
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 08 May 2019
Sec. Plant Pathogen Interactions
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00570
This article is part of the Research TopicEvolutionary Trajectories in Plant-Associated Pseudomonas and Xanthomonas StrainsView all 12 articles
The Pseudomonas syringae complex comprises different genetic groups that include strains from both agricultural and environmental habitats. This complex group has been used for decades as a “hodgepodge,” including many taxonomically related species. More than 60 pathovars of P. syringae have been described based on distinct host ranges and disease symptoms they cause. These pathovars cause disease relying on an array of virulence mechanisms. However, P. syringae pv. syringae (Pss) is the most polyphagous bacterium in the P. syringae complex, based on its wide host range, that primarily affects woody and herbaceous host plants. In early 1990s, bacterial apical necrosis (BAN) of mango trees, a critical disease elicited by Pss in Southern Spain was described for the first time. Pss exhibits important epiphytic traits and virulence factors, which may promote its survival and pathogenicity in mango trees and in other plant hosts. Over more than two decades, Pss strains isolated from mango trees have been comprehensively investigated to elucidate the mechanisms that governs their epiphytic and pathogenic lifestyles. In particular, the vast majority of Pss strains isolated from mango trees produce an antimetabolite toxin, called mangotoxin, whose leading role in virulence has been clearly demonstrated. Moreover, phenotypic, genetic and phylogenetic approaches support that Pss strains producers of BAN symptoms on mango trees all belong to a single phylotype within phylogroup 2, are adapted to the mango host, and produce mangotoxin. Remarkably, a genome sequencing project of the Pss model strain UMAF0158 revealed the presence of other factors that may play major roles in its different lifestyles, such as the presence of two different type III secretion systems, two type VI secretion systems and an operon for cellulose biosynthesis. The role of cellulose in increasing mango leaf colonization and biofilm formation, and impairing virulence of Pss, suggests that cellulose may play a pivotal role with regards to the balance of its different lifestyles. In addition, 62-kb plasmids belonging to the pPT23A-family of plasmids (PFPs) have been strongly associated with Pss strains that inhabit mango trees. Further, complete sequence and comparative genomic analyses revealed major roles of PFPs in detoxification of copper compounds and ultraviolet radiation resistance, both improving the epiphytic lifestyle of Pss on mango surfaces. Hence, in this review we summarize the research that has been conducted on Pss by our research group to elucidate the molecular mechanisms that underpin the epiphytic and pathogenic lifestyle on mango trees. Finally, future directions in this particular plant–pathogen story are discussed.
Pseudomonas syringae complex has been traditionally used as a taxonomic hodgepodge that currently includes 15 recognized bacterial species and more than 60 different pathovars of the sensu stricto species P. syringae (Gomila et al., 2017). The taxonomy of the P. syringae complex has been widely discussed over the last 40 years, yet still remains a controversial group. The classification of this group is defined based on host range and symptomatology, dividing P. syringae species into pathogenic varieties known as pathovars (Dye et al., 1980; Young, 2010). The pathovar-based classification is widely accepted even today, but does not reveal the genetic relationships between pathovars. Initial genomic studies were based on DNA-DNA hybridization methods (Palleroni et al., 1972; Pecknold and Grogan, 1973; Denny et al., 1988; Gardan et al., 1992; Janse et al., 1996). Gardan et al. (1999) described nine discrete genomospecies classification groups that have been widely accepted until recently. Phylogenetic approaches based on multilocus sequence typing analysis (MLST) have had a significant impact on P. syringae classification (Sarkar and Guttman, 2004; Hwang et al., 2005; Almeida et al., 2010; Bull et al., 2011; Berge et al., 2014). Although the classification proposed by Berge et al. (2014) is generally accepted, a recent study using comparative genomics of the whole genome sequences of this species proposed the delineation of phylogenomic P. syringae complex and confirmed, as one might expect, that a high proportion of strains were misclassified (Gomila et al., 2017). Significantly, different P. syringae strains isolated from different sources (i.e., snow, irrigation water, and a diseased crop) have been identified as belonging to the same evolutionary lineage (Monteil et al., 2016). This fact suggests that the evolutionary history of the plant pathogen P. syringae is linked to the water cycle, which promoted the colonization of agricultural and non-agricultural habitats (Morris et al., 2008).
Pseudomonas syringae species possess a great diversity of virulence factors, such as a type III secretion system (T3SS) and its effector repertoires, toxic compounds, exopolysaccharides, ice nucleation activity, cell-wall-degrading enzymes and plant hormones, that make it the model phytopathogenic bacterium for understanding plant–pathogen interactions. Additionally, adaptation mechanisms to its plant hosts and microbial evolution have more recently become of great interest to many research groups (Xin et al., 2018). In particular, Pseudomonas syringae pv. syringae (Pss), has been described as the most polyphagous bacterium into the P. syringae complex due to its broad host range (Kennelly et al., 2007). Pss strains isolated from mango trees were identified as the causative agent of bacterial apical necrosis (BAN) disease of mango trees, which is the most limiting factor for mango crop in the Mediterranean region (Cazorla et al., 1998). A novel antimetabolite toxin called “mangotoxin” was reported to be intimately associated with all Pss strains isolated from mango trees, and with a few Pss strains from other hosts (Arrebola et al., 2003). The presence of different variants of copper resistance genes, as well as ultraviolet resistance determinants, were found to be associated with 62-kb plasmids belonging to the pPT23A family plasmids (PFPs) (Cazorla et al., 2002, 2008; Gutiérrez-Barranquero et al., 2013b). In addition, several studies have attempted to unravel the biosynthesis pathway and the regulatory mechanisms of mangotoxin production (Arrebola et al., 2007; Arrebola et al., 2012; Carrión et al., 2012, 2014). A molecular evolutionary approach using mangotoxin biosynthetic operon gene cluster, revealed that this operon was specifically distributed within the P. syringae Genomospecies 1, and which was acquired only once during evolution (Carrión et al., 2013). Moreover, a diversity survey of Pss strains isolated from mango trees was performed using phenotypic, genetic and phylogenetic approaches based on MLST analysis (Gutiérrez-Barranquero et al., 2013a) in order to understand the epidemiology of BAN disease. This study strongly indicated that Pss isolated from mango trees were forming a single phylotype inside the Pss species, characterized mainly by its adaptation to the mango host and by the production of mangotoxin. Subsequently, and due to the genome sequencing project of the model strain Pss UMAF0158 (Martínez-García et al., 2015), a gene cluster involved in the production of cellulose was discovered (Arrebola et al., 2015). This study demonstrated that cellulose was an important exopolysaccharide (EPS) to attach to the mango surface that could also act as a switch modulating the transition from epiphytic to pathogenic phases of Pss on the mango host. Finally, a PFPs sequencing project determined the importance of the 62-kb plasmids in improving the epiphytic survival of Pss strains isolated from mango trees (Gutiérrez-Barranquero et al., 2017a).
Therefore, this review summarizes the work that has been conducted on Pss strains isolated from mango trees over more than two decades of research. This phytopathogenic bacterium has arisen as a particular pathogen developing important features that modulate their epiphytic and pathogenic lifestyle phases on the mango tree surface.
Mango crops (Mangifera indica L.) are present in many tropical and subtropical regions and represent one of the most important subtropical fruit crops distributed worldwide (Galán-Saúco, 2015). This crop was established in Southern Spain in Malaga in the early 1980s. The pace of the planting of this crop was relatively high over the last few years, expanding from 800 hectares (ha) in 2004 to 4500 ha in 2016 in Spain, of which more than 2,000 ha are in full production (Gutiérrez-Barranquero et al., 2017b). Very recent data claim that there are more than 6,000 ha, of which more than 3,000 are currently in full production, which would break the historical record of more than 30,000 tons of mango fruit harvested (Anonymous, 2018) August. Thus, the mango crop has been considered one of the most promising crops in Southern Spain, mainly in the tropical coastal areas of Malaga and Granada. As new crops are deployed in new regions, there might be spill-over effects and the emergence of new diseases. The commercial viability of this crop has been threatened by different bacterial and fungal plant pathogens (Bradbury, 1986; Gagnevin and Pruvost, 2001; Gutiérrez-Barranquero et al., 2019). In Southern Spain, the fungal pathogen Fusarium mangiferae which causes mango malformation disease (Crespo et al., 2012) and Pss the causal agent of BAN disease (Cazorla et al., 1998) are the most severe phytopathogens causing important economic losses. The main symptomatology associated with BAN disease, the isolation and identification of Pss as the causal agent of BAN disease, and the control methods specifically tested to limit and prevent Pss infections are discussed in detail below.
The mango crop develops well at temperatures between 20 and 25°C, reaching a dormancy period when the temperature is below 15°C (Samson, 1986; Galán-Saúco, 2015). Thus, cool temperatures and wet periods play an important role in favoring the development of BAN symptoms, which has also been described in other infections caused by P. syringae in other woody hosts (Kennelly et al., 2007). Rain or dew are essential for inoculum dissemination to other buds and leaves, and wind exposure facilitates BAN development by causing microinjuries (Cazorla et al., 1998). BAN disease on mango trees is characterized by rapidly expanding necrotic spots on buds and leaves from October–November. January–February are the coolest and rainiest months in Southern Spain, giving rise to the highest incidence of necrotic symptoms, which is consistent with the period with the largest Pss population on mango trees (Cazorla et al., 1998). Additionally, at this time the symptoms can extend from buds through the leaf petiole to reach the leaves and stems. Typically, lesions on leaves start as interveinal, angular, water-soaked spots that may coalesce, becoming black and slightly raised. Importantly, favorable weather conditions for the pathogen that are maintained throughout the winter and even into the spring season can promote the appearance of wood necrosis on branches to such a degree that, in extreme cases, this can lead to the death of the tree. These symptoms are quite similar to those described for blossom blast of pear and stone fruits (English et al., 1980). Additionally, a white milky gum exudate can also be observed. Necrotic symptoms affecting flower panicles are less frequently observed but can become very apparent in years with severe attacks. These symptoms cause the most severe economic losses due to decreases in fruit yield (Cazorla et al., 1998). The typical symptoms of BAN disease of mango trees are summarized in Figure 1.

Figure 1. Typical symptoms of bacterial apical necrosis (BAN) disease on mango trees. (A) Healthy mango tree. (B) Mango tree affected by BAN disease. (C) Healthy mango apical bud. (D) Typical gum exudes on mango apical bud. (E) Initial necrotic spots on mango apical bud. (F) Severe necrosis of mango apical bud. (G) Necrotic symptoms progression from apical bud to leaves through the petiole. (H) Dead mango apical bud and surrounded leaves. (I) Flower panicles. Yellow arrow: healthy mango flower panicle; red arrow: necrosis on mango flower panicle.
The phytopathogenic bacterium P. syringae has the ability to survive as an epiphyte on plant surfaces. During its epiphytic phase, P. syringae has to cope with different abiotic factors by using different mechanisms (Sundin and Jacobs, 1999; Yu et al., 1999; Lindow and Brandl, 2003), which allow it to achieve large population sizes before starting an infection process (Hirano and Upper, 2000). Although P. syringae can elicit disease symptoms in a wide variety of woody and herbaceous plants, P. syringae has been considered a weak pathogen because the infection process on their plant hosts can be strongly improved by frost damage or mechanical injury. Thus, P. syringae can elicit disease outbreaks in temperate regions distributed worldwide in important crops, causing significant yield losses (Kennelly et al., 2007). Since the early 1990s, necrotic symptoms have been observed in apical buds, leaves and stems in mango trees in Southern Spain and Portugal (Cazorla et al., 1998). In years with severe attacks, which correlate with cool and wet winters, necrotic symptoms were more evident in the mango tree canopy and could cause a reduction of 30–50% in mango fruit production (Gutiérrez-Barranquero et al., 2012). Preliminary isolation from the edge of necrotic tissues of mango trees revealed that over 90% of bacterial isolates recovered were fluorescent Pseudomonas. Similar necrotic symptoms have been reported in many other woody hosts infected by Pss, such as peaches (Endert and Ritchie, 1984), citrus (Mirik et al., 2005; Ivanović et al., 2017), cherry (Sundin et al., 1989; Wenneker et al., 2013), almond (Lindow and Connell, 1984), apple (Mansvelt and Hattingh, 1986; Gasic et al., 2018) and pear (Montesinos and Vilardell, 1991; Xu et al., 2008). Different biochemical and physiological characteristics suggested the tentative identification of P. syringae. Furthermore, the presence of ice nucleation activity (INA), a virulence trait well-documented in P. syringae to be used by the bacterium to cause micro-wounds on the plant surface to provide an entry way to the plant (Hirano and Upper, 1995; Hwang et al., 2005), was found in bacterial isolates following a protocol previously described by Cazorla et al. (1995). The production of lipodepsipeptidic toxins typically associated with P. syringae, such as syringomycin and syringopeptins, were also confirmed in bacterial isolates from mango (Gross and DeVay, 1977; Ballio et al., 1991; Arrebola et al., 2003). All the results obtained conclusively confirmed that the bacterial isolates associated with necrotic symptoms in mango trees belonged to the P. syringae species (Cazorla et al., 1992, 1998). P. syringae is a highly heterogeneous species comprising more than 60 pathovars (Young, 2010). To determine which pathovar was the causal agent of necrotic symptoms, different pathogenicity tests were performed in tomato and lilac plants, immature lemon and pear fruits, and bean pods (Lelliott and Stead, 1987). All P. syringae strains assayed induced typical symptoms in all plant hosts of Pss. Once the bacterial strains associated with necrotic symptoms in mango trees were identified, a pathogenicity test in adult mango plants was carried out in order to fulfill Koch’s postulates. Two different experiments under field conditions were performed using 2-year-old mango plants growing in pots. Buds and stems were inoculated with 10 μl of bacterial suspensions using a microsyringe. Necrotic symptoms developed in the inoculated mango trees, and the incidence and severity of necrotic symptoms that occurred in each experiment (i.e., different years) were different, indicating the importance of the weather conditions in symptom development, as has been previously observed for P. syringae in other hosts (Hirano and Upper, 2000). The subsequent re-isolation from the necrotic lesions artificially reproduced in mango tissues and the subsequent identification confirmed that Pss was the causal agent of bacterial apical necrosis (BAN) of mangos (Cazorla et al., 1998).
Therefore, the life cycle of Pss on mango trees is clearly divided first, in an epiphytic phase, in which Pss has to survive and grow under harsh environmental conditions, and second, in a pathogenic phase to produce BAN symptomatology. In both phases, different genetic traits are expressed to either, improve survival or to enhance an infection process (Figure 2).
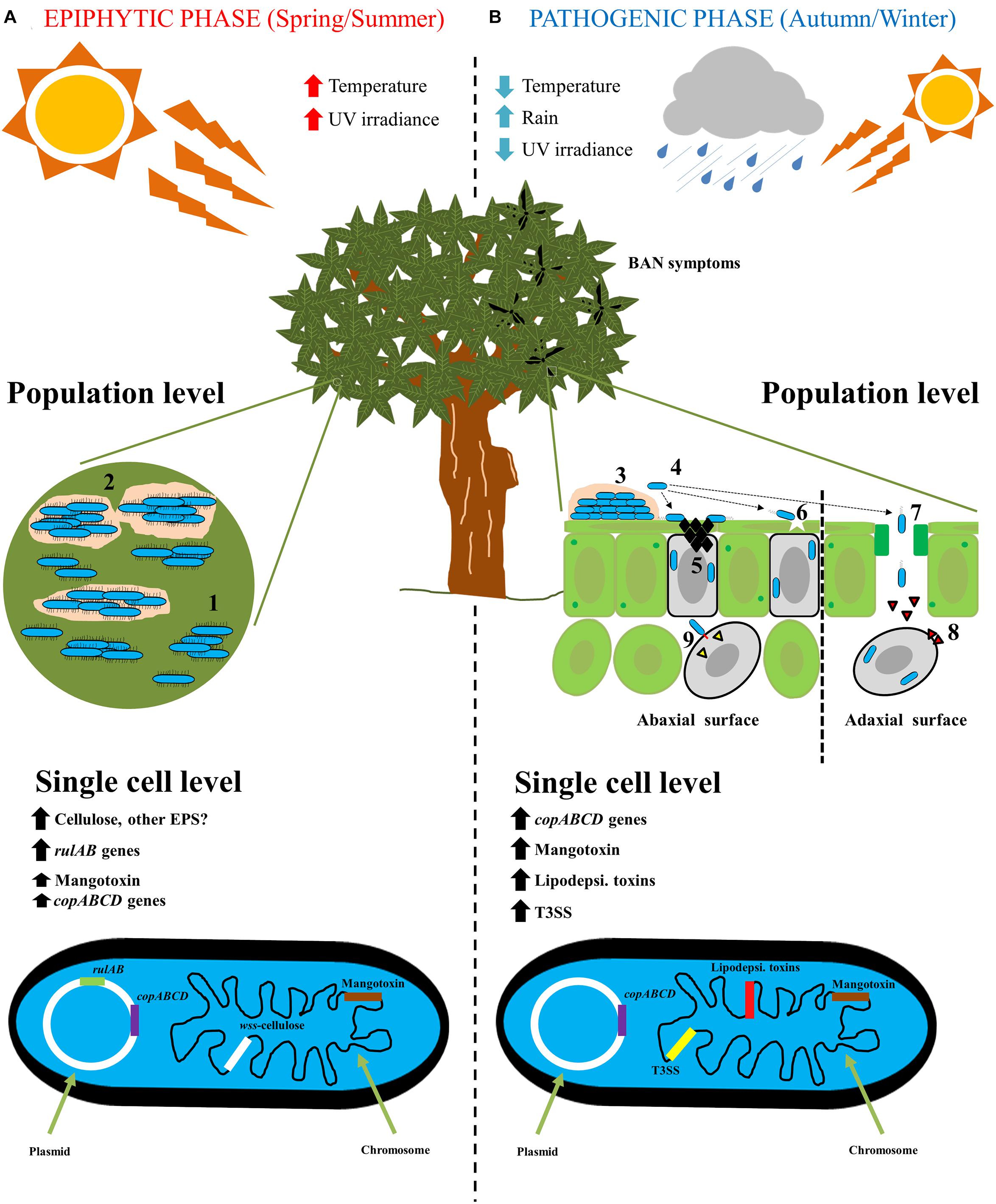
Figure 2. The life cycle of Pseudomonas syringae pv. syringae on mango trees. (A) Epiphytic phase of P. syringae pv. syringae on mango trees is developing mainly in spring/summer seasons, where high temperature and high UV radiation are present. At population level, P. syringae pv. syringae is present mainly on the buds and leaves surfaces forming microcolonies (1), that will subsequently form a mature biofilm with the biosynthesis of an extracellular matrix (2). At single cell level, rulAB operon encoded by 62-kb PFP plasmid involved in resistance to UV radiation, and wss operon present at the chromosome and involved in the biosynthesis of cellulose, are both highly expressed. On the contrary, copABCD operon encoded by 62-kb PFP plasmid involved in copper resistance, and mbo operon located at the chromosome and involved in mangotoxin biosynthesis, are less expressed. (B) Pathogenic phase of P. syringae pv. syringae on mango trees arise primarily in autumn/winter seasons, where low temperatures, low UV radiation and high rainfall are present. At population level, the infection process on mango leaves and buds is the following: (3) epiphytic survival and biofilm formation; (4) biofilm disassembly and bacterial migration; (5) Ice nucleation activity to damage mango surfaces; (6) bacterial entry into cells by microinjuries; (7) Bacterial entry into cells through stomata; (8) Release of phytotoxins; and (9) Release of type III effectors by using the type III secretion system. At single cell level, firstly, copABCD operon involved in detoxification of copper compounds is highly expressed in response to copper treatment applications by farmers. Then, all genes that encode virulence factors are highly expressed (Mangotoxin, lipodepsipeptidic toxins and type III secretion system and its effectors) to elicit the typical BAN disease symptoms.
Management of woody plant diseases caused by P. syringae, and particularly those provoked by Pss, are a major concern for growers worldwide due to the broad host range. Sprays of copper compounds have been used for decades as standard bactericides to combat many bacterial diseases, but their use is subject to a number of constraints (Kennelly et al., 2007). The most common treatment for controlling BAN disease in Southern Spain is the spraying of a copper compound with a film-forming mode of action known as Bordeaux mixture (BM). However, different copper-based compounds fail to protect against BAN. Unfortunately, continuous treatments with copper sprays can lead to many problems. The efficacy of copper treatments for the control of bacterial diseases is often limited, largely due to the selection of copper-resistant strains; this has previously been described for Pss strains isolated from mango trees (Cazorla et al., 2002). Another serious problem associated with the excessive usage of copper is that copper is a major heavy metal contaminant that accumulates in soil from different sources (Wang, 1997; Xiong, 1998; Kabata-Pendias, 2001). Copper has demonstrated toxicity to roots and young shoots and leaves (Kairu et al., 1985; Alva and Graham, 1991; Iannotta et al., 2007), and has sustained bioaccumulation effects (Xiong and Wang, 2005). Finally, the European Union has introduced legislation limiting the use of copper compounds in regulation No. 473/2002 (Anonymous, 2002). For all of these reasons, there has been an urgent need expressed by growers and extension services to search for alternative treatments to copper compounds that may be effective for the control of BAN disease. In this context, Cazorla et al. (2006) evaluated the capacity of several different control treatments to cope with BAN disease in mango crops. In addition, the mechanisms of action of the different treatments were examined, analyzing their effect on Pss population levels. The treatments assayed in this work included BM, fosetyl-Al, gibberellic acid, acibenzolar-S-methyl, silicon gel (soluble potassium silicate 34%) and combined treatments (Cazorla et al., 2006). Interestingly, treatments reduced symptoms but did not reduce the size of the pathogen population, suggesting a non-bactericidal mode of action of these compounds. After evaluation of the different treatments, this study concluded that the best treatment to control BAN disease was conventional copper-based treatment BM. However, there were promising effects showed by other assayed treatments against BAN disease, indicating that a few of them could be interesting alternatives to traditional chemical control (Cazorla et al., 2006). The silicon gel was highly relevant, because its reduction of necrotic symptoms in apical buds was similar to the levels obtained with BM; it also has potential for use in organic farming.
Due to the limitations concerning the use of copper compounds, together with the increasing demand for organic crops, have led to in-depth analysis of different alternative treatments to combat plant diseases. Particularly, Gutiérrez-Barranquero et al. (2012) performed a study where they analyzed different alternative treatments, including the silicon gel that previously showed potential to control BAN disease. In this study after different scale trials (small, semi-commercial, and commercial), confirmed the efficacy of silicon gel to control BAN disease, reducing the occurrence of necrotic symptoms at a similar level to the conventional treatment BM. Moreover, mango growers directly observed the effectiveness of silicon gel, and thus, this treatment has been registered for commercial use in mango crops in Spain as a phytostrengthener compatible with organic farming (Gutiérrez-Barranquero et al., 2012). Interestingly, silicon gel failed to reduce the bacterial population in mango tress, suggesting a film-forming mode of action acting as a physical barrier to avoid the entry of the pathogen, as it was previously reported for BM (Becerra, 1995). A similar mode of action has been previously described for silicon protective effects in other plant hosts against fungal and bacterial pathogens (Diogo and Wydra, 2007; Guével et al., 2007; Sun et al., 2010). However, other putative modes of action for silicon gel cannot be ruled out, as might be the induction of systemic resistance (ISR) (Bélanger et al., 2003; Rodrigues et al., 2003; Rodgers-Gray and Shaw, 2004; Fauteux et al., 2005) and to enhance cell wall lignification (Kim et al., 2002).
Plant surfaces are hostile and dynamic environments for plant-associated bacteria due to rapidly changing climatic conditions (Lindow and Brandl, 2003). P. syringae is an epiphytic bacterium and an opportunistic plant pathogen that needs to survive on plant surfaces (Hirano and Upper, 2000). Before initiating infection, P. syringae has to face environmental abiotic stressors via different survival mechanisms (Sundin and Jacobs, 1999; Yu et al., 1999; Lindow and Brandl, 2003). The life cycle of Pss on mango plant surfaces (as depicted in Figure 2) involves an epiphytic phase mainly during the spring and summer seasons, that subsequently leads to an infection process during the autumn and winter seasons, when the weather conditions are favorable for the disease development (Cazorla et al., 1998).
Pseudomonas syringae pv. syringae isolated from mango trees has therefore developed different strategies to survive on the mango plant surface. Where present, the 62 Kb PFP plasmids exhibit a key role (Cazorla et al., 2002, 2008; Arrebola et al., 2009; Gutiérrez-Barranquero et al., 2013b, 2017a). Recently, other important genes located on the chromosomal genetic material have been described as having a primary role in adhesion and subsequent biofilm formation on mango plant surfaces (Arrebola et al., 2015).
Plasmids have been reported to be one of the most important sources for bacterial evolution, due to their ability to acquire foreign DNA and be rapidly transmitted among bacteria via the horizontal gene transfer process (Vivian et al., 2001; Norman et al., 2009). Plasmids are part of the flexible genome and represent a portion of the genome that does not contribute to basic survival functions. However, plasmids encompass important genes that can improve the ecological fitness of their bacterial hosts (Medini et al., 2005; Sundin, 2007) and improve virulence mechanisms (Jackson et al., 1999; Arnold et al., 2001). The PFPs are a family of native plasmids that appear to be indigenous to P. syringae. All PFP plasmids share a major replication protein, gene repA (Sesma et al., 1998, 2000). Apart from specific genes involved in self-maintenance and replication processes of PFPs, different genes implicated in virulence and/or ecological fitness are encoded. In particular, copper- and ultraviolet radiation-resistance genes are two of the most widely distributed genes in this family of plasmids, which play a fundamental role in epiphytic survival (Sundin, 2007).
As mentioned previously, the use of copper compounds has been strongly associated with agriculture (Lamichhane et al., 2018). The extensive use of copper by growers led to an increase in the dosage and frequency of applications, giving rise the emergence of copper-resistant strains, a concerning issue that is very common among plant pathogenic bacteria, such as P. syringae (Sundin et al., 1989; Andersen et al., 1991; Sundin and Bender, 1993; Scheck and Pscheidt, 1998). In Southern Spain, different copper compounds have been largely used to control BAN disease in mango trees, as well as other plant diseases. This suggests that the selection of copper-resistant strains could be a major reason for further control failures with copper bactericides. The copABCD operon is the most common genetic determinant associated with copper resistance in P. syringae and has been reportedly associated with conjugative native PFP plasmids (Bender and Cooksey, 1986; Cooksey, 1987; Lim and Cooksey, 1993; Sundin and Bender, 1996). The copABCD operon encoded by a 35-kb plasmid from P. syringae pv. tomato was the first of these genes to be sequenced (Mellano and Cooksey, 1988). Based on this background, a study was performed to analyze the role of the copABCD operon in copper treatment tolerance, as well as its association with PFP plasmids in Pss strains isolated from mango trees (Cazorla et al., 2002). The presence of the copABCD operon and its association with PFPs plasmids was further analyzed. Over 75% of the copper-resistant strains, harbored 62-kb plasmids that showed a hybridization signal by Southern blot analysis with the copABCD probe obtained from P. syringae pv. tomato PT23 (Bender and Cooksey, 1987). The copABCD operon is also encoded, albeit to a lesser extent, in the chromosome, as well as in 120- and 45-kb plasmids. This observation suggested that different variants of copper resistance determinants could be found in Pss mango populations, as has previously been reported in other Pss populations (Sundin and Bender, 1993; Rogers et al., 1994). These data were also supported by 62-kb plasmids restriction profiles, identifying different restriction profiles in both copper-resistant and copper-sensitive plasmids. Moreover, in order to determine whether those plasmids were conjugative and also the main determinants of copper resistance, mating experiments proved that those plasmids were conjugative and were involved in the copper resistance phenotype. The presence of copper-resistant conjugative plasmids could be considered the main cause of control strategy failures when treating with copper bactericides. Thus, field experiments where copper treatments were applied to mango trees once per month, from September to June, were analyzed to assess the emergence of copper-resistant strains. It was clearly demonstrated that excessive usage of copper in mango trees to control BAN disease promoted an increase in copper-resistant strains, which could be mainly due to the ability of these plasmids with be transmitted by conjugative processes (Cazorla et al., 2002).
Subsequently, based on a PFPs sequencing project that included strains that harbored different variants of copper-resistance determinants (Gutiérrez-Barranquero et al., 2017a), it was shown that the presence of a novel genetic structure in Pss UMAF0081 strain isolated from mango increased copper-resistance phenotypes. This novel genetic structure encoded the cusCBA genes (detoxifying monovalent cations of silver and copper) and copG, a putative metal-transporting P-type ATPase, both inserted within the copABCD operon (Gutiérrez-Barranquero et al., 2013b). Furthermore, the novel genetic structure was found in another strain of Pss analyzed in this study (Pss 6–9 strain isolated from sweet cherry), and was also present in another two strains from the database that belonged to different pathovars (ATCC1128 pv. tabaci and NCPPB1108 pv. tomato). This structure encompassed 15 genes that were more than 17 kb in size, according to data that was recently updated (Gutiérrez-Barranquero et al., 2017a). To determine whether those extra genes were responsible for the increase in copper resistance, the minimal inhibitory concentrations of copper and other heavy metals were investigated. A collection of Pss strains isolated from mangos and others hosts, two strains from different pathovars, a transconjugant strain obtained previously (Cazorla et al., 2002, FF5-km + 62-kb 0081 plasmid), and two Pss FF5 transformants that harbored copG and cusCBA were independently evaluated. It was observed that the transconjugant strain showed the same MIC value for copper as the original 0081 strain; the transformed strains also had increased their MIC values in comparison with the copper-sensitive parental FF5 strain (Sundin and Bender, 1993). A growth curve performed in minimal medium supplemented with 0.8 mM of copper sulfate clearly demonstrated that copG and cusCBA were responsible for the increase in copper resistance. The role of cusCBA in detoxifying heavy metals has been previously reported in Cupriavidus metallidurans (Mergeay et al., 2003; Von Rozycki and Nies, 2009), Escherichia coli (Franke et al., 2003) and Pseudomonas putida KT2440 (Cánovas et al., 2003; Leedjärv et al., 2008). Finally, qRT-PCR experiments were performed to analyze the expression profiles of copG and cusA in the presence or absence of 0.8 mM copper sulfate. The results showed that the expression levels of cusA and copG increased 13- and 100-fold, respectively, in the presence of copper, and the expression of cusA was 3-fold higher than copG. These results confirmed the previous results obtained in the MIC and growth curve experiments, supporting the hypothesis that the novel rearrangement of three different genetic determinants into a conjugative plasmid increases copper resistance in P. syringae (Gutiérrez-Barranquero et al., 2013b). Thus, the presence of different copper-resistance structures associated primarily with 62-Kb PFPs plasmids has been demonstrated in Pss strains isolated from mango trees. However, little is known concerning the dynamics of maintenance or preference of the different types of 62-kb plasmids in Pss mango populations.
UV radiation affects bacterial communities that are intimately associated with plant surfaces; to overcome this growth-limiting environmental stress, different mechanisms have been developed (Beattie and Lindow, 1995; Sundin and Jacobs, 1999; Jacobs and Sundin, 2001). Among the different mechanisms described for avoiding UV damage, the presence of DNA repair mechanisms, such as rulAB operon encoded by PFP plasmids, are the most relevant in Pss (Sundin et al., 1996; Sesma et al., 1998; Sundin and Murillo, 1999; Sundin et al., 2000). In Southern Spain, mango crops are exposed to high UV radiation, especially in the spring and summer seasons. These highly restrictive solar radiation conditions suggest that a similar rulAB-like operon could play an indispensable role in the epiphytic survival of Pss associated with mango trees. As noted above, there was a high incidence of 62-kb plasmids associated with Pss isolated from mango trees that belong to the PFP family, which were also strongly associated with copper resistance phenotype (Cazorla et al., 2002). In this sense, Cazorla et al. (2008) analyzed the presence of the rulAB-like operon and its role in UV radiation tolerance in the 62-Kb PFP plasmids. Over 62% of the strains analyzed harbored a 62-kb plasmid. Additionally, it was observed that the Pss strains harboring 62-kb plasmids, rather than those lacking plasmids or having a different plasmid, were more tolerant to UVC exposure and were able to maintain higher population levels in vitro. However, the UVC wavelengths do not naturally reach the earth’s surface; thus, its impact on ecological fitness is low (Kim and Sundin, 2000). Subsequently, two different exposures conditions of UVA+B (high irradiation and similar radiation in a summer day in Southern Spain) were tested, and in both conditions, the role of plasmids in UVA+B tolerance was demonstrated. This result reinforced the importance of 62-kb plasmids in epiphytic survival of Pss isolated from mango trees in Southern Spain. Finally, the role of 62-kb plasmids in UV tolerance was tested in vivo on mango leaf surfaces, evaluating different conditions (leaves in sunny and shady areas, and adaxial and abaxial parts of the leaves). Once again, a greater surviving population of Pss was observed in the strains harboring 62-kb PFP plasmids, although this difference was only notable in the adaxial side of leaves exposed to direct sunlight radiation (Cazorla et al., 2008). Therefore, it has been clearly demonstrated the rulAB+ Pss strains shown an advantage regarding their epiphytical fitness, and thus this operon plays a relevant role in growth and dispersion of Pss on mango surfaces during its harsh epiphytic phase suffered in Southern Spain. This competitive advantage may be promoting the selection and the dispersion of these plasmids among the mango microbiome.
Exopolysaccharides (EPS) have been reported to play essential roles in plant colonization and epiphytic survival of plant-associated bacteria (Pfeilmeier et al., 2016), including P. syringae (Yu et al., 1999). Different EPS have been associated with different functions of P. syringae during the epiphytic phase on the plant surface, as well as with its pathogenic lifestyle. Alginate is one of the most-studied EPS in P. syringae, and its involvement in osmotic stress tolerance, epiphytic survival, and virulence has been well-established (Yu et al., 1999; Freeman et al., 2013). Although the role of alginate and levan are not directly related to biofilm formation (Laue et al., 2006), their role in the initial stages of adhesion prior to biofilm development cannot be ignored (Yu et al., 1999). In addition, the putative role of levan as a nutrient source in mature biofilms, as well as its activity as a barrier blocking the recognition by the plant during pathogenesis, have been proposed (Kasapis et al., 1994; Laue et al., 2006). Cellulose is an important EPS that is well-documented in many bacterial species (Römling and Galperin, 2015). It is an integral part of extracellular matrix components of biofilms, mainly in environmental and pathogenic Pseudomonas (Ude et al., 2006; Römling et al., 2013). It is noteworthy that cellulose also exhibits major roles in the modulation of virulence mechanisms in both human and plant pathogenic bacteria (Römling et al., 2013). Based on a “genome mining” approach using the complete genome sequence of the model strain Pss UMAF0158 (Martínez-García et al., 2015), an orthologous gene cluster to the operon wss of Pseudomonas fluorescens SBW25 involved in cellulose biosynthesis was identified in the chromosome (Rainey and Travisano, 1998; Spiers et al., 2002). This gene cluster is organized as an operon, and encompasses 14,642 bp that encodes nine genes with putative functions associated with cellulose production and acetylation. Additionally, the evolutionary history of this gene cluster revealed that it was present in both pathogenic and non-pathogenic Pseudomonas. In addition, the flanking regions of the cellulose gene cluster were consistent between Pss UMAF0158 and other P. syringae cellulose-producing strains, suggesting an identical chromosome location.
Epiphytic colonization by P. fluorescens SBW25 and its survival on plant surfaces is primarily due to cellulose overproduction by the wss operon (Gal et al., 2003; Spiers et al., 2003). The role of cellulose in biofilm formation of P. syringae pv. tomato DC300 has also been shown (Pérez-Mendoza et al., 2014). To determine the role of cellulose in the lifecycle of Pss isolated from mangos, insertional mutants in the biosynthetic genes of the wss cluster, wssB and wssE (Römling, 2002), were constructed and proved to be impaired in cellulose production. Furthermore, a cellulose-overproducing strain was obtained via the transformation of Pss UMAF0158 with plasmid pVS61-WsR19 that contained wspR19 from P. fluorescens SBW25 (Ude et al., 2006). Scanning electronic microscopy on mango buds and tomato leaves revealed the formation of microcolonies of the wild-type and overproducing strains immersed in the extracellular matrix, but not for the cellulose-defective mutants. Furthermore, adhesion experiments on mango leaves revealed that the amount of bacteria recovered were higher in the wild-type and overproducing strains, in respect to the wss mutants. In contrast, although growth curves on minimal medium for the different strains exhibited similar patterns, the incidence (number of necrotic points developed) and the severity (necrotic area developed) on tomato leaflets were higher in the wss mutants, lower in the wild-type, and practically abolished in the cellulose overproducing strain. The competitive index approach analysis supported these results, showing that the competitiveness of the overproducing strain was decreased during the plant infection experiments (Arrebola et al., 2015). It is evident that cellulose plays a primary dual role between epiphytic and pathogenic lifestyle of Pss on mango tree surfaces, which suggests that this trait is maintained on Pss mango populations for mango leaf and bud colonization and adaptation. Mechanisms of the regulation of cellulose biosynthesis by Pss isolated from mango trees has not yet been determined, but some clues have been discovered in related Pseudomonas. The second messenger c-di-GMP controls cellulose biosynthesis in P. fluorescens SBW25 (Spiers et al., 2002) and regulates the switch between the static and motile phases in many different bacterial species (Römling et al., 2013). More recently, the transcriptional regulator AmrZ has been reported to be a key regulator in the biosynthesis of cellulose in P. syringae pv. tomato DC3000 (Prada-Ramírez et al., 2016). The regulon of the AmrZ transcriptional regulator includes putative c-di-GMP proteins such as AdcA and MorA; thus, AmrZ could be directly involved in cellulose biosynthesis by modulating the available pool of c-di-GMP.
As described by Salmond (1994), a virulence factor could be any molecule present on the bacterial cell surface or released from the cell that could influence the growth of the pathogen in plants, enhancing infection and subsequent disease development. Plant pathogenic bacteria have developed many different and specific virulence strategies to infect successfully their plant hosts. The identification, characterization and dissection of the modes of action of different virulence factors is complex, despite the efforts of many research groups (Mansfield et al., 2012; Pfeilmeier et al., 2016). Whereas the traits that confer P. syringae pathogenicity are numerous and well-studied, the mechanisms underlying susceptibility of mango are unknown. The lack of balance in our understanding of the mechanisms involved (well understood for the pathogen, poorly understood for the host) make us to focus in the role of the pathogen during the interaction with the host. P. syringae, in particular, and Pss strains isolated from mango specifically, shows a broad and sophisticated armament of different virulence factors (Ichinose et al., 2013), among which bacterial toxins are one of the most studied in depth.
Bacterial toxins are important virulence factors of P. syringae (Mitchell, 1991) and have been described to be involved in the development of chlorotic and necrotic disease symptoms in its plant hosts (Volksch and Weingart, 1998; Scholz-Schroeder et al., 2001). Lipodepsipeptidic toxins, such as syringomycins and syringopeptins, have been strongly associated with several pathovars of P. syringae and are mainly related with the production of necrotic symptoms (Gross and DeVay, 1977; Ballio et al., 1991; Adetuyi et al., 1995; Vassilev et al., 1996; Bender et al., 1999; Scholz-Schroeder et al., 2001). Pss strains isolated from mango trees were found to produce syringomycin by using growth inhibition tests toward Geotrichum candidum (Gross and DeVay, 1977) and Rhodotorula pilimanae (Iacobellis et al., 1992), and the detection of a specific gene involved in its biosynthesis was done by a PCR protocol (Sorensen et al., 1998). In addition, Pss strains isolated from mango trees were also found to produce syringopeptins by using a grown inhibition bioassay of Bacillus megaterium (Lavermicocca et al., 1997). Another group of important toxins described in several pathovars of P. syringae are the so-called “antimetabolite toxins.” This group of toxins blocks the function of enzymes involved in the biosynthetic pathways of crucial amino acids, as well as the biosynthesis of polyamine (Bender et al., 1999; Arrebola et al., 2011a,b). These toxins produce chlorotic symptoms in plant tissue due to the accumulation of different intermediates (Patil et al., 1972; Turner and Debbage, 1982; Bachmann et al., 1998). The best-known antimetabolite toxins produced by different pathovars of P. syringae are tabtoxin, phaseolotoxin, and the recently identified mangotoxin (Arrebola et al., 2003). Mangotoxin was initially identified to be produced mainly by Pss strains isolated from mango trees, although its production was also reported in a few Pss strains from other hosts (Arrebola et al., 2003). The biosynthesis pathway of mangotoxin, its regulation, and the role that this toxin plays in the different lifestyles of Pss-mango interactions are discussed extensively in the next section.
Mangotoxin is the most recent antimetabolite toxin discovered and was first described to be mainly produced by Pss strains isolated from mango trees. This toxin was called “mangotoxin” due to the plant host (mango tree) from which most of the Pss strains mangotoxin producers were isolated (Arrebola et al., 2003). As mentioned above, antimetabolite toxins block enzymes functions involved in the biosynthetic pathways of crucial amino acids and the biosynthesis of polyamine (Bender et al., 1999; Arrebola et al., 2011a,b). The toxic activity of mangotoxin is reversed by the addition of ornithine, and thus, its target enzyme was identified as ornithine N-acetyl transferase (OAT) (Arrebola et al., 2003). In Figure 3A, a schematic representation of the arginine-glutamine and polyamine biosynthesis pathways shows the target enzymes of the different antimetabolite toxins including mangotoxin. In order to decipher the chemical structure of mangotoxin a physicochemical characterization was performed initially using cell-free filtrates revealing that mangotoxin is a small secreted molecule of a hydrophilic nature smaller than 3 kDa in size, extremely resistant to high pH and high temperature, but sensitive to protease treatments. The analysis of a Tn5 defective mutant in mangotoxin production (UMAF0158-3aE10) and the wild-type strain Pss UMAF0158 by using High-performance liquid chromatography (HPLC), revealed a specific peak associated with mangotoxin activity (Arrebola et al., 2003). Another chemical separation techniques such hydrophilic interaction liquid chromatography (HILIC) and ion Exchange chromatography (FPLC) have been also applied to decode the mangotoxin structure (data not published). However, the efforts conducted to unravel the chemical structure of mangotoxin have been in vain to date, largely due to its high chemical instability.
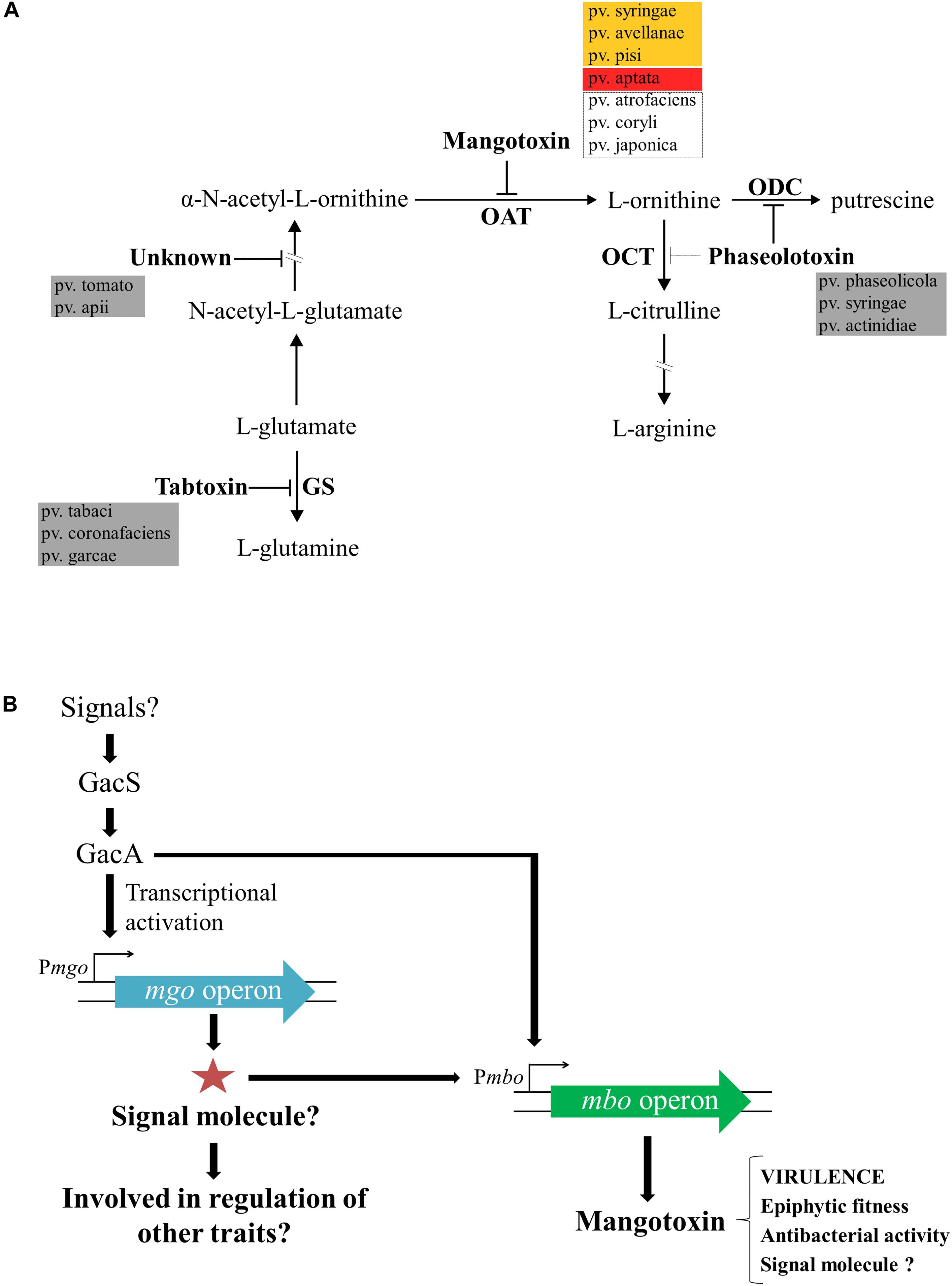
Figure 3. Enzymatic target of mangotoxin and its biosynthesis regulation. (A) Representative scheme of the arginine-glutamine and polyamine biosynthesis pathways. The enzymatic targets of the different antimetabolite toxins, including mangotoxin, and the different pathovars involved in the production of the different antimetabolite toxins are depicted. OAT, ornithine N-acetyltransferase; OCT, ornithine carbamoyltransferase; ODC, ornithine decarboxylase, and GS, glutamine synthetase. Orange box: pathovars positives for the presence of mbo genes and positives for mangotoxin production; red box: pathovar positive for the presence of mbo genes, but negative for mangotoxin production; dotted lines box: pathovars positive for the presence of mbo genes, but not experimentally tested for mangotoxin production. (B) Mangotoxin biosynthesis regulation model. GacS/GacA two-component regulatory system regulates directly or indirectly the transcription of the mgo operon. Mgo operon could synthetize a positive regulator (signal molecule) to activate the mbo operon transcription. The mbo operon produces mangotoxin, which acts primarily as a virulence factor, although, other functions have been described.
To understand the molecular basis of mangotoxin production, three mutants impaired in mangotoxin production obtained from a genomic library (Pss UMAF0158-3γH1, -6γF6, and -5αC5) that displayed growth characteristics and production of lipodepsipeptidic toxins similar to wild-type strain UMAF0158 (Arrebola et al., 2007) were studied in depth. The insertion in the mutant UMAF0158-6γF6 was located in a DNA region that showed high similarity with an non-ribosomal peptide synthetase (NRPS) present in Pss B728a, P. syringae pv. tomato DC3000 and P. syringae pv. phaseolicola 1448A. This orf called mgoA gene has a size of 3447 bp, and the amino acids sequence of this protein was composed of an activation module with conserved domains typical for NRPS (Stein and Vater, 1996; Marahiel et al., 1997). The role of mgoA in virulence of Pss was demonstrated in tomato leaflets, showing this mutant a lower disease incidence than the wild-type. Therefore, the NRPS gene mgoA was confirmed to be involved in mangotoxin biosynthesis and, also, in virulence (Arrebola et al., 2007). Furthermore, three additional genes were detected together with the mgoA gene and were designated mgoB, mgoC, mgoA, and mgoD, in accordance with the mangotoxin generating operon. Insertional mutants in mgoC, mgoA, and mgoD, had altered mangotoxin production. Additionally, by using RT-PCR all mgo genes were co-transcribed together, forming a single polycistronic mRNA and thus forming an operon. Complementation experiments with the mgo operon restored the ability of the mutants to produce mangotoxin, and therefore, these results confirmed strongly that the mgo operon was necessary for mangotoxin production (Arrebola et al., 2007, 2012). The mgo operon has been found to be well-distributed in the majority of Pseudomonas species, including different pathovars of P. syringae (Lindeberg et al., 2008; Vallet-Gely et al., 2010). A homologous gene cluster to the mgo operon, pvf, has been proposed to be encoded in Pseudomonas entomophila as a regulator of virulence factors (Vallet-Gely et al., 2010). Recently, the family of pyrazine N-oxides (PNOs), including a novel dihydropyrazine N,N′-dioxide metabolite, were identified to be produced by the pvf gene cluster in P. entomophila, suggesting that these molecules could be involved in Pseudomonas signaling and virulence (Kretsch et al., 2018). In addition, fragin biosynthesis, the main antifungal compound produced by Burkholderia cenocepacia H111 is under the control of valdiazen, a novel quorum-sensing signaling molecule produced by a gene cluster homologous to the mgo and pvf operons (Jenul et al., 2018). Although the structure of the putative signaling molecule produced by the mgo operon in Pss isolated from mango trees remains unknown, its function as a regulator of biosynthesis of mangotoxin, and likely other secondary metabolites, is quite feasible.
Interestingly, another two Tn5 mutants abolished in mangotoxin production (UMAF01585aC5 and UMAF0158-4βA2), and thus, affected in virulence (tested in virulence assay in tomato leaflets) were studied in depth because they did not show homology with the genome sequences of Pss B728a, P. syringae pv. tomato DC3000 or P. syringae pv. phaseolicola 1448A. The involvement of mangotoxin in the epiphytic survival of Pss strains isolated from mango was demonstrated by Arrebola et al. (2009). Epiphytic survival experiments on tomato leaflets revealed that there was no difference between the wild-type Pss UMAF0158 and both mutants. Nevertheless, when the bacteria were co-inoculated together the wild-type with each of the mutants individually a slight but significant decrease was observed in the mutants, and the difference reached almost one order of magnitude. Thus, in addition to its virulence function, mangotoxin could also play a role in improving the ecological fitness of Pss strains isolated from mango trees. Furthermore, the screening of both mutant insertions in the genomic library showed that both were in a cluster of six genes present in wild-type strain Pss UMAF0158 and not in Pss B728a, P. syringae pv. tomato DC3000 or P. syringae pv. phaseolicola 1448A. Complementation experiments restored the ability of both mutants to produce mangotoxin (Carrión et al., 2012). These six genes were named mboA, B, C, D, E, and F in accordance with the mangotoxin biosynthetic operon and experiments based on RT-PCR and Northern blot analysis confirmed that these six genes were co-transcribed as a single polycistronic mRNA molecule confirming that these genes were forming an operon. Furthermore, site directed insertional mutations performed in each gene have shown a complete abolition of mangotoxin production in mboA, B, C, and D gene mutants and altered phenotypes in mboE and F gene mutants. Transformation experiments with pLAC-AF (pBBR1-MCS5 + mboA-F), a plasmid that contains the six mbo genes in different non-producing Pseudomonas strain genetic backgrounds, resulted in mangotoxin producers. Therefore, all experiments strongly confirmed that the mbo genes were essential for full production of mangotoxin.
Unambiguously, Carrión et al. (2014) demonstrated that the regulation of mangotoxin production was under the control of both gacS/gacA and mgo genes and additionally, that mgo genes were regulated by gacS/gacA genes. Tn5 mutants that were all defective in mangotoxin production (mgoA mutant, mboD mutant, mboB mutant, gacS mutant, and gacA mutant) were used to unravel the regulation of the mangotoxin biosynthetic pathway. Transcriptional analysis by qRT-PCR showed that expression levels of the mboA, C, and E genes were significantly lower in the gacA and mgoA mutants than in the wild-type; however, the mgo and mbo mutants did not affect the transcription levels of the gacS/gacA genes. These results suggested that the gacS/gacA system controls the regulation of both mgo and mbo operons and downstream the mgo operon controlled the regulation of the mbo operon, and thus controlling the mangotoxin production. Promoter fusion experiments using the mbo promoter showed high levels of β-galactosidase activity in the wild-type, whereas the expression was significantly lower in mgoA, gacA, and gacS mutants, supporting the results obtained previously. Taken together, a model for the regulation of mangotoxin production has been proposed (Figure 3B) (Carrión et al., 2014). In this model, it is proposed that mgo molecules could serve as signaling molecules, as has been previously described in similar bacteria, and may be involved in the regulation of other virulence traits in Pss strains isolated from mango trees. Moreover, other functions in addition to virulence have been described for mangotoxin, and its putative role as a signaling molecule has been hypothesized.
A diversity survey using different approaches (genetic, phenotypic, and phylogenetic) showed that Pss strains isolated from mango trees formed a single phylotype into the pathovar syringae associated with the mango host, producers of mangotoxin and distributed worldwide in areas where mango is grown and BAN is a relevant disease (Gutiérrez-Barranquero et al., 2013a). Despite of Pss strains isolated from mango are more similar among them in comparison with other Pss isolated from others hosts and other pathovars, phenotypic (including virulence degree) and genetic variability has been observed (Gutiérrez-Barranquero et al., 2013a). Then, in order to determine the evolutionary history of the mbo operon, a phylogenetic analysis using the housekeeping genes rpoD and gyrB grouped all strains belonging to the Genomospecies 1 together but separated in three different clusters. Two of these clusters were associated with the presence of the mbo operon (Carrión et al., 2013). Group I mbo+ was mainly composed of strains from the pathovar syringae, mainly isolated from woody hosts, but predominantly from mango trees, group which correspond with the single phylotype of Pss strains associated with mango trees described by Gutiérrez-Barranquero et al. (2013a). Group II mbo+ was composed of five different pathovars of P. syringae isolated from herbaceous and woody plants (aptata, avellanae, japonica, pisi, and syringae) and group III mainly composed by the pathovar syringae that was negative for the presence of mbo genes. Interestingly, group III (the group that lacked the mbo operon) diverged before the separation of groups I and II. These results suggested that the mbo operon was acquired by groups I and II in only one or two acquisition events after their separation from group III. Thus, this work strongly suggested that the mbo operon was horizontally acquired only once during the evolution of the P. syringae complex shown to be specifically distributed within the P. syringae Genomospecies 1 (Carrión et al., 2013). In the last few years, the databases have suffered a veritable explosion regarding the number of P. syringae genome sequences available (Baltrus et al., 2011; Thakur et al., 2016), which also contributed to a novel classification of the P. syringae complex in 13 different phylogroups (Berge et al., 2014). A more in depth phylogenetic analysis has been performed including 150 strains of the P. syringae complex belonging to the phylogenetic groups 1, 2, 3, 4, 5, 6, 7, and 11 (Figure 4). Inside the phylogenetic group 2, where the Pss strains isolated from mango are present, it is possible to observe the differentiation of three main groups, similar to those previously reported by Carrión et al. (2013). Group I mbo+ was mostly composed of pathovar syringae, mainly isolated from mango trees, corresponding with the single phylotype described. Group II mbo+ was composed of the 5 pathovars previously identified in this group. However, this new analysis included two more pathovars into this group (pathovar atrofaciens and coryli). Finally, a third group was composed mainly by the pathovar syringae that was negative for the presence of mbo genes. This new phylogenetic analysis confirms the previous assumption that Pss strains isolated from mango are forming a single phylotype inside the Genomospecies1-phylogenetic group 2.
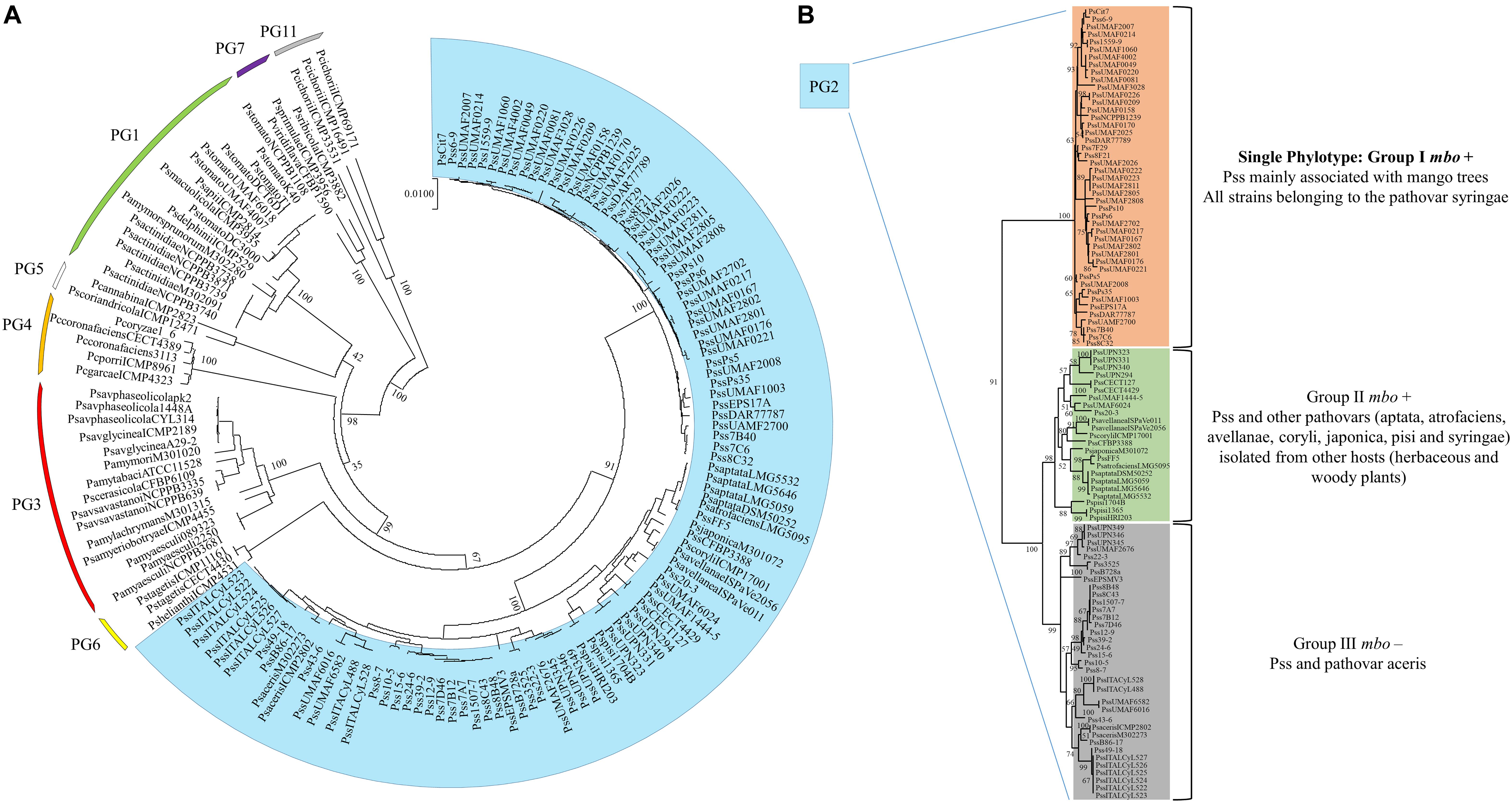
Figure 4. Multilocus sequence typing analysis of strains belonging to the P. syringae complex. (A) The neighbor-joining tree was constructed with combined partial sequences of rpoD and gyrB housekeeping genes using MEGA 7 software. Bootstrap values (1,000 repetitions) are shown on branches and evolutionary distances are in units of nucleotide substitutions per site. One hundred and fifty strains belonging to the phylogenetic groups 1, 2, 3, 4, 5, 6, 7, and 11 of the P. syringae complex are depicted in the circular phylogenetic tree. Marked in blue are represented the strains belonging to the phylogenetic group 2, where the P. syringae pv. syringae strains isolated from mango are found. (B) Exclusive representation of the phylogenetic group 2. Three main groups are defined regarding the presence or not of the mbo genes necessary for mangotoxin production. The topology was similar among phylogenetic trees produced by the maximum-parsimony and maximum-likelihood methods. Supplementary Table S1 provides the phylogenetic groups, the host of isolation and the accession numbers of the DNA sequences used for each strain represented in this phylogenetic analysis.
Pseudomonas syringae infections tend to be favored by cool and wet conditions due to its ability to induce ice nuclei formation at warm, subfreezing temperatures (-2 to -4°C) (Lindow et al., 1982; Hirano and Upper, 1995). Ice nucleation activity (INA), is considered an important virulence factor wide spread throughout P. syringae complex that plays a major role in the early stages of infections causing wounds that can facilitate disease particularly in woody plant species (Lindow et al., 1982; O’Brien and Lindow, 1988; Hwang et al., 2005; Lamichhane et al., 2014). In this sense, Cazorla et al. (1995) developed a simple and alternative multiple-tube test that showed an increase in detection sensitivity of active ice nuclei forming bacteria relative to the traditional drop-freezing methods (Lindow et al., 1978). This method revealed that all Pss strains isolated from mango trees were positive for INA detection. Although the INA virulence factor could be important at the initial stages of BAN development, the low probability of occurrence of frost in mango-producing areas makes its role in virulence largely anecdotal.
The most-studied and well-characterized virulence factor associated with P. syringae is the T3SS (Lindeberg et al., 2012). The T3SS is a complex nanomolecular machinery used by P. syringae and many other plant and animal pathogens to inject effector proteins into host plant cells to subvert the plant immune system and induce disease development (Lindeberg et al., 2012). While the T3SS is the most-studied virulence factor in P. syringae-plant interactions (Collmer et al., 2000; Oh et al., 2010; Cunnac et al., 2011; Lo et al., 2017), the role that this secretion system might play in the development of BAN disease has not been examined in depth to date. At this stage, a genome sequencing project performed on the Pss model strain isolated from mango trees, UMAF0158, revealed the presence of two different T3SSs (Martínez-García et al., 2015). The first T3SS (T3SS-1) is similar to the Hrp-1 T3SS family (Egan et al., 2014) found in different pathovars of P. syringae (Lindeberg et al., 2012) and represents the canonical T3SS widely distributed in pathogenic P. syringae strains (Block and Alfano, 2011; Lindeberg et al., 2012). Pss strains isolated from mango trees were able to induce a hypersensitivity response (HR) in tobacco plants (Cazorla et al., 1998). The capability of P. syringae to provoke a HR in non-host plants is dependent on a functional T3SS (Huang et al., 1992). Thus, in Pss UMAF0158, a simple deletion mutant constructed in the hrpL gene (UMAF0158ΔhrpL) (an alternative sigma factor that binds to the hrp box promoter sequence of the T3SS genes that upregulates their expression) confirmed the involvement of T3SS-1 in HR development (Martínez-García et al., 2015). The role of this particular T3SS in overall virulence has been widely recorded in Pss B728a, P. syringae pv. tomato DC3000, and many others (Schechter et al., 2006; Vinatzer et al., 2006; Kvitko et al., 2009; Lee et al., 2012).
Additionally, bioinformatics analysis highlighted the presence of an additional T3SS (called T3SS-2) in the chromosome of Pss UMAF0158 (Martínez-García et al., 2015) that was also found in different strains from different pathovars (Reinhardt et al., 2009; Studholme et al., 2009; Clarke et al., 2010; Matas et al., 2014). This T3SS-2 shows high similarity to the rhizobial-like T3SS Rhc of the Rhizobiales family (Gazi et al., 2012; Egan et al., 2014). A typical hrp box promoter regulatory sequences of the HrpL regulon found preceding the genes of the typical T3SS (Fouts et al., 2002) was missed in the T3SS-2. As it has been demonstrated in other P. syringae strains, the T3SS-2 is dispensable for pathogenicity, although a possible role in plant surface colonization or interaction with insects cannot be ruled out (Lindgren et al., 1986; Clarke et al., 2010; Pérez-Martínez et al., 2010; Silby et al., 2011). Different specific mutants in the T3SS-1, T3SS-2, and in combination in both systems constructed in Pss UMAF0158 did not revealed the function of the T3SS-2 in Pss isolated from mango trees, which remains unknown to date (Martínez-García et al., 2015).
Due to the release of the Plant-bacteria Interaction FActors Resource (PIFAR), an open-access web-based resource for genetic factors involved in bacterial interactions with plant–hosts1 (Martínez-García et al., 2016), the detection of type 3 effectors (T3Es) has become more accurate than the method previously selected to identify T3Es in Pss UMAF0158 (Martínez-García et al., 2015). By using PIFAR tool, 15 putative T3Es have been identified in Pss UMAF0158, 4 T3Es more than the 11 previously identified. A Venn diagram analysis of the core T3Es, comparing the Pss UMAF0158 genome with the genome sequencing of three Pss strains (B728a, HS191, and B301D) and P. syringae Cit7 all belonging to Genomospecies 1 (Gardan et al., 1999) and Phylogenetic Group 2 (Berge et al., 2014), has been performed (Figure 5A). In addition, the presence or absence of the different T3Es present in these four strains are depicted (Figure 5B). hopA1, hopAX1, hopAZ1, and hopBK1 are the unique T3Es shared by Pss UMAF0158 with several other strains (Ps Cit7 and Pss HS191). On the other hand, hopA1, hopAX1, and hopBK1 have been found in other pathovars of P. syringae belonging to different phylogroups (Berge et al., 2014). Remarkably, the effector hopAX1 appears to be mainly associated with a few strains of different pathovars all belonging to the Genomospecies 1-Phylogenetic Group 2 (pv. aptata, pv. pisi, pv. aceris, and pv. syringae). Dillion et al. (2019) have recently described the high-specificity of hopAX1 T3E in the Phylogenetic Group 2.
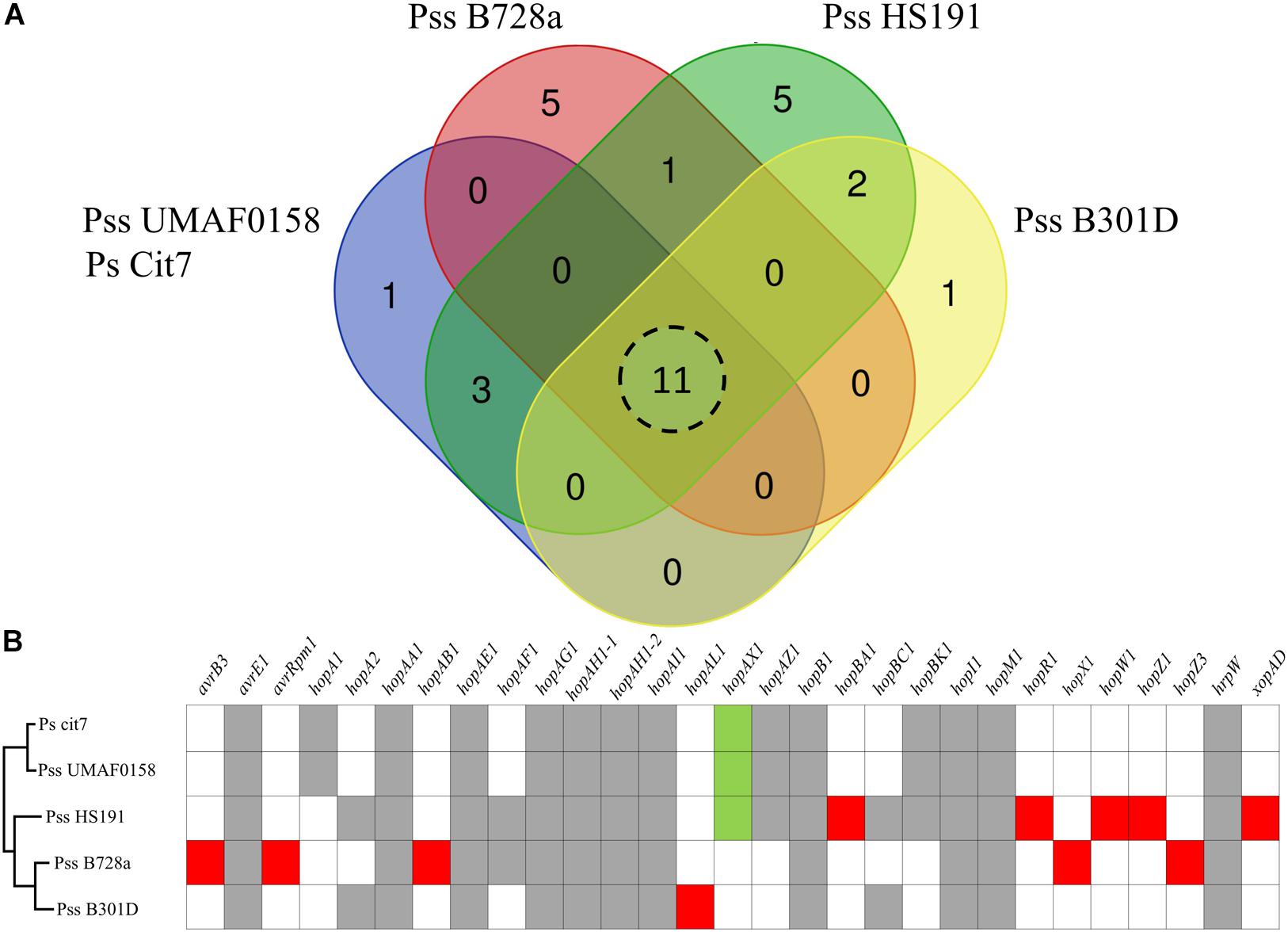
Figure 5. Type III effectors repertoire. (A) Venn diagram comparing the putative type III effectors presence in selected strains of P. syringae including the model strain P. syringae pv. syringae isolated from mango UMAF0158. Eleven type III effectors shared by all the strains analyzed. (B) Presence of specific type III effectors. Gray boxes indicate type III effectors presence in all the strains analyzed. Red boxes indicate type III effectors specific for each strain analyzed. Green boxes represents the hopAX1effector, an effector present in UMAF0158 that shows a high-specificity for a few strains of different pathovars exclusively belonging to the Genomospecies 1-Phylogenetic Group 2.
High-Throughput Sequencing technologies (HTS) has had a large impact on plant pathology and other research areas. In recent years, there has been substantial growth regarding genome sequencing of bacterial plant pathogens (Studholme et al., 2011) that can provide a strong basis for a better understanding of plant–microbe interactions that 1 day will contribute to the eradication of plant diseases. P. syringae is the model plant pathogen par excellence most often used worldwide to dissect plant–pathogen interactions (Baltrus et al., 2017). From the first genome sequenced of the model strain P. syringae pv. tomato DC 3000, the current landscape has changed markedly, with many groups interested in P. syringae comparative genomics and evolution (Lovell et al., 2009; Green et al., 2010; Baltrus et al., 2011; McCann et al., 2013; Thakur et al., 2016; Hulin et al., 2018). Currently, the complete genome sequences of 29 P. syringae strains, along with more than 400 draft genome sequences, are included in the NCBI database2,3. To date, there is only one complete genome sequenced of Pss strains isolated from mango trees (chromosome + 62-kb PFP plasmid), which was performed in the model strain Pss UMAF0158 (Martínez-García et al., 2015). This work revealed a high degree of conservation with other Pseudomonas from the P. syringae complex; however, different genetic factors were identified for their potential involvement in the epiphytic or pathogenic lifestyle, and these factors have been described in depth in this review. Among these factors, the most important were the presence of the mbo operon (mangotoxin biosynthetic operon), the presence of the wss operon (operon involved in cellulose biosynthesis), the additional type III-like rhizobial secretion system, the additional type VI secretion system, and a particular T3E repertoire.
Recently, a PFP sequencing project that includes 4 62-kb PFP plasmids from different strains of Pss strains isolated from mango trees was carried out (Gutiérrez-Barranquero et al., 2017a). In this work, it was revealed that the main functions of 62-kb plasmids of Pss strains isolated from mango trees were related to the increase in UV radiation and copper treatment tolerance. The backbone of the different plasmids regarding the genes involved in the maintenance, replication and conjugation was similar and showed a high degree of synteny. Interestingly, these plasmids were included in the previously described subgroup B (Ma et al., 2007), sharing more than the repA gene (replicase gene shared by all PFPs plasmids; Sundin, 2007). In addition, a novel genetic structure likely related to a cell-to-cell communication signaling system appeared in those plasmids upstream of the type IV secretion system, suggesting that the conjugation process could be under the regulation of this signaling mechanism (Gutiérrez-Barranquero et al., 2017a). On the other hand, there is a relatively low degree of homology in remaining genes found in each 62-kb PFP plasmids.
The enormous efforts that have been carried out over the last two decades have led us to gain more in-depth understanding of the P. syringae pv. syringae-mango host interactions. Pss causes important economic losses in mango crop production in the Mediterranean region. Pss strains isolated from mango trees form a single phylotype within the pathovar syringae and exhibit important factors that contribute to the epiphytic-pathogenic phase establishment on the mango plant, revealing a deep interaction between the pathogenic microbe and the host plant. It is worthy to note that the traits in P. syringae that are involved in pathogenic and epiphytic lifestyles have been studied in depth, but particularly, the mechanisms underlying the association of Pss with the mango host are little known. Thus, the major traits analyzed in depth in this review would help Pss to interact successfully with mango trees, but some of them are also useful in the interaction of other P. syringae strains with other plant hosts. Mangotoxin is the main virulence factor of this particular group of bacteria, and although much attention has been paid to it, the structure of this toxic molecule remains elusive. In addition, the possible role of mangotoxin as a signaling molecule modulating specific gene expression has been hypothesized. Further experiments are currently being carried out to confirm this hypothesis. Additionally, another important virulence factor not well-studied is the T3SS. In Pss isolated from mango trees, an extra copy of the T3SS is present. However, despite the efforts made, its role in the ecology of Pss remains unknown. Another relevant factor recently discovered in Pss strains isolated from mango trees is the presence of a cellulose biosynthetic gene cluster. The cellulose gene cluster has been described as involved in adhesion and biofilm formation development in Pss on the mango leaf surfaces. This gene cluster is only present in a few strains of P. syringae but is present in all Pss strains isolated from mango trees, suggesting that it is a crucial factor in the adaptation to the mango host. Its role in modulating epiphytic and pathogenic phases on mango surfaces has also been addressed. In addition, 62-kb PFP plasmids have been shown to play a key role in epiphytic survival of Pss on mangos, harboring UV and copper resistance determinants, among others. This long-lasting interaction among Pss and mango led us to search for effective control methods to allow farmers to deal with BAN symptoms. The efficacy of the alternative treatment silicon gel compared to the spray of copper compound BM has been demonstrated, and silicon gel has finally been registered for its commercial use in mango crops in Spain to combat BAN disease.
Given all of this, the future directions of this research are actually being targeted in two aims: (1) to unravel signaling mechanisms of Pss in interactions with other bacterial members of the mango microbiome by analysis of the transcript-level expression using in vitro and in vivo approaches; and (2) comparative genomics and evolutionary history analysis. In spite of the massive development of genomic sequencing technologies, there is a lack of information regarding genomic data from Pss strains isolated from mango trees. Thus, a great effort is currently being carried out to perform a major genome sequencing project involving a number of different strains to unravel the evolutionary processes that have occurred in mango populations from different geographical regions, separated in time. Phylogenetic and evolutionary approaches will open new windows of research that allow us to better understand why this phytopathogenic bacterium is so peculiar.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by grants from CICE-Junta de Andalucía, Proyecto de Excelencia (P12-AGR-1473) co-financed by FEDER (EU). JG-B was supported by a Postdoctoral Fellowship from the Research Own Plan of the University of Malaga “Ayuda de Incorporación de Doctores 2017”.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are very grateful to all the people who were directly involved in the development of this research: José María Farré, José María Hermoso, Emilio Guirado, David Sarmiento, Alejandro Pérez-García, Juan C. Codina, Eva Arrebola, Victor J. Carrión, and Jesús Murillo. We would like to thank SAT 2803 TROPS and all collaborating farmers. We also extend a special thanks to Irene Linares for her collaboration and technical support. This work is especially dedicated to the memory of our colleague Juan A. Torés Montosa, who sadly passed away in July 2018. He was one of the original researchers responsible for the discovery of BAN disease, and the development of this research line in our laboratory.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00570/full#supplementary-material
Adetuyi, F. C., Isogai, A., Di Giorgio, D., Ballio, A., and Takemoto, J. Y. (1995). Saprophytic Pseudomonas syringae strain M1 of wheat produces cyclic lipodepsipeptides. FEMS Microbiol. Lett. 131, 63–67. doi: 10.1111/j.1574-6968.1995.tb07755.x
Almeida, N. F., Yan, S., Cai, R., Clarke, C. R., Morris, C. E., Schaad, N. W., et al. (2010). PAMDB, a multilocus sequence typing and analysis database and website for plant associated microbes. Phytopathology 100, 208–215. doi: 10.1094/PHYTO-100-3-0208
Andersen, G. L., Menkissoglou, O., and Lindow, S. E. (1991). Occurrence and properties of copper-tolerant strains of Pseudomonas syringae isolated from fruit trees in California. Phytopathology 81, 648–656. doi: 10.1094/Phyto-81-648
Anonymous (2002). Commission Regulation (EC) No 472/2002. Official Journal of the European Communities. Available at: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2002:075:0018:0020:EN:PDF
Anonymous (2018). Available at: https://www.diariosur.es/economia/agroalimentacion/cosecha-mango-batira-20180806211309-nt.html
Arnold, D. L., Jackson, R. W., Fillingham, A. J., Goss, S. C., Taylor, J. D., Mansfield, J. W., et al. (2001). Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv.pisi. Microbiology 147, 1171–1182. doi: 10.1099/00221287-147-5-1171
Arrebola, E., Carrión, V. J., Cazorla, F. M., Pérez-García, A., Murillo, J., and de Vicente, A. (2012). Characterisation of the mgo operon in Pseudomonas syringae pv. syringae UMAF0158 that is required for mangotoxin production. BMC Microbiol. 12:10. doi: 10.1186/1471-2180-12-10
Arrebola, E., Carrión, V. J., Gutiérrez-Barranquero, J. A., Pérez-García, A., Rodríguez-Palenzuela, P., Cazorla, F. M., et al. (2015). Cellulose production in Pseudomonas syringae pv. syringae: a compromise between epiphytic and pathogenic lifestyles. FEMS Microbiol. Ecol. 91:fiv071. doi: 10.1093/femsec/fiv071
Arrebola, E., Cazorla, F. M., Codina, J. C., Gutiérrez-Barranquero, J. A., Pérez-García, A., and de Vicente, A. (2009). Contribution of mangotoxin to the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Int. Microbiol. 12, 87–95. doi: 10.2436/20.1501.01.85
Arrebola, E., Cazorla, F. M., Durán, V. E., Rivera, E., Olea, F., Codina, J. C., et al. (2003). Mangotoxin: a novel antimetabolite toxin produced by Pseudomonas syringae inhibiting ornithine/ arginine biosynthesis. Physiol. Mol. Plant Pathol. 63, 117–127. doi: 10.1016/j.pmpp.2003.11.003
Arrebola, E., Cazorla, F. M., Pérez-García, A., and de Vicente, A. (2011a). Chemical and metabolic aspects of antimetabolite toxins produced by Pseudomonas syringae pathovars. Toxins 3, 1089–1110. doi: 10.3390/toxins3091089
Arrebola, E., Cazorla, F. M., Pérez-García, A., and de Vicente, A. (2011b). Genes involved in the production of antimetabolite toxins by Pseudomonas syringae pathovars. Genes 2, 640–660. doi: 10.3390/genes2030640
Arrebola, E., Cazorla, F. M., Romero, D., Pérez-García, A., and de Vicente, A. (2007). A nonribosomal peptide synthetase gene (mgoA) of Pseudomonas syringae pv. syringae is involved in mangotoxin biosynthesis and is required for full virulence. Mol. Plant Microbe Interact. 20, 500–509. doi: 10.1094/MPMI-20-5-0500
Bachmann, A. S., Matile, P., and Slusarenko, A. J. (1998). Inhibition of ornithine decarboxylase activity by phaseolotoxin: Implications for symptom production in halo blight of French bean. Physiol. Mol. Plant Pathol. 53, 287–299. doi: 10.1006/pmpp.1998.0183
Ballio, A., Barra, D., Bossa, F., Collina, A., Grgurina, I., Marino, G., et al. (1991). Syringopeptins, new phytotoxic lipodepsipeptides of Pseudomonas syringae pv. syringae. FEBS Lett. 291, 109–112. doi: 10.1016/0014-5793(91)81115-O
Baltrus, D. A., McCann, H. C., and Guttman, D. S. (2017). Evolution, genomics and epidemiology of Pseudomonas syringae: challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 18, 152–168. doi: 10.1111/mpp.12506
Baltrus, D. A., Nishimura, M. T., Romanchuk, A., Chang, J. H., Mukhtar, M. S., Cherkis, K., et al. (2011). Dynamic evolution of pathogenicity revealed by sequencing and comparative genomics of 19 Pseudomonas syringae isolates. PLoS Pathog. 7:e1002132. doi: 10.1371/journal.ppat.1002132
Beattie, G., and Lindow, S. E. (1995). The secret life of foliar bacterial pathogens on leaves. Annu. Rev. Phytopathol. 33, 145–172. doi: 10.1146/annurev.py.33.090195.001045
Becerra, L. (1995). “Enfermedades del cultivo del mango,” in Producción del Mango en México, eds I. Mata and R. Mosqueda (México: Noriega editores), 84–86.
Bélanger, R. R., Benhamou, N., and Menzies, J. G. (2003). Cytological evidence of an active role of silicon in wheat resistance to powdery mildew (Blumeria graminis f. sp. tritici). Phytopathology 93, 402–412. doi: 10.1094/PHYTO.2003.93.4.402
Bender, C., and Cooksey, D. (1986). Indigenous plasmids in Pseudomonas syringae pv. tomato: conjugative transfer and role in copper resistance. J. Bacteriol. 165, 534–541. doi: 10.1128/jb.165.2.534-541.1986
Bender, C. L., Alarcón-Chaidez, F., and Gross, D. C. (1999). Pseudomonas syringae phytotoxins: mode of action, regulation, and biosynthesis by peptide and polyketide synthetases. Microbiol. Mol. Biol. Rev. 63, 266–292.
Bender, C. L., and Cooksey, D. (1987). Molecular cloning of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 169, 470–474. doi: 10.1128/jb.169.2.470-474.1987
Berge, O., Monteil, C. L., Bartoli, C., Chandeysson, C., Guilbaud, C., Sands, D. C., et al. (2014). A user’s guide to a data base of the diversity of Pseudomonas syringae and its application to classifying strains in this phylogenetic complex. PLoS One 9:e105547. doi: 10.1371/journal.pone.0105547
Block, A., and Alfano, J. R. (2011). Plant targets for Pseudomonas syringae type III effectors: virulence targets or guarded decoys? Curr. Opin. Microbiol. 14, 39–46. doi: 10.1016/j.mib.2010.12.011
Bradbury, J. F. (1986). Guide to Plant Pathogenic Bacteria. Slough: CAB International Mycological Institute.
Bull, C. T., Clarke, C. R., Cai, R., Vinatzer, B. A., Jardini, T. M., and Koike, S. T. (2011). Multilocus sequence typing of Pseudomonas syringae sensu lato confirms previously described genomospecies and permits rapid identification of P. syringae pv. coriandricola and P. syringae pv. apii causing bacterial leaf spot on parsley. Phytopathology 101, 847–858. doi: 10.1094/PHYTO-11-10-0318
Cánovas, D., Cases, I., and de Lorenzo, V. (2003). Heavy metal tolerance and metal homeostasis in Pseudomonas putida as revealed by complete genome analysis. Environ. Microbiol. 5, 1242–1256. doi: 10.1111/j.1462-2920.2003.00463.x
Carrión, V. J., Arrebola, E., Cazorla, F. M., Murillo, J., and de Vicente, A. (2012). The mbo operon is specific and essential for biosynthesis of mangotoxin in Pseudomonas syringae. PLoS One 7:e36709. doi: 10.1371/journal.pone.0036709
Carrión, V. J., Gutiérrez-Barranquero, J. A., Arrebola, E., Bardaji, L., Codina, J. C., de Vicente, A., et al. (2013). The mangotoxin biosynthetic operon (mbo) is specifically distributed within Pseudomonas syringae Genomospecies 1 and was acquired only once during evolution. Appl. Environ. Microbiol. 79, 756–767. doi: 10.1128/AEM.03007-12
Carrión, V. J., van der Voort, M., Arrebola, E., Gutiérrez-Barranquero, J. A., de Vicente, A., Raaijmakers, J. M., et al. (2014). Mangotoxin production of Pseudomonas syringae pv. syringae is regulated by MgoA. BMC Microbiol. 14:46. doi: 10.1186/1471-2180-14-46
Cazorla, F. M., Arrebola, E., Olea, F., Velasco, L., Hermoso, J. M., Pérez- García, A., et al. (2006). Field evaluation of treatments for the control of the bacterial apical necrosis of mango (Mangifera indica) caused by Pseudomonas syringae pv. syringae. Eur. J. Plant Pathol. 116, 279–288. doi: 10.1007/s10658-006-9059-7
Cazorla, F. M., Arrebola, E., Sesma, A., Pérez-García, A., Codina, J. C., Murillo, J., et al. (2002). Copper resistance in Pseudomonas syringae strains isolated from mango is encoded mainly by plasmids. Phytopathology 92, 909–916. doi: 10.1094/PHYTO.2002.92.8.909
Cazorla, F. M., Codina, J. C., Abad, C., Arrebola, E., Torés, J. A., Murillo, J., et al. (2008). 62-kb plasmids harboring rulAB homologues confer UV-tolerance and epiphytic fitness to Pseudomonas syringae pv. syringae mango isolates. Microbiol. Ecol. 56, 283–291. doi: 10.1007/s00248-007-9346-7
Cazorla, F. M., Olalla, L., Torés, J. A., Pérez-García, A., Codina, J. C., and de Vicente, A. (1995). A method for estimation of population densities of ice nucleating active Pseudomonas syringae in buds and leaves of mango. J. Appl. Bacteriol. 79, 341–346. doi: 10.1111/j.1365-2672.1995.tb03146.x
Cazorla, F. M., Torés, J. A., de Vicente, A., and Farré, J. M. (1992). Procesos necróticos de mango: aislamiento de Pseudomonas syringae. Bol. Cultivos Trop. 92:8.
Cazorla, F. M., Torés, J. A., Olalla, L., Pérez-García, A., Farré, J. M., and de Vicente, A. (1998). Bacterial apical necrosis of mango in Southern Spain: a disease caused by Pseudomonas syringae pv. syringae. Phytopathology 88, 614–620. doi: 10.1094/PHYTO.1998.88.7.614
Clarke, C. R., Cai, R., Studholme, D. J., Guttman, D. S., and Vinatzer, B. A. (2010). Pseudomonas syringae strains naturally lacking the classical P. syringae hrp/hrc locus are common leaf colonizers equipped with an atypical type III secretion system. Mol. Plant Microbe Interact. 23, 198–210. doi: 10.1094/MPMI-23-2-0198
Collmer, A., Badel, J. L., Charkowski, A. O., Deng, W.-L., Fouts, D. E., Ramos, A. R., et al. (2000). Pseudomonas syringae Hrp type III secretion system and effector proteins. Proc. Natl. Acad. Sci. U.S.A. 97, 8770–8777. doi: 10.1073/pnas.97.16.8770
Cooksey, D. A. (1987). Characterization of a copper resistance plasmid conserved in copper-resistant strains of Pseudomonas syringae pv. tomato. Appl. Environ. Microbiol. 53, 454–456.
Crespo, M., Cazorla, F. M., Hermoso, J. M., Guirado, E., Maymon, M., Torés, J. A., et al. (2012). First report of mango malformation disease caused by Fusarium mangiferae in Spain. Plant Dis. 96, 286–287. doi: 10.1094/PDIS-07-11-0599
Cunnac, S., Chakravarthy, S., Kvitko, B. H., Russell, A. B., Martin, G. B., and Collmer, A. (2011). Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. U.S.A. 108, 2975–2980. doi: 10.1073/pnas.1013031108
Denny, T. P., Gilmour, M. N., and Selander, R. K. (1988). Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J. Gen. Microbiol. 134, 1949–1960. doi: 10.1099/00221287-134-7-1949
Dillion, M. M., Thakur, S., Almeida, R. N. D., Wang, P. W., Weir, B. S., and Guttman, D. S. (2019). Recombination of ecologically and evolutionarily significant loci maintains genetic cohesion in the Pseudomonas syringae species complex. Genome Biol. 20:3. doi: 10.1186/s13059-018-1606-y
Diogo, R., and Wydra, K. (2007). Silicon-induced basal resistance in tomato against Ralstonia solanacearum is related to modification of pectic cell wall polysaccharide structure. Physiol. Mol. Plant Pathol. 70, 120–129. doi: 10.1016/j.pmpp.2007.07.008
Dye, D. W., Bradbury, J. F., Goto, M., Hayward, A. C., Lelliott, R. A., and Schroth, M. N. (1980). International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotypes. Rev. Plant Pathol. 59, 153–168.
Egan, F., Barret, M., and O’Gara, F. (2014). The SPI-1-like type III secretion system: more roles tan you think. Front. Plant Sci. 5:34. doi: 10.3389/fpls.2014.00034
Endert, E., and Ritchie, D. F. (1984). Overwintering and survival of Pseudomonas syringae pv. syringae and symptom development in peach trees. Plant Dis. 68, 468–470. doi: 10.1094/PD-69-468
English, H., DeVay, J. E., and Ogawa, J. M. (1980). Bacterial Canker and Blast of Deciduous Fruits. Geneva: WHO, 2155.
Fauteux, F., Rémus-Bore, W., Menzies, J. G., and Bélanger, R. R. (2005). Silicon and plant disease resistance against pathogenic fungi. FEMS Microbiol. Lett. 249, 1–6. doi: 10.1016/j.femsle.2005.06.034
Fouts, D. E., Abramovitch, R. B., Alfano, J. R., Baldo, A. M., Buell, C. R., Cartinhour, S., et al. (2002). Genome wide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. U.S.A. 99, 2275–2280. doi: 10.1073/pnas.032514099
Franke, S., Grass, G., Rensing, C., and Nies, D. H. (2003). Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185, 3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003
Freeman, B. C., Chen, C., Yu, X., Nielsen, L., Peterson, K., and Beattie, G. A. (2013). Physiological and transcriptional responses to osmotic stress of two Pseudomonas syringae strains that differ in epiphytic fitness and osmotolerance. J. Bacteriol. 195, 4742–4752. doi: 10.1128/JB.00787-13
Gagnevin, L., and Pruvost, O. (2001). Epidemiology and control of mango bacterial black spot. Plant Dis. 85, 928–935. doi: 10.1094/PDIS.2001.85.9.928
Gal, M., Preston, G. M., Massey, R. C., Spiers, A. J., and Rainey, P. B. (2003). Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Molec. Ecol. 12, 3109–3121. doi: 10.1046/j.1365-294X.2003.01953.x
Galán-Saúco, V. (2015). Current situation and future prospects of worldwide mango production and market. Acta Hortic. 1066, 69–84. doi: 10.17660/ActaHortic.2015.1066.7
Gardan, L., Bollet, C., Abu Ghorrah, M., Grimont, F., and Grimont, P. A. D. (1992). DNA relatedness among the pathovar strains of Pseudomonas syringae subsp. savastanoi Janse (1982) and proposal of Pseudomonas savastanoi sp. nov. Int. J. Syst. Bacteriol. 42, 606–612. doi: 10.1099/00207713-42-4-606
Gardan, L., Shafik, H., Belouin, S., Broch, R., Grimont, F., and Grimont, P. A. D. (1999). DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959). Int. J. Syst. Bacteriol. 49, 469–478. doi: 10.1099/00207713-49-2-469
Gasic, K., Pavlovic, Z., Santander, R., Meredith, C., and Aćimović, S. (2018). First report of Pseudomonas syringae pv. syringae associated with bacterial blossom blast on apple (Malus pumila) in USA. Plant Dis. 102:1848. doi: 10.1094/PDIS-01-18-0184-PDN
Gazi, A. D., Sarris, P. F., Fadouloglou, V. E., Charova, S. N., Mathioudakis, N., Panopoulos, N., et al. (2012). Phylogenetic analysis of a gene cluster encoding an additional, rhizobial-like type III secretion system that is narrowly distributed among Pseudomonas syringae strains. BMC Microbiol. 12:188. doi: 10.1186/1471-2180-12-188
Gomila, M., Busquets, A., Mulet, M., García-Valdés, E., and Lalucat, J. (2017). Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analysis. Front. Microbiol. 8:2422. doi: 10.3389/fmicb.2017.02422
Green, S., Studholme, D. J., Laue, B. E., Dorati, F., Lovell, H., Arnold, D., et al. (2010). Comparative genome analysis provides insights into the evolution and adaptation of Pseudomonas syringae pv. aesculi on Aesculus hippocastanum. PLoS One 5:e10224. doi: 10.1371/journal.pone.0010224
Gross, D. C., and DeVay, J. E. (1977). Production and purification of syringomycin, a phytotoxin produced by Pseudomonas syringae. Physiol. Plant. Pathol. 11, 13–28. doi: 10.1016/0048-4059(77)90083-2
Guével, M. H., Menzies, J. G., and Bélanger, R. R. (2007). Effect of root and foliar applications of soluble silicon on powdery mildew control and growth of wheat plants. Eur. J. Plant Pathol. 119, 429–436. doi: 10.1007/s10658-007-9181-1
Gutiérrez-Barranquero, J. A., Arrebola, E., Bonilla, N., Sarmiento, D., Cazorla, F. M., and de Vicente, A. (2012). Environmentally friendly treatment alternatives to Bordeaux mixture for controlling bacterial apical necrosis (BAN) of mango. Plant Pathol. 61, 665–676. doi: 10.1111/j.1365-3059.2011.02559.x
Gutiérrez-Barranquero, J. A., Cazorla, F. M., de Vicente, A., and Sundin, G. W. (2017a). Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family. BMC Genomics 18:365. doi: 10.1186/s12864-017-3763-x
Gutiérrez-Barranquero, J. A., Cazorla, F. M., Guirado, E., Sarmiento, D., Torés, J. A., and de Vicente, A. (2017b). Cómo manejar la necrosis apical y la malformación, las dos principales enfermedades que afectan al mango en España. Phytoma 287, 16–21.
Gutiérrez-Barranquero, J. A., Cazorla, F. M., Torés, J. A., and de Vicente, A. (2019). Pantoea agglomerans as a new etiological agent of a bacterial necrotic disease of mango trees. Phytopathology 109, 17–26. doi: 10.1094/PHYTO-06-18-0186-R
Gutiérrez-Barranquero, J. A., Carrión, V. J., Murillo, J., Arrebola, E., Arnold, D. L., and Cazorla, F. M. (2013a). A Pseudomonas syringae diversity survey reveals a differentiated phylotype of the pathovar syringae associated with the mango host and mangotoxin production. Phytopathology 103, 1115–1129. doi: 10.1094/PHYTO-04-13-0093-R
Gutiérrez-Barranquero, J. A., de Vicente, A., Carrión, V. J., Sundin, G. W., and Cazorla, F. M. (2013b). Recruitment and rearrangement of three different genetic determinants into a conjugative plasmid increase copper resistance in Pseudomonas syringae. Appl. Environ. Microbiol. 79, 1028–1033. doi: 10.1128/AEM.02644-12
Hirano, S. S., and Upper, C. D. (1995). “Ecology of ice nucleation-active bacteria,” in Biological Ice Nucleation and Its Applications, eds R. E. Lee, G. J. Warren, and L. V. Gusta (St. Paul, MN: APS Press), 41–61.
Hirano, S. S., and Upper, C. D. (2000). Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 64, 624–653. doi: 10.1128/MMBR.64.3.624-653.2000
Huang, H. C., He, S. Y., Bauer, D. W., and Collmer, A. (1992). The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J. Bacteriol. 174, 6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992
Hulin, M. T., Armitage, A. D., Vicente, J. G., Holub, E. B., Baxter, L., Bates, H. J., et al. (2018). Comparative genomics of Pseudomonas syringae reveals convergent gene gain and loss associated with specialization onto cherry (Prunus avium). New Phytol. 219, 672–696. doi: 10.1111/nph.15182
Hwang, M. S., Morgan, R. L., Sarkar, S. F., Wang, P. W., and Guttman, D. S. (2005). Phylogenetic characterization of virulence and resistance phenotypes of Pseudomonas syringae. Appl. Environ. Microbiol. 71, 5182–5191. doi: 10.1128/AEM.71.9.5182-5191.2005
Iacobellis, N. S., Lavermicocca, P., Grgurina, I., Simmaco, M., and Ballio, A. (1992). Phytotoxic properties of Pseudomonas syringae pv. syringae toxins. Physiol. Mol. Plant Pathol. 40, 107–116. doi: 10.1016/0885-5765(92)90038-W
Iannotta, N., Belfiore, T., Brandmaya, P., Noce, M. E., and Scalercio, S. (2007). Evaluation of the impact on entomocoenosis of active agents allowed in organic olive farming against Bactrocera oleae (Gmelin, 1790). J. Environ. Sci. Health B 42, 783–788. doi: 10.1080/03601230701551020
Ichinose, Y., Taguchi, F., and Mukaihara, T. (2013). Pathogenicity and virulence factors of Pseudomonas syringae. J. Gen. Plant Pathol. 79, 285–296. doi: 10.1007/s10327-013-0452-8
Ivanović, Ž, Perović, T., Popović, T., Blagojević, J., Trkulja, N., and Hrnčić, S. (2017). Characterization of Pseudomonas syringae pv. syringae, causal agent of citrus blast of mandarin in Montenegro. Plant Pathol. J. 33, 21–33. doi: 10.5423/PPJ.OA.08.2016.0161
Jackson, R. W., Athanassopoulos, E., Tsiamis, G., Mansfield, J. W., Sesma, A., Arnold, D. L., et al. (1999). Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc. Natl. Acad. Sci. U.S.A. 96, 10875–10880. doi: 10.1073/pnas.96.19.10875
Jacobs, J. L., and Sundin, G. W. (2001). Effect of solar UV-B radiation on a phyllosphere bacterial community. Appl. Environ. Microbiol. 67, 5488–5496. doi: 10.1128/AEM.67.12.5488-5496.2001
Janse, J. D., Rossi, P., Angelucci, L., Scortichini, M., Derks, J. H. J., Akkermans, A. D. L., et al. (1996). Reclassification of Pseudomonas syringae pv. avellanae as Pseudomonas avellanae (spec. nov.), the bacterium causing canker of hazelnut (Corylus avellana L.). Syst. Appl. Microbiol. 19, 589–595. doi: 10.1016/S0723-2020(96)80030-0
Jenul, C., Sieber, S., Daeppen, C., Mathew, A., Lardi, M., Pessi, G., et al. (2018). Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 9:1297. doi: 10.1038/s41467-018-03690-2
Kairu, G. M., Nyangena, C. M. S., and Crosse, J. E. (1985). The effect of copper sprays on bacterial blight and coffee berry disease in Kenya. Plant Pathol. 34, 207–213. doi: 10.1111/j.1365-3059.1985.tb01351.x
Kasapis, S., Morris, E. R., Gross, M., and Rudolph, K. (1994). Solution properties of levan polysaccharide from Pseudomonas-syringae pv phaseolicola, and its possible primary role as a blocker of recognition during pathogenesis. Carbohydr. Polym. 23, 55–64. doi: 10.1016/0144-8617(94)90090-6
Kennelly, M. M., Cazorla, F. M., de Vicente, A., Ramos, C., and Sundin, G. W. (2007). Pseudomonas syringae diseases of fruit trees. Progress toward understanding and control. Plant Dis. 91, 4–17. doi: 10.1094/PD-91-0004
Kim, J. J., and Sundin, G. W. (2000). Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J. Bacteriol. 182, 6137–6144. doi: 10.1128/JB.182.21.6137-6144.2000
Kim, S. G., Kim, K. W., Park, E. W., and Choi, D. (2002). Silicon-induced cell wall fortification of rice leaves: a possible cellular mechanism of enhanced host resistance to blast. Phytopathology 92, 1095–1103. doi: 10.1094/PHYTO.2002.92.10.1095
Kretsch, A. M., Morgan, G. L., Tyrrell, J., Mevers, E., Vallet-Gély, I., and Li, B. (2018). Discovery of (dihydro)pyrazine N-oxides via genome mining in Pseudomonas. Org. Lett. 20, 4791–4795. doi: 10.1021/acs.orglett.8b01944
Kvitko, B. H., Park, D. H., Velásquez, A. C., Wei, C. F., Russell, A. B., Martin, G. B., et al. (2009). Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 5:e1000388. doi: 10.1371/journal.ppat.1000388
Lamichhane, J. R., Osdaghi, E., Behlau, F., Köhl, J., Jones, J. B., and Aubertot, J.-N. (2018). Thirteen decades of antimicrobial copper compounds applied in agriculture. A review. Agron. Sustain. Dev. 38:28. doi: 10.1007/s13593-018-0503-9
Lamichhane, J. R., Varvaro, L., Parisi, L., Audergon, J.-M., and Morris, C. E. (2014). Disease and frost damage of woody plants caused by Pseudomonas syringe: seeing the forest for the trees. Adv. Agron. 126, 235–295. doi: 10.1016/B978-0-12-800132-5.00004-3
Laue, H., Schenk, A., Li, H., Lambertsen, L., Neu, T. R., Molin, S., et al. (2006). Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology 152, 2909–2918. doi: 10.1099/mic.0.28875-0
Lavermicocca, P., Iacobellis, N., Simmaco, M., and Graniti, A. (1997). Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins. Physiol. Mol. Plant Pathol. 50, 129–140. doi: 10.1006/pmpp.1996.0078
Lee, J., Teitzel, G. M., Munkvold, K., del Pozo, O., Martin, G. B., Michelmore, R. W., et al. (2012). Type III secretion and effectors shape the survival and growth pattern of Pseudomonas syringae on leaf surfaces. Plant Physiol. 158, 1803–1818. doi: 10.1104/pp.111.190686
Leedjärv, A., Ivask, A., and Virta, M. (2008). Interplay of different transporters in the mediation of divalent heavy metal resistance in Pseudomonas putida KT2440. J. Bacteriol. 190, 2680–2689. doi: 10.1128/JB.01494-07
Lelliott, R. A., and Stead, D. E. (1987). Methods in Plant Pathology, Vol. 2: Methods for the Diagnosis of Bacterial Diseases of Plants. Oxford: Blackwell Scientific Publications.
Lim, C. K., and Cooksey, D. A. (1993). Characterization of chromosomal homologs of the plasmid-borne copper resistance operon of Pseudomonas syringae. J. Bacteriol. 175, 4492–4498. doi: 10.1128/jb.175.14.4492-4498.1993
Lindeberg, M., Cunnac, S., and Collmer, A. (2012). Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20, 199–208. doi: 10.1016/j.tim.2012.01.003
Lindeberg, M., Myers, C. R., Collmer, A., and Schneider, D. J. (2008). Roadmap to new virulence determinants in Pseudomonas syringae: insights from comparative genomics and genome organization. Mol. Plant Microbe Interac. 21, 685–700. doi: 10.1094/MPMI-21-6-0685
Lindgren, P. B., Peet, R. C., and Panopoulos, N. J. (1986). Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J. Bacteriol. 168, 512–522. doi: 10.1128/jb.168.2.512-522.1986
Lindow, S. E., Arny, D. C., and Upper, C. D. (1978). Erwinia herbicola: a bacterial ice nucleus active in increasing frost injury to corn. Phytopathology 68, 523–527.
Lindow, S. E., Arny, D. C., and Upper, C. D. (1982). Bacterial ice nucleation: a factor in frost injury to plants. Plant physiol. 70, 1084–1089. doi: 10.1104/pp.70.4.1084
Lindow, S. E., and Brandl, M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003
Lindow, S. E., and Connell, J. H. (1984). Reduction of frost injury to almond by control of ice nucleation active bacteria. J. Am. Soc. Hort. Sci. 109, 48–53.
Lo, T., Koulena, N., Seto, D., Guttman, D. S., and Desveaux, D. (2017). The HopF family of Pseudomonas syringae type III secreted effectors. Mol. Plant Pathol. 18, 457–468. doi: 10.1111/mpp.12412
Lovell, H. C., Mansfield, J. W., Godfrey, S. A. C., Jackson, R. W., Hancock, J. T., and Arnold, D. L. (2009). Bacterial evolution by genomic island transfer occurs via DNA transformation in planta. Curr. Biol. 19, 1586–1590. doi: 10.1016/j.cub.2009.08.018
Ma, Z., Smith, J. J., Zhao, Y., Jackson, R. W., Arnold, D. L., Murillo, J., et al. (2007). Phylogenetic analysis of the pPT23A plasmid family of Pseudomonas syringae. Appl. Environ. Microbiol. 73, 1287–1295. doi: 10.1128/AEM.01923-06
Mansfield, J. W., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., Ronald, P., et al. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
Mansvelt, E. L., and Hattingh, M. J. (1986). Bacterial blister bark and blight of fruit spurs of apple in South Africa caused by Pseudomonas syringae pv. syringae. Plant Dis. 70, 403–405. doi: 10.1094/PD-70-403
Marahiel, M. A., Stachelhaus, T., and Mootz, H. D. (1997). Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97, 2651–2674. doi: 10.1021/cr960029e
Martínez-García, P. M., López-Solanilla, E., Ramos, C., and Rodríguez-Palenzuela, P. (2016). Prediction of bacterial associations with plants using a supervised machine-learning approach. Environ. Microbiol. 18, 4847–4861. doi: 10.1111/1462-2920.13389
Martínez-García, P. M., Rodríguez-Palenzuela, P., Arrebola, E., Carrión, V. J., Gutiérrez-Barranquero, J. A., Pérez-García, A., et al. (2015). Bioinformatics analysis of the complete genome sequence of the mango tree pathogen Pseudomonas syringae pv. syringae UMAF0158 reveals traits relevant to virulence and epiphytic lifestyle. PLoS One 10:e0136101. doi: 10.1371/journal.pone.0136101
Matas, I. M., Castañeda-Ojeda, M. P., Aragón, I. M., Antúnez-Lamas, M., Murillo, J., Rodríguez-Palenzuela, P., et al. (2014). Translocation and functional analysis of Pseudomonas savastanoi pv. savastanoi NCPPB 3335 type III secretion system effectors reveals two novel effector families of the Pseudomonas syringae complex. Mol. Plant Microbe Interact. 27, 424–436. doi: 10.1094/MPMI-07-13-0206-R
McCann, H. C., Rikkerink, E. H. A., Bertels, F., Fiers, M., Lu, A., Rees-George, J., et al. (2013). Genomic analysis of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae provides insight into the origins of an emergent plant disease. PLoS Pathog. 9:e1003503. doi: 10.1371/journal.ppat.1003503
Medini, D., Donati, C., Tettelin, H., Masignani, V., and Rappouli, R. (2005). The microbial pangenome. Curr. Opin. Genet. Dev. 15, 589–594. doi: 10.1016/j.gde.2005.09.006
Mellano, M. A., and Cooksey, D. A. (1988). Nucleotide sequence and organization of copper resistance genes from Pseudomonas syringae pv. tomato. J. Bacteriol. 170, 2879–2883. doi: 10.1128/jb.170.6.2879-2883.1988
Mergeay, M., Monchy, S., Vallaeys, T., Auquier, V., Benotmane, A., Bertin, P., et al. (2003). Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes. FEMS Microbiol. Rev. 27, 385–410. doi: 10.1016/S0168-6445(03)00045-7
Mirik, M., Baloglu, S., Aysan, Y., Cetinkaya-Yildiz, R., Kusek, M., and Sahin, F. (2005). First outbreak and occurrence of citrus blast disease, caused by Pseudomonas syringae pv. syringae, on orange and mandarin trees in Turkey. Plant Pathol. 54:238. doi: 10.1111/j.1365-3059.2005.01134.x
Mitchell, R. E. (1991). Implications of toxins in the ecology and evolution of plant pathogenic microorganisms: bacteria. Experientia 47, 791–803. doi: 10.1007/BF01922459
Monteil, C. L., Yahara, K., Studholme, D. J., Mageiros, L., Méric, G., Swingle, B., et al. (2016). Population-genomic insights into emergence, crop adaptation and dissemination of Pseudomonas syringae pathogens. Microb. Genom. 2:e000089. doi: 10.1099/mgen.0.000089
Montesinos, E., and Vilardell, P. (1991). Relationships among population levels of Pseudomonas syringae, amount of ice nuclei, and incidence of blast dormant flower in commercial pear orchards in Catalunya. Spain. Phytopathology 81, 113–119. doi: 10.1094/Phyto-81-113
Morris, C. E., Sands, D. C., Vinatzer, B. A., Glaux, C., Guilbaud, C., Buffière, A., et al. (2008). The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2, 321–334. doi: 10.1038/ismej.2007.113
Norman, A., Hansen, L. H., and Sørensen, S. J. (2009). Conjugative plasmids: vessels of the communal gene pool. Philos. Trans. R. Soc. Lond. B 364, 2275–2289. doi: 10.1098/rstb.2009.0037
O’Brien, R. D., and Lindow, S. E. (1988). Effect of plant species and environmental conditions on ice nucleation activity of Pseudomonas syringae on leaves. Appl. Environ. Microbiol. 54, 2281–2286.
Oh, H. S., Park, D. H., and Collmer, A. (2010). Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity. Mol. Plant Microbe Interact. 23, 727–739. doi: 10.1094/MPMI-23-6-0727
Palleroni, N. J., Ballard, R. W., Ralston, E., and Doudoroff, M. (1972). Deoxyribonucleic acid homologies among some Pseudomonas species. J. Bacteriol. 110, 1–11.
Patil, S. S., Tam, L. Q., and Sakai, W. S. (1972). Mode of action of the toxin from Pseudomonas phaseolicola. I. Toxin specificity, chlorosis and ornithine accumulation. Plant Physiol. 49, 803–807. doi: 10.1104/pp.49.5.803
Pecknold, P. C., and Grogan, R. G. (1973). Deoxyribonucleic acid homology groups among phytopathogenic Pseudomonas species. Int. J. Syst. Bacteriol. 23, 111–121. doi: 10.1099/00207713-23-2-111
Pérez-Martínez, I., Rodríguez-Moreno, L., Lambertsen, L., Matas, I. M., Murillo, J., Tegli, S., et al. (2010). Fate of a Pseudomonas savastanoi pv. savastanoi type III secretion system mutant in olive plants (Olea europaea L.). Appl. Environ. Microbiol. 76, 3611–3619. doi: 10.1128/AEM.00133-10
Pérez-Mendoza, D., Aragón, I. M., Prada-Ramírez, H. A., Romero-Jiménez, L., Ramos, C., Gallegos, M. T., et al. (2014). Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS One 9:e91645. doi: 10.1371/journal.pone.0091645
Pfeilmeier, S., Caly, D. L., and Malone, J. G. (2016). Bacterial pathogenesis of plants: future challenges from a microbial perspective: challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 17, 1298–1313. doi: 10.1111/mpp.12427
Prada-Ramírez, H. A., Pérez-Mendoza, D., Felipe, A., Martínez-Granero, F., Rivilla, R., Sanjuán, J., et al. (2016). AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000. Mol. Microbiol. 99, 960–977. doi: 10.1111/mmi.13278
Rainey, P. B., and Travisano, M. (1998). Adaptive radiation in a heterogeneous environment. Nature 394, 69–72. doi: 10.1038/27900
Reinhardt, J. A., Baltrus, D. A., Nishimura, M. T., Jeck, W. R., Jones, C. D., and Dangl, J. L. (2009). De novo assembly using low coverage short read sequence data from the rice pathogen Pseudomonas syringae pv. oryzae. Genome Res. 19, 294–305. doi: 10.1101/gr.083311.108
Rodgers-Gray, B. S., and Shaw, M. W. (2004). Effects of straw and silicon soil amendments on some foliar and stem-base diseases in pot-grown winter wheat. Plant Pathol. 53, 733–740. doi: 10.1111/j.1365-3059.2004.01102.x
Rodrigues, F. A., Benhamou, N., Datnoff, L. E., Jones, J. B., and Bélanger, R. R. (2003). Ultrastuctural and cytochemical aspects of silicon mediated rice blast resistance. Phytopathology 93, 535–546. doi: 10.1094/PHYTO.2003.93.5.535
Rogers, J. S., Clark, E., Cirvilleri, G., and Lindow, S. E. (1994). Cloning and characterization of genes conferring copper resistance in epiphytic ice nucleation-active Pseudomonas syringae strains. Phytopathology 84, 891–897. doi: 10.1094/Phyto-84-891
Römling, U. (2002). Molecular biology of cellulose production in bacteria. Res. Microbiol. 153, 205–212. doi: 10.1016/S0923-2508(02)01316-5
Römling, U., and Galperin, M. Y. (2015). Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 23, 545–557. doi: 10.1016/j.tim.2015.05.005
Römling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Salmond, G. P. (1994). Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32, 181–200. doi: 10.1146/annurev.py.32.090194.001145
Sarkar, S. F., and Guttman, D. S. (2004). Evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl. Environ. Microbiol. 70, 1999–2012. doi: 10.1128/AEM.70.4.1999-2012.2004
Schechter, L. M., Vencato, M., Jordan, K. L., Schneider, S. E., Schneider, D. J., and Collmer, A. (2006). Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant Microbe Interact. 19, 1180–1192. doi: 10.1094/MPMI-19-1180
Scheck, H. J., and Pscheidt, J. W. (1998). Effect of copper bactericides on copper-resistant and-sensitive strains of Pseudomonas syringae pv. syringae. Plant Dis. 82, 397–406. doi: 10.1094/PDIS.1998.82.4.397
Scholz-Schroeder, B. K., Hutchison, M. L., Grgurina, I., and Gross, D. C. (2001). The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant Microbe Interact. 14, 336–348. doi: 10.1094/MPMI.2001.14.3.336
Sesma, A., Murillo, J., and Sundin, G. W. (2000). Phylogeny of the replication regions of pPT23A-like plasmids from Pseudomonas syringae. Microbiology 146, 2375–2384. doi: 10.1099/00221287-146-10-2375
Sesma, A., Sundin, G. W., and Murillo, J. (1998). Closely related plasmid replicons in the phytopathogen Pseudomonas syringae show a mosaic organization of the replication region and an altered incompatibility behavior. Appl. Environ. Microbiol. 64, 3948–3953.
Silby, M. W., Winstanley, C., Godfrey, S. A. C., Levy, S. B., and Jackson, R. W. (2011). Pseudomonas genomes: diverse and adaptable. FEMS Microbiol. Rev. 35, 652–680. doi: 10.1111/j.1574-6976.2011.00269.x
Sorensen, K. N., Kim, K. H., and Takemoto, J. Y. (1998). PCR detection of cyclic lipodepsinonapeptide-producing Pseudomonas syringae pv. syringae and similarity of strains. Appl. Environ. Microbiol. 64, 226–230.
Spiers, A. J., Bohannon, J., Gehrig, S., and Rainey, P. B. (2003). Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50, 15–27. doi: 10.1046/j.1365-2958.2003.03670.x
Spiers, A. J., Kahn, S. G., Travisano, M., Bohannon, J., and Rainey, P. B. (2002). Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics 161, 33–46.
Stein, T., and Vater, J. (1996). Amino acid activation and polymerization at modular multienzymes in nonribosomal peptide biosynthesis. Amino Acids 10, 201–227. doi: 10.1007/BF00807324
Studholme, D. J., Ibanez, S. G., MacLean, D., Dangl, J. L., Chang, J. H., and Rathjen, J. P. (2009). A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics 10:395. doi: 10.1186/1471-2164-10-395
Studholme, D. J., Rachel, H., and Glover, N. B. (2011). Application of high-throughput DNA sequencing in phytopathology. Annu. Rev. Phytopathol. 49, 87–105. doi: 10.1146/annurev-phyto-072910-095408
Sun, W., Zhang, J., Fan, Q., Xue, G., Li, Z., and Liang, Y. (2010). Silicon enhanced resistance to rice blast is attributed to silicon-mediated defence resistance and its role as physical barrier. Eur. J. Plant Pathol. 128, 39–49. doi: 10.1007/s10658-010-9625-x
Sundin, G. W. (2007). Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu. Rev. Phytopathol. 45, 129–151. doi: 10.1146/annurev.phyto.45.062806.094317
Sundin, G. W., and Bender, C. L. (1993). Ecological and genetic analysis of copper and streptomycin resistance in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 59, 1018–1024.
Sundin, G. W., and Bender, C. L. (1996). Molecular analysis of closely related copper- and streptomycin-resistance plasmids in Pseudomonas syringae pv. syringae. Plasmid 35, 98–107. doi: 10.1006/plas.1996.0012
Sundin, G. W., and Jacobs, J. L. (1999). Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.). Microb. Ecol. 38, 27–38. doi: 10.1007/s002489900152
Sundin, G. W., Jacobs, J. L., and Murillo, J. (2000). Sequence diversity of rulA among natural isolates of Pseudomonas syringae and effect on function of rulAB-mediated UV radiation tolerance. Appl. Environ. Microbiol. 66, 5167–5173. doi: 10.1128/AEM.66.12.5167-5173.2000
Sundin, G. W., Jones, A. L., and Fulbright, D. W. (1989). Copper resistance in Pseudomonas syringae pv. syringae from cherry orchards and its associated transfer in vitro and in planta with a plasmid. Phytopathology 79, 861–865. doi: 10.1094/Phyto-79-861
Sundin, G. W., Kidambi, S. P., Ulrich, M., and Bender, C. L. (1996). Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene 177, 77–81. doi: 10.1016/0378-1119(96)00273-9
Sundin, G. W., and Murillo, J. (1999). Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290-320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ. Microbiol. 1, 75–88. doi: 10.1046/j.1462-2920.1999.00008.x
Thakur, S., Weir, B. S., and Guttman, D. S. (2016). Phytopathogen genome announcement: draft genome sequences of 62 Pseudomonas syringae type and pathotype strains. Mol. Plant Microbe Interac. 29, 243–246. doi: 10.1094/MPMI-01-16-0013-TA
Turner, J. G., and Debbage, J. M. (1982). Tabtoxin-induced symptoms are associated with accumulation of ammonia formed during photorespiration. Physiol. Plant Pathol. 20, 223–233. doi: 10.1016/0048-4059(82)90087-X
Ude, S., Arnold, D. A., Moon, C. D., Timmis-Wilson, T., and Spiers, A. J. (2006). Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 8, 1997–2011. doi: 10.1111/j.1462-2920.2006.01080.x
Vallet-Gely, I., Opota, O., Boniface, A., Novikov, A., and Lemaitre, B. (2010). A secondary metabolite acting as a signalling molecule controls Pseudomonas entomophila virulence. Cell. Microbiol. 12, 1666–1679. doi: 10.1111/j.1462-5822.2010.01501.x
Vassilev, V., Lavermicocca, P., Di Giorgio, C., and Iacobellis, N. (1996). Production of syringomycins and syringopeptins by Pseudomonas syringae pv. atrofaciens. Plant Pathol. 45, 316–322. doi: 10.1046/j.1365-3059.1996.d01-126.x
Vinatzer, B. A., Teitzel, G. M., Lee, M. W., Jelenska, J., Hotton, S., Fairfax, K., et al. (2006). The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol. Microbiol. 62, 26–44. doi: 10.1111/j.1365-2958.2006.05350.x
Vivian, A., Murillo, J., and Jackson, R. W. (2001). The roles of plasmids in phytopathogenic bacteria: mobile arsenals? Microbiology 147, 763–780. doi: 10.1099/00221287-147-4-763
Volksch, B., and Weingart, H. (1998). Toxin production by pathovars of Pseudomonas syringae and their antagonistic activities against epiphytic microorganisms. J. Basic Microbiol. 38, 135–145.
Von Rozycki, T., and Nies, D. H. (2009). Cupriavidus metallidurans: evolution of a metal-resistant bacterium. Antonie Van Leeuwenhoek 96, 115–139. doi: 10.1007/s10482-008-9284-5
Wang, M. J. (1997). Land application of sewage sludge in China. Sci. Total Environ. 197, 149–160. doi: 10.1016/S0048-9697(97)05426-0
Wenneker, M., Meijer, H., Maas, F. M., de Bruine, A., Vink, P., and Pham, K. (2013). Bacterial canker of plum trees (prunus domestica), caused by Pseudomonas syringae pathovars, in the netherlands. Acta Hortic. 985, 235–239. doi: 10.17660/ActaHortic.2013.985.30
Xin, X.-F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Xiong, Z. (1998). Heavy metal contamination of urban soils and plants in relation to traffic inWuhan City, China. Toxicol. Environ. Chem. 65, 31–39. doi: 10.1080/02772249809358555
Xiong, Z., and Wang, H. (2005). Copper toxicity and bioaccumulation in Chinese cabbage (Brassica pekinensis Rupr.). Environ. Toxicol. 20, 188–194. doi: 10.1002/tox.20094
Xu, L. H., Xie, G. L., Li, B., Zhu, B., Xu, F. S., and Qian, J. (2008). First report of pear blossom blast caused by Pseudomonas syringae pv. syringae in China. Plant Dis. 92:832. doi: 10.1094/PDIS-92-5-0832C
Young, J. M. (2010). Taxonomy of Pseudomonas syringae. J. Plant Pathol. 92, S5–S14. doi: 10.4454/jpp.v92i1sup.2501
Keywords: Pseudomonas syringae pv. syringae, mango tree, epiphytic fitness, virulence strategies, mangotoxin, pPT23A family plasmid, ultraviolet radiation and copper resistance
Citation: Gutiérrez-Barranquero JA, Cazorla FM and de Vicente A (2019) Pseudomonas syringae pv. syringae Associated With Mango Trees, a Particular Pathogen Within the “Hodgepodge” of the Pseudomonas syringae Complex. Front. Plant Sci. 10:570. doi: 10.3389/fpls.2019.00570
Received: 18 December 2018; Accepted: 15 April 2019;
Published: 08 May 2019.
Edited by:
Olivier Pruvost, UMR Peuplements Végétaux et Bio-Agresseurs en Milieu Tropical (CIRAD), FranceReviewed by:
Boris Alexander Vinatzer, Virginia Tech, United StatesCopyright © 2019 Gutiérrez-Barranquero, Cazorla and de Vicente. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonio de Vicente, YWRldmljZW50ZUB1bWEuZXM=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.