
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 10 January 2020
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01664
 Tao Kang1,2,3†
Tao Kang1,2,3† Chun-Yan Yu1,4†
Chun-Yan Yu1,4† Yue Liu4,5
Yue Liu4,5 Wei-Meng Song2,3
Wei-Meng Song2,3 Yan Bao2,3
Yan Bao2,3 Xiao-Tong Guo1,4
Xiao-Tong Guo1,4 Bei Li1,2,4*
Bei Li1,2,4* Hong-Xia Zhang1,2,4*
Hong-Xia Zhang1,2,4*Plants in the juvenile state are more tolerant to adverse conditions. Constitutive expression of MicroRNA156 (miR156) prolonged the juvenile phase and increased resistance to abiotic stress, but also affected the architecture of transgenic plants. In this study, we investigated the possibility of subtle manipulation of miR156 expression in flowering plants, with the goal to increase tolerance to abiotic stress without altering the normal growth and development of transgenic plants. Transgenic tobacco plants expressing ZmmiR156 from maize were generated, driven either by the cauliflower mosaic virus (CaMV) 35S promoter or the stress-inducible ZmRab17 promoter. Expression of ZmmiR156 led to improved drought and salt tolerance in both 35S::MIR156 and Rab17::MIR156 transgenic plants, as shown by more vigorous growth, greater biomass production and higher antioxidant enzyme expression after a long period of drought or salt treatment, when compared to wild type and transgenic vector control plants. However, constitutive expression of ZmmiR156 also resulted in retarded growth, increased branching and delayed flowering of transgenic plants. These undesirable developmental changes could be mitigated by using the stress-inducible ZmRab17 promoter. Furthermore, under drought or salt stress conditions, expression of ZmmiR156 reduced the transcript level of NtSPL2 and NtSPL9, the genes potentially targeted by ZmmiR156, as well as that of CP1, CP2, and SAG12, the senescence-associated genes in tobacco. Collectively, our results indicate that ZmmiR156 can be temporally manipulated for the genetic improvement of plants resistant to various abiotic stresses.
Unlike animals, plants are sessile organisms that cannot avoid or flee from adverse conditions. Therefore, they have developed a series of strategies to cope with or tolerate different environmental stresses. Drought and salt are two of the most serious environmental factors which have severely restricted the production of agricultural crops worldwide (Rivero et al., 2007). To date, a number of drought and salt stress-related genes and pathways have been isolated and identified in different plant species (Zhang and Blumwald, 2001; Xiong et al., 2002; Yamaguchi-Shinozaki and Shinozaki, 2006; Yang et al., 2008).
In higher plants, MicroRNA156 (miR156), a class of 20–24 nucleotide non-coding regulatory RNAs derived from hairpin precursors by the Dicer-like enzymes, plays crucial roles in transcriptional and post-transcriptional gene regulation via the silencing mechanism (Carrington and Ambros, 2003; Bartel, 2004; Baulcombe, 2004; He and Hannon, 2004). As one of the conserved miRNA families in higher plants, miR156 is highly expressed in seedlings and decreased during the juvenile-to-adult transition (Wang et al., 2011). Together with its target SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) family, miR156 has acted as a versatile toolbox in regulating various developmental processes of plants (Wang and Wang, 2015). In Arabidopsis and poplar, the miRNA156/157 recognition element in the 3′ UTR of SPL3 prevented early flowering by translational inhibition in seedlings (Gandikota et al., 2007). The miRNA regulation cascades (miR156-SPL13-miR172-SNZ) also played a crucial role during the post-germination developmental stages (Martin et al., 2010). Overexpression of miR156 delayed the phase transitions from juvenile to adult and adult to reproductive, and caused shortened plastochron and leaf overproduction in transgenic plants (Wu and Poethig, 2006; Schwarz et al., 2008; Wang et al., 2008). In maize, enhanced expression of miR156b/c dramatically increased the branch numbers of transgenic plants (Chuck et al., 2007). In rice transgenic expression of miR156 enhanced branch/tiller numbers of transgenic plants (Jiao et al., 2010). In poplar, overexpression of miR156 reduced the expression of miR156-targeted SPL genes, and prolonged the juvenile phase of transgenic poplar (Wang et al., 2011). Recently, overexpression of GmmiR156b in soybean improved shoot architecture and significantly increased the numbers of long branches, nodes and pods, leading to improved grain yield in transgenic plants (Sun et al., 2019).
In addition to its developmental functions, miRNAs also play crucial roles in plant response to environmental stress. In Arabidopsis, miR156 isoforms were induced by heat stress (HS) and promoted the expression of HS-responsive genes (Stief et al., 2014). In barley, Hvu-miR156, Hvu-miR166, Hvu-miR171, and Hvu-miR408 were responsive to drought stress (Kantar et al., 2010). In maize, miRNA399b, miRNA156, and Zma-miR3 were induced by low phosphorus stress (Zhang et al., 2012). Transgenic Arabidopsis plants expressing soybean GmmiR172c exhibited increased resistance to drought and salt stresses (Li et al., 2016). Expression of miRNA156 in Arabidopsis and rice led to improved tolerance to NaCl and osmotic stresses in transgenic plants (Cui et al., 2014). In alfalfa, expression of miR156 reduced plant height and increased biomass, branch number and time to complete vegetative growth under both normal and salt stress conditions (Arshad et al., 2017). Similarly, in chickpea, overexpression of miR408 led to reduced plant height and increased drought tolerance (Hajyzadeh et al., 2015).
In this study, we explored whether subtly manipulating the expression of ZmmiR156 from maize (Zea mays) in tobacco would improve the tolerance to drought and salt stresses but cause no significant effects on the regular growth and development of transgenic plants. Our results demonstrate that, by temporally regulating the expression of ZmmiR156 with a stress-inducible promoter, transgenic plants resistant to both drought and salt stresses with no significant morphological changes can be generated.
Tobacco plants (Nicotiana tabacum cv. xanthi) were grown on MS medium (Murashige and Skoog, 1962) with 2% (w/v) sucrose and 0.8% (w/v) agar with cool white fluorescent light (~200 μmol/m2/s) under a 12 h light/12 h dark photoperiod at 21–25°C/15–18°C (day/night). Plants in soil were grown in a greenhouse under a 14 h photoperiod, comprised of natural daylight supplemented with lamps (120–150 µEm2/s) at 21–25°C/15–18°C (day/night). The sequenced maize (Zea mays L.) cultivar B73 was grown in greenhouse as described previously (Li et al., 2013).
For expression pattern analyses of ZmmiR156 in maize plants, juvenile (1-month-old) and old (4-month-old) maize plants or roots of 2-week-old seedlings treated with 15% PEG, 100 mM NaCl or 50 μM ABA were used. Total RNA was extracted from different tissues with RNAiso Plus and RNAiso-mate for Plant Tissue (TaKaRa, Dalian, China), and treated with RNase-free DNase (Promega, Shanghai, China).
The ZmmiR156 sequence was cloned from B73 (GenBank accession No. EF541486.1). The 764-bp pre-mature miR156 sequence was inserted into a modified pCAMBIA3301 vector via the Bam HI and Spe I restriction sites, under the control of two copies of the cauliflower mosaic virus (CaMV) 35S promoter or the drought- and ABA-inducible maize Rab17 promoter. The resultant constructs, as well as the modified pCAMBIA3301 vector, were respectively introduced into Agrobacterium tumefaciens strain EHA105 for tobacco transformation. Leaves of N. tabacum cv. xanthi were transformed as described previously (Yang et al., 2008). Independently regenerated transgenic shoots were propagated and transplanted into soil for homozygous seed production. Transgenic plants transformed with the modified pCAMBIA3301 vector were used as vector control.
Sequence alignment of mature NtmiR156 and ZmmiR156 was performed on Clustal omega website (http://www.ebi.ac.uk/Tools/msa/clustalo). Phylogenetic trees were generated using the MEGA 7.0 and drawn using the neighbor joining method. The accession numbers for AtSPLs, NtSPLs, and ZmSPLs are shown in Supplementary Table S1.
First-strand cDNA was synthesized using the ReverTra Ace Kit (TOYOBO, Osaka, Japan) following the manufacturer’s instruction. A total amount of 2 µg RNA was subjected to reverse transcription reaction using ReverTra Ace (Vazyme, Shanghai, China) at 50°C for 30 min. The resultant cDNA was then used for PCR amplification with gene-specific primers. ZmActin and NtActin were employed as internal controls, respectively. Quantitative real-time PCR (qRT-PCR) was performed with the SYBR Green Real-time PCR Master Mix (Vazyme, Shanghai, China) and monitored in real time with the CFX Connect (BIO-RAD, Shanghai, China). All the primers used in this research are listed in Supplementary Table S2.
For drought stress analyses, 4-week-old seedlings for 35S::MIR156#1 and 35S::MIR156#2 and 6-week-old seedlings for Rab17::MIR156#1 and Rab17::MIR156#7 were withheld from watering for another four weeks. For salt stress treatment, 2-month-old 35S::MIR156#1 and 35S::MIR156#2 and 6-week-old Rab17::MIR156#1 and Rab17::MIR156#7 plants grown in greenhouse were watered with 1/8 concentration of MS salt solution supplemented with or without 200 mM NaCl bi-weekly for 6 weeks.
To measure the content of MDA, leaf samples were ground in 5 ml of 0.1% trichloroacetic acid (TCA) and mixed with 5 ml of 0.5% thiobarbituric acid. The samples were then boiled for 10 min, cooled to room temperature and centrifuged at 12,000 g. The supernatant was analyzed by monitoring the difference in absorbance at A532 and A600. Proline content was determined as described previously (Bates and Walden, 1973).
Leaf disks were stained with nitroblue tetrazolium (NBT) as described previously (Fryer et al., 2002). To measure chlorophyll fluorescence concentration, the fourth new grown leaf counted from the bottom of each plant was taken and photographed at the end of the relative treatment. Chlorophyll content was determined as described by Lichtenthaler (1987).
For stoma observation, epidermal strips were peeled from the leaves of two-month-old wild type, vector control and transgenic plants. The samples were incubated in a solution containing 10 mM KCl, 10 mM MES-Tris, and 50 µM CaCl2 (pH 6.15), and exposed to light (100 µmol m−2 s−1) for 3 h. Subsequently, 0 µM or 50 µM ABA was added to the solution. After 2 h, samples were examined under 20× and 40× magnification using a Nikon microscope. The number of stomata was counted, and the widths and lengths of stomatal apertures were measured. Mean ratios of stomatal density and width to length ± SE of three independent experiments (n = 30–50) were calculated.
All the data were normalized and all samples were normally distributed with homogeneity of variance. Error bars represent the SDs from three biological replicates. Student’s t-test was used for statistical analyses. The tests were one-tailed.
To understand the biological functions of miR156 in plant responses to abiotic stresses, the full length sequence of ZmmiR156 was isolated from maize. Consistent with previous reports that miR156 is highly conserved in plants, ZmmiR156 shares very high sequence identity with miR156s in tobacco (Axtell and Bowman, 2008; Supplementary Figure S1A). Since the miR156/SPLs regulatory module was conserved in diverse plant species, we also conducted phylogenetic analyses of SPLs from Arabidopsis, tobacco and maize using MEGA 7.0 (Chuck et al., 2007; Wang et al., 2011; Wang et al., 2014; Supplementary Figure S1B).
To see whether ZmmiR156 is responsive to abiotic stress, we performed qRT-PCR analyses. Under normal growth condition, ZmmiR156 was ubiquitously expressed in different tissues and organs including roots, leaves, and stems, with a relatively higher expression in juvenile roots and leaves (Supplementary Figure S2A). Upon treatments with 15% PEG, 100 mM NaCl or 50 μM ABA, ZmmiR156 transcript level increased after 1 h and then decreased after 6 h (Supplementary Figures S2B–D). These results demonstrate that ZmmiR156 is responsive to different abiotic stresses and ABA.
To explore whether it is feasible to improve the resistance to different abiotic stresses without changing the architecture of transgenic plants, transgenic tobacco plants with constitutive or stress-induced ZmmiR156 expression were generated. ZmmiR156 sequence was cloned into the pCAMBIA-3301 vector, under the control of either two copies of the cauliflower mosaic virus 35S (2×35S) promoter or the stress-inducible ZmRab17 promoter, and introduced into the genome of tobacco (N. tabacum cv. xanthi) by A. tumefaciens-mediated transformation (Supplementary Figures S3A, B). Fifty-two independently regenerated transgenic lines (T0 generation) were obtained, and twenty lines were confirmed by PCR for the integration of ZmmiR156. The expression level of ZmmiR156 in different transgenic lines was examined by qRT-PCR. Two homozygous 35S::ZmmiR156 transgenic lines which showed high expression of ZmmiR156 (35S::MIR156#1, 35S::MIR156#2) and two homozygous Rab17::MIR156 transgenic lines which showed very low expression of ZmmiR156 (Rab17::MIR156#1, Rab17::MIR156#7) were selected for further stress resistance and morphological analyses (Supplementary Figures S3C, D).
We first assessed the resistance of ZmmiR156-expressing plants to osmotic or salt stress at the early developing stages of seedlings. Seeds of wild type, vector control and the four transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1 and Rab17::MIR156#7 were sown on MS medium supplemented with 7.5% PEG, 200 mM mannitol or 150 mM NaCl. The growth of wild type, vector control and transgenic seedlings were approximately the same when cultured on normal MS medium (Figures 1A–F). However, in the presence of 7.5% PEG, 200 mM mannitol or 150 mM NaCl, growth of wild type, vector control and transgenic seedlings was all impaired, although the growth of transgenic seedlings was less severely affected (Figures 1A–F). Transgenic seedlings produced longer roots and greater biomass than the wild type and vector control seedlings (Figures 1A–F). These results indicate that the expression of ZmmiR156 increased the osmotic and salt tolerance in tobacco.
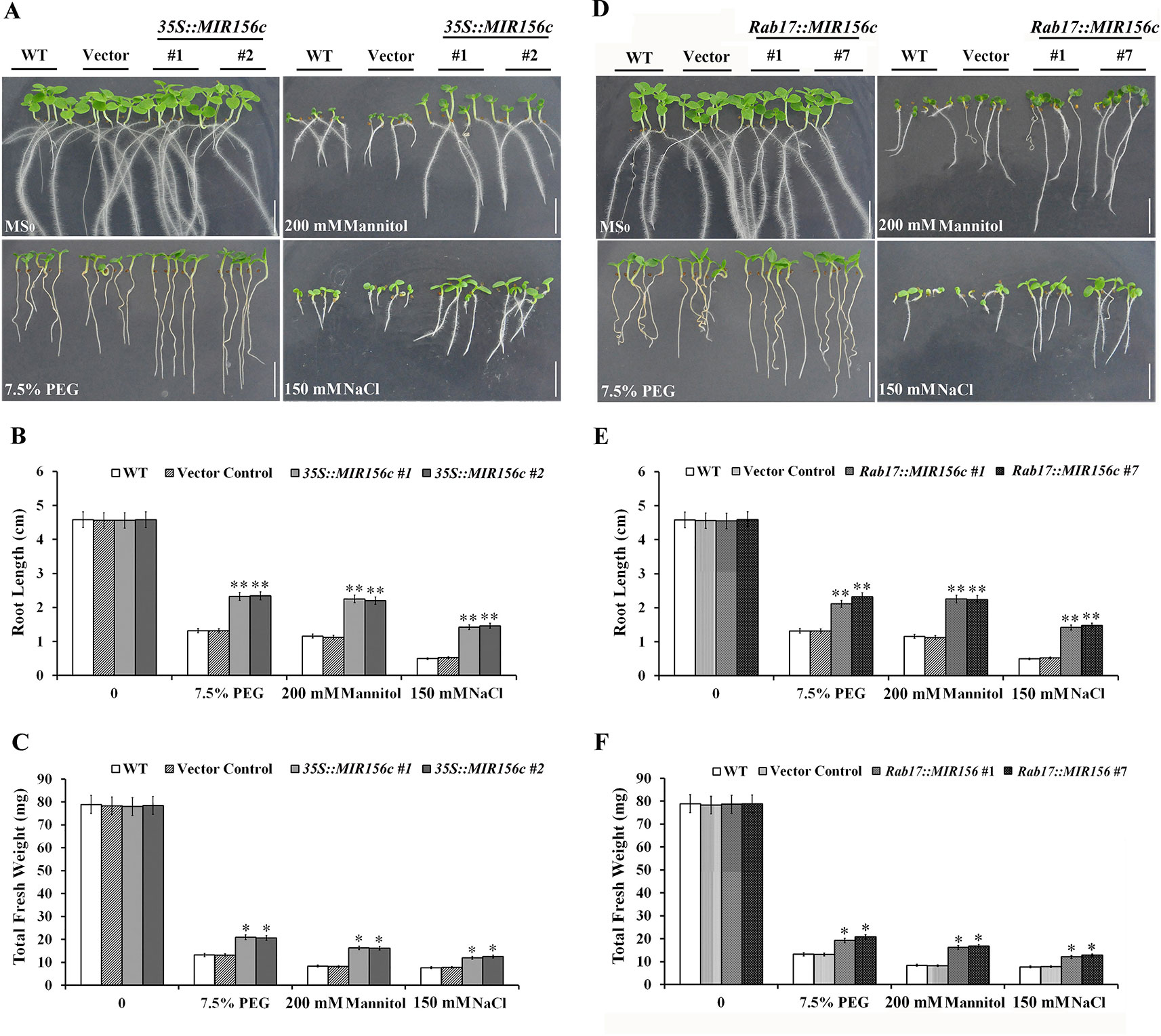
Figure 1 Osmotic and salt stress analyses of wild type (WT), vector control, and both 35S::MIR156, and Rab17::MIR156 transgenic seedlings. Seeds of WT, vector control and transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1, and Rab17::MIR156#7 were sown on MS medium supplemented with 7.5% PEG, 200 mM mannitol, or 150 mM NaCl and vertically cultured for two weeks. (A) Phenotypes of WT, vector control and 35S::MIR156 transgenic seedlings. (B, C) Root lengths and fresh weights of seedlings in (A). (D) Phenotypes of WT, vector control and Rab17::MIR156 transgenic seedlings. (E, F) Root lengths and fresh weights of seedlings in (D). Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at *0.01 < P < 0.05 and **P < 0.01. Scale bar = 1 cm.
To further test the effect of drought on the growth of wild type and ZmmiR156-expressing plants, seedlings grown in soil in greenhouse were withheld from watering for 4 weeks. To minimize the growth retardation caused by the constitutive expression of ZmmiR156, 4-week-old seedlings of 35S::MIR156#1 and 35S::MIR156#2, instead of 6-week-old seedlings of Rab17::MIR156#1 and Rab17::MIR156#7, were used for drought treatment. Expression of ZmmiR156 did not cause any significant morphological changes since wild type, vector control, and both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants all grow normally under well-watered condition (Figures 2A and 3A). After 4 weeks of water withholding, the growth of wild type and vector control plants was dramatically retarded, whereas the growth of both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants was less severely retarded (Figures 2B and 3B). The plant heights and fresh weights of transgenic plants were remarkably higher compared to the wild type and vector control plants (Figures 2C, D, and 3C, D).
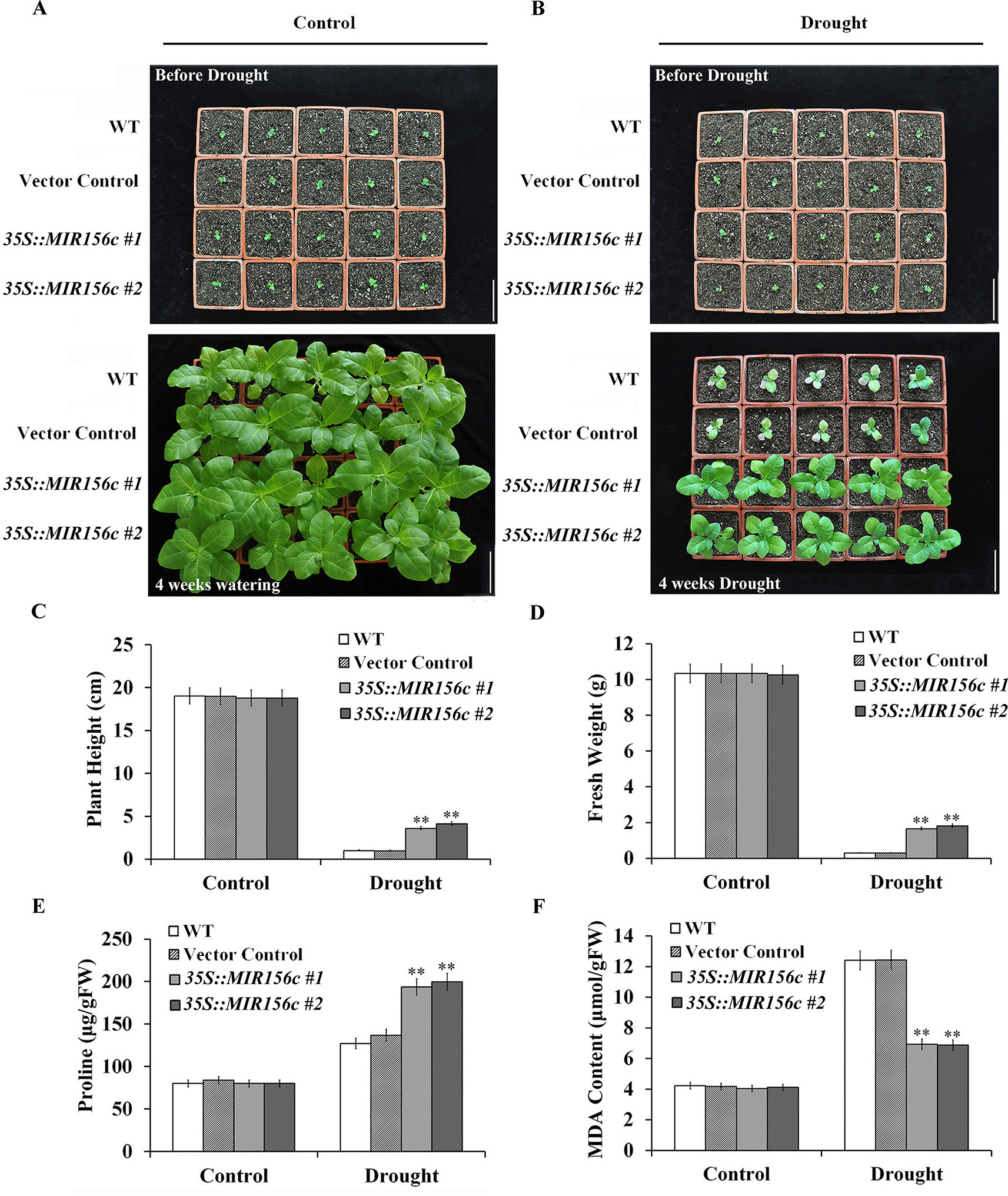
Figure 2 Drought tolerance, plant heights, fresh weights, and leaf proline and MDA content analyses of wild type (WT), vector control and 35S::MIR156 transgenic plants grown in greenhouse. Four-week-old seedlings of WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 were withheld from watering for four weeks. (A) Phenotypes of well-watered seedlings and plants at the beginning and end of experiment, respectively. (B) Phenotypes of drought-treated seedlings and plants at the beginning and end of experiment, respectively. (C–F) Plant heights, fresh weights, and leaf proline and MDA contents of plants in (A) and (B) at the end of experiment. Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at **P < 0.01. Scale bar = 10 cm.
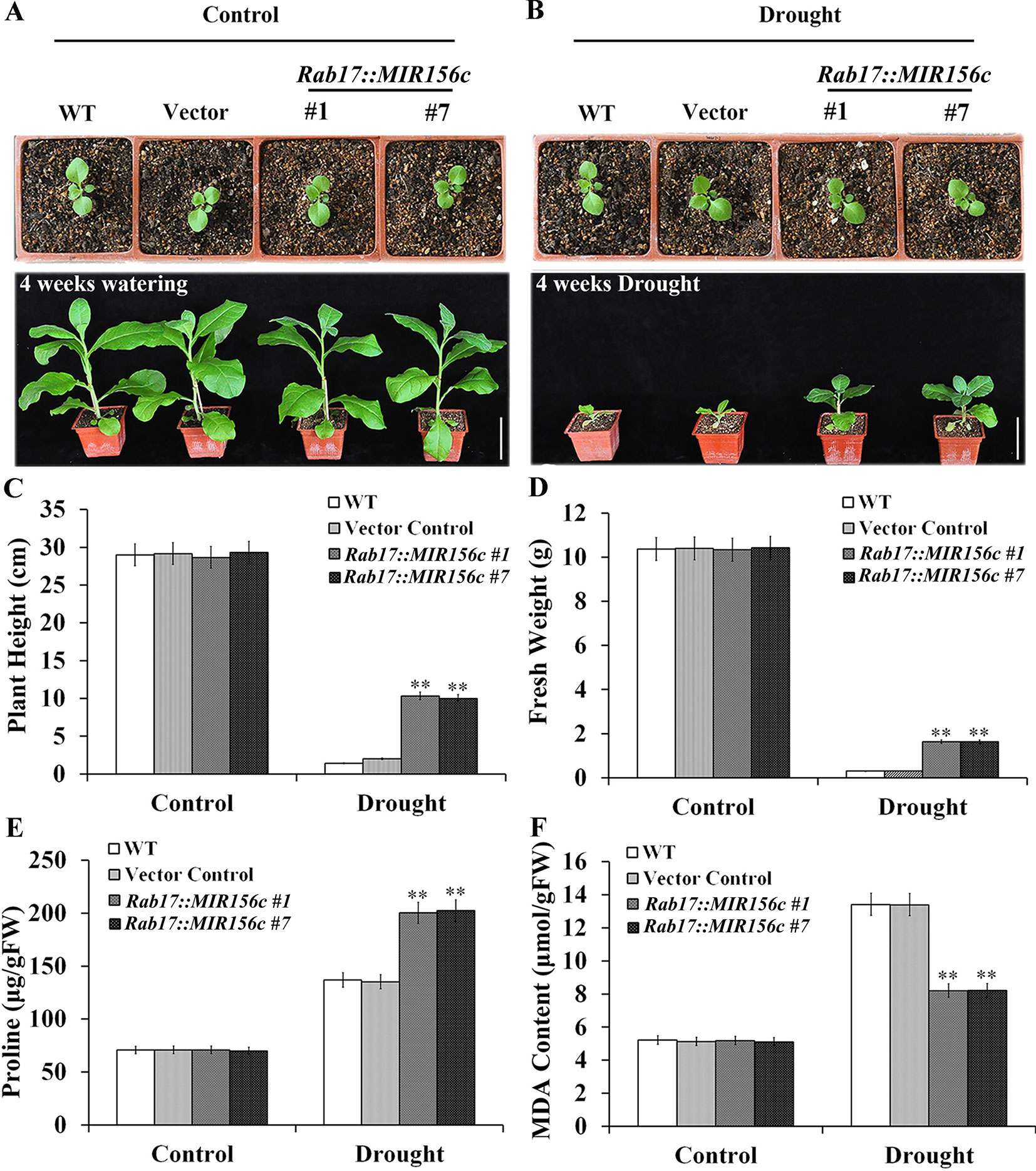
Figure 3 Drought tolerance, plant heights, fresh weights, and leaf proline and MDA content analyses of wild type (WT), vector control and Rab17::MIR156 transgenic plants grown in greenhouse. Six-week-old seedlings of WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 were withheld from watering for four weeks. (A) Phenotypes of well-watered seedlings and plants at the beginning and end of experiment, respectively. (B) Phenotypes of drought treated seedlings and plants at the beginning and end of experiment, respectively. (C–F) Plant heights, fresh weights, and leaf proline and MDA contents of plants in (A) and (B) at the end of experiment. Data are shown as mean ± SD from three replicates. Asterisks indicate significant differences from the corresponding control values at **P < 0.01. Scale bar = 10 cm.
Since miR156 plays a pivotal role in regulating the transition of plants from vegetative growth to reproductive phase, we further examined the effect of salt stress on the growth of adult plants expressing ZmmiR156. Two-month-old transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants or 6-week-old transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 seedlings grown in greenhouse were watered with 1/8 concentration of MS salt solution supplemented with or without 200 mM NaCl bi-weekly for 6 weeks. Under normal growth condition, constitutive expression of ZmmiR156 led to retarded growth, promoted branching and delayed flowering in 35S::MIR156 transgenic plants (Figure 4A; Supplementary Figures S4A, B, and S5A, B). The plant heights and fresh weights of transgenic lines 35S::MIR156#1 and 35S::MIR156#2 were drastically lower than those of the wild type and vector control plants (Figures 4B, C). However, stress-induced expression of ZmmiR156 only caused very minor growth changes in Rab17::MIR156 transgenic plants (Figure 4F; Supplementary Figures S4A, B, and S5A, B). The plant heights and fresh weights of transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 were approximately the same as those of the wild type and vector control plants, although flowering time was slightly delayed (Figures 4G, H; Supplementary Figures S4A, B, and S5A, B). Upon treatment with 200 mM NaCl, both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants showed improved tolerance to salt stress (Figures 4A, F). The plant heights and fresh weights of transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1 and Rab17::MIR156#7 were significantly higher than those of the wild type and relative vector control plants (Figures 4B, C, G, H).
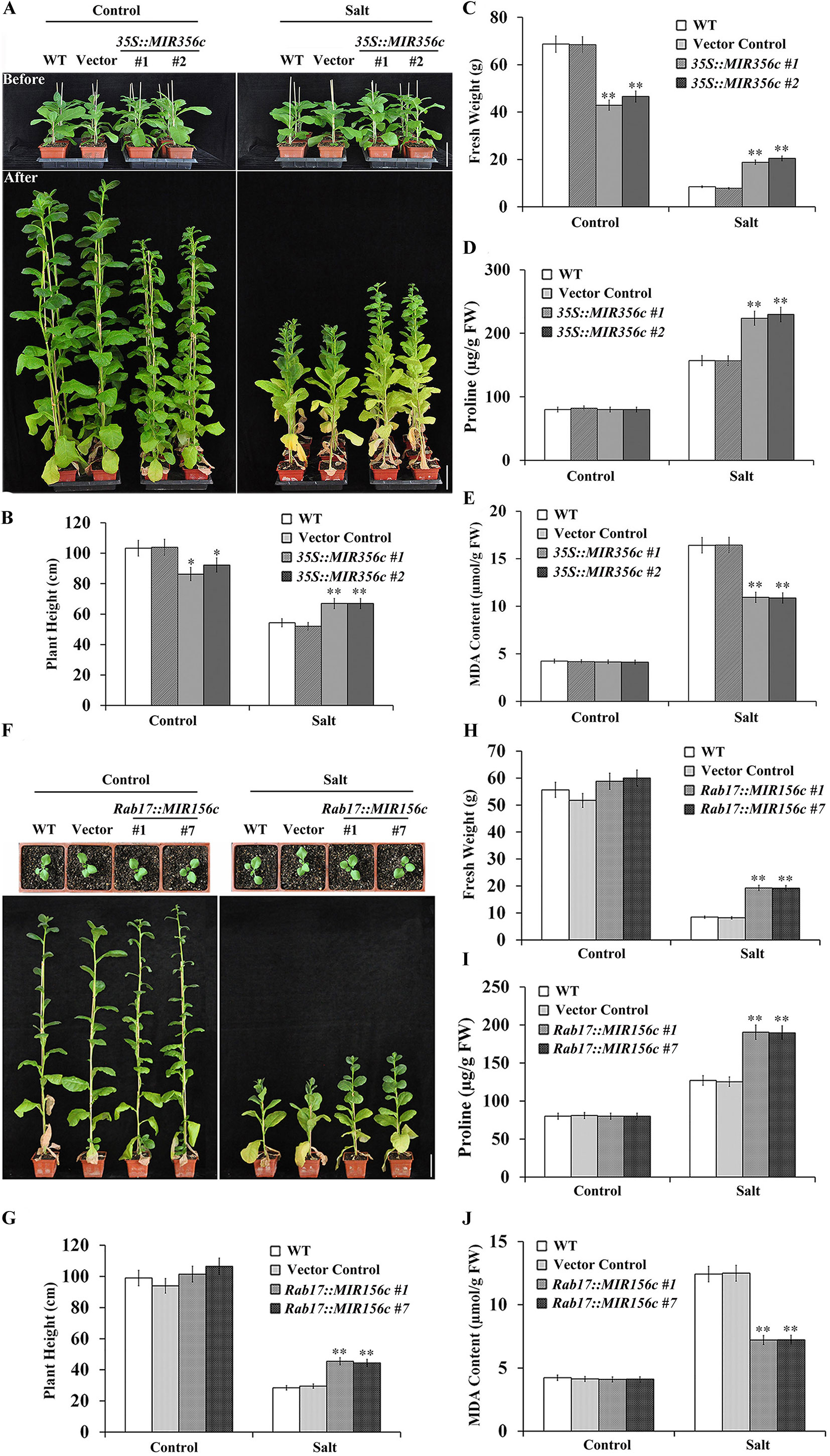
Figure 4 Salt tolerance, plant heights, fresh weights, leaf proline and MDA content analyses of wild type (WT), vector control, and both 35S::MIR156 and Rab17::MIR156 transgenic plants grown in greenhouse. (A) Phenotypes of plants before and after the salt treatment. Two-month-old plants of WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 were treated with 200 mM NaCl for six weeks. (B–E) Plant heights, fresh weights, and leaf proline and MDA content of plants in (A) at the end of experiment. (F) Phenotypes of plants before and after the salt treatment. Six-week-old plants of WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 were treated with 200 mM NaCl for six weeks. (G–J) Plant heights, fresh weights, and leaf proline and MDA content of plants in (F) at the end of experiment. Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at **P < 0.01. Scale bar = 10 cm.
Since drought and salt stresses also exert osmotic stress to plant cells, we further explored whether expression of ZmmiR156 confers elevated resistance to oxidative stress on transgenic plants. It is well known that proline accumulates in response to drought or salt stress in both prokaryotic and eukaryotic organisms (Schobert 1997,). We examined the proline content in the leaves of wild type, vector control and transgenic plants grown under both normal and stress conditions. We observed that although proline accumulation increased in all plants upon the treatment of drought or salt stress, proline concentration was significantly higher in transgenic plants compared to that in the wild type and vector control plants (Figures 2E, 3E, and 4D, I).
Lipid hydroperoxidation is used as an indicator of cell damage caused by oxidative stress (Yoshimura et al., 2004). We measured the lipid hydroperoxide production rate changes induced by oxidative stress in the leaves of wild type, vector control and transgenic plants grown under both normal and stress conditions by determining the content of MDA. Under either drought or salt stress condition, MDA content increased in all plants, but was prominently lower in transgenic plants than that in wild type and vector control plants (Figures 2F, 3F, and 4E, J). Although the expression of ascorbate peroxidase (APX), catalase (CAT) and superoxide dismutase (SOD) was approximately the same under normal conditions, a higher expression was observed in transgenic plants under both drought and salt stress conditions (Supplementary Figures S6A–F and S7A–F).
We further performed nitroblue tetrazolium (NBT; free-radical stain) staining with the leaf disks of wild type, vector control, and 35S::ZmmiR156 and Rab17::MIR156 transgenic plants grown under both normal and stress conditions. Compared to the leaf disks of wild type and vector control plants, leaf disks of 35S::ZmmiR156 and Rab17::MIR156 transgenic plants showed a stronger tolerance to the damage caused by oxidative stress (Figures 5A, B, E, F). Upon treatment of drought or salt stress, there was significant chlorophyll loss in wild type and vector control plants but only a marginal decrease in 35S::ZmmiR156 and Rab17::MIR156 transgenic plants (Figures 5C, D, G, H). Collectively, these results imply that expression of ZmmiR156 improved resistance to the oxidative stress-induced membrane hydroperoxidation in transgenic tobacco plants.
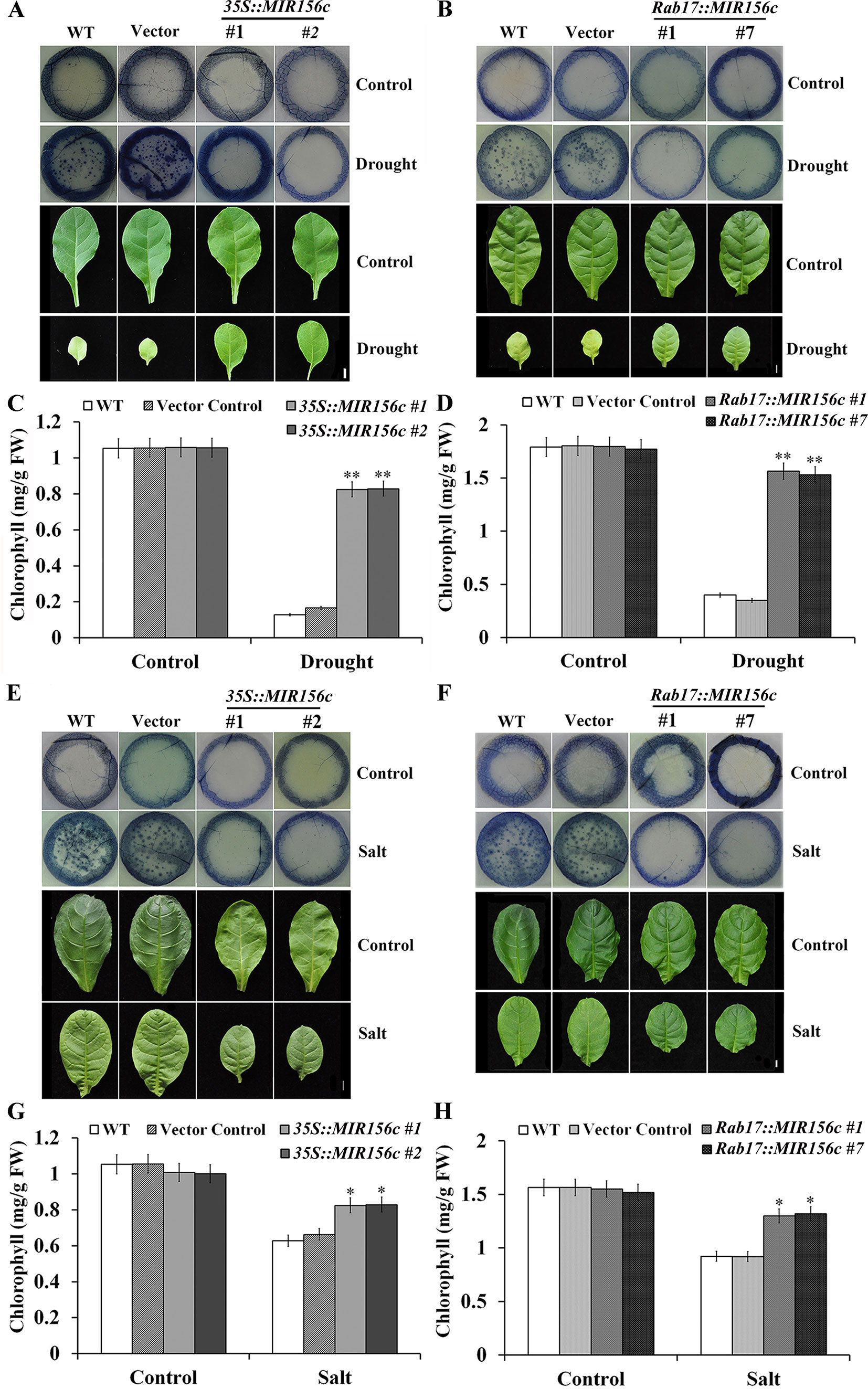
Figure 5 Nitroblue tetrazolium (NBT) staining and chlorophyll content assays of wild type (WT), vector control, and both 35S::MIR156 and Rab17::MIR156 transgenic plants after drought or salt treatment. (A) Leaf disk NBT staining of WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants after the drought treatment. (B) Leaf disk NBT staining of WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants after the drought treatment. (C, D) Leaf disk chlorophyll content of WT, vector control and transgenic plants in (A) and (B), respectively. (E) Leaf disk NBT staining of WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants after the salt treatment. (F) Leaf disk NBT staining of WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants after the salt treatment. (G, H) Leaf disk chlorophyll contents of WT, vector control and transgenic plants in (E) and (F), respectively. Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at values at *0.01 < P < 0.05 and **P < 0.01. Scale bar = 1 cm.
To see if ZmmiR156 expression has been properly manipulated in transgenic tobacco plants, we first investigated the transcript level of ZmmiR156 in wild type, vector control, and 35S::ZmmiR156 and Rab17::MIR156 transgenic plants. Under either normal or stress condition, no ZmmiR156 expression was detected in wild type and vector control plants, whereas constitutive ZmmiR156 expression was observed in transgenic lines 35S::MIR156#1 and 35S::MIR156#2 (Figures 6A, D). Under normal growth condition, although a very low expression of ZmmiR156 was also observed in transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7, the transcription of ZmmiR156 was strongly induced by drought or salt stress (Figures 6B, E). We then examined the relative expression levels of NtSPL2 and NtSPL9, the two SPL family genes potentially targeted by ZmmiR156 in tobacco. Consistent with the expression of ZmmiR156, NtSPL2 and NtSPL9 in transgenic lines 35S::MIR156#1 and 35S::MIR156#2 were drastically down-regulated under both normal and stress conditions, but in transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7, NtSPL2 and NtSPL9 were significantly down-regulated only under drought or salt stress conditions (Figures 6C, F).
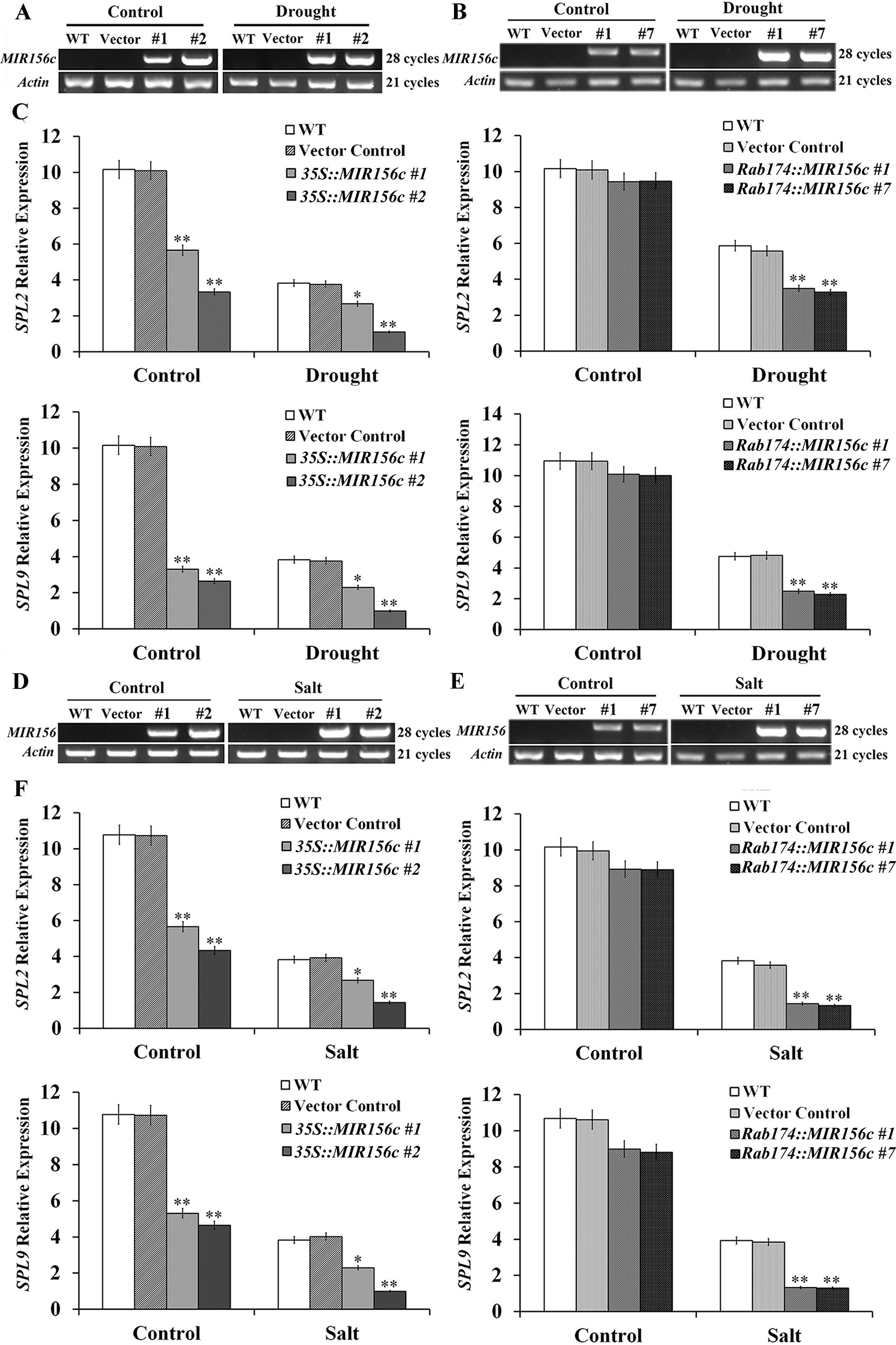
Figure 6 ZmmiR156 and SPL gene expression in wild type (WT), vector control, and both 35S::MIR156 and Rab17::MIR156 transgenic plants after drought or salt treatment. (A) RT-PCR analyses of ZmmiR156 expression in WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants grown under normal and drought stress conditions. (B) RT-PCR analyses of ZmmiR156 expression in WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants grown under normal and drought stress conditions. (C) Quantitative real-time PCR analyses of SPL gene expression in WT, vector control, and transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1, and Rab17::MIR156#7 plants grown under normal and drought stress conditions. (D) RT-PCR analyses of ZmmiR156 expression in WT, vector control, and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants grown under normal and salt stress conditions. (E) RT-PCR analyses of ZmmiR156 expression in WT, vector control, and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants grown under normal and salt stress conditions. (F) Quantitative real-time PCR analyses of SPL gene expression in WT, vector control, and transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1, and Rab17::MIR156#7 plants grown under normal and salt stress conditions. The NtActin gene was employed as an internal control. Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at *P < 0.05, **P < 0.01.
Senescence is a process of growth and recession at cellular, tissue, organ or whole plant level which is influenced by both internal and external factors (Lim et al., 2007). In this process, the relative expression of a number of genes, especially transcription factors and their downstream target genes involved in metabolism and signal perception, will be up- or down-regulated. To date, senescence-associated genes (SAGs) from different plant species have been isolated, including Arabidopsis, rice and tobacco (Getu et al., 2006; Lim et al., 2007; Ewa et al., 2009; Branka et al., 2015). We compared the expression of Cysteine protease 1 (CP1), Cysteine protease 2 (CP2) and SAG12 in the leaves of wild type, vector control, and 35S::ZmmiR156 and Rab17::MIR156 transgenic plants grown under normal and stress conditions. Under normal growth condition, although no significant difference was seen in the expression level of CP1, possibly due to its low expression, the expression of CP2 and SAG12 was significantly down-regulated in transgenic lines 35S::MIR156#1 and 35S::MIR156#2 (Figures 7A, C). No significant difference in expression was observed in transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 (Figures 7B, D). Upon treatment with drought or salt stress, expression of CP1, CP2, and SAG12 was up-regulated in all the plants, with less significant up-regulation in transgenic lines 35S::MIR156#1, 35S::MIR156#2, Rab17::MIR156#1, and Rab17::MIR156#7 (Figures 7A–D).
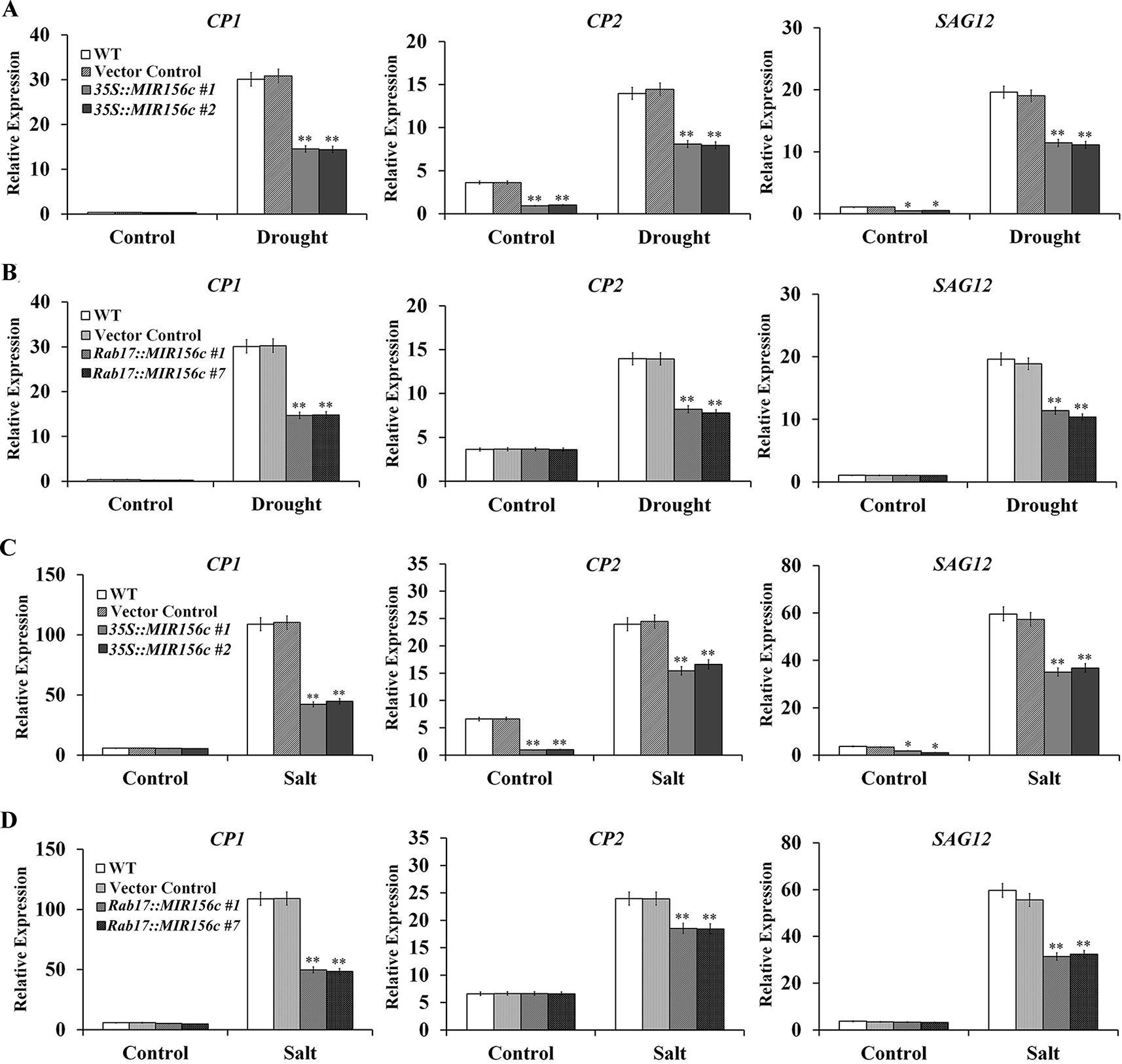
Figure 7 Senescence-associated gene expression in wild type (WT), vector control, and both 35S::MIR156 and Rab17::MIR156 transgenic plants after drought or salt treatment. (A) Quantitative real-time PCR analyses of CP1, CP2, and SAG12 expression in WT, vector control and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants grown under normal and drought stress conditions. (B) Quantitative real-time PCR analyses of CP1, CP2, and SAG12 expression in WT, vector control and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants grown under normal and drought stress conditions. (C) Quantitative real-time PCR analyses of CP1, CP2, and SAG12 expression in WT, vector control and transgenic lines 35S::MIR156#1 and 35S::MIR156#2 plants grown under normal and salt stress conditions. (D) Quantitative real-time PCR analyses of CP1, CP2, and SAG12 expression in WT, vector control and transgenic lines Rab17::MIR156#1 and Rab17::MIR156#7 plants grown under normal and salt stress conditions. The NtActin gene was employed as an internal control. Data are shown as mean ± SD from three biological replicates. Asterisks indicate significant differences from the corresponding control values at **P < 0.01.
To deal with harmful environmental conditions such as drought and salt stress, plants have developed various systems to incorporate the reprogramming of gene expressions (Zhu, 2002; Chinnusamy et al., 2004). By high-throughput small RNA deep sequencing, a number of drought- and salt-responsive miRNAs involved in these regulatory networks have been identified in different plant species (Hobert, 2008; Yu et al., 2008; Jian et al., 2016; Saminathan et al., 2016). Among them, miR156 has been shown to have an important function in plant response to drought and salinity stresses (Sun et al., 2012; Wang et al., 2013; Bhardwaj et al., 2014).
In addition to its function in plant response to adverse stresses, miR156 also plays a crucial role in plant growth and development. Previously, constitutive expression of miR156 improved the tolerance to abiotic stress, but also affected the regular growth and development of transgenic plants (Wu and Poethig, 2006; Chuck et al., 2007; Schwarz et al., 2008; Wang et al., 2008; Jiao et al., 2010; Wang et al., 2011; Xie et al., 2012; Sun et al., 2019). We aimed to investigate the feasibility of improving resistance to abiotic stress without altering the regular growth and development of transgenic plants by genetically manipulating the expression pattern of miR156. Therefore, two kinds of transgenic tobacco plants with either constitutive or stress-induced expression of ZmmiR156 were generated, and their resistance to drought and salt stress as well as their growth features were compared under both normal and abiotic stress conditions (Supplementary Figures S3A–D).
Based on the assumption that senescence is a form of cell death programming activated by drought, isopentenyltransferase (IPT) has been expressed in tobacco and successfully improved the tolerance to drought stress by delaying the senescence of transgenic plants (Rivero et al., 2007). Similarly, constitutive expression of miR156 in Arabidopsis and rice prolonged the juvenile phase, and improved the resistance of transgenic plants to salt and mannitol stresses (Wu and Poethig, 2006; Wang et al., 2011; Cui et al., 2014). However, detailed analyses on the growth and resistance to abiotic stress at different developmental stages were not carried out. In this work, the relative growth of wild type, vector control, and both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants at seedling, juvenile and adult stages grown under normal and stress conditions were compared. Consistent with the previously reported transgenic Arabidopsis and rice, both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants showed improved tolerance to salt and mannitol stresses at the early seedling development stages, as indicated by the more vigorous growth of roots and shoots compared to the wild type and relative vector control seedlings (Figures 1A–F). Similar results were also observed in transgenic tobacco plants at juvenile and adult stages grown in greenhouse (Figures 2A–D, 3A–D, and 4A–C, F–H).
As a hydroxyl radical scavenger, proline makes a significant contribution to the adjustment to osmotic stress and the protection of macromolecules during drought and salt stresses (Hong et al., 2000). Upon treatment with drought or salt stress, proline accumulated to a significantly higher level in both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants than in wild type and vector control plants (Figures 2E, 3E, and 4D, I). This is consistent with previous reports that transgenic salt tolerant plants accumulated more proline (Nanjo et al., 1999; Zhang and Blumwald, 2001; Zhang et al., 2016). Therefore, the augmented proline accumulation in transgenic plants may have helped protect the activity of antioxidative enzymes and as a result alleviated the adverse impacts imposed by drought and salt on transgenic plants.
One of the major causes of the adverse impacts of drought and salt is the generation of reactive oxygen species (ROS) from chloroplast and mitochondrial metabolism induced by abiotic stress, which causes membrane damage and electrolyte leakage (Apse and Blumwald, 2002). Upon treatment with drought or salt stress, a significant increase of MDA content was observed in wild type and vector control plants (Figures 2F, 3F, and 4E, J). In addition, a higher chlorophyll content and antioxidant enzyme expression was also observed in both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants (Figures 5A–H, Supplementary Figures S6A–F, and 7A–F). Therefore, we postulated that under adverse growth conditions, expression of ZmmiR156 helped protect cell membrane integrity in transgenic plants. This hypothesis was also supported by the observations of transgenic tobacco expressing inositol polyphosphate 6-/3-kinase AtIpk2β and heat shock protein LeHSP21 (Yang et al., 2008; Zhang et al., 2016).
At both seedling and juvenile stages, no significant phenotype changes were observed between wild type, vector control, and both 35S::ZmmiR156 and Rab17::MIR156 transgenic plants under normal growth condition (Figures 1A–F, 2A, C, D, and 3A, C, D). However, at adult and reproductive stages, transgenic tobacco plants with constitutive expression of ZmmiR156 showed slower growth, delayed flowering and increased branching compared to the wild type, vector control and transgenic tobacco plants with stress-induced expression of ZmmiR156 (Figures 4A, C, F, G, H; Supplementary Figures S4A, B, and S5A, B). This is consistent with the biological role of ZmmiR156 in plant growth and development. Similar morphological changes were also observed in Arabidopsis, rice, maize, soybean, and aspen with increased expression of miR156, and transgenic chickpea constitutively expressing miR408 (Wu and Poethig, 2006; Jiao et al., 2010; Wang et al., 2011; Xie et al., 2012; Cui et al., 2014; Sun et al., 2019).
As a key channel to exchange gas with the outside environment, the stoma regulates the photosynthesis, transpiration and water use of plants (Chaerle et al., 2005). Environmental cues, such as light intensity and quality, water status, temperature and atmospheric carbon dioxide concentration, as well as endogenous signals, control the development, density and aperture of stomata in plants (Hetherington and Woodward, 2003; Saibo et al., 2003). In addition to the altered growth and development, transgenic plants with constitutive expression of ZmmiR156 also showed decreased leaf stomata number (Supplementary Figures S8A, B). A nearly 66% reduction, leading to a slower water loss, was observed in 35S::MIR156 transgenic plants compared to the wild type, vector control and Rab17::MIR156 transgenic plants (Supplementary Figures S8C, D). Further treatment with ABA showed no significant difference in stomatal aperture in all of the plants (Supplementary Figure S9A, B). We then evaluated the ABA sensitivity of wild type, vector control and 35S::MIR156 transgenic plants during germination and early seedling development. Seeds were sown on MS medium containing different concentrations of ABA. Again, no significant difference was observed after two weeks, as indicated by root lengths and total fresh weights, suggesting that ZmmiR156 may have improved tolerance to abiotic stress in an ABA-independent manner (Supplementary Figures S10A–C).
Leaf senescence involves the expression of a specific set of SAGs. Since constitutive expression of ZmmiR156 prolonged the juvenile phase of transgenic plants, we compared the expression profiles of SPL and SAG genes in both 35S::MIR156 and Rab17::MIR156 transgenic plants. Constitutive expression of ZmmiR156 reduced the expression of miR156-targeted SPL and SAG genes under both normal and stress conditions, whereas stress-induced expression of ZmmiR156 reduced the expression of miR156-targeted SPL and SAG genes only under drought or salt stress conditions (Figures 6C, F, and 7A–D). Similar strategy was also used to improve the salt, drought and freezing tolerance of transgenic Arabidopsis, wheat and barley plants expressing DREB transcription factor genes, driven with the stress inducible rd29A or rab17 promoter (Kasuga et al., 1999; Morran et al., 2011). Taken together, our results demonstrate that ZmmiR156 expression can be temporally engineered to improve plant resistance to abiotic stress without causing any significant effects on the regular growth and development of transgenic plants.
All datasets generated for this study are included in the article/Supplementary Material.
TK, C-YY, YL, X-TG, BL, W-MS, and YB performed the experiments and analyzed the data. TK, H-XZ, and BL conceived the study. TK, C-YY, H-XZ, and BL wrote the manuscript. All authors read and agreed at the last version of the manuscript.
This work was supported by the following grants: the National Key R & D Program of China (2016YFD0600106); The Modern Agricultural Industry Technology System Innovation Team of Shandong Province of China (SDAIT-02-05); The Shandong Agricultural Elite Variety Project (2019LZGC010); The Natural Science Foundation of Shandong Province of China (ZR2016CB48); The National Key Program on Transgenic Research (2018ZX08020002-003-004); The National Natural Science Foundation of China (31700524, 31870576); and The Science and Technology Develop Project in Yantai (2018XSCC041).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Ms. Jessie Zhang (Faculty of Health Sciences, McMaster University, Canada) and Prof. Danielle E. Vindua (College of Food Engineering, Ludong University, China) for the critical reading and editing of this paper.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01664/full#supplementary-material
Apse, M. P., Blumwald, E. (2002). Engineering salt tolerance in plants. Curr. Opin. Biotechnol. 13, 146–150. doi: 10.1016/S0958-1669(02)00298-7
Arshad, M., Gruber, M. Y., Wall, K., Hannoufa, A. (2017). An insight into microRNA156 Role in salinity stress responses of alfalfa. Front. Plant Sci. 8, 356. doi: 10.3389/fpls.2017.00356
Axtell, M. J., Bowman, J. L. (2008). Evolution of plant microRNAs and their targets. Trends Plant Sci. 13, 343–349. doi: 10.1016/j.tplants.2008.03.009
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297. doi: 10.1016/S0092-8674(04)00045-5
Bates, L. S., Walden, R. P. (1973). Rapid determination of free proline for water stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bhardwaj, A. R., Joshi, G., Pandey, R., Kukreja, B., Goel, S., Jagannath, A. (2014). A genome-wide perspective of miRNA in response to high temperature, salinity and drought stresses in brassica juncea (Czern) L. PloS One 9, e92456. doi: 10.1371/journal.pone.0092456
Branka, U., Dušica, J., Ana, S., Václav, M., Petre, I. D., Snežana, B. (2015). Characterization of natural leaf senescence in tobacco (Nicotiana tabacum) plants grown in vitro. Protoplasma 253, 259–275. doi: 10.1007/s00709-015-0802-9
Carrington, J. C., Ambros, V. (2003). Role of MicroRNAs in plant and animal development. Science 301, 336–338. doi: 10.1126/science.1085242
Chaerle, L., Saibo, N., Straeten, D. V. D. (2005). Tuning the pores: towards engineering plants for improved water use efficiency. Trends Biotechnol. 23, 308–315. doi: 10.1016/j.tibtech.2005.04.005
Chinnusamy, V., Schumaker, K., Zhu, J. K. (2004). Molecular genetic perspectives on cross-talk and specificity in abiotic stress signaling in plants. J. Exp. Bot. 55, 225–236. doi: 10.1093/jxb/erh005
Chuck, G., Cigan, A. M., Saeteurn, K., Hake, S. (2007). The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat. Genet. 39, 544–549. doi: 10.1038/ng2001
Cui, L. G., Shan, J. X., Shi, M., Gao, J. P., Lin, H. X. (2014). The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 80, 1108–1117. doi: 10.1111/tpj.12712
Ewa, N., Lisa, P., Christine, D., Piotr, R., Zbigniew, M., Karin, K. (2009). Spatial patterns of senescence and development-dependent distribution of reactive oxygen species in tobacco (Nicotiana tabacum) leaves. J. Plant Physiol. 166, 1057–1068. doi: 10.1016/j.jplph.2008.12.014
Fryer, M. J., Oxborough, K., Mullineaux, P. M., Baker, N. R. (2002). Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 53, 1249–1254. doi: 10.1093/jxb/53.3721249
Gandikota, M., Birkenbihl, R. P., Höhmann, S., Cardon, G. H., Saedler, H., Huijser, P. (2007). The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 49, 683–693. doi: 10.1111/j.1365-313X.2006.02983.x
Getu, B., Christine, H. F., Karl, J. K. (2006). Two new cysteine proteinases with specific expression patterns in mature and senescent tobacco (Nicotiana tabacum L.) leaves. J. Exp. Bot. 57, 1431–1443. doi: 10.1093/jxb/erj123
Hajyzadeh, M., Turktas, M., Khawar, K. M., Unver, T. (2015). miR408 overexpression causes increased drought tolerance in chickpea. Gene 555, 186–193. doi: 10.1016/j.gene.2014.11.002
He, L., Hannon, G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531. doi: 10.1038/nrg1379
Hetherington, A. M., Woodward, F. I. (2003). The role of stomata in sensing and driving environmental change. Nature 424, 901–908. doi: 10.1038/nature01843
Hobert, O. (2008). Gene regulation by transcription factors and MicroRNAs. Science 319, 1785–1786. doi: 10.1126/science.1151651
Hong, Z. L., Lakkineni, K., Zhang, Z. M., Verma, D. P. S. (2000). Removal of feedback inhibition of DELTA1-pyrroline-5-carboxylate synthetase results in increased proline accumulation and protection of plants from osmotic stress. Plant Physiol. 122, 1129–1136. doi: 10.1104/pp.122.41129
Jian, H. J., Wang, J., Wang, T. Y., Wei, L. J., Li, J. N., Liu, L. Z. (2016). Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front. Plant Sci. 7, 658. doi: 10.3389/fpls.2016.00658
Jiao, Y. Q., Wang, Y. H., Xue, D. W., Wang, J., Yan, M. X., Liu, G. F., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kantar, M., Unver, T., Budak, H. (2010). Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct. Integr. Genom. 10, 493–507. doi: 10.1007/s10142-010-0181-4
Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17 (3), 287–291. doi: 10.1038/7036
Li, B., Liu, H., Zhang, Y., Kang, T., Zhang, L., Tong, J. H., et al. (2013). Constitutive expression of cell-wall invertase genes increase grain yield and starch content in maize. Plant Biotechnol. J. 11, 1080–1091. doi: 10.1111/pbi.12102
Li, W., Wang, T., Zhang, Y., Li, Y. (2016). Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 67, 175–194. doi: 10.1093/jxb/erv450
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Lim, P. O., Kim, H. J., Nam, H. G. (2007). Leaf senescence. Annu. Rev. Plant Biol. 58, 115–136. doi: 10.1146/annurev.arplant.57.032905.105316
Martin, R. C., Asahina, M., Liu, P. P., Kristof, J. R., Coppersmith, J. L., Pluskota, W. E., et al. (2010). The microRNA156 and microRNA172 gene regulation cascades at post-germinative stages in Arabidopsis. Seed Sci. Res. 20, 79–87. doi: 10.1017/S0960258510000085
Morran, S., Eini, O., Pyvovarenko, T., Parent, B., Singh, R., Ismagul, A., et al. (2011). Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. doi: 10.1111/j.1467-7652.2010.00547.x
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant 15, 473–495. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nanjo, T., Kobayashi, M., Yoshiba, Y., Kakubari, Y., Yamaguchi-Shinozaki, K., Shinozaki, K. (1999). Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett. 461, 205–210. doi: 10.1016/S0014-5793(99)01451-9
Rivero, R. M., Kojima, M., Gepstein, A., Sakakibara, H., Mittler, R., Gepstein, S., et al. (2007). Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl. Acad. Sci. U.S.A. 104, 19631–19636. doi: 10.1073/pnas.0709453104
Saibo, N. J., Vriezen, W. H., Beemster, G. T. S., Straeten, D. V. D. (2003). Growth and stomata development of Arabidopsis hypocotyls are controlled by gibberellins and modulated by ethylene and auxins. Plant J. 33, 989–1000. doi: 10.1046/j.1365-313X.2003.01684.x
Saminathan, T., Bodunrin, A., Singh, N. V., Devarajan, R., Nimmakayala, P., Jeff, M., et al. (2016). Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 16, 122. doi: 10.1186/s12870-016-0807-3
Schobert, B. (1997). Is there an osmotic regulatory mechanism in algae and higher plants? J. Theor. Biol. 68, 17–26. doi: 10.1016/0022-5193(77)90224-7
Schwarz, S., Grande, A. V., Bujdoso, N., Saedler, H., Huijser, P. (2008). The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 67, 183–195. doi: 10.1007/s11103-008-9310-z
Stief, A., Altmann, S., Hoffmann, K., Pant, B. D., Scheible, W. R., Baurle, I. (2014). Arabidopsis miR156 regulates tolerance to recurring environmental stress through SPL transcription factors. Plant Cell 26, 1792–1807. doi: 10.1105/tpc.114.123851
Sun, G., Stewart, C. N., Jr., Xiao, P., Zhang, B. H. (2012). MicroRNA expression analysis in the cellulosic biofuel crop switchgrass (Panicum virgatum) under abiotic stress. PloS One 7, e32017. doi: 10.1371/journal.pone.0032017
Sun, Z., Su, C., Yun, J., Jiang, Q., Wang, L., Wang, Y., et al. (2019). Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol. J. 17, 50–62. doi: 10.1111/pbi.12946
Wang, H., Wang, H. Y. (2015). The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol. Plant 8, 677–688. doi: 10.1016/j.molp.2015.01.008
Wang, J. W., Schwab, R., Czech, B., Mica, E., Weigel, D. (2008). Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20, 1231–1243. doi: 10.1105/tpc.108.058180
Wang, J. W., Park, M. Y., Wang, L. J., Koo, Y., Chen, X. Y., Weigel, D., et al. (2011). MiRNA control of vegetativephase change in trees. PloS Genet. 7, e1002012. doi: 10.1371/journal.pgen.1002012
Wang, M., Wang, Q. L., Zhang, B. L. (2013). Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530, 26–32. doi: 10.1016/j.gene.2013.08.009
Wang, Y. L., Wu, F. J., Bai, J. J., He, Y. K. (2014). BrpSPL9 (Brassicarapa ssp pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 12, 312–321. doi: 10.1111/pbi.12138
Wu, G., Poethig, R. S. (2006). Temporal regulation of shoot development in Arabidopsis thaliana by Mir156 and its target SPL3. Development 133, 3539–3547. doi: 10.1242/dev.02521
Xie, K., Shen, J., Hou, X., Yao, J., Li, X., Xiao, J., et al. (2012). Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 158, 1382–1394. doi: 10.1104/pp.111.190488
Xiong, L., Schumaker, K. S., Zhu, J. K. (2002). Functional and phylogenetic analysis of a DREB/CBF-like gene in perennial ryegrass (Lolium perenne L.). Planta 224, 878–888. doi: 10.1007/s00425-006-0273-5
Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, L., Tang, R. J., Zhu, J. Q., Liu, H., Mueller-Roeber, B., Xia, H. J., et al. (2008). Enhancement of stress tolerance in transgenic tobacco plants constitutively expressing AtIpk2β, an inositol polyphosphate 6-/3-kinase from Arabidopsis thaliana. Plant Mol. Biol. 66, 329–343. doi: 10.1007/s11103-007-9267-3
Yoshimura, K., Miyao, K., Gaber, A., Takeda, T., Kanaboshi, H., Miyasaka, H., et al. (2004). Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 37, 21–33. doi: 10.1046/j.1365-313X.2003.01930.x
Yu, X. P., Lin, J., Zack, D. J., Mendell, J. T., Qian, J. (2008). Analysis of regulatory network topology reveals functionally distinct classes of microRNAs. Nucleic Acids Res. 36, 6494–6503. doi: 10.1093/nar/gkn712
Zhang, H. X., Blumwald, E. (2001). Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat. Biotechnol. 19, 765–768. doi: 10.1038/90824
Zhang, Z., Lin, H., Shen, Y., Gao, J., Xiang, K., Liu, L., et al. (2012). Cloning and characterization of miRNAs from maize seedling roots under low phosphorus stress. Mol. Biol. Rep. 39, 8137–8146. doi: 10.1007/s11033-012-1661-5
Zhang, J., Chen, H. Y., Wang, H. H., Li, B., Yi, Y. J., Kong, F. J., et al. (2016). Constitutive expression of a tomato small heat shock protein gene LeHSP21 improves tolerance to high temperature stress by enhancing antioxidation capacity in tobacco. Plant Mol. Biol. Rep. 34, 399–409. doi: 10.1007/s11105-015-0925-3
Keywords: drought, salt, ZmmiR156, SPL, transgenic plants
Citation: Kang T, Yu C-Y, Liu Y, Song W-M, Bao Y, Guo X-T, Li B and Zhang H-X (2020) Subtly Manipulated Expression of ZmmiR156 in Tobacco Improves Drought and Salt Tolerance Without Changing the Architecture of Transgenic Plants. Front. Plant Sci. 10:1664. doi: 10.3389/fpls.2019.01664
Received: 31 August 2019; Accepted: 26 November 2019;
Published: 10 January 2020.
Edited by:
Dean E. Riechers, University of Illinois at Urbana-Champaign, United StatesReviewed by:
Yoshiharu Y. Yamamoto, Gifu University, JapanCopyright © 2020 Kang, Yu, Liu, Song, Bao, Guo, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bei Li, YmVpbGlAc2licy5hYy5jbg==; Hong-Xia Zhang, aHh6aGFuZ0BzaWJzLmFjLmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.