- Plant Breeding Institute, Agrar und Ernährungswissenschaftliche Fakultät, Christian-Albrechts-University of Kiel, Kiel, Germany
Understanding phosphate uptake and storage is interesting to optimize the plant performance to phosphorus fluctuations. Phytic acid (PA) is the major source of inorganic phosphorus (Pi) in plants. Genetic analyses of PA pathway transporter genes (BnMRP5) and their functional characterization might provide clues in better utilizing the available phosphate resources. Furthermore, the failure to assimilate PA by monogastric animals results in its excess accumulation in manure, which ultimately causes groundwater eutrophication. As a first step toward breeding low PA mutants in oilseed rape (Brassica napus L.), we identified knockout mutants in PA biosynthesis and transporter genes. The obtained M3 single mutants of Bn.MRP5.A10 and Bn.MRP5.C09 were combined by crossing to produce double mutants. Simultaneously, crosses were performed with the non-mutagenized EMS donor genotype to reduce the background mutation load. Double mutants identified from the F2 progeny of direct M3 crosses and BC1 plants showed 15% reduction in PA contents with no significant differences in Pi. We are discussing the function of BnMRP5 paralogs and the benefits for breeding Bnmrp5 mutants in respect to low PA, yield, and stress tolerances.
Introduction
Phosphorus is an essential macronutrient required for plant development. In plants, numerous transporters such as PHT, SPX-MFS, and phosphate antiporters facilitate the uptake of phosphorus from the soil and its mobilization across various tissues (Kopriva and Chu, 2018). The up taken phosphorus is utilized in various metabolic processes and is finally stored in seed vacuoles in the form of phytic acid (PA; also referred as inositol hexakis phosphate), which is readily available for seedling development. The PA stored in vacuoles is referred to as phytin, which is a complex salt of divalent ions. In dry mature seeds of various plant species, up to 80% of total phosphorus is stored as PA (Raboy et al., 2000). Therefore, low PA mutants are desirable for a reduced application of external phosphorus in the form of fertilizers. Furthermore, they could help to decrease the dependency on non-renewable rock phosphates (Lott et al., 2007). In this regard, PA biosynthetic genes have been knocked out in different plant species to obtain low PA mutants with simultaneous increase of inorganic phosphorus (Pi) (Sparvoli and Cominelli, 2015). In some cases, the knockout of genes involved in PA biosynthesis has proven to have adverse effects on plant performances such as poor seed set, delayed germination, and low yield (Shi et al., 2007). Therefore, targeting the transporters that are accumulating PA in seeds gained more attention.
A multi-drug resistant protein (MRP5), acting as a tonoplast transporter was shown to have high affinity to PA (Nagy et al., 2009). MRP5 is a member of the ATP binding cassette (ATP) super family and belongs to the ABCC subfamily (Kang et al., 2011). The protein has two transmembrane domains and two nucleotide-binding domains encasing the walker A, B, and C motifs (Verrier et al., 2008). It has been shown that in Arabidopsis the transport of PA is strictly dependent on ATP (Nagy et al., 2009) whereas in rice knockout of SULTR-like phosphorus distribution transporter (SPDT) genes lead to 25–32% reduced PA contents in seeds (Yamaji et al., 2016). This indicates that the vacuolar loading of phosphorus in seeds is not limited to the MRP5 protein but can also be achieved by other transporters. Furthermore, the accumulation of PA in the vacuoles depends on the availability of Pi, as it is highly important to maintain the ratio of vacuolar to cytoplasmic Pi for overall phosphorus homeostasis (Pratt et al., 2009).
Oilseed rape (Brassica napus L.) is the third most important oil crop in the world and its seed meal is rich in proteins and amino acids (Gacek et al., 2018). However, anti-nutritive compounds such as glucosinolates and tannins together with a high fiber content impede its use as a valuable protein source for human and animal feed. Also, PA has an anti-nutritive effect because it is a strong chelator of positive ions like magnesium, potassium, zinc, and iron, thereby reducing their bioavailability in monogastric animals. Moreover, it contributes to phosphate pollution because PA cannot be digested due to lack of phytases thereby leading to high P concentrations in the manure. Several knockout and knockdown mutants of PA transporter genes were identified in cereal crops such as maize, rice, wheat, and barley. Their seed PA contents were reduced by 32–90% with a simultaneous increase of Pi (Dorsch et al., 2003; Shi et al., 2007; Xu et al., 2009; Panzeri et al., 2011; Bhati et al., 2016). In addition, MRP mutants were identified from common bean and soybean, which also showed a substantial reduction of PA contents by up to 90% (Wilcox et al., 2000; Shi et al., 2007; Campion et al., 2009).
We expected that MRP genes display a similar function in rapeseed. Therefore, we aimed to select MRP mutants with reduced PA seed storage. We identified MRP orthologs in the rapeseed genome and used the sequence information to select EMS mutants by a novel TILLING by Sequencing (TbyS) approach (Sashidhar et al., 2019). Only double mutants showed a significant reduction in seed PA content. These mutants will be important for breeding low PA rapeseed varieties.
Materials and Methods
Plant Material and Growth Conditions
The rapeseed winter type variety Express 617 was used to study PA and Pi accumulation in developing seeds. Plants were grown in a greenhouse in 11 cm × 11 cm pots at 22°C under long day conditions (16 h of light and 8 h of dark). No external phosphorus was added to the soil. Three weeks old plants were vernalized for 8 weeks at 4°C (16 h light and 8 h dark) and after 8 weeks plants were transferred back to 22°C (16 h light and 8 h dark) until harvest. At each time point, five plants were harvested. During flowering, young buds were emasculated and hand-pollinated with Express 617 pollen to mark the day of pollination. For PA and Pi extractions, developing seeds were collected 15, 25, 35, 45, and 55 days after pollination (DAP) and frozen in liquid nitrogen. Samples were kept at −80°C until extraction.
M3 seeds were obtained from the breeding company Norddeutsche Pflanzenzucht Hans-Georg Lembke KG (Hohenlieth, Germany). M3 plants homozygous for the Bn.MRP5.A10 allele (M3 seed code 170482) were crossed with homozygous Bn.MRP5.C05 mutants (M3 seed code 170481) to produce F1 plants. Simultaneously, homozygous M3 plants from each family were crossed with non-mutagenized Express 617 plants to obtain F1 offspring with reduced background mutation load. All F1 plants were selfed to produce F2 seeds (Figure 1).
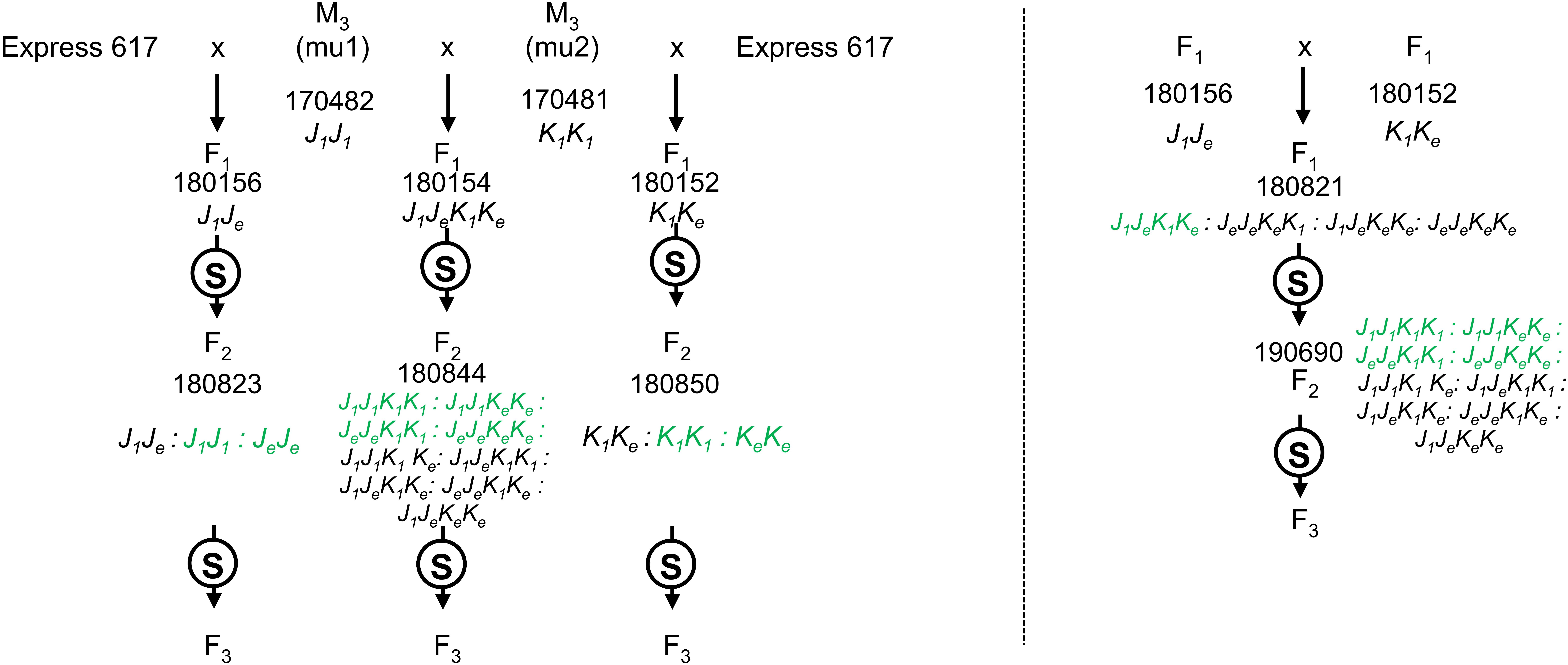
Figure 1. Crossing scheme for the production of BnMRP5 single and double mutants. The seed codes are written above each genotype (see Table 3 for respective allele codes). The genotypes highlighted in green were selfed to produce F3 seeds, which were used for phenotyping; “S”: selfing; Bn.MRP5.A10 and Bn.MRP5.C05 mutations are indicated as mu1 and mu2, respectively.
Identification of MRP5 Genes and Mutant Screening
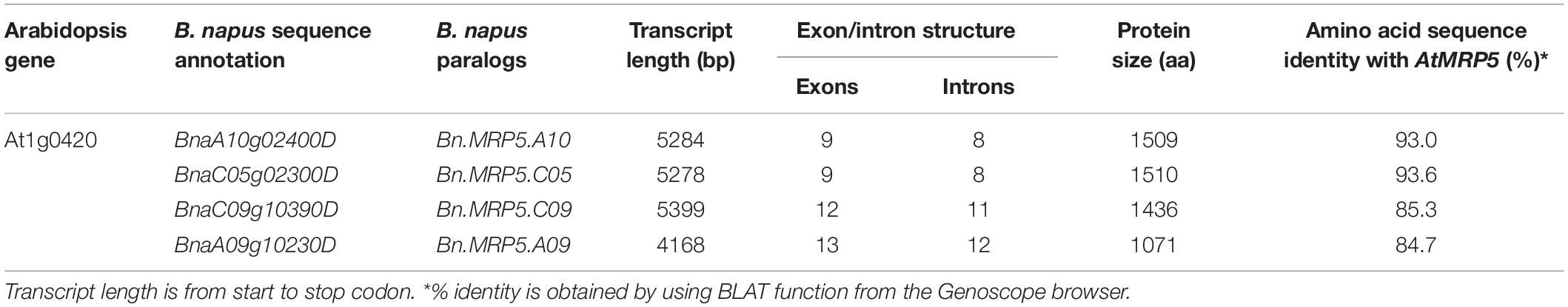
In Arabidopsis, the MRP5 protein is encoded by one gene (At1g0420), which was used for retrieving all paralogous genes from B. napus using the Genoscope browser1. Genes having high sequence identity to the Arabidopsis genes were considered as true paralogous genes. Protein domains were identified using the pFam database search2. All hits were confirmed using TAIR3, NCBI annotations4 and BRAD databases5.
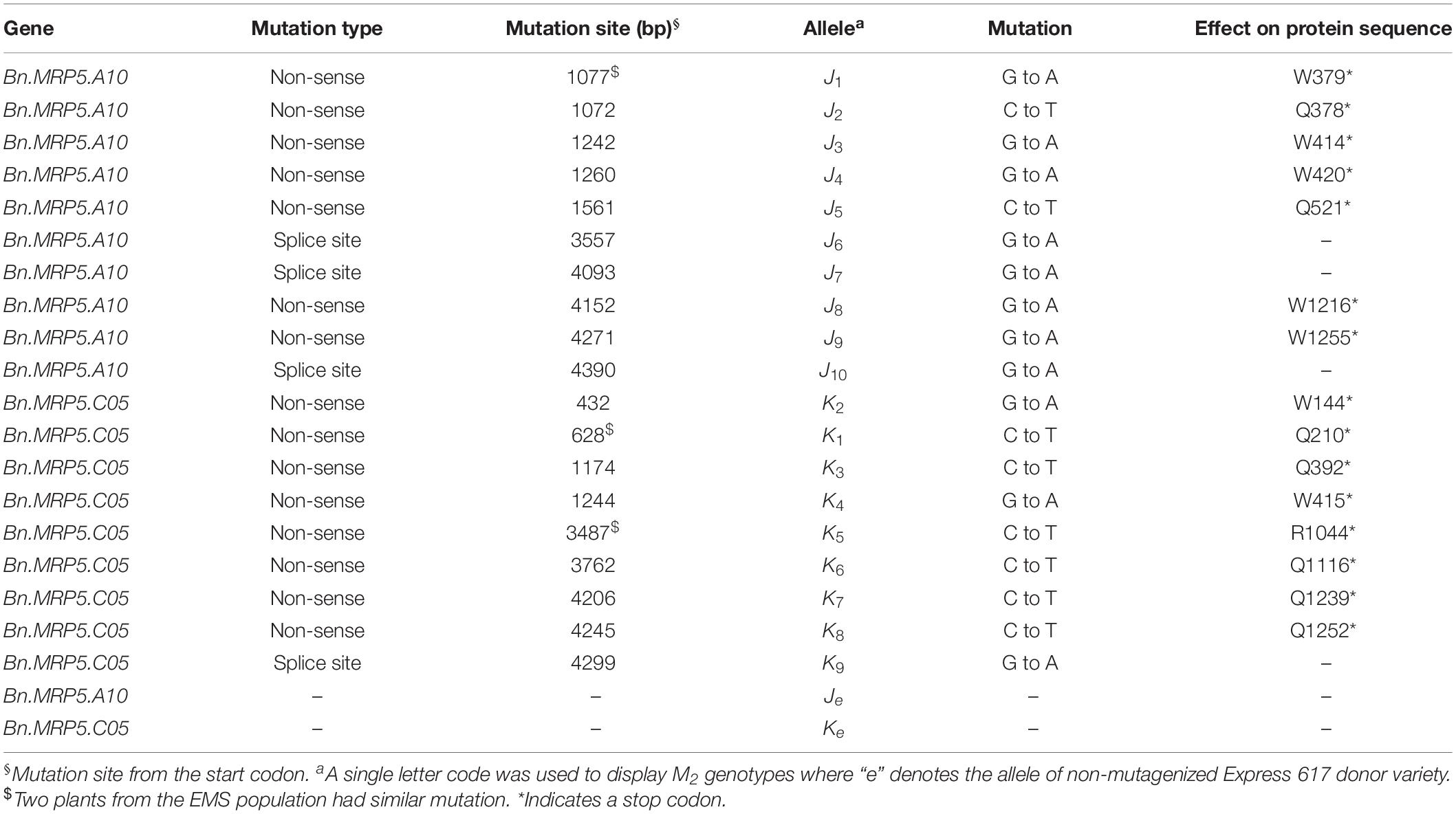
We have used TbyS for screening the mutations as described in Sashidhar et al. (2019). Two BnMRP5 paralogs were chosen for identifying the putative loss of function mutations. Four primer pairs were used to screen amplicons 1414–1509 bp in size. Sanger sequencing was used to confirm the mutations in the M2 generation with the help of paralog specific primers.
Mutation frequencies (F) were calculated based on the mutations per M1 plant (Harloff et al., 2012):
Nucleic Acid Isolation, PCR, and RT-qPCR
For genotyping experiments, DNA was isolated from the leaves using the CTAB protocol (Rogers and Bendich, 1985) with minor modifications. PCR was performed using DNA aliquots of 2 μl as a template for amplifying the genes (92°C: 3 min, [92°C: 30 s, 58–63°C: 30 s, 72°C: 65 s] 36x, 72°C: 5 min). Amplicon lengths were verified on 1% agarose gels and the presence of mutations was confirmed by Sanger sequencing (Table 1).
For expression analysis, the winter type line Express 617 was grown in the greenhouse as described above. Five plants were used for expression analysis. Immature flower buds were emasculated followed by hand pollination to ensure the day of pollination. Seed samples of 50 mg were harvested at 15, 25, 35, and 45 DAP and were shock frozen in liquid nitrogen. RNA was isolated using the peqGOLD RNA isolation kit (PEQLAB Biotechnologie GmbH, Erlangen, Germany) according to the manufacturer’s protocol. DNase digestion was performed after RNA isolation using the peqGOLD DNase I Digest Kit (PEQLAB Biotechnologie GmbH, Erlangen, Germany) to remove genomic DNA. cDNA first strand synthesis was carried out using a first strand cDNA synthesis kit (ThermoFisher Scientific Inc., Waltham, MA, United States) according to the manufacturer’s protocol. RT-qPCR was performed using Platinum® SYBR® Green qPCR SuperMix-UDG with ROX (Invitrogen, Karlsruhe, Germany) on a CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, United States) in a volume of 20 μl. BnActin2 was used as a reference gene for normalization. Relative expression was calculated using the ΔCt method (Table 1).
Inorganic Phosphorus and Phytic Acid Measurements
Dry seeds of 200 mg were ground into fine powder and PA was extracted in three replicates of 50 mg according to Matthäus et al. (1995) with minor modifications. PA was measured by HPLC according to Rounds and Nielsen (1993) calibrating with a PA standard (Sigma P-8810). For determining Pi contents, 50 μl of the purified column extracts was mixed with 500 μl of coloring reagent [10% w/v of ascorbic acid (Roth-Art.Nr.3666.1) and 5% w/v of ammonium molybdate (Roth, Art. No.3525.2)] adjusted to 1.5 ml with double distilled water and incubated at 40°C for 1 h. The samples were measured against a reagent blank buffer in a spectrophotometer at 655 nm. A calibration curve ranging from 0 to 100 nmol was used for both PA and Pi measurements with standards for Pi and PA, respectively.
Statistical Analysis
Data obtained from PA and Pi measurements were used for statistical analysis. Significance was calculated using ANOVA in R 3.6.0 and the MulticomView package and post hoc test was performed by using Tukey’s multiple comparison test; p = 0.05. Standard deviation was calculated between five biological samples along with their three technical repeats.
Results
Dynamics of Phytic Acid and Inorganic Phosphorous Accumulation in Winter Type Rapeseed
Seed samples from 15, 25, 35, 45, and 55 DAP were used to determine the dynamic changes of PA and Pi in developing seeds. We observed that PA was not measurable at 15 DAP, but it accumulated significantly from 15 to 35 DAP. In contrast, Pi contents decreased significantly from 15 to 35 DAP. The coordinated decrease of Pi and increase of PA might indicate that most of the Pi is assimilated to PA between 35 and 45 DAP (Figure 2).
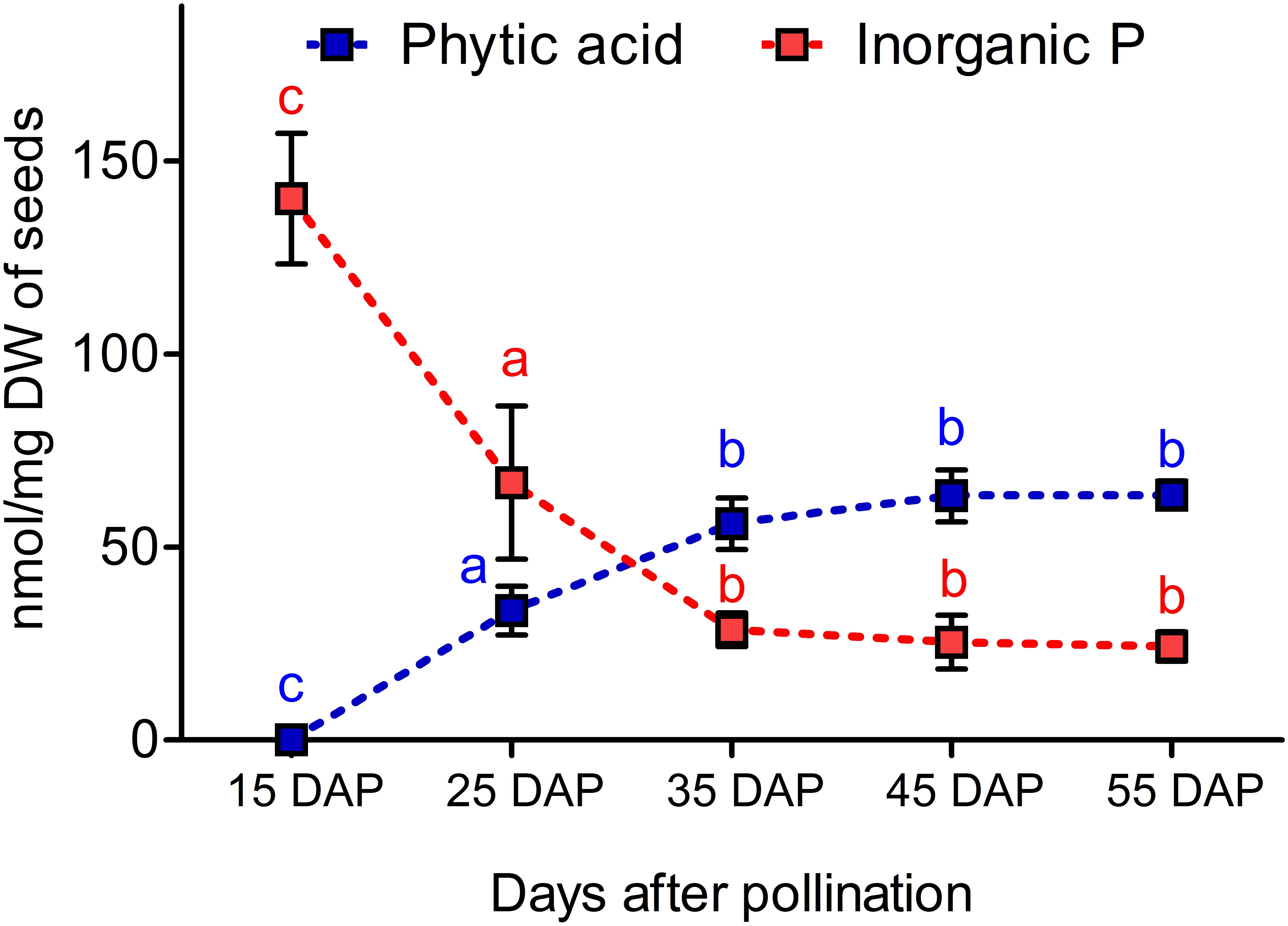
Figure 2. Dynamics of phytic acid (PA) and inorganic phosphorus (Pi) accumulation during seed maturation in the winter type Express 617. Plants were grown in a greenhouse under 16 h of light and 8 h of dark at 22°C. Five plants were used as biological replicates for PA and Pi measurements. Error bars denote the SD among five biological samples along with three technical repeats. Statistical significance was calculated using ANOVA and grouping was done using a Tuckey test. Different letters indicate statistical significance.
Searching the Rapeseed Genome for MRP5 Orthologs
We used the Arabidopsis AtMRP5 sequence as query to search the Brassica Genoscope database. Four rapeseed sequences were identified as putative paralogous genes of AtMRP5 (Table 2). All genes showed Walker A, B, and C motifs, which were encased by the transmembrane domain (Supplementary Figure S1). The BLAT results from Genoscope browser showed that Bn.MRP5.A09 had a lower homology and also a shorter protein sequence when compared to the three other proteins. Upon protein alignment with AtMRP5, we found that Bn.MRP5.A09 lacked most of the transmembrane domain regions, indicating that this protein might lack a membrane transport function for PA. Therefore, we excluded Bn.MRP5.A09 from our analyses (Supplementary Figure S2).
Messenger RNA abundance was measured in developing seeds of Express 617 and Ct values were normalized to the housekeeping gene BnActin2. The transcriptional activities of all three genes strongly increased between 15 and 25 DAP. They peaked between 25 and 35 DAP and decreased 45 DAP (Figure 3). Finally, the two highest expressed paralogs (Bn.MRP5.A10, Bn.MRP5.C05) were used for mutation screening in the EMS mutant population.
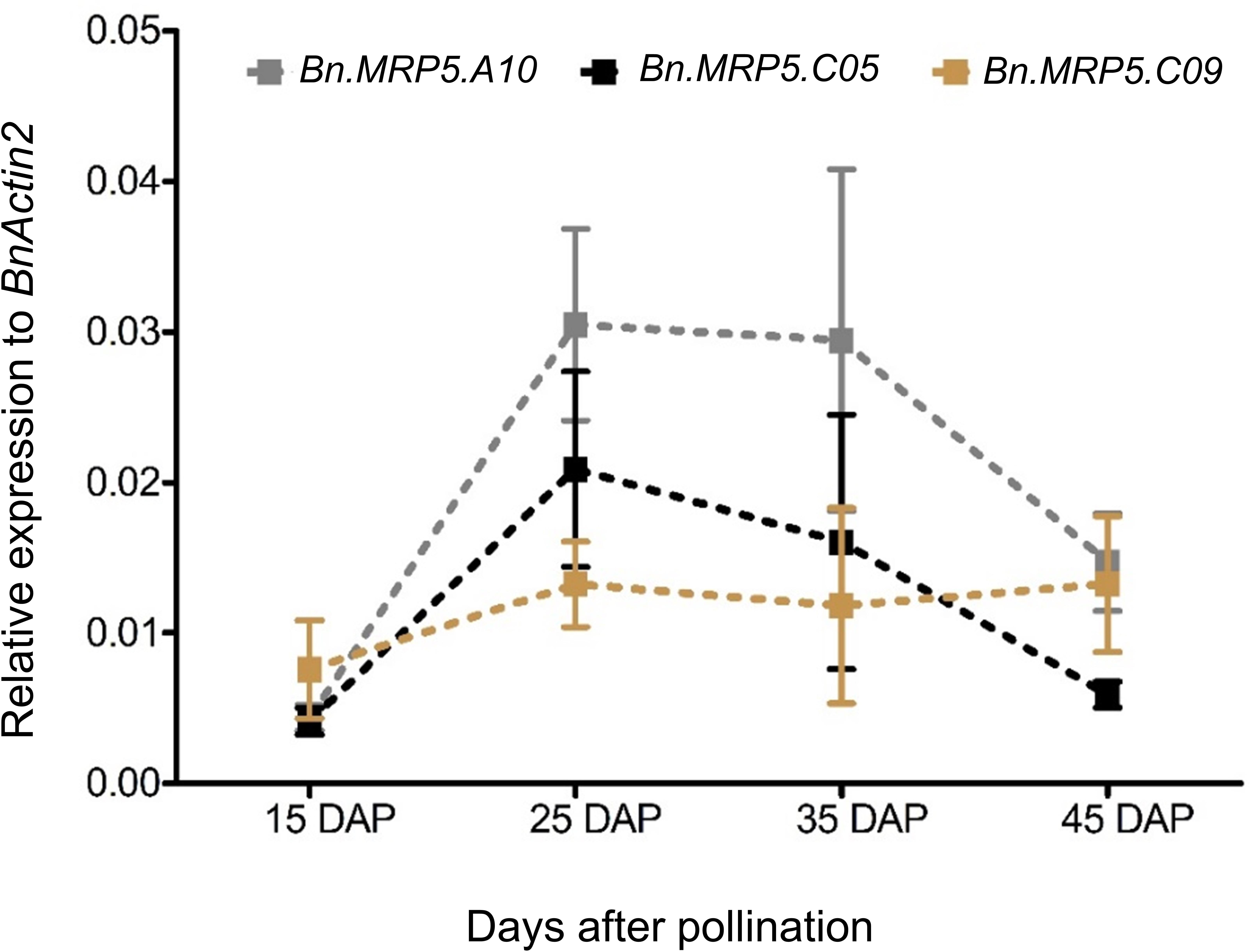
Figure 3. Expression profiles of three BnMRP5 genes in Express 617 at seed filling stages. Plants were grown at 16 h of light and 8 h of dark at 22°C. Seed samples from four developmental stages of Express 617 were harvested at 15, 25, 35, and 45 DAP. The expression values were normalized to the housekeeping gene BnActin2. Error bars indicate SD of five biological samples along with three technical repeats.
Identification of EMS Mutants and Production of Double Mutants
We applied a new TbyS protocol to search for EMS mutants in the Express 617 mutant population (Sashidhar et al., 2019). We screened for mutations within the translated regions of Bn.MRP5.A10 (5284 bp) and Bn.MRP5.C05 (5278 bp). The amplicons used for TbyS covered up to 60% of the coding regions of Bn.MRP5.A10 and Bn.MRP5.C05, including the Walker A and C motifs of the genes (Figure 4). As a result, eight stop codon and three splice site mutations were found in Bn.MRP5.A10 while 10 stop codon and one splice site mutation were found in Bn.MRP5.C05 (Table 3). All mutations were confirmed by Sanger sequencing confirming the reliability of the TbyS strategy (Sashidhar et al., 2019). For ease of understanding, mutations in Bn.MRP5.A10 and Bn.MRP5.C05 were assigned with the allele code J and K, respectively. For phenotyping, we chose the alleles J1 and K1, as these mutations were at the beginning of the gene and the translation should result in a loss of all the important protein domains (Table 3 and Figure 4).
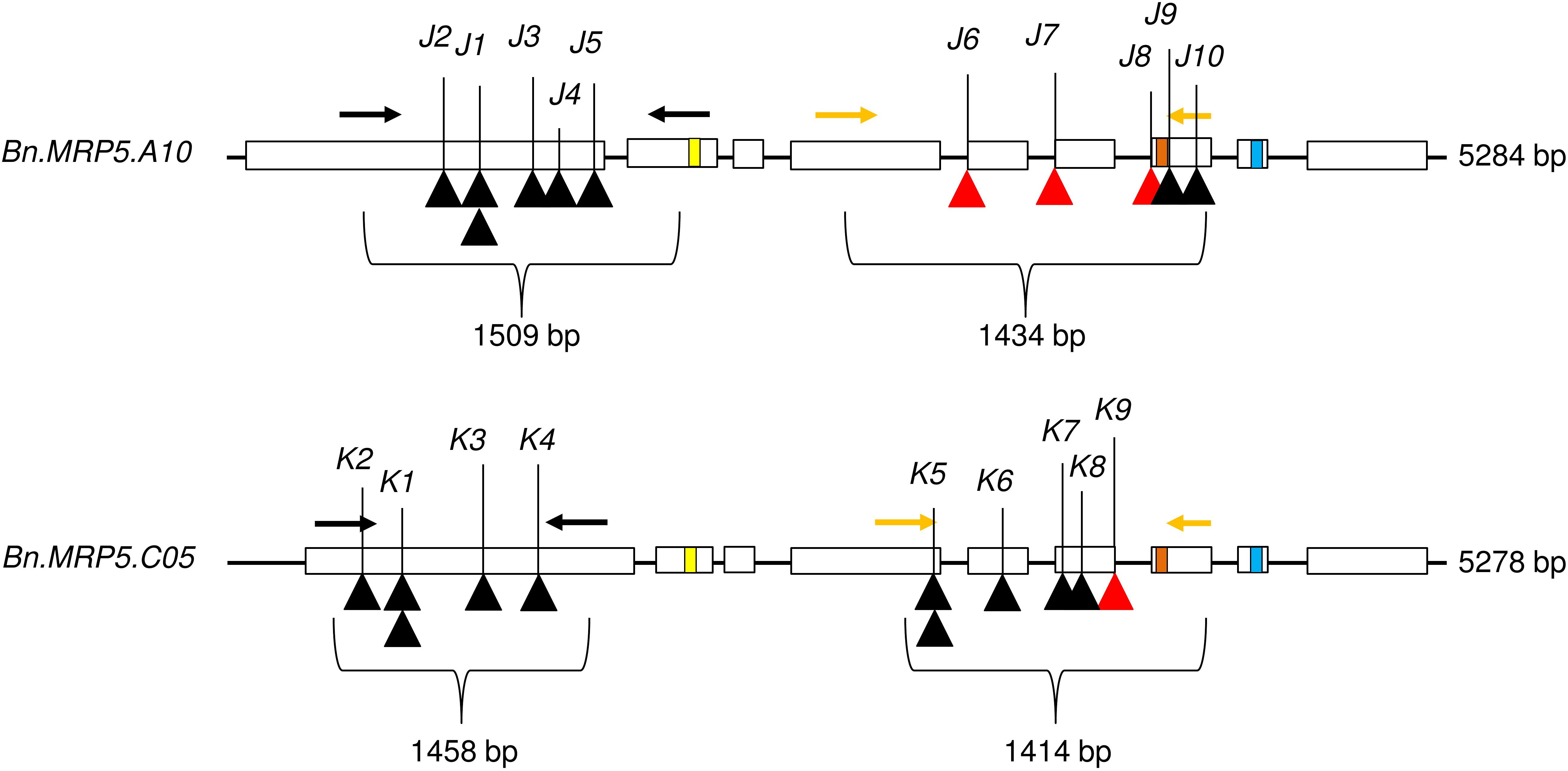
Figure 4. Exon–intron structure showing the positions of the mutations. Exons are shown as white boxes. Primer pairs used for amplicon sequencing are shown as black and yellow arrows. C motif, Walker A, and Walker B motif are shown as yellow, orange, and blue boxes, respectively. The C motif is also known as ABC signature motif. Stop codon and splice site mutations are shown as black and red triangles, respectively. The mutant alleles are indicated by respective numbers (see Table 3).
It was known from previous studies that single mutants in rapeseed often do not display a new phenotype due to gene redundancy. Moreover, EMS mutants suffer from a huge number of background mutations, which severely hampers the phenotypic evaluation (Braatz et al., 2018; Shah et al., 2018). Therefore, we initiated a crossing program to produce double mutants and backcross generations with reduced mutation load. Homozygous M3 mutants were crossed with non-mutated Express 617 to obtain F1 seeds. Then, the F1 plants carrying mutations in both the selected copies were crossed with each other (Figure 1) and F2 populations were generated to be used for PA measurements. In the F2 generation, segregation ratios were as expected for mono (3:1) and digenic (15:1) segregation (Table 4).
Bnmrp5 Double Mutants With Reduced Phytic Acid Seed Content
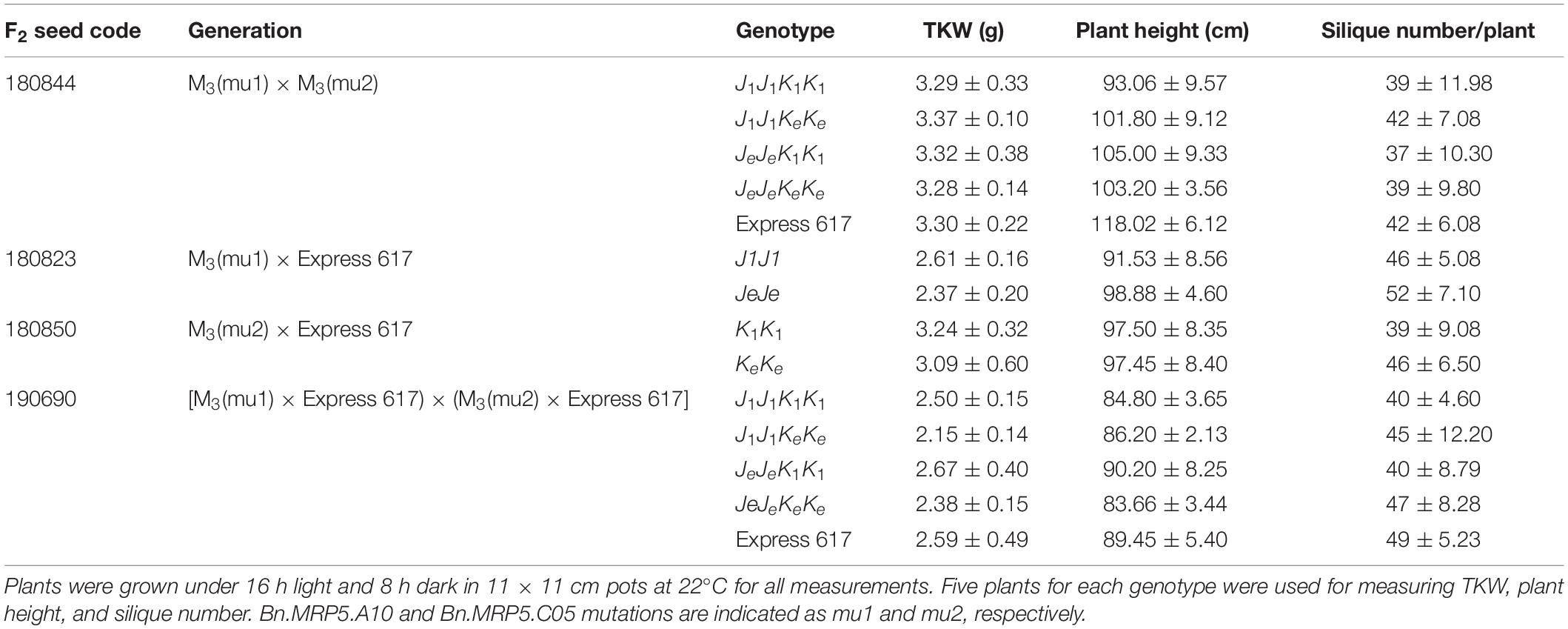
We measured the effect of single and double mutations with full and reduced mutation load. Plants from M3, F2, and BC1 generations were grown in the greenhouse. Express 617 and mutant offspring of Express 617 (suffix “e”) were used as controls. Single mutants from the two F2 families, 180823 (J1J1) and 180850 (K1K1), showed no significant reduction in PA contents when compared to both Express 617 controls (Figure 5). However, double mutants obtained from F2 family 180844 (J1J1K1K1) showed significant reduction in comparison to the mutant control plants carrying homozygous Express 617 alleles (JeJeKeKe) but not to non-mutagenized Express 617 (Figure 6). This could be explained due to a high mutation load. Therefore, we expected clear phenotypic effects in the double mutants backcrossed with Express 617 (see Figure 1). As a result, we observed a significant PA reduction in backcrossed double mutants from the F2 population 190690 in comparison to the internal control plants (JeJeKeKe) as well as to the Express 617 plants. We concluded that a reduction of 15% was due to the underlying mutant alleles and not due to the background mutation load. Indeed, a similar reduction was observed in the double mutants of the 180844 family when compared to internal control plants (JeJeKeKe), indicating that the observed reduction of 15% PA is due to the two mutated alleles. In addition, we measured the Pi contents in matured seeds of the 190690 progeny to see if the reduced PA content had any effect on Pi accumulation. We did not find a significant increase in double mutants as compared to control plants (Figure 6). Besides, plant height, silique number per plant, and thousand-kernel weight were not significantly different in the mutants, indicating that the low PA was not affecting the overall plant performance (Table 5).
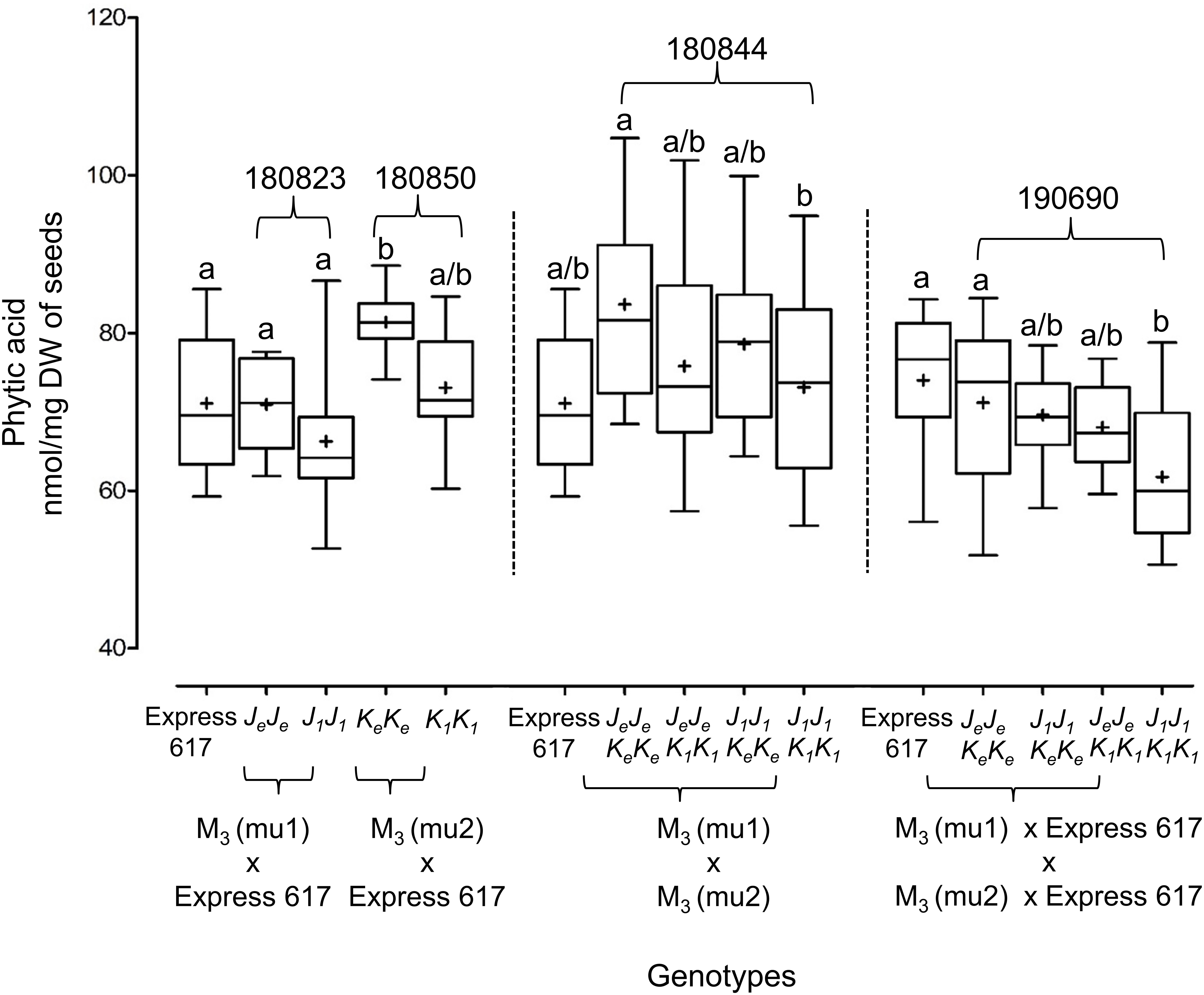
Figure 5. Phenotypic analysis of Bnmrp5 mutants. Box plot of phytic acid contents in seeds of Bnmrp5 mutants. Statistical analysis was performed using ANOVA test at p < 0.05 and grouping was done using the Tukey test. Same letters indicate no significant differences. The non-mutated Express 617 alleles are indicated by “e.” Bn.MRP5.A10 and Bn.MRP5.C05 mutations are indicated as mu1 and mu2, respectively. Seed codes of F2 families are given above the brackets. Means are indicated by “+.” Allele codes are given in Table 3.
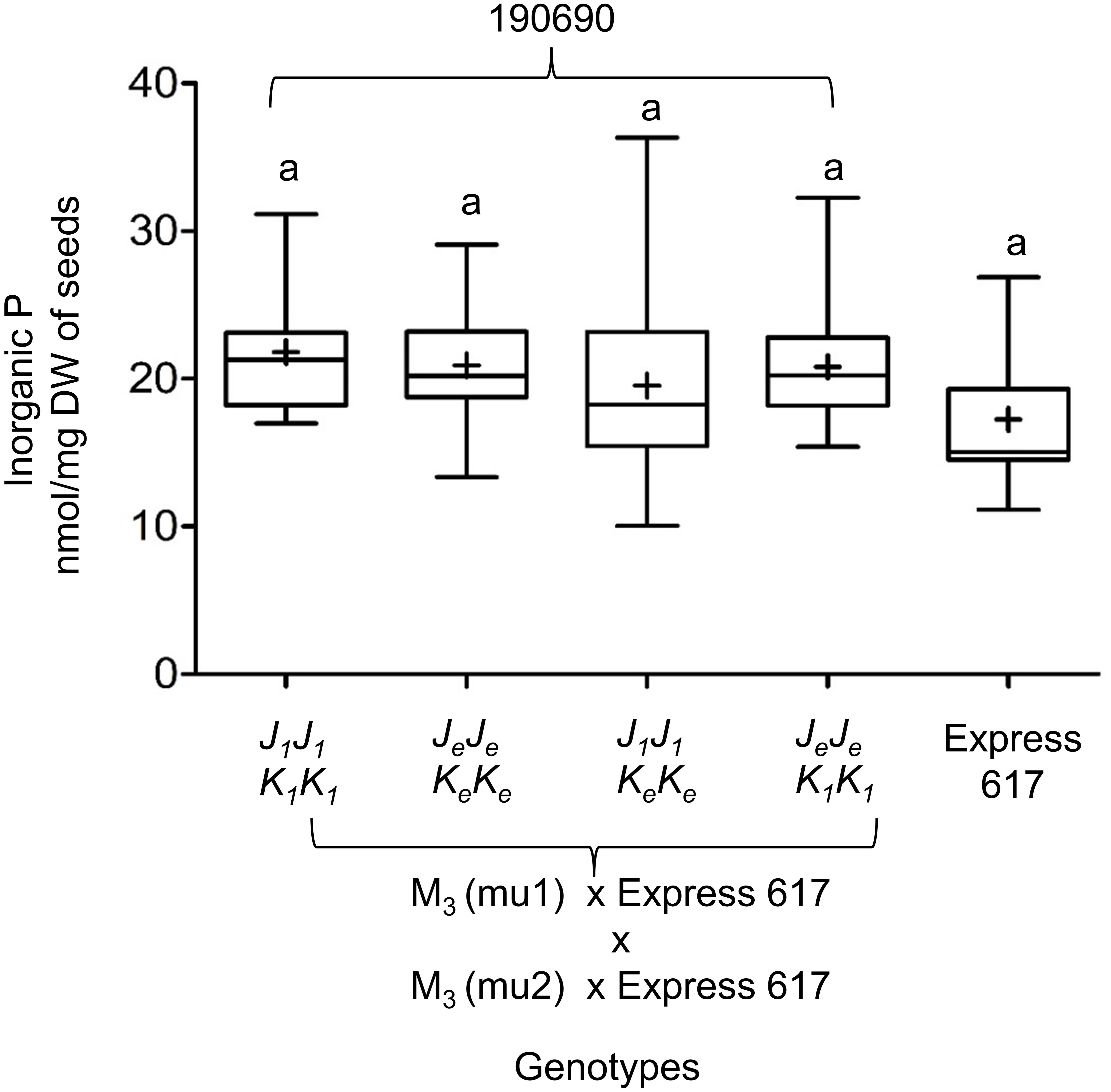
Figure 6. Inorganic P (Pi) measurements in Bnmrp5 double mutants of the F2 family 190690. The boxplot represents Pi measurements in five plants for each genotype from matured seeds. Of each plant, three technical replicates were taken. Means are indicated by “+.” Statistical analysis was performed using ANOVA test at p < 0.05 and grouping was done using the Tukey test. Same letters indicate no significant differences between the samples. The non-mutated Express 617 alleles are indicated by “e.” Bn.MRP5.A10 and Bn.MRP5.C05 mutations are indicated as mu1 and mu2, respectively Allele codes are given in Table 3.
Discussion
In this study, we describe low PA mutants in oilseed rape by targeting BnMRP5 PA transporter genes. Three paralogs were expressed during seed development, two showed a maximum expression between 25 and 35 DAP similar to the expression of MRP5 genes in siliques of Arabidopsis (AtMRP5, Kim and Tai, 2011) and common bean (PvMRP1, measured 6 and 24 days after flowering (Panzeri et al., 2011). The expression pattern of the paralogs was in line with the PA and Pi accumulation pattern from Express 617. Shortly after 35 DAP, PA storage reached its maximum and Pi depletion its minimum suggesting a major incorporation of Pi into PA until 35 DAP.
The double mutants obtained displayed up to 15% reduced PA contents. One of the major concerns regarding EMS mutants is their high mutation load. This problem became obvious when comparing primary (non-backcrossed) mutants and double mutants with their non-mutagenized parent Express 617. Therefore, we could not observe significant differences between primary double mutants (J1J1K1K1, F2 population 180844) and Express 617. However, double mutants once backcrossed with Express 617 (F2 population 190690) had significantly reduced PA contents compared to their mutant siblings (JeJeKeKe) and compared to Express 617.
We expected that the two knockout mutants resulted in a truncated protein, where the Walker A, B, and C motifs were completely lost, thereby leading to a failure to transport PA. However, the reduction in PA content was somewhat lower as in other studies with maize, rice, wheat, barley, Arabidopsis, soybean, and common bean where reductions between 32 and 90% have been reported (Sparvoli and Cominelli, 2015). One reason for a relatively lower reduction in our study could be that the third gene might partially contribute to the compensation of the phenotypic effect in the double mutant. In rice it has been shown that a mutation in OsPLDα1 caused a reduction of PA in seeds, however, in the mutant other PLD genes were upregulated, indicating that the other PLD proteins can in part compensate the mutation in OsPLDα1 (Khan et al., 2019). This genetic compensation response has first been described after mutant mRNA degradation for genes with sequence similarities to the mutated genes in zebrafish (El-Brolosy et al., 2019). Whether this applies also for the mutation of the two MRP5 genes we have studied will be an interesting direction for further studies.
In polyploids, the simultaneous knockout of multiple genes is required to obtain a phenotypic effect (Wang et al., 2014; Del Pozo and Ramirez-Parra, 2015). In wheat, a reduction in PA content by “only” 22% was observed when all three paralogous genes of TaABCC5 were downregulated by RNAi (Bhati et al., 2016). Therefore, this study and our study might indicate the existence of alternative transporters in polyploids, which might compensate for the loss of knockout mutations in MRP5 proteins. A study in common bean has shown that loss of PvMRP1 was compensated by PvMRP2. However, compensation was not observed in seeds but in leaf tissues (Cominelli et al., 2018). In addition, reduced PA seed contents in knockout mutants of OsSPDT in rice, an SPDT (Yamaji et al., 2016), suggest its contribution to PA loading in seeds (Sacchi and Nocito, 2019). Since, loss of function mutants in PA transporters were never identified before in oilseed rape, the hypothesis that other MRP transporters compensate the knockout effects need to be further validated for instance by additional mutations in the third paralog Bn.MRP5.C09. Moreover, they can be crossed with bn2-pgk2 mutants which we have been recently identified (Sashidhar et al., 2019). Like the bnmrp5 mutants, bn2-pgk2 mutants did not show any trade off effects regarding agronomically important characters. Therefore, creating multiple mutants in both gene families could be a promising option to further reduce PA seed contents in oilseed rape.
Knock out mutants of MRP5 in Arabidopsis showed a major reduction in PA contents. Therefore, we assumed that this might also be the case in BnMRP5 mutants of the close relative B. napus. PA transporter mutants in various species do have higher inositol kinases (Vip/VIH) referred to as inositol hexakis phosphate kinases (Desai et al., 2014), which synthesize InsP7 (also referred as PP-InsP5) and InsP8 (also referred as 1,5 PP2-InsP4; Supplementary Figure S3). For instance, the knockout mutants of atmrp5 and zmmrp4 showed an increased accumulation of InsP7. Loss of function in MRP5 might therefore lead to an increased concentration of InsP6 (or PA) in the cytosol, which can be used as a substrate by the inositol hexakis phosphate kinases to produce InsP7 or InsP8, resulting in a lower PA storage rate. Accumulating InsP7 and InsP8 might be beneficial to plants as they possess energy-rich pyrophosphate moieties having a comparable free energy of hydrolysis to ATP (Hand and Honek, 2007; Desai et al., 2014). It was shown in Arabidopsis that InsP7/InsP8 binds to SPX domains, which are crucial in phosphate sensing and in regulating Pi homeostasis (Jung et al., 2018; Ried et al., 2019). Thereby with an increased accumulation of such molecules, the yield capacity can be improved and also fertilizer demands might be reduced (Williams et al., 2015). Finally, inositol pyrophosphates are regulating the jasmonic acid-based defense mechanisms, which might be beneficial for an improved growth under stress conditions (Laha et al., 2015).
AtMRP5 Arabidopsis mutants have an elevated drought tolerance due to a reduced ABA sensitivity resulting in stomatal closure (Gaedeke et al., 2001; Klein et al., 2003). In atmrp5 mutants, failure to transport PA into vacuoles resulted in reduced K+ uptake into the guard cells by inhibiting K+ inward rectifying channels (Nagy et al., 2009). The stomatal closure might reduce water loss from leaves and might foster plants to cope with drought stress and it was shown that atmpr5 mutants showed reduced water uptake and survived longer than the wild type plants under low water levels (Klein et al., 2003). However, as we observed only 15% reduction of PA, a complete knockout of BnMRP5 genes in future might pave the way for a comprehensive analysis of BnMRP5 genes under drought stress. In conclusion, the mutants presented here are important for breeding low PA rapeseed varieties. Moreover, they can be utilized in exploring phosphorous sensing and homeostasis mechanisms.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
NS planned, performed, and analyzed the experiments and wrote the manuscript. HH and CJ designed the study, supervised the experiments, and revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by a grant from the German Research Foundation, DFG (Grant No. JU 205/26-1).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Siegbert Melzer for his valuable discussions and Nirosha Karunarathna in sampling the seed material for the accumulation data. We also thank the Institute of Clinical Molecular Biology in Kiel for providing Sanger sequencing as supported in part by the DFG Clusters of Excellence “Precision Medicine in Chronic Inflammation” and “ROOTS” and we thank T. Naujoks, Dr. D. Langfeldt, and Dr. B. Löscher for technical support. We thank Dipl. Biol. Jens Hermann and Prof. Dr. Wolfgang Bilger from the Department of Ecophysiology of Plants at the CAU Kiel for HPLC measurements.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00603/full#supplementary-material
Footnotes
- ^ http://www.genoscope.cns.fr/brassicanapus/
- ^ http://pfam.xfam.org/
- ^ https://www.arabidopsis.org/servlets/Search?type=gene&action=new_search
- ^ https://www.ncbi.nlm.nih.gov/gene
- ^ http://brassicadb.org/brad/
References
Bhati, K. K., Alok, A., Kumar, A., Kaur, J., Tiwari, S., and Pandey, A. K. (2016). Silencing of ABCC13 transporter in wheat reveals its involvement in grain development, phytic acid accumulation and lateral root formation. J. Exp. Bot. 67, 4379–4389. doi: 10.1093/jxb/erw224
Braatz, J., Harloff, H. J., Emrani, N., Elisha, C., Heepe, L., Stanislav, N., et al. (2018). The effect of INDEHISCENT point mutations on silique shatter resistance in oilseed rape (Brassica napus). Theoret. Appl. Genet. 131, 959–971. doi: 10.1007/s00122-018-3051-4
Campion, B., Sparvoli, F., Doria, E., Tagliabue, G., Galasso, I., Fileppi, M., et al. (2009). Isolation and characterisation of an lpa (low phytic acid) mutant in common bean (Phaseolus vulgaris L.). TAG Theoret. Appl. Genet. 118, 1211–1221. doi: 10.1007/s00122-009-0975-8
Cominelli, E., Confalonieri, M., Carlessi, M., Cortinovis, G., Daminati, M. G., Porch, T. G., et al. (2018). Phytic acid transport in Phaseolus vulgaris: a new low phytic acid mutant in the PvMRP1 gene and study of the PvMRPs promoters in two different plant systems. Plant Sci. 270, 1–12. doi: 10.1016/j.plantsci.2018.02.003
Del Pozo, J. C., and Ramirez-Parra, E. (2015). Whole genome duplications in plants: an overview from Arabidopsis. J. Exp. Bot. 66, 6991–7003. doi: 10.1093/jxb/erv432
Desai, M., Rangarajan, P., Donahue, J. L., Williams, S. P., Land, E. S., Mandal, M. K., et al. (2014). Two inositol hexakisphosphate kinases drive inositol pyrophosphate synthesis in plants. Plant J. 80, 642–653. doi: 10.1111/tpj.12669
Dorsch, J., Cook, A., Young, K., Anderson, J., Bauman, A., Volkmann, C., et al. (2003). Seed phosphorus and inositol phosphate phenotype of barley low phytic acid genotypes. Phytochemistry 62, 691–706. doi: 10.1016/S0031-9422(02)00610-6
El-Brolosy, M. A., Kontarakis, Z., Rossi, A., Kuenne, C., Günther, S., Fukuda, N., et al. (2019). Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197.
Gacek, K., Bartkowiak-Broda, I., and Batley, J. (2018). Genetic and molecular regulation of seed storage proteins (SSPs) to improve protein nutritional value of oilseed rape (Brassica napus L.) Seeds. Front. Plant Sci. 9:890. doi: 10.3389/fpls.2018.00890
Gaedeke, N., Klein, M., Kolukisaoglu, U., Forestier, C., Müller, A., Ansorge, M., et al. (2001). The Arabidopsis thaliana ABC transporter AtMRP5 controls root development and stomata movement. EMBO J. 20, 1875–1887. doi: 10.1093/emboj/20.8.1875
Hand, C. E., and Honek, J. F. (2007). Phosphate transfer from inositol pyrophosphates InsP5PP and InsP4(PP)2: a semi-empirical investigation. Bioorg. Med. Chem. Lett. 17, 183–188. doi: 10.1016/j.bmcl.2006.09.066
Harloff, H. J., Lemcke, S., Mittasch, J., Frolov, A., Wu, J. G., Dreyer, F., et al. (2012). A mutation screening platform for rapeseed (Brassica napus L.) and the detection of sinapine biosynthesis mutants. TAG Theoret. Appl. Genet. 124, 957–969. doi: 10.1007/s00122-011-1760-z
Jung, J.-Y., Ried, M. K., Hothorn, M., and Poirier, Y. (2018). Control of plant phosphate homeostasis by inositol pyrophosphates and the SPX domain. Curr. Opin. Biotechnol. 49, 156–162. doi: 10.1016/j.copbio.2017.08.012
Kang, J., Park, J., Choi, H., Burla, B., Kretzschmar, T., Lee, Y., et al. (2011). Plant ABC Transporters. Arabidopsis Book 9:e0153. doi: 10.1199/tab.0153
Khan, M., Basnet, R., Islam, S., and Shu, Q. (2019). Mutational analysis of OsPLDα1 reveals its involvement in phytic acid biosynthesis in rice grains. J. Agric. Food Chem. 67, 11436–11443. doi: 10.1021/acs.jafc.9b05052
Kim, S. I., and Tai, T. H. (2011). Identification of genes necessary for wild-type levels of seed phytic acid in Arabidopsis thaliana using a reverse genetics approach. Mol. Genet. Genomics 286, 119–133. doi: 10.1007/s00438-011-0631-2
Klein, M., Perfus-Barbeoch, L., Frelet, A., Gaedeke, N., Reinhardt, D., Mueller-Roeber, B., et al. (2003). The plant multidrug resistance ABC transporter AtMRP5 is involved in guard cell hormonal signaling and water use. Plant J. 33, 119–129. doi: 10.1046/j.1365-313X.2003.016012.x
Kopriva, S., and Chu, C. (2018). Are we ready to improve phosphorus homeostasis in rice? J. Exp. Bot. 69, 3515–3522. doi: 10.1093/jxb/ery163
Laha, D., Johnen, P., Azevedo, C., Dynowski, M., Weiß, M., Capolicchio, S., et al. (2015). VIH2 regulates the synthesis of inositol pyrophosphate InsP8 and Jasmonate-dependent defenses in Arabidopsis. Plant Cell 27:1082. doi: 10.1105/tpc.114.135160
Liu, J., Huang, S., Sun, M., Liu, S., Liu, Y., Wang, W., et al. (2012). An improved allele-specific PCR primer design method for SNP marker analysis and its application. Plant Methods 8:34. doi: 10.1186/1746-4811-8-34
Lott, J. N. A., Ockenden, I., Raboy, V., and Batten, G. D. (2007). Phytic acid and phosphorus in crop seeds and fruits: a global estimate. Seed Sci. Res. 10, 11–33. doi: 10.1017/S0960258500000039
Matthäus, B., Lösingand, R., and Fiebig, H.-J. (1995). Determination of inositol phosphates IP3 – IP6 in rapeseed and rapeseed meal by an HPLC method, Part 2: investigations of rapeseed and rapeseed meal and comparison with other methods. Lipid/Fett 97, 372–374. doi: 10.1002/lipi.19950971004
Nagy, R., Grob, H., Weder, B., Green, P., Klein, M., Frelet-Barrand, A., et al. (2009). The Arabidopsis ATP-binding cassette protein AtMRP5/AtABCC5 is a high affinity inositol hexakisphosphate transporter involved in guard cell signaling and phytate storage. J. Biol. Chem. 284, 33614–33622. doi: 10.1074/jbc.M109.030247
Panzeri, D., Cassani, E., Doria, E., Tagliabue, G., Forti, L., Campion, B., et al. (2011). A defective ABC transporter of the MRP family, responsible for the bean lpa1 mutation, affects the regulation of the phytic acid pathway, reduces seed myo-inositol and alters ABA sensitivity. New Phytol. 191, 70–83. doi: 10.1111/j.1469-8137.2011.03666.x
Pratt, J., Boisson, A.-M., Gout, E., Bligny, R., Douce, R., and Aubert, S. (2009). Phosphate (Pi) starvation effect on the cytosolic Pi concentration and Pi exchanges across the tonoplast in plant cells: an in vivo 31P-nuclear magnetic resonance study using methylphosphonate as a Pi analog. Plant Physiol. 151, 1646–1657. doi: 10.1104/pp.109.144626
Raboy, V., Gerbasi, P. F., Young, K. A., Stoneberg, S. D., Pickett, S. G., Bauman, A. T., et al. (2000). Origin and seed phenotype of maize low phytic acid 1 and low phytic acid 2-1. Plant Physiol. 124:355. doi: 10.1104/pp.124.1.355
Ried, M. K., Wild, R., Zhu, J., Broger, L., Harmel, R. K., Hothorn, L. A., et al. (2019). Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. BioRxiv [Preprint], doi: 10.1101/2019.12.13.875393
Rogers, S. O., and Bendich, A. J. (1985). Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Mol. Biol. 5, 69–76. doi: 10.1007/BF00020088
Rounds, M. A., and Nielsen, S. S. (1993). Anion-exchange high-performance liquid chromatography with post-column detection for the analysis of phytic acid and other inositol phosphates. J. Chromatogr. A 653, 148–152. doi: 10.1016/0021-9673(93)80404-V
Sacchi, G. A., and Nocito, F. F. (2019). Plant sulfate transporters in the low phytic acid network: some educated guesses. Plants 8:616. doi: 10.3390/plants8120616
Sashidhar, N., Harloff, H.-J., and Jung, C. (2019). Identification of phytic acid mutants in oilseed rape (Brassica napus) by large scale screening of mutant populations through amplicon sequencing. New Phytol. 225, 2022–2034. doi: 10.1111/nph.16281
Sashidhar, N., Harloff, H. J., Potgieter, L., and Jung, C. (2020). Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 1–10. doi: 10.1111/pbi.13380
Shah, S., Karunarathna, N. L., Jung, C., and Emrani, N. (2018). An APETALA1 ortholog affects plant architecture and seed yield component in oilseed rape (Brassica napus L.). BMC Plant Biol. 18:380. doi: 10.1186/s12870-018-1606-9
Shi, J., Wang, H., Schellin, K., Li, B., Faller, M., Stoop, J. M., et al. (2007). Embryo-specific silencing of a transporter reduces phytic acid content of maize and soybean seeds. Nat. Biotechnol. 25, 930–937. doi: 10.1038/nbt1322
Sparvoli, F., and Cominelli, E. (2015). Seed biofortification and phytic acid reduction: a conflict of interest for the plant? Plants 4, 728–755. doi: 10.3390/plants4040728
Verrier, P. J., Bird, D., Burla, B., Dassa, E., Forestier, C., Geisler, M., et al. (2008). Plant ABC proteins – a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. doi: 10.1016/j.tplants.2008.02.001
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Wilcox, J., Premachandra, G., Young, K., and Raboy, V. (2000). Isolation of high seed inorganic P, Low-Phytate soybean mutants. Crop Sci. 40, 1601–1605. doi: 10.2135/cropsci2000.4061601x
Williams, S. P., Gillaspy, G. E., and Perera, I. Y. (2015). Biosynthesis and possible functions of inositol pyrophosphates in plants. Front. Plant Sci. 6:67. doi: 10.3389/fpls.2015.00067
Xu, X. H., Zhao, H. J., Liu, Q. L., Frank, T., Engel, K. H., An, G., et al. (2009). Mutations of the multi-drug resistance-associated protein ABC transporter gene 5 result in reduction of phytic acid in rice seeds. Theoret. Appl. Genet. 119, 75–83. doi: 10.1007/s00122-009-1018-1
Keywords: ATP binding cassette, Brassica napus, BnMRP5, lpa, phosphorous, TILLING, EMS, mutant
Citation: Sashidhar N, Harloff HJ and Jung C (2020) Knockout of MULTI-DRUG RESISTANT PROTEIN 5 Genes Lead to Low Phytic Acid Contents in Oilseed Rape. Front. Plant Sci. 11:603. doi: 10.3389/fpls.2020.00603
Received: 24 February 2020; Accepted: 21 April 2020;
Published: 26 May 2020.
Edited by:
Nicolas Rispail, Spanish National Research Council, SpainReviewed by:
Qingyao Shu, Zhejiang University, ChinaFrancesca Sparvoli, Italian National Research Council, Italy
Copyright © 2020 Sashidhar, Harloff and Jung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Jung, Yy5qdW5nQHBsYW50YnJlZWRpbmcudW5pLWtpZWwuZGU=
 Niharika Sashidhar
Niharika Sashidhar Hans J. Harloff
Hans J. Harloff Christian Jung
Christian Jung