., Weckwerth, W. (2008). Integration of metabolomic and proteomic phenotypes: analysis of data covariance dissects starch and RFO metabolism from low and high temperature compensation response in Arabidopsis thaliana. Mol. Cell. Proteomics 7, 1725–1736. doi: 10.1074/mcp.M700273-MCP200
Wishart, D. S., Feunang, Y. D., Marcu, A., Guo, A. C., Liang, K., Vázquez-Fresno, R., et al. (2018). HMDB 4.0: The human metabolome database for. Nucleic Acids Res. 46, D608–D617. doi: 10.1093/nar/gkx1089
Xue, A. G., Warkentin, T. D., Greeniaus, M. T., Zimmer, R. C. (1996). Genotypic variability in seedborne infection of field pea by Mycosphaerella pinodes and its relation to foliar disease severity. Can. J. Plant Pathol. 18, 370–374. doi: 10.1080/07060669609500590
Young, T., Cameron, D. D., Phoenix, G. K. (2015). Using AMF inoculum to improve the nutritional status of Prunella vulgaris plants in green roof substrate during establishment. Urban. For. Urban Green. 14, 959–967. doi: 10.1016/j.ufug.2015.08.012
Zhang, W., Chang, J., Lei, Z., Huhman, D., Sumner, L. W., Zhao, P. X. (2014). MET-COFEA: a liquid chromatography/mass spectrometry data processing platform for metabolite compound feature extraction and annotation. Anal.Chem 86, 6245–6253. doi: 10.1021/ac501162k
Zhang, W., Lei, Z., Huhman, D., Sumner, L. W., Zhao, P. X. (2015). MET-XAlign: a metabolite cross-alignment tool for LC/MS-based comparative metabolomics. Anal. Chem. 87, 9114–9119. doi: 10.1021/acs.analchem.5b01324
Zhu, Y.-G., Smith, F. A., Smith, S. E. (2003). Phosphorus efficiencies and responses of barley (Hordeum vulgare L.) to arbuscular mycorrhizal fungi grown in highly calcareous soil. Mycorrhiza 13, 93–100. doi: 10.1007/s00572-002-0205-6
Zimmer, R. C., Sabourin, D. (1986). Determining Resistance Reactions of Field Pea Cultivars at the Seedling Stage to Mycosphaerella pinodes. Phytopathology 76, 878–881. doi: 10.1094/Phyto-76-878
Keywords: seed metabolism, Pisum sativum, mycorrhizal colonization, secondary metabolites, plant stress
Citation: Ranjbar Sistani N, Desalegn G, Kaul H-P and Wienkoop S (2020) Seed Metabolism and Pathogen Resistance Enhancement in Pisum sativum During Colonization of Arbuscular Mycorrhizal Fungi: An Integrative Metabolomics-Proteomics Approach. Front. Plant Sci. 11:872. doi: 10.3389/fpls.2020.00872
Received: 09 March 2020; Accepted: 28 May 2020;
Published: 12 June 2020.
Edited by:
Mingxun Chen, Northwest A and F University, ChinaReviewed by:
Kei Hiruma, Nara Institute of Science and Technology (NAIST), JapanCopyright © 2020 Ranjbar Sistani, Desalegn, Kaul and Wienkoop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stefanie Wienkoop, c3RlZmFuaWUud2llbmtvb3BAdW5pdmllLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.