
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 July 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00890
This article is part of the Research Topic Understanding the Molecular Mechanisms of Plant Responses to Abiotic Stress View all 25 articles
The NAC proteins form one of the largest families of plant-specific transcription factors (TFs) and play essential roles in developmental processes and stress responses. In this study, we characterized a NAC domain transcription factor, OoNAC72, from a legume Oxytropis ochrocephala. OoNAC72 was proved to be localized in the nuclei in tobacco lower epidermal cells and had transcriptional activation activity in yeast, confirming its transcription activity. OoNAC72 expression could be induced by drought, salinity and exogenous abscisic acid (ABA) in O. ochrocephala seedlings. Furthermore, over-expression of OoNAC72 driven by CaMV35S promoter in Arabidopsis resulted in ABA hypersensitivity and enhanced tolerance to drought and salt stresses during seed germination and post-germinative growth periods. In addition, over-expression of OoNAC72 enhanced the expression of stress-responsive genes such as RD29A, RD29B, RD26, LEA14, ANACOR19, ZAT10, PP2CA, and NCED3. These results highlight the important regulatory role of OoNAC72 in multiple abiotic stress tolerance, and may provide an underlying reason for the spread of O. ochrocephala.
Oxytropis ochrocephala Bunge, one of the toxic Oxytropis locoweeds, distributed widely among Northwest China, where the plant often suffered from stress environment such as drought, high soil salinity and low temperature. In natural grassland plant, O. ochrocephala can rapidly replace local forages grass species because of its unpalatability and strong biotic stress tolerance. Grasslands infested by O. ochrocephala lead to tremendous losses to livestock husbandry, as well as great damage to the grassland ecological equilibrium (Zhao et al., 2013; He et al., 2015). However, existing research on O. ochrocephala mainly focused on surveys, allelopathy and toxicological studies of distribution (Tulsiani et al., 1988; Zhao et al., 2013), and have not yet investigated its resistance mechanism.
In general, plants often experience some harsh environments during growth and development. As a result, plants must respond to those stresses by regulating the resistance-related genes. Transcription factors play an extremely important role in the process of stresses response by activating or inhibiting the expression of the downstream target genes. In plants, several families of stress-responsive transcription factors have been functionally characterized under stress regulation networks, such as NAC, bZIP, WRKY, MYB/MYC, and AP2/ERF (Hénanff et al., 2013). NAC transcription factors, one of the largest plant-specific transcription factor families, play an important role in plant growth and stress response, and have become a hot spot in the research of gene regulation (Olsen et al., 2005b; Zheng et al., 2009). NAC transcriptional factors are derived from three kinds of genes containing particular domains of NAM (no apical meristem), ATAF (Arabidopsis transcription activation factor) and CUC (cup-shaped cotyledon) (Souer and Al, 1996; Aida et al., 1997).
NAC transcription factors are key regulators of plant resistance to stress by ABA-dependent or ABA-independent pathways (Puranik et al., 2012). Currently, the NAC transcription factor family have been systematically screened and analyzed in various plants, such as Arabidopsis (138), rice (158), wheat (134), canola, cotton, banana, and soybean (Hegedus et al., 2003; Ooka et al., 2003; Yujie et al., 2008; Meng et al., 2009; Tran et al., 2009; Tang et al., 2012; Jia et al., 2014; Jinpu et al., 2014; Tak et al., 2017). In Arabidopsis, Miki et al. (2010) found that RD26/ANAC072 was significantly induced by drought, high salt and ABA, and the rd26 mutant was not sensitive to exogenous ABA, revealing positive regulation by ABA signaling under drought stress. Similar study showed that ANAC096 also exhibits ABA-dependent signaling and regulates the response of transgenic Arabidopsis to osmotic stress (Xu et al., 2013). Moreover, ANAC096 was reported to have a synergistic relationship with ABRE binding factor and increased plant stress resistance (Xu et al., 2013). In rice, the expression of ONAC022 was up-regulated by various stresses (Hong et al., 2016). Transgenic rice plants overexpressing OsNAC5 and OsNAC6 enhanced dehydration, high salinity and disease tolerances (Nakashima et al., 2007; Takasaki et al., 2010; Jeong et al., 2013). The overexpression of OsNAC9 altered root architecture of rice plants, enhancing drought resistance and grain yield under field conditions (Redillas et al., 2012). In wheat, TaNAC29 and TaNAC2 can be up-regulated by different abiotic stresses, and transgenic Arabidopsis plants overexpressing these genes improved salt and drought tolerance (Mao et al., 2012, 2014; Huang et al., 2015; Huang and Wang, 2016). Additionally, in soybean, Tran et al., 2009 screened and cloned 31 soybean NAC genes, and found that nine of them were induced by drought stress. Pinheiro et al. (2009) found that the expression of GmNAC2/3/4 was significantly induced by osmotic pressure, and GmNAC3/4 was simultaneously induced by ABA, JA, and salt. Although a growing number of studies have shown that NAC transcription factors play a critical regulatory role in a variety of stress-responsive signaling pathways in higher plants, the biological function of O. ochrocephala NAC transcription factor is still unknown.
In this study, an abiotic stress-related NAC family gene OoNAC72 from O. ochrocephala were screened and characterized, and then the subcellular localization and transcriptional activation activities of OoNAC72 protein were further verified. The expression patterns of OoNAC72 in response to polyethylene glycol (PEG), salt and exogenous ABA treatments were also determined by the quantitative real-time PCR (qRT-PCR). Moreover, transgenic Arabidopsis plants over-expressing OoNAC72 (OoNAC72-OX) were measured for phenotypic and physiological characteristics under drought and salt stress conditions. Through this study, we aim to gain a more in-depth and comprehensive understanding of the OoNAC72 structure and its function. The results may provide a new insight into the mechanism for the rapid spread of O. ochrocephala.
Mature O. ochrocephala seeds were collected from Haiyuan, Ningxia Province (36°29′49″N 105°36′49″E 2171 mH) in July 2014. Seeds after collection were pretreated with 98% H2SO4 for 6∼9 min, then they were washed in distilled water 4∼6 times and germinated on wet filter papers for 3 days in the dark on petri dishes. For hormone treatments, 3-week-old seedlings grown in a greenhouse were treated by spraying with 100 mM gibberellin (GA), 100 mM ethephon (ETH) and 100 mM abscisic acid (ABA) with equal volume of solution containing only 0.1% ethanol and distilled water as controls. For high salinity and drought treatments, roots of O. ochrocephala seedlings were soaked in 150 mM NaCl and 20% PEG-6000, respectively. O. ochrocephala seedlings treated with various chemicals and stress elicitors along with control plants were sampled at 0, 1, 3, 6, 12, 24, and 48 h post-treatment (hpt). All samples were frozen in liquid nitrogen and stored at −80°C for RNA extraction. Three independent biological replications were performed for each experiment.
Extraction of O. ochrocephala total RNA was performed with the Trizol Reagent (TIANGEN, Beijing, China) according to the manufacturer’s instructions. Quality and integrity of total RNA was assessed by 1.0% agarose gel electrophoresis. RNA purity and concentration were determined on a NanoDropTM 2000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). In order to perform RT-PCR and qRT-PCR, the first-strand cDNA was synthesized by reverse transcription using 3 μg total RNA in a 10 μl reaction volume according to the manufacturer’s instructions using the transcription kit (Thermo Fisher Scientific, Waltham, MA, United States). The cDNA was diluted 10-fold with nuclease-free water for RT-PCR and qRT-PCR.
The sequence of OoNAC72 was obtained by our research group from the O. ochrocephala’s transcriptome sequencing data (He et al., 2015). Using the specific primers, we amplified the ORF of OoNAC72 (Supplementary Table S1). The PCR condition was as follows: 3 min at 95°C; 34 cycles of 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C; and then 10 min at 72°C. The resulting PCR products were cloned to the pGEM-T Easy Vector (TaKaRa, Beijing, China) and sequenced by Sangon Biotech Co., Ltd., (Shanghai, China). Multiple sequence alignment of OoNAC72 with NAC TFs in other species was performed with DNAMAN 8.0. A phylogenetic tree was constructed using a neighbor-joining (NJ) method with 1000 bootstrap replicates in MEGA 5.0.
Expression profiles of OoNAC72 after different treatments were determined by qRT-PCR analyses with a pair of primers amplifying a 101-bp fragment (Supplementary Table S2). To ensure gene-specific amplification, the primers were used to amplify the OoNAC72 gene by regular PCR and sequenced. For qRT-PCR analyses, reactions were conducted following the method of Zhuang et al. (2015a). O. ochrocephala Histone H3 (KR733680.1) and Actin101 (KR822225.1) were used as the internal references (He et al., 2015). OoNAC72 expression level was calculated using the relative 2–ΔΔCt method. Three replications were performed for each experiment.
The coding sequence without the stop codon of OoNAC72 was transferred into the pCAMBI1302-eGFP vector (Invitrogen, United States) to generate a pCAMBI1302-OoNAC72-eGFP fusion protein using a pair of primers containing Bgl II or Spe I site (Supplementary Table S3). The re-combinational construct pCAMBI1302-OoNAC72-eGFP and pCAMBI1302-eGFP (control vector) were infiltrated into the leaves of 6-week-old Nicotiana benthamiana, respectively (Sheludko et al., 2007). After transformation for 36–60 h, the expression location of OoNAC72:eGFP fusion protein was observed using confocal laser scanning microscopy (CLSM, Olympus FV1000, Olympus Optical Company Ltd., Japan). Laser scanning confocal microscope was used to detect the eGFP (excitation: 488 nm, emission: 510 nm) fluorescence signal. eGFP images, The 40,6-diamidino-2-phenylindole (DAPI) (excitation: 405 nm, emission: 461 nm) nuclear stain was used to determine the location of nucleus. Images were acquired with the software FV10-ASW 4.2 Viewer. Each digital image was recorded with the same camera settings and was not further processed.
For transactivation analysis of OoNAC72 in yeast cell, the yeast strain AH109 (Clontech) was transformed with the appropriate bait vectors. The full-length coding sequence without the stop codon of OoNAC72 was amplified using three pairs of primers with Smal I - and Pst I sites (Supplementary Table S3). The PCR products were digested with Smal I and Pst I and then were cloned into the GAL4 binding domain vector pGBKT7 according to the manufacture’s protocol (Clontech). Empty pGBKT7 vector was used as a negative control and the pGAL4 vector was used as a positive control. Transformed yeast cells were transformed onto SD medium (SD / -Trp, SD/ -Trp-His-Ade, SD / -Trp-His-Ade / X-α-gal) to compare their survival. Plates were incubated at 28°C for 3 days before photographing.
Although O. ochrocephala is widely distributed as a leguminous plant on the grassland, its current culture system with a complete growth cycle in the laboratory has not been reported, which makes the gene function research become extremely difficult with O. ochrocephala as a host plant. Therefore, OoNAC72 was transferred into the model plant Arabidopsis to reveal its potential biological functions. The full-length coding sequence without the stop codon of OoNAC72 was cloned into pCAMBI1302-OoNAC72-eGFP and transformed into Arabidopsis thaliana Columbia-0 (WT) plants according to the floral dip method using Agrobacterium tumefaciens strain GV3101 (Clough and Bent, 1998). The positive transgenic lines were screened on homomycin (50 mg/L) plates, and further identified by genomic DNA PCR, and the OoNAC72 expression level in leaves of each transgenic line was examined by qRT-PCR. The homozygous lines of T3 generation plants were used for study.
Two representative transgenic lines overexpressing OoNAC72-OX lines and wild type (WT) Arabidopsis plants were selected for abiotic stress tolerance assays. For the analysis of germination rate, surface-sterilized seeds were sown on 1/2 MS solid medium supplemented with 100 and 150 mM NaCl, 200 and 250 mM mannitol, and 1 and 3 μM ABA, respectively. The seeds were first vernalized at 4°C for 3 days in dark, and then were incubated at 22°C with 16 h light / 8 h dark cycle. The germination rates of seeds were calculated when the green cotyledons emerged. For seedling root length experiment, 5-day-old seedlings cultivated on 1/2 MS solid medium were transferred onto 1/2 MS solid medium supplemented with 100 and 150 mM NaCl, 200, 250 mM mannitol or 1, 3 μM ABA for vertical culture (Yuriko et al., 2013). The length of primary roots of each subset were measured after 14 days of the treatments.
For evaluation of stresses tolerance at the vegetative growth stage, 3-week-old WT and OoNAC72-OX lines grown in soil under non-stress conditions were irrigated continuously with 150 mM NaCl for 30 days. For drought tolerance assay, 40-day-old plants were continuously dehydrated until the leaves withered, and then were rehydrated. Dehydration rate of detached leaves and stomatal conductance were determined on the 10th day after the onset of drought stress. To further evaluate the response mechanism of OoNAC72-OX lines to ABA-mediated drought, the leaves of 40-day-old seedlings of the WT and OoNAC72-OX lines were sheared with 10 μM ABA for 3 h (Huang and Wang, 2016; Mao et al., 2016), and then the stomatal conductance was measured under light microscopy.
Three-week-old OoNAC72-OX seedlings and WT plants grown in soil were transferred to 1/2 MS liquid medium supplemented with 20% PEG-6000 and incubated at greenhouse for 2 days. Fresh Arabidopsis samples harvested at different time stages during development were immediately frozen for ABA quantification by the ABA immunoassay kit (Yang et al., 2001; Yanjuan et al., 2012).
To further investigate the molecular mechanism of stress tolerance, the expression levels of marker genes were detected in the WT and OoNAC72-OX plants. Total RNAs of the whole plants from 14-day-old WT and OoNAC72-OX plant seedlings grown on 1/2 MS solid medium supplemented with 150 mM NaCl and 200 mM mannitol were extracted with the Trizol Reagent. The qRT-PCR was performed using specific primers (Supplementary Table S4) for the expression levels of marker genes, and Actin8 was employed as a reference control.
To determine water loss rate, leaves were harvested from 4-week-old seedlings of the WT and OoNAC72-OX plants and dehydrated on the dry filter paper (22–25°C, humidity 45–60%) for weighing at designated time points. Images were captured at 0 and 3 h after the treatment. The water loss rate was calculated based on the initial fresh weight of the leaves. Proline and malondialdehyde (MDA) content, SOD and POD activities were measured according to Wang et al. (2010) and Moore (1998). Proline content was measured according to Bates et al. (1973). At least 20 seedlings were employed for physiological indices analysis in each sample.
Data were presented as means ± SD of at least three independent replicates from one representative experiment. Analysis of significant difference was performed by Duncan’s multiple range tests in the ANOVA program of SPSS (IBM SPSS 22), taking *P < 0.05, ∗∗P < 0.01 as critical value.
A 1301 bp cDNA containing an ORF of 996 bp (65–1060) was cloned from the transcriptome of O. ochrocephala. This ORF coded a protein of 331 amino acids with a theoretical molecular weight of 37.49 kDa. The conserved domain analysis revealed that its N-terminal region had a highly conserved NAC domain (amino acid 98–220), which consisted of five subdomains A–E (Supplementary Figure S1). Whereas its C-terminal region had no significant similarity to any other members of the NAC family. BLASTP analysis revealed that this protein shared the highest similarity (70%) to AtNAC72 (XP_016170502.1) in Arabidopsis. Further phylogenetic analysis confirmed that the relatedness of the predicted protein to AtNAC72 was highly homologous with those of Medicago truncatula and Cicer arietinum (Supplementary Figure S2). Therefore, this O. ochrocephala gene was designated as OoNAC72 (MH142381).
The sequence analysis showed that OoNAC72 possessed a conserved nuclear localization signal (NLS, positions 74–88, 117–129 aa) (Supplementary Figure S1). Meanwhile, analysis by ProtComp v.9.0 indicated a high likelihood of nuclear localization for OoNAC72 protein. To confirm this prediction, an expression cassette fusing OoNAC72 with the eGFP protein was constructed. Then the fused protein was expressed transiently in Nicotiana benthamiana while the pCAMBI1302-eGFP functioned as a control. Fluorescence microscopy revealed that the OoNAC72-eGFP fusion protein was exclusively localized in the nucleus in the transformed cells, whereas the control eGFP was uniformly distributed throughout the cell (Figure 1). These results further confirmed that the OoNAC72 protein was a nuclear-localized protein.
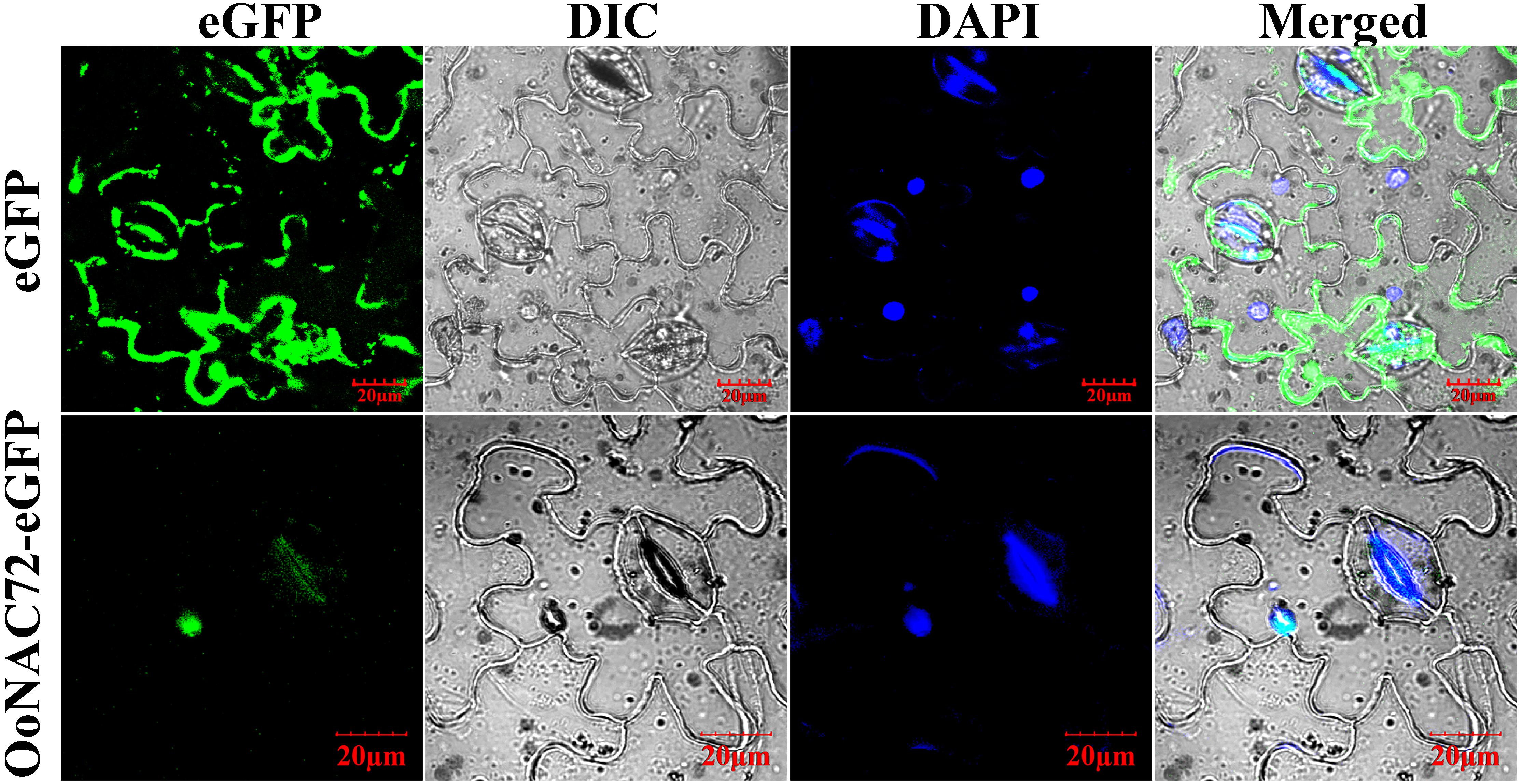
Figure 1. Subcellular localization of the OoNAC72 protein in leaf epidermal cells of Nicotiana benthamiana. eGFP and OoNAC72-eGFP constructs were separately expressed instantly in leaf epidermal cells of Nicotiana benthamiana and observed under a laser scanning confocal microscope after agroinfiltration for 48 h. DAPI images indicate nuclear staining. eGFP images, DAPI stained images, differential interference contrast images (DIC) and merged images were taken. Bars = 20 μm.
The result of transcriptional activity analysis of OoNAC72 was illustrated in Figure 2. The vector pGBKT7 fused with OoNAC72 was used as the experimental set, and the empty pGBKT7 vector and pGAL4 were hired as the negative and positive controls, respectively (Figure 2A). All of the transformants grew well on selective SD/-Trp medium (Figure 2B), indicating that the three vectors were successfully transformed into the yeast cells. The GAL4-binding domain-OoNAC72 construct and pGAL4 grew well on SD/-Trp-His-Ade medium, while the transformants containing the pGBKT7 vector did not grow on the same medium (Figure 2C). The results of α-galactosidase activity assays showed that transformants containing GAL4-binding domain-OoNAC72 construct and pGAL4 appeared blue in color on SD/-Trp-His-Ade medium containing 5-bromo-4-chloro-3-indoxyl-α-D-galactopyranoside (X-α-Gal) (Figure 2D). These results indicated that OoNAC72 had transcriptional activity in yeast.
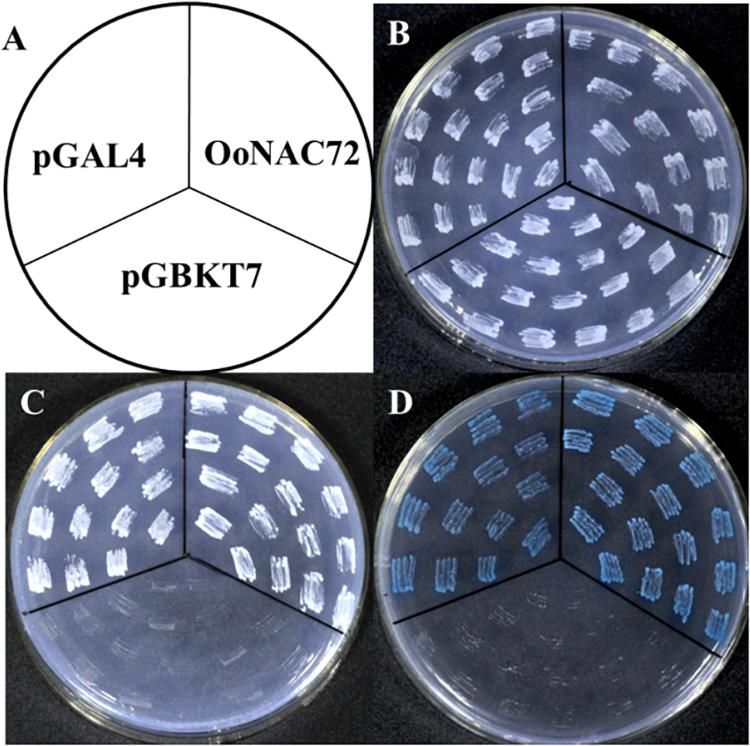
Figure 2. Transcriptional activation assay of the OoNAC72 in yeast cell. (A) Schematic Diagram. The numbers on the left indicate the last residues of polypeptides. Vectors pGBKT7 and pGAL4 were used as negative and positive control, respectively. All plasmids were transformed into the yeast strain AH109 (B–D). Transformed yeasts were dripped on the SD/-Trp, SD/-Trp-His-Ade, and SD/-Trp-His-x-gal after being cultured for 3 days in the growth chamber.
Quantitative real-time PCR was used to evaluate the expression patterns of OoNAC72 during dehydration (20% PEG-6000), salt (150 mM NaCl) and three hormone treatments (100 mM ABA, ETH, and GA) (Figure 3). When the seedlings were treated with dehydration, OoNAC72 mRNA abundance was slightly induced at 1 h, followed by progressive elevation until reaching the peak value at 3 h, which showed an approximately 11.9-fold increase relative to the initial level (Figure 3A). Treatment with 150 mM NaCl led to a quick accumulation of OoNAC72 mRNA level, which progressed until a maximum level was reached at 6 h, approximately 12.3-fold increase relative to the initial level (Figure 3A). Moreover, for the ABA treatment, the expression levels of OoNAC72 increased rapidly and reached the maximum level at 3 h, being 41.8-fold greater than the control (0 h), while there were no significant changes when treated with ETH and GA (Figure 3B). All the results suggested that OoNAC72 may respond to stress in O. ochrocephala by participating in ABA-dependent signal transduction pathways.
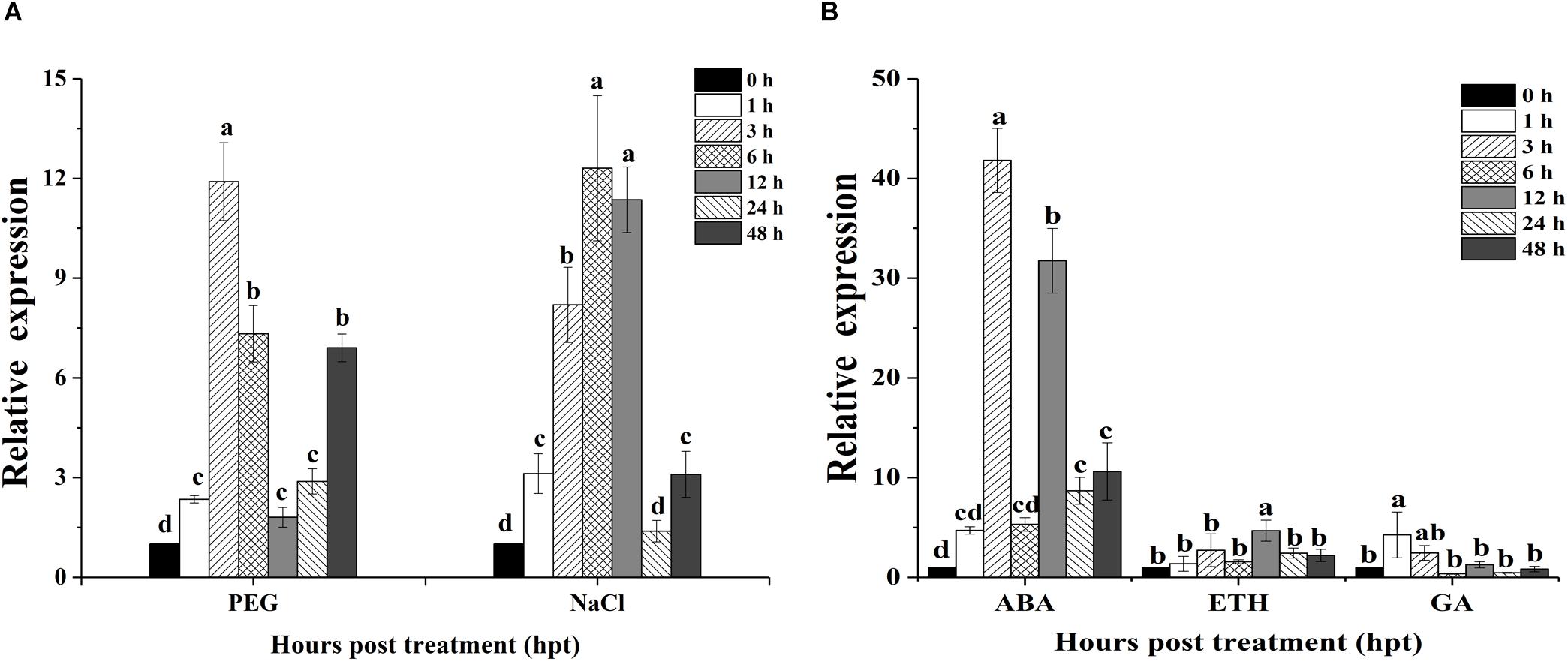
Figure 3. Expression patterns of OoNAC72 in response to stress treatments. (A) Expression patterns under abiotic treatments: 20% PEG and NaCl. (B) Expression patterns under exogenous hormone treatments: ABA, ETH and GA. Three-week-old seedlings were exposed to stresses as described in section Materials and Methods. The 2−ΔΔct method was used to measure the relative expression levels of the target genes in stressed and non-stressed leaves. Means are generated from three independent measurements; Error bars indicate SD, and Different letters on the histograms indicate significantly different values (*P < 0.05; ∗∗P < 0.01 by Duncan’s test) compared to the non-treatment controls.
To explore the function of OoNAC72 during the tolerance to abiotic stress, transgenic Arabidopsis over-expressing plants driven by the CaMV35S promoter were generated. We analyzed the growth status of two OoNAC72-OX transgenic lines (OX1, OX2) from the screening transgenic positive seedlings. Compared with wild-type plants, OoNAC72-OX transgenic lines (OX1, OX2) showed rosette leaves during the vegetative growth stage, and the number of leaves was significantly increased (Figures 4A,B,D); At the reproductive stage, OoNAC72-OX transgenic lines (OX1, OX2) showed significant delayed bolting and flowering (Figures 4C,E). OoNAC72-OX transgenic lines (OX1, OX2) showed no significant difference in leaf number, flowering plant height, dry and wet weight (Figures 4D–G). These findings suggested that OoNAC72 was an important regulator of plant development.
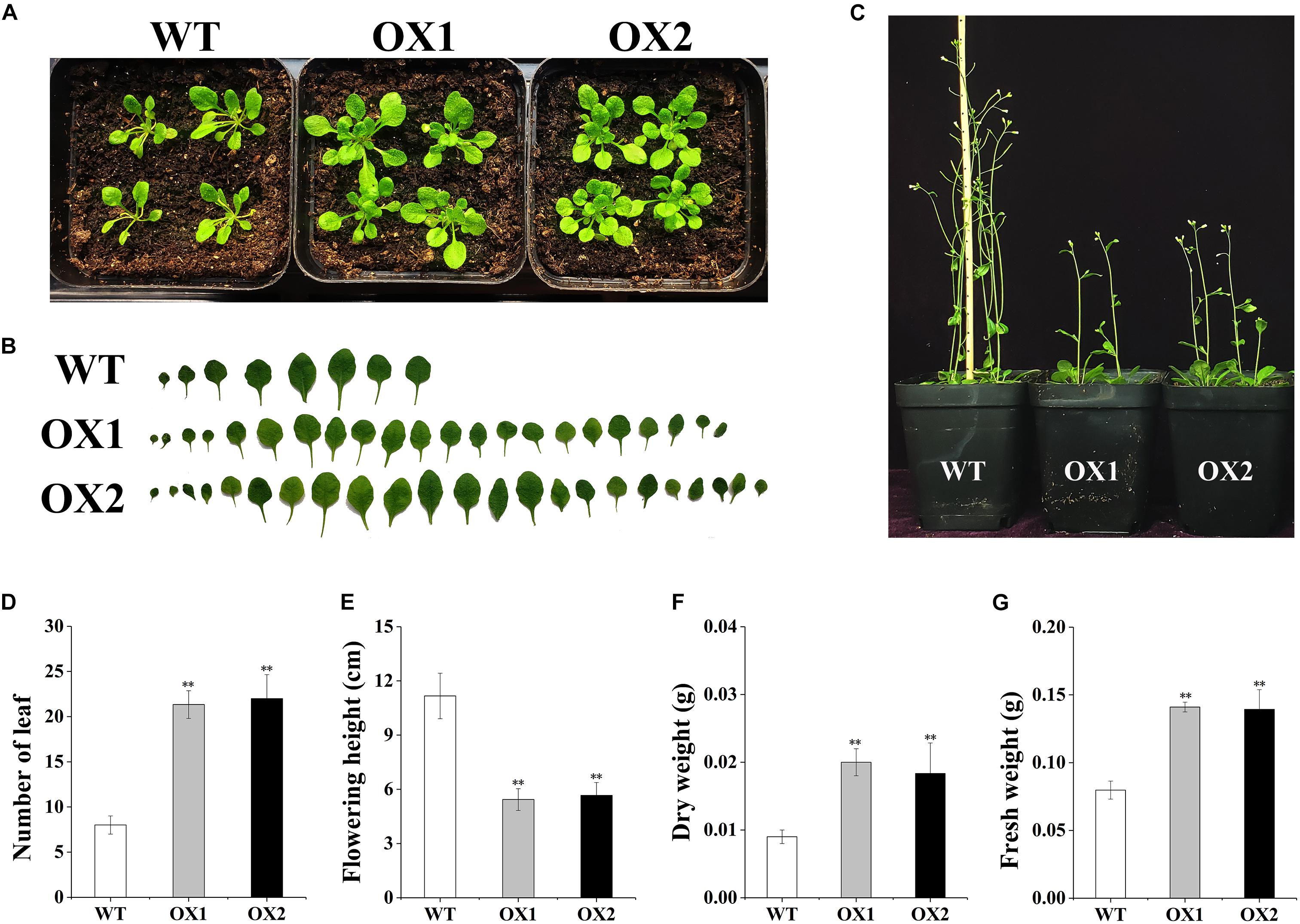
Figure 4. The Comparison of OoNAC72-OX and wild plants. (A) The plant phenotype of WT and OoNAC72-OX (OX1, OX2) plants grown under LD conditions after 3 weeks. (B) Schematic diagram of leaves phenotypic analyses of WT and OoNAC72-OX (OX1, OX2). Results from one of five biological replicates were shown. (C) The plant height of WT and OoNAC72-OX plants grown under LD conditions after 6 weeks. (D–G) Statistical analysis of WT and OoNAC72-OX (OX1, OX2) in leaf number, flowering plant height, dry and wet weight. Wet and dry weight in 3 weeks old seedlings test results. Data are represented as mean ± SD of at least three independent replications. Asterisk indicates significant difference (*P < 0.05; ∗∗P < 0.01) between transgenic lines and WT.
To further investigate mechanisms of hypersensitivity to abiotic stress in OoNAC72-OX plants, we evaluated the stress tolerance of transgenic (OX1 and OX2 lines) and WT Arabidopsis in germination and root growth in seedlings (Figure 5). WT and OoNAC72-OX lines were no significant difference on 1/2 MS medium, whereas OX1 and OX2 lines better growth and longer root length under 150 mM NaCl, 200 mM NaCl, 200 mM mannitol and 250 mM mannitol stresses. These results suggested that OoNAC72-OX plants had improved tolerance to salt and drought stresses during seed germination and post-germinative growth periods.
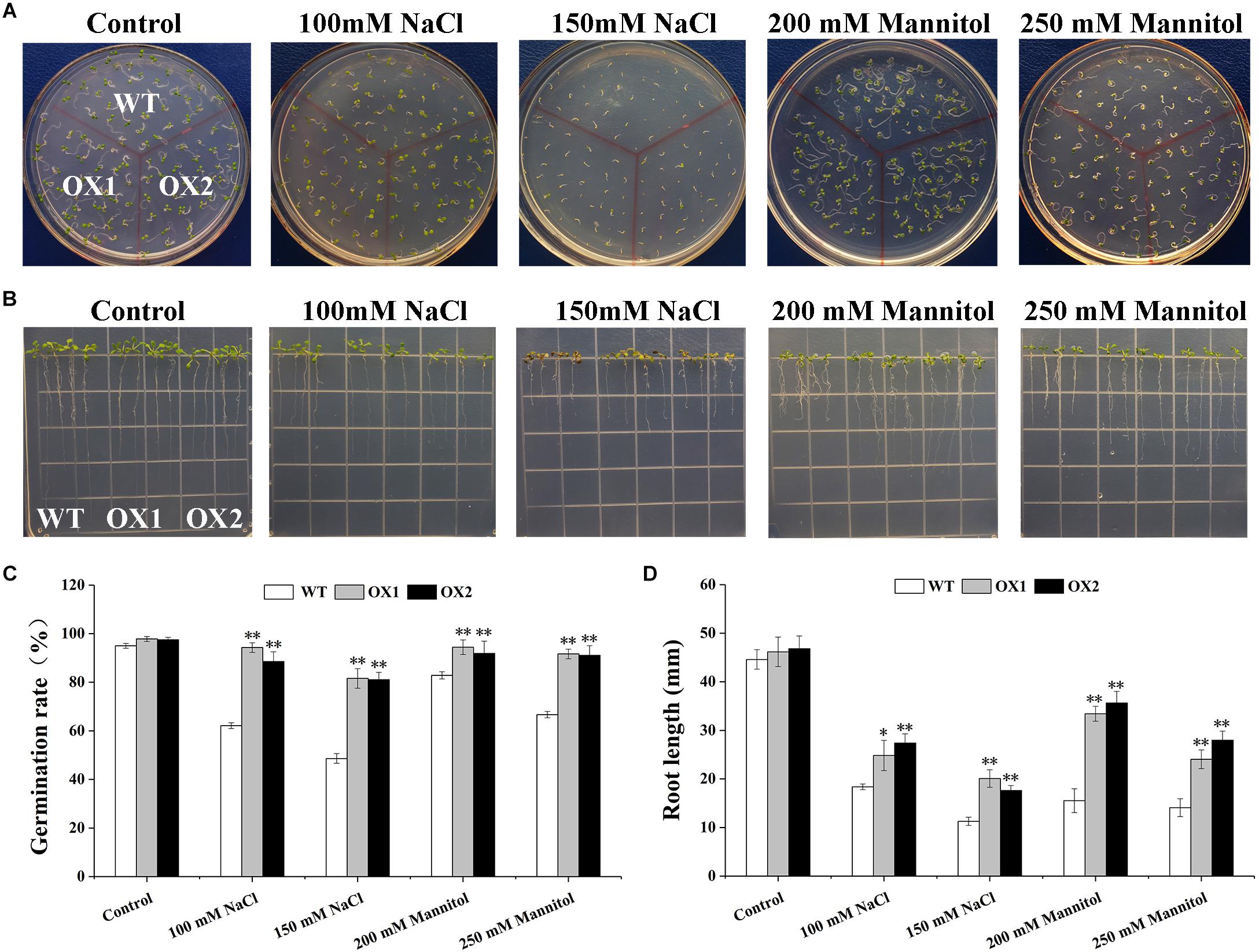
Figure 5. Comparison of the germination rate and root length between OoNAC72-OX and WT Arabidopsis plants. (A) Seed germination assay between OoNAC72-OX and WT Arabidopsis plants. The surface-sterilized seeds were sowed on 1/2 MS solid medium supplemented with 100 mM NaCl, 150 mM NaCl, 200 mM mannitol and 250 mM mannitol incubated at 22°C. (B) Root length assay between OoNAC72-OX and WT Arabidopsis plants. Five-day-old seedlings were cultured vertically on 1/2 MS solid medium supplemented with 100 mM NaCl, 150 mM NaCl, 200 mM mannitol and 250 mM mannitol for 7 days. (C,D) Effects of salt and drought stress treatments on germination rate and root length. Data are represented as mean ± SD of at least three independent replications. Asterisk indicates significant difference (*P < 0.05; ∗∗P < 0.01) between transgenic lines and WT.
To further elucidate the possible involvement of OoNAC72 in the response to drought and salt, we compared drought and salt tolerance of the OoNAC72-OX lines and WT plants at the vegetative growth stage (Figure 6). After 20 days of 150 mM NaCl stress treatment, the phenotypes of the WT and OX1 and OX2 lines began to show significant differences, in which 87.5% of the WT died, while the majority of OX1 and OX2 lines remained green,whereas OX1 and OX2 showed significantly higher survival rate 75 and 62.5% (Figures 6A,C). In order to detect the drought resistance of OoNAC72-OX lines, WT and OoNAC72-OX lines were simultaneously withholding water for 35 and 5 days after rehydration, the phenotypes of the WT and OoNAC72-OX lines began to show significant differences, the leaves of WT lines completely wilted and the plants survival rate was 3.33%. However, OX1 and OX2 lines survival rate were 87.5% (Figures 6B,C). Furthermore, there were no significant differences in proline and MDA content, POD and SOD activities between WT and OX1 and OX2 lines under normal condition (Figures 6D–G). Whereas, under salt and drought stresses, OX1 and OX2 lines exhibitedhigher levels of proline content, POD and SOD activities, and lower MDA level compared with those of WT (Figures 6D–G). Phenotypic analysis showed that the OoNAC72-OX plants had the highest drought and salt tolerance. These data demonstrated that the OoNAC72-OX plants exhibited increased tolerance to salt and drought stresses, thus we speculated OoNAC72 plays a critical role in O. ochrocephala response to salt and drought stresses.
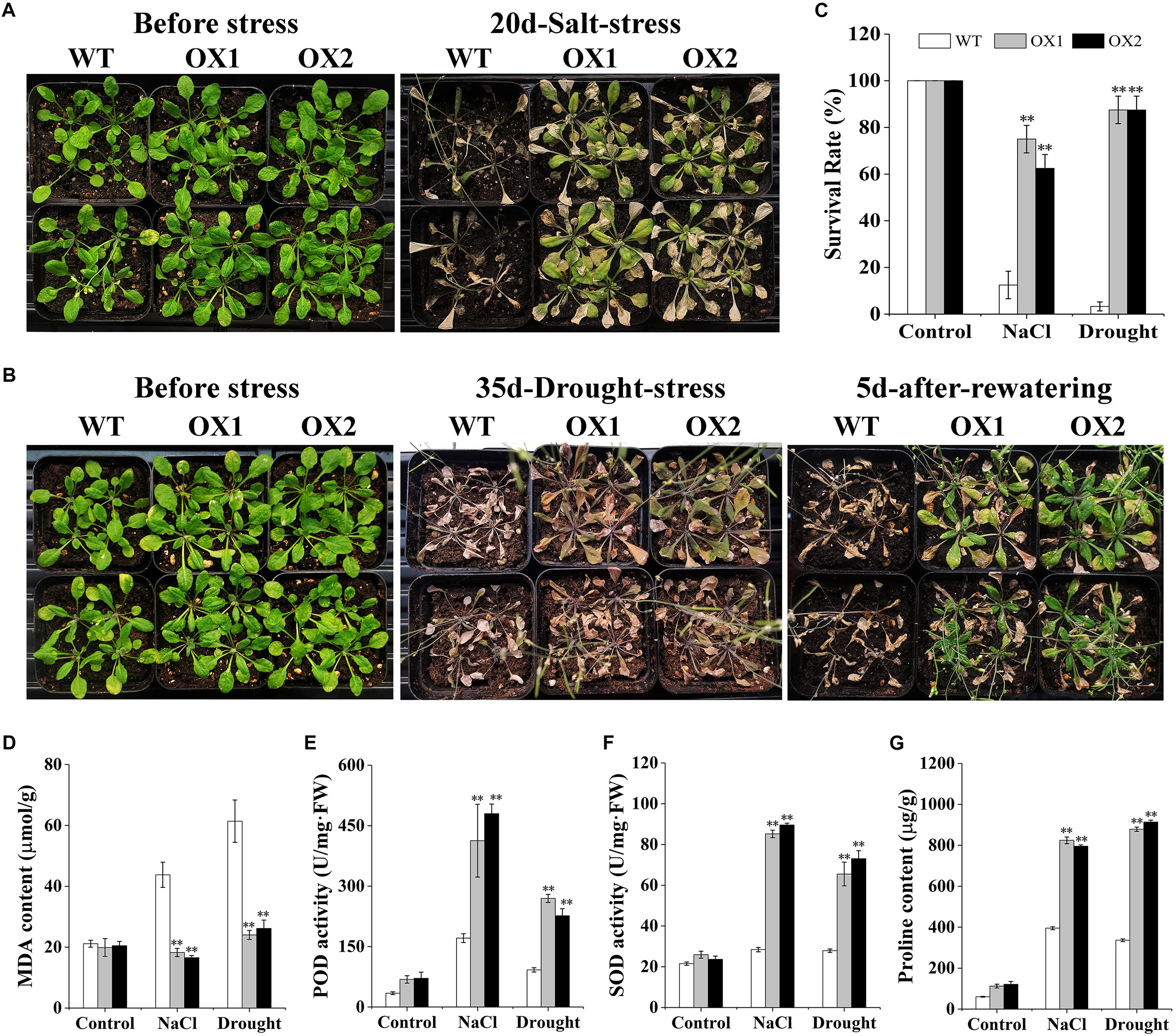
Figure 6. Dehydration and salt stress resistance analysis of transgenic lines. (A,B) Performance of OoNAC72-OX Arabidopsis plants under salt and drought stresses. Seedlings were grown without water for 35 days and re-watered for 5 days. (C) Survival rate after recovery from drought and salt stress. Each experiment comprises 20 plants. (D–G) Proline and MDA content, SOD and POD activities in WT and OoNAC72-OX lines after drought and salt stress. Each data point is the mean of three biological replicates. Error bars indicate SD, and asterisks indicate a significant difference (*P < 0.05; ∗∗P < 0.01) compared with WT.
To further investigate drought sensitivity of the OoNAC72-OX plants, the rate of nature water loss and stomatal apertures of leaves from the 25-day-old soil-grown WT and OoNAC72-OX plants were detected (Figure 7). After 3 h of air drying, the leaves of OX1 and OX2 were slightly curled, while the WT plants were severely curled (Figure 7A). Additionally, OoNAC72-OX lines showed lower water loss rate compared with WT plants (Figure 7B), indicating that over expression of OoNAC72 had increased water retention capacity in Arabidopsis. After 10 days of drought control, stomatal apertures index of the transgenic and WT lines all appeared significant changes. OX1 and OX2 plants decreased from 0.40 and 0.49 to 0.09 and 0.08, which was significantly smaller than WT (from 0.46 to 0.27) (Figures 7C,D). WT and OoNAC72-OX plants were treated with 20% PEG-6000 simulated drought stress. ABA was detected at different time points. The ABA content of these OoNAC72-OX lines were significantly higher than that of the WT (Supplementary Figure S3). Those results indicated that OoNAC72-OX plants may reduce the loss of water by regulating the stomata closure to improve drought resistance.
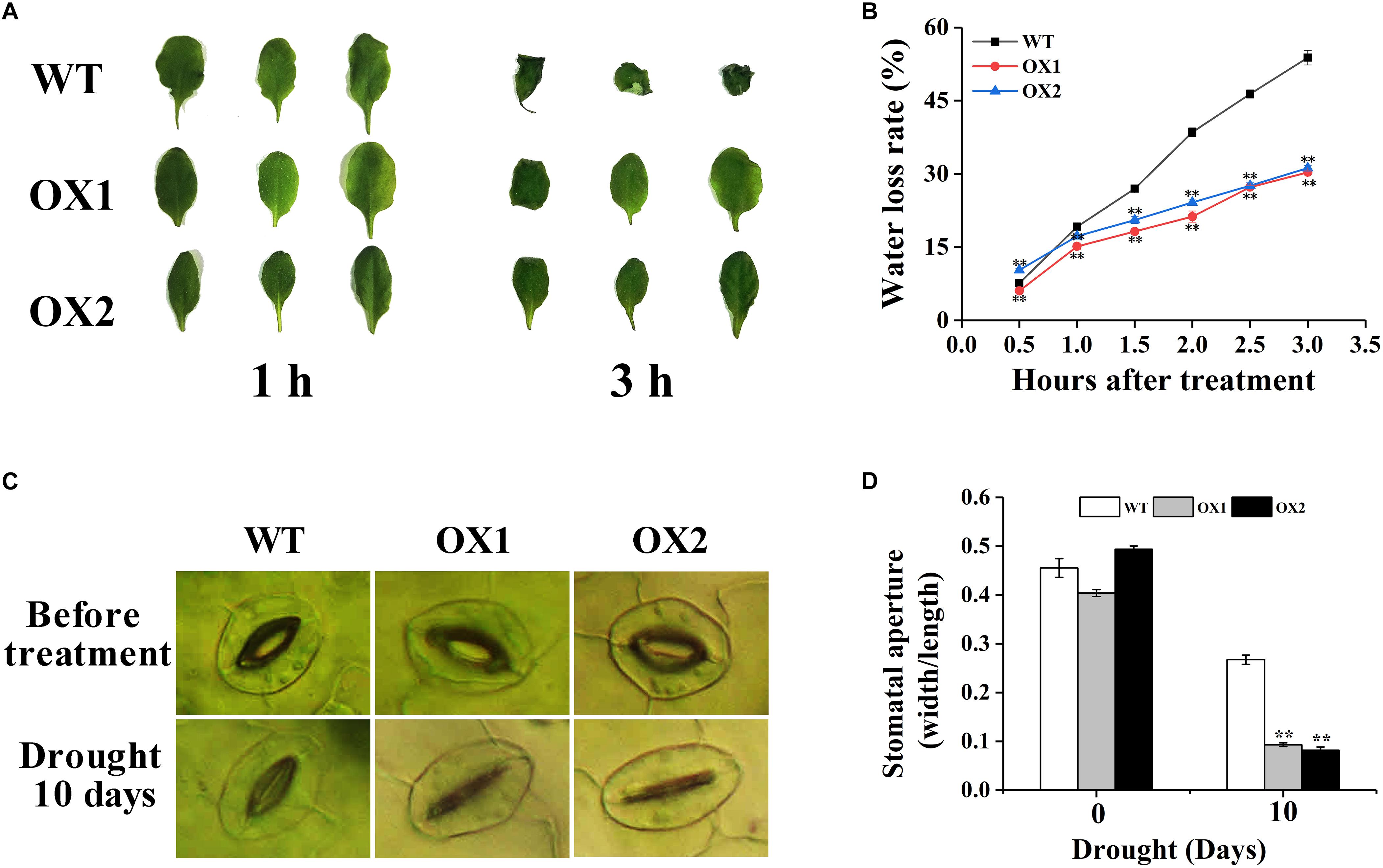
Figure 7. OoNAC72-OX plants exhibit reduced tolerance to water deficiency. (A) Phenotypes of detached leaves under water deficiency treatment for 1 and 3 h. (B) Water loss from detached rosettes of WT and OoNAC72-OX plants. Values are means ± SD of three independent experiments. (C) Stomatal apertures of WT and OoNAC72-OX plants. Scale bars = 30 μm. (D) Stomatal aperture width/length ratios under dehydration conditions in 10 days. The stomatal aperture was measured by microscope. Values are means ± SD (n = 50) in assays. Asterisks indicate statistically significant differences from WT. *P < 0.05, ∗∗P < 0.01.
To explore the underlying basis of this phenotype caused by drought or salt stress in the transgenic plants, eight stress-responsive genes: RD29A (Nakashima et al., 2006), RD29B (Nakashima et al., 2006), RD26 (Fujita et al., 2004), LEA14 (Jia et al., 2014), ANACOR19 (Tran et al., 2004), ZAT10 (Mittler et al., 2002), PP2CA (Yoshida and Hirayama, 2006) and NCED3 (Iuchi, 2001) were selected for expression pattern assays (Figure 8). Under normal condition, the expression levels of eight genes showed no significant difference between the OoNAC72-OX lines and WT. However, the expression levels of the eight genes in the OX1 and OX2 lines were significantly higher than those in WT plants under salt and drought conditions.
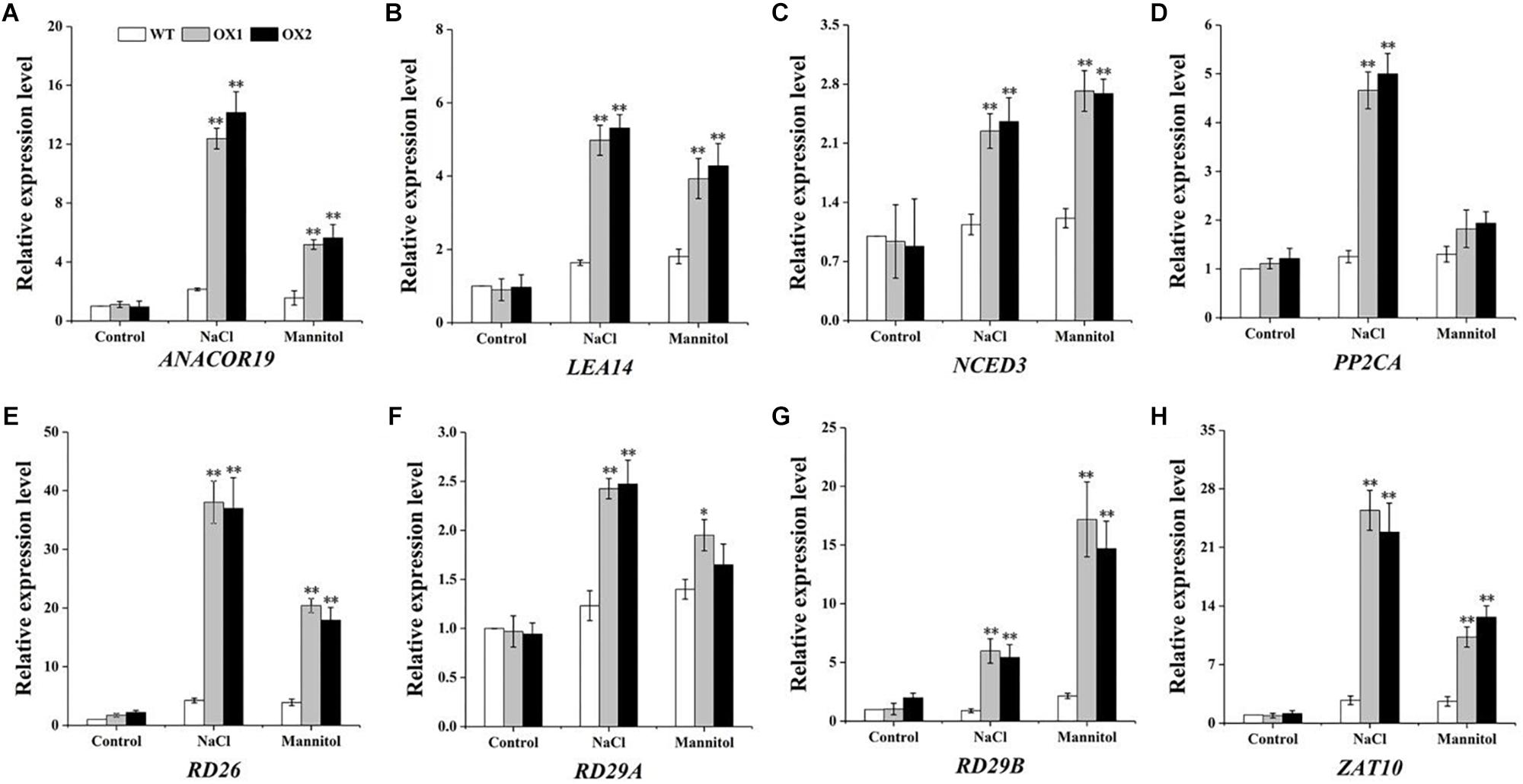
Figure 8. Expression levels of the stress-associated genes in WT and OoNAC72-OX plants. The 14-day-old seedlings grown on 1/2 MS medium were subsequently submerged in 1/2 MS cultures supplemented with 150 mM NaCl and 200 mM mannitol for 7 days. (A–H) Transcript levels of stress-associated genes were determined against with ACTIN8. Data are means ± SD of three biological replicates. Asterisks indicates insignificant differences from WT (*P < 0.05; ∗∗P < 0.01).
The ABA sensitivity of the OoNAC72-OX lines was assessed by analyses of seed germination and seedling growth (Figure 9). In the absence of ABA, no obvious difference was observed between transgenic and WT plants under normal growing conditions. However, when supplied with 1 or 3 μM ABA, seeds germination and seedling root length of the OX1 and OX2 lines were significantly inhibited compared with the WT lines (Figures 9A–D). Moreover, when 40-day-old plants were treated with 10 μM ABA, the stomatal apertures of both wild-type and OoNAC72-OX lines all happened to change (Figure 9E). The stomatal apertures index of WT plants decreased from 0.47 to 0.25, while the OX1 and OX2 lines dropped from 0.48 and 0.52 to 0.12 and 0.15 (Figure 9F). It indicated that the OoNAC72-OX lines were more rapid and variable in stomatal conductance. These results indicated that overexpression of OoNAC72 gene led to increased ABA sensitivity, which resulted in retarded growth of transgenic plants.
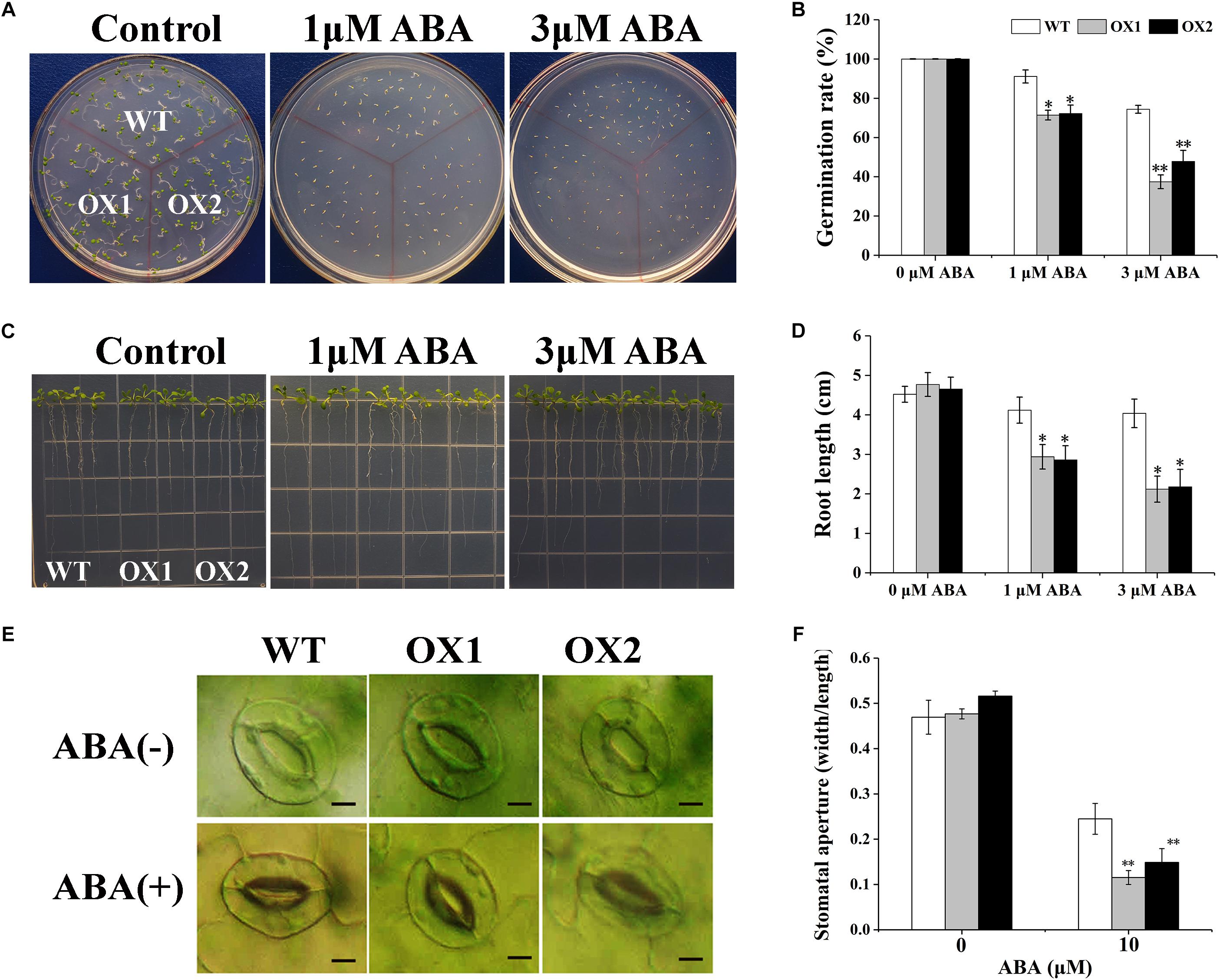
Figure 9. Responses of OoNAC72-OX plants to ABA. (A,B) Seed germination assay between OoNAC72-OX and WT Arabidopsis plants. The surface-sterilized seeds were sowed on 1/2 MS solid medium supplemented with 1 and 3 μM ABA incubated at 22°C. (C,D) Root length assay between OoNAC72-OX and WT Arabidopsis plants. Five-day-old seedlings were cultured vertically on 1/2 MS solid medium supplemented with 1 and 3 μM ABA for 7 days. (E,F) Stomatal closure of WT and OoNAC72-OX plants with 10 μM of ABA. Values are means ± SD (n = 50) in assays. Asterisks indicate statistically significant differences from WT. *P < 0.05, ∗∗P < 0.01.
O. ochrocephala is a perennial poisonous plant widely distributed in harsh environments, and livestock generally do not eat it when edible pasture is relatively abundant. In recent years, due to the emergence of extreme climatic and human overgrazing, O. ochrocephala has spread rapidly and become one of the main poisonous weeds in Western China. Studies have shown that O. ochrocephala is rich in protein and mineral elements and can be used as a good forage forb for livestock after detoxification (Shen and Mo, 2017). At the same time, as a highly resistant plant, O. ochrocephala has important ecological value in wind break and sand fixation, as well as biodiversity maintenance. In view of these characteristics, we focus on O. ochrocephala to elucidate the survival mechanism in adversity, and ultimately to provide a theoretical basis for its development and utilization.
In the present study, we characterized the OoNAC72 gene, a novel stress-related member of the NAC gene family in O. ochrocephala. OoNAC72 protein contains a typical NAC conserved domain located in the N-terminal region, which can be divided into five subdomains. As the transcriptional regulatory domain, the C-terminus is extremely variable. This result is in agreement with previous reports on other NAC TFs domains, such as PbeNAC1, ZmNAC55, CmNAC1 (Mao et al., 2016; Cao et al., 2017; Jin et al., 2017). Phylogenetic analysis confirmed the OoNAC72 had the highest homology with the two legumes, Medicago truncatula and Cicer arietinum. Subcellular localization and transactivation analysis jointly revealed that OoNAC72 was a transcription factor, which was consistent with previous research (Mao et al., 2016). Therefore, OoNAC72 act as a transcriptional activator, depending on interactions with other transcription factors or conformational changes. These results speculate that OoNAC72 is a novel stress-related member of the NAC gene family in O. ochrocephala.
In previous researches, many NAC genes had been identified as an important part of the progress of complex signal transduction when plants were subjected to various stresses, especially abiotic stress such as drought and salt stress (Puranik et al., 2012). In our study, we found that OoNAC72 can be induced by high salt, drought and ABA treatment, whereas not induced by GA and ETH. we speculated that OoNAC72 may respond to stress in O. ochrocephala by ABA-dependent signal transduction pathways. These abiotic stresses induced expression pattern of OoNAC72 was supported by earlier studies, such as in pumpkin (Cao et al., 2017), Pyrus betulifolia (Jin et al., 2017), tomato (Zhu et al., 2014), chickpea (Yu et al., 2014, 2016) and Tamarix hispida (Wang et al., 2017).
Numerous reports have demonstrated that NAC TFs are involved in a number of biological processes, such as regulating the growth of plant cells (Hiroaki et al., 2010), seed development (Sperotto et al., 2009), embryonic development, fiber formation and development, cell differentiation and leaf senescence (Youn-Sung et al., 2007). In our study, we found that OoNAC72-OX (OX1, OX2, OX3) lines showed significant rosette leaves, delayed twitches and flowering compared with WT plants. However, the mechanism resulting from overexpression of OoNAC72 in regulating this phenotype is still unknown and waiting for further research.
Overexpression or deletion mutation is the primary way to study the function of NAC transcription factors in adversity stress. In this study, several physiological changes of OoNAC72-OX lines seem to be involved in abiotic stress-resistant and molecular mechanisms. Firstly, in terms of phenotype, OoNAC72-OX lines appeared improved tolerance to salt and drought stresses during seed germination, post-germinative growth periods and vegetative growth stage. Consistent with the results of OoNAC72, CarNAC4-transgenic plants exhibited enhanced drought and salt tolerance than the WT plant, which were strongly demonstrated by both morphological and physiological changes (Yu et al., 2016). Secondly, at the physiological and biochemical levels, proline and MDA content, POD and SOD activities in plants overexpressing OoNAC72 have been significantly induced under different stress conditions. Thirdly, at the molecular level, our data also demonstrated that over-expression of OoNAC72 enhanced the expression of stress-responsive genes such as RD29A, RD29B, RD26, LEA14, ANACOR19, ZAT10, PP2CA, and NCED8. Previous studies have confirmed that NAC proteins could bind to NACRS containing the core sequence “CGT[A/G]” to regulate gene expression through ABA dependent pathway for various stress response (Duval et al., 2002; Olsen et al., 2005a; Takasaki et al., 2010; Jensen et al., 2013). However, the genome of O. ochrocephala has not been sequenced and the OoNAC72 promoter sequence could not be cloned, and so it is impossible to predict these cis-acting elements associated with stress in the promoter, and resistance mechanisms need to be further investigated.
Abscisic acid is a very important signaling molecule that respond to many adverse environmental stresses such as high salt, drought and extreme temperatures (Agarwal and Jha, 2010; Fujita et al., 2011). In plants, higher ABA sensitivity may irritate the stomata to maintain moisture and enhance drought resistance. In our study, ABA content of WT and OoNAC72-OX plants treated with 20% PEG-6000 simulating drought stress showed that ABA content of the OoNAC72-OX lines was significantly higher than that of the WT (Supplementary Figure S3). Furthermore, we found that the stomata in Arabidopsis overexpressing OoNAC72 had different degrees of closure after drought stress, indirectly mapping the relationship between drought stress and the ABA pathway. Similar results were found in Arabidopsis and maize (Ping-Li et al., 2007; Mao et al., 2016). Overexpression of KUP6 (K+ uptake transporter 6) in Arabidopsis increased the sensitivity of ABA to drought stress through faster stomatal closure, thereby improving the tolerance of transgenic plants to drought stress (Yuriko et al., 2013). Genes such as PP2Cs, SnRK2s, NAC and WRKY played a vital role in ABA signaling pathways induced by drought stress (Kim, 2014). Therefore, we supposed that OoNAC72 may enhance the drought resistance of transgenic Arabidopsis by participating in the ABA pathway.
Under stress conditions, plants reduce the damage caused by stress-induced ROS by enhancing their antioxidant defense system (Oracz et al., 2009; Suzuki et al., 2012). MDA is one of the important parameters to measure the degree of oxidative damage in plant cells. While pro contributes to osmotic adjustment to effectively enhance the antioxidant system and reduce peroxidative damage (Zhuang et al., 2015a). In our study, transgenic plants produced less MDA and more pro under stress than wild-type plants, revealing that OoNAC72 overexpression enhanced the body’s regulation of feedback regulatory substances.
Under drought stress, ABA acts as an upstream signaling of NO and they form cross-signal pathways co-regulate a balance between ROS (H2O2) and NO production (Zhuang et al., 2015b). Our resistance assays showed that the ABA contents, and SOD and POD activities were significantly induced by OoNAC72 under drought stresses condition. The increase in ABA content leads to an increase in the concentration of NO for activating NO signaling pathway. On the one hand, the low concentration of NO interacts with H2O2 to stimulate the antioxidant defense system. On the other hand, the increase of SOD and POD activity further enhances plant’s ability to scavenge excessive H2O2. The synergy between NO signal and ROS signal promotes the dynamic balance between NO and H2O2, and ultimately improves plant resistance tolerance (Miller et al., 2010). These results indicate that showing the positive regulation of OoNAC72 on ABA signaling pathway under drought stress.
Plant NAC transcription factors control diverse biological processes, such as differentiation, development and abiotic stress responses. In this study, we identified a gene encoding NAC72-type transcription factor from Oxytropis ochrocephala, and characterized its role. The nuclear localization and transcriptional activity of this protein indicate the possible function of OoNAC72 as a transcription factor. In addition, expression of OoNAC72 transcript was shown to be up-regulated in response to abiotic stresses and exogenous ABA. Furthermore, analyses of transgenic Arabidopsis expressing OoNAC72 also supported the involvement of OoNAC72 in drought and salinity responses as well as in the regulation of ABA-dependent processes.
HG and YF conceived and designed the study. HG, XL, and FN performed the experiments. HG, XL, and QZ analyzed the data and wrote the manuscript. FN, QZ, NF, DC, and DM reviewed the manuscript. WH, BG, YW, and YF contributed reagents, materials, and fund support.
This work was supported by Special Fund for Agro-scientific Research in The Public Interest (201203062), Key Research and Development Plan Project of Shaanxi Province (2018ZDXM-SF-016) and Opening Foundation of Key Laboratory of Resource Biology and Biotechnology in Western China (Northwest University), Ministry of Education (ZSK2016003) to YW, Bureau of Science and Technology in Shaanxi Province, Xi’an City, Beilin District (GX1702) to BG, Natural Science Foundation of Shanxi Province of China (2018JQ3029) and Natural Science Foundation of China (31702159) to YF, Key Research and Development Plan Project of Shaanxi Province (2018SF-034) and Scientific Research from Shaanxi Provincial Department of Education (17JK0243) to NF.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the reviewers for their constructive suggestions. We are also particularly grateful to other members of our research group for their helpful comments to improve the article.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00890/full#supplementary-material
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biol. Plant. 54, 201–212. doi: 10.1007/s10535-010-0038-7
Aida, M., Ishida, T., Fukaki, H., Fujisawa, H., and Tasaka, M. (1997). Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9, 841–857. doi: 10.1105/tpc.9.6.841
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1016/j.dental.2010.07.006
Cao, H., Wang, L., Nawaz, M. A., Niu, M., Sun, J., Xie, J., et al. (2017). Ectopic expression of pumpkin NAC transcription factor CmNAC1 improves multiple abiotic stress tolerance in Arabidopsis. Front. Plant Sci. 8:2052. doi: 10.3389/fpls.2017.02052
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Duval, M., Hsieh, T. F., Kim, S. Y., and Thomas, T. L. (2002). Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol. Biol. 50, 237–248.
Fujita, M., Fujita, Y., Maruyama, K., Seki, M., Hiratsu, K., Ohme-Takagi, M., et al. (2004). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313x.2004.02171.x
Fujita, Y., Fujita, M., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2011). ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 124, 509–525. doi: 10.1007/s10265-011-0412-3
He, W., Zhuang, H., Fu, Y., Guo, L., Guo, B., Guo, L., et al. (2015). De novo transcriptome assembly of a chinese locoweed (Oxytropis ochrocephala) species provides insights into genes associated with drought, salinity, and cold tolerance. Front. Plant Sci. 6:1086. doi: 10.3389/fpls.2015.01086
Hegedus, D. Y. M., Baldwin, D., Gruber, M., Sharpe, A., Parkin, I., Whitwill, S., et al. (2003). Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol. Biol. 53, 383–397. doi: 10.1023/b:plan.0000006944.61384.11
Hénanff, G. L., Profizi, C., Courteaux, B., Rabenoelina, F., Gérard, C., Clément, C., et al. (2013). Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 64, 4877–4893. doi: 10.1093/jxb/ert277
Hiroaki, K., Taizo, M., Yoshibumi, K., Tamao, S., and Atsushi, K. (2010). Overexpression of the NAC transcription factor family gene ANAC036 results in a dwarf phenotype in Arabidopsis thaliana. J. Plant Physiol. 167, 571–577. doi: 10.1016/j.jplph.2009.11.004
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Huang, Q., and Wang, Y. (2016). Overexpression of TaNAC2D displays opposite responses to abiotic stresses between seedling and mature stage of transgenic Arabidopsis. Front. Plant Sci. 7:1754. doi: 10.3389/fpls.2016.01754
Huang, Q., Wang, Y., Li, B., Chang, J., Chen, M., Li, K., et al. (2015). TaNAC29, a NAC transcription factor from wheat, enhances salt and drought tolerance in transgenic Arabidopsis. BMC Plant Biol. 15:268. doi: 10.1186/s12870-015-0644-9
Iuchi, S. (2001). Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 27, 325–333. doi: 10.1046/j.1365-313x.2001.01096.x
Jensen, M. K., Lindemose, S., Masi, F. D., Reimer, J. J., Nielsen, M., Perera, V., et al. (2013). ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio. 3:321. doi: 10.1016/j.fob.2013.07.006
Jeong, J. S., Kim, Y. S., Redillas, M. C., Jang, G., Jung, H., Bang, S. W., et al. (2013). OsNAC5 overexpression enlarges root diameter in rice plants leading to enhanced drought tolerance and increased grain yield in the field. Plant Biotechnol. J. 11, 101–114. doi: 10.1111/pbi.12011
Jia, F., Qi, S., Hui, L., Pu, L., Li, P., Wu, C., et al. (2014). Overexpression of late embryogenesis abundant 14 enhances Arabidopsis salt stress tolerance. Biochem. Biophys. Res. Commun. 454, 505–511. doi: 10.1016/j.bbrc.2014.10.136
Jin, C., Li, K. Q., Xu, X. Y., Zhang, H. P., Chen, H. X., Chen, Y. H., et al. (2017). A novel NAC transcription factor, PbeNAC1, of Pyrus betulifolia confers cold and drought tolerance via interacting with PbeDREBs and activating the expression of stress-responsive genes. Front. Plant Sci. 8:1049. doi: 10.3389/fpls.2017.01049
Jinpu, J., He, Z., Lei, K., Ge, G., and Jingchu, L. (2014). PlantTFDB 3.0: a portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 42, 1182–1187. doi: 10.1093/nar/gkt1016
Kim, T. H. (2014). Mechanism of ABA signal transduction: agricultural highlights for improving drought tolerance. J. Plant Biol. 57, 1–8. doi: 10.1007/s12374-014-0901-8
Mao, X., Chen, S., Li, A., Zhai, C., and Jing, R. (2014). Novel NAC transcription factor TaNAC67 confers enhanced multi-abiotic stress tolerances in Arabidopsis. PLoS One 9:e84359. doi: 10.1371/journal.pone.0084359
Mao, X., Zhang, H., Qian, X., Li, A., Zhao, G., and Jing, R. (2012). TaNAC2, a NAC-type wheat transcription factor conferring enhanced multiple abiotic stress tolerances in Arabidopsis. J. Exp. Bot. 63, 2933–2946. doi: 10.1093/jxb/err462
Mao, H., Yu, L., Han, R., Li, Z., and Liu, H. (2016). ZmNAC55, a maize stress-responsive NAC transcription factor, confers drought resistance in transgenic Arabidopsis. Plant Physiol. Biochem. 105, 55–66. doi: 10.1016/j.plaphy.2016.04.018
Meng, C., Caiping, C., Zhang, T., and Guo, W. (2009). Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 176, 352–359. doi: 10.1016/j.plantsci.2008.12.003
Miki, F., Yasunari, F., Kyonoshin, M., Motoaki, S., Keiichiro, H., Masaru, O. T., et al. (2010). A dehydration-induced NAC protein, RD26, is involved in a novel ABA-dependent stress-signaling pathway. Plant J. 39, 863–876. doi: 10.1111/j.1365-313x.2004.02171.x
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mittler, R., Kim, Y. S., Song, L., Coutu, J., Coutu, A., Ciftci-Yilmaz, S., et al. (2002). Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 580, 6537–6542. doi: 10.1016/j.febslet.2006.11.002
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005a). DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 169, 785–797. doi: 10.1016/j.plantsci.2005.05.035
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005b). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Ooka, H., Satoh, K., Doi, K., Nagata, T., Otomo, Y., Murakami, K., et al. (2003). Comprehensive analysis of NAC family genes in oryza sativa and Arabidopsis thaliana. DNA Res. 10:239. doi: 10.1093/dnares/10.6.239
Oracz, K., Elmaaroufbouteau, H., Kranner, I., Bogatek, R., Corbineau, F., and Bailly, C. (2009). The mechanisms involved in seed dormancy alleviation by hydrogen cyanide unravel the role of reactive oxygen species as key factors of cellular signaling during germination. Plant Physiol. 150, 494–505. doi: 10.1104/pp.109.138107
Ping-Li, L., Nai-Zhi, C., Rui, A., Zhao, S., Bi-Shu, Q., Fei, R., et al. (2007). A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63:289. doi: 10.1007/s11103-006-9089-8
Pinheiro, G. L., Marques, C. S., Costa, M. D. B. L., Reis, P. A. B., Alves, M. S., Carvalho, C. M., et al. (2009). Complete inventory of soybean NAC transcription factors: sequence conservation and expression analysis uncover their distinct roles in stress response. Gene 444, 10–23. doi: 10.1016/j.gene.2009.05.012
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Redillas, M. C., Jeong, J. S., Kim, Y. S., Jung, H., Bang, S. W., Choi, Y. D., et al. (2012). The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 10, 792–805. doi: 10.1111/j.1467-7652.2012.00697.x
Sheludko, Y. V., Sindarovska, Y. R., Gerasymenko, I. M., Bannikova, M. A., and Kuchuk, N. V. (2007). Comparison of several Nicotiana species as hosts for high-scale Agrobacterium-mediated transient expression. Biotechnol. Bieng. 96, 608–614. doi: 10.1002/bit.21075
Shen, M. H., and Mo, C. H. (2017). Comparison of nutritions and swainsonine content in different regions of Oxytropis ochrocephala. Anim. Husband. Vet. Med. 49, 39–42.
Souer, E., and Al, E. (1996). The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85, 159–170. doi: 10.1016/s0092-8674(00)81093-4
Sperotto, R. A., Ricachenevsky, F. K., Duarte, G. L., Tatiana, B., Lopes, K. L., Sperb, E. R., et al. (2009). Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230, 985–1002. doi: 10.1007/s00425-009-1000-9
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Tak, H., Negi, S., and Ganapathi, T. R. (2017). Banana NAC transcription factor MusaNAC042 is positively associated with drought and salinity tolerance. Protoplasma 254, 803–816. doi: 10.1007/s00709-016-0991-x
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284, 173–183. doi: 10.1007/s00438-010-0557-0
Tang, Y., Liu, M., Gao, S., Zhang, Z., Zhao, X., Zhao, C., et al. (2012). Molecular characterization of novel TaNAC genes in wheat and overexpression of TaNAC2a confers drought tolerance in tobacco. Physiol. Plant 144, 210–224. doi: 10.1111/j.1399-3054.2011.01539.x
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Tran, L. S., Quach, T. N., Guttikonda, S. K., Aldrich, D. L., Kumar, R., Neelakandan, A., et al. (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics 281, 647–664. doi: 10.1007/s00438-009-0436-8
Tulsiani, D. R., Broquist, H. P., James, L. F., and Touster, O. (1988). Production of hybrid glycoproteins and accumulation of oligosaccharides in the brain of sheep and pigs administered swainsonine or locoweed. Arch. Biochem. Biophys. 264, 607–617. doi: 10.1016/0003-9861(88)90327-x
Wang, L., Li, Z., Lu, M., and Wang, Y. (2017). ThNAC13, a NAC transcription factor from Tamarix hispida, confers salt and osmotic stress tolerance to transgenic Tamarix and Arabidopsis. Front. Plant Sci. 8:635. doi: 10.3389/fpls.2017.00635
Wang, Y., Gao, C., Liang, Y., Wang, C., Yang, C., and Liu, G. (2010). A novel bZIP gene from Tamarix hispida mediates physiological responses to salt stress in tobacco plants. J. Plant Physiol. 167, 222–230. doi: 10.1016/j.jplph.2009.09.008
Xu, Z. Y., Kim, S. Y., Hyeon do, Y., Kim, D. H., Dong, T., Park, Y., et al. (2013). The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. doi: 10.1105/tpc.113.119099
Yang, J., Zhang, J., Wang, Z., Zhu, Q., and Wang, W. (2001). Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 127, 315–323. doi: 10.1104/pp.127.1.315
Yanjuan, J., Liang, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Yoshida, T., and Hirayama, T. (2006). ABA-hypersensitive germination3 encodes a protein phosphatase 2C (AtPP2CA) that strongly regulates abscisic acid signaling during germination among Arabidopsis protein phosphatase 2Cs. Plant Physiol. 140, 115–126. doi: 10.1104/pp.105.070128
Youn-Sung, K., Sang-Gyu, K., Jung-Eun, P., Hye-Young, P., Mi-Hye, L., Nam-Hai, C., et al. (2007). A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell 18, 3132–3144. doi: 10.1105/tpc.106.043018
Yu, X., Liu, Y., Wang, S., Tao, Y., Wang, Z., Shu, Y., et al. (2016). CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 35, 613–627. doi: 10.1007/s00299-015-1907-5
Yu, X., Peng, H., Liu, Y., Zhang, Y., Shu, Y., Chen, Q., et al. (2014). CarNAC2, a novel NAC transcription factor in chickpea (Cicer arietinum L.), is associated with drought-response and various developmental processes in transgenic Arabidopsis. J. Plant Biol. 57, 55–66. doi: 10.1007/s12374-013-0457-z
Yujie, F., Jun, Y., Kabin, X., Weibo, X., and Lizhong, X. (2008). Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 547–563. doi: 10.1007/s00438-008-0386-6
Yuriko, O., Naoko, A., Taishi, U., Shogo, K., Keita, N., Hidenori, T., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25, 609–624. doi: 10.1105/tpc.112.105700
Zhao, M., Gao, X., Wang, J., He, X., and Han, B. (2013). A review of the most economically important poisonous plants to the livestock industry on temperate grasslands of China. J. Appl. Toxicol. 33, 9–17. doi: 10.1002/jat.2789
Zheng, X., Bo, C., Lu, G., and Han, B. (2009). Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 379, 985–989. doi: 10.1016/j.bbrc.2008.12.163
Zhu, M., Chen, G., Zhang, J., Zhang, Y., Xie, Q., Zhao, Z., et al. (2014). The abiotic stress-responsive NAC-type transcription factor SlNAC4 regulates salt and drought tolerance and stress-related genes in tomato (Solanum lycopersicum). Plant Cell Rep. 33, 1851–1863. doi: 10.1007/s00299-014-1662-z
Zhuang, H., Fu, Y., He, W., Wang, L., and Wei, Y. (2015a). Selection of appropriate reference genes for quantitative real-time PCR in Oxytropis ochrocephala Bunge using transcriptome datasets under abiotic stress treatments. Front. Plant Sci. 6:475. doi: 10.3389/fpls.2015.00475
Keywords: O. ochrocephala, NAC transcription factor, drought stress, salt stress, ABA hypersensitivity, transgenic Arabidopsis
Citation: Guan H, Liu X, Niu F, Zhao Q, Fan N, Cao D, Meng D, He W, Guo B, Wei Y and Fu Y (2019) OoNAC72, a NAC-Type Oxytropis ochrocephala Transcription Factor, Conferring Enhanced Drought and Salt Stress Tolerance in Arabidopsis. Front. Plant Sci. 10:890. doi: 10.3389/fpls.2019.00890
Received: 06 January 2019; Accepted: 21 June 2019;
Published: 11 July 2019.
Edited by:
Motoaki Seki, RIKEN, JapanReviewed by:
Yong Hwa Cheong, Sunchon National University, South KoreaCopyright © 2019 Guan, Liu, Niu, Zhao, Fan, Cao, Meng, He, Guo, Wei and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yahui Wei, d2VpeWFodWlAbnd1LmVkdS5jbg==; Yanping Fu, ZnV5YW5waW5nQG53dS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.