
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 September 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01059
The type III secretion system (T3SS) in many Gram-negative bacterial pathogens is regarded as the most critical virulence determinant and an attractive target for novel anti-virulence drugs. In this study, we constructed a T3SS secretion reporter containing the β-lactamase gene fused with a signal peptide sequence of the T3SS effector gene, and established a high-throughput screening system for T3SS inhibitors in the plant pathogenic bacterium Acidovorax citrulli. From a library of 12,000 chemical compounds, we identified a series of benzyloxy carbonimidoyl dicyanide (BCD) derivatives that effectively blocked T3SS-dependent β-lactamase secretion. Substitution of halogens or nitro groups at the para-position on the benzene ring contributed to an increased inhibitory activity. One representative compound, BCD03 (3,4-dichloro-benzyloxy carbonimidoyl dicyanide), dramatically reduced pathogenicity of A. citrulli on melon seedlings, and attenuated hypersensitive responses in the non-host Nicotiana tabacum caused by pathogenic bacteria A. citrulli, Xanthomonas oryzae pv. oryzae and Pseudomonas syringae pv. tomato at sub-MIC concentrations. Western blotting assay further confirmed that BCD03 inhibited effector secretion from the above bacteria via T3SS in the liquid medium. Taken together, our data suggest that BCD derivatives act as novel inhibitors of T3SS in multiple plant bacterial pathogens.
Antibiotic resistance is becoming a major concern in agricultural industries. Traditional antibiotics, which target growth and survival, induce high selective pressure on pathogenic bacteria. One promising way to reduce bacterial resistance is to target virulence factors, rather than growth (Rasko and Sperandio, 2010). The type III secretion system (T3SS) is highly conserved among Gram-negative bacteria and is directly related to pathogenicity during host cell invasion. The needle-like T3SS penetrates the cell membrane where it injects T3SS effectors (T3Es), suppressing host immunity and promoting invasion and dissemination (Cornelis, 2006). T3SS is not necessary for bacterial survival, making it an attractive target for novel antibacterial drugs (Büttner, 2012). To date, a number of chemical compounds acting as T3SS inhibitors have been identified in a wide range of important plant and animal bacterial pathogens, such as Erwinia, Pseudomonas, Xanthomonas, Escherichia, Yersinia and Salmonella species (Felise et al., 2008; Wang et al., 2011; Jessen et al., 2013; Yang et al., 2014; Anantharajah et al., 2016; Fan et al., 2017).
Most of the known T3SS inhibitors were screened out by using transcriptional reporters that fused T3SS-related genes to the genes encoding reporter proteins like luciferase, phospholipase, and β-lactamase. (Kauppi et al., 2003; Li et al., 2005; Felise et al., 2008; Pan et al., 2009; Duncan et al., 2014). A few screenings were performed by using translational or translocational reporters (Kim et al., 2009; Pan et al., 2009; Swietnicki et al., 2011). The known inhibitors, which include alicylates, benzoates, thiazoles, phenolic compounds and specific antibodies, affect T3SS functions by directly targeting T3SS injectisome (Lynch et al., 2010; Jessen et al., 2013; Bowlin et al., 2014), regulating T3SS gene expression (Yang et al., 2014; Marsden et al., 2015; Fan et al., 2017), or through other indirect interactions (Wang et al., 2011; Bowlin et al., 2014; Faure et al., 2014).
In many plant bacterial pathogens, T3SS and T3Es are essential virulence factors during infection. T3SSs in plant bacteria fall into two groups, hypersensitive reaction and pathogenicity (Hrp) groups I and II, based on their nucleotides, operon structures, and regulatory systems (Alfano and Collmer, 1996; Cornelis, 2006; Nilles and Condry, 2017). Representatives of group I include Pseudomonas syringae and Erwinia amylovora, while Ralstonia solanacearum and Xanthomonas oryzae belong in group II (Tampakaki et al., 2010; Diepold and Armitage, 2015). Although these two groups share similar hrc (HR and conserved) genes, some of their hrp genes appear completely different. One significant difference is that group I T3SSs are activated by HrpL, an ECF family sigma factor activated by HrpS to promote gene expression (Lidell and Hutcheson, 1994; Alfano and Collmer, 1996), while group II T3SSs are regulated by AraC family transcriptional regulators, HrpB in R. solanacearum and HrpX in X. oryzae and Acidovorax citrulli. HrpB and hrpX are activated by the OmpR-type transcriptional regulator HrpG, a response regulator protein of a two-component signal transduction system, which induces expression of T3SS (Wengelnik and Bonas, 1996; Wengelnik et al., 1996; Tampakaki et al., 2010). However, expression of T3SS is not constitutive. Transcription of T3SS genes in both Hrp groups is repressed in full nutrient medium, but induced in planta or in medium mimicking the intracellular space of plants (Bonas, 1994; Nilles and Condry, 2017). Induction of T3SS expression tends to involve low-nutrient culture media; for example, XVM2 and XOM2 medium for X. oryzae (pH 6.5 and 5 mM Mg2+), 1/4 M63 medium for R. solanacearum (1 mM Mg2+and 3.9 mM Fe2+) and MG medium for P. syringae (pH 7.0 and 2 mM Mg2+) (Keane et al., 1970; Boucher et al., 1985; Tsuge et al., 2002). Meanwhile, with bacteria such as A. citrulli, T3SS expression is induced in modified lysogenic broth (LB) medium (pH5.8, 10 mM Mg2+) (Zhang et al., 2018).
A. citrulli is the causal agent of bacterial fruit blotch in cucurbit crops such as watermelons (Citrullus lanatus) and melons (Cucumis melo) (Schaad et al., 2008). T3SS is required for pathogenicity and hypersensitive response (HR) induction in host and non-host plants, respectively (Bahar and Burdman, 2010; Johnson et al., 2011). In this study, we established a high-throughput screening system for T3SS inhibitors in A. citrulli strain MH21 and successfully screened a large number of chemical compounds under the optimized T3SS-inducing conditions. Chemicals showing inhibitory activity were further tested using different plant bacterial pathogens to explore their effective target spectrum.
The bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli was grown in LB medium at 37°C, A. citrulli MH21 and its derivatives were grown in LB medium at 28°C, X. oryzae pv. oryzae PXO99A and its T3SS mutant were grown in M210 medium (0.8% casein enzymatic hydrolysate, 0.5% sucrose, 0.4% yeast extract, 17.2 mM K2HPO4, 1.2 mM MgSO4·7H2O) at 28°C. XOM2 medium (0.18% D-(+) xylose, 670 μM L-methionine, 10 mM sodium L-(+) glutamate, 14.7 mM KH2PO4, 40 μM MnSO4, 240 μM Fe(III)-EDTA and 5 mM MgCl2, pH adjusted to 6.5 with KOH) was used for hrp-inducing (Tsuge et al., 2002). P. syringae pv. tomato DC3000 and its T3SS mutant were grown in King’s B medium at 28°C. MG medium (1% Mannitol, 0.2% L-glutamic acid, 3.6 mM KH2PO4, 3.4 mM NaCl and 1.6mM MgSO4·7H2O, pH adjusted to 7.0 with NaOH) was used for hrp-inducing (Keane et al., 1970). The following antibiotics were used when required: 50 μg/mL ampicillin (Ap), 50 μg/mL streptomycin (Sm), 25 μg/mL spectinomycin (Sp), 50 μg/mL kanamycin (Kan), 10 μg/mL gentamycin (Gm), and 50 μg/mL rifampicin (Rif).
Chemical compounds used for T3SS inhibitor screening were provided by Shenyang Sinochem Agrochemicals R&D Co. Ltd (Shenyang, China) and were dissolved in dimethyl sulfoxide (DMSO).
To construct the T3E secretion reporter pZAC-3502sig-penAC, the penAC (GenBank accession MK576102) fragment without its 57-bp 5’ Type II secretion system (T2SS) signal peptide sequence was PCR amplified from MH21 using primers penAC-F and penAC-R (Table S1). A 180-bp T3SS-dependent signal peptide sequence from the predicted effector Aave_3502, a homologue of P. syringae effector HopAO1 (Bretz et al., 2003; Underwood et al., 2007), was PCR amplified using primers Aave-3502sig-F and Aave-3502sig-R (Table S1) then in-frame fused to the 5’end of the penAC fragment digested with EcoRI. The modified β-lactamase sequence was cloned into pZLQ (Luo and Farrand, 1999) under control of the Ptrc promoter. The reporter plasmid was introduced from E. coli into A. citrulli MH21 via mating with an E. coli strain carrying the helper plasmid (pRK600) (Finan et al., 1986). The pZAC-3502-FLAG plasmid containing the FLAG-tagged hopAO1 gene under the Ptrc promoter was generated and introduced into A. citrulli MH21 for Western blotting analysis. The X. oryzae pv. oryzae carrying plasmid pHZY-avrXa27 (Ji et al., 2014) and P. syringae pv. tomato DC3000 carrying plasmid pCPP5372-avrPto-HA (Wei et al., 2015) were also used for Western blotting analysis.
A. citrulli MH21 reporter strain containing the pZAC-3502sig-penAC plasmid was grown overnight in LB medium at 28°C then the bacterial cells were harvested by centrifugation. After washing twice with sterilized distilled water (sdH2O), the bacterial cells were resuspended in sdH2O to OD600 = 1.0 and inoculated into LB medium (pH 5.8) supplemented with 10 mM MgCl2 (Zhang et al., 2018) in a 96-well microplate at a ratio of 4%. Test compounds were added at a final concentration of 25 μg/mL. An identical concentration of DMSO was used as a control. Nitrocefin (TOKU-E, WA, USA) was added at a concentration of 250 μg/mL when the bacteria grew to OD600 = 0.4–0.6 (20 h) followed by incubation for 15 min. Color measurements of the medium (OD500) were then taken using a Microplate Reader (SpectraMax i3x; Molecular Devices, Sunnyvale, CA). The screening experiment was performed with three technical replicates.
Bacterial strain MH21 harboring the pZAC-3502sig-penAC plasmid was prepared as above in a 96-well microplate. The MEC assay was carried out by diluting benzyloxy carbonimidoyl dicyanide (BCD) derivatives at a final concentration of 70, 60, 50, 40, 30, 20, 10, 5, 2.5, or 1.25 μg/mL, respectively. Nitrocefin was then added and the OD500 was determined as described above. The molarity of chemical compounds was converted based on the MECs and molecular weights (MWs). IC50 of these compounds was calculated based on the OD500 values and molarity (Sebaugh, 2011). Experiments were performed twice with three biological replicates.
A MIC assay was carried out using strains MH21, PXO99A and DC3000 and their T3SS mutants. Bacteria were cultured in their own favorable medium until OD600 reached 1.0 then inoculated in 96-well microplates with the same medium at a ratio of 1:1,000, supplemented with BCD derivatives at a final concentration of 200, 100, 50, 25, 12.5, 6.25 or 3.125 μg/mL, respectively. The microplates were placed at 28 ºC and OD600 was measured using a microplate reader after 48 h incubation. The experiments were performed twice with three biological replicates.
Germinated seeds of melon (Cucumis melo INV192) were dipped in MH21 bacterial suspension at 3 × 108 CFU, supplemented with 25 or 50 μg/mL BCD03 for 20 min. ΔhrcC mutant strain was used as a control. Cotyledons of the seedlings were collected 12 days after sowing and disease severity was assessed on a scale of 1 to 9: 1, no symptoms; 3, small, necrotic lesions on cotyledons, <20% necrotic cotyledon; 5, necrotic lesions with chlorosis on cotyledons, 20–50% necrotic cotyledon; 7, large spreading lesions, >50% of cotyledon necrotic; 9, >90% necrosis of the cotyledon or a dead plant (Hopkins and Thompson, 2002). The experiments were repeated twice with three biological replicates. Each replicate contained 5 seedlings.
Nicotiana tabacum seedlings were grown under greenhouse conditions for 4–5 weeks. Bacterial cells were then harvested from precultured medium and suspended in 10 mM MgCl2. Leaves of N. tabacum were subsequently inoculated with bacterial strains at 3 × 108 cfu/mL (MH21, PXO99A and their T3SS mutants) or 3 × 106 cfu/mL (DC3000 and its T3SS mutant) by infiltration with a blunt syringe and assayed 18 h post inoculation (Alfano et al., 1996). BCD03 was added at a final concentration of 25 μg/mL 2 h before infiltration. BCD03 alone was used as a control. The experiment was performed with three biological replicates.
Strains MH21, PXO99A and DC3000 harboring reporter plasmids pZAC-3502-FLAG, pHZY-avrXa27 and pCPP5372-avrPto-HA, respectively, were cultured in their own hrp-inducing medium supplemented with 25 μg/mL T3SS inhibitor BCD03 (or DMSO as a control) at 28ºC for 8 h. Bacterial cultures were diluted to OD600 = 0.5 and centrifuged at 4000 × g for 10 min then the supernatant was filter sterilized, mixed with cold 100% trichloroacetic acid to a final concentration of 15% and incubated overnight at 4ºC. Samples were then centrifuged again at 6000 × g for 30 min at 4ºC, the supernatant was discarded, and the protein pellet was washed twice with 2 mL cold acetone. The bacterial pellet was resuspended in 50 µl phosphate buffered saline (PBS) and boiled for 10 min then HopAO1 and AvrXA27 were detected by Western blotting using a primary anti-FLAG mouse monoclonal antibody (Transgen Biotech, Beijing, China), diluted 1:2,000, and a goat anti-mouse IgG (H+L) secondary antibody conjugated to HRP (Invitrogen, Beijing, China), diluted 1:5,000. AvrPto was detected by Western blotting using a primary anti-HA mouse monoclonal antibody conjugated to HRP (MBL, Beijing, China) and diluted 1:5,000. Antibodies used in this study were all diluted in 40 ml PBS buffer supplemented with 0.5% tween 20 and 1% skimmed milk. Immunodetected HopAO1-FLAG, AvrXA27-FLAG, and AvrPto-HA were developed using Immobilon ECL (Beyotime, Shanghai, China) and membranes were photographed using the ChemiDoc Imaging System (BIO-RAD, Beijing, China).
Statistical analyses were performed using IBM SPSS Statistics 17.0 (SPSS, Chicago, IL, USA) or GraphPad 7.0 (GraphPad Software, La Jolla, CA, USA) as specified.
We constructed a T3E secretion reporter plasmid containing a T3E signal peptide sequence fused with the β-lactamase gene in A. citrulli MH21 to screen for T3SS inhibitors. The β-lactamase gene was used as a reporter because it hydrolyzes nitrocefin and gives a clear color change readout from yellow to red, fulfilling the purpose of high through-put screening. However, wild-type A. citrulli MH21 is naturally resistant to ampicillin and hydrolyzes nitrocefin, indicating the presence of a gene with β-lactamase activity on its chromosome (Figure S1A). Genome sequence analysis subsequently revealed a potential β-lactamase gene named penAC in MH21, which had 39.8% identity and 83.0% coverage with the β-lactamase gene penP in Bacillus subtilis (GenBank accession BG11016). Further phylogenetic analysis (Figure S2) and conserved structure analysis (Table S2) revealed that PenAC belongs to β-lactamase class A (Ambler, 1980; Matagne et al., 1999; Eiamphungporn et al., 2018). In addition, mutation of penAC in A. citrulli MH21 led to loss of ampicillin resistance (Figures S1A, B), suggesting that PenAC is indeed a β-lactamase. We therefore constructed pZAC-3502sig-penAC, a reporter plasmid containing the Ptrc promoter to drive a recombined β-lactamase gene penAC. The original N-terminal T2SS signal peptide of penAC was replaced by the T3SS-signal peptide of effector gene Aave_3502, a homologue of the P. syringae effector HopAO1. (Figure 1A). The mutant MH21ΔpenAC was then used as a host for the reporter plasmid pZAC-3502sig-penAC to remove the background β-lactamase activity. A double gene mutant, MH21ΔpenACΔhrcC, was also constructed as a T3SS-negative control (Figure S1C).
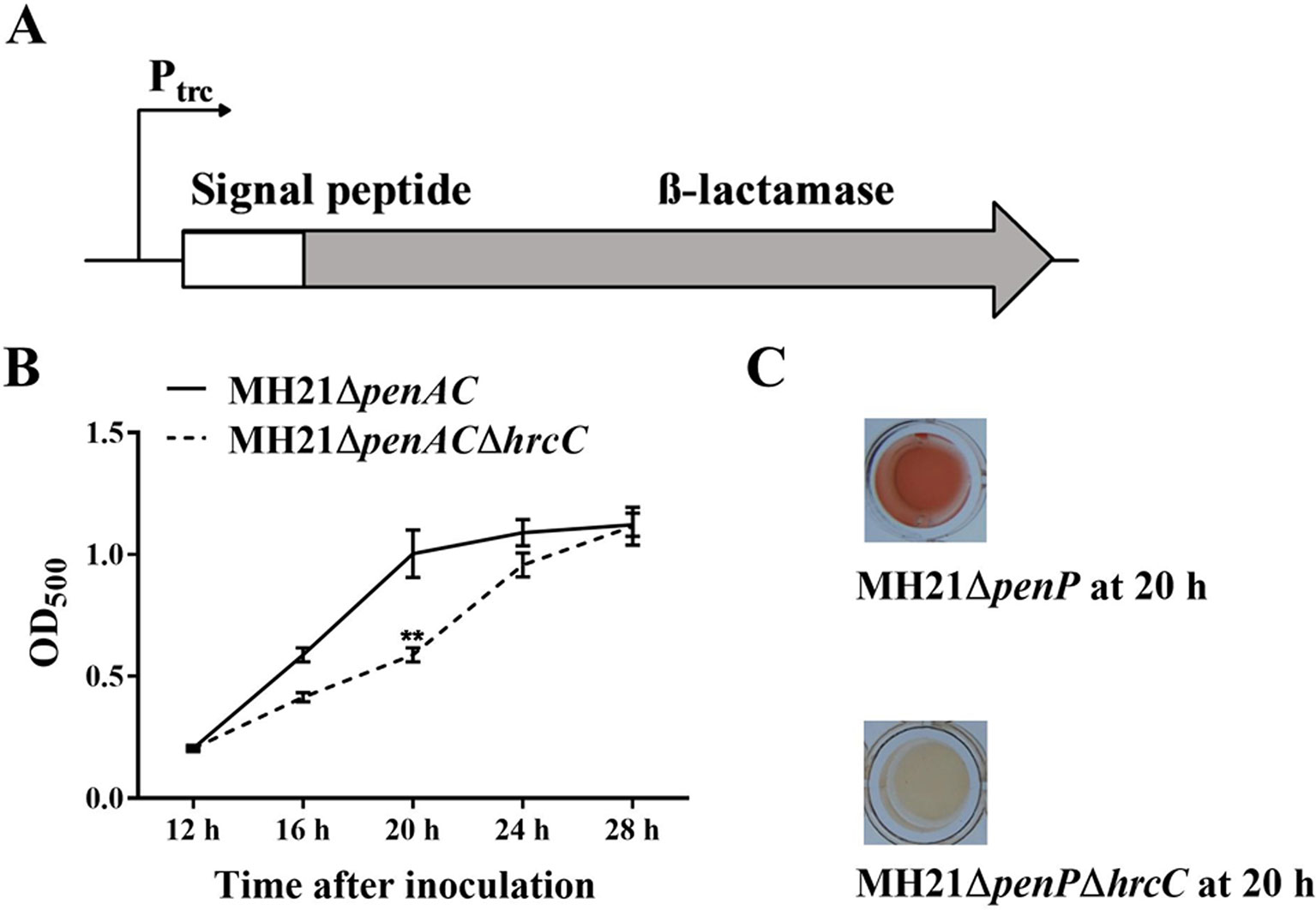
Figure 1 Reporter structure and suitable growth state of reporter strain Acidovorax citrulli MH21∆penAC (pZAC-3502sig-penAC). (A) Reporter structure on plasmid pZAC-3502sig-penAC. The Ptrc promoter driven β-lactamase gene was fused with a signal peptide sequence of the T3SS effector gene (Aave_3502, a hopAO1 homologous gene). (B) Extracellular β-lactamase activity of MH21∆penAC and its T3SS mutant MH21∆penAC∆hrcC cultured in hrp-inducing medium. β-lactamase activity was measured spectrophotometrically by determining the OD500. Experiments were performed twice with three biological replicates. Statistically significant differences (Student’s t-test) were observed at 20 h post inoculation. P-value < 0.05 (**). (C) The reaction mixture containing the reporter plasmid in the T3SS mutant MH21∆penAC∆hrcC remained yellow, but changed into red in mixture containing the T3SS wild-type strain MH21∆penAC.
When cultured in T3SS-inducing LB medium (pH 5.8, 10 mM MgCl2), MH21ΔpenAC containing the reporter plasmid pZAC-3502sig-penAC secreted β-lactamase in a T3SS-dependent manner. Extracellular β-lactamase activity was therefore used to indicate the effect of each inhibitor. Because over-cultured bacteria release undesired β-lactamase via cell lysis, we identified the most suitable bacterial state for reporter detection by comparing extracellular β-lactamase activity between the wild-type MH21 and its T3SS mutant throughout growth. As a result, no growth differences were observed between MH21 mutants ΔpenAC and ΔpenACΔhrcC carrying pZAC-3502sig-penAC when cultured in T3SS-inducing medium for 28 h. However, extracellular β-lactamase activity was significantly higher with MH21ΔpenAC than the T3SS mutant MH21ΔpenACΔhrcC at 20 h (OD600 = 0.4– 0.6) post inoculation (Figures 1B, C), indicating secretion of β-lactamase via T3SS. These culture conditions were therefore used to screen the T3SS inhibitors.
More than 12,000 compounds at a final concentration of 25 µg/mL were screened in 96-well microplates using the above method (Figure 2A). A series of BCD derivatives prevented the color change from yellow to red in the reaction mixture, indicating potential T3SS inhibitory activity. These compounds shared the same BCD backbone as a number of substituents, including halogen, hydro, nitro, and methoxycarbonimidoyl dicyanide (MD), on its benzene ring (Figure 2B). The MICs of the BCD derivatives was always higher than 50 µg/mL, and with BCD03, BCD10, and BCD12 was as high as 100 µg/mL (Table S3). These findings indicate specific inhibition of T3SS by these compounds. The BCD derivatives were further tested with the wild- type MH21, which produced T2SS-dependent β-lactamase PenAC using the system described above. Accordingly, no color variation was observed, indicating that the compounds did not directly inhibit β-lactamase activity (data not shown).
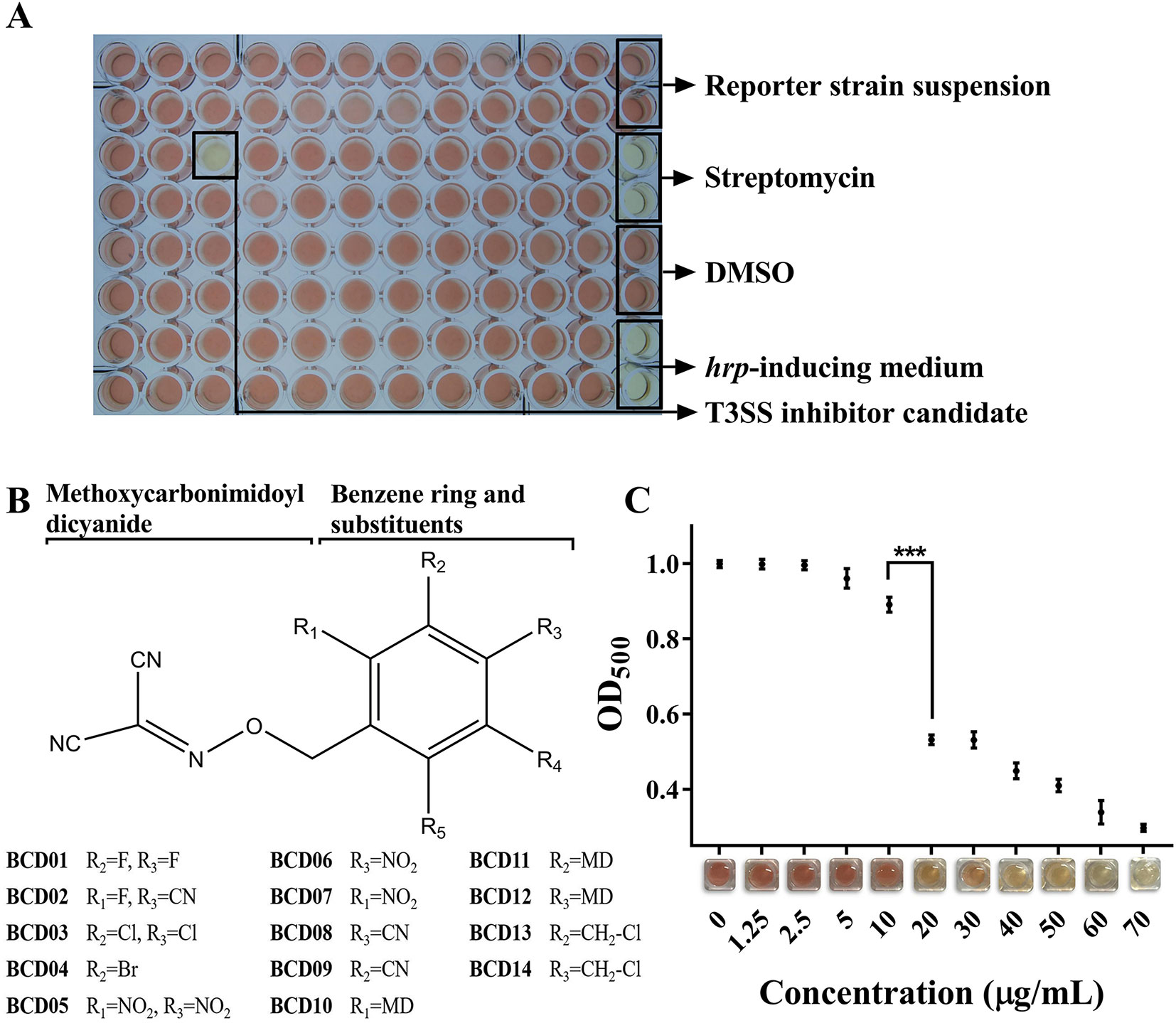
Figure 2 Benzyloxy carbonimidoyl dicyanide derivatives (BCDs)-induced inhibition of T3SS function in Acidovorax citrulli MH21. (A) High-throughput screening in a 96-well microplate. Each well contained the reporter strain MH21∆penAC harboring pZAC-3502sig-penAC in 200 µL hrp-inducing medium (LB with 10 mM MgCl2, pH 5.8) supplemented with candidate chemicals and nitrocefin. Control groups were arranged in the rightmost row, including reaction mixture without candidate chemicals, reaction mixture with the antibiotic streptomycin, reaction mixture with solvent DMSO, and hrp-inducing medium only. (B) Chemical structures of BCD derivatives that had an inhibitory effect on T3E reporter secretion. The benzene ring substituted by one BCD is stabilized as a core. R1-R5 substitutions on the benzene ring were evaluated. (C) Inhibitory effect of different concentrations of BCD03 on T3SS function. The readout of OD500 decreased as the concentration of BCD03 increased, as shown by the color variation from red to yellow. Experiments were performed twice with three biological replicates. Statistically significant differences (Student’s t-test) were observed between 10 and 20 µg/mL. P-value < 0.01 (***).
The MECs of effective inhibitors of T3SS from A. citrulli MH21 were subsequently measured at concentrations below their MICs. BCD01, BCD02, BCD03, BCD04, and BCD05 had relatively low MEC (Table 2). Moreover, using BCD03 as an example, the color of the reaction changed from red to yellow with increasing addition, indicating a dosage effect of the inhibition on the function of T3SS (Figure 2C).
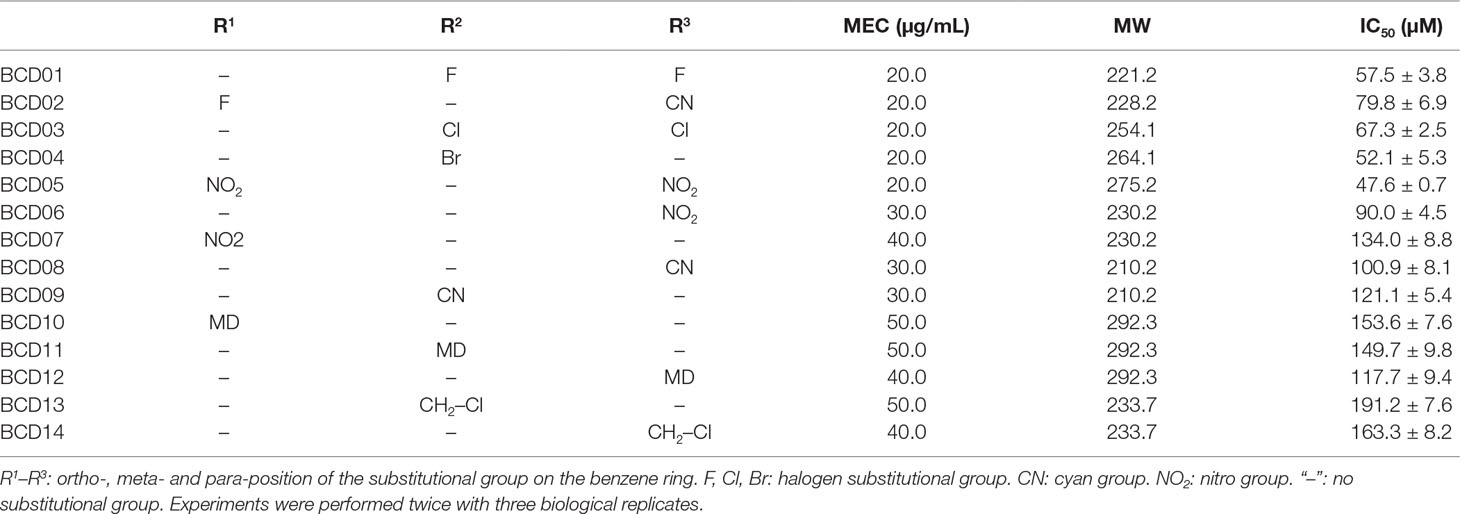
Table 2 Derivatives of benzyloxy carbonimidoyl dicyanide (BCD) with their minimum effective concentrations (MEC), molecular weights (MW) and half maximal inhibitory concentrations (IC50).
Benzene substituted by halogens as in BCD01, BCD02 (containing an additional cyan group), BCD03 and BCD04, had relatively low IC50 values that mean the high T3SS-inhibitory activities. BCD13 and BCD14 also possessed a halogen substitution, but not directly on the benzene, resulting in a decreased inhibitory activity. Of the three nitro-substituted BCDs (BCD05, BCD06, and BCD07), BCD05, which had two nitro substitutes at both a para- and ortho-position, had the highest inhibitory activity, suggesting that two substitutions are better than one. The increased inhibitory activity of BCD06 compared to BCD07 further suggests a para-position substitution is also beneficial under single substitutional situation. A similar position-activity relationship was observed with BCD10-12, whereby ortho-, meta- and para-position substitutions resulted in IC50 values of 153.6, 149.7, and 117.7 μM, respectively. However, cyan substitutions at para- and meta-positions did not result in any significant differences between BCD08 and BCD09. Overall, with the single substitution, para-position of the compounds resulted in the highest T3SS inhibitory activity, and the replacement of halogens or nitro substitutional groups resulted in an increase of T3SS inhibitory effect.
The HR-inducing capacities of bacteria belonging to Hrp groups I (P. syringae pv. tomato DC3000) and II (X. oryzae pv., oryzae PXO99A, and A. citrulli MH21), and their T3SS mutants were examined in N. tabacum in the presence of BCD03. MICs of BCD03 in strains DC3000, PXO99A, and MH21 were 200, 50 and 100 µg/mL, respectively (Figure S3). BCD03 was therefore used at the sub-MIC concentration of 25 µg/mL for all the above bacteria. HR symptoms were observed on leaves of N. tabacum 18 h after inoculation with wild-type bacteria (MH21, PXO99A, and DC3000), but were dramatically suppressed when BCD03 was added (Figure 3A). No cell death symptoms developed on leaves treated with the T3SS mutants or BCD03 alone (Figure 3A). These results suggest that BCD03 inhibits T3SS-based HR induction in bacteria of at least three genera belonging to two Hrp groups.
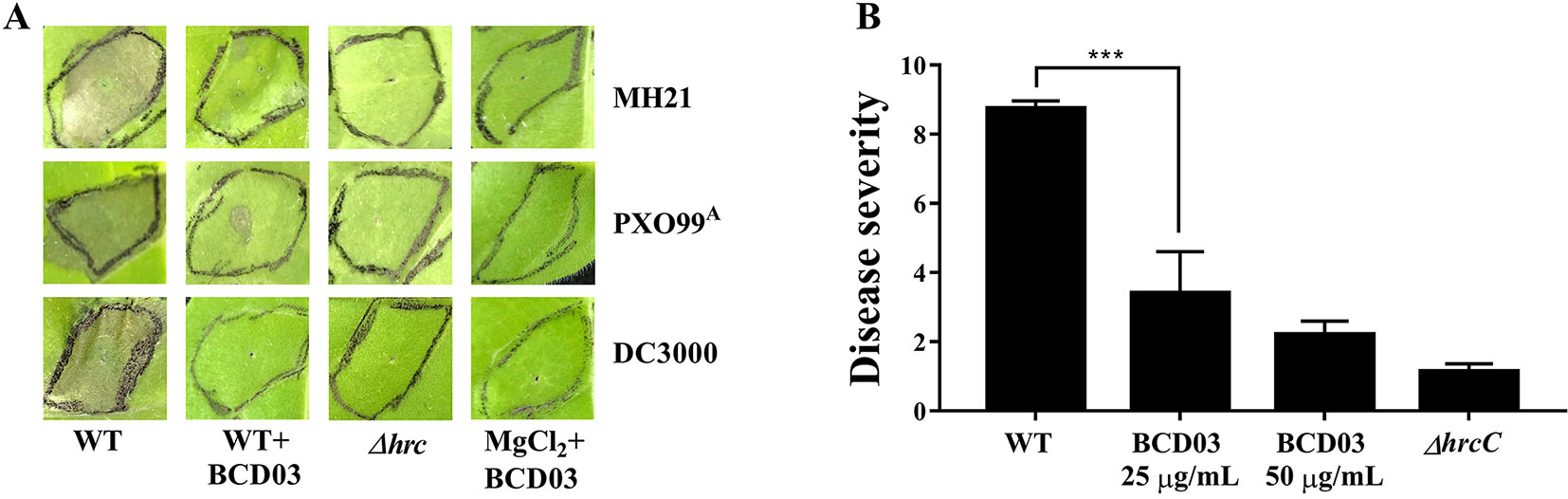
Figure 3 (A) BCD03 suppression of hypersensitive responses in Nicotiana tabacum triggered by pathogenic bacteria. A. citrulli MH21, X. oryzae pv. oryzae PXO99A, P. syringae pv. tomato DC3000 and their T3SS mutants, with or without BCD03 (25 µg/mL added 2 h before infiltration), were infiltrated into the leaves (3 × 108 cfu/mL for MH21 and PXO99A, 3 × 106 cfu/ml for DC3000) then the response was photographed at 18 h post inoculation. The control group was treated with 10 mM MgCl2 mix with 25 µg/mL BCD03. Photos from one of three biological replicate experiments are shown. (B) Effect of BCD03 on infection of A. citrulli MH21 on melon seedlings. Germinated melon seeds were dipped in bacterial suspension at 3 x 108 CFU, supplemented with 0, 25 or 50 µg/mL BCD03, respectively. Disease severity was investigated 12 days after sowing. Disease ratings are defined using a scale-based symptom appearance as described in Methods section. The experiments were repeated twice with three biological replicates. Each replicate contained 5 seedlings. Statistically significant differences (Two-way ANOVA) were observed between wild type treated groups and other groups. P-value < 0.01 (***).
To test the inhibitory activity of BCD03 on bacterial pathogenicity, the wild type A. citrulli MH21 with or without BCD03 at its sub-MICs was inoculated on the germinated melon seeds before sowing. Disease investigation at seedling stage showed a sever disease caused by the wild type MH21, but not its T3SS mutant ΔhrcC, indicating the critical role of T3SS in bacterial infection (Figure 3B). A dramatic decrease of disease severity was observed on seedlings treated with the wild type MH21 and BCD03 at concentrations 25 µg/mL and 50 µg/mL (Figure 3B). As a result, BCD03 reduced the T3SS-dependent pathogenicity of A. citrulli MH21 on its host plant at sub-MICs.
When cultured in hrp-inducing media, plant pathogenic bacteria deliver T3Es to extracellular spaces via T3SS. We therefore examined the inhibitory effect of BCD03 on T3SS-dependent effector secretion in three different bacteria, A. citrulli MH21, X. oryzae PXO99A, and P. syringae DC3000. All three wild-type bacteria and their T3SS mutants produced T3Es inside the cell, but only those of the wild-type bacteria were detectable in intracellular medium by Western blotting analysis, indicating the requirement of T3SS for effector secretion (Figure 4). BCD03 treatment suppressed secretion of effector HopAO1 and most of effector AvrXa27 in the Hrp group II bacterial strains MH21 and PXO99A, respectively (Figures 4A, B). Similarly, secretion of effector AvrPto was suppressed in Hrp group I strain DC3000 (Figure 4C). It is remarkable that intracellular production of T3Es in all three bacteria was not reduced, which suggests that only secretion but not expression of T3Es was inhibited by BCD03. Overall, the Western blotting results suggest that BCD03 has a broad-spectrum inhibitory effect on plant pathogen bacterial T3SSs.
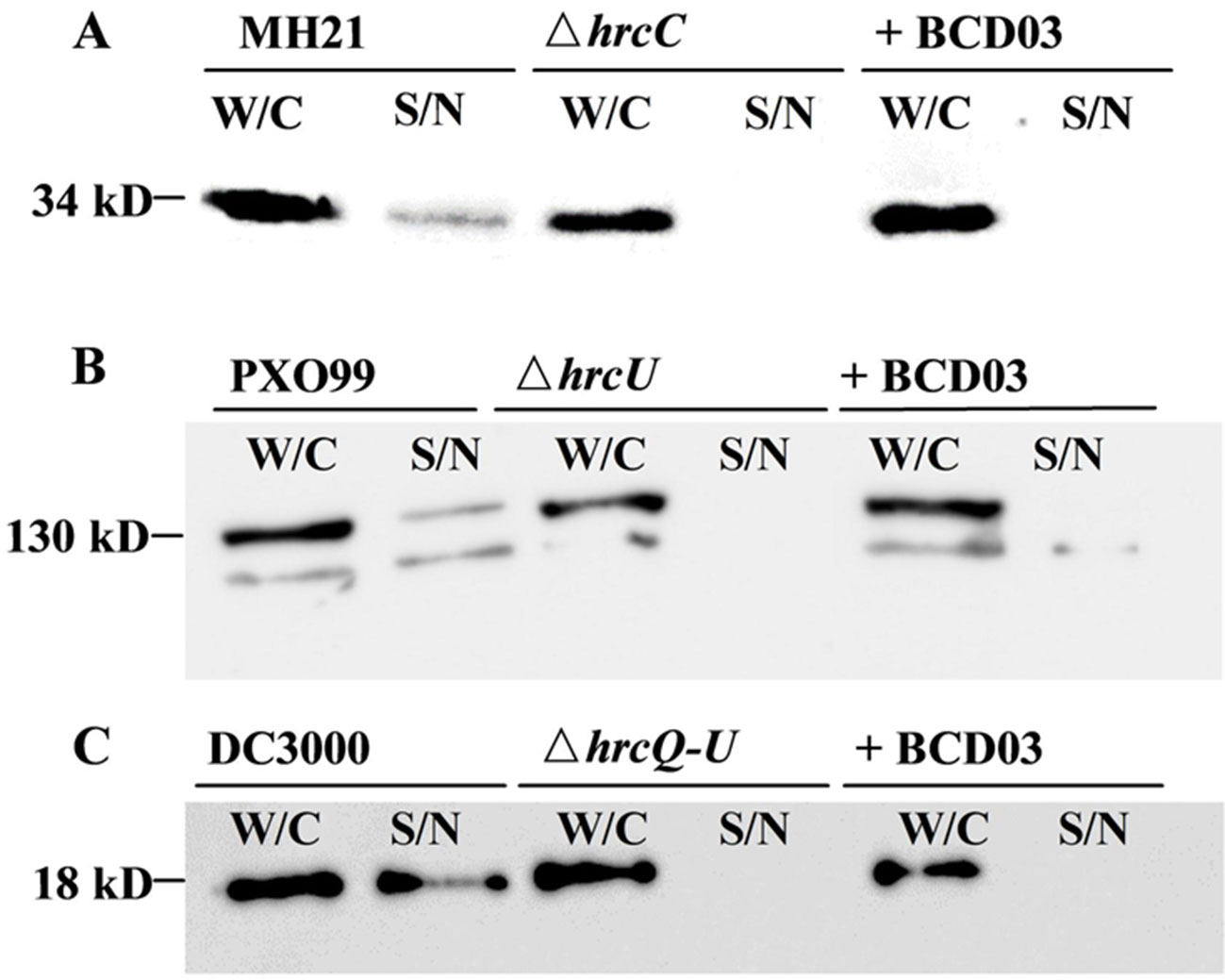
Figure 4 BCD03 inhibition of effector secretion via bacterial T3SS. (A) A. citrulli MH21 bearing the hopAO1-FLAG construct. (B) X. oryzae pv. oryzae PXO99A bearing the avrXa27-FLAG construct. (C) P. syringae pv. tomato DC3000 bearing the avrPto-HA construct. All bacteria were grown for 8 h in their own hrp-inducing media with or without BCD03 at a finial concentration of 25 µg/mL. Whole cell extracts (W/C) and supernatants (S/N) were separated by centrifugation, and their secreted proteins were detected with anti-FLAG or anti-HA antibodies.
In this study, we developed a high-throughput system based on a T3E secretion reporter to screen bacterial T3SS inhibitors, which is distinguished from most previous studies using transcriptional reporters of T3SS structure genes, regulatory genes or T3E genes (Kauppi et al., 2003; Gauthier et al., 2005; Li et al., 2005; Nordfelth et al., 2005; Pan et al., 2007; Felise et al., 2008; Pan et al., 2009; Aiello et al., 2010; Enquist et al., 2012; Fan et al., 2017). Inhibitors obtained from previous screening commonly affect transcription of T3SS structural gene clusters and T3E genes (Gauthier et al., 2005; Lee et al., 2007; Arnoldo et al., 2008; Kim et al., 2009; Garrity-Ryan et al., 2010; Grier et al., 2010; Swietnicki et al., 2011; Yamazaki et al., 2012; Kim et al., 2014; Williams et al., 2015; Marsden et al., 2016), while potential inhibitors targeting post-transcriptional regulation or effector secretion were missed. In this study, we constructed a T3E secretion reporter for inhibitor screening since effector secretion is the end result of T3SS expression, which is delicately regulated by a complex network, including transcriptional and post-transcriptional regulation of T3SS components and regulatory genes, post-translational modification of T3SS structural proteins and assembly of the needle or pilus structure, as well as interactions between T3Es and T3SS structural proteins (Keyser et al., 2008; Harmon et al., 2010; Kimura et al., 2011; Warrener et al., 2014). A similar screening system was developed in Yersinia using β-lactamase fused with the T3E YopE, revealing a group of compounds that specifically inhibited translocation of effectors into the host cells without altering the synthesis and secretion of T3Es (Harmon et al., 2010). Revealing T3SS inhibitors with this kind of action model is difficult by using transcriptional reporters, highlighting the importance of secretion reporters in large-scale screening.
Because our screening system detected T3SS-induced extracellular activity of β-lactamase, cell growth had to be tightly controlled to avoid β-lactamase leakage during bacterial cell lysis. With A. citrulli MH21, a narrow window of time was revealed when the bacteria were grown in suitable medium for 20 h (OD600 = 0.4–0.6), during which extracellular β-lactamase activity was significantly higher in the wild-type strain than its T3SS mutant. This difference was less obvious with further incubation, suggesting that β-lactamase release occurs following cell lysis in the T3SS mutant (Figure 1B). β-lactamase was used as the reporter because of its high sensitivity, ease of operation and clear color change readout from yellow to red; however, special attention is required when using it in T3SS inhibitor screening. β-lactamase is not applicable when screening dark-colored compounds, as it can disturb the color readout of the reaction mixture. In addition, the positive compounds obtained using β-lactamase reporter need to be re-tested to avoid false positive results caused by potential lactamase-inhibitory activity.
Structure–function analysis of the BCD derivatives revealed that the single substitution at para-position on the benzene ring had the greatest effect on T3SS inhibition. It is likely that para-position substitution resulted in a stronger binding ability with the targets, or was similar to its competitors. However, although halogen substitutions of the BCD derivatives resulted in an increased T3SS inhibitory effect, it remains unknown whether other substitutions such as hydroxy, alkyl, and sulfo, also play a role in T3SS inhibition, suggesting the possible existence of other highly-efficient BCD derivatives.
The BCD compound BCD03 inhibited T3SS of plant pathogenic bacteria in distinct Hrp groups, suggesting that it targets a common component shared by T3SSs in different bacteria. Some broad-spectrum T3SS inhibitors have already been reported, such as salicylidene, acylhydrazides, and thiazolidinones, both of which affect T3SS in multiple bacterial species (Aiello et al., 2010; Duncan et al., 2012). Salicylidene acylhydrazides were first found using Yersinia as the screening model, and later proved effective against Chlamydia, Pseudomonas, Escherichia, Salmonella and Shigella bacteria (Kauppi et al., 2003; Gauthier et al., 2005; Muschiol et al., 2006; Hudson et al., 2007; Veenendaal et al., 2009). Similarly, thiazolidinone inhibits the pathogenicity of Yersinia by inhibiting the formation of T3SS injectisome, preventing effector secretion of Francisella by targeting the type IV secretion system (Felise et al., 2008). Moreover, thiazolidinone was the first T3SS inhibitor reported to inhibit the plant pathogen P. syringae (Felise et al., 2008). Accordingly, the inhibitory effect of BCD derivatives on animal pathogenic bacteria is an attractive subject of future study.
The discovery of T3SS inhibitors has become a significant topic in recent years, and a number of inhibitors with different action models have subsequently been examined. Salicylidene acylhydrazides derivatives have diverse inhibitory mechanisms. For example, INP0007 blocks the translocation of Yop T3Es and protects HeLa cells from Yersinia infection (Nordfelth et al., 2005), while INP1750 binds to YscN, the ATPase of Yersinia, resulting in the suppression of T3Es secretion (Swietnicki et al., 2011). Meanwhile, other inhibitors specifically target T3Es. For example, exosin, which was identified in P. aeruginosa, directly blocks the ADP-ribosyltransferase activity of T3E ExoS, while pseudolipasin A suppresses the phospholipase A2 activity of T3E ExoU (Lee et al., 2007; Arnoldo et al., 2008). Other compounds have also been screened as T3SS inhibitors, such as caminoside A, aurodox, and o-coumaric acid; however, their molecular targets remain unknown (Linington et al., 2002; Kimura et al., 2011; Fan et al., 2017). The BCD derivatives identified in this study had a significant inhibitory effect on T3Es secretion in several plant pathogenic bacteria and showed the ability to suppress the pathogenicity of A. citrulli on its natural host. To express sufficient T3Es for Western blotting analysis, the T3E gene hopAO1 in A. citrulli and avrXa27 gene in X. oryzae were placed under control of constitutive expression promoters, making it hard to distinguish between transcriptional differences by comparing Western Blotting bands. Meanwhile, expression of the T3Es gene avrPto under its own T3SS-inducing promoter had no obvious influence by BCD03 in terms of transcription of the reporter gene. It enhances our speculation that the BCD derivatives directly blocks T3E secretion through the T3SS complex. Our future study is to identify the potential targets of the BCD compounds in bacteria, which may help us explain their T3SS inhibitory mechanism and determine their applicability.
All datasets for this study are included in the manuscript and the Supplementary Files.
LC, Y-NM, J-LL and L-QZ designed the research; J-LL and L-QZ supervised the study; LC and Y-NM performed all the experiments and analyzed data. Y-NM and L-QZ wrote the paper. All authors revised the manuscript and approved the final version for submission.
This work was funded by the Key Project of Chinese National Programs for Fundamental Research and Development (973 Program) (2015CB150605), National Key Research and Development Program (2017YFD0201106), Special Fund for Agro-scientific Research in the Public Interest (201503109), and the National Natural Science Foundation of China (31572045, 31872020).
Authors Liang Chen, Nai-Guo Si, Jun-Li Liu were employed by company Shenyang Sinochem Agrochemicals R&D Co., Ltd, Shenyang, China. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to thank Dr. Hailei Wei, Chinese Academy of Agricultural Sciences, for providing the plasmid pCPP5372-avrPto-HA and strains DC3000 and DC3000ΔhrcQ-U; and Dr. Gongyou Chen, Shanghai Jiao Tong University, for providing the plasmid pHZY-avrXa27 and strains PXO99A and PXO99AΔhrcU.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01059/full#supplementary-material
Aiello, D., Williams, J. D., Majgier-Baranowska, H., Patel, I., Peet, N. P., Huang, J., et al. (2010). Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob. Agents Chemother. 54, 1988–1999. doi: 10.1128/AAC.01598-09
Alfano, J. R., Bauer, D. W., Milos, T. M., Collmer, A. (1996). Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19, 715–728. doi: 10.1046/j.1365-2958.1996.415946.x
Alfano, J. R., Collmer, A. (1996). Bacterial pathogens in plants: life up against the wall. Plant Cell 8, 1683. doi: 10.2307/3870222
Ambler, R. P. (1980). The structure of β-lactamases. Philos. Trans. R. Soc. B-Biol. Sci. 289, 321–331. doi: 10.1098/rstb.1980.0049
Anantharajah, A., Mingeot-Leclercq, M.-P., Van Bambeke, F. (2016). Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol. Sci. 37, 734–749. doi: 10.1016/j.tips.2016.05.011
Arnoldo, A., Curak, J., Kittanakom, S., Chevelev, I., Lee, V. T., Sahebol-Amri, M., et al. (2008). Identification of small molecule inhibitors of Pseudomonas aeruginosa exoenzyme S using a yeast phenotypic screen. PLoS. Genet. 4, e1000005. doi: 10.1371/journal.pgen.1000005
Büttner, D. (2012). Protein export according to schedule: architecture, assembly, and regulation of type III secretion systems from plant-and animal-pathogenic bacteria. Microbiol. Mol. Biol. Rev. 76, 262–310. doi: 10.1128/MMBR.05017-11
Badel, J. L., Shimizu, R., Oh, H.-S., Collmer, A. (2006). A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant-Microbe Interact. 19, 99–111. doi: 10.1094/MPMI-19-0099
Bahar, O., Burdman, S. (2010). Bacterial fruit blotch: a threat to the cucurbit industry. Isr. J. Plant Sci. 58, 19–31. doi: 10.1560/IJPS.58.1.19
Bonas, U. (1994). “Hrp genes of phytopathogenic bacteria,” in Bacterial pathogenesis of plants and animals. (Berlin, Heidelberg: Springer), 79–96. doi: 10.1007/978-3-642-78624-2_4
Boucher, C. A., Barberis, P. A., Demery, D. A. (1985). Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5-induced avirulent mutants. Microbiology 131, 2449–2457. doi: 10.1099/00221287-131-9-2449
Bowlin, N. O., Williams, J. D., Knoten, C. A., Torhan, M. C., Tashjian, T. F., Li, B., et al. (2014). Mutations in the P. aeruginosa needle protein gene pscF confer resistance to phenoxyacetamide inhibitors of the type III secretion system. Antimicrob. Agents Chemother. 58, 02795–02713. doi: 10.1128/AAC.02795-13
Bretz, J. R., Mock, N. M., Charity, J. C., Zeyad, S., Baker, C. J., Hutcheson, S. W. (2003). A translocated protein tyrosine phosphatase of Pseudomonas syringae pv. tomato DC3000 modulates plant defence response to infection. Mol. Microbiol. 49, 389–400. doi: 10.1046/j.1365-2958.2003.03616.x
Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., et al. (2003). The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000 . Proc. Natl. Acad. Sci. 100, 10181–10186. doi: 10.1073/pnas.1731982100
Cornelis, G. R. (2006). The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811. doi: 10.1038/nrmicro1526
Diepold, A., Armitage, J. P. (2015). Type III secretion systems: the bacterial flagellum and the injectisome. Philos. Trans. R. Soc. B-Biol. Sci. 370, 20150020. doi: 10.1098/rstb.2015.0020
Duncan, M. C., Linington, R. G., Auerbuch, V. (2012). Chemical inhibitors of the type three secretion system: disarming bacterial pathogens. Antimicrob. Agents Chemother. 56, 5433–5441. doi: 10.1128/AAC.00975-12
Duncan, M. C., Wong, W. R., Dupzyk, A. J., Bray, W. M., Linington, R. G., Auerbuch, V. (2014). An NF-κB-based high-throughput screen identifies piericidins as inhibitors of the Yersinia pseudotuberculosis type III secretion system. Antimicrob. Agents Chemother. 58, 1118–1126. doi: 10.1128/AAC.02025-13
Eiamphungporn, W., Schaduangrat, N., Malik, A., Nantasenamat, C. (2018). Tackling the antibiotic resistance caused by class A β-Lactamases through the use of β-Lactamase inhibitory protein. Int. J. Mol. Sci. 19, 2222. doi: 10.3390/ijms19082222
Enquist, P.-A., Gylfe, Hägglund, U., Lindström, P., Norberg-Scherman, H., Sundin, C., et al. (2012). Derivatives of 8-hydroxyquinoline—antibacterial agents that target intra-and extracellular Gram-negative pathogens. Bioorg. Med. Chem. Lett. 22, 3550–3553. doi: 10.1016/j.bmcl.2012.03.096
Fan, S., Tian, F., Li, J., Hutchins, W., Chen, H., Yang, F., et al. (2017). Identification of phenolic compounds that suppress the virulence of Xanthomonas oryzae on rice via the type III secretion system. Mol. Plant Pathol. 18, 555–568. doi: 10.1111/mpp.12415
Faure, E., Mear, J.-B., Faure, K., Normand, S., Couturier-Maillard, A., Grandjean, T., et al. (2014). Pseudomonas aeruginosa type-3 secretion system dampens host defense by exploiting the NLRC4-coupled inflammasome. Am. J. Respir. Crit. Care Med. 189, 799–811. doi: 10.1164/rccm.201307-1358OC
Felise, H. B., Nguyen, H. V., Pfuetzner, R. A., Barry, K. C., Jackson, S. R., Blanc, M.-P., et al. (2008). An inhibitor of gram-negative bacterial virulence protein secretion. Cell host Microbe. 4, 325–336. doi: 10.1016/j.chom.2008.08.001
Finan, T. M., Kunkel, B., De Vos, G. F., Signer, E. R. (1986). Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167, 66–72. doi: 10.1128/jb.167.1.66-72.1986
Garrity-Ryan, L. K., Kim, O. K., Balada-Llasat, J.-M., Bartlett, V. J., Verma, A. K., Fisher, M. L., et al. (2010). Small molecule inhibitors of LcrF, a Yersinia pseudotuberculosis transcription factor, attenuate virulence and limit infection in a murine pneumonia model. Infect. Immun. 78, 4683–4690. doi: 10.1128/IAI.01305-09
Gauthier, A., Robertson, M. L., Lowden, M., Ibarra, J. A., Puente, J. L., Finlay, B. B. (2005). Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49, 4101–4109. doi: 10.1128/AAC.49.10.4101-4109.2005
Grier, M. C., Garrity-Ryan, L. K., Bartlett, V. J., Klausner, K. A., Donovan, P. J., Dudley, C., et al. (2010). N-Hydroxybenzimidazole inhibitors of ExsA MAR transcription factor in Pseudomonas aeruginosa: In vitro anti-virulence activity and metabolic stability. Bioorg. Med. Chem. Lett. 20, 3380–3383. doi: 10.1016/j.bmcl.2010.04.014
Harmon, D. E., Davis, A. J., Castillo, C., Mecsas, J. (2010). Identification and characterization of small-molecule inhibitors of Yop translocation in Yersinia pseudotuberculosis. Antimicrob. Agents Chemother. 54, 3241–3254. doi: 10.1128/AAC.00364-10
Hopkins, C. M., White, F., Choi, S., Guo, A., Leach, J. (1992). Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol. Plant-Microbe Interact. 5, 451–459. doi: 10.1094/MPMI-5-451
Hopkins, D., Thompson, C. (2002). Evaluation of Citrullus sp. germ plasm for resistance to Acidovorax avenae subsp. citrulli. Plant Dis. 86, 61–64. doi: 10.1094/PDIS.2002.86.1.61
Hudson, D. L., Layton, A. N., Field, T. R., Bowen, A. J., Wolf-Watz, H., Elofsson, M., et al. (2007). Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51, 2631–2635. doi: 10.1128/AAC.01492-06
Jessen, D. L., Bradley, D. S., Nilles, M. L. (2013). A type III secretion system inhibitor targets YopD while revealing differential regulation of secretion in calcium blind mutants of Yersinia pestis. Antimicrob. Agents Chemother. 58, 01170–01113. doi: 10.1128/AAC.01170-13
Ji, Z.-Y., Xiong, L., Zou, L.-F., Li, Y.-R., Ma, W.-X., Liu, L., et al. (2014). AvrXa7-Xa7 mediated defense in rice can be suppressed by transcriptional activator-like effectors TAL6 and TAL11a from Xanthomonas oryzae pv. oryzicola. Mol. Plant-Microbe Interact. 27, 983–995. doi: 10.1094/MPMI-09-13-0279-R
Johnson, K., Minsavage, G., Le, T., Jones, J., Walcott, R. (2011). Efficacy of a nonpathogenic Acidovorax citrulli strain as a biocontrol seed treatment for bacterial fruit blotch of cucurbits. Plant Dis. 95, 697–704. doi: 10.1094/PDIS-09-10-0660
Kauppi, A. M., Nordfelth, R., Uvell, H., Wolf-Watz, H., Elofsson, M. (2003). Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10, 241–249. doi: 10.1016/S1074-5521(03)00046-2
Keane, P., Kerr, A., New, P. (1970). Crown gall of stone fruit II. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23, 585–596. doi: 10.1071/BI9700585
Keyser, P., Elofsson, M., Rosell, S., Wolf-Watz, H. (2008). Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264, 17–29. doi: 10.1111/j.1365-2796.2008.01941.x
Kim, D., Baek, J., Song, J., Byeon, H., Min, H., Min, K. H. (2014). Identification of arylsulfonamides as ExoU inhibitors. Bioorg. Med. Chem. Lett. 24, 3823–3825. doi: 10.1016/j.bmcl.2014.06.064
Kim, O. K., Garrity-Ryan, L. K., Bartlett, V. J., Grier, M. C., Verma, A. K., Medjanis, G., et al. (2009). N-hydroxybenzimidazole inhibitors of the transcription factor LcrF in Yersinia: novel antivirulence agents. J. Med. Chem. 52, 5626–5634. doi: 10.1021/jm9006577
Kimura, K., Iwatsuki, M., Nagai, T., Matsumoto, A., Takahashi, Y., Shiomi, K., et al. (2011). A small-molecule inhibitor of the bacterial type III secretion system protects against in vivo infection with Citrobacter rodentium. J. Antibiot. 64, 197. doi: 10.1038/ja.2010.155
Kovach, M., Phillips, R., Elzer, P., Peterson, K. (1994). pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16, 800–802. doi: 10.1002/bip.360340510
Lee, V. T., Pukatzki, S., Sato, H., Kikawada, E., Kazimirova, A. A., Huang, J., et al. (2007). Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 75, 1089–1098. doi: 10.1128/IAI.01184-06
Li, X., Kang, F.-A., Macielag, M. (2005). “Triazine compounds as inhibitors of bacterial type III protein secretion systems”. Google Patents.
Lidell, M., Hutcheson, S. (1994). Characterization of the hrpJ and hrpU operons of Pseudomonas syringae pv. syringae Pss61: similarity with components of enteric bacteria involved in flagellar biogenesis and demonstration of their role in HarpinPss secretion. Mol. Plant-Microbe Interact. 7, 488–497. doi: 10.1094/MPMI-7-0488
Linington, R. G., Robertson, M., Gauthier, A., Finlay, B. B., Van Soest, R., Andersen, R. J. (2002). Caminoside A, an antimicrobial glycolipid isolated from the marine sponge Caminus sphaeroconia. Org. Lett. 4, 4089–4092. doi: 10.1021/ol0268337
Liu, X., Zou, H., Zou, L., Chen, G. (2010). Site-directed mutagenesis and functional analysis of hrcU gene from rice pathogen Xanthomonas oryzae pv. oryzae. Chin. J. Rice Sci. 24, 348–352. doi: 10.3724/SP.J.1011.2010.01385
Luo, Z.-Q., Farrand, S. K. (1999). Signal-dependent DNA binding and functional domains of the quorum-sensing activator TraR as identified by repressor activity. Proc. Natl. Acad. Sci. 96, 9009–9014. doi: 10.1073/pnas.96.16.9009
Lynch, S. V., Flanagan, J. L., Sawa, T., Fang, A., Baek, M. S., Rubio-Mills, A., et al. (2010). Polymorphisms in the Pseudomonas aeruginosa type III secretion protein, PcrV — implications for anti-PcrV immunotherapy. Microb. Pathog. 48, 197–204. doi: 10.1016/j.micpath.2010.02.008
Marsden, A. E., King, J. M., Spies, M. A., Kim, O. K., Yahr, T. L. (2015). Inhibition of Pseudomonas aeruginosa ExsA DNA-binding activity by N-hydroxybenzimidazoles. Antimicrob. Agents Chemother. 60, 02242–02215. doi: 10.1128/AAC.02242-15
Marsden, A. E., King, J. M., Spies, M. A., Kim, O. K., Yahr, T. L. (2016). Inhibition of Pseudomonas aeruginosa ExsA DNA-binding activity by N-hydroxybenzimidazoles. Antimicrob. Agents Chemother. 60, 766–776. doi: 10.1128/AAC.02242-15
Matagne, A., Dubus, A., Galleni, M., Frère, J.-M. (1999). The β-lactamase cycle: a tale of selective pressure and bacterial ingenuity. Nat. Prod. Rep. 16, 1–19. doi: 10.1039/a705983c
Muschiol, S., Bailey, L., Gylfe, Å., Sundin, C., Hultenby, K., Bergström, S., et al. (2006). A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. 103, 14566–14571. doi: 10.1073/pnas.0606412103
Nilles, M. L., Condry, D. L. J. (2017). Type 3 Secretion Systems. (New York, NY: Springer). doi: 10.1007/978-1-4939-6649-3
Nordfelth, R., Kauppi, A. M., Norberg, H., Wolf-Watz, H., Elofsson, M. (2005). Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73, 3104–3114. doi: 10.1128/IAI.73.5.3104-3114.2005
Pan, N., Goguen, J., Lee, C. (2007). “High throughput screening for small-molecule inhibitors of type III secretion in Yersinia pestis,” in The Genus Yersinia. (New York, NY: Springer) 367–375. doi: 10.1007/978-0-387-72124-8_34
Pan, N. J., Brady, M. J., Leong, J. M., Goguen, J. D. (2009). Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53, 385–392. doi: 10.1128/AAC.00670-08
Rasko, D. A., Sperandio, V. (2010). Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117. doi: 10.1038/nrd3013
Ren, Z., Jiang, W., Ni, X., Lin, M., Zhang, W., Tian, G., et al. (2014). Multiplication of Acidovorax citrulli in planta during infection of melon seedlings requires the ability to synthesize leucine. Plant Pathol. 63, 784–791. doi: 10.1111/ppa.12156
Schaad, N. W., Postnikova, E., Sechler, A., Claflin, L. E., Vidaver, A. K., Jones, J. B., et al. (2008). Reclassification of subspecies of Acidovorax avenae as A. Avenae (Manns 1905) emend., A. cattleyae (Pavarino, 1911) comb. nov., A. citrulli (Schaad et al., 1978) comb. nov., and proposal of A. oryzae sp. nov. Syst. Appl. Microbiol. 31, 434–446. doi: 10.1016/j.syapm.2008.09.003
Sebaugh, J. (2011). Guidelines for accurate EC50/IC50 estimation. Pharm. Stat. 10, 128–134. doi: 10.1002/pst.426
Swietnicki, W., Carmany, D., Retford, M., Guelta, M., Dorsey, R., Bozue, J., et al. (2011). Identification of small-molecule inhibitors of Yersinia pestis type III secretion system YscN ATPase. PLoS One 6, e19716. doi: 10.1371/journal.pone.0019716
Tampakaki, A. P., Skandalis, N., Gazi, A. D., Bastaki, M. N., Panagiotis, F, S., Charova, S. N., et al. (2010). Playing the “Harp”: evolution of our understanding of hrp/hrc genes. Annu. Rev. Phytopathol. 48, 347–370. doi: 10.1146/annurev-phyto-073009-114407
Tsuge, S., Furutani, A., Fukunaka, R., Takashi, O., Tsuno, K., Ochiai, H., et al. (2002). Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J. Gen. Plant Pathol. 68, 363–371. doi: 10.1007/PL00013104
Underwood, W., Zhang, S., He, S. Y. (2007). The Pseudomonas syringae type III effector tyrosine phosphatase HopAO1 suppresses innate immunity in Arabidopsis thaliana. Plant J. 52, 658–672. doi: 10.1111/j.1365-313X.2007.03262.x
Veenendaal, A. K., Sundin, C., Blocker, A. J. (2009). Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. Bacteriol. 191, 563–570. doi: 10.1128/JB.01004-08
Wang, D., Zetterstrom, C. E., Gabrielsen, M., Beckham, K. S., Tree, J. J., Macdonald, S. E., et al. (2011). Identification of bacterial target proteins for the salicylidene acylhydrazide class of virulence blocking compounds. J. Biol. Chem. M111, 233858. doi: 10.1074/jbc.M111.233858
Warrener, P., Varkey, R., Bonnell, J. C., Digiandomenico, A., Camara, M., Cook, K., et al. (2014). A novel anti-PcrV antibody providing enhanced protection against Pseudomonas aeruginosa in multiple animal infection models. Antimicrob. Agents Chemother. 58, 4384–4391. doi: 10.1128/AAC.02643-14
Wei, H.-L., Chakravarthy, S., Mathieu, J., Helmann, T. C., Stodghill, P., Swingle, B., et al. (2015). Pseudomonas syringae pv. tomato DC3000 type III secretion effector polymutants reveal an interplay between HopAD1 and AvrPtoB. Cell host Microbe 17, 752–762. doi: 10.1016/j.chom.2015.05.007
Wengelnik, K., Bonas, U. (1996). HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178, 3462–3469. doi: 10.1111/j.1365-2672.1996.tb03274.x
Wengelnik, K., Van Den Ackerveken, G., Bonas, U. (1996). HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria ls homologous to two-component response regulators. Mol. Plant-Microbe Interact. 9, 704–712. doi: 10.1094/MPMI-9-0704
Williams, J. D., Torhan, M. C., Neelagiri, V. R., Brown, C., Bowlin, N. O., Di, M., et al. (2015). Synthesis and structure–activity relationships of novel phenoxyacetamide inhibitors of the Pseudomonas aeruginosa type III secretion system (T3SS). Bioorg. Med. Chem. 23, 1027–1043. doi: 10.1016/j.bmc.2015.01.011
Yamazaki, A., Li, J., Zeng, Q., Khokhani, D., Hutchins, W. C., Yost, A. C., et al. (2012). Derivatives of plant phenolic compound affect the type III secretion system of Pseudomonas aeruginosa via a GacS-GacA two-component signal transduction system. Antimicrob. Agents Chemother. 56, 36–43. doi: 10.1128/AAC.00732-11
Yang, F., Korban, S. S., Pusey, P. L., Elofsson, M., Sundin, G. W., Zhao, Y. (2014). Small-molecule inhibitors suppress the expression of both type III secretion and amylovoran biosynthesis genes in Erwinia amylovora. Mol. Plant Pathol. 15, 44–57. doi: 10.1111/mpp.12064
Keywords: Acidovorax, Xanthomonas, Pseudomonas, type III secretion system, hypersensitive response, virulence inhibitor, benzyloxy carbonimidoyl dicyanide
Citation: Ma Y-N, Chen L, Si N-G, Jiang W-J, Zhou Z-G, Liu J-L and Zhang L-Q (2019) Identification of Benzyloxy Carbonimidoyl Dicyanide Derivatives as Novel Type III Secretion System Inhibitors via High-Throughput Screening. Front. Plant Sci. 10:1059. doi: 10.3389/fpls.2019.01059
Received: 15 March 2019; Accepted: 05 August 2019;
Published: 05 September 2019.
Edited by:
Gail Preston, University of Oxford, United KingdomReviewed by:
Hai-Lei Wei, Institute of Agricultural Resources and Regional Planning (CAAS), ChinaCopyright © 2019 Ma, Chen, Si, Jiang, Zhou, Liu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Qun Zhang, emhhbmdscUBjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.