
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 April 2019
Sec. Plant Cell Biology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00516
This article is part of the Research Topic The Maintenance of Genome Integrity in Plants: Novel Challenges in Basic and Applied Research View all 11 articles
Plant DNA is damaged by exposure to solar radiation, which includes ultraviolet (UV) rays. UV damaged DNA is repaired either by photolyases, using visible light energy, or by nucleotide excision repair (NER), also known as dark repair. NER consists of two subpathways: global genomic repair (GGR), which repairs untranscribed DNA throughout the genome, and transcription-coupled repair (TCR), which repairs transcribed DNA. In mammals, CSA, CSB, UVSSA, USP7, and TFIIS have been implicated in TCR. Arabidopsis homologs of CSA (AtCSA-1/2) and CSB (CHR8) have previously been shown to contribute to UV tolerance. Here we examine the role of Arabidopsis homologs of UVSSA, USP7 (UBP12/13), and TFIIS (RDO2) in UV tolerance. We find that loss of function alleles of UVSSA, UBP12, and RDO2 exhibit increased UV sensitivity in both seedlings and adults. UV sensitivity in atcsa-1, uvssa, and ubp12 mutants is specific to dark conditions, consistent with a role in NER. Interestingly, chr8 mutants exhibit UV sensitivity in both light and dark conditions, suggesting that the Arabidopsis CSB homolog may play a role in both NER and light repair. Overall our results indicate a conserved role for UVSSA, USP7 (UBP12), and TFIIS (RDO2) in TCR.
Unable to move, plants must adapt to their surroundings. An important and unavoidable component of a plant’s environment is solar radiation, which includes both beneficial visible light and damaging ultraviolet (UV) rays. UV radiation harms a variety of cellular components including DNA. UV damaged DNA, primarily pyrimidine photodimers, is repaired by photolyases, using the energy from visible light (light repair), and by nucleotide excision repair (NER) (dark repair) (Pang and Hays, 1991; Molinier, 2017).
Nucleotide excision repair is a conserved multistep pathway involving damage recognition, strand unwinding, excision, repair synthesis, and ligation. Damage recognition is via one of two NER sub-pathways. Global genomic repair (GGR) identifies UV damage in DNA throughout the genome, while transcription coupled repair (TCR) initiates repair of transcribed strands. TCR has been well studied in humans, where deficiencies in this process can result in Cockayne Syndrome and UV-sensitive syndrome (Gregersen and Svejstrup, 2018). UV damaged DNA arrests progression of RNA polymerase II (RNAP), resulting in stabilization of RNAP – Cockayne Syndrome B (CSB) interaction. CSB then recruits the Cockayne Syndrome A (CSA)-DDB1-cullin 4 complex, which ubiquitinates CSB, followed by UV Stimulated Scaffold protein A (UVSSA) and Ubiquitin Specific Peptidase 7 (USP7), which stabilize CSB. Subsequently, core NER components, such as TFIIH and the XPG and XPF endonucleases, are recruited, and resulting in damage excision and repair. Re-initiation of transcription following repair is thought to involve the TFIIS elongation factor (Geijer and Marteijn, 2018).
In plants, UV damage in transcribed strands is preferentially repaired, and this process is regulated by the circadian clock (Fidantsef and Britt, 2012; Oztas et al., 2018). The Arabidopsis homologs of CSA, CSB, USP7, and TFIIS have previously been identified and described. Arabidopsis thaliana has two CSA homologs, AtCSA-1/ CSAat1A (At1g27840) and AtCSA-2/ CSAat1B (At1g19750) (Kunz et al., 2005). Despite the fact that these two proteins are 92% identical, they are both required for tolerance to UV and MMS and repair of transcribed strands. The CSA homologs interact with DDB1A, localize to the nucleus, and form heterotetramers (Biedermann and Hellmann, 2010; Zhang et al., 2010). The Arabidopsis CSB homolog is SWI2/SNF2 protein Chromatin Remodeling 8 (CHR8, At2g18760) (Kunz et al., 2005; Singh et al., 2010). CHR8 RNAi lines result in UV sensitivity, but do not exhibit ionizing radiation or intrachromosomal recombination rate phenotypes, consistent with a role in NER (Shaked et al., 2006). UBP12 (At5g06600) and UBP13 (At3g11910) are the Arabidopsis USP7 homologs and have been implicated in plant immunity, flowering, seed, and root development, as well as jasmonate signaling (Ewan et al., 2011; Cui et al., 2013; Derkacheva et al., 2016; Jeong et al., 2017; An et al., 2018). The Arabidopsis TFIIS homolog is Reduced Dormancy 2 (RDO2, At2g38560), which is required for regulation of seed dormancy by Delay of Germination 1 (DOG1) (Léon-Kloosterziel et al., 1996; Grasser et al., 2009; Liu et al., 2011; Mortensen and Grasser, 2014). RDO2/TFIIS has also been implicated in mRNA processing in plants, including in response to light (Dolata et al., 2015; Antosz et al., 2017; Godoy Herz et al., 2019). In this study we identify the Arabidopsis UVSSA homolog and examine the roles of UVSSA, UBP12/13, and RDO2 in UV tolerance.
Gymnosperm UVSSA homologs were accessed via the PLAZA gymnosperm site1 (Proost et al., 2015) while all other homologs were identified via KEGG (Kyoto Encyclopedia of Genes and Genomes2). UVSSA amino acid sequences were aligned in CLUSTAL Omega (Sievers et al., 2011) using the default settings and saved in NEXUS format for phylogenetic analysis. The aligned amino acid sequences were then analyzed by maximum parsimony as implemented in PAUP∗ version 4.0b8/4.0d78 using the default settings unless otherwise specified (Swofford, 2002). One million maximum parsimony heuristic search replicates were performed with random sequence addition, tree bisection and reconnection branch swapping on only the best trees, multiple trees saved at each step, and retention of all best trees. In addition, 1 million random sequence addition fast addition bootstrap search replicates were performed with retention of all groups consistent with 50% bootstrap consensus.
The following T-DNA alleles were used in this study: SALK_030558 (AtCSA-1) (Lee et al., 2010), SALK_000799 and SAIL_273_G11 (CHR8), SAIL_58_C12 and SALK_061538 (UVSSA), GABI_742C10 (UBP12) (Cui et al., 2013), and SALK_027259 (RDO2) (Grasser et al., 2009; Liu et al., 2011). Col-0 was used as the wild type control for the SALK and GABI lines (Alonso et al., 2003; Kleinboelting et al., 2012), while Col-3 was used as the control for the SAIL lines (Sessions et al., 2002). All plant material was obtained from the Arabidopsis Biological Resource Center (ABRC) (Columbus, OH, United States) or the Nottingham Arabidopsis Stock Centre (NASC) (Nottingham, Loughborough, United Kingdom). Alleles were genotyped with the primers listed in Supplementary Table S1 along with T-DNA specific primers LBb1.3: ATTTTGCCGATTTCGGAAC (SALK lines), LB3SAIL: TAGCATCTGAATTTCATAACCAATCTCGATACAC (SAIL lines), and GK_8409: ATATTGACCATCATACTCATTGC (GABI line). For plant growth, seeds were sterilized and plated on Linsmaier and Skoog (LS) media (Caisson, Smithfield, UT, United States) with 0.6% sucrose and 0.8% Phytoblend (Caisson). After 2–3 days of stratification at 4°C, plates were moved to an incubator with fluorescent bulbs (100 μM photons m-2 s-1) and grown under long day conditions (16 h light/8 h dark) at 20°C and 50% relative humidity. For adult growth, 14 day old plants were transplanted into soil (Sunshine mix no. 1, Sun Gro, Bellevue, WA, United States) and grown under the same conditions.
Ribonucleic acid was extracted from approximately fifty 7-day-old seedlings per genotype with the RNeasy plant mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions including a DNase treatment. RNA was quantified with a Nanodrop spectrophotometer (Thermo Scientific) and 1 μg used to synthesize cDNA, using the Maxima First Strand cDNA synthesis kit (Fermentas, Waltham, MA, United States). For semi-quantitative RT-PCR, CHR8, UVSSA, AtCSA-1, and UBP12 were amplified for 30 cycles and RDO2 for 26 cycles using the primers indicated (Supplementary Table S1) and the Actin loading control amplified for 22 cycles. For quantitative real time PCR, cDNA was diluted 40 fold and PCR performed using SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, United States), a CFX Connect Real time PCR detection system (Bio-Rad), and the primers listed in Supplementary Table S1. EF1α (At5g60390) (Jain et al., 2006; Hossain et al., 2012) was used to normalize sample loading and three technical replicates were analyzed per sample.
The following data was collected from plants transplanted to soil: flowering time (day the first bud is detected), rosette diameter at 4 weeks, number of shoots and silique length at 6 weeks.
Seeds were plated, stratified, and grown vertically in the conditions above for 3 days, then seedlings irradiated with 1000 J m-2 UV-C (corresponding to 65 s exposure to shortwave UV lamp XX-15S, UVP/LLC, Upland, CA, United States). Plates were rotated 90° and incubated in either long day or dark conditions for the indicated number of days, then scanned. Image J was used to measure root and hypocotyl length.
For adult UV assays, 21 day old plants in soil were irradiated with 500 J m-2 UV-C, incubated in the dark for 3 days, then returned to long day conditions. Three days later, individual leaves were scored as either undamaged (green) or damaged (yellow or brown), and % damaged leaves (number of damaged leaves/total leaves) was calculated for all plants.
All experiments were performed at least twice and representative experiments shown. Two-tailed student’s t-tests (p ≤ 0.05) were used to assess statistical significance.
In this study we identify the Arabidopsis UVSSA homolog. Arabidopsis UVSSA (encoded by At3g61800) is 39% and 28% identical to rice and human UVSSA, respectively. Clear UVSSA homologs are found throughout the animal and plant kingdoms including angiosperms, gymnosperms, ferns, and moss. One million maximum parsimony phylogenetic search replicates for UVSSA homolog amino acid sequences recovered a single most parsimonious tree (score 2561) (Figure 1) that is topologically congruent with well supported hypotheses of plant evolutionary history (Morris et al., 2018). Conserved domains in UVSSA proteins include ENTH/VHS in the N terminus and DUF2043 in the C terminus (Figure 2). ENTH/VHS domains are multi-helical with an alpha-alpha 2-layered structural fold, while DUF2043 is an approximately 100 amino acid long UVSSA-specific domain, which includes three conserved cysteines and a CP(y/l)HG motif (Marchler-Bauer et al., 2017). AtUVSSA has a potential bipartite NLS in the C terminus and SUBAcon predicts nuclear localization (score 0.994) (De Castro et al., 2006; Hooper et al., 2014), consistent with a role in DNA repair.
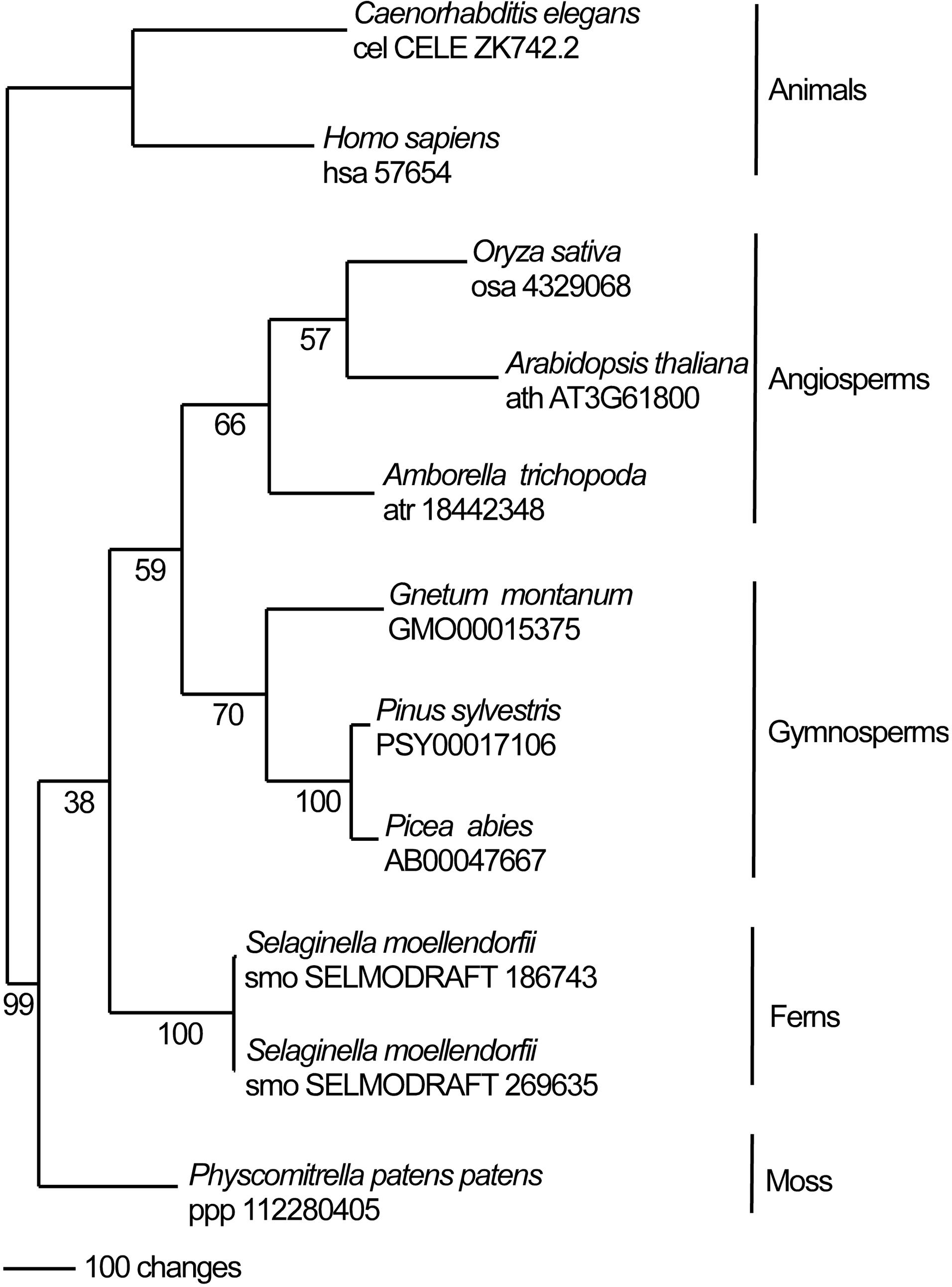
Figure 1. The single most parsimonious phylogenic tree (score 2561) based on analysis of amino acid sequences of UVSSA homologs. Maximum parsimony bootstrap support is indicated under each node. Gymnosperm UVSSA homologs are from the PLAZA gymnosperm site (https://bioinformatics.psb.ugent.be/plaza/versions/gymno-plaza/) (Proost et al., 2015) while all other homologs were accessed via KEGG (Kyoto Encyclopedia of Genes and Genomes) (http://www.kegg.jp/).
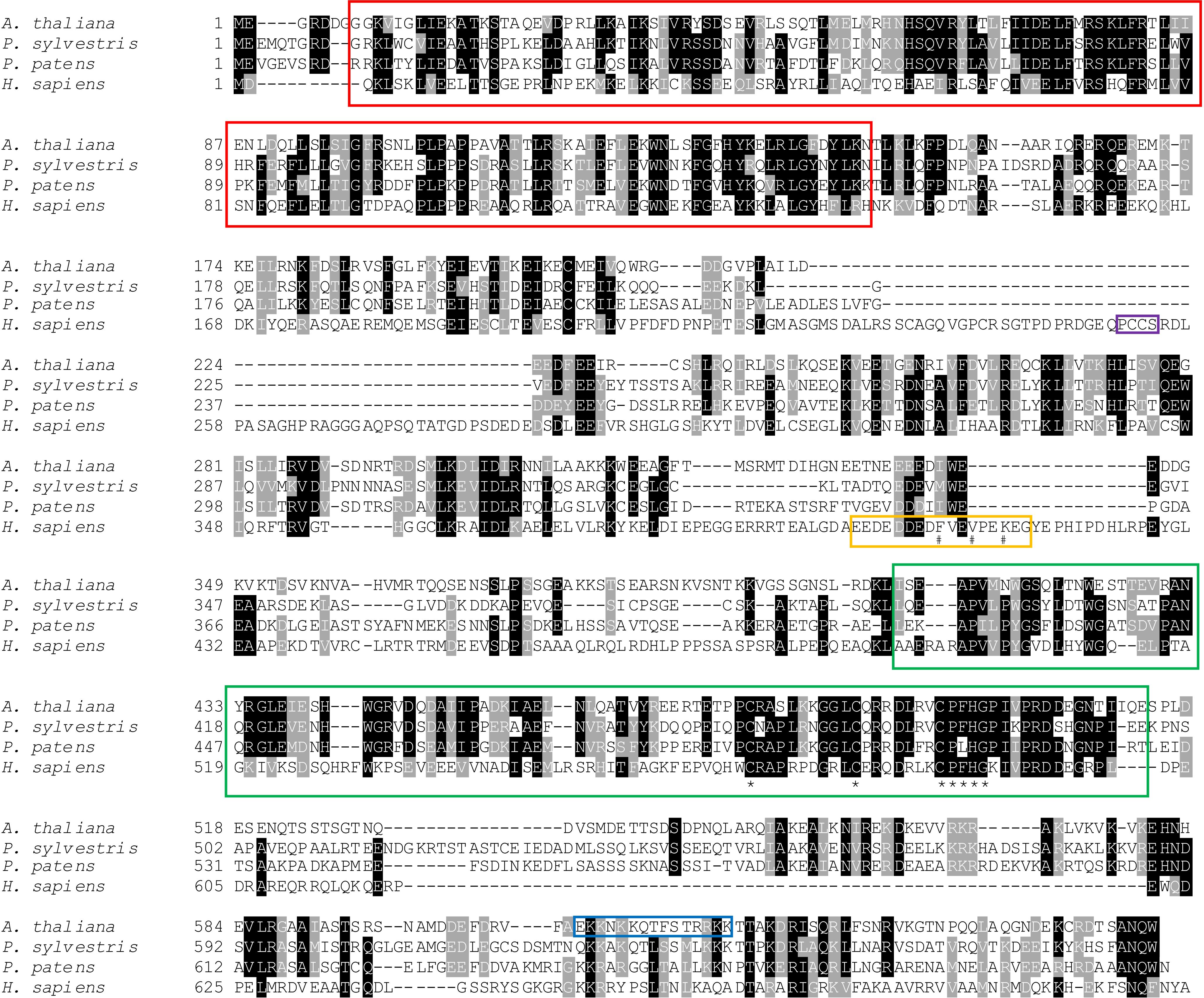
Figure 2. Amino acid alignment of UVSSA from representative angiosperm (Arabidopsis thaliana), gymnosperm (Pinus sylvestris), moss (Physcomitrella patens patens), and animal (Homo sapiens) species. Sequences were aligned using NCBI COBALT (Papadopoulos and Agarwala, 2007) and formatted using Boxshade. Amino acids showing identity (black) and similarity (gray) are indicated. Conserved ENTH/VHS (red) and DUF2043 (green) domains are boxed, with asterisks indicating conserved cysteines and CP(y/l)HG motif in the DUF2043 domain (Marchler-Bauer et al., 2017). The blue box indicates potential bipartite NLS in Arabidopsis UVSSA identified using Prosite (De Castro et al., 2006). Regions required for interaction of human UVSSA with USP7 (purple) and TFIIH (yellow) are shown and key residues indicated with #s (Higa et al., 2016, 2018; Okuda et al., 2017).
Public gene expression data was examined for Arabidopsis UVSSA and the other TCR gene homologs: AtCSA-1, CHR8 (CSB homolog), UBP12 and UBP13 (USP7 homologs), and RDO2 (TFIIS homolog). With respect to absolute levels of expression (Supplementary Figure S1A), UBP12, UBP13, and RDO2 are expressed throughout the plant, consistent with the broad role of these genes in development (Grasser et al., 2009; Cui et al., 2013; Derkacheva et al., 2016), while AtCSA-1, CHR8, and UVSSA are expressed at lower levels (Schmid et al., 2005). With respect to relative levels of expression (Supplementary Figure S1B), CHR8 and UVSSA are enriched in mature pollen, while RDO2, AtCSA-1, CHR8, and UVSSA are up-regulated more than two-fold in dry seed, perhaps contributing to maintenance of seed genome integrity (Waterworth et al., 2015).
Public expression data was also examined to determine the effect of potentially mutagenic stress on expression of these genes. CHR8 was found to be upregulated by genotoxic stress induced by bleomycin and mitomycin C treatment, consistent with previous reports (Molinier et al., 2005), in both the shoot and root, but the other genes were not, while UV-B treatment did not result in major changes to the levels of any of the genes (Supplementary Figure S2; Kilian et al., 2007).
In order to examine the role of these genes in Arabidopsis UV tolerance, T-DNA insertion mutants were obtained. Previously described alleles of AtCSA-1 (SALK_030558) (Lee et al., 2010) and RDO2 (SALK_027259) (Grasser et al., 2009; Liu et al., 2011) were utilized. UBP12 allele GABI_742C10 (ubp12-2) has previously been shown to result in reduced levels of both UBP12 and UBP13, thus acts as a weak double mutant (Cui et al., 2013). In previous studies RNAi lines of CHR8 were shown to exhibit UV sensitivity (Shaked et al., 2006). Here we examine two T-DNA alleles of CHR8, chr8-1 (SALK_000799) and chr8-2 (SAIL_273_G11) (Figure 3A). For UVSSA, two T-DNA alleles were examined, uvssa-1 (SAIL_58_C12), located 38 bp upstream of the start codon, and uvssa-2 (SALK_061538), located in the first intron past the start codon.
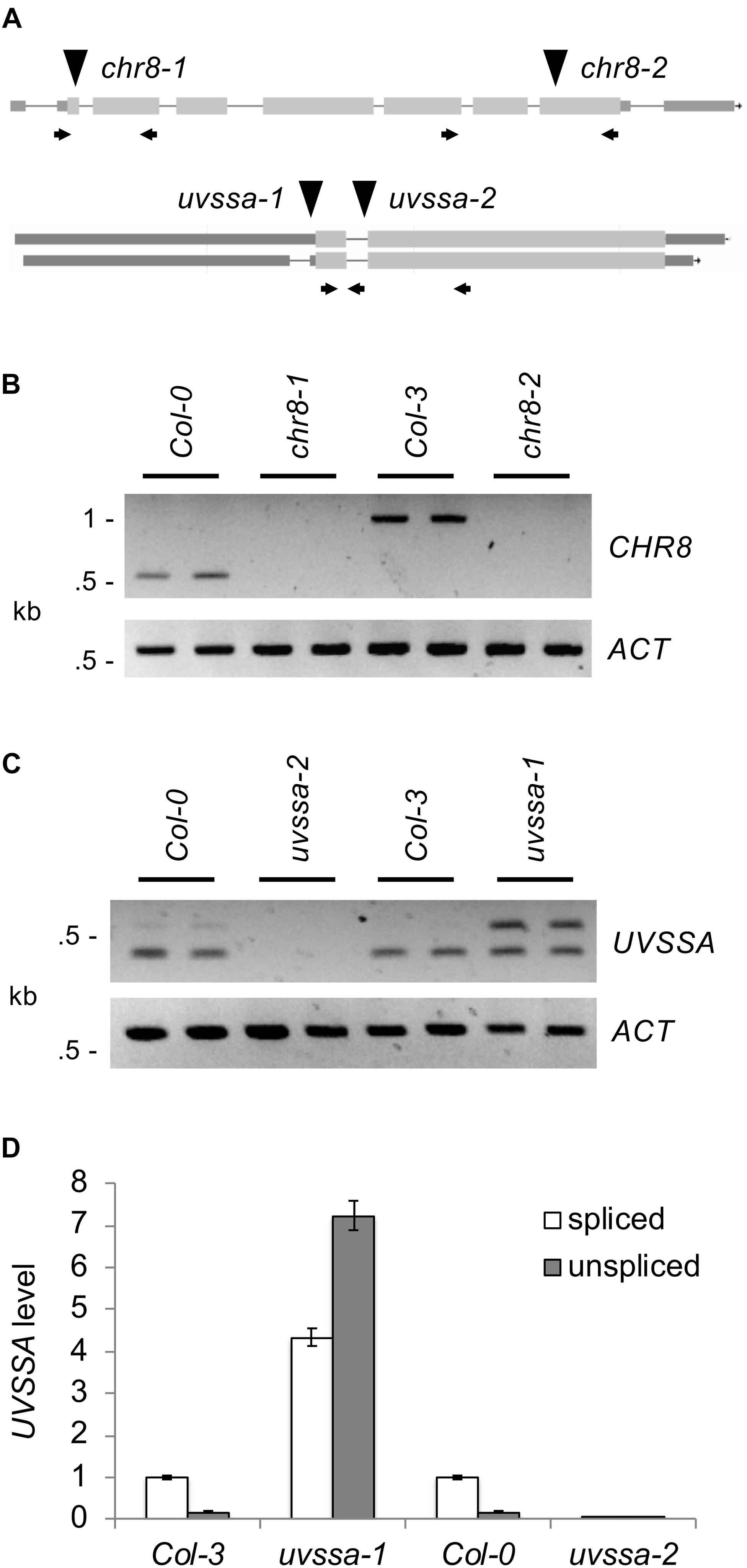
Figure 3. Analysis of CHR8 and UVSSA alleles. (A) Map of CHR8 (At2g18760) and UVSSA (At3g61800) genes. Lines indicate introns and boxes exons, with coding regions shaded light gray and untranslated regions dark gray. For CHR8, transcript At2g18760.4 is shown because it is the best match to light grown seedling RNA-Seq data in JBrowse (Buels et al., 2016). For UVSSA, both known transcripts, At3g61800.1 (above) and At3g61800.2 (below), are shown. The location of T-DNA alleles chr8-1 (SALK_000799), chr8-2 (SAIL_273_G11), uvssa-1 (SAIL_58_C12), and uvssa-2 (SALK_061538) are indicated with triangles. Arrows indicate primers used in RNA analysis. (B) Semi-quantitative RT-PCR analysis of CHR8 and ACTIN expression in chr8-1 and chr8-2 and their respective wild-type backgrounds, using CHR8 primers flanking each allele. (C) Semi-quantitative RT-PCR analysis of UVSSA and ACTIN expression in uvssa-1 and uvssa-2 and their respective wild-type backgrounds, using UVSSA primers in the first and second coding exons. For (B,C), two technical replicates per sample are shown. (D) qPCR analysis of spliced (primers in first coding exon and spanning exon 1–2 junction) and unspliced (primers in first coding exon and following intron) UVSSA level. UVSSA level was normalized using EF1α and expressed as relative to Col-3 spliced product level. Error bars indicate SE of three technical replicates.
We examined the effect of these alleles on gene expression using semi-quantitative RT-PCR. Primers flanking the chr8-1 and chr8-2 insertion sites detected no CHR8 transcript, indicating these are null alleles (Figure 3B). Semi-quantitative RT-PCR with T-DNA insertion flanking primers also confirmed loss of transcript in the atcsa-1, ubp12, and rdo2 lines (Supplementary Figure S3). For UVSSA, we utilized primers in the first and second coding exons, since the effect of T-DNA insertion on coding sequences was our primary concern. uvssa-2 results in a null allele, but in uvssa-1 both the predicted band and a larger band were detected (Figure 3C). The size of the larger band was consistent with that of the unspliced transcript, so we hypothesized that uvssa-1 insertion affected intron splicing [note the uvssa-1 samples did not result in larger gDNA-size bands of CHR8, thus were not gDNA contaminated (data not shown)]. Real-time qPCR with an intron-specific primer was used to quantify the effect of the uvssa-1 allele on splicing, and large amounts of the unspliced product were detected (Figure 3D). Due to the presence of an in frame stop codon in the intron, this transcript results in a truncated 77 amino acid product. uvssa-1 also resulted in increased levels of correctly spliced UVSSA. Thus uvssa-1 would be predicted to result in increased levels of both full length and truncated UVSSA.
Mutant alleles of the TCR genes were grown in long day conditions with their respective controls and their developmental phenotypes examined. ubp12-2 mutants exhibited decreased rosette size, early flowering (days), and decreased apical dominance (increased number of shoots) (Supplementary Figure S4), consistent with previously described phenotypes (Cui et al., 2013; Derkacheva et al., 2016). The other mutant alleles did not exhibit any developmental phenotypes with the exception of a slight increase in apical dominance in chr8-2. RDO2 mutants have been described as early flowering (Grasser et al., 2009), however, additional analysis indicates this phenotype is observed with respect to number of leaves, rather than number of days, at flowering (Mortensen and Grasser, 2014), consistent with our results.
The UV tolerance of the mutant alleles of the TCR genes was then assessed. Since TCR is a sub-pathway of NER, or dark repair, we assessed UV tolerance in seedlings following dark incubation after UV treatment. As previously described (Shaked et al., 2006; Biedermann and Hellmann, 2010; Zhang et al., 2010), AtCSA-1 and CHR8 (CSB) loss of function resulted in increased UV sensitivity in the dark (Figures 4A–C and Supplementary Figures S5A,B). The UVSSA loss of function allele, uvssa-2, also resulted in increased UV sensitivity in the dark (Figure 4E). The uvssa-1 allele, which results in increased levels of both truncated and full length UVSSA, did not exhibit either increased or decreased UV tolerance following 2 or 3 days of dark incubation (Figure 4D and Supplementary Figure S5C). ubp12-2 also exhibited increased UV sensitivity in the dark (Figure 4F and Supplementary Figure S5D). rdo2 exhibited increased dark UV sensitivity in hypocotyls (but not roots) after 2 days of incubation, but not after 3 days (Figure 4G and Supplementary Figure S5E). We also examined UV sensitivity in adult plants following dark incubation and found that, as in seedlings, atcsa-1, chr8, uvssa-2, ubp12, and rdo2 mutants exhibit UV sensitivity, while uvssa-1 does not (Figure 5).
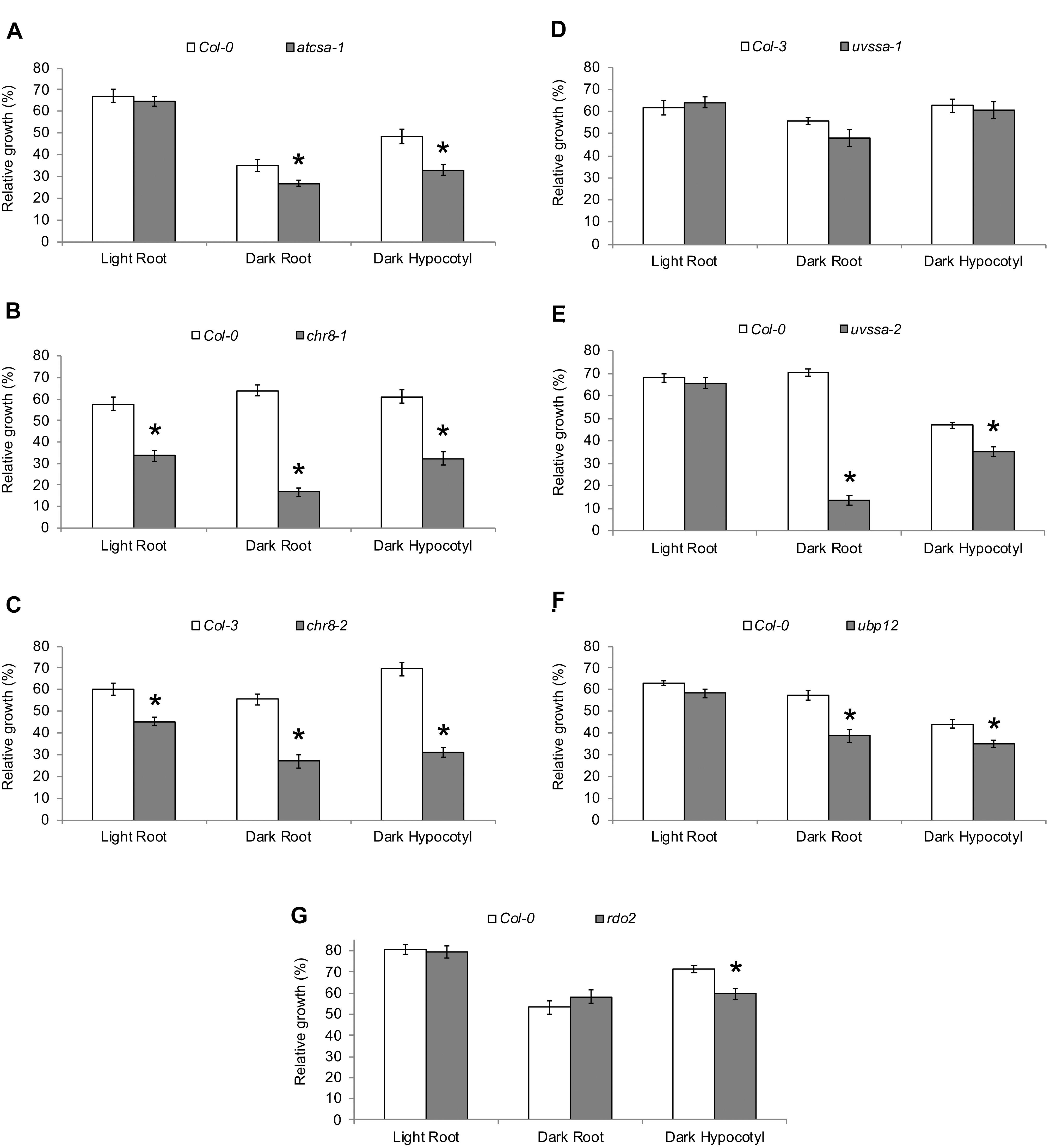
Figure 4. UV tolerance of mutants in TCR genes. Relative growth of roots and hypocotyls of (A) atcsa-1, (B) chr8-1, (C) chr8-2, (D) uvssa-1, (E) uvssa-2, (F) ubp12, and (G) rdo2 after 1000 J m-2 UV treatment, followed by 2 days of long-day (light) or dark incubation. Data are expressed as length relative to unirradiated control of the same genotype. Values are means ± SE (n = 20), ∗p ≤ 0.05 of mutants vs wild type.
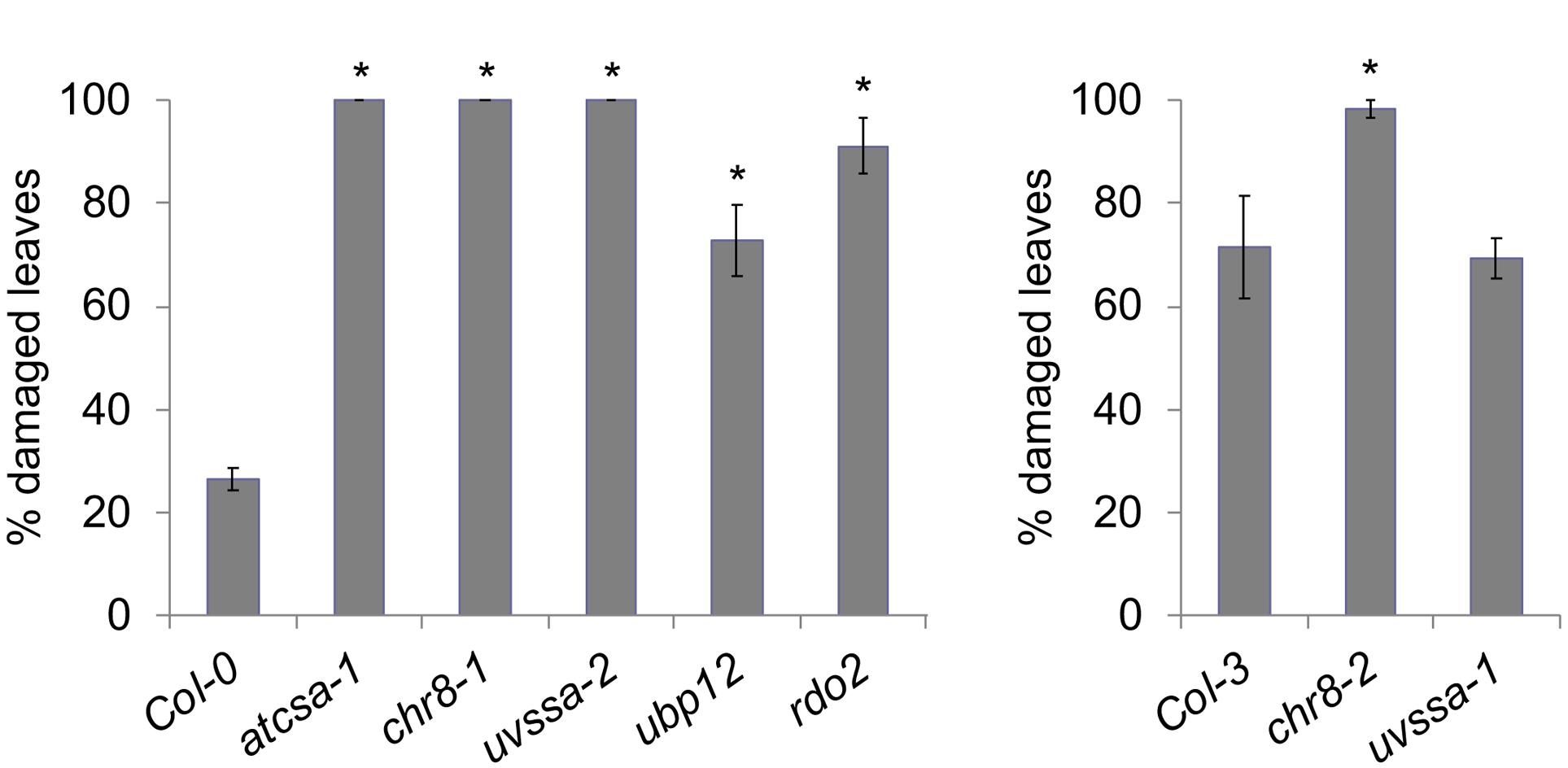
Figure 5. UV tolerance in adult plants. Percentage damaged leaves after 500 J m-2 UV treatment, followed by 3 days of dark incubation. Values are means ± SE (n = 6), ∗p ≤ 0.05 of mutants vs respective wild type.
To examine the specificity of the UV sensitivity of these alleles, they were also incubated in light (long day) following UV treatment. atcsa-1, uvssa-2, and ubp12 were not UV sensitive in the light (Figures 4A,E,F), consistent with the dark specific role of NER. Surprisingly, both chr8 alleles displayed UV sensitivity following light incubation (Figures 4B,C), exhibiting the expected dose dependence, with the more severely truncated chr8-1 allele demonstrating a stronger root phenotype in both light and dark. This result suggests that CHR8 plays a role in light repair, distinct from the other components of the TCR pathway.
In this study, we examined the UV sensitivity of mutant alleles of Arabidopsis homologs of genes implicated in mammalian TCR. As previously reported, we find atcsa-1 mutants exhibit increased dark specific UV sensitivity (Biedermann and Hellmann, 2010). Our atcsa-1 dark root phenotype is not as strong as that of mutants in other TCR components such as CSB/CHR8 and UVSSA, this may be due to redundancy with AtCSA-2/CSAat1B.
The Arabidopsis homolog of mammalian CSB [also known as Excision Repair Cross-Complementing 6 (ERCC6)] and yeast Rad26 is CHR8 (Kunz et al., 2005; Singh et al., 2010). In this study, we utilized CHR8 T-DNA lines and observed increased UV sensitivity following dark incubation, consistent with previous studies using CHR8 RNAi lines (Shaked et al., 2006). Also, unique among the TCR mutants we examined, chr8 alleles exhibited increased UV sensitivity following light incubation. Mammalian CSB has been implicated in regulation of transcription and base excision repair in addition to TCR (Stevnsner et al., 2008; Boetefuer et al., 2018), so one of these roles may contribute to the chr8 light UV sensitivity phenotype.
In humans, mutation of UVSSA results in defective TCR and UV sensitive syndrome (Cleaver, 2012). Loss of the C. elegans UVSSA homolog also results in increased UV sensitivity (Babu and Schumacher, 2016). While UVSSA is conserved throughout the animal kingdom (Nakazawa et al., 2012), it is absent from Drosophila. However, Drosophila also lack CSA and CSB homologs, and do not appear to perform TCR (Sekelsky, 2017). Yeast also lack UVSSA, although both S. cerevisiae and S. pombe have CSB homologs and perform TCR (Li and Li, 2017; Xu et al., 2017). Here we show that UVSSA is found throughout the plant kingdom, with conserved ENTH/VHS and DUF2043 domains. Recently, the region corresponding to amino acid 400–415 of human UVSSA was found to be well conserved in animals and required for TFIIH interaction (Okuda et al., 2017). Although this region is still acidic in plants, it not well conserved with human UVSSA and plants lack F408 and V411, which are required for TFIIH interaction and TCR in humans (Okuda et al., 2017), as well as K414, which is mono-ubiquitinated (Higa et al., 2018). In addition, residues 251–254 of human UVSSA have been shown to be required for USP7 interaction, CSB stability, and TCR (Higa et al., 2016), yet this sequence is also not conserved in plants. Nonetheless our data show that lack of UVSSA results in dark specific UV sensitivity in Arabidopsis, consistent with a role in NER.
Arabidopsis USP7 homologs UBP12 and UBP13, like other ubiquitin specific proteases, play important roles in plant development and environmental response (Zhou et al., 2017). UBP12/13 interact with LHP1 and deubiquitinate RGFR1 and MYC2 (Derkacheva et al., 2016; Jeong et al., 2017; An et al., 2018). Our results here indicate that UBP12 (and UBP13) are involved in UV tolerance, suggesting they may also deubiquitinate UVSSA and CSB, as has been proposed for mammalian USP7 (Geijer and Marteijn, 2018). UBP12 and UBP13 act redundantly, and double null alleles are inviable due to pollen defects (Ewan et al., 2011; Derkacheva et al., 2016). Here we use an allele of UBP12, ubp12-2, which also results in a partial decrease in UBP13 level, and resulting in a weak double mutant (Cui et al., 2013). However, because this is a weak (non-null) double mutant, we may be underestimating the role of UBP12/13 in UV tolerance.
In mammals, in addition to acting during transcript elongation, TFIIS has been shown to facilitate transcription re-initiation following RNAP arrest, and is recruited to the stalled polymerase in a CSB and CSA dependent manner (Donahue et al., 1994; Kalogeraki et al., 2005; Fousteri et al., 2006; Dutta et al., 2015). In yeast, loss of TFIIS only results in increased UV sensitivity in a GGR-deficient background, however, the same is true of CSB homolog Rad26 (Wong and Ingles, 2001). In mammals, reduction of TFIIS resulted in reduced RNA synthesis recovery, but had no effect on UV sensitivity (Jensen and Mullenders, 2010). In this study we detected a UV sensitive phenotype in TFIIS deficient Arabidopsis (rdo2), however, it was milder than observed for the other TCR mutants and not detectable 3 days after seedling UV treatment. Interestingly, the UV sensitive phenotype of both rdo2 and atcsa-1 was stronger in hypocotyls than in roots, at 2 days than at 3 days, and in adults than in seedlings, suggesting the role of these genes in UV tolerance may vary with tissue, time, and phenotype assessed (growth versus tissue death).
In this study, we have identified the Arabidopsis UVSSA homolog and shown that Arabidopsis UVSSA, USP7 (UBP12/13), and TFIIS (RDO2) homologs contribute to UV tolerance, along with CSA and CSB (CHR8) homologs, suggesting conservation in the mechanisms of TCR.
All datasets for this study are included in the manuscript and the Supplementary Files.
WAK, AS, and DS performed the experiments. JM conducted the phylogenetic analysis. DS wrote the first draft of the manuscript. All authors contributed to revised manuscript and approved the final version, designed the experiments and analyzed the data.
This study was supported by the Natural Sciences and Engineering Research Council of Canada.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We are grateful to Janelle Lazaro and Linda Al Rayes for technical assistance and Triparna Lahari for helpful discussion. Arabidopsis seeds were obtained from the Arabidopsis Biological Resource Center and the Nottingham Arabidopsis Stock Centre.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00516/full#supplementary-material
CHR8, chromatin remodeling 8; CSA/B, Cockayne syndrome A/B; GGR, global genomic repair; NER, nucleotide excision repair; RDO2, reduced dormancy 2; RNAP, RNA polymerase II; TCR, transcription coupled repair; TFIIS, transcription elongation factor IIS; UBP and USP, ubiquitin specific protease; UV, ultraviolet irradiation; UVSSA, UV stimulated scaffold protein A.
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
An, Z., Liu, Y., Ou, Y., Li, J., Zhang, B., Sun, D., et al. (2018). Regulation of the stability of RGF1 receptor by the ubiquitin-specific proteases UBP12/UBP13 is critical for root meristem maintenance. Proc. Natl. Acad. Sci. U.S.A. 115, 1123–1128. doi: 10.1073/pnas.1714177115
Antosz, W., Pfab, A., Ehrnsberger, H. F., Holzinger, P., Köllen, K., Mortensen, S. A., et al. (2017). The composition of the Arabidopsis RNA Polymerase II transcript elongation complex reveals the interplay between elongation and mRNA processing factors. Plant Cell 29, 854–870. doi: 10.1105/tpc.16.00735
Babu, V., and Schumacher, B. (2016). A C. elegans homolog for the UV-hypersensitivity syndrome disease gene UVSSA. DNA Repair 41, 8–15. doi: 10.1016/j.dnarep.2016.03.008
Biedermann, S., and Hellmann, H. (2010). The DDB1a interacting proteins ATCSA-1 and DDB2 are critical factors for UV-B tolerance and genomic integrity in Arabidopsis thaliana. Plant J. 62, 404–415. doi: 10.1111/j.1365-313X.2010.04157.x
Boetefuer, E. L., Lake, R. J., and Fan, H. Y. (2018). Mechanistic insights into the regulation of transcription and transcription-coupled DNA repair by Cockayne syndrome protein B. Nucleic Acids Res. 46, 7471–7479. doi: 10.1093/nar/gky660
Buels, R., Yao, E., Diesh, C. M., Hayes, R. D., Munoz-Torres, M., Helt, G., et al. (2016). JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 17:66. doi: 10.1186/s13059-016-0924-1
Cleaver, J. E. (2012). Photosensitivity syndrome brings to light a new transcription-coupled DNA repair cofactor. Nat. Genet. 44, 477–478. doi: 10.1038/ng.2255
Cui, X., Lu, F., Li, Y., Xue, Y., Kang, Y., Zhang, S., et al. (2013). Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol. 162, 897–906. doi: 10.1104/pp.112.213009
De Castro, E., Sigrist, C. J. A., Gattiker, A., Bulliard, V., Langendijk-Genevaux, P. S., Gasteiger, E., et al. (2006). ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365.
Derkacheva, M., Liu, S., Figueiredo, D. D., Gentry, M., Mozgova, I., Nanni, P., et al. (2016). H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat. Plants 2:16126. doi: 10.1038/nplants.2016.126
Dolata, J., Guo, Y., Kołowerzo, A., Smoliñski, D., Brzyżek, G., Jarmołowski, A., et al. (2015). NTR1 is required for transcription elongation checkpoints at alternative exons in Arabidopsis. EMBO J. 34, 544–558. doi: 10.15252/embj.201489478
Donahue, B. A., Yin, S., Taylor, J. S., Reines, D., and Hanawalt, P. C. (1994). Transcript cleavage by RNA polymerase II arrested by a cyclobutane pyrimidine dimer in the DNA template. Proc. Natl. Acad. Sci. U.S.A. 91, 8502–8506. doi: 10.1073/pnas.91.18.8502
Dutta, A., Babbarwal, V., Fu, J., Brunke-Reese, D., Libert, D. M., Willis, J., et al. (2015). Ccr4-Not and TFIIS function cooperatively to rescue arrested RNA Polymerase II. Mol. Cell. Biol. 35, 1915–1925. doi: 10.1128/MCB.00044-15
Ewan, R., Pangestuti, R., Thornber, S., Craig, A., Carr, C., O’Donnell, L., et al. (2011). Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol. 191, 92–106. doi: 10.1111/j.1469-8137.2011.03672.x
Fidantsef, A. L., and Britt, A. B. (2012). Preferential repair of the transcribed DNA strand in plants. Front. Plant Sci. 2:105. doi: 10.3389/fpls.2011.00105
Fousteri, M., Vermeulen, W., van Zeeland, A. A., and Mullenders, L. H. (2006). Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23, 471–482. doi: 10.1016/j.molcel.2006.06.029
Geijer, M. E., and Marteijn, J. A. (2018). What happens at the lesion does not stay at the lesion: transcription-coupled nucleotide excision repair and the effects of DNA damage on transcription in cis and trans. DNA Repair 71, 56–68. doi: 10.1016/j.dnarep.2018.08.007
Godoy Herz, M. A., Kubaczka, M. G., Brzyżek, G., Servi, L., Krzyszton, M., Simpson, C., et al. (2019). Light regulates plant alternative splicing through the control of transcriptional elongation. Mol. Cell 73, 1066.e–1074.e. doi: 10.1016/j.molcel.2018.12.005
Grasser, M., Kane, C. M., Merkle, T., Melzer, M., Emmersen, J., and Grasser, K. D. (2009). Transcript elongation factor TFIIS is involved in Arabidopsis seed dormancy. J. Mol. Biol. 386, 598–611. doi: 10.1016/j.jmb.2008.12.066
Gregersen, L. H., and Svejstrup, J. Q. (2018). The cellular response to transcription-blocking DNA damage. Trends Biochem. Sci. 43, 327–341. doi: 10.1016/j.tibs.2018.02.010
Higa, M., Tanaka, K., and Saijo, M. (2018). Inhibition of UVSSA ubiquitination suppresses transcription-coupled nucleotide excision repair deficiency caused by dissociation from USP7. FEBS J. 285, 965–976. doi: 10.1111/febs.14382
Higa, M., Zhang, X., Tanaka, K., and Saijo, M. (2016). Stabilization of ultraviolet (UV)-stimulated scaffold protein A by interaction with ubiquitin-specific peptidase 7 Is essential for transcription-coupled nucleotide excision repair. J. Biol. Chem. 291, 13771–13779. doi: 10.1074/jbc.M116.724658
Hooper, C. M., Tanz, S. K., Castleden, I. R., Vacher, M. A., Small, I. D., and Millar, A. H. (2014). SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30, 3356–3364. doi: 10.1093/bioinformatics/btu550
Hossain, Z., Amyot, L., McGarvey, B., Gruber, M., Jung, J., and Hannoufa, A. (2012). The translation elongation factor eEF-1Bβ1 is involved in cell wall biosynthesis and plant development in Arabidopsis thaliana. PLoS One 7:e30425. doi: 10.1371/journal.pone.0030425
Jain, M., Nijhawan, A., Tyagi, A. K., and Khurana, J. P. (2006). Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 345, 646–651. doi: 10.1016/j.bbrc.2006.04.140
Jensen, A., and Mullenders, L. H. (2010). Transcription factor IIS impacts UV-inhibited transcription. DNA Repair 9, 1142–1150. doi: 10.1016/j.dnarep.2010.08.002
Jeong, J. S., Jung, C., Seo, J. S., Kim, J. K., and Chua, N. H. (2017). The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29, 1406–1424. doi: 10.1105/tpc.17.00216
Kalogeraki, V. S., Tornaletti, S., Cooper, P. K., and Hanawalt, P. C. (2005). Comparative TFIIS-mediated transcript cleavage by mammalian RNA polymerase II arrested at a lesion in different transcription systems. DNA Repair 4, 1075–1087. doi: 10.1016/j.dnarep.2005.05.007
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313x.2007.03052.x
Kleinboelting, N., Huep, G., Kloetgen, A., Viehoever, P., and Weisshaar, B. (2012). GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 40, D1211–D1215. doi: 10.1093/nar/gkr1047
Kunz, B. A., Anderson, H. J., Osmond, M. J., and Vonarx, E. J. (2005). Components of nucleotide excision repair and DNA damage tolerance in Arabidopsis thaliana. Environ. Mol. Mutagen. 45, 115–127.
Lee, J. H., Yoon, H. J., Terzaghi, W., Martinez, C., Dai, M., Li, J., et al. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22, 1716–1732. doi: 10.1105/tpc.109.073783
Léon-Kloosterziel, K. M., van de Bunt, G. A., Zeevaart, J. A., and Koornneef, M. (1996). Arabidopsis mutants with a reduced seed dormancy. Plant Physiol. 110, 233–240. doi: 10.1104/pp.110.1.233
Li, W., and Li, S. (2017). Facilitators and repressors of transcription-coupled DNA repair in Saccharomyces cerevisiae. Photochem. Photobiol. 93, 259–267. doi: 10.1111/php.12655
Liu, Y., Geyer, R., van Zanten, M., Carles, A., Li, Y., Hörold, A., et al. (2011). Identification of the Arabidopsis REDUCED DORMANCY 2 gene uncovers a role for the polymerase associated factor 1 complex in seed dormancy. PLoS One 6:e22241. doi: 10.1371/journal.pone.0022241
Marchler-Bauer, A., Bo, Y., Han, L., He, J., Lanczycki, C. J., Lu, S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45, D200–D203. doi: 10.1093/nar/gkw1129
Molinier, J. (2017). Genome and epigenome surveillance processes underlying UV exposure in plants. Genes 8:e316. doi: 10.3390/genes8110316
Molinier, J., Oakeley, E. J., Niederhauser, O., Kovalchuk, I., and Hohn, B. (2005). Dynamic response of plant genome to ultraviolet radiation and other genotoxic stresses. Mutat. Res. 571, 235–247. doi: 10.1016/j.mrfmmm.2004.09.016
Morris, J. L., Puttick, M. N., Clark, J. W., Edwards, D., Kenrick, P., Pressel, S., et al. (2018). The timescale of early land plant evolution. Proc. Natl. Acad. Sci. U.S.A. 115, E2274–E2283. doi: 10.1073/pnas.1719588115
Mortensen, S. A., and Grasser, K. D. (2014). The seed dormancy defect of Arabidopsis mutants lacking the transcript elongation factor TFIIS is caused by reduced expression of the DOG1 gene. FEBS Lett. 588, 47–51. doi: 10.1016/j.febslet.2013.10.047
Nakazawa, Y., Sasaki, K., Mitsutake, N., Matsuse, M., Shimada, M., Nardo, T., et al. (2012). Mutations in UVSSA cause UV-sensitive syndrome and impair RNA polymerase IIo processing in transcription-coupled nucleotide-excision repair. Nat. Genet. 44, 586–592. doi: 10.1038/ng.2229
Okuda, M., Nakazawa, Y., Guo, C., Ogi, T., and Nishimura, Y. (2017). Common TFIIH recruitment mechanism in global genome and transcription-coupled repair subpathways. Nucleic Acids Res. 45, 13043–13055. doi: 10.1093/nar/gkx970
Oztas, O., Selby, C. P., Sancar, A., and Adebali, O. (2018). Genome-wide excision repair in Arabidopsis is coupled to transcription and reflects circadian gene expression patterns. Nat. Commun. 9:1503. doi: 10.1038/s41467-018-03922-5
Pang, Q., and Hays, J. B. (1991). UV-B-Inducible and temperature-sensitive photoreactivation of cyclobutane pyrimidine dimers in Arabidopsis thaliana. Plant Physiol. 95, 536–543. doi: 10.1104/pp.95.2.536
Papadopoulos, J. S., and Agarwala, R. (2007). COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics 23, 1073–1079. doi: 10.1093/bioinformatics/btm076
Proost, S., Van Bel, M., Vaneechoutte, D., Van de Peer, Y., Inzé, D., Mueller-Roeber, B., et al. (2015). PLAZA 3.0: an access point for plant comparative genomics. Nucleic Acids Res. 43, D974–D981. doi: 10.1093/nar/gku986
Schmid, M., Davison, T. S., Henz, S. R., Pape, U. J., Demar, M., Vingron, M., et al. (2005). A gene expression map of Arabidopsis thaliana development. Nat. Genet. 37, 501–506.
Sekelsky, J. (2017). DNA repair in Drosophila: mutagens, models, and missing genes. Genetics 205, 471–490. doi: 10.1534/genetics.116.186759
Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. doi: 10.1105/tpc.004630
Shaked, H., Avivi-Ragolsky, N., and Levy, A. A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994. doi: 10.1534/genetics.105.051664
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Singh, S. K., Roy, S., Choudhury, S. R., and Sengupta, D. N. (2010). DNA repair and recombination in higher plants: insights from comparative genomics of Arabidopsis and rice. BMC Genomics 11:443. doi: 10.1186/1471-2164-11-443
Stevnsner, T., Muftuoglu, M., Aamann, M. D., and Bohr, V. A. (2008). The role of cockayne syndrome group B (CSB) protein in base excision repair and aging. Mech. Age. Dev. 129, 441–448. doi: 10.1016/j.mad.2008.04.009
Swofford, D. L. (2002). PAUP∗. Phylogenetic Analysis Using Parsimony (∗and Other Methods). Sunderland, MA: Sinauer Associates. doi: 10.1016/j.mad.2008.04.009
Waterworth, W. M., Bray, C. M., and West, C. E. (2015). The importance of safeguarding genome integrity in germination and seed longevity. J. Exp. Bot. 66, 3549–3558. doi: 10.1093/jxb/erv080
Wong, J. M., and Ingles, C. J. (2001). A compromised yeast RNA polymerase II enhances UV sensitivity in the absence of global genome nucleotide excision repair. Mol. Gen. Genet. 264, 842–851. doi: 10.1007/s004380000374
Xu, J., Lahiri, I., Wang, W., Wier, A., Cianfrocco, M. A., Chong, J., et al. (2017). Structural basis for the initiation of eukaryotic transcription-coupled DNA repair. Nature 551, 653–657. doi: 10.1038/nature24658
Zhang, C., Guo, H., Zhang, J., Guo, G., Schumaker, K. S., and Guo, Y. (2010). Arabidopsis cockayne syndrome A-like proteins 1A and 1B form a complex with CULLIN4 and damage DNA binding protein 1A and regulate the response to UV irradiation. Plant Cell 22, 2353–2369. doi: 10.1105/tpc.110.073973
Keywords: Arabidopsis, transcription coupled repair, UV, CSA, CSB, UVSSA, UBP7, TFIIS
Citation: Al Khateeb WM, Sher AA, Marcus JM and Schroeder DF (2019) UVSSA, UBP12, and RDO2/TFIIS Contribute to Arabidopsis UV Tolerance. Front. Plant Sci. 10:516. doi: 10.3389/fpls.2019.00516
Received: 26 February 2019; Accepted: 03 April 2019;
Published: 24 April 2019.
Edited by:
Alma Balestrazzi, University of Pavia, ItalyReviewed by:
Michael G. Kemp, Wright State University, United StatesCopyright © 2019 Al Khateeb, Sher, Marcus and Schroeder. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dana F. Schroeder, RGFuYS5TY2hyb2VkZXJAdW1hbml0b2JhLmNh
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.