- 1Division of Applied Life Science (BK21plus program), Plant Molecular Biology and Biotechnology Research Center, Gyeongsang National University, Jinju, South Korea
- 2Department of Biotechnology, Bacha Khan University, Charsadda, Pakistan
- 3Department of Biomedical Science and Engineering, Konkuk University, Seoul, South Korea
- 4Institute of Glocal Disease Control, Konkuk University, Seoul, South Korea
High salt stress caused by ionic and osmotic stressors eventually results in the suppression of plant growth and a reduction in crop productivity. In our previous reports, we isolated the endophytic bacterium Bacillus oryzicola YC7007 from the rhizosphere of rice (Oryza sativa L.), which promoted plant growth and development and suppressed bacterial disease in rice by inducing systemic resistance and antibiotic production. In this study, Arabidopsis thaliana seedlings under salt stress that were bacterized with YC7007 displayed an increase in the number of lateral roots and greater fresh weight relative to that of the control seedlings. The chlorophyll content of the bacterized seedlings was increased when compared with that of untreated seedlings. The accumulation of salt-induced malondialdehyde and Na+ in seedlings was inhibited by their co-cultivation with YC7007. The expression of stress-related genes in the shoots and roots of seedlings was induced by YC7007 inoculation under salt stress conditions. Interestingly, YC7007-mediated salt tolerance requires SOS1, a plasma membrane-localized Na+/H+ antiporter, given that plant growth in sos2-1 and sos3-1 mutants was promoted under salt-stress conditions, whereas that of sos1-1 mutants was not. In addition, inoculation with YC7007 in upland-crops, such as radish and cabbage, increased the number of lateral roots and the fresh weight of seedlings under salt-stress conditions. Our results suggest that B. oryzicola YC7007 enhanced plant tolerance to salt stress via the SOS1-dependent salt signaling pathway, resulting in the normal growth of salt-stressed plants.
Introduction
Agricultural productivity cannot meet the requirements of an exploding world population despite distinct progress in farm management and agronomy; in addition, climate change affects agriculture and increases the risk of food insecurity (Nelson et al., 2014). In particular, soil salinity is one of the most representative environmental stressors and constitutes an important agriculture problem (Flowers, 2004; Van Oosten et al., 2017). Salinity has been shown to reduce the content of leaf chlorophyll, resulting in impaired photosynthesis under abiotic stress conditions (Chaves et al., 2009; Zhang et al., 2009a). Salt stress seriously damages photosynthesis as well as plant growth and development by resulting in water deficits, oxidant imbalances, cell-division impairment, membrane degeneration, ion injury, and osmotic stress (Abdel Latef et al., 2017; Abdel Latef et al., 2018; Abdel Latef et al., 2019a; Abdel Latef et al., 2019b). Plants have developed various mechanisms to avoid salt stress by employing compatible solutes which reduce sodium (Na+) uptake while regulating stomatal closure, shoot branching, and lateral root formation to conserve moisture (Acosta-Motos et al., 2017; Zhang et al., 2018). The expression of stress-response genes is regulated upon the initiation of unfavorable stress conditions (Shinozaki and Yamaguchi-Shinozaki, 2007; Hernández, 2019).
Plants heavily rely on two active defense mechanisms that involve ion transporters that either intensely exclude Na+ ions out of the cells or lock the ions into the vacuole (Zhu, 2002). The disintegration of ion homeostasis is the deleterious effect observed in plants that are exposed to salt stress (Munns and Tester, 2008). The salt overly sensitive (SOS) pathway is a key regulatory system that retains ion homeostasis during salt stress and sos mutants exhibit hypersensitivity to salt (Zhu, 2002). The SOS pathway involving the SOS1, SOS2, and SOS3 proteins, is essential for the maintenance of ion homeostasis (Zhu, 2002). Intercellular Na+ elevates intracellular Ca2+ levels, and these Ca2+ ions modulate sodium ion homeostasis together with the SOS proteins (Ji et al., 2013). Upon binding with Ca2+, SOS3 interacts with and activates SOS2 by releasing its self-inhibition. Then, the SOS3-SOS2 complex phosphorylates the plasma membrane protein SOS1, which effluxes Na+, reduces Na+ toxicity, and whose function is crucial to protect the root meristem (Ji et al., 2013). The high-affinity K+ transporter 1 (HKT1) functions as a major regulator that maintains the Na+ and K+ balance by regaining Na+ from the xylem in shoots (Rus et al., 2001).
Recently, it has been reported that plant tolerance can be enhanced by exogenous biostimulants, such as plant growth-promoting rhizobacteria (PGPR), to protect the plants against salt stress (Upadhyay and Singh, 2015; Habib et al., 2016; Li et al., 2017; Zou et al., 2018). Endophytic bacteria that are considered as PGPR have been applied to a broad range of agricultural crops to improve seed germination while increasing plant biomass and productivity (Gray and Smith, 2005; Paré et al., 2011; Bhattacharyya and Jha, 2012; Chung et al., 2015). The majority of PGPR include various strains of Agrobacterium, Azospirillum, Bacillus, Pseudomonas, and Rhizobium species (Hamdia et al., 2004; Bharti et al., 2013; Ahmad et al., 2014). These PGPR benefit plant growth through diverse mechanisms including nitrogen fixation; phosphate solubilization; and the production of 1-aminocyclopropane-1-carboxylate (ACC), deaminase, exopolysaccharide (EPS), and phytohormones, such as auxin, indole-3-acetic acid (IAA), abscisic acid (ABA), gibberellin (GA), and cytokinin (Cowan et al., 1999; Gyaneshwar et al., 2002; Mayak et al., 2004; Ryu et al., 2004; Figueiredo et al., 2008; Zhang et al., 2008; Yang et al., 2009).
Bacillus subtilis in Arabidopsis thaliana and Rhizobium and Pseudomonas in Zea mays increase plant tolerance to salt stress by regulating proline contents and ion homeostasis (Chen et al., 2007; Bano and Fatima, 2009). Lettuce leaves of Lactuca sativa inoculated with ACC-producing PGPR Pseudomonas mendocina exhibited enhanced plant tolerance to salt stress by increasing the uptake of nutrients and increasing the activity of antioxidant enzymes, such as peroxidase and catalase (Kohler et al., 2009). Brahmi (Bacopa monnieri) inoculated with EPS-producing PGPR Bacillus pumilus (STR2) and Exiguobacterium oxidotolerans (STR36) contained higher proline levels and lower lipid peroxidation levels in a saline environment (Bharti et al., 2013). The other multifarious bacteria, including Pantoea, Paenibacillus, Burkholderia, Achromobacter, Microbacterium, Methylobacterium, Variovorax, and Enterobacter species have also been reported as PGPR and mediate plant tolerance to various abiotic stressors, such as light, cold, heat, drought, salt, and oxygen (Choudhary, 2012; Kasotia and Choudhary, 2014).
Thus, the application of salt-tolerant PGPR to salt-hypersensitive crops, such as tomato, red pepper, maize, mung bean, and lettuce, might ensure plant growth and increase productivity during salt-stress conditions (Bano and Fatima, 2009; Tank and Saraf, 2010; Siddikee et al., 2011; Ahmad et al., 2012; Shukla et al., 2012; Ahmad et al., 2014). We previously reported the isolation and identification of an endophytic bacterium, Bacillus oryzicola YC7007, from the rice (Oryza sativa L.) rhizosphere (Chung et al., 2015). The YC7007 bacteria was found to promote plant growth and act as a biocontrol against fungal and bacterial rice pathogens through induced systemic resistance (ISR) and antibiotic production (Chung et al., 2015; Hossain et al., 2016). In this study, we investigated whether the PGPR B. oryzicola YC7007 ameliorates the adverse effects of high salt stress on the growth of Arabidopsis thaliana plants. The application of YC7007 may constitute an efficacious and sustainable approach for the agricultural management and protection of crops under salt-stress conditions.
Materials and Methods
Cultivation of the Endophytic Bacterium Bacillus oryzicola YC7007
The endophytic bacterium B. oryzicola YC7007 isolated from rice roots was used in the present study (Chung et al., 2015). The YC7007 strain was kept on tryptic soy agar (TSA; Difco Laboratories, Detroit, MI, USA) medium at 28°C for 24 h. The YC7007 cells were cultured in 1/10 X tryptic soy broth (TSB; Difco Laboratories, Detroit, MI, USA) at 28°C for 24 h and were adjusted to a 1 X 109 colony forming unit (CFU)/ml in a 10 mM MgSO4 solution after centrifugation and washing in sterile distilled water for further experiments.
Plant Materials and Growth Conditions
The ecotypes of Arabidopsis thaliana used in this study include Landsberg (Ler, Landsberg erecta), Wassilewskija (Ws-0), Colombia (Col-0), and a derivative of the Columbia C24 ecotype (TAIR, https://www.arabidopsis.org/). The sos1-1, sos2-1, and sos3-1 mutants are described in Zhu et al. (1998). Arabidopsis thaliana seeds were surface-sterilized with 70% ethanol for 1 min followed by soaking for 3 min in 10% sodium hypochlorite and then washed with distilled water. The seeds were sown on 1/2 X Murashige and Skoog (MS) medium supplemented with 1.5% sucrose under short day conditions (8 h light / 16 h dark) at 23 °C. Surface-sterilized seeds from radish, Raphanus sativus, and cabbage, Brassica campestris ssp. pekinensis, were sown on 1/2 X MS medium that contained 1.5% sucrose under short day conditions at 28 °C.
Treatment With Bacillus Oryzicola YC7007 and Salt
A total of 20 μl of the bacterial suspension of YC7007 that was adjusted to 109 CFU/ml in 10 mM MgSO4 was used for the treatments. For horizontal growing, 5-day-old seedlings (10 seedlings per plate) were transferred onto one side of specialized plastic petri dishes containing a center partition (I plates; Fisher Scientific). Both sides of the petri dished contained 1/2 X MS with 1% sucrose and NaCl (0, 20, 40, 60, 80, or 100 mM). The treated plants were inoculated with either 50 µl of the bacterial suspension or 10 mM MgSO4 buffer at the center of the petri dish site opposite to the seedlings and grown for 14 days after incubation (DAI). For vertical growth, seedlings (10 seedlings per plate) were arranged in a line on one side of the petri dish. The other side of the petri dish was inoculated with a bacterial suspension at a distance of 5 cm from the seedling root tip on the opposite side of the petri dish. Plates were sealed with parafilm and placed in a growth chamber. Our experiments were performed independently at least 4 or 5 times with 6, 10, or 12 seedlings for each experiment.
The salt overly sensitive sos1, sos2, and sos3 mutant seedlings were arranged in square plates (4 seedlings each) on one side of specialized square plastic petri dishes containing 1/2 X MS with 1% sucrose and either 30 or 50 mM NaCl and treated with 100 µl of the bacterial suspension at a distance of 5 cm from the primary root tip. The experiments were performed with six replicates with twelve seedlings per replicate.
Analysis of Root Development
The root development parameters of horizontally grown seedlings were measured 14 days after inoculation (DAG). The growth of vertically grown seedlings was determined at 8 DAG. The fresh weight of the root and shoots was determined with an analytical balance. Primary root length was measured with a ruler and lateral root number was also determined immediately after harvesting. The dry weight of the seedlings was measured after being oven dried for 3 days at 80°C. The experiments were performed with five replicates with ten seedlings per replicate.
Proton Concentration Measurements
The measurements of pH in the growth medium was determined by pH indicator (Zhang et al., 2009b). To quantify acidification of the plant growth medium, 4-day-old Arabidopsis (ecotype Col-0) seedlings were grown in 1/2 X MS medium with YC7007 or dH2O as a control for 5 days, then transferred to 1/2 X MS medium without agar. Before and after 24 h of roots or bacteria acidification, the pH value in growth medium was determined by a pH meter (Orion Star™ A211 pH Benchtop Meter; Thermo Scientific, Waltham, MA USA). The pH values without plant or YC7007 were used as blank controls for calculation of the proton concentration. Root proton release (unit: mol/g of fresh weight/h) was determined as (10–final pH – 10–initial pH)*0.01/g of fresh weight of roots/24 h. The experiments were performed with three replicates of ten seedlings per replicate.
Measurement of Chlorophyll Content
Leaf chlorophyll content was determined according to the methodology of Xie et al. (2009). 5 mg of tissue was pulverized with 1 ml 80% acetone and the absorbance of its supernatant was measured at 645 nm and 663 nm. Total chlorophyll content was calculated using the following formula:
The experiments performed with three replicates with twenty seedlings per replicate.
Measurement of Malondialdehyde
The oxidative stress biomarker malondialdehyde (MDA) was measured spectrophotometrically. Ground leaf tissue (0.1 mg) was mixed with 2 ml of 0.1% trichloroacetic acid (TCA) and centrifuged at 8,000 × g for 10 min. The supernatant was mixed with 20% TCA and 0.5% thiobarbituric acid (TBA) and incubated at 95°C for 30 min and then cooled immediately in an ice bath. After centrifugation, the absorbance of its supernatant was measured at 450 nm, 532 nm, and 600 nm to estimate the total leaf MDA content. The experiments were performed in triplicate with twenty seedlings per replicate.
Visualization of Intracellular Na+ Ions and Ion Content Measurements
To visualize and measure the sodium ion content in the roots after NaCl treatment, we followed the methodology of Choi et al. (2011). Seven-day-old seedlings were treated with 200 mM NaCl and B. oryzicola YC7007 for 24 h. Then, the roots were stained with 20 μM CoroNa-Green in 0.02% pluoric acid for 3 h, and the fluorescence signals were observed using a GFP filter (excitation, 488 nm; emission, 510 nm) on a confocal laser-scanning microscope (Olympus FV1000; Olympus, Tokyo, Japan). Ten-day-old seedlings (n= 20) grown on 1/2 MS medium were transferred to the same medium supplemented with 200 mM NaCl and YC7007 for 6 h. The Na+ ion content was determined using inductively coupled plasma (ICP) spectrometry. The experiments were performed with three replicates of twenty seedlings per replicate.
Analysis of Quantitative Real-Time PCR
Total RNA was isolated using a RNeasy Kit (Qiagen, Valencia, CA, USA) according to the instructions of the manufacturer and treated with DNase I (Qiagen, Valencia, CA, USA) to remove genomic DNA contamination. Total RNA (2 µg) was used for the first strand of cDNA synthesis using a cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) and subjected to qRT-PCR analysis. The primers used in the qRT-PCR analysis are described in Supplementary Table 1. QuantiMix SYBR (PhileKorea, Daejeon, Korea) was used for PCR reactions. The PCR conditions were 95°C for 10 min, 60 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The relative expression levels of all samples were automatically calculated and analyzed three times using CFX Manager Software (Bio-Rad, Hercules, CA, USA). The expression of A. thaliana TUBULIN2 was used as the endogenous control. The qRT-PCR experiments were performed in triplicate. The specific primers of the stress-response genes used are listed in Supplementary Table S1.
Statistical Analysis
The statistical analyses including Student’s t test were performed by Excel 2010 software. The qRT-PCR analyses were performed three-independent experiments, the average values of 2−ΔCT were used to determine the differences, and the data were expressed as means ± sd. A significant difference was considered at 0.01 < p-value ≤ 0.05 and p-value < 0.01.
Results
Bacillus oryzicola YC7007 Promoted the Growth of All Arabidopsis Ecotypes
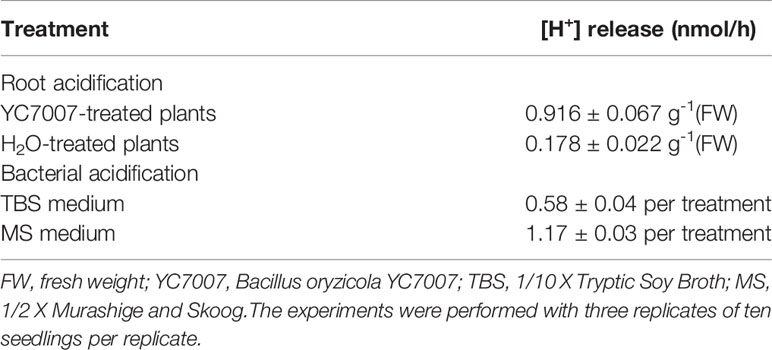
To investigate whether the YC7007 strain promotes plant growth, we used four ecotypes of Arabidopsis. At 11 DAG, the plant size of the seedlings co-cultivated with YC7007 was slightly greater than that of the non-treated control plants (Figure 1). The fresh weight of the seedling shoots and roots treated with YC7007 for 11 days was measured (Figure 1B). The shoot fresh weight of seedlings in the YC7007 treatment increased significantly by 37.7–43.4% when compared to that of the control seedlings (Figure 1B, left). The fresh weight of the roots in the YC7007 treatment also increased by 25.4% in Ler, 51.5% in Ws-0, 35.8% in Col-0, and 21% in C24 ecotypes compared to that of the control seedlings (Figure 1B, right).
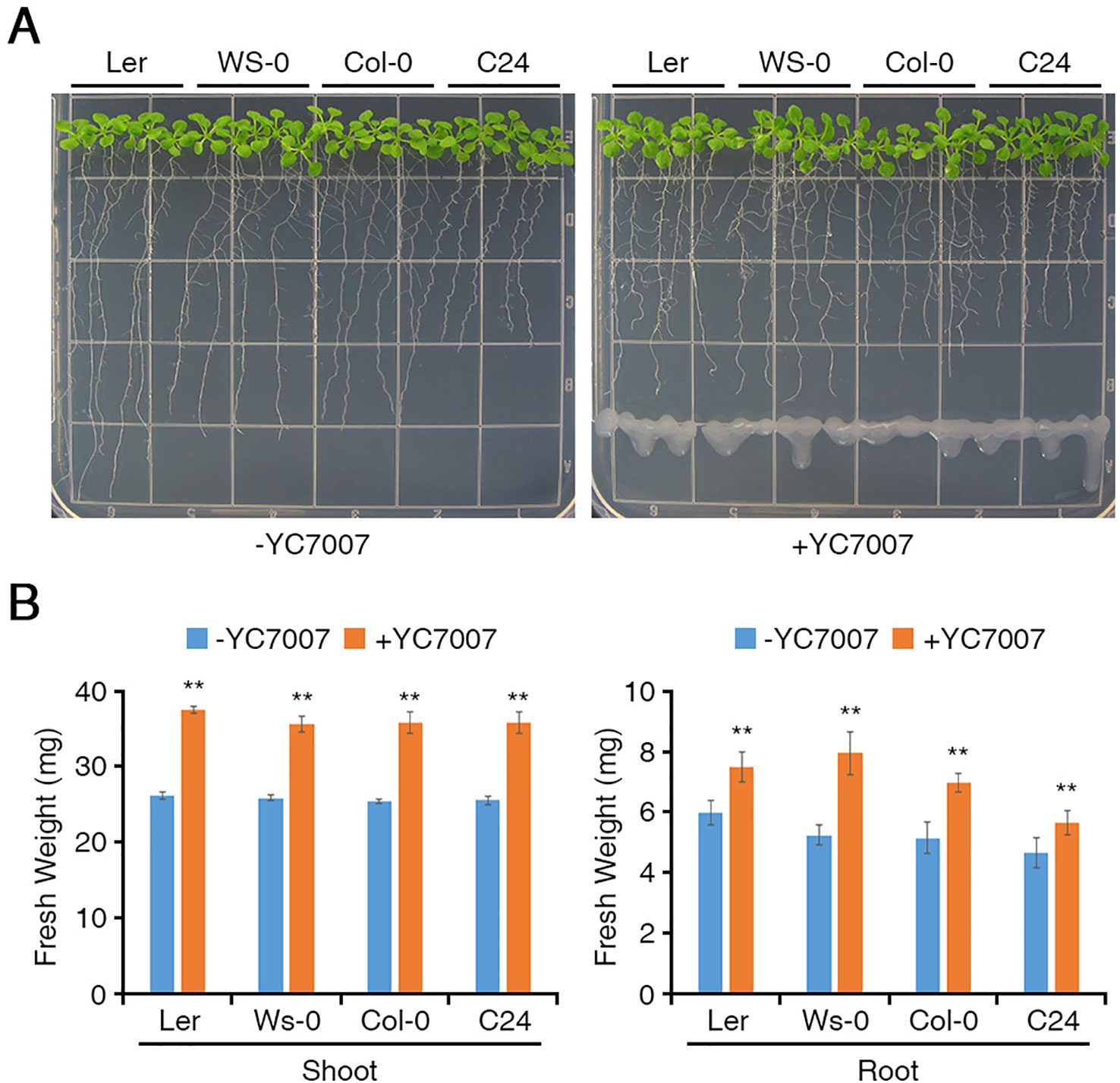
Figure 1 Comparison of plant growth among Arabidopsis Ler, Ws-0, Col-0, and C24 by an endophytic Bacillus oryzicola YC7007. Ler, Ws-0, Col-0 and C24 ecotype Arabidopsis plants were grown on 1/2 MS agar plate supplied with or without B. oryzicola YC7007. Suspension of B. oryzicola YC7007 in 10 mM MgSO4 was streak on the medium plate (+YC7007). The 10 mM MgSO4 solution was used as a control (Mock; -YC7007). (A) Photos were taken after 11 Days-After-Germination (DAG). (B) Fresh weight of shoot (left) and root (right), excising from the seedlings 11 DAG after B. oryzicola YC7007 treatment, were measured. The values indicated means ± SE of n = 3 replicates of 10 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (**, p-value < 0.01; Student’s t-test).
The volatile organic compounds (VOCs) of PGPR induced acidification of the rhizosphere (Zhang et al., 2009b). To test whether VOCs of YC7007 was involved in rhizosphere acidification, we performed quantitative measurement of [H+] release in the growth medium before and after YC7007 treatment (Table 1). The values of pH in bacterial growth medium (TBS) and plant growth medium (MS) decreased by YC7007 (Table 1). 4-day-old A. thaliana ecotype Col-0 seedlings were initially grown in medium with exposure to H2O or YC7007 for 5 days. The [H+] release ability in root was enhanced approximately 5-fold with exposure to YC7007 than H2O (Table 1). These results suggested that YC7007 effectively increased rhizosphere acidification through by promoting root [H+] release.
Bacillus oryzicola YC7007 Enhanced Plant Tolerance to Salt Stress
To test whether YC7007 has a protective effect in conditions of salt stress, we performed plant growth assays using A. thaliana ecotype Col-0 plants. At 14 DAI, the fresh weight of the shoots from the seedlings grown on salt MS media decreased approximately 37.8% compared to that of the untreated control shoots (Figure 2). Interestingly, the shoots from seedlings treated with YC7007 under salt stress conditions exhibited a 2-fold higher weight than untreated seedlings (Figure 2).
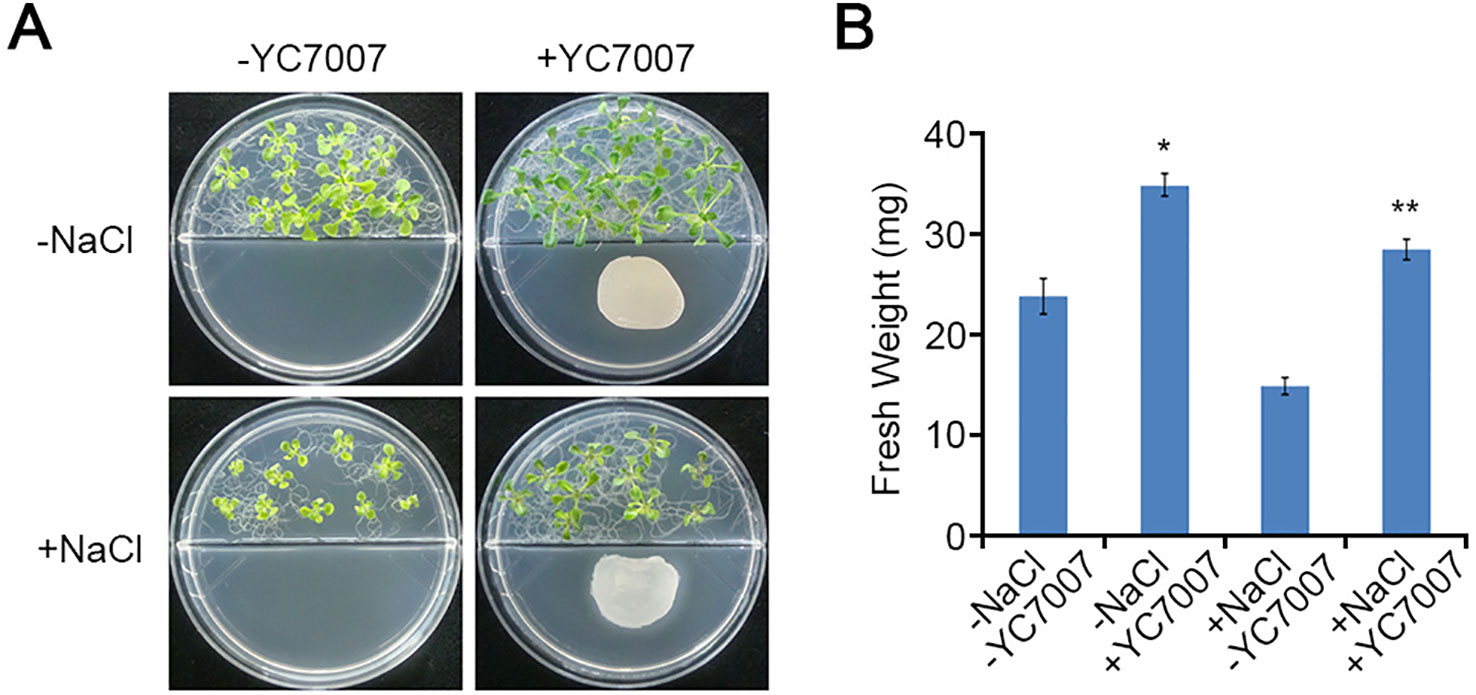
Figure 2 Effect of plant growth by B. oryzicola YC7007 during salt stress. Col-0 ecotype Arabidopsis plants were grown on 1/2 MS agar plate supplied with both 100 mM NaCl and B. oryzicola YC7007. Suspension of B. oryzicola YC7007 in 10 mM MgSO4 was streak on the medium plate with NaCl for co-cultivation. The 10 mM MgSO4 solution was used as a control (Mock; -YC7007). (A) Photos were taken after 14 Days-After-Inoculation (DAI). (B) Fresh weight of shoot, excising from the seedlings 14 DAI after both NaCl and B. oryzicola YC7007 treatment, were measured. The values indicated means ± SE of n = 5 replicates of 10 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (*, 0.01 < p-value ≤ 0.05; **, p-value < 0.01; Student’s t-test).
To investigate the effects of YC7007 on plant root development under salt-stress conditions, we performed root growth assays. The weight of the roots was reduced by about 35.2% by the salt treatment when compared to that of the untreated control. The inoculation of YC7007 for 14 days increased the weight of salt-treated seedlings under salt stress 2.2-fold (Figures 3A, B). In addition, co-cultivation with YC7007 increased the length of the primary root and the number of lateral roots about 1.7- and 2.5-fold, respectively, under conditions of salt stress (Figures 3C, D). These results suggested that PGPR YC7007 enhanced plant tolerance to salt stress through the promotion of shoot and root development.
Figure 3 Effect of root development by B. oryzicola YC7007 during salt stress.Four-day-old seedlings of Col-0 ecotype Arabidopsis plants were transferred and vertically grown on 1/2 MS agar plate supplied with both 100 mM NaCl and B. oryzicola YC7007. (A) Roots of plants were photographed after 14 Days-After-Inoculation (DAI). Bar = 10 mm. (B) Fresh weight of roots, excising from the seedlings 14 DAI after both NaCl and B. oryzicola YC7007 treatment, were measured. (C, D) Changes in length of primary root (C) and number of lateral root (D) were analyzed in (A). The values indicated means ± SE of n = 5 replicates of 10 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (**, p-value < 0.01; Student’s t-test).
Bacillus oryzicola YC7007 Increased the Content of Chlorophyll and MDA Under Salt Stress
The A. thaliana Col-0 shoots from seeds cultivated on the 1/2 X MS containing 1.5% sucrose and streaked with YC7007 exhibited 20% more chlorophyll than untreated control plants at 14 DAG (Figure 4A). Under salt stress, the inoculation of YC7007 increased chlorophyll content by 45.4% compared to that of the control plants (Figure 4A). These results suggest that YC7007 promotes photosynthesis through the accumulation of chlorophyll under normal conditions and ensures its ability upon conditions of salt stress.
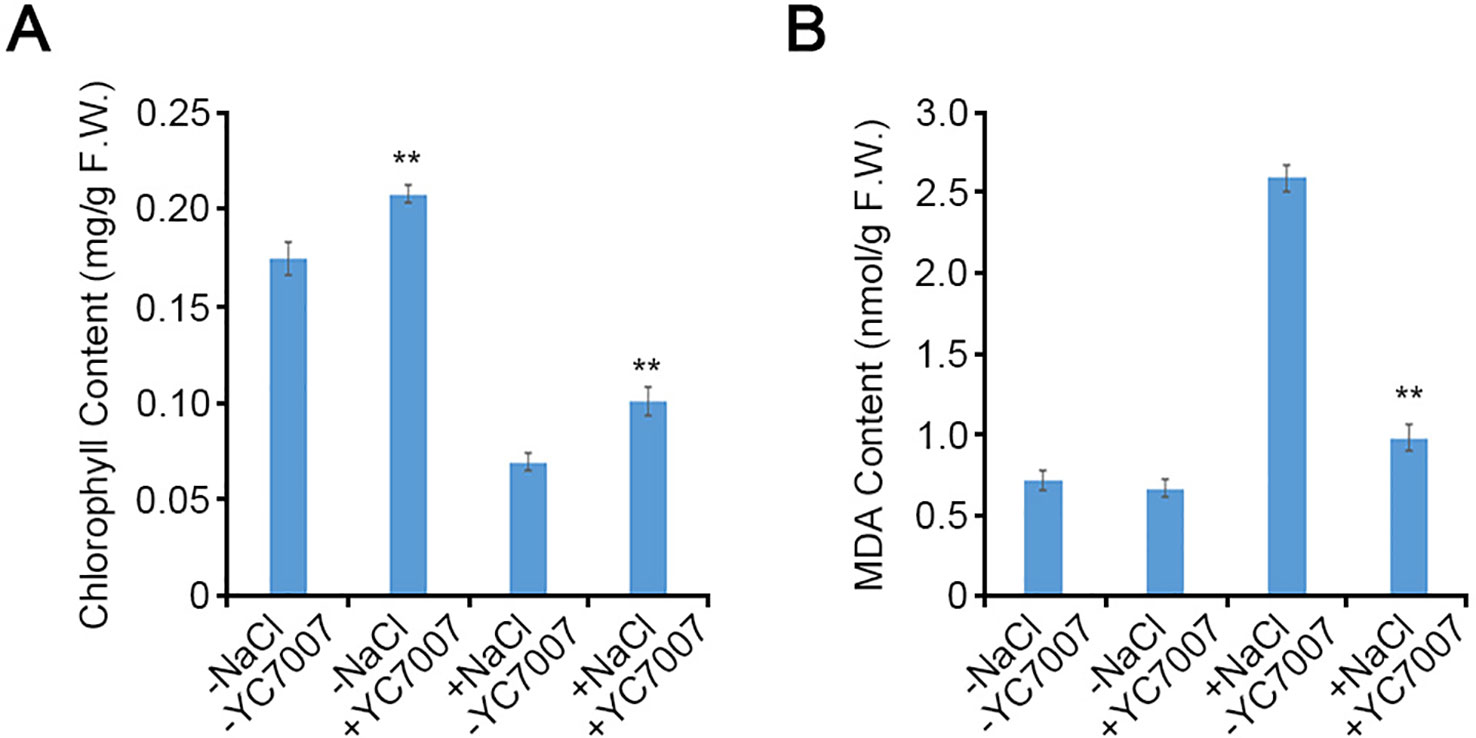
Figure 4 Quantitative comparison of chlorophyll and malondialdehyde (MDA) contents by B. oryzicola YC7007 during salt stress. Col-0 ecotype Arabidopsis plants were grown on 1/2 MS agar plate supplied with both 100 mM NaCl and B. oryzicola YC7007 after 14 Days-After-Germination (DAG). (A) 10 mg shoots from both NaCl- and B. oryzicola YC7007-treated seedlings were pulverized with acetone solution. The absorbance of samples was analyzed at wavelength of 645 nm and 663 nm. Total chlorophyll content was calculated using the following formula, as mg/g chlorophyll = (7.15 × A663 + 18.71 × A645) / 1000 / fresh weight of shoots. The values indicated means ± SE of n = 3 replicates of 10 seedlings for each experiment. (B) The MDA content in fresh weight of leaves was analyzed and compared between NaCl and B. oryzicola YC7007 treatment. The values indicated means ± SE of n = 3 replicates of 5 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (**, p-value < 0.01; Student’s t-test).
To investigate the change in MDA content due to treatment with YC7007 under salt-stress conditions, we measured a biomarker of lipid peroxidation (Kong et al., 2016). Even though there was no difference in MDA content under normal conditions, in the presence of salt, the MDA content of seedling shoots co-cultivated with YC7007 at 14 DAG decreased by about 62.3% compared to that of the untreated seedlings (Figure 4B). These results suggest that PGPR YC7007 improved plant tolerance to membrane oxidative damage under salt-stress conditions.
Bacillus oryzicola YC7007 Maintained Na+ Homeostasis
In the absence of salt, YC7007 treatment significantly increased the concentration of Na+ ion by about 5.4- and 10.4-fold in the shoots and roots, respectively (Figures 5A, B). However, in the presence of salt, YC7007 decreased Na+ ion content by about 20.9% and 38.9% in the shoots and roots, respectively (Figures 5A, B).
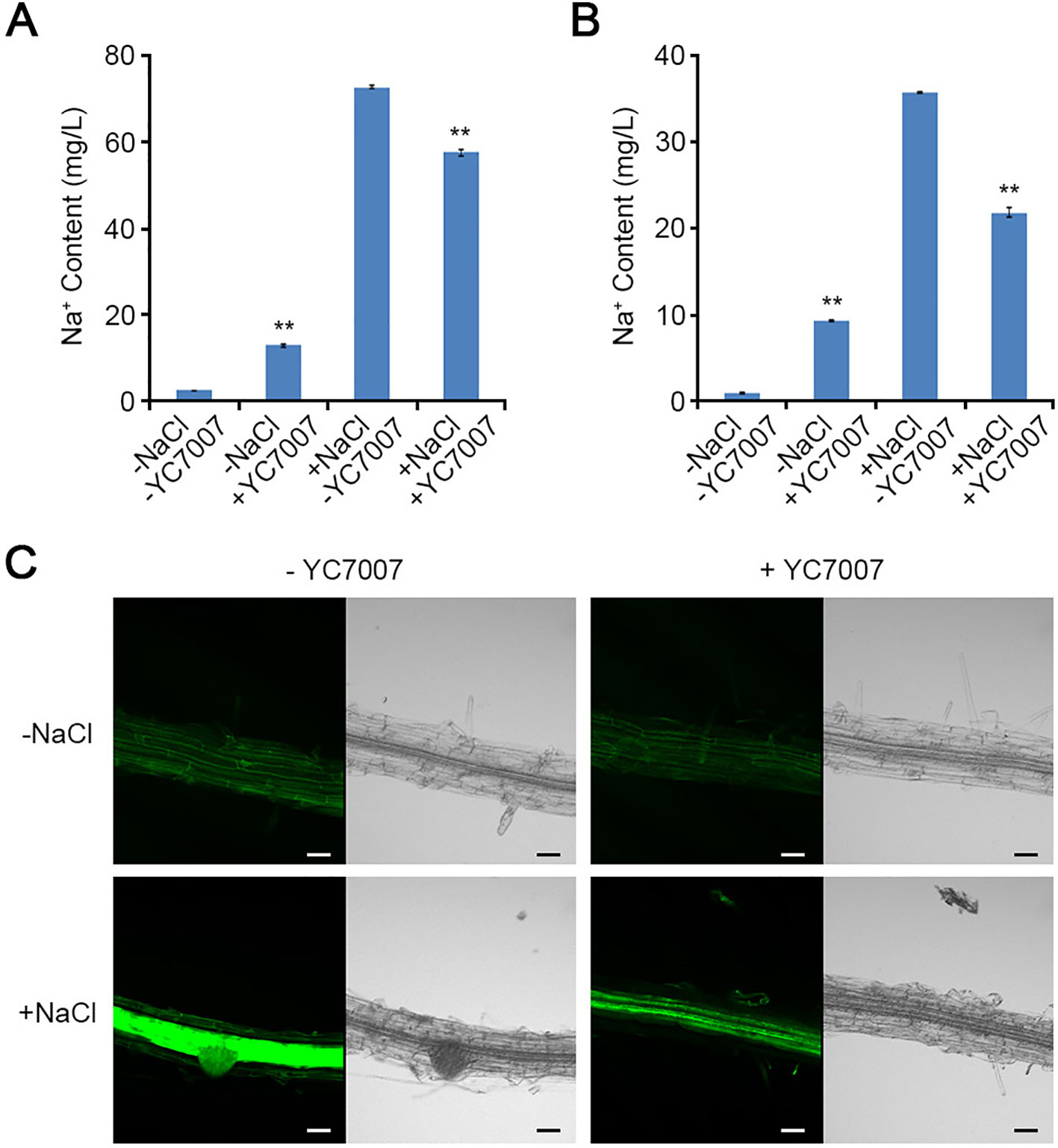
Figure 5 Effect of intracellular Na+ accumulation by B. oryzicola YC7007 during salt stress. (A, B) 10-day-old seedlings of Col-0 ecotype Arabidopsis plants were transferred 1/2 MS liquid medium supplied with both 200 mM NaCl and B. oryzicola YC7007. After 6 h, intracellular Na+ contents in shoots (A) and root (B) of Col-0 plants were measured by inductively coupled plasma (ICP) spectrometry. The values indicated means ± SE of n = 3 replicates of 10 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (**, p-value < 0.01; Student's t-test). (C) Visualization of intracellular Na+ by CoroNa-Green staining. 7-day-old seedlings of Col-0 ecotype Arabidopsis plants were transferred on filter papers supplied with both 200 mM NaCl and B. oryzicola YC7007. After 24 h, seedlings were stained with CoroNa-Green, and then visualized by confocal fluorescence microscopy. A representative image were photographed. Bar = 20 μm.
To visually confirm the effect of YC7007 in the control of Na+ ion content, a CoroNa-Green staining assay was performed. The dyes used indicators intracellular Na+ ions (Choi et al., 2011). The intensity of the fluorescent signal was reduced by YC7007 inoculation. However, the green fluorescence signals from the salt-treated roots without YC7007 were much stronger than those of YC7007-inoculated roots (Figure 5C). Taken together, these results indicate that YC7007 improved salt tolerance by controlling Na+ ion homeostasis A. thaliana.
The Expression of Stress-Response Genes was Increased by Bacillus oryzicola YC7007 Under Salt-Stress Conditions
Under normal conditions, YC7007 enhanced the transcriptional expression of RD29A, RD22, KIN1, and ERD1 in the roots (Figure 6A) and RD29A in the shoots (Figure 6B). Under salt-stress conditions, most stress-response genes (RD29A, RD29B, RD20, RD22, KIN1, and ERD1) were expressed strongly in the shoots and roots of seedlings co-cultivated with YC7007. These results indicated that YC7007 led to the up-regulation of stress-response genes under both natural and salt-stress conditions.
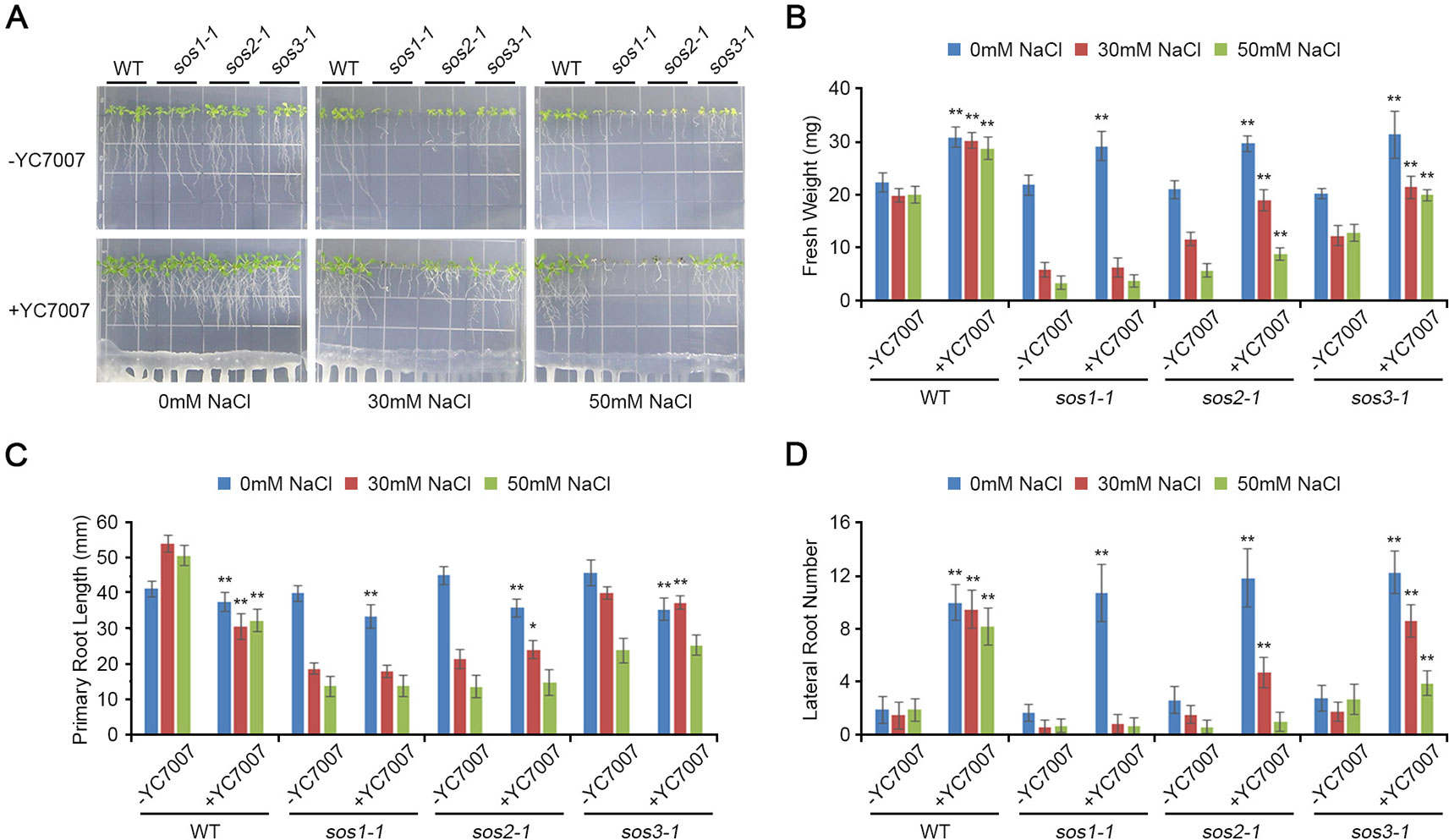
Figure 6 Comparison of plant growth among Arabidopsis sos mutants (sos1-1, sos2-1, and sos3-1) by B. oryzicola YC7007 during salt stress. Four-day-old seedlings of wild-type (WT), sos1-1, sos2-1, and sos3-1 plants were transferred and vertically grown on 1/2 MS agar plate supplied with both different concentration (0 mM, 30 mM, and 50 mM) of NaCl and B. oryzicola YC7007. (A) Roots of plants were photographed after 14 Days-After-Inoculation (DAI). Bar = 10 mm. (B) Fresh weight of shoots, excising from the seedlings 14 DAI after both different concentration of NaCl and B. oryzicola YC7007 treatment, were measured. (C, D) Changes in length of primary root (C) and number of lateral root (D) were analyzed in (A). The values indicated means ± SE of n = 4 replicates of 6 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (*, 0.01 < p-value ≤ 0.05; **, p-value < 0.01; Student’s t-test).
Bacillus oryzicola YC7007 was Involved in the SOS-Dependent Pathway of the Induction of Salt Tolerance
At 14 DAI, YC7007 increased the fresh weight and lateral root number of sos mutants but not primary root length under normal conditions (Figure 7). Upon conditions of salt stress, YC7007 alleviated the salt hypersensitivity of only sos2-1 and sos3-1 mutants, as reflected by an increase in their fresh weight and number of roots (Figure 7). Interestingly, the sos1-1 mutants were not affected by co-cultivation with YC7007 (Figure 7). These data suggest that YC7007 might be involved in salt tolerance through SOS1-dependent signaling. The phenotype of sos1-1 mutant to salt stress was more hypersensitive than that of sos2-1 and sos3-1 mutants (Figure 6) (Qiu et al., 2002; Qiu et al., 2004). The salt tolerance of sos2-1 and sos3-1 mutants by YC7007 were increased, suggesting that the functions of two genes in the same pathway to regulate by YC7007, unlike SOS1 function.
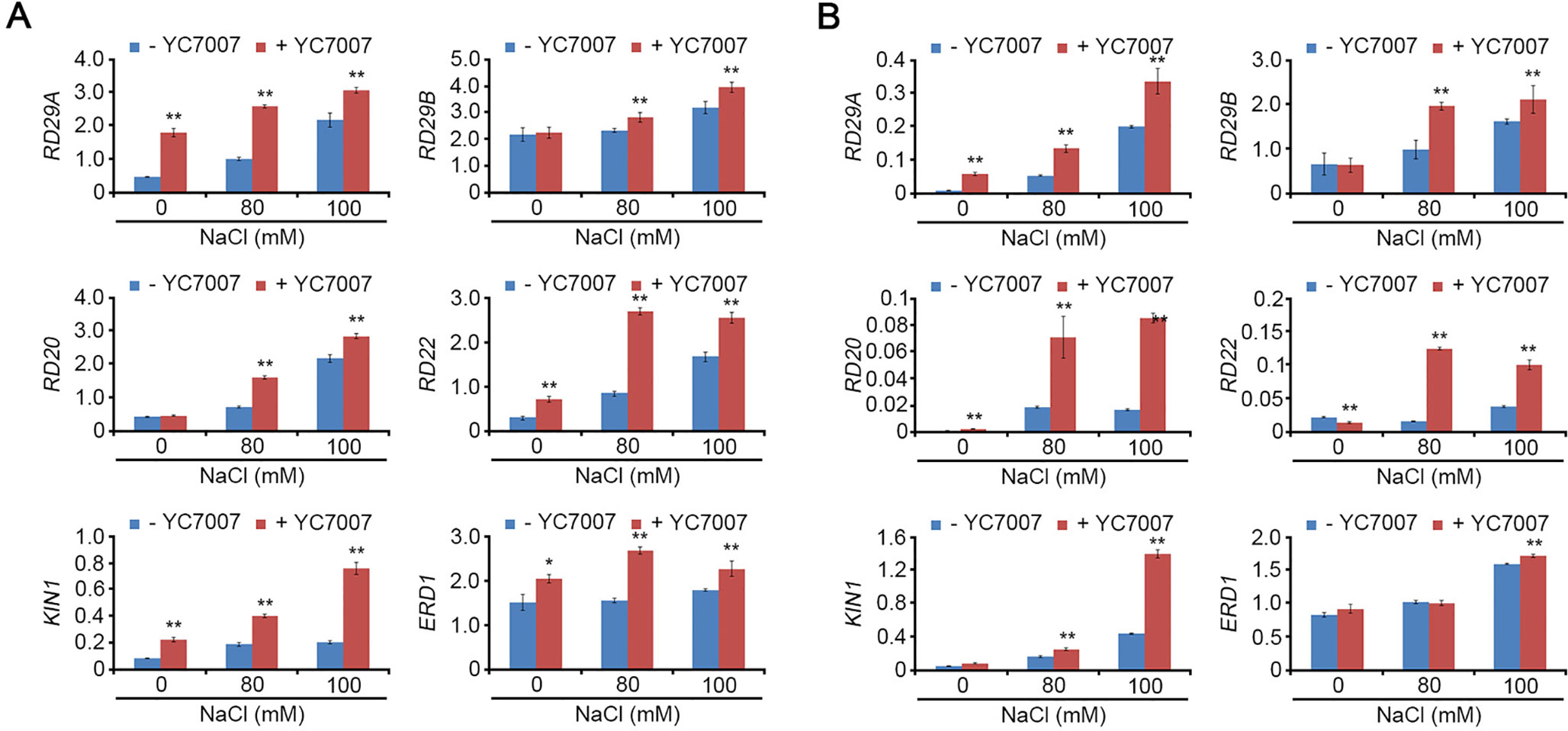
Figure 7 Effect of transcriptional expression of stress-responsive genes by B. oryzicola YC7007 during salt stress. 10-day-old seedlings of Col-0 ecotype Arabidopsis plants were transferred on 1/2 MS liquid medium supplied with both different concentration (0 mM, 80 mM, and 100 mM) of NaCl and B. oryzicola YC7007 for 6 h. Transcript levels of stress-responsive genes were measured by qRT-PCR in total RNA extracted from shoot (A) and root (B) of treated seedlings. Expression of TUBULIN2 was used for normalization. Bars represent mean ± SD of three biological replicates with three technical replicates each. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (*, 0.01 < p-value ≤ 0.05; **, p-value < 0.01; Student’s t-test).
Bacillus oryzicola YC7007 Enhanced Salt Tolerance in Vegetable Crops
To understand the effect of YC7007 on vegetable crops in relation to salt tolerance, we evaluated radish and cabbage plants in our assays. At 15 DAG, the plant growth of the two crops was enhanced given the observed increase in fresh weight due to co-cultivation with YC7007 under both normal and salt-stress conditions (Figure 8). Notably, even under conditions of high salt stress (e.g., 150 mM NaCl), the inoculation of YC7007 increased the fresh weight of both radish and cabbage seedlings by about 2.34- and 2.37-fold, respectively, compared to that of the untreated control plants. These results indicate that YC7007 enhanced the plant growth, plant development, and salt tolerance of vegetables under salt-stress conditions.
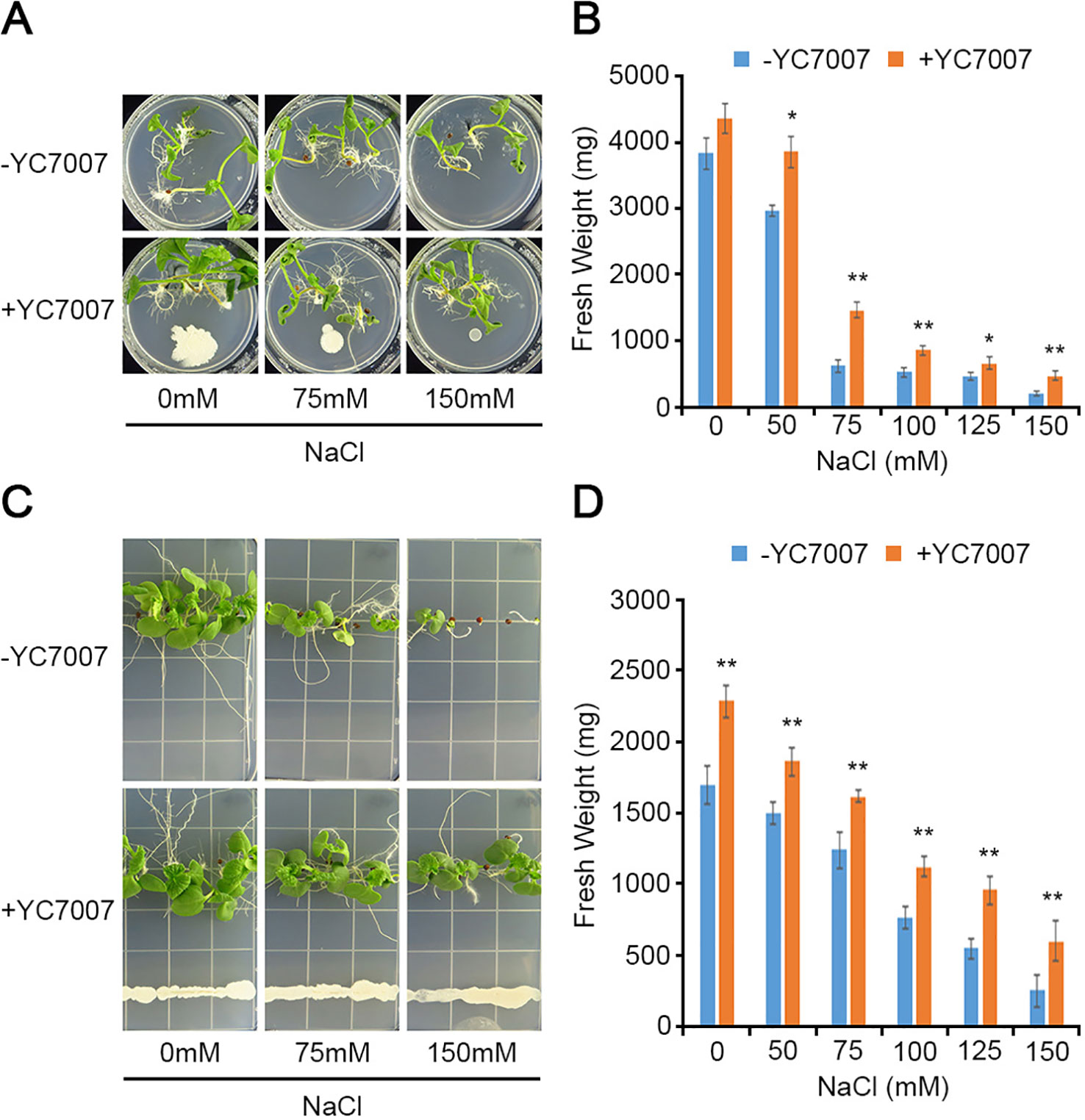
Figure 8 Comparison of plant growth among upland-crops, such as radish and cabbage, by B. oryzicola YC7007 during salt stress. The radish (Raphanus sativus var.; A, B) and cabbage (Brassica campestris ssp. Pekinensis; C, D) plants were grown on 1/2 MS agar plate supplied with both different concentration (0 mM, 75 mM, and 150 mM) of NaCl and B. oryzicola YC7007. (A, C) Radish (A) and cabbage (C) plants were photographed after 15 Days-After-Inoculation (DAI). (B, D) After both indicated different concentration of NaCl and B. oryzicola YC7007 treatment for 15 DAI, fresh weight of radish (B) and cabbage (D) seedlings were measured. The values indicated means ± SE of n = 3 replicates of 6 seedlings for each experiment. Asterisks represent significant differences from the untreated B. oryzicola YC7007 (-YC7007) (*, 0.01 < p-value ≤ 0.05; **, p-value < 0.01; Student’s t-test).
Discussion
The treatment of endophytic B. oryzicola YC7007 significantly promoted the growth and development of A. thaliana seedlings of all the tested ecotypes on 1/2 MS media given the observed increase in shoot and root weight. This bacterial strain has been previously found to promote rice growth while showing biological control activity against important seed-borne diseases (Chung et al., 2015). Some endophytic bacteria can be used as biocontrol agents for plant pathogens, insect, nematodes, and biostimulants to enhance plant growth and productivity (Santoyo et al., 2016). Furthermore, in this study we show that the B. oryzicola YC7007 strain enhanced the growth and the salt-stress tolerance of A. thaliana seedlings grown on 1/2 MS medium supplied with 100 mM NaCl given the increase in primary and lateral root development (Figures 1–3; Chung et al., 2015).
Various PGPR of different genera have been reported to induce salt-stress tolerance through the production of phytohormones, such as ACC deaminase, IAA, ABA, GA, and cytokinin (Nadeem et al., 2010; Numan et al., 2018). Plant growth-promoting rhizobacteria, such as B. subtilis, Rhizobium, and Pseudomonas species, have been found to increase plant tolerance to salt stress through the regulation of proline content and ion homeostasis in A. thaliana and Z. mays (Chen et al., 2007; Bano and Fatima, 2009). To reduce the impact of salt stress in plant, endophytic bacteria induce the accumulation of osmolytes and anti-oxidant compounds such as proline, glycine betaine, and anti-oxidant enzymes (Vaishnav et al., 2019). Rice inoculated with P. pseudoalcaligenes and Bacillus pumilus enhance salinity with higher level of glycine betaline (Jha et al., 2011). Volatile organic compounds (VOC) emitted from PGPR, which enter the atmosphere as vapors due to significantly high vapor pressure and low molecular weight (Vespermann et al., 2007; Dimkpa et al., 2009). VOCs from B. subtilis GB03 can stimulate many different hormonal signals which includes auxin, brassinosteroid cytokinin, GA and salicylic acid (SA) in Arabidopsis (Ryu et al., 2004). In addition, upon salt stress VOCs enhanced the plant tolerance of Arabidopsis to salt by regulating tissue-specific HKT1 expression, repressed in roots while upregulated in the shoots (Ryu et al., 2004; Zhang et al., 2008). Here, our study showed that the YC7007 strain enhanced plant growth and salinity tolerance (Figure 2). These salt stress responses alter the root architecture, especially the development of primary and lateral roots, by regulating cell division and differentiation (Vacheron et al., 2013; Etesami and Maheshwari, 2018; El-Esawi et al., 2018). The YC7007 strain has also been found to increase the fresh root weight, primary root length, and the number of lateral roots in A. thaliana plants under salt stress (Figure 3). Thus, YC7007 appears to influence the root system architecture via root developmental changes, which might enhance plant tolerance to salt stress. The changes in cellular root levels by YC7007 resulted in root formation, which was probably due to the production of VOCs emitted from YC7007 in response to salt stress as the bacterial cells did not touch the plant roots.
Plant growth-promoting rhizobacteria-mediated salt tolerance appears to include ABA- and SOS-mediated pathways and detoxification via the up-regulated expression of ABA-signaling cascade genes as well as the genes of antioxidant enzymes and osmolytes (Bharti et al., 2016). In our study, the salt sensitivity of sos mutants was examined under salt-stress conditions and co-cultivation with YC7007 to understand SOS-mediated salt tolerance. The YC7007 strain reduced salt accumulation both in the shoots and roots of A. thaliana seedlings (Figure 5). The salt hypersensitivity of the sos1 mutants was not affected by YC7007, while sos2 and sos3 mutants exhibited increased salt-stress tolerance, suggesting that SOS1 is required for Na+ exclusion even in the presence of PGPR (Figure 7). The level of salt-induced MDA during salt stress was reduced by YC7007, which resulted oxidative damage (Figure 4). Moreover, genes involved in stress responses, such as RD29A, RD29B, KIN1, and ERD1, were expressed more in bacteria-treated plants under conditions of salt stress (Figure 6). The Na+/H+ transporter SOS1 was concerned in Na+ exclusion to salt stress response via alkalinization of intracellular pH and H+ efflux in sos1 mutant (Guo et al., 2009). Our observations demonstrated that YC7007 was involved in rhizosphere acidification (Table 1). Taken together, YC7007 seems to induce salt tolerance via the SOS1-dependent signaling pathway under salt-stress conditions.
Each plant species has a different sensitivity to salt stress, although plant growth is eventually interrupted by high concentrations of salt in soil (Shannon and Grieve, 1999; Parida and Das, 2005). Many PGPR have been reported to enhance plant growth by enhancing stress tolerance in various crops, such as tomato, pepper, canola, bean, and lettuce during exposure to salt stress (Barassi et al., 2006). In particular, YC7007 enhanced the stress tolerance of radish and cabbage seedlings under conditions of salt stress, reflected in the observed increase in fresh weight under multiple salt-stress conditions (Figure 8).
We functionally characterized the role of B. oryzicola YC7007 in the SOS1-dependent signaling pathway involved in plant tolerance to salt stress. Our molecular, biological, and biochemical results suggest that B. oryzicola YC7007 regulates chlorophyll and MDA content while maintaining intracellular Na+ ion concentrations to enhance stress tolerance under conditions of salt stress. The physiological functions of the endophytic bacteria YC7007 are essential to understanding bacterial interactions that enhance plant growth and systemic tolerance to salt stress in plants as well as plant-bacteria communications. A clear understanding of the communication between PGPR and plants is more important than ever to ensure crop productivity and the development of economical crop management systems in agriculture.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
YC and DB designed the experiments and wrote the manuscript. DB, MR, AK, and HP performed the experiments. MK and D-JY discussed the experimental results and commented on the manuscript.
Funding
This study was supported by grant from the Next-Generation BioGreen21 Program (grant No. PJ 01104901 to YC and PJ01104902 to DB), Rural Development Administration Republic of Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01646/full#supplementary-material
References
Abdel Latef, A. A. H., Srivastava, A. K., Saber, H., Alwaleed, E. A., Tran, L. P. (2017). Sargassum muticum and Jania rubens regulate amino acid metabolism to improve growth and alleviate salinity in chickpea. Sci. Rep. 7 (1), 10537. doi: 10.1038/s41598-017-07692-w
Abdel Latef, A. A. H., Srivastava, A. K., Abdel-sadek, M. S., Kordrostam, M., Tran, L. P. (2018). Titanium dioxide nanoparticles improve growth and enhance tolerance of broad bean plants under saline conditions. LandDegrad. Develop. 29 (4), 1065–1073. doi: 10.1002/ldr.2780
Abdel Latef, A. A. H., Mostofa, M. G., Rahman, M. M., Abdel-Farid, I. B., Tran, L. P. (2019a). Extracts from yeast and carrot roots enhance maize performance under seawater-induced salt stress by altering physio-biochemical characteristics of stressed plants. J. Plant Growth Regulat. 38 (3), 966–979. doi: 10.1007/s00344-018-9906-8
Abdel Latef, A. A. H., Kordrostami, M., Zakir, A., Zaki, H., Saleh, O. M. (2019b). Eustress with H2O2 facilitates plant growth by improving tolerance to salt stress in two wheat cultivars. Plants (Basel) 8 (9), E303. doi: 10.3390/plants8090303
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sánchez-Blanco, M. J., Hernández, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy. 7 (1), 1–18. doi: 10.3390/agronomy7010018
Ahmad, M., Zahir, Z. A., Asghar, H. N., Arshad, M. (2012). The combined application of rhizobial strains and plant growth promoting rhizobacteria improves growth and productivity of mung bean (Vigna radiata L.) under salt-stressed conditions. Ann. Microbiol. 62 (3), 1321–1330. doi: 10.1007/s13213-011-0380-9
Ahmad, M., Zahir, Z. A., Nazli, F., Akram, F., Arshad, M., Khalid, M. (2014). Effectiveness of halo-tolerant, auxin producing Pseudomonas and Rhizobium strains to improve osmotic stress tolerance in mung bean (Vigna radiata L.). Braz. J. Microbiol. 44 (4), 1341–1348. doi: 10.1590/S1517-83822013000400045
Bano, A., Fatima, M. (2009). Salt tolerance in Zea mays (L.) following inoculation with Rhizobium and Pseudomonas. Biol. Fertil. Soils 45, 405–413. doi: 10.1007/s00374-008-0344-9
Barassi, C. A., Ayrault, G., Creus, C. M., Sueldo, R. J., Sobero, M. T. (2006). Seed inoculation with Azospirillum mitigates NaCl effects on lettuce. Sci. Hortic. 109, 8–14. doi: 10.1016/j.scienta.2006.02.025
Bharti, N., Yadav, D., Barnawal, D., Maji, D., Kalra, A. (2013). Exiguobacterium oxidotolerans, a halotolerant plant growth promoting rhizobacteria, improves yield and content of secondary metabolites in Bacopa monnieri (L.) Pennell under primary and secondary salt stress. World J. Microbiol. Biotechnol. 29 (2), 379–387. doi: 10.1007/s11274-012-1192-1
Bharti, N., Pandey, S. S., Barnawal, D., Patel, V. K., Kalra, A. (2016). Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci. Rep. 6, 34768. doi: 10.1038/srep34768
Bhattacharyya, P. N., Jha, D. K. (2012). Plant growth-promoting rhizobacteria (PGPR): emergence in agriculture. World J. Microbiol. Biotechnol. 28 (4), 1327–1350. doi: 10.1007/s11274-011-0979-9
Chaves, M. M., Flexas, J., Pinheiro, C. (2009). Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann. Bot. 103 (4), 551–560. doi: 10.1093/aob/mcn125
Chen, M., Wei, H., Cao, J., Liu, R., Wang, Y., Zheng, C. (2007). Expression of Bacillus subtilis proBA genes and reduction of feedback inhibition of proline synthesis increases proline production and confers osmotolerance in transgenic Arabdopsis. J. Biochem. Mol. Biol. 40, 396403. doi: 10.5483/BMBRep.2007.40.3.396
Choi, W., Baek, D., Oh, D. H., Park, J., Hong, H., Kim, W. Y., et al. (2011). NKS1, Na(+)- and K(+)-sensitive 1, regulates ion homeostasis in an SOS-independent pathway in Arabidopsis. Phytochemistry. 72 (4–5), 330–336. doi: 10.1016/j.phytochem.2010.12.005
Choudhary, D. K. (2012). Microbial rescue to plant under habitat-imposed abiotic and biotic stresses. Appl. Microbiol. Biotechnol. 96 (5), 1137–1155. doi: 10.1007/s00253-012-4429-x
Chung, E. J., Hossain, M. T., Khan, A., Kim, K. H., Jeon, C. O., Chung, Y. R. (2015). Bacillus oryzicola sp. nov., an Endophytic Bacterium Isolated from the Roots of Rice with Antimicrobial, Plant Growth Promoting, and Systemic Resistance Inducing Activities in Rice. Plant Pathol. J. 31 (2), 152–164. doi: 10.5423/PPJ.OA.12.2014.0136
Cowan, A. K., Cairns, A. L. P., Bartels-Rahm, B. (1999). Regulation of abscisic acid metabolism: towards a metabolic basis for abscisic acid-cytokinin antagonism. J. Exp. Bot. 50 (334), 595–603. doi: 10.1093/jxb/50.334.595
Dimkpa, C., Weinand, T., Asch, F. (2009). Plant rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
El-Esawi, M. A., Alaraidh, I. A., Alsahli, A. A., Alamri, S. A., Ali, H. M., Alayafi, A. A. (2018). Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress-responsive genes expression. Plant Physiol. Biochem. 132, 375–384. doi: 10.1016/j.plaphy.2018.09.026
Etesami, H., Maheshwari, D. K. (2018). Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 156, 225–246. doi: 10.1016/j.ecoenv.2018.03.013
Figueiredo, M. V. B., Burity, H. A., Martínez, C. R., Chanway, C. P. (2008). Alleviation of drought stress in the common bean (Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil Ecol. 40 (1), 182–188. doi: 10.1016/j.apsoil.2008.04.005
Flowers, T. J. (2004). Improving crop salt tolerance. J. Exp. Bot. 55 (396), 307–319. doi: 10.1093/jxb/erh003
Gray, E. J., Smith, D. L. (2005). Intracellular and extracellular PGPR: commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol. Biochem. 37, 395–412. doi: 10.1016/j.soilbio.2004.08.030
Guo, K. M., Babourina, O., Rengel, Z. (2009). Na(+)/H(+) antiporter activity of the SOS1 gene: lifetime imaging analysis and electrophysiological studies on Arabidopsis seedlings. Physiol. Plant. 137 (2), 155–165. doi: 10.1111/j.1399-3054.2009.01274.x
Gyaneshwar, P., Kumar, G. N., Parekh, L. J., Poole, P. S. (2002). Role of soil microorganisms in improving P nutrition of plants. Plant Soil. 245 (1), 133–143. doi: 10.1007/978-94-017-1570-6_15
Habib, S. H., Kausar, H., Saud, H. M. (2016). Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ROS-scavenging enzymes. Biomed. Res. Int. 2016, 6284547. doi: 10.1155/2016/6284547
Hamdia, A. B. E., Shaddad, M. A. K., Doaa, M. M. (2004). Mechanisms of salt tolerance and interactive effects of Azospirillum brasilense inoculation on maize cultivars grown under salt stress conditions. Plant Growth Regul. 44 (2), 165–174. doi: 10.1023/B:GROW.0000049414.03099.9b
Hernández, J. A. (2019). Salinity tolerance in plants: trends and perspectives. Int. J. Mol. Sci. 20 (10), E2408. doi: 10.3390/ijms20102408
Hossain, M. T., Khan, A., Chung, E. J., Rashid, M. H., Chung, Y. R. (2016). Biological control of rice bakanae by an endophytic Bacillus oryzicola YC7007. Plant Pathol. J. 32 (3), 228–241. doi: 10.5423/PPJ.OA.10.2015.0218
Jha, Y., Subramanian, R. B., Patel, S. (2011). Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiologiae Plantarum. 33 (3), 797–802. doi: 10.1007/s11738-010-0604-9
Ji, H., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., Li, X. (2013). The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6 (2), 275–286. doi: 10.1093/mp/sst017
Kasotia, A., Choudhary, D. K. (2014). “Role of endophytic microbes in mitigation of abiotic stress in plants,” in Emerging Technologies and Management of Crop Stress Tolerance. Chapter 4, vol. 2., 97–108. doi: 10.1016/B978-0-12-800875-1.00004-1
Kohler, J., Hernández, J. A., Caravaca, F., Roldán, A. (2009). Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ. Exp. Bot. 65 (2–3), 245–252. doi: 10.1016/j.envexpbot.2008.09.008
Kong, W., Liu, F., Zhang, C., Zhang, J., Feng, H. (2016). Non-destructive determination of Malondialdehyde (MDA) distribution in oilseed rape leaves by laboratory scale NIR hyperspectral imaging. Sci. Rep. 6, 35393. doi: 10.1038/srep35393
Li, L., Li, L., Wang, X., Zhu, P., Wu, H., Qi, S. (2017). Plant growth-promoting endophyte Piriformospora indica alleviates salinity stress in Medicago truncatula. Plant Physiol. Biochem. 119, 211–223. doi: 10.1016/j.plaphy.2017.08.029
Mayak, S., Tirosh, T., Glick, B. R. (2004). Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 42 (6), 565–572. doi: 10.1016/j.plaphy.2004.05.009
Munns, R., Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Nadeem, S. M., Zahir, Z. A., Naveed, M., Ashraf, M. (2010). Microbial ACC-deaminase: prospects and applications for inducing salt tolerance in plants. Crit. Rev. Plant Sci. 29 (6), 360–393. doi: 10.1080/07352689.2010.524518
Nelson, G. C., Valin, H., Sands, R. D., Havlík, P., Ahammad, H., Deryng, D., et al. (2014). Climate change effects on agriculture: economic responses to biophysical shocks. Proc. Natl. Acad. Sci. U. S. A. 111 (9), 3274–3279. doi: 10.1073/pnas.1222465110
Numan, M., Bashir, S., Khan, Y., Mumtaz, R., Shinwari, Z. K., Khan, A. L., et al. (2018). Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol. Res. 209, 21–32. doi: 10.1016/j.micres.2018.02.003
Paré, P. W., Zhang, H. M., Aziz, M., Xie, X. T., Kim, M. S., Shen, X. (2011). Beneficial rhizobacteria induce plant growth: mapping signaling networks in Arabidopsis. Biocommun. Soil Microorg. Soil Biol. 23, 403–412. doi: 10.1007/978-3-642-14512-4_15
Parida, A. K., Das, A. B. (2005). Salt tolerance and salinity effects on plants: a review. Ecotoxicol. Environ. Saf. 60, 324–349. doi: 10.1016/j.ecoenv.2004.06.010
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U. S. A. 99 (12), 8436–8441. doi: 10.1073/pnas.122224699
Qiu, Q. S., Guo, Y., Quintero, F. J., Pardo, J. M., Schumaker, K. S., Zhu, J. K. (2004). Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the salt-overly-sensitive (SOS) pathway. J. Biol. Chem. 279 (1), 207–215. doi: 10.1074/jbc.M307982200
Rus, A., Yokoi, S., Sharkhuu, A., Reddy, M., Lee, B. H., Matsumoto, T. K., et al. (2001). AtHKT1 is a salt tolerance determinant that controls Na(+) entry into plant roots. Proc. Natl. Acad. Sci. U. S. A. 98 (24), 14150–14155. doi: 10.1073/pnas.241501798
Ryu, C. M., Farag, M. A., Hu, C. H., Reddy, M. S., Kloepper, J. W., Paré, P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134 (3), 1017–1026. doi: 10.1104/pp.103.026583
Santoyo, G., Moreno-Hagelsieb, G., Orozco-Mosqueda Mdel, C., Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Shannon, M. C., Grieve, C. M. (1999). Tolerance of vegetable crops to salinity. Sci. Hortic. 78, 5–38. doi: 10.1016/S0304-4238(98)00189-7
Shinozaki, K., Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007 58 (2), 221–227. doi: 10.1093/jxb/erl164
Shukla, P. S., Agarwal, P. K., Jha, B. (2012). Improved salinity tolerance of Arachis hypogaea (L.) by the interaction of halotolerant plant-growth-promoting rhizobacteria. J. Plant Growth Regul. 31 (2), 195–206. doi: 10.1007/s00344-011-9231-y
Siddikee, M. A., Glick, B. R., Chauhan, P. S., Yim, W. J., Sa, T. (2011). Enhancement of growth and salt tolerance of red pepper seedlings (Capsicum annuum L.) by regulating stress ethylene synthesis with halotolerant bacteria containing 1-aminocyclopropane-1-carboxylic acid deaminase activity. Plant Physiol. Biochem. 49 (4), 427–434. doi: 10.1016/j.plaphy.2011.01.015
Tank, N., Saraf, M. (2010). Salinity-resistant plant growth promoting rhizobacteria ameliorates sodium chloride stress on tomato plants. J. Plant Interact. 5 (1), 51–58. doi: 10.1080/17429140903125848
Upadhyay, S. K., Singh, D. P. (2015). Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. (Stuttg). 17 (1), 288–293. doi: 10.1111/plb.12173
Vacheron, J., Desbrosses, G., Bouffaud, M. L., Touraine, B., Moënne-Loccoz, Y., Muller, D., et al. (2013). Plant growth-promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 356. doi: 10.3389/fpls.2013.00356
Vaishnav, A., Shukla, A. K., Sharma, A., Kumar, R., Choudhary, D. K. (2019). Endophytic bacteria in plant salt stress tolerance: current and future prospects. J. Plant Growth Regul. 38 (2), 650–668. doi: 10.1007/s00344-018-9880-1
Van Oosten, M. J., Silletti, S., Guida, G., Cirillo, V., Di Stasio, E., Carillo, P., et al. (2017). A benzimidazole proton pump inhibitor increases growth and tolerance to salt stress in tomato. Front. Plant Sci. 8, 1220. doi: 10.3389/fpls.2017.01220
Vespermann, A., Kai, M., Piechulla, B. (2007). Rhizobacterial volatiles affect the growth of fungi and Arabidopsis thaliana. Appl. Environ. Microbiol. 73 (17), 5639–5641. doi: 10.1128/AEM.01078-07
Xie, X., Zhang, H., Paré, P. W. (2009). Sustained growth promotion in Arabidopsis with long-term exposure to the beneficial soil bacterium Bacillus subtilis (GB03). Plant Signal Behav. 4 (10), 948–953. doi: 10.4161/psb.4.10.9709
Yang, J., Kloepper, J. W., Ryu, C. M. (2009). Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14 (1), 1–4. doi: 10.1016/j.tplants.2008.10.004
Zhang, H., Kim, M. S., Sun, Y., Dowd, S. E., Shi, H., Paré, P. W. (2008). Soil bacteria confer plant salt tolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant Microbe Interact. 21 (6), 737–744. doi: 10.1094/MPMI-21-6-0737
Zhang, R. H., Li, J., Guo, S. R., Tezuka, T. (2009a). Effects of exogenous putrescine on gas-exchange characteristics and chlorophyll fluorescence of NaCl-stressed cucumber seedlings. Photosynth. Res. 100 (3), 155–162. doi: 10.1007/s11120-009-9441-3
Zhang, H., Sun, Y., Xie, X., Kim, M. S., Dowd, S. E., Paré, P. W. (2009b). A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 58 (4), 568–577. doi: 10.1111/j.1365-313X.2009.03803.x
Zhang, Y., Lv, Y., Jahan, N., Chen, G., Ren, D., Guo, L. (2018). Sensing of abiotic stress and ionic stress responses in plants. Int. J. Mol. Sci. 19 (11), E3298. doi: 10.3390/ijms19113298
Zhu, J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. doi: 10.1146/annurev.arplant.53.091401.143329
Zhu, J. K., Liu, J., Xiong, L. (1998). Genetic analysis of salt tolerance in arabidopsis. Evidence for a critical role of potassium nutrition. Plant Cell. 10 (7), 1181–1191. doi: 10.1105/tpc.10.7.1181
Keywords: Bacillus oryzicola, plant growth-promoting rhizobacteria, salt-stress tolerance, SOS pathway, malondialdehyde, sodium ion
Citation: Baek D, Rokibuzzaman M, Khan A, Kim MC, Park HJ, Yun D-j and Chung YR (2020) Plant-Growth Promoting Bacillus oryzicola YC7007 Modulates Stress-Response Gene Expression and Provides Protection From Salt Stress. Front. Plant Sci. 10:1646. doi: 10.3389/fpls.2019.01646
Received: 13 September 2019; Accepted: 22 November 2019;
Published: 09 January 2020.
Edited by:
Huiming Zhang, Shanghai Institutes for Biological Sciences (CAS), ChinaReviewed by:
Arafat Abdel Hamed Abdel Latef, South Valley University, EgyptKazuo Nakashima, Japan International Research Center for Agricultural Sciences, Japan
Copyright © 2020 Baek, Rokibuzzaman, Khan, Kim, Park, Yun and Chung. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Young Ryun Chung, eXJjaHVuZ0BnbnUuYWMua3I=
 Dongwon Baek
Dongwon Baek Mohammad Rokibuzzaman
Mohammad Rokibuzzaman Ajmal Khan2
Ajmal Khan2 Min Chul Kim
Min Chul Kim Hee Jin Park
Hee Jin Park Dae-jin Yun
Dae-jin Yun Young Ryun Chung
Young Ryun Chung