
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 16 May 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00630
The abscisic acid (ABA)-responsive element binding factors (ABFs) play important regulatory roles in multiple abiotic stresses responses. However, information on the stress tolerance functions of ABF genes in sweetpotato (Ipomoea batatas [L.] Lam) remains limited. In the present study, we isolated and functionally characterized the sweetpotato IbABF4 gene, which encodes an abiotic stress-inducible basic leucine zipper (bZIP) transcription factor. Sequence analysis showed that the IbABF4 protein contains a typical bZIP domain and five conserved Ser/Thr kinase phosphorylation sites (RXXS/T). The IbABF4 gene was constitutively expressed in leaf, petiole, stem, and root, with the highest expression in storage root body. Expression of IbABF4 was induced by ABA and several environmental stresses including drought, salt, and heat shock. The IbABF4 protein localized to the nucleus, exhibited transcriptional activation activity, and showed binding to the cis-acting ABA-responsive element (ABRE) in vitro. Overexpression of IbABF4 in Arabidopsis thaliana not only increased ABA sensitivity but also enhanced drought and salt stress tolerance. Furthermore, transgenic sweetpotato plants (hereafter referred to as SA plants) overexpressing IbABF4, generated in this study, exhibited increased tolerance to drought, salt, and oxidative stresses on the whole plant level. This phenotype was associated with higher photosynthetic efficiency and lower malondialdehyde and hydrogen peroxide content. Levels of endogenous ABA content and ABA/stress-responsive gene expression were significantly upregulated in transgenic Arabidopsis and sweetpotato plants compared with wild-type plants under drought stress. Our results suggest that the expression of IbABF4 in Arabidopsis and sweetpotato enhances tolerance to multiple abiotic stresses through the ABA signaling pathway.
Abiotic stresses such as drought, salinity, and high temperature severely affect plant growth, development, and productivity (Moore et al., 2009). Understanding the mechanisms of abiotic stress tolerance in plants is a crucial topic of environmental research (Bartels and Sunkar, 2005). In response to adverse environment conditions, plants have evolved a number of defense mechanisms involving transcription factors (TFs) that bind to conserved cis-acting elements in target gene promoters, thus activating their expression and leading to enhanced stress tolerance (Fujita et al., 2005). Numerous stress-responsive TFs, such as those belonging to the basic leucine zipper (bZIP), WRKY, MYB, basic helix-loop-helix (bHLH), and NAC families, have been well-characterized through genetic, molecular, and biochemical analyses (Wang et al., 2003; Golldack et al., 2011). Transgenic plants overexpressing these TFs show improved tolerance to different environmental stresses (Hossain M. et al., 2010).
The bZIP proteins, containing a basic DNA-binding region and a leucine zipper dimerization motif, comprise one of the largest and diverse TF families (Latchman, 1993). Recent studies show that bZIP TFs are involved in diverse biological processes such as abiotic and biotic stress responses, seed germination, flower and seed development, and hormone and sugar signaling (Thurow et al., 2005; Muszynski et al., 2006; Lindemose et al., 2013). In Arabidopsis thaliana, the bZIP TFs are classified into 10 groups, based on sequence similarity of the basic region and additional conserved motifs (Corrêa et al., 2008). The abscisic acid (ABA)-responsive element (ABRE)-binding factors (ABFs), also known as ABA-responsive element binding (AREB) proteins, belong to Group A of the bZIP protein family. The consensus core sequence (C/T)ACGTGGC has been identified as a major cis-acting regulatory element (Hirayama and Shinozaki, 2010; Yoshida et al., 2010). The ABFs regulate the expression of ABA and other stress-responsive genes by binding to the ABREs in their promoter regions (Busk and Pagès, 1998; Hyungin et al., 2000). Five conserved Ser/Thr kinase phosphorylation sites (RXXS/T) are characteristic of the abiotic stress-responsive AtABFs/AREB proteins. Amino-acid residue (S26, S86, S94, T135) of four RXXS/T phosphorylation target sites in the N-terminal three conserved regions can be phosphorylated by SnRK2-type protein kinases and further activate AREB1 protein (Furihata et al., 2006; Fujii et al., 2009). Furihata et al. (2006) also confirmed that additional kinases exist for the phosphorylation of AREB1b in response to salt and/or osmotic stresses. The ABF genes have been extensively investigated in a number of plant species, such as Arabidopsis, rice (Oryza sativa), wheat (Triticum aestivum), barley (Hordeum vulgare), potato (Solanum tuberosum), and tomato (Solanum lycopersicum) (Hyungin et al., 2000; Casaretto and Ho, 2005; Kobayashi et al., 2008; Hossain M.A. et al., 2010; Tsaihung et al., 2010; García et al., 2012). Most ABF genes such as ABF2/AREB1, ABF4/AREB2, and ABF3 are highly induced by ABA, drought, and salt treatments (Uno et al., 2000; Furihata et al., 2006; Yoshida et al., 2010). Furthermore, overexpression of ABFs increases abiotic stress tolerance in several plant species (Hossain M.A. et al., 2010; Roychoudhury et al., 2013). Transgenic Arabidopsis plants overexpressing ABF3 and ABF4 exhibit improved drought tolerance via the up-regulation of several ABA and other stress-responsive genes (Kang et al., 2002). Overexpressing AtABF3 in alfalfa (Medicago sativa) reduces the transpiration rate and accumulation of reactive oxygen species, and increases tolerance to drought, salt, and oxidative stresses (Wang Z. et al., 2016).
Sweetpotato (Ipomoea batatas [L.] Lam) is the seventh most important food crop on the basis of the annual starch production worldwide (Pradhan et al., 2015), and has the potential to be commercially utilized as a health food as it is rich in antioxidants, dietary fiber, and minerals. Furthermore, sweetpotato is widely used as a source of starch and bioethanol (Madzlan et al., 2012; Duvernay et al., 2013). However, pests, viral diseases, and various environmental stresses, such as drought and extreme temperature, generally limit the production of sweetpotato in many areas worldwide (Lebot, 2010). Therefore, understanding the mechanisms of adaptation to adverse environmental conditions in sweetpotato plants is key for the development of stress tolerant cultivars.
In this study, we isolated the sweetpotato IbABF4 gene, which encodes an abiotic stress-inducible bZIP TF, and characterized its expression in different tissues and in response to ABA and several environmental stresses. The transactivation and ABRE-binding ability of IbABF4 was tested using a yeast one-hybrid assay and electrophoretic mobility shift assays (EMSAs). We also generated transgenic Arabidopsis and sweetpotato plants overexpressing IbABF4. The tolerance of these transgenic plants to drought, salt, and oxidative stresses was investigated at the seed and whole plant levels. Our results indicate that IbABF4 is a positive transcriptional regulator of the abiotic stress response, and overexpression of IbABF4 significantly increases the tolerance of transgenic plants to multiple abiotic stresses.
Sweetpotato cultivar Xushu 18 and Arabidopsis thaliana ecotype Colombia-0 (Col-0) were used in this study. Col-0 plants, used as the wild-type (WT), were grown in a growth chamber at 22 ± 1°C under 16 h light/8 h dark photoperiod. Sweetpotato plants were propagated by cuttings, which were grown at 25 ± 1°C under a 16 h light/8 h dark photoperiod. To analyze gene expression, 3-week-old sweetpotato plants were cultured in half-strength Hoagland nutrient solution supplemented with 10 μM ABA, 25% polyethylene glycol (PEG) 8000 (drought stress), 350 mM NaCl (salt stress), respectively, and subjected to 47°C (heat stress) treatments. The third leaf from the top of the plants was sampled at 0, 3, 6, 12, 24, and 48 h for further quantitative real-time PCR analysis. Plant tissues (leaves, petioles, stems, fibrous roots, pencil roots, proximal end of storage roots, storage root bodes, and distal end of storage roots) were separated from the 10-week-old sweetpotato plants under no environmental stress for tissue-specific gene expression analysis.
Total RNA of sweetpotato cultivar Xushu18 was used for cDNA synthesis. Full-length cDNA of the IbABF4 gene was isolated by reverse transcription PCR (RT-PCR) using sequence-specific primers (Supplementary Table S1) and cloned into the pGWB5 vector using the gateway method (Curtis and Grossniklaus, 2003). In this vector, the IbABF4 gene was fused to the green fluorescent protein (GFP) gene under the control of the cauliflower mosaic virus (CaMV) 35S promoter. The resulting 35S::IbABF4-GFP plasmid was used for subcellular localization analysis and Agrobacterium-mediated transformation.
To determine the transcriptional activation activity of IbABF4, full-length or partial (N- or C-terminal) coding sequences (Figure 3B) of IbABF4 cDNA were cloned in the pDEST32 vector containing the GAL4 DNA-binding domain (pBD) using the gateway method. This generated three in-frame protein fusion constructs, pBD::IbABF4, pBD::IbABF4ΔN, and pBD::IbABF4ΔC.
To conduct EMSAs, full-length IbABF4 cDNA was cloned into the pDEST15 vector containing the glutathione S-transferase (GST) tag using the gateway method, thus generating the GST::IbABF4 plasmid.
Total RNA and cDNA templates were obtained as described previously (Jin et al., 2017). All qRT-PCR analyses were performed in triplicate on a CFX Connect Real-Time PCR Detection system (Bio-Rad, Berkeley, CA, United States) using gene-specific primers (Supplementary Table S1) and Ever-Green PCR master mix kit (BioFact, Daejeon, South Korea), according to the manufacturer’s instructions. Relative expression levels of genes were calculated using the 2–ΔΔCT method.
The fusion construct 35S::IbABF4-GFP was transformed into Agrobacterium tumefaciens strain GV3101, which was then infiltrated into tobacco (Nicotiana benthamiana) leaves to obtain transient expression of IbABF4. After 3 days of culture, the infiltrated parts of leaves were cut and immersed in 4′,6-diamidino-2-phenylindole (DAPI) solution for nuclear staining and subjected to fluorescent signal detection under a confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany) with proper filter sets (Ke et al., 2017).
Three fusion constructs pBD::IbABF4, pBD::IbABF4ΔN, and pBD::IbABF4ΔC, as well as pBD (pDEST32 empty vector; negative control) and pGAL4 (positive control) vectors were transformed individually into the yeast strain PJ69-4A containing His3 and LacZ reporter genes. The transformed yeast cells were cultured on synthetic defined (SD) plates containing media lacking either only leucine (SD/Leu-) or Leu and histidine (SD/Leu-/His-). The transactivation activity of each protein was evaluated by the filter lift assay and quantitative assays of β-galactosidase activity using O-nitrophenyl-β-d-galactopyranoside (ONPG) as a substrate. All procedures were performed according to the Yeast Protocols Handbook (Clontech, United States).
The GST::IbABF4 fusion protein was expressed in Escherichia coli strain BL21 and purified as described previously (Jin et al., 2017). Optimal conditions for protein expression were created by adding 0.4 mM isopropyl-β-d-thiogalactoside (IPTG) and by incubating the E. coli cells at 20°C for 12 h. Oligonucleotide sets containing the ABRE repeat motif (5′-GGACA GCTGGC GGGACACGTGGC GGGACACGTGGC G-3′) were annealed by boiling for 5 min and then labeled with [γ-32P]-ATP by adding T4 Polynucleotide Kinase (Promega, Madison, WI, United States). A 30 μL mixture of a labeled probe (0.5 μg) and purified GST-IbABF4 or GST protein (10 μg) was incubated in binding buffer (200 mM HEPES, 5 mM DTT, 1 mM EDTA, 50 mM KCl, and 20 pmol of poly dI-dC) at room temperature for 30 min. Subsequently, the reaction mixture was loaded on an 8% native polyacrylamide gel and visualized by autoradiography. Competition experiments were carried out by incubating increasing amounts of unlabeled competitor probe with purified GST-IbABF4 fusion protein before the addition of labeled probes.
To overexpress the IbABF4 gene in sweetpotato cultivar Xushu 18, IbABF4 expression plasmids were introduced into A. tumefaciens strain EHA105 using the freeze-thaw method (Höfgen and Willmitzer, 1988) and then transformed into embryogenic calli of Xushu 18 plants via Agrobacterium-mediated transformation, as described previously (Lim et al., 2004; Kang et al., 2017). The transformed calli were selected on Murashige and Skoog (MS) medium containing 400 mg L–1 cefotaxime and 25 mg L–1 hygromycin. To generate transgenic Arabidopsis plants overexpressing IbABF4, the 35S::IbABF4-GFP recombinant plasmid was introduced in A. tumefaciens strain GV3101 and then transformed into Col-0 plants by the floral dip method (Clough and Bent, 1998). Homozygous T3 lines were used for further analysis.
Transgenic sweetpotato and Arabidopsis lines were confirmed and selected by genotyping and qRT-PCR analysis of hygromycin resistant plants. Primers used for PCR and qRT-PCR analyses are listed in Supplementary Table S1.
To perform cotyledon greening analyses, surface-sterilized Col-0 and T3 seeds were sown on half-strength MS (1/2 MS) medium supplemented with or without 0.6 μM ABA, 200 mM mannitol, or 125 mM NaCl. Greening of seedlings was determined at 1 week post-germination. To perform root growth assays, 5-days-old Arabidopsis seedlings were transferred to 1/2 MS medium containing 300 mM mannitol or 125 mM NaCl to induce drought or salt stress, respectively. Root length and fresh weight were measured after 10 days. To determine the drought tolerance of adult plants, irrigation of 3-week-old plants was withheld for 10 days, followed by re-watering. After the plants were photographed, contents of malondialdehyde (MDA), ABA and hydrogen peroxide (H2O2) and expression levels of stress-responsive genes were measured. Gene-specific primer sequences are listed in Supplementary Table S1.
To assess the stress tolerance of sweetpotato plants, 5-week-old plants of the WT and two transgenic lines were subjected to drought (without irrigation for 17 days and recovery for 2 days) or salt (irrigation with 200 mM NaCl solution every 3 days for 6 days) stress treatments. After photographing the plants, the third leaf from the top of the plants was used for the analysis of photosynthetic efficiency (Fv/Fm) of photosystem II (PSII) and measurement of ABA, MDA, and H2O2 contents. In plants subjected to drought stress, ABA content (at 3 and 6 days) and expression pattern of stress-responsive marker genes (at 6 days) were also measured. To determine the effect of oxidative stress on transgenic sweetpotato (hereafter referred to as SA) and WT plants, leaf disks (1 cm diameter) were excised from the third or fourth healthy leaf from the top of 5-week-old plants and submerged in 5 mL medium containing 2 μM methyl viologen (MV). The mixture was incubated in the dark for 12 h, followed by continuous light at 25°C. The analysis of relative membrane permeability (at 0, 12, 24, 36, and 48 h) and 3,3′-diaminobenzidine (DAB) staining (at 48 h) were performed according to the methods described by Jin et al. (2017) and Kang et al. (2017), respectively. All experiments were repeated three times.
The Fv/Fm values were measured using a portable chlorophyll fluorescence meter (Handy pEA, Hansatech, England) after 30 min of dark adaptation. The ABA content of SA and WT plants was measured using an enzyme-linked immunosorbent assay (ELISA), as described previously (Yang et al., 2001). The MDA content of plants was measured using the modified thiobarbituric acid (TBA) method described by Wang et al. (2009). Values of specific and non-specific absorbance of extracts were measured at 532 and 600 nm, respectively. Non-specific absorbance was then subtracted from the specific absorbance. The H2O2 content of plants was assessed using the xylenol orange method, as described previously (Bindschedler et al., 2001; Wang et al., 2009). The H2O2 content was expressed as μmol of H2O2 per gram of fresh weight of plant tissue.
All experimental assays were repeated at least three times. Significant differences between means were determined by analysis of variance (ANOVA and Tukey–Kramer test) at P < 0.01.
The full-length cDNA sequence of IbABF4 isolated from Xushu 18 was 1,272 bp in length, with an open reading frame of 423 amino acids (GenBank Accession No. MK503986). Analysis of the deduced amino acid residues using the SMART program1 revealed that the IbABF4 protein contained a typical bZIP domain (N-X7-R/K-X9-L-X6-L-X6-L), including a basic DNA-binding domain and a leucine zipper domain (Figure 1A). Five conserved Ser/Thr kinase phosphorylation sites (RXXS/T) were also observed in IbABF4 (data not shown). Furthermore, alignment and phylogenetic analysis revealed that IbABF4 was the most closely related to the ABF4 homologs of tomato and Nicotiana sylvestris (Figure 1B).
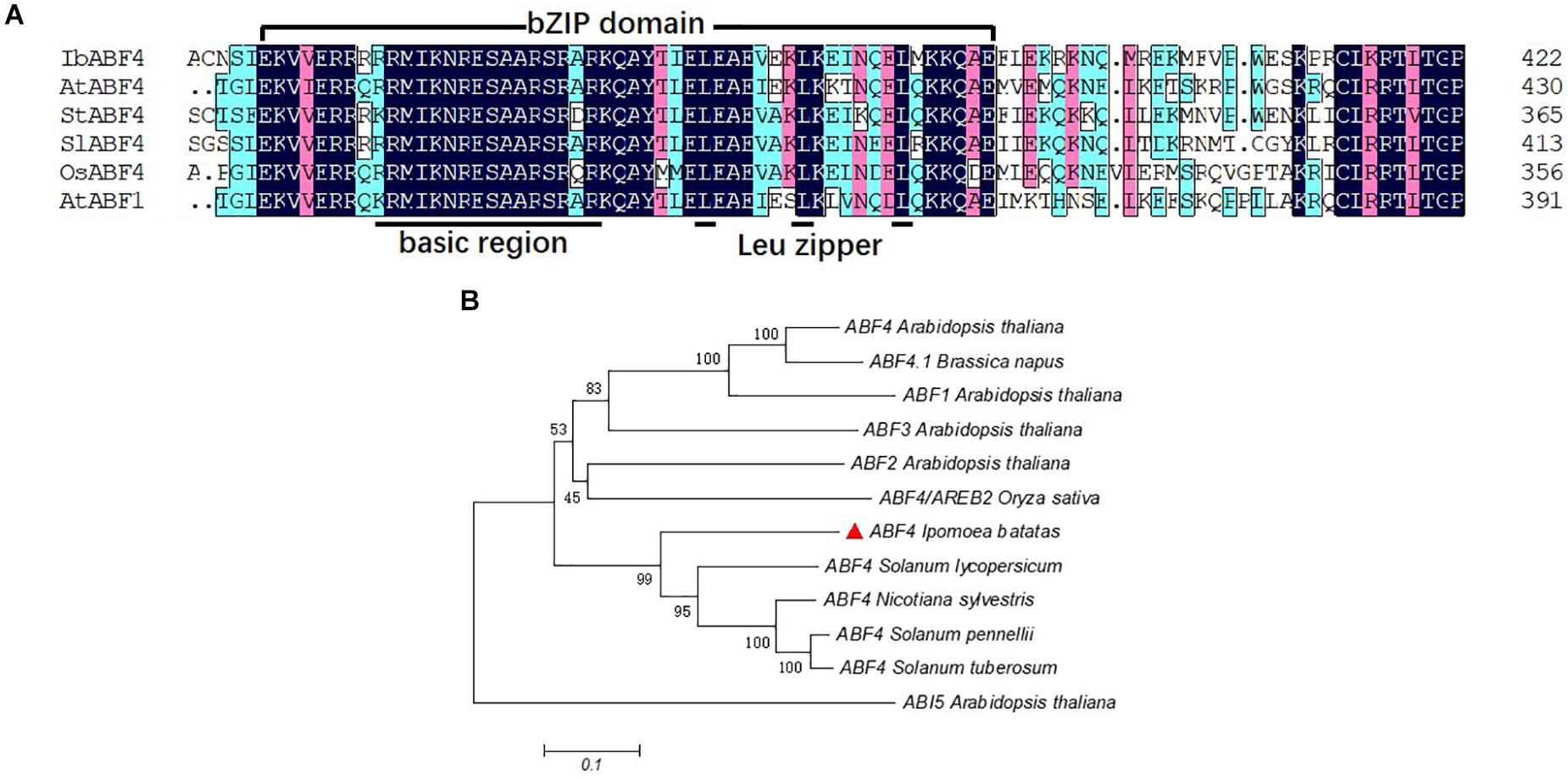
Figure 1. Multiple sequence alignment of the highly conserved bZIP domains and phylogenetic analysis of selected ABF-related proteins. (A) Multiple sequence alignment of the conserved bZIP domains of selected ABF-related proteins. Color-coding indicates sequence similarity, with black indicating the highest sequence similarity, pink indicating lower similarity, and blue indicating the least similarity. (B) Phylogenetic analysis of IbABF4 homologs in plants.
Tissue-specific expression levels of IbABF4 were measured by qRT-PCR (Figure 2A). IbABF4 was expressed in all analyzed tissues, with higher expression in the storage root, especially storage root body, compared with other tissues. The relative expression level of IbABF4 in sweetpotato plants exposed to ABA, PEG, NaCl, and heat was also measured by qRT-PCR (Figure 2B). In plants exposed to 10 μM ABA, the relative expression level of IbABF4 started increasing at 3 h, reaching a peak at 6 h, and then declining over the following 12 h. At the peak time point, the expression of IbABF4 in ABA-treated plants was 2.7-fold higher than that in untreated control plants. The expression of IbABF4 was also strongly induced by 25% PEG, 350 mM NaCl, and 47°C heat shock treatments, with expression patterns similar to those observed in ABA-treated plants. Under dehydration, salinity, and heat stresses, the expression of IbABF4 was the highest at 3 h; at this time point, IbABF4 expression in stress-treated plants was 3.5-, 3-, and 3.5-fold higher than that in control plants, respectively. Taken together, these results suggest that IbABF4 is induced by ABA, dehydration, salinity, and high temperature.
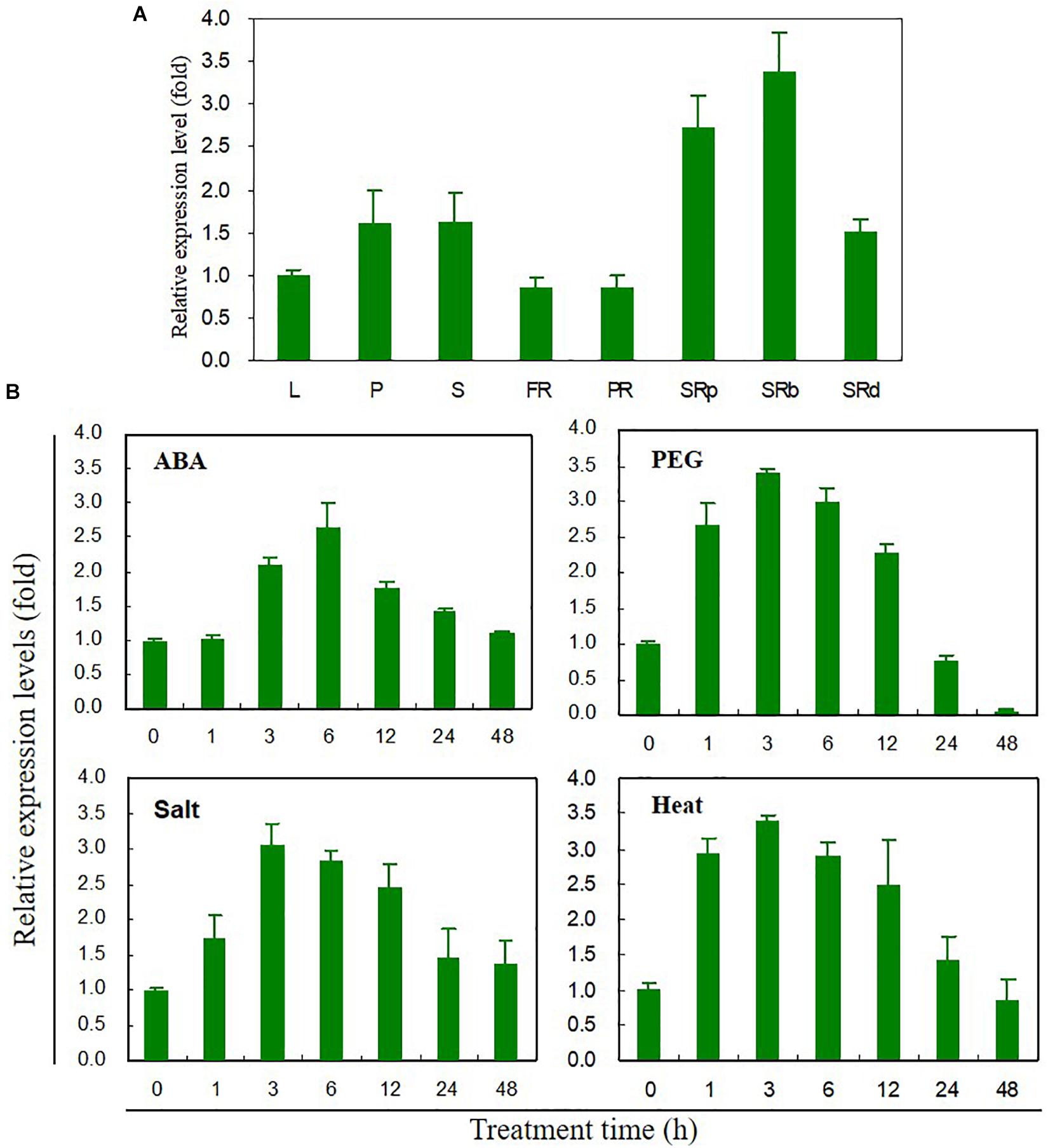
Figure 2. Expression profiles of IbABF4 in various tissues and under various abiotic stress conditions. (A) Tissue-specific expression pattern of IbABF4. L, leaf; P, petiole; S, stem; FR, fibrous root; PR, pencil root; SRp, proximal end of storage root; SRb, storage root body; SRd, distal end of storage root. (B) Expression pattern of IbABF4 in response to various abiotic stresses. Three-week-old sweetpotato plants grown in half-strength Hoagland nutrient solution were subjected to ABA (10 μM), dehydration (25% PEG8000), high salt (350 mM NaCl), and heat shock (47°C) treatments. Expression levels of genes were determined in the third fully expanded intact leaf (from the top) by qRT-PCR analysis. Expression levels of genes were normalized relative to the IbActin gene (internal control). Data represent mean ± standard deviation (SD) of three biological replicates.
We determined the subcellular localization of IbABF4 by transiently expressing the 35S::IbABF4-GFP plasmid in tobacco leaf epidermal cells. Co-localization of green fluorescence with the DAPI signal indicated that the IbABF4 protein was localized to the nucleus (Figure 3A).
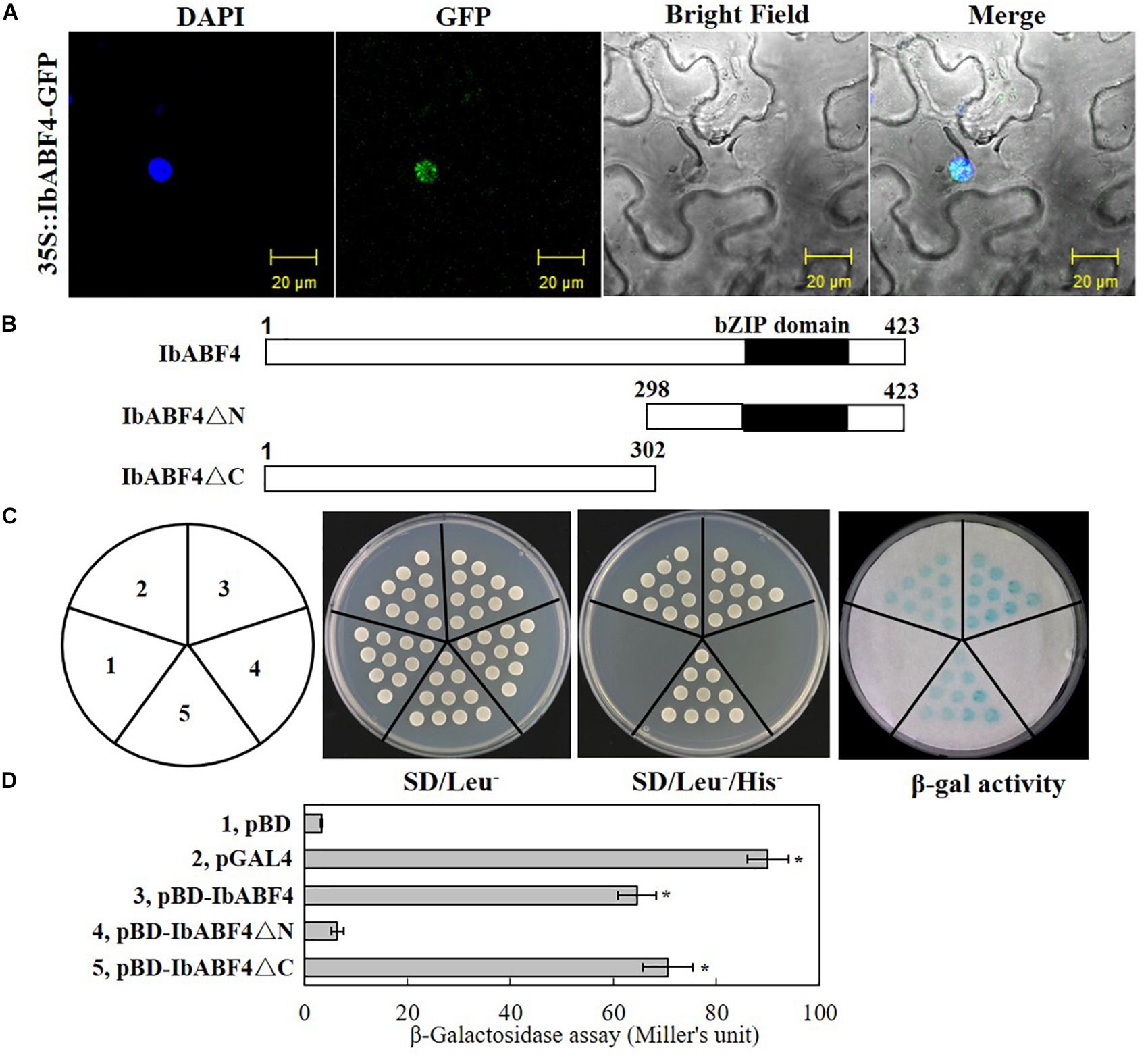
Figure 3. Subcellular localization and transactivation assay of the IbABF4 protein. (A) Subcellular localization of IbABF4. (B) Vectors used in the yeast one-hybrid assay. (C) Transactivation activity assay of the IbABF4 protein. The pGAL4 and pBD empty vectors were used as positive and negative controls, respectively, in yeast one-hybrid assay. (D) Relative quantitative assay of β-galactosidase activity (expressed in Miller units). Data represent mean ± SD of three independent biological replicates. Asterisks indicate significant differences between pBD and other vectors at P < 0.01.
We further determined the transcriptional activation capability of IbABF4 protein using the yeast one-hybrid assay. Full-length and N- and C-terminal fragments (IbABF4ΔC and IbABF4ΔN, respectively) of the IbABF4 protein were cloned into the yeast expression vector pBD containing the GAL4 DNA BD (Figure 3B). The pGAL4 and pBD empty vectors were used as positive and negative controls, respectively. All transformants grew well on SD/Leu- plates (Figure 3C). Yeast cells harboring pGAL4, pBD::IbABF4, and pBD::IbABF4ΔC grew normally on SD/Leu-/His- medium and showed β-galactosidase activity, whereas yeast cells containing pBD and pBD::IbABF4ΔN failed to grow on the SD/Leu-/His- medium and did not show β-galactosidase activity. These results revealed that IbABF4 possesses transcriptional activation capability, dependent on its N-terminal bZIP region. The results of the relative quantitative assay of β-galactosidase activity, using ONPG as a substrate, were consistent with those of the yeast one-hybrid assay (Figure 3D).
The cis-acting ABRE, which is often present in the promoters of ABA and other stress-responsive genes, is the binding site of the ABF/AREB proteins. To determine whether IbABF4 binds to the cis-acting ABRE in vitro, we performed EMSA using recombinant GST-IbABF4 fusion protein and 32P-labeled DNA containing three repeats of the ABRE core sequence (Figure 4A). Expression of GST-IbABF4 in E. coli BL21 cells was induced by the addition of IPTG, and the recombinant fusion protein was successfully purified by affinity chromatography. Results of the EMSA showed a shift in electrophoretic mobility of the 32P-labeled ABRE probe (Figure 4B). The binding activity of GST-IbABF4 increased with an increase in the amount of protein (Figure 4B, lanes 3 and 4). By contrast, GST alone did not bind to the 32P-labeled ABRE probe (Figure 4B, lane 2). The addition of increasing amounts of unlabeled ABRE probe prior to the addition of labeled probes (Figure 4C, lanes 3, 4, and 5) gradually abolished the binding. These results indicate that IbABF4 specifically binds to the cis-acting ABRE in vitro.
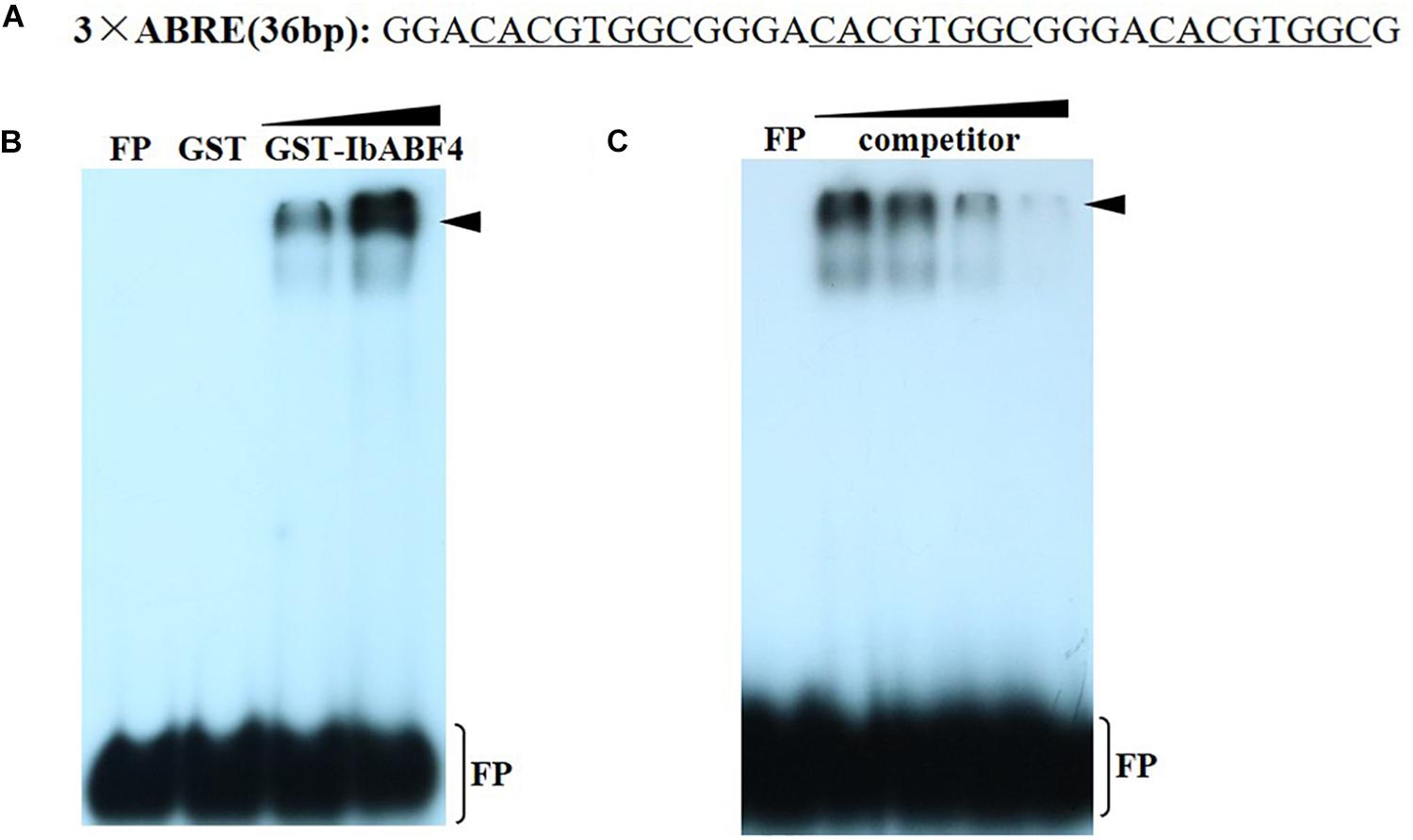
Figure 4. Binding of IbABF4 to the cis-acting ABRE in EMSA. (A) Oligonucleotide probes containing the ABRE repeat motif. The underlined letters indicate the ABRE core sequence. (B) Analysis of binding specificity of IbABF4. FP indicates free probe. EMSA was performed using 32P-labeled ABRE probe with GST protein (lane 2) or GST-IbABF4. Triangles indicate increasing amounts of GST-IbABF4 protein (5 and 10 μg) used for DNA-binding analysis. (C) Competition analysis of unlabeled probe in EMSA. Triangles indicate increasing amounts of unlabeled probe. GST-IbABF4 protein was preincubated with 1-, 20-, 40-, and 80-fold molar excess of ABRE before the addition of probe.
To characterize the function of IbABF4 in ABA, drought, and salt stress tolerance during seed germination, two independent T3 homozygous lines (OE9 and OE13) of Arabidopsis overexpressing IbABF4 were selected (data not shown) for further analysis; WT Col-0 plants were used as a control. Under normal growth conditions, no differences were observed in seed germination phenotype (Figure 5A) and germination rate (Figure 5B) between OE and WT plants. However, OE seeds were more sensitive to exogenously applied ABA than WT seeds during germination. The OE seeds showed significant improvement in germination rate upon exposure to mannitol and NaCl compared with WT seeds. These results suggest that IbABF4 increases the level of ABA sensitivity and drought and salt stress tolerance in plants.
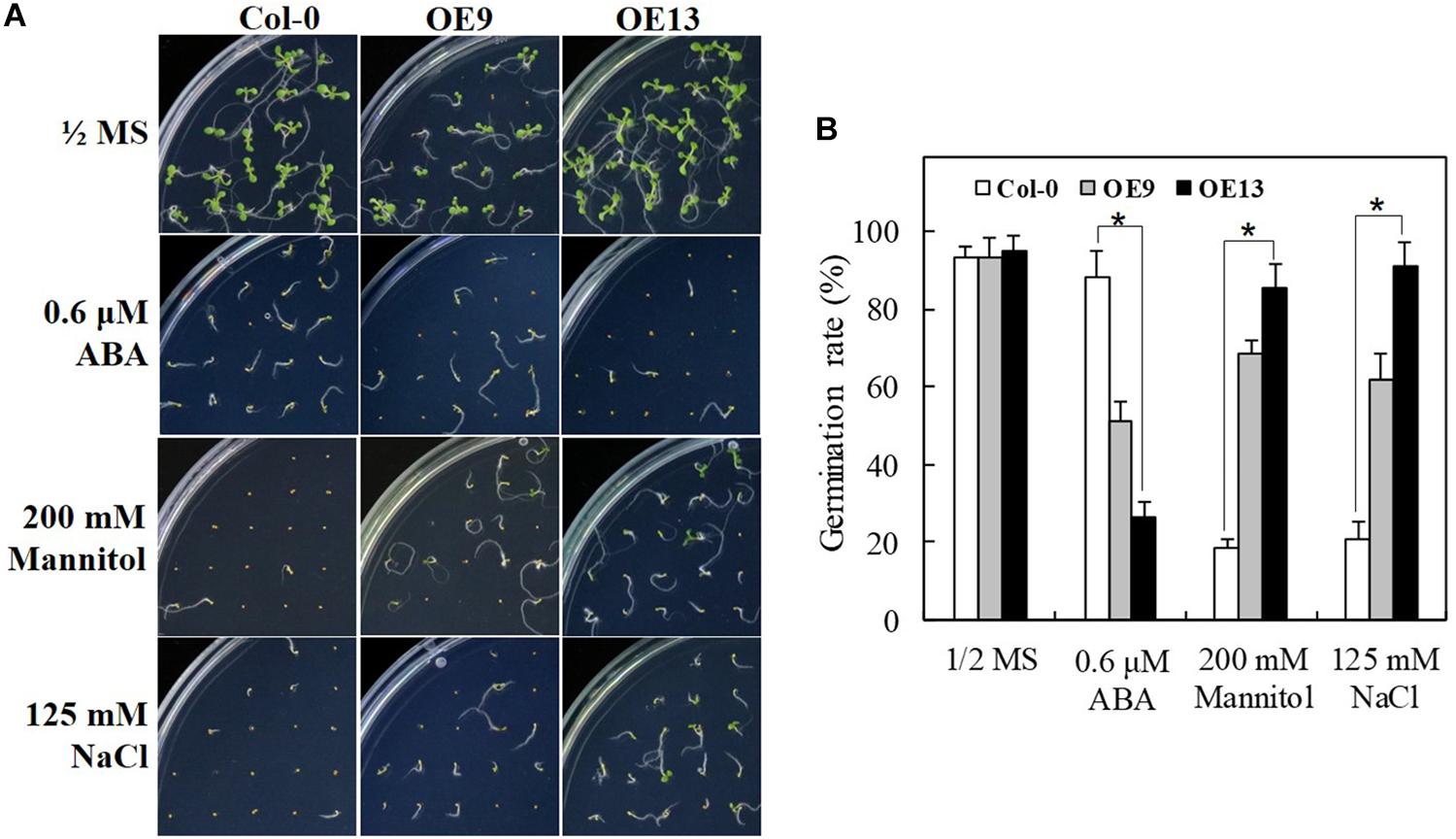
Figure 5. Seed germination assay of transgenic Arabidopsis lines overexpressing IbABF4 under ABA, drought, and salt stresses. (A) Seed germination phenotype of transgenic and WT Col-0 seeds on 1/2 MS medium supplemented with or without (control) 0.6 μM ABA, 200 mM mannitol, or 125 mM NaCl. (B) Germination rate of transgenic and Col-0 seeds under ABA, drought, and salt stresses. A total of 100 seeds were used for the calculation of germination rate in each treatment. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences at P < 0.01.
To gain further insights into the possible roles of IbABF4 in abiotic stress tolerance at post-germination, we investigated the growth phenotype of transgenic Arabidopsis seedlings overexpressing IbABF4 under drought and salt stresses. Under normal growth conditions, no difference was observed in growth phenotype (Figure 6A), root length (Figure 6B), and fresh weight (Figure 6C) between OE plants and WT. By contrast, under drought and salt stresses imposed by 1/2 MS medium containing 300 mM mannitol or 125 mM NaCl, respectively, OE plants exhibited significantly greater root elongation and fresh weight than WT plants. In the presence of 300 mM mannitol, root length was 4.7 cm in OE9 and OE13 lines but only 3.7 cm in WT seedlings. These results indicate that constitutive overexpression of IbABF4 enhances drought and salt tolerance of transgenic Arabidopsis plants.
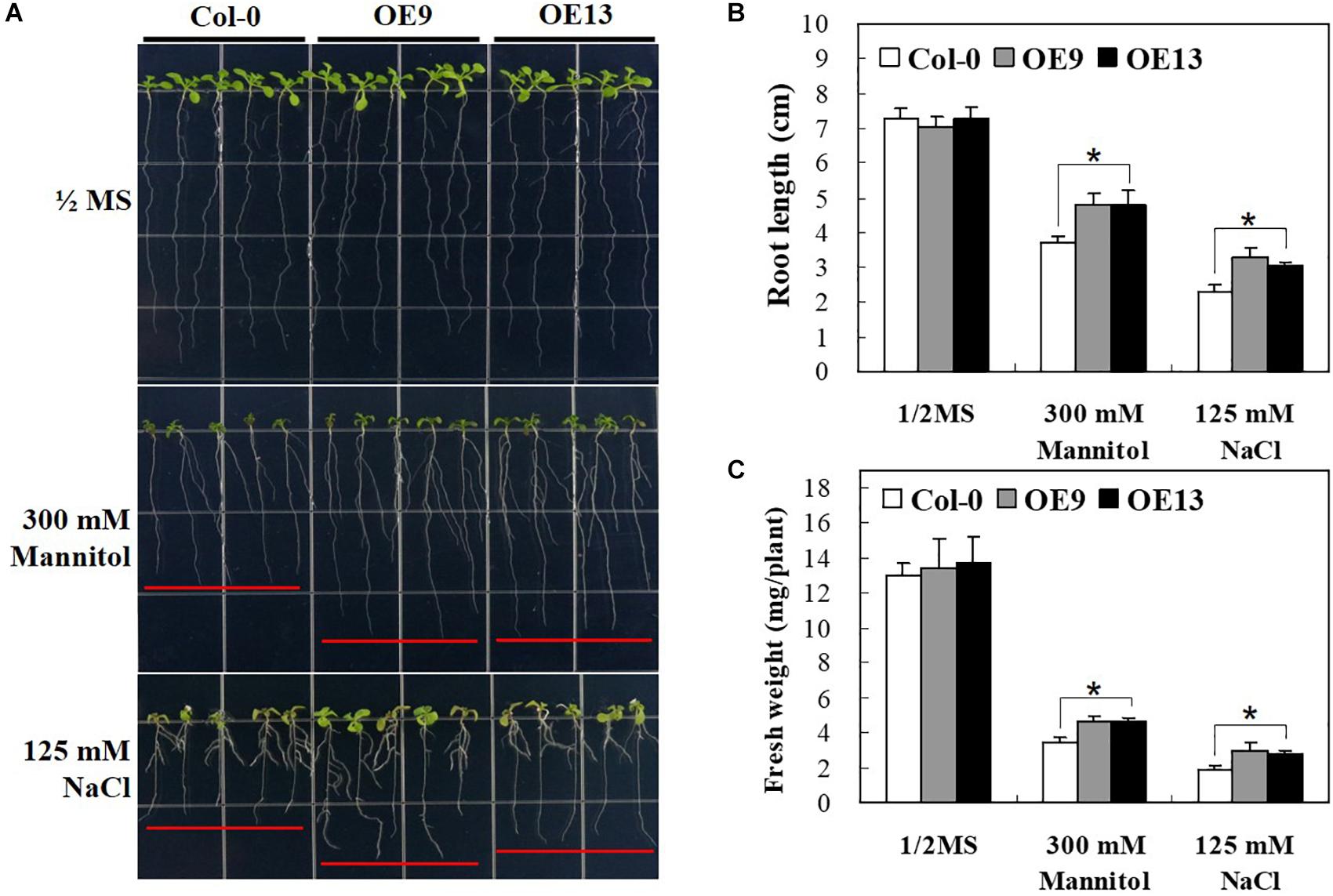
Figure 6. Post-germination growth assay of transgenic Arabidopsis plants overexpressing IbABF4 under drought and salt stresses. (A) Growth of transgenic Arabidopsis plants on 1/2 MS medium supplemented with or without (control) 300 mM mannitol, or 125 mM NaCl. (B) Root length of Arabidopsis plants subjected to drought and salt stresses. (C) Fresh weight of Arabidopsis plants exposed to drought and salt stresses. A total of 40 seedlings were used in each treatment. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences at P < 0.01.
We evaluated the drought tolerance of transgenic and WT Arabidopsis plants grown in soil. No significant differences were detected between transgenic and WT plants under normal growth conditions. However, when irrigation was withheld for 10 days, most of the leaves of WT plants turned yellow, shrunk in size, and ultimately died. By contrast, plants of transgenic Arabidopsis lines OE9 and OE13 only displayed slight wilting but grew well and recovered successfully following re-watering for 1 day (Figure 7A). The MDA and H2O2 contents of both transgenic and WT plants gradually increased at 3 and 6 days of drought treatment, although transgenic lines exhibited lower MDA and H2O2 contents than WT plants (Figures 7B,C). The ABA content of OE plants was higher than that of WT plants at 3 and 6 days of drought stress (Figure 7D). Furthermore, the expression of stress-responsive genes, including AtRD29A, AtRD29B, and AtCOR47, was highly induced by drought stress, with significantly higher expression in transgenic Arabidopsis plants than in WT Col-0 plants (Figure 7E). Taken together, these data suggest that IbABF4 is a functional homolog of Arabidopsis ABF4, which activates stress-responsive genes such as AtRD29A, AtRD29B, and AtCOR47, thus enhancing drought tolerance.
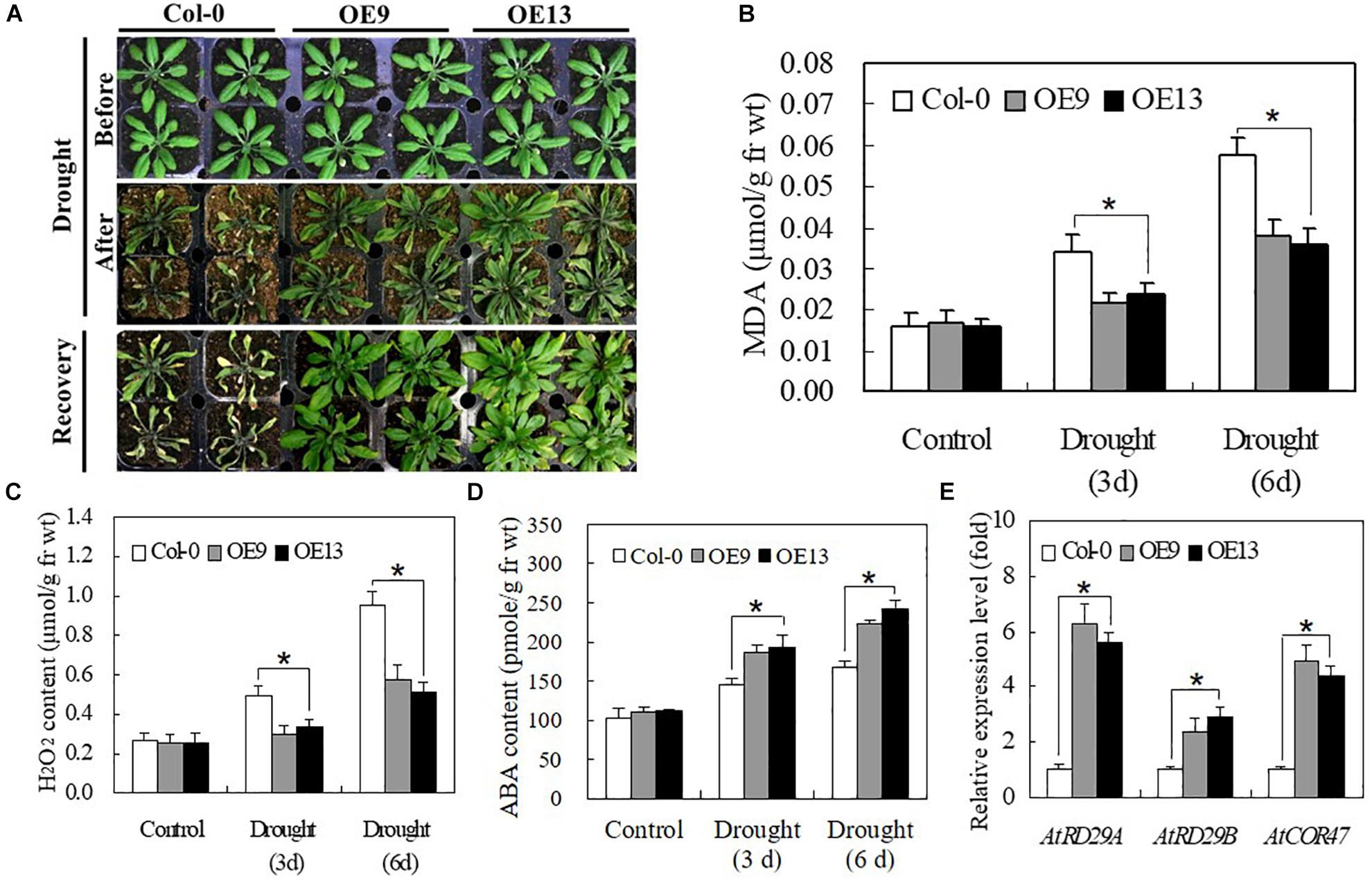
Figure 7. Overexpression of IbABF4 in Arabidopsis confers drought tolerance. (A) Drought tolerance phenotypes of 3-week-old transgenic Arabidopsis (OE9 and OE13) lines and Col-0 plants grown in soil without irrigation for 10 days, followed by recovery for 1 day. Contents of MDA (B), H2O2 (C) and ABA (D) in leaves of Col-0 and OE plants grown without irrigation for 3 and 6 days. (E) Transcript levels of stress-responsive marker genes in OE9, OE13, and Col-0 plants. The AtActin gene was used as an internal control. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences at P < 0.01.
The results of transgenic Arabidopsis plants encouraged us to generate transgenic sweetpotato plants with improved abiotic stress tolerance through the overexpression of IbABF4. The IbABF4 cDNA was cloned in the pGWB5 vector under the control of CaMV 35S promoter (Figure 8A), and transgenic sweetpotato plants were generated by Agrobacterium-mediated transformation. A total of 15 independent hygromycin resistant sweetpotato lines were confirmed by PCR amplification of genomic DNA (Figure 8B). Two transgenic lines (SA3 and SA4) exhibiting high IbABF4 expression were identified by qRT-PCR for further characterization (Figure 8C).
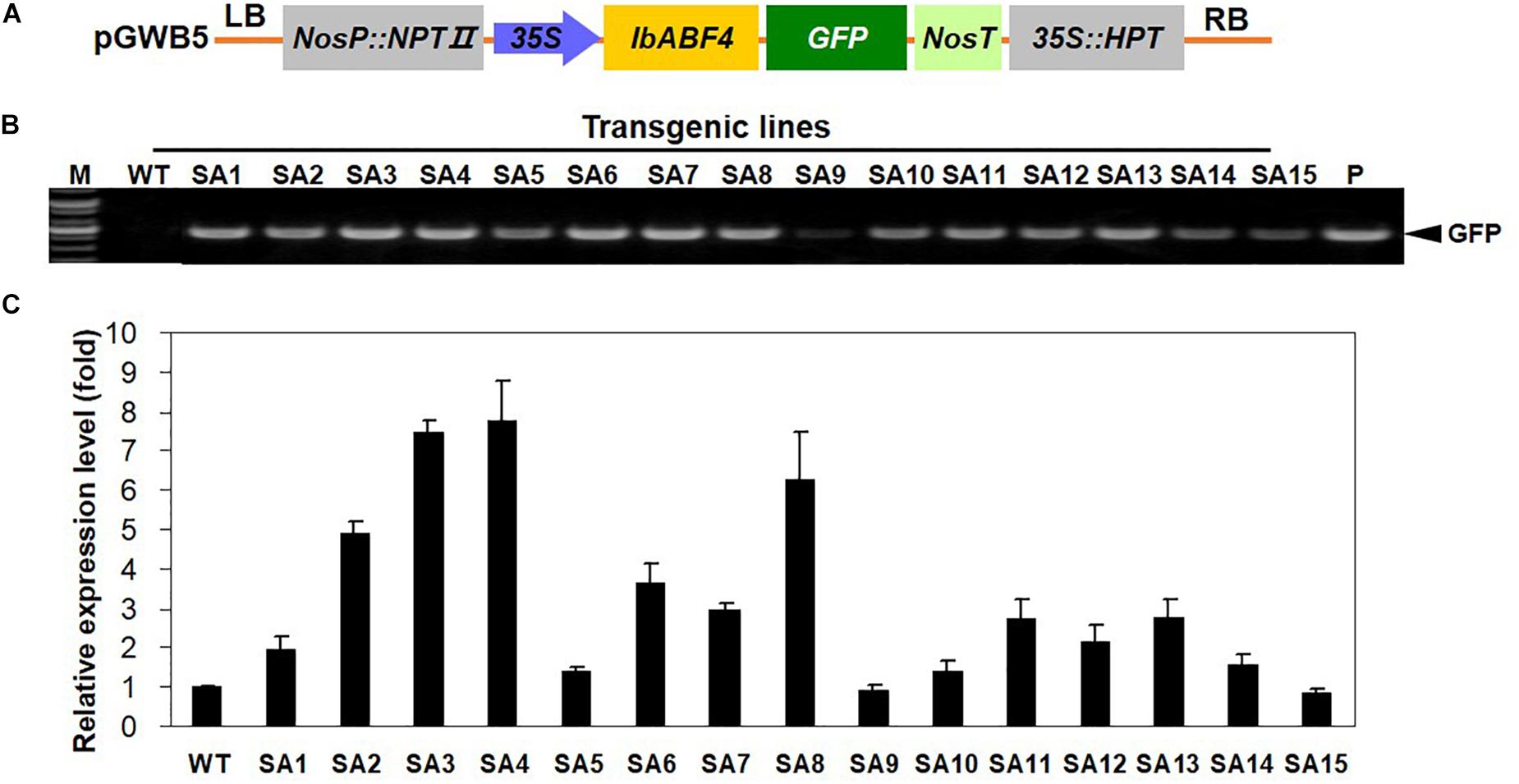
Figure 8. Generation of SA plants overexpressing IbABF4. (A) Schematic representation of the constructs used for sweetpotato transformation. (B) PCR analysis of SA plants using GFP-specific primers. M, DNA size markers; P, positive control. (C) qRT-PCR analysis of SA plants using IbABF4-specific primers. The IbActin gene was used as an internal control. Data represent mean ± SD of three biological replicates.
To evaluate the effect of IbABF4 overexpression in sweetpotato on drought stress tolerance, we compared the growth phenotypes of SA and WT plants subjected to water deficiency for 17 days, followed by recovery for 2 days. Under drought stress conditions, SA plants showed slight wilting but grew well and recovered successfully, whereas WT plants displayed severe wilting at 17 days and slight recovery after re-watering for 2 days (Figure 9A). The SA plants maintained higher photosynthetic efficiency (Fv/Fm) of PSII throughout the water deficiency and recovery periods than WT plants (Figure 9B). In both WT and SA plants, MDA and H2O2 contents increased after 17 days of continuous drought stress and then decreased after 2 days of recovery. The WT plants consistently exhibited a higher level of MDA and H2O2 contents than SA plants throughout the drought stress and recovery periods (Figures 9C,D). Taken together, these results suggest that IbABF4 overexpression in sweetpotato enhances drought tolerance.
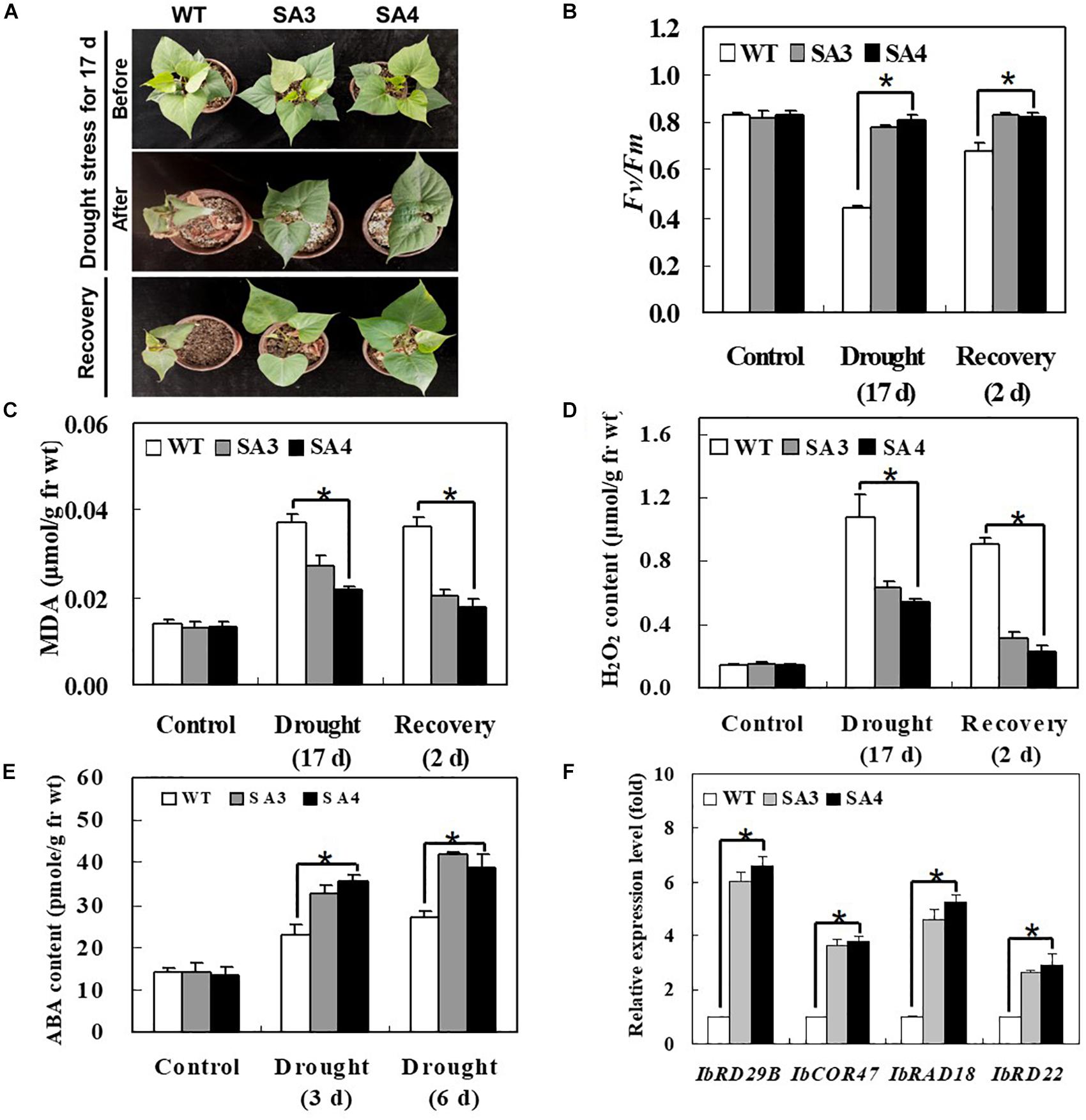
Figure 9. Phenotypic and physiological characterization of SA plants under drought stress. (A) Visible damage in the leaves of sweetpotato plants after drought stress treatment for 17 days, followed by recovery for 2 days. (B) Photosynthetic efficiency (Fv/Fm) of PSII. Contents of MDA (C) and H2O2 (D) in leaves of 5-week-old WT and SA plants subjected to drought stress for 17 days, followed by recovery for 2 days. (E) ABA content of leaves of 5-week-old WT and SA plants subjected to drought stress for 3 and 6 days. (F) Transcript levels of stress-responsive marker genes in WT and SA plants determined by qRT-PCR. Samples of WT plants were collected prior to drought stress treatment, and transcript levels of genes were normalized relative to IbActin transcripts. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences between WT and SA plants at P < 0.01.
We also examined the content of ABA content and expression pattern of several stress-responsive marker genes in SA and WT plants subjected to drought stress. The ABA content of SA plants was higher than that of WT plants at 3 and 6 days of drought stress (Figure 9E). The SA plants also exhibited significantly higher expression levels of ABA/stress-responsive marker genes, including IbRD29B, IbCOR47, IbRAB18, and IbRD22, than WT plants (Figure 9F). Increased levels of ABA content and stress-responsive marker gene expression in SA plants likely explain their enhanced drought tolerance.
To determine whether IbABF4 overexpression in sweetpotato confers salt stress tolerance, 5-week-old WT and SA plants were irrigated with 200 mM NaCl solution every 3 days for 6 days. Under normal growth conditions, no phenotypic differences were observed between WT and SA plants. However, under salt stress, WT plants exhibited more severe wilting and chlorosis than SA plants (Figure 10A). The SA plants also showed higher Fv/Fm values than WT plants at 6 days of salt stress treatment (Figure 10B). Furthermore, under salt stress conditions, contents of MDA and H2O2 in WT plants were significantly higher than those in SA plants (Figures 10C,D). The ABA content of SA plants was higher than that of WT plants at 6 days of salt stress (Figure 10E).
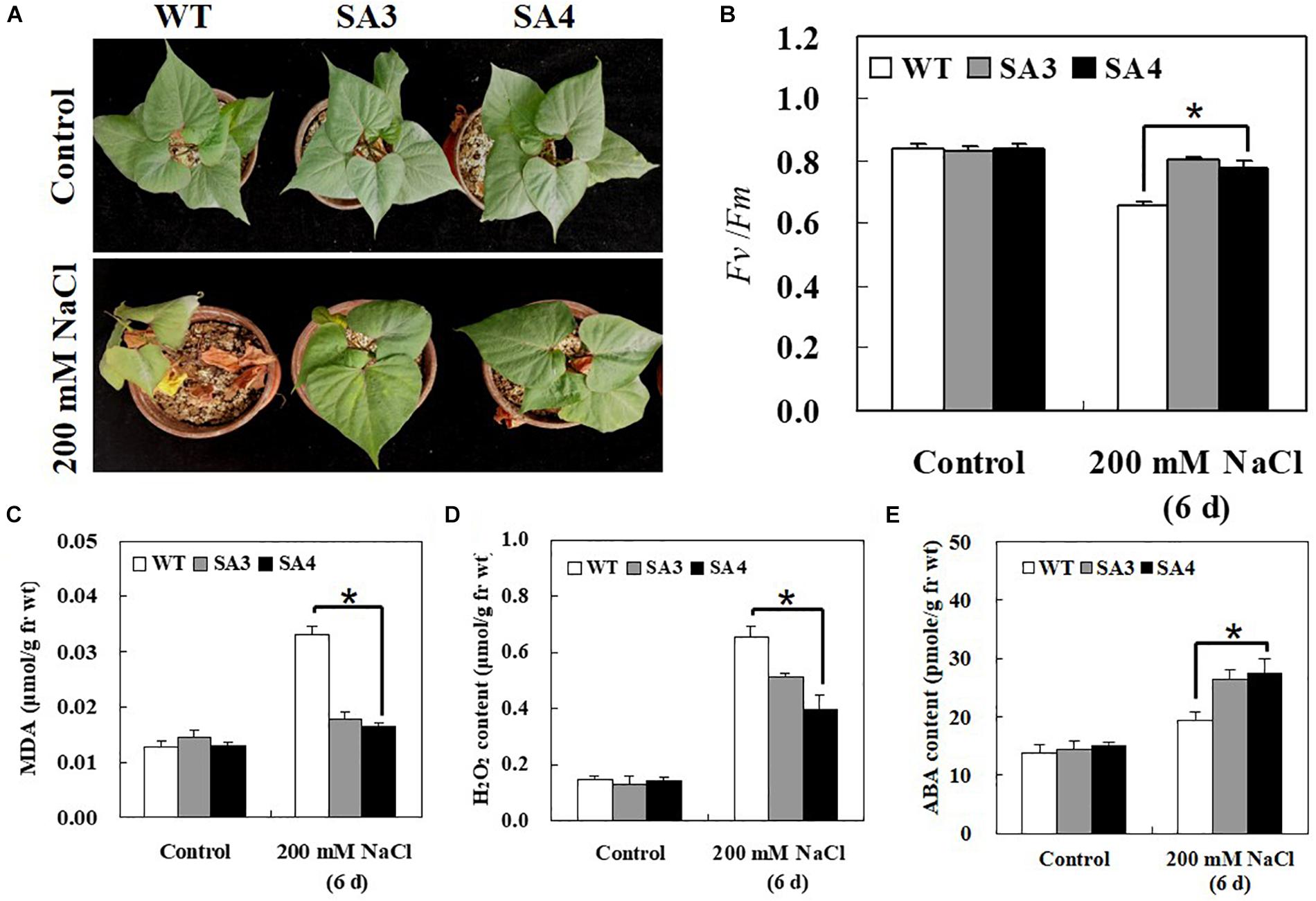
Figure 10. Phenotypic and physiological characterization of SA plants under salt stress. (A) Visible damage in the leaves of sweetpotato plants after 6 days of salt stress treatment. (B) PSII photosynthetic efficiency (Fv/Fm). Contents of MDA (C), H2O2 (D), and ABA (E) in leaves of 5-week-old WT and SA plants subjected to salt stress treatment for 6 days. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences between WT and SA plants at P < 0.01.
A leaf disk senescence assay was performed to investigate the oxidative stress tolerance of SA plants. When treated with MV, WT plants showed symptoms of injury including rapid senescence and chlorosis, whereas SA plants only exhibited slight damage (Figure 11A). Moreover, SA plants showed significantly lower relative membrane permeability than WT plants (Figure 11B). We also investigated the level of H2O2 accumulation following MV-induced oxidative stress by DAB staining. Detached leaves of SA plants exhibited reduced DAB staining intensity than those of WT plants after MV treatment (Figure 11C), indicating lower levels of H2O2 in SA leaves. Overall, we conclude that overexpression of IbABF4 confers tolerance to salt stress and MV-induced oxidative stress in sweetpotato.
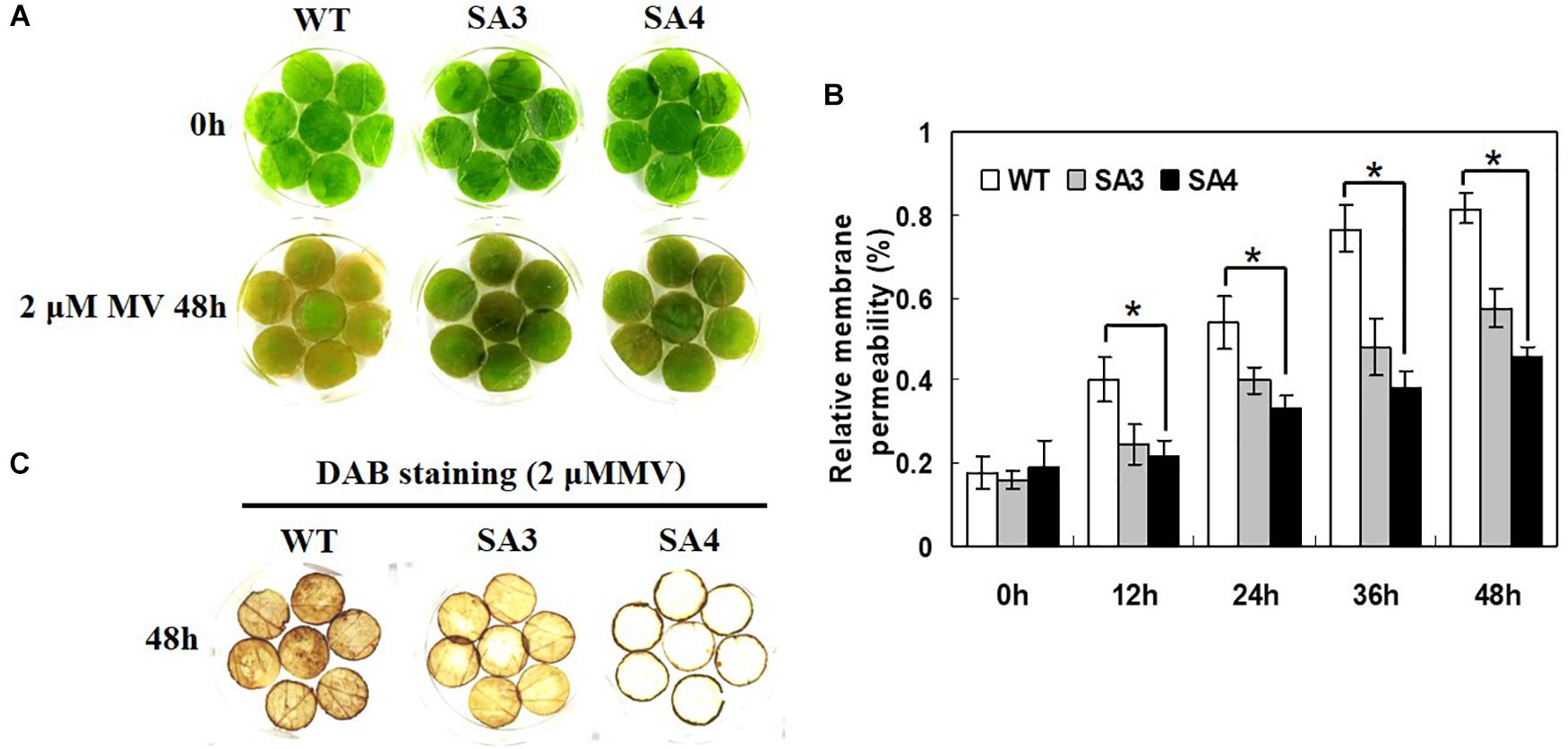
Figure 11. Oxidative stress tolerance of SA plants. (A) Visible damage in detached leaves of WT and SA plants treated with 2 μM MV. (B) Relative membrane permeability of detached leaves treated with 2 μM MV at 48 h. Data represent mean ± SD of three biological replicates. Asterisks indicate significant differences between WT and SA plants at P < 0.01. (C) Analysis of H2O2 accumulation in detached leaves using DAB staining. The third or fourth fully expanded leaves were collected from the top of 5-week-old WT and SA plants.
Sweetpotato is one of the most important food crops, as it is rich in nutrients and useful as a source of starch and bioethanol (Duvernay et al., 2013; Pradhan et al., 2015). The production of sweetpotato is limited by a variety of abiotic stresses, resulting in significant yield losses (Lebot, 2010). Therefore, the expression and function of genes regulating abiotic stress response in sweetpotato are of considerable interest. Several sweetpotato genes including IbOr (Kim et al., 2013; Goo et al., 2015; Kang et al., 2017), IbCBF3 (Jin et al., 2017), IbPsbP (Kang et al., 2017), IbCHY-β (Kang et al., 2017), IbLCY-β (Kim et al., 2014), IbMPK3, and IbMPK6 (Kim et al., 2016) have been cloned and characterized in our laboratory. Although many genes encoding bZIP TFs are known to play key roles in abiotic stress response, only a few such genes have been reported in sweetpotato. In this study, we demonstrated that a Group A bZIP encoding gene, IbABF4/IbABRE2, is a component of the ABA signaling pathway involved in adaptation to drought, salt, and oxidative stresses.
Based on the basic DNA-binding domain containing an invariant motif (N-X7-R/K) and a leucine zipper domain (L-X6-L-X6-L), bZIP TFs are classified into 10 groups in Arabidopsis (Jakoby et al., 2002) and 13 groups in rice (Corrêa et al., 2008). The ABF/AREB proteins belong to Group A of bZIP TFs. Five conserved Ser/Thr kinase phosphorylation sites (RXXS/T) are characteristic of the abiotic stress-responsive AtAREB/ABFs (Furihata et al., 2006; Fujii et al., 2009). In the present study, we showed that like other ABFs, IbABF4 contains the typical bZIP domain (Figure 1A) and five conserved Ser/Thr kinase phosphorylation sites. Amino acid sequence alignments demonstrated that IbABF4 is similar to other ABFs including AtABF4, OsABF4, StABF4, SlABF4, and AtABF1. Together with phylogenetic analysis (Figure 1B), these findings suggest that IbABF4 is a functional homolog of plant ABF4 proteins responsive to abiotic stresses.
The expression of several ABA/stress-responsive genes in plants is regulated by cis-acting elements that function as important molecular switches in the transcriptional regulation of a dynamic network of genes (Yamaguchishinozaki and Shinozaki, 2005, 2006). Subcellular localization experiments in this study indicated that IbABF4 functions as a transcriptional regulator in the nucleus (Figure 3A). The results of yeast one-hybrid assay revealed that IbABF4 possesses transcriptional activation capability dependent on its N-terminal bZIP domain (Figures 3C,D). Our results are consistent with those of previous yeast one-hybrid assays of OsABF1 (Hossain M. et al., 2010), OsABF2 (Hossain M.A. et al., 2010), OsbZIP23 (Xiang et al., 2008), and TaAREB3 (Wang J. et al., 2016). Furthermore, the results of EMSAs indicate that IbABF4 specifically binds to the cis-acting ABRE with the core sequence CACGTGGC in vitro (Figure 4). Similar observations on AtABF1, SlAREB, TabZIP60, TaAREB3, and TabZIP14-B are reported in yeast one-hybrid assay and EMSAs (Hyungin et al., 2000; Tsaihung et al., 2010; Zhang et al., 2015, 2017; Wang J. et al., 2016). In conclusion, these data suggest that IbABF4 is a transcriptional activator that specifically binds to the cis-acting ABRE in the promoters of downstream stress-responsive genes, thus activating their expression.
Most ABF genes such as ABF2/AREB1, ABF4/AREB2, and ABF3 are highly induced by ABA, drought, and salinity treatments in plant tissues (Uno et al., 2000; Furihata et al., 2006; Yoshida et al., 2010). Overexpression of ABFs increases abiotic stress tolerance in several plant species (Roychoudhury et al., 2013). In this study, transcript levels and response times of IbABF4 to ABA, PEG, salt, and heat shock treatments indicated that IbABF4 is highly sensitive to environmental stresses (Figure 2B). Thus, our data indicate that IbABF4 plays an important role in the response to abiotic stresses. Compared with WT Col-0 seeds, transgenic Arabidopsis seeds overexpressing IbABF4 exhibited a significantly lower germination rate when treated with exogenous ABA and an improved germination rate when treated with mannitol or NaCl (Figure 5). Additionally, Arabidopsis OE plants exhibited significantly greater root length and fresh weight than Col-0 plants under drought and salt stress treatments (Figure 6). Furthermore, lower MDA and H2O2 contents of transgenic Arabidopsis plants than WT plants indicate that transgenic plants have greater tolerance to salt and dehydration stresses (Figure 7). Moreover, we successfully generated SA plants overexpressing IbABF4 (Figure 8). Under drought and salt stresses, SA plants exhibited lower MDA and H2O2 contents than WT plants (Figures 9C,D, 10C,D), suggesting that IbABF4 overexpression increases antioxidant activity in sweetpotato. The reduced Fv/Fm values of WT plants indicated that the PSII reaction center was more severely damaged in WT plants than in SA plants (Figures 9B, 10B). Under MV-induced oxidative stress, SA plants showed only slight damage compared with WT plants (Figure 11). Together, these results suggest that IbABF4 overexpression improves the germination rate and post-germination growth of transgenic lines under drought, salt, and oxidative stress conditions. Our results are consistent with a previous study of Kim et al. (2004) showing that Arabidopsis abf3 and abf4 mutants display defects in response to ABA, salt, and dehydration stresses.
In the present study, endogenous ABA contents were higher in transgenic plants than in WT plants under drought stress (Figures 7D, 9E). Moreover, transcript levels of several stress-responsive genes including RD29A, RD29B, COR47, RAB18, and RD22 were upregulated in transgenic Arabidopsis and sweetpotato plants overexpressing IbABF4 under drought stress (Figures 7E, 9F). All stress-responsive genes harbor ABREs in their promoter regions (Fujita et al., 2005; Yamaguchishinozaki and Shinozaki, 2006). These results support our conclusion that IbABF4 mediates responses to drought stress via the ABA signaling pathway. The results of EMSA suggest that IbABF4 interacts with ABREs in the promoters of ABA-responsive genes, thus enhancing the expression level of these genes and resulting in greater drought tolerance (Agarwal and Jha, 2010). These genes are also induced by salt stress and ABA (Nakashima et al., 2009; Tsaihung et al., 2010). Increased expression levels of these genes may, at least in part, explain the improved drought and salt stress tolerance of transgenic plants. There is evidence for cross-talk between ABA signals and H2O2 signaling in plant tissue (Saxena et al., 2016; Phillips and Ludidi, 2017). In present study, transgenic Arabidopsis and sweetpotato plants overexpressing IbABF4 showed increasing ABA and H2O2 contents under drought and salt treatments than normal growth condition (Figures 7C,D, 9D,E, 10D,E). We deduced that H2O2 may function as an intermediate in ABA signaling. MAPK cascade is activated by the increasing H2O2 which in turn is mediated by the hormones like ABA (Saxena et al., 2016). Subsequently, ABA-activated SnRK2-type protein kinases may phosphorylate the Ser/Thr residues of RXXS/T sites and activate IbABF4 protein similar to AtAREB1 (Furihata et al., 2006). The lower level of H2O2 contents in transgenic Arabidopsis and sweetpotato plants than WT under stresses treatments (Figures 7C, 9D, 10D) may be caused by over expression of various downstream antioxidants such as CAT and APX mediated by IbABF4 protein. However, the ABA and H2O2 signaling pathways in IbABF4 protein response to abiotic stress still need further investigation. The higher level of H2O2 contents in WT plants caused corresponding increases in lipid peroxidation (Figures 7B, 9C, 10C).
We report that the IbABF4 TF specifically binds to the cis-regulatory ABRE. Overexpression of IbABF4 in Arabidopsis resulted in enhanced tolerance to drought and salt stresses. Moreover, SA plants confirmed the role of IbABF4 in tolerance to drought, salt, and oxidative stresses. Agronomic traits of sweetpotato plants overexpressing IbABF4 need further characterization in the field. Overall, the results of this study broaden our understanding of sweetpotato bZIP TFs and suggest an excellent candidate gene for the improvement of stress tolerance in crop plants.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
WW, HK, and S-SK conceived and designed the experiments. WW, XQ, YY, HK, XJ, and HY performed the experiments. WW, XQ, YY, and HK analyzed the data. S-SK contributed to reagents, materials, and analysis tools. WW, XQ, and S-SK wrote the manuscript .
This work was supported by the Basic Research Program of Shanxi Province (201801D121204), Cultivate Project of National Science Foundation of Shanxi Agricultural University (2017GPY06), Science Innovation Foundation of Shanxi Agricultural University (2018yz003), Doctoral Research Grant of Shanxi Agricultural University (XB2009002), and KRIBB initiative program.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00630/full#supplementary-material
Agarwal, P. K., and Jha, B. (2010). Transcription factors in plants and ABA dependent and independent abiotic stress signaling. Biol. Plant 54, 201–212. doi: 10.1007/s10535-010-0038-7
Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24, 23–58. doi: 10.1080/07352680590910410
Bindschedler, L. V., Minibayeva, F., Gardner, S. L., Gerrish, C., Davies, D. R., and Bolwell, G. P. (2001). Early signalling events in the apoplastic oxidative burst in suspension cultured French bean cells involve cAMP and Ca2+. New Phytol. 151, 185–194. doi: 10.1046/j.1469-8137.2001.00170.x
Busk, P. K., and Pagès, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. doi: 10.1023/A:1006058700720
Casaretto, J. A., and Ho, T. H. (2005). Transcriptional regulation by abscisic acid in barley (Hordeum vulgare L.) seeds involves autoregulation of the transcription factor HvABI5. Plant Mol. Biol. 57, 21–34. doi: 10.1007/s11103-004-6520-x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation ofArabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Corrêa, L. G., Riaño-Pachón, D. M., Schrago, C. G., Dos Santos, R. V., Mueller-Roeber, B., and Vincentz, M. (2008). The role of bZIP transcription factors in green plant evolution: adaptive features emerging from four founder genes. PLoS One 3:e2944. doi: 10.1371/journal.pone.0002944
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
Duvernay, W. H., Chinn, M. S., and Yencho, G. C. (2013). Hydrolysis and fermentation of sweetpotatoes for production of fermentable sugars and ethanol. Ind. Crop. Prod. 42, 527–537. doi: 10.1016/j.indcrop.2012.06.028
Fujii, H., Chinnusamy, V., Rodrigues, A., Rubio, S., Antoni, R., Park, S. Y., et al. (2009). In vitro reconstitution of an abscisic acid signalling pathway. Nature 462, 660–664. doi: 10.1038/nature08599
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17, 3470–3488. doi: 10.1105/tpc.105.035659
Furihata, T., Maruyama, K., Fujita, Y., Umezawa, T., Yoshida, R., Shinozaki, K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993. doi: 10.1073/pnas.0505667103
García, M. N. M., Giammaria, V., Grandellis, C., Télleziñón, M. T., Ulloa, R. M., and Capiati, D. A. (2012). Characterization of StABF1, a stress-responsive bZIP transcription factor from Solanum tuberosum L. that is phosphorylated by StCDPK2 in vitro. Planta 235, 761–778. doi: 10.1007/s00425-011-1540-7
Golldack, D., Lüking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Goo, Y. M., Han, E. H., Jeong, J. C., Kwak, S. S., Yu, J., Kim, Y. H., et al. (2015). Overexpression of the sweet potato IbOr gene results in the increased accumulation of carotenoid and confers tolerance to environmental stresses in transgenic potato. C. R. Biol. 338, 12–20. doi: 10.1016/j.crvi.2014.10.006
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. Cell Mol. Biol. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Höfgen, R., and Willmitzer, L. (1988). Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 16:9877. doi: 10.1093/nar/16.20.9877
Hossain, M., Lee, Y., Cho, J., Ahn, C., Lee, S., Jeon, J., et al. (2010). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72, 557–566. doi: 10.1007/s11103-009-9592-9
Hossain, M. A., Cho, J. I., Han, M., Ahn, C. H., Jeon, J. S., An, G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. doi: 10.1016/j.jplph.2010.05.008
Hyungin, C., Hong, J. H., Jinok, H., Kang, J. Y., and Sooyoung, K. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jin, R., Kim, B. H., Ji, C. Y., Kim, H. S., Li, H. M., Ma, D. F., et al. (2017). Overexpressing IbCBF3 increases low temperature and drought stress tolerance in transgenic sweetpotato. Plant Physiol. Biochem. 118, 45–54. doi: 10.1016/j.plaphy.2017.06.002
Kang, J. Y., Choi, H. I., Im, M. Y., and Kim, S. Y. (2002). Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14, 343–357. doi: 10.1105/tpc.010362
Kang, L., Kim, H. S., Kwon, Y. S., Ke, Q. B., Ji, C. Y., Park, S. C., et al. (2017). IbOr regulates photosynthesis under heat stress by stabilizing IbPsbP in sweetpotato. Front. Plant Sci. 8:989. doi: 10.3389/fpls.2017.00989
Ke, Q. B., Kim, H. S., Wang, Z., Ji, C. Y., Jeong, J. C., Lee, H. S., et al. (2017). Down-regulation of GIGANTEA-like genes increases plant growth and salt stress tolerance in poplar. Plant Biotechnol. J. 15, 331–343. doi: 10.1111/pbi.12628
Kim, H. S., Park, S. C., Ji, C. Y., Park, S., Jeong, J. C., Lee, H. S., et al. (2016). Molecular characterization of biotic and abiotic stress-responsive MAP kinase genes, IbMPK3 and IbMPK6, in sweetpotato. Plant Physiol. Biochem. 108, 37–48. doi: 10.1016/j.plaphy.2016.06.036
Kim, S., Kang, J. Y., Cho, D. I., Ji, H. P., and Kim, S. Y. (2004). ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 40, 75–87. doi: 10.1111/j.1365-313X.2004.02192.x
Kim, S. H., Ahn, Y. O., Ahn, M. J., Jeong, J. C., Lee, H. S., and Kwak, S. S. (2013). Cloning and characterization of an Orange gene that increases carotenoid accumulation and salt stress tolerance in transgenic sweetpotato cultures. Plant Physiol. Biochem. 70, 445–454. doi: 10.1016/j.plaphy.2013.06.011
Kim, S. H., Jeong, J. C., Park, S., Bae, J. Y., Ahn, M. J., Lee, H. S., et al. (2014). Down-regulation of sweetpotato lycopene β-cyclase gene enhances tolerance to abiotic stress in transgenic calli. Mol. Biol. Rep. 41, 8137–8148. doi: 10.1007/s11033-014-3714-4
Kobayashi, F., Maeta, E., Terashima, A., and Takumi, S. (2008). Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol. Plant. 134, 74–86. doi: 10.1111/j.1399-3054.2008.01107.x
Latchman, D. S. (1993). Transcription factors: an overview. Int. J. Exp. Pathol. 74, 417–422. doi: 10.1016/S1357-2725(97)00085-X
Lebot, V. (2010). “Sweet potato,” in Root and Tuber Crops, ed. J. E. Bradshaw (New York, NY: Springer Science+Business Media), 97–125.
Lim, S., Yang, K. S., Kwon, S. Y., Paek, K. Y., Kwak, S. S., and Lee, H. S. (2004). Agrobacterium-mediated genetic transformation and plant regeneration of sweetpotato (Ipomoea batatas). J. Plant Biotechnol. 31, 267–271. doi: 10.5010/jpb.2004.31.4.267
Lindemose, S., O’shea, C., Jensen, M. K., and Skriver, K. (2013). Structure, function and networks of transcription factors involved in abiotic stress responses. Int. J. Mol. Sci. 14, 5842–5878. doi: 10.3390/ijms14035842
Madzlan, K., Hasnisa, H., Sabeetha, S., and Dayana, M. N. (2012). Extraction of starch and enzymatic production of high amylose starch from sweetpotato (Ipomea batatas) var. Telong. J. Trop. Agric. Food Sci. 222, 203–210. doi: 10.1016/j.jconrel.2015.12.011
Moore, J. P., Le, N. T., Brandt, W. F., Driouich, A., and Farrant, J. M. (2009). Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 14, 110–117. doi: 10.1016/j.tplants.2008.11.007
Muszynski, M. G., Dam, T., Li, B., Shirbroun, D. M., Hou, Z., Bruggemann, E., et al. (2006). delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 142, 1523–1536. doi: 10.1104/pp.106.088815
Nakashima, K., Ito, Y., and Yamaguchishinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Phillips, K., and Ludidi, N. (2017). Drought and exogenous abscisic acid alter hydrogen peroxide accumulation and differentially regulate the expression of two maize RD22-like genes. Sci. Rep. 7:8821. doi: 10.1038/s41598-017-08976-x
Pradhan, D. M. P., Mukherjee, A., George, J., Chakrabarti, S. K., Vimala, B., Naskar, S. K., et al. (2015). High starch, beta carotene and anthocyanin rich sweet potato: ascent to future food and nutrition security in coastal and backward areas. Int. J. Trop. Agric. 10, 9–22. doi: 10.4067/S0718-22012014000100002
Roychoudhury, A., Paul, S., and Basu, S. (2013). Cross-talk between abscisic acid-dependent and abscisic acid-independent pathways during abiotic stress. Plant Cell Rep. 32, 985–1006. doi: 10.1007/s00299-013-1414-5
Saxena, I., Srikanth, S., and Chen, Z. (2016). Cross talk between H2O2 and interacting signal molecules under plant stress response. Front. Plant Sci. 7:570. doi: 10.3389/fpls.2016.00570
Thurow, C., Schiermeyer, A., Krawczyk, S., Butterbrodt, T., Nickolov, K., and Gatz, C. (2005). Tobacco bZIP transcription factor TGA2.2 and related factor TGA2.1 have distinct roles in plant defense responses and plant development. Plant J. 44, 100–113. doi: 10.1111/j.1365-313X.2005.02513.x
Tsaihung, H., Li, C. W., Su, R. C., Cheng, C. P., Yichien, T., and Chan, M. T. (2010). A tomato bZIP transcription factor, SLAREB, is involved in water deficit and salt stress response. Planta 231, 1459–1473. doi: 10.1007/s00425-010-1147-4
Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc. Natl. Acad. Sci. U.S.A. 97, 11632–11637. doi: 10.1073/pnas.190309197
Wang, J., Li, Q., Mao, X., Li, A., and Jing, R. (2016). Wheat transcription factor TaAREB3 participates in drought and freezing tolerances in Arabidopsis. Int. J. Biol. Sci. 12, 257–269. doi: 10.7150/ijbs.13538
Wang, Z., Su, G., Li, M., Ke, Q., Kim, S. Y., Li, H., et al. (2016). Overexpressing arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol. Biochem. 109, 199–208. doi: 10.1016/j.plaphy.2016.09.020
Wang, W., Vinocur, B., and Altman, A. (2003). Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218, 1–14. doi: 10.1007/s00425-003-1105-5
Wang, W. B., Kim, Y. H., Lee, H. S., Kim, K., Deng, X. P., and Kwak, S. S. (2009). Analysis of antioxidant enzyme activity during germination of alfalfa under salt and drought stresses. Plant Physiol. Biochem. 47, 570–577. doi: 10.1016/j.plaphy.2009.02.009
Xiang, Y., Tang, N., Du, H., Ye, H., and Xiong, L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Yamaguchishinozaki, K., and Shinozaki, K. (2005). Organization of cis-acting regulatory elements in osmotic- and cold-stress-responsive promoters. Trends Plant Sci. 10, 88–94. doi: 10.1016/j.tplants.2004.12.012
Yamaguchishinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, J., Zhang, J., Wang, Z., Zhu, Q., and Wang, W. (2001). Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 127, 315–323. doi: 10.1104/pp.127.1.315
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Zhang, L., Zhang, L., Xia, C., Gao, L., Hao, C., Zhao, G., et al. (2017). A novel wheat C-bZIP gene, TabZIP14-B, participates in salt and freezing tolerance in transgenic plants. Front. Plant Sci. 8:710. doi: 10.3389/fpls.2017.00710
Keywords: abiotic stress, drought tolerance, salt tolerance, IbABF4, sweetpotato
Citation: Wang W, Qiu X, Yang Y, Kim HS, Jia X, Yu H and Kwak S-S (2019) Sweetpotato bZIP Transcription Factor IbABF4 Confers Tolerance to Multiple Abiotic Stresses. Front. Plant Sci. 10:630. doi: 10.3389/fpls.2019.00630
Received: 21 February 2019; Accepted: 26 April 2019;
Published: 16 May 2019.
Edited by:
Omar Borsani, Universidad de la República, UruguayReviewed by:
Yong Hwa Cheong, Sunchon National University, South KoreaCopyright © 2019 Wang, Qiu, Yang, Kim, Jia, Yu and Kwak. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sang-Soo Kwak, c3Nrd2FrQGtyaWJiLnJlLmty
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.