
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 04 November 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01402
γ-glutamyl transferases/transpeptidases (E.C. 2.3.2.2, GGTs) are involved in the catabolism of many compounds that are conjugated to glutathione (GSH), which have a variety of roles. GSH can act as storage and transport vehicle for reduced sulfur; it is involved in the detoxification of xenobiotics and also acts as a redox buffer by utilizing its thiol residue to protect against reactive oxygen species, which accumulate in response to biotic and abiotic stress. Furthermore, many distinctive flavor and aroma compounds in Sauvignon blanc wines originate from odorless C5- and C6-GSH conjugates or their GGT catabolized derivatives. These precursors are then processed into their volatile forms by yeast during fermentation. In many plant species, two or more isoforms of GGTs exist that target GSH-conjugates to either the apoplast or the vacuole. A bioinformatics approach identified multiple GGT candidates in grapevine (Vitis vinifera). However, only a single candidate, VvGGT3, has all the conserved residues needed for GGT activity. This is intriguing given the variety of roles of GSH and GGTs in plant cells. Characterization of VvGGT3 from cv. Sauvignon blanc was then undertaken. The VvGGT3 transcript is present in roots, leaves, inflorescences, and tendril and at equal abundance in the skin, pulp, and seed of mature berries and shows steady accumulation over the course of whole berry development. In addition, the VvGGT3 transcript in whole berries is upregulated upon Botrytis cinerea infection as well as mechanical damage to leaf tissue. VvGGT3-GFP fusion proteins transiently over-expressed in onion cells were used to study subcellular localization. To confirm VvGGT3 activity and localization in vivo, the fluorescent γ-glutamyl-7-amido-4-methylcoumarin substrate was added to Nicotiana benthamiana leaves transiently over-expressing VvGGT3. In combination, these results suggest that the functional VvGGT3 is associated with membrane-like structures. This is not consistent with its closely related functionally characterized GGTs from Arabidopsis, radish and garlic.
In plants, glutathione (GSH, γ-Glu-Cys-Gly) is involved in a multitude of cellular processes. It is often conjugated by glutathione S-transferases (GSTs) to a wide range of chemicals, both derived from cellular metabolism and externally occurring xenobiotics (Rouhier et al., 2008). These GSH-conjugates are mostly targeted to the vacuole before being metabolized further (Edwards et al., 2000). The initial step of GSH-conjugate catabolism is carried out by γ-glutamyl transferases/transpeptidases (GGTs; EC 2.3.2.2), that promote the cleavage of the γ-glutamyl moiety of GSH. Following which, the remaining Cys-Gly-conjugated dipeptide is attacked by a proposed Cys-Gly dipeptidase to release Gly, with Cys remaining linked to the conjugate (Taniguchi and Ikeda, 2006; Castellano and Merlino, 2013; Masi et al., 2015).
In mammalian systems, where GGTs have been most extensively studied, GGTs are found to be N-terminally bound to the plasma membrane with the catalytic site facing the extracellular space. Following translation, mammalian as well as Escherichia coli GGTs undergo an autoproteolytic cleavage that is dependent on a conserved threonine residue (Hashimoto et al., 1995; Okada et al., 2006; Taniguchi and Ikeda, 2006) into a large and a small subunit. These subunits then self-assemble into functional heterodimers (Tate and Meister, 1978; Masi et al., 2007; Castellano and Merlino, 2013). GGT transcription is regulated in a tissue-specific manner and is highly represented in organs with secretory and absorptive functions such as the kidney and liver. GGTs play a role in maintaining cellular redox homeostasis by regulating the cellular levels of GSH (Castellano and Merlino, 2012).
In plants, there has been comparatively little research focus on GGTs. However, GGTs from a range of model as well as non-model plant species have begun to be functionally characterized (Nakano et al., 2006b; Yoshimoto et al., 2015). In Arabidopsis thaliana (Arabidopsis), where plant GGTs have been most extensively studied, four GGT genes have been identified, of which three are functional (AtGGT1, AtGGT2, and AtGGT4) (Grzam et al., 2007; Martin et al., 2007; Ohkama-Ohtsu et al., 2007a; Tolin et al., 2013). AtGGT3 lacks segments of exons which possess catalytic activity and is most likely a pseudogene (Ohkama-Ohtsu et al., 2009). AtGGT1 and AtGGT2 are located in the apoplast, ionically-bound to the cell wall and/or plasma membrane, as they could only be liberated with high-salt treatments (Ohkama-Ohtsu et al., 2007a; Giaretta et al., 2017). Both are proposed to be involved in the prevention of oxidative stress by degrading the oxidized form of glutathione (GSSG) (Ohkama-Ohtsu et al., 2009; Noctor et al., 2012). AtGGT1 is expressed throughout the plant with expression predominantly found in leaves and the vascular system, whereas AtGGT2 is not transcribed in leaves but is specifically transcribed in GSH sink tissues such as seeds, pollen, and trichomes, AtGGT2 is also weakly transcribed in roots (Martin et al., 2007; Destro et al., 2011; Giaretta et al., 2017). In Atggt1 knock-out mutants, levels of GSSG in the apoplastic fraction from leaves are 7-fold higher than in wild type plants (Ohkama-Ohtsu et al., 2007a) indicating the need for AtGGT1 to metabolize GSSG. Phenotypically, Atggt1 plants show yellowing in the older leaves. This is proposed to be caused by oxidative stress as indicated by higher lipid peroxidation in the mutant leaves (Martin et al., 2007; Ohkama-Ohtsu et al., 2007a). In a separate Atggt1 knock-out study, Tolin et al. (2013) found both an increase in GSH content and a drop in the Cys-Gly content when compared to wild type plants, the authors accredit this to a loss of apoplastic GGT activity.
In Atggt1/2 double knock-out mutants generated via RNAi, no significant changes to the overall levels of GSH were observed. However, the double mutant plants exhibited a reduction in seed yield and also produced fewer trichomes (where GSH levels were high) (Giaretta et al., 2017). AtGGT4 is expressed in vacuoles and is involved in the degradation of GSH-conjugated xenobiotics (Grzam et al., 2007; Ohkama-Ohtsu et al., 2007b). It is divergent from the other three GGTs sharing only about 50% sequence identity with AtGGT1 and is prevalent in the roots, where it accounts for 50% of total GGT enzyme activity (Grzam et al., 2007; Destro et al., 2011).
There are three functional GGT genes in radish, RsGGT1 (BAC45233.1), RsGGT2 (BAC56855.1), and RsGGT3 (BAD22536.1). These genes have been used to assist in the classification of plant GGTs, which fall into two groups based on their solubility (either soluble or bound) (Nakano et al., 2006a; Nakano et al., 2006b). In addition, they have been used to classify GGTs based molecular structures, these include the monomeric GGT and heterodimeric GGT, both of which present different substrate specificities (Nakano et al., 2006a; Nakano et al., 2006b). Similarly, three functional GGT genes are present in garlic, AsGGT1 (BAQ21911.1), AsGGT2 (BAQ21912.1), and AsGGT3 (BAQ21913.1) (Yoshimoto et al., 2015). AsGGT2 localizes to the vacuole, whereas AsGGT1 and AsGGT3 do not appear to possess a transit peptide targeting the protein for localization to intracellular organelles. In addition, the kinetic affinities differ based on both the γ-glutamyl donor substrate and the AsGGT enzyme used to catalyze the substrate (Yoshimoto et al., 2015).
New Zealand Sauvignon blanc wines have distinctive flavor and aroma, which are in part due to the volatile thiols, 3-mercaptohexan-1-ol (3MH, passion fruit) and its derivative 3-mercaptohexyl-acetate (3MHA, boxwood) (Lund et al., 2009). Other volatile thiols present in Sauvignon blanc wines include 4-mercapto-4-methyl-2-pentanone (4MMP, boxwood) and 4-mercapto-4-methyl-2-pentanol (4MMPOH, citrus zest) (Darriet et al., 1995; Tominaga et al., 1998; des Gachons et al., 2000). These C5 and C6 based compounds have been discovered in free form in most tropical fruits, but not in grape berries, where they are found as odorless GSH, Cys-Gly, and Cys- coupled precursors (Pena-Gallego et al., 2012). A pathway for the biosynthesis of the volatile thiol 3MH has been proposed by Kobayashi et al. (2011), involving the conjugation of C6 hexenal to GSH to produce 3MH-GSH (3MH-S-glut). The breakdown of 3MH-S-glut forming other recognized precursors is proposed to involve the action of GGT to produce 3MH-S-Cys-Gly, as well as additional enzymes that include carboxypeptidases to produce 3MH-S-Cys. It is now considered that these volatile precursors are released by the action of yeasts during the alcoholic fermentation of grape (Pena-Gallego et al., 2012).
Pathogen infections are thought to stimulate stress response pathways via the production of pathogen-associated molecular patterns and/or elicitors during the fungal infection (Thibon et al., 2009; Thibon et al., 2011). These responses, in turn, upregulate the production of compounds that are able to be conjugated by GSH and grape berries infected with Botrytis cinerea have much higher levels of 3MH-S-Cys (Thibon et al., 2009; Thibon et al., 2011). Kobayashi et al. (2011) provided data on the involvement of GSTs and GGTs in the formation of volatile thiol precursors in grapes. However, other than transcript and GGT enzyme activity data in grape berries exposed to heat and UV-C stress, no information on the wider roles for grape GGT currently exists.
Grapevine appears to contain only a single GGT gene, possessing all the key conserved residues needed for GGT activity (annotated as VvGGT3, NCBI accession XP_002280190.1). This raises many questions about the role of this enzyme compared to other plants, where at least two GGTs exist, and specifically with respect to its role in the formation of important flavor and aroma precursors in grape berries. Consequently, we set out to functionally characterize VvGGT3 from V. vinifera L. cv. Sauvignon blanc. We explored subcellular localization as well as VvGGT3 transcript abundance in a range of tissues, including whole grape berry development and whole berries infected with Botrytis.
V. vinifera L. cv. Sauvignon blanc whole berries for gene expression studies, as described in Podolyan et al. (2010), were collected at 20, 30, 50, 60, 80, 90, 100, and 110 days after anthesis (daa). Collection was during development in the 2006, 2007, 2008, and 2009 seasons from the Booker vineyard, Brancott Estate, Marlborough, New Zealand (41°.56′ South, 173°.85′ East). Berries harvested at 110 daa had soluble solid levels of between 21 and 23 °Brix and were considered fully ripe. For each time point, the phenological stages mostly represented E-L 30, 31, 32, 35, 36, 36, 37, and 38, respectively (Coombe, 1995). Root tip (∼15 mm), leaf (∼225 mm2), E-L 15 inflorescence (Coombe, 1995), and tendril (∼20 mm), as described in Tashiro et al. (2016), were also collected from the Booker vineyard during dormancy in 2010 and grown as single node cuttings in the Lincoln University glasshouse nursery, Canterbury, New Zealand (43°.38′ South, 172°.27′ East) in 2011. Young leaves (∼2.5–3 cm wide) used for wounding responses were also collected from the Lincoln University glasshouse nursery in 2013. Skin, pulp and seed fractions were separated from mature whole Sauvignon blanc grape berries at 21 °Brix and B. cinerea-infected berries were collected from the Lincoln University research vineyard in the 2012 and 2014 seasons, respectively. Uninfected berries on the same bunch had a soluble solids content of 21 °Brix, mostly representing E-L 38 berries (Coombe, 1995). All samples were collected in biological triplicates and were snap-frozen in liquid nitrogen then stored at -80°C until use. The LAB genotype of Nicotiana benthamiana (Bally et al., 2018) was used in transient transformation experiments. These plants were grown in a controlled environment room at Lincoln University until 4 weeks of age, at constant temperature of 25°C with a 16/8 h photoperiod and a light intensity of 180 µM · m−2 · s−1. Onion biolistic transient transformation experiments were carried out using red onion (Allium cepa L., family Amaryllidaceae) bulbs purchased locally.
The triplicate samples of the various tissue types were ground in liquid nitrogen and ∼100 mg was processed by the Spectrum Plant Total RNA kit (Sigma-Aldrich). The resultant total RNA, ∼300 ng, was reverse transcribed by PrimeScript RT reagent Kit (Perfect Real Time, TaKaRa Bio Inc.) in a 10 µL reaction and then diluted to a final volume of 200 µL, as described in Tashiro et al. (2016). For later stage berries (100 and 110 daa), ∼200 mg of ground tissue was used and supplemented with a proportional increase in buffers. For seed tissue, 5% w/v Polyvinylpolypyrrolidone (Sigma-Aldrich) was added to the lysis buffer.
The VvGGT3 open reading frame was amplified from Sauvignon blanc grape berry cDNA using VvGGT3 gateway forward and VvGGT3 reverse primers (Supplementary Table 1). Amplification was carried out using PrimeStar HS DNA polymerase (TaKaRa Bio Inc.) according to manufacturer's instructions. The PCR amplification was conducted using the following conditions; 30 cycles comprising a denaturation step at 98°C for 10 s, annealing at 55°C for 5 s and extension at 72°C for 2 min. The VvGGT3 PCR fragment was subsequently cloned into pENTR/D-TOPO (ThermoFisher), according to manufacturer's instructions. DNA isolated from individual clones was sequenced to confirm the identity of VvGGT3 and this sequence has been deposited to NCBI (VvGGT3_ORF; MN101215.3).
VvGGT3 was re-amplified with VvGGT3 gateway forward and the VvGGT3 reverse-STOP primers (Supplementary Table 1) and this amplicon was subcloned into pB7FWG2 (Karimi et al., 2002) for subcellular localization. A shorter VvGGTN75 fragment was also subcloned into pB7FWG2 using the VvGGT3 (N75) gateway forward and VvGGT3 (N75) reverse primers (Supplementary Table 1). To explore in vivo VvGGT3 enzymatic activity, VvGGT3 was subcloned into pEAQ-HT-DEST2 (Sainsbury et al., 2009) using Gateway cloning as per manufacturer's instructions (ThermoFisher). Each of the binary-destination vectors containing VvGGT3 was checked for integrity by sequencing and finally transferred into Agrobacterium tumefaciens GV3101 (MP90) by electroporation, using a MicroPulser (BioRad), with the Agrobacterium setting.
Transient transformation of the inner epidermis of excised bulb leaves of red onion and N. benthamiana leaves with VvGGT3 and positive GFP control constructs was carried out according to Wiltshire et al. (2017) and Dumin et al. (2018), respectively. A positive eGFP control construct was obtained by liberating 3×eGFP-HygR as a SfiI restriction fragment from pUMA647 (Scherer et al., 2006). This fragment was then placed under the transcriptional control of the CaMv35S promoter in pBS-SK (Artemio Mendoza, unpublished data). For transient transformations of onion inner epidermal cells, purified plasmid constructs were deposited onto gold particles and used to bombard the inner epidermis of excised bulb leaves using an particle inflow gun (Kiwi Scientific, Levin, New Zealand) as described in Wiltshire et al. (2017). Leaf pieces were incubated for 24 h prior to imaging. GFP fluorescence images were obtained using confocal microscopy (Zeiss LSM510, Axio imager, with a LCI plan-neofluar 25×0.8 Imm corr DIC objective). Images were captured using the fluorescence contrast method with an excitation wavelength of 488 nm, an emission wavelength of 538 nm and a pinhole of 3.86 AU/78 µm.
Transient transformation of N. benthamiana leaves was performed according to Dumin et al. (2018). Briefly, overnight cultures of transformed Agrobacterium were centrifuged, the pellets washed with half-strength Murashige and Skoog medium (Duchefa Biochemie), re-suspended to an OD600 of 0.2 and acetosyringone (Sigma-Aldrich) added to a final concentration of 100 µM. The re-suspended cultures were subsequently incubated at room temperature on a rocking platform for at least 1 h prior to being used for transient agro-infiltrations. Infiltrated leaves were left on the plant for either: 3 days prior to excision for GFP analysis at 30% laser power, 501–551 nm (green color) wavelengths, using the TCS SP5 confocal microscope (Leica Microsystems), as described in Dumin et al. (2018) or 5 days for in vivo activity. At least 10 individual infiltrations were prepared for each experiment and representative images presented.
The metabolism of a fluorescent γ-GGT substrate was carried out as per Storozhenko et al. (2002). After 5 days post agro-infiltration, leaf discs from at least 10 independent infiltrations were cut and vacuum infiltrated twice for 2 min (Lizamore and Winefield, 2015) with a solution containing 0.1 M Tris-HCl [pH 7.5], 1 mM γ-glutamyl-7-amido-4-methylcoumarin (Biosynth), and 40 mM glycylglycine (Sigma-Aldrich). The leaf discs were then briefly rinsed in molecular grade water and immediately mounted onto microscope slides before being examined at 10x magnification using a DM IRB inverted fluorescent microscope (Leica), with excitation at 340–380 nm and emission at 425 nm. Images were captured using SPOT Software v4.6 (Diagnostic Instruments Inc.). Negative controls consisted of leaves transiently transformed with Agrobacterium GV3101 (MP90) without a transgene and treated in exactly the same manner as described.
Quantitative reverse-transcriptase PCR (RT-qPCR) was performed on cDNA synthesized as described earlier on the triplicate samples. Assays were designed and executed according to MIQE guidelines (Bustin et al., 2009). Each individual PCR reaction was carried out with triplicate technical replication for each biological replicate. VvActin and VvEF1α were described as being suitable normalization factors in grape berry (Reid et al., 2006). We thus used VvActin as described in Reid et al. (2006) and an optimized VvEF1α qPCR primer set, as described Tashiro et al. (2016), as normalization factors. Relative expression assays were run using the Eco Real-Time PCR System (Illumina) and resulting data analyzed with the supplied Illumina Eco software. Final reaction volumes were 10 µL and are as described in Philips et al. (2019), with primer sequences presented in (Supplementary Table 1). All liquid handling steps were performed using an Eppendorf epMotion 5070 liquid handling robot. PCR efficiency, standard curves, and no-template controls were determined and confirmed as described in Tashiro et al. (2016). The qRT-PCR conditions consisted of; 95°C for 5 min, followed by 2-step PCR of 40 cycles comprising of 95°C for 5 s and 60°C for 30 s, with fluorescence being recorded during the 60°C extension step. Following this, one cycle of melt curve analysis was performed for each amplified product as per default Illumina Eco settings.
Two multiple alignments were performed on the selection of protein sequences retrieved (Table 1) using the default ClustalW settings within Geneious 10.1.3 (Biomatters Ltd.) and consisted of the BLOSUM62 cost matrix, gap open cost of 10 and gap extend cost of 0.1. The first alignment included full length plant GGTs, where V77 of VvGGT3 is the first largely conserved (97.7%) residue. The second alignment excluded the N-terminal leader and targeting sequences by removal of residues prior to this position. Cladograms were then generated from both the full length and truncated alignments using the default tree builder feature within Geneious 10.1.3 (Biomatters Ltd.); these include Jukes-Cantor as the genetic distance model and the tree built with the neighbor-joining method with no outgroup. The multiple protein sequence alignments of both the full length and truncated GGTs are presented in Supplementary Data Sheet 2 and 3.
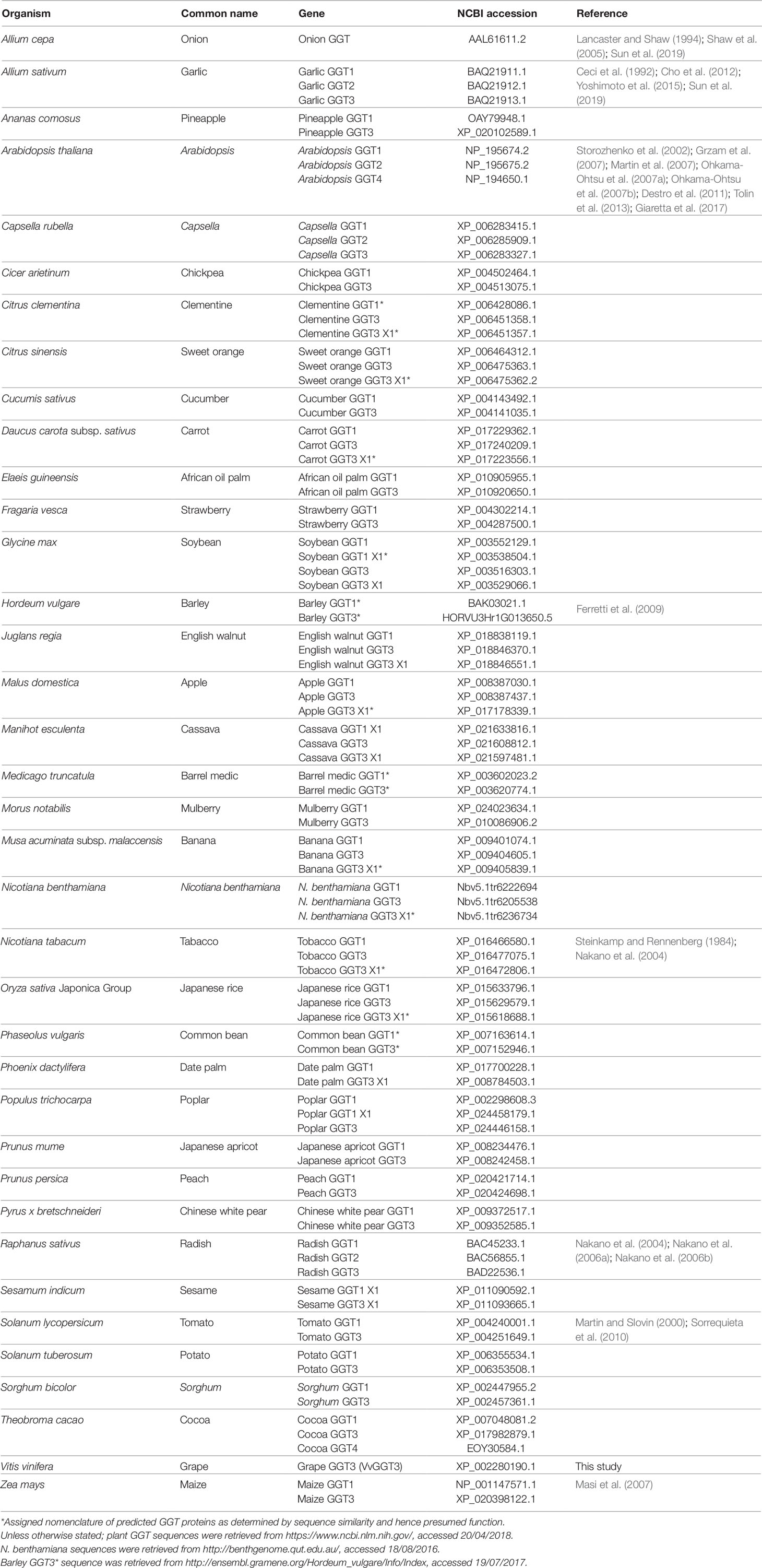
Table 1 List of plant GGT enzymes and their accessions used to generate the phylogenetic tree presented in Figure 1. GGTs associated with a reference have been completely or partially characterized.
Post-hoc analysis of the mean relative abundance of VvGGT3 in the various tissue types and treatments was performed by the Tukey-Kramer test after ANOVA using GenStat v15 (VSN International, Ltd.). Means significantly different at P < 0.05 are indicated by different letters above each bar in the graphs.
Initial mining of the 12X grape genome reference sequence deposited on NCBI, accessed 22/11/2016 revealed a single GGT sequence, (XP_002280190.1, designated as VvGGT3), which is unlike most other plant species where multiple genes encode GGT isoforms (Table 1). These GGT sequences from planta, retrieved from online repositories, are either published and characterized or currently putative. Since then, mining the latest 12X.v2 genome (http://genomes.cribi.unipd.it/grape/, accessed 30/08/2019) revealed three GGT candidates. The first on chromosome 11 has three splice variants (VIT_211s0016g02830.1,.2, and.3), encoding putative GGTs at 626, 475, and 557 amino acids (AA) in length, respectively. The other potential GGTs, VIT_212s0142g00530, located on chromosome 12, and VIT_201s0146g00200, located on chromosome 1, are 406 and 174 AA in length, respectively. In addition, mining UniProt (accessed 30/08/2019), reveals two potential GGTs. The first, designated D7TBD5 is identical in sequence to XP_002280190.1 and VIT_211s0016g02830.1. The second, A0A438IHM7, is slightly larger at 683 AA in length.
Generation of a phylogenetic tree with the deduced VvGGT3 amino acid sequence (XP_002280190.1) with a selection of deduced plant GGT amino acid sequences revealed that when two or more genes for GGT were present in a species that they grouped into two distinct subgroups (Figure 1). In accordance to the functional work carried out in Arabidopsis and garlic, these two subgroups possess GGTs that target either the apoplast or the vacuole (Yoshimoto et al., 2015; Giaretta et al., 2017). Based on the phylogenetic analysis, VvGGT3 groups with Arabidopsis GGT4 (AtGGT4) and garlic GGT2 (AsGGT2) both of which have been functionally shown to be located in the vacuole (Grzam et al., 2007; Yoshimoto et al., 2015). Additionally, radish has three at least GGT enzymes (Nakano et al., 2006b) of which RsGGT3 is the closest match to VvGGT3 (Figure 1). When over-expressed in tobacco, RsGGT3 unlike RsGGT1 and RsGGT2 which are most likely bound to cell walls was able to be purified from the soluble fraction (Nakano et al., 2006b), suggesting that it too like AtGGT4 may be localized to the vacuole (Ohkama-Ohtsu et al., 2009). Removal of the putative N-terminal leader/targeting peptides and reconstruction of the phylogenetic tree does not substantially affect the sequence relationships observed (Supplementary Figure 1).
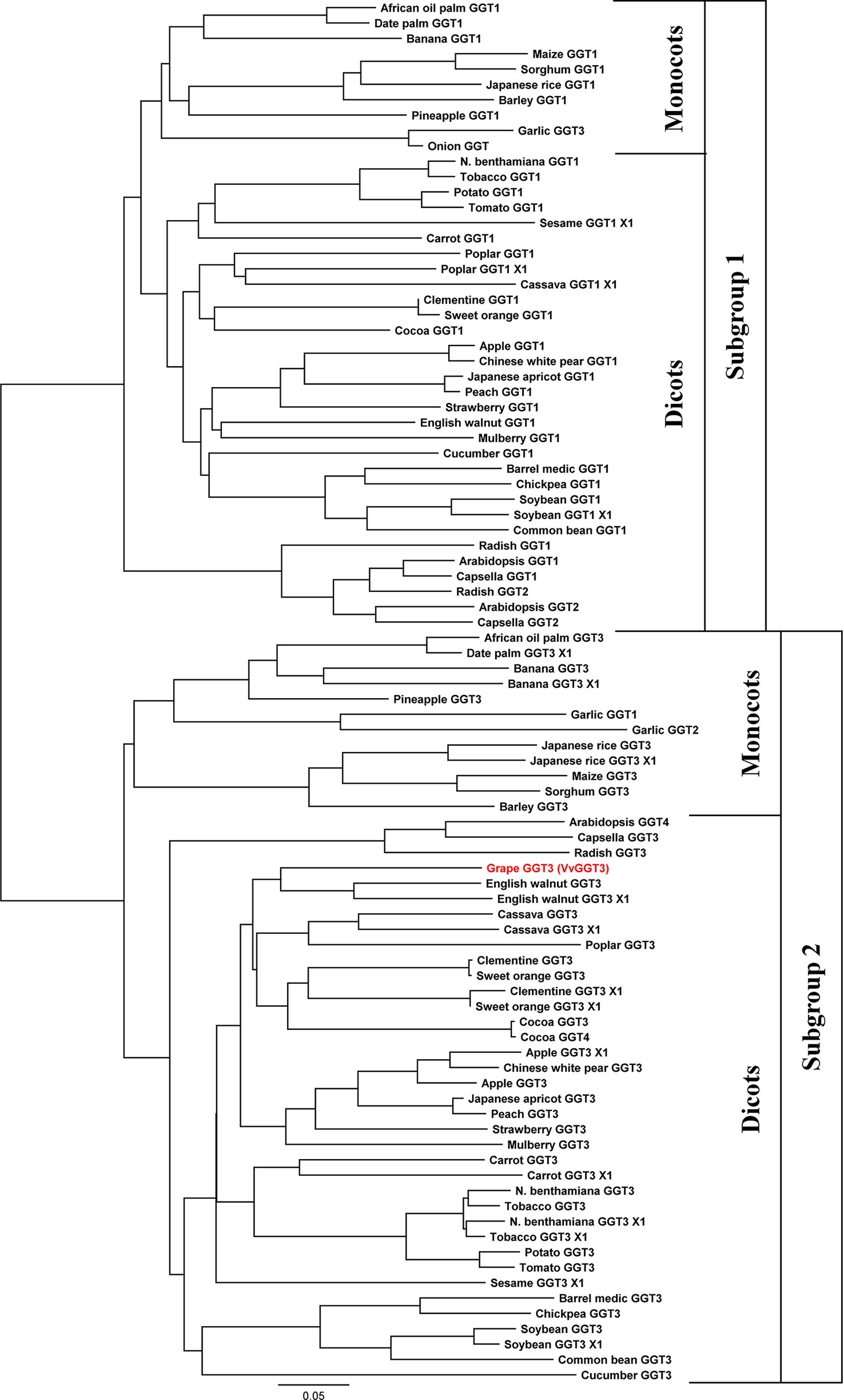
Figure 1 Phylogeny of selected plant γ-glutamyl transferases/transpeptidases (GGTs). The accession numbers of these GGTs are presented in Table 1. GGTs group into two distinctive subgroups, then further into the monocot and dicot clades. Subgroup 1 has GGTs, which are thought to prevent oxidative stress by degrading the oxidized form of glutathione (GSSG) and the breakdown of extracellular GSH in the apoplast. Subgroup 2 has GGTs which are thought to degrade GSH-conjugates in the vacuole. The GGT from grape, VvGGT3, highlighted in red, falls under subgroup 2. Multiple sequence alignments of deduced protein sequence that were used in this analysis can be found in Supplementary Data Sheet 2.
Generation of a phylogenetic tree with the other potential Vitis GGT candidates was then undertaken (Supplementary Figure 2). As expected, the splice variants, VIT_211s0016g02830.2, .3, and UniProt A0A438IHM7 clustered together with VvGGT3 (XP_002280190.1). Surprisingly, none of the candidates grouped to subgroup 1. Candidate GGT genes, VIT_212s0142g00530 and VIT_201s0146g00200, were the most divergent when compared to the other plant GGT sequences (Supplementary Figure 2).
A subset of the functionally characterized plant GGTs, along with Escherichia coli, human, and rat GGTs, where key residues have been characterized, were then aligned with the potential Vitis GGT candidates (Supplementary Figure 3). Compared to other GGTs, the UniProt A0A438IHM7 candidate has a longer N-terminus sequence, most likely from mis-annotation of the genome. Key residues, essential for GGT activity are conserved in bacterial, mammalian, and plant GGT sequences (Supplementary Figure 3). These residues include: R107 and D423 in human, which are involved in the binding of substrates (Taniguchi and Ikeda, 2006); the catalytic nucleophile, T391 in E. coli, and the residue involved in stabilizing the nucleophile, T409 (Okada et al., 2006); residues S451 and S452 in human, which are involved in enzyme catalysis (Okada et al., 2006; Taniguchi and Ikeda, 2006); and G483 and G484 in E. coli, which comprise the GGT oxyanion hole (Okada et al., 2006). Of all the VvGGT isoforms, only the R107 residue in human is conserved, apart from splice variant VIT_211s0016g02830.3, where it is missing. The VIT_212s0142g00530 and VIT_201s0146g00200 candidates, do not possess the remaining conserved residues. Furthermore, they are also missing the region that has been described as the GGT molecular signature, [T-[STA]-H-x-[ST]-[LIVMA]-x(4)-G-[SN]-x-V-[STA]-x-T-x-T-[LIVM]-[NE]-x(1,2)-[FY]-G] (Ferretti et al., 2009) (Supplementary Figure 3). As the VIT_212s0142g00530 and VIT_201s0146g00200 GGT candidates are deficient in the key GGT residues, molecular signature and lack the length of other plant GGTs, they are not considered to be genes possessing "classical" GGT activity.
Separation of the PCR amplification of VvGGT3 from Sauvignon blanc berry cDNA as described in Materials and Methods, produced a single distinct band, which was sequenced to be the transcript encoding the 626 AA variant (XP_002280190.1 and VIT_211s0016g02830.1). The nucleotide sequence in cv. Sauvignon blanc has been deposited to NCBI (MN101215.3). Splice variant VIT_211s0016g02830.2, is missing the first conserved residue, R107 (in human) needed of GGT substrate binding (Taniguchi and Ikeda, 2006). Whereas, splice variant VIT_211s0016g02830.3 does not have conserved residues at its C-terminus, when compared to the selection of other plant GGT sequences (Supplementary Figure 3). It is therefore likely that these splice variants are mis-annotated. Due to the aforementioned reasons and as we were unable to experimentally detect the other splice variants, we consider cv. Sauvignon blanc as possessing a sole GGT gene.
Determination of the biochemical function of VvGGT3 using recombinant protein from over-expression experiments in E. coli was not successful due to difficulties in obtaining soluble protein. Similar difficulties have also been previously reported (Kobayashi et al., 2011), and have been partly attributed to the propensity for the recombinant protein to remain insoluble and self-cleave (Suzuki and Kumagai, 2002; Kobayashi et al., 2011). We, therefore, opted to determine whether VvGGT3 encoded an active GGT by using a plant-specific recombinant protein vector, pEAQ-HT-DEST2, (Sainsbury et al., 2009; Peyret and Lomonossoff, 2013) by transient over-expression of the VvGGT3 open reading frame in N. benthamiana leaves. Attempts to purify the resulting 6xHis tagged protein from the leaves also proved difficult. Finally, we decided to determine if functional recombinant VvGGT3 protein was being produced in the transiently transformed N. benthamiana leaves by infiltrating a quenched GGT fluorescent substrate (γ-glutamyl-7-amido-4-methylcoumarin) and directly observing the enzymatic release of fluorescent 7-amido-4-methylcoumarin (Figure 2A). Controls were included to confirm that the observed fluorescence was due to VvGGT3-induced cleavage and not due to either auto-degradation of the substrate, or due to autofluorescence, or due to the infiltration of Agrobacterium GV3101 (MP90) resulting in pathogen-induced expression of endogenous N. benthamiana GGTs. These consisted of untransformed leaves infiltrated with γ-glutamyl-7-amido-4-methylcoumarin (Figure 2B), VvGGT3 transformed leaves, which were subsequently infiltrated with molecular grade water (Figure 2C) and leaves infiltrated with untransformed Agrobacterium GV3101 (MP90) and subsequently infiltrated with the substrate (Figure 2D), respectively. In each case, the controls did not show the same accumulation of fluorescence as seen in Figure 2A. These results demonstrates that VvGGT3 encodes a functional GGT when transiently over-expressed in N. benthamiana leaves and that the location of the enzymatic activity did not appear to localize to the vacuole as predicted based on sequence similarity, but rather in an undetermined location associated with cellular membrane structures.
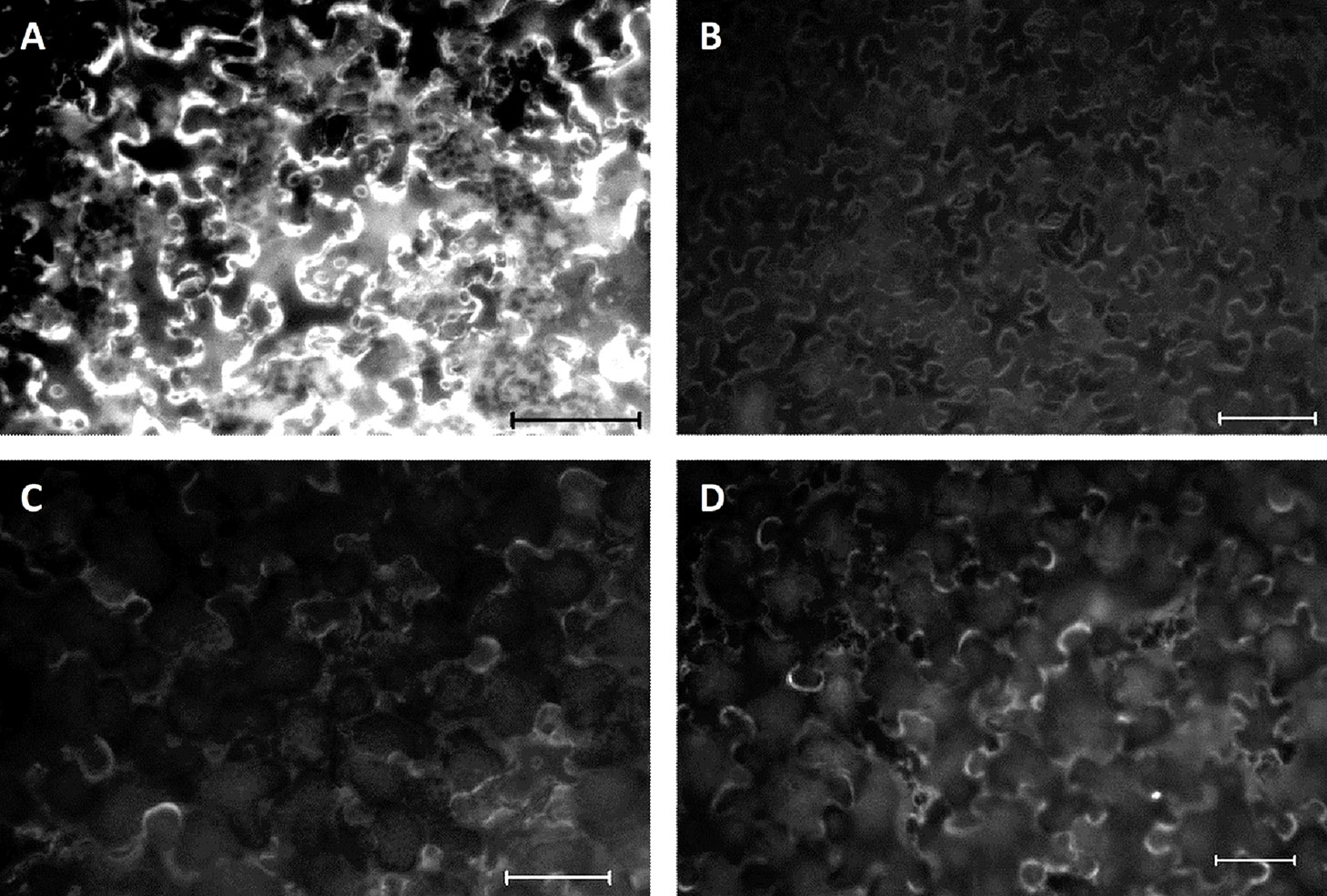
Figure 2 Fluorescent images of Nicotiana benthamiana leaf discs infiltrated with the over-expressing VvGGT3 construct five days prior to the addition of the γ-glutamyl-7-amido-4-methylcoumarin substrate. Images are representative of the result obtained from at least 10 independent infiltrations. (A) Leaves transiently transformed with the VvGGT3 over-expressing construct and subsequently infiltrated with the substrate. (B) Untransformed leaves at the same age infiltrated with the substrate. (C) Leaves transiently transformed with the VvGGT3 over-expressing construct and subsequently infiltrated with molecular grade water. (D) Leaves transiently transformed with Agrobacterium tumefaciens GV3101 (MP90) without a transgene and infiltrated with the substrate. Scale bars = 100 μm.
To further resolve the subcellular localization of VvGGT3, we transiently transformed onion epidermal cells with a VvGGT3-GFP full-length translational fusion construct. The VvGGT3-GFP fusion protein was found to be associated with what appears to be membrane structures or the tonoplast (Figure 3, top row). A 3D rendered video of the z-stacked confocal image captured in the top row of (Figure 3) demonstrates that the VvGGT3-GFP fusion protein does not localize to the vacuole (Supplementary Video 1). The pattern of accumulation is also consistent with that observed for the in vivo enzymatic activity (Figure 2), indicating that the site of enzyme activity is not within the vacuole as predicted by VvGGT3 subgrouping with AtGGT4 and AsGGT2.
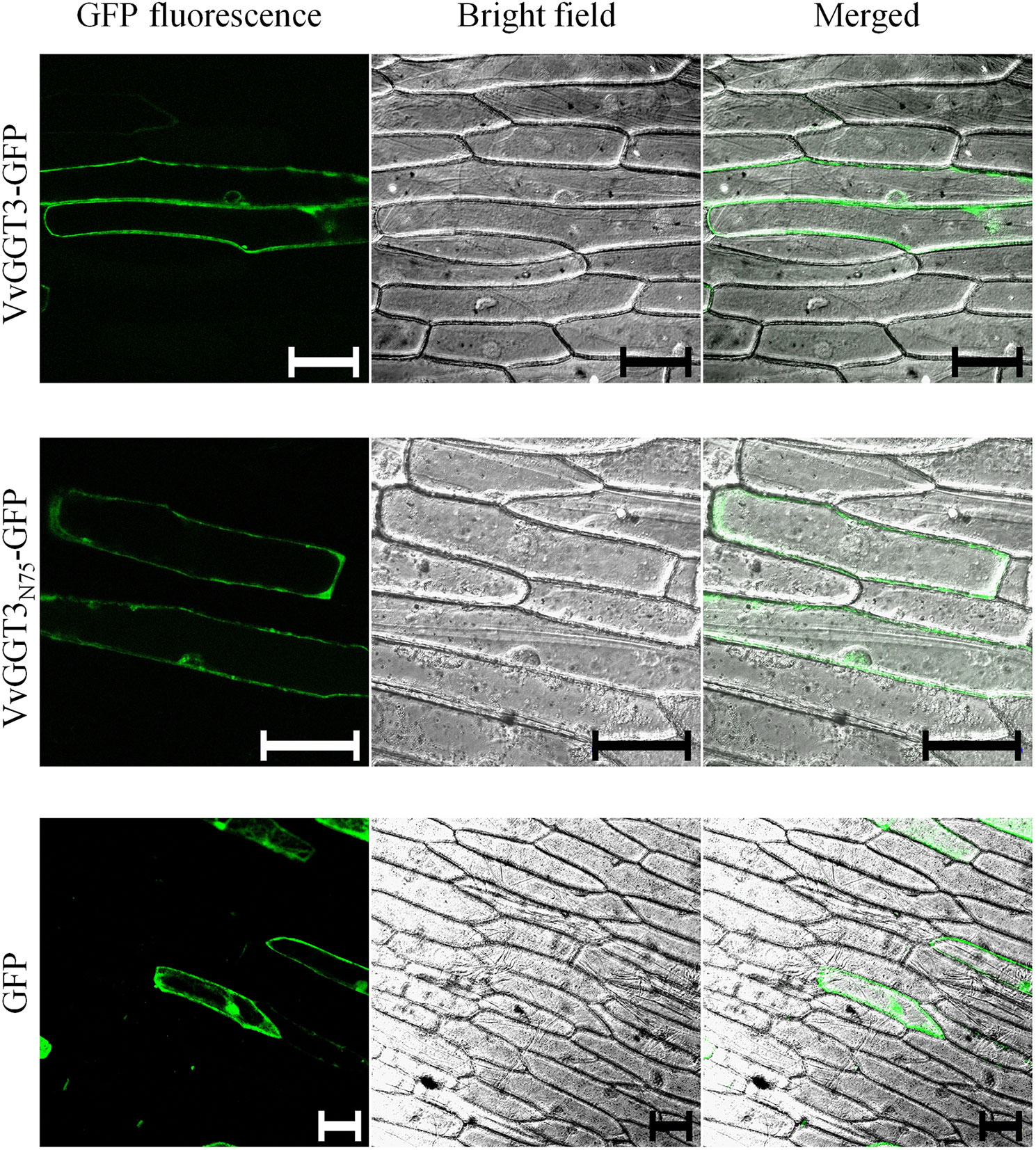
Figure 3 Confocal images of onion cells transiently transformed with either: the VvGGT3-GFP fusion protein (top row), VvGGT3N75-GFP fusion protein (middle row), or GFP alone (bottom row). The merged images demonstrate that neither the full length (VvGGT3) nor the first 75 AA of the protein (VvGGT3N75) localize to the vacuole as predicted by phylogenetic analysis. The free GFP localizes to the nucleus and cell membranes as described in Ohkama-Ohtsu et al. (2007b). The images presented are representative of cells obtained from epidermal peels from three independent biolistic events on three individual bulb leaf sections bulb for each construct. Each peel was inspected to compare cells from at least three different fields of view. Two independent experiments carried out. Scale bars = 100 µm.
In garlic, weak fluorescent signals have been detected when full-length AsGGTs or the first 300 AA residues have been fused to GFP and the authors suggest that this may be in part due to the incorrect processing of the AsGGT-GFP fusion (Yoshimoto et al., 2015). A stronger fluorescent signal was then produced when the authors instead used the first 100 AA from AsGGTs for their localization experiments (Yoshimoto et al., 2015). To test if the full-length VvGGT3-GFP fusion was unstable or incorrectly processed, we fused the first 75 AA of VvGGT3 (VvGGT3N75) to GFP and found the same localization patterns (Figure 3, middle row). We then transiently transformed onion epidermal cells with free GFP. Free GFP was shown to localize to the nucleus and cell membranes, as demonstrated in Ohkama-Ohtsu et al. (2007b), which is different to that observed by the VvGGT-GFP fusion constructs. We also captured similar localization patterns upon transiently transformation in N. benthamiana leaf cells (Supplementary Figure 4). In addition, the fluorescence of the VvGGT3-GFP fusion constructs are in accordance with the computational programs WoLF PSORT (Horton et al., 2007) and PSORT (Nakai and Horton, 1999), both predicting VvGGT3 to be localized to membranous compartments.
There has been the suggestion that plant GGTs are regulated in response to stress, such as oxidative stresses in Arabidopsis, (Ohkama-Ohtsu et al., 2009), γ-irradiation to garlic bulbs after harvest (Ceci et al., 1992), and UV-C and temperature exposure to grape berries (Kobayashi et al., 2011). With only a single GGT isoform identified in grapevine and considering the numerous roles that GGTs are proposed to play within plants, we were interested to determine the patterns of VvGGT3 transcript accumulation. In particular, due to the proposed roles of VvGGT3 in the formation of important flavor and aroma precursors in grape berries, we were very interested to discover the patterns of transcript accumulation between berry fractions, across berry development, and in response to exposure of tissues to biotic and abiotic stress treatments.
The VvGGT3 transcript was found to be expressed in all tissues tested (roots, leaf, inflorescences, and tendril, and in the skin, pulp, and seed fractions of mature grape berries (Figure 4A). The transcript abundance of VvGGT3 was the lowest in the tendril, followed by the root, with a 1.7- and 1.8-fold increase in leaf and inflorescence compared to the tendril, respectively (Figure 4A). Focusing on the relative VvGGT3 abundance in the skin, pulp, and seed fractions of mature grape berries, we observed similar levels of transcript in all three berry tissues, with these all being over 7-fold higher than the tendril (Figure 4A).
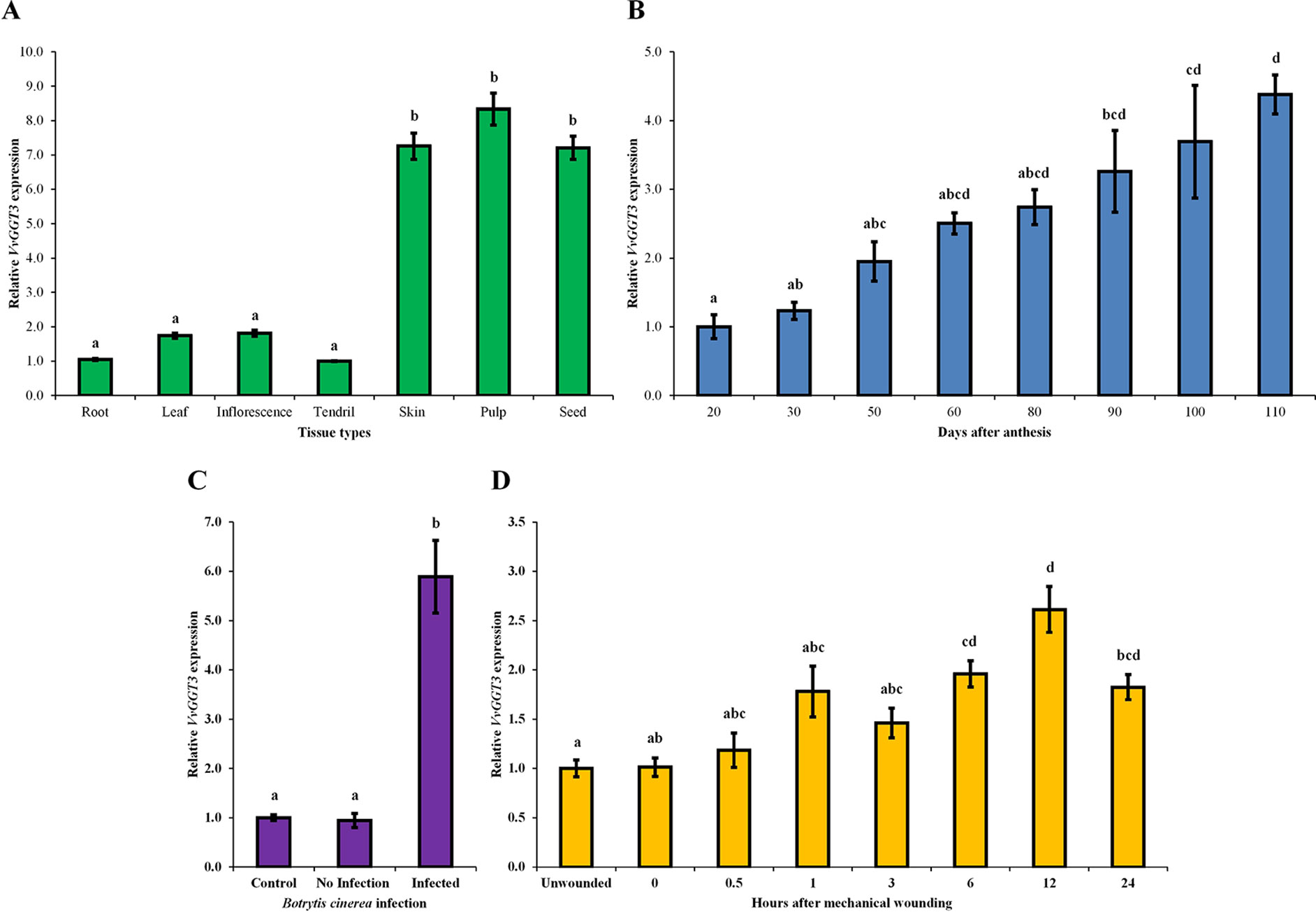
Figure 4 The mean relative abundance of VvGGT3 in a range of Sauvignon blanc tissues and stress treatments. All data is derived from biological replicates and technical triplicates were performed for each RT-qPCR gene targets. (A) The mean relative abundance in root (root tip, ∼15 mm), leaf (∼225 mm2), E-L 15 inflorescence (Coombe, 1995), and tendril (∼20 mm), and the skin, pulp, and seed fraction of mature berries [21°Brix, mostly E-L 38 berries (Coombe, 1995)], the expression level of tendril was set to 1. (B) The mean relative abundance in grape berries throughout development was studied in the 2006, 2007, 2008, and 2009 growing seasons. For each time point, the phenological stages mostly represented E-L 30, 31, 32, 35, 36, 36, 37, and 38, respectively (Coombe, 1995). Veraison in each season took place approximately 50 to 60 days after anthesis (daa). The level at 20 daa was set to 1 (n = 4 biological replicates, n = 3 biological replicates for 20 and 30 daa). (C) The mean relative abundance in whole grape berries from bunches prior to harvest [21 °Brix, mostly E-L 38 berries (Coombe, 1995)] infected with B. cinerea. Berries that showed no sign of infection were considered the control and set to 1, berries of healthy appearance adjacent to infected berries were considered not infected (no infection), and grape berries that showed signs of infection but were not fully infected (infected) were analyzed (Supplementary Figure 6). (D) The mean relative abundance over 24 h in mechanically damaged young grape leaves (∼2.5–3 cm wide). The level of unwounded leaves was set to 1. For all assays (A–D), the geometrical means of VvActin and VvEF1α as reference genes were used as a normalization factors. Different letters indicate statistically significant differences, P < 0.05, ANOVA Tukey-Kramer, n = 3 biological replicates for graphs in (A), (C), and (D), means ± SEM.
Across whole berry development, VvGGT3 accumulated continuously, peaking at harvest (110 days after anthesis—berries between 21 and 23 ˚Brix; Figure 4B). Across the individual seasons, we saw a relatively consistent pattern of accumulation, indicating that while there are clearly some inter-seasonal variations, VvGGT3 transcript accumulation is largely under developmental regulation (Supplementary Figure 5).
VvGGT3 has a proposed role in the formation of precursors of key varietal aromas in wine (Kobayashi et al., 2011). These compounds are considered to be derived from the lipoxygenase/hydroperoxide lyase mediated breakdown of linolenic and linoleic C18 fatty acids to produce C6 hexanal and then its derivatives (Capone and Jeffery, 2011; Kobayashi et al., 2011; Thibon et al., 2011; Capone et al., 2012). These derivatives are then thought to be conjugated to GSH by GSTs and are potential substrates for GGT activity within the vacuole of the berry. Levels of C6-GSH, C6-Cys-Gly, and C6-Cys accumulation in grape berries and juice have been shown to increase when the berries have been subjected to pathogens such as B. cinerea and/or wound stress (Capone and Jeffery, 2011; Kobayashi et al., 2011; Thibon et al., 2011). Therefore, we investigated whether either of these stresses might also impact VvGGT3 gene transcription in Sauvignon blanc. To test the impact of Botrytis infection on VvGGT3 transcript accumulation, we isolated berries from infected bunches that had (i) no sign of infection, (ii) berries on infected bunches immediately adjacent to infected berries but did not show sign of infection, and (iii) berries that showed signs of Botrytis infection, but were not fully infected (Supplementary Figure 6). As expected, Figure 4C clearly shows a correlation between Botrytis infection and a strong 5.8-fold upregulation of the VvGGT3 transcript. To test the impact of wounding on VvGGT3 transcription, we mechanically wounded leaves and collected leaf samples at regular intervals up to 24 h. In comparison to Botrytis infection, there is only a mild (∼2.0-fold) increase in the relative transcript abundance of VvGGT3 compared to the unwounded control leaves at 6 h post wounding. This mild increase remains apparent at 12 h (∼2.6-fold) and at 24 h (∼1.8-fold), which are statistically different (P < 0.05) to that of the unwounded control leaves (Figure 4D).
Analysis of the 12X.v2 V. vinifera reference genome revealed three potential GGT isoforms, with one isoform having three splice variants. After aligning these potential isoforms to functionally characterized GGTs from a range of species, candidates VIT_212s0142g00530 and VIT_201s0146g00200 do not possess conserved residues (Okada et al., 2006; Taniguchi and Ikeda, 2006) or the GGT molecular signature, [T-[STA]-H-x-[ST]-[LIVMA]-x(4)-G-[SN]-x-V-[STA]-x-T-x-T-[LIVM]-[NE]-x(1,2)-[FY]-G] (Ferretti et al., 2009) that are characteristic of GGT enzymes (Supplementary Figure 3). In addition, splice variant VIT_211s0016g02830.2, is missing the first conserved residue, R107 (in human) needed of GGT substrate binding (Taniguchi and Ikeda, 2006). Whereas, splice variant VIT_211s0016g02830.3 does not have conserved residues at its C-terminus, when compared to the selection of other plant GGT sequences (Supplementary Figure 3). It is therefore likely that these splice variants are mis-annotated. The remaining splice variant VIT_211s0016g02830.1, is identical in length to the reference XP_002280190.1. We isolated this GGT isoform from Sauvignon blanc and also found it to be the same length, we also find SNPs that are distinctive to this cultivar and have deposited this sequence to NCBI (MN101215.3). Thus, we only find a single GGT isoform in V. vinifera, which is in contrast to most other plants where two or more GGT isoforms exist (Table 1), tending to group into two distinct subgroups. Based on the characterized GGTs from Arabidopsis and garlic, these subgroups could potentially be GGTs that are either targeted to the vacuole or apoplast (Figure 1).
Given the range of roles that GGT enzymes are proposed to play in the maintenance of cellular activities, possession of only a single isoform in grapevine is intriguing. Phylogenetic comparison of the VvGGT3 putative peptide sequences with other plant GGT sequences shows that VvGGT3 associates in the subgroup containing the characterized AtGGT4 and AsGGT2, both of which localize to vacuoles and have presumed roles in the catabolism of GSH conjugated to a wide range of small molecules (Grzam et al., 2007; Yoshimoto et al., 2015). This is consistent with the proposed role of VvGGT3 in the formation of precursors of volatile thiols (e.g. C6-Cys-Gly, C6-Cys). However, it also raises questions as to how other roles proposed for GGTs in grapevine, in particular, maintenance of cellular redox potential via modulation of cellular GSH levels, are achieved? It is possible that an alternative GSH degradation pathway exists that is present in the cytoplasm rather than the apoplast or vacuole which utilizes γ-glutamyl cyclotransferases (GGCT) and 5-oxoprolinase (5OPase) rather than GGT (Ohkama-Ohtsu et al., 2008). As the cleavage of the γ-glutamyl bond is only known to be carried out by GGT, our results suggest that the range of activities proposed for GGT in grapevine is carried out by this single VvGGT3 isoform.
Due to difficulties in generating stable transgenic lines in V. vinifera (Sauvignon blanc), we were unable to investigate the impact of a Vvggt3 knockout in grape. Therefore, we opted to study whether recombinant VvGGT3 possessed GGT activity by transient over-expression in N. benthamiana leaves and then supplying the leaf sections the quenched substrate γ-glutamyl-7-amido-4-methylcoumarin. As determined by the GGT-mediated cleavage of the γ-glutamyl moiety and release of fluorescent 7-amido-4-methylcoumarin, VvGGT3 is indeed functional and the substrate does not accumulate in the vacuole (Figure 2A).
To investigate the localization of VvGGT3, we generated a genetic construct designed to produce a C-terminal translational fusion of GFP with the full-length VvGGT3 protein. When this fusion protein construct was transiently transformed into onion epidermal cells (Figure 3, Supplementary Video 1) and N. benthamiana leaves (Supplementary Figure 4), fluorescence of the VvGGT3-GFP fusion protein was not detected in the vacuole, but was rather associated with membranes. This result complements the location of the GGT-cleaved 7-amido-4-methylcoumarin substrate seen in Figure 2A. Close inspection of the GFP images indicates that the localization may be in the cytoplasm or also associated with the tonoplast but is clearly not in the vacuole.
In garlic, weak fluorescent signals were detected when the full-length AsGGTs or the first 300 AA residues were used in GFP-fusion constructs and the authors suggest that this may be in part due to the incorrect processing of the AsGGT-GFP fusion (Yoshimoto et al., 2015). Similarly, in Arabidopsis, no fluorescence was detected when the full-length AtGGT4 or the first 300 AA residues were used in GFP-fusion constructs (Ohkama-Ohtsu et al., 2007b). Fluorescence was detected when the first 100 AA were instead used in localization experiments (Ohkama-Ohtsu et al., 2007b; Yoshimoto et al., 2015). To determine whether the full-length VvGGT3 sequence was unstable or incorrectly processed, we fused the first 75 amino acids of VvGGT3 (VvGGT3N75) to GFP and found the same localization patterns. This result indicates that there was no difference in the stability or processing of either the full length (VvGGT3-GFP) or truncated (VvGGT3N75-GFP) fusions (Figure 3).
The apoplast generally has an acidic pH (<6.5) and GFP fusion proteins have been reported to show poor to variable fluorescence in this compartment (Schulte et al., 2006). This is due to reversible protonation of GFP leading to quenching of fluorescence when subjected to a pH range between pH 7 and 5 (Schulte et al., 2006). If the pH drops below pH 5, then an irreversible conformational change of GFP takes place leading to protein instability (Schulte et al., 2006). Thus, the strong fluorescence we have observed with the transient expression of the VvGGT3-GFP fusion in onion epidermal cells argues against an apoplastic location for the fusion protein. However, further experiments are needed to conclusively determine the subcellular localization of VvGGT3 and the apoplast cannot be dismissed.
Despite the bioinformatic prediction of the vacuolar localization for VvGGT3, the localization of the VvGGT3-GFP fusion protein as well as the corresponding enzymatic activity in N. benthamiana suggest that VvGGT3 is unlikely to be involved in the processing of GSH-conjugated metabolites in the vacuoles of grapevine cells. In addition, due to the strong GFP signal of the VvGGT3-GFP fusion protein, the enzyme is also unlikely to be involved in the apoplastic recycling of GSSH. The presumed absence of a GGT in grape vacuoles also raises important questions as to the subcellular localization of enzymatic activities responsible for green leaf volatile and green leaf volatile conjugate formation in grapevine. In particular, these data raise questions as to the possible mechanisms for the production of the Cys-Gly and Cys-precursors of the volatile C6-thiolated metabolites that are important for Sauvignon blanc flavors and aromas. There is little data on the physical storage location of these precursors in grapevine and data suggest that it may be possible that the removal of the γ-Glu from GSSH conjugated metabolites may occur (leaving a Cys-Gly conjugate) in the cytoplasm prior to transport of these conjugated metabolites into the vacuole. There are alternate recycling pathways for GSSH reported in plants that involve GGCT and 5OPase (Ohkama-Ohtsu et al., 2008). In Arabidopsis, this recycling pathway has been shown to be the main metabolic flux route for recycling of GSSH compared to the GGT route (Ohkama-Ohtsu et al., 2008). Thus, there is a possibility that the metabolism of both apoplastic GSSH and GSSH conjugates in the vacuole may preferentially use this alternative pathway in grapevine. At this time we cannot rule out these results are an artifact of the transient expression of these gene constructs in a heterologous species and consequently further work in grapevine would be required to determine the validity of this data.
Our main interest in VvGGT3 is its potential role in the formation of important precursors of the volatile thiol aromas (3MH and 3MHA) in grape berries that are characteristic of Sauvignon blanc wines. Consequently, we explored the pattern of VvGGT3 transcript accumulation across tissues and with a focus on grape berries. It is clear that VvGGT3 is expressed in every tissue tested (Figure 4) and thus, a sole GGT in grape is consistent with a presumed wide range of roles GGTs play in cellular metabolism, regulation, and recovery of GSH as well as acting on glutathionylated xenobiotics. Exploration of the patterns of transcript accumulation in mature berry tissues revealed a fairly even distribution of transcript abundance between the skin, pulp, and seed fractions (Figure 4A). However, the levels of VvGGT3 transcript seen to be accumulating in mature berry pulp is surprising given the debate as to the varying levels of cell viability recorded for the berry pulp across a range of grape varietals (Tilbrook and Tyerman, 2008; Fuentes et al., 2010), which would otherwise result in global transcript reduction. Therefore, while we are able to detect significant levels of VvGGT3 transcript in the total RNA isolated from mature berry pulp, we are not able to comment on the levels of active protein in these tissues. GGTs are known to accumulate in certain storage tissues such as seed and bulbs (Balakrishna and Prabhune, 2014; Sun et al., 2019). When stored at cool temperatures below 20°C, GGT activity has been detected in the bulbs of garlic and onion. This activity increases over time and can be detected up to eight months after storage (Hanum et al., 1995; Li et al., 2008). Similarly, in Arabidopsis seed, amongst the AtGGT members, AtGGT4, which based on sequence alignment is the closest match to VvGGT3, has the highest expression (Martin et al., 2007). Thus, taken together, it is not surprising that the expression of VvGGT3 in a storage tissue such as the seed is high when compared to the other tissues tested.
In tomato fruit (cv. Alisa Craig and Rutgers), GGT activity was shown to increase during ripening, reviewed in Sorrequieta et al. (2010). Similarly, across whole berry development, VvGGT3 demonstrates a general pattern of increasing in transcript abundance (Figure 4B). Grape berries feature a distinctive stage during the development known as veraison, where significant changes at a physiological level take place. These changes include, softening of the berry, accumulation of sugars, reduction in acids and changes in color in red varieties that signal the onset of ripening. Veraison is also associated with an oxidative burst and changes in cell structure, where cell division stops and berry enlargement occurs, mainly due to cell expansion (Coombe, 1992; Podolyan et al., 2010). Such physiological changes at veraison (50–60 days after anthesis) could account for the 2- to 2.5-fold increase in transcript abundance of VvGGT3 (Figure 4B). In considering the accumulation of the VvGGT3 transcript across four seasons, we clearly notice some inter-seasonal variation at each time point, which are likely to be due to differing climates (Supplementary Figure 5). However, the pattern of accumulation is generally stable, indicating that the main drivers for VvGGT3 transcription are related to berry development and ripening. This pattern of accumulation is similar to patterns of transcript abundance for other genes involved in volatile thiol precursor formation [namely VvLOX, VvHPL, and VvGST (Podolyan et al., 2010; Kobayashi et al., 2011; Zhu et al., 2012)].
In the context of flavors and aroma, wine produced from grapes infected by B. cinerea have a considerably higher concentration of volatile thiols (especially 3MH), reviewed by Coetzee and du Toit (2012). This is due in part to the berry's increase in the precursors such as 3MH-S-Cys (Thibon et al., 2009). This precursor is most likely formed after GGT and carboxypeptidase enzymes have cleaved Glu and Gly from the GSH-conjugate. Evidence of the role GGT plays in this formation was demonstrated by des Gachons et al. (2002), who showed 3MH-S-Cys to be liberated from 3MH-S-glut in grape must after passing the must through a 4B sepharose column with immobilized copper (II) and the GGT enzyme.
B. cinerea is not found to be directly responsible for this precursor formation but most likely stimulates the grape metabolic pathway involved in its formation due to the release of the fungal mebabolites (Thibon et al., 2011). In stressed plants, the lipoxygenase-hydroperoxide lyase pathway releases C6 volatiles and other reactive species, reviewed by Bate and Rothstein (1998); Thibon et al. (2011). More specifically, in grape berries, the lipoxygenase, VvLOXC and VvLOXO, transcripts are upregulated when infected by B. cinerea (Podolyan et al., 2010), this in turn may induce the detoxification pathway as the C6 compounds produced are not only toxic to the pathogen but at a critical level can become toxic to the plant itself (Coetzee and du Toit, 2012). Thus, to decrease toxicity, these compounds can be conjugated to GSH via the GST enzymes. In grape berry skins, the transcript of VvGST1, VvGST3, and VvGST4 are found to be upregulated when infected with the downy mildew pathogen, which in turn leads to an increase in the concentrations of GSH-conjugates (Kobayashi et al., 2011). In addition, the VvGGT3 transcript was upregulated 1.7-fold (Kobayashi et al., 2011) and is likely to be involved in the breakdown of the GSH-conjugates, which include the flavor and aroma precursor compounds as demonstrated by a higher production of the 3MH-S-Cys precursor (Kobayashi et al., 2011). Similarly, in our study, while a general pattern of developmentally regulated VvGGT3 transcript accumulation is evident, we also show a strong 5.8-fold pathogen related increase for VvGGT3 in mature grape berries (Figure 4C). However, we see only small levels of transcript accumulation in response to wounding of young grape leaves. Collectively, these data point to an important role for pathogen infection rather than wounding in modulating the levels of these important flavor and aroma precursors in grapevine.
Based on a bioinformatics search in V. vinifera, we have functionally characterized a sole GGT enzyme, VvGGT3. Our substrate localization and VvGGT3-GFP fusion results indicate that VvGGT3 does not localize to the vacuole as predicted by phylogenetic analysis. Rather, it is localized to membrane structures. This raises important questions about the subcellular localization for production transport and storage of C6 volatiles and their conjugates in grapevine. The VvGGT3 transcript was detected in all tissue tested and was most abundant in the skin, pulp, and seed fractions of mature berries. In whole berries, VvGGT3 appears to be under developmental control as the transcript steadily increases over time. We observed VvGGT3 transcript upregulation in whole berries infected with B. cinerea and in mechanically wounded leaves, which is not surprising given the production of GGT-cleavable volatiles upon biotic and abiotic stresses. The sole GGT member in grapevine is intriguing given the range of roles GGTs play in planta, the findings presented here may be foundational for other crop species where only a single GGT may exist.
The VvGGT3 sequence from Sauvignon blanc can be found in GenBank accession MN101215.3.
JP and CW designed the experiments. JP performed experiments and analyzed all data. WD performed leaf wounding and tissue type qRT-PCR experiments. CW performed GFP localization experiments in onion cells. CW supervised the experiments. JP and CW wrote the original draft. JP and CW reviewed and edited the manuscript.
This work was supported by Lincoln University and the Ministry of Business, Innovation and Employment (previously the New Zealand Foundation for Research, Science and Technology), contract number UOAX0404.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Dr. Andriy Podolyan (Lincoln University) for providing the whole berry developmental series samples. We thank both Dr. David Collings and Mr Manfred Ingelfeld (Biomolecular Interaction Centre and the University of Canterbury) for their assistance with the confocal microscopy in N. benthamiana leaves. We also thank Dr. Artemio Mendoza for the triple-GFP vector construct used in the onion biolistic experiments. We would also like to thank both reviewers for their invaluable comments, which significantly improved this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01402/full#supplementary-material
Balakrishna, S., Prabhune, A. A. (2014). Gamma-glutamyl transferases: a structural, mechanistic and physiological perspective. Front. Biol. 9 (1), 51–65. doi: 10.1007/s11515-014-1288-0
Bally, J., Jung, H., Mortimer, C., Naim, F., Philips, J. G., Hellens, R., et al. (2018). The rise and rise of Nicotiana benthamiana: a plant for all reasons. Annu. Rev. Phytopathol. 56 (1), 405–426. doi: 10.1146/annurev-phyto-080417-050141
Bate, N. J., Rothstein, S. J. (1998). C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J. 16 (5), 561–569. doi: 10.1046/j.1365-313x.1998.00324.x
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., et al. (2009). The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 (4), 611–622. doi: 10.1373/clinchem.2008.112797
Capone, D. L., Black, C. A., Jeffery, D. W. (2012). Effects on 3-mercaptohexan-1-ol precursor concentrations from prolonged storage of Sauvignon blanc grapes prior to crushing and pressing. J. Agric. Food. Chem. 60 (13), 3515–3523. doi: 10.1021/jf300054h
Capone, D. L., Jeffery, D. W. (2011). Effects of transporting and processing Sauvignon blanc grapes on 3-mercaptohexan-1-ol precursor concentrations. J. Agric. Food. Chem. 59 (9), 4659–4667. doi: 10.1021/jf200119z
Castellano, I., Merlino, A. (2012). γ-Glutamyltranspeptidases: sequence, structure, biochemical properties, and biotechnological applications. Cell. Mol. Life Sci. 69 (20), 3381–3394. doi: 10.1007/s00018-012-0988-3
Castellano, I., Merlino, A. (2013). Gamma-glutamyl transpeptidases: structure and function. Heidelberg: Springer. doi: 10.1007/978-3-0348-0682-4
Ceci, L. N., Curzio, O. A., Pomilio, A. B. (1992). Effects of irradiation and storage on the γ-glutamyl transpeptidase activity of garlic bulbs cv ‘Red’. J. Sci. Food Agric. 59 (4), 505–510. doi: 10.1002/jsfa.2740590413
Cho, J., Park, M., Choi, D., Lee, S. K. (2012). Cloning and expression of γ-glutamyl transpeptidase and its relationship to greening in crushed garlic (Allium sativum) cloves. J. Sci. Food Agric. 92 (2), 253–257. doi: 10.1002/jsfa.4610
Coetzee, C., du Toit, W. J. (2012). A comprehensive review on Sauvignon blanc aroma with a focus on certain positive volatile thiols. Food Res. Int. 45 (1), 287–298. doi: 10.1016/j.foodres.2011.09.017
Coombe, B. G. (1992). Research on development and ripening of the grape berry. Am. J. Enol. and Vitic. 43 (1), 101–110.
Coombe, B. G. (1995). Growth stages of the grapevine: adoption of a system for identifying grapevine growth stages. Aust. J. Grape Wine Res 1 (2), 104–110. doi: 10.1111/j.1755-0238.1995.tb00086.x
Darriet, P., Tominaga, T., Lavigne, V., Boidron, J.-N., Dubourdieu, D. (1995). Identification of a powerful aromatic component of Vitis vinifera L. var. Sauvignon wines: 4-mercapto-4-methylpentan-2-one. Flavour Fragance J 10 (6), 385–392. doi: 10.1002/ffj.2730100610
des Gachons, C. P., Tominaga, T., Dubourdieu, D. (2000). Measuring the aromatic potential of Vitis vinifera L. cv. Sauvignon blanc grapes by assaying S-cysteine conjugates, precursors of the volatile thiols responsible for their varietal aroma. J. Agric. Food. Chem. 48 (8), 3387–3391. doi: 10.1021/jf990979b
des Gachons, C. P., Tominaga, T., Dubourdieu, D. (2002). Sulfur aroma precursor present in S-glutathione conjugate form: Identification of S-3-(Hexan-1-ol)-glutathione in must from Vitis vinifera L. cv. Sauvignon blanc. J. Agric. Food. Chem. 50 (14), 4076–4079. doi: 10.1021/jf020002y
Destro, T., Prasad, D., Martignago, D., Lliso Bernet, I., Trentin, A. R., Renu, I. K., et al. (2011). Compensatory expression and substrate inducibility of γ-glutamyl transferase GGT2 isoform in Arabidopsis thaliana. J. Exp. Bot. 62 (2), 805–814. doi: 10.1093/jxb/erq316
Dumin, W., Rostas, M., Winefield, C. (2018). Identification and functional characterisation of an allene oxide synthase from grapevine (Vitis vinifera L. Sauvignon blanc). Mol. Biol. Rep.45 (3), 263–277. doi: 10.1007/s11033-018-4159-y
Edwards, R., Dixon, D. P., Walbot, V. (2000). Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 5 (5), 193–198. doi: 10.1016/S1360-1385(00)01601-0
Ferretti, M., Destro, T., Tosatto, S. C. E., La Rocca, N., Rascio, N., Masi, A. (2009). Gamma-glutamyl transferase in the cell wall participates in extracellular glutathione salvage from the root apoplast. New Phytol. 181 (1), 115–126. doi: 10.1111/j.1469-8137.2008.02653.x
Fuentes, S., Sullivan, W., Tilbrook, J., Tyerman, S. (2010). A novel analysis of grapevine berry tissue demonstrates a variety-dependent correlation between tissue vitality and berry shrivel. Aust. J. Grape Wine Res 16 (2), 327–336. doi: 10.1111/j.1755-0238.2010.00095.x
Giaretta, S., Prasad, D., Forieri, I., Vamerali, T., Trentin, A. R., Wirtz, M., et al. (2017). Apoplastic gamma-glutamyl transferase activity encoded by GGT1 and GGT2 is important for vegetative and generative development. Plant Physiol. Biochem. 115, 44–56. doi: 10.1016/j.plaphy.2017.03.007
Grzam, A., Martin, M. N., Hell, R., Meyer, A. J. (2007). γ-Glutamyl transpeptidase GGT4 initiates vacuolar degradation of glutathione S-conjugates in Arabidopsis. FEBS Lett. 581 (17), 3131–3138. doi: 10.1016/j.febslet.2007.05.071
Hanum, T., Sinha, N. K., Cash, J. N. (1995). Characteristics of γ-glutamyl transpeptidase and alliinase of onion and their effects on the enhancement of pyruvate formation in onion macerates. J. Food Biochem. 19 (1), 51–65. doi: 10.1111/j.1745-4514.1995.tb00520.x
Hashimoto, W., Suzuki, H., Yamamoto, K., Kumagai, H. (1995). Effect of site-directed mutations on processing and activity of γ-glutamyltranspeptidase of Escherichia coli K-12. J. Biochem. 118 (1), 75–80. doi: 10.1093/oxfordjournals.jbchem.a124894
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (Web Server issue), W585–W587. doi: 10.1093/nar/gkm259
Karimi, M., Inzé, D., Depicker, A. (2002). GATEWAY™ vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7 (5), 193–195. doi: 10.1016/S1360-1385(02)02251-3
Kobayashi, H., Takase, H., Suzuki, Y., Tanzawa, F., Takata, R., Fujita, K., et al. (2011). Environmental stress enhances biosynthesis of flavor precursors, S-3-(hexan-1-ol)-glutathione and S-3-(hexan-1-ol)-L-cysteine, in grapevine through glutathione S-transferase activation. J. Exp. Bot. 62 (3), 1325–1336. doi: 10.1093/jxb/erq376
Lancaster, J. E., Shaw, M. L. (1994). Characterization of purified γ-glutamyl transpeptidase in onions: Evidence for in vivo role as a peptidase. Phytochemistry 36 (6), 1351–1358. doi: 10.1016/S0031-9422(00)89723-X
Li, L., Hu, D., Jiang, Y., Chen, F., Hu, X., Zhao, G. (2008). Relationship between gamma-glutamyl transpeptidase activity and garlic greening, as controlled by temperature. J. Agric. Food. Chem. 56 (3), 941–945. doi: 10.1021/jf072470j
Lizamore, D., Winefield, C. (2015). The addition of an organosilicone surfactant to Agrobacterium suspensions enables efficient transient transformation of in vitro grapevine leaf tissue at ambient pressure. Plant Cell Tissue Organ Culture 120 (2), 607–615. doi: 10.1007/s11240-014-0627-9
Lund, C. M., Thompson, M. K., Benkwitz, F., Wohler, M. W., Triggs, C. M., Gardner, R., et al. (2009). New Zealand Sauvignon blanc distinct flavor characteristics: Sensory, chemical, and consumer aspects. Am. J. Enol. and Vitic. 60 (1), 1–12.
Martin, M. N., Saladores, P. H., Lambert, E., Hudson, A. O., Leustek, T. (2007). Localization of members of the γ-glutamyl transpeptidase family identifies sites of glutathione and glutathione S-conjugate hydrolysis. Plant Physiol. 144 (4), 1715–1732. doi: 10.1104/pp.106.094409
Martin, M. N., Slovin, J. P. (2000). Purified γ-glutamyl transpeptidases from tomato exhibit high affinity for glutathione and glutathione S-conjugates. Plant Physiol. 122 (4), 1417–1426. doi: 10.1104/pp.122.4.1417
Masi, A., Destro, T., Turetta, L., Varotto, S., Caporale, G., Ferretti, M. (2007). Localization of gamma-glutamyl transferase activity and protein in Zea mays organs and tissues. J. Plant Physiol. 164 (11), 1527–1535. doi: 10.1016/j.jplph.2006.07.016
Masi, A., Trentin, A. R., Agrawal, G. K., Rakwal, R. (2015). Gamma-glutamyl cycle in plants: a bridge connecting the environment to the plant cell? Front. Plant Sci. 6 (252), 4. doi: 10.3389/fpls.2015.00252
Nakai, K., Horton, P. (1999). PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24 (1), 34–35. doi: 10.1016/S0968-0004(98)01336-X
Nakano, Y., Okawa, S., Yamauchi, T., Koizumi, Y., Sekiya, J. (2004). Occurrence of two forms of γ-glutamyltransferases in radish plant. Plant Biotechnol. 21 (3), 243–246. doi: 10.5511/plantbiotechnology.21.243
Nakano, Y., Okawa, S., Yamauchi, T., Koizumi, Y., Sekiya, J. (2006a). Purification and properties of soluble and bound γ- glutamyltransferases from radish cotyledon. Biosci. Biotechnol., Biochem 70 (2), 369–376. doi: 10.1271/bbb.70.369
Nakano, Y., Prieto, R., Hara, M., Sekiya, J. (2006b). Cloning and molecular analysis of radish (Raphanus sativus L.) cDNAs encoding heterodimeric γ-glutamyltransferases. Plant Biotechnol. 23 (4), 419–424. doi: 10.5511/plantbiotechnology.23.419
Noctor, G., Mhamdi, A., Chaouch, S., Han, Y., Neukermans, J., Marquez-Garcia, B., et al. (2012). Glutathione in plants: an integrated overview. Plant Cell Environ. 35 (2), 454–484. doi: 10.1111/j.1365-3040.2011.02400.x
Ohkama-Ohtsu, N., Fukuyama, K., Oliver, D. J., (2009). “Roles of γ-glutamyl transpeptidase and γ-glutamyl cyclotransferase in glutathione and glutathione-conjugate metabolism in plants,” in Advances in Botanical Research,” in Oxidative Stress and Redox Regulation in Plants. Ed. Jacquot, J. P. (London: Academic Press Ltd-Elsevier Science Ltd), 87–113. doi: 10.1016/s0065-2296(10)52004-4
Ohkama-Ohtsu, N., Oikawa, A., Zhao, P., Xiang, C., Saito, K., Oliver, D. J. (2008). A gamma-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol. 148 (3), 1603–1613. doi: 10.1104/pp.108.125716
Ohkama-Ohtsu, N., Radwan, S., Peterson, A., Zhao, P., Badr, A. F., Xiang, C., et al. (2007a). Characterization of the extracellular γ-glutamyl transpeptidases, GGT1 and GGT2, in Arabidopsis. Plant J. 49 (5), 865–877. doi: 10.1111/j.1365-313X.2006.03004.x
Ohkama-Ohtsu, N., Zhao, P., Xiang, C., Oliver, D. J. (2007b). Glutathione conjugates in the vacuole are degraded by γ-glutamyl transpeptidase GGT3 in Arabidopsis. Plant J. 49 (5), 878–888. doi: 10.1111/j.1365-313X.2006.03005.x
Okada, T., Suzuki, H., Wada, K., Kumagai, H., Fukuyama, K. (2006). Crystal structures of g-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate. Proc. Natl. Acad. Sci. 103 (17), 6471–6476. doi: 10.1073/pnas.0511020103
Pena-Gallego, A., Hernandez-Orte, P., Cacho, J., Ferreira, V. (2012). S-cysteinylated and S-glutathionylated thiol precursors in grapes. A review. Food Chem. 131 (1), 1–13. doi: 10.1016/j.foodchem.2011.07.079
Peyret, H., Lomonossoff, G. P. (2013). The pEAQ vector series: the easy and quick way to produce recombinant proteins in plants. Plant Mol. Biol. 83 (1), 51–58. doi: 10.1007/s11103-013-0036-1
Philips, J. G., Dudley, K. J., Waterhouse, P. M., Hellens, R. P. (2019). The rapid methylation of T-DNAs upon Agrobacterium inoculation in plant leaves. Front. Plant Sci. 10 (312), 17. doi: 10.3389/fpls.2019.00312
Podolyan, A., White, J., Jordan, B., Winefield, C. (2010). Identification of the lipoxygenase gene family from Vitis vinifera and biochemical characterisation of two 13-lipoxygenases expressed in grape berries of Sauvignon Blanc. Funct. Plant Biol. 37 (8), 767–784. doi: 10.1071/FP09271
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6 (1), 27. doi: 10.1186/1471-2229-6-27
Rouhier, N., Lemaire, S. D., Jacquot, J.-P. (2008). The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59 (1), 143–166. doi: 10.1146/annurev.arplant.59.032607.092811
Sainsbury, F., Thuenemann, E. C., Lomonossoff, G. P. (2009). pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol. J. 7 (7), 682–693. doi: 10.1111/j.1467-7652.2009.00434.x
Scherer, M., Heimel, K., Starke, V., Kämper, J. (2006). The Clp1 protein is required for clamp formation and pathogenic development of Ustilago maydis. Plant Cell 18 (9), 2388–2401. doi: 10.1105/tpc.106.043521
Schulte, A., Lorenzen, I., Böttcher, M., Plieth, C. (2006). A novel fluorescent pH probe for expression in plants. Plant Methods 2 (1), 7. doi: 10.1186/1746-4811-2-7
Shaw, M. L., Pither-Joyce, M. D., McCallum, J. A. (2005). Purification and cloning of a γ-glutamyl transpeptidase from onion (Allium cepa). Phytochemistry 66 (5), 515–522. doi: 10.1016/j.phytochem.2005.01.017
Sorrequieta, A., Ferraro, G., Boggio, S. B., Valle, E. M. (2010). Free amino acid production during tomato fruit ripening: a focus on l-glutamate. Amino Acids 38 (5), 1523–1532. doi: 10.1007/s00726-009-0373-1
Steinkamp, R., Rennenberg, H. (1984). γ-Glutamyl-transpeptidase in tobacco suspension cultures: catalytic properties and subcellular localization. Physiol. Plant. 61 (2), 251–256. doi: 10.1111/j.1399-3054.1984.tb05905.x
Storozhenko, S., Belles-Boix, E., Babiychuk, E., Herouart, D., Davey, M. W., Slooten, L., et al. (2002). γ-glutamyl transpeptidase in transgenic tobacco plants. cellular localization, processing, and biochemical properties. Plant Physiol. 128 (3), 1109–1119. doi: 10.1104/pp.010887
Sun, Y., Hu, J., Wang, W., Zhang, B., Shen, Y. (2019). Characterization of γ-glutamyltranspeptidases from dormant garlic and onion bulbs. Food Sci. Nutr. 7 (2), 499–505. doi: 10.1002/fsn3.820
Suzuki, H., Kumagai, H. (2002). Autocatalytic processing of γ-glutamyltranspeptidase. J. Biol. Chem. 277 (45), 43536–43543. doi: 10.1074/jbc.M207680200
Taniguchi, N., Ikeda, Y. (2006). “γ‐Glutamyl Transpeptidase: Catalytic Mechanism and Gene Expression” in Advances in Enzymology - and Related Areas of Molecular Biology. Purich, DL, editor. Wiley. doi: 10.1002/9780470123188.ch7
Tashiro, R. M., Philips, J. G., Winefield, C. S. (2016). Identification of suitable grapevine reference genes for qRT-PCR derived from heterologous species. Mol. Genet. Genomic 291 (1), 483–492. doi: 10.1007/s00438-015-1081-z
Tate, S. S., Meister, A. (1978). Serine-borate complex as a transition-state inhibitor of gamma-glutamyl transpeptidase. Proc. Natl. Acad. Sci. 75 (10), 4806–4809. doi: 10.1073/pnas.75.10.4806
Thibon, C., Cluzet, S., Merillon, J. M., Darriet, P., Dubourdieu, D. (2011). 3-sulfanylhexanol precursor biogenesis in grapevine cells: The stimulating effect of Botrytis cinerea. J. Agric. Food. Chem. 59 (4), 1344–1351. doi: 10.1021/jf103915y
Thibon, C., Dubourdieu, D., Darriet, P., Tominaga, T. (2009). Impact of noble rot on the aroma precursor of 3-sulfanylhexanol content in Vitis vinifera L. cv Sauvignon blanc and Semillon grape juice. Food Chem. 114 (4), 1359–1364. doi: 10.1016/j.foodchem.2008.11.016
Tilbrook, J., Tyerman, S. D. (2008). Cell death in grape berries: varietal differences linked to xylem pressure and berry weight loss. Funct. Plant Biol. 35 (3), 173–184. doi: 10.1071/FP07278
Tolin, S., Arrigoni, G., Trentin, A. R., Veljovic-Jovanovic, S., Pivato, M., Zechman, B., et al. (2013). Biochemical and quantitative proteomics investigations in Arabidopsis ggt1 mutant leaves reveal a role for the gamma-glutamyl cycle in plant's adaptation to environment. Proteomics 13 (12-13), 2031–2045. doi: 10.1002/pmic.201200479
Tominaga, T., Furrer, A., Henry, R., Dubourdieu, D. (1998). Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Fragance J 13 (3), 159–162. doi: 10.1002/(SICI)1099-1026(199805/06)13:3<159::AID-FFJ709>3.0.CO;2-7
Wiltshire, E. J., Eady, C. C., Collings, D. A. (2017). Induction of anthocyanin in the inner epidermis of red onion leaves by environmental stimuli and transient expression of transcription factors. Plant Cell Rep. 36 (6), 987–1000. doi: 10.1007/s00299-017-2132-1
Yoshimoto, N., Yabe, A., Sugino, Y., Murakami, S., Sai-ngam, N., Sumi, S.-i., et al. (2015). Garlic γ-glutamyl transpeptidases that catalyze deglutamylation of biosynthetic intermediate of alliin. Front. Plant Sci. 5 (758), 11. doi: 10.3389/fpls.2014.00758
Keywords: glutathione, γ-glutamyl transpeptidase, grape berry development, GGT, volatile thiol, New Zealand Sauvignon blanc
Citation: Philips JG, Dumin W and Winefield C (2019) Functional Characterization of the Grapevine γ-Glutamyl Transferase/Transpeptidase (E.C. 2.3.2.2) Gene Family Reveals a Single Functional Gene Whose Encoded Protein Product Is Not Located in Either the Vacuole or Apoplast. Front. Plant Sci. 10:1402. doi: 10.3389/fpls.2019.01402
Received: 16 July 2019; Accepted: 10 October 2019;
Published: 04 November 2019.
Edited by:
Michael James Considine, University of Western Australia, AustraliaReviewed by:
Naoko Ohkama-Ohtsu, Tokyo University of Agriculture and Technology, JapanCopyright © 2019 Philips, Dumin and Winefield. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher Winefield, Christopher.winefield@lincoln.ac.nz
†Present address: Joshua G. Philips, Queensland University of Technology, Brisbane, QLD, Australia
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.