- 1Department of Biology, University of Florida, Gainesville, FL, United States
- 2Genetics Institute, University of Florida, Gainesville, FL, United States
- 3Section on Molecular Medicine, Department of Internal Medicine, Center for Precision Medicine, Wake Forest School of Medicine, Winston-Salem, NC, United States
- 4Plant Molecular and Cellular Biology, University of Florida, Gainesville, FL, United States
- 5Interdisciplinary Center for Biotechnology Research, University of Florida, Gainesville, FL, United States
Glucosinolates (GLSs) are a well-defined group of specialized metabolites, and like any other plant specialized metabolites, their presence does not directly affect the plant survival in terms of growth and development. However, specialized metabolites are essential to combat environmental stresses, such as pathogens and herbivores. GLSs naturally occur in many pungent plants in the order of Brassicales. To date, more than 200 different GLS structures have been characterized and their distribution differs from species to species. GLSs co-exist with classical and atypical myrosinases, which can hydrolyze GLS into an unstable aglycone thiohydroximate-O-sulfonate, which rearranges to produce different degradation products. GLSs, myrosinases, myrosinase interacting proteins, and GLS degradation products constitute the GLS-myrosinase (GM) system (“mustard oil bomb”). This review discusses the cellular and subcellular organization of the GM system, its chemodiversity, and functions in different cell types. Although there are many studies on the functions of GLSs and/or myrosinases at the tissue and whole plant levels, very few studies have focused on different single cell types. Single cell type studies will help to reveal specific functions that are missed at the tissue and organismal level. This review aims to highlight (1) recent progress in cellular and subcellular compartmentation of GLSs, myrosinases, and myrosinase interacting proteins; (2) molecular and biochemical diversity of GLSs and myrosinases; and (3) myrosinase interaction with its interacting proteins, and how it regulates the degradation of GLSs and thus the biological functions (e.g., plant defense against pathogens). Future prospects may include targeted approaches for engineering/breeding of plants and crops in the cell type-specific manner toward enhanced plant defense and nutrition.
Introduction
One of the most extensively studied classes of anti-herbivore chemical defenses in plants is glucosinolates (GLSs), a group of sulfur-rich, amino acid-derived metabolites combining a β-d-glucopyranose residue linked via a sulfur atom to an N-hydroxyimino sulfate ester, which are plant-derived natural products (Halkier and Gershenzon, 2006; Halkier, 2016). GLSs are widely distributed in the order Brassicales, which includes vegetables (cabbage, cauliflower, and broccoli), spice plants supplying condiments (mustard, horseradish, and wasabi), and reference species, Arabidopsis thaliana (Fahey et al., 2001; Reichelt et al., 2002). Upon insect feeding or mechanical disruption, GLSs are hydrolyzed by myrosinases (thioglucoside glucohydrolase, TGG, EC 3.2.1.147) into unstable thiohydroximate-O-sulfonates, which rearrange to form different hydrolytic products such as isothiocyanates (ITCs), nitriles, and other by-products depending on the nature of the GLS side chain and the reaction conditions, such as iron, pH, and presence of myrosinase interacting proteins (Chen and Andreasson, 2001; Wittstock et al., 2016a). This GLS-myrosinase (GM) system is popularly known as “mustard oil bomb” (Lüthy and Matile, 1984; Ratzka et al., 2002). Myrosin cells (an idioblast cell type accumulating TGGs) are involved in plant defense by hydrolyzing GLSs into toxic volatiles such as ITCs or nitriles (Wittstock et al., 2003). TGGs are known to be present in all A. thaliana organs and were reported in A. thaliana and B. napus phloem parenchyma as well as in guard cells (Andréasson et al., 2001; Thangstad et al., 2004). In general, GLSs are enriched in “S-cells” that are found in Arabidopsis flower stalks and occur close to myrosin cells (Koroleva et al., 2000; Andréasson et al., 2001).
The spatial distribution of GLSs was demonstrated in A. thaliana leaves by constructing ion intensity maps from matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectra, where major GLSs were found to be more abundant in tissues of the midvein and the periphery of the leaf than the inner lamina (Shroff et al., 2008). Although this study concluded that GLSs are not abundant on A. thaliana leaf surfaces, the authors could not obtain information on the cell type distribution of GLSs in leaves. Moreover, all the genes in the GLS biosynthetic pathways have been identified, and it is somewhat known where GLSs are stored (Koroleva et al., 2000; Andréasson et al., 2001), but it has remained elusive where GLSs are specifically produced at the subcellular, cellular, and tissue levels (Rask et al., 2000; Nintemann et al., 2017). Neither is it clear about the cellular and subcellular compartmentation of different myrosinases and their interacting proteins, which include myrosinase-binding proteins (MBPs), myrosinase-associated proteins (MyAPs), and different specifier proteins.
In the following sections, we discuss various aspects of the GM system based on current knowledge, starting from the cellular control of enzymes, cell type, and subcellular organization, to uniqueness of myrosinases and myrosinase interacting proteins covering a range of small molecule and macromolecular interactions of the “mustard oil bomb.”
The Glucosinolate-Myrosinase System and Cellular Control of Enzyme Reactions
As found in the order of Brassicales, including important crops (e.g., mustard, oilseed rape, radish, broccoli, and cabbage), GLSs co-exist with myrosinases. When tissue damage occurs, the “mustard oil bomb” is detonated and GLSs are hydrolyzed and converted to different degradation products with a variety of biological activities (Rask et al., 2000; Halkier and Gershenzon, 2006; Yan and Chen, 2007; Bednarek et al., 2009; Clay et al., 2009; Halkier, 2016; Wittstock et al., 2016a). For example, these degradation products play important roles in plant defense against pathogens and herbivores, as well as serve as attractants to specialists (Rask et al., 2000; Barth and Jander, 2006; Clay et al., 2009; Wittstock et al., 2016a). Several of these degradation products are involved in plant nutrition (Holmes, 1980; Armengaud et al., 2004) and growth regulation (Hasegawa et al., 2000; Hull et al., 2000; Mikkelsen et al., 2000). In plant metabolism, it is important that enzymes and substrates are under tight regulation, which is more relevant for toxic compounds, as these chemical defenses are derived from specialized metabolites. There are several ways of regulation: (1) coarse control through biosynthesis; (2) fine control of enzyme activity through protein interaction and allosteric regulation; and (3) substrate and enzyme compartmentalization (Sweetlove and Fernie, 2013). While the regulation is well studied in primary metabolism (e.g., photosynthesis and respiration), it is not clear in many of the specialized metabolic processes such as GLS metabolism. Furthermore, protein-protein interactions are intrinsic to virtually every cellular process and have been extensively studied in animals and yeast (Uetz et al., 2000; Gavin et al., 2002; Ho et al., 2002; Li et al., 2004; Huttlin et al., 2017). In plants, this area has lagged behind in spite of recent progress (Hosseinpour et al., 2012; Zhang et al., 2016; Jiang et al., 2018). Vast majority of the studies did not go beyond identifying physical interactions to the point of functional analysis. Figure 1 shows the potential molecular interactions of the GM system in the context of cell type-specific metabolisms.
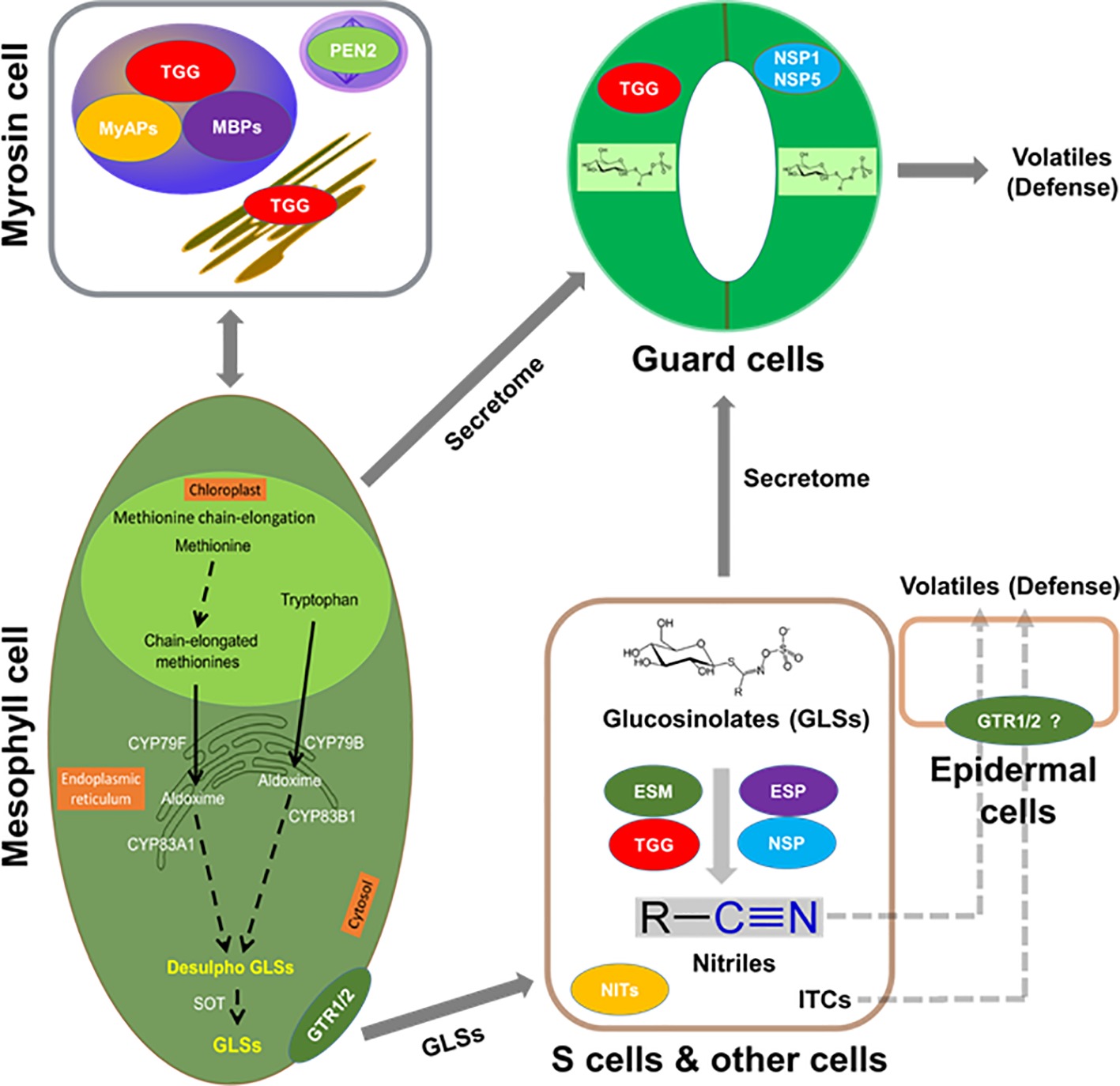
Figure 1. Putative interactions between myrosinases (TGGs), myrosinase interacting proteins, GLSs, and volatiles in the context of cell type compartmentation. A myrosin cell shows vacuolar localization of TGGs, myrosinase-binding proteins (MBPs), and myrosinase-associated proteins (MyAPs); peroxisomal localization of penetration (PEN2); and ER localization of TGG. Transporters that are specific to GLSs such as NRT1/PTR glucosinolate transporter (GTR) 1, GTR2 or non-specific transporters could be aiding in their transport to site of accumulation such as S-cells or guard cells. Importantly, these cells may have the capability of de novo biosynthesis of GLSs. In addition, the presence of epithiospecifier modifier (ESM, MyAP-like), epithiospecifier (ESP), and nitrile specifier (NSP) 1, NSP5, etc. may lead to the breakdown of GLSs to nitriles and isothiocyanates (ITCs) for roles in cell type-specific signaling and defense against pathogen and herbivores. GLSs, TGGs, and ESP were found in the S-cells, and the presence of ESM and NSP is indicative of other cell types.
Cell Type-Specific Cellular and Subcellular Organization of the “Mustard Oil Bomb”
Myrosinase is located in myrosin cells, which are scattered cells in radicles, stems, leaves, petioles, seeds, and seedlings of several species (Husebye et al., 2002). A cell-specific localization was found in radicles and cotyledons of the maturing embryo resembling the pattern of the myrosin cells (Bones et al., 1991). Most GLSs are constitutively present in all Arabidopsis tissues (Petersen et al., 2002; Brown et al., 2003). The key steps in the biosynthesis of the different types of GLSs are localized in distinct cells in separate as well as overlapping vascular tissues (Nintemann et al., 2018). The presence of GLS biosynthetic enzymes in parenchyma cells of the vasculature may assign new defense-related functions to these cell types (Nintemann et al., 2018). To date, the cellular and subcellular compartmentation of the “mustard oil bomb” (Lüthy and Matile, 1984) is not completely clear and is rather contradictory. For instance, in Arabidopsis flower stalks, GLSs were found in the elongated sulfur-rich “S-cells” situated between phloem and endodermis (Koroleva et al., 2000; Husebye et al., 2002). However, the myrosinase TGG1 was found to be abundant in guard cells, whereas TGG1 and TGG2 were localized to the phloem-associated cells close to the “S-cells” (Husebye et al., 2002; Thangstad et al., 2004; Barth and Jander, 2006). Thus, it appears that myrosinases and their substrates were physically separated in the plant tissues. However, such an arrangement may not be the case as a recent proteomics study located the myrosinases in “S-cells” (Koroleva and Cramer, 2011). In Brassica juncea seedlings, myrosinase was found to co-localize with GLSs in aleurone-type cells (Kelly et al., 1998). In Arabidopsis suspension cells, both myrosinases and GLSs were present (Alvarez et al., 2008). Such diverse co-localization results may indicate that myrosinases and GLSs are spatially separated at the subcellular levels. Alternatively, they could be in the same compartment with tight control of myrosinase activities. GLSs were found in vacuoles rich in ascorbic acid (Grob and Matile, 1979), which plays a role to inhibit myrosinase at high concentration and activate myrosinase at low concentration. This dual regulation supports the potential co-localization of GLSs and myrosinases in the same subcellular compartment.
Recent metabolomics data have confirmed the presence of GLSs in guard cells (Geng et al., 2016; Zhu and Assmann, 2017) and revealed the changes in GLS metabolism in guard cells upon treatment with CO2 (Geng et al., 2016) and ABA (Zhu and Assmann, 2017). The first indication of the role of GLS metabolism in stomatal movement was obtained through analysis of the effect of ABA on stomatal movement of the Arabidopsis myrosinase mutant tgg1 (Zhao et al., 2008). Subsequently, additional reverse genetics studies corroborated the role of GLS metabolism in stomatal movement (Islam et al., 2009; Zhu et al., 2014). Furthermore, stomatal closure was induced by pharmacological treatments with different GLS hydrolysis products (Khokon et al., 2011; Sobahan et al., 2015). However, these products and the amounts used are of synthetic origin and abundance. It is not known what degradation products are produced and how much in vivo, which GLSs and myrosinases [TGGs and/or Penetration 2 (PEN2)] are involved, and how protein interactions regulate the GLS breakdown in guard cells.
The Arabidopsis cyp79b2/cyp79b3 mutants are known to produce mostly aliphatic GLSs (Zhao et al., 2002; Chen et al., 2003; Grubb and Abel, 2006; Khokon et al., 2011; Sobahan et al., 2015), while the myb28/myb29 mutants are known to produce mostly indolic GLSs (Hirai et al., 2007; Beekwilder et al., 2008). Furthermore, the tgg1/tgg2 double mutant showed undetectable myrosinase activity, and damage-induced breakdown of endogenous GLSs was not from aliphatic GLSs and was greatly slowed for indole GLSs (Barth and Jander, 2006). Moreover, the tgg1/tgg2 mutant lacking the foliar myrosinases was compromised in activation of their GLS defense. Another mutant, atvam3 mutant showed abnormal distribution of myrosin cells and overproduction of TGG1 and TGG2 (Ueda et al., 2006). Thus, beyond TGGs, MYB28, MYB29, AtVAM, CYP79s, and other biosynthetic genes all affect GLS deposition levels and possibly cell type specificity of the GM system. To understand the regulation and correlation of these proteins, we used GeneMANIA software (Warde-Farley et al., 2010) and predicted the association of the known genes involved in GLS metabolism (from our selected gene list in Supplementary Table S1). This software further added putative proteins with similar functions and potential involvement in the GM system (Figure 2).
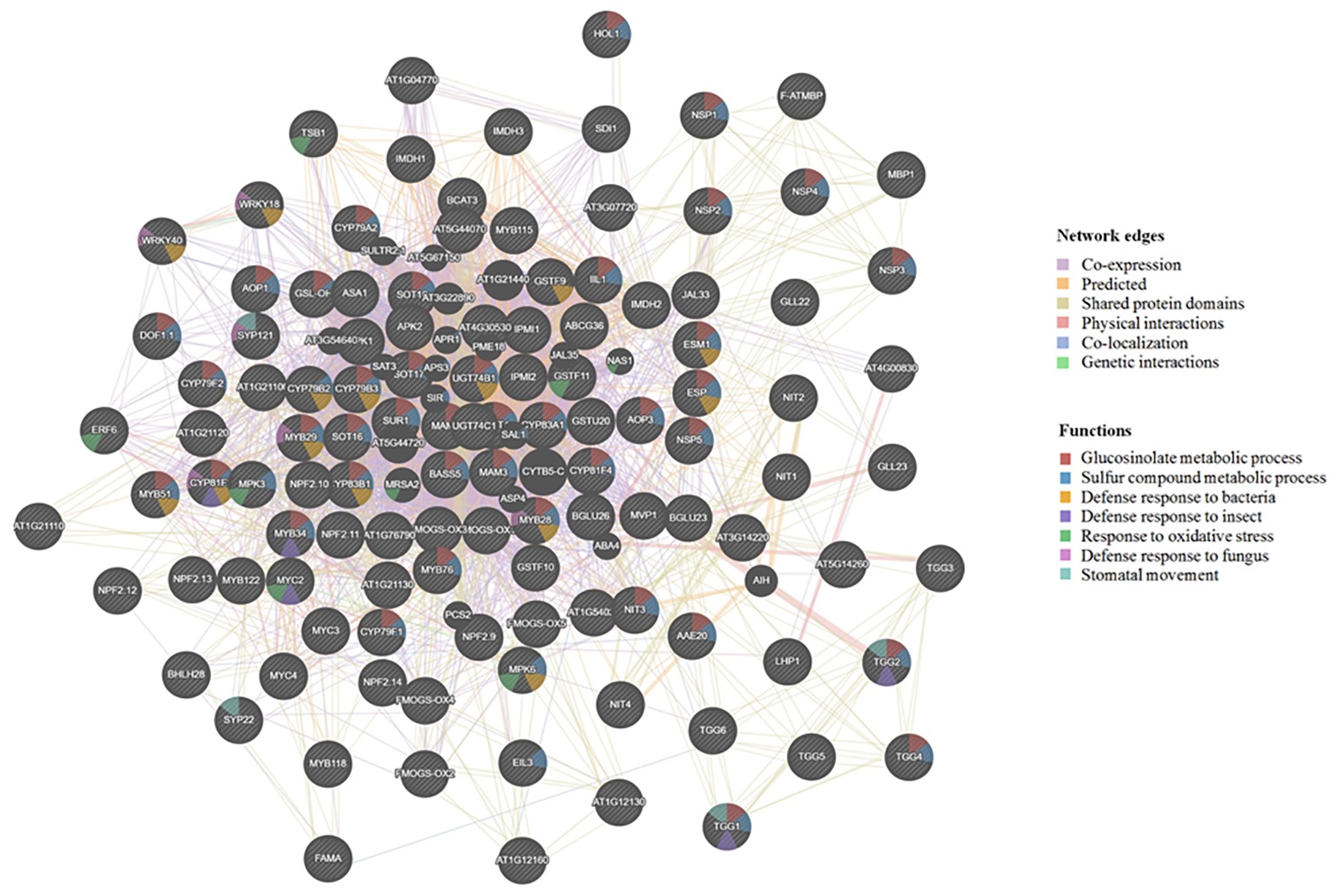
Figure 2. Visualization of functional prediction of protein networks in the glucosinolate-myrosinase (GM) system using GeneMANIA (http://genemania.org/). The protein names are indicated inside the nodes, and the links between the nodes indicate the network edges in which the proteins are connected. The color of the edges represents evidence for the connection, which includes co-expression (purple), predicted (yellow), shared protein domains (beige), physical interactions (pink), co-localization (blue), and genetic interactions (green). As to functions associated with each protein, the color code inside the nodes indicates GLS metabolic process (red), sulfur compound metabolic process (blue), defense response to bacterium (yellow), defense response to insect (purple), response to oxidative stress (green), defense response to fungus (pink), and stomatal movement (light blue).
In B. napus leaves, myrosinases are localized in mesophyll cells and phloem cells (Chen and Andreasson, 2001) and were mainly stored in protein-rich vacuolar structures of myrosin cells (Rask et al., 2000; Ueda et al., 2006). There is also a report of the presence of myrosinase as cytosolic enzymes bound to intracellular membranes (Lüthy and Matile, 1984). The knowledge of the localization of myrosinases and interacting proteins was advanced by vacuolar proteomics. Myrosinases, TGG1 and TGG2, and myrosinase-associated protein (MyAP) 1 were identified in the vacuoles. In the early leaf developmental stages, TGG1 is more abundant than TGG2, whereas in fully expanded leaves, both TGG1 and TGG2 levels show increased accumulation. Concurrently, MyAP1 levels are increasingly abundant. We have previously observed such regulation of myrosinase expression, which correlated with GLS turnover (Petersen et al., 2002). The co-localization of myrosinase and MyAP1 and the concurrent expression during development lead to the hypothesis that the vacuolar myrosinases may be active and MyAPs may interact with myrosinase to play a role in GLS hydrolysis. For example, MyAPs may facilitate ITC production (Zhang et al., 2006). Indeed, immunogold analysis of leaf sections showed the presence of TGG1 and TGG2 in the same vacuoles (Ueda et al., 2006). An independent vacuolar proteomics study also identified these proteins (Carter et al., 2004). In addition, two more MyAPs (At1g54000 and At1g54010) and three myrosinase-binding proteins (MBPs) (At1g52040, At3g16470, and At2g39330) were localized in the vacuoles (Carter et al., 2004). TGG1 and TGG2 were also found in the endoplasmic reticulum (ER), ER bodies, and transvacuolar strands, and this localization is dependent on MyAP1 (MVP1). Mutation of the MyAP1 clearly altered the subcellular localization profiles of the green fluorescent protein (GFP)-tagged TGG1 and TGG2 (Agee et al., 2010). Interestingly, the myrosinase PEN2 (hydrolyzing indole GLSs and shown to function in plant defense (Bednarek et al., 2009; Clay et al., 2009; Millet et al., 2010; Fan et al., 2011; Johansson et al., 2014; Frerigmann et al., 2016; Luti et al., 2016; Xu et al., 2016; Vilakazi et al., 2017)) is targeted to peroxisomes and the outer mitochondrial membrane (Fuchs et al., 2016). In addition to MyAPs and MBPs, specifier proteins including epithiospecifier modifier (ESM, MyAP-like), epithiospecifier protein (ESP), nitrile specifier protein (NSP), and thiocyanate forming protein (TFP) may affect the outcome of GLS degradation (Lambrix et al., 2001; Burow et al., 2006; Zhang et al., 2006; Wittstock et al., 2016a,b ; Backenköhler et al., 2018). ESP was found to be in “S-cells” and in guard cells with NSP1 and NSP5 (Burow et al., 2007; Zhao et al., 2008). The functions of these MyAPs, MBPs, and specifier proteins in “S-cells” and guard cells and their interactions with myrosinases in different cell types are not known.
To understand the subcellular organization of the GM system, we compared the proteins and pathways involved in GLS biosynthesis, degradation, and transport using available and/or predicted subcellular localization information. Figure 3 and Supplementary Table S1 provide an overview of the GM system at subcellular level based on available literature and analysis using different protein localization tools: (1) Plant-mPLoc1 (Chou and Shen, 2007, 2008, 2010); (2) TAIR2 (with annotation based on literature); (3) Eplant3 using SUBA (Subcellular Localisation Database for Arabidopsis) with annotation based on subcellular proteomics and/or protein fluorescence microscopy; (4) TargetP4 based on the N-terminal targeting sequences (chloroplast transit peptide (cTP), mitochondrial targeting peptide (mTP), or secretory pathway signal peptide (SP) (Emanuelsson et al., 2000) [with a reliability score of 1–5 (1 being most reliable and 5 least reliable)]; (5) LocTree5 using support vector machines for localization prediction (in the form of expected accuracy); and (6) ngLOC6 using Bayesian method for prediction of localization. As shown in Figure 3, most GM system proteins were found to be in the cytoplasm followed by nucleus, where the transcriptional regulators were localized. All the cytochrome P450s involved in GLS biosynthesis and modification were localized to endoplasmic reticulum, and other GLS biosynthesis-related proteins were in the chloroplast and cytoplasm. Glucosinolate transporters (GTR1 and GTR2) and nitrate transporters (NRT1.6, NRT1.7, and NRT1.9) were found to be in plasma membrane. It is not known how glucosinolates are transported into vacuoles. PEN2 and BZO1 were localized in peroxisomes. No GM system proteins were found on Golgi apparatus. Out of the 114 GM system proteins used in this study (Supplementary Table S1), 65 proteins had experimental evidence of localization, 26 were predicted using the software tools (at least three tools with consistent result), and 23 proteins could not be conclusively localized.
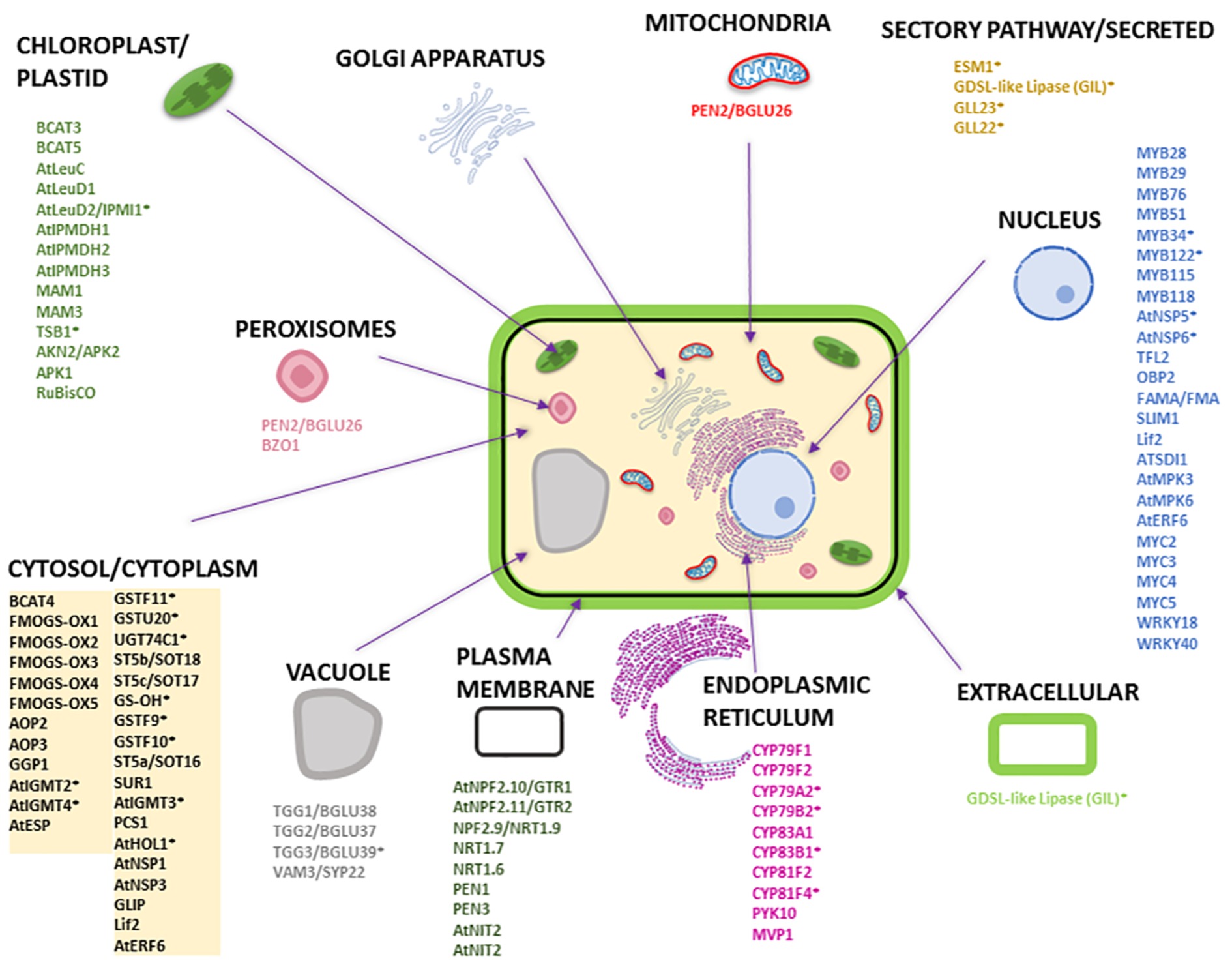
Figure 3. Subcellular localization of Arabidopsis proteins involved in GLS biosynthesis, degradation, and transport. The subcellular organelles and their localized proteins are given in similar color. Most of the subcellular localizations are based on literature evidence. The names of the proteins with an asterisk are predicted localization information (i.e., for proteins that do not have localization information in literature) that are based on consensus from at least three software tools used. The prediction tools used were Eplant, TargetP, LocTree, ngLOC, Plant-mPLoc, and TAIR. Please refer to Supplementary Table S1 for detailed information.
Distinct Molecular and Biochemical Properties of Myrosinases
Myrosinases are classified into two types, typical (classical) and atypical myrosinases. The crystal structure of a classical myrosinase shows that the protein folds into an (β/α)8 barrel structure (Burmeister et al., 1997). In the active site, a Glu (E) residue is involved in nucleophilic attack to initiate the release of an aglycone (thiohydroximate-O-sulfonate) and form a glucosyl-enzyme intermediate. Another Gln (Q) residue enables the hydrolysis of this intermediate with assistance from water and ascorbate. Classical myrosinases (with QE catalytic residues) use ascorbate as a cofactor and proton donor to facilitate the release of bound glucose (Burmeister et al., 1997; Wittstock and Burow, 2010; Bhat and Vyas, 2019). In contrast, atypical myrosinases have two catalytic Glu residues (EE), which function as acid/base catalyst in the active site. They do not require ascorbate. In addition, atypical myrosinases have two basic amino acid residues at different positions (+6 and +7) for glucosinolate binding compared to +0 position arginine residue of classical myrosinases (Wittstock and Burow, 2010; Nakano et al., 2017; Shirakawa and Hara-Nishimura, 2018; Bhat and Vyas, 2019). Classical myrosinases are glycosylated, activated by low concentrations of ascorbate, and accepted GLSs as the only substrates (Chen and Halkier, 1999; Chen and Andreasson, 2001). In contrast, atypical myrosinases, such as PEN2 and PYK10, can hydrolyze indole GLSs and also use O-glucosides as substrates (Bednarek et al., 2009; Nakano et al., 2017). Although myrosinase does not use acylated GLSs and desulpho-GLSs as substrates, it may accept a wide range of GLS substrates (Chen and Halkier, 1999; Rask et al., 2000; Barth and Jander, 2006). Myrosinases from B. napus and Crambe abyssinica degrade different GLS at different rates (James and Rossiter, 1991; Finiguerra et al., 2001). However, the mechanism underlying this substrate specificity is not established. Myrosinases in B. napus are encoded by >29 genes in three subfamilies, denoted as MA, MB, and MC. The MA myrosinases occur as dimers, while MB and MC myrosinases exist in complexes with MBPs and/or MyAPs (Lenman et al., 1990; Rask et al., 2000). By heterologous expression in yeast, we have previously produced a functional free form myrosinase Myr1 from the MB subfamily (Chen and Halkier, 1999). The activity of this Myr1 suggests that MBPs and MyAPs are not absolutely necessary for myrosinase activity, but raises questions on the functions of MBPs and MyAPs and their interactions with myrosinases.
Bioinformatic analysis of the Arabidopsis genome revealed the presence of six myrosinase genes TGG1-TGG6 (Xu et al., 2004). TGG1 and TGG2 are expressed in leaves (Xue et al., 1995; Husebye et al., 2002; Thangstad et al., 2004; Barth and Jander, 2006; Ueda et al., 2006) and flowers (Ruan et al., 1998; Barth and Jander, 2006), while TGG4 and TGG5 are specifically expressed in roots (Zimmermann et al., 2004). TGG3 and TGG6 are pseudogenes (Husebye et al., 2002; Zhang et al., 2002). Although TGG1 and TGG2 appear to display a low degree of substrate specificity, the activities of TGG1 and TGG2 have been correlated with the feeding preference and growth of different generalist and specialist insects (Barth and Jander, 2006). Interestingly, overexpression of TGG1 and TGG2 leads to accumulation of several GLS degradation products, including 5-methylhexanenitrile, heptanenitrile, 1-isothiocyanato-3-methylbutane, 1-isothiocyanato-4-methylpentane, and 1-isothiocyanato-3-methylhexane. Based on the degradation product profile, possible endogenous substrates for the two TGGs include 4-methylthiobutylglucosinolate, 4-methylpentylglucosinolate and 3-methylbutylglucosinolates (Ueda et al., 2006). Investigating endogenous substrates of different classical and atypical myrosinases is an important future direction.
In leaves, TGG1 was found to be abundant in guard cells, while TGG2 appeared only present in phloem-associated cells (Barth and Jander, 2006; Zhao et al., 2008). Considering the presence of GLSs in guard cells (Geng et al., 2016; Zhu and Assmann, 2017), how the GM system plays a role in guard cell functions (e.g., stomatal immunity) is an interesting question. Clearly, mutation of the TGG1 and/or TGG2 genes affected the guard cell size, stomatal aperture, and leaf metabolites, such as fatty acids, glucosinolates, and indole compounds (Ahuja et al., 2016). Another proteomic study of trichome and epidermal pavement cells did not identify the TGG1 protein in the samples (Wienkoop et al., 2004). However, a single cell type study in trichomes found the presence of gene encoding transcription factors of aliphatic GLS (MYB28, MYB29 and MYB76) and indole GLS (MYB34, MYB51 and MYB122), indicating that trichomes have biosynthetic genes for the GM system (Frerigmann et al., 2012), but nothing was suggested about myrosinases activity or expression. Given the defense roles of guard cells and trichomes, characterization of the GM systems in these special cell types is of great importance to understand the molecular mechanisms underlying the cell type-specific functions, e.g., defense against pathogen invasion.
Complex Formation Between Myrosinase and Its Interacting Proteins
As described earlier, myrosinase interacting proteins include MBP, MyAP, and specifier proteins (ESM, ESP, NSP, and TFP). The first six MBPs identified in B. napus range in size from 30 to 110 kDa (Taipalensuu et al., 1996; Chisholm et al., 2000). All MBPs contain jacalin-like repeats (Chisholm et al., 2000; Andréasson et al., 2001). Jacalin-related proteins share the domain structure of plant lectins and are upregulated by phytohormones (e.g., jasmonic acid, salicylic acid, and ethylene) and pathogens (Taipalensuu et al., 1996; Geshi and Brandt, 1998; Xiang et al., 2011; Vilakazi et al., 2017). Recently, identification of bacterial lipopolysaccharide interacting proteins in Arabidopsis revealed myrosinases, TGG1 and TGG2, and a MBP (Vilakazi et al., 2017). It remains unclear how the MBP levels are regulated and whether MBPs directly interact and affect myrosinase activity and specificity. In B. napus seeds, MBPs are present in most cells but not in the myrosin cells (Rask et al., 2000; Ueda et al., 2006). During germination, MBPs are co-localized with myrosinases in cotyledons, suggesting that preformed myrosinase complexes do exist (Geshi and Brandt, 1998; Eriksson et al., 2002). Using basic local alignment search tool (BLAST) to interrogate the Arabidopsis genome reveals >30 putative MBPs. MBP1 and MBP2 are like lectin jacalins and plant aggregating factors. MBP1 and MBP2 are abundantly expressed in immature flowers, and the pattern is similar to that of myrosinase TGG1 (Capella et al., 2001). MBP expression and myrosinase activity are affected in the coi1 mutant, which is insensitive to jasmonate (Capella et al., 2001). Depletion of MBPs does not alter the cellular distribution of myrosinases but prevents myrosinases from forming complexes (Eriksson et al., 2002). Thus, the functions of MBPs are not fully understood. Interestingly, most NSPs possess jacalin-like domains and are MBP-like (Kuchernig et al., 2012). The jacalin-like domain may interact with the glycans of myrosinases to potentially affect GLS degradation. However, experimental evidence is lacking. NSPs were shown to enhance simple nitrile formation (He et al., 2009; Kissen and Bones, 2009; Chen et al., 2015; Wittstock et al., 2016b). Recently, iron was shown to be a centrally bound cofactor of ESP, TFP, and NSP involved in glucosinolate breakdown. In addition, NSP active site has fewer restrictions to the aglycone conformation than ESP and TFP. This may explain why NSP facilitates simple nitrile production, but not production of epithionitrile and thiocyanate that may need exact positioning of the aglycone thiolate relative to the side chain (Backenköhler et al., 2018). In addition to MBPs, MyAPs form complexes with myrosinases in B. napus (Taipalensuu et al., 1996). In Arabidopsis, TGG2 was pulled down with MyAP1 in leaf extracts (Agee et al., 2010). MyAPs display high similarity to GDSL lipases, which have a motif of Gly, Asp, Ser, and Leu residues in the active site. The Arabidopsis genome contains >80 genes encoding GDSL lipases, typically with a GDSL-like motif, a catalytic triad of Ser, Asp and His residues, and a lipase signature sequence GxSxxxxG (Brick et al., 1995). The possible lipase activity of MyAP suggests a potential role of MyAP in releasing acyl groups from acylated GLSs, thereby making them available for myrosinase hydrolysis. Arabidopsis contains acylated GLSs in seeds, but B. napus does not contain acylated GLSs; thus, MyAP in B. napus may have other functions. A recent study shows that overexpression of B. napus MyAP1 led to enhanced plant defense against a fungal pathogen Sclerotinia sclerotiorum (Wu et al., 2017). A MyAP-like ESM was found to favor ITC production and protect Arabidopsis from herbivory (Zhang et al., 2006). However, whether this system involves myrosinase complex formation is still not known. In some plants, ESPs are involved in GLS hydrolysis (Foo et al., 2000; Burow et al., 2006). Hydrolysis of alkenyl GLSs in the presence of ESP leads to the formation of nitriles or epithionitriles, instead of isothiocyanates (Zabala Mde et al., 2005; Burow et al., 2006). Because ESPs can alter the course of hydrolysis, they are important in determining plant herbivore choice and host resistance (Lambrix et al., 2001). Furthermore, this suggests that ESP is situated close to the active site so that it could promptly convert the unstable aglycone to nitriles. Although kinetic studies have showed that ESP acts as a non-competitive inhibitor of myrosinase (MacLeod and Rossiter, 1985), no stable interaction between ESP and myrosinase has been reported (Burow et al., 2006). Like nitrile formation, the production of thiocyanate was found to be associated with TFP. For detailed description of myrosinase specifier proteins, please refer to a recent review (Wittstock et al., 2016a). In summary, several other groups of proteins may interact with myrosinases and function to affect how GLSs are degraded, leading to the formation of different metabolic products. Systematic studies to characterize the interaction of these proteins with myrosinases are needed to elucidate their specific functions.
Directions for Future Research and Conclusions
Currently, the cellular and subcellular location of myrosinases, GLSs, and their interacting proteins, i.e., the GM system, are far from established. Given >100 cell types in plants and >5,500 species of GLS producers, it would be a challenge to capture all the species-specific and cell type-specific information of the “mustard oil bomb.” In addition, with temporal accumulation and expression patterns of metabolites and enzymes involved typically in case of specialized metabolites, these eventual pictures could be very complex. Using cell type-specific genetic manipulations (e.g., GFP fusion and CRISPR), one can envision to capture the cell type-specific expression patterns and functional role of the glucosinolate biosynthetic proteins, myrosinases, and myrosinase interacting proteins. There exist large gaps in the knowledge base of the GM system, e.g., myrosinase interacting proteins in terms of their interactions, co-localizations, regulations, and functions in specific cell types. Furthermore, the developmental staged appearance and regulation of the proteins and metabolites are not clear. Moreover, the reported interactions of myrosinases and other proteins could be very much cell type-specific or subcellular localized, which is not well studied till date. Without resolving the cell type specificity of the proteins and metabolites, it would be very challenging to draw mechanistic conclusions on the specific roles of the enzymes, interactors, transporters, and the metabolites from tissue- and whole plant-based data where the information are averaged out (Dai and Chen, 2012; Misra et al., 2014).
In the future, efforts need to focus on large-scale speedy preparations of organelles and subcellular fractions (e.g., vacuoles, peroxisomes, and chloroplasts) in a time-dependent manner to capture the dynamics of protein interactions and GLS metabolism. It is obviously challenging to prepare and enrich plant cell types (e.g., the “S-cells”) in copious amounts for more system-wide experiments such as transcriptomics, proteomics, and metabolomics and to obtain preparations at a given time and for a specific treatment. With the recent development of single-cell omics tools (Misra et al., 2014; Efroni and Birnbaum, 2016; Doerr, 2019), such large-scale molecular characterization of different single cells is within sight and will greatly enhance the understanding of the chemodiversity of the GM system at the single-cell resolution.
Author Contributions
SChh made the list of protein localization, Figures 2, 3, generated the reference list, and edited the manuscript. BM wrote one-third of the manuscript draft, made Figure 1 draft, and edited the manuscript. NT assisted with Supplementary Table S1 analysis, reference list, and edited the manuscript. SChe designed the manuscript, wrote two-third of the text, provided guidance to students, and finalized the manuscript for submission.
Funding
The authors would like to acknowledge funding from the National Science Foundation MCB 1758820 to SChe. A NIH supported SF2UF Bridge Program (to Dr. David Julian) and a NSF REU grant 1560049 (to Dr. Ramesh Katam) have provided support to NT.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00618/full#supplementary-material
Footnotes
1. http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/
2. https://www.arabidopsis.org/tools/bulk/protein/index.jsp
3. https://bar.utoronto.ca/eplant/
4. http://www.cbs.dtu.dk/services/TargetP/
References
Agee, A. E., Surpin, M., Sohn, E. J., Girke, T., Rosado, A., Kram, B. W., et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152, 120–132. doi: 10.1104/pp.109.145078
Ahuja, I., de Vos, R. C., Rohloff, J., Stoopen, G. M., Halle, K. K., Ahmad, S. J., et al. (2016). Arabidopsis myrosinases link the glucosinolate-myrosinase system and the cuticle. Sci. Rep. 6:38990. doi: 10.1038/srep38990
Alvarez, S., He, Y., and Chen, S. (2008). Comparative investigations of the glucosinolate–myrosinase system in Arabidopsis suspension cells and hypocotyls. Plant Cell Physiol. 49, 324–333. doi: 10.1093/pcp/pcn007
Andréasson, E., Jørgensen, L. B., Höglund, A. S., Rask, L., and Meijer, J. (2001). Different myrosinase and idioblast distribution in Arabidopsis and Brassica napus. Plant Physiol. 127, 1750–1763. doi: 10.1104/pp.010334
Armengaud, P., Breitling, R., and Amtmann, A. (2004). The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling. Plant Physiol. 136, 2556–2576. doi: 10.1104/pp.104.046482
Backenköhler, A., Eisenschmidt, D., Schneegans, N., Strieker, M., Brandt, W., and Wittstock, U. (2018). Iron is a centrally bound cofactor of specifier proteins involved in glucosinolate breakdown. PLoS One 13:e0205755. doi: 10.1371/journal.pone.0205755
Barth, C., and Jander, G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46, 549–562. doi: 10.1111/j.1365-313X.2006.02716.x
Bednarek, P., Piślewska-Bednarek, M., Svatoš, A., Schneider, B., Doubský, J., Mansurova, M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106. doi: 10.1126/science.1163732
Beekwilder, J., Van Leeuwen, W., Van Dam, N. M., Bertossi, M., Grandi, V., Mizzi, L., et al. (2008). The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS One 3:e2068. doi: 10.1371/journal.pone.0002068
Bhat, R., and Vyas, D. (2019). Myrosinase: insights on structural, catalytic, regulatory, and environmental interactions. Crit. Rev. Biotechnol. 39, 508–523. doi: 10.1080/07388551.2019.1576024
Bones, A. M., Thangstad, O. P., Haugen, O. A., and Espevik, T. (1991). Fate of myrosin cells: characterization of monoclonal antibodies against myrosinase. J. Exp. Bot. 42, 1541–1550. doi: 10.1093/jxb/42.12.1541
Brick, D. J., Brumlik, M. J., Buckley, J. T., Cao, J. X., Davies, P. C., Misra, S., et al. (1995). A new family of lipolytic plant enzymes with members in rice, Arabidopsis and maize. FEBS Lett. 377, 475–480. doi: 10.1016/0014-5793(95)01405-5
Brown, P. D., Tokuhisa, J. G., Reichelt, M., and Gershenzon, J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481. doi: 10.1016/S0031-9422(02)00549-6
Burmeister, W. P., Cottaz, S., Driguez, H., Iori, R., Palmieri, S., and Henrissat, B. (1997). The crystal structures of Sinapis alba myrosinase and a covalent glycosyl–enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 5, 663–676. doi: 10.1016/S0969-2126(97)00221-9
Burow, M., Markert, J., Gershenzon, J., and Wittstock, U. (2006). Comparative biochemical characterization of nitrile-forming proteins from plants and insects that alter myrosinase-catalysed hydrolysis of glucosinolates. FEBS J. 273, 2432–2446. doi: 10.1111/j.1742-4658.2006.05252.x
Burow, M., Rice, M., Hause, B., Gershenzon, J., and Wittstock, U. (2007). Cell-and tissue-specific localization and regulation of the epithiospecifier protein in Arabidopsis thaliana. Plant Mol. Biol. 64, 173–185. doi: 10.1007/s11103-007-9143-1
Capella, A., Menossi, M., Arruda, P., and Benedetti, C. (2001). COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Planta 213, 691–699. doi: 10.1007/s004250100548
Carter, C., Pan, S., Zouhar, J., Avila, E. L., Girke, T., and Raikhel, N. V. (2004). The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16, 3285–3303. doi: 10.1105/tpc.104.027078
Chen, S., and Andreasson, E. (2001). Update on glucosinolate metabolism and transport. Plant Physiol. Biochem. 39, 743–758. doi: 10.1016/S0981-9428(01)01301-8
Chen, Y., Fei, M., Wang, Y., Chen, S., and Yan, X. (2015). Proteomic investigation of glucosinolate systematically changes in Arabidopsis rosette leaves to exogenous methyl jasmonate. Plant Biosyst. 149, 346–353. doi: 10.1080/11263504.2013.819044
Chen, S., Glawischnig, E., Jørgensen, K., Naur, P., Jørgensen, B., Olsen, C. E., et al. (2003). CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J. 33, 923–937. doi: 10.1046/j.1365-313X.2003.01679.x
Chen, S., and Halkier, B. A. (1999). Functional expression and characterization of the myrosinase MYR1 from Brassica napus in Saccharomyces cerevisiae. Protein Expr. Purif. 17, 414–420. doi: 10.1006/prep.1999.1158
Chisholm, S. T., Mahajan, S. K., Whitham, S. A., Yamamoto, M. L., and Carrington, J. C. (2000). Cloning of the Arabidopsis RTM1 gene, which controls restriction of long-distance movement of tobacco etch virus. Proc. Natl. Acad. Sci. USA 97, 489–494. doi: 10.1073/pnas.97.1.489
Chou, K. C., and Shen, H. B. (2007). Large-scale plant protein subcellular location prediction. J. Cell. Biochem. 100, 665–678. doi: 10.1002/jcb.21096
Chou, K. C., and Shen, H. B. (2008). Cell-PLoc: a package of web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 3:153. doi: 10.1038/nprot.2007.494
Chou, K. C., and Shen, H. B. (2010). Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS One 5:e11335. doi: 10.1371/journal.pone.0011335
Clay, N. K., Adio, A. M., Denoux, C., Jander, G., and Ausubel, F. M. (2009). Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323, 95–101. doi: 10.1126/science.1164627
Dai, S., and Chen, S. (2012). Single-cell-type proteomics toward a holistic understanding of plant functions. Mol. Cell. Proteomics 11, 1622–1630. doi: 10.1074/mcp.R112.021550
Efroni, I., and Birnbaum, K. D. (2016). The potential of single-cell profiling in plants. Genome Biol. 17:65. doi: 10.1186/s13059-016-0931-2
Emanuelsson, O., Nielsen, H., Brunak, S., and Von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. doi: 10.1006/jmbi.2000.3903
Eriksson, S., Andréasson, E., Ekbom, B., Granér, G., Pontoppidan, B., Taipalensuu, J., et al. (2002). Complex formation of myrosinase isoenzymes in oilseed rape seeds are dependent on the presence of myrosinase-binding proteins. Plant Physiol. 129, 1592–1599. doi: 10.1104/pp.003285
Fahey, J. W., Zalcmann, A. T., and Talalay, P. (2001). The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51. doi: 10.1016/S0031-9422(00)00316-2
Fan, J., Crooks, C., Creissen, G., Hill, L., Fairhurst, S., Doerner, P., et al. (2011). Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331, 1185–1188. doi: 10.1126/science.1199707
Finiguerra, M. G., Iori, R., and Palmieri, S. (2001). Soluble and total myrosinase activity in defatted Crambe abyssinica meal. J. Agric. Food Chem. 49, 840–845. doi: 10.1021/jf000917h
Foo, H. L., Grønning, L. M., Goodenough, L., Bones, A. M., Danielsen, B. E., Whiting, D. A., et al. (2000). Purification and characterisation of epithiospecifier protein from Brassica napus: enzymic intramolecular sulphur addition within alkenyl thiohydroximates derived from alkenyl glucosinolate hydrolysis. FEBS Lett. 468, 243–246. doi: 10.1016/S0014-5793(00)01176-5
Frerigmann, H., Böttcher, C., Baatout, D., and Gigolashvili, T. (2012). Glucosinolates are produced in trichomes of Arabidopsis thaliana. Front. Plant Sci. 3:242. doi: 10.3389/fpls.2012.00242
Frerigmann, H., Piślewska-Bednarek, M., Sánchez-Vallet, A., Molina, A., Glawischnig, E., Gigolashvili, T., et al. (2016). Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 9, 682–695. doi: 10.1016/j.molp.2016.01.006
Fuchs, R., Kopischke, M., Klapprodt, C., Hause, G., Meyer, A. J., Schwarzländer, M., et al. (2016). Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28, 130–145. doi: 10.1105/tpc.15.00887
Gavin, A. C., Bösche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. doi: 10.1038/415141a
Geng, S., Misra, B. B., De Armas, E., Huhman, D. V., Alborn, H. T., Sumner, L. W., et al. (2016). Jasmonate-mediated stomatal closure under elevated CO2 revealed by time-resolved metabolomics. Plant J. 88, 947–962. doi: 10.1111/tpj.13296
Geshi, N., and Brandt, A. (1998). Two jasmonate-inducible myrosinase-binding proteins from Brassica napus L. seedlings with homology to jacalin. Planta 204, 295–304. doi: 10.1007/s004250050259
Grob, K., and Matile, P. H. (1979). Vacuolar location of glucosinolates in horseradish root cells. Plant Sci. Lett. 14, 327–335. doi: 10.1016/S0304-4211(79)90281-5
Grubb, C. D., and Abel, S. (2006). Glucosinolate metabolism and its control. Trends Plant Sci. 11, 89–100. doi: 10.1016/j.tplants.2005.12.006
Halkier, B. A. (2016). “General introduction to glucosinolates” in Advances in Botanical Research. Vol. 80, ed. S. Kopriva (Cambridge, MA, USA: Academic Press), 1–14.
Halkier, B. A., and Gershenzon, J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333. doi: 10.1146/annurev.arplant.57.032905.105228
Hasegawa, T., Yamada, K., Kosemura, S., Yamamura, S., and Hasegawa, K. (2000). Phototropic stimulation induces the conversion of glucosinolate to phototropism-regulating substances of radish hypocotyls. Phytochemistry 54, 275–279. doi: 10.1016/S0031-9422(00)00080-7
He, Y., Mawhinney, T. P., Preuss, M. L., Schroeder, A. C., Chen, B., Abraham, L., et al. (2009). A redox-active isopropylmalate dehydrogenase functions in the biosynthesis of glucosinolates and leucine in Arabidopsis. Plant J. 60, 679–690. doi: 10.1111/j.1365-313X.2009.03990.x
Hirai, M. Y., Sugiyama, K., Sawada, Y., Tohge, T., Obayashi, T., Suzuki, A., et al. (2007). Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA 104, 6478–6483. doi: 10.1073/pnas.0611629104
Ho, Y., Gruhler, A., Heilbut, A., Bader, G. D., Moore, L., Adams, S. L., et al. (2002). Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183. doi: 10.1038/415180a
Holmes, M. R. J. (1980). Nutrition of the oilseed rape crop. (London, UK: Applied Science Publishers).
Hosseinpour, B., HajiHoseini, V., Kashfi, R., Ebrahimie, E., and Hemmatzadeh, F. (2012). Protein interaction network of Arabidopsis thaliana female gametophyte development identifies novel proteins and relations. PLoS One 7:e49931. doi: 10.1371/journal.pone.0049931
Hull, A. K., Vij, R., and Celenza, J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97, 2379–2384. doi: 10.1073/pnas.040569997
Husebye, H., Chadchawan, S., Winge, P., Thangstad, O. P., and Bones, A. M. (2002). Guard cell-and phloem idioblast-specific expression of thioglucoside glucohydrolase 1 (myrosinase) in Arabidopsis. Plant Physiol. 128, 1180–1188. doi: 10.1104/pp.010925
Huttlin, E. L., Bruckner, R. J., Paulo, J. A., Cannon, J. R., Ting, L., Baltier, K., et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. doi: 10.1038/nature22366
Islam, M. M., Tani, C., Watanabe-Sugimoto, M., Uraji, M., Jahan, M. S., Masuda, C., et al. (2009). Myrosinases, TGG1 and TGG2, redundantly function in ABA and MeJA signaling in Arabidopsis guard cells. Plant Cell Physiol. 50, 1171–1175. doi: 10.1093/pcp/pcp066
James, D. C., and Rossiter, J. T. (1991). Development and characteristics of myrosinase in Brassica napus during early seedling growth. Physiol. Plant. 82, 163–170. doi: 10.1111/j.1399-3054.1991.tb00076.x
Jiang, M., Niu, C., Cao, J., Ni, D. A., and Chu, Z. (2018). In silico-prediction of protein–protein interactions network about MAPKs and PP2Cs reveals a novel docking site variants in Brachypodium distachyon. Sci. Rep. 8:15083. doi: 10.1038/s41598-018-33428-5
Johansson, O. N., Fantozzi, E., Fahlberg, P., Nilsson, A. K., Buhot, N., Tör, M., et al. (2014). Role of the penetration-resistance genes PEN 1, PEN 2 and PEN 3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J. 79, 466–476. doi: 10.1111/tpj.12571
Kelly, P. J., Bones, A., and Rossiter, J. T. (1998). Sub-cellular immunolocalization of the glucosinolate sinigrin in seedlings of Brassica juncea. Planta 206, 370–377. doi: 10.1007/s004250050412
Khokon, M. A. R., Jahan, M. S., Rahman, T., Hossain, M. A., Muroyama, D., Minami, I., et al. (2011). Allyl isothiocyanate (AITC) induces stomatal closure in Arabidopsis. Plant Cell Environ. 34, 1900–1906. doi: 10.1111/j.1365-3040.2011.02385.x
Kissen, R., and Bones, A. M. (2009). Nitrile-specifier proteins involved in glucosinolate hydrolysis in Arabidopsis thaliana. J. Biol. Chem. 284, 12057–12070. doi: 10.1074/jbc.M807500200
Koroleva, O. A., and Cramer, R. (2011). Single-cell proteomic analysis of glucosinolate-rich S-cells in Arabidopsis thaliana. Methods 54, 413–423. doi: 10.1016/j.ymeth.2011.06.005
Koroleva, O. A., Davies, A., Deeken, R., Thorpe, M. R., Tomos, A. D., and Hedrich, R. (2000). Identification of a new glucosinolate-rich cell type in Arabidopsis flower stalk. Plant Physiol. 124, 599–608. doi: 10.1104/pp.124.2.599
Kuchernig, J. C., Burow, M., and Wittstock, U. (2012). Evolution of specifier proteins in glucosinolate-containing plants. BMC Evol. Biol. 12:127. doi: 10.1186/1471-2148-12-127
Lambrix, V., Reichelt, M., Mitchell-Olds, T., Kliebenstein, D. J., and Gershenzon, J. (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13, 2793–2807. doi: 10.1105/tpc.13.12.2793
Lenman, M., Rödin, J., Josefsson, L. G., and Rask, L. (1990). Immunological characterization of rapeseed myrosinase. Eur. J. Biochem. 194, 747–753. doi: 10.1111/j.1432-1033.1990.tb19465.x
Li, S., Armstrong, C. M., Bertin, N., Ge, H., Milstein, S., Boxem, M., et al. (2004). A map of the interactome network of the metazoan C. elegans. Science 303, 540–543. doi: 10.1126/science.1091403
Lüthy, B., and Matile, P. (1984). The mustard oil bomb: rectified analysis of the subcellular organisation of the myrosinase system. Biochem. Physiol. Pflanz. 179, 5–12. doi: 10.1016/S0015-3796(84)80059-1
Luti, S., Caselli, A., Taiti, C., Bazihizina, N., Gonnelli, C., Mancuso, S., et al. (2016). PAMP activity of cerato-platanin during plant interaction: an-omic approach. Int. J. Mol. Sci. 17:866. doi: 10.3390/ijms17060866
MacLeod, A. J., and Rossiter, J. T. (1985). The occurrence and activity of epithiospecifier protein in some cruciferae seeds. Phytochemistry 24, 1895–1898. doi: 10.1016/S0031-9422(00)83087-3
Mikkelsen, M. D., Hansen, C. H., Wittstock, U., and Halkier, B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717. doi: 10.1074/jbc.M001667200
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Misra, B. B., Assmann, S. M., and Chen, S. (2014). Plant single cell and single cell-type metabolomics. Trends Plant Sci. 19, 637–646. doi: 10.1016/j.tplants.2014.05.005
Nakano, R. T., Piślewska-Bednarek, M., Yamada, K., Edger, P. P., Miyahara, M., Kondo, M., et al. (2017). PYK10 myrosinase reveals a functional coordination between endoplasmic reticulum bodies and glucosinolates in Arabidopsis thaliana. Plant J. 89, 204–220. doi: 10.1111/tpj.13377
Nintemann, S. J., Hunziker, P., Andersen, T. G., Schulz, A., Burow, M., and Halkier, B. A. (2018). Localization of the glucosinolate biosynthetic enzymes reveals distinct spatial patterns for the biosynthesis of indole and aliphatic glucosinolates. Physiol. Plant. 163, 138–154. doi: 10.1111/ppl.12672
Nintemann, S. J., Vik, D., Svozil, J., Bak, M., Baerenfaller, K., Burow, M., et al. (2017). Unravelling protein-protein interaction networks linked to aliphatic and indole glucosinolate biosynthetic pathways in Arabidopsis. Front. Plant Sci. 8:2028. doi: 10.3389/fpls.2017.02028
Petersen, B., Chen, S., Hansen, C., Olsen, C., and Halkier, B. (2002). Composition and content of glucosinolates in developing Arabidopsis thaliana. Planta 214, 562–571. doi: 10.1007/s004250100659
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–114. doi: 10.1023/A:1006380021658
Ratzka, A., Vogel, H., Kliebenstein, D. J., Mitchell-Olds, T., and Kroymann, J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 99, 11223–11228. doi: 10.1073/pnas.172112899
Reichelt, M., Brown, P. D., Schneider, B., Oldham, N. J., Stauber, E., Tokuhisa, J., et al. (2002). Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59, 663–671. doi: 10.1016/S0031-9422(02)00014-6
Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. doi: 10.1046/j.1365-313X.1998.00254.x
Shirakawa, M., and Hara-Nishimura, I. (2018). Specialized vacuoles of myrosin cells: chemical defense strategy in Brassicales plants. Plant Cell Physiol. 59, 1309–1316. doi: 10.1093/pcp/pcy082
Shroff, R., Vergara, F., Muck, A., Svatoš, A., and Gershenzon, J. (2008). Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 105, 6196–6201. doi: 10.1073/pnas.0711730105
Sobahan, M. A., Akter, N., Okuma, E., Uraji, M., Ye, W., Mori, I. C., et al. (2015). Allyl isothiocyanate induces stomatal closure in Vicia faba. Biosci. Biotechnol. Biochem. 79, 1737–1742. doi: 10.1080/09168451.2015.1045827
Sweetlove, L. J., and Fernie, A. R. (2013). The spatial organization of metabolism within the plant cell. Annu. Rev. Plant Biol. 64, 723–746. doi: 10.1146/annurev-arplant-050312-120233
Taipalensuu, J., Falk, A., and Rask, L. (1996). A wound-and methyl jasmonate-inducible transcript coding for a myrosinase-associated protein with similarities to an early nodulin. Plant Physiol. 110, 483–491. doi: 10.1104/pp.110.2.483
Thangstad, O. P., Gilde, B., Chadchawan, S., Seem, M., Husebye, H., Bradley, D., et al. (2004). Cell specific, cross-species expression of myrosinases in Brassica napus, Arabidopsis thaliana and Nicotiana tabacum. Plant Mol. Biol. 54, 597–611. doi: 10.1023/B:PLAN.0000038272.99590.10
Ueda, H., Nishiyama, C., Shimada, T., Koumoto, Y., Hayashi, Y., Kondo, M., et al. (2006). AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol. 47, 164–175. doi: 10.1093/pcp/pci232
Uetz, P., Giot, L., Cagney, G., Mansfield, T. A., Judson, R. S., Knight, J. R., et al. (2000). A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. doi: 10.1038/35001009
Vilakazi, C. S., Dubery, I. A., and Piater, L. A. (2017). Identification of lipopolysaccharide-interacting plasma membrane-type proteins in Arabidopsis thaliana. Plant Physiol. Biochem. 111, 155–165. doi: 10.1016/j.plaphy.2016.11.025
Warde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., et al. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38(suppl. 2), W214–W220. doi: 10.1093/nar/gkq537
Wienkoop, S., Zoeller, D., Ebert, B., Simon-Rosin, U., Fisahn, J., Glinski, M., et al. (2004). Cell-specific protein profiling in Arabidopsis thaliana trichomes: identification of trichome-located proteins involved in sulfur metabolism and detoxification. Phytochemistry 65, 1641–1649. doi: 10.1016/j.phytochem.2004.03.026
Wittstock, U., and Burow, M. (2010). Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8:e0134. doi: 10.1199/tab.0134
Wittstock, U., Kliebenstein, D. J., Lambrix, V., Reichelt, M., and Gershenzon, J. (2003). “Chapter five-Glucosinolate hydrolysis and its impact on generalist and specialist insect herbivores” in Recent Advances in Phytochemistry. Vol. 37, ed. J. T. Romeo (London, UK: Elsevier), 101–125.
Wittstock, U., Kurzbach, E., Herfurth, A.-M., and Stauber, E. J. (2016a). “Chapter six-Glucosinolate breakdown” in Advances in Botanical Research. Vol. 80, ed. S. Kopriva (Cambridge, MA, USA: Academic Press), 126–159.
Wittstock, U., Meier, K., Dörr, F., and Ravindran, B. M. (2016b). NSP-dependent simple nitrile formation dominates upon breakdown of major aliphatic glucosinolates in roots, seeds, and seedlings of Arabidopsis thaliana columbia-0. Front. Plant Sci. 7:1821. doi: 10.3389/fpls.2016.01821
Wu, L., Yu, M., Holowachuk, J., Sharpe, A., Lydiate, D., Hegedus, D., et al. (2017). Overexpression of Brassica napus myrosinase-associated protein 1 improved Sclerotinia sclerotiorum tolerance in Arabidopsis thaliana. Can. J. Plant Sci. 97, 842–851. doi: 10.1139/cjps-2016-0255
Xiang, X., Song, M., Zhaoyan, W., Tong, J., Zhang, L., Xiao, L., et al. (2011). A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J. Exp. Bot. 10, 1–13. doi: 10.1093/jxb/err226
Xu, Z., Escamilla-Treviño, L., Zeng, L., Lalgondar, M., Bevan, D., Winkel, B., et al. (2004). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367. doi: 10.1007/s11103-004-0790-1
Xu, J., Meng, J., Meng, X., Zhao, Y., Liu, J., Sun, T., et al. (2016). Pathogen-responsive MPK3 and MPK6 reprogram the biosynthesis of indole glucosinolates and their derivatives in Arabidopsis immunity. Plant Cell 28, 1144–1162. doi: 10.1105/tpc.15.00871
Xue, J., Jørgensen, M., Pihlgren, U., and Rask, L. (1995). The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol. Biol. 27, 911–922. doi: 10.1007/BF00037019
Yan, X., and Chen, S. (2007). Regulation of plant glucosinolate metabolism. Planta 226, 1343–1352. doi: 10.1007/s00425-007-0627-7
Zabala Mde, T., Grant, M., Bones, A. M., Bennett, R., Lim, Y. S., Kissen, R., et al. (2005). Characterisation of recombinant epithiospecifier protein and its over-expression in Arabidopsis thaliana. Phytochemistry 66, 859–867. doi: 10.1016/j.phytochem.2005.02.026
Zhang, F., Liu, S., Li, L., Zuo, K., Zhao, L., and Zhang, L. (2016). Genome-wide inference of protein-protein interaction networks identifies crosstalk in abscisic acid signaling. Plant Physiol. 171, 1511–1522. doi: 10.1104/pp.16.00057
Zhang, Z., Ober, J. A., and Kliebenstein, D. J. (2006). The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18, 1524–1536. doi: 10.1105/tpc.105.039602
Zhang, J., Pontoppidan, B., Xue, J., Rask, L., and Meijer, J. (2002). The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant. 115, 25–34. doi: 10.1034/j.1399-3054.2002.1150103.x
Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. doi: 10.1101/gad.1035402
Zhao, Z., Zhang, W., Stanley, B. A., and Assmann, S. M. (2008). Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell 20, 3210–3226. doi: 10.1105/tpc.108.063263
Zhu, M., and Assmann, S. M. (2017). Metabolic signatures in response to abscisic acid (ABA) treatment in Brassica napus guard cells revealed by metabolomics. Sci. Rep. 7:12875. doi: 10.1038/s41598-017-13166-w
Zhu, M., Zhu, N., Song, W. Y., Harmon, A. C., Assmann, S. M., and Chen, S. (2014). Thiol-based redox proteins in abscisic acid and methyl jasmonate signaling in Brassica napus guard cells. Plant J. 78, 491–515. doi: 10.1111/tpj.12490
Keywords: glucosinolate, myrosinase, cell type, metabolism, protein-protein interaction
Citation: Chhajed S, Misra BB, Tello N and Chen S (2019) Chemodiversity of the Glucosinolate-Myrosinase System at the Single Cell Type Resolution. Front. Plant Sci. 10:618. doi: 10.3389/fpls.2019.00618
Edited by:
Kazuki Saito, RIKEN Center for Sustainable Resource Science (CSRS), JapanReviewed by:
Ryosuke Sugiyama, RIKEN Center for Sustainable Resource Science (CSRS), JapanFranziska S. Hanschen, Leibniz-Institut für Gemüse-und Zierpflanzenbau (IGZ), Germany
Copyright © 2019 Chhajed, Misra, Tello and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sixue Chen, c2NoZW5AdWZsLmVkdQ==
†These authors have contributed equally to this work
 Shweta Chhajed
Shweta Chhajed Biswapriya B. Misra
Biswapriya B. Misra Nathalia Tello
Nathalia Tello Sixue Chen
Sixue Chen