
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 28 February 2020
Sec. Plant Systematics and Evolution
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00202
This article is part of the Research Topic Origins and Domestication of the Grape View all 8 articles
Wild grapevine, Vitis vinifera L. subsp. sylvestris (Gmelin, Hegi) is spontaneous to Europe and common in Tuscany. In this study, wild grapevines were identified in 22 populations from eight locations in Tuscan Maremma (Grosseto and Siena province). The plants were propagated by cuttings, collected in a vineyard, genotyped by nuclear simple sequence repeats (SSRs), chloroplast SSRs and single nucleotide polymorphisms (SNPs), and compared to locally cultivated varieties (Vitis vinifera L. subsp. sativa) and to non-vinifera and non-vitis genotypes. The identity analysis revealed that some individuals were redundant genotypes, suggesting natural vegetative propagation. In addition, four of the supposed V.v. sylvestris were in fact naturalized V.v. sativa. The majority of putative sylvestris genotypes had chlorotype A, while the remainder had chlorotype D, as the majority of Vitis vinifera subsp. sativa cultivated in Italy. Some of the recovered sylvestris genotypes appeared to be natural crosses with cultivated grapevine varieties in Tuscany, and their chlorotype suggests a higher pollen flow from sativa to the sylvestris genotypes than in the opposite direction. In addition, other genotypes appeared to be crosses within sylvestris, sylvestris-sativa or sylvestris-sylvestris siblings, or equivalent relationships. These relationships suggest a noticeably level of sexual reproductive activities among sylvestris and sylvestris-sativa genotypes. A cluster and structure analysis clearly differentiated the true sylvestris from the sativa, and the non-vinifera or non-vitis genotypes, and also highlighted a possible introgression of sylvestris into some Italian and French cultivated varieties. The results therefore suggest that, in addition to the primary ancient center of domestication from the Near East to Central Asia, the introgression among cultivated and wild grapevine occurred in other centers of diversification along the migration routes, contributing to the domestication processes, and suggesting that these processes are still ongoing despite the reduction in populations of sylvestris. The results also highlight that the GrapeReSeq 18K Vitis genotyping chip are suitable for non-vitis genotyping and that the range of SNPs heterozygosity in sylvestris appears to be up to 6% less and does not overlap the heterozygosity range of sativa genotypes.
The domesticated grapevine (Vitis vinifera L. subsp. sativa) is the most cultivated fruit crop within the genus Vitis. Grapevine domestication occurred about 8000 years ago during the Neolithic Age, in the Near East and the area of northern Mesopotamia and central Asian countries, from the wild grapevine Vitis vinifera L. subsp. sylvestris (Levadoux, 1956; McGovern, 2003; This et al., 2006; Forni, 2012; Bacilieri et al., 2013). The domesticated grapevines were disseminated from the primary domestication center toward Mesopotamia, the Balkans and the east Mediterranean Basin (at the end of the fifth millenium BC), and toward Sicily and western Europe. They were introduced to central Europe during the first millenium BC (Forni, 2012).
Negrul (1946) divided the domesticated grapevine cultivars into three proles (pontica, orientalis, and occidentalis) depending on the geographical distribution and morphological and ecological differences. The proles orientalis include grapevine varieties from eastern Georgia, Armenia, and Azerbaijan, the former Soviet republics in Central Asia to the Near East. This proles has two sub-proles: caspica (the ancient wine grape and Muscat varieties) and the antasiatica (table and raisin grape cultivars of a more recent origin). The proles pontica comprise the varieties from Georgia to the Balkans, divided into two sub-proles: georgica and balkanica, respectively. The proles occidentalis include the varieties from Western Europe.
Despite the grapevine diffusion from east to west after its first domestication, recent studies on DNA molecular markers provide some evidence of genetic characteristics and introgression from local sylvestris individuals in cultivated accessions (Myles et al., 2011). A significant gene flow between sativa and sylvestris has been observed in regions where these two taxa came into contact (Di Vecchi-Staraz et al., 2008), and additional domestication events that gave rise to the cultivated grapevine have been suggested in Sardinia (Grassi et al., 2006) and also in Spain and France (Riaz et al., 2018). However, the degree to which local sylvestris contributed to the domestication of Western European sativa cultivars has not yet been well defined.
The wild grapevine is spontaneous from Central Asia to the Mediterranean Basin (Zohary and Hopf, 2000) and is typically found in riparian ravines where it has access to water and can climb into the tree canopies. There is a considerable variety of forms among wild grapevine germplasm, and it can present biotypes of botanical or farming interest. In addition, the wild grapevine is an important source of genetic variation as a source of resilience in breeding programs for the improvement of cultivated vines as well as for dealing with climate change and the increasing demand for sustainable viticulture (Coleman et al., 2009; Ellstrand et al., 2010; Popescu et al., 2013; Meléndez et al., 2016; Marrano et al., 2018).
However, due to the ongoing restriction of its natural habitat, this sub-species is now at risk of extinction. The wild grapevine is undergoing a dramatic regression together with a significant increase in naturalized cultivated accessions, thus endangering the endurance of the wild grapevine in natural ecosystems (Meléndez et al., 2016; Riaz et al., 2018).
Previous investigations have reported that wild vines are widespread in Italy and particularly in Tuscan Maremma (Biagini et al., 2014). This paper presents the results of a project on the safeguarding, genotyping and phylogenetic study of Vitis vinifera subsp. sylvestris vines in the Tuscan Maremma.
About 150 vines of putative Vitis vinifera subsp. sylvestris (Gmelin) Hegi were identified in 22 populations (the distances between populations was over 5 km) from eight locations in the provinces of Grosseto and Siena (Figure 1). Most of the vines were identified in typical wild grapevine environments, such as land and woods with a high degree of humidity and the presence of elm, poplar and oak, in coastline zones and valley floors mainly with alluvial and colluvial lands. However, some were collected from locations at the edges of areas that had once been cultivated.
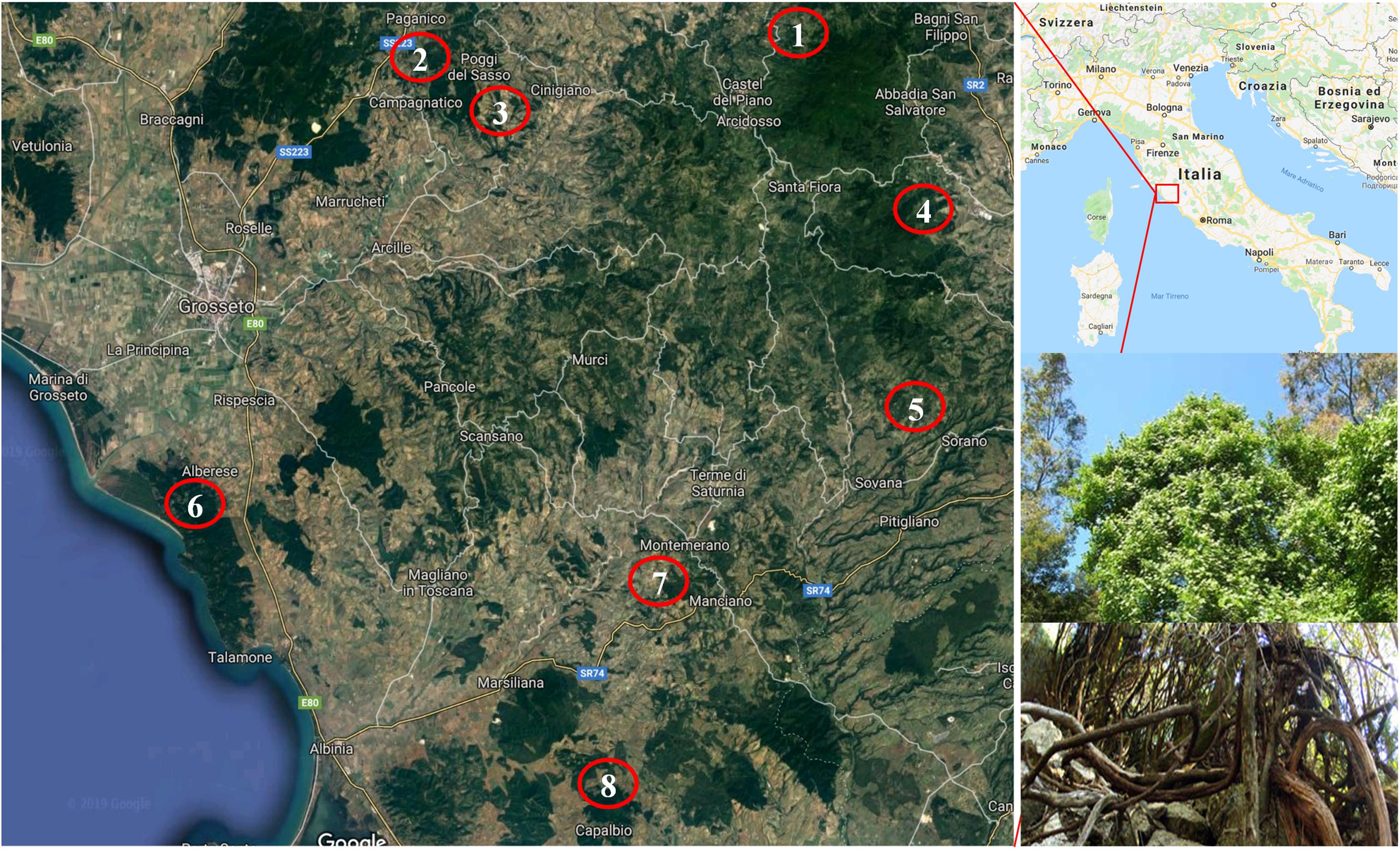
Figure 1. Location of the identification of putative Vitis vinifera subsp. sylvestris. (1) Castiglione D’Orcia; (2) Paganico; (3) Cinigiano; (4) Castell’Azzara; (5) Sorano; (6) Alberese; (7) Manciano; (8) Capalbio. In the two inserts in the right bottom: details of the top and bottom sections of a putative Vitis vinifera subsp. sylvestris vine.
The identification of the wild individual was based mainly on the morphology of leaves: mature leaves with open petiole sinus and upper lateral sinus, absence of prostrate hairs and a low density of erect hairs on the lower side of the blade, and teeth with both sides straight. However, vines with dubious sylvestris leaf morphology were also sampled to assess the presence of naturalized sativa genotypes and sylvestris-sativa crosses. When possible, flower sex was also verified in situ by selecting diecious plants.
Of the plants identified, 82 were sampled for DNA polymorphism analysis, ranging from 1 to 21 per population (Table 1) and vegetatively propagated by cuttings. The self-rooted plants (usually five for each accession) were planted in a collection vineyard located on the estate of Castello di Collemassari in the Montecucco Sangiovese DOCG (Grosseto Province, Tuscany south coastal area), where flower sex was identified as suggested in a previous study (Stout, 1921; Caporali et al., 2003) and berry color was identified in accordance with OIV descriptor 225 (OIV, 2009).
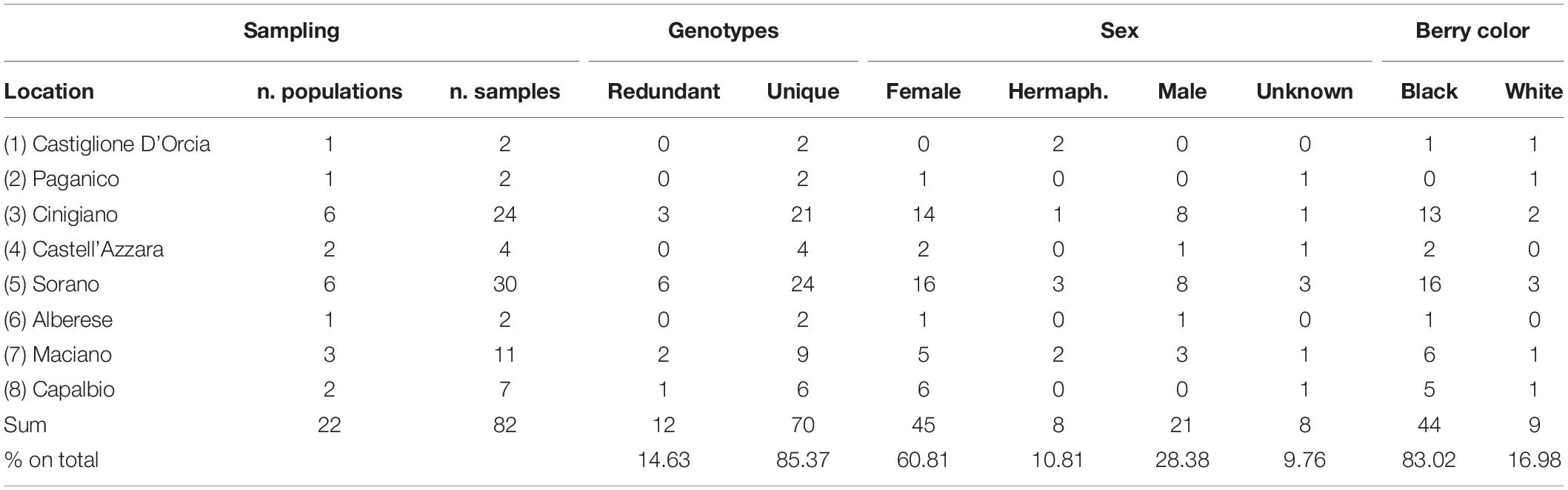
Table 1. Location, number of populations per location, number of samples per population, genotypes, sex and berry color of collected putative Vitis vinifera subsp. sylvestris.
Other 77 genotypes were sampled for DNA polymorphism analysis from the vineyard collection of the Department of Agriculture, Food and Environment of University of Pisa (Supplementary Table S1): 70 Vitis vinifera subsp. sativa genotypes cultivated in Tuscany, including local and international reference varieties introduced in Tuscany more than 50 years ago; ‘Isabel’, a cross between Vitis vinifera and the American genotype Vitis labrusca (D’Onofrio et al., 2016); two American Vitis species used as parents of the most widespread rootstocks (Vitis riparia and Vitis rupestris) belonging to the subgenus Euvitis such as Vitis vinifera; a Vitis rotundifolia (another American species, but belonging to the subgenus Muscadinia); and three Vitaceae not belonging to the genus Vitis (Cissus rhombifolia; Cayratia japonica; and Leea guineensis) obtained from the Karlsruher Institut für Technologie (KIT) Botanisches Institut.
DNA was extracted from young growing leaves (about 2 cm ∅) according to the protocol reported by Mulcahy et al. (1993), and modified as indicated by D’Onofrio et al. (2010).
The 82 collected putative Vitis vinifera subsp. sylvestris accessions and the other 77 genotypes were genotyped at 14 nuclear simple sequence repeats (SSRs) loci. To facilitate comparisons with other genotypes in Vitis databases, a core set of microsatellites selected from the GrapeGen06 research European Project was included in the analysis: VVS2 (Thomas and Scott, 1993), VVMD5, VVMD7, VVMD28, and VVMD32 (Bowers et al., 1996), VVMD25, VVMD27 (Bowers and Meredith, 1997), VrZAG62 and VrZAG79 (Sefc et al., 1999). The additional five microsatellite loci were: VVMD6 (Bowers et al., 1996), VVMD17, VVMD21, and VVMD24 (Bowers et al., 1999), VMC1b11b (Zyprian and Topfer, 2005).
PCR amplification was conducted in a 20 μl reaction mixture containing 10 ng of genomic DNA, 0.5 U Go Taq Flexi DNA polymerase (Promega, Madison, WI, United States), 1x Go Taq Flexi Colorless PCR buffer (Promega), 1.5 mM MgCl2, 0.5 μM of each primer and 200 μM of each deoxynucleoside triphosphate. A primer of each couple was fluorescently labeled with dye phosphoramidites (6-FAM, HEX, NED, and PET).
PCRs were carried out using a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, United States), initially set for 4 min at 94°C, followed by 39 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 5 min. The separation and sizing of alleles were performed on an ABI 310 Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany) and analyzed with GeneScan v. 2.1, as reported by D’Onofrio et al. (2016). In each PCR run, ‘Sangiovese’ microsatellites were amplified as a reference to standardize the microsatellite profiles, as implemented in the Italian Vitis Database (D’Onofrio and Scalabrelli, 2010)1.
To determine the chlorotype of the 82 collected putative Vitis vinifera subsp. sylvestris, 9 chloroplast SSRs were analyzed: ccmp3, ccmp5, and ccmp10 (Weising and Gardner, 1999); ccSSR-5, ccSSR-9 and ccSSR-14 (Chung and Staub, 2003); NTCP-8 (Bryan et al., 1999); ccSSR-23b and VVCP67629 (Péros et al., 2010). PCR amplification was carried out in a 20 μl reaction mixture containing 10 ng of genomic DNA, 0.5 U Go Taq Flexi DNA polymerase (Promega, Madison, WI, United States), 1x Go Taq Flexi Colorless PCR buffer (Promega), 1.5 mM MgCl2, 0.5 μM of each primer and 200 μM of each deoxynucleoside triphosphate. A primer of each couple was fluorescently labeled with a dye phosphoramidites (6-FAM, HEX, NED, and PET).
PCRs were carried out using a MyCycler Thermal Cycler (Bio-Rad, Hercules, CA, United States), initially set for 4 min at 94°C, followed by 35 cycles of 94°C for 1 min, 50°C for 30 s (at 55°C for 30 s for the ccmp5 locus), and 72°C for 1 min, with a final extension at 72°C for 5 min. Allele separation and sizing were performed on a ABI 310 Genetic Analyzer (Applied Biosystems, Weiterstadt, Germany) and analyzed with GeneScan v. 2.1, as reported by D’Onofrio et al. (2016).
Of the non-redundant sylvestris genotypes, 37 were selected based on the results of the SSRs polymorphism analysis. These sylvestris genotypes, along with the other 77 genotypes (Supplementary Table S1) were then subjected to SNPs analysis by the Grapevine Illumina Infinium SNP chip developed by the GrapeReSeq Consortium (GrapeReSeq 18K Vitis genotyping chip), and analyzed by Traits Genetics GmbH (Gatersleben, Germany).
As the dataset includes highly distant genotypes with a high percentage of failed SNPs, the markers were selected using three different procedures: (i) by the missing rate per genotype (<0.1), missingness test (the maximum number of missing SNP states per SNP site, GENO > 0.2) and minor allele frequency (MAF < 0.05) test (matrix SNPs-A) performed by Plink software (Purcell et al., 2007); (ii) or applying the same missingness test and minor allele frequency test including all the genotypes (matrix SNPs-B) even with a high rate of missing markers (>0.1); or (iii) simply selecting only the totally successful SNPs in all the genotypes (matrix SNPs-C).
Redundant sylvestris genotypes were identified in base to SSR analysis by CERVUS 3.0.3 (Kalinowski et al., 2007). GenAlEx 6.503 (Peakall and Smouse, 2006, 2012) was used to calculate the following parameters for genetic diversity on the SSRs profile of non-redundant sylvestris genotypes, the 70 Vitis vinifera subsp. sativa, and the 4 Vitis non-vinifera genotypes: the number of alleles (Na), the effective number of alleles (Ne), observed heterozygosity (Ho), expected heterozygosity (He) (Nei, 1973); and the fixation index (F), also referred to as the inbreeding coefficient.
Allelic richness (AR) and private allelic richness (PAR) for each population were estimated using the rarefaction method, which compensates for differences in sample size (i.e., rarefied allelic richness) among populations as implemented in HP-Rare 1.1 (Kalinowski, 2005). GENEPOP v. 4.7.2 (Rousset, 2008) was used to estimate the effective number of migrants per generation (Nm) among the grapevine populations and between the two subspecies using the private allele method (Barton and Slatkin, 1986), and Weir and Cockerham’s F-statistics (Weir and Cockerham, 1984) per each population (FST).
Identity analyses were conducted using CERVUS for all the accessions in the Italian Vitis Database2 at 9 loci, or at 14 loci for the genotypes uploaded by the Department of Agriculture, Food and Agro-environmental, University of Pisa, and at 9 loci in the European Vitis Database3.
A preliminary analysis of parent-offspring relationships was conducted among the non-redundant sylvestris genotypes, the seventy Vitis vinifera subsp. sativa and the four Vitis non-vinifera genotypes, by identity-by-state (IBS) on nuclear SSRs using CERVUS. The parent-offspring, sibling or equivalent relationships were therefore checked on the SNPs-A data set by the identity-by-descent (IBD) coefficients calculated using PLINK (Purcell et al., 2007). The two IBD coefficients inferring the probabilities that a pair shares one or two alleles IBD (Z1, Z2, respectively), and relatedness value (PI_HAT) were calculated for all pair-wise comparisons. The expected values of Z1, Z2, and PI_HAT were, respectively, 1, 0, and 0.5 for parent-offspring (PO); 0.5, 0.25, and 0.5 for full-siblings (FS); and 0.5, 0, and 0.25 for second-degree relatives. The values for known pedigree relationships included in the set of 70 sativa genotypes were used to evaluate IBD and relatedness coefficients, thus establishing threshold values.
The allele sharing distance between each pair of individuals was calculated using PEAS software (Xu et al., 2010) and UPGMA clustering (Sokal and Michener, 1958) performed using MEGA 6 (Tamura et al., 2013). Poppr 2.8.3 (Kamvar et al., 2015) implemented in R 3.6.1 software (R Core Team, 2019) was also used to calculate neighbor-joining unrooted phylogenetic trees based on the Nei distance (Nei, 1972), which was then circle plotted by MEGA 6.
A Bayesian clustering was derived on both nuclear SSRs and SNPs using STRUCTURE (Pritchard et al., 2000) in its revised 2.3.4 version (Jul 2012). An admixture model and independent allelic frequencies were used to analyze the dataset without prior population information. To estimate the cluster number (K) of ancestral genetic groups and the ancestry membership proportion of each individual in these clusters, the algorithm was run 10 times for each K value from 1 to 10. The optimum number of clusters (K) was chosen based on the second order rate of change in the log probability of data between successive K values (Evanno et al., 2005).
Of the collected accessions 28.38% had male flowers, 71.62 had female-hermaphrodite flowers, while 9.76% were not classified. Among the female-hermaphrodites, 83.02% had black berries and 16.98% had white berries (Table 1).
The nuclear SSRs profiles and the chlorotypes are reported in Supplementary Table S1: the SSRs analysis failed for the three non-Vitis genotypes.
Of the 82 putative sylvestris accessions analyzed, 12 (14.63%) were redundant genotypes and were identified in 4 of the 22 populations sampled (Table 1).
The number of nuclear SSR alleles in the 70 putative Vitis vinifera subsp. sylvestris and the 70 grapevine genotypes cultivated in Tuscany (Vitis vinifera subsp. sativa) ranged from 6 for VVMD17 to 19 for VVMD28 and VMC1b11, with a mean value of 13.143 alleles per locus (Table 2). The number of effective alleles ranged from 2.318 for VVMD21 to 7.849 for VVMD28, and the mean overall value was 4.988. The He ranged from 0.569 for VVMD21 to 0.873 for VVMD28, with an average of 0.781. The Ho ranged from 0.486 (VVMD21) to 0.854 (VVS2) and the mean value was 0.726. The mean F value for the dataset was 0.073, with the lowest of −0.015 for VVS2 and the highest of 0.180 for VVMD17.
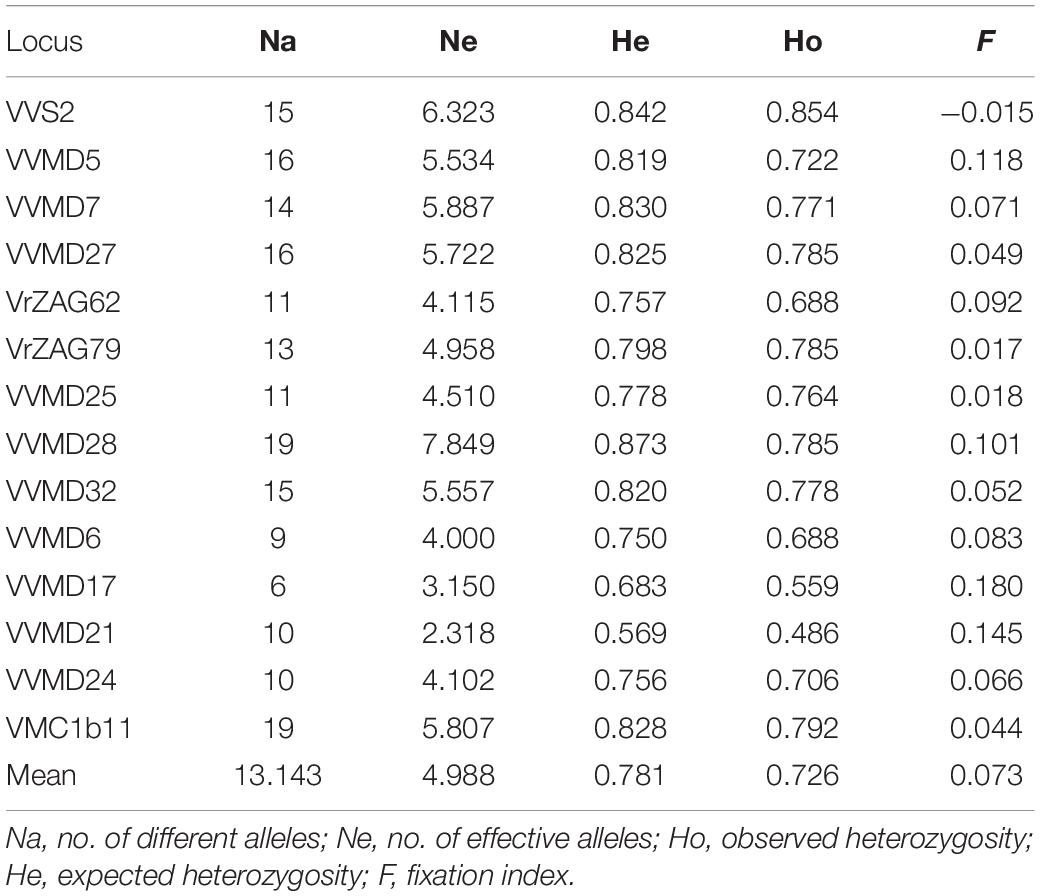
Table 2. Nuclear SSRs genetic diversity indices of 70 putative Vitis vinifera subsp. sylvestris and 70 cultivated grapevine (Vitis vinifera subsp. sativa) genotypes from Tuscany.
Concerning the genetic diversity among populations (Table 3A), the number of alleles per locus (Na) was identical in both sativa and sylvestris populations (9.714), but lower for the non-vinifera population (5.643). On the other hand, the number of effective alleles per locus (Ne) was higher in the sativa population (5.040) than in sylvestris (3.789), and the value of the non-vinifera population was in the middle (4.789). There was a similar trend for observed and expected heterozygosity. The allelic richness in sativa was higher than in sylvestris, while the private allele richness was higher in sylvestris: sylvestris genotypes had 10 private alleles in 6 of the 14 SSRs loci analyzed which could be useful in distinguishing sylvestris from sativa genotypes. The mean Shannon Information Index (I) for the wild accessions (1.581) was lower than for sativa genotypes (1.775) and the non-vinifera population (1.603). The inbreeding coefficient within individuals (F) was negative for sativa (−0.016) and positive for sylvestris (0.075) and non-vinifera genotypes (0.130).
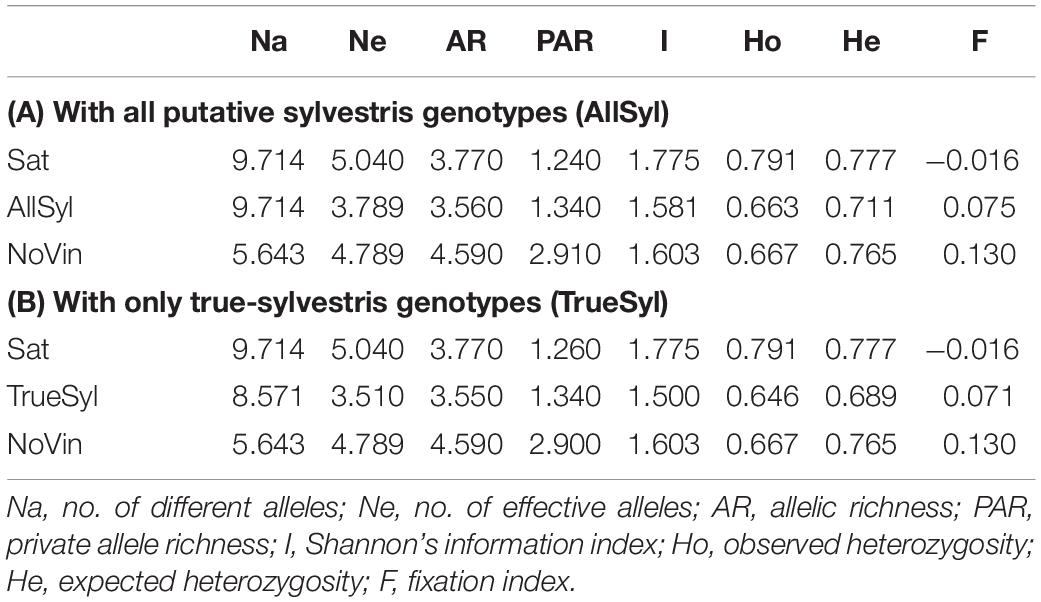
Table 3. (A) Nuclear SSRs genetic diversity estimates for each analyzed population: 70 Vitis vinifera subsp. sativa (Sat), 70 Vitis vinifera subsp. sylvestris (AllSyl), 4 non-vinifera (NoVin). (B) Then, the analysis war repeated including only the 61 true-sylvestris genotypes (TrueSyl).
The heterozygosity (Ho) was also lower in sylvestris than the sativa population (0.663 and 0.791, respectively) as was the expected heterozygosity (He) (0.711 and 0.777, respectively). The value of Ne, I, Ho, He, and F were in agreement with previously observed values in sylvestris from central Italy (Biagini et al., 2014).
The effective number of migrants per generation (Nm) after correction for sample size was 1.133 (data not shown) among the three populations (sativa, sylvestris, non-vinifera). On the other hand between the sativa and sylvestris populations, it was 3.224, and again much lower (0.379) between sativa and non-vinifera and between sylvestris and non-vinifera (0.444) populations (Table 4A). The Weir and Cockerham F-statistics (Weir and Cockerham, 1984) FST between sativa and sylvestris populations was 0.067, while between sativa and non-vinifera, it was 0.106 and between sylvestris and non-vinifera populations, 0.148 (Table 4A).
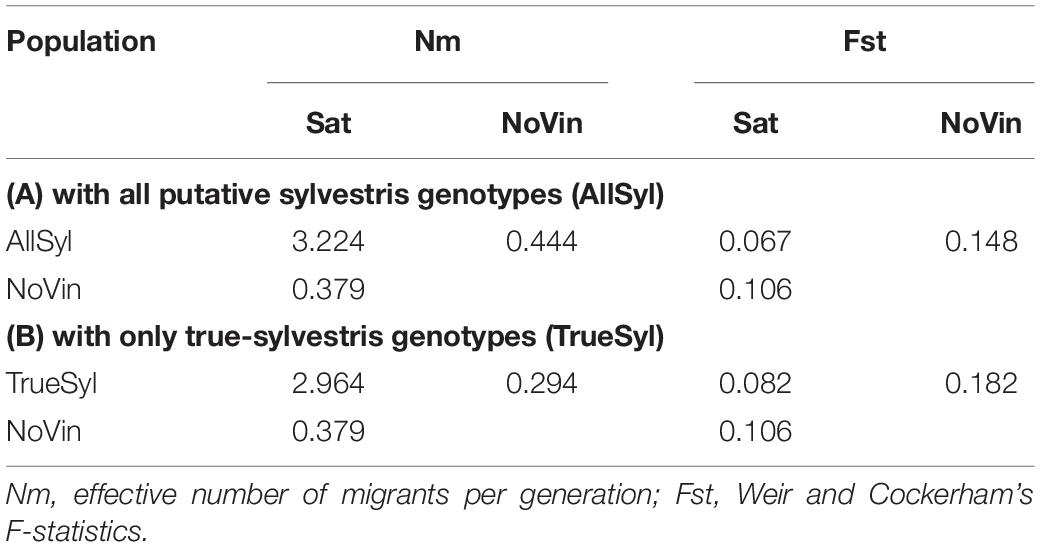
Table 4. Effective number of migrants per generation (Nm) and Weir and Cockerham’s F-statistics among the grapevine populations. (A) 70 Vitis vinifera subsp. sativa (Sat), 70 Vitis vinifera subsp. sylvestris (AllSyl), 4 non-vinifera (NoVin); (B) 70 Vitis vinifera subsp. sativa (Sat), 61 true-sylvestris genotypes (TrueSyl), 4 non-vinifera (NoVin).
The analysis of chloroplast microsatellites revealed three chlorotypes: A, B and D, as indicated by Arroyo-García et al. (2006) (Supplementary Table S1). In agreement with previous studies (Arroyo-García et al., 2006; Grassi et al., 2006; Cunha et al., 2009; Butorac et al., 2018), the majority of the identified genotypes of putative sylvestris had chlorotype A (51 samples; 72.86%), while 18 samples (25.71%) had chlorotype D as in the majority of the Vitis vinifera subsp. sativa cultivated in Italy. Only one genotype (1.43%) had chlorotype B, which has not previously been identified in any Italian sylvestris, but only in a few Italian cultivated varieties (Arroyo-García et al., 2006).
Of the 18071 SNPs in the array, 623 (3.4%) completely failed and were thus removed from the matrix when calculating the subsequent statistics. The percentage of other failed markers ranged from 0.57 to 78.54%. It was mainly related to the taxonomic groups, increasing gradually with the genetic distance from Vitis vinifera, and was similar among subsp. sativa and putative subsp. sylvestris genotypes (Figure 2). However, the heterozygosity ranged from 23.9 to 47.15%, in an inverse trend to percentage of failed markers, and was lower in sylvestris (36.90–45.20) than in sativa (41.91–47.15) genotypes (Supplementary Table S2). Among the sativa, the French genotypes typically have a slightly higher level of heterozygosity than other cultivated genotypes, particularly ‘Sauvignon blanc’ and ‘Pinot noir’ (Supplementary Table S2).
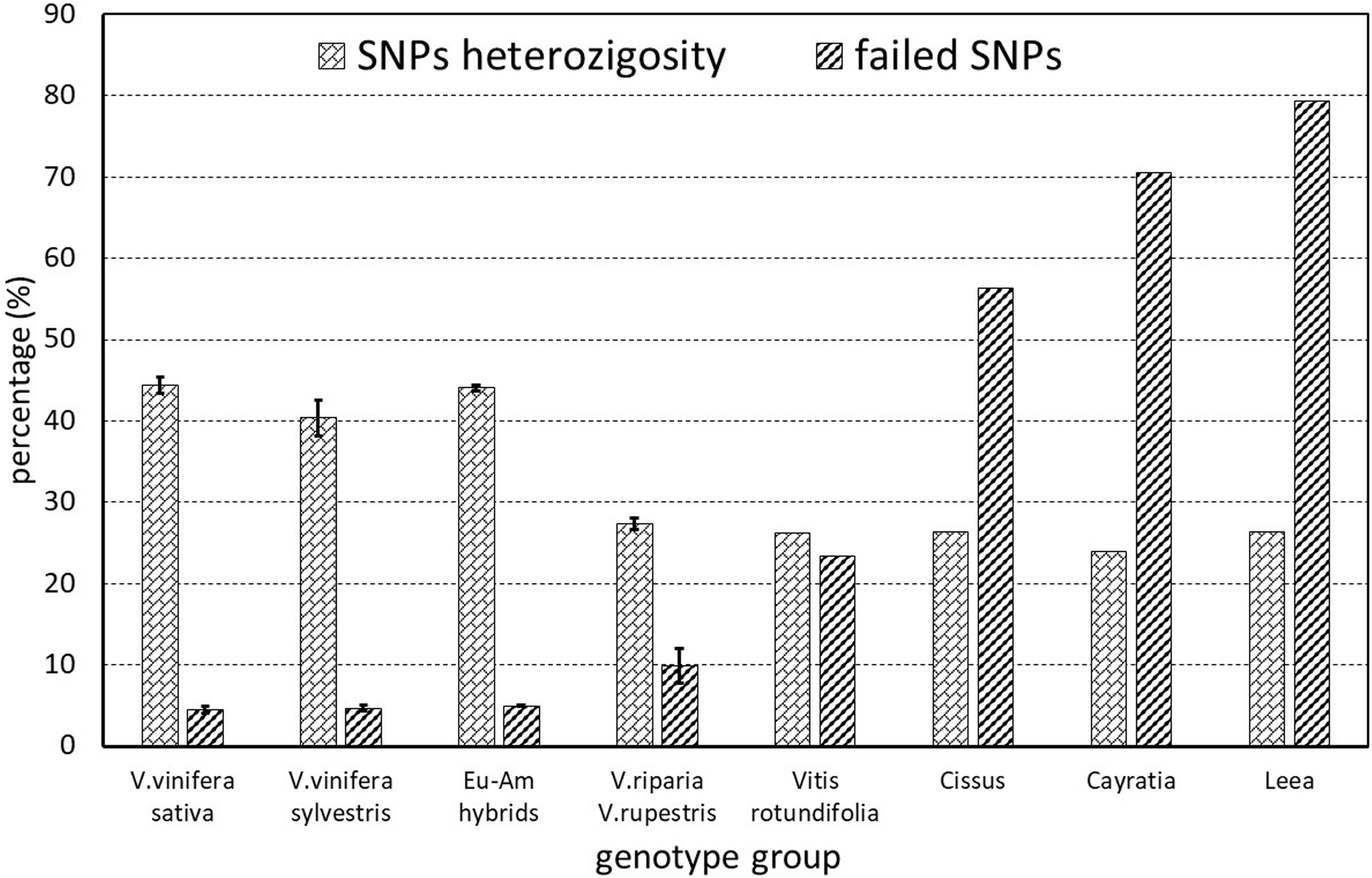
Figure 2. SNPs missing rate and heterozygosity of the various genetic groups analyzed. The European–American groups (Eu-Am hybrids) include the hybrid ‘Isabel’ and the hybrid ‘Seibel 128’ identified among the putative sylvestris.
The SNP quality check excluded four genotypes with a missing rate over 10% (Vitis rotundifolia and the three non-Vitis genotypes) and revealed that 267 SNPs failed the missingness test (the maximum number of missing SNP states per SNP site, GENO > 0.2) and 4751 SNPs failed the minor allele frequency test (MAF < 0.05). Therefore, after frequency and genotyping pruning, there were 13154 SNPs in the dataset (SNPs-A).
Including all the 114 genotypes, even together with the Leea guineensis with a missing rate of 78.54%, 313 SNPs failed the missingness test (the maximum number of missing SNP states per SNP site, GENO > 0.2) and 4792 SNPs failed the minor allele frequency test (MAF < 0.05), and 13080 SNPs passed the tests (SNPs-B). Finally, a third matrix (SNPs-C) included only the SNPs (1070) that were successful in the whole set of 114 genotypes (Supplementary Table S3).
The identity analysis using the SSRs profiles of genotypes in the Italian Vitis Database4 and the European Vitis Database5 indicated that three accessions matched with previously characterized genotypes (Supplementary Table S1).
The genotype Syl_55, identified in the Sorano area (Sorano1), which was hermaphrodite and had white berries, was the Italian variety ‘Bellone’ (Bellone ITA388-G012 EVD; European Vitis Database). The Syl_60 recovered from the same population (Sorano1) matched the table grapevine ‘Regina dei vigneti’ (FRA139-1078Mtp1, European Vitis Database), and again being hermaphrodite with white berries confirmed this.
Another genotype Syl_3 was identified at Campiglia D’Orcia, in a flat area near a stream named Formone, which was hermaphrodite and had black berries, and matched the European-American Vitis interspecific crossing ‘Seibel 128’ (Seibel 128 FRA139-5026Mtp4, European Vitis Database).
The parent-offspring analysis performed by identity-by-state (IBS) on nuclear SSRs and by identity-by-descent (IBD) on the SNP profile indicated that some accessions were possibly natural sativa-sylvestris crosses, while others might be crosses between sylvestris (Supplementary Table S1). The IBD analysis was performed on the SNPs-A data set with a threshold of 0.462 PI_HAT identified as the lowest pair-wise relationship (Cabernet sauvignon-Sauvignon blanc) among the known PO relationships in the set of the 70 sativa genotypes.
Of the 67 non-redundant genotypes not matching known genotypes, six (8.95%) appeared to be parent-offspring of grapevine varieties cultivated in Tuscany (Supplementary Table S1).
The accession Syl_26 identified in the area of Cinigiano at Poggi del Sasso, which was hermaphrodite with black berries, appeared to be a natural cross of ‘Sangiovese’. Another natural cross of ‘Sangiovese’ was the accession Syl_16. This genotype was identified in a population in the area of Manciano and had hermaphrodite flowers and black berries. The genotype Syl_10, identified at Castell’Azzara, which was female and had black berries, appeared to be a natural cross of ‘Trebbiano toscano.’ Again, this relationship was confirmed by both SSRs and SNPs. The Syl_69, from the Sorano area, which had female flowers and produces black grapes, was a cross with another genotype of the same population, the Syl_55 identified as ‘Bellone.’ Another genotype (Syl_56) from the same area (Sorano) was a natural cross of the hybrid ‘Isabel’. This accession had female flowers and produces black grapes.
From an analysis exclusively on nuclear SSRs, the genotype Syl_27, with female flowers and white berries from Cinigiano, was identified as a natural cross of ‘Berzemino/Negretta toscana’, a local Tuscany variety reported in the Italian Vitis Database6.
The analysis of only the SSRs profile, and not SNPs because of the lack of profiles, indicated a parent-offspring relationship between the sylvestris genotypes Syl_49 and Syl_63 from Sorano, and between Syl_54 and Syl_61 also from Sorano (Supplementary Table S1).
In addition, the IBD analysis suggested a full-sibling relationship between Syl_4 and Syl_7 from Capalbio, and between Syl_48 and Syl_49 from Sorano area (Supplementary Table S1).
The second-degree relationship between Syl_26 and Syl_16, as well as their second-degree relationship with ‘Ciliegiolo,’ a well-known offspring of ‘Sangiovese,’ confirmed that both these accessions were ‘Sangiovese’ crosses. In addition, the second-degree relationship between Syl_10 and ‘San Lorenzo’ (Sat_4), a local Tuscan offspring of the ‘Trebbiano toscano’ variety, confirmed this parent-offspring relation with ‘Trebbiano toscano.’
Interestingly, the IBD analysis of highlighted some second-degree relationships among ‘Petit manseng’ (Sat_38), ‘Petit verdot’ (Sat_48), ‘Greco bianco’ (Sat_7) and some sylvestris.
Three UPGMA tree-A, tree-B, and tree-C, were generated by the allele sharing distance between each pair of individuals on selected SNPs in SNPs-A, SNPs-B, and SNPs-C matrices, respectively (Figure 3), using the PEAS and MEGA packages.
In all UPGMA trees there was a clear separation of three main clusters: a cluster with non-vinifera genotypes, another with the majority of sylvestris genotypes, and the last including almost all sativa genotypes, non-true-sylvestris genotypes and putative sylvestris-sativa crosses.
In the tree-A obtained from the matrix of the selected SNPs through the missingness and minor allele frequency test on genotypes with a low rate of missing markers (<10%) and without Vitis rotundifola and non-vitis genotypes, the non-vinifera cluster was between the sylvestris and sativa clusters. While, in tree-B, which was made up of the matrices including the Vitis rotundifola and non-vitis genotypes with a high missing rate (over 10%), the cluster of sylvestris genotypes was between the non-vinifera and sativa clusters. In both these trees, the arrangement of genotypes within sylvestris and sativa clusters was exactly the same. In the sativa cluster, there are two main sub-clusters: one including most of the Italian sativa and Muscat varieties, and another grouping most of the French genotypes and some Italian genotypes. In tree-C, although generated from a low number of SNPs (1070), the cluster of sylvestris was between the non-vinifera and sativa genotypes, but the arrangement of genotypes within sylvestris and sativa clusters has some differences.
In all trees, the sylvestris accessions identified as the European–American Vitis hybrid ‘Seibel 128’ (Syl_3), and the accession identified as a full-sibling of the European-American Vitis hybrid ‘Isabel’ (Syl_56) were in the non-vinifera cluster. In addition, the accessions Syl_55 and Syl_60, matching the cultivated varieties ‘Bellone’ and ‘Regina dei vigneti,’ respectively, were properly in the sativa cluster.
The Syl_26 and Syl_16 were always in the sub-cluster with ‘Sangiovese,’ confirming these accessions as crosses of ‘Sangiovese’ (Sat_31), the most widely diffused grapevine in Tuscany. Similarly, the Syl_10 was in the sub-cluster including ‘Trebbiano toscano’ (Sat_44), confirming its origin as a cross of this important variety that is widespread in Tuscany. In addition, Syl_69 was among the sativa genotypes, near Syl_55 identified as ‘Bellone,’ thus confirming the parent-offspring relationship between these two genotypes.
In tree-C, the non-vitis genotypes were rightly in the non-vinifera cluster, and unlike the other two trees, the sativa genotypes Sat_38, Sat_48 and Sat_7 (respectively ‘Petit Manseng,’ ‘Petit Verdot’ and ‘Greco bianco’) were in the cluster of sylvestris genotypes, in agreement with the IBD second-degree relationships.
Similarly to the UPGMA trees, three unrooted NJ-dendrograms (NJ-A, NJ-B, and NJ-C, respectively) were generated from SNPs-A, SNPs-B, and SNPs-C matrices by neighbor-joining (NJ) cluster analysis based on the pair-wise Nei’s distance (Figure 4). All three NJ-dendrograms clearly differentiated the three main clusters: sylvestris cluster, sativa cluster, and the cluster with non-vinifera genotypes. The cluster of non-vinifera accessions was among the sativa and sylvestris clusters in the NJ-B dendrogram but among sativa genotypes in NJ-A and NJ-C dendrograms.
In agreement with the UPGMA trees, in all the NJ-dendrograms, the two naturalized sativa (Syl_55 and Syl_60) and the four sativa-sylvestris (Syl_10, Syl_16, Syl_26, and Syl_69) crosses were all in the sativa cluster. In addition, the non-true sylvestris accessions Syl_3 and Syl_56 related to the hybrid ‘Isabel’ were in the non-vinifera cluster in NJ-B and NJ-C dendrograms as in all UPGMA trees, while in NJ-A, Syl_3 was confirmed in the non-vinifera cluster but Syl_56 appeared in the sylvestris cluster. In all NJ dendrograms, the sativa genotypes ‘Petit manseng’ (Sat_38), ‘Petit verdot’ (Sat_48) and ‘Greco bianco’ (Sat_7) were in the sylvestris cluster, as in the UPGMA tree-C and in agreement with the IBD second-degree relationships.
The structure analysis was performed on both nuclear SSR markers (barplot-A) and SNP matrices (barplot-B, barplot-C, barplot-D from SNPs-A, SNPs-B, SNPs-C matrices, respectively) (Figure 5).
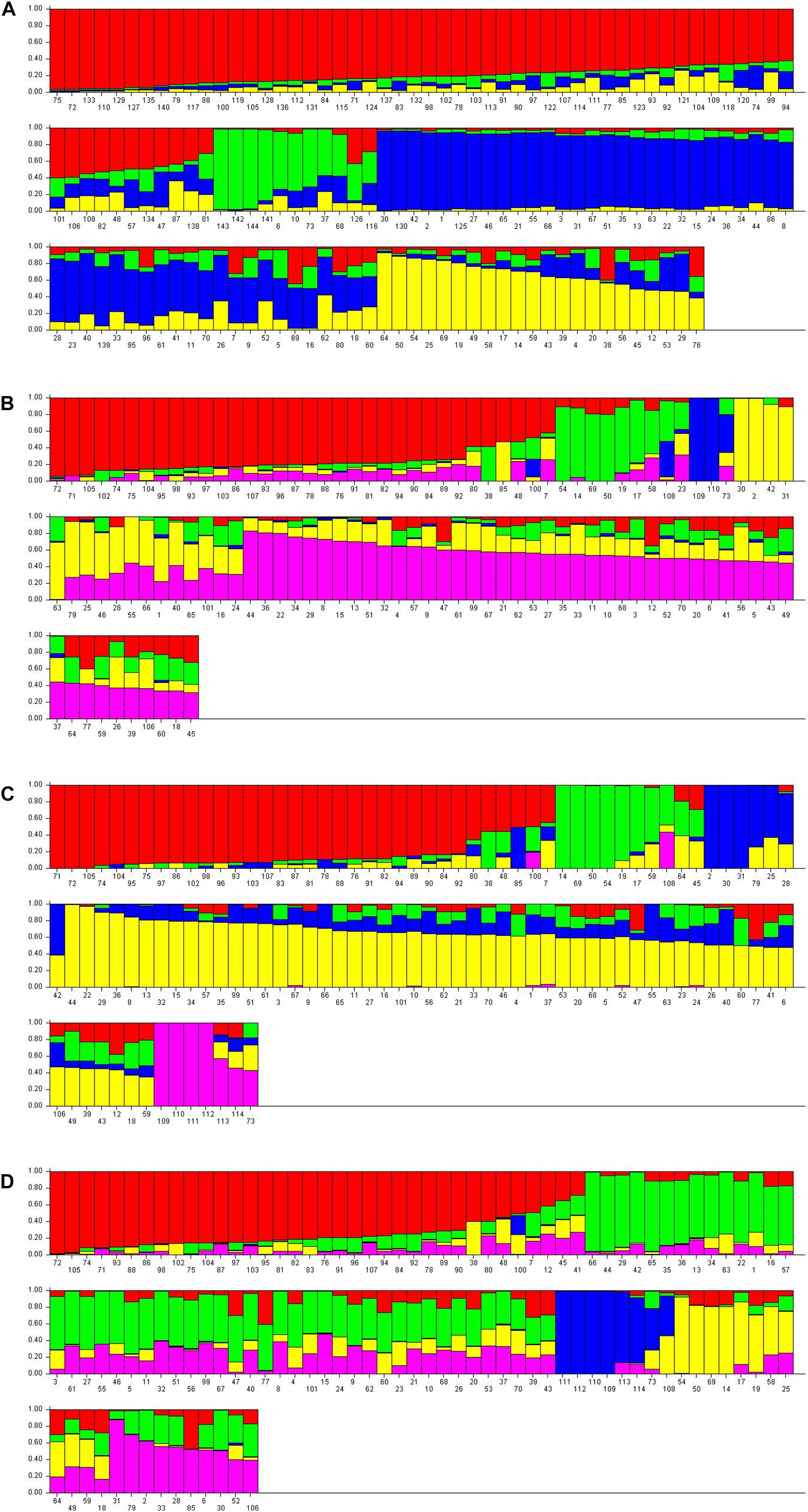
Figure 5. Barplot displaying the admixture proportions of wild, cultivated grapevine and non-vinifera genotypes as estimated by STRUCTURE analysis. (A) 144 genotypes, 14 SSR loci, K = 4; (B) 110 genotypes, 13154 SNPs, K = 5; (C) 114 genotypes, 13080 SNPs, K = 5; (D) 114 genotypes, 1070 SNPs, K = 5.
Analysis on the SSRs of 144 genotypes (70 sativa, 70 putative sylvestris, three non-vinifera, Vitis rotundifolia, but not the three non-Vitis genotypes because the SSRs failed) revealed four populations (Figure 5A): non-vinifera genotypes, true-sylvestris, most Italian sativa and Muscat varieties, and most French sativa and some Italian sativa varieties (Figure 5). In agreement with the cluster analysis, the Italian sativa population included the two naturalized sativa (Syl_55 and Syl_60; accessions 125 and 130, respectively) and the four sativa-sylvestris crosses (Syl_10, Syl_16, Syl_26 and Syl_69; accessions 80, 86, 96, and 139, respectively), but also the genotype Syl_19 (accession 89) not analyzed by SNPs. Another sylvestris genotype (Syl_6; accession 76), not analyzed by SNPs, was in the French population, and another (Syl_46; accession 116) was in the cluster of non-vinifera genotypes. The presence of ‘Petit verdot’ (Sat_48; accession 48) in the population of sylvestris was also confirmed, but not the varieties ‘Petit manseng’ (Sat_38; accession 38) and ‘Greco bianco’ (Sat_7; accession 7), which were respectively assigned to the French sativa and Italian sativa cluster. In addition, the local Tuscan varieties ‘Colorino’ (Sat_47; accession 47) and ‘Schiava gentile’ (Sat_57; accession 57) appeared among sylvestris.
The structure analysis on all the three SNP matrices always revealed five populations: non-vinifera, sylvestris, Italian sativa, French sativa, and a population including all Muscat varieties, the ‘Sangiovese’ and a few other Tuscan varieties. The barplot-B and C, from SNP-A and SNPs-B matrices, appeared very similar (Figures 5B,C), and in agreement with UPGMA tree-C and all NJ dendrograms, the varieties ‘Petit manseng’ (Sat_38; accession 38), ‘Petit verdot’ (Syl_48, accession 48) and Greco bianco’ (Sat_7; accession 7) were included in the sylvestris population. The naturalized sativa genotypes (Syl_55 and Syl_60; accessions 99 and 101, respectively) and most of the sativa offspring were in the sativa population, as well as the naturalized hybrid ‘Seibel 128’ (Syl_3; accession 73), and the full-siblings of the hybrid ‘Isabel’ (Syl_56; accession 100) were in the non-vinifera population. Looking at the ancestry of the single genotypes, it seems that barplot-C better separated the sylvestris and sativa ancestries.
The barplot-D generated from the matrix SNPc-C including the non-vitis genotypes but with only the 1070 SNPs that were successful in all genotypes, confirmed the same 5 populations (Figure 5D). All the non-vinifera and non-vitis genotypes were in the same cluster together the naturalized hybrid ‘Seibel 128’ (Syl_3; accession 73), while the descendant of the hybrid ‘Isabel’ (Syl_56; accession 100) was in the sativa population despite a high non-vinifera ancestry. In addition to ‘Petit manseng’ (Sat_38; accession 38), ‘Petit verdot’ (Syl_48, accession 48) and ‘Greco bianco’ (Sat_7; accession 7), also the sativa varieties ‘Cabernet franc’ (Sat_12; accession 12), ‘Syrah (Sat_45; accession 45) and ‘Roussane bianco’ (Sat_41; accession 41) were in the sylvestris population although their sylvestris ancestry is less than 50%.
The genotyping analysis of the nuclear and chloroplast SSRs and SNPs polymorphism of putative wild vines identified and recovered in southern Tuscany highlights the difficulty of identifying true Vitis vinifera subsp. sylvestris.
The presence of redundant genotypes and their high percentage in 6 out of 22 sampled populations, and the identification of redundant plants over 20 m from each other, suggest not only the possible extent of such plants, but also the possibility of natural vegetative propagation.
The identity analysis of the main Vitis databases revealed that 3 of the 70 non-redundant putative wild genotypes are known genotypes: two are Vitis vinifera subsp. sativa varieties (‘Bellone’ and ‘Regina dei vigneti’) and one is the European-American Vitis hybrid ‘Seibel 128.’ Two of these (‘Bellone’ and ‘Regina dei vigneti’) were collected from the same population in the Sorano area, while the other (‘Seibel 128’) was identified in a population about 20 km away (Castiglione D’Orcia). This locality is not currently an intensive wine viticulture area, but the known naturalized genotypes identified suggest that this was previously a cultivation area of allochthonous grapevine varieties, which was later abandoned, and consequently the cultivated grapevine genotypes acquired a naturalized appearance, as previously also observed for other sativa genotypes around Europe (Riaz et al., 2018). Their identity is supported by the high level of sativa ancestry of the genotypes identified as ‘Bellone’ and ‘Regina dei vigneti’ (Supplementary Table S2), and their position in the cluster of sativa in all the UPGMA-trees and NJ-dendrograms. Other evidence includes the high non-vinifera ancestry of the genotypes identified as ‘Seibel 128’ and their position in the cluster of non-vinifera in all the UPGMA-trees and NJ-dendrograms.
Six (8.95%) of the 67 putative sylvetris genotypes studied appeared to be natural crosses of the most widespread grapevine varieties in Tuscany (‘Sangiovese’ and ‘Trebbiano toscano’), of a minor local variety (‘Berzemino’), of the non-true sylvestris matching ‘Bellone,’ and of the European-American Vitis hybrid ‘Isabel.’ These results suggest a previous pollen flow between wild and cultivated grapevine proven by the analysis of seeds collected from sylvestris grapevines (Di Vecchi-Staraz et al., 2008). Four of the six crosses observed had chlorotype A, like most of the sylvestris genotypes, and only one had chlorotype D, like most cultivated sativa varieties in Italy. The progeny usually inherits the mother’s chlorotype, thus the higher number of crosses with chlorotype A suggest a higher pollen flow from sativa to the sylvestris genotypes compared to the opposite direction. This is also supported by the probable higher quantity of sativa than sylvetris pollen in highly intensive viticulture regions such as Tuscany. However, seedlings from wild plants have a higher probability of growing in woods than those from cultivated plants in areas with worked soil. The two pairs of sylvestris genotypes with a parent-offspring relationship (Syl_49-Syl_63, Syl_54-Syl_61) and the two pairs with a full-sibling relationship (Syl_4-Syl_7, Syl_48-Syl_49) had chlorotype A and a high sylvestris ancestry, confirming their true sylvestris nature.
All the above described parent-offspring and full-siblings highlight sexual reproductive activities among sylvestris and sylvestris-sativa genotypes.
The population of sativa had a higher number of SSRs effective alleles and allele richness than the sylvestris population (Table 3A), highlighting a higher diversity among sativa genotypes cultivated in Tuscany, coming from western to eastern Europe, compared to sylvestris genotypes collected from a few isolated populations in a relative small area (about 100 km2) in southern Tuscany. These differences increased with the exclusion from the sylvestris population of accessions that were non-true-sylvestris according to the parentage analysis (Table 3B).
The expected and observed heterozygosity calculated on SSR markers appeared higher in sativa than in sylvestris population, in agreement with previous results (Riaz et al., 2018). In addition, the Ho value of the sativa population appeared to be slightly higher than the He values, while the reverse was true for the sylvestris accessions. These differences correspond with the positive F values in sylvestris, which suggests a high level of genetic relationship among the individuals from the same wild populations (Table 3).
With the exclusion from the sylvestris population of accessions that were non-true-sylvestris the FST between sativa and sylvestris populations increased, while the effective number of migrants per generation decreased (Table 4). The FST indicated that the overall level of genetic differentiation between sativa and sylvestris genotypes was moderate, in agreement with previous results (Marrano et al., 2018; Riaz et al., 2018). However it was higher between sativa and non-vinifera genotypes and even higher between sylvestris and non-vinifera genotypes (Table 4B).
All these results are in agreement with previous studies attributing the low level of diversity among sylvestris and the absence of an inter-sylvestris population gene-flow to the small size and the isolation of the sylvestris populations, and the high level of diversity within cultivated genotypes to the sexual crossing occurring during the grapevine domestication (This et al., 2006; Di Vecchi-Staraz et al., 2008; Vitti et al., 2013; Marrano et al., 2017; Riaz et al., 2018).
Also the SNPs heterozygosity was higher in the sativa grapevine compared to sylvestris, supporting previous observations based on SNPs analysis (Lijavetzky et al., 2007; Emanuelli et al., 2013; Marrano et al., 2017). With the exclusion of the accessions indicated as non-true-sylvestris, the SNPs heterozygosity of sylvestris further decreased to a range of 37–41% (Supplementary Table S2). Thus, the heterozygosity range of true-sylvestris appears to be up to 6% less and does not overlap with that of sativa genotypes. Consequently, the SNPs heterozygosity revealed by the GrapeReSeq 18K Vitis genotyping chip could be a useful additional method for separating sylvestris and sativa genotypes, together other SNPs genomic signatures that are able to differentiate between these two sub-taxa (Marrano et al., 2018).
Therefore, although a higher level of heterozygosity is expected in sylvestris because of its obligate out-crossing nature compared to sativa, the results indicate a lower level of diversity in sylvestris than sativa, and a high level of genetic relationship among individuals from the same wild populations. This is in agreement with other studies which highlight that man-made and natural geographical barriers can also lead to the isolation of wild populations in their native habitat, and could lead to significant inbreeding, reduced gene flow within and among different geographic groups (Bacilieri et al., 2013; Emanuelli et al., 2013; Imazio et al., 2013; Riaz et al., 2018).
The UPGMA-trees and structure barplots from SNPs were generally more accurate than the SNPs NJ-dendrograms in the separation of populations. All the UPGMA-trees and barplots from SNPs clearly separate the true sylvestris genotypes from sativa and non-vinifera genotypes, while in the NJ-dendrograms, the cluster of non-vinifera genotypes is mainly among sativa genotypes. Among the sativa population, the UPGMA-trees clearly separate French genotypes (representing the proles occidentalis) from Italian and Muscat genotypes. The barplot from the structure analysis also appeared more informative than the UPGMA-trees because it further separated the Italian sativa (representing the proles pontica) from Muscat genotypes (representing the proles orientalis subpr. caspica), in agreement with Marrano et al. (2015). The UPGMA-tree and barplot obtained from the SNPs-B matrix of 13080 SNPs selected by the missingness (GENO > 0.2) and minor allele frequency (MAF < 0.05) tests including the non-vinifera genotypes with a high rate of failed SNPs (>10%), appeared to be slightly more informative and probable than the tree generated by the matrix of the 13154 SNPs selected only on genotypes with a low rate of failed SNPs (<10%).
With regard to the single genotypes, all the UPGMA-trees, NJ-dendrograms and barplot, correctly assigned the naturalized sativa (Syl_55 and Syl_60), as well as the naturalized European-American Vitis interspecific hybrid ‘Seibel 128’ expectedly in the non-vinifera cluster. The UPGMA-tree generated by the matrix SNPs-C, including only the 1070 SNPs that were successful in all 114 genotypes genotyped by SNPs, although generally less accurate, placed the French varieties ‘Petit Manseng’ and ‘Petit Verdot’ and the Italian variety ‘Greco bianco’ among sylvestris. This was in agreement with all the NJ-dendrograms, all the barplots from SNPs, and also in agreement with the evidence of a second-degree relationship of these genotypes with sylvestris as indicated by IBD analysis. In addition, in the barplot generated by the SNPs-C matrix also ‘Cabernet franc’, ‘Syrah and ‘Roussane bianco’ are among the sylvestris.
The barplot from SSRs (Figure 5A) appears to be highly informative despite the low number of markers (14 loci) compared to the high number of SNPs, although it is less accurate in the separation of genotypes at the edges of the populations. Although it correctly separates the true sylvestris from naturalized sativa and hybrids, only ‘Petit verdot’ is in the cluster of sylvestris, while ‘Petit manseng’ and ‘Greco bianco’ are among the sativa genotypes, even though showing a visible sylvestris ancestry in the barplot. However the Tuscan sativa varieties ‘Colorino’ and ‘Schiava gentile’ are assigned to the population of sylvestris.
These results suggest an introgression of sylvestris into important cultivated sativa varieties, supporting the previous hypothesis that in the areas of grapevine cultivation, the wild germplasm contributed to creating some of the current cultivated varieties (De Andrés et al., 2012; Riaz et al., 2018). The high sylvestris ancestry in ‘Petit Verdot’ has been previously highlighted, together with another five French (Arvine Petite, Cot, Chenin Blanc, Pinot Meunier and Sauvignon Blanc) and five Spanish (Albariño, Caiño Blanco, Ferrón, Maturana, Ondarrabi Betlza) varieties. This research adds the ‘Petit manseng’ to this group of varieties, and possibly also ‘Cabernet franc’ and ‘Syrah, strengthening the hypothesis of a second center of domestication of cultivated grape in Western Europe, in addition to the primary center of domestication in Transcaucasia (Riaz et al., 2018).
The introgression of sylvestris in the Italian cultivated variety ‘Greco bianco,’ but possibly also in the Tuscany local varieties ‘Colorino,’ ‘Schiava gentile’ and ‘Roussane bianco,’ highlights an additional center of domestication in Tuscany.
The identification of a hybrid between a sylvestris and the European-American hybrid ‘Isabel’ and a putative sylvestris with a high non-vinifera ancestry also support a genetic flux among non-vinifera and sylvestris genotypes. Thus, there is a general flux among the various taxa of grapevine in intensive viticulture areas.
The results of this research highlight a higher pollen flow from sativa to sylvestris genotypes than in the opposite direction, and thus a significant introgression of cultivated genotypes into wild grapevines. The evidence of introgression of sylvestris genotypes into cultivated varieties, although at a much lower rate than in the opposite direction, also supports previous research suggesting separate domestication events in Western Europe, in addition to the primary ancient center of domestication from the Near East to Central Asia.
It is therefore plausible that crosses among sylvestris and sativa occurred in the numerous centers of diversification along the migration routes, as well as crosses among sativa genotypes, and that these processes are still ongoing despite the reduction in sylvestris populations.
Finally, the genotype (Syl_26) identified as a cross between ‘Sangiovese’ and a sylvestris, called ‘Femmina Poggi,’ produces grapes of an acceptable quality which are suitable for the production of an interesting wine with a strong black color and intense fruity and spicy notes.
The datasets generated for this study can be found in NCBI and a full list of urls and accession can be found in Supplementary Table S1.
The author confirms being the sole contributor of this work and has approved it for publication.
Project supported by University of Pisa, Fondazione Bertarelli and ColleMassari s.p.a.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author would like to thank Prof. Giancarlo Scalabrelli, collecting sylvestris in Tuscany and for having initiated this project.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00202/full#supplementary-material
TABLE S1 | SSRs allelic profile and chlorotype of 70 putative Vitis vinifera subsp. sylvestris collected in southern Tuscany, 70 Vitis vinifera subsp. sativa, 3 Vitis non-vinifera and 4 non-Vitis genotypes.
TABLE S2 | SNPs missing rate and heterozygosity of single genotypes and various populations (the European–American groups include the hybrid ‘Isabel’ and the hybrid ‘Seibel 128’ identified among the putative sylvestris); SNPs ancestry values for genetic groups inferred by STRUCTURE analysis on 13080 SNPs (SNPs-B matrix) including 37 sylvestris, 70 sativa, 3 non-vinifera, and 4 non-Vitis reference genotypes.
TABLE S3 | SNPs matrix of 114 grapevine genotypes: 70 Vitis vinifera subsp. sativa, 37 Vitis vinifera subsp. sylvestris, 4 Vitis non-vinifera, 3 non-Vitis.
Arroyo-García, R., Ruiz-García, L., Bolling, L., Ocete, R., López, M. A., Arnold, C., et al. (2006). Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 15, 3707–3714. doi: 10.1111/j.1365-294x.2006.03049.x
Bacilieri, R., Lacombe, T., Le Cunff, L., Di Vecchi-Staraz, M., Laucou, V., Genna, B., et al. (2013). Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 13:25. doi: 10.1186/1471-2229-13-25
Barton, N. H., and Slatkin, M. A. (1986). quasi-equilibrium theory of the distribution of rare alleles in a subdivided population. Heredity 56, 409–415. doi: 10.1038/hdy.1986.63
Biagini, B., de Lorenzis, G., Imazio, S., Failla, O., and Scienza, A. (2014). Italian wild grapevine (Vitis vinifera subsp. sylvestris) population: insights into eco-geographical aspects and genetic structure. Tree Genet. Genome 10, 1369–1385. doi: 10.1007/s11295-014-0767-4
Bowers, J. E., Dangl, G. S., and Meredith, C. P. (1999). Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Vitic. 50, 243–246.
Bowers, J. E., Dangl, G. S., Vignani, R., and Meredith, C. P. (1996). Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L). Genome 39, 628–636.
Bowers, J. E., and Meredith, C. P. (1997). The parentage of a classic wine grape, Cabernet Sauvignon. Nat. Genet. 16, 84–87. doi: 10.1038/ng0597-84
Bryan, G. J., McNicoll, J., Ramsay, G., Meyers, R. C., and De Jong, W. S. (1999). Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theor. Appl. Genet. 99, 859–867. doi: 10.1007/s001220051306
Butorac, L., Hancevic, K., Luksic, K., Skvorc, Z., Leko, M., Maul, E., et al. (2018). Assessment of wild grapevine (Vitis vinifera ssp. sylvestris) chlorotypes and accompanying woody species in the Eastern Adriatic region. PLoS One 13:e0199495. doi: 10.1371/journal.pone.0199495
Caporali, E., Spada, A., Marziani, G., Failla, Q., and Scienza, A. (2003). The arrest of development ofabortive reproductive organs in the unisexual flower of Vitis vinifera ssp. silvestris. Sex. Plant Reprod. 15, 291–300. doi: 10.1007/s00497-003-0169-5
Chung, S. M., and Staub, J. E. (2003). The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theor. Appl. Genet. 107, 757–767. doi: 10.1007/s00122-003-1311-3
Coleman, C., Copetti, D., Cipriani, G., Hoffmann, S., Kozma, P., Kovács, L., et al. (2009). The powdery mildew resistance gene REN1 cosegregates with an NBS-LRR gene cluster in two central Asian grapevines. BMC Genet. 10:89. doi: 10.1186/1471-2156-10-89
Cunha, J., Teixeira Santos, M., Carneiro, L. C., Fevereiro, P., and Eiras-Dias, J. E. (2009). Portuguese traditional grapevine cultivars and wild vines (Vitis vinifera L.) share morphological and genetic traits. Genet. Resour. Crop Evol. 56, 975–989. doi: 10.1007/s10722-009-9416-4
De Andrés, M. T., Benito, A., Pérez-Rivera, G., Ocete, R., Lopez, M. A., Gaforio, L., et al. (2012). Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol. Ecol. 21, 800–816. doi: 10.1111/j.1365-294X.2011.05395.x
Di Vecchi-Staraz, M., Laucou, V., Bruno, G., Lacombe, T., Gerber, S., Bourse, T., et al. (2008). Low level of pollen-mediated gene flow from cultivated to wild grapevine: consequences for the evolution of the endangered subspecies Vitis vinifera L. subsp silvestris. J. Hered. 99, 45–55. doi: 10.1093/jhered/esn084
D’Onofrio, C., De Lorenzis, G., Giordani, T., Natali, L., Cavallini, A., and Scalabrelli, G. (2010). Retrotransposon-based molecular markers for grapevine species and cultivars identification. Tree Genet. Genomes 6, 451–466. doi: 10.1007/s11295-009-0263-4
D’Onofrio, C., Fausto, C., Matarese, F., Materazzi, A., Scalabrelli, G., Fiorani, F., et al. (2016). Genotyping of grapevine varieties from garfagnana (Northern Tuscany): evidence of a historical centre of diversity. Am. J. Enol. Vitic. 67, 120–126. doi: 10.5344/ajev.2015.15046
D’Onofrio, C., and Scalabrelli, G. (2010). The software for a universal grapevine database. Acta Hort. 1046, 665–672. doi: 10.1094/PDIS-94-3-0373A
Ellstrand, N. C., Heredia, S. M., Leak-Garcia, J. A., Heraty, J. M., Burger, J. C., Yao, L., et al. (2010). Crops gone wild: evolution of weeds and invasive from domesticated ancestors. Evol. Appl. 3, 494–504. doi: 10.1111/j.1752-4571.2010.00140.x
Emanuelli, F., Lorenzi, S., Grzeskowiak, L., Catalano, V., Stefanini, M., Troggio, M., et al. (2013). Genetic diversity and population structure assessed by SSR and SNP markers in large germplasm collection of grape. BMC Plant Biol. 13:39. doi: 10.1186/1471-2229-13-39
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi: 10.1111/j.1365-294x.2005.02553.x
Forni, G. (2012). “The origin of “old world” viticulture,” in Caucasus and Northern Black Sea Region Ampelography, eds D. Maghradze, L. Rustioni, A. Scienza, J. Turok, and O. Failla, (Quedlinburg: JKI-Julius Kühn-Institut) 27–38.
Grassi, F., Labra, M., Imazio, S., Ocete Rubio, R., Failla, O., Scienza, A., et al. (2006). Phylogeographical structure and conservation genetics of wild grapevine. Conserv. Genet. 7, 837–845. doi: 10.1007/s10592-006-9118-9
Imazio, S. A., Maghradze, D., Lorenzis, G., Bacilieri, R., Laucou, V., This, P., et al. (2013). From the cradle of grapevine domestication: molecular overview and description of Georgian grapevine (Vitis vinifera L.) germplasm. Tree Genet. Genomes 9, 641–658. doi: 10.1007/s11295-013-0597-9
Kalinowski, S. T. (2005). Hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 5, 187–189. doi: 10.1111/j.1471-8286.2004.00845.x
Kalinowski, S. T., Taper, M. L., and Marshall, T. C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. doi: 10.1111/j.1365-294x.2007.03089.x
Kamvar, Z. N., Brooks, J. C., and Grünwald, N. J. (2015). Novel R tools for analysis of genomewide population genetic data with emphasis on clonality. Front. Genet. 6:208. doi: 10.3389/fgene.2015.00208
Levadoux, L. (1956). Les populations sauvages et cultivées de Vitis vinifera L. Ann. Amellior Plantes 1, 59–118.
Lijavetzky, D., Cabezas, J. A., Ibanez, A., Rodriguez, V., and Martinez-Zapater, J. M. (2007). High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genomics 8:424. doi: 10.1186/1471-2164-8-424
Marrano, A., Birolo, G., Prazzoli, M. A., Lorenzi, S., Valle, G., and Grando, S. (2017). SNP-discovery by RAD-Sequencing in a germplasm collection of wild and cultivated grapevines (V. vinifera L.). PLoS One 12:e0170655. doi: 10.1371/journal.pone.0170655
Marrano, A., Grzeskowiak, L., Moreno Sanz, P., Lorenzi, S., Prazzoli, M. L., Arzumanov, A., et al. (2015). Genetic diversity and relationships in the grapevine germplasm collection from Central Asia. Vitis 54, 233–237.
Marrano, A., Micheletti, D., Lorenzi, S., Neale, D., and Grando, M. S. (2018). Genomic signatures of different adaptations to environmental stimuli between wild and cultivated Vitis vinifera L. Hortic. Res. 5:34. doi: 10.1038/s41438-018-0041-2
McGovern, P. E. (2003). Ancient Wine: The Search for the Origins of Viniculture, 1st Edn. Princeton, NJ: Princeton University Press.
Meléndez, E., Puras, P., García, J. L., Cantos, M., Gómez-Rodríguez, J. A., Íñiguez, M., et al. (2016). Evolution of wild and feral vines from the Ega river gallery forest (Basque country and Navarra, Spain) from 1995 to 2015. J. Int. Sci. Vigne Vin 50, 65–75.
Mulcahy, D., Cresti, M., Sansavini, S., Douglas, C., Linskens, H.-F., Bergamini-Mulcahy, G., et al. (1993). The use of random amplified polymorphic DNAs to fingerprint apple genotypes. Sci. Hortic. 54, 89–96. doi: 10.1016/0304-4238(93)90057-w
Myles, S., Boyko, A. R., Owens, C. L., Brown, P. J., Grassi, F., Aradhya, M. K., et al. (2011). Genetic structure and domestication history of the grape. Proc. Natl. Acad. Sci. U.S.A. 108, 3457–3458.
Negrul, A. M. (1946). “Origin of cultivated grapevine and its classification,” in Ampelography of the Soviet Union, ed. A. M. Frolov-Bagreev, (Moscow: Pishchepromizdat), 159–216.
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. U.S.A. 70, 3321–3323. doi: 10.1073/pnas.70.12.3321
Peakall, R., and Smouse, P. E. (2006). GenAlEx 6: genetic analysis in excel. population genetic software for teaching and research. Mol. Ecol. Notes 6, 288–295. doi: 10.1111/j.1471-8286.2005.01155.x
Peakall, R., and Smouse, P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research – an update. Bioinformatics 28, 2537–2539. doi: 10.1093/bioinformatics/bts460
Péros, J. P., Berger, G., Portemont, A., Boursiquot, J. M., and Lacombe, T. (2010). Genetic variation and biogeography of the disjunct Vitis subgenus (Vitaceae). J. Biogeogr. 38, 471–486. doi: 10.1111/j.1365-2699.2010.02410.x
Popescu, C. F., Dejeu, L. C., and Ocete, R. R. (2013). Preliminary characterization of wild grapevine populations (Vitis vinifera ssp. sylvestris) grown along the Danube River. Not. Bot. Horti Agrobot. Cluj Napoca 41, 472–477.
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959.
Purcell, S., Neale, B., Todd-Brown, K., Thomas, L., Ferreira, M. A. R., Bender, D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575. doi: 10.1086/519795
R Core Team, (2019). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Riaz, S., De Lorenzi, G., Velasco, D., Koehmstedt, A., Maghradze, D., Bobokashvili, Z., et al. (2018). Genetic diversity analysis of cultivated and wild grapevine (Vitis vinifera L.) accessions around the Mediterranean basin and Central Asia. BMC Plant Biol. 18:137. doi: 10.1186/s12870-018-1351-0
Rousset, F. (2008). Genepop’007: a complete reimplementation of the genepop software for windows and Linux. Mol. Ecol. Resour. 8, 103–106. doi: 10.1111/j.1471-8286.2007.01931.x
Sefc, K. M., Regner, F., Turetschek, E., Glöss, J., and Steinkellner, H. (1999). Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 42, 367–373. doi: 10.1139/g98-168
Sokal, R. R., and Michener, C. D. (1958). A statistical method for evaluating systematic relationships [J]. Univ. Kans. Sci. Bull. 28, 1409–1438.
Stout, A. B. (1921). Types of flowers and intersexes in grapes with reference to fruit development. N. Y. Agric. Exp. Station Tech. Bull. 82, 1–16.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kuma, R. S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
This, P., Lacombe, T., and Thomas, M. R. (2006). Historical origins and genetic diversity of wine grapes. Trends Genet. 22, 511–519. doi: 10.1016/j.tig.2006.07.008
Thomas, M. R., and Scott, N. S. (1993). Microsatellite repeats in grapevine reveal DNA polymorphism when analysed as sequence tagged sites (STSs). Theor. Appl. Genet. 86, 985–990. doi: 10.1007/bf00211051
Vitti, J. J., Grossman, S. R., and Sabeti, P. C. (2013). Detecting natural selection in genomic data. Annu. Rev. Genet. 47, 97–120. doi: 10.1146/annurev-genet-111212-133526
Weir, B. S., and Cockerham, C. (1984). Estimating F-statistics for the analysis of population genetics. Evolution 38, 1358–1370.
Weising, K., and Gardner, R. C. (1999). A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledoneous angiosperms. Genome 42, 9–19. doi: 10.1139/g98-104
Xu, S., Gupta, S., and Jin, L. (2010). PEAS V1.0: a package for elementary analysis of SNP data. Mol. Ecol. Resour. 10, 1085–1088. doi: 10.1111/j.1755-0998.2010.02862.x
Zohary, D., and Hopf, M. (2000). Domestication of Plants in the Old World, 3rd Edn. New York, NY: Oxford University Press.
Keywords: chlorotype, domestication, GrapeReSeq 18K Vitis genotyping chip, lambrusque, microsatellite, spontaneous crosses, Vitis vinifera subsp. sylvestris, Vitaceae
Citation: D’Onofrio C (2020) Introgression Among Cultivated and Wild Grapevine in Tuscany. Front. Plant Sci. 11:202. doi: 10.3389/fpls.2020.00202
Received: 14 October 2019; Accepted: 11 February 2020;
Published: 28 February 2020.
Edited by:
Rosa Arroyo-Garcia, Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (INIA), SpainReviewed by:
Javier Ibáñez, Institute of Vine and Wine Sciences (ICVV), SpainCopyright © 2020 D’Onofrio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Claudio D’Onofrio, Y2xhdWRpby5kb25vZnJpb0B1bmlwaS5pdA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.