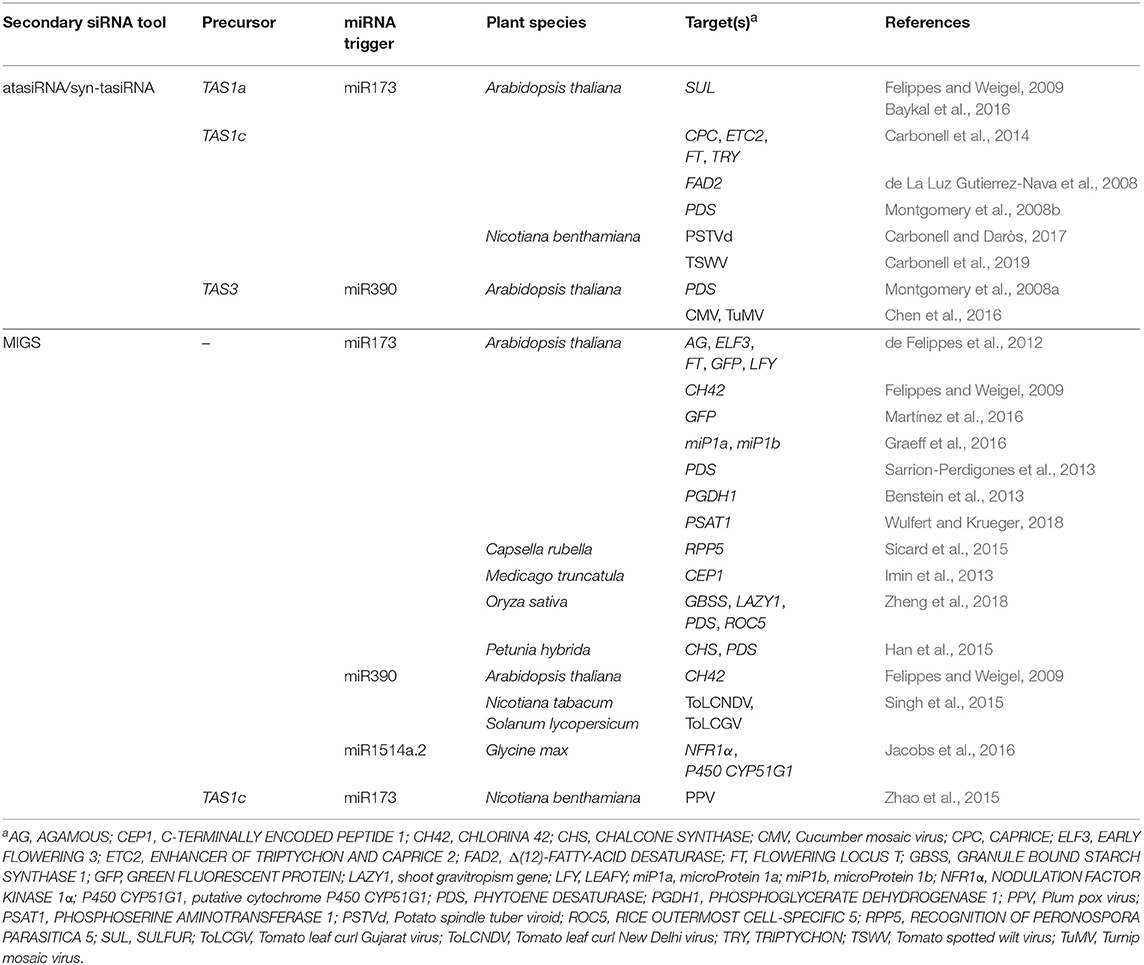
- Instituto de Biología Molecular y Celular de Plantas, Consejo Superior de Investigaciones Científicas-Universitat Politècnica de València, Valencia, Spain
Plant Secondary Small Interfering RNAs
In plants, RNA silencing regulates key biological processes such as development, response to stress, genome integrity, and antiviral resistance. RNA silencing functions through diverse classes of small RNAs (sRNAs) that associate with ARGONAUTE (AGO) proteins to repress highly sequence-complementary target transcripts (Baulcombe, 2004).
Small interfering RNAs (siRNAs) are a class of sRNAs arising from double-stranded RNA (dsRNA) precursors. Secondary siRNAs are those siRNAs whose dsRNA precursor synthesis is triggered by an upstream sRNA-guided transcript cleavage event followed by RNA-dependent RNA polymerase (RDR) activity (for a recent review see de Felippes, 2019). Many secondary siRNAs are produced in 21-nucleotide (nt) register with the sRNA-guided cleavage site by successive Dicer-Like (DCL) processing and are therefore called phased secondary siRNAs (phasiRNAs). In contrast, only a subset of secondary siRNAs act in trans to repress one or more targets distinct from their locus of origin. These siRNAs are called trans-acting siRNAs (tasiRNAs), most of which are also phased (Axtell, 2013).
Classes of Secondary siRNA-based Silencing Tools
While secondary siRNAs may be ultimately generated in classic RNA interference (RNAi) approaches such as virus induced gene silencing (VIGS) or hairpin-based silencing after the initial targeting of transgene-derived (primary) siRNAs (Ossowski et al., 2008), only two classes of silencing tools operate primarily through the action of secondary siRNAs. These are (i) artificial or synthetic tasiRNAs (atasiRNAs and syn-tasiRNAs, respectively, both terms are accepted), and (ii) miRNA-induced gene silencing (MIGS). Both classes of secondary siRNA-based tools have been extensively used in plants to induce selective gene silencing in basic gene function studies or to improve agronomic traits.
atasiRNA/syn-tasiRNAs are expressed from transgenes including engineered TAS precursors in which a region corresponding to various endogenous tasiRNAs is substituted by a fragment containing multiple atasiRNA/syn-tasiRNA sequences (Figure 1A). In Arabidopsis thaliana (Arabidopsis), modified TAS transcripts are cleaved by a specific microRNA (miRNA)/AGO complex (e.g., miR173/AGO1 and miR390/AGO7 cleave TAS1- and TAS3-based precursors, respectively), and one of the cleavage products is converted by RDR6 to dsRNA, which is processed by DCL4 into phased tasiRNA duplexes in 21-nt register with the miRNA cleavage site. atasiRNA/syn-tasiRNA guide strands, typically designed to contain an AGO1-preferred 5' U, are incorporated into AGO1 to direct silencing of one or multiple transcripts at one or multiple sites (Figure 1A). Importantly, the multiplexing of several atasiRNAs/syn-tasiRNAs in a single construct allows for the efficient and simultaneous multitargeting of various sequence-related or unrelated genes. Moreover, as for artificial miRNAs (amiRNAs), atasiRNAs/syn-tasiRNAs can be computationally designed with user-friendly web tools such as P-SAMS (http://p-sams.carringtonlab.org/) (Fahlgren et al., 2016) to be highly specific and prevent the so-called off-target effects characteristic of other RNAi approaches. For instance, P-SAMS designs artificial sRNAs that contain (i) an AGO1-preferred 5' U, (ii) a C in position 19 to generate a star strand with an AGO1 non-preferred 5' G thus avoiding competition for AGO1 loading, and (iii) an intentional mismatch with the target transcript at position 21 to limit possible transitivity effects (Carbonell, 2017). Initially, atasiRNAs/syn-tasiRNAs were used to efficiently repress one or multiple endogenous genes in gene function studies in Arabidopsis (de La Luz Gutierrez-Nava et al., 2008; Montgomery et al., 2008a,b; Carbonell et al., 2014) (Table 1). More recently, atasiRNAs/syn-tasiRNAs have emerged as an effective approach to induce resistance against viruses and viroids in several plant species (Chen et al., 2016; Carbonell and Daròs, 2017; Carbonell et al., 2019) (Table 1), and, more broadly, as a promising tool for plant biology study and crop improvement (for a review see Zhang, 2014).
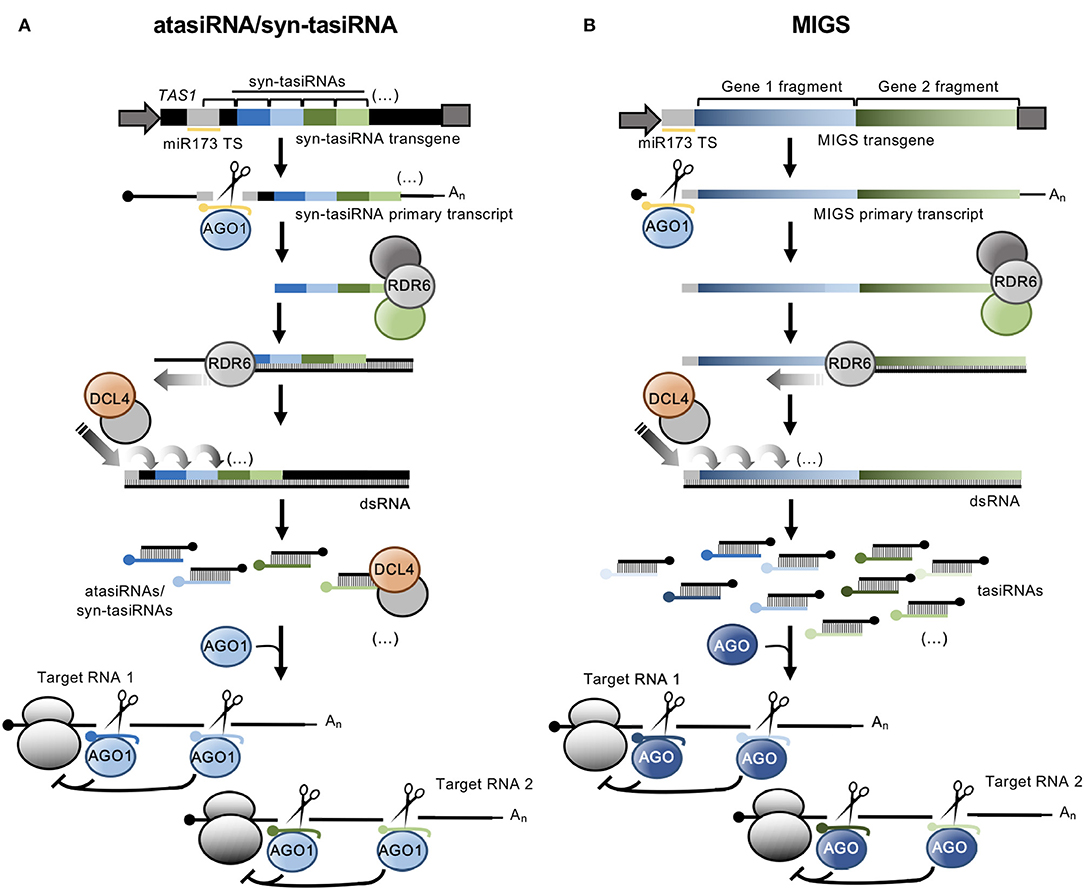
Figure 1. Secondary siRNA-based silencing tools in plants. (A). The atasiRNA/syn-tasiRNA pathway. TAS1 precursor sequence is in black, with trigger miR173 target site (TS) in light gray. AtasiRNA/syn-tasiRNA sequences targeting RNA 1 are shown in dark and light blue; atasiRNA/syn-tasiRNA sequences targeting RNA 2 are shown in dark and light green; miR173 sequence is shown in yellow. Promoter and terminator sequences are shown with a dark gray arrow and box, respectively. Participating proteins are represented with colored ovals. (B) The MIGS pathway. Sequences corresponding to gene fragments 1 and 2 are in diverse blue and green tonalities, respectively. Other details are as in (A).
MIGS was named (de Felippes et al., 2012) a few years later than was first reported (Montgomery et al., 2008b; Felippes and Weigel, 2009). Initial MIGS transgenes included one or more fragments from one or more target genes fused downstream of miR173 target site (Figure 1B) (de Felippes et al., 2012). miR173, as other 22 nt miRNAs, possesses the ability of triggering the production of phasiRNAs from target transcripts (Chen et al., 2010; Cuperus et al., 2010). In MIGS, miR173/AGO1-guided cleavage of the MIGS primary transcript triggers RDR6-dependent synthesis of dsRNA which is subsequently processed by DCL4 to release phased tasiRNAs that lead to the efficient silencing of target genes (Figure 1B). Interestingly, MIGS can also be triggered by other 22-nt miRNAs such as miR1514a.2 (Jacobs et al., 2016), or by miR390 (Felippes and Weigel, 2009; Singh et al., 2015), a 21-nt miRNA with unique properties for triggering tasiRNA formation from TAS3 transcripts (Montgomery et al., 2008a) (Table 1). Because miR173 is present only in Arabidopsis and closely-related species, miR173 co-expression with MIGS transgenes is necessary to trigger tasiRNA biogenesis in non-Arabidopsis species as reported in Medicago truncatula (Imin et al., 2013), Petunia (Han et al., 2015), soybean (Jacobs et al., 2016), and rice (Zheng et al., 2018). Despite having been widely used in gene function studies and also to confer antiviral resistance (Table 1), the MIGS approach presents a significant risk of off-target effects due to (i) the large number of tasiRNAs generated from the MIGS construct (similarly to those from classic RNAi constructs), (ii) the generation of out-of-phase siRNAs from MIGS constructs that can accumulate to high levels as observed in Petunia (Han et al., 2015), and (iii) the possibility that MIGS-derived tasiRNAs induce transitivity as reported (Han et al., 2015). Finally, loading of MIGS-derived tasiRNA into particular AGOs cannot be controlled, and for instance only a subset of these tasiRNAs, typically those with a 5' U, will be loaded into AGO1 while others may be loaded into different AGOs or degraded.
Appropriate Terminology to Refer to Secondary siRNA-based Silencing Tools
After having reviewed the literature, it seems necessary to make some brief remarks to improve the proper and consistent use of the terminology related to these secondary siRNA-based tools. For example, the production of tasiRNAs from a transgene including a gene fragment fused downstream to a miRNA trigger target site should always be referred to as MIGS, and not to as atasiRNA/syn-tasiRNA as observed in several works (Singh et al., 2015; Zhao et al., 2015; Guo et al., 2016), even if the MIGS cassette is inserted into a TAS precursor (Zhao et al., 2015; Guo et al., 2016). Also, secondary siRNAs generated from MIGS transgenes should be referred as phasiRNAs or, even better, as tasiRNAs as they are expected to act in trans to target the desired target gene(s), but not as atasiRNAs or syn-tasiRNAs as reported (Singh et al., 2015; Zhao et al., 2015). I suggest to use the term “artificial” or “synthetic” when referring to those transgene-derived tasiRNAs that are incorporated into precursors (TAS or others) as individual 21-nt guide sequences that may have been designed computationally to be highly specific in silencing the corresponding target transcript(s), to be preferentially and selectively loaded by AGO1 or to avoid transitivity effects.
Conclusions and Future Perspectives
Still in the genome editing era dominated by CRISPR/CAS9-based technologies, we anticipate that secondary sRNA-based silencing tools will continue to be broadly used because of their unique features in allowing (i) highly specific silencing (e.g., atasiRNAs/syn-tasiRNAs), (ii) the study of genes whose complete knock-out induces lethality, (iii) multitargeting, as well as the targeting of duplicated genes, antisense transcripts and individual isoforms, and (iv) the spatio-temporal control of silencing when transgene expression is regulated with tissue specific or inducible promoters. Moreover, as gene knock-down tools, it might be possible to develop secondary siRNA-based strategies for the fine-tuning regulation of secondary siRNA activity to induce the desired degree of target gene silencing.
The atasiRNA/syn-tasiRNA approach seems especially attractive due to its multiplexing capability and high specificity, as well as for the availability of high-throughput cloning strategies and automated design tools for the simple generation of atasiRNA/syn-tasiRNA constructs (Carbonell et al., 2014; Carbonell, 2019). In particular, antiviral atasiRNAs/syn-tasiRNAs designed to target multiple sites in viral RNAs should induce a more effective and durable resistance compared to single target site targeting approaches such as amiRNAs, as the possibility that the virus mutates all target sites to break the resistance seems highly improbable. Still, a deeper knowledge of the basic mechanisms governing secondary siRNA biogenesis, mode of action, and targeting efficacy is needed to further refine these secondary siRNA-based silencing tools in view of accelerating studies of gene function and crop improvement.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
This work was supported by grants RYC-2017-21648 and RTI2018-095118-A-100 from the Ministerio de Ciencia, Innovación y Universidades (MCIU, Spain), Agencia Estatal de Investigación (AEI, Spain), and Fondo Europeo de Desarrollo Regional (FEDER, European Union) to AC.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
AC acknowledge support of the publication fee by the CSIC Open Access Publication Support Initiative through its Unit of Information Resources for Research (URICI).
References
Axtell, M. J. (2013). Classification and comparison of small RNAs from plants. Annu. Rev. Plant Biol. 64, 137–159. doi: 10.1146/annurev-arplant-050312-120043
Baykal, U., Liu, H., Chen, X., Nguyen, H. T., and Zhang, Z. J. (2016). Novel constructs for efficient cloning of sRNA-encoding DNA and uniform silencing of plant genes employing artificial trans-acting small interfering RNA. Plant Cell Rep. 35, 2137–2150. doi: 10.1007/s00299-016-2024-9
Benstein, R. M., Ludewig, K., Wulfert, S., Wittek, S., Gigolashvili, T., Frerigmann, H., et al. (2013). Arabidopsis phosphoglycerate dehydrogenase1 of the phosphoserine pathway is essential for development and required for ammonium assimilation and tryptophan biosynthesis. Plant Cell 25, 5011–5029. doi: 10.1105/tpc.113.118992
Carbonell, A. (2017). “Artificial small RNA-based strategies for effective and specific gene silencing in plants,” in Plant Gene Silencing: Mechanisms and Applications, ed T. Dalmay (CABI Publishing), 110–127. doi: 10.1079/9781780647678.0110
Carbonell, A. (2019). Design and high-throughput generation of artificial small RNA constructs for plants. Methods Mol. Biol. 1932, 247–260. doi: 10.1007/978-1-4939-9042-9_19
Carbonell, A., and Daròs, J. A. (2017). Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol. Plant Pathol. 18, 746–753. doi: 10.1111/mpp.12529
Carbonell, A., López, C., and Daròs, J. A. (2019). Fast-forward identification of highly effective artificial small RNAs against different tomato spotted wilt virus isolates. Mol. Plant Microbe Interact. 32, 142–156. doi: 10.1094/MPMI-05-18-0117-TA
Carbonell, A., Takeda, A., Fahlgren, N., Johnson, S. C., Cuperus, J. T., and Carrington, J. C. (2014). New generation of artificial MicroRNA and synthetic trans-acting small interfering RNA vectors for efficient gene silencing in Arabidopsis. Plant Physiol. 165, 15–29. doi: 10.1104/pp.113.234989
Chen, H. M., Chen, L. T., Patel, K., Li, Y. H., Baulcombe, D. C., and Wu, S. H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 107, 15269–15274. doi: 10.1073/pnas.1001738107
Chen, L., Cheng, X., Cai, J., Zhan, L., Wu, X., Liu, Q., et al. (2016). Multiple virus resistance using artificial trans-acting siRNAs. J. Virol. Methods 228, 16–20. doi: 10.1016/j.jviromet.2015.11.004
Cuperus, J. T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R. T., Takeda, A., et al. (2010). Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003. doi: 10.1038/nsmb.1866
de Felippes, F. F. (2019). Gene regulation mediated by microRNA-triggered secondary small RNAs in plants. Plants 8:112. doi: 10.3390/plants8050112
de Felippes, F. F., Wang, J. W., and Weigel, D. (2012). MIGS: miRNA-induced gene silencing. Plant J. 70, 541–547. doi: 10.1111/j.1365-313X.2011.04896.x
de La Luz Gutiérrez-Nava, M., Aukerman, M. J., Sakai, H., Tingey, S. V., and Williams, R. W. (2008). Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol. 147, 543–551. doi: 10.1104/pp.108.118307
Fahlgren, N., Hill, S. T., Carrington, J. C., and Carbonell, A. (2016). P-SAMS: a web site for plant artificial microRNA and synthetic trans-acting small interfering RNA design. Bioinformatics 32, 157–158. doi: 10.1093/bioinformatics/btv534
Felippes, F. F., and Weigel, D. (2009). Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 10, 264–270. doi: 10.1038/embor.2008.247
Graeff, M., Straub, D., Eguen, T., Dolde, U., Rodrigues, V., Brandt, R., et al. (2016). MicroProtein-mediated recruitment of CONSTANS into a TOPLESS trimeric complex represses flowering in Arabidopsis. PLoS Genet. 12:e1005959. doi: 10.1371/journal.pgen.1005959
Guo, Q., Liu, Q., Smith, N. A., Liang, G., and Wang, M. B. (2016). RNA silencing in plants: mechanisms, technologies and applications in horticultural crops. Curr. Genomics 17, 476–489. doi: 10.2174/1389202917666160520103117
Han, Y., Zhang, B., Qin, X., Li, M., and Guo, Y. (2015). Investigation of a miRNA-induced gene silencing technique in petunia reveals alterations in miR173 precursor processing and the accumulation of secondary siRNAs from endogenous genes. PLoS ONE 10:e0144909. doi: 10.1371/journal.pone.0144909
Imin, N., Mohd-Radzman, N. A., Ogilvie, H. A., and Djordjevic, M. A. (2013). The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 64, 5395–5409. doi: 10.1093/jxb/ert369
Jacobs, T. B., Lawler, N. J., Lafayette, P. R., Vodkin, L. O., and Parrott, W. A. (2016). Simple gene silencing using the trans-acting siRNA pathway. Plant Biotechnol. J. 14, 117–127. doi: 10.1111/pbi.12362
Martínez, G., Panda, K., Köhler, C., and Slotkin, R. K. (2016). Silencing in sperm cells is directed by RNA movement from the surrounding nurse cell. Nat Plants 2:16030. doi: 10.1038/nplants.2016.30
Montgomery, T. A., Howell, M. D., Cuperus, J. T., Li, D., Hansen, J. E., Alexander, A. L., et al. (2008a). Specificity of ARGONAUTE7-miR390 interaction anddual functionality in TAS3 trans-acting siRNA formation. Cell 133, 128–141. doi: 10.1016/j.cell.2008.02.033
Montgomery, T. A., Yoo, S. J., Fahlgren, N., Gilbert, S. D., Howell, M. D., Sullivan, C. M., et al. (2008b). AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl. Acad. Sci. U.S.A. 105, 20055–20062. doi: 10.1073/pnas.0810241105
Ossowski, S., Schwab, R., and Weigel, D. (2008). Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 53, 674–690. doi: 10.1111/j.1365-313X.2007.03328.x
Sarrion-Perdigones, A., Vazquez-Vilar, M., Palací, J., Castelijns, B., Forment, J., Ziarsolo, P., et al. (2013). GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 162, 1618–1631. doi: 10.1104/pp.113.217661
Sicard, A., Kappel, C., Josephs, E. B., Lee, Y. W., Marona, C., Stinchcombe, J. R., et al. (2015). Divergent sorting of a balanced ancestral polymorphism underlies the establishment of gene-flow barriers in Capsella. Nat. Commun. 6:7960. doi: 10.1038/ncomms8960
Singh, A., Taneja, J., Dasgupta, I., and Mukherjee, S. K. (2015). Development of plants resistant to tomato geminiviruses using artificial trans-acting small interfering RNA. Mol. Plant Pathol. 16, 724–734. doi: 10.1111/mpp.12229
Wulfert, S., and Krueger, S. (2018). Phosphoserine aminotransferase1 is part of the phosphorylated pathways for serine biosynthesis and essential for light and sugar-dependent growth promotion. Front. Plant Sci. 9:1712. doi: 10.3389/fpls.2018.01712
Zhang, Z. J. (2014). Artificial trans-acting small interfering RNA: a tool for plant biology study and crop improvements. Planta 239, 1139–1146. doi: 10.1007/s00425-014-2054-x
Zhao, M., San León, D., Mesel, F., García, J. A., and Simón-Mateo, C. (2015). Assorted processing of synthetic trans-acting siRNAs and its activity in antiviral resistance. PLoS ONE 10:e0132281. doi: 10.1371/journal.pone.0132281
Keywords: RNA silencing, secondary siRNA, tasiRNA, atasiRNA, syn-tasiRNA, MIGS
Citation: Carbonell A (2019) Secondary Small Interfering RNA-Based Silencing Tools in Plants: An Update. Front. Plant Sci. 10:687. doi: 10.3389/fpls.2019.00687
Received: 28 March 2019; Accepted: 07 May 2019;
Published: 28 May 2019.
Edited by:
Andrea Chini, Centro Nacional de Biotecnología (CNB), SpainReviewed by:
German Martinez, Swedish University of Agricultural Sciences, SwedenBlake Meyers, Donald Danforth Plant Science Center, United States
Copyright © 2019 Carbonell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alberto Carbonell, YWNhcmJvbmVsbEBpYm1jcC51cHYuZXM=
 Alberto Carbonell
Alberto Carbonell