
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 03 July 2019
Sec. Plant Membrane Traffic and Transport
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00851
This article is part of the Research TopicRecent Advances in Understanding Plant Hormone TransportersView all 13 articles
Auxin is a crucial phytohormone involved in multiple plant developmental processes. Spatiotemporal regulation of auxin levels is necessary to achieve development of organs in the proper place and at the proper time. These levels can be regulated by conversion of auxin [indole 3-acetic acid (IAA)] from its conjugated forms and its precursors. Indole 3-butyric acid (IBA) is an auxin precursor that is converted to IAA in a peroxisomal β-oxidation process. In Arabidopsis, altered IBA-to-IAA conversion leads to multiple plant defects, indicating that IBA contributes to auxin homeostasis in critical ways. Like IAA, IBA and its conjugates can be transported in plants, yet many IBA carriers still need to be identified. In this review, we discuss IBA transporters identified in Arabidopsis thus far, including the pleiotropic drug resistance (PDR) members of the G subfamily of ATP-binding cassette transporter (ABCG) family, the TRANSPORTER OF IBA1 (TOB1) member of the major facilitator superfamily (MFS) family and hypothesize other potential IBA carriers involved in plant development.
Multiple pathways coordinate plant development; many of which require auxin for their effect on development. Auxin is a well-studied plant hormone that is important for multiple plant developmental processes with major roles in cell division, differentiation, and elongation (Ljung, 2013; reviewed in Zhao, 2010). Endogenous active auxins include indole acetic acid (IAA), phenyl acetic acid (PAA), and 4-chloroindole-3-acetic acid (4-Cl-IAA) (Cook, 2019; reviewed in Korasick et al., 2013). IAA is widely considered to be the predominant form of active auxin within the plant and likely contributes to the majority of auxin activity in many plants.
Attaining auxin maxima in specific tissues is essential for organogenesis and for environmental responses. Mechanisms that contribute to the regulated distribution of auxin include polar auxin transport and IAA metabolism. The polarized localization of IAA transporters regulates auxin movement in specific directions, whereas local auxin metabolism provides the IAA for this transport and is also critical for establishing auxin maxima. These metabolism and transport mechanisms for IAA likely act in concert (Brumos et al., 2018; Morffy and Strader, 2018).
Auxin is primarily synthesized through a two-step tryptophan-dependent auxin biosynthesis pathway catalyzed by the tryptophan aminotransferase of Arabidopsis (TAA) and YUCCA (flavin monooxygenase enzyme) families of enzymes (reviewed in Zhao, 2012). TAA family enzymes convert the aromatic amino acid tryptophan into indole-3-pyruvic acid (IPyA); this is subsequently converted into IAA by YUCCA family members. Higher order taa and higher order yucca mutants display drastic developmental phenotypic defects (Zhao et al., 2001; Cheng et al., 2006, 2007; Stepanova et al., 2008; Tao et al., 2008), suggesting functional redundancy within these families and also revealing critical roles for de novo auxin biosynthesis in plant development.
In addition to de novo auxin biosynthesis, IAA can be released from conjugates with sugars, amino acids, and the chain-lengthened precursor indole-3-butyric acid (IBA) (reviewed in Korasick et al., 2013; Frick and Strader, 2018). Several amide-linked IAA conjugates have been identified in plants, including IAA-Ala and IAA-Leu, which can be hydrolyzed to release free IAA in Arabidopsis, maize, and several other plant species (Bartel and Fink, 1995; LeClere et al., 2002; Campanella et al., 2003a,b, 2008; Rampey et al., 2004). IAA stored in the form of an ester-linked IAA-sugar conjugate is hydrolyzed to release active IAA in both monocots and dicots (Jakubowska et al., 1993; Jakubowska and Kowalczyk, 2005; Campanella et al., 2008). The auxin precursor IBA is converted in to active IAA through a β-oxidation process in the peroxisome (Strader et al., 2011).
IBA was initially assumed to be a synthetic auxinic compound that was primarily used as a rooting media agent. In marigold, tomato, buckwheat, pea, bean, sunflower, and few other plant species, exogenous IBA induces root elongation, leaf epinasty, and stem bending (Zimmerman and Wilcoxon, 1935). Although IBA was once thought to be a synthetic auxin, it was later detected as an endogenous compound in potato peelings using paper chromatography (Blommaert, 1954). Endogenous IBA has been detected in multiple plant species, including Arabidopsis, tobacco, pea, and maize (reviewed in Korasick et al., 2013). This widespread occurrence of IBA suggests that this molecule may play a conserved role across species.
Although IBA has been identified as an endogenous compound in multiple plant species, it is often present at low levels and is difficult to detect. Indeed, the presence of IBA as an endogenous molecule has been questioned in one study, due to an inability to detect it using gas chromatography-mass spectrometry and liquid chromatography-mass spectrometry in Arabidopsis, poplar, and wheat (Novak et al., 2012). In studies in which endogenous IBA has been detected, its levels are often found at a lower level than IAA (Sutter and Cohen, 1992; Ludwig-Müller et al., 1993). Among these studies, differences in IBA detection could reflect (1) differences in metabolite extraction techniques, (2) detection limits of the technology being used, and/or (3) differences in IBA accumulation in plants grown under distinct laboratory conditions. IBA contribution to overall auxin level varies among species and even among accessions within a single species, suggesting differential IBA metabolism in different plants (Ludwig-Müller and Epstein, 1991; Ludwig-Müller et al., 1993). Resolving these differences in the future will be key to understanding the prevalence of IBA contributions to the auxin pool.
IBA is structurally similar to IAA except for the side chain, in which IAA has two carbons and IBA has a four-carbon side chain (Figure 1). IAA binds to the TIR1/AFB-Aux/IAA (TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX PROTEIN-Auxin/INDOLE-3-ACETIC ACID) co-receptor complex to initiate downstream auxin-responsive gene expression (reviewed in Lavy and Estelle, 2016). Because of its lengthened side chain, IBA is unable to stimulate formation of the auxin co-receptor complex (Uzunova et al., 2016). Thus, physiological effects of IBA treatment are likely caused by IBA-derived IAA and not by the IBA molecule itself.
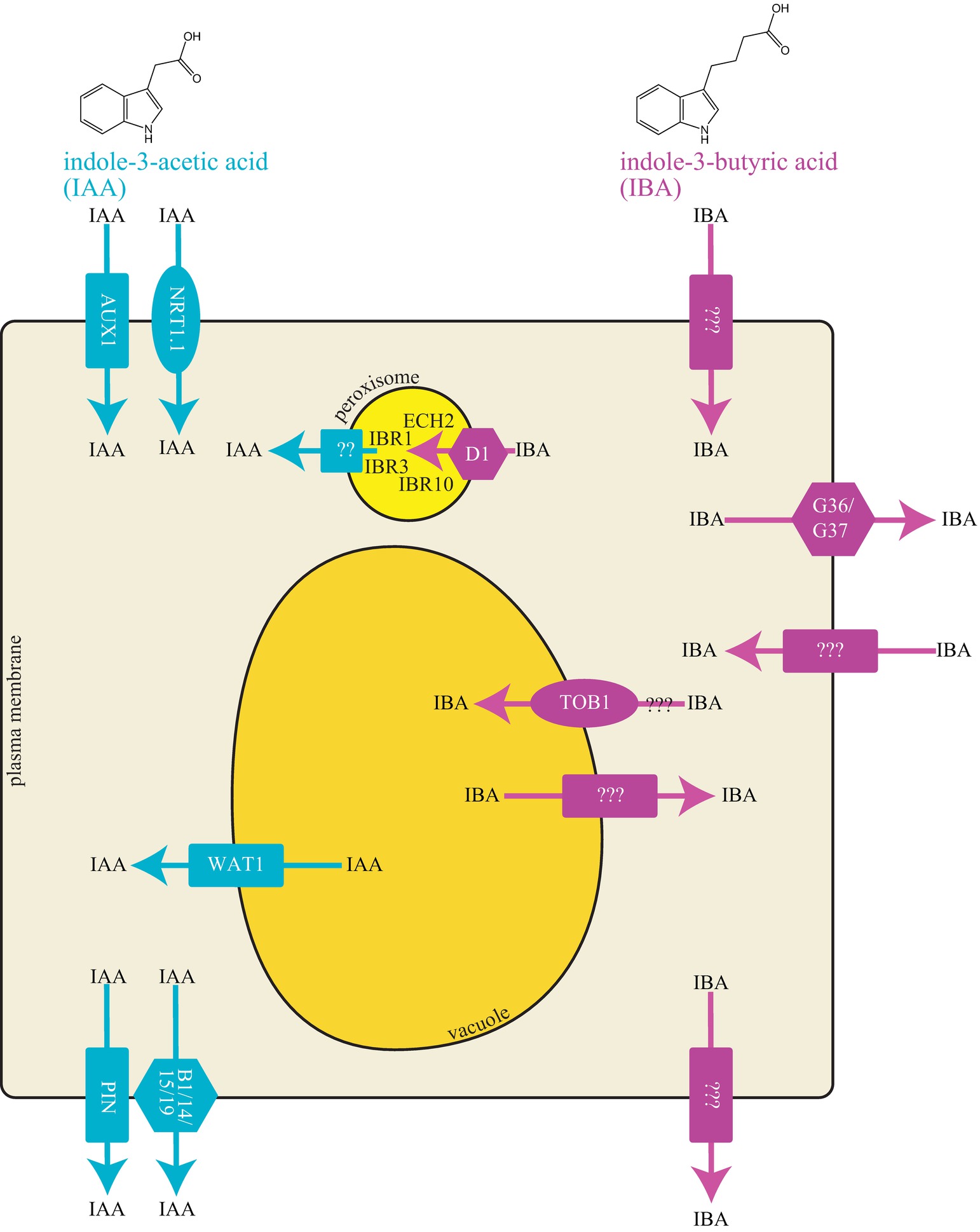
Figure 1. Cellular model of IAA and IBA transporters. IAA and IBA are indolic compounds with two and four carbon side chains, respectively. Graphical representation of a cell with different transport proteins involved in transport of IAA and IBA.
The major mechanism by which IBA influences plant development is through the reduction of its carbon side chain to convert in to IAA (reviewed in Strader and Bartel, 2011). IBA-to-IAA conversion occurs through a fatty acid β-oxidation process housed in the peroxisome (Zolman et al., 2000; Strader et al., 2010). Enzymes specifically involved in catalyzing IBA conversion include the short-chain dehydrogenase/reductase indole-3-butyric acid response 1 (IBR1; Zolman et al., 2007), the acyl-coA dehydrogenase/oxidase-like IBR3 (Zolman et al., 2007), the predicted enoyl coA hydratase IBR10 (Zolman et al., 2008), and enoyl-COA hydratase2 (ECH2; Strader et al., 2011). In addition, generalist enzymes such as PED1 (3-ketoacyl COA thiolase) (Zolman et al., 2000) and ACX enzymes (Adham et al., 2005) may also act in the conversion of IBA to IAA. Peroxisomal fatty acid β-oxidation of oil bodies is required in Arabidopsis to fuel growth prior to photosynthesis. Thus, dark-grown seedlings defective in fatty acid β-oxidation display reduced growth unless the media is supplemented with an exogenous carbon source such as sucrose (Zolman et al., 2000). Because mutants defective in IBR1, IBR3, IBR10, or ECH2 grow normally in the dark in the absence of sucrose for growth (Zolman et al., 2007, 2008; Strader et al., 2011), these enzymes are unlikely to be involved in fatty acid β-oxidation; however, it is possible they are involved in β-oxidation of substrates in addition to IBA. Conversely, mutants defective in PED1 or ACX enzymes are resistant to IBA and also require an exogenous carbon source to fuel dark-grown growth (Zolman et al., 2000; Adham et al., 2005), suggesting that these are in involved in both IBA and fatty acid β-oxidation.
Mutants defective in IBA-to-IAA conversion enzymes display multiple plant developmental defects, including reduced cotyledon expansion, reduced apical hook curvature, reduced lateral root formation, and smaller root apical meristems, along with decreased levels of free IAA (Zolman et al., 2008; Strader et al., 2010, 2011). IBA treatment fails to stimulate lateral root organogenesis in mutants defective in IBA conversion enzymes (Strader et al., 2011), suggesting that IBA cannot function directly to stimulate lateral root production but rather acts through its conversion to IAA (Strader et al., 2011). Further, the chemical naxillin stimulates lateral root production through stimulation of IBA-to-IAA conversion in Arabidopsis (De Rybel et al., 2012), confirming strong roles for IBA contribution to the pool of active auxin to regulate production of lateral roots.
Similar to IAA conjugates, both amide- and ester-linked conjugates of IBA have been detected in plants (Ludwig-Müller et al., 1993; Tognetti et al., 2010; Liu et al., 2012; Sherp et al., 2018). Overexpression of UGT74E2, encoding a glucosyltransferase, results in reduced IBA levels and elevated IBA-glucose conjugate levels in Arabidopsis (Tognetti et al., 2010), suggesting that UGT74E2 catalyzes conjugation of IBA to glucose. Further, Arabidopsis plants overexpressing UGT74E2 display increased shoot branching and improved abiotic stress tolerance (Tognetti et al., 2010), consistent with the possibility that altered IBA homeostasis affects plant growth and stress response. In these experiments, it is unclear whether depletion of the IBA molecule itself or depletion of IBA-derived IAA affects growth and stress responses when UGT74E2 is overexpressed. In addition to conjugation to glucose, IBA is also conjugated to amino acids (reviewed in Korasick et al., 2013). The Arabidopsis GH3-15 acyl acid amido synthetase enzyme specifically conjugates IBA to aspartate; however, whether this conjugated form is for IBA storage or for degradation is unknown (Sherp et al., 2018). Overall, these data suggest that homeostasis of IBA and its conjugates play important roles in maintaining IAA levels.
Polar auxin transport regulates IAA distribution via specific cellular carriers. Similar to IAA, IBA is moved by transporters (reviewed in Strader and Bartel, 2011; Michniewicz et al., 2014). Because IBA is structurally similar to IAA (Figure 1), the question arises whether IAA transporters can also transport IBA. IAA is transported directionally with the aid of tissue-specific influx and efflux proteins (reviewed in Vieten et al., 2007; Peer et al., 2011). Transporters involved in polar IAA transport include both uptake and efflux carriers. IAA uptake is carried out by members of the amino acid permease-like AUXIN RESISTANT1 (AUX1) family. Two transporter families mediate cellular IAA efflux: the family of PIN-FORMED (PIN) proteins and the MULTIDRUG RESISTANCE/GLYCOPROTEIN (PGP) class of ATP-binding cassette (ABC) transporters (reviewed in Vieten et al., 2007; Petrášek and Friml, 2009). No examined IAA transporter appears to mediate IBA transport (see sections below) suggesting the presence of IBA specific transporters in Arabidopsis.
Long-distance IBA transport was first suggested based on evidence that localized IBA treatment affected distal developmental events in marigold, tomato, sweet pea, and few other species (Zimmerman and Wilcoxon, 1935). Long-distance transport of radiolabeled IBA allowed tracking of IBA movement in cleopatra mandarin (Epstein and Sagee, 1992) and Arabidopsis (Figure 1; Ludwig-Müller et al., 1995; Rashotte et al., 2003). However, a major caveat to these tracking experiments is that the identity of the tracked molecule is unknown; thus, movement of [3H]IBA is indistinguishable from movement of [3H]IBA-conjugates or [3H]IAA derived from [3H]IBA. To overcome this limitation, analytical methods have been used to determine the identity of IBA-derived molecules in transport assays (Ruzicka et al., 2010; Liu et al., 2012). Application of [3H]-IBA to Arabidopsis root columella cells results in high-performance liquid chromatography-based detection of [3H]-IAA in root tissue 4 mm above the application site after a 2-h incubation (Ruzicka et al., 2010), suggesting that most of the transported molecule in this assay was IBA-derived IAA. Further, gas chromatography-mass spectrometry (GC/MS) methods were used to determine transported molecules across the Arabidopsis hypocotyl and inflorescence stem after heavy IBA application; multiple IBA-derived molecules were transported through these tissues, including IAA and ester-linked IBA (Liu et al., 2012). By comparison, movement of heavy IBA through these tissues occurred at lower levels, suggesting that, in Arabidopsis, IBA metabolites are transported long distances more efficiently than the IBA molecule itself (Liu et al., 2012). Thus, it seems that much of the long-distance transport of “IBA” may be of IBA conjugates, rather than the IBA molecule itself. Further, no identified IBA transporter has been shown to have roles in long-distance IBA transport.
IBA uptake is a saturable process (Ludwig-Müller et al., 1995; Rashotte et al., 2003), suggesting that IBA uptake is mediated by carriers rather than by simple diffusion. IBA transport is unaltered in the aux1 mutant in both long-distance (Rashotte et al., 2003) and root tip (Strader and Bartel, 2009) transport assays. Thus, IBA is likely not a substrate for AUX1; however, it remains possible that it may be a substrate for other members of the AUX1 family. In particular, LIKE AUX1 (LAX3) appears to display some affinity toward IBA when heterologously expressed in Xenopus oocytes (Swarup et al., 2008), suggesting that this transporter may use IBA, in addition to IAA, as a substrate.
IBA efflux appears to be mediated by carriers distinct from IAA efflux carriers. Application of the polar auxin transport inhibitors 1-N-napthylphthalamic acid (NPA) or 2,3,5-triiodobenzoic acid (TIBA) fail to block IBA efflux, whereas they have a dramatic effect on IAA transport (Rashotte et al., 2003; Liu et al., 2012), suggesting independent efflux mechanisms for these two molecules. Further, heterologously expressed PIN2, PIN7, ABCB1, and ABCB19 display no IBA efflux activity (Ruzicka et al., 2010). Thus, plants seem to use distinct efflux carriers for IBA and IAA. Indeed, several transporters that use IBA, but not IAA, as a substrate have been described. Using genetic and molecular approaches in Arabidopsis, IBA transporters including PXA1/ABCD1, ABCG36, ABCG37, and TOB1 have been identified, and their roles are elaborated in the following text.
IBA to IAA conversion occurs in the peroxisome; thus, an intracellular carrier is necessary for movement of IBA into peroxisomes. The peroxisomal ABC transporter PEROXISOMAL ABC TRANSPORTER1/ABCD1/PXA1 appears to be the influx carrier involved in transporting IBA for IAA conversion through the β-oxidation process (Figure 1; Zolman et al., 2001b). The loss-of-function pxa1 mutant displays resistance to the long-chain auxin precursor IBA, but wild-type sensitivity to the active auxin IAA (Zolman et al., 2001b), due to a reduction in IBA-to-IAA conversion (Strader et al., 2010). PXA1 likely transports compounds in addition to IBA for peroxisomal β-oxidation, such as fatty acid and jasmonic acid (Zolman et al., 2001a; Linka et al., 2008; Kunz et al., 2009). The peroxisomal transporter required for efflux of IBA-derived IAA from the peroxisome is unknown.
The ABCG36 and ABCG37 members of the pleiotropic drug resistance (PDR) subclade of the ABCG family of ATP-binding cassette transporters (Strader et al., 2008; Strader and Bartel, 2009; Ruzicka et al., 2010) are required for efflux of IBA from the root. Mutants defective in ABCG36 were found in a screen for altered IBA sensitivity (Strader and Bartel, 2009). Mutants defective in ABCG37 were identified in screens for altered sensitivity to 2,4-dichlorophenoxyacetic acid (2,4-D) (Ito and Gray, 2006), for altered sensitivity to polar auxin transport inhibitors (Ruzicka et al., 2010), or for altered sensitivity to IBA (Strader et al., 2008). Excised root tips from abcg36 and abcg37 loss-of-function mutants hyperaccumulate [3H]-IBA, but not [3H]-IAA, suggesting that the ABCG36 and ABCG37 transporters act to efflux IBA (Figure 1; Strader et al., 2008; Strader and Bartel, 2009; Ruzicka et al., 2010). The hyperaccumulation of [3H]-IBA, combined with the localization of ABCG36 and ABCG37 to the outer polar domain of root epidermal and lateral root cap cells (Stein et al., 2006; Strader and Bartel, 2009; Ruzicka et al., 2010), suggest that these transporters move IBA out of the root.
To determine whether ABCG37 directly transports IBA, it was expressed in Schizosaccharomyces pombe (see Table 1), in which ABCG37-expressing cells accumulated less [3H]-IBA compared to control cells, with no change in [3H]-IAA accumulation (Ruzicka et al., 2010), suggesting that ABCG37 directly transports IBA. Although ABCG36 has not been evaluated in a heterologous system for efflux activity, the IBA hyperaccumulation in the abcg36 mutant (Strader and Bartel, 2009) suggests that ABCG36 could be functionally similar to ABCG37 in IBA transport. Further, the abcg36 abcg37 double mutant displays additive IBA hypersensitivity and [3H]-IBA hyperaccumulation in comparison to either the abcg36 or abcg37 single mutants, suggesting overlapping functions (Ruzicka et al., 2010).
ABCG37 and ABCG36 appear to transport substrates in addition to IBA. PDR proteins typically have broad range of substrates (reviewed in van den Brule and Smart, 2002; Verrier et al., 2008). ABCG37 displays broad substrate specificity, including several auxinic compounds such as the synthetic auxin 2,4-D (Ito and Gray, 2006; Strader et al., 2008), 2,4-dichlorophenoxy butyric acid (2,4-DB) (Strader et al., 2008; Ruzicka et al., 2010), and the auxin transport inhibitor (NPA) (Ito and Gray, 2006). ABCG36 likely transports cadmium (Kim et al., 2007) and the indolic defense compound 4-methoxyindol-3-yl-methanol (Matern et al., 2019).
ABCG36 and ABCG37 play roles in distinct areas of plant development. In particular, the abcg36 mutant displays an increased number of lateral roots, larger cotyledons, and increased root hair lengths (Strader and Bartel, 2009), consistent with elevated auxin levels in these tissues. ABCG36 and ABCG37 localize to the outward face of epidermal cells in leaves (ABCG36) and roots (ABCG36 and 37) (Strader and Bartel, 2009; Langowski et al., 2010; Ruzicka et al., 2010), suggesting that these transporters exude IBA into the environment. This activity may be crucial for maintaining IBA levels and auxin homeostasis, as abcg36 and abcg37 mutant display phenotypes consistent with elevated auxin accumulation. Further, blocking IBA-to-IAA conversion in the abcg36 mutant suppresses these phenotypes (Strader et al., 2011), suggesting that IBA conversion is necessary for the developmental defects in abcg36. In roots, ABCG36 and ABCG37 are polarly localized to the outward-facing plasma membrane of root epidermal and lateral root cap cells (Strader and Bartel, 2009; Langowski et al., 2010; Ruzicka et al., 2010) and may exude IBA into the rhizosphere, which could have potential implications on the microbial diversity.
TRANSPORTER OF IBA1 (TOB1) was identified in a forward genetics screen for suppression of abcg36 IBA hypersensitivity (Michniewicz et al., 2019). TOB1 belongs to the NTR1 PTR FAMILY (NPF), a part of the larger group of major facilitator superfamily (MFS) transporters (Leran et al., 2014). The tob1 loss-of-function mutant displays mild resistance to 2,4-DB and IBA and wild-type sensitivity to IAA and 2,4-D in root elongation assays (Michniewicz et al., 2019). Indeed, transport assays using excised Arabidopsis root tips reveal reduced accumulation of [3H]-IBA and no difference in [3H]-IAA accumulation in tob1 compared to wild type root tips (Michniewicz et al., 2019). Further, TOB1 directly transports IBA in heterologous systems (see Table 1) such as yeast and Xenopus oocytes (Michniewicz et al., 2019). In addition, TOB1 transports nitrate (He et al., 2017; Michniewicz et al., 2019). In oocytes, nitrate serves as a better substrate than IBA, although IBA can compete with nitrate uptake in TOB1-expressing oocytes (Michniewicz et al., 2019).
TOB1 localizes to the vacuolar membrane and is primarily expressed in cells adjacent to the lateral root primordia and in the lateral root cap cells of the primary root (Michniewicz et al., 2019). The tob1 loss-of-function mutants display increased numbers of lateral roots and altered root system architecture (Michniewicz et al., 2019), suggesting that TOB1 normally limits production of lateral roots, possibly by sequestering IBA to the vacuole to limit its contributions to the pool of active auxin in these tissues. TOB1 is a direct target of the cytokinin reponse regulator ARR10 (Zubo et al., 2017) and TOB1 expression is induced by cytokinin treatment (Michniewicz et al., 2019). Further, the tob1 mutant is resistant to the inhibitory effects of cytokinin on lateral root production (Michniewicz et al., 2019), suggesting that cytokinin’s inhibitory effects on lateral root production are at least partially mediated by moving IBA into the vacuole. These data suggest a model in which cytokinin regulates TOB1 expression to regulate IBA contributions to auxin homeostasis during lateral root development.
Thus far, four proteins, PXA1 (Zolman et al., 2001b), ABCG36 (Strader and Bartel, 2009), ABCG37 (Strader et al., 2008; Ruzicka et al., 2010), and TOB1 (Michniewicz et al., 2019) have been implicated in IBA transport. Identification of these transporters has demonstrated the significance of IBA transporters in regulating aspects of development. Although progress has been made in identification of IBA transporters and IBA metabolic enzymes, much remains to be uncovered. Identifying additional transporters and determining their substrates (either IBA or IBA conjugates), combined with a detailed knowledge of IBA metabolism, will shine more light into understanding the plant developmental pathways.
Remaining questions on IBA transport:
•How and where is IBA made and metabolized? IBA may be derived from IAA in maize (Ludwig-Müller and Epstein, 1991; Ludwig-Müller et al., 1995a) and Arabidopsis (Ludwig-Muller, 2007). Yet, enzymes involved in IBA biosynthesis remain unidentified. IBA-derived auxin appears to be important in very specific aspects of plant development; it seems an inefficient system to create IBA from IAA, just to metabolize it to IAA again. Because IBA is important for early seedling growth, a stage in which peroxisomal activity is high in metabolizing storage oils, and because IBA β-oxidation releases not only free IAA but also acetyl-CoA, it may be possible that IBA is used in this scenario to not only provide auxin, but also energy, to drive growth. In addition, much of the transported IBA is in the form of IBA ester-linked conjugates (likely conjugated to sugars) (Liu et al., 2012). Whether IBA conjugates are made in one tissue; then transported to another for metabolism remains an open question. A greater understanding of tissue-type expression of IBA conversion enzymes and IBA transporters will be beneficial in understanding this system.
•Roles for additional ABCG or TOB proteins in IBA transport. Known IBA transporters belong to the PDR clade of the ATP-binding cassette (ABC) family and of the NPF clade of the major facilitator superfamily. Additional members of these clades may be involved in mediating IBA transport. In particular, mutants defective in ABCG29/PDR1 and ABCG33/PDR5 display hypersensitivity to the synthetic IBA mimic 2,4-DB (Michniewicz et al., 2014), consistent with the possibility that, similar to ABCG36 and ABCG37 family members, ABCG29 and ABCG33 transport IBA. In addition, TOB1 also has six closely related paralogs, which could function, similar to TOB1, in transporting IBA (Michniewicz et al., 2019). Functional characterization of these close paralogs could reveal potential IBA carriers. To determine whether these are bona fide IBA transporters, IBA transport activity must be demonstrated in a heterologous system (Table 1) such as yeast, Xenopus oocytes, insect cells, or mammalian cells (Haferkamp and Linka, 2012). There is a possibility that TOB1 paralogs (Michniewicz et al., 2019) could transport IBA in distinct tissues of Arabidopsis and promote the development of different tissues. Characterization of these members in addition to other potential transporters would aid in understanding the role of IBA transporters function at spatial levels to indirectly regulate IAA levels.
•IBA uptake carriers. IBA uptake is a rate-limited process (Rashotte et al., 2003), suggesting that its import into the cell is mediated by a carrier, rather than the effect of diffusion. Thus far, no IBA uptake carriers have been reported. A potential candidate for an uptake carrier is RESISTANT TO IBA1 (RIB1). The semi-dominant mutant rib1 displays IBA resistance (Poupart and Waddell, 2000) and altered IBA transport (Poupart et al., 2005), consistent with the possibility that RIB1 mediates IBA uptake. Identification of the underlying mutation and molecular characterization of the gene product will allow for understanding the role of RIB1 in IBA transport.
•Directionality of TOB1 transport. The reduced 3H-IBA accumulation in tob1 mutants combined with developmental phenotypes consistent with increased IBA contributions to the auxin pool suggests that TOB1 sequesters IBA in to vacuole (Michniewicz et al., 2019). Conversely, tob1 resistance to the inhibitory effects of exogenous IBA on root elongation and electrophysiology assays are consistent with the possibility that TOB1 moves IBA out of the vacuole (Michniewicz et al., 2019). These contradictory pieces of data may reflect concentration-dependent differences in TOB1 transport direction. Thus, the direction of IBA movement by TOB1 remains unclear (Figure 1).
•Identity of transported molecules. Although it is clear that PDR9 and TOB1 directly transport IBA based on heterologous transport assays, it is not clear whether IBA itself or IBA-derived molecules are being tracked in other assays. Liu et al. (2012) elegantly used heavy IBA and mass spec analysis to determine that, in Arabidopsis hypocotyl tissues, most of the transported material was IBA-derived IAA or IBA conjugates. Likewise, in Ruzicka et al. (2010), [3H]-IBA applied to the columella was IBA-derived [3H]-IAA when sampled 4 mm from the root tip. Clearly, analytical methods need to be combined with transport assays for both IBA and IAA. Indeed, transport assays in heterologous systems, such as Xenopus oocytes, can be done on a time scale for which metabolism of molecules is not a factor.
•IAA efflux carrier in peroxisome. IBA-to-IAA conversion occurs in the peroxisome (reviewed in Figure 1; Strader and Bartel, 2011). However, the transporter involved in movement of IBA-derived IAA from the peroxisome into the cytoplasm is unidentified.
•Long distance IBA efflux carriers. Although long-distance transport has been speculated to exist based on radiotracer experiments, it seems that much of this tracked material was IBA metabolites, rather than free IBA. Certainly, none of the identified IBA transporters are involved in long-distance IBA transport. At this point, transporters involved in the long-distance transport of IBA or IBA conjugates remain elusive. Further, developmental roles for transport of IBA or IBA remain unknown; thus, it is possible that this mechanism is not a contributor to physiologically relevant processes.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was supported by the National Science Foundation (IOS-1453750 to LS), the NSF Center for Engineering Mechanobiology (CMMI-1548571 to LS), and the National Institutes of Health (R01 GM112898 to LS).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Adham, A. R., Zolman, B. K., Millius, A., and Bartel, B. (2005). Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in beta-oxidation. Plant J. 41, 859–874. doi: 10.1111/j.1365-313X.2005.02343.x
Bartel, B., and Fink, G. R. (1995). ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268, 1745–1748. doi: 10.1126/science.7792599
Blommaert, K. L. J. (1954). Growth and inhibiting substances in relation to the rest period of the potato tuber. Nature 174, 970–972. doi: 10.1038/174970b0
Brumos, J., Robles, L. M., Yun, J., Vu, T. C., Jackson, S., Alonso, J. M., et al. (2018). Local auxin biosynthesis is a key regulator of plant development. Dev. Cell 47, 306–318.e305. doi: 10.1016/j.devcel.2018.09.022
Campanella, J. J., Bakllamaja, V., Restieri, T., Vomacka, M., Herron, J., Patterson, M., et al. (2003a). Isolation of an ILR1 auxin conjugate hydrolase homolog from Arabidopsis suecica. Plant Growth Regul. 39, 175–181. doi: 10.1023/A:1022528318188
Campanella, J. J., Larko, D., and Smalley, J. (2003b). A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp. Funct. Genomics 4, 584–600. doi: 10.1002/cfg.340
Campanella, J. J., Smith, S. M., Leibu, D., Wexler, S., and Ludwig-Müller, J. (2008). The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J. Plant Growth Regul. 27, 26–38. doi: 10.1007/s00344-007-9027-2
Carrier, D. J., Bakar, N. T. A., Swarup, R., Callaghan, R., Napier, R. M., Bennett, M. J., et al. (2008). The binding of auxin to the Arabidopsis auxin influx transporter AUX1. Plant Physiol. 148, 529–535. doi: 10.1104/pp.108.122044
Cheng, Y., Dai, X., and Zhao, Y. (2006). Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 20, 1790–1799. doi: 10.1101/gad.1415106
Cheng, Y., Dai, X., and Zhao, Y. (2007). Auxin synthesized by the YUCCA flavin monooxygenases is essential for embryogenesis and leaf formation in Arabidopsis. Plant Cell 19, 2430–2439. doi: 10.1105/tpc.107.053009
Cook, S. D. (2019). An historical review of phenylacetic acid. Plant Cell Physiol. 60, 243–254. doi: 10.1093/pcp/pcz004
De Rybel, B., Audenaert, D., Xuan, W., Overvoorde, P., Strader, L. C., Kepinski, S., et al. (2012). A role for the root cap in root branching revealed by the non-auxin probe naxillin. Nat. Chem. Biol. 8, 798–805. doi: 10.1038/nchembio.1044
Epstein, E., and Sagee, O. (1992). Effect of ethylene treatment on transport and metabolism of indole-3-butyric acid in citrus leaf midribs. Plant Growth Regul. 11, 357–362. doi: 10.1007/BF00130642
Frick, E. M., and Strader, L. C. (2018). Roles for IBA-derived auxin in plant development. J. Exp. Bot. 69, 169–177. doi: 10.1093/jxb/erx298
Haferkamp, I., and Linka, N. (2012). Functional expression and characterisation of membrane transport proteins. Plant Biol. 14, 675–690. doi: 10.1111/j.1438-8677.2012.00591.x
He, Y. N., Peng, J. S., Cai, Y., Liu, D. F., Guan, Y., Yi, H. Y., et al. (2017). Tonoplast-localized nitrate uptake transporters involved in vacuolar nitrate efflux and reallocation in Arabidopsis. Sci. Rep. 7:6417. doi: 10.1038/s41598-017-06744-5
Ito, H., and Gray, W. M. (2006). A gain-of-function mutation in the Arabidopsis pleiotropic drug resistance transporter PDR9 confers resistance to auxinic herbicides. Plant Physiol. 142, 63–74. doi: 10.1104/pp.106.084533
Jakubowska, A., and Kowalczyk, S. (2005). A specific enzyme hydrolyzing 6-O(4-O)-indole-3-ylacetyl-beta-D-glucose in immature kernels of Zea mays. J. Plant Physiol. 162, 207–213. doi: 10.1016/j.jplph.2004.05.015
Jakubowska, A., Kowalczyk, S., and Leźnicki, A. J. (1993). Enzymatic hydrolysis of 4-O and 6-O-Indol-3-Ylacetyl-β-D-Glucose in plant tissues. J. Plant Physiol. 142, 61–66. doi: 10.1016/S0176-1617(11)80108-2
Kim, D. Y., Bovet, L., Maeshima, M., Martinoia, E., and Lee, Y. (2007). The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 50, 207–218. doi: 10.1111/j.1365-313X.2007.03044.x
Korasick, D. A., Enders, T. A., and Strader, L. C. (2013). Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555. doi: 10.1093/jxb/ert080
Kunz, H.-H., Scharnewski, M., Feussner, K., Feussner, I., Flügge, U.-I., Fulda, M., et al. (2009). The ABC transporter PXA1 and peroxisomal β-oxidation are vital for metabolism in mature leaves of Arabidopsis during extended darkness. Plant Cell 21, 2733–2749. doi: 10.1105/tpc.108.064857
Kuromori, T., Miyaji, T., Yabuuchi, H., Shimizu, H., Sugimoto, E., Kamiya, A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 107, 2361–2366. doi: 10.1073/pnas.0912516107
Langowski, L., Ruzicka, K., Naramoto, S., Kleine-Vehn, J., and Friml, J. (2010). Trafficking to the outer polar domain defines the root-soil interface. Curr. Biol. 20, 904–908. doi: 10.1016/j.cub.2010.03.059
Lavy, M., and Estelle, M. (2016). Mechanisms of auxin signaling. Development 143, 3226–3229. doi: 10.1242/dev.131870
LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P., and Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 277, 20446–20452. doi: 10.1074/jbc.M111955200
Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N., Daniel-Vedele, F., et al. (2014). A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 19, 5–9. doi: 10.1016/j.tplants.2013.08.008
Linka, N., Theodoulou, F. L., Haslam, R. P., Linka, M., Napier, J. A., Neuhaus, H. E., et al. (2008). Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana. Plant Cell 20, 3241–3257. doi: 10.1105/tpc.108.062042
Liu, X., Barkawi, L., Gardner, G., and Cohen, J. D. (2012). Transport of indole-3-butyric acid and indole-3-acetic acid in Arabidopsis hypocotyls using stable isotope labeling. Plant Physiol. 158, 1988–2000. doi: 10.1104/pp.111.191288
Ljung, K. (2013). Auxin metabolism and homeostasis during plant development. Development 140, 943–950. doi: 10.1242/dev.086363
Ludwig-Muller, J. (2007). Indole-3-butyric acid synthesis in ecotypes and mutants of Arabidopsis thaliana under different growth conditions. J. Plant Physiol. 164, 47–59. doi: 10.1016/j.jplph.2005.10.008
Ludwig-Müller, J., and Epstein, E. (1991). Occurrence and in vivo biosynthesis of indole-3-butyric acid in corn (Zea mays L.). Plant Physiol. 97, 765–770. doi: 10.1104/pp.97.2.765
Ludwig-Müller, J., Hilgenberg, W., and Epstein, E. (1995a). The in vitro biosynthesis of indole-3-butyric acid in maize. Phytochemistry 40, 61–68. doi: 10.1016/0031-9422(95)00259-A
Ludwig-Müller, J., Raisig, A., and Hilgenberg, W. (1995). Uptake and transport of indole-3-butyric acid in Arabidopsis thaliana: comparison with other natural and synthetic Auxins. J. Plant Physiol. 147, 351–354. doi: 10.1016/S0176-1617(11)82166-8
Ludwig-Müller, J., Sass, S., Sutter, E. G., Wodner, M., and Epstein, E. (1993). Indole-3-butyric acid in Arabidopsis thaliana. Plant Growth Regul. 13, 179–187. doi: 10.1007/BF00024260
Matern, A., Böttcher, C., Eschen-Lippold, L., Westermann, B., Smolka, U., Döll, S., et al. (2019). A substrate of the ABC transporter PEN3 stimulates bacterial flagellin (flg22)-induced callose deposition in Arabidopsis thaliana. J. Biol. Chem. 294, 6857–6870. doi: 10.1074/jbc.RA119.007676
Michniewicz, M., Ho, C., Enders, T., Floro, E., Gunther, L. K., Damodoran, S., et al. (2019). Transporter of IBA1 links auxin and cytokinin to influence root architecture. Available at: https://ssrncom/abstract=3339905 (Accessed February 26, 2019).
Michniewicz, M., Powers, S. K., and Strader, L. C. (2014). “IBA transport by PDR proteins” in Plant ABC transporters. ed. M. Geisler (Cham: Springer International Publishing), 313–331.
Morffy, N. J., and Strader, L. C. (2018). Locally sourced: auxin biosynthesis and transport in the root meristem. Dev. Cell 47, 262–264. doi: 10.1016/j.devcel.2018.10.018
Novak, O., Henykova, E., Sairanen, I., Kowalczyk, M., Pospisil, T., and Ljung, K. (2012). Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523–536. doi: 10.1111/j.1365-313X.2012.05085.x
Peer, W. A., Blakeslee, J. J., Yang, H., and Murphy, A. S. (2011). Seven things we think we know about auxin transport. Mol. Plant 4, 487–504. doi: 10.1093/mp/ssr034
Petrášek, J., and Friml, J. (2009). Auxin transport routes in plant development. Development 136, 2675–2688. doi: 10.1242/dev.030353
Poupart, J., Rashotte, A. M., Muday, G. K., and Waddell, C. S. (2005). The rib1 mutant of Arabidopsis has alterations in indole-3-butyric acid transport, hypocotyl elongation, and root architecture. Plant Physiol. 139, 1460–1471. doi: 10.1104/pp.105.067967
Poupart, J., and Waddell, C. S. (2000). The rib1 mutant is resistant to indole-3-butyric acid, an endogenous auxin in Arabidopsis. Plant Physiol. 124, 1739–1751. doi: 10.1104/pp.124.4.1739
Rampey, R. A., LeClere, S., Kowalczyk, M., Ljung, K., Sandberg, G., and Bartel, B. (2004). A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988. doi: 10.1104/pp.104.039677
Rashotte, A. M., Poupart, J., Waddell, C. S., and Muday, G. K. (2003). Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 133, 761–772. doi: 10.1104/pp.103.022582
Ruzicka, K., Strader, L. C., Bailly, A., Yang, H., Blakeslee, J., Langowski, L., et al. (2010). Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proc. Natl. Acad. Sci. USA 107, 10749–10753. doi: 10.1073/pnas.1005878107
Sherp, A. M., Westfall, C. S., Alvarez, S., and Jez, J. M. (2018). Arabidopsis thaliana GH3.15 acyl acid amido synthetase has a highly specific substrate preference for the auxin precursor indole-3-butyric acid. J. Biol. Chem. 293, 4277–4288. doi: 10.1074/jbc.RA118.002006
Stein, M., Dittgen, J., Sánchez-Rodríguez, C., Hou, B.-H., Molina, A., Schulze-Lefert, P., et al. (2006). Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell 18, 731–746. doi: 10.1105/tpc.105.038372
Stepanova, A. N., Robertson-Hoyt, J., Yun, J., Benavente, L. M., Xie, D. Y., Dolezal, K., et al. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133, 177–191. doi: 10.1016/j.cell.2008.01.047
Strader, L. C., and Bartel, B. (2009). The Arabidopsis pleiotropic drug resistance8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21, 1992–2007. doi: 10.1105/tpc.109.065821
Strader, L. C., and Bartel, B. (2011). Transport and metabolism of the endogenous auxin precursor indole-3-butyric acid. Mol. Plant 4, 477–486. doi: 10.1093/mp/ssr006
Strader, L. C., Culler, A. H., Cohen, J. D., and Bartel, B. (2010). Conversion of endogenous indole-3-butyric acid to indole-3-acetic acid drives cell expansion in Arabidopsis seedlings. Plant Physiol. 153, 1577–1586. doi: 10.1104/pp.110.157461
Strader, L. C., Monroe-Augustus, M., Rogers, K. C., Lin, G. L., and Bartel, B. (2008). Arabidopsis IBA response5 suppressors separate responses to various hormones. Genetics 180, 2019–2031. doi: 10.1534/genetics.108.091512
Strader, L. C., Wheeler, D. L., Christensen, S. E., Berens, J. C., Cohen, J. D., Rampey, R. A., et al. (2011). Multiple facets of Arabidopsis seedling development require indole-3-butyric acid-derived auxin. Plant Cell 23, 984–999. doi: 10.1105/tpc.111.083071
Sutter, E. G., and Cohen, J. D. (1992). Measurement of indolebutyric acid in plant tissues by isotope dilution gas chromatography-mass spectrometry analysis. Plant Physiol. 99, 1719–1722. doi: 10.1104/pp.99.4.1719
Swarup, K., Benkova, E., Swarup, R., Casimiro, I., Peret, B., Yang, Y., et al. (2008). The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 10, 946–954. doi: 10.1038/ncb1754
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Tognetti, V. B., Van Aken, O., Morreel, K., Vandenbroucke, K., van de Cotte, B., De Clercq, I., et al. (2010). Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22, 2660–2679. doi: 10.1105/tpc.109.071316
Uzunova, V. V., Quareshy, M., Del Genio, C. I., and Napier, R. M. (2016). Tomographic docking suggests the mechanism of auxin receptor TIR1 selectivity. Open Biol. 6:160139. doi: 10.1098/rsob.160139
van den Brule, S., and Smart, C. C. (2002). The plant PDR family of ABC transporters. Planta 216, 95–106. doi: 10.1007/s00425-002-0889-z
Verrier, P. J., Bird, D., Burla, B., Dassa, E., Forestier, C., Geisler, M., et al. (2008). Plant ABC proteins–a unified nomenclature and updated inventory. Trends Plant Sci. 13, 151–159. doi: 10.1016/j.tplants.2008.02.001
Vieten, A., Sauer, M., Brewer, P. B., and Friml, J. (2007). Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 12, 160–168. doi: 10.1016/j.tplants.2007.03.006
Yang, H., and Murphy, A. S. (2009). Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 59, 179–191. doi: 10.1111/j.1365-313X.2009.03856.x
Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 61, 49–64. doi: 10.1146/annurev-arplant-042809-112308
Zhao, Y. (2012). Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol. Plant 5, 334–338. doi: 10.1093/mp/ssr104
Zhao, Y., Christensen, S. K., Fankhauser, C., Cashman, J. R., Cohen, J. D., Weigel, D., et al. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291, 306–309. doi: 10.1126/science.291.5502.306
Zimmerman, P. W., and Wilcoxon, F. (1935). Several chemical growth substances which cause initiation of roots and other responses in plant. Contrib. Boyce Thompson Inst. 7, 209–229.
Zolman, B. K., Martinez, N., Millius, A., Adham, A. R., and Bartel, B. (2008). Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180, 237–251. doi: 10.1534/genetics.108.090399
Zolman, B. K., Monroe-Augustus, M., Thompson, B., Hawes, J. W., Krukenberg, K. A., Matsuda, S. P., et al. (2001a). chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. J. Biol. Chem. 276, 31037–31046. doi: 10.1074/jbc.M104679200
Zolman, B. K., Nyberg, M., and Bartel, B. (2007). IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol. Biol. 64, 59–72. doi: 10.1007/s11103-007-9134-2
Zolman, B. K., Silva, I. D., and Bartel, B. (2001b). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid beta-oxidation. Plant Physiol. 127, 1266–1278. doi: 10.1104/pp.010550
Zolman, B. K., Yoder, A., and Bartel, B. (2000). Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156, 1323–1337.
Zourelidou, M., Absmanner, B., Weller, B., Barbosa, I. C. R., Willige, B. C., Fastner, A., et al. (2014). Auxin efflux by PIN-FORMED proteins is activated by two different protein kinases D6 PROTEIN KINASE and PINOID. elife 3:e02860. doi: 10.7554/eLife.02860
Zubo, Y. O., Blakley, I. C., Yamburenko, M. V., Worthen, J. M., Street, I. H., Franco-Zorrilla, J. M., et al. (2017). Cytokinin induces genome-wide binding of the type-B response regulator ARR10 to regulate growth and development in Arabidopsis. Proc. Natl. Acad. Sci. USA 114, E5995–E6004. doi: 10.1073/pnas.1620749114
Keywords: auxin, indole-3-butyric acid, phytohormone, ATP-binding cassette transporter, TRANSPORTER OF IBA1, transporters
Citation: Damodaran S and Strader LC (2019) Indole 3-Butyric Acid Metabolism and Transport in Arabidopsis thaliana. Front. Plant Sci. 10:851. doi: 10.3389/fpls.2019.00851
Received: 31 March 2019; Accepted: 13 June 2019;
Published: 03 July 2019.
Edited by:
Markus Geisler, Université de Fribourg, SwitzerlandReviewed by:
Jutta Ludwig-Müller, Dresden University of Technology, GermanyCopyright © 2019 Damodaran and Strader. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lucia C.Strader, c3RyYWRlckB3dXN0bC5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.