- 1College of Agriculture, Ludong University, Yantai, China
- 2Key Laboratory of Molecular Module-Based Breeding of High Yield and Abiotic Resistant Plants in Universities of Shandong, Ludong University, Yantai, China
- 3National Key Facility for Crop Gene Resources and Genetic Improvement, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 4Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
Timing of flowering is crucial for the transformation from vegetative to reproductive growth in the important food crop, wheat (Triticum aestivum L.). The circadian clock is a central part of photoperiod regulation, with Pseudo-Response Regulators (PRRs) representing key components of circadian networks. However, little is known about the effects of PRR family members on yield-related traits in crop plants. In this study, we identified polymorphisms and haplotypes of TaPRR1, demonstrating that natural variations in TaPRR1 are associated with significant differences in yield-related traits including heading date, plant height and thousand grain weight. TaPRR1-6A-Hapla showed an earlier heading date, advanced by 0.9 to 1.7%. TaPRR1-6B-Hapla and TaPRR1-6D-Hapla displayed reduced plant height and increased thousand grain weight of up to 13.3 to 26.4% and 6.3 to 17.3%, respectively. Subcellular localization and transcriptional activity analysis showed that TaPRR1 is a nuclear localization protein with transcriptional activity controlled by an IR domain. The expression profiles of TaPRR1 genes over a 48-h period were characterized by circadian rhythms, which had two peaks under both short- and long- day conditions. In addition, geographical distribution analysis indicated higher distribution frequencies of TaPRR1-6A-Hapla, TaPRR1-6B-Haplb, and TaPRR1-6D-Haplb in different agro-ecological production zones. Furthermore, analysis of molecular variance of the distribution frequency of TaPRR1 haplotypes suggested significant differences in haplotype distribution frequency between landraces and modern cultivars. Our study provides a basis for in-depth understanding of TaPRR1 function on yield-related traits in wheat, as well as establishing theoretical guidance for wheat molecular marker-assisted breeding.
Introduction
Wheat (Triticum aestivum L., AABBDD) is one of the most important food crops and sources of calories for humans. During growth, the timing of flowering (heading date in cereal crops) is key for the transformation of wheat from vegetative to reproductive growth. Photoperiod response genes not only affect the growth phase, but also play an important role in the adaptability and crop yield of wheat. In the photoperiod pathway, the photoreceptor senses a light signal and transmits it to the circadian clock, which generates biological rhythms and exports them to downstream-regulated genes. The circadian clock is a central part of photoperiod regulation, the spontaneous mechanisms of biological rhythms with a cycle of nearly 24 h (Niinuma et al., 2007). It is an internal time-keeping mechanism that provides an adaptive advantage by enabling organisms to anticipate daily changes and orchestrate biological processes accordingly. Circadian regulated Pseudo-Response Regulators (PRRs) are key components of circadian networks in plants (Farré and Liu, 2013). PRRs were first described in Arabidopsis and have been demonstrated to play a key role in the regulation of the circadian oscillator and clock output processes (Makino et al., 2000; Strayer et al., 2000; Harmer, 2009).
PRR1 (Pseudo-response regulator 1), also known as TOC1 (TIMING OF CAB EXPRESSION 1), is a member of the PRR gene family. The Arabidopsis PRR gene family consists of five members (PRR9, PRR7, PRR5, PRR3, and PRR1), all of which are regulated by the circadian clock, but each of which peaks at a different time of day (Matsushika et al., 2000). PRR1 together with two MYB transcription factors, CCA1 (CIRCADIAN CLOCK-ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL), constitute the main feedback loop of the central oscillator. Reciprocal transcriptional regulation between PRR1, CCA1, and LHY was proposed to be essential for circadian rhythmicity, with PRR1 repressing the transcription of CCA1 and LHY (Harmer et al., 2000; Alabadí et al., 2001; Gendron et al., 2012; Huang et al., 2012). Arabidopsis prr1-1 exhibits a shortened period phenotype in multiple rhythms and altered circadian clock regulation of multiple outputs during development (Somers et al., 1998; Strayer et al., 2000). Silencing or overexpression of PRR1 abolishes photoperiod rhythmicity and changes the light sensitivity in the control of hypocotyl elongation, indicating that PRR1 has dual functionality, not only in the integration of light signals to control circadian rhythm, but also in the photomorphogenic response (Más et al., 2003).
PRR gene family members also play an important role in wheat. Ppd-1 (TaPRR37), the famous gene conferring photoperiod insensitivity in “Green Revolution” cultivars, belongs to the PRR gene family and has made a significant contribution to wheat yield improvement (Laurie, 1997; Worland and Snape, 2001; Turner et al., 2005; Beales et al., 2007). Genetic and epigenetic variations in the Ppd-1 gene have been used to develop specific molecular markers for polymorphic genetic loci and classify Ppd-1 haplotypes. Genetic variations in the Ppd-1 gene not only affect the photoperiod sensitivity of wheat, but are also associated with important agronomic traits, including plant height and thousand grain weight (Beales et al., 2007; Wilhelm et al., 2009; Guo et al., 2010; Seki et al., 2011; Díaz et al., 2012; Nishida et al., 2013; Sun et al., 2014; Arjona et al., 2018). TaPRR73 is a paralog of Ppd-1 known to affect heading date and plant height in wheat. Overexpression of TaPRR73 accelerates flowering under long photoperiods (Zhang et al., 2016). TaPRR1 has also been reported in wheat. MicroRNA tae-miR408 functions in wheat heading time by mediating the expression of TaPRR1, one of its targets. Transgenic plants with knockdown of TaPRR1 and overexpression of tae-miR408 show an early heading phenotype (Zhao et al., 2016). Although some progress has been made in functional studies of wheat PRR family members, studies defining the influence of the circadian clock core member TaPRR1 on yield-related traits, as well as functional polymorphic sites, characteristics of haplotype classification and geographical distribution, and the development and application of molecular markers, are still relatively lacking.
In our current study, we further analyzed the functions of the circadian clock gene TaPRR1. We discovered and evaluated haplotypes of the wheat TaPRR1 gene. Association analysis using natural populations in multiple environments indicated that TaPRR1 is significantly associated with important agronomic traits, including plant height, thousand grain weight, and heading time. Geographical distribution analysis indicated that TaPRR1 haplotypes show distinctive characteristics in different agro-ecological production zones. Furthermore, assessment of the distribution frequency of the different haplotypes suggested that TaPRR1 genes have undergone selection during the wheat breeding process. Our study provides not only a theoretical basis for understanding the functional mechanisms of the wheat PRR gene family, but also some theoretical guidance for wheat molecular marker-assisted breeding.
Materials and Methods
Plant Material
Hexaploid wheat (T. aestivum) cultivar “Chinese Spring” (CS) and diploid varieties T. urartu UR102 (AA), Aegilops Tauschii Y215 (DD), and A. Speltoides Y2022 (SS) were used for TaPRR1 gene cloning. Twenty-eight accessions with wide variations in heading date and yield-related traits (Supplementary Table S1) were used to detect naturally occurring variations in TaPRR1 gene sequences. A total of 177 wheat accessions, including 129 accessions from the Chinese wheat Mini Core Collection (MCC) (Hao et al., 2011), 40 germplasm resources preserved in the Chinese Academy of Agricultural Sciences (CAAS, Beijing, China), and eight synthetic hexaploid wheat lines, were screened for TaPRR1 genotype and used for association analysis of TaPRR1 (Supplementary Table S2). The above materials were separately planted in Luoyang (111.6°E, 33.8°N), Xinxiang (113.8°E, 35.2°N), Changping (116.2°E, 40.2°N), Beijing (116.2°E, 39.5°N), Bashang (115.7°E, 41.7°N), Jiaozuo (113.2°E, 35.1°N), and Shunyi (116.5°E, 40.1°N) during the years 2006–2015. The characteristics of these locations are described in Supplementary Table S3. Six plants showing even growth were selected for each genotype for investigating agronomic traits including effective tiller number (ETN), plant height (PH), spike length (SL), total number of spikelets per spike (TNSS), number of grains per spike (NGS), thousand grain weight (TGW), and heading date (HD). For geographic distribution analysis, a total of 177 wheat accessions spanning a wide geographical area were evaluated (Supplementary Table S2). The Chinese accessions comprised landraces and modern wheat cultivars derived from the 10 major agroecological wheat regions (Jin, 1983) and were selected from the Chinese wheat MCC (Hao et al., 2011). Accessions chosen from Asia, Europe, North America, South America, and Oceania were representative accessions with a rich diversity of photoperiod responses selected from both the MCC and CAAS. For haplotype frequencies analysis, a total of 124 wheat accessions, including 60 modern cultivars, 56 landraces, and eight synthetic hexaploid wheat lines, were investigated (Supplementary Table S4).
Subcellular Localization
Coding sequences of TaPRR1-D1 were cloned into p2GWF7 using Gateway cloning technology (Invitrogen, United States) to generate the TaPRR1-D1-GFP construct (Karimi et al., 2002). Similarly, the coding sequence of OsMADS15 was cloned into p2GWR7 to generate OsMADS15-RFP as a nuclear marker. TaPRR1-D1-GFP and OsMADS15-RFP constructs were co-transformed into rice protoplasts as described previously (Bart et al., 2006). After 20–24 h of incubation at 28°C, green fluorescent protein (GFP) and red fluorescent protein (RFP) fluorescence were observed using a confocal laser-scanning microscope (Carl Zeiss LAM510, Germany).
Transactivation Activity Assay
TaPRR1-D1 proteins with no functional domain (1–29 aa), CheY-like REC (receiver) domain (1–176 aa), IR domain (186–431 aa), CCT domain (426–520 aa), IR-CCT domain (186–520 aa), and full length TaPRR1-D1 (1–520 aa) were subcloned into pGBKT7 (Clontech) to generate DNA binding domain fusion constructs. For transactivation activity assay, vectors carrying the target segment were transformed into yeast strain AH109 (Clontech) according to the manufacturer’s protocols. Yeast cells transformed with the indicated vectors were diluted and then dropped onto tryptophan-negative (–Trp) and tryptophan-, histidine- negative (–Trp/–His) synthetic dropout (SD) media, respectively. Plates were incubated at 30°C until colonies appeared.
Molecular Marker Development for TaPRR1
Three cleaved amplified polymorphic sequence (CAPS) markers, P78, P272, and P321, for TaPRR1-A1 were designed to identify polymorphisms in the 5′UTR, intron 1 and exon 2, respectively. Marker P78 was used to detect a single nucleotide polymorphism (SNP) locus in the 5′UTR. This assay used nested PCR with two pairs of primers. Products generated by PCR using the first primer pair, P78-1F and P78-1R, were used as templates for amplification with the second pair, P78-2F and P78-2R. PCR products were then digested by Bsp1286I (GDGCH/C) at 37°C for 3 h. If the product could not be digested by Bsp1286I, base A was located at −78 bp upstream of the initiation codon ATG. If the product could be digested into two fragments (114 bp and 294 bp), base G was located at this SNP locus. Using the same method, markers P272 and P321 were designed to detect polymorphisms in intron 1 and exon 2, respectively. Two allele-specific PCR (AS-PCR) markers, P1055 and P1230, were developed to detect variations in TaPRR1-B1. The CAPs marker P1941 was designed to identify polymorphism in the TaPRR1-D1 promoter region. This assay also used nested PCR with two pairs of primers, P1941-1F/1R and 2F/2R. The PCR product was digested by BtsCI (GGATGNN/) at 50°C for 3 h. If the product could not be digested by BtsCI, base G was located at −1941 bp upstream of ATG. If the product could be digested into two fragments (116 bp and 315 bp), base A was located at this SNP locus (Supplementary Table S5). All PCR primers are listed in Supplementary Table S6.
Expression Analysis
The expression experiment was conducted under short-day (9 h: 15 h, light: dark) and long-day (15 h: 9 h, light: dark) conditions with samples collected every 3 h during a 48-h period from 14-day-old seedlings of wheat cultivars “Chinese Spring” (CS, Hapla/Haplb/Haplb), “Yanzhan1” (YZ1, Hapla/Hapla/Hapla), and “Hussar” (Hapla/Haplb/Hapla). Four plants were pooled at each time-point and three biologically independent replications were performed for each genotype. Total RNA was extracted using RNAiso Plus (Takara, Ohtsu, Shiga, Japan). DNA was removed by digestion with DNase I (Fermentas, Ontario, Canada), and first-strand cDNA was synthesized using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Invitrogen). Quantitative reverse transcription PCR (RT-qPCR) was conducted using SYBR® Premix Ex TaqTM (Takara) on an ABI PRISM 7300 (Applied Biosystems, Foster City, CA, United States). Expression levels of TaPRR1 genes were standardized against that of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and quantitative data were analyzed using the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Phylogenetic Analysis
Amino acid sequences of the PRR gene family were obtained from the Protein database1. Multiple sequence alignment was performed using ClustalW. A neighbor-joining tree was reconstructed from alignment of 15 wheat PRR protein sequences using MEGA 6.06 with 1,000 bootstrap replicates and the Jones-Taylor-Thornton (JTT) model (Tamura et al., 2013).
PCR Primers, Sequence Analysis and Statistical Analysis
Primers were designed using Primer Premier 5.0 software (Supplementary Table S6). Sequence alignment and SNP detection were carried out using DNASTAR Lasergene 7.1.0 (DNASTAR). Cis-regulatory elements were predicted using PLACE (Higo et al., 1999). Significance tests were performed using SPSS Statistics 18.0. Tukey’s test was used to determine statistical differences by one-way ANOVA. Association analysis was performed by retrieving genotype data spanning 1-Mb regions upstream and downstream of TaPRR1 using the “Axiom Wheat 660K Genotyping Array” scanning 681 natural population (Supplementary Table S7)2. The phenotype data was collected in Changping (116.2°E, 40.2°N) during the years 2012–2013. Sequence variants with minor allele frequency (MAF) ≥0.05 were retained, and their associations with yield-related traits were tested using a general linear model that accounts for familial relatedness in Tassel 5.0 (Bradbury et al., 2007). Pairwise linkage disequilibrium analysis was performed using LDheatmap packages (Shin et al., 2006). Analysis of molecular variance (AMOVA) was performed using ARLEQUIN version 3.53 with an F-statistic to assign the genetic variation to either divergence among or variation within groups. Bisulfite sequencing data of DNA methylation levels of TaPRR1 in two-week-old CS leaf tissue at the three-leaf stage was available under project ID SRP133674 (Appels et al., 2018). Histone modification levels of TaPRR1 in 10-day-old in vitro seedlings were available under SRA study PRJNA420988 (SRP126222) (Appels et al., 2018). The integrated and visualized data was downloaded from The Triticeae Multi-omics Center website4.
Results
Isolation of TaPRR1 Homeologs and Phylogenetic Analysis of the PRR Gene Family in Wheat
Primer pair TaPRR1-F1/R1 specific for full-length TaPRR1 was designed and used to amplify three homeologs of TaPRR1 genes (TaPRR1-A1: TraesCS6A02G227900; TaPRR1-B1: TraesCS6B02G253900; and TaPRR1-D1: TraesCS6D02G207100) from hexaploid wheat cultivar “Chinese Spring” (T. aestivum, AABBDD). The full-length TaPRR1 genes of diploid A, B, and D genome donors UR102 (T. urartu, AA), Y2022 (A. speltoides, SS), and Y215 (A. tauschii, DD) were also obtained. Comparison of gene sequences from hexaploid and diploid accessions showed that different sequences from CS correspond to A, B, and D genomes, respectively (Supplementary Table S8).
TaPRR1 is a member of the PRR gene family. To explore the genetic relationships among PRR gene family members, we reconstructed a neighbor-joining tree (Figure 1A). The results showed that wheat PRR proteins could be divided into two clusters, which were further divided into five groups, each containing three homeologs from the wheat A, B, and D subgenomes. TaPRR1 showed the closest relationship with TaPRR95 and TaPRR59. Ppd-1 (TaPRR37) and TaPRR73 clustered in another branch of the tree. It is noteworthy that TaPRR1 and other PRR gene family members all contain two conserved domains, a REC domain and a CCT domain. The REC domain is a signal receiver domain and usually found N-terminal to a DNA binding effector domain (Marchler-Bauer et al., 2017). The CCT (CONSTANS, CO-like, and TOC1) domain is found in a number of plant proteins. It is about 45 amino acids long and contains a putative nuclear localization signal within the second half of the CCT motif (Marchler-Bauer et al., 2017). Remarkably the DBP (Duffy-antigen binding protein) domain is unique to the Ppd-D1 gene.
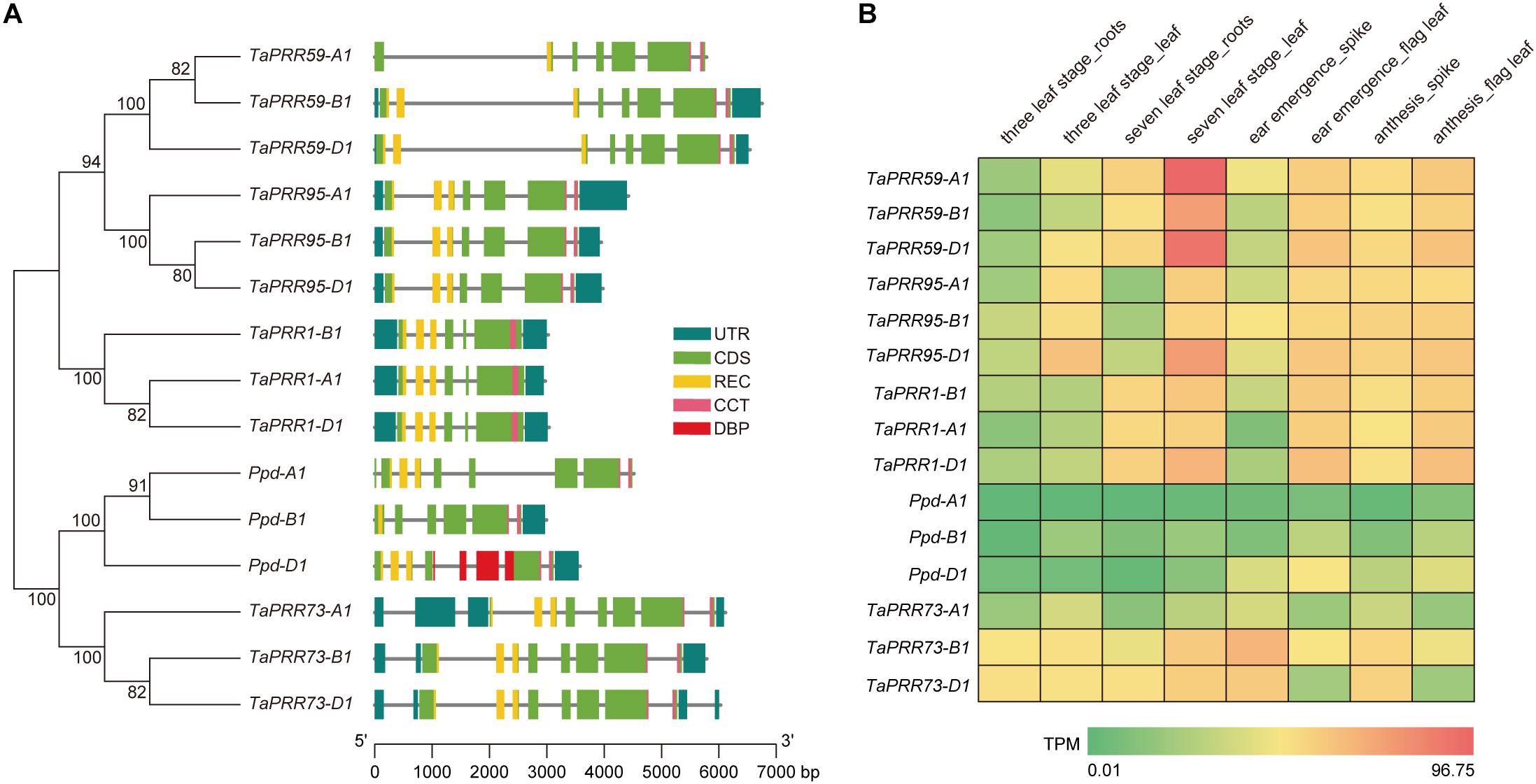
Figure 1. Phylogenetic relationships and expression profiles of the PRR gene family in wheat. (A) Phylogenetic analysis of the PRR gene family in wheat. A neighbor-joining tree was reconstructed from alignment of 15 wheat PRR protein sequences using MEGA 6.06 and the Jones-Taylor-Thornton model. Numbers above the branches represent bootstrap support based on 1,000 bootstrap replicates. Gene structures of TaPRR genes. UTR and CDS are represented by blue and green rectangles, respectively. Conserved domains of the proteins are also marked on the gene structure. REC, CCT, and DBP domains are represented by yellow, purples, and red respectively. Genes and their constituent parts can be inferred using the scalebar at the bottom. (B) Heat map of in silico expression profiles of PRR gene family members in different organs at different growth stages of “Chinese Spring”. The expression data were generated from the expVIP database (http://www.wheat-expression.com/). The color scale represents TPM (Transcripts Per Kilobase of exon model per Million mapped reads) value. Green indicates low levels of transcript abundance, while red indicates high levels.
Using expression data from the expVIP database5, we drew a heat map of PRR expression profiles at different developmental stages in different tissues (Figure 1B). The spatiotemporal expression patterns of TaPRR1 were similar to those of TaPRR59 and TaPRR95, but different from those of Ppd-1 and TaPRR73. The expression level of TaPRR1 in leaves (TPM value 25.32, 31.67, 43.47 for TaPRR1-A1, -B1, -D1 at 7-leaf stage) was higher than that of roots (TPM value 18.75, 21.46, 25.30 for TaPRR1-A1, -B1, -D1 at 7-leaf stage). During the reproductive growth period, the expression level of TaPRR1 in flag leaves (TPM value 28.21, 28.90, 37.14 for TaPRR1-A1, -B1, -D1 at ear emergence stage) was higher than that of spikes (TPM value 2.26, 7.20, 5.13 for TaPRR1-A1, -B1, -D1 at ear emergence stage). The spatiotemporal expression patterns of TaPRR1-A1, -B1, and -D1 were consistent. The expression level of TaPRR1-D1 was higher than those of TaPRR1-A1 and -B1 (Figure 1B).
Subcellular Localization and Transcriptional Activity Analysis of TaPRR1
TaPRR1 contains a CheY-like REC domain at its N-terminus and a CCT motif at the C-terminus. The CCT motif contains a putative nuclear localization signal. Based on coding regions of the D subgenome member, we determined the subcellular localization of TaPRR1 by transient expression (Figure 2A). A nuclear marker protein, OsMADS15, fused with mCherry, was used as a positive control. The subcellular localization results showed overlapping GFP and RFP fluorescence signals in the nuclei, indicating that TaPRR1 is localized in the nucleus.
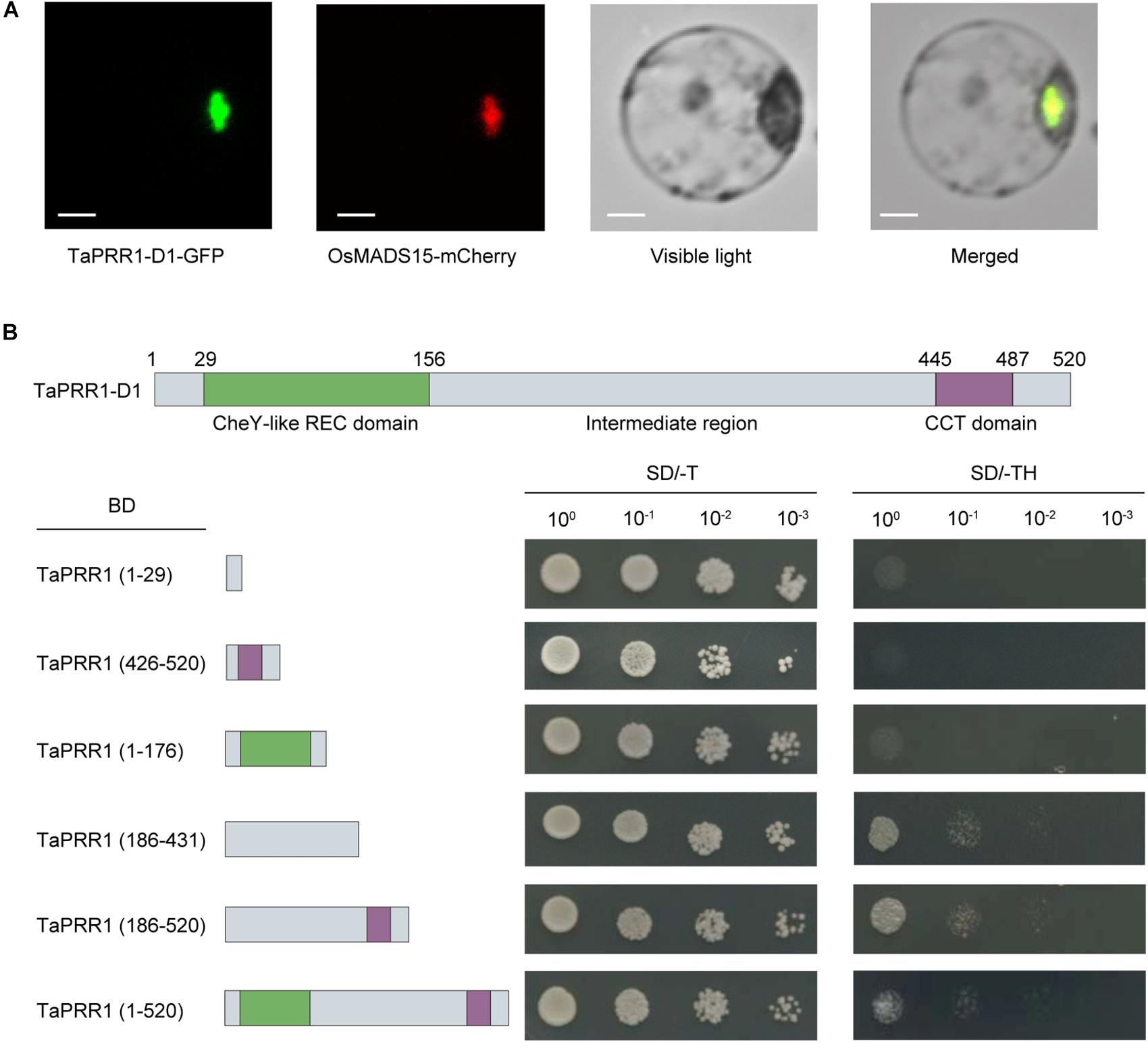
Figure 2. Subcellular localization and transcriptional activity analysis of TaPRR1. (A) Subcellular localization of TaPRR1. A fusion protein (TaPRR1-D1-GFP) and control (OsMADS15-RFP) were introduced into rice protoplasts. GFP and RFP fluorescence were observed using a confocal laser-scanning microscope. A nuclear marker protein, OsMADS15, fused with mCherry, was used as a positive control. Scale bars = 5 μm. (B) Transactivation activity assay. TaPRR1-D1 proteins with no functional domain (1–29 aa), CheY-like REC domain (1–176 aa), IR domain (186–431 aa), CCT domain (426–520 aa), IR-CCT domain (186–520 aa), and full length TaPRR1-D1 (1–520 aa) were assembled into pGBKT7 vectors. Yeast cells transformed with the indicated vectors were diluted and then dropped onto tryptophan- negative (–Trp) and tryptophan-, histidine- negative (–Trp/–His) synthetic dropout (SD) media, respectively.
To further investigate whether TaPRR1 has transcriptional activation activity, we carried out a transcriptional activation assay based on coding regions of TaPRR1-D1. The results showed that the constructs fusing the full length or IR (intermediate region) domain of TaPRR1-D1 with BD (GAL4 binding domain) resulted in expression of reporter genes (Figure 2B). We observed no activation signal from fusion proteins containing only the CheY-like REC or CCT domain. These results suggested that TaPRR1 has transcriptional activity in its IR motif, which is similar to Arabidopsis PRR1 (Gendron et al., 2012). Taken together, these results demonstrated that TaPRR1 is a nuclear-localized protein with transcriptional activity controlled by the IR domain.
Haplotype Analysis of TaPRR1 and Molecular Marker Development
To analyze the polymorphism of TaPRR1 and explore the relationship between natural variations of TaPRR1 and agronomic traits, we analyzed the entire genomic fragment of TaPRR1 in 28 cultivars with wide variations in HD and yield-related traits (Supplementary Table S1). Two haplotypes of TaPRR1-A1 were characterized by five variations, all of which were SNPs and located upstream of the coding region, intron 1, exon 2, and exon 6, respectively (Figure 3A). Notably, SNP3-SNP5 were located in the exon region, but only SNP4 caused amino acid changes (from E to K). Three cleaved amplified polymorphic sequence (CAPS) marker were developed based on SNP1-SNP3 to identify the two haplotypes simultaneously (Figure 3A). For TaPRR1-B1, three haplotypes were identified, characterized by three SNPs located within intron 4 and intron 5, respectively. Two allele-specific PCR (AS-PCR) markers, P1055 and P1230, were developed to detect the variations (Figure 3B). For TaPRR1-D1, there was no nucleotide variation in the coding region. We only found a SNP located 1,941 bp upstream of the coding region, which resulted in two haplotypes of TaPRR1-D1. The CAPs marker P1941 was developed based on this polymorphism for diagnosing the TaPRR1-D1 alleles of more accessions (Figure 3C). It is noteworthy that an ARR1-binding element (GGATT)6 was found at this SNP site and a single-base substitution mutation caused variations of this site, which might block its function.
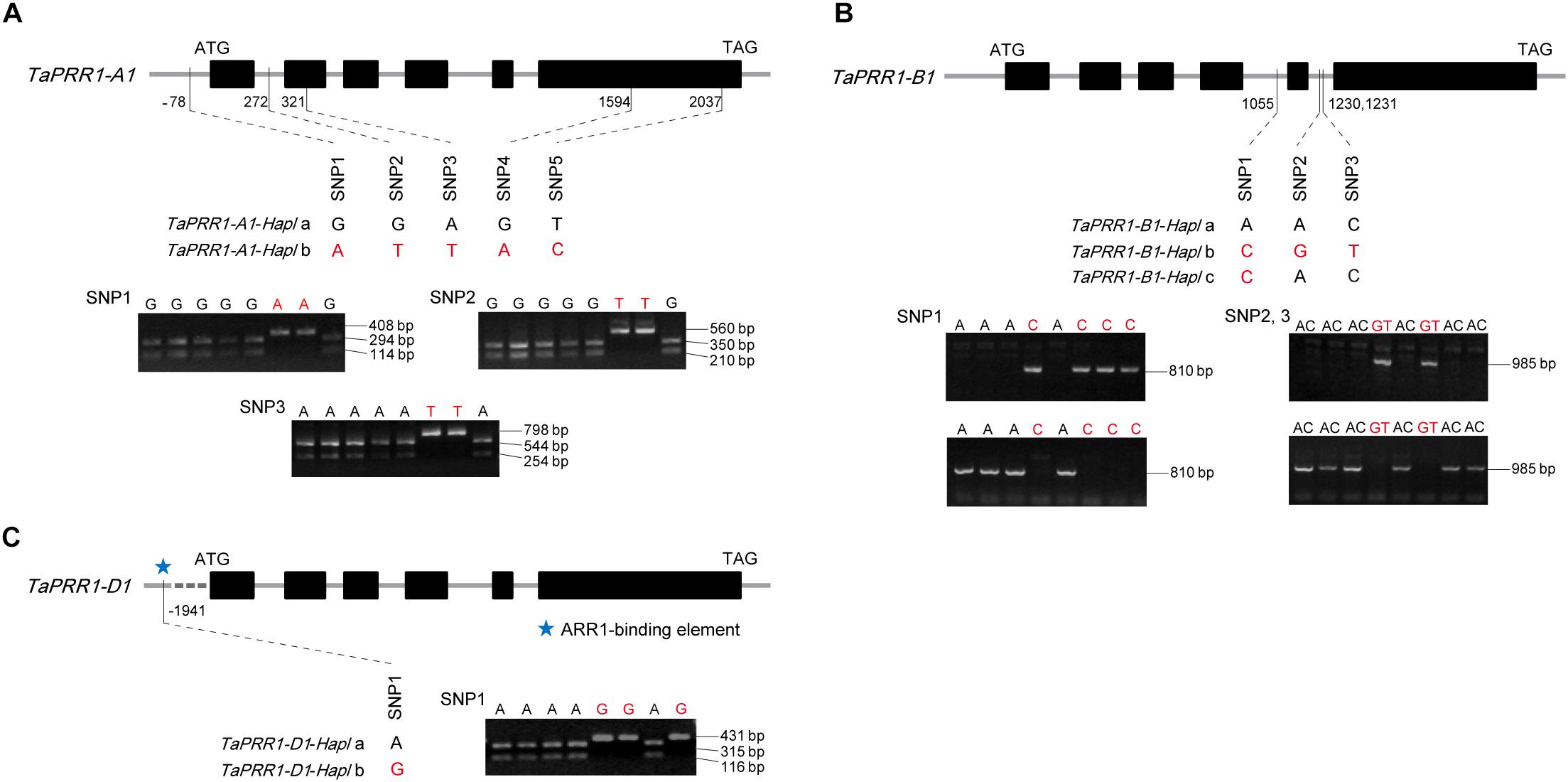
Figure 3. Gene structure, haplotypes, and molecular markers of TaPRR1. Taller rectangles represent the coding regions; shorter rectangles represent the candidate promoter, 5′ untranslated region (UTR), introns, and 3′UTR. (A) Haplotypes and molecular markers of TaPRR1-A1. Two haplotypes were identified for TaPRR1-A1. Three CAPS markers were developed based on SNP1-SNP3 with restriction endonucleases Bsp1286 I (SNP1 and SNP2) and BtgI (SNP3). (B) Haplotypes and molecular markers of TaPRR1-B1. Three haplotypes were identified for TaPRR1-B1, characterized by three SNPs located within intron 4 and intron 5. Two allele-specific PCR markers were developed to detect the variations of TaPRR1-B1. (C) Haplotypes and molecular markers of TaPRR1-D1. Two haplotypes were identified for TaPRR1-D1. A CAPS marker was developed based on SNP1 with restriction endonuclease BtsCI. The sizes of PCR products are shown on the right.
TaPRR1 Exhibited a Diurnal Rhythm Expression Pattern
We analyzed the expression patterns of TaPRR1 genes over a 48-h period in common wheat cultivars grown under short- and long- day conditions (Figure 4). Three representative cultivars, “Chinese Spring” (CS, Hapla/Haplb/Haplb), “Yanzhan1” (YZ1, Hapla/Hapla/Hapla), and “Hussar” (Hapla/Haplb/Hapla) were selected for expression tests. Overall, the expression patterns of TaPRR1 were characterized by diurnal rhythms, which had peaks at around 9 h after dawn and 18 h after dawn under both short- and long-day conditions. Specifically, “Hussar” showed different expression patterns compared to CS and YZ1. The first peak was higher than the second peak in CS and YZ1, whereas the second peak was higher in TaPRR1-B1 and TaPRR1-D1 of “Hussar” under short-day conditions. The peak appeared a few hours later in “Hussar” compared to CS and YZ1 under long-day conditions. The genotypes of TaPRR1-B1 and -D1 in “Hussar” were different from those in CS and YZ1, which may be the cause of the expression differentiation. In future studies, it will be of great significance to verify cis-regulatory elements/trans-transcriptional regulators to elucidate the detailed molecular mechanisms regulating TaPRR1 expression.
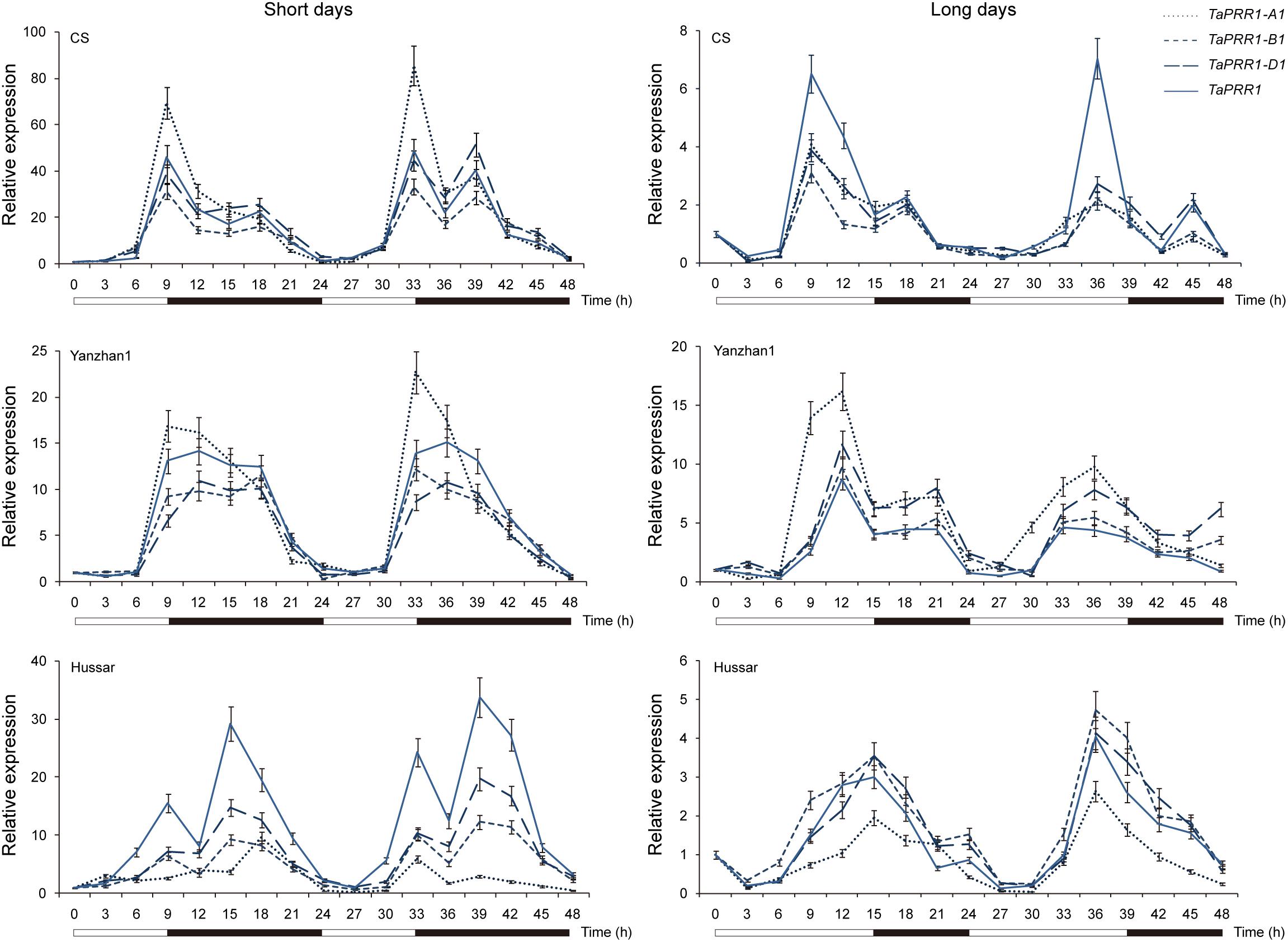
Figure 4. Relative expression levels of TaPRR1 under short days (9 h light/15 h dark) and long days (15 h light/9 h dark) by quantitative RT-PCR. RNAs were sampled at 3-h intervals over a 48-h period. Means ± SD from three replicated experiments are shown. Open bars, daytime; closed bars, nighttime.
Association of TaPRR1 Haplotypes With Agronomic Traits
In order to investigate the association of TaPRR1 haplotypes with yield-related traits, we performed an association analysis of each haplotype with seven yield traits using 177 accessions spanning 10 major agroecological wheat regions (Supplementary Table S2). Association analysis showed the homeolog-specific functions of TaPRR1 (Figure 5). For TaPRR1-A1, there were weak associations between TaPRR1-A1 and HD (seven environments), as well as SL (seven environments) (Figure 5A). However, TaPRR1-B1 was significantly associated with PH in all environments except E5 (18 environments) and weakly associated with TGW (10 environments) (Figure 5B). Similarly, TaPRR1-D1 was strongly associated with PH in all environments (19 environments) and weakly associated with TGW (10 environments) (Figure 5C).
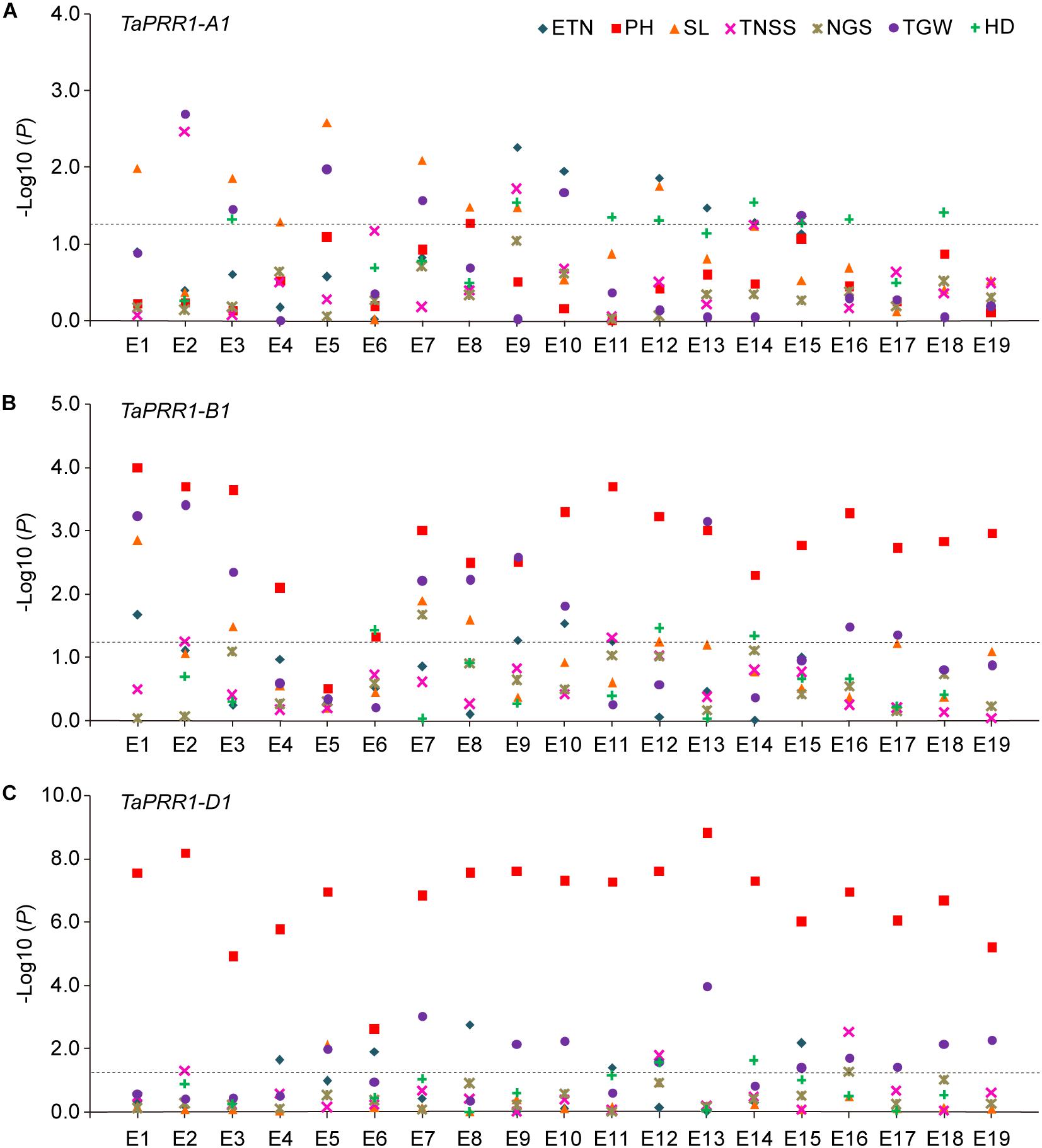
Figure 5. Association analysis of TaPRR1 homeologs. (A) Association analysis of TaPRR1-A1. (B) Association analysis of TaPRR1-B1. (C) Association analysis of TaPRR1-D1. ETN, effective tiller number; PH, plant height; SL, spike length; TNSS, total number of spikelets per spike; NGS, number of grains per spike; TGW, thousand grain weight; HD, heading date. E1 to E19 indicate environments. Negative log10-transformed P values are plotted. Significance tests were performed using SPSS Statistics 18.0. Tukey’s test was used to determine statistical differences by one-way ANOVA. Black horizontal dotted line indicates the threshold value for significant associations (P < 0.05).
We compared the phenotype variations of different TaPRR1 haplotypes for the above agronomic traits. In general, for TaPRR1-A1, varieties with Hapla showed earlier HD. It is noteworthy that Haplb showed a longer SL compared to Hapla (Supplementary Figure S1A). For TaPRR1-B1 and -D1, accessions with Hapla showed more favorable phenotypic traits, including shorter PH and higher TGW (Supplementary Figures S1B,C). Specifically, for TaPRR1-A1, the HD of Hapla was about 1.83 to 3.45 days ahead, advanced by 0.9 to 1.7%. The SL of Haplb increased by about 0.79 to 1.79 cm, representing an increase of 8.2 to 16.9% compared to Hapla. For TaPRR1-B1 and -D1, Hapla reduced PH by about 12.37 to 23.60 and 12.89 to 30.26 cm, respectively, representing a decrease of 13.3 to 19.7% and 14.1 to 26.4% compared to Haplb. Hapla also increased TGW by about 3.17 to 6.85 and 2.28 to 5.78 g, representing an increase of 9.1 to 17.3% and 6.3 to 14.8%, respectively, compared to Haplb.
To further confirm the association between natural variations of TaPRR1 and yield-related traits, we performed an association analysis by retrieving genotype data spanning 1-Mb regions upstream and downstream of TaPRR1 using the “Axiom Wheat 660K Genotyping Array” scanning 681 natural population (Supplementary Table S7). For TaPRR1-A1, there were 16 SNP markers spanning the physical position from 428,384,625 to 430,343,495 bp. Overall, TaPRR1-A1 and 11 other SNPs exhibited significant association with HD and SL (P < 0.05) (Supplementary Table S9). TaPRR1-A1 was in strong linkage disequilibrium (r2 > 0.5) with other significant variants, creating a linkage disequilibrium block flanking TaPRR1-A1 (Figure 6A). Similar to TaPRR1-A1, TaPRR1-B1 showed significant association with PH and TKW (P < 0.0001) (Supplementary Table S9) and was in high linkage disequilibrium with other significant SNPs (r2 > 0.5) (Figure 6B). For TaPRR1-D1, there were 35 SNP markers (Supplementary Table S9) and TaPRR1-D1 was significantly associated with PH and TKW (P < 0.0001) (Figure 6C). It was also noticeable that the region of TaPRR1-D1 coincided with several significantly associated SNPs (Figure 6C). Thus, TaPRR1 possibly confers pleiotropic effects on agronomic traits.
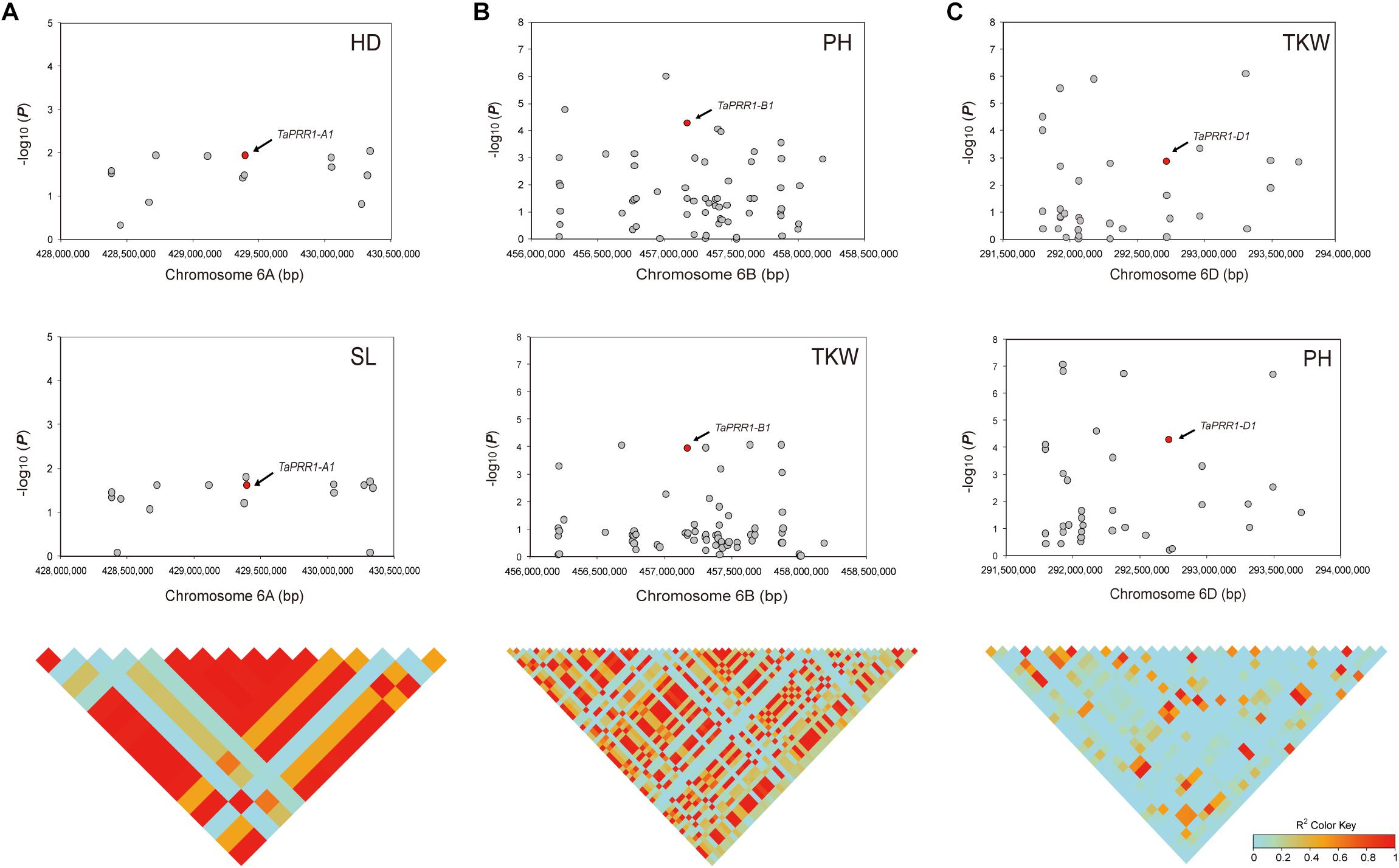
Figure 6. Association analysis of the 2-Mb region for TaPRR1 in a panel of 681 natural population. (A) Association studies were performed for TaPRR1-A1 with HD and SL. (B) Association studies were performed for TaPRR1-B1 with PH and TGW. (C) Association studies were performed for TaPRR1-D1 with PH and TGW. The lower diagram shows a triangle matrix of pairwise linkage disequilibrium. The intensity of red shading indicates the level of linkage disequilibrium (r2) between variants.
Geographical Distribution Characteristics of TaPRR1 Haplotypes
In order to determine the distribution characteristics of TaPRR1, we investigated the distribution frequency of different TaPRR1 haplotypes using 177 accessions from five continents including 15 countries.
The Chinese wheat planting areas were divided into ten major agro-ecological production zones according to climatic characteristics, topography and soil type, planting system, and cultivation characteristics (Jin, 1983). We investigated the distribution frequencies of different TaPRR1 haplotypes and found different distribution characteristics (Figure 7). For TaPRR1-A1, Hapla accounted for a larger proportion than Haplb in all ten ecological areas, which suggests Hapla varieties are photoperiod insensitive and can adapt to a wide range of geographical conditions (Figure 7A). For TaPRR1-B1, Haplb accounted for the highest proportion, followed by Hapla and Haplc in all ten ecological areas except SCW (South China Winter Wheat Region, where Haplc accounted for the largest ratio) (Figure 7B). The cause of the above phenomenon may be that SCW is located in a low-latitude area with comparatively shorter day lengths as well as high temperatures in summer than other areas, and Haplc is more suitable and adaptable for this climate compared to other haplotypes. For TaPRR1-D1, the proportion of Haplb was higher than that of Hapla (Figure 7C). It is worth noting that the haplotypes with the highest proportions in the B and D subgenomes do not carry favorable alleles. Therefore, the discovery of favorable alleles provides valuable theoretical reference for molecular breeding of wheat in these regions.
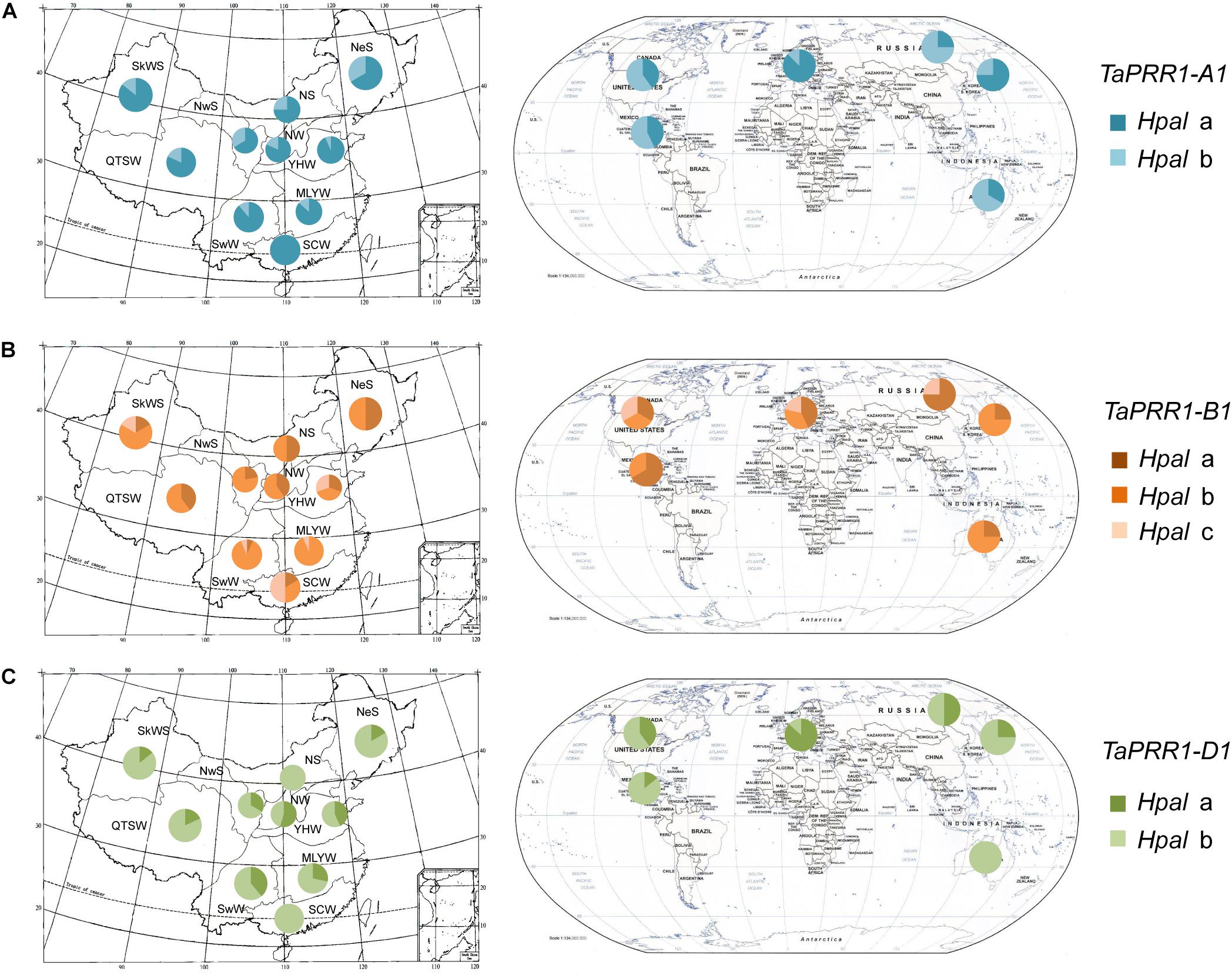
Figure 7. Geographic distributions of TaPRR1 haplotypes. (A) Geographic distributions of TaPRR1-A1. (B) Geographic distributions of TaPRR1-B1. (C) Geographic distributions of TaPRR1-D1. The Chinese wheat planting areas were divided into ten major agro-ecological production zones (SkWS, Sinkiang Winter-Spring Wheat Region; QTSW, Qinghai-Tibet Spring-Winter Wheat Region; NeS, Northeastern Spring Wheat Region; NwS, Northwestern Spring Wheat Region; NS, Northern Spring Wheat Region; NW, Northern Winter Wheat Region; YHW, Yellow and Huai River Winter Wheat Region; MLYW, Middle and Low Yangtze Valley Winter Wheat Region; SwW, Southwestern Winter Wheat Region; and SCW, South China Winter Wheat Region). Other areas included Asia, Europe, North America, South America, and Oceania. Pie charts represent the proportional distribution of different haplotypes.
The distribution characteristics of different TaPRR1 haplotypes in other countries were different from those in China (Figure 7). For TaPRR1-A1, Hapla accounted for a larger proportion than Haplb in Asia and Europe, whereas other regions were dominated by Haplb. For TaPRR1-B1, Haplb accounted for the highest proportion in Asia and Oceania. For TaPRR1-D1, it is noteworthy that Hapla was predominant in Europe, which was opposite to situations in other regions.
Differentiation of Genetic Variations of TaPRR1 Between Landraces and Modern Cultivars
To determine the differentiation degree of genetic variation of TaPRR1, we analyzed the distribution frequencies of haplotypes of TaPRR1-A1, -B1, and -D1 in landraces, modern cultivars, and synthetic hexaploid wheat lines (Supplementary Table S4). Haplotype network analysis showed that genetic variations in TaPRR1 were different between landraces and modern cultivars (Figure 8). However, no differentiation existed between synthetic hexaploid wheat and landraces/modern cultivars. For TaPRR1-A1, Hapla was predominant in landraces. However, the percentage of Haplb increased from 5.1% in landraces to 23.3% in modern cultivars (Figure 8A). For TaPRR1-B1, the percentage of Hapla, which represents a favorable haplotype able to reduce PH and increase TGW, was increased from 7.3% in landraces to 42.9% in modern cultivars (Figure 8B). Similarly, the percentage of TaPRR1-D1-Hapla was greatly increased in modern cultivars (5.1% in landraces versus 58.6% in modern cultivars) (Figure 8C). AMOVA revealed significant differences in haplotype distribution frequency between landraces and modern cultivars in TaPRR1-A1 (Fst = 0.07; P < 0.05), TaPRR1-B1 (Fst = 0.21; P < 0.05), and TaPRR1-D1 (Fst = 0.43; P < 0.05), respectively (Supplementary Table S10). According to the Fst value, we speculated that TaPRR1-A1 showed a moderate degree of genetic differentiation whereas TaPRR1-B1 and -D1 showed a high degree of genetic differentiation between landraces and modern cultivars.
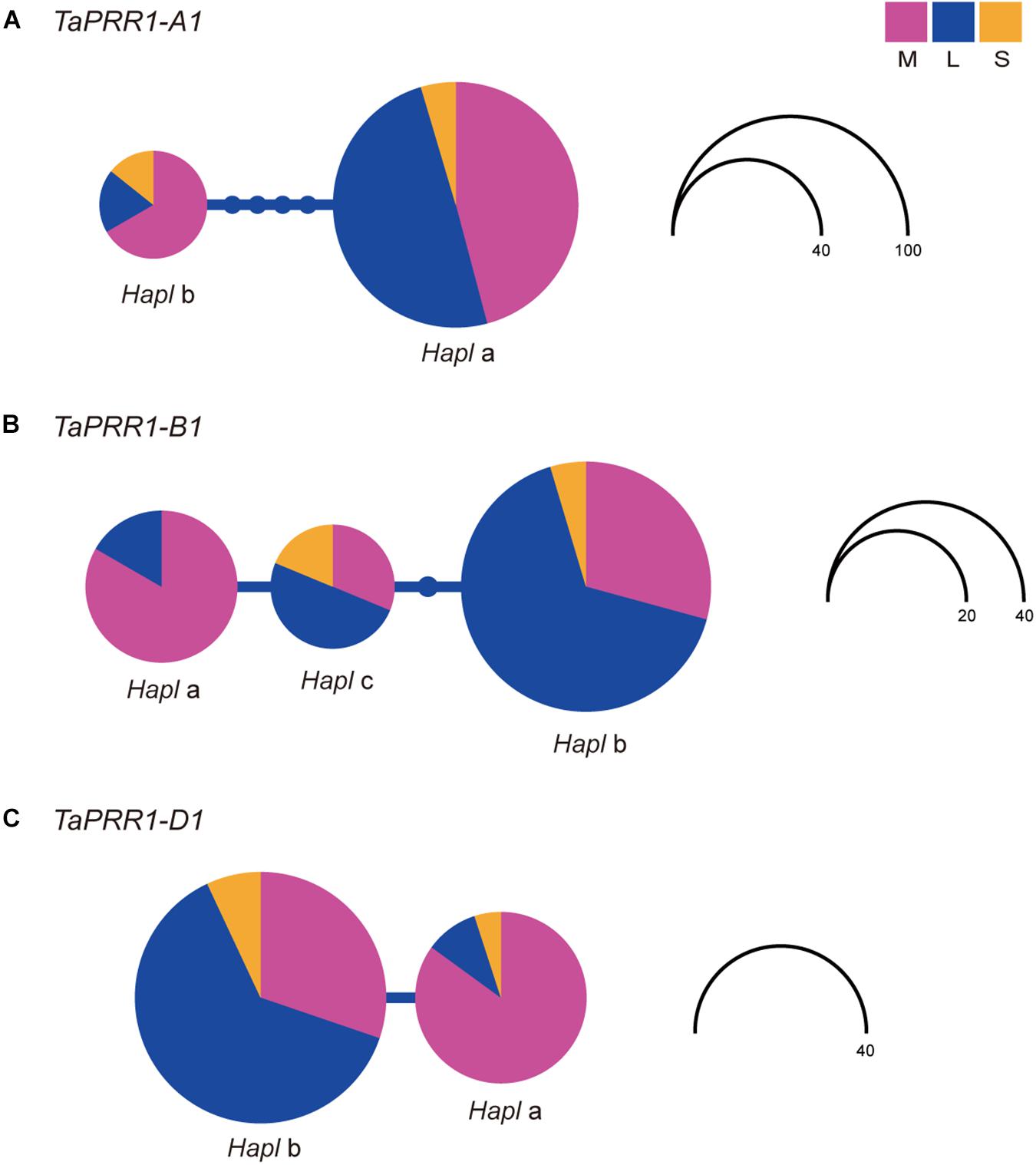
Figure 8. Haplotype network of TaPRR1-A1, -B1, and -D1. Coding regions of TaPRR1-A1, -B1, and -D1 plus 2kb upstream of the coding region of TaPRR1-D1 were used for haplotype analysis. A haplotype network was drawn using haplotype viewer (http://www.cibiv.at/~greg/haploviewer). One circle represents a haplotype. The node above the line indicates the number of base substitutions needed to get from one haplotype to another. Circle size represents the number of haplotypes. Different colors represent different subgroups. Purple, blue, and orange pie charts represent modern cultivars, landraces, and synthetic hexaploid wheat, respectively. Haplotype networks of TaPRR1-A1, -B1, and -D1 are shown in (A), (B), and (C), respectively.
Discussion
Wheat is a widely cultivated crop that has formed diversified ecological types adapted to different conditions worldwide (Worland and Snape, 2001). With the continuous enrichment of wheat resources as well as extensive exchange of new cultivars across the world, the ecological types of wheat are becoming more diversified. Developmental characteristics affect the regional distribution, introduction range, and utilization of wheat, and form an important basis for research into wheat cultivation and adaptation. Wheat is a long-day crop and its flowering is accelerated by a long photoperiod. Under short days, heading is delayed in photoperiod-sensitive cultivars (Klaimi and Qualset, 1973). Mutation of common hexaploid wheat into photoperiod-insensitive accessions enables it to adapt to different environmental changes (Worland et al., 1998).
Homeolog-Specific Functions of TaPRR1
The circadian clock regulates diverse aspects of plant growth and development; it not only confers daily rhythms in growth and metabolism, but also interacts with signaling pathways involved in plant responses to the environment (Harmer, 2009). PRR1 is a key component of the plant circadian clock, and the maintenance of its rhythmic expression is essential for proper functioning of the circadian clock (Somers et al., 1998; Más, 2008). In the present study, we analyzed polymorphisms in TaPRR1 among diverse wheat germplasm. Based on the observed polymorphisms, we identified two, three, and two haplotypes for TaPRR1-A1, TaPRR1-B1, and TaPRR1-D1, respectively. Association analyses supported an association between haplotypes of TaPRR1 and agronomic traits under multiple environmental conditions. However, it is noteworthy that the functional strength of TaPRR1-A1, TaPRR1-B1, and TaPRR1-D1 was different. Specifically, TaPRR1-A1 was related to the photoperiod sensitivity of wheat; accessions with Hapla had an earlier HD and showed a photoperiod-insensitive phenotype. TaPRR1-A1 was also weakly associated with SL. Different from the characteristics controlled by TaPRR1-A1, TaPRR1-B1, and -D1 had similar functions and were significantly associated with PH and TGW. However, TaPRR1-D1 showed a stronger function than TaPRR1-B1. Interestingly, different from TaPRR1-A1, neither TaPRR1-B1 nor -D1 was associated with heading time. Therefore, although TaPRR1 belongs to the circadian clock genes, its function is not limited to the regulation of heading stage in wheat, but also affects other yield-related traits. In addition, TaPRR1 is reported to regulate flag leaf angle, which is significantly correlated with the photosynthetic ability of a plant (Zhao et al., 2016). Moreover, it is worth noting that there are several previously reported yield-related quantitative trait loci (QTLs) (Wang et al., 2012; Edae et al., 2014; Zou et al., 2017) in the region of TaPRR1. Therefore, TaPRR1 might co-locate with these QTLs and confer complex pleiotropic effects on agronomic traits.
Significance of the PRR Gene Family in Wheat
Although wheat PRR family members vary in length, they all contain two conserved domains, a REC domain at the N-terminus and a CCT motif at the C-terminus. PRRs have conserved protein structure, suggesting that they might have similar biological functions. PRR gene family members in Arabidopsis function as central oscillators of the circadian clock. PRR3 is a vascular regulator of PRR1 stability, which perturbs PRR1 interaction with ZTL (ZEITLUPE) that targets PRR1 for proteasome-dependent degradation (Para et al., 2007). PRR9, PRR7, and PRR5 act as transcriptional repressors of CCA1 and LHY, constituting the molecular mechanism accounting for the role of these proteins in the feedback loop of the circadian clock (Nakamichi et al., 2010). In wheat, PRR family members, including “Green Revolution” gene Ppd-1 (PRR37) and its homologous gene TaPRR73 have been studied extensively (Beales et al., 2007; Wilhelm et al., 2009; Guo et al., 2010; Seki et al., 2011; Díaz et al., 2012; Nishida et al., 2013; Sun et al., 2014; Zhang et al., 2016; Arjona et al., 2018). However, research on TaPRR1, the core member of the circadian clock, has lagged behind. Both this study and previous studies have shown that TaPRR1 affects heading time of wheat. In our study, we also found that TaPRR1 affects agronomic traits including PH, TGW, and SL. It is worth noting that in the majority of agro-ecological production zones of China, varieties with favorable alleles of TaPRR1 did not prevail, although the favorable haplotypes were positively selected during the wheat breeding process. Undoubtedly, the discovery of favorable alleles of TaPRR1 and the development of molecular markers are of great significance for marker-assisted breeding of wheat. In addition, a pair of homologous genes, TaPRR59 and TaPRR95, is also worth studying. Phylogenetic analysis showed that TaPRR1 has a close relationship with TaPRR95 and TaPRR59, and spatiotemporal expression analysis showed that these genes also have similar expression patterns. Considering the important effects of TaPRR1 on agronomic traits, TaPRR95 and TaPRR59 may be worth further investigation in the future.
Epigenetic Modification Characteristics of TaPRR1
Previous studies have shown that TaPRR1 is regulated by tae-miR408, a form of epigenetic regulation (Zhao et al., 2016). Interestingly, Ppd-B1 was also reported to be regulated by epigenetic modification (DNA methylation), and DNA methylation affects the transcriptional levels of this gene (Sun et al., 2014). To further determine whether TaPRR1 is regulated by other epigenetic factors, we retrieved DNA methylation and histone modification data for the TaPRR1 gene (Appels et al., 2018) from The Triticeae Multi-omics Center website. DNA methylation represents an important epigenetic mechanism of gene regulation and is generally associated with the repression of gene transcription (Chan et al., 2005; Reddington et al., 2013). Histone methylation is an important post-translational modification. In general, H3K4me3 modification is usually enriched in the activated promoter region and relates to the activation of gene expression whereas H3K27me3 is associated with transcriptional inhibition (Kouzarides, 2007; Vaillant and Paszkowski, 2007). According to ChIP-seq data, the DNA methylation level of the gene body region of TaPRR1 was lower than that of the promoter region (Supplementary Figure S2). Interestingly, TaPRR1-A1, TaPRR1-B1, and TaPRR1-D1 had distinct DNA methylation patterns. The methylation level of TaPRR1-A1 was the highest whereas that of TaPRR1-B1 was the lowest. In terms of histone modification, the degree of TaPRR1 histone modification on different subgenomes was roughly the same (Supplementary Figure S2). H3K4me3 was mainly concentrated near the transcriptional start site, whereas H3K27me3 was enriched over the entire genic region. Therefore, epigenetic modification, especially DNA methylation, might play an important role in the regulation of TaPRR1, which is also a key research object for the future.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
HS, ZG, and JJ designed the research. HS, WZ, YW, FC, and CZ performed the experiments. HS, YW, and LG analyzed the data. HS and ZG wrote the manuscript. JJ revised the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the National Natural Science Foundation for Young Scholars of China (Grant No. 31801348), the Natural Science Foundation of Shandong Province, China (Grant Nos. ZR2019PC003 and ZR2018PC016), the National Natural Science Foundation of China (Grant Nos. 31501308, 31871612, and 31701505), and Major Research plan of Yantai (Grant No. 2017ZH052).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yannan Liu (School of Life Sciences, Peking University) for critical reading and comments on our manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00285/full#supplementary-material
Footnotes
- ^ https://www.uniprot.org/
- ^ http://wheat.pw.usda.gov/ggpages/topics/Wheat660_SNP_array_developed_by_CAAS.pdf
- ^ http://cmpg.unibe.ch
- ^ http://202.194.139.32/
- ^ http://www.wheat-expression.com/
- ^ https://www.dna.affrc.go.jp/PLACE/
References
Alabadí, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Más, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Appels, R., Eversole, K., Feuillet, C., Keller, B., Rogers, J., Stein, N., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated refernce genome. Science 361:eaar7191. doi: 10.1126/science.aar7191
Arjona, J. M., Royo, C., Dreisigacker, S., Ammar, K., and Villegas, D. (2018). Effect of Ppd-A1 and Ppd-B1 allelic variants on grain number and thousand kernel weight of durum wheat and their impact on final grain yield. Front. Plant Sci. 9:888. doi: 10.3389/fpls.2018.00888
Bart, R., Chern, M., Park, C. J., Bartley, L., and Ronald, P. C. (2006). A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2:13. doi: 10.1186/1746-4811-2-13
Beales, J., Turner, A., Griffiths, S., Snape, J. W., and Laurie, D. A. (2007). A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. doi: 10.1007/s00122-007-0603-4
Bradbury, P. J., Zhang, Z., Kroon, D. E., Casstevens, T. M., Ramdoss, Y., and Buckler, E. S. (2007). TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. doi: 10.1093/bioinformatics/btm308
Chan, S. W., Henderson, I. R., and Jacobsen, S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 6, 351–360. doi: 10.1038/nrg1601
Díaz, A., Zikhali, M., Turner, A. S., Isaac, P., and Laurie, D. A. (2012). Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7:e33234. doi: 10.1371/journal.pone.0033234
Edae, E. A., Byrne, P. F., Haley, S. D., Lopes, M. S., and Reynolds, M. P. (2014). Genome-wide association mapping of yield and yield components of spring wheat undercontrasting moisture regimes. Theor. Appl. Genet. 127, 791–807. doi: 10.1007/s00122-013-2257-8
Farré, E. M., and Liu, T. (2013). The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr. Opin. Plant Biol. 16, 621–629. doi: 10.1016/j.pbi.2013.06.015
Gendron, J. M., Pruneda-Paz, J. L., Doherty, C. J., Gross, A. M., Kang, S. E., and Kay, S. A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. U.S.A. 109, 3167–3172. doi: 10.1073/pnas.1200355109
Guo, Z., Song, Y., Zhou, R., Ren, Z., and Jia, J. (2010). Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 185, 841–851. doi: 10.1111/j.1469-8137.2009.03099.x
Hao, C., Wang, L., Ge, H., Dong, Y., and Zhang, X. (2011). Genetic diversity and linkage disequilibrium in Chinese bread wheat (Triticum aestivum L.) revealed by SSR markers. PLoS One 6:e17279. doi: 10.1371/journal.pone.0017279
Harmer, S. L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60, 357–377. doi: 10.1146/annurev.arplant.043008.092054
Harmer, S. L., Hogenesch, J. B., Straume, M., Chang, H. S., Han, B., Zhu, T., et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113. doi: 10.1126/science.290.5499.2110
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Huang, W., Pérez-García, P., Pokhilko, A., Millar, A. J., Antoshechkin, I., Riechmann, J. L., et al. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336, 75–79. doi: 10.1126/science.1219075
Jin, S. (1983). Chinese Wheat Varieties and Their Genealogies. Beijing: Agricultural publishing house.
Karimi, M., Inzé, D., and Depicker, A. (2002). GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7, 193–195. doi: 10.1016/S1360-1385(02)02251-3
Klaimi, Y. Y., and Qualset, C. O. (1973). Genetics of heading time in wheat (Triticum aestivum L.). I. The inheritance of photoperiodic response. Genetics 74, 139–156.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Laurie, D. A. (1997). Comparative genetics of flowering time in cereals. Plant Mol. Biol. 35, 167–177. doi: 10.1007/978-94-011-5794-0_16
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Makino, S., Kiba, T., Imamura, A., Hanaki, N., Nakamura, A., Suzuki, T., et al. (2000). Genes encoding pseudo-response regulators: insight into His-to-Asp phosphorelay and circadian rhythm in Arabidopsis thaliana. Plant Cell Physiol. 41, 791–803. doi: 10.1093/pcp/41.6.791
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Más, P. (2008). Circadian clock function in Arabidopsis thaliana: time beyond transcription. Trends Cell Biol. 18, 273–281. doi: 10.1016/j.tcb.2008.03.005
Más, P., Alabadí, D., Yanovsky, M. J., Oyama, T., and Kay, S. A. (2003). Dual role of TOC1 in the control of circadian and photomorphogenic responses in Arabidopsis. Plant Cell 15, 223–236. doi: 10.1105/tpc.006734
Matsushika, A., Makino, S., Kojima, M., and Mizuno, T. (2000). Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 41, 1002–1012. doi: 10.1093/pcp/pcd043
Nakamichi, N., Kiba, T., Henriques, R., Mizuno, T., Chua, N. H., and Sakakibara, H. (2010). PSEUDO-RESPONSE REGULATORS 9,7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22, 594–605. doi: 10.1105/tpc.109.072892
Niinuma, K., Nakagawa, M., Calvino, M., and Mizoguchi, T. (2007). Dance of plants with circadian clock. Plant Biotechnol. 24, 87–97. doi: 10.5511/plantbiotechnology.24.87
Nishida, H., Yoshida, T., Kawakami, K., Fujita, M., Long, B., Akashi, Y., et al. (2013). Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time. Mol. Breed. 31, 27–37. doi: 10.1007/s11032-012-9765-0
Para, A., Farré, E. M., Imaizumi, T., Pruneda-Paz, J. L., Harmon, F. G., and Kay, S. A. (2007). PRR3 is a vascular regulator of TOC1 Stability in the Arabidopsis circadian clock. Plant Cell 19, 3462–3473. doi: 10.1105/tpc.107.054775
Reddington, J. P., Pennings, S., and Meehan, R. R. (2013). Non-canonical functions of the DNA methylome in gene regulation. Biochem. J. 451, 13–23. doi: 10.1042/BJ20121585
Seki, M., Chono, M., Matsunaka, H., Fujita, M., Oda, S., Kubo, K., et al. (2011). Distribution of photoperiod-insensitive alleles Ppd-B1a and Ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed. Sci. 61, 405–412. doi: 10.1270/jsbbs.61.405
Shin, J. H., Blay, S., McNeney, B., and Graham, J. (2006). LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 16:3.
Somers, D. E., Webb, A. A., Pearson, M., and Kay, S. A. (1998). The short-period mutant toc1-1 alters circadian clock regulation of multiple outputs throughout development in Arabidopsis thaliana. Development 125, 485–494.
Strayer, C., Oyama, T., Schultz, T. F., Raman, R., Somers, D. E., Más, P., et al. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289, 768–771. doi: 10.1126/science.289.5480.768
Sun, H., Guo, Z., Gao, L., Zhao, G., Zhang, W., Zhou, R., et al. (2014). DNA methylation pattern of Photoperiod-B1 is associated with photoperiod insensitivity in wheat (Triticum aestivum). New Phytol. 204, 682–692. doi: 10.1111/nph.12948
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Turner, A., Beales, J., Faure, S., Dunford, R. P., and Laurie, D. A. (2005). The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310, 1031–1034. doi: 10.1126/science.1117619
Vaillant, I., and Paszkowski, J. (2007). Role of histone and DNA methylation in gene regulation. Curr. Opin. Plant Biol. 10, 528–533. doi: 10.1016/j.pbi.2007.06.008
Wang, L., Ge, H., Hao, C., Dong, Y., and Zhang, X. (2012). Identifying loci influencing 1,000-kernel weight in wheat by microsatellite screening for evidence of selection during breeding. PLoS One 7:e29432. doi: 10.1371/journal.pone.0029432
Wilhelm, E. P., Turner, A. S., and Laurie, D. A. (2009). Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.). Theor. Appl. Genet. 118, 285–294. doi: 10.1007/s00122-008-0898-9
Worland, A. J., Börner, A., Korzun, V., Li, W. M., Petrovíc, S., and Sayers, E. J. (1998). The influence of photoperiod genes to the adaptability of European winter wheats. Euphytica 100, 385–394. doi: 10.1007/978-94-011-4896-2_67
Worland, T., and Snape, J. W. (2001). “Genetic basis of worldwide wheat varietal improvement,” in The World Wheat Book: A History of Wheat Breeding, eds A. P. Bonjean and W. J. Angus (Paris: Lavoisier Publishing), 59–100.
Zhang, W., Zhao, G., Gao, L., Kong, X., Guo, Z., Wu, B., et al. (2016). Functional studies of heading date-related gene TaPRR73, a paralog of Ppd1 in common wheat. Front. Plant Sci. 7:772. doi: 10.3389/fpls.2016.00772
Zhao, X. Y., Hong, P., Wu, J. Y., Chen, X. B., Ye, X. G., Pan, Y. Y., et al. (2016). The tae-miR408-mediated control of TaTOC1 genes transcription is required for the regulation of heading time in wheat. Plant Physiol. 170, 1578–1594. doi: 10.1104/pp.15.01216
Keywords: TaPRR1, haplotype, circadian rhythm, association analysis, yield-related traits, genetic differentiation
Citation: Sun H, Zhang W, Wu Y, Gao L, Cui F, Zhao C, Guo Z and Jia J (2020) The Circadian Clock Gene, TaPRR1, Is Associated With Yield-Related Traits in Wheat (Triticum aestivum L.). Front. Plant Sci. 11:285. doi: 10.3389/fpls.2020.00285
Received: 11 January 2020; Accepted: 25 February 2020;
Published: 12 March 2020.
Edited by:
Guijun Yan, The University of Western Australia, AustraliaReviewed by:
Jian Ma, Sichuan Agricultural University, ChinaShahidul Islam, Murdoch University, Australia
Copyright © 2020 Sun, Zhang, Wu, Gao, Cui, Zhao, Guo and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Han Sun, c3VuaGFuQGxkdS5lZHUuY24=; Zhiai Guo, Z3VvemhpYWlAaWJjYXMuYWMuY24=; Jizeng Jia, amlhaml6ZW5nQGNhYXMuY24=
†These authors have contributed equally to this work
 Han Sun
Han Sun Wenping Zhang
Wenping Zhang Yongzhen Wu
Yongzhen Wu Lifeng Gao
Lifeng Gao Fa Cui
Fa Cui Chunhua Zhao
Chunhua Zhao Zhiai Guo
Zhiai Guo Jizeng Jia3*
Jizeng Jia3*