
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 26 February 2020
Sec. Plant Breeding
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00214
This article is part of the Research Topic Breeding Forage and Grain Legumes for Sustainable Protein Production View all 15 articles
 Ying Zhao1†
Ying Zhao1† Yifan Cui1†
Yifan Cui1† Shiyu Huang1
Shiyu Huang1 Jingyao Yu1
Jingyao Yu1 Xinyu Wang1
Xinyu Wang1 Dawei Xin1
Dawei Xin1 Xin Li2
Xin Li2 Yonghui Liu1
Yonghui Liu1 Yuxin Dai1
Yuxin Dai1 Zhaoming Qi1*
Zhaoming Qi1* Qingshan Chen1*
Qingshan Chen1*Glucose-6-phosphate dehydrogenase (G6PDH) is known as a critical enzyme responsible for nicotinamide adenine dinucleotide phosphate (NADPH) generation in the pentose phosphate pathway (PPP), and has an essential function in modulating redox homeostasis and stress responsiveness. In the present work, we characterized the nine members of the G6PDH gene family in soybean. Phylogenic analysis and transit peptide prediction showed that these soybean G6PDHs are divided into plastidic (P) and cytosolic (Cy) isoforms. The subcellular locations of five GmG6PDHs were further verified by confocal microscopy in Arabidopsis mesophyll protoplasts. The respective GmG6PDH genes had distinct expression patterns in various soybean tissues and at different times during seed development. Among them, the Cy-G6PDHs were strongly expressed in roots, developing seeds and nodules, while the transcripts of P-G6PDHs were mainly detected in green tissues. In addition, the activities and transcripts of GmG6PDHs were dramatically stimulated by different stress treatments, including salt, osmotic and alkali. Notably, the expression levels of a cytosolic isoform (GmG6PDH2) were extraordinarily high under salt stress and correlated well with the G6PDH enzyme activities, possibly implying a crucial factor for soybean responses to salinity. Enzymatic assay of recombinant GmG6PDH2 proteins expressed in Escherichia coli showed that the enzyme encoded by GmG6PDH2 had functional NADP+-dependent G6PDH activity. Further analysis indicated overexpression of GmG6PDH2 gene could significantly enhance the resistance of transgenic soybean to salt stress by coordinating with the redox states of ascorbic acid and glutathione pool to suppress reactive oxygen species generation. Together, these results indicate that GmG6PDH2 might be the major isoform for NADPH production in PPP, which is involved in the modulation of cellular AsA-GSH cycle to prevent the oxidative damage induced by high salinity.
The pentose phosphate pathway (PPP) is a pivotal carbohydrate metabolic pathway that acts as a key role in plant development and stress responses (Krüger et al., 2011; Caretto et al., 2015). The PPP is one of the major sources of nicotinamide adenine dinucleotide phosphate (NADPH), which is the principal reducing molecule used in many metabolic pathways, such as nitrogen assimilation and amino acids synthesis (Devi et al., 2007; Sharkey and Weise, 2016). The two dehydrogenases in the PPP pathway, 6-phosphogluconate dehydrogenase (6PGDH, EC 1.1.1.44) and glucose-6- phosphate dehydrogenase (G6PDH, EC 1.1.1.49), utilize NADP+ as a cofactor to generate NADPH during the conversion of glucose-6-phosphate (G6P) to pentoses (Wood, 1986). The step catalyzed by G6PDH enzyme is known as the vital reaction in the PPP due to its strict control of the NADPH/NADP+ redox balance.
Genes encoding G6PDH have been cloned and characterized from some plants including Oryza sativa (Zhang et al., 2013), Populus suaveolens (Lin et al., 2005), Arabidopsis thaliana (Wakao and Benning, 2005), Solanum tuberosum (Wendt et al., 2000; Hauschild and von Schaewen, 2003), Hordeum vulgare (Esposito et al., 2001; Cardi et al., 2015), and Triticum aestivum (Nemoto and Sasakuma, 2000), and their involvement in plant development has been reported. Cytosolic (Cy) and plastidic (P) isoforms have been certified for plant G6PDHs based on their subcellular localization (Cardi et al., 2013; Castiglia et al., 2015). In addition, the P-G6PDHs are divided into two types, P1-G6PDH and P2-G6PDH, which can be distinguished by diverse gene expression profiles as well as specific biochemical characteristics, indicating the different functions of each isoform in plant metabolism (Cardi et al., 2016). In Arabidopsis, there are six G6PDHs targeted to different subcellular compartments: two cytosolic NADP+-dependent isoforms encoded by AtG6PDH5 and AtG6PDH6 genes respectively, and four plastidic NADP+-dependent isoforms encoded by AtG6PDH1, AtG6PDH2, AtG6PDH3, and AtG6PDH4 genes respectively (Wakao and Benning, 2005). Previous studies have shown that knocking down the cytosolic G6PDH in Arabidopsis may inhibit the seed oil accumulation, suggesting that Cy-G6PDH is crucial for regulating the oil biosynthesis during seed development (Wakao et al., 2008). Furthermore, the plastidic isoforms are proved to be essential in providing reducing power (NADPH) for enzymes involved in ammonium assimilation and nitrate reduction (Esposito et al., 2004; Esposito, 2016).
In addition to their pivotal role in developmental processes, the key functions of plant G6PDHs in responses to different types of environmental stresses have been widely proven, including salinity (Wang et al., 2008), cold (Lin et al., 2013), drought (Landi et al., 2016), and heat (Gong et al., 2012). It has been demonstrated that the cytosolic G6PDH is the major contributor to the total cellular G6PDH activity in plants (Castiglia et al., 2015), which seems to be a significant factor for the outcome of abiotic stress responses (Esposito et al., 2001; Honjoh et al., 2007). As a main example, overexpression of a kinetically engineered G6PDH in cytosol enhanced both biotic (defense reactions) and abiotic (drought) stress tolerance of transgenic tobacco through inhibiting NADPH oxidases induced reactive oxygen species (ROS) by improving NADPH provision during early oxidative bursts (Scharte et al., 2009). Besides, overexpression of a cytosolic PsG6PDH gene from Populus suaveolens confers an increased cold tolerance in transgenic tobacco by elevating the activity of antioxidative enzymes, such as peroxidase and superoxide dismutase, and decreasing the level of membrane lipid peroxidation (Lin et al., 2005, 2013). Also, the enhanced cytosolic G6PDH activities would contribute to the improvement of drought tolerance in soybean roots, with the involvement of ABA-dependent signaling pathway (Liu et al., 2013; Wang et al., 2016).
The characteristics of G6PDHs with respect to salt resistance have been validated by several researchers (Liu et al., 2007; Wang et al., 2008; Sang et al., 2018). It has been shown that the oxidative burst is counteracted, more or less, by the activities and expression of G6PDH isoforms upon salt stress (Valderrama et al., 2006; Liu et al., 2007). The Cy-G6PDH isoforms in Arabidopsis are essential in the provision of NADPH to maintain the cellular redox homeostasis via the phosphorylation of Thr467 by glycogen synthase kinase 3 under high salinity condition (Dal Santo et al., 2012); and this process is identified as associated with a sugar-signaling molecule (Lejay et al., 2008). Northern hybridization revealed a noticeable increase of TaG6PDH transcripts within 12 h of exposure of wheat seedlings to salinity treatment (150 mM NaCl), and the high transcript level was maintained for several hours (Nemoto and Sasakuma, 2000, 2002). In addition, the G6PDH enzyme in rice is the key in sustaining ROS homeostasis by regulating the coordination state of G6PDH activity and NAPDH oxidase under salt stress, however, the molecular metabolism have not been investigated (Zhang et al., 2013).
Although the biological functions of G6PDH in stress responses have been described in several model plants, few information are known about soybean. Here, we characterized nine members of the G6PDH gene family in soybean. The cellular locations of GmG6PDHs were initially predicted by transit peptide analysis and subsequently verified by transient expression of GFP-tagged GmG6PDH fusion proteins in Arabidopsis protoplasts. We also determined the transcriptional profiles of GmG6PDHs in distinct organs and under various abiotic stress using qRT-PCR and high-throughput sequencing data analyses. Most notably, one cytosolic isoform (GmG6PDH2) had apparent transcriptional response to salt stress and did well correlate with the activity of G6PDH enzyme, possibly implying a prominent role for this isoform in response to salinity. The prokaryotic expression of GmG6PDH2 in Escherichia coli demonstrated that this gene encoded an active G6PDH enzyme. In addition, overexpression of GmG6PDH2 in soybean hair roots increased the salt tolerance in transgenic soybean seedlings, with higher AsA/DHA (ascorbic acid/dehydroascorbate), GSH/GSSG (reduced/oxidized glutathione) ratios, lower levels of ROS and lipid peroxidation. These findings indicate that the soybean G6PDHs participate in plant growth and stress responses, of which the cytosolic GmG6PDH2 is the main isoform in regulating the cellular redox pool and defending against oxidative stress.
To obtain all G6PDHs from the soybean genome, a systematic BLASTP search was carried out against the soybean genetics and genomics database (SoyBase1) using the published A. thaliana G6PDHs as queries. The protein sequences of putative soybean G6PDH family members with an E-value of <10–10 and a sequence identity threshold > 90% were downloaded. The candidate genes were further verified by SMART2 and Pfam3 databases to confirm the presence of a C-terminal NADP-dependent G6PD domain (PF02781) and an N-terminal NADP+-binding domain (PF00479). Information about the genetic characteristics of GmG6PDHs, including coding sequence lengths, chromosome locations and protein lengths were collected from the SoyBase. The isoelectric point and molecular mass were determined on ExPASy server4. Transit peptides and subcellular localization were predicted using TargetP 2.05 and CELLO 2.56 (Yu et al., 2004).
The full-length proteins of G6PDHs from Glycine max (GmG6PDHs), Zea mays (ZmG6PDHs), Oryza sativa (OsG6PDHs), Phaseolus vulgaris (PvG6PDHs), Medicago truncatula (MtG6PDHs), Sorghum bicolor (SbG6PDHs), Brachypodium distachyon (BdG6PDHs), and A. thaliana (AtG6PDHs) were used for building a phylogenetic tree using MEGA 5.0 software based on neighbor-joining method with the default parameter values (Tamura et al., 2011). The gene structures of G6PDHs were affirmed using the GSDS database7 by aligning the coding regions with their corresponding genomic regions. The genomic sequences and coding sequences of G6PDH genes in G. max and A. thaliana were obtained from the soybean genetics and genomics database and NCBI database. The syntenic blocks among G. max, Z. mays, A. thaliana, O. sativa, P. vulgaris, M. truncatula, B. distachyon, and S. bicolor G6PDHs were identified from the plant genome duplication database (PGDD8) (Lee et al., 2012). The gene ID and other information of the G6PDHs used in this study were available in Supplementary Table S1.
To investigate the critical cis-acting elements in the promoter of GmG6PDH genes, 2.0 kb upstream of the position of the ATG codon in these genes were obtained from the soybean genetics and genomics database9. The cis-acting regulatory DNA elements were predicted from the PlantCARE database10 and presented by the IBS 2.0 (Liu et al., 2015).
The entire coding region of five GmG6PDH genes were amplified from the seeds of soybean cultivar “SN14” (provided by the Soybean Breeding Research Center of Northeast Agricultural University, Haerbin, China) by reverse transcription- polymerase chain reaction (RT-PCR) with the high-fidelity KOD-Plus-DNA polymerase (TOYOBO, Osaka, Japan). These genes were further constructed into pBI121 vector, which both contain a CaMV35S promoter and green fluorescent protein (GFP) tag. The gene-specific primers used for cloning the putative G6PDH genes were shown in Supplementary Table S2. The fusion proteins pBI121- GmG6PDHs:GFP or positive control (empty vector) were temporarily expressed in Arabidopsis mesophyll protoplasts, which were isolated from the leaves of 14 days-old seedlings grown under a weak light condition to minimize the chloroplast autofluorescence. The subcellular location of GmG6PDH-GFP proteins was monitored 14 h after polyethylene glycol (PEG)-mediated protoplast transfection protocol (Yoo et al., 2007). Confocal laser-scanning microscopy (LSM 710, Carl Zeiss, Jena, Germany) with a 488-nm argon ion laser (for GFP excitation) was used to visualize and localize GFP-tagged proteins. The excitation/emission wavelength were as follows for GFP (488 nm/507 to 535 nm) and chlorophyll autofluorescence (610 nm/650 to 750 nm).
The transcriptional patterns of GmG6PDHs in multiple tissues via high-throughput sequencing data from Phytozome database11, including leaves, root, root hairs, shoot apical meristem, nodules, stem, seed, pod, and flower tissues. The results are shown as heat maps with hierarchical clustering using the software TBtools 0.665 (Chen et al., 2018) and the values were log2-transformed with normalization. To analyze the transcriptional profiles of the GmG6PDHs in different stages of seed development, total RNA was extracted from soybean seeds at 4, 7, 14, 30, 50, 80, 110, and 120 days after flowering (DAF). The expression level of GmG6PDHs in developing seeds at 4DAF was used as a calibrator. To examine the transcriptional profiling of GmG6PDHs under various abiotic stresses, soybean seedlings at the second trifoliolate stage were subjected to salt stress induced by 150 mM NaCl, alkali stress induced by 100 mM NaHCO3, and osmotic stress induced by 20% (w/v) PEG (with a molecular weight of 6000 g/M) or 200 mM mannitol solutions. The osmotic potential of 20% PEG6000 and 200 mM mannitol was −0.53 and −0.50 MPa, respectively. Total RNA was extracted from leaf samples at 0, 6, and 12 h after the above treatments. The transcripts of GmG6PDHs in soybean leaf under normal environment condition were used as a calibrator. GmGAPDH and GmACTIN were used as internal reference. Each quantitative real time-polymerase chain reaction (qRT-PCR) reaction was performed in triplicate (technical replicates) on three biological replicates and the transcriptional level of GmG6PDHs was calculated based on the 2–ΔΔct method. All the primers used for qRT-PCR were available in Supplementary Table S2.
The CDS of GmG6PDH2 with the XhoI and NcoI sites was inserted into the prokaryotic expression vector pET32a (+). The recombinant plasmid pET32a- GmG6PDH2 was transformed into E. coli Rosetta strain to produce the putative recombinants. The positive clone was sequenced and cultivated in liquid LB medium supplemented with 1 mM IPTG at 37°C for 4 h to induce the expression of GmG6PDH2. The recombinant proteins were wall-broken by ultrasonic wave with a power output of 250 W for 10 min and then harvested by centrifuging at 10,000 g for 15 min. The supernatant was detected by 12% SDS-PAGE and collected for enzymatic activities assay. The total protein concentration was monitored by Bradford Protein Assay Kit purchased from Solarbio Science and Technology (Beijing, China). The kinetic parameters of GmG6PDH2 recombinant proteins with regard to the glucose-6-phosphate (G6P) and NAPD+ was determined using Eadie–Hofstee plot (Wakao and Benning, 2005).
G6PDH activity assays were performed as described by Wakao and Benning (2005), with slight modifications. The soybean roots (0.2 g) were extracted in 5 mL extraction buffer containing 50 mM 2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid -Tris(hydroxymethyl)aminomethane (Hepes-Tris) buffer (pH 7.8), 1 mM EDTA, 3 mM MgCl2 and 1 mM phenylmethylsulfonyl fluoride. The G6PDH activities were measured with regard to the oxidation of G6P by NADP+. The total reaction mixture was reduced to 1 mL with 0.5 mM NADPNa2, 0.5 mM D-glucose-6-phosphate disodium salt, 3.3 mM MgCl2, 50 mM Hepes-Tris (pH 7.8) and an appropriate amount of enzyme extracts (Wakao and Benning, 2005). Hepes-Tris buffer was made as follows: 0.5M Hepes was titrated to pH 7.8 with about 1M Tris, and then diluted 10-fold to give 50 mM Hepes.
The plasmid of pBI121-GmG6PDH2:GFP was transformed by electroporation into Agrobacterium rhizogenes strain K599, which was used to transform soybean hypocotyls. Soybean transformation in “SN14” hypocotyls and hairy root induction were performed as reported previously (Tóth et al., 2016). Soybean plants infected with the A. rhizogenes strain K599 were considered as control hairy roots. The transgenic lines were screened by PCR amplification and enzyme activity assay as described (Pan et al., 2016), and the non-transgenic hair roots were removed from the seedlings. Transgenic lines with similar-length hairy roots were selected and treated with 1/2 Hoagland solution containing 0 or 100 mM NaCl for 5 days, respectively. The root fresh weight and maximum root length of transgenic soybean plants were researched after 5 day of salt treatment. More than 10 independent hairy root lines were analyzed in this work to check the effects of GmG6PDH2 over expression on salinity stress responses.
The metabolites contents of AsA-GSH cycle, including AsA and GSH, and their oxidized forms, DHA and GSSG were determined using Ascorbic Acid or Glutathione Colorimetric Assay Kit purchased from Solarbio Science and Technology (Beijing, China), as per the manufacturer’s protocol. The assays of hydrogen peroxide (H2O2) content were conducted according to the method published previously (Velikova et al., 2000). The membrane damage was determined with regard to thiobarbituric acid- reactive substances (TBARS) content, a product of lipid peroxidation (Hodges et al., 1999). Briefly, the soybean root samples (0.5 g) were extracted with 10 mL 0.1% (w/v) trichloroacetic acid (TCA), and then the homogenate was centrifuged at 10,000 g for 10 min at 4°C. The supernatant was used for the determination of H2O2 and TBARS contents. The total reaction volume of H2O2 assay was 2 mL containing 0.5 mL of the supernatant, 1 mL 1M potassium iodide and 0.5 mL 10 mM potassium phosphate buffer (pH 7.0). The intensity was measured at 390 nm. The total reaction volume of TBARS assay was 2 mL containing 0.5 mL of the supernatant and 1.5 mL 0.5% (w/v) thiobarbital acid in 15% TCA. The absorbancy of supernatant was read at both 532 and 600 nm.
All experiments were performed with at least three biological replicates. Values are presented as mean ± SD. The significance of the data was evaluated using Student’s t-test with SPSS statistics 22.0 software. The significance level was set at P < 0.05.
In this study, full-length proteins and conserved domains of six glucose-6-phosphate dehydrogenases (G6PDHs) in A. thaliana were used as BLAST queries against the soybean genetics and genomics database12. A total of nine G6PDH genes were originally obtained in the soybean genome, which were designated as GmG6PDH1-9 (Table 1). Full-length coding sequences of GmG6PDH 1-9 ranged from 1560 to 1839 bp, and encoded nine putative proteins with 518 to 612 amino acid residues. The protein isoelectric points and molecular mass of the nine GmG6PDHs varied from 5.80/59.3 to 8.76/68.8 kDa, respectively (Table 1). Online software, CELLO 2.5 and TargetP 1.1, were used to assess the existence of a predicted N-terminal transit peptide (TP), which indicated the localization of GmG6PDH2, 4 and 6 in the cytosol, and others in the plastid (Table 1).
To examine the classification and evolutionary history of soybean G6PDHs, the full-length protein sequences of GmG6PDHs were aligned with homologous G6PDHs from S. bicolor (StG6PDH1-4), A. thaliana (AtG6PDH1-6), O. sativa (OsG6PDH1-5), Z. mays (ZmG6PDH1-6), B. distachyon (BdG6PDH1-5), M. truncatula (MtG6PDH1-7), and P. vulgaris (PvG6PDH1-5), and a phylogenetic tree was constructed. As shown in Figure 1A, the phylogenic analysis suggested that the different plant G6PDHs could be clearly classified into two major clades (I and II). Clade I corresponded to plastidic (P) isoforms containing four Arabidopsis P-G6PDHs (AtG6PDH1-4) (Wakao and Benning, 2005; Née et al., 2009). Clade I was further segmented into three classes (a, b, and c), in which GmG6PDH3 and GmG6PDH8 were subdivided into class a, along with an Arabidopsis P1 isoform (AtG6PDH1); GmG6PDH5, 7 and two Arabidopsis P2 isoforms (AtG6PDH2, 3) fall into class b; GmG6PDH1, 9 and an inactive-G6PDH isoform (AtG6PDH4) were clustered within class c. Moreover, clade II corresponded to the cytosolic (Cy) isoforms, including GmG6PDH2, 4 and 6, together with two Arabidopsis Cy-G6PDHs (AtG6PDH5, 6) (Wakao et al., 2008). The phylogenetic clades were in accordance with the in silico prediction of GmG6PDHs. As expected, the GmG6PDH isoforms were more closely related to its homolog from P. vulgaris in each cluster, which all belonged to the leguminous family (Figure 1A).
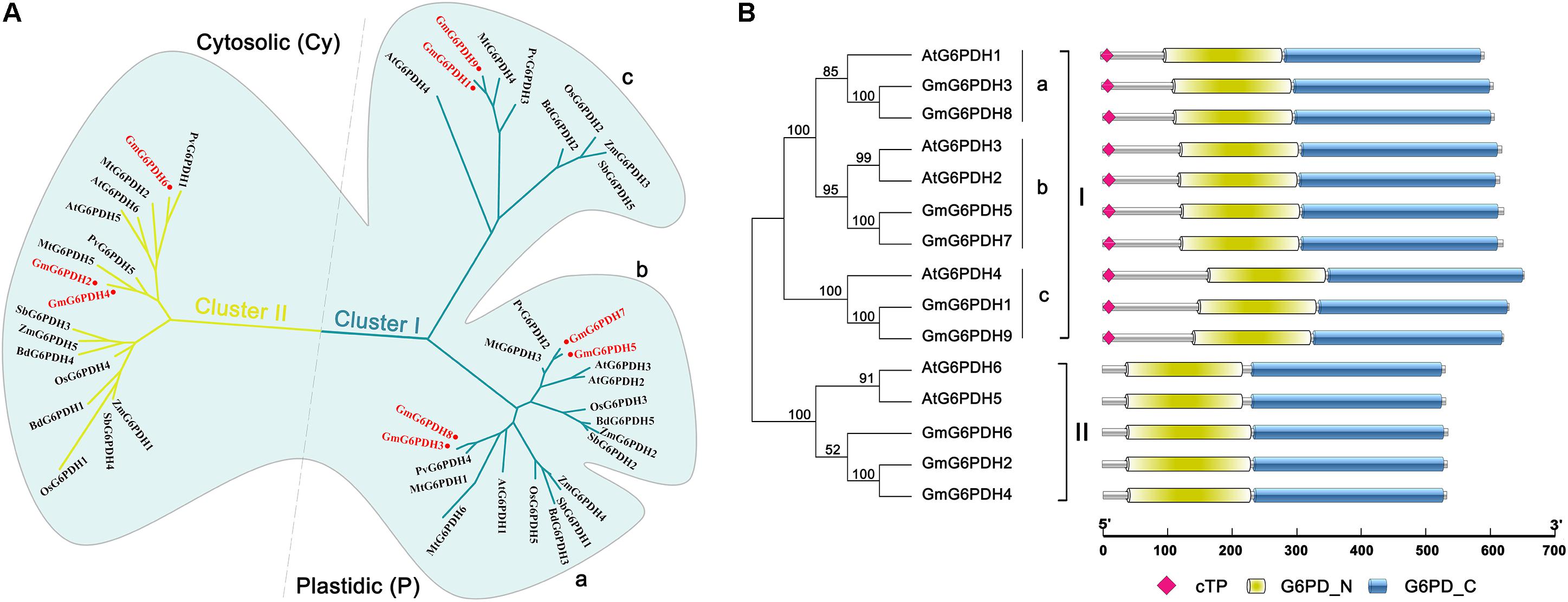
Figure 1. Phylogenic and protein domain analysis of GmG6PDHs. (A) Phylogenetic tree of G6PDH proteins from Glycine max (red circles), Arabidopsis thaliana, Zea mays, Oryza sativa, Phaseolus vulgaris, Medicago truncatula, Brachypodium Distachyon, and Sorghum bicolor. (B) The bi-domain structure of the soybean and Arabidopsis G6PDH proteins. Predicted chloroplast transit peptide (cTP) is depicted as colored diamonds.
Structural analysis of translated proteins for GmG6PDH1-9 demonstrated that the GmG6PDHs exhibited a bi-domain protein structure similar to the reported A. thaliana G6PDH proteins, consisting of an N-terminal NADP+-binding domain (PF00479) and a C-terminal G6PD domain (PF02781) (Figure 1B). Two motifs, NADP+-binding motif (NEFVIRLQP) and substrate-binding motif (RIDHYLGKE), were highly conserved in all the GmG6PDH proteins (Supplementary Figure S1). These protein sequences also contained a conserved Rossman fold (GXXGDLA) domain (Supplementary Figure S1). Based on the subcellular localization prediction, phylogenetic and protein structural analyses, GmG6PDHs were separated into two types: three cytosolic NADP+-G6PDH isoforms (GmG6PDH2, 4 and 6), and six plastidic NADP+-G6PDH isoforms (GmG6PDH1, 3, 5, 7, 8, and 9). The specific localization feature as represented by soybean G6PDH genes revealed the relatedness of distinct function for G6PDH isoforms in each clade with their evolutionary process.
A synteny analysis among G6PDHs from G. max, P. vulgaris, Z. mays, O. sativa, A. thaliana, M. truncatula, B. distachyon, and S. bicolor was performed in the present study to gain some insight into the potential function of GmG6PDHs. As shown in Figure 2A, the nine GmG6PDHs were scattered along seven out of twenty soybean chromosomes and each of the seven chromosomes comprised one to three GmG6PDHs. Moreover, a total of 30 orthologous pairs of G6PDHs were found in the above eight species (Supplementary Figure S2 and Supplementary Table S3). The GmG6PDHs had syntenic relationship only with PvG6PDHs, OsG6PDHs, SbG6PDHs and BdG6PDHs, including four orthologous gene pairs between G. max and O. sativa or S. bicolor, five orthologous gene pairs between G. max and B. distachyon, and 12 orthologous gene pairs between G. max and P. vulgaris (Figure 2A). Besides, five paralogous G6PDH gene pairs were confirmed in soybean genome, and the paralogous gene pairs were apt to be found among the members in the same subfamily (Figure 2A). To obtain further details about the structural diversity of GmG6PDH genes, we subsequently compared the localization and size of exon/intron among GmG6PDHs and AtG6PDHs. As shown in Figure 2B, G6PDH genes belonged to the same cluster shared the similar exon–intron structures, particularly in relation to exon numbers. For instance, G6PDH genes in cluster I possess 8–12 exons, and G6PDH genes in cluster II exhibited an equal number of exons (15) and nearly identical exons length. These results indicated that the GmG6PDH genes were highly conserved in gene sequence and exon–intron organization within each phylogenic group.
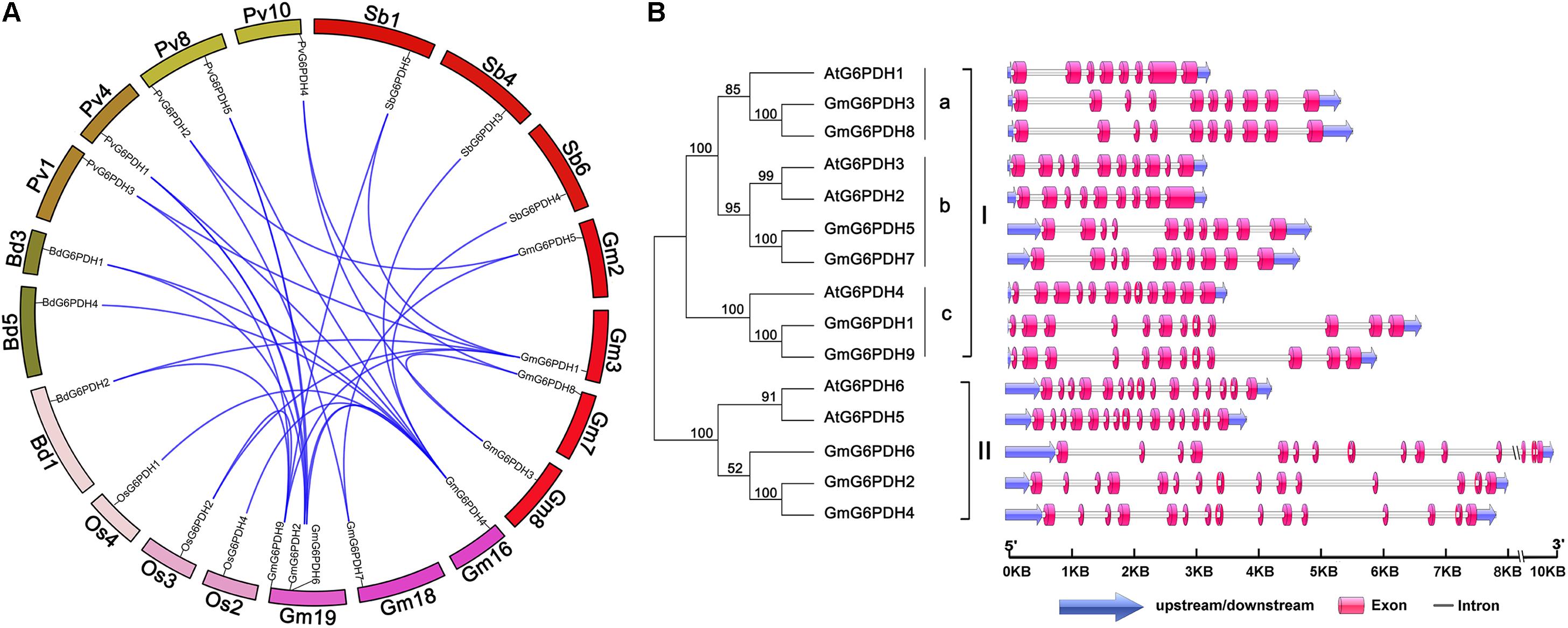
Figure 2. Syntenic and exon–intron structural analysis of GmG6PDH family genes. (A) Syntenic analysis of soybean G6PDHs with the corresponding genes in B. distachyon, P. vulgaris, S. bicolor, and Oryza sativa. The chromosome of the above species is shown as a circle. The colored curves indicate the collinear region of the G6PDHs. (B) The exon–intron organization of AtG6PDHs and GmG6PDHs. Untranslated regions (UTRs) are indicated by blue arrow. Exons and introns are visualized as colored boxes and gray lines respectively.
To identify putative cis-elements involved in GmG6PDHs transcriptional regulation, a 2.0 kb promoter region upstream from the ATG translation start codon of each GmG6PDH gene was analyzed. As shown in Figure 3, most GmG6PDH genes displayed the existence of some stress responsive cis-elements, such as ARE, a cis-regulatory element involved in anoxic-inducibility, was found in all GmG6PDH genes, except for GmG6PDH3. Beyond that, MBS, a cis-element involved in drought responsiveness, was found in GmG6PDH2, GmG6PDH4, GmGPDH6, and GmGPDH7 genes; TC-rich repeats, a cis-element associated to stress and defense responsiveness, was observed in GmG6PDH8; and LTR (cold-responsive element) was seen in GmG6PDH5 and GmG6PDH9 genes (Figure 3). Meanwhile, all GmG6PDH promoter sequences contained one or more cis-elements involved in response to multiple hormones, such as auxin-responsive element (TGA), ABA-responsive element (ABRE), and SA-responsive element (TCA). Remarkably, GCN4, a cis-acting element required for endosperm expression was observed in GmG6PDH6. Bioinformatics analyses of cis-elements suggest GmG6PDHs may be pivotal in mediating stress responses as well as plant growth.
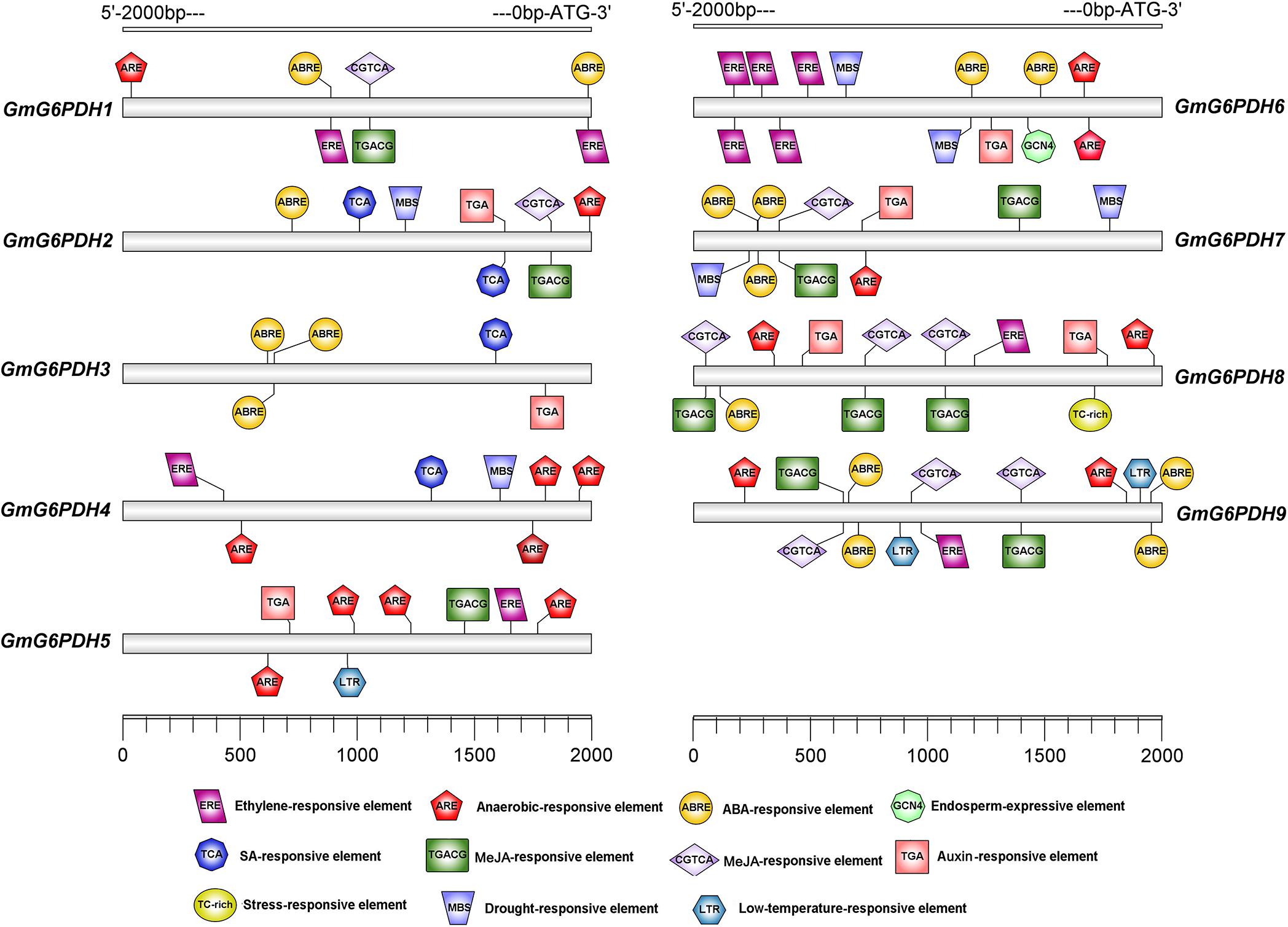
Figure 3. Cis-elements prediction in the 2.0 kb promoter region upstream from the start codon of GmG6PDHs. The relative positions of cis-elements in each GmG6PDH gene are marked by different-colored boxes.
The cellular location of G6PDH proteins is closely linked to and indicates their functions. To certify the subcellular localization of GmG6PDHs, the coding sequences of five GmG6PDHs (GmG6PDH1, 2, 5, 8, and 9) were successfully cloned, verified and submitted to GenBank with the following accession numbers: GmG6PDH1 (MN339553), GmG6PDH2 (MN339554), GmG6PDH5 (MN339555), GmG6PDH8 (MN339556), and GmG6PDH9 (MN339557). We then fused the coding region of five GmG6PDHs in-frame with the N-terminus of the GFP coding region. The positive control (35S: GFP) and GFP-tagged GmG6PDH proteins were transiently expressed in Arabidopsis mesophyll protoplasts. The free GFP protein was evenly distributed throughout the cell except for chloroplast and vacuole (Figure 4), while the GmG6PDH1, 5, 8 and 9 were specifically targeted to the chloroplast. Moreover, the GmG6PDH2 fusion protein was merely detected in the cytosol, suggesting that it encoded a cytoplasmic protein (Figure 4). These results were in line with the previous online prediction.
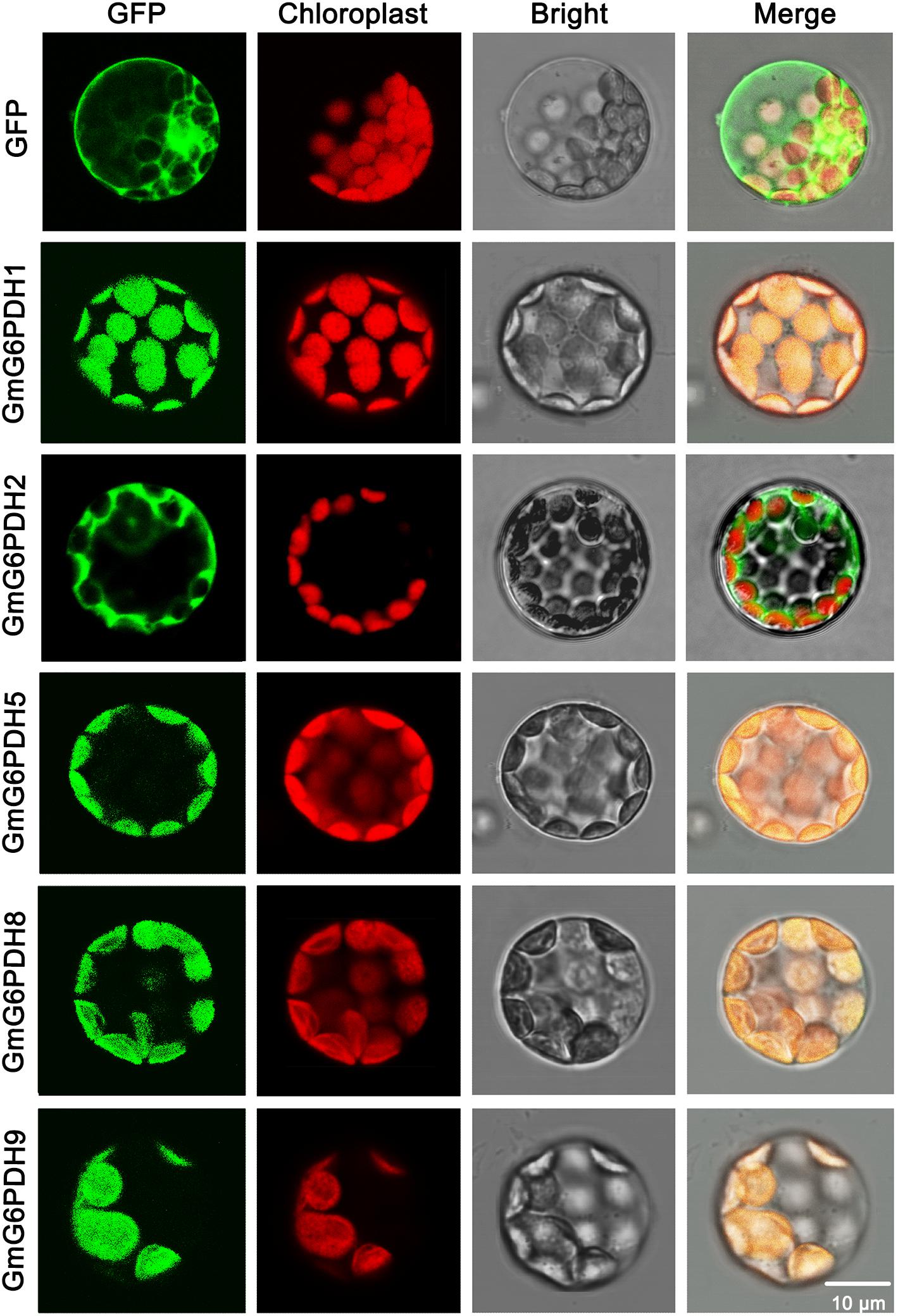
Figure 4. Subcellular localization analysis of GmG6PDHs by transiently expressing five GmG6PDH-GFP fusion proteins in Arabidopsis mesophyll protoplasts. Confocal micrographs showing the subcellular localization of GmG6PDH1, GmG6PDH2, GmG6PDH5, GmG6PDH6, and GmG6PDH9 proteins. Merged pictures of the GFP fluorescence (first panels), the chloroplast autofluorescence (second panels) and the corresponding bright field (third panels) are shown in the fourth panels.
In succession, we evaluated the transcriptional patterns of GmG6PDHs in multiple tissues via high-throughput sequencing data from Phytozome database, including leaves, root, root hairs, shoot apical meristem, nodules, stem, seed, pod, and flower tissues. The transcripts of nine GmG6PDHs could be observed in all the tissues tested, but the transcriptional profiles were different between the cytosolic (GmG6PDH2, 4, and 6) and plastidial (GmG6PDH1, 3, 5, 7, 8, and 9) isoforms (Figure 5A). The cytosolic GmG6PDHs were strongly expressed in roots, pod and nodules. Nonetheless, the transcripts of plastidial GmGPDH3, 5, and 9 genes were primarily seen in leaves, and GmG6PDH7 gene was mainly expressed in pod. The transcriptional levels of GmG6PDH1 and 8 genes were comparatively low in all nine tissues compared with other genes. Furthermore, the GmG6PDHs were classed into two major groups (I and II) based upon their expression patterns across all nine tissues, which is correlated with the phylogenetic clades for GmG6PDHs (Figure 5A). Within each subgroup, most of these genes exhibited similar expression profiles. Notably, the average relative mRNA levels of cytosolic GmG6PDHs in group II were much higher than that of plastidic GmG6PDHs in group I. The tissue-specific expression characteristics of GmG6PDHs reflected their versatile functions in multiple aspects of soybean growth and development.
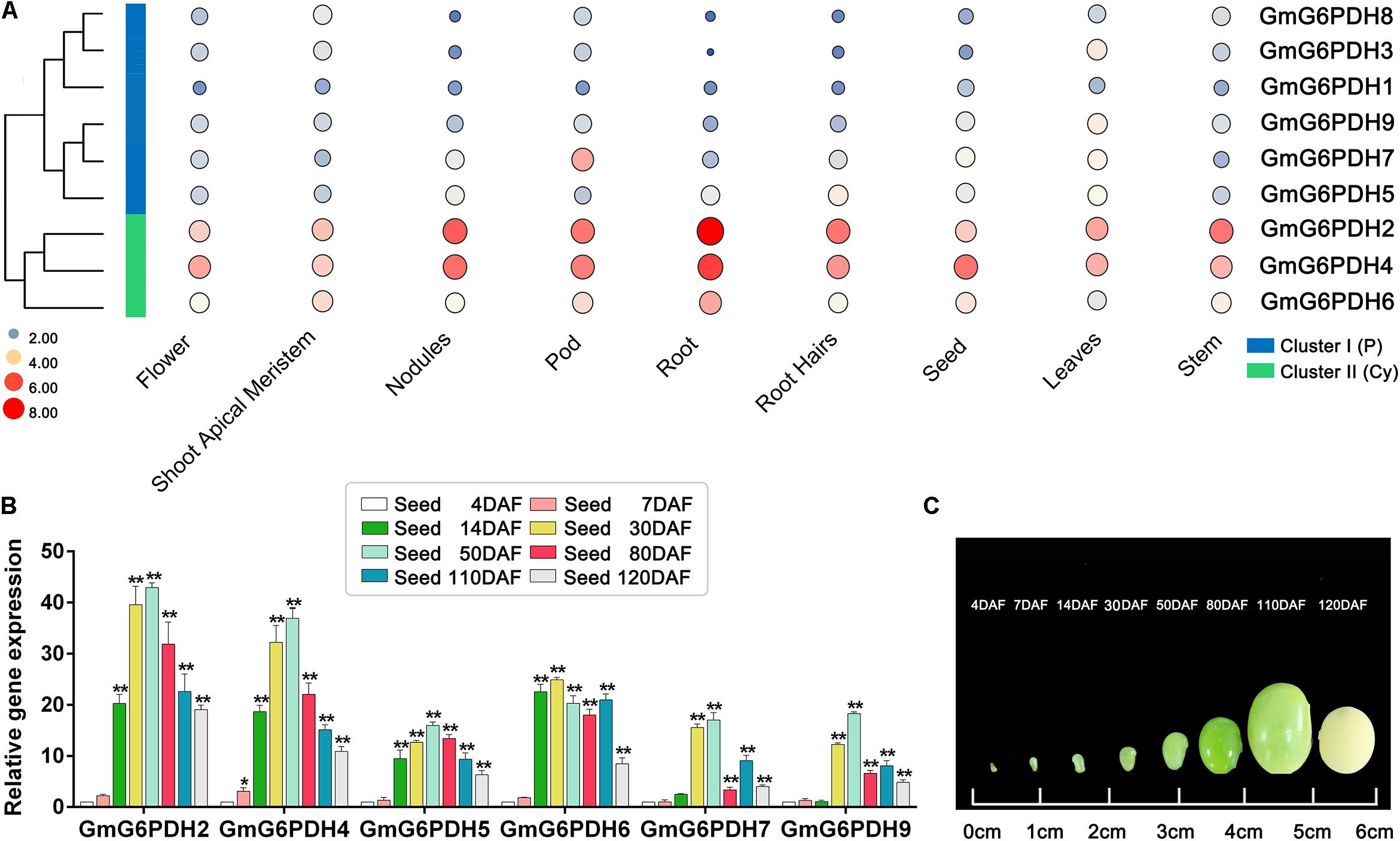
Figure 5. Expression profiles of GmG6PDHs in multiple tissues and developmental stages. (A) Expression patterns cluster analysis of GmG6PDHs involved in tissue development. The transcripts of GmG6PDH genes in various tissues are investigated from Phytozome database. The results are shown as heat maps. The color scale represents log2 expression values, with red denoting high-level transcription and blue denoting low-level transcription. The size of the circle also indicates the transcription level, with a larger circle indicating high-abundance transcripts. (B) The transcript profiling and (C) physiological phenotype of GmG6PDHs in developing seeds at 4, 7, 14, 30, 50, 80, 110, and 120 days after flowering (DAF). The transcripts of GmG6PDHs in developing seeds at 4 DAF were served as the internal reference. Three biological replicates for each tissue were collected. Asterisks above bars denote a statistically significant difference by Student’s t-test (*P < 0.05, **P < 0.01).
In addition, it has been reported that G6PDH enzymes are required for the accumulation of lipid and starch in developing seeds (Wakao et al., 2008). Therefore, to explore the potential function of GmG6PDHs during soybean seed development, the transcriptional patterns of GmG6PDHs in developing seeds at different stages was further affirmed by qRT-PCR. All the tested genes remained extremely high expression levels during early-middle seed development (14, 30, and 50 DAF) and low expression levels during the late maturation stage of seed development (110 and 120 DAF) (Figure 5B). The maximum level of GmG6PDH2, 4, 5, 7, and 9 genes expression occurred in developing seeds at 50 DAF, while the expression level of GmG6PDH6 was peaked in developing seeds at 30 DAF. Among the tested genes, the transcript abundance of cytosolic GmG6PDH2 gene was the highest at several stages of seed development (30, 50, 80, 110, and 120 DAF).
It is well-known that G6PDH genes are important for stress adaptation in several model plants (Wakao and Benning, 2005; Long et al., 2016). Also, the promoter analysis of soybean G6PDHs revealed a number of potential cis-elements and transcription binding motifs (MBS, ABRE, ARE, and TCA-elements) involved in responsiveness to abiotic stresses, such as salt and drought stresses frequently encountered in our area (northeast China). Hence, to further understand how GmG6PDHs respond to abiotic stresses, we analyzed the transcriptional profiles of GmG6PDHs under alkali (100 mM NaHCO3), salt (150 mM NaCl), and osmotic (20% PEG or 200 mM mannitol) stresses. As shown in Figure 6, there were distinct differences among cytosolic and plastidial GmG6PDH isoforms based on expression patterns under various stress conditions. The expression levels of cytosolic GmG6PDHs (GmG6PDH2, 4, and 6) were extraordinarily high upon salt stress, of which the transcript expression of GmG6PDH2 was highly inducible (reaching upto 100-fold) under the salinity stress treatment for 6 h. The plastidic forms GmG6PDH1, 3, 7, and 8 were also upregulated under salinity stress of 6 h (about 30–40-fold), while the average mRNA levels of P-G6PDHs was much lower than that of Cy-G6PDHs. Also, the cytosolic GmG6PDHs were dramatically induced at the early stage of PEG treatment, and the transcripts of GmG6PDH6 at 6 h of treatment were much higher than that of other genes. Furthermore, most of GmG6PDH genes were apparently up-regulated under alkaline treatment, particularly the plastidial GmG6PDHs, which had obviously high level of transcription at 6 h. In contrast, the cytosolic GmG6PDHs showed the maximum transcriptional level at 12 h of the same treatment. Likewise, the plastidial GmG6PDHs were originally stimulated after 6 h of mannitol treatment, maintaining comparatively high abundant transcripts throughout the entire treatment period, while the cytosolic GmG6PDHs were markedly up-regulated after 12 h (Figure 6).
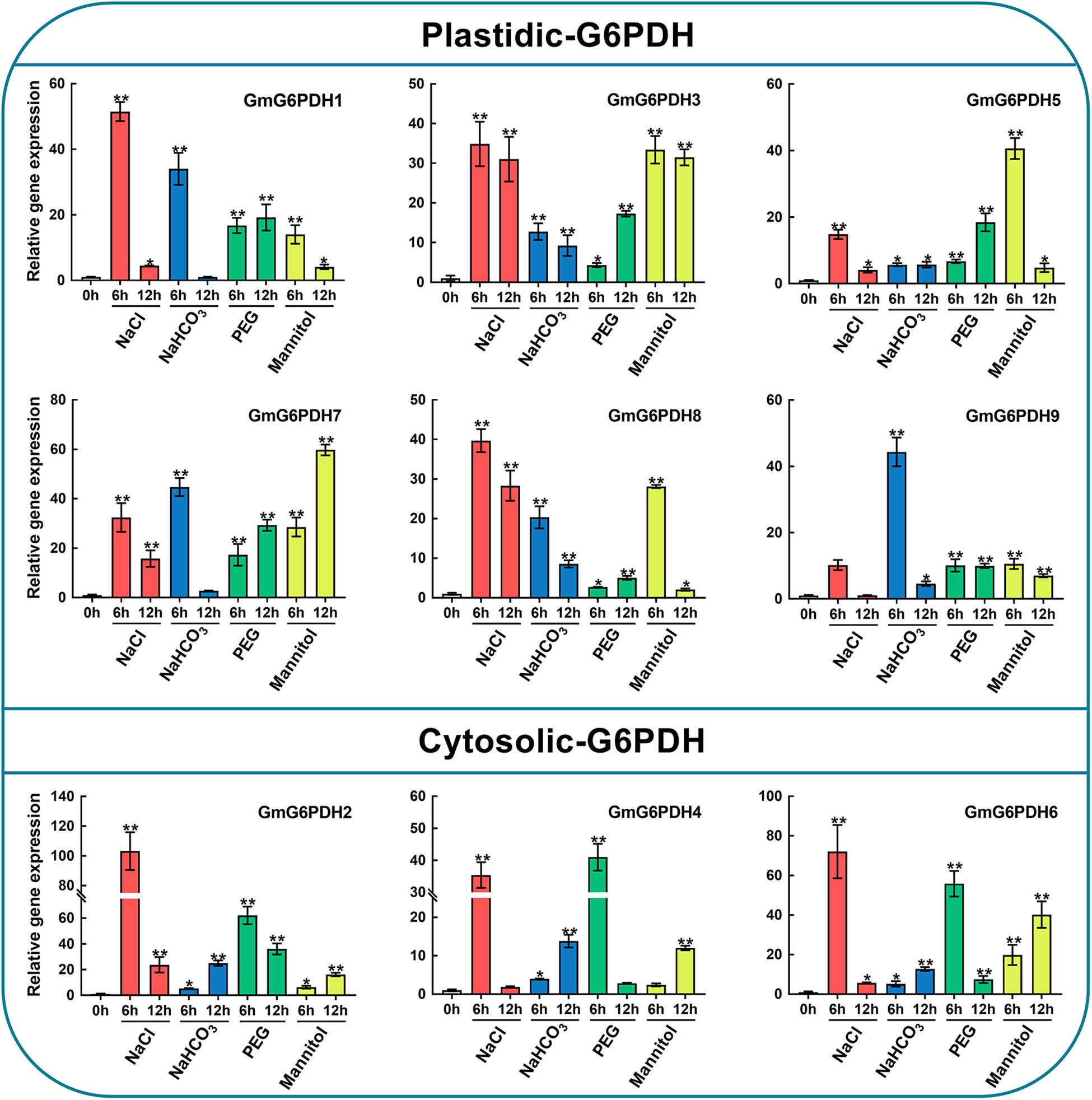
Figure 6. Expression profiles of GmG6PDHs in leaves of soybean plants subjected to 150 mM NaCl, 100 mM NaHCO3, 20% PEG, 200 mM mannitol or water (control) for 0, 6, and 12 h, respectively. The expression of GmG6PDHs in non-stress conditions was used as a calibrator. Three independent biological replicates were carried out and the results of qRT-PCR were analyzed using the 2–ΔΔct method. Asterisks above bars denote a statistically significant difference by Student’s t-test (*P < 0.05, **P < 0.01).
Similarly, the enzymatic assay revealed that the soybean G6PDH activity was significantly increased under NaCl, NaHCO3, PEG and mannitol treatments (Figure 7A). The G6PDH activities under the above treatments were approximately 1.5- to 4.2 -fold higher than that under the normal condition. What’s more, salt treatment exhibited an even higher stimulatory influence on G6PDH activity than other treatments, which causes the rapid increases of the enzyme activity within 6 h after salt stress (Figure 7A). During drought treatment, the G6PDH activity first peaked at 6 h and was raised again during the 12–24 h time period. The level of the G6PDH activity increased and then decreased during alkali treatment, and peaked at 12 h, while the G6PDH activity exhibited a continuous increase during osmotic treatment. Meanwhile, correlation analysis revealed the trend in G6PDH activity under abiotic stresses was well-consistent with gene expression of cytosolic GmG6PDH2, which indicated that it to encode the major G6PDH isoform involved in response to abiotic stresses (Figure 7B).
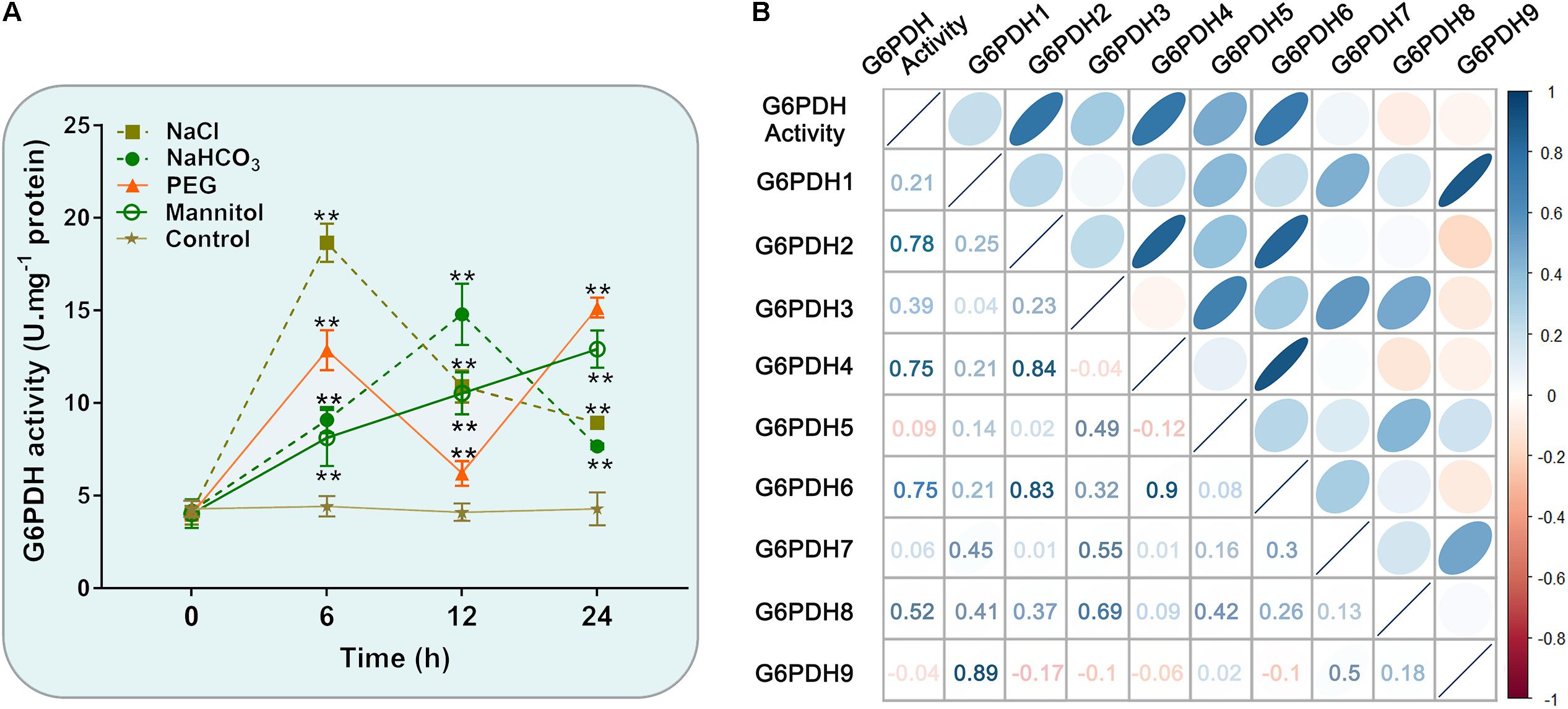
Figure 7. Enzyme activity profiles of G6PDH in soybean under different abiotic stress treatments. (A) Analysis of G6PDH activity in leaves of soybean plants subjected to 150 mM NaCl, 100 mM NaHCO3, 20% PEG, 200 mM mannitol or water (control) for 0, 6, 12, and 24 h, respectively. Data represent the means ± SD of three biological replicates. Asterisks above bars denote a statistically significant difference from the control group by Student’s t-test (*P < 0.05; **P < 0.01). (B) Correlation coefficient between G6PDH enzymatic activity and the expression levels of GmG6PDHs. Each ellipse chart represents the correlation coefficient between any two traits. The color and slope of the ellipse represent magnitude of correlation. The ellipses of negative correlations are displayed in red and positive correlations in blue.
To deeply comprehend the resistance functions of GmG6PDH2 gene, its catalytic characteristics were first determined by extracting the crude proteins from E. coli cells expression plasmid of His-tagged GmG6PDH2. An expected molecular mass of GmG6PDH2-His fusion protein (consisting of histidine marker and target gene) was identified by SDS/PAGE (Figure 8A). The recombinant GmG6PDH2 protein was analyzed for its enzyme kinetic with regard to both substrates: glucose-6-phosphate (G6P) and NADP+. The kinetic parameters were calculated by using Eadie–Hofstee data plots, in which the Km of G6P and NADP+ was estimated as 2.14 and 0.033 mM respectively, and the Vmax of G6P and NADP+ was estimated as 0.67 and 0.62 umol⋅min–1⋅mg–1 protein respectively (Figures 8B,C). These results demonstrated that the protein encoded by GmG6PDH2 had the functional NADP+-dependent G6PDH enzyme activity.
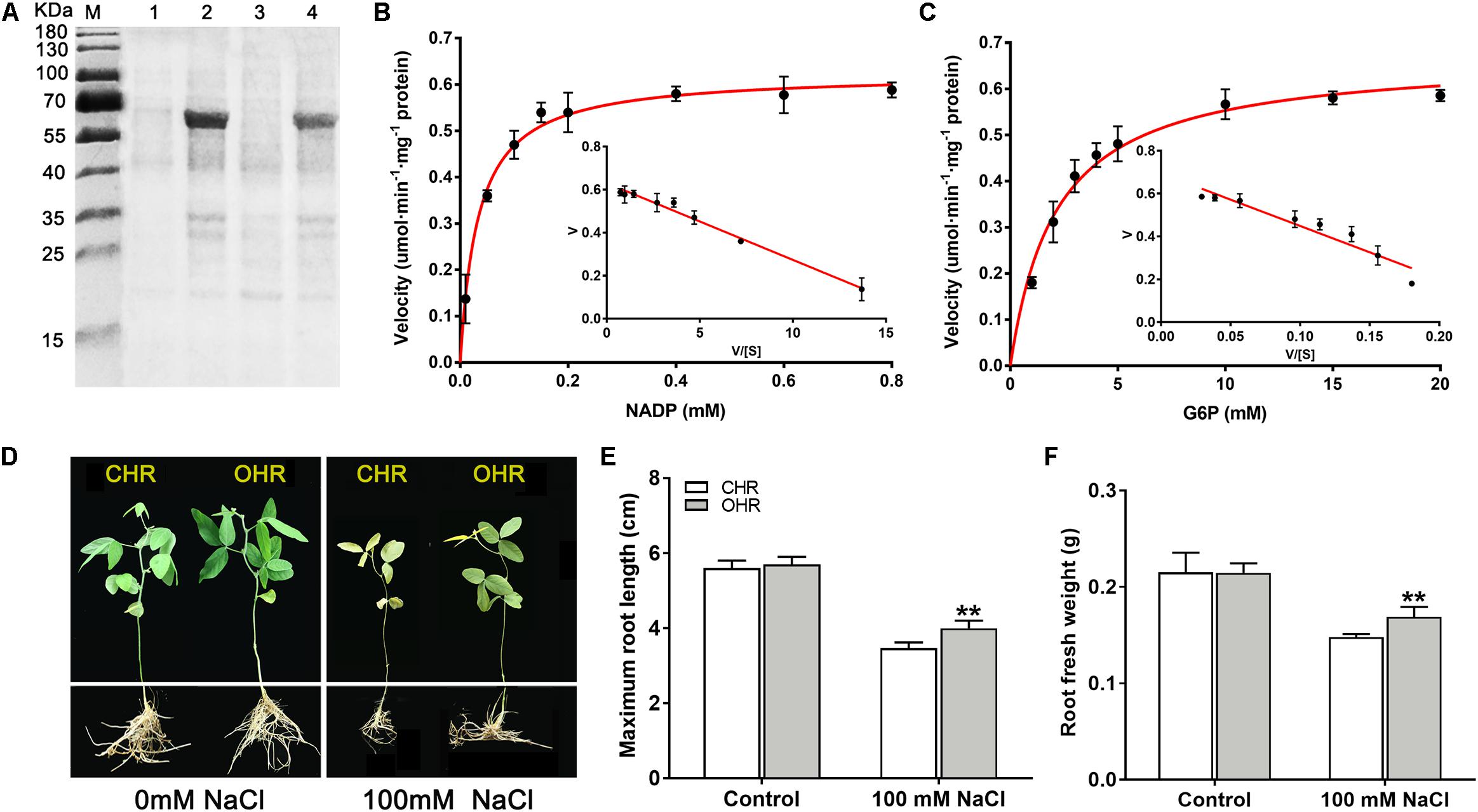
Figure 8. Overexpression of cytosolic NADP+-dependent isoform (GmG6PDH2) confers salt tolerance in transgenic soybean. (A) Coomassie-stained 12% SDS-PAGE of crude extracts of Escherichia coli cells harboring pET32a-GmG6PDH2 without (Lanes 1, 3) and with IPTG induction (Lanes 2, 4). The kinetic properties of GmG6PDH2 with regard to (B) NADP+ and (C) glucose-6-phosphate by Eadie–Hofstee plot. (D) Performance of soybean plants harboring GmG6PDH2-overexpressing hairy roots (OHR) and control hairy roots (CHR) treated with 0 or 100 mM NaCl for 5 days. CHR plants are generated after the infection of Agrobacterium rhizogenes. Comparisons of (E) maximum root length and (F) root fresh weight of CHR and OHR plants subjected to 0 or 100 mM NaCl for 5 days. More than 10 hair root lines were investigated and shown as means ± SD. Asterisks above bars denote a statistically significant difference from the CHR plants by Student’s t-test (*P < 0.05, **P < 0.01).
Next, to better elucidate how GmG6PDH2 gene contribute to the response to high salinity, GmG6PDH2-overexpressing soybean hairy roots (OHRs) were generated by Agrobacterium rhizogenes-mediated gene transformation, of which 10 positive transgenic plants were verified by PCR detection (Supplementary Figure S3). The physiological activity test signified that the G6PDH enzyme activity in GmG6PDH2-OHR lines were 1.7- to 2.5-fold higher than that in the control hair roots (CHR), which suggested that GmG6PDH2 gene was expressed successfully and functioned with G6PDH activities in OHR lines (Supplementary Figure S3). To look into the effects of GmG6PDH2 overexpression on salt tolerance at soybean seedling stage, 4-week-old CHR and OHR plants were transferred to 1/2 Hoagland solution supplemented with 0 or 100 mM NaCl for 5 days. As demonstrated in Figure 8D, the CHR seedlings exhibited a severe growth-inhibitory phenotype, with a partial leaf shriveling phenomenon, whereas GmG6PDH2-OHR plants displayed significantly improved resistance to salinity stress, as reflected by a higher root fresh weight and root length (Figures 8E,F).
It has been reported that the plant G6PDHs perform significant functions in maintaining the cellular redox balance (Krüger et al., 2011; Cardi et al., 2016). To validate whether overexpression of cytosolic GmG6PDH2 gene could affect the redox fluctuation, the redox states of AsA and GSH were monitored in soybean transgenic hair roots. The levels of reduced and oxidized AsA and GSH did not change among the CHR and OHR plants under standard growing conditions (Figures 9A,B). Nevertheless, the GmG6PDH2-OHR plants accumulated higher contents of AsA and GSH, and substantially lower GSH and GSSG contents than the CHR plants, thereby resulting in much higher ratios of AsA/DHA and GSH/GSSG. These results illustrated that the function of GmG6PDH2 gene in salt tolerance could partly ascribe to the improved regeneration of the reduced form of AsA and GSH.
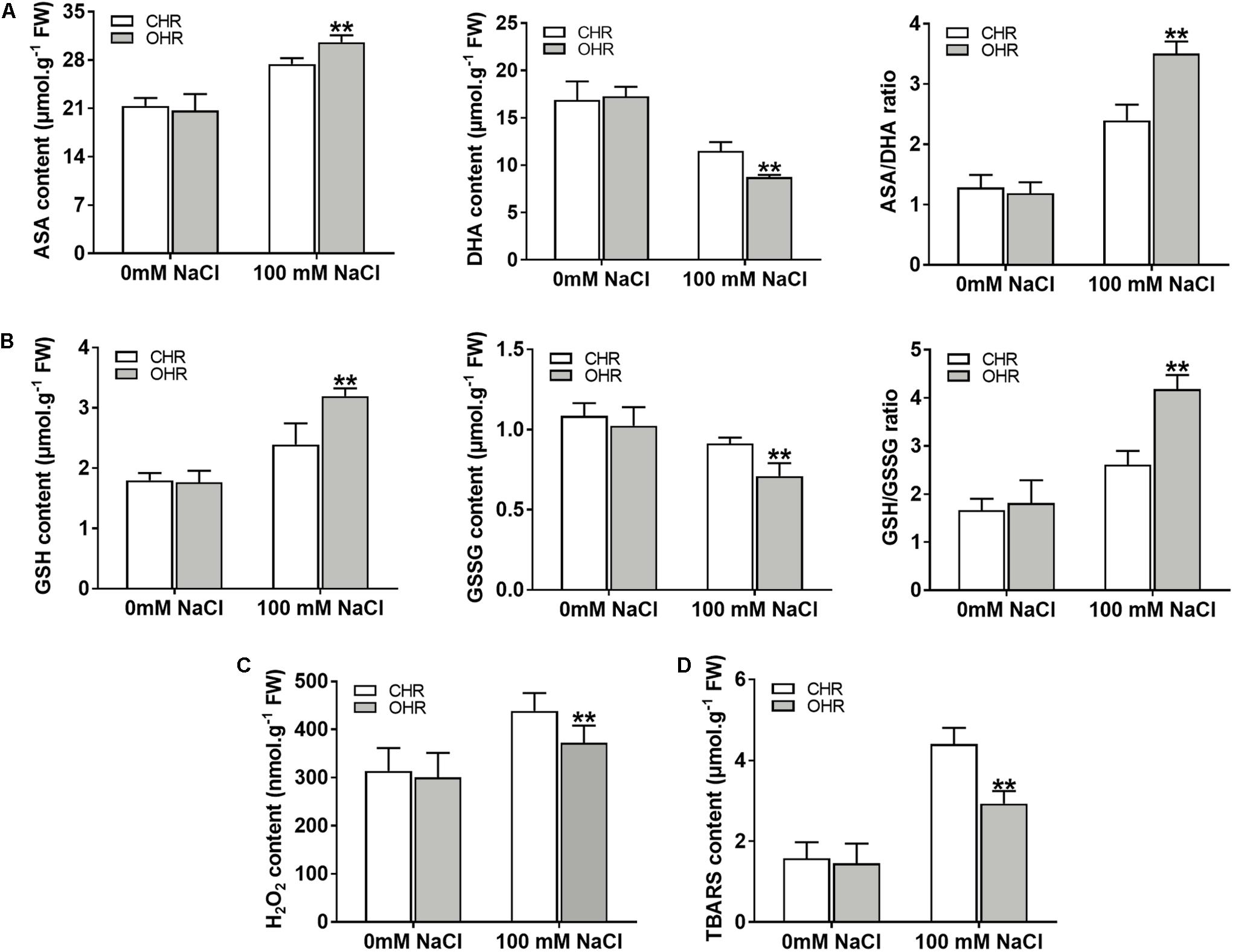
Figure 9. Overexpression of GmG6PDH2 improves the ROS-scavenging capacity under high salinity condition. (A) The contents of AsA and DHA, and the ratio of AsA/DHA; (B) The contents of GSH and GSSG, and the ratio of GSH/GSSG; (C) H2O2 and (D) TBARS levels in CHR and OHR plants subjected to 0 or 100 mM NaCl for 5 days. More than 10 independent transgenic hairy root lines were analyzed and shown as means ± SD. Asterisks above bars denote a statistically significant difference from the CHR plants by Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
In plant stress reactions, the AsA-GSH cycle has been considered as a powerful ROS scavenging pathway, exclusively eliminating the cellular hydrogen peroxide (H2O2) through utilizing the AsA and GSH (Fotopoulos et al., 2010; Li et al., 2010). Thus, the remarkable changes in reduced and oxidized forms of ASA and GSH in GmG6PDH2 transgenic lines reminded us to examine the H2O2 contents upon salt stress. As expected, the contents of H2O2 in GmG6PDH2-OHR lines were remarkably lower than that in the CHR plants, supporting an important role for GmG6PDH2 in ROS detoxification under salt stress (Figure 9C). Moreover, the TBARS levels, as an indicator of lipid peroxidation, were evidently lower in OHR plants than that in the CHR plants under salt stress (Figure 9D). In conclusion, these results suggested that overexpression of cytosolic GmG6PDH2 gene alleviated the stress-induced ROS accumulation through the regulation of the redox status of AsA and GSH, and consequently minimize the cell membrane damages under salt stress.
Soybean as the important economic food crop is one of the major sources of plant protein and oil for mankind, and its growth is largely affected by different abiotic stresses (Ghosh and Islam, 2016). A great number of researches have shown that G6PDHs play a critical role in plant growth and abiotic stress responses (Esposito, 2016; Hýsková et al., 2017). Although G6PDH genes have been cloned from several model organisms such tobacco (Scharte et al., 2009; Silva et al., 2018), Arabidopsis (Wakao and Benning, 2005), barley (Cardi et al., 2013; Caretto et al., 2015), wheat (Nemoto and Sasakuma, 2000) and tomato (Landi et al., 2016), there is scarce information about their biological functions in soybean. In this study, we identified nine G6PDH gene family members from the soybean genome, named as GmG6PDH1-9 (Table 1). Similar to other reported G6PDHs, all the GmG6PDH proteins contained the typical and necessary protein domains (PF00479, PF02781) (Figure 1B). Meanwhile, GmG6PDH genes have two highly conserved motifs in their translated protein sequences: NADP+-binding motif (NEFVIRLQP) and substrate- binding motif (RIDHYLGKE) (Supplementary Figure S1), illustrating the proteins encoded by GmG6PDH1-9 have enzyme catalytic activities (Hauschild and von Schaewen, 2003). According to the putative subcellular location, GmG6PDHs were segmented into two types: three cytosolic (Cy) isoforms (GmG6PDH2, 4, and 6), six plastidic (P) isoforms (GmG6PDH1, 3, 5, 7, 8, and 9), which was in accord with previous reports (Esposito et al., 2001; Wakao and Benning, 2005). Phylogeny analyses also resulted in the same classification of GmG6PDHs by dividing them into two clades (I and II), with clade I corresponding to P-G6PDHs and clade II corresponding to Cy-G6PDHs (Figure 1A). These results indicated that the divergence of G6PDHs into Cy and P types may be to ensure the plant adaptations to changing conditions.
All the GmG6PDHs were found having a close relationship with the corresponding genes in common bean, which is in agreement with their genetic relationships, suggesting possible functional conservation (Figure 1B). Additionally, we found that GmG6PDHs had collinear relationships with S. bicolor, O. sativa, P. vulgaris and B. distachyon G6PDHs, illustrating that GmG6PDHs might have occurred before the divergence of sorghum, rice, common bean and purple false brome grass lineages (Figure 2). The predicted of subcellular localization of plant G6PDHs are basically assigned dependent on the presence of signal peptide (Yu et al., 2004). In the present work, we assessed the subcellular localization of five soybean G6PDHs (GmG6PDH1, 2, 5, 8, and 9) by the transient expression of GFP-tagged recombinant proteins in Arabidopsis mesophyll protoplasts (Figure 4). The merged image of chloroplast autofluorescence and GFP indicated that the GmG6PDH1, 5, 8, and 9 fusion proteins were clearly located in the chloroplast, while the GmG6PDH2 protein was specially targeted to cytosol (Figure 4). The results correlated with the in silico prediction and phylogenetic clades for GmG6PDHs. The earlier identified AtG6PDH proteins were predicted to be cytosol or plastid-localized isoforms because of the absence or presence of pronounced targeting signal and transmembrane domains (Wakao and Benning, 2005), but a clear experimental evidence was lacking.
Using quantitative real-time qRT-PCR and high-throughput sequencing data analyses, we examined the transcriptional profiling of GmG6PDHs in different soybean tissues and at different times during seed development. Our data showed differences in the expression level of GmG6PDHs in distinct tissues. To be specific, the plastidic GmG6PDHs exhibited a tissue-specific transcript profiling with the high mRNA levels in green tissue, such as leaves and pods (Figure 5A), consistent with the expression profiles of their orthologous genes from Hevea brasiliensis, A. thaliana, and Nicotiana tabacum (Wakao and Benning, 2005; Gong et al., 2012; Long et al., 2016). By contrast, the expression analysis of cytosolic GmG6PDH genes revealed relatively high-abundant transcripts in developing seeds, roots and nodules (Figures 5A,B). In Arabidopsis, the Cy-G6PDH isoforms are also proved to be crucial in the metabolism of developing seeds (Wakao et al., 2008), suggesting that the cytosolic G6PDH genes mainly function in sorts of sink tissues. The tissue-specific expression characteristics indicate that GmG6PDHs may play versatile physiological roles in soybean development (Figures 5A–C).
In addition to regulating plant growth, the key functions of plant G6PDHs in stress- response mechanisms have also been widely proven (Esposito, 2016). The promoter regions of the GmG6PDHs contained lots of cis-acting elements that possibly participated in plant responses to drought and salt stresses, such as MBS, ABRE, ARE, and TCA-elements (Figure 3) (Liu et al., 2017). By qRT-PCR and enzyme activity assays, we further examined the physiological and transcriptional responses of GmG6PDHs to different stress conditions, including salt, alkali and osmotic, and significant induction in GmG6PDHs was observed under the above treatments, especially under salt stress (Figures 6, 7). In addition, the different osmotic stresses induced by PEG6000 and mannitol have different effects on the transcription and expression of GmG6PDHs, probably due to the inhibitory effects of PEG on root oxygen availability (Guo et al., 2018). It has also been reported that the activities and transcripts of AtG6PDHs (Wakao and Benning, 2005), HbG6PDHs (Long et al., 2016), ScG6PDHs (Yang et al., 2014), and PsG6PDHs (Lin et al., 2005) are markedly stimulated by adverse environmental conditions, like oxygen availability, salinity, cold, and dehydration. Furthermore, the subcellular location of GmG6PDHs seemed to have certain impacts on their stress responses, as represented by the higher average mRNA expression levels of cytosolic GmGPDH2, 4 and 6 in comparison with plastidic GmG6PDHs under various stresses. Meanwhile, it is worthwhile to note that a cytosolic isoform (GmG6PDH2) respond faster and more vigorously to salinity stress than other genes, reaching its maximum mRNA level (about 100-fold) within 6 h of salinity treatment. The average mRNA abundances of GmGPDH2 under salt stress was much higher than that of other genes, and its expression patterns correlated well with the activity of G6PDH enzyme (Figure 7), implying a major role in response to salinity. Same results have been available earlier in other plants: a cytosolic G6PDH encoded by WESR5 are proved to be important for early salt responding in wheat (Nemoto and Sasakuma, 2000, 2002); and a cytosolic ScG6PDH also plays a positive role in salt-induced responses in sugarcane, but the corresponding functional verification is lacking (Yang et al., 2014).
In this study, the underlying molecular metabolism of the cytosolic GmG6PDH2 in mediating salinity adaption was further studied. Enzymatic properties analysis of recombinant GmG6PDH2 proteins expressed in E. coli (Figure 8A) showed that the protein encoded by GmG6PDH2 gene had substrate affinity (KmG6P of 2.138 mM) (Figure 8C), and a good agreement was achieved compared to the enzymatic characteristics available in previously published G6PDHs (Hauschild and von Schaewen, 2003; Scharte et al., 2009). In addition, overexpression of GmG6PDH2 dramatically enhanced the salt tolerance in transgenic soybean plants, as reflected by a noticeable increase in root length and root fresh weight (Figures 8E,F). Cytosolic G6PDHs have been previously proved to participate in the modulation of cellular redox states by supply for NADPH (Xu et al., 2003; Valderrama et al., 2006). In the present work, an apparent elevation in the levels of antioxidants (AsA and GSH) as well as the ratios of AsA/DHA and GSH/GSSG were seen in GmG6PDH2-OHR plants related to the CHR plants (Figures 9A,B). These data validate that GmG6PDH2 plays a pivotal role in modulating the changes of metabolite contents and the redox state of AsA and GSH pool under salinity condition, which was consistent with previous results (Krüger et al., 2011; Wang et al., 2016). It is known that the AsA-GSH cycle has a main function of eliminating H2O2, a potentially harmful ROS (Fotopoulos et al., 2010; Li et al., 2010). In agreement with this, we found that the levels of H2O2 were obviously lower in GmG6PDH2 OHRs than in control plants (Figure 9C). In addition, the cellular membrane damage in GmG6PDH2-OHR plants was less severe as manifested by the reduction in TBARS content under salt stress (Figure 9D), suggesting that overexpression of GmG6PDH2 alleviate the salt-induced ROS accumulation and consequently minimize the membrane lipid peroxidation. In summary, the cytosolic GmG6PDH2 gene has profound effects on salt tolerance by increasing components of the AsA-GSH cycle which subsequently may act to prevent oxidative stress caused by salinity.
Nine soybean G6PDH genes were characterized in the soybean genome. Based on the subcellular localization and phylogenetic analysis, GmG6PDHs were divided into plastidic (P) and cytosolic (Cy) isoforms. The GmG6PDH genes had distinct expression patterns under various abiotic stresses, reflected the potential functional distinction of each isoform. Of the nine GmG6PDH genes, the cytosol-localized GPDH (GmG6PDH2) gene had an apparent transcriptional response to salinity, and the expression of GmG6PDH2 showed a high correlation with the G6PDH activity, suggesting a principal participant in response to salinity. Further study indicated that GmG6PDH2 gene encode active G6PDH enzyme, which was coordinated with AsA-GSH cycle to maintain the redox state of AsA and GSH and consequently minimize the NaCl-induced oxidative damages.
All datasets generated for this study are included in the article/Supplementary Material.
YZ and YC designed and conceived the experiments. YZ, YC, SH, and JY performed the experiments. XW, DX, XL, YL, and YD analyzed the data and interpreted the results. YZ prepared the manuscript. ZQ and QC conceived the experiments and revised the manuscript. All authors agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved and approved the final version to be published.
This study was financially supported by EUCLEG (727312; 2017YFE0111000), National Key R&D Program of China (2016YFD0100500, 2016YFD0100300, and 2016YFD0100201-21), Post-Doctoral Project of Northeast Agricultural University (NEAUBH-19002), National Natural Science Foundation of China (31701449, 31971968, 31971899, and 31501332), Natural Science Foundation of Heilongjiang (QC2017013), Special Financial Aid to Post-doctor Research Fellow in Heilongjiang (LBH-TZ1714), Heilongjiang Academy of Agricultural Sciences Funds (2019YYYF 019), International Post-doctoral Exchange Fellowship Program of China Post-doctoral Council (20180004), China Post-Doctoral Project (2015M581419), Heilongjiang Funds for Distinguished Young Scientists (JC2016004 and JC2017006), Dongnongxue zhe Project (to QC), Backbone of Young Talent scholar Project (to ZQ, 18XG01) of Northeast Agricultural University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Jinhui Wang (Northeast Agricultural University, China) for providing us soybean seeds.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00214/full#supplementary-material
Cardi, M., Castiglia, D., Ferrara, M., Guerriero, G., Chiurazzi, M., and Esposito, S. (2015). The effects of salt stress cause a diversion of basal metabolism in barley roots: possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 86, 44–54. doi: 10.1016/j.plaphy.2014.11.001
Cardi, M., Chibani, K., Castiglia, D., Cafasso, D., Pizzo, E., Rouhier, N., et al. (2013). Overexpression, purification and enzymatic characterization of a recombinant plastidial glucose-6-phosphate dehydrogenase from barley (Hordeum vulgare cv. Nure) roots. Plant Physiol. Biochem. 73, 266–273. doi: 10.1016/j.plaphy.2013.10.008
Cardi, M., Zaffagnini, M., De Lillo, A., Castiglia, D., Chibani, K., Gualberto, J. M., et al. (2016). Plastidic P2 glucose-6P dehydrogenase from poplar is modulated by thioredoxin m-type: distinct roles of cysteine residues in redox regulation and NADPH inhibition. Plant Sci. 252, 257–266. doi: 10.1016/j.plantsci.2016.08.003
Caretto, S., Linsalata, V., Colella, G., Mita, G., and Lattanzio, V. (2015). Carbon fluxes between primary metabolism and phenolic pathway in plant tissues under stress. Int. J. Mol. Sci. 16, 26378–26394. doi: 10.3390/ijms161125967
Castiglia, D., Cardi, M., Landi, S., Cafasso, D., and Esposito, S. (2015). Expression and characterization of a cytosolic glucose-6-phosphate dehydrogenase isoform from barley (Hordeum vulgare) roots. Protein Expr. Purif. 112, 8–14. doi: 10.1016/j.pep.2015.03.016
Chen, C., Chen, H., He, Y., and Xia, R. (2018). TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv [Preprint]. doi: 10.1101/289660
Dal Santo, S., Stampfl, H., Krasensky, J., Kempa, S., Gibon, Y., Petutschnig, E., et al. (2012). Stress-induced GSK3 regulates the redox stress response by phosphorylating glucose-6- phosphate dehydrogenase in Arabidopsis. Plant Cell 24, 3380–3392. doi: 10.1105/tpc.112.101279
Devi, R., Munjral, N., Gupta, A. K., and Kaur, N. (2007). Cadmium induced changes in carbohydrate status and enzymes of carbohydrate metabolism, glycolysis and pentose phosphate pathway in pea. Environ. Exp. Bot. 61, 167–174. doi: 10.1016/j.envexpbot.2007.05.006
Esposito, S. (2016). Nitrogen assimilation, abiotic stress and glucose 6-phosphate dehydrogenase: the full circle of reductants. Plants 5:E24. doi: 10.3390/plants5020024
Esposito, S., Carfagna, S., Massaro, G., Vona, V., and Rigano, V. D. M. (2001). Glucose-6-phosphate dehydrogenase in barley roots: kinetic properties and localisation of the isoforms. Planta 212, 627–634. doi: 10.1007/s004250000443
Esposito, S., Guerriero, G., Vona, V., Di Martino Rigano, V., Carfagna, S., and Rigano, C. (2004). Glutamate synthase activities and protein changes in relation to nitrogen nutrition in barley: the dependence on different plastidic glucose-6P dehydrogenase isoforms. J. Bot. 56, 55–64.
Fotopoulos, V., Ziogas, V., Tanou, G., and Molassiotis, A. (2010). “Involvement of AsA/DHA and GSH/GSSG ratios in gene and protein expression and in the activation of defence mechanisms under abiotic stress conditions,” in Ascorbate-Glutathione Pathway and Stress Tolerance in Plants (Berlin: Springer), 265–302. doi: 10.1007/978-90-481-9404-9_10
Ghosh, A., and Islam, T. (2016). Genome-wide analysis and expression profiling of glyoxalase gene families in soybean (Glycine max) indicate their development and abiotic stress specific response. BMC Plant Biol. 16:87. doi: 10.1186/s12870-016-0773-9
Gong, H., Chen, G., Li, F., Wang, X., Hu, Y., and Bi, Y. (2012). Involvement of G6PDH in heat stress tolerance in the calli from Przewalskia tangutica and Nicotiana tabacum. Biol. Plant. 56, 422–430. doi: 10.1007/s10535-012-0072-8
Guo, Y. Y., Tian, S. S., Liu, S. S., Wang, W. Q., and Sui, N. (2018). Energy dissipation and antioxidant enzyme system protect photosystem II of sweet sorghum under drought stress. Photosynthetica 56, 861–872. doi: 10.1007/s11099-017-0741-0
Hauschild, R., and von Schaewen, A. (2003). Differential regulation of glucose-6-phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol. 133, 47–62. doi: 10.1104/pp.103.025676
Hodges, D. M., DeLong, J. M., Forney, C. F., and Prange, R. K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207, 604–611. doi: 10.1007/s004250050524
Honjoh, K., Machida, T., Hagisako, T., Suga, K., Yonekura, M., Shimizu, H., et al. (2007). Molecular cloning and characterization of a cDNA for low-temperature inducible cytosolic glucose 6-phosphate dehydrogenase gene from Chlorella vulgaris and expression of the gene in Saccharomyces cerevisiae. Plant Sci. 172, 649–658. doi: 10.1016/j.plantsci.2006.12.004
Hýsková, V., Pliskova, V., Červený, V., and Ryšlavá, H. (2017). NADP-dependent enzymes are involved in response to salt and hypoosmotic stress in cucumber plants. Gen. Physiol Biophys. 36, 247–258. doi: 10.4149/gpb_2016053
Krüger, A., Grüning, N. M., Wamelink, M. M., Kerick, M., Kirpy, A., Parkhomchuk, D., et al. (2011). The pentose phosphate pathway is a metabolic redox sensor and regulates transcription during the antioxidant response. Antioxid. Redox Signal. 15, 311–324. doi: 10.1089/ars.2010.3797
Landi, S., Nurcato, R., De Lillo, A., Lentini, M., Grillo, S., and Esposito, S. (2016). Glucose-6- phosphate dehydrogenase plays a central role in the response of tomato (Solanum lycopersicum) plants to short and long-term drought. Plant Physiol. Biochemi. 105, 79–89. doi: 10.1016/j.plaphy.2016.04.013
Lee, T. H., Tang, H., Wang, X., and Paterson, A. H. (2012). PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res. 41, D1152–D1158. doi: 10.1093/nar/gks1104
Lejay, L., Wirth, J., Pervent, M., Cross, J. M. F., Tillard, P., and Gojon, A. (2008). Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 146, 2036–2053. doi: 10.1104/pp.107.114710
Li, Y., Liu, Y., and Zhang, J. (2010). Advances in the research on the AsA-GSH cycle in horticultural crops. Front. Agricu. China 4:84. doi: 10.1007/s11703-009-0089-8
Lin, Y., Lin, S., Guo, H., Zhang, Z., and Chen, X. (2013). Functional analysis of PsG6PDH, a cytosolic glucose-6-phosphate dehydrogenase gene from Populus suaveolens, and its contribution to cold tolerance improvement in tobacco plants. Biotechnol Lett. 35, 1509–1518. doi: 10.1007/s10529-013-1226-2
Lin, Y. Z., Lin, S. Z., Zhang, W., Zhang, Q., Zhang, Z. Y., Guo, H., et al. (2005). Cloning and sequence analysis of a glucose-6-phosphate dehydrogenase gene PsG6PDH from freezing-tolerant Populus suaveolens. For. Stud. China 7, 1–6. doi: 10.1007/s11632-005-0048-2
Liu, J., Wang, X., Hu, Y., Hu, W., and Bi, Y. (2013). Glucose-6-phosphate dehydrogenase plays a pivotal role in tolerance to drought stress in soybean roots. Plant Cell Rep. 32, 415–429. doi: 10.1007/s00299-012-1374-1
Liu, W., Xie, Y., Ma, J., Luo, X., Nie, P., Zuo, Z., et al. (2015). IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. doi: 10.1093/bioinformatics/btv362
Liu, Y., Wu, R., Wan, Q., Xie, G., and Bi, Y. (2007). Glucose-6-phosphate dehydrogenase plays a pivotal role in nitric oxide-involved defense against oxidative stress under salt stress in red kidney bean roots. Plant Cell Physiol. 48, 511–522. doi: 10.1093/pcp/pcm020
Liu, Z., Ge, X., Yang, Z., Zhang, C., Zhao, G., Chen, E., et al. (2017). Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.). BMC Genet. 18:54. doi: 10.1186/s12863-017-0517-3
Long, X., He, B., Fang, Y., and Tang, C. (2016). Identification and characterization of the glucose-6- phosphate dehydrogenase gene family in the para rubber tree. Hevea brasiliensis. Front. Plant Sci. 7:215. doi: 10.3389/fpls.2016.00215
Née, G., Zaffagnini, M., Trost, P., and Issakidis-Bourguet, E. (2009). Redox regulation of chloroplastic glucose-6-phosphate dehydrogenase: a new role for f-type thioredoxin. FEBS Lett. 583, 2827–2832. doi: 10.1016/j.febslet.2009.07.035
Nemoto, Y., and Sasakuma, T. (2000). Specific expression of glucose-6-phosphate dehydrogenase (G6PDH) gene by salt stress in wheat (Triticum aestivum L.). Plant Sci. 158, 53–60. doi: 10.1016/s0168-9452(00)00305-8
Nemoto, Y., and Sasakuma, T. (2002). Differential stress responses of early salt-stress responding genes in common wheat. Phytochemistry 61, 129–133. doi: 10.1016/s0031-9422(02)00228-5
Pan, W. J., Tao, J. J., Cheng, T., Bian, X. H., Wei, W., Zhang, W. K., et al. (2016). Soybean miR172a improves salt tolerance and can function as a long-distance signal. Mol. Plant 9, 1337–1340. doi: 10.1016/j.molp.2016.05.010
Sang, S., Xie, L., Cui, X., Wang, N., Gao, M., and Wang, Z. (2018). Glucose-6-phosphate dehydrogenase is required for hpa1xoo (harpin protein fragment)-mediated salt stress tolerance in transgenic Arabidopsis thaliana. Pak. J. Bot. 50, 841–845.
Scharte, J., Schön, H., Tjaden, Z., Weis, E., and von Schaewen, A. (2009). Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc. Natl. Acad. S. U.S.A. 106, 8061–8066. doi: 10.1073/pnas.0812902106
Sharkey, T. D., and Weise, S. E. (2016). The glucose 6-phosphate shunt around the Calvin-Benson cycle. J. Exp. Bot. 67, 4067–4077. doi: 10.1093/jxb/erv484
Silva, F. L. B., Vieira, L. G. E., Ribas, A. F., Moro, A. L., Neris, D. M., and Pacheco, A. C. (2018). Proline accumulation induces the production of total phenolics in transgenic tobacco plants under water deficit without increasing the G6PDH activity. Theor. Exp. Plant Physiol. 30, 251–260. doi: 10.1007/s40626-018-0119-0
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tóth, K., Batek, J., and Stacey, G. (2016). Generation of soybean (Glycine max) transient transgenic roots. Curr. Protoc. Plant Biol. 1, 1–13. doi: 10.1002/cppb.20017
Valderrama, R., Corpas, F. J., Carreras, A., Gómez-Rodríguez, M. V., Chaki, M., Pedrajas, J. R., et al. (2006). The dehydrogenase-mediated recycling of NADPH is a key antioxidant system against salt-induced oxidative stress in olive plants. PlantCell Environ. 29, 1449–1459. doi: 10.1111/j.1365-3040.2006.01530.x
Velikova, V., Yordanov, I., and Edreva, A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants: protective role of exogenous polyamines. Plant Sci. 151, 59–66. doi: 10.1016/s0168-9452(99)00197-1
Wakao, S., Andre, C., and Benning, C. (2008). Functional analyses of cytosolic glucose-6-phosphate dehydrogenases and their contribution to seed oil accumulation in Arabidopsis. Plant Physiol. 146, 277–288. doi: 10.1104/pp.107.108423
Wakao, S., and Benning, C. (2005). Genome-wide analysis of glucose-6-phosphate dehydrogenases in Arabidopsis. Plant J. 41, 243–256. doi: 10.1111/j.1365-313x.2004.02293.x
Wang, H., Yang, L., Li, Y., Hou, J., Huang, J., and Liang, W. (2016). Involvement of ABA-and H2O2-dependent cytosolic glucose-6-phosphate dehydrogenase in maintaining redox homeostasis in soybean roots under drought stress. Plant Physiol. Biochem. 107, 126–136. doi: 10.1016/j.plaphy.2016.05.040
Wang, X., Ma, Y., Huang, C., Li, J., Wan, Q., and Bi, Y. (2008). Involvement of glucose-6-phosphate dehydrogenase in reduced glutathione maintenance and hydrogen peroxide signal under salt stress. Plant Signal. Behav. 3, 394–395. doi: 10.4161/psb.3.6.5404
Wendt, U. K., Wenderoth, I., Tegeler, A., and Von Schaewen, A. (2000). Molecular characterization of a novel glucose-6-phosphate dehydrogenase from potato (Solanum tuberosum L.). Plant J. 23, 723–733. doi: 10.1046/j.1365-313x.2000.00840.x
Wood, T. (1986). Physiological functions of the pentose phosphate pathway. Cell Biochem. Funct. 4, 241–247. doi: 10.1002/cbf.290040403
Xu, J., Maki, D., and Stapleton, S. R. (2003). Mediation of cadmium-induced oxidative damage and glucose-6-phosphate dehydrogenase expression through glutathione depletion. J. Biochem. Mo,. Toxicol. 17, 67–75. doi: 10.1002/jbt.10062
Yang, Y., Fu, Z., Su, Y., Zhang, X., Li, G., Guo, J., et al. (2014). A cytosolic glucose-6-phosphate dehydrogenase gene, ScG6PDH, plays a positive role in response to various abiotic stresses in sugarcane. Sci. Rep. 4:7090. doi: 10.1038/srep07090
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2:1565. doi: 10.1038/nprot.2007.199
Yu, C. S., Lin, C. J., and Hwang, J. K. (2004). Predicting subcellular localization of proteins for Gram- negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Keywords: glucose-6-phosphate dehydrogenase, expression, enzyme activity, salt stress, transgenic plants, soybean (Glycine max L.)
Citation: Zhao Y, Cui Y, Huang S, Yu J, Wang X, Xin D, Li X, Liu Y, Dai Y, Qi Z and Chen Q (2020) Genome-Wide Analysis of the Glucose-6-Phosphate Dehydrogenase Family in Soybean and Functional Identification of GmG6PDH2 Involvement in Salt Stress. Front. Plant Sci. 11:214. doi: 10.3389/fpls.2020.00214
Received: 22 November 2019; Accepted: 12 February 2020;
Published: 26 February 2020.
Edited by:
Thomas Henry Noel Ellis, John Innes Centre, United KingdomReviewed by:
Alberto A. Iglesias, National University of the Littoral, ArgentinaCopyright © 2020 Zhao, Cui, Huang, Yu, Wang, Xin, Li, Liu, Dai, Qi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhaoming Qi, cWl6aGFvbWluZzE4NjBAMTI2LmNvbQ==; Qingshan Chen, cXNoY2hlbkAxMjYuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.