- 1School of Life Sciences, Chongqing University, Chongqing, China
- 2School of Chemistry and Chemical Engineering, Chongqing University of Science and Technology, Chongqing, China
- 3Department of Cellular and Structural Biology, UT Health San Antonio, San Antonio, TX, United States
- 4Zhengzhou Research Base, State Key Laboratory of Cotton Biology, Zhengzhou University, Zhengzhou, China
- 5State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
Melatonin functions as a plant hormone/regulator in the regulation of growth and development. However, the underlying mechanisms are still unclear. In this study, we found that a high dose of melatonin inhibited hypocotyl elongation in a dose-dependent manner in Arabidopsis. An expression profile analysis showed that hypocotyl growth inhibition by melatonin was involved in reprograming the expression of cell elongation genes and brassinosteroid (BRs) biosynthetic genes. Furthermore, similar to BR biosynthetic inhibitor brassinazole (BRZ), a high concentration of melatonin upregulated BR-biosynthetic genes and downregulated BR-induced genes involved in cell elongation, while melatonin was inefficient in brassinazole-resistant mutants like the bzr1-1D and bes1-D in hypocotyl inhibition. The comparative expression profile analysis showed an opposite expression mode in the co-regulated genes between melatonin and BZR1 or melatonin and brassinolide (BL). Additionally, exogenous BL rescued the repressive phenotype of BR biosynthesis-deficient mutant like det2-1 even in the presence of high-dose melatonin, but not BR receptor mutant bri1-5 or signal transduction mutant bin2-1. A biochemical analysis further confirmed that melatonin reduced endogenous BR levels in a dose-dependent manner in Arabidopsis. Taken together, these results indicate that melatonin inhibits BR biosynthesis but does not block BR signaling in the inhibition of hypocotyl elongation and extends insights on the role of melatonin in cross-talking with plant hormone signaling.
Introduction
Melatonin, chemically N-acetyl-5-methoxytryptamine, is a low-molecular-weight substance found in living organisms ranging from bacteria to animals, with pleiotropic biological activities (Hardeland et al., 2006; Hardeland et al., 2011; Reiter et al., 2017). In plants, melatonin has a wide range of physiological functions such as regulating plant stress tolerance, senescence retardation, and pathogen defense. Additionally, melatonin also participates in the processes of root growth and architecture, flowering, seed germination, and fruit ripening (Arnao and Hernández-Ruiz, 2014; Arnao and Hernández-Ruiz, 2015; Wei et al., 2015; Nawaz et al., 2016; Kanwar et al., 2018). Regarding growth regulation, melatonin has been found to promote growth in shoots, roots, and also explants. Melatonin reportedly stimulated root generation and cotyledon expansion, induced new lateral and adventitious roots, and also extended the coleoptiles in some monocot plants (Murch et al., 2001; Hernandez-Ruiz et al., 2005; Pelagio-Flores et al., 2013; Sarropoulou et al., 2012; Hernandez et al., 2015; Erland et al., 2018). Melatonin also acts as a growth promoter inducing active growth of hypocotyls in etiolated seedlings of lupin at micromolar concentrations while displaying an inhibitory effect at 100 μM (Hernandez-Ruiz et al., 2004). Generally, growth inhibition by melatonin only occurs at a high concentration (> 100 μM). High-concentration melatonin repressed root growth and reduced biomass, as well as retarded plant leaf growth by reducing both cell size and cell proliferation (Wang et al., 2017; Lin et al., 2018). Although melatonin plays an important role in the regulation of plant growth and development, the underlying mechanism is still unclear and remains to be elucidated.
In early studies, melatonin was considered primarily as an auxin-like effector in regulating of plant growth. However, to date, the accumulated data suggested that melatonin functions as a new plant hormone or regulator with versatile and complex activities in which it participates in the regulation of gene expression related to plant hormones including auxin, cytokinin, gibberellins (GAs), abscisic acid, ethylene, jasmonic acid, and salicylic acid (Arnao and Hernández-Ruiz, 2018; Arnao and Hernández-Ruiz, 2019). Melatonin showed effects on inducing expression of phytohormone-related genes, such as biosynthesis, catabolism, receptors, and transcription factors, and also influencing the endogenous plant hormone levels and their physiological actions. A recent investigation showed that a drop in melatonin by knockdown of the key enzyme gene like the SNAT2 (serotonin N-acetyltransferase 2) and TDC (tryptophan decarboxylase), involved in melatonin biosynthesis, resulted in downregulation of the expression of DWARF4 gene and reduction on endogenous brassinosteroid (BR) levels as well as a semi-dwarf phenotype in rice (Hwang and Back, 2018; Lee and Back, 2018), implying that there exists a close relationship between melatonin and the BR signaling pathway.
BRs, a class of plant-specific steroid hormones, play vital roles in regulating plant cell growth and morphogenesis, particularly in hypocotyl cell elongation, as the BR biosynthetic mutants, such as de-etiolated 2 (det2), are defective in hypocotyl elongation and shows a dwarf phenotype (Li et al., 1996; Fujioka and Sakurai 1997; Clouse and Sasse, 1998). In BR signaling, BRs are perceived by the BR receptor BR-insensitive 1 (BRI1), which initiates a signaling cascade to inactivate the negative regulator BR insensitive 2 (BIN2), thereby activating the downstream transcription factors brassinazole-resistant 1 (BZR1) and bri1-EMS suppressor 1 (BES1), two key transcription factors mediating BR responses (Li and Chory, 1997; Li and Nam, 2002; Xu et al., 2008; Wang et al., 2011; Wang et al., 2012; Zhu et al., 2013). BZR1 and BES1 play central roles in BR-regulated gene expression and plant development. Activated BZR1 and BES1 accumulate in the nucleus and promote stem elongation through the transcriptional activation of their target genes, such as PER1, IAA19, small auxin upregulated RNA (SAUR), and Expansin A8 (EXPA8), involved in cell elongation (Sun et al., 2010; Yu et al., 2011; Bai et al., 2012; Wang et al., 2012, Nolan et al., 2017), whereas negatively regulated by feedback suppression of expression of BR-biosynthetic genes like DWF4, CPD, and BR6OX2 (Mathur et al., 1998; Asami et al., 2001; Wang et al., 2002; Fujioka and Yokota, 2003; He et al., 2005).
In this study, we found that melatonin inhibited hypocotyl elongation and reprogramed expression of cell elongation genes and BR-biosynthetic genes in a dose-dependent manner in Arabidopsis. Through a transcriptome analysis in combination with gene expression analysis, we also found that melatonin inhibited hypocotyl elongation involved in repressing BR pathway. Additionally, we demonstrated that melatonin inhibited BR biosynthesis but did not block BR signaling in the regulation of hypocotyl elongation in Arabidopsis. Our findings provide new insights into the function of melatonin in the regulation of plant hormone signaling pathways.
Materials and Methods
Plant Materials and Growth Conditions
The wild-type (WT) Arabidopsis Columbia (Col-0) ecotype was used unless otherwise indicated. The mutants used in this study include BR-related mutants det2-1 (Chory et al., 1991), bri1-5 (Noguchi et al., 1999), bin2-1 (Peng et al., 2008), bzr1-1D (Wang et al., 2002), and bes1-D (Yin et al., 2002), in which bri1-5 and bes1-D are in Wassilewskija (Ws) and Ertkheim-2 (En2) backgrounds, respectively, and the others are in Col-0 background. Seeds were surface sterilized with 70% (v/v) ethanol for 2 min, followed by 10% (v/v) bleach solution for 5 min, and rinsed with sterile water five times. The seeds were then placed on 1/2 strength Murashige and Skoog (MS) medium (pH = 5.8) containing 1% sucrose and 0.8% agar. Seeds on medium were grown in growth chambers with 22°C and 16-h/8-h light/dark cycle settings unless indicated otherwise.
Hormone or Inhibitor Treatments and Hypocotyl Elongation Assays
As for the observation of hypocotyl experiments, seeds were incubated 1/2MS medium with various concentrations of melatonin (MEL; Sigma, M5250) or 0.1% dimethyl sulfoxide (DMSO; solvent control) for 7 days or more in the light conditions. For another treatment, 4-day-old seedlings were transferred on medium plates supplemented with melatonin, brassinazole (BRZ; Sigma, SML1406) dissolved in DMSO, brassinolide (BL; Sigma, B1439), indole acetic acid (IAA; Sigma, I5148), gibberellin (GA3, Sigma, G7645), or the combination for treatment in darkness for another 3 days. BL, IAA, and GA3 were dissolved in 80% ethanol. After the indicated time of incubation, about 40 seedlings were laid horizontally on an agar plate and photographed. Hypocotyl length was measured using the ImageJ software (http://imagej.nih.gov/ij/) from digitally computed images of seedlings.
RNA-Seq Analysis
RNA-Sequencing was carried out by BGI Life Tech Co. Ltd. (Wuhan, China). Four-day-old seedlings were transferred on 1/2MS plate containing 1 mM of melatonin or 0.1% DMSO (control). After 3 days of incubation in a dark condition, about 100 seedlings were collected as one replicate, and a total of three replicates were used for each treatment. Total RNA was isolated using RNeasy Plant Mini Kit (Tiangen, China) following the manufacturer’s protocol. The cDNA libraries were constructed, and the library quality was determined by using Bioanalyzer 2100 (Agilent), subsequently sequenced using the BGISEQ-500 platform (BGI). Clean reads obtained from the raw reads were obtained by removing the adapter and low-quality sequences and then mapped to the annotated genome sequence of Arabidopsis in The Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/) by using HISAT and Bowtie2 software (Langmead and Salzberg, 2012; Kim et al., 2015). Transcript abundance was also normalized by transforming the data to reads per kilobase of exon model per million mapped reads (FPKM) method, and DEGseq method was used to identify differentially expressed genes (DEGs) (Wang et al., 2010). A twofold difference in expression levels with an adjusted p-value ≤ 0.001 (q-value ≤ 0.001) was set as the threshold for the determination of the significant changes (Storey and Tibshirani, 2003). Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were carried out with the web tool DAVID bioinformatics resources 6.8 (http://david.abcc.ncifcrf.gov/home.jsp) (Huang da et al., 2009a; Huang da et al., 2009b) and KOBAS (http://kobas.cbi.pku.edu.cn/home.do) (Kanehisa et al., 2008; Kanehisa et al., 2016). All RNA-Seq data are available in Supplementary Table S1.
Comparative Analysis of RNA-Seq Data
RNA-Seq was compared with the public online data from published articles. DEGs with 1.5-fold difference were set to keep the consistence between the data. Venn diagrams for expression differences were generated online (http://bioinfogp.cnb.csic.es/tools/venny/). Expression levels of overlapping DEGs were exhibited by heatmaps created by using the MeV4.9.0 software (Saeed et al., 2003). The overlapping DEGs are listed in Supplementary Table S2–S3.
RNA Isolation and Real-Time PCR (qRT-PCR) Analysis
Total RNA was isolated by using the RNAprep Pure Plant Kit (TianGen, DP432), and reverse transcription was performed using the PrimeScript™ RT Reagent Kit (Takara, RR047A) following the manufacturer’s instructions. Real-time polymerase chain reaction (qRT-PCR) was performed on a Bio-Rad CFX96 System (Bio-Rad) using the TB Green™ Premix Ex Taq™ (Takara, RR420A). ACTIN2 was used as internal control for BR-related experiments. Primer sequences used for qRT-PCR analyses are listed in Supplementary Table S4.
Bioactive BRs Determination by ELISA
Ten-day-old seedlings grown on 1/2MS, including various concentrations of melatonin (100, 500, and 1,000 μM), 1 μM of BRZ, or 0.1% DMSO (control), were collected for determining BR levels. Bioactive BR contents were quantified by enzyme-linked immunosorbent assay (ELISA) as described previously (Hwang and Back, 2018).
Data Processing
The presented results were obtained from three independent experiments. Data were expressed as means ± standard deviations (SDs) of 40 plants (n ≥ 40) with each measurement performed in triplicate. The data were evaluated by a one-way analysis of variance (ANOVA), followed by Student tests at a significance level of p < 0.05 (*) or extremely significant level of p < 0.01 (**).
Accession Numbers
Information for the main genes involving in this study is available in The TAIR (http://www.arabidopsis.org/) under the following accession numbers: Paclobutrazol resistance1 (PRE1) (AT5G39860), IAA19 (AT3G15540), ACS5 (AT5G65800), SAUR15 (AT4G38850), SAUR16 (AT4G38860), EXPA8 (AT2G40610), DWF4 (AT3G50660), CPD (AT5G05690), BR6OX2 (AT3G30180), and ACTIN2 (AT3G18780).
Results
Melatonin Inhibits Hypocotyl Elongation of Arabidopsis
Melatonin as a plant growth regulator promotes or inhibits plant growth depending on the concentration of melatonin. To understand how melatonin affects plant growth, a series of concentrations of melatonin (0, 1, 10, 100, 500, and 1,000 μM) were set to assess its effects on seedling growth. After 10 days of germination, coincident with previous report, seedlings grown on 1/2MS plates with high-dose melatonin showed obvious growth arrest with smaller cotyledon and true leaves compared with those in solvent control (Supplementary Figure S1A). Melatonin reportedly promotes hypocotyl growth in etiolated lupin (Lupinus albus) at a micromolar concentration level, while it has an inhibitory effect at high concentrations (Hernandez-Ruiz et al., 2004). Similarly, our observation showed that the Arabidopsis seedlings also displayed a repressed hypocotyl and cell length by high-dose melatonin in a dose-dependent manner (Supplementary Figures S1B, C). Specifically, seedlings had displayed slight response to low-dose melatonin (1 and 10 μM); however, when concentrations of melatonin were increased (100, 500, and 1,000 μM), seedlings showed considerable reduction in hypocotyl length, which declined to about 80%, 60%, and 40%, respectively, compared with that of solvent control (Supplementary Figure S1D). To extend these findings and confirm inhibition in cell elongation of high dose of melatonin, we transferred 4 days seedlings to 1/2MS plates with various concentrations of melatonin (100, 500, and 1,000 μM) and incubation in a dark condition to better observe cell elongation. After 3 days of incubation, similar results were observed in which seedlings showed obvious reduction in hypocotyl and cell length as with increasing dose of melatonin in comparison with solvent control (Figures 1A–C).
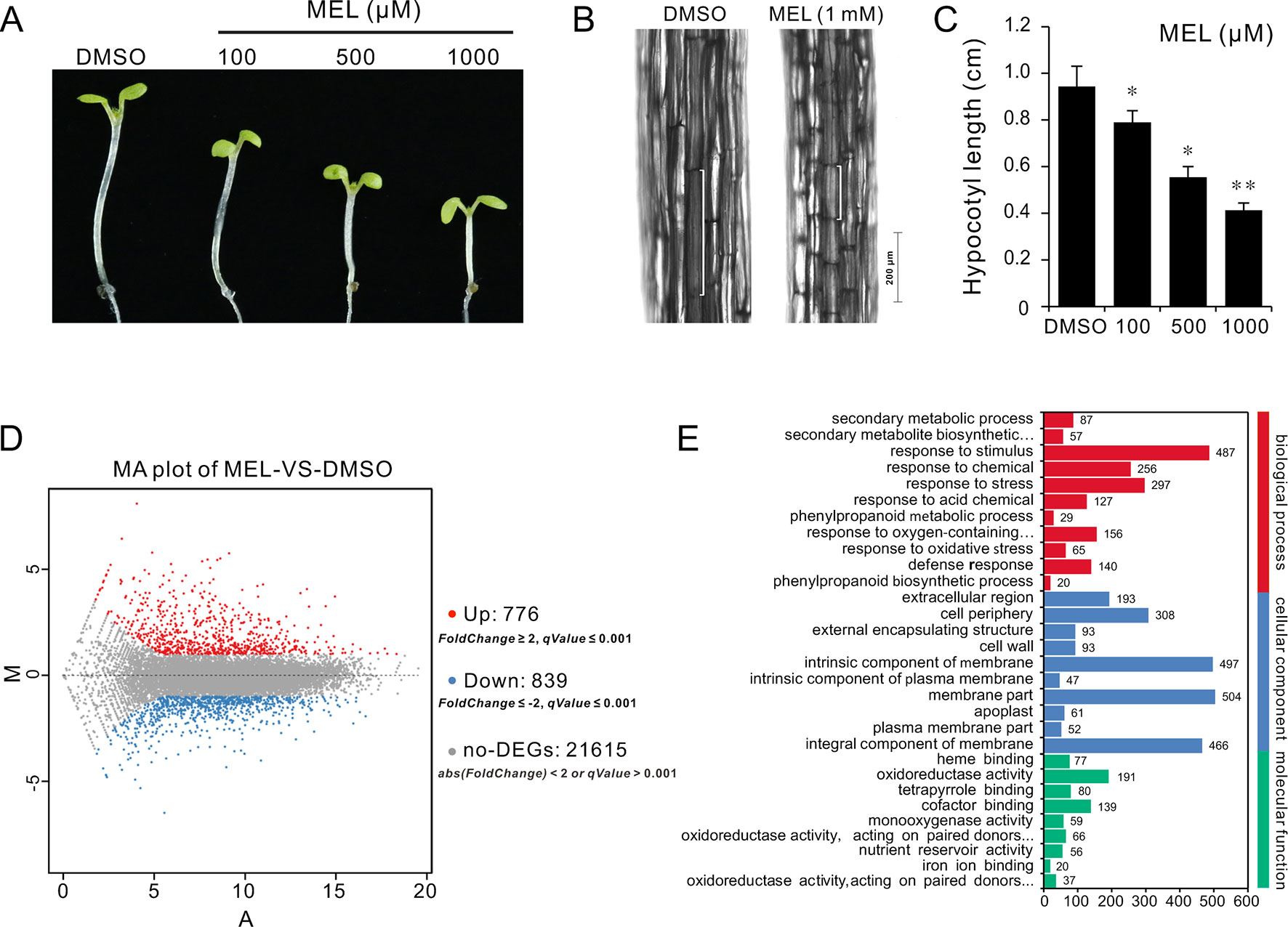
Figure 1 RNA-Seq of analysis genes affected by melatonin. Seedlings of four DAGs were transferred on 1/2MS plate containing different concentrations of melatonin (100, 500, and 1,000 µM) or 0.1% DMSO (solvent control). After 3 days of incubation in a dark condition, seedlings from 1 mM of melatonin and DMSO treatment were used for gene expression profile analysis by RNA-Seq. (A,B) Melatonin inhibits hypocotyl and cell elongation in a dose-dependent manner. (C) Hypocotyl length of seedlings. Error bars indicate the SEM (n ≥ 40). *p < 0.05, **p < 0.01 (Student’s t test). (D) DEGs affected by 1 mM of melatonin. (E) The top 30 of the most enriched GO terms (q-value < 0.01). DAG, days after germination; DMSO, dimethyl sulfoxide; GO, gene ontology; MS, Murashige and Skoog; SEM, standard error of the mean.
Melatonin Reprograms Cell Elongation Gene Expression
To investigate the mechanism of hypocotyl elongation inhibition caused by melatonin, we performed a whole-genome expression profiling analysis between melatonin and DMSO treatment (DMSO as the solvent) by RNA sequencing. RNA-Seq results showed that there were 1,615 DEGs (7.47% of all unigenes), consisting of 776 upregulated DEGs and 839 downregulated DEGs (Figure 1D, Supplementary Table S1). The GO enrichment analysis showed that the DEGs were categorized into 68 significantly (q-value < 0.01) enriched GO terms including 17 terms of cellular component in which typical terms in respect to cell elongation, such as “cell wall” and “plant-type cell wall,” were included (Supplementary Table S5). In the top 30 of the most enriched GO terms, there were 11 terms involved in cellular component (Figure 1E). As far as the number of genes are concerned, many DEGs were enriched in cellular component ontology following the terms of “membrane part (504),” “intrinsic components of membrane (497),” and “integral component of membrane (466),” etc. (Figure 1E). Furthermore, the KEGG pathway analysis showed that the majority of DEGs were mainly distributed on 26 significantly enriched KEGG pathways (q-value < 0.05), such as “Phenylpropanoid biosynthesis,” “Biosynthesis of secondary metabolites,” and “Metabolic pathways” (Figure 2A, Supplementary Table S6). In addition, we found that “brassinosteroid biosynthesis” pathway displayed in the top 20 of most significantly enriched KEGG pathways (Figure 2A), and BR biosynthesis-related genes, such as three cytochrome P450 superfamily genes (CYP724A1, CYP90D1, and BR6OX2) and one abietane diterpene oxidase gene ADTO1 (Fujioka and Yokota, 2003), showed differential expression (Figure 2B, Table 1), suggesting that melatonin affects the BR metabolism.
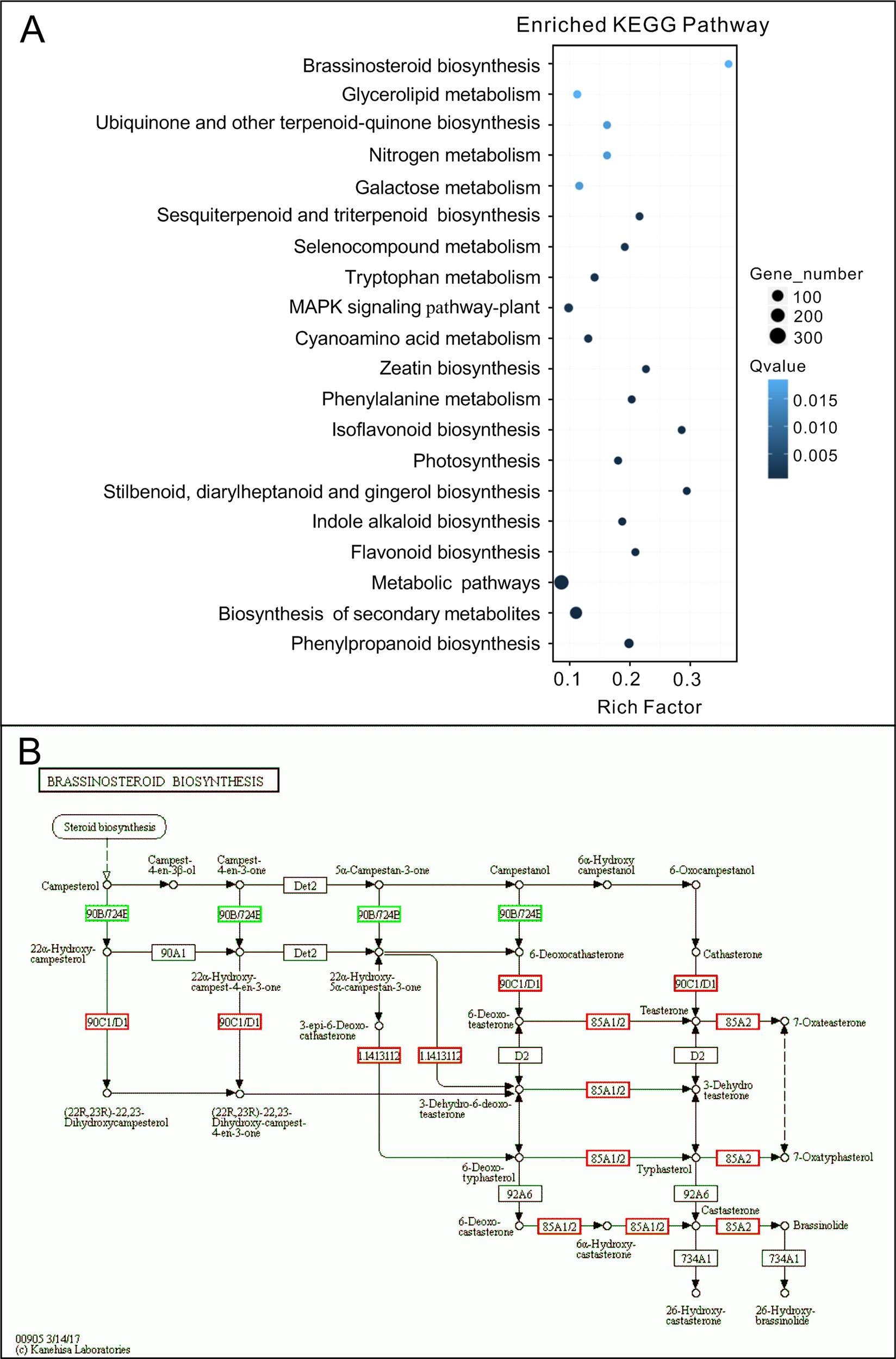
Figure 2 KEGG analysis of DEGs affected by melatonin. (A) The top 20 of the most significantly enriched KEGG pathways (q-value < 0.05). (B) Expression changes of genes in the brassinosteroid biosynthesis pathway. Red boxes, upregulated genes; green boxes, downregulated genes. KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.

Table 1 Differentially expressed genes involved in brassinosteroid biosynthesis and cell elongation.
A further analysis showed that there were 52 DEGs in respect to cell elongation/expansion. Among these DEGs, most of them (38 out of 51) were downregulated and only 13 DEGs were upregulated (Table 1). In addition, SAUR genes involved in auxin response and functions on the regulation of cell elongation and cell expansion (Knauss et al., 2003; Jain and Khurana, 2009; Ren and Gray, 2015) accounted for 37% (19 out of 51) and displayed consistent downregulated expression (Table 1). In these SAURs, most of them, such as SAUR15, SAUR16, and SAUR19 to SAUR24, have also been proven to respond to BR signaling in the regulation of hypocotyl growth (Favero et al., 2017). Furthermore, the BR-induced genes as well the marker genes like the PRE1 and indole-3-acetic acid inducible 19 (IAA19) also displayed decreasing transcript levels (Table 1). These results suggest that a high concentration of melatonin represses hypocotyl elongation in relation to reprograming cell elongation/expansion genes expression likely through regulating BR metabolism.
Melatonin Regulates BR-Related Genes Expression
To investigate the regulatory relationship between melatonin and BR signaling, we screened the BR biosynthesis-related genes and representative BR-induced genes involved in cell elongation in the total detected genes of RNA-Seq data. Except of CYP724A1, CYP90D1, BR6OX2, and ADTO1, the key rate-limiting enzyme gene CPD in BR biosynthesis also displayed differential upregulated expression at 1.5-fold change threshold (Supplementary Table S1). Conversely, in addition to PER1 and IAA19, BRs-induced as well as marker genes regulating cell elongation, like the SAUR15, SAUR16, and EXPA8, exhibited consistent downregulated expression with a 1.5-fold change threshold. Furthermore, ACS5, usually taken as a marker gene in response to BRs, exhibited reduced transcript level as well (Table 1, Supplementary Table S1). We next performed qRT-PCR to validate the expression levels of these BR-biosynthetic and BR-induced genes in the seedlings after being treated with various concentrations of melatonin. Except of the two genes CYP724A1 and ADTO1, which showed smooth expression levels, a consistent trend of expression was obtained with these detected genes (Figure 3A). BR-biosynthetic genes (DWF4, CPD, CYP90D1, and BR6OX2) showed dramatic enhancement with increasing concentrations of melatonin, especially those of DWF4 and BR6OX2 (Figure 3A). On the contrary, the BR-induced genes (PER1, IAA19, ACS5, SAUR15, SAUR16, and EXPA8) displayed an identical decline in transcript level caused by increasing dose of melatonin (Figure 3B). Additionally, we also detected the transcript levels of BZR1 and BES1, two key transcription factors in BR signaling, which promote cell elongation through transcriptional activation of BR-induced genes, such as PER1, IAA19, SAURs, and EXPA8, whereas promote feedback suppression in the expression of BR-biosynthetic genes (like the DWF4, CPD, and BR6OX2) (Sun et al., 2010; Yu et al., 2011; Bai et al., 2012; Wang et al., 2012; Nolan et al., 2017). Consistent with BR-induced genes, BZR1 and BES1 also showed downregulated trend in transcript levels with increasing dose of melatonin (Figure 3C). These results are consistent with the gene expression of the inhibition of BR signaling, implying that a high dose of melatonin represses BR signaling pathway in the regulation of hypocotyl elongation.
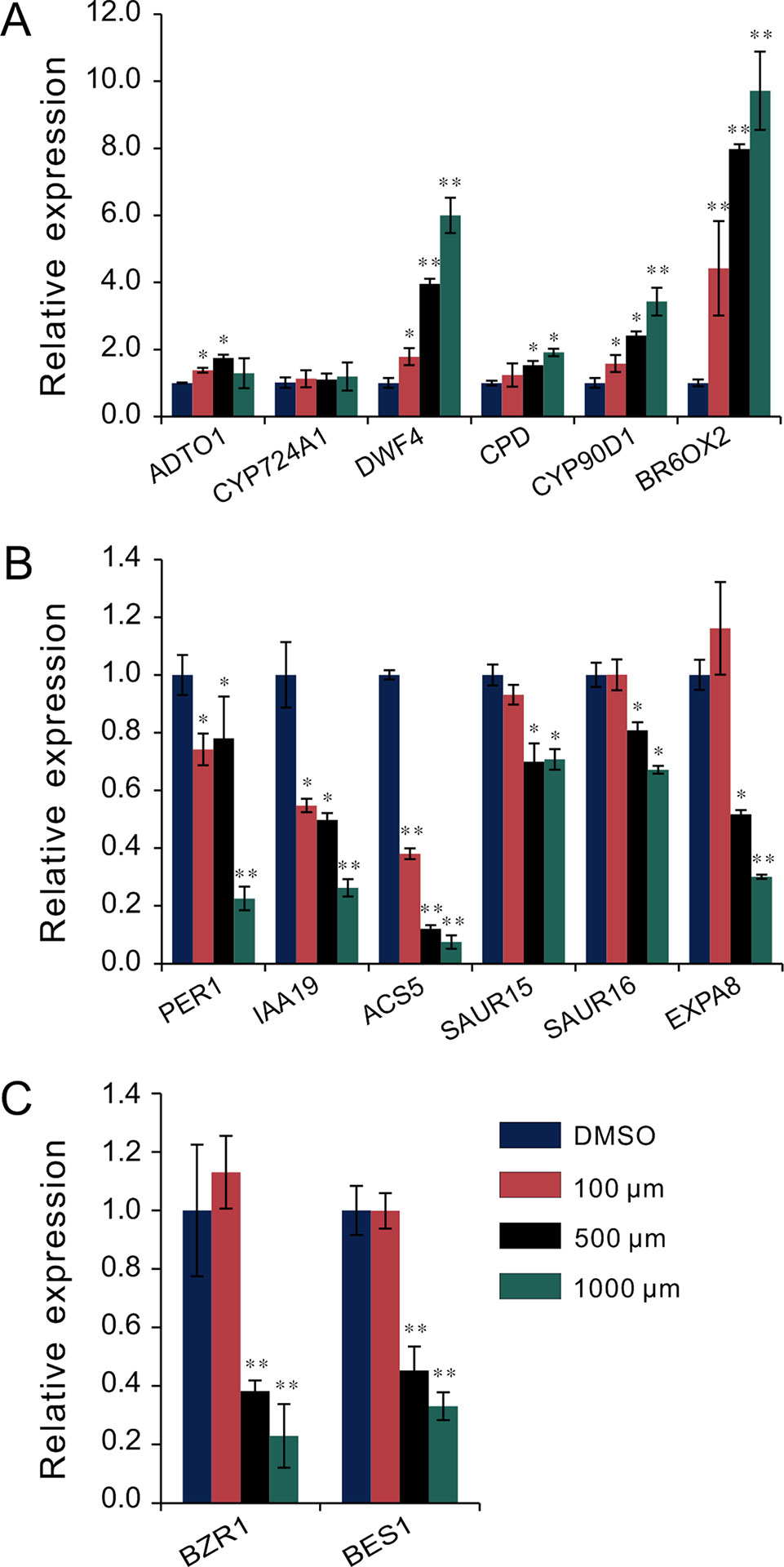
Figure 3 qRT-PCR analysis of BR-related gene expression under melatonin treatment. Seedlings were grown in light for 4 days and then transferred to 1/2MS plates with various concentrations of melatonin in a dark condition for another 3 days. Expression levels were normalized to an internal control ACTIN2. (A) qRT-PCR analysis of BR-biosynthetic genes. (B) qRT-PCR analysis of BR-induced genes involved in cell elongation. (C) qRT-PCR analysis of the two key transcription factors, BZR1 and BES1. Data are represented as means ± SD (n = 3), *p < 0.05, **p < 0.01 (Student’s t test). BR, brassinosteroid; MS, Murashige and Skoog; qRT-PCR, real-time polymerase chain reaction.
Melatonin Inhibits Hypocotyl Elongation by Repressing BR Signaling
Based on the above observations, we speculated that melatonin inhibits hypocotyl likely by direct repression of BR signaling. This idea was supported by the melatonin sensitivity experiment performed on bzr1-1D and bes1-D, which are BR constitutive active form mutants of BZR1 and BES1, respectively. Similar with the BR biosynthetic inhibitor BRZ, melatonin markedly (p < 0.01) inhibited hypocotyl elongation of WT but showed slight (p < 0.05) suppression on bzr1-1D and had no impact on hypocotyl elongation of bes1-D (Figures 4A, B). Furthermore, an expression analysis also showed the same tendency in downregulation of BR-induced genes (PRE1, IAA19, ACS5, EXP8, SAUR15, and SUAR16) between melatonin and BRZ (Figure 4C), suggesting that high concentrations of melatonin are equivalent to the BRZ in the inhibition of hypocotyl elongation. We also analyzed the expression of these genes in bzr1-1D seedlings after being treated with melatonin and found that these genes were poorly affected by melatonin as compared to those of WT control (Figure 4D). In addition, the upregulation of DWF4 and BR6OX2 caused by melatonin was partly suppressed in bzr1-1D (Figure 4E).
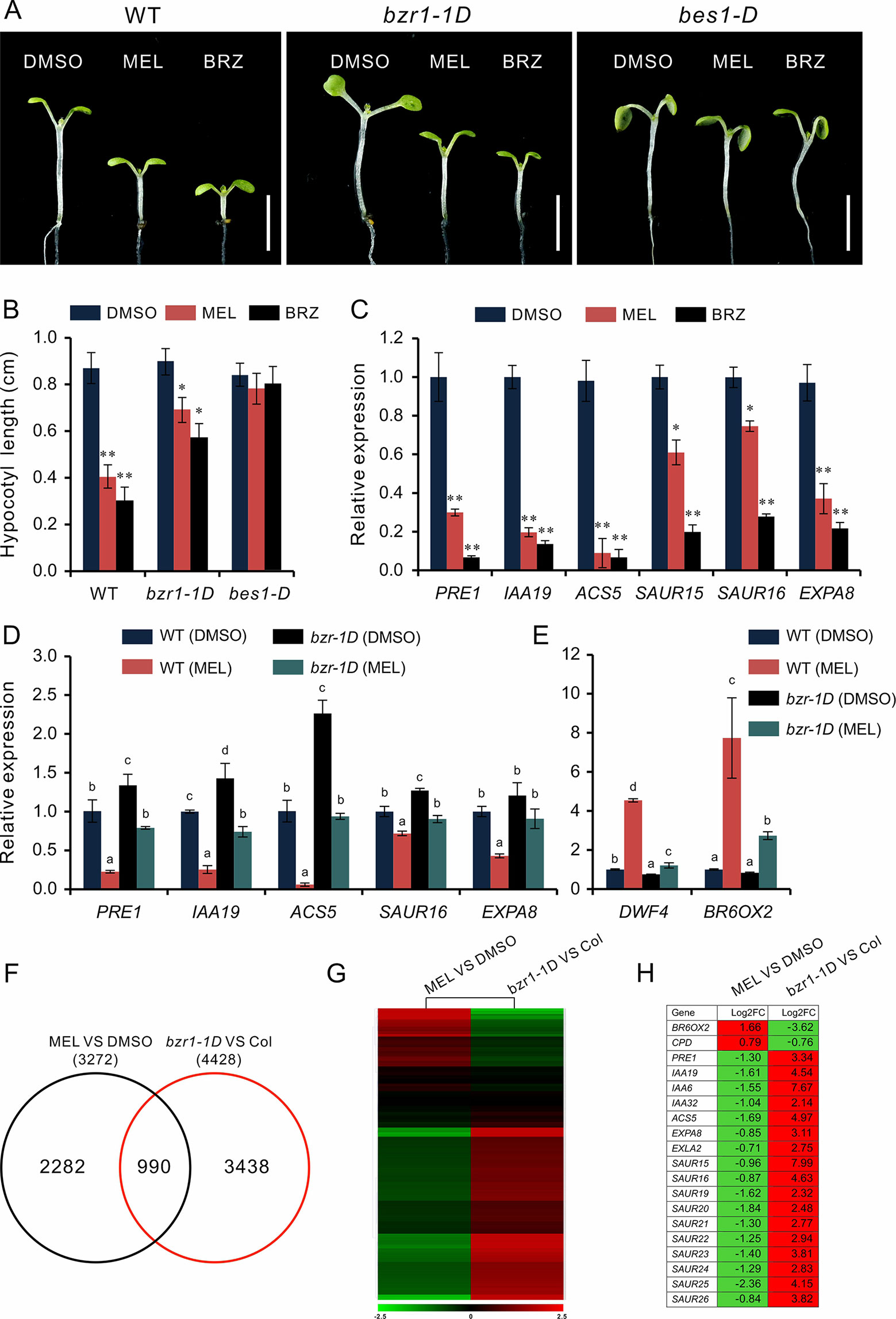
Figure 4 High-dose melatonin represses BR signaling. (A, B) Four-day-old seedlings of WT, bzr1-1D, and bes1-D were transferred on medium containing 1 mM of melatonin (MEL), 1 µM brassinazole (BRZ), or 0.1% DMSO (solvent control) for 3 days in darkness. Scale bar represents 0.5 cm. *p < 0.05, **p < 0.01 (Student’s t test). Error bars indicate the SEM (n ≥ 40). (C) qRT-PCR analyses of expression of BR-related genes in WT seedlings under treatment of MEL, BRZ, and DMSO (control). Data are represented as means ± SD (n = 3), *p < 0.05, **p < 0.01 (Student’s t test). (D, E) Expression level analysis of BR-related genes in both WT and bzr1-1D after being treated with melatonin. Significant differences (p < 0.05) are denoted by different lowercase letters. (F) Venn diagram displaying the overlapping genes regulated by melatonin and BZR1. (G, H) Hierarchical cluster analysis of 990 overlapping genes and the representative genes in BR signaling are listed. Red and green colors in the heatmap represent induced and repressed genes, respectively. BR, brassinosteroid; DMSO, dimethyl sulfoxide; qRT-PCR, real-time polymerase chain reaction; SEM, standard error of the mean; WT, wild type.
We then carried out a comparative transcription analysis between the data of MEL versus DMSO and that of bzr1-1D versus Col, which was obtained from public RNA-Seq data online (Supplementary Table S2) (Tian et al., 2018). We found that of 3,272 DEGs regulated by melatonin, 990 genes (30%) were shared by the DEGs (4,428) in bzr1-D versus Col group (Figure 4F). Heatmap showed an opposite pattern of expression among these overlapping genes, in which 705 genes (71%) were affected in the opposite way (Figure 4G). In particular, the marker genes of BR signaling, such as BR-biosynthetic genes (BR6OX2 and CPD) and BR-induced gens (PER1, IAA19, EXPA8, SAURs, etc) involved in cell elongation, also displayed an opposite expression trend (Figure 4H), suggesting that the inhibition of hypocotyl elongation caused by melatonin involves the repression of BR signaling.
BL Antagonizes Melatonin in the Regulation of Hypocotyl Elongation
Exogenous BL is usually used to rescue dwarf hypocotyl of BR biosynthesis synthesis mutants (Wang et al., 2002; De Rybel et al., 2009). To test the relationship between the melatonin and BR pathways, we transferred the seedlings to melatonin-enriched medium supplemented with various concentrations of BL. The results showed that hypocotyl inhibition caused by high dose of melatonin was gradually restored by additional BL, but not IAA or GA3 (Figures 5A, B, and Supplementary Figure S2). Consistent with the retrieval of bzr1-1D and bes1-D on hypocotyl repression caused by melatonin (Figure 4A), the inhibitory effects of 1 mM of melatonin on hypocotyl were almost eliminated by 1 μM BL (Figures 5A, B). Furthermore, an expression analysis of BR-related genes showed that BL antagonized the high dose of melatonin in the regulation of the expression of BR-related genes. Concretely, BL increased the expression of BR-induced genes (PRE1, IAA19, ACS5, SAUR15, and SUAR16) but reduced the expression of BR-biosynthetic genes (DAWF4, CPD, and BR6OX2), even in the presence of 1 mM of melatonin (Figure 5C), implying that melatonin affects BR biosynthesis.
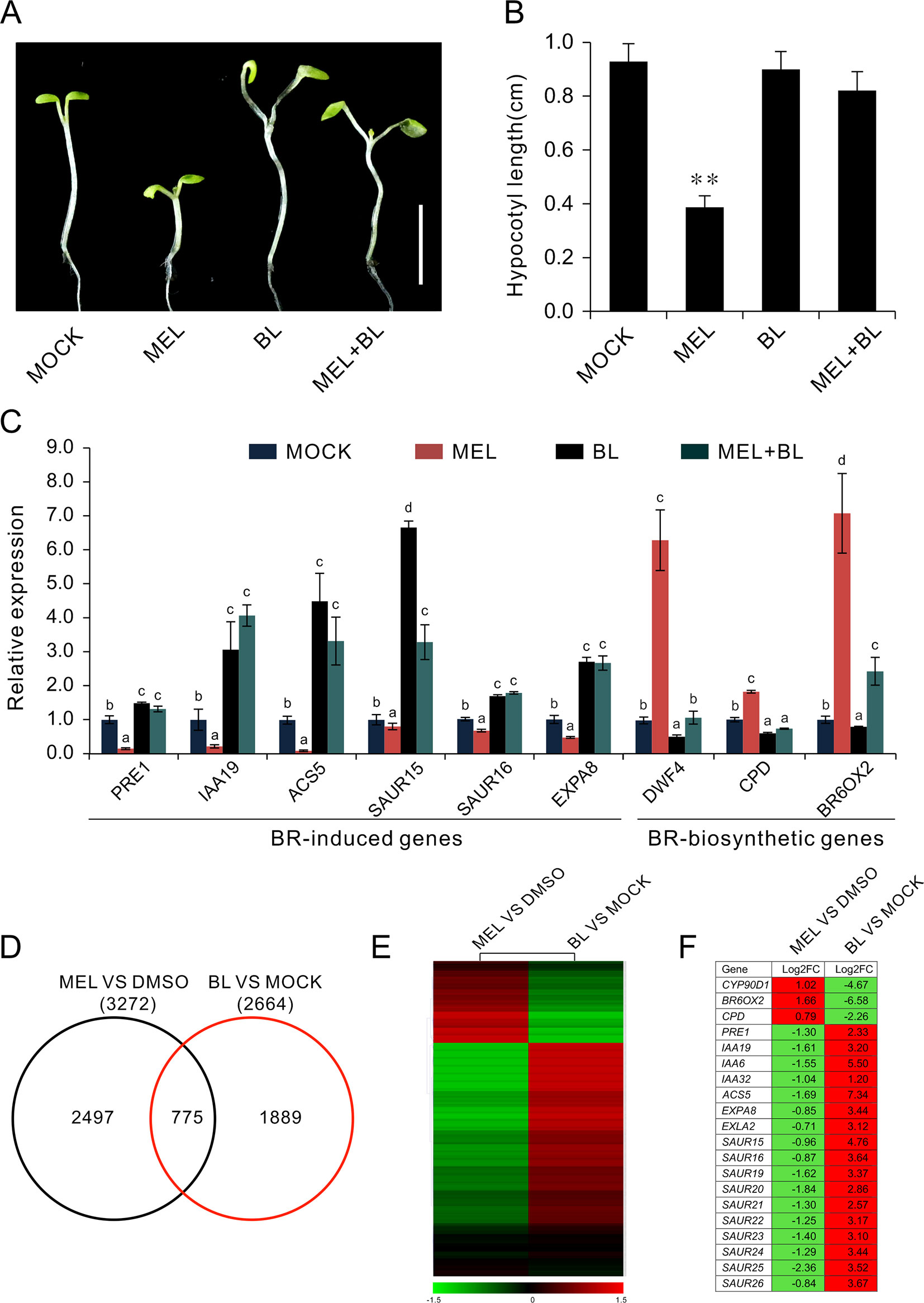
Figure 5 BL antagonizes melatonin in the regulation of hypocotyl elongation. (A) Additional BL restores hypocotyl elongation against a high concentration of melatonin. Four-day-old seedlings grown on 1/2MS under light were transferred on medium containing 1 mM of melatonin with indicated concentration of BL for another 3 days in darkness. Scale bar represents 0.5 cm. (B) The corresponding hypocotyl length.*p < 0.05, **p < 0.01 (Student’s t test). Error bars indicate the SEM (n ≥ 40). (C) qRT-PCR analyses showing the expression of BR-related genes. Significant differences (p < 0.05) are denoted by different lowercase letters. (D) Venn diagram displaying the overlapping genes induced by melatonin and BL. (E, F) Hierarchical cluster analysis of 775 overlapping genes and the representative genes in BR signaling are listed. Red and green colors in the heatmap represent induced and repressed genes, respectively. BL, brassinolide; MS, Murashige and Skoog; qRT-PCR, real-time polymerase chain reaction; SEM, standard error of the mean.
To further explore the regulatory relationship between melatonin and BR signaling, we compared the 3,272 DEGs (1.5-fold change) affected by melatonin (MEL versus DMSO) with the 2,664 DEGs induced by BL (BL versus mock) from public RNA-Seq data (Supplemental Table S3) (Oh et al., 2014) and found that of 3,272 melatonin regulated DEGs, and 775 genes (24%) were also affected by BL (Figure 5D, Supplementary Table S3). Among these co-regulated genes, 544 genes (70%) in MEL versus DMSO group were downregulated, whereas in BL versus mock group, most of the genes (492 out of 775) were upregulated (Supplementary Table S3). Heatmap also showed an opposite regulated mode where 597 (77%) genes were affected in an opposite way between melatonin and BL (Figure 5E, Supplementary Table S3). In addition, some representative genes such as BR-biosynthetic genes (CYP90D1, BR6OX2, and CPD) and BR-induced genes (PER1, IAA19, ACS5, EXPA8, SAUR15, SAUR16, etc.) of the co-regulated genes exhibited reverse expression mode (Figure 5F). In addition, similar results were observed when genes affected by melatonin (MEL versus DMSO) were compared with the genes induced by BL (BL versus mock) and the genes regulated in bzr1-1D (bzr1-1D versus Col) (Supplementary Figure S3).
Melatonin Inhibits BR Biosynthesis
To explore how melatonin acts on BR signaling, the effects of melatonin on BR-deficient mutants were also investigated, such as BR synthesis mutant det2-1, the BR receptor mutant bir1-5, and BR-insensitive 2 mutant bin 2-1. The results showed that exogenous melatonin had little effect on hypocotyl elongation of these BR-deficient mutants compared with the control whether they were under normal light or dark conditions (Figure 6A). It has been report that additional BL can repress a BR-deficient phenotype of BR synthesis mutants (such as det2, cpd, and dwarf4), but not to BR receptor mutants or other mutants involved in BR signal transduction (Wang et al., 2002; De Rybel et al., 2009). Consistent with previous reports, exogenous BL dramatically promoted hypocotyl elongation of det2-1 but not bri1-5 and bin2-1, even in presence of a high dose of melatonin (Figures 6A–C). Furthermore, we measured the bioactive BR contents by ELISA method. Consistent with the hypocotyl inhibition, a high concentration of melatonin reduced endogenous BR levels to a similar extent with biosynthetic inhibitor BRZ (Figures 7A–C). These results further confirmed that a high dose of melatonin repressed BR biosynthesis in the inhibition of hypocotyl elongation.
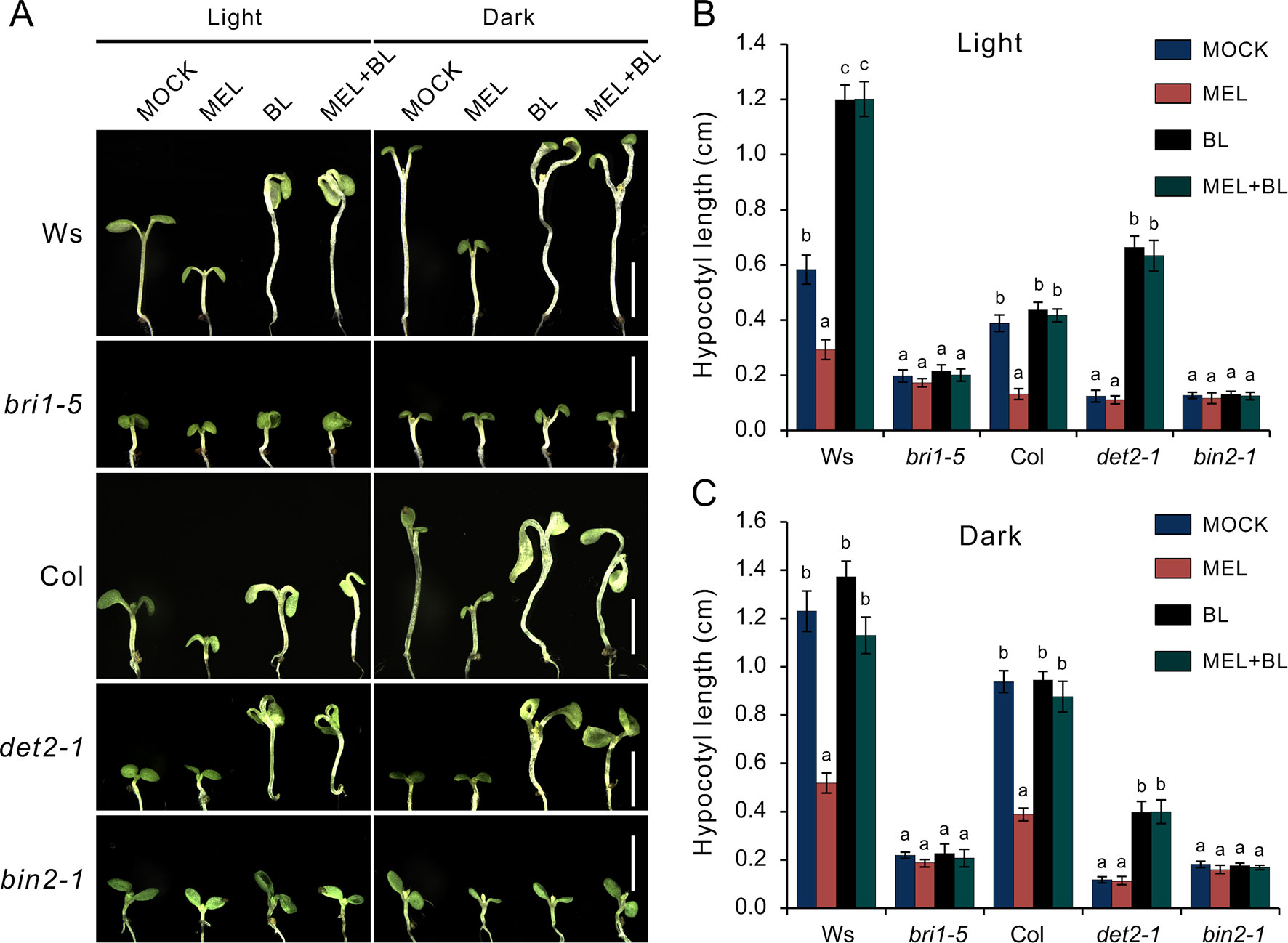
Figure 6 Melatonin involves in repressing BR biosynthesis. (A) Phenotype of BR-related mutants like the det2-1, bri1-5, and bin2-1 response to 1 mM of melatonin, 1 µM of BL or their combination, and mock solvent as the control. Scale bar represents 0.5 cm. (B, C) Hypocotyl length of the corresponding seedlings. Error bars indicate the SEM (n ≥ 40). Significant differences (p < 0.05) are denoted by different lowercase letters. BL, brassinolide; BR, brassinosteroid; SEM, standard error of the mean.
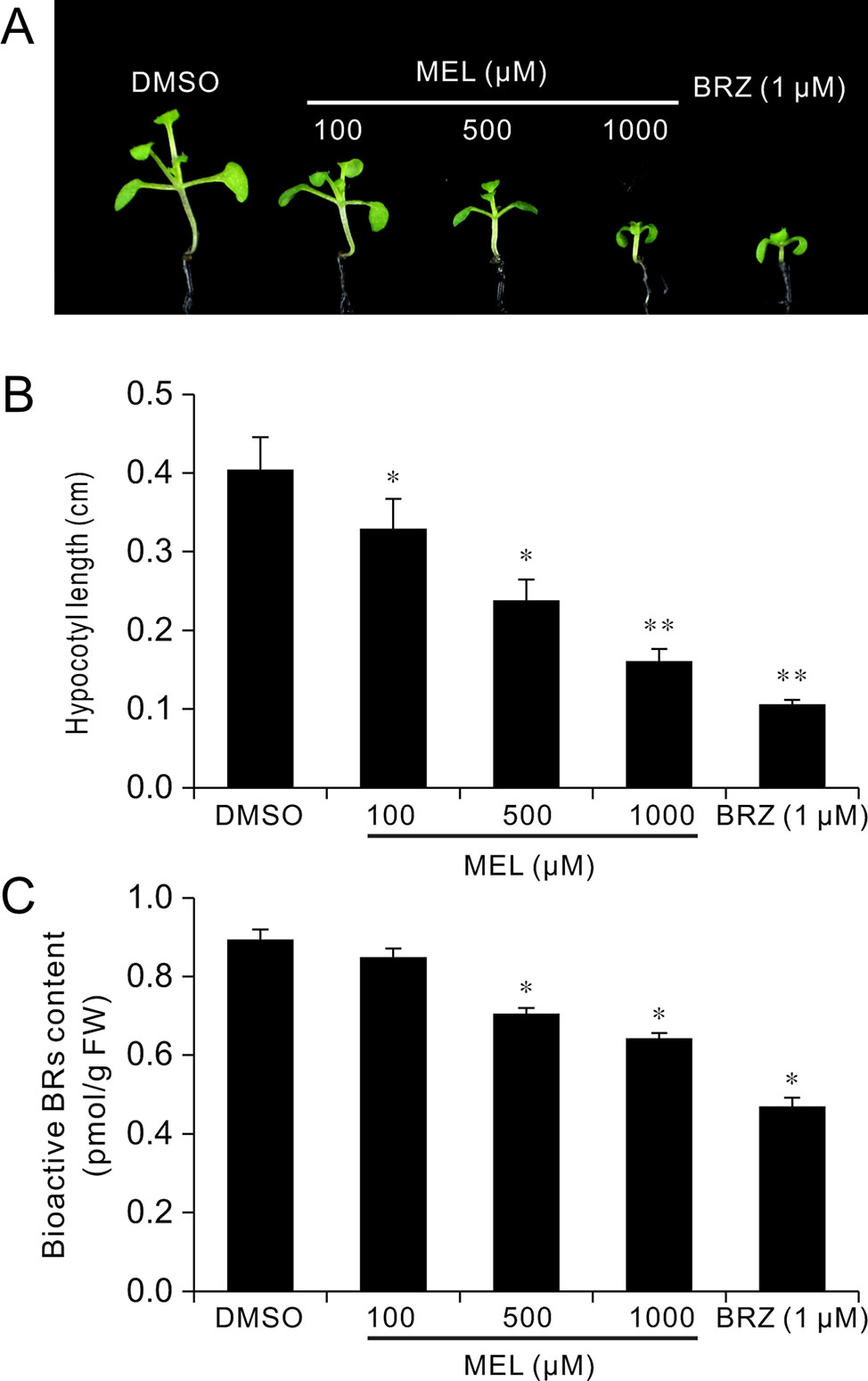
Figure 7 Melatonin reduces endogenous BR levels. Seedlings 12 DAG from 1/2MS amended with melatonin, BRZ, or DMSO (control) were collected for the determination of BR levels by ELISA method. (A) Phenotype of seedlings from melatonin, BRZ, or DMSO treatments. (B) Hypocotyl length of the corresponding seedlings. *p < 0.05, **p < 0.01 (Student’s t test). Error bars indicate the SEM (n ≥ 40). (C) Endogenous bioactive BR content. Data are represented as means ± SD (n = 3), *p < 0.05 (Student’s t test). BR, brassinosteroid; BRZ, brassinazole; DAG, days after germination; DMSO, dimethyl sulfoxide; ELISA, enzyme-linked immunosorbent assay; MS, Murashige and Skoog; SEM, standard error of the mean.
Discussion
Melatonin functions as a pleiotropic regulator in the regulation of plant growth, development, and multiple stress responses (Hardeland et al., 2011; Arnao and Hernández-Ruiz, 2014; Arnao and Hernández-Ruiz, 2015; Erland et al., 2015; Nawaz et al., 2016; Kanwar et al., 2018). In terms of dosage melatonin, the working concentration of melatonin is usually considerably higher than that of other plant hormones in the study of plant growth and development. For example, in the research of how melatonin regulates Arabidopsis root system architecture, the maximum concentration of melatonin (600 μM) was used (Pelagio-Flores et al., 2013); in another study, melatonin regulates the floral transition in Arabidopsis, and up to 1 mM of melatonin was applied (Shi et al., 2016). In addition, a high concentration of melatonin (1 mM) was used to investigate its inhibitory effects on leaf development in Arabidopsis (Wang et al., 2017). Growth retardations caused by a relatively high concentration of exogenous melatonin have been reported in many studies. It has been reported that melatonin at a high concentration shows an inhibitory effect on hypocotyl elongation of etiolated seedlings in lupin (Hernandez-Ruiz et al., 2004). A recent study showed us that high-concentration melatonin restrained root growth and reduced biomass, as well as retarded plant leaf growth by reducing both cell size and cell proliferation (Wang et al., 2017; Lin et al., 2018). There were many investigations showing that high-dose melatonin accompanied inhibitory effects; however, the molecular mechanism remains largely unknown. In present study, we found that a high dose of melatonin inhibited hypocotyl and cell elongation (Supplementary Figure S1, Figures 1A–C) and mechanistically revealed that hypocotyl inhibition of melatonin is involved in the repression of BR biosynthesis in Arabidopsis.
Our data revealed that there were 1,615 DEGs affected by melatonin, in which many genes involved in cell elongation/expansion were downregulated (Figure 1D, Table 1, Supplementary Table S1). This also explains why a high dose of melatonin inhibits hypocotyl elongation. There are several literatures described that melatonin participates in regulating gene expression in respect with plant hormones including auxin, cytokinin, GAs, abscisic acid, ethylene, jasmonic acid, and salicylic acid (Arnao and Hernández-Ruiz, 2018; Arnao and Hernández-Ruiz, 2019). Melatonin showed effects on inducing expression of phytohormone related-genes such as biosynthesis, catabolism, receptors, and transcription factors (Arnao and Hernández-Ruiz, 2018; Arnao and Hernández-Ruiz, 2019). Our KEGG pathway analysis also found that “brassinosteroid biosynthesis” pathway listed in the top 20 of most significantly enriched KEGG pathways and BR biosynthesis-related genes showed differential expression (Figures 2A, B, Table 1), suggesting that melatonin affects the BR metabolism. It is well known that BRs play a predominant role in the regulation of cell elongation; thus, we speculated that hypocotyl inhibition of melatonin is likely to be involved in the regulation of BR signaling. Our results further showed that melatonin modulated BR-related genes expression, such as downregulating BR-induced genes involved in cell elongation (PRE1, IAA19, and SAURs) but upregulating BR-biosynthetic genes (DAWF4, CPD, and BR6OX2) (Figures 3A, B, Table 1). In BR signaling, BRs activated and stabilized the two key transcription factors, BZR1 and BES1, to promote cell elongation through transcriptional activation of their target genes involved in cell elongation, including PER1, IAA19, SAURs, and EXPA8 (Sun et al., 2010; Yu et al., 2011; Bai et al., 2012; Wang et al., 2012, Nolan et al., 2017). Therefore, it is easy to understand that constitutive active forms of mutants bzr1-1D and bes1-D showed insensitivity to BRZ as well as a high dose of melatonin (Figures 4A–D). Through a comparative expression analysis, we further confirmed that melatonin represses BR signaling in regulating hypocotyl elongation (Figures 4F–H and 5D–F and Supplementary Figure S3).
We also observed that an inhibitory effect on hypocotyl of melatonin was eliminated by exogenous BL (Figures 5A–C, Supplementary Figure S2), implying that melatonin functions in BR biosynthesis, because an exogenous application of BL could recover hypocotyl elongation of a BR-deficient phenotype of BR synthesis mutants (det2, cpd, dwarf4, etc.), but not to BR receptor mutants (bri1-5) or other mutants (bin2-1) involved in BR signal transduction (Wang et al., 2002; De Rybel et al., 2009). In the present study, we also observed that even in the presence of high concentrations of melatonin, BL reversed hypocotyl inhibition of WT and det2-1 mutant seedlings, but not bri1-5 or bin2-1 mutants (Figures 6A–C). These observations further confirmed that melatonin suppresses BR biosynthesis in the regulation of hypocotyl elongation. In addition, the results of quantification of bioactive BR contents under treatment with melatonin also supported our conclusions (Figures 7A–C). On the other hand, the recent investigations reported that rice seedlings with melatonin deficiency by knockdown of the enzyme gene required for melatonin biosynthesis showed a semi-dwarf phenotype and reduction of DWARF4 transcripts and reduction of endogenous BR levels (Hwang and Back, 2018; Lee and Back, 2018), suggesting that the lack of melatonin synthesis is likely related to BR suppression. In the present study, contradicting results were obtained likely due to the difference in species, because of no BR-deficient phenotype; for example, a de-etiolated phenotype in the dark condition and a short hypocotyl in the light condition were observed in melatonin biosynthesis-deficient mutants such as snat1 (Lee and Back, 2018).
BRs and GAs are two primary hormones regulating plant cell elongation/expansion; additionally, auxin also plays an important role in regulating cell elongation (Depuydt and Hardtke, 2011; Bai et al., 2012). It has been reported that melatonin upregulated GA biosynthesis genes such as GA20ox and GA3ox in cucumber seedlings in response to saline stress and resulted in a high level of active GAs such as GA3 and GA4 in the salt-inhibited germination process (Zhang et al., 2013; Zhang et al., 2014). Recently, melatonin was reported to negatively regulate auxin biosynthesis and the auxin response in Arabidopsis (Wang et al., 2016). Together, the above findings imply that melatonin inhibited hypocotyl elongation likely involved in the regulation of GA or auxin signaling. The expression profile analysis also showed a certain amount of auxin response genes, such as SAURs and IAAs, in which some are co-regulated by BRs and auxin, involved in the regulation of cell elongation (Table 1). However, our results showed that just BL, but not IAA or GA3, could save the hypocotyl elongation (Supplementary Figure S2), suggesting that BRs, not auxin and GAs, play a predominant role in mediating the inhibition of hypocotyl elongation caused by a high concentration of melatonin.
Based on our results, we propose that melatonin decreases endogenous BR content through repressing BR biosynthesis, which leads to inactivation of BR signaling and reduction of transcriptional activity of BZR1/BES1 and then downregulation of the expression of BR-induced genes (PRE1, IAA19, EXPA8 and SAURs) involved in hypocotyl cell elongation and upregulation of the expression of BR-biosynthetic genes (DAWF4, CPD, CYP90D1, and BR6OX2) by negative feedback regulation. In summary, our findings reveal that melatonin inhibits hypocotyl elongation through repression of the BR biosynthesis in Arabidopsis; however, the mechanism of action needs further study to investigate how melatonin acts in the biosynthesis of BRs.
Data Availability
All datasets for this study are included in the manuscript and the Supplementary Files.
Author Contributions
FX and FZ designed the experiments; FX, ZW, and LF performed the experiments; KD, YS, LW, and GQ analyzed the data; FX and MR wrote the manuscript; and RR, ZY, and FL revised the manuscript and provided valuable advice.
Funding
This work was supported by the following grants: the National Natural Science Foundation of China (Nos. 31801913 and 31801271), the Major Program of Joint Funds (Henan, Grant No. U1804231), the China Postdoctoral Science Foundation (No. 2017M622958), and the Fundamental and Frontier Research Project of Chongqing (No. cstc2016jcyjA0822).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhiyong Wang (Carnegie Institution for Science, Stanford, CA, USA) and Honghui Lin (Sichuan University, Chengdu, Sichuan, China) for generously providing the BR-related Arabidopsis mutant seeds.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01082/full#supplementary-material
Supplementary Figure S1 | Exogenous melatonin inhibits seedling growth in Arabidopsis.
Supplementary Figure S2 | Additional BL rescued hypocotyl elongation under melatonin treatment
Supplementary Figure S3 | Comparative expression spectrum analysis
Supplementary Table S1 | Differentially expressed genes between melatonin treatment and DMSO control (MEL VS DMSO).
Supplementary Table S2 | Comparative expression spectrum analysis between MEL VS DMSO and bzr1-1D VS Col.
Supplementary Table S3 | Comparative expression spectrum analysis between MEL VS DMSO and BL VS Mock.
Supplementary Table S4 | Primers used for qRT-PCR.
Supplementary Table S5 | Enrichment of GO terms
Supplementary Table S6 | Enrichment of KEGG pathways
References
Arnao, M. B., Hernández-Ruiz, J. (2014). Melatonin: plant growth regulator and/or biostimulator during stress? Trends Plant Sci. 19, 789–797. doi: 10.1016/j.tplants.2014.07.006
Arnao, M. B., Hernández-Ruiz, J. (2015). Functions of melatonin in plants: a review. J. Pineal Res. 59, 133–150. doi: 10.1111/jpi.12253
Arnao, M. B., Hernández-Ruiz, J. (2018). Melatonin and its relationship to plant hormones. Ann. Bot. 121, 195–207. doi: 10.1093/aob/mcx114
Arnao, M. B., Hernández-Ruiz, J. (2019). Melatonin: a new plant hormone and/or a plant master regulator? Trends Plant Sci. 24, 38–48. doi: 10.1016/j.tplants.2018.10.010
Asami, T., Mizutani, M., Fujioka, S., Goda, H., Min, Y. K., Shimada, Y., et al. (2001). Selective interaction of triazole derivatives with DWF4, a cytochrome P450 monooxygenase of the brassinosteroid biosynthetic pathway, correlates with brassinosteroid deficiency in planta. J. Biol. Chem. 276, 25687–25691. doi: 10.1074/jbc.M103524200
Bai, M. Y., Shang, J. X., Oh, E., Fan, M., Bai, Y., Zentella, R., et al. (2012). Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 14, 810–817. doi: 10.1038/ncb2546
Chory, J., Nagpal, P., Peto, C. A. (1991). Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. Plant Cell 3, 445–459. doi: 10.1105/tpc.3.5.445
Clouse, S. D., Sasse, J. M. (1998). BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Mol. Biol. 49, 427–451. doi: 10.1146/annurev.arplant.49.1.427
De Rybel, B., Audenaert, D., Vert, G., Rozhon, W., Mayerhofer, J., Peelman, F., et al. (2009). Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 16, 594–604. doi: 10.1016/j.chembiol.2009.04.008
Depuydt, S., Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21, R365–R373. doi: 10.1016/j.cub.2011.03.013
Erland, L. A., Murch, S. J., Reiter, R. J., Saxena, P. K. (2015). A new balancing act: the many roles of melatonin and serotonin in plant growth and development. Plant Signal. Behav. 10, e1096469. doi: 10.1080/15592324.2015.1096469
Erland, L. A. E., Shukla, M. R., Singh, A. S., Murch, S. J., Saxena, P. K. (2018). Melatonin and serotonin: mediators in the symphony of plant morphogenesis. J. Pineal Res. 64, e12452. doi: 10.1111/jpi.12452
Favero, D. S., Le, K. N., Neff, M. M. (2017). Brassinosteroid signaling converges with SUPPRESSOR OF PHYTOCHROME B4-#3 to influence the expression of SMALL AUXIN UP RNA genes and hypocotyl growth. Plant J. 89, 1133–1145. doi: 10.1111/tpj.13451
Fujioka, S., Sakurai, A. (1997). Brassinosteroids. Nat. Prod. Rep. 14, 1–10. doi: 10.1039/np9971400001
Fujioka, S., Yokota, T. (2003). Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164. doi: 10.1146/annurev.arplant.54.031902.134921
Hardeland, R., Pandi-Perumal, S. R., Cardinali, D. P. (2006). Melatonin. Int. J. Biochem. Cell Biol. 38, 313–316. doi: 10.1016/j.biocel.2005.08.020
Hardeland, R., Cardinali, D. P., Srinivasan, V., Spence, D. W., Brown, G. M., Pandi-Perumal, S. R. (2011). Melatonin—a pleiotropic, orchestrating regulator molecule. Prog. Neurobiol. 93, 350–384. doi: 10.1016/j.pneurobio.2010.12.004
He, J. X., Gendron, J. M., Sun, Y., Gampala, S. S., Gendron, N., Sun, C. Q., et al. (2005). BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307, 1634–1638. doi: 10.1126/science.1107580
Hernandez, I. G., Gomez, F. J., Cerutti, S., Arana, M. V., Silva, M. F. (2015). Melatonin in Arabidopsis thaliana acts as plant growth regulator at low concentrations and preserves seed viability at high concentrations. Plant Physiol. Biochem. 94, 191–196. doi: 10.1016/j.plaphy.2015.06.011
Hernandez-Ruiz, J., Cano, A., Arnao, M. B. (2004). Melatonin: a growth-stimulating compound present in lupin tissues. Planta 220, 140–144. doi: 10.1007/s00425-004-1317-3
Hernandez-Ruiz, J., Cano, A., Arnao, M. B. (2005). Melatonin acts as a growth-stimulating compound in some monocot species. J. Pineal Res. 39, 137–142. doi: 10.1111/j.1600-079X.2005.00226.x
Huang da, W., Sherman, B. T., Lempicki, R. A. (2009a). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. doi: 10.1038/nprot.2008.211
Huang da, W., Sherman, B. T., Lempicki, R. A. (2009b). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13. doi: 10.1093/nar/gkn923
Hwang, O. J., Back, K. (2018). Melatonin is involved in skotomorphogenesis by regulating brassinosteroids biosynthesis in rice plants. J. Pineal Res. 65, e12495. doi: 10.1111/jpi.12495
Jain, M., Khurana, J. P. (2009). Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 276, 3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M., Tanabe, M. (2016). KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–D462. doi: 10.1093/nar/gkv1070
Kanehisa, M., Araki, M., Goto, S., Hattori, M., Hirakawa, M., Itoh, M., et al. (2008). KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484. doi: 10.1093/nar/gkm882
Kanwar, M. K., Yu, J., Zhou, J. (2018). Phytomelatonin: recent advances and future prospects. J. Pineal Res. 65, e12526. doi: 10.1111/jpi.12526
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Knauss, S., Rohrmeier, T., Lehle, L. (2003). The auxin-induced maize gene ZmSAUR2 encodes a short-lived nuclear protein expressed in elongating tissues. J. Biol. Chem. 278, 23936–23943. doi: 10.1074/jbc.M212585200
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lee, K., Back, K. (2018). Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 66, e12537. doi: 10.1111/jpi.12537
Li, J., Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. doi: 10.1016/S0092-8674(00)80357-8
Li, J., Nam, K. H. (2002). Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. doi: 10.1126/science.1065769
Li, J., Nagpal, P., Vitart, V., McMorris, T. C., Chory, J. (1996). A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401. doi: 10.1126/science.272.5260.398
Lin, L., Li, J., Chen, F., Liao, M., Tang, Y., Liang, D., et al. (2018). Effects of melatonin on the growth and cadmium characteristics of Cyphomandra betacea seedlings. Environ. Monit. Assess. 190, 119. doi: 10.1007/s10661-018-6481-1
Mathur, J., Molnar, G., Fujioka, S., Takatsuto, S., Sakurai, A., Yokota, T., et al. (1998). Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 14, 593–602. doi: 10.1046/j.1365-313X.1998.00158.x
Murch, S. J., Campbell, S. S. B., Saxena, P. K. (2001). The role of serotonin and melatonin in plant morphogenesis: regulation of auxin-induced root organogenesis in in vitro-cultured explants of St. John’s wort (Hypericum perforatum L.). In Vitro Cell. Dev. Biol. Plant 37, 786–793. doi: 10.1007/s11627-001-0130-y
Nawaz, M. A., Huang, Y., Bie, Z., Ahmed, W., Reiter, R. J., Niu, M., et al. (2016). Corrigendum: Melatonin: current status and future perspectives in plant science. Front. Plant Sci. 7, 714. doi: 10.3389/fpls.2015.01230
Noguchi, T., Fujioka, S., Choe, S., Takatsuto, S., Yoshida, S., Yuan, H., et al. (1999). Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 121, 743–752. doi: 10.1104/pp.121.3.743
Nolan, T., Liu, S., Guo, H., Li, L., Schnable, P., Yin, Y. (2017). Identification of brassinosteroid target genes by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) and RNA-sequencing. Methods Mol. Biol. 1564, 63–79. doi: 10.1007/978-1-4939-6813-8_7
Oh, E., Zhu, J. Y., Bai, M. Y., Arenhart, R. A., Sun, Y., Wang, Z. Y. (2014). Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 27, 3. doi: 10.7554/eLife.03031
Pelagio-Flores, R., Munoz-Parra, E., Ortiz-Castro, R., Lopez-Bucio, J. (2013). Melatonin regulates Arabidopsis root system architecture likely acting independently of auxin signaling. J. Pineal Res. 53, 279–288. doi: 10.1111/j.1600-079X.2012.00996.x
Peng, P., Yan, Z., Zhu, Y., Li, J. (2008). Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 1, 338–346. doi: 10.1093/mp/ssn001
Reiter, R. J., Rosales-Corral, S., Tan, D. X., Jou, M. J., Galano, A., Xu, B. (2017). Melatonin as a mitochondria-targeted antioxidant: one of evolution’s best ideas. Cell. Mol. Life Sci. 74, 3863–3881. doi: 10.1007/s00018-017-2609-7
Ren, H., Gray, W. M. (2015). SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol. Plant 8, 1153–1164. doi: 10.1016/j.molp.2015.05.003
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34, 374–378. doi: 10.2144/03342mt01
Sarropoulou, V. N., Therios, I. N., Dimassi-Theriou, K. N. (2012). Melatonin promotes adventitious root regeneration in in vitro shoot tip explants of the commercial sweet cherry rootstocks CAB-6P (Prunus cerasus L.), Gisela 6 (P. cerasus x P. canescens), and MxM 60 (P. avium x P. mahaleb). J. Pineal Res. 52, 38–46. doi: 10.1111/j.1600-079X.2011.00914.x
Shi, H., Wei, Y., Wang, Q., Reiter, R.J., He, C. (2016). Melatonin mediates the stabilization of DELLA proteins to repress the floral transition in Arabidopsis. J. Pineal Res. 60, 373–379. doi: 10.1111/jpi.12320
Storey, J. D., Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Sun, Y., Fan, X. Y., Cao, D. M., Tang, W., He, K., Zhu, J. Y., et al. (2010). Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 19, 765–777. doi: 10.1016/j.devcel.2010.10.010
Tian, Y., Fan, M., Qin, Z., Lv, H., Wang, M., Zhang, Z., et al. (2018). Hydrogen peroxide positively regulates brassinosteroid signaling through oxidation of the BRASSINAZOLE-RESISTANT1 transcription factor. Nat. Commun. 9, 1063. doi: 10.1038/s41467-018-03463-x
Wang, H., Yang, C., Zhang, C., Wang, N., Lu, D., Wang, J., et al. (2011). Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834. doi: 10.1016/j.devcel.2011.08.018
Wang, L., Feng, Z., Wang, X., Wang, X., Zhang, X. (2010). DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138. doi: 10.1093/bioinformatics/btp612
Wang, Q., An, B., Shi, H., Luo, H., He, C. (2017). High concentration of melatonin regulates leaf development by suppressing cell proliferation and endoreduplication in Arabidopsis. Int. J. Mol. Sci. 18, E991. doi: 10.3390/ijms18050991
Wang, Q., An, B., Wei, Y., Reiter, R. J., Shi, H., Luo, H., et al. (2016). Melatonin regulates root meristem by repressing auxin synthesis and polar auxin transport in Arabidopsis. Front. Plant Sci. 7, 1882. doi: 10.3389/fpls.2016.01882
Wang, Z. Y., Bai, M. Y., Oh, E., Zhu, J. Y. (2012). Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724. doi: 10.1146/annurev-genet-102209-163450
Wang, Z. Y., Nakano, T., Gendron, J., He, J., Chen, M., Vafeados, D., et al. (2002). Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2, 505–513. doi: 10.1016/S1534-5807(02)00153-3
Wei, W., Li, Q. T., Chu, Y. N., Reiter, R. J., Yu, X. M., Zhu, D. H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Xu, W., Huang, J., Li, B., Li, J., Wang, Y. (2008). Is kinase activity essential for biological functions of BRI1? Cell Res. 18, 472–478. doi: 10.1038/cr.2008.36
Yin, Y., Wang, Z. Y., Mora-Garcia, S., Li, J., Yoshida, S., Asami, T., et al. (2002). BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. doi: 10.1016/S0092-8674(02)00721-3
Yu, X., Li, L., Zola, J., Aluru, M., Ye, H., Foudree, A., et al. (2011). A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646. doi: 10.1111/j.1365-313X.2010.04449.x
Zhang, N., Zhao, B., Zhang, H. J., Weeda, S., Yang, C., Yang, Z. C., et al. (2013). Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 54, 15–23. doi: 10.1111/j.1600-079X.2012.01015.x
Zhang, H. J., Zhang, N., Yang, R. C., Wang, L., Sun, Q. Q., Li, D. B., et al. (2014). Melatonin promotes seed germination under high salinity by regulating antioxidant systems, ABA and GA(4) interaction in cucumber (Cucumis sativus L.). J. Pineal Res. 57, 269–279. doi: 10.1111/jpi.12167
Keywords: melatonin, hypocotyl inhibition, BR biosynthesis, gene expression, Arabidopsis
Citation: Xiong F, Zhuo F, Reiter RJ, Wang L, Wei Z, Deng K, Song Y, Qanmber G, Feng L, Yang Z, Li F and Ren M (2019) Hypocotyl Elongation Inhibition of Melatonin Is Involved in Repressing Brassinosteroid Biosynthesis in Arabidopsis. Front. Plant Sci. 10:1082. doi: 10.3389/fpls.2019.01082
Received: 24 May 2019; Accepted: 08 August 2019;
Published: 26 September 2019.
Edited by:
Peter Ulvskov, University of Copenhagen, DenmarkReviewed by:
Jozef Mravec, University of Copenhagen, DenmarkMaren Müller, University of Barcelona, Spain
Copyright © 2019 Xiong, Zhuo, Reiter, Wang, Wei, Deng, Song, Qanmber, Feng, Yang, Li and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuguang Li, bGlmdWd1YW5nQGNhYXMuY24=; Maozhi Ren, cmVubWFvemhpQGNxdS5lZHUuY24=
 Fangjie Xiong
Fangjie Xiong Fengping Zhuo1,2
Fengping Zhuo1,2 Russel J. Reiter
Russel J. Reiter Zuoren Yang
Zuoren Yang Maozhi Ren
Maozhi Ren