- Molecular Plant Pathology, Faculty of Science, Swammerdam Institute for Life Sciences, University of Amsterdam, Amsterdam, Netherlands
Interactions between plants and the root-colonizing fungus Fusarium oxysporum (Fo) can be neutral, beneficial, or detrimental for the host. Fo is infamous for its ability to cause wilt, root-, and foot-rot in many plant species, including many agronomically important crops. However, Fo also has another face; as a root endophyte, it can reduce disease caused by vascular pathogens such as Verticillium dahliae and pathogenic Fo strains. Fo also confers protection to root pathogens like Pythium ultimum, but typically not to pathogens attacking above-ground tissues such as Botrytis cinerea or Phytophthora capsici. Endophytes confer biocontrol either directly by interacting with pathogens via mycoparasitism, antibiosis, or by competition for nutrients or root niches, or indirectly by inducing resistance mechanisms in the host. Fo endophytes such as Fo47 and CS-20 differ from Fo pathogens in their effector gene content, host colonization mechanism, location in the plant, and induced host-responses. Whereas endophytic strains trigger localized cell death in the root cortex, and transiently induce immune signaling and papilla formation, these responses are largely suppressed by pathogenic Fo strains. The ability of pathogenic strains to compromise immune signaling and cell death is likely attributable to their host-specific effector repertoire. The lower number of effector genes in endophytes as compared to pathogens provides a means to distinguish them from each other. Co-inoculation of a biocontrol-conferring Fo and a pathogenic Fo strain on tomato reduces disease, and although the pathogen still colonizes the xylem vessels this has surprisingly little effect on the xylem sap proteome composition. In this tripartite interaction the accumulation of just two PR proteins, NP24 (a PR-5) and a β-glucanase, was affected. The Fo-induced resistance response in tomato appears to be distinct from induced systemic resistance (ISR) or systemic acquired resistance (SAR), as the phytohormones jasmonate, ethylene, and salicylic acid are not required. In this review, we summarize our molecular understanding of Fo-induced resistance in a model and identify caveats in our knowledge.
Introduction
The Fusarium oxysporum species complex embraces a variety of strains ubiquitously present in soils. Most of these strains are saprotrophs and despite their ability to colonize plant roots the majority represents commensal endophytes not affecting plant fitness (Bao et al., 2004). Some F. oxysporum (Fo) strains, such as Fo47 and CS-20, are actually beneficial to the host and can provide protection against root pathogens (Table 1). Biocontrol-conferring Fo strains, such as Fo47, have been identified in vascular wilt-disease suppressive soils (Alabouvette, 1986). Identification of the causal microbes in wilt suppressive soils is typically done by sterilizing the soil following subsequent re-inoculation with the original microbes and screening for isolates that restore the suppressive effect against Fusarium wilt (Tamietti et al., 1993).
Fusarium wilt is one of the major diseases caused by pathogenic Fo strains. Wilts are a major threat for agriculture (Fisher et al., 2012) and Fo ranks among the 10 most devastating fungal plant pathogens worldwide (Dean et al., 2012). Besides wilt disease some strains can also cause foot- or root-rot resulting in serious yield losses in affected crops (Michielse and Rep, 2009). Fo produces micro- and macroconidia and chlamydospores that can remain viable in infected soils for decades, thereby frustrating crop rotation schemes (Nelson, 1981). Pathogenicity of Fo is host-specific, as typically strains infecting one plant species do not cause disease in others. Based on this host-specificity, pathogenic strains have been classified into so-called formae speciales (ff.spp.), of which over 100 have currently been described (Armstrong and Armstrong, 1981). An explanation for the emergence of host-specific pathogenic strains may be the extensive use of monocultures with limited crop rotation serving as breeding grounds for pathogens (Xiong et al., 2016). The evolved Fo pathogens can give rise to devastating crop losses, Fusarium wilt disease of banana, caused by Fo f.sp. cubense, being a prime example (García-Bastidas et al., 2014; Ordoñez et al., 2016).
To control wilt diseases different strategies are currently being employed in agriculture. One of these is chemical control, which includes broad-spectrum biocides like methyl bromide, benomyl, or carbendazim applied before planting. These chemicals can prevent infection, but do not cure a plant once infected. A caveat of these compounds is that they also affect beneficial soil microbiota and some accumulate in the food chain and for this reason many of these products are, or will be, banned (Lopez-Aranda et al., 2016). Heat sterilization of soils overcomes some of these drawbacks, but has the disadvantage that it is non-selective and it can have undesired effects on soil quality (Mahmood et al., 2014). Use of resistant plant varieties, e.g. plants carrying resistance genes is currently the most effective in terms of economy, ecology, and disease control. However, genetically encoded resistance is seldom durable and sooner or later new races emerge that overcome resistance in a never-ending arms race between Fo and its host (Takken and Rep, 2010; de Sain and Rep, 2015). Furthermore, Fo resistance genes are not available in the germplasm of all crops or they cannot be introgressed by breeding (Ploetz, 2015).
The limitations of the current approaches of wilt disease control urges the need to develop alternatives. An interesting alternative strategy is the use of beneficial Fo strains that confer biocontrol and thereby reduce disease incidence. A major advantage of biocontrol is the relatively broad-spectrum- and non-race specific protection conferred by endophytic strains (Table 1). A limitation is that the protection provided by these biological agents is highly variable and not consistent between seasons, crops, or fields. As illustration, even in greenhouse trials using tomato plants artificially co-inoculated with a pathogenic and a biocontrol Fo strain significantly different degrees of protection where observed in subsequent years (Fuchs et al., 1999). Furthermore, biocontrol observed under controlled lab conditions is not necessarily scalable to field conditions. For example, controlled soil co-inoculation of asparagus with Fo47 and Fo f.sp. asparagi (Foa) resulted in partial disease protection under lab conditions, but application of Fo47 in Foa-infested greenhouses did not reduce wilt disease (Blok et al., 1997).
A better understanding of the molecular mechanisms underlying biocontrol conferred by endophytic Fo strains may help to unleash the full potential that these organisms harbor to control disease conferred by their brothers in crime. In this review we mostly focus on two endophytic Fo strains, Fo47 and CS-20, as these are the best studied strains. We assess the differences between pathogenic and endophytic strains at their root colonization behavior, at the genome level and the responses they trigger in plants. Endophyte-mediated biocontrol consists of two components. The first is based on a direct activity on the pathogenic strain via parasitism and antibiosis (Benhamou et al., 2002; Le Floch et al., 2009) or by competing for nutrients or root niches. Several excellent reviews are available describing these non-plant mediated processes (Fravel et al., 2003; Alabouvette et al., 2009; Vos et al., 2014; Latz et al., 2018). In this review we focus on the other component of biocontrol, the indirect plant-mediated resistance response triggered by Fo endophytes, called endophyte-mediated resistance (EMR).
F. oxysporum Confers Biocontrol in Various Plant Species Against Root Pathogens
The ability of a large variety of endophytic Fo strains to confer biocontrol has been reported in many independent studies implying that it is a generic feature for Fo (Table 1). This idea is supported by a study in which over 200 different non-pathogenic Fo strains isolated from a tomato field were able to confer biocontrol in tomato, albeit to various degrees (Bao et al., 2004). Another observation is that Fo-based biocontrol is effective in a wide variety of plant species including both monocot and dicot species. This suggests that biocontrol is an ancient property as these families diverged over 200 million years ago (Wolfe et al., 1989). A number of oomycete-caused diseases can also be suppressed by Fo. For instance, Fo47 is reported to reduce disease incidence caused by Pythium oligandrum in tomato (Le Floch et al., 2009), Pythium ultimum in cucumber (Benhamou et al., 2002) and Phytophthora capsici in pepper (Veloso and Díaz, 2012). A common property among these pathogens is that they infect roots, but unlike most pathogenic Fo strains not all colonize the vasculature, implying that EMR is not vasculature-specific.
Two studies report on Fo-induced biocontrol that is not exclusively targeted against a root pathogen (Table 1). Pre-inoculation of watermelon roots with Fo f.sp. cucumerinum (Foc) (a pathogen on cucumber) reduced lesion sizes of Colletotrichum lagenarium infected leaves (Biles and Martyn, 1989). The other example details enhanced tolerance to Botrytis cinerea in pepper plants pre-inoculated with Fol (a tomato pathogen) (Díaz et al., 2005). Based on our literature survey Fo-induced EMR appears to be mostly root-confined.
The Root Colonization Pattern of F. oxysporum Pathogens Differs From That of Endophytes
Root colonization by Fo endophytes and pathogens has been extensively studied. In this chapter, we compare the root colonization process of Fo endophytes with that of pathogens. In our comparison we include colonization of ‘incompatible interactions’ in which a pathogenic Fo strain colonizes a resistant host, which does not result in disease emergence.
Spore Germination
The first stage of the colonization process starts with Fo spores or hyphae that grow in the vicinity of a root. Addition of sugar to the soil induces chlamydospore germination of pathogenic Fol and Fo f.sp. basilici (Larkin and Fravel, 1999). In natural settings root exudates presumably provide these carbohydrates as exudates from different crops enhance germination of microconidia of Fol and Fo f.sp. radicis-lycopersici (Forl) pathogens (Steinkellner et al., 2005). Hyphal exudations may also play a role in conidia germination as Fo uses autocrine pheromone signaling to control germination in a conidial-density dependent manner (Vitale et al., 2019). Some root pathogens effectively grow towards roots using chemotropism (Yao and Allen, 2006). While earlier studies found no evidence of chemotaxis toward tomato roots by Fo47 or the pathogens Fol or Forl (Steinberg et al., 1999; Olivain et al., 2006) a recent study showed that peroxidases secreted by tomato roots elicit Fol chemotropism towards roots (Turrà et al., 2015; Nordzieke et al., 2019). Altogether, it seems that root exudations trigger spore germination and induces directional mycelial growth. At this pre-colonization stage, no differences are noticeable between endophytic and pathogenic Fo strains.
Host Colonization
Upon germination, both Fo endophytes and pathogens colonize the root surfaces of host and non-host plants. Contact with the root triggers hyphal branching, after which Fo produces hyphal swellings to invade the root. The fungal hyphae enter plant roots via wounds, cracks in the epidermis, lateral root emergence points, or by direct penetration of the root tip depending on the Fo strain and plant species involved. Hyphae reach the vascular stele via the apoplast of the root cortex. In some cases, intracellular growth is noticeable along with local host cell-death, a phenomenon observed more often among non-pathogenic strains (He et al., 2002; Olivain et al., 2003; Humbert et al., 2015; Gordon, 2017). Both pathogenic and non-pathogenic strains colonize the root cortex (Gordon, 2017), but although the initial colonization pattern is similar, the extent and pattern of colonization differs during later stages. The amount of biomass of a pathogenic strain in the root is typically higher than that of an endophyte. This difference is already apparent at early stages. At 48 h post-inoculation (hpi) higher amounts of fungal biomass for the Fo40 pathogen were detected in roots of soybean plants than for the endophytic Fo36 (Lanubile et al., 2015). A similar difference was reported for other systems, like the interaction between tomato and Fo47 or Forl. Two weeks post-inoculation Fo47 biomass was 10-fold less than that of the pathogen (Validov et al., 2011). These observations imply that in early stages of the interaction Fo endophytes are less efficient root colonizers than pathogens.
Besides the amount of fungal biomass also the root colonization pattern differs between Fo pathogens and endophytes. Typically, only pathogenic strains are able to reach the xylem vessels from where they colonize above-ground tissues. The induced occlusions of the xylem vessels, aimed to restrict pathogen progress, results in the classical wilting symptoms of infected plants (Gordon, 2017). A well-studied example is the interaction between pea roots, Fo47, and the pathogen Fo f.sp. pisi. Whereas Fo47 colonization is restricted to the root surface and outermost cell layers of the cortex, the pathogen massively invades the deeper root tissues including the vasculature (Benhamou and Garand, 2001). A similar pattern is seen upon Fo colonization of tomato. When grew in hydroponics, Fo47 and Fol8 both efficiently colonized the surface of tomato tap roots following attachment of the microconidia to the root hair zone. Subsequently, both strains grew towards the elongation zone until they reached the root apex. Whereas Fol8 intensively colonized the deeper root tissues and eventually reached the vasculature, the endophyte was confined to the epidermis and cortex (Nahalkova et al., 2008). In line with these observations, Fo47 proteins were not identified in xylem sap of Fo47-inoculated soil-grown tomato plants, whereas Fol proteins were detected in sap of Fol-infected plants (de Lamo et al., 2018). This difference indicates that even at later stages of root colonization Fo47 does not reach the vasculature. Contrarily, Fo47 was reported to colonize xylem vessels of eucalyptus (Salerno et al., 2000) and the Fo endophyte CS-20 was found to colonize xylem vessels of cucumber (Pu et al., 2014). An explanation for the vascular presence of the latter two endophytic strains could be the clipping of the roots prior to inoculation, providing direct vascular access to the endophyte.
In incompatible interactions, the first stages of the root colonization pattern are similar to that of a compatible interaction. The pathogen colonizes the root cortex, but in contrast to a purely endophytic strain, the pathogen frequently reaches the vasculature of a resistant host. Examples of vascular colonization of a resistant host are chickpea and tomato roots inoculated by either avirulent Fo f.sp. ciceris or Fol (Mes et al., 2000; Jimenez-Fernandez et al., 2013; van der Does et al., 2018). Recently vascular colonization of tomato plants carrying three different classes of resistance (R) gene types by Fol was compared. Although the plasma membrane-localized immune receptors (I and I-3) restricted colonization to a larger extent than the intracellular receptor (I-2), vascular colonization was observed in all cases (van der Does et al., 2018). The amount of fungal biomass in a resistant plant, however, is low and fungal proteins cannot be detected in the xylem sap of infected plants (de Lamo et al., 2018). These findings are in line with the low number of hyphae observed in vessels of a resistant tomato variety (Mes et al., 2000). Inoculation of resistant cabbage roots with Fo f.sp. conglutinans also resulted in marginal colonization of the vasculature and fungal proteins were not identified in the xylem sap either (Pu et al., 2016).
The general pattern is that Fo endophytes, similar to endophytes such as Serendita indica (Jacobs et al., 2011) and arbuscular mycorrhizal fungi (Gadkar et al., 2001), are mostly root surface- and cortex-colonizers. Extensive colonization of the root cortex and vasculature is typically restricted to pathogens, a property that correlates with enhanced secretion of cell wall-degrading enzymes by these strains (Jonkers et al., 2009). Pathogenic strains are also able to enter and, to a limited extent, colonize the vasculature of a resistant host. Upon (co-)inoculation of an endophyte and a pathogen both strains coincide at the same root tissues during the early stages of the interaction, but become spatially separated when the pathogen invades the vascular bundle. Therefore, disease protection induced by Fo endophytes at these later stages is likely plant-mediated.
F. oxysporum Endophytism and Pathogenicity Are Genetically Determined by the Fungus–Host Combination
Bioassays can reveal whether a strain is pathogenic on a specific host, or on a specific variety of that host, but these assays cannot establish whether a strain is non-pathogenic. Given the narrow host-range of pathogenic Fo strains (mostly restricted to single plant species) it would be necessary to inoculate a particular strain on all possible plant species and varieties to conclude that it is likely a non-pathogen in case of a negative outcome. Giving the impracticality of such an approach, and the limitation of classical taxonomic features, there has been ample focus on identifying molecular features that could be used to distinguish ff.spp. and to diagnose pathogenic isolates and discriminate them from non-pathogenic strains.
Phylogenetic analyses of the Fo species complex using conserved gene sequences such as those encoding elongation factor 1α typically result in phylogenetic trees in which Fo pathogens and endophytes are distributed together over different clades (Wong and Jeffries, 2006; Ellis et al., 2014; Pinaria et al., 2015). Likewise, trees based on genomic markers such as restriction fragment length polymorphism of the ribosomal intergenic spacer regions, or on mating type results in trees in which the ff. spp. are polyphyletic and cluster together with Fo endophytes in various clades (Alves-Santos et al., 1999; Abo et al., 2005; Ellis et al., 2014; Nirmaladevi et al., 2016). Even a multiple-sequence alignment of 441 conserved core genes from various Fo genomes did not result in a tree that enabled differentiating ff. spp. or allowed unambiguous identification of non-pathogenic strains (van Dam et al., 2018).
A decade ago, however, it was reported that the presence of a lineage-specific chromosome determines pathogenicity of Fol toward tomato (Ma et al., 2010). Horizontal transfer of a pathogenicity chromosome from Fol to Fo47 turned the endophyte into a tomato pathogen (Ma et al., 2010). Subsequent studies revealed that chromosome transfer from a cucurbit-infecting Fo strain could transform Fo47 into a cucurbit pathogen (van Dam et al., 2017). Vice versa, loss of a dispensable pathogenicity chromosome from a Fol strain resulted in loss of pathogenicity (Vlaardingerbroek et al., 2016). Hence, pathogenicity appears to correlate with the presence of a pathogenicity chromosome. These pathogenicity chromosomes differ from core chromosomes by a high content of transposable elements and a low gene density (Ma et al., 2010). Genetic analysis of Fol revealed that its pathogenicity chromosome carries the genes encoding the putative host-specific virulence proteins (effectors) that the fungus secretes in the tomato xylem sap following infection (Schmidt et al., 2013). Some of these Secreted In Xylem, or SIX, proteins such as SIX1 (Avr3) (Rep et al., 2004), SIX3 (Avr2) (Di et al., 2017b), SIX4 (Avr1) (Houterman et al., 2008) and SIX6 (Gawehns et al., 2014) are genuine effectors and contribute to fungal virulence on tomato. Many of these effectors were found to be specific for the tomato-infecting strain, providing the means to identify this pathogen based on its effector profile. Based on the features of these Fol SIX effector genes an effector prediction pipeline could be constructed in which putative effector genes in Fo can be identified based on: 1) a relatively small size (>25 aa and <300 bp), 2) presence of a signal peptide for secretion, and 3) proximity to a “miniature impala” transposable element (van Dam et al., 2016). Analyses of predicted Fo effectoromes revealed that these are shared between strains from a f.sp. infecting the same host, while they are divergent for those infecting other plant species, thereby allowing distinction of ff.spp. based on their effector profiles (Lievens et al., 2009; van Dam et al., 2016; van Dam et al., 2018). Hence, in contrast to phylogenetic analyses, Fo effectorome exploration proves a powerful tool to predict pathogenicity and the potential plant host for a given strain. Whether a potential pathogen is indeed able to cause disease ultimately depends on the corresponding genotype of the host. If the host carries a resistance gene recognizing a specific effector of the pathogen this may result in activation of gene-for-gene-based resistance response restricting host colonization (Flor, 1956; Jones and Dangl, 2006). For instance, the Fol effector proteins Avr1, Avr2, or Avr3 are recognized by the tomato resistance proteins I, I-2, or I-3 (Simons et al., 1998; Catanzariti et al., 2015; Catanzariti et al., 2017) resulting in the activation of a resistance response in plants carrying these genes (Houterman et al., 2008). Similarly, the effector protein AvrFom2 from the melon pathogen Fo f.sp. melonis can be recognized by the melon resistance protein Fom2 thereby conferring avirulence to the fungus (Risser et al., 1976; Schmidt et al., 2016).
Non-pathogenic strains share a set of conserved putative effector genes with pathogenic strains, but typically carry much fewer effector candidates and no or few host-specific effectors (van Dam et al., 2016). This notable difference provides a means to distinguish potential pathogens from non-pathogens by the number of candidate effectors they carry. It is tempting to speculate that the candidate effectorome of non-pathogens determines their capacity to colonize roots and confer EMR. Unfortunately, little is known of the role of effectors for Fo endophytes. One putative effector, CS20EP, of the EMR-conferring CS-20 strain was reported to trigger a defense response in tomato against Fol when applied prior to inoculation with the pathogen (Shcherbakova et al., 2015). The protein was identified in the culture filtrate of in vitro-grown fungus. However, whether the CS20EP gene is actually expressed during host root colonization awaits future study, as does its role in EMR, for which a knockout strain should be assessed. Altogether, the predicted effector profile from a Fo strain allows its classification as a likely endophyte or as a putative (a)virulent pathogen on a given host. The increasing number of Fo genomes becoming available allows f.sp.-specific effector candidates to be identified and to more precisely predict host-specific pathogenicity of a given strain. Functional analysis of these effectors, and identification of their host targets, could provide new leads to combat pathogens (Gawehns et al., 2013).
The Timing and Amplitude of Root Responses Upon Colonization by Endophytic or Pathogenic F. oxysporum Differ
Plant roots are typically exposed to a highly diverse soil microbiota (Hacquard et al., 2017). Plants recognize microorganisms via microbe-associated molecular patterns (MAMPs) that are present in both pathogens and non-pathogens (Henry et al., 2012). Well-known fungal MAMPs are chitin (Kaku et al., 2006) and ß-glucan (Cheong and Hahn, 1991). MAMP recognition is mediated by pattern recognition receptors (PRRs) located at the cell surface (Macho and Zipfel, 2014) such as the CERK1 chitin receptor of Arabidopsis (Miya et al., 2007). Forward genetics in Arabidopsis identified the receptor-like kinase MIK2 as a potential PRR and as a crucial component to recognize and respond to MAMPs from Fo (Coleman et al., 2019). PRRs are mainly expressed in root zones vulnerable to pathogen entry resulting in a heterogenic and tissue-specific responsiveness to different MAMPs (Chuberre et al., 2018). Responsiveness to chitin, for instance, is mostly confined to the mature zone and other parts of the root system are relatively insensitive to this MAMP and do not mount immune responses upon exposure to chitin (Millet et al., 2010; Coleman et al., 2019). This heterogeneity could explain why Fol typically does not penetrate mature root zones (Mes et al., 2000). In the responsive zones MAMP recognition results in activation of pattern-triggered immunity (PTI), which confers resistance to a wide variety of potential pathogens (Jones and Dangl, 2006; Bigeard et al., 2015).
Activation of PTI induces a variety of early signaling responses, such as a cellular Ca2+ and H+ influx resulting in extracellular alkalinization, production of reactive oxygen species (ROS), and phosphorylation of mitogen-associated protein kinases (MAPKs) (Bigeard et al., 2015; Chuberre et al., 2018). A number of PTI-associated signaling responses that have been monitored in cell cultures upon Fo treatment are depicted in Figure 1. Cell cultures have been instrumental to study early plant responses to Fo (Olivain et al., 2003; Humbert et al., 2015). Flax cell cultures exposed to germinated microconidia of Fo47 show a stronger extracellular alkalinization response than cells treated with pathogenic Fo f.sp. lini (Foln) (Figure 1) (Olivain et al., 2003). Also the Ca2+ influx was higher upon Fo47 exposure than to Foln application (Olivain et al., 2003). Ca2+ influx activates calmodulin (CaM), and in cucumber roots the CaM signal transduction pathway was more strongly induced upon CS-20 colonization than when treated with pathogenic Foc (Pu et al., 2014), implying a weaker PTI induction by pathogenic strains.
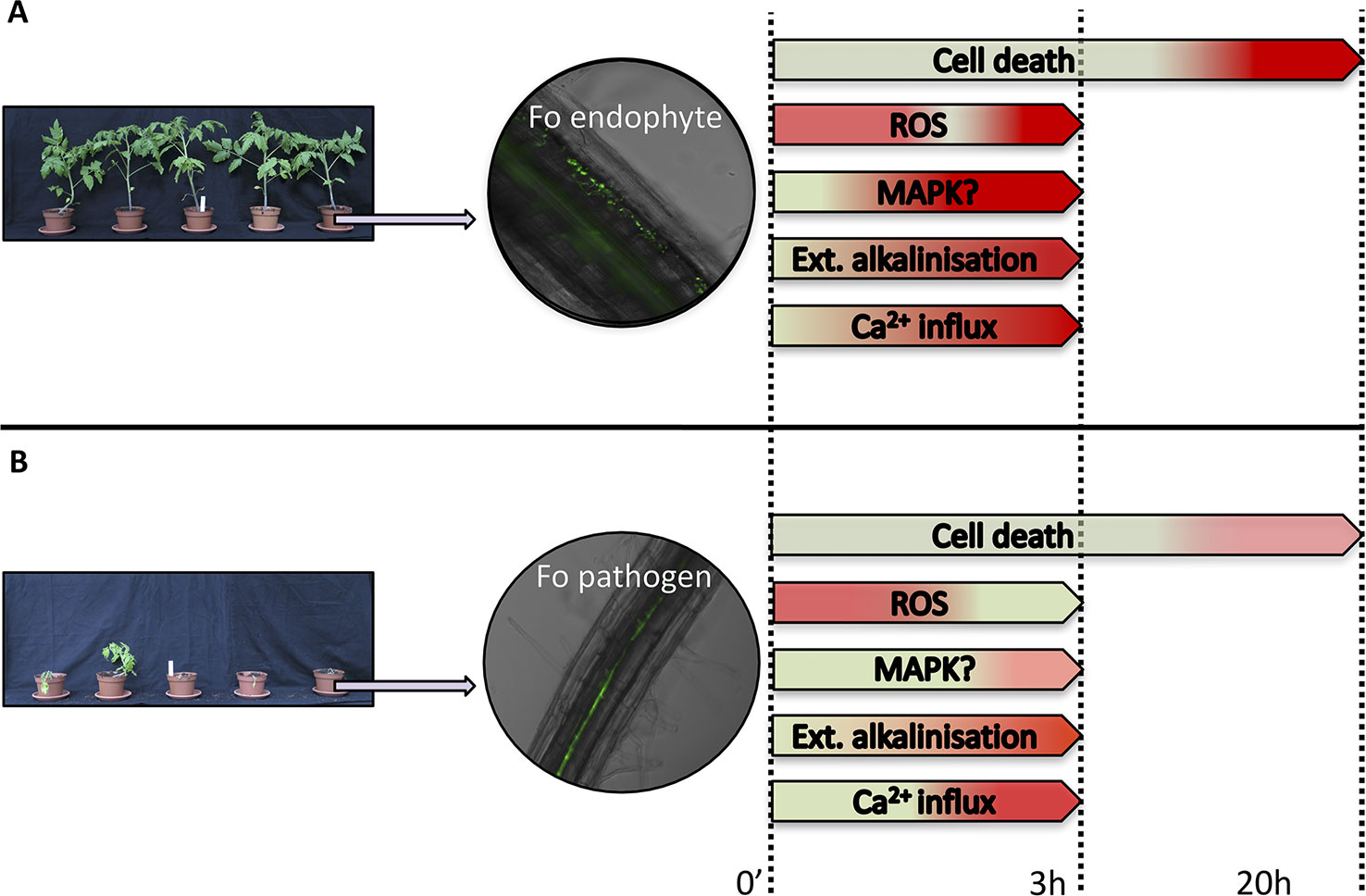
Figure 1 Schematic representation of plant responses upon Fusarium oxysporum (Fo) inoculation. Responses of plants following exposure to a Fo endophyte (A) or a pathogen (B). Pictures on the left show representative phenotypes of tomato plants upon inoculation with either an endophytic or a pathogenic Fo strain. Middle panels show root colonization by GFP-labeled Fo strains visualised by fluorescence microscopy. The right panel summarizes early signaling responses upon Fo exposure to plant cell cultures (Olivain et al., 2003; Humbert et al., 2015). The response amplitudes are color-coded from green (lowest) to red (highest).
ROS, besides being signaling molecules, have direct toxic effects on microbes (O'Brien et al., 2012) and can induce cell death thereby limiting progression of biotrophic pathogens (Mur et al., 2008). Within minutes Fo47 and Foln induce a similar early ROS burst in flax cells (Olivain et al., 2003). Fo47, however, also triggers a second more vigorous burst 3 h post-exposure, which is absent upon Foln treatment. Fo47 also induced more cell death than Foln especially at 14 hpi. A similar observation was made using tomato cell cultures incubated with germinated microconidia of Fo47 or Fol (Humbert et al., 2015). Analogously, inoculation of non-pathogenic Fo that triggers EMR against pathogenic Foa in asparagus induced a cell death response (≈10% cell death) in roots while no cell death was detected when Foa alone was inoculated (He et al., 2002). Transcriptome analysis of soybean roots infected by pathogenic Fo pathogen revealed upregulation of several MAPKs at a relative late stage (72 hpi) of infection, while none was induced by an endophytic strain (Lanubile et al., 2015). Whether MAPKs are differentially phosphorylated in an interaction between roots and Fo endophytes or pathogens remains a question for future study. Taken together, whereas both endophytes and pathogenic Fo strains trigger early PTI signaling, these responses are typically less pronounced in the presence of the latter, suggestive of stronger immune suppression by pathogenic strains.
An effective PTI response results in a transient, local and systemic transcriptional reprogramming of the host (Boller and Felix, 2009; Millet et al., 2010; Bigeard et al., 2015; Chuberre et al., 2018). For instance, in Fo47-inoculated pepper roots a transient expression of a PR-1 protein, a chitinase and a sesquiterpene cyclase (involved in capsidiol synthesis) was observed at 48 hpi, after which expression returned to basal levels at 120 hpi (Veloso and Díaz, 2012). In tomato roots, Fo47 and Fol did not differentially affect expression of a set of PR marker genes when monitored at 48, 72, or 96 hpi: two chitinases (CHI9 and CHI3), two glucanases (GLUB and GLUA), a lypoxygenase (LOXD) and PR-1a (Aimé et al., 2013). However, during later stages of infection at 6 to 22 days post-inoculation (dpi), Fo47, unlike Fol, did not trigger accumulation of PR transcripts (Aimé et al., 2008). In contrast, in cucumber roots CS-20 did induce major transcriptional changes, and at 72 hpi there was a strong induction of PR3, LOX1, PAL1, and NPR1 and of CaMs, CsCam7 and CsCam12 being the strongest induced. At the same time point pathogenic Foc induced NPR1 and to a lesser extent PR3 and PAL1 expression (Pu et al., 2014). RNA-seq analysis of soybean roots inoculated with endophytic or pathogenic Fo revealed that the latter induced more, and stronger, transcriptional changes at 72 and 96 hpi (Lanubile et al., 2015). The literature is ambiguous regarding transcriptional reprogramming in plant–Fo interactions, which might originate from dissimilarities in experimental setup, sampling time, and/or plant-endophyte combination (Table 1). Together the data shows that transcriptional reprogramming following Fo endophyte colonization varies depending on the strain, but typically is transient and returns to basal levels within days. The observation that CS-20 affects transcriptional responses more strongly than Fo47 correlates with CS-20 being a more potent EMR-inducer (Larkin and Fravel, 1999). Xylem sap proteome analysis of susceptible tomato plants showed a significant change in abundance of up to 92% of the identified proteins at 2 weeks post-inoculation of Fol (Gawehns et al., 2015; de Lamo et al., 2018). Among the proteins showing the highest induction are the PR proteins PR-1 and PR-10. Contrarily, at the same time point no significant changes were detected in the proteome of Fo47-inoculated plants as compared to mock treatment (de Lamo et al., 2018). In summary, the transcriptional reprogramming in response to Fo endophytes is confined to the first days of the interaction, while pathogenic strains induce changes mostly during later stages when disease symptoms emerge. At these later stages, major changes are also detected in the xylem sap proteome of diseased tomato plants.
PTI is hypothesized to result in establishment of physicochemical barriers such as callose depositions at the cell walls and exudation of phytoalexins aimed at restricting microbial invasion (Millet et al., 2010). In Fo47-inoculated pea roots, host cell-wall penetration attempts by the endophyte appear constrained by callose-containing papillae depositions (Benhamou and Garand, 2001). Similar observations have been made in Fo–cucumber (Benhamou et al., 2002), Fo–flax (Olivain et al., 2003), and Fo–tomato interactions (Le Floch et al., 2009). Fo47 also induced accumulation of the phenolic compound caffeic acid in pepper roots at 48 hpi (Veloso et al., 2016), whereas in tomato roots CS-20 induced accumulation of ferulic acid at 72 hpi (Panina et al., 2007). Both compounds have in vitro antimicrobial activity to Verticillium dahliae (Veloso et al., 2016). Pea roots colonized by Fo47 respond by formation of an osmiophilic compound coating the secondary wall and the pit membranes of the vessel lumen (Benhamou and Garand, 2001). Inoculation with pathogenic Fo f.sp. pisi did not trigger these types of responses in pea (Benhamou and Garand, 2001).
Taken together, both Fo endophytes and pathogens trigger local PTI responses but these appear suppressed/evaded by the latter, likely by the secretion of host-specific effectors. Indeed, Fo effectors have been identified that suppress PTI, a prime example being Fol Avr2 that suppresses ROS production, callose deposition, MAPK phosphorylation, and growth-inhibition upon MAMP application (Di et al., 2017b). Recently, a chitin deacetylase (PDA1) has been found to be required for pathogenicity of Fo f.sp. vasinfectum to cotton (Gao et al., 2019). This provides evidence of a PTI avoidance strategy as de-acetylation of chitin converts it into chitosan, which is a poor inducer of PTI (Gao et al., 2019). Another strategy to evade PTI activation is masking fungal MAMPs. LysM-containing effectors in Cladosporium fulvum are involved in chitin-binding, thereby preventing their perception by the host (Bolton et al., 2008). LysM domain-containing effector genes are also present in Fo genomes (de Jonge et al., 2010; de Sain and Rep, 2015) and a LysM-containing protein secreted by Fol has been identified in tomato xylem sap (Gawehns et al., 2015; de Lamo et al., 2018). However, further research should clarify whether its role in pathogenicity is similar to that of C. fulvum. In summary, successful suppression of PTI by Fo pathogens seems required to cause disease and strains unable to do so, e.g. because they lack the proper host-specific effectors, do not cause disease and exert endophytic lifestyles.
EMR Involves Localized Cell Death and Accumulation of Specific PR Proteins in the Xylem Sap
The molecular and physiological changes in roots during EMR have been studied in some detail and were mostly focused on changes in transcriptome, metabolome, and xylem sap proteome. Whereas Fo endophytes typically trigger an early, minor, and transient change in gene expression, pathogenic strains induce a major and later (days) transcriptional reprogramming during the onset and development of disease (see The Timing and Amplitude of Root Responses Upon Colonization by Endophytic or Pathogenic F. oxysporum Differ). In tri-partite interactions surprisingly little changes in gene expression have been reported. One study of Fo47-inoculated tomato roots challenged with Fol revealed induction of transcripts encoding an acidic extracellular chitinase (CHI3), an acidic extracellular ß-1,3-glucanase (GLUA), and PR-1a 48 h after inoculation (Aimé et al., 2013).
Metabolomic studies revealed that pre-treatment of pepper plants with Fo47 2 days prior to V. dahliae inoculation enhanced the accumulation (at 8 and 24 hpi) of a phenolic acid, chlorogenic acid, in the roots in response to the latter (Veloso et al., 2016). Phenolic acids are involved in fortification of cell walls when cross-linked to cell wall polymers by a ROS-catalyzed process (McLusky et al., 1999; Bubna et al., 2011; O'Brien et al., 2012). Phenolics, together with callose, ROS, peroxidases, and structural proteins form the major constituents of the papillae depositions that are proposed to block cell entry of Fo (Underwood, 2012). In agreement, pre-treatment of cucumber roots with Fo47 resulted in more papillae depositions preventing P. ultimum to penetrate host cells (Benhamou et al., 2002). Another physiological aspect of EMR is the endophyte-induced host cell death during early stages of colonization. This phenomenon seems to be common among non-pathogenic strains as Fo endophytes typically induce host cell death in the root cortex to a larger extent than Fo pathogens during early stages of infection (He et al., 2002; Olivain et al., 2003; Humbert et al., 2015; Gordon, 2017). Noteworthy, in a mutagenesis screen of different Fo endophytes, those losing their ability to trigger biocontrol also showed a reduced induction of host cell death in cell cultures despite retaining its host colonization capabilities (Trouvelot et al., 2002; L'Haridon et al., 2007; Alabouvette et al., 2009).
Pathogenic Fo strains show reduced vasculature colonization upon EMR induction, which might be caused by a change in the xylem sap proteome. To address this hypothesis the xylem sap proteome of tomato plants inoculated with Fo47 and/or Fol was compared (de Lamo et al., 2018). Of the 388 quantifiable proteins, the abundance of only two proteins was strongly increased in the tri-partite interaction as compared to the mock controls. Accumulation of these two proteins, a β-glucanase and NP24, was induced 45- and 33-fold respectively as compared to the control. β-Glucanases exert its antimicrobial activity by hydrolyzing glucan molecules, one of the most abundant polysaccharides in fungal cell walls (Stintzi et al., 1993). Furthermore, the released ß-1,6-glucans act as fungus- and oomycete-specific MAMPs triggering host immune responses (Fesel and Zuccaro, 2016). NP24 is a member of the PR-5 family that includes osmotin and thaumatin-like proteins (Stintzi et al., 1993; Liu et al., 2010). PR-5 proteins exert in planta antimicrobial activity against the pathogens Phytophthora infestans (Woloshuk et al., 1991), P. capsici, and Fo (Mani et al., 2012) by disrupting their plasma membrane integrity via the formation of pores (Vigers et al., 1992). In addition, some PR-5 proteins exert ß-1,3-glucanase activity that could contribute to their antimicrobial activity (Grenier et al., 1999; Menu-Bouaouiche et al., 2003). Besides a direct effect on the pathogen, overexpression of a plum PR-5 in Arabidopsis activated the production of the phytoalexin camalexin (El-kereamy et al., 2011). The correlation between EMR and NP24 abundance is intriguing, as the only differentially accumulated protein in the xylem sap of resistant tomato plants inoculated with an avirulent Fol strain is also a PR-5 family member. Accumulation of this xylem sap-specific PR-5x protein was induced 158-fold upon inoculation of the avirulent pathogen. In a compatible interaction the abundance of the protein also increased, but to a much lower extent (de Lamo et al., 2018). The finding that PR-5 isoforms also specifically accumulate in xylem sap of susceptible and resistant Brassica oleracea infected with Fo f.sp. conglutinans (Pu et al., 2016) further indicates a role for these proteins in controlling the proliferation of pathogenic Fo strains in the vasculature. The observation that pathogenicity-compromised Fol strains in which specific effectors are deleted trigger an >200-fold induction of NP24 in the xylem sap provides additional support for this hypothesis (Gawehns et al., 2015). How these Fol effectors affect accumulation of PR-5 isoforms in tomato is unknown, but various plant pathogens, including V. dahliae (Zhang et al., 2019), Blumeria graminis (Pennington et al., 2016), and B. cinerea (Gonzalez et al., 2017), secrete effectors that directly target PR-5 proteins, stressing their importance in plant fungal interactions.
Altogether, Fo-based EMR seems to be a root-mediated response that triggers, among other responses, specific accumulation of xylem sap-localized PR-5 and ß-glucanase proteins and secretion of phenolic compounds that together with ROS are involved in cell wall lignification and callose depositions. Furthermore, host cell death induced by Fo endophytes correlates with the induction of an effective EMR response.
EMR is Distinct From Induced Systemic Resistance and Systemic Acquired Resistance Responses
Many studies attribute Fo-induced resistance response in plants as the main contributor to biocontrol (Table 1). Split-root systems, in which the Fo endophyte is spatially separated from the pathogen, have shown that EMR can act systemically in root tissues (Kroon et al., 1991; Fuchs et al., 1997; Duijff et al., 1998; Larkin and Fravel, 1999; Kaur and Singh, 2007; Pantelides et al., 2009; Aïcha et al., 2014). The mechanism that transduce this signal to distant root tissues is unknown. Root colonization by endophytes such as S. indica or Trichoderma spp. triggers an induced systemic resistance response (ISR) that relies on the phytohormones jasmonic acid (JA) and ethylene (ET) (Shoresh et al., 2010; Franken, 2012). Other microbes, especially avirulent pathogens, can trigger a salicylic acid (SA)-dependent immune response, which results in systemic acquired resistance (SAR) (Pieterse et al., 2014). Both systemic responses prime the plant to respond faster and stronger to subsequent pathogen attack, thereby reducing it susceptibility to foliage-attacking pathogens (Durrant and Dong, 2004; Fu and Dong, 2013; Pieterse et al., 2014). The observation that Fo endophytes typically do not confer protection to pathogens attacking above-ground tissues (Table 1) raises the question whether EMR mechanistically differs from ISR or SAR. Only two reports describe Fo-induced resistance to a foliar pathogens (Biles and Martyn, 1989; Díaz et al., 2005). It was reported that a Fol strain that is pathogenic on tomato reduced susceptibility to B. cinerea in pepper (Díaz et al., 2005). Pre-treatment with 1-methylcyclopropene, an inhibitor of ET perception, compromised this Fol-induced plant protection to the fungus. The involvement of ET in this response implies that the non-host pathogen Fol can trigger ISR in pepper. Remarkably, Fo47 inoculation did not confer protection against B. cinerea in the same experimental setup, although this strain triggered EMR (Veloso and Díaz, 2012), suggesting that Fol triggers both. The other example details the cucumber-pathogen Foc that induced systemic responses in aerial tissues in watermelon (Biles and Martyn, 1989). It will be interesting to investigate whether both pathogenic Fo strains carry effectors that are recognized by these non-host plants responsible for triggering a systemic ISR-type immune response. These examples imply that non-host pathogenic Fo strains can induce both ISR and EMR, while purely endophytic strains trigger only the latter response.
Whereas tomato mutants compromised in SA signaling are hypersensitive to Fusarium wilt disease, an increased tolerance was observed in mutants affected in ET biosynthesis or perception (Di et al., 2017a). In contrast, susceptibility of tomato mutants deficient in JA biosynthesis towards Fol was unaffected, showing that these three phytohormones have distinct roles in the interaction between tomato and pathogenic Fo (Di et al., 2017a). The interaction between these phytohormones and Fo is complex and differs for different pathosystems (Di et al., 2016). To elucidate the role of these defense phytohormones in EMR, Constantin and co-workers analyzed Fo47-induced immune responses in wild-type tomato plants and in mutants compromised in ET, JA or SA signaling (Constantin et al., 2019). Expression of ET marker genes (Pti4 and ETR4) was not induced in a tri-partite tomato–Fol–Fo47 interaction suggesting that ET is not involved in EMR (Constantin et al., 2019). Indeed, EMR was intact in tomato lines affected in either their ability to sense- (never-ripe mutant) or produce ET (transgenic lines constitutive expressing ACC deaminase) (Constantin et al., 2019). Also tomato plants with a defect in JA biosynthesis (def1) were still capable of mounting EMR upon co-inoculation with Fo47 and Fol. These findings make involvement of ISR in EMR unlikely, as this response requires intact ET/JA signaling pathways (Pieterse et al., 2014). Likewise, SAR, which requires SA, appears not to be involved as tomato lines compromised in SA accumulation (expressing NahG) exert a functional EMR response against Fol (Constantin et al., 2019). Together, these findings support a model in which EMR induced by Fo47 is distinct from ISR and SAR, as these responses require either JA/ET or SA and result in induced resistance in shoots, unlike EMR that is mostly root confined.
Discussion
Based on the data presented we propose a mechanistic model on how Fo-induced EMR prevents disease. Figure 2 illustrates an early (≈ two dpi) interaction between a root and Fo. Both pathogenic and non-pathogenic Fo strains colonize the root epidermis and cortex. Whereas pathogenic Fo strains effectively compromise immune signaling by secreting effector proteins (Figure 2A) endophytes are unable to do so and trigger immune activation (Figure 2B). The transient induction of immune signaling confines the non-pathogenic fungus to the root cortex and restricts its growth by preventing entry into host cells by the formation of papillae and cell wall fortifications. Together these responses prevent the fungus from reaching the vasculature and causing disease. Localized cell death induced by Fo endophytes (He et al., 2002; Alabouvette et al., 2009) appears to be involved in the induction of EMR, because Fo mutants that lost their ability to induce cell death are also unable to trigger EMR even though they can still colonize the roots (Alabouvette et al., 2009). Endophytes such as Harpophora oryzae or the phylogenetically distant basidiomycete S. indica, are also known to trigger localized cell death upon root colonization (Deshmukh et al., 2006; Su et al., 2013). It is tempting to speculate that induction of host cell death by endophytes may be a generic property required for EMR induction. Possibly cell death primes, or potentiates, immune responses to an extent that they can no longer be mitigated by the effectors secreted by the pathogen. The potentiated immune responses restrict pathogen development in a tri-partite interactions and results in a reduced xylem sap colonization (Figure 2C). Although pathogenic Fo strains colonize the vasculature in tri-partite interactions their proliferation is reduced as are the disease symptoms. We speculate that the reduced ability to colonize the vasculature is in part due to the increased abundance of PR-5 protein family members and plant-produced ß-glucanases. Assessing the biocontrol properties of Fo in plants in which these genes are knocked-out can put this hypothesis to the test.
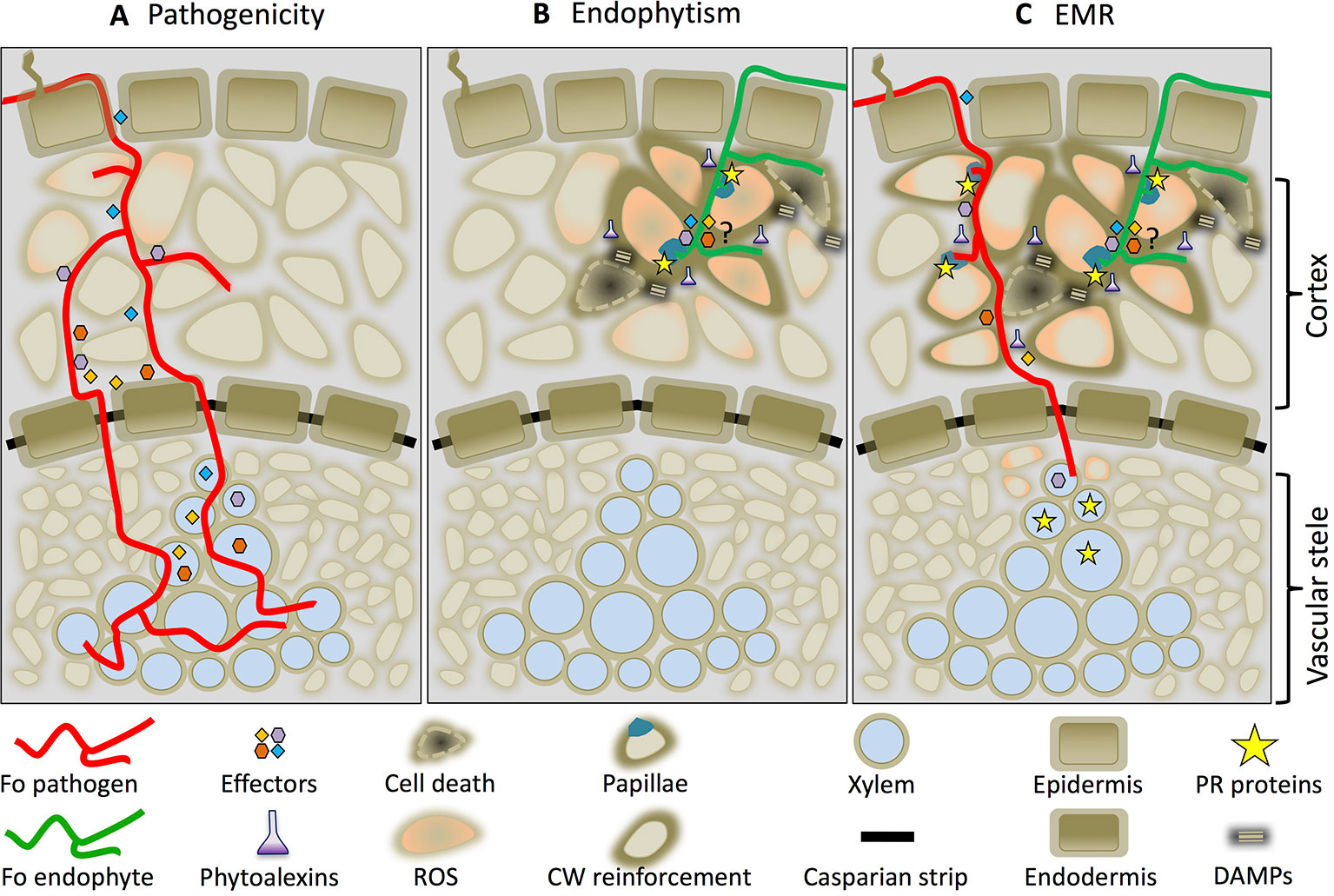
Figure 2 Endophyte-mediated resistance (EMR) working model. Cross-section of a root colonized by a Fo pathogen (red line) (A), an endophyte (green line) (B), or by both in a tri-partite interaction in which EMR is triggered (C). The drawings depict an interaction around 2 days after inoculation.
Conclusion
Although being studied for over more than three decades the mechanism underlying EMR remains elusive. Understanding this inducible defense mechanism, which confers protection against root-invading vascular pathogens, holds potential for improved control of wilt diseases without affecting the conventional defense pathways. Future studies focusing on the nature of the systemic signal, the role of secondary metabolites, PR protein production, and papillae formation in tri-partite interactions will be instrumental to get a better understanding of the mechanism underlying EMR. Elucidating the relation between localized host cell death and EMR will reveal whether damage merely amplifies, or is essential, to trigger this immune response. Studying the potential role of host-specific and generic effector candidates in modulating EMR will increase our understanding of the endophytic side of the interaction, possibly allowing selection of endophytic strains conferring robust biocontrol in agricultural settings. A concern is that horizontal chromosome transfer from pathogenic Fo strains to the applied Fo endophytes could turn the latter into pathogens. Whether chromosome transfer occurs in natural setting should be investigated before agricultural application.
Author Contributions
FL and FT conceived and wrote the manuscript.
Funding
FL and FT are supported by the BestPass project. This project is funded by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 676480 (International Training Network BestPass). FT also is supported by the NWO-Earth and Life Sciences funded VICI project No. 865.14.003.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Martijn Rep and Ben Cornelissen for their feedback on the manuscript, to Maria Constantin and Babette Vlieger for providing the microscopy pictures used in Figure 1, and to both reviewers for their constructive, critical, and detailed comments.
References
Abo, K., Klein, K. K., Edel-Hermann, V., Gautheron, N., Traore, D., Steinberg, C. (2005). High genetic diversity among strains of Fusarium oxysporum f. sp. vasinfectum from cotton in Ivory Coast. Phytopathology 95 (12), 1391–1396. doi: 10.1094/PHYTO-95-1391
Aïcha, B., Nisserine, H. K., Abdeslem, S. M., Hadjira, B., Lalia, B. S., Mebrouk, K., et al. (2014). Antagonist effect of nonpathogenic Fusarium oxysporum strain Fo47 on Fusarium oxysporum f. sp. radicis-lycopersici the cause of fusarium crown and root rot in tomato cultivation. Adv. Environ. Biol. 8 (10), 50–56.
Aimé, S., Cordier, C., Alabouvette, C., Olivain, C. (2008). Comparative analysis of PR gene expression in tomato inoculated with virulent Fusarium oxysporum f. sp. lycopersici and the biocontrol strain F. oxysporum Fo47. Physiol. Mol. Plant P. 73 (1-3), 9–15. doi: 10.1016/j.pmpp.2008.10.001
Aimé, S., Alabouvette, C., Steinberg, C., Olivain, C. (2013). The endophytic strain Fusarium oxysporum Fo47: a good candidate for priming the defense responses in tomato roots. Mol. Plant Microbe Interact. 26 (8), 918–926. doi: 10.1094/MPMI-12-12-0290-R
Alabouvette, C., Olivain, C., Migheli, Q., Steinberg, C. (2009). Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 184 (3), 529–544. doi: 10.1111/j.1469-8137.2009.03014.x
Alabouvette, C. (1986). Fusarium-wilt suppressive soils from the châteaurenard region: review of a 10-year study. Agronomie 6 (3), 273–284. doi: 10.1051/agro:19860307
Alves-Santos, F. M., Benito, E. P., Eslava, A. P., Diaz-Minguez, J. M. (1999). Genetic diversity of Fusarium oxysporum strains from common bean fields in Spain. Appl. Environ. Microb. 65 (8), 3335–3340. doi: 10.1128/AEM.65.8.3335-3340.1999
Armstrong, G. M., Armstrong, J. K. (1981). another approach to race classification of Fusarium oxysporum f. sp. pisi. Phytopathology 71 (5), 474–478. doi: 10.1094/Phyto-71-474
Bao, J., Fravel, D., Lazarovits, G., Chellemi, D., van Berkum, P., O'Neill, N. (2004). Biocontrol genotypes of Fusarium oxysporum from tomato fields in Florida. Phytoparasitica 32 (1), 9–20.
Benhamou, N., Garand, C. (2001). Cytological analysis of defense-related mechanisms induced in pea root tissues in response to colonization by nonpathogenic Fusarium oxysporum Fo47. Phytopathology 91 (8), 730–740. doi: 10.1094/PHYTO.2001.91.8.730
Benhamou, N., Garand, C., Goulet, A. (2002). Ability of nonpathogenic Fusarium oxysporum strain fo47 to induce resistance against Pythium ultimum infection in cucumber. Appl. Environ. Microbiol. 68 (8), 4044–4060. doi: 10.1128/AEM.68.8.4044-4060.2002
Bigeard, J., Colcombet, J., Hirt, H. (2015). Signaling mechanisms in Pattern-Triggered Immunity (PTI). Mol. Plant 8 (4), 521–539. doi: 10.1016/j.molp.2014.12.022
Biles, C. L., Martyn, R. D. (1989). Local and systemic resistance induced in watermelons by formae speciales of Fusarium oxysporum. Phytopathology 79 (8), 856–860. doi: 10.1094/Phyto-79-856
Blok, W. J., Zwankhuizen, M. J., Bollen, G. J. (1997). Biological control of Fusarium oxysporum f.sp. asparagi by applying non-pathogenic isolates of F. oxysporum. Biocontrol Sci. Techn. 7 (4), 527–541. doi: 10.1080/09583159730596
Boller, T., Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bolton, M. D., van Esse, H. P., Vossen, J. H., de Jonge, R., Stergiopoulos, I., Stulemeijer, I. J. E., et al. (2008). The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol. Microbiol. 69 (1), 119–136. doi: 10.1111/j.1365-2958.2008.06270.x
Bolwerk, A., Lagopodi, A. L., Lugtenberg, B. J. J., Bloemberg, G. V. (2005). Visualization of interactions between a pathogenic and a beneficial Fusarium strain during biocontrol of tomato foot and root rot. Mol. Plant Microbe 18 (7), 710–721. doi: 10.1094/MPMI-18-0710
Bubna, G. A., Lima, R. B., Zanardo, D. Y. L., dos Santos, W. D., Ferrarese, M. D. L., Ferrarese, O. (2011). Exogenous caffeic acid inhibits the growth and enhances the lignification of the roots of soybean (Glycine max). J. Plant Physiol. 168 (14), 1627–1633. doi: 10.1016/j.jplph.2011.03.005
Catanzariti, A. M., Lim, G. T. T., Jones, D. A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207 (1), 106–118. doi: 10.1111/nph.13348
Catanzariti, A. M., Do, H. T. T., Bru, P., de Sain, M., Thatcher, L. F., Rep, M., et al. (2017). The tomato/gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 89 (6), 1195–1209. doi: 10.1111/tpj.13458
Cheong, J. J., Hahn, M. G. (1991). A specific, high-affinity binding site for the hepta-beta-glucoside elicitor exists in soybean membranes. Plant Cell 3 (2), 137–147. doi: 10.1105/tpc.3.2.137
Chuberre, C., Plancot, B., Driouich, A., Moore, J. P., Bardor, M., Gugi, B., et al. (2018). Plant immunity is compartmentalized and specialized in roots. Front. Plant Sci. 9, 1692. doi: 10.3389/fpls.2018.01692
Coleman, A. D., Raasch, L., Maroschek, J., Ranf, S., Hückelhoven, R. (2019). The Arabidopsis leucine-rich repeat receptor kinase MIK2 is a crucial component of pattern-triggered immunity responses to Fusarium fungi. bioRxiv 720037. doi: 10.1101/720037
Constantin, M. E., de Lamo, F. J., Vlieger, B. V., Rep, M., Takken, F. L. W. (2019). Endophyte-mediated resistance in tomato to Fusarium oxysporum is independent of ET, JA, and SA. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00979
Díaz, J., Silvar, C., Varela, M. M., Bernal, A., Merino, F. (2005). Fusarium confers protection against several mycelial pathogens of pepper plants. Plant Pathol. 54, 773–780. doi: 10.1111/j.1365-3059.2005.01285.x
de Jonge, R., van Esse, H. P., Kombrink, A., Shinya, T., Desaki, Y., Bours, R., et al. (2010). Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329 (5994), 953–955. doi: 10.1126/science.1190859
de Lamo, F. J., Constantin, M. E., Fresno, D. H., Boeren, S., Rep, M., Takken, F. L. W. (2018). Xylem sap proteomics reveals distinct differences between R Gene- and endophyte-mediated resistance against fusarium wilt disease in tomato. Front. Microbiol. 9, 2977. doi: 10.3389/fmicb.2018.02977
de Sain, M., Rep, M. (2015). The role of pathogen-secreted proteins in fungal vascular wilt diseases. Int. J. Mol. Sci. 16 (10), 23970–23993. doi: 10.3390/ijms161023970
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13 (4), 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Deshmukh, S., Hueckelhoven, R., Schaefer, P., Imani, J., Sharma, M., Weiss, M., et al. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. PNAS 103 (49), 18450–18457. doi: 10.1073/pnas.0605697103
Di, X., Takken, F. L. W., Tintor, N. (2016). How phytohormones shape interactions between plants and the soil-borne fungus fusarium oxysporum. Front. Plant Sci. 7, 170.
Di, X., Gomila, J., Takken, F. L. W. (2017a). Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Mol. Plant Pathol. 18 (7), 1024–1035. doi: 10.1111/mpp.12559
Di, X. T., Cao, L. X., Hughes, R. K., Tintor, N., Banfield, M. J., Takken, F. L. W. (2017b). Structure-function analysis of the Fusarium oxysporum Avr2 effector allows uncoupling of its immune-suppressing activity from recognition. New Phytol. 216 (3), 897–914. doi: 10.1111/nph.14733
Duijff, B. J., Pouhair, D., Olivain, C., Alabouvette, C., Lemanceau, P. (1998). Implication of systemic induced resistance in the suppression of fusarium wilt of tomato by Pseudomonas fluorescens WCS417r and by nonpathogenic Fusarium oxysporum Fo47. Eur. J. Plant Pathol. 104 (9), 903–910. doi: 10.1023/A:1008626212305
Duijff, B. J., Recorbet, G., Bakker, P. A. H. M., Loper, J. E., Lemanceau, P. (1999). Microbial antagonism at the root level is involved in the suppression of Fusarium wilt by the combination of nonpathogenic Fusarium oxysporum Fo47 and Pseudomonas putida WCS358. Phytopathology 89 (11), 1073–1079. doi: 10.1094/PHYTO.1999.89.11.1073
Durrant, W. E., Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
El-kereamy, A., El-sharkawy, I., Ramamoorthy, R., Taheri, A., Errampalli, D., Kumar, P., et al. (2011). Prunus domestica pathogenesis-related protein-5 activates the defense response pathway and enhances the resistance to fungal infection. PloS One 6 (3), e17973. doi: 10.1371/journal.pone.0017973
Ellis, M. L., Jimenez, D. R. C., Leandro, L. F., Munkvold, G. P. (2014). Genotypic and phenotypic characterization of fungi in the Fusarium oxysporum species complex from soybean roots. Phytopathology 104 (12), 1329–1339. doi: 10.1094/PHYTO-02-14-0043-R
Elmer, W. H. (2004). Combining nonpathogenic strains of Fusarium oxysporum with sodium chloride to suppress fusarium crown rot of asparagus in replanted fields. Plant Pathol. 53 (6), 751–758. doi: 10.1111/j.1365-3059.2004.01096.x
Fesel, P. H., Zuccaro, A. (2016). Beta-glucan: crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 90, 53–60. doi: 10.1016/j.fgb.2015.12.004
Fisher, M. C., Henk, D. A., Briggs, C. J., Brownstein, J. S., Madoff, L. C., McCraw, S. L., et al. (2012). Emerging fungal threats to animal, plant and ecosystem health. Nature 484 (7393), 186–194. doi: 10.1038/nature10947
Flor, H. H. (1956). The complementary genic systems in flax and flax rust. Adv. Genet. 8, 29–54. doi: 10.1016/S0065-2660(08)60498-8
Franken, P. (2012). The plant strengthening root endophyte Piriformospora indica: potential application and the biology behind. Appl. Microbiol. Biot. 96 (6), 1455–1464. doi: 10.1007/s00253-012-4506-1
Fravel, D. R., Larkin, R. P. (2002). Reduction of fusarium wilt of hydroponically grown basil by Fusarium oxysporum strain CS-20. Crop Prot. 21 (7), 539–543. doi: 10.1016/S0261-2194(01)00143-0
Fravel, D. R., Olivain, C., Alabouvette, C. (2003). Fusarium oxysporum and its biocontrol. New Phytol. 157, 493–502. doi: 10.1046/j.1469-8137.2003.00700.x
Fu, Z. Q., Dong, X. N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fuchs, J. G., Moënne-Loccoz, Y., Défago, G. (1997). Nonpathogenic Fusarium oxysporum strain Fo47 induces resistance to Fusarium wilt in tomato. Plant Dis. 81 (5), 492–496. doi: 10.1094/PDIS.1997.81.5.492
Fuchs, J. G., Moënne-Loccoz, Y., Défago, G. (1999). Ability of nonpathogenic Fusarium oxysporum Fo47 to protect tomato against Fusarium wilt. Biol. Control 14 (2), 105–110. doi: 10.1006/bcon.1998.0664
Gadkar, V., David-Schwartz, R., Kunik, T., Kapulnik, Y. (2001). Arbuscular mycorrhizal fungal colonization. Factors involved in host recognition. Plant Physiol. 127 (4), 1493–1499. doi: 10.1104/pp.010783
Gao, F., Zhang, B. S., Zhao, J. H., Huang, J. F., Jia, P. S., Wang, S., et al. (2019). Deacetylation of chitin oligomers increases virulence in soil-borne fungal pathogens. Nat. Plants 5 (11), 1167–1176. doi: 10.1038/s41477-019-0527-4
García-Bastidas, F., Ordoñez, N., Konkol, J., Al-Qasim, M., Naser, Z., Abdelwali, M., et al. (2014). First report of Fusarium oxysporum f. sp. cubense tropical race 4 associated with panama disease of banana outside Southeast Asia. Plant Dis. 98 (5), 694–694. doi: 10.1094/PDIS-09-13-0954-PDN
Gawehns, F., Cornelissen, B. J. C., Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microb. Biotechnol. 6 (3), 223–229. doi: 10.1111/1751-7915.12023
Gawehns, F., Houterman, P. M., Ichou, F. A., Michielse, C. B., Hijdra, M., Cornelissen, B. J. C., et al. (2014). The Fusarium oxysporum effector six6 contributes to virulence and suppresses I-2-mediated cell death. MPMI 27 (4), 336–348. doi: 10.1094/MPMI-11-13-0330-R
Gawehns, F., Ma, L., Bruning, O., Houterman, P. M., Boeren, S., Cornelissen, B. J. C., et al. (2015). The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 6, 967. doi: 10.3389/fpls.2015.00967
Gonzalez, M., Brito, N., Gonzalez, C. (2017). The Botrytis cinerea elicitor protein BcIEB1 interacts with the tobacco PR5-family protein osmotin and protects the fungus against its antifungal activity. New Phytol. 215 (1), 397–410. doi: 10.1111/nph.14588
Gordon, T. R. (2017). Fusarium oxysporum and the Fusarium wilt syndrome. Annu. Rev. Phytopathol. 55, 23–39. doi: 10.1146/annurev-phyto-080615-095919
Grenier, J., Potvin, C., Trudel, J., Asselin, A. (1999). Some thaumatin-like proteins hydrolyse polymeric beta-1,3-glucans. Plant J. 19 (4), 473–480. doi: 10.1046/j.1365-313X.1999.00551.x
Hacquard, S., Spaepen, S., Garrido-Oter, R., Schulze-Lefert, P. (2017). Interplay between innate immunity and the plant microbiota. Annu. Rev. Phytopathol. 55, 565–589. doi: 10.1146/annurev-phyto-080516-035623
He, C. Y., Hsiang, T., Wolyn, D. J. (2002). Induction of systemic disease resistance and pathogen defence responses in Asparagus officinalis inoculated with nonpathogenic strains of Fusarium oxysporum. Plant Pathol. 51 (2), 225–230. doi: 10.1046/j.1365-3059.2002.00682.x
Henry, G., Thonart, P., Ongena, M. (2012). PAMPs, MAMPs, DAMPs and others: an update on the diversity of plant immunity elicitors. Biotechnol. Agron. Soc. 16 (2), 257–268.
Houterman, P. M., Cornelissen, B. J., Rep, M. (2008). Suppression of plant resistance gene-based immunity by a fungal effector. PloS Pathog. 4 (5), e1000061. doi: 10.1371/journal.ppat.1000061
Huertas-Gonzalez, M. D., Ruiz-Roldan, M. C., Di Pietro, A., Roncero, M. I. G. (1998). Cross protection provides evidence for race-specific avirulence factors in Fusarium oxysporum. Physiol. Mol. Plant P. 54 (3-4), 63–72. doi: 10.1006/pmpp.1998.0185
Humbert, C., Aimé, S., Alabouvette, C., Steinberg, C., Olivain, C. (2015). Remodelling of actin cytoskeleton in tomato cells in response to inoculation with a biocontrol strain of Fusarium oxysporum in comparison to a pathogenic strain. Plant Pathol. 64 (6), 1366–1374. doi: 10.1111/ppa.12375
Jacobs, S., Zechmann, B., Molitor, A., Trujillo, M., Petutschnig, E., Lipka, V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the Fungus Piriformospora indica. Plant Physiol. 157 (1), 531–531. doi: 10.1104/pp.111.900420
Jimenez-Fernandez, D., Landa, B. B., Kang, S., Jimenez-Diaz, R. M., Navas-Cortes, J. A. (2013). Quantitative and microscopic assessment of compatible and incompatible interactions between chickpea cultivars and Fusarium oxysporum f. sp. ciceris races. PloS One 8 (4), e61360. doi: 10.1371/journal.pone.0061360
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444 (7117), 323–329. doi: 10.1038/nature05286
Jonkers, W., Rodrigues, C. D., Rep, M. (2009). Impaired colonization and infection of tomato roots by the Δfrp1 mutant of Fusarium oxysporum correlates with reduced CWDE gene expression. Mol. Plant Microbe Interact. 22 (5), 507–518. doi: 10.1094/MPMI-22-5-0507
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. PNAS 103 (29), 11086–11091. doi: 10.1073/pnas.0508882103
Kaur, R., Singh, R. S. (2007). Study of induced systemic resistance in Cicer arietinum L. due to nonpathogenic Fusarium oxysporum using a modified split root technique. J. Phytopathol. 155 (11-12), 694–698. doi: 10.1111/j.1439-0434.2007.01300.x
Kroon, B. A. M., Scheffer, R. J., Elgersma, D. M. (1991). Induced resistance in tomato plants against Fusarium wilt invoked by Fusarium oxysporum f.sp. dianthi. Neth. J. Plant Pathol. 97 (6), 401–408. doi: 10.1007/BF03041387
L'Haridon, F., Aime, S., Alabouvette, C., Olivain, C. (2007). Lack of biocontrol capacity in a non-pathogenic mutant of Fusarium oxysporum f.sp. melonis. Eur. J. Plant Pathol. 118 (3), 239–246. doi: 10.1007/s10658-007-9139-3
Lanubile, A., Muppirala, U. K., Severin, A. J., Marocco, A., Munkvold, G. P. (2015). Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genomics 16. doi: 10.1186/s12864-015-2318-2
Larkin, R. P., Fravel, D. R. (1999). Mechanisms of action and dose-response relationships governing biological control of fusarium wilt of tomato by nonpathogenic Fusarium spp. Phytopathology 89 (12), 1152–1161. doi: 10.1094/PHYTO.1999.89.12.1152
Latz, M. A. C., Jensen, B., Collinge, D. B., Jørgensen, H. J. L. (2018). Endophytic fungi as biocontrol agents: elucidating mechanisms in disease suppression. Plant Ecol. Divers. 11 (5-6), 555–567. doi: 10.1080/17550874.2018.1534146
Le Floch, G., Vallance, J., Benhamou, N., Rey, P. (2009). Combining the oomycete Pythium oligandrum with two other antagonistic fungi: Root relationships and tomato grey mold biocontrol. Biol. Control 50 (3), 288–298. doi: 10.1016/j.biocontrol.2009.04.013
Lievens, B., Houterman, P. M., Rep, M. (2009). Effector gene screening allows unambiguous identification of Fusarium oxysporum f.sp lycopersici races and discrimination from other formae speciales. FEMS Microbiol. Lett. 300 (2), 201–215. doi: 10.1111/j.1574-6968.2009.01783.x
Liu, J. J., Sturrock, R., Ekramoddoullah, A. K. M. (2010). The superfamily of thaumatin-like proteins: its origin, evolution, and expression towards biological function. Plant Cell Rep. 29 (5), 419–436. doi: 10.1007/s00299-010-0826-8
Lopez-Aranda, J. M., Dominguez, P., Miranda, L., de los Santos, B., Talavera, M., Daugovish, O., et al. (2016). Fumigant use forsStrawberry production in Europe: the current landscape and solutions. Int. J. Fruit Sci. 16, 1–15. doi: 10.1080/15538362.2016.1199995
Ma, L. J., van der Does, H. C., Borkovich, K. A., Coleman, J. J., Daboussi, M. J., Di Pietro, A., et al. (2010). Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464 (7287), 367–373. doi: 10.1038/nature08850
Macho, A. P., Zipfel, C. (2014). Plant PRRs and the activation of innate immune signaling. Mol. Cell 54 (2), 263–272. doi: 10.1016/j.molcel.2014.03.028
Mahmood, T., Mehnaz, S., Fleischmann, F., Ali, R., Hashmi, Z. H., Iqbal, Z. (2014). Soil sterilization effects on root growth and formation of rhizosheaths in wheat seedlings. Pedobiologia 57 (3), 123–130. doi: 10.1016/j.pedobi.2013.12.005
Mani, T., Sivakumar, K. C., Manjula, S. (2012). Expression and functional analysis of two osmotin (PR5) isoforms with differential antifungal activity from Piper colubrinum: prediction of structure-function relationship by bioinformatics approach. Mol. Biotechnol. 52 (3), 251–261. doi: 10.1007/s12033-011-9489-0
McLusky, S. R., Bennett, M. H., Beale, M. H., Lewis, M. J., Gaskin, P., Mansfield, J. W. (1999). Cell wall alterations and localized accumulation of feruloyl-3 ‘-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J. 17 (5), 523–534. doi: 10.1046/j.1365-313X.1999.00403.x
Menu-Bouaouiche, L., Vriet, C., Peumans, W. J., Barre, A., Van Damme, E. J. M., Rouge, P. (2003). A molecular basis for the endo-beta 1,3-glucanase activity of the thaumatin-like proteins from edible fruits. Biochimie 85 (1-2), 123–131. doi: 10.1016/S0300-9084(03)00058-0
Mes, J. J., van Doorn, A. A., Wijbrandi, J., Simons, G., Cornelissen, B. J. C., Haring, M. A. (2000). Expression of the Fusarium resistance gene I-2 colocalizes with the site of fungal containment. Plant J. 23 (2), 183–193. doi: 10.1046/j.1365-313x.2000.00765.x
Michielse, C. B., Rep, M. (2009). Pathogen profile update: Fusarium oxysporum. Mol. Plant Pathol. 10 (3), 311–324. doi: 10.1111/j.1364-3703.2009.00538.x
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22 (3), 973–990. doi: 10.1105/tpc.109.069658
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. PNAS 104 (49), 19613–19618. doi: 10.1073/pnas.0705147104
Mur, L. A. J., Kenton, P., Lloyd, A. J., Ougham, H., Prats, E. (2008). The hypersensitive response; the centenary is upon us but how much do we know? J. Exp. Bot. 59 (3), 501–520. doi: 10.1093/jxb/erm239
Nahalkova, J., Fatehi, J., Olivain, C., Alabouvette, C. (2008). Tomato root colonization by fluorescent-tagged pathogenic and protective strains of Fusarium oxysporum in hydroponic culture differs from root colonization in soil. FEMS Microbiol. Lett. 286 (2), 152–157. doi: 10.1111/j.1574-6968.2008.01241.x
Nel, B., Steinberg, C., Labuschagne, N., Viljoen, A. (2006). The potential of nonpathogenic Fusarium oxysporum and other biological control organisms for suppressing fusarium wilt of banana. Plant Pathol. 55 (2), 217–223. doi: 10.1111/j.1365-3059.2006.01344.x
Nelson, P. E. (1981). Chapter 3 - Life cycle and epidemiology of Fusarium oxysporum. Fungal Wilt. Dis. Plants 51–80. doi: 10.1016/B978-0-12-464450-2.50008-5
Nirmaladevi, D., Venkataramana, M., Srivastava, R. K., Uppalapati, S. R., Gupta, V. K., Yli-Mattila, T., et al. (2016). Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Sci. Rep. 6. doi: 10.1038/srep21367
Nordzieke, D. E., Fernandes, T. R., El Ghalid, M., Turrà, D., Di Pietro, A. (2019). NADPH oxidase regulates chemotropic growth of the fungal pathogen Fusarium oxysporum towards the host plant. New Phytol. 1600–1612. doi: 10.1111/nph.16085
O'Brien, J. A., Daudi, A., Butt, V. S., Bolwell, G. P. (2012). Reactive oxygen species and their role in plant defence and cell wall metabolism. Planta 236 (3), 765–779. doi: 10.1007/s00425-012-1696-9
Olivain, C., Trouvelot, S., Binet, M. N., Cordier, C., Pugin, A., Alabouvette, C. (2003). Colonization of flax roots and early physiological responses of flax cells inoculated with pathogenic and nonpathogenic strains of Fusarium oxysporum. Appl. Environ. Microbiol. 69 (9), 5453–5462. doi: 10.1128/AEM.69.9.5453-5462.2003
Olivain, C., Humbert, C., Nahalkova, J., Fatehi, J., L'Haridon, F., Alabouvette, C. (2006). Colonization of tomato root by pathogenic and nonpathogenic Fusarium oxysporum strains inoculated together and separately into the soil. Appl. Environ. Microbiol. 72 (2), 1523–1531. doi: 10.1128/AEM.72.2.1523-1531.2006
Ordoñez, L. N., García-Bastidas, F., Laghari, H. B., Akkary, M. Y., Harfouche, E. N., al Awar, ,. B. N., et al. (2016). First report of Fusarium oxysporum f. sp. cubense tropical race 4 causing panama disease in Cavendish bananas in Pakistan and Lebanon. Plant Dis. 100 (1), 209–210. doi: 10.1094/PDIS-12-14-1356-PDN
Panina, Y., Fravel, D. R., Baker, C. J., Shcherbakova, L. A. (2007). Biocontrol and plant pathogenic Fusarium oxysporum-induced changes in phenolic compounds in tomato leaves and roots. J. Phytopathol. 155 (7-8), 475–481. doi: 10.1111/j.1439-0434.2007.01260.x
Pantelides, I. S., Tjamos, S. E., Striglis, I. A., Chatzipavhdis, I., Paplomatals, E. J. (2009). Mode of action of a non-pathogenic Fusarium oxysporum strain against Verticillium dahliae using Real Time QPCR analysis and biomarker transformation. Biol. Control 50 (1), 30–36. doi: 10.1016/j.biocontrol.2009.01.010
Pennington, H. G., Gheorghe, D. M., Damerum, A., Pliego, C., Spanu, P. D., Cramer, R., et al. (2016). Interactions between the powdery mildew effector BEC1054 and barley proteins identify candidate host targets. J. Proteome Res. 15 (3), 826–839. doi: 10.1021/acs.jproteome.5b00732
Pieterse, C. M. J., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C. M., Bakker, P. A. H. M. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Pinaria, A. G., Laurence, M. H., Burgess, L. W., Liew, E. C. Y. (2015). Phylogeny and origin of Fusarium oxysporum f. sp. vanillae in Indonesia. Plant Pathol. 64 (6), 1358–1365. doi: 10.1111/ppa.12365
Ploetz, R. C. (2015). Fusarium Wilt of Banana. Phytopathology 105 (12), 1512–1521. doi: 10.1094/PHYTO-04-15-0101-RVW
Pu, X., Xie, B., Li, P., Mao, Z., Ling, J., Shen, H., et al. (2014). Analysis of the defence-related mechanism in cucumber seedlings in relation to root colonization by nonpathogenic Fusarium oxysporum CS-20. FEMS Microbiol. Lett. 355 (2), 142–151. doi: 10.1111/1574-6968.12461
Pu, Z., Ino, Y., Kimura, Y., Tago, A., Shimizu, M., Natsume, S., et al. (2016). Changes in the Proteome of Xylem Sap in Brassica oleracea in Response to Fusarium oxysporum Stress. Front. Plant Sci. 7, 31. doi: 10.3389/fpls.2016.00031
Rep, M., van der Does, H. C., Meijer, M., van Wijk, R., Houterman, P. M., Dekker, H. L., et al. (2004). A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol. Microbiol. 53 (5), 1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x
Risser, G., Banihashemi, Z., Davis, D. W. (1976). Proposed Nomenclature of Fusarium oxysporum f.sp. melonis races and resistance genes in Cucumis melo. Phytopathology 66 (9), 1105–1106. doi: 10.1094/Phyto-66-1105
Salerno, M. I., Gianinazzi, S., Gianinazzi-Pearson, V. (2000). Effects on growth and comparison of root tissue colonization patterns of Eucalyptus viminalis by pathogenic and nonpathogenic strains of Fusarium oxysporum. New Phytol. 146 (2), 317–324. doi: 10.1046/j.1469-8137.2000.00629.x
Schmidt, S. M., Houterman, P. M., Schreiver, I., Ma, L. S., Amyotte, S., Chellappan, B., et al. (2013). MITEs in the promoters of effector genes allow prediction of novel virulence genes in Fusarium oxysporum. BMC Genomics 14. doi: 10.1186/1471-2164-14-119
Schmidt, S. M., Lukasiewicz, J., Farrer, R., van Dam, P., Bertoldo, C., Rep, M. (2016). Comparative genomics of Fusarium oxysporum f. sp. melonis reveals the secreted protein recognized by the Fom-2 resistance gene in melon. New Phytol. 209 (1), 307–318. doi: 10.1111/nph.13584
Shcherbakova, L. A., Odintsova, T. I., Stakheev, A. A., Fravel, D. R., Zavriev, S. K. (2015). Identification of a novel small cysteine-rich protein in the fraction from the Biocontrol Fusarium oxysporum strain CS-20 that mitigates fusarium wilt symptoms and triggers defense responses in tomato. Front. Plant Sci. 6, 1207. doi: 10.3389/fpls.2015.01207
Shoresh, M., Harman, G. E., Mastouri, F. (2010). Induced systemic resistance and plant responses to Fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43. doi: 10.1146/annurev-phyto-073009-114450
Simons, G., Groenendijk, J., Wijbrandi, J., Reijans, M., Groenen, J., Diergaarde, P., et al. (1998). Dissection of the Fusarium I2 gene cluster in tomato reveals Six homologs and one active gene copy. Plant Cell 10, 1055–1068. doi: 10.1105/tpc.10.6.105510.2307/3870690
Steinberg, C., Whipps, J. M., Wood, D., Fenlon, J., Alabouvette, C. (1999). Mycelial development of Fusarium oxysporum in the vicinity of tomato roots. Mycol. Res. 103, 769–778. doi: 10.1017/S0953756298007710
Steinkellner, S., Mammerler, R., Vierheilig, H. (2005). Microconidia germination of the tomato pathogen Fusarium oxysporum in the presence of root exudates. J. Plant Interact. 1 (1), 23–30. doi: 10.1080/17429140500134334
Stintzi, A., Heitz, T., Prasad, V., Wiedemannmerdinoglu, S., Kauffmann, S., Geoffroy, P., et al. (1993). Plant pathogenesis-related proteins and their role in defense against pathogens. Biochimie 75 (8), 687–706. doi: 10.1016/0300-9084(93)90100-7
Su, Z. Z., Mao, L. J., Li, N., Feng, X. X., Yuan, Z. L., Wang, L. W., et al. (2013). Evidence for biotrophic lifestyle and biocontrol potential of dark septate endophyte Harpophora oryzae to rice blast disease. PloS One 8 (4). doi: 10.1371/journal.pone.0061332
Takken, F. L. W., Rep, M. (2010). The arms race between tomato and Fusarium oxysporum. Mol. Plant Pathol. 11 (2), 309–314. doi: 10.1111/j.1364-3703.2009.00605.x
Tamietti, G., Ferraris, L., Matta, A., Gentile, I. A. (1993). Physiological responses of tomato plants grown in Fusarium suppressive soil. J. Phytopathol. 138 (1), 66–76. doi: 10.1111/j.1439-0434.1993.tb01361.x
Trouvelot, S., Olivain, C., Recorbet, G., Migheli, Q., Alabouvette, C. (2002). Recovery of Fusarium oxysporum Fo47 mutants affected in their biocontrol activity after transposition of the Fot1 element. Phytopathology 92 (9), 936–945. doi: 10.1094/PHYTO.2002.92.9.936
Turrà, D., El Ghalid, M., Rossi, F., Di Pietro, A. (2015). Fungal pathogen uses sex pheromone receptor for chemotropic sensing of host plant signals. Nature 527 (7579), 521–52+. doi: 10.1038/nature15516
Underwood, W. (2012). The plant cell wall: a dynamic barrier against pathogen invasion. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00085
Validov, S. Z., Kamilova, F. D., Lugtenberg, B. J. (2011). Monitoring of pathogenic and non-pathogenic Fusarium oxysporum strains during tomato plant infection. Microb. Biotechnol. 4 (1), 82–88. doi: 10.1111/j.1751-7915.2010.00214.x
van Dam, P., Fokkens, L., Schmidt, S. M., Linmans, J. H. J., Kistler, H. C., Ma, L. J., et al. (2016). Effector profiles distinguish formae speciales of Fusarium oxysporum. Environ. Microbiol. 18 (11), 4087–4102. doi: 10.1111/1462-2920.13445
van Dam, P., Fokkens, L., Ayukawa, Y., van der Gragt, M., ter Horst, A., Brankovics, B., et al. (2017). A mobile pathogenicity chromosome in Fusarium oxysporum for infection of multiple cucurbit species. Sci. Rep. 7. doi: 10.1038/s41598-017-07995-y
van Dam, P., de Sain, M., ter Horst, A., van der Gragt, M., Rep, M. (2018). Use of comparative genomics-based markers for discrimination of host specificity in Fusarium oxysporum. Appl. Environ. Microb. 84 (1), 55–65. doi: 10.1128/AEM.01868-17
van der Does, H. C., Constantin, M. E., Houterman, P. M., Takken, F. L. W., Cornelissen, B. J. C., Haring, M. A., et al. (2018). Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur. J. Plant Pathol. doi: 10.1007/s10658-018-1596-3
Veloso, J., Díaz, J. (2012). Fusarium oxysporum Fo47 confers protection to pepper plants against Verticillium dahliae and Phytophthora capsici, and induces the expression of defence genes. Plant Pathol. 61, 281–288. doi: 10.1111/j.1365-3059.2011.02516.x
Veloso, J., Alabouvette, C., Olivain, C., Flors, V., Pastor, V., García, T., et al. (2016). Modes of action of the protective strain Fo47 in controlling verticillium wilt of pepper. Plant Pathol. 65 (6), 997–1007. doi: 10.1111/ppa.12477
Vigers, A. J., Wiedemann, S., Roberts, W. K., Legrand, M., Selitrennikoff, C. P., Fritig, B. (1992). Thaumatin-Like Pathogenesis-Related Proteins Are Antifungal. Plant Sci. 83 (2), 155–161. doi: 10.1016/0168-9452(92)90074-V
Vitale, S., Di Pietro, A., Turrà, D. (2019). Autocrine pheromone signalling regulates community behaviour in the fungal pathogen Fusarium oxysporum. Nat. Microbiol. 1443–1449. doi: 10.1038/s41564-019-0456-z
Vlaardingerbroek, I., Beerens, B., Schmidt, S. M., Cornelissen, B. J. C., Rep, M. (2016). Dispensable chromosomes in Fusarium oxysporum f. sp. lycopersici. Mol. Plant Pathol. 17 (9), 1455–1466. doi: 10.1111/mpp.12440
Vos, C. M., Yang, Y., De Coninck, B., Cammue, B. P. A. (2014). Fungal (-like) biocontrol organisms in tomato disease control. Biol. Control 74, 65–81. doi: 10.1016/j.biocontrol.2014.04.004
Wolfe, K. H., Gouy, M. L., Yang, Y. W., Sharp, P. M., Li, W. H. (1989). Date of the monocot dicot divergence estimated from chloroplast DNA-Sequence Data. PNAS 86 (16), 6201–6205. doi: 10.1073/pnas.86.16.6201
Woloshuk, C. P., Meulenhoff, J. S., Sela-Buurlage, M., van den Elzen, P. J., Cornelissen, B. J. C. (1991). Pathogen-induced proteins with inhibitory activity toward Phytophthora infestans. Plant Cell 3 (6), 619–628. doi: 10.1105/tpc.3.6.61910.2307/3869190
Wong, J. Y., Jeffries, P. (2006). Diversity of pathogenic Fusarium populations associated with asparagus roots in decline soils in Spain and the UK. Plant Pathol. 55 (3), 331–342. doi: 10.1111/j.1365-3059.2006.01360.x
Xiong, W., Zhao, Q. Y., Xue, C., Xun, W. B., Zhao, J., Wu, H. S., et al. (2016). Comparison of fungal community in black pepper-vanilla and vanilla monoculture systems associated with vanilla Fusarium wilt disease. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00117
Yao, J., Allen, C. (2006). Chemotaxis is required for virulence and competitive fitness of the bacterial wilt pathogen Ralstonia solanacearum. J. Bacteriol. 188 (10), 3697–3708. doi: 10.1128/JB.188.10.3697-3708.2006
Zhang, J., Chen, J., Jia, R. M., Ma, Q., Zong, Z. F., Wang, Y. (2018). Suppression of plant wilt diseases by nonpathogenic Fusarium oxysporum Fo47 combined with actinomycete strains. Biocontrol Sci. Techn. 28 (6), 562–573. doi: 10.1080/09583157.2018.1468996
Keywords: Fusarium, endophyte, induced resistance, biocontrol, PR protein
Citation: de Lamo FJ and Takken FLW (2020) Biocontrol by Fusarium oxysporum Using Endophyte-Mediated Resistance. Front. Plant Sci. 11:37. doi: 10.3389/fpls.2020.00037
Received: 25 September 2019; Accepted: 13 January 2020;
Published: 06 February 2020.
Edited by:
Kalliope K. Papadopoulou, University of Thessaly, GreeceReviewed by:
Víctor Flors, University of Jaume I, SpainMarta Berrocal-Lobo, Polytechnic University of Madrid, Spain
Copyright © 2020 de Lamo and Takken. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Frank L. W. Takken, F.L.W.Takken@uva.nl
 Francisco J. de Lamo
Francisco J. de Lamo Frank L. W. Takken
Frank L. W. Takken