- 1U.S. Department of Agriculture – Agricultural Research Service, North Carolina State University, Raleigh, NC, United States
- 2Department of Crop Genetics, John Innes Centre, Norwich, United Kingdom
Important advances have been made in understanding the relationship of necrotrophic effectors (NE) and host sensitivity (Snn) genes in the Parastagonospora nodorum-wheat pathosystem. Yet much remains to be learned about the role of these interactions in determining wheat resistance levels in the field, and there is mixed evidence on whether breeding programs have selected against Snn genes due to their role in conferring susceptibility. SNB occurs ubiquitously in the U.S. Atlantic seaboard, and the environment is especially well suited to field studies of resistance to natural P. nodorum populations, as there are no other important wheat leaf blights. Insights into the nature of SNB resistance have been gleaned from multi-year data on phenotypes and markers in cultivars representative of the region’s germplasm. In this perspective article, we review the evidence that in this eastern region of the U.S., wheat cultivars have durable quantitative SNB resistance and Snn–NE interactions are of limited importance. This conclusion is discussed in light of the relevant available information from other parts of the world.
Introduction
Septoria nodorum blotch (SNB) of wheat, caused by the ascomycete fungus Parastagonospora nodorum, has a global distribution, although severe epidemics appear to be confined mainly to Australia, parts of northern Europe and the U.S., and parts of north Asia. In some areas such as the United Kingdom, SNB has become less significant than formerly as a result of successful breeding for resistance combined with reductions in atmospheric sulphur dioxide [discussed by (Arraiano et al., 2009); see also (Brown, 2015) and (Chandramohan and Shaw, 2013)]. In the U.S., SNB is a recurrent disease of wheat in several geographic regions (Figure 1). It co-occurs with Septoria tritici blotch in the western, moist areas of the Pacific Northwest; the upper Plains states of North Dakota, South Dakota, and Minnesota; and the states adjacent to the Great Lakes (Shaner and Buechley, 1995; Cowger et al., 2002; Figure 1). The only U.S. region where P. nodorum is the sole important leaf blotch pathogen of wheat is the eastern seaboard, making that environment ideal for field studies of SNB resistance and epidemiology (Cowger and Murphy, 2007).
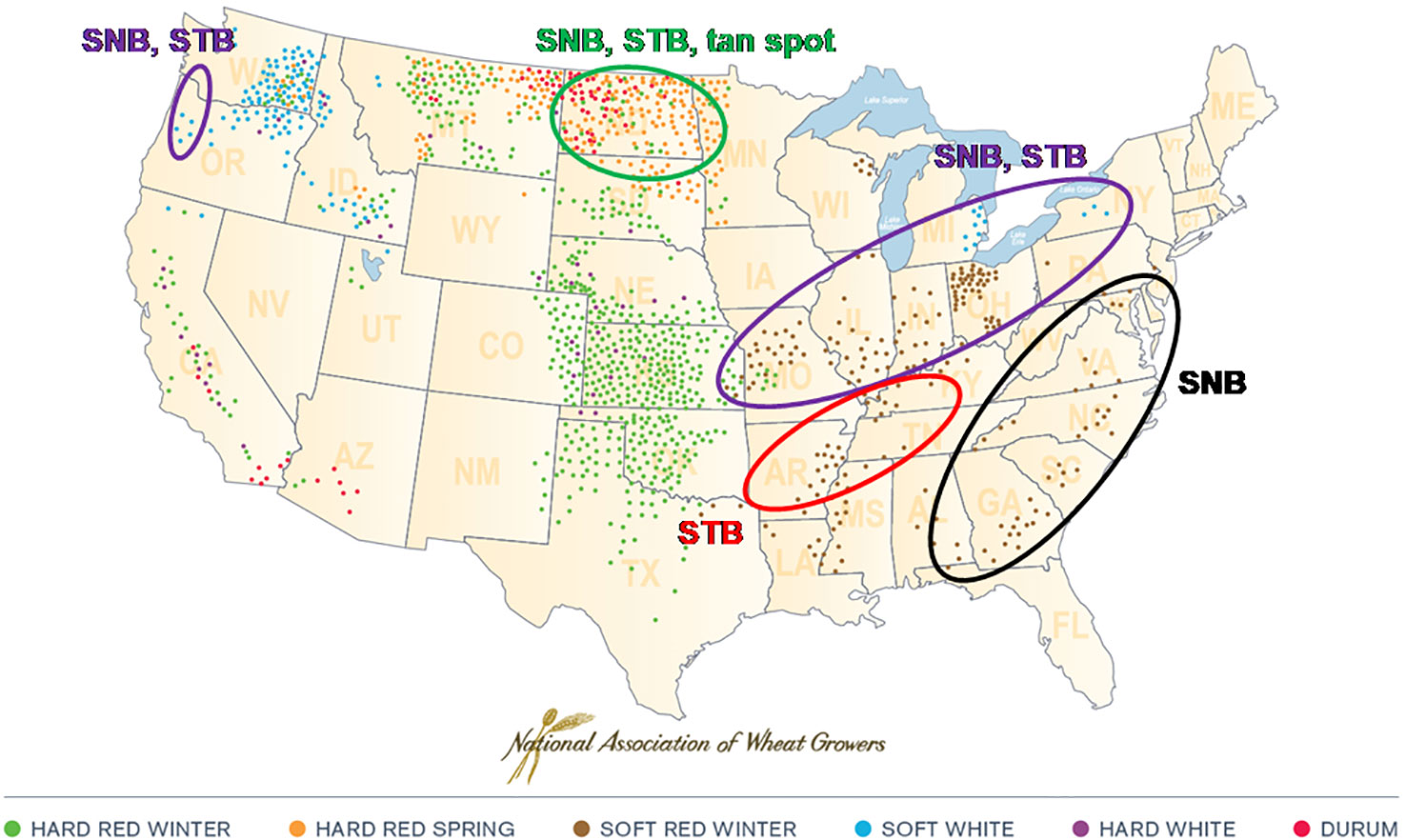
Figure 1 Septoria nodorum blotch (SNB), caused by Parastagonospora nodorum, co-occurs with Septoria tritici blotch (STB) (caused by Mycosphaerella graminicola) in the western, moist areas of the Pacific Northwest; the upper Plains states of North Dakota, South Dakota, and Minnesota; and the states adjacent to the Great Lakes. It is the only important wheat leaf blotch in the eastern seaboard states from Florida to Delaware. (Tan spot caused by Pyrenophora tritici-repentis).
SNB can cause significant yield losses in the U.S. and other countries (Shah and Bergstrom, 1993; Bhathal et al., 2003). In the southeastern U.S., SNB has been a frequent and sometimes damaging disease of winter wheat for many years (Nelson et al., 1974; Allingham and Jackson, 1981; Cunfer, 1998). Soft red winter wheat (SRWW) is the main market class of wheat bred and grown in the eastern U.S. (Figure 1), and the third largest wheat market class in the nation by production volume (after hard red spring and hard red winter wheats) (USDA Economic Research Service, 2019). Annual production of U.S. SRWW ranged between 6.5 million to 15.5 million tons (2.4 million and 5.7 million bushels) during the decade 2010 to 2019 (NASS, 2019). Given its low to medium levels of protein and soft endosperm, the primary uses of SRWW are in flatbreads, crackers, cakes, pastries and animal feed. Resistance to SNB is an ongoing breeding objective for SRWW (Cowger and Murphy, 2007).
Snn–NE Interactions and Resistance
Wheat has nine named sensitivity (Snn) genes that interact with matching necrotrophic effectors (NE) of P. nodorum (Peters-Haugrud et al., 2019). The Snn genes have been found on wheat chromosomes 1AS, 1BS, 2DS, 2DL, 4BL, 5BS, 5DS, 5BL, and 6AL. These genes, which were detected as QTL using bi-parental mapping populations, interact with pathogen NE to increase the severity of SNB in what is known as an “inverse gene-for-gene” interaction. In wheat-P. nodorum and some other necrotrophic pathosystems, some sensitivity genes are apparently R-genes of the type that normally elicits resistance upon pathogen recognition, but have been hijacked by necrotrophic pathogens to elicit susceptibility (Nagy and Bennetzen, 2008; Faris et al., 2010; Gilbert and Wolpert, 2013; Shi et al., 2016). The NE are small secreted proteins, generally in the 10–30 kD range, that elicit programmed cell death when recognized by the host (Friesen and Faris, 2010). In the wheat-P. nodorum pathosystem, the interactions of Snn genes and NE may be incremental, unlike a classic gene-for-gene interaction; i.e., possessing two sensitivity genes can cause more susceptibility than possessing one, if the matching pathogen effectors are present (Chu et al., 2010; Abeysekara et al., 2012).
Of the wheat NE sensitivity genes investigated to date, Tsn1 appears to be the one with the largest positive effect on susceptibility (Liu et al., 2006). Tsn1 has features typical of pathogen recognition genes, including serine/threonine protein kinase (S/TPK) and nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains (Faris et al., 2010). SnToxA is the NE that is recognized by Tsn1, while SnTox1 is recognized by Snn1 and SnTox3 by Snn3.
In an international comparison of NE frequencies and haplotypes, SnToxA was usually absent in the majority of P. nodorum isolates, depending on the country of origin, including in 75% of P. nodorum isolates from both spring and winter regions of the U.S. (Stukenbrock and McDonald, 2007; McDonald et al., 2013). An exception was Australia, but the evidence there appears to be mixed. Of 73 Australian P. nodorum isolates, 97% possessed SnToxA (Stukenbrock and McDonald, 2007; McDonald et al., 2013); however, that collection was from a single field and the Tsn1 status of the wheat cultivar was unknown (Oliver et al., 2009). As over half of sampled wheat cultivars grown in Western Australia were sensitive to SnToxA, it appeared that breeders there had not selected against Tsn1, possibly because neither sensitivity to SnToxA nor to SnTox3 was significantly associated with cultivar resistance level as measured via artificial inoculation with a mixture of current isolates (Oliver et al., 2009; Waters et al., 2011) By contrast, effector sensitivity tests on Vavilov wheat collection accessions showed that since 1940, breeders in Russia and Kazakhstan had selected directly or indirectly against Tsn1 and also against Snn1, although not against Snn3 (Phan et al., 2018).
The nine known P. nodorum NE and matching wheat sensitivity genes have been identified and characterized mainly using hard red spring or durum wheat lines and P. nodorum isolates from the upper Midwest state of North Dakota (references in Peters-Haugrud et al., 2019). It was recently discovered that the upper Midwest and eastern U.S. P. nodorum populations differ dramatically in the frequency of SnToxA (Richards et al., 2019). Specifically, Population 1 as examined by Richards et al. (2019) consisted of 105 P. nodorum isolates from hard spring and winter wheat and durum wheat in North Dakota, South Dakota, and Minnesota, while Population 2 included 67 isolates from the soft winter wheat states of the eastern U.S., stretching from Texas and Arkansas northeast to New York and south to Georgia. The frequencies of SnToxA were 96 and 4% in Population 1 and Population 2, respectively. That is, SnToxA was almost universal in the upper Plains isolates, and almost entirely absent in the eastern soft wheat isolates. The populations did not differ greatly for the other analyzed NEs: SnTox1 occurred at frequencies of 100 and 88% in Populations 1 and 2, respectively, and SnTox3 at frequencies of 62 and 52%, respectively. These results are consistent with a smaller previous study showing that SnToxA and SnTox3 were present only in a minority of isolates from the eastern U.S. P. nodorum population (15 and 39%, respectively), but SnTox1 was present in 74% (Crook et al., 2012).
On the host side, a sample of 26 wheat genotypes from eastern U.S. wheat breeding programs was surveyed to determine the presence of the Tsn1, Snn1, and Snn3 sensitivity genes (Bertucci et al., 2014). Tsn1 was not found in any of 12 eastern U.S. soft winter lines. However, the gene was present in 6 of 11 hard winter wheat cultivars, particularly those with origins in the southern Plains states of Nebraska, Kansas, Oklahoma and Texas, that had been utilized for hardness and other traits in a new program to breed bread wheats for the eastern U.S. market. Further, the sensitivity gene Snn3 was present in 7 of the 12 soft wheat cultivars, and 6 of the 11 hard wheat cultivars, while Snn1 was absent from all lines. In North America, Snn1 is thought to be less frequent in hexaploid than in durum wheat germplasm (J. Faris, personal communication.) The evidence from these studies suggests that variation in susceptibility of SRWW could be affected by Snn3, but not by Tsn1 and probably not by Snn1. From the small sample of hard winter wheats with origins in the central-southern Great Plains (Nebraska, Kansas, Oklahoma, and Texas), it appears that Tsn1 may play a role in susceptibility there.
The hypothesis that Tsn1 is rare in eastern U.S. soft wheat germplasm was reinforced by examining data from available eastern wheat germplasm. A dominant Kompetitive Allele-Specific Polymerase chain reaction (KASP) marker (primer sequences Tsn1_AL1 5ʹ-GAAGGTCGGAGTCAACGGATTCTATTCGTAATCGTGCCTTCCGG-3ʹ and Tsn1_C1 5ʹ-CTGCCCTTCACTTAGCCTGTCAC-3ʹ) was developed based on the published Tsn1 gene sequence (Faris et al., 2010). Frequency of Tsn1 was assessed using DNA of a panel of 330 eastern wheat lines that included ten 19th century landraces, 188 cultivars developed from 1915 to 2011, and 132 elite experimental lines. Tsn1 was detected in only 15 samples, including seven soft wheat cultivars and two experimental lines. Six of the 15 eastern hard winter wheat breeding lines in the panel possessed the gene. None of the soft winter wheat landraces had Tsn1 and there was only one Tsn1-positive line developed prior to 1950, suggesting that Tsn1 was either rare or absent in foundational germplasm of the eastern soft wheat growing region.
Although this panel was not an exhaustive sampling of historical germplasm, this conclusion is supported by the low frequency of Tsn1 in later cultivar releases. Starting in 2013, the Tsn1 marker was routinely analyzed on advanced wheat experimental lines and released varieties serving as checks in the elite Uniform Eastern and Uniform Southern SRWW nurseries operated by the U.S. Department of Agriculture-Agricultural Research Service. Entries in these nurseries have a high likelihood of subsequent release as commercial varieties. Through 2019, Tsn1 marker data were available for 350 lines, which came from public and private breeding programs in states throughout the eastern U.S. soft wheat area, from Texas to Michigan and eastward. Only 9% carried the Tsn1 marker. With the exception of cv. Bess (PI 642794), the 32 Tsn1-bearing genotypes were all experimental lines, and it is unknown how many were subsequently released for commercial use.
Taken together, the information suggests that Tsn1 probably occurs at relatively high frequency in wheat cultivars planted in North Dakota, South Dakota, and Minnesota, given the high frequency of SnToxA in the corresponding P. nodorum population, and thus Tsn1 may contribute to variation in SNB susceptibility in wheat crops in that region. Tsn1 may also be common in hard winter wheats from central states farther south, such as Oklahoma and Kansas. However, Tsn1 is rare in eastern U.S. commercial SRWW cultivars. It appears to have been nearly absent from the foundational germplasm of that market class, and although the gene was subsequently introduced into SRWW, it has remained rare, perhaps due to phenotypic selection for SNB resistance. Other sensitivity genes, in particular Snn3, could be more widely present in eastern U.S. wheat production.
The efficacy of such phenotypic selection is best illustrated in the U.K. There, breeding for SNB resistance succeeded in reducing the disease to inconsequential levels (Scott and Benedikz, 1977; Scott et al., 1982; Brown, 2015) despite the absence of relevant information about Snn genes until a recent survey. In a panel of 480 northwest European varieties, of which approximately 330 were from the U.K., sensitivity to SnTox3 was more common than that to SnTox1, while sensitivity to SnToxA was rare (Downie et al., 2018). The relative contributions of Snn genes and other, minor genes to susceptibility remain to be investigated in the U.K.
In contrast with the findings of Downie et al. (2018), SnToxA sensitivity was more common in Scandinavian spring wheats: among 157 genotypes, the majority Norwegian and Swedish and the rest from CIMMYT, sensitivity to SnToxA was present in 45% (Ruud et al., 2018). In keeping with this relatively high frequency of SnToxA sensitivity in Scandinavian spring wheats, 69% of a sample of Norwegian P. nodorum isolates had the SnToxA gene; the frequencies of SnTox1 and SnTox3 were 53 and 76%, respectively (Ruud et al., 2018). The higher SnToxA frequency is the main difference with the only other survey to date of NE in western European P. nodorum: there, frequencies were reported of about 12% SnToxA, 85% SnTox1, and 65% SnTox3 in samples mainly from Switzerland, Sweden, and Denmark (McDonald et al., 2013).
Ruud et al. (2018) found some association between field susceptibility and sensitivity to SnToxA, but not to either of the other NE. This is to date the only evidence that Snn and NE frequencies are factors explaining SNB resistance in European wheat production. In other words, it appears that SnToxA-Tsn1 interactions are important in Norway, likely because the prevalence of Tsn1 in spring wheat germplasm has selected for a relatively high frequency of SnToxA in the pathogen population. There is no evidence that SnToxA-Tsn1 interactions or other Snn–NE interactions are important in other, primarily winter-wheat producing regions of western Europe.
Available information from Australia also leads to the conclusion that NE sensitivity is not closely related to disease phenotype in the field. Neither individual sensitivity to SnToxA, to SnTox1, or to SnTox3 was consistently significantly associated with field disease resistance scores of representative Australian wheat cultivars in more than one year (Tan et al., 2014). A P. nodorum strain lacking SnToxA, SnTox1, and SnTox3 was found to produce one or more novel NE in culture filtrates, but there was no clear relationship between sensitivity to that new NE and SNB susceptibility in 46 tested cultivars with varying degrees of resistance (Tan et al., 2015). Only by pooling scores of sensitivity to SnToxA, SnTox1, and SnTox3 and the novel NE did the authors detect a significant association between NE sensitivity and field disease score in the 46 cultivars, and even then a quarter of the more susceptible lines had low pooled NE sensitivity and about 40% of more resistant lines had high pooled NE sensitivity (Tan et al., 2015).
Given the above, what is known about the genetics of wheat resistance to SNB? The Tsn1-SnToxA interaction can confer susceptibility in the field in regions where Tsn1 has not been selected against. However, contrary to what is sometimes stated [e.g., (Waters et al., 2011; Tan et al., 2015; Phan et al., 2018)], there is no reason to suppose that in general, wheat-P. nodorum interactions are completely or even mainly defined by Snn–NE relationships, or that NE are required for pathogenesis. Resistance is polygenic, involving genes with minor effects, and foliar and glume infections are controlled by genes that segregate independently (Fried and Meister, 1987; Bostwick et al., 1993; Francki, 2013; Francki et al., 2017). While some of these minor interactions may involve NE and Snn genes, whether or not they have been characterized, others may well not.
Capitalizing on Long-Term Screening Data
Resistance to SNB is quantitative, and we sought evidence on its durability in commercial wheat production in the eastern U.S. If Snn–NE interactions were common in this region, one might expect cultivars would decline in quantitative resistance (QR) as common Snn genes exerted a selective effect by increasing the frequency of matching NE. To evaluate this hypothesis, we took advantage of data from a long-term resistance screening nursery to assess durability and to conduct a genome-wide association study (GWAS); both analyses are described below.
Between 2009 and 2018, 2,161 wheat genotypes (the vast majority SRWW) were phenotyped for SNB severity in the Eastern SNB Nursery of the USDA, the goal of which is to increase SNB resistance in the germplasm of participating breeding programs. Replicated field trials were performed in 16 year* location environments in North Carolina; in most years there were two locations, but in a few years there was only one due to crop or disease failure. Most genotypes were advanced experimental lines from 21 breeding programs in the central and eastern U.S., and were in the nursery fewer than 3 years; however, some commercial varieties were tested longer.
Genotypes were grown in plots consisting of two adjacent 1.3-meter rows, with two replicate plots per location. Each year, wheat straw infected by the natural P. nodorum population of North Carolina was spread evenly on the plots at Zadoks growth stage 25-29 (Zadoks et al., 1974; prior to stem elongation) to provide inoculum (Cowger and Murphy, 2007). The pathogen population in the nursery was therefore expected to broadly reflect the composition of that in North Carolina generally. During heading and anthesis, large-droplet irrigation was applied at some sites to enhance disease development and ensure inoculum dispersal to all upper plant parts, minimizing infection escape due to tall stature. Disease severity was rated using a 1–9 scale on a whole-plot basis at Zadoks growth stage 75–77 (medium to late milk stage of grain filling) for foliar and glume symptoms separately. Earlier-maturing lines were scored earlier than late-maturing lines, and no significant relationship was detected between SNB severity and heading date. Significant differences among genotypes were observed (https://www.ars.usda.gov/southeast-area/raleigh-nc/plant-science-research/docs/nursery-reports/page-6/).
Statistical Evidence of Durability of Eastern US Wheat QR to SNB
Large longitudinal datasets on varieties in official trials over years or decades are an underutilized resource that can provide insight into advances in breeding crop varieties for diverse traits (Brown et al., 2019; Figure 1 in Cowger and Brown, 2019). In the present case, to assess durability of QR to SNB, a linear mixed model was fitted to whole-plot SNB severity data from 19 SRWW cultivars that were tested at least 7 years each in the Eastern SNB Nursery trials, and one genotype (P26R61) that appeared 5 years in the trials and was known to possess QR to multiple diseases (Cowger and Brown, 2019, Figure 2). Variety and the linear effect of year were fixed effects and the interaction of variety and year was a random effect. While mean levels of foliar and glume SNB, estimated across all cultivars, increased significantly in the nursery over the period due to improved screening methodology such as irrigation (P < 0.002), there was only weak evidence for variation in the rate at which foliar SNB severity changed on cultivars with respect to each other (P = 0.06), and no such evidence with regard to glume SNB, demonstrating the durability of QR in this germplasm [for further details, see (Cowger and Brown, 2019)]. This degree of durability would not be expected if a large proportion of resistance were controlled by genes involved in specific interactions with P. nodorum genotypes, such as those for NE sensitivity.
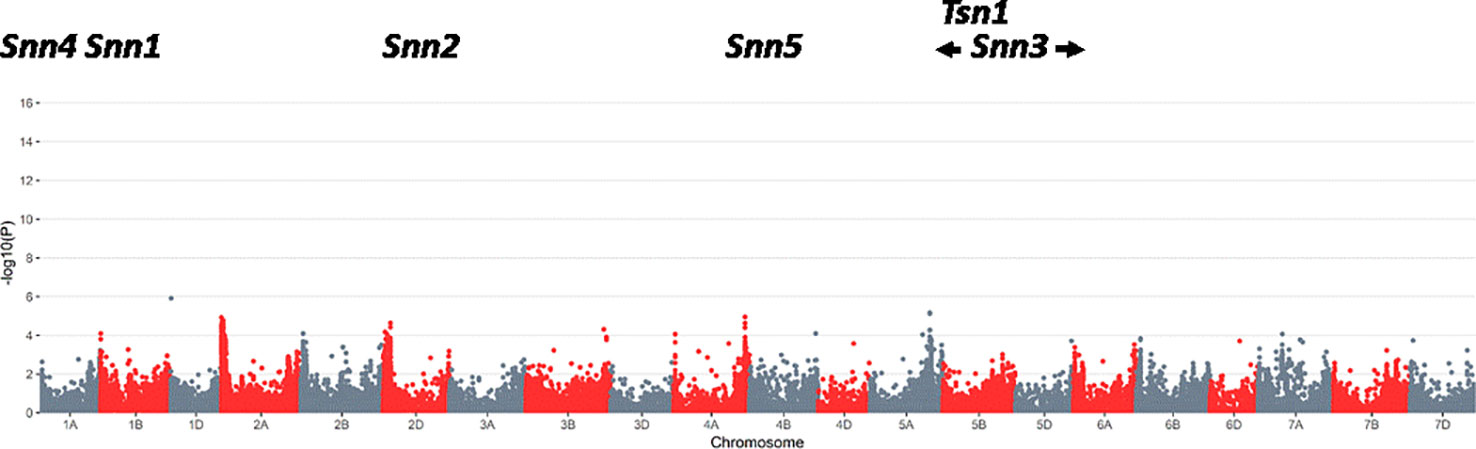
Figure 2 Results of a genome wide association study using SNPs and foliar severity of Septoria nodorum blotch on 1,231 wheat lines from the eastern U.S. during the period 2009 to 2018. No significant quantitative trait loci were detected for foliar resistance (false discovery rate threshold of 0.05), whereas a significant QTL was separately detected for glume resistance (data not shown; report in preparation).
Genome-Wide Association Study Reveals No Significant Loci for Foliar QR
A mixed-model GWAS was conducted on 1,231 of the lines phenotyped in the Eastern SNB Nursery from 2009 to 2018 for which genotyping-by-sequencing (GBS) data were available. The 1,231 lines were mainly from two regional nurseries: the Gulf Atlantic Wheat Nursery (GAWN), which comprises wheat lines from southern soft wheat states, and the Mason-Dixon Nursery, which includes wheat lines from the more northerly soft wheat states of Kentucky, Virginia, North Carolina and Maryland.
The GBS data included 54,856 SNPs. Briefly, genomic DNA was isolated from seedlings at the two-leaf stage. GBS library preparation was performed on genomic DNA according to Poland et al. (2012). The Burrows-Wheeler Aligner (Li and Durbin, 2009) v0.7.17 was used to align single-end Illumina reads to the Chinese Spring IWGSC RefSeq v1.0 wheat reference genome (International Wheat Genome Sequencing Consortium, 2018). Variant identification, SNP filtering and imputation were as described in Ward et al. (2019). The first five genotypic principal components, calculated for the SNP matrix using EIGENSTRAT (Price et al., 2006), were used to model and account for population structure, i.e., relatedness of genotypes. The Genome Wide Complex Trait Analysis (GCTA) software (Yang et al., 2011) was used to perform single-locus mixed linear model GWAS and to estimate the proportion of phenotypic variance explained by all SNPs jointly.
The GWAS revealed no significant QTL for foliar resistance to P. nodorum, including no significant effects in chromosomal regions near the Tsn1, Snn1, or Snn3 loci [at a false discovery rate (Benjamini and Hochberg, 1995) threshold of 0.05; Figure 2]. This was consistent with the low frequencies of Tsn1 and SnToxA found in SRWW and the southeastern P. nodorum population, respectively. It was notable that Snn3 did not contribute significantly to foliar resistance in the GWAS, despite the intermediate frequencies of Snn3 and SnTox3 in the host and pathogen populations. This suggests that SNB resistance in the study area is primarily determined by genes other than those conferring NE sensitivity, although it is possible that as-yet undetected Snn–NE pairs are playing very small roles.
It should be noted that a GWAS using the natural P. nodorum population is different from one designed to detect genes when only a fraction of either host or pathogen diversity is present. In other words, the use of one or a few isolates would give greater power to detect resistance loci that are effective against a specific set of pathogen genotypes, depending also on the host genes being tested. However, such a GWAS would also have to be done under controlled conditions to avoid confounding inoculum. The results would not be reflective of field performance of tested genotypes, whereas the present question was precisely whether Snn-SnTox interactions significantly influence field performance of regionally representative germplasm in the presence of the full range of pathogen diversity. In fact, the same GWAS that did not detect foliar QTL did detect a new major QTL for glume resistance, and a separate report on the novelty of that locus is in preparation. The foliar QR to SNB in eastern U.S. wheat germplasm appears to be governed by numerous loci, each with a small effect, consistent with previous studies in this region (Nelson and Gates, 1982; Bostwick et al., 1993), and with durable resistance to other foliar diseases of cereals (Brown, 2015).
Conclusion
The above evidence leads to two conclusions:
(1) Regional/national differences in SnToxA frequency are likely to mirror the frequency of Tsn1 in the wheat germplasm that is widely deployed in each area. Globally, evidence on whether breeders have selected against Tsn1 or other Snn genes is mixed and regionally specific. In commercial wheat production environments that are conducive to SNB, extensive use of Tsn1 in widely planted cultivars appears to have selected a high frequency of SnToxA in the P. nodorum population. Also, it may be that due to germplasm histories, spring wheat areas are more apt to have high Tsn1 and SnToxA frequencies than winter wheat areas, provided conditions are conducive to SNB. At the same time, SnToxA appears to exact a fitness cost, as it has a low frequency in the pathogen population where Tsn1 is selected against in current wheat varieties.
In the U.S., it appears there are important regional differences with regard to Tsn1. Eastern SRWW breeding programs started with a germplasm base in which Tsn1 was all but absent, and had apparently selected against the gene even before marker data became available in 2013, in part because SNB symptoms have been more ubiquitous, frequent and unconfounded by other leaf blotches than in other U.S. regions. Hard wheat breeding programs in the central U.S. states have apparently retained Tsn1, perhaps because it is a deliberately selected resistance gene and/or is linked to important agronomic traits such as hardness, and because phenotypic selection is hampered by infrequent occurrence of SNB epidemics.
(2) Snn–NE interactions do not appear to be an important factor in the QR observed in eastern U.S. soft wheat germplasm. The QR in this germplasm is evidently stable and relatively durable, and there is no evidence that the pathogen population is being strongly selected for aggressiveness as one might expect if Snn genes were exerting a substantial influence. Surprisingly, while Snn3 has been detected in soft wheat germplasm, the GWAS yielded no evidence that Snn3 by itself was important in explaining resistance variation, similar to findings in Norway and Australia. It is possible that as-yet undetected NE and Snn genes are playing a role in the host-pathogen relationship in the eastern U.S. It is also possible that as in other crop diseases (Cowger and Brown, 2019), this QR is governed by many genes with small effects that are not directly involved in Snn–NE interactions. A field screening program that utilizes the natural P. nodorum population of an area is an essential tool for selecting against susceptibility, whether Snn-linked or not.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
CC contributed field data, BW conducted GWAS, GB-G contributed marker data, and JB performed statistical analysis. CC led manuscript production and all authors contributed text and edited manuscript.
Funding
The research of CC, BW, and GB-G is funded by the United States Department of Agriculture-Agricultural Research Service (USDA-ARS), an equal opportunity provider and employer. The research of JB is supported by the Biotechnology & Biological Sciences Research Council Plant Health strategic programme.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Urmila Adhikari, Tyler Cole, Michael Elliott, Kimberly Howell, Keith Merrill, and Jared Smith for technical assistance.
Abbreviations
NE, necrotrophic effectors; GWAS, genome-wide association study; GBS, genotyping by sequencing; Snn gene, host sensitivity gene; QR, quantitative resistance; QTL, quantitative trait locus; SNP, single-nucleotide polymorphism; SNB, Septoria nodorum blotch; SRWW, soft red winter wheat; USDA, United States Department of Agriculture.
References
Abeysekara, N. S., Faris, J. D., Chao, S., McClean, P. E., Friesen, T. L. (2012). Whole-genome QTL analysis of Stagonospora nodorum blotch resistance and validation of the SnTox4–Snn4 interaction in hexaploid wheat. Phytopathology 102, 94–104. doi: 10.1094/PHYTO-02-11-0040
Allingham, E. A., Jackson, L. F. (1981). Variation in pathogenicity, virulence, and aggressiveness of Septoria nodorum in Florida. Phytopathology 71, 1080–1085. doi: 10.1094/Phyto-71-1080
Arraiano, L. S., Balaam, N., Fenwick, P. M., Chapman, C., Feuerhelm, D., Howell, P., et al. (2009). Contributions of disease resistance and escape to the control of septoria tritici blotch of wheat. Plant Pathol. 58, 910–922. doi: 10.1111/j.1365-3059.2009.02118.x
Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Bertucci, M., Brown-Guedira, G., Murphy, J. P., Cowger, C. (2014). Genes conferring sensitivity to Stagonospora nodorum necrotrophic effectors in Stagonospora nodorum blotch-susceptible U.S. wheat cultivars. Plant Dis. 98, 746–753. doi: 10.1094/PDIS-08-13-0820-RE
Bhathal, J. S., Loughman, R., Speijers, J. (2003). Yield reduction in wheat in relation to leaf disease from yellow (tan) spot and Septoria nodorum blotch. Eur. J. Plant Pathol. 109, 435–443. doi: 10.1023/A:1024277420773
Bostwick, D. E., Ohm, H. W., Shaner, G. (1993). Inheritance of septoria glume blotch resistance in wheat. Crop Sci. 33, 439–443. doi: 10.2135/cropsci1993.0011183X003300030005x
Brown, J. K. M., Beeby, R., Penfield, S. (2019). Yield instability of winter oilseed rape modulated by early winter temperature. Sci. Rep. 9, 6953. doi: 10.1038/s41598-019-43461-7
Brown, J. K. M. (2015). Durable resistance of crops: a Darwinian perspective. Annu. Rev. Phytopathol. 53, 513–539. doi: 10.1146/annurev-phyto-102313-045914
Chandramohan, P., Shaw, M. W. (2013). Sulphate and sulphurous acid alter the relative susceptibility of wheat to Phaeosphaeria nodorum and Mycosphaerella graminicola. Plant Pathol. 62, 1342–1349. doi: 10.1111/ppa.12052
Chu, C.-G., Faris, J. D., Xu, S. S., Friesen, T. L. (2010). Genetic analysis of disease susceptibility contributed by the compatible Tsn1–SnToxA and Snn1–SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120, 1451–1459. doi: 10.1007/s00122-010-1267-z
Cowger, C., Brown, J. K. M. (2019). Durability of quantitative resistance in crops: greater than we know? Annu. Rev. Phytopathol. 57, 253–277. doi: 10.1146/annurev-phyto-082718-100016
Cowger, C., Murphy, J. P. (2007). Artificial inoculation of wheat for selecting resistance to Stagonospora nodorum blotch. Plant Dis. 91, 539–545. doi: 10.1094/PDIS-91-5-0539
Cowger, C., McDonald, B. A., Mundt, C. C. (2002). Frequency of sexual reproduction by Mycosphaerella graminicola on partially resistant wheat cultivars. Phytopathology 92, 1175–1181. doi: 10.1094/PHYTO.2002.92.11.1175
Crook, A. D., Friesen, T. L., Liu, Z. H., Ojiambo, P. S., Cowger, C. (2012). Novel necrotrophic effectors from Stagonospora nodorum and corresponding host sensitivities in winter wheat germplasm in the southeastern United States. Phytopathology 102, 498–505. doi: 10.1094/PHYTO-08-11-0238
Cunfer, B. M. (1998). Seasonal availability of inoculum of Stagnospora nodorum in the field in the southeastern USA. Cereal Res. Commun. 26, 259–263. doi: 10.1007/BF03543497
Downie, R. C., Bouvet, L., Furuki, E., Gosman, N., Gardner, K. A., Mackay, I. J., et al. (2018). Assessing European wheat sensitivities to Parastagonospora nodorum necrotrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00881
Faris, J. D., Zhang, Z., Lu, H., Lu, S., Reddy, L., Cloutier, S., et al. (2010). A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. U. S. A. 107, 13544–13549. doi: 10.1073/pnas.1004090107
Francki, M. G., Walker, E., Li, D. A., Forrest, K. (2017). High-density SNP mapping reveals closely linked QTL for resistance to Stagonospora nodorum blotch (SNB) in flag leaf and glume of hexaploid wheat. Genome 61, 145–149. doi: 10.1139/gen-2017-0203
Francki, M. G. (2013). Improving Stagonospora nodorum resistance in wheat: a review. Crop Sci. 53, 355–365. doi: 10.2135/cropsci2012.06.0347
Fried, P. M., Meister, E. (1987). Inheritance of leaf and head resistance of winter wheat to Septoria nodorum in a diallel cross. Phytopathology 77, 1371–1375. doi: 10.1094/Phyto-77-1371
Friesen, T. L., Faris, J. D. (2010). Characterization of the wheat-Stagonospora nodorum disease system: what is the molecular basis of this quantitative necrotrophic disease interaction? Can. J. Plant Pathol. 32, 20–28. doi: 10.1080/07060661003620896
Gilbert, B. M., Wolpert, T. J. (2013). Characterization of the LOV1-mediated, victorin-induced, cell-death response with virus-induced gene silencing. Mol. Plant-Microbe Interact. 26, 903–917. doi: 10.1094/MPMI-01-13-0014-R
International Wheat Genome Sequencing Consortium, Appels, R., Eversole, K., Stein, N., Feuillet, C., Keller, B., et al. (2018). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361 (6403), eaar7191. doi: 10.1126/science.aar7191
Li, H., Durbin, R. (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, Z., Friesen, T. L., Ling, H., Meinhardt, S. W., Oliver, R. P., Rasmussen, J. B., et al. (2006). The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49, 1265–1273. doi: 10.1139/g06-088
McDonald, M. C., Oliver, R. P., Friesen, T. L., Brunner, P. C., McDonald, B. A. (2013). Global diversity and distribution of three necrotrophic effectors in Phaeosphaeria nodorum and related species. New Phytol. 199, 241–251. doi: 10.1111/nph.12257
Nagy, E. D., Bennetzen, J. L. (2008). Pathogen corruption and site-directed recombination at a plant disease resistance gene cluster. Genome Res. 18, 1918–1923. doi: 10.1101/gr.078766.108
NASS, National Agricultural Statistics Service (2019). U.S. and state data. United States Department of Agriculture, http://www.nass.usda.gov/index.asp.
Nelson, L. R., Gates, C. E. (1982). Genetics of host plant resistance of wheat to Septoria nodorum. Crop Sci. 22, 771–773. doi: 10.2135/cropsci1982.0011183X002200040017x
Nelson, L. R., Morey, D. D., Brown, A. R. (1974). Wheat cultivar responses to severe glume blotch in Georgia. Plant Dis. Rep. 58, 21–23.
Oliver, R. P., Rybak, K., Solomon, P. S., Ferguson-Hunt, M. (2009). Prevalence of ToxA-sensitive alleles of the wheat gene Tsn1 in Australian and Chinese wheat cultivars. Crop Pasture Sci. 60, 348–352. doi: 10.1071/CP08259
Peters-Haugrud, A. R., Zhang, Z., Richards, J. K., Friesen, T. L., Faris, J. D. (2019). Genetics of variable disease expression conferred by inverse gene-for-gene interactions in the wheat-Parastagonospora nodorum pathosystem. Plant Physiol. 180, 420–434 pp.00149.02019. doi: 10.1104/pp.19.00149
Phan, H. T. T., Rybak, K., Bertazzoni, S., Furuki, E., Dinglasan, E., Hickey, L. T., et al. (2018). Novel sources of resistance to Septoria nodorum blotch in the Vavilov wheat collection identified by genome-wide association studies. Theor. Appl. Genet. 131, 1223–1238. doi: 10.1007/s00122-018-3073-y
Poland, J. A., Brown, P. J., Sorrells, M. E., Jannink, J.-L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PloS One 7, e32253. doi: 10.1371/journal.pone.0032253
Price, A., Patterson, N., Plenge, R. M., Weinblatt, M. E., Shadick, N. A., Reich, D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. doi: 10.1038/ng1847
Richards, J. K., Stukenbrock, E. H., Carpenter, J., Liu, Z., Cowger, C., Faris, J. D., et al. (2019). Local adaptation drives the diversification of effectors in the fungal wheat pathogen Parastagonospora nodorum in the United States. PloS Genet. 15(10), e1008223. doi: 10.1371/journal.pgen.1008223
Ruud, A. K., Dieseth, J. A., Lillemo, M. (2018). Effects of three Parastagonospora nodorum necrotrophic effectors on spring wheat under Norwegian field conditions. Crop Sci. 58, 159–168. doi: 10.2135/cropsci2017.05.0281
Scott, P. R., Benedikz, P. W. (1977). Field techniques for assessing reaction of winter wheat cultivars to Septoria nodorum. Ann. Appl. Biol. 85, 345–358. doi: 10.1111/j.1744-7348.1977.tb01920.x
Scott, P. R., Benedikz, P. W., Cox, C. W. (1982). A genetic study on the relationship between height, time of ear emergence, and resistance to Septoria nodorum in wheat. Plant Pathol. 31, 45–60. doi: 10.1111/j.1365-3059.1982.tb02811.x
Shah, D. A., Bergstrom, G. C. (1993). Assessment of seedborne Stagonospora nodorum in New York soft white winter wheat. Plant Dis. 77, 468–471. doi: 10.1094/PD-77-0468
Shaner, G., Buechley, G. (1995). Epidemiology of leaf blotch of soft red winter wheat caused by Septoria tritici and Stagonospora nodorum. Plant Dis. 79, 928–938. doi: 10.1094/PD-79-0928
Shi, G., Zhang, Z., Friesen, T. L., Raats, D., Fahima, T., Brueggeman, R. S., et al. (2016). The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2, e1600822–e1600822. doi: 10.1126/sciadv.1600822
Stukenbrock, E. H., McDonald, B. A. (2007). Geographical variation and positive diversifying selection in the host-specific toxin SnToxA. Mol. Plant Pathol. 8, 321–332. doi: 10.1111/j.1364-3703.2007.00396.x
Tan, K.-C., Waters, O. D. C., Rybak, K., Antoni, E., Furuki, E., Oliver, R. P. (2014). Sensitivity to three Parastagonospora nodorum necrotrophic effectors in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci. 65, 150–158. doi: 10.1071/CP13443
Tan, K.-C., Phan, H. T. T., Rybak, K., John, E., Chooi, Y. H., Solomon, P. S., et al. (2015). Functional redundancy of necrotrophic effectors - consequences for exploitation for breeding. Front. Plant Sci. 6, 501–501. doi: 10.3389/fpls.2015.00501
USDA Economic Research Service (2019). Wheat sector at a glance. U.S. Department of Agriculture. https://www.ers.usda.gov/topics/crops/wheat/wheat-sector-at-a-glance/#classes. Last updated August 20, 2019.
Ward, B. P., Brown-Guedira, G., Kolb, F. L., Van Sanford, D. A., Tyagi, P., Sneller, C. H., et al. (2019). Genome-wide association studies for yield-related traits in soft red winter wheat grown in Virginia. PloS One. 14(2), e0208217. doi: 10.1371/journal.pone.0208217
Waters, O. D. C., Lichtenzveig, J., Rybak, K., Friesen, T. L., Oliver, R. P. (2011). Prevalence and importance of sensitivity to the Stagonospora nodorum necrotrophic effector SnTox3 in current Western Australian wheat cultivars. Crop Pasture Sci. 62, 556–562. doi: 10.1071/CP11004
Yang, J., Lee, S. H., Goddard, M. E., Visscher, P. M. (2011). GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82. doi: 10.1016/j.ajhg.2010.11.011
Keywords: Parastagonospora nodorum, Septoria nodorum blotch, quantitative resistance, genotyping by sequencing, genome-wide association study, necrotrophic effectors, necrotrophic effector sensitivity genes
Citation: Cowger C, Ward B, Brown-Guedira G and Brown JKM (2020) Role of Effector-Sensitivity Gene Interactions and Durability of Quantitative Resistance to Septoria Nodorum Blotch in Eastern U.S. Wheat. Front. Plant Sci. 11:155. doi: 10.3389/fpls.2020.00155
Received: 30 October 2019; Accepted: 31 January 2020;
Published: 06 March 2020.
Edited by:
Morten Lillemo, Norwegian University of Life Sciences, NorwayReviewed by:
Thomas Miedaner, University of Hohenheim, GermanyMarja Johaana Jalli, Natural Resources Institute Finland (Luke), Finland
Copyright © 2020 Cowger, Ward, Brown-Guedira and Brown. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christina Cowger, Q2hyaXN0aW5hLkNvd2dlckB1c2RhLmdvdg==
 Christina Cowger
Christina Cowger Brian Ward
Brian Ward Gina Brown-Guedira
Gina Brown-Guedira James K. M. Brown
James K. M. Brown