
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 13 August 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.01010
This article is part of the Research Topic New Insights into Salinity Sensing, Signaling and Adaptation in Plants View all 26 articles
Mitochondria play a central role in the energy metabolism of plants. At the same time, they provide energy for plant stress responses. We here report a first view on the mitochondrial Oxidative Phosphorylation (OXPHOS) system of the halophile (salt tolerant) plant Cakile maritima. Mitochondria were purified from suspension cultures of C. maritima and for comparison of Arabidopsis thaliana, a closely related glycophyte (salt sensitive) plant. Mitochondria were treated with digitonin and solubilized protein complexes were analyzed by 2D Blue native/SDS polyacrylamide gel electrophoresis. The OXPHOS systems of the two compared plants exhibit some distinct differences. C. maritima mitochondria include a very abundant respiratory supercomplex composed of monomeric complex I and dimeric complex III. At the same time the complexes II and IV are of reduced abundance. The stability of the OXPHOS complexes was investigated by combined salt and temperature treatments of isolated mitochondria. ATP synthase (complex V) is of increased stability in C. maritima. Also, the I + III2 supercomplex is present in high abundance during stress treatments. These results give insights into the mitochondrial contribution to the plant salt stress response.
Halophile plants have extraordinary competence to live on soils with high contents of salt. This aptitude is based on various physiological properties, like active salt secretion from cells, inner-cellular accumulation of salt in the vacuole and the biosynthesis of compatible osmolytes like proline or glycine betaine (Munns and Gilliham, 2015). In general, increased inner-cellular salt concentrations can cause severe damages, e.g., denaturation of proteins and formation of reactive oxygen species (ROS). However, halophile plants have particular capabilities to cope with these circumstances. For instance, they may have enhanced intra-cellular levels of reductants, such as ascorbate, glutathione or NADPH, which counteract increased ROS formation (Ismail and Horie, 2017). At the same time, enhanced levels of heat stress proteins can stabilize the native structures of proteins.
Plant life in the presence of increased salt levels requires extra energy in the form of ATP (Jacoby et al., 2011, 2018; Bose et al., 2017). ATP is needed for actively secreting salt ions from cells or accumulating salt ions in the vacuole (Nikalje et al., 2018). The corresponding transport processes against the respective concentration gradients are based on proton gradients across the plasma membrane or the tonoplast, which are generated by the act of membrane-bound proton-ATPases (Munns et al., 2016). Furthermore, the biosynthesis of compatible osmolytes, reductants and heat-stress proteins requires additional ATP. In plants, ATP is mainly produced by oxidative phosphorylation (OXPHOS) in the mitochondria and photophosphorylation (PHOTOPHOS) in the chloroplasts. However, PHOTOPHOS only takes place in green cells (it is absent in roots and some organs of flowers) and only operates at daytime. OXPHOS therefore is of outstanding importance for halophyte physiology.
Prerequisite for mitochondrial ATP formation is the respiratory electron transfer chain and the ATP synthase complex (complex V). The respiratory chain is composed of four protein complexes, the NADH dehydrogenase complex (complex I), the succinate dehydrogenase complex (complex II), the cytochrome c reductase complex (complex III), and the cytochrome c oxidase complex (complex IV). Furthermore, cytochrome c, a small monomeric protein, and the lipid ubiquinone are required for the respiratory electron transport. Additionally, in plants and some other groups of organisms, extra enzymes can participate in respiratory electron transport, e.g., the alternative oxidase (AOX) or alternative NADH dehydrogenases (Millar et al., 2011; Schertl and Braun, 2014). As such, the respiratory electron transfer chain is branched, which offers additional physiological functions, but at the same requires some extra levels of regulation.
To our knowledge, the molecular features of the OXPHOS system in halophytes have not been characterized so far. Due to extra energy requirement, we hypothesize that the system should have efficient operation modes. Furthermore, high salt and simultaneously, in some environments, high temperatures might require increased structural stability of the involved protein complexes.
Here, we report the molecular characterization of the OXPHOS system of Cakile maritima, a halophile plant. In parallel, the OXPHOS system of the model plant Arabidopsis thaliana, a glycophyte (salt sensitive) plant, which also belongs to the Brassicaceae family of angiosperms, is characterized for comparison. For optimal comparability, non-green suspension cell cultures were established for both species and used for parallel mitochondrial isolations. Using two-dimensional Blue native/SDS polyacrylamide gel electrophoresis in combination with differential fluorophore-based labeling of proteins, we here provide insights into the composition and stability of the OXPHOS system of a halophile plant.
Seeds of A. thaliana (Columbia 0 ecotype) and C. maritima (Raoued ecotype) were sterilized by treatment with 70% ethanol (4 min under shaking) and 6% sodium hypochlorite solution (4 min under shaking). After washing the seeds five times with sterile distilled H2O they were plated on solid Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) supplemented with 0.8% agar. Young A. thaliana and C. maritima plants (age approximately 6 days) grown under sterile conditions were dissected into small pieces with a diameter of about 3 mm. Obtained plant pieces were laid out on solid B5-medium and cultivated in the dark for 2–3 weeks for callus generation.
Arabidopsis thaliana and C. maritima cell suspension cultures were established as outlined in May and Leaver (1993): Calli were transferred into 500 mL Erlenmeyer flasks containing 100 mL medium composed of 0.3% (w/v) Gamborg B5 medium supplemented with 3% (w/v) sucrose, 0.01% (w/v) 2,4-D and 0.001% (w/v) kinetin. Cultivation took place at 24°C, at darkness and continuous shaking at 100 rpm. The medium was renewed every 7 days until the generation of approximately 3 g of cell material per Erlenmeyer flask. Afterward, cells were distributed to several flasks. Cell cultures were maintained by transferring about 1.5 g of cells to new medium after 7 days (yielding about 3 g of cells prior to the next round of transferring cells to new medium). Starting material for mitochondria isolations were suspension cells from about 10 Erlenmeyer flasks at the end of a subculturing round (day 7; approximately 30 g of cells in total).
Mitochondria were isolated from A. thaliana and C. maritima suspension cell cultures as described by Werhahn et al. (2001): The cell cultures (about 30 g of cells per species) were filtered through two layers of Miracloth and homogenized at 4°C in Disruption buffer [450 mM sucrose, 15 mM MOPS, 1.5 mM EGTA, 0.6% (w/v) PVP40, 2% (w/v) BSA, 10 mM sodium ascorbate, 10 mM cysteine, and 0.2 mM PMSF, pH 7.4]. Cells were ground three times using a Waring blender (1 × 15 s at high speed, 2 × 15 s at low speed, 30–60 s intervals in between). The obtained homogenate was then centrifuged twice at 2,700 × g for 5 min (organelles in supernatant), once at 8,300 × g for 5 min (organelles in supernatant), and once at 17,000 × g for 10 min (organelles in pellet). Mitochondria were then resuspended in Wash buffer containing 0.3 M sucrose, 10 mM MOPS, 1 mM EGTA, and 0.2 mM PMSF (pH 7.4) and carefully dispersed using two strokes in a Teflon homogenizer. The resulting suspension was transferred on top of Percoll gradients [3 gradients per species; 18, 23, and 40% Percoll prepared in Gradient buffer (0.3 M sucrose, 10 mM MOPS, pH 7.4)]. After ultracentrifugation for 90 min at 70,000 × g, the mitochondria were collected from the 23/40% interphase of the gradients. Percoll was removed by three rounds of resuspending the mitochondria in Resuspension buffer (0.4 mM mannitol, 1 mM EGTA, 10 mM Tricine, and 0.2 mM PMSF, pH 7.2)/re-collecting them by centrifugation at 14,500 × g for 10 min. Final mitochondrial pellets were resuspended in Resuspension buffer (weight corresponding to 10× the weight of the mitochondrial pellet). Organelle suspensions were finally divided into aliquots of 100 μL and directly used for gel electrophoresis (see below) or shock frozen and stored at −80°C.
Isolated mitochondria of A. thaliana or C. maritima (aliquots of 100 μL corresponding to 10 mg mitochondria) were sedimented by centrifugation for 10 min at 14,300 × g. Resulting pellets were re-suspended in 100 μL of Digitonin solubilization buffer, pH 7.4 [30 mM HEPES, 150 mM potassium acetate, 10% (v/v) glycerol, and 5% (w/v) digitonin]. For salt-treatment, mitochondria were resuspended in Digitonin solubilization buffer (see above) supplemented with 300 mM NaCl. Suspensions were kept on ice for 20 min or incubated for 2 or 5 min at 50°C. After incubation on ice or at 50°C, insoluble material was removed from the suspensions by centrifugation for 10 min at full speed and 4°C. Obtained supernatants were supplemented with 5 μL of Coomassie-blue solution [750 mM aminocaproic acid, 5% (w/v) Coomassie-blue 250 G].
Fractions were directly loaded onto a Blue native (BN) gel. 2D BN/SDS–PAGE was performed as outlined by Wittig et al. (2006). Blue native separation of protein complexes was carried out in gradient gels of 4.5–16% (w/v) polyacrylamide. SDS–PAGE for second gel dimension was carried out in a separation gel [constant polyacrylamide concentration of 16.5% (w/v)], which was overlaid with a 10% (w/v) spacer gel. After completion of the electrophoretic runs, gels were fixed for 2 h [fixing solution: 15% (v/v) ethanol, 10% (v/v) acetic acid] and stained according to the Coomassie-blue colloidal protocol [staining solution: 5% (w/v) Coomassie-blue, 2% ortho phosphoric acid, and 10% (w/v) ammonia sulfate] as described by Neuhoff et al. (1985, 1990).
All comparative proteome analyses were based on at least four independent experiments (biological controls) and data evaluation using the Delta 2D software package, version 4.3 (Decodon, Greifswald, Germany) according to Berth et al. (2007) and Lorenz et al. (2014).
Fluorescent differential gel electrophoresis (DIGE) in combination with 2D Blue native/SDS–PAGE was carried out as outlined in Heinemeyer et al. (2009). CyDyes were obtained from GE Healthcare (Munich, Germany). 100 μL of the mitochondrial solutions prepared from A. thaliana and C. maritima (see above) were solubilized using Digitonin solubilization buffer (see above) and subsequently incubated with either Cy5 or Cy3 for 10 min. The reaction was stopped by addition of 1 μL Lysine solution (10 mM lysine). The two fractions were finally mixed, supplemented with Coomassie-blue buffer and proteins were separated by 2D Blue native/SDS–PAGE as given above. For details see Heinemeyer et al. (2009).
Suspension cell cultures are an ideal system for investigating house-keeping functions of organisms because they can be maintained at very defined conditions. Suspension cell cultures for A. thaliana were first established 25 years ago (May and Leaver, 1993). Their physiological properties have been carefully investigated (Davy de Virville et al., 1994, 1998). Meanwhile, numerous studies have been carried out for investigating the basic functions of cells in A. thaliana (e.g., Kruft et al., 2001; Lee et al., 2008). Successful establishment of a suspension cell culture for C. maritima was only reported 5 years ago (Ben Hamed et al., 2014). Halophyte behavior of the cells is maintained in the cell culture (Ben Hamed-Laouti et al., 2016). If compared to a suspension cell culture from A. thaliana, the cell death rate upon treatment with 400 mM NaCl was much reduced. Indeed, C. maritima suspension cells have remarkable properties for NaCl exclusion. At the same time, they have increased inner-cellular ascorbate levels (Ben Hamed-Laouti et al., 2016). We conclude that cell cultures are a suitable starting material for investigating protein complex stabilities in A. thaliana and C. maritima.
For investigating the OXPHOS system, suspension cell cultures for A. thaliana and C. maritima were established simultaneously as outlined in Figure 1. After subculturing for 7 days, about 30 g cells per species were used as starting material for parallel mitochondrial isolations by differential centrifugation and Percoll density gradient centrifugation. The protocol used for preparing mitochondria from A. thaliana suspension cell cultures has been evaluated previously and shown to generate very pure organelle fractions (purity > 95%; Senkler et al., 2017). The yield of a typical organelle preparation was about 150 mg mitochondria (pellet weight) per 30 g of A. thaliana or C. maritima cells. Mitochondrial fractions of both species were divided into aliquots corresponding to about 1 mg mitochondrial protein, shock-frozen in liquid nitrogen and stored at −80°C.
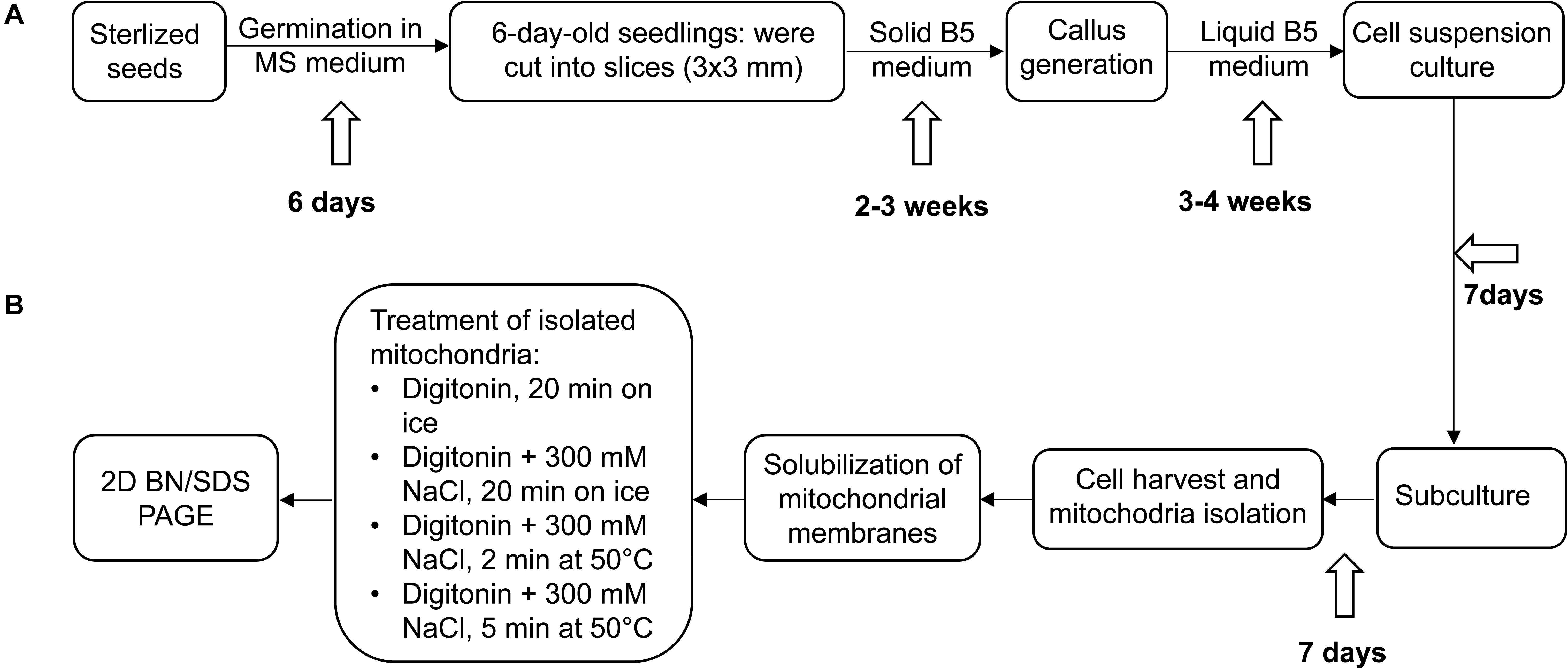
Figure 1. Experimental outline. (A) Establishment of Arabidopsis thaliana and Cakile maritima cell suspension cultures, (B) further processing of cell cultures, mitochondria isolation and solubilization. For details see Materials and methods section.
The mitochondrial OXPHOS system can be nicely characterized by Blue native gel electrophoresis (Schägger and von Jagow, 1991). Mitochondrial membranes are carefully dissolved by a mild non-ionic detergent and solubilized protein complexes incubated with a Coomassie-blue solution for careful introducing negative charge into protein complexes. Subsequently, protein complexes are separated by electrophoresis in polyacrylamide gradient gels. The native gel dimension can be combined with SDS–PAGE in orthogonal direction to separate subunits of protein complexes (Schägger and von Jagow, 1991). Mitochondrial fractions from A. thaliana and C. maritima were solubilized by digitonin (5 mg/mg mitochondrial protein) and supplemented with Coomassie-blue solution. The result of a typical 2D Blue native/SDS–PAGE analysis is shown in Figure 2.
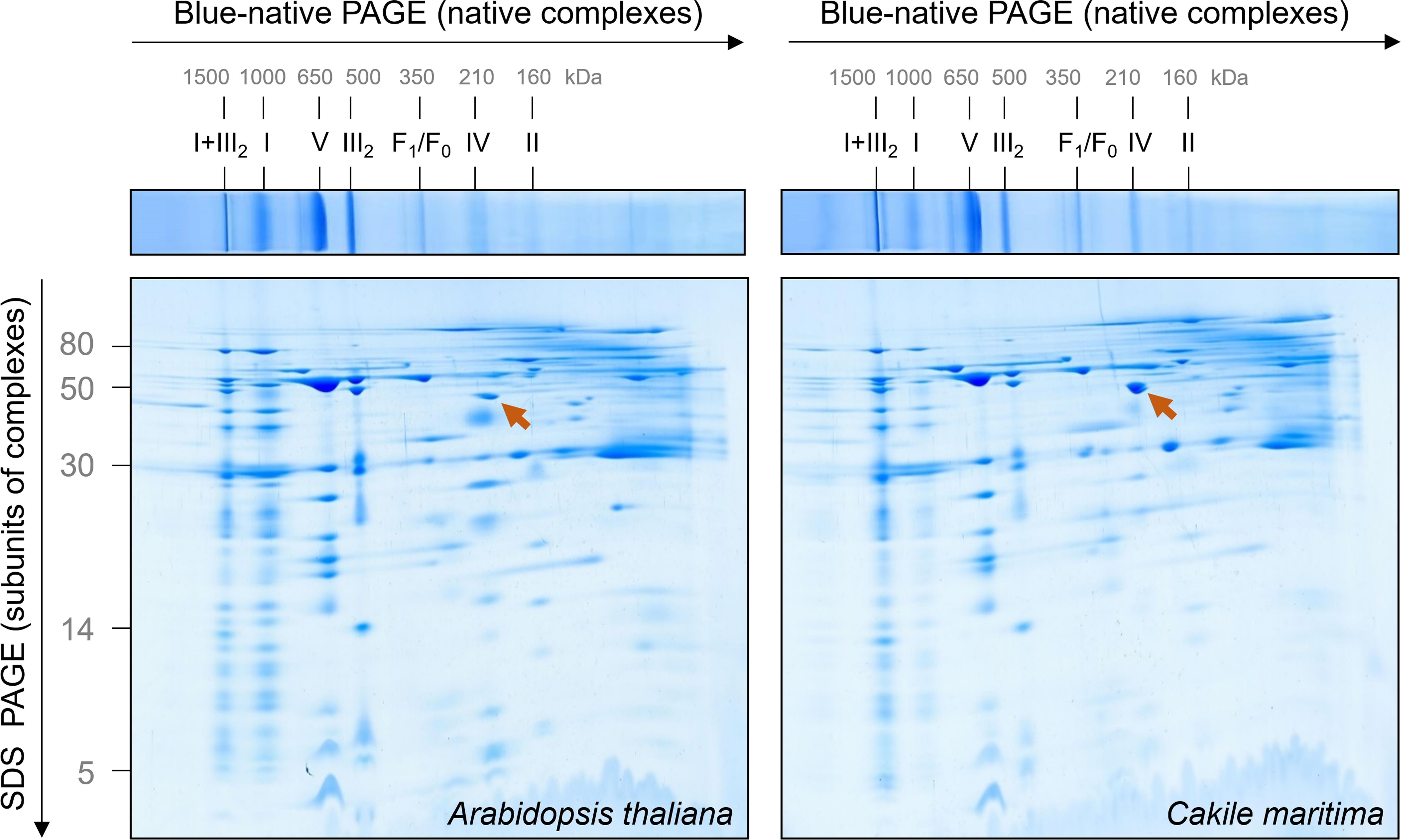
Figure 2. Comparative analysis of the mitochondrial proteomes of Arabidopsis thaliana and Cakile maritima by Blue native/SDS–PAGE. Mitochondria purified from cell suspension cultures were solubilized by digitonin. Subsequently, protein fractions were supplemented with Coomassie-blue and separated by 2D Blue native/SDS–PAGE. After the second gel dimension, gels were Coomassie stained. Molecular masses of standard protein complexes are given above, molecular masses of monomeric proteins to the left of the 2D gel (in kDa). Identities of the protein complexes are given above the BN gel. Designations: I + III2, supercomplex composed of complex I and dimeric complex III; I, complex I; V, complex V (ATP synthase); III2, dimeric complex III; F1, F1 part of ATP synthase; F0, F0 part of ATP synthase; IV, complex IV; II, complex II. The arrow points to glutamate dehydrogenase.
The OXPHOS system of A. thaliana mitochondria has been extensively characterized by 2D Blue native/SDS–PAGE in combination with mass spectrometry (Klodmann et al., 2011). A GelMap of the Arabidopsis mitochondrial proteome from cell culture is presented at the GelMap portal1. This data background allows evaluating the 2D gels shown in Figure 2. Overall, the OXPHOS systems in C. maritima and A. thaliana are highly similar, which can be expected because both species are from the same family. However, some features clearly differ: (i) Nearly all complex I forms a respiratory supercomplex with dimeric complex III in C. maritima; at identical conditions, only about 50% of complex I is associated with dimeric complex III in A. thaliana. (ii) The complexes II and IV are of decreased abundance in C. maritima. (iii) Besides differences related to the OXPHOS system, C. maritima contains high amounts of the glutamate dehydrogenase complex.
Cakile maritima and A. thaliana are closely related organisms, but the amino acid sequences of their subunits forming part of OXPHOS complexes differ slightly (precise information will become clear upon knowledge of the genome sequence of C. maritima). To visually compare molecular masses of subunits of the two species, mitochondrial fractions were differentially labeled with CyDye fluorophores, mixed, and co-electrophoresed by 2D Blue native/SDS–PAGE. On the resulting overly image, C. maritima proteins are red and A. thaliana proteins green (Figure 3). If proteins exactly match in size and abundance, the spots on the resulting 2D gel are visible in yellow. Several of the subunits of the complexes III and V are yellow, indicating similar subunit abundances and sizes. However, distinct subunits are visible as pairs of spots in red + green, indicating differences in subunit size between the two compared species. Subunits of the I + III2 supercomplex are more abundant in C. maritima (red) and subunits of monomeric complex I, complex II and complex IV in A. thaliana (green), supporting our results obtained by Coomassie-staining of the 2D Blue native/SDS gels shown in Figure 2.
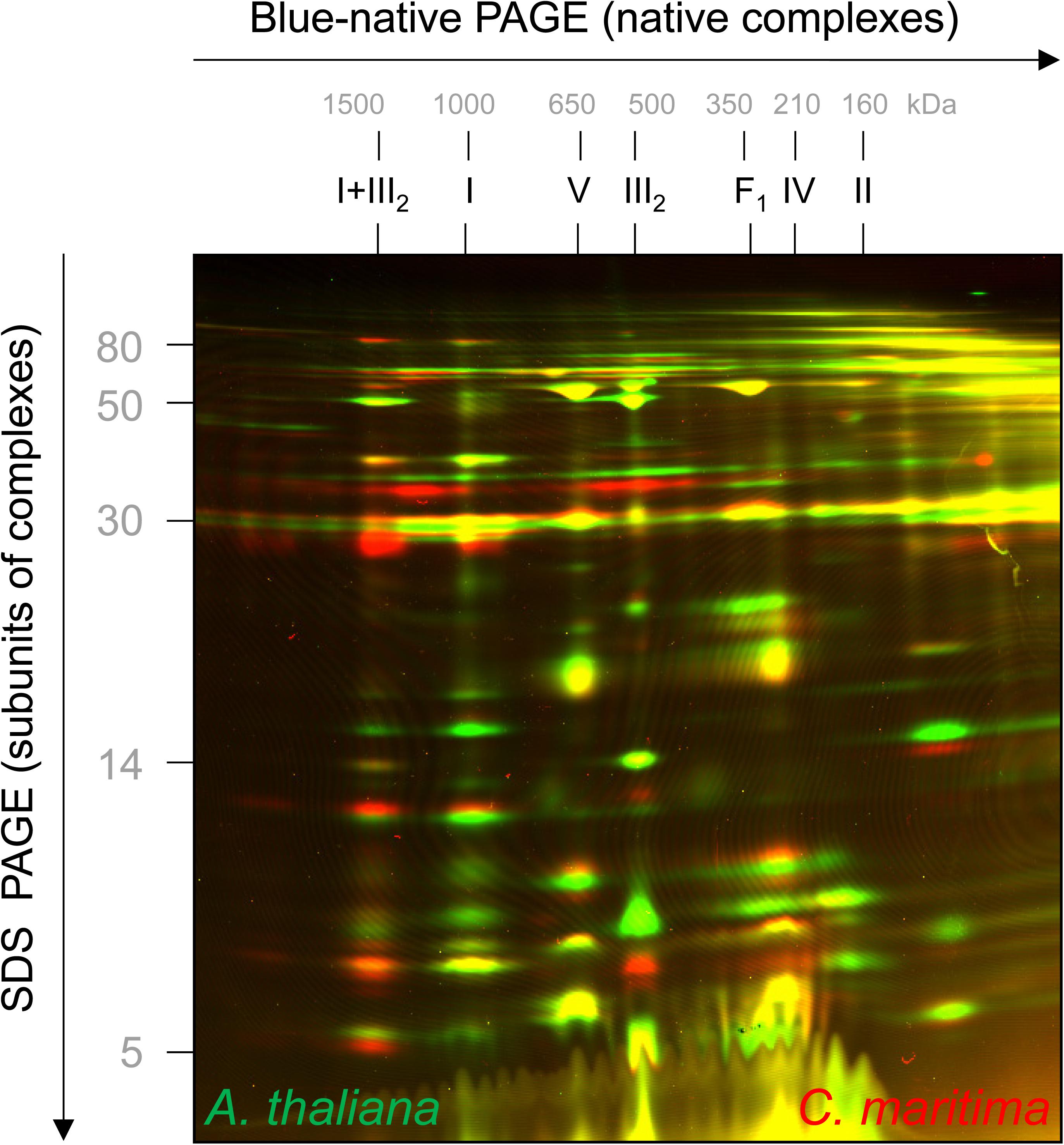
Figure 3. 2D fluorescence differential gel electrophoresis analysis of mixed mitochondrial fractions of A. thaliana and C. maritima. Proteins of A. thaliana and C. maritima mitochondrial fractions were labeled with different CyDyes (Cy3 for A. thaliana and Cy5 for C. maritima) and separated by 2D Blue native/SDS–PAGE. Visualization was carried out by laser scanning at the respective wavelengths using the Typhoon laser scanner (GE Healthcare, Munich, Germany). Identities of the resolved protein complexes are given above the 2D gel (for designations see legend of Figure 2). On the resulting overlay image, proteins of A. thaliana are seen in green and those of C. maritima in red. If proteins exactly overlap, they are seen in yellow.
We used the Raoued ecotype of C. maritima and the Col-O ecotype of A. thaliana for our investigations. Raoued is from the Mediterranean coast of Tunisia (20 km to the north of Tunis) while Col-O probably originated from central Germany (see Nordborg et al., 2005 and Somssich, 2018 for discussion). The two ecotypes are adapted to differing environments. Raoued not only is exposed to salty soils but also to increased average temperatures when compared to the Col-O ecotype of A. thaliana. What are the molecular adaptations of the two ecotypes compared in our study? We hypothesize that high salt and simultaneously high temperatures may have promoted evolution of protein complexes of increased stability in C. maritima ecotype Raoued. This was tested by directly exposing mitochondrial protein fractions of A. thaliana and C. maritima to salt and temperature followed by 2D Blue native/SDS–PAGE evaluation. Mitochondrial protein fractions of A. thaliana and C. maritima (1 mg of mitochondrial protein in 100 μL of Digitonin solubilization buffer; see Materials and methods) were treated with different NaCl concentrations and temperatures for defined periods of time. Conditions were systematically optimized for defining boundary values with respect to protein complex stability (data not shown). The following treatments were finally used: (i) 0 mM NaCl at 0°C for 20 min (= control), (ii) 300 mM NaCl at 0°C for 20 min, (iii) 300 mM NaCl at 50°C for 2 min, afterward 0°C for 18 min, (iv) 300 mM NaCl at 50°C for 15 min, afterward 0°C for 15 min. All samples were analyzed by 2D Blue native/SDS–PAGE and proteins were visualized by Coomassie-blue staining (Figure 4).
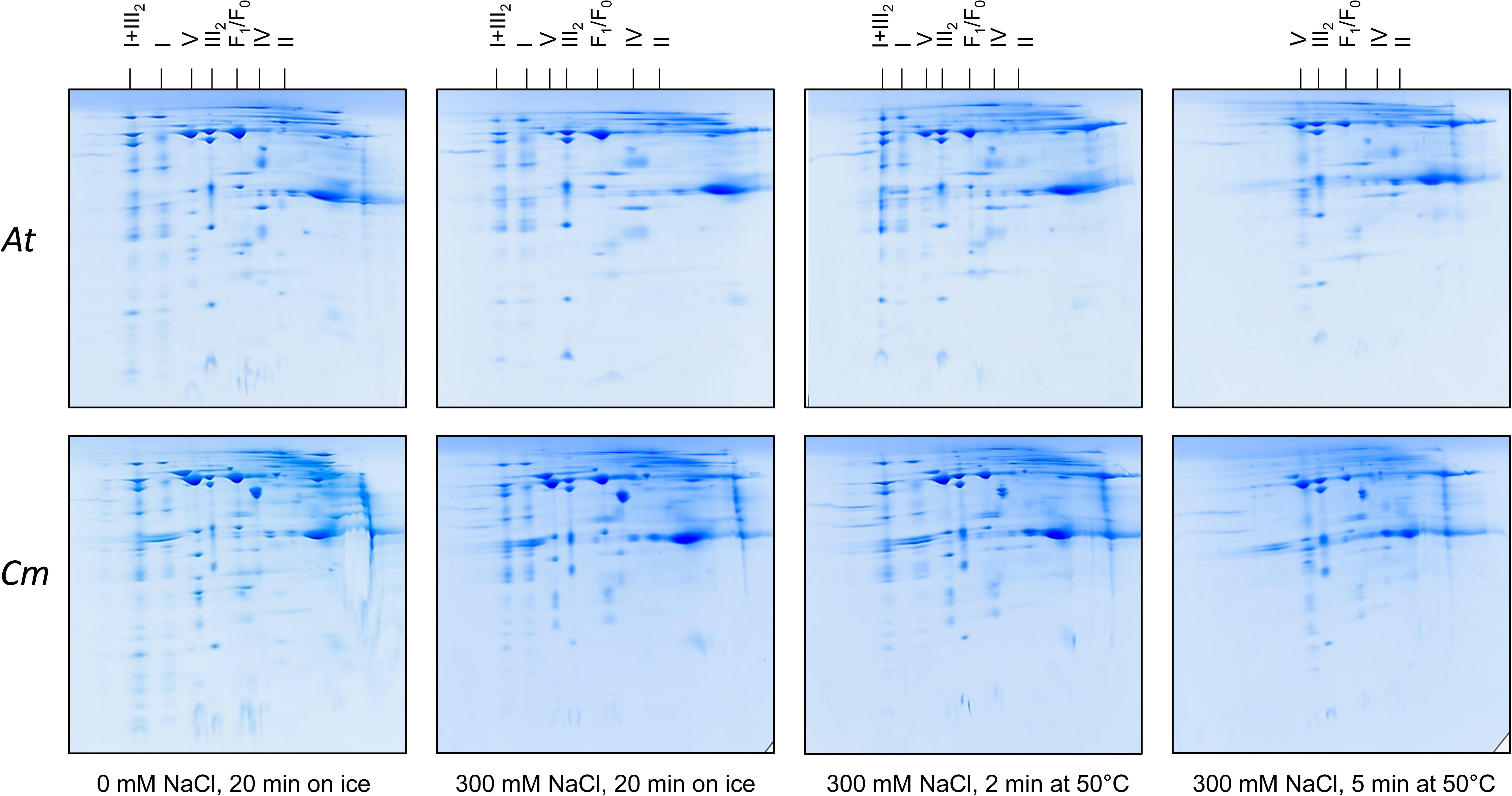
Figure 4. Salt and heat stability of the mitochondrial OXPHOS complexes from A. thaliana (At) and C. maritima (Cm). Isolated mitochondria were treated as indicated below the 2D gels. Subsequently, mitochondria were solubilized by digitonin and mitochondrial proteins separated by 2D Blue native/SDS–PAGE. Identities of the resolved protein complexes are given above the 2D gels. For designations see legend of Figure 2.
In A. thaliana, ATP synthase (complex V) abundance on the 2D gels significantly drops in fractions treated with 300 mM NaCl (Figure 4). Also, complex II of A. thaliana clearly is destabilized by 300 mM NaCl (Figure 4). Complexes I and IV as well as the I + III2 supercomplex of A. thaliana are stable in 300 mM salt, even if treated for 2 min at 50°C. In contrast, all three complexes completely disappear on the 2D gels upon treatment for 5 min at 50°C (Figure 4). Dimeric complex III proved to be the most stable OXPHOS complex in A. thaliana. It is not even affected at 300 mM NaCl and treatment at 50°C for 5 min (Figure 4).
In C. maritima, salt and temperature treatments led to very similar effects. However, some differences with respect to A. thaliana were visible: (i) Complex V is more stable in C. maritima upon salt treatment. (ii) Amount of dimeric complex III even increases upon combined salt and heat treatment. This result probably reflects dissociation of the I + III2 supercomplex (which is more abundant in C. maritima) into monomeric complex I and dimeric complex III. (iii) Besides the OXPHOS complexes, the glutamate dehydrogenase complex is of increased abundance in C. maritima mitochondria. It is clearly affected by salt and temperature. However, a small percentage of this complex was stable even at the harshest conditions. All experiments illustrated in Figure 4 were repeated several times (three biological controls and several technical controls) and quantitatively evaluated using the Delta 2D software package (Supplementary Figures 1, 3). A statistical evaluation of the results has been exemplarily carried out for the ATP synthase complex (Supplementary Figure 2).
This study is dedicated to the OXPHOS system of C. maritima. As expected, the OXPHOS system resembles that of the closely related model plant A. thaliana, which has been characterized extensively (Eubel et al., 2003; Klodmann et al., 2011; Senkler et al., 2017). In Arabidopsis, complex I consists of about 49 subunits, complex II of 8 subunits, dimeric complex III of 2 × 10 subunits, complex IV of about 13 and ATP synthase of about 15 subunits. In C. maritima, we could not detect any differences with respect to subunit numbers of the five OXPHOS complexes. However, several OXPHOS subunits have slightly varying molecular masses (Figure 3). Furthermore, the stoichiometry of the OXPHOS complexes differs between A. thaliana and C. maritima (Figure 2). In C. maritima, the I + III2 supercomplex is more abundant (Figure 2). This may promote efficient electron transfer from complex I to complex III. Furthermore, it also may have a positive effect on the stability of the monomeric complexes. However, the precise physiological role of respiratory supercomplexes is still a matter of debate (see Hirst, 2018 for discussion). In contrast, complexes II and IV seem to be of reduced abundance in C. maritima. This could affect the capacity for electron insertion into the respiratory chain (ETC)/reduction of oxygen to water by the ETC. The latter effect could be compensated by AOX (this enzyme is difficult to detect on BN/SDS gels). Finally, glutamate dehydrogenase is quite prominent in C. maritima. Halophile plants usually have much increased capacities for proline biosynthesis during salt stress as well as proline catabolism upon salt stress release. Glutamate dehydrogenase is involved in the mitochondrial proline degradation pathway.
Salt and temperature might affect protein and protein complex stability. Indeed, it is known that species living in very hot environments have protein complexes of high stability (Graziano and Merlino, 2014). For this reason, structural analyses of proteins and protein complexes using x-ray crystallography or single particle cryo electron microscopy often is performed with protein fractions isolated from thermophilic bacteria. Also, prerequisite of the polymerase chain reaction (PCR) is a heat-stable DNA polymerase like present is thermophilic bacteria. Due to its natural environment, we hypothezised hat C. maritima may have more stable OXPHOS complexes than the glycophyte plant A. thaliana. However, the stability of the OXPHOS complexes turned out to be similar. Indeed, the natural environments of these ecotypes do not differ drastically. At the same time, some differences were observed. The ATP synthase complex (complex V) was slightly more stable in C. maritima. This complex is in the very center of mitochondrial ATP production. Furthermore, the I + III2 supercomplex is very abundant in C. maritima. Only at the harshest treatment condition (300 mM NaCl and 50°C for 5 min), this supercomplex dissociates. As a result, the amount of dimeric complex III increases, while the complex I monomer is degraded. Physiological experiments using isolated mitochondria from A. thaliana and C. maritima should be employed next to further compare mitochondrial functions in these two species in the presence and absence of stress factors like salt. This should give further insights into the mitochondrial contribution to the salt stress response in plants.
Available data are presented in the manuscript and the Supplementary Material.
NF performed all the experiments. NF, SH, and TMH carried out the Delta2D evaluations. H-PB and AD designed and supervised the project. H-PB and NF wrote the manuscript.
This study was supported by the German Academic Exchange Service (DAAD) in the framework of the “Change by Exchange”-program (program line 4), title of the project: “Proteomics and Halophyte Stress Tolerance” (ID 57247769). The publication of this manuscript was funded by the Open Access fund of Leibniz Universität Hannover.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Christa Ruppelt and Dagmar Lewejohann for the support in cell culture maintenance and mitochondria isolations. We also thank Dr. Holger Eubel for critically reading the manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01010/full#supplementary-material
Ben Hamed, I., Biligui, B., Arbelet-Bonnin, D., Abdelly, C., Ben Hamed, K., and Bouteau, F. (2014). Establishment of a cell suspension culture of the halophyte Cakile maritima. Adv. Hort. Sci. 28, 43–48.
Ben Hamed-Laouti, I., Arbelet-Bonnin, D., De Bont, L., Biligui, B., Gakière, B., Abdelly, C., et al. (2016). Comparison of NaCl-induced programmed cell death in the obligate halophyte Cakile maritima and the glycophyte Arabidospis thaliana. Plant Sci. 247, 49–59. doi: 10.1016/j.plantsci.2016.03.003
Berth, M., Moser, F. M., Kolbe, M., and Bernhardt, J. (2007). The state of the art in the analysis of two-dimensional gel electrophoresis images. Appl. Microbiol. Biotechnol. 76, 1223–1243. doi: 10.1007/s00253-007-1128-0
Bose, J., Munns, R., Shabala, S., Gilliham, M., Pogson, B., and Tyerman, S. D. (2017). Chloroplast function and ion regulation in plants growing on saline soils: lessons from halophytes. J. Exp. Bot. 68, 3129–3143. doi: 10.1093/jxb/erx142
Davy de Virville, J., Aaron, I., Alin, M. F., and Moreau, F. (1994). Isolation and properties of mitochondria from Arabidopsis thaliana cell suspension cultures. Plant Physiol. Biochem. 32, 159–166.
Davy de Virville, J., Alin, M. F., Aaron, Y., Remy, R., Guillot-Salomon, T., and Cantrel, C. (1998). Changes in functional properties of mitochondria during growth cycle of Arabidopsis thaliana cell suspension cultures. Plant Physiol. Biochem. 36, 347–356. doi: 10.1016/s0981-9428(98)80076-4
Eubel, H., Jänsch, L., and Braun, H. P. (2003). New insights into the respiratory chain of plant mitochondria. Supercomplexes and a unique composition of complex II. Plant Physiol. 133, 274–286. doi: 10.1104/pp.103.024620
Graziano, G., and Merlino, A. (2014). Molecular bases of protein halotolerance. Biochim. Biophys. Acta 1844, 850–858. doi: 10.1016/j.bbapap.2014.02.018
Heinemeyer, J., Scheibe, B., Schmitz, U. K., and Braun, H. P. (2009). Blue native DIGE as a tool for comparative analyses of protein complexes. J. Proteom. 72, 539–544. doi: 10.1016/j.jprot.2008.12.008
Hirst, J. (2018). Open questions: respiratory chain supercomplexes - why are they there and what do they do? BMC Biol. 16:111. doi: 10.1186/s12915-018-0577-5
Ismail, A. M., and Horie, T. (2017). Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 68, 405–434. doi: 10.1146/annurev-arplant-042916-040936
Jacoby, R. P., Millar, A. H., and Taylor, N. L. (2018). Mitochondrial biochemistry: stress responses and roles in stress alleviation. Plant Mitochondria 50, 227–268. doi: 10.1111/1567-1364.12218
Jacoby, R. P., Taylor, N. L., and Millar, A. H. (2011). The role of mitochondrial respiration in salinity tolerance. Trends Plant Sci. 16, 614–623. doi: 10.1016/j.tplants.2011.08.002
Klodmann, J., Senkler, M., Rode, C., and Braun, H. P. (2011). Defining the protein complex proteome of plant mitochondria. Plant Physiol. 157, 587–598. doi: 10.1104/pp.111.182352
Kruft, V., Eubel, H., Jänsch, L., Werhahn, W., and Braun, H. P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. doi: 10.1104/pp.127.4.1694
Lee, C. P., Eubel, H., O’Toole, N., and Millar, A. H. (2008). Heterogeneity of the mitochondrial proteome for photosynthetic and non-photosynthetic Arabidopsis metabolism. Mol. Cell Proteom. 7, 1297–1316. doi: 10.1074/mcp.M700535-MCP200
Lorenz, C., Rolletschek, H., Sunderhaus, S., and Braun, H. P. (2014). Brassica napus seed endosperm metabolism and signaling in a dead end tissue. J. Proteom. 108, 382–426. doi: 10.1016/j.jprot.2014.05.024
May, M. J., and Leaver, C. J. (1993). Oxidative stimulation of glutathione synthesis in Arabidopsis thaliana suspension cultures. Plant Physiol. 103, 621–627. doi: 10.1104/pp.103.2.621
Millar, A. H., Whelan, J., Soole, K. L., and Day, D. A. (2011). Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. doi: 10.1146/annurev-arplant-042110-103857
Munns, R., and Gilliham, M. (2015). Salinity tolerance of crops–what is the cost? New Phytol. 208, 668–673. doi: 10.1111/nph.13519
Munns, R., James, R. A., Gilliham, M., Flowers, T. J., and Colmer, T. D. (2016). Tissue tolerance: an essential but elusive trait for salt-tolerant crops. Funct. Plant Biol. 43, 1103–1113.
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Neuhoff, V., Stamm, R., and Eibl, H. (1985). Clear background and highly sensitive protein staining with coomassie blue dyes in polyacrylamide gels: a systematic analysis. Electrophoresis 6, 427–448. doi: 10.1002/elps.1150060905
Neuhoff, V., Stamm, R., Pardowitz, I., Arold, N., Ehrhardt, W., and Taube, D. (1990). Essential problems in quantification of proteins following colloidal staining with coomassie brilliant blue dyes in polyacrylamide gels, and their solution. Electrophoresis 11, 101–117. doi: 10.1002/elps.1150110202
Nikalje, G. C., Srivastava, A. K., Pandey, G. K., and Suprasanna, P. (2018). Halophytes in biosaline agriculture: mechanism, utilization, and value addition. Land Degrad. Dev. 29, 1081–1095. doi: 10.1002/ldr.2819
Nordborg, M., Hu, T. T., Ishino, Y., Jhaveri, J., Toomajian, C., Zheng, H., et al. (2005). The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol. 3:e196. doi: 10.1371/journal.pbio.0030196
Schägger, H., and von Jagow, G. (1991). Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199, 223–231. doi: 10.1016/0003-2697(91)90094-a
Schertl, P., and Braun, H. P. (2014). Respiratory electron transfer pathways in plant mitochondria. Front. Plant Sci. 5:163. doi: 10.3389/fpls.2014.00163
Senkler, J., Senkler, M., Eubel, H., Hildebrandt, T., Lengwenus, C., Schertl, P., et al. (2017). The mitochondrial complexome of Arabidopsis thaliana. Plant J. 89, 1079–1092. doi: 10.1111/tpj.13448
Somssich, M. (2018). A short history of Arabidopsis thaliana (L.) Heynh. Columbia-0. Peer J. 6:e26931v4.
Werhahn, W., Niemeyer, A., Jänsch, L., Kruft, V., Schmitz, U. K., and Braun, H. P. (2001). Purification and characterization of the preprotein translocase of the outer mitochondrial membrane from Arabidopsis thaliana: identification of multiple forms of TOM20. Plant Physiol. 125, 943–954. doi: 10.1104/pp.125.2.943
Keywords: halophyte, mitochondria, respiratory chain, oxidative phosphorylation, mitochondrial ATP synthase, Blue native PAGE, Arabidopsis thaliana, Cakile maritima
Citation: Farhat N, Hichri S, Hildebrandt TM, Debez A and Braun H-P (2019) Composition and Stability of the Oxidative Phosphorylation System in the Halophile Plant Cakile maritima. Front. Plant Sci. 10:1010. doi: 10.3389/fpls.2019.01010
Received: 27 April 2019; Accepted: 18 July 2019;
Published: 13 August 2019.
Edited by:
Jayakumar Bose, The University of Adelaide, AustraliaReviewed by:
Nicolas L. Taylor, The University of Western Australia, AustraliaCopyright © 2019 Farhat, Hichri, Hildebrandt, Debez and Braun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nèjia Farhat, bmVqaWFmYXJoYXRAZ21haWwuY29t; Hans-Peter Braun, YnJhdW5AZ2VuZXRpay51bmktaGFubm92ZXIuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.