
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 11 April 2019
Sec. Plant Abiotic Stress
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00468
This article is part of the Research Topic Construction of Regulatory Networks in Plant Abiotic Stress Responses View all 4 articles
Autophagy, a highly conserved process in eukaryotes that involves vacuolar degradation of intracellular components and decomposition of damaged or toxic constituents, is induced by endogenous reactive oxygen species (ROS) accumulation, endoplasmic reticulum stress, and other factors. In plants, the role of autophagy in the induction of programmed cell death (PCD) is still unclear. Here, we show that ROS contribute to the regulation of PCD during waterlogging (which results in oxygen depletion) via autophagy. In wild-type roots, waterlogging induces the transcription of hypoxia-responsive genes and respiratory burst oxidase homolog (RBOH)-mediated ROS production. It also altered the transcription level of alternative oxidase1a and the activity level of antioxidant enzymes. Moreover, waterlogging increased the transcription levels of autophagy-related (ATG) genes and the number of autophagosomes. Autophagy first occurred in the root stele, and then autophagosomes appeared at other locations in the root. In rboh mutants, upregulation of autophagosomes was less pronounced than in the wild type upon waterlogging. However, the accumulation of ROS and the level of cell death in the roots of atg mutants were higher than those in the wild type after waterlogging. In conclusion, our results suggest that autophagy induced in Arabidopsis roots during waterlogging has an attenuating effect on PCD in the roots.
HIGHLIGHTS
– This study examined the impact of waterlogging-induced autophagy on programmed cell death in Arabidopsis roots.
Flooding, which results in soil waterlogging, and in many situations complete submergence, is a major issue for plant survival in many regions of the world. When waterlogging occurs, restricted gas exchange between the soil and atmosphere and altered respiratory processes of soil organisms contribute to hypoxia and anoxia (Mühlenbock et al., 2007). Under hypoxic or anoxic conditions, ethanol produced by anaerobic respiration and reactive oxygen species (ROS) leakage from mitochondria affect root growth and function. To cope with a shortage of oxygen, some gramineous plants form lysigenous aerenchyma in their roots as a consequence of cell death of cortical cells (Jackson and Armstrong, 1999; Evans, 2003).
Previous studies of Arabidopsis thaliana have shown that the expression of genes encoding enzymes related to glucose metabolism, glycolysis, and fermentation increases in response to hypoxia (Liu F. et al., 2005; Loreti et al., 2005; van Dongen et al., 2009). Under hypoxic conditions, the transition of plants from aerobic respiration to lactic acid fermentation increases the level of expression of the lactate dehydrogenase (LDH) gene (Kreuzwieser et al., 2009). Because the accumulation of lactic acid leads to the acidification of the cytoplasm, the activity of LDH is reduced, allowing alcohol fermentation (Kreuzwieser and Rennenberg, 2014). The activities of enzymes required for alcohol fermentation, which include pyruvate decarboxylase (PDC) and alcohol dehydrogenase (ADH), are higher in waterlogged roots (Atkinson et al., 2008). Sucrose synthase (SUS), which participates in sucrose metabolism, is of great importance to surviving hypoxia (Santaniello et al., 2014). A specific subfamily of transcription factors, Group VII ethylene response factors (ERF-VIIs), plays important roles in regulating the activation of fermentation-related genes (Bailey-Serres and Voesenek, 2008). In plants, ERF-VII genes have been found to play a role in the oxygen-sensing mechanism through the N-end rule pathway of proteolysis (Gibbs et al., 2011; Licausi et al., 2011). Under hypoxia, ERF-VIIs are released and activate the expression of hypoxia-responsive genes, including the fermentative genes PDC1 and ADH (Gasch et al., 2016).
Well-known ROS molecules in plants include ozone, singlet oxygen, the superoxide anion, hydrogen peroxide (H2O2), and hydroxyl radicals. ROS signaling, especially that of H2O2, plays a key role in plant adaptation to low-oxygen conditions (Dat et al., 2004; Bailey-Serres and Voesenek, 2008; Sauter, 2013; Pucciariello and Perata, 2017). H2O2 is produced by the double electron reduction of molecular oxygen catalyzed by the respiratory burst oxidase homologs (RBOHs), which is a plant homolog of mammalian nicotinamide adenine dinucleotide phosphate (NADPH) oxidase in the plasma membrane (Torres et al., 1998). The Arabidopsis genome has 10 RBOH genes, and each homolog has a specific role in different biological processes (Foreman et al., 2003). The expression of the RBOH gene contributes to the production and signaling of RBOH-dependent ROS in the immune response of A. thaliana (Morales et al., 2016). However, the functions of RBOHs under hypoxic or anoxic conditions are poorly understood.
In the plant mitochondrial electron transport chain (ETC), there are two terminal oxidases, cytochrome oxidase and alternative oxidase (AOX). AOX is an integral protein of the inner mitochondrial membrane and catalyses the alternative respiratory pathway (Feng et al., 2013). Under environmental stresses, electrons produced by the respiratory oxidation of NADPH can flow through the alternative respiratory pathway instead of the usual cytochrome respiratory pathway in higher plants. This can limit the excessive production of ROS and nitric oxide and maintain the redox balance of plant cells (Maxwell et al., 1999; Giraud et al., 2008; Smith et al., 2009; Cvetkovska and Vanlerberghe, 2012). Enzymatic mechanisms to scavenge overproduced ROS in plants include superoxide dismutase (SOD), ascorbate peroxidase (APX), and catalase (CAT). The main function of SOD is dismutation of superoxide to H2O2, which is detoxified to H2O and O2 by APX and CAT (Apel and Hirt, 2004).
Autophagy is a highly conserved degradation process that transfers intracellular components to vacuoles or lysosomes under environmental stress or at certain developmental stages (Klionsky and Ohsumi, 1999; Klionsky, 2005, 2007). In plants, two major autophagic pathways have been described, microautophagy and macroautophagy (hereafter, autophagy) (Bassham et al., 2006). During autophagy, damaged or toxic constituents are engulfed in a double membrane vesicle called an autophagosome, which fuses with the vacuole to deliver the autophagic body into the vacuolar lumen for further break-down (Li and Vierstra, 2012; Liu and Bassham, 2012). Autophagy is active at very low levels during plant growth and development but is highly inducible in response to a variety of abiotic stresses, such as oxidative stress, salt stress, osmotic stress, and heat stress (Zhou et al., 2014). Autophagy-defective (atg) mutants or transgenic plants are more hypersensitive to ROS, ethanol, and submergence conditions than wild-type plants (Chen et al., 2015). In mammals, ROS is an important stimulator of autophagy induction, whereas autophagy reduces the accumulation of ROS through the removal of damaged organelles (Li et al., 2015). The interaction between ROS and autophagy has also been reported in plants recently. Autophagy is induced after treatment of Arabidopsis plants with H2O2 or ROS-producing agent methylviologen (MV). After treatment with MV, RNAi-ATG18a seedlings accumulated more oxidized proteins and were more sensitive to oxidative stress (Xiong et al., 2007).
Programmed cell death (PCD) is a planned, step-by-step, active suicide process that selectively eliminates unnecessary or damaged cells in eukaryotes (Gadjev et al., 2008). PCD is a genetically controlled form of cell death, both in the course of development and in response to osmotic, thermal, and oxidative stresses and in defenses against pathogens. Like animal cell apoptosis, PCD in plants shares common morphological and biochemical features, including cell contraction, cytoplasmic contraction, cytochrome c release from mitochondria, excessive production of ROS, chromatin condensation and aggregation, and DNA fragmentation. Recent studies have shown that multifarious PCD processes are accompanied by autophagy (Liu Y. et al., 2005). An interesting view has developed in recent issues of Trends in Plant Science, autophagy plays a unique role in the initiation, execution of, and protection against cell death (Minina et al., 2014), suggesting that the morphological features and mechanisms of autophagy may be different in living cells and dying cells. Therefore, we plan to perform further work to determine the exact role of autophagy in the induction of PCD in plant cells.
Recently, identification of autophagy-related (ATG) genes and research on the autophagy pathway have made considerable progress (Bassham et al., 2006; Liu and Bassham, 2012). Several studies have demonstrated the pro-survival role of autophagy in plant cells under environmental stresses (Doelling et al., 2002; Xiong et al., 2007; Shin et al., 2009) including submergence (Chen et al., 2015). We further investigated the impact of waterlogging-induced autophagy on PCD in Arabidopsis roots. Here, we report that ROS-dependent autophagy has an attenuating effect on PCD in waterlogged Arabidopsis roots.
The strains of A. thaliana used in this study were all Columbia-0 (Col-0) ecotype. A. thaliana mutants atg2 (SALK_076727), atg5 (SAIL_129_B07), atg7 (SAIL_11_H07), atg10 (SALK_084434C), and rboh (rbohd, CS9555; rbohf, CS9557 and rbohdf, CS9558) were obtained from the Arabidopsis Information Resources Centre. Seeds of plants expressing 35Spro:GFP-ATG8e and ATG8epro:GUS (Chen et al., 2015) were kindly provided by Prof. Shi Xiao (Sun Yat-sen University). All Arabidopsis seeds were surface sterilized in 20% bleach [1% (v/v) NaClO] and 0.01% Triton X-100 for 12 min, washed five times with sterile water, and stored at 4°C for 3 days. Then, the seeds were sown on plates containing half-strength MS medium with 1% sucrose and 0.8% (w/v) agar. The plates were vertically placed in growth chambers with a 16-h-light/8-h-dark cycle at 22°C.
All waterlogging treatments were performed in three independent biological replicates. Waterlogging treatment was carried out as described previously, with minor modifications (Wang et al., 2016). Briefly, 1-week-old seedlings were transferred to pots containing a stagnant liquid MS medium with 1% sucrose. The stagnant solution contained 0.2% (w/v) dissolved agar to prevent convective movement and was deoxygenated (dissolved O2, <0.5 mg/L) by flushing with N2 gas (Wiengweera et al., 1997; Yamauchi et al., 2017). The plant roots were completely immersed in stagnant solution, and leaves protruded 0.5 cm above the surface of the solution. The plants were placed in chambers at 22°C under a 16-h photoperiod. In order to calculate root length and survival rates after waterlogging, more than 20 plants of each genotype were waterlogged. Survival rates were based on the ability to produce new leaves and continue to grow.
Acid dyes, i.e., monodansylcadaverine (MDC) and GFP-ATG8 fusion proteins, are often used to detect autophagy in plants (Contento et al., 2005). Seedlings (1 weeks old) expressing GFP-ATG8e fusion protein were waterlogged with stagnant solution for the indicated times. GFP fluorescence was observed and photographed using a Leica SP8 laser scanning confocal microscope (Leica, Germany), and the excitation and emission wavelengths were 488 and 507 nm, respectively.
Monodansylcadaverine staining was carried out as described previously (Contento et al., 2005). Briefly, seedlings (1 weeks old) were waterlogged for the indicated times and subsequently incubated in 0.05 mM MDC (Sigma) in PBS for 10 min, followed by three washes with PBS at room temperature. Seedling were observed and photographed with a Leica SP8 laser scanning confocal microscope using a DAPI-specific filter. The excitation and emission wavelengths for MDC were 345 and 455 nm, respectively.
Each sample consisted of three independent biological replicates. Three roots per experiment were analyzed microscopically. Five optical sections per root were counted. The thickness of a section is 1 μm. The GFP-ATG8e-labeled and MDC staining punctate structures were counted using the Adobe Photoshop software1.
We used diaminobenzidine (DAB) staining to detect H2O2 in situ as described previously, with minor modifications (Yang et al., 2014). Seedlings (1 weeks old) were incubated in 0.5 mg/mL DAB (Sigma) dissolved in 50 mM Tris–HCl (pH 5.0) for 2 h. The seedlings were rapidly washed three times with water and observed on a differential interference contrast microscope (Nikon 80i Eclipse). The quantitative measurement of staining intensity was carried out using Image pro plus 6.0.
Nitroblue tetrazolium (NBT) staining was performed to detect superoxide anion as described previously (Dunand et al., 2007). Briefly, seedlings (1 weeks old) were steeped in 2 mM NBT in 20 mM phosphate buffer (pH 6.1) for 15 min. The seedlings were transferred to distilled water and observed on a differential interference contrast microscope (Nikon 80i Eclipse). The quantitative measurement of staining intensity was carried out using Image pro plus 6.0.
For 2′,7′-dichlorofluorescin diacetate (DCFH-DA) staining to detect ROS (Soh, 2006; Kalyanaraman et al., 2012), 1-week-old seedlings were incubated at 37°C in 10 μM staining solution for 30 min. The DCFH-DA (sigma) was diluted from a 10 mM stock in ethanol using PBS. The stained seedlings were rapidly washed with PBS three times and were observed on a Leica SP8 laser scanning confocal microscope with an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The quantitative measurement of fluorescence intensity was carried out using Image pro plus 6.0.
The content of H2O2 was measured as described previously (Patterson et al., 1984). The content of superoxide anion was determined according to hydroxylamine method (Elstner and Heupel, 1976).
For the GUS staining assay, 1-week-old seedlings were soaked in GUS assay buffer [0.1 M phosphate buffer (pH 7.0), 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, 0.1% Triton X-100, and 0.5 mg/mL X-Gluc] at 37°C in the dark for 9 min and were then incubated in 75% ethanol. Seedlings were photographed with a differential interference contrast microscope (Nikon 80i Eclipse).
One-week-old Arabidopsis roots (1.0 g) prepared for enzyme activity were homogenized in an extraction solution containing 50 mM Na2HPO4–NaH2PO4 buffer, 0.2 mM EDTA, and 2% insoluble polyvinylpyrrolidone. The homogenate was centrifuged at 12,000 rpm for 20 min and the supernatant was used to determine enzyme activity. The activity of SOD, APX, and CAT was determined by using SOD assay kit, APX assay kit, and CAT assay kit purchased from Nanjing Jiancheng Bioengineering Institute according to manufacturer’s directions (Hussain et al., 2016).
Total RNA was extracted from 1-week-old Arabidopsis roots using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The isolated RNA was reverse transcribed using the PrimeScript RT reagent kit with gDNA Eraser (Takara). Reverse transcription quantitative PCR (qRT-PCR) was performed with SYBR Premix Ex Taq II (Takara) using a StepOne Plus real-time PCR system (Applied Biosystems). The program for qRT-PCR was 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. Each reaction had three biological and three technical replicates. ACTIN2 was used as the internal control and transcription levels were calculated according to the 2-ΔΔCt formula. Information of genes included in this study are listed in Supplementary Table S1. Primers used for qRT-PCR analysis are listed in Supplementary Table S2.
The general procedures of preparing conventional samples for transmission electron microscopy (TEM), described previously, were followed with minor modifications (Xu et al., 2013). Briefly, middle areas of root sections (1 mm3) were cut from seedlings (1 weeks old) and fixed immediately in 2.0% paraformaldehyde and 2.5% glutaraldehyde in PBS overnight at 4°C. The tissues were rinsed three times using PBS, post-fixed in 1% osmium tetroxide for 4 h, rinsed three times using PBS again, dehydrated in a graded acetone series, and embedded in SPI-PON 812 resin (SPI Supplies). For TEM, the ultrathin sections were contrasted with uranyl acetate and lead citrate and were observed directly under the TEM (Hitachi H-7650, Japan) at 80 kV. For statistical analysis of the autophagic structures, five roots for per experiments and three cells from every root/root tissue were analyzed microscopically.
Total protein was extracted from 1-week-old Arabidopsis roots using an ice-cold RIPA lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS) supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF; Roche). The homogenates were placed on ice for 30 min and were then centrifuged at 4°C for 15 min at 12,000 rpm. The supernatant was transferred to a new microfuge tube before electrophoresis.
For the immunoblot analysis, total proteins were separated by 15% SDS–PAGE and were then electroblotted to a nitrocellulose membrane (Biosharp). Anti-AOX (agersera), anti-Actin (sigma), anti-ATG8a (Abcam), and anti-GFP (Abcam) antibodies were used in the immunoblotting analysis. Quantification of the protein signal was performed using Image pro plus 6.0 software.
For phenotypic analysis, 1-week-old Arabidopsis roots under normal or waterlogged conditions were stained using 10 μg/mL propidium iodide (PI; Sigma) and were observed on a Leica SP8 laser scanning confocal microscope with an excitation wavelength of 535 nm and an emission wavelength of 615 nm.
For FDA staining, 1-week-old Arabidopsis roots under normal or waterlogged conditions were stained with 5 ug/mL Fluorescein diacetate (FDA; Sigma) for 5 min and were washed three times using sterile water. The fluorescence signal was observed on a Leica SP8 laser scanning confocal microscope with an excitation wavelength of 488 nm and an emission wavelength of 525 nm. The quantitative measurement of fluorescence intensity was carried out using Image pro plus 6.0.
For trypan blue staining, 1-week-old seedlings were placed at room temperature in trypan blue staining buffer (0.4% solution in PBS) for 10 min, followed by washing three times using PBS. Seedlings were photographed with a differential interference contrast microscope (Nikon 80i Eclipse).
For DAPI staining, 1-week-old Arabidopsis roots under normal or waterlogged conditions were stained in PBS containing 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI; Sigma) for 30 min and washed three times with PBS. The fluorescence signal was observed on a Leica SP8 laser scanning confocal microscope with an excitation wavelength of 358 nm and an emission wavelength of 461 nm.
A TUNEL assay was performed using an In Situ Cell Death Detection Kit (Roche, Germany) according to the manufacturer’s instructions with a few modifications. In brief, 1-week-old Arabidopsis roots were fixed in 4% (w/v) paraformaldehyde in PBS (pH 7.4) for 20 min. Then, the fixed seedlings were washed for 30 min with PBS before immersing in permeabilization solution (0.1% Triton X-100, 0.1% sodium citrate) for 2 min. After washing the samples three times with PBS, 50 μL aliquot of TUNEL reaction mixture was then added to the sample, and it was incubated for 1 h (37°C) in a humidified atmosphere in the dark. After being rinsed three times with PBS, photographs were taken under a Leica SP8 laser scanning confocal microscope with an excitation wavelength of 488 nm and an emission wavelength of 530 nm.
All the experiments were run in triplicate or more unless otherwise indicated, and the results reported in this study are presented as the mean ± SD. Five roots were used for every kind of experiment (ROS assays, GUS assays, and cell death detection). Data were analyzed by a two-tailed Student’s t-test using GraphPad Prism 7.0. The significance levels are ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001.
To validate the system of waterlogging, we analyzed the relative expression of the hypoxia-responsive genes ADH1, PDC1, PDC2, SUS1, SUS4, LDH, hemoglobin 1 (HB1), hypoxia-responsive unknown protein 43 (HUP43), and LOB domain-containing protein 41 (LBD41) (Hunt et al., 2002; Mustroph et al., 2010) in wild-type roots at 0, 4, 8, 12, and 24 h after waterlogging. The expression of the hypoxia-responsive genes ADH1, PDC1, SUS1, HB1, HUP43, and LBD41 started to increase within 4 h and peaked at 4 h (Supplementary Figure S1), and expression of PDC2, SUS4, and LDH peaked at 8 h under waterlogged conditions (Supplementary Figure S1). These results demonstrate that the system is appropriate to study the hypoxia response of waterlogged Arabidopsis roots.
We previously showed that accumulation of ROS in wheat root cells was induced by waterlogging (Xu et al., 2013). To investigate whether ROS are involved in the hypoxic response of waterlogged A. thaliana, we investigated the generation of ROS in wild-type roots subjected to waterlogging. DAB, NBT, and DCFH-DA staining were used to detect H2O2, superoxide anion, and ROS in situ. Pronounced accumulation of H2O2, superoxide anion (Figure 1A), and ROS (Figure 1B) was detected in the meristematic and elongation zones of the primary root under waterlogging conditions. In contrast, plants grown under normal conditions had relatively low ROS levels (Figures 1A–D). We also found that the ROS in the root stele seemed to be higher. To further confirm this, we determined the level of H2O2 and superoxide anion in wild-type roots under waterlogging conditions. The content of H2O2 in Arabidopsis roots was significantly upregulated within 24 h (Figure 1E). The content of superoxide anion started to increase within 4 h and peaked at 12 h (Figure 1F). These findings suggest that waterlogging induced accumulation of ROS.
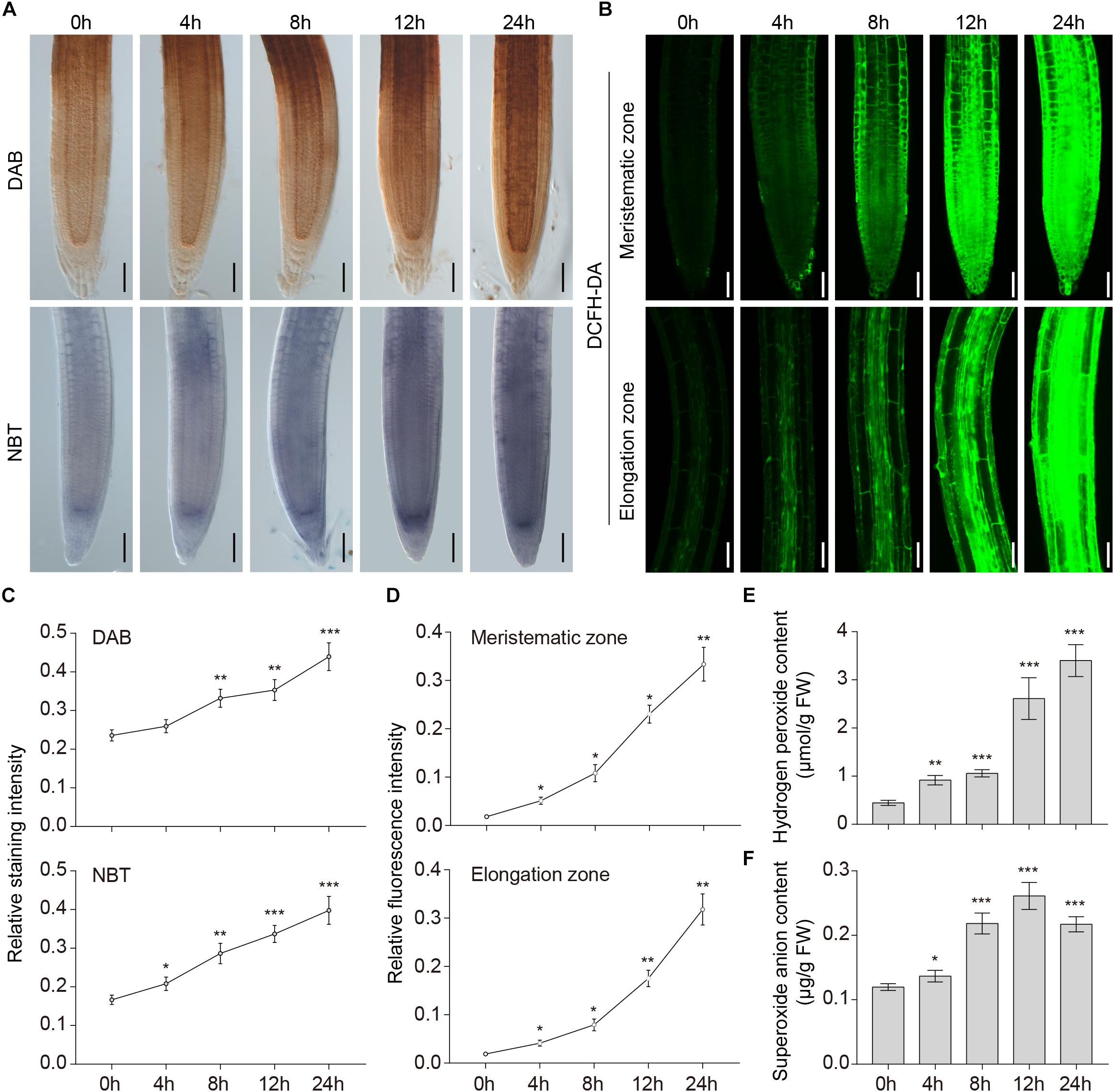
Figure 1. Accumulation of ROS induced by waterlogging. (A) DAB staining for H2O2 and NBT staining for superoxide anion in the primary root of wild-type Arabidopsis after waterlogging treatment. Bars = 50 μm. (B) DCFH-DA staining for ROS in the primary root of wild-type Arabidopsis after waterlogging treatment. Bars = 50 μm. (C–D) Relative staining intensities calculated from (A–B). (E) H2O2 and (F) superoxide anion contents in wild-type roots after waterlogging treatment. All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
RBOHD and RBOHF are the most in-depth studied enzymatic ROS-generating systems, and reports of their participation in various plant processes have increased considerably in recent years. To obtain genetic evidence of the response of ROS to waterlogged conditions, we obtained rbohd, rbohf, and rbohdf mutants and characterized their responses to waterlogging. The results showed that waterlogging markedly suppressed the growth of all plants. Of note, the double mutants and the two single mutants grew more slowly than the wild type (Supplementary Figure S2A). The root length of rbohdf was clearly less than that of wild type, rbohd and rbohf after exposure to waterlogged conditions (Supplementary Figure S2B). Moreover, we used DAB and NBT staining to detect ROS accumulation in roots of wild type and rboh mutants. DAB and NBT staining results showed that the root tips of rbohd, rbohf, and rbohdf mutants accumulated less H2O2 and superoxide anion than those of the wild type after waterlogging (Supplementary Figures S2C–E). Together, these results indicate that both RBOHD and RBOHF are required for waterlogging induced ROS generation.
The fact that AOX can use reductant in excess of either the Cyt pathway capacity or the rate of ATP use suggests that AOX may play an important role in reducing the generation of ROS (Umbach et al., 2005). To test whether waterlogging affected the expression of the AOX protein, total endogenous protein extracted from wild-type roots was immunoblotted with specific monoclonal anti-AOX antibody. The levels of the AOX protein increased upon waterlogging and accumulated to the highest level at 8 h (Figure 2A). As shown in Figure 2B, the gray values of the total AOX protein and the control are significantly different after treatment for 4, 8, and 12 h.
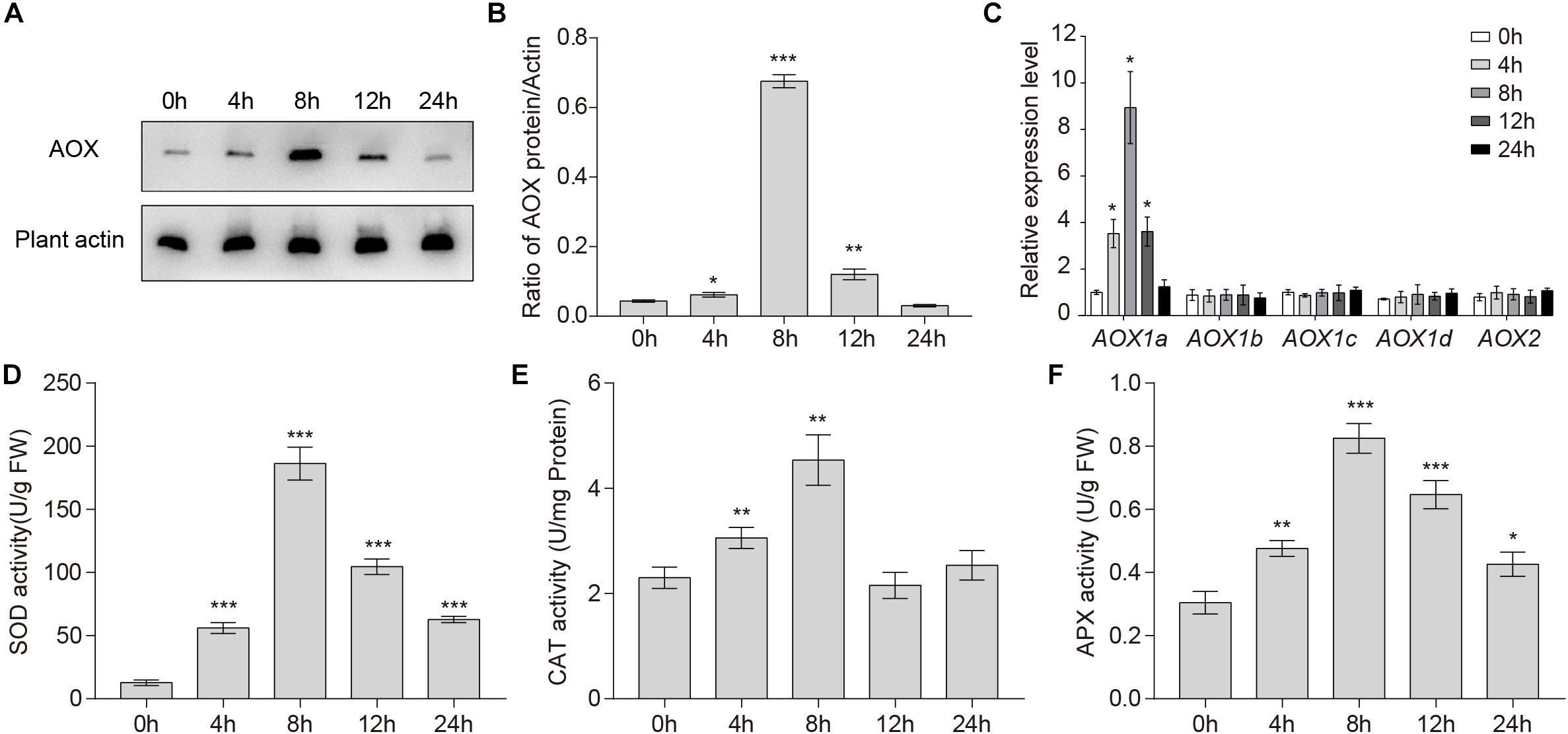
Figure 2. Changes in AOX1a expression levels and antioxidant enzymes activities under waterlogged conditions. (A) Western blot analysis of the level of total AOX proteins in wild-type roots under waterlogged conditions. Plant actin below the blots to indicate the amount of protein loaded per lane. (B) Quantitative analyses of the relative gray values of the western blotting. (C) qRT-PCR analyses showing the relative expression of AOX genes (AOX1a, AOX1b, AOX1c, AOX1d, and AOX2) upon waterlogging. (D) SOD, (E) CAT, and (F) APX activities in wild-type roots after waterlogging treatment. All of the experiments were repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
In A. thaliana, there are five AOX genes: AOX1a, AOX1b, AOX1c, AOX1d, and AOX2 (Giraud et al., 2008). Because the expression of these five AOX genes in response to various stresses is complex, we investigated their transcription levels in wild-type roots after waterlogging. qRT-PCR analyses showed that only the transcription level of AOX1a was significantly increased. The transcription level of AOX1a started to increase within 4 h, and peaked at 8 h (Figure 2C), which was consistent with the change in the AOX total protein level (Figure 2A). In conclusion, waterlogging may be affecting only the expression of AOX1a.
To counteract oxidative stress in plant cells, three enzymes, SOD, APX, and CAT, are crucial. To further investigate enzymatic mechanisms that scavenge overproduced ROS, we measured the activities of SOD, APX, and CAT in wild-type roots during waterlogging. The activities of SOD, CAT, and APX in Arabidopsis roots were significantly upregulated within 24 h (Figures 2D–F). The activities of SOD, CAT, and APX started to increase within 4 h and peaked at 8 h (Figures 2D–F). Together, these findings indicate that activities of antioxidant enzymes were enhanced under waterlogged conditions.
To investigate the role of waterlogging in autophagosome formation in plant cells, we used Arabidopsis plants expressing the GFP-ATG8e fusion protein as a marker (Contento et al., 2005). Seedlings expressing GFP-ATG8e were waterlogged for 4 and 8 h and GFP fluorescence was analyzed using confocal microscopy. In contrast to the root cells of the control, there were more GFP-ATG8e labeled dots and ring-like structures in wild-type root tips and mature root cells subjected to waterlogging (Figures 3A,B). At the beginning of autophagy, GFP-ATG8e that appeared in the shape of dots formed autophagosomes with other autophagy-associated proteins and subsequently fused with the vacuole. After entering the vacuole, GFP-ATG8e was sheared into free GFP and ATG8 hydrolysis fragments by acidic hydrolases. Since the GFP protein is very stable under acidic conditions, it is possible to determine the intensity of autophagy by determining the degradation of GFP-ATG8e (Ichimura et al., 2004). Thus, the appearance of free GFP can be used to monitor the extent of autophagy. The waterlogging treatment enhanced the degradation of GFP-ATG8e, as measured by the accumulation of free GFP, in comparison to the control (Figure 3C).
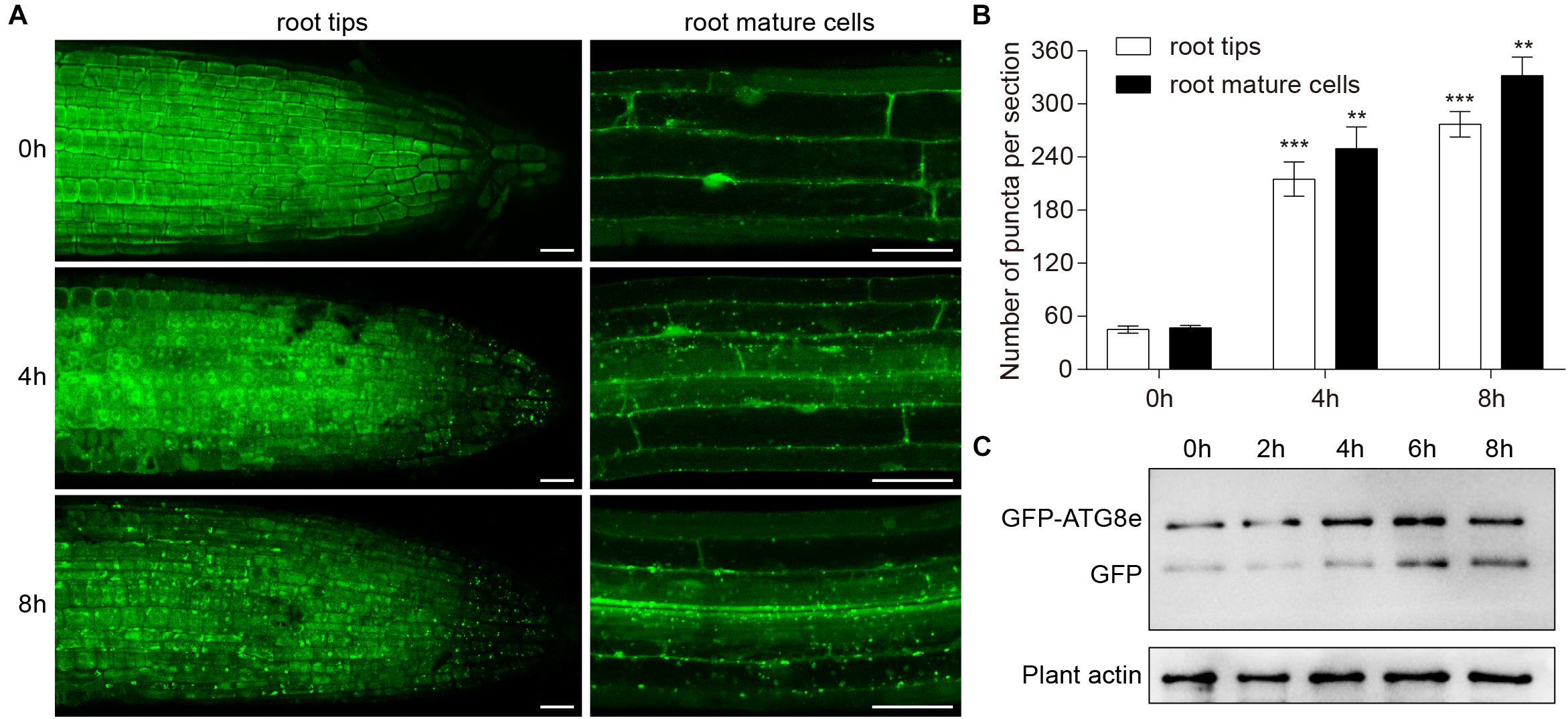
Figure 3. Induction of autophagy by waterlogging. (A) GFP-ATG8e-labeled dots and ring-like structures accumulate in wild-type roots after waterlogging. Bars = 50 μm. (B) Numbers of puncta per root section in the root tips and mature root cells in (A). (C) Detection of free GFP generated from transgenic lines expressing GFP-ATG8e after waterlogging. Plant actin below the blots to indicate the amount of protein loaded per lane. All of the experiments were repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
To investigate autophagy characteristics upon waterlogging, we examined ATG8e expression in Arabidopsis roots using seedlings expressing GFP-ATG8e and ATG8epro-GUS. PI staining was used to distinguish the structure of Arabidopsis roots. In contrast to the control, there were more GFP-ATG8e-labeled dots and ring-like structures initially observed in the root stele cells after 4 h of waterlogging followed by epidermis of the root after 8 h of waterlogging (Figure 4A). After waterlogging, the appearance of GFP-labeled autophagosome-like structures was significantly increased in the wild-type root stele cells (Figure 4B). GUS staining revealed that the expression of ATG8e initially intense in the root stele cells after 4 h of waterlogging followed by other parts of the root after 8 h of waterlogging (Figure 4C). This result shows that waterlogging-activated autophagy initially occurs in the root stele. To further confirm this, we used TEM to examine ultrastructure of Arabidopsis root. Analysis of cross sections by TEM revealed autophagy-related structures increased in the root stele after 4 and 8 h of waterlogging treatment and in epidermis of the root after 8 h of waterlogging treatment (Supplementary Figure S3), whereas no obvious difference was detected in other parts of the root (Supplementary Figure S3).
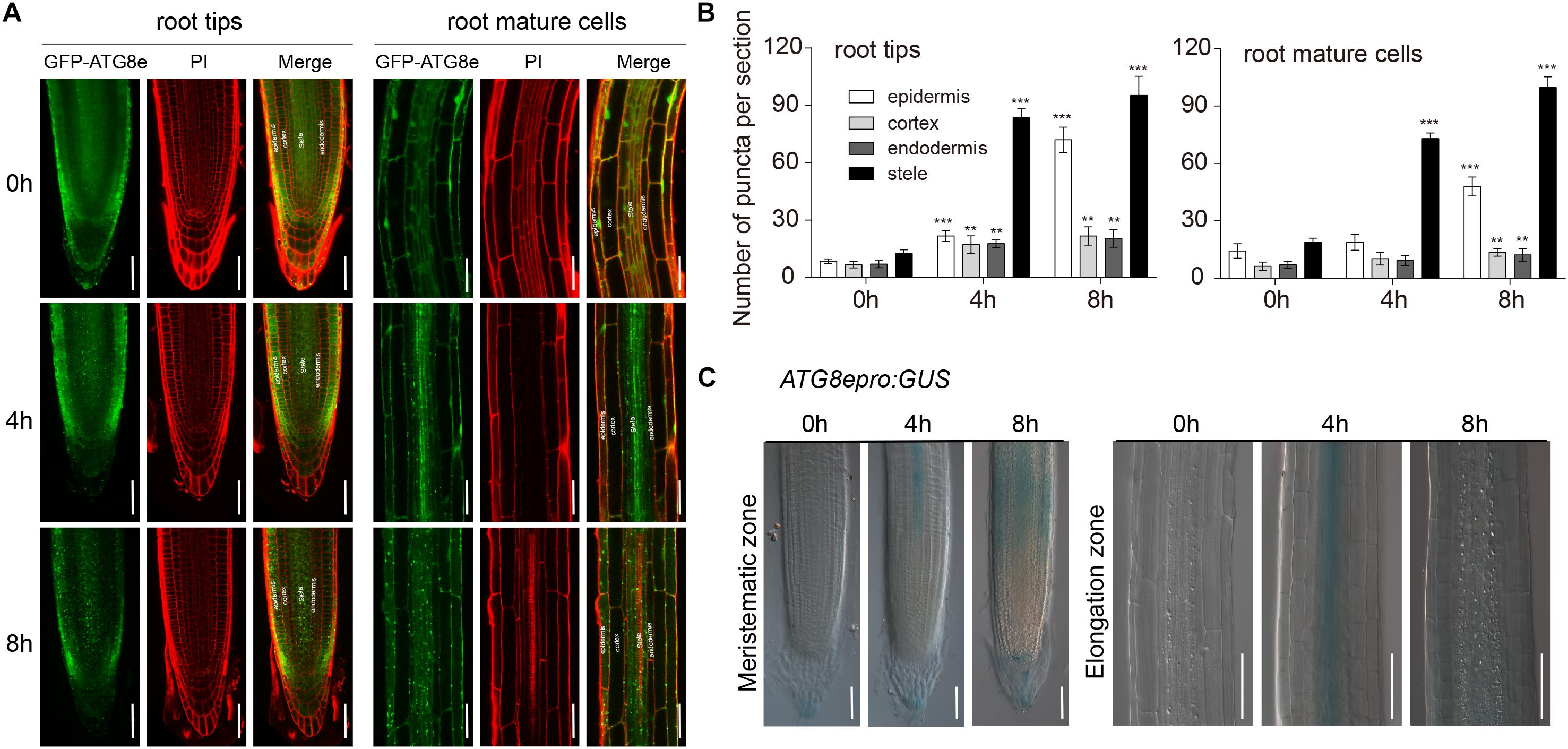
Figure 4. Autophagy occurs first in the root stele. (A) Confocal images of primary roots expressing GFP-ATG8e. Roots were counterstained with PI. Bars = 50 μm. (B) Numbers of puncta per root section in the root tips and mature root cells of wild type in (A). (C) The expression of the ATG8epro:GUS reporter was monitored in wild-type roots. Bars = 50 μm. All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
Previous studies have implied the function of autophagy under stress conditions (Bassham et al., 2006; Han et al., 2011). To investigate the role of autophagy in the response to waterlogging, we transferred 1-week-old atg mutant plants (atg2, atg5, atg7, and atg10), alongside wild-type plants, to hypoxic conditions induced by waterlogging. We analyzed the atg mutants that showed enhanced hypersensitivity to waterlogging, waterlogging + 1% alcohol, and waterlogging + 1% perhydrol (Supplementary Figure S4A). As shown in Supplementary Figure S4B, the primary roots of atg mutants were significantly shorter than those of that wild type after waterlogging and the waterlogging + 1% alcohol treatment. During the course of waterlogging + 1% perhydrol stress, most of the wild-type plants survived, but more than half of the atg mutants died (Supplementary Figure S4C).
To further evaluate the function of autophagy in the waterlogging response, we transferred 1-week-old wild-type and atg mutant seedlings (atg5 and atg7) to waterlogged conditions, and then MDC staining was used to observe autophagosomes. Accumulation of MDC-stained autophagosomes was observed in root tips and mature root cells of wild-type plants subjected to waterlogging (Supplementary Figure S4D), whereas the number of autophagosomes was significantly decreased in the atg mutant root cells after waterlogging (Supplementary Figure S4E).
Under waterlogged incubation conditions, accumulation of ROS and autophagy are induced. To investigate the relationship between ROS and autophagy, qRT-PCR was used to quantify the expression levels of ATG genes in the wild-type and rboh mutants exposed to waterlogging. In comparison to the wild type, the expression of ATG genes was significantly downregulated in the rboh mutants (Figure 5A). To further confirm the qRT-PCR data, we observed autophagosomes of rboh mutants following waterlogging using MDC staining. Upon waterlogging exposure, the appearance of autophagosome-like structures was decreased in the rbohd and rbohf mutants in comparison with the wild type (Figures 5B,C). We also used TEM to monitor the autophagic activity after waterlogging. The results showed that lower levels of autophagic activity were detected in the rboh mutants than in wild type after waterlogging (Supplementary Figures S5A,B). These data demonstrated that the occurrence of autophagy is significantly decreased in rboh mutants after waterlogging.
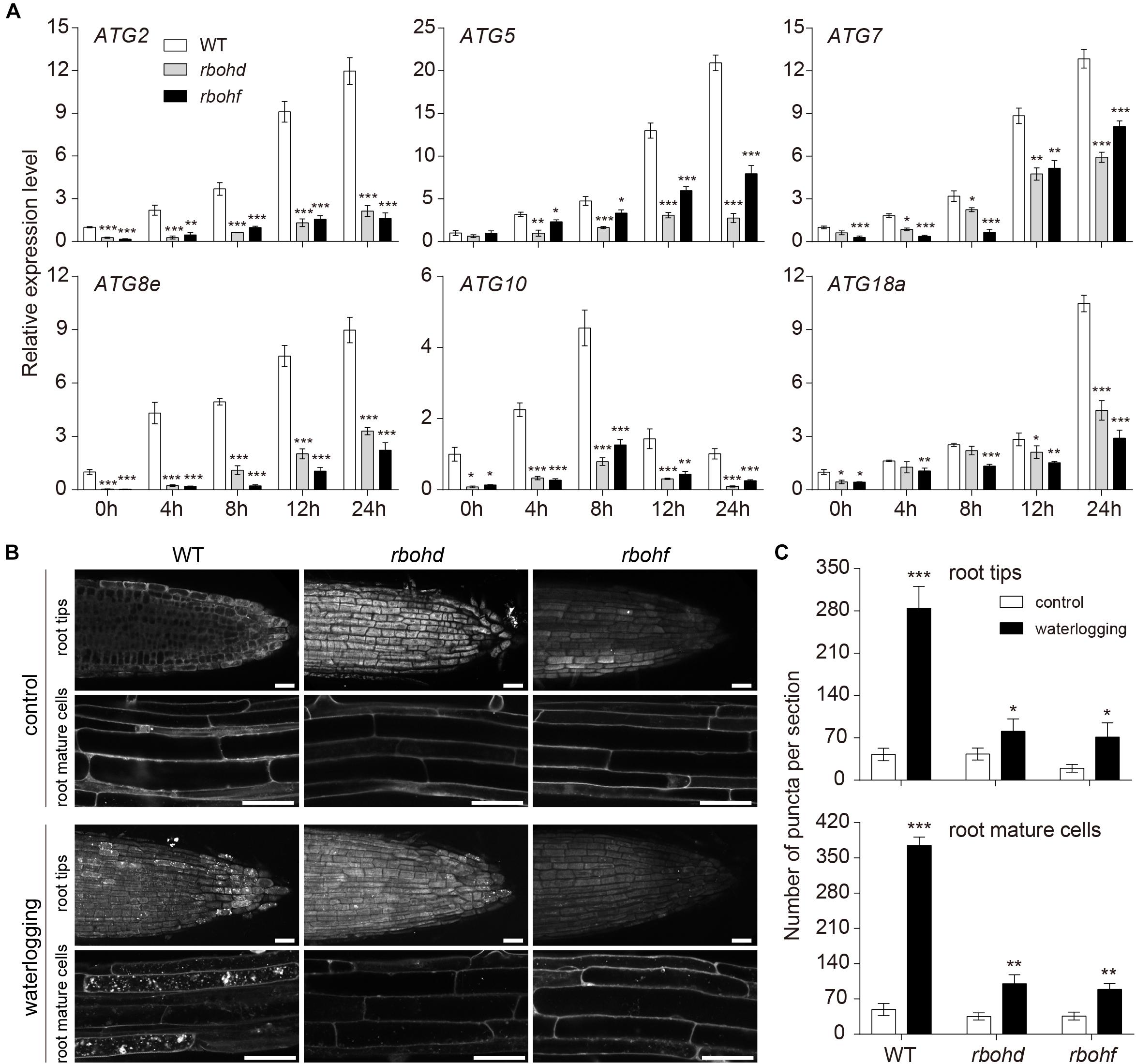
Figure 5. The level of autophagy attenuated in rboh mutants after waterlogging. (A) Relative transcription levels of autophagy-related genes (ATG2, ATG5, ATG7, ATG8e, ATG10, and ATG18a) in the wild-type and rboh mutants upon waterlogging. (B) Confocal microscopy of MDC-stained wild-type, rbohd, and rbohf seedlings after waterlogging. Bars = 50 μm. (C) Quantification of autophagosome number in the root tips and mature root cells of wild-type, rbohd, and rbohf under waterlogged conditions as in (B). All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
In order to further investigate the interrelation between ROS and autophagy, we analyzed the mRNA expression level of biosynthesis and scavenging genes of ROS in the wild-type and atg mutants upon waterlogging. In comparison to the wild type, the transcription level of RBOHD and RBOHF was notably increased in the atg mutants at 0 and 4 h of waterlogging (Figure 6A). However, the expressions of SOD, CAT1, APX1, and APX2, which constitute the enzymatic scavenging system to eliminate ROS in plants, were significantly reduced in the atg mutants in comparison to the wild type (Figure 6A). In that case, the atg mutant plants exhibited excessive accumulation of ROS compared with the wild type under waterlogging stress. In order to further verify higher levels of ROS in atg mutants, we transferred 1-week-old atg mutant plants, alongside wild-type plants, to DCFH-DA dye. DCFH-DA staining showed higher accumulation of ROS in atg mutant plants under waterlogging conditions, in comparison to wild type (Supplementary Figures S6A,B). In comparing the DAB and NBT staining in root tips of the wild-type and atg mutants, we found the H2O2 and superoxide anion levels were both greatly elevated in the atg mutants after waterlogging treatment, compared with those of the wild type (Figures 6B–D). Waterlogging treatment significantly enhanced the production of ROS in both wild-type and atg mutants (Figures 6B–D and Supplementary Figure S6). In summary, accumulation of ROS is increased in atg mutants after waterlogging.
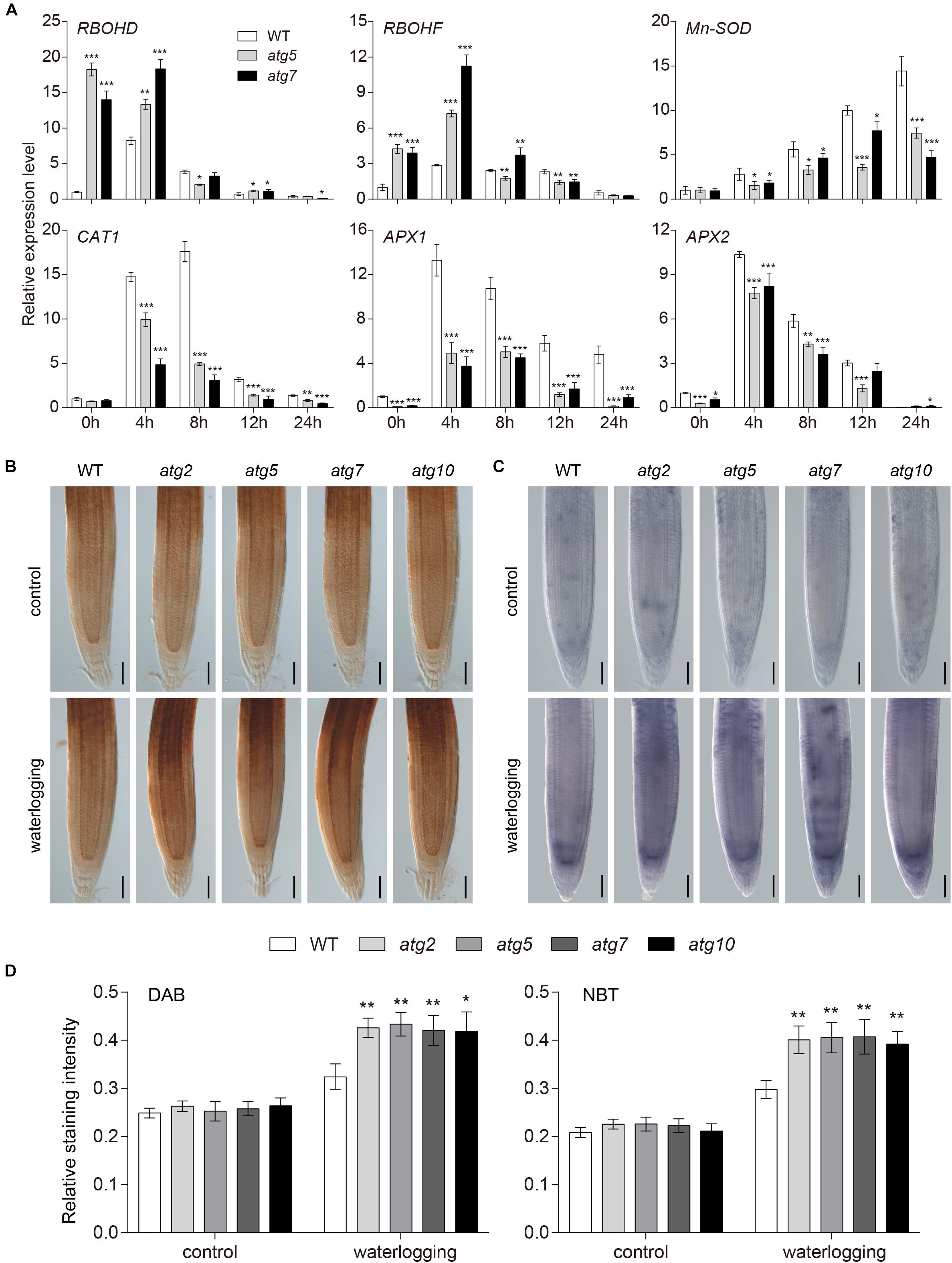
Figure 6. Accumulation of ROS increased in atg mutants after waterlogging. (A) Relative expression levels of genes encoding enzymes involved in production or reduction of ROS in the wild-type and atg mutants after waterlogging treatment. (B) DAB staining for H2O2 and (C) NBT staining for superoxide anion in primary root of wild-type and atg mutants after waterlogging treatment. Bars = 50 μm. (D) Relative staining intensities calculated from (B–C). All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
Arabidopsis hypocotyls form lysigenous aerenchyma as a consequence of cell death of secondary xylem during hypoxia (Mühlenbock et al., 2007). To investigate the PCD process upon waterlogging, 1-week-old wild-type seedlings were treated with PI, which cannot pass through a live cell membrane, but can cross a damaged cell membrane and stain the nucleus. PI staining showed that cell death started at 24 h in the primary root, whereas under normal conditions it was hardly detected (Figure 7A). We used FDA staining to show the effects of waterlogging on cell viability in the meristematic and elongation zones of wild-type roots after waterlogging for 24 and 48 h. Arabidopsis plants in the control showed strong FDA fluorescence, whereas weak fluorescence was detected under waterlogging conditions (Figures 7B,C). Typical characteristic of PCD is the occurrence of morphological changes in the nucleus and the cleavage of genomic DNA at internucleosomal sites by endogenous nucleases. To further determine whether the root cell death induced by waterlogging is a kind of PCD, we used DAPI staining and TEM to show the effects of waterlogging on the chromatin condensation of wild-type roots. Confocal images and TEM images showed that a marked increase in DAPI fluorescence and condensed and moon-shaped nuclei were detected under waterlogging conditions (Figure 7D). We also investigated the internucleosomal fragmentation of the DNA by TUNEL assay. TUNEL-positive signals were detected in the meristematic zone to the elongation zone from root tips after waterlogging treatment (Figure 7E). These results indicate that waterlogging induced cell death in Arabidopsis roots is a kind of PCD.
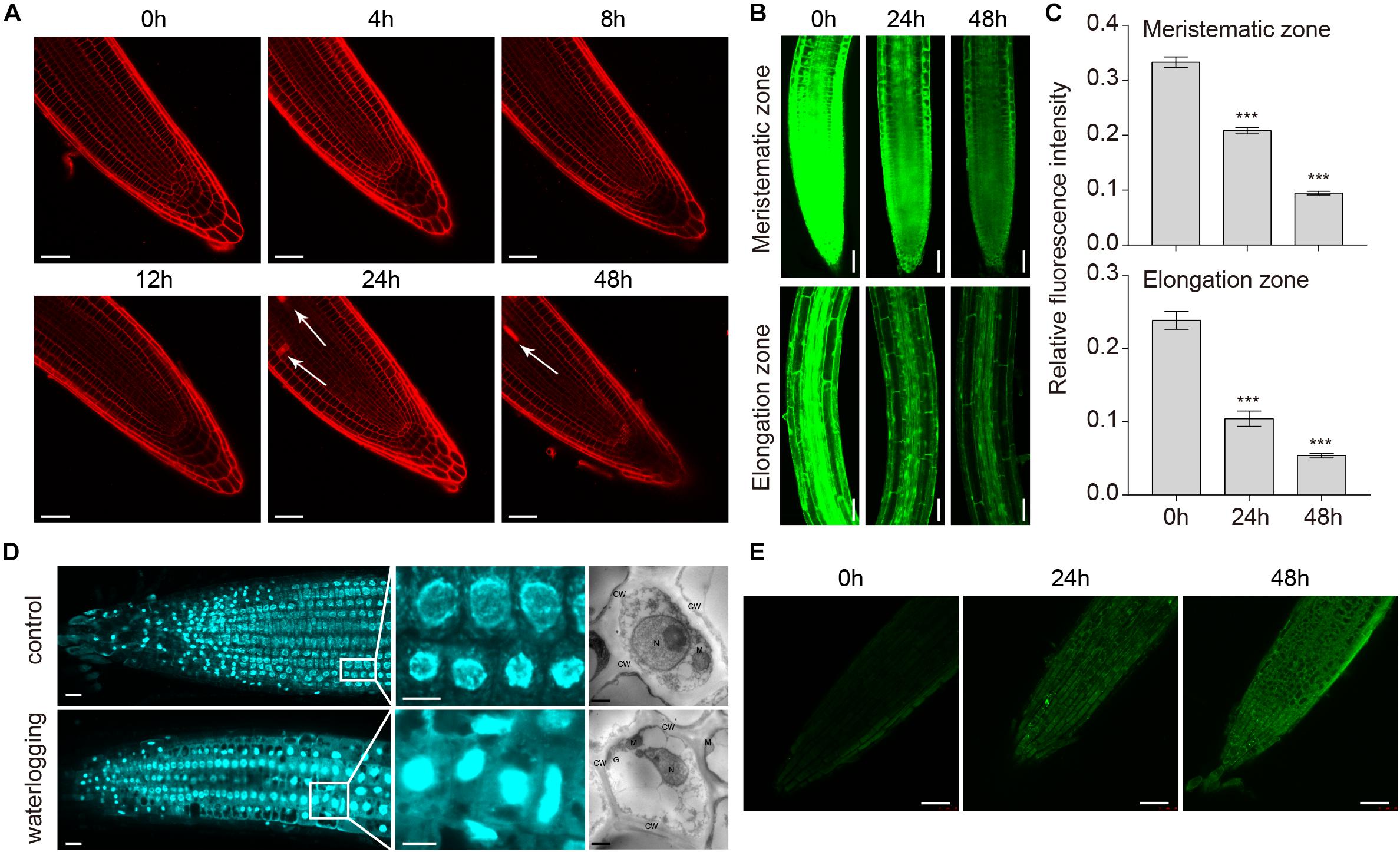
Figure 7. Programmed cell death in roots of A. thaliana induced by waterlogging. (A) Confocal images of wild-type roots stained with propidium iodide (PI). Arrows indicate the cell death. Bars = 50 μm. (B) Confocal images of the detection of cell viability in primary roots. Bars = 50 μm. (C) Meristematic and elongation zones of wild type of FDA staining intensity are quantified by Image pro plus 6.0 software. (D) Confocal images and TEM images of the detection of nucleus in primary roots with or without waterlogging treatment. CW, cell wall; M, mitochondrion; N, nucleus; G, Golgi body. Bars = 50 μm. (E) TUNEL assay of DNA fragmentation (stained in green) in root tip cells of wild-type seedlings. Bars = 50 μm. All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
To examine whether autophagy is also involved in the PCD process in Arabidopsis roots, 1-week-old atg mutant plants were waterlogged. FDA staining was used to observe cell viability of waterlogged wild-type and atg mutants. The fluorescence intensity of atg mutants was obviously weaker than that of the wild type in the waterlogged treatment (Figure 8A). However, the fluorescence intensity of wild-type and atg mutants grown under normal conditions was fairly similar (Figure 8B). Waterlogging triggered cell death, as indicated by the blue color, in root of the wild-type and atg mutants (Figure 8C). In comparison to the wild type and controls, much higher levels of cell death were observed in the roots of atg mutants after waterlogging (Figure 8C). To further evaluate the role of autophagy in PCD, we examined cell death in Arabidopsis root subjected to waterlogging. In comparison to the wild type, cell death was significantly increased in the atg mutants after exposure to waterlogging. And cell death was mainly concentrated in root stele (Figure 8D). In conclusion, autophagy has effect on PCD in Arabidopsis roots during waterlogging.
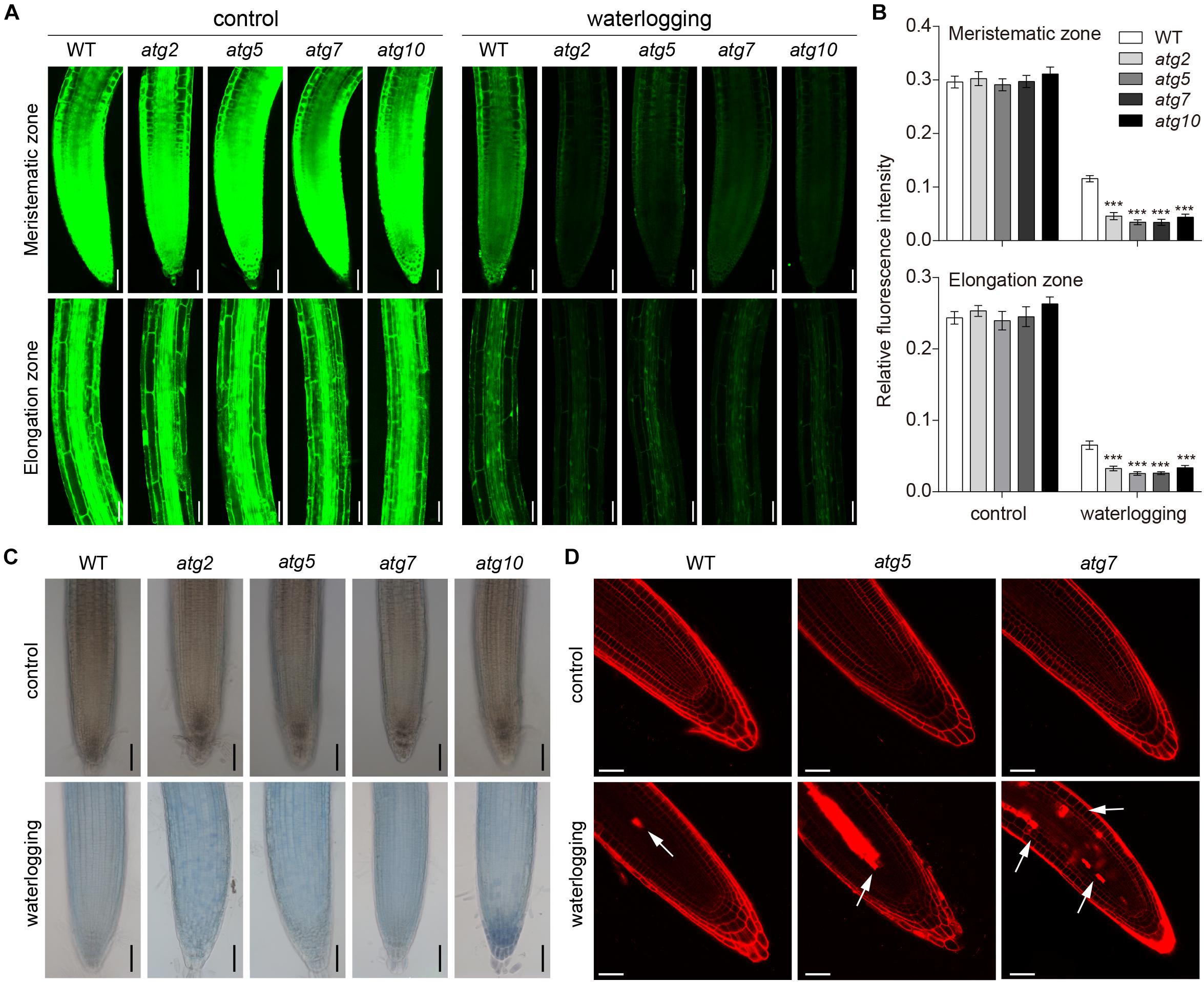
Figure 8. Autophagy regulates the PCD process upon waterlogging. (A) Confocal images of the detection of cell viability in primary roots of atg mutants in comparison with wild type. Bars = 50 μm. (B) Meristematic and elongation zones of wild-type and atg mutants of FDA staining intensity are quantified by Image pro plus 6.0 software. (C) Trypan blue staining cell death in root of wild-type and atg mutants without treatment or waterlogged for 8 h. Bars = 50 μm. (D) Cell death of the root of wild-type and atg mutants visualized by PI staining. Arrows indicate the cell death. Bars = 50 μm. All of the experiments have been repeated at least three times. Data shown are the mean ± SD (n = 3). ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001 by Student’s t-test.
Autophagy, which etymologically means “to eat oneself,” is a highly conserved process that involves vacuolar degradation of damaged or toxic constituents and is induced by ROS accumulation and other factors (Liu et al., 2009). Although autophagy was first described in plants a number of years ago, in the past few years, the study of autophagy molecular mechanisms and regulatory pathways has flourished. In this paper, we demonstrate the effect of waterlogging-induced autophagy on PCD in Arabidopsis roots. First, the levels of ROS were found to increase and the activities of SOD, CAT, and APX were significantly increased to cope with ROS under waterlogged conditions (Figures 1, 2). Second, waterlogging-induced autophagy was observed in the stele of Arabidopsis roots under waterlogged conditions (Figures 3, 4). Third, atg mutant plants displayed enhanced sensitivity to waterlogging stress (Supplementary Figure S4). Fourth, ROS signaling contributed to autophagy, and accumulation of ROS decreased in Arabidopsis roots under waterlogged conditions as a result of autophagy (Figures 5, 6 and Supplementary Figures S5, S6). Finally, autophagy-defective mutants showed much higher levels of cell death in comparison to the wild type (Figures 7, 8). Together, this evidence suggests that autophagy is important for A. thaliana survival during PCD induced by waterlogging.
In higher plants, ROS are considered to be an important signaling molecules that are produced by different mechanisms through electron transport in chloroplasts and mitochondria or by enzymes in the extracellular space and peroxisomes (Wrzaczek et al., 2013). Plant NADPH oxidase, also known as RBOH, is a major source of ROS in plant–pathogen interactions (Torres and Dangl, 2005). It plays an important role in plant response to abiotic and biological stresses, regulation of plant growth and development, and ROS signal transduction (Gechev et al., 2006; Suzuki et al., 2011). The observations that rbohd, rbohf, and rbohdf mutants accumulated less H2O2 and superoxide anion than wild type after waterlogging (Supplementary Figures S2C–E) and waterlogging suppressed the growth of rbohd, rbohf, and rbohdf mutants (Supplementary Figures S2A,B) are a confirmation of the data previously published (Chen et al., 2015). This suggests that RBOHD and RBOHF are required for ROS accumulation in waterlogged Arabidopsis roots. The ROS signaling pathway includes the rapid generation and removal of various forms of ROS to maintain the homeostasis of ROS levels (Mittler et al., 2011). However, the dynamic balance between production and clearance of ROS may be destroyed by certain biotic and abiotic stresses (Apel and Hirt, 2004). Under oxidative stress induced by ROS accumulation, autophagy is involved in reducing oxidative damage and ROS by scavenging damaged organelles, and compared with the wild type, there is a large amount of oxidatively damaged protein accumulation in autophagic mutant plants (Xiong et al., 2007). Consistent with previous observations (Chen et al., 2015), our quantitative PCR analysis showed that the transcription levels of RBOHD and RBOHF were significantly elevated and antioxidant enzyme genes were significantly decreased in the waterlogged atg mutant roots (Figure 6A). Previous studies have shown that atg mutants accumulated more H2O2 than wild type upon submergence (Chen et al., 2015). In our study, NBT and DCFH-DA staining showed higher accumulation of superoxide anion and ROS in atg mutant plants under waterlogging conditions, in comparison to wild type (Figures 6B–D and Supplementary Figure S6). While ROS levels were increased in the wild-type and atg mutants, there were significant differences among genotypes (Supplementary Figures S4A–C), suggesting that ROS are likely to be scavenged as a result of autophagy under waterlogged conditions. During animal starvation stress, ROS activates autophagy by directly regulating the ATG4 protein (Scherz-Shouval et al., 2007). Autophagy can be induced by different signaling pathways under different abiotic stresses in Arabidopsis, such as autophagy induced by starvation and salt stress, which depends on the activity of NADPH oxidase, in contrast to osmotic stress-induced autophagy (Liu et al., 2009). We observed that the expression of ATG genes was markedly reduced in the rboh mutants in comparison to the wild type (Figure 5A). The waterlogging-induced appearance of autophagosome-like structures was decreased in the rbohd and rbohf mutants in comparison with the wild type (Figures 5B,C and Supplementary Figure S5). This is consistent with the few autophagosome-like structures that were observed in the rboh mutants after submergence (Chen et al., 2015). This suggests that ROS participate in the process of autophagy.
Alternative oxidase helps to remove excess ROS in response to abiotic stress (Umbach et al., 2005). Recent studies have shown that AOX1a-OE plants pretreated with 1-(aminocarbonyl)-1-cyclopropanecarboxylic acid (ACC) exhibited higher whereas aox1a showed lower autophagic activity than wild-type plants. ATG8d- and ATG18h-silenced plants had significantly lower levels of AOX protein (Zhu et al., 2018). Applying exogenous H2O2 and its scavenger DMTU to AOX1a-OE and aox1a plants, respectively, found the autophagic activity reduced in AOX1a-OE and increased in aox1a plants under drought in response to ACC (Zhu et al., 2018). We found that the expression levels of the AOX protein increased and accumulated to the highest level at 8 h under waterlogging (Figure 2A). Furthermore, it found that only the transcription level of AOX1a was significantly increased and it peaked at 8 h (Figure 2B). We also found there were more autophagic structures initially observed in the root stele cells after 4 h of waterlogging followed by epidermis of the root after 8 h of waterlogging (Figures 4A,B). AOX1a-dependent ROS signaling maybe a key factor for the induction of autophagy upon waterlogging. Besides, the activities of antioxidant enzymes started to increase within 4 h and peaked at 8 h (Figures 2D–F). Together, waterlogging may induce autophagy through the regulation of ROS by antioxidant enzymes and AOX1a. All these need further experiments to prove.
Programmed cell death, an autonomous and orderly form of cell death controlled by genes, occurs during plant development or in response to biotic and abiotic stresses (Gadjev et al., 2008). As a signal molecule, ROS can not only promote the process of PCD but also inhibit it (Patel et al., 2006). In recent years, an increasing number of reports suggest that abiotic stresses are associated with the appearance of ROS, which are involved in the induction of autophagy. However, the role of autophagy in PCD induction remains elusive. A dual role of autophagy in plant PCD (autophagy acts as either an initiator or executioner of cell death) has been proposed (Minina et al., 2014). Autophagy plays an essential role in plant innate immunity and negatively regulates PCD (Liu Y. et al., 2005). Previous studies have shown that PCD phenotypes may require the change of SA signaling in the autophagy defective mutants after submergence (Chen et al., 2015). Disruptions of ATG10 enhance PCD compared to the wild type (Phillips et al., 2008). Enhanced trypan blue staining in atg5 and atg7 mutants were observed in UV-B-exposed leaves (Izumi et al., 2017). Compared with previous studies, this study focused only on the root of 1 weeks old Arabidopsis. Our findings from trypan blue and PI staining analyses showed that cell death was significantly upregulated in atg mutants upon waterlogging (Figures 8C,D). We also used FDA staining to observe the cell viability of waterlogged wild-type and atg mutant plants. The fluorescence intensity of the atg mutants was obviously weaker than that of the wild type after waterlogging (Figure 8A), whereas that of plants grown under normal conditions was fairly similar (Figure 8B). Furthermore, we found that waterlogging-induced root cell death is a kind of PCD by observing nuclear morphological changes and internucleosomal fragmentation of the DNA (Figures 7D–E). Thus, we propose that PCD may be affected by waterlogging induced autophagy. Most interestingly, this study also found that waterlogging induced ROS generation, autophagy and PCD process mainly occurred in the root stele. Recent studies have shown that the expression of HRE2 which is a member of oxygen-sensing subgroup VII AP2/ERFs was increased in the root stele at hypoxic conditions (Eysholdt-Derzsó and Sauter, 2017). It is probably due to oxygen-sensing subgroup VII AP2/ERFs mainly expressed in stele, leading to ROS generation, autophagy, and PCD process mainly occurred in the root stele.
Based on the results presented above, a model for effect of waterlogging-induced autophagy on PCD process in Arabidopsis roots can be proposed (Figure 9). In waterlogged wild-type roots, continued respiration and constrained gas influx can rapidly induce hypoxia. During the PCD process in waterlogged Arabidopsis roots, the amount of ROS is increased by enhancement of RBOH. In addition, the activities of SOD, APX, and CAT are upregulated to alleviate the generation of ROS under waterlogged conditions. ROS results in a significant increase in the transcription level of ATG genes and the formation of autophagosomes and autophagy, which in turn reduces the levels of ROS. Autophagy may induce the expression of antioxidant enzymes and inhibit the expression of RBOH. Finally, autophagy alleviates PCD in Arabidopsis roots. By contrast, autophagy deficiency of the atg mutants disrupts the induction of antioxidant enzymes expression and the inhibition of RBOH expression by autophagy. Autophagy deficiency also disrupts the interaction between ROS and autophagy, which may alleviate PCD. In that case, excessive accumulation of ROS increased PCD in atg mutant roots.
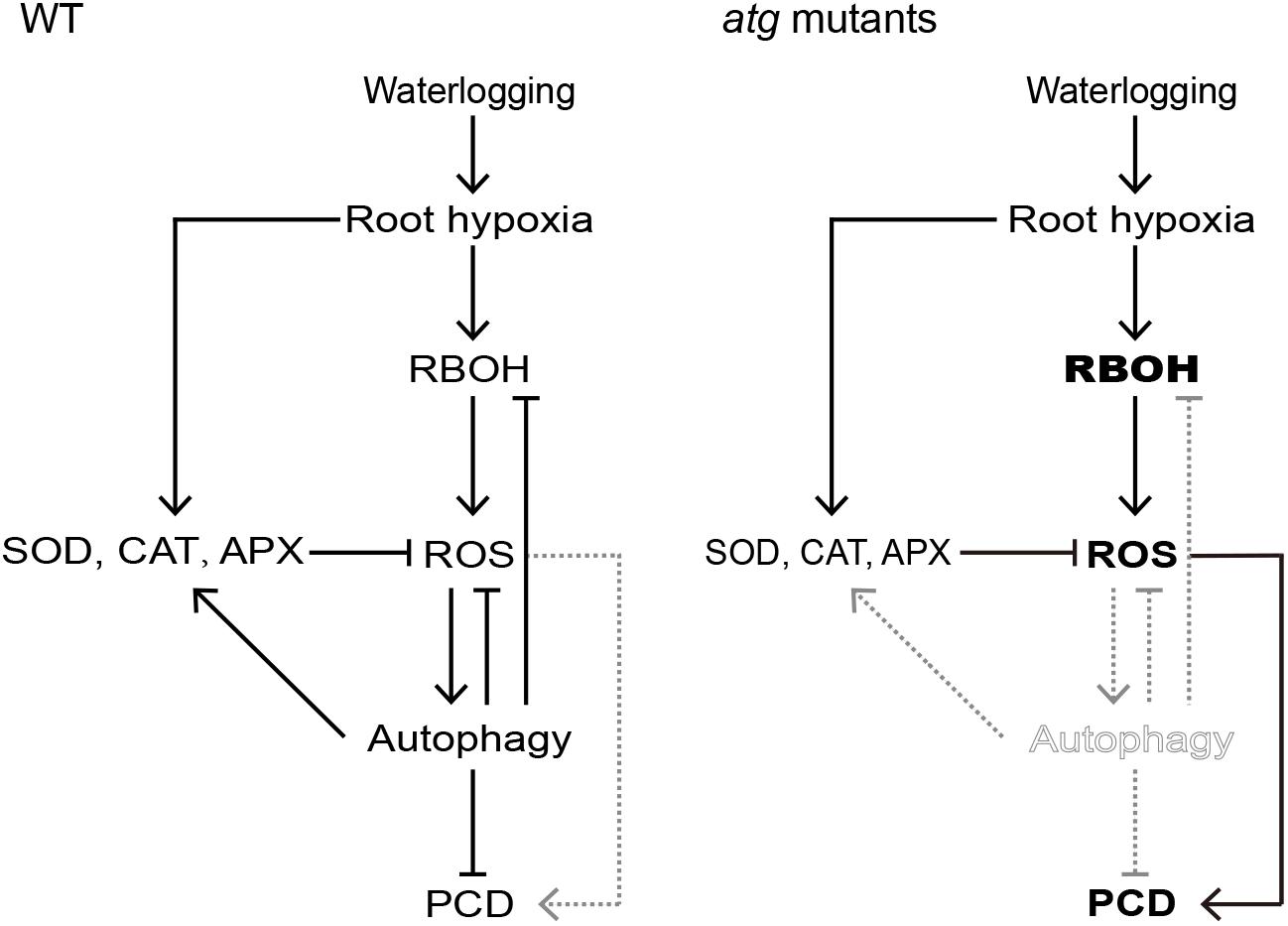
Figure 9. Proposed working model for signaling pathways that regulate PCD in Arabidopsis roots. In waterlogged wild-type roots, hypoxia induces both RBOH expression (leading to an increase in ROS), as well as increasing the expression and activities of ROS-scavenging enzymes CAT, APX, and SOD. The RBOH-dependent increase in ROS activates autophagy which alleviates PCD. However, autophagy deficiency of the atg mutants disrupts the induction of antioxidant enzymes expression and the inhibition of RBOH expression by autophagy. In that case, excessive accumulation of ROS contributes to PCD in atg mutant roots.
BG and ZZ conceived and designed the experiments, and revised the manuscript. BG performed the experiments and wrote the manuscript. ZL and DL performed the part of experiments. All authors discussed the results and revised the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Shi Xiao (Sun Yat-sen University) for GFP-ATG8e and ATG8epro:GUS seeds. We also thank the National Natural Science Foundation of China for its support (Nos. 31471428 and 31871530). We would like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00468/full#supplementary-material
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Atkinson, C. J., Harrison-Murray, R. S., and Taylor, J. M. (2008). Rapid flood-induced stomatal closure accompanies xylem sap transportation of root-derived acetaldehyde and ethanol in Forsythia. Environ. Exp. Bot. 64, 196–205. doi: 10.1016/j.envexpbot.2008.02.001
Bailey-Serres, J., and Voesenek, L. A. C. J. (2008). Flooding stress: acclimations and genetic diversity. Annu. Rev. Plant Biol. 59, 313–339. doi: 10.1146/annurev.arplant.59.032607.092752
Bassham, D. C., Laporte, M., Marty, F., Moriyasu, Y., Ohsumi, Y., Olsen, L. J., et al. (2006). Autophagy in development and stress responses of plants. Autophagy 2, 2–11. doi: 10.4161/auto.2092
Chen, L., Liao, B., Qi, H., Xie, L. J., Huang, L., Tan, W. J., et al. (2015). Autophagy contributes to regulation of the hypoxia response during submergence in Arabidopsis thaliana. Autophagy 11, 2233–2246. doi: 10.1080/15548627.2015.1112483
Contento, A. L., Xiong, Y., and Bassham, D. C. (2005). Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J. 42, 598–608. doi: 10.1111/j.1365-313X.2005.02396.x
Cvetkovska, M., and Vanlerberghe, G. C. (2012). Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol. 195, 32–39. doi: 10.1111/j.1469-8137.2012.04166.x
Dat, J. F., Capelli, N., Folzer, H., Bourgeade, P., and Badot, P. M. (2004). Sensing and signalling during plant flooding. Plant Physiol. Biochem. 42, 273–282. doi: 10.1016/j.plaphy.2004.02.003
Doelling, J. H., Walker, J. M., Friedman, E. M., Thompson, A. R., and Vierstra, R. D. (2002). The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 277, 33105–33114. doi: 10.1074/jbc.M204630200
Dunand, C., Crevecoeur, M., and Penel, C. (2007). Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: possible interaction with peroxidases. New Phytol. 174, 332–341. doi: 10.1111/j.1469-8137.2007.01995.x
Elstner, E. F., and Heupel, A. (1976). Inhibition of nitrite formation from hydroxylammonium chloride: a simple assay for superoxide dismutase. Anal. Biochem. 70, 616–620. doi: 10.1016/0003-2697(76)90488-7
Evans, D. E. (2003). Aerenchyma formation. New Phytol. 161, 35–49. doi: 10.1046/j.1469-8137.2003.00907.x
Eysholdt-Derzsó, E., and Sauter, M. (2017). Root bending is antagonistically affected by hypoxia and ERF-mediated transcription via auxin signaling. Plant Physiol. 175, 412–423. doi: 10.1104/pp.17.00555
Feng, H., Guan, D., Sun, K., Wang, Y., Zhang, T., and Wang, R. (2013). Expression and signal regulation of the alternative oxidase genes under abiotic stresses. Acta Biochim. Biophys. Sin. 45, 985–994. doi: 10.1093/abbs/gmt094
Foreman, J., Demidchik, V., Bothwell, J. H., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Gadjev, I., Stone, J. M., and Gechev, T. S. (2008). Chapter 3: programmed cell death in plants. Int. Rev. Cell Mol. Biol. 270, 87–144. doi: 10.1007/978-1-60761-702-0_18
Gasch, P., Fundinger, M., Müller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28, 160–180. doi: 10.1105/tpc.15.00866
Gechev, T. S., Van, B. F., Stone, J. M., Denev, I., and Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. doi: 10.1002/bies.20493
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Giraud, E., Ho, L. H., Clifton, R., Carroll, A., Estavillo, G., Tan, Y. F., et al. (2008). The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol. 147, 595–610. doi: 10.1104/pp.107.115121
Han, S., Yu, B., Wang, Y., and Liu, Y. (2011). Role of plant autophagy in stress response. Protein Cell 2, 784–791. doi: 10.1007/s13238-011-1104-4
Hunt, P. W., Klok, E. J., Trevaskis, B., Watts, R. A., Ellis, M. H., Peacock, W. J., et al. (2002). Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 17197–17202. doi: 10.1073/pnas.212648799
Hussain, S., Khan, F., Hussain, H. A., and Nie, L. (2016). Physiological and biochemical mechanisms of seed priming-induced chilling tolerance in rice cultivars. Front. Plant Sci. 7:116. doi: 10.3389/fpls.2016.00116
Ichimura, Y., Imamura, Y., Emoto, K., Umeda, M., Noda, T., and Ohsumi, Y. (2004). In vivo and in vitro reconstitution of Atg8 conjugation essential for autophagy. J. Biol. Chem. 279, 40584–40592. doi: 10.1074/jbc.M405860200
Izumi, M., Ishida, H., Nakamura, S., and Hidema, J. (2017). Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 29, 377–394. doi: 10.1105/tpc.16.00637
Jackson, M. B., and Armstrong, W. (1999). Formation of aerenchyma and the processes of plant ventilation in relation to soil flooding and submergence. Plant Biol. 1, 274–287. doi: 10.1111/j.1438-8677.1999.tb00253.x
Kalyanaraman, B., Darley-Usmar, V., Davies, K. J. A., Dennery, P. A., Forman, H. J., Grisham, M. B., et al. (2012). Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic. Biol. Med. 52, 1–6. doi: 10.1016/j.freeradbiomed.2011.09.030
Klionsky, D. J. (2005). The molecular machinery of autophagy: unanswered questions. J. Cell Sci. 118, 7–18. doi: 10.1242/jcs.01620
Klionsky, D. J. (2007). Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 8, 931–937. doi: 10.1038/nrm2245
Klionsky, D. J., and Ohsumi, Y. (1999). Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 1–32. doi: 10.1146/annurev.cellbio.15.1.1
Kreuzwieser, J., Hauberg, J., Howell, K. A., Carroll, A., Rennenberg, H., Millar, A. H., et al. (2009). Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiol. 149, 461–473. doi: 10.1104/pp.108.125989
Kreuzwieser, J., and Rennenberg, H. (2014). Molecular and physiological responses of trees to waterlogging stress. Plant Cell Environ. 37, 2245–2259. doi: 10.1111/pce.12310
Li, F., and Vierstra, R. D. (2012). Autophagy: a multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 17, 526–537. doi: 10.1016/j.tplants.2012.05.006
Li, L., Tan, J., Miao, Y., Lei, P., and Zhang, Q. (2015). ROS and autophagy: interactions and molecular regulatory mechanisms. Cell Mol. Neurobiol. 35, 615–621. doi: 10.1007/s10571-015-0166-x
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Liu, F., VanToai, T., Moy, L. P., Bock, G., Linford, L. D., and Quackenbush, J. (2005). Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol. 137, 1115–1129. doi: 10.1104/pp.104.055475
Liu, Y., and Bassham, D. C. (2012). Autophagy: pathways for self-eating in plant cells. Annu. Rev. Plant Biol. 63, 215–237. doi: 10.1146/annurev-arplant-042811-105441
Liu, Y., Schiff, M., Czymmek, K., Tallóczy, Z., Levine, B., and Dinesh-Kumar, S. P. (2005). Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577. doi: 10.1016/j.cell.2005.03.007
Liu, Y., Xiong, Y., and Bassham, D. C. (2009). Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5, 954–963. doi: 10.4161/auto.5.7.9290
Loreti, E., Poggi, A., Novi, G., Alpi, A., and Perata, P. (2005). A genome-wide analysis of the effects of sucrose on gene expression in Arabidopsis seedlings under anoxia. Plant Physiol. 137, 1130–1138. doi: 10.1104/pp.104.057299
Maxwell, D. P., Wang, Y., and Mcintosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U.S.A. 96, 8271–8276. doi: 10.1073/pnas.96.14.8271
Minina, E. A., Bozhkov, P. V., and Hofius, D. (2014). Autophagy as initiator or executioner of cell death. Trends Plant Sci. 19, 692–697. doi: 10.1016/j.tplants.2014.07.007
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Morales, J., Kadota, Y., Zipfel, C., Molina, A., and Torres, M. A. (2016). The Arabidopsis NADPH oxidases RbohD and RbohF display differential expression patterns and contributions during plant immunity. J. Exp. Bot. 67, 1663–1676. doi: 10.1093/jxb/erv558
Mühlenbock, P., Plaszczyca, M., Plaszczyca, M., Mellerowicz, E., and Karpinski, S. (2007). Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 19, 3819–3830. doi: 10.1105/tpc.106.048843
Mustroph, A., Lee, S. C., Oosumi, T., Zanetti, M. E., Yang, H., Ma, K., et al. (2010). Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152, 1484–1500. doi: 10.1104/pp.109.151845
Patel, S., Caplan, J., and Dineshkumar, S. P. (2006). Autophagy in the control of programmed cell death. Curr. Opin. Plant Biol. 9, 391–396. doi: 10.1016/j.pbi.2006.05.007
Patterson, B. D., MacRae, E. A., and Ferguson, I. B. (1984). Estimation of hydrogen peroxide in plant extracts using titanium (IV). Anal. Biochem. 139, 487–492. doi: 10.1016/0003-2697(84)90039-3
Phillips, A. R., Suttangkakul, A., and Vierstra, R. D. (2008). The ATG12-Conjugating Enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 178, 1339–1353. doi: 10.1534/genetics.107.086199
Pucciariello, C., and Perata, P. (2017). New insights into reactive oxygen species and nitric oxide signalling under low oxygen in plants. Plant Cell Environ. 40, 473–482. doi: 10.1111/pce.12715
Santaniello, A., Loreti, E., Gonzali, S., Novi, G., and Perata, P. (2014). A reassessment of the role of sucrose synthase in the hypoxic sucrose-ethanol transition in Arabidopsis. Plant Cell Environ. 37, 2294–2302. doi: 10.1111/pce.12363
Sauter, M. (2013). Root responses to flooding. Curr. Opin. Plant Biol. 16, 282–286. doi: 10.1016/j.pbi.2013.03.013
Scherz-Shouval, R., Shvets, E., Fass, E., Shorer, H., Gil, L., and Elazar, Z. (2007). Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760. doi: 10.1038/sj.emboj.7601623
Shin, J. H., Yoshimoto, K., Ohsumi, Y., Jeon, J., and An, G. (2009). OsATG10b, an autophagosome component, is needed for cell survival against oxidative stresses in rice. Mol. Cells 27, 67–74. doi: 10.1007/s10059-009-0006-2
Smith, C. A., Melino, V. J., Sweetman, C., and Soole, K. L. (2009). Manipulation of alternative oxidase can influence salt tolerance in Arabidopsis thaliana. Physiol. Plant. 137, 459–472. doi: 10.1111/j.1399-3054.2009.01305.x
Soh, N. (2006). Recent advances in fluorescent probes for the detection of reactive oxygen species. Anal. Bioanal. Chem. 386, 532–543. doi: 10.1007/s00216-006-0366-9
Suzuki, N., Miller, G., Morales, J., Shulaev, V., Torres, M. A., and Mittler, R. (2011). Respiratory burst oxidases: the engines of ROS signalling. Curr. Opin. Plant Biol. 14, 691–699. doi: 10.1016/j.pbi.2011.07.014
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91 phox). Plant J. 14, 365–370. doi: 10.1046/j.1365-313X.1998.00136.x
Umbach, A. L., Fiorani, F., and Siedow, J. N. (2005). Characterization of transformed Arabidopsis with altered alternative oxidase levels and analysis of effects on reactive oxygen species in tissue. Plant Physiol. 139, 1806–1820. doi: 10.1104/pp.105.070763
van Dongen, J. T., Fröhlich, A., Ramírez-Aguilar, S. J., Schauer, N., Fernie, A. R., Erban, A., et al. (2009). Transcript and metabolite profiling of the adaptive response to mild decreases in oxygen concentration in the roots of Arabidopsis plants. Ann. Bot. 103, 269–280. doi: 10.1093/aob/mcn126
Wang, F., Chen, Z. H., Liu, X., Colmer, T. D., Zhou, M., and Shabala, S. (2016). Tissue-specific root ion profiling reveals essential roles of the CAX and ACA calcium transport systems in response to hypoxia in Arabidopsis. J. Exp. Bot. 67, 3747–3762. doi: 10.1093/jxb/erw034
Wiengweera, A., Greenway, H., and Thomson, C. J. (1997). The use of agar nutrient solution to simulate lack of convection in waterlogged soils. Ann. Bot. 80, 115–123. doi: 10.1006/anbo.1996.0405
Wrzaczek, M., Brosche, M., and Kangasjarvi, J. (2013). ROS signaling loops - production, perception, regulation. Curr. Opin. Plant Biol. 16, 575–582. doi: 10.1016/j.pbi.2013.07.002
Xiong, Y., Contento, A. L., Nguyen, P. Q., and Bassham, D. C. (2007). Degradation of oxidized proteins by autophagy during oxidative stress in Arabidopsis. Plant Physiol. 143, 291–299. doi: 10.1104/pp.106.092106
Xu, Q. T., Yang, L., Zhou, Z. Q., Mei, F. Z., Qu, L. H., and Zhou, G. S. (2013). Process of aerenchyma formation and reactive oxygen species induced by waterlogging in wheat seminal roots. Planta 238, 969–982. doi: 10.1007/s00425-013-1947-4
Yamauchi, T., Yoshioka, M., Fukazawa, A., Mori, H., Nishizawa, N. K., Tsutsumi, N., et al. (2017). An NADPH oxidase RBOH functions in rice roots during lysigenous aerenchyma formation under oxygen-deficient conditions. Plant Cell 29, 775–790. doi: 10.1105/tpc.16.00976
Yang, L., Zhang, J., He, J., Qin, Y. Y., Hua, D. P., Duan, Y., et al. (2014). ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 10:e1004791. doi: 10.1371/journal.pgen.1004791
Zhou, J., Wang, J., Yu, J. Q., and Chen, Z. (2014). Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 5:174. doi: 10.3389/fpls.2014.00174
Keywords: reactive oxygen species, respiratory burst oxidase homolog, autophagy, programmed cell death, waterlogging
Citation: Guan B, Lin Z, Liu D, Li C, Zhou Z, Mei F, Li J and Deng X (2019) Effect of Waterlogging-Induced Autophagy on Programmed Cell Death in Arabidopsis Roots. Front. Plant Sci. 10:468. doi: 10.3389/fpls.2019.00468
Received: 01 December 2018; Accepted: 28 March 2019;
Published: 11 April 2019.
Edited by:
Jin-Song Zhang, Institute of Genetics and Developmental Biology (CAS), ChinaReviewed by:
Olga V. Voitsekhovskaja, Komarov Botanical Institute (RAS), RussiaCopyright © 2019 Guan, Lin, Liu, Li, Zhou, Mei, Li and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhuqing Zhou, emhvdXpodXFpbmdAbWFpbC5oemF1LmVkdS5jbg==
†Present address: Bin Guan, National Key Laboratory of Plant Molecular Genetics, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai, China
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.