- Institute of Biochemical Plant Pathology, Helmholtz Zentrum München – German Research Center for Environmental Health, Munich, Germany
Nitric oxide (NO) is a key signaling molecule in all kingdoms. In plants, NO is involved in the regulation of various processes of growth and development as well as biotic and abiotic stress response. It mainly acts by modifying protein cysteine or tyrosine residues or by interacting with protein bound transition metals. Thereby, the modification of cysteine residues known as protein S-nitrosation is the predominant mechanism for transduction of NO bioactivity. Histone acetylation on N-terminal lysine residues is a very important epigenetic regulatory mechanism. The transfer of acetyl groups from acetyl-coenzyme A on histone lysine residues is catalyzed by histone acetyltransferases. This modification neutralizes the positive charge of the lysine residue and results in a loose structure of the chromatin accessible for the transcriptional machinery. Histone deacetylases, in contrast, remove the acetyl group of histone tails resulting in condensed chromatin with reduced gene expression activity. In plants, the histone acetylation level is regulated by S-nitrosation. NO inhibits HDA complexes resulting in enhanced histone acetylation and promoting a supportive chromatin state for expression of genes. Moreover, methylation of histone tails and DNA are important epigenetic modifications, too. Interestingly, methyltransferases and demethylases are described as targets for redox molecules in several biological systems suggesting that these types of chromatin modifications are also regulated by NO. In this review article, we will focus on redox-regulation of histone acetylation/methylation and DNA methylation in plants, discuss the consequences on the structural level and give an overview where NO can act to modulate chromatin structure.
Sources and Intracellular Localization of Nitric Oxide
In plants, NO is formed either by reductive or oxidative pathways. In mammals, three cell-specific NO synthases (NOS) oxidize arginine to citrulline, thereby releasing NO. Although NOS-like activities have been measured in chloroplasts and peroxisomes of higher plants (He et al., 2004; Corpas and Barroso, 2018), NO synthase has only been identified in the algae (Foresi et al., 2010). Other possible substrates for oxidative NO production involve polyamines and hydroxylamine (Groß et al., 2013; Farnese et al., 2016). Reduction of nitrite to NO constitutes the reductive route of NO production (Rockel et al., 2002; Yamamoto-Katou et al., 2006; Srivastava et al., 2009). Usually, nitrate reductase catalyzes the reduction of nitrate to nitrite. However, under low oxygen conditions, light and high nitrite levels, nitrite can be reduced to NO (Rockel et al., 2002; Planchet et al., 2005). Finally, enzyme-independent reduction of nitrite has been described in apoplast under acidic conditions (Bethke et al., 2004). Intracellular sources of NO are located in various compartments, including cytosol, peroxisomes, mitochondria, and chloroplasts (summarized in Groß et al., 2013; Farnese et al., 2016). Nuclear NO production is not described in plants. However, thiol reducing systems like thioredoxins and glutaredoxins as well as reducing molecules such as glutathione were found in the nucleus, suggesting that thiol modifications occur in this compartment (Delorme-Hinoux et al., 2016; Martins et al., 2018). Because of its lipophilic character, NO can easily cross the nuclear membrane or enter via nuclear pores (Toledo and Augusto, 2012; Lancaster, 2015). Moreover, NO can be transferred into the nucleus via S-nitrosylated proteins or S-nitrosylated low molecular weight thiols, such as S-nitrosoglutathione (GSNO) or S-nitrosocysteine. S-Nitrosylated nuclear proteins have been identified using the biotin switch technique, which labels S-nitrosylated proteins with a biotin linker allowing detection, purification, and identification of these proteins (Chaki et al., 2015). In mammals, nuclear translocation of S-nitrosylated proteins is described for gylceralaldehyd-3-phosphat-dehydrogenase and chloride intracellular channel protein CLIC4 (Hara et al., 2005; Malik et al., 2010). Nuclear localization of gylceralaldehyd-3-phosphat-dehydrogenase has been characterized in Arabidopsis (Holtgrefe et al., 2008; Vescovi et al., 2013; Aroca et al., 2017).
Physiological Function and Biochemistry of Nitric Oxide
The chemical properties of nitric oxide (NO) make it highly multifunctional. Whereas some studies report toxic and harmful action of NO species, such as cell death (Pedroso et al., 2000), damage of proteins, membranes, and nucleic acids, or photosynthetic inhibition (Yamasaki, 2000), others demonstrate protective and/or signaling function of NO species. In fact, the dual function of NO is often dependent on its concentration and environment. Based on its functions, NO has been proposed as a stress-responding agent. It can counteract toxic processes induced by ROS (Beligni and Lamattina, 1998; Sun et al., 2007). It was shown that NO is involved in abiotic stress responses such as salinity, drought, UV-B radiation, temperature, and heavy metal toxicity (Mata and Lamattina, 2001; Tian et al., 2007). The role of NO in biotic stress is essential. It plays a key role in disease resistance against Pseudomonas syringae in Arabidopsis leaves, and is required for SAR induction in tobacco (Delledonne et al., 1998; Hong et al., 2008). Moreover, NO participates in plant development and physiological processes such as germination, gravitropism, root development, and flowering (Correa-Aragunde et al., 2004; He et al., 2004; Zhang et al., 2005). Although there is no doubt that NO is crucial for plant development and survival, the mechanism by which NO activates signaling function and the genes underlying this process remain to be elucidated.
NO chemical properties contribute to its role in signal transduction in a living cell (Toledo and Augusto, 2012; Lancaster, 2015). It can rapidly undergo multiple chemical reactions with enzymes, transcription factors, second messengers, or chromatin modifiers (Yu et al., 2014; Kovacs et al., 2016a). NO and its related species are able to modulate protein activities and biological function through covalent post-translational modifications (PTM) by binding to the metal centers of proteins and by affecting their cysteine and tyrosine residues. Tyrosine nitration is a post-translational modification that arises through the binding of a NO2 into ortho carbons of aromatic ring of tyrosine residues that leads to the formation of 3-nitrotyrosine (Mata-Pérez et al., 2016; Kolbert et al., 2017). In a direct reaction termed metal nitrosylation, NO binds to transition metals, resulting in formation of metal nitrosyl complexes. In this way, activity and function of proteins can be regulated. Well studied targets for NO interaction are iron-sulfur clusters, as well as heme groups and zinc ions of proteins (Astier et al., 2010).
Examples of NO binding to iron present in heme proteins have also been observed in plants. It was suggested that two major H2O2-scavenging enzymes in tobacco, ascorbate peroxidase, and catalase are reversible inhibited by NO donors through the formation of an iron-nitrosyl complex (Clark et al., 2000). Plant hemoglobins were also identified as a target for NO. It was shown that Arabidopsis nonsymbiotic hemoglobin AHb1 binds NO and oxidizes it to nitrate, suggesting a role of hemoglobins in detoxification of NO (Perazzolli et al., 2004; Kuruthukulangarakoola et al., 2017). S-Nitrosation is the most studied redox-based post-translational modification. This modification results in the formation of S-nitrosothiols (SNO). S-Nitrosation enables a living organism to directly respond to environmental stimulus through the regulation of protein activity, protein-protein interaction, or protein localization (Hara et al., 2006; Yun et al., 2011). The release of the NO moiety from proteins and therefore the control of SNO homeostasis in a cell is maintained by two enzymes: GSNOR reductase (GSNOR), which metabolizes GSNO to a mixture of intermediates, and thioredoxins, which mediate denitrosylation (López-Sánchez et al., 2008; Cañas et al., 2012; Kneeshaw et al., 2014). Furthermore, reduced glutathione (GSH) alone is able to denitrosylate S-nitrosylated proteins. For instance, physiological levels of GSH rapidly removed the NO moiety of S-nitrosylated GAPDH resulting in the reduced and active form of GAPDH (Zaffagnini et al., 2013). For this GSH-dependent protein denitrosylation, the GSH/GSNO ratio is of relevance, but not the GSH/GSSG ratio. Although a high number of candidates for S-nitrosation were identified, only a few of them were experimentally confirmed and their functions in response to NO demonstrated (Astier et al., 2012). Most of the studies are based on biotin switch technique, where S-nitrosated cysteines are labeled with a biotinylating agent, allowing easy detection by immunoblotting using anti-biotin antibodies or purification using streptavidin matrix.
Interestingly, S-nitrosation of transcription factors can affect their function. For instance, S-nitrosation of Cys53 of the Arabidopsis R2R3-MYB2 transcription factor inhibited its DNA-binding activity (Serpa et al., 2007). Similarly, S-nitrosation of Cys49 and Cys53 of MYB30 results are structural changes negatively affecting DNA affinity of this transcription factor (Tavares et al., 2014). In contrast, S-nitrosation of TGA1, a transcription factor involved in the activation of pathogenesis-related (PR) genes, promotes the binding to the as1-element of the PR1 promoter (Lindermayr et al., 2010). Besides the regulatory function of NO on transcription factors, NO can control gene transcription also by affecting chromatin structure and/or DNA accessibility.
Regulation of Histone Acetylation/Methylation and DNA Methylation by Nitric Oxide
On the genome level (nucleotide sequence), cells of multicellular organisms are identical. However, each cell differs from others through the differences in gene expression pattern that might occur in a temporally and spatially-dependent manner. Genes might be silenced/switched off and switched on again only when they are required. Such an activation/inactivation can be regulated through a direct control of regulatory elements on gene promoters. Moreover, over the last few decades, it was found that affecting the accessibility of the DNA by modification of the chromatin structure, is also a key regulator of transcription.
Genetic material of all eukaryotic organisms has to be packed into the nucleus to prevent it from becoming damaged. Since the length of eukaryotic DNA is far greater than the diameter of a nucleus, it has to be organized in a very tightly packaged structure, known as chromatin. The core subunit of chromatin is an octamer, which is composed of two copies of the histone proteins, H2A, H2B, H3, and H4, which are positively charged and enable an electrostatic interaction with negatively charged DNA. 145–147 bp of DNA are wrapped around a histone complex forming repeating nucleosomal units, which are connected with each other by short DNA fragments called “linker DNA.” Linker histone H1 is located between the nucleosomes and stabilizes chromatin structure, resulting in highly condensed 30 nm fibers (Luger et al., 2012).
Chromatin structure in eukaryotic organisms is very dynamic, and can be changed during growth and development and in response to environmental stimuli. Chromatin marks are able to induce chromatin remodeling and therefore to control important molecular processes such as gene transcription, replication, repair, and recombination (Bannister and Kouzarides, 2011). DNA methylation and histone modifications are the key mediators of epigenetic modifications. DNA methylation is usually associated with long-term silencing of genes, whereas histone modifications contribute to both activation and repression of gene transcription and can be removed after several cell cycles (Jaenisch and Bird, 2003; Minard et al., 2009).
Histone modifications play an important role in the regulation of chromatin structure and in subsequent gene transcription. N-terminal histone tails, which are exposed outside the nucleosome may interact with neighboring nucleosomes and therefore manipulate the chromatin structure (Bannister and Kouzarides, 2011). Histone tails can undergo different posttranslational modifications such as acetylation, methylation, phosphorylation, and ubiquitination. These can act alone or in combination, resulting in different molecular changes that effect DNA accessibility.
Nitric Oxide Inhibits Histone Deacetylases
Histone acetylation plays a key role in regulation of gene transcription (Servet et al., 2010; Shen et al., 2015). This modification is very dynamic and is catalyzed by two families of enzymes: histone acetyltransferases (HATs) and histone deacetylases (HDAs). The transfer of acetyl groups from acetyl-coenzyme A on histone lysine residues is catalyzed by histone acetyltransferases. This modification neutralizes the positive charge of the lysine residue and reduces the interaction between histones and the negatively charged DNA (Schiedel and Conway, 2018). This results in a loose chromatin structure accessible for DNA binding proteins. Histone deacetylases, in contrast, remove the acetyl group of histone tails resulting in condensed chromatin with reduced gene expression activity (Hollender and Liu, 2008; Luo et al., 2012). Therefore, histone acetylation is usually associated with gene transcription. For instance, differential acetylation at H3K9 and H3K27 and phosphorylation at H3S28 between end-of-night and end-of-day correlates with changes in diurnal transcript levels of core clock genes in Arabidopsis (Baerenfaller et al., 2016). In poplar, expression of carbonic anhydrase, pyruvate orthophosphate dikinase, phosphoenolpyruvate carboxykinase, and phosphoenolpyruvate carboxylase correlates with acetylation of H3K9 and H4K5 at their promoter regions (Li et al., 2017). Moreover, deacetylation of the flowering gene AGL19 represses its transcription (Kim et al., 2013). However, there are also examples where enhanced acetylation of nearby regulatory elements and coding sequences does not generally result in higher transcription of the corresponding gene (Mengel et al., 2017). For example, comparison of ChIP-seq and transcript data of genes displaying GSNO-regulated H3K9/14 ac demonstrated that the mRNA levels of more than 60% of these genes remained unchanged (Mengel et al., 2017), concluding that histone acetylation is indeed making DNA accessible, but does not per se leads directly to gene transcription.
There is increasing evidence that the catalytic activity of at least some HDAs is regulated by redox modifications, which are involved in the regulation of unwinding and wrapping of chromatin. Until now, most studies of redox regulation by HDAs have been done in human and animal cells. It was reported that HDA2 in neurons gets S-nitrosated upon NO signaling triggered by brain-derived neurotrophic factor (Nott et al., 2008). S-Nitrosation of HDA2 results in chromatin acetylation and activation of gene expression that are involved in neuronal development. Notably, S-nitrosation does not affect the enzymatic activity of HDA2, but stimulates its release from chromatin. Influence of NO on HDA2 was confirmed, when redox-sensitive cysteines were mutated to alanine preventing dissociation of HDA2 from chromatin (Nott et al., 2008). S-Nitrosation of HDA2 was also demonstrated in muscles of dystrophin-deficient MDX mice (Colussi et al., 2008). The catalytic activity of this enzyme was impaired by NO in vivo and in vitro. Additionally, protein activity in the presence of NO was also measured in purified Escherichia coli produced HDA1, HDA2, and HDA3. Recombinant HDA2 was highly sensitive to NO donors, and a slight reduction of protein activity was measured in HDA1 that was not caused by S-nitrosation (Colussi et al., 2008). Human HDA6 and HDA8 were also identified as potential targets for NO (Feng et al., 2001; Okuda et al., 2015). Endogenous HDA6 was identified as target for S-nitrosation using the biotin switch assay (Okuda et al., 2015). S-Nitrosation of HDA6 inhibited its catalytic activity and increased the level of acetylated alpha-tubulin suggesting that HDA6 plays a crucial regulatory function in acetylation of proteins others than histones (Okuda et al., 2015). HDA8 is S-nitrosated by GSNO in vitro (Feng et al., 2001). Moreover, the protein activity was significantly reduced by GSNO and another NO donor, S-nitrosocysteine, in time- and concentration-dependent manner. Interestingly, application of the NO donor sodium nitroprusside (SNP) to HDA8 had no effect on the catalytic activity of this protein, indicating that a special structural interaction is required for transferring NO (Feng et al., 2001). NO-dependent inhibition of gene expression was measured in human umbilical vein endothelial cells (Illi et al., 2008). It was demonstrated that upon NO production, protein phosphatase becomes activated and associates with a histone deacetylase complex pCamkIV/HDAs, promoting its dephosphorylation. This process leads to the shuttling of HDA4 and HDA5 (members of pCamkIV/HDAs complex) to the nucleus and deacetylation of histones. As a consequence, c-fos gene expression is inhibited. c-fos encodes for a protein with a basic leucine zipper region for dimerization and DNA-binding and a C-terminal transactivation domain (Illi et al., 2008). It is involved in important cellular events, including cell proliferation, differentiation and survival. Under non-stressed conditions, when the NO level in the cell is low, HDA4 and HDA5 remain in the cytosol, allowing hyperacetylation of chromatin (Illi et al., 2008). If similar mechanisms exist in plants as well still has to be investigated.
In Arabidopsis there are 18 members of HDAs, which are divided into 3 families: RPD3-like, HD-tuins, and sirtuins (Hollender and Liu, 2008; König et al., 2014; Shen et al., 2015; Bourque et al., 2016). The first family is the largest one and is composed of 12 putative members (HDA2, HDA5-10, HDA14-15, HDA17-19), which, based on their structure, can be further divided into 3 subclasses. This family of HDAs is homologous to yeast reduced potassium deficiency 3 (RPD3) proteins that are present across all eukaryotes (Hollender and Liu, 2008). All members of this family contain a specific deacetylase domain that is required for their catalytic activity. It should be highlighted that this class of HDAs is able to deacetylate more targets than just histones. Lysine acetylome profiling uncovered 91 acetylated proteins in Arabidopsis leaves after the treatment with deacetylase inhibitors apicidin and trichostatin A. Of these, only 14 were histone-like proteins (Hartl et al., 2017). The second family is plant-specific and contains the HD-tuins (HD2). These type of proteins was originally found in maize. The amino acid sequence of HD-tuins is related to cis-trans prolyl isomerases, which are present in other eukaryotes (Aravind and Koonin, 1998; Bourque et al., 2016). HD2s are structurally distinct from RPD3-like members, but display a sequence similarity with FK506-binding proteins. In total, four members of HD-tuins have been identified in Arabidopsis: HDT1 (HD2A), HDT2 (HD2B), HDT3 (HD2C), and HDT4 (HD2D). These consist of an N-terminal domain that has a conserved pentapeptide MEFWG region, which is part of a gene repression activity (Bourque et al., 2016). This region is followed by a high-charged acidic motif that is rich in glutamic and/or aspartic acid and a variable C-terminal region (Dangl et al., 2001). Moreover, HDT1 and HDT3 possess a zinc-finger motif that probably is involved in protein-protein interaction and DNA-binding (Bourque et al., 2016). The third family of plant HDAs is represented by sirtuins (SIR2-like proteins), which are homologs to yeast silent information regulator 2 (SIR2) (König et al., 2014; Bheda et al., 2016). These HDAs are unique because they require a NAD cofactor for their function and unlike RPD3 proteins, they are not inhibited by trichostatin A or sodium butyrate. Moreover, sirtuins use a wide variety of substrates beyond histones (König et al., 2014; Bheda et al., 2016).
Similar as in humans/animals redox molecules modulate histone acetylation in plants, too. Two members of the plant RPD-3 like family (HDA9 and HDA19) are sensitive to oxidation; however, the physiological function of this modification is still not understood (Liu et al., 2015). Treatment of Arabidopsis seedlings with the physiological NO-donor GSNO increased the abundance of several histone 3 and histone 4 acetylation marks. Presence of the NO scavenger 2-4-carboxyphenyl-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) strongly diminished the abundance of these histone mark (Mengel et al., 2017). Since, GSNO and S-nitroso-N-acetyl-DL-penicillamine (SNAP) reversibly reduced total HDA activity both in vitro and in vivo, the increased acetylation was likely caused by NO-dependent inhibition of HDA activity. Moreover, the major plant defense hormone salicylic acid, inhibited HDA activity and increased histone acetylation by inducing endogenous NO production. Additionally, genome-wide NO-dependent H3K9/14 ac profiling in Arabidopsis seedlings identified NO-regulated histone acetylation of genes involved in plant defense response and abiotic stress response. This includes, for example, genes encoding for TIR class nucleotide-binding site-leucine-rich repeat (TIR-NBS-LRR) class disease resistance proteins and the transcription factors WRKY27, WRKY53, TGA2 and TGA5 (Mengel et al., 2017). Plant proteins belonging to the nucleotide-binding site-leucine-rich repeat (NBS-LRR) family are used for pathogen detection. These proteins detect pathogen-associated proteins, such as the effector molecules responsible for virulence. The TIR class of plant NBS-LRR proteins contains an additional amino-terminal domain homolog to the Toll and interleukin 1 receptors (DeYoung and Innes, 2006). WRKY transcription factors are key players in modulating the transcriptome during plant defense response. WRKY27 controls the expression of genes involved in nitrogen metabolism and NO production and negatively influences symptom development of Ralstonia solanacearum in Arabidopsis (Mukhtar et al., 2008). WRKY53 acts in a transcription factor signaling network mediating together with the EPITHIOSPECIFYING SENESCENCE REGULATOR a negative crosstalk between pathogen resistance and senescence, most likely controlled by the equilibrium between jasmonic acid and salicylic acid (Miao and Zentgraf (2007). TGA2 or TGA5 simultaneously bind to the TGACG motif of the Pathogenesis-related1 promoter activating expression of this defense gene (Zhang et al., 2003; Hussain et al., 2018). In sum, NO regulates histone acetylation by modifying and inhibiting HDA complexes. This results in hyperacetylation of specific genes enabling their transcription. This might be an important mechanism operating in the plant stress response and facilitating expression of stress-related genes (Figure 1; Mengel et al., 2017).
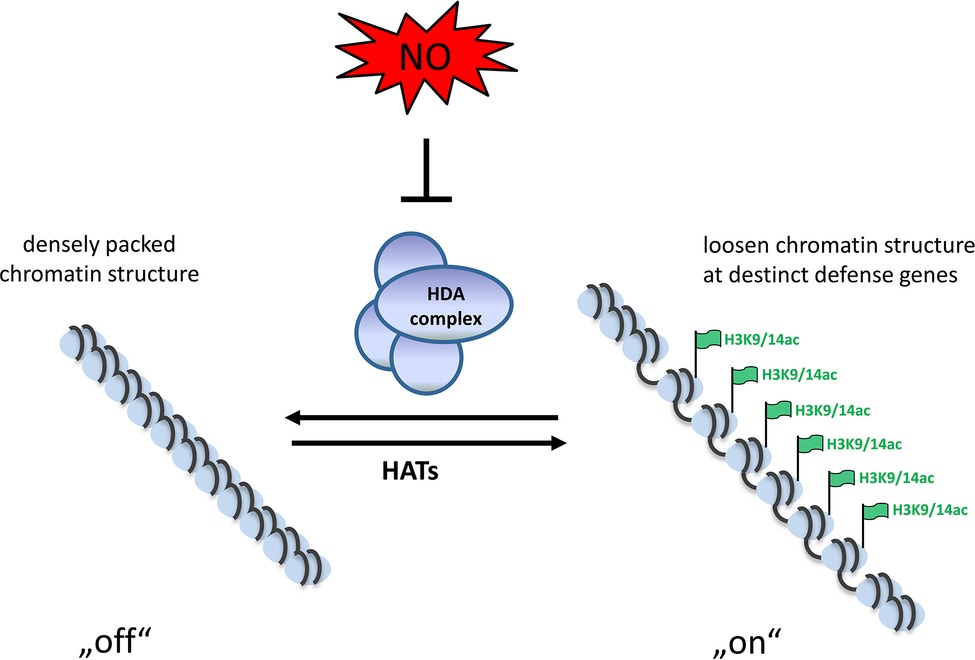
Figure 1. Schematic model of NO-induced chromatin modulation. Due to the activity of HDAs the chromatin is densely packed and genes are not transcribed. Upon formation of NO, the HDA-complexes become inhibited by S-nitrosation, leading to acetylation of the chromatin. This loosen chromatin structure allows transcription in tight interplay with activating transcription factors.
All members of the RPD3-superfamily contain several cysteine residues and in many cases, S-nitrosation of protein cysteine residues is conserved across the kingdoms. Human HDA2 is S-nitrosated at Cys262 and Cys274, which are located close to the catalytic center (Nott et al., 2008, 2013). A comparison of the amino acid sequence of human HDA2 and HDAs from different plant species revealed that HDA6 and HDA19 are closely related to human HDA2. The HDA domains of HDA6, HDA19 and HDA2 are highly similar (ca. 65–70% identity) whereas the C-terminal parts of the sequences are divergent (Figure 2). The highly conserved part contains the HDA domain including six highly conserved cysteine residues (Cys112, Cys163, Cys273, Cys285, Cys296, and Cys323 of Arabidopsis HDA6). Additionally, Cys325 of Arabidopsis HDA6 is highly conserved within the plant HDa6 and HDA19. The cysteine residues, which are targeted by NO in human HDA2 (Cys262 and Cys274) are located within the region, which is conserved in plant HDA6 and HDA19 (Figure 2). Moreover, structural modeling of the HDA domain of Glycine max HDA6 and HDA19 (HDA6 68.12% sequence identity to HDA2, Glycine max HDA19 69.81% sequence identity to human HDA2) based on the available crystal structure of HDA2 revealed a strikingly similar 3D-fold of these proteins, where these two conserved cysteines are located close to the substrate binding site at the same positions (Figure 3). This makes plant HDA6 and HDA19 promising candidates for NO-affected/regulated nuclear HDA isoform(s). In sum, NO-dependent regulation of plant HDAs can be considered as a key mechanism in regulation of histone acetylation and gene expression.
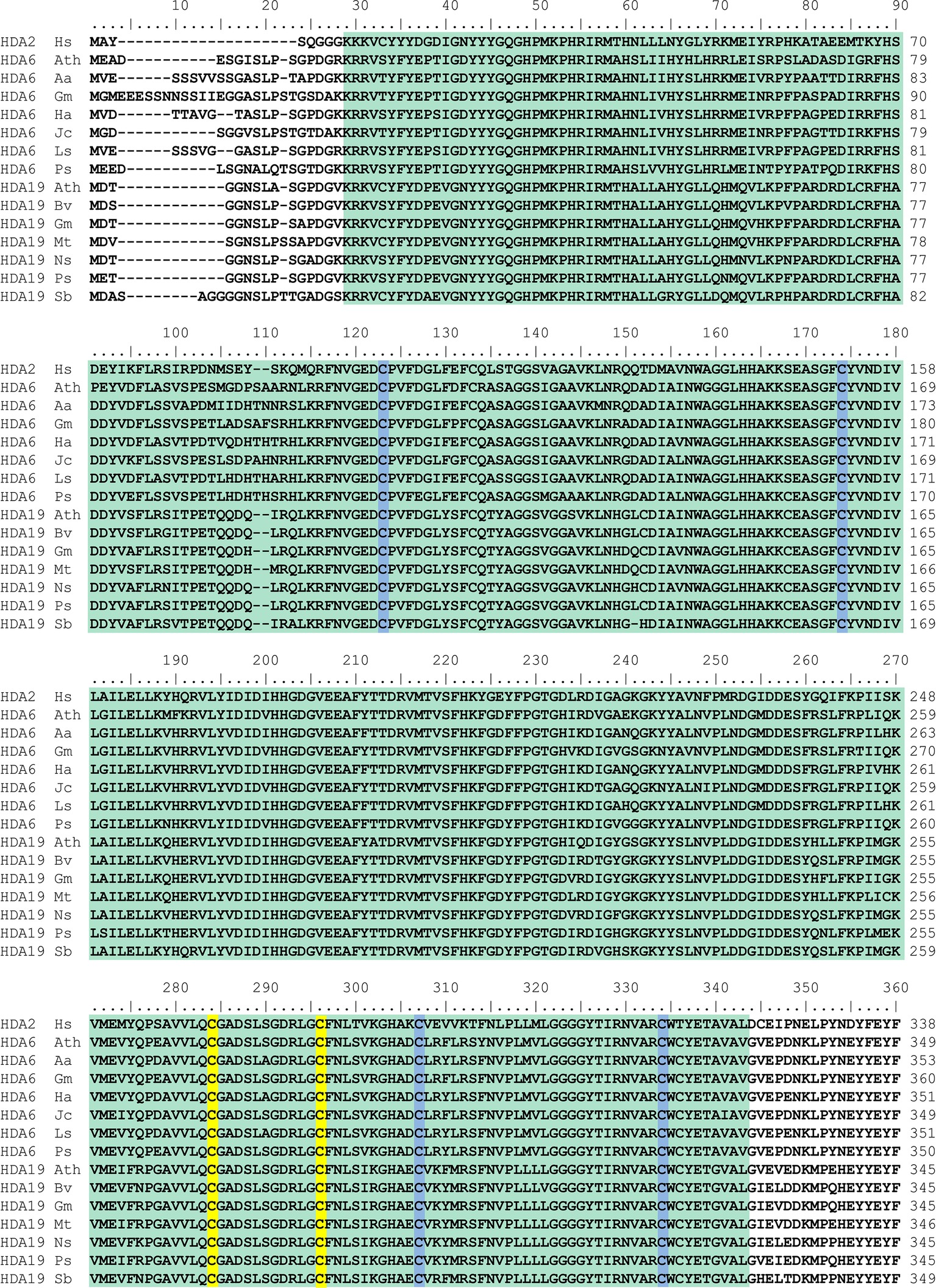
Figure 2. Alignment of the amino acid sequences of the HDA domain of human HDA2 and different plant histone deacetylase 6 and 19 proteins. HDA amino acid sequences were aligned using Clustal W. The HDA domain is depicted in green. Cysteine residues of human HDA2 which are targets for S-nitrosation and the corresponding cysteine residues of plant HDAs are highlighted in yellow. Other conserved cysteine residues are marked in blue. Hs, Homo sapiens NP_001518.3; Ath, Arabidospsis thaliana AED97705.1 (HDA6) and O22446.2 (HDA19); Aa, Artemisia annua PWA92260.1; Gm, Glycine max XP_003525556.1 (HDA6) and XP_003543935.1 (HDA19); Ha, Helianthus annuus XP_021978414.1; Jc, Jatropha curcas XP_012079994.1; Ls, Lactuca sativa XP_023740973.1; Ps, Papaver somniferum XP_026387130.1 (HDA6) and XP_026455725.1 (HDA19); Bv, Beta vulgaris XP_010690952.1; Mt., Medicago truncatula XP_013462369.1; Ns, Nicotiana sylvestris XP_009770456.1; Sb, Sorghum bicolor XP_002438614.1.
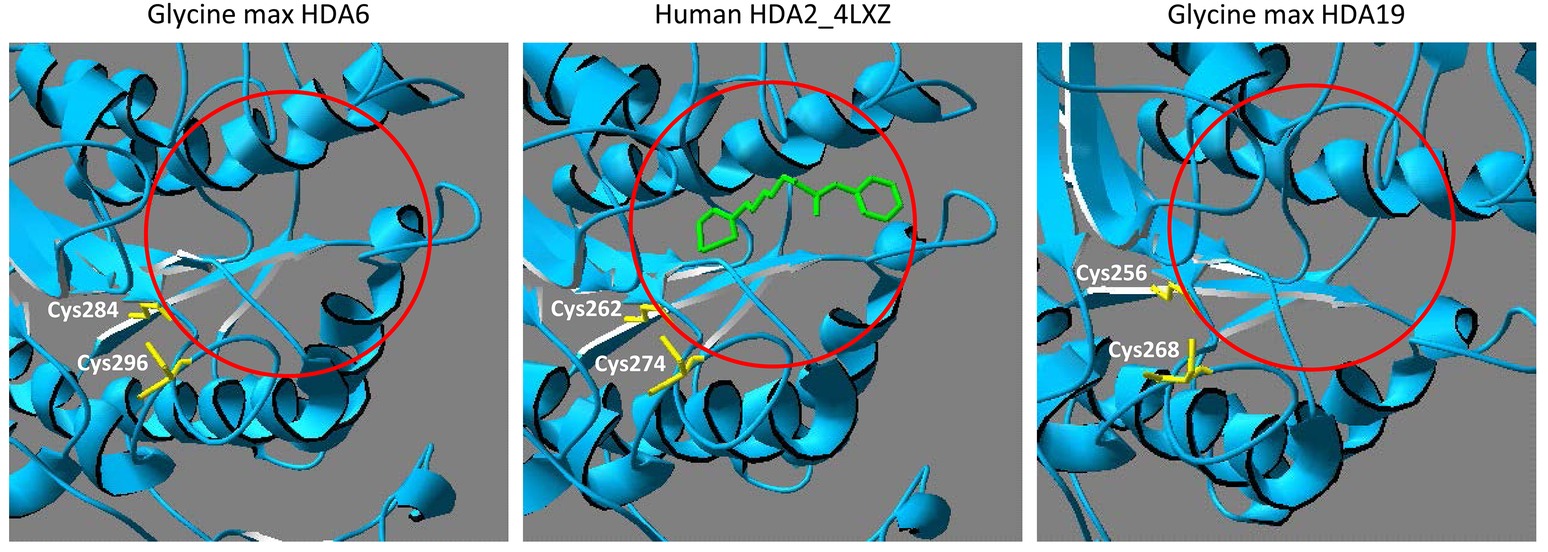
Figure 3. Comparison of HDA6, HDA19 and human HDA2 substrate binding site. Structural comparison between human HDA2 and Glycine max HDA6 and HDA19. The HDA domains of G. max HDA6 (amino acids 29–397, Uniprot entry I1MTD8) and of G. max HDA19 (amino acids 3–371, Uniprot entry A0A0R0H2W2) were modeled using the SwissProt Modeling server with human HDA2 as template (PDB entry 4LXZ). The histone deacetylase inhibitor octanedioic acid hydroxyamide phenylamide is highlighted in green and shows the location of the active site (mark with a red circle). Cysteine residues, which are located next to the active site are marked in yellow. These cysteine residues are targets for S-nitrosation in human HDA2.
Nitric Oxide Induces Expression of Demethylases and Methyltransferases and/or Affects Their Activities
Reports about the effect of NO on protein or DNA methylation in plants are rare. However, transcriptional changes in response to NO have been analyzed intensively using different techniques, for instance, cDNA-amplified fragment length polymorphism (Polverari et al., 2003), microarray (Huang et al., 2002), real-time PCR (Parani et al., 2004), and RNA-seq (Begara-Morales et al., 2014). The results of these different types of transcriptome studies demonstrated that NO is inducing a large set of genes involved in plant signal transduction, transport, defense and cell death, primary and secondary metabolism, and reactive oxygen species production and degradation. NO might regulate these genes by interacting directly with transcription factors or by regulating components of signal transduction cascades (Serpa et al., 2007; Lindermayr et al., 2010; Tavares et al., 2014; Imran et al., 2018). However, the accessibility of DNA is also an important regulatory mechanism in context of gene transcription. The accessibility of DNA can be regulated either by modification of histone tails (mainly acetylation and methylation) or by methylation of DNA. Interestingly, NO alters the expression of several methyltransferases and demethylases suggesting a regulatory role of NO in DNA and/or histone methylation (Ahlfors et al., 2009; Gibbs et al., 2014; Shi et al., 2014; Hussain et al., 2016; Kovacs et al., 2016a; Imran et al., 2018). For instance, CysNO and SNP treatment of leaves induced H3K27me3 Jumonji domain-containing histone demethylase 13 (JMJ13) expression (Ahlfors et al., 2009; Hussain et al., 2016) acting as a temperature- and photoperiod- dependent flowering repressor (Zheng et al., 2019). Enhanced endogenous levels of NO or GSNO, due to overexpression of rat neuronal NO synthase (nNOS) or knockout of GSNOR, respectively, results in downregulation of JMJ30 (Shi et al., 2014; Kovacs et al., 2016a), which demethylates H3K36me2/3, regulates period length in the circadian clock (Lu et al., 2011), and is involved in the control of flowering time (Yan et al., 2014). Transcriptomic analysis of NO-deficient noa1-2, nia1nia2, and nia1nia2noa1-2 mutants also revealed that enzymes involved in epigenetic methylation processes are differentially expressed. For instance, chromomethylase 2 (CMT2), responsible for CHH methylation at pericentromeric heterochromatin (Stroud et al., 2013), is downregulated in each mutant (Gibbs et al., 2014). Additionally, DNA METHYLTRANSFERASE 1 (MET1) maintaining CG methylation is upregulated in noa1-2, but downregulated in nia1nia2. Further, enzymes involved in the active DNA demethylation system such as REPRESSOR of SILENCING1 (ROS1) and DEMETER-like protein 2 (DML2) (Furner and Matzke, 2011) are differently expressed in these NO-deficient mutants. Interestingly, several protein arginine methyltransferases (PRMTs) are upregulated in NO-deficient plants, for example, PRMT1a, PRMT1b, PRMT3, PRMT10, PRMT5 in noa1-2; PRMT1b, PRMT3, PRMT4B in nia1nia2; and PRMT1b, PRMT3, PRMT4B, PRMT10, PRMT5 in nia1nia2noa1-2 (Gibbs et al., 2014). PRMT1b, upregulated in all three NO-deficient mutants, methylates H4R3 and non-histone proteins such as the RNA methyltransferase fibrillarin 2 (Yan et al., 2007). Another example is PRMT5, which catalyze symmetric dimethylation of H4R3 in vitro and is essential for proper pre-mRNA splicing (Deng et al., 2010). PRMT5 is upregulated in noa1-2 and nia1nia2noa1-2 (Gibbs et al., 2014) and positively regulated by S-nitrosylation during stress responses (Hu et al., 2017). Regarding the late flowering phenotype of these NO-deficient mutants, it is worth mentioning that the histone demethylases JMJ18 is downregulated in each mutant. JMJ18 is a H3K4 demethylase controlling flowering time (Yang et al., 2012).
NO-dependent changes in DNA-methylation have been described in rice plants exposed to 0.5 mM NO donor sodium nitroprusside (Ou et al., 2015). The treatment resulted in stress symptoms and complete growth inhibition accompanied by hypomethylation of genomic DNA predominantly at CHG sites. As a consequence, transcription of a number of genes and transposable elements was activated and expression of several genes involved in chromatin remodeling and DNA methylation homeostasis, for example, chromomethylase 3, deficient in DNA methylation 1a and 1b, and DEMETER, was disturbed (Ou et al., 2015). In these cases, DNA methylation might be regulated via differential expression of DNA-methyl modifiers. However, modulation of their activity by NO-based post-translational modifications cannot be excluded.
Recently, it was shown that NO regulates protein methylation during stress responses in Arabidopsis plants (Hu et al., 2017). The authors demonstrated that S-nitrosation of protein arginine methyltransferase 5 (PRMT5), an enzyme catalyzing symmetric demethylation of protein arginine residues, activates its enzyme activity leading to proper splicing-specific pre-mRNA of stress-related genes (Hu et al., 2017). Although this mechanism does not evolve alteration of the chromatin structure, other studies showed that PRMT5 is a highly conserved type II protein Arg methyltransferase, which amongst others interacts with and methylates histones (Hu et al., 2017). In the prmt5-1 mutant methylation of several proteins in the range of a molecular mass of 14 kD, including histone H4 and several core components of small nuclear ribonucleoprotein of the spliceosome, was barely detectable, while this phenotype could be rescued by a PRMT5 transgene (Hu et al., 2017). Therefore, it cannot be excluded that S-nitrosation of PRMT5 also affects chromatin structure by methylating defined histone Arg residues.
In mammals, NO exposure results in decease in global 5-methylcytosine and activation of transcriptional response. This might be linked to decreased expression of DNA methyltransferases Dnmt1 and Dnmt3a. Moreover, significant changes in the methylation level of H3K9, H3K27, H3K36, and H4K20 were observed in presence of NO. These changes are well-known as important modulators of gene transcription (Vasudevan et al., 2015) and are mainly a consequence of inhibition of KDM3A/JmjC histone demethylase activities and/or increased expression of a set of histone de-methylases (KDM1, KDM3A, KDM3B, KDM4A, KDM4B, KDM4C, KDM4D, and KDM7A). JmjC histone demethylases catalyze demethylation of mono-, di-, and trimethylated lysine residues by an oxidative, Fe(II)-dependent mechanism. NO directly inhibits the KDM3A demethylase activity by forming a nitrosyl-iron complex in the active site (Hickok et al., 2013) and exposure of mammalian cells to NO resulted in a significant increase in H3K9me2, the preferred substrate for KDM3A. Furthermore, exposure of embryonic stem cells to DETA-NO caused increased H3K4me1/2/3 and H3K9me3 methylation (Mora-Castilla et al., 2010) and concentration- and time-dependent accumulation of H3K9me2 upon DETA-NO treatment of human breast carcinoma cells was reported (Hickok et al., 2013).
Eukaryotic JmjC genes are separated in 14 subfamilies: Lysine-Specific Demethylase (KDM) 3, KDM5, JMJD6, and Putative-Lysine-Specific Demethylase (PKDM) 11, and PKDM13 subfamilies present in plants, animals, and fungi. Other subfamilies are detected only in plants and animals (PKDM12) or in animals and fungi but not in plants (KDM2 and KDM4). PKDM7-9 are plant-specific groups. The existence of Jumonji C-containing histone demethylases in plants suggested that at least some might be regulated by NO, similar the ones described in mammals.
Nitric Oxide Regulates Histone and DNA-Methylation on Metabolite Level
Methylation of histones and DNA can be regulated on two main levels–on the level of methyltransferases and de-methylases, which are catalyzing the methylation/de-methylation reactions, and on metabolite level. S-Adenosylmethionine (SAM) is the major methyl group donor in the cell. DNA and histones are subject to methylation by specific SAM-dependent methyltransferases. Each transfer of a methyl-group generates S-adenosylhomocysteine (SAH), which is cleaved into homocysteine and adenosine (Ado) by S-adenosylhomocysteine hydrolase (SAHH) (Kovacs et al., 2016b). It is known already for a long time that the equilibrium of this reversible reaction favoring SAH synthesis (Palmer and Abeles, 1976) is driven toward hydrolysis of SAH due to removal of its products (Poulton and Butt, 1976). Methionine synthase converts homocysteine to methionine, which is in turn adenylated to SAM by SAM synthetase, whereas Ado is metabolized in the Ado salvage cycle. The ratio of SAM and SAH is considered as important regulator of cellular methylation processes.
For example, Zhou et al. (2013) reported the importance of a balanced SAM/SAH ratio for DNA and H3K9me2 methylation. The defect of folate polyglutamylation caused by mutation of folylpolyglutamate synthase affects the folate-mediated one-carbon metabolism resulting in increased SAH level, reduced SAM/SAH ratio and continuatively in a reduced DNA methylation and H3K9me2. A connection between the folate cycle, DNA methylation and redox homeostasis is also reported by Groth et al. (2016). Mutation in methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase (MTHFD1) results in loss of DNA methylation. The authors assume that reduced MTHFD1 function disturbs the cellular redox state, which might affect enzyme activities of the methylation cycle and/or methylation/demethylation reactions.
Other studies highlighted the importance of SAHH activity toward chromatin modifications (reviewed in Pikaard and Mittelsten Scheid (2014) and Vriet et al. (2015)): Mutation of the AtSAHH1 gene resulted in reduced cytosine methylation and release of transcriptional gene silencing (Rocha et al., 2005; Mull et al., 2006; Jordan et al., 2007). Moreover, silencing of SAHH expression in tobacco plants lead to loss of DNA methylation in repetitive elements (Tanaka et al., 1997). Furthermore, in Arabidopsis levels of DNA and histone methylation at endogenous repeats are reduced after treatment with the SAHH inhibitor dihydroxypropyladenine (Baubec et al., 2010).
There are several hints that the supply of SAM and the removal of SAH—the by-product inhibitor of transmethyl reactions—are at least partly regulated by NO, since the activities of key enzymes of the methylation cycle seems to be modulated by NO-dependent posttranslational modifications. In several independent proteomic studies, cobalamin-independent methionine synthase, SAMS, and SAHH were identified as targets for S-nitrosation (Lindermayr et al., 2005; Abat and Deswal, 2009; Puyaubert et al., 2014; Hu et al., 2015). In Arabidopsis, different SAMS isoforms are differentially inhibited by protein S-nitrosation (Lindermayr et al., 2006). While isoform SAMS1 is reversibly inhibited by GSNO, the activity of the isoforms SAMS2 or SAMS3 is not affected. Responsible for the inhibition of SAMS1 is S-nitrosation of Cys114, which is located nearby the catalytic center as part of the active site loop (Lindermayr et al., 2006). In mammals, a similar regulatory mechanism of SAMS activity is described. Here two SAMS isoforms are present. While the activity of SAMS1A is reversibly inhibited by NO, SAMS2A activity is not affected (Pérez-Mato et al., 1999).
In Arabidopsis, two genes encode for SAHH, but only SAHH1 seems to play a role in DNA-methylation processes (Rocha et al., 2005; Vriet et al., 2015). S-Nitrosation of Arabidopsis SAHH1 upon cold stress was reported, but the physiological function of S-nitrosated SAHH1 is not yet investigated (Puyaubert et al., 2014; Puyaubert and Baudouin, 2014). Beside S-nitrosation, tyrosine nitration has been observed in sunflower (Helianthus annuus L.) SAHH. This modification decreased the catalytic activity of SAHH (Chaki et al., 2009). The results of all these different studies suggest that NO plays an important regulatory role in allocation of the major methyl-group donor SAM and the removal of the methyltransferase inhibitor SAH, which consequently affects histone and DNA methylation.
In sum, there are different levels, where NO can affect chromatin modulation, for instance transcription and activity of chromatin modifiers or the supply of methyl group donors or methylation inhibitors (summarized in Figure 4). Since NO is an important signaling molecule in plant growth and development and in plant stress response all these different mechanisms discussed above allows NO to regulate physiological processes. The most important future challenges are the identification and/or verification of NO-regulated chromatin modifiers responsible for histone modifications and DNA methylation and the characterization of the mode of action of NO on these proteins. Moreover, the corresponding histone marks and the chromatin regions controlled by NO-regulated chromatin modifiers have to be identified. Such analysis would also include possible interaction between histone marks and DNA-methylation. Additionally, these NO-dependent chromatin modifications have to be analyzed in a physiological context to complement the picture of NO signaling function in plants.
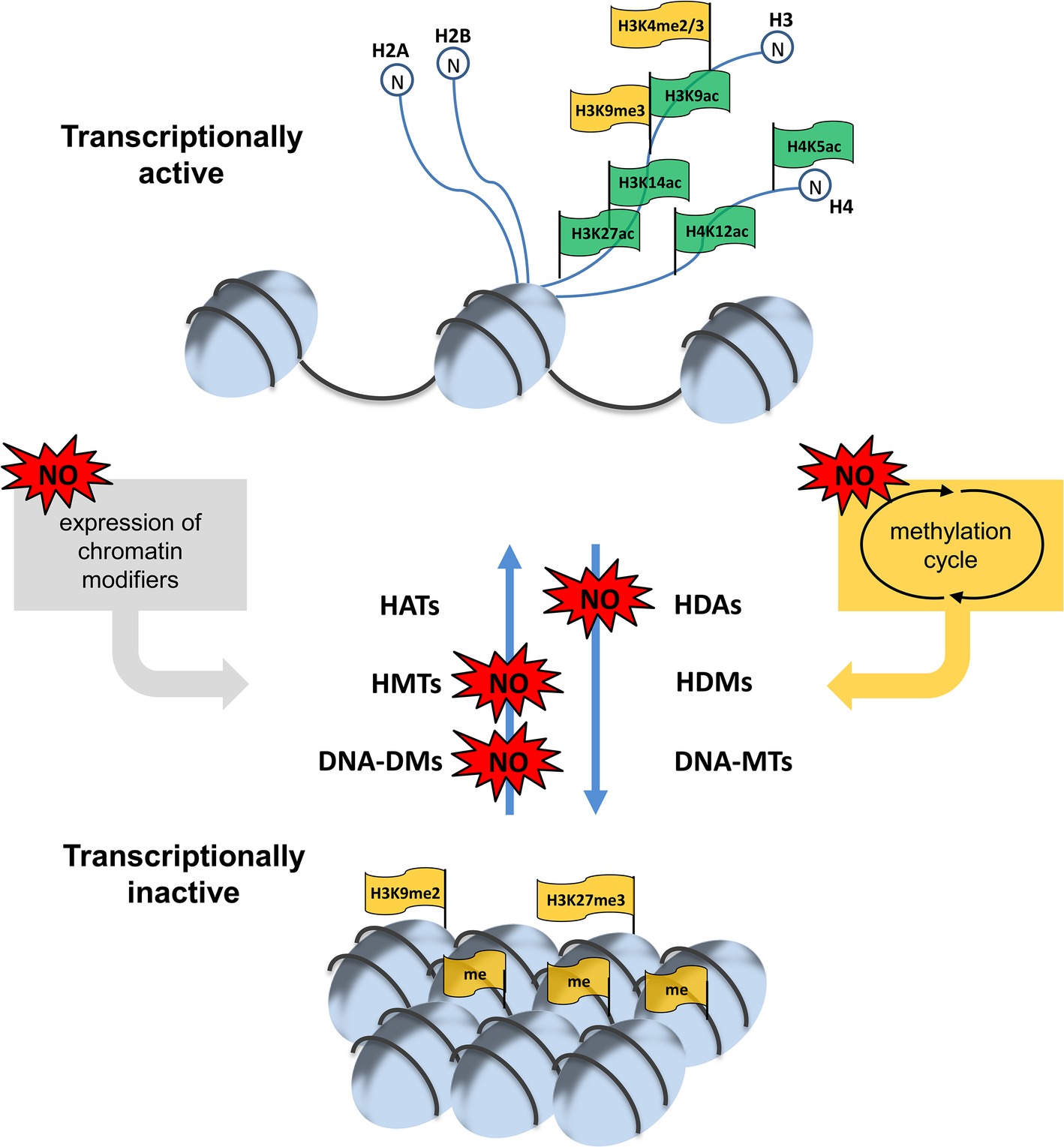
Figure 4. NO-dependent regulation of chromatin modulation. Histone acetylation/methylation and DNA-methylation is controlled by different sets of acetylases/deacetylases (HATs/HDAs) and methyl transferases/demethylases (HMT/HDMs and DNA-MTs/DNA-DMs). NO can regulate the expression of some of these chromatin modifiers as well as their activity. Moreover, NO can affect the supply of the methyl group donor SAM and the level of the methyltransferase inhibitor SAH by altering the activity of enzymes of the methylation cycle and/or connected pathways. For more details see this paper.
Finally, the gained knowledge could be subjected to genetic/epigenetic engineering or classical breeding to improve plant traits permanently.
Author Contributions
AA-K, ER and CL wrote the review article. CL coordinated the completion of the article.
Funding
This work was supported by the Bundesministerium fürBildung und Forschung.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abat, J. K., and Deswal, R. (2009). Differential modulation of S-nitrosoproteome of Brassica juncea by low temperature: change in S-nitrosylation of Rubisco is responsible for the inactivation of its carboxylase activity. Proteomics 9, 4368–4380. doi: 10.1002/pmic.200800985
Ahlfors, R., Brosché, M., Kollist, H., and Kangasjärvi, J. (2009). Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 58, 1–12. doi: 10.1111/j.1365-313X.2008.03756.x
Aravind, L., and Koonin, E. V. (1998). Second Family of Histone Deacetylases. Science 280:1167. doi: 10.1126/science.280.5367.1167a
Aroca, A., Schneider, M., Scheibe, R., Gotor, C., and Romero, L. C. (2017). Hydrogen sulfide regulates the cytosolic/nuclear partitioning of glyceraldehyde-3-phosphate dehydrogenase by enhancing its nuclear localization. Plant Cell Physiol. 58, 983–992. doi: 10.1093/pcp/pcx056
Astier, C. P. J., Besson-Bard, A., Wawer, I., Rasul, D. W. S., Jeandroz, S., and Dat, J. F. (2010). Nitric oxide signalling in plants: cross-talk with Ca2+, protein kinases and reactive oxygen species. Annu. Plant Rev. 42, 147–170. doi: 10.1002/9781119312994.apr0454
Astier, J., Kulik, A., Koen, E., Besson-Bard, A., Bourque, S., Jeandroz, S., et al. (2012). Protein S-nitrosylation: what’s going on in plants? Free Radic. Biol. Med. 53, 1101–1110. doi: 10.1016/j.freeradbiomed.2012.06.032
Baerenfaller, K., Shu, H., Hirsch-Hoffmann, M., Fütterer, J., Opitz, L., Rehrauer, H., et al. (2016). Diurnal changes in the histone H3 signature H3K9ac|H3K27ac|H3S28p are associated with diurnal gene expression in Arabidopsis. Plant Cell Environ. 39, 2557–2569. doi: 10.1111/pce.12811
Bannister, A. J., and Kouzarides, T. (2011). Regulation of chromatin by histone modifications. Cell Res. 21, 381–395. doi: 10.1038/cr.2011.22
Baubec, T., Dinh, H. Q., Pecinka, A., Rakic, B., Rozhon, W., Wohlrab, B., et al. (2010). Cooperation of multiple chromatin modifications can generate unanticipated stability of epigenetic states in Arabidopsis. Plant Cell 22, 34–47. doi: 10.1105/tpc.109.072819
Begara-Morales, J. C., Sánchez-Calvo, B., Luque, F., Leyva-Pérez, M. O., Leterrier, M., Corpas, F. J., et al. (2014). Differential transcriptomic analysis by RNA-Seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 55, 1080–1095. doi: 10.1093/pcp/pcu044
Beligni, L., and Lamattina, M. (1998). Nitric oxide counteracts cytotoxic processes mediated by reactive oxygen species in plant tissues. Planta 208, 337–344.
Bethke, P. C., Badger, M. R., and Jones, R. L. (2004). Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16, 332–341. doi: 10.1105/tpc.017822
Bheda, P., Jing, H., Wolberger, C., and Lin, H. (2016). The substrate specificity of sirtuins. Annu. Rev. Biochem. 85, 405–429. doi: 10.1146/annurev-biochem-060815-014537
Bourque, S., Jeandroz, S., Grandperret, V., Lehotai, N., Aimé, S., Soltis, D. E., et al. (2016). The evolution of HD2 proteins in green plants. Trends Plant Sci. 21, 1008–1016. doi: 10.1016/j.tplants.2016.10.001
Cañas, A., López-Sánchez, L. M., Valverde-Estepa, A., Mengel, A., Hernández, V., Fuentes, E., et al. (2012). Maintenance of S-nitrosothiol homeostasis plays an important role in growth suppression of estrogen receptor-positive breast tumors. Breast Cancer Res. 14:R153. doi: 10.1186/bcr3366
Chaki, M., Shekariesfahlan, A., Ageeva, A., Mengel, A., von Toerne, C., Durner, J., et al. (2015). Identification of nuclear target proteins for S-nitrosylation in pathogen-treated Arabidopsis thaliana cell cultures. Plant Sci. 238, 115–126. doi: 10.1016/j.plantsci.2015.06.011
Chaki, M., Valderrama, R., Fernández-Ocaña, A. M., Carreras, A., López-Jaramillo, J., Luque, F., et al. (2009). Protein targets of tyrosine nitration in sunflower (Helianthus annuus L.) hypocotyls. J. Exp. Bot. 60, 4221–4234. doi: 10.1093/jxb/erp263
Clark, D. F. K., Durner, J., and Navarre, D. A. (2000). Nitric oxide inhibition of tobacco catalase and ascorbate peroxidase. Mol. Plant-Microbe Interact. 13, 1380–1384. doi: 10.1094/MPMI.2000.13.12.1380
Colussi, C., Mozzetta, C., Gurtner, A., Illi, B., Rosati, J., Straino, S., et al. (2008). HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. USA 105, 19183–19187. doi: 10.1073/pnas.0805514105
Corpas, F. J., and Barroso, J. B. (2018). Peroxisomal plant metabolism–an update on nitric oxide, Ca2+ and the NADPH recycling network. J. Cell Sci. 131. pii: jcs202978. doi: 10.1242/jcs.202978
Correa-Aragunde, N., Lamattina, L., and Graziano, M. (2004). Nitric oxide plays a central role in determining lateral root development in tomato. Planta 218, 900–905. doi: 10.1007/s00425-003-1172-7
Dangl, M., Brosch, G., Haas, H., Loidl, P., and Lusser, A. (2001). Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213, 280–285. doi: 10.1007/s004250000506
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
Delorme-Hinoux, V., Bangash, S. A. K., Meyer, K. J., and Reichheld, J.-P. (2016). Nuclear thiol redox systems in plants. Plant Sci. 243, 84–95. doi: 10.1016/j.plantsci.2015.12.002
Deng, X., Gu, L., Liu, C., Lu, T., Lu, F., Lu, Z., et al. (2010). Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc. Natl. Acad. Sci. USA 107, 19114–19119. doi: 10.1073/pnas.1009669107
DeYoung, B. J., and Innes, R. W. (2006). Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 7, 1243–1249. doi: 10.1038/ni1410
Farnese, F. S., Menezes-Silva, P. E., Gusman, G. S., and Oliveira, J. A. (2016). When Bad Guys Become Good Ones: The Key Role of Reactive Oxygen Species and Nitric Oxide in the Plant Responses to Abiotic Stress. Front Plant Sci. 7:471. doi: 10.3389/fpls.2016.00471
Feng, J. H., Jing, F. B., Fang, H., Gu, L. C., and Xu, W. F. (2001). Expression, purification, and S-nitrosylation of recombinant histone deacetylase 8 in Escherichia coli. Biosci. Trends 5, 17–22. doi: 10.5582/bst.2011.v5.1.17
Foresi, N., Correa-Aragunde, N., Parisi, G., Caló, G., Salerno, G., and Lamattina, L. (2010). Characterization of a Nitric Oxide Synthase from the Plant Kingdom: NO Generation from the Green Alga Ostreococcus tauri Is Light Irradiance and Growth Phase Dependent. Plant Cell 22, 3816–3830. doi: 10.1105/tpc.109.073510
Furner, I. J., and Matzke, M. (2011). Methylation and demethylation of the Arabidopsis genome. Curr. Opin. Plant Biol. 14, 137–141. doi: 10.1016/j.pbi.2010.11.004
Gibbs, D. J., MdIsa, N., Movahedi, M., Lozano-Juste, J., Mendiondo, G. M., Berckhan, S., et al. (2014). Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53, 369–379. doi: 10.1016/j.molcel.2013.12.020
Groß, F., Durner, J., and Gaupels, F. (2013). Nitric oxide, antioxidants and prooxidants in plant defence responses. Front. Plant Sci. 4:419. doi: 10.3389/fpls.2013.00419
Groth, M., Moissiard, G., Wirtz, M., Wang, H., Garcia-Salinas, C., Ramos-Parra, P. A., et al. (2016). MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 7:11640. doi: 10.1038/ncomms11640
Hara, M. R., Agrawal, N., Kim, S., Cascio, M. B., Fujimuro, M., Ozeki, Y., et al. (2005). S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674. doi: 10.1038/ncb1268
Hara, M. R., Thomas, B., Cascio, M. B., Bae, B. I., Hester, L. D., Dawson, V. L., et al. (2006). Neuroprotection by pharmacologic blockade of the GAPDH death cascade. Proc. Natl. Acad. Sci. USA 103, 3887–3889. doi: 10.1073/pnas.0511321103
Hartl, M., Füßl, M., Boersema, P. J., Jost, J. O., Kramer, K., Bakirbas, A., et al. (2017). Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 23, 949–964. doi: 10.15252/msb.20177819
He, Y., Tang, R. H., Hao, Y., Stevens, R. D., Cook, C. W., Ahn, S. M., et al. (2004). Nitric oxide represses the Arabidopsis floral transition. Science 305, 1968–1971. doi: 10.1126/science.1098837
Hickok, J. R., Vasudevan, D., Antholine, W. E., and Thomas, D. D. (2013). Nitric oxide modifies global histone methylation by inhibiting Jumonji C domain-containing demethylases. J. Biol. Chem. 288, 16004–16015. doi: 10.1074/jbc.M112.432294
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Holtgrefe, S., Gohlke, J., Starmann, J., Druce, S., Klocke, S., Altmann, B., et al. (2008). Regulation of plant cytosolic glyceraldehyde 3-phosphate dehydrogenase isoforms by thiol modifications. Physiol. Plant. 133, 211–228. doi: 10.1111/j.1399-3054.2008.01066.x
Hong, J. K., Yun, B. W., Kang, J. G., Raja, M. U., Kwon, E., Sorhagen, K., et al. (2008). Nitric oxide function and signalling in plant disease resistance. J. Exp. Bot. 59, 147–154. doi: 10.1093/jxb/erm244
Hu, J., Huang, X., Chen, L., Sun, X., Lu, C., Zhang, L., et al. (2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 167, 1731–1746. doi: 10.1104/pp.15.00026
Hu, J., Yang, H., Mu, J., Lu, T., Peng, J., Deng, X., et al. (2017). Nitric oxide regulates protein methylation during stress responses in plants. Mol. Cell 67, 702–710. doi: 10.1016/j.molcel.2017.06.031
Huang, X., von Rad, U., and Durner, J. (2002). Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215, 914–923. doi: 10.1007/s00425-002-0828-z
Hussain, A., Mun, B.-G., Imran, Q. M., Lee, S.-U., Adamu, T. A., Shahid, M., et al. (2016). Nitric oxide mediated transcriptome profiling reveals activation of multiple regulatory pathways in Arabidopsis thaliana. Front. Plant Sci. 7, 1–18. doi: 10.3389/fpls.2016.00975
Hussain, R. M. F., Sheikh, A. H., Haider, I., Quareshy, M., and Linthorst, H. J. M. (2018). Arabidopsis WRKY50 and TGA transcription factors synergistically activate expression of PR1. Front. Plant Sci. 9:930. doi: 10.3389/fpls.2018.00930
Illi, B., Dello Russo, C., Colussi, C., Rosati, J., Pallaoro, M., Spallotta, F., et al. (2008). Nitric oxide modulates chromatin folding in human endothelial cells via protein phosphatase 2A activation and class II histone deacetylases nuclear shuttling. Circ. Res. 102, 51–58. doi: 10.1161/CIRCRESAHA.107.157305
Imran, Q. M., Hussain, A., Lee, S. U., Mun, B. G., Falak, N., Loake, G. J., et al. (2018). Transcriptome profile of NO-induced Arabidopsis transcription factor genes suggests their putative regulatory role in multiple biological processes. Sci. Rep. 15:771. doi: 10.1038/s41598-017-18850-5
Jaenisch, R., and Bird, A. (2003). Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254. doi: 10.1038/ng1089
Jordan, N. D., West, J. P., Bottley, A., Sheikh, M., and Furner, I. (2007). Transcript profiling of the hypomethylated hog1 mutant of Arabidopsis. Plant Mol. Biol. 65, 571–586. doi: 10.1007/s11103-007-9221-4
Kim, W., Latrasse, D., Servet, C., and Zhou, D. X. (2013). Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 432, 394–398. doi: 10.1016/j.bbrc.2012.11.102
Kneeshaw, S., Gelineau, S., Tada, Y., Loake, G. J., and Spoel, S. H. (2014). Selective protein denitrosylation activity of thioredoxin-h5 modulates plant immunity. Mol. Cell 56, 153–162. doi: 10.1016/j.molcel.2014.08.003
Kolbert, Z., Feigl, G., Borde, A., Molnar, A., and Erdei, L. (2017). Protein tyrosine nitration in plants: present knowledge, computational prediction and future perspectives. Plant Physiol. Biochem. 113, 56–63. doi: 10.1016/j.plaphy.2017.01.028
König, A. C., Hartl, M., Pham, P. A., Laxa, M., Boersema, P. J., Orwat, A., et al. (2014). The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 164, 1401–1414. doi: 10.1104/pp.113.232496
Kovacs, I., Ageeva, A., König, E.-E., and Lindermayr, C. (2016b). S-Nitrosylation of nuclear proteins: new pathways in regulation of gene expression. Adv. Bot. Res. 77, 15–39. doi: 10.1016/bs.abr.2015.10.003
Kovacs, I., Holzmeister, C., Wirtz, M., Geerlof, A., Fröhlich, T., Römling, G., et al. (2016a). ROS-mediated inhibition of S-nitrosoglutathione reductase contributes to the activation of anti-oxidative mechanisms. Front. Plant Sci. 7, 1–17. doi: 10.3389/fpls.2016.01669
Kuruthukulangarakoola, G. T., Zhang, J., Albert, A., Winkler, B., Lang, H., Buegger, F., et al. (2017). Nitric oxide-fixation by non-symbiotic haemoglobin proteins in Arabidopsis thaliana under N-limited conditions. Plant Cell Environ. 40, 36–50. doi: 10.1111/pce.12773
Li, Y., Dong, X. M., Jin, F., Shen, Z., Chao, Q., and Wang, B. C. (2017). Histone Acetylation Modifications Affect Tissue-Dependent Expression of Poplar Homologs of C4 Photosynthetic Enzyme Genes. Front Plant Sci. 8:950. doi: 10.3389/fpls.2017.00950
Lancaster, J. R. Jr. (2015). Nitric oxide: a brief overview of chemical and physical properties relevant to therapeutic applications. Future Sci. OA 1, 1–5. doi: 10.4155/fso.15.59
Lindermayr, C., Saalbach, G., Bahnweg, G., and Durner, J. (2006). Differential inhibition of Arabidopsis methionine adenosyltransferases by protein S-nitrosylation. J. Biol. Chem. 281, 4285–4291. doi: 10.1074/jbc.M511635200
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Lindermayr, C., Sell, S., Müller, B., Leister, D., and Durner, J. (2010). Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22, 2894–2907. doi: 10.1105/tpc.109.066464
Liu, P., Zhang, H., Yu, B., Xiong, L., and Xia, Y. (2015). Proteomic identification of early salicylate- and flg22-responsive redox-sensitive proteins in Arabidopsis. Sci. Rep. 5:8625. doi: 10.1038/srep08625
López-Sánchez, L. M., Corrales, F. J., González, R., Ferrín, G., Muñoz-Castañeda, J. R., Ranchal, I., et al. (2008). Alteration of S-nitrosothiol homeostasis and targets for protein S-nitrosation in human hepatocytes. Proteomics 8, 4709–4720. doi: 10.1002/pmic.200700313
Lu, S. X., Knowles, S. M., Webb, C. J., Celaya, R. B., Cha, C., Siu, J. P., et al. (2011). The Jumonji C domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol. 155, 906–915. doi: 10.1104/pp.110.167015
Luger, K., Dechassa, M. L., and Tremethick, D. J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13, 436–447. doi: 10.1038/nrm3382
Luo, M., Yu, C. W., Chen, F. F., Zhao, L., Tian, G., Liu, X., et al. (2012). Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 8:e1003114. doi: 10.1371/journal.pgen.1003114
Malik, M., Shukla, A., Amin, P., Niedelman, W., Lee, J., Jividen, K., et al. (2010). S-nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4. J. Biol. Chem. 285, 23818–23828. doi: 10.1074/jbc.M109.091611
Martins, L., Trujillo-Hernandez, J. A., and Reichheld, J. P. (2018). Thiol based redox signaling in plant nucleus. Front. Plant Sci. 9:705. doi: 10.3389/fpls.2018.00705
Mata, C. G., and Lamattina, L. (2001). Nitric oxide induces stomatal closure and enhances the adaptive plant responses against drought stress. Plant Physiol. 126, 1196–1204. doi: 10.1104/pp.126.3.1196
Mata-Pérez, C., Begara-Morales, J. C., Chaki, M., Sánchez-Calvo, B., Valderrama, R., Padilla, M. N., et al. (2016). Protein tyrosine nitration during development and abiotic stress response in plants. Front. Plant Sci. 7:1699. eCollection. doi: 10.3389/fpls.2016.01699
Mengel, A., Ageeva, A., Georgii, E., Bernhardt, J., Wu, K., Durner, J., et al. (2017). Nitric oxide modulates histone acetylation at stress genes by inhibition of histone deacetylases. Plant Physiol. 173, 1434–1452. doi: 10.1104/pp.16.01734
Miao, Y., and Zentgraf, U. (2007). The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 19, 819–830. doi: 10.1105/tpc.106.042705
Minard, M. E., Jain, A. K., and Barton, M. C. (2009). Analysis of epigenetic alterations to chromatin during development. Genesis 47, 559–572. doi: 10.1002/dvg.20534
Mora-Castilla, S., Tejedo, J. R., Hmadcha, A., Cahuana, G. M., Martín, F., Soria, B., et al. (2010). Nitric oxide repression of Nanog promotes mouse embryonic stem cell differentiation. Cell Death Differ. 17, 1025–1033. doi: 10.1038/cdd.2009.204
Mukhtar, M. S., Deslandes, L., Auriac, M. C., Marco, Y., and Somssich, I. E. (2008). The Arabidopsis transcription factor WRKY27 influences wilt disease symptom development caused by Ralstonia solanacearum. Plant J. 56, 935–947. doi: 10.1111/j.1365-313X.2008.03651.x.
Mull, L., Ebbs, M. L., and Bender, J. (2006). A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics 174, 1161–1171. doi: 10.1534/genetics.106.063974
Nott, A., Nitarska, J., Veenvliet, J. V., Schacke, S., Derijck, A. A., Sirko, P., et al. (2013). S-nitrosylation of HDAC2 regulates the expression of the chromatin-remodeling factor BRM during radial neuron migration. Proc. Natl. Acad. Sci. USA 110, 3113–3118. doi: 10.1073/pnas.1218126110
Nott, A., Watson, P. M., Robinson, J. D., Crepaldi, L., and Riccio, A. (2008). S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature 455, 411–415. doi: 10.1038/nature07238
Okuda, K., Ito, A., and Uehara, T. (2015). Regulation of histone deacetylase 6 activity via S-nitrosylation. Biol. Pharm. Bull. 38, 1434–1437. doi: 10.1248/bpb.b15-00364
Ou, X., Zhuang, T., Yin, W., Miao, Y., Wang, B., Zhang, Y., et al. (2015). DNA methylation changes induced in rice by exposure to high concentrations of the nitric oxide modulator, sodium nitroprusside. Plant Mol. Biol. Report. 33, 1428–1440. doi: 10.1007/s11105-014-0843-9
Palmer, J. L., and Abeles, R. H. (1976). Mechanism for enzymatic thioether formation. Mechanism of action of S-adenosylhomocysteinase. J. Biol. Chem. 251, 5817–5819.
Parani, M., Rudrabhatla, S., Myers, R., Weirich, H., Smith, B., Leaman, D. W., et al. (2004). Microarray analysis of nitric oxide responsive transcripts in Arabidopsis. Plant Biotechnol. J. 2, 359–366. doi: 10.1111/j.1467-7652.2004.00085.x
Pedroso, M. C., Magalhaes, J. R., and Durzan, D. (2000). Nitric oxide induces cell death in Taxus cells. Plant Sci. 157, 173–180. doi: 10.1016/S0168-9452(00)00278-8
Perazzolli, M., Dominici, P., Romero-Puertas, M. C., Zago, E., Zeier, J., Sonoda, M., et al. (2004). Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. Plant Cell 16, 2785–2794. doi: 10.1105/tpc.104.025379
Pérez-Mato, I., Castro, C., Ruiz, F. A., Corrales, F. J., and Mato, J. M. (1999). Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J. Biol. Chem. 274, 17075–17079. doi: 10.1074/jbc.274.24.17075
Pikaard, C. S., and Mittelsten Scheid, O. (2014). Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6:a019315. doi: 10.1101/cshperspect.a019315
Planchet, E., Jagadis Gupta, K., Sonoda, M., and Kaiser, W. M. (2005). Nitric oxide emission from tobacco leaves and cell suspensions: rate limiting factors and evidence for the involvement of mitochondrial electron transport. Plant J. 41, 732–743. doi: 10.1111/j.1365-313X.2005.02335.x
Polverari, A., Molesini, B., Pezzotti, M., Buonaurio, R., Marte, M., and Delledonne, M. (2003). Nitric oxide-mediated transcriptional changes in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 16, 1094–1105. doi: 10.1094/MPMI.2003.16.12.1094
Poulton, J. E., and Butt, V. S. (1976). Purification and properties of S-adenosyl-L-homocysteine hydrolase from leaves of spinach beet. Arch. Biochem. Biophys. 172, 135–142. doi: 10.1016/0003-9861(76)90058-8
Puyaubert, J., and Baudouin, E. (2014). New clues for a cold case: nitric oxide response to low temperature. Plant Cell Environ. 37, 2623–2630. doi: 10.1111/pce.12329
Puyaubert, J., Fares, A., Reze, N., Peltier, J. B., and Baudouin, E. (2014). Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 215–216, 150–156. doi: 10.1016/j.plantsci.2013.10.014
Rocha, P. S., Sheikh, M., Melchiorre, R., Fagard, M., Boutet, S., Loach, R., et al. (2005). The arabidopsis homology-dependent gene silencing1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell 17, 404–417. doi: 10.1105/tpc.104.028332
Rockel, P., Strube, F., Rockel, A., Wildt, J., and Kaiser, W. M. (2002). Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J. Exp. Bot. 53, 103–110. doi: 10.1093/jexbot/53.366.103
Schiedel, M., and Conway, S. J. (2018). Small molecules as tools to study the chemical epigenetics of lysine acetylation. Curr. Opin. Chem. Biol. 45, 166–178. doi: 10.1016/j.cbpa.2018.06.015
Serpa, V., Vernal, J., Lamattina, L., Grotewold, E., Cassia, R., and Terenzi, H. (2007). Inhibition of AtMYB2 DNA-binding by nitric oxide involves cysteine S-nitrosylation. Biochem. Biophys. Res. Commun. 361, 1048–1053. doi: 10.1016/j.bbrc.2007.07.133
Servet, C., Conde, S. N., and Zhou, D. X. (2010). Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 3, 670–677. doi: 10.1093/mp/ssq018
Shen, Y., Wei, W., and Zhou, D. X. (2015). Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 20, 614–621. doi: 10.1016/j.tplants.2015.07.005
Shi, H., Ye, T., Zhu, J. K., and Chan, Z. (2014). Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J. Exp. Bot. 65, 4119–4131. doi: 10.1093/jxb/eru184
Srivastava, N., Gonugunta, V. K., Puli, M. R., and Raghavendra, A. S. (2009). Nitric oxide production occurs downstream of reactive oxygen species in guard cells during stomatal closure induced by chitosan in abaxial epidermis of Pisum sativum. Planta 229, 757–765. doi: 10.1007/s00425-008-0855-5
Stroud, H., Do, T., Du, J., Zhong, X., Feng, S., Johnson, L., et al. (2013). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol. 21, 64–72. doi: 10.1038/nsmb.2735
Sun, B., Jing, Y., Chen, K., Song, L., Chen, F., and Zhang, L. (2007). Protective effect of nitric oxide on iron deficiency-induced oxidative stress in maize (Zea mays). J. Plant Physiol. 164, 536–543. doi: 10.1038/nsmb.2735
Tanaka, H., Masuta, C., Uehara, K., Kataoka, J., Koiwai, A., and Noma, M. (1997). Morphological changes and hypomethylation of DNA in transgenic tobacco expressing antisense RNA of the S-adenosyl-L-homocysteine hydrolase gene. Plant Mol. Biol. 35, 981–986. doi: 10.1023/A:1005896711321
Tavares, C. P., Vernal, J., Delena, R. A., Lamattina, L., Cassia, R., and Terenzi, H. (2014). S-nitrosylation influences the structure and DNA binding activity of AtMYB30 transcription factor from Arabidopsis thaliana. Biochim. Biophys. Acta Proteins Proteomics 1844, 810–817. doi: 10.1016/j.bbapap.2014.02.015
Tian, Q.-Y., Sun, D.-H., Zhao, M.-G., and Zhang, W.-H. (2007). Inhibition of nitric oxide synthase (NOS) underlies aluminum-induced inhibition of root elongation in Hibiscus moscheutos. New Phytol. 174, 322–331. doi: 10.1111/j.1469-8137.2007.02005.x
Toledo, J. C. Jr., and Augusto, O. (2012). Connecting the chemical and biological properties of nitric oxide. Chem. Res. Toxicol. 25, 975–989. doi: 10.1021/tx300042g
Vasudevan, D., Hickok, J. R., Bovee, R. C., Pham, V., Mantell, L. L., Bahroos, N., et al. (2015). Nitric oxide regulates gene expression in cancers by controlling histone posttranslational modifications. Cancer Res. 75, 5299–5308. doi: 10.1158/0008-5472.CAN-15-1582
Vescovi, M., Zaffagnini, M., Festa, M., Trost, P., Lo Schiavo, F., and Costa, A. (2013). Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiol. 162, 333–346. doi: 10.1104/pp.113.215194
Vriet, C., Hennig, L., and Laloi, C. (2015). Stress-induced chromatin changes in plants: of memories, metabolites and crop improvement. Cell. Mol. Life Sci. 72, 1261–1273. doi: 10.1007/s00018-014-1792-z
Yamamoto-Katou, A., Katou, S., Yoshioka, H., Doke, N., and Kawakita, K. (2006). Nitrate reductase is responsible for elicitin-induced nitric oxide production in Nicotiana benthamiana. Plant Cell Physiol. 47, 726–735. doi: 10.1093/pcp/pcj044
Yamasaki, H. (2000). Nitrite-dependent nitric oxide production pathway: implications for involvement of active nitrogen species in photoinhibition in vivo. Philos. Trans. R. Soc. B 355, 1477–1488. doi: 10.1098/rstb.2000.0708
Yan, Y., Shen, L., Chen, Y., Bao, S., Thong, Z., and Yu, H. (2014). A MYB-domain protein EFM mediates flowering responses to environmental cues in Arabidopsis. Dev. Cell 30, 437–448. doi: 10.1016/j.devcel.2014.07.004
Yan, D., Zhang, Y., Niu, L., Yuan, Y., and Cao, X. (2007). Identification and characterization of two closely related histone H4 arginine 3 methyltransferases in Arabidopsis thaliana. Biochem. J. 408, 113–121. doi: 10.1042/BJ20070786
Yang, H., Han, Z., Cao, Y., Fan, D., Li, H., Mo, H., et al. (2012). A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in Arabidopsis via the repression of FLC expression. PLoS Genet. 8:e1002664. doi: 10.1371/journal.pgen.1002664
Yu, M., Lamattina, L., Spoel, S. H., and Loake, G. J. (2014). Nitric oxide function in plant biology: a redox cue in deconvolution. New Phytol. 202, 1142–1156. doi: 10.1111/nph.12739
Yun, B. W., Feechan, A., Yin, M., Saidi, N. B., Le Bihan, T., Yu, M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478, 264–268. doi: 10.1038/nature10427
Zaffagnini, M., Morisse, S., Bedhomme, M., Marchand, C. H., Festa, M., Rouhier, N., et al. (2013). Mechanisms of nitrosylation and denitrosylation of cytoplasmic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana. J. Biol. Chem. 288, 22777–22789. doi: 10.1074/jbc.M113.475467
Zhang, H., Shen, W. B., Zhang, W., and Wu, L. L. (2005). A rapid response of beta-amylase to nitric oxide but not gibberellin in wheat seeds during the early stage of germination. Planta 220, 708–716. doi: 10.1007/s00425-004-1390-7
Zhang, Y., Tessaro, M. J., Lassner, M., and Lia, X. (2003). Knockout analysis of arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential rolesin systemic acquired resistance. Plant Cell 15, 2647–2653. doi: 10.1105/tpc.014894
Zheng, S., Hu, H., Ren, H., Yang, Z., Qiu, Q., Qi, W., et al. (2019). The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat. Commun. 10, 1–11. doi: 10.1038/s41467-019-09310-x
Keywords: nitric oxide, redox-modification, S-nitrosation, chromatin modulation, acetylation, methylation
Citation: Ageeva-Kieferle A, Rudolf EE and Lindermayr C (2019) Redox-Dependent Chromatin Remodeling: A New Function of Nitric Oxide as Architect of Chromatin Structure in Plants. Front. Plant Sci. 10:625. doi: 10.3389/fpls.2019.00625
Edited by:
Jean-Benoit Charron, McGill University, CanadaReviewed by:
Jean-Philippe Reichheld, Centre National de la Recherche Scientifique (CNRS), FranceGiorgio Perrella, ENEA – Centro Ricerche Trisaia, Italy
Copyright © 2019 Ageeva-Kieferle, Rudolf and Lindermayr. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christian Lindermayr, bGluZGVybWF5ckBoZWxtaG9sdHotbXVlbmNoZW4uZGU=
 Alexandra Ageeva-Kieferle
Alexandra Ageeva-Kieferle Christian Lindermayr
Christian Lindermayr