- 1Center for Viticulture and Enology, College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China
- 2Key Laboratory of Viticulture and Enology, Ministry of Agricultural and Rural Affairs, Beijing, China
Norisoprenoids are important aromatic volatiles contributing to the pleasant floral/fruity odor in grapes and wine. They are produced from carotenoids through the cleavage of carotenoid cleavage dioxygenases (CCDs). However, the underlying mechanisms regulating VvCCD expression remain poorly understood. In this study, we showed that VvCCD4b expression was positively correlated with the accumulation of β-damascenone, β-ionone, 6-methyl-5-hepten-2-one, geranylacetone, dihydroedulan I, and total norisoprenoids in developing grapes in two vintages from two regions. VvCCD4b was found to be principally expressed in flowers, mature leaves, and berries. Abscisic acid strongly induced the expression of this gene. Additionally, the present study preliminarily indicated that the activity of the VvCCD4b promoter was dropped under 37°C treatment and also responded to the illumination change. VvCCD4b was expressed in parallel with VvMADS4 in developing grape berries. The latter is a MADS family transcription factor and nucleus-localized protein that was captured by yeast one-hybrid. A dual-luciferase reporter assay in tobacco leaves revealed that VvMADS4 downregulated the activity of the VvCCD4b promoter. VvMADS4 overexpression in grape calli and Vitis quinquangularis Rehd. leaves repressed the VvCCD4b expression. In summary, this work demonstrates that VvCCD4b expression is positively correlated with the accumulation of norisoprenoids, and VvMADS4 is a potential negative regulator of VvCCD4b. Our results provide a new perspective for understanding the regulation of VvCCD4b expression and norisoprenoid accumulation in grapes.
Introduction
Norisoprenoids are volatile C9, C10, C11, and C13 molecules with a low odor perception threshold, generating the floral and fruity scents in grape berries and wine. The content of monoterpenes is extremely low in neutral grape varieties and their corresponding wines (e.g., Vitis vinifera L. cv. Cabernet Sauvignon, Merlot, Syrah, and Chardonnay). In contrast, norisoprenoids are more important than other terpenoids in forming the characteristic aromas of these varieties, because the norisoprenoid concentrations tend to be higher than their thresholds (Mateo and Jiménez, 2000). Indeed, several norisoprenoids are the primary contributors to aroma in red and white grapes and wines (Xu et al., 2015; Gao et al., 2016; Asproudi et al., 2018; Chen W. K. et al., 2018). For example, β-damascenone exhibits a complex smell of “cooked apple,” “floral,” and “quince” (Kotseridis et al., 1999), whereas β-ionone was described as “violet,” “woody,” and “raspberry” (Aznar et al., 2001). Aged Riesling wines have a floral profile of “kerosene” and “petrol” owing to the presence of 1,1,5-trimethyl-1,2-dihydronaphthalene (TDN) (Simpson, 1978; Simpson and Miller, 1983).
Carotenoids are the precursors of norisoprenoids, and they are synthesized via the methyl-erythritol-phosphate (MEP) pathway in plastids (Dunlevy and Kalua, 2009). Major enzymes driving biochemical reactions in this pathway include deoxyxylulose 5-phosphate synthase (DXS), deoxyxylulose 5-phosphate reductoisomerase (DXR), and 1-hydroxy-2-methyl-2-(E)-butenyl-4-PP reductase (HDR) (Cunningham and Gantt, 2000; Dunlevy and Kalua, 2009). Besides, carotenoid cleavage dioxygenases (CCDs) are critical enzymes that catalyze the generation of norisoprenoids (apocarotenoids) by cleaving the conjugate double bond of carotenoids (Fleischmann et al., 2003; Castillo et al., 2005; Baldermann et al., 2010; Campbell et al., 2010; Chiou et al., 2010; Ledger et al., 2010; Brandi et al., 2011; Frusciante et al., 2014). In Arabidopsis thaliana, AtCCD1 and AtCCD4 cleave carotenoids to produce norisoprenoid compounds. The CCD1 and CCD4 homologs from some other plants were also reported to possess these biochemical functions, although they show different preferences toward the substrates and the cleavage sites (Fleischmann et al., 2003; Castillo et al., 2005; Baldermann et al., 2010; Campbell et al., 2010; Chiou et al., 2010; Ledger et al., 2010; Brandi et al., 2011; Adami et al., 2013; Ma et al., 2013; Frusciante et al., 2014; Bai et al., 2015). CCD4 usually has more than one isoform in plants, involving the coloration of petals or the generation of floral/fruity aroma by cleaving carotenoids. In Chrysanthemum morifolium, CmCCD4a, which mostly expressed in petals, cleaves the carotenoid, leading to the formation of a white flower (Ohmiya et al., 2006). CsCCD4c cleaves β-carotene at different sites to produce β-ionone and β-cyclocitral; meanwhile, this enzyme also uses lutein, neoxanthin, and violaxanthin as substrates (Rubio-Moraga et al., 2014). In grapevines, VvCCD1, VvCCD4a, and VvCCD4b have been biochemically characterized. VvCCD1 isolated from “Shiraz” can cleave zeaxanthin to generate 3-hydroxy-β-ionone (Mathieu et al., 2005). Overexpressing VvCCD1, VvCCD4a, and VvCCD4b from V. vinifera L. cv “Pinotage” in carotenoid-generating Escherichia coli strains revealed that they all can cleave lycopene and ε-carotene to form 6-methyl-5-hepten-2-one (MHO) and β-ionone, respectively (Lashbrooke et al., 2013). VvCCD1 was the only enzyme capable of cleaving β-carotene and incapable of cleaving neurosporene. In another experiment, VvCCD1 and VvCCD4b from “Cabernet Sauvignon” were expressed in recombinant Saccharomyces cerevisiae cells that were able to produce carotenoids; both enzymes were found to cleave lycopene and β-carotene (Meng et al., 2019). The three genes are expressed lowly in young grape berries but increased with ripening (Lashbrooke et al., 2013).
Individual norisoprenoid has a distinct accumulation pattern. In “Pinot noir” grapes, total β-damascenone, vitispirane, and TDN increased during ripening, whereas total α-ionone and β-ionone decreased (Yuan and Qian, 2016). In “Cabernet Sauvignon” grapes, the contents of MHO and geranylacetone were nearly identical between the two producing regions in some stages of development (Chen et al., 2017). Actually, environmental factors markedly influence norisoprenoid production in grape berries. Light exposure and water deficit increase the concentration of norisoprenoids (Bindon et al., 2007; Kwasniewski et al., 2010; Feng et al., 2015). The three VvCCD genes present some differences in the spatial and temporal expression patterns. VvCCD4b is expressed predominantly in mature grape berries (Lashbrooke et al., 2013). Our previous research also observed that norisoprenoid concentration was correlated with the transcript level of VvCCD4b in developing “Cabernet Sauvignon” grape berries (Chen et al., 2017). However, the regulation of VvCCD4b expression is poorly understood.
In plants, cis- and trans-acting elements are largely responsible for regulating gene transcription. Cis-acting elements in the promoter region determine temporal and spatial gene expression patterns, as well as the manner in which these patterns respond to stress. CmCCD4a-5 in C. morifolium and AtCCD7 in A. thaliana are petal specific (Imai et al., 2013) and vascular tissue specific (Liang et al., 2011), respectively. In Malus, drought, waterlogging, and methyl jasmonate treatments all decreased CCD7 promoter activity (Yue et al., 2015).
Trans-acting elements, especially transcription factors, bind to the cis-acting regulatory regions and influence gene expression. Major transcription factor families in plants include WRKY, MYB, NAC, bZIP, and MADS, and they participate in numerous functions such as stress response, metabolism, and hormonal induction. However, we know little about transcription factors that regulate CCD expression. A recent study in Citrus sinensis found that CsMADS6 activated CsCCD1 expression (Lu et al., 2018). Additionally, OfWRKY3 and OfERF61 in Osmanthus fragrans Lour. stimulate OfCCD4 expression, which results in carotenoid cleavage and influences β-ionone synthesis in sweet osmanthus petals (Han et al., 2016, 2019). However, until now, there has been no report involving the transcriptional regulation of VvCCDs in grapes.
Thus, in this study, we examined the relationship between CCD expression and norisoprenoid accumulation in “Cabernet Sauvignon” grapes of two vintages from two regions with very different climates. We also assessed the temporal and spatial expression patterns of VvCCD4b and the responses of its promoter to temperature, light, and abscisic acid (ABA) treatments. Furthermore, we identified a transcription factor potentially regulating the VvCCD4b expression. The findings help clarify the mechanisms underlying norisoprenoid biosynthesis in grape berries.
Materials and Methods
Plant Materials
This study used “Cabernet Sauvignon” in 2013 and 2016 vintages from the Changli (CL) and Gaotai (GT) regions. These two regions are located in Hebei Province of Northeastern China and Gansu Province of Northwestern China, respectively, and are important wine-producing zones with a monsoon climate and continental climate, respectively. Details on regional climate, vineyards, grapevine cultivation, and sampling method are described in our previous publications (Xu et al., 2015; Chen et al., 2017). Grapes were sampled according to the E-L system (Coombe, 1995). In 2013, berries were sampled 4 (E-L31), 6 (E-L34), 8 (E-L35), 10 (E-L36), 12 (E-L37), and 14 (E-L38) weeks after flowering (WAF) in GT and 6 (E-L31), 8 (E-L34), 10 (E-L35), 12 (E-L36), 14 (E-L37), and 16 (E-L38) WAF in CL. In 2016, GT samples were collected 4 (E-L31), 7 (E-L35), 14 (E-L37), and 16 (E-L38) WAF, and CL samples were collected 6 (E-L31), 9 (E-L35), 16 (E-L37), and 20 (E-L38) WAF. Three biological replicates were performed per sampling. Each replicate randomly collected 300 berries without physical damage or disease, from 150 clusters (Xu et al., 2015). Grape berries with 2-mm pedicels were placed into a plastic bag and then a foam box and transported into the nearest laboratory within 2 h (Wen et al., 2015). At the laboratory, they were frozen in liquid nitrogen and stored at −80°C.
For spatial and temporal expression analyzes, stems, flowers, young leaves (10 days of age), mature leaves (1 month of age), tendrils, and roots were sampled at the full-bloom stage, and grape berries were gathered at 5, 15, 63, 70, 80, and 98 days after flowering following the E-L system from E-L29 to E-L38 (Coombe, 1995) in 2019. All the samples were collected from the self-rooted “Cabernet Sauvignon” grapevines at the Chateau SunGod Great Wall (Huailai, Hebei Province). Except for roots, other tissues were sampled from nine grapevines, which were divided into three groups (three grapevines each group) corresponding to three biological replicates. As for individual tissue, material from one biological replicate was pooled and frozen in liquid nitrogen. They were powdered prior to use. The roots were dug out from the underground parts of another nine grapevines in the same vineyard, which were also divided into three biological replicates. The land is privately owned, and no protected species were sampled (manager: Qing-quan Yu, e-mail: eXVxcUBjb2Zjby5jb20=).
The “Cabernet Sauvignon” grape calli were induced from pulp cells. Callus incubation followed the method described by Wang H. et al. (2015). Wild-type calli were cultured on B5 plates (3.21 g/L B5 basic medium, 30 g/L sucrose, 2.5 g/L acid-hydrolyzed casein, 0.2 mg/L KT, 0.1 mg/L NAA, and 3.0 g/L plant gel; pH 5.9–6.0) at 25°C in the dark. Transgenic calli were cultured on B5 plates with 5 mg/L hygromycin. Both kinds of calli were subcultured every 20–25 days.
Wild-type A. thaliana (ecotype Columbia) and tobacco (Nicotiana benthamiana) were grown in soil in a greenhouse under a 16 h/8 h light/dark cycle at 23°C.
Detection of Norisoprenoid Volatiles
Sample pretreatment was performed according to the published method (Lan et al., 2016), with some modifications. Briefly, pedicels and seeds were removed from 100 g grape berries; the remainder was powdered and then blended with 1 g polyvinylpolypyrrolidone and 0.5 g D-gluconic acid lactone under liquid nitrogen. After being macerated at 4°C for 4 h, the clear juice was collected through centrifugation at 6,000 × g for 10 min at 4°C. The juice of 5 ml, added with 1 g NaCl and 10 μl internal standard (4-methyl-2-pentanol), was used to detect the free-form volatile compounds using a headspace solid-phase microextraction (HS-SPME) gas chromatography–mass spectrometer (GC-MS). Another 4 ml juice was used to extract glycosidically bound volatile compounds using Cleanert PEP-SPE resin. The extract was dissolved in 10 ml citric acid/sodium citrate buffer (0.2 M, pH 2.5), moved averagely into two 20 ml vials containing 1 g NaCl, and then acid-hydrolyzed to release volatile aglycone (Feng et al., 2015). The vials were tightly capped and incubated for 1 h in a 99°C water bath. After cooling to room temperature, 10 μl of internal standard (4-methyl-2-pentanol) was added to each vial for the detection of volatile aglycones using HS-SPME GC-MS. Two technical replicates of the free-form or bound-form detection were performed per biological replicate. Norisoprenoid compounds were identified and quantified according to the published methods (Xu et al., 2015). The standards β-damascenone, β-ionone, geranylacetone, and MHO were used to establish standard curves in this research. The total concentration (free form + bound form) of each compound was used for further analysis in this study. And the norisoprenoid concentration was expressed as micrograms per berry. β-Damascenone-Z and β-damascenone-E were individually identified and quantified, and their concentrations, named β-damascenone, were added together for k-means analysis and correlation analysis.
Total RNA Extraction, Reverse Transcription, Quantitative Real-Time PCR, and Reverse Transcription PCR
RNA was extracted from different samples using different amounts. For a single RNA extraction, at least 10 frozen grape berries, half a leaf, or two clumps of 1 cm diameter calli were needed. At least 0.3 g of other samples (stems, roots, flowers, etc.) was needed once. All the frozen samples were ground in liquid nitrogen. Total RNA extraction of grape samples used the SpectrumTM Plant Total RNA Kit (Sigma-Aldrich, St. Louis, MO, United States), and the other materials used E.Z.N.A.® Plant RNA Kit (Omega, Norcross, GA, United States). Both extraction processes used on-column DNase I (Promega, Durham, United Kingdom). The quality and concentration of the obtained RNA were detected by agarose gel electrophoresis using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, MA, United States). RNA for further analysis required showing clear and bright bands in the agarose gel; the value of OD260/OD230 was more than 1.8, and the value of OD260/OD280 was between 1.8 and 2.1. First-strand cDNA was synthesized from 1 μg total RNA in a 20 μl reaction mixture following the protocol of HiScript® II Q RT SuperMix for qPCR + gDNA wiper (Vazyme, Nanjing, China).
Quantitative real-time PCR (qRT-PCR) was performed with 2 μl of cDNA as the template using ChamQ Universal SYBR qPCR Master Mix (Vazyme); the Ubiquitin gene was used as the reference. The number of PCRs per gene comprised at least three biological replicates and three technical runs of each replicate (at least nine values). The biological replicate number was noted in the experimental methods. Thermocycling conditions (Wen et al., 2015) and analysis methods (Ruijter et al., 2009) were described previously. The dissolution curve illustrated the specificity of the primers. The size of the amplicon was examined by agarose gel electrophoresis, and its nucleotide sequence was confirmed by sequencing.
Reverse-transcription PCR (RT-PCR) was performed, referencing the published methods with some modification (Chang-ho et al., 2015; Tanabe et al., 2015). 2 × Taq PCR MasterMix (KT201) (Tiangen Biotech, Beijing, China) was used for PCR in which a 25 μl reaction mixture contained 1 μl of cDNA template, 2 μl RT-PCR primers, and 9.5 μl ddH2O. The internal control was AtActin8 (Tanabe et al., 2015). DNA polymerase was first activated at 94°C for 5 min, and PCR was run for 25 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C, followed by a final extension step for 5 min at 72°C. Products were visualized through agarose gel electrophoresis. And ImageJ software (NIH, Bethesda, United States) was used to measure the bright intensity of bands on an agarose gel. The relative brightness of the target gene was normalized by that of AtActin8.
The primer sequences, primer concentrations, and length of the amplicons are listed in Supplementary Table S1.
Cloning, Sequence Analysis, and Activity Assay of the VvCCD4b Promoter
The region ∼1.0 kb at the upstream of start codon ATG was regarded as the VvCCD4b promoter (PCCD4b), based on the genomic sequence of “Pinotage1” Genomic DNA from “Cabernet Sauvignon” grape berries was obtained using a plant genome extraction kit (Bioteke, Beijing, China). The fragment of PCCD4b was amplified using a gene-specific primer pair, promoter-F/R, designed by Primer Premier 5.0 (Premier Biosoft, United States), and then inserted into a T-vector (Tsingke, Beijing, China) for sequencing. The cis-acting elements on PCCD4b were predicted using PLACE2 and PlantCARE3. The transcription initiation site was identified using TSSP4.
To verify the promoter activity, we introduced PCCD4b into a modified pCAMBIA 1300-LUC vector carrying a luciferin (LUC) reporter gene (Shang et al., 2010) using homologous recombination (Clontech, Mountain View, CA, United States) with the KpnI and SmaI restriction sites. The recombinant vector was transferred into Agrobacterium tumefaciens strain GV3101 using the freeze–thaw method (Hofgen and Willmitzer, 1988). Detailed methods on tobacco (N. benthamiana) infection and LUC detection were described by Sun et al. (2015).
Construction of Transgenic A. thaliana
A pCAMBIA 1381-PCCD4b-β-glucuronidase (GUS) plasmid was constructed by inserting PCCD4b using homologs recombination with SmaI and SalI into pCAMBIA 1381 (CambiaLabs) carrying the GUS gene. The construct was transferred into GV3101 and then transformed into A. thaliana using the floral-dip method (Clough and Bent, 1998). Transgenic Arabidopsis plants were selected from Murashige–Skoog (MS) medium plates (4.43 g/L MS, 30 g/L sucrose, and 8 g/L agar; pH 5.9–6.0) with 50 mg/L hygromycin, cultured in the greenhouse with a 16-h/8-h light/dark cycle at 23°C for 2 weeks. Then the healthy seedlings were transplanted into soil, also cultured in the same greenhouse.
Abiotic Treatments of Transgenic A. thaliana, Grapevine Calli, and Grape Berries
Four-week-old T3-generation transgenic A. thaliana individuals (from Construction of Transgenic A. thaliana) were divided into four groups and placed in climate chambers for 16 h under the following conditions: 25°C (control), 37°C, 30°C, or 10°C. A different line of transgenic 4-week-old seedlings was subjected to illumination treatments [4,240 (control), 6,630, 1,315, and 0 lx] in a climate chamber for 16 h. Three biological replicates were maintained. Leaves were sampled immediately after the treatments to detect GUS expression through RT-PCR. Histochemical staining was also performed to determine GUS activity in plants as described previously (Cho and Cosgrove, 2000).
“Cabernet Sauvignon” grape calli were grown on B5 medium plates containing different ABA concentrations (0, 0.4, 0.8, 1.0, and 2.0 mg/L) for 25 days and then sampled for RNA extraction. Five clumps of callus were placed on one plate, and one concentration treatment used three plates, serving as three biological replicates.
ABA spraying treatment was also performed on Chateau SunGod Great Wall plants in 2019. The three center rows of a “Cabernet Sauvignon” vineyard, containing an average of 30 plants per row in a north–south orientation, were selected. Six vines were chosen randomly and divided into two groups in each row. One group was sprayed with 1 g/L ABA and 0.05% Tween 20 when 5% of the berries reached veraison (began coloration). Another group, as the control, was sprayed with 0.05% Tween 20 only. The sampling was carried out every 2 days until 10 days post-spraying. Approximately 100 berries were collected for each biological replicate per sampling and used for RNA extraction.
Yeast One-Hybrid Assay
A yeast one-hybrid (Y1H) assay was performed by using the Matchmaker Gold Yeast One-Hybrid Library Screening System (Clontech). The short fragment of the VvCCD4b promoter (205–893 bp upstream of ATG of VvCCD4b) was inserted into pAbAi to construct the bait vector pAbAi-PVvCCD4b. The bait vector was linearized and transformed into S. cerevisiae Y1HGold to create bait strain. A cDNA library of “Cabernet Sauvignon” was constructed by Clontech. The cDNA library was transformed into the bait strain and screened using synthetic dropout medium (SD) lacking leucine (–Leu) with aureobasidin A (AbA). The prey fragments from the positive colonies were identified by DNA sequencing (Sangon) using the primer pair of pGADT7-F/R and blasted by NCBI.
Dual-Luciferase Activity Assay in a Transient Expression System
Dual-luciferase activity was assayed in a tobacco transient expression system. PCCD4b was subcloned into a pGreen II 0800 double-reporter vector, and the full-length coding sequence (CDS) of transcription factor genes were subcloned into a pCAMBIA 1301 vector as effectors. Recombinant reporter vector and individual effector vector were transferred into A. tumefaciens EHA105. The two A. tumefaciens strains were mixed at a 1:1 (v:v) ratio and injected into tobacco leaves (Voinnet et al., 2003). Dual-luciferase activity was measured using a Dual-Luciferase® Reporter Assay System (Promega). Six biological replicates were used in one independent experiment. An independent experiment of each transcription factor was repeated for two to four times.
Subcellular Localization of VvMADS4
The CDS of VvMADS4 (NM_001281185.1) without the stop codon was amplified, sequenced, and inserted into pEZS-NL to express a green fluorescent protein (GFP) fusion protein. The recombinant plasmid was transformed into onion epidermal cells using a gene gun (Bio-Rad, Hercules, CA, United States). Cells were incubated in the dark at 25°C for 16 h and then co-incubated with 50 mg/L 4’,6-diamidino-2-phenylindole (DAPI) for 20 min as a nucleus localization marker. The samples stained with DAPI were observed under a confocal microscope (Nikon A1, Tokyo, Japan). An empty vector was not used as a control because it could not properly express GFP5.
VvMADS4 Overexpression in Grape Calli and Leaves
To verify the role of VvMADS4 in controlling VvCCD4b expression, we overexpressed VvMADS4 in grapevine calli. After inserting VvMADS4 CDS into pCXSN, the recombinant plasmid was transformed into GV3101. Calli transformation was performed using an A. tumefaciens-mediated method (Van Eck et al., 2006) with some modifications. Each callus was submerged in a medium containing A. tumefaciens strains and gently shaken for 6 min. After the medium was dried off, the calli were cultivated on a plate containing a sterile filter paper and T1 medium (B5 medium with 100 μmol/L acetosyringone and 100 mg/L DL-dithiothreitol; pH 5.9–6.0). Cultivation occurred in the dark for 3 days. The calli were subsequently washed using sterile water and sterile water with 400 mg/L cefalexin and 400 mg/L carboxymycin in turn for three times. The calli were then dried and cultivated in the dark on a plate containing T2 medium (B5 medium with 100 mg/L DL-dithiothreitol, 400 mg/L cefalexin, 400 mg/L carboxymycin, and 5 mg/L hygromycin). Concentrations of cefalexin and carboxymycin in the solid medium were gradually decreased until new calli appeared. Transgenic calli were identified by hygromycin gene detection and target gene expression quantitation.
Transient VvMADS4 overexpression using Vitis quinquangularis Rehd leaves was also performed to further verify the function of VvMADS4 on the regulation of VvCCD4b expression. V. quinquangularis Rehd leaves contain little wax and are easily infiltrated by bacterial solution. The expression construct pCAMBIA 1301-VvMADS4 was transferred into GV3101, and empty pCAMBIA 1301 was used as the control. The transformation method followed the procedure described by Xu et al. (2010), with some modifications. A single colony of the GV3101 strain containing the plasmid was inoculated into 5 ml LB liquid medium (50 mg/L rifampicin and 50 mg/L kanamycin) and cultured at 28°C and 220 rpm for 20 h. Then, 300 μl bacterial liquid was added to 30 ml LB liquid medium (50 mg/L rifampicin and 50 mg/L kanamycin) and cultured at 28°C and 220 rpm until OD600 reached 0.5–0.6. Subsequently, the culture was centrifuged at 4°C and 4,000 × g for 10 min and then resuspended with induction buffer (2.132 g/L MES, 2.033 g/L MgCl2⋅6H2O, 5 g/L sucrose, and 0.039 g/L acetosyringone; pH 5.9–6.0) until OD600 reached 0.3–0.4. After the bacterial suspension was inoculated at 25°C for 3 h, the leaves were submerged into the suspension and subjected to vacuum (−0.8 MPa) for 20 min to induce infiltration. After 3 days of incubation at 25°C in the dark, the leaves were used for gene expression analysis. This experiment was repeated twice, each time with three biological replicates at least and one leaf per replicate.
Statistical Analysis
Data are presented as means ± SD (standard deviation). Pearson’s correlation and one-way analysis of variance (ANOVA) were conducted using SPSS for Windows version 20.0 (SPSS Inc., United States). String diagrams, bar charts, and box plots were created in OriginPro 9.0 (OriginLab, Northampton, MA, United States). k-means and heatmaps were generated using k-mean in the “base” and “pheatmap” packages in R software. Pearson’s correlation analysis of VvCCD4b expression and norisoprenoid accumulation was performed after data normalization. The concentration of individual norisoprenoid compound was normalized by dividing the concentration by the maximum value of all samples (over 2 years from both regions) to enable the compounds to be compared, regardless of a wide range of concentrations. The relative expression of VvCCDs was also normalized using the same method. Pearson’s correlation analysis was performed using the normalized relative expression of VvCCDs and the normalized concentration of norisoprenoids during the entire development stage over 2 years from both regions (Xu et al., 2015). Pearson’s correlation was also estimated between the expression of VvCCD4b and candidate transcription factor genes identified by Y1H. The gene expression data were from the RNA-seq dataset6. The transcript abundance of each gene was calculated using the FPKM (fragments per kilobase per million fragments mapped) method. The information about the experiment protocol, data analysis of RNA-seq, and submission to the NCBI Gene Expression Omnibus was recorded in the publication of our research group (Sun et al., 2019). In this study, we selected the data of the control group at the E-L29, E-L31, E-L35, E-L36, E-L37, and E-L38 stages of berry development for Pearson’s correlation analysis.
Results
Correlation Between VvCCDs Expression and Norisoprenoid Content
We quantified 15 free-form and 6 glycosidically bound-form norisoprenoid compounds (Supplementary Table S2). The total concentration (free form plus bound form) of individual norisoprenoid compound is shown in Supplementary Table S3. The k-means analysis divided the norisoprenoids into three clusters based on the total concentration changes during berry development across 2 years (for individual compounds per cluster, see Figure 1C). The compounds in the first cluster accumulated continuously as the grapes matured. The compound concentration in the second cluster peaked between E-L35 and E-L36 before decreasing gradually. Finally, the third cluster maintained a steady concentration, except for the GT grapes in 2013 (Figure 1A). Correspondingly, the VvCCD expression patterns differed across development (Figure 1B). VvCCD1 expression was higher during early development than during maturation. VvCCD4a expression was very low before maturation but increased sharply from E-L36 to E-L37 until ripening. VvCCD4b transcription level in ripening grapes was higher than that in green berries. Pearson’s correlation analysis revealed that the accumulation of 2,2,6-trimethylcyclohexanone (TCH) and β-cyclocitral was positively correlated only with VvCCD1 expression. The accumulation of dihydroedulan I, geranylacetone, and β-damascenone, in cluster 1, highly paralleled with VvCCD4b expression but opposed to VvCCD1 expression. MHO and β-ionone, in cluster 3, were positively correlated with VvCCD4a and VvCCD4b expression (Figure 1C). Notably, VvCCD4b expression was significantly correlated with the total concentration of norisoprenoids, indicating that VvCCD4b is a key enzyme in norisoprenoid production during grape berry development.
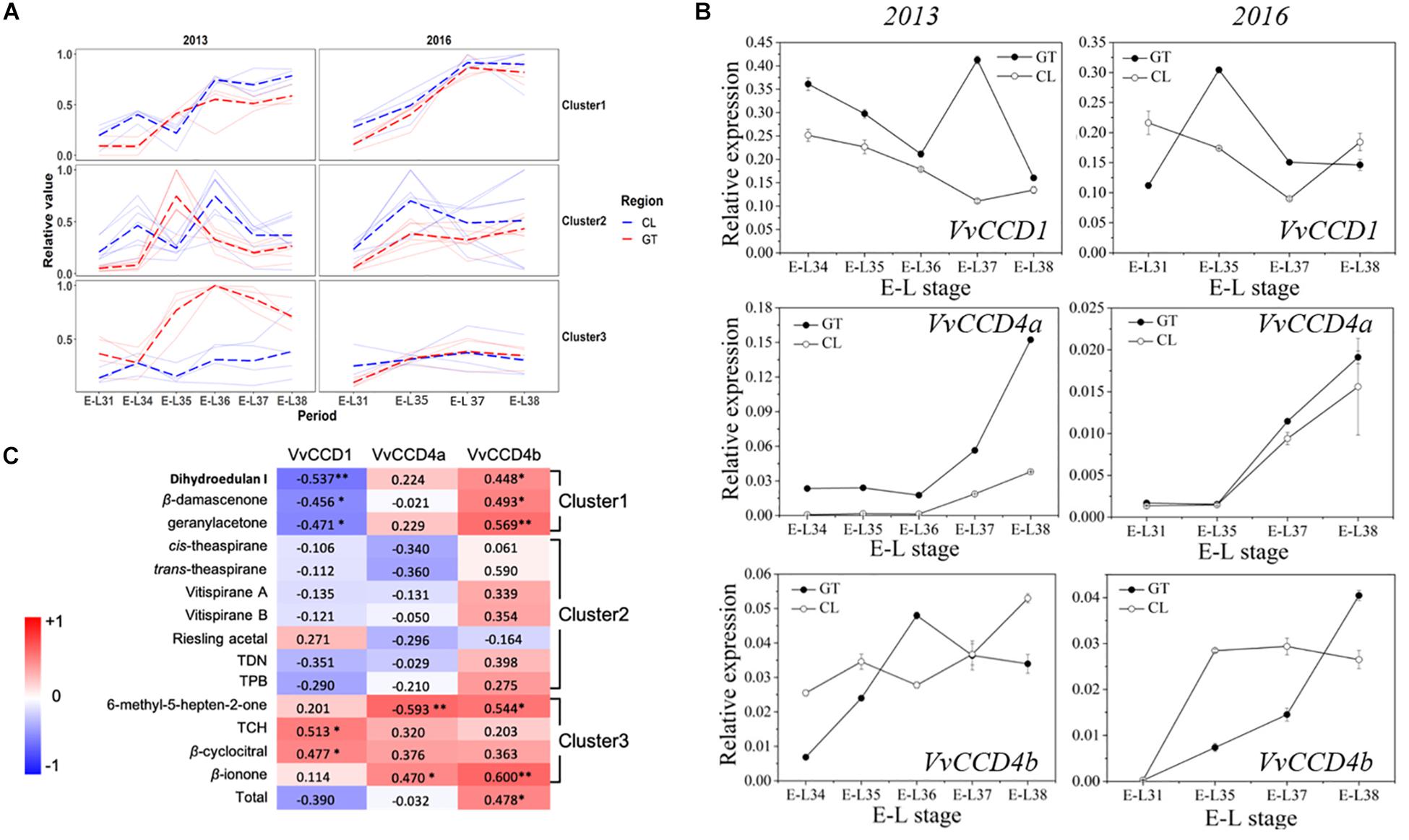
Figure 1. The concentrations of norisoprenoids and the expression levels of VvCCDs in grape berries from two regions in 2 years. (A) The analysis of k-means clustering for 2013 and 2016 vintages. (B) The expression of VvCCDs in “Cabernet Sauvignon” grape berries from two regions in 2 years. The data are expressed as means ± SD of three replications. (C) Heatmap of Pearson’s correlation results. The correlation is assessed based on the normalized concentration of individual norisoprenoid volatile and the normalized expression level of VvCCDs during the development period. The data on the heatmap are correlation coefficients. ∗∗Significant correlation at P < 0.01 (two-sided test). ∗Significant correlation at P < 0.05 (two-sided test). The red block indicates positive correlation, and the blue block indicates negative correlation. The clusters were corresponding to the results of the k-means analysis. TDN: 1,1,5–trimethyl–1,2–dihydronaphthalene; TCH: 2,2,6-trimethylcyclohexanone; TPB: (E)-1-(2,3,6-trimethylphenyl) buta-1,3-diene.
Temporal and Spatial VvCCD4b Expression Patterns
The temporal and spatial expression pattern of VvCCD4b was examined by qRT-PCR. VvCCD4b showed low expression levels in the root, stem, and tendril and high expression levels in the flower, mature leaf, and mature fruit (Figure 2). The expression of VvCCD4b increased with the age of leaves and the maturation of grape berries. Combining the results of Figures 1B, 2, we found that there is an upregulation of VvCCD4b expression at the E-L35 stage.
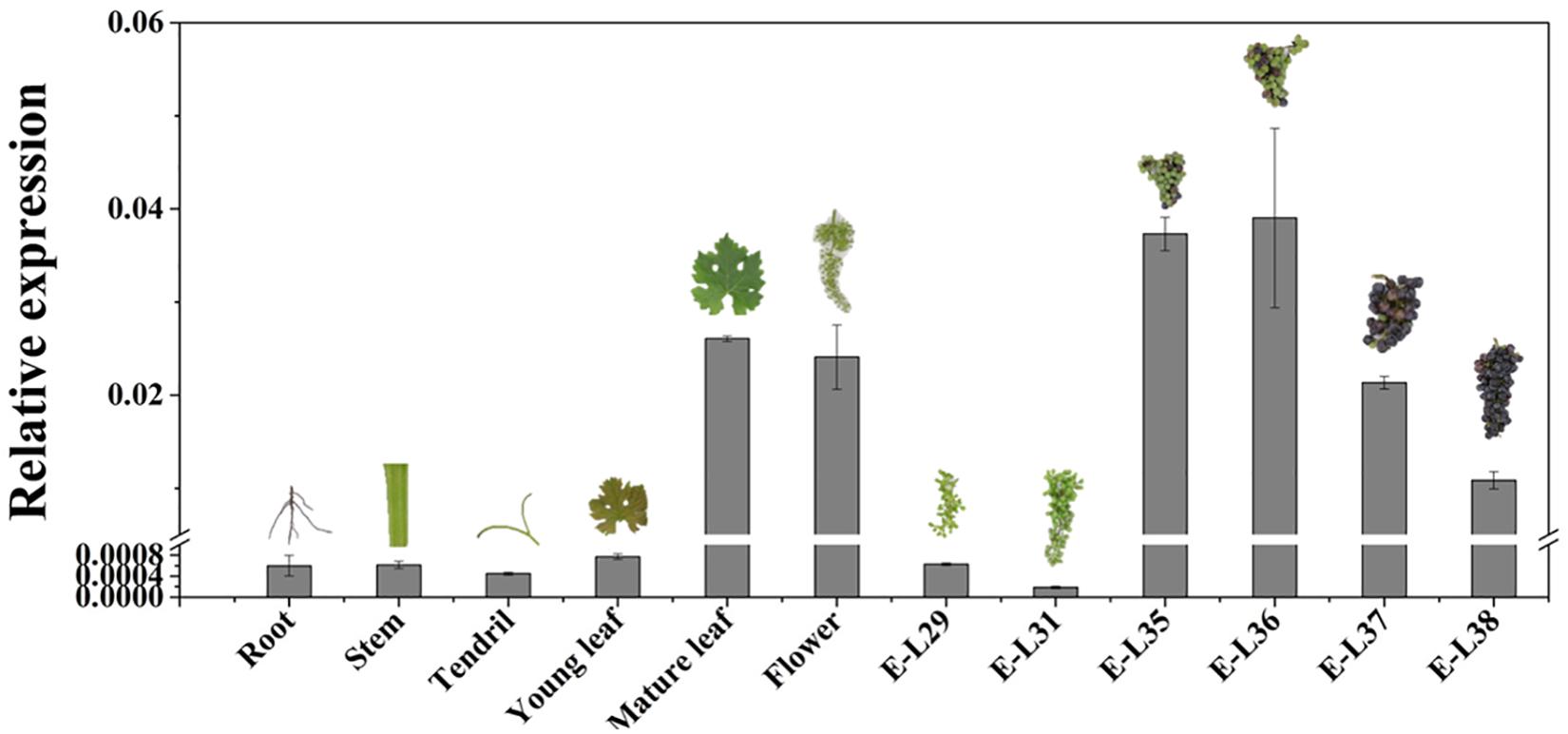
Figure 2. Temporal and spatial expression of VvCCD4b in various organs of “Cabernet Sauvignon” grapevine. The data are expressed as means ± SD of three replications.
Response of VvCCD4b Promoter to Light and Temperature
To research the cis-acting elements on the VvCCD4b promoter and its responses to stresses, we cloned a 1,057-bp VvCCD4b promoter from the DNA of “Cabernet Sauvignon.” The promoter shared 99% similarity with the sequence from “Pinot noir” (Supplementary Figure S1). The transcription initiation site, located 70 bp upstream of the start codon (ATG), was set as position + 1 (Figure 3A). A putative TATA-box (TCATTATAAAA) and a CAAT-box (CAAAT) that are necessary for transcription were found at positions −23 to −33 and −93 to −97, respectively. We also found several putative environmental stress-responsive or hormone-responsive cis-acting regulatory elements in the VvCCD4b promoter. Box 4, GT1 motif, I-box, and G-box are involved in light responsiveness. Ethylene-responsive element (ERE), TCA element, and ABA-responsive element (ABRE) are associated with responses to ethylene, salicylic acid, and ABA, respectively. W-box, MYC binding site, CCA1 binding site, box L-like, and CArG motif are all important binding sites for transcription factors (Supplementary Table S4).
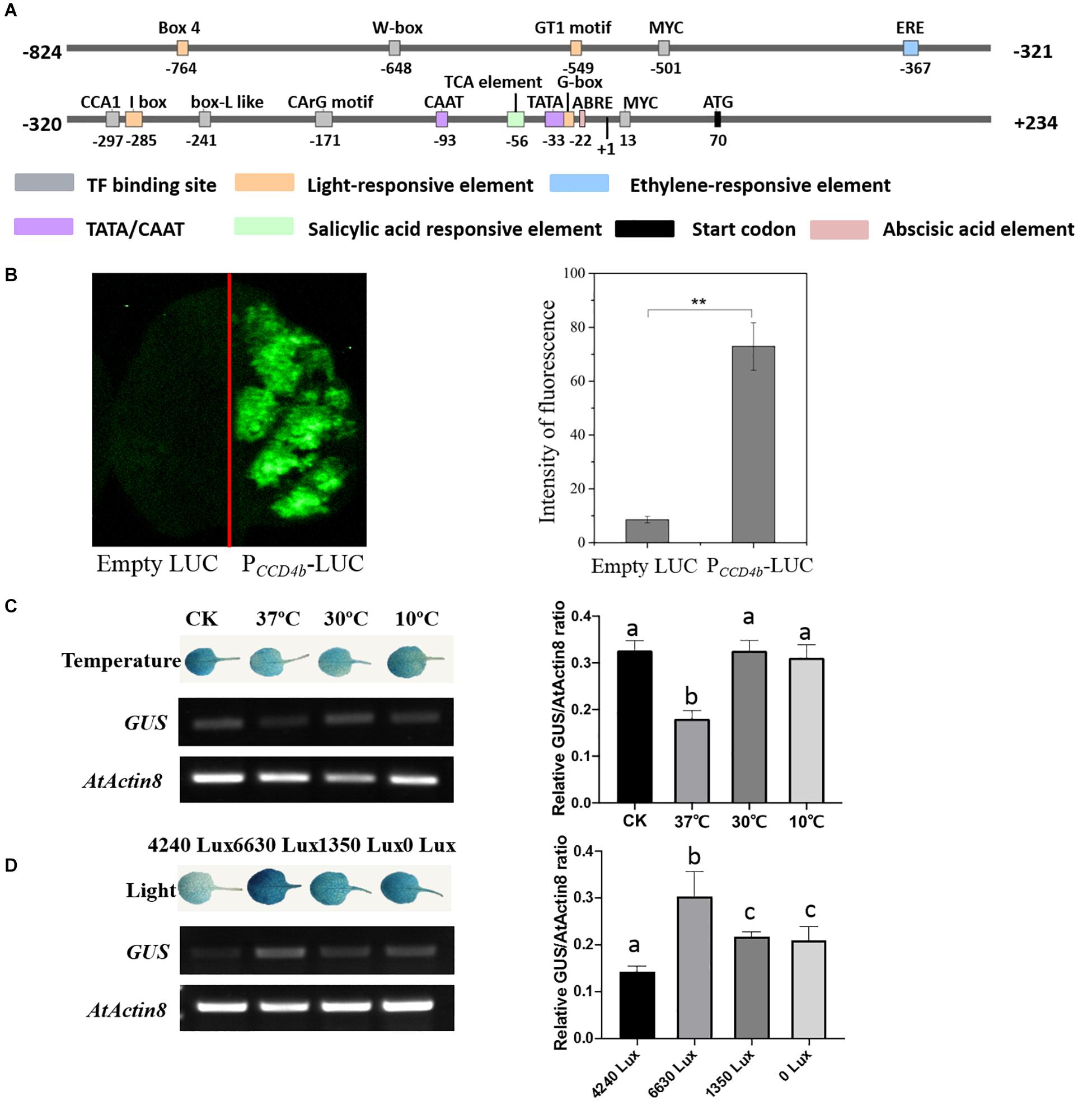
Figure 3. Analysis of cis-acting elements and examination of VvCCD4b promoter activity. (A) The map of the VvCCD4b promoter region deduced by the PlantCARE and PLACE programs. (B) Transient expression of the LUC gene driven by the VvCCD4b promoter. Tobacco leaves were transformed with equal amounts of empty LUC plasmid (left side) and PCCD4b-LUC (right side). Left panel: fluorescence imaging. Right panel: intensity of fluorescence analyzed by ImageJ. Asterisks indicate significant differences relative to the control by one-way ANOVA test (**P < 0.01). (C,D) Effects of temperatures or illuminations on the expression of the GUS gene driven by the VvCCD4b promoter in the transgenic Arabidopsis leaves. AtActin8 was used as a control for the RT-PCR. Amplified products were electrophoresed on 1.5% (w/v) agarose gels. The relative GUS/AtActin8 ratio is quantified by ImageJ software according to the brightness of GUS and AtActin8 bands in agarose gels. Data are expressed as means ± SD of three biological replicates. Lowercase letters indicate significant differences analyzed by one-way ANOVA tests. The original figures of the agarose gels in (C,D) were shown in Supplementary Figures S2A,B, respectively. GUS staining patterns and agarose gel results of two additional biological replicates are shown in Supplementary Figures S2C,D, respectively.
Then, PCCD4b-LUC was transformed into tobacco leaves to identify the activity of the VvCCD4b promoter. The transformation of tobacco leaves with a construct containing VvCCD4b-promoter-driven LUC yielded strong fluorescence (Figure 3B), whereas fluorescence was absent under control conditions (empty vector). These findings indicate that the VvCCD4b promoter has activity to drive LUC expression.
To test the responses of the VvCCD4b promoter to temperature and light treatment, we subcloned PCCD4b into the pCAMBIA 1381 vector to drive the GUS gene and then transformed it into A. thaliana. The expression levels of GUS and AtActin8 in transgenic Arabidopsis were assessed by RT-PCR, and the relative GUS/AtActin8 ratio was analyzed using the brightness of bands on the agarose gel. The result revealed that GUS transcript abundance was lower at 37°C than that at 25°C (the control group), whereas the treatments at 10 and 30°C had no significant effect on the GUS expression (Figure 3C). This indicated that the activity of the VvCCD4b promoter was repressed by extremely high temperatures. Additionally, strong illumination, weak illumination, and dark treatment all promoted GUS transcript accumulation, especially strong light stimulation (Figure 3D). The investigation illustrated that the VvCCD4b promoter could respond to illumination changes.
VvCCD4b Response to ABA Treatments in Calli and Grapes
Given that an upregulation of VvCCD4b expression was observed at E-L35 (the onset of berry ripening) (Figure 1B) and that an ABRE element was present in the VvCCD4b promoter, we hypothesized that the grape ripening hormone ABA may be involved in the induction of VvCCD4b expression. To test this hypothesis, we treated the grape calli with ABA of different concentrations for 25 days. The results verified that ABA upregulated VvCCD4b expression, with the latter increasing with increasing ABA (Figure 4A).
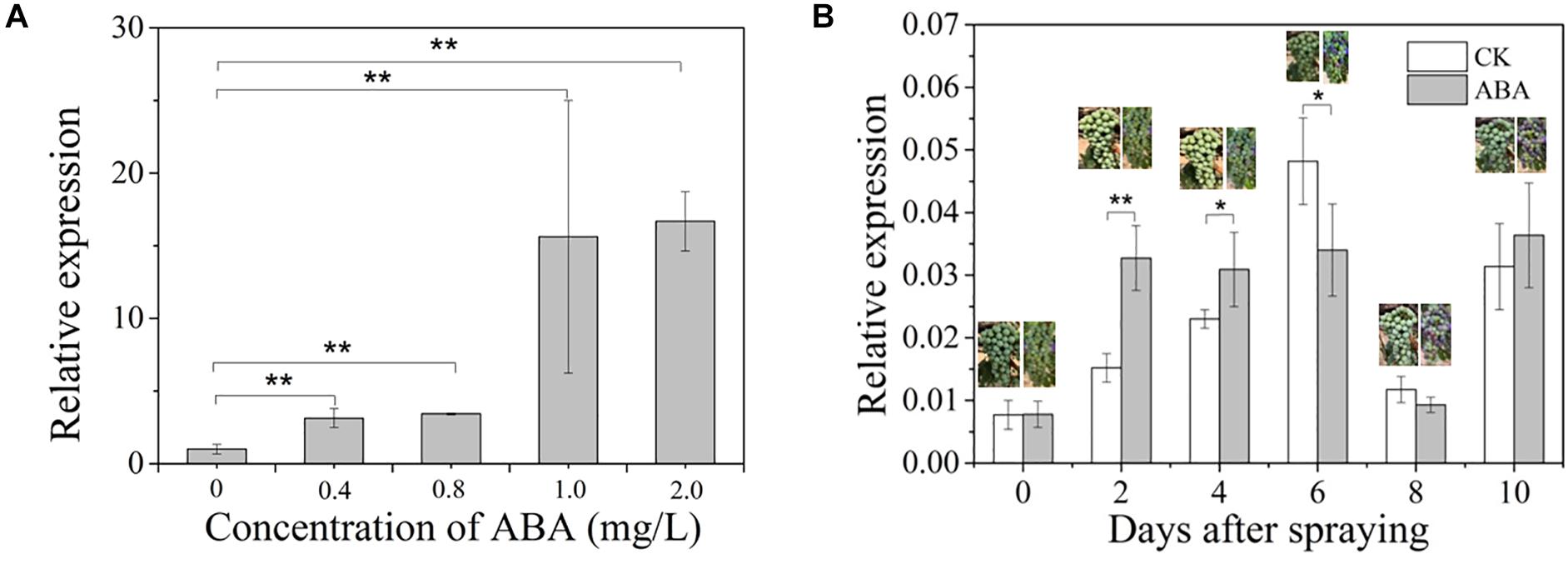
Figure 4. Effects of exogenous ABA treatment on VvCCD4b expression. (A) The relative expression of VvCCD4b in calli treated by ABA with different concentrations. (B) The relative expression of VvCCD4b in grape berries after being sprayed by ABA or Tween 20 solution. The data are expressed as means ± SD of three replications in both figures. Asterisks indicate significant differences relative to the control by one-way ANOVA test (*P < 0.05; **P < 0.01).
Afterward, we sprayed ABA solution on grape berries at the onset of ripening (5% berry coloration) to test if ABA had a similar effect on VvCCD4b expression in developing grape berries. The result showed that immediately after ABA spraying, the coloration of berries was accelerated and that VvCCD4b expression was upregulated compared to that in the control; however, this effect disappeared gradually. VvCCD4b expression amount in the treated group was about 2-fold and 1.34-fold greater than that in the control at 2 and 4 days after ABA spraying, respectively (Figure 4B). On Day 6 post-spraying, the expression level of VvCCD4b in the treated group was lower than that in the control. Subsequently, VvCCD4b transcript abundance ceased to differ between the ABA-treated group and the control. Both experiments of grape calli and berries suggested that VvCCD4b expression is induced by ABA, explaining the upregulation of VvCCD4b expression at E-L35.
Identification of Transcription Factors Regulating VvCCD4b Expression
A Y1H screening was performed to search candidate transcription factors possibly regulating VvCCD4b expression. The short fragment of the VvCCD4b promoter was used as a bait to screen the cDNA library of “Cabernet Sauvignon.” Nine candidate transcription factors were captured (Supplementary Table S5). Except for VvPCL1, which failed to be cloned successfully, the CDS of the other transcription factor genes was all obtained and subcloned into the pCAMBIA 1301 vector. Following that, a dual-luciferase reporter assay in a tobacco leaf transient expression system was carried out to test their influences on the activity of the VvCCD4b promoter (Figure 5 and Supplementary Figure S3). At least two independent experiments all demonstrated that VvbZIP53, VvMYB4, VvWRKY40-like, VvGATA24, and VvbHLH47 could not activate the VvCCD4b promoter, whereas VvMYBCS1 and VvMYB1R1 could suppress the activity of the VvCCD4b promoter only once among three independent experiments. Only VvMADS4 always showed a consistent effect on the activity of the VvCCD4b promoter in the four independent experiments with six biological replicates per experiment. Compared with the control group (pGreen-PCCD4b + pCAMBIA 1301), the relative LUC/REN ratio was significantly lower when the LUC-containing construction was co-expressed with pCAMBIA 1301-VvMADS4, indicating that VvMADS4 decreased VvCCD4b promoter activity (Figure 5). The co-expression analysis between VvCCD4b and the above nine transcription factors was conducted using RNA-seq data across the whole development period of grape berry7. The VvMYBCS1, VvMYB1R1, VvWRKY40-like, VvbZIP53, and VvMYB4 were all negatively correlated with the VvCCD4b expression, and Pearson’s coefficient values were −0.66, −0.78, −0.68, −0.76, and −0.54, respectively. VvMADS4, VvbHLH47, VvGATA24, and VvPCL1 were positively correlated with VvCCD4b expression, and Pearson’s coefficient values were 0.63, 0.20, 0.41, and 0.58, respectively. VvMADS4 co-expressed with VvCCD4b with the highest coefficient. Moreover, the VvCCD4b promoter included a CArG box cis-acting element, required by MADS binding (Figure 3A). Therefore, the biological functions of VvMADS4 were further researched.
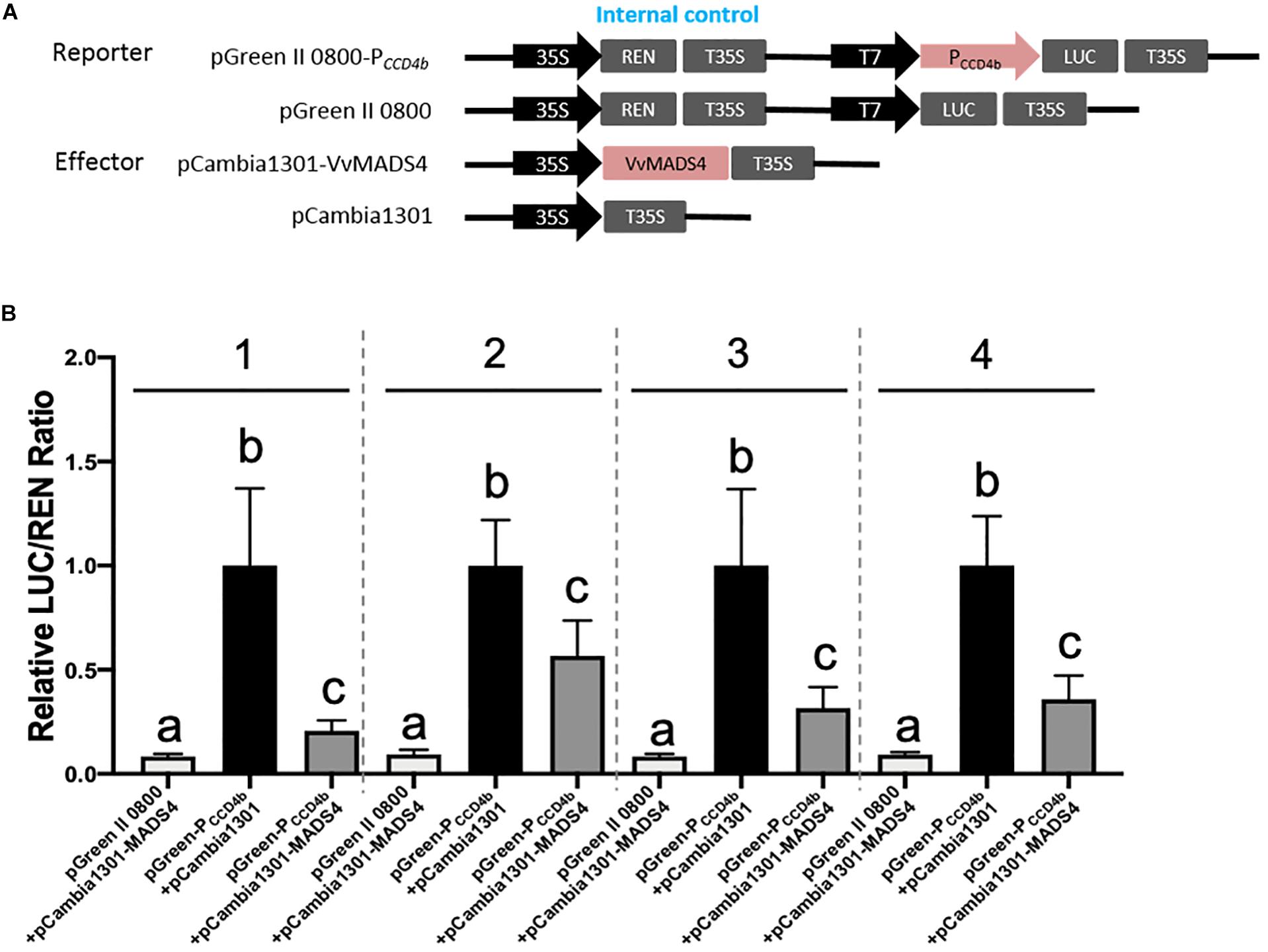
Figure 5. Regulation of VvMADS4 on the activity of the VvCCD4b promoter. (A) Schematic diagrams of vectors used for the dual-luciferase assay. The pGreen II 0800, empty, pGreen-PCCD4b, and reporter vectors contained the VvCCD4b promoter fused to LUC. The pCAMBIA 1301, empty, pCAMBIA 1301-VvMADS4, and overexpression vectors contained VvMADS4. (B) The dual-luciferase assay was performed using a tobacco transient expression system. pGreen II 0800 + pCAMBIA 1301-VvMADS4 and pGreen-PCCD4b + pCAMBIA 1301 are controls. Four independent experiments were performed, and the numbers above the bar chart indicate the number of independent experiments. Each independent experiment had six biological replicates. The data are expressed as the means ± SD from six biological replicates. Lowercase letters indicate significant differences among the controls and the experimental group by one-way ANOVA test in each independent experiment (P < 0.05).
The temporal and spatial expression assays showed that VvMADS4 was mainly expressed in grape flowers and berries (Figure 6A). VvMADS4 expression increased as the grape berries developed until E-L36, after which point, it began decreasing.
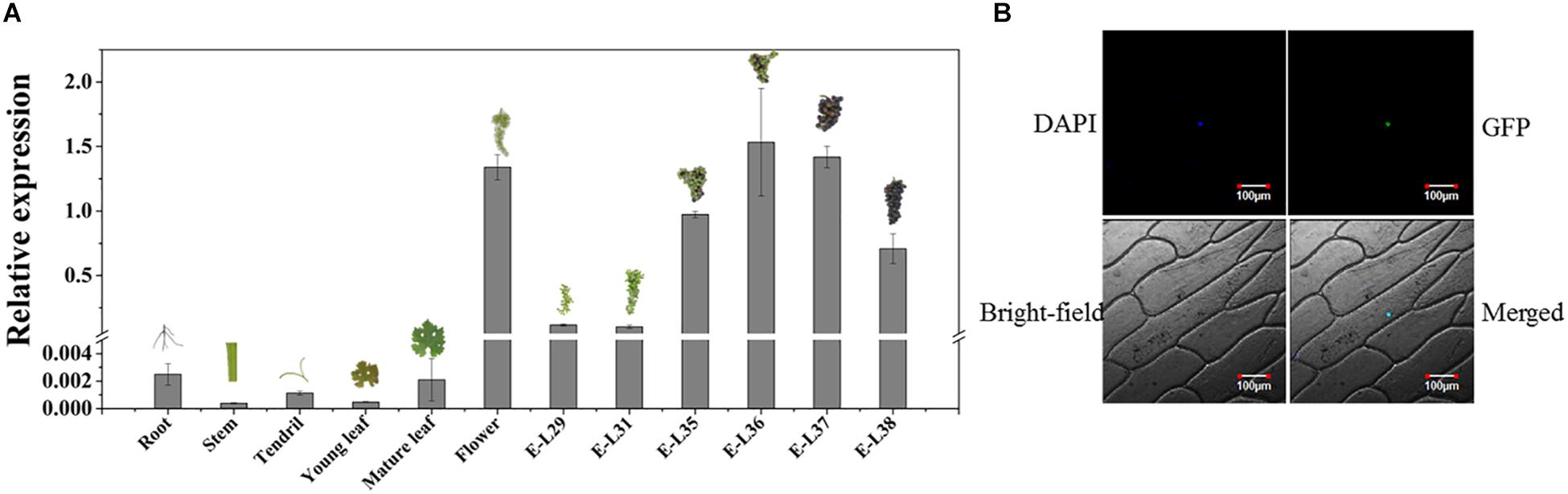
Figure 6. Characterization of VvMADS4. (A) Temporal and spatial expression patterns of VvMADS4. The data are expressed as means ± SD of three replications. (B) Subcellular localization of VvMADS4. DAPI is a nuclear-localized marker.
Subcellular localization of VvMADS4 in onion epidermal cells revealed that VvMADS-GFP was only present in the nucleus, confirming its role as a transcription factor (Figure 6B).
To elucidate the regulatory effect of VvMADS4 on VvCCD4b transcription in the homologous system, we overexpressed VvMADS4 stably in grape calli and transiently in V. quinquangularis leaves. It was observed that transgenic calli expressed the Hygromycin gene, whereas wild-type calli did not, demonstrating that the CaMV 35S promoter-driven VvMADS4 gene (35:VvMADS4) was successfully transformed into the calli (Figure 7A). In comparison to that in the wild-type calli, overexpression of VvMADS4 resulted in the reduction of VvCCD4b and VvCCD4a expression amounts (Figure 7B). Additionally, VvMADS4 was transiently overexpressed in V. quinquangularis leaves, and an empty vector was also transformed as the control. We observed significant downregulation of VvCCD4b in VvMADS4-overexpressed leaves (Figure 7C). Both of the results suggest that VvMADS4 overexpression negatively regulates VvCCD4b transcription.
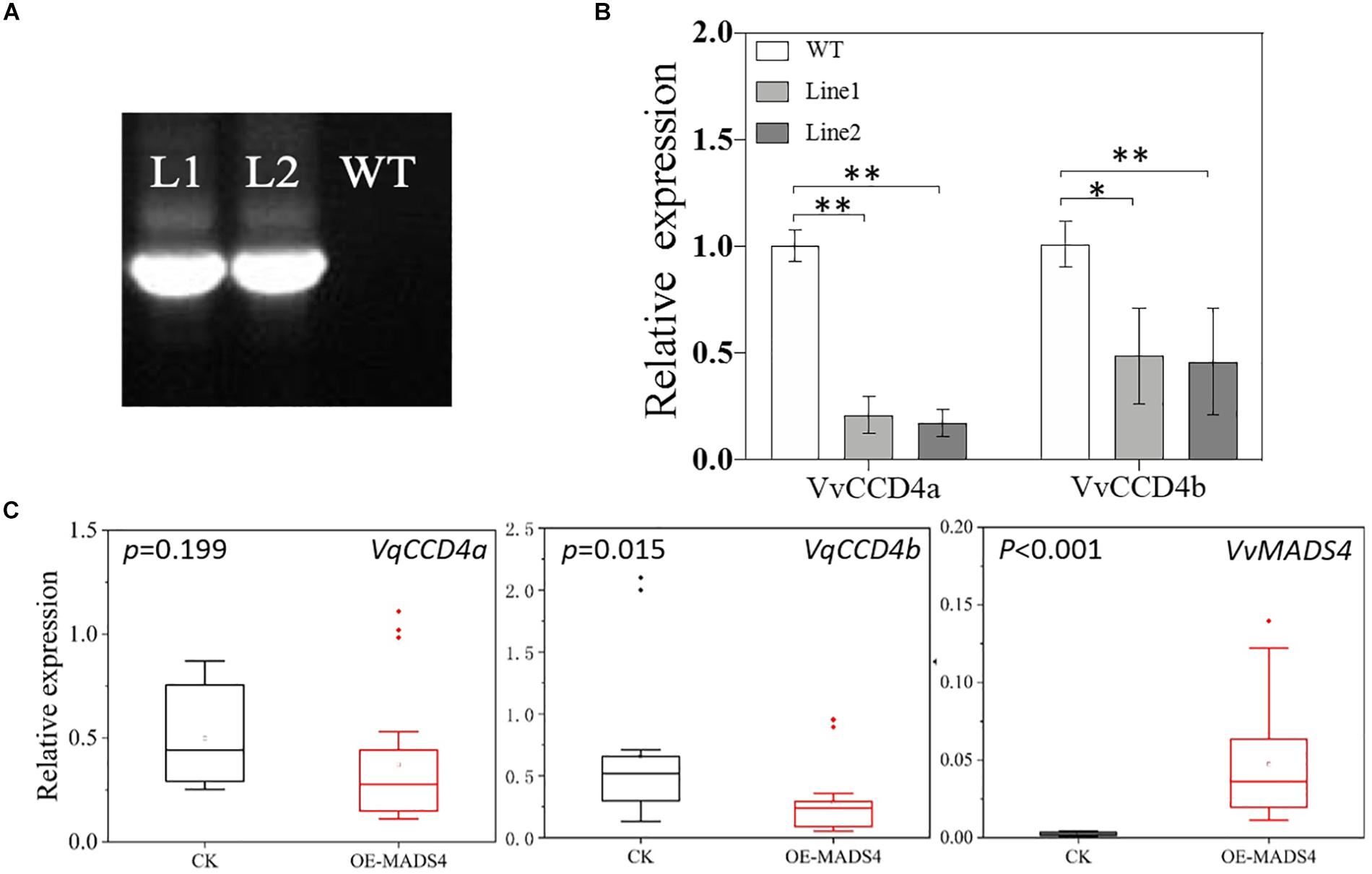
Figure 7. Transcriptional regulation of VvMADS4 on the VvCCD4b gene. (A) Hygromycin gene expression detected by PCR and analyzed by gel electrophoresis. The original figure is shown in Supplementary Figure S4. (B) Relative expression levels of VvCCD4s in transgenic grape calli carrying the VvMADS4 sequence. The name of the pertinent gene is indicated on each panel. Data are expressed as means ± SD of three replicates. Asterisks indicate significant differences relative to the control by one-way ANOVA test (∗P < 0.05; ∗∗P < 0.01). (C) Relative expression levels of VqCCD4s in VvMADS4 transiently overexpressed V. quinquangularis leaves. At least seven biological replicates were carried out, and a one-way ANOVA test was performed (∗P < 0.05; ∗∗P < 0.01).
Discussion
Importance of VvCCD4b in Grape Berries
CCDs are instrumental in norisoprenoid production because they cleave double bonds at specific sites in carotenoid molecules; in vitro and in vivo recombinant enzymatic experiments have yielded strong evidence for this (Rubio et al., 2008; Huang et al., 2009; Ma et al., 2013; Rodrigo et al., 2013; Rubio-Moraga et al., 2014; Bruno et al., 2015), as have in planta experiments (Campbell et al., 2010; Lashbrooke et al., 2013; Zhang et al., 2015). Overexpression experiments in carotenoid-accumulating E. coli or S. cerevisiae verified that VvCCD1 cleaves zeaxanthin, lycopene, ε-carotene, and β-carotene to yield 3-hydroxy-β-ionone, MHO, geranylacetone, β-ionone, and β-cyclocitral, respectively (Mathieu et al., 2005; Lashbrooke et al., 2013; Meng et al., 2019). Another study noted that overexpressing or silencing VvCCD1 in transgenic grapevine did not influence leaf norisoprenoid levels (Lashbrooke et al., 2013). However, overexpressing VvCCD4a and VvCCD4b in carotenoid-accumulating E. coli revealed that VvCCD4a and VvCCD4b could cleave neurosporene, lycopene, and ε-carotene to generate geranylacetone, MHO, and α-ionone, respectively (Lashbrooke et al., 2013). Overexpressing VvCCD4b in β-carotene-accumulating S. cerevisiae resulted in the formation of β-ionone and β-cyclocitral (Meng et al., 2019). However, there is no in planta evidence regarding the function of VvCCD4a and VvCCD4b.
In this study, we demonstrated that VvCCD4b expression is significantly correlated with the accumulation of norisoprenoids, particularly MHO, β-ionone, and geranylacetone. These three compounds are all direct products of VvCCD4b interacting with carotenoids (Lashbrooke et al., 2013; Meng et al., 2019). Together, our findings and previous data strongly indicate that VvCCD4b is a critical enzyme affecting norisoprenoid production in grape berries.
Development-Dependent VvCCD4b Expression
Although CCDs in different species are homologous, their expression patterns are species specific. In Malus domestica, MdCCD4b is mainly expressed in flowers but not in fruits or buds (Chen H. et al., 2018). CCD4b of Solanum lycopersicum is expressed in all tissues but is the most prominent in mature leaves and the least prominent in fruit (Wei et al., 2015). In C. sinensis, CsCCD4b1 is expressed only in petals and fruit peel. In the latter, CsCCD4b1 expression is the lowest at the green stage, peaks at the late breaker stage, and decreases until ripening (Rodrigo et al., 2013). In potato, CCD4 is primarily expressed in leaves and flowers, with much lower expression in stems, tubers, and roots (Campbell et al., 2010). CCD4 of Rosa × damascena is predominantly expressed in flowers; it exhibits very low expression in leaves, stems, and roots (Huang et al., 2009).
Here, our study reveals that VvCCD4b is abundantly expressed in mature leaves, flowers, and ripening berries, similar to what was observed in “Pinotage” (Lashbrooke et al., 2013). Notably, VvCCD4b expression was induced at the beginning of the coloration stage (E-L35) compared to the earlier stages. The timing of the expression, combined with our results from the ABA spraying of grapes, indicates that ABA upregulates VvCCD4b expression. The present findings corroborate the previous report in apples where the researchers observe that ABA induces the expression of MdCCD4c, MdCCD7b, and MdCCD8a (Chen H. et al., 2018). Likewise, ABA also increases CCD4 expression in soybean (Wang et al., 2011). Combining these above observations, we suggest that the VvCCD4b expression promoted by ABA might be related to the ABRE element on the VvCCD4b promoter.
CCDs have distinct expression patterns and divergent functions. The cis-acting elements are not well conserved among CCD promoters in numerous plants, such as A. thaliana, Brassica rapa, Crocus sativus, Medicago truncatula, Oryza sativa, Populus trichocarpa, Sorghum bicolor, S. lycopersicum, and V. vinifera (Ahrazem et al., 2010). The present study preliminarily indicates that the activity of the VvCCD4b promoter is dropped in response to the 37°C treatment and that it also responds to the illumination change. This finding diverges from those of some previous reports. In soybean, cold and heat treatments both increase CCD4 expression, though cold treatment for 6 and 12 h did decrease the expression (Wang et al., 2011). Cold and heat treatments also upregulate CsCCD4c expression in C. sativus (Rubio-Moraga et al., 2014). Exposure to red, blue, and white light strongly decreases SbCCD4 in Scutellaria baicalensis (Tuan et al., 2017), whereas darkness treatment decreases CCD4b1 expression in clementines, but not in Navelina oranges (Lado et al., 2019).
Considering the finding of the VvCCD4b promoter responding to different light and temperature conditions, we tried to dissect why the expression patterns of VvCCD4b diverged during the ripening stage in different regions and different vintages. In this study, the GT and CL regions are characterized by a temperate continental arid climate and a temperate continental monsoon climate. Under both climates, diurnal temperature difference and sunshine are responsively altered by rainfall. In our grape-producing regions, extensive field investigations also support the fact that grape berry quality is strongly affected by seasonal rainfall. The researchers also reported that the accumulation patterns of β-damascenone and TDN were correlated with precipitation and humidity and that the expression pattern of VvCCD4b was also influenced by water deficit (Xu et al., 2015; Savoi et al., 2016; Chen et al., 2017). Only the responses of VvCCD4b promoter activity to light and temperature treatments are insufficient to interpret the relevance of VvCCD4b expression in the production of norisoprenoids in grapes. The rainfall and water status are also important factors. More experiments need to be conducted.
Functional Characterization of VvMADS4
In this study, we found that VvMADS4 is a potential transcription factor negatively regulating VvCCD4b expression. MADS transcription factors regulate fruit ripening (Ito et al., 2008), vegetative organ development (Guo et al., 2017; Li et al., 2019), flowering time (Jeon et al., 2000; Alter et al., 2016), floral meristem and organ identity (Thompson et al., 2009), stress tolerance (Guo et al., 2016), and metabolism (Lu et al., 2018; Zha et al., 2019). MADS proteins are divided into Type I and Type II based on conserved motifs and exon count. Type II (also known as MIKC) includes MADS (M-), intervening (I-), keratin-like (K-), and C-terminal (C-) domains (Theißen et al., 1996; Díaz-Riquelme et al., 2009). MIKC-type genes are classified as MIKCC- and MIKC∗- based on the I domain (Henschel et al., 2002). MIKCC members can be further subdivided into A, B, C, D, and E classes according to their function in flower organogenesis (Theißen and Saedler, 2001). VvMADS4 belongs to the E-class and is homologous with AtSEP3 from Arabidopsis (Boss et al., 2002; Wang L. et al., 2015; Grimplet et al., 2016). AtSEP3 interacts with other MADS to influence flower development and organ identity (Immink et al., 2009; Melzer et al., 2009; Wu et al., 2012). Specifically, AtSEP3 binds to LEAFY to activate B- and C-class genes (Krizek and Fletcher, 2005; Liu et al., 2009). This pair also directly activates AP3, AG, SEP1-4, and AP1 (Kaufmann et al., 2009) and is part of the positive feedback loop that maintains ABCE gene expression (Liu and Mara, 2010). AGAMOUS-like24 (AGL24), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), and SHORT VEGETATIVE PHASE (SVP) all repress AtSEP3 expression (Gregis et al., 2008; Kaufmann et al., 2009). SEPs are involved in regulating fruit ripening in fleshy fruits. For example, the best-known ripening-related gene in tomato is Rin, a SEP4-like gene and MADS family member (Vrebalov et al., 2002). Likewise, FaMADS9, a member of the SEP1/2 subfamily, modulates strawberry receptacle, achene, and petal development (Seymour et al., 2011). MaMADS2, a SEP3 homolog in banana, increases ethylene production; however, the un-ripening phenotype is still not complemented (Elitzur et al., 2010). Similarly, in the VvMADS4-transformed tomato rin mutant, the un-ripening phenotype is not complemented (Mellway and Lund, 2013). Collectively, all of these results indicate that MADS transcription factors participate in fruit ripening development by interacting with each other.
MADS proteins contact DNA by inserting amino acid residues of the α-helix in the N-terminus into the major groove or into the minor groove of the DNA (Pellegrini et al., 1995; Huang et al., 2000; Santelli and Richmond, 2000). There is a conserved arginine residue at the third amino acid position (R3) in the N-terminal arm of the MADS domain, which is directed into the minor groove of the DNA (Pellegrini et al., 1995; Huang et al., 2000; Santelli and Richmond, 2000). Using a lysine or an alanine residue to replace the R3 will reduce the DNA-binding affinity (Käppel et al., 2018). The VvMADS4 binding domain that interacts with the VvCCD4b promoter was not identified in this study. Although VvMADS4 was firstly screened out by a Y1H screening assay, when we verified their interaction again by the Y1H system using the VvCCD4b promoter sequence as the bait, the unexpected result was attained that VvMADS4 could not bind with the promoter (Supplementary Figure S5). Further study found that the VvMADS4 could bind to the three-tandem CArG box sequence, which was designed with reference to the sequence of CArG box on the VvCCD4b promoter. The number of binding sites may influence the binding efficiency. SEP3 specifically binds to CArG box [CC(A/T)6GG], whereas CArG box [CAAATTTAAG] was not consistently found in the VvCCD4b promoter, which may be also a reason for the low binding efficiency. MADS proteins usually bind to the CArG motif of the target gene DNA as dimers or multiple complex (Schwarzsommer et al., 1992; Shore and Sharrocks, 1996). However, not all the CArG elements combine with MADS proteins. Previous researchers found that the promoter of VERDANDI (VDD) contained three different CArG elements and that SEEDSTICK (STK) and SEP3 preferred to bind to CArG box 1 and CArG box 3 in the VDD promoter; as a result, STK-SEP3 protein–protein cooperative interactions form a loop between CArG boxes 1 and 3. CArG box 2 was ignored in the normal condition. But, when there was a mutant of CArG box 1 or CArG box 3, the multiple complex of STK and SEP3 could bind to CArG box 2 and another normal CArG box 1 or 3. When both CArG box 1 and CArG box 3 were mutant, the STK–SEP3 interactions could not bind to CArG box 2 (Matias-Hernandez et al., 2010; Mendes et al., 2013). So it is thought that the preference of the CArG box sequence and the number of CArG box are all important for MADS binding with DNA. SEP always coordinates with AG, SEP, and AP1. Proteins that interact with VvMADS4 (VvSEP3) in grape berries at veraison have been screened previously (Mellway and Lund, 2013). VvMADS4 can form a binary complex with VvAP3.2, VvAG1, VvAG2, VvSEP3, and VvSEP4. It can also form a ternary complex with VvAG1 and VvAG1; VvAG1 and VvAG2; or VvAG1 and VvAG3. In transgenic tomatoes, VvSEP4, VvAG1, and VvAG2 are all involved in regulating carotenoid metabolism. In our study, a dual-luciferase activity assay in Arabidopsis protoplast also revealed that VvMADS4 upregulated VvCCD4b promoter activity, which is opposite to the results in the tobacco and grape systems (Supplementary Figure S6). Therefore, we propose that VvMADS4 regulates VvCCD4b expression by coordinating with other proteins, in addition to binding directly to the VvCCD4b promoter. Future studies should directly test this hypothesis.
In summary, we verified that VvCCD4b expression was positively correlated with norisoprenoid accumulation in developing grape berries; we also investigated the responses of the VvCCD4b promoter to high temperatures and different illuminations. Furthermore, this study indicated that VvCCD4b expression was induced by ABA and that VvMADS4, a nucleus-localized transcription factor, possessed a potential function in downregulating VvCCD4b expression. Both VvMADS4 and VvCCD4b were mainly expressed in flowers and ripening berries undergoing isoprene metabolism. Our findings may be valuable for efforts to improve grape fragrance by manipulating the norisoprenoid content. Moreover, this study provides insight into VvCCD4b expression regulation. In the future, we aim to screen for potential co-regulators of VvCCD4b that interact with VvMADS4. We will also try to establish VvMADS4 transgenic grapevines, which should allow us to clarify the transcription factor’s effects on norisoprenoid accumulation.
Data Availability Statement
This study used publicly available datasets, available at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129916.
Author Contributions
NM, YG, and YW performed the research and analyzed data. KY, JC, and X-YL perfected the research scheme. C-QD and Q-HP guided the research. NM wrote the paper. All the authors critically revised the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, Grant Nos. 31471834 and 31772278.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Dapeng Zhang (Tsinghua University) for helping us in using the Multifunction Reader. We thank Prof. Daqi Fu, Prof. Hongliang Zhu, and Assoc. Prof. Xiuqin Wang (China Agricultural University) for giving the pCXSN, pGreen II 0800, and pCAMBIA 1301 vectors and calli of grape as gifts.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00483/full#supplementary-material
Footnotes
- ^ http://genomes.cribi.unipd.it/grape/
- ^ https://www.dna.affrc.go.jp/PLACE/?action=newplace
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ http://linux1.softberry.com/berry.phtml?topic=tssp&group=programs&subgroup=promoter
- ^ https://deepgreen.dpb.carnegiescience.edu/cell%20imaging%20site%20/html/vectors.html
- ^ https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129916
- ^ https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE129916
References
Adami, M., De Franceschi, P., Brandi, F., Liverani, A., Giovannini, D., Rosati, C., et al. (2013). Identifying a Carotenoid Cleavage Dioxygenase (ccd4) gene controlling yellow/white fruit flesh color of peach. Plant Mol. Biol. Report. 31, 1166–1175. doi: 10.1007/s11105-013-0628-6
Ahrazem, O., Trapero, A., Gómez, M. D., Rubio-Moraga, A., and Gómez-Gómez, L. (2010). Genomic analysis and gene structure of the plant carotenoid dioxygenase 4 family: A deeper study in Crocus sativus and its allies. Genomics 96, 239–250. doi: 10.1016/j.ygeno.2010.07.003
Alter, P., Bircheneder, S., Zhou, L. Z., Schlüter, U., Gahrtz, M., Sonnewald, U., et al. (2016). Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiol. 172, 389–404. doi: 10.1104/pp.16.00285
Asproudi, A., Ferrandino, A., Bonello, F., Vaudano, E., Pollon, M., and Petrozziello, M. (2018). Key norisoprenoid compounds in wines from early-harvested grapes in view of climate change. Food Chem. 268, 143–152. doi: 10.1016/j.foodchem.2018.06.069
Aznar, M., López, R., Cacho, J. F., and Ferreira, V. (2001). Identification and quantification of impact Odorants of aged red wines from Rioja. GC-Olfactometry, Quantitative GC-MS, and odor evaluation of HPLC fractions. J. Agric Food Chem. 49, 2924–2929. doi: 10.1021/jf001372u
Bai, S., Tuan, P. A., Tatsuki, M., Yaegaki, H., Ohmiya, A., Yamamizo, C., et al. (2015). Knockdown of Carotenoid Cleavage Dioxygenase 4 (CCD4) via virus-induced gene silencing confers yellow coloration in peach fruit: evaluation of gene function related to fruit traits. Plant Mol. Biol. Report. 34, 257–264. doi: 10.1007/s11105-015-0920-8
Baldermann, S., Kato, M., Kurosawa, M., Kurobayashi, Y., Fujita, A., Fleischmann, P., et al. (2010). Functional characterization of a carotenoid cleavage dioxygenase 1 and its relation to the carotenoid accumulation and volatile emission during the floral development of Osmanthus fragrans Lour. J. Exp. Bot. 61, 2967–2977. doi: 10.1093/jxb/erq123
Bindon, K. A., Dry, P. R., and Loveys, B. R. (2007). Influence of plant water status on the production of C13- norisoprenoid precursors in Vitis vinifera L. cv. cabernet sauvignon grape berries. J. Agric. Food Chem. 55, 4493–4500. doi: 10.1021/jf063331p
Boss, P. K., Sensi, E., Hua, C., Davies, C., and Thomas, M. R. (2002). Cloning and characterisation of grapevine (Vitis vinifera L.) MADS-box genes expressed during inflorescence and berry development. Plant Sci. 162, 887–895. doi: 10.1016/S0168-9452(02)00034-1
Brandi, F., Bar, E., Mourgues, F., Horvath, G., Turcsi, E., Giuliano, G., et al. (2011). Study of “Redhaven” peach and its white-fleshed mutant suggests a key role of CCD4 carotenoid dioxygenase in carotenoid and norisoprenoid volatile metabolism. BMC Plant Biol. 11:24. doi: 10.1186/1471-2229-11-24
Bruno, M., Beyer, P., and Al-Babili, S. (2015). The potato carotenoid cleavage dioxygenase 4 catalyzes a single cleavage of beta-ionone ring-containing carotenes and non-epoxidated xanthophylls. Arch. Biochem. Biophys. 572, 126–133. doi: 10.1016/j.abb.2015.02.011
Campbell, R., Ducreux, L. J., Morris, W. L., Morris, J. A., Suttle, J. C., Ramsay, G., et al. (2010). The metabolic and developmental roles of carotenoid cleavage dioxygenase4 from potato. Plant Physiol. 154, 656–664. doi: 10.1104/pp.110.158733
Castillo, R., Fernandez, J. A., and Gomez-Gomez, L. (2005). Implications of carotenoid biosynthetic genes in apocarotenoid formation during the stigma development of Crocus sativus and its closer relatives. Plant Physiol. 139, 674–689. doi: 10.1104/pp.105.067827
Chang-ho, E., Seong-U, K., and In-J, K. (2015). The promoter from the Citrus unshiu carotenoid isomerase gene directs differential GUS expression in transgenic Arabidopsis. Mol. Breed. 35:116. doi: 10.1007/s11032-015-0310-9
Chen, H., Zuo, X., Shao, H., Fan, S., Ma, J., Zhang, D., et al. (2018). Genome-wide analysis of carotenoid cleavage oxygenase genes and their responses to various phytohormones and abiotic stresses in apple (Malus domestica). Plant Physiol. Biochem. 123, 81–93. doi: 10.1016/j.plaphy.2017.12.001
Chen, W. K., Wang, Y., Gao, X. T., Yang, X. H., He, F., Duan, C. Q., et al. (2018). Flavonoid and aromatic profiles of two Vitis vinifera L. teinturier grape cultivars. Aust. J. Grape Wine Res. 24, 379–389. doi: 10.1111/ajgw.12336
Chen, W. K., Yu, K. J., Liu, B., Lan, Y., Sun, R. Z., Li, Q., et al. (2017). Comparison of transcriptional expression patterns of carotenoid metabolism in ‘Cabernet Sauvignon’ grapes from two regions with distinct climate. J. Plant Physiol. 213, 75–86. doi: 10.1016/j.jplph.2017.03.001
Chiou, C. Y., Pan, H. A., Chuang, Y. N., and Yeh, K. W. (2010). Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232, 937–948. doi: 10.1007/s00425-010-1222-x
Cho, H. T., and Cosgrove, D. J. (2000). Altered expression of expansin modulates leaf growth and pedicel abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 97, 9783–9788. doi: 10.1073/pnas.160276997
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313X.1998.00343.x
Coombe, B. G. (1995). Growth stages of the grapevine: adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res. 1, 104–110. doi: 10.1111/j.1755-0238.1995.tb00086.x
Cunningham, F. X., and Gantt, E. (2000). Identification of multi-gene families encoding isopentenyl diphosphate isomerase in plants by heterologous complementation in Escherichia coli. Plant Cell Physiol. 41, 119–123. doi: 10.1093/pcp/41.1.119
Díaz-Riquelme, J., Lijavetzky, D., Martínez-Zapater, J. M., and Carmona, M. J. (2009). Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiol. 149, 354–369. doi: 10.1104/pp.108.131052
Dunlevy, J. D., and Kalua, C. M. (2009). “The production of flavour & aroma compounds in grape berries,” in Grapevine Molecular Physiology & Biotechnology, Ed. K. A. Roubelakis-Angelakis, (Dordrecht: Springer), 293–240.
Elitzur, T., Vrebalov, J., Giovannoni, J. J., Goldschmidt, E. E., and Friedman, H. (2010). The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J. Exp. Bot. 61, 1523–1535. doi: 10.1093/jxb/erq017
Feng, H., Yuan, F., Skinkis, P. A., and Qian, M. C. (2015). Influence of cluster zone leaf removal on Pinot noir grape chemical and volatile composition. Food Chem. 173, 414–423. doi: 10.1016/j.foodchem.2014.09.149
Fleischmann, P., Watanabe, N., and Winterhalter, P. (2003). Enzymatic carotenoid cleavage in star fruit (Averrhoa carambola). Phytochemistry 63, 131–137. doi: 10.1016/s0031-9422(02)00657-x
Frusciante, S., Diretto, G., Bruno, M., Ferrante, P., Pietrella, M., Prado-Cabrero, A., et al. (2014). Novel carotenoid cleavage dioxygenase catalyzes the first dedicated step in saffron crocin biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, 12246–12251. doi: 10.1073/pnas.1404629111
Gao, Y., Li, X. X., Han, M. M., Yang, X. F., Li, Z., Wang, J., et al. (2016). Rain-Shelter cultivation modifies carbon allocation in the polyphenolic and volatile metabolism of Vitis vinifera L. chardonnay grapes. PLoS One 11:e0156117. doi: 10.1371/journal.pone.0156117
Gregis, V., Sessa, A., Colombo, L., and Kater, M. M. (2008). AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis. Plant J. 56, 891–902. doi: 10.1111/j.1365-313X.2008.03648.x
Grimplet, J., Martinez-Zapater, J. M., and Carmona, M. J. (2016). Structural and functional annotation of the MADS-box transcription factor family in grapevine. BMC Genomics 17:80. doi: 10.1186/s12864-016-2398-7
Guo, X., Chen, G., Cui, B., Gao, Q., Guo, J. E., Li, A., et al. (2016). Solanum lycopersicum agamous-like MADS-box protein AGL15-like gene, SlMBP11, confers salt stress tolerance. Mol. Breed. 36, 1–15. doi: 10.1007/s11032-016-0544-1
Guo, X., Chen, G., Naeem, M., Yu, X., Tang, B., Li, A., et al. (2017). The MADS-box gene SlMBP11 regulates plant architecture and affects reproductive development in tomato plants. Plant Sci. 258, 90–101. doi: 10.1016/j.plantsci.2017.02.005
Han, Y., Wang, H., Wang, X., Li, K., Dong, M., Li, Y., et al. (2019). Mechanism of floral scent production in Osmanthus fragrans and the production and regulation of its key floral constituents, β-ionone and linalool. Hortic. Res. 6:106. doi: 10.1038/s41438-019-0189-4
Han, Y., Wu, M., Cao, L., Yuan, W., Dong, M., Wang, X., et al. (2016). Characterization of OfWRKY3, a transcription factor that positively regulates the carotenoid cleavage dioxygenase gene OfCCD4 in Osmanthus fragrans. Plant Mol. Biol. 91, 485–496. doi: 10.1007/s11103-016-0483-6
Henschel, K., Kofuji, R., Hasebe, M., Saedler, H., Münster, T., and Theißen, G. (2002). Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol. Biol. Evol. 19, 801–814. doi: 10.1093/oxfordjournals.molbev.a004137
Hofgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16:9877. doi: 10.1093/nar/16.20.9877
Huang, F. C., Molnár, P., and Schwab, W. (2009). Cloning and functional characterization of carotenoid cleavage dioxygenase 4 genes. J. Exp. Bot. 60, 3011–3022. doi: 10.1093/jxb/erp137
Huang, K., Louis, J. M., Donaldson, L., Lim, F., Sharrocks, A. D., and Clore, G. M. (2000). Solution structure of the MEF2A–DNA complex: structural basis for the modulation of DNA bending and specificity by MADS-box transcription factors. EMBO J. 19, 2615–2628. doi: 10.1093/emboj/19.11.2615
Imai, A., Takahashi, S., Nakayama, K., and Satoh, H. (2013). The promoter of the carotenoid cleavage dioxygenase 4a-5 gene of Chrysanthemum morifolium (CmCCD4a-5) drives petal-specific transcription of a conjugated gene in the developing flower. J. Plant Physiol. 170, 1295–1299. doi: 10.1016/j.jplph.2013.04.001
Immink, R. G. H., Tonaco, I. A. N., de Folter, S., Shchennikova, A., van Dijk, A. D. J., Busscher-Lange, J., et al. (2009). SEPALLATA3: The “glue” for MADS box transcription factor complex formation. Genome Biol. 10:R24. doi: 10.1186/gb-2009-10-2-r24
Ito, Y., Kitagawa, M., Ihashi, N., Yabe, K., Kimbara, J., Yasuda, J., et al. (2008). DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 55, 212–223. doi: 10.1111/j.1365-313X.2008.03491.x
Jeon, J. S., Jang, S., Lee, S., Nam, J., Kim, C., Lee, S. H., et al. (2000). leafy hull sterile 1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12, 871–884. doi: 10.1105/tpc.12.6.871
Käppel, S., Melzer, R., Rümpler, F., Gafert, C., and Theißen, G. (2018). The floral homeotic protein SEPALLATA3 recognizes target DNA sequences by shape readout involving a conserved arginine residue in the MADS-domain. Plant J. 95, 341–357. doi: 10.1111/tpj.13954
Kaufmann, K., Muiño, J. M., Jauregui, R., Airoldi, C. A., Smaczniak, C., Krajewski, P., et al. (2009). Target genes of the MADS transcription factor sepallata3: Integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7:e90. doi: 10.1371/journal.pbio.1000090
Kotseridis, Y., Baumes, R. L., Bertrand, A., and Skouroumounis, G. K. (1999). Quantitative determination of β-ionone in red wines and grapes of Bordeaux using a stable isotope dilution assay. J. Chromatogr. A 848, 317–325. doi: 10.1016/S0021-9673(99)00422-7
Krizek, B. A., and Fletcher, J. C. (2005). Molecular mechanisms of flower development: An armchair guide. Nat. Rev. Genet. 6, 688–698. doi: 10.1038/nrg1675
Kwasniewski, M. T., Vanden Heuvel, J. E., Pan, B. S., and Sacks, G. L. (2010). Timing of cluster light environment manipulation during grape development affects C13 norisoprenoid and carotenoid concentrations in Riesling. J. Agric. Food Chem. 58, 6841–6849. doi: 10.1021/jf904555p
Lado, J., Alós, E., Manzi, M., Cronje, P. J. R., Gómez-Cadenas, A., Rodrigo, M. J., et al. (2019). Light regulation of carotenoid biosynthesis in the peel of mandarin and sweet orange fruits. Front. Plant Sci. 10:1288. doi: 10.3389/fpls.2019.01288
Lan, Y. B., Qian, X., Yang, Z. J., Xiang, X. F., Yang, W. X., Liu, T., et al. (2016). Striking changes in volatile profiles at sub-zero temperatures during over-ripening of “Beibinghong” grapes in Northeastern China. Food Chem. 212, 172–182. doi: 10.1016/j.foodchem.2016.05.143
Lashbrooke, J. G., Young, P. R., Dockrall, S. J., Vasanth, K., and Vivier, M. A. (2013). Functional characterisation of three members of the Vitis vinifera L. carotenoid cleavage dioxygenase gene family. BMC Plant Biol. 13:156. doi: 10.1186/1471-2229-13-156
Ledger, S. E., Janssen, B. J., Karunairetnam, S., Wang, T., and Snowden, K. C. (2010). Modified CAROTENOID CLEAVAGE DIOXYGENASE8 expression correlates with altered branching in kiwifruit (Actinidia chinensis). New Phytol. 188, 803–813. doi: 10.1111/j.1469-8137.2010.03394.x
Li, A., Chen, G., Yu, X., Zhu, Z., Zhang, L., Zhou, S., et al. (2019). The tomato MADS-box gene SlMBP9 negatively regulates lateral root formation and apical dominance by reducing auxin biosynthesis and transport. Plant Cell Rep. 38, 951–963. doi: 10.1007/s00299-019-02417-x
Liang, Y. S., Jeon, Y. A., Lim, S. H., Kim, J. K., Lee, J. Y., Kim, Y. M., et al. (2011). Vascular-specific activity of the Arabidopsis carotenoid cleavage dioxygenase 7 gene promoter. Plant Cell Rep. 30, 973–980. doi: 10.1007/s00299-010-0999-1
Liu, C., Xi, W., Shen, L., Tan, C., and Yu, H. (2009). Regulation of floral patterning by flowering time genes. Dev. Cell 16, 711–722. doi: 10.1016/j.devcel.2009.03.011
Liu, Z., and Mara, C. (2010). Regulatory mechanisms for floral homeotic gene expression. Semin. Cell Dev. Biol. 21, 80–86. doi: 10.1016/j.semcdb.2009.11.012
Lu, S., Zhang, Y., Zhu, K., Yang, W., Ye, J., Chai, L., et al. (2018). The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 176, 2657–2676. doi: 10.1104/pp.17.01830
Ma, G., Zhang, L., Matsuta, A., Matsutani, K., Yamawaki, K., Yahata, M., et al. (2013). Enzymatic formation of β-citraurin from β-cryptoxanthin and zeaxanthin by carotenoid cleavage dioxygenase4 in the flavedo of citrus fruit. Plant Physiol. 163, 682–695. doi: 10.1104/pp.113.223297
Mateo, J. J., and Jiménez, M. (2000). Monoterpenes in grape juice and wines. J. Chromatogr. A 881, 557–567. doi: 10.1016/S0021-9673(99)01342-4
Mathieu, S., Terrier, N., Procureur, J., Bigey, F., and Günata, Z. (2005). A carotenoid cleavage dioxygenase from Vitis vinifera L.: Functional characterization and expression during grape berry development in relation to C13-norisoprenoid accumulation. J. Exp. Bot. 56, 2721–2731. doi: 10.1093/jxb/eri265
Matias-Hernandez, L., Battaglia, R., Galbiati, F., Rubes, M., Eichenberger, C., Grossniklaus, U., et al. (2010). VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22, 1702–1715. doi: 10.1105/tpc.109.068627
Mellway, R. D., and Lund, S. T. (2013). Interaction analysis of grapevine MIKCc-type MADS transcription factors and heterologous expression of putative véraison regulators in tomato. J. Plant Physiol. 170, 1424–1433. doi: 10.1016/j.jplph.2013.05.010
Melzer, R., Verelst, W., and Theißen, G. (2009). The class E floral homeotic protein SEPALLATA3 is sufficient to loop DNA in ‘floral quartet’-like complexes in vitro. Nucleic Acids Res. 37, 144–157. doi: 10.1093/nar/gkn900
Mendes, M. A., Guerra, R. F., Berns, M. C., Manzo, C., Masiero, S., Finzi, L., et al. (2013). MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 25, 2560–2572. doi: 10.1105/tpc.112.108688
Meng, N., Yan, G. L., Zhang, D., Li, X. Y., Duan, C. Q., and Pan, Q. H. (2019). Characterization of two Vitis vinifera carotenoid cleavage dioxygenases by heterologous expression in Saccharomyces cerevisiae. Mol. Biol. Rep. 46, 6311–6323. doi: 10.1007/s11033-019-05072-3
Ohmiya, A., Kishimoto, S., Aida, R., Yoshioka, S., and Sumitomo, K. (2006). Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 142, 1193–1201. doi: 10.1104/pp.106.087130
Pellegrini, L., Tan, S., and Richmond, T. J. (1995). Structure of serum response factor core bound to DNA. Nature 376, 490–498. doi: 10.1038/376490a0
Rodrigo, M. J., Alquézar, B., Alós, E., Medina, V., Carmona, L., Bruno, M., et al. (2013). A novel carotenoid cleavage activity involved in the biosynthesis of Citrus fruit-specific apocarotenoid pigments. J. Exp. Bot. 64, 4461–4478. doi: 10.1093/jxb/ert260
Rubio, A., Rambla, J. L., Santaella, M., Gomez, M. D., Orzaez, D., Granell, A., et al. (2008). Cytosolic and plastoglobule-targeted carotenoid dioxygenases from Crocus sativus are both involved in beta-ionone release. J. Biol. Chem. 283, 24816–24825. doi: 10.1074/jbc.M804000200
Rubio-Moraga, A., Rambla, J. L., Fernández-De-Carmen, A., Trapero-Mozos, A., Ahrazem, O., Orzáez, D., et al. (2014). New target carotenoids for CCD4 enzymes are revealed with the characterization of a novel stress-induced carotenoid cleavage dioxygenase gene from Crocus sativus. Plant Mol. Biol. 86, 555–569. doi: 10.1007/s11103-014-0250-5
Ruijter, J. M., Ramakers, C., Hoogaars, W. M., Karlen, Y., Bakker, O., van den Hoff, M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37:e45. doi: 10.1093/nar/gkp045
Santelli, E., and Richmond, T. J. (2000). Crystal structure of MEF2A core bound to DNA at 1.5 Å resolution11Edited by J. Karn. J. Mol. Biol. 297, 437–449. doi: 10.1006/jmbi.2000.3568
Savoi, S., Wong, D. C. J., Arapitsas, P., Miculan, M., Bucchetti, B., Peterlunger, E., et al. (2016). Transcriptome and metabolite profiling reveals that prolonged drought modulates the phenylpropanoid and terpenoid pathway in white grapes (Vitis vinifera L.). BMC Plant Biol. 16:760. doi: 10.1186/s12870-016-0760-1
Schwarzsommer, Z., Hue, I., Huijser, P., Flor, P. J., Hansen, R., Tetens, F., et al. (1992). Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 11, 251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x
Seymour, G. B., Ryder, C. D., Cevik, V., Hammond, J. P., Popovich, A., King, G. J., et al. (2011). A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria×ananassa Duch.) fruit, a non-climacteric tissue. J. Exp. Bot. 62, 1179–1188. doi: 10.1093/jxb/erq360
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shore, P., and Sharrocks, A. D. (1996). MADS-Box Family of Transcription Factors BT - EJB Reviews, eds P. Christen, and E. Hofmann, (Berlin: Springer Berlin Heidelberg), 87–99. doi: 10.1007/978-3-642-85252-7_7
Simpson, R. F. (1978). 1,1,6-Trimethyl-1,2-dihydronaphthalene - important contributor to bottle aged bouquet of wine. Chem. Indus. 1, 37–37.
Sun, R. Z., Cheng, G., Li, Q., Zhu, Y. R., Zhang, X., Wang, Y., et al. (2019). Comparative physiological, metabolomic, and transcriptomic analyses reveal developmental stage-dependent effects of cluster bagging on phenolic metabolism in Cabernet Sauvignon grape berries. BMC Plant Biol. 19:583. doi: 10.1186/s12870-019-2186-z
Sun, R. Z., Pan, Q. H., Duan, C. Q., and Wang, J. (2015). Light response and potential interacting proteins of a grape flavonoid 3’-hydroxylase gene promoter. Plant Physiol. Biochem. 97, 70–81. doi: 10.1016/j.plaphy.2015.09.016
Tanabe, N., Tamoi, M., and Shigeoka, S. (2015). The sweet potato RbcS gene (IbRbcS1) promoter confers high-level and green tissue-specific expression of the GUS reporter gene in transgenic Arabidopsis. Gene 567, 244–250. doi: 10.1016/j.gene.2015.05.006
Theißen, G., Kim, J. T., and Saedler, H. (1996). Classification and phylogeny of the MADS-Box multigene family suggest defined roles of MADS-Box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/pl00006110
Theißen, G., and Saedler, H. (2001). Plant biology: Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Thompson, B. E., Bartling, L., Whipple, C., Hall, D. H., Sakai, H., Schmidt, R., et al. (2009). Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 21, 2578–2590. doi: 10.1105/tpc.109.067751
Tuan, P. A., Park, C. H., Park, W. T., Kim, Y. B., Kim, Y. J., Chung, S. O., et al. (2017). Expression levels of carotenoid biosynthetic genes and carotenoid production in the callus of scutellaria baicalensis exposed to white, blue, and red light-emitting diodes. Appl. Biol. Chem. 60, 591–596. doi: 10.1007/s13765-017-0314-8
Van Eck, J., Kirk, D. D., and Walmsley, A. M. (2006). Tomato (Lycopersicum esculentum). Methods Mol. Biol. 343, 459–473.
Voinnet, O., Rivas, S., Mestre, P., and Baulcombe, D. (2003). Retracted: An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 33, 949–956. doi: 10.1046/j.1365-313X.2003.01676.x
Vrebalov, J., Ruezinsky, D., Padmanabhan, V., White, R., Medrano, D., Drake, R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346. doi: 10.1126/science.1068181
Wang, H., Wang, W., Zhan, J., Huang, W., and Xu, H. (2015). An efficient PEG-mediated transient gene expression system in grape protoplasts and its application in subcellular localization studies of flavonoids biosynthesis enzymes. Sci. Hortic. 191, 82–89. doi: 10.1016/j.scienta.2015.04.039
Wang, L., Yin, X., Cheng, C., Wang, H., Guo, R., Xu, X., et al. (2015). Evolutionary and expression analysis of a MADS-box gene superfamily involved in ovule development of seeded and seedless grapevines. Mol. Genet. Genomics 290, 825–846. doi: 10.1007/s00438-014-0961-y
Wang, R. K., Lu, J. J., Xing, G. N., Gai, J. Y., and Zhao, T. J. (2011). Molecular evolution of two consecutive carotenoid cleavage dioxygenase genes in strigolactone biosynthesis in plants. Genet. Mol. Res. 10, 3664–3673. doi: 10.4238/2011.December.2.2
Wei, Y., Wan, H., Wu, Z., Wang, R., Ruan, M., Ye, Q., et al. (2015). A comprehensive analysis of carotenoid cleavage dioxygenases genes in solanum lycopersicum. Plant Mol. Biol. Rep. 34, 512–523. doi: 10.1007/s11105-015-0943-1
Wen, Y. Q., Zhong, G. Y., Gao, Y., Lan, Y. B., Duan, C. Q., and Pan, Q. H. (2015). Using the combined analysis of transcripts and metabolites to propose key genes for differential terpene accumulation across two regions. BMC Plant Biol. 15:240. doi: 10.1186/s12870-015-0631-1
Wu, M. F., Sang, Y., Bezhani, S., Yamaguchi, N., Han, S. K., Li, Z., et al. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U.S.A. 109, 3576–3581. doi: 10.1073/pnas.1113409109
Xu, W., Yu, Y., Ding, J., Hua, Z., and Wang, Y. (2010). Characterization of a novel stilbene synthase promoter involved in pathogen- and stress-inducible expression from Chinese wild Vitis pseudoreticulata. Planta 231, 475–487. doi: 10.1007/s00425-009-1062-8
Xu, X. Q., Liu, B., Zhu, B. Q., Lan, Y., Gao, Y., Wang, D., et al. (2015). Differences in volatile profiles of Cabernet Sauvignon grapes grown in two distinct regions of China and their responses to weather conditions. Plant Physiol. Biochem. 89, 123–133. doi: 10.1016/j.plaphy.2015.02.020
Yuan, F., and Qian, M. C. (2016). Development of C13-norisoprenoids, carotenoids and other volatile compounds in Vitis vinifera L. Cv. Pinot noir grapes. Food Chem. 192, 633–641. doi: 10.1016/j.foodchem.2015.07.050
Yue, Z., Liu, H., and Ma, F. (2015). The Malus carotenoid cleavage dioxygenase 7 is involved in stress response and regulated by basic pentacysteine 1. Sci. Hortic. 192, 264–270. doi: 10.1016/j.scienta.2015.06.027
Zha, K., Xie, H., Ge, M., Wang, Z., Wang, Y., Si, W., et al. (2019). Expression of Maize MADS transcription factor ZmES22 negatively modulates starch accumulation in rice endosperm. Int. J. Mol. Sci. 20:483. doi: 10.3390/ijms20030483
Keywords: carotenoid cleavage dioxygenase 4b, VvMADS4, norisoprenoids, Vitis vinifera L., expression and regulation
Citation: Meng N, Wei Y, Gao Y, Yu K, Cheng J, Li X-Y, Duan C-Q and Pan Q-H (2020) Characterization of Transcriptional Expression and Regulation of Carotenoid Cleavage Dioxygenase 4b in Grapes. Front. Plant Sci. 11:483. doi: 10.3389/fpls.2020.00483
Received: 06 January 2020; Accepted: 31 March 2020;
Published: 08 May 2020.
Edited by:
Stephan Pollmann, Center for Plant Biotechnology and Genomics, National Institute of Agricultural and Food Research and Technology, SpainReviewed by:
Christine Böttcher, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaYuejin Wang, Northwest A&F University, China
Copyright © 2020 Meng, Wei, Gao, Yu, Cheng, Li, Duan and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiu-Hong Pan, cGFucWhAY2F1LmVkdS5jbg==
 Nan Meng
Nan Meng Yi Wei1,2
Yi Wei1,2 Qiu-Hong Pan
Qiu-Hong Pan