
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 15 April 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00397
Ovules are female reproductive organs of angiosperms, containing sporophytic integuments and gametophytic embryo sacs. After fertilization, embryo sacs develop into embryos and endosperm whereas integuments into seed coat. Ovule development is regulated by transcription factors (TF) whose expression is often controlled by microRNAs. Mutations of Arabidopsis DICER-LIKE 1 (DCL1), a microRNA processing protein, caused defective ovule development and reduced female fertility. However, it was not clear whether other microRNA processing proteins participate in this process and how defective ovule development influenced female fertility. We report that mutations of HUA ENHANCER1 (HEN1) and HYPONASTIC LEAVES 1 (HYL1) interfered with integument growth. The sporophytic defect caused abnormal embryo sac development and inability of mutant ovules to attract pollen tubes, leading to reduced female fertility. We show that the role of HEN1 in integument growth is cell-autonomous. Although AUXIN RESPONSE FACTOR 6 (ARF6) and ARF8 were ectopically expressed in mutant ovules, consistent with the reduction of microRNA167 in hen1, introducing arf6;arf8 did not suppress ovule defects of hen1, suggesting the involvement of more microRNAs in this process. Results presented indicate that the microRNA processing machinery is critical for ovule development and seed production through multiple microRNAs and their targets.
Ovule development is critical for seed yield and plant reproduction. A mature ovule is composed of sporophytic cells, i.e., integuments, and a female gametophyte (FG), called embryo sac (Schneitz et al., 1995, 1997; Christensen et al., 1997; Drews et al., 1998; Drews and Yadegari, 2002; Shi and Yang, 2011). The embryo sac forms embryo and endosperm whereas integuments develop into seed coat after fertilization. During ovule development, megasporogenesis establishes a proximal-distal polarity while megagametogenesis ensures the production of FG (Christensen et al., 1997; Drews et al., 1998; Drews and Yadegari, 2002). A critical event during megagametogenesis in Arabidopsis is the asymmetric growth of outer integuments, which finally envelops the embryo sac and leads to the anatrophy of mature ovules (Schneitz et al., 1995, 1997; Drews et al., 1998; Drews and Yadegari, 2002). Arabidopsis mutants defective in the asymmetric growth of outer integuments often shows defective formation of FG (Bencivenga et al., 2011; Chevalier et al., 2011; Wang et al., 2016), suggesting sporophytic control of gametophytic development.
A number of transcriptional factors (TFs) regulate integument growth (Colombo et al., 2008). Mutations of INNER NO OUTER (INO), ABERRANT TESTA SHAPE (ATS), or AINTEGUMENTA (ANT) compromised the initiation and growth of integuments, resulting in a loss of the outer-inner integument organization (Elliott et al., 1996; Klucher et al., 1996; Villanueva et al., 1999; McAbee et al., 2006). Genes encoding a few homeodomain proteins, such as BELL1 (BEL1) (Reiser et al., 1995), PHABULOSA (PHB) (Sieber et al., 2004), and WUSHEL (WUS) (Gross-Hardt et al., 2002; Lieber et al., 2011), play roles in integument growth and ovule development. Except for the positive regulators of integument growth, ectopic expression of AUXIN RESPONSE FACTOR6 (ARF6) and ARF8 resulted in the arrest of integument growth (Wu et al., 2006), suggesting that there are negative regulators in this process.
Transcription factors are major targets of microRNAs (miRNAs), which are small RNAs of 20- to 24-nucleotide (nt), produced from pre-miRNA-encoding genes, and sequence-specific regulators of gene expression. Because of key roles of miRNAs in plant growth and responses to environment, their expression, processing, and turnover are tightly regulated (Dong et al., 2008; Rogers and Chen, 2013). Mutations at miRNA processing genes often result in reduced fertility (Lu and Fedoroff, 2000; Schauer et al., 2002; Olmedo-Monfil et al., 2010). Arabidopsis DICER-LIKE 1 (DCL1) is a miRNA processing protein (Kurihara and Watanabe, 2004). A mutant of DCL1, short integuments (sin1/dcl1-7), was identified from a chemical mutagenesis (Robinson-Beers et al., 1992). sin1/dcl1-7 is defective in the asymmetric growth of integuments (Robinson-Beers et al., 1992) during ovule developmental stage 3-I when a functional megaspore (FM) is formed (Schneitz et al., 1995, 1997). Efficient and precise processing of pri-miRNAs requires the interaction between DCL1 and HYPONASTIC LEAVES 1 (HYL1) (Kurihara et al., 2006), a dsRNA-binding protein (Vazquez et al., 2004; Dong et al., 2008). HUA ENHANCER1 (HEN1), a multidomain AdoMet-dependent 2′-O-methyltransferase critical for miRNA biogenesis (Chen et al., 2002; Yu et al., 2005, 2010; Baranauske et al., 2015), act in the same pathway as DCL1 and HYL1 (Yang et al., 2010; Baranauske et al., 2015). Whether they participate in ovule development and how their mutations influence female fertility are not clear.
We report here that hen1-8, a hypomorphic mutant of HEN1 (Chen et al., 2002; Yu et al., 2005, 2010), and hyl1-2, a null mutant of HYL1 (Han et al., 2004; Kurihara et al., 2006; Dong et al., 2008), are defective in ovule development. Mutant ovules failed to have asymmetric integument growth, leading to abnormal embryo sac development, compromised pollen tube guidance, and thus reduced female fertility. Downregulating HEN1 specifically in outer integuments phenocopied hen1-8, suggesting a cell-autonomous action. Ectopic expression of ARF6 and ARF8 and distorted auxin maximum in hen1-8 ovules are consistent with the reduction of miRNA167, whose processing relies on HEN1 (Yu et al., 2010; Ren et al., 2012). However, introducing the arf6;arf8 double mutant did not suppress ovule defects of hen1-8, suggesting the involvement of more microRNAs in this process.
Arabidopsis mutants lines, including hen1-8 (Yu et al., 2010), hen1-2 (Chen et al., 2002), hyl1-2 (Vazquez et al., 2004), arf6-1 (Okushima et al., 2005), arf8-3 (Nagpal et al., 2005), the transgenic line ProLAT52:GUS (Li et al., 2013), DR5:GFP (Ulmasov et al., 1997), PIN1:GFP (Benkova et al., 2003), and ProES1:NLS-YFP (Bencivenga et al., 2012) were described previously. Col-0 ecotype or Landsberg erecta (Ler) was used as the wild type as noted. Arabidopsis plants were grown as described (Zhou et al., 2013). In brief, the seeds were surface-sterilized and then sown on half-strength Murashige and Skoog (MS) basal medium with vitamins (Phytotechlab, SPS0519160A). Plates were placed under 4°C in darkness for 3 days before being moved to a growth chamber with a 16-h-light/8-h-dark cycle at 21°C. One week later, the seedlings were transferred to a 3:1 mix of nutrient soil: vermiculite under same conditions with growth chamber. Plant growth, transformation, and selection were as described (Zhou et al., 2013).
The artificial miRNA construct targeting HEN1 (amiR-HEN1) was designed with the primers ZP7781/ZP7782/ZP7783/ZP7784 using WMD3-Designer. The amiR-HEN1 was cloned into pROKII-GFP to generate ProLAT52:amiR-HEN1. Later, ProLat52 was replaced by ProINO to generate ProINO:amiR-HEN1. The RNAi-HEN1 fragment (2297bp to 2599bp of HEN1 coding sequence) was amplified with the primer pair ZP6753/ZP6754. The resultant PCR products were sub-cloned into the RNAi vector pTCK303 (Guo et al., 2010) to obtain the ProUBQ10:RNAi-HEN1 construct. Later, ProUBQ10 was replaced by ProINO to generate ProINO:RNAi-HEN1. ProHEN1 was cloned into pENTR/SD/D-TOPO (Invitrogen) with the primer pair ZP5140/ZP5173, including a 1847 bp sequence upstream of HEN1 start codon. The entry vector was used in a LR reaction with the destination vector pMD163 (Curtis and Grossniklaus, 2003) to generate ProHEN1:GUS. All primers are listed in Supplementary Table S1.
Genotyping PCRs for arf6-1 and arf8-3 were performed using following primer pairs: ZP308/ZP309 and ZP306/ZP307 for the wild copy, ZP1/ZP309 and ZP7546/ZP307 for the mutant copy of ARF6 and ARF8, respectively. Total RNAs were extracted from mature ovules using a Qiagen RNeasy plant mini kit. For qPCRs of ARF6 and ARF8 in ovules, oligo(dT)-primed cDNAs were synthesized using a FastQuant RT Kit (TIAN GEN, Cat#KR106-02). Internal controls were as described (Zhou et al., 2013). qPCRs were performed with three biological replicates. Primers used in qPCRs were ZP201/ZP202 for TUBLIN2, ZP687/ZP688 for GAPDH, ZP7207/ZP7208 for ARF6, ZP7209/ZP7210 for ARF8, and ZP9325/ZP9326 for HEN1. All primers are listed in Supplementary Table S1.
RNA in situ hybridization was performed as previously described (Zhou et al., 2013). In brief, the emasculate pistils were fixed in 4% Paraformaldehyde solution (aladdin) at 4°C overnight. Then the fixed tissues were embedded in Paraplast (Sigma-Aldrich) after dehydration and were then sectioned at 8 μm. RNA probes of ARF6 and ARF8 were amplified with the primer pairs ZP8093/8094 and ZP8095/8096, respectively. The sense and antisense probes were modified in vitro with digoxigenin-UTP by SP6 or T7 RNA polymerases (Roche), respectively. Sections were hybridized with 1.5 ng/μL probes at 42°C overnight in a hybridization solution that contained formamide. Hybridization signals were detected by antidigoxigenin antibody (Anti-Digoxigenin-Ap Fab fragments; Roche). The samples were observed using an Olympus BX53 microscope. All primers are listed in Supplementary Table S1.
Pollen tube in vivo growth by histochemical GUS staining of ProLAT52:GUS-pollinated pistils and aniline blue staining were performed as described (Li et al., 2013). Whole-mount ovule clearing and CLSM of ovules were performed as described (Wang et al., 2016; Liu et al., 2019). Flowers at stage 12 were emasculated and left to grow for 12–16 h before pollination assays.
Lysotracker red staining was used to show cell silhouettes as described (Wang et al., 2016). CLSM of fluorescence materials was performed with a LSM880 (Zeiss) with the excitation and emission wavelengths set to 488 nm/505–550 nm for YFP and GFP signals and 561 nm/600 nm for RFP signals, respectively.
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are: AT4G20910 for HEN1; AT1G09700 for HYL1; AT3G22886 for miRNA167; AT1G30330 for ARF6; AT5G37020 for ARF8.
To determine what caused the reduced fertility in hen1-8 (Chen et al., 2002; Yu et al., 2005, 2010), we performed the following experiments. First, we observed white and wrinkled ovules dispersed among developing seeds in the maturing siliques of hen1-8 plants, but not in those of wild type or of hen1-8/ + (Figures 1A,B), indicating that the reduced fertility is sporophytic. Indeed, segregation ratio by reciprocal crosses indicated that both the male and female gametophytes of hen1-8 were transmitted normally (Supplementary Table S2). Pollen development of hen1-8 is also comparable to that of wild type (Supplementary Figure S1). By dissecting siliques from crosses between wild type and hen1-8, we observed reduced fertility only when hen1-8 was used as the female parent (Figures 1A,B), suggesting that the reduced fertility of hen1-8 was due to sporophytic female defects.
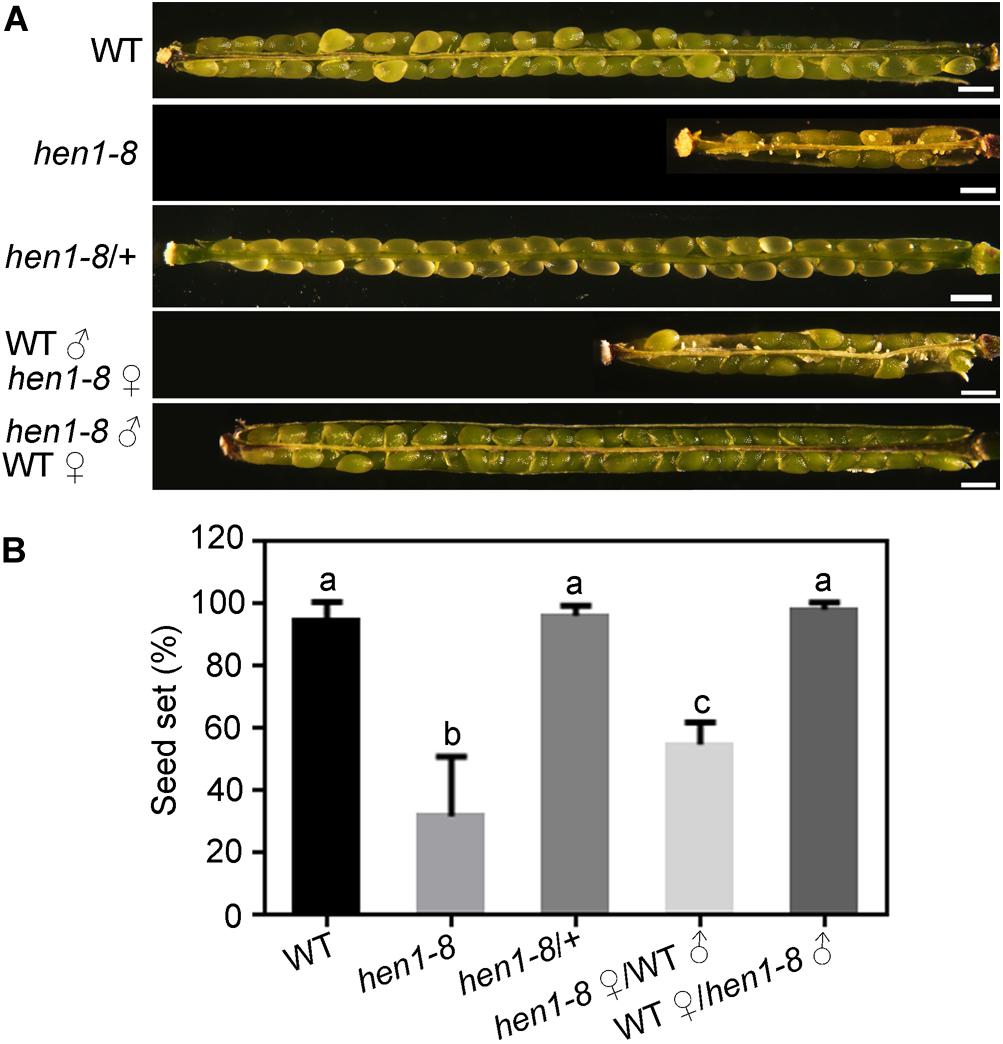
Figure 1. hen1-8 is significantly reduced in female fertility. (A) Representative seed set. For some, two overlapping high-magnification images were taken for one silique and were then overlaid with Photoshop (Adobe Systems) to show the whole silique. (B) Quantitative analysis of seed set. Results are means ± standard deviation (SD, n = 15). Different numbers indicate significantly different groups (One-Way ANOVA, Tukey’s multiple comparisons test, P < 0.05). Bars = 500 μm.
To determine the reason for sporophytic female defects that caused reduced fertility in hen1-8, we examined the morphology of mature ovules by scanning electron micrographs (SEMs) and whole-mount ovule clearing (Wang et al., 2016). At maturation, wild-type ovules showed a typical anatropy with the micropyle proximal to the funiculus (Figures 2A,B). An embryo sac was clearly seen in the mature ovule of wild type (Figure 2E). By contrast, micropyle structure was not discernible in a portion of hen1-8 ovules (Figures 2C,R). Instead, a bulge, likely a deformed embryo sac, was exposed (Figures 2D,G,H). These results suggested that ovule development is compromised in hen1-8.
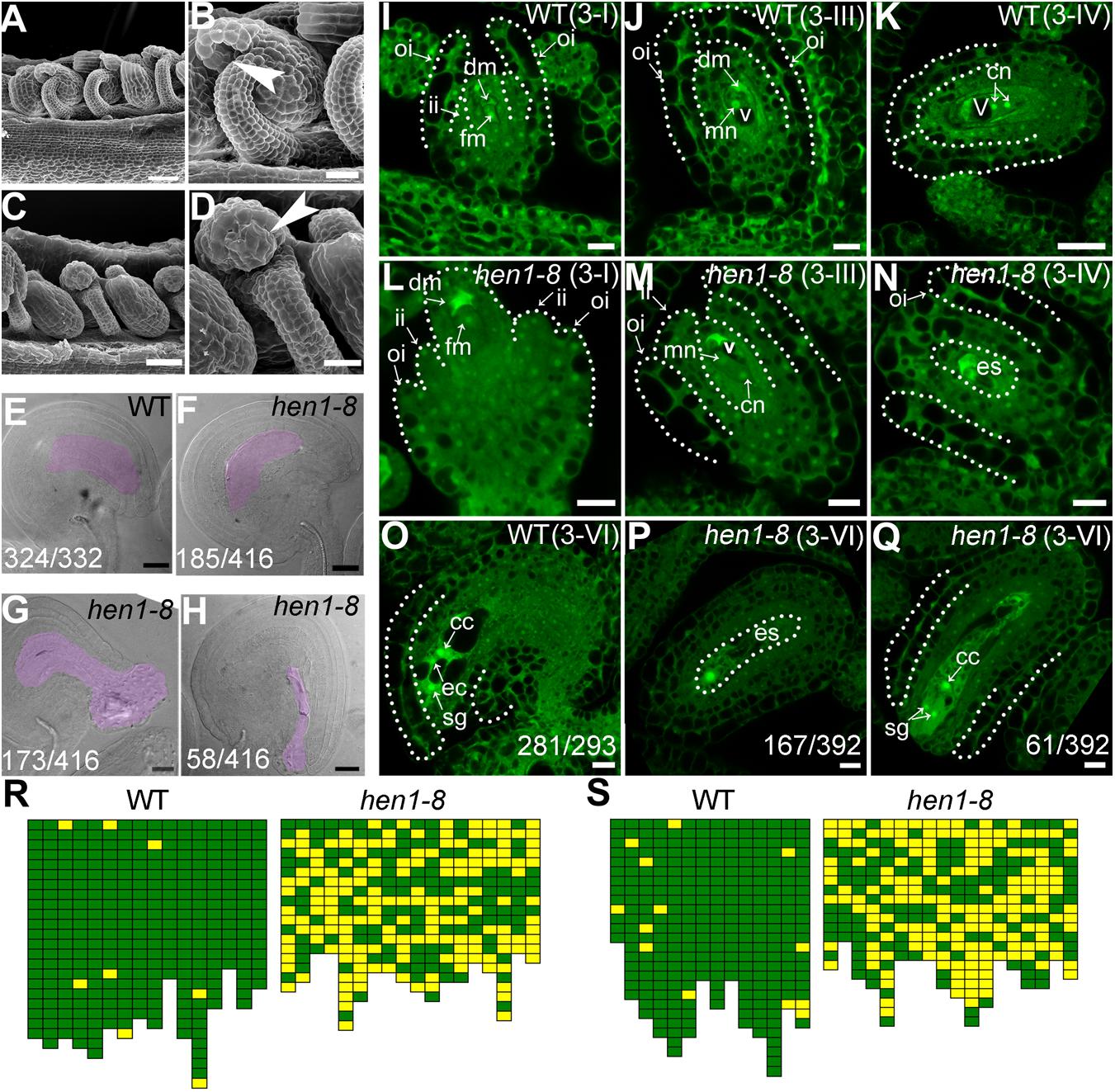
Figure 2. hen1-8 is defective in ovule development. (A–D) Scanning electron micrographs (SEMs) of mature ovules from wild type (A,B) or from hen1-8 (C,D). (B) and (D) are close-ups of (A) and (C), respectively. Arrows in (B,D) point at the micropyle. (E–H) Whole-mount ovule clearing of wild type (E) or hen1-8 (F–H). The ovule in (F) or in (G–H) represents normal or abnormal types, respectively. Embryo sacs are highlighted with lilac. (I–Q) Confocal laser scanning microscopy (CLSM) of wild-type (I–K,O) or hen1-8 (L–N,P,Q) ovules at stage 3-I (I,L), 3-III (J,M), 3-IV (K,N), or 3-VI (O–Q). Ovules were stained with PI and mid-optical sections are shown. For (I–Q), only hen1-8 ovules with visible nuclei were documented. For (E–H) and (O–Q), numbers on the bottom of each image indicate displayed ovules/total ovules examined. Cc, central cell; cn, chalazal nucleus; dm, degenerating megaspore; ec, egg cell; es, embryo sac; fm, functional megaspore; ii, inner integument; oi, outer integument; mn, micropylar nucleus; sg, synergid cell; v, vacuole. (R–S) Quantitative analysis of ovule development by ovule clearing (R) or by optical sections (S). Each pistil examined was represented by two neighboring columns; the number of cubes in each column indicates the number of countable ovules; normal and abnormal ovules (R) or embryo sacs (S) are displayed in green and yellow, respectively. Bars = 50 μm for (A,C); 20 μm for (B,D,E–H); 10 μm for (I–J,L–Q); 20 μm for (K).
To determine at which stage the hen1-8 ovules started to be defective, we performed confocal laser scanning microscopy (CLSM) of developing ovules. At early stages, i.e., before the meiosis of megaspore mother cell (MMC), hen1-8 and wild type are comparable although HEN1 is expressed in ovules throughout development (Supplementary Figure S2). However, at stage 3-I when the outer integuments of wild type started rapid and asymmetric growth, extending above the inner integuments (Figure 2I), the growth of hen1-8 outer integuments was delayed, hardly reaching the length of the inner integuments (Figure 2L). At this stage, functional megaspore (FM) was formed both in wild type and in hen1-8 (Figures 2I,L). In wild type, from stage 3-III to maturation, the outer integuments continued extended growth, finally enclosing the inner integuments (Figure 2J,K). Every mature ovules of wild type contains an embryo sac with a central cell, an egg cell and two synergid cells (Figure 2O). By contrast, the outer integuments of hen1-8 failed to enclose the inner structure (Figures 2M,N). At maturation, these ovules contain embryo sacs with abnormal cellular structures such that only one nucleus was visible (Figures 2P,Q,S).
To provide further evidence that HEN1 is the causative gene for the observed ovule defects in hen1-8, we performed additional experiments. Because hen1-2 is an allelic HEN1 mutant in Landsberg errecta (Ler) that contains exactly the same site mutation as in hen1-8 (Yu et al., 2010), we first examined ovule development of hen1-2 by CLSM. Indeed, hen1-2 showed the same ovule defects as those of hen1-8 (Supplementary Figure S3). Next, we crossed hen1-8 and hen1-2 and examined ovules of the F1 progenies. Ovules of the F1 progenies from the cross showed exactly the same defects (Supplementary Figure S4). These results indicated defective outer integument growth affected embryo sac development when HEN1 is mutated.
CLSM of ProES1:NLS-YFP;hen1-8 in which a nucleus-targeted YFP was driven by an embryo sac-specific promoter (Pagnussat et al., 2009), often showed one nucleus, sometimes no nucleus at all, in the embryo sac in contrast to the eight nuclei structure in wild-type ovules (Supplementary Figure S5), suggesting that embryo sac development was compromised due to sporophytic defects in hen1-8. This result is also consistent with those obtained by optical section of mature hen1-8 ovules (Figure 2).
Because the embryo sac within an ovule attracts pollen tubes for fertilization (Higashiyama and Yang, 2017), abnormal embryo sacs due to defective integument growth might be the reason for reduced female fertility in hen1-8 (Figure 1). To test this hypothesis, wild-type or hen1-8 pistils were emasculated, and hand-pollinated with ProLAT52:GUS pollen and pollen tube attraction at 12 h after pollination (HAP) was examined by histochemical GUS staining. In contrast to wild type in which almost all ovules were targeted by a pollen tube, as indicated by a blue blob inside embryo sacs (Figures 3A,C), over half of hen1-8 ovules failed to attract a pollen tube (Figures 3B,D). By aniline blue staining of pistils at 48 HAP, we determined that most wild-type ovules were fertilized as indicated by size increase (Figure 3E). By contrast, around half of hen1-8 ovules were not targeted by pollen tubes and were not fertilized (Figures 3F,G). Therefore, we concluded that defective embryo sac development in hen1-8 resulted in its reduced female fertility.
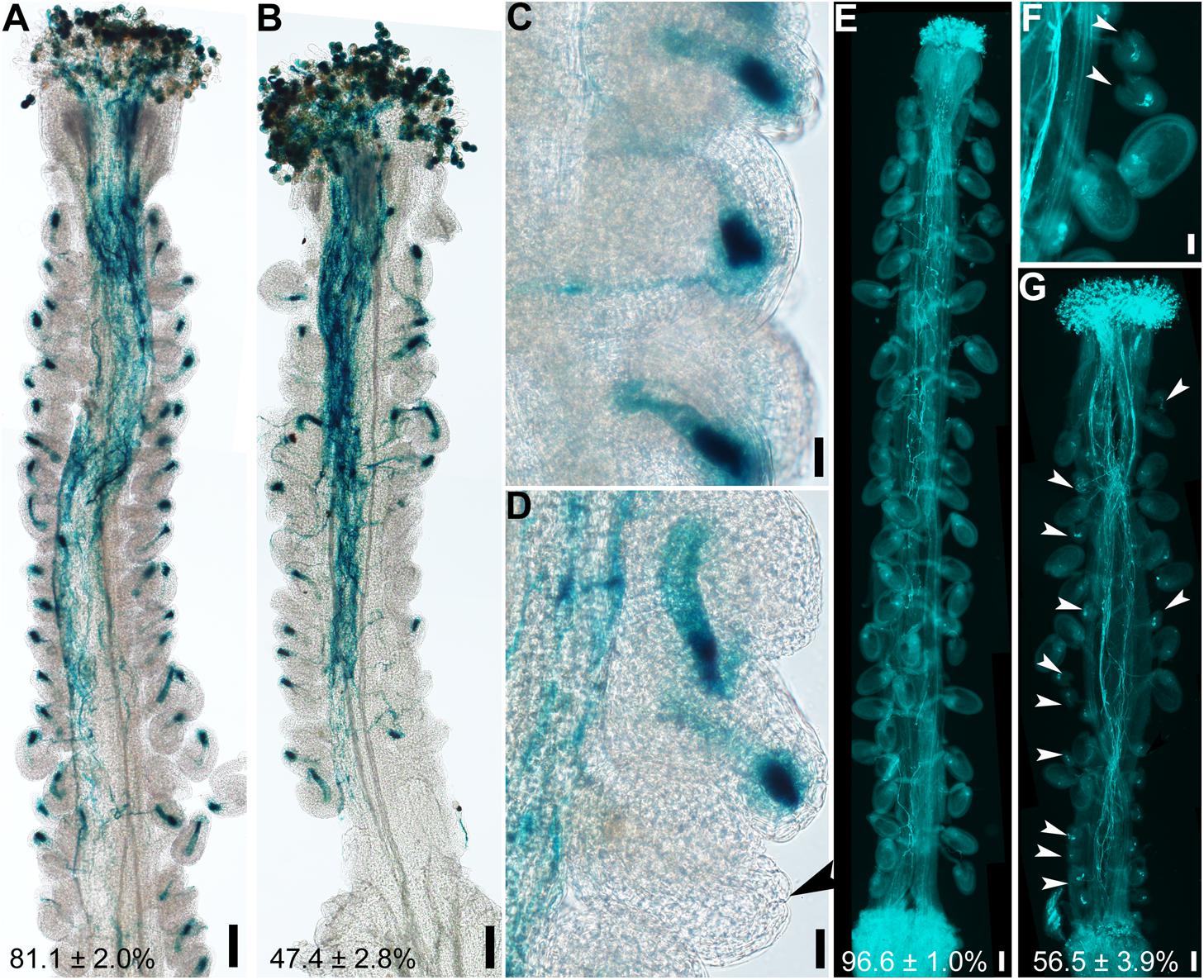
Figure 3. hen1-8 ovules are compromised in pollen tube guidance. (A–D) Histochemical GUS staining of wild-type (A,C) or hen1-8 (B,D) pistils at 12 h after pollination (HAP) with ProLAT52:GUS pollen. Arrowheads point at an ovule that failed to attract pollen. (E–G) Aniline blue staining of wild-type (E) or hen1-8 (F–G) pistils at 48 HAP with wild-type pollen. Arrowheads point at ovules that did not develop as a result of fertilization failure. Two to three overlapping high-magnification images were taken for one pistil and overlaid with Photoshop (Adobe) to show the whole pistil (A,B,E,G). Numbers at the bottom of (A), (B), (C), (D) are quantification of targeted ovules out of total ovules. Results are means ± SD (n = 15). Bars = 100 μm for (A,B,E,G), 20 μm for (C,D,F).
Ovule defects of hen1-8 were likely due to compromised miRNA processing because HEN1 is critical for the processing of various miRNAs (Yu et al., 2005, 2010; Zhao et al., 2012) and sin1/dcl1-7 showed a similar phenotype (Robinson-Beers et al., 1992). To provide further evidence that the miRNA processing pathway was critical for ovule development, we also examined hyl1-2 (Vazquez et al., 2004), a null mutant of HYL1 whose severely reduced fertility was restored to the wild-type level by exogenous HYL1 (Lian et al., 2013). Female gametophytes of hyl1-2 transmitted comparably to those of wild type (Xiong et al., 2020), indicating that HYL1 is not required for the development of female gametophytes. However, the homozygous hyl1-2 showed a significantly reduced seed set due to sporophytic female defects (Figures 4A–E). By ovule whole-mount analysis (Figures 4F–G), optical sections (Figures 4H–I), and SEM analysis (Figures 4L–M), we demonstrated that hyl1-2 was defective in ovule development due to the growth arrest of outer integuments. Because of the defects, hyl1-2 ovules showed a reduced ability to attract pollen tubes compared with those of wild type (Figures 4J–K), leading to significantly reduced female fertility (Figures 4B,D,E).
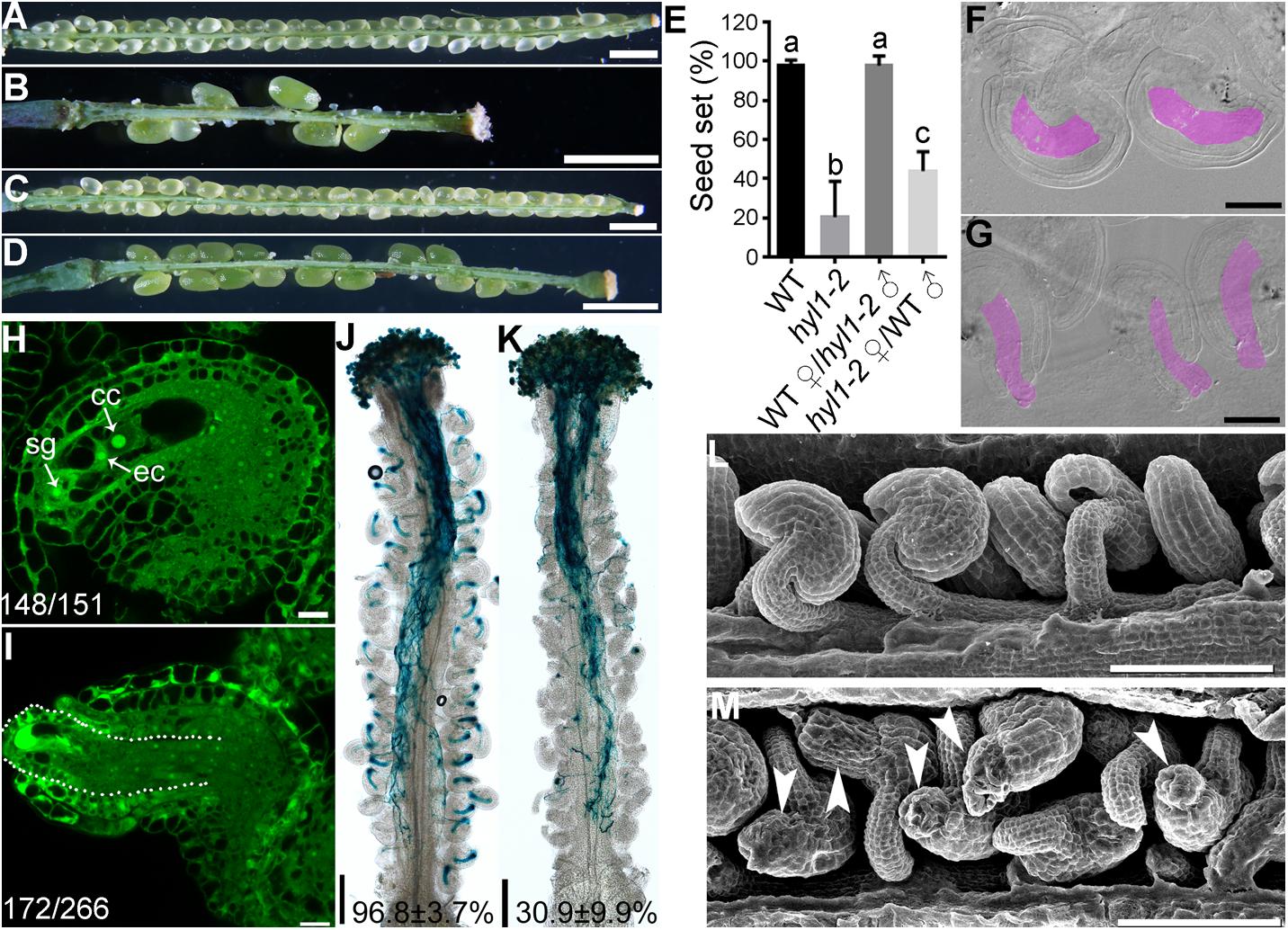
Figure 4. hyl1-2 is defective in ovule development, similar to hen1-8. (A–D) Representative silique of wild type (A), hyl1-2 (B), wild type pollinated with hyl1-2 pollen (C), or hyl1-2 pollinated with wild-type pollen (D). (E) Quantitative analysis of seed set. Results are means ± SD (n = 15). Different numbers indicate significantly different groups (One-Way ANOVA, Tukey’s multiple comparisons test, P < 0.05). (F,G) Whole-mount ovule clearing of wild type (F) or hyl1-2 (G). Embryo sacs are highlighted with lilac. (H,I) CLSM of wild-type (H) or hyl1-2 (I) ovules at stage 3-VI. Ovules were stained with PI and mid-optical sections are shown. Numbers on the bottom of each image indicate displayed ovules/total ovules examined. cc, central cell; ec, egg cell; sg, synergid cell. (J,K) Histochemical GUS staining of wild-type (J) or hyl1-2 (K) pistils at 12 HAP with ProLAT52:GUS pollen. Two to three overlapping high-magnification images were taken for one pistil and overlaid with Photoshop (Adobe) to show the whole pistil. Numbers at the bottom are quantification of targeted ovules out of total ovules. Results are means ± SD (n = 15). (L,M) SEMs of mature ovules from wild type (L) or from hyl1-2 (M). Arrowheads point at ovules with protruding embryo sac. Bars = 1 mm for (A–D); 50 μm for (F,G); 10 μm for (H–I); 200 μm for (J,K); 100 μm for (L,M).
hen1-8 shows vegetative growth retardation (Chen et al., 2002), which could have an impact in female fertility. To exclude the possibility that ovule developmental defect of hen1-8 was resulted from its reduced vegetative growth, we attempted to downregulate the expression of HEN1 specifically in outer integuments by using the outer integument-specific promoter ProINO (Wang et al., 2016). A dozen of transgenic lines containing either ProINO:amiR-HEN1 (artificial microRNA-HEN1) were generated. The transgenic plants were comparable to that of wild type regarding the growth of vegetative tissues (Supplementary Figure S6), consistent with the use of outer-integument-specific promoter. However, seed set of the transgenic plants was compromised (Figures 5B,C,F). We examined two independent transgenic lines representing mildly or severely affected types by CLSM. In the line of ProINO:amiR-HEN1 in which seed set was reduced by 50% (Figures 5B,F), half of the mature ovules showed abnormal number of nuclei in their embryo sacs while the integuments were morphologically indistinguishable from those of wild type (Figures 5G,H). In the line where there was hardly any seed set (Figures 5C,F), most mature ovules had no discernible outer integuments or nucleus structure in their embryo sacs (Figures 5I,J).
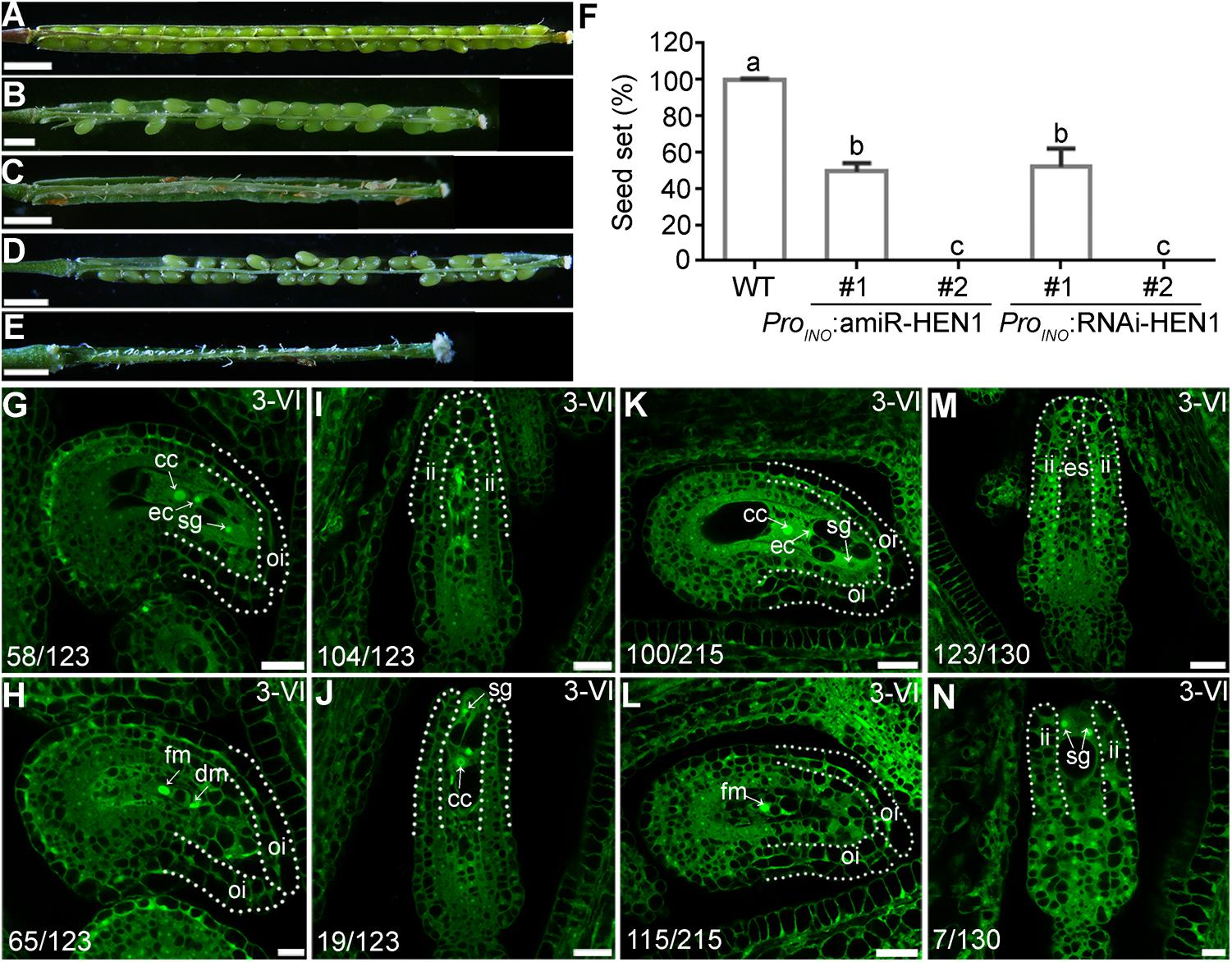
Figure 5. Downregulating HEN1 in outer integuments mimicked hen1-8 in ovule development. (A–E) Representative silique from wild type (A), two lines of ProINO:amiR-HEN1 (B,C), and two lines of ProINO:HEN1-RNAi (D,E) transgenic plants. For some pistils, two to three overlapping high-magnification images were taken and overlaid with Photoshop (Adobe). (F) Quantification of seed set. Results are means ± SD (n = 15). Different numbers indicate significantly different groups (One-Way ANOVA, Tukey’s multiple comparisons test, P < 0.05). (G–N) CLSM of representative ovules at stage 3-VI from the #1 (G,H) or #2 (I,J) line of ProINO:amiR-HEN1, and #1 (K,L) or #2 (M,N) line of ProINO:RNAi-HEN1. Numbers on the bottom of each image indicate displayed ovules/total ovules examined. Dotted lines illustrate either outer integuments (oi) or inner integuments (ii). Cc, central cell; ec, egg cell; sg, synergid. Bars = 1 mm for (A,C,D); 500 μm for (B,E); 20 μm for (G,I,K,M,J,L); 10 μm for (H, N).
Because the specificity of ProINO used to downregulating HEN1 (Wang et al., 2016) and the constitutive expression of HEN1 in ovules (Supplementary Figure S2), we could not verify the downregulation of HEN1 in the ProINO:amiR-HEN1 transgenic plants by quantitative real-time PCRs (qPCRs). Instead, we performed two experiments to support that HEN1 mediates integument growth in a cell-autonomous way. First, we generated Pro35S:amiR-HEN1 transgenic plants and examined the transcript abundance of HEN1 in transgenic seedlings by qPCRs. In randomly selected two Pro35S:amiR-HEN1 independent lines, HEN1 abundance was significantly reduced compared to that in wild type (Supplementary Figure S7), suggesting that the amiR-HEN1 expression did reduce the mRNA level of HEN1. Consistently, the transgenic plants were shorter and smaller than wild-type plants (Supplementary Figure S7). Second, we used a RNA interference (RNAi) approach instead of amiR to downregulate HEN1 specifically in outer integuments. The ProINO:HEN1-RNAi transgenic plants phenocopied ProINO:amiR-HEN1 in reduced seed set (Figures 5D,E,F) and defective ovule development (Figures 5K–N). These results suggested that HEN1 mediates outer integument growth in a cell-autonomous way.
Auxin is a determinant factor in ovule development (Benkova et al., 2003; Bencivenga et al., 2012; Ceccato et al., 2013). Both auxin receptors and response factors are regulated by miRNAs whose processing depends on the DCL1-HEN1-HYL1 pathway (Navarro et al., 2006; Gutierrez et al., 2009; Ren et al., 2012; Zhao et al., 2012). Therefore, we examined auxin responses by introducing DR5:GFP (Ulmasov et al., 1997) into hen1-8 and examining GFP distribution. In wild type, GFP signals were detected only in the epidermal cell layer of the nucellus at stage 2-III when both outer and inner integuments were initiated (Figure 6A) and at stage 3-I when outer integuments underwent rapid growth to establish ovule anatrophy (Figure 6B). At maturation, GFP signals were hardly visible in ovules except in the vascular tissues of the funiculus (Figure 6C). The GFP distribution of hen1-8 was similar to, albeit weaker than, that of wild type at early stages (Figure 6D) and at maturation in morphologically normal hen1-8 ovules (Figure 6F). However, at stage 3-I, auxin maximum was expanded from the nucellus to the developing female gametophytes of hen1-8 ovules (Figure 6E), suggesting a spatially disturbed auxin response.
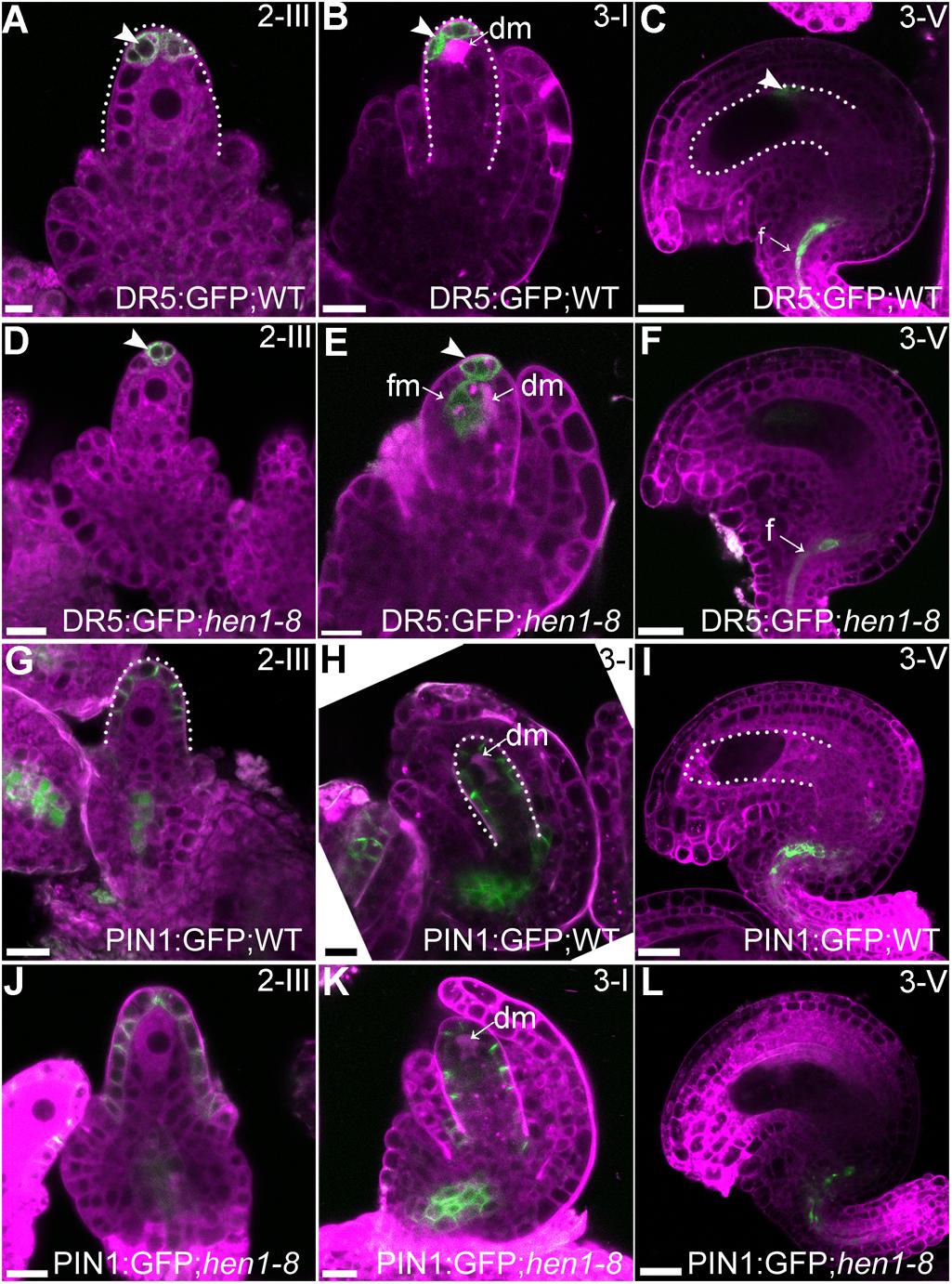
Figure 6. Auxin responses but not the asymmetric PIN1 localization were compromised in hen1-8 during ovule development. (A–F) DR5:GFP in wild-type (A–C) or in hen1-8 (D–F) ovules at different stages by CLSM. Arrowheads indicate DR5 signals. (G–L) PIN1:GFP in wild-type (G–I) or in hen1-8 (J–L) ovules at different stages by CLSM. Ovules were stained with lysotracker red (magenta). dm: degenerating megaspore. Bars = 10 μm.
Because PIN1 is the key auxin efflux carrier responsible for auxin distribution during ovule development (Ceccato et al., 2013), we also generated the PIN1:GFP;hen1-8 plants to examine its distribution. As reported previously (Ceccato et al., 2013; Wang et al., 2016), PIN1 was asymmetrically distributed at the epidermal cells of the nucellus during ovule development (Figures 6G–H) and restricted to the funiculus at maturation (Figure 6I). No difference of PIN1 distribution was observed between wild type and hen1-8 (Figures 6J–L), indicating that compromised auxin maximum in hen1-8 ovules was likely resulted from signaling rather than auxin transport.
Mutations of DCL1 (Robinson-Beers et al., 1992), HEN1, and HYL1 resulted in defective ovule development, suggesting a role of miRNAs in this process. Among miRNAs whose accumulation relies on HEN1 (Ren et al., 2012; Zhao et al., 2012), miRNA167 was demonstrated a positive regulator for ovule development by suppressing the expression of ARF6 and ARF8 (Wu et al., 2006; Yao et al., 2019). Indeed, ovule development of the mir167 mutants was largely recovered by introducing either arf6 or arf8 (Yao et al., 2019).
To test the possibility that ectopic expression of ARF6 and ARF8 resulted in the arrest of integuments in hen1-8, we performed the following experiments. First, we examined the expression of ARF6 and ARF8 in hen1-8 ovules by RNA in situ hybridization. As reported previously (Wu et al., 2006), ARF6 (Figures 7A,B) and ARF8 (Figures 7E–F) were highly expressed in the funiculus during ovule development in wild type. By contrast, signals of either ARF6 (Figures 7C,D) or ARF8 (Figures 7G,H) were detected in whole ovules of hen1-8, indicating its ectopic expression. Second, by quantitative RT-PCR (qRT-PCR), we could verify that transcript abundance of both ARF6 and ARF8 was significantly increased in hen1-8 ovules (Figures 7I,J). Third, we introduced the mutants of ARF6 and ARF8, i.e., arf6-1 and arf8-3 respectively, into hen1-8 and analyzed the resultant hierarchy mutants. Introducing arf6-1 or arf8-3 alone into hen1-8 did not affect vegetative growth whereas the arf6-1;arf8-3;hen1-8 showed a severe growth retardation (Supplementary Figure S8). Close examination of mature ovules from different genotypes showed that either arf6-1, or arf8-3, or the arf6-1;arf8-3 double mutant could not restore ovule developmental defects of hen1-8 (Supplementary Figure S9), suggesting that ectopic expression of ARF6 and ARF8 was not the reason for the arrest of hen1-8 integuments.
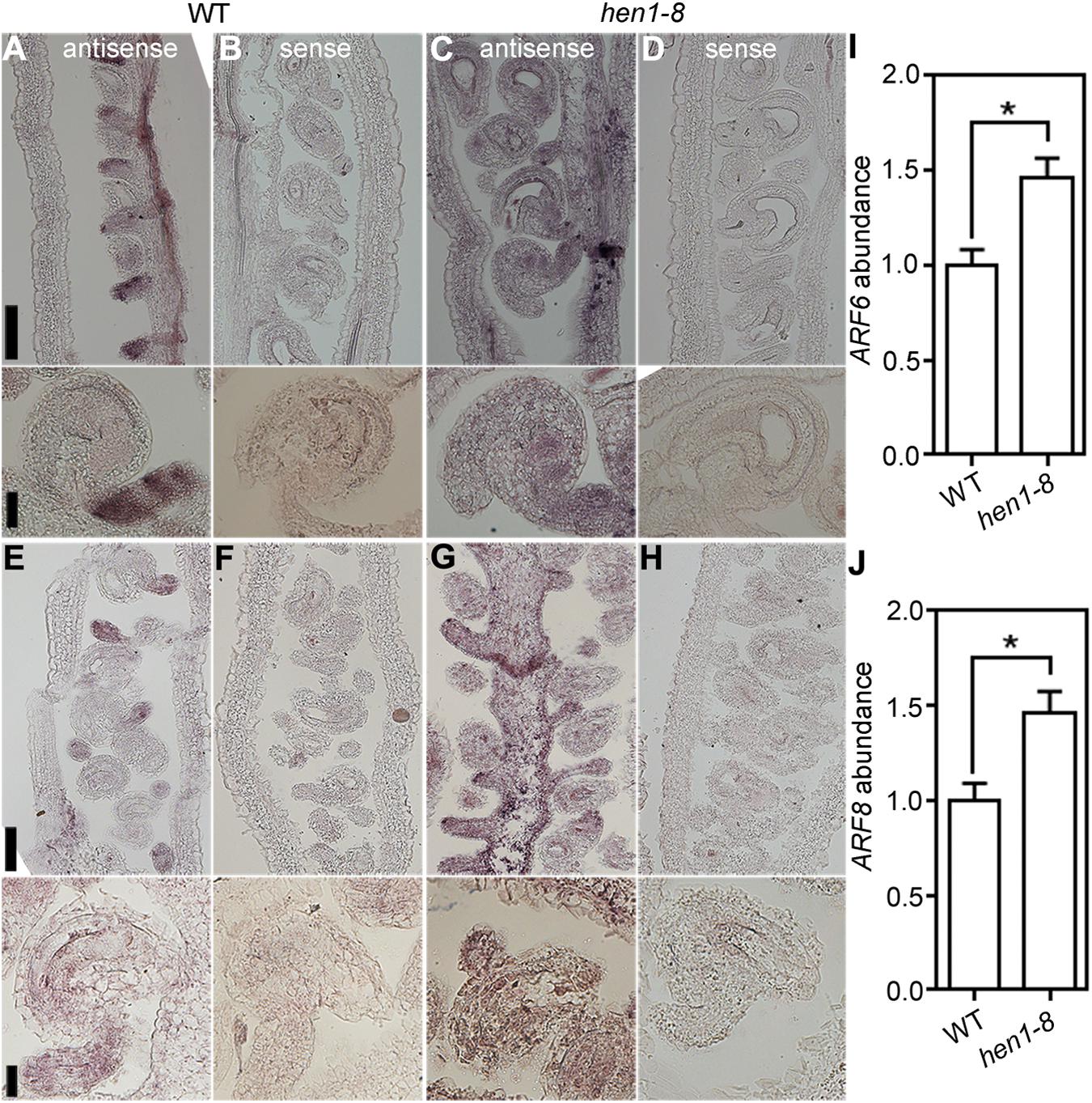
Figure 7. ARF6 and ARF8 are ectopically expressed in hen1-8 ovules. (A–D) Stage 3-IV wild-type (A,B) or hen1-8 (C,D) ovules labeled by the ARF6 antisense probe (A,C) or sense probe (B,D). (E–H) Stage 3-IV wild-type (E,F) or hen1-8 (G,H) ovule labeled by the ARF8 antisense probe (E,G) or sense probe (F,H). Funiculus of wild-type ovules whereas integuments of hen1-8 ovules show strong signals by the antisense probes. (I,J) Relative ARF6 (I) or ARF8 (J) abundance in mature ovules of wild type or hen1-8 by qRT-PCRs. GAPDH and TUBULIN2 were used as internal controls. Results shown are means ± SE (n = 3). Asterisks indicate significant difference (t-test, P < 0.01). Bars = 50 μm for whole views of pistils and 20 μm for close-ups of ovules.
We demonstrate here that mutations of HEN1 and HYL1 resulted in the arrest of integument growth during ovule development, similar to those of DCL1. Because female gametophyte of hen1-8 (Supplementary Table S2) and hyl1-2 (Xiong et al., 2020) transmits comparably that of wild type, the embryo sac defects observed in hen1-8 and hyl1-2, possibly also in sin1/dcl1-7 (Robinson-Beers et al., 1992), were resulted from sporophytic defects through intercellular signaling (Bencivenga et al., 2011), as being reported (Wang et al., 2016; Liu et al., 2019). Embryo sacs send signals to guide directional pollen tube growth (Higashiyama and Yang, 2017). It is therefore understandably that hen1-8 or hyl1-2 ovules showed a significantly reduced ability to attract pollen tubes (Figures 3, 4). hen1-8 is a hypomorphic allele (Yu et al., 2010) whereas hyl1-2 is a null allele (Han et al., 2004; Kurihara et al., 2006; Dong et al., 2008), which explains the different severity of ovule defects (Figures 1, 4).
A recent study reported that the mutations of HYL1, DCL1, or HEN1 caused a reduced number of pollen and megaspore mother cells (Oliver et al., 2017). Our results strongly suggested that fertility reduction in hen1-8 was due to abnormal ovule development (Figure 3). First, hen1-8 as the pollen donor to wild type resulted in a full seed set (Figure 1), indicating normal pollen function. Second, pollen development is comparable between hen1-8 and wild type, despite the relatively small size of hen1-8 anthers (Supplementary Figure S1). Third, the heterozygous hen1-8 mutant also produced full seed set (Figure 1), arguing against female gametophytic defects. Indeed, reciprocal crosses between wild type and the heterozygous hen1-8 indicated that both male and female transmission of hen1-8 are normal (Supplementary Table S1). The discrepancy between the previous report (Oliver et al., 2017) and ours might be due to the fact that hen1-8 and hen1-2 are weaker alleles of HEN1.
Despite that the hen1-8 plants showed sub-optical vegetative growth (Chen et al., 2002; Ren et al., 2012), we believe that HEN1 works in a cell-autonomous way to regulate the asymmetric growth of outer integuments. Downregulating HEN1 specifically in outer integuments was sufficient to cause ovule defect similar to, even more severe than, that of hen1-8 (Figure 5), without affecting vegetative growth (Supplementary Figure S6). Because the cell-specific feature of these transgenic lines, it is difficult, if possible, to examine the reduction of HEN1 transcript abundance. To make it additionally difficult, in the more severely affected RNAi or amiR lines, the growth of outer integuments was arrested very early on. But the two different constructs used to downregulating HEN1 in outer integuments gave the same results, strongly supporting a cell autonomous role of HEN1 in outer integuments.
The accumulation of miRNA167 was significantly reduced in hen1 mutants (Yu et al., 2010; Ren et al., 2012). Consistently, ARF6 and ARF8, major targets of miRNA167 (Nagpal et al., 2005; Wu et al., 2006; Yao et al., 2019; Zheng et al., 2019), were ectopically expressed in hen1-8 (Figure 7). However, introducing arf6 or arf8 did not suppress developmental defects of hen1-8 ovules (Supplementary Figure S9). The inability is unlikely to have caused by substantially compromised growth of the arf6-1;arf8-3;hen1-8 triple mutant since introducing either arf6-1 or arf8-3 didn’t aggravate the growth of hen1-8 but yet was not able to rescue its defects (Supplementary Figure S8). A more likely possibility is that more miRNAs downstream of HEN1 play roles in this process. Auxin maximum was altered in developing ovules of hen1-8 such that DR5 signals were expanded to the developing female gametophytes in hen1-8 rather than restricted to the nucellus as in wild type (Figure 6). Because the sporophytic integuments affect FG development (Bencivenga et al., 2011; Wang et al., 2016; Liu et al., 2019), these results indicated that auxin signaling in integuments was compromised in hen1-8. Indeed, genes encoding auxin receptors are also targets of miRNAs (Navarro et al., 2006; Gutierrez et al., 2009). A genome-wide small RNA sequencing will be useful to identify miRNAs that are expressed in integuments and whose reduced levels result in the arrest of integument growth in mutants of the DCL1-HEN1-HYL1 pathway.
All datasets generated for this study are included in the article/Supplementary Material.
S-JW and SC performed all the experiments with the assistance of R-MZ and C-YD. SL and YZ conceived and supervised the project and secured the funding. S-JW, SL, and YZ analyzed the data. YZ wrote the article with input from all authors.
This work was supported by Natural Science Foundation of China (31871422 and 31771558 to SL, 31970332 and 31625003 to YZ). YZ’s laboratory is partially supported by Tai-Shan Scholar Program by Shandong Provincial Government.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Xian Sheng Zhang for DR5:GFP and PIN1:GFP; Prof. Guo-Dong Ren for the kind gift of hen1-8 and hen1-2; Prof. Yijun Qi for hyl1-2.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00397/full#supplementary-material
Baranauske, S., Mickute, M., Plotnikova, A., Finke, A., Venclovas, C., Klimasauskas, S., et al. (2015). Functional mapping of the plant small RNA methyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins. Nucleic Acids Res. 43, 2802–2812. doi: 10.1093/nar/gkv102
Bencivenga, S., Colombo, L., and Masiero, S. (2011). Cross talk between the sporophyte and the megagametophyte during ovule development. Sex Plant Reprod. 24, 113–121. doi: 10.1007/s00497-011-0162-3
Bencivenga, S., Simonini, S., Benkova, E., and Colombo, L. (2012). The transcription factors BEL1 and SPL are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Benkova, E., Michniewicz, M., Sauer, M., Teichmann, T., Seifertova, D., Jurgens, G., et al. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115, 591–602. doi: 10.1016/s0092-8674(03)00924-3
Ceccato, L., Masiero, S., Sinha Roy, D., Bencivenga, S., Roig-Villanova, I., Ditengou, F. A., et al. (2013). Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS One 8:e66148. doi: 10.1371/journal.pone.0066148
Chen, X., Liu, J., Cheng, Y., and Jia, D. (2002). HEN1 functions pleiotropically in Arabidopsis development and acts in C function in the flower. Development 129, 1085–1094.
Chevalier, E., Loubert-Hudon, A., Zimmerman, E. L., and Matton, D. P. (2011). Cell-cell communication and signalling pathways within the ovule: from its inception to fertilization. New Phytol. 192, 13–28. doi: 10.1111/j.1469-8137.2011.03836.x
Christensen, C. A., King, E. J., Jordan, J. R., and Drews, G. N. (1997). Megagametogenesis in Arabidopsis wild type and the Gf mutant. Sex Plant Reprod. 10, 49–64. doi: 10.1007/s004970050067
Colombo, L., Battaglia, R., and Kater, M. M. (2008). Arabidopsis ovule development and its evolutionary conservation. Trends Plant Sci. 13, 444–450. doi: 10.1016/j.tplants.2008.04.011
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Dong, Z., Han, M. H., and Fedoroff, N. (2008). The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 9970–9975. doi: 10.1073/pnas.0803356105
Drews, G. N., Lee, D., and Christensen, C. A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. doi: 10.1105/tpc.10.1.5
Drews, G. N., and Yadegari, R. (2002). Development and function of the angiosperm female gametophyte. Annu. Rev. Genet. 36, 99–124. doi: 10.1146/annurev.genet.36.040102.131941
Elliott, R. C., Betzner, A. S., Huttner, E., Oakes, M. P., Tucker, W. Q., Gerentes, D., et al. (1996). AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell 8, 155–168. doi: 10.1105/tpc.8.2.155
Gross-Hardt, R., Lenhard, M., and Laux, T. (2002). WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 16, 1129–1138. doi: 10.1101/gad.225202
Guo, J., Wang, F., Song, J., Sun, W., and Zhang, X. S. (2010). The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231, 293–303. doi: 10.1007/s00425-009-1051-y
Gutierrez, L., Bussell, J. D., Pacurar, D. I., Schwambach, J., Pacurar, M., and Bellini, C. (2009). Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 21, 3119–3132. doi: 10.1105/tpc.108.064758
Han, M. H., Goud, S., Song, L., and Fedoroff, N. (2004). The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl. Acad. Sci. U.S.A. 101, 1093–1098. doi: 10.1073/pnas.0307969100
Higashiyama, T., and Yang, W. C. (2017). Gametophytic pollen tube guidance: attractant peptides, gametic controls, and receptors. Plant Physiol. 173, 112–121. doi: 10.1104/pp.16.01571
Klucher, K. M., Chow, H., Reiser, L., and Fischer, R. L. (1996). The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell 8, 137–153. doi: 10.1105/tpc.8.2.137
Kurihara, Y., Takashi, Y., and Watanabe, Y. (2006). The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA 12, 206–212. doi: 10.1261/rna.2146906
Kurihara, Y., and Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 101, 12753–12758. doi: 10.1073/pnas.0403115101
Li, S., Ge, F. R., Xu, M., Zhao, X. Y., Huang, G. Q., Zhou, L. Z., et al. (2013). Arabidopsis COBRA-LIKE 10, a GPI-anchored protein, mediates directional growth of pollen tubes. Plant J. 74, 486–497. doi: 10.1111/tpj.12139
Lian, H., Li, X., Liu, Z., and He, Y. (2013). HYL1 is required for establishment of stamen architecture with four microsporangia in Arabidopsis. J. Exp. Bot. 64, 3397–3410. doi: 10.1093/jxb/ert178
Lieber, D., Lora, J., Schrempp, S., Lenhard, M., and Laux, T. (2011). Arabidopsis WIH1 and WIH2 genes act in the transition from somatic to reproductive cell fate. Curr. Biol. 21, 1009–1017. doi: 10.1016/j.cub.2011.05.015
Liu, H. H., Xiong, F., Duan, C. Y., Wu, Y. N., Zhang, Y., and Li, S. (2019). Importin β4 mediates nuclear import of GRF-interacting factors to control ovule development in Arabidopsis. Plant Physiol. 179, 1080–1092. doi: 10.1104/pp.18.01135
Lu, C., and Fedoroff, N. (2000). A mutation in the Arabidopsis HYL1 gene encoding a dsRNA binding protein affects responses to abscisic acid, auxin, and cytokinin. Plant Cell 12, 2351–2366.
McAbee, J. M., Hill, T. A., Skinner, D. J., Izhaki, A., Hauser, B. A., Meister, R. J., et al. (2006). ABERRANT TESTA SHAPE encodes a KANADI family member, linking polarity determination to separation and growth of Arabidopsis ovule integuments. Plant J. 46, 522–531. doi: 10.1111/j.1365-313X.2006.02717.x
Nagpal, P., Ellis, C. M., Weber, H., Ploense, S. E., Barkawi, L. S., Guilfoyle, T. J., et al. (2005). Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development 132, 4107–4118. doi: 10.1242/dev.01955
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Oliver, C., Pradillo, M., Jover-Gil, S., Cunado, N., Ponce, M. R., and Santos, J. L. (2017). Loss of function of Arabidopsis microRNA-machinery genes impairs fertility, and has effects on homologous recombination and meiotic chromatin dynamics. Sci. Rep. 7:9280. doi: 10.1038/s41598-017-07702-x
Olmedo-Monfil, V., Duran-Figueroa, N., Arteaga-Vazquez, M., Demesa-Arevalo, E., Autran, D., Grimanelli, D., et al. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. doi: 10.1038/nature08828
Pagnussat, G. C., Alandete-Saez, M., Bowman, J. L., and Sundaresan, V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689. doi: 10.1126/science.1167324
Reiser, L., Modrusan, Z., Margossian, L., Samach, A., Ohad, N., Haughn, G. W., et al. (1995). The BELL1 gene encodes a homeodomain protein involved in pattern formation in the Arabidopsis ovule primordium. Cell 83, 735–742. doi: 10.1016/0092-8674(95)90186-8
Ren, G., Chen, X., and Yu, B. (2012). Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr. Biol. 22, 695–700. doi: 10.1016/j.cub.2012.02.052
Robinson-Beers, K., Pruitt, R. E., and Gasser, C. S. (1992). Ovule development in wild-type Arabidopsis and two female-sterile mutants. Plant Cell 4, 1237–1249. doi: 10.1105/tpc.4.10.1237
Rogers, K., and Chen, X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25, 2383–2399. doi: 10.1105/tpc.113.113159
Schauer, S. E., Jacobsen, S. E., Meinke, D. W., and Ray, A. (2002). DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 7, 487–491. doi: 10.1016/s1360-1385(02)02355-5
Schneitz, K., Hulskamp, M., Kopczak, S. D., and Pruitt, R. E. (1997). Dissection of sexual organ ontogenesis: a genetic analysis of ovule development in Arabidopsis thaliana. Development 124, 1367–1376.
Schneitz, K., Hülskamp, M., and Pruitt, R. E. (1995). Wild-type ovule development in Arabidopsis thaliana: a light microscope study of cleared whole-mount tissue. Plant J. 7, 731–749. doi: 10.1046/j.1365-313X.1995.07050731.x
Shi, D. Q., and Yang, W. C. (2011). Ovule development in Arabidopsis: progress and challenge. Curr. Opin. Plant Biol. 14, 74–80. doi: 10.1016/j.pbi.2010.09.001
Sieber, P., Gheyselinck, J., Gross-Hardt, R., Laux, T., Grossniklaus, U., and Schneitz, K. (2004). Pattern formation during early ovule development in Arabidopsis thaliana. Dev. Biol. 273, 321–334. doi: 10.1016/j.ydbio.2004.05.037
Ulmasov, T., Murfett, J., Hagen, G., and Guilfoyle, T. J. (1997). Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9, 1963–1971. doi: 10.1105/tpc.9.11.1963
Vazquez, F., Gasciolli, V., Crete, P., and Vaucheret, H. (2004). The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 14, 346–351. doi: 10.1016/j.cub.2004.01.035
Villanueva, J. M., Broadhvest, J., Hauser, B. A., Meister, R. J., Schneitz, K., and Gasser, C. S. (1999). INNER NO OUTER regulates abaxial- adaxial patterning in Arabidopsis ovules. Genes Dev. 13, 3160–3169. doi: 10.1101/gad.13.23.3160
Wang, J. G., Feng, C., Liu, H. H., Ge, F. R., Li, S., Li, H. J., et al. (2016). HAPLESS13-mediated trafficking of STRUBBELIG is critical for ovule development in Arabidopsis. PLoS Genet. 12:e1006269. doi: 10.1371/journal.pgen.1006269
Wu, M. F., Tian, Q., and Reed, J. W. (2006). Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 133, 4211–4218. doi: 10.1242/dev.02602
Xiong, F., Duan, C. Y., Liu, H. H., Wu, J. H., Zhang, Z. H., Li, S., et al. (2020). Arabidopsis KETCH1 is critical for the nuclear accumulation of ribosomal proteins and gametogenesis. Plant Cell (in press). doi: 10.1105/tpc.19.00791
Yang, S. W., Chen, H. Y., Yang, J., Machida, S., Chua, N. H., and Yuan, Y. A. (2010). Structure of Arabidopsis HYPONASTIC LEAVES1 and its molecular implications for miRNA processing. Structure 18, 594–605. doi: 10.1016/j.str.2010.02.006
Yao, X., Chen, J., Zhou, J., Yu, H., Ge, C., Zhang, M., et al. (2019). An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 180, 453–464. doi: 10.1104/pp.19.00127
Yu, B., Bi, L., Zhai, J., Agarwal, M., Li, S., Wu, Q., et al. (2010). siRNAs compete with miRNAs for methylation by HEN1 in Arabidopsis. Nucleic Acids Res. 38, 5844–5850. doi: 10.1093/nar/gkq348
Yu, B., Yang, Z., Li, J., Minakhina, S., Yang, M., Padgett, R. W., et al. (2005). Methylation as a crucial step in plant microRNA biogenesis. Science 307, 932–935. doi: 10.1126/science.1107130
Zhao, Y., Mo, B., and Chen, X. (2012). Mechanisms that impact microRNA stability in plants. RNA Biol. 9, 1218–1223. doi: 10.4161/rna.22034
Zheng, L., Nagpal, P., Villarino, G., Trinidad, B., Bird, L., Huang, Y., et al. (2019). miR167 limits anther growth to potentiate anther dehiscence. Development 146:dev174375. doi: 10.1242/dev.174375
Keywords: integument, fertility, microRNA, HYL1, female gametophytes
Citation: Wei S-J, Chai S, Zhu R-M, Duan C-Y, Zhang Y and Li S (2020) HUA ENHANCER1 Mediates Ovule Development. Front. Plant Sci. 11:397. doi: 10.3389/fpls.2020.00397
Received: 07 February 2020; Accepted: 19 March 2020;
Published: 15 April 2020.
Edited by:
Yuling Jiao, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, ChinaReviewed by:
Wen-hui Lin, Shanghai Jiao Tong University, ChinaCopyright © 2020 Wei, Chai, Zhu, Duan, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhang, eXpoYW5nQHNkYXUuZWR1LmNu; Sha Li, c2hhbGlAc2RhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.