
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Plant Sci. , 11 February 2020
Sec. Plant Development and EvoDevo
Volume 11 - 2020 | https://doi.org/10.3389/fpls.2020.00055
This article is part of the Research Topic Linking Stomatal Development and Physiology: From Stomatal Models to Non-Model Species and Crops View all 15 articles
Gas exchange between the plant and the atmosphere takes place through stomatal pores formed by paired guard cells. Grasses develop a unique stomatal structure that consists of two dumbbell-shaped guard cells flanked by lateral subsidiary cells. These structures confer a very efficient gas exchange capacity, which may have contributed to the evolutionary success of grasses. Recent works have identified orthologues of Arabidopsis MUTE in three grass species: BdMUTE in Brachypodium distachyon, BZU2/ZmMUTE in maize, and OsMUTE in rice. These genes induce the recruitment of subsidiary cells, and it appears to rely upon the ability of intercellular movement, from the guard mother cell to subsidiary mother cells, of the proteins encoded by them. Unexpectedly, this function of these grass MUTE genes contrasts with that of Arabidopsis MUTE, which promotes guard mother cell identity. These MUTE orthologues also appear to control guard mother cell fate progression, with the action of BdMUTE being less severe than those of BZU2/ZmMUTE and OsMUTE. The emerging picture unravels that grass MUTE genes have not only diverged, due to neo-functionalization, from Arabidopsis MUTE, but also among them.
Plants colonized land more than 400 million years ago (Edwards et al., 1998; Berry et al., 2010). One of the key innovations that enabled this to be possible was the development of a waxy cuticle to prevent water loss from the plant surface (Berry et al., 2010). The appearance of this impermeable layer coincides with the presence of stomatal pores, thus allowing the uptake of carbon dioxide to perform photosynthesis with a minimal loss of water vapor (Edwards et al., 1998; Berry et al., 2010). These microscopic innovations, bordered by a pair of kidney-shaped guard cells (GCs), are conserved across all land plants except liverworts and some mosses and hornworts (Chater et al., 2017; Renzaglia et al., 2017). Although to date no other structures has replaced the stoma, its shape, and its relationship with other epidermal cells have changed over time. Grasses, which develop a unique stomatal structure consisting of two dumbbell-shaped GCs flanked by two lateral subsidiary cells (SCs) (Stebbins and Shah, 1960; Rudall et al., 2017; Hepworth et al., 2018; Nunes et al., 2019), are a beautiful example of these changes. In addition, several works comparing stomatal responses between grasses and species with different stomatal morphology suggest that the stomatal complexes of grasses increase stomatal responsiveness with large and rapid GC movements (Franks and Farquhar, 2007; Bertolino et al., 2019 and references therein). Moreover, it has even been proposed that this developmental innovation has contributed, at least in part, to the extraordinary evolutionary success of this plant group (Kellogg, 2001; Hetherington and Woodward, 2003; Chen et al., 2017).
In the leaves of grasses, stomatal development occurs only in some epidermal cell files and it proceeds acropetally, with early stages of this process taking place in the basal regions of the leaf and stomata developing later in the distal ones (Stebbins and Shah, 1960). The development of four-celled stomatal complexes takes place through a simple and invariant pattern of cell divisions (Stebbins and Shah, 1960; Serna, 2011; Hepworth et al., 2018; Nunes et al., 2019; Figure 1A). They initiate with an asymmetric cell division from a protodermal cells leading to a smaller guard mother cell (GMC) and a larger sister cell. Before GMC division, cells from files in either side of newly formed GMC acquire subsidiary mother cell (SMC) identity and divide asymmetrically. The smaller cells resulting from these divisions, which are always placed next to the GMC, differentiate as SCs. Following SCs recruitment, the GMC divides symmetrically, with the cell division plane being parallel to the main axis of leaf growth, and it yields the paired GCs. This cell division pattern differs from that taking place in Arabidopsis (Serna and Fenoll, 2000; Bergmann and Sack, 2007; Figure 1A). First, in Arabidopsis, stomatal precursors, named meristemoids, are self-renewing cells. They can undergo several rounds of cell division in an inward spiral, regenerating themselves in each division, before assuming GMC identity. In contrast, in grasses, an asymmetric division directly gives rise to the stomatal precursor. Thus, meristemoids appear to be absent in this plant group. Second, the GMC of Arabidopsis does not recruit SCs. In addition, while grasses form dumbbell-shaped GCs, eudicots and most monocots develop kidney-shaped GCs pairs (Stebbins and Shah, 1960).
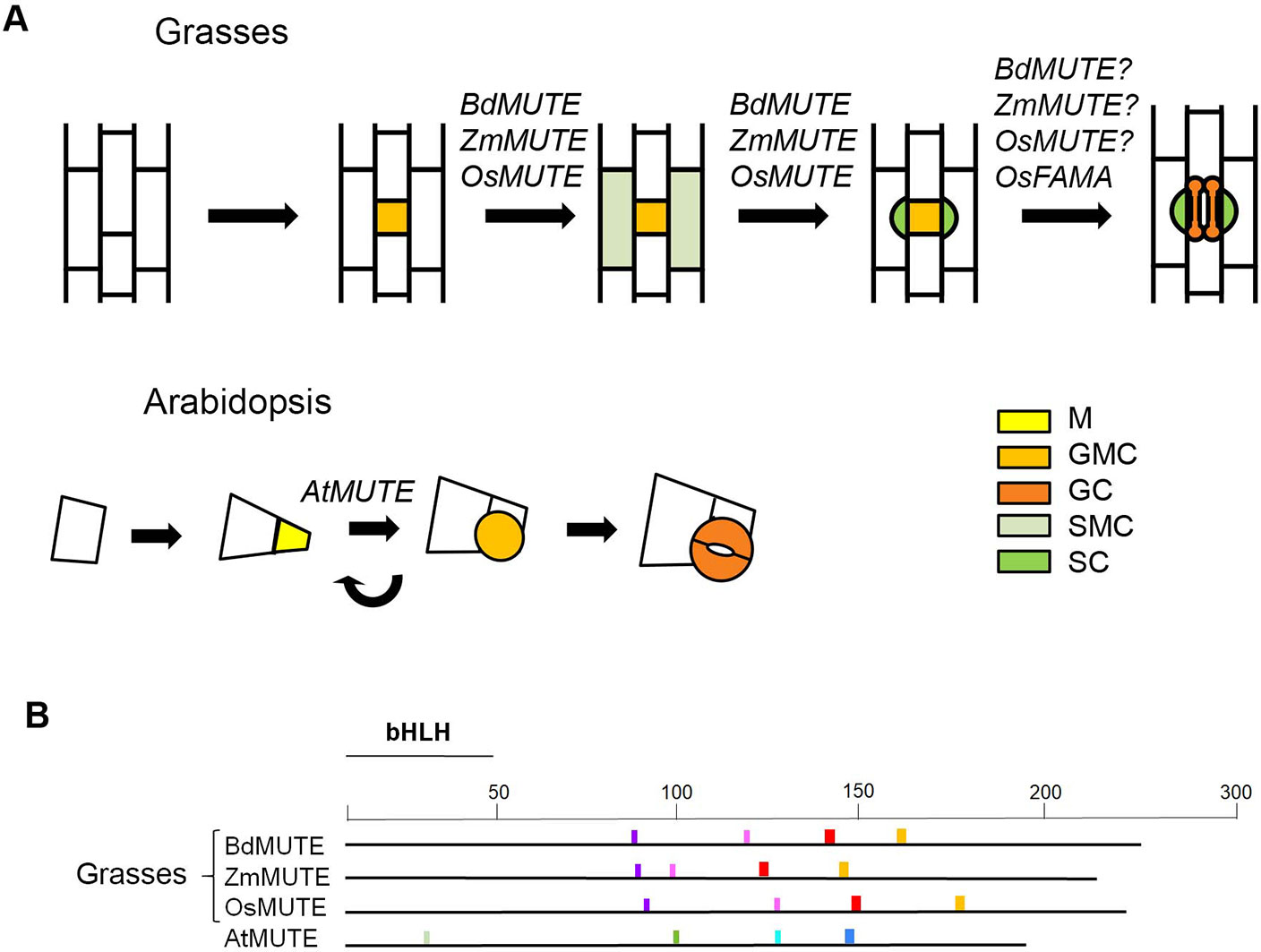
Figure 1 Role of MUTE orthologues of grasses and AtMUTE during stomatal development. (A) Stomatal development in grasses starts with an asymmetric division that produces a guard mother cell (GMC). Before GMC division, cells from files on either side of the GMC assume subsidiary mother cell (SMC) identity. SMCs then divide asymmetrically to produce subsidiary cells (SCs) in direct contact with the GMC. Only when the GMC is flanked by the two SCs, a symmetric cell division produces the two dumbbell-shaped guard cells (GCs). Grass MUTE genes specify SMC identity. They also appear to control the fate of the GMC, with the action of BdMUTE being less severe than those of ZmMUTE and OsMUTE. OsFAMA regulates the last stage of stomatal development. In Arabidopsis, a protodermal cell divides asymmetrically to produce a meristemiod (M) and a larger pavement cell. Ms usually reiterate asymmetric division several times, in an inward spiral, until they assume GMC identity. The GMC divides symmetrically to produce the two kidney-shaped GCs. AtMUTE regulates the transition of the M to GMC. (B) Schematic diagram of potential mobility motifs in the MUTE protein sequences. Conserved motifs in grass MUTE proteins could promote the intercellular movement, from the GMC to SMC, of these transcriptional factors. In contrast, the motifs conserved in AtMUTE could prevent its intercellular movement. The different-coloured boxes represent different motifs. These motifs are conserved among grasses but not in eudicots or vice versa, or are different between grasses and eudicots. Arabidopsis motifs are shown as an example of eudicot ones. The position of the bHLH domain is indicated. GC, guard cell; GMC, guard mother cell; M, meristemoid. SMC, subsidiary mother cell; SC, subsidiary cell.
The recruitment of SCs in grasses is preceded by a process of polarization of the SMC that is very well known in maize (Serna, 2015; Apostolakos et al., 2018; Figure 2). This begins with the accumulation of the SCAR/WAVE regulatory complex (WRC) at the cell surface of the SMC, specifically at the site of GMC contact (Facette et al., 2015). Unknown signals emanating from GMC activate PANs receptors, which also, in a WRC-dependent manner (Facette et al., 2015), accumulate at the SMC/GMC contact site (Cartwright et al., 2009; Zhang et al., 2012). Then PANs recruit and activate ROPs (Humphries et al., 2011), which activate the WRC complex (Facette et al., 2015). Finally, activated WRC activates the ARP2/3 complex giving rise to a dense T-actin and inducing the migration of the nucleus toward the GMC (Deeks and Hussey, 2005).
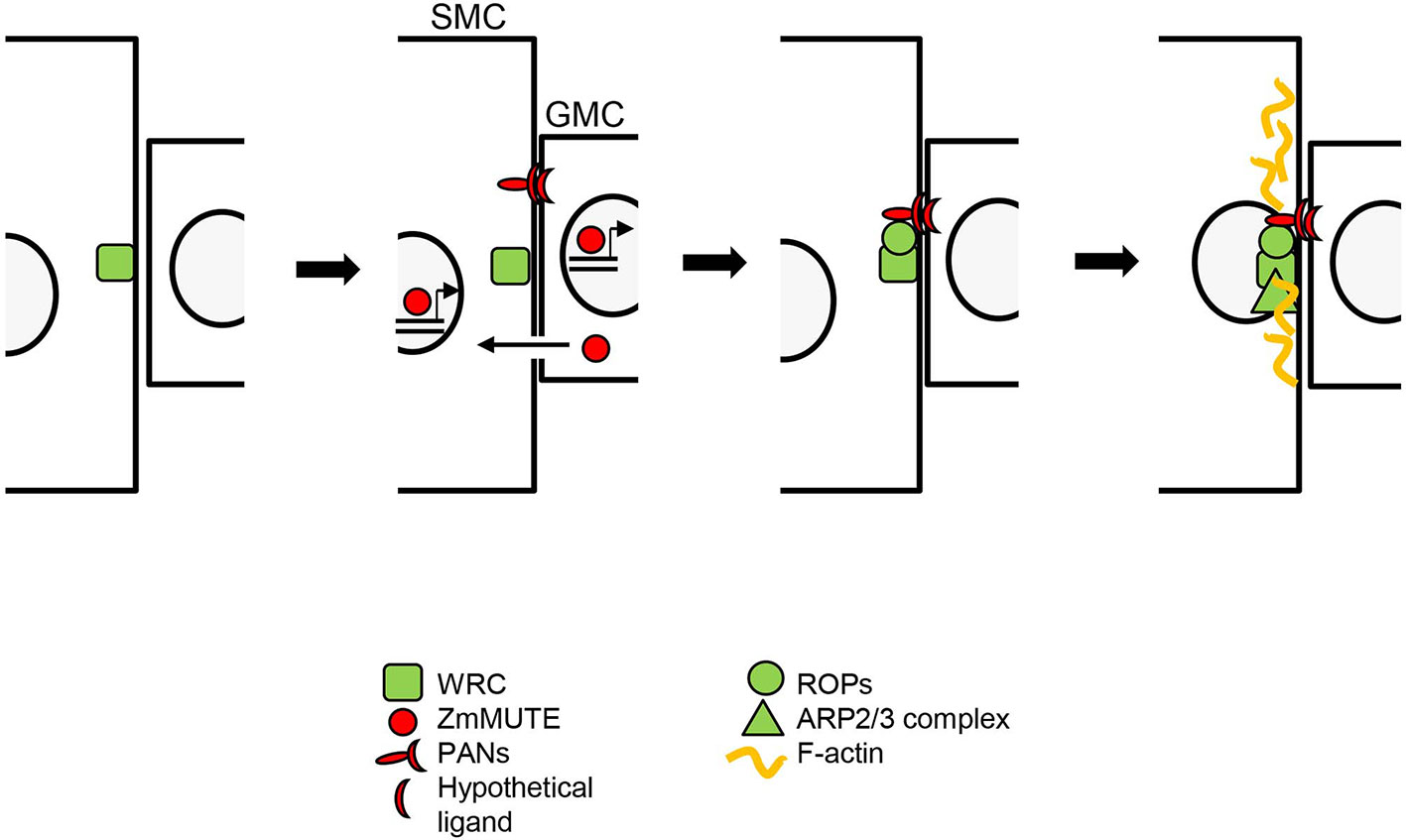
Figure 2 ZmMUTE role during the polarization of the subsidiary mother cell (SMC) in maize. The WAVE regulatory complex (WRC) complex polarizes to the cell surface of the SMC, at the site of guard mother cell (GMC) contact. ZmMUTE moves from the GMC to SMCs, where it binds to PANs promoters and promotes their induction. PANs accumulate, in a WRC-dependent manner, at the SMC/GMC contact site. PANs then recruit and activate ROPs. Activated ROPs physically interact and activate the WRC, which activates the ARP2/3 complex. Finally, ARP2/3 activity produces a dense F-actin patch and promotes nuclear migration towards the GMC in an actin-dependent manner. ZmMUTE may also exert a cell-autonomous role inducing, directly or indirectly, the expression of the hypothetical ligands of PANs. GMC, guard mother cell. SMC, subsidiary mother cell.
Over the last twenty-five years, the isolation and characterization of numerous genes have illuminated our understanding of stomatal development in Arabidopsis. In this model plant, three basic helix-loop-helix (bHLH) transcription factors, SPEECHLESS (SPCH), MUTE, and FAMA sequentially specify stomatal lineage identity, regulate the transition from meristemoids to GMCs, and promote GCs differentiation, respectively (Ohashi-Ito and Bergmann, 2006; MacAlister et al., 2007; Pillitteri et al., 2007). The function of these transcriptional factors requires heterodimerization with the functionally redundant bHLH proteins ICE1 or SCREAM2 (Kanaoka et al., 2008). With origins which predate the divergence of the mosses and hornworts, these bHLH proteins diverged prior to the monocot-dicot split (Chater et al., 2017; Hepworth et al., 2018, and references therein). This divergence enabled the emergence of new protein functions, which are essential for the unique stomatal development of grasses (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019).
Recent works have uncovered the role of three orthologues of Arabidopsis MUTE during stomatal development in three grass species (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019): BdMUTE of Brachypodium distachyon, BZU2/ZmMUTE of Zea mays (maize) and OsMUTE of Oryza sativa (rice). The role of these genes, regulating the formation of SCs (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019), differs from that of AtMUTE. These orthologues of MUTE also seem to regulate the identity of the GMC, with the action of BdMUTE in this process being less severe. Taken together, these recent discoveries suggest not only that MUTE orthologues of grasses have diverged from AtMUTE, but also that MUTE genes of domesticated grasses studied thus far have diverged comparatively to BdMUTE.
In Brachypodium, maize and rice, like in other grass species, stomatal complexes comprise a pair of dumbbell-shaped GCs associated with two SCs (Campbell, 1881; Stebbins and Shah, 1960; Shoemaker and Srivastava, 1973; Kamiya et al., 2003; Raissig et al., 2017). Interestingly, the mutants subsidiary cell identity defective (sid) of B. distachyon, bizui2 (bzu2) of maize and c-osmute of rice, in addition to having impaired other steps of the stomatal developmental, fail to recruit SCs (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019). These mutants have alterations in their orthologues of Arabidopsis MUTE (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019): sid in BdMUTE, bzu2-1 in BZU2/ZmMUTE and c-osmute in OsMUTE. The function of these MUTE orthologues contrasts with that of AtMUTE, which triggers GMC formation only in Arabidopsis (MacAlister et al., 2007; Pillitteri et al., 2007; Table 1).
How do these grass orthologues of Arabidopsis MUTE recruit SCs? The expression of the yellow fluorescent protein (YFP) under the control of the BdMUTE promoter showed that its induction is restricted to GMCs and GCs (Raissig et al., 2017). However, analysis of transgenic plants expressing the protein encoded by YFP-BdMUTE construct driven by the BdSPCH2 promoter showed that BdMUTE locates not only in GMCs but also in SMCs (Raissig et al., 2017). Considering that the BdSPCH2 promoter is active only in the stomatal lineage cells (Raissig et al., 2016), Raissig et al. (2017) inferred that BdMUTE protein moves from the GMC to epidermal cells of neighboring files. In consonance, successful complementation experiments of sid mutants with a fusion of the BdMUTE promoter to the YFP-BdMUTE construct lights up not only GMCs and young GCs, but also SMCs and young SCs (Raissig et al., 2017). In rice, YFP-OsMUTE expression driven by the OsMUTE promoter, whose induction in the developing four-celled complex is restricted to GMCs (Liu et al., 2009; Wang et al., 2019), lights up also GMCs and SMCs (Wang et al., 2019). This indicates that OsMUTE, like BdMUTE, also moves from the GMC to epidermal cells of neighboring files (Wang et al., 2019). Maize expressing YFP-ZmMUTE driven by the ZmMUTE promoter illuminates also GMCs, young GCs and SMCs (Wang et al., 2019). Assuming that the cellular localization of the ZmMUTE promoter induction is restricted to GMCs and young GCs, ZmMUTE would also move from the GMC to epidermal cells of neighboring files. Indeed, ZmMUTE protein is also able to move from the GMCs to SMCs in rice, and to epidermal cells adjacent to the stoma in Arabidopsis (Wang et al., 2019). These experiments strongly suggest that the recruitment of SCs in grasses depends on the intercellular movement of the grass MUTE proteins. Interestingly, the overexpression of BdMUTE driven by the Ubi constitutive promoter produces not only lateral, but also polar SCs (Raissig et al., 2017). This emphasizes the relationship between SCs recruitment and intercellular movement of MUTE orthologues in grasses. However, direct proof conclusively validating that the SCs formation relies upon the grass MUTE intercellular movement is lacking.
Multiple studies have shown that transcriptional factors move among cells via plasmodesmata (Han et al., 2014). GMCs of Brachypodium are symplastically connected with surrounding epidermal cells (Raissig et al., 2017). Therefore, BdMUTE may also move from the GMC to cells of neighboring files through plasmodesmata. But, what allows this protein to move laterally but not radially? Plasmodesmata continuously adjust their permeability in response to multiple cues (Sager and Lee, 2018). In addition, it is known that the control of this permeability is essential to the proper segregations of cell fate determinants during stomatal development in Arabidopsis (Guseman et al, 2010; Kong et al., 2012). Therefore, the lateral mobility of BdMUTE, and the unique design of the grass stomatal complexes, could depend on the restriction of the permeability of the plasmodesmata that symplastically connect cells of the same row. Future research should include delving in this direction.
MUTE orthologues of grasses move from the GMC to the cells of neighboring files and this is linked with the formation of SCs. But, what do they do there? Wang and co-workers (2019) examined the role of ZmMUTE, specifically with regard to the regulation of SMC polarization. They found that cells adjacent to stomata, placed in neighboring epidermal files, of the bzu2-1 mutant, in contrast to the wild type, do not show enrichment of F-actin patches at the GMC contact sites or polarization of their nuclei (Wang et al., 2019). This indicates that SMC polarization is not cell-autonomous, and that ZmMUTE regulates this process (Table 1). The bzu2-1 mutation downregulates the transcription of both PAN1 and PAN2, indicating that ZmMUTE transcriptional factor positively regulates PAN1 and PAN2 expression (Wang et al., 2019). Because PAN1, whose polarization at the SMC/GMC interface requires PAN2 (Cartwright et al., 2009; Zhang et al., 2012), recruits and activates ROPs (Humphries et al., 2011), ZmMUTE must induce PANs expression before ROPs polarization (Figure 2). Interestingly, while the bzu2-1 mutant is almost devoid of SCs (Wang et al., 2019), most of the SCs of pan1 and pan2 mutants show no defects and most probably they derive from normal asymmetric cell divisions (Gallagher and Smith, 2000; Cartwright et al., 2009; Zhang et al., 2012; Facette et al., 2015). Therefore, ZmMUTE, or its downstream transcriptional factors, controls the expression not only of PANs, but also of other unknown genes to induce the recruitment of SCs. Among these genes could be those that encode for the hypothetical ligands of PANs, and his discovery would reveal one of the best-kept secrets of SMC polarization. If this is so, and assuming that these ligands emanate from the GMC, ZmMUTE may have both cell-autonomous and non-cell-autonomous functions (Figure 2).
Yeast one-hybrid and EMSA experiments showed that ZmMUTE binds to the E-box P1 and P2 motifs of the PAN1 and PAN2 promoters respectively (Wang et al., 2019). This suggests that the action of ZmMUTE on the expression of these genes may be direct. This does not rule out that ZmMUTE could also indirectly affect the activity of PAN1 and PAN2 promoters through upregulation of positive regulators and/or downregulation of repressors of PANs expression. For example, the transcriptional factor Glis3, directly and indirectly, regulates the expression of the insulin gene (Yang et al., 2009). The ZmMUTE protein does not bind the E-box P3 motif of the PAN2 promoter in vitro, but ChlP-qPCR data indicate that it binds this motif in vivo (Wang et al., 2019). This suggests that the ZmMUTE protein physically interacts with other proteins to activate the E-box P3 of the PAN2 promoter (Wang et al., 2019). Interestingly, yeast two-hybrid and bimolecular fluorescence complementation assays have just showed that its orthologue of rice, OsMUTE, interacts with OsICE1 and OsICE2 (Wu et al., 2019). Therefore, ZmMUTE may interact with their homologs to regulate the E-box P3 of the PAN2 promoter.
BdMUTE, ZmMUTE and OsMUTE conserve the motifs that could promote their intercellular movement or lack those that could prevent such movement (Raissig et al., 2017; Xu et al., 2018; Figure 1B). These proteins move from the GMC to epidermal cells of neighboring files, where they may specify SMC identity to recruit SCs (Raissig et al., 2017; Wang et al., 2019; Wu et al., 2019). In contrast, the Arabidopsis MUTE protein, whose gene is expressed in GMCs (MacAlister et al., 2007; Pillitteri et al., 2007), does not move among cells (Wang et al., 2019). As expected, AtMUTE does not have the conserved mobility motifs of grass MUTE proteins, but those conserved in eudicots (Raissig et al., 2017; Figure 1B). In accordance, the recruitment of SCs does not take place in Arabidopsis. The role of AtMUTE is restricted to control the meristemoid to GMC transition (MacAlister et al., 2007; Pillitteri et al., 2007).
The fact that the YFP-BdMUTE construct, driven by the GMC-specific AtMUTE promoter (MacAlister et al., 2007; Pillitteri et al., 2007), illuminates not only stomatal precursors but also adjacent epidermal cells in Arabidopsis (Raissig et al., 2017; Wang et al., 2019), underlines the very likely importance of the mobility motifs in the protein movement. Interestingly, this construct does not induce the recruitment of SCs in Arabidopsis (Raissig et al., 2017; Wang et al., 2019), highlighting that AtMUTE and BdMUTE have diverged. AtMUTEp : YFP-ZmMUTE in Arabidopsis also lights up GMCs and neighboring epidermal cells (Wang et al., 2019). Given that OsMUTE conserve the motifs that could promote its movement or lack those that could prevent it (Raissig et al., 2017), it is expected that this protein also moves from stomatal precursors to neighboring epidermal cells in Arabidopsis. OsMUTE and ZmMUTE expressed under the control of the AtMUTE promoter partially complement the defects of Arabidopsis mute-1 by inducing the formation of stomata from some stomatal precursors (Liu et al., 2009), but, like BdMUTE, they do not induce the recruitment of SCs (Figure 4 in Liu et al., 2009). Although OsMUTE and ZmMUTE, and perhaps BdMUTE, retain the function of inducing stomata formation, they are unable to induce the recruitment of SCs in Arabidopsis. This underlies that these proteins have diverged from the AtMUTE protein acquiring of a new function: the recruitment of SCs.
Although the three orthologues of MUTE regulate the formation of SCs, their function during stomatal development is not identical (Table 1). The bzu2-1 mutant forms GMCs but displays defects in their divisions, undergoing excessive, randomly oriented and/or asymmetric divisions (Wang et al., 2019). This gives rise to short columns of elongated cells instead of stomata, which results in a slower transpiration rate and in a decreased photosynthetic activity (Wang et al., 2019). c-osmute exhibits also columns of undifferentiated cells, produced by misoriented and/or asymmetric cell divisions (Wu et al., 2019). Morphologically, the phenotype of the c-osmute mutant is reminiscent of the bzu2-1 one, and the physiology of c-osmute mutants must also be dramatically affected. Certainly, both mutants exhibit a lethal phenotype at the seedling stage (Wang et al., 2019; Wu et al., 2019).
The sid mutant is fully viable and fertile, although its stomatal physiology is also affected (Raissig et al., 2017). Like the bzu2-1 and c-osmute mutants, the sid mutant also undergoes misoriented GMC divisions (Raissig et al., 2017). However, in contrast to these mutants, about 70% of the GMCs of this mutant develop dicot-like two celled stomata (Raissig et al., 2017). Therefore, while bzu2-1 and c-osmute mutants exhibit a fully penetrant phenotype affecting the division of the GMC (Wang et al., 2019; Wu et al., 2019), many of GMC divisions of the sid mutant are normal (Raissig et al., 2017). The molecular nature triggering the lack of a fully penetrant phenotype in sid is unknown, and to delve into this question is one of the most exciting future directions. OsFAMA also controls GC morphogenesis, with c-osmute exhibiting stomata with swollen GCs (Wu et al., 2019). This mutant also exhibits a fraction of swollen SCs, suggesting that, in addition to OsFAMA, other genes regulate SC differentiation (Wu et al., 2019). Among these unknown genes could be OsMUTE.
The defects induced by mutations in the grass MUTE orthologues in the maintenance of the GMC identity could reflect a mechanism of cellular signaling from the SMC towards the GMC to induce stomatal formation. It has been proposed that, prior SC formation, high levels of a grass peptide similar to AtEPF1/2 may cause GMC arrest (Hughes et al., 2017; Hepworth et al., 2018), perhaps through the suppression of grass MUTE orthologues activity specifically in GMCs. Grass MUTE activity in SMC would allow SC formation. Then, signals from SCs may activate grass MUTE orthologues in GMCs, perhaps by reducing grass EPF1/2 production, to promote stomatal formation. Agree with this, 1) GMCs do not progress to become stomata until SC formation, and 2) barley overexpressing HvEPF1 exhibits arrested GMCs (Hughes et al., 2017). It is then likely that grass MUTE genes, in addition to having a non-cell-autonomous role specifying the SMC fate, have a cell-autonomous one triggering the progression of the GMC fate. The complementation of the Arabidopsis mute-1 mutant phenotype, inducing stomatal development from some stomatal precursors, with at least the OsMUTE and ZmMUTE genes (Liu et al., 2009), also supports the cell-autonomous role of MUTE orthologues regulating GMC fate.
Several works comparing physiological stomatal behaviors among species with different stomatal complexes suggest that those of grasses are more efficient (Grantz and Zeiger, 1986; Franks and Farquhar, 2007; Vico et al., 2011; Merilo et al., 2014; McAusland et al., 2016; Haworth et al., 2018). The isolation of the sid mutant, the first grass mutant to date that disrupts the two main attributes of the grass stomatal complexes, the presence of dumbbell-shaped GCs and the recruitment of SCs (Raissig et al., 2017), underscores the important role of this innovative morphology in the stomatal function (Nunes et al., 2019). The maximum area of the open pore in the sid mutant, and its gas exchange capacity, were noticeably smaller than those in the wild type, even when stomatal opening was induced by the toxin fusicoccin (Raissig et al., 2017). The sid mutant also exhibited slower stomatal movements to fluctuating light conditions, and its stomata could not open as wide compared with the wild type (Raissig et al., 2017). Consequently, sid mutants produced less biomass than the wild types (Raissig et al., 2017). These results link the morphology of the stomatal complexes with its impact on gas exchange and biomass production in the wild grass Brachypodium, and strongly they suggest that this relationship may extend to the remaining grass species.
The improvement of stomatal function in grasses could have contributed to their expansion and diversification, 30 to 45 million years ago, when a progressive and global aridification took place (Kellogg, 2001; Hetherington and Woodward, 2003; Chen et al., 2017). The inability of the sid mutant to open widely its pores indicates that grass stomatal complexes are associated with greater stomatal openings and conductance. Interestingly, species with greater maximum stomatal conductance exhibit higher sensitivity to closure during drought (Henry et al., 2019). Under a global drought, a more sensitive stomatal closure could have allowed to capture carbon dioxide without losing too much water, thus favoring the successful diversification of this plant group. Certainly, Poaceae, with around 12,000 species, includes almost a quarter of all monocots of the planet, and it is one of the largest families of flowering plants. Curiously, the enrichment of species in genera of monocotyledons is associated with geographical variables, like larger ranges and lower elevations, rather than with biological attributes (Tang et al., 2016). It is likely that the success of the grasses lies partly in their morphology, including their unique stomatal complexes, and partly in the places they have occupied.
The development of the unique grass stomatal complex is a great advantage, which may have contributed to the expansion of this plant group. Undoubtedly, MUTE orthologues of grasses provide a starting point to unravel not only the mechanism underlying stomatal complexes formation, but also the evolution of this essential trait. MUTE orthologues of grasses have not only functionally diverged, due to neo-functionalization, from AtMUTE, but also among them, with BdMUTE exhibiting divergence from ZmMUTE and OsMUTE. Certainly, protein phylogenetic analysis of bHLH regulators of stomatal development supports this view (Wu et al., 2019). The comparison of the grass MUTE function between domesticated plants and their wild relatives, will allow us to know if the agricultural practices have driven the divergence of these genes. Because grass stomatal complexes have largely contributed to the adaptive success in hotter and drier environment, delving into the function of these genes will also provide useful genetic tools for producing plants with better tolerance to drought caused by climate change.
LS wrote the article and designed the figures.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Apostolakos, P., Livanos, P., Giannoutsou, E., Panteris, E., Galatis, B. (2018). The intracellular and intercellular cross-talk during subsidiary cell formation in Zea mays: existing and novel components orchestrating cell polarization and asymmetric division. Ann. Bot. 122, 679–696. doi: 10.1093/aob/mcx193
Bergmann, D. C., Sack, F. D. (2007). Stomatal development. Annu. Rev. Plant Biol. 58, 163–181. doi: 10.1146/annurev.arplant.58.032806.104023
Berry, J. A., Beerling, D. J., Franks, P. J. (2010). Stomata: key players in the earth system, past and present. Curr. Opin. Plant Biol. 13, 233–240. doi: 10.1016/j.pbi.2010.04.013
Bertolino, L. T., Caine, R. S., Gray, J. E. (2019). Impact of stomatal density and morphology on water-use efficiency in a changing world. Front. Plant Sci. 10, 225. doi: 10.3389/fpls.2019.00225
Campbell, D. H. (1881). On the development of the stomata of Tradescantia and Indian corn. Amer. Nat. 15, 761–766. doi: 10.1086/272919
Cartwright, H. N., Humphries, J. A., Smith, L. G. (2009). PAN1: a receptor-like protein that promotes polarization of an asymmetric cell division in maize. Science 323, 649–651. doi: 10.1126/science.1161686
Chater, C. C. C., Caine, R. S., Fleming, A. J., Gray, J. E. (2017). Origins and evolution of stomatal development. Plant Physiol. 174, 624–638. doi: 10.1104/pp.17.00183
Chen, Z. H., Chen, G., Dai, F., Wang, Y., Hills, A., Ruan, Y. L., et al. (2017). Molecular evolution of grass stomata. Trends Plant Sci. 22, 124–139. doi: 10.1016/j.tplants.2016.09.005
Deeks, M. J., Hussey, P. J. (2005). Arp2/3 and SCAR: plants move to the fore. Nat. Rev. Mol. Cell. Biol. 6, 954–964. doi: 10.1038/nrm1765
Edwards, D., Kerp, H., Hass, H. (1998). Stomata in early land plants: an anatomical and ecophysiological approach. J. Exp. Bot. 49, 255–278. doi: 10.1093/jxb/49.Special_Issue.255
Facette, M. R., Park, Y., Sutimantanapi, D., Luo, A., Cartwright, H. N., Yang, B., et al. (2015). The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat. Plants 1, 14024. doi: 10.1038/nplants.2014.24
Franks, P. J., Farquhar, G. D. (2007). The mechanical diversity of stomata and its significance in gas-exchange control. Plant Physiol. 143, 78–87. doi: 10.1104/pp.106.089367
Gallagher, K., Smith, L. G. (2000). Roles for polarity and nuclear determinants in specifying daughter cell fates after an asymmetric cell division in the maize leaf. Curr. Biol. 10, 1229–1232. doi: 10.1016/S0960-9822(00)00730-2
Grantz, D. A., Zeiger, E. (1986). Stomatal responses to light and leaf-air water vapor pressure difference show similar kinetics in sugarcane and soybean. Plant Physiol. 81, 865–868. doi: 10.1104/pp.81.3.865
Guseman, J. M., Lee, J. S., Bogenschutz, N. L., Peterson, K. M., Virata, R. E., Xie, B., et al. (2010). Dysregulation of cell-to-cell connectivity and stomatal patterning by loss-of-function mutation in Arabidopsis chorus (glucan synthase-like 8). Development 137, 1731–1741. doi: 10.1242/dev.049197
Han, X., Kumar, D., Chen, H., Wu, S., Kim, J.-Y. (2014). Transcription factor-mediated cell-to-cell signalling in plants. J. Exp. Bot. 65, 1737–1749. doi: 10.1093/jxb/ert422
Haworth, M., Scutt, C. P., Douthe, C., Marino, G., Gomes, M. T. G., Loreto, F., et al. (2018). Allocation of the epidermis to stomata relates to stomatal physiological control: stomatal factors involved in the evolutionary diversification of the angiosperms and development of amphistomaty. Environ. Exp. Bot. 151, 55–63. doi: 10.1016/j.envexpbot.2018.04.010
Henry, C., John, G. P., Pan, R., Bartlett, M. K., Fletcher, L. R., Scoffoni, C., et al. (2019). A stomatal safety-efficiency trade-off constrains responses to leaf dehydration. Nat. Commun. 10, 3398. doi: 10.1038/s41467-019-11006-1
Hepworth, C., Caine, R. S., Harrison, E. L., Sloan, J., Gray, J. E. (2018). Stomatal development: focusing on the grasses. Curr. Opin. Plant Biol. 41, 1–7. doi: 10.1016/j.pbi.2017.07.009
Hetherington, A. M., Woodward, F. I. (2003). The role of stomata in sensing and driving environmental change. Nature 424, 901–908. doi: 10.1038/nature01843
Hughes, J., Hepworth, C., Dutton, C., Dunn, J. A., Hunt, L., Stephens, J., et al. (2017). Reducing stomatal density in barley improves drought tolerance without impacting on yield. Plant Physiol. 174, 776–787. doi: 10.1104/pp.16.01844
Humphries, J. A., Vejlupkova, Z., Luo, A., Meeley, R. B., Sylvester, A. W., Fowler, J. E., et al. (2011). ROP GTPases act with the receptor-like protein PAN1 to polarize asymmetric cell division in maize. Plant Cell 23, 2273–2284. doi: 10.1105/tpc.111.085597
Kamiya, N., Itoh, J. I., Morikami, A., Nagato, Y., Matsuoka, M. (2003). The SCARECROW gene’s role in asymmetric cell divisions in rice plants. Plant J. 36, 45–54. doi: 10.1046/j.1365-313X.2003.01856.x
Kanaoka, M. M., Pillitteri, L. J., Fujii, H., Yoshida, Y., Bogenschutz, N. L., Takabayashi, J., et al. (2008). SCREAM/ICE1 and SCREAM2 specify three cell-state transitional steps leading to arabidopsis stomatal differentiation. Plant Cell. 20, 1775–1785. doi: 10.1105/tpc.108.060848
Kellogg, E. A. (2001). Evolutionary history of the grasses. Plant Physiol. 125, 1198–1205. doi: 10.1104/pp.125.3.1198
Kong, D., Karve, R., Willet, A., Chen, M.-K., Oden, J., Shpak, E. D. (2012). Regulation of plasmodesmatal permeability and stomatal patterning by the glycosyltransferase-like protein KOBITO1. Plant Physiol. 159, 156–168. doi: 10.1104/pp.112.194563
Liu, T., Ohashi-Ito, K., Bergmann, D. C. (2009). Orthologues of Arabidopsis thaliana stomatal bHLH genes and regulation of stomatal development in grasses. Development 136, 2265–2276. doi: 10.1242/dev.032938
MacAlister, C. A., Ohashi-Ito, K., Bergmann, D. C. (2007). Transcription factor control of asymmetric cell divisions that establish the stomatal lineage. Nature 445, 537–540. doi: 10.1038/nature05491
McAusland, L., Vialet-Chabrand, S., Davey, P., Baker, N. R., Brendel, O., Lawson, T. (2016). Effects of kinetics of light-induced stomatal responses on photosynthesis and water-use efficiency. New Phytol. 211, 1209–1220. doi: 10.1111/nph.14000
Merilo, E., Jõesaar, I., Brosché, M., Kollist, H. (2014). To open or to close: species-specific stomatal responses to simultaneously applied opposing environmental factors. New Phytol. 202, 499–508. doi: 10.1111/nph.12667
Nunes, T. D. G., Zhang, D., Raissig, M. T. (2019). Form, development and function of grass stomata. Plant J. doi: 10.1111/tpj.14552
Ohashi-Ito, K., Bergmann, D. C. (2006). Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 18, 2493–2505. doi: 10.1105/tpc.106.046136
Pillitteri, L. J., Sloan, D. B., Bogenschutz, N. L., Torii., K. U. (2007). Termination of asymmetric cell division and differentiation of stomata. Nature 445, 501–505. doi: 10.1038/nature05467
Raissig, M. T., Abrash, E., Bettadapur, A., Vogel, J. P., Bergmann, D. C. (2016). Grasses use an alternatively wired bHLH transcription factor network to establish stomatal identity. Proc. Natl. Acad. Sci. U. S. A. 113, 8326–8331. doi: 10.1073/pnas.1606728113
Raissig, M. T., Matos, J. L., Gil, M. X. A., Kornfeld, A., Bettadapur, A., Abrash, E., et al. (2017). Mobile MUTE specifies subsidiary cells to build physiologically improved grass stomata. Science 355, 1215–1218. doi: 10.1126/science.aal3254
Renzaglia, K. S., Villarreal, J. C., Piatkowski, B. T., Lucas, J. R., Merced, A. (2017). Hornwort stomata: architecture and fate shared with 400-million-year-old fossil plants without leaves. Plant Physiol. 174, 788–797. doi: 10.1104/pp.17.00156
Rudall, P. J., Chen, E. D., Cullen, E. (2017). Evolution and development of monocot stomata. Am. J. Bot. 104, 1122–1140. doi: 10.3732/ajb.1700086
Sager, R. E., Lee, J. Y. (2018). Plasmodesmata at a glance. J. Cell Sci. 131, pii: jcs209346. doi: 10.1242/jcs.209346
Serna, L., Fenoll, C. (2000). Stomatal development in Arabidopsis: how to make a functional pattern. Trends Plant Sci. 5, 458–460. doi: 10.1016/S1360-1385(00)01782-9
Serna, L. (2011). Stomatal development in Arabidopsis and grasses: differences and commonalities. Int. J. Dev. Biol. 55, 5–10. doi: 10.1387/ijdb.103094ls
Serna, L. (2015). Development: Early events in asymmetric division. Nat. Plants 1, 15008. doi: 10.1038/nplants.2015.8
Shoemaker, E. M., Srivastava, L. M. (1973). The mechanics of stomatal opening in corn (Zea mays L.) leaves. J. Theor. Biol. 42, 219–225. doi: 10.1016/0022-5193(73)90086-6
Stebbins, G. L., Shah, S. S. (1960). Developmental studies of cell differentiation in the epidermis of monocotyledons. II. Cytological features of stomatal development in the Gramineae. Dev. Biol. 2, 477–500. doi: 10.1016/0012-1606(60)90050-6
Tang, C. Q., Orme, C. D. L., Bunnefeld, L., Jones, F. A., Powell, S., Chase, M. W., et al. (2016). Global monocot diversification: geography explains variation in species richness better than environment or biology. Bot. J. Linn. Soc. 83, 1–15. doi: 10.1111/boj.12497
Vico, G., Manzoni, S., Palmroth, S., Katul, G. (2011). Effects of stomatal delays on the economics of leaf gas exchange under intermittent light regimes. New Phytol. 192, 640–652. doi: 10.1111/j.1469-8137.2011.03847.x
Wang, H., Guo, S., Qiao, X., Guo, J., Li, Z., Zhou, Y., et al. (2019). BZU2/ZmMUTE controls symmetrical division of guard mother cell and specifies neighbor cell fate in maize. PloS Genet. 15, e1008377. doi: 10.1371/journal.pgen.1008377
Wu, Z., Chen, L., Yu, Q., Zhou, W., Gou, X., Li, J., et al. (2019). Multiple transcriptional factors control stomata development in rice. New Phytol. 223, 220–232. doi: 10.1111/nph.15766
Xu, M., Chen, F., Qi, S., Zhang, L., Wu, S. (2018). Loss or duplication of key regulatory genes coincides with environmental adaptation of the stomatal complex in Nymphaea colorata and Kalanchoe laxiflora. Hortic. Res. 5, 42. doi: 10.1038/s41438-018-0048-8
Yang, Y., Chang, B. H., Samson, S. L., Li, M. V., Chan, L. (2009). The Krüppel-like zinc finger protein Glis3 directly and indirectly activates insulin gene transcription. Nucleic Acids Res. 37, 2529–2538. doi: 10.1093/nar/gkp122
Zhang, X., Facette, M., Humphries, J. A., Shen, Z., Park, Y., Sutimantanapi, D., et al. (2012). Identification of PAN2 by quantitative proteomics as a leucine-rich repeat-receptor-like kinase acting upstream of PAN1 to polarize cell division in maize. Plant Cell 24, 4577–4589. doi: 10.1105/tpc.112.104125
Keywords: grasses, MUTE, orthologues, polarization, stomata, subsidiary cells
Citation: Serna L (2020) The Role of Grass MUTE Orthologues During Stomatal Development. Front. Plant Sci. 11:55. doi: 10.3389/fpls.2020.00055
Received: 05 November 2019; Accepted: 15 January 2020;
Published: 11 February 2020.
Edited by:
Michael T. Raissig, Heidelberg University, GermanyReviewed by:
Chun-Peng Song, Henan University, ChinaCopyright © 2020 Serna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laura Serna, bGF1cmEuc2VybmFAdWNsbS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.