
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 07 February 2020
Sec. Plant Systematics and Evolution
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01731
The Eriobotrya-Rhaphiolepis (ER) clade consists of about 46 species distributed in East and Southeast Asia. Although Eriobotrya and Rhaphiolepis have been supported to form a clade, the monophyly of Eriobotrya and Rhaphiolepis at the genus level has never been well tested and their phylogenetic positions in Maleae still remain uncertain. This study aims to reconstruct a robust phylogeny of the ER clade in the framework of Maleae with a broad taxon sampling and clarify the phylogenetic relationship between Eriobotrya and Rhaphiolepis. This study employed sequences of the whole plastome (WP) and entire nuclear ribosomal DNA (nrDNA) repeats assembled from the genome skimming approach and included 83 samples representing 76 species in 32 genera of Rosaceae, especially Maleae. The Maximum Likelihood (ML) and Bayesian Analysis (BI) based on three datasets, i.e., WP, coding sequences of plastome (CDS), and nrDNA, strongly supported the paraphyly of Eriobotrya, within which Rhaphiolepis was nested. Our plastid tree supported the sister relationship between the ER clade and Heteromeles, and the nrDNA tree, however, did not resolve the phylogenetic placement of the ER clade in Maleae. Strong incongruence between the plastid and the nuclear trees is most likely explained by hybridization events, which may have played an important role in the evolutionary history of the ER clade. Molecular, morphological, and geographic evidence all supports the merge of Eriobotrya with Rhaphiolepis, which has the nomenclatural priority. We herein transferred 36 taxa of Eriobotrya to Rhaphiolepis. We also proposed a new name, Rhaphiolepis loquata B.B.Liu & J.Wen, for the economically important loquat, as the specific epithet “japonica” was pre-occupied in Rhaphiolepis.
Rosaceae is a widely distributed and economically important plant family currently classified into three subfamilies: Rosoideae, Dryadoideae, and Amygdaloideae (Morgan et al., 1994; Potter et al., 2007). The apple tribe Maleae consists of more than 1,000 species, mainly distributed in the temperate regions of the Northern Hemisphere with several genera extending to subtropical and tropical Southeast Asia and South America (Phipps et al., 1990; Lo and Donoghue, 2012). The apple tribe includes a number of commercially important fruits, such as apples (Malus domestica (Suckow) Borkh.), pears (Pyrus L. spp.), loquats (Eriobotrya japonica (Thunb.) Lindl.), as well as some ornamentals, e.g. serviceberries (Amelanchier Medik. spp.), chokeberries (Aronia Medik. spp.), and photinias (Photinia Lindl. spp.). Due to the economic significance, Maleae has intrigued taxonomists, botanists, horticulturists, and agriculturists for several hundred years. Furthermore, the phylogenetic position of the Eriobotrya Lindl.-Rhaphiolepis Lindl. complex within Maleae has been uncertain, perhaps due to the low sequence divergence, hybridizations, and the extensive extinctions of its close relatives (Campbell et al., 2007; Liu et al., 2019). The phylogenetic relationships among the members of Maleae have never been resolved confidently using either the morphological characteristics (Phipps et al., 1991; Aldasoro et al., 2005) or the limited plastid and/or nuclear sequences (Campbell et al., 1995; Verbylaitė et al., 2006; Campbell et al., 2007; Li et al., 2012; Lo and Donoghue, 2012; Sun et al., 2018).
The generic delimitation in Maleae has been notoriously difficult, which may be due to the low sequence divergence resulted from ancient, rapid radiations (Wolfe and Wehr, 1988; Campbell et al., 2007) and/or frequent hybridizations (Robertson et al., 1991; Lo and Donoghue, 2012; Liu et al., 2019). As the one of the few genera of Maleae largely distributed in subtropical and tropical regions (Robertson et al., 1991), Eriobotrya consists of ca. 15–20 species ranging from the Himalayas throughout continental southeast Asia to Japan and the islands of western Malesia (Kalkman, 2004). The well-known fruit loquat, Eriobotrya japonica, has been widely cultivated all over the world, making Eriobotrya of great economical importance. Characterized by persistent sepals and craspedodromous lateral veins of leaves, Eriobotrya has been treated to be distinct from Rhaphiolepis in all taxonomic literature and floras (Vidal, 1968; Vidal, 1970; Robertson et al., 1991; Kalkman, 1993; Lu et al., 2003; Kalkman, 2004). The monophyly of the Eriobotrya-Rhaphiolepis complex has been strongly supported by recent molecular phylogenetic studies based on limited taxon sampling, however, its phylogenetic placement was still controversial (Campbell et al., 2007; Li et al., 2009; Li et al., 2012; Lo and Donoghue, 2012; Liu et al., 2019; Xiang et al., 2017; Yang et al., 2012; Zhang et al., 2017). In addition, with the synapomorphies of the proportionally large seed with rounded or wide-elliptic cross-section and the lack of endosperm (Aldasoro et al., 2005; Rohrer et al., 1991), Eriobotrya and Rhaphiolepis were morphologically distinct from other members of Maleae. Although the nuclear Adh topology indicated the nonmonophyly of Eriobotrya with Rhaphiolepis nested within it, Yang et al. (2012) did not provide any discussions on the taxonomic implications.
As a rapid and cost-effective method in phylogenomics, genome skimming (i.e. low-coverage genome shotgun sequencing) could generate a large number of phylogenetically informative sites from the whole chloroplast genome, partial mitochondrial genome, and entire nuclear ribosomal DNA (nrDNA) repeats (Straub et al., 2012; Zhang et al, 2015; Zhang et al, 2017; Zimmer and Wen, 2015). Strongly supported incongruence between the maternally inherited plastomes and nuclear markers has been utilized to address polyploidy and reticulate evolution (Bock et al., 2014; McKain et al., 2018; Liu et al., 2019). Recently Liu et al. (2019) have successfully clarified the phylogenetic relationships between Photinia and its morphologically similar allies in the framework of Maleae using whole chloroplast genomes and nuclear ribosomal DNA (nrDNA) repeats assembled from genome skimming approach. The case study showed great potential in resolving the phylogenetic relationships in Maleae using genome skimming data. These datasets have been successfully applied to reconstruct the phylogenetic history of various plant lineages (e.g., in Bock et al., 2014; Zhang et al., 2015; Dillenberger et al., 2018; Mabry and Simpson, 2018; Thomson et al, 2018; Wen et al., 2018; Liu et al., 2019).
The main objectives of the present study were to clarify the phylogenetic placements of Eriobotrya and Rhaphiolepis with a broader sampling using sequences of the whole chloroplast genomes and entire nrDNA repeats via genome skimming. We aim to (1) explore the phylogenetic position of the Eriobotrya-Rhaphiolepis complex; (2) test the monophyly of Eriobotrya and clarify its phylogenetic relationship with Rhaphiolepis; and (3) discuss the taxonomic implications of the new phylogenetic results.
We used 83 chloroplast genomes and 68 nrDNA repeats for this analysis, in which 37 samples were sequenced for this study, 31 samples were from our previous study (Liu et al., 2019), and 15 accessions of plastomes were from GenBank. These 83 samples of plastomes represented 76 species and 32 genera which represented nearly all the genera recognized currently in Maleae expect Chamaemeles Lindl. As the core ingroup, nine species of Rhaphiolepis and 12 species of Eriobotrya were included in the phylogenetic analysis, representing all the morphological and geographic diversity (Figure 1). In order to resolve the phylogenetic positions of the Eriobotrya-Rhaphiolepis complex, we extensively sampled its closely related genera in the plastid tree, i.e., Cotoneaster Medik., Heteromeles M.Roem., and Photinia, as well as its closely related genus Stranvaesia Lindl. in the nrDNA tree (Liu et al., 2019). We also sampled 67 accessions of plastomes from 30 genera of Maleae as the ingroup, i.e., Amelanchier, Aronia, Chaenomeles Lindl., Cormus Spach, Cotoneaster, Crataegus L., Cydonia Mill., Dichotomanthes Kurz, Docynia Decne., Eriolobus (DC.) M.Roem., Hesperomeles Lindl., Heteromeles, Kageneckia Ruiz & Pav., Lindleya Kunth, Malacomeles (Decne.) Decne., Malus Mill., Mespilus L., Osteomeles Lindl., Peraphyllum Nutt., Phippsiomeles B.B.Liu & J.Wen, Photinia, Pourthiaea Decne., Pseudocydonia (C.K.Schneid.) C.K.Schneid., Pyracantha M.Roem., Pyrus, Sorbus L., Stranvaesia, Torminalis Medik., and Vauquelinia Corrêa ex Bonpl. The most closely related genus of Maleae, Gillenia was selected as the outgroup. As for the nrDNA analysis, 68 accessions representing 67 species and 29 genera were used herein.
The total genomic DNAs sequenced for this study were extracted either from silica-gel dried leaves or from the specimens in herbarium PE and US (Supplementary Table 1). The DNAs of the samples with species names marked with α (Supplementary Table 1) were extracted with a modified CTAB (mCTAB) method (Li et al., 2013b) in the lab of the State Key Laboratory of Systematic and Evolutionary Botany, CAS, China. Library preparations were done at Novogene, Beijing, China using NEBNext® Ultra™ II DNA Library Prep Kit and the libraries were sequenced on the NovaSeq 6000 Sequencing System. The DNAs of the samples with β (Supplementary Table 1) were extracted using Qiagen DNeasy® plant mini-kit (Qiagen Gmbh, Hilden, Germany) following the manufacturer's protocol and the libraries were prepared with the same kit in the Laboratories of Analytical Biology (LAB), National Museum of Natural History (NMNH), Smithsonian Institution (SI), USA. The pooled libraries were shipped to Novogene in UC Davis Sequencing Centre, Davis, California, USA on the Illumina HiSeq 2500 instrument. Paired-end reads of 2 × 150 bp were generated on both sequencing platforms.
We removed the adaptors introduced by Illumina sequencing using cutadapt 2.4 (Martin, 2011) with AGATCGGAAGAGC as the forward and the reverse adaptor. The results were checked for quality control with FastQC 0.11.8 (Andrews, 2018). NOVOPlasty 3.6 (Dierckxsens et al., 2016) was then used to assemble the plastomes de novo with the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (rbcL) gene sequence as the seed. Twenty-two circular plastomes (59.5%) of the 37 samples were assembled with NOVOPlasty. The remaining 15 samples were assembled using Zhang et al. (2015)'s successive method combining mapping-based and de novo assembly. For these 15 samples, the raw data generated from the Illumina HiSeq runs were trimmed using Trimmomatic v.0.39 (Bolger et al., 2014), removing bases below PHRED 15 within a sliding window of four bases, keeping only reads of 36 bases or longer. The results were also checked by FastQC for quality control. Zhang's successive assembly used a multistep approach, including reference-guided assembly and de novo assembly to obtain a high-quality plastome. The reference-guided assemblies were performed by Bowtie2 2.3.5.1 (Langmead and Salzberg, 2012) using only the forward and reverse paired reads generated by Trimmomatic v.0.39 (Bolger et al., 2014). We used the closely related plastomes generated from the previous step of this study or the GenBank as the reference genome. After obtaining a complete plastome for one species, we used it as the reference to assemble its closely related samples (see Zhang et al., 2015). The resulting plastomes were used as subsequent references. De novo assemblies were constructed by SPAdes 3.13.1 with careful error correction and K-mer length of 21, 33, 55, 77 (Nurk et al., 2013). To correct errors and ambiguities resulting from each approach, the scaffolds obtained by de novo assembly, in combination with the contigs generated from NOVOPlasty were mapped to the plastome from the reference-guided assembly. Through the combined effort using NOVOPlasty and Zhang et al. (2015)'s method, we assembled high-quality plastomes for all samples.
We also used Zhang et al. (2015)'s method to assemble the entire nrDNA repeats. The raw data were trimmed by Trimmomatic v.0.39 (Bolger et al., 2014). We used the previously assembled nrDNA sequences (Eriobotrya cavaleriei (H.Lév.) Rehder [GenBank accession no. MN215982]and E. deflexa (Hemsl.) Nakai [GenBank accession no. MN215978]) to capture and map the reads to reference by Bowtie2 2.3.5.1 (Langmead and Salzberg, 2012). The scaffolds generated by SPAdes 3.13.1 (Nurk et al., 2013) in assembling the plastome were mapped to the consensus sequences, and the gaps and ambiguous sites generated by the reference method were corrected. If the quality of nrDNA sequences from de novo assembly was not good, we used a different reference to assemble the sequences again with the above procedure. Generally, we obtained high-quality nrDNA sequences using this successive approach that combined the strengths of reference-guided and de novo approaches.
The assembled plastid genomes and nrDNA repeats were annotated using Geneious Prime (Kearse et al., 2012) with a closely related and well-annotated sequence downloaded from NCBI as a reference (KT633951), and the results of automated annotation were checked manually. The coding sequences of plastome were translated into proteins for checking the start and stop codons manually in Geneious Prime. With the two reverse complementary repeats in the plastome of Rosaceae, the boundary of the large-single copy (LSC), small-single copy (SSC), and inverted-repeats (IRs) for each plastome were verified by the Find Repeats in Geneious Prime. Eighty-three plastomes were aligned by MAFFT v.7.409 (Nakamura et al., 2018) with default parameters. To reduce the systematic errors produced by poor alignment, we used the trimAL v.1.2 (Capella-Gutiérrez et al., 2009) with a heuristic method to decide on the best-automated method to trim the alignment of the whole plastome (WP). All 79 coding sequences (CDSs) of each plastome were extracted separately by Geneious Prime (Kearse et al., 2012), each coding sequence was aligned by MAFFT as specified above, and the alignment of each gene was concatenated by AMAS (Borowiec, 2016). Sixty-eighty nrDNA sequences were also aligned by MAFFT as specified above, and each region of nrDNA was extracted separately by Geneious Prime (Kearse et al., 2012), and then concatenated by AMAS (Borowiec, 2016). The gene map of Eriobotrya japonica and Rhaphiolepis indica (L.) Lindl. ex Ker Gawl. were generated by OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al., 2019).
We first performed independent phylogenetic analyses for each dataset (WP and nrDNA) obtained via genome skimming using the Maximum Likelihood (ML) and Bayesian Inference (BI). As the sequence of two IR copies was completely or nearly identical, only one copy of inverted repeat (IR) region was included for the whole plastome (WP) analyses. We used all the 79 coding sequences (CDSs) of each plastome extracted separately by Geneious Prime for phylogenetic analyses. For nrDNA sequences, the intergenic spacer (IGS) and external transcribed spacer (ETS) regions were difficult to align, only part of ETS and the complete sequences of 18S, ITS1, 5.8S, ITS2, and 26S in nrDNA were used for the phylogenetic analyses.
The best-fit partitioning schemes and nucleotide substitution models for the coding sequences of whole plastome and nrDNA dataset were estimated using PartitionFinder2 (Stamatakis, 2006; Lanfear et al., 2016) for unpartitioned whole plastome or partitioned coding sequences (CDS) of plastomes and nrDNA sequences. Under the corrected Akaike information criterion (AICc) and linked branch lengths, the PartitionFinder2 were performed by greedy (Lanfear et al., 2012) and rcluster (Lanfear et al., 2014) algorithm option for WP, plastid CDS, and nrDNA datasets, respectively, with prior defined data blocks by codon positions of each protein-coding genes and all models. The partitioning schemes and evolutionary model for each subset were used for the downstream ML and BI analyses. The ML tree was inferred by IQ-TREE v.1.6.9 (Nguyen et al., 2015) with 1000 bootstrap replicates using UFBoot2 (Hoang et al., 2017) and collapsing near zero branches option. The BI was performed with MrBayes 3.2.7 (Ronquist et al., 2012). The Markov Chain Monte Carlo (MCMC) analyses were run for 10,000,000 generations. The stationarity was regarded to be reached when the average standard deviation of split frequencies remained below 0.01. Trees were sampled at every 1,000 generations, and the first 25% of samples were discarded as burn-in. The remaining trees were used to build a 50% majority-rule consensus tree. The ML and BI trees were visualized using Geneious Prime (Kearse et al., 2012).
The chloroplast genome of Rhaphiolepis indica (the type species of Rhaphiolepis) was 159,466 bp in length with a classic quadripartite structure that comprised of inverted repeat's pairs (Figure 2), and that of Eriobotrya japonica (type species of Eriobotrya) had the same structure as R. indica with a length of 159,156 bp (Supplementary Figure 1). They contained the same number of coding sequences (79), tRNAs (37), and rRNAs (8). No significant rearrangements or gene losses were found in the other species of Eriobotrya and Rhaphiolepis.
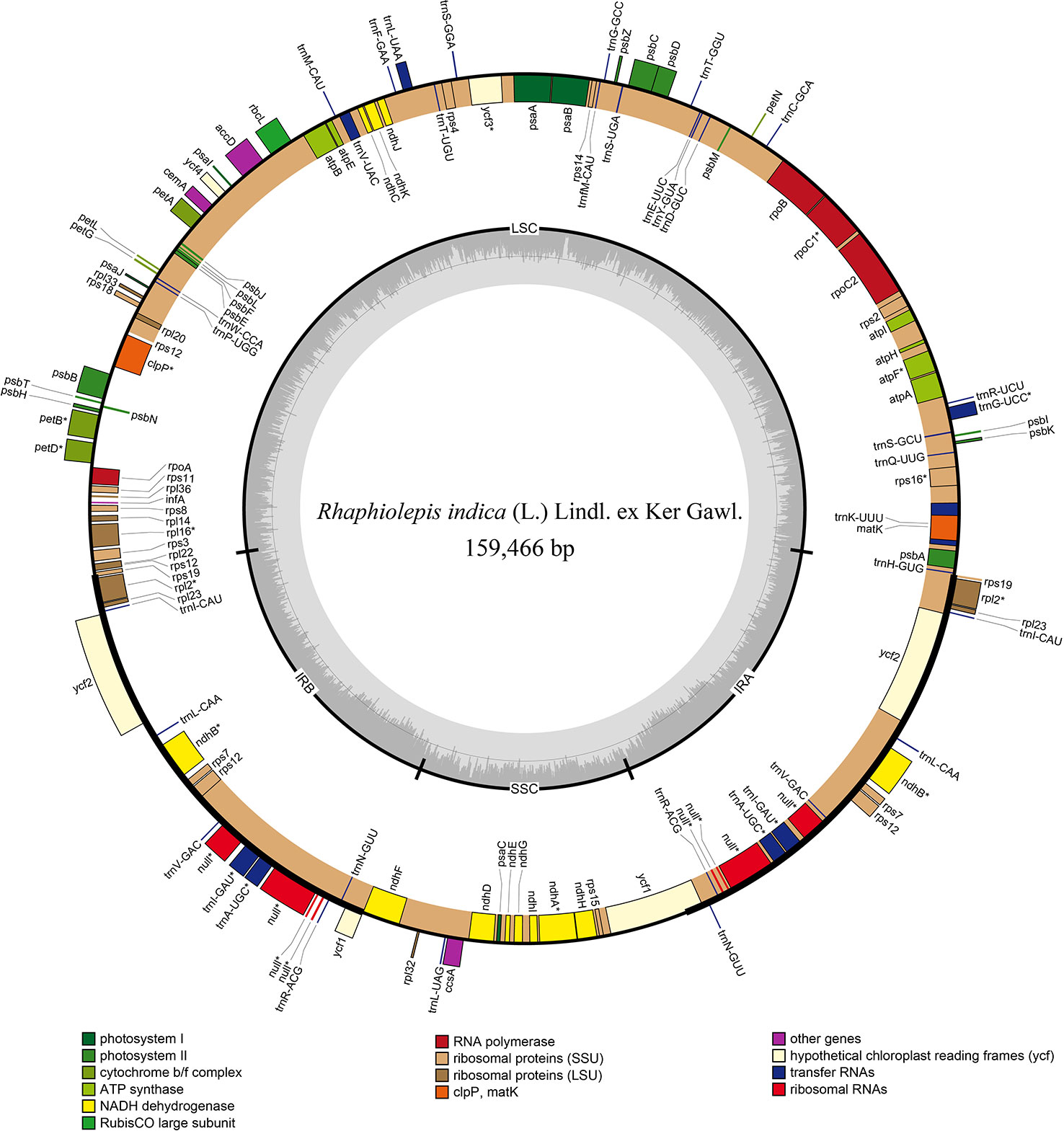
Figure 2 Gene map of the chloroplast genome of Rhaphiolepis indica (L.) Lindl. ex Ker Gawl. The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belong to the different functional group are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions.
The aligned matrix of the 83 whole chloroplast genomes was 129,168 bp in length with the pool alignment trimmed by trimAL (Capella-Gutiérrez et al., 2009). The best-fit model of nucleotide substitutions for the ML analysis was TVM+I+G calculated by PartitionFinder, and that for the BI analysis was GTR+Γ+I. The 79 concatenated CDS sequences from 83 plastomes were generated an aligned matrix of 67,961 bp in length, which was split into 25 sets of sites (aka data blocks) with the same best scheme for each data block for the subsequent ML and BI analysis. The ML and BI analysis of WP dataset resulted in five strongly supported clades within Maleae (Figure 3): clade A (Kageneckia and Lindleya), clade B (only Vauquelinia), clade C (only Pyracantha), clade D (Amelanchier, Crataegus, Hesperomeles, Malacomeles, and Peraphyllum), and two large clades (E and F). These five major clades were also recovered by the phylogenetic analysis of CDS (Supplementary Figure 2). Two tropical American genera (Kageneckia and Lindleya) formed clade A, which constituted the first diverged major clade of Maleae. Clade A was sister to the North American genus Vauquelinia (clade B), and then together they were sister to the core Maleae (Figure 3 and Supplementary Figure 2). As the basalmost group of the core Maleae, Pyracantha (clade C) was sister to a large clade, including clades D, E, and F. Most of the members of clade D were from the New World, and those of clades E and F were from Eurasia except that Aronia and Heteromeles are from North America. The two major Eurasian clades (E and F) were sister to each other, and then together sister to the New World clade D (Figure 3 and Supplementary Figure 2).
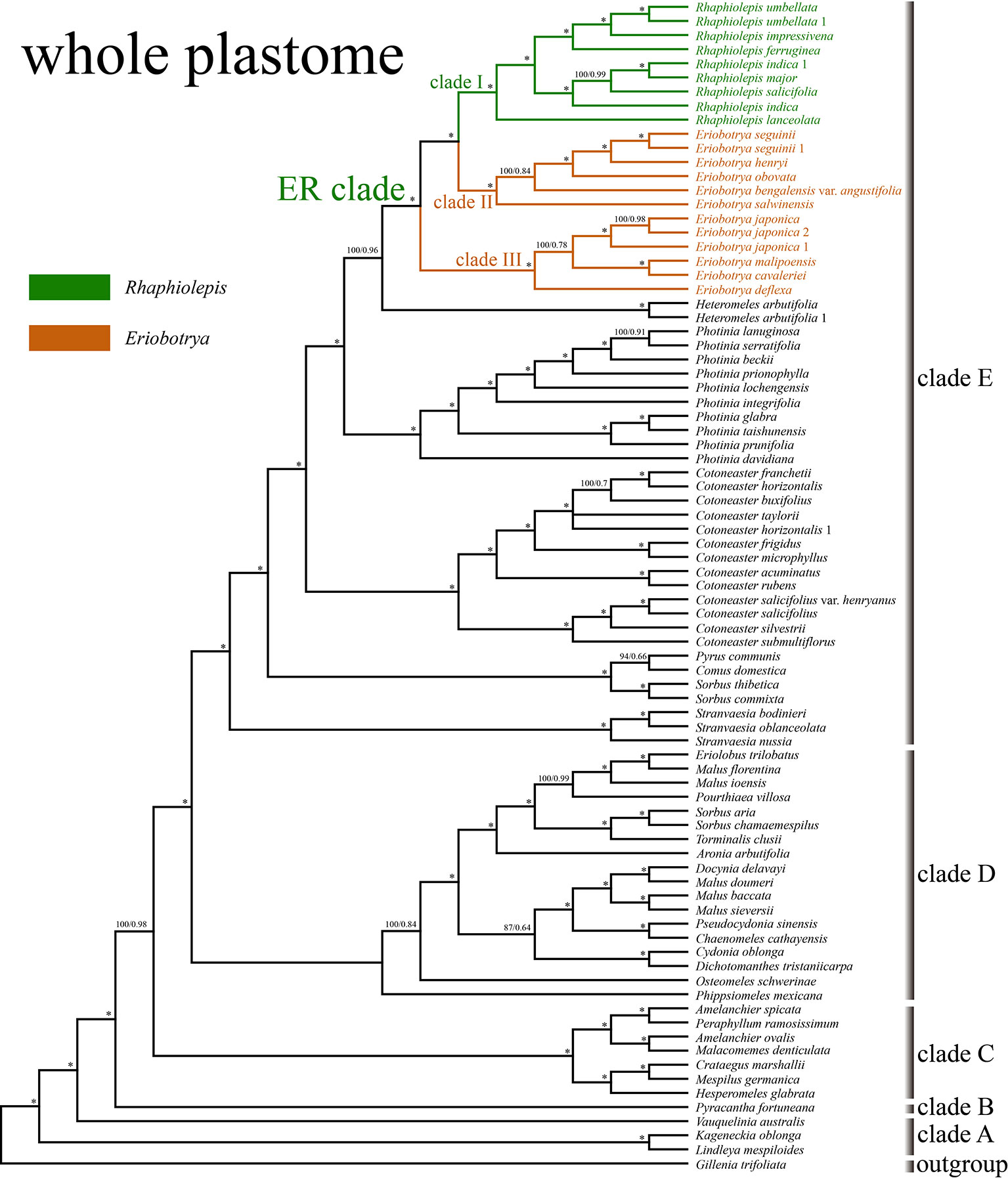
Figure 3 The phylogenetic relationships between Eriobotrya and Rhaphiolepis in the framework of Maleae resolved by Bayesian inference of the whole plastid genome (WP). Numbers associated with the branches are ML bootstrap value (BS) and BI posterior probabilities (PP), and asterisks (*) indicate bootstrap support/posterior probability of 100/1.00.
Samples of Eriobotrya and Rhaphiolepis were strongly supported to form a monophyletic group (BS = 100, PP = 1) by the WP and CDS trees (Figure 3 and Supplementary Figure 2). Although Rhaphiolepis was recovered as a monophyletic group (clade I) with strong support (WP: BS = 99, PP = 1; CDS: BS = 96, PP = 1), Eriobotrya was divided into two clades, clades II & III, with the Rhaphiolepis clade (I) sister to clade II (E. henryi Nakai, E. obovata W.W.Sm., E. salwinensis Hand.-Mazz., and E. seguinii (H.Lév.) Cardot ex Guillaumin), and together they were sister to clade III (E. cavaleriei, E. deflexa, E. japonica, and E. malipoensis K.C.Kuan). Eriobotrya was also paraphyletic in these two trees (Figure 3 and Supplementary Figure 2).
The concatenated nrDNA data (six regions, i.e. partial ETS, 18S, ITS1, 5.8S, ITS2, and 26S) generated an aligned matrix of 6,361 bp in length. Each region of nrDNA was treated as a data block, and these six sets of sites were partitioned into five subsets with each corresponding models of molecular evolution for the subsequent ML and BI analyses. The ML and BI analyses had nearly the same topology (Figure 4). The ER clade was well-supported in the nrDNA tree (BS = 100, PP = 1), however, its phylogenetic position was not resolved (Figure 4). Two species of Eriobotrya (E. henryi and E. seguinii) were strongly supported to be sister to all Rhaphiolepis species sampled here (BS = 90, PP = 0.96), and then together they were sister to a clade of the other species of Eriobotrya (E. cavaleriei, E. deflexa, E. japonica, E. malipoensis, E. obovata, and E. salwinensis) (BS = 100, PP = 1). The topology of the ER clade generated from the nrDNA dataset showed strong conflicts with that from the plastome dataset (Figure 5).
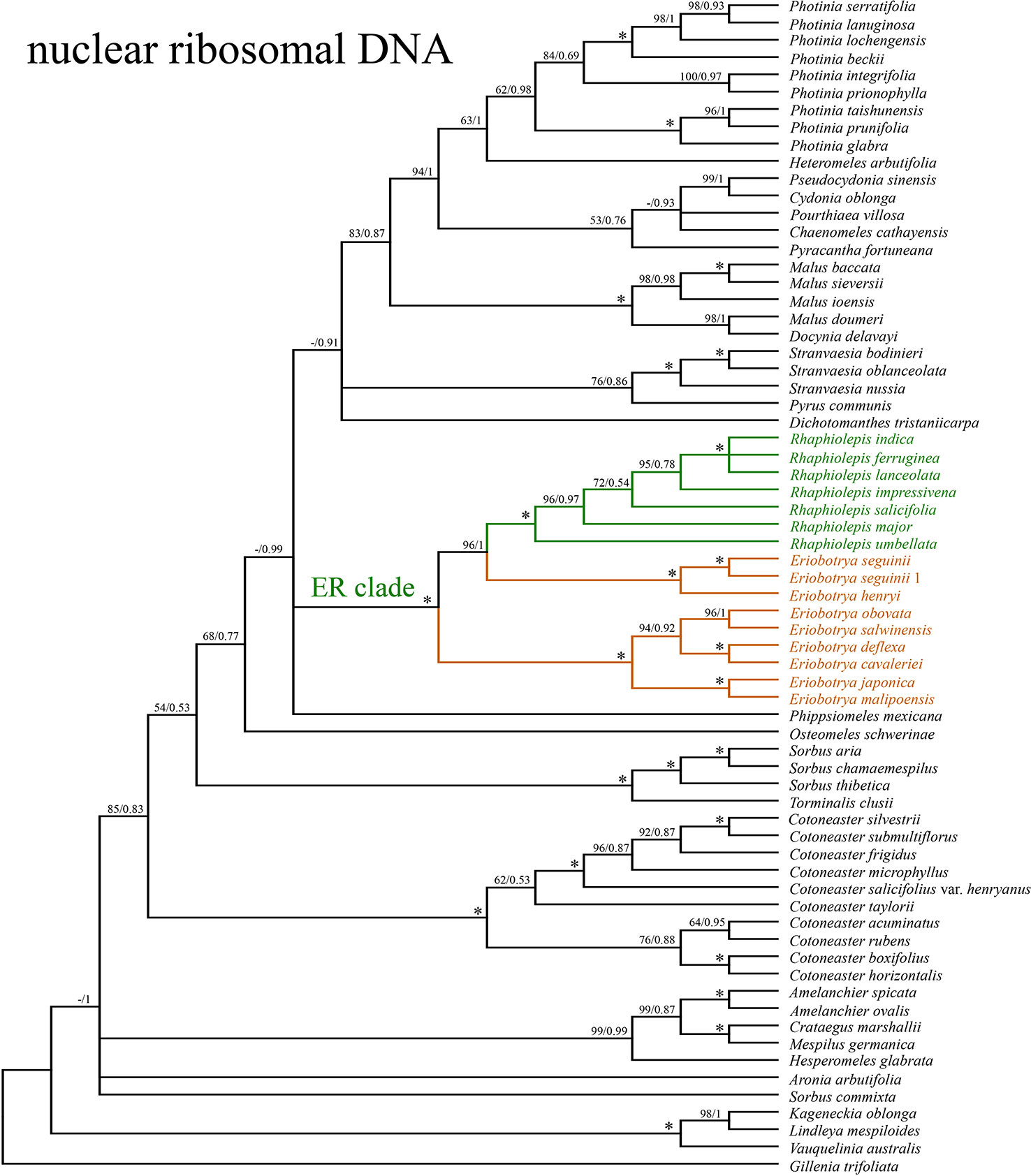
Figure 4 The phylogenetic relationships between Eriobotrya and Rhaphiolepis in the framework of Maleae resolved by Bayesian inference of the sequences of the nuclear ribosomal DNA (nrDNA) repeats. Numbers associated with the branches are ML bootstrap value (BS) and BI posterior probabilities (PP), and asterisks (*) indicate bootstrap support/posterior probability of 100/1.00.
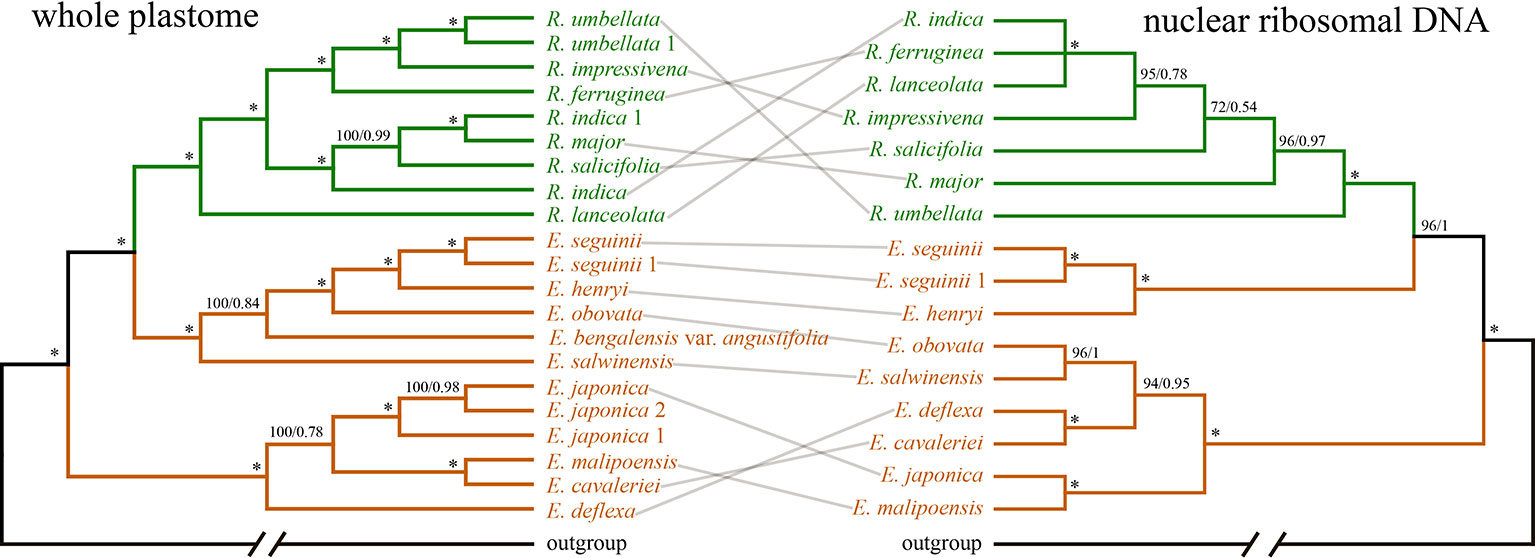
Figure 5 COMPARISONS of the phylogenies based on Bayesian inference of the whole plastid genome (WP: left) and nuclear ribosomal DNA (nrDNA) repeats (right). Branches are colored by genera. Numbers associated with the branches are ML bootstrap value (BS) and BI posterior probabilities (PP), and asterisks (*) indicate bootstrap support/posterior probability of 100/1.00.
Our phylogenetic results based on the whole chloroplast genome and entire nrDNA repeats strongly supported the monophyly of the Eriobotrya-Rhaphiolepis complex (Figure 3 and Supplementary Figure 2). The ER clade was first reported by Campbell et al. (2007) based on phylogenetic analyses using six cpDNA regions, and this relationship was supported by subsequent studies using chloroplast regions or even plastomes, but with limited taxon sampling (Lo and Donoghue, 2012; Zhang et al.,2017; Sun et al., 2018; Liu et al., 2019). In addition, Eriobotrya and Rhaphiolepis have also been supported to form a clade based on nuclear data, e.g., GBSSI-1A and nrITS plus GBSSI-2B sequences (Campbell et al., 2007), as well as nuclear Adh sequences (Yang et al., 2012). Although the topology of [Eriobotrya, (Rhaphiolepis, Vauquelinia)] was reported by Campbell et al. (1995) using nrITS and a small portion of the 5.8S sequences, all the subsequent studies based on the nrITS sequences supported the close relationship between Eriobotrya and Rhaphiolepis (Campbell et al., 2007; Li et al., 2009; Li et al., 2012; Lo and Donoghue, 2012) instead of between Rhaphiolepis and Vauquelinia (Campbell et al., 1995). Our nrDNA tree showed a distant relationship between Vauquelinia and the ER clade. In addition, Vauquelinia can be easily distinguished from the members of the ER clade morphologically by the former's dry capsular fruits, winged seeds with endosperms, and different wood ray anatomy (Zhang, 1992). Morphologically the ER clade is supported by two synapomorphies: the rounded or widely elliptic cross-section of seeds and the absence of endosperm (Aldasoro et al., 2005).
The ML and BI analyses based on the WP and CDS dataset strongly supported the sister relationship between the ER clade and Heteromeles, then together they were sister to Photinia. Previous studies based on the limited chloroplast regions or genomes have shown the uncertain placement of the ER clade. The clade may be sister to the [(Aria, Heteromeles) Cotoneaster] clade (Campbell et al., 2007), or Cotoneaster (Lo and Donoghue, 2012), or the [(Cotoneaster, Stranvaesia), (Heteromeles, Photinia)] clade (Sun et al., 2018), or the [Photinia (Cotoneaster, Heteromeles)] clade (Zhang et al., 2017; Liu et al., 2019). Such uncertainties may be due to the limited markers and/or samples used in the previous studies, or extinctions of the closest relatives of the ER clade. The ER clade was found to be closely related to the small North American genus Heteromeles based on our plastome data (Figure 3 and Supplementary Figure 2). But we will test this relationship in our further analyses of Maleae.
By contrast, the ML and BI analyses based on the nrDNA dataset did not resolve the phylogenetic position of the ER clade. The ML result supported that the ER clade was sister to a large clade that includes Chaenomeles, Cydonia, Dichotomanthes, Docynia, Heteromeles, Malus, Phippsiomeles, Photinia, Pourthiaea, Pseudocydonia, Pyracantha, Pyrus, and Stranvaesia (Figure 4). The BI result did not resolve the relationships among the ER clade, the Central American Phippsiomeles, and the remaining genera (Chaenomeles, Cydonia, Dichotomanthes, Docynia, Heteromeles, Malus, Photinia, Pourthiaea, Pseudocydonia, Pyracantha, Pyrus, and Stranvaesia). Campbell et al. (1995) recovered the ER clade as well as Vauquelinia as forming the basalmost clade of Maleae using ITS sequences, and this result was supported by the cladistic analysis using 16 morphological and anatomical characters (Aldasoro et al., 2005). While the ER clade was placed either as sister to Pyrus using GBSSI-1A sequences or as a weakly supported sister group with Chaenomeles using GBSSI-2B and nrITS plus GBSSI-2B sequences (Campbell et al., 2007). The ER clade was moderately supported to be sister to Osteomeles and Stranvaesia using entire nrDNA repeats (Liu et al., 2019). Based on the transcriptome data, Xiang et al. (2017) placed the ER clade as sister to a large clade that includes Chaenomeles, Cydonia, Docynia, Eriolobus, Malus, Photinia, Pseudocydonia, Pyrus, Sorbus, and Stranvaesia, and this relationship does not conflict with our result. The ER clade is largely distributed in the warm temperate to subtropical and tropical regions of China, Indochina and the Malesian region, whereas most other Asian Maleae including its closely related genera are well developed in cool temperate and warm temperate regions, extending to subtropical regions of the Northern Hemisphere. Fossils of the ER clade were reported from northern China and northeastern Siberia in the Miocene when the earth was warmer (Hsu and Chaney, 1940; Baikovskaja, 1974).
The ER clade can be easily distinguished from other genera of Maleae. They have evergreen, coriaceous leaves with unlobed, entire to finely or coarsely serrate margins (Robertson et al., 1992), and semi-inferior to inferior ovaries (Rohrer et al., 1991; and its seed morphology is unique with the lack of endosperm and proportionally larger in size with a rounded or wide-elliptic cross-section (Rohrer et al., 1991; Aldasoro et al., 2005). The clade was placed as sister to all pome-bearing members of Maleae based on morphological characters (Aldasoro et al., 2005), supporting its isolated position. The incongruent positions of the ER clade based on our plastome and the nrDNA data may also point to the potential impact of hybridization. So the ER clade remains an enigmatic position within Maleae.
Our results strongly supported the paraphyly of Eriobotrya, with Rhaphiolepis nested within it, based on the plastome as well as the nrDNA datasets. The paraphyly of Eriobotrya was never reported in any previous studies. Yang et al. (2012) sampled only one species of Rhaphiolepis (R. indica) in their preliminary phylogenetic analyses of Eriobotrya using the nuclear Adh sequences. Rhaphiolepis indica was shown to be sister to a subclade of Eriobotrya (E. bengalensis Hook.f., E. prinoides Rehder & E.H.Wilson f. angustifolia J.E.Vidal, E. henryi, and E. seguinii), and this clade was then sister to another subclade of nine Eriobotrya species, showing the biphyly of Eriobotrya, with an extremely limited sampling of Rhaphiolepis. Yang et al. (2012) never discussed the nonmonophyly of Eriobotrya nor its taxonomic implications. Due to the general limited taxon sampling of Eriobotrya and Rhaphiolepis, most previous studies only emphasized the close relationships between these two genera (Campbell et al., 2007; Lo and Donoghue, 2012; Xiang et al., 2017; Yang et al., 2017; Zhang et al., 2017), but their data were insufficient in addressing the phylogenetic relationships between the two genera.
Our study represents the first phylogenetic analysis that was designed to test the monophyly of Eriobotrya and Rhaphiolepis with the taxon sampling representing their respective morphological and geographic diversity. The paraphyly of Eribototrya revealed in our plastid and nrDNA supports the merge of Eriobotrya and Rhaphiolepis into one genus. Eriobotrya was thought to be easily distinguished from Rhaphiolepis by the former's persistent sepals (vs. caducous sepals in Rhaphiolepis) and the excurrent lateral veins of leaves (vs. curved lateral veins in Rhaphiolepis). However, these two characters were not always stable to be used to distinguish Eriobotrya and Rhaphiolepis. For example, based on our field observations and herbarium studies in PE and US, the sepals of Eriobotrya henryi are obviously caducous (Figure 6A) and the lateral veins of leaves in E. henryi and E. seguinii are curved (Figures 6B, C), furthermore, the lateral veins of Rhaphiolepis sometimes terminate at the leaf margins (Figure 6D). Treating the ER clade as one genus is also supported by two synapomorphies: the presence of rounded or widely elliptic seeds and the absence of endosperm). Geographically, these two genera overlap broadly in East and Southeast Asia (Figure 1). The phylogenetic, morphological and geographic evidence all supports merging Eriobotrya into Rhaphiolepis, which has the nomenclatural priority. Nevertheless, describing the clade II of Figures 3 as a new genus may seem a likely alternative solution to ensure the monophyly of each genus, however, the phylogenetic incongruence (e.g., between E. obovata and E. salwinensis) in the chloroplast and nuclear trees is consistent with the extensive gene flow between clades II and III (see below). It thus seems not a good taxonomic solution to segregate clade II of Eriobotrya into a separate genus.

Figure 6 The caducous sepals (A, Eriobotrya henryi from Yunnan, China); curved lateral veins of leaves (camptodroumous) in Eriobotrya (B, E. henryi A[bardoce 00063057] and C, E. seguinii); the lateral veins terminate at the leaf margin (craspedodromous) in Rhaphiolepis ferruginea F.P.Metcalf (Guangxi, China). Photo credits: (A), Jian Huang; (D), Bin-Bin Liu.
We detected topological incongruence within the expanded Rhaphiolepis between the chloroplast and nrDNA trees (Figure 5). Processes that might explain the incongruence between chloroplast and nuclear phylogenies include incomplete lineage sorting, allopolyploidy, and hybridization (Rieseberg and Soltis, 1991; Mckinnon et al., 2001; Zou and Ge, 2008). Allopolyploidy has never been reported in Eriobotrya except for the cultivars of E. japonica (Mehra et al., 1973; Singhal et al., 1990; Chen, 1993; Liang et al., 1999; Liang et al., 2001; Yahata et al., 2005; Zhang et al., 2012; Li et al., 2013a), and this mechanism may be excluded for explaining the conflicts between the plastid and nrDNA trees. Lineage sorting could also result in incongruence between chloroplast and nuclear topologies, however, it is very difficult to distinguish hybridization from incomplete lineage sorting (Joly et al., 2009). Hybridization has been shown to be very common in Maleae (Robertson et al., 1991; Lo and Donoghue, 2012; Liu et al., 2019), and artificial hybrids were reported even between Eriobotrya and Rhaphiolepis (Coombes and Robertson, 2008). The conflicts shown in our results likely reflect frequent hybridization events in the evolutionary history of the expanded Rhaphiolepis. The extent and impact of hybridizations in Maleae will need to be further analyzed using next-generation sequencing and genomic tools (Zimmer and Wen, 2015). We will discuss the evolutionary events, involving hybridization, chloroplast capture, introgression, and/or allopolyploidy that occurred in the expanded Rhaphiolepis with a comprehensive sampling in a follow-up paper.
Rhaphiolepis Lindl., Bot. Reg. 6: t. 468 (1820)≡Rhaphiolepis Poir., Dict. Sci. Nat., ed. 2. [F. Cuvier] 45: 314. 1827. Type: Rhaphiolepis indica (L.) Lindl. ex Ker Gawl.≡Crataegus indica L. = Eriobotrya Lindl., Trans. Linn. Soc. London 13: 96, 102. 1821. syn. nov.
Trees, small trees, or shrubs, unarmed, evergreen. Leaves simple, penninerved; stipules on the extreme base of petiole, free, rarely intrapetiolarly connate. Petiole present, venation craspedodromous or camptodromous, margin serrate or entire; stipules caducous, subulate. Inflorescences in terminal racemes, panicles, or compound racemes, many flowered. Hypanthium campanulate, cupular, tubular, or obconical, the free part inside lined with an intrastaminal disk, open at top. Sepals 5, persistent or caducous. Petals 5, white, yellow, or pink, obovate or orbicular, base clawed. Stamens 15–20(-40). Ovary inferior, carpels 2–5, ventrally and laterally connate (in upper part ventrally free), completely connate with each other and dorsally adnate to the hypanthium, the hairy apex exposed; ovules normally 2 per carpel, rarely more; styles 2–5, connate at base and often pubescent; stigma truncate. Fruit a pome, yellowish, yellowish red, brown, dark purplish-brown, bluish, or purplish-black, subglobose, globose, or obovate, fleshy or dry, flesh mostly of hypanthial origin, sclereids absent or present, endocarp (core) thin, membranous. Seeds 1–3, large, with a thin but firm testa; endosperm absent, cotyledons thick. 2n = 34.
About 46 species (Vidal, 1965; Vidal, 1968; Vidal, 1970; Kalkman, 1973; Lu et al., 2003; Kalkman, 2004) in East & Southeast Asia and the Himalayas, south to Borneo and Sumatra.
Below we transfer all taxa of Eriobotrya to Rhaphiolepis and make the nomenclatural changes.
1. Rhaphiolepis angustissima (Hook.f.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya angustissima Hook.f., Fl. Brit. India [J. D. Hooker] 2(5): 372. 1878≡Pyrus angustissima (Hook.f.) M.F.Fay & Christenh., Global Fl. 4: 95. 2018.
Distribution: India (Mt. Khasia); South Vietnam.
2. Rhaphiolepis balgooyi (K.M.Wong & Ent) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya balgooyi K.M.Wong & Ent, Pl. Ecol. Evol. 147(1): 136. 2014.
Distribution: Malaysia (Borneo on Mt. Kinabalu and Mt. Tambuyukon).
3. Rhaphiolepis bengalensis (Roxb.) B.B.Liu & J.Wen, comb. nov.≡Mespilus bengalensis Roxb., Hort. Bengal. 38; Fl. Ind. (ed. 1832) 2: 510. 1832≡Eriobotrya bengalensis Hook.f., Fl. Brit. India [J. D. Hooker] 2(5): 371. 1878.
= Alsodeia grandis Miq., Fl. Ned. Ind., Eerste Bijv. 3: 391. 1861.
= Eriobotrya tinctoria Kurz, Prelim. Rep. For. Veg. Pegu, App. B. 48. 1875, in clavi.
Distribution: widely distributed from East Himalaya (Sikkim, Assam) through Bangladesh (Chittagong) to Myanmar, Laos, Cambodia, S. Vietnam, Malay Peninsula, Sumatra, and Borneo.
4. Rhaphiolepis cavaleriei (H.Lév.) B.B.Liu & J.Wen, comb. nov.≡Hiptage cavaleriei H.Lév., Repert. Spec. Nov. Regni Veg. 10: 372. 1912≡Eriobotrya cavaleriei (H.Lév.) Rehder, J. Arnold Arbor. 13: 307. 1932≡Pyrus athenae M.F.Fay & Christenh., Global Fl. 4: 96. 2018.
= Eriobotrya brackloi Hand.-Mazz., Anz. Akad. Wiss. Wien, Math.-Naturwiss. Kl. 59: 102. 1922≡Eriobotrya cavaleriei (H.Lév.) Rehder var. brackloi (Hand.-Mazz.) Rehder, J. Arnold Arbor. 13(3): 308. 1932.
= Eriobotrya brackloi Hand.-Mazz. var. atrichophylla Hand.-Mazz., Anz. Akad. Wiss. Wien, Math.-Naturwiss. Kl. 59: 103. 1922.
= Eriobotrya grandiflora Rehder & E.H.Wilson, Pl. Wilson. (Sargent) 1(2): 193. 1912≡Eriobotrya deflexa (Hemsl.) Nakai var. grandiflora (Rehder & E.H.Wilson) Nakai, J. Arnold Arbor. 5(2): 72. 1924.
Distribution: China (Fujian, Guangdong, Guangxi, Guizhou, Hubei, Hunan, Jiangxi, Sichuan); North Vietnam (Hòa Bình, Lao Cai).
5. Rhaphiolepis condaoensis (X.F.Gao, Idrees & T.V.Do) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya condaoensis X.F.Gao, Idrees & T.V.Do, Phytotaxa 365(3): 290. 2018.
Distribution: Southeast Vietnam (Ba Ria-Vung Tau: Con Dao National Park).
6. Rhaphiolepis × daduheensis (H.Z.Zhang ex W.B.Liao, Q.Fan & M.Y.Ding) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya × daduheensis H.Z.Zhang ex W.B.Liao, Q.Fan & M.Y.Ding, Phytotaxa 212(1): 97. 2015.
Distribution: As a putative natural hybrid between Rhaphiolepis loquata (= Eriobotrya japonica) and R. prinoides (= E. prinoides), this species is restricted to Daduhe River Basin in Sichuan, China (Ding et al., 2015).
7. Rhaphiolepis deflexa (Hemsl.) B.B.Liu & J.Wen, comb. nov.≡Photinia deflexa Hemsl., in Ann. Bot. ix. 153. 1895≡Eriobotrya deflexa (Hemsl.) Nakai, Bot. Mag. (Tokyo) 30: 18, in adnot. 1916.
= Photinia buisanensis Hayata, Icon. Pl. Formosan. 3: 100. 1913≡Eriobotrya deflexa (Hemsl.) Nakai f. buisanensis (Hayata) Nakai, Bot. Mag. (Tokyo) 30(349): 18. 1916≡Eriobotrya buisanensis (Hayata) Kaneh., Formosan Trees 218. 1918≡Eriobotrya deflexa Nakai var. buisanensis (Hayata) Hayata, Catal. Governm. Herb. Formos. 246. 1930≡Eriobotrya buisanensis (Hayata) Makino & Nemoto, Fl. Japan., ed. 2 (Makino & Nemoto) 464. 1931.
= Eriobotrya deflexa Nakai var. koshunensis Kaneh. & Sasaki, Catal. Gov't Herb. Formosa 246. 1930≡Eriobotrya deflexa Nakai f. koshunensis (Kaneh. & Sasaki) H.L.Li, Lloydia 14(4): 232. 1951.
Distribution: China (Guangdong, Hainan, Taiwan); Vietnam (Nha Trang).
8. Rhaphiolepis dubia (Lindl.) B.B.Liu & J.Wen, comb. nov.≡Photinia dubia Lindl., Trans. Linn. Soc. London 13(1): 104, t. 10. 1821≡Eriobotrya dubia (Lindl.) Decne., in Nouv. Arch. Mus. Par. Ser. I, x. 145. 1874.
= Mespilus tinctoria D.Don, Prodr. Fl. Nepal. 238. 1825.
Distribution: Bhutan; India (Sikkim); Myanmar (Kachin, Mandalay, Shan); Nepal.
9. Rhaphiolepis elliptica (Lindl.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya elliptica Lindl., Trans. Linn. Soc. London 13(1): 102. 1821≡Cotoneaster ellipticus (Lindl.) Hort ex Loudon, Encyc. Pl. 1208≡Pyrus elliptica (Lindl.) M.F.Fay & Christenh., Global Fl. 4: 102. 2018.
= Mespilus cuila Buch.-Ham. ex D.Don, Prodr. Fl. Nepal. 238. 1825.
Distribution: China (Tibet); Nepal (Narainhetty).
10. Rhaphiolepis elliptica (Lindl.) B.B.Liu & J.Wen var. petelotii (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya elliptica Lindl. var. petelotii J.E.Vidal, Adansonia sér. 2, 5: 552. 1965.
Distribution: N Vietnam (Lao Cai).
11. Rhaphiolepis fulvicoma (Chun ex W.B.Liao, F.F.Li & D.F.Cui) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya fulvicoma Chun ex W.B.Liao, F.F.Li & D.F.Cui, Ann. Bot. Fenn. 49(4): 264. 2012.
Distribution: China (Guangdong).
12. Rhaphiolepis glabrescens (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya glabrescens J.E.Vidal, Adansonia sér. 2, 5: 554. 1965≡Pyrus serpentae M.F.Fay & Christenh., Global Fl. 4: 121. 2018.
Distribution: North Myanmar (Triangle, Centre Ouest, Khai Yang).
13. Rhaphiolepis glabrescens (J.E.Vidal) B.B.Liu & J.Wen var. victoriensis (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya glabrescens J.E.Vidal var. victoriensis J.E.Vidal, Adansonia sér. 2, 5: 555. 1965.
Distribution: North Mayanmar (Centre Ouest: Mt Victoria).
14. Rhaphiolepis henryi (Nakai) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya henryi Nakai, J. Arnold Arbor. 5: 70. 1924≡Pyrus henryi (Nakai) M.F.Fay & Christenh., Global Fl. 4:106. 2018.
Distribution: China (Guizhou, Yunnan); Myanmar (Pyin Oo Lwin).
15. Rhaphiolepis hookeriana (Decne.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya hookeriana Decne., Nouv. Arch. Mus. Par. Ser. I 10:146. 1874≡Pyrus hookeriana (Decne.) M.F.Fay & Christenh., Global Fl. 4: 107. 2018.
Distribution: Bhutan; India (Sikkim).
16. Rhaphiolepis herae (M.F.Fay & Christenh.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya latifolia Hook.f., Fl. Brit. India [J. D. Hooker] 2(5): 370. 1878≡Pyrus herae M.F.Fay & Christenh., Global Fl. 4: 106. 2018.
Distribution: Myanmar (Kayin, Taninthayi).
17. Rhaphiolepis longifolia (Decne.) B.B.Liu & J.Wen, comb. nov.≡Photinia longifolia Decne., in Nouv. Arch. Mus. Par. Ser. I 10: 142. 1874≡Eriobotrya longifolia (Decne.) Hook.f., Fl. Brit. India [J. D. Hooker] 2(5): 370. 1878.
Distribution: Bangladesh (East Bengal).
18. Rhaphiolepis loquata B.B.Liu & J.Wen, nom. nov.≡Mespilus japonica Thunb., Fl. Jap. (Thunberg) 206. 1784≡Eriobotrya japonica (Thunb.) Lindl., Trans. Linn. Soc. London 13: 102. 1821≡Photinia japonica Benth. & Hook.f. ex Asch. & Schweinf., Mém. Inst. Égypt. [Illustr. Fl. Egypt.] 73. 1887.
= Crataegus bibas Lour., Fl. Cochinch. 1: 319. 1790≡Pyrus bibas (Lour.) M.F.Fay & Christenh., Global Fl. 4: 98. 2018.
Distribution: Native in Chongqing (Nanchuan) and Hubei (Yichang) of China. As an economically important fruit, this species has been widely cultivated in central & south China, as well as in Japan, Korea, India, and some countries in Southeast Asia.
Note: The species epithet has been pre-occupied by Rhaphiolepis japonica Siebold & Zucc. (Siebold and Zuccarini, 1841), so a new name is needed for this taxon (Turland et al., 2018). The epithet “loquata” is derived from the English name loquat.
19. Rhaphiolepis macrocarpa (Kurz) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya macrocarpa Kurz, J. Asiat. Soc. Bengal, Pt. 2, Nat. Hist. 41(4): 306. 1872.
Distribution: Myanmar (Bago, Mandalay).
20. Rhaphiolepis malipoensis (K.C.Kuan) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya malipoensis K.C.Kuan, Acta Phytotax. Sin. 8(3): 231. 1963≡Pyrus malipoensis (K.C.Kuan) M.F.Fay & Christenh., Global Fl. 4: 111. 2018.
Distribution: China (SE Yunnan).
21. Rhaphiolepis merguiensis (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya merguiensis J.E.Vidal, Adansonia sér. 2, 5: 563. 1965.≡Pyrus merguiensis (J.E.Vidal) M.F.Fay & Christenh., Global Fl. 4: 112. 2018.
Distribution: Myanmar (Mergui Archipelago, Taninthayi).
22. Rhaphiolepis oblongifolia (Merr. & Rolfe) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya oblongifolia Merr. & Rolfe, Philipp. J. Sci., C 3: 102. 1908.
Distribution: the Philippines (Mindanao).
23. Rhaphiolepis obovata (W.W.Sm.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya obovata W.W.Sm., Notes Roy. Bot. Gard. Edinburgh 10: 29. 1917≡Pyrus obovata (W.W.Sm.) M.F.Fay & Christenh., Global Fl. 4: 114. 2018.
Distribution: China (C Yunnan).
24. Rhaphiolepis petiolata (Hook.f.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya petiolata Hook.f., Fl. Brit. India [J. D. Hooker] 2(5): 370. 1878≡Pyrus petiolata (Hook.f.) M.F.Fay & Christenh., Global Fl. 4: 115. 2018.
Distribution: Bangladesh (Chittagong); Bhutan; India (Khasia, Sikkim); Myanmar (Chin).
25. Rhaphiolepis platyphylla (Merr.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya platyphylla Merr., Brittonia 4(1): 80. 1941≡Pyrus platyphylla (Merr.) M.F.Fay & Christenh., Global Fl. 4: 116. 2018.
Distribution: Myanmar (Kachin).
26. Rhaphiolepis poilanei (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya poilanei J.E.Vidal, Adansonia sér. 2, 5: 557. 1965≡Pyrus poilanei (J.E.Vidal) M.F.Fay & Christenh., Global Fl. 4: 116. 2018.
Distribution: Vietnam (Haut-Donnai).
27. Rhaphiolepis prinoides (Rehder & E.H.Wilson) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya prinoides Rehder & E.H.Wilson, Pl. Wilson. (Sargent) 1(2): 194. 1912≡Pyrus prinoides (Rehder & E.H.Wilson) M.F.Fay & Christenh., Global Fl. 4: 116. 2018.
Distribution: China (Sichuan, Yunnan); Laos.
28. Rhaphiolepis prinoides (Rehder & E.H.Wilson) B.B.Liu & J.Wen var. laotica (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya prinoides Rehder & E.H.Wilson var. laotica J.E.Vidal, Adansonia sér. 2, 5: 573. 1965.
Distribution: Laos (Xièng Khouang).
29. Rhaphiolepis salwinensis (Hand.-Mazz.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya salwinensis Hand.-Mazz., Symb. Sin. Pt. 7(3): 475. 1933≡Pyrus salwinensis (Hand.-Mazz.) M.F.Fay & Christenh., Global Fl. 4: 120. 2018.
Distribution: China (NE Yunnan; Tibet); India; Myanmar.
30. Rhaphiolepis seguinii (H.Lév.) B.B.Liu & J.Wen, comb. nov.≡Symplocos seguinii H.Lév., Repert. Spec. Nov. Regni Veg. 10: 431. 1912≡Eriobotrya seguinii (H.Lév.) Cardot ex Guillaumin, Bull. Soc. Bot. France 71: 287, in obs. 1924.
= Eriobotrya pseudorhaphiolepis Cardot, Notul. Syst. (Paris) 3: 371. 1918.
Distribution: China (SW Guizhou, SE Yunnan).
31. Rhaphiolepis serrata (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya serrata J.E.Vidal, Adansonia sér. 2, 5: 558. 1965≡Pyrus serrata (J.E.Vidal) M.F.Fay & Christenh., Global Fl. 4: 121. 2018.
Distribution: China (Guangxi, Yunnan); Laos (Xièng Khouang).
32. Rhaphiolepis stipularis (Craib) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya stipularis Craib, Bull. Misc. Inform. Kew 1929(4): 109. 1929≡Pyrus stipularis (Craib) M.F.Fay & Christenh., Global Fl. 4: 122. 2018.
Distribution: Cambodia; Thailand (Satun).
33. Rhaphiolepis tengyuehensis (W.W.Sm.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya tengyuehensis W.W.Sm., Notes Roy. Bot. Gard. Edinburgh 10: 30. 1917≡Pyrus tengyuehensis (W.W.Sm.) M.F.Fay & Christenh., Global Fl. 4: 123. 2018.
Distribution: China (NW Yunnan; Tibet); Myanmar (Kachin).
34. Rhaphiolepis wardii (C.E.C.Fisch.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya wardii C.E.C.Fisch., Bull. Misc. Inform. Kew 1929(6): 205. 1929≡Pyrus alabaster M.F.Fay & Christenh., Global Fl. 4: 94. 2018.
Distribution: Myanmar (Kachin, North Triangle).
35. Rhaphiolepis williamtelliana (M.F.Fay & Christenh.) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya fragrans Champ., Hooker's J. Bot. Kew Gard. Misc. 4: 80. 1852≡Pyrus williamtelliana M.F.Fay & Christenh., Global Fl. 4: 126. 2018.
Distribution: China (Guangdong, Guangxi, Hainan, Hongkong, Tibet); Vietnam.
36. Rhaphiolepis williamtelliana (M.F.Fay & Christenh.) B.B.Liu & J.Wen var. furfuracea (J.E.Vidal) B.B.Liu & J.Wen, comb. nov.≡Eriobotrya fragrans Champ. ex Benth. var. furfuracea J.E.Vidal, Adansonia sér. 2, 5: 557. 1965.
Distribution: Vietnam (Nha Trang).
Our phylogenetic analyses of Maleae using sequences of the whole chloroplast genomes and entire nrDNA repeats have strongly supported the monophyly of the Eriobotrya-Rhaphiolepis clade and the paraphyly of Eriobotrya, in which Rhaphiolepis was nested within it. Molecular, morphological, and geographic evidence supports merging these two genera into one genus, Rhaphiolepis. We herein have transferred taxa currently recognized in Eriobotrya to Rhaphiolepis, making 35 new combinations and a new name, Rhaphiolepis loquata, in this paper.
Publicly available datasets were analyzed in this study. This data can be found here: MN577874, MN577892, MN577875, MN577893, MN577873, MN577895, MN577863, MN577894, MN577872, MN577880, MN577877, MN577881, MN577882, MN577883, MN577884, MN577885, MN068252, MN068266, MN068248, MN577878, MN577889, MN577879, MN577890, MN577888, MN577891, MN577865, MN577871, MN577870, MN577866, MN577886, MN577864, MN577867, MN577887, MN577876, MN577868, MN068250, MN577908, MN577928, MN577909, MN577929, MN577907, MN577931, MN577896, MN577930, MN577905, MN577914, MN577911, MN577915, MN577916, MN577918, MN577919, MN577920, MN577923, MN577932, MN577906, MN577912, MN577925, MN577913, MN577926, MN577924, MN577927, MN577898, MN577904, MN577903, MN577899, MN577921, MN577897, MN577900, MN577922, MN577910, MN577901, MN577917.
B-BL, G-NL, D-YH, and JW designed the study. B-BL and G-NL conducted the experiments, analyzed the data, and drafted the manuscript. JW and D-YH provided suggestions on structuring the paper and the main points of the discussion, and revised the manuscript. All authors approved the final manuscript.
This research is supported by the National Natural Science Foundation of China (Grant No 31620103902).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
We thank John Wiersema (US) for his advice on nomenclature and Chao Xu (PE) for assistance with experiments. Our thanks also go to Xin-Tang Ma (PE), Xiao-Hua Jin (PE), You-Sheng Chen (SCBG), Hui-Min Li (NAS), and Ting Wang (HHBG) for the sample collections and Jian Huang (HITBC) for supplying the photo of Eriobotrya henryi. The computational analyses were conducted on the Smithsonian Institution High Performance Cluster (SI/HPC, “Hydra”).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01731/full#supplementary-material
Supplementary Figure 1 | Gene map of the chloroplast genome of Rhaphiolepis loquata (Eriobotrya japonica). The genes inside and outside of the circle are transcribed in the clockwise and counterclockwise directions, respectively. Genes belonging to the different functional group are shown in different colors. The thick lines indicate the extent of the inverted repeats (IRa and IRb) that separate the genomes into small single-copy (SSC) and large single-copy (LSC) regions.
Supplementary Figure 2 | The phylogenetic relationships between Eriobotrya and Rhaphiolepis in the framework of Maleae resolved by Bayesian inference of the coding sequences (CDS) of chloroplast genome. Numbers associated with the branches are ML bootstrap value (BS) and BI posterior probabilities (PP), and asterisks (*) indicate 100/1 support.
Supplementary Table 1 | Accessions of Maleae used in this study. † and ‡ indicate that the samples were extracted from silica-gel dried leaves and herbarium specimens, respectively.
Aldasoro, J. J., Aedo, C., Navarro, C. (2005). Phylogenetic and phytogeographical relationships in Maloideae (Rosaceae) based on morphological and anatomical characters. Blumea 50, 3–32. doi: 10.3767/000651905X623256
Andrews, S. (2018). FastQC: A quality control tool for high throughput sequence data, Available at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed September 18, 2019).
Baikovskaja, T. N. (1974). Upper Miocene flora of the Southern Primorski Kray (Leningrad: Acad. Sci. SSSR). [in Russian].
Bock, D. G., Kane, N. C., Ebert, D. P., Rieseberg, L. H. (2014). Genome skimming reveals the origin of the Jerusalem Artichoke tuber crop species: neither from Jerusalem nor an artichoke. New Phytol. 201, 1021–1030. doi: 10.1111/nph.12560
Bolger, A. M., Lohse, M., Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Borowiec, M. L. (2016). AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ 4, e1660. doi: 10.7717/peerj.1660
Campbell, C. S., Donoghue, M. J., Baldwin, B. G., Wojciechowski, M. F. (1995). Phylogenetic relationships in Maloideae (Rosaceae): evidence from sequences of the internal transcribed spacers of nuclear ribosomal DNA and its congruence with morphology. Am. J. Bot. 82, 903–918. doi: 10.1002/j.1537-2197.1995.tb15707.x
Campbell, C. S., Evans, R. C., Morgan, D. R., Dickinson, T. A., Arsenault, M. P. (2007). Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): Limited resolution of a complex evolutionary history. Plant Syst. Evol. 266, 119–145. doi: 10.1007/s00606-007-0545-y
Capella-Gutiérrez, S., Silla-Martínez, J. M., Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, R. Y. (1993). Chromosome atlas of Chinese principal economic plants. Volume 1. Chromosome atlas of Chinese fruit trees and their close wild relatives (Beijing: Science Press).
Coombes, A. J., Robertson, K. R. (2008). An intergeneric hybrid between Eriobotrya and Rhaphiolepis. Plantsman 7, 162–165.
Dierckxsens, N., Mardulyn, P., Smits, G. (2016). NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. doi: 10.1093/nar/gkw955
Dillenberger, M. S., Wei, N., Tennessen, J. A., Ashman, T. L., Liston, A. (2018). Plastid genomes reveal recurrent formation of allopolyploid Fragaria. Am. J. Bot. 105, 862–874. doi: 10.1002/ajb2.1085
Greiner, S., Lehwark, P., Bock, R. (2019). OrganellarGenomeDRAW (OGDRAW) version 1.3. 1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47, W59–W64. doi: 10.1093/nar/gkz238
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q., Vinh, L. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hsu, J., Chaney, R. W. (1940). A Miocene flora from Shantung province, China. Contr. Paleontol. Carnegie Inst. Wash. 507, 1–147.
Joly, S., McLenachan, P. A., Lockhart, P. J. (2009). A statistical approach for distinguishing hybridization and incomplete lineage sorting. Am. Nat. 174, E54–E70. doi: 10.1086/600082
Kalkman, C. (1993). “Rosaceae,” in Flora Malesiana series I-Spermatophyta Flowering Plants. Eds. de Wilde, W. J. J. O., Nooteboom, H. P., Kalkman, C. (Leiden: Rijksherbarium), 227–351.
Kalkman, C. (2004). “Rosaceae,” in The Families and Genera of Vascular Plants. VI Flowering Plants-Dicotyledons Celastrales, Oxalidales, Rosales, Cornales, Ericales. Eds. Kubitzki, K. (Berlin: Springer), 343–386.
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analyses of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Lanfear, R., Calcott, B., Ho, S. Y. W., Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Lanfear, R., Calcott, B., Kainer, D., Mayer, C., Stamatakis, A. (2014). Selecting optimal partitioning schemes for phylogenomic datasets. BMC Evol. Biol. 14, 82. doi: 10.1186/1471-2148-14-82
Lanfear, R., Frandsen, P. B., Wright, A. M., Senfeld, T., Calcott, B. (2016). PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 34, 772–773. doi: 10.1093/molbev/msw260
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, P., Lin, S. Q., Yang, X. H., Hu, G. B., Jiang, Y. M. (2009). Molecular phylogeny of Eriobotrya Lindl. (loquat) inferred from internal transcribed spacer sequences of nuclear ribosome. Pakistan J. Bot. 41, 185–193.
Li, Q. Y., Guo, W., Liao, W. B., Macklin, J. A., Li, J. H. (2012). Generic limits of Pyrinae: insights from nuclear ribosomal DNA sequences. Bot. Stud. 53, 151–164.
Li, G. F., Liang, G. L., Lin, S. Q. (2013a). A study on karyotype analysis of genus Eriobotrya and its application to systematic taxonomy (in Chinese). Acta Hortic. Sin. 40, 1465–1474.
Li, J. L., Wang, S., Yu, J., Wang, L., Zhou, S. L. (2013b). A modified CTAB protocol for plant DNA extraction (in Chinese). Chin. Bull. Bot. 48, 72–78. doi: 10.3724/SP.J.1259.2013.00072
Liang, G. L., Ren, Z. C., Yan, Y., Huang, H., Wu, C. Q. (1999). Chromosome variation in 8 loquat varieties in Sichuan Province (in Chinese). Acta Hortic. Sin. 26, 71–76.
Liang, G. L., Ren, Z. C., Yan, Y., Li, X. L., Jiang, N. G. (2001). Variation and evolution of genus Eriobotrya in Sichuan (in Chinese). Adv. In Chromosome Sci. 1, 311–316.
Liu, B. B., Hong, D. Y., Zhou, S. L., Xu, C., Dong, W. P., Johnson, G., et al. (2019). Phylogenomic analyses of the Photinia complex support the recognition of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Rosaceae). J. Syst. Evol. 57, 678–694. doi: 10.1111/jse.12542
Lo, E. Y. Y., Donoghue, M. J. (2012). Expanded phylogenetic and dating analyses of the apples and their relatives (Pyreae, Rosaceae). Mol. Phylogenet. Evol. 63, 230–243. doi: 10.1016/j.ympev.2011.10.005
Lu, L. T., Gu, C. Z., Li, C. L., Alexander, C., Bartholomew, B., Brach, A. R., et al. (2003). “Rosaceae,” in Flora of China Volume 9 (Pittosporaceae through Connaraceae). Eds. Wu, Z. Y., Raven, P. H., Hong, D. Y. (Beijing: Science Press; St. Louis: Missouri Botanical Garden Press), 46–434.
Mabry, M. E., Simpson, M. G. (2018). Evaluating the monophyly and biogeography of Cryptantha (Boraginaceae). Syst. Bot. 43, 53–76. doi: 10.1600/036364418X696978
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/ej.17.1.200
Mckain, M. R., Johnson, M. G., Uribe-Convers, S., Eaton, D., Yang, Y. (2018). Practical considerations for plant phylogenomics. Appl. Plant Sci. 6 (3), e1038. doi: 10.1002/aps3.1038
McKinnon, G. E., Vaillancourt, R. E., Jackson, H. D., Potts, B. M. (2001). Chloroplast sharing in the Tasmanian eucalypts. Evolution 55, 703–711. doi: 10.1554/0014-3820(2001)055[0703:CSITTE]2.0.CO;2
Mehra, P. N., Sareen, T. S., Hans., A. S. (1973). Cytology of some woody species of Rosaceae from the Himalayas. Silvae Genet. 22, 188–190.
Morgan, D. R., Soltis, D. E., Robertson, K. R. (1994). Systematic and evolutionary implications of rbcl sequence variation in Rosaceae. Am. J. Bot. 81, 890–903. doi: 10.2307/2445770
Nakamura, T., Yamada, K. D., Tomii, K., Katoh, K. (2018). Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34, 2490–2492. doi: 10.1093/bioinformatics/bty121
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., Bui Quang, M. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). “Assembling genomes and mini-metagenomes from highly chimeric reads,” in Research in Computational Molecular Biology. Eds. Deng, M., Jiang, R., Sun, F., Zhang, X. (Berlin, Heidelberg: Springer), 158–170.
Phipps, J. B., Robertson, K. R., Smith, P. G., Rohrer, J. R. (1990). A checklist of the subfamily Maloideae (Rosaceae). Canad. J. Bot. 68, 2209–2269. doi: 10.1139/b90-288
Phipps, J. B., Robertson, K. R., Rohrer, J. R., Smith, P. G. (1991). Origins and evolution of subfam. Maloideae (Rosaceae). Syst. Bot. 16, 303–332. doi: 10.2307/2419283
Potter, D., Eriksson, T., Evans, R. C., Oh, S., Smedmark, J. E. E., Morgan, D. R., et al. (2007). Phylogeny and classification of Rosaceae. Plant Syst. Evol. 266, 5–43. doi: 10.1007/s00606-007-0539-9
Rohrer, J. R., Robertson, K. R., Phipps, J. B. (1991). Variation in structure among fruits of Maloideae (Rosaceae). Am. J. Bot. 78, 1617–1635. doi: 10.2307/2444843
Rohrer, J. R., Robertson, K. R., Phipps, J. B. (1994). Floral morphology of Maloideae (Rosaceae) and its systematic relevance. Am. J. Bot. 81, 574–581. doi: 10.2307/2445732
Rieseberg, L. H., Soltis, D. (1991). Phylogenetic consequences of cytoplasmic gene flow in plants. Evol. Trends Pl. 5, 65–84.
Robertson, K. R., Phipps, J. B., Rohrer, J. R., Smith, P. G. (1991). A synopsis of genera in Maloideae (Rosaceae). Syst. Bot. 16, 376–394. doi: 10.2307/2419287
Robertson, K. R., Phipps, J. B., Rohrer, J. R. (1992). Summary of leaves in the genera of Maloideae (Rosaceae). Ann. Missouri Bot. Gard. 79, 81–94. doi: 10.2307/2399811
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Siebold, P. F. von, Zuccarini, J. G. (1841). Flora Japonica; sive, Plantae Quas in Imperio Japonico Collegit, Descripsit, ex Parte in Ipsis Locis Pingendas Curavit. Sectio Prima Continens Plantas Ornatui vel Usui Inservientes (Leiden: Lugduni Batavorum).
Singhal, V., Gill, B., Sidhu, M. (1990). Cytology of woody members of Rosaceae. Proc. Indian Acad. Sci. (Plant Sci.) 100, 17–21. doi: 10.1007/BF03053464
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Straub, S. C., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., Liston, A. (2012). Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 99, 349–364. doi: 10.3732/ajb.1100335
Sun, J. H., Shi, S., Li, J. L., Yu, J., Wang, L., Yang, X. Y., et al. (2018). Phylogeny of Maleae (Rosaceae) based on multiple chloroplast regions: implications to genera circumscription. BioMed. Res. Int. 2018, 7627191. doi: 10.1155/2018/7627191
Thomson, A. M., Vargas, O. M., Dick, C. W. (2018). Complete plastome sequences from Bertholletia excelsa and 23 related species yield informative markers for Lecythidaceae. Appl. Plant Sci. 6, e1151. doi: 10.1002/aps3.1151
Turland, N. J., Wiersema, J. H., Barrie, F. R., Greuter, W., Hawksworth, D. L., Herendeen, P. S., et al. (2018). International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. Regnum Vegetabile 159 (Glashütten: Koeltz Botanical Books). doi: 10.12705/Code.2018
Verbylaitė, R., Ford-Lloyd, B., Newbury, J. (2006). The phylogeny of woody Maloideae (Rosaceae) using chloroplast trnL-trnF sequence data. Biologija 1, 60–63.
Vidal, J. E. (1965). Notes sur quelques Rosacées asiatiques (III). Révision du genre Eriobotrya (Pomoideae). Adansonia 5, 537–580.
Vidal, J. E. (1968). Flore du Cambodge du Laos et du Vietnam (Paris: National Museum of Natural History).
Vidal, J. E. (1970). “Rosaceae,” in Flora of Thailand. Eds. Larsen, K. (Bangkok: Forest Herbarium, Royal Forest Department), 31–74.
Wen, J., Harris, A. J., Kalburgi, Y., Zhang, N., Xu, Y., Zheng, W., et al. (2018). Chloroplast phylogenomics of the New World grape species (Vitis, Vitaceae). J. Syst. Evol. 56, 297–308. doi: 10.1111/jse.12447
Wolfe, J. A., Wehr, W. (1988). Rosaceous Chamaebatiaria-like foliage from the Paleogene of western North America. Aliso 12, 177–200. doi: 10.5642/aliso.19881201.14
Xiang, Y. Z., Huang, C. H., Hu, Y., Wen, J., Li, S. S., Yi, T. S., et al. (2017). Evolution of Rosaceae fruit types based on nuclear phylogeny in the context of geological times and genome duplication. Mol. Biol. Evol. 34, 262–281.
Yahata, S., Sato, S., Ohara, H., Matsui, H. (2005). Differences of morphological and fruit bearing characteristics among the different ploidy plants and triploid production by crossings between diploid and triploid plants in loquat [Eriobotrya japonica]. Hortic. Res. (Japan) 4, 379–384. doi: 10.2503/hrj.4.379
Yang, X. H., Li, P., Lin, S. Q., Hu, G. B., He, X. L. (2012). A preliminary phylogeny study of the Eriobotrya based on the nrDNA Adh sequences. Not. Bot. Horti. Agrobot. Cluj-Na. 40, 233–237. doi: 10.15835/nbha4027997
Yang, X. H., Najafabadi, S. K., Shahid, M. Q., Zhang, Z. K., Jing, Y., Wei, W. L., et al. (2017). Genetic relationships among Eriobotrya species revealed by genome-wide RAD sequence data. Ecol. Evol. 7, 2861–2867. doi: 10.1002/ece3.2902
Zhang, Z. K., Wang, Y. Q., Lin, S. Q., Du, K. (2012). Ploidy identification of loquats for genome sequencing project by flow cytometry. J. Fruit Sci. 29, 498–504.
Zhang, N., Wen, J., Zimmer, E. A. (2015). Congruent deep relationships in the grape family (Vitaceae) based on sequences of chloroplast genomes and mitochondrial genes via genome skimming. PloS One 10, e0144701. doi: 10.1371/journal.pone.0144701
Zhang, S. D., Jin, J. J., Chen, S. Y., Chase, M. W., Soltis, D. E., Li, H. T., et al. (2017). Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 214, 1355–1367. doi: 10.1111/nph.14461
Zimmer, E. A., Wen, J. (2015). Using nuclear gene data for plant phylogenetics: progress and prospects II. Next-gen approaches. J. Syst. Evol. 53, 371–379. doi: 10.1111/jse.12174
Keywords: chloroplast genome, Eriobotrya, hybridization, Maleae, nrDNA, Rhaphiolepis, Rosaceae
Citation: Liu B-B, Liu G-N, Hong D-Y and Wen J (2020) Eriobotrya Belongs to Rhaphiolepis (Maleae, Rosaceae): Evidence From Chloroplast Genome and Nuclear Ribosomal DNA Data. Front. Plant Sci. 10:1731. doi: 10.3389/fpls.2019.01731
Received: 14 October 2019; Accepted: 09 December 2019;
Published: 07 February 2020.
Edited by:
Matthew Aaron Gitzendanner, University of Florida, United StatesReviewed by:
Núria Garcia-Jacas, Instituto Botánico de Barcelona (IBB), SpainCopyright © 2020 Liu, Liu, Hong and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Wen, d2VuakBzaS5lZHU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.