- 1Key Laboratory of Horticulture Science for Southern Mountains Regions of Ministry of Education, College of Horticulture and Landscape Architecture, Southwest University, Chongqing, China
- 2State Cultivation Base of Crop Stress Biology for Southern Mountainous Land of Southwest University, Academy of Agricultural Sciences of Southwest University, Chongqing, China
- 3College of Forestry and Landscape Architecture, South China Agricultural University, Guangzhou, China
Double-flower Eriobotrya japonica, of which one phenotype is homeotic transformation of sepals into petals, is a new germplasm for revealing the molecular mechanisms underlying the floral organ transformation. Herein, we analyzed the sequence, expression pattern and functional characterization of EjPI, which encoded a B-class floral homeotic protein referred to as PISTILLATA ortholog, from genetically cognate single-flower and double-flower E. japonica. Phylogenetic analysis suggested that the EjPI gene was assigned to the rosids PI/GLO lineage. Analysis of protein sequence alignments showed that EjPI has typical domains of M, I, K, and C, and includes a distinctive PI motif at the C-terminal region. Compared with asterids PI/GLO lineage, the K1 and K3 subdomains of EjPI both contain a single amino acid difference. Subcellular localization of EjPI was determined to be in the nucleus. Expression pattern analysis revealed that EjPI expressed not only in petals, filament, and anther in single-flower E. japonica, but also in petaloid sepals in double-flower E. japonica. Meanwhile, there were high correlation between EjPI transcript level and petaloid area within a sepal. Furthermore, 35S::EjPI transgenic wild-type Arabidopsis caused the homeotic transformation of the first whorl sepals into petaloid sepals. Ectopic expression of EjPI in transgenic pi-1 mutant Arabidopsis rescued normal petals and stamens. These results suggest expression pattern of EjPI is associated with the formation of petaloid sepal. Our study provides the potential application of EjPI for biotechnical engineering to create petaloid sepals or regulate floral organ identity in angiosperms.
Introduction
Structural diversification of flower organs has often been considered to be function requirements of floral pollination/ecology biology (Endress, 2006). Understanding the underlying mechanisms for the diversification of floral organs has long been a challenge in angiosperms (Schwarz-Sommer et al., 1990; Frohlich, 2006; Irish, 2010). However, the most well-known ABCE-model maintains that each whorl organ identity is determined in a combinational way of four class homeotic proteins, termed A, B, C, and E class proteins (Bowman et al., 1991; Coen and Meyerowitz, 1991; Theißen, 2001; Theissen and Saedler, 2001; Litt and Kramer, 2010). According to this model, sepal identity is specified by the combinational A and E class proteins; petal formation is regulated by the combination of A, B, and E class proteins; stamen identity is specified by the combinational B, C, and E class proteins; carpel formation is regulated by the combination of C and E class proteins. All the A, B, C, and E proteins, except for APETALA2, belong to MIKC-type MADS-box transcription factors (Irish, 2010).
The MIKC-type MADS-box transcription factors are identified originally as floral homeotic proteins, and exhibit the characteristic domains from N- to C-terminus: a MADS (M), an intervening (I), a keratin-like (K), and a C-terminal (C) domains (Purugganan et al., 1995; Theißen et al., 1996; Theissen et al., 2000; Kaufmann et al., 2005; Litt and Kramer, 2010). Among these domains, the M domain, which is the most highly conserved region, contributes to the dimerization and nuclear localization (Gramzow and Theissen, 2010). The I domain, a relatively weakly conserved region, is also important for the DNA-binding dimer formation (Kaufmann et al., 2005). By contrast, the relatively conserved K domain, which allows a coiled-coil secondary structure by the formation of three-segment amphipathic helices, is involved in the formation of multimeric complex and protein dimerization (Yang and Jack, 2004; Puranik et al., 2014). Finally, the C domain, which is quite variable, contributes to the transcriptional activation and the formation of multimeric complex (Kaufmann et al., 2005; Theißen and Gramzow, 2016). Sequence differences of the MADS-box proteins from different flowering plant species has been used to clarify the evolution and diversification of floral organ identity (Theißen et al., 2016).
PISTILLATA (PI) homologues, which encode floral homeotic B-function MADS-box transcription factors, play a crucial role in the specification of petal and stamen identities in angiosperms (Goto and Meyerowitz, 1994; Hernandez-Hernandez et al., 2007; Whipple et al., 2007; Irish, 2009; Theißen et al., 2016). Molecular evolution of PI lineages indicates that the PI homologues are generated from a major duplication event of an ancestral gene, and their encoding proteins include a highly conserved PI motif in most angiosperms (Kramer et al., 1998). Therefore, functional diversification or conservation of PI orthologous genes need to be focused after the duplication events in different clades of angiosperms. The altered expressional patterns of PI orthologs in some angiosperm shaped floral organ diversification (Soltis et al., 2007; Viaene et al., 2009; Chen et al., 2012; Hofer et al., 2012; Sasaki et al., 2014; Jing et al., 2015; Dodsworth, 2017). However, the expression pattern and functional roles of PI ortholog in Eriobotrya remain unclear.
Double flower is one of the earliest documented examples of floral mutants (Meyerowitz et al., 1989). In many land plants, double-flower cultivars are selected as ornamentals and provide resources for elucidating the genetic difference between normal and double-flower phenotypes (Dubois et al., 2010; Galimba et al., 2012). Compared with the single-flower phenotype in angiosperms, extra petals of double flower are from homeotic transformation of the first whorl sepals or the third whorl stamens. At present, the formation of double flower in few species has been reported and mainly focus on the transformation of stamens into petaloid organ. Recent studies indicated that the transformation from stamen to petal in double flowers is associated with expression patterns of C-class genes (Dubois et al., 2010; Galimba et al., 2012; Liu et al., 2013; Ma et al., 2018). However, the regulatory mechanisms underlying the transformation from sepals to petals need further research.
Eriobotrya japonica, a Chinese originated evergreen tree, belongs to the family Rosaceae and is cultivated broadly in tropical and subtropical regions (Lin et al., 1999). The single-flower E. japonica has four normal floral whorls, which include four sepals in the first whorl, four petals in the second whorl, numerous stamens in the third whorl and five carpels in the fourth whorl (Figures 1A, D). However, the double-flower E. japonica, a recently discovered natural variation, has four floral whorls including four homeotic conversional petaloid organ from sepals in the first whorl, (Figures 1B–F), four petals in the second whorl, numerous stamens in the third whorl and five carpels in the fourth whorl. In this study, we isolated and identified EjPI gene, a PI ortholog, from genetically cognate single-flower and double-flower E. japonica. Analyses of protein sequence alignment and phylogenetic tree showed that EjPI is a typical B-class MADS-box genes and assigned to the rosids PI/GLO lineage. Expression pattern analysis suggested that EjPI expressed not only in petals, filament and anther in single-flower E. japonica, but also in petaloid sepals in double-flower E. japonica. Meanwhile, the expression level of EjPI was highly correlated with petaloid area within a sepal. The 35S::EjPI transgenic wild-type Arabidopsis caused the first whorl sepals replaced into petaloid sepals. Ectopic expression of the EjPI in homozygous pi-1 mutant Arabidopsis rescued normal petals and stamens. These results reveal that expression pattern of EjPI is associated with the formation of petaloid sepal in double-flower E. japonica. Our study contributes to better understand the roles of EjPI for homeotic transformation of sepals into petaloid organs in double-flower E. japonica.
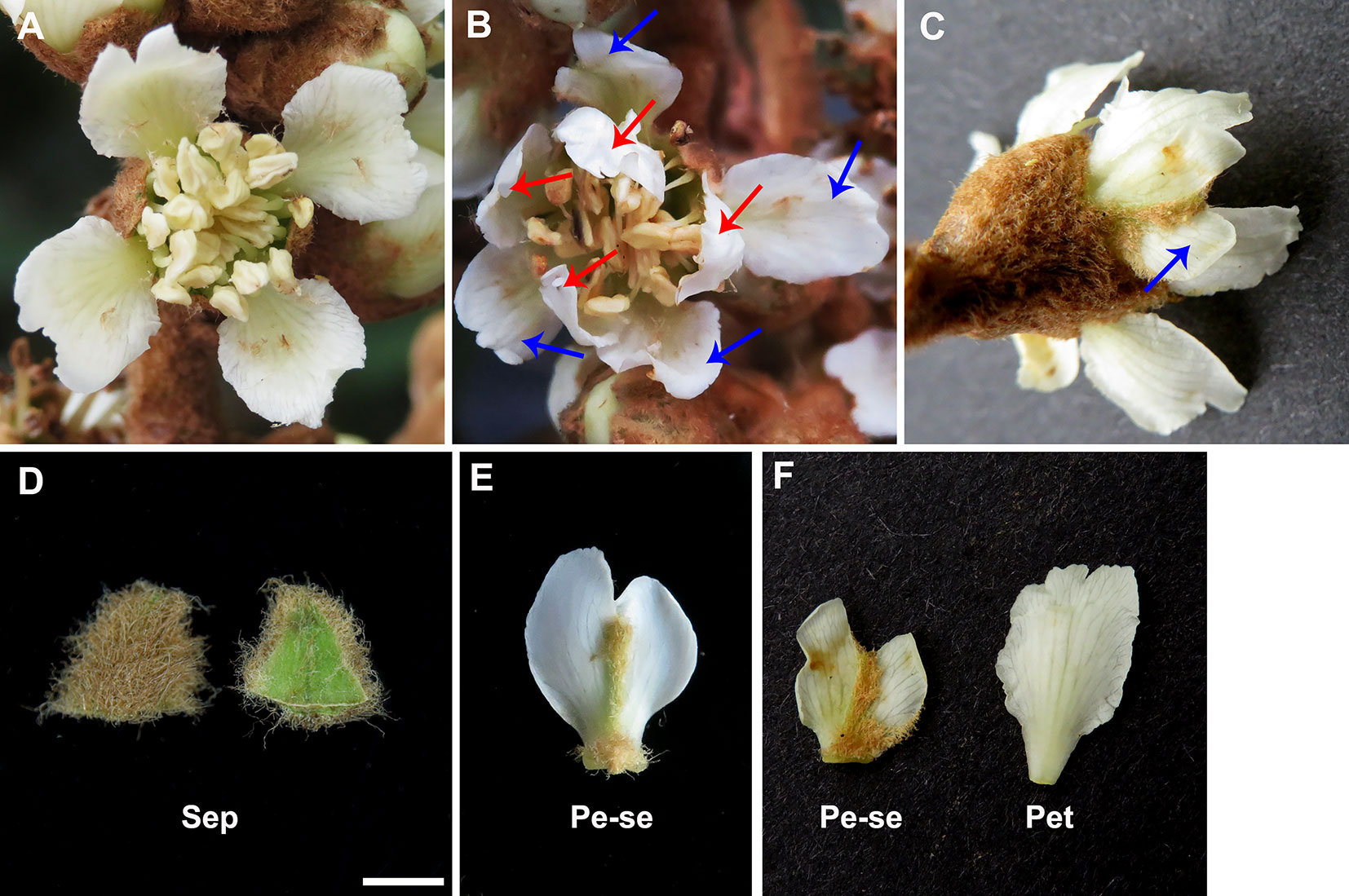
Figure 1 Comparative morphological observation in the single-flower and double-flower of E. japonica. (A) Single-flower E. japonica; (B) Double-flower E. japonica, showing homeotic conversional petaloid sepals from sepals in the first whorl (blue arrows) and petals in the second whorl (red arrows). (C) Petaloid sepals (blue arrow) in double-flower E. japonica. (D) Sepal. (E) Petaloid sepal. (F) Comparison of petaloid sepal and petal. Sep, sepal; Pe-se, petaloid sepals; Pet, petal.
Materials and Methods
Plant Materials
At different development stages, flower buds from the single-flower and double-flower E. japonica were collected from an experimental farm of Southwest University (Chongqing, China). Sepals, petals, filaments, anthers, and carpels were sampled from single-flower E. japonica. The petaloid sepals, petals, filaments, anthers, and carpels from double-flower E. japonica were collected, and immediately frozen in liquid nitrogen. The seeds of pi-1 mutant Arabidopsis (Landsberg erecta, CS77) were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA).
Isolation of EjPI in E. japonica
Total RNA from floral buds of single-flower and double-flower E. japonica was extracted using EASYspin plant RNA Extraction kit (RN09, Aidlab, China). The 3′ rapid amplification of cDNA ends (RACE) of EjPI was conducted from the DNase I-treated RNA using a 3′-full RACE Core Set Version 2.0 kit (Takara, Japan). The primers of 3′ RACE, 3REjPI1, and 3REjPI2 were designed according to the conserved sequences of PI orthologs from related species in Rosaceae, such as Kerria japonica (GenBank accession number MH161203), Pyrus pyrifolia (KP164019), Prunus pseudocerasus (KM243373), and Malus domestica (AB081092). Then, first PCR was conducted using the gene-specific primer 3REjPI1. The nested PCR was conducted using the gene specific primer 3REjPI2. Furthermore, the primers of 5′ RACE, 5REjPI1, and 5REjPI2 were designed based on the obtained sequences of 3′ RACE. The 5′ partial cDNA of EjPI was isolated using the SMARTer RACE 5′/3′ kit (Takara, Japan). Then, first PCR was conducted using the gene specific primer 5REjPI1. The nested PCR was conducted using the gene specific primer 5REjPI2. To further verify the full-length cDNA sequence of EjPI, PCR was conducted using the primers of FLEjPIF and FLEjPIR. PCR parameters was at 94°C denaturation step for 5 min, followed by 35 cycles of 50 s at 94°C, 50 s annealing at 57°C and 50 s extension at 72°C, with a final extension period of 72°C for 10 min. These primers of PCR are shown in Table S1.
Sequence Alignments and Phylogenetic Analysis
The BLAST analysis of deduced amino acid sequences of EjPI was performed on the Genbank database. Multiple A, B, C, and E class proteins were selected for alignment from various angiosperm lineages. Accession numbers of these proteins are listed in the Table S2. Amino acid sequences of these proteins, which contain the M, I, K, and C domains, were aligned using a ClustalW program (Thompson et al., 1994). A phylogenetic tree was constructed using MEGA 5.0 software (Kumar et al., 2008; Tamura et al., 2011) and the method described by Jing et al. (2015). Parameters of the phylogenetic tree were the bootstrap of 1,000 replicates, substitution model of Jones–Taylor–Thornton (Jones et al., 1992), uniform rates, nearest neighbor interchange, and the complete deletion of gaps/missing data. Lower than 50% of the values at each node was hidden.
Subcellular Localization, Semi-Quantitative Reverse Transcription PCR and Quantitative Real-Time PCR (qRT-PCR)
Subcellular localization of EjPI was detected using the modified pCAMBIA 1300 vector (Liu et al., 2017) and Agrobacterium-mediated transient transformation in Nicotiana benthamiana leaves. Then, fluorescence signals of green fluorescent protein (GFP) were observed via fluorescent microscopy using an Observer DP80 (Olympus, Japan). As a control, the vector expressing GFP alone was used.
Total RNA concentrations of sepals, petaloid sepals, petals, filament, anther, and carpels were assayed using a Nanodrop 2000 Spectrophotometer (Thermo Scientific, USA). To synthesize first-strand cDNA, 2 µg of the DNase I-treated RNA was used with oligo (dT)-18 adaptor primer and M-MLV reverse transcriptase (Takara, Japan). Then, semi-quantitative RT-PCR was conducted using 2 µl of the cDNA, the forward primer of RTEjPIF and the reverse primer of RTEjPIR. The PCR products from each reaction were determined using a 1% agarose gel electrophoresis. The experiments were assayed for three independent biological replicates for each sample. As an internal control, the ACTIN gene of E. japonica was used with the specific primers RTEjactinF and RTEjactinR (Shan et al., 2008). These primers of semi-quantitative PCR are shown in Table S3.
Flower organ samples (200 mg) were collected, respectively from sepals, petals, and different petaloid sepals, i.e., ~34%, ~45%, ~65%, and ~86% petaloid area within a sepal. Three independent biological replicates were collected for each sample. Then, total RNA from sepals, petaloid sepals and petals, was extracted individually and treated with RNase-free DNase I (Takara, Japan). To generate the first-strand cDNA, 2 µg of total RNA were used using PrimeScript RT reagent Kit with gDNA Eraser (Takara, Japan). Then, cDNA was added in a 20 µL PCR reaction with the primers of QEjPIF and QEjPIR. The qRT-PCR were assayed using the SYBR green I (Takara, Japan) and CFX96 Touch Real-time PCR Detection System (Bio-Rad, USA). The reaction mixture was cycled using the previous parameters described by Jing et al. (2015). As an internal control, the E. japonica β-actin was used to normalize small differences in template amounts with the primers qEjactinF and qEjactinR (Shan et al., 2008). These primers of qRT-PCR are shown in Table S3. Three biological replicates for each sample were conducted. The relative quantification of gene expression level was determined by the 2-ΔΔCT method (Livak and Schmittgen, 2001).
Vectors Construction and Arabidopsis Transformation
Coding sequences of EjPI was cloned into a pBI121 vector (BD Biosciences Clontech, USA) using restriction enzymes of XbaI and SmaI (Takara, Japan). The 35S::EjPI construct was transformed into heterozygous pi-1 mutant Arabidopsis lines via the Agrobacterium tumefaciens strain GV3101-90 using the floral-dip method described by Clough and Bent (1998). The seeds of transgenic Arabidopsis lines were selected using the previous method described by Jing et al. (2015). Then, the seedlings were transplanted in soil. The 35S::EjPI transgenic lines were detected by PCR and qRT-PCR. Genotype of transgenic Arabidopsis lines were identified using the primers of PI-1MF and PI-1MR designed by the dCAPS Finder program (Neff et al., 2002; Lamb and Irish, 2003; Jing et al., 2015). After genotyping, transgenic Arabidopsis lines of wild-type and homozygous pi-1 mutant were observed. For qRT-PCR analysis of transgenic Arabidopsis lines, three biological replicates were performed using the primers of QEjPIF and QEjPIR, qApiF and qApiR. The Actin gene of Arabidopsis was used to normalize small differences in template amounts with the primers of qAactinF and qAactinR (Zhang et al., 2014). The primers of these PCR are shown in Table S3.
Analysis of Scanning Electron Microscopy
Epidermal cells of floral organ from different Arabidopsis lines were fixed in 2.5% glutaraldehyde solution at 4°C for 48 h. The materials were dehydrated in a graded ethanol series and introduced at a critical point into the liquid CO2. The dried samples were coated with gold-palladium using a Hitachi E-1010 sputter Coater (Hitachi, Japan). Epidermal cells of the samples were observed using a FEI-Quanta 200F scanning electron microscope (FEI Company, Hillsboro, USA).
Results
Isolation and Sequence Analyses of EjPI
To isolate the EjPI sequence, we performed RACE technique to obtain full-length cDNA of EjPI from E. japonica flower bud (Figure S1). The EjPI cDNA sequence was 988 base pairs (bp) including a 61-bp of 5′ untranslated region (UTR), 648-bp open reading frame and 279-bp 3′ UTR with a poly-A tail at 3′-end (Figure S2). Isoelectric points and molecular weight of EjPI protein is 8.69 and 25.04 kD (Table S4), respectively. Accession number of the sequence was MK913362 in the GenBank database.
Analysis of Phylogenetic Tree and Protein Sequence Alignments
Analysis of phylogenetic tree showed that the EjPI gene is assigned to the rosids PI/GLO lineage (Figure 2). Conceptual translation reveals that EjPI encode 215 amino acids (aa) including a 59-aa M domain, a 27-aa I domain, an 82-aa K domain, and a 47-aa C domain from N- to C-terminus (Figure 3). Among these domains, the M, I, and K domains were conserved in these aligned PI orthologous proteins. The K domain contains K1 (87-108), K2 (121-135), and K3 (143-169) subdomains with (abcdefg)n heptad repeats, which could potentially mediate protein interaction (Yang et al., 2003; Yang and Jack, 2004). In comparison to K domain of asterids PI/GLO lineage, K1 subdomain contains substitution of polar neutral amino acid (Asn-92) to acidic amino acid (Asp-92/Glu-92), and K3 subdomain contains substitution of basic amino acid (Lys-149/His-149) to polar neutral amino acid (Asn-149) in rosids PI/GLO lineage, respectively. However, the C domain, which includes a distinctive PI motif, showed more variable than the other domains (Figure 3).
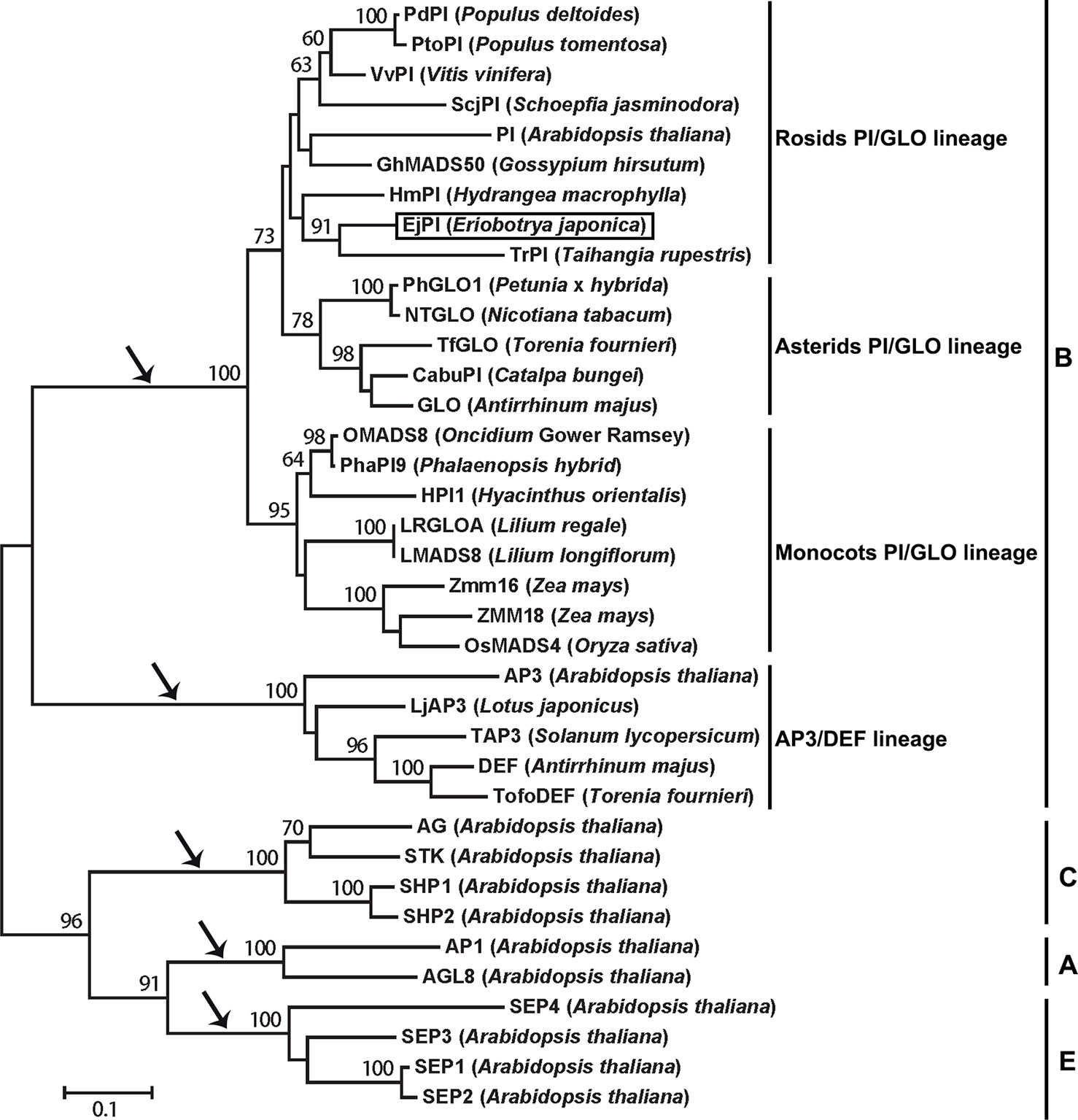
Figure 2 Phylogenetic analysis of PI/GLO-like MADS-box proteins. The EjPI protein sequence is blasted with twenty-six B-class proteins from other angiosperms, with two A-class proteins, four C-class proteins and four E-class proteins as out group. Black arrows show that the gene lineages are obtained through gene duplication. EjPI protein is marked. PI, PISTILLATA.
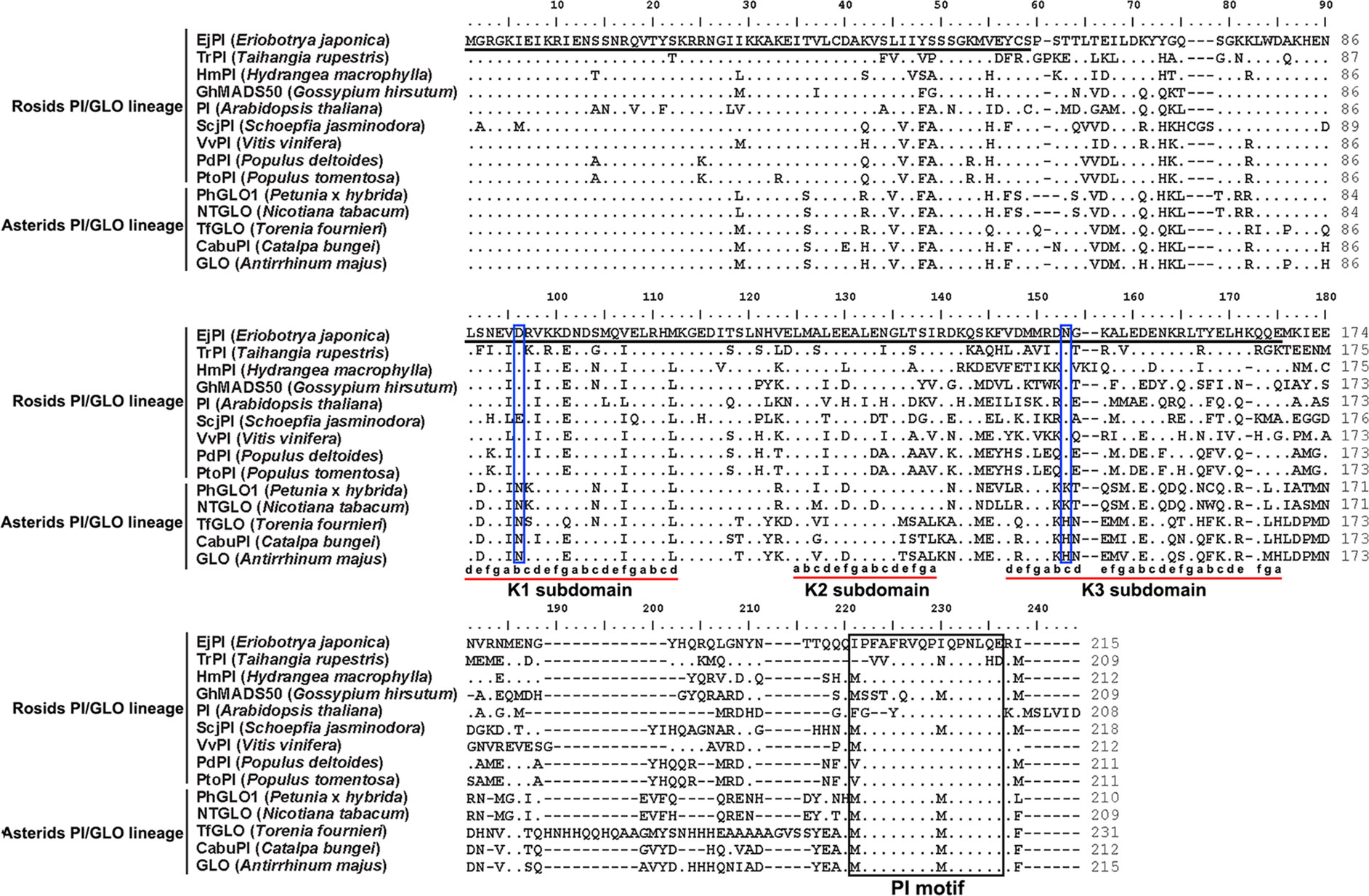
Figure 3 Sequence comparisons of EjPI and the other PI/GLO orthologous proteins. First underlined region represents the MADS domain. Second underlined region represents the K domain. The PI-motif is boxed. Dots indicate the amino acid residues identical to EjPI. Dashes are introduced into the sequences to improve the alignment. The K domain contains K1, K2, and K3 subdomains with (abcdefg)n heptad repeats (Yang et al., 2003), which are also underlined. Meanwhile, Asp-92/Glu-92 and Asn-149 in K1 and K3 subdomains in rosids PI/GLO lineage are boxed.
Subcellular Localization and Spatial Expression of EjPI
To observe the subcellular localization of EjPI, EjPI-GFP fusion proteins transiently expressed in leaf epidermal cells in N. benthamiana. Fluorescence from 35S::GFP was as control and detected in both the nucleus and cytoplasm, but the fluorescence from 35S::EjPI-GFP was detected only in the nucleus (Figure 4).
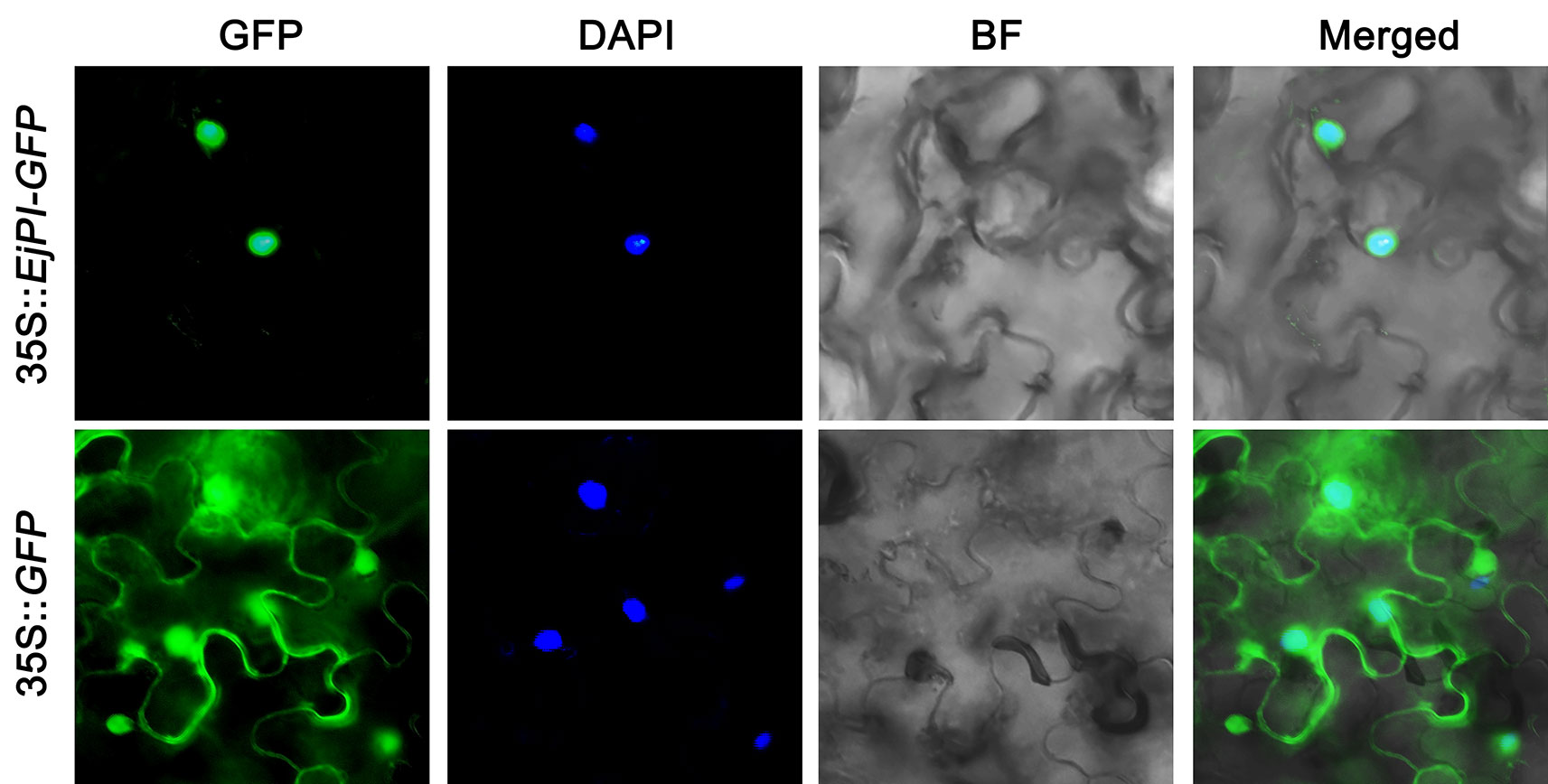
Figure 4 Subcellular localization of EjPI. GFP, GFP fluorescence; 4,6-diamidino-2-phenylindole (DAPI) staining shows nuclear localization; BF, bright-field; Merged, merged image of GFP and DAPI. GFP, green fluorescent protein.
To analyze spatial expression pattern of EjPI, the semi-quantitative RT-PCR were performed in single-flower and double-flower E. japonica. In flower buds of single-flower E. japonica, EjPI was transcribed only in the petals, filaments and anther, but not in sepals and carpels (Figure 5). However, in the flower buds of double-flower E. japonica, EjPI was transcribed in petaloid sepals, petals, filaments, and anthers (Figure 5).
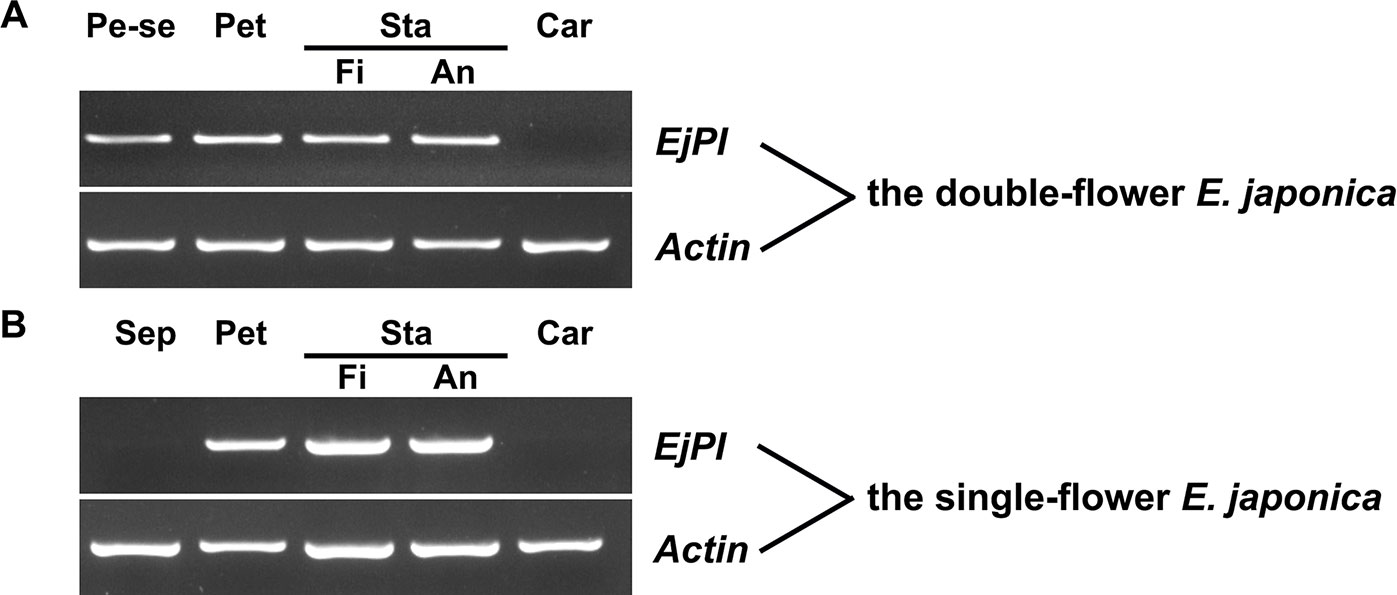
Figure 5 Spatial expression of EjPI in single-flower and double-flower E. japonica by semi-quantitative RT-PCR. (A) Spatial expression of EjPI in double-flower E. japonica. (B) Spatial expression of EjPI in single-flower E. japonica. Sep, sepals; Pe-se, petaloid sepals; Pet, petals; Sta, stamens; Car, carpels; Fi, filaments; An, anthers.
We analyzed EjPI expression levels in petaloid sepals of different areas within the sepals. The expression level of EjPI in larger petaloid area within a sepal was significantly higher than that in small petaloid area (Figure 6). The correlationship between petaloid area and EjPI transcript level was further investigated. There were high correlation coefficients between EjPI transcript level and petaloid area (Pearson’s correlation coefficient of 0.977, Table S5), indicated that increased EjPI expression level causes the increased petaloid area within a sepal.
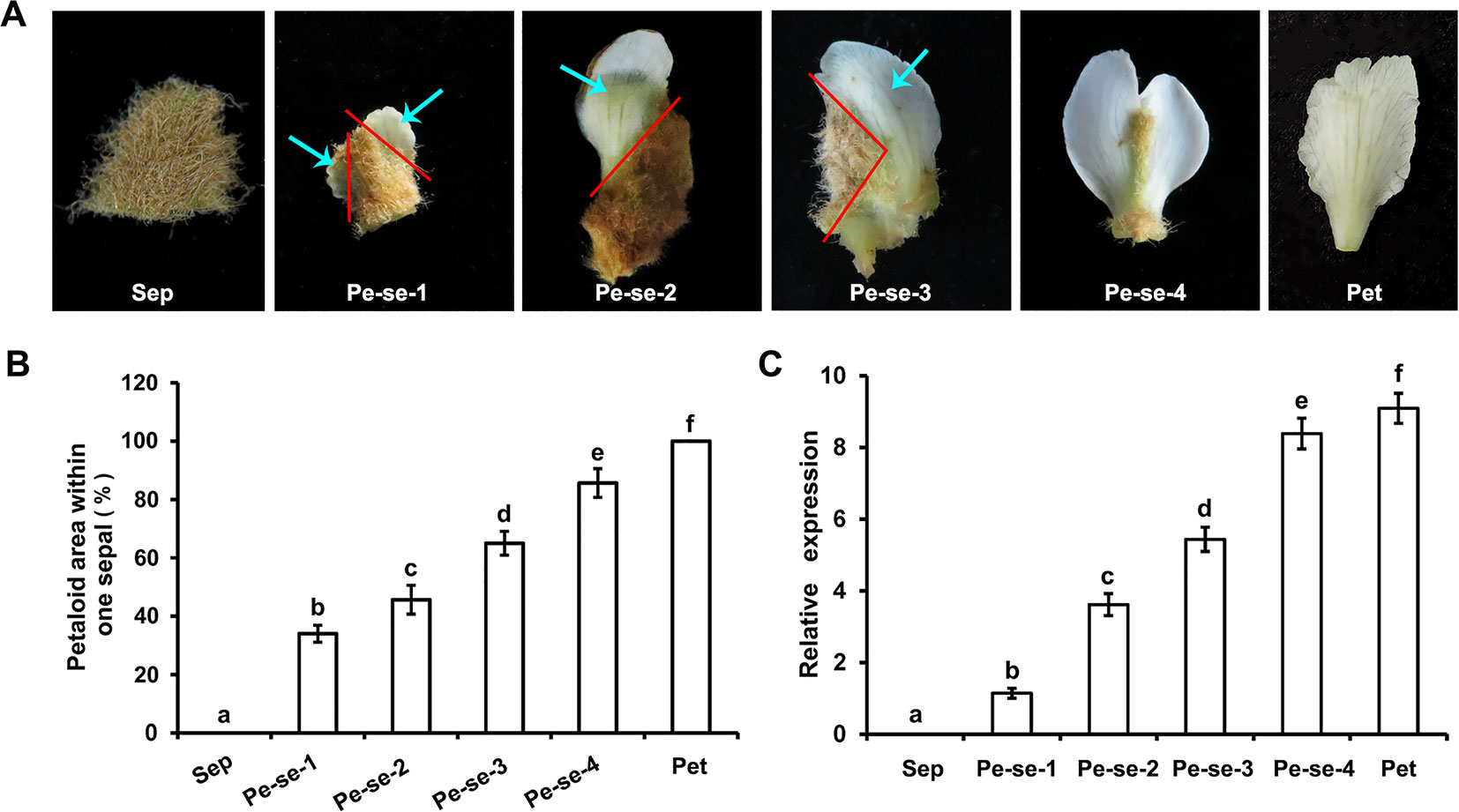
Figure 6 Relative expression levels of EjPI in petaloid sepals in double-flower E. japonica by qRT-PCR. (A) Different types of petaloid sepals. (B) Petaloid area within one sepal. (C) Relative expression levels of EjPI in petaloid sepals. Sep, sepals; Pe-se, petaloid sepals; Pet, petals. Error bars indicate the standard deviation of three biological replicates. Different letters indicate significant differences (P < 0.05).
Functional Analyses of EjPI in Transgenic Arabidopsis
The pi-1 Arabidopsis, which is a strong allele leading to a truncated protein product, results in the phenotype of full B-class loss-of-function. Flower of wild-type Arabidopsis has four sepals in the first whorl, four petals in the second whorl, six stamens in the third whorl and a carpel in the fourth whorl (Figure 7A). However, in homozygous pi-1 mutant Arabidopsis, the second whorl petals are transformed into sepals and the third whorl stamens are replaced into a carpel (Figure 9A).
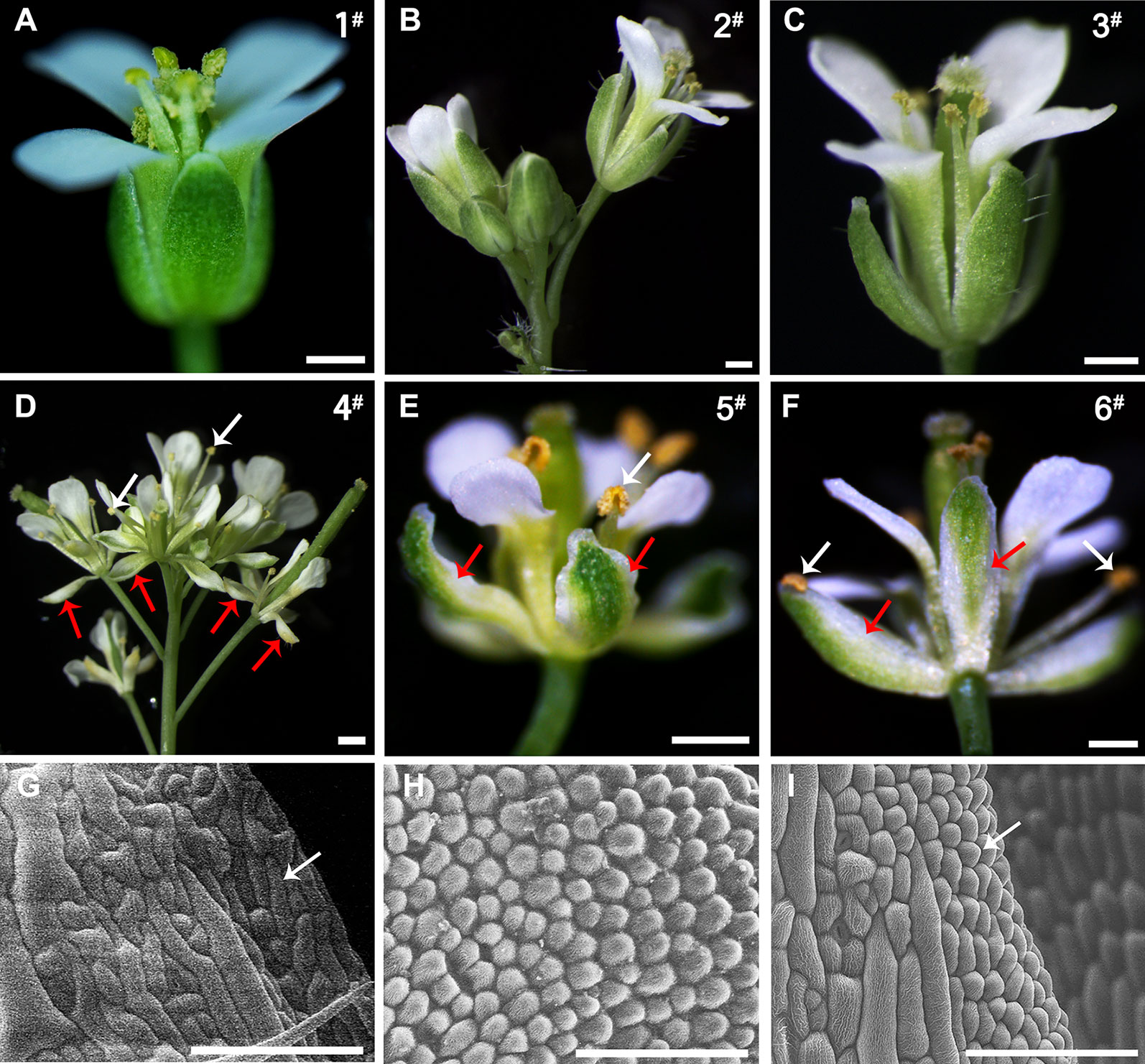
Figure 7 Comparison of the phenotypes of the wild-type and 35S::EjPI transgenic wild-type lines. (A) Flower of the wild-type Arabidopsis, showing tightly closed sepals and stamens (line 1#). (B) Inflorescence of the transgenic wild-type Arabidopsis with the pBI121 vector only (negative control, line 2#). (C) Flower of the transgenic wild-type Arabidopsis with the pBI121 vector only, showing no phenotypic alteration (line 3#). (D) Inflorescence of 35S::EjPI, showing completely separating petaloid sepals (red arrows) and opening stamens (white arrows) (line 4#). (E) Flower of 35S::EjPI transgenic wild-type Arabidopsis, showing green/white petaloid sepals in the first whorl (red arrows) (line 5#). (F) Flower of 35S::EjPI transgenic wild-type Arabidopsis, showing completely separating petaloid sepals (red arrows) and opening stamens (white arrows) (line 6#). (G) Cell shapes of adaxial surface in a wild-type Arabidopsis sepals, showing irregular cell margin; (H) Cell shapes of adaxial surface in petals in wild-type Arabidopsis; (I) Cell shapes of adaxial surface in petaloid sepals in 35S::EjPI transgenic wild-type lines, showing a petaloid margins (white arrow). Bars = 500 µm in (A), (B), (C), (D), (E), and (F) and Bars = 100 µm in (G), (H), and (I).
To analyze the functional characterization of EjPI, overexpression and complementation assays were conducted by ectopic expression of EjPI into wild-type and homozygous pi-1 mutant Arabidopsis plants, respectively. We obtained thirty-one 35S::EjPI transgenic wild-type Arabidopsis lines. Among them, compared with untransformed wild-type plants, ten lines (32.26%) were phenotypically indistinguishable, but 21 remaining transgenic lines (67.74%) showed identical phenotypic alterations. In these transgenic lines with altered phenotypes, green/white petaloid sepals were produced in the first whorl (Figures 7D–F). Meanwhile, the first whorl sepals of Arabidopsis plants of wild-type and transgenic wild-type with the pBI121 vector were tightly closed even after pollination (Figures 7A–C). However, the first whorl petaloid sepals of 35S::EjPI transgenic wild-type lines opened immediately and separated completely after flower opening (Figures 7D–F).
Epidermis cellular shapes of adaxial side from the first whorl floral parts were further examined. Adaxial epidermis cellular shapes of sepal margin in a wild-type Arabidopsis were irregular (Figure 7G). Epidermis cellular shapes of the second whorl petals exhibited cone-shaped (Figure 7H). However, cell shapes of white portion of the petaloid sepals in transgenic wild-type Arabidopsis exhibited to be morphologically distinct from the first whorl sepal epidermis in wild-type plants, but similar to the epidermal cells of the second whorl petals (Figure 7I). Furthermore, the expression levels of EjPI in transgenic wild-type lines were confirmed by qRT-PCR (Figure S3).
Twenty-seven 35S::EjPI transgenic homozygous pi-1 mutant Arabidopsis lines were obtained by PCR analysis (Figure 8). Compared with untransformed homozygous pi-1 lines, nineteen of these lines (70.37%) showed altered phenotypes in flower organ structure, but eight remaining transgenic plants (29.63%) were phenotypically indistinguishable. In these 35S::EjPI transgenic lines with altered phenotypes, eleven plants produced shortened petals in the second whorl and runtish stamens in the third whorl (Figure 9C). Eight transgenic homozygous pi-1 lines produced normal petals in the second whorl and completely rescued stamens in the third whorl (Figure 9D). The expression levels of EjPI in transgenic lines were further confirmed by qRT-PCR. The Pearson correlation between EjPI expression level and phenotype alteration was significant in transgenic lines (P-value < 0.01) (Table S6). Meanwhile, EjPI expression levels of transgenic pi-1 lines with normal petals and stamens were significantly higher than those of transgenic lines with runtish petals and stamens (Figure S4).
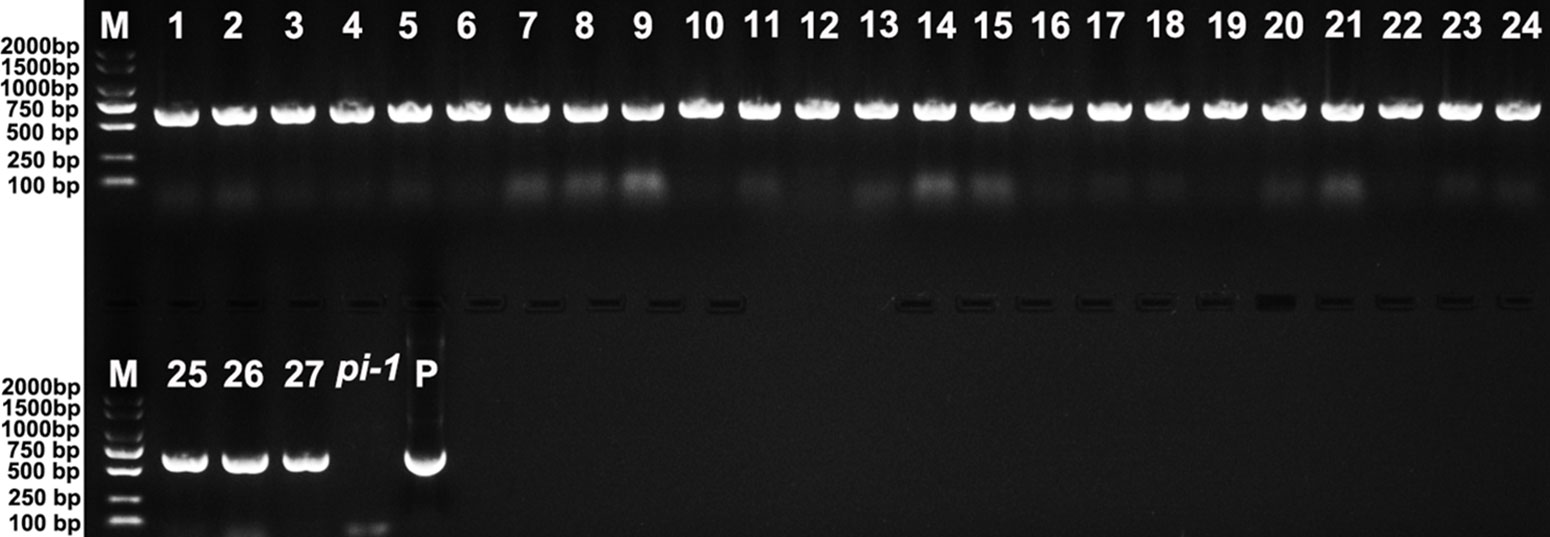
Figure 8 PCR analysis of 35S::EjPI transgenic homozygous pi-1 mutant lines. Lane M: DL2000 DNA marker. Lane 1-27: the PCR with DNA of 35S::EjPI transgenic homozygous pi-1 Arabidopsis as templates. Lane pi-1: the PCR with DNA of pi-1 line containing the pBI121 vector only as a template. Lane P: the PCR with plasmid containing EjPI full-length CDS as a template.

Figure 9 Phenotypic comparison of the homozygous pi-1 mutant and 35S::EjPI transgenic homozygous pi-1 Arabidopsis. (A) Flower of a homozygous pi-1 mutant Arabidopsis (line 1#). (B) Flower of the transgenic homozygous pi-1 mutant with the pBI121 vector only (negative control) (line 2#). (C) The flower of 35S::EjPI transgenic homozygous pi-1 Arabidopsis, showing shortened petals (red arrows) and runtish stamens (white arrows) (line 3#). (D) Flower of 35S::EjPI transgenic homozygous pi-1 Arabidopsis, showing normal petals (red arrows) and completely rescuing stamens (white arrows) (line 4#). Bars = 500 µm.
Discussion
Characterization of key regulatory genes in model plants provides the opportunity to uncover the functional roles of orthologous genes in other plants. In model plants, such as Arabidopsis and Antirrhinum, PI homologues encode floral homeotic B-function MADS-box transcription factors and regulate petal and stamen identities (Trobner et al., 1992; Goto and Meyerowitz, 1994). In this study, we identified the sequence, phylogenetic evolution, and expression patterns of EjPI from single-flower and double-flower E. japonica. Meanwhile, ectopic expression of EjPI in Arabidopsis plants of wild-type and pi-1 mutant was further conducted to confirm the functional roles in regulating floral organ identities.
Protein sequence and phylogenetic analysis showed EjPI protein was assigned to the rosids euPI lineage containing a highly-conserved M domain, an I domain, a less-conserved K domain, a highly divergent C-terminal domain and a distinctive PI motif at the C-terminal region. Therefore, EjPI encodes a typical class B-function MADS-box transcription factor according to the structural features of MADS-box proteins (Kim et al., 2004; Kaufmann et al., 2005). However, K1 and K3 subdomains both contain a single amino acid difference between rosids and asterids PI/GLO lineage. This difference may be related to the divergence of rosids and asterids in the core eudicot. Furthermore, the subcellular localization of EjPI was detected in the nucleus; this is consistent with the nuclear localization of PI in Arabidopsis (Mcgonigle et al., 1996).
In our study, EjPI was expressed in the petals, filament and anther in the flower buds of E. japonica. Spatial expression of EjPI matched well with that in some core eudicots, such as Arabidopsis thaliana (Goto and Meyerowitz, 1994), Taihangia rupestris (Lu et al., 2010), Antirrhinum majus (Trobner et al., 1992), Torenia fournieri (Sasaki et al., 2014), Nicotiana tabacum (Hansen et al., 1993), and Catalpa bungei (Jing et al., 2015). However, expression pattern of EjPI was different from that of the PI orthologs in some eudicots and monocots. In basal eudicot Magnolia wufengensis, MawuPI was mainly expressed in the first and second whorl inner tepals and the third whorl stamens (Liu et al., 2018). In Fagopyrum esculentum, FaesPI was expressed only in stamens (Fang et al., 2015). In monocot Oncidium Gower Ramsey, OMADS8 expression was restricted in all floral organs, such as sepals, petals, lips, stamen, and carpel (Chang et al., 2010).
Beside expression in the petals, filaments and anthers, EjPI was also transcribed in petaloid sepals in double-flower E. japonica. Furthermore, there was a high correlation between the expression level of EjPI and petaloid area within a sepal in double-flowers E. japonica. Previously, compared with the sepal in Papaver somniferum, higher expression level of PapsPI was detected in the petaloid sepals (Singh et al., 2014). In the flowers of monocot such as Zingiberales, Commelinales, Alismatales, and Liliales, expanded expression of class B genes into the first floral whorl was correlated with the formation of petaloid organs (Kanno et al., 2007; Hsu et al., 2015; Kanno, 2015; Theißen et al., 2016). These results suggested that altering expression pattern of PI orthologous genes caused the transformation from sepal to petaloid organ in flowers. However, in some basal angiosperms, expression patterns of PI orthologues in petaloid organs are variability, which can be found in the relative expression level of PI vs paleoAP3. The expression pattern differences of PI orthologues in petaloid organs between E. japonica and basal angiosperms might be due to functional diversification within PI and AP3 gene lineages during the course of angiosperm evolution.
Functional analysis suggested that ectopic expression of EjPI in transgenic wild-type Arabidopsis caused the first whorl sepals replaced into petaloid sepals. Similar phenotypes of transgenic wild-type Arabidopsis were observed in the ectopic expression of PI homologues from rosids species such as Arabidopsis (Krizek and Meyerowitz, 1996; Lamb and Irish, 2003), and asterids species such as C. bungei (Jing et al., 2015), T. fournieri (Sasaki et al., 2010) and monocots such as Lilium longiflorum (Chen et al., 2012), Agapanthus praecox (Nakamura et al., 2005) and Cymbidium faberi Rolfe (Fei and Liu, 2019). For instance, ectopic expression of PI caused the transformation of the first whorl sepals to petaloid organs in transgenic wild-type Arabidopsis (Lamb et al., 2003). Ectopic expression of CabuPI in transgenic wild-type Arabidopsis produced homeotic conversion of sepals into petaloid sepals in the first floral whorl (Jing et al., 2015). In T. fournieri, TfGLO expression in sepals exhibited a petaloid sepal phenotype (Sasaki et al., 2010; Sasaki et al., 2014).
Further functional complementation assay showed that EjPI could substitute the endogenous PI gene in pi-1 mutant Arabidopsis and rescue the identity of petals and stamens. The phenotype differences of EjPI between transgenic wild-type and pi-1 Arabidopsis may due to an effect of pi genotype. However, transgenic phenotypes of EjPI in pi-1 Arabidopsis differed from those of PI orthologs from Magnoliaceae such as MAwuPI (Liu et al., 2018), and from monocots such as LMADS8/9 of L. longiflorum (Chen et al., 2012) and CyfaPI of C. faberi (Fei and Liu, 2019). Ectopic expression of these monocots PI orthologs only partially rescued petal formation in pi mutant Arabidopsis.
Conclusions
In our study, EjPI was isolated and its expression pattern and functional characterization were analyzed. The relative expression level of EjPI in larger petaloid area within a sepal was significantly higher than that in small petaloid area. Ectopic expression of EjPI in transgenic wild-type Arabidopsis caused a petaloid sepal phenotype in the first floral whorl. These data revealed that expression pattern and function of EjPI are associated with the formation of petaloid sepals in double-flower E. japonica. This improves our knowledge of PI orthologous genes in E. japonica, and provides the potential application of EjPI for biotechnical engineering to create petaloid sepals in angiosperm. Meanwhile, we did not find the difference of amino acid sequences of EjPI between the single-flower and double-flower E. japonica on distinct phenotype. Therefore, future work should compare the difference in EjPI promoter between the single-flower and double-flower E. japonica.
Data Availability Statement
The datasets generated for this study can be found in the MK913362 in the GenBank database.
Author Contributions
YX and MS conducted the experiments and drafted the manuscript. WC and RH carried out the analyses of qRT-PCR and endogenous hormones. DJ, DW, SW, QL, and HD contributed to the data analysis. QG and GL provided plant tissues, laboratory facilities, and project supervision. All authors approved the final draft of the manuscript.
Funding
This work was financially supported by National Key Research and Development Program of China (No. 2019YFD1000200), Natural Science Foundation of China (No. 31800600), Fundamental Research Funds for the Central Universities (XDJK2019AA001 and XDJK2011D011), Key Projects of Chongqing Science and Technology Commission (cstc2018jscx-mszdX0054).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01685/full#supplementary-material
References
Bowman, J. L., Smyth, D. R., Meyerowitz, E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1–20.
Chang, Y. Y., Kao, N. H., Li, J. Y., Hsu, W. H., Liang, Y. L., Wu, J. W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152, 837–853. doi: 10.1104/pp.109.147116
Chen, M. K., Hsieh, W. P., Yang, C. H. (2012). Functional analysis reveals the possible role of the C-terminal sequences and PI motif in the function of lily (Lilium longiflorum) PISTILLATA (PI) orthologues. J. Exp. Bot. 63, 941–961. doi: 10.1093/jxb/err323
Clough, S. J., Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Coen, E. S., Meyerowitz, E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37. doi: 10.1038/353031a0
Dodsworth, S. (2017). Petal, Sepal, or Tepal? B-Genes and Monocot Flowers. Trends Plant Sci. 22, 8–10. doi: 10.1016/j.tplants.2016.11.006
Dubois, A., Raymond, O., Maene, M., Baudino, S., Langlade, N. B., Boltz, V., et al. (2010). Tinkering with the C-function: a molecular frame for the selection of double flowers in cultivated roses. PloS One 5, e9288. doi: 10.1371/journal.pone.0009288
Endress, P. K. (2006). Angiosperm floral evolution: morphological developmental framework. Adv. In .Botanical Res. 44, 1–61. doi: 10.1016/S0065-2296(06)44001-5
Fang, Z. W., Li, X. P., Li, X. F., Liu, Z. X. (2015). FaesPI, a Fagopyrum esculentum PISTILLATA ortholog, is involved only in stamen development. J. Plant Biol. 58, 102–109. doi: 10.1007/s12374-014-0390-9
Fei, Y., Liu, Z. X. (2019). Isolation and characterization of the PISTILLATA ortholog gene from Cymbidium faberi Rolfe. Agronomy 9, 425. doi: 10.3390/agronomy9080425
Frohlich, M. W. (2006). Recent developments regarding the evolutionary origin of flowers. Adv. In .Botanical Res. 44, 63–127. doi: 10.1016/S0065-2296(06)44002-7
Galimba, K. D., Tolkin, T. R., Sullivan, A. M., Melzer, R., Theißen, G., Di Stilio, V. S. (2012). Loss of deeply conserved C-class floral homeotic gene function and C-and E-class protein interaction in a double-flowered ranunculid mutant. Proc. Natl. Acad. Sci. 109, E2267–E2275. doi: 10.1073/pnas.1203686109
Goto, K., Meyerowitz, E. M. (1994). Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8, 1548–1560. doi: 10.1101/gad.8.13.1548
Gramzow, L., Theissen, G. (2010). A hitchhiker’s guide to the MADS world of plants. Genome Biol. 11, 214. doi: 10.1186/gb-2010-11-6-214
Hansen, G., Estruch, J. J., Sommer, H., Spena, A. (1993). NTGLO: a tobacco homologue of the GLOBOSA floral homeotic gene of Antirrhinum majus: cDNA sequence and expression pattern. Mol. Gen. Genet. 239, 310–312. doi: 10.1007/bf00281633
Hernandez-Hernandez, T., Martinez-Castilla, L. P., Alvarez-Buylla, E. R. (2007). Functional diversification of B MADS-box homeotic regulators of flower development: Adaptive evolution in protein-protein interaction domains after major gene duplication events. Mol. Biol. Evol. 24, 465–481. doi: 10.1093/molbev/msl182
Hofer, K. A., Ruonala, R., Albert, V. A. (2012). The double-corolla phenotype in the Hawaiian lobelioid genus Clermontia involves ectopic expression of PISTILLATA B-function MADS box gene homologs. Evodevo 3, 26–26. doi: 10.1186/2041-9139-3-26
Hsu, H. F., Hsu, W. H., Lee, Y. I., Mao, W. T., Yang, J. Y., Li, J. Y., et al. (2015). Model for perianth formation in orchids. Nat. Plants 1, 15046. doi: 10.1038/nplants.2015.46
Irish, V. F. (2009). Evolution of petal identity. J. Exp. Bot. 60, 2517–2527. doi: 10.1093/jxb/erp159
Irish, V. F. (2010). The flowering of Arabidopsis flower development. Plant J. 61, 1014–1028. doi: 10.1111/j.1365-313x.2009.04065.x
Jing, D., Xia, Y., Chen, F., Wang, Z., Zhang, S., Wang, J. (2015). Ectopic expression of a Catalpa bungei (Bignoniaceae) PISTILLATA homologue rescues the petal and stamen identities in Arabidopsis pi-1 mutant. Plant Sci. 231, 40–51. doi: 10.1016/j.plantsci.2014.11.004
Jones, D. T., Taylor, W. R., Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kanno, A., Nakada, M., Akita, Y., Hirai, M. (2007). Class B gene expression and the modified ABC model in nongrass monocots. ScientificWorldJournal 7, 268–279. doi: 10.1100/tsw.2007.86
Kanno, A. (2015). Molecular mechanism regulating floral architecture in monocotyledonous ornamental plants. Hortic. J. 85, 8–22.
Kaufmann, K., Melzer, R., Theissen, G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347, 183–198. doi: 10.1016/j.gene.2004.12.014
Kim, S., Yoo, M. J., Albert, V. A., Farris, J. S., Soltis, P. S., Soltis, D. E. (2004). Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91, 2102–2118. doi: 10.3732/ajb.91.12.2102
Kramer, E. M., Dorit, R. L., Irish, V. F. (1998). Molecular evolution of genes controlling petal and stamen development: duplication and divergence within the APETALA3 and PISTILLATA MADS-box gene lineages. Genetics 149, 765–783.
Krizek, B. A., Meyerowitz, E. M. (1996). Mapping the protein regions responsible for the functional specificities of the Arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. U.S.A. 93 (9), 4063–4070. doi: 10.1073/pnas.93.9.4063
Kumar, S., Nei, M., Dudley, J., Tamura, K. (2008). MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 9, 299–306. doi: 10.1093/bib/bbn017
Lamb, R. S., Irish, V. F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. U.S.A. 100, 6558–6563. doi: 10.1073/pnas.0631708100
Lin, S., Sharpe, R. H., Janick, J. (1999). “Loquat: botany and horticulture,” in Horticultural Reviews John Wiley & Sons, Inc: 233–276.
Litt, A., Kramer, E. M. (2010). The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 21, 129–137. doi: 10.1016/j.semcdb.2009.11.019
Liu, Z., Zhang, D., Liu, D., Li, F., Lu, H. (2013). Exon skipping of AGAMOUS homolog PrseAG in developing double flowers of Prunus lannesiana (Rosaceae). Plant Cell Rep. 32, 227–237. doi: 10.1007/s00299-012-1357-2
Liu, H., Huang, R., Ma, J., Sui, S., Guo, Y., Liu, D., et al. (2017). Two C3H type zinc finger protein genes, CpCZF1 and CpCZF2, from Chimonanthus praecox affect stamen development in Arabidopsis. Genes 8, E199. doi: 10.3390/genes8080199
Liu, W., Shen, X., Liang, H., Wang, Y., Chen, F. (2018). Isolation and Functional Analysis of PISTILLATA Homolog From Magnolia wufengensis. Front. In Plant Sci. 9, 1743. doi: 10.3389/fpls.2018.01743.
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S., Fan, Y., Liu, L., Liu, S., Zhang, W., Meng, Z. (2010). Ectopic expression of TrPI, a Taihangia rupestris (Rosaceae) PI ortholog, causes modifications of vegetative architecture in Arabidopsis. J. Plant Physiol. 167, 1613–1621. doi: 10.1016/j.jplph.2010.06.028
Ma, J., Shen, X., Liu, Z., Zhang, D., Liu, W., Liang, H., et al. (2018). Isolation and Characterization of AGAMOUS-Like Genes Associated With Double-Flower Morphogenesis in Kerria japonica (Rosaceae). Front. Plant Sci. 9, 959. doi: 10.3389/fpls.2018.00959
Mcgonigle, B., Bouhidel, K., Irish, V. F. (1996). Nuclear localization of the Arabidopsis APETALA3 and PISTILLATA homeotic gene products depends on their simultaneous expression. Genes Dev. 10, 1812. doi: 10.1101/gad.10.14.1812
Meyerowitz, E. M., Smyth, D. R., Bowman, J. L. (1989). Abnormal flowers and pattern formation in floral. Development 106, 209–217.
Nakamura, T., Fukuda, T., Nakano, M., Hasebe, M., Kameya, T., Kanno, A. (2005). The modified ABC model explains the development of the petaloid perianth of Agapanthus praecox ssp. orientalis (Agapanthaceae) flowers. Plant Mol. Biol. 58, 435–445. doi: 10.1007/s11103-005-5218-z
Neff, M. M., Turk, E., Kalishman, M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18, 613–615. doi: 10.1016/s0168-9525(02)02820-2
Puranik, S., Acajjaoui, S., Conn, S., Costa, L., Conn, V., Vial, A., et al. (2014). Structural basis for the oligomerization of the MADS domain transcription factor SEPALLATA3 in Arabidopsis. Plant Cell 26, 3603–3615. doi: 10.1105/tpc.114.127910
Purugganan, M. D., Rounsley, S. D., Schmidt, R. J., Yanofsky, M. F. (1995). Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140, 345–356.
Sasaki, K., Aida, R., Yamaguchi, H., Shikata, M., Niki, T., Nishijima, T., et al. (2010). Functional divergence within class B MADS-box genes TfGLO and TfDEF in Torenia fournieri Lind. Mol. Genet. Genomics 284, 399–414. doi: 10.1007/s00438-010-0574-z
Sasaki, K., Yamaguchi, H., Nakayama, M., Aida, R., Ohtsubo, N. (2014). Co-modification of class B genes TfDEF and TfGLO in Torenia fournieri Lind. alters both flower morphology and inflorescence architecture. Plant Mol. Biol. 86, 319–334. doi: 10.1007/s11103-014-0231-8
Schwarz-Sommer, Z., Huijser, P., Nacken, W., Saedler, H., Sommer, H. (1990). Genetic control of flower development by homeotic genes in Antirrhinum majus. Science 250, 931. doi: 10.1126/science.250.4983.931
Shan, L. L., Li, X., Wang, P., Cai, C., Zhang, B., Sun, C. D., et al. (2008). Characterization of cDNAs associated with lignification and their expression profiles in loquat fruit with different lignin accumulation. Planta 227, 1243–1254. doi: 10.1007/s00425-008-0696-2
Singh, S. K., Shukla, A. K., Dhawan, O. P., Shasany, A. K. (2014). Recessive loci Pps-1 and OM differentially regulate PISTILLATA-1 and APETALA3-1 expression for sepal and petal development in Papaver somniferum. PloS One 9, e101272. doi: 10.1371/journal.pone.0101272
Soltis, D. E., Ma, H., Frohlich, M. W., Soltis, P. S., Albert, V. A., Oppenheimer, D. G., et al. (2007). The floral genome: an evolutionary history of gene duplication and shifting patterns of gene expression. Trends Plant Sci. 12, 358–367. doi: 10.1016/j.tplants.2007.06.012
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Theißen, G., Gramzow, L. (2016). Structure and evolution of plant MADS domain transcription factors in Plant Transcription Factors. Elsevier Inc. 127–138. doi: 10.1016/B978-0-12-800854-6.00008-7
Theißen, G., Kim, J. T., Saedler, H. (1996). Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J. Mol. Evol. 43, 484–516. doi: 10.1007/bf02337521
Theißen, G., Melzer, R., Rümpler, F. (2016). MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143, 3259–3271.
Theißen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4, 75–85. doi: 10.1016/s1369-5266(00)00139-4
Theissen, G., Saedler, H. (2001). Plant biology. Floral quartets. Nature 409, 469–471. doi: 10.1038/35054172
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149. doi: 10.1023/A:1006332105728
Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Trobner, W., Ramirez, L., Motte, P., Hue, I., Huijser, P., Lonnig, W. E., et al. (1992). GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 11, 4693–4704.
Viaene, T., Vekemans, D., Irish, V. F., Geeraerts, A., Huysmans, S., Janssens, S., et al. (2009). Pistillata–duplications as a mode for floral diversification in (Basal) asterids. Mol. Biol. Evol. 26, 2627–2645. doi: 10.1093/molbev/msp181
Whipple, C. J., Zanis, M. J., Kellogg, E. A., Schmidt, R. J. (2007). Conservation of B class gene expression in the second whorl of a basal grass and outgroups links the origin of lodicules and petals. Proc. Natl. Acad. Sci. U. S. A. 104, 1081–1086. doi: 10.1073/pnas.0606434104
Yang, Y., Jack, T. (2004). Defining subdomains of the K domain important for protein–protein interactions of plant MADS proteins. Plant Mol. Biol. 55, 45–59. doi: 10.1007/s11103-004-0416-7
Yang, Y., Xiang, H., Jack, T. (2003). pistillata-5, an Arabidopsis B class mutant with strong defects in petal but not in stamen development. Plant J. 33, 177–188. doi: 10.1046/j.1365-313x.2003.01603.x
Keywords: Eriobotrya japonica, double-flower, PISTILLATA, MADS-box gene, expression pattern, ectopic expression
Citation: Xia Y, Shi M, Chen W, Hu R, Jing D, Wu D, Wang S, Li Q, Deng H, Guo Q and Liang G (2020) Expression Pattern and Functional Characterization of PISTILLATA Ortholog Associated With the Formation of Petaloid Sepals in Double-Flower Eriobotrya japonica (Rosaceae). Front. Plant Sci. 10:1685. doi: 10.3389/fpls.2019.01685
Received: 27 August 2019; Accepted: 29 November 2019;
Published: 17 January 2020.
Edited by:
Zhongchi Liu, University of Maryland, College Park, United StatesReviewed by:
Hengfu Yin, Chinese Academy of Forestry, ChinaShaohua Zeng, Chinese Academy of Sciences, China
Copyright © 2020 Xia, Shi, Chen, Hu, Jing, Wu, Wang, Li, Deng, Guo and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qigao Guo, cWdndW9AMTI2LmNvbQ==; Guolu Liang, bGlhbmdnbEBzd3UuZWR1LmNu
†These authors have contributed equally to this work
 Yan Xia
Yan Xia Min Shi1,2†
Min Shi1,2† Weiwei Chen
Weiwei Chen Danlong Jing
Danlong Jing Qigao Guo
Qigao Guo Guolu Liang
Guolu Liang