
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 10 October 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01258
This article is part of the Research Topic Orchid Genomics and Developmental Biology View all 13 articles
Orchids comprise one of the largest, most highly evolved angiosperm families, and form an extremely peculiar group of plants. Various orchids are available through traditional breeding and micro-propagation since they are valuable as potted plants and/or cut flowers in horticultural markets. The flowering of orchids is generally influenced by environmental signals such as temperature and endogenous developmental programs controlled by genetic factors as is usual in many flowering plant species. The process of floral transition is connected to the flower developmental programs that include floral meristem maintenance and floral organ specification. Thanks to advances in molecular and genetic technologies, the understanding of the molecular mechanisms underlying orchid floral transition and flower developmental processes have been widened, especially in several commercially important orchids such as Phalaenopsis, Dendrobium and Oncidium. In this review, we consolidate recent progress in research on the floral transition and flower development of orchids emphasizing representative genes and genetic networks, and also introduce a few successful cases of manipulation of orchid flowering/flower development through the application of molecular breeding or biotechnology tools.
The orchid family is one of the largest families of angiosperms. The flowers of Orchidaceae exhibit a high degree of speciation, with wide variations in floral features such as morphology, color, size and fragrance for attraction of pollinators (Peakall, 2007). Orchids are traded worldwide as cut flowers and potted flowering plants. The flowering of orchids can generally be divided into two steps: floral transition and flower development. In floral transition, juvenility, ambient to cool temperature and/or photoperiod are crucial in determining the time that orchids initiate flowering with respect to ontogeny and season. Although molecular and genetic approaches have made it possible to shed some light on the mechanisms underlying floral transition in model plants such as Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) (He, 2009; Sun et al., 2014), orchid flowering is still not well-understood. Flower formation is initiated by activities of genes known as flowering time genes which regulate the conversion of the vegetative meristem to the floral meristem. Then, floral meristem identity genes regulate the formation of the flower. Once flowering initiates, cadastral genes which govern the whorl formation in flowers are expressed, and the structures of the whorls and their occurrence at the right position are regulated by homeotic genes. Although extreme variation in flower morphology is found across the angiosperms, four types of relatively simple floral organs can be distinguished, namely sepals, petals, stamens and carpels (Theissen and Melzer, 2007; Heijmans et al., 2012). Based mainly on studies with Arabidopsis, the ABCDE model explains the establishment and maintenance of floral organ specification through an interactive network of MADS-box transcription factors. Herein, we review recent research on floral transition and flower development of a few representative orchid species from the perspective of molecular genetics. Some successful cases of the use of biotechnological tools to modulate orchid floral traits are also highlighted.
The Orchidaceae plants are widely distributed across the globe, thus the mechanisms of flowering control among different genera may have developed according to their natural habitats. Apostasioideae, Cypripedioideae, Vanilloideae, Epidendroideae and Orchidoideae are the subfamilies of Orchidaceae. Among these five subfamilies, the Epidendroideae is the largest containing more than 500 genera with around 20,000 species (Freudenstein and Chase, 2015). Different orchids of the Epidendroideae require different conditions of ambient temperature and photoperiod to induce flowering (Hsiao et al., 2011).
Most orchids take several years to finish the juvenile stage. Phalaenopsis usually reach maturity after three to five leaves. Usually, the floral spike (inflorescence) emerges from the axillary buds of the fourth node below the apical leaf whereas other axillary buds are maintained in a dormant state during the flowering season (Sakanishi et al., 1980). The mechanism of differential commitment of distinct axillary buds for flowering is unknown. At the beginning of flowering, the axillary buds are enlarged and then protrude from the base of a leaf, which takes about 3–4 weeks. After that, the bud is elongated to become a floral spike.
The flowering of several orchids is influenced by change in ambient temperature. The flowering of Phalaenopsis is promoted by low ambient temperature, usually less than 26 °C (Blanchard and Runkle, 2006); and can be reversed if the ambient temperature is elevated. The flowering activity of Dendrobium is also promoted by low ambient temperature (Campos and Kerbauy, 2004); however, flowering of some hybrid cultivars of Dendrobium– such as Dendrobium Chao Praya Smile and Dendrobium Madame Thong-In, are promoted by high ambient temperature. The flowering of Miltoniopsis and Zygopetalum are promoted by cool temperature 11–14 °C (Lopez et al., 2003; Lopez and Runkle, 2006). For the Oncidium, another popular orchid, change in ambient temperature enhances or is neutral to their flowering activity. For Cypripedium species, vernalization below 5 °C or subzero temperature is required for flowering (Zhang et al., 2014).
In the model plant Arabidopsis, flowering activity is delayed by low ambient temperature (16 °C) but promoted by vernalization. In low ambient temperature, an alternatively spliced form of FLOWERING LOCUS M (FLM), FLM-β associates with SUPPRESSOR OF VEGETATIVE PHASE (SVP) to suppress the floral integrator gene FLOWERING LOCUS T (FT) (Lee et al., 2013). The expression of FLM is down-regulated via alternative splicing coupled with nonsense-mediated decay (AS-NMD) in response to elevated temperatures (Sureshkumar et al., 2016), indicating that FLM plays an important role in flowering initiation by modulating thermo-sensitivity. The FT expression is also suppressed by a MADS-box gene FLOWERING LOCUS C (FLC). Constant expression of FLC prevents the winter-annual Arabidopsis from floral transition before winter and this repression is released by the vernalization pathway signaling (He, 2009). The vernalization pathway suppresses the FLC activity via the trimethylation of histone 3 lysine 27 (H3K9me3) of FLC chromatin. Vernalization is an obligatory requirement for the flowering of lily (Lilium longiflorum). In high ambient temperature, a fully developed floral bud is present inside the bulb. A period of prolonged cold temperature in winter is required for the switch of shoot apical meristem (SAM) to inflorescence meristem (IM). The candidate genes involved in vernalization response in lily have been elucidated using transcriptome analysis (Villacorta-Martin et al., 2015; Li et al., 2016) and several orthologs of Arabidopsis flowering genes were identified. Among them, SVP and VERNALIZATION 1 (VRN1) were down- and up-regulated respectively in floral buds when the bulbs were treated with low ambient temperature. Furthermore, ectopic expression of lily SVP or VRN1 in Arabidopsis resulted in delayed or earlier flowering, respectively.
Understanding how ambient temperature regulates the floral transition of orchids is valuable not only for the horticultural market but also for the comprehension of flowering regulation in different plant species. Using molecular genomics tools to investigate flowering in popular ornamental orchids has made it possible to identify many flowering-related genes; however, how such genes regulate flowering in an ambient temperature-dependent manner is still not clear.
In Arabidopsis and rice, photoperiod is crucial for flowering control (He, 2009; Sun et al., 2014; Song, 2016). Under long-day conditions, Arabidopsis FT expression is activated in vascular tissues of leaves in a circadian rhythmic manner by the transcriptional regulator CONSTANS (CO) whose expression and activity are controlled by light signaling pathways and the circadian clock (Song, 2016). In rice, two orthologs of FT, Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T (RFT) are mainly responsible for floral induction under short-days (SD) and LD conditions, respectively (Sun et al., 2014).
Floral initiation has been shown to be regulated by photoperiod in a few orchids. In Doritis pulcherrima (now Phalaenopsis pulcherrima), under the 30 °C/20 °C (day/night) condition, floral spikes are initiated more efficiently by 9-h light than 12-h light treatment (Wang et al., 2003). In Miltoniopsis orchids, SD incubation at 23 °C before shifting to cool temperature (11–14 °C) facilitates flowering (Lopez and Runkle, 2006). However, during cool temperature treatment, different photoperiods have no significant effect on flowering suggesting that ambient temperature may play a major role in flowering of Miltoniopsis. On the other hand, the formation of floral spikes in Psygmorchis pusilla is positively correlated with the increase in day length implying that P. pusilla is a quantitative LD plant (Vaz et al., 2004). The effect of photoperiod on flowering seems to be various among different orchid species which have great diversity in adaptation. Furthermore, most orchids are native to tropical areas where day length does not change dramatically throughout the year. Thus, it is reasonable to anticipate that photoperiod has limited effects on flowering of orchids.
The effects of phytohormones on orchid flowering have been studied (Goh and Yang, 1978). A synthetic cytokinin, 6-benzylaminopurine (BA), stimulated the flowering of monopodial (e.g., Phalaenopsis) and sympodial (e.g., Dendrobium) orchids whereas auxin suppressed the BA effect. The positive effect of BA on flowering is likely to be enhanced when combined with gibberellic acid (GA3) although GA3 applied alone does not have an influence on floral induction (Hew and Clifford, 1993). In Phalaenopsis and Doritaenopsis, plants sprayed with BA produced visible inflorescences 3 to 9 days earlier than those without BA treatment although the application of BA could not replace the inductive low temperature in Phalaenopsis (Blanchard and Runkle, 2008). Interestingly, even though GAs do not induce flowering, optimum levels of endogenous GAs in the flowering shoot tips are required for flower development in Phalaenopsis. Indeed, injection of GAs can rescue the blockage of flower development under high temperature (Su et al., 2001). The existence of abscisic acid (ABA) in different tissues of Phalaenopsis has been examined. Relatively higher level of free ABA has been detected in dormant axillary buds whereas free or bound forms of ABA were not found in floral shoots (Wang et al., 2002). In addition, exogenous application of ABA to the stem of Phalaenopsis inhibited the formation of floral spikes even under inductive low ambient temperature conditions implying that ABA plays an inhibitory role in orchid floral transition.
Taken together, developmental maturity and ambient temperature are likely the main endogenous and environmental factors, respectively, controlling orchid flowering together with the combined action of phytohormones. Many orchids are epiphytic plants, meaning they are adapted to face frequent periods of nutrient and water scarcity. Although this kind of orchid uses the crassulacean acid metabolism (CAM) as the strategy for carbon fixation like other CAM plants such as succulents and pineapples to adapt themselves to arid conditions (Silvera et al., 2009), flowering consumes a lot of energy. Therefore, the right timing of bud commitment for floral induction with the best physiological situation is crucial for the maintenance of the species through successful sexual propagation. In addition, most orchids have particular pollinators in the wild environment; the timing of floral transition should be consistent with the appearance of their pollinators. Thus, sensing ambient temperature seems to be a good strategy to achieve this aim if their pollinators only appear in a particular season.
Based on expression analyses and functional studies with heterologous expression systems, candidate genes for flowering regulatory networks in orchids have been reported. Recently, genome-wide analyses have also been taken into consideration for the identification of Orchidaceae-specific and/or species-specific key genes controlling flowering/flower development of orchids (Huang et al., 2016; Lin et al., 2016; Wen et al., 2017).
P. aphrodite FT1 (PaFT1) has been isolated and functionally characterized (Jang et al., 2015). Flowering of P. aphrodite is induced by low ambient temperature (≤25 °C) but prohibited by high ambient temperature (≥28 °C). Moreover, photoperiod has no significant influence on flowering of P. aphrodite. Expression patterns of PaFT1 reflect the flowering behavior of the orchid; its flowering is induced by low ambient temperature but not by different light regimes. Reduced expression level of PaFT1 by virus-induced gene silencing (VIGS) methods resulted in delayed flowering under inductive low ambient temperature, while ectopic expression of PaFT1 suppressed the late flowering phenotype caused by the induced expression of SVP and active FRIGIDA (FRI), a FLC activator, in Arabidopsis. Moreover, PaFT1 is able to physically interact with PaFD reminiscent of the Arabidopsis FT-FD module (He, 2009). LEAFY (LFY) is another floral integrator in Arabidopsis (He, 2009). The LFY gene of P. aphrodite (PhapLFY) has also been characterized (Jang, 2015). Expression of PhapLFY under the control of Arabidopsis LFY promoter rescued the abnormal floral structure of Arabidopsis lfy mutant and caused early heading by overexpression in rice. Recently, a CO-like gene, PhalCOL has been identified in P. hybrida (Wedding Promenade) (Zhang et al., 2011). PhalCOL has significant sequence similarity to CO of Arabidopsis. Overexpression of PhalCOL in tobacco caused an early-flowering phenotype suggesting the functional convergence of CO genes in flowering among different plant species. To discover candidate genes responsible for the flowering control of P. aphrodite, transcription profiles in axillary buds of plants treated with low and high ambient temperature were analyzed and compared to each other (Huang et al., 2016). The result showed that, in addition to FT, LFY, APETALA1 (AP1) and OVEREXPRESSION OF CONSTANS 1 (SOC1) homologs, genes involved in GA biosynthesis were also up-regulated by low ambient temperature. Another expression analysis using Phalaenopsis Fortune Saltzman found that transcripts of KNOX1, R2R3-like MYB, adenosine kinase 2, S-adenosylmethionine synthetase, dihydroflavonol 4-reductase, and naringenin 3-dioxygenase were accumulated at a higher level in spikes grown under warm day/cool night (28 °C/21 °C) compared with those grown at daily warm temperature (28 °C/26 °C) under natural light conditions (Li et al., 2014).
Flowering of D. nobile is promoted by low ambient temperature (Campos and Kerbauy, 2004). Orthologs of FT and MOTHER OF FT AND TFL1 (MFT), a homolog of FT, have been identified in D. nobile Lindl. (Li et al., 2012). Expression of DnFT is up-regulated in leaves but down-regulated in axillary buds under low temperature treatment (12 °C/9 °C, day/night) whereas DnMFT expression is not affected by the low temperature treatment. Transgenic Arabidopsis ectopically expressing DnFT exhibited early flowering. In addition, an ortholog of SOC1, DOSOC1 has been identified from Dendrobium Chao Praya Smile (Ding et al., 2013). Increased expression of DOSOC1 was detected during floral transition. Moreover, overexpression of DOSOC1 in both Arabidopsis and Dendrobium resulted in early-flowering phenotypes. DOAP1, an ortholog of AP1 has also been identified and characterized from the orchid (Sawettalake et al., 2017). The role of DOAP1 is similar to that of AP1 in Arabidopsis. Overexpression of DOAP1 rescued the floral defect of ap1 mutant and resulted in early flowering in wild-type Arabidopsis. Differential gene expression in the SAM during in vitro transition from vegetative to reproductive growth has been investigated in Dendrobium Madame Thong-In (Yu and Goh, 2000a). Several transcription factors, including a MADS-box gene of the AP1/AGL2 family, a class I KNOX gene and a homolog of the DrosophilaSEVEN-UP gene were differentially expressed in vegetative and transitional SAM. The KNOX gene plays an important role in the function of SAM, and encodes a KNOTTED1-like homeobox (Knox) protein later designated as DOH1 (Dendrobium Orchid Homeobox 1) (Yu et al., 2000). In tissue culture conditions, the expression of DOH1 was gradually increased in the apical meristem during vegetative growth whereas it was decreased during the progress of reproductive growth. Overexpression of antisense DOH1 resulted in early flowering in Dendrobium. On the other hand, DOMADS1, a MADS-box gene of the AP1/AGL9 superfamily, was identified from the cDNA library of transitional SAM (Yu and Goh, 2000b). Expression of DOMADS1 was up-regulated in transgenic plants harboring p35S: antisense DOH1 suggesting that DOH1 is a possible upstream repressor of DOMADS1 in the flowering control of Dendrobium orchids. Recently, transcriptomes of D. nobile were analyzed under cold (4 °C) or exogenous cytokinin (thidiazuron) treatment (Wen et al., 2017). The results showed that SOC1, LFY and AP1 genes were induced by both low temperature and thidiazuron treatment whereas DnVRN1 and FT were induced only by cold treatment. Also, some marker genes for the GA signaling pathway were up-regulated under both low temperature and thidiazuron treatments. Further investigation is needed to uncover the cytokinin-GA signaling network in the inductive temperature condition underlying the floral transition in Dendrobium.
High ambient temperature (30 °C) accelerates the flowering of Oncidium. The flowering response to changes in ambient temperature in Oncidium is opposite to that of Phalaenopsis. The flowering promoter FT and repressor TERMINAL FLOWER 1 (TFL1) both encode proteins belonging to phosphatidylethanolamine-binding protein (PEBP) family and have also been identified in Oncidium Gower Ramsey (Hou and Yang, 2009). OnFT was expressed in axillary buds, leaves, pseudo-bulb and flowers while OnTFL1 was only expressed in axillary buds and pseudo-bulb. The expression of OnFT was regulated by photoperiod, with highest expression from the 8th to 12th hour of the light period and lowest expression at dawn. However, expression of OnTFL1 is not influenced by photoperiod. Ectopic expression of OnFT resulted in an early-flowering phenotype in Arabidopsis, and late-flowering of Arabidopsis ft-1 mutant was rescued by OnFT overexpression (Hou and Yang, 2009). On the contrary, ectopic expression of OnTFL1 in Arabidopsis displayed late flowering. OMADS1, a homolog of ArabidopsisAGL6 has also been identified in Oncidium Gower Ramsey (Hsu et al., 2003). OMADS1 transcripts were detectable in the apical meristem and floral organs, and ectopic expression of OMADS1 in Arabidopsis caused early flowering with up-regulation of FT, SOC1, LFY and AP1. Overexpression of OMADS1 in Oncidium also resulted in precocious flowering (Hsu et al., 2003). Recently, ascorbate (AsA) content has been shown to play an important role in floral transition in response to thermal stress (30 °C over a 14-day period) in Oncidium Gower Ramsey (Chin et al., 2014). Under thermal stress conditions, the level of reactive oxygen species (ROS; e.g., H2O2) was significantly increased and the AsA redox ratio [AsA to dehydroascorbate (DHA, the oxidized form of AsA)] was decreased with prominent up-regulation of cytosolic ascorbate peroxidase (cytAPX1) expression. The oxidation of AsA to DHA by ascorbate peroxidase is the key reaction to remove H2O2. The report by Chin and colleagues suggests that the AsA/DHA redox ratio may act as one of the endogenous signals that induce the flowering of Oncidium in response to high ambient temperature. Furthermore, it has been shown that reduced GSH redox ratio caused by down-regulation of GSH metabolism-related genes such as glutathione reductase (GR1), γ-glutamylcysteine synthase (GSH1) and glutathione synthase (GSH2) was linked to the decrease in the AsA redox ratio for flowering of Oncidium orchid (Chin et al., 2016).
DhFVE, a Dortiaenopsis ortholog of Arabidopsis FVE, which is a component of the autonomous flowering pathway, has been identified (Sun et al., 2012). Flowering of Dortiaenopsis (now Phalaenopsis) is promoted by low ambient temperature (22 °C/18 °C, day/night) (Luo et al., 2011) and the expression of DhFVE is increased in the stem during floral transition. In addition, EARLY FLOWERING 4 (EFL4) family genes, DhEFL2, DhEFL3 and DhEFL4 have been identified in Dortiaenopsis (Chen et al., 2015). Arabidopsis EFL4 is known to affect flowering through photoperiod perception and circadian regulation (Doyle et al., 2002). Ectopic expression of DhEFL2, DhEFL3 or DhEFL4 delayed flowering of Arabidopsis. Moreover, GIGANTEA (GI), an upstream activator of CO (Sawa et al., 2007), has been identified as DhGI1 in Dortiaenopsis hybrid (Luo et al., 2011). Expression of DhGI1 is up-regulated by low temperature and may play a role in flowering initiation in Dortiaenopsis hybrid. Recently, many putative flowering genes have been identified in Cymbidium and Erycina through transcriptome analyses. However, their functions still remain to be examined (Li et al., 2013; Lin et al., 2016).
The regulatory networks of floral transition in different orchids may be divergent since the required inductive conditions are not all the same. In addition, the genetic tools for orchid research are still limited; therefore, it is time-consuming and also difficult to reveal the molecular mechanisms of flowering control in orchids. The popular ornamental orchid P. aphrodite has a particular requirement for floral transition, i.e., low ambient temperature, which makes this orchid an interesting target for a case-study of flowering regulation in orchids. Furthermore, a transformation system has been established in orchids including P. aphrodite (Hsing et al., 2016). Thus, the molecular mechanism of flowering regulation could be investigated more thoroughly.
The hypothetical gene regulatory network in flowering of P. aphrodite is illustrated in Figure 1 based on the published results (data adapted from research on orchids). The expression of PaFT1 is induced by low ambient temperature, and PaFT1 interacts with PaFD to possibly activate the downstream genes required for floral induction. Reflecting the Arabidopsis model, PaFT1 protein is likely transported from leaves to axillary buds to induce spiking in Phalaenopsis orchid. If the PaFT1 moves to dormant/juvenile buds, there must be a repressive mechanism against PaFT1 action or other unknown floral co-activators are still absent in dormant/juvenile buds. However, we cannot exclude the possibility that the pavement for the PaFT1 movement to the dormant/juvenile buds is not available. Based on the transcriptomic analyses of Phalaenopsis orchids, KNOX1, SOC1 and FVE genes are induced by low ambient temperature (Sun et al., 2012; Li et al., 2014; Huang et al., 2016) although Dendrobium KNOX1 gene is down-regulated by low ambient temperature (Yu and Goh, 2000a; Yu et al., 2000). Thus, the role of KNOX1 in floral transition of Phalaenopsis needs to be further examined. In Phalaenopsis, SOC1 may activate LFY and AP1 during floral transition as is the case in Arabidopsis. Recently, FVE has been suggested to be an upstream activator of SOC1 in P. aphrodite (Koh et al., 2018). The CO in Phalaenopsis may also activate FT irrespective of photoperiod (Zhang et al., 2011; Kaewphalug et al., 2017). In addition, ELF genes in Phalaenopsis may repress the CO activity (Doyle et al., 2002; Chen et al., 2015). Moreover, the GI-FLAVIN-BINDING, KELCH REPEAT, F BOX 1 (FKF1) and CYCLING DOF FACTOR (CDF) may also regulate the CO activity in Phalaenopsis as is also the case in Arabidopsis (Song, 2016). Since SVP and FLM are critical flowering regulators responding to changes in ambient temperature in Arabidopsis, it is also possible to anticipate that the two orthologs of SVP and FLM also act in floral induction of Phalaenopsis in various ways. Actually, SVP and FLM repress the expression of FT at low ambient temperature in Arabidopsis (Lee et al., 2013). Thus, the working mechanism of those orthologs in Phalaenopsis might be distinct from that of Arabidopsis. In addition, further studies are required to reveal the mechanisms underlying phytohormone-dependent flowering pathways linked to changes in ambient temperature in orchids. Even taking these findings together, it can be seen that there is a long way to go to achieve a better understanding of the flowering regulatory network in P. aphrodite. As one of the shortcuts to reach this goal, high throughput analyses can be applied to identify candidates involved in the floral transition of Phalaenopsis. Also, collection of information on flowering networks in various plant species would be very useful in the interpretation of large-scale experimental results such as omics data. Most importantly, forward or reverse genetic studies are required for the functional confirmation of candidate genes in orchid flowering. The homologs of flowering genes in a few orchid species are listed in Table S1.
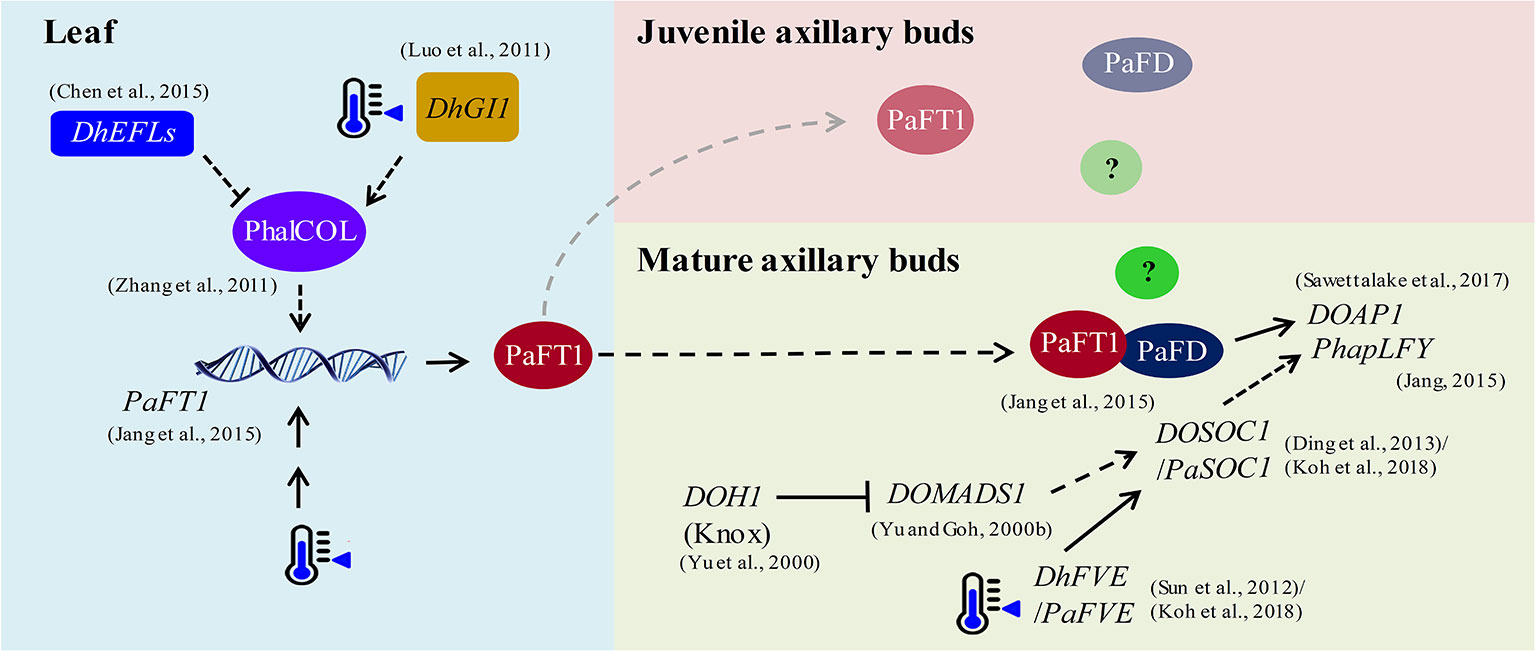
Figure 1 The hypothetical flowering regulation in P. aphrodite under low ambient temperature. Low ambient temperature drives PaFT1 activity in leaves and then PaFT1 protein is transported selectively to mature axillary buds or both mature and juvenile axillary buds. A complex formed by the interaction between PaFT1 and PaFD may activate downstream genes such as AP1 required for floral meristem identity. An example of unknown factors for flowering in the meristem is presented by a green circle with a question mark inside. In juvenile axillary buds, some flowering activators required for floral induction may be absent. PaFT1 can also be activated by CO pathway and the CO activity—PhalCOL (Phalaenopsis CO Like), here—which is likely to be regulated by EFL and GI genes. This regulation should not be dependent on photoperiod. Low ambient temperature also activates PaFVE. Consequently, PaSOC1 is activated by the PaFVE. Moreover, the reduced expression of a KNOTTED1-like homeobox (Knox) gene may lead to the activation of SOC1 through other MADS box genes such as DOMADS1. The orchid LFY genes such as PhapLFY can also be induced by PaSOC1. The solid lines represent pathways that have been reported in orchids and the dotted lines indicate hypothetical regulations based on studies using other plant species. References for each regulation are presented in the figure.
Floral pattern formation is one of the significant features of angiosperms, the mechanisms of which can be explained using the ABC or ABCDE model (Figure 2A). The identity of various floral organs is determined by MADS-box transcription factors or their complexes during flower development. According to the ABC model, in Arabidopsis, sepals in the first whorl are characterized by the expression of A-class genes; petals in whorl 2 are characterized by co-expression of A- and B-class genes, stamens in whorl 3 are characterized by co-expression of B- and C-class genes, and carpels in whorl 4 are determined when C-class genes are solely expressed (Jack, 2001; Theissen, 2001). The ABC model has been extended to the ABCDE model with additional functions of D- and E-class genes in floral organ identity determination (Theissen and Melzer, 2007; Heijmans et al., 2012). D-class genes are involved in ovule development while E-class genes are involved in the identity of all floral verticils. Most genes in the ABCDE model are MADS-box genes. AP1 and AP2 (the only non-MADS-box genes) are A-class genes; AP3 and PISTILLATA (PI) are B-class genes. AGAMOUS (AG) is a C-class gene; SEEDSTICK (STK) is a D-class gene. The D-class genes were first identified in Petunia, namely FLORAL BINDING PROTEIN 7 (FBP7) and FBP11. Finally, SEPALLATA 1 (SEP1), SEP2, SEP3 and SEP4 formally known as AGAMOUS-LIKE 2 (AGL2), AGL4, AGL9 and AGL3, respectively, are characterized as E-class genes. According to the floral quartet model, MADS-box proteins form whorl-specific tetrameric complexes during floral organ determination. The tetrameric transcriptional factor complexes recognize specific cis-regulatory elements termed CArG-boxes (CC-AT rich-GG), and different complexes are postulated to function in controlling expression of different target genes (Jetha et al., 2014). Recently, a reliable and efficient two-stage approach has been developed for angiosperms MADS-Box genes classification using machine learning methods to overcome errors in phylogenetic tree construction (Chen et al., 2019).
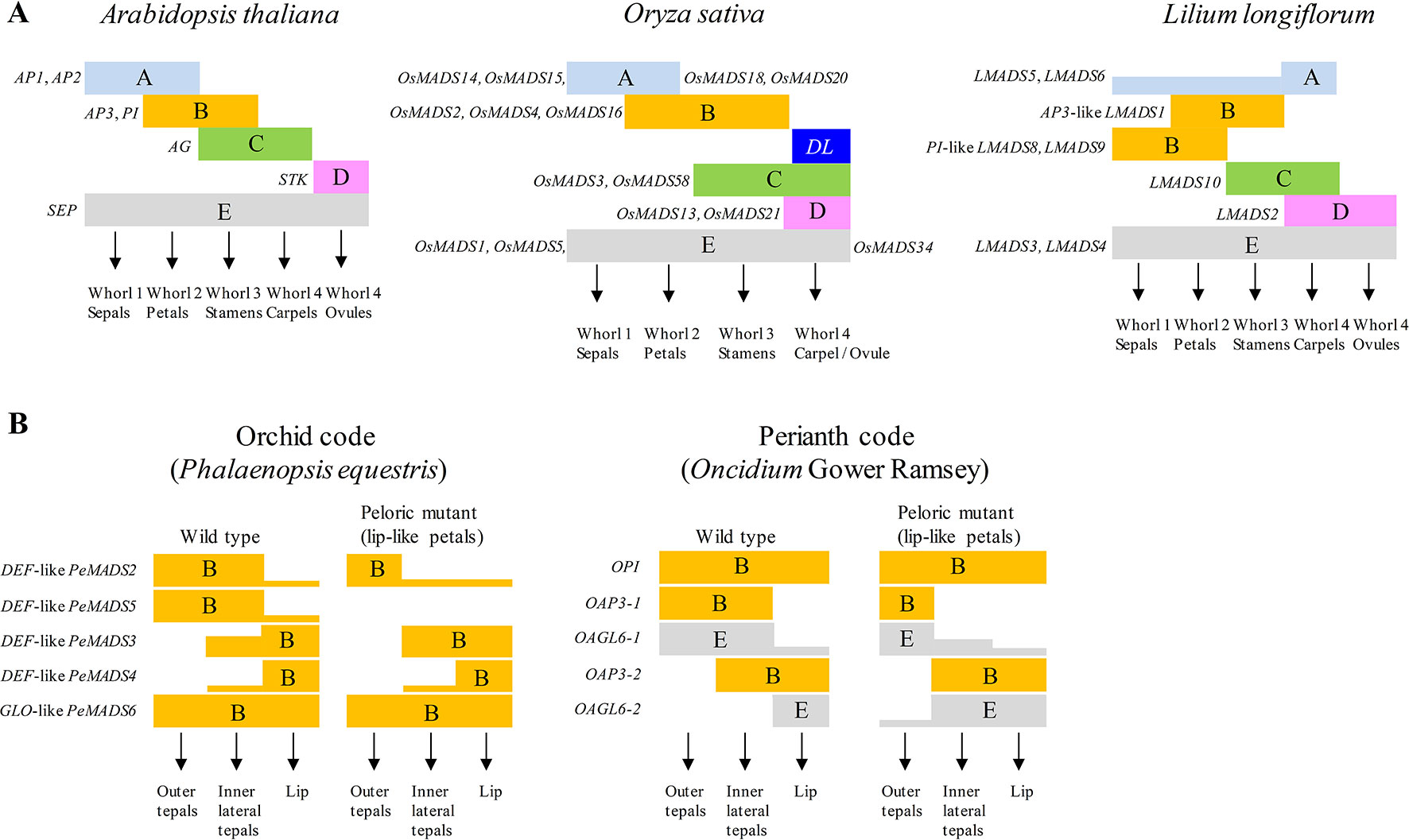
Figure 2 The models of floral development in Arabidopsis, rice, lily and orchids. (A) The ABCDE model explains the floral organ identity in Arabidopsis, rice and lily. The floral development genes are grouped into different classes from A to E. Combinational expressions of different classes of genes determine the formation of different floral organs. Representative genes of different classes are indicated in the figures. DL, DROOPING LEAF. (B) The orchid code and perianth code (P code) explain the formation of perianth of orchids. In the orchid code, combinational expressions of B-class genes from different phylogenetic clades determine the morphology of orchid perianth. The expression patterns of B-class genes changed in peloric mutant are compared with that of wild type. In the P code model, combinational expressions of different B and E-class genes determine the formation sepal/petal or lip of Oncidium orchids.
The MADS-box genes involved in the floral development in monocot plants such as rice, maize (Zea mays), wheat (Triticum aestivum), barley (Hordeum vulgare) and lily have also been elucidated (Callens et al., 2018). MADS-box protein structure is conserved between diverse plants. Comparing the expressions and functions of the ABCDE MADS-box genes among different monocot plants provides the opportunity to elucidate their roles in determining the floral development during evolution. In rice, the A-class gene, the FUL-like (FRUITFULL locus of Arabidopsis) OsMADS14, OsMADS15, OsMADS18 and OsMADS20, are expressed to determine the whorl 1 structures of lemma and palea; co-expression of A and B-class genes, DEF-like (DEFICIENS: an Antirrhinum B-class gene) SUPERWOMEN1 (SPW1 or OsMADS16) and GLO-like (GLOBOSA: another Antirrhinum B-class gene) OsMADS2 and OsMADS4, determines the whorl 2 structure of lodicules (Yoshida and Nagato, 2011); co-expression of B and C-class genes, OsMADS3 and OsMADS58, determines the whorl 3 structure of stamens. Another C function gene DROOPING LEAF (DL) have been identified in carpel specification and floral meristem determinacy (Shige-Hiro et al., 2019). C and D-class genes, OsMADS13 and OsMADS21, are expressed to determine the whorl 4 structure of the carpel and ovule, respectively. In addition, the E-class genes, OsMADS1, OsMADS5 and OsMADS34, are involved in the development of all four whorl structures (Figure 2A).
The lily has a floral character most similar to orchids, with almost identical sepals and petals. Flowers from the lily family Liliaceae have three sepals in whorl 1, three petals in whorl 2, six stamens in whorl 3 and three fused carpels in whorl 4. Three A-class genes from lily, LMADS5, LMADS6 and LMADS7 have been analyzed (Figure 2A) (Chen et al., 2008). They were all expressed in the vegetative stem and inflorescence meristem. With regard to the floral organs, LMADS5 and LMADS6 are mainly expressed in the carpel, whereas LMADS7 is only expressed in the vegetative stem and inflorescence meristem. Early flowering phenotype and homeotic conversions of sepals to carpel-like and of petals to stamen-like structure were observed when they were ectopically expressed in Arabidopsis. For the B-class gene, mRNA of AP3-like LMADS1 could be detected in all four whorls but the protein was detected only in the petal and stamen (Tzeng and Yang, 2001). The PI-like LMADS8 and LMADS9 are expressed in whorl 1 and 2 during all stages of development and are expressed in the stamen only in young flower buds (Chen et al., 2012a). The ectopic expression of LMADS8 and LMADS9 partially converted the sepals into petaloid structures in Arabidopsis. The PI-like gene LFGLOA and LFGLOB identified from L. x formolongi are also expressed in the first, second and third whorl of lily (Akita et al., 2008). Several C-class genes have been identified from different lilies including LMADS10, LFAG1 and LLAG1 (Benedito et al., 2004; Akita et al., 2008; Hsu et al., 2010). They are expressed in whorl 3 and 4, and ectopic expression of C-class genes in Arabidopsis resulted in carpel-like septal and stamen-like petal phenotypes. In addition, a double-flowered cultivar “Aphrodite” of L. x formolongi with petal-like stamen may be caused by reduction in the expression of LFAG1 in whorl 3 (Akita et al., 2008). The lily D-class gene, LMADS2 completely accumulated in the carpel and more specifically in the ovules (Tzeng et al., 2002). Ectopic expression of LMADS2 in Arabidopsis converted the sepals into a stamen-like structure in whorl 2. LMADS3 and LMADS4 have been identified as E-class genes in lily (Tzeng et al., 2003). Both LMADS3 and LMADS4 expression are expressed in the inflorescence meristem, floral buds and all four whorls of the flower organ. Ectopic expression of LMADS3 in Arabidopsis showed extremely early flowering, which was associated with up-regulation of FT, SOC1, LUMINIDEPENDENS and flower meristem identity gene LFY and AP1. Taken together, the different expression patterns of A- and B-class genes from that of the dicot plants such as Arabidopsis suggest a modified ABCDE model for flower development in lily.
The structure of the orchid flower has a zygomorphic nature in contrast to most plant groups leading to precise interaction with the pollinator (Cubas, 2004). Orchid flowers generally contain an outer whorl with three sepals, an inner whorl with three petals, and a single column in the center. The sepals in most orchids are enlarged and look like petals. Two of the petals are displayed in a normal shape and one becomes a highly specialized structure called the lip or labellum. The sepals and petals are usually called tepals since they are very similar in appearance. The column, also called the gynostemium, is the reproductive organ of orchids, which combines the gynoecium (female organ) and androecium (male organ). On the top of the column is the male anther which contains packets of pollen called pollinia. Many floral MADS-box genes have been identified from different orchids. According to their expression patterns and putative roles in determining the different floral organs, advanced models of flower development for orchids have been proposed. Also, these studies offer an opportunity to understand the relationships between the diverse array of floral MADS-box genes and flower development.
A- and E-class genes belong to the same AP1/AGL9 superfamily group. A-function genes come from the SQUA-like (SQUAMOSA: an Antirrhinum A-class gene) subgroup, which can be divided into FUL/AGL8-like and euAP1-like clades. E-function genes come from the SEP-like subgroup containing AGL9-like and AGL2/3/4-like clades. AGL6 is a member of the AP1/AGL9 group between the SQUA-like and SEP-like subgroups (Purugganan et al., 1995; Chang et al., 2009). Two AP1-like genes, ORAP11 and ORAP13, were isolated and characterized in Phalaenopsis Formosa Rose. Both genes are highly expressed during the early stages of floral buds and vegetative organs (Chen et al., 2007). At later stages of flower development, ORAP11 is expressed only in the column and ORAP13 expression is absent from all of the floral organs. PaAP1-1 identified from P. aphrodite is expressed in the inner whorls of the pollinia and pedicel whereas PaAP1-2 is expressed in the pedicel only (Su et al., 2013). This expression pattern suggests that PaAP1-1 and PaAP1-2 play a role in the development of the pollinia and gynoecium rather than being involved in the perianth formation like that in Arabidopsis. Furthermore, PaAP2-5 is mainly expressed in whorl 1 and 2, PaAP2-7 is specifically expressed in pollinia, and PaAP2-11 is expressed at a low level among all floral organs. Novel functions of homeotic genes may be acquired based on the diversified expression in Phalaenopsis through evolutionary processes. Two AP1/FUL-like genes in Phalaenopsis hybrid “Athens,” PhaMADS1 and PhaMADS2 are highly expressed in the ovary before pollination, but a low level of expression is detected in the perianth and gynostemium (Acri-Nunes-Miranda and Mondragon-Palomino, 2014). DOMADS2 identified from Dendrobium Madam Thong-in, and DthyrFL1, DthyrFL2 and DthyrFL3 from D. thyrsiflorum have been characterized as SQUA-like and FUL-like genes, respectively (Yu and Goh, 2000b; Skipper et al., 2005). The expression of DOMADS2 could be detected in the SAM during floral transition but it was limited to the column at the later stages of flower development. Low level expression of DthyrFL1, DthyrFL2 and DthyrFL3 was observed in the vegetative tissues, but higher levels were detected in the inflorescences and ovules. The AP1 homolog of Oncidium Gower Ramsey, OMADS10 is expressed in vegetative leaves, lip and carpel of mature flowers (Chang et al., 2009). An AP2-like gene of D. crumenatum, DcOAP2 is expressed in all floral organs (Xu et al., 2010). Weak expression of three AP1-like genes, EpMADS10, EpMADS11 and EpMADS12 from Erycina pusilla has also been observed in all floral organs (Dirks-Mulder et al., 2017). An AP2-like gene of C. ensifolium, CeAP2, is expressed mainly in sepals and petals with negative regulation by miR172 (Yang et al., 2015).
B-class genes in Orchidaceae have been surveyed and have 11 species characterized into four AP3- and two PI-duplicated homologs (Pan et al., 2011). PI homologs are uniformly expressed in all floral whorls whereas different clades of AP3 homologs may have different expression patterns within floral organs. In P. equestris, four DEF-like genes, PeMADS2, PeMADS3, PeMADS4 and PeMADS5 have been identified. (Tsai et al., 2004). They are expressed in floral organs with distinct patterns. PeMADS2 and PeMADS5 are expressed in sepals, petals, lip and column; PeMADS3 is expressed in petals, lip and column, whereas PeMADS4 is expressed in lip and column only. In peloric (lip-like petal) mutant, expression of PeMADS5 is absent from all floral organs, and expression of PeMADS4 is extended to the lip-like petal. This indicates that, among the four DEF-like genes, PeMADS4 may determine the lip formation in Phalaenopsis. In addition, these four DEF-like MADS genes are able to interact with a GLO-like gene PeMADS6 (Tsai et al., 2005; Tsai et al., 2008). Ectopic expression of PeMADS6 caused petaloid sepals in Arabidopsis flowers. Furthermore, these heterodimers could bind the CArG cis-element. A single copy of PI-like gene of Phalaenopsis hybrid, PhPI10 has been also characterized (Guo et al., 2008). Expression of PhPI10 is restricted to the lip of flowers. Several AP3-like genes PaAP3-1, PaAP3-2, PaAP3-3 and PaAP3-4 were identified in P. aphrodite and their expression was preferentially detected in sepals, petals and the lip/column (Su et al., 2013). Another PI-like gene, PaPI, has also been identified but its transcripts are detectable in all floral organs. In Dendrobium crumenatum, DcOAP3A/B and DcOPI were identified as AP3-like and PI-like genes, respectively (Xu et al., 2006). Transcripts of DcOAP3A and DcOPI are accumulated in all floral organs and their proteins form heterodimers. Abnormal flowers containing petaloid sepals were produced when DcOPI was overexpressed in Arabidopsis. In addition, the phenotypic alteration of B-function mutants has also been observed by overexpressing DcOAP3-SRDX repressor fusion construct implying a dominant negative effect on the B-function via the hetero-dimerization with its interacting partners. The Oncidium Gower Ramsey AP3-like gene, OMADS3 is also expressed in all floral organs (Hsu and Yang, 2002). Another study characterized the B-class MADS-box genes of Oncidium including the AP3-like OMADS3, OMADS5 and OMADS9, and PI-like OMADS8 (Chang et al., 2010). OMADS5 is expressed in sepals and petals; OMADS9 is expressed in petals and lip; OMADS8 is expressed in all four floral organs like OMADS3. In lip-like petals and lip-like sepals of peloric mutant flowers, expression of OMADS5 is down-regulated suggesting that OMADS5 negatively regulates the lip formation. Recently, the perianth formation in Cymbidium goeringii was determined through the complete analysis of expression levels of B-class genes along with co-expression of A-class and E-class genes (Xiang et al., 2018). CgDEF1 is expressed in sepals and petals but not lip; CgDEF3 and CgDEF4 are highly expressed in lip and lip-like petals whereas CgDEF2 is expressed in all floral organs. Several B-class genes have been isolated from Habenaria (Kim et al., 2007), Orchis (Salemme et al., 2011) and Erycina (Dirks-Mulder et al., 2017). HrDEF expression is exhibited in the petals and column whereas that of HrGLO1 and HrGLO2 is detected in all floral organs. In the petaloid-sepal mutant, expression of HrDEF was extended to the petaloid sepals suggesting that distinctive expression of HrDEF determines the differentiation of sepals and petals of H. radiata flowers. In Orchis italica, the PI/GLO-like genes OrcPI and OrcPI2 are expressed in all floral organs of immature floral buds but their expression is restricted to the lip in the mature flower. In E. pusilla, EpMADS13, EpMADS14 and EpMADS15 are AP3-like genes while EpMADS16 is a PI-like gene. Their expression is detectable in almost all floral organs but has diverse patterns. In Rhynchostylis gigantea, RgAP3 an AP3-like and RgPI a PI-like genes were cloned and expression levels of RgAP3 were noticed only in the petal and sepal, and RgPI was expressed in every part of the floral organs (Zhang et al., 2013).
The reproductive organ, the gynostemium, is another unique floral structure in orchids in addition to the labellum. In the ABCDE model, C-class genes are important for the development of stamens and carpels, and D-class genes are required for ovule development. Both C- and D-class genes belong to the AG subfamily of MADS-box genes (Theissen et al., 2000; Kramer et al., 2004). In orchids, C- and D-class genes involved in the development of the gynostemium and ovule have been also identified. In P. equestris, PeMADS1 and PeMADS7 were characterized as C- and D-class genes, respectively (Chen et al., 2012b). Spatial expression analyses of PeMADS1 and PeMADS7 demonstrated that they are specifically expressed in the gynostemium of the flower. Development of ovules is initiated after pollination in orchids and PeMADS1 and PeMADS7 transcripts are accumulated in the ovules after pollination. Moreover, expression of PeMADS1 could be detected in petals of gynostemium-like petal (gylp) mutant suggesting that PeMADS1 functions in gynostemium development. PeMADS1 and PeMADS7 could form a homodimer or heterodimer via the PeMADS8, an E-class protein. Ectopic expression analyses showed that PeMADS1 could rescue the phenotype of AG mutant, and PeMADS7 in Arabidopsis produced characteristic phenotypes of the D-class gene family without homeotic conversions. Another two studies also characterized C- and D-class genes in Phalaenopsis Hatsuyuki and Phalaenopsis Athens including AG-like PhlAG1, PhaMADS8, PhaMADS10 and STK-like PhlAG2, PhaMADS9 (Song et al., 2006; Acri-Nunes-Miranda and Mondragon-Palomino, 2014). Expressions of PhlAG1 and PhlAG2 could be detected in the lip, column and ovule, and PhaMADS8, PhaMADS9 and PhaMADS10 were specifically expressed in the gynostemium and ovary. Expression of PhaMADS9 but not PhaMADS8 and PhaMADS10 is increased in the gynostemium of peloric mutants suggesting that the expression level of STK-like gene is crucial for gynostemium development. The C- and D-class genes in D. crumenatum and D. thyrsiflorum were also characterized; DcOAG1 and DthyrAG1 are C-class genes; DcOAG2 and DthyrAG2 are D-class genes (Skipper et al., 2006; Xu et al., 2006). DcOAG1 is expressed in all floral organs while DcOAG2 is mainly expressed in the ovary and in the envelope cells of pollinia. Overexpression of DcOAG1 in Arabidopsis transformed the sepals and petals into carpel-like and stamen-like structures, respectively. DthyrAG1 and DthyrAG2 are expressed in the inflorescences and ovules after pollination, and DthyrAG2 is believed to play more important roles in later stages of ovule development in D. thyrsiflorum since its expression was higher than that of DthyrAG1 in ovules. The C- and D-class genes in Oncidium Gower Ramsey, namely OMADS4 and OMADS2 respectively, were also characterized (Hsu et al., 2010). OMADS4 is expressed in the stamens and carpels whereas expression of OMADS2 is restricted to the stigmatic cavity and ovary of carpels. In addition, yeast two-hybrid analyses showed that OMADS4 and OMADS2 are able to form homodimers by themselves or form heterodimers with each other. Ectopic expression of OMADS4 and OMADS2 caused only early or moderately early flowering in Arabidopsis without homeotic conversion of floral organs. C- and D-class genes characterized in Cymbidium (Wang et al., 2011b), Orchis (Salemme et al., 2013) and Erycina (Dirks-Mulder et al., 2017), include C-class genes CeMADS1, CeMADS2, OitaAG, EpMADS20, EpMADS21 and EpMADS22, and D-class genes OitaSTK and EpMADS23. CeMADS1 is only expressed in the column but CeMADS2 is expressed in all floral organs. In the multitepal mutant whose male and female reproductive organs are replaced by a newly emerged flower, expression of CeMADS1 is lost, suggesting that CeMADS1 is associated with the development of the gynostemium. O. italica OitaAG and OitaSTK are also expressed in the reproductive organs in both the early and late stages. Among the E. pusilla MADS-box genes, EpMADS20 is expressed in all floral organs while others, such as EpMADS21, EpMADS22 and EpMADS23 are mainly expressed in the gynostemium.
E-class genes show unique interaction with other floral organ identity genes to determine all floral organs. Arabidopsis has four SEP genes: SEP1, SEP2, SEP3, and SEP4. The flowers of a triple mutant of sep1 sep2 sep3 consist entirely of sepal-like organs (Pelaz et al., 2000) while the quadruple mutant of sep1 sep2 sep3 sep4 produces leaf-like organs instead of floral organs (Ditta et al., 2004). Moreover, simultaneously reduced expression of four rice SEP-like genes (OsMADS1, OsMADS5, OsMADS7 and OsMADS8) caused the conversion of all floral organs except the lemma into leaf-like structures (Cui et al., 2010). In orchids, a number of genes belonging to the E-class have been characterized at the molecular level. A SEP3-like gene, OM1 was the first E-class gene identified, from the mature flower of Aranda Deborah, and its expression was identified in sepals and petals (Lu et al., 1993). In Phalaenopsis hybrid Athens, three SEP-like genes PhaMADS4, PhaMADS5 and PhaMADS7 have been identified and these are expressed in the sepal, petal and labellum (Acri-Nunes-Miranda and Mondragon-Palomino, 2014). In P. aphrodite, PaAGL6-1 is expressed in the lip, whereas the PaAGL6-2 is expressed throughout all floral organs (Su et al., 2013). Four SEP-like genes PeSEP1, PeSEP2, PeSEP3 and PeSEP4 were characterized from P. equestris (Pan et al., 2014). These PeSEP genes are expressed in all floral organs and VIGS of PeSEP3 resulted in production of leaf-like tapels. Down-regulation of PeSEP2 expression alone by VIGS has minor effects, but silencing of both PeSEP2 and PeSEP3 expression caused reduced expression of B-class genes such as PeMADS2, PeMADS3, PeMADS4, PeMADS5 and PeMADS6 suggesting an association between PeSEP functions and B-class gene expression. DOMADS1 and DOMADS3 from Dendrobium Madame Thong-In are homologous genes of SEP3 and SEP4, respectively, and DcOSEP1 from D. crumenatum is a homolog of SEP3 (Yu and Goh, 2000b; Xu et al., 2006). Expressions of these genes are constantly activated during floral transition and continue into the mature floral stage. Four E-class genes belonging to the AP1/AGL9 superfamily, OMADS6, OMADS7, OMADS10 and OMADS11 have been identified from Oncidium Gower Ramsey (Chang et al., 2009). OMADS6 is expressed in all floral organs except the stamen, and expression patterns of OMADS7 and OMADS11 are similar to that of OMADS6. Unlike OMADS6, OMADS10 is expressed in vegetative leaves although its transcripts are specifically accumulated in the lip and carpels of mature flowers. Moreover, overexpression of OMADS6, OMADS7 and OMADS11 resulted in extremely early flowering but OMADS10 overexpression caused moderately early flowering in Arabidopsis. Several AGL6-like genes including EpMADS3, EpMADS4 and EpMADS5 have been identified in E. pusilla. They are expressed in floral organs with various expression levels in distinct floral organs suggesting that multiple AGL6-like genes may also contribute to the development of floral organs (Dirks-Mulder et al., 2017). Seven E-class genes CgSEP1, 2, 3, 4 and CgAGL6-1, -2, -3 were characterized in C. goeringii (Xiang et al., 2018). The expression level of CgSEP1 was increased in the peloric mutant lips and decreased in the peloric mutant sepals. High expression levels of CgSEP2 and CgAGL6-1 could be detected in the sepals, but rarely in the lips and columns of the wild-type and the peloric mutant. An increased expression level of CgSEP3 was noticed in peloric mutant lip-like petals and lips, but expression level of CgSEP4 was decreased in the peloric mutant lip-like petals and lips. The expression level of CgAGL6-2 is increased in all floral organs of the wild-type and peloric mutant, but increased level of CgAGL6-3 could only be detected the in lip and lip-like petals of wild-type and peloric mutant. Based on expression patterns and phenotypic alterations caused by the E-class genes, it is likely that E-class genes are required for petal, stamen, and carpel formation in both dicot and monocot plants. Of note, a recent report has shown that the greenish flower phenotype of a mutant orchid cultivar in Habenaria radiata is due to the absence of SEP (HrSEP-1) function (Mitoma and Kanno, 2018). The characterized genes of flower development in orchids are listed in Table S2 (Aceto and Gaudio, 2011; Mondragon-Palomino, 2013; Teixeira da Silva et al., 2014).
Genetic models related to the regulatory patterning formation of actinomorphic flowers in Arabidopsis and Antirrhinum are well characterized. However, orchid flowers usually have zygomorphic symmetry with a prominent, well-differentiated labellum in the inner tepals, and it is morphologically distinct from tepals of other angiosperms (Salemme et al., 2011). The perianth formation in orchids cannot be solely explained by the ABCDE model. Many divergent genes with novel functions are responsible for the regulation of perianth formation in orchids. The model “Orchid Code” has been proposed to explain how the various expression levels of duplicated AP3/DEF-like genes regulate the perianth morphogenesis in orchids (Figure 2B) (Mondragon-Palomino and Theissen, 2008; Mondragon-Palomino and Theissen, 2009). In this model, duplicated DEF-like paralogous are grouped into four clades phylogenetically and their expression patterns determine the formation of different flower organs. The development of outer tepals (sepal) is regulated by the co-expression of clade 1 (PeMADS2-like) and clade 2 (PeMADS5-like or OMADS3-like) genes whereas the two lateral petals are regulated by the co-expression of clade 1, clade 2 and clade 3 (PeMADS3-like) genes. The lip structure is regulated by the co-expression of clade 1, clade 2, clade 3 and clade 4 (PeMADS4-like) genes. In contrast, GLO-like genes form a single clade and express in all four floral whorls. In other words, the divergent DEF-like gene is the key that leads to the morphological diversity of the flowers in orchids. Later, the orchid code model was refined by extensive analysis of DEF-like genes in different subfamilies of orchids (Mondragon-Palomino and Theissen, 2011). According to this refined orchid code, four clades of DEF-like genes are all expressed in lateral petal and lip. However, lower expression level of clade 3 and clade 4 is displayed in lateral petal whereas lower expression level of clade 1 and clade 2 genes is displayed in the lip. Moreover, a combination of four clade genes with differential expression level may also contribute to the developments of gynostemium and ovary.
Development of different floral organs of orchids has also been proposed by a homeotic orchid tepal (HOT) model (Pan et al., 2011). Twenty-four AP3-like genes were identified from 11 species of orchids and grouped into four clades (PeMADS3-like, PeMADS4-like, PeMADS2-like and PeMADS5-like). Similar to the orchid code model, the sepal organ expresses PeMADS2-like and PeMADS5-like genes whereas the lateral petal expresses PeMADS2-like, PeMADS5-like and PeMADS3-like genes. In the lip organ, all four clade genes are expressed. In addition, the HOT model also addresses the temporal change of PeMADS3-like and PeMADS4-like expressions in different stages of inflorescence development. In the early floral organ primordial stage, they are expressed in all whorl organs. In the late floral organ primordial and floral bud stages, PeMADS3-like genes are restricted in petal, lip and column whereas the PeMADS4-like genes are restricted in the lip and column. Combinations of PI-like genes and other MADS-box genes also determine the identity of lip and column.
Another study also based on the expression analysis proposed a “Perianth code” (P code) hypothesis to explain the floral identity of orchids (Figure 2B) (Hsu et al., 2015). Two clades of AP3-like genes (OAP3-1 and AP3-2) in Oncidium associate with OPI and different clades of OAGL6-like genes (E-class). Different heterotetradmers determine different organs. The SP (sepal/petal) complex (OAP3-1/OAGL6-1/OAGL6-1/OPI) specifies the sepal/petal formation whereas the L (lip) complex (OAP3-2/OAGL6-2/OAGL6-2/OPI) is exclusively required for lip formation. The P code model suggests that the diverse E-class genes are also important for development of lip structure of orchids. Diverse roles of E-class genes in floral identity have also been suggested in lily (Tzeng et al., 2003). Two AGL2-like genes, LMADS3 and LMADS4, were identified in lily, and they were expressed in all four whorls of floral organs. Ectopic expression of LMADS3 but bot LMADS4 in Arabidopsis resulted in reduced plant size, early flowering and loss of floral determinacy. In H. radiata, two E-class genes (HrSEP-1 and HeSEP-2) were characterized recently (Mitoma and Kanno, 2018). The expression of HrSEP-1 is lost in a mutant cultivar “Ryokusei” that has greenish petals and a smaller lip in whorl 2, and several septaloid organs and a ventral column in whorls 3 and 4. Moreover, the expression of HrSEP-2 is up-regulated in Ryokusei. Alternative expressions of B-class genes by silencing of E-class genes have also been revealed in Phalaenopsis (Pan et al., 2014). This indicates a feedback regulation between the floral identity genes may also play an important role in the development of floral organs.
All these models specify that the diverse roles of duplicated AP3/DEF-like and other floral identify genes are important for specialized flower morphogenesis, especially for the lip development of orchid flowers. This idea is supported by the recent study of whole-genome sequencing of Apostasia shenzhenica, a genus of Apostasioideae, which is regarded as being phylogenetically plesiomorphic in the orchid family (Zhang et al., 2017). The Apostasia has a radially symmetrical (actinomorphic) flower without lip and complex column structures. Comparing A. shenzhenica to other subfamilies of Orchidaceae, A. shenzhenica seems to have fewer B-class AP3 genes.
Conventional breeding efforts were initiated more than one century ago and helped propel the growth of the ornamental orchid market. In order to obtain profitable horticultural traits, successful breeding usually proceeds by preserving or selecting advantageous parents or progenies. Application of a well-controlled cultivation management system is another important factor for successful commercial orchid farming. In the worldwide market, hundreds and thousands of successful cultivars have been generated using the conventional methods through enthusiastic professional and amateur breeders. The discovery of native tetraploid Phalaenopsis orchids that have better flower shapes and color intensities initiated the creation of numerous significant commercial hybrids through polyploidy breeding. Over the last two decades, a successful program to convert 20 diploid Phalaenopsis species to tetraploid has been developed by Chen et al. (2011). This program facilitated a breeding process that feeds the needs of the nursery business. Polyploidization can typically be achieved by introduction of colchicine in many agricultural or horticultural crops including orchids (Griesbach, 1981; Caperta et al., 2006; Azmi et al., 2016; Tuwo and Indrianto, 2016). One of the most successful product series, “Big White Flower,” is comprised of tetraploid Phalaenopsis hybrids developed through polyploidization with the parents Phalaenopsis Doris. Among them, P. Sogo Yukidian ‘V3’ has been very popular in the market. At present, it is still one of the most profitable hybrids in this group. Chen et al. concluded that the importance of polyploidy to the improvement of orchid cultivars may rely on the increase in the number of sets of genes (Chen et al., 2011). The accumulation of additive genetic and heterotic effects of these genes could potentially improve the varieties possible. Currently, polyploidy is still a major technique that drives orchid breeding programs to commercial success. An alternative strategy for polyploid breeding is the phenomenon of unreduced gametes. By analyzing sporad types, Bolaños-Villegas et al. observed a certain percentage of dyads (2n gametes) and suggested the pollination from individual of these dyad may produce polyploid progeny as revealed in other plants (Bolaños-Villegas et al., 2008).
In the 1970s, the creation of recombination DNA molecules brought about revolutionary genetic engineering, also called genetic transformation, which constitutes the direct manipulation of an organism’s genome using biotechnology. It is a set of technologies that is used to change genetic makeup, including the transfer of genes within and across species boundaries to produce improved or novel traits. In 2013, the blue Phalaenopsis orchids created by the research team of Professor Mii at Chiba University, Japan were exhibited at the 11th Asian Pacific Orchid Conference. These true-blue orchids were genetically transformed using a flavonoid 3’,5’-hydroxylase gene derived from Commelina communis which was incorporated into the Phalaenopsis genome and expressed in the flowers to produce delphinidin, a blue anthocyanin pigment that is also in the flowers of delphinium (larkspur). In 2016, the white Oncidium orchids created by a research group from the National Taiwan University, Taiwan were exhibited at the Taiwan International Orchid Show. The genetically engineered white orchids were transformed by a flower specific promotor driving the carotenoid cleavage dioxygenase gene that degrades carotenoids, which led to white petals.
Similar to many other crops, two major transformation systems were successfully used to transport foreign genes into orchid genomes: particle bombardment and Agrobacterium-mediated transformation systems. The early genetic transformation studies in orchids were confined to the biolistic-mediated transformation which needed a so-called gene gun to deliver the target genes (Kuehnle and Sugii, 1992; Chia et al., 1994). The first successful genetic transformation reported using the Agrobacterium-mediated method was the transfer of gus gene into Phalaenopsis orchid (Belarmino and Mii, 2000). Many years later, this system also successfully transformed other genera of orchids such as Dendrobium, Cymbidium and Oncidium (Yu et al., 2001; Liau et al., 2003; Chin et al., 2007). Target explants for transformation using protocorms, protocorm-like bodies (PLB) and calluses have been reported so far (Belarmino and Mii, 2000; Mishiba et al., 2005; Chin et al., 2007). Usually, the genetic transformation efficiency has been low for orchid plants and the transformation process is also time-consuming. The general slow growth rate could mean the entire transformation process could take up to 2 or 3 years to reach the stage of two-leaf young plantlets. Recently, a protocol using protocorms that could shorten the transformation process to 8 months was reported for Phalaenopsis orchids (Hsing et al., 2016). It will be worth investigating whether a similar time frame can be achieved in other orchid genera. Genome editing, a new and revolutionary genetic engineering technology, is an approach in which a specific target DNA sequence of the genome is altered by adding, removing, or replacing DNA bases in a highly precise manner (Gaj et al., 2013). One particular tool, the CRISPR/Cas system has been developed at an accelerated pace. The application of the CRISPR/Cas system to create engineered crops or plants has yielded fruitful results for further investigation. The technology development and application have been widely reviewed (Bortesi and Fischer, 2015; Yin et al., 2017). One crucial criterion to precisely edit orchid genomes is to rely heavily on the availability of the whole genome sequence information. We predict that the combination of the genome editing technology with bioinformatics analyses will create revolutionary breeding programs for orchid research and development in the future.
For annual plants such as Arabidopsis, rice and wheat, precise control of flowering time in response to environmental cues is necessary for successful reproduction. For perennial plants like orchids, the flowering network may not be so complicated since vernalization is not required and the effect of photoperiod is limited. The changes in ambient temperature may play a more important role in the regulation of floral transition in orchids. Recently, regulations of microRNAs and anti-florigens such as TFL1 have been shown to be involved in the flowering control of A. alpina, a perennial relative of A. thaliana (Wang et al., 2011a; Bergonzi et al., 2013). Although further investigation is required to discover whether orchid TFL1 also plays a negative role in flowering in orchids, it is no exaggeration to say that TFL1 may regulate the dormancy of juvenile axillary buds in the flowering season of orchids. Furthermore, regulation of PHYTOCHROME INTERACTING FACTOR 4 (PIF4) via phytohormone pathways and sugar metabolism may also be involved (Kumar et al., 2012; Bolouri Moghaddam and Van den Ende, 2013). Flower development studies revealed that multiplicated floral homeotic genes (Table S2) are the basis of the specific morphology of orchids (Figure 2). However, how the high order complex of these proteins regulates floral organ identity needs to be further investigated. In addition, the feedback regulation between different floral homeotic genes needs further investigation to explain the floral development of orchids in detail. The A and E-class genes are involved in both floral initiation and flower development. It will be interesting to investigate whether A and E-class genes regulate the expressions of other MADS-box genes. Finally, further advances in functional studies on key genes for flowering/flower development may rely on a breakthrough in orchid transformation technology which leads to more efficient results. Recently, the genome sequences of several orchids have been determined. Understanding of the flowering control and flower development of the CAM-using plants may provide an insightful view into plant evolution.
All the authors wrote and reviewed the manuscript.
This work was supported in part by grants from World Vegetable Center Korea Office (WKO #10000379) and core donors to the World Vegetable Center: Republic of China (ROC), UK aid from the UK government, United States Agency for International Development (USAID), Australian Center for International Agricultural Research (ACIAR), Germany, Thailand, Philippines, Korea and Japan and, a grant from the Ministry of Science and Technology, Taiwan (MOST 106-2321-B-020-002-) to F-C. C. and a grant from Council of Agriculture, Taiwan (COA 106AS-8.6.3-FD-Z1(1)) to F-C. C.
The authors are grateful to Miranda Loney for English editing.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01258/full#supplementary-material
Aceto, S., Gaudio, L. (2011). The MADS and the beauty: genes involved in the development of orchid flowers. Curr. Genomics 12 (5), 342–356. doi: 10.2174/138920211796429754
Acri-Nunes-Miranda, R., Mondragon-Palomino, M. (2014). Expression of paralogous SEP-, FUL-, AG- and STK-like MADS-box genes in wild-type and peloric Phalaenopsis flowers. Front. Plant Sci. 5, 76. doi: 10.3389/fpls.2014.00076
Akita, Y., Horikawa, Y., Kanno, A. (2008). Comparative analysis of floral MADS-box genes between wild-type and a putative homeotic mutant in lily. J. Hortic. Sci. Biotechnol. 83 (4), 453–461. doi: 10.1080/14620316.2008.11512406
Azmi, T. K. K., Sukma, D., Aziz, S. A., Syukur, M. (2016). Polyploidy induction of moth orchid (Phalaenopsis amabilis (L.) Blume) by colchicine treatment on pollinated flowers. J. Agric. Sci. 11 (2), 62. doi: 10.4038/jas.v11i2.8118
Belarmino, M. M., Mii, M. (2000). Agrobacterium-mediated genetic transformation of a Phalaenopsis orchid. Plant Cell Rep. 19, 435–442. doi: 10.1007/s002990050752
Benedito, V. A., Visser, P. B., van Tuyl, J. M., Angenent, G. C., de Vries, S. C., Krens, F. A. (2004). Ectopic expression of LLAG1, an AGAMOUS homologue from lily (Lilium longiflorum Thunb.) causes floral homeotic modifications in Arabidopsis. J. Exp. Bot. 55 (401), 1391–1399. doi: 10.1093/jxb/erh156
Bergonzi, S., Albani, M. C., Ver Loren van Themaat, E., Nordstrom, K. J., Wang, R., Schneeberger, K., et al. (2013). Mechanisms of age-dependent response to winter temperature in perennial flowering of Arabis alpina. Science 340 (6136), 1094–1097. doi: 10.1126/science.1234116
Blanchard, M. G., Runkle, E. S. (2006). Temperature during the day, but not during the night, controls flowering of Phalaenopsis orchids. J. Exp. Bot. 57 (15), 4043–4049. doi: 10.1093/jxb/erl176
Blanchard, M. G., Runkle, E. S. (2008). Benzyladenine promotes flowering in Doritaenopsis and Phalaenopsis orchids. J. Plant Growth Regul. 27 (2), 141–150. doi: 10.1007/s00344-008-9040-0
Bolaños-Villegas, P., Chin, S.-W., Chen, F.-C. (2008). Meiotic chromosome behavior and capsule setting in Doritaenopsis hybrids. J. Am. Soc. Hortic. Sci. 133, 107–116. doi: 10.21273/JASHS.133.1.107
Bolouri Moghaddam, M. R., Van den Ende, W. (2013). Sugars, the clock and transition to flowering. Front. Plant Sci. 4, 22. doi: 10.3389/fpls.2013.00022
Bortesi, L., Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33 (1), 41–52. doi: 10.1016/j.biotechadv.2014.12.006
Callens, C., Tucker, M. R., Zhang, D., Wilson, Z. A. (2018). Dissecting the role of MADS-box genes in monocot floral development and diversity. J. Exp. Bot. 69 (10), 2435–2459. doi: 10.1093/jxb/ery086
Campos, K. O., Kerbauy, G. B. (2004). Thermoperiodic effect on flowering and endogenous hormonal status in Dendrobium (Orchidaceae). J. Plant Physiol. 161 (12), 1385–1387. doi: 10.1016/j.jplph.2004.07.008
Caperta, A. D., Delgado, M., Ressurreicao, F., Meister, A., Jones, R. N., Viegas, W., et al. (2006). Colchicine-induced polyploidization depends on tubulin polymerization in c-metaphase cells. Protoplasma 227 (2–4), 147–153. doi: 10.1007/s00709-005-0137-z
Chang, Y. Y., Chiu, Y. F., Wu, J. W., Yang, C. H. (2009). Four orchid (Oncidium Gower Ramsey) AP1/AGL9-like MADS box genes show novel expression patterns and cause different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 50 (8), 1425–1438. doi: 10.1093/pcp/pcp087
Chang, Y. Y., Kao, N. H., Li, J. Y., Hsu, W. H., Liang, Y. L., Wu, J. W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152 (2), 837–853. doi: 10.1104/pp.109.147116
Chao, Y. T., Yen, S. H., Yeh, J. H., Chen, W. C., Shih, M. C. (2017). Orchidstra 2.0—A transcriptomics resource for the orchid family. Plant Cell Physiol. 58 (1), e9. doi: 10.1093/pcp/pcw220
Chen, D., Guo, B., Hexige, S., Zhang, T., Shen, D., Ming, F. (2007). SQUA-like genes in the orchid Phalaenopsis are expressed in both vegetative and reproductive tissues. Planta 226 (2), 369–380. doi: 10.1007/s00425-007-0488-0
Chen, M. K., Hsieh, W. P., Yang, C. H. (2012a). Functional analysis reveals the possible role of the C-terminal sequences and PI motif in the function of lily (Lilium longiflorum) PISTILLATA (PI) orthologues. J. Exp. Bot. 63 (2), 941–961. doi: 10.1093/jxb/err323
Chen, M. K., Lin, I. C., Yang, C. H. (2008). Functional analysis of three lily (Lilium longiflorum) APETALA1-like MADS box genes in regulating floral transition and formation. Plant Cell Physiol. 49 (5), 704–717. doi: 10.1093/pcp/pcn046
Chen, W.-H., Kao, Y.-L., Tang, C.-Y., Jean, G.-T. (2011). “Endopolyploidy in Phalaenopsis orchids and its application in polyploid breeding,” in Orchid Biotechnology II. Eds. Chen, W. H., Chen, H. H. (Singapore: World Scientific), 25–48. doi: 10.1142/9789814327930_0002
Chen, W., Qin, Q., Zhang, C., Zheng, Y., Wang, C., Zhou, M., et al. (2015). DhEFL2, 3 and 4, the three EARLY FLOWERING4-like genes in a Doritaenopsis hybrid regulate floral transition. Plant Cell Rep. 34 (12), 2027–2041. doi: 10.1007/s00299-015-1848-z
Chen, Y. T., Chang, C. C., Chen, C. W., Kuan, C. C., Wei, Y. C. (2019). MADS-box gene classification in Angiosperms by clustering and machine learning approaches. Front. Genet. 9, 707. doi: 10.3389/fgene.2018.00707
Chen, Y. Y., Lee, P. F., Hsiao, Y. Y., Wu, W. L., Pan, Z. J., Lee, Y. I., et al. (2012b). C- and D-class MADS-box genes from Phalaenopsis equestris (Orchidaceae) display functions in gynostemium and ovule development. Plant Cell Physiol. 53 (6), 1053–1067. doi: 10.1093/pcp/pcs048
Chia, T.-F., Chan, Y.-S., Chua, N.-H. (1994). The firefly luciferase gene as a non-invasive reporter for Dendrobium transformation. The Plant Journal 6, 441–446. doi: 10.1046/j.1365-313X.1994.06030441.x
Chin, D. C., Hsieh, C. C., Lin, H. Y., Yeh, K. W. (2016). A low glutathione redox state couples with a decreased ascorbate redox ratio to accelerate flowering in Oncidium orchid. Plant Cell Physiol. 57 (2), 423–436. doi: 10.1093/pcp/pcv206
Chin, D. C., Shen, C. H., SenthilKumar, R., Yeh, K. W. (2014). Prolonged exposure to elevated temperature induces floral transition via up-regulation of cytosolic ascorbate peroxidase 1 and subsequent reduction of the ascorbate redox ratio in Oncidium hybrid orchid. Plant Cell Physiol. 55 (12), 2164–2176. doi: 10.1093/pcp/pcu146
Chin, D. P., Mishiba, K., Mii, M. (2007). Agrobacterium-mediated transformation of protocorm-like bodies in Cymbidium. Plant Cell Rep. 26 (6), 735–743. doi: 10.1007/s00299-006-0284-5
Cubas, P. (2004). Floral zygomorphy, the recurring evolution of a successful trait. Bioessays 26 (11), 1175–1184. doi: 10.1002/bies.20119
Cui, R., Han, J., Zhao, S., Su, K., Wu, F., Du, X., et al. (2010). Functional conservation and diversification of class E floral homeotic genes in rice (Oryza sativa). Plant J. 61, 767–781. doi: 10.1111/j.1365-313X.2009.04101.x
Ding, L., Wang, Y., Yu, H. (2013). Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao parya smile. Plant Cell Physiol. 54 (4), 595–608. doi: 10.1093/pcp/pct026
Dirks-Mulder, A., Butot, R., van Schaik, P., Wijnands, J. W., van den Berg, R., Krol, L., et al. (2017). Exploring the evolutionary origin of floral organs of Erycina pusilla, an emerging orchid model system. BMC Evol. Biol. 17 (1), 89. doi: 10.1186/s12862-017-0938-7
Ditta, G., Pinyopich, A., Robles, P., Pelaz, S., Yanofsky, M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14, 1935–1940. doi: 10.1016/j.cub.2004.10.028
Doyle, M. R., Davis, S. J., Bastow, R. M., McWatters, H. G., Kozma-Bognar, L., Nagy, F., et al. (2002). The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 419 (6902), 74–77. doi: 10.1038/nature00954
Freudenstein, J. V., Chase, M. W. (2015). Phylogenetic relationships in Epidendroideae (Orchidaceae), one of the great flowering plant radiations: progressive specialization and diversification. Ann. Bot. 115 (4), 665–681. doi: 10.1093/aob/mcu253
Gaj, T., Gersbach, C. A., Barbas, C. F. (2013). ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31 (7), 397–405. doi: 10.1016/j.tibtech.2013.04.004
Goh, C. J., Yang, A. L. (1978). Effects of growth regulators and decapitation on flowering of Dendrobium orchid hybrids. Plant Sci. Lett. 12, 278–292. doi: 10.1016/0304-4211(78)90080-9
Griesbach, R. J. (1981). Colchicine-induced polyploidy in Phanalenopsis orchid. Plant Cell Tissue Organ Cult. 1, 103–107. doi: 10.1007/BF02318909
Guo, B., Zhang, T., Shi, J., Chen, D., Shen, D., Ming, F. (2008). Cloning and characterization of a novel PI-like MADS-box gene in Phalaenopsis orchid. DNA Seq. 19 (3), 332–339. doi: 10.1080/10425170701606193
He, Y. (2009). Control of the transition to flowering by chromatin modifications. Mol. Plant. 2 (4), 554–564. doi: 10.1093/mp/ssp005
Heijmans, K., Ament, K., Rijpkema, A. S., Zethof, J., Wolters-Arts, M., Gerats, T., et al. (2012). Redefining C and D in the petunia ABC. Plant Cell 24 (6), 2305–2317. doi: 10.1105/tpc.112.097030
Hew, C. S., Clifford, P. E. (1993). Plant growth regulator and orchid cut-flower industry. Plant Growth Regul. 13, 231–239. doi: 10.1007/BF00024843
Hou, C. J., Yang, C. H. (2009). Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant Cell Physiol. 50 (8), 1544–1557. doi: 10.1093/pcp/pcp099
Hsiao, Y. Y., Pan, Z. J., Hsu, C. C., Yang, Y. P., Hsu, Y. C., Chuang, Y. C., et al. (2011). Research on orchid biology and biotechnology. Plant Cell Physiol. 52 (9), 1467–1486. doi: 10.1093/pcp/pcr100
Hsing, H. X., Lin, Y. J., Tong, C. G., Li, M. J., Chen, Y. J., Ko, S. S. (2016). Efficient and heritable transformation of Phalaenopsis orchids. Bot. Stud. 57 (1), 30. doi: 10.1186/s40529-016-0146-6
Hsu, H.-F., Hsu, W.-H., Lee, Y.-I., Mao, W.-T., Yang, J.-Y., Li, J.-Y., et al. (2015). Model for perianth formation in orchids. Nat. Plant 1, 1–8. doi: 10.1038/nplants.2015.46
Hsu, H. F., Hsieh, W. P., Chen, M. K., Chang, Y. Y., Yang, C. H. (2010). C/D class MADS box genes from two monocots, orchid (Oncidium Gower Ramsey) and lily (Lilium longiflorum), exhibit different effects on floral transition and formation in Arabidopsis thaliana. Plant Cell Physiol. 51 (6), 1029–1045. doi: 10.1093/pcp/pcq052
Hsu, H. F., Huang, C. H., Chou, L. T., Yang, C. H. (2003). Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol. 44 (8), 783–794. doi: 10.1093/pcp/pcg099
Hsu, H. F., Yang, C. H. (2002). An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol. 43 (10), 1198–1209. doi: 10.1093/pcp/pcf143
Huang, J. Z., Lin, C. P., Cheng, T. C., Huang, Y. W., Tsai, Y. J., Cheng, S. Y., et al. (2016). The genome and transcriptome of Phalaenopsis yield insights into floral organ development and flowering regulation. PeerJ 4, e2017. doi: 10.7717/peerj.2017
Jack, T. (2001). Plant development going MADS. Plant Mol. Biol. 46 (5), 515–520. doi: 10.1023/A:1010689126632
Jang, S. (2015). Functional characterization of PhapLEAFY, a FLORICAULA/LEAFY Ortholog in Phalaenopsis aphrodite. Plant Cell Physiol. 56 (11), 2234–2247. doi: 10.1093/pcp/pcv130
Jang, S., Choi, S. C., Li, H. Y., An, G., Schmelzer, E. (2015). Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS One 10 (8), e0134987. doi: 10.1371/journal.pone.0134987
Jetha, K., Theissen, G., Melzer, R. (2014). Arabidopsis SEPALLATA proteins differ in cooperative DNA-binding during the formation of floral quartet-like complexes. Nucleic Acids Res. 42 (17), 10927–10942. doi: 10.1093/nar/gku755
Kaewphalug, W., Huehne, P. S., Sriboonlert, A. (2017). Characterization of a CONSTANS-like gene from Pigeon orchid (Dendrobium crumenatum Swartz) and its expression under different photoperiod conditions. Hortic. J. 86 (2), 252–262. doi: 10.2503/hortj.MI-123
Kim, S.-Y., Yun, P.-Y., Fukuda, T., Ochiai, T., Yokoyama, J., Kameya, T., et al. (2007). Expression of a DEFICIENS-like gene correlates with the differentiation between sepal and petal in the orchid, Habenaria radiata (Orchidaceae). Plant Sci. 172 (2), 319–326. doi: 10.1016/j.plantsci.2006.09.009
Koh, K. W., Lee, S. H., Chen, H. K., Chang, C. Y., Chan, M. T. (2018). Phalaenopsis flowering locus VE regulates floral organ maturation. Plant Cell Rep. 37 (3), 467–482. doi: 10.1007/s00299-017-2243-8
Kramer, E. M., Jaramillo, M. A., Di Stilio, V. S. (2004). Patterns of gene duplication and functional evolution during the diversification of the AGAMOUS subfamily of MADS box genes in angiosperms. Genetics 166 (2), 1011–1023. doi: 10.1534/genetics.166.2.1011
Kuehnle, A. R., Sugii, N. (1992). Transformation of Dendrobium orchid using particle bombardment of protocorms. Plant Cell Rep. 11 (9), 484–488. doi: 10.1007/BF00232696
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alos, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484 (7393), 242–245. doi: 10.1038/nature10928
Lee, J. H., Ryu, H. S., Chung, K. S., Pose, D., Kim, S., Schmid, M., et al. (2013). Regulation of temperature-responsive flowering by MADS-box transcription factor repressors. Science 342 (6158), 628–632. doi: 10.1126/science.1241097
Li, D. M., Lu, F. B., Zhu, G. F., Sun, Y. B., Xu, Y. C., Jiang, M. D., et al. (2014). Identification of warm day and cool night conditions induced flowering-related genes in a Phalaenopsis orchid hybrid by suppression subtractive hybridization. Genet. Mol. Res. 13 (3), 7037–7051. doi: 10.4238/2014.February.14.7
Li, R., Wang, A., Sun, S., Liang, S., Wang, X., Ye, Q., et al. (2012). Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell Tissue Organ Cult. (PCTOC) 111 (2), 143–151. doi: 10.1007/s11240-012-0178-x
Li, W., Liu, X., Lu, Y. (2016). Transcriptome comparison reveals key candidate genes in response to vernalization of Oriental lily. BMC Genomics 17, 664. doi: 10.1186/s12864-016-2955-0
Li, X., Luo, J., Yan, T., Xiang, L., Jin, F., Qin, D., et al. (2013). Deep sequencing-based analysis of the Cymbidium ensifolium floral transcriptome. PLoS One 8 (12), e85480. doi: 10.1371/journal.pone.0085480
Liau, C. H., You, S. J., Prasad, V., Hsiao, H. H., Lu, J. C., Yang, N. S., et al. (2003). Agrobacterium tumefaciens-mediated transformation of an Oncidium orchid. Plant Cell Rep. 21 (10), 993–998. doi: 10.1007/s00299-003-0614-9
Lin, C. S., Hsu, C. T., Liao, D. C., Chang, W. J., Chou, M. L., Huang, Y. T., et al. (2016). Transcriptome-wide analysis of the MADS-box gene family in the orchid Erycina pusilla. Plant Biotechnol. J. 14 (1), 284–298. doi: 10.1111/pbi.12383
Lopez, R. G., Runkle, E. S. (2006). Temperature and photoperiod regulate flowering of potted Miltoniopsis Orchids. Hortscience 41, 593–597. doi: 10.21273/HORTSCI.41.3.593
Lopez, R. G., Runkle, E. S., Heins, R. D., Whitman, C. M. (2003). Temperature and photoperiodic effects on growth and flowering of Zygopetalum redvale ‘Fire Kiss’ orchid. Acta Hortic. 624, 155–162. doi: 10.17660/ActaHortic.2003.624.20
Lu, Z. X., Wu, M., Loh, C. S., Yeong, C. Y., Goh, C. J. (1993). Nucleotide sequence of a flower-specific MADS box cDNA clone from orchid. Plant Mol. Biol. 23 (4), 901–904. doi: 10.1007/BF00021545
Luo, X., Zhang, C., Sun, X., Qin, Q., Zhou, M., Paek, K. Y., et al. (2011). Isolation and characterization of a Doritaenopsis hybrid GIGANTEA gene, which possibly involved in inflorescence initiation at low temperatures. Korean J. Hortic. Sci. Technol. 29, 135–143.
Mishiba, K., Chin, D. P., Mii, M. (2005). Agrobacterium-mediated transformation of Phalaenopsis by targeting protocorms at an early stage after germination. Plant Cell Rep. 24 (5), 297–303. doi: 10.1007/s00299-005-0938-8
Mitoma, M., Kanno, A. (2018). The greenish flower phenotype of Habenaria radiata (Orchidaceae) is caused by a mutation in the SEPALLATA-Like MADS-Box gene HrSEP-1. Front. Plant Sci. 9, 831. doi: 10.3389/fpls.2018.00831
Mondragon-Palomino, M. (2013). Perspectives on MADS-box expression during orchid flower evolution and development. Front. Plant Sci. 4, 377. doi: 10.3389/fpls.2013.00377
Mondragon-Palomino, M., Theissen, G. (2008). MADS about the evolution of orchid flowers. Trends Plant Sci. 13 (2), 51–59. doi: 10.1016/j.tplants.2007.11.007
Mondragon-Palomino, M., Theissen, G. (2009). Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann. Bot. 104 (3), 583–594. doi: 10.1093/aob/mcn258
Mondragon-Palomino, M., Theissen, G. (2011). Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the ‘orchid code’. Plant J. 66 (6), 1008–1019. doi: 10.1111/j.1365-313X.2011.04560.x
Pan, Z. J., Chen, Y. Y., Du, J. S., Chen, Y. Y., Chung, M. C., Tsai, W. C., et al. (2014). Flower development of Phalaenopsis orchid involves functionally divergent SEPALLATA-like genes. New Phytol. 202 (3), 1024–1042. doi: 10.1111/nph.12723
Pan, Z. J., Cheng, C. C., Tsai, W. C., Chung, M. C., Chen, W. H., Hu, J. M., et al. (2011). The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol. 52 (9), 1515–1531. doi: 10.1093/pcp/pcr092
Peakall, R. (2007). Speciation in the Orchidaceae: confronting the challenges. Mol. Ecol. 16 (14), 2834–2837. doi: 10.1111/j.1365-294X.2007.03311.x
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E., Yanofsky, M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203. doi: 10.1038/35012103
Purugganan, M. D., Rounsley, S. D., Schmidt, R. J., Yanofsky, M. F. (1995). Molecular evolution of flower development: diversification of the plant MADS-box regulatory gene family. Genetics 140 (1), 345–356.
Sakanishi, Y., Imanishi, H., Ishida, G. (1980). Effect of temperature on growth and flowering of Phalaenopsis amabilis. Bull. Univ. Osaka Prefect. Ser. B 32, 1–9.
Salemme, M., Sica, M., Gaudio, L., Aceto, S. (2011). Expression pattern of two paralogs of the PI/GLO-like locus during Orchis italica (Orchidaceae, Orchidinae) flower development. Dev. Genes Evol. 221 (4), 241–246. doi: 10.1007/s00427-011-0372-6
Salemme, M., Sica, M., Gaudio, L., Aceto, S. (2013). The OitaAG and OitaSTK genes of the orchid Orchis italica: a comparative analysis with other C- and D-class MADS-box genes. Mol. Biol. Rep. 40 (5), 3523–3535. doi: 10.1007/s11033-012-2426-x
Sawa, M., Nusinow, D. A., Kay, S. A., Imaizumi, T. (2007). FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 318 (5848), 261–265. doi: 10.1126/science.1146994
Sawettalake, N., Bunnag, S., Wang, Y., Shen, L., Yu, H. (2017). DOAP1 promotes flowering in the orchid Dendrobium Chao Praya Smile. Front. Plant Sci. 8, 400. doi: 10.3389/fpls.2017.00400
Shige-Hiro, S., Yukiko, Y., Suzuha, O., Wakana, T., Hiro-, Y. H. (2019). Rice flower development revisited: regulation of carpel specification and flower meristem determinacy. Plant Cell Physiol. 60 (6), 1284–1295. doi: 10.1093/pcp/pcz020
Silvera, K., Santiago, L. S., Cushman, J. C., Winter, K. (2009). Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 149 (4), 1838–1847. doi: 10.1104/pp.108.132555
Skipper, M., Johansen, L. B., Pedersen, K. B., Frederiksen, S., Johansen, B. B. (2006). Cloning and transcription analysis of an AGAMOUS- and SEEDSTICK ortholog in the orchid Dendrobium thyrsiflorum (Reichb. f.). Gene 366 (2), 266–274. doi: 10.1016/j.gene.2005.08.014
Skipper, M., Pedersen, K. B., Johansen, L. B., Frederiksen, S., Irish, V. F., Johansen, B. B. (2005). Identification and quantification of expression levels of three FRUITFULL-like MADS-box genes from the orchid Dendrobium thyrsiflorum (Reichb. f.). Plant Sci. 169 (3), 579–586. doi: 10.1016/j.plantsci.2005.04.011
Song, I. J., Nakamura, T., Fukuda, T., Yokoyama, J., Ito, T., Ichikawa, H., et al. (2006). Spatiotemporal expression of duplicate AGAMOUS orthologues during floral development in Phalaenopsis. Dev. Genes Evol. 216 (6), 301–313. doi: 10.1007/s00427-005-0057-0
Song, Y. H. (2016). The effect of fluctuations in photoperiod and ambient temperature on the timing of flowering: time to move on natural environmental conditions. Mol. Cells 39 (10), 715–721. doi: 10.14348/molcells.2016.0237
Su, C. L., Chen, W. C., Lee, A. Y., Chen, C. Y., Chang, Y. C., Chao, Y. T., et al. (2013). A modified ABCDE model of flowering in orchids based on gene expression profiling studies of the moth orchid Phalaenopsis aphrodite. PLoS One 8 (11), e80462. doi: 10.1371/journal.pone.0080462
Su, W. R., Chen, W. S., Koshioka, M., Mander, L. N., Hung, L. S. (2001). Changes in gibberellin levels in the flowering shoot of Phalaenopsis hybrida under high temperature conditions when flower development is blocked. Plant Physiol. Biochem. 39, 45–50. doi: 10.1016/S0981-9428(00)01218-3
Sun, C., Chen, D., Fang, J., Wang, P., Deng, X., Chu, C. (2014). Understanding the genetic and epigenetic architecture in complex network of rice flowering pathways. Protein Cell 5 (12), 889–898. doi: 10.1007/s13238-014-0068-6
Sun, X., Qin, Q., Zhang, J., Zhang, C., Zhou, M., Paek, K. Y., et al. (2012). Isolation and characterization of the FVE gene of a Doritaenopsis hybrid involved in the regulation of flowering. Plant Growth Regul. 68 (1), 77–86. doi: 10.1007/s10725-012-9695-1
Sureshkumar, S., Dent, C., Seleznev, A., Tasset, C., Balasubramanian, S. (2016). Nonsense-mediated mRNA decay modulates FLM-dependent thermosensory flowering response in Arabidopsis. Nat. Plants 2 (5), 16055. doi: 10.1038/nplants.2016.55
Teixeira da Silva, J. A., Aceto, S., Liu, W., Yu, H., Kanno, A. (2014). Genetic control of flower development, color and senescence of Dendrobium orchids. Sci. Hortic. 175, 74–86. doi: 10.1016/j.scienta.2014.05.008
Theissen, G. (2001). Development of floral organ identity: stories from the MADS house. Curr. Opin. Plant Biol. 4 (1), 75–85. doi: 10.1016/S1369-5266(00)00139-4
Theissen, G., Becker, A., Di Rosa, A., Kanno, A., Kim, J. T., Munster, T., et al. (2000). A short history of MADS-box genes in plants. Plant Mol. Biol. 42 (1), 115–149. doi: 10.1023/A:1006332105728
Theissen, G., Melzer, R. (2007). Molecular mechanisms underlying origin and diversification of the angiosperm flower. Ann. Bot. 100 (3), 603–619. doi: 10.1093/aob/mcm143
Tsai, W. C., Kuoh, C. S., Chuang, M. H., Chen, W. H., Chen, H. H. (2004). Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol. 45 (7), 831–844. doi: 10.1093/pcp/pch095
Tsai, W. C., Lee, P. F., Chen, H. I., Hsiao, Y. Y., Wei, W. J., Pan, Z. J., et al. (2005). PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol. 46 (7), 1125–1139. doi: 10.1093/pcp/pci125
Tsai, W. C., Pan, Z. J., Hsiao, Y. Y., Jeng, M. F., Wu, T. F., Chen, W. H., et al. (2008). Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol. 49 (5), 814–824. doi: 10.1093/pcp/pcn059
Tuwo, M., Indrianto, A. (2016). Improvement of orchid Vanda hybrid (Vanda limbata Blume X Vanda tricolor Lindl. var. suavis) by colchicines treatment in vitro. Modern Appl. Sci. 10 (11), 83. doi: 10.5539/mas.v10n11p83
Tzeng, T. Y., Chen, H. Y., Yang, C. H. (2002). Ectopic expression of carpel-specific MADS box genes from lily and lisianthus causes similar homeotic conversion of sepal and petal in Arabidopsis. Plant Physiol. 130 (4), 1827–1836. doi: 10.1104/pp.007948
Tzeng, T. Y., Hsiao, C. C., Chi, P. J., Yang, C. H. (2003). Two lily SEPALLATA-like genes cause different effects on floral formation and floral transition in Arabidopsis. Plant Physiol. 133 (3), 1091–1101. doi: 10.1104/pp.103.026997
Tzeng, T. Y., Yang, C. H. (2001). A MADS box gene from lily (Lilium longiflorum) is sufficient to generate dominant negative mutation by interacting with PISTILLATA (PI) in Arabidopsis thaliana. Plant Cell Physiol. 42 (10), 1156–1168. doi: 10.1093/pcp/pce151
Vaz, A. P., Figueiredo-Ribeiro Rd Rde, C., Kerbauy, G. B. (2004). Photoperiod and temperature effects on in vitro growth and flowering of P. pusilla, an epiphytic orchid. Plant Physiol. Biochem. 42 (5), 411–415. doi: 10.1016/j.plaphy.2004.03.008
Villacorta-Martin, C., Nunez de Caceres Gonzalez, F. F., de Haan, J., Huijben, K., Passarinho, P., Lugassi-Ben Hamo, M., et al. (2015). Whole transcriptome profiling of the vernalization process in Lilium longiflorum (cultivar White Heaven) bulbs. BMC Genomics 16, 550. doi: 10.1186/s12864-015-1675-1
Wang, R., Albani, M. C., Vincent, C., Bergonzi, S., Luan, M., Bai, Y., et al. (2011a). Aa TFL1 confers an age-dependent response to vernalization in perennial Arabis alpina. Plant Cell 23 (4), 1307–1321. doi: 10.1105/tpc.111.083451
Wang, S. Y., Lee, P. F., Lee, Y. I., Hsiao, Y. Y., Chen, Y. Y., Pan, Z. J., et al. (2011b). Duplicated C-class MADS-box genes reveal distinct roles in gynostemium development in Cymbidium ensifolium (Orchidaceae). Plant Cell Physiol. 52 (3), 563–577. doi: 10.1093/pcp/pcr015
Wang, W.-Y., Chen, W.-S., Chen, W.-H., Huang, L.-S., Chang, P.-S. (2002). Influence of abscisic acid on flowering in Phalaenopsis hybrida. Plant Physiol. Biochem. 40, 97–100. doi: 10.1016/S0981-9428(01)01339-0
Wang, W.-Y., Chen, W.-S., Huang, K.-L., Huang, L.-S., Chen, W.-H., Su, W.-R. (2003). The effects of daylength on protein synthesis and flowering in Doritis pulcherrima. Sci. Hortic. 97, 49–56. doi: 10.1016/S0304-4238(02)00128-0
Wen, Z., Guo, W., Li, J., Lin, H., He, C., Liu, Y., et al. (2017). Comparative transcriptomic analysis of vernalization- and cytokinin-induced floral transition in Dendrobium nobile. Sci. Rep. 7, 45748. doi: 10.1038/srep45748
Xiang, L., Chen, Y., Chen, L., Fu, X., Zhao, K., Zhang, J., et al. (2018). B and E MADS-box genes determine the perianth formation in Cymbidium goeringii Rchb.f. Physiol. Plant 162 (3), 353–369. doi: 10.1111/ppl.12647
Xu, Y., Teo, L. L., Zhou, J., Kumar, P. P., Yu, H. (2006). Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J. 46 (1), 54–68. doi: 10.1111/j.1365-313X.2006.02669.x
Xu, Y., Yu, H., Kumar, P. P. (2010). Characterization of floral organ identity genes of the orchid Dendrobium crumenatum. AsPac J. Mol. Biol. Biotechnol. 18, 185–187.
Yang, F. X., Zhu, G. F., Wang, Z., Liu, H. L., Huang, D. (2015). A putative miR172-targeted CeAPETALA2-like gene is involved in floral patterning regulation of the orchid Cymbidium ensifolium. Genet. Mol. Res. 14 (4), 12049–12061. doi: 10.4238/2015.October.5.18
Yin, K., Gao, C., Qiu, J. L. (2017). Progress and prospects in plant genome editing. Nat. Plants 3, 17107. doi: 10.1038/nplants.2017.107
Yoshida, H., Nagato, Y. (2011). Flower development in rice. J. Exp. Bot. 62.14, 4719–4730. doi: 10.1093/jxb/err272
Yu, H., Goh, C. J. (2000a). Differential gene expression during floral transition in an orchid hybrid Dendrobium Madame Thong-In. Plant Cell Rep. 19, 926–931. doi: 10.1007/s002990000227
Yu, H., Goh, C. J. (2000b). Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiol. 123 (4), 1325–1336. doi: 10.1104/pp.123.4.1325
Yu, H., Yang, S. H., Goh, C. J. (2000). DOH1, a class 1 knox gene, is required for maintenance of the basic plant architecture and floral transition in orchid. Plant Cell 12 (11), 2143–2160. doi: 10.1105/tpc.12.11.2143
Yu, H., Yang, S. H., Goh, C. J. (2001). Agrobacterium -mediated transformation of a Dendrobium orchid with the class 1 knox gene DOH1. Plant Cell Rep. 20 (4), 301–305. doi: 10.1007/s002990100334
Zhang, G. Q., Liu, K. W., Li, Z., Lohaus, R., Hsiao, Y. Y., Niu, S. C., et al. (2017). The Apostasia genome and the evolution of orchids. Nature 549 (7672), 379–383. doi: 10.1038/nature23897
Zhang, J.-X., Wu, K.-L. W., Tian, L.-N., Zeng, S.-J., Duan, J. D. (2011). Cloning and characterization of a novel CONSTANS-like gene from Phalaenopsis hybrida. Acta Physiol. Plant 33, 409–417. doi: 10.1007/s11738-010-0560-4
Zhang, J., Li, Z., Xu, L. (2013). Cloning and expression analysis of B type MADS-box genes involving in floral development from Rhynchostylis gigantean. Mol. Plant Breed. 11, 570–574. doi: 10.3969/mpb.011.000570
Keywords: floral transition, flower development, molecular genetics, orchids, orchid biotechnology, transgenic orchids
Citation: Wang S-L, Viswanath KK, Tong C-G, An HR, Jang S and Chen F-C (2019) Floral Induction and Flower Development of Orchids. Front. Plant Sci. 10:1258. doi: 10.3389/fpls.2019.01258
Received: 09 May 2019; Accepted: 10 September 2019;
Published: 10 October 2019.
Edited by:
Jen-Tsung Chen, National University of Kaohsiung, TaiwanReviewed by:
Lee Jeong Hwan, Chonbuk National University, South KoreaCopyright © 2019 Wang, Viswanath, Tong, An, Jang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seonghoe Jang, c2Vvbmdob2UuamFuZ0B3b3JsZHZlZy5vcmc=; Fure-Chyi Chen, ZmNoZW5AbWFpbC5ucHVzdC5lZHUudHc=
†ORCID: Seonghoe Jang, orcid.org/0000-0001-5018-3480
Fure-Chyi Chen, orcid.org/0000-0001-5475-9248
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.