
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 10 February 2020
Sec. Plant Breeding
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01781
This article is part of the Research Topic Translational Research for Cucurbit Molecular Breeding: Traits, Markers, and Genes View all 22 articles
 Zhihua Cheng1
Zhihua Cheng1 Shibin Zhuo1
Shibin Zhuo1 Xiaofeng Liu1
Xiaofeng Liu1 Gen Che1
Gen Che1 Zhongyi Wang1
Zhongyi Wang1 Ran Gu1
Ran Gu1 Junjun Shen1
Junjun Shen1 Weiyuan Song1
Weiyuan Song1 Zhaoyang Zhou1
Zhaoyang Zhou1 Deguo Han2*
Deguo Han2* Xiaolan Zhang1*
Xiaolan Zhang1*Cucumber is an important vegetable crop bearing fleshy pepo fruit harvested immature. Fruits left unpicked in time during summer production, as well as unfavorable environmental conditions during post-harvest shelf, will cause cucumber fruits to turn yellow and ripen, and thus impair the market value. Identification of maturity-related genes is of great agricultural and economic importance for cucumber production. Here, we isolated and characterized a MADS-box gene, Cucumis sativus SHATTERPROOF (CsSHP) in cucumber. Expression analysis indicated that CsSHP was specifically enriched in reproductive organs including stamens and carpels. Ectopic expression of CsSHP was unable to rescue the indehiscence silique phenotype of shp1 shp2 mutant plant in Arabidopsis. Instead, overexpression of CsSHP resulted in early flowering, precocious phenotypes, and capelloid organs in wild-type Arabidopsis. Biochemical analysis indicated that CsSHP directly interacted with cucumber SEPALLATA (SEP) proteins. CsSHP expression increased significantly during the yellowing stage of cucumber ripening, and was induced by exogenous application of abscisic acid (ABA). Therefore, CsSHP may participate in fruit maturation through the ABA pathway and floral organ specification via interaction with CsSEPs to form protein complex in cucumber.
The fruit is a major evolutionary success in angiosperms which is essential for plant sexual reproduction and environmental adaptation (Fourquin and Ferrandiz, 2012). The main functions of fruits are to protect and nourish the developing seeds, and to act as a seed dispersal agent (van der Knaap et al., 2014). Angiosperms have evolved different types of fruit to meet the need of diverse dispersal strategies, such as the dry dehiscent fruit in Arabidopsis thaliana opens through dehiscence zones to release seeds, whereas the fleshy fruit in tomato attracts frugivorous animals to disperse seeds by means of bright color and pleasant aromas upon ripening (Roeder and Yanofsky, 2006; Ferrandiz and Fourquin, 2014). Moreover, fruits are the commercial organs for many agricultural crops and play important roles for human diet and health, thus the fruit has been under strong selective pressure during crop domestication (van der Knaap et al., 2014).
The fruit is generally developed from the ovary, which is an important component of the gynoecium. Gynoecium is located in the center of the flower, and surrounded sequentially as whorls by stamens, petals, and sepals, respectively (Robles and Pelaz, 2005). According to the ABC model, the sepal is specified by A function genes, the petal is determined by A+B function genes, the stamen is controlled by the B+C function genes, and the carpel is specified by the C class of genes (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994; Fourquin and Ferrandiz, 2012). In Arabidopsis, the AGAMOUS (AG) gene is the C class of gene that determines the carpel identity, specifies stamen identity with B-function genes, inhibits A-function genes and controls floral meristem determinacy (Bowman et al., 1989; Bowman et al., 1991; Mizukami and Ma, 1992; Mizukami and Ma, 1995). Subsequent studies showed that SEPALLATA (SEP) genes, expressing in four floral whorls, act as co-factors with ABC homeotic genes in specifying all types of floral organs (Theissen and Saedler, 2001; Favaro et al., 2003; Robles and Pelaz, 2005; Ruelens et al., 2017). Strikingly, all the genes involved in floral organ specification in Arabidopsis are from MADS-box transcription factor family excepting the gene APETALA2 (Jofuku et al., 1994; Dreni and Kater, 2014). MADS-box genes are reported to be the key players in organ morphogenesis throughout the plant life cycle, with a typical MADS domain and a K-box domain in their protein structure (Smaczniak et al., 2012).
There are two MADS-box genes, the SHATTERPROOF1 (SHP1) and SHP2, in the AG subfamily of Arabidopsis acting as the major regulators directing dehiscence zone differentiation and stimulating lignification of adjacent cells in siliques (Liljegren et al., 2000). In the double mutant shp1 shp2 plant, the mature siliques were unable to dehisce due to failure of dehiscence zone formation (Liljegren et al., 2000). Constitutive expression of SHP genes led to small fruits with overlignified valves (Liljegren et al., 2000). In other dry dehiscent fruits such as Nicotiana benthamiana (NbSHP), Glycine max (GmAGL1), and Medicago, the SHP gene promotes lignin accumulation in fruit pods to ensure cracking upon maturation (Liljegren et al., 2000; Fourquin and Ferrandiz, 2012; Fourquin et al., 2013; Chi et al., 2017). In the fleshy berry fruit tomato (Giovannoni, 2007), the SHP1/2 ortholog TAGL1 participates in fruit expansion and promotes fruit ripening. Knockdown of TAGL1 resulted in yellow orange fruit with decreased carotenoids and thin pericarps. Overexpression of TAGL1 led to enlarged sepals and overaccumulation of lycopene, supporting the roles of TAGL1 in fruit ripening (Itkin et al., 2009; Vrebalov et al., 2009; Gimenez et al., 2010). Similarly, in the fleshy false fruit of strawberry, FaSHP was shown to promote maturation as well (Itkin et al., 2009; Vrebalov et al., 2009; Daminato et al., 2013). In citrus, CsMADS6 (the ortholog of SHP1/2) positively modulated carotenoid metabolism by directly regulating the expression of carotenogenic genes, suggesting an active role in fruit ripening (Lu et al., 2018). In addition to the functions in fruit opening and ripening, SHP genes play important roles in floral organ determination (Pinyopich et al., 2003; Vrebalov et al., 2009; Chi et al., 2017). In Arabidopsis, a redundant roles of SHP1/2 and AG were found to promote carpel development, and overexpression of SHP2 was sufficient to rescue the stamen and carpel phenotypes in the ag mutant (stamens were replaced with petals and carpels were replaced by new abnormal flowers) (Bowman et al., 1989; Pinyopich et al., 2003). Transient knockdown of NbSHP in tobacco exhibited unfused pistils, and increased number of styles and stigmas (Fourquin and Ferrandiz, 2012). Ectopic expression of soybean GmAGL1 in Arabidopsis resulted in petal-free flowers (Chi et al., 2017). Overexpression of grape Vvmads1 (the ortholog of SHP1/2) in tobacco led to carpelloid sepals and stamenoid petals (Boss et al., 2001).
Cucumber (Cucumis sativus L.) is a world-wide cultivated vegetable crop bearing fleshy pepo fruit, which is developed from three-syncarpous inferior ovary (Che and Zhang, 2019). The fruit of North China cucumber can be generally divided into five developmental stages: immature green [0–9 days after anthesis days after anthesis (DAA)], breaker (12–15 DAA), turning (18–24 DAA), fully ripe (27–30 DAA), and senescence (35–43 DAA) (Leng et al., 2014). Cucumber fruits are consumed freshly or processed into pickles, and typically are harvested immature (about 7~14 DAA) (Weng et al., 2015). During summer production of cucumber, fruits left unpicked in time are prone to turn yellow and ripen on plants, which will greatly impair the commercial value and result in economic loss (Wang et al., 2013). Meanwhile, during cucumber post-harvest storage and transportation, unfavorable environmental conditions will cause immature fruits a series of senescence phenomena such as yellowing, accumulation of citrate, and tissue softening (Mainardi et al., 2006), which will adversely affect the market value. Therefore, identification of maturity-related genes is of great agricultural and economic importance for inhibiting fruit ripening on plants and delaying senescence of postharvest cucumbers.
As a non-climacteric fruit, it is abscisic acid (ABA), not ethylene, that was shown to promote fruit ripening on the cucumber plant (Nilsson, 2005; Hurr et al., 2009; Wang et al., 2013). To isolate putative fruit ripening related genes, we cloned a MADS-box gene CsSHP (the ortholog of SHP) in cucumber. CsSHP expression was highly enriched in reproductive organs of cucumber and were positively correlated with ABA accumulation during fruit maturation. Overexpression of CsSHP caused early flowering, precocious phenotypes, and ectopic capelloid organs in Arabidopsis. Biochemical analysis indicated that CsSHP directly interact with CsSEPs at protein level. Thus, CsSHP may participate in fruit maturation through the ABA pathway and floral organ specification via interaction with CsSEPs, which provides a possible target for genetic manipulation of fruit maturation progression to meet the different market demands in cucumber.
Cucumber (Cucumis sativus L.) inbred lines R1461 (Chinese long type) was used in this study, and grown in the experimental field of China Agricultural University at Beijing under standard greenhouse conditions. The A. thaliana Columbia (Col) ecotype and the double mutant shp1 shp2, and relevant transgenic plants were grown in soil at 22°C under 16 h/8 h light/dark condition in a growth chamber.
A 711-bp fragment containing the complete CsSHP coding sequence was amplified from the young female buds using gene-specific primers (Supplementary Table S2). The gene structure of CsSHP was analyzed using online software GSDS 2.0 (http://gsds.cbi.pku.edu.cn/). Protein sequences of SHPs and other MADS-box genes from diverse plant species were obtained using the protein BLAST search (NCBI blast: https://blast.ncbi.nlm.nih.gov/Blast.cgi). Multiple sequence alignment was performed using CLUSTALW in MEGA5. The phylogenetic tree was generated using the neighbor-joining method in MEGA5 with 1,000 bootstrap replicates. The GenBank accession numbers for related proteins are listed in Supplementary Table S1.
The young leaves, stems, tendrils, male flower buds, female flower buds, male flowers, female flowers, ovaries at anthesis, fruits at different developmental stages from cucumber, as well as the inflorescences of Arabidopsis were frozen in the liquid nitrogen and stored at −80°C until use. Total RNA was extracted with TRIzol reagent as described in the manufacturer's instructions (Waryoung, China, http://www.huayueyang.com/), and cDNA was synthesized using FastQuant RT Kit (Tiangen, China, http://www.tiangen.com/). Quantitative real-time PCR (qRT-PCR) was performed using TB Green™ Premix Ex Taq™ (Takara, Japan, http://www.takarabiomed.com.cn/) on the Applied Biosystems 7500 RT-qPCR system. Ubiquitin extension protein (UBI CsaV3_5G031430) and ACTIN2 (AT3G18780) were used as the internal reference genes in cucumber and Arabidopsis, respectively (Wan et al., 2010). The relative expression was calculated according to the comparative cycle threshold (CT) method (Schmittgen and Livak, 2008). Three biological replicates and three technical replicates were performed for each gene. The primer information is listed in Supplementary Table S2.
The 25-day-old shoot tips, male and female flower buds of R1461 were fixed with 3.7% formalin-acetic acid-alcohol (FAA) fixative. Sampling and recognition of flower buds at different developmental stages were performed as described (Bai et al., 2004). Sense and antisense probes were amplified with gene-specific primers containing SP6 and T7 RNA polymerase binding sites, respectively. Sample fixation, sectioning and hybridization were performed as described previously (Zhang et al., 2013). The primer information is listed in Supplementary Table S2.
β-glucuronidase (GUS) assay was performed according to the protocol as described previously (Liu et al., 2016). The Arabidopsis inflorescences and fruits at different developmental stage were fixed and incubated in GUS-staining solution at 37°C for 24–48 h until dyed blue. Stained samples were then cleared with 70% ethanol and observed by anatomic microscope (Leica DFC450, Germany). The experiment was repeated three times.
The full-length CDS of CsSHP was fused into the pSUPER1300 vector to generate a CsSHP-GFP in-frame fusion protein. The empty pSUPER1300 vector was used as a positive control. Agrobacterium tumefaciens with the vectors were injected into the abaxial side of tobacco leaves (4–6 weeks old) by syringe as described previously (Schutze et al., 2009). After 48 h of infiltration, the fluorescence of the expressed GFP proteins was detected and captured under 488 nm excitation wavelength from Argon laser of fluorescence microscope (Leica sp5, Germany). The wavelength range for GFP is 495–545 nm.
The binary vector pBI121 and pSUPER1300 were used for ectopic expression of CsSHP in Arabidopsis Col and shp1 shp2, respectively. The CsSHP CDS was cloned into pBI121 and pSUPER1300 through XbaI and SmaI cleavages sites, and HindIII and KpnI cleavages sites to generate CsSHP overexpression constructs, respectively. Col and shp1 shp2 plants were transformed by A. tumefaciens containing the pBI121 and pSUPER1300 recombinant construct through the floral dip method, respectively (Clough and Bent, 1998). Transgenic seeds were germinated on solid Murashige and Skoog (MS) medium with 50mg/L kanamycin and 25mg/L hygromycin, respectively. Resistant seedlings were transferred to soil and further verified by PCR and qRT-PCR. Three biological replicates and three technical replicates were performed for each gene. T2 transgenic plants were chosen for further phenotypic analysis and data statistics. The primers used for vector construction and transgene identification are given in Supplementary Table S2.
About 0.3 g cucumber ovaries at different developmental stages were used as samples. The content of indole-3-acetic acid (IAA), zeatin riboside (ZR), abscisic acid (ABA) were measured using enzyme-linked immunosorbent assays according to methods previously described (Sun et al., 2016). Three biological replicates were performed.
Three groups, each consisting of 20 cucumber ovaries at 4 days before anthesis, were used as samples. The first group was sprayed with a solution of 200 mg/L ABA in 0.5% (v/v) Tween 20, the second group was sprayed with a solution of 50 mg/L IAA in 0.5% (v/v) Tween 20, and the third group was sprayed only with 0.5% (v/v) Tween 20 as control (Daminato et al., 2013). Three ovaries from each group were collected after 0, 1, 3, 12, and 24 h treatments. RNA of the collected ovaries was extracted, and the CsSHP expression was evaluated by qRT-PCR.
Full-length CDSs of IND, SPT, CsSHP, CUM10, CsSEP2, CsSEP3, and CsSEP4 were cloned into pGADT7 (prey vector) or pGBKT7 (bait vector). All constructs were verified by sequencing and then transformed into yeast strain AH109. The yeast two-hybrid assays were conducted and protein interactions were analyzed on selective medium lacking Leu, Trp, His, and Ade (Ding et al., 2015). The primers for yeast two-hybrid assays are listed in Supplementary Table S2.
CsSEP2, CsSEP3, CsSEP4, and CUM10 full-length CDSs without stop codon were cloned in pCAMBIA1300-nLUC, and CsSHP full-length CDS with stop codon in pCAMBIA1300-cLUC. A. tumefaciens GV3101 strain carrying the above constructs was mixed in proportion and resuspended, then injected into tobacco (N. benthamiana) leaves by syringe. The interactions of the expressed fusion proteins were indicated by reconstituted LUC enzyme after 2–3 days of infiltration, and images were obtained using a chemiluminescent imaging system (Tanon 5200, China) as described (Chen et al., 2008). The primers for vector construction are given in Supplementary Table S2.
To isolate the SHP gene in cucumber, we performed a BLAST search in National Center for Biotechnology Information (NCBI) database, and found the cucumber protein (NP_001292697.1) displays the highest sequence homology (59.7% identity) to Arabidopsis SHP1. A reciprocal BLAST search was performed in The Arabidopsis Information Resource (TAIR) and cucurbit genomics database (CuGenDB), and the NP_001292697.1 (CsaV3_6G015770.1) was confirmed to be the SHP homolog in cucumber, therefore, we named it as CsSHP hereinafter. There were two SHP genes (SHP1 and SHP2) in Arabidopsis, while only one SHP in cucumber. The genomic sequence of CsSHP is 8,591 bp, which is much longer than that of SHP1 (4,058 bp) and SHP2 (3,759 bp) in Arabidopsis. CsSHP is predicted to contain seven exons and six introns, with the first and second intron being particularly long (3,005 and 3,232 bp, respectively) (Figure 1A). The second intron of AG/PLE genes contains several conserved motifs and has been shown to be essential for fulfilling their proper functions (Causier et al., 2009). The extremely large first and second introns in CsSHP may imply the more complex functional regulation than its Arabidopsis counterpart.
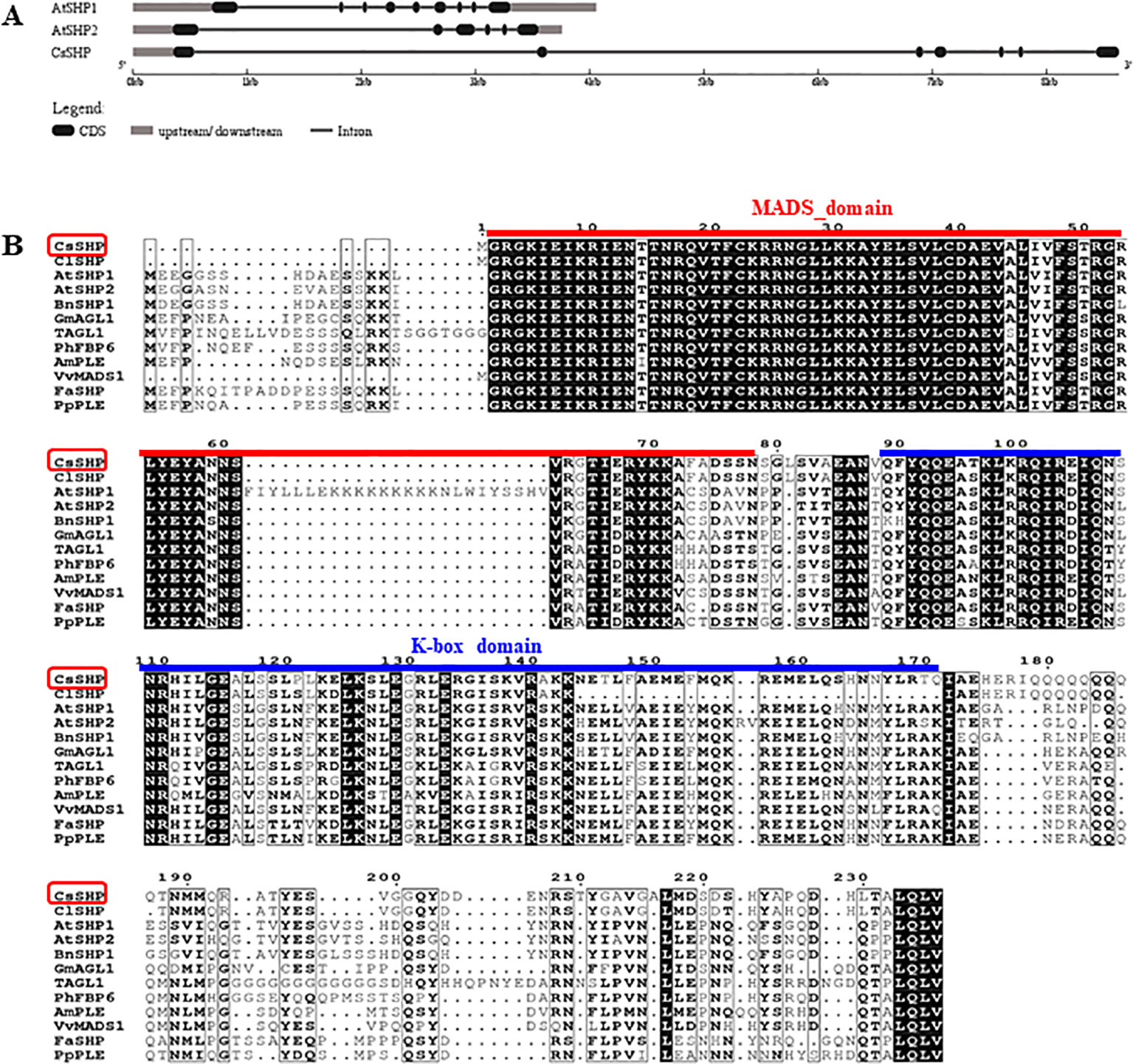
Figure 1 Gene structure and sequence alignment. (A) Gene structure analysis of SHP1, SHP2, and CsSHP. (B) Multiple sequence alignment of CsSHP and orthologs from related plant species. The red and blue lines indicate the conserved MADS domain and K-box domain, respectively.
The full-length coding sequence (CDS) of CsSHP was obtained from the female bud of cucumber inbred line R1461, which encodes a protein of 237 amino acids with a calculated molecular mass of 27.18 kD (Supplementary S1). A multiple sequence alignment of CsSHP and its homologs from other species indicated that these proteins contained the conserved MADS domain and K-box domain (Figure 1B). Phylogenetic analysis indicated that CsSHP is very close to ClSHP in watermelon, which was clustered with other known SHP proteins and located in the PLENA (PLE) lineages of AG subfamily, while CUM1 (the ortholog of AG in cucumber) was clustered with other AG proteins (Figure 2) (Kater et al., 1998).
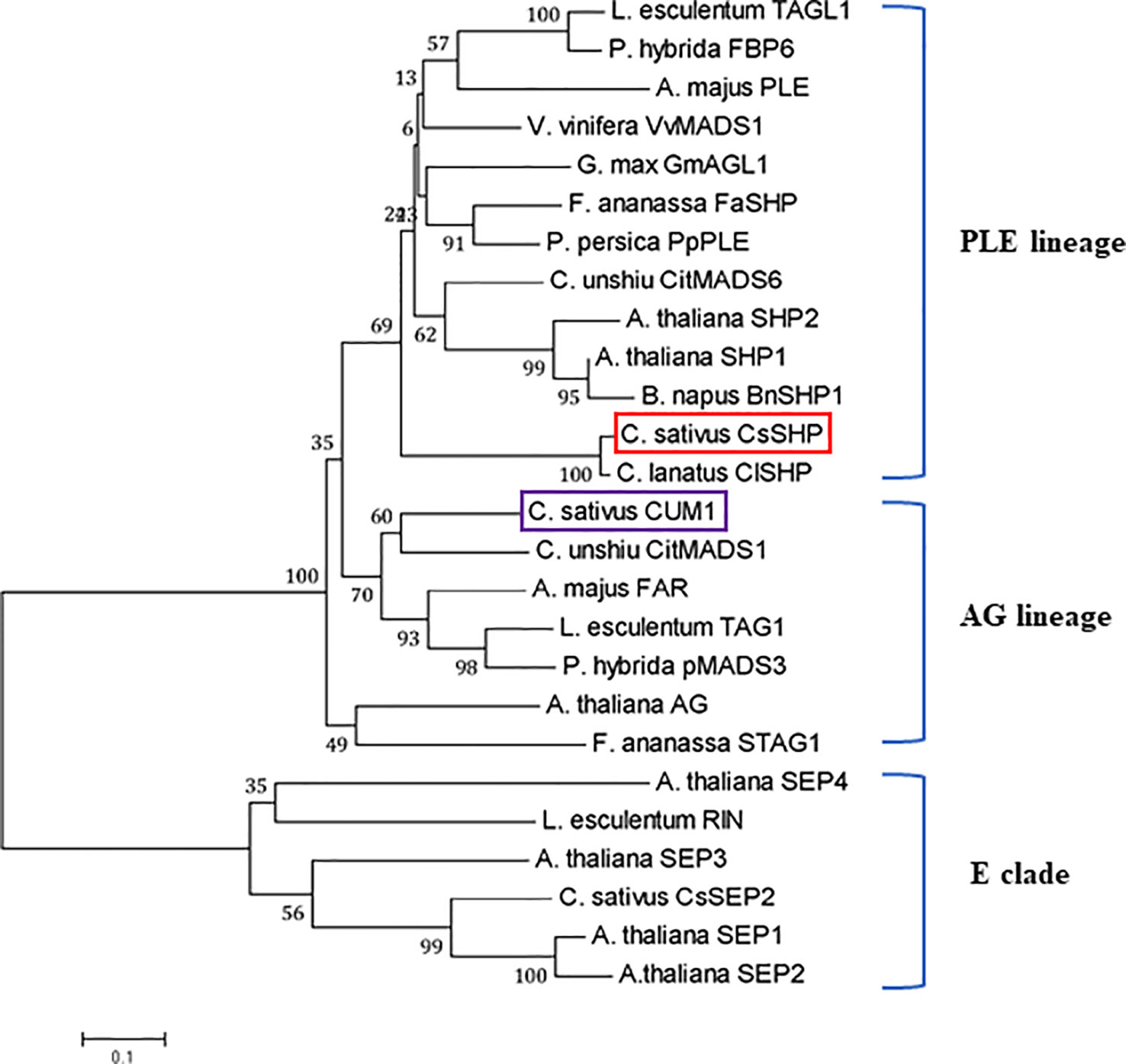
Figure 2 Phylogenetic analysis of CsSHP and its homologous proteins. The scale bar represents 0.1 substitutions per site. The red and purple squares indicate CsSHP and CUM1, respectively. E lineage proteins were used as outgroups.
To explore the expression pattern of CsSHP, qRT-PCR was performed in different cucumber organs including young leaves, stems, tendrils, male buds, female buds, male flowers at anthesis, female flowers, and ovaries at anthesis. Transcripts of CsSHP were highly accumulated in reproductive organs such as male flowers, female flowers, and ovaries, but with low levels in vegetative organs including leaves, stems, and tendrils (Figure 3A). In the four floral organs at anthesis, CsSHP was specifically expressed in stamens of male flowers and stigma of female flowers, while very low levels in sepals and petals (Figure 3B).
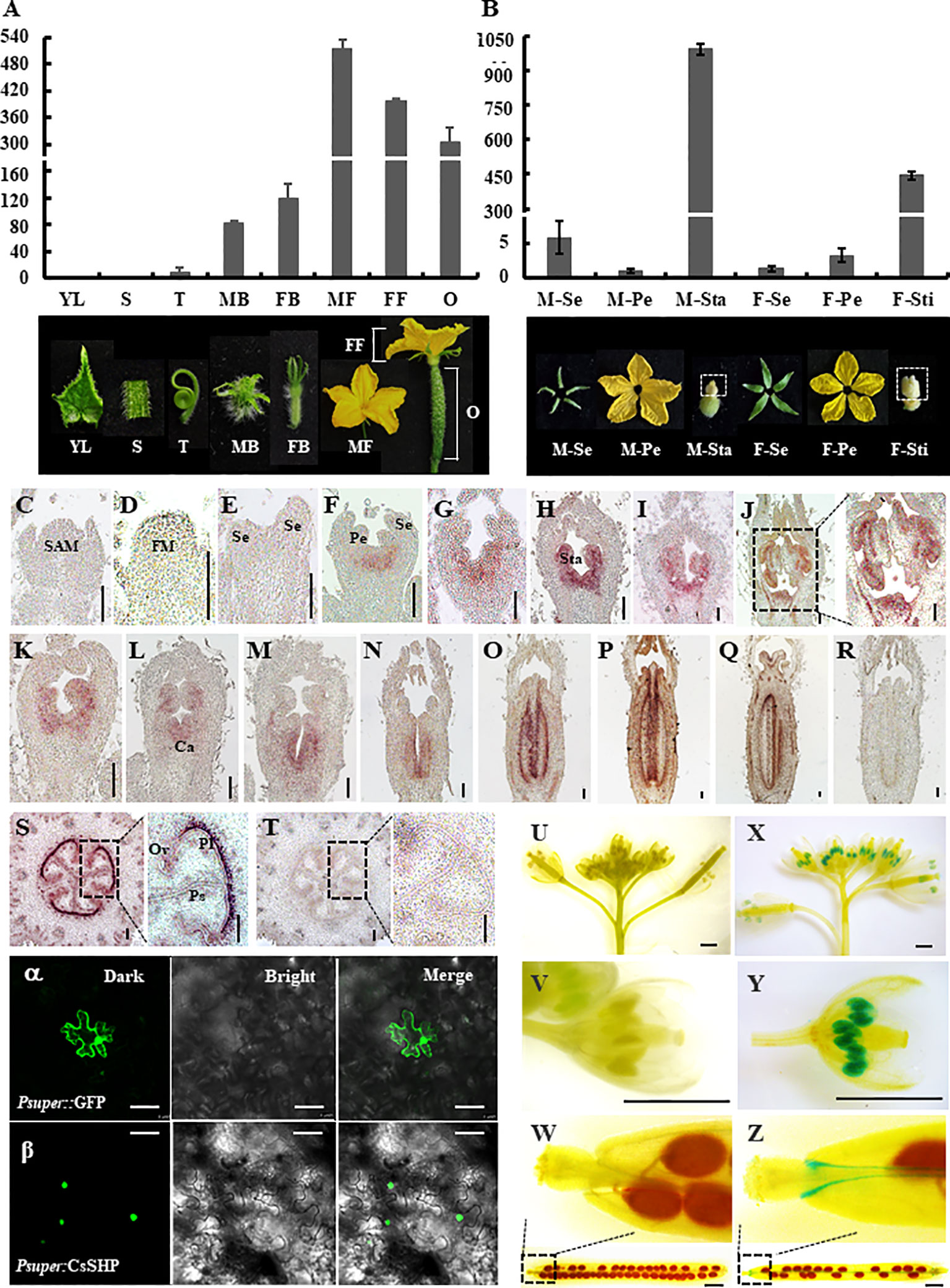
Figure 3 Expression analysis and subcellular localization of CsSHP. (A) quantitative real-time (qRT)-PCR analysis of CsSHP expression in different cucumber organs. YL, young leaves; S, stems; T, tendrils; MB, male buds; FB, female buds; MF, male flowers at anthesis; FF, female flowers at anthesis; O, ovaries at anthesis. Three biological replicates and three technical replicates were performed. (B) qRT-PCR analysis of CsSHP expression in different floral organs at anthesis. M-Se, sepals from male flowers; M-Pe, petals from male flowers; M-Sta, stamens from male flowers; F-Se, sepals from female flowers; F-Pe, petals from female flowers; F-Sti, stigmas from female flowers. Three biological replicates and three technical replicates were performed. Pictures of the corresponding organs are displayed at the bottom. (C–T) In situ hybridization of CsSHP in young organs of cucumber. Scale bar = 100 μm. (C) Shoot apical meristem (SAM). (D–H) Longitudinal sections of male flower buds at stages 1–5 in inbred line R1461. (I, J) Male flower buds at stages 8 and 9 in R1461 line. (K–Q) Longitudinal sections of female flower buds at different developmental stages in inbred line R1461. (S) Transection of ovary 10 days before anthesis. (R, T) The sense CsSHP probe was hybridized as a negative control. FM, floral meristem; Se, sepal; Pe, petal; Sta, stamen; Ca, carpel; Pl, placenta; Ps, pseudoseptum; Ov, ovule. (U–Z) β-Glucuronidase (GUS) signals of CsSHP in Arabidopsis. Scale bar = 1 mm. (U–W) Negative control plants showed no GUS signal. (X, Y) GUS signals were highly enriched in stamens. (Z) GUS signals were detected in valve margin of silique. The experiment was repeated three times. (α, β) Subcellular localization of CsSHP. GFP signals were localized in the nucleus. GFP driven by the pSUPER promoter was used as a positive control. GFP is shown in green. The left, middle, and right panels represent pictures taken under dark field, bright field, and merge views, respectively. Scale bar = 50 μm.
A previous study showed that SHP1/2 in Arabidopsis was specifically expressed in gynoecia, but not in stamens (Pinyopich et al., 2003; Colombo et al., 2010). To further detect the spatial and temporal expression pattern of CsSHP, in situ hybridization was applied to the shoot apex, ovaries, and flower buds at different developmental stages in cucumber. In the shoot apical meristem (SAM), floral meristem, and sepal primordia, CsSHP transcripts were undetectable (Figures 3C–E). CsSHP transcripts were first found in the initiating stamen primordia at stage 3 (Figure 3F), then CsSHP was expressed at stamen and carpel primordia from stage 4 to 6 (Figures 3G, H), and maintained its expression in developing stamens and degenerate carpels in male flowers (Figures 3I, J). In female flower buds, CsSHP was highly expressed in carpel primordia and decreasingly expressed in degenerate stamens (Figures 3K–N). Since stage 8, CsSHP transcripts were specifically detected at stigmas, placenta, and ovule primordia (Figures 3O–Q). No signal was detected upon hybridized with the sense probe control (Figure 3R). Transverse sections of ovaries showed significant enrichment of CsSHP signals at placenta, pseudoseptum, and ovules (Figures 3S, T).
To visualize the expression pattern at whole plant level, transgenic Arabidopsis lines expressing β-glucuronidase (GUS) driven by the 1,716 bp CsSHP promoter fragment were generated. Unlike the specific expression of SHP2 throughout the developing gynoecium in Arabidopsis (Colombo et al., 2010), GUS signal of CsSHP was found strongly in stamens as well as the valve margin of siliques, but not in developing gynoecium (Figures 3U–Z).
To explore the subcellular localization, CsSHP was fused with GFP under the control of a pSUPER promoter, and transiently expressed in tobacco leaves. Confocal green fluorescence imaging revealed that CsSHP-GFP fusion protein was located to the nucleus, whereas free GFP was distributed throughout the cell (Figures 3A, B).
In Arabidopsis, the mature siliques in the double mutant shp1 shp2 plant were unable to dehisce due to failure of dehiscence zone formation (Liljegren et al., 2000). To explore the function of CsSHP, we first transformed CsSHP driven by the pSUPER promoter into shp1 shp2 mutant plants. A total of 10 transgenic plants were obtained and the T2 plants were used for further characterization. Our data showed that ectopic expression of CsSHP was unable to rescue the indehiscence phenotype of shp1 shp2 mutant plant, but instead resulted in early flowering in Arabidopsis (Supplementary Figures S1A, B). Next, we transformed the CsSHP driven by the cauliflower mosaic virus 35S (CaMV 35S) into wild-type Arabidopsis (35S::CsSHP/Col). A total of 29 transgenic lines were generated. Based on CsSHP expression levels, three representative lines (#41, #45, #53) (T2) were chosen for further phenotypic analysis and data statistics. Compared to the wild-type control (Col), the expression of CsSHP was dramatically increased in the transgenic lines (Supplementary Figure S2A). Overexpression of CsSHP resulted in early flowering (Figure 4A). Quantification analysis indicated that the days to bolting was 27.2 ± 2.1 in Col, while that of overexpression lines varied from 21.5 ± 1.3 to 23.3 ± 1.0 (Figure 4B). Similarly, the number of rosette leaves upon bolting, as well as the days to the 1st flower opening were significantly reduced in the transgenic lines (Supplementary Figures S2B, C). Moreover, ectopic expression of CsSHP accelerated the progression of reproductive growth in Arabidopsis. Under the same conditions, wild-type siliques in the main inflorescence were still green, while many of those in the transgenic plants had turned yellow and or even cracked (Figure 4C). Data statistics showed that the days to 1st silique yellowing and the days from anthesis to silique cracking were significantly shorter in transgenic lines than Col plants (Figures 4D, E).
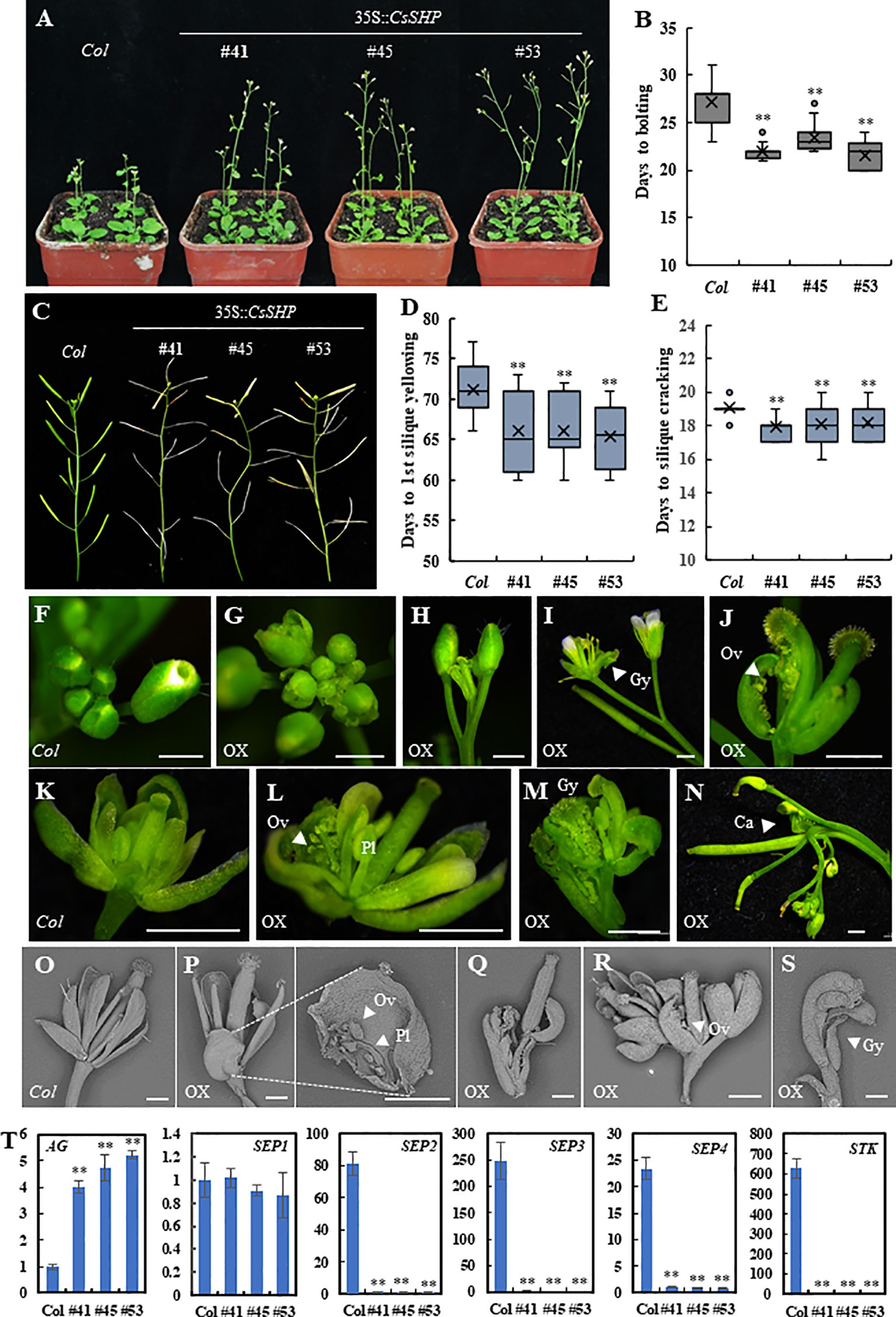
Figure 4 Phenotypic characterization of 35S::CsSHP transgenic plants in Arabidopsis. (A) Representative images of 35S::CsSHP transgenic plants indicated early flowering in Arabidopsis. (B) Box-plot of the days to bolting in Col and 35S::CsSHP transgenic lines. (C) Overexpression of CsSHP led to precocious phenotype in Arabidopsis. (D, E) Quantification of the days to yellowing of the 1st silique (D) and the days from anthesis to silique cracking (E). The data were the average of 20 plants for each line. (F–S) Overexpression of CsSHP caused defected floral organ development in Arabidopsis. (F–N) Anatomic micrographs of flowers and inflorescences. Scale bar = 1 mm. (O–S) Scanning electron micrographs of flowers and siliques. Scale bar = 500 μm. (T) The expression levels of AG, SEP1, SEP2, SEP3, SEP4, and STK in 35S::CsSHP Arabidopsis flowers. Gy, gynoecium; Ov, ovule; Pl, placenta; Ca, carpel **, t-test (p < 0.01).
Much more dramatic changes were found in flower organ development in the 35S::CsSHP transgenic lines. As compared to the wild-type control, some flower buds in transgenic plants were precociously opened (Figures 4F, G), flower patterning was disrupted, and the inflorescence meristem prematurely terminated (Figures 4H, I). For each individual flower, the flower patterning of sepal, petal, stamen, carpel was disturbed upon CsSHP overexpression. Some flowers in transgenic lines lack petals and stamens, consisting of only gynoecia and sepals, in which the gynoecia formed a longitudinal cleft to expose the placenta and ovules, and sepals were carpelloid (Figure 4J). Sometimes, one sepal of the flower was replaced by an ectopic gynoecium containing placenta tissue and ovules (Figures 4K, L). In more severe cases, a misshapen flower with carpelloid sepals grew from the base of another abnormal silique (Figure 4M), or an inflorescence grow out from the base of an ectopic carpel bearing ovules, and almost all flowers on the inflorescences were abnormal (Figure 4N). Scanning electron microscopy images showed that wild-type sepals were long and narrow, while that of the transgenic plants were short and round, with ectopic placenta and ovules in the inner side of the sepal (Figures 4O, P). Abnormal flowers with only gynoecium and carpelloid sepal grew from the inner base of another gynoecium or another flower (Figures 4Q–S). Statistic data showed that average of 51.6% plants displayed abnormal flowers in the transgenic lines, in which 8% of flowers were defected, as compared to none in the wild-type control (Supplementary Figure S2D).
Based on the carpelloid organ phenotype, a set of carpel related genes including AG, SEP1/2/3/4, and STK were chosen for expression analysis (Pelaz et al., 2000; Favaro et al., 2003). As compared to the wild-type control, the expression of AG was significantly elevated, whereas that of SEP2, SEP3, SEP4, and STK was dramatically decreased in the CsSHP transgenic plants (Figure 4T).
In addition, unlike the stretching and flat wild-type leaves, there was a considerable proportion of crooked rosette and cauline leaves curling upwards and inward in transgenic lines (Supplementary Figure S3).
SEP1/2/3/4 are a class of organ-identity genes that are required for development of sepals, petals, stamens, and carpels in Arabidopsis (Pelaz et al., 2000), and SEP proteins are thought to act as a “bridge” allowing the formation of higher order complex with the floral organ identity MADS-box factors (Favaro et al., 2003). In vitro and in vivo evidence was provided for the existence of SEP, STK, and/or AG and SHP protein complexes in promoting the Arabidopsis ovule identity (Mendes et al., 2013). In cucumber, constitutive expression of CUM1 (AG ortholog) resulted in sepals transformed into carpelloid structures, and petals reduced significantly in size or completely absent (Kater et al., 2001). CUM10 (STK ortholog) mediates floral organ identity in cucumber, and ectopic expression of CUM10 resulted in partial transformation of petals into antheroid structures in petunia (Kater et al., 1998). CsSEP2 was shown to participate in floral organ development in cucumber, since abolishment of the transcriptional activity of CsSEP2 led to increased floral organ size with disturbed floral patterning (Wang et al., 2016). To explore the existence of possible interactions in cucumber, a yeast two-hybrid assay was performed between CsSHP and cucumber homologs of AG, SEPs, and STK. Our data showed that CsSHP displayed strong interactions with CsSEP2, CsSEP3, CsSEP4, and a weak interaction with CUM10, while CsSHP could not interact with CUM1, neither with itself to form homodimer (Figure 5A). To verify the protein interactions in vivo, a LUC complementation imaging assay was performed in the abaxial leaf epidermis of tobacco (Figure 5B). The reconstituted LUC enzyme can be detected by luminometer from combinations of CsSHP with CsSEP2, CsSEP3, or CsSEP4, but not from combination of CsSHP with CUM1 or CUM10, indicating that CsSHP interacts with CsSEPs to form multimeric protein complex in cucumber.
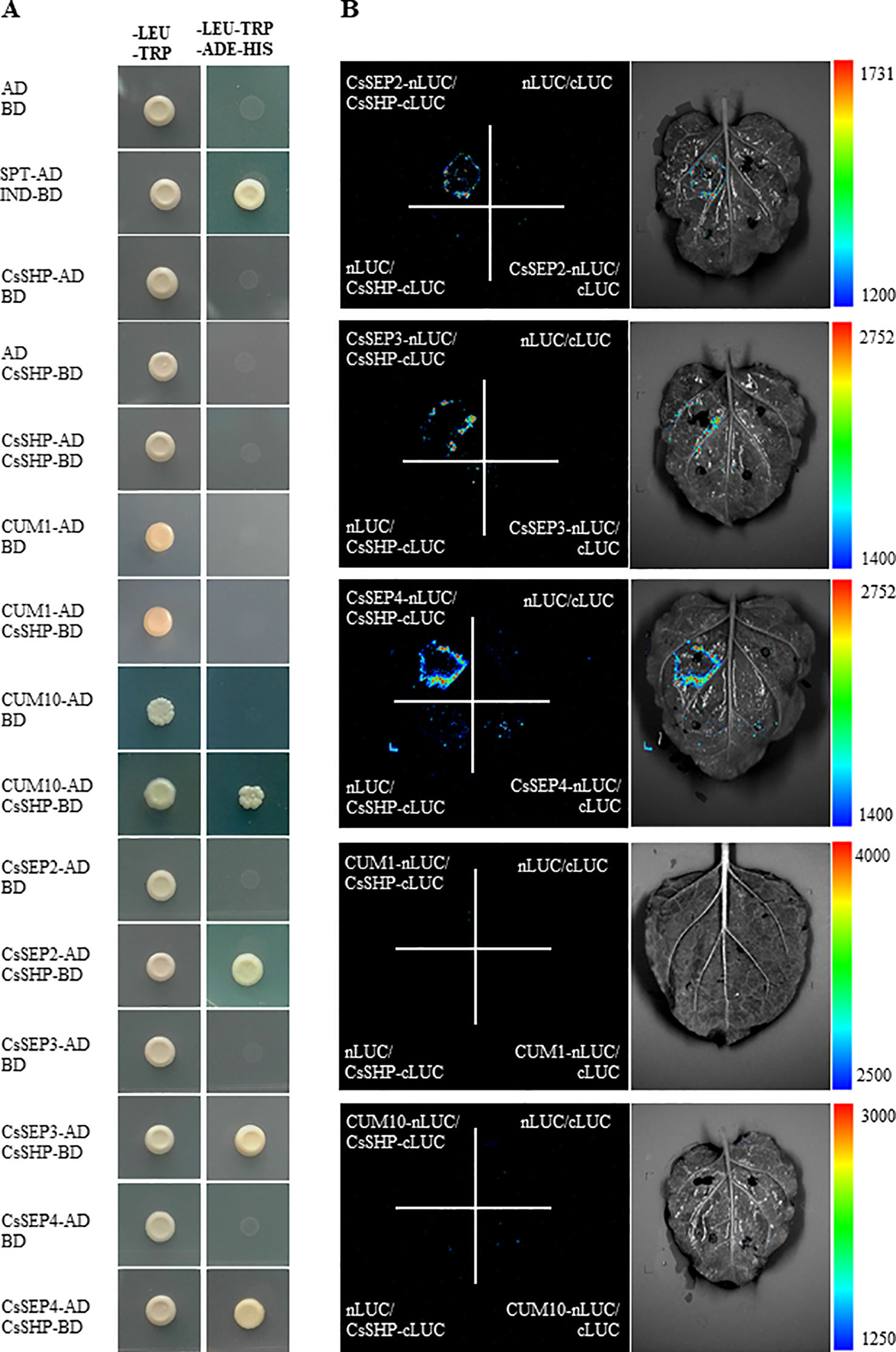
Figure 5 Physical interactions of CsSHP with CsSEP2, CsSEP3, and CsSEP4. (A) Yeast two-hybrid assay. The combination of SPT-AD and IND-BD was used as a positive control. (B) Luciferase (LUC) complementation imaging analysis.
In climacteric tomato, TAGL1 was highly expressed in flowers at anthesis and in fruits at red ripe stage (Gimenez et al., 2010). Similarly, during the development of non-climacteric citrus fruits, the transcript levels of CsMADS6 increased gradually, peaking at 180 DAA (breaker stage) and declining at 220 DAA (Lu et al., 2018). To understand whether CsSHP expression is correlated with fruit maturation in cucumber, qRT-PCR analysis was performed in cucumber fruit at fourteen developmental points. As shown in Figure 6A, the CsSHP expression showed a slight downward trend from 0.3 cm flower buds (14 days before anthesis) to commercially mature fruits (nine DAA). From 9 to 15 DAA, CsSHP transcripts increased threefold during the 6 days, and the highest accumulation of transcripts was detected at 21 DAA when the pericarp began to yellow. After 21 DAA, CsSHP expression gradually declined as fruit maturation progression and the pericarp became more and more yellow.
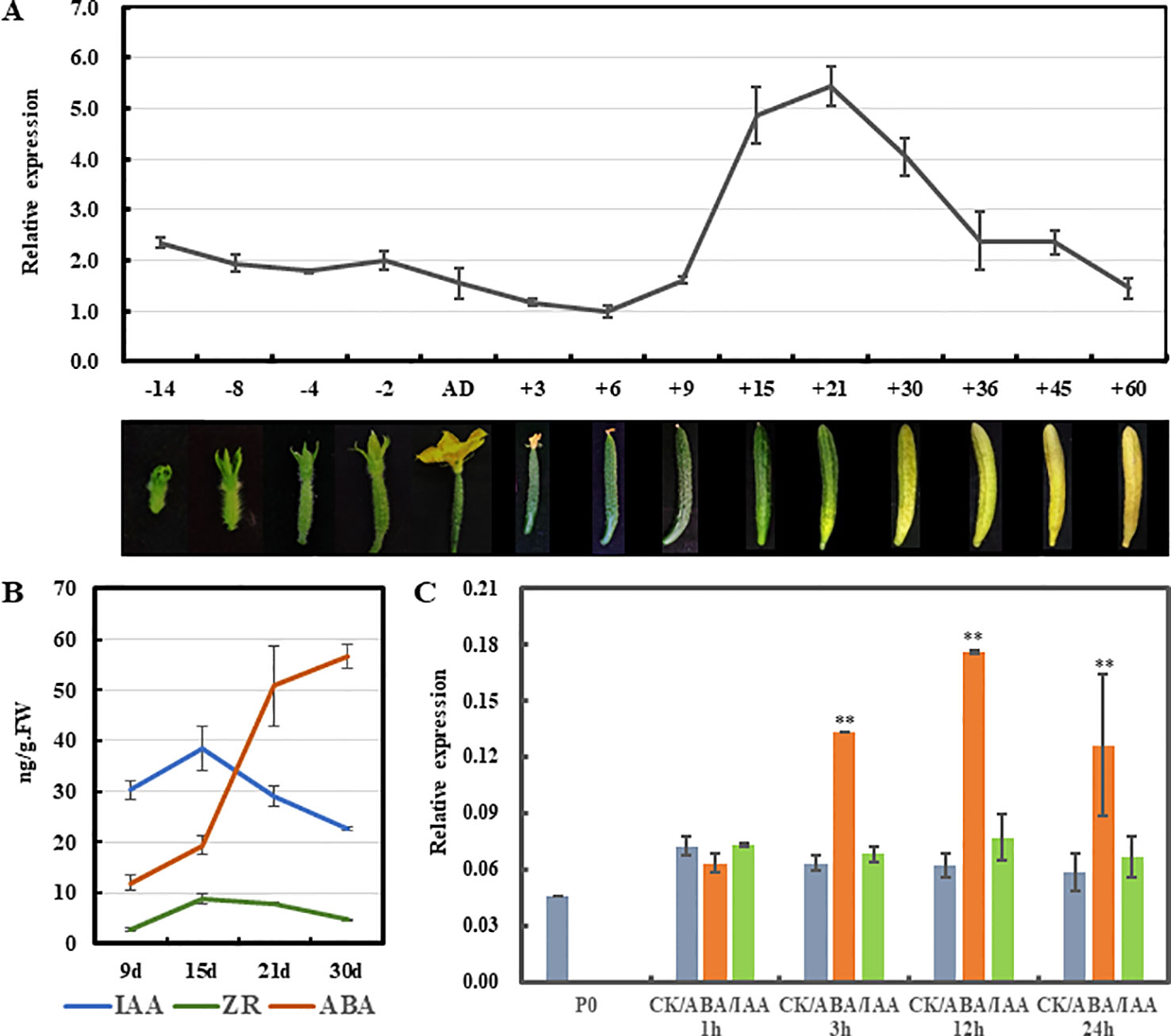
Figure 6 Expression analysis of CsSHP during fruit maturation in cucumber. (A) CsSHP expression in fruits at different developmental stages in inbred line R1461. Fruit pictures of the corresponding developmental stages are displayed at the bottom in different proportions. (B) Measurements of three endogenous hormones in fruit from 9 to 30 days after anthesis (DAA). Three biological replicates were performed. (C) Expression response of CsSHP to abscisic acid (ABA) or indole-3-acetic acid (IAA) treatment in cucumber ovaries of 4 days before anthesis. Three biological replicates and three technical replicates were performed **, t-test (p < 0.01).
Cucumber is classified as a non-climacteric fruit, and endogenous ethylene displayed no influence on the postharvest yellowing of cucumber (Nilsson, 2005). However, abscisic acid (ABA) was shown to play important and direct roles in regulating cucumber fruit development and ripening. Exogenous ABA application at the turning stage promoted fruit ripening in cucumber (Wang et al., 2013). Therefore, the content of ABA, as well as the indole-3-acetic acid (IAA) and zeatin riboside (ZR), was measured in fruits of the variety R1461 from 9 to 30 DAA, when CsSHP expression changed drastically. Our data showed that the content of ABA increased continuously from 9 to 30 DAA as fruit ripening progression, with a dramatic upregulation from 15 to 30 DAA, while that of IAA and ZR displayed a mild increase from 9 to 15 DAA, and then decreased (Figure 6B), confirming that ABA may play key roles in mediating fruit ripening in cucumber. Given that the CsSHP expression and ABA level showed similar trend as fruit maturation, we next examined CsSHP response to ABA treatment in ovaries of 4 days before anthesis. Our data indicated that CsSHP transcription was significantly induced after ABA treatment for 3 hours, and remained at 2–3 times upregulation to 24 h (Figure 6C), suggesting that CsSHP may be involved in fruit maturation in cucumber through the ABA pathway.
MADS-box genes are critical transcription factors, participating in virtually all aspects of plant development, especially during flower and fruit development (Smaczniak et al., 2012; Wang et al., 2015). Previous study revealed the expression pattern of 43 MADS-box genes in cucumber (Hu and Liu, 2012). CUM1, CUM10, CUM26, CsAP3, and CsSEP2 are currently known cucumber MADS-box genes that play essential roles in flower and fruit development (Kater et al., 1998; Kater et al., 2001; Sun et al., 2016; Wang et al., 2016), whereas there are still a considerable portion of MADS-box genes need to be functionally characterized. In this study, we identified the CsSHP gene in cucumber and revealed the roles in fruit maturation and floral organ determination.
CsSHP is a member of MADS-box family and has the conserved MADS domain and K-box domain. The MADS domain is responsible for binding downstream target DNA, and the K-box domain acts on protein-protein interactions (Shore and Sharrocks, 1995; Yang et al., 2003). The CDS length of CsSHP and SHP1/2 is almost equivalent, while the genomic length of CsSHP gene is about twice that of SHP1/2, due to extremely long first and second introns (Figure 1A). Expression analyses by RT-PCR and in situ hybridization showed that CsSHP was specifically expressed in reproductive tissues including stamens of male flowers, stigma of female flowers, and ovaries (Figure 3). Such expression is similar to that of TAGL1 in tomato and NbSHP in tobacco, but different from SHP2 in Arabidopsis, which is specifically expressed in gynoecium and absent in stamen (Vrebalov et al., 2009; Gimenez et al., 2010; Fourquin and Ferrandiz, 2012). Interestingly, GUS signals driven by CsSHP promoter were detected only in stamens and valve margin in Arabidopsis, but not in developing gynoecium (Figure 3). There are two possible reasons for such discrepancy. One is the different flower structure in Arabidopsis (complete flower) and cucumber (unisexual flower), and the other is that the promoter sequence used for driving the GUS signal was unable to confer the correct expression pattern of CsSHP. Considering that the second intron of AG/PLE genes contain multiple regulatory elements that determine the proper spatial and temporal expression (Liu and Liu, 2008; Gu et al., 2018), it is more plausible that the expression pattern of CsSHP is coordinately controlled by its promoter and introns.
Overexpression of CsSHP in wild-type Arabidopsis resulted on ectopic carpelloid organs with the characteristics of gynoecium: stigma-like tissue at the top, placenta tissue in the middle, and infertile ovules along the valve margin (Figure 4). The inflorescence or flower patterning were often terminated in an ectopic carpel, showing a phenotype of “flower in carpel” or “inflorescence in carpel” (Figure 4). Although some phenotypes like early flowering, curly leaves and prematurely open flower buds, are similar to those previously described upon overexpression of SHP orthologs, such as Arabidopsis SHP1/2 and tomato TAGL1, in Arabidopsis (Favaro et al., 2003; Vrebalov et al., 2009), the carpelloid phenotype caused by CsSHP overexpression is more severe. For example, 35S::SHP1/2 in Arabidopsis showed a transformation of petals toward stamens and a partial conversion of sepals toward carpels, with stigmatic papillae on it (Favaro et al., 2003). However, in the Arabidopsis inflorescences of 35S::CsSHP plants, ectopic carpels may grow anywhere of the flower (Figure 4). The carpelloid phenotype in 35S::CsSHP transgenic plants suggested the function of CsSHP in specifying carpel identity in cucumber. Consistently, the expression of AG was significantly elevated, while that of STK was almost abolished in the transgenic flowers (Figure 4), which may due to the ectopic carpelloid organs with infertile ovules. Interestingly, the expression of SEP2, SEP3 and SEP4 was dramatically decreased in the 35S::CsSHP transgenic flowers (Figure 4). However, without a time course analysis in CsSHP inducible lines, we were unable to differentiate the expression change of CsSEP2/3/4 is a cause or a result of floral patterning defect. Biochemical data showed that CsSHP interacts with CsSEP2/3/4 at protein level (Figure 5). Loss of CsSEP2 function led to disturbed flower patterning with enlarged floral organs in cucumber (Wang et al., 2016). Therefore, we hypothesized that CsSHP may specify carpel identity in cucumber through interacting with CsSEP2 to form protein complex. In fact, SHP homologs were shown to interact with SEP-like proteins in Arabidopsis, tomato, and soybean (Favaro et al., 2003; Leseberg et al., 2008; Mendes et al., 2013; Chi et al., 2017), indicating the interactions between SHP and SEPs are relatively conserved among species.
Cucumber fruit is harvested immature and consumed freshly or as processed pickles. Fruit ripening is important for seed maturation, but has adverse effect on cucumber production and post-harvest shelf life. Unlike the dry pod in Arabidopsis, cucumber fruit has no cracking characteristics, and fruit ripening involved in changes of color, texture, and aroma (Seymour et al., 2008). As a non-climacteric fruit, ethylene has no influence on the postharvest yellowing of cucumber (Nilsson, 2005), but instead, ABA application at the turning stage promoted fruit ripening (Wang et al., 2013). Here, we found that ectopic expression of CsSHP resulted in early flowering and accelerated ripening in Arabidopsis (Figure 4). Moreover, CsSHP expression is significantly increased during the fruit yellowing from 9 to 21 DAA, concomitant with the dramatic increase of ABA level (Figure 6), implying the positive roles of CsSHP in cucumber ripening. Further, CsSHP expression was found to be induced upon exogenous ABA application (Figure 6D). Hence, it is plausible to speculate that high level of ABA from commodity fruit stage (nine DAA) induces CsSHP expression, which positively modulates carotenoid metabolism and ripening-related genes, just like AGL1 in tomato and CsMADS6 in citrus (Vrebalov et al., 2009; Lu et al., 2018), to promote fruit yellowing and fruit ripening in cucumber. Further studies are needed to test above hypothesis using transgenic cucumbers through CRISPR/Cas9 techniques (Hu et al., 2017).
All datasets generated for this study are included in the article/Supplementary Material.
ZC and XZ conceived this project. ZC, SZ, XL, GC, ZW, RG, JS, WS performed the experiments, ZC and XZ wrote the manuscript, XL, ZZ and DH contributed to critical discussions. All authors read and approved the final version.
This study was supported by the National Key Research and Development Program [2018YFD1000800], National Natural Science Foundation of China [31572132] and [31772315], and the Construction of Beijing Science and Technology Innovation and Service Capacity in Top Subjects [CEFFPXM2019_014207_000032].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to Dr Wei Shi for technical assistance with the LUC activity assay.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01781/full#supplementary-material
Bai, S. L., Peng, Y. B., Cui, J. X., Gu, H. T., Xu, L. Y., Li, Y. Q., et al. (2004). Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. doi: 10.1007/s00425-004-1342-2
Boss, P. K., Vivier, M., Matsumoto, S., Dry, I. B., Thomas, M. R. (2001). A cDNA from grapevine (Vitis vinifera L.), which shows homology to AGAMOUS and SHATTERPROOF, is not only expressed in flowers but also throughout berry development. Plant Mol. Biol. 45, 541–553. doi: 10.1023/A:1010634132156
Bowman, J. L., Smyth, D. R., Meyerowitz, E. M. (1989). Genes directing flower development in Arabidopsis. Plant Cell. 1, 37–52. doi: 10.1105/tpc.19.00276
Bowman, J. L., Drews, G. N., Meyerowitz, E. M. (1991). Expression of the Arabidopsis floral homeotic gene agamous is restricted to specific cell-types late in flower development. Plant Cell. 3, 749–758. doi: 10.1105/tpc.3.8.749
Causier, B., Bradley, D., Cook, H., Davies, B. (2009). Conserved intragenic elements were critical for the evolution of the floral C-function. Plant J. 58, 41–52. doi: 10.1111/j.1365-313X.2008.03759.x
Che, G., Zhang, X. (2019). Molecular basis of cucumber fruit domestication. Curr. Opin. Plant Biol. 47, 38–46. doi: 10.1016/j.pbi.2018.08.006
Chen, H. M., Zou, Y., Shang, Y. L., Lin, H. Q., Wang, Y. J., Cai, R., et al. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146, 368–376. doi: 10.1104/pp.107.111740
Chi, Y. J., Wang, T. T., Xu, G. L., Yang, H., Zeng, X. R., Shen, Y. X., et al. (2017). GmAGL1, a MADS-Box gene from soybean, is involved in floral organ identity and fruit dehiscence. Front. Plant Sci. 8, 175. doi: 10.3389/fpls.2017.00175
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls -genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Colombo, M., Brambilla, V., Marcheselli, R., Caporali, E., Kater, M. M., Colombo, L. (2010). A new role for the SHATTERPROOF genes during Arabidopsis gynoecium development. Dev. Biol. 337, 294–302. doi: 10.1016/j.ydbio.2009.10.043
Daminato, M., Guzzo, F., Casadoro, G. (2013). A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 64, 3775–3786. doi: 10.1093/jxb/ert214
Ding, L., Yan, S. S., Jiang, L., Zhao, W. S., Ning, K., Zhao, J. Y., et al. (2015). HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. Plos Genet. 11, e1005479. doi: 10.1371/journal.pgen.1005479
Dreni, L., Kater, M. M. (2014). MADS reloaded: evolution of the AGAMOUS subfamily genes. New Phytol. 201, 717–732. doi: 10.1111/nph.12555
Favaro, R., Pinyopich, A., Battaglia, R., Kooiker, M., Borghi, L., Ditta, G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell. 15, 2603–2611. doi: 10.1105/tpc.015123
Ferrandiz, C., Fourquin, C. (2014). Role of the FUL-SHP network in the evolution of fruit morphology and function. J. Exp. Bot. 65, 4505–4513. doi: 10.1093/jxb/ert479
Fourquin, C., Ferrandiz, C. (2012). Functional analyses of AGAMOUS family members in Nicotiana benthamiana clarify the evolution of early and late roles of C-function genes in eudicots. Plant J. 71, 990–1001. doi: 10.1111/j.1365-313X.2012.05046.x
Fourquin, C., del Cerro, C., Victoria, F. C., Vialette-Guiraud, A., de Oliveira, A. C., Ferrandiz, C. (2013). A change in SHATTERPROOF protein lies at the origin of a fruit morphological novelty and a new strategy for seed dispersal in Medicago Genus. Plant Physiol. 162, 907–917. doi: 10.1104/pp.113.217570
Gimenez, E., Pineda, B., Capel, J., Anton, M. T., Atares, A., Perez-Martin, F., et al. (2010). Functional Analysis of the arlequin mutant corroborates the essential role of the ARLEQUIN/TAGL1 gene during reproductive development of tomato. Plos One 5, e14427. doi: 10.1371/journal.pone.0014427
Giovannoni, J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10, 283–289. doi: 10.1016/j.pbi.2007.04.008
Gu, R., Liu, X. F., Zhao, W. S., Yan, S. S., Sun, L. H., Wu, B. N., et al. (2018). Functional characterization of the promoter and second intron of CUM1 during flower development in Cucumber (Cucumis sativus L.). Hortic. Plant J. 4, 103–110. doi: 10.1016/j.hpj.2018.03.004
Hu, L. F., Liu, S. Q. (2012). Genome-wide analysis of the MADS-box gene family in cucumber. Genome 55, 245–256. doi: 10.1139/g2012-009
Hu, B. W., Li, D. W., Liu, X., Qi, J. J., Gao, D. L., Zhao, S. Q., et al. (2017). Engineering non-transgenic gynoecious cucumber using an improved transformation protocol and optimized CRISPR/Cas9 System. Mol. Plant. 10, 1575–1578. doi: 10.1016/j.molp.2017.09.005
Hurr, B. M., Huber, D. J., Vallejos, C. E., Talcott, S. T. (2009). Developmentally dependent responses of detached cucumber (Cucumis sativus L.) fruit to exogenous ethylene. Postharv. Biol. Tec. 52, 207–215. doi: 10.1016/j.postharvbio.2008.12.006
Itkin, M., Seybold, H., Breitel, D., Rogachev, I., Meir, S., Aharoni, A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60, 1081–1095. doi: 10.1111/j.1365-313X.2009.04064.x
Jofuku, K. D., Denboer, B. G. W., Vanmontagu, M., Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene Apetala2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Kater, M. M., Colombo, L., Franken, J., Busscher, M., Masiero, S., Campagne, M. M. V., et al. (1998). Multiple AGAMOUS homologs from cucumber and petunia differ in their ability to induce reproductive organ fate. Plant Cell 10, 171–182. doi: 10.1105/tpc.10.2.171
Kater, M. M., Franken, J., Carney, K. J., Colombo, L., Angenent, G. C. (2001). Sex determination in the monoecious species cucumber is confined to specific floral whorls. Plant Cell 13, 481–493. doi: 10.1105/tpc.13.3.481
Leng, P., Yuan, B., Guo, Y. D. (2014). The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 65, 4577–4588. doi: 10.1093/jxb/eru204
Leseberg, C. H., Eissler, C. L., Wang, X., Johns, M. A., Duvall, M. R., Mao, L. (2008). Interaction study of MADS-domain proteins in tomato. J. Exp. Bot. 59, 2253–2265. doi: 10.1093/jxb/ern094
Liljegren, S. J., Ditta, G. S., Eshed, H. Y., Savidge, B., Bowman, J. L., Yanofsky, M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404, 766–770. doi: 10.1038/35008089
Liu, Z. R., Liu, Z. C. (2008). The second intron of AGAMOUS drives carpel- and stamen-specific expression sufficient to induce complete sterility in Arabidopsis. Plant Cell Rep. 27, 855–863. doi: 10.1007/s00299-008-0511-3
Liu, B., Liu, X. W., Yang, S., Chen, C. H., Xue, S. D., Cai, Y. L., et al. (2016). Silencing of the gibberellin receptor homolog, CsGID1a, affects locule formation in cucumber (Cucumis sativus) fruit. New Phytol. 210, 551–563. doi: 10.1111/nph.13801
Lu, S. W., Zhang, Y., Zhu, K. J., Yang, W., Ye, J. L., Chai, L. J., et al. (2018). The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 176, 2657–2676. doi: 10.1104/pp.17.01830
Mainardi, J. A., Purgatto, E., Vieira, A., Jr., Bastos, W. A., Cordenunsi, B. R., Oliveira do Nascimento, J. R., et al. (2006). Effects of ethylene and 1-methylcyclopropene (1-MCP) on gene expression and activity profile of alpha-1,4-glucan-phosphorylase during banana ripening. J. Agric. Food Chem. 54, 7294–7299. doi: 10.1021/jf061180k
Mendes, M. A., Guerra, R. F., Berns, M. C., Manzo, C., Masiero, S., Finzi, L., et al. (2013). MADS domain transcription factors mediate short-range DNA looping that is essential for target gene expression in Arabidopsis. Plant Cell 25, 2560–2572. doi: 10.1105/tpc.112.108688
Mizukami, Y., Ma, H. (1992). Ectopic expression of the floral homeotic gene agamous in transgenic Arabidopsis plants alters floral organ identity. Cell 71, 119–131. doi: 10.1016/0092-8674(92)90271-d
Mizukami, Y., Ma, H. (1995). Separation of ag function in floral meristem determinacy from that in reproductive organ identity by expressing antisense ag RNA. Plant Mol. Biol. 28, 767–784. doi: 10.1007/BF00042064
Nilsson, T. (2005). Effects of ethylene and 1-MCP on ripening and senescence of European seedless cucumbers. Postharv. Biol. Tec. 36, 113–125. doi: 10.1016/j.postharvbio.2004.11.008
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Pinyopich, A., Ditta, G. S., Savidge, B., Liljegren, S. J., Baumann, E., Wisman, E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424, 85–88. doi: 10.1038/nature01741
Robles, P., Pelaz, S. (2005). Flower and fruit development in Arabidopsis thaliana. Int. J. Dev. Biol. 49, 633–643. doi: 10.1387/ijdb.052020pr
Roeder, A. H., Yanofsky, M. F. (2006). Fruit development in Arabidopsis, Arabidopsis Book. vol. 4, e0075. doi: 10.1199/tab.0075
Ruelens, P., Zhang, Z. C., van Mourik, H., Maere, S., Kaufmann, K., Geuten, K. (2017). The origin of floral organ identity quartets. Plant Cell 29, 229–242. doi: 10.1105/tpc.16.00366
Schmittgen, T. D., Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108. doi: 10.1038/nprot.2008.73
Schutze, K., Harter, K., Chaban, C. (2009). Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol. 479, 189–202. doi: 10.1007/978-1-59745-289-2_12
Seymour, G., Poole, M., Manning, K., King, G.J. (2008). Genetics and epigenetics of fruit development and ripening. Curr. Opin. Plant Biol. 11, 58–63. doi: 10.1016/j.pbi.2007.09.003
Shore, P., Sharrocks, A. D. (1995). The MADs-Box family of transcription factors. Eur. J. Biochem. 229, 1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x
Smaczniak, C., Immink, R. G. H., Angenent, G. C., Kaufmann, K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098. doi: 10.1242/dev.074674
Sun, J. J., Li, F., Wang, D. H., Liu, X. F., Li, X., Liu, N., et al. (2016). CsAP3: a cucumber homolog to Arabidopsis APETALA3 with novel characteristics. Front. Plant Sci. 7, 1181. doi: 10.3389/fpls.2016.01181
Theissen, G., Saedler, H. (2001). Plant biology - floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
van der Knaap, E., Chakrabarti, M., Chu, Y. H., Clevenger, J. P., Illa-Berenguer, E., Huang, Z. J., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5, 227. doi: 10.3389/fpls.2014.00227
Vrebalov, J., Pan, I. L., Arroyo, A. J. M., McQuinn, R., Chung, M., Poole, M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 21, 3041–3062. doi: 10.1105/tpc.109.066936
Wan, H. J., Zhao, Z. G., Qian, C. T., Sui, Y. H., Malik, A. A., Chen, J. F. (2010). Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399, 257–261. doi: 10.1016/j.ab.2009.12.008
Wang, Y. P., Wang, Y., Ji, K., Dai, S. J., Hu, Y., Sun, L., et al. (2013). The role of abscisic acid in regulating cucumber fruit development and ripening and its transcriptional regulation. Plant Physiol. Bioch. 64, 70–79. doi: 10.1016/j.plaphy.2012.12.015
Wang, H. S., Yu, C., Fan, P. P., Bao, B. F., Li, T., Zhu, Z. J. (2015). Identification of two cucumber putative silicon transporter genes in Cucumis sativus. J. Plant Growth Regul. 34, 332–338. doi: 10.1007/s00344-014-9466-5
Wang, X., Gao, D. L., Sun, J. J., Liu, M., Lun, Y. Y., Zheng, J. S., et al. (2016). An exon skipping in a SEPALLATA-Like gene is associated with perturbed floral and fruits development in cucumber. J. Integr. Plant Biol. 58, 766–771. doi: 10.1111/jipb.12472
Weigel, Meyerowitz (1994). The ABCs of floral homeotic genes. Cell 79, 180–180. doi: 10.1016/0092-8674(94)90291-7
Weng, Y., Colle, M., Wang, Y., Yang, L., Rubinstein, M., Sherman, A., et al. (2015). QTL mapping in multiple populations and development stages reveals dynamic quantitative trait loci for fruit size in cucumbers of different market classes. Theor. Appl. Genet. 128, 1747–1763. doi: 10.1007/s00122-015-2544-7
Yang, Y. Z., Fanning, L., Jack, T. (2003). The K domain mediates heterodimerization of the Arabidopsis floral organ identity proteins, APETALA3 and PISTILLATA. Plant J. 33, 47–59. doi: 10.1046/j.0960-7412.2003.01473.x
Zhang, X. L., Zhou, Y., Ding, L., Wu, Z. G., Liu, R. Y., Meyerowitz, E. M. (2013). Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101. doi: 10.1105/tpc.112.107854
Keywords: cucumber, Cucumis sativus SHATTERPROOF, floral organ identity, fruit maturation, abscisic acid
Citation: Cheng Z, Zhuo S, Liu X, Che G, Wang Z, Gu R, Shen J, Song W, Zhou Z, Han D and Zhang X (2020) The MADS-Box Gene CsSHP Participates in Fruit Maturation and Floral Organ Development in Cucumber. Front. Plant Sci. 10:1781. doi: 10.3389/fpls.2019.01781
Received: 03 October 2019; Accepted: 20 December 2019;
Published: 10 February 2020.
Edited by:
Rafael Lozano, University of Almeria, SpainReviewed by:
Muriel Quinet, Catholic University of Louvain, BelgiumCopyright © 2020 Cheng, Zhuo, Liu, Che, Wang, Gu, Shen, Song, Zhou, Han and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deguo Han, ZGVndW9oYW5AbmVhdS5lZHUuY24=; Xiaolan Zhang, emh4aWFvbGFuQGNhdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.