
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 30 January 2020
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01709
This article is part of the Research Topic Orchid Genomics and Developmental Biology View all 13 articles
 Pei-Han Lai1†
Pei-Han Lai1† Li-Min Huang1†
Li-Min Huang1† Zhao-Jun Pan2
Zhao-Jun Pan2 Wann-Neng Jane3
Wann-Neng Jane3 Mei-Chu Chung3
Mei-Chu Chung3 Wen-Huei Chen4
Wen-Huei Chen4 Hong-Hwa Chen1,4,5*
Hong-Hwa Chen1,4,5*Phalaenopsis orchids have a spectacular floral morphology with a highly evolved lip that offers a landing platform for pollinators. The typical morphological orchid lip features are essential for the special pollination mechanism of Phalaenopsis flowers. Previously, we found that in the lip, a member of the AP2/EREBP protein family was highly expressed. Here, we further confirmed its high expression and characterized its function during lip development. Phylogenetic analysis showed that AP2/EREBP belongs to the Va2 subgroup of ERF transcription factors. We named it PeERF1. We found that PeERF1 was only expressed at stage 5, as flowers opened. This coincided with both thickening of the cuticle and development of nanoridges. We performed knockdown expression of PeERF1 using CymMV-based virus-induced gene silencing in either the AP2 conserved domain, producing PeERF1_AP2-silenced plants, or the SHN specific domain, producing PeERF1_SHN-silenced plants. Using cryo-SEM, we found that the number of nanoridges was reduced only in the PeERF1_AP2-silenced group. This change was found on both the abaxial and adaxial surfaces of the central lip lobe. Expression of PeERF1 was reduced significantly in PeERF1_AP2-silenced plants. In cutin biosynthesis genes, expression of both PeCYP86A2 and PeDCR was significantly decreased in both groups. The expression of PeCYP77A4 was reduced significantly only in the PeERF1_AP2-silenced plants. Although PeGPAT expression was reduced in both silenced plants, but to a lesser degree. The expression of PeERF1 was significantly reduced in the petal-like lip of a big-lip variant. PeCYP77A4 and PeGPAT in the lip were also reduced, but PeDCR was not. Furthermore, heterologous overexpression of PeERF1 in the genus Arabidopsis produced leaves that were shiny on the adaxial surface. Taken together, our results show that in Phalaenopsis orchids PeERF1 plays an important role in formation of nanoridges during lip epidermis development.
Phalaenopsis orchids are renowned for their unique and elegant floral morphology and long florescence duration. Recently, they have become the model Orchidaceae research plants. Two databases for genetic information have been established, OrchidBase 3.0 and Orchidstra 2.0 (Fu et al., 2011; Su et al., 2013a; Tsai et al., 2013; Chao et al., 2017). The floral morphology of Phalaenopsis orchids includes three sepals, three petals, and one column. The column is formed by fusion of the style and a part of the androecium. The outer two perianth whorls are typically petaloid and are referred to as tepals. Instead of producing three uniform petals, Phalaenopsis flowers have a highly evolved, modified, and resupinated inner medium petal, the lip (Rudall and Bateman, 2002; Tsai et al., 2004; Tsai and Chen, 2006; Tsai et al., 2008; Pan et al., 2011; Pan et al., 2014). This lip is understood to play an important role both in pollination and evolution (Robinson and Burns-Balogh, 1982; Cozzolino and Widmer, 2005; Mondragón-Palomino and Theißen, 2008), as it provides a platform for pollinators.
Lip morphogenesis consists of five stages, from the embedded stage 1 to the open flower of stage 5. There is no division of the lip at stage 1. However, in stage 2, the lip divides quickly into three distinct parts. There are two lateral lobes, one central lobe, and one callus. The split lip forms a tunnel-like structure in the mature flower (Figure 1A, Li), a feature related to evolved pollination strategies (Cozzolino and Widmer, 2005). Although Phalaenopsis orchids exhibit unique floral morphological features, Cryo-scanning electron microscopy (Cryo-SEM) has revealed that the perianth lip epidermis has a unique functional morphology not found in sepals or petals (Pan et al., 2011; Hsieh et al., 2013a; Hsieh et al., 2013b; Pan et al., 2014; Hsu et al., 2015b). Heavy and dense nanoridges cover the lip epidermis. These undulated nanostructures, also called cuticular folds, are assumed to contain cuticular lipids (Koch et al., 2008; Koch et al., 2009a; Koch et al., 2009b).
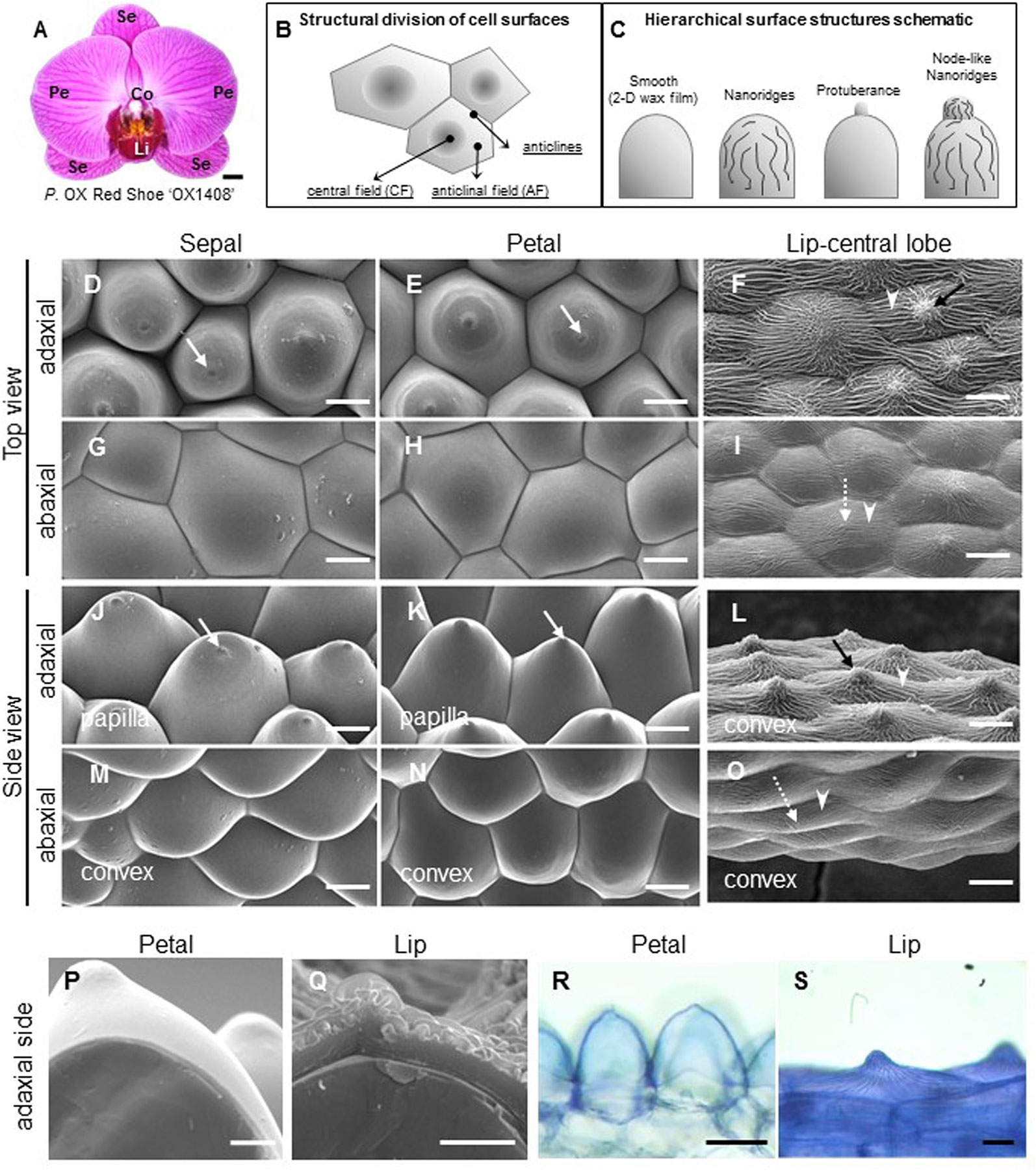
Figure 1 Ultrastructure of floral epidermal cells in the cultivar P. OX Red Shoes “OX1408.” (A) Floral morphology. Scale bar, 5 cm. Se, sepal; Pe, petal; Li, lip; Co, column. (A) Schematic diagrams of the cell structural division of a single cell surface. The central field (CF) is in the center middle of the cell surface and the anticline field (AF) is between the middle area and the boundary of the cell. Anticlines directly connected to the cell area (modified after Koch et al., 2009a). (A) Schematic diagrams of epidermis that built up the cell hierarchical structures with 2-D smooth wax films or decorated with protuberance, nanoridges, and node-like nanoridges. (D–O) Cyro-SEM top view and side view of epidermal cells of perianth organs. White arrows indicate a protuberance on the top of the cell. Black arrows indicate node-like nanoridges in the CF of the cell. White arrowheads indicate parallel nanoridges in the AF of the cell. Scale bars, 30 μm. (P–Q) Cross section of adaxial epidermal cells of petal and lip. Scale bars, 10 μm. (R–S) Lipophilic dye “Sudan Black B” staining on the adaxial epidermal cells of petal and lip. Scale bars, 100 μm.
Diverse perianth and floral epidermis adaptations have evolved in both eudicots and monocots. In many flowering plants, the epidermal surfaces of sepals and petals display a range of patterns in combination with diverse micro- and nanostructures (Kay et al., 1981; Whitney et al., 2009; Whitney et al., 2011b; Kourounioti et al., 2013). More than 75% of petal epidermal cells of angiosperms are conical or papillate, usually on the adaxial side where potential pollinators would be found (Kay et al., 1981; Whitney et al., 2009; Whitney et al., 2011a; Whitney et al., 2011b). Moreover, in many plants, sepal and petal epidermal cells are covered with various density and orientation of nanoridges (Jeffree, 2006). These structures on the surface of sepal and petal epidermal cells are believed to attract pollinators and enhance pollination success through visual signals (Whitney et al., 2009; Whitney et al., 2011b; Kourounioti et al., 2013; Moyroud et al., 2017) and act as tactile signals affecting pollinator movement (Prüm et al., 2011; Rands et al., 2011; Prüm et al., 2012; Prüm et al., 2013; Adachi et al., 2015). Moreover, cell surface cuticle structures can strengthen cells and thereby function many ways in plant development as well as survival and defense in unfavorable environments, such as under biotic or abiotic stress (e.g., dehydration, pathogens, UV light, frost, and insect attacks) (Koch et al., 2008; Koch et al., 2009a; Koch et al., 2009b).
The first identified transcription factors (TFs) that regulate cuticle biosynthesis are SHINEs/WAX INDUCERs (SHNs/WINs), members of the V group of the ethylene responsive factor (ERF) subfamily of the apetala2/ethylene response element binding protein (AP2/EREBP) TF family (Aharoni et al., 2004; Broun et al., 2004; Nakano et al., 2006). In Arabidopsis SHINE gain-of-function mutant (shn) and plants overexpressing AtWIN1/SHN1, AtSHN2, or AtSHN3 have shiny leaves and increased accumulation of epidermal wax on the top of leaves as compared with wild type (Aharoni et al., 2004; Broun et al., 2004). By co-silencing all three AtSHN clade members, SHNs redundantly regulate the formation of petal surface nanoridges and also cell elongation, adhesion, and separation (Shi et al., 2011). Recently, increased number of SHN-like TFs that belong to the ERF-V group have been identified and they exhibit various functions during plant physiological processes.
The ERF-V group includes two subgroups: Va and Vb. The Va subgroup contains two conserved motifs of the conserved middle motif (CMV-1) and C-terminal motif (CMV-2), which the Vb subgroup does not contain (Nakano et al., 2006). The ERF-Va subgroup is further divided into two subgroups, Va1 and Va2, containing a complete or incomplete CMV-1 motif, respectively (Nakano et al., 2014). Functional characterization of genes in the Va1 subgroup from several plants indicates that they are involved in cuticle development. Arabidopsis AtSHNs regulate cuticle formation (Aharoni et al., 2004; Broun et al., 2004; Shi et al., 2011).
Similar research has been done with several plant species. In barley it was found that HvNud is involved in the lipid biosynthesis of the grain surface, which produces hulled caryopses (Taketa et al., 2008). Tomato SlSHINE3 is involved in cutin metabolism of fleshy fruit epidermal cells for patterning the epidermal surface (Shi et al., 2013) and SlSHN1 is involved in wax accumulation of leaf epidermal cells, which enhances drought tolerance (Al-Abdallat et al., 2014). Rice OsAP2/ERF-”N-22” is involved in wax biosynthesis and also enhances drought resistance (Mawlong et al., 2014), while wheat TdSHN1 is involved in the cuticle formation of leaf surfaces (Jäger et al., 2015). Eucalyptus EgrSHN1 and EgrSHN2 are involved in cell wall biosynthesis of flowers (Marques et al., 2013). Hence, the complete CMV-1 and CMV-2 motifs of ERF-Va1 genes are deemed the SHINE domains. SHINE domains are considered to be important in cuticle development. The subgroup Va1 is identified as the SHINE clade (Aharoni et al., 2004).
In contrast, the Va2 subgroup, with an incomplete CMV-1 motif, is involved in various physiological processes: Arabidopsis At5g25190 is induced by 1-aminocyclopropane-1-carboxylicacid (ACC) and salt (NaCl) and was named the ethylene- and salt-inducible ERF gene (AtESE3) (Zhang et al., 2011), but its overexpression confers no typical SHINE phenotype (Aharoni et al., 2004). Tomato LeERF1 regulates fruit ripening and softening (Li et al., 2007). Populus PtaERF003 is involved in lateral root formation (Trupiano et al., 2013). Eucalyptus Egr33m and Egr40m are involved in wood cell wall biosynthesis (Marques et al., 2013). Tomato SlERF52 regulates flower pedicel abscission (Nakano et al., 2014). Whereas, NvERF045 in berries regulates berry ripening and is also involved in cuticle development (Leida et al., 2016). Moreover, the Vb subgroup At5g19790 does not contain CMV-1 and CMV-2 motifs and is important in low potassium signaling (Kim et al., 2012).
Gene associated with cutin biosynthesis for epidermal nanoridge formation include CYP86A and CYP77A, members of a cytochrome P450 family, glycerol-3-phosphate acyltransferase 6 (GPAT), and defective in cuticular ridges (DCR) (Kannangara et al., 2007; Li-Beisson et al., 2009; Panikashvili et al., 2009; Shi et al., 2011; Shi et al., 2013; Petit et al., 2016; Mazurek et al., 2017). GPAT6 and CYP77A6 are for the formation of floral cutin in Arabidopsis thaliana (Li-Beisson et al., 2009). CYP86A4 has been reported as one of the downstream target genes SHN (Shi et al., 2011; Shi et al., 2013). It has been shown that DCR-deficient plants have defective cuticle formation with altered epidermal cell differentiation (Panikashvili et al., 2009). This defective formation in reproductive and vegetative tissues was correlated with low abundance of 9(10),16-dihydroxyhexadecanoic acid in the cutin polymer of DCR (At5g23940)-deficient plants (Panikashvili et al., 2009).
We previously identified several unigenes dominantly expressed in the Phalaenopsis lip (Hsiao et al., 2013). Among them, one member of the AP2 family, P. equestris ethylene responsive factor 1 (PeERF1), was found to be most similar to the SHINE clade homolog At5g25190.
Here, to extend our understanding of the function of PeERF1 in Phalaenopsis orchids, we analyzed its spatial and temporal gene expression; downregulated PeERF1 expression by using CymMV-based virus-induced gene silencing (VIGS) in orchids; and examined heterologous overexpression of PeERF1 in Arabidopsis for comparison. The abnormal phenotypes of nanoridge sculpture patterns of lip epidermal cells were observed in the somaclonal variant, P. “Join Big foot TH365” containing enlarged petal-like lip mutants. We further investigated the relationship of putative Phalaenopsis orthologs of known cutin biosynthetic genes with the cuticle formation in Phalaenopsis lips. These genes include two cytochrome P450s, PeCYP86A2 and PeCYP77A4, and two putative acyltransferases, PeGPAT and PeDCR. We hope that these results will contribute to the understanding of transcriptional regulation of late-stage orchid lip formation during floral morphogenesis.
We conducted gene spatial expression analysis on specimens of Phalaenopsis equestris obtained from the Taiwan Sugar Corp. (Tainan, Taiwan), and both gene temporal expression and VIGS experiments on the commercial cultivar P. OX Red Shoes “OX1408” obtained from Oxen Biotechnology Corp. (Tainan, Taiwan).
The flower buds of P. OX Red Shoes “OX1408” were divided into five stages by size (Figure 3C). A single raceme spike inflorescence embraces 8–10 flowers. The smallest flower bud is embedded in the tip of an inflorescence and is stage 1 (< 0.5 cm), followed by successive stage 2 (0.5–1 cm), stage 3 (1–2 cm), stage 4 (2–3 cm), and blooming flowers are stage 5 (floral diameter of 12 cm) (Figure 3C). For comparison, we used wild-type flower and big-lip variant flower (petal-like lip) of P. hybrid “Join Big foot” (P. Yu Pin Easter Island x P. I- Hsin Diamond “Join White of Love”). These were provided by Join Orchids Incorporation (Tainan, Taiwan). For VIGS experiments, plants were kept in the greenhouse at the Tainan District Agricultural Research and Extension Station, Council of Agriculture, under a controlled temperature of 27°C/22°C (day/night). Other mature orchid plants were maintained in the greenhouse at National Cheng Kung University under natural light and controlled temperature from 23°C to 27°C.
We examined changes in cellular morphology from the 1st to the 8th blooming flowers of silenced plants (stage 5, floral diameter of 12 cm) using Cryo-SEM. Sample preparation and Cryo-SEM examination follows previous research (Hsieh et al., 2013a; Pan et al., 2014). Fresh samples were dissected and loaded on the stub, which was subsequently frozen with liquid nitrogen slush, and then quickly transferred to a sample preparation chamber at -160°C for 5 min. After that time the temperature was raised to -85°C and sublimed for 15 min. Samples were then coated with platinum (Pt) at -130°C and transferred to the cryo-stage in an SEM chamber and observed at -160°C using Cryo scanning electron microscope (FEI Quanta 200 SEM/Quorum Cryo System PP2000TR FEI) with 20 kV.
Images were taken under a Cryo stage at < -160°C. We measured the nanoridge area on 40 flowers, 5 flowers from each of 8 plants using ImageJ (http://rsb.info.nih.gov/ij/). Mean data were compared by Duncan’s multiple-range test, using SPSS v17.
Using TRIsure reagent (Bioline, UK) total RNA was extracted and then treated with RNase-free DNaseI (Invitrogen, USA) to remove residual DNA. We cloned the full-length cDNA of PeERF1 (accession no. MG948436) using a SMART rapid amplification of cDNA ends (RACE) kit (Clontech, USA). We randomly selected 6 to 8 positive clones for sequencing. For gene expression analysis, quantitative real-time RT-PCR (qRT-PCR) was performed in triplicate and repeated independently three times as previously described (Hsu et al., 2015a). Primers for all the PCR and qRT-PCR experiments are in Supplementary Table S1.
For qRT-PCR, the cDNA template was mixed with 2X SYBR Green PCR master mix (Applied Biosystems, Norwalk, CT, USA) in an ABI 7300 instrument (Applied Biosystems) with three biological replicates. For gene quantification, qRT-PCR was performed at stage 5 flowers of PeERF1-silenced plants in triplicate, and repeated in three silenced plants independently. For PCR reaction, each sample was analyzed in triplicate. Reactions involved incubation at 50°C for 2 min, then 95°C for 10 min, and thermal cycling for 40 cycles (95°C for 15 s and 60°C for 1 min). The relative quantification was calculated according to the manufacturer’s instructions (Applied Biosystems). To control the integrity of RNA and normalize target RNA copy numbers in gene-silenced and mock-treated flowers, the housekeeping gene PeActin4 (AY134752) was recruited as an internal control for normalization (Chen et al., 2005).
Multiple sequence alignment was generated by using AlignX (Vector NTI advance 11, Invitrogen). The protein sequences of SHN homologous TFs were obtained from the National Center for Biotechnology Information (NCBI) databases (http://www.ncbi.nlm.nih.gov/), and accession numbers are as follows: A. thaliana AtSHN1 (NP_172988), AtSHN2 (NP_196680), AtSHN3 (NP_851073), At5g25190 (NP_197901.1), and At5g19790 (NP_197480.1); tomato (Solanum lycopersicum) SlSHN1 (XP_004235965), SlSHN3 (XP_004240977), and SlERF52 (BAO18577); tomato (Lycopersicon esculentum) LeERF1 (AAL75809); berry (Vitis vinifera) VvERF045 (ANT73695); barley (Hordeum vulgare L.) HvNud (BAG12386); wheat (Triticum aestivum L.) TdSHN1 (ANY98960); rice (Oryza sativa) OsAP2/ERF-”N-22” (ACU44657), and Populus (P. tremulax P. alba) PtaERF003 (Potri.018G021900). Eucalyptus grandis SHN homologous TFs can be accessed in the Phytozome database (http://www.phytozome.net/cgi-bin/gbrowse/eucalyptus/): EgrSHN1 (Eucgr.C04221.1), EgrSHN2 (Eucgr.C01178.1), Egr33m (Eucgr.C02719.1), and Egr40m (Eucgr.C03947.1) (Marques et al., 2013). These sequences were used to construct phylogenetic trees by using MEGA5.0 (Tamura et al., 2011). Phylogenetic relationships were inferred by the neighbor-joining method and evolutionary distances were computed by the Poisson correction method. Bootstrap values were calculated with 1,000 replicates.
VIGS experiments with PeERF1 were performed as previously described (Hsieh et al., 2013a; Pan et al., 2014); 142-nt and 175-nt fragments for the conserved AP2 domain and the specific incomplete “SHINE domains,” i.e., CMV-1 and CMV-2 motifs of PeERF1, respectively, were constructed into the pCymMV-Gateway plasmid (Lu et al., 2007). The constructed VIGS-silencing plasmids for producing the PeERF1_AP2-silenced and PeERF1_SHN-silenced plants were named pCymMVGateway-PeERF1_AP2 and pCymMV-Gateway-PeERF1_SHN, respectively. These plasmids were transformed into Agrobacterium (strain EHA105). For infiltration, Agrobacterium tumefaciens strain EHA105 containing pCymMVGateway-PeERF1_AP2 or mMV-Gateway-PeERF1_SHN were grown overnight at 28°C to OD600 = 1. After centrifugation, bacterial cell pellets were resuspended by adding 300 μl MS medium containing 100 μM acetosyringone and allowed to stand at room temperature for 0.5 h. Two methods of Agro-infiltration were used: inflorescence injection and leaf injection. For inflorescence injection, suspensions were injected into the stalk of the raceme with eight internodes and one visible floral bud (extruding out of its bract) by use of a 1-ml syringe with a needle in all silencing treatments. The raceme stalk usually emerges from the stem between the third and fourth leaves. For leaf injection, suspensions were injected into the leaf directly above the emerging inflorescence. Mock-treated plants were recruited as the negative control. They were handled the same and contained an empty vector of a Cymbidium mosaic virus infectious clone with a Gateway system vector. Transformed EHA105 was injected into both inflorescence spikes and the leaf directly above the emerging inflorescence. For VIGS, eight independent PeERF1_AP2-silenced and eight PeERF1_ SHN-silenced plants as well as eight mock control plants were generated, and repeated twice independently. qRT-PCR was used to examine the knockdown expression of cuticle biosynthesis-related genes in the 5th floral buds (stage 3, length of 1–2 cm) after agro-infiltration in triplicate and repeated three times independently; Cryo-SEM was used to examine the changes in cellular morphology from the 5th to the 8th blooming flowers of silenced plants (stage 5, floral diameter of 12 cm).
A. thaliana ecotype Columbia was used for transformation experiments as previously described (Chen et al., 2012). Full-length cDNA of PeERF1 was cloned into the pBI121 vector under the control of the constitutive Cauliflower mosaic virus (CaMV) 35S promoter, and the resulted plasmid was named pBI121-PeERF1. We then introduced pBI121-PeERF1 plasmid into Agrobacterium tumefaciens (strain GV3101) and transformed into wild-type Arabidopsis by the floral dip method (Clough and Bent, 1998). In total, 60 kanamycin-resistant T1 seedlings were obtained and grown at 23°C in a growth chamber under long-day conditions (16-h light/8-h dark). A total of 457 T2 heterozygous seedlings were obtained with a segregation ratio of 3:1 as analyzed by chi-square test. For gene expression analysis, RNA samples of T1 heterozygous plants were extracted to confirm the successful expression by qRT-PCR in triplicate and repeated three times independently. The primers were listed in Supplementary Table S1.
To investigate the detailed ultrastructure of orchid floral epidermal cells, we first examined the floral epidermal morphology of a native species, P. aphrodite subsp. formosana (Supplementary Figure S1 ) and the commercial cultivar, P. OX Red Shoe “OX1408” (Figure 1A) by using Cryo-SEM. The schematic diagrams of different regions and hierarchical structures on the cell surface of Phalaenopsis flowers are shown in Figures 1B, C. The “central field” (CF) and “anticlinal field” (AF) represent the inner and outer parts of cells, respectively, and the boundaries of two perpendicular cell walls are “anticlines” (Figure 1B). The epidermal cells of the Phalaenopsis orchid perianth have a variety of cell morphology, including convex or papilla cell shapes. The cell surface of the perianth epidermal cells can be smooth, covered with 2-D wax films or decorated with protuberances, nanoridges, or other node-like nanoridges (Figure 1C).
The adaxial epidermal cells of sepals and petals in P. OX Red Shoe “OX1408” flowers featured a papilla cell shape with a protuberance on the top (Figures 1D, E, J, K, white arrow), and the abaxial epidermal cells of sepals and petals had a convex cell shape with smooth 2-D wax films (Figures 1G, H, M, N). In contrast, heavy and dense nanoridges were found on the lip epidermis. The lip adaxial epidermal cells had convex cell shape with the appearance of node-like nanoridges in the CF (Figures 1F, L, black arrow) and parallel and radial nanoridges in the AF (Figures 1F, L, white arrowhead) of cell surfaces. The lip abaxial epidermal cells had a convex cell shape with parallel nanoridges in the CF (Figures 1I, O, white dashed arrow) and AF of cell surfaces (Figures 1I, O, white arrowhead). The same heavy nanoridges were also observed on the lip epidermis of P. aphrodite subsp. formosana (Supplementary Figure S1D).
Cross sections of petal and lip adaxial epidermal cells showed markedly different thickness of cuticles (Figures 1P, Q). The petal adaxial epidermal cells were covered with smooth 2-D wax films (Figure 1P) as compared with the complex and heavy nanoridges on the lip adaxial epidermal cells (Figure 1Q). Histochemistry staining with the lipophilic dye “Sudan Black B” revealed a lipid layer on the petal cuticle and lip adaxial epidermal cells (Figures 1R, S).
We analyzed and confirmed the spatial expression patterns of PeERF1 in both vegetative (root, leaf, and stalk) and reproductive organs (pedicle, bud, sepal, petal, lip, and column) of P. equestris (Figures 2A, B) by qRT-PCR. PeERF1 was highly expressed in reproductive organs (pedicle and flower bud) with lower expression in roots, and very low expression in leaf and stalk tissue. As expected, PeERF1 was highly expressed in the lip and column, with threefold expression in lip as compared with sepals and petals (Figure 2C).
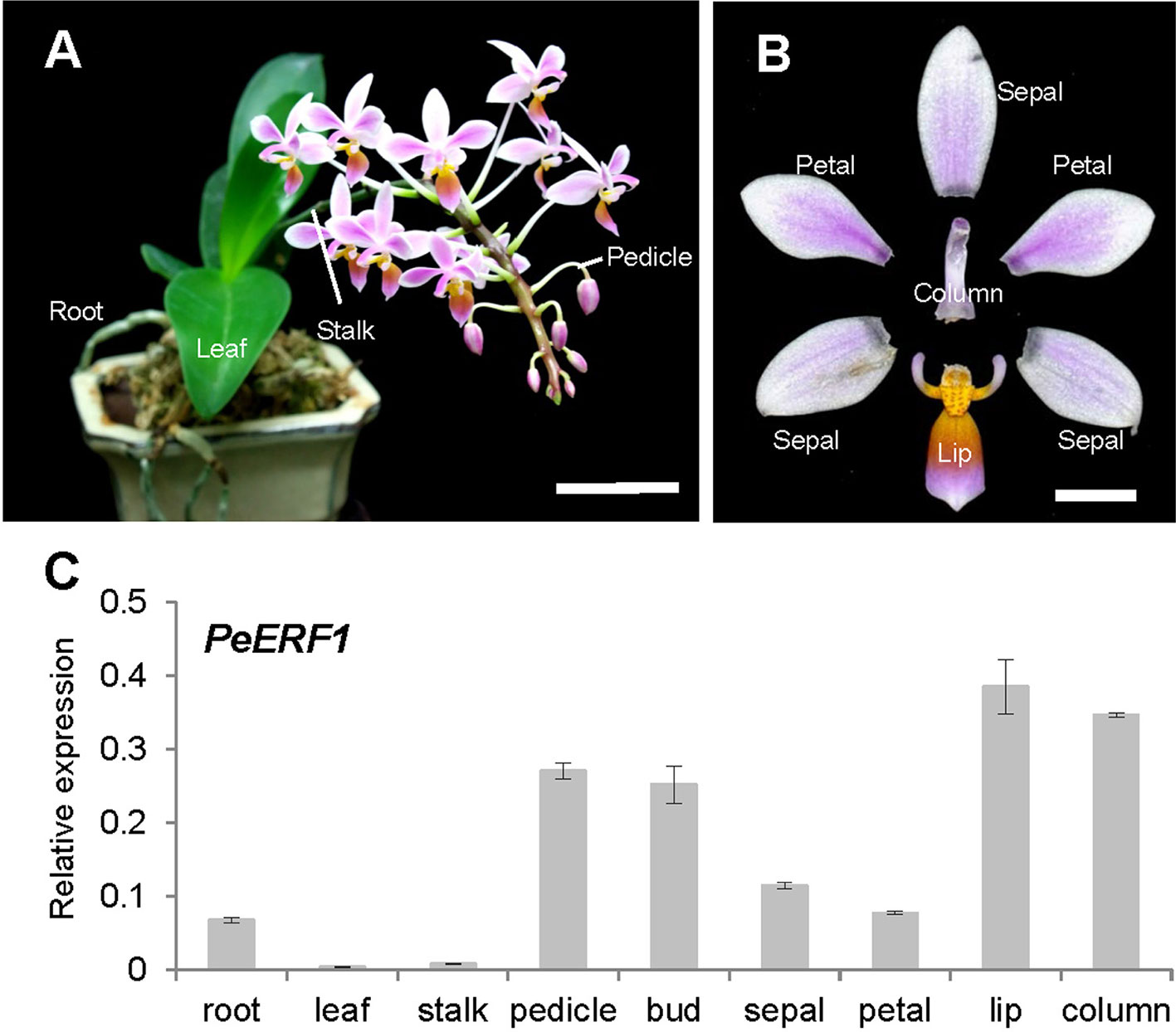
Figure 2 Spatial expression patterns of PeERF1 in the native species P. equestris. (A) Various vegetative and reproductive tissues were analyzed, including roots, leaves, pedicles, and stalks. Scale bar, 5 cm. (B) Flower organs were analyzed, including sepals, petals, lip, and column. Scale bar, 1 cm. (C) Spatial expression patterns of PeERF1 in various organs. Total RNA were extracted from variant tissues in three independent plants. Three technical repeats were performed for each sample. Data are mean ± SD. Numbers above the bars are expression levels after normalization with the internal control (PeActin4) (Chen et al., 2005).
Temporal expression of PeERF1 during lip morphogenesis was examined in five stages of lip development (Figures 3A–C). PeERF1 showed low and increasing expression from stage 1 to stage 4, with a sharp increase at stage 5 (Figure 3D).
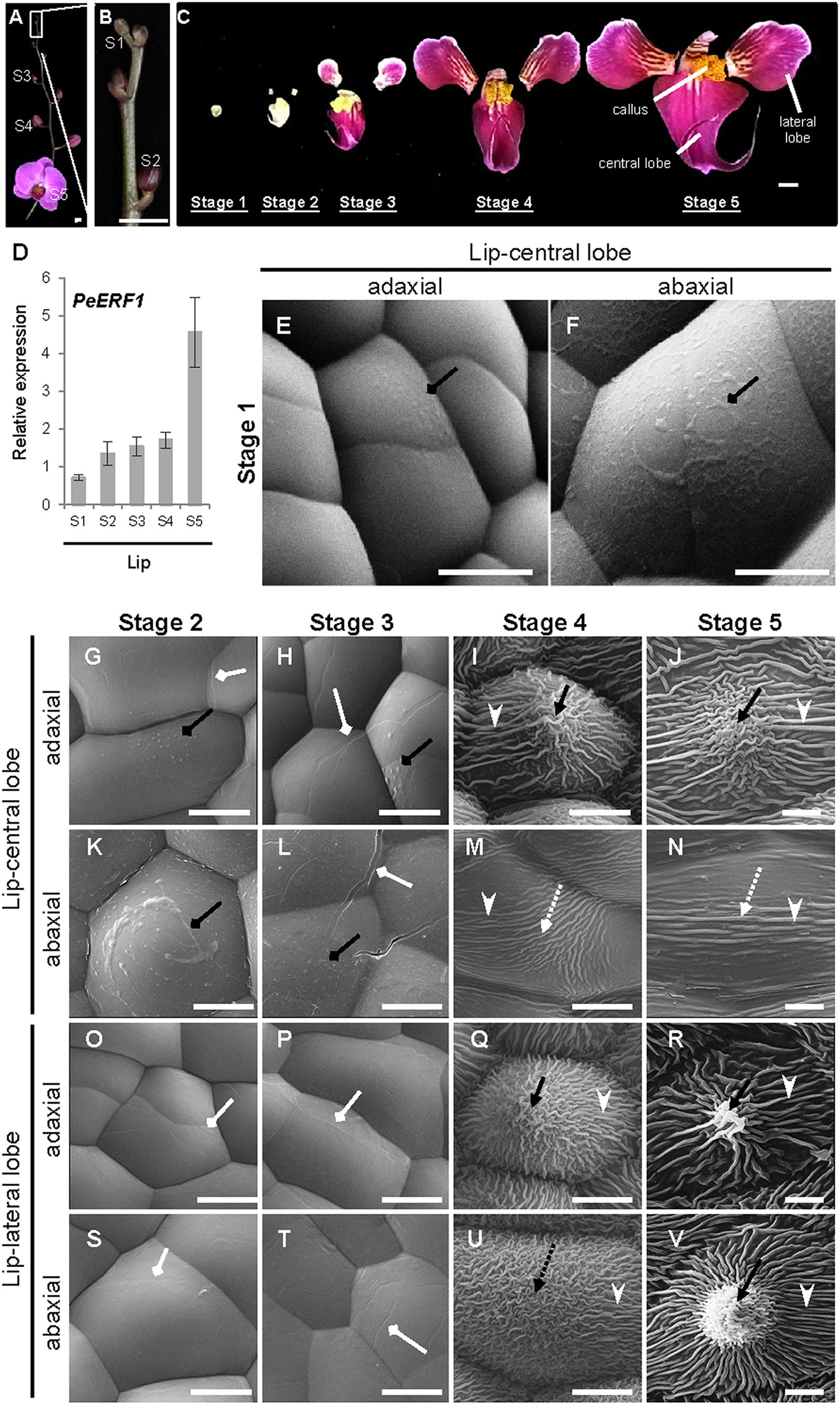
Figure 3 Temporal expression patterns of PeERF1 and the ultrastructure of lip epidermal cells during various developmental stages of P. OX Red Shoes “OX1408” flowers. (A–C) Various stages of lip development during floral morphogenesis. S1, stage 1, flower bud (0–0.5 cm); S2, stage 2, flower bud (0.5–1 cm); S3, stage 3, flower bud (1–2 cm); S4, stage 4, flower bud (2–3 cm); S5, stage 5, flowering. Scale bars, 2 cm (A–B) and 1 cm (C). (D) Temporal expression patterns of PeERF1 at various lip developmental stages. Total RNA were extracted from the various lip developmental stages in three independent plants. Three technical repeats were performed for each sample. Data are mean ± SD. Numbers above the bars are expression levels after normalization with the internal control (PeActin4). (E–V) Cyro-SEM of adaxial and abaxial sites of epidermal cells of different parts of lip organs during developmental stages. Black rhombus-head lines indicate the secreted bubble-like preliminary cuticle components. White rhombus-head lines indicate the traces of preliminary nanoridges. Black arrows indicate node-like nanoridges in the CF of the cell. Black dashed arrows indicate irregular nanoridges in the CF of the cell. White arrowheads indicate parallel nanoridges in the AF of the cell. Scale bars, 10 μm.
The ultrastructure of the lip epidermis during various developmental stages was examined under Cryo-SEM. Both adaxial and abaxial surfaces of epidermal cells in the lip-central lobe showed secreted bubble-like preliminary cuticle components and heavy cuticle layers from stages 1 to 3 (Figures 3E, F, G, H, K, L, black rhombus-head line). Traces of preliminary nanoridges started to form on the lip epidermis from stages 2 to 3 (Figures 3G, H, L, O, P, S, T, white rhombus-head line). These then thickened and the number of nanoridges increased from stages 4 to 5 (Figures 3I, J, M, N, Q, R, U, V).
The final mature forms of nanoridges varied in different parts of the lip. The adaxial epidermal cells of the central lobe showed anode-like nanoridges in the CF (Figure 3J, black arrow), and parallel and radial nanoridges in the AF (Figure 3J, white arrowhead). Similar nanoridges on adaxial epidermal cells of the central lobe were also found on the adaxial and abaxial epidermal cells of lateral lobes (Figure 3R, V). In contrast, the abaxial epidermal cells of the central lobe showed looser parallel nanoridges in the CF (Figure 3N, white dashed arrow) and AF (Figure 3N, white arrowhead). Thus, the typical lip epidermal features containing nanoridges are important markers of the lip morphological identity. These results suggest that PeERF1 may have a correlation with nanoridge formation during lip morphogenesis.
Phylogenetic analysis showed that PeERF1 belongs to the Va2 subgroup of the ERF subfamily of the AP2/EREBP family (Figure 4A), and the sequence alignment indicated that PeERF1 contains incomplete “SHINE domains” (CMV-1 and CMV-2 motifs) (Figure 4B). Of note, the protein sequences of incomplete “SHINE domains” of PeERF1 are distinguished from other members of the ERF-Va2 subgroup, which results in a separate branch from the other ERF-Va2 members and closer to the ERF-Va1 subgroup (Figure 4).
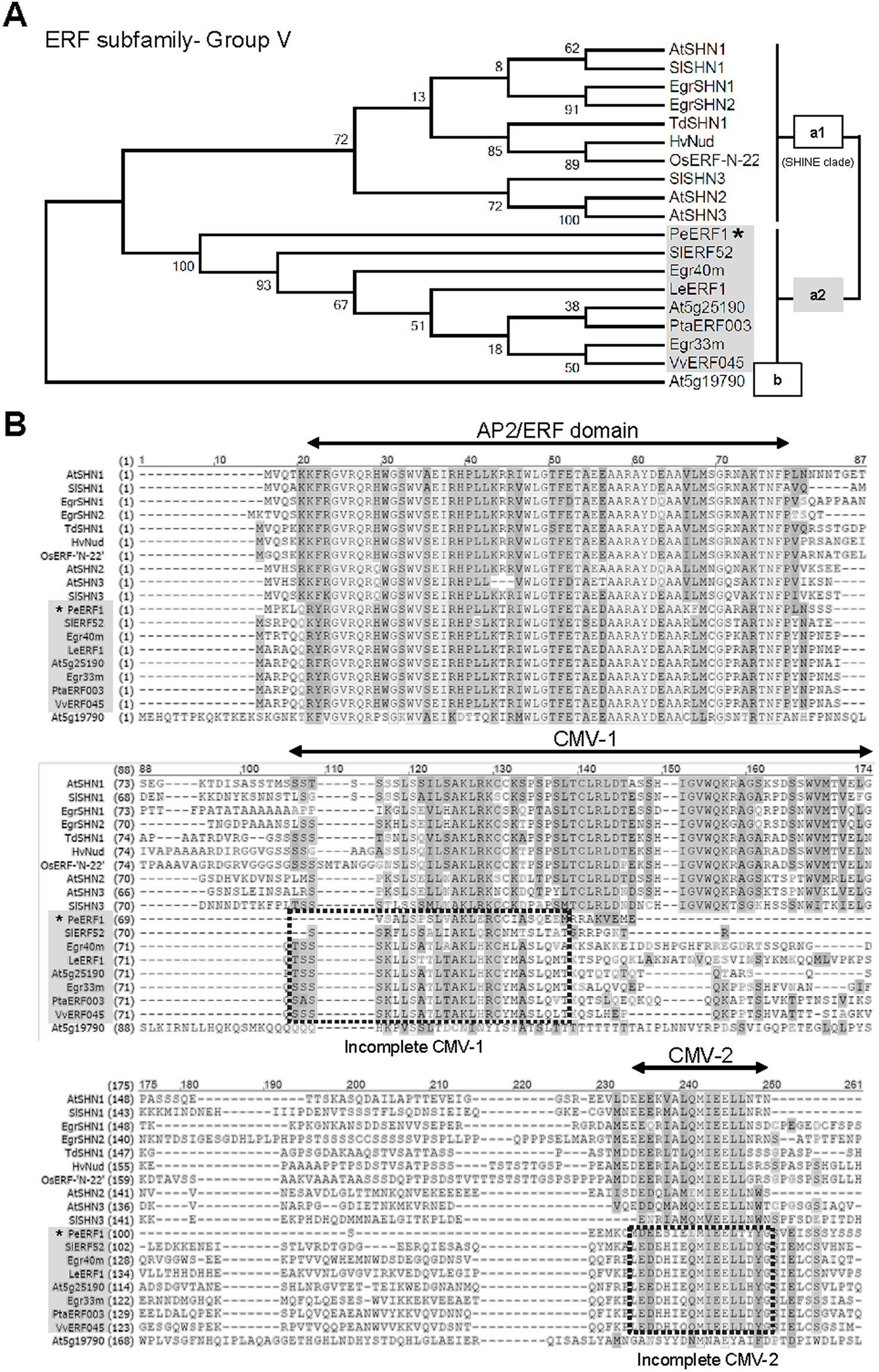
Figure 4 Phylogenetic analysis and sequence alignment of PeERF1. (A) Phylogenetic analysis of PeERF1 with published ERF-V group proteins. Bootstrap values were calculated with 1,000 replicates. PeERF1 is highlighted with a star (*). (B) Multiple alignment of amino acid sequences of PeERF1 and ERF-V group proteins. According to the existence of “SHINE domains” (CMV-1 and CMV-2 motifs), the ERF-V group was classified into two groups: Va group, with SHINE domains, and Vb group, without SHINE domains. In addition, the Va group was further divided into two subgroups: Va1 subgroup, with complete SHINE domains (deemed as “SHINE clade”), and Va2 subgroup, with incomplete SHINE domains.
To assess the role of PeERF1 in lip cuticle development, CymMV-based VIGS was used to silence its expression with a 142-nt conserved AP2 domain (139–281 nt) and the 175-nt incomplete “SHINE domains” (346–521 nt) in P. OX Red Shoes “OX1408.” This resulted in plants that were PeERF1_AP2-silenced and PeERF1_SHN-silenced (Supplementary Figure S2A). Using Cryo-SEM we examined the perianth epidermis micro-morphology from PeERF1_AP2-silenced plants. Lip epidermis nanoridges showed continuously altered distribution of structures within one plant from the 1st to the 7th flowers. We also found more severely altered phenotypes, with looser, thinner, uneven, and hollowed distribution of nanoridge structures especially on the lip epidermis of the 6th and the 7th flowers (Supplementary Figure S3). Flowers on these silenced plants showed no obvious difference in floral morphology as compared with mock-treated plants (Supplementary Figures S2B–D).
The most severe phenotype was observed on the 7th blooming flowers in PeERF1_AP2-silenced plants (Figure S3D). This was concomitant with the expression of PeERF1 at late stage of lip morphogenesis. We then further examined the cell surface on the lip epidermis of the 7th blooming flowers of both types of silenced plants (Figures 5A–L, top view; Supplementary Figure S4, side view). Looser and fewer nanoridges in the AF of adaxial and abaxial surfaces of central lobe epidermal cells were observed as compared with mock-treated plants (Figures 5A–F, white arrowhead). In addition, the AF showed uneven distribution of parallel and irregular nanoridges, which was hollow by reducing the coverage of nanoridges on adaxial and abaxial surfaces of lateral lobe epidermal cells (Figures 5H, I, K, L, white arrowhead; Figure 5M). Moreover, PeERF1_AP2-silenced plants showed unique, thicker, and extended nanoridges across anticlines of both the adaxial and abaxial surface of lip-lateral lobe epidermal cells (Figures 5H, K, black spherical-head line).
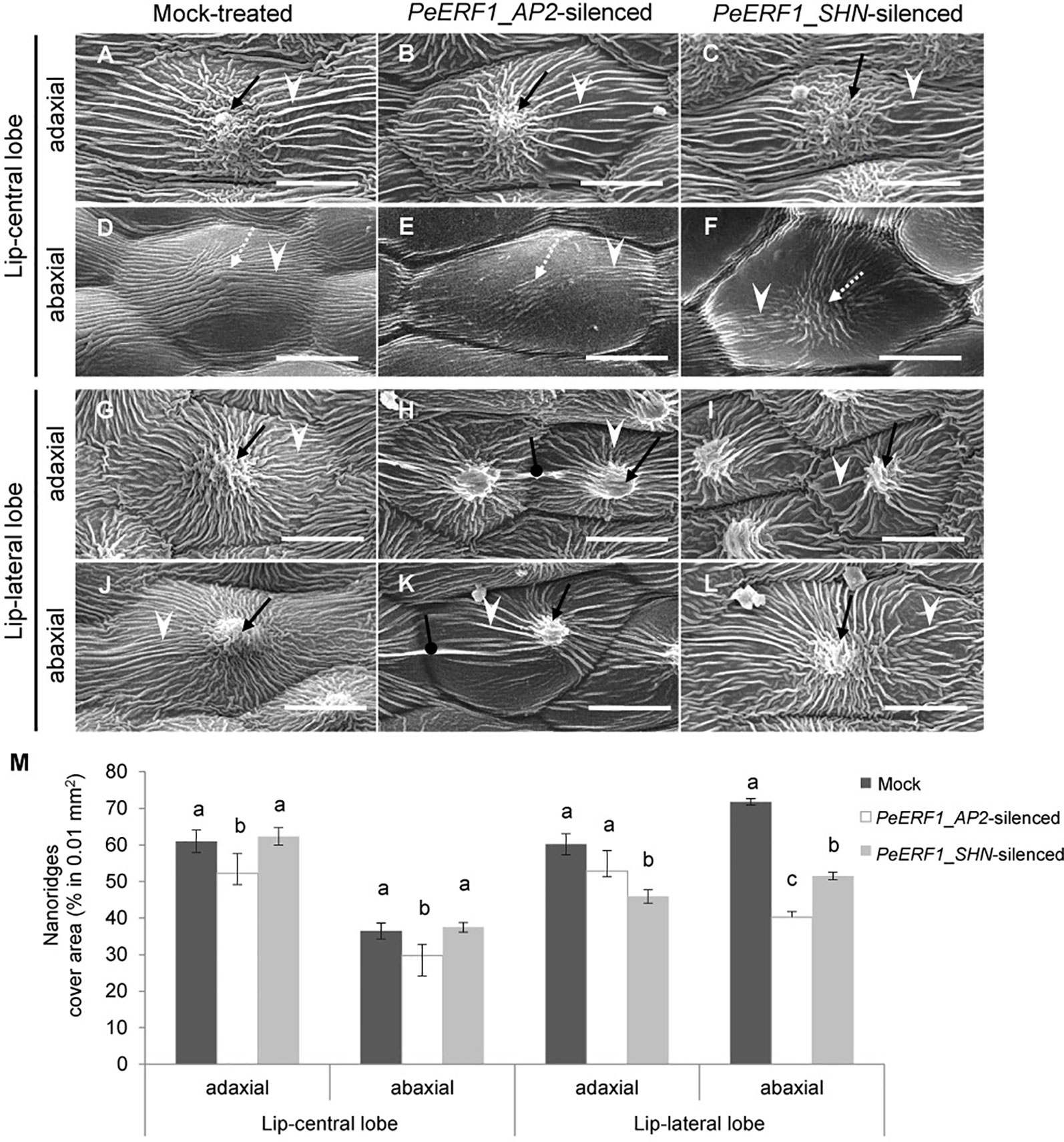
Figure 5 Nanoridge characteristics of ultrastructure of lip epidermal cells from 7th blooming flowers of PeERF1-silenced P. OX Red Shoes “OX1408.” (A–L) Cryo-SEM was used to examine the changes in cellular morphology from the 5th to the 8th blooming flowers of silenced plants (stage 5, floral diameter of 12 cm). Top view of adaxial and abaxial sites of lip central and lateral lobe epidermis in mock-treated, PeERF1_AP2-silenced, and PeERF1_SHN-silenced plants. Black arrows indicate node-like nanoridges in the CF of the cell. White dashed arrows indicate parallel nanoridges in the CF of the cell. White arrowheads indicate parallel nanoridges in the AF of the cell. Black spherical-head line indicates thicker and extended nanoridges across anticlines of cells. Scale bars, 30 μm. (M) The coverage of nanoridges on the top of adaxial and abaxial lip epidermal cells of mock-treated and PeERF1-silenced flowers. For Cryo-SEM examination, eight plants were injected with control, specific domain and conserved domain separately, and repeated twice. Data are mean ± SD (n = 15); the same letters above the bars indicate no statistical difference by Duncan's multiple range test (P < 0.05).
In contrast, the sepals and petals of all blooming flowers of PeERF1-silenced plants otherwise showed no obvious phenotypic changes in the adaxial and abaxial surface of epidermal cells (Supplementary Figure S5). PeERF1 and cutin biosynthesis genes were downregulated in both silenced plants; however, these cells do not have cuticle on their cell surface (Supplementary Figure S6).
Both PeERF1-silenced plants showed downregulation of PeERF1 (Figures 6A and S6A). However, the transcript level of PeSHN1 was much lower than that of PeERF1 (Figures 6A, B). We speculate that the downregulation of PeSHN1 was due to off-target effect since there are 71.8% and 47.1% identity between the AP2 and SHN domains of PeERF1 and PeSHN1 genes, respectively (Supplementary Figure S7). However, it is also possible that transcription of PeSHN1 is regulated by PeERF1 to some extent (Figures 6A, B and Supplementary Figure S6B).
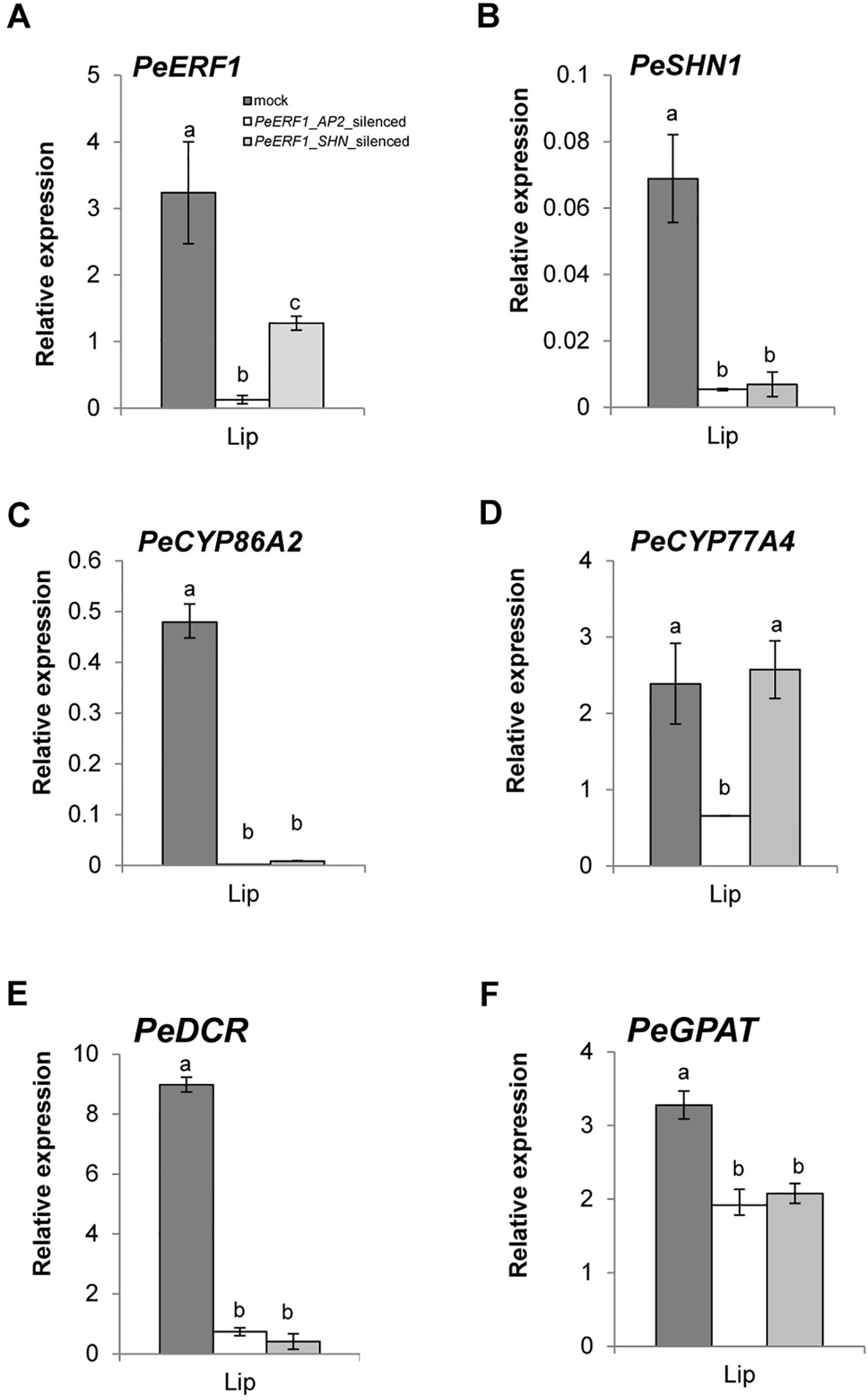
Figure 6 Expression patterns of PeERF1 and cuticle-associated genes in PeERF1-silenced plants. Transcript level of PeERF1 (A), PeSHN1 (B), and cutin metabolism-related genes (PeCYP86A2, PeCYP77A4, PeGPAT, and PeDCR) (C–F) in mock-treated and PeERF1-silenced flowers. Total RNA of sepal, petal, and lip were extracted from the 5th floral bud (stage 3, length of 1–2 cm) at 30 days post-inoculation. Three technical repeats and three biological repeats were performed for gene expression analysis in the PeERF1_silenced plants. Data are mean ± SD; the same letters above the bars indicate no statistical difference by Duncan's multiple range test (P < 0.05).
Next, we examined whether the expression of cuticle biosynthesis genes was altered upon the reduction of PeERF1 expression in the VIGS plants. Expression of PeERF1 in the lip was reduced significantly in PeERF1_AP2-silenced plants, but less so in PeERF1_SHN-silenced plants. PeCYP86A2 and PeCYP77A4, PeGPAT, PeGPAT, and PeDCR are putative Phalaenopsis orthologs of known cutin biosynthetic genes, which contribute to the cuticle formation in Phalaenopsis lips. Similar to the reduced expression of PeERF1 in the PeERF1_SHN-silenced plants, the expressions of PeCYP86A2, PeDCR, and PeCYP77A4 were reduced significantly in PeERF1_AP2-silenced plants (Figures 6C, D). The expressions of PeCYP86A2 and PeDCR were reduced, while the expression of PeCYP77A4 was not affected in the PeERF1_SHN-silenced plants (Figures 6C, D). The expression of PeGPAT was reduced in both PeERF1_AP2-silenced and PeERF1_SHN-silenced plants, but to a less extent (Figure 6F). Therefore, both PeCYP77A4 and PeGPAT were found to be crucial for cuticle formation, and their expressions were regulated by other TFs in addition to PeERF1 or PeSHN1.
From population of the P. hybrid “Join Big foot” grown from seedlings we selected two plants each with wild-type flower (normal split-lip organ) and with the big-lip enlarged petal-like lip flower. We observed these in order to confirm the association of PeERF1 and cuticle formation during lip morphogenesis (Figure 7A). Cryo-SEM revealed that cell morphology and ultrastructure were severely changed in the petal-like lip epidermal cells of the big-lip variant as compared with wild type (Figures 7B–Q). In wild type, the epidermal cells of split-lip organs showed an abnormal, uneven, and hollowed distribution of parallel nanoridges in the AF of cells (Figures 7B, D, F, H). In the big-lip variant, all adaxial and abaxial surfaces of the lip epidermal cells displayed similar characteristics of petal adaxial epidermal cells with a papilla cell shape with smooth 2-D wax films and a protuberance on the top of the cell (Figures 7C, E, G, I, K, M, O, Q; Supplementary Figures S8 and S9). The phenotypic observations were done in three big-foot variant plants. Lip morphology was altered to various levels, from severe to mild, yet the phenotypic change for nanoridge formation was the same (data not shown).
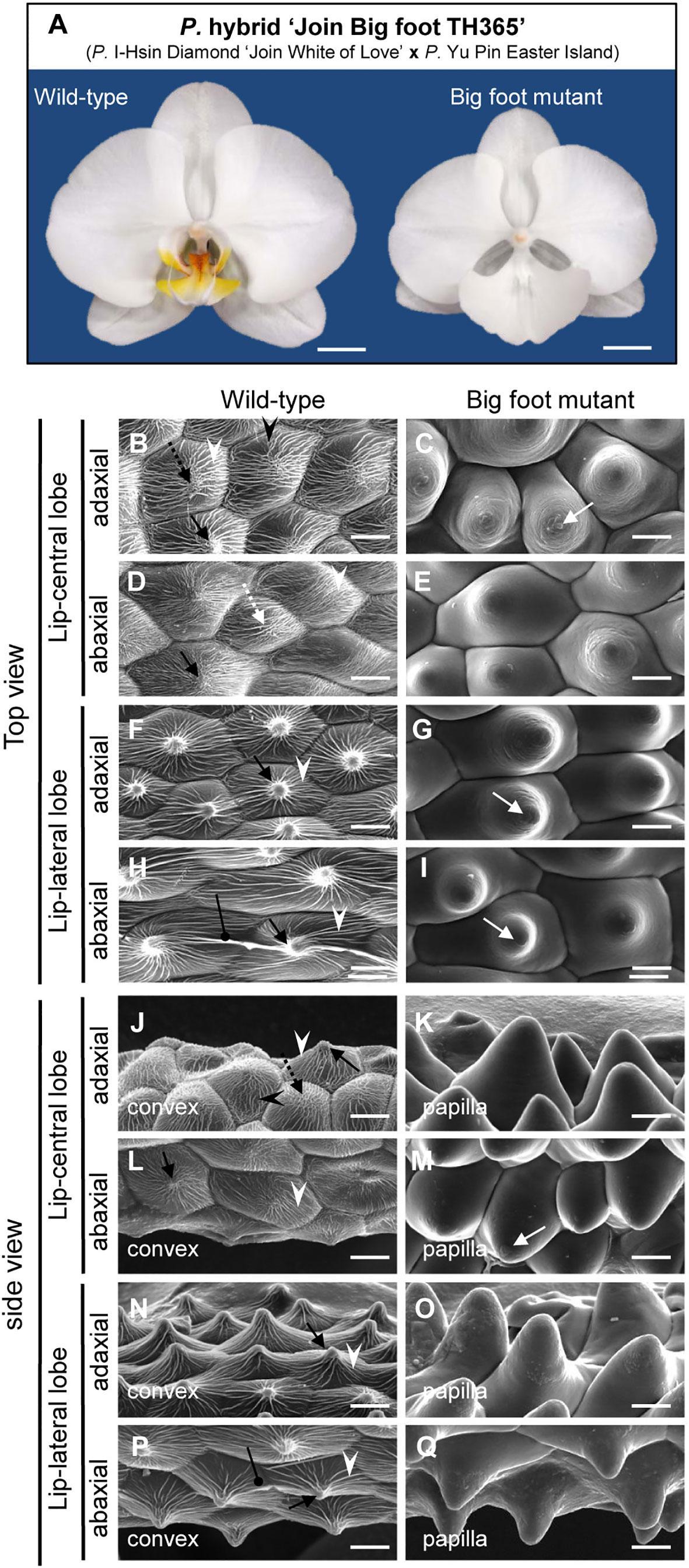
Figure 7 Floral morphology and ultrastructure of lip epidermal cells of somaclonal variants with normal lip or enlarged petal-like lip mutants. (A) The flowers of somaclonal variants of P. hybrid “Join Big foot TH365” (P. I-Hsin Diamond “Join White of Love” x P. Yu Pin Easter Island). The lip morphology of “big foot mutant” flower showed morphological conversions to petal-like structure. Scale bars, 2 cm. (B–Q) Top view and side view of adaxial and abaxial sites of lip central and lateral lobe epidermis in “wild-type” and “big foot mutant” of somaclonal variants of P. “Join Big foot TH365” flowers. White arrows indicate a protuberance on the top of the cell. Black arrows indicate node-like nanoridges in the CF of the cell. White dashed arrows indicate parallel nanoridges in the CF of the cell. Black dashed arrows indicate irregular nanoridges in the CF of the cell. White arrowheads indicate parallel nanoridges in the AF of the cell. Black arrowheads indicate irregular nanoridges in the AF of the cell. Black spherical-head line indicates thicker and extended nanoridges across anticlines of the cells. Different cell shape types of floral epidermal cells are in the lower left corner of each panel. Scale bars, 30 μm.
To further understand the molecular mechanisms of petal-like lip formation, we investigated gene expression of cutin biosynthesis genes PeERF1, PeSHN1 (PeCYP86A2, PeCYP77A4, PeDCR, and PeGPAT) (Figure 8), as well as expression of floral morphogenesis genes (B-class [DEFICIENS (DEF)/APETALA3 (AP3)-like (PeMADS2-5) and GLOBOSA (GLO)/PISTILLATA (PI)-like (PeMADS6)], AGAMOUS-LIKE6a (PeAGL6a), and E-class [SEPALLATA1-4 (PeSEP1-4)] MADS box genes) (Supplementary Figure S10).
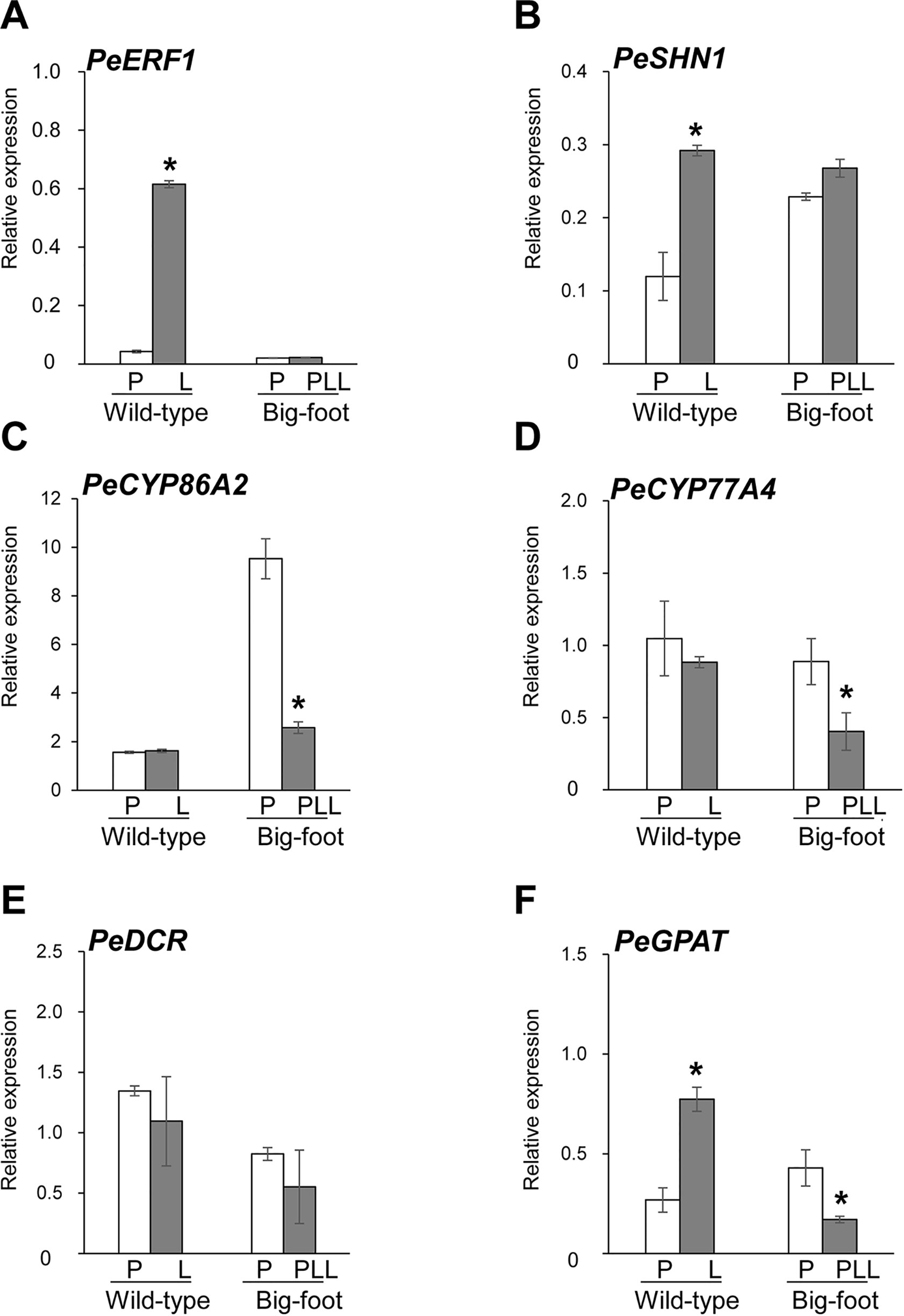
Figure 8 Expression patterns of PeERF1, PeSHN1, cutin-biosynthetic genes in petal and lip of “wild-type” and “big foot mutant” flowers of P. hybrid “Join Big foot TH365.” Transcript level of genes related to lip epidermis development PeERF1 (A), PeSHN1 (B), and cutin metabolism-related genes (PeCYP86A2, PeCYP77A4, PeGPAT, and PeDCR) (C–F) in flowers of “wild-type” and “big foot mutant” of P. “Join Big foot TH365” were examined. P, L, and PPL were represented as petal, lip, and petal-like lip, respectively. mRNA of petal and lip were extracted from the 2nd floral bud in three independent plants of big foot mutant and wild type. Three technical repeats were performed for each sample. Data are mean ± SD. Numbers above the bars are expression levels after normalization with internal control (PeActin4). *P < 0.05 by one-tailed t-test.
Expression of PeERF1 was significantly reduced in the lip of the big-foot variant as compared to the wild type. In addition, the expressions of PeCYP77A4 and PeGPAT were reduced in the big-foot variant lip compared to that of the wild-type plant, while expression levels of PeCYP86A2 and PeDCR were not significantly different between the big-foot variant and the wild type. Interestingly, expression of PeSHN1 was nearly unaffected for petal-like lip in big-foot variant as compared to that of lip in wild-type plant. These results suggest that PeERF1, PeCYP77A4, and PeGPAT were involved in the nanoridge formation in the orchid lip.
For MADS box genes, highest expressions of PeMADS3, 4 and 6, and PeAGL6a were detected in the lip of “wild-type” flowers, whereas PeMADS2 expressed higher in the petals (Figure 8A). In contrast, only PeMADS2 and PeMADS3 expressed higher in petal-like lip compared to the petal in the big-foot variant (Supplementary Figure S10). Intriguingly, we found little or no difference in transcriptional levels of PeMADS4~6 between the petal-like lip and the petals of the big-foot variant (Supplementary Figure S10).
To further characterize the biological function of PeERF1, we performed ectopic overexpression of PeERF1 under the control of CaMV 35S promoter by Agrobacterium-mediated transformation in Arabidopsis. Two batches of overexpression were performed. In the first batch, a total of 60 T1 transgenic lines were generated, and in the second batch of overexpression, a total of 48 transgenic lines were generated. Most of overexpressing lines showed SHINE phenotypes. Three overexpressing lines (L24, L37, and L52) from the first batch and three overexpression lines (P12, P13, and P15) from the second batch were used for further analysis (Figure 9). These plants have enhanced brilliant, shiny green color and curved-down edges rosette leaves as compared to wild-type plants (Figures 9A–C). For seedlings of 35S:PeERF1 transgenic lines, the adaxial surface of the second pair of true leaves show a shiny surface with few trichomes as compared with the wild type (Figures 9D, E). At the cellular level, the adaxial surfaces of rosette leaf epidermal cells of 35S:PeERF1 transgenic plants showed ectopic nanoridge formation and wax deposition on the surface, in contrast to the wild-type leaf with a smooth surface (Figures 9F–H, black arrow).
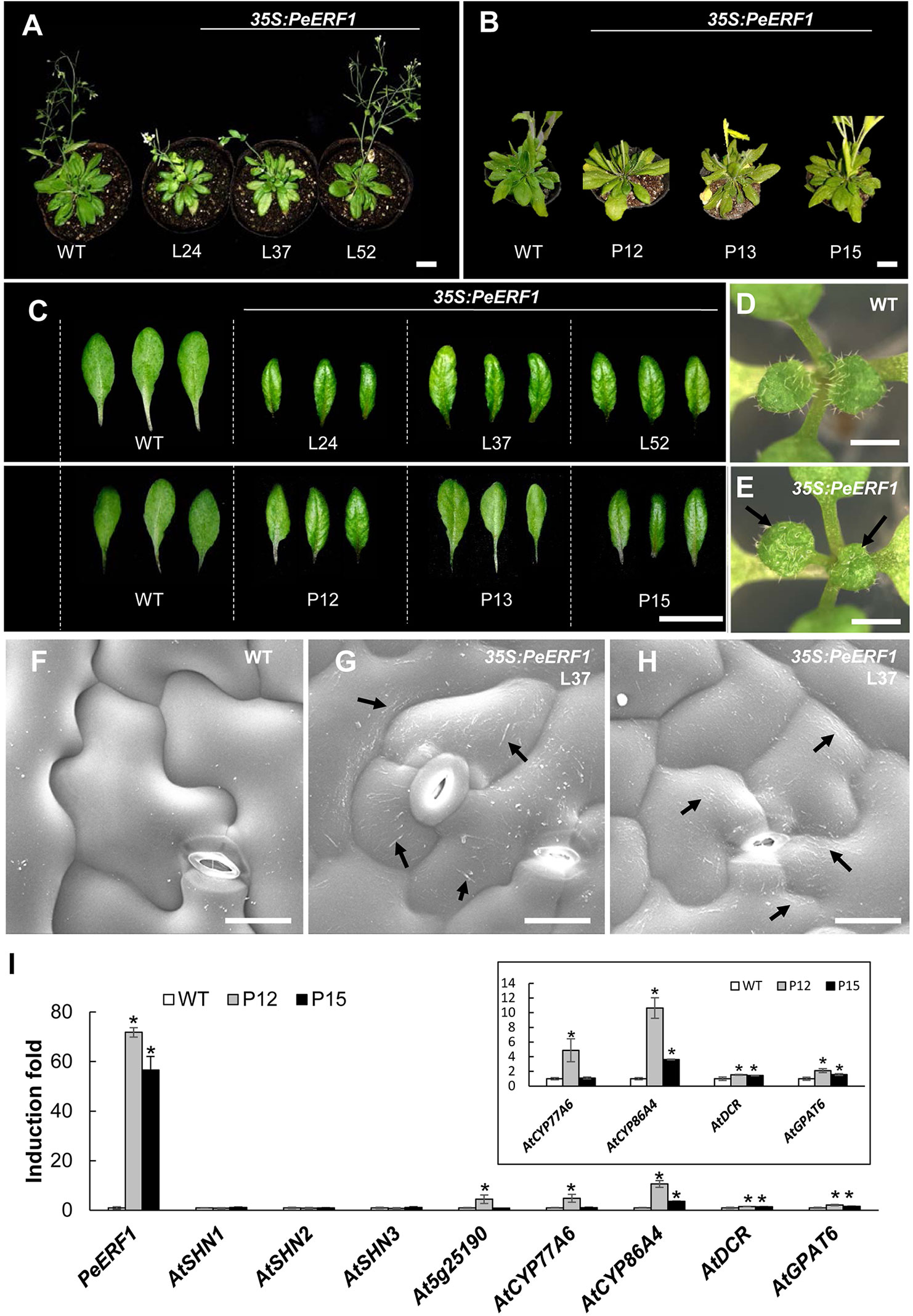
Figure 9 Phenotype analysis and expression patterns of transgenic Arabidopsis plants ectopically expressing PeERF1. (A) Wild-type and 35S:PeERF1 transgenic lines (the first batch, L24, L37, and L52) at 45 days old, and (B) wild-type and 35S:PeERF1 transgenic lines (the second batch, P12, P13, and P15) at 60 days old. Scale bars, 5 cm. (C) Shiny and curved-down edges of the rosette leaves of 45-day-old wild-type and 35S:PeERF1 transgenic plants of L24, L37 and L52, and 60-day-old wild-type and 35S:PeERF1 transgenic plants of P12, P13, and P15. Scale bars, 2 cm. (D–E) Seedlings of wild-type and 35S:PeERF1 transgenic plants. Black arrow indicates the top view of the second pair of true leaves with a shiny surface. (F–H) Micrograph images of adaxial surface of rosette leaf epidermal cells of wild-type and 35S:PeERF1 transgenic line 37. Black arrow indicates ectopic wax deposition in the AF and anticlines of the cell. Scale bars, 10 μm. (I) Expression patterns of PeERF1 and Arabidopsis cuticle-associated genes (AtSHN1-3, At5g25190, AtCYP86A4, AtCYP77A6, AtGPAT6, and AtDCR6) in wild type and 35S:PeERF1 overexpressing lines P12 and P15 were determined by qRT-PCR and normalized to the expression level of the Actin gene as an internal control. Fold induction of PeERF1 and 8 genes associated with cuticle biosynthesis in 35S:PeERF1 transgenic plants and wild-type plants, as compared to control. Asterisks were used to indicate statistically significant difference compared with wild-type plants. Three technical replicates were performed for each overexpression line and repeated in two different overexpression lines independently. Data are mean ± SD. *P < 0.05 by one-tailed t-test.
In addition, qRT-PCR was performed to examine the gene expression in the two transgenic lines with obvious phenotype, P12 and P15 from the second batch of overexpression. Ectopic expression of PeERF1 was accompanied by upregulation of cuticle biosynthesis genes, including AtCYP86A4, AtDCR, and AtGPAT6 in two independent lines (Figure 9I). Furthermore, the ectopic overexpression of PeERF1 in Arabidopsis did not disturb the expression of the three endogenous AtSHNs (AtSHN1~AtSHN3) as well as the PeERF1 orthologous gene At5g25190 in all ectopic overexpression lines except the expression of At5g25190 in the line P12 (Figure 9I). These results suggest that PeERF1, with similar SHINE phenotypes, might be assigned to one of the SHINE-like TFs and can be ectopically overexpressed and functionally involved in leaf cuticle development in Arabidopsis.
ERF-Va2 subgroup genes are not known to have a typical SHINE phenotype; rather, they are involved in various developmental and physiological processes (Aharoni et al., 2004; Li et al., 2007; Marques et al., 2013; Trupiano et al., 2013; Nakano et al., 2014; Leida et al., 2016). Recently, berry VvERF045 SHINE domains were found to be involved in berry ripening and epidermal cuticle development. Transgenic grapevine lines overexpressing VviERF045 show stunted growth, discolored, and smaller leaves, with reduced gene expression for epidermal wax decoration and wax biosynthesis. This indicates that VviERF045 is a potential repressor in epidermis patterning and cuticle development (Leida et al., 2016).
In this study, PeERF1_AP2-silenced plants showed significantly reduced expressions of PeERF1 and PeSHN1 with loose and uneven nanoridges on the lip epidermal surface, accompanied by drastically reduced expression of cutin biosynthesis genes including PeCYP86A2, PeCYP77A4, and PeDCR. Expression of PeGPAT, on the other hand, was affected only to a less extent (Figure 6). These results suggest that both PeERF1 and PeSHN1 are important for cuticle formation. Yet the expression level of PeSHN1 was much lower than that of PeERF1. Similar to the reduced expression of PeERF1 in the PeERF1_SHN-silenced plants, the expressions of PeCYP86A2, PeDCR, and PeCYP77A4 were reduced significantly in PeERF1_AP2-silenced plants. The expression of PeCYP77A4 was not affected in the PeERF1_SHN-silenced plants (Figure 6D). The expression of PeGPAT was reduced in both PeERF1_AP2-silenced and PeERF1-SHN plants, but to a lesser extent (Figure 6F). Therefore, it appears that PeCYP77A4 and PeGPAT are regulated by PeERF1 and/or PeSHN1 and downregulation of these genes might explain the cuticle formation defects in the PeERF1-silenced plants. Based on our observations, it is also suggested that proper expression of PeCYP77A4 and PeGPAT might involve additional TF(s). Thus, our results suggest that PeERF1 as a SHN-like homolog is a potential activator in the lip epidermal cell patterning of Phalaenopsis flowers. Furthermore, transgenic Arabidopsis lines overexpressing PeERF1 showed typical SHINE phenotypes similar to AtSHN-overexpressing Arabidopsis (Figures 9A–C). The PeERF1 overexpressing plants enhanced the expression of AtCYP86A4, AtDCR, and AtGPAT6 without disturbance of the expression of endogenous AtSHNs and the homologous gene At5g25190 except the expression of At5g25190 in the line P12 was increased (Figure 9I). With the fact that At5g25190 does not have the function for cuticle formation (Aharoni et al., 2004), the phenotype observed in the overexpressor P12 was due to the PeERF1 per se. Therefore, our results suggest that PeERF1, as a SHINE-like TF, increase the Arabidopsis cuticle-associated genes and result the shiny surface of rosette leaves.
Recent studies have indicated the importance of CMV-1 and CMV-2 motifs of SHINE domains. A highly conserved valine (V) residue in the complete CMV-1 motif in barley has been found to be associated with lipid biosynthesis of grain (Taketa et al., 2008). When V is changed to aspartic acid (D) in the complete CMV-1 motif, instead of the typical hulled caryopsis in barley, a naked one results. This is associated with lipid formation of caryopsis and hull adhesion (Taketa et al., 2008). In addition, the existence of a C-terminal 30-amino-acid region of the CMV-2 motif is also important for transcriptional activation of SlERF52 (Nakano et al., 2014). The protein sequences of PeERF1 were more distinguished from other members of the ERF-Va2 subgroup (Figure 4). We found that PeERF1 has a serine (S) residue at position 5 of the incomplete CMV-2 motif, whereas several members of the Va2 subgroup have a histidine (H) residue at the same position (Figure 4B).
Studies of SHINE genes modulating cuticle permeability and epidermal cell patterning have revealed the requirement of the downstream synthesis of cutin polyesters (Kannangara et al., 2007; Li-Beisson et al., 2009; Panikashvili et al., 2009; Shi et al., 2011; Shi et al., 2013; Petit et al., 2016; Mazurek et al., 2017). A recent model was formulated of the association between cutin biosynthesis and nanoridge formation of the petal cuticle (Mazurek et al., 2017). However, although several SHN putative downstream target genes related to cuticle formation have been reported, SHN TFs do not bind directly to most of their presumed targets. In fact, they require an interacting partner for SHN-mediated target regulation (Kannangara et al., 2007). So far, only the promoter regions of CYP86A cytochrome P450s (AtCYP86A4, AtCYP86A7, and SlCYP86A69) and GSDL-motif lipases (AtRXF26) have been found to be activated by SHNs in Arabidopsis and tomato (Shi et al., 2011; Shi et al., 2013).
When PeERF1 was silenced, we found associated downregulation of cutin biosynthesis gene expressions and reduced numbers of nanoridges on lip abaxial and adaxial surfaces. However, even though the expressions of PeCYP86A2 and PeDCR were significantly reduced, that of PeGPAT was only mildly reduced in lip epidermis in both PeERF1-silenced plants (Figure 6). Thus, PeCYP86A2 and PeDCR may be downstream genes of PeSHN1, and PeCYP77A4 a downstream gene of PeERF1. PeGPAT was regulated by PeERF1, PeSHN1, and non-SHN like TFs. Hence, PeERF1 as a SHN-like TF is involved in decorating the floral organ epidermal surface by regulating downstream cutin biosynthesis genes.
Recently, the roles of B- and E-class MADS-box genes were revealed for orchid tepal development (Tsai et al., 2004; Tsai et al., 2005; Lu et al., 2007; Hsieh et al., 2013a; Su et al., 2013b; Pan et al., 2014; Hsu et al., 2015b; Huang et al., 2015; Huang et al., 2016). We have shown that knock-down expression of E-class PeSEP1-4 genes reduced the expression of PeERF1. PeSEP1-silenced plants had a changed ultrastructure in terms of nanoridge formation, of the floral epidermal cells (Pan et al., 2014). This result suggests that PeERF1 could be a downstream gene of E-class MADS box genes.
In this study, we found that the expression of PeERF1 was reduced to almost zero in the big-foot somaclonal variants with a petal-like lip, along with the loss of gene expression of B-class (PeMADS4, PeMADS5, and PeMADS6) and E-class (PeAGL6a) MADS box genes (Supplementary Figure S10). These results suggest that the developmental program of lip morphogenesis is very complex, and PeERF1 is a downstream target gene of B- and E-class MADS box genes and is involved in lip morphogenesis.
Our previous VIGS results of MADS box silencing data showed the highest silencing efficiency at 4–7 weeks post-silencing with the first four flowers blooming after viral inoculation (Hsieh et al., 2013a; Hsieh et al., 2013b; Pan et al., 2014; Hsu et al., 2015a). This indicates that MADS box genes are induced in early floral morphogenesis.
PeSHN1 as an SHN ortholog contained the complete SHINE domains and was assigned to the ERF-Va1 subgroup (SHINE clade) (Figure 4). However, PeERF1 was expressed at late stage lip morphogenesis so that VIGS phenotype was more distinct on the late emerged floral buds. The identity of PeSHN1 with PeERF1_AP2-silenced and PeERF1_SHN-silenced regions were 71.8% and 47.1%, respectively. Yet, no contiguous matches were longer than 11 nt for the PeSHN1 coding region and the VIGS fragment (Supplementary Figure S7). A nucleotide identity of less than 11 nt on target mRNA has previously been shown to reduce the chances of silencing induction (Senthil-Kumar and Mysore, 2011; Hsieh et al., 2013a). The off-target effects of PeSHN1 silencing that occurred in all PeERF1-silenced plants may be due to sequence similarity. The combined effects of specific silencing of PeERF1 and off-target silencing of PeSHN1 together resulted in a change of nanoridge formation in lip epidermal cell patterning in the PeERF1-silenced plants, yet PeSHN1 expressed much lower than PeERF1 in the Phalaenopsis orchids.
To contribute to successful sexual reproduction in higher plants, the perianth of flower creates various cues, such as tactile, visual, and olfactory signals, to reward or not reward (deceive) pollinators. In Orchidaceae, approximately one-third of orchid species have a deceit pollination strategy by using general floral signals without rewarding pollinators with nectar or pollen (Ackerman, 1986; Nilsson, 1992; Jersáková et al., 2006). Although numerous Phalaenopsis species are scentless, with a diversity of colorful perianths, floral morphology, and floral scents, bees are the major pollinators of Phalaenopsis flowers via food-deceptive pollination (Roman Kaiser, 1993; Hsiao et al., 2006; Tsai et al., 2008).
In this study, we examined the floral epidermal cell surfaces of Phalaenopsis species (Figure 1; Supplementary Figure S1); the tissues of sepals and petals had a papillae shape with a protuberance on the smooth epidermal surface, whereas the lip harbored numerous nanoridges on the epidermal surface. The biological function of nanoridges on the floral organs for pollinator attraction has been linked with the unique visual and tactile signals they produce (Whitney et al., 2009; Prüm et al., 2011; Rands et al., 2011; Whitney et al., 2011b; Prüm et al., 2012; Kourounioti et al., 2013; Prüm et al., 2013; Adachi et al., 2015; Moyroud et al., 2017).
We propose three steps of a special deceit pollination strategy in the scentless Phalaenopsis species. First, the papillae cell shape on the sepal and petal epidermis creates big brilliant visual cues to attract pollinators from a distance. Then, the convex cell shape with heavy nanoridges on the top of the lip epidermal cells generates more “flashy” visual cues (similar to guide lights on an aircraft runway) and direct pollinators to land on the lip instead of sepals and petals. Finally, the distinctive split lip with central lobes and lateral lobes creates a tunnel-like structure, and the nanoridges on the lip epidermis generate tactile cues that help pollinators walk and explore the area. During this process, because of the heavy and dense nanoridges on the top of the lip epidermis, the pollinators may slip toward the column and attach to the pollinia. While visiting subsequent flowers, successful pollination may occur when attached pollinia are placed into the stigmatic cavity underneath the column. The slippery quality of nanoridges for beetles has been reported (Prüm et al., 2011), and the slip-and-fall pollination mechanism related to the ultrastructural characterization of the floral lip was also shown for Gongora bufonia (Orchidaceae) (Adachi et al., 2015).
In conclusion, our results demonstrate that PeERF1, as an SHN-like TF, was involved in lip epidermal cell morphological formation at the last flowering stage by regulating lip nanoridge development in Phalaenopsis flowers. In addition, the heavy nanoridges on the lip epidermis, as typical lip features, may be essential for the pollination mechanism of Phalaenopsis. This study gives a better understanding of the transcriptional regulation of the late stage development of lip morphogenesis and the special pollination mechanism of Phalaenopsis.
The datasets generated for this study can be found in the PeERF1, MG948436; PeSHN1, XM_020736987.1; PeCYP86A2, XM_020732683.1; PeCYP77A4, XM_020725159.1; PeGPAT, XM_020727266.1; PeDCR, XM_020725429.1.
P-HL, W-HC, and H-HC conceived the research plans. P-HL performed most of the experiments, analyzed the data, and wrote the article with contributions from all the authors. L-MH assisted in the identification and expression analysis of cutin biosynthesis genes. Z-JP assisted with the performance of VIGS experiments. W-NJ and M-CC performed Cryo-SEM analysis and provided service for Cryo-scanning electron microscope.
This work was supported by grant no. MOST-107-2313-B-006-003-MY3 from the Ministry of Science and Technology, Taiwan.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Ming-Hsien Hsieh (Tainan District Agricultural Research and Extension Station, Council of Agriculture, Taiwan) for providing the greenhouse and for assistance with operating the VIGS experiment, and Dr. Shau-Ting Chiu (Biology Department, National Museum of Natural Science, Taichung, Taiwan) for helpful discussion of floral epidermal morphology. We also thank Oxen Biotechnology Corp. (Tainan, Taiwan) and Join Orchids Corp. (Tainan, Taiwan) for providing the plant materials for of P. OX Red Shoes “OX1408” and P. “Join Big foot TH365,” respectively.
AF, anticlinal field; CF, central field; Cryo-SEM, Cryo-scanning electron microscopy; ERF, ethylene responsive factor; RT-PCR, reverse transcription–polymerase chain reaction; SHN/WIN, SHINE/WAX INDUCER; TFs, transcription factors; VIGS, virus-induced gene silencing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01709/full#supplementary-material
Ackerman, J. D. (1986). Mechanisms and evolution of food-deceptive pollination systems in orchids. Lindleyana 1, 108–113.
Adachi, S. A., Machado, S. R., Guimarães, E. (2015). Structural and ultrastructural characterization of the floral lip in Gongora bufonia (Orchidaceae): understanding the slip-and-fall pollination mechanism. Botany 93, 759–768. doi: 10.1139/cjb-2015-0114
Aharoni, A., Dixit, S., Jetter, R., Thoenes, E., van Arkel, G., Pereira, A. (2004). The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16, 2463–2480. doi: 10.1105/tpc.104.022897
Al-Abdallat, A. M., Al-Debei, H. S., Ayad, J. Y., Hasan, S. (2014). Over-expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in tomato. Int. J. Mol. Sci. 15, 19499–19515. doi: 10.3390/ijms151119499
Broun, P., Poindexter, P., Osborne, E., Jiang, C.-Z., Riechmann, J. L. (2004). WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 4706–4711. doi: 10.1073/pnas.0305574101
Chao, Y. T., Yen, S. H., Yeh, J. H., Chen, W. C., Shih, M. C. (2017). Orchidstra 2.0-A Transcriptomics Resource for the Orchid Family. Plant Cell Physiol. 58, e9–e9. doi: 10.1093/pcp/pcw220
Chen, Y. H., Tsai, Y. J., Huang, J. Z., Chen, F. C. (2005). Transcription analysis of peloric mutants of Phalaenopsis orchids derived from tissue culture. Cell Res. 15, 639–657. doi: 10.1038/sj.cr.7290334
Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J., Lee, Y. I., et al. (2012). C-and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiol. 53, 1053–1067. doi: 10.1093/pcp/pcs048
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Cozzolino, S., Widmer, A. (2005). Orchid diversity: an evolutionary consequence of deception? Trends In Ecol. Evol. 20, 487–494. doi: 10.1016/j.tree.2005.06.004
Fu, C. H., Chen, Y. W., Hsiao, Y. Y., Pan, Z. J., Liu, Z. J., Huang, Y. M., et al. (2011). OrchidBase: a collection of sequences of the transcriptome derived from orchids. Plant Cell Physiol. 52, 238–243. doi: 10.1093/pcp/pcq201
Hsiao, Y. Y., Tsai, W. C., Kuoh, C. S., Huang, T. H., Wang, H. C., Wu, T. S., et al. (2006). Comparison of transcripts in Phalaenopsis bellina and Phalaenopsis equestris (Orchidaceae) flowers to deduce monoterpene biosynthesis pathway. BMC Plant Biol. 6, 14. doi: 10.1186/1471-2229-6-14
Hsiao, Y. Y., Huang, T. H., Fu, C. H., Huang, S. C., Chen, Y. J., Huang, Y. M., et al. (2013). Transcriptomic analysis of floral organs from Phalaenopsis orchid by using oligonucleotide microarray. Gene 518, 91–100. doi: 10.1016/j.gene.2012.11.069
Hsieh, M. H., Lu, H. C., Pan, Z. J., Yeh, H. H., Wang, S. S., Chen, W. H., et al. (2013a). Optimizing virus-induced gene silencing efficiency with Cymbidium mosaic virus in Phalaenopsis flower. Plant Sci. 201, 25–41. doi: 10.1016/j.plantsci.2012.11.003
Hsieh, M. H., Pan, Z. J., Lai, P. H., Lu, H. C., Yeh, H. H., Hsu, C. C., et al. (2013b). Virus-induced gene silencing unravels multiple transcription factors involved in floral growth and development in Phalaenopsis orchids. J. Exp. Bot. 64, 3869–3884. doi: 10.1093/jxb/ert218
Hsu, C. C., Chen, Y. Y., Tsai, W. C., Chen, W. H., Chen, H. H. (2015a). Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 168, 175–191. doi: 10.1104/pp.114.254599
Hsu, H. F., Hsu, W. H., Lee, Y. I., Mao, W. T., Yang, J. Y., Li, J. Y., et al. (2015b). Model for perianth formation in orchids. Nat. Plants 1, 15046. doi: 10.1038/nplants.2015.46
Huang, J. Z., Lin, C. P., Cheng, T. C., Chang, B. C. H., Cheng, S. Y., Chen, Y. W., et al. (2015). A de novo floral transcriptome reveals clues into Phalaenopsis orchid flower development. PloS One 10, e0123474. doi: 10.1371/journal.pone.0123474
Huang, J. Z., Lin, C. P., Cheng, T. C., Huang, Y. W., Tsai, Y. J., Cheng, S. Y., et al. (2016). The genome and transcriptome of Phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ J. Life Environ. Sci. 4, e2017. doi: 10.7717/peerj.2017
Jäger, K., Miskó, A., Fábián, A., Deák, C., Kiss-Bába, E., Polgári, D., et al. (2015). Expression of a WIN/SHN-type regulator from wheat triggers disorganized proliferation in the Arabidopsis leaf cuticle. Biol. Plantarum 59, 29–36. doi: 10.1007/s10535-014-0471-0
Jeffree, C. E. (2006). Annual Plant Reviews Volume 23: Biology of the Plant Cuticle ed Riederer, M., Müller, C. (Oxford, UK: Blackwell Publishing), 11–125.
Jersáková, J., Johnson, S. D., Kindlmann, P. (2006). Mechanisms and evolution of deceptive pollination in orchids. Biol. Rev. Cambridge Philos. Soc. 81, 219–235. doi: 10.1017/S1464793105006986
Kannangara, R., Branigan, C., Liu, Y., Penfield, T., Rao, V., Mouille, G. G., et al. (2007). The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19, 1278–1294. doi: 10.1105/tpc.106.047076
Kay, Q. O. N., Daoud, H. S., Stirton, C. H. (1981). Pigment distribution, light reflection and cell structure in petals. Bot. J. Linn. Soc. 83, 57–83. doi: 10.1111/j.1095-8339.1981.tb00129.x
Kim, M. J., Ruzicka, D., Shin, R., Schachtman, D. P. (2012). The Arabidopsis AP2/ERF transcription factor RAP2. 11 modulates plant response to low-potassium conditions. Mol. Plant 5, 1042–1057. doi: 10.1093/mp/sss003
Koch, K., Bhushan, B., Barthlott, W. (2008). Diversity of structure, morphology and wetting of plant surfaces. Soft Mater. 4, 1943–1963. doi: 10.1039/B804854A
Koch, K., Bhushan, B., Barthlott, W. (2009a). Multifunctional surface structures of plants: an inspiration for biomimetics. Prog. In Mater. Sci. 54, 137–178. doi: 10.1016/j.pmatsci.2008.07.003
Koch, K., Bohn, H. F., Barthlott, W. (2009b). Hierarchically sculptured plant surfaces and superhydrophobicity. Langmuir 25, 14116–14120. doi: 10.1021/la9017322
Kourounioti, R. L. A., Band, L. R., Fozard, J. A., Hampstead, A., Lovrics, A., Moyroud, E., et al. (2013). Buckling as an origin of ordered cuticular patterns in flower petals. J. R. Soc. Interface 10, 20120847. doi: 10.1098/rsif.2012.0847
Leida, C., Dal Rì, A., Dalla Costa, L., Gómez, M. D., Pompili, V., Sonego, P., et al. (2016). Insights into the role of the berry-specific ethylene responsive factor VviERF045. Front. In Plant Sci. 7, 1793. doi: 10.3389/fpls.2016.01793
Li, Y., Zhu, B., Xu, W., Zhu, H., Chen, A., Xie, Y., et al. (2007). LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 26, 1999–2008. doi: 10.1007/s00299-007-0394-8
Li-Beisson, Y., Pollard, M., Sauveplane, V., Pinot, F., Ohlrogge, J., Beisson, F. (2009). Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc. Natl. Acad. Sci. U.S.A. 106, 22008–22013. doi: 10.1073/pnas.0909090106
Lu, H. C., Chen, H. H., Tsai, W. C., Chen, W. H., Su, H. J., Chang, C. N., et al. (2007). Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 143, 558–569. doi: 10.1104/pp.106.092742
Marques, W. L., Salazar, M. M., Camargo, E. L. O., Lepikson-Neto, J., Tiburcio, R. A., do Nascimento, L. C., et al. (2013). Identification of four Eucalyptus genes potentially involved in cell wall biosynthesis and evolutionarily related to SHINE transcription factors. Plant Growth Regul. 69, 203–208. doi: 10.1007/s10725-012-9754-7
Mawlong, I., Ali, K., Kurup, D., Yadav, S., Tyagi, A. (2014). Isolation and characterization of an AP2/ERF-type drought stress inducible transcription factor encoding gene from rice. J. Plant Biochem. Biotechnol. 23, 42–51. doi: 10.1007/s13562-012-0185-3
Mazurek, S., Garroum, I., Daraspe, J., De Bellis, D., Olsson, V., Mucciolo, A., et al. (2017). Connecting the molecular structure of cutin to ultrastructure and physical properties of the cuticle in petals of Arabidopsis. Plant Physiol. 173, 1146–1163. doi: 10.1104/pp.16.01637
Mondragón-Palomino, M., Theißen, G. N. (2008). MADS about the evolution of orchid flowers. Trends Plant Sci. 13, 51–59. doi: 10.1016/j.tplants.2007.11.007
Moyroud, E., Wenzel, T., Middleton, R., Rudall, P. J., Banks, H., Reed, A., et al. (2017). Disorder in convergent floral nanostructures enhances signalling to bees. Nature 550, 469. doi: 10.1038/nature24285
Nakano, T., Suzuki, K., Fujimura, T., Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nakano, T., Fujisawa, M., Shima, Y., Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nilsson, L. A. (1992). Orchid pollination biology. Trends In Ecol. Evol. 7, 255–259. doi: 10.1016/0169-5347(92)90170-G
Pan, Z. J., Cheng, C. C., Tsai, W. C., Chung, M. C., Chen, W. H., Hu, J. M., et al. (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 52, 1515–1531. doi: 10.1093/pcp/pcr092
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C., et al. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202, 1024–1042. doi: 10.1111/nph.12723
Panikashvili, D., Shi, J. X., Schreiber, L., Aharoni, A. (2009). The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol. 151, 1773–1789. doi: 10.1104/pp.109.143388
Petit, J., Bres, C., Mauxion, J. P., Tai, F. W., Martin, L. B., Fich, E. A., et al. (2016). The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 171, 894–913. doi: 10.1104/pp.16.00409
Prüm, B., Seidel, R., Bohn, H. F., Speck, T. (2011). Plant surfaces with cuticular folds are slippery for beetles. J. R. Soc. Interface, rsif20110202. doi: 10.1098/rsif.2011.0202
Prüm, B., Seidel, R., Bohn, H. F., Speck, T. (2012). Impact of cell shape in hierarchically structured plant surfaces on the attachment of male Colorado potato beetles (Leptinotarsa decemlineata). Beilstein J. Nanotechnol. 3, 57. doi: 10.3762/bjnano.3.7
Prüm, B., Bohn, H. F., Seidel, R., Rubach, S., Speck, T. (2013). Plant surfaces with cuticular folds and their replicas: influence of microstructuring and surface chemistry on the attachment of a leaf beetle. Acta Biomater. 9, 6360–6368. doi: 10.1016/j.actbio.2013.01.030
Rands, S. A., Glover, B. J., Whitney, H. M. (2011). Floral epidermal structure and flower orientation: getting to grips with awkward flowers. Arthropod-Plant Interact. 5, 279–285. doi: 10.1007/s11829-011-9146-3
Robinson, H., Burns-Balogh, P. (1982). Evidence for a primitively epiphytic habit in Orchidaceae. Syst. Bot. 7, 353–358. doi: 10.2307/2418670
Roman Kaiser, G. R. (1993). The scent of orchids: olfactory and chemical investigations (Dübendorf, Switzerland, and Elsevier, Amsterdam: Amer Orchid Society).
Rudall, P. J., Bateman, R. M. (2002). Roles of synorganisation, zygomorphy and heterotopy in floral evolution: the gynostemium and labellum of orchids and other lilioid monocots. Biol. Rev. Cambridge Philos. Soc. 77, 403–441. doi: 10.1017/S1464793102005936
Senthil-Kumar, M., Mysore, K. S. (2011). New dimensions for VIGS in plant functional genomics. Trends In Plant Sci. 16, 656–665. doi: 10.1016/j.tplants.2011.08.006
Shi, J. X., Malitsky, S., De Oliveira, S., Branigan, C., Franke, R. B., Schreiber, L., et al. (2011). SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PloS Genet. 7, e1001388. doi: 10.1371/journal.pgen.1001388
Shi, J. X., Adato, A., Alkan, N., He, Y., Lashbrooke, J., Matas, A. J., et al. (2013). The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol. 197, 468–480. doi: 10.1111/nph.12032
Su, C. L., Chao, Y. T., Yen, S. H., Chen, C. Y., Chen, W. C., Chang, Y. C. A., et al. (2013a). Orchidstra: an integrated orchid functional genomics database. Plant Cell Physiol. 54, e11–e11. doi: 10.1093/pcp/pct004
Su, C. L., Chen, W. C., Lee, A. Y., Chen, C. Y., Chang, Y. C. A., Chao, Y. T., et al. (2013b). A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PloS One 8, e80462. doi: 10.1371/journal.pone.0080462
Taketa, S., Amano, S., Tsujino, Y., Sato, T., Saisho, D., Kakeda, K., et al. (2008). Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc. Natl. Acad. Sci. U.S.A. 105, 4062–4067. doi: 10.1073/pnas.0711034105
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 10, 2731–9. doi: 10.1093/molbev/msr121.
Trupiano, D., Yordanov, Y., Regan, S., Meilan, R., Tschaplinski, T., Scippa, G. S., et al. (2013). Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 238, 271–282. doi: 10.1007/s00425-013-1890-4
Tsai, W. C., Chen, H. H. (2006). The orchid MADS-box genes controlling floral morphogenesis. Sci. World J. 6, 1933–1944. doi: 10.1100/tsw.2006.321
Tsai, W. C., Kuoh, C. S., Chuang, M. H., Chen, W. H., Chen, H. H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 45, 831–844. doi: 10.1093/pcp/pch095
Tsai, W. C., Lee, P. F., Chen, H. I., Hsiao, Y. Y., Wei, W. J., Pan, Z. J., et al. (2005). PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 46, 1125–1139. doi: 10.1093/pcp/pci125
Tsai, W. C., Hsiao, Y. Y., Pan, Z. J., Hsu, C. C., Yang, Y. P., Chen, W. H., et al. (2008). Molecular biology of orchid flowers: With emphasis on Phalaenopsis. Adv. In Bot. Res. 47, 99–145. doi: 10.1016/S0065-2296(08)00003-7
Tsai, W. C., Fu, C. H., Hsiao, Y. Y., Huang, Y. M., Chen, L. J., Wang, M., et al. (2013). OrchidBase 2.0: comprehensive collection of Orchidaceae floral transcriptomes. Plant Cell Physiol. 54, e7–e7. doi: 10.1093/pcp/pcs187
Whitney, H. M., Kolle, M., Andrew, P., Chittka, L., Steiner, U., Glover, B. J. (2009). Floral iridescence, produced by diffractive optics, acts as a cue for animal pollinators. Science 323, 130–133. doi: 10.1126/science.1166256
Whitney, H. M., Bennett, K. M. V., Dorling, M., Sandbach, L., Prince, D., Chittka, L., et al. (2011a). Why do so many petals have conical epidermal cells? Ann. Bot. 108, 609–616. doi: 10.1093/aob/mcr065
Whitney, H. M., Glover, B. J., Walker, R., Ellis, A. G. (2011b). The contribution of epidermal structure to flower colour in the South African flora. Curtis’s Bot. Mag. 28, 349–371. doi: 10.1111/j.1467-8748.2011.01762.x
Keywords: AP2/EREBP, cutin biosynthesis genes, lip, nanoridge, orchid, Phalaenopsis, transcription factor
Citation: Lai P-H, Huang L-M, Pan Z-J, Jane W-N, Chung M-C, Chen W-H and Chen H-H (2020) PeERF1, a SHINE-Like Transcription Factor, Is Involved in Nanoridge Development on Lip Epidermis of Phalaenopsis Flowers. Front. Plant Sci. 10:1709. doi: 10.3389/fpls.2019.01709
Received: 23 July 2019; Accepted: 04 December 2019;
Published: 30 January 2020.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Hirokazu Tanaka, Meiji University, JapanCopyright © 2020 Lai, Huang, Pan, Jane, Chung, Chen and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Hwa Chen, aGhjaGVuQG1haWwubmNrdS5lZHUudHc=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.