
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 19 May 2020
Sec. Plant Metabolism and Chemodiversity
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00532
In plants, gibberellins (GAs) play important roles in regulating growth and development. Early studies revealed the large chemodiversity of gibberellins in plants, but only GA1, GA3, GA4, and GA7 show biological activity that controls plant development. However, the elucidation of the GA metabolic network at the molecular level has lagged far behind the chemical discovery of GAs. Recent advances in downstream GA biosynthesis (after GA12 formation) suggest that species-specific gibberellin modifications were acquired during flowering plant evolution. Here, we summarize the current knowledge of GA metabolism in flowering plants and the physiological functions of GA deactivation, with a focus on GA 13 hydroxylation. The potential applications of GA synthetic biology for plant development are also discussed.
Gibberellins (GAs), a type of 6-5-6-5 tetracyclic diterpenoid, are essential phytohormones for plant growth and development. The more than 130 GAs discovered to date have been classified into two groups based on their number of carbon atoms: C19-GAs (with one carboxylic group at the C-7 position, e.g., GA20 and GA9 in Figure 1) and C20-GAs (with two carboxylic groups at the C-7 and C-19 positions, respectively, e.g., GA12 and GA53 in Figure 1). GAs have also been classified into two groups based on other chemical structural criteria: 13-H GAs (hydrogen at the C-13 position) and 13-OH GAs (hydroxyl group at the C-13 position). Plant biologists consider GA4, GA1 (also known as 13-OH GA4), GA7, and GA3 (also known as 13-OH GA7) to be the only common bioactive GAs in flowering plants. The bioactivity of GA1 in plants is ∼1000-fold lower than that of GA4 (Eriksson et al., 2006).
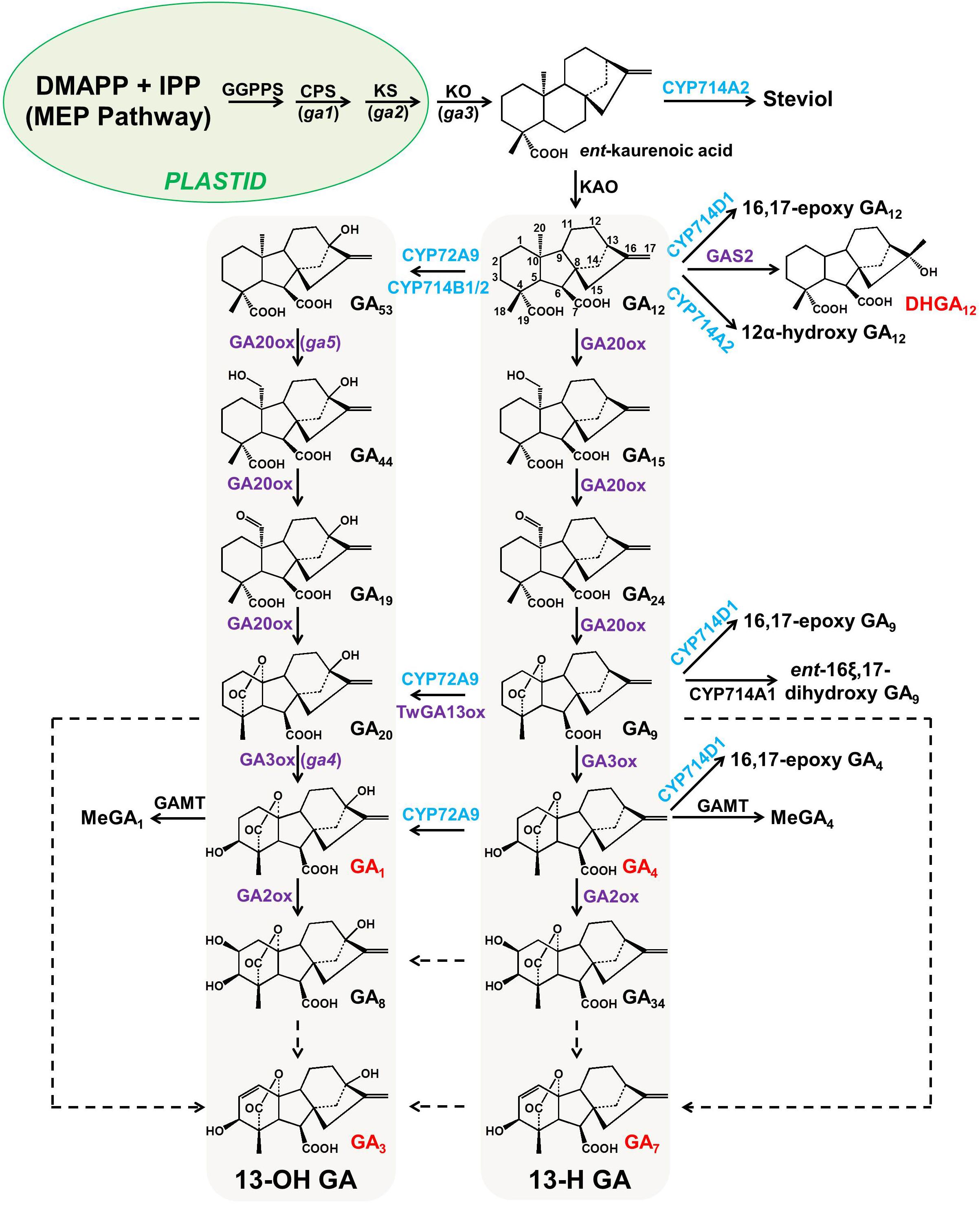
Figure 1. Simplified GA metabolic pathway in flowering plants. All enzymes mapped to the GA metabolic pathway, except GA13ox from Tripterygium wilfordii, were verified with enzymatic assays and the chemical profiling of transgenic plants. P450s (ER membrane-associated proteins) in this pathway are highlighted in blue, while 2-oxoglutarate-dependent dioxygenases (2-ODDs, cytosolic proteins) are highlighted in purple. All dashed lines indicate enzymatic a step that has not yet been characterized at the genetic level in any flowering plant. The genes/enzymes characterized from Arabidopsis and rice are marked in black and blue, respectively. The carbon backbone of GA12 is labeled with numbers, and the bioactive GAs (GA1, GA3, GA4, GA7, and DHGA12) are marked in red. The Arabidopsis GA-deficient mutants (ga1 to ga5) used for map-based cloning are shown in parentheses next to the corresponding genes. CPS, ent-copalyl diphosphate synthase; GAMT, GA methyltransferase; GAS2, gain of function in ABA-modulated seed germination 2; GGPPS, geranylgeranyl diphosphate synthase; KAO, ent-kaurenoic acid oxidase; KO, ent-kaurene oxidase; KS, ent-kaurenoic acid synthase; MEP, 2-C-methyl-D-erythritol 4-phosphate.
The GA metabolic pathway at the molecular level has been extensively investigated using both forward and reverse genetics strategies. Both strategies used on GA metabolism started with the dwarfism phenotype caused by bioactive GA deficiency (e.g., so-called GA-sensitive mutants). A forward genetics strategy, in which a mutant population is scored by the dwarf phenotype change followed by map-based cloning of the causal genes, has been applied to GA biosynthesis gene identification in Arabidopsis, from GGPP to GA4, for a long time. Thus far, five GA-sensitive Arabidopsis mutants (ga1 to ga5) have been used to determine the causal genes: GA1 encodes CPS (ent-copalyl diphosphate synthase; EC 5.5.1.13), GA2 encodes KS (ent-kaurene synthase; EC 4.2.3.19), GA3 encodes KO (ent-kaurene oxidase; EC 1.14.13.79), and GA4 and GA5 encode GA 3β-hydroxylase and GA 20-oxidase respectively (Yamaguchi, 2008; Hedden and Sponsel, 2015). As there are 10 GGPPS (geranylgeranyl diphosphate synthase; EC 2.5.1.29) or GGPPS-like genes in the Arabidopsis genome, it is not clear which GGPPS gene(s) is responsible for GA biosynthesis. A recent study demonstrated that GGPPSL1, 2, 3, 4, and 11 are bona fide GGPPS enzymes in Arabidopsis (Nagel et al., 2015; Wang et al., 2016). Combined with the subcellular localization (GGPPSL2 and 11 are plastidial proteins) and tissue specificity (GGPPSL11 showed a much higher level than GGPPSL2 in all tested Arabidopsis tissues) information, it can be inferred that GGPPSL11 (At4g36810) is likely the key player in providing GGPP for GA biosynthesis, although GGPPSL11 also functioned as a GGPP provider for chlorophyll and carotenoid biosynthesis in Arabidopsis (Beck et al., 2013; Vranova et al., 2013; Chen et al., 2015; Wang et al., 2016). The knock-down mutant of GGPPSL11 (SALK_140601, T-NDA inserted at the promoter region) showed a dwarf phenotype and pale-green leaves, while knock-out mutants of GGPPSL11 showed a seedling-lethal phenotype (Chen et al., 2015). Notably, endogenous GAs were not measured in any tissue of either GGPPSL2 or GGPPSL11 mutants.
Thanks to the work of phytochemists, 136 naturally occurring GAs have been identified thus far (Hedden, 2019). However, no novel GAs have been reported in plants for a long time. Recently, Liu et al. (2019) reported a novel gibberellin, DHGA12 (chemical structure shown in Figure 1), which possesses a C20 skeleton structure and can promote seed germination and seedling establishment in Arabidopsis. DHGA12, like other bioactive GAs (e.g., GA1, GA3, GA4, and GA7), has the ability to bind the GA receptor (GID1), although it has a lower affinity to GID1 than GA4. GAS2, a 2-oxoglutarate-dependent dioxygenase (2-ODD), was also identified to catalyze GA12 into DHGA12 (Liu et al., 2019). However, the ABA (abscisic acid) level was significantly reduced in GAS2 overexpression lines, which complicates the explanation of how GAS2 promotes seed germination. Does the increase in DHGA12 (twofold) compensate for the decrease of GA4 (10-fold) in the GAS2 overexpression lines? Furthermore, DHGA12 or GA4 could only partially rescue the phenotype of cotyledon greening and hypocotyl elongation in gas2 mutants, indicating that there were other factors involved in these processes. Given the promiscuity of plant 2-ODDs (Mitchell and Weng, 2019), it seems that the activity of GAS2 is not restricted to GA12 oxidation to form DHGA12 but that it acts on other unknown substrates. The possibility of whether GAS2 is also involved in ABA inactivation via oxidation remains to be clarified. Comprehensive metabolite analysis of GAS2 transgenic lines will provide valuable evidence to elucidate the GAS2 functions in Arabidopsis.
As shown in Figure 1, it remains unknown how the other two bioactive GAs, GA3, and GA7, are biosynthesized in plants. GA4-desaturase (GA4-DES), which also belongs to the 2-ODD family and which converts GA4 to GA7 (Figure 1), has been functionally identified from Gibberella fujikuroi (Tudzynski et al., 2003; Bhattacharya et al., 2012). The overexpression of GA4-DES in plants resulted in a 20-fold higher concentration of GA3 along with an ∼70-fold reduction in GA4. Moreover, the resulting transgenic plants showed a substantial growth increase, which reflected the increase in the total bioactivity of GA in planta (Bhattacharya et al., 2012). However, the functional homologs of fungal GA4-DES have not been characterized from plants to date. The Peter Hedden group recently demonstrated that two GA 3-oxidases from southern wild cucumber (Marah macrocarpus), MmGA3ox1 and MmGA3ox2, were involved in GA3/GA7 production (from GA9 and/or GA20) (Ward et al., 2010). Notably, these results do not exclude the possibility that plants evolved other enzyme systems (a case of convergent evolution) to produce GA3/GA7, which merits further investigation.
Previous studies have shown that not only GA biosynthesis but also GA catabolism (deactivation) play important roles in GA bioactivity homeostasis in planta. Thus far, three types of GA deactivation enzymes and their encoding genes have been identified, including GA 2-oxidase (belonging to the 2-ODD family; seven GA2ox genes in Arabidopsis and ten GA2ox genes in rice) (Rieu et al., 2008), GA methyltransferase (GAMT1/2 from Arabidopsis) (Varbanova et al., 2007), and GA 16,17-oxidase (CYP714D1/EUI from rice) (Zhu et al., 2006). Phylogenetic analysis shows that there are three CYP714 subclades (CYP714B, C, and D) in the rice genome (Figure 2A). It is plausible to speculate that other members of the CYP714 family might be involved in GA deactivation in plants. Recently, reverse genetic studies in rice have suggested a dominant role for CYP714B1 and CYP714B2 in reducing GA activity through 13-hydroxylation of GA12 to form GA53 (Magome et al., 2013). Interestingly, both cyp714d1 mutant and cyp714b1/cyp714b2 double mutant rice display an elongated uppermost internode, where these three P450s are highly expressed, at the heading stage. The measurement of endogenous GAs showed that the levels of 13-OH GAs were decreased, while the levels of the 13-H GAs were increased in the cyp714b1/cyp714b2 double mutant. However, there are no members of the CYP714D and CYP714B subfamily in Arabidopsis. The closest analogs to CYP714D1 and CYP714B1/B2 in Arabidopsis are CYP714A1 and CYP714A2 (Figure 2A). Both are GA deactivation enzymes, at least in vitro: CYP714A1 catalyzes the conversion of GA12 into 16-carboxylated GA12, while CYP714A2 catalyzes the conversion of ent-kaurenoic acid into steviol (ent-13-hydroxy kaurenoic acid) (Zhang et al., 2011; Nomura et al., 2013). In contrast to their rice counterparts, knockdown of both CYP714A1 and CYP714A2 led to increased bioactive GA4 and no change in GA1 when compared to those in wild-type plants. Together with the wide occurrence of GA1 in flowering plants, these results indicate that CYP714A1 and CYP714A2 are not the main contributors to GA 13-hydroxylation and that other genes are responsible for 13-OH GA production in Arabidopsis.
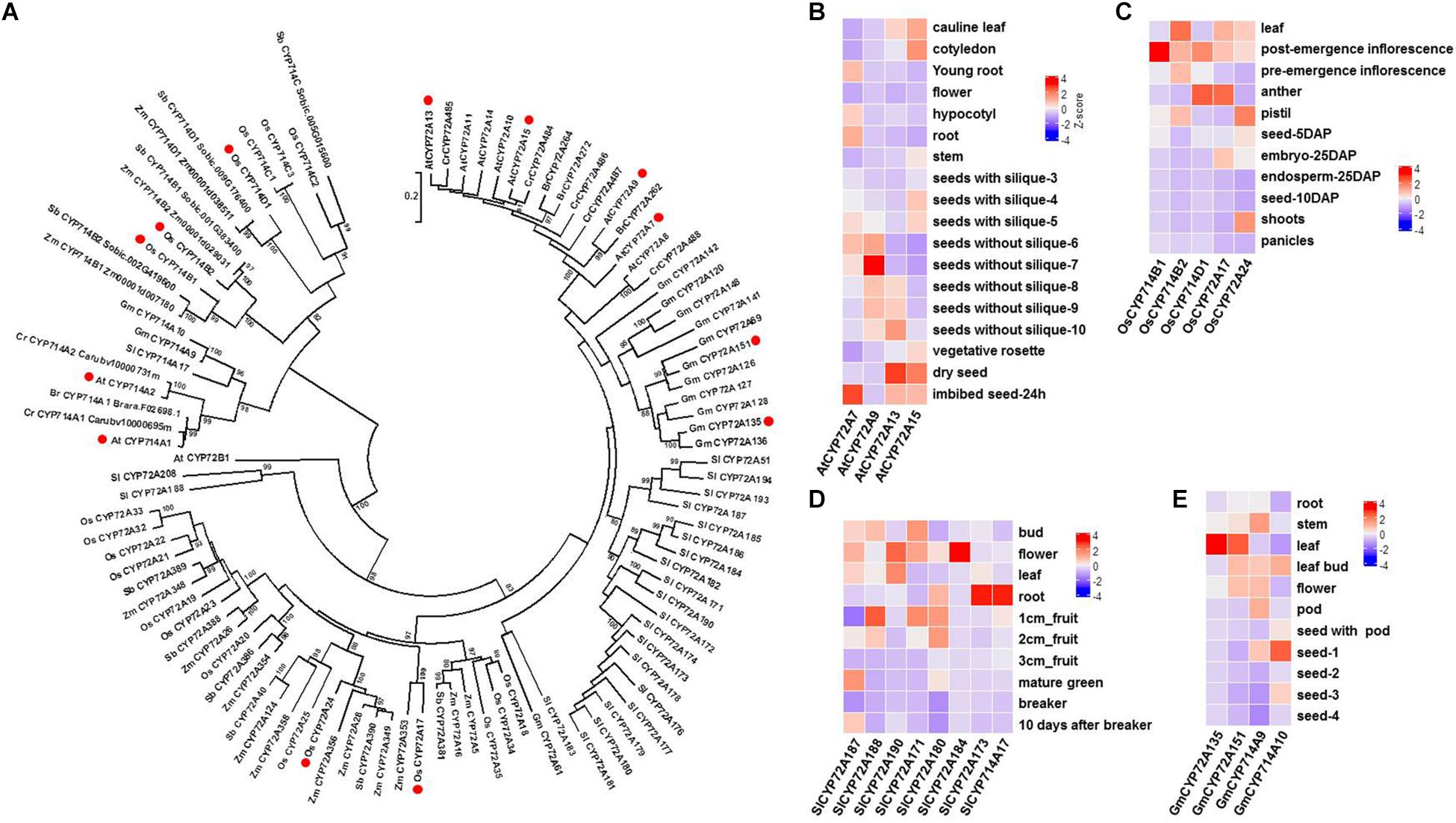
Figure 2. Characterization of GA oxidases encoded by P450 genes in flowering plants. (A). Phylogenetic analysis of plant GA oxidases encoded by P450 genes using MEGA 6.0 (Tamura et al., 2013). A total of 102 P450 proteins (members of the CYP72A and CYP714 subfamilies) were obtained from 8 representative species in plant evolutionary history. P450 proteins with GA oxidase activity are marked with red circles. Bootstrap values (based on 500 replicates) >80% are shown for corresponding nodes. At, Arabidopsis thaliana; Br, Brassica rapa; Cr, Capsella rubella; Gm, Glycine max; Os, Oryza sativa; Sb, Sorghum bicolor; Sl, Solanum lycopersicum; Zm, Zea mays. (B) Tissue specificity of four CYP72A genes in Arabidopsis. Data were extracted from the gene expression map of Arabidopsis development (http://arabidopsis.org). (C) Tissue specificity of three CYP714 genes and two CYP72A genes in rice. RNA-Seq (FPKM) data were extracted from the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu). (D) Tissue specificity of seven CYP72A genes and one CYP714A gene in tomato plants. Data were extracted from the Tomato Genome Consortium (Sato et al., 2012). (E) Tissue specificity of two CYP72A genes and two CYP714A genes in soybean plants. Data were extracted from Shen et al. (Shen et al., 2014).
More recently, CYP72A9 (encoded by At3g14630) was reported to catalyze the 13-H GAs (GA12, GA9, GA4) into 13-OH GAs (GA53, GA20, GA1), which is inconsistent with the traditional view that GA4 and GA1 are biosynthesized in parallel by GA20ox and GA3ox in flowering plants, from GA12 and GA53, respectively (Figure 1; He et al., 2019). It was unexpected to discover that CYP72A9 encoded GA 13-hydroxylase, as CYP72A9 was located in one geranylfarnesyl pyrophosphate synthase (C25)-sesterterpene synthase-P450 (GFPPS-sesterTPS-P450) gene cluster. Eight tandem duplicated CYP72As (CYP72A7, At3g14610; A8, At3g14620; A9, At3g14630; A10, At3g14640; A11, At3g14650; A13, At3g14660; A14, At3g14680; A15, At3g14690) are located together at the same chromosome region. These eight CYP72As were first thought to be involved in sesterterpene (C25) biosynthesis (Wang et al., 2016; Shao et al., 2017; Chen Q. et al., 2019). Plants overexpressing CYP72A9 exhibited semi-dwarfism, which was caused by a significant reduction in GA4 levels. Moreover, CYP72A9 was expressed predominantly in developing seeds in Arabidopsis, and cyp72a9 mutants showed a deficiency of GA1 but an increase of GA4 in the siliques. CYP72A9 was further proven to be involved in primary seed dormancy (He et al., 2019). The other three Arabidopsis CYP72A members (A7, A13, and A15) also showed GA 13-hydroxylase activity, but overexpression plants did not show a semi-dwarf phenotype, which merits further investigation of the physiological functions of these genes in planta. Homologs of CYP72A9 in other Brassicaceae plants, such as CYP72A262 in Brassica rapa and CYP72A484 in Capsella rubella, also have GA 13-hydroxylase activity, although with slightly different substrate preferences (He et al., 2019). It appears that the recruitment of GA deactivation enzymes shows different tissue specificity in different plant species, at least at the family level: CYP714D1 and CYP714B1/B2 are highly expressed in the uppermost internode of rice (Poaceae family), while all GA deactivation genes (CYP72A9, GAMT1/2, and CYP714A1/A2) are predominantly expressed in the developing siliques/seeds of Arabidopsis (Brassicaceae family). As both CYP714 and CYP72A are widely distributed in flowering plants, it will be interesting to determine the role of each member of both P450 subfamilies in contributing to GA homeostasis in different plant species (Figure 2). The tissue specificity of potential GA genes from the CYP714 and CYP72A subfamilies also suggested the functional divergence of GA deactivation in different plant species (the genes from Arabidopsis, rice, soybean, and tomato are shown in Figures 2B–E). For example, CYP72A135 has been demonstrated to catalyze the conversion of GA9 to GA20 (He et al., 2019), and CYP72A135 was predominantly expressed in leaves in soybean plants (Figure 2D). Thus, further investigation is needed to test whether CYP72A135 is involved in leaf development in soybean.
Zhang et al. recently reported another type of GA13-hydroxylase, TwGA13ox, from Tripterygium wilfordii (Celastraceae family). TwGA13ox, in contrast to the P450s characterized in rice and Arabidopsis, is a 2-ODD enzyme, like GA20ox, GA3ox, and GA2ox (Figure 1; Zhang et al., 2019). TwGA13ox was highly expressed in root phloem and could be induced by methyl jasmonate (MeJA) treatment. TwGA13ox specifically catalyzes the conversion of GA9 to GA20 in vitro but does not catalyze the formation of GA1 from GA4. It is noteworthy that the more detailed biochemical and physiological function of TwGA13ox has not yet been determined in Tripterygium wilfordii. Additionally, GA13-hydroxylase activity of the 2-ODD enzyme has previously been reported in cucumber plants (Cucumis sativus L.) (Lange et al., 2013). However, it is unlikely that 2-ODD enzymes are the main contributor to 13-OH GA production in flowering plants, as the above-mentioned P450s are responsible for 13-OH GA production in rice and Arabidopsis, which has been shown at the biochemical and genetic levels.
Due to the very low level of endogenous GAs, accurate quantitative measurement plays an important role in elucidating the GA metabolic pathway in plants. Currently, triple quadrupole MS (mass spectrometry) with MRM (multi-reaction monitoring) is the most frequently used operation mode for the quantitative analysis of target GAs (Pan et al., 2010; Urbanova et al., 2013; Chu et al., 2017). In most cases, the GAs shown in Figure 1 can be quantitatively measured using the LC-QQQ-MS/MS technique combined with GA enrichment using SPE (solid-phase extraction) sorbents. For the qualitative discovery and identification of unknown GAs, MS, and MS/MS full scans are commonly used to obtain more structural information. Sometimes, high-resolution MS, such as Q-TOF (quadrupole time-of-flight), Orbitrap, or FT-ICR (Fourier transform ion cyclotron resonance), is necessary to derive the molecular formulae of the precursor and product ions. The purification of a novel GA from plants and the elucidation of its absolute chemical structure is always a time-consuming and laborious procedure.
Since GAs play important roles in regulating plant growth and development, the GA genes described here represent a promising toolkit with which synthetic biologists can adjust bioactive GA levels to manipulate plant traits. The most famous GA gene used for crop improvement is the “Green Revolution” gene in rice: the “dwarfing” rice caused by the knock-out of Os20ox2 (semi-dwarfing allele, sd-1 mutant) showed high resistance to lodging stress and significantly increased the yield (to support the heavy grain of the high-yielding varieties), in combination with the application of large amounts of fertilizer and pesticides (Sasaki et al., 2002; Spielmeyer et al., 2002; Hedden, 2003). In Arabidopsis, a previous study showed that overexpression of AtCPS (At4g02780) in Arabidopsis leads to an increase in ent-kaurene/ent-kaurenoic acid and GA12 but does not affect the level of bioactive GAs or cause a phenotype change (Fleet et al., 2003). Moreover, the overexpression of AtGA20ox (At4g02780) in Arabidopsis (Col-0) leads to an increase in bioactive GAs and the GA-overproduction phenotype (Huang et al., 1998). Notably, there were higher concentrations of GA1 and GA20 in AtGA20ox OE plants than in wild-type seedlings, whereas the concentration of GA4 remained approximately the same. The constitutive promoters, coupled with additional copies of the GA genes, were applied in all of the above-mentioned cases in Arabidopsis. All these results also confirmed that GA genes are sensitive to endogenous bioactive GA changes: GA biosynthesis genes are usually upregulated when GA4 decreases, while GA catabolism genes are downregulated under the same conditions, and vice versa (He et al., 2019). Synthetic biology could thus be used to replace native promoters with new tunable promoters that active/deactivate in response to endogenous bioactive GA and even other internal/external signals (Kassaw et al., 2018). At the enzyme level, the targeted near-saturated mutagenesis of GA genes, especially those downstream of GA12 formation (multiple copies with different tissue-specificity), using CRISPR-based editing could be applied to fine-tune bioactive GA levels to achieve ideal plant traits (Chen K. L. et al., 2019; Eshed and Lippman, 2019). After quantitatively characterizing these GA components in planta, other GA-related factors, such as signal transduction and interactions with other phytohormones, etc., will be considered and re-designed to drive GA synthetic biology forward.
GW and JH conceptualized the content of the manuscript. JH wrote the first draft of the manuscript. GW finalized the manuscript with contributions from JH, PX, XM, and JC.
This work was financially supported by the National Key R&D Program of China (Grant No. 2018YFA0900600), the National Natural Science Foundation of China (Grant Nos. 31970315 and 31770398), the Key R&D Program of Shandong Province (Grant No. 2019JZZY020610), and the State Key Laboratory of Plant Genomics of China (SKLPG2016A-13).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Beck, G., Coman, D., Herren, E., Ruiz-Sola, M., Rodriguez-Concepcion, M., Gruissem, W., et al. (2013). Characterization of the GGPP synthase gene family in Arabidopsis thaliana. Plant. Mol. Biol. 82, 393–416. doi: 10.1007/s11103-013-0070-z
Bhattacharya, A., Kourmpetli, S., Ward, D. A., Thomas, S. G., Gong, F., Powers, S. J., et al. (2012). Characterization of the fungal gibberellin desaturase as a 2-oxoglutarate-dependent dioxygenase and its utilization for enhancing plant growth. Plant Physiol. 160, 837–845. doi: 10.1104/pp.112.201756
Chen, K. L., Wang, Y. P., Zhang, R., Zhang, H. W., and Gao, C. X. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Chen, Q., Jiang, T., Liu, Y., Liu, H., Zhao, T., Liu, Z., et al. (2019). Recently duplicated sesterterpene (C25) gene clusters in Arabidopsis thaliana modulate root microbiota. Sci. China Life Sci. 62, 947–958. doi: 10.1007/s11427-019-9521-2
Chen, Q., Fan, D., and Wang, G. (2015). Heteromeric geranyl(geranyl) diphosphate synthase is involved in monoterpene biosynthesis in Arabidopsis flowers. Mol. Plant 8, 1434–1437. doi: 10.1016/j.molp.2015.05.001
Chu, J., Fang, S., Xin, P., Guo, Z., and Chen, Y. (2017). “Quantitative analysis of plant hormones based on LC-MS/MS,” in Hormone Metabolism and Signaling in Plants, eds J. Li, C. Li, and S. M. Smith (Cambridge, MA: Academic Press), 471–537.
Eriksson, S., Bohlenius, H., Moritz, T., and Nilsson, O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18, 2172–2181. doi: 10.1105/tpc.106.042317
Eshed, Y., and Lippman, Z. B. (2019). Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 366:eaax0025. doi: 10.1126/science.aax0025
Fleet, C. M., Yamaguchi, S., Hanada, A., Kawaide, H., David, C. J., Kamiya, Y., et al. (2003). Overexpression of AtCPS and AtKS in Arabidopsis confers increased ent-kaurene production but no increase in bioactive gibberellins. Plant Physiol. 132, 830–839. doi: 10.1104/pp.103.021725
He, J., Chen, Q. W., Xin, P. Y., Yuan, J., Ma, Y. H., Wang, X. M., et al. (2019). CYP72A enzymes catalyse 13-hydroxylation of gibberellins. Nat. Plants 5, 1057–1065. doi: 10.1038/s41477-019-0511-z
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/s0168-9525(02)00009-4
Hedden, P. (2019). A novel gibberellin promotes seedling establishment. Nat. Plants 5, 459–460. doi: 10.1038/s41477-019-0427-7
Hedden, P., and Sponsel, V. (2015). A century of gibberellin research. J. Plant Growth Regul. 34, 740–760. doi: 10.1007/s00344-015-9546-1
Huang, S. S., Raman, A. S., Ream, J. E., Fujiwara, H., Cerny, R. E., and Brown, S. M. (1998). Overexpression of 20-oxidase confers a gibberellin overproduction phenotype in Arabidopsis. Plant Physiol. 118, 773–781. doi: 10.1104/pp.118.3.773
Kassaw, T. K., Donayre-Torres, A. J., Antunes, M. S., Morey, K. J., and Medford, J. I. (2018). Engineering synthetic regulatory circuits in plants. Plant Sci. 273, 13–22. doi: 10.1016/j.plantsci.2018.04.005
Lange, M. J. P., Liebrandt, A., Arnold, L., Chmielewska, S. M., Felsberger, A., Freier, E., et al. (2013). Functional characterization of gibberellin oxidases from cucumber, Cucumis sativus L. Phytochemistry 90, 62–69. doi: 10.1016/j.phytochem.2013.02.006
Liu, H., Guo, S., Lu, M., Zhang, Y., Li, J., Wang, W., et al. (2019). Biosynthesis of DHGA12 and its roles in Arabidopsis seedling establishment. Nat. Commun. 10:1768. doi: 10.1038/s41467-019-09467-5
Magome, H., Nomura, T., Hanada, A., Takeda-Kamiya, N., Ohnishi, T., Shinma, Y., et al. (2013). CYP714B1 and CYP714B2 encode gibberellin 13-oxidases that reduce gibberellin activity in rice. Proc. Natl. Acad. Sci. U.S.A. 110, 1947–1952. doi: 10.1073/pnas.1215788110
Mitchell, A. J., and Weng, J. K. (2019). Unleashing the synthetic power of plant oxygenases: from mechanism to application. Plant Physiol. 179, 813–829. doi: 10.1104/pp.18.01223
Nagel, R., Bernholz, C., Vranová, E., Košuth, J., Bergau, N., Ludwig, S., et al. (2015). Arabidopsis thaliana isoprenyl diphosphate synthases produce the C25 intermediate geranylfarnesyl diphosphate. Plant J. 84, 847–859. doi: 10.1111/tpj.13064
Nomura, T., Magome, H., Hanada, A., Takeda-Kamiya, N., Mander, L. N., Kamiya, Y., et al. (2013). Functional analysis of Arabidopsis CYP714A1 and CYP714A2 reveals that they are distinct gibberellin modification enzymes. Plant Cell Physiol. 54, 1837–1851. doi: 10.1093/pcp/pct125
Pan, X., Welti, R., and Wang, X. (2010). Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat. Protoc. 5, 986–992. doi: 10.1038/nprot.2010.37
Rieu, I., Eriksson, S., Powers, S. J., Gong, F., Griffiths, J., Woolley, L., et al. (2008). Genetic analysis reveals that C-19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20, 2420–2436. doi: 10.1105/tpc.108.058818
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). Green revolution: a mutant gibberellin-synthesis gene in rice new insight into the rice variant that helped to avert famine over thirty years ago. Nature 416, 701–702. doi: 10.1038/416701a
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Shao, J., Chen, Q. W., Lv, H. J., He, J., Liu, Z. F., Lu, Y. N., et al. (2017). (+)-Thalianatriene and (-)-retigeranin B catalyzed by sesterterpene synthases from Arabidopsis thaliana. Org. Lett. 19, 1816–1819. doi: 10.1021/acs.orglett.7b00586
Shen, Y. T., Zhou, Z. K., Wang, Z., Li, W. Y., Fang, C., Wu, M., et al. (2014). Global dissection of alternative splicing in paleopolyploid soybean. Plant Cell 26, 996–1008. doi: 10.1105/tpc.114.122739
Spielmeyer, W., Ellis, M. H., and Chandler, P. M. (2002). Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. U.S.A. 99, 9043–9048. doi: 10.1073/pnas.132266399
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tudzynski, B., Mihlan, M., Rojas, M. C., Linnemannstons, P., Gaskin, P., and Hedden, P. (2003). Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi - des and P450-3 encode GA(4) desaturase and the 13-hydroxylase, respectively. J. Biol. Chem. 278, 28635–28643. doi: 10.1074/jbc.M301927200
Urbanova, T., Tarkowska, D., Novak, O., Hedden, P., and Strnad, M. (2013). Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 112, 85–94. doi: 10.1016/j.talanta.2013.03.068
Varbanova, M., Yamaguchi, S., Yang, Y., McKelvey, K., Hanada, A., Borochov, R., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45. doi: 10.1105/tpc.106.044602
Vranova, E., Coman, D., and Gruissem, W. (2013). Network analysis of the MVA and MEP pathways for isoprenoid synthesis. Annu. Rev. Plant Biol. 64, 665–700. doi: 10.1146/annurev-arplant-050312-120116
Wang, C. Y., Chen, Q. W., Fan, D. J., Li, J. X., Wang, G. D., and Zhang, P. (2016). Structural analyses of short-chain prenyltransferases identify an evolutionarily conserved GFPPS clade in Brassicaceae plants. Mol. Plant 9, 195–204. doi: 10.1016/j.molp.2015.10.010
Ward, D. A., MacMillan, J., Gong, F., Phillips, A. L., and Hedden, P. (2010). Gibberellin 3-oxidases in developing embryos of the southern wild cucumber. Marah macrocarpus. Phytochemistry 71, 2010–2018. doi: 10.1016/j.phytochem.2010.09.015
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Zhang, Y., Su, P., Wu, X., Zhou, J., Zhao, Y., Hu, T., et al. (2019). The gibberellin 13-oxidase that specifically converts gibberellin A9 to A20 in Tripterygium wilfordii is a 2-oxoglutarate-dependent dioxygenase. Planta 250, 1613–1620. doi: 10.1007/s00425-019-03240-0
Zhang, Y., Zhang, B., Yan, D., Dong, W., Yang, W., Li, Q., et al. (2011). Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 67, 342–353. doi: 10.1111/j.1365-313X.2011.04596.x
Keywords: gibberellins, plant development, metabolism, hydroxylation, oxidation
Citation: He J, Xin P, Ma X, Chu J and Wang G (2020) Gibberellin Metabolism in Flowering Plants: An Update and Perspectives. Front. Plant Sci. 11:532. doi: 10.3389/fpls.2020.00532
Received: 07 February 2020; Accepted: 08 April 2020;
Published: 19 May 2020.
Edited by:
Deyu Xie, North Carolina State University, United StatesReviewed by:
Raimund Nagel, Institute of Biology, University of Leipzig, GermanyCopyright © 2020 He, Xin, Ma, Chu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, Z2R3YW5nQGdlbmV0aWNzLmFjLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.