
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 06 March 2020
Sec. Plant Physiology
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00140
 Hagit Zer1
Hagit Zer1 Hila Mizrahi1
Hila Mizrahi1 Nikol Malchenko1
Nikol Malchenko1 Tamar Avin-Wittenberg1
Tamar Avin-Wittenberg1 Liron Klipcan2*
Liron Klipcan2* Oren Ostersetzer-Biran1*
Oren Ostersetzer-Biran1*Plants produce a myriad of specialized (secondary) metabolites that are highly diverse chemically, and exhibit distinct biological functions. Here, we focus on meta-tyrosine (m-tyrosine), a non-proteinogenic byproduct that is often formed by a direct oxidation of phenylalanine (Phe). Some plant species (e.g., Euphorbia myrsinites and Festuca rubra) produce and accumulate high levels of m-tyrosine in their root-tips via enzymatic pathways. Upon its release to soil, the Phe-analog, m-tyrosine, affects early post-germination development (i.e., altered root development, cotyledon or leaf chlorosis, and retarded growth) of nearby plant life. However, the molecular basis of m-tyrosine-mediated (phyto)toxicity remains, to date, insufficiently understood and are still awaiting their functional characterization. It is anticipated that upon its uptake, m-tyrosine impairs key metabolic processes, or affects essential cellular activities in the plant. Here, we provide evidences that the phytotoxic effects of m-tyrosine involve two distinct molecular pathways. These include reduced steady state levels of several amino acids, and in particularly altered biosynthesis of the phenylalanine (Phe), an essential α-amino acid, which is also required for the folding and activities of proteins. In addition, proteomic studies indicate that m-tyrosine is misincorporated in place of Phe, mainly into the plant organellar proteomes. These data are supported by analyses of adt mutants, which are affected in Phe-metabolism, as well as of var2 mutants, which lack FtsH2, a major component of the chloroplast FtsH proteolytic machinery, which show higher sensitivity to m-tyrosine. Plants treated with m-tyrosine show organellar biogenesis defects, reduced respiration and photosynthetic activities and growth and developmental defect phenotypes.
The chemical diversity of terrestrial plants is truly exceptional. Plants are estimated to produce hundreds of thousands of different metabolites, probably the largest number among all other species (see e.g., Moghe and Last, 2015; Wurtzel and Kutchan, 2016; Obata, 2019). Such an exceptional diversity may be a consequence of the large diversification and rapid evolution of specialized metabolic pathways in plants since they occupied the terrestrial environment, about 500 million years ago (Morris et al., 2018). Phytochemicals produced by land-plants are also of great economic and ecological importance, as herbicidal compounds to control weeds.
Phytochemicals, as other natural compounds, are classified into two main categories. Primary metabolites that are associated with essential cellular functions, e.g., nucleotides, amino acids, fatty acids, sugars, and organic acids, which are typically present in all organisms and cells. In addition, organisms also produce numerous specialized (secondary) metabolites that are not essential to basic physiological functions, but otherwise play important roles during specific growth and developmental stages, and aid in adapting and adjusting the developmental need of the plant with specific physiological or environmental signals (Witzany, 2006; Tissier et al., 2014; Moghe and Last, 2015; Wurtzel and Kutchan, 2016; Li et al., 2018). Specialized compounds produced in plants were shown to have key roles in defense mechanisms, in inter- or intracellular signaling, coloring, regulation of primary metabolism, as well as in allelochemisrty (i.e., biomolecules produced by one organism that have a physiological effect on another species when released to the environment) (Rizvi and Rizvi, 1992).
Plants have complex relationships with other organisms, which involve physical and chemical interactions with their surrounding (reviewed by e.g., van Loon, 2016; Bennett and Klironomos, 2018; Mhlongo et al., 2018). These are indicated, for example, by complex interactions between different plants and microorganisms in the soil. Growing roots also need to compete with their neighboring plants, and at the same time to attract beneficial microorganisms to supply them with minerals and nutrients, e.g., root nodules in legumes, where bacterial symbionts fix atmospheric nitrogen (van Loon, 2016; Bennett and Klironomos, 2018; Mhlongo et al., 2018). Accordingly, many chemical signals are exchanged between plants and their neighboring organisms in the soil (Mhlongo et al., 2018). Our study focuses on meta-tyrosine (m-tyrosine, or m-Tyr), an oxidized byproduct of the aromatic amino acid phenylalanine, which inhibits early post-germination and seedling growth (Bertin et al., 2007; Bertin et al., 2009).
At various conditions and especially under stresses, the production of tyrosine isomers para- (i.e., the native amino acid), meta-, and ortho-tyrosine, can spontaneously occur by the oxidation of the benzyl ring of phenylalanine (Mager and Berends, 1974). A few plant species, including Euphorbia myrsinites (donkey-tail spurge) and Festuca rubra (red fescue) synthesize enzymatically the isomer meta-tyrosine (m-tyrosine) in their root tip tissues, and then secrete it to the soil to inhibit nearby plant-life (Mothes et al., 1964; Bertin et al., 2007). In E. myrsinites, m-tyrosine is produced via a transamination of m-hydroxyphenylpyruvate (Mothes et al., 1964), whereas the biosynthesis of m-tyrosine in F. rubra is mediated directly through the hydroxylation of phenylalanine (Bertin et al., 2007; Huang et al., 2012). These data indicate that different plants utilize m-tyrosine as a phytotoxic allelochemical using distinct metabolic pathways, activities which likely arose independently during the evolution of land plants (Huang et al., 2012). While m-tyrosine is highly toxic to plants, its structural related isomers, o- and p-tyrosine, have no or only little effects on the germination, growth, or development of land plants (Bertin et al., 2007).
The molecular basis for m-tyrosine toxicity has not been resolved yet. We show, by biochemical and genetic approaches, that m-tyrosine affects early post-germination development of Arabidopsis plantlets by altering the biosynthesis of various amino acids, and in particular the aromatic amino acid Phenylalanine (Phe). Proteomic analyses of young Arabidopsis seedlings, grown in the absence or presence of m-tyrosine, strongly support that m-tyrosine is misincorporated instead of Phe to various organellar (i.e., mitochondrial and plastidial) proteins, an activity which is likely mediated by a dually-localized organellar phenylalanine-transfer RNA (tRNA) synthetase (PheRS) enzyme (Duchêne et al., 2005).
Plants growth and analyses generally followed the procedures described in (Sultan et al., 2016). Arabidopsis thaliana ecotype Columbia (Col-0) seeds were obtained from the ABRC center, at Ohio State University (Columbus, OH). Arabidopsis mutants of different arogenate dehydratase encoding genes (ADT1, At1g11790; ADT3, At2g27820; ADT4, At3g44720; ADT5, At5g22630; ADT6, At1g08250) (Chen et al., 2016; Höhner et al., 2018), were generously provided by Prof. Jirong Huang (Shanghai Normal University), while mutants in FtsH2 (At2g30950, also denoted as var2) (Takechi et al., 2000; Sakamoto et al., 2003; Zaltsman et al., 2005) were generously given by Prof. Zach Adam (The Hebrew University). We further analyzed the Arabidopsis atg7-2 (At5g45900; GK-655B06) and atg5-1 (At5g17290; SAIL-129B079) mutant-lines (Hofius et al., 2009; Yoshimoto et al., 2009; Avin-Wittenberg et al., 2015), which are affected in autophagy. Prior to their germination, the seeds of wild-type and mutant lines were surface sterilized by a vapor-phase method, using a 50 ml sodium hypochlorite (bleach, 6%) solution supplemented with 1.5 ml HCl (37%) solution. The sterilized seeds were sown on Murashige and Skoog (MS)-agar plates, incubated in the dark for 2 days at 4°C, and then transferred to controlled temperature (22°C) and humidity (50%) growth chamber (Percival Scientific, Perry, IA, USA), under short day conditions (8-h light, 250 µE·m-2·s-1 and 16-h dark).
For the analysis of plant morphology, plant tissues (i.e., leaves and roots) where obtained from 5-day-old Arabidopsis plants grown on MS-plates in the presence or absence of 10 μM m-tyrosine. The morphologies of mitochondria and plastids were established by transmission electron microscopy (TEM) of ultrathin plant sections, using Tecnai 12 TEM 100 kV (Phillips, Eindhoven, the Netherlands) microscope equipped with MegaView II CCD camera and Analysis® version 3.0 software (SoftImaging System GmbH, Münstar, Germany), at the Bio-Imaging unit of the Institute of Life Sciences (The Hebrew University of Jerusalem). The relative densities (i.e., pixel intensities) of thylakoid grana membrane stacks and the average surface area of mitochondria have been manually evaluated from TEM images of ultrathin sections of 5-day-old plantlets grown in the absence or presence of m-tyrosine, using the ImageJ software (Version 1.52a) (Jensen, 2013). Student's t-test was performed to determine significant differences (P ≤ 0.05).
Crude organellar proteins were prepared essentially as described previously (Pineau et al., 2008; Shevtsov et al., 2018). In brief, organellar membrane extracts were obtained from 200 mg Arabidopsis seedlings (5 days-old), grown in the presence or absence of 10 μM m-tyrosine supplemented to the growth media. The seedlings were then homogenized in 2 ml of 75 mM MOPS-KOH, pH 7.6, 0.6 M sucrose, 4 mM ethylenediaminetetraacetic acid (EDTA), 0.2% polyvinylpyrrolidone-40, 8 mM cysteine, 0.2% bovine serum albumin (BSA), and protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany). Protein concentration was determined by the Bradford method (Bio-Rad, Catalog no. 5000201), according to the manufacturer's protocol. For immunoassays, crude membrane fraction were suspended in sample loading buffer (Laemmli, 1970) and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (at a constant 100 V). Following electrophoresis, the proteins were transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Catalog no. 1620177), essentially as described previously (Eubel et al., 2005), and incubated overnight at 4°C with various antibodies (Table S1). Detection was carried out by chemiluminescence assay after incubation with an appropriate horseradish peroxidase (HRP)-conjugated secondary antibody.
Blue native (BN)-PAGE of organellar membranous complexes was performed according to the methods described previously (Pineau et al., 2008; Shevtsov et al., 2018). Crude organellar membranes were solubilized with n-dodecyl-ß-maltoside [DDM; 1.5% (w/v)] and loaded onto a native 4 to 16% linear gradient gel. For immunoblotting of non-denaturing PAGE, the proteins were transferred from the gel onto a PVDF membrane (Bio-Rad, Catalog no. 1620177). The membranes were then incubated with specific primary antibodies (Supplemental Table S1), and detection was carried out by chemiluminescence assay after incubation with horseradish peroxidase (HRP)-conjugated “secondary” antibodies.
Following the extraction of total protein from 5-day-old Arabidopsis seedlings and crude organellar preparations (Pineau et al., 2008; Shevtsov et al., 2018), total proteins were obtained by the borate/ammonium acetate method (Maayan et al., 2008). For this purpose, plant tissues were homogenized in the presence of polyvinylpolypyrrolidone (PVPP). The homogenate was added to microfuge tubes containing 400 ml ice-cold protein extraction buffer [50 mM Na-borate, 50 mM ascorbic acid, 1.25% (w/v) sodium dodecyl sulfate (SDS), 12.5 mM β-mercaptoethanol, pH 9.0] and the protease inhibitor cocktail “complete Mini” from Roche Diagnostics GmbH (Mannheim, Germany). Proteins were recovered by centrifugation (25,000 g) in the presence of three volumes of ice-cold 0.1 M ammonium acetate in methanol buffer (NH4-OAc-MeOH), following (80% v/v) acetone precipitation. The protein pellet was resuspended with 25 mM Tris-HCl pH 8.0, 10 mM dithiothreitol (DTT), 2% SDS buffer solution. Protein concentration was determined according to the Bradford method, with BSA used as a standard. Twenty-five micrograms of protein was alkylated with 55 mM iodoacetamide (Sigma Chem. Corp. St. Louis, MO) for 30 min at room-temperature in the dark. Removal of SDS followed by digestion with trypsin (Promega Corp., Madison, WS) were performed using the S-Trap microspin column kit (ProtiFi, LLC, Huntington, NY), according to the manufacturer protocol. The tryptic peptides were then desalted, as previously described (Rappsilber et al., 2007). A total of 1.5 µg of peptides from each sample were injected into the mass spectrometer (Q Exactive Plus mass spectrometer, Thermo Fisher Scientific, USA). The data were analyzed using the PEAKS proteomics software, version 8 (Bioinformatics Solutions Inc., Waterloo, ON, Canada), with specified Phe to Tyr replacements (i.e., the mass of m-tyrosine is equal to Tyr) (see Table S3).
The extraction of Arabidopsis plant free amino acids (and various other metabolites) was performed according to a previously described procedure with some modifications by (Roessner-Tunali et al., 2003). In general, 50 mg Arabidopsis seedlings (5-day-old) were frozen in liquid nitrogen and homogenized in 700 µl of MeOH, in the presence of 30 µl ribitol (0.2 mg/ml in DDW) as an internal standard. Extraction, derivatization, standard addition, and sample injection by gas chromatography–mass spectrometry (GC-MS) were performed essentially as described previously (Lisec et al., 2006). The GC-MS system comprised of the an Agilent 7693 Autosampler, Agilent J&W DB-35ms column, Agilent 7200B gas chromatograph, quadrupole time-of-flight MS with removable electron ionization source. Amino acids were identified in comparison to commercial standard compounds purchased from Sigma. Chromatograms and mass spectra were evaluated using MassHunter Data Analysis by Agilent, Quantitative and Qualitative Analysis (TOF). Calculations of the relative free amino acid levels were evaluated by calculating the relative pick area of each amino acid in untreated versus m-tyrosine grown seedlings. For detailed parameters see Supplemental Dataset 1 (https://figshare.com/articles/Frontiers_in_Plant_Sciences_MS_data/11627211).
Specialized phytochemical compounds are highly diverged chemically, and are expected to arise in plants to aid with key biological functions, such as in surviving and communicating with other organisms in complex environmental niches. F. rubra, a grass species that is widespread across the northern hemisphere, releases to the soil biochemicals that inhibit the growth of nearby plant-life. NMR-spectroscopy led to the identification of m-tyrosine as the key phytotoxic compound within root exudates of F. rubra plants (Bertin et al., 2007). Plants treated with exogenous m-tyrosine are characterized by shortened roots and reduced biomass phenotypes [Bertin et al. (2007) and Supplemental Figure S1]. The phytotoxic effects of m-tyrosine seem to be mediated by mechanisms that are not related to auxin (Bertin et al., 2007). Our analyses further indicate that Arabidopsis seedlings germinated in the presence of m-tyrosine (i.e., 0 to 320 μM), show post-germination developmental defects, and strong effects on root growth (calculated IC50 value of 2.365 μM; i.e., concentrations required to achieve 50% reduction of Arabidopsis root growth) (Supplemental Figure S1 and Table 1).
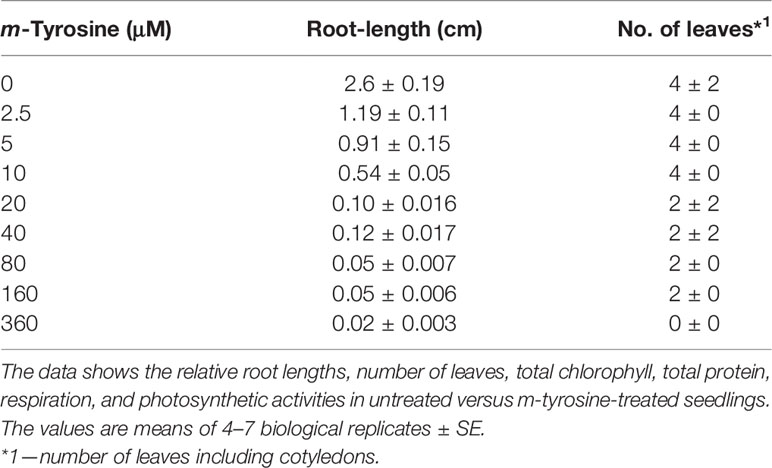
Table 1 Effects of m-tyrosine on root length and leaf number of 5-day-old Arabidopsis thaliana (col-0) seedlings.
The molecular basis for m-tyrosine mediated phytotoxicity is largely unclear in plants. Given its chemical properties, we speculate that m-tyrosine might be incorporated to the plant proteome and/or interferes with cellular metabolism. To address this, we analyzed the relative accumulation of free amino acids in 5-day-old Arabidopsis thaliana seedlings grown in the absence (control, −m-Tyr) or presence of 10 μM m-tyrosine (+m-Tyr) by GC-MS analysis (see Figure 1, and Supplemental Table S2 and S3). The accumulation of various amino acids was evaluated relatively to the control (non-treated) plants grown under the same growth conditions (see Table S3).
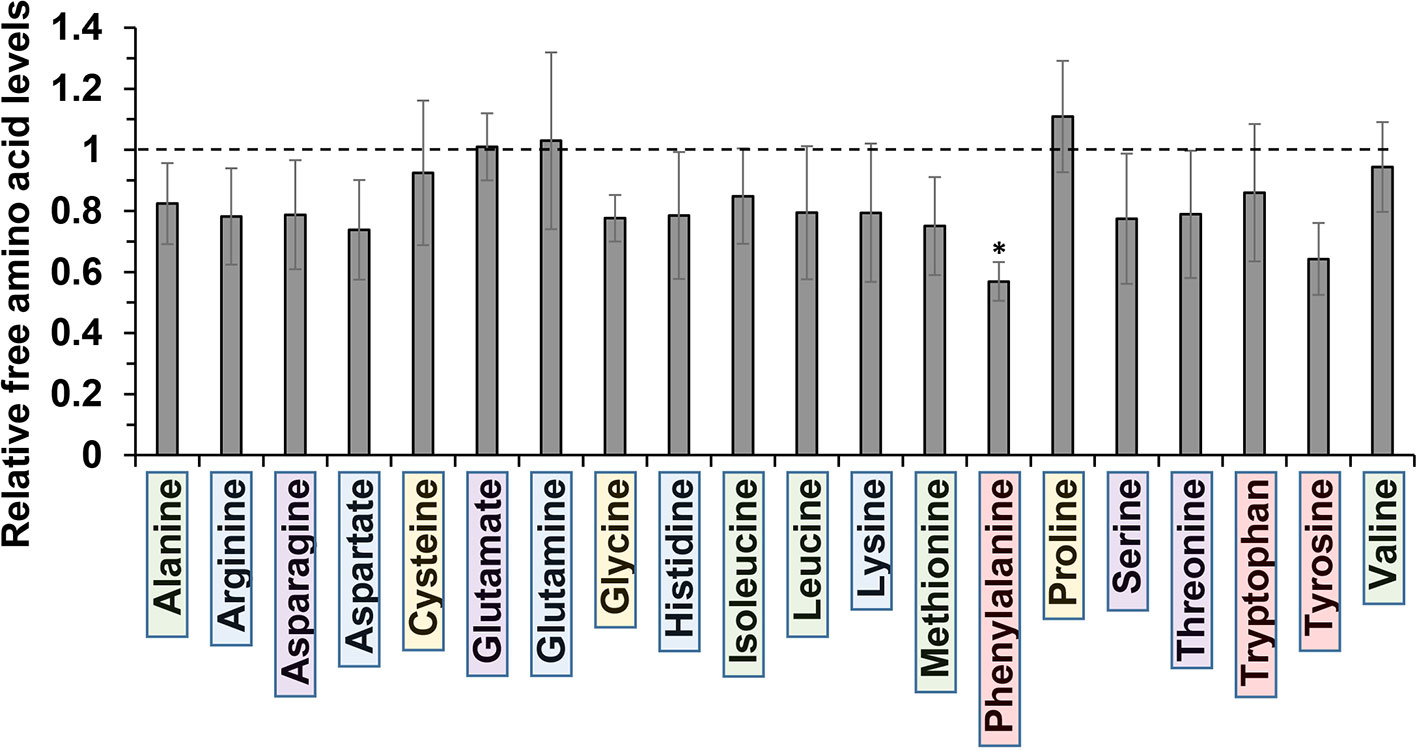
Figure 1 Relative change in free amino acid levels in 5-day-old Arabidopsis thaliana seedlings grown in the presence or absence of 10 μM m-tyrosine. Arabidopsis seeds were germinated in Murashige and Skoog (MS)-agar plates in the absence or presence of 10 μM m-tyrosine (m-Tyr). Amino acids, extracted from 50 mg of 5-day-old Arabidopsis seedlings, were identified in comparison to commercial standard compounds from each treatment (i.e., control and m-Tyr-grown seedlings), in the presence of ribitol (0.2 mg/ml) as an internal standard. Extraction, derivatization, standard addition, and sample injection were performed essentially as described previously (Lisec et al., 2006). Calculations of the relative free amino acid levels were evaluated from the relative pick area of each amino acid in untreated versus m-tyrosine-treated seedlings. The values are means of four biological replicates, while error bars indicate one standard deviation (see Table S2). Color boxes indicate to functional groups: in green hydrophobic side chains, blue for electrically charged side chains, purple for polar (uncharged) side chains, red for aromatic residues, and yellow indicates special side chains.
Under “normal” growth conditions (see Materials and Methods), the steady-state levels of various amino acids, including the nonpolar (hydrophobic) Ala, Ile, Leu, Met, and Val, the uncharged Ser and Thr, positively charged Arg, His, and Lys, the negatively charged Asp, as well as the aromatic residues Trp and Tyr, were all reduced to some extent (i.e., from 7 to 26%) in the m-tyrosine germinated seedlings (Figure 1, and Table S2). However, only Phe showed a statistically significant reduction (43.1 ± 6.3%) in the treated plants (see Figure 1, and Table S2). The nonpolar amino acid Pro, which is known to accumulate under different stress conditions (Szabados and Savouré, 2010), was found to be somewhat higher (110.9 ± 18.2%) in plants germinated in the presence of m-tyrosine, whereas the steady-state levels of the negative charged Glu (1.01 ± 0.14%) and the uncharged Gln (103.0 ± 2.9%) amino acids were not significantly affected by the addition of m-tyrosine to the growth media (Figure 1, Table S2).
Growing plants in the presence of exogenous amino acids was previously shown to partially restore the growth and developmental defect phenotypes of Arabidopsis seedlings treated with m-tyrosine (Bertin et al., 2007). Root growth inhibition by 3 μM m-tyrosine was counteracted by 14 out of 21 different amino acids given at 40 μM concentrations (Bertin et al., 2007). However, when the amino acids were administered to the growth media together with a higher concentration (i.e., 10 μM) of m-tyrosine, only Phe showed a notable “rescue” effect on Arabidopsis root growth inhibition (Figure 2).
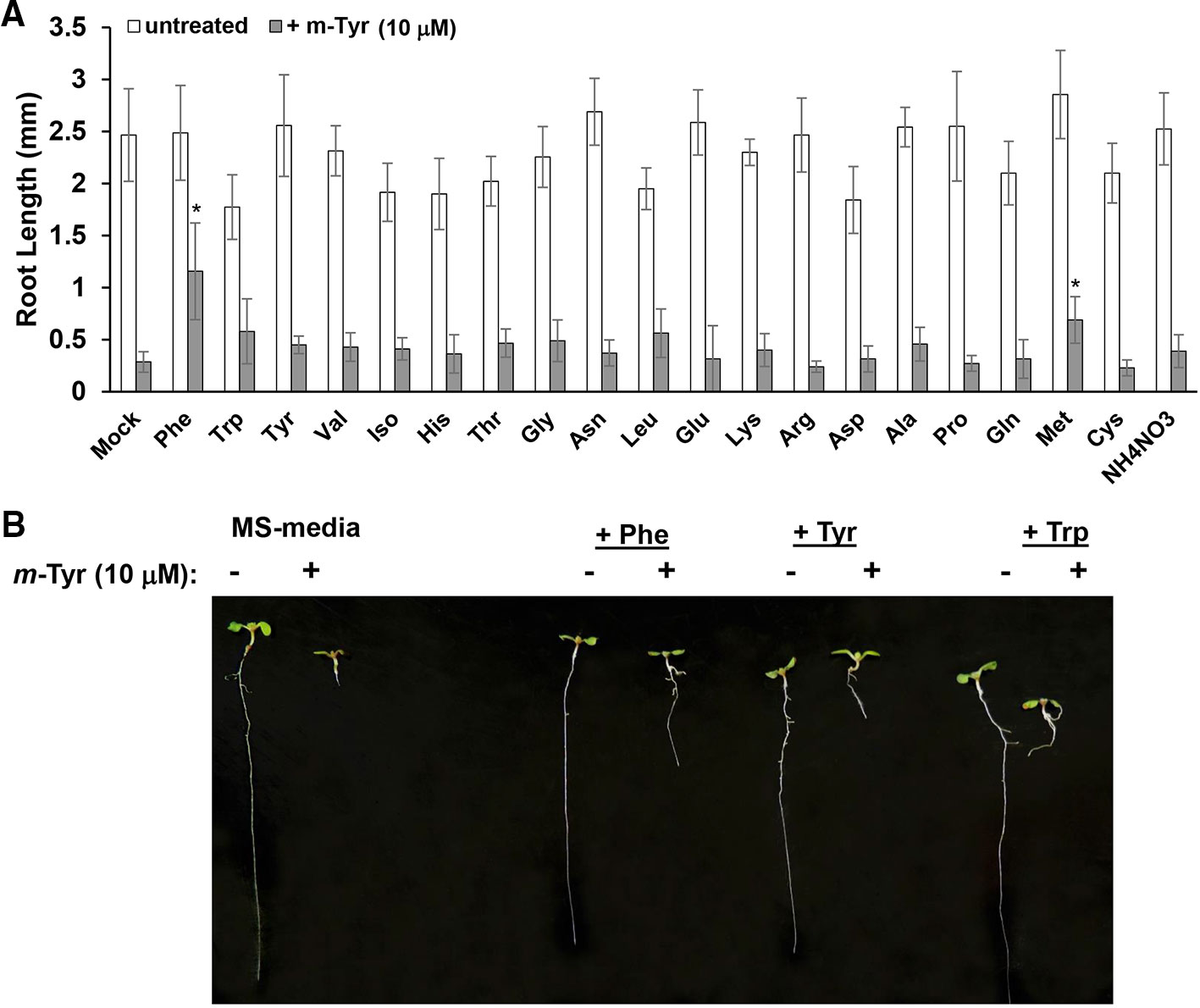
Figure 2 The effect of exogenous amino acids on m-tyrosine mediated toxicity. (A) Five-day-old Arabidopsis seeds were germinated in Murashige and Skoog (MS)-agar plates containing various amino acids (40 μM), in the absence or presence of 10 μM m-tyrosine. (B) Measurements of the root lengths in 5-day-old Arabidopsis seedlings. Bar in (panel A) represents 1.0 cm. The values are means of three biological replicates with about 25 seedlings in each treatment. Error bars indicate one standard deviation. Asterisks in (panel B) indicate a significant difference from 10 μM m-Tyr grown plants (Student's T-test, P ≤ 0.05).
The biosynthesis of Phe in various organisms involves a series of enzymatic reactions, which convert prephanate (a product of the shikimate pathway) into Phe, from either phenylpyruvate or arogenate (the later serves as the primarily route for Phe synthesis in angiosperms) (Cho et al., 2007; Tzin and Galili, 2010). In Arabidopsis, arogenate dehydratase (ADT) catalyzes the final step in Phe biosynthesis (i.e., decarboxylation of arogenate), a reaction that takes place within the plastids (Tohge et al., 2013). Arabidopsis contains six closely related ADT genes, which differ in their organ locations and contribution to Phe accumulation (Höhner et al., 2018). Arabidopsis ADT-deficient mutants show altered photosynthetic rates, where stronger phenotypes are seen in lines that had deficiencies in multiple ADT isoforms (Höhner et al., 2018). These data are in accordance with the genetic and biochemical analyses, as well as with the expression patterns of different ADT genes in Arabidopsis, which indicate that different ADT isoforms in land plants are redundant in Phe-biosynthesis (Chen et al., 2016; Höhner et al., 2018). Arabidopsis mutants that accumulate lower levels of free-Phe are expected to show higher sensitivity to m-tyrosine toxicity, due to altered Phe metabolism (Figure 1 and Table S2) and/or increased misincorporation of the Phe-analog into the plant proteome. Analysis of single, double, triple, and quadruple knockout lines (Chen et al., 2016; Höhner et al., 2018) (generously provided by Prof. Jirong Huang, Shanghai Normal University) indicated that triple adt3/4/5 and quadruple adt3/4/5/6 mutants are more sensitive to m-tyrosine, further supporting a functional redundancy between different ADT paralogs in Arabidopsis thaliana plants (Figure 3 and S2). In the absence of m-tyrosine, the root sizes were similar in 5-day-old wild-type plants and adt mutant-lines (i.e., about 3 cm in length; Figures 3A, B). Yet, when the seeds were germinated in the presence of 2.5 or 5 μM m-tyrosine, root development was more strongly affected in the triple adt3/4/5 (81% inhibition at 2.5 μM and 89% at 5 μM m-Tyr) or quadruple adt3/4/5/6 mutants (90% inhibition at 2.5 μM and 89% at 5 μM m-Tyr) than in wild-type plants or single adt mutant lines (i.e., 61~75% inhibition at 2.5 μM and 82~84% at 5 μM m-Tyr) (Figures 3B, D). Accordingly, the m-tyrosine concentration required to achieve 50% reduction of root growth of adt3/4/5/6 mutant-line was notably lower than that of wild-type plants (i.e., IC50 values 0.761 and 2.365 μM, respectively) (Figures 3C and S1).
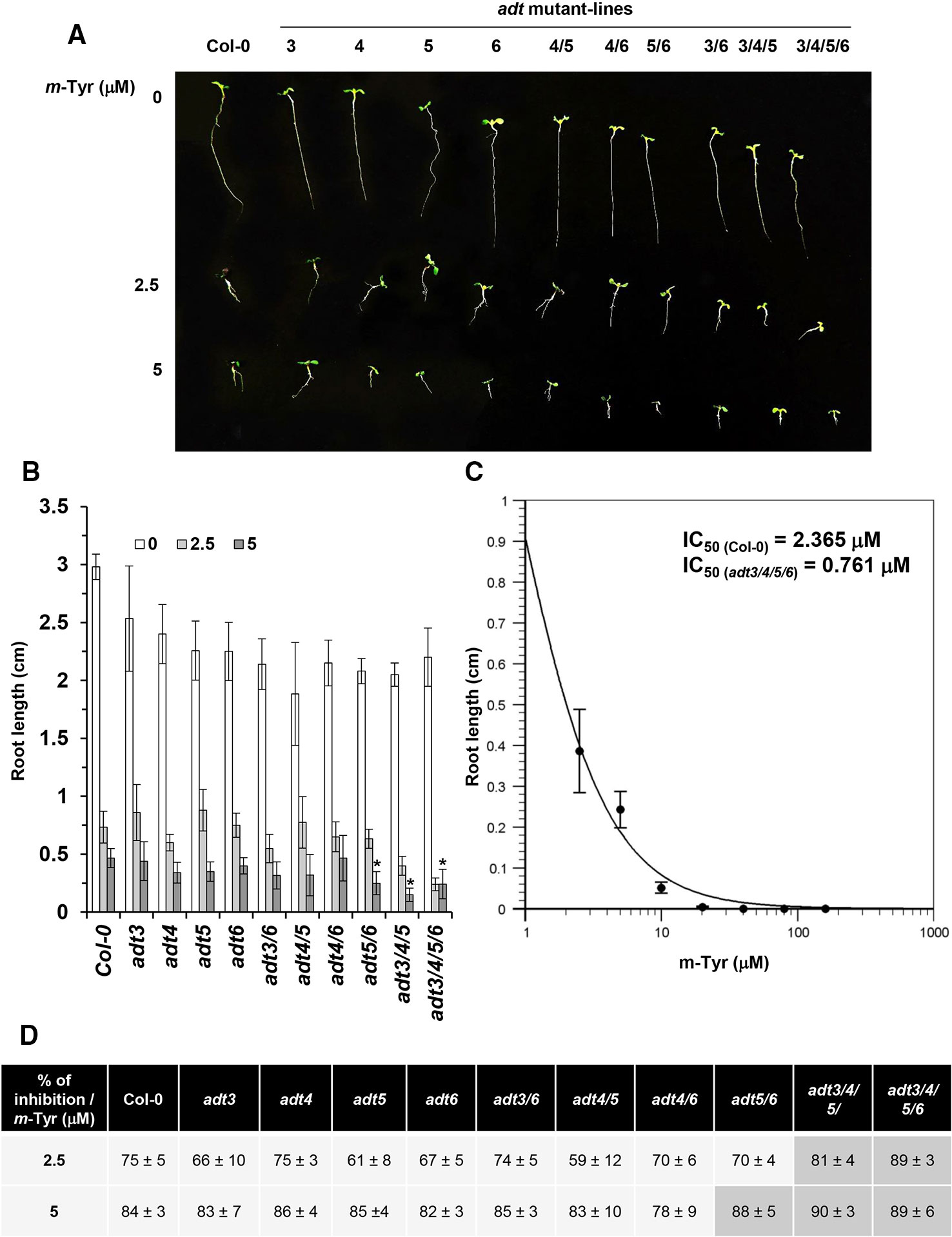
Figure 3 The effect of m-tyrosine on Arabidopsis wild-type and single, double, triple, or quadruple adt knockout mutant-lines. (A) Seeds of Arabidopsis wild-type (Col-0) and homozygous adt mutant-lines (Chen et al., 2016; Höhner et al., 2018) (generously provided by Prof. Jirong Huang, Shanghai Normal University) were germinated in Murashige and Skoog (MS)-agar plates in the absence or presence of different concentration of m-tyrosine (i.e., 0, 2.5, and 5 μM), as indicated in the panel. The figure show 5-day-old seedlings. (B) Measurements of the root lengths in 5-day-old Arabidopsis wild-type (Col-0) plants and adt mutants. (C) IC50 of the quadruple adt3/4/5/6 mutant was calculated using the ‘Quest-Graph™ IC50 calculator' (https://www.aatbio.com/tools/ic50-calculator). (D) The effect of m-tyrosine (i.e., % of inhibition) on the relative root length ratios in 5-day-old wild-type (Col-0) plants and adt mutants. Bar in (panel A) represents 1.0 cm. The values are means of three biological replicates with about 25 seedlings in each treatment. Error bars indicate one standard deviation. Asterisk in (panel B) indicates a significant difference from 5 μM m-Tyr grown plants (Student's T-test, P ≤ 0.05).
Next, we examined the effects of different Phe concentrations (i.e., 20, 40, and 80 μM) on root growth of Arabidopsis thaliana wild-type (Col-0) and adt3/4/5/6 mutants grown in the absence or presence of 10 μM m-tyrosine (Figure S2A). Increased concentrations of Phe partially rescued the early seedlings establishment and root developmental defect phenotypes by m-tyrosine (Figure S2A). At concentrations above 40 μM phenylalanine, the m-tyrosine mediated altered post-germination development and inhibited root development were restored in both the wild-type and adt3/4/5/6 mutant (Figure S2A). To a lesser degree, the effect of m-tyrosine on root growth was also counteracted by the addition of Phe in other adt mutant-lines (Figure S2B).
Arabidopsis seedlings germinated in the presence of m-tyrosine (i.e., above 20~40 μM) had yellowish to white cotyledons (Figure S1), with reduced chlorophyll content (Table 2), thus suggesting that the seedlings are defective in chloroplast development. We observed that the m-tyrosine-associated phenotypes can be partially restored by the addition of sucrose to the growth media (Figure S3, + sucrose), further suggesting that m-tyrosine affects organellar biogenesis during early seedlings establishment and root development. Microscopic analysis of young (i.e., 5-day-old) Arabidopsis plantlets treated with 10 μM m-tyrosine showed altered chloroplast morphologies, with less grana lamella (Figure 4). The relative densities of the thylakoid grana stacks in m-tyrosine treated plants were found to be notably lower (0.56 ± 0.07 times) than those of Col-0 plants grown under the same conditions in the absence of m-tyrosine (1.00 ± 0.07, Table 2). We further noticed the appearance of many plastoglobuli (PG) in the m-tyrosine treated plants (Figures 4D–F). These lipoprotein particles, which are commonly observed in colored plastids (i.e., chromoplasts), seem particularly prominent in plants affected in chloroplast development (Austin et al., 2006).
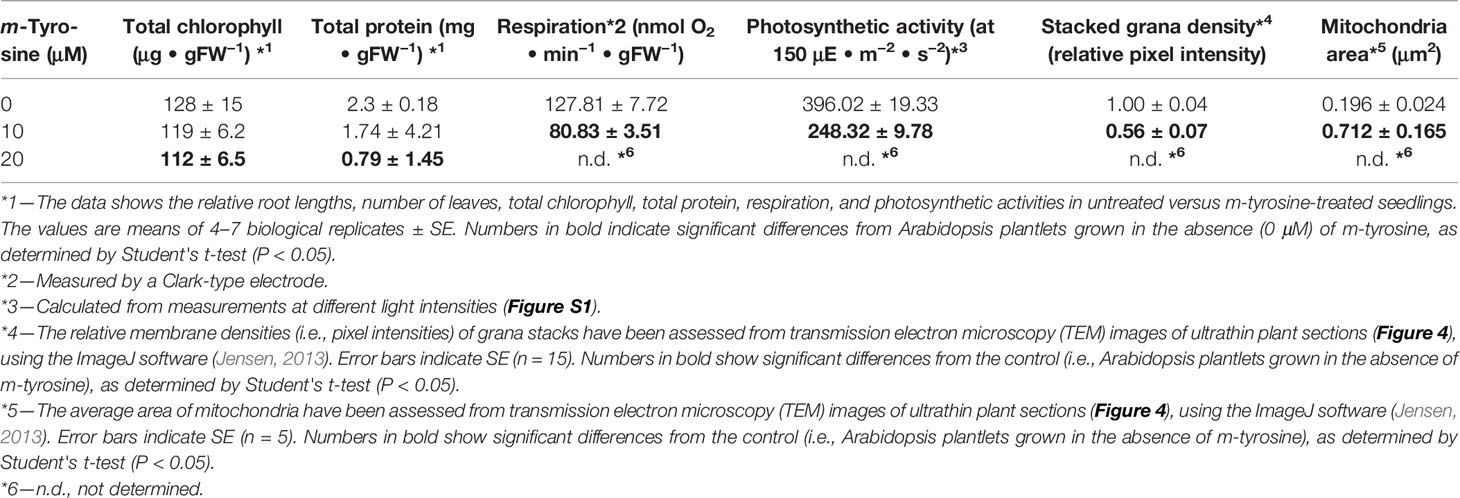
Table 2 Effects of m-tyrosine on protein levels and organellar activities of 5-day-old Arabidopsis thaliana (Col-0) seedlings.
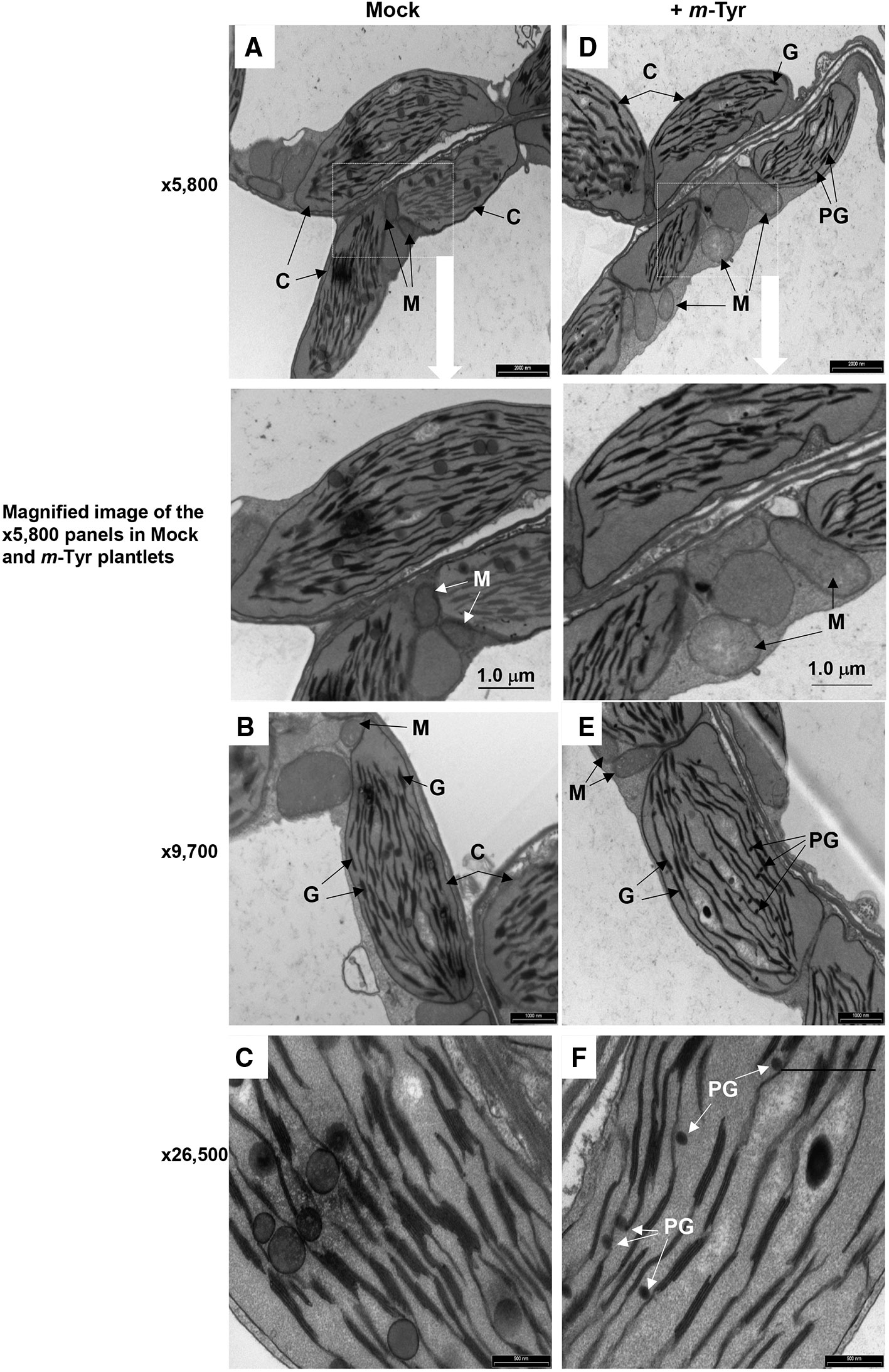
Figure 4 The effect of m-tyrosine on organellar morphologies in Arabidopsis plants. Representative transmission electron micrographs of ultrathin sections from the hypocotyl tissue of 5-day-old Arabidopsis seedlings seeded on Murashige and Skoog (MS)-agar plates in the absence (panels A–C) or presence of 10 μM m-tyrosine (panels D–F). Mitochondria are labeled by “M,” “C” indicates to chloroplasts, “G” refers to grana stacks, while plastoglobuli are labeled as “PG.” Bars represent 2 µm in panels A, D, 1.0 µm in panels B, E, and 0.5 µm in panels C, F, as indicated in each panel.
In addition to altered chloroplast biogenesis defects, we also noticed mild alterations in mitochondria structure in the presence of m-tyrosine (i.e., 10 μM). While the electron micrographs of mitochondria of the non-treated plantlets showed characteristic internal cristae formation, as dense folds of the inner-membrane sections (Figures 4A–C), many mitochondria in m-tyrosine grown seedlings seemed larger (about 3.6x in organellar area, Table 2), with reduced inner mitochondrial membrane electron density and less cristae organization (Figures 4D–F). Similar organellar morphologies were also noticed in various Arabidopsis mutants affected in mitochondria gene expression (see e.g., Keren et al., 2009; Keren et al., 2012; Zmudjak et al., 2013; Cohen et al., 2014).
To determine whether the photosynthetic and respiratory activities were altered in plants grown in the presence of m-tyrosine, we monitored the O2-evolution rates of 5-day-old seedlings in the dark, using a Clark-type electrode (Table 2 and Figure S4). In the dark, the average O2-uptake rate of untreated plants was 127.81 ± 7.72 nmol O2 ·min−1 ·gFW−1. Arabidopsis seedlings grown in the presence of 10 μM m-tyrosine showed lower respiratory activities (i.e., 80.83 ± 3.51 nmol O2 ·min−1 ·gFW−1) than those of the control (i.e., untreated) plants (Table 2 and Figure S4). The photosynthetic activities of control and m-tyrosine treated plants were examined by monitoring the O2-evolution rates at different light intensities (0–1,000 μmol photons · m−2 ·s−2). The light compensation point was similar in control plants and seedlings grown in the presence of m-tyrosine (12.4 ± 3.7 and 11.2 ± 5.3 µmol ·m−2 ·s−1) (Figure S4). Yet, noticeable differences were observed in light saturation curves between treated (+ m-Tyr) and untreated (−m-Tyr) plants.
Comparative measurements of light saturation curves of photosynthesis (i.e., O2-evolution rates, using a Clark-type electrode) were performed with untreated (Figure S4, open boxes) and 5-day-old seedlings grown in the presence of 10 μM m-tyrosine (Figure S4, filled boxes). The characteristics parameters of the photosynthetic activities were evaluated by fitting the net photosynthesis data to the equation (MacIntyre et al., 2002). The maximal photosynthetic yield value (Pmax) of the control plants was 428.5 nmol O2 ·min−1 ·gFW−1, while the Pmax value of m-tyrosine treated plants was notably lower (i.e., 318.4 nmol O2 ·min−1 ·gFW−1). Yet, a lower light intensity was required for the saturation of photosynthesis in the m-tyrosine grown plants (i.e., 50.9 µmol ·m−2 ·s−1, compare with 102.3 µmol ·m−2 ·s−1 in the control plants). The data also suggested that m-tyrosine treated seedlings experienced photoinhibition under lower light intensities, as seen by a decline in O2-evolution rates in light intensities higher than ~600 µmol photons · m−2 · s−1 (Figure S4), while the control plants did not show reduced photosynthetic activities in light intensities above ~1,000 µmol photons ·m−2 ·s−1. Photoinhibition in the m-tyrosine treated plants was further apparent by their higher β values (i.e., 0.05198), in comparison to those evaluated for the untreated plants (i.e., β values = 0.009976). These data are indicating that the photosynthetic and respiration activities were reduced in Arabidopsis plantlets treated with m-tyrosine.
The accumulation of organellar proteins was analyzed in 5-day-old Arabidopsis seedlings treated with 10 or 20 μM m-tyrosine. Total protein, extracted from control (−m-Tyr) and m-tyrosine treated plantlets, was analyzed by (12%) SDS-PAGE, following immunoblotting with antibodies raised against various organellar proteins (Figure 5A and Table S1). The steady-state levels of the mitochondrial NAD9, CA2, and COX2 (i.e., between 30 up to 55% reductions), the plastidial PsbC protein (about 64% lower), and to a lesser extent the plastidial PsaA subunit (~25% lower), were all found to be reduced in seedlings germinated in the presence of m-tyrosine (Figures 5B, C). The accumulations of AtpA, RISP, VDAC, and PsbD subunits were not significantly affected by the addition of m-tyrosine to the growth media (Figures 5B, C). The rotenone-insensitive NADH dehydrogenases (NDBs) and alternative oxidases (AOXs) are induced in a coordinated manner under respiratory chain dysfunction and oxidative stress conditions (Clifton et al., 2005; Garmier et al., 2008; Rasmusson et al., 2008; Yoshida and Noguchi, 2009; Keren et al., 2012). The signals of AOX and NDB2 were both found to be somewhat higher (about 2.5 and 7.2 times higher, respectively) in the 5-day-old m-tyrosine treated Arabidopsis plantlets (Figures 5B, C).
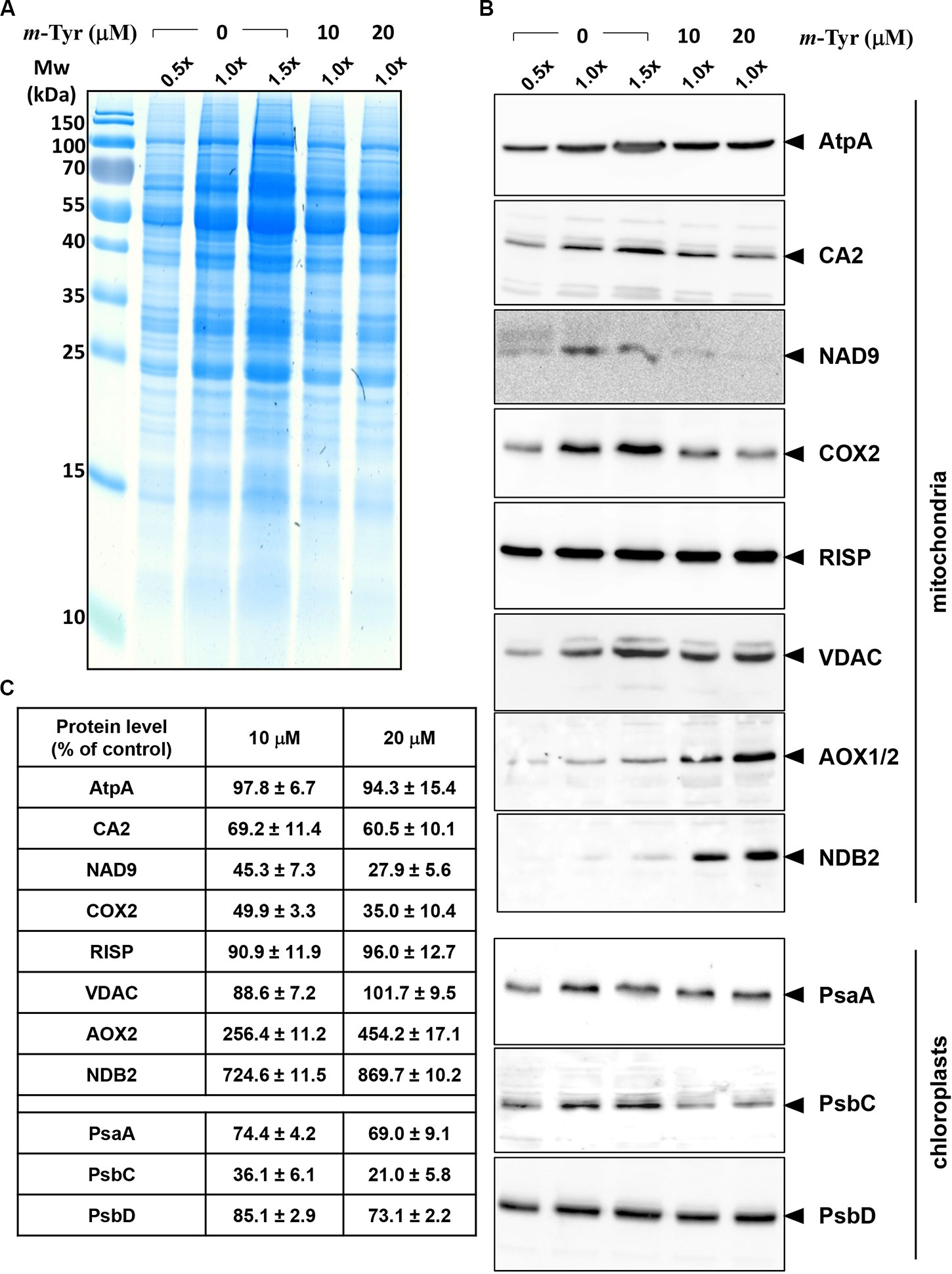
Figure 5 Relative accumulation of organellar proteins in 5-day-old Arabidopsis plants grown in the absence or presence of m-tyrosine. Immunoblot analyses of 5-day-old Arabidopsis thaliana (Col-0) plants grown in the absence or presence of m-tyrosine. For the quantification of the relative abundances of organellar proteins in the plants, different amounts of total proteins extracted from control (untreated) and m-tyrosine-grown seedlings were loaded and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After electrophoresis, the gel was stained with Coomassie Blue (A), or transferred to a PVDF membrane for immunobloting (B). The blots were probed with polyclonal antibodies raised to different organellar proteins (see Table S1), as indicated in the right panel. Detection was carried out by chemiluminescence assays after incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. These include the mitochondrial proteins, ATP-synthase subunit 1 (AtpA), subunit CA2 of complex I (γ-type carbonic anhydrase-like subunit 2) (Perales et al., 2005), and NADH dehydrogenase subunit 9 (NAD9) (Lamattina et al., 1993), the Rieske iron-sulfur protein (RISP) of complex III (cytochrome c reductase) (Carrie et al., 2010), cytochrome oxidase subunit 2 (COX2) of complex IV (C-IV), alternative oxidase (AOX1a) (Elthon et al., 1989), the external NADH-dehydrogenase subunit 2 (NDB2) (Carrie et al., 2008), and the voltage-dependent anion channel (VDAC, or Porin), and the plastidial Photosystem I P700 chlorophyll a apoprotein A1 (PsaA), photosystem II CP43 reaction center protein (PsbC), and photosystem II D2 protein (PsbD). Arrows in the blots indicate to AtpA (55 kDa), CA2 (28 kDa), NAD9 (23 kDa), COX2 (30 kDa), RISP (23 kDa), VDAC (29 kDa), AOX1/2 (36 kDa), NDB2 (50 kDa), PsaA (52 kDa), PsbC (43 kDa), PsbD (39 kDa). Hybridization signals were analyzed by chemiluminescence assays after incubation with HRP-conjugated secondary antibody. The intensities of protein signals in panel B were evaluated using the ImageJ software (Jensen, 2013). The data shows the relative accumulation of each protein in untreated versus m-tyrosine-treated seedlings (panel C). The values are means of three biological replicates ± SE.
To investigate the effects of m-tyrosine on the accumulation of native organellar complexes, crude membranous fractions were obtained from 5-day-old seedlings germinated in the absence or presence of 10 μM m-tyrosine. The membranous proteins were separated under native conditions by blue native-polyacrylamide gel electrophoresis (BN-PAGE), and then subjected to immunoblot analyses with various antibodies (Table S1), as indicated in Figure 6. Arrows indicate to the native CI, CIII, VIV, CV, and PSI complexes. Under the gel electrophoretic conditions (Pineau et al., 2008; Shevtsov et al., 2018), we noticed to reduced steady-state levels of the respiratory complexes CI (47.5%), CIII (36.7%), CIV (59.9%), and CV (26.4%) upon m-tyrosine treatment (Figure 6). Reduced CI activity was also apparent by “in-gel” activity assays (Figure 6A). Similarly, immunoblots with PsaA indicated to reduced accumulation in photosystem I (PI) in seedlings grown in the presence of 10 μM m-tyrosine (Figure 6B). The immunoassays with anti-CA2, COX2, and anti-PsaA antibodies further indicated the presence of additional protein bands (Figure 6B, marked with asterisks). We speculate that the lower molecular weight bands in the CA2 and COX2 blots (i.e., about 800 and 150 kDa, respectively) may correspond to a partially assembled sub-CI and CIV particles, respectively (Meyer et al., 2011), whereas the upper-band (~500 kDa) in the PsaA blot may correspond to an oligomeric state of PSI (Salomon and Keren, 2011).
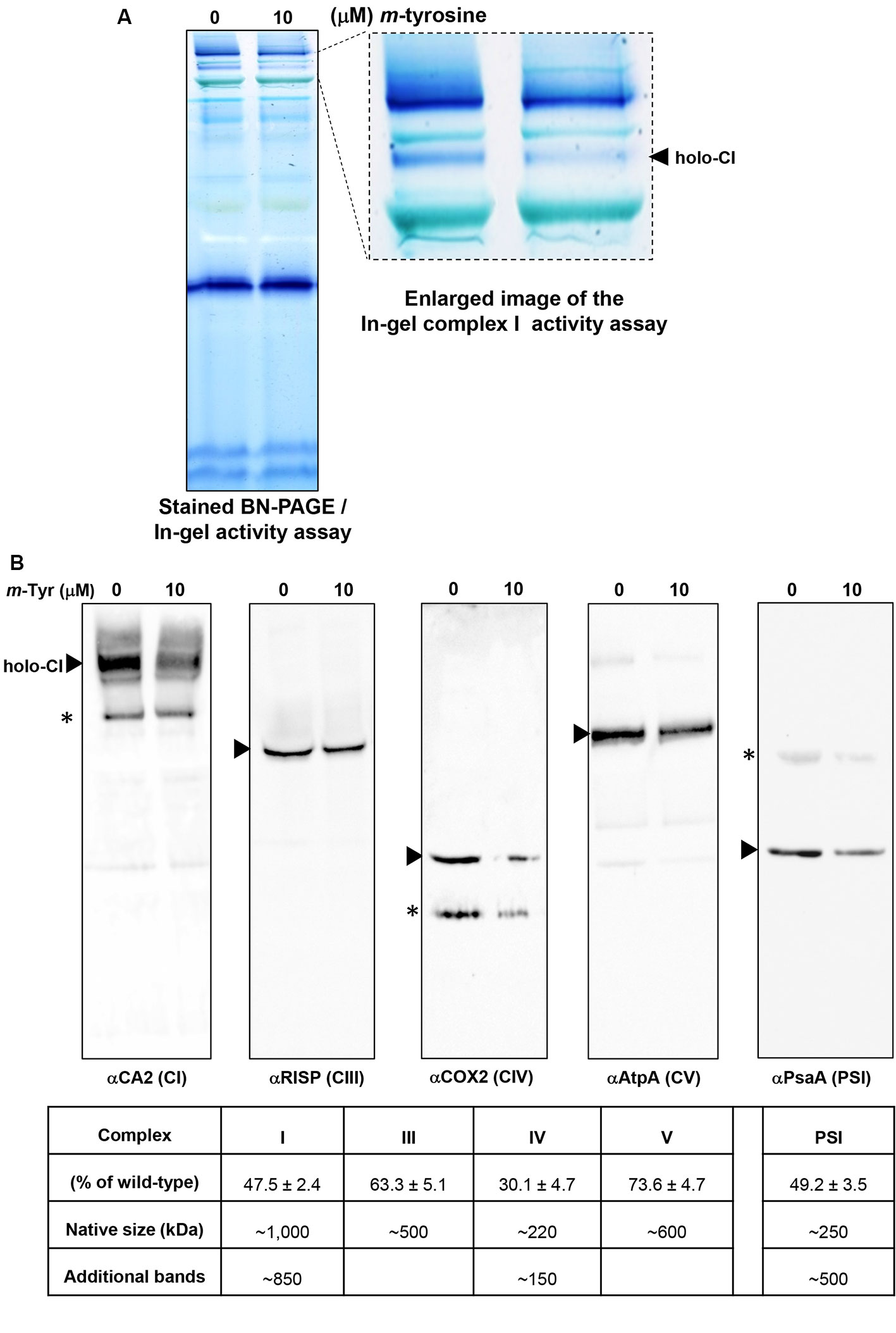
Figure 6 Relative accumulation of native organellar complexes in 5-day-old Arabidopsis plants grown in the absence or presence of m-tyrosine. Blue native-polyacrylamide gel electrophoresis (BN-PAGE) of crude organellar preparations was performed according to the method described in Pineau et al. (2008). Equal amounts of crude membranous fractions (equivalent to 50 mg fresh-weight seedlings), were obtained from 5-day-old Arabidopsis seedlings, solubilized with 1.5% n-dodecyl-β-d-maltoside (DDM), and the organellar complexes were resolved by BN-PAGE. (Panel A) shows stained PAGE, followed by in-gel activity assay of complex I, according to the method described in Eubel et al. (2005). For immunodetections (panel B), proteins were transferred from the native gels onto a polyvinylidene difluoride (PVDF) membrane (see Materials and Methods) and then probed with specific antibodies (see Table S1), as indicated below each blot. Arrows indicate to the native mitochondrial complexes I (~1,000 kDa), III (dimer, ~500 kDa), IV (~220 kDa), V (~600 kDa), and the plastidial PSI complex (~250 kDa). Asterisks in the COX2 and PsaA panels indicate the presence of lower or higher (respectively) molecular weight particles of unknown origins. Hybridization signals were analyzed by chemiluminescence assays after incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. The intensities of protein signals in (panel B) were evaluated using the ImageJ software (Jensen, 2013). The data shows the relative accumulation of each protein in untreated versus m-tyrosine-treated seedlings. The data shows the relative accumulation of each protein in untreated versus m-tyrosine-treated seedlings (panel C). The values are means of three biological replicates ± SE.
The Phe-analog, m-tyrosine, affects early seedlings development and radicle elongation of plants (Figures S1 and S3, and Bertin et al., 2007). Growing Arabidopsis seedlings in the presence of m-tyrosine results with a reduction in the level of free-Phe (Figure 1). Accordingly, adt mutants are more severely affected by m-tyrosine (Figures 3 and S2), whereas the adt2-1D mutant plant-line that accumulates higher levels of free-Phe (Cho et al., 2007) seems less susceptible to m-tyrosine toxicity (Huang et al., 2010). Similarly, the addition of exogenous Phe can partially restore the growth and developmental defect phenotypes associated with m-tyrosine (Figure 2, and Bertin et al., 2007). Here, we further show that m-tyrosine alters the biogenesis and activities of both the mitochondria and chloroplasts during early developmental stages (Figures 4, 6, S1, and S3, Table 2). It remains possible, therefore, that the developmental defect phenotypes cause by m-tyrosine may relate to its misincorporation to the mitochondrial and plastidial proteomes.
To analyze whether m-tyrosine is incorporated into Arabidopsis proteins, we performed MS-based proteomic analyses of 5-day-old plantlets grown in the absence or presence of m-tyrosine (Supplemental Table S3, doi: 10.6084/m9.figshare.11627211). The proteomic analyses indicated many Phe → m-Tyr/Tyr exchanges (i.e., due to their identical masses it is not possible to distinguish between the native amino acid Tyr and its structural related isomers, o- and p- and m-tyrosine in the MS data) in various proteins of Arabidopsis seedlings grown in the presence of m-tyrosine, which were not observed in plants grown in the absence of the analog (i.e., under the same growth conditions). Table 3 summarizes the MS-based data of four independent proteomic analyses of 5-day-old seedlings grown in the absence (control) or presence of 10 μM and 20 μM m-tyrosine. The frequencies of Phe mistranslation is dose-dependent, as the misincorporation frequencies increase about two folds when plants were treated with 20 μM instead of 10 μM m-tyrosine (Table 3). In accordance with previous observations suggesting that organellar PheRSs might be more prone to misacylation of m-tyrosine (Klipcan et al., 2009; Klipcan et al., 2010), the proteomic data indicated that Phe → m-Tyr/Tyr exchanges occur more frequently during the synthesis of organellar proteins, and in particularly of those translated within the chloroplasts (Table 3 and Supplementary Data File 1).
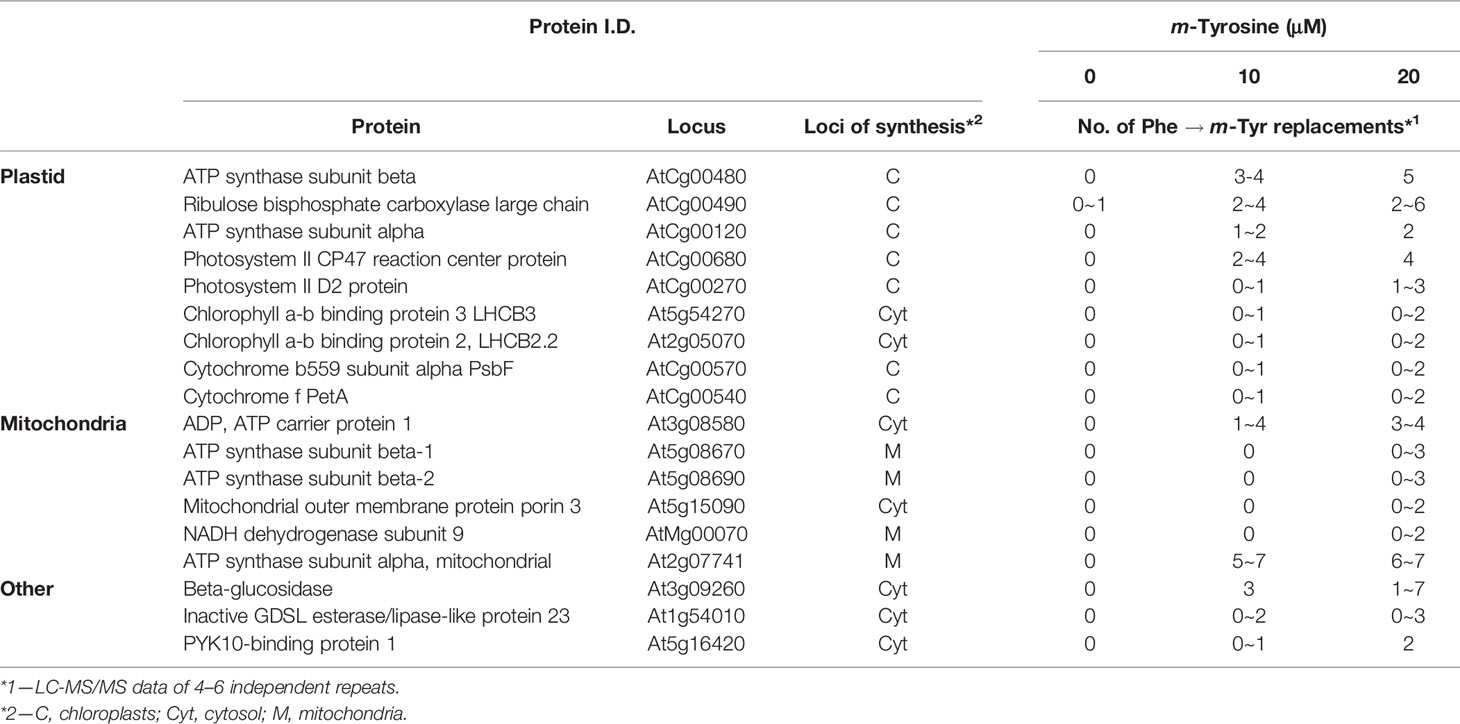
Table 3 Summary of four independent proteome analyses of enriched organellar fractions obtained from 5-day-old seedlings grown in the absence or presence of m-tyrosine.
Our results (Table 3) indicate that m-tyrosine can be delivered into the proteomes of plants, an abnormal cellular condition that can be associated with increased protein turnover (Rodgers et al., 2002). FtsH is a membrane-bound ATP-dependent zinc metalloprotease complex, which has been characterized in bacteria and organelles of eukaryotic cells (Kato and Sakamoto, 2018 and references therein). While Escherichia coli harbors a single FtsH gene, Arabidopsis plants encode 12 related FtsHs that play key roles in protein quality control, as removal of damaged and abnormal proteins (Ostersetzer and Adam, 1997; Adam and Ostersetzer, 2001; Zaltsman et al., 2005; Sakamoto, 2006). Nine FtsHs reside in the plastids, whereas the other three paralogs are located within the mitochondria (Kato and Sakamoto, 2018). In chloroplasts, FtsH is made of different hetero-hexamer complexes (Moldavski et al., 2012). FtsH2 (At2g30950, also denoted as VAR2) is the most abundant isoform within the plastids. var2 mutants show aberrant leaf development with severe variegation phenotypes, which are related to altered chloroplast development (Chen et al., 2000; Takechi et al., 2000; Sakamoto et al., 2003).
Based on these data we speculated that var2 mutants might show higher sensitivity to m-tyrosine, due to the accumulation of abnormal and damaged proteins within the plastids. To examine this hypothesis, we analyzed the root development and early seedling establishment of Arabidopsis wild-type (Col-0) and var2 mutants (Takechi et al., 2000; Sakamoto et al., 2003; Zaltsman et al., 2005), germinated in the absence or presence of various concentrations of m-tyrosine. The data indicated that var2 mutants seem to be more susceptible to inhibition by m-tyrosine (Figure 7). In particular, the Phe-analog, m-tyrosine, had a strong effect on chloroplast biogenesis of var2 plants, as evident by the cotyledon chlorosis (Figure 7A) and reduced chlorophyll content (Figure 7B). Reduced leaf pigmentation in var2 was already observed at m-tyrosine concentrations ≥ 1 μM, while no chlorophyll was detected when the mutants were grown in m-tyrosine concentrations above 5.0 μM (Figure 7).
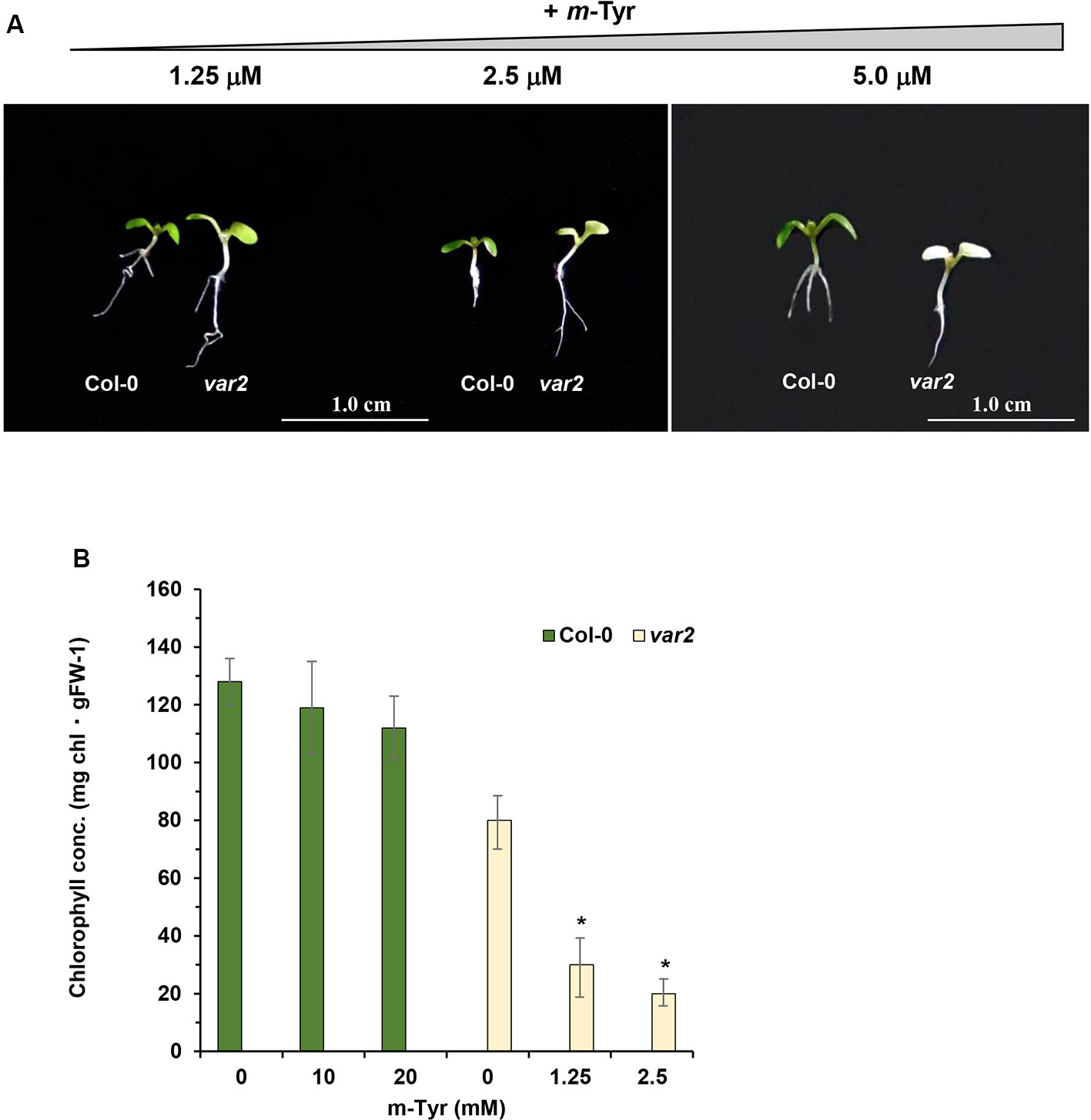
Figure 7 The effect of m-tyrosine on Arabidopsis wild-type (Col-0) and var2 knockout mutant-line. (A) Seeds of Arabidopsis wild-type (Col-0) and homozygous var2 mutant-line (Takechi et al., 2000; Sakamoto et al., 2003; Zaltsman et al., 2005) (generously provided by the group of Prof. Zach Adam, The Hebrew University) were germinated in Murashige and Skoog (MS)-agar plates in the absence or presence of different concentration of m-tyrosine. The values are means of three biological replicates with about 25 seedlings, as indicated in the panel. The figure shows 5-day-old seedlings. Bars represent 1 cm in each panel. The values are means of three biological replicates with about 25 seedlings (i.e., 5-day-old). Error bars indicate one standard deviation. Asterisk in (panel B) indicates a significant difference from var2 mutants control plants (Student's T-test, P ≤ 0.05).
Autophagy is an essential process in eukaryotic cells that involves a control degradation of large substrates including protein aggregates, pathogens, and organelles (reviewed by e.g., Avin-Wittenberg et al., 2018). Forward genetics led to the identification of many autophagy-related loci in Arabidopsis, denoted as ATG genes (Young et al., 2019). Among these genes are ATG5 (At5g17290) and ATG7 (At5g45900), which their mutants show a full inhibition of autophagy (Barros et al., 2017 and references herein). When Arabidopsis wild-type plants (Col-0) and atg mutants (Hofius et al., 2009; Yoshimoto et al., 2009; Avin-Wittenberg et al., 2015) were germinated in the absence or presence of various concentrations of m-tyrosine (Figure S5A), the atg mutant-lines were found to be more susceptible to high concentrations of m-tyrosine (i.e., > 10 μM). Yet, the phenotypes of the autophagy mutants were undistinguishable from those of wild-type (Col-0) plants at lower concentrations of the Phe-analog (Figure S5B). These results seem to be correlated with the organellar biogenesis defects of Arabidopsis plantlets germinated in the presence of m-tyrosine (Figures 4, S1, and S3, Table 2).
m-Tyrosine is a non-proteinogenic analog of the aromatic amino acids phenylalanine and tyrosine, which acts as a natural herbicide, affecting the post-germination development and early establishment of nearby plant life (Bertin et al., 2007). Published data indicates that m-tyrosine is significantly more toxic to plants than its structural related isomers, o- and p-tyrosine (Bertin et al., 2007). Our data provide further insights into m-tyrosine-mediated phytotoxicity. Under different conditions, and more particularly under oxidative stress, m-tyrosine is accumulating to higher levels in the cell (Ipson and Fisher, 2016). m-tyrosine is directly generated by hydroxyl radical attack of phenylalanine when levels of reactive oxygen species are elevated (Rodgers and Shiozawa, 2008), or might be produced enzymatically, as indicated in a few plant species (Mothes et al., 1964; Bertin et al., 2007; Huang et al., 2012). In F. rubra, m-tyrosine biosynthesis occurs through the hydroxylation of phenylalanine in the root tips. The allelochemical is then released to the biosphere, were it inhibits the post-germination development of nearby plants (Huang et al., 2012).
Arabidopsis seedlings germinated in the presence of m-tyrosine show slow seedlings establishment, altered root elongation, and reduced biomass (Figures 3, S1–S3, and S5, and Bertin et al., 2007). The severity of these phenotypes are correlated with the levels of m-tyrosine in the growth media, with calculated IC50 value of 2.365 μM (Figure S1). At m-tyrosine concentrations higher than 40~80 μM, the plants show post-germination arrest of embryo development (Figure S1). Apparently, while m-tyrosine had no obvious effect on the photosynthetic activity of lettuce seedlings (Bertin et al., 2007), at concentrations above 40 μM Arabidopsis seedlings showed signs of leaf (i.e., cotyledon) chlorosis (Figure S1). Analyses of 5 day-old seedlings grown in the absence or presence of 10 μM m-tyrosine indicated to reduced chlorophyll production (Table 2), altered organellar biogenesis (Figure 4) and reduced photosynthetic activities (Table 2 and Figure S4). Our data also indicate to a reduction in oxygen consumption when the plants are grown in the presence of m-tyrosine (Table 2). Respiration in some CI-deficient Arabidopsis mutants can be upregulated, a phenomenon that has been associated with the induction of the alternative respiratory pathway, mediated by non-phosphorylating NAD(P)H dehydrogenases (NDs) and alternative oxidases (AOXs) (see e.g., Kuhn et al., 2015). We speculate that although AOX and ND2 are upregulated in m-tyrosine treated plants, the high energy demand (i.e., ATP production) during germination and early seedling metabolism necessitates the electron transport via the canonical respirasome mediated pathway. Accordingly, in different plant species, the affinity of AOX is significantly lower compared to that of COX to O2 (see e.g., van Dongen et al., 2011). The m-tyrosine-mediated defect phenotypes can be partially rescued by the addition of sucrose to the growth media (Figure S3), in a similar manner to the effects of sugar on Arabidopsis mutants affected in chloroplast or mitochondria biogenesis (see e.g., Keren et al., 2012; Van Dingenen et al., 2016).
Based on the experimental results and published data we consider two different cellular pathways in which m-tyrosine inhibits the early establishment of Arabidopsis plants. Together, the analyses of the steady-state levels of free amino acids in control and m-tyrosine-grown seedlings (Figure 1 and Table S2), the effects of various amino acids added to the growth media on m-tyrosine toxicity (Figure 2), the higher-sensitivity of adt mutants to m-tyrosine (Figures 3 and S2) and lower sensitivity of adt2-1D mutant which accumulates higher levels of Phe (Cho et al., 2007), strongly suggest that m-tyrosine affects the biosynthesis of phenylalanine in early-developing Arabidopsis seedlings. Another molecular mechanism by which m-tyrosine can affect Arabidopsis vitality involves the misincorporation of this aromatic non-proteinogenic analog into the plant proteome instead of Phe, and in particular to various organellar proteins. These are evident by proteomic data (Table 3 and Supplemental Table S3, doi: 10.6084/m9.figshare.11627211), altered organellar biogenesis and functions (Table 2, Figures 4–6 and S4), the hypersensitivity of var2 mutants to m-tyrosine (Figure 7), as well as previous data indicating that tRNAPhe can be charged with m-tyrosine, in vitro (Gurer-Orhan et al., 2006; Klipcan et al., 2009).
Published data (Huang et al., 2010; Höhner et al., 2018) and our own results (Figures 2 and S2) strongly suggest that m-tyrosine toxicity is affected by the (bio-) availability of Phe. Accordingly, exogenous Phe can partially rescue the growth defect phenotypes mediated by m-tyrosine (Figure 2). Seedlings grown in the presence of the amino acid methionine (Met) are also less susceptible to inhibition by m-tyrosine (Figure 2, and Bertin et al., 2007). Therefore, it remains possible that Met plays a non-redundant role through indirectly affecting Phe metabolism, or by scavenging reactive oxygen species (ROS) that may accumulate to higher levels in the m-tyrosine treated plants. Methionine may also act as catalytic antioxidants, protecting protein where they reside, as well as other molecules within the cells (Luo and Levine, 2009). Accordingly, Arabidopsis seedlings germinated in the presence of m-tyrosine show altered organellar activities (Table 2 and Figure S4) and accumulate high levels of AOX (Figure 5).
An effect of m-tyrosine on Phe biosynthesis is further supported by genetic analyses. Arogenate dehydratase (ADT) is a key enzyme in Phe biosynthesis, converting arogenate into Phe. ADT activity in plants is positively regulated by Tyr and negatively regulated by Phe (Jung et al., 1986; Siehl and Conn, 1988). While Arabidopsis adt mutants show a hipper sensitivity to m-tyrosine (Figures 3 and S2), the adt2-1D mutant-line that accumulates higher levels of Phe, most likely due to a disruption in the feedback inhibition (i.e., allosteric effects) of the ADT enzyme (Cho et al., 2007), are less susceptible to m-tyrosine toxicity (Huang et al., 2010). Accordingly, analyses of the steady-state levels of free amino-acids indicate that the phytotoxic effects of m-tyrosine correlate with altered amino acid metabolism, in particularly phenylalanine (Figure 1 and Table S2). Taken together, these data suggest that m-tyrosine reduces the biosynthesis of Phe, probably by directly affecting the activity of ADT. However, these assumptions need to be validated experimentally.
Another molecular mechanism, by which m-tyrosine affects plants, may involve the incorporation of unnatural chemical groups (i.e., non-canonical amino acids) into proteins (Smith and Fowden, 1968), which are expected to affect the activities of proteins and enzymes, or to interfere with the functions of native side chains. Proteomic analyses of plants germinated in the absence or presence of m-tyrosine indicate that this analog is misincorporated into the plant proteome (Table 3, and Supplemental Table S3, doi: 10.6084/m9.figshare.11627211). These data are also in agreement with earlier reports suggesting that m-tyrosine may be wrongly delivered into proteins in bacteria (Aronson and Wermus, 1965) and animals (Gurer-Orhan et al., 2006). In Arabidopsis, the majority of the Phe → m-Tyr/Tyr replacements are identified in proteins that are expressed within the organelles, in particular plastidial ones (Table 3). Furthermore, analyses of mutant plants affected in FtsH2, a major component of the chloroplast protein quality control system (Kato and Sakamoto, 2018), resulted with an increased sensitivity to m-tyrosine, an effect that is expected to be associated with the accumulation of damaged and abnormal organellar proteins (Ostersetzer and Adam, 1997; Adam and Ostersetzer, 2001; Sakamoto, 2006). Also, mutant plants affected in autophagy are more severely affected in seedling establishment and show strong cotyledon bleaching at m-tyrosine concentrations above 10 μM (Figure S5), although the effects at high m-tyrosine could rather relate to general stress responses rather than to a specific cellular damage.
How can m-tyrosine be incorporated into the plant proteome? Aminoacyl tRNA synthases (aaRSs) ensure the integrity of the translation of the genetic code, by covalently attaching an appropriate amino acid to the corresponding nucleic acid adaptor tRNA molecule. Although aaRS are known to be highly specific, mistakes in the recognition may still occur, due to stereo-chemical similarities shared by some native amino acids and their non-proteinogenic analogs (Moghal et al., 2014; Chaliotis et al., 2016; Mohler and Ibba, 2017). A key repair mechanism to ensure the acetylation of the correct amino acid involves an intrinsic editing activity, by which misacylated tRNAs can be hydrolyzed. In plants, the attachment of phenylalanine to tRNAPhe is catalyzed by two major forms of PheRSs. These include i) a heterotetrameric (2·αβ) form, which is structurally related to the bacterial 2·αβ enzymes and acts in the cytosol, and ii) a degenerated monomeric form (contains fragments of α and β), which dually functions in the plastid and mitochondria of plants (Duchêne et al., 2005; Klipcan et al., 2010).
The aromatic amino acids Phe, Tyr, and their cognate non-proteinogenic m-tyrosine analog, are distinguished by a single hydroxyl group at the aromatic ring, and thus discrimination between these molecules may not always be accurate (Roy et al., 2004; Kotik-Kogan et al., 2005; Ling et al., 2007; Klipcan et al., 2009; Moghal et al., 2014; Popp et al., 2015). To maintain accuracy during translation, the PheRS enzyme possess an editing domain that hydrolyzes misacylated tRNAPhe (Roy et al., 2004; Kotik-Kogan et al., 2005; Ling et al., 2007). Structural and biochemical assays suggest that bacterial and both eukaryotic PheRS forms can load m-tyrosine into tRNAPhe (Klipcan et al., 2009; Moghal et al., 2016). However, only the bacterial enzyme seems to be able to efficiently hydrolyze the misacylated m-tyrosine (Klipcan et al., 2009). In the cytosolic PheRS form of eukaryotes, discrimination against m-tyrosine mainly occurs during the aminoacylation step (Klipcan et al., 2009). Structural and biochemical studies suggest that organellar PheRS is not able to discriminate against m-tyrosine and lacks editing activity (Klipcan et al., 2009; Klipcan et al., 2010). It is, therefore, expected that the dually-localized organellar PheRS enzyme will be more prone to errors, opening up the rout for delivery of misacylated tRNAPhe-m-tyrosine to the organellar ribosomes.
Currently, we are not able to provide with a definitive answer. Plants are highly susceptible to m-tyrosine [Figures S1 and S3, and (Bertin et al., 2007)], with IC50 (i.e., root growth inhibition) at the low micromolar range (Figure S1), about 100~1,000 times more toxic to plants than to bacterial (Smith et al., 1964; Aronson and Wermus, 1965), yeast (Bullwinkle et al., 2014), or animal cells (Gurer-Orhan et al., 2006). Phe is an essential α-amino acid that is produced through the shikimate pathway, which is present in bacteria, fungi, and plants, as well as in some protozoa, but not in Animalia who need to obtain aromatic amino acids from their diet. While animals cannot produce Phe, it was previously shown that m-tyrosine can be biodegraded in several microorganisms, which can utilize this metabolite as the sole source of carbon, nitrogen, and energy (Khan et al., 2013 and references therein). In addition, the levels of free Phe, seem to be notably higher in animal cells (about 55 to 60 μM in human cells) then in plants (i.e., Arabidopsis, ~15 μM) (Kaufman, 1999; Voll et al., 2004).
As indicated above, there are also differences in the way that m-tyrosine can be delivered into the proteomes of different organisms. While prokaryotes encode a single heterotetrameric PheRS that is able to efficiently hydrolyze the misacylated m-tyrosine (Klipcan et al., 2009), eukaryotes possess two PheRS isoforms, one of which resides within the organelles (Klipcan et al., 2009). In plants, the organellar PheRS isoform resides in both the plastids and mitochondria (Duchêne et al., 2005). Plastid biogenesis seems in particularly amenable to m-tyrosine toxicity. This is apparent by altered chloroplast morphologies, reduced grana stacking, the presence of many plastoglobuli (Figure 4) and lower photosynthetic rates (Table 2 and Figure S4), associated with Arabidopsis plantlets germinated in the presence of m-tyrosine, as well as the fact that many polypeptides that undergo Phe → (m-)Tyr exchanges are identified as plastidial proteins (Table 3). These may relate to the high complexity of the translational apparatus in plant chloroplasts (Zoschke and Bock, 2018). We speculate that the effects of m-tyrosine on Phe biosynthesis together with the ability of the organellar PheRS to deliver the non-proteinogenic analog to both mitochondrial and plastidial proteins makes plants highly sensitive to m-tyrosine.
In summary, this work is founded on earlier reports that indicate that m-tyrosine is highly toxic to different plant species. Based on the data we consider two related molecular mechanisms by which m-tyrosine can affect angiosperm's root development, radicle elongation, and early establishment. These involve (a) a direct interference in the metabolism of various amino acids, and in particular Phe, and (b) the misincorporation of the Phe-analogue to some cytosolic and mainly the plant organellar proteins. The correct transfer of information from the genome to proteins is pivotal for the development and physiology of plants, as well as for other organisms. The specificity of aminoacyl-tRNA synthetases (aaRSs) is key for ensuring the proper decoding of the genetic information into proteins. Distinguishing between closely related amino acids and their analogs by aaRSs is not always accurate, leading to errors in tRNA loading and hence for the translation of aberrant polypeptides. When a non-cognate amino acid is activated, some aaRSs employ an editing mechanism leading to the hydrolysis of the misacylated tRNA molecule. Biochemical and structural studies suggest that organellar PheRS forms lack editing activity. The two effects (i.e., altered Phe biosynthesis and increased misincorporation of m-tyrosine) are related to one another, i.e., reduced availability of the natural amino acid Phe would ultimately result an increased misincorporation of its analogous compound, m-tyrosine, in particularly into organellar proteins (Klipcan et al., 2009). The effects of m-tyrosine are in particularly prevalent during early development, this in accordance with mutant plants affected in organellar biogenesis, which also show early growth retardation, inhibition of root development, and altered seedling establishment phenotypes (see e.g., Keren et al., 2012; Van Dingenen et al., 2016).
The datasets generated for this study can be found in the https://doi.org/10.6084/m9.figshare.11627211.
HZ: Experimental design, plant growth and analysis, analyses of the protein profiles, BN-PAGE assays. NM: Assisted in growth and analyses: HM and TA-W: GC-MS analyses of plant extracts. LK: Assisted with experimental design, co-corresponding author. OO-B: Principal Investigator, MS preparation and corresponding authors.
Research at the TA-W group is funded by the Israel Science Foundation (ISF grant no. 1899/16). This work was supported by grants from the ‘Israel Ministry of Agriculture, Nitsan Fund' (No. 20-01-0162) to LK and OO-B and the ‘Israel Science Foundation' (ISF grant no. 741/15) to OO-B.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Mr. Yuval Kolodny for his help with the analyses of photosynthetic measurements, the Arabidopsis biological resource center for providing wild-type Arabidopsis thaliana var Columbia (Col-0) seeds, Prof. Zach Adam for providing Arabidopsis var2 mutant-lines, and Prof. Jirong Huang (Shanghai Normal University) for providing us Arabidopsis adt mutant lines.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00140/full#supplementary-material
Adam, Z., Ostersetzer, O. (2001). Degradation of unassembled and damaged thylakoid proteins. Biochem. Soc. Trans. 29, 427–430. doi: 10.1042/bst0290427
Aronson, J. N., Wermus, G. R. (1965). Effects of m-tyrosine on growth and sporulation of bacillus species. J. Bacteriol. 90 (1), 38–46. doi: 10.1128/JB.90.1.38-46.1965
Austin, J. R., Frost, E., Vidi, P.-A., Kessler, F., Staehelin, L. A. (2006). Plastoglobules are lipoprotein subcompartments of the chloroplast that are permanently coupled to thylakoid membranes and contain biosynthetic enzymes. Plant Cell 18 (7), 1693–1703. doi: 10.1105/tpc.105.039859
Avin-Wittenberg, T., Bajdzienko, K., Wittenberg, G., Alseekh, S., Tohge, T., Bock, R., et al. (2015). Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. Plant Cell 27 (2), 306–322. doi: 10.1105/tpc.114.134205
Avin-Wittenberg, T., Baluška, F., Bozhkov, P. V., Elander, P. H., Fernie, A. R., Galili, G., et al. (2018). Autophagy-related approaches for improving nutrient use efficiency and crop yield protection. J. Exp. Bot. 69 (6), 1335–1353. doi: 10.1093/jxb/ery069
Barros, J. A. S., Cavalcanti, J. H. F., Medeiros, D. B., Nunes-Nesi, A., Avin-Wittenberg, T., Fernie, A. R., et al. (2017). Autophagy deficiency compromises alternative pathways of respiration following energy deprivation in Arabidopsis thaliana. Plant Physiol. 175 (1), 62–76. doi: 10.1104/pp.16.01576
Bennett, J. A., Klironomos, J. (2018). Mechanisms of plant-soil feedback: interactions among biotic and abiotic drivers. New Phytol. 222 (1), 91–96. doi: 10.1111/nph.15603
Bertin, C., Weston, L. A., Huang, T., Jander, G., Owens, T., Meinwald, J., et al. (2007). Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proc. Natl. Acad. Sci. 104 (43), 16964–16969. doi: 10.1073/pnas.0707198104
Bertin, C., Harmon, R., Akaogi, M., Weidenhamer, J. D., Weston, L. A. (2009). Assessment of the phytotoxic potential of m-tyrosine in laboratory soil bioassays. J. Chem. Ecol. 35 (11), 1288–1294. doi: 10.1007/s10886-009-9707-4
Bullwinkle, T. J., Reynolds, N. M., Raina, M., Moghal, A., Matsa, E., Rajkovic, A., et al. (2014). Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. Elife 3, e02501, 1–16. doi: 10.7554/eLife.02501
Carrie, C., Murcha, M. W., Kuehn, K., Duncan, O., Barthet, M., Smith, P. M., et al. (2008). Type II NAD(P)H dehydrogenases are targeted to mitochondria and chloroplasts or peroxisomes in Arabidopsis thaliana. FEBS Lett. 582 (20), 3073–3079. doi: 10.1016/j.febslet.2008.07.061
Carrie, C., Giraud, E., Duncan, O., Xu, L., Wang, Y., Huang, S. B., et al. (2010). Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 285 (46), 36138–36148. doi: 10.1074/jbc.M110.121202
Chaliotis, A., Vlastaridis, P., Mossialos, D., Ibba, M., Becker, H. D., Stathopoulos, C., et al. (2016). The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 45 (3), 1059–1068. doi: 10.1093/nar/gkw1182
Chen, M., Choi, Y., Voytas, D. F., Rodermel, S. (2000). Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J. 22 (4), 303–313. doi: 10.1046/j.1365-313x.2000.00738.x
Chen, Q., Man, C., Li, D., Tan, H., Xie, Y., Huang, J. (2016). Arogenate dehydratase isoforms differentially regulate anthocyanin biosynthesis in Arabidopsis thaliana. Mol. Plant 9 (12), 1609–1619. doi: 10.1016/j.molp.2016.09.010
Cho, M.-H., Corea, O. R. A., Yang, H., Bedgar, D. L., Laskar, D. D., Anterola, A. M., et al. (2007). Phenylalanine biosynthesis in Arabidopsis thaliana: identification and characterization of arogenate dehydratases. J. Biol. Chem. 282 (42), 30827–30835. doi: 10.1074/jbc.M702662200
Clifton, R., Lister, R., Parker, K., Sappl, P., Elhafez, D., Millar, A., et al. (2005). Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58 (2), 193–212. doi: 10.1007/s11103-005-5514-7
Cohen, S., Zmudjak, M., Colas des Francs-Small, C., Malik, S., Shaya, F., Keren, I., et al. (2014). nMAT4, a maturase factor required for nad1 pre-mRNA processing and maturation, is essential for holocomplex I biogenesis in Arabidopsis mitochondria. Plant J. 78 (2), 253–268. doi: 10.1111/tpj.12466
Duchêne, A.-M., Giritch, A., Hoffmann, B., Cognat, V., Lancelin, D., Peeters, N. M., et al. (2005). Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 102 (45), 16484–16489. doi: 10.1073/pnas.0504682102
Elthon, T. E., Nickels, R. L., McIntosh, L. (1989). Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol. 89 (4), 1311–1317. doi: 10.1104/pp.89.4.1311
Eubel, H., Braun, H.-P., Millar, A. H. (2005). Blue-native PAGE in plants: a tool in analysis of protein-protein interactions. Plant Methods 1 (1), 11–11. doi: 10.1186/1746-4811-1-11
Garmier, M., Carroll, A. J., Delannoy, E., Vallet, C., Day, D. A., Small, I. D., et al. (2008). Complex I dysfunction redirects cellular and mitochondrial metabolism in Arabidopsis. Plant Physiol. 148 (3), 1324–1341. doi: 10.1104/pp.108.125880
Gurer-Orhan, H., Ercal, N., Mare, S., Pennathur, S., Orhan, H., Heinecke, J. W. (2006). Misincorporation of free m-tyrosine into cellular proteins: a potential cytotoxic mechanism for oxidized amino acids. Biochem. J. 395 (2), 277–284. doi: 10.1042/BJ20051964
Höhner, R., Marques, J. V., Ito, T., Amakura, Y., Budgeon, A. D., Weitz, K., et al. (2018). Reduced arogenate dehydratase expression: ramifications for photosynthesis and metabolism. Plant Physiol. 177 (1), 115–131. doi: 10.1104/pp.17.01766
Hofius, D., Schultz-Larsen, T., Joensen, J., Tsitsigiannis, D. I., Petersen, N. H., Mattsson, O., et al. (2009). Autophagic components contribute to hypersensitive cell death in Arabidopsis. Cell 137 (4), 773–783. doi: 10.1016/j.cell.2009.02.036
Huang, T., Tohge, T., Lytovchenko, A., Fernie, A. R., Jander, G. (2010). Pleiotropic physiological consequences of feedback-insensitive phenylalanine biosynthesis in Arabidopsis thaliana. Plant J. 63 (5), 823–835. doi: 10.1111/j.1365-313X.2010.04287.x
Huang, T., Rehak, L., Jander, G. (2012). meta-Tyrosine in Festuca rubra ssp. commutata (Chewings fescue) is synthesized by hydroxylation of phenylalanine. Phytochemistry 75, 60–66. doi: 10.1016/j.phytochem.2011.09.018
Ipson, B. R., Fisher, A. L. (2016). Roles of the tyrosine isomers meta-tyrosine and ortho-tyrosine in oxidative stress. Ageing Res. Rev. 27, 93–107. doi: 10.1016/j.arr.2016.03.005
Jensen, E. C. (2013). Quantitative analysis of histological staining and fluorescence using ImageJ. Anat. Rec. 296 (3), 378–381. doi: 10.1002/ar.22641
Jung, E., Zamir, L. O., Jensen, R. A. (1986). Chloroplasts of higher plants synthesize L-phenylalanine via L-arogenate. Proc. Natl. Acad. Sci. 83 (19), 7231–7235. doi: 10.1073/pnas.83.19.7231
Kato, Y., Sakamoto, W. (2018). FtsH protease in the thylakoid membrane: physiological functions and the regulation of protease activity. Front. Plant Sci. 9, 855–855. doi: 10.3389/fpls.2018.00855
Kaufman, S. (1999). A model of human phenylalanine metabolism in normal subjects and in phenylketonuric patients. Proc. Natl. Acad. Sci. 96 (6), 3160–3164. doi: 10.1073/pnas.96.6.3160
Keren, I., Bezawork-Geleta, A., Kolton, M., Maayan, I., Belausov, E., Levy, M., et al. (2009). AtnMat2, a nuclear-encoded maturase required for splicing of group-II introns in Arabidopsis mitochondria. RNA 15 (12), 2299–2311. doi: 10.1261/rna.1776409
Keren, I., Tal, L., Colas des Francs-Small, C., Araújo, W. L., Shevtsov, S., Shaya, F., et al. (2012). nMAT1, a nuclear-encoded maturase involved in the trans-splicing of nad1 intron 1, is essential for mitochondrial complex I assembly and function. Plant J. 71 (3), 413–426. doi: 10.1111/j.1365-313X.2012.04998.x
Khan, F., Kumari, M., Cameotra, S. S. (2013). Biodegradation of the allelopathic chemical m-tyrosine by Bacillus aquimaris SSC5 involves the homogentisate central pathway. PloS One 8 (10), e75928–e75928. doi: 10.1371/journal.pone.0075928
Klipcan, L., Moor, N., Kessler, N., Safro, M. G. (2009). Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc. Natl. Acad. Sci. 106 (27), 11045–11048. doi: 10.1073/pnas.0905212106
Klipcan, L., Finarov, I., Moor, N., Safro, M. G. (2010). Structural aspects of phenylalanylation and quality control in three major forms of phenylalanyl-tRNA synthetase. J. Amino Acids 2010983503, 1–7. doi: 10.4061/2010/983503
Kotik-Kogan, O., Moor, N., Tworowski, D., Safro, M. (2005). Structural basis for discrimination of L-phenylalanine from L-tyrosine by phenylalanyl-tRNA synthetase. Structure 13 (12), 1799–1807. doi: 10.1016/j.str.2005.08.013
Kuhn, K., Obata, T., Feher, K., Bock, R., Fernie, A. R., Meyer, E. H. (2015). Complete mitochondrial complex I deficiency induces an up-regulation of respiratory fluxes that is abolished by traces of functional complex I. Plant Physiol. 168 (4), 1537–1549. doi: 10.1104/pp.15.00589
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Lamattina, L., Gonzalez, D., Gualberto, J., Grienenberger, J.-M. (1993). Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217 (3), 831–838. doi: 10.1111/j.1432-1033.1993.tb18311.x
Li, J., Schuman, M. C., Halitschke, R., Li, X., Guo, H., Grabe, V., et al. (2018). The decoration of specialized metabolites influences stylar development. eLife 7, e38611. doi: 10.7554/eLife.38611
Ling, J., Yadavalli, S. S., Ibba, M. (2007). Phenylalanyl-tRNA synthetase editing defects result in efficient mistranslation of phenylalanine codons as tyrosine. RNA 13 (11), 1881–1886. doi: 10.1261/rna.684107
Lisec, J., Schauer, N., Kopka, J., Willmitzer, L., Fernie, A. R. (2006). Gas chromatography mass spectrometry–based metabolite profiling in plants. Nat. Protoc. 1, 387–396. doi: 10.1038/nprot.2006.59
Luo, S., Levine, R. L. (2009). Methionine in proteins defends against oxidative stress. FASEB J. 23 (2), 464–472. doi: 10.1096/fj.08-118414
Maayan, I., Shaya, F., Ratner, K., Mani, Y., Lavee, S., Avidan, B., et al. (2008). Photosynthetic activity during olive (Olea europaea) leaf development correlates with plastid biogenesis and Rubisco levels. Physiol. Plant. 134 (3), 547–558. doi: 10.1111/j.1399-3054.2008.01150.x
MacIntyre, H. L., Kana, T. M., Anning, T., Geider, R. J. (2002). Photoacclimation of photosynthesis irradiance response curves and photosynthetic pigments in microalgae and cyanobacteria. J. Phycol. 38 (1), 17–38. doi: 10.1046/j.1529-8817.2002.00094.x
Mager, H., Berends, W. (1974). Activation and transfer of oxygen—IX: nonenzymic hydroxylation of phenylalanine by model systems of dihydroalloxazine/O2 dihydroalloxazine/H2O2 and alloxazinium cation/H2O. Tetrahedron 30 (7), 917–927. doi: 10.1002/recl.19720910913
Meyer, E. H., Solheim, C., Tanz, S. K., Bonnard, G., Millar, A. H. (2011). Insights into the composition and assembly of the membrane arm of plant complex I through analysis of subcomplexes in Arabidopsis mutant lines. J. Biol. Chem. 286 (29), 26081–26092. doi: 10.1074/jbc.M110.209601
Mhlongo, M. I., Piater, L. A., Madala, N. E., Labuschagne, N., Dubery, I. A. (2018). The chemistry of plant–microbe interactions in the rhizosphere and the potential for metabolomics to reveal signaling related to defense priming and induced systemic resistance. Front. Plant Sci. 9 (112), 1–17. doi: 10.3389/fpls.2018.00112
Moghal, A., Mohler, K., Ibba, M. (2014). Mistranslation of the genetic code. FEBS Lett. 588 (23), 4305–4310. doi: 10.1016/j.febslet.2014.08.035
Moghal, A., Hwang, L., Faull, K., Ibba, M. (2016). Multiple quality control pathways limit non-protein amino acid use by yeast cytoplasmic phenylalanyl-tRNA synthetase. J. Biol. Chem. 291 (30), 15796–15805. doi: 10.1074/jbc.M116.726828
Moghe, G. D., Last, R. L. (2015). Something old, something new: conserved enzymes and the evolution of novelty in plant specialized metabolism. Plant Physiol. 169 (3), 1512–1523. doi: 10.1104/pp.15.00994
Mohler, K., Ibba, M. (2017). Translational fidelity and mistranslation in the cellular response to stress. Nat. Microbiol. 2 (9), 1–9. doi: 10.1038/nmicrobiol.2017.117
Moldavski, O., Levin-Kravets, O., Ziv, T., Adam, Z., Prag, G. (2012). The hetero-hexameric nature of a chloroplast AAA+ FtsH protease contributes to its thermodynamic stability. PloS One 7 (4), e36008. doi: 10.1371/journal.pone.0036008
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. 115 (10), E2274–E2283. doi: 10.1073/pnas.1719588115
Mothes, K., Schütte, H. R., Müller, P., Ardenne, M. V., Tümmler, R. (1964). m-tyrosin, eine neue aminosäure aus dem milchsaft von euphorbia myrsinites l. Z. Naturforsch. B 19 (12), 1161–1162. doi: 10.1515/znb-1964-1219
Obata, T. (2019). Metabolons in plant primary and secondary metabolism. Phytochemistry 60 (7), 651–674. doi: 10.1007/s11101-019-09619-x
Ostersetzer, O., Adam, Z. (1997). Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell 9 (6), 957–965. doi: 10.1105/tpc.9.6.957
Perales, M., Eubel, H., Heinemeyer, J., Colaneri, A., Zabaleta, E., Braun, H.-P. (2005). Disruption of a nuclear gene encoding a mitochondrial gamma carbonic anhydrase reduces complex I and supercomplex I+III2 levels and alters mitochondrial physiology in Arabidopsis. J. Mol. Biol. 350 (2), 263–277. doi: 10.1016/j.jmb.2005.04.062
Pineau, B., Layoune, O., Danon, A., De Paepe, R. (2008). L-galactono-1,4-lactone dehydrogenase is required for the accumulation of plant respiratory complex I. J. Biol. Chem. 283 (47), 32500–32505. doi: 10.1074/jbc.M805320200
Popp, O., Larraillet, V., Kettenberger, H., Gorr, I. H., Hilger, M., Lipsmeier, F., et al. (2015). Molecular polygamy: the promiscuity of l-phenylalanyl-tRNA-synthetase triggers misincorporation of meta- and ortho-tyrosine in monoclonal antibodies expressed by Chinese hamster ovary cells. Biotechnol. Bioeng. 112 (6), 1187–1199. doi: 10.1002/bit.25528
Rappsilber, J., Mann, M., Ishihama, Y. (2007). Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906. doi: 10.1038/nprot.2007.261
Rasmusson, A. G., Geisler, D. A., Møller, I. M. (2008). The multiplicity of dehydrogenases in the electron transport chain of plant mitochondria. Mitochondrion 8 (1), 47–60. doi: 10.1016/j.mito.2007.10.004
Rizvi, S. J. H., Rizvi, V. (1992). Allelopathy: Basic and Applied Aspects (New York: Chapman and Hall), 480.
Rodgers, K. J., Shiozawa, N. (2008). Misincorporation of amino acid analogues into proteins by biosynthesis. Int. J. Biochem. Cell Biol. 40 (8), 1452–1466. doi: 10.1016/j.biocel.2008.01.009
Rodgers, K. J., Wang, H., Fu, S., Dean, R. T. (2002). Biosynthetic incorporation of oxidized amino acids into proteins and their cellular proteolysis. Free Radical Biol. Med. 32 (8), 766–775. doi: 10.1016/s0891-5849(02)00768-2
Roessner-Tunali, U., Hegemann, B., Lytovchenko, A., Carrari, F., Bruedigam, C., Granot, D., et al. (2003). Metabolic profiling of transgenic tomato plants overexpressing hexokinase reveals that the influence of hexose phosphorylation diminishes during fruit development. Plant Physiol. 133 (1), 84–99. doi: 10.1104/pp.103.023572
Roy, H., Ling, J., Irnov, M., Ibba, M. (2004). Post-transfer editing in vitro and in vivo by the beta subunit of phenylalanyl-tRNA synthetase. EMBO J. 23 (23), 4639–4648. doi: 10.1038/sj.emboj.7600474
Sakamoto, W., Zaltsman, A., Adam, Z., Takahashi, Y. (2003). Coordinated regulation and complex formation of YELLOW VARIEGATED1 and YELLOW VARIEGATED2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 (12), 2843–2855. doi: 10.1105/tpc.017319
Sakamoto, W. (2006). Protein degradation machineries in plastids. Annu. Rev. Plant Biol. 57 (1), 599–621. doi: 10.1146/annurev.arplant.57.032905.105401
Salomon, E., Keren, N. (2011). Manganese limitation induces changes in the activity and in the organization of photosynthetic complexes in the cyanobacterium Synechocystis sp. Strain PCC 6803. Plant Physiol. 155 (1), 571–579. doi: 10.1104/pp.110.164269
Shevtsov, S., Nevo-Dinur, K., Faigon, L., Sultan, L. D., Zmudjak, M., Markovits, M., et al. (2018). Control of organelle gene expression by the mitochondrial transcription termination factor mTERF22 in Arabidopsis thaliana plants. PloS One 13 (7), e0201631. doi: 10.1371/journal.pone.0201631
Siehl, D. L., Conn, E. E. (1988). Kinetic and regulatory properties of arogenate dehydratase in seedlings of Sorghum bicolor (L.) Moench. Arch. Biochem. Biophys. 260 (2), 822–829. doi: 10.1016/0003-9861(88)90513-9
Smith, I. K., Fowden, L. (1968). Studies on the specificities of the phenylalanyl- and tyrosyl-sRNA synthetases from plants. Phytochemistry 7 (7), 1064–1075. doi: 10.1016/S0031-9422(00)88252-7
Smith, L. C., Ravel, J. M., Lax, S. R., Shive, W. (1964). The effects of phenylalanine and tyrosine analogs on the synthesis and activity of 3-deoxy-d-arabino-heptulosonic acid 7-phosphate synthetases. Arch. Biochem. Biophys. 105, 424–430. doi: 10.1016/0003-9861(64)90026-8
Sultan, L. D., Mileshina, D., Grewe, F., Rolle, K., Abudraham, S., Głodowicz, P., et al. (2016). The reverse-transcriptase/RNA-maturase protein MatR is required for the splicing of various group II introns in Brassicaceae mitochondria. Plant Cell 28 (11), 2805–2829. doi: 10.1105/tpc.16.00398
Szabados, L., Savouré, A. (2010). Proline: a multifunctional amino acid. Trends Plant Sci. 15 (2), 89–97. doi: 10.1016/j.tplants.2009.11.009
Takechi, K., Sodmergen, Murata, M., Motoyoshi, F., Sakamoto, W. (2000). The yellow variegated (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol. 41 (12), 1334–1346. doi: 10.1093/pcp/pcd067
Tissier, A., Ziegler, J., Vogt, T. (2014). “Specialized Plant Metabolites: Diversity and Biosynthesis,” in Ecological Biochemistry. Eds. Krauss, G., Nies, D. H. (Hoboken, New Jersey, USA: Wiley-Blackwell). doi: 10.1002/9783527686063.ch2
Tohge, T., Watanabe, M., Hoefgen, R., Fernie, A. (2013). Shikimate and Phenylalanine Biosynthesis in the Green Lineage. Front. Plant Sci. 4 (62), 1–13. doi: 10.3389/fpls.2013.00062
Tzin, V., Galili, G. (2010). New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant 3 (6), 956–972. doi: 10.1093/mp/ssq048
Van Dingenen, J., Blomme, J., Gonzalez, N., Inzé, D. (2016). Plants grow with a little help from their organelle friends. J. Exp. Bot. 67 (22), 6267–6281. doi: 10.1093/jxb/erw399
van Dongen, J. T., Gupta, K. J., Ramirez-Aguilar, S. J., Araujo, W. L., Nunes-Nesi, A., Fernie, A. R. (2011). Regulation of respiration in plants: a role for alternative metabolic pathways. J. Plant Physiol. 168 (12), 1434–1443. doi: 10.1016/j.jplph.2010.11.004
van Loon, L. C. (2016). The Intelligent Behavior of Plants. Trends Plant Sci. 21 (4), 286–294. doi: 10.1016/j.tplants.2015.11.009
Voll, L. M., Allaire, E. E., Fiene, G., Weber, A. P. M. (2004). The Arabidopsis phenylalanine insensitive growth mutant exhibits a deregulated amino acid metabolism. Plant Physiol. 136 (2), 3058–3069. doi: 10.1104/pp.104.047506
Witzany, G. (2006). Plant communication from biosemiotic perspective: differences in abiotic and biotic signal perception determine content arrangement of response behavior. context determines meaning of meta-, inter- and intraorganismic plant signaling. Plant Signaling Behav. 1 (4), 169–178. doi: 10.4161/psb.1.4.3163
Wurtzel, E. T., Kutchan, T. M. (2016). Plant metabolism, the diverse chemistry set of the future. Science 353 (6305), 1232–1236. doi: 10.1126/science.aad2062
Yoshida, K., Noguchi, K. (2009). Differential gene expression profiles of the mitochondrial respiratory components in illuminated Arabidopsis leaves. Plant Cell Physiol. 50 (8), 1449–1462. doi: 10.1093/pcp/pcp090
Yoshimoto, K., Jikumaru, Y., Kamiya, Y., Kusano, M., Consonni, C., Panstruga, R., et al. (2009). Autophagy negatively regulates cell death by controlling NPR1-dependent Salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21 (9), 2914–2927. doi: 10.1105/tpc.109.068635
Young, P. G., Passalacqua, M. J., Chappell, K., Llinas, R. J., Bartel, B. (2019). A facile forward-genetic screen for Arabidopsis autophagy mutants reveals twenty-one loss-of-function mutations disrupting six ATG genes. Autophagy 15 (6), 941–959. doi: 10.1080/15548627.2019.1569915
Zaltsman, A., Ori, N., Adam, Z. (2005). Two types of FtsH protease subunits are required for chloroplast biogenesis and photosystem II repair in Arabidopsis. Plant Cell 17 (10), 2782–2790. doi: 10.1105/tpc.105.035071
Zmudjak, M., Colas des Francs-Small, C., Keren, I., Shaya, F., Belausov, E., Small, I., et al. (2013). mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 199 (2), 379–394. doi: 10.1111/nph.12282
Keywords: m-tyrosine, phenylalanine-tRNA synthetase, mitochondria, chloroplasts, translation, Arabidopsis thaliana
Citation: Zer H, Mizrahi H, Malchenko N, Avin-Wittenberg T, Klipcan L and Ostersetzer-Biran O (2020) The Phytotoxicity of Meta-Tyrosine Is Associated With Altered Phenylalanine Metabolism and Misincorporation of This Non-Proteinogenic Phe-Analog to the Plant's Proteome. Front. Plant Sci. 11:140. doi: 10.3389/fpls.2020.00140
Received: 19 November 2019; Accepted: 29 January 2020;
Published: 06 March 2020.
Edited by:
Philippe Giegé, Centre National de la Recherche Scientifique (CNRS), FranceReviewed by:
Markus Schwarzländer, University of Münster, GermanyCopyright © 2020 Zer, Mizrahi, Malchenko, Avin-Wittenberg, Klipcan and Ostersetzer-Biran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liron Klipcan, bGlyb25rQHZvbGNhbmkuYWdyaS5nb3YuaWw=; Oren Ostersetzer-Biran, b3Jlbi5vc3RlcnNldHplckBtYWlsLmh1amkuYWMuaWw=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.