
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 26 September 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01172
This article is part of the Research TopicProceedings of iMMM 2019 – International Molecular Mycorrhiza MeetingView all 14 articles
Phosphorus (P), zinc (Zn), and iron (Fe) are three essential elements for plant survival, and severe deficiencies in these nutrients lead to growth retardation and crop yield reduction. This review synthesizes recent progress on how plants coordinate the acquisition and signaling of Pi, Zn, and Fe from surrounding environments and which genes are involved in these Pi–Zn–Fe interactions with the aim of better understanding of the cross-talk between these macronutrient and micronutrient homeostasis in plants. In addition, identification of genes important for interactions between Pi, Zn, and/or Fe transport and signaling is a useful target for breeders for improvement in plant nutrient acquisition. Furthermore, to understand these processes in arbuscular mycorrhizal plants, the preliminary examination of interactions between Pi, Zn, and Fe homeostasis in some relevant crop species has been performed at the physiological level and is summarized in this article. In conclusion, the development of integrative study of cross-talks between Pi, Zn, and Fe signaling pathway in mycorrhizal plants will be essential for sustainable agriculture all around the world.
Inorganic phosphate (Pi), zinc (Zn), and iron (Fe) are three essential macronutrient and micronutrients for the survival and development of all living organisms including mycorrhizal plants and edible crops (Westheimer, 1987; Briat et al., 1995; Marschner, 1995; Salgueiro et al., 2000). These three mineral elements are relatively inaccessible to plants and crops because of their low solubility and relative immobilization in the agricultural soils (Lopez et al., 2000; Hirsch et al., 2006). Crops are therefore subjected to Pi, Zn, and Fe deficiencies, which can adversely impact multiple metabolic processes in cells. Nevertheless, plants have evolved a number of strategies to cope with low Pi, Zn, and Fe availabilities, including development of a mycorrhizal symbiosis (Karandashov and Bucher, 2005; Smith and Read, 2008), conversion of metabolism, remodeling of root morphology, secretion of root exudates, and induction of the high-affinity transport systems.
In recent decades, the effects of Pi, Zn, and Fe deficiencies on crop yield and quality have become a global concern due to the issues of food availability and malnutrition (Abelson, 1999; Neset and Cordell, 2012; Shahzad et al., 2014). To guarantee the sustainable food source for the growing population, worldwide agriculture has become dependent on the massive use of Pi, Zn, and Fe fertilizers for improving crop yield and quality. Nevertheless, this strategy has adverse long-term economic and ecological impacts. Development of sustainable agricultural practices will require crops with improved Pi, Zn, and Fe nutrition in order to reduce the application of these fertilizers. The novel plant genotypes with high-efficiency nutrient use are genetically desired in an appropriate way to fit the lower input into the environment. Despite the importance of these issues, the biological interactions between P, Zn, and Fe elements still remain incompletely studied, and our understanding is limited of how various signaling pathways are induced in response to nutrient availability and how these changes are integrated with relation to other nutrients (Briat et al., 2015). On the other hand, some key genes involved in the acquisition and distribution of macronutrient and micronutrients in nonmycorrhizal and mycorrhizal plants have been identified (Javot et al., 2007a; Gojon et al., 2009; Pilon et al., 2009; Giovannetti et al., 2014; Watts-Williams and Cavagnaro, 2018), and their expression in response to nutrient status has started to be elucidated (Schachtman and Shin, 2007; Giehl et al., 2009; Liu et al., 2009; Hindt and Guerinot, 2012).
Approximately 72% of terrestrial vascular plant species are capable of establishing symbiotic mutualistic associations with obligate biotrophic soil-borne arbuscular mycorrhizal fungi (AMF) from the phylum Glomeromycota (Remy et al., 1994; Schüßler et al., 2001; Bonfante, 2018). The endosymbiotic associations between plants and AMF, namely, arbuscular mycorrhizas (AM), are widespread in terrestrial ecosystems (Parniske, 2008). In AM symbiosis, the fungal symbiont provides mineral nutrients to the plant and in return obtains sugars and lipids (Smith and Read, 2008; Jiang et al., 2017), and thus, this symbiosis has significant contribution to plant productivity and ecosystem function (van der Heijden et al., 1998).
AM symbiosis not only is capable of significantly improving the acquisition of macronutrients such as Pi, N, and S to host plant (Ames et al., 1983; Smith et al., 2003; Smith and Read, 2008; Allen and Shachar, 2009; Leigh et al., 2009; Sieh et al., 2013) but also facilitates the uptake and translocation of micronutrients such as Zn and Fe in the soil–AMF–plant continuum (Caris et al., 1998; Liu et al., 2000; Chen et al., 2003; Farzaneh et al., 2011; González et al., 2016). The acquisition of Pi, N, and S in AM symbiosis through a specific symbiotic uptake pathway has been extensively described (Rausch et al., 2001; Harrison et al., 2002; Javot et al., 2007b; Chen et al., 2007; Guether et al., 2009; Yang et al., 2012; Giovannetti et al., 2014; Volpe et al., 2016). However, very few studies have been undertaken to uncover the molecular mechanisms underlying the uptake and homeostasis of Zn and Fe from AM fungus Rhizophagus irregularis to the plant (González et al., 2005; Tamayo et al., 2014; Tamayo et al., 2018), and the impact of this symbiosis on Zn and Fe homeostasis in plant is far from being understood (Chorianopoulou et al., 2015; Watts-Williams et al., 2015). A very recent study has revealed the involvement of AM-modified ZmNAS1, ZmNAS3, and ZmYS1 genes in the regulation of Fe homeostasis in mycorrhizal maize through sulfate deficiency signaling (Chorianopoulou et al., 2015), suggesting the existence of a cross-talk between S and Fe homeostasis in mycorrhizal symbiosis. Nevertheless, the molecular basis of the double or tripartite interactions between Pi, Zn, and Fe homeostasis in AM symbiosis is still lacking in mycorrhizal plants. Therefore, it is of biological significance to decipher the mechanisms of coordinating the Pi, Zn, and Fe deficiency signaling in AM symbiosis and consequently profit mycorrhizal plant growth and fitness during multiple Pi–Zn–Fe deficiency stresses.
In such context, the aim of this review is to summarize current knowledge on cross-talk between Pi and Zn, Pi and Fe, Zn and Fe, and tripartite Pi–Zn–Fe homeostasis in both nonmycorrhizal and mycorrhizal plants. Additionally, Pi (or Fe) nutrition is also affected by the interaction between Zn and Fe (or Pi) in plants, such as Arabidopsis and rice. The MYB transcription factor (TF) PHR1 acting as a potential integrator of Pi, Zn, and Fe nutrient signals to regulate mineral nutrition in plants is discussed. Moreover, a novel role of the OsPHO1;1 in Fe transport through integrating Pi and Zn deficiency signaling is proposed, and these complicated nutritional interactions are presented, with a focus on the emerging roles of nutrient transporters in mycorrhizal plants.
In plants, Pi, Zn, and Fe are acquired at the root periphery in the form of free ions (Guerinot, 2000; Curie et al., 2001; Vert et al., 2002; Nussaume et al., 2011; Milner et al., 2013), and the uptake and translocation of these minerals in plants involve multiple and complex transport systems.
The Pi is taken up at the root system via the high-affinity Pi:H+ symporters belonging to members of the PHT1 family (Schachtman et al., 1998; Bucher, 2007; Nussaume et al., 2011). The Arabidopsis, rice, soybean, and tomato genomes harbor 9, 13, 14, and 8 members of the PHT1 family, respectively (Paszkowski et al., 2002; Poirier and Bucher, 2002; Fan et al., 2013; Chen et al., 2014). Some of these Pht1 genes are predominantly expressed in roots, and the encoded proteins function as high-affinity Pi uptake transporters (Muchhal et al., 1996; Shin et al., 2004; Remy et al., 2012; Sun et al., 2012). Nevertheless, the transcripts of Pht1 genes are also detected in shoots (including vegetative and reproductive tissues), implicating their role beyond Pi uptake at the root surface (Mudge et al., 2002; Nagarajan et al., 2011; Chen et al., 2014). In Arabidopsis, five out of nine Pht1 members have been functionally characterized by genetic approaches. Earlier work reported that both AtPT1 and AtPT4 transporters contributed to Pi uptake in Arabidopsis thaliana under both low and high Pi levels (Shin et al., 2004). However, the double mutant pht1;1Δpht1;4Δ showed a more pronounced reduction in Pi acquisition relative to wild-type from both low and high Pi environments, suggesting redundant functions of these two Pi transporters (Shin et al., 2004). Nevertheless, Nagarajan et al. (2011) showed that the AtPT5 could mobilize Pi between the source and sink organs for Pi homeostasis in A. thaliana. Recently, it was demonstrated that AtPT8 and AtPT9 transporters act in a redundant manner during Pi uptake in Arabidopsis seedlings during Pi starvation (Remy et al., 2012). These results indicated the compensatory effects of root Pi uptake and shoot Pi accumulation between the four Arabidopsis Pi transporters AtPT1, AtPT4, AtPT8, and AtPT9 during Pi deficiency. In rice (Oryza sativa), a total of 13 members of the PHT1 family have been identified (Goff et al., 2002), and 10 out of 13 genes had been well characterized in O. sativa by reverse genetics. The constitutively expressed OsPT1 mediates Pi translocation in shoots and also induces root hair growth in rice during Pi-repletion (Sun et al., 2012). Ai et al. (2009) demonstrated that OsPT2 was transcriptionally induced in roots under Pi deficiency and functioned in Pi translocation in rice, while OsPT3 mediated Pi uptake, translocation, and remobilization in rice under extremely low Pi regimes (Chang et al., 2019). OsPT4 not only facilitated Pi mobilization but also played a pivotal role in embryo development (Zhang F et al., 2015), whereas OsPT6 displayed a broad role in Pi acquisition and translocation throughout the plant (Ai et al., 2009). It was observed that the high-affinity Pi transporter gene OsPT8 was involved in Pi homeostasis in rice (Jia et al., 2011). However, Wang et al. (2014b) found that OsPT9 and OsPT10 redundantly functioned in Pi uptake under both low and high Pi conditions. OsPT11 and OsPT13 were exclusively induced in arbusculated cells and non-redundantly regulated the arbuscular mycorrhizal symbiosis in rice (Paszkowski et al., 2002; Yang et al., 2012). The current understanding of the Pi transport activities of Pht1 transporters and their complex regulation in plants has been well documented and intensively summarized in multiple reviews in recent years (Poirier and Bucher, 2002; Bayle et al., 2011; Lin et al., 2013; Chen et al., 2015; Poirier and Jung, 2015; Gu et al., 2016).
For Zn2+ acquisition in roots, transmembrane transporters belonging to the ZIP (ZRT and IRT-like protein) family are considered to be the primary Zn2+ uptake transporters, which have been identified in both dicotyledons and monocotyledons (Eng et al., 1998; Maser et al., 2001; López et al., 2004; Palmer and Guerinot, 2009; Lee et al., 2010; Tiong et al., 2014). Some ZIP family transporters preferentially localize to the plasma membrane of root epidermal cells and deletion, or overexpression of these genes results in plants that accumulate less or more Zn2+ than do wild-type plants, respectively. This is indicative of their roles in Zn2+ acquisition at root–soil interface (Lee et al., 2010; Milner et al., 2012). In Arabidopsis, the plasma membrane-localized AtIRT1, belonging to the ZIP gene family, is involved in Zn2+ uptake at root epidermal cells (Henriques et al., 2002; Vert et al., 2002; Barberon et al., 2011). The well-characterized ZIP gene IRT3 is transcriptionally induced in response to Zn2+ deficiency and confers increased shoot Zn2+ accumulation when overexpressed in Arabidopsis (van de Mortel et al., 2006; Sinclair and Krämer, 2012). Moreover, AtIRT3 is localized to the plasma membrane where it transports Zn2+ across the plasma membrane into the cell (Lin et al., 2009). In rice, the node-localized transporter, OsZIP3, is responsible for unloading Zn2+ from the xylem as well as Zn2+ distribution to the developing tissues (Sasaki et al., 2015), whereas the OsZIP4 located in the phloem cells acts as a Zn2+ transporter that may be responsible for Zn2+ translocation within plant (Ishimaru et al., 2005). Other Arabidopsis and rice ZIP family members involved in Zn2+ uptake and homeostasis are barely known, and therefore, further works need to determine their precise roles in plants.
Iron (Fe) from the soils enters the root cells through two distinct strategies (Figure 1), according to non-Graminacea plants (strategy I) and Graminacea plants (strategy II). In strategy I plants, the ferric iron (Fe3+) is reduced in the ferrous iron (Fe2+) prior to uptake into the root epidermal cells (Morrissey and Guerinot, 2009; Conte and Walker, 2011). For example, in Arabidopsis, under Fe deficiency, the FIT and bHLH TFs, bHLH38 and bHLH39, are activated in roots and bind to the promoters of the iron-responsive genes. Subsequently, the induction of ferric reductase oxidase 2 (FRO2) and IRT1 activity is co-regulated in response to iron deficiency through the reduction-based strategy I for iron uptake (Figure 1A), while iron in rhizosphere is firstly solubilized by the activated H+-ATPase AHA2 and is then reduced from ferric (Fe3+) to ferrous (Fe2+) iron by the reductase FRO2 (Ivanov et al., 2012). Fe2+ is then imported into the root cell by the metal transporter IRT1 (Vert et al., 2002).
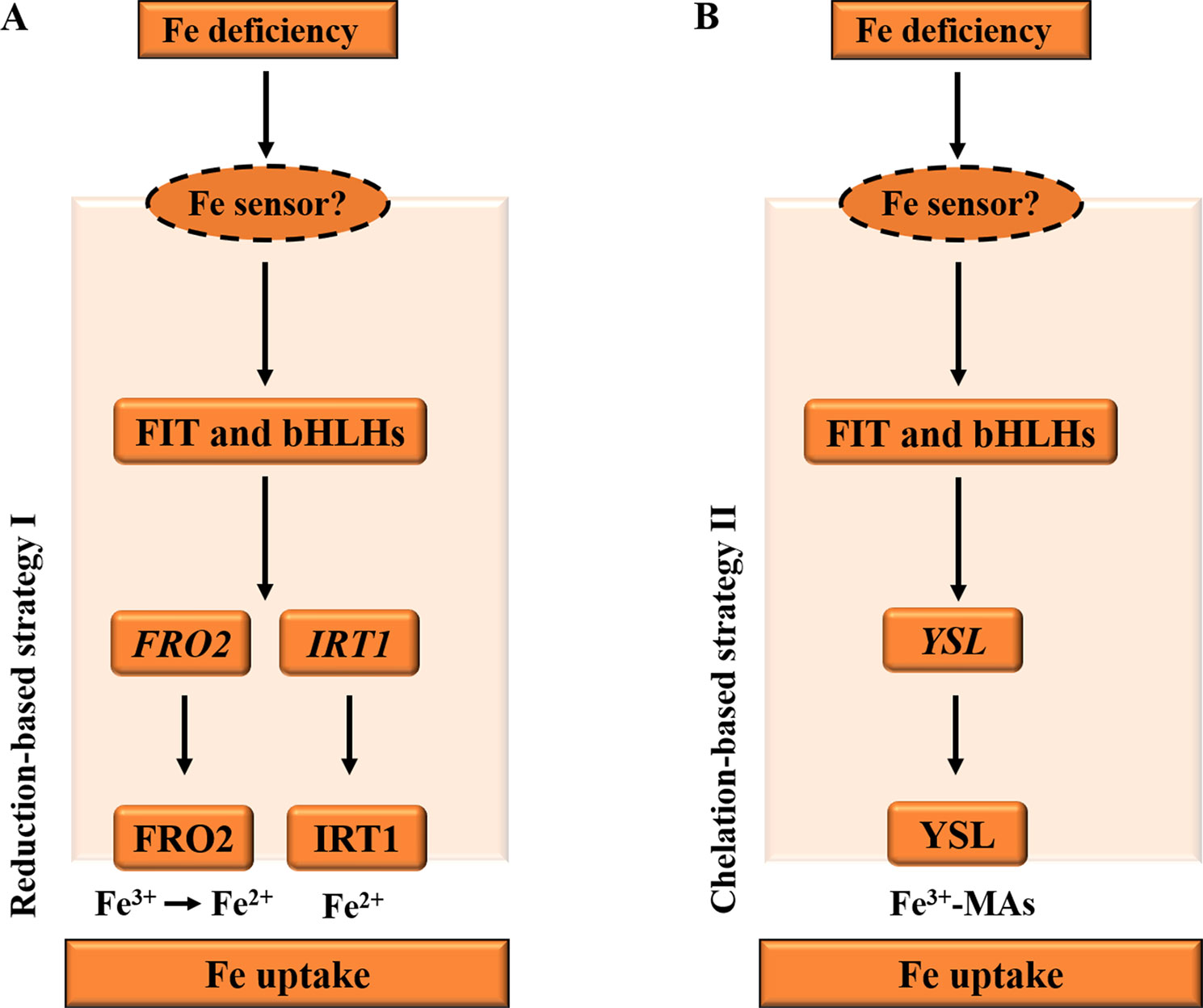
Figure 1 Diagrams illustrating the iron deficiency response in Arabidopsis and Graminacea plants. (A) Under Fe deficiency, in Arabidopsis roots, the FIT and bHLH transcription factors, such as bHLH38 and bHLH39 in Arabidopsis, are activated by an unknown PM iron sensor in order to bind to the promoters of the iron-responsive genes. Subsequently, the induction of FRO2 and IRT1 activity is co-regulated in response to iron deficiency through the reduction-based strategy I for iron uptake. Iron is firstly reduced from ferric (Fe3+) to ferrous (Fe2+) iron by the reductase FRO2. Fe2+ is then imported into the root cell by the metal transporter IRT1. (B) Under Fe deficiency, the FIT and bHLH transcription factors are activated to induce the strategy II Fe uptake system in Graminacea plant roots. Fe3+–MAs are transported into the root cell by the YSL. PM, plasma membrane; YSL, yellow stripe like; IRT1, iron-regulated transporter 1. The arrows refer to the positive interactions, while the question marks indicate the unknown iron sensor.
In strategy II plants, the ferric iron (Fe3+) is first chelated by interaction with mugineic acids (MAs) (Figure 1B), and then these Fe3+–MA complexes are taken up into root cells by plasma membrane-localized transporter proteins (Curie et al., 2001). For example, in maize, under iron deficiency, MAs are synthesized in root cells by nicotianamine synthase (NAS), NA aminotransferase (NAAT), and deoxymugineic acid synthase (DMAS); and MAs are secreted into rhizosphere by transporter of MA family phytosiderophores1 (TOM1) (Li et al., 2018). Then, Fe3+–MAs are transported into root cells by the transmembrane transporter yellow stripe 1-like (YSL) (Curie et al., 2001).
After their uptake at the root surface, these minerals can be transported to the vacuoles. Alternatively, Pi, Zn2+, and iron can undergo symplastic journey towards the root xylem for movement upward to the aerial tissues. For Pi, phosphate exporters PHO1 and PHO1;H1 have been identified as important components in the long-distance transfer of Pi from roots to shoots (Poirier et al., 1991; Hamburger et al., 2002; Stefanovic et al., 2011; Kisko et al., 2015).
For Zn2+, two plasma membrane transporters AtHMA2 and AtHMA4 belonging to P1B-ATPase subfamily played key roles in Zn loading into the xylem and root-to-shoot translocation of Zn2+ in Arabidopsis (Hussain et al., 2004; Verret et al., 2004; Hanikenne et al., 2008; Wong et al., 2009). NA had been proposed to form stable complexes with Zn and to play an important role in Zn2+ movement in the xylem and phloem (Stephan and Scholz, 1993). NAS genes were induced under Zn2+ deficiency (Wintz et al., 2003) and were functionally involved in the intercellular movement and long-distance transport of Zn2+ in A. thaliana (Takahashi et al., 2003). Overexpression of AhNAS2 gene in roots contributed to Zn2+ hyperaccumulation of Arabidopsis halleri (Deinlein et al., 2012). Interestingly, the constitutive expression of NAS genes from other plant species caused an increase in Zn2+ translocation and accumulation in polished rice grains (Masuda et al., 2009; Lee et al., 2011), illustrating the significant importance of the NAS proteins in the Zn2+ translocation in plants. The major facilitator superfamily (MFS) transporter (Pao et al., 1998), zinc-induced facilitator 1 (ZIF1), was shown to contribute to Zn2+ tolerance in Arabidopsis (Haydon and Cobbett, 2007), and tonoplast-localized ZIF1 proteins have been implicated in vacuolar Zn2+ sequestration (Arrivault et al., 2006; Kawachi et al., 2009). Under Zn2+ deficiency, the vacuole-stored Zn2+ was remobilized (Lanquar et al., 2004) to the cytosol. Natural resistance-associated macrophage protein (NRAMP) family members played roles in heavy metal transport in plants (Belouchi et al., 1995; Thomine et al., 2000). Arabidopsis NRAMP4 localized to the vacuolar membrane and associated with Zn2+ remobilization (Lanquar et al., 2004). Whether other members of the NRAMP family could contribute to Zn2+ remobilization in plants remains unknown.
For Fe, many transporters and soluble proteins responsible for Fe long-distance transfer and distribution have been characterized in recent years (Morrissey and Guerinot, 2009; Kobayashi and Nishizawa, 2012). In Arabidopsis, the AtFRD3 protein, which is a member of the multidrug and toxic compound extrusion (MATE) family, functions during efflux of citrate into xylem and is responsible for Fe long-distance transport from root xylem to shoots (Green and Rogers, 2004; Durrett et al., 2007; Magalhaes et al., 2007), whereas the rice OsFRDL1, the ortholog of FRD3, maintains the Fe3+ levels in the xylem sap (Yokosho et al., 2009). YSL transporters play a significant role in the transportation and distribution of Fe through the phloem (Curie et al., 2009) and are also involved in loading Fe from old leaves to flowers and developing seeds (Kobayashi and Nishizawa, 2012). Moreover, in rice, OsYSL2 and OsYSL15 may coordinate the long-distance Fe transport from root to shoot to seed (Koike et al., 2004; Inoue et al., 2009). In addition to YSLs, the iron transport protein ITP, which is an Fe-binding dehydrin in the phloem sap, helps promote Fe3+ mobility within the phloem of Ricinus communis (Kruger et al., 2002). Plant NAS genes are also required for long-distance Fe transport. For instance, in Arabidopsis, AtNAS2 and AtNAS4 may be involved in Fe translocation from roots to shoots (Klatte et al., 2009). Interestingly, the rice OsIRT1 are highly expressed in the companion cells of phloem under Fe deficiency (Ishimaru et al., 2006), and it is possible that the corresponding encoding protein OsIRT1 could transport Fe2+ into the phloem prior to being chelated by NA. More recently, the involvement of OsPHO1;1 in the Fe loading into the root xylem has been reported, where it may affect overaccumulation of Fe in roots of the Ospho1;1 mutant under Pi and Zn deficiency (Saenchai et al., 2016).
Considerable advances have been made in studying the molecular mechanisms underlying Pi, Zn, and Fe sensing and signaling in plants in recent decades (Abel et al., 2002; Chiou and Lin, 2011; Assuncão et al., 2013; Kobayashi and Nishizawa, 2014; Zhang et al., 2014). Nevertheless, the cross-talks between these signaling pathways integrating the tripartite interaction among Pi, Zn, and Fe homeostasis remains poorly understood (Briat et al., 2015; Saenchai et al., 2016). How Pi homeostasis is regulated in plants has already been documented in numerous studies, and plant Pi sensing seems to be conserved in flowering plants, with several signaling networks having been proposed (Rouached et al., 2010; Wang et al., 2014a). The defined mechanism is the systemic Pi signaling cascade, which contains the MYB TF PHR1, the miRNA399, and the ubiquitin E2 conjugase PHO2 components (Bari et al., 2006; Pant et al., 2008). Generally, the well-characterized PHR1–PHO2–miRNA399 signaling pathway controls the expression of most Pi starvation-induced (PSI) genes in plants (Figure 2). In response to low Pi, miRNA399 is transcriptionally induced by PHR1 activity (Figure 2A) and then mediates shoot-to-root Pi signaling via the phloem, where it targets the mRNA of PHO2 (Lin et al., 2008; Pant et al., 2008). The inhibition of PHO2 leads to an increase in the PHR1-dependent expression of root Pi transporters that include the members of PHT1 and PHO1, and hence an increase in Pi acquisition in roots and Pi translocation to shoots (Bari et al., 2006; Lin et al., 2008). Under high Pi conditions, miRNA399 is down-regulated due to the inhibition of PHR1 activity (Figure 2B), and the PHO2–miR399 pathway in roots is dysfunctional through target mimicry between miR399 and PHR1-dependent IPS1 (Franco et al., 2007). Target genes of PHR1 are also reduced at transcriptional level, and PHO2 protein is activated to facilitate the degradation of Pi transporters.
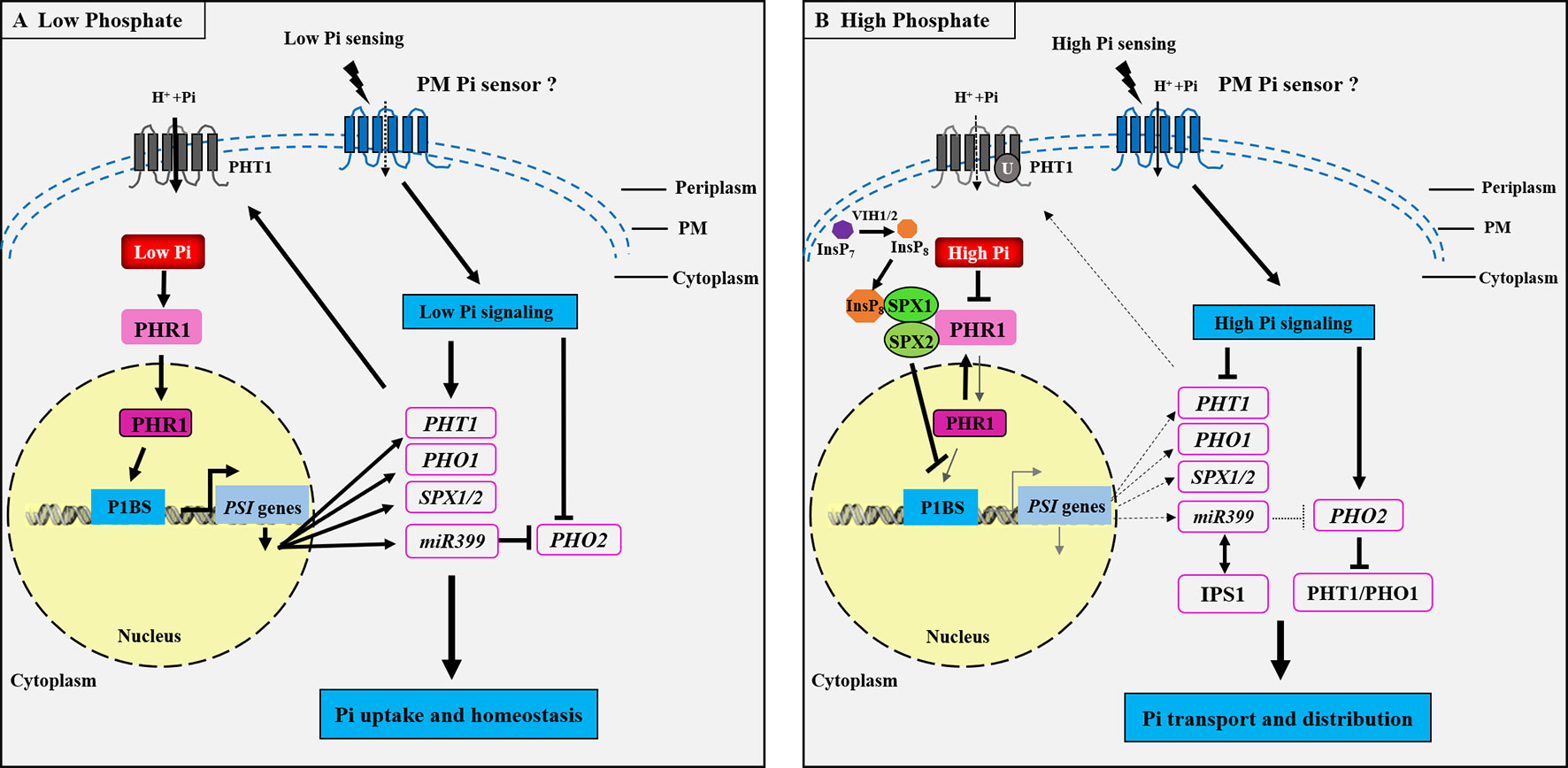
Figure 2 Schematic representation of the phosphate (Pi) signaling pathway essential for plant adaptation to low Pi concentration. Under Pi deficiency (A), a set of phosphate starvation-induced (PSI) genes are transcriptionally activated through binding of the transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1) to the cis-element (P1BS) present in the promoter region of the PSI genes, and subsequently PHT1 and PHO1 mRNAs are induced to be necessary for Pi uptake and translocation in roots. The SPX1/2 and miR399 genes are also activated by PHR1 during Pi starvation. miR399 inhibits the ubiquitin E2 conjugase PHO2 in order to maintain the PHT1 protein activity at the PM. It could be proposed that the Pi signaling is activated for sensing external low Pi through an unknown PM Pi sensor, which induces the low Pi responsive genes PHT1, PHO1, SPX1/2, and miR399, whereas the PHO2 is repressed, thus activating the Pi regulatory pathway to modulate Pi uptake and homeostasis. Under high Pi concentration (B), the Pi signaling pathway is repressed, the diphosphoinositol pentakisphosphate kinases VIH1 and VIH2 function redundantly to synthesize InsP8, and InsP8 can directly bind to the SPX domain of SPX1 and is essential for the interaction between SPX1/2 and PHR1. This interaction leads to the inhibition of PHR1 binding to the cis-element P1BS present in the promoter region of the PSI genes. Thus, the PSI genes, including PHT1, PHO1, SPX1/2, and miR399, are transcriptionally repressed, while the PHO2 is activated to be responsible of the ubiquitination of PHT1 and PHO1 proteins to promote Pi transporters degradation. IPS1 encodes a non-coding RNA and enables post-transcriptional regulation under high Pi through RNA mimicry. IPS1-miR399 matching thus results in the inhibition of the miR399 activity to target PHO2. It is also predicted that there may exist an unknown PM Pi sensor responsible for high Pi sensing. PM, plasma membrane. The arrows and flat-ended lines refer to the positive and negative interactions, respectively. The dotted arrows represent the repression processes.
Recently, the SPX domain-containing proteins have been proposed to function as the intracellular Pi sensors for sensing cellular Pi levels and controlling Pi homeostasis in both monocotyledonous and dicotyledonous plants. In Arabidopsis, the PHR1-dependent AtSPX1 gene is transcriptionally induced under Pi deficiency (Figure 2A), while the AtSPX1 protein can interact with the AtPHR1 at the protein level under Pi sufficiency, inhibiting AtPHR1 binding to P1BS cis-element (GNATATNC) (Puga et al., 2014). Similarly, in rice, OsSPX1 and OsSPX2 inhibit Pi deficiency response through interaction with OsPHR2 in a Pi-dependent manner (Wang et al., 2014a), involvement of SPX proteins in the Pi sensing, and signaling mechanisms in plants (Figure 2B). Very recently, it has demonstrated that both the diphosphoinositol pentakisphosphate kinases (PPIP5K) VIH1 and VIH2 function redundantly to synthesize the inositol pyrophosphate (InsP8) (see Figure 2B), and InsP8 can directly bind to the intracellular Pi sensor SPX1 to control Pi homeostasis in Arabidopsis during Pi repletion (Dong et al., 2019). This study revealed that InsP8 acts as an intracellular phosphate signal in plants. The next major challenge in this field is to unmask the extracellular Pi sensor sensing.
In plants, how Zn-deficient signal is sensed, relayed, and integrated into a signal response remains elusive. Nevertheless, a first working model of Zn deficiency signaling has been proposed by Assuncão et al. (2013). Two bZIP TFs, bZIP19/23, have been identified in Zn homeostasis via the regulation of target genes, including the members of ZIP family (Guerinot, 2000; Assuncão et al., 2010a) for root Zn transport and NAS2/4 for NA synthesis (Assuncão et al., 2010b), since the shoots appear to the first organ to sense the Zn deficiency and then transmit the signal to the roots where these ZIP transporters are activated (Assuncão et al., 2013). This observation has led to the proposal of the existence of unknown long-distance Zn deficiency signaling molecules. Additionally, a ZDRE element (RTGTCGACAY) is present in the promoter regions of both the ZIP and NAS genes in response to Zn deficiency (Assuncão et al., 2010; Assuncão et al., 2010b).
How the Fe status of plant is sensed and how this signal is transmitted to the transcriptional networks for Fe acquisition and response are currently areas of great interest in the field of Fe homeostasis in plants (Briat et al., 2015). A major goal is to find a master Fe sensor controlling Fe homeostasis in plants (Hindt and Guerinot, 2012). Some degree of progress towards these aims has been achieved by exploiting members of the basic helix-loop-helix (bHLH) TF family (Hindt and Guerinot, 2012; Ivanov et al., 2012; Kobayashi and Nishizawa, 2012; Moran et al., 2014). Also, hemerythrin motif-containing RING and zinc-finger proteins HRZ1/2 and its ortholog E3 ligase BTS that have been recently characterized in both the monocotyledonous and dicotyledonous plants, respectively (Kobayashi and Nishizawa, 2014). The tomato FIT is referred as to FER (Ling et al., 2002; Brumbarova and Bauer, 2005), and FER mutant fit-1 repressed about 50% of Fe deficiency-induced genes in roots (Colangelo and Guerinot, 2004). A second PYE bHLH protein is exclusively induced in roots under Fe deficiency. The pye-1 mutant line is sensitive to low Fe. In pye-1 mutant, three Fe transport-related genes, NAS4, FRO3, and ZIF1, are strongly induced under Fe deficiency and are identified as the targets of PYE (Long et al., 2010). In addition, Arabidopsis bHLH104 and ILR3 play crucial roles in the regulation of Fe deficiency responses through targeting other bHLH genes and PYE expression (Zhang et al., 2015). The overexpression lines of rice iron-related TF 2 (OsIRO2), ortholog of Arabidopsis bHLH38/39, showed both enhanced Fe uptake and transportation to seeds (Ogo et al., 2007; Ogo et al., 2011). Furthermore, a PYE homologous protein OsIRO3 is induced under Fe deficiency, whereas it is a negative regulator of Fe deficiency responses due to the hypersensitivity to Fe deficiency and the inhibition of genes up-regulated by Fe deficiency (Zheng et al., 2010). Recently, rice bHLH133 was identified to play an important role in the regulation of Fe translocation from roots to shoots (Wang et al., 2013). On the other hand, BTS and its orthologs HRZ1/2 could negatively regulate Fe acquisition, accumulation of Fe, and tolerance to Fe deficiency in rice HRZ1/2 mutants (Kobayashi et al., 2013).
Cross-talks between macronutrient and micronutrients in plants have long been recognized, and these interactions are understood to some extent. Hence, we here emphasize the interactions between Pi, Zn, and iron (Fe) homeostasis at the physiological and molecular levels. The interaction between two nutrients homeostasis has been observed in crop species.
The interaction between Pi and Zn homeostasis in plants is relatively well understood. Pi deficiency results in overaccumulation of Zn in shoots, and inversely, Zn deficiency leads to overaccumulation of Pi in the aerial part of plants (Reed, 1946; Cakmak and Marschner, 1986; Huang et al., 2000; Bouain et al., 2014a; Khan et al., 2014; Ova et al., 2015). In addition to the well-known antagonistic effect of Pi and Zn nutrition in plants, there is some evidence of similar physiological interactions between Pi and Fe nutrition (Zheng et al., 2009), and between Zn and Fe nutrition (Haydon et al., 2012) as well. Pi acquisition in both roots and shoots is promoted under Fe deficiency, and conversely, Pi deficiency significantly increases Fe availability within the plants (Misson et al., 2005; Hirsch et al., 2006; Ward et al., 2008; Zheng et al., 2009; Briat et al., 2015). Fe deficiency leads to an accumulation of Zn, while an excess Zn causes physiological Fe deficiency (Haydon et al., 2012; Shanmugam et al., 2012; Briat et al., 2015).
In plants, the intricate cross-talks between the homeostasis of macronutrients and micronutrients have recently become clear (Briat et al., 2015), and evidence of a complex tripartite interaction between Pi, Fe, and Zn nutrients for maintenance of Pi homeostasis in Arabidopsis has been described (Rai et al., 2015). In addition, Saenchai et al. (2016) have also provided evidence that iron transport in rice is regulated by integration of Pi and Zn deficiencies, highlighting the presence of tripartite cross-talk between Pi, Zn, and Fe homeostasis for better plant survival and fitness (Figure 3).
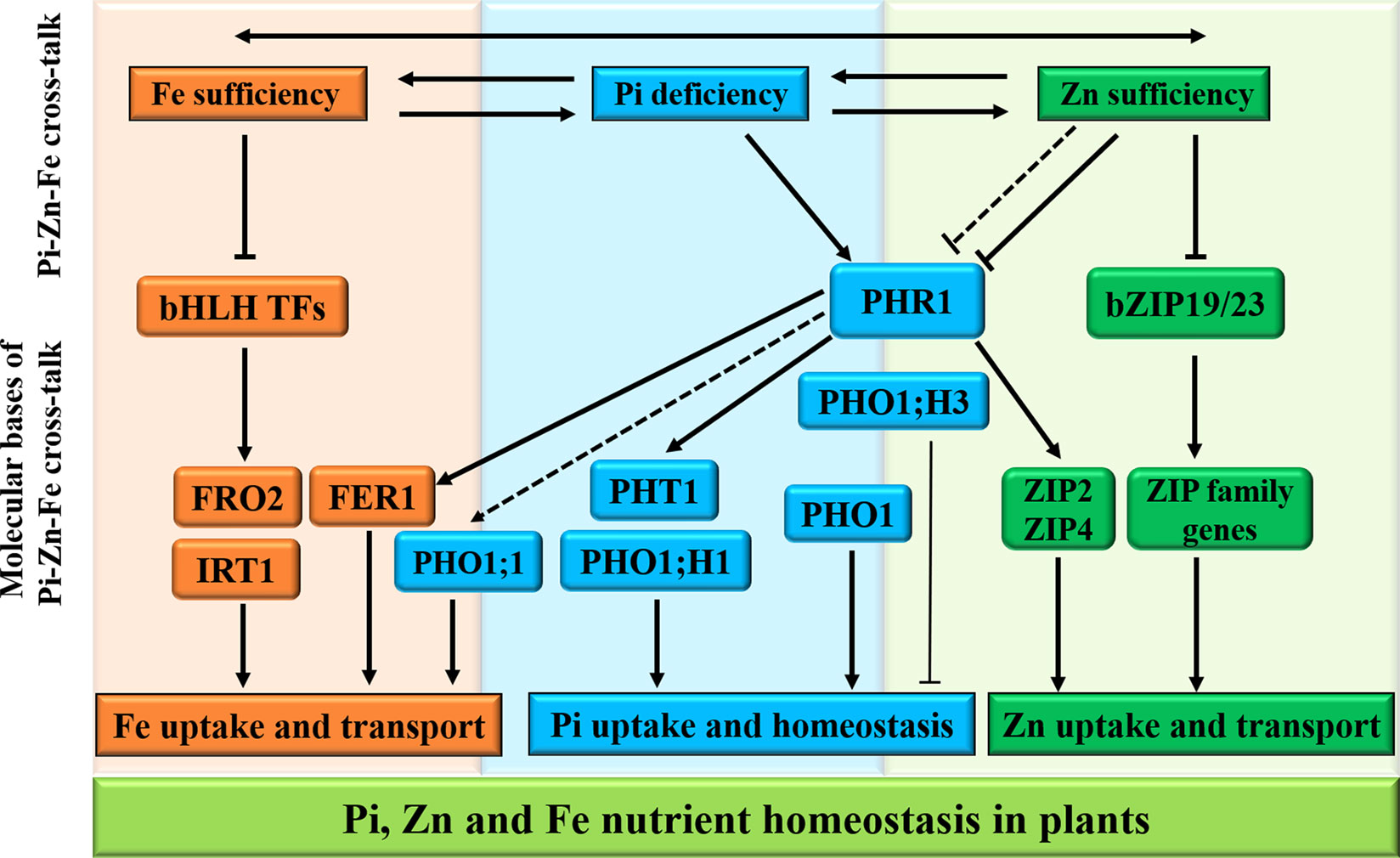
Figure 3 Schematic representation of Pi, Zn, and Fe homeostasis interactions in plants. The cross-talks between phosphate (Pi), zinc (Zn), and iron (Fe) nutrients are shown at the physiological level by two-way arrows. For the molecular bases of the Pi–Zn–Fe cross-talks, the PHR1 acts as a potential integrator of Pi, Zn, and Fe nutrient signals in plants. Firstly, the PHR1 was defined as a key regulator of the expression of Pi transporters PHT1 and PHO1; H1 through the PHR1–miR399–PHO2 pathway (see Figure 2). Secondly, ZIP2 and ZIP4 genes, belonging to plant ZIP gene family, are transcriptionally induced via the activated PHR1 transcription factor binding to the P1BS (GNATATNC) sequences found in the promoter regions of their genes. Under Zn sufficiency, the bZIP19/23 transcription factors are inactivated, and bZIP19/23-mediated Zn regulatory pathways repress the plant ZIP gene family transporters in order to regulate Zn homeostasis. On the other hand, PHO1;H3, which is transcriptionally down-regulated by high Zn supply, and the PHR1 and PHO1 proteins contribute to the Pi–Zn nutrient homeostasis cross-talk. In addition, the transcriptional activation of some genes involved in maintaining Fe homeostasis is also shown to be PHR1-dependent manner, including the FER1 gene encoding the Fe storage protein ferritin, and the PHO1;1 gene encoding Pi transporter. The arrows and flat-ended lines indicate the positive and negative interactions, respectively.
Although the cross-talks between Pi, Zn, and Fe homeostasis have been reported in many plant species (Briat et al., 2015), the molecular basis and biological significance of these nutritional interactions remain thus far largely unknown. It can be first achieved through transcriptomic and genetic analyses of Pi-, Zn-, or Fe-deficient plants (Hammond et al., 2003; Wu et al., 2003; Misson et al., 2005; van de Mortel et al., 2006; Zheng et al., 2009; Bustos et al., 2010; Thibaud et al., 2010; Rouached et al., 2011b; Pineau et al., 2012; Khan et al., 2014; Moran et al., 2014; Rai et al., 2015; Saenchai et al., 2016).
Zn deficiency activates the transcription of numerous Pi-related genes (van de Mortel et al., 2006), while Pi deficiency up-regulates the expression of genes involved in Zn and Fe homeostasis (Misson et al., 2005; Bustos et al., 2010). More recently, several reports have proposed that PHR1, PHO1 and PHO1;H3 are coordinatively involved in the homeostasis between Pi and Zn in Arabidopsis (Bouain et al., 2014b; Khan et al., 2014; Kisko et al., 2015), reinforcing the interaction between Pi and Zn signaling at the molecular level (Figure 3).
In the absence of Pi, plants induce the expression of genes in response to sufficient Fe, whereas Pi-starvation plants reduce the transcripts of genes in response to Fe deficiency (Misson et al., 2005; Müller et al., 2007; Thibaud et al., 2010). Reciprocally, Fe deficiency alters the transcription of Pi-related genes (Zheng et al., 2009; Moran et al., 2014). Genome-wide analysis further reveals 547 and 579 overlapping genes regulated by both Pi and Fe deficiency in rice and Arabidopsis roots, respectively (Zheng et al., 2009; Li and Lan, 2015). In these cases, the expression of FER1 gene encoding Fe storage protein ferritin is in response to Pi starvation mediating by PHR1 (Figure 3) and Fe excess (Petit et al., 2001; Bournier et al., 2013), and NAS3 and YSL8 genes responsible for Fe homeostasis are also induced upon Pi starvation in plants (Bustos et al., 2010). However, the IRT1/2, FRO3/6, and NAS1 genes are repressed in response to Fe deficiency in Pi-deficient plants. Recently, it has reported that the Arabidopsis phr1 × phl1 double mutant influenced Fe distribution and Fe-related gene expression (Bournier et al., 2013; Briat et al., 2015), suggesting that PHR1 and PHL1 may integrate Fe and Pi nutrient signals. The high-affinity copper transport protein COPT2 acts as a key player in the interaction between Pi and Fe deficiency signaling in Arabidopsis (Perea et al., 2013). COPT2 may play a dual role under Fe deficiency. It participates in copper uptake and distribution in Fe-limited roots to minimize iron loss. On the other hand, loss of COPT2 function exacerbates Pi starvation responses in Arabidopsis plants. These findings open new approaches to mitigate iron deficiency in crop species.
For Zn and Fe cross-talk, transcriptomic analysis indicates that many Zn uptake- and homeostasis-related genes are up-regulated in Fe-deficient soybean root and leaf (Moran et al., 2014), including those encoding six members of the ZIP gene family, IRT1, the NAS2, and NRAMP3. Similarly, the Fe deficiency responsive AtIRT1 gene (Figure 3) identified could be a key player in the coordination between Zn- and Fe-deficient signaling in Arabidopsis (Connolly et al., 2002; Vert et al., 2002; Briat et al., 2015). Furthermore, the vacuolar membrane protein encoding genes MTP3, HMA3, and ZIF essential for Zn tolerance are up-regulated in response to Fe deficiency or Zn excess (Becher et al., 2003; Arrivault et al., 2006; van de Mortel et al., 2006; Haydon and Cobbett, 2007; Haydon et al., 2012). A recent study has confirmed that the MATE transporter gene FRD3 is involved in cross-talk between Zn and Fe homeostasis for the tolerance to Zn excess in Arabidopsis (Pineau et al., 2012), highlighting the complexity of cross-talk between these signaling pathways to regulate Fe deficiency and Zn excess.
Several recent reports have started to discuss the complex tripartite cross-talks among Pi, Zn, and Fe (Briat et al., 2015; Rai et al., 2015). Pi nutrition is affected by the interaction between Zn and Fe in plants. The MYB TF PHR1 apparently acts as a common regulator of Pi, Zn, and Fe homeostasis (Figure 3) and functions as a general integrator of multiple nutrition signals (Briat et al., 2015). Firstly, PHR1 was defined as a key regulator for the expression of Pi transporters PHT1 and PHO1;H1 through PHR1–miR399–PHO2 pathway. Secondly, PHR1 seems to be a regulator of the ZIP transporters ZIP2 and ZIP4 for Zn mobilization. In addition, the transcriptional activation of some genes involved in maintaining Fe homeostasis is also shown to be PHR1-dependent manner, including the FER1 gene encoding the Fe storage protein ferritin, and the PHO1;1 gene encoding Pi transporter. Saenchai et al. (2016) have reported the OsPHO1;1 is involved in the coordination between Fe transport and Pi–Zn deficiency signaling in rice. Nevertheless, fundamental aspects regulating the cross-talk between Pi, Zn, and Fe deficiency signaling and the regulation of nutritional homeostasis in plants remain to be discovered.
In the last several decades, the cross-talk between Pi and Zn nutrient homeostasis has been well recognized at the physiological level in many mycorrhizal plants (Reed, 1946; Cakmak and Marschner, 1986; Huang et al., 2000; Watts-Williams et al., 2014; Watts-Williams et al., 2017; Nafady and Elgharably, 2018). High Pi treatment substantially decreased Zn concentration in wheat shoots and grain when these plants were grown in native soils (Ova et al., 2015), and these data also revealed that the negative effect of increasing Pi application on root Zn accumulation and shoot Zn distribution in wheat is dependent on mycorrhization. Furthermore, Zhang et al. (2016) proposed that Pi treatment decreased the Zn concentration in wheat, and they also found that Zn concentration in roots and shoots of maize decreased with increasing Pi supply, and root Zn accumulation exhibits the Pi-induced Zn deficiency during mycorrhization (Zhang et al., 2017), because Pi treatment inhibits colonization resulting in impaired mycorrhizal uptake pathway and then affects the Zn uptake and tissue Zn status of host plants (Loneragan and Webb, 1993; Marschner, 2012; Watts-Williams et al., 2013). The negative relationship between Pi application and the grain Zn status was also confirmed in field studies (Ryan et al., 2008; Zhang et al., 2012). Conversely, under AM conditions, Pi content in shoots of Medicago truncatula was greatly reduced when excess Zn was applied in soil (Watts-Williams et al., 2017). Interestingly, an experiment with lettuce plants grown under excessive Zn levels showed that Zn content in mycorrhizal lettuce was greatly reduced when the nutrient solution contained low Pi concentration (Konieczny and Kowalska, 2017). This is indicative of the “protective effect” of arbuscular mycorrhiza, where host plants acquire much less Zn from the Zn excess soils (Chen et al., 2003; Watts-Williams et al., 2013; Christie et al., 2004). Altogether, the interaction between Pi–Zn nutrients during AM symbiosis can be concluded as follows: Crops grown with sufficient Pi decrease Zn in the roots and/or shoots of crops, and inversely, excess Zn reduces Pi in the shoots. However, the underlying molecular mechanisms of the Pi–Zn interaction in mycorrhizal symbiosis are still unclear, and only a few reports discuss the molecular basis of these interactions (Cakmak and Marschner, 1986; Zhu et al., 2001; Watts-Williams et al., 2013; Watts-Williams et al., 2017). Future studies are required to elucidate the molecular basis of the interactions between Pi and Zn nutrient homeostasis during AM symbiosis.
The antagonistic physiological and molecular interactions between Pi and Fe nutrition have been established in model systems such as Arabidopsis and rice (Hirsch et al., 2006; Ward et al., 2008; Zheng et al., 2009; Jain et al., 2013; Rai et al., 2015), but very little information is available on their interactions in mycorrhizal plants.
A couple of studies performed in some edible crop species uncovered the existence of a negative relationship between Pi and Fe uptake in mycorrhizal plants (Azcón et al., 2003; Ferrol et al., 2016; Hoseinzade et al., 2016; Nafady and Elgharably, 2018). Under low Pi supply, the acquisition of Fe increases in mycorrhizal plants (Watts-Williams and Cavagnaro, 2014; Ferrol et al., 2016), and conversely, host plants decrease the Fe accumulation under high Pi conditions during AM symbiosis. Interestingly, Fe content of the straw was greatly increased with low Pi supply during AM symbiosis (Hoseinzade et al., 2016), indicating that mycorrhized rice has reduced Fe nutrient transported to shoots at high Pi status. Very recently, Nafady and Elgharably (2018) reported a similar negative effect of Fe content when maize was treatment with Pi fertilizers. These studies have demonstrated the negative effects of high Pi application in soil on Fe accumulation in mycorrhizal plants (Azcón et al., 2003). Further, the studies have showed the effect of high Pi application on the uptake and transport of Fe nutrition in both rice and maize during AM symbiosis, which could result in the appearance of iron deficiency symptoms under low Fe conditions. However, the effect of Fe treatments on Pi nutrition has not been investigated so far during mycorrhization. The molecular bases of the cross-talk between Pi and Fe in mycorrhizal plants need to be further explored.
Zinc interacts with some micronutrients such as Fe and copper (Cu) in plants (Poshtmasari et al., 2008; Jain et al., 2013). The cross-talk between the effects of Zn rates on Fe accumulation and translocation has been partially studied in several mycorrhizal plants. Zinc treatment resulted in Fe accumulation in soybean roots under arbuscular mycorrhizal conditions but inhibited Fe translocation from roots to shoots (Ibiang et al., 2017), indicating the cross-talk in Zn and Fe status within the whole soybean during AM symbiosis. However, excess Zn increased root to fruit Fe translocation during AM symbiosis in tomato plants (Ibiang et al., 2018), whereas excess Zn could also lead to a decrease in Fe concentration in mycorrhizal roots. These studies performed under AM conditions have revealed that the physiological antagonistic interaction between Zn and Fe nutrients occurred in roots or shoots depending on the host-plant species. Zn status may therefore affect Fe uptake and transport mechanisms in mycorrhizal plants. These studies have indicated the effect of Zn treatment on the accumulation and homeostasis of Fe nutrition in mycorrhizal plants. However, the effect of Fe availability on Zn nutrition in mycorrhizal plants has not been studied yet, and little information is available on this issue. From the nutritional aspect, there exists a competition between Zn and Fe elements; host plants require coordinate Zn–Fe homeostasis to avoid ion imbalances. Under excess Zn, mycorrhizal plants will decrease the overaccumulation of Fe in shoots prone to Fe starvation. Few studies have identified the potential molecular components involved, and no key genes have been characterized so far acting in the phenomenon. Therefore, the molecular bases of the Zn–Fe interactions in mycorrhizal plants remain largely unknown, and the evidence for the molecular basis of the Zn–Fe co-regulation that mediates the adaptation of a mycorrhizal plant to Zn and Fe availability should be provided in future studies. In particular, the potential genes are involved in the cross-talk between the Zn and Fe homeostasis during AM symbiosis. For instance, the expression of the zinc- and iron-regulated transporter-like proteins (ZRT, IRT-like proteins, referred as to ZIP family) encoding genes in roots and shoots is induced at the transcriptional level by Zn and/or Fe availability (Pedas et al., 2009; Li et al., 2013; Fu et al., 2017), indicating that these ZIP genes may control the uptake and homeostasis of Zn and Fe in mycorrhizal plant species (Grotz et al., 1998; Hall and Williams, 2003).
The above studies provide new insights on genes involved in the potential regulation of nutrient homeostasis in conditions when an individual element is limiting. However, recent research indicated that plant survival is affected by a complex cross-talk between Pi, Zn, and Fe homeostasis (Briat et al., 2015). Interestingly, Saenchai et al. (2016) reported that OsPHO1;1 was transcriptionally up-regulated in response to Pi–Zn–Fe combined stresses and involved in Fe transport and integrative Pi–Zn deficiency signaling in rice, providing a genetic basis for tripartite Pi–Zn–Fe signaling cross-talks in plants. However, how the members of the plant PHO1-type Pi transporter family function as key linkers in the cross-talks between Pi–Zn–Fe signaling during AM symbiosis has not been elucidated. Although the cross-talks between these nutrients have been touched upon in some model plant studies (Misson et al., 2005; Zheng et al., 2009; Saenchai et al., 2016), the molecular mechanisms of the tripartite interactions during AM symbiosis are still lacking.
Over the last seven decades, large numbers of studies have focused on how to interpret the potential mechanisms for phosphorus uptake and signaling at molecular and cellular levels in Arabidopsis or rice. The combination of molecular and cellular biology, multiple “omics” approaches, and reverse genetics has resulted in the characterization of many important genes that control Pi accumulation and homeostasis in Arabidopsis and rice in response to Pi limitation. However, Pi is well known to interact with some micronutrients such as Zn and Fe in plants (Bouain et al., 2014a; Briat et al., 2015). Future research will need to undertake an integrative study to uncover the defined mechanisms by which plants coordinate the Pi, Zn, and Fe deficiency signaling in order to enhance their fitness during multiple Pi, Zn, and Fe deficiency stresses. In such a context, the principal aim of this review is to broaden the current understanding of the cross-talk between the Pi and Zn, Pi and Fe, Zn and Fe, and Pi–Zn–Fe homeostasis in nonmycorrhizal and mycorrhizal plants. In addition, the identification of important genes regulating the interactions between Pi, Zn, and/or Fe transport and signaling in plants, particularly in crop species, will help breeders develop new strategies for nutrient management, and taking into account the interactions between plants and their AM fungal symbionts. In conclusion, the development of the integrative study of cross-talk between Pi, Zn, and Fe signaling pathway will be of great interest and essential for sustainable agricultural development all around the world.
MT and XX conceived and designed this study. XX, WH, and XF wrote the manuscript. HC proposed related theories and assisted with the interpretation of some references. All authors have read, edited, and approved the current version of the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 31800092), the Natural Science Foundation of Guangdong Province in China (grant no. 2018A030313141), the Key Projects of Guangzhou Science and Technology Plan (grant no. 201904020022), and the High-level Talent Start Funding Project of South China Agricultural University (grant no. 218066).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful for the critical revision on the draft manuscript by the three reviewers. The authors are also grateful for the critical proofreading on the draft manuscript by Prof. Erik Nielsen (The Plant Cell, Guest editor) and also thank the Associated Researchers Junwei Liu and Jianyong An (Huazhong Agricultural University, Wuhan, China) and Dr. Nianwu Tang (Kunming Institute of Botany, Chinese Academy of Sciences) for the professional reading and valuable discussion on this manuscript.
Abel, S., Ticconi, C. A., Delatorre, C. A. (2002). Phosphate sensing in higher plants. Physiol. Plant. 115, 1–8. doi: 10.1034/j.1399-3054.2002.1150101.x
Ai, P. H., Sun, S. B., Zhao, J. N., Fan, X. R., Xin, W. J., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Allen, J. W., Shachar, H. Y. (2009). Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol. 149, 549–560. doi: 10.1104/pp.108.129866
Ames, R. N., Reid, C. P. P., Porter, L. K., Cambardella, C. (1983). Hyphal uptake and transport of nitrogen from two 15N-labelled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol. 95, 381–396. doi: 10.1111/j.1469-8137.1983.tb03506.x
Arrivault, S., Senger, T., Krämer, U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46, 861–879. doi: 10.1111/j.1365-313X.2006.02746.x
Assuncão, A. G., Herrero, E., Lin, Y. F., Huettel, B., Talukdar, S., Smacznia, C., et al. (2010a). Arabidopsis thaliana transcription factors bZIP19 and bZIP23 regulate the adaptation to zinc deficiency. Proc. Natl. Acad. Sci. U. S. A. 107, 10296–10301. doi: 10.1073/pnas.1004788107
Assuncão, A. G., Persson, D. P., Husted, S., Schjorring, J. K., Alexander, R. D., Aarts, M. G. (2013). Model of how plants sense zinc deficiency. Metallomics. 5, 1110–1116. doi: 10.1039/c3mt00070b
Assuncão, A. G., Schat, H., Aarts, M. G. (2010b). Regulation of the adaptation to zinc deficiency in plants. Plant Signal. Behav. 5, 1553–1555. doi: 10.4161/psb.5.12.13469
Azcón, R., Ambrosano, E., Charest, C. (2003). Nutrient acquisition in mycorrhizal lettuce plants under different phosphorus and nitrogen concentration. Plant Sci. 165, 1137–1145. doi: 10.1016/S0168-9452(03)00322-4
Barberon, M., Zelazny, E., Robert, S., Conejero, G., Curie, C., Friml, J., et al. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U. S. A. 108, E450–E458 doi: 10.1073/pnas.1100659108
Bari, R., Datt, P., Stitt, M., Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. doi: 10.1104/pp.106.079707
Bayle, V., Arrighi, J. F., Creff, A., Nespoulous, C., Vialaret, J., Rossignol, M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell. 23, 1523–1535. doi: 10.1105/tpc.110.081067
Becher, M., Talke, I. N., Krall, L., Krämer, U. (2003). Cross-species microarray transcript profiling reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37, 251–268. doi: 10.1046/j.1365-313X.2003.01959.x
Belouchi, A., Cellier, M., Kwan, T., Saini, H. S., Leroux, G., Gros, P. (1995). The macrophage-specific membrane protein Nramp controlling natural resistance to infections in mice has homologues expressed in the root system of plants. Plant Mol. Biol. 29, 1181–1196. doi: 10.1007/BF00020461
Bereczky, Z., Wang, H. Y., Schubert, V., Ganal, M., Bauer, P. (2003). Differential regulation of NRAMP and IRT metal transporter genes in wild type and iron uptake mutants of tomato. J. Biol. Chem. 278, 24697–24704. doi: 10.1074/jbc.M301365200
Bonfante, P. (2018). The future has roots in the past: the ideas and scientists that shaped mycorrhizal research. New Phytol. 220, 982–995. doi: 10.1111/nph.15397
Bournier, M., Tissot, N., Mari, S., Boucherez, J., Lacombe, E., Briat, J. F., et al. (2013). Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288, 22670–22680. doi: 10.1074/jbc.M113.482281
Bouain, N., Kisko, M., Rouached, A., Dauzat, M., Lacombe, B., Belgaroui, N., et al. (2014a). Phosphate/zinc interaction analysis in two lettuce varieties reveals contrasting effects on biomass, photosynthesis, and dynamics of Pi transport. BioMed. Res. Int. 2014, 548254. doi: 10.1155/2014/548254
Bouain, N., Shahzad, Z., Rouached, A., Khan, G. A., Berthomieu, P., Abdelly, C., et al. (2014b). Phosphate and zinc transport and signalling in plants: toward a better understanding of their homeostasis interaction. J. Exp. Bot. 65, 5725–5741. doi: 10.1093/jxb/eru314
Briat, J. F., Fobis, L. I., Grignon, N., Lobreaux, S., Pascal, N., Savino, G., et al. (1995). Cellular and molecular aspects of iron metabolism in plants. Biol. Cell. 84, 69–81. doi: 10.1016/0248-4900(96)81320-7
Briat, J., Rouached, H., Tissot, N., Gaymard, F., Dubos, C. (2015). Integration of P, S, Fe and Zn nutrition signals in Arabidopsis thaliana: potential involvement of PHOSPHATE STARVATION RESPONSE 1 (PHR1). Front. Plant Sci. 6, 290. doi: 10.3389/fpls.2015.00290
Brumbarova, T., Bauer, P. (2005). Iron-mediated control of the basic helix-loop-helix protein FER, a regulator of iron uptake in tomato. Plant Physiol. 137, 1018–1026. doi: 10.1104/pp.104.054270
Bucher, M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173, 11–26. doi: 10.1111/j.1469-8137.2006.01935.x
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez, P. J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PloS Genet. 6, e1001102. doi: 10.1371/journal.pgen.1001102
Cakmak, I., Marschner, H. (1986). Mechanism of phosphorus induced zinc deficiency in cotton. I. Zinc deficiency-enhanced uptake rate of phosphorus. Physiol. Plant. 68, 483–490. doi: 10.1111/j.1399-3054.1986.tb03386.x
Caris, C., Hördt, W., Hawkins, H. J., Römheld, V., George, E. (1998). Studies of iron transport by arbuscular mycorrhizal hyphae from soil to peanut and sorghum plants. Mycorrhiza. 8, 35–39. doi: 10.1007/s005720050208
Chang, M. X., Gu, M., Xia, Y. W., Dai, X. L., Dai, C. R., Zhang, J., et al. (2019). OsPHT1;3 mediates uptake, translocation, and remobilization of phosphate under extremely low phosphate regimes. Plant Physiol. 179, 656–670. doi: 10.1104/pp.18.01097
Chen, A., Chen, X., Wang, H., Liao, D., Gu, M., Qu, H., et al. (2014). Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 14, 61. doi: 10.1186/1471-2229-14-61
Chen, A., Hu, J., Sun, S., Xu, G. (2007). Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol. 173, 817–831. doi: 10.1111/j.1469-8137.2006.01962.x
Chen, B. D., Li, X. L., Tao, H. Q., Christie, P., Wong, M. H. (2003). The role of arbuscular mycorrhiza in zinc uptake by red clover growing in a calcareous soil spiked with various quantities of zinc. Chemosphere 50, 839–846. doi: 10.1016/S0045-6535(02)00228-X
Chen, J. Y., Wang, Y. F., Wang, F., Yang, J., Gao, M. X., Li, C. Y., et al. (2015). The rice CK2 kinase regulates trafficking of phosphate transporters in response to phosphate levels. Plant Cell. 27, 711–723. doi: 10.1105/tpc.114.135335
Chiou, T. J., Lin, S. I. (2011). Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 62, 185–206. doi: 10.1146/annurev-arplant-042110-103849
Chorianopoulou, S. N., Saridis, Y. I., Dimou, M., Katinakis, P., Bouranis, D. L. (2015). Arbuscular mycorrhizal symbiosis alters the expression patterns of three key iron homeostasis genes, ZmNAS1, ZmNAS3, and ZmYS1, in S deprived maize plants. Front. Plant Sci. 6, 257. doi: 10.3389/fpls.2015.00257
Christie, P., Li, X. L., Chen, B. D. (2004). Arbuscular mycorrhiza can depress translocation of zinc to shoots of host plants in soils moderately polluted with zinc. Plant Soil. 261, 209–217. doi: 10.1023/B:PLSO.0000035542.79345.1b
Colangelo, E. P., Guerinot, M. L. (2004). The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 16, 3400–3412. doi: 10.1105/tpc.104.024315
Connolly, E. L., Fett, J. P., Guerinot, M. L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell. 14, 1347–1357. doi: 10.1105/tpc.001263
Conte, S. S., Walker, E. L. (2011). Transporters contributing to iron trafficking in plants. Mol. Plant 4, 464–476. doi: 10.1093/mp/ssr015
Curie, C., Cassin, G., Couch, D., Divol, F., Higuchi, K., Le Jean, M., et al. (2009). Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103, 1–11. doi: 10.1093/aob/mcn207
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
Deinlein, U., Weber, M., Schmidt, H., Rensch, S., Trampczynska, A., Hansen, T. H., et al. (2012). Elevated nicotianamine levels in Arabidopsis halleri roots play a key role in Zn hyperaccumulation. Plant Cell. 24, 708–723. doi: 10.1105/tpc.111.095000
Dong, J., Ma, G., Sui, L., Wei, M., Satheesh, V., Zhang, R., et al. (2019). Inositol pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant. S1674–2052(19)30263–1. doi: 10.1016/j.molp.2019.08.002
Durrett, T. P., Gassmann, W., Rogers, E. E. (2007). The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 144, 197–205. doi: 10.1104/pp.107.097162
Eng, B. H., Guerinot, M. L., Eide, D., Saier, M. H. J. (1998). Sequence analyses and phylogenetic characterization of the ZIP family of metal ion transport proteins. J. Membr. Biol. 166, 1–7. doi: 10.1007/s002329900442
Fan, C., Wang, X., Hu, R., Wang, Y., Xiao, C., Jiang, Y., et al. (2013). The pattern of phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol. 13, 48. doi: 10.1186/1471-2229-13-48
Farzaneh, M., Vierheilig, H., Loessl, A. (2011). Arbuscular mycorrhiza enhances nutrient uptake in chickpea. Plant Soil Environ. 57, 465–470. doi: 10.17221/133/2011-PSE
Ferrol, N., Tamayo, E., Vargas, P. (2016). The heavy metal paradox in arbuscular mycorrhizas: from mechanisms to biotechnological applications. J. Exp. Bot. 22, 6253–6265. doi: 10.1093/jxb/erw403
Franco, Z. J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio, S. I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Fu, X., Zhou, X., Xing, F., Ling, L., Chun, C., Cao, L., et al. (2017). Genome-wide identification, cloning and functional analysis of the zinc/iron-regulated transporter-like protein (ZIP) gene family in trifoliate orange (Poncirus trifoliata L. Raf.). Front. Plant Sci. 8, 588. doi: 10.3389/fpls.2017.00588
Giehl, R. F., Meda, A. R., Von, W. N. (2009). Moving up, down, and everywhere: signaling of micronutrients in plants. Curr. Opin. Plant Biol. 12, 320–327. doi: 10.1016/j.pbi.2009.04.006
Giovannetti, M., Tolosano, M., Volpe, V., Kopriva, S., Bonfante, P. (2014). Identification and functional characterization of a sulfate transporter induced by both sulfur starvation and mycorrhiza formation in Lotus japonicus. New Phytol. 204, 609–619. doi: 10.1111/nph.12949
Green, L. S., Rogers, E. E. (2004). FRD3 controls iron localization in Arabidopsis. Plant Physiol. 136, 2523–2531. doi: 10.1104/pp.104.045633
Goff, S. A., Ricke, D., Lan, T. H., Presting, G., Wang, R., Dunn, M., et al. (2002). A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296, 92–100. doi: 10.1126/science.1068275
Gojon, A., Nacry, P., Davidian, J. C. (2009). Root uptake regulation: a central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 12, 328–338. doi: 10.1016/j.pbi.2009.04.015
González, G. M., Azcón, A. C., Mooney, M., Valderas, A., MacDiarmid, C. W., Eide, D. J., et al. (2005). Characterization of a Glomus intraradices gene encoding a putative Zn transporter of the cation diffusion facilitator family. Fungal Genet. Biol. 42, 130–140. doi: 10.1016/j.fgb.2004.10.007
González, G. M., Escudero, V., Saez, A., Tejada, J. M. (2016). Transition metal transport in plants and associated endosymbionts: arbuscular mycorrhizal fungi and rhizobia. Front. Plant Sci. 7, 1088. doi: 10.3389/fpls.2016.01088
Grotz, N., Fox, T., Connolly, E., Park, W., Guerinot, M. L., Eide, D. (1998). Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. U. S. A. 95, 7220–7224. doi: 10.1073/pnas.95.12.7220
Guerinot, M. L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465, 190–198. doi: 10.1016/S0005-2736(00)00138-3
Guether, M., Neuhauser, B., Balestrini, R., Dynowsk, M., Ludewig, U., Bonfante, P. (2009). A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 150, 73–83. doi: 10.1104/pp.109.136390
Gu, M., Chen, A., Sun, S., Xu, G. (2016). Complex regulation of plant phosphate transporters and the gap between molecular mechanisms and practical application: what is missing? Mol. Plant 9, 396–416. doi: 10.1016/j.molp.2015.12.012
Hall, J. L., Williams, L. E. (2003). Transition metal transporters in plants. J. Exp. Bot. 54, 2601–2613. doi: 10.1093/jxb/erg303
Hamburger, D., Rezzonico, E., MacDonald, C. P. J., Somerville, C., Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 14, 889–902. doi: 10.1105/tpc.000745
Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. doi: 10.1104/pp.103.020941
Hanikenne, M., Talke, I. N., Haydon, M. J., Lanz, C., Nolte, A., Motte, P., et al. (2008). Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 453, 391–395. doi: 10.1038/nature06877
Harrison, M. J., Dewbre, G. R., Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 14, 2413–2429. doi: 10.1105/tpc.004861
Haydon, M. J., Cobbett, C. S. (2007). A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143, 1705–1719. doi: 10.1104/pp.106.092015
Haydon, M. J., Kawachi, M., Wirtz, M., Hillmer, S., Hell, R., Kramer, U. (2012). Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24, 724–737. doi: 10.1105/tpc.111.095042
Henriques, R., Jasik, J., Klein, M., Martinoia, E., Feller, U., Schell, J., et al. (2002). Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol. Biol. 50, 587–597. doi: 10.1023/A:1019942200164
Hindt, M. N., Guerinot, M. L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823, 1521–1530. doi: 10.1016/j.bbamcr.2012.03.010
Hirsch, J., Marin, E., Floriani, M., Chiarenza, S., Richaud, P., Nussaume, L., et al. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie. 88, 1767–1771. doi: 10.1016/j.biochi.2006.05.007
Hoseinzade, H., Ardakani, M. R., Shahdi, A., Rahmani, H. A., Noormohammadi, G., Miransari, M. (2016). Rice (Oryza sativa L.) nutrient management using mycorrhizal fungi and endophytic Herbaspirillum seropedicae. J. Integr. Agric. 6, 1385–1394. doi: 10.1016/S2095-3119(15)61241-2
Huang, C., Barker, S. J., Langridge, P., Smith, F. W., Graham, R. D. (2000). Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 124, 415–422. doi: 10.1104/pp.124.1.415
Hussain, D., Haydon, M. J., Wang, Y., Wong, E., Sherson, S. M., Young, J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell. 16, 1327–1339. doi: 10.1105/tpc.020487
Ibiang, Y. B., Mitsumoto, H., Sakamoto, K. (2017). Bradyrhizobia and arbuscular mycorrhizal fungi modulate manganese, iron, phosphorus, and polyphenols in soybean (Glycine max (L.) Merr.) under excess zinc. Environ. Exp. Bot. 137, 1–13. doi: 10.1016/j.envexpbot.2017.01.011
Ibiang, Y. B., Innami, H., Sakamoto, K. (2018). Effect of excess zinc and arbuscular mycorrhizal fungus on bioproduction and trace element nutrition of tomato (Solanum lycopersicum L. cv. Micro-Tom). Soil Sci. Plant Nutr. 3, 342–351. doi: 10.1080/00380768.2018.1425103
Inoue, H., Kobayashi, T., Nozoye, T., Takahashi, M., Kakei, Y., Suzuki, K., et al. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479. doi: 10.1074/jbc.M806042200
Ishimaru, Y., Suzuki, M., Kobayashi, T., Takahashi, M., Nakanishi, H., Mori, S., et al. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56, 3207–3214. doi: 10.1093/jxb/eri317
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Ivanov, R., Brumbarova, T., Bauer, P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant 5, 27–42. doi: 10.1093/mp/ssr065
Jain, A., Sinilal, B., Dhandapani, G., Meagher, R. B., Sahi, S. V. (2013). Effects of deficiency and excess of zinc on morphophysiological traits and spatiotemporal regulation of zinc-responsive genes reveal incidence of cross talk between micro- and macronutrients. Environ. Sci. Technol. 47, 5327–5335. doi: 10.1021/es400113y
Javot, H., Penmetsa, R. V., Terzaghi, N., Cook, D. R., Harrison, M. J. (2007a). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U. S. A. 104, 1720–1725. doi: 10.1073/pnas.0608136104
Javot, H., Pumplin, N., Harrison, M. J. (2007b). Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322. doi: 10.1111/j.1365-3040.2006.01617.x
Jia, H., Ren, H., Gu, M., Zhao, J., Sun, S., Zhang, X., et al. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175. doi: 10.1104/pp.111.175240
Jiang, Y., Wang, W., Xie, Q., Liu, N., Liu, L., Wang, D., et al. (2017). Plants transfer lipids to sustain colonization by mutualistic mycorrhizal and parasitic fungi. Science 356, 1172–1175. doi: 10.1126/science.aam9970
Karandashov, V., Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29. doi: 10.1016/j.tplants.2004.12.003
Kawachi, M., Kobae, Y., Mori, H., Tomioka, R., Lee, Y., Maeshima, M. (2009). A mutant strain Arabidopsis thaliana that lacks vacuolar membrane zinc transporter MTP1 revealed the latent tolerance to excessive zinc. Plant Cell Physiol. 50, 1156–1170. doi: 10.1093/pcp/pcp067
Khan, G. A., Bouraine, S., Wege, S., Li, Y., De, C. M., Berthomieu, P., et al. (2014). Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 65, 871–884. doi: 10.1093/jxb/ert444
Kisko, M., Bouain, N., Rouached, A., Choudhary, S. P., Rouached, H. (2015). Molecular mechanisms of phosphate and zinc signaling crosstalk in plants: phosphate and zinc loading into root xylem in Arabidopsis. Environ. Exp. Bot. 114, 57–64. doi: 10.1016/j.envexpbot.2014.05.013
Klatte, M., Schuler, M., Wirtz, M., Fink-Straube, C., Hell, R., Bauer, P. (2009). The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiol. 150, 257–271. doi: 10.1104/pp.109.136374
Kobayashi, T., Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kobayashi, T., Nishizawa, N. K. (2014). Iron sensors and signals in response to iron deficiency. Plant Sci. 224, 36–43. doi: 10.1016/j.plantsci.2014.04.002
Kobayashi, T., Nagasaka, S., Senoura, T., Itai, R. N., Nakanishi, H., Nishizawa, N. K. (2013). Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 4, 2792. doi: 10.1038/ncomms3792
Koike, S., Inoue, H., Mizuno, D., Takahashi, M., Nakanishi, H., Mori, S., et al. (2004). OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 39, 415–424. doi: 10.1111/j.1365-313X.2004.02146.x
Konieczny, A., Kowalska, I. (2017). Effect of arbuscular mycorrhizal fungi on the content of zinc in lettuce grown at two phosphorus levels and an elevated zinc level in a nutrient solution. J. Elementol. 2, 761–772. doi: 10.5601/jelem.2016.21.4.1335
Kruger, C., Berkowitz, O., Stephan, U. W., Hell, R. (2002). A metal-binding member of the late embryogenesis abundant protein family transports iron in the phloem of Ricinus communis L. J. Biol. Chem. 277, 25062–25069. doi: 10.1074/jbc.M201896200
Lanquar, V., Lelièvre, F., Barbier-Brygoo, H., Thomine, S. (2004). Regulation and function of AtNRAMP4 metal transporter protein. Soil Sci. Plant Nutr. 50, 477–484. doi: 10.1080/00380768.2004.10408587
Lee, S., Kim, S. A., Lee, J., Guerinot, M. L., An, G. (2010). Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells. 29, 551–558. doi: 10.1007/s10059-010-0069-0
Lee, S., Persson, D. P., Hansen, T. H., Husted, S., Schjoerring, J. K., Kim, Y. S., et al. (2011). Bio-available zinc in rice seeds is increased by activation tagging of nicotianamine synthase. Plant Biotechnol. J. 9, 865–873. doi: 10.1111/j.1467-7652.2011.00606.x
Leigh, J., Hodge, A., Fitter, A. H. (2009). Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol. 181, 199–207. doi: 10.1111/j.1469-8137.2008.02630.x
Lin, S. I., Chiang, S. F., Lin, W. Y., Chen, J. W., Tseng, C. Y., Wu, P. C., et al. (2008). Regulatory network of microRNA399 and PHO2 by systemic signaling. Plant Physiol. 147, 732–746. doi: 10.1104/pp.108.116269
Lin, Y. F., Liang, H. M., Yang, S. Y., Boch, A., Clemens, S., Chen, C. C., et al. (2009). Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 182, 392–404. doi: 10.1111/j.1469-8137.2009.02766.x
Li, S., Zhou, X., Chen, J., Chen, R. (2018). Is there a strategy I iron uptake mechanism in maize? Plant Signal. Behav. 3, 13 (4), e1161877. doi: 10.1080/15592324.2016.1161877
Li, S., Zhou, X., Huang, Y., Zhu, L., Zhang, S., Zhao, Y., et al. (2013). Identification and characterization of the zinc-regulated transporters, iron-regulated transporter-like protein (ZIP) gene family in maize. BMC Plant Biol. 13, 114. doi: 10.1186/1471-2229-13-114
Li, W., Lan, P. (2015). Genome-wide analysis of overlapping genes regulated by iron deficiency and phosphate starvation reveals new interactions in Arabidopsis roots. BMC. Res. Notes. 12, 555. doi: 10.1186/s13104-015-1524-y
Lin, W. Y., Huang, T. K., Chiou, T. J. (2013). NITROGEN LIMITATION ADAPTATION, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 25, 4061–4074. doi: 10.1105/tpc.113.116012
Ling, H. Q., Bauer, P., Bereczky, Z., Keller, B., Ganal, M. (2002). The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc. Natl. Acad. Sci. U. S. A. 99, 13938–13943. doi: 10.1073/pnas.212448699
Liu, A., Hamel, C., Hamilton, R. I., Ma, B. L., Smith, B. L. (2000). Acquisition of Cu, Zn, Mn and Fe by mycorrhizal maize (Zea mays L.) grown in soil at different P and micronutrient levels. Mycorrhiza. 9, 331–336. doi: 10.1007/s005720050277
Liu, T. Y., Chang, C. Y., Chiou, T. J. (2009). The long-distance signaling of mineral macronutrients. Curr. Opin. Plant Biol. 12, 312–319. doi: 10.1016/j.pbi.2009.04.004
Loneragan, J. F., Webb, M. J. (1993). “Interactions between zinc and other nutrients affecting the growth of plants,” in Zinc in soils and plants. Ed. Robson, A. D. (Dordrecht: Springer Netherlands), 119–134. doi: 10.1007/978-94-011-0878-2_9
Long, T. A., Tsukagoshi, H., Busch, W., Lahner, B., Salt, D. E., Benfey, P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell. 22, 2219–2236. doi: 10.1105/tpc.110.074096
Lopez, B. J., De, L. V. O., Guevara, G. A., Herrera, L. (2000). Enhanced phosphorus uptake in transgenic tobacco plants that overproduce citrate. Nat. Biotechnol. 18, 450–453. doi: 10.1038/74531
López, M. A., Ellis, D. R., Grusak, M. A. (2004). Identification and characterization of several new members of the ZIP family of metal ion transporters in Medicago truncatula. Plant Mol. Biol. 54, 583–596. doi: 10.1023/B:PLAN.0000038271.96019.aa
Magalhaes, J. V., Liu, J., Guimarães, C. T., Lana, U. G., Alves, V. M., Wang, Y. H., et al. (2007). A gene in the multidrug and toxic compound extrusion (MATE) family confers aluminum tolerance in sorghum. Nat Genet. 39, 1156–1161. doi: 10.1038/ng2074
Marschner, P. (2012). Marschner’s mineral nutrition of higher plants. 3rd. USA: Boston: Academic Press, Elsevier.
Mäser, P., Thomine, S., Schroeder, J. I., Ward, J. M., Hirschi, K., Sze, H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. doi: 10.1104/pp.126.4.1646
Masuda, H., Usuda, K., Kobayashi, T., Ishimaru, Y., Kakei, Y., Takahashi, M., et al. (2009). Overexpression of the barley nicotianamine synthase gene HvNAS1 increases iron and zinc concentrations in rice grains. Rice 2, 155–166. doi: 10.1007/s12284-009-9031-1
Milner, M. J., Seamon, J., Craft, E., Kochian, L. (2013). Transport properties of members of the ZIP family in plants and their role in Zn and Mn homeostasis. J. Exp. Bot. 64, 369–603. doi: 10.1093/jxb/ers315
Milner, M. J., Craft, E., Yamaji, N., Koyama, E., Ma, J. F., Kochian, L. V. (2012). Characterization of the high affinity Zn transporter from Noccaea caerulescens, NcZNT1, and dissection of its promoter for its role in Zn uptake and hyperaccumulation. New Phytol. 195, 113–123. doi: 10.1111/j.1469-8137.2012.04144.x
Misson, J., Raghothama, K. G., Jain, A., Jouhet, J., Block, M. A., Bligny, R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U. S. A. 102, 11934–11939. doi: 10.1073/pnas.0505266102
Moran, L. A., Peiffer, G. A., Yin, T., Whitham, S. A., Cook, D., Shoemaker, R. C., et al. (2014). Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genomics 15, 702. doi: 10.1186/1471-2164-15-702
Morrissey, J., Guerinot, M. L. (2009). Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109, 4553–4567. doi: 10.1002/chin.201005266
Muchhal, U. S., Pardo, J. M., Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U. S. A. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Mudge, S. R., Rae, A. L., Diatloff, E., Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Müller, R., Morant, M., Jarmer, H., Nilsson, L., Hamborg, N. T. H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143, 156–171. doi: 10.1104/pp.106.090167
Nafady, N. A., Elgharably, A. (2018). Mycorrhizal symbiosis and phosphorus fertilization effects on Zea mays growth and heavy metals uptake. Int. J. Phytorem. 20, 869–875. doi: 10.1080/15226514.2018.1438358
Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G., Smith, A. P. (2011). Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 156, 1149–1163. doi: 10.1104/pp.111.174805
Neset, T. S., Cordell, D. (2012). Global phosphorus scarcity: identifying synergies for a sustainable future. J. Sci. Food Agric. 92, 2–6. doi: 10.1002/jsfa.4650
Nussaume, L., Kanno, S., Javot, H, M. E., Pochon, N., Ayadi, A., Nakanishi, T. M., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2, 83. doi: 10.3389/fpls.2011.00083
Ogo, Y., Itai, R. N., Kobayashi, T., Aung, M. S., Nakanishi, H., Nishizawa, N. K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75, 593–605. doi: 10.1007/s11103-011-9752-6
Ogo, Y., Itai, R. N., Nakanishi, H., Kobayashi, T., Takahashi, M., Mori, S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377. doi: 10.1111/j.1365-313X.2007.03149.x
Ova, E. A., Kutman, U. B., Ozturk, L., Cakmak, I. (2015). High phosphorus supplies reduced zinc concentration of wheat in native soil but not in autoclaved soil or nutrient solution. Plant Soil 393, 147–162. doi: 10.1007/s11104-015-2483-8
Palmer, C. M., Guerinot, M. L. (2009). Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340. doi: 10.1038/nchembio.166
Pant, B. D., Buhtz, A., Kehr, J., Scheible, W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53, 731–738. doi: 10.1111/j.1365-313X.2007.03363.x
Pao, S. S., Paulsen, I. T., Saier, M. H. J. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34. doi: 10.9783/9781512806809-002
Parniske, M. (2008). Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev. Microbiol. 6, 763–775. doi: 10.1038/nrmicro1987
Paszkowski, U., Kroken, S., Roux, C., Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U. S. A. 99, 13324–13329. doi: 10.1073/pnas.202474599
Pedas, P., Schjoerring, J. K., Husted, S. (2009). Identification and characterization of zinc-starvation-induced ZIP transporters from barley roots. Plant Physiol. Biochem. 47, 377–383. doi: 10.1016/j.plaphy.2009.01.006
Perea, G. A., Garcia, M. A., Andrés, N., Vera, S. F., Pérez, A. M. A., Puig, S., et al. (2013). Arabidopsis copper transport protein COPT2 participates in the cross talk between iron deficiency responses and low-phosphate signaling. Plant Physiol. 162, 180–194. doi: 10.1104/pp.112.212407
Petit, J. M., Briat, J. F., Lobréaux, S. (2001). Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 359, 575–582. doi: 10.1042/bj3590575
Pilon, M., Cohu, C. M., Ravet, K., Abdel, G. S. E., Gaymard, F. (2009). Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 12, 347–357. doi: 10.1016/j.pbi.2009.04.011
Pineau, C., Loubet, S., Lefoulon, C., Chalies, C., Fizames, C., Lacombe, B., et al. (2012). Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet. 8, e1003120. doi: 10.1371/journal.pgen.1003120
Poirier, Y., Bucher, M. (2002). Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book. 1, e0024. doi: 10.1199/tab.0024
Poirier, Y., Jung, J. Y. (2015). Phosphate transporters. Annu. Plant Rev. 48, 125–159. doi: 10.1002/9781118958841.ch5
Poirier, Y., Thoma, S., Somerville, C., Schiefelbein, J. (1991). Mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol. 97, 1087–1093. doi: 10.1104/pp.97.3.1087
Poshtmasari, H. K., Bahmanyar, M. A., Pirdashti, H., Shad, M. A. (2008). Effects of Zn rates and application forms on protein and some micronutrients accumulation in common bean (Phaseolus vulgaris L.). Pak J Biol Sci. 11, 1042–1046.
Puga, M. I., Mateos, I., Charukesi, R., Wang, Z. Y., Franco, Z. J. M., De, L. L., et al. (2014). SPX1 is a phosphate-dependent inhibitor of PHOSPHATE STARVATION RESPONSE 1 in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 111, 14947–14952. doi: 10.1073/pnas.1404654111
Rai, V., Sanagala, R., Sinilal, B., Yadav, S., Sarkar, A. K., Dantu, P. K., et al. (2015). Iron availability affects phosphate deficiency-mediated responses, and evidence of cross-talk with auxin and zinc in Arabidopsis. Plant Cell Physiol. 56, 1107–1123. doi: 10.1093/pcp/pcv035
Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M., Leggewie, G., et al. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414, 462–470. doi: 10.1038/35106601
Reed, H. (1946). Effects of zinc deficiency on phosphate metabolism of the tomato plant. Am. J. Bot. 33, 778–784. doi: 10.1002/j.1537-2197.1946.tb12940.x
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sa, C. I., Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Remy, W., Taylor, T. N., Hass, H., Kerp, H. (1994). Four hundred million-year-old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U. S. A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Rouached, H., Arpat, A. B., Poirier, Y. (2010). Regulation of phosphate starvation responses in plants: signaling players and cross-talks. Mol. Plant 3, 288–299. doi: 10.1093/mp/ssp120
Rouached, H., Stefanovic, A., Secco, D., Bulak Arpat, A., Gout, E., Bligny, R., et al. (2011b). Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 65, 557–570. doi: 10.1111/j.1365-313X.2010.04442.x
Ryan, M. H., Mcinerney, J. K., Record, I. R., Angus, J. F. (2008). Zinc bioavailability in wheat grain in relation to phosphorus fertiliser, crop sequence and mycorrhizal fungi. J. Sci. Food Agric. 88, 1208–1216. doi: 10.1002/jsfa.3200
Salgueiro, M., Zubillaga, M., Lysionek, A., Sarabia, M. I., Caro, R., De, P. T., et al. (2000). Zinc as an essential micronutrient: a review. Nutr. Res. 20, 737–755. doi: 10.1016/S0271-5317(00)00163-9
Saenchai, C., Bouain, N., Kisko, M., Prom, C., Doumas, P., Rouached, H. (2016). The involvement of OsPHO1;1 in the regulation of iron transport through integration of phosphate and zinc deficiency signaling. Front. Plant Sci. 7, 396. doi: 10.3389/fpls.2016.00396
Sasaki, A., Yamaji, N., Mitani-Ueno, N., Kashino, M., Ma, J. F. (2015). A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 84, 374–384. doi: 10.1111/tpj.13005
Schachtman, D. P., Reid, R. J., Ayling, S. M. (1998). Phosphorus uptake by plants: from soil to cell. Plant Physiol. 116, 447–453. doi: 10.1104/pp.116.2.447
Schachtman, D. P., Shin, R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58, 47–69. doi: 10.1146/annurev.arplant.58.032806.103750
Schüßler, A., Schwarzott, D., Walker, C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421. doi: 10.1017/S0953756201005196
Shahzad, Z., Rouached, H., Rakha, A. (2014). Combating mineral malnutrition through iron and zinc biofortification of cereals. Compr. Rev. Food Sci. F. 13, 329–346. doi: 10.1111/1541-4337.12063
Shanmugam, V., Tsednee, M., Yeh, K. C. (2012). ZINC TOLERANCE INDUCED BY IRON 1 reveals the importance of glutathione in the cross-homeostasis between zinc and iron in Arabidopsis thaliana. Plant J. 69, 1006–1017. doi: 10.1111/j.1365-313X.2011.04850.x
Shin, H., Shin, H. S., Dewbre, G. R., Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Sieh, D., Watanabe, M., Devers, E. A., Brueckner, F., Hoefgen, R., Krajinski, F. (2013). The arbuscular mycorrhizal symbiosis influences sulfur starvation responses of Medicago truncatula. New Phytol. 197, 606–616. doi: 10.1111/nph.12034
Sinclair, S. A., Krämer, U. (2012). The zinc homeostasis network of land plants. Biochim. Biophys. Acta 1823, 1553–1567. doi: 10.1016/j.bbamcr.2012.05.016
Smith, S. E., Smith, F. A., Jakobsen, I. (2003). Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 133, 16–20. doi: 10.1104/pp.103.024380
Stefanovic, A., Arpat, A. B., Bligny, R., Gout, E., Vidoudez, C., Bensimon, M., et al. (2011). Overexpression of PHO1 in Arabidopsis leaves reveals its role in mediating phosphate efflux. Plant J. 66, 689–699. doi: 10.1111/j.1365-313X.2011.04532.x
Stephan, U. W., Scholz, G. (1993). Nicotianamine: mediator of transport of iron and heavy metals in the phloem? Physiol. Plant. 88, 522–529. doi: 10.1111/j.1399-3054.1993.tb01367.x
Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X., Ai, P., et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159, 1571–1581. doi: 10.1104/pp.112.196345
Takahashi, M., Terada, Y., Nakai, I., Yoshimura, E., Mori, S., Nishizawa, N. K. (2003). Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell. 15, 1263–1280. doi: 10.1105/tpc.010256
Tamayo, E., Gómez-Gallego, T., Azcón-Aguilar, C., Ferrol, N. (2014). Genome-wide analysis of copper, iron and zinc transporters in the arbuscular mycorrhizal fungus Rhizophagus irregularis. Front. Plant Sci. 5, 547. doi: 10.3389/fpls.2014.00547
Tamayo, E., Knight, S. A. B., Valderas, A., Dancis, A., Ferrol, N. (2018). The arbuscular mycorrhizal fungus Rhizophagus irregularis uses a reductive iron assimilation pathway for high-affinity iron uptake. Environ Microbiol. 20, 1857–1872. doi: 10.1111/1462-2920.14121
Thibaud, M. C., Arrighi, J. F., Bayle, V., Chiarenza, S., Creff, A., Bustos, R., et al. (2010). Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64, 775–789. doi: 10.1111/j.1365-313X.2010.04375.x
Thomine, S., Wang, R., Ward, J. M., Crawford, N. M., Schroeder, J. I. (2000). Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc. Natl. Acad. Sci. U. S. A. 97, 4991–4996. doi: 10.1073/pnas.97.9.4991
Tiong, J., McDonald, G. K., Genc, Y., Pedas, P., Hayes, J. E., Toubia, J., et al. (2014). HvZIP7 mediates zinc accumulation in barley (Hordeum vulgare) at moderately high zinc supply. New Phytol. 201, 131–143. doi: 10.1111/nph.12468
van de Mortel, J. E., Almar, V. L., Schat, H., Kwekkeboom, J., Coughlan, S., Moerland, P. D., et al. (2006). Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142, 1127–1147. doi: 10.1104/pp.106.082073
van der Heijden, M. G. A., Klironomos, J. N., Ursic, M., Moutoglis, P., Streitwolf, R., Boller, T., et al. (1998). Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 396, 69–72. doi: 10.1038/23932
Verret, F., Gravot, A., Auroy, P., Leonhardt, N., David, P., Nussaume, L., et al. (2004). Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 576, 306–312. doi: 10.1016/j.febslet.2004.09.023
Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 14, 1223–1233. doi: 10.1105/tpc.001388
Volpe, V., Giovannetti, M., Sun, X. G., Fiorilli, V., Bonfante, P. (2016). The phosphate transporters LjPT4 and MtPT4 mediate early root responses to phosphate status in nonmycorrhizal roots. Plant Cell Environ. 39, 660–671. doi: 10.1111/pce.12659
Wang, L., Ying, Y., Narsai, R., Ye, L., Zheng, L., Tian, J., et al. (2013). Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ. 36, 224–236. doi: 10.1111/j.1365-3040.2012.02569.x
Wang, Z., Ruan, W., Shi, J., Zhang, L., Xiang, D., Yang, C., et al. (2014a). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U. S. A. 111, 14953–14958. doi: 10.1073/pnas.1404680111
Wang, X., Wang, Y., Piñeros, M. A., Wang, Z., Wang, W., Li, C., et al. (2014b). Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37, 1159–1170. doi: 10.1111/pce.12224
Ward, J. T., Lahner, B., Yakubova, E., Salt, D. E., Raghothama, K. G. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147, 1181–1191. doi: 10.1104/pp.108.118562
Watts-Williams, S. J., Cavagnaro, T. R. (2018). Arbuscular mycorrhizal fungi increase grain zinc concentration and modify the expression of root ZIP transporter genes in a modern barley (Hordeum vulgare) cultivar. Plant Sci. 247, 163–170. doi: 10.1016/j.plantsci.2018.05.015
Watts-Williams, S. J., Cavagnaro, T. R. (2014). Nutrient interactions and arbuscular mycorrhizas: a meta-analysis of a mycorrhiza-defective mutant and wild-type tomato genotype pair. Plant Soil 2, 79–92. doi: 10.1007/s11104-014-2140-7
Watts-Williams, S. J., Smith, F. A., McLaughlin, M. J., Patti, A. F., Cavagnaro, T. R. (2015). How important is the mycorrhizal pathway for plant Zn uptake? Plant Soil. 390, 157–166. doi: 10.1007/s11104-014-2374-4
Watts-Williams, S. J., Tyerman, S. D., Cavagnaro, T. R. (2017). The dual benefit of arbuscular mycorrhizal fungi under soil zinc deficiency and toxicity: linking plant physiology and gene expression. Plant Soil 420, 375–388. doi: 10.1007/s11104-017-3409-4
Watts-Williams, S. J., Patti, A., Cavagnaro, T. R. (2013). Arbuscular mycorrhizas are beneficial under both deficient and toxic soil zinc conditions. Plant Soil 371, 299–312. doi: 10.1007/s11104-013-1670-8
Westheimer, F. H. (1987). Why nature chose phosphates. Science 235, 1173–1178. doi: 10.1126/science.2434996
Wintz, H., Fox, T., Wu, Y., Feng, V., Chen, W., Chang, H., et al. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278, 47644–47653. doi: 10.1074/jbc.M309338200
Wong, C. K., Jarvis, R. S., Sherson, S. M., Cobbett, C. S. (2009). Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana. New Phytol. 181, 79–88. doi: 10.1111/j.1469-8137.2008.02637.x
Wu, P., Ma, L., Hou, X., Wang, M., Wu, Y., Liu, F., et al. (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132, 1260–1271. doi: 10.1104/pp.103.021022
Xue, Y. F., Xia, H. Y., Christie, P., Zhang, Z., Li, L., Tang, C. X. (2016). Crop acquisition of phosphorus, iron and zinc from soil in cereal/legume intercropping systems: a critical review. Ann. Bot. 3, 363–377. doi: 10.1093/aob/mcv182
Yang, S. Y., Grønlund, M., Jakobsen, I., Grotemeyer, M. S., Rentsch, D., Miyao, A., et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell. 24, 4236–4251. doi: 10.1105/tpc.112.104901
Yokosho, K., Yamaji, N., Ueno, D., Mitani, N., Ma, J. F. (2009). OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 149, 297–305. doi: 10.1104/pp.108.128132
Zhang, F., Sun, Y., Pei, W., Jain, A., Sun, R., Cao, Y., et al. (2015). Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82, 556–569. doi: 10.1111/tpj.12804
Zhang, J., Liu, B., Li, M., Feng, D., Jin, H., Wang, P., et al. (2015). The bHLH transcription factor bhlh104 interacts with IAA-LEUCINE RESISTANT3 and modulates iron homeostasis in Arabidopsis. Plant Cell. 27, 787–805. doi: 10.1105/tpc.114.132704
Zhang, W., Chen, X. X., Liu, Y. M., Liu, D. Y., Chen, X. P., Zou, C. Q. (2017). Zinc uptake by roots and accumulation in maize plants as affected by phosphorus application and arbuscular mycorrhizal colonization. Plant Soil 413, 59–71. doi: 10.1007/s11104-017-3213-1
Zhang, W., Liu, D. Y., Liu, Y. M., Cui, Z. L., Chen, X. P., Zou, C. Q. (2016). Zinc uptake and accumulation in winter wheat relative to changes in root morphology and mycorrhizal colonization following varying phosphorus application on calcareous soil. Field Crop Res. 197, 74–82. doi: 10.1016/j.fcr.2016.08.010
Zhang, Y. Q., Deng, Y., Chen, R. Y., Cui, Z. L., Chen, X. P., Yost, R., Zhang, F.S., et al. (2012). The reduction in zinc concentration of wheat grain upon increased phosphorus-fertilization and its mitigation by foliar zinc application. Plant Soil. 361, 143–152. doi: 10.1007/s11104-012-1238-z
Zhang, Z., Liao, H., Lucas, W. J. (2014). Molecular mechanisms underlying phosphate sensing, signaling, and adaption in plants. J. Integr. Plant Biol. 56, 192–220. doi: 10.1111/jipb.12163
Zheng, L., Huang, F., Narsai, R., Wu, J., Giraud, E., He, F., et al. (2009). Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151, 262–274. doi: 10.1104/pp.109.141051
Zheng, L., Ying, Y., Wang, L., Wang, F., Whelan, J., Shou, H. (2010). Identification of a novel iron regulated basic helix-loop-helix protein involved in Fe homeostasis in Oryza sativa. BMC Plant Biol. 10, 166. doi: 10.1186/1471-2229-10-166
Keywords: phosphorus, zinc, iron, Pi–Zn–Fe interactions, arbuscular mycorrhizal plants
Citation: Xie X, Hu W, Fan X, Chen H and Tang M (2019) Interactions Between Phosphorus, Zinc, and Iron Homeostasis in Nonmycorrhizal and Mycorrhizal Plants. Front. Plant Sci. 10:1172. doi: 10.3389/fpls.2019.01172
Received: 17 March 2019; Accepted: 27 August 2019;
Published: 26 September 2019.
Edited by:
Valentina Fiorilli, University of Turin, ItalyReviewed by:
Nuria Ferrol, Experimental Station of Zaidín (EEZ), SpainCopyright © 2019 Xie, Hu, Fan, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianan Xie, eGlleGlhbmFuODgzNDIwM0AxMjYuY29t; Ming Tang, dGFuZ21pbmd5bEAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.