
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 28 February 2020
Sec. Plant Cell Biology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00164
Autophagy is an intracellular process that facilitates the bulk degradation of cytoplasmic materials by the vacuole or lysosome in eukaryotes. This conserved process is achieved through the coordination of different autophagy-related genes (ATGs). Autophagy is essential for recycling cytoplasmic material and eliminating damaged or dysfunctional cell constituents, such as proteins, aggregates or even entire organelles. Plant autophagy is necessary for maintaining cellular homeostasis under normal conditions and is upregulated during abiotic and biotic stress to prolong cell life. In this review, we present recent advances on our understanding of the molecular mechanisms of autophagy in plants and how autophagy contributes to plant development and plants’ adaptation to the environment.
Autophagy, which means self-eating, is found universally in all eukaryotic cells. It is an important process for degrading proteins and organelles, via the vacuole in yeast and plants or through the lysosome in animals, to facilitate intracellular recycling (Marshall and Vierstra, 2018). In plants, autophagy is continuously maintained at a basal level during growth and development to ensure homeostasis, but is upregulated under environmental stresses to aid plant survival (Wang et al., 2018a). Three major types of autophagy have been described in plants: microautophagy, macroautophagy, and mega-autophagy. Among these, macroautophagy has been the best studied (van Doorn and Papini, 2013).
During microautophagy, cytosolic proteins or entire organelles are congregated together near the vacuole and become encapsulated by an invagination or protrusion of the vacuolar membrane. This forms an intravacuolar vesicle called an autophagic body, which is released to the vacuolar lumen by membrane scission and is degraded (van Doorn and Papini, 2013; Marshall and Vierstra, 2018). In contrast, in macroautophagy, a cup-shaped double-membrane structure, named the phagophore, is formed at the phagophore assembly site (PAS). Some studies show that the phagophore arises from endoplasmic reticulum (ER), mitochondria and plasma membrane or from ER-mitochondria contact sites (Hamasaki et al., 2013; Le Bars et al., 2014; Zhuang et al., 2017). The phagophore elongates and encircles the cytosolic substances to form double membrane-bound autophagosomes that subsequently fuse with the vacuole to release the internal vesicle for degradation (Liu and Bassham, 2012). The third route of autophagy, which is only found in plants, is mega-autophagy and occurs concomitantly with programmed cell death (PCD). Large amounts of hydrolases are released into the cytoplasm from the vacuole, resulting in large-scale degradation of cellular components including cytoplasm, all organelles, the plasma membrane and part of the cell wall (Hara-Nishimura and Hatsugai, 2011; van Doorn and Papini, 2013). Unlike both microautophagy and macroautophagy that recycle macromolecular constituents back to the cytosol from the vacuole, mega-autophagy is an extreme form of massive degradation resulting in cell death.
In addition to the conserved microautophagy and macroautophagy pathways, yeast also have a cytoplasm-to-vacuole targeting (Cvt) pathway that is biosynthetic and occurs constitutively to deliver precursors of resident hydrolases to the vacuole. The Cvt pathway operates under a similar mechanism to macroautophagy (Klionsky and Emr, 2000). In mammals, chaperone-mediated autophagy is the third pathway used for transferring substrate proteins directly to the lysosome without the use of separate vesicles (Dice, 2007). However, no evidence has been shown that the Cvt pathway nor chaperone-mediated autophagy exist in plants.
Macroautophagy is characterized by the presence of autophagosomes with a double-membrane envelope that form at the PAS and will eventually fuse with the vacuole. A series of autophagy-related genes (ATGs) are responsible for the initiation and formation of autophagosomes. Although autophagy has long been regarded as a process of nonselective degradation of cellular structures (general autophagy), plentiful evidence indicates that autophagy is also a highly selective mechanism to target a wide range of superfluous or damaged components as part of cellular quality control and stress responses (selective autophagy) (Yoshimoto and Ohsumi, 2018). Many ATG proteins involved in general autophagy are conserved in yeast, plants, and animals, indicating that eukaryotes may share similar autophagy mechanisms. In contrast, the selective autophagy pathways rely on a variety of selective autophagy receptors. Although the selective autophagy receptors are not conserved, their modes of action and regulatory mechanisms are similar across eukaryotes (Farré and Subramani, 2016). Various autophagy pathways have specific roles during plant development and stress response. In this review, we summarize recent advances in understanding the molecular mechanisms underlying autophagy in plants and the important roles that autophagy plays in plant development and stress responses.
Although autophagosomes were first observed in mammalian cells in the 1950s (de Duve et al., 1955), the molecular mechanisms of autophagy have been primarily revealed from research in yeast, then extended to animal and plant (Tsukada and Ohsumi, 1993; Ohsumi, 2001; Marshall and Vierstra, 2018). These great progresses expand our understanding to higher eukaryotes in recent decades.
Macroautophagy, often simply called autophagy, has been well characterized (Figure 1). The autophagy process can be divided into distinct stages: induction, cargo recognition, phagophore formation, phagophore expansion and closure, and autophagosome fusion and breakdown (Masclaux-Daubresse et al., 2017). More than 40 ATG genes have been characterized in yeast. Homologs of many ATGs have been characterized in animals and plants (Tang and Bassham, 2018). At present, about 40 ATGs have been identified in Arabidopsis thaliana, most of which are homologous with yeast ATGs (Chung, 2019). The ATG proteins can be divided into four core functional groups: 1) the ATG1/ATG13 kinase complex that initiates autophagosome formation in response to nutrient limitation; 2) the autophagy-specific class III phosphatidylinositol (PI) 3-kinase complex; 3) the ATG9 complex which promotes phagophore expansion; and 4) the ATG8/ATG12 ubiquitin-like conjugation systems which act during phagophore expansion and maturation (Kim et al., 2012; Wang et al., 2018a).
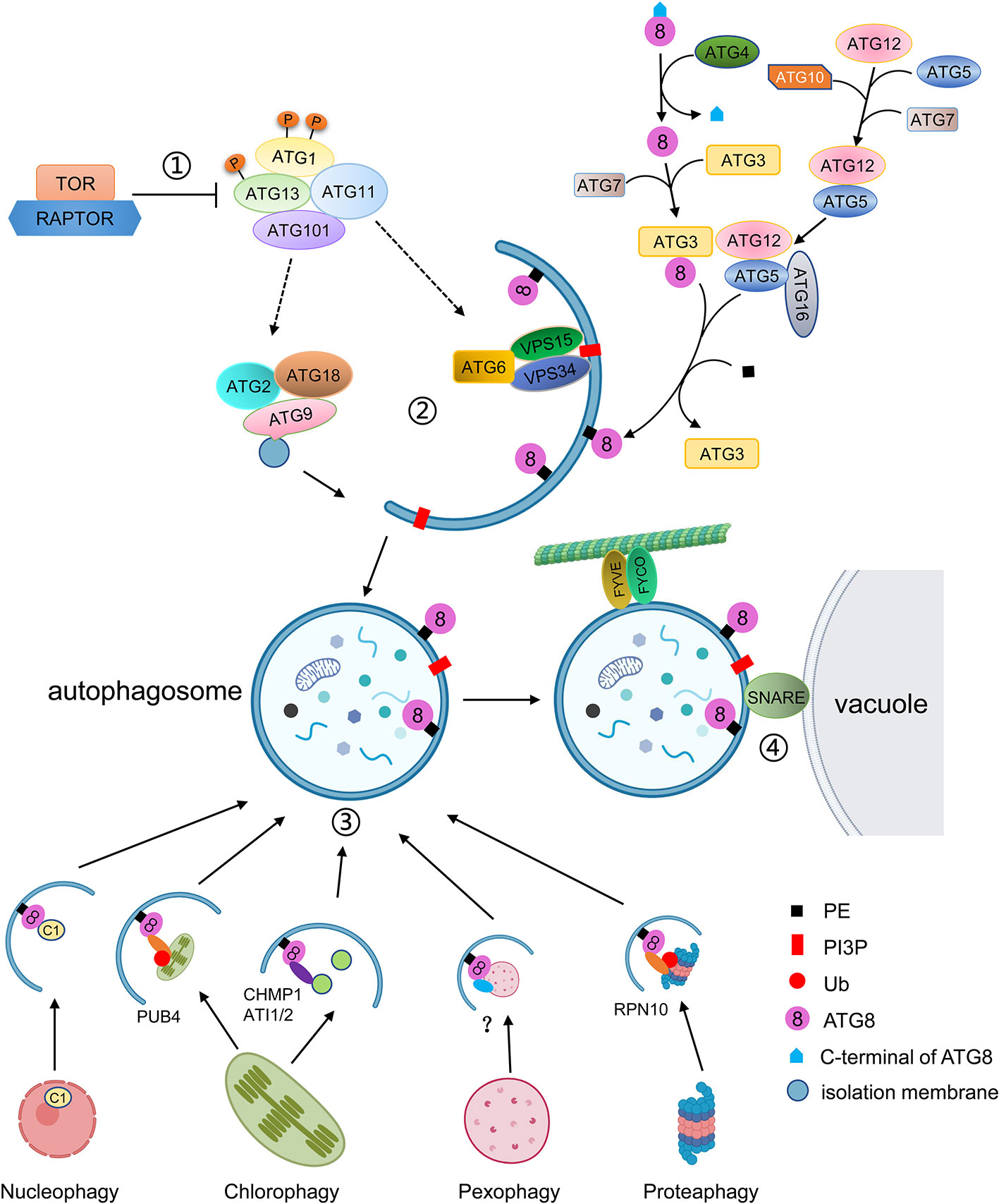
Figure 1 Schematic diagram of the autophagy process. ① Autophagy is regulated by TOR kinase. Inhibition of TOR results in activation of constitutive autophagy, while overexpression of TOR blocks autophagy. Autophagy is initiated by the association of ATG13 and ATG1, together with ATG11 and ATG101, into an active complex. ② Autophagosome formation includes membrane delivery, vesicle nucleation and phagophore expansion and closure. ATG9, with ATG2 and ATG18, is involved in the delivery of lipids to the expanding phagophore. The VPS34 lipid kinase complex generates PI3P decoration, which is accompanied by the conjugation of ATG8 to PE. ATG8 firstly is matured by cleavage of its C-terminal by ATG4, and then is conjugated to PE by E2-like ATG3 and the E3-like ATG12-ATG5-ATG16 complex. ATG8-PE localizes to the autophagosomal membrane for phagophore expansion. ③ Selective autophagy is mediated by interaction of ATG8 with specific autophagic receptors through the AIM domain. Different cell components can be recognized by specific autophagic receptors. For instance, the C1 protein is translocated from the nucleus to the cytoplasm and binds to ATG8 for degradation. The whole chloroplast is ubiquitulated by PUB4 and binds to ATG8. The fragments of chloroplasts are recognized by CHMO1 or ATI1/2 for degradation by RCBs or ATI1/2 decorated plastid bodies. In plants, peroxisomes to be degraded are probably recognized by PEX6 or PEX10, and the ubiquitylated proteasome is recognized by RPN10. ④ The mature autophagosome is transported to the vacuole with the help of FYVE and FYCO proteins and fused with the vacuole to be degraded.
Autophagy is initiated by the down-regulation of the TOR (Target of Rapamycin) complex as a consequence of various stimuli, such as developmental and nutritional signals (Marshall and Vierstra, 2018). TOR is an essential serine/threonine kinase belonging to the phosphatidylinositol kinase-related kinase family and negatively regulates autophagy (Díaz-Troya et al., 2008). In yeast, TOR1/2, KOG1 (Kontroller of growth 1), LST8 (Lethal with Sec13 protein 8), and TCO89 (89-kDa subunit of Tor complex 1) form TOR complex 1 (TORC1) to inhibit autophagy (Díaz-Troya et al., 2008; Cebollero and Reggiori, 2009; He and Klionsky, 2009). It has been reported that the TORC1 equivalent also exists in plants (Robaglia et al., 2012). In Arabidopsis, TOR is encoded by a single gene, which is essential for embryo development (Menand et al., 2002). The Arabidopsis RAPTOR family is homologous to yeast KOG1 and functions as the target recognition cofactor of TOR. The RAPTOR family consists of RAPTOR1/Raptor1B and RAPTOR2/Raptor1A. Disruption of RAPTOR1/Raptor1B causes repression of embryo development and post-embryonic plant growth, whereas disruption of RAPTOR2/Raptor1A does not affect plant development and growth (Anderson et al., 2005; Deprost et al., 2005). In Arabidopsis, two putative homologs of LST8 have been identified; however, no TCO89 homologs have yet been found (Díaz-Troya et al., 2008; Moreau et al., 2012). As regulatory associated proteins, the LST8 homologs may be involved in plant autophagy by regulating TOR activity, but this needs to be confirmed. In plants, active TOR inhibits autophagy under normal conditions, whereas inactive TOR promotes autophagy under nutrient starvation or during treatment with chemical inhibitors such as rapamycin or AZD8055 (Ren et al., 2012; Li et al., 2015b).
Arabidopsis SnRK1 (SNF1/AMPK-related kinase 1), the homolog of SNF1/AMPK, which can inhibit TOR activity in yeast and mammals, is a heterotrimeric complex consisting of a catalytic α subunit and regulatory β and γ subunits (Smeekens et al., 2010). KIN10 is one of the three isoforms of the SnRK1 catalytic subunit involved in autophagy. Autophagy is blocked in mutants lacking KIN10 under nutrient deficiency and salt, oxidative, and ER stress conditions, whereas overexpression of KIN10 induces autophagy under abiotic stresses (Chen et al., 2017; Soto-Burgos and Bassham, 2017). Under nutrient deprivation, KIN10 promotes the phosphorylation of ATG1 and the TOR complex subunit RAPTOR to inhibit TOR activity and initiate autophagy (Nukarinen et al., 2016; Chen et al., 2017).
In Arabidopsis, under normal conditions, TOR is active and hyperphosphorylates ATG13. Highly phosphorylated ATG13 has a low binding capacity for ATG1, resulting in low activity of ATG1 and maintenance of basal levels of autophagy. In Arabidopsis, ATG1 and ATG13, which along with other two accessory subunits ATG11 and ATG101, are assembled into an active complex to initiate autophagy (Suttangkakul et al., 2011; Li and Vierstra, 2014). However, whether the assembly of the ATG1-ATG13 complex is regulated by nutrient availability is still under debate.
In Arabidopsis, the activation of the ATG1-ATG13 complex stimulates the downstream steps of autophagy including membrane delivery, vesicle nucleation, and phagophore expansion and closure (Li and Vierstra, 2012; Marshall and Vierstra, 2018). ATG9, along with ATG2 and ATG18, is involved in the formation of the isolation membrane at the PAS and the delivery of lipids to the expanding phagophore (Zhuang et al., 2017). In yeast and mammals, atg9 mutants fail to form autophagosomes, but in Arabidopsis, the deletion of ATG9 results in expanding autophagosome-related tubules connected to the ER (Orsi et al., 2012; Yamamoto et al., 2012; Zhuang et al., 2017). Also, the sequence of AtATG9 shows limited homology with those of yeast and mammals, suggesting that AtATG9 may function in a plant-specific manner during autophagosome formation (Zhuang et al., 2018).
In yeast, the Atg2–Atg18 complex is localized to the PAS in a phosphatidylinositol-3-phosphate (PI3P)-dependent manner with Atg2 associating with the ER to form the isolation membrane (Kotani et al., 2018). In yeast, PI3P, a small membrane lipid, is generated by the class III PI3K (phosphatidylinositol-3-kinase) complex I, consisting of Vps34, Vps15, Vps30/Atg6, and Atg14 (Kihara et al., 2001). During autophagy, PI3K complex I localizes to the PAS to recruit downstream proteins through its product, PI3P. Loss of Arabidopsis VPS34, ATG6/VPS30, and VPS15, cause lethality due to defects in pollen germination and development (Fujiki et al., 2007; Lee et al., 2008; Xu et al., 2011). Heterozygous Arabidopsis vps34 and vps15 mutants display abnormal vacuole morphology (Lee et al., 2008). In tobacco cell culture, PI3K inhibitors such as wortmannin, LY294002, and 3-methyladenine (3-MA) impair the formation of autophagosomes and block the accumulation of autolysosomes, indicating that PI3K plays a crucial role in autophagy (Takatsuka et al., 2004).
Meanwhile, a ubiquitin-like conjugation pathway is required for the insertion of ATG8-phosphatidylethanolamine (PE) into the membrane of the expanding phagophore (Marshall and Vierstra, 2018). In yeast, Atg8 is initially synthesized as a long, inactive precursor, which is subsequently cleaved by the cysteine protease Atg4 to expose a glycine residue on the C-terminus (Ohsumi, 2001). The mature ATG8 can be activated through binding of ATP-dependent E1 (ubiquitin-activating enzyme)-like ATG7 to its newly exposed glycine. ATG8 is subsequently donated to the E2 (ubiquitin-conjugating enzyme)-like ATG3 and finally conjugated to PE by the E3 (ubiquitin ligase)-like ATG12-ATG5-ATG16 complex. This complex is formed by another ubiquitin-fold protein, ATG12, that is conjugated to ATG5 through the sequential actions of ATG7 and the ATG12-specific E2 conjugating enzyme ATG10. ATG12-ATG5 then connects to the dimeric ATG16 protein to tether to the phagophore, allowing for the lipidation of ATG8 with PE (Hanada et al., 2007; Chung et al., 2010).
The ATG8 ubiquitin-like conjugation pathway was also been reported to exist in Arabidopsis. However, unlike in yeast, Arabidopsis has nine homologs of ATG8 (ATG8a-ATG8i), two homologs of ATG4 (ATG4a and ATG4b), and two homologs of ATG12 (ATG12a and ATG12b) (Doelling et al., 2002; Hanaoka et al., 2002). The Arabidopsis ATG8 genes have different expression patterns, indicating that they may have distinct functions (Sláviková et al., 2005). In Arabidopsis, the ATG4s can cleave the C-terminus of ATG8, as its homolog does in yeast. Moreover, the atg4a atg4b double mutant displays autophagy defects, evidenced by early senescence and decreased silique production, indicating that ATG4s are essential for plant development (Yoshimoto et al., 2004). In Arabidopsis, the atg12a atg12b double mutant shows premature senescence, sensitivity to nutrient starvation, and lack of autophagic bodies, whereas the single atg12a or atg12b mutants do not. The accumulation of the ATG12-ATG5 conjugate was reduced in either atg12a or atg12b single mutants, and ATG8-PEs were not detected in atg12a atg12b, indicating that the ATG12-ATG5 conjugate is essential for ATG8-PE conjugations (Chung et al., 2010). Mutation of ATG5, ATG7 or ATG10 results in plants which are hypersensitive to nitrogen and carbon starvation (Chung et al., 2010). Like atg12, the atg10-1 and atg5-1 mutants fail to form autophagic bodies in the vacuole (Phillips et al., 2008).
The ATG8-PE conjugates, which are located in both the inner and outer autophagosome membranes, aid phagophore expansion and vesicle closure to form autophagosomes and recognize autophagic cargoes through ATG8-interacting proteins (Johansen and Lamark, 2011; Liu and Bassham, 2012). In Arabidopsis, a membrane associated protein SH3P2 (SH3 domain-containing protein 2) translocates to the PAS during autophagy (Zhuang et al., 2013). SH3P2 not only interacts with ATG8 but also binds to PI3P and associates with the PI3K complex to facilitate membrane elongation and autophagosome closure (Zhuang et al., 2013).
In mammals, when the autophagosome is sealed, FYCO1 (FYVE and coiled-coil domain containing 1) binds to LC3/ATG8 and PI3P on the outer autophagosome membrane to promote autophagosome movements through microtubule plus end-directed transport (Pankiv et al., 2010). In Arabidopsis, co-sedimentation and co-localization assays showed that ATG8 can bind to microtubules in vivo, suggesting microtubules may be involved in relocation of autophagosomes to the vacuole (Ketelaar et al., 2004).
Correct targeting of autophagosomes to the vacuole requires SNAREs (soluble NSF attachment protein receptors) (Surpin et al., 2003). In Arabidopsis, under nutrient starvation conditions, loss of VTI12, a VTI1-type v-SNARE (vesicle-SNARE) located on the target membrane, results in failure of the autophagosomes to enter the vacuole, indicating that VTI12 plays an important role in autophagosome fusion with the tonoplast (Surpin et al., 2003).
Autophagosome fusion and degradation require the ESCRT (Endosomal Sorting Complex Required for Transport) machinery. Recently, some of the components involved in this process have been identified. Arabidopsis AMSH3 (associated molecule with the SH3 domain of STAM3), a deubiquitinating enzyme, interacts with the ESCRT-III subunit VPS2.1 (vacuolar protein sorting 2.1) and is essential for autophagosome trafficking to the vacuole (Katsiarimpa et al., 2013). A plant-specific ESCRT component, FREE1 (FYVE domain protein required for endosomal sorting 1), was reported to directly interact with the autophagy regulator SH3P2 and to play a role in autophagosome-vacuole fusion in Arabidopsis (Zhuang et al., 2013; Gao et al., 2015). Arabidopsis CFS1 (Cell death related endosomal FYVE/SYLF protein 1), a FYVE and SYLF domain-containing protein unique to plants that co-localizes and interacts with PI3P and the ESCRT-I component ELCH, is enriched in the autophagosome membrane and participates in autophagosome turnover (Sutipatanasomboon et al., 2017).
After the autophagosome and vacuole are fused, the internal vesicle, called the autophagic body, is released into the vacuole and degraded by a series of resident hydrolases (Marshall and Vierstra, 2018). ATG8-PE on the inner autophagosome membrane is also degraded in the vacuole, while the ATG8-PE attached to the outer autophagosome membrane is cleaved by ATG4 to release ATG8 from PE for recycling (Yoshimoto et al., 2004).
Autophagy occurs during all developmental stages of plants. It has become increasingly clear that selective autophagy also exists to specifically remove certain cellular components. The specificity of autophagy is mediated by interactions between ATG8 and specific autophagic receptors containing an ATG8-interacting motif (AIM) (Marshall et al., 2015; Farré and Subramani, 2016; Maqbool et al., 2016). Autophagic receptors mediate selective autophagy of specific cellular components, such as various organelles, protein aggregates, and even invading pathogens (Marshall and Vierstra, 2018). Selective autophagy endows plants with the ability to specifically regulate cell activity according to their physiological state and external environment.
In plants, the quality control machinery, in which the ubiquitin-proteasome system (UPS) plays an important role, is utilized to degrade defective proteins and maintain the integrity of the cellular proteome (Balchin et al., 2016). Selective autophagy also plays an important role in the clearance of damaged protein complexes, such as proteasomes (Yoon and Chung, 2019). Proteaphagy was first described in plants and is activated by two pathways: one is induced by nitrogen starvation and controlled by ATG1, the other is activated by proteasome inhibitors such as MG132 in an ATG1-independent manner (Marshall et al., 2015). When the proteasome is blocked by inhibitors, the proteasome complex is ubiquitylated and recognized by proteaphagy receptors, RPN10 in Arabidopsis, to initiate the assembly of autophagosomes (Marshall et al., 2015). RPN10 is a ubiquitin-binding proteasome subunit which can bind the ubiquitylated proteasome through a UIM (Ubiquitin-interacting motif) and ATG8 via another UIM-related sequence (Marshall et al., 2019). In the Arabidopsis rpn10 mutant, inhibitor-induced proteaphagy is blocked (Marshall et al., 2015).
Nonfunctional proteins are also degraded by selective autophagy in the form of aggregates, termed aggrephagy, with ubiquitin-chains acting as the marker for the degradation (Yoon and Chung, 2019). p62/SEQUESTOSOME 1 (SQSTM1) and Neighbor of BRCA 1 (NBR1) in mammals and Cue5 in yeast are aggrephagy receptors which simultaneously bind ATG8 via an AIM and poly-ubiquitin chains via a ubiquitin-binding domain (Pankiv et al., 2007; Lu et al., 2014). A homolog of NBR1 has been found in plants. Plant NBR1 contains an N-terminal PB1 (Phox and Bem1p) domain and binds to ubiquitin and ATG8 simultaneously, suggesting conserved mechanisms of aggrephagy in mammals/yeast and plants (Svenning et al., 2011). In Arabidopsis undergoing heat stress, mutation of NBR1 results in the accumulation of ubiquitylated insoluble proteins (Zhou et al., 2013). Also, heat stress can induce the association of NBR1 and ATG8 with accumulated cytoplasmic protein aggregates, suggesting that NBR1 is a plant aggrephagy receptor essential for maintaining proteostasis (Jung et al., 2020).
The chloroplast is a unique organelle found only in plants and algae, which mainly performs the functions of material and energy metabolism through photosynthesis and signal transduction (Jarvis and López-Juez, 2013). Damaged or redundant chloroplasts are degraded through selective autophagy, called chlorophagy. For example, under darkness or in senescent leaves, chloroplasts become redundant and are transported to the vacuole for recycling via an autophagy-dependent process (Wada et al., 2009). Similarly, chloroplasts damaged by strong light or ultraviolet-B are also subjected to degradation through autophagy (Izumi et al., 2017). Chlorophagy happens in many ways, including the whole chloroplast, the Rubisco containing body (RCB), the ATI1-GFP labels plastid-associated body (ATI-PS body), and the small starch granule-like structures pathways (Izumi et al., 2019; Nakamura et al., 2019; Zhuang and Jiang, 2019).
The whole chloroplast pathway includes the ubiquitylation of damaged chloroplasts by the cytosolic ubiquitin ligase PUB4 (Plant U-BOX Protein 4). The ubiquitylated chloroplasts are then encapsulated into ATG8-decorated autophagic vesicles and delivered to the vacuole (Izumi et al., 2017). In contrast, during the early stages of senescence, small double-membrane spherical structures called RCBs, containing stromal proteins such as rubisco but not the chloroplast envelope nor thylakoid proteins, bud from chloroplasts (Ishida et al., 2008). The RCBs may arise from the fission of stroma-filled tubules that protrude from the chloroplast, in which the ESCRT component CHMP1 (Charged Multivesicular Body 1) may play an important role, and are degraded in the vacuole through autophagy (Spitzer et al., 2015; Marshall and Vierstra, 2018). A third pathway of chlorophagy involves plant-specific proteins, ATI1 and ATI2 (ATG8-interacting protein 1 and 2), which localize to the ER- and plastid-derived bodies (Michaeli et al., 2014). These ATI1-decorated plastid bodies contain plastid membrane proteins from the outer envelope or thylakoids and are delivered to the vacuole through the autophagy machinery (Michaeli et al., 2014). Although both come from plastids, RCBs and ATI1-decorated bodies contain different plastid proteins, suggesting two distinct pathways, and function in different developmental stages or environmental conditions. Starch granules exist in the chloroplasts and serve as a carbon reservoir (Malinova et al., 2018). But a small starch granule-like structure (SSGL) is also observed in the cytosol and is sequestered into the autophagic bodies (Wang et al., 2013), indicating that the SSGLs derived from chloroplasts may be degraded by autophagy. Consistent with this idea, silencing of ATG6 can reduce the number of vacuole-localized SSGLs, suggesting that the degradation of SSGLs is ATG-dependent (Wang et al., 2013).
Plant peroxisomes are ubiquitous organelles housing various metabolic reactions including the glyoxylate cycle, photorespiration, and the β-oxidation of fatty acids (Su et al., 2019). The types of metabolic reactions occurring in peroxisomes change according to the plant developmental stage (Young and Bartel, 2016). For example, peroxisomes are enriched in enzymes of the glyoxylate cycle as they are necessary for seed germination in early developmental stage (Hu et al., 2012). However, in green seedlings, these enzymes are eliminated and replaced by those required for photorespiration (Lingard et al., 2009). It has been demonstrated that this remodeling process requires the involvement of selective autophagy of peroxisomes, termed pexophagy (Bartel et al., 2014).
Peroxisomes are the major site of reactive oxygen species (ROS) production, making them vulnerable to oxidative damage. The peroxisomes also contain multiple antioxidative enzymes to remove excess ROS, such as catalase that specifically degrades H2O2 (Mhamdi et al., 2012; Su et al., 2018). Peroxisomal catalase detoxifies H2O2 by converting it to water and molecular oxygen, but catalase itself is also susceptible to damage by H2O2 (Anand et al., 2009). Aggregates of inactive catalase accumulate in peroxisomes of autophagy-deficient seedlings, and atg2 mutants show clustered, damaged peroxisomes, indicating that damaged peroxisomes are degraded by pexophagy (Shibata et al., 2013; Yoshimoto et al., 2014). Moreover, other atg mutants (atg5, atg7, atg10, and atg12) display increased accumulation and aggregation of peroxisomes in guard cells. Additionally, these mutants display increased ROS levels and impaired stomatal opening, indicating that autophagy regulates guard cell ROS homeostasis and stomal opening (Yamauchi et al., 2019).
Pexophagy seems to occur at a higher rate than other types of selective autophagy and is tissue-dependent. For example, pexophagy is active in above ground parts of the plant but not in the roots, possibly because of the higher photorespiratory activity in green tissue (Yoshimoto et al., 2014). However, it has recently been shown that pexophagy in roots is involved in the regulation of glucose-mediated root meristem activity through mediation of auxin biosynthesis (Huang et al., 2019a). In the roots of wild-type plants, high levels of glucose induce ROS accumulation. On the one hand, ROS oxidize active IAA (indole-3-acetic acid). On the other hand, constitutive pexophagy is enhanced by high levels of ROS to attenuate the synthesis of IAA, in turn reducing root meristem activity (Huang et al., 2019a). However, in atg5 and atg7, the autophagy deficiency disrupts transmission of the high-glucose signal to the peroxisomes, enhancing the activity of IAA and the root meristem, resulting in longer primary roots than in wild type under high glucose conditions (Huang et al., 2019a).
How peroxisomes are marked for degradation is not yet clear in plants. In yeast, the AIM-containing pexophagy receptor (Atg30 in Pichia pastoris and Atg36 in Saccharomyces cerevisiae) binds to Pex3, a peroxisome membrane protein, to recruit the autophagic machinery by interacting with Atg8 and Atg11 (Farré et al., 2013). In Arabidopsis, two peroxisomal membrane proteins, PEX6 and PEX10, have been shown to interact with ATG8 via an AIM, suggesting that they may participate in the initiation of pexophagy (Xie et al., 2016).
As the central organelle of the eukaryotic cell, the nucleus plays a key role in maintaining genomic integrity and controlling gene expression. Cells require a strategy to eliminate undesirable nuclear proteins and components when suffering stress or pathogen infection. The mechanisms of selective autophagy of nuclear components including nuclear envelope components, DNA, RNA and nucleoli, termed nucleophagy, are conserved from yeast to mammals (Klionsky et al., 2008; Papandreou and Tavernarakis, 2019). However, for a long time, no evidence demonstrated that nuclear autophagy occurs in plants. Recent research has shown that the accumulation of a geminivirus nuclear protein C1 induces autophagy. ATG8h interacts with C1 and translocates it from the nucleus to the cytoplasm, through the XPO1-mediated nuclear export-dependent pathway (Li et al., 2019a). Mutation of the AIM in C1 abolishes its interaction with ATG8. Degradation of C1 is blocked by autophagy inhibitors and the deletion of ATG8h, ATG5, or ATG7 (Li et al., 2019a). This discovery was the first to reveal that autophagy is involved in the degradation of nuclear proteins in plants.
Moreover, the BRI1-EMS Suppressor 1 (BES1), a transcription factor which positively regulates brassinosteroid signaling, is ubiquitylated and interacts with DSK2A to be degraded in a DSK2- and core ATG-dependent manner. DSK2A is an autophagy adaptor which contains a ubiquitin-like domain, two AIMs, and a ubiquitin-associated domain (Nolan et al., 2017). Binding of DSK2 to ATG8 is regulated by BIN2 kinase, which phosphorylates DSK2 near the AIM domains to enhance its binding ability to ATG8 (Nolan et al., 2017). However, it is not clear whether BES1 is degraded before or after it enters the nucleus.
In addition to the above described selective autophagy pathways, there are also other types of selective autophagy found in plants cells. For example, in Arabidopsis, a selective autophagy pathway involved in rRNA turnover which is dependent on ATG5 has been reported (Floyd et al., 2015). In addition, it has been shown recently that mammalian NUFIP1 is a ribophagy receptor required for the selective autophagy of ribosomes during starvation conditions (Wyant et al., 2018). A homolog of mammalian NUFIP1 exists in Arabidopsis, but further investigation is needed to show whether Arabidopsis NUFIP1 is also involved in ribophagy (Rodor et al., 2011).
Recently, a new class of ATG8 interactors has been described that harbor a UIM-like domain to interact with ATG8 in plants, yeast, and humans (Marshall et al., 2019). Therefore, it is likely that new selective autophagy pathways may be discovered in the near future. These various kinds of selective autophagy pathways ensure that plant cells can specifically remove useless or damaged cell components efficiently to maintain cell viability and enable plant survival during environmental stresses.
Autophagy is a highly conserved process which plays significant roles in controlling overall plant development, metabolism, senescence, and stress responses (Janse van Rensburg et al., 2019). Under normal growth conditions, basal levels of autophagy are essential for maintaining cellular homeostasis. Although most atg mutants are not fatal and are able to complete their life cycle, they do show clear growth defects. The Arabidopsis atg4, atg5, atg7, atg10, and atg12 mutants all display hypersensitization to nitrogen and carbon starvation and premature senescence compared with wild type (Phillips et al., 2008; Chung et al., 2010). Under nitrogen starvation, atg5, atg7, and atg10 seedlings show severely slowed leaf emergence and expansion as well as chlorotic leaves. Similar defects are seen with the atg4, atg5, atg7, and atg12 mutants under carbon starvation (Phillips et al., 2008; Chung et al., 2010). In contrast, in Arabidopsis, overexpression of ATG5 and ATG7 stimulates the lipidation of ATG8, autophagosome formation, and autophagic flux, concomitant with increased growth, seed set, and seed oil content (Minina et al., 2018). Overexpression of ATG8 in Arabidopsis not only increases autophagosome number and promotes autophagic activity, but also improves nitrogen remobilization efficiency and grain filling (Chen et al., 2019b). These results indicate that increased autophagy improves plant productivity.
In flowering plants, the tapetum provides nutrients and lipids to developing microspores and pollen grains where autophagy is proposed to play an important role. Autophagosome structures and vacuole-enclosed lipid bodies have been observed in post-meiotic tapetum cells during pollen development. These structures disappeared in the autophagy defective mutant Osatg7. Moreover, the mutant fails to accumulate lipid and starch in pollen grains and shows complete male sterility, indicating that autophagy is induced in tapetum cells and is essential for another development in rice (Kurusu et al., 2014). Using fluorescent protein-tagged AtATG8 as a marker, it has been shown that the number of autophagosomes increases rapidly at the uninucleate stages in the tapetal cells during anther development (Hanamata et al., 2019). In the anthers of the Osatg7 mutant, the endogenous levels of active-forms of gibberellins (GAs) and cytokinin, trans-zeatin, are significantly lower than that in wild type, indicating that autophagy may regulate phytohormone metabolism during rice anther development (Kurusu et al., 2017).
During seed development in Arabidopsis, almost all ATG genes are upregulated in siliques (Di Berardino et al., 2018). ATG8f is strongly expressed in the phloem companion cells of pericarps and in the embryo. Moreover, GFP-ATG8 labelled autophagosomes have been observed in seed embryos (Di Berardino et al., 2018). In maize, many ATG genes are induced in the endosperm, and the ATG8-PE adducts accumulate in the maize endosperm beginning 18 days after pollination (DAP) with continuing increases until 30 DAP, indicating that autophagy is involved in endosperm development (Chung et al., 2009). The rice Osatg7-1 mutant also produces smaller seeds with a chalky appearance and lower starch content. During seed maturation of Osatg7-1, the starch degradation pathways are activated abnormally in the endosperm, indicating that autophagy plays critical roles in metabolic regulation in the endosperm (Sera et al., 2019).
Additionally, autophagy can regulate the nutrient supply during seed development (Chen et al., 2019a). In atg5-1, iron translocation from vegetative organs to seeds is severely decreased during seed formation. It was also shown that the translocations of zinc and manganese to the seeds are also dependent on autophagy (Pottier et al., 2019). In Arabidopsis atg mutants, the amount of 12S globulins in seeds is decreased, but the amount of 12S globulin precursors are increased, suggesting that autophagy is involved in delivering the precursors to the protein storage vacuoles (PSVs), where the precursors are processed into their mature forms (Di Berardino et al., 2018).
Lipids in membranous organelles are important substrates for energy production and as cellular structural materials. Before being used directly for β-oxidation, fatty acids are first stored in lipid droplets (LDs) in the form of triacylglycerol (TAG) (Fan et al., 2014; Fan et al., 2017). The LDs are hydrolyzed by lipolysis to supply the cell with fatty acids. In mammalian cells, the degradation of LDs is carried out through a selective autophagy pathway, termed lipophagy (Wang, 2016; Zechner et al., 2017). In comparison with yeast and mammals, the mechanisms of lipophagy have been less well characterized in plants (van Zutphen et al., 2014; Jaishy and Abel, 2016).
During pollen maturation, LDs containing TAGs in the tapetum are necessary as a supply of lipid components (Kurusu et al., 2014). In tapetum cells of rice, LDs enclosed in vacuoles have been detected, and LD-like structures were more abundant in the cytoplasm of Osatg7 and Osatg9 mutants than that of wild type, indicating that LDs in plant may also be degraded through lipophagy (Kurusu et al., 2014). Furthermore, lipidomic analysis has shown impairment of phosphatidylcholine (PC) editing and lipid desaturation in the anthers of these mutants during pollen maturation, indicating the involvement of autophagy in the regulation of lipid metabolism during plant development (Hanamata et al., 2014; Kurusu et al., 2014). In Arabidopsis, the autophagy of cellular organelles can provide a source of fatty acids for TAG synthesis under normal and starvation conditions, indicating that autophagy can promote TAG synthesis. Under normal conditions, TAGs stored in LDs are hydrolyzed by SDP1 (sugar-dependent 1), a lipase responsible for the initiation of TAG catabolism (Eastmond, 2006). But under nutrient starvation, lipophagy is induced to degrade the LDs for energy production (Fan et al., 2019). In the atg5 mutant, peroxisomal and ER proteins involved in very long chain fatty acid synthesis and β-oxidation are up-regulated, and the concentrations of sphingolipids, phospholipids and galactolipids are changed, indicating that lipid metabolism is severely affected in autophagy mutants and that autophagy can play a role in the control of plant lipid metabolism in addition to regulating TAG synthesis and LD degradation (Havé et al., 2019).
The regulation of primary and secondary root development is important for the establishment of root systems. It has been recently demonstrated that autophagy is involved in the regulation of plant root development. Under phosphate starvation, phosphorylation and activation of PUB9 (U Box/Armadillo Repeat-Containing E3 ligase) by ARK2 (S-Domain receptor Kinase) is responsible for ubiquitination of either AUX/IAA (auxin/indole-3-acetic acid) proteins or other repressors of auxin accumulation. These auxin repressors are subsequently selectively targeted to the autophagosome for degradation, releasing ARFs (auxin response factors) or other signals to promote auxin accumulation and lateral root development. The inhibition of autophagy by 3-MA leads to disruption of both lateral root growth and auxin accumulation in the roots under phosphate starvation, suggesting autophagy plays an important role in root growth (Deb et al., 2014; Sankaranarayanan and Samuel, 2015). In untreated Arabidopsis, overexpression of AtATG8f results in plants with fewer lateral roots than control plants, despite similar primary root lengths. The application of zeatin results in reduced length of primary roots and reduced number of lateral roots in both plants. However, the extent of inhibition was remarkable in length of the primary roots but similar in number of the lateral roots in overexpression of AtATG8f plants. Together these data indicate that ATG8 affects the cytokinin-mediated regulation of root architecture (Slavikova et al., 2008).
Glucose is a key nutrient signal regulating root meristem activity in plants. It has recently been found that autophagy is involved in glucose-regulated root meristem maintenance (Pacifici et al., 2015; Huang et al., 2019a). Under high glucose conditions, accumulation of ROS oxidizes active IAA and impairs root meristem activity and growth. However, in atg mutants, the transmission of the high-glucose signal to the peroxisomes is disrupted, alleviating ROS-oxidized IAA, causing increased root growth (Huang et al., 2019a). These data demonstrate that constitutive autophagy regulates the production of ROS and IAA by the peroxisome to modulate root meristem activity. Autophagy was also found to play a key role in the hydrotropic curvature of A. thaliana roots. Autophagosomes accumulate in the root curvature of wild type seedlings after being transferred to a water potential gradient stress system called Normal Medium-Water Stress Medium (NM-WSM). In contrast, atg2, atg5, atg8b, atg8i, and atg9 mutants do not show hydrotropic curvature in the NM-WSM system. During the hydrotropic response, H2O2 also accumulates in the root curvature at a similar rate as the autophagosomes (Jiménez-Nopala et al., 2018), suggesting that oxidative stress, and specifically H2O2, induced during the hydrotropic response in NM-WSM regulates autophagy.
In Populus trichocarpa, ATG genes and the ATG8 protein are expressed at different stages during root or stem primary and secondary development. Ultrastructural observations have revealed that autophagosomes accumulate during differentiation of xylem in roots (Wojciechowska et al., 2019). These results exhibit autophagy is involved in root morphogenesis.
The involvement of autophagy during senescence has been supported by the observation that many ATG genes are upregulated in older leaves. In Arabidopsis, 15 ATG genes are upregulated during senescence (Breeze et al., 2011). In maize, ATG8 lipidation, which is a marker for higher levels of autophagy activity, is highly accumulated in the yellowing area of senescent leaves (Chung et al., 2009). ATG11, which links the ATG1/ATG13 complex to autophagic membranes, also plays a critical role in senescence-induced mitophagy in Arabidopsis. The senescence-induced breakdown of mitochondria-resident proteins and mitochondrial vesicles occurs via an autophagic process requiring ATG11 and other ATG components (Li et al., 2014). In atg mutants, defective autophagy results in early senescence and PCD irrespective of nutrient conditions. The salicylic acid (SA) signaling pathways are required for this senescence/PCD phenotype (Yoshimoto et al., 2009). Application of a SA agonist cannot induce the senescence/PCD phenotype in atg npr1 mutants, indicating that induction of this phenotype in atg mutants requires the SA signal transducer NPR1 (nonexpressor of pathogenesis-related genes 1). Autophagy is induced by a SA agonist via NPR1, thus indicating that autophagy can operate as a negative feedback regulator of SA signaling to limit senescence and PCD (Yoshimoto et al., 2009). In conclusion, autophagy can be induced at different developmental stages to perform different physiological functions in plants.
As immovable organisms, plants need to cope with various environmental changes, including nutrient starvation, salt, drought, and heat stresses (Han et al., 2014; Shen et al., 2014; Qi et al., 2018; Hilker and Schmülling, 2019). To respond appropriately to these stresses, plants have evolved various distinct signaling and regulatory mechanisms (Hilker and Schmülling, 2019). It has been shown that autophagy is involved in removing damaged proteins and cellular components produced in response to environmental stresses (Wang et al., 2018b). Furthermore, Autophagy is induced by various abiotic stresses and autophagy-defective mutants are hypersensitive to these stress conditions (Wang et al., 2018b). For example, macroautophagy-defective RNAi-AtATG18a plants are more sensitive than wild type plants to methyl viologen, a ROS inducer, and accumulate more oxidized proteins (Xiong et al., 2007). In this section, we review the roles of autophagy in plant stress response and the underlying mechanisms.
In response to starvation, autophagy is upregulated to facilitate the recycling of cellular material and remobilization of nutrients. The evidence that autophagy is involved in nitrogen remobilization from leaves into seeds comes from research using 15NO3−. It was shown that 15N remobilization is decreased significantly in ATG18a RNAi, atg5, and atg9 mutants compared with wild type plants under nitrogen starvation (Guiboileau et al., 2012). Moreover, more ammonium, amino acids, and proteins accumulate in the rosette leaves of these atg mutants than in wild type (Guiboileau et al., 2013; Chen et al., 2019b). In maize, the growth rate and seed yield of the atg12 mutant is decreased significantly in low nitrogen conditions but not in sufficient nitrogen conditions. However, even under nitrogen rich conditions, the seed yield and 15N reallocation into the seeds are much lower in atg12 than in wild type (Li et al., 2015a). Recently, a study illustrated that the overexpression of ATG18a in apple results in increased tolerance to nitrogen starvation due to increased autophagy under these conditions (Sun et al., 2018). Additionally, under carbon starvation, the Arabidopsis atg5 and atg7 mutants exhibit delayed growth, reduced amino acid levels, increased respiration, and decreased flux to net protein synthesis. These results highlight the importance of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings (Avin-Wittenberg et al., 2015).
Heat stress is one of the most dangerous threats for plant growth, causing the misfolding and denaturing of normal proteins, which can be degraded through autophagy (Zhang et al., 2010; Fu et al., 2015). It has been recently shown that the NBR1-mediated selective autophagy pathway is involved in the degradation of denatured or otherwise damaged proteins caused by heat stress (Zhou et al., 2013). The nbr1 mutant shows reduced heat tolerance and increased accumulation of insoluble, detergent-resistant and highly ubiquitinated proteins under heat stress, implying that the autophagy adaptor NBR1 targets the ubiquitinated protein aggregates for degradation under stress conditions (Zhou et al., 2013). As further evidence that autophagy is involved in the heat stress response, the expression of ATG genes and the accumulation of autophagosomes are induced by heat stress (Wang et al., 2015). Moreover, it was shown that ATG8 coimmunoprecipitates with different classes of heat shock proteins (HSP90s, HSP101, and small HSP17.6) in plants. Defective autophagy causes accumulation of these HSPs, suggesting that autophagy promotes the degradation of HSPs and unfolded proteins (Sedaghatmehr et al., 2019). Additionally, in Arabidopsis, high-temperature stress (30°C) promotes autophagy in both anther wall cells and microspores in developing anthers of plants, while atg mutants (atg2-1, atg5-1, atg7-2, and atg10-1) have visibly impaired pollen development and anther dehiscence, suggesting that autophagy functions in tapetum degeneration and pollen development during high-temperature stress (Dündar et al., 2019).
Drought and salt are two common environmental stresses that plants may encounter (Golldack et al., 2014; Zheng et al., 2017; Zhang et al., 2018). The involvement of autophagy in response to drought stress was first elucidated in Arabidopsis, indicated by the upregulation of ATG18a during drought conditions (Liu et al., 2009). In Arabidopsis, the atg5, atg7, and ATG18a RNAi mutants display hypersensitization to drought stress (Zhou et al., 2013). On the other hand, overexpression of ATG18a in apple results in higher autophagy activity and increased drought tolerance, indicating the key role for autophagy in responses to drought (Sun et al., 2018). Recently, Medicago truncatula dehydrin MtCAS31 (cold acclimation-specific 31) was found to interact with both ATG8a and a negative drought stress regulator MtPIP2;7 to improve drought tolerance by facilitating autophagic degradation of MtPIP2;7, demonstrating that MtCAS31 functions as a positive regulator of drought-induced autophagy (Li et al., 2019b). Consistent with this idea, the Arabidopsis atg2 and atg7 mutants contain more oxidized proteins and are hypersensitive to both salt and osmotic stresses (Luo et al., 2017). Under salt stress, autophagy is required for Na+ sequestration in the central vacuole in root cortex cells (Luo et al., 2017). Interestingly, autophagy induction by nutrient starvation and salt stress, but not osmotic stress, can be blocked by NADPH oxidase inhibitors (Liu et al., 2009). These results suggest that autophagy is also involved in the oxidative stress response and can be induced by NADPH oxidase-dependent or -independent pathways.
TOR kinase is a major switch in controlling autophagy, and the phenotypes of TOR knockout and knockdown mutant lines indicate that TOR plays important roles in nutrient stress responses in plants (Menand et al., 2002; Liu and Bassham, 2010). Recently, it was shown that ATG13 in plants can be phosphorylated directly by TOR (Van Leene et al., 2019), suggesting that autophagy may be involved in stress response in plants through similar regulatory mechanisms as in yeast and mammals. TOR activity is also regulated by SnRK1, which is repressed by sugars and activated under energy-deficient conditions, such as darkness or under biotic and abiotic stresses (Baena-González et al., 2007). Thus, during responses to environmental stresses, TOR activity is regulated by SnRKs, subsequently, regulating the autophagy process. Interestingly, under prolonged fixed-carbon starvation, the active KIN10 subunit of SnRK1 phosphorylates ATG6 to activate the PI3K complex, which bypasses the requirement of the ATG1 kinase complex for autophagy initiation (Huang et al., 2019a; Huang et al., 2019b) (Figure 2).
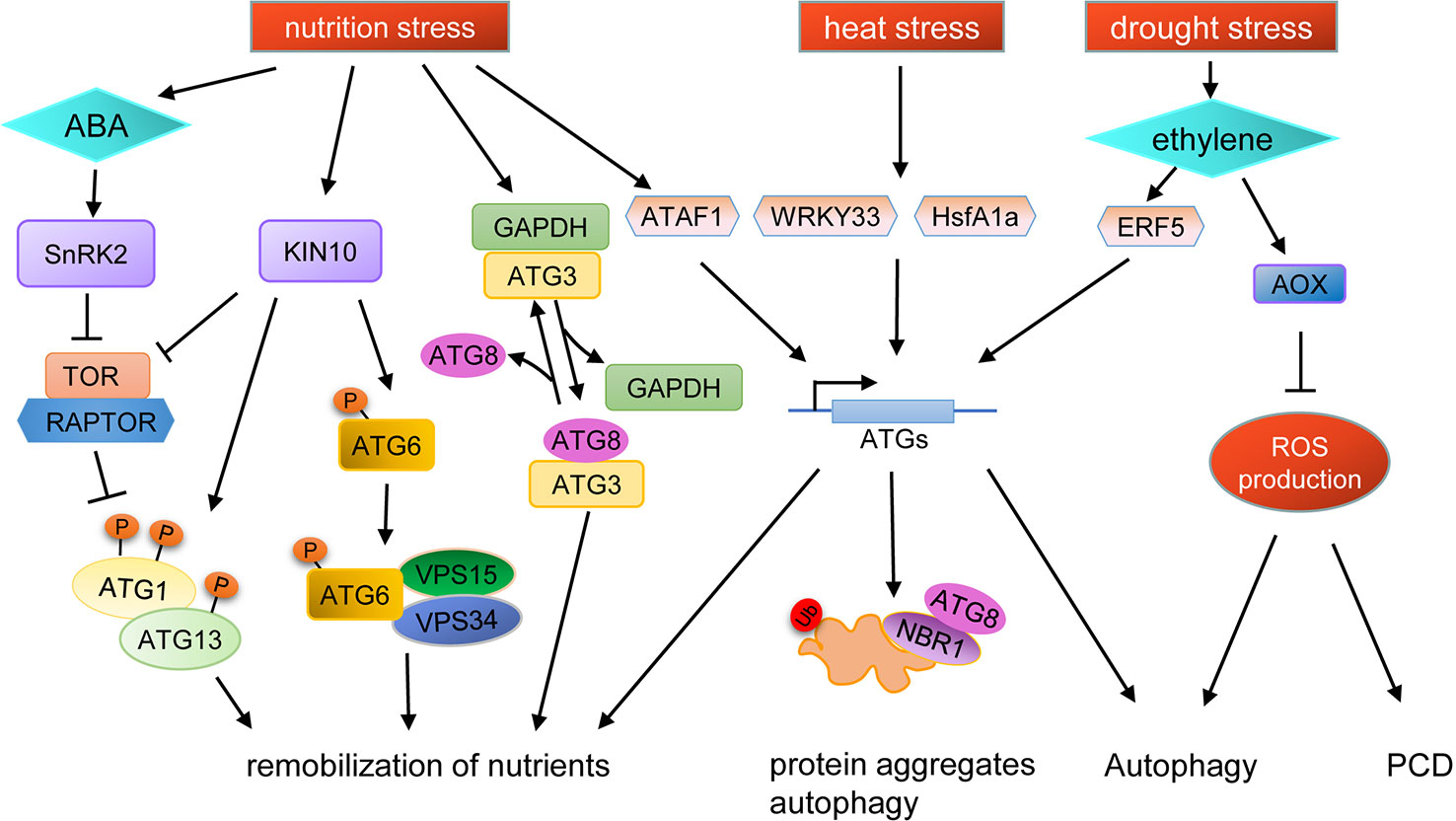
Figure 2 Schematic diagram of autophagy regulation under various stresses. KIN10, a catalytic subunit of SnRK1, is activated by stresses to phosphorylate TOR-interacting proteins, in turn inhibiting TOR kinase activity and activating ATG1. Additionally, active KIN10 can phosphorylate ATG6 and activate the PI3K complex to induce autophagy by an ATG1 kinase independent pathway. Under nutrient starvation stress, SnRK2 is activated by ABA to inhibit the activity of the TOR complex, and the expression of ATG genes are induced by the transcription factor ATAF1, resulting in the promotion of autophagy. Nutrient starvation stress can also cause the dissociation of GAPDH and ATG3, thus promoting the binding of ATG8 and ATG3. The expression of ATG genes are induced by WRKY33 and HsfA1a under heat stress to promote the degradation of protein aggregates through autophagy. Autophagy can also be induced by drought stress in an ethylene-dependent manner. Moreover, autophagy is also regulated by ROS levels during drought stress, in which AOX plays an important role.
Another regulatory pathway of autophagy in plants is mediated by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Masclaux-Daubresse et al., 2017). In tobacco, cytosolic GAPDHs (GAPCs) interact with ATG3, resulting in the inhibition of the ATG3-ATG8 interaction, thereby preventing ATG3-dependent ATG8-PE conjugation (Han et al., 2015). In Arabidopsis, the gapc1 mutant shows enhanced autophagy compared to wild type (Henry et al., 2015).
Autophagy is also controlled at the transcriptional level (Wang et al., 2019a). In yeast and mammals, Rph1/KDM4, a histone demethylase protein, is a repressor of the transcription of several ATG genes and thus of autophagy induction. Under nitrogen starvation conditions, Rph1/KDM4 can be phosphorylated by Rim15 to relieve the inhibition of ATG gene expression (Bernard et al., 2015). In Arabidopsis, under carbon starvation, a NAC transcription factor ATAF1 (Arabidopsis transcription activation factor 1) induces ATG gene expression to promote the carbon starvation response (Garapati et al., 2015). Under heat stress, downregulation of WRKY33 expression in tomato plants causes an increase in sensitivity to heat stress and reduces the expression of several ATG genes, suggesting that WRKY33 participates in the heat stress response through transcriptional regulation of autophagy (Zhou et al., 2014). Moreover, in tomato, the expression of ATG10 and ATG18f are regulated by HsfA1a (heat shock transcription factor A1a) to activate autophagy under drought stress (Wang et al., 2015).
Many kinds of abiotic stresses can induce ROS production, which can act as a signal molecule for stress response. ROS is produced in many compartments of plant cells, such as the plasma membrane, mitochondria, chloroplasts, and peroxisomes (Mhamdi et al., 2012). Recent studies have shown that ROS acts as a link between autophagy and abiotic stress (Figure 2). For example, plasma membrane-associated NADPH oxidase, an important source of ROS, is necessary for plant tolerance to submergence and autophagy activation (Chen et al., 2015). In Solanum lycopersicum, mitochondrial alternative oxidase (AOX) may regulate autophagy by controlling ROS production during drought stress (Zhu et al., 2018). ROS production can be limited by AOX, and overexpression of AOX results in increased drought tolerance in tomato, concomitant with increased autophagy activity during drought (Selinski et al., 2018; Zhu et al., 2018). In AOX1a-RNAi plants, ethylene-induced drought tolerance is disrupted and the level of autophagy is decreased, accompanied by higher levels of H2O2, which results in rapid programmed cell death (PCD). Pharmacological scavenging of H2O2 in the aox19 mutant induces autophagosome accumulation under drought stress, indicating that AOX-dependent ROS signaling is critical in triggering autophagy (Zhu et al., 2018). In summary, AOX can reduce the ROS burst caused by drought and subsequently, the low ROS levels promote autophagy to aid drought tolerance in tomato (Zhu et al., 2018).
ROS can modulate autophagy by causing damage to cellular components (Signorelli et al., 2019). Upon treating Arabidopsis roots with exogenous H2O2, autophagy is induced to degrade oxidized proteins (Xiong et al., 2007). In Chlamydomonas, a deficiency in carotenoid synthesis triggers autophagy in the light, but not in the dark, indicating that ROS produced in the chloroplastic electron chain are able to induce autophagy (Pérez-Pérez et al., 2012). Redox status can also modulate autophagy by regulating ATG4 activity. In Arabidopsis, cleavage of ATG8f by ATG4 is dramatically decreased with increasing concentrations of H2O2, but ATG4 activity can be restored with the addition of the reducing agent DTT (dithiothreitol) (Woo et al., 2014). Similarly, in Chlamydomonas reinhardtii, DTT treatment promotes monomeric and active ATG4, whereas NF (norflurazon) treatment results in oxidation and aggregation of ATG4, rendering it inactive. In the C. reinhardtii carotenoid-less mutant lts1-204 that lacks phytoene synthase, a shift from dark to light conditions leads to increased ROS levels and induction of autophagy. Moreover, in the lts1-204 mutant, the abundance of oligomeric ATG4 is significantly increased, concomitant with accumulation of lipidated ATG8. Together, these data indicate that redox status regulates autophagy through the oxidation and inactivation of ATG4 under stress conditions (Pérez-Pérez et al., 2016).
Autophagy in turn contributes to the antioxidant system through degradation of oxidative damaged organelles, such as mitochondria, chloroplasts, and peroxisomes (Signorelli et al., 2019). Consistent with the role of autophagy in oxidative stress, atg2 and atg5 mutants exhibit enhanced accumulation of H2O2 (Yoshimoto et al., 2009). Additionally, silencing of ATG18a in Arabidopsis results in increased oxidative damage and subsequently, hypersensitivity to oxidative stress (Yoshimoto et al., 2009; Minibayeva et al., 2012). In autophagy-deficient seedlings, inactive catalases accumulate in clustered peroxisomes, implying that pexophagy actively clears damaged peroxisomes (Shibata et al., 2013). ROS are key signaling molecules for promoting stomatal closure in response to diverse environmental stresses. The disruption of other ATGs, such as ATG2, ATG5, ATG7, ATG10 or ATG12, promotes the aggregation of peroxisomes in guard cells. These mutants exhibit reduced activity of the ROS scavenger catalase, resulting in impairment of stomatal opening in response to light and low CO2 concentrations (Yamauchi et al., 2019). In summary, autophagy and ROS production are two important cellular physiological processes that work cooperatively during the plant’s response to abiotic stresses, enabling plants to cope with various environmental challenges.
Phytohormones play important roles in the regulation of plant development and environmental stress response. The atg mutants defective in autophagy have been used to uncover the connection between the phytohormone pathway and autophagy (Signorelli et al., 2019) (Figure 2). Arabidopsis TSPO (Tryptophan-rich Sensory Protein) can respond to osmotic and salt stresses and is induced by abscisic acid (ABA) to directly bind to ATG8 in an AIM-dependent manner. In wild-type, ABA-induced TSPO is degraded quickly, while in the atg5 mutant, degradation of ABA-induced TSPO is inhibited. Moreover, ABA-induced TSPO is stabilized in the presence of autophagic inhibitors, such as 3-MA, and the vacuolar Cys protease inhibitor E64d. Thus, these data indicate that autophagy is involved in the ABA-mediated stress response (Guillaumot et al., 2009; Vanhee et al., 2011). In addition, ATG8-interacting protein 1 and 2 (ATI1 and ATI2) are involved in ABA-mediated germination (Avin-Wittenberg et al., 2012; Honig et al., 2012). Under unstressed conditions, TOR kinase phosphorylates PYL ABA receptors, disrupting PYL association with ABA and PP2C phosphatase effectors, leading to inactivation of SnRK2 kinases (Wang et al., 2018b). Under stress conditions, SnRK2 is activated by ABA to phosphorylate RAPTOR, a component of the TOR complex, resulting in the dissociation and inhibition of the TOR complex (Wang et al., 2018b; Salem et al., 2018). Therefore, as an endogenous messenger during abiotic stresses, ABA is capable of inducing autophagy in multiple ways.
In addition to ABA, the abundance of Brassinazole-resistant 1 (BZR1) is controlled by TOR signaling to maintain growth under carbon starvation conditions in Arabidopsis, suggesting crosstalk between autophagy and BR signaling (Zhang et al., 2016). ATG gene expression and autophagosome formation are decreased in the bzr1 mutant but are enhanced in BZR1 overexpressing plants after BR treatment. In the atg2 and atg6 mutants, initiation of BR-induced autophagy is disrupted. Additionally, silencing of BZR1 decreases resistance to nitrogen starvation in tomato (Wang et al., 2019b). As previously described above, BES1 is degraded by a selective autophagy pathway mediated by DSK2 under drought and starvation stresses (Nolan et al., 2017), indicating that BR signaling is also involved in autophagy regulation during stress response.
Ethylene is also a stress response hormone. In atg5 and atg9 mutants, a subset of ethylene signaling genes are induced, such as ETR2 (ethylene response 2) and CTR1 (constitutive triple response 1), suggesting that ethylene is overproduced in the atg mutants (Masclaux-Daubresse et al., 2014). Moreover, under drought stress, ERF5 (ethylene response factor 5) binds to the tomato promoters of ATG8d and ATG18h to induce autophagy (Zhu et al., 2018). These results suggest that ethylene signaling may also be involved in the regulation of autophagy induced by stresses, although the underlying mechanism requires further investigation.
Autophagy plays an important role in recycling cytosolic material and maintaining cellular homeostasis during growth, development and response to diverse stresses. In recent years, considerable efforts have focused on revealing the mechanisms of autophagy, with great achievements being made in recent decades. Many of the protein components and molecular mechanisms involved in autophagy have been identified and key regulatory factors have also been discovered, such as the TOR complex (Liu and Bassham, 2010) and SnRK1 (Soto-Burgos and Bassham, 2017). Studies on the roles of autophagy in plant stress tolerance have enabled us to understand the positive significance of autophagy on plants.
However, most studies have only described the phenomenon that autophagy is involved in plant development and stress response, leading to several important unanswered questions about the underlying molecular mechanisms that remain to be further investigated. For example, how do cells choose the right autophagy pathways at the proper time when adjusting their life state and how are these autophagy pathways related to each other? During plant development, different tissues undergo different physiological processes, and selective autophagy must cooperate with other regulatory pathways to promote normal development. Therefore, the appropriate switch of autophagy and its coordination with other pathways are crucial processes that require further investigation. Additionally, many of the receptors for selective autophagy are unknown. Thus, it is of great importance to identify and characterize the adaptors/receptors or other new components that mediate the various selective autophagy pathways. Lastly, how the autophagy machinery is activated in response to biotic and abiotic stresses of plants, and how it works cooperatively with other regulatory pathways in response to stress still remain to be elucidated. The study of these scientific issues will enable us to better understand the important physiological process of autophagy.
TS, XL, PW and CM designed the concept and wrote the manuscript. MY, YZ and QS contributed to revision of the manuscript. All authors read and approved the submitted version.
This study was funded by the National Natural Science Foundation of China (31670073, 31600204, 31770290 and 31970301), the Key Technology Research and Development Program of Shandong (2018GSF121037 and 2018GNC113010), and the China Postdoctoral Science Foundation (2017M612333).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Anand, P., Kwak, Y., Simha, R., Donaldson, R. P. (2009). Hydrogen peroxide induced oxidation of peroxisomal malate synthase and catalase. Arch. Biochem. Biophys. 491, 25–31. doi: 10.1016/j.abb.2009.09.019
Anderson, G. H., Veit, B., Hanson, M. R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3, 12. doi: 10.1186/1741-7007-3-12
Avin-Wittenberg, T., Michaeli, S., Honig, A., Galili, G. (2012). ATI1, a newly identified atg8-interacting protein, binds two different Atg8 homologs. Plant Signal. Behav. 7, 685–687. doi: 10.4161/psb.20030
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Bock, R., et al. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cel. 27, 306–322. doi: 10.1105/tpc.114.134205
Baena-González, E., Rolland, F., Thevelein, J. M., Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Balchin, D., Hayer-Hartl, M., Hartl, F. U. (2016). In vivo aspects of protein folding and quality control. Science 353, aac4354. doi: 10.1126/science.aac4354
Bartel, B., Farmer, L. M., Rinaldi, M. A., Young, P. G., Danan, C. H., Burkhart, S. E. (2014). Mutation of the Arabidopsis LON2 peroxisomal protease enhances pexophagy. Autophagy 10, 518–519. doi: 10.4161/auto.27565
Bernard, A., Jin, M., González-Rodríguez, P., Füllgrabe, J., Delorme-Axford, E., Backues, S.K., et al. (2015). Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr. Biol. 25, 546–555. doi: 10.1016/j.cub.2014.12.049
Breeze, E., Harrison, E., McHattie, S., Hughes, L., Hickman, R., Hill, C., et al. (2011). High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23, 873–894. doi: 10.1105/tpc.111.083345
Cebollero, E., Reggiori, F. (2009). Regulation of autophagy in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1793, 1413–1421. doi: 10.1016/j.bbamcr.2009.01.008
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W. J., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Chen, L., Su, Z. Z., Huang, L., Xia, F. N., Qi, H., Xie, L. J., et al. (2017). The AMP-activated protein kinase KIN10 is involved in the regulation of autophagy in Arabidopsis. Front. Plant Sci. 8, 1201. doi: 10.3389/fpls.2017.01201
Chen, Q., Shinozaki, D., Luo, J., Pottier, M., Havé, M., Marmagne, A., et al. (2019a). Autophagy and nutrients management in plants. Cells 8, pii: E1426. doi: 10.3390/cells8111426
Chen, Q., Soulay, F., Saudemont, B., Elmayan, T., Marmagne, A., Masclaux-Daubresse, C. L. (2019b). Overexpression of ATG8 in Arabidopsis stimulates autophagic activity and increases nitrogen remobilization efficiency and grain filling. Plant Cell Physiol. 60, 343–352. doi: 10.1093/pcp/pcy214
Chung, T., Suttangkakul, A., Vierstra, R. D. (2009). The ATG autophagic conjugation system in maize: ATG transcripts and abundance of the ATG8-lipid adduct are regulated by development and nutrient availability. Plant Physiol. 149, 220–234. doi: 10.1104/pp.108.126714
Chung, T., Phillips, A. R., Vierstra, R. D. (2010). ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12a and ATG12b loci. Plant J. 62, 483–493. doi: 10.1111/j.1365-313X.2010.04166.x
Chung, T. (2019). How phosphoinositides shape autophagy in plant cells. Plant Sci. 281, 146–158. doi: 10.1016/j.plantsci.2019.01.017
Díaz-Troya, S., Pérez-Pérez, M. E., Florencio, F. J., Crespo, J. L. (2008). The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy 4, 851–865. doi: 10.4161/auto.6555
Dündar, G., Shao, Z., Higashitani, N., Kikuta, M., Izumi, M., Higashitani, A. (2019). Autophagy mitigates high-temperature injury in pollen development of Arabidopsis thaliana. Dev. Biol. 456, 190–200. doi: 10.1016/j.ydbio.2019.08.018
de Duve, C., Pressman, B. C., Gianetto, R., Wattiaux, R., Appelmans, F. (1955). Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem. J. 60, 604–617. doi: 10.1042/bj0600604
Deb, S., Sankaranarayanan, S., Wewala, G., Widdup, E., Samuel, M. A. (2014). The S-Domain receptor kinase Arabidopsis receptor kinase2 and the U box/armadillo repeat-containing E3 ubiquitin ligase9 module mediates lateral root development under phosphate starvation in Arabidopsis. Plant Physiol. 165, 1647–1656. doi: 10.1104/pp.114.244376
Deprost, D., Truong, H. N., Robaglia, C., Meyer, C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326, 844–850. doi: 10.1016/j.bbrc.2004.11.117
Di Berardino, J., Marmagne, A., Berger, A., Yoshimoto, K., Cueff, G., Chardon, F., et al. (2018). Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 69, 1403–1414. doi: 10.1093/jxb/ery012
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Eastmond, P. J. (2006). SUGAR-DEPENDENT1 encodes a patatin domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18, 665–675. doi: 10.1105/tpc.105.040543
Fan, J., Yan, C., Roston, R., Shanklin, J., Xu, C. (2014). Arabidopsis lipins, PDAT1 acyltransferase, and SDP1 triacylglycerol lipase synergistically direct fatty acids toward β-oxidation, thereby maintaining membrane lipid homeostasis. Plant Cell 26, 4119–4134. doi: 10.1105/tpc.114.130377
Fan, J., Yu, L., Xu, C. (2017). A central role for triacylglycerol in membrane lipid breakdown, fatty acid β-oxidation, and plant survival under extended darkness. Plant Physiol. 174, 1517–1530. doi: 10.1104/pp.17.00653
Fan, J., Yu, L., Xu, C. (2019). Dual role for autophagy in lipid metabolism in Arabidopsis. Plant Cell 31, 1598–1613. doi: 10.1105/tpc.19.00170
Farré, J. C., Subramani, S. (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat. Rev. Mol. Cell Biol. 17, 537–552. doi: 10.1038/nrm.2016.74
Farré, J. C., Burkenroad, A., Burnett, S. F., Subramani, S. (2013). Phosphorylation of mitophagy and pexophagy receptors co-ordinates their interaction with Atg8 and Atg11. EMBO Rep. 14, 441–449. doi: 10.1038/embor.2013.40
Floyd, B. E., Morriss, S. C., MacIntosh, G. C., Bassham, D. C. (2015). Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11, 2199–2212. doi: 10.1080/15548627.2015.1106664
Fu, C., Zhang, J. X., Liu, X. X., Yang, W. W., Yu, H. B., Liu, J. (2015). AtFes1A is essential for highly efficient molecular chaperone function in Arabidopsis. J Plant Biol. 58, 366–373. doi: 10.1007/s12374-015-0181-y
Fujiki, Y., Yoshimoto, K., Ohsumi, Y. (2007). An Arabidopsis homolog of yeast ATG6 is essential for pollen germination. Plant Physiol. 143, 1132–1139. doi: 10.1104/pp.106.093864
Gao, C., Zhuang, X., Cui, Y., Fu, X., He, Y., Zhao, Q., et al. (2015). Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. U. S. A. 112, 1886–1891. doi: 10.1073/pnas.1421271112
Garapati, P., Feil, R., Lunn, J. E., Van Dijck, P., Balazadeh, S., MuellerRoeber, B. (2015). Transcription factor Arabidopsis activating factor1 integrates carbon starvation responses with trehalose metabolism. Plant Physiol. 69, 379–390. doi: 10.1104/pp.15.00917
Golldack, D., Li, C., Mohan, H., Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5, 151. doi: 10.3389/fpls.2014.00151
Guiboileau, A., Yoshimoto, K., Soulay, F., Bataillé, M. P., Avice, J. C., Masclaux-Daubresse, C. (2012). Autophagy machinery controls nitrogen remobilization at the whole-plant level under both limiting and ample nitrate conditions in Arabidopsis. New Phytol. 194, 732–740. doi: 10.1111/j.1469-8137.2012.04084.x
Guiboileau, A., Avila-Ospina, L., Yoshimoto, K., Soulay, F., Azzopardi, M., Marmagne, A., et al. (2013). Physiological and metabolic consequences of autophagy defisciency for the management of nitrogen and protein resources in Arabidopsis leaves depending on nitrate availability. New Phytol. 199, 683–694. doi: 10.1111/nph.12307
Guillaumot, D., Guillon, S., Morsomme, P., Batoko, H. (2009). ABA, porphyrins and plant TSPO-related protein. Plant Signal. Behav. 4, 1087–1090. doi: 10.4161/psb.4.11.9796
Hamasaki, M., Furuta, N., Matsuda, A., Nezu, A., Yamamoto, A., Fujita, N., et al. (2013). Autophagosomes form at ER-mitochondria contact sites. Nature 495, 389–393. doi: 10.1038/nature11910
Han, G., Wang, M., Yuan, F., Sui, N., Song, J., Wang, B. (2014). The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 86, 237–253. doi: 10.1007/s11103-014-0226-5
Han, S., Wang, Y., Zheng, X., Jia, Q., Zhao, J., Bai, F., et al. (2015). Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in Nicotiana benthamiana. Plant Cell 27, 1316–1331. doi: 10.1105/tpc.114.134692
Hanada, T., Noda, N. N., Satomi, Y., Ichimura, Y., Fujioka, Y., Takao., T., et al. (2007). The ATG12-ATG5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302. doi: 10.1074/jbc.C700195200
Hanamata, S., Kurusu, T., Kuchitsu, K. (2014). Roles of autophagy in male reproductive development in plants. Front. Plant Sci. 5, 457. doi: 10.3389/fpls.2014.00457
Hanamata, S., Sawada, J., Toh, B., Ono, S., Ogawa, K., Fukunaga, T., et al. (2019). Monitoring autophagy in rice tapetal cells during pollen maturation. Plant Biotechnol. 36, 99–105. doi: 10.5511/plantbiotechnology.19.0417a
Hanaoka, H., Noda, T., Shirano, Y., Kato, T., Hayashi, H., Shibata, D., et al. (2002). Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiol. 129, 1181–1193. doi: 10.1104/pp.011024
Hara-Nishimura, I., Hatsugai, N. (2011). The role of vacuole in plant cell death. Cell Death Differ. 18, 1298–1304. doi: 10.1038/cdd.2011.70
Havé, M., Luo, J., Tellier, F., Balliau, T., Cueff, G., Chardon, F. (2019). Proteomic and lipidomic analyses of the Arabidopsis atg5 autophagy mutant reveal major changes in endoplasmic reticulum and peroxisome metabolisms and in lipid composition. New Phytol. 223, 1461–1477. doi: 10.1111/nph.15913
He, C., Klionsky, D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. doi: 10.1146/annurev-genet-102808-114910
Henry, E., Fung, N., Liu, J., Drakakaki, G., Coaker, G. (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PloS Genet. 11, e1005199. doi: 10.1371/journal.pgen.1005199
Hilker, M., Schmülling, T. (2019). Stress priming, memory, and signalling in plants. Plant Cell Environ. 42, 753–761. doi: 10.1111/pce.13526
Honig, A., Avin-Wittenberg, T., Ufaz, S., Galili, G. (2012). A new type of compartment, defined by plant-specific Atg8-interacting proteins, is induced upon exposure of Arabidopsis plants to carbon starvation. Plant Cell 24, 288–303. doi: 10.1105/tpc.111.093112
Hu, J., Baker, A., Bartel, B., Linka, N., Mullen, R. T., Reumann, S., et al. (2012). Plant peroxisomes: biogenesis and function. Plant Cell 24, 2279–2303. doi: 10.1105/tpc.112.096586
Huang, L., Yu, L. J., Zhang, X., Fan, B., Wang, F. Z., Dai, Y. S., et al. (2019a). Autophagy regulates glucose-mediated root meristem activity by modulating ROS production in Arabidopsis. Autophagy 15, 407–422. doi: 10.1080/15548627.2018.1520547
Huang, X., Zheng, C., Liu, F., Yang, C., Zheng, P., Lu, X., et al. (2019b). Genetic analyses of the Arabidopsis ATG1 kinase complex reveal both kinase-dependent and independent autophagic routes during fixed-carbon starvation. Plant Cell 31, 2973–2995. doi: 10.1105/tpc.19.00066
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., et al. (2008). Mobilization of rubisco and stromalocalized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Ishida, H., Nakamura, S., Hidema, J. (2017). Entire photo-damaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29, 377–394. doi: 10.1105/tpc.16.00637
Izumi, M., Nakamura, S., Li, N. (2019). Autophagic turnover of chloroplasts: its roles and regulatory mechanisms in response to sugar starvation. Front. Plant Sci. 10, 280. doi: 10.3389/fpls.2019.00280
Jaishy, B., Abel, E. D. (2016). Lipids, lysosomes, and autophagy. J. Lipid Res. 57, 1619–1635. doi: 10.1194/jlr.R067520
Janse van Rensburg, H. C., Van den Ende, W., Signorelli, S. (2019). Autophagy in plants: both a puppet and a puppet master of sugars. Front. Plant Sci. 10, 14. doi: 10.3389/fpls.2019.00014
Jarvis, P., López-Juez, E. (2013). Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. doi: 10.1038/nrm3702
Jiménez-Nopala, G., Salgado-Escobar, A. E., Cevallos-Porta, D., Cárdenas, L., Sepúlveda-Jiménez, G., Cassab, G., et al. (2018). Autophagy mediates hydrotropic response in Arabidopsis thaliana roots. Plant Sci. 272, 1–13. doi: 10.1016/j.plantsci.2018.03.026
Johansen, T., Lamark, T. (2011). Selective autophagy mediated by autophagic adapter proteins. Autophagy 7, 279–296. doi: 10.4161/auto.7.3.14487
Jung, H., Lee, H. N., Marshall, R. S., Lomax, A. W., Yoon, M. J., Kim, J., et al. (2020). Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 71, 73–89. doi: 10.1093/jxb/erz404.
Katsiarimpa, A., Kalinowska, K., Anzenberger, F., Weis, C., Ostertag, M., Tsutsumi, C., et al. (2013). The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25, 2236–2252. doi: 10.1105/tpc.113.113399
Ketelaar, T., Voss, C., Dimmock, S. A., Thumm, M., Hussey, P. J. (2004). Arabidopsis homologues of the autophagy protein Atg8 are a novel family of microtubule binding proteins. FEBS Lett. 567, 302–306. doi: 10.1016/j.febslet.2004.04.088
Kihara, A., Noda, T., Ishihara, N., Ohsumi, Y. (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 152, 519–530. doi: 10.1083/jcb.152.3.519
Kim, S. H., Kwon, C., Lee, J. H., Chung, T. (2012). Genes for plant autophagy: functions and interactions. Mol. Cells 34, 413–423. doi: 10.1007/s10059-012-0098-y
Klionsky, D. J., Emr, S. D. (2000). Autophagy as a regulated pathway of cellular degradation. Science 290, 1717–1721. doi: 10.1126/science.290.5497.1717
Klionsky, D. J., Abeliovich, H., Agostinis, P., Agrawal, D. K., Aliev., G., Askew, D. S., et al. (2008). Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4, 151–175. doi: 10.4161/auto.5338
Kotani, T., Kirisako, H., Koizumi, M., Ohsumi, Y., Nakatogawa, H. (2018). The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. U. S. A. 115, 10363–10368. doi: 10.1073/pnas.1806727115
Kurusu, T., Koyano, T., Hanamata, S., Kubo, T., Noguchi, Y., Yagi, C., et al. (2014). OsATG7 is required for autophagy-dependent lipid metabolism in rice postmeiotic anther development. Autophagy 10, 878–888. doi: 10.4161/auto.28279
Kurusu, T., Koyano, T., Kitahata, N., Kojima, M., Hanamata, S., Sakakibara, H., et al. (2017). Autophagy-mediated regulation of phytohormone metabolism during rice anther development. Plant Signal. Behav. 12, e1365211. doi: 10.1080/15592324.2017.1365211
Le Bars, R., Marion, J., Le Borgne, R., Satiat-Jeunemaitre, B., Bianchi, M. W. (2014). ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat. Commun. 5, 4121. doi: 10.1038/ncomms5121
Lee, Y., Kim, E. S., Choi, Y., Hwang, I., Staiger, C. J., Chung, Y. Y., et al. (2008). The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol. 147, 1886–1897. doi: 10.1104/pp.108.121590
Li, F., Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Li, F., Vierstra, R. D. (2014). Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy 10, 1466–1467. doi: 10.4161/auto.29320
Li, F., Chung, T., Vierstra, R. D. (2014). AUTOPHAGY-RELATED11 plays a critical role in general autophagy and senescence-induced mitophagy in Arabidopsis. Plant Cell 26, 788–807. doi: 10.1105/tpc.113.120014
Li, F., Chung, T., Pennington, J. G., Federico, M. L., Kaeppler, H. F., Kaeppler, S. M., et al. (2015a). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389–1408. doi: 10.1105/tpc.15.00158
Li, L., Song, Y., Wang, K., Dong, P., Zhang, X., Li, F., et al. (2015b). TOR-inhibitor insensitive-1 (TRIN1) regulates cotyledons greening in Arabidopsis. Front. Plant Sci. 6, 861. doi: 10.3389/fpls.2015.00861
Li, F., Zhang, M., Zhang, C., Zhou, X. (2019a). Nuclear autophagy degrades a geminivirus nuclear protein to restrict viral infection in Solanaceous plants. New Phytol. 225, 1746–1761. doi: 10.1111/nph.16268
Li, X., Liu, Q., Feng, H., Deng, J., Zhang, R., Wen, J., et al. (2019b). Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy 30, 1–16. doi: 10.1080/15548627.2019.1643656
Lingard, M. J., Monroe-Augustus, M., Bartel, B. (2009). Peroxisome-associated matrix protein degradation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 106, 4561–4566. doi: 10.1073/pnas.0811329106
Liu, Y. M., Bassham, D. C. (2010). TOR is a negative regulator of autophagy in Arabidopsis thaliana. PloS One 5, e11883. doi: 10.1371/journal.pone.0011883
Liu, Y., Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Xiong, Y., Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Lu, K., Psakhye, I., Jentsch, S. (2014). Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563. doi: 10.1016/j.cell.2014.05.048
Luo, L., Zhang, P., Zhu, R., Fu, J., Su, J., Zheng, J., et al. (2017). Autophagy is rapidly induced by salt stress and is required for salt tolerance in Arabidopsis. Front. Plant Sci. 8, 1459. doi: 10.3389/fpls.2017.01459
Malinova, I., Qasim, H. M., Brust, H., Fettke, J. (2018). Parameters of starch granule genesis in chloroplasts of Arabidopsis thaliana. Front. Plant Sci. 9, 761. doi: 10.3389/fpls.2018.00761
Maqbool, A., Hughes, R. K., Dagdas, Y. F., Tregidgo, N., Zess, E., Belhaj, K., et al. (2016). Structural basis of host autophagy-related protein 8 (ATG8) binding by the irish potato famine pathogen effector protein PexRD54. J. Biol. Chem. 291, 20270–20282. doi: 10.1074/jbc.M116.744995
Marshall, R. S., Vierstra, R. D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173–208. doi: 10.1146/annurev-arplant-042817-040606
Marshall, R. S., Li, F., Gemperline, D. C., Book, A. J., Vierstra, R. D. (2015). Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 58, 1053–1066. doi: 10.1016/j.molcel.2015.04.023
Marshall, R. S., Hua, Z., Mali, S., McLoughlin, F., Vierstra, R. D. (2019). ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell 177, 766–781. doi: 10.1016/j.cell.2019.02.009
Masclaux-Daubresse, C., Clément, G., Anne, P., Routaboul, J. M., Guiboileau, A., Soulay, F., et al. (2014). Stitching together the multiple dimensions of autophagy using metabolomics and transcriptomics reveals impacts on metabolism, development, and plant responses to the environment in Arabidopsis. Plant Cell 26, 1857–1877. doi: 10.1105/tpc.114.124677
Masclaux-Daubresse, C., Chen, Q., Havé, M. (2017). Regulation of nutrient recycling via autophagy. Curr. Opin. Plant Biol. 39, 8–17. doi: 10.1016/j.pbi.2017.05.001
Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., et al. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. U. S. A. 99, 6422–6427. doi: 10.1073/pnas.092141899
Mhamdi, A., Noctor, G., Baker, A. (2012). Plant catalases: peroxisomal redox guardians. Arch. Biochem. Biophys. 525, 181–194. doi: 10.1016/j.abb.2012.04.015
Michaeli, S., Honig, A., Levanony, H., Peled-Zehavi, H., Galili, G. (2014). Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 26, 4084–4101. doi: 10.1105/tpc.114.129999
Minibayeva, F., Dmitrieva, S., Ponomareva, A., Ryabovol, V. (2012). Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol. Biochem. 59, 11–19. doi: 10.1016/j.plaphy.2012.02.013
Minina, E. A., Moschou, P. N., Vetukuri, R. R., Sanchez-Vera, V., Cardoso, C., Liu, Q., et al. (2018). Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 69, 1415–1432. doi: 10.1093/jxb/ery010
Moreau, M., Azzopardi, M., Clément, G., Dobrenel, T., Marchive, C., Renne, C., et al. (2012). Mutations in the Arabidopsis homolog of LST8/GβL, a partner of the target of rapamycin kinase, impair plant growth, flowering, and metabolic adaptation to long days. Plant Cell 24, 463–481. doi: 10.1105/tpc.111.091306
Nakamura, S., Hidema., J., Sakamoto, W., Ishida, H., Izumi, M. (2019). Selective elimination of membrane-damaged chloroplasts via microautophagy. Plant Physiol. 177, 1007–1026. doi: 10.1104/pp.18.00444
Nolan, T. M., Brennan, B., Yang, M., Chen, J., Zhang, M., Li, Z., et al. (2017). Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 41, 33–46. doi: 10.1016/j.devcel.2017.03.013
Nukarinen, E., Nägele, T., Pedrotti, L., Wurzinger, B., Mair, A., Landgraf, R., et al. (2016). Quantitative phosphoproteomics reveals the role of the AMPK plant ortholog SnRK1 as a metabolic master regulator under energy deprivation. Sci. Rep. 6, 31697. doi: 10.1038/srep31697
Ohsumi, Y. (2001). Molecular dissection of autophagy: two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2, 211–216. doi: 10.1038/35056522
Orsi, A., Razi, M., Dooley, H. C., Robinson, D., Weston, A. E., Collinson, L. M., et al. (2012). Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 23, 1860–1873. doi: 10.1091/mbc.e11-09-0746
Pérez-Pérez, M. E., Couso, I., Crespo, J. L. (2012). Carotenoid deficiency triggers autophagy in the model green alga Chlamydomonas reinhardtii. Autophagy 8, 376–388. doi: 10.4161/auto.18864
Pérez-Pérez, M. E., Lemaire, S. D., Crespo, J. L. (2016). Control of autophagy in Chlamydomonas is mediated through redox-dependent inactivation of the ATG4 protease. Plant Physiol. 172, 2219–2234. doi: 10.1104/pp.16.01582
Pacifici, E., Polverari, L., Sabatini, S. (2015). Plant hormone cross-talk: the pivot of root growth. J. Exp. Bot. 66, 1113–1121. doi: 10.1093/jxb/eru534
Pankiv, S., Clausen, T. H., Lamark, T., Brech, A., Bruun, J. A., Outzen, H., et al. (2007). p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282, 24131–24145. doi: 10.1074/jbc.M702824200
Pankiv, S., Alemu, E. A., Brech, A., Bruun, J. A., Lamark, T., Overvatn, A., et al. (2010). FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J. Cell Biol. 188, 253–269. doi: 10.1083/jcb.200907015
Papandreou, M. E., Tavernarakis, N. (2019). Nucleophagy: from homeostasis to disease. Cell Death Differ. 26, 630–639. doi: 10.1038/s41418-018-0266-5
Phillips, A. R., Suttangkakul, A., Vierstra, R. D. (2008). The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Pottier, M., Dumont, J., Masclaux-Daubresse, C., Thomine, S. (2019). Autophagy is essential for optimal translocation of iron to seeds in Arabidopsis. J. Exp. Bot. 70, 859–869. doi: 10.1093/jxb/ery388
Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J., Zhu, J. K., et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 60, 805–826. doi: 10.1111/jipb.12654
Ren, M., Venglat, P., Qiu, S., Feng, L., Cao, Y., Wang, E., et al. (2012). Target of rapamycin signaling regulates metabolism, growth, and life span in Arabidopsis. Plant Cell 24, 4850–4874. doi: 10.1105/tpc.112.107144
Robaglia, C., Thomas, M., Meyer, C. (2012). Sensing nutrient and energy status by SnRK1 and TOR kinases. Curr. Opin. Plant Biol. 15, 301–307. doi: 10.1016/j.pbi.2012.01.012
Rodor, J., Jobet, E., Bizarro, J., Vignols, F., Carles, C., Suzuki, T., et al. (2011). AtNUFIP, an essential protein for plant development, reveals the impact of snoRNA gene organisation on the assembly of snoRNPs and rRNA methylation in Arabidopsis thaliana. Plant J. 65, 807–819. doi: 10.1111/j.1365-313X.2010.04468.x
Salem, M. A., Li, Y., Bajdzienko, K., Fisahn, J., Watanabe, M., Hoefgen, R., et al. (2018). RAPTOR controls developmental growth transitions by altering the hormonal and metabolic balance. Plant Physiol. 177, 565–593. doi: 10.1104/pp.17.01711
Sankaranarayanan, S., Samuel, M. A. (2015). A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal. Behav. 10, e989749. doi: 10.4161/15592324.2014.989749
Sedaghatmehr, M., Thirumalaikumar, V. P., Kamranfar, I., Marmagne, A., Masclaux-Daubresse, C., Balazadeh, S. (2019). A regulatory role of autophagy for resetting the memory of heat stress in plants. Plant Cell Environ. 42, 1054–1064. doi: 10.1111/pce.13426
Selinski, J., Scheibe, R., Day, D. A., Whelan, J. (2018). Alternative oxidase is positive for plant performance. Trends Plant Sci. 23, 588–597. doi: 10.1016/j.tplants.2018.03.012
Sera, Y., Hanamata, S., Sakamoto, S., Ono, S., Kaneko, K., Mitsui, Y., et al. (2019). Essential roles of autophagy in metabolic regulation in endosperm development during rice seed maturation. Sci. Rep. 9, 18544. doi: 10.1038/s41598-019-54361-1
Shen, X., Wang, Z., Song, X., Xu, J., Jiang, C., Zhao, Y., et al. (2014). Transcriptomic profiling revealed an important role of cell wall remodeling and ethylene signaling pathway during salt acclimation in Arabidopsis. Plant Mol. Biol. 86, 303–317. doi: 10.1007/s11103-014-0230-9
Shibata, M., Oikawa, K., Yoshimoto, K., Kondo, M., Mano, S., Yamada, K., et al. (2013). Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell 25, 4967–4983. doi: 10.1105/tpc.113.116947
Signorelli, S., Tarkowski, Ł.P., Van den Ende, W., Bassham, D. C. (2019). Linking autophagy to abiotic and biotic stress responses. Trends Plant Sci. 24, 413–430. doi: 10.1016/j.tplants.2019.02.001
Sláviková, S., Shy, G., Yao, Y., Glozman, R., Levanony, H., Pietrokovski, S., et al. (2005). The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 56, 2839–2849. doi: 10.1093/jxb/eri276
Slavikova, S., Ufaz, S., Avin-Wittenberg, T., Levanony, H., Galili, G. (2008). An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 59, 4029–4043. doi: 10.1093/jxb/ern244
Smeekens, S., Ma, J., Hanson, J., Rolland, F. (2010). Sugar signals and molecular networks controlling plant growth. Curr. Opin. Plant Biol. 13, 274–279. doi: 10.1016/j.pbi.2009.12.002
Soto-Burgos, J., Bassham, D. C. (2017). SnRK1 activates autophagy via the TOR signaling pathway in Arabidopsis thaliana. PloS One 12, e0182591. doi: 10.1371/journal.pone.0182591
Spitzer, C., Li, F., Buono, R., Roschzttardtz, H., Chung, T., Zhang., M., et al. (2015). The endosomal protein CHARGED MULTI-VESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 27, 391–402. doi: 10.1105/tpc.114.135939
Su, T., Wang, P., Li, H., Zhao, Y., Lu, Y., Dai, P., et al. (2018). The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 60, 591–607. doi: 10.1111/jipb.12649
Su, T., Li, W., Wang, P., Ma, C. (2019). Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front. Plant Sci. 10, 705. doi: 10.3389/fpls.2019.00705
Sun, X., Jia, X., Huo, L., Che, R., Gong, X., Wang, P., et al. (2018). MdATG18a overexpression improves tolerance to nitrogen deficiency and regulates anthocyanin accumulation through increased autophagy in transgenic apple. Plant Cell Environ. 41, 469– 480. doi: 10.1111/pce.13110
Surpin, M., Zheng, H., Morita, M. T., Saito, C., Avila, E., Blakeslee, J. J., et al. (2003). The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15, 2885–2899. doi: 10.1105/tpc.016121
Sutipatanasomboon, A., Herberth, S., Alwood, E. G., Häweker, H., Müller, B., Shahriari, M., et al. (2017). Disruption of the plant-specific CFS1 gene impairs autophagosome turnover and triggers EDS1-dependent cell death. Sci. Rep. 2017 7, 8677. doi: 10.1038/s41598-017-08577-8
Suttangkakul, A., Li, F., Chung, T., Vierstra, R. D. (2011). The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 23, 3761–3779. doi: 10.1105/tpc.111.090993
Svenning, S., Lamark, T., Krause, K., Johansen, T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993–1010. doi: 10.4161/auto.7.9.16389
Takatsuka, C., Inoue, Y., Matsuoka, K., Moriyasu, Y. (2004). 3-Methyladenine inhibits autophagy in tobacco culture cells under sucrose starvation conditions. Plant Cell Physiol. 45, 265–274. doi: 10.1093/pcp/pch031
Tang, J., Bassham, D. C. (2018). Autophagy in crop plants: what’s new beyond Arabidopsis? Open Biol. 8, 180162. doi: 10.1098/rsob.180162
Tsukada, M., Ohsumi, Y. (1993). Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333, 169–174. doi: 10.1016/0014-5793(93)80398-E
van Doorn, W. G., Papini, A. (2013). Ultrastructure of autophagy in plant cells: a review. Autophagy 9, 1922–1936. doi: 10.4161/auto.26275
Van Leene, J., Han, C., Gadeyne, A., Eeckhout, D., Matthijs, C., Cannoot, B., et al. (2019). Capturing the phosphorylation and protein interaction landscape of the plant TOR kinase. Nat. Plants 5, 316–327. doi: 10.1038/s41477-019-0378-z
van Zutphen, T., Todde, V., de Boer, R., Kreim, M., Hofbauer, H. F., Wolinski, H., et al. (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25, 290–301. doi: 10.1091/mbc.e13-08-0448
Vanhee, C., Zapotoczny, G., Masquelier, D., Ghislain, M., Batoko, H. (2011). The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805. doi: 10.1105/tpc.110.081570
Wada, S., Ishida, H., Izumi, M., Yoshimoto, K., Ohsumi, Y., Mae, T., et al. (2009). Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 149, 885–893. doi: 10.1104/pp.108.130013
Wang, Y., Yu, B. J., Zhao, J. P., Guo, J. B., Li, Y., Han, S. J., et al. (2013). Autophagy contributes to leaf starch degradation. Plant Cell 25, 1383–1399. doi: 10.1105/tpc.112.108993
Wang, Y., Cai, S., Yin, L., Shi, K., Xia, X., Zhou, Y., et al. (2015). Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 11, 2033–2047. doi: 10.1080/15548627.2015.1098798
Wang, P., Mugume, Y., Bassham, D. C. (2018a). New advances in autophagy in plants: regulation, selectivity and function. Semin. Cell Dev. Biol. 80, 113–122. doi: 10.1016/j.semcdb.2017.07.018
Wang, P., Zhao, Y., Li, Z., Hsu, C. C., Liu, X., Fu, L., et al. (2018b). Reciprocal regulation of the TOR kinase and ABA receptor balances plant growth and stress response. Mol. Cell 69, 100–112. doi: 10.1016/j.molcel.2017.12.002
Wang, P., Nolan, T. M., Yin, Y., Bassham, D. C. (2019a). Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 25, 1–17. doi: 10.1080/15548627.2019.1598753
Wang, Y., Cao, J. J., Wang, K. X., Xia, X. J., Shi, K., Zhou, Y. H. (2019b). BZR1 mediates brassinosteroid-induced autophagy and nitrogen starvation in tomato. Plant Physiol. 179, 671–685. doi: 10.1104/pp.18.01028
Wang, C. W. (2016). Lipid droplets, lipophagy, and beyond. Biochim. Biophys. Acta 1861, 793–805. doi: 10.1016/j.bbalip.2015.12.010
Wojciechowska, N., Smugarzewska, I., Marzec-Schmidt, K., Zarzyńska-Nowak, A., Bagniewska-Zadworna, A. (2019). Occurrence of autophagy during pioneer root and stem development in Populus trichocarpa. Planta 250, 1789–1801. doi: 10.1007/s00425-019-03265-5
Woo, J., Park, E., Dinesh-Kumar, S. P. (2014). Differential processing of Arabidopsis ubiquitin-like Atg8 autophagy proteins by Atg4 cysteine proteases. Proc. Natl. Acad. Sci. U. S. A. 111, 863–868. doi: 10.1073/pnas.1318207111
Wyant, G. A., Abu-Remaileh, M., Frenkel, E. M., Laqtom, N. N., Dharamdasani, V., Lewis, C. A., et al. (2018). NUFIP1 is a ribosome receptor for starvation-induced ribophagy. Science 360, 751–758. doi: 10.1126/science.aar2663
Xie, Q., Tzfadia, O., Levy, M., Weithorn, E., Peled-Zehavi, H., Van Parys, T., et al. (2016). hfAIM: a reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy 12, 876–887. doi: 10.1080/15548627.2016.1147668
Xiong, Y., Contento, A. L., Nguyen, P. Q., Bassham, D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Xu, N., Gao, X. Q., Zhao, X. Y., Zhu, D. Z., Zhou, L. Z., Zhang, X. S. (2011). Arabidopsis VPS15 is essential for pollen development and germination through modulating phosphatidylinositol-3-phosphate formation. Plant Mol. Biol. 77, 251–260. doi: 10.1007/s11103-011-9806-9
Yamamoto, H., Kakuta, S., Watanabe, T. M., Kitamura, A., Sekito, T., Kondo-Kakuta, C., et al. (2012). Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J. Cell Biol. 198, 219–233. doi: 10.1083/jcb.201202061
Yamauchi, S., Mano, S., Oikawa, K., Hikino, K., Teshima, K. M., Kimori, Y., et al. (2019). Autophagy controls reactive oxygen species homeostasis in guard cells that is essential for stomatal opening. Proc. Natl. Acad. Sci. U. S. A. 116, 19187–19192. doi: 10.1073/pnas.1910886116
Yoon, S. H., Chung, T. (2019). Protein and RNA quality control by autophagy in plant cells. Mol. Cells 42, 285–291. doi: 10.14348/molcells.2019.0011
Yoshimoto, K., Ohsumi, Y. (2018). Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol. 59, 1337–1344. doi: 10.1093/pcp/pcy112
Yoshimoto, K., Hanaoka, H., Sato, S., Kato, T., Tabata, S., Noda, T., et al. (2004). Processing of ATG8s, ubiquitinlike proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 16, 2967–2983. doi: 10.1105/tpc.104.025395
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21, 2914–2927. doi: 10.1105/tpc.109.068635
Yoshimoto, K., Shibata, M., Kondo, M., Oikawa, K., Sato, M., Toyooka, K., et al. (2014). Organ-specific quality control of plant peroxisomes is mediated by autophagy. J. Cell Sci. 12, 1161–1168. doi: 10.1242/jcs.139709
Young, P. G., Bartel, B. (2016). Pexophagy and peroxisomal protein turnover in plants. Biochim. Biophys. Acta 1863, 999–1005. doi: 10.1016/j.bbamcr.2015.09.005
Zechner, R., Madeo, F., Kratky, D. (2017). Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat. Rev. Mol. Cell Biol. 18, 671–684. doi: 10.1038/nrm.2017.76
Zhang, J. X., Wang, C., Yang, C. Y., Wang, J. Y., Chen, L., Bao, X. M., et al. (2010). The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. Plant J. 62, 539–548. doi: 10.1111/j.1365-313X.2010.04173.x
Zhang, Z., Zhu, J. Y., Roh, J., Marchive, C., Kim, S. K., Meyer, C., et al. (2016). TOR signaling promotes accumulation of BZR1 to balance growth with carbon availability in Arabidopsis. Curr. Biol. 26, 1854–1860. doi: 10.1016/j.cub.2016.05.005
Zhang, H., Li, Y., Zhu, J. K. (2018). Developing naturally stress resistant crops for a sustainable agriculture. Nat. Plants 4, 989–996. doi: 10.1038/s41477-018-0309-4
Zheng, Y., Liao, C., Zhao, S., Wang, C., Guo, Y. (2017). The glycosyltransferase QUA1 regulates chloroplast-associated calcium signaling during salt and drought stress in Arabidopsis. Plant Cell Physiol. 58, 329–341. doi: 10.1093/pcp/pcw192
Zhou, J., Wang, J., Cheng, Y., Chi, Y. J., Fan, B., Yu, J. Q., et al. (2013). NBR1-mediated selective autophagy targets insoluble ubiquitinated protein aggregates in plant stress responses. PloS Genet. 9, e1003196. doi: 10.1371/journal.pgen.1003196
Zhou, J., Wang, J., Yu, J. Q., Chen, Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5, 174. doi: 10.3389/fpls.2014.00174
Zhu, T., Zou, L., Li, Y., Yao, X., Xu, F., Deng, X., et al. (2018). Mitochondrial alternative oxidase-dependent autophagy involved in ethylene-mediated drought tolerance in Solanum lycopersicum. Plant Biotechnol. J. 16, 2063–2076. doi: 10.1111/pbi.12939
Zhuang, X., Jiang, L. (2019). Chloroplast degradation: multiple routes into the vacuole. Front. Plant Sci. 10, 359. doi: 10.3389/fpls.2019.00359
Zhuang, X., Wang, H., Lam, S. K., Gao, C., Wang, X., Cai, Y., et al. (2013). A BAR-domain protein SH3P2, which binds to phosphatidylinositol-3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 25, 4596–4615. doi: 10.1105/tpc.113.118307
Zhuang, X., Chung, K. P., Cui, Y., Lin, W., Gao, C., Kang, B. H., et al. (2017). ATG9 regulates autophagosome progression from the endoplasmic reticulum in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 114, E426–E435. doi: 10.1073/pnas.1616299114
Keywords: autophagy, selective autophagy, receptor, stress, plant
Citation: Su T, Li X, Yang M, Shao Q, Zhao Y, Ma C and Wang P (2020) Autophagy: An Intracellular Degradation Pathway Regulating Plant Survival and Stress Response. Front. Plant Sci. 11:164. doi: 10.3389/fpls.2020.00164
Received: 15 November 2019; Accepted: 03 February 2020;
Published: 28 February 2020.
Edited by:
Diane C. Bassham, Iowa State University, United StatesReviewed by:
Taijoon Chung, Pusan National University, South KoreaCopyright © 2020 Su, Li, Yang, Shao, Zhao, Ma and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changle Ma, bWFjaGFuZ2xlQHNkbnUuZWR1LmNu; Pingping Wang, cGluZ3Bpbmcud2FuZ0BzZG51LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.