
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 22 October 2019
Sec. Plant Metabolism and Chemodiversity
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01147
Calcium-dependent protein kinases (CDPKs) are recognized as important calcium (Ca2+) sensors in signal transduction and play multiple roles in plant growth and developmental processes, as well as in response to various environmental stresses. However, little information is available about the CDPK family in the green microalga Chlamydomonas reinhardtii. In this study, 15 CrCDPK genes were identified in C. reinhardtii genome, and their functions in nitrogen (N) deficiency-induced oil accumulation were analyzed. Our results showed that all CrCDPK proteins harbored the typical elongation factor (EF)-hand Ca2+-binding and protein kinase domains. Phylogenetic analysis revealed that these CrCDPKs were clustered into one group together with a subclade of several CPKs from Arabidopsis and rice, clearly separating from the remaining AtCPKs and OsCPKs. These genes were located in 10 chromosomes and one scaffold of C. reinhardtii and contained 6–17 exons. RNA sequencing and quantitative reverse transcription (qRT)-PCR assays indicated that most of these CrCDPKs were significantly induced by N deficiency and salt stress. Lanthanum chloride (LaCl3), a plasma membrane Ca2+ channel blocker, limited oil accumulation in C. reinhardtii under N-deficient conditions, suggesting that Ca2+ was involved in N deficiency-induced oil accumulation. Furthermore, RNA interference (RNAi) silencing analyses demonstrated that six CrCDPKs played positive roles and three CrCDPKs played negative roles in N deficiency-induced oil accumulation in C. reinhardtii.
Calcium ion (Ca2+), a ubiquitous second messenger in cell signaling, plays essential roles in many physiological processes, such as plant growth, development, and abiotic and biotic stresses (Hetherington and Brownlee, 2004). Various extracellular stimuli can cause transient and minor changes in cytosolic Ca2+ concentration, which can be first sensed and decoded by several Ca2+ sensors, triggering appropriate downstream responses in plant cells (Lenzoni et al., 2018). In general, three classes of Ca2+ sensors, namely, calcineurin B-like (CBL), calmodulin (CaM) and CaM-like proteins, and Ca2+-dependent protein kinases (CDPKs or CPKs), have been identified (Boudsocq and Sheen, 2013). However, only CDPKs can directly transduce Ca2+ signal into downstream phosphorylation cascades by a single gene product, which arise from their special structures (Hu et al., 2016b). The CDPK family harbors four functional domains, namely, the variable N-terminal domain, highly conserved Ser/Thr kinase domain, autoinhibitory junction domain, and CaM-like domain. The N-terminal domain contains potential myristoylation or palmitoylation sites for membrane association, and the Ser/Thr kinase domain is required for substrate phosphorylation. The autoinhibitory junction domain modulates the kinase activity of CDPKs via a pseudosubstrate mechanism, and the CaM-like domain contains four elongation factor (EF)-hand motifs for binding to Ca2+ (Liese and Romeis, 2013; Schulz et al., 2013; Liu et al., 2018). Ca2+ binding to EF-hands induces a conformational change in CDPK, resulting in the refolding and exposure of the active site of the kinase domain and thus activating the CDPK to phosphorylate an array of substrates.
A large number of CDPKs have been widely found in plants, green algae, and certain protozoans but not in bacteria, fungi, yeast, and animals (Batistič and Kudla, 2012; Hamel et al., 2014). For instance, 34 and 31 CDPKs have been identified in Arabidopsis and rice genomes, respectively (Hrabak et al., 2003; Ray et al., 2007). In addition, there are 20 CDPKs in wheat genome (Li et al., 2008), 23 in potato (Gromadka et al., 2018), 27 in cassava (Hu et al., 2016a), 29 in tomato (Hu et al., 2016b), 30 in poplar (Zuo et al., 2013), 40 in maize (Kong et al., 2013), and 41 in cotton (Liu et al., 2014). The biological functions of CDPKs, including pollen tube development (AtCPK2/AtCPK11/AtCPK17/AtCPK20/AtCPK24/AtCPK34) (Gutermuth et al., 2013; Zhao et al., 2013), response to environmental stresses (AtCPK1/AtCPK3/AtCPK4/AtCPK5/AtCPK6/AtCPK8/AtCPK10/AtCPK11/AtCPK13/AtCPK21) (Choi et al., 2005; Mori et al., 2006; Zhu et al., 2007; Boudsocq et al., 2010; Franz et al., 2011; Zhou et al., 2015), and plant growth regulation (AtCPK12/AtCPK28) (Zhao et al., 2011; Matschi et al., 2013), have been well characterized in Arabidopsis. Several rice CDPKs have been demonstrated to respond to cold (OsCPK7/OsCPK13/OsCPK17/OsCPK24) (Saijo et al., 2000; Almadanim et al., 2018; Liu et al., 2018), drought, and salt stresses (OsCPK4/OsCPK9/OsCPK12/OsCPK13/OsCPK21) (Asano et al., 2011; Asano et al., 2012; Campo et al., 2014; Wei et al., 2014). By contrast, information about the roles of CDPKs derived from other plant species is still scarce, apart from a few CDPKs that are involved in tolerance to abiotic stresses (ZmCPK1, VaCPK20, BnaCPK2, and StCDPK4/5) (Kobayashi et al., 2007; Dubrovina et al., 2015; Weckwerth et al., 2015; Wang et al., 2018b).
Microalgae are a large and diverse group of photosynthetic eukaryotes with a simple cellular structure, which range from unicellular to multicellular forms. In general, the lipid metabolic pathways in microalgae were inferred from those of plants wherein the lipid synthesis have been well characterized. However, the recent studies have shown that there were many differences between algal species and higher plants. For instance, cells usually contain betaine lipid diacylglyceryl-(N,N,N-trimethyl)homoserine (DGTS) instead of phosphatidylcholine (PtdCho) in Chlamydomonas reinhardtii (Liu and Benning, 2013). With the development of next-generation sequencing technology, more and more algal genes involved in lipid metabolism were identified, and the gene functions were annotated to different extent in microalgae. The lipid biochemistry of eukaryotic algae has been reviewed by Li-Beisson et al. (2019). Stress conditions, especially nitrogen (N) deficiency, can induce oil droplet biosynthesis, mainly in the form of triacylglycerol (TAG), which accumulates in cells to generate lipid droplets that store carbon and energy in many microalgae species, such as C. reinhardtii (Hu et al., 2008). During N deprivation, massive metabolic alterations were observed in C. reinhardtii. Wase et al. (2014) reported that, compared with non-stressed cultures, metabolites and proteins involved in citrate [tricarboxylic acid (TCA)] cycles, glycolysis, and starch and lipid metabolism were elevated, and the enzymes of the glyoxylate cycle, the Calvin cycle, and photosynthesis were reduced in response to N starvation. These metabolic changes play important roles in the supply of carbon skeletons and energy for lipid storage under N-deficient conditions. Moreover, citrate accumulated during nitrogen deprivation, which may be exported for use in fatty acid (FA) synthesis. Similar results were obtained by Park et al. (2015). Once N deficiency-induced TAG synthesis was initiated in the cells, carbon metabolism shifted from a gluconeogenic state to a glycolytic state. In addition, the transcriptional levels of genes related to pyruvate and acetyl coenzyme A (acetyl-CoA) synthesis steadily increased during N starvation. Pyruvate and acetyl-CoA are two central intermediates linking glycolysis and TCA cycles to lipid metabolism, respectively.
N deficiency-induced microalgal oil accumulation always associated with reduced biomass yield, which was a large obstacle for algal-based biodiesel production. Currently, an alternative strategy has been developed to select small-molecule chemical inducers to trigger TAG accumulation without terminating cell growth. A total of 243 compounds that clustered into five distinct structural scaffolds were effective in multiple algal species including Chlamydomonas, Chlorella sorokiniana, Chlorella vulgaris, and Tetrachlorella alternans (Wase et al., 2017), and 34 molecules were also identified to act as inducers of TAG accumulation in Phaeodactylum tricornutum (Conte et al., 2018). Possible targets for these compounds were distributed in various metabolic steps of carbon and lipid metabolism such as lipase inhibition, acyltransferase activity modulation, and channeling of acetyl-CoA to FA biosynthesis (Wase et al., 2018).
Even though many efforts have been made to exploit lipid metabolism in microalgae, its molecular mechanisms remain unclear and controversial. Most evidence shows that CDPKs act as signaling hubs in plant stress signaling and development (Schulz et al., 2013), suggesting their contribution to stress-related TAG accumulation in microalgae. In this study, we identified 15 CDPK genes on the basis of a systematic genome-wide analysis in C. reinhardtii and analyzed their protein motifs, phylogenetic relationship, chromosome localization, gene structure, expression profiles under various abiotic stresses, and the functions of CrCDPKs in N deficiency-induced oil accumulation. RNAi silencing of CrCDPKs resulted in a significant decrease or increase in the oil content of C. reinhardtii under N deficiency.
The BLASTP program at an expectation value (E) of 10−10 was performed in Phytozome V11.0 C. reinhardtii v5.5 database (https://phytozome.jgi.doe.gov/) by using the published CDPK proteins (Arabidopsis thaliana and Oryza sativa) as query sequences (Table S1). In addition, another way to identify the CDPK genes was searching the database using the keyword “calcium-dependent protein kinase kinase.” Only non-redundant full-length sequences containing EF-hand motifs were considered. Conserved protein motifs were predicted using ScanProsite (http://prosite.expasy.org/) and SMART (http://smart.embl-heidelberg.de/). The protein domains were visualized by the Interactive Tree of Life (iTOL) (http://itol2.embl.de/). The molecular weights and isoelectric points of proteins were predicted using ExPASy (http://web.expasy.org/compute_pi/). Myristoylation sites were predicted by NMT—the MYR predictor script (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm).
Multiple sequence alignments were performed with Clustal X 2.0, and a phylogenetic tree was constructed on the basis of the neighbor-joining method using MEGA 6.0 software with 1,000 bootstrap replicates. The chromosomal locations of 15 CrCDPK genes were drafted using MapInspect 1.0. The structures of CrCDPK genes were depicted by using the Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/)
C. reinhardtii CC425 (cw15 arg2) was purchased from the Chlamydomonas Genetic Center of Duke University. For RNA-Seq analysis, the wild-type CC425 was first grown in Tris–acetate–phosphate (TAP) medium for 3 days to reach a growth plateau and then collected by centrifugation (3,000 g). After being washed twice with sterile water, the algal cells were inoculated in high salt minimal (HSM) medium that is deficient in N, phosphorus (P), and iron (Fe) and contains 100 mM of NaCl (salt stress). They were placed in an incubator shaker maintained at 230 rpm and 24 °C and exposed to continuous illumination at a light intensity of 150 μmol·m−2·s−1. The cultured cells that reached a growth plateau were inoculated in normal HSM medium and were divided into two groups: one group was used as control, and the other group was placed under a temperature of 16 °C (cold stress). Samples were collected after 24 h for RNA-Seq analysis. N deficiency-treated algal cells were also collected after 0, 2, 12, 24, 48, and 72 h for quantitative reverse transcription (qRT)-PCR.
RNA-Seq analysis was performed on an Illumina HiSeq 2000 platform in GENE DENOVO, Guangzhou, China (http://www.genedenovo.com), as described by Li et al. (2017). Raw reads were first filtered by removing the adapter sequences, low-quality sequences, and reads with more than 10% unknown nucleotides. The obtained clean reads were mapped to the Ribosomal Database Project with Bowtie (Langmead et al., 2009), and the reads that belonged to ribosomal DNA (rRNA) were removed. The remaining clean reads were mapped to the reference genome of C. reinhardtii by TopHat2 (Kim et al., 2013). The transcriptome was assembled with Cufflinks (Trapnell et al., 2010), and Cuffmerge was used to merge the assembly of two replicate samples. The gene expression level was calculated and normalized by using the FPKM (fragments per kilobase of transcript per million mapped reads) method. The edgeR package (http://www.r-project.org/) was used to identify differentially expressed genes across samples. The false discovery rate (FDR) was used to determine the p-value threshold in multiple tests. Genes with FDR ≤0.05 and an absolute value of log2 change fold ≥1 were regarded as significant differentially expressed genes.
The fold change value of each stress treatment compared with the control was normalized to complete hierarchical clustering analysis, and the heatmap was drawn by using the R3.5.1 software. For qRT-PCR, total RNA was extracted using TRIzol Reagent (Takara, Japan) and subjected to RNase-free DNase (RQ1, Promega, USA) for digestion and purification. qRT-PCR analyses were performed as described by Li et al. (2017). The gene-specific primers of qRT-PCR are listed in Table S2. The experiment was repeated three times, and statistical analysis was performed by using SPSS software with independent-samples t-test.
LaCl3 was purchased from Sigma (St. Louis, MO). C. reinhardtii CC425 cells grown on TAP agar plate were inoculated into 100-ml Erlenmeyer flasks containing 50 ml of TAP media for 3 days to reach a growth plateau and were collected by centrifugation at 3,000 g. After being washed twice with sterile water, the cultured cells were inoculated into P- and N-and-P-deficient HSM media with various concentrations of LaCl3 (0, 1, 5, 10, 25, 50, 75, 100, 150, 200, 500, and 1,000 μM). Moreover, 0 μM of LaCl3-treated cells was used as control. All cultures were maintained in an incubator shaker at 230 rpm and 24 °C and exposed to continuous illumination at a light intensity of 150 μmol·m−2s−1. Samples for cell viability assay and measurement of chlorophyll and oil contents were collected after 24, 48, and 72 h. For cell viability assay, the algal cells were directly stained with 0.01% (w/v) fluorescein diacetate (FDA) (final concentration) for 2 min, and flow cytometry using CyFlow Cube 6 was employed to count cells. A total of 104 cells were counted per sample. Cellular viability was shown as the percentages of FDA-stained live cells to total 104 cell numbers.
The Nile Red fluorescence method was performed to determine the oil content as described by Li et al. (2012a). The algal cells were directly stained with 0.1 mg/ml (final concentration) Nile Red (dissolved in DMSO) for 10 min, and fluorescence was measured on a GloMax-Multi Detection System (Promega, USA) with excitation and emission wavelengths of 470 and 570 nm, respectively. The fluorescence value was calculated by using the equation FD (470/570) = (A2 − A1), where A2 and A1 are the fluorescence values of algal cells after and before staining with Nile Red, respectively. To establish the relationship of fluorescence values of samples with their neutral lipid contents, a standard curve was drawn by preparing different concentrations of Triolein (Sigma, USA) and detecting their fluorescence value after staining with Nile Red. The lipid content of algal cells was calculated using the following formula: lipid content (μg/106 cell) = [0.0004 × FD (470/570) − 0.0038] × 50/cell density. Chlorophyll content was measured as described by Li et al. (2012b). Statistical analysis was performed using the SPSS software by one-way ANOVA and Duncan’s multiple range tests, and the experiment was repeated three times.
To construct the RNAi vectors, a fragment of the C. reinhardtii 18S gene was amplified with primers 5′-CGAACTTCTGCGAAAGCAT-3′ and 5′-TCAGCCTTGCGACCATACT-3′ and inserted into pMD18T to obtain pMD18T-18S. The fragments from non-conservative domain-encoding regions of 15 CrCDPK genes were amplified by PCR using C. reinhardtii complementary DNA (cDNA) as template. The fragments were digested with KpnI/BamHI and HindIII/SalI and subsequently inserted into the corresponding cloning sites of pMD18T-18S to form vector pMD18T-CrCDPKF-18S-CrCDPKR, which contained an inverted repeat sequence of CrCDPK (CrCDPK IR). The vector pMD18T-CrCDPKF-18S-CrCDPKR was double digested with KpnI and HindIII to obtain CrCDPK IR. Finally, the fragment of CrCDPK IR was inserted as a blunt-end fragment into EcoRI digested pMaa7 IR/XIR to give pMaa7 IR/CrCDPK IR.
The transformation of C. reinhardtii strain CC425 was performed by the glass bead procedure as described by Kindle (1990). The algal cells were cultured in TAP medium to a cell density of 2 × 106 cells/ml, collected by centrifugation (3,000 g, 5 min), and resuspended in TAP medium to a cell density of 2 × 108 cells/ml. Approximately 10 μg of plasmid DNA was added in a mixture containing 400 μl of algal cells and 300 mg of sterile glass beads. The reaction was mixed for 20 s on a bench-top vortex and allowed to recover for 2 days before plating at selective conditions. Maa7 IR/XIR and Maa7 IR/CrCDPK IR transformants were selected on TAP agar medium containing 1.5 mM of l-tryptophan, 5 μg/ml of paromomycin, and 5 μM of indole analog 5-fluoroindole (5-FI) as described by Rohr et al. (2004). Plates were incubated under dim light (approximately 50 μmol·m−2·s−1). Isolated transgenic strains were kept under constant selective pressure to prevent the loss of integrated IR transgenes. The diagram of RNAi vector Maa7 IR/XIR is shown in Figure S1. Because the IR corresponding to the target gene is internal to the arms of the Maa7 IR, efficient Maa7 gene silencing may result in an efficiently suppressed target gene. Only the transgenic algal strains exhibiting Maa7 and target gene silencing can grow on TAP medium with 5-FI and tryptophan because the Maa7 gene encodes tryptophan synthase β-subunit, which can convert 5-FI into the toxic tryptophan 5-FI. Therefore, Maa7-silencing transgenic algal cells are resistant to 5-FI.
For oil content measurement, each transformant was inoculated into HSM agar plates containing 0.5 mM of ammonium. After 12 days of cultivation, transgenic lines were transferred into a 96-well plate containing 200 μl of sterile water, and the oil content was measured as mentioned above. The cell density was determined by measuring the optical density of samples at 490 nm (OD490). To produce the standard curve, a series of C. reinhardtii samples of different cell densities were collected, and their OD490 values and cell densities were determined gravimetrically after plotting the standard curve of OD490 versus cell density (cell/ml). Samples were diluted by using appropriate ratios to ensure that the measured OD490 values were in the range of 0.15–0.75, if applicable. Cell density was calculated by using the following formula: cell density (cell/ml) = 2 × 107 × OD490-669662 (R2 = 0.9996). C. reinhardtii transformed with the empty vector Maa7 IR/XIR (Maa7) was used as control. A total of 192 transgenic lines were selected to determine the oil content of each CrCDPK gene, and the boxplot was drawn by R3.5.1 software. Statistical analysis was performed using the SPSS software by means of one-way ANOVA and Duncan’s multiple range tests.
CrCDPK3-, CrCDPK5-, and CrCDPK7-RNAi-silencing algal transformants were selected for log-phase culture. Maa7 empty vector transgenic lines were used as control. Ten transgenic lines were used for each CrCDPK gene. Maa7 and RNAi strains grown on a TAP agar plate were inoculated into 100-ml Erlenmeyer flasks containing 50 ml of HSM media with 0.5 mM of ammonium (N-limited). All cultures were placed in an incubator shaker maintained at 230 rpm and 24 °C and exposed to continuous illumination at a light intensity of 150 μmol·m−2·s−1. Samples for cell density, chlorophyll content, and oil content measurement were collected after 1, 2, 3, 4, and 5 days. Chlorophyll content was measured as described by Li et al. (2012b). Statistical analysis was performed using SPSS software by means of one-way ANOVA and Duncan’s multiple range tests.
BLAST searches of the C. reinhardtii genome were performed using Arabidopsis and rice CDPK gene sequences as queries. A total of 15 non-redundant CrCDPK genes were identified and designated as CrCDPK1–CrCDPK15 (Table 1), all of which had domains typical for the CDPK family, which include a variable N-terminal domain, Ser/Thr kinase domain, autoinhibitory junction domain, and CaM-like domain (Figure 1). All CrCDPKs have four EF-hand motifs except for CrCDPK10, which lacked two EF-hands in the CaM-like domain (Figure 1). Eight CrCDPKs contain the predicted myristoylation sites at their N-terminus (Table 1). The open reading frame length of CrCDPKs ranged from 1,455 bp (CrCDPK9) to 5,940 bp (CrCDPK7), which encoded polypeptides in the range of 484 to 1,979 aa with predicted protein molecular masses from 54 to 198.9 kDa (Table 1). As shown in the table, the predicted isoelectric points of CrCDPKs ranged from 5.23 to 8.61. All of them tended to be acidic, except for CrCDPK11 (8.61) and CrCDPK6 (7.26).
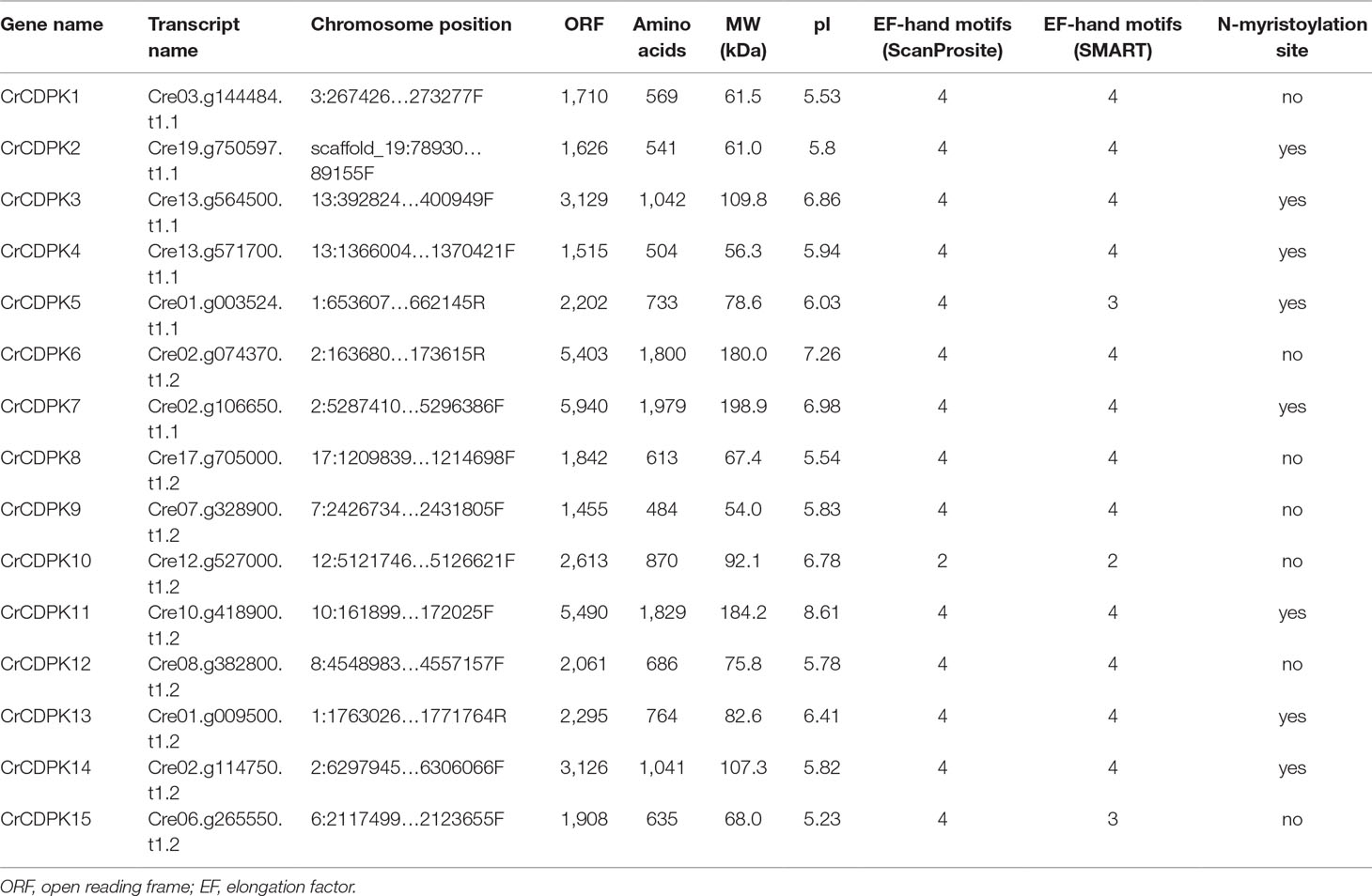
Table 1 List of the 15 CDPK genes identified in Chlamydomonas reinhardtii and their sequence characteristics.
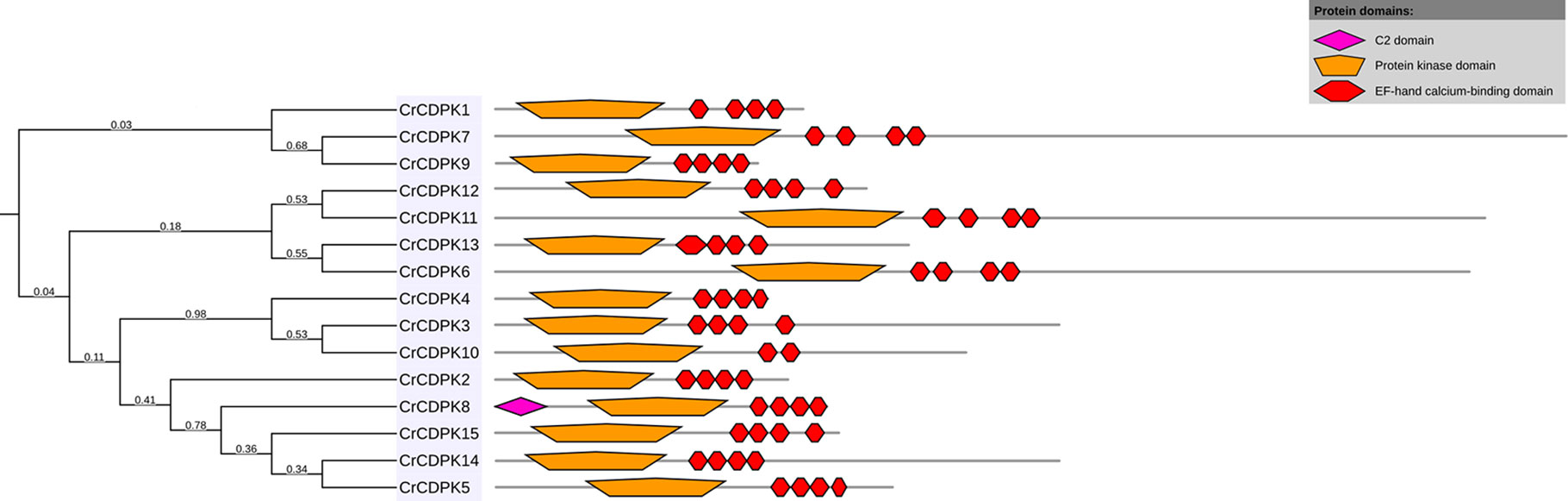
Figure 1 Conserved motif composition of CrCDPKs. The pink, orange, and red boxes indicate the C2, protein kinase, and elongation factor (EF)-hand calcium-binding domains.
To evaluate the evolutionary relationships of CDPK family members among C. reinhardtii, Arabidopsis, and rice, 80 full-length amino acid sequences, including 15 CrCDPKs, 34 AtCPKs, and 31 OsCPKs, were used to generate a phylogenetic tree. The results showed that these CDPKs could be divided into three different clusters (I, II, and III) (Figure 2). Cluster I, the largest group, contained 18 AtCPKs and 19 OsCPKs. Cluster II included 13 AtCPKs and 8 OsCPKs. Interestingly, all CrCDPKs were assigned to Cluster III with a subclade of three AtCPKs and four OsCPKs. Additionally, CrCDPK12 had high similarity with OsCPK31 (Figure 2). The CrCDPK gene family of Cluster III was clearly separated from the other two clusters with a strong bootstrap support, suggesting that this family probably came from a CDPK ancestor with Arabidopsis and rice but had a distant relationship with them.
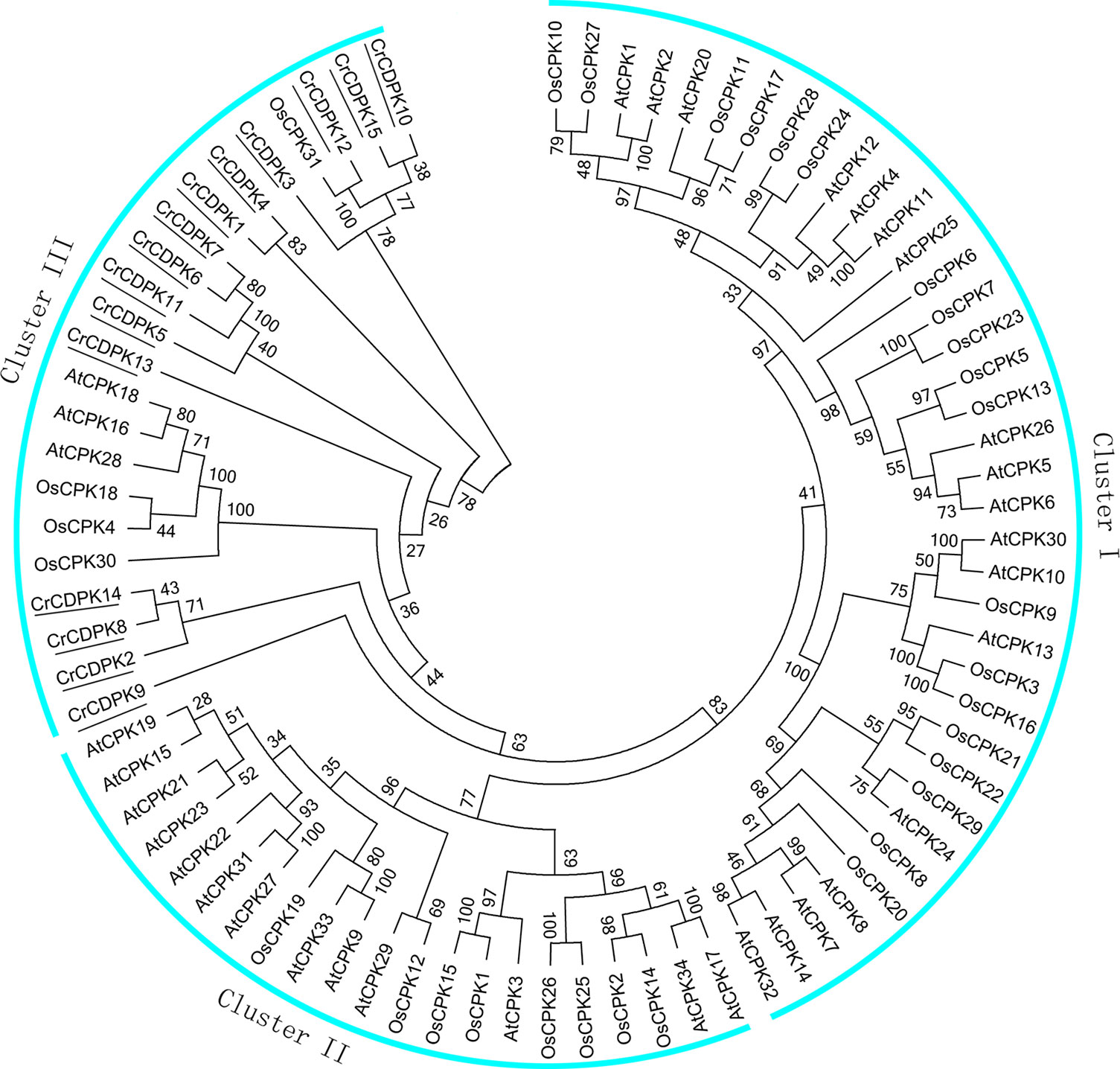
Figure 2 Phylogenetic tree of calcium-dependent protein kinase (CDPK) genes in Chlamydomonas reinhardtii, Arabidopsis, and rice. The phylogenetic tree was created according to the neighbor-joining method using Clustal X 2.0 and MEGA 6.0 software with 1,000 bootstraps on the basis of the amino acid sequence of 15 CrCDPKs (indicated with a black line), 34 AtCPKs, and 31 OsCPKs.
The chromosomal localization of CDPKs indicated that 14 CrCDPKs were distributed among 10 chromosomes in C. reinhardtii and one CrCDPK was located on scaffold_19 (Figure S2). Seven chromosomes had one unique CDPK gene each. CrCDPK1, CrCDPK15, CrCDPK9, CrCDPK12, CrCDPK11, CrCDPK10, and CrCDPK8 were located on chromosomes 3, 6, 7, 8, 10, 12, and 17, respectively. Chromosomes 1 and 13 had two CDPK genes each. Chromosome 1 carried CrCDPK5 and CrCDPK13, whereas chromosome 13 carried CrCDPK3 and CrCDPK4. CrCDPK6, CrCDPK7, and CrCDPK14 were mapped on chromosome 2 (Figure S2). CDPK genes were widely distributed in the genome, implying a multiple-gene function of this family in C. reinhardtii. In addition, the CrCDPK gene cluster was not identified in the C. reinhardtii genome, suggesting that a few duplication events occurred in this gene family.
The exon and intron structures of CrCDPK genes were visualized by using GSDS. As shown in Figure 3, all CrCDPKs contained 9–17 exons except for CrCDPK10, which had only six exons. CrCDPK14 had a maximum of 17 exon regions. CrCDPK4 and CrCDPK9 both contained nine exons, and CrCDPK5, CrCDPK7, and CrCDPK8 contained 12 exons. Fourteen exons were contained in five genes, namely, CrCDPK1, CrCDPK3, CrCDPK12, CrCDPK13, and CrCDPK15. CrCDPK2 and CrCDPK11 contained 15 exons, and CrCDPK6 contained 16 exons.
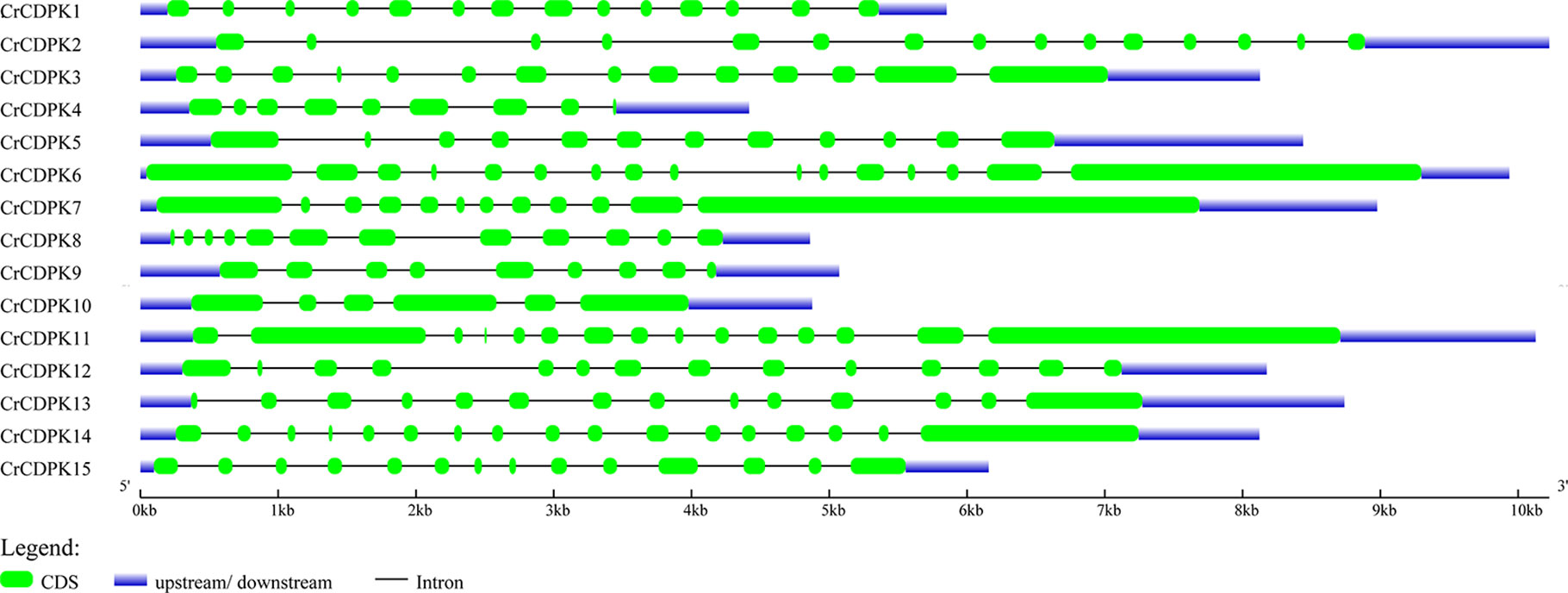
Figure 3 Gene structure of the CDPK gene family in Chlamydomonas reinhardtii. Blue and green bars represent untranslated regions (UTRs) and exons, respectively, and black strings represent introns.
To evaluate the expression profiles of the CrCDPK genes under stress conditions, algal cells treated with N, P, and Fe deficiencies and cold and salt stresses for 24 h were collected for RNA sequencing. We normalized the fold changes (stress treatment/control) of CrCDPKs and subjected them to hierarchical cluster analysis. As shown in Figure 4, except CrCDPK15, the expression profiles of CDPK genes can be divided into seven groups, wherein each group contained two genes. CrCDPK1 and CrCDPK10, CrCDPK4 and CrCDPK12, CrCDPK8 and CrCDPK9, CrCDPK7 and CrCDPK11, CrCDPK2 and CrCDPK5, CrCDPK3 and CrCDPK14, and CrCDPK6 and CrCDPK13 shared similar expression patterns under various stress conditions.
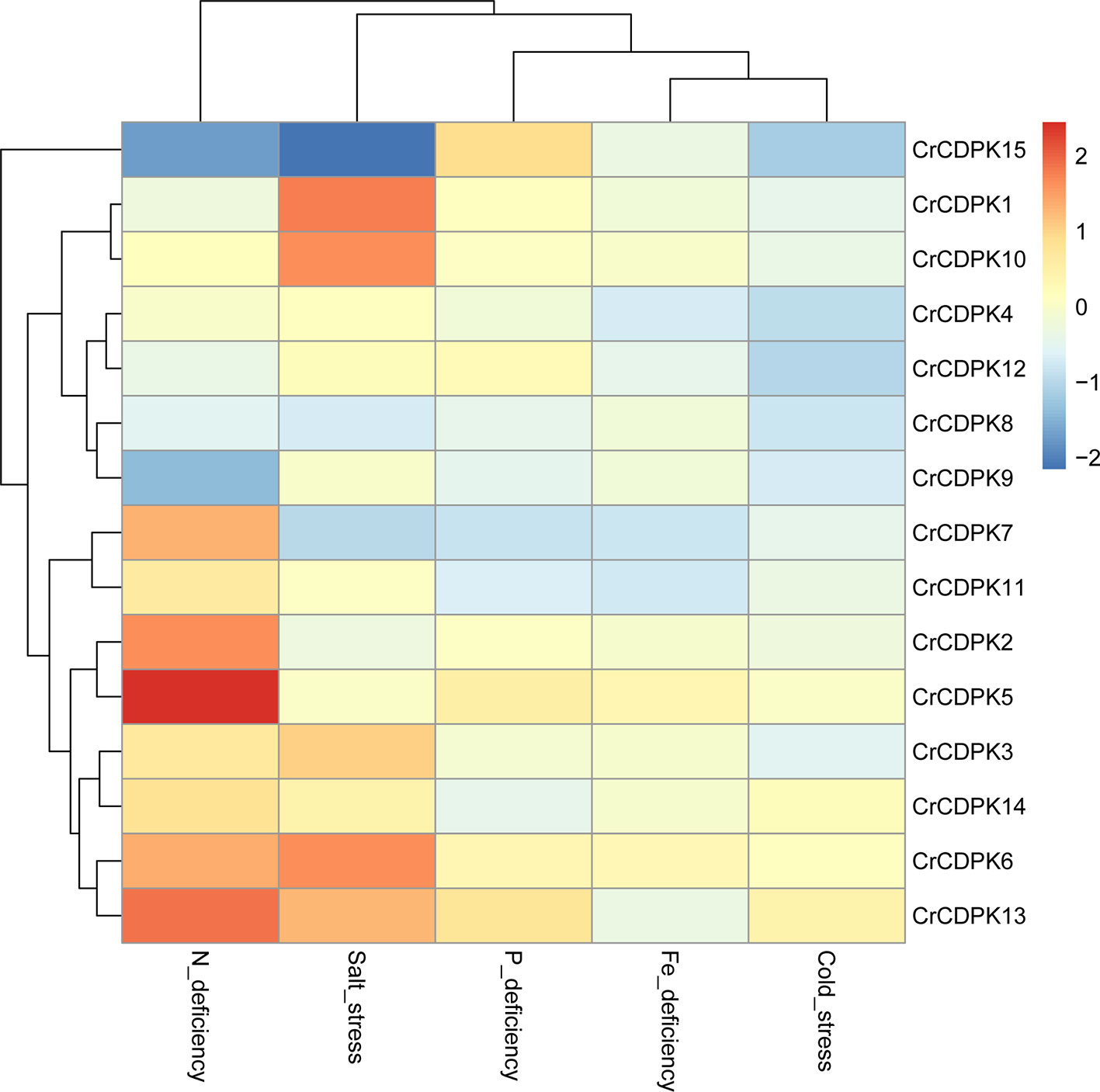
Figure 4 Expression profiles of 15 CrCDPK genes under nitrogen (N), phosphorus (P), and iron (Fe) deficiencies and cold and salt stresses. The algal cells were treated with various abiotic stresses and collected after 24 h for RNA-Seq. “Cold_stress” means that algal cells were exposed to 16 °C. The fold change value of each stress treatment compared with the control was normalized to complete hierarchical clustering analysis. Each CrCDPK gene was represented in a single row, and log2 (fold change) of each treatment was represented in a single column. Red and blue indicated higher and lower gene expression levels of CrCDPKs, respectively, in response to stress treatments compare with those of the control. The key is shown at the right side of the heat map.
N deficiency and salt stress activated more differentially expressed CrCDPK genes than the other treatments (Figure 4). The log2 (fold change) values of CrCDPK genes under various stress conditions are shown in Table 2. The algal cells grown in normal conditions were treated as control. When FDR ≤0.05 and |log2 (fold change)| ≥1 were applied as filter conditions, five genes (CrCDPK2, CrCDPK5, CrCDPK6, CrCDPK7, and CrCDPK13) and two genes (CrCDPK9 and CrCDPK15) were upregulated and downregulated, respectively, by N deficiency. In the case of salt stress, five genes (CrCDPK1, CrCDPK3, CrCDPK6, CrCDPK10, and CrCDPK13) were upregulated, and one gene (CrCDPK15) was downregulated. Moreover, CrCDPK6 and CrCDPK13 were upregulated, and CrCDPK15 was downregulated by both N deficiency and salt stress treatment. By contrast, P and Fe deficiencies and cold stress caused a slight upregulation or downregulation of CrCDPK genes.
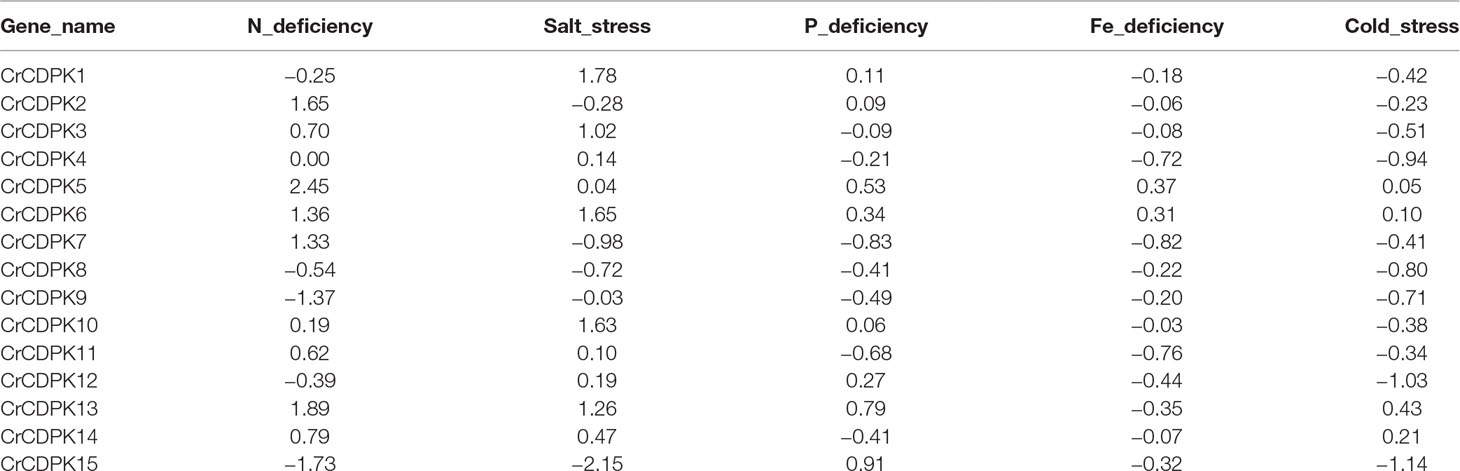
Table 2 The log2 (fold change) value of 15 CrCDPKs in various stress treatments compared to the control.
To learn more about the expression profiles of CrCDPK genes under N-deficient conditions, the algal cells were sampled after 2, 12, 24, 48, and 72 h of N-deficient treatment, and the untreated cells were used as control. The expression patterns of CrCDPKs in C. reinhardtii were examined through qRT-PCR (Figure 5). Compared with the control, all CrCDPK genes were found to be not significantly differentially expressed after 2 h. The transcript level of CrCDPK6 was remarkably increased after 12 h and reached its peak, which was approximately 600 times higher than that of control (Figure 5). The expression levels of seven CrCDPK genes were significantly induced by N deficiency after 24 h. Among them, four CrCDPKs (CrCDPK3, CrCDPK7, CrCDPK11, and CrCDPK12) showed similar expression patterns, which peaked after 24 h. CrCDPK1 and CrCDPK15 were dramatically upregulated after 24 h of N deficiency but significantly downregulated after 48 and 72 h. CrCDPK5 was remarkably activated after 24 h and continually expressed up to 72 h (Figure 5). Interestingly, three genes (CrCDPK2, CrCDPK8, and CrCDPK13) showed lower expression levels than the control after 48 h, and two genes (CrCDPK4 and CrCDPK14) were significantly upregulated after 72 h (Figure 5). The expression profile of the CrCDPK gene family suggested that most CrCDPKs showed earlier expression and/or higher transcription abundance under N-deficient conditions.
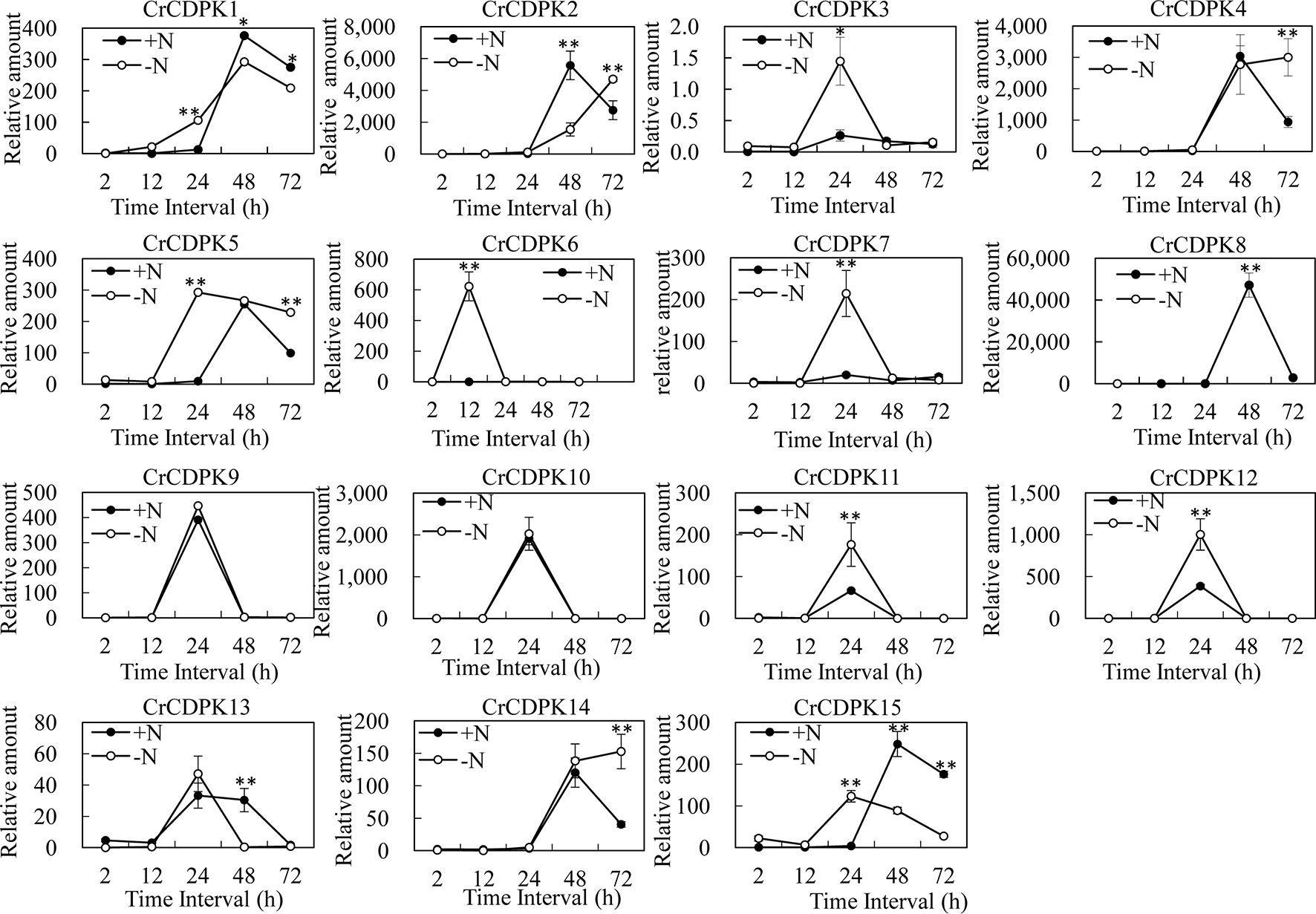
Figure 5 Expression profiles of 15 CrCDPK genes under normal and nitrogen-deficient conditions. Two-step cultivation was used for nitrogen deficiency treatment. The algal cells were first grown in Tris–acetate–phosphate (TAP) medium for 3 days to reach a growth plateau and collected by centrifugation (3,000 g). Finally, the cultured cells were divided into two groups: one group was inoculated into high salt minimal (HSM) medium without nitrogen (HSM-N), and the other was inoculated into normal HSM that was used as control. The samples for quantitative reverse transcription (qRT)-PCR were collected after 0, 2, 12, 24, 48, and 72 h. All expression level values were normalized to the value of the 18S gene level. The data shown are means ( ± SD, n = 3). Significance is indicated as *p < 0.05 and **p < 0.01 (t-test). This experiment was repeated three times with similar results.
To determine whether the Ca2+ signaling pathway played an important role in N deficiency-induced oil accumulation in C. reinhardtii, LaCl3, a general plasma membrane Ca2+ blocker, was used to prevent the Ca2+ influx in N deficiency-treated algal cells. Because phosphate and La3+ formed precipitates, the medium for N deficiency treatment (HSM-N) did not contain P. Algal cells were grown in TAP media for 3 days, collected by centrifugation, and inoculated into HSM-P and HSM-N-P media with various concentrations of LaCl3 (1, 5, 10, 25, 50, 75, 150, 100, 200, 500, and 1,000 μM). At different treatment times, cellular viability, chlorophyll content, and oil content of the samples were measured. Algal cells treated with P or N and P deficiency and 0 μM of LaCl3 were used as control. The results showed that compared with those in untreated algal cells, the oil contents in C. reinhardtii cells dramatically decreased after treating with over 10 μM of LaCl3 for 48 h under P or N and P starvation (Figure 6A). Algal cells treated with 10–50 μM of LaCl3 displayed lower oil content than those treated with other concentrations, suggesting that 10–50 μM LaCl3 is toxic to algal cells (Figure 6A). Because LaCl3 toxicity is not dosage dependent (Figure S3), the cell viabilities did not significantly change under 75–500 μM of LaCl3 and N and P starvation. Although the oil content significantly decreased, results suggested that Ca2+ contributed to N and P deficiency-induced oil accumulation in C. reinhardtii. LaCl3 had a significant effect on the chlorophyll content of C. reinhardtii (Figure 6B). After the application of over 50 μM of LaCl3, chlorophyll (a + b) contents were significantly decreased under P-deficient conditions. By contrast, treatment with over 10 μM of LaCl3 induced a dramatic decline in the chlorophyll content of algal cells grown in N-and-P-deficient media.
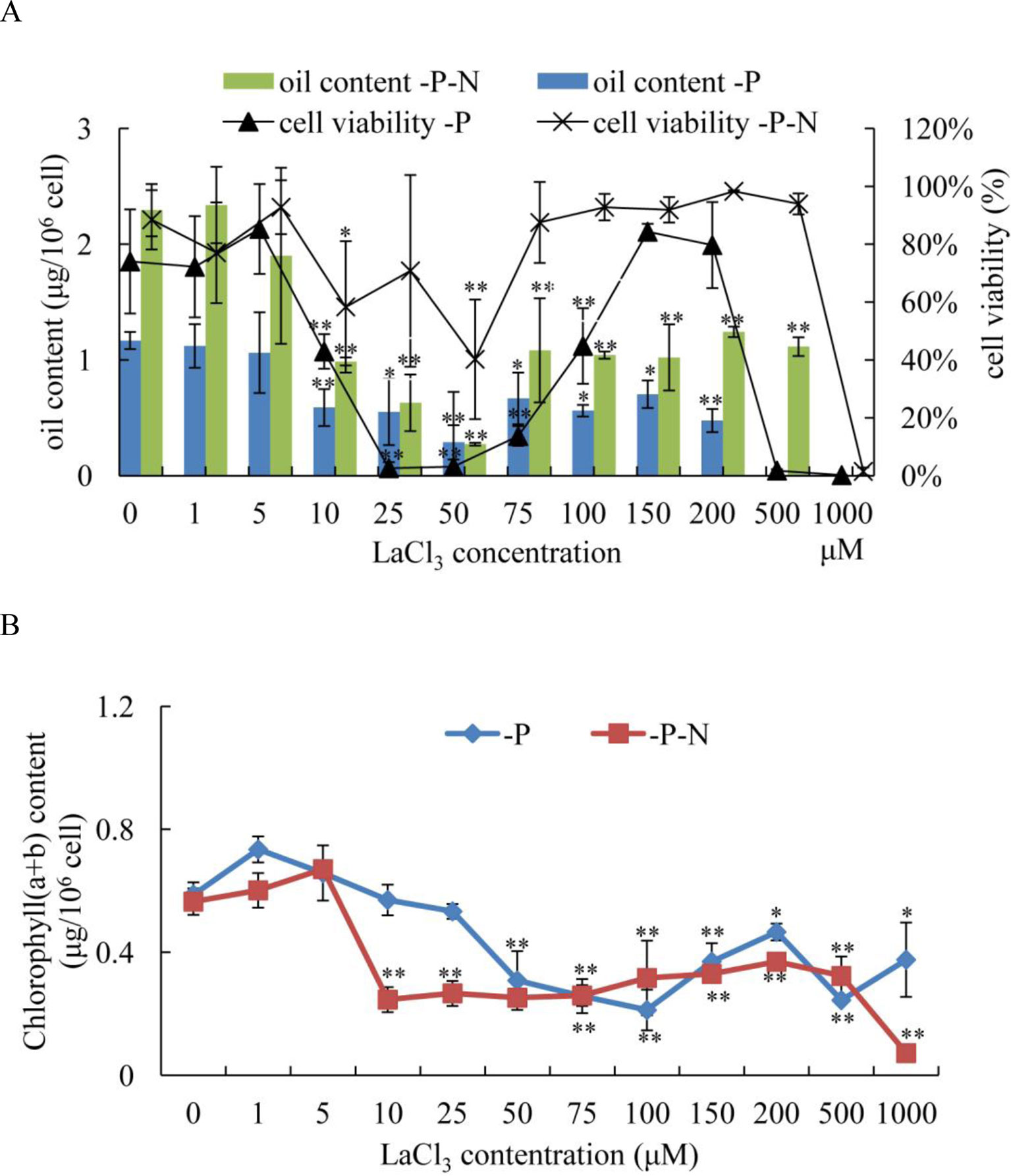
Figure 6 Effects of lanthanum chloride (LaCl3) on oil accumulation (A) and chlorophyll content (B) of Chlamydomonas reinhardtii cells for 48 h under P- and N-and-P-deficient conditions. (A) Algal cells were grown in high salt minimal (HSM) media for 3 days to reach a growth plateau and collected by centrifugation. Finally, the cultured cells were inoculated into P- and N-and-P-deficient HSM media but with various concentrations of LaCl3 (0, 1, 5, 10, 25, 50, 75, 100, 150, 200, 500, and 1,000 µM). The oil contents and cell viability of the samples were measured at 48 h after treatment with different concentrations of LaCl3. Fluorescein diacetate [FDA; 0.01% (w/v)]-stained cells were counted by flow cytometry using CyFlow Cube 6. Cellular viability was presented as percentages of FDA-stained live cells to total 104 cell numbers. “0 µM” means LaCl3-untreated cells (control). Data shown are means ( ± SD, n = 3). Asterisks indicate statistically significant differences compared with the corresponding control (Duncan’s multiple range test: *p < 0.05, **p < 0.01). This experiment was repeated three times with similar results.
To understand the function of CrCDPK genes in N deficiency-induced oil accumulation in C. reinhardtii, RNAi-mediated gene silencing was performed for all 15 CrCDPKs. A total of 192 transgenic lines were generated in each case, and their oil contents were determined under N-deficient conditions. Transgenic cells containing RNAi plasmid Maa7 IR/XIR were used as control. The results are shown in Figure 7. Compared with those of the control, the oil contents of algal cells transformed with CrCDPK1, CrCDPK3, CrCDPK5, CrCDPK11, CrCDPK12, and CrCDPK15 significantly decreased after N deficiency treatment, indicating that these genes play positive roles in N deficiency-induced oil accumulation in C. reinhardtii. By contrast, transgenic lines carrying the small interfering RNA (siRNA) against CrCDPK4, CrCDPK6, and CrCDPK7 exhibited a dramatic increase in oil content (Figure 7), implying that these three genes are negatively involved in oil accumulation under N-deficient conditions in C. reinhardtii. No significant changes were found in the cell densities between the RNAi strains and control (Figure S4). Additionally, no obvious difference was observed between the control and CrCDPK2-, CrCDPK8-, CrCDPK9-, CrCDPK10-, CrCDPK13-, and CrCDPK14-RNAi-silencing transgenic lines (Figure 7), suggesting that these genes are not involved in oil accumulation or that other members of CrCDPK act redundantly.
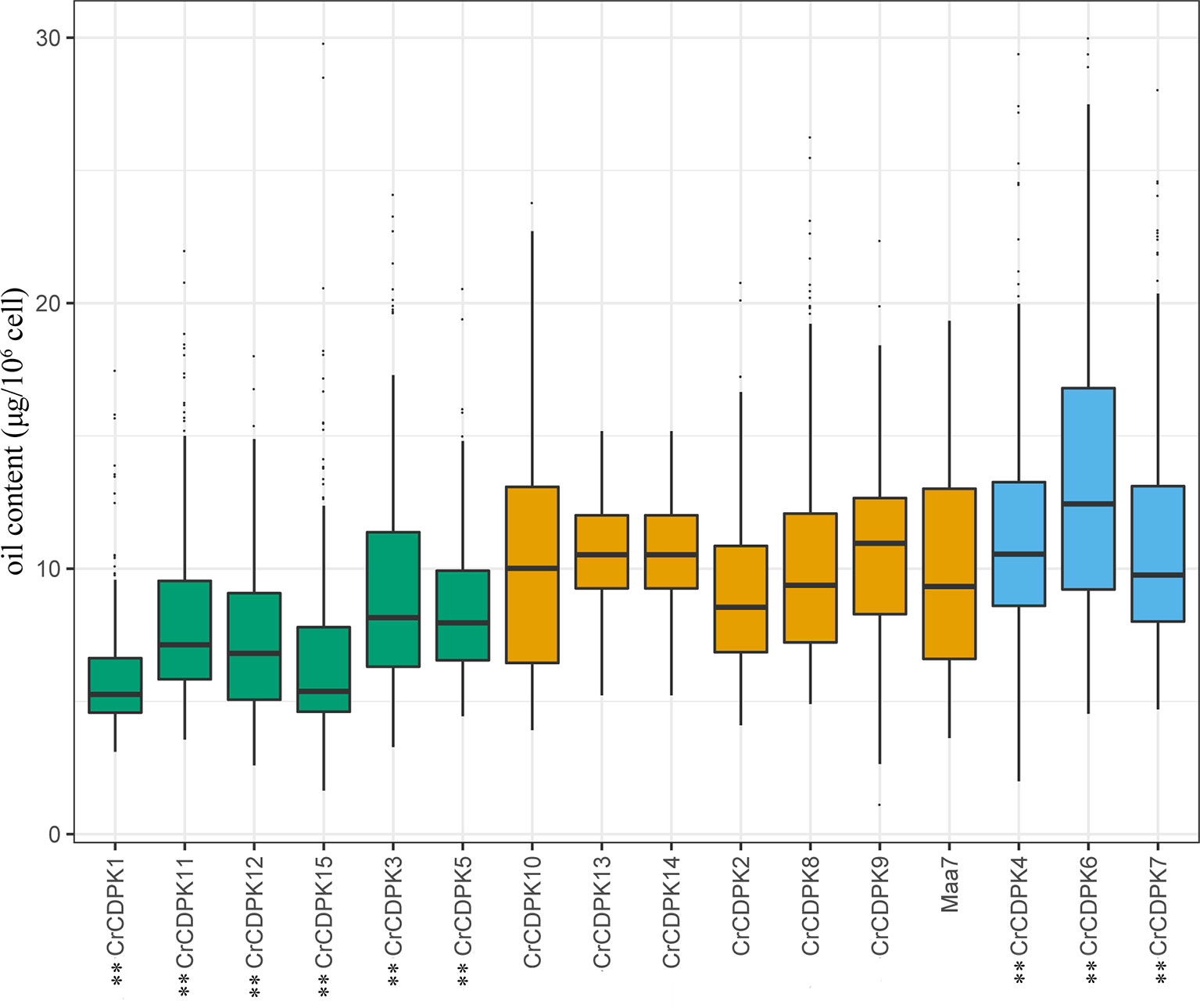
Figure 7 The oil contents of CrCDPK-silenced cells of Chlamydomonas reinhardtii grown under N-deficient conditions. CrCDPK RNA interference (RNAi)-silencing transgenic lines were inoculated into Tris–acetate–phosphate (TAP) agar medium containing 0.5 mM of ammonium and transferred into a 96-well plate containing sterile water for the measurement of oil content after 12 days of cultivation. Boxplots display medians with first and third quartiles, whereas whiskers indicate the smallest and largest non-outlier observations (n = 192). Maa7 is Chlamydomonas reinhardtii transformed with the empty vector Maa7/XIR, which was used as control. An independent-samples t-test was performed for statistical analysis. Significance is indicated as **p < 0.01.
To validate the oil changes in RNAi strains, CrCDPK3-, CrCDPK5-, and CrCDPK7-RNAi-silencing algal transformants were selected for log-phase culture. Maa7 empty vector transgenic lines were used as control. The results are shown in Figure 8. Under N-limited conditions, the cell density gradually increased from 1 to 5 days (Figure 8A), and the chlorophyll content sharply increased after 4 days and then decreased rapidly after 5 days (Figure 8C). No significant changes were observed in the cell density and chlorophyll content between the RNAi strains and control except for CrCDPK5-RNAi cells, which showed more cell density than the control. On the contrary, compared with the control, CrCDPK5-RNAi cells showed a significant decline in oil content from 3 to 5 days of N-limited cultivation, and CrCDPK3-RNAi cells exhibited lower oil content after 4 days. On the other hand, the oil content of CrCDPK7-RNAi was significantly increased after 5 days of cultivation (Figure 8B). This result was consistent with those obtained above.
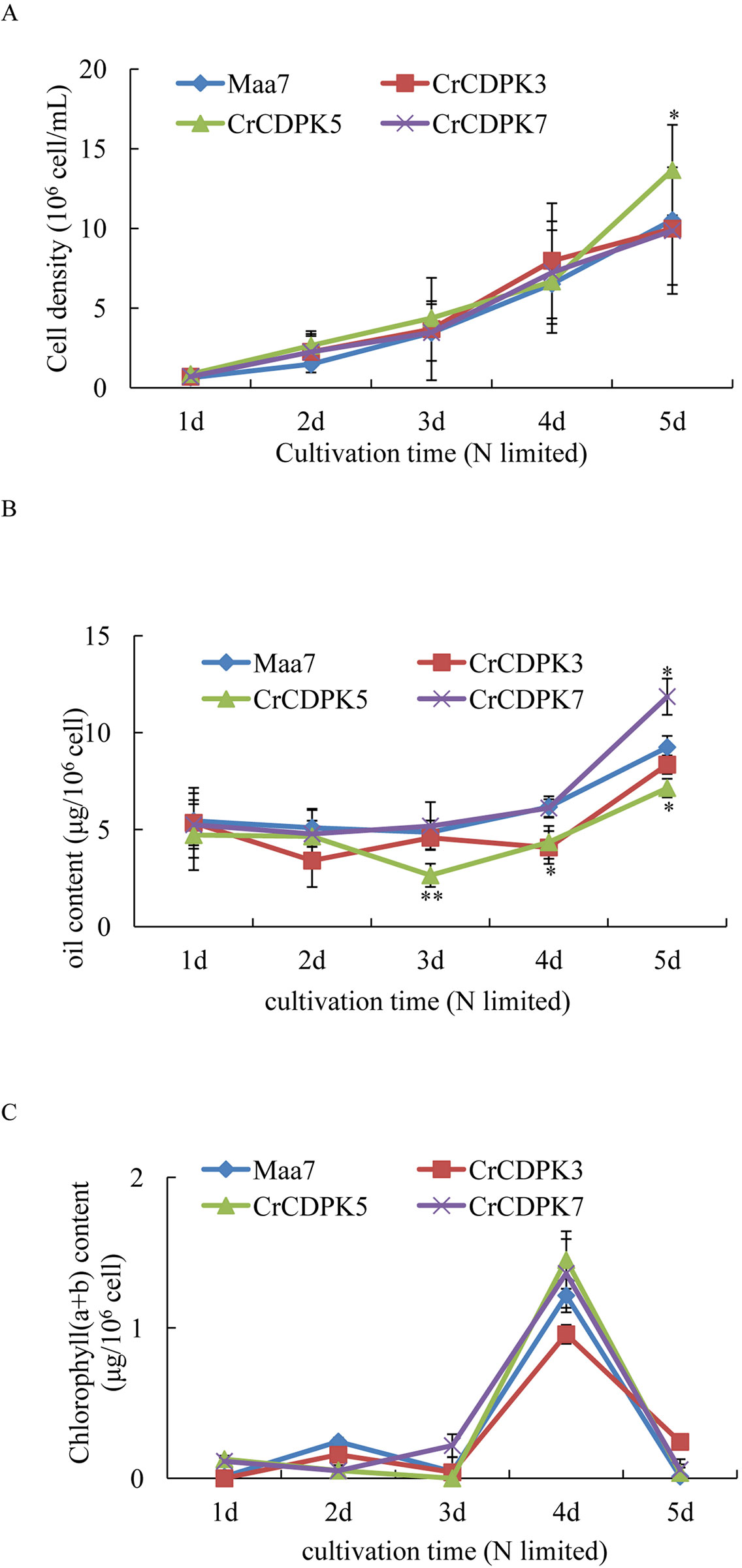
Figure 8 Cell density (A), oil content (B), and chlorophyll content (C) of CrCDPK3-, CrCDPK5-, and CrCDPK7-RNA interference (RNAi) strains. Maa7- and CrCDPK-RNAi strains grown on a Tris–acetate–phosphate (TAP) agar plate were inoculated into 100-ml Erlenmeyer flasks containing 50 ml of high salt minimal (HSM) media with 0.5 mM of ammonium (N-limited). Samples for cell density, chlorophyll content, and oil content measurement were collected after 1, 2, 3, 4, and 5 days. Statistical analysis was performed using the SPSS software by means of one-way ANOVA and Duncan’s multiple range tests. Data shown are means ( ± SD, n = 10). Asterisks indicate statistically significant differences compared with the corresponding control (Duncan’s multiple range test: *p < 0.05, **p < 0.01). This experiment was repeated three times with similar results.
Microalgae tend to accumulate oils in response to different environmental stresses with a slow growth rate. Several genetic strategies have been employed to obtain strains with high oil content and a rapid growth rate. However, they are inefficient due to the limited knowledge about the molecular mechanisms of stress-induced oil biosynthesis in microalgae. On the other hand, Ca2+ signaling is a core regulator of cellular responses to miscellaneous biotic and abiotic stresses (Dodd et al., 2010) and plays an important role in oil biosynthesis in mammals. Ca2+ imbalance may result in dysfunctions of adipose tissue in mammals. For example, a rise in cytosolic Ca2+ can promote TAG accumulation and storage in mice (Arruda and Hotamisligil, 2015). Lipid droplets may also accumulate in the liver, heart, and skeletal muscles of mice deficient in store-operated Ca2+ entry, which is a ubiquitous Ca2+ influx pathway (Maus et al., 2017). However, little information is available on the Ca2+ signaling pathway involved in stress-induced oil biosynthesis in microalgae. In the present study, LaCl3 at concentrations of 75 μM or higher has important consequences for N and P deficiency-induced oil accumulation in C. reinhardtii. After the application of LaCl3, the oil content of algal cells was significantly decreased compared to that of the control (Figure 6). As a plasma membrane calcium channel blocker, LaCl3 prevented Ca2+ influx via inhibiting Ca2+-ATPase activity. The results indicated that changes in Ca2+ homeostasis directly affected stress-induced oil accumulation in C. reinhardtii, in which Ca2+ is an important contributing factor.
Several Ca2+ sensors, including CaM and CaM-like proteins, CBL, and CDPKs, have been identified in organisms. However, only CDPKs display both Ca2+ sensing and protein kinase domains within a single protein. Thus, they can directly translate Ca2+ signals into downstream protein phosphorylation without protein partners (Boudsocq and Sheen, 2013). In recent years, CDPKs have attracted increasing attention due to their special feature. In this study, we investigated the CDPK gene family in the C. reinhardtii genome by using Arabidopsis and rice CDPK sequences as queries. A total of 15 non-redundant CrCDPK genes were identified from C. reinhardtii (Table 1), which was consistent with the results obtained by Hamel et al. (2014). All CrCDPK genes harbor a typical structure containing protein kinase and Ca2+ binding domains in C. reinhardtii (Figure 1), and all of them contain four EF-hand motifs except for CrCDPK10, which only has two EF-hands. In other plant species, such as tomato and potato, similar results were obtained: some CDPK genes have less than four EF-hands (Gromadka et al., 2018; Hu et al., 2016a; Hu et al., 2016b). Boudsocq et al. (2012) reported that the AtCPK25 gene is Ca2+ independent owing to the absence of functional EF-hands in Arabidopsis. The qRT-PCR results also showed that the expression level of CrCDPK10 under N-deficient conditions was quite similar with that under normal conditions (Figure 5), implying that CrCDPK10 probably has weak Ca2+-binding affinities and is insensitive to Ca2+ in C. reinhardtii.
In addition, eight CrCDPK genes were predicted to have myristoylation sites that play an essential role in membrane targeting and protein–protein interactions. On the basis of their phylogenetic relationships, all CrCDPKs were clustered into a distinct group along with a subclade of AtCPK16/AtCPK18/AtCPK28 and OsCPK4/OsCPK18/OsCPK30, whereas the remaining AtCPKs and OsCPKs were split into two other groups (Figure 2). The AtCPK28 gene responds to osmotic stress and controls stem elongation and vascular development in Arabidopsis (Matschi et al., 2013; Gao et al., 2014). OsCPK4 functions as a positive modulator of various abiotic stress and negatively regulates innate immunity in rice (Wang et al., 2018a). Because these known-function genes are involved in various physiological processes, the functions of CrCDPK genes are difficult to predict and remain unclear in C. reinhardtii.
To investigate the expression patterns of CrCDPK genes in response to abiotic stress, we determined their transcript levels in C. reinhardtii against N, P, and Fe deficiencies and salt and cold stresses. Interestingly, the expression profiles of CrCDPKs were paired except for CrCDPK15 and varied greatly between treatments. About half of CrCDPKs were significantly induced by N deficiency or salt stress. By contrast, CrCDPK genes were insensitive to P and Fe deficiencies and cold stress because no significant changes were observed in their messenger RNA (mRNA) levels under such conditions (Figure 4). Many salt stress-responsive CDPK genes have been identified in higher plants in previous studies. For instance, OsCPK12 positively regulates salt tolerance by reducing the accumulation of reactive oxygen species (Asano et al., 2012). Recently, CDPKs have been demonstrated to be related to N deficiency. The transcription levels of three OtCDPKs were upregulated by N depletion in marine green algae Ostreococcus tauri (Caló et al., 2017), and the mRNA levels of CDPK1 and CDPK3 from C. reinhardtii showed over 10-fold increase after 1 h of N starvation, followed by a decrease from 3 to 18 h (Motiwalla et al., 2014). These two C. reinhardtii CDPKs corresponded to CrCDPK8 and CrCDPK9 in our study. With the combination of the results of RNA sequencing and qRT-PCR analysis, CrCDPK8 was significantly downregulated by N deficiency after 48 h (Figure 5), and CrCDPK9 was significantly downregulated after 24 h (Figure 4).
As mentioned above, oil droplet biogenesis was prevented by over 75 μM of LaCl3 in C. reinhardtii (Figure 6), revealing that Ca2+ has an important regulatory role in N deficiency-induced oil accumulation. Ma et al. (2011) reported that TAG is dose-dependently decreased with increasing dietary calcium levels, and TAG transport protein is downregulated in hamster. Thus, the relationship between calcium and oil biosynthesis is still unclear. In higher plants, several CDPKs have been found to be associated with oil bodies in some oilseeds, such as sandalwood, cotton, and soybean, indicating that CDPK and Ca2+ may have a regulatory role during oil accumulation or oil body biogenesis (Anil et al., 2003). In the present study, to understand the functions of CrCDPKs in oil biosynthesis in C. reinhardtii, RNAi gene silencing was performed for all 15 CrCDPK genes. The transgenic cells containing the empty vector Maa7 XR/XIR were used as control. Six CrCDPK-silencing transgenic lines, especially CrCDPK1, exhibited lower levels of oil content than the control under N-deficient conditions (Figure 7). Oil content was significantly decreased by 39.02% after the CrCDPK1 gene was silenced in C. reinhardtii. Notably, the results of qRT-PCR showed that all of these six CrCDPK genes were upregulated after 24 h in response to N deficiency (Figure 5). The data indicated that these six CrCDPK genes, which include CrCDPK1, CrCDPK3, CrCDPK5, CrCDPK11, CrCDPK12, and CrCDPK15, play positive roles in oil accumulation in C. reinhardtii under N-deficient conditions. The other three CrCDPKs (CrCDPK4, CrCDPK6, and CrCDPK7) showed higher levels of oil content than the control (Figure 7), suggesting that they play negative roles in oil accumulation. In addition, according to their expression patterns examined by qRT-PCR (Figure 5), they may be involved in the different stages of oil biosynthesis in N-deficient cells in C. reinhardtii because CrCDPK4 was upregulated after 72 h, CrCDPK6 after 12 h, and CrCDPK7 after 24 h. The oil contents of the remaining six CrCDPK-silencing transgenic cells showed no significant changes compared with those of the control. This result was consistent with the observation that they were all insensitive to N deficiency at transcript levels (Figure 5).
In conclusion, there were at least 15 members of the CDPK gene family in the C. reinhardtii genome, and they all harbored the typical conserved domains, namely, the EF-hand Ca2+-binding domain and protein kinase domain. Phylogenetic analysis showed that these CrCDPKs were clustered into the same group, separating from most Arabidopsis and rice CDPK genes. The CrCDPK genes were widely distributed in the genome, and all of them contained 6–17 exons. Most CrCDPKs were transcriptionally insensitive to P and Fe deficiencies and cold stress after 24 h treatment, but half of them were significantly upregulated/downregulated by N deficiency and salt stress. Under N-deficient conditions, oil droplets disappeared in C. reinhardtii cells after application of over 100 μM of LaCl3. When the 15 CrCDPK genes were silenced in C. reinhardtii, six genes displayed lower levels of oil content, and three genes showed higher levels than the control. Correspondingly, nine CrCDPK genes were significantly upregulated by N deficiency at different times.
The data generated in this study can be found in the NCBI using accession numbers SRX7003584, SRX7003583, SRX7003582, SRX7003581, SRX7003580.
YL and XD designed the experiments, analyzed the data, and wrote the manuscript. YL, XF, HD, JL, and WZ conducted the experiments.
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31770272) and Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No. 1630052018001, No. 1630052016009, and No. 19CXTD-32).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01147/full#supplementary-material.
Almadanim, M. C., Gonçalves, N. M., Rosa, M. T. G., Alexandre, B. M., Cordeiro, A. M., Rodrigues, M., et al. (2018). The rice cold-responsive calcium-dependent protein kinase OsCPK17 is regulated by alternative splicing and post-translational modifications. BBA-Mol. Cell Res. 1865, 231. doi: 10.1016/j.bbamcr.2017.10.010
Anil, V. S., Harmon, A. C., Rao Sankara, K. (2003). Temporal association of Ca2+ dependent protein kinase with oil bodies during seed development in Santalum album L.: its biochemical characterization and significance. Plant Cell Physiol. 44 (4), 367–376. doi: 10.1093/pcp/pcg046
Arruda, A. P., Hotamisligil, G. S. (2015). Calcium homeostasis and organelle function in the pathogenesis of obesity and diabetes. Cell Metab. 22, 381–397. doi: 10.1016/j.cmet.2015.06.010
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi: 10.1007/s11103-010-9717-1
Asano, T., Hayashi, N., Kobayashi, M., Aoki, N., Miyao, A., Mitsuhara, I., et al. (2012). A rice calcium-dependent protein kinase OsCPK12 oppositely modulates salt-stress tolerance and blast disease resistance. Plant J. 69, 26–36. doi: 10.1111/j.1365-313X.2011.04766.x
Batistič, O., Kudla, J. (2012). Analysis of calcium signaling pathways in plants. BBA-Gen. Subj. 1820, 1238–1293. doi: 10.1016/j.bbagen.2011.10.012
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18 (1), 30–40. doi: 10.1016/j.tplants.2012.08.008
Boudsocq, M., Droillard, M. J., Regad, L., Laurière, C. (2012). Characterization of Arabidopsis calcium-dependent protein kinases: activated or not by calcium? Biochem. J. 447, 291–299. doi: 10.1042/BJ20112072
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 464, 418–422. doi: 10.1038/nature08794
Caló, G., Scheidegger, D., Martínez-Noël, G. M. A., Salerno, G. L. (2017). Ancient signal for nitrogen status sensing in the green lineage: functional evidence of CDPK repertoire in Ostreococcus tauri. Plant Physiol. Bioch. 118, 377–384. doi: 10.1016/j.plaphy.2017.07.009
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Choi, H. I., Park, H. J., Park, J. H., Kim, S., Im, M. Y., Seo, H. H., et al. (2005). Arabidopsis calcium-dependent protein kinase AtCPK32 interacts with ABF4, transcriptional regulator of abscisic acid-responsive gene expression, and modulates its activity. Plant Physiol. 139, 1750–1761. doi: 10.1104/pp.105.069757
Conte, M., Lupette, J., Seddiki, K., Meï, C., Dolch, L. J., Gros, V., et al. (2018). Screening for biologically annotated drugs that triggerw triacylglycerol accumulation in the diatom Phaeodactylum. Plant Physiol. 177 (2), 532–552. doi: 10.1104/pp.17.01804
Dodd, A. N., Kudla, J., Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Dubrovina, A. S., Kiselev, K. V., Khristenko, V. S., Aleynova, O. A. (2015). VaCPK20, a calcium-dependent protein kinase gene of wild grapevine Vitis amurensis Rupr., mediates cold and drought stress tolerance. J. Plant Physiol. 185, 1–12. doi: 10.1016/j.jplph.2015.05.020
Franz, S., Ehlert, B., Liese, A., Kurth, J., Cazalé, A. C., Romeis, T. (2011). Calcium-dependent protein kinase CPK21 functions in abiotic stress response in Arabidopsis thaliana. Mol. Plant 4, 83–96. doi: 10.1093/mp/ssq064
Gao, A., Wu, Q., Zhang, Y., Miao, Y., Song, C. (2014). Arabidopsis calcium-dependent protein kinase CPK28 is potentially involved in the response to osmotic stress. Chin. Sci. Bull. 59, 1113–1122. doi: 10.1007/s11434-013-0062-z
Gromadka, R., Cieśla, J., Olszak, K., Szczegielnniak, J., Muszyńska, G., Polkowska-Kowalczyk, L. (2018). Genome-wide analysis and expression profiling of calcium dependent protein kinases in potato (Solanum tuberosum). Plant Growth Regul. 84, 303–315. doi: 10.1007/s10725-017-0341-9
Gutermuth, T., Lassig, R., Portes, M. T., Maierhofer, T., Romeis, T., Borst, J. W., et al. (2013). Pollen tube growth regulation by free anions depends on the interaction between the anion channel SLAH3 and calcium-dependent protein kinases CPK2 and CPK20. Plant Cell 25, 4525–4543. doi: 10.1105/tpc.113.118463
Hamel, L. P., Sheen, J., Séguin, A. (2014). Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89. doi: 10.1016/j.tplants.2013.10.009
Hetherington, A. M., Brownlee, C. (2004). The generation of Ca2+ signals in plants. Annu. Rev. Plant Biol. 55, 401–427. doi: 10.1146/annurev.arplant.55.031903.141624
Hrabak, E. M., Chan, C. W. M., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Hu, Q., Sommerfeld, M., Jarvis, E., Ghirardi, M., Posewitz, M., Seibert, M., et al. (2008). Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 54, 621–639. doi: 10.1111/j.1365-313X.2008.03492.x
Hu, W., Hou, X., Xia, Z., Yan, Y., Wei, Y., Wang, L., et al. (2016a). Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava. Mol. Genet. Genomics 291, 241–253. doi: 10.1007/s00438-015-1103-x
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016b). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant Plant Sci. 7, 469. doi: 10.3389/fpls.2016.00469
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. R36: Genome Biol. 14 (4). doi: 10.1186/gb-2013-14-4-r36
Kindle, K. L. (1990). High frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 87, 1228–1232. doi: 10.1016/S0076-6879(98)97005-7
Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M., Shimamoto, K., et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. Plant Cell 19, 1065–1080. doi: 10.1105/tpc.106.048884
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomic. 14, 433. doi: 10.1186/1471-2164-14-433
Langmead, B., Trapnell, C., Pop, M., Salzberg, S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. R25: Genome Biol. 10 (3). doi: 10.1186/gb-2009-10-3-r25
Lenzoni, G., Liu, J., Knight, M. R. (2018). Predicting plant immunity gene expression by identifying the decoding mechanism of calcium signatures. New Phytol. 217, 1598–1609. doi: 10.1111/nph.14924
Li, A. L., Zhu, Y. F., Tan, X. M., Wang, X., Wei, B., Guo, H., et al. (2008). Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Mol. Biol. 66, 429–443. doi: 10.1007/s11103-007-9281-5
Li, Y., Fei, X., Deng, X. (2012a). Novel molecular insights into nitrogen starvation-induced triacylglycerols accumulation revealed by differential gene expression analysis in green algae Micractinium pusillum. Biomass. Bioenerg. 42, 199–211. doi: 10.1016/j.biombioe.2012.03.010
Li, Y. J., Deng, X. D., Yang, J. H., Fei, X. W. (2012b). A putative protein kinase Femu9p contributes to the iron deficiency-inducible expression of FOX1 gene in Chlamydomonas reinhardtii. Aust. J. Crop Sci. 6, 1724–1731.
Li, Y., Fei, X., Wu, X., Deng, X. (2017). Iron deficiency response gene Femu2 plays a positive roles in protecting Chlamydomonas reinhardtii against salt stress. BBA-Gen. Subj. 1861, 3345–3354. doi: 10.1016/j.bbagen.2016.08.017
Li-Beisson, Y., Thelen, J. J., Fedosejevs, E., Harwood, J. L. (2019). The lipid biochemistry of eukaryotic algae. Prog. Lipid Res. 74, 31–68. doi: 10.1016/j.plipres.2019.01.003
Liese, A., Romeis, T. (2013). Biochemical regulation of in vivo function of plant calcium-dependent protein kinases (CDPK). Biochim. Biophys. Acta. 1833, 1582–1589. doi: 10.1016/j.bbamcr.2012.10.024
Liu, B., Benning, C. (2013). Lipid metabolism in microalgae distinguishes itself. Curr. Opin. Biotech. 24, 300–309. doi: 10.1016/j.copbio.2012.08.008
Liu, W., Li, W., He, Q., Daud, M., Chen, J., Zhu, S. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. Plos One 9, e98189. doi: 10.1371/journal.pone.0098189
Liu, Y., Xu, C., Zhu, Y., Zhang, L., Chen, T., Zhou, F., et al. (2018). The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 60, 173–188. doi: 10.1111/jipb.12614
Ma, K. Y., Yang, N., Jiao, R., Peng, C., Lei, G., Huang, Y., et al. (2011). Dietary calcium decreased plasma cholesterol by down-regulation of intestinal Niemann-Pick C1 like 1 and microsomal triacylglycerol transport protein and up-regulation of CYP7A1 and ABCG 5/8 in hamsters. Mol. Nutr. Food Res. 55, 247–258. doi: 10.1002/mnfr.201000161
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., Romeis, T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73 (3), 883–896. doi: 10.1111/tpj.12090
Maus, M., Cuk, M., Patel, B., Lian, J., Oumet, M., Kaufmann, U., et al. (2017). Store-operated Ca2+ entry controls induction of lipolysis and the transcriptional reprogramming to lipid metabolism. Cell Metab. 25, 698–712. doi: 10.1016/j.cmet.2016.12.021
Mori, I. C., Murata, Y., Yang, Y., Munemasa, S., Wang, Y. F., Andreoli, S., et al. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4, e327. doi: 10.1371/journal.pbio.0040327
Motiwalla, M. J., Sequeira, M. P., D’Souza, J. S. (2014). Two calcium-dependent protein kinases from Chlamydomonas reinhardtii are transcriptionally regulated by nutrient starvation. Plant Signal Behav. 2014 9, e27969. doi: 10.4161/psb.27969
Park, J. J., Wang, H., Gargouri, M., Deshpande, R. R., Skepper, J. N., Holguin, F. O., et al. (2015). The response of Chlamydomonas reinhardtii to nitrogen deprivation: a systems biology analysis. Plant J. 81 (4), 611–624. doi: 10.1111/tpj.12747
Ray, S., Agarwal, P., Arora, R., Kapoor, S., Tyagi, A. K. (2007). Expression analysis of calcium-dependent protein kinase gene family during reproductive development and abiotic stress conditions in rice (Oryza sativa L. ssp. indica). Mol. Genet. Genomics 278, 493–505. doi: 10.1007/s00438-007-0267-4
Rohr, J., Sarkar, N., Balenger, S., Jeong, B., Cerutti, H. (2004). Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 40, 611–621. doi: 10.1111/j.1365-313X.2004.02227.x
Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. doi: 10.1046/j.1365-313x.2000.00787.x
Schulz, P., Herde, M., Romeis, T. (2013). Calcium-dependent protein kinases: hubs in plant stress signaling and development. Plant Physiol. 163, 523–530. doi: 10.1104/pp.113.222539
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Barenm, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28 (5), 511–515. doi: 10.1038/nbt.1621
Wang, J., Wang, S., Hu, K., Yang, J., Xin, X., Zhou, W., et al. (2018a). The kinase OsCPK4 regulates a buffering mechanism that fine-tunes innate immunity. Plant Physiol. 176, 1835–1849. doi: 10.1104/pp.17.01024
Wang, W., Zhang, H., Wei, X., Yang, L., Yang, B., Zhang, L., et al. (2018b). Functional characterization of calcium-dependent protein kinase (CPK)2 gene from oilseed rape (Brassica napus L.) in regulating reactive oxygen species signaling and cell death control. Gene 651, 49–56. doi: 10.1016/j.gene.2018.02.006
Wase, N., Black, P., DiRusso, C. (2018). Innovations in improving lipid production: algal chemical genetics. Prog. Lipid Res. 71, 101–123. doi: 10.1016/j.plipres.2018.07.001
Wase, N., Black, P. N., Stanley, B. A., DiRusso, C. C. (2014). Integrated quantitative analysis of nitrogen stress response in Chlamydomonas reinhardtii using metabolite and protein profiling. J. Proteome Res. 13 (3), 1373–1396. doi: 10.1021/pr400952z
Wase, N., Tu, B., Allen, J. W., Black, P. N., DiRusso, C. C. (2017). Identification and metabolite profiling of chemical activators of lipid accumulation in green algae. Plant Physiol. 174 (4), 2146–2165. doi: 10.1104/pp.17.00433
Weckwerth, P., Ehlert, B., Romeis, T. (2015). ZmCPK1, a calcium-independent kinase member of the Zea mays CDPK gene family, functions as a negative regulator in cold stress signaling. Plant Cell Environ. 38, 544–558. doi: 10.1111/pce.12414
Wei, S., Hu, W., Deng, X., Zhang, Y., Liu, X., Zhao, X., et al. (2014). A rice calcium dependent protein kinase OsCPK9 positively regulates drought stress tolerance and spikelet fertility. BMC Plant Biol. 14, 133. doi: 10.1186/1471-2229-14-133
Zhao, L. N., Shen, L. K., Zhang, W. Z., Zhang, W., Wang, Y., Wu, W. H. (2013). Ca2+-dependent protein kinase 11 and 24 modulate the activity of the inward rectifying K+ channels in Arabidopsis pollen tubes. Plant Cell 25, 649–661. doi: 10.1105/tpc.112.103184
Zhao, R., Sun, H. L., Mei, C., Wang, X. J., Yan, L., Liu, R., et al. (2011). The Arabidopsis Ca2+-dependent protein kinase CPK12 negatively regulates abscisic acid signaling in seed germination and post-germination growth. New Phytol. 192, 61–73. doi: 10.1111/j.1469-8137.2011.03793.x
Zhou, J. J., Li, X. D., Ratnasekera, D., Wang, C., Liu, X. X., Song, L. F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. The Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Zhu, S. Y., Yu, X. C., Wang, X. J., Zhao, R., Li, Y., Fan, R. C., et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19, 3019–3036. doi: 10.1105/tpc.107.050666
Keywords: Chlamydomonas reinhardtii, Ca2+-dependent protein kinases, expression pattern, gene function, nitrogen deficiency, oil accumulation
Citation: Li Y, Fei X, Dai H, Li J, Zhu W and Deng X (2019) Genome-Wide Identification of Calcium-Dependent Protein Kinases in Chlamydomonas reinhardtii and Functional Analyses in Nitrogen Deficiency-Induced Oil Accumulation. Front. Plant Sci. 10:1147. doi: 10.3389/fpls.2019.01147
Received: 19 March 2019; Accepted: 22 August 2019;
Published: 22 October 2019.
Edited by:
Basil J. Nikolau, Iowa State University, United StatesReviewed by:
Nishikant Wase, University of Nebraska-Lincoln, United StatesCopyright © 2019 Li, Fei, Dai, Li, Zhu and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaodong Deng, ZGVuZ3hpYW9kb25nQGl0YmIub3JnLmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.