
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 05 April 2019
Sec. Plant Development and EvoDevo
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00362
Phospholipases (PLs) are lipid-hydrolyzing enzymes known to have diverse signaling roles during plant abiotic and biotic stress responses. They catalyze lipid remodeling, which is required to generate rapid responses of plants to environmental cues. Moreover, they produce second messenger molecules, such as phosphatidic acid (PA) and thus trigger or modulate signaling cascades that lead to changes in gene expression. The roles of phospholipases in plant abiotic and biotic stress responses have been intensively studied. Nevertheless, emerging evidence suggests that they also make significant contributions to plants’ cellular and developmental processes. In this mini review, we summarized recent advances in the study of the cellular and developmental roles of phospholipases in plants.
Phospholipids are major components of biological membranes and have important signaling functions (Hou et al., 2016). Phospholipases (PLs) represent a ubiquitous family of proteins that cleave various bonds in phospholipids to maintain membrane lipid homeostasis and stability (Wang et al., 2012). Plant PLs are divided into three major classes: A, C, and D. These classes are distinguished by the type of catalytic reactions they perform. Phospholipases A (PLAs) hydrolyze acyl groups from the sn-1 and sn-2 positions of phospholipids and produce free fatty acids and lysophospholipids. Phospholipases C (PLCs) catalyze the hydrolysis of the entire phosphoryl head group, cleaving the glycerophosphate bond and thus yielding diacylglycerol and the phosphorylated head group. Phospholipases D (PLDs) cleave the phosphodiester (phosphorus-oxygen) bond in the phospholipid, yielding phosphatidic acid (PA; Chen et al., 2011). The hydrolytic activity of PLs is often accompanied by the production of lipid second messengers, which mediate plant responses to external stimuli. Notably, PLs have the ability to bind and modulate other regulatory proteins, including the subunits of G-protein complexes and sphingosine kinase (Mishra et al., 2006; Guo et al., 2012; Pandey, 2016). Their role in membrane lipid remodeling and the subsequent regulation of membrane physical properties makes PLs and their hydrolytic products key regulators of endomembrane organization. In addition, PLs also control the organization and dynamics of the cytoskeleton (Zhang et al., 2012; Pleskot et al., 2013). They are involved in jasmonic acid (JA) biosynthesis (Ishiguro et al., 2001) and also have notable impact on plant responses to such hormones as abscisic acid (ABA; Mishra et al., 2006; Peters et al., 2010; Uraji et al., 2012), auxins (Scherer et al., 2012), and cytokinins (Repp et al., 2004). The interplay of PLs with hormones strongly regulates plant developmental processes, as indicated by the phenotypes of mutants with deficient PL expression (e.g., Labusch et al., 2013). Through all of these mechanisms, PLs are involved in plant responses to drought (Sang et al., 2001), chilling (Huo et al., 2016), heat (Krčková et al., 2015), salt (Arisz and Munnik, 2011), osmotic pressure (Hong et al., 2008), heavy metal stress (Pejchar et al., 2015), phosphorus deficiency (Cruz-Ramírez et al., 2006; Li et al., 2006b), and pathogens (Zhao, 2015). Last but not least, PLs modulate plant developmental processes, such as pollen tube growth (Potocký et al., 2003; Kim et al., 2011), embryogenesis, and root and leaf development (Wimalasekera et al., 2010), as well as gravitropism (Lee, 2003).
The heterogeneity of PLs indicates that they carry specific developmental functions with different degrees of redundancy within or across particular PL subfamilies. The involvement of PLs in the abiotic and biotic stress responses of plants has been frequently and comprehensively reviewed in recent studies (Wang, 2014; Hong et al., 2016), while their role in plant developmental processes is largely underestimated. This mini review aimed to summarize the recent advances in the study of the roles of PLs in plant developmental processes, which have been mostly related to their cellular functions.
PLAs are a multigene family of enzymes that are subdivided into PLA1s, secretory PLAs (PLA2s) and patatin-like PLAs (pPLAs). They show affinities for diverse phospholipid substrates, including phosphatidylcholine (PC), monogalactosyldiacylglycerol, digalactosyldiacylglycerol, and triacylglycerol (Chen et al., 2013).
PLA1 enzymes are calcium-independent and catalyze the hydrolysis of acyl groups from the sn-1 position of phospholipids (Chen et al., 2013). Their developmental expression and localization is very elusive. PLA1Iα1 is targeted to the chloroplasts and is expressed specifically in the stamens before flower opening (Ishiguro et al., 2001). PLA1III is expressed at high levels in young seedlings and localized to the mitochondria (Figure 2; Seo et al., 2011). Some members of this subfamily are expressed at high levels in the pollen (PLA1Iα1, PLA1Iα2, PLA1IIα, and PLA1IIβ), petals and sepals (PLA1Iγ3), seed coats (PLA1Iα2), inflorescence stems (PLA1Iα1, PLA1Iα2, PLA1Iβ1, and PLA1IIβ), and xylem of the hypocotyl stele (PLA1Iγ3) in Arabidopsis (Figure 1). They show increased expression during young rosette development (PLA1Iα2), silique maturation, and senescence (PLA1Iβ1; Genvestigator). These findings indicate that these PLA1s have prominent roles in generative organ development (Figure 1). For example, processes such as anther dehiscence, pollen maturation, and flower opening are controlled by the chloroplast-associated PLA1Iα1 (Figure 2), which was proposed to prefer PC as a substrate (Ishiguro et al., 2001). It was shown experimentally that this regulation is mediated by the participation of PLA1Iα1 in chloroplast-dependent JA biosynthesis (Figure 3A). Notably, PLAs catalyze the release of α-linolenic acid, which is a precursor of JA, from membrane phospholipids (Wasternack and Song, 2016). This supports the role of PLAs in JA-controlled developmental processes. Remarkably, PLA1Iα1 is not only linked to JA, as its expression is also dependent on AUXIN RESPONSE FACTOR 6 and 8 (Tabata et al., 2010), ubiquitin ligase DAF (Peng et al., 2013), and the floral homeotic gene AGAMOUS (Ito et al., 2007). Nevertheless, PLA1 functions are not restricted only to generative organ development. The vacuolar membrane-localized PA-PLA1 (also known as SHOOT GRAVITROPISM 2; SGR2) is important in the regulation of shoot gravitropism. This role is linked to vacuole biogenesis, a process that is inevitably involved in amyloplast redistribution in the shoot endodermis (Kato et al., 2002; Morita et al., 2002). Recently, a similar function of PA-PLA1 was discovered in the zygote, where it controls the polar vacuole positioning to ensure asymmetrical cell division (Kimata et al., 2019).
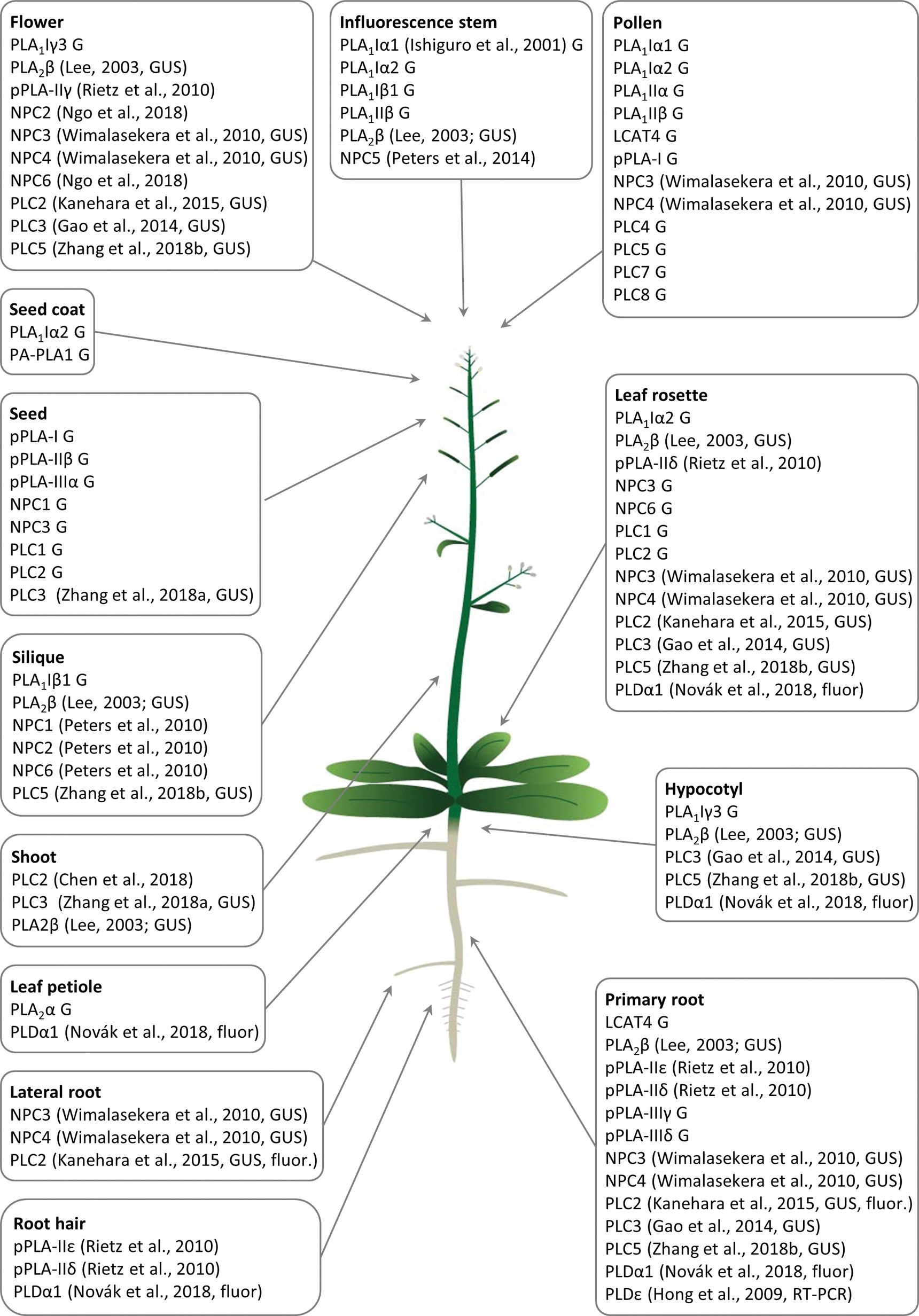
Figure 1. Overview of Arabidopsis tissue- and organ-specific expression patterns and enrichment of diverse phospholipases important for the development of respective plant organs and tissues. G means that the gene expression data were retrieved from the Genvestigator database, GUS means that the expression data represented the results of promoter-GUS reporter gene assays, and fluor means that the expression data represented microscopic observations of fluorescently tagged phospholipases. Citations to the relevant original articles are provided.
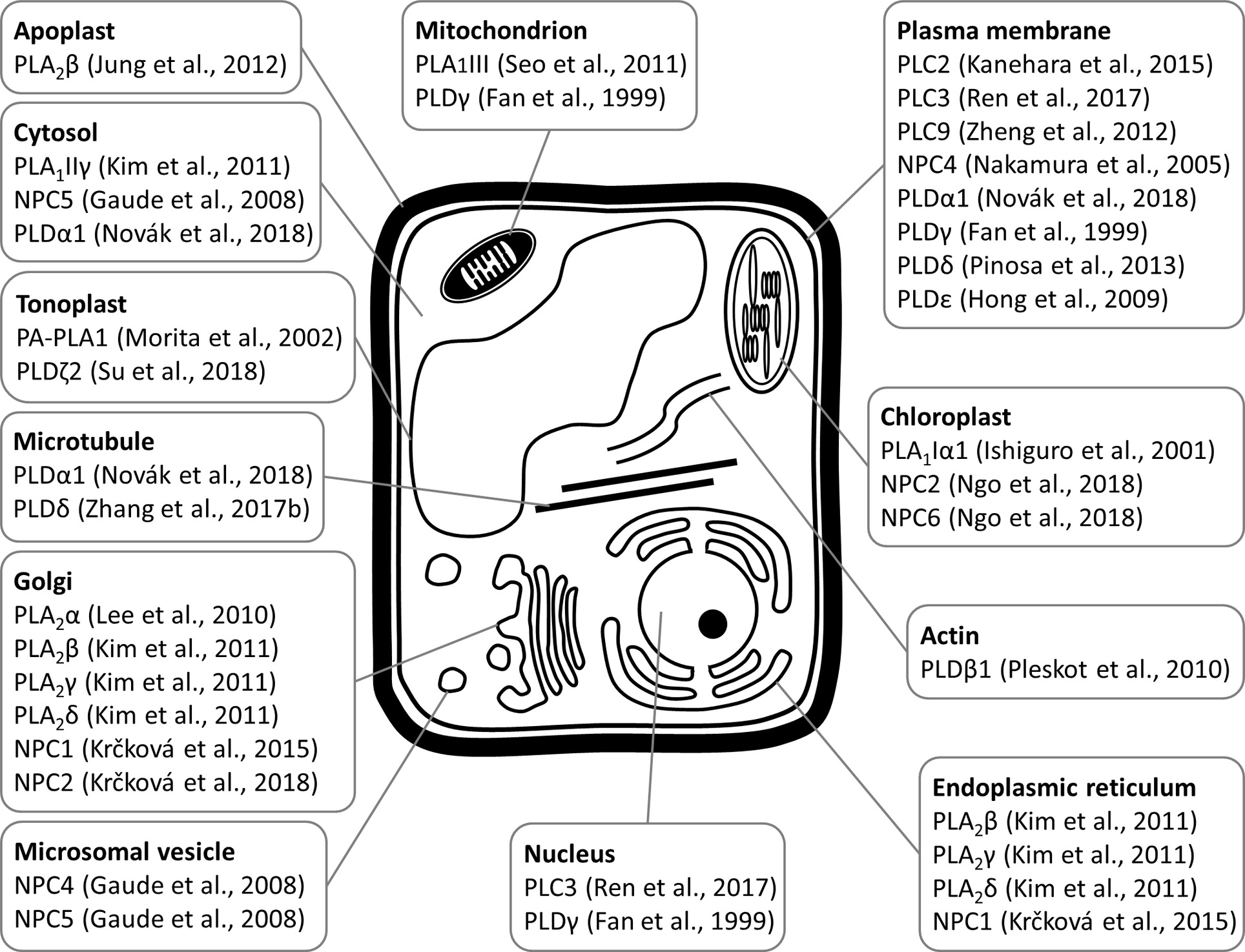
Figure 2. Schematic overview of subcellular localizations of plant phospholipases to diverse subcellular compartments and organelles, as revealed by experimental approaches with relevant citations to the original articles.
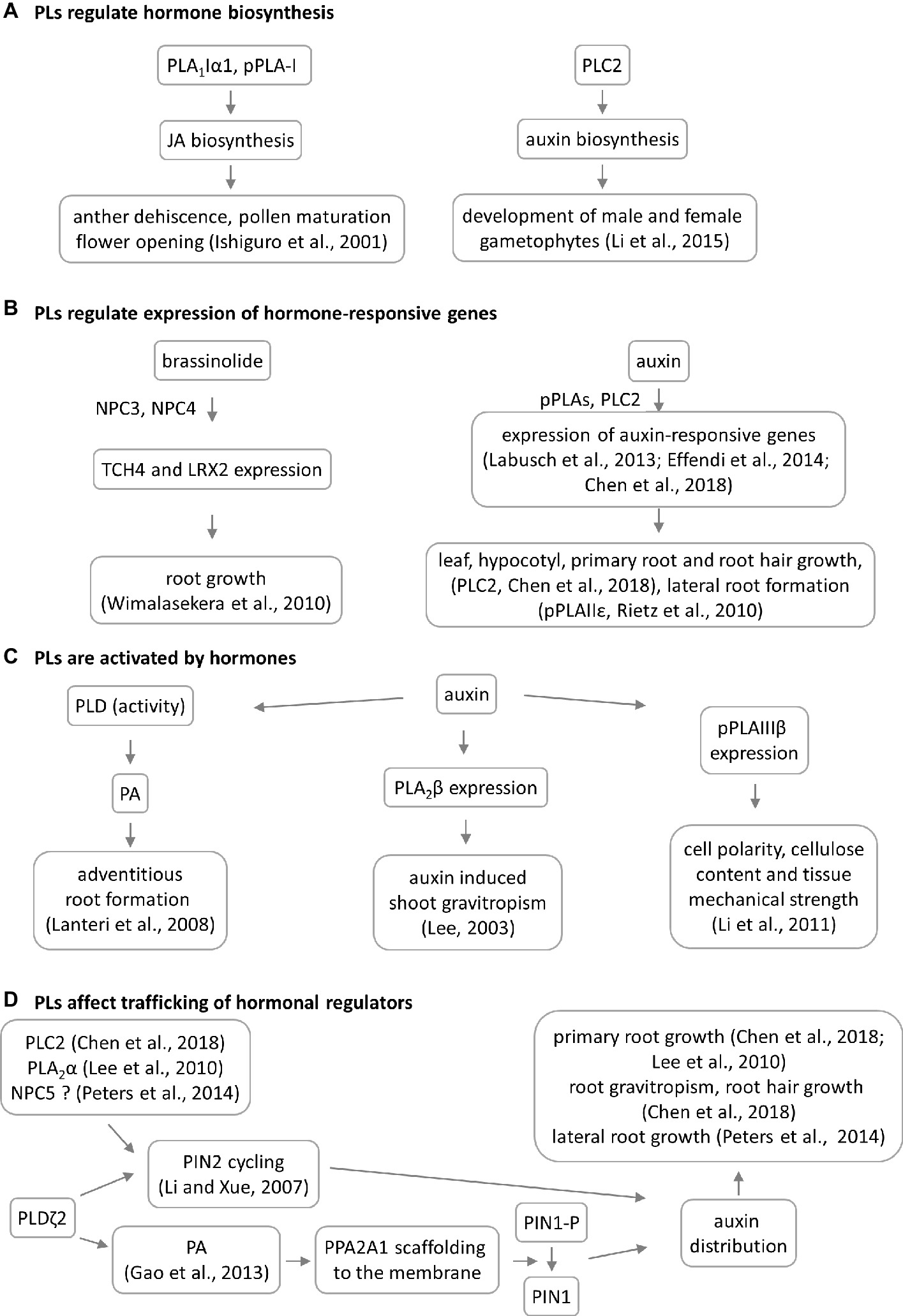
Figure 3. Overview of crosstalk between phospholipases (PLs) and hormones during plant development. Four models of PL–hormone interaction are proposed. (A) PLs are involved in jasmonic acid and auxin biosynthesis. This has consequences in reproductive organ development. (B) PLs are regulating the hormone-induced gene expression having impact on root and shoot development, TCH4 = xyloglucan endotransglycosylase 4, LRX2 = leucine-rich extensin 1. (C) PLs are activated by hormones leading to developmental phenotypes. In some cases, the PL mutation does not lead to phenotypic manifestation. (D) Particular PLs have impact on the auxin distribution through 2 different mechanisms. NPC5 promotes lateral root formation likely through regulating auxin transport and distribution (Peters et al., 2014), which occurs via PIN2 cycling.
The gene FSE1 encodes a PL-like protein that is homologous to the PA-preferring PLA1s and controls the synthesis of galactolipids in the rice endosperm. It is important for seed and endosperm development (Long et al., 2018). Furthermore, PLA1s may also have root-specific functions. A pepper PLA1 is expressed selectively in young, growing roots and regulates growth rates and root and leaf tissue patterning. The authors of the previous study (Seo et al., 2008) that examined it proposed that this PLA1 may be involved in cell cycle control via the signaling functions of its hydrolytic products. Recently, a beneficial biotechnological application of this PLA1 was presented, as the yields of transgenic rice plants overexpressing pepper PLA1 were increased relative to normal plants (Park et al., 2016).
Similar to PLA1s, only limited information exists on the developmental expression of most PLA2s. These are also called secretory PLAs and catalyze phospholipid hydrolysis at the sn-2 position. PLA2s are expressed at high levels in pollen (lecithin-cholesterol acyltransferase-like 4; LCAT4), leaf petioles (PLA2α), the root pericycle (LCAT4; Figure 1), and flowers (PLA2γ; Bahn et al., 2003). They produce lysophospholipids (Lee et al., 2005; Chen et al., 2013) capable of inducing local positive membrane curvature (Boutté and Moreau, 2014), which is important for endomembrane morphogenesis. PLA2α, PLA2β, PLA2γ, and PLA2δ are localized in the Golgi apparatus and endoplasmic reticulum (ER; Lee et al., 2010; Kim et al., 2011), while PLA2β is secreted into the apoplast (Bahn et al., 2003; Lee, 2003). Specifically, PLA2β is gradually translocated to the apoplast during leaf maturation (Figure 2; Jung et al., 2012).
PLA2β controls cell elongation in the leaf petioles and inflorescence stems (Lee, 2003). It is also implicated in shoot (but not root) gravitropism via an auxin-dependent mechanism, which is seemingly different from that of the abovementioned PA-PLA1 (Figure 3; Lee, 2003; Kimata et al., 2019). The interplay between PLA2s and auxin was previously explained mechanistically. It was found that PLA2α and the product of its lipid-hydrolyzing activity, lysophosphatidyl ethanolamine (LPE), are both required for the trafficking of PINs (auxin efflux transporters) to the plasma membrane (PM). This is required for proper root growth in Arabidopsis (Figure 3D; Lee et al., 2010). Interestingly, PLA2-mediated production of LPE is also important for the proper organization of the endomembrane system during pollen development, germination, and elongation (Kim et al., 2011). This finding enriched the list of important signaling lipids involved in determining polar tip growth.
Patatin-related PLAs (pPLAs) hydrolyze a broad range of phospholipids at both the sn-1 and sn-2 positions and contain a patatin-catalytic domain. In addition, they can also use galactolipids as substrates. The pPLAs in Arabidopsis are classified into three subfamilies (pPLA-I, pPLA-II, and pPLA-III) (Scherer et al., 2010; Li and Wang, 2014), although the individual members of these subfamilies might possess different substrate specificities (Rietz et al., 2010). Arabidopsis pPLA-IIε and pPLA-IIδ are expressed at high levels in the roots and root hairs, and pPLA-IIδ is expressed also in the leaves and cotyledons. Expression of pPLA-IIγ is restricted to the generative organs (Rietz et al., 2010). Other pPLAs are expressed at high levels in the pollen (pPLA-I), seeds (pPLA-I, pPLA-IIβ, and pPLA-IIIα), and roots (pPLA-IIIγ and pPLA-IIIδ; Figure 1). The overexpression of pPLA-IIIδ leads to stiff inflorescence stems, thicker leaves, shorter siliques, larger seeds, more rounded flowers, and delayed plant growth (Huang, 2001).
There is solid evidence of the occurrence of crosstalk between pPLAs and plant hormones. While pPLA-I is involved in jasmonic acid biosynthesis, pPLA-IIδ and pPLAIIIβ expression is strongly induced by auxin (Rietz et al., 2010; Li et al., 2011). Interestingly, it was also discovered that pPLAs are important for the expression of early auxin-responsive genes in Arabidopsis (Figure 3B; Labusch et al., 2013; Effendi et al., 2014). The knock-out mutants pplaIIIβ and pplaIIε do not exhibit auxin-related phenotypes under normal growth conditions (Rietz et al., 2010; Li et al., 2011), while the pplaIIIδ mutant consistently showed higher sensitivity to auxin (Labusch et al., 2013). Crosstalk between pPLAs and auxin likely affects cell polarity, cellulose content, and tissue mechanical strength in Arabidopsis, rice, and ginseng (Figure 3C; Li et al., 2011; Liu et al., 2015; Jang and Lee, 2019). Since cellulose deposition is tightly controlled by microtubules, it would be extremely challenging to further investigate the interplay between pPLAIIIβ-induced lipid remodeling, cellulose deposition, and microtubule dynamics. This assumption is also substantiated by the fact that pPLAIIIδ overexpression inhibited longitudinal growth but promoted transverse growth, in most organs of Arabidopsis and Brassica napus (Dong et al., 2014). Growth polarity appears to be under the control of auxin and ethylene, and these plant hormones are involved in growth reprogramming through modifications of microtubule orientation governed by katanin (Lindeboom et al., 2013; Sassi et al., 2014; Luptovčiak et al., 2017). Proof of this concept was provided by the differential proteomic analysis of pPLAIIIδ-overexpressing plants, which demonstrated the impact of this protein on the abundance of the microtubule-associated protein MAP18, which modulates directional cell growth (Zheng et al., 2014).
The phospholipid-hydrolyzing activity of PLCs on the phosphodiester bond results in the production of diacylglycerol (DAG) and head group (Singh et al., 2015; Hong et al., 2016). DAG may then be converted into PA by DAG kinase (Arisz et al., 2009), so the developmental impacts of PLCs might also be assigned to their product, PA. This was previously tested by a pharmacological approach employing PLC and PLD inhibitors (den Hartog et al., 2001). Nevertheless, PLC- and PLD-mediated PA production pathways may differ in their functions, as exemplified by their effects on pollen tube growth. Pharmacological inhibition of both PLC and PLD caused pollen tube growth to cease, while cellular processes essential for polarized growth exhibited different sensitivities to PLC and PLD inhibitors. Vacuole organization was heavily altered by PLC inhibitor, while actin dynamics was affected specifically by pharmacological inhibition of PLD (Pleskot et al., 2012). PLCs in plants can be divided into three groups according to their substrate specificity and cellular functions. Non-specific PLCs (NPCs; in Arabidopsis these were designated as NPC1-NPC6) act on common phospholipids, such as PC and PE (Pokotylo et al., 2013). Phosphoinositide-specific PLCs (PI-PLCs; in Arabidopsis designated as PLC1-PLC9) hydrolyze phosphoinositides, such as phosphatidylinositol 4,5-bisphosphate (PIP2) (Tasma et al., 2008; Heilmann, 2016). Glycosylphosphatidylinositol (GPI)-specific PLCs cleave GPI anchors from proteins (Hong et al., 2016). To our knowledge, no PL with GPI-PLC activity has been characterized in plants so far.
Previous studies reported that NPC1, NPC2, and NPC6 were expressed at high levels in siliques, while NPC3 and NPC4 were expressed at high levels in roots and inflorescences. NPC5 expression is similar to that of NPC3 and NPC4, although its expression is absent in roots (Peters et al., 2010, 2014; Wimalasekera et al., 2010). The expression of NPC2 and NPC6 is developmentally regulated, and at later stages, they show rather limited expression in vegetative tissues but pronounced expression in reproductive organs (Ngo et al., 2018). NPC3 and NPC4 are expressed in the young anthers, particularly in pollen sac tissues (Wimalasekera et al., 2010). According to the Genevestigator database, non-specific PLs (NPCs) are ubiquitously expressed in Arabidopsis and show high levels of expression in embryos (NPC2 and NPC3) and in the xylem (NPC3). They are also expressed during seed (NPC1 and NPC3) and rosette development (NPC3 and NPC6; Figure 1). Subcellularly, they localize mainly to membranous compartments (Figure 2). They have been found in the endoplasmic reticulum (NPC1; Krčková et al., 2015) and Golgi apparatus (NPC1 and NPC2; Krčková et al., 2015, 2018; Figure 2). In the leaves, NPC2 and NPC6 were recently localized in the plastids (Ngo et al., 2018). NPC4 and NPC5 are commonly localized in the microsomal fraction, and NPC5 in the cytosol, while NPC4 also associates with the plasma membrane (Figure 2; Nakamura et al., 2005; Gaude et al., 2008; Peters et al., 2014).
Experimental data show that crosstalk between NPCs and plant hormones may control gametophyte development and root growth and affect tissue mechanical strength. Consistently with expression patterns, NPC2 and NPC6 (hydrolyzing PC and PE) are redundantly required for male and female gametophyte development in Arabidopsis (Ngo et al., 2018). Gametophyte development also requires the expression of the PI-dependent PLC2, suggesting that some PLs may act together in this process (Figure 1). Other NPCs, such as NPC3 and NPC4, are inducible by auxin and control primary root growth and lateral root initiation under brassinolide regulation (Figure 3B; Wimalasekera et al., 2010) and root hair growth during Pi starvation (Su et al., 2018). In addition, NPC4 positively modulates ABA responses during seed germination and root elongation (Peters et al., 2010), while NPC5 positively regulates lateral root formation, perhaps by promoting auxin signaling (Peters et al., 2014).
According to expression studies based on the technique using the promoter-ß-glucuronidase (GUS), PI-PLCs, including PLC2, PLC3, and PLC5, are expressed throughout the whole plant but are mainly restricted to the vasculature. PLC2 and PLC5, unlike PLC3, show increased expression in the root tips, but all three PLCs are expressed at high levels in the reproductive organs (Kanehara et al., 2015; Zhang et al., 2018a,b). Specifically, PLC2 shows tissue-specific expression in the embryo sac during female gametophyte development, including in the maternal tissues of the ovule at the chalazal pole and in the embryo dermatogen at the late globular stage (di Fino et al., 2017).
In accordance with the abovementioned experimental results, Genevestigator data confirmed the high expression levels of PI-PLCs in the pollen (PLC4, PLC5, PLC7, and PLC8), embryos (PLC7), and phloem (PLC3). PLC2, which is expressed at the highest levels among the PLCs, shows elevated expression together with that of PLC1 during rosette development and seed formation (Figure 1). In agreement with this finding, PLC2 represents a major PL catalyzing PIP2 hydrolysis (Kanehara et al., 2015). PI-PLCs (PLC2, PLC3, and PLC9) localize to the PM (Zheng et al., 2012; Kanehara et al., 2015; di Fino et al., 2017; Ren et al., 2017), while PLC3 is localized in the nucleus (Figure 2; Ren et al., 2017).
Genetic depletion of PI-PLCs leads, in some cases, to generally reduced plant growth and a dwarf phenotype (Table S1). This is obvious for PLC2 (Kanehara et al., 2015; Chen et al., 2018) and PLC5 (Zhang et al., 2018b), while deficiencies in PLC3 (Zhang et al., 2018a) and PLC4 (Xia et al., 2017) show either very slight growth retardation or a wild type-like phenotype. Arabidopsis PLC2 is implicated in auxin biosynthesis, which controls the development of male and female gametophytes (Figure 3A; Li et al., 2015, di Fino et al., 2017) and roots and shoots (Figure 1; Kanehara et al., 2015, Chen et al., 2018), as well as the early stages of embryogenesis (di Fino et al., 2017). In addition, PLC2 influences also the polar distribution of auxin efflux carrier PIN2, which thus affects root hair formation and gravitropism (Figure 3D; Chen et al., 2018). These roles are likely linked to the known roles of phosphoinositides in membrane transport (Noack and Jaillais, 2017). The mode of action of Arabidopsis PLC3 is different from that of PLC2, and plc3 knock-down mutants show no defects in their fertility and generative organ development. PLC3 controls lateral root development and seed germination, and it promotes ABA signaling (Zhang et al., 2018a). In addition, PLC3 functions redundantly with PLC9 in Arabidopsis thermotolerance (Gao et al., 2014). Importantly, PLCs (due to their phosphoinositide-hydrolyzing and PA-producing activity) control developmental processes that depend on membrane transport, such as the polar tip growth of root hairs and pollen tubes (Dowd et al., 2006; Helling et al., 2006; Pleskot et al., 2012; Hong et al., 2016). Some authors have suggested that they are also associated with the actin cytoskeleton (Huang and Crain, 2009; Andreeva et al., 2010) and thus serve as proteins connecting the actin cytoskeleton and membrane transport. Finally, pharmacological experiments revealed that PLCs and PLDs are determinants of asymmetrical cell divisions but not the cell division plane orientation, during stomatal development in maize (Apostolakos et al., 2008).
PLDs catalyze the hydrolysis of structural glycerophospholipids (e.g., PC, PE, and phosphatidylglycerol), producing PA and free soluble head groups (e.g., choline or ethanolamine) (Selvy et al., 2011; Hong et al., 2016). Plant PLDs can be subdivided into six classes: α (α1, α2, and α3), β (β1 and β2), γ (γ1, γ2, and γ3), δ, ε, and ζ (ζ1 and ζ2) (Hong et al., 2016). According to published data, they are mostly localized to the PM (PLDγ, PLDε, and PLDδ; Figure 2; Fan et al., 1999; Hong et al., 2009; Pinosa et al., 2013), tonoplast (PLDζ2; Su et al., 2018), nucleus and mitochondria (PLDγ; Fan et al., 1999), and cytoplasm (PLDα1; Novák et al., 2018; Figure 2). PLDα1 is capable of relocalization to the PM under conditions of salt stress (Figure 2; Novák et al., 2018). PLDζ2 is localized to the tonoplast, and upon phosphate deprivation, it accumulates in domains in the tonoplast preferentially positioned close to the mitochondria and chloroplasts (Su et al., 2018).
The expression patterns of PLDs are developmentally controlled (Fan et al., 1999; Novák et al., 2018) and tissue-dependent (Li and Xue, 2007; Hong et al., 2008; Yamaguchi et al., 2009).
Since PA is a direct product of the phospholipid-hydrolyzing activity of PLDs, many developmental and stress-related roles may be assigned to PA production. PA binds to a diverse array of substrates, including kinases (e.g., mitogen-activated protein kinase 6, MPK6; Yu et al., 2010), enzymes (e.g., glyceraldehyde 3-phosphate dehydrogenase; Kim et al., 2013), and proteins regulating the cytoskeleton (e.g., microtubule-associated protein 65–1, MAP65–1; Zhang et al., 2012) and signaling (e.g., 14–3-3 proteins; Camoni et al., 2012). PA can recruit regulatory proteins to membranes and affect diverse developmental processes (Gao et al., 2013). In addition, PLDs bind and modulate the functions of important signaling and structural proteins, such as components of the G-protein complex (Mishra et al., 2006; Roy Choudhury and Pandey, 2016), sphingosine kinase (Guo et al., 2012), MPK3 (Vadovič et al., 2019), and PLD-regulated protein1 (Ufer et al., 2017). Single T-DNA insertional mutants of PLD isoforms (PLDα and PLDδ) exhibited reduced production of PA (Hong et al., 2008), while a more drastic reduction in PA production was observed by carrying out the simultaneous mutation of both of these PLD isoforms (Bargmann et al., 2009; Uraji et al., 2012). These reports suggest that the total PA pool is generated by the additive actions of several PLDs, as well as by PI-specific PLCs. Interestingly, the reduction in PA production was much more pronounced under stress, which likely explains why Arabidopsis with mutant PLDs possess rather conditional phenotypes (Table S1; Li et al., 2006a; Zhao et al., 2011; Pandey, 2016).
Several of the developmental roles of PLDs depend on stress and hormonal stimuli (Table S1) because PLDs are activated in response to stress factors and hormones, which are accompanied by PA accumulation (Lanteri et al., 2008; Figure 3C). PLDα1 is important for the ABA-mediated regulation of seed germination, which depends on G-protein signaling (Roy Choudhury and Pandey, 2016). A direct physical interaction was recently reported between MPK3 and PLDα1, showing that their genetic interaction hinders the ABA-dependent inhibition of seed germination (Vadovič et al., 2019). PLDs and PA control the distribution of auxin by at least two different mechanisms. In the first of these, PLDζ2-generated PA interacts with and recruits the scaffolding of the A1 subunit of protein phosphatase 2A to the membrane, which is followed by the dephosphorylation of the auxin efflux carrier PIN1 in Arabidopsis (Gao et al., 2013). The second mechanism is connected to the well-known ability of PLDs to regulate membrane transport (described below). PLDζ2 is required for the recycling of PIN2-containing vesicles, as well as for polar auxin transport and distribution (Figure 3D; Li and Xue, 2007).
PLDs are regularly involved in membrane transport, which together with the activity of the cytoskeleton is a major process required for plant development. PLDα1 is required for the homeostasis of proteins involved in vesicular trafficking, membrane fusions, and ER-PM contact sites (Takáč et al., 2019b). PLD-generated PA can regulate membrane transport by causing the direct modification of membrane curvature or by recruiting important regulatory proteins (Donaldson, 2009). These proteins positively affect protein internalization (Li and Xue, 2007; Antonescu et al., 2010) and vesicle fusion (Roth, 2008). PLDα1 was also shown to interact with the adaptor-protein 2 complex, which controls the uptake and sorting of the proteins internalized by clathrin-mediated endocytosis (Yamaoka et al., 2013). This is consistent with the enhanced accumulation of PLDα1-YFP in the cortical cytoplasmic layer of Arabidopsis cells (Novák et al., 2018). Interestingly, the ARC1 E3 ubiquitin ligase-dependent ubiquitination of Brassica napus PLDα1 leads to the inhibition of multivesicular body (MVB) exocytosis in stigmas, resulting in self-incompatibility (Scandola and Samuel, 2019).
Further, PLDs and PA may regulate the nuclear localization of transcription factors important for stress responses or plant development (Yao et al., 2013; Janda et al., 2015). After the binding of the PA generated by PLDζ1 to the WER (WEREWOLF) protein, WER is translocated to the nucleus, which hinders root hair formation and elongation (Yao et al., 2013). PLDζ2 controls root and root hair elongation under phosphate-limited conditions. The exact mechanism is not known, although lipid remodeling is thought to have an important role in this regulation (Su et al., 2018).
A recent proteomic study suggested there is an integrative role of PLDα1 in chloroplast protein import and processing, as well as in plastid and cytosolic translation during chloroplast biogenesis in Arabidopsis (Takáč et al., 2019a). Nevertheless, the molecular mechanism linking PLDα1, which is absent in the chloroplasts, to translation and chloroplast biogenesis remains unknown. One possible link could arise from the ability of ABA to modulate the scaffold protein RACK1, which can regulate translation (Guo et al., 2011) and interact with PLDα1 (our unpublished results).
PLDs are also important regulators of cytoskeletal organization in plant cells (Pleskot et al., 2013). PLDβ1, PLDβ2, PLDγ1, PLDγ2, and PLDγ3 contain an actin-binding domain in their amino acid sequences (Hong et al., 2016). It was previously shown experimentally that tobacco PLDβ1 interacts with actin, and its enzymatic activity is enhanced by F-actin and inhibited by G-actin (Pleskot et al., 2010). On the other hand, reductions in the levels of PLD-produced PA leads to perturbed actin organization, which is mediated by the binding of PA to an actin-capping protein (Huang, 2006). In addition to actin, PLDs also bind to microtubules (Dhonukshe et al., 2003; Zhang et al., 2017b; Angelini et al., 2018; Figure 2), and MAP65-1 was found to be a direct target of PA that is important for microtubule regulation. In this way, PLDα1-derived PA controls microtubule organization during salt stress and enhances the tolerance of Arabidopsis to this stress (Zhang et al., 2012). This type of regulation is strictly limited to stressful conditions because the organization of cortical and mitotic microtubules in the pldα1 mutant does not differ from that in the wild type. We recently reported that PLDα1 decorates cortical microtubules and links them with clathrin (Figure 2; Novák et al., 2018), perhaps forming a putative scaffold for the molecular players involved in the endocytic machinery. Surprisingly, a tight connection of PLDα1 with cytokinetic microtubules was also revealed, especially at the trailing (inner) edge of the enlarging phragmoplast. In addition, PLDα1 was found in the preprophase band and mitotic spindle. Thus, PLDα1 likely contributes to vesicle trafficking events connected with the delivery of membranous material to the newly forming cell plate (Novák et al., 2018). However, the possibility cannot be excluded that this PLDα1-microtubule interaction determines the activation status of PLDα1, since PLDα1 is activated by treatment with cytoskeletal inhibitors (Zhang et al., 2017a).
PLs have been intensively studied for more than two decades due to their ability to modulate plant stress responses. Our review points to their important roles in plant development, which are tightly interlinked with the cellular regulation and subcellular localization of PLs. Generally, PLs show overlapping functions and cooperatively control diverse developmental processes independently of their biochemical properties (Figure 1, Table S1). The developmental roles of PLs are regulated in time-, tissue- (Figure 1), and subcellular localization-dependent manners (Figure 2). The expression patterns of PLs are tightly controlled by signaling and phytohormone-dependent mechanisms. The mechanisms of PL-hormone signaling crosstalk are also diverse. PLs may regulate hormonal biosynthesis or promote the transcription of hormone-responsive genes. Moreover, such crosstalk affects the polar distributions of master regulators of hormonal signaling (Figure 3). In general, crosstalk between PLs and hormones seems to be essential to plant growth and development.
Nevertheless, little is known about the molecular regulation of the expression and activity of PLs. For example, NPC4 might be controlled transcriptionally by the homeodomain protein ALFIN-LIKE6 (Chandrika et al., 2013). However, its expression might also be controlled by post-translational modifications, such as phosphorylation (Rietz et al., 2010; Umezawa et al., 2013) or ubiquitination (Scandola and Samuel, 2019). Finally, the tissue- or cell-specific expression patterns of PLs, along with the proper subcellular localization and function of PLs and their enzymatic products, appear to control diverse developmental processes in plants. Therefore, in the future, efforts should be devoted to the more detailed elucidation of spatiotemporal patterns in the expression and subcellular localization of PLs during plant development. Recently, major advances were achieved in the determination of the developmental and subcellular localization of PLDα1 using advanced light sheet and super-resolution microscopy (Novák et al., 2018). In conclusion, more systematic studies have to be conducted to clarify the molecular mechanisms that control the expression, subcellular localization, and developmental functions of PLs.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was financially supported by Grant No. 16-22044S from the Czech Science Foundation GAČR and by ERDF Project “Plants as a tool for sustainable global development” (No. CZ.02.1.01/0.0/0.0/16_019/0000827).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00362/full#supplementary-material
Andreeva, Z., Barton, D., Armour, W. J., Li, M. Y., Liao, L. F., McKellar, H. L., et al. (2010). Inhibition of phospholipase C disrupts cytoskeletal organization and gravitropic growth in Arabidopsis roots. Planta 232, 1263–1279. doi: 10.1007/s00425-010-1256-0
Angelini, J., Vosolsobě, S., Skůpa, P., Ho, A. Y. Y., Bellinvia, E., Valentová, O., et al. (2018). Phospholipase Dδ assists to cortical microtubule recovery after salt stress. Protoplasma 255, 1195–1204. doi: 10.1007/s00709-018-1204-6
Antonescu, C. N., Danuser, G., and Schmid, S. L. (2010). Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol. Biol. Cell 21, 2944–2952. doi: 10.1091/mbc.E10-05-0421
Apostolakos, P., Panteris, E., and Galatis, B. (2008). The involvement of phospholipases C and D in the asymmetric division of subsidiary cell mother cells of Zea mays. Cell Motil. Cytoskeleton 65, 863–875. doi: 10.1002/cm.20308
Arisz, S. A., and Munnik, T. (2011). The salt stress-induced LPA response in Chlamydomonas is produced via PLA 2 hydrolysis of DGK-generated phosphatidic acid. J. Lipid Res. 52, 2012–2020. doi: 10.1194/jlr.M016873
Arisz, S. A., Testerink, C., and Munnik, T. (2009). Plant PA signaling via diacylglycerol kinase. Biochim. Biophys. Acta 1791, 869–875. doi: 10.1016/j.bbalip.2009.04.006
Bahn, S. C., Lee, H. Y., Kim, H. J., Ryu, S. B., and Shin, J. S. (2003). Characterization of Arabidopsis secretory phospholipase A2-gamma cDNA and its enzymatic properties. FEBS Lett. 553, 113–118. doi: 10.1016/S0014-5793(03)00982-7
Bargmann, B. O. R., Laxalt, A. M., Riet, B. t., van Schooten, B., Merquiol, E., Testerink, C., et al. (2009). Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant Cell Physiol. 50, 78–89. doi: 10.1093/pcp/pcn173
Boutté, Y., and Moreau, P. (2014). Modulation of endomembranes morphodynamics in the secretory/retrograde pathways depends on lipid diversity. Curr. Opin. Plant Biol. 22, 22–29. doi: 10.1016/j.pbi.2014.08.004
Camoni, L., Di Lucente, C., Pallucca, R., Visconti, S., and Aducci, P. (2012). Binding of phosphatidic acid to 14-3-3 proteins hampers their ability to activate the plant plasma membrane H+-ATPase. IUBMB Life 64, 710–716. doi: 10.1002/iub.1058
Chandrika, N. N. P., Sundaravelpandian, K., Yu, S.-M., and Schmidt, W. (2013). ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol. 198, 709–720. doi: 10.1111/nph.12194
Chen, G., Greer, M. S., and Weselake, R. J. (2013). Plant phospholipase A: advances in molecular biology, biochemistry, and cellular function. Biomol. Concepts 4, 527–532. doi: 10.1515/bmc-2013-0011
Chen, G., Snyder, C. L., Greer, M. S., and Weselake, R. J. (2011). Biology and biochemistry of plant phospholipases. Crit. Rev. Plant Sci. 30, 239–258. doi: 10.1080/07352689.2011.572033
Chen, X., Li, L., Xu, B., Zhao, S., Lu, P., He, Y., et al. (2018). Phosphatidylinositol-specific phospholipase C2 functions in auxin-modulated root development. Plant Cell Environ. in press. doi: 10.1111/pce.13492
Cruz-Ramírez, A., Oropeza-Aburto, A., Razo-Hernández, F., Ramírez-Chávez, E., and Herrera-Estrella, L. (2006). Phospholipase DZ2 plays an important role in extraplastidic galactolipid biosynthesis and phosphate recycling in Arabidopsis roots. Proc. Natl. Acad. Sci. U.S.A. 103, 6765–6770. doi: 10.1073/pnas.0600863103
den Hartog, M., Musgrave, A., and Munnik, T. (2001). Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: a role for phospholipase C and D in root hair deformation. Plant J. 25, 55–65. doi: 10.1046/j.1365-313x.2001.00931.x
Dhonukshe, P., Laxalt, A. M., Goedhart, J., Gadella, T. W. J., and Munnik, T. (2003). Phospholipase d activation correlates with microtubule reorganization in living plant cells. Plant Cell 15, 2666–2679. doi: 10.1105/tpc.014977
di Fino, L. M., D’Ambrosio, J. M., Tejos, R., van Wijk, R., Lamattina, L., Munnik, T., et al. (2017). Arabidopsis phosphatidylinositol-phospholipase C2 (PLC2) is required for female gametogenesis and embryo development. Planta 245, 717–728. doi: 10.1007/s00425-016-2634-z
Donaldson, J. G. (2009). Phospholipase D in endocytosis and endosomal recycling pathways. Biochim. Biophys. Acta 1791, 845–849. doi: 10.1016/j.bbalip.2009.05.011
Dong, Y., Li, M., Zhang, P., Wang, X., Fan, C., and Zhou, Y. (2014). Patatin-related phospholipase pPLAIIIδ influences auxin-responsive cell morphology and organ size in Arabidopsis and Brassica napus. BMC Plant Biol. 14:332. doi: 10.1186/s12870-014-0332-1
Dowd, P. E., Coursol, S., Skirpan, A. L., Kao, T. H., and Gilroy, S. (2006). Petunia phospholipase c1 is involved in pollen tube growth. Plant Cell 18, 1438–1453. doi: 10.1105/tpc.106.041582
Effendi, Y., Radatz, K., Labusch, C., Rietz, S., Wimalasekera, R., Helizon, H., et al. (2014). Mutants of phospholipase A (pPLA-I) have a red light and auxin phenotype: phospholipase A pPLA-I in auxin and light signalling. Plant Cell Environ. 37, 1626–1640. doi: 10.1111/pce.12278
Fan, L., Zheng, S., Cui, D., and Wang, X. (1999). Subcellular distribution and tissue expression of phospholipase Dalpha, Dbeta, and Dgamma in Arabidopsis. Plant Physiol. 119, 1371–1378. doi: 10.1104/pp.119.4.1371
Gao, H.-B., Chu, Y.-J., and Xue, H.-W. (2013). Phosphatidic acid (PA) binds PP2AA1 to regulate PP2A activity and PIN1 polar localization. Mol. Plant 6, 1692–1702. doi: 10.1093/mp/sst076
Gao, K., Liu, Y.-L., Li, B., Zhou, R.-G., Sun, D.-Y., and Zheng, S.-Z. (2014). Arabidopsis thaliana phosphoinositide-specific phospholipase C isoform 3 (AtPLC3) and AtPLC9 have an additive effect on thermotolerance. Plant Cell Physiol. 55, 1873–1883. doi: 10.1093/pcp/pcu116
Gaude, N., Nakamura, Y., Scheible, W.-R., Ohta, H., and Dörmann, P. (2008). Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. Plant J. 56, 28–39. doi: 10.1111/j.1365-313X.2008.03582.x
Guo, J., Wang, S., Valerius, O., Hall, H., Zeng, Q., Li, J. F., et al. (2011). Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol. 155, 370–383. doi: 10.1104/pp.110.160663
Guo, L., Mishra, G., Markham, J. E., Li, M., Tawfall, A., Welti, R., et al. (2012). Connections between sphingosine kinase and phospholipase D in the abscisic acid signaling pathway in Arabidopsis. J. Biol. Chem. 287, 8286–8296. doi: 10.1074/jbc.M111.274274
Heilmann, I. (2016). Phosphoinositide signaling in plant development. Development 143, 2044–2055. doi: 10.1242/dev.136432
Helling, D., Possart, A., Cottier, S., Klahre, U., and Kost, B. (2006). Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18, 3519–3534. doi: 10.1105/tpc.106.047373
Hong, Y., Devaiah, S. P., Bahn, S. C., Thamasandra, B. N., Li, M., Welti, R., et al. (2009). Phospholipase Dε and phosphatidic acid enhance Arabidopsis nitrogen signaling and growth. Plant J. 58, 376–387. doi: 10.1111/j.1365-313X.2009.03788.x
Hong, Y., Pan, X., Welti, R., and Wang, X. (2008). Phospholipase D 3 is involved in the hyperosmotic response in Arabidopsis. Plant Cell 20, 803–816. doi: 10.1105/tpc.107.056390
Hong, Y., Zhao, J., Guo, L., Kim, S.-C., Deng, X., Wang, G., et al. (2016). Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 62, 55–74. doi: 10.1016/j.plipres.2016.01.002
Hou, Q., Ufer, G., and Bartels, D. (2016). Lipid signalling in plant responses to abiotic stress: lipid signalling in plant responses to abiotic stress. Plant Cell Environ. 39, 1029–1048. doi: 10.1111/pce.12666
Huang, S. (2001). Cloning of an Arabidopsis patatin-like gene, STURDY, by activation T-DNA tagging. Plant Physiol. 125, 573–584. doi: 10.1104/pp.125.2.573
Huang, S. (2006). Heterodimeric capping protein from Arabidopsis is regulated by phosphatidic acid. Mol. Biol. Cell 17, 1946–1958. doi: 10.1091/mbc.E05-09-0840
Huang, C. H., and Crain, R. C. (2009). Phosphoinositide-specific phospholipase C in oat roots: association with the actin cytoskeleton. Planta 230, 925–933. doi: 10.1007/s00425-009-0990-7
Huo, C., Zhang, B., Wang, H., Wang, F., Liu, M., Gao, Y., et al. (2016). Comparative study of early cold-regulated proteins by two-dimensional difference gel electrophoresis reveals a key role for phospholipase Dα1 in mediating cold acclimation signaling pathway in rice. Mol. Cell. Proteomics 15, 1397–1411. doi: 10.1074/mcp.M115.049759
Ishiguro, S., Kawai-Oda, A., Ueda, J., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. doi: 10.1105/tpc.13.10.2191
Ito, T., Ng, K. H., Lim, T. S., Yu, H., and Meyerowitz, E. M. (2007). The homeotic protein AGAMOUS controls late stamen development by regulating a jasmonate biosynthetic gene in Arabidopsis. Plant Cell 19, 3516–3529. doi: 10.1105/tpc.107.055467
Janda, M., Šašek, V., Chmelařová, H., Andrejch, J., Nováková, M., Hajšlová, J., et al. (2015). Phospholipase D affects translocation of NPR1 to the nucleus in Arabidopsis thaliana. Front. Plant Sci. 6:59. doi: 10.3389/fpls.2015.00059
Jang, J. H., and Lee, O. R. (2019). Overexpression of ginseng patatin-related phospholipase pPLAIIIβ alters the polarity of cell growth and decreases lignin content in Arabidopsis. J. Ginseng Res. in press. doi: 10.1016/j.jgr.2019.01.004
Jung, J., Kumar, K., Lee, H. Y., Park, Y.-I., Cho, H.-T., and Ryu, S. B. (2012). Translocation of phospholipase A2α to apoplasts is modulated by developmental stages and bacterial infection in Arabidopsis. Front. Plant Sci. 3:126. doi: 10.3389/fpls.2012.00126
Kanehara, K., Yu, C.-Y., Cho, Y., Cheong, W.-F., Torta, F., Shui, G., et al. (2015). Arabidopsis AtPLC2 is a primary phosphoinositide-specific phospholipase C in phosphoinositide metabolism and the endoplasmic reticulum stress response. PLoS Genet. 11:e1005511. doi: 10.1371/journal.pgen.1005511
Kato, T., Morita, M. T., Fukaki, H., Yamauchi, Y., Uehara, M., Niihama, M., et al. (2002). SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14, 33–46. doi: 10.1105/tpc.010215
Kim, H. J., Ok, S. H., Bahn, S. C., Jang, J., Oh, S. A., Park, S. K., et al. (2011). Endoplasmic reticulum– and golgi-localized phospholipase A2 plays critical roles in Arabidopsis pollen development and germination. Plant Cell 23, 94–110. doi: 10.1105/tpc.110.074799
Kim, S. C., Guo, L., and Wang, X. (2013). Phosphatidic acid binds to cytosolic glyceraldehyde-3-phosphate dehydrogenase and promotes its cleavage in Arabidopsis. J. Biol. Chem. 288, 11834–11844. doi: 10.1074/jbc.M112.427229
Kimata, Y., Kato, T., Higaki, T., Kurihara, D., Yamada, T., Segami, S., et al. (2019). Polar vacuolar distribution is essential for accurate asymmetric division of Arabidopsis zygotes. Proc. Natl. Acad. Sci. U. S. A. 116, 2338–2343. doi: 10.1073/pnas.1814160116
Krčková, Z., Brouzdová, J., Daněk, M., Kocourková, D., Rainteau, D., Ruelland, E., et al. (2015). Arabidopsis non-specific phospholipase C1: characterization and its involvement in response to heat stress. Front. Plant Sci. 6:928. doi: 10.3389/fpls.2015.00928
Krčková, Z., Kocourková, D., Daněk, M., Brouzdová, J., Pejchar, P., Janda, M., et al. (2018). The Arabidopsis thaliana non-specific phospholipase C2 is involved in the response to Pseudomonas syringae attack. Ann. Bot. 121, 297–310. doi: 10.1093/aob/mcx160
Labusch, C., Shishova, M., Effendi, Y., Li, M., Wang, X., and Scherer, G. F. E. (2013). Patterns and timing in expression of early auxin-induced genes imply involvement of phospholipases A (pPLAs) in the regulation of auxin responses. Mol. Plant 6, 1473–1486. doi: 10.1093/mp/sst053
Lanteri, M. L., Laxalt, A. M., and Lamattina, L. (2008). Nitric oxide triggers phosphatidic acid accumulation via phospholipase D during auxin-induced adventitious root formation in cucumber. Plant Physiol. 147, 188–198. doi: 10.1104/pp.107.111815
Lee, H. Y. (2003). Secretory low molecular weight phospholipase A2 plays important roles in cell elongation and shoot gravitropism in Arabidopsis. Plant Cell 15, 1990–2002. doi: 10.1105/tpc.014423
Lee, H. Y., Bahn, S. C., Shin, J. S., Hwang, I., Back, K., Doelling, J. H., et al. (2005). Multiple forms of secretory phospholipase A2 in plants. Prog. Lipid Res. 44, 52–67. doi: 10.1016/j.plipres.2004.10.002
Lee, O. R., Kim, S. J., Kim, H. J., Hong, J. K., Ryu, S. B., Lee, S. H., et al. (2010). Phospholipase A(2) is required for PIN-FORMED protein trafficking to the plasma membrane in the Arabidopsis root. Plant Cell 22, 1812–1825. doi: 10.1105/tpc.110.074211
Li, G., and Xue, H.-W. (2007). Arabidopsis PLD 2 regulates vesicle trafficking and is required for auxin response. Plant Cell 19, 281–295. doi: 10.1105/tpc.106.041426
Li, L., He, Y., Wang, Y., Zhao, S., Chen, X., Ye, T., et al. (2015). Arabidopsis PLC2 is involved in auxin-modulated reproductive development. Plant J. 84, 504–515. doi: 10.1111/tpj.13016
Li, M., Bahn, S. C., Guo, L., Musgrave, W., Berg, H., Welti, R., et al. (2011). Patatin-related phospholipase pPLAIII-induced changes in lipid metabolism alter cellulose content and cell elongation in Arabidopsis. Plant Cell 23, 1107–1123. doi: 10.1105/tpc.110.081240
Li, M., Qin, C., Welti, R., and Wang, X. (2006a). Double knockouts of phospholipases Dzeta1 and Dzeta2 in Arabidopsis affect root elongation during phosphate-limited growth but do not affect root hair patterning. Plant Physiol. 140, 761–770. doi: 10.1104/pp.105.070995
Li, M., and Wang, X. (2014). “pPLA: patatin-related phospholipase as with multiple biological functions” in Phospholipases in plant signaling. ed. X. Wang (Berlin, Heidelberg: Springer Berlin Heidelberg), 93–108.
Li, M., Welti, R., and Wang, X. (2006b). Quantitative profiling of Arabidopsis polar glycerolipids in response to phosphorus starvation. Roles of phospholipases D zeta1 and D zeta2 in phosphatidylcholine hydrolysis and digalactosyldiacylglycerol accumulation in phosphorus-starved plants. Plant Physiol. 142, 750–761. doi: 10.1104/pp.106.085647
Lindeboom, J. J., Nakamura, M., Hibbel, A., Shundyak, K., Gutierrez, R., Ketelaar, T., et al. (2013). A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science 342:1245533. doi: 10.1126/science.1245533
Liu, G., Zhang, K., Ai, J., Deng, X., Hong, Y., and wang, X. (2015). Patatin-related phospholipase A, pPLAIIIα, modulates the longitudinal growth of vegetative tissues and seeds in rice. J. Exp. Bot. 66, 6945–6955. doi: 10.1093/jxb/erv402
Long, W., Wang, Y., Zhu, S., Jing, W., Wang, Y., Ren, Y., et al. (2018). FLOURY SHRUNKEN ENDOSPERM1 connects phospholipid metabolism and amyloplast development in rice. Plant Physiol. 177, 698–712. doi: 10.1104/pp.17.01826
Luptovčiak, I., Komis, G., Takáč, T., Ovečka, M., and Šamaj, J. (2017). Katanin: a sword cutting microtubules for cellular, developmental, and physiological purposes. Front. Plant Sci. 8:1982. doi: 10.3389/fpls.2017.01982
Mishra, G., Zhang, W., Deng, F., Zhao, J., and Wang, X. (2006). A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science 312, 264–266. doi: 10.1126/science.1123769
Morita, M. T., Kato, T., Nagafusa, K., Saito, C., Ueda, T., Nakano, A., et al. (2002). Involvement of the vacuoles of the endodermis in the early process of shoot gravitropism in Arabidopsis. Plant Cell 14, 47–56. doi: 10.1105/tpc.010216
Nakamura, Y., Awai, K., Masuda, T., Yoshioka, Y., Takamiya, K., and Ohta, H. (2005). A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. J. Biol. Chem. 280, 7469–7476. doi: 10.1074/jbc.M408799200
Ngo, A. H., Lin, Y.-C., Liu, Y., Gutbrod, K., Peisker, H., Dörmann, P., et al. (2018). A pair of nonspecific phospholipases C, NPC2 and NPC6, are involved in gametophyte development and glycerolipid metabolism in Arabidopsis. New Phytol. 219, 163–175. doi: 10.1111/nph.15147
Noack, L. C., and Jaillais, Y. (2017). Precision targeting by phosphoinositides: how PIs direct endomembrane trafficking in plants. Curr. Opin. Plant Biol. 40, 22–33. doi: 10.1016/j.pbi.2017.06.017
Novák, D., Vadovič, P., Ovečka, M., Šamajová, O., Komis, G., Colcombet, J., et al. (2018). Gene expression pattern and protein localization of Arabidopsis phospholipase D alpha 1 revealed by advanced light-sheet and super-resolution microscopy. Front. Plant Sci. 9:371. doi: 10.3389/fpls.2018.00371
Pandey, S. (2016). Phospholipases as GTPase activity accelerating proteins (GAPs) in plants. Plant Signal. Behav. 11:e1176821. doi: 10.1080/15592324.2016.1176821
Park, K. Y., Kim, E. Y., Seo, Y. S., and Kim, W. T. (2016). Constitutive expression of CaPLA1 conferred enhanced growth and grain yield in transgenic rice plants. Plant Mol. Biol. 90, 517–532. doi: 10.1007/s11103-016-0440-4
Pejchar, P., Potocký, M., Krčková, Z., Brouzdová, J., Daněk, M., and Martinec, J. (2015). Non-specific phospholipase C4 mediates response to aluminum toxicity in Arabidopsis thaliana. Front. Plant Sci. 6:66. doi: 10.3389/fpls.2015.00066
Peng, Y. J., Shih, C. F., Yang, J. Y., Tan, C. M., Hsu, W. H., Huang, Y. P., et al. (2013). A RING-type E3 ligase controls anther dehiscence by activating the jasmonate biosynthetic pathway gene DEFECTIVE IN ANTHER DEHISCENCE1 in Arabidopsis. Plant J. 74, 310–327. doi: 10.1111/tpj.12122
Peters, C., Kim, S. C., Devaiah, S., Li, M., and Wang, X. (2014). Non-specific phospholipase C5 and diacylglycerol promote lateral root development under mild salt stress in Arabidopsis. Plant Cell Environ. 37, 2002–2013. doi: 10.1111/pce.12334
Peters, C., Li, M., Narasimhan, R., Roth, M., Welti, R., and Wang, X. (2010). Nonspecific phospholipase C NPC4 promotes responses to abscisic acid and tolerance to hyperosmotic stress in Arabidopsis. Plant Cell 22, 2642–2659. doi: 10.1105/tpc.109.071720
Pinosa, F., Buhot, N., Kwaaitaal, M., Fahlberg, P., Thordal-Christensen, H., Ellerstrom, M., et al. (2013). Arabidopsis phospholipase D is involved in basal defense and nonhost resistance to powdery mildew fungi. Plant Physiol. 163, 896–906. doi: 10.1104/pp.113.223503
Pleskot, R., Li, J., Žárský, V., Potocký, M., and Staiger, C. J. (2013). Regulation of cytoskeletal dynamics by phospholipase D and phosphatidic acid. Trends Plant Sci. 18, 496–504. doi: 10.1016/j.tplants.2013.04.005
Pleskot, R., Pejchar, P., Bezvoda, R., Lichtscheidl, I. K., Wolters-Arts, M., Marc, J., et al. (2012). Turnover of phosphatidic acid through distinct signaling pathways affects multiple aspects of pollen tube growth in tobacco. Front. Plant Sci. 3:54. doi: 10.3389/fpls.2012.00054
Pleskot, R., Potocký, M., Pejchar, P., Linek, J., Bezvoda, R., Martinec, J., et al. (2010). Mutual regulation of plant phospholipase D and the actin cytoskeleton. Plant J. 62, 494–507. doi: 10.1111/j.1365-313X.2010.04168.x
Pokotylo, I., Pejchar, P., Potocký, M., Kocourková, D., Krčková, Z., Ruelland, E., et al. (2013). The plant non-specific phospholipase C gene family. Novel competitors in lipid signalling. Prog. Lipid Res. 52, 62–79. doi: 10.1016/j.plipres.2012.09.001
Potocký, M., Eliás, M., Profotová, B., Novotná, Z., Valentová, O., and Zárský, V. (2003). Phosphatidic acid produced by phospholipase D is required for tobacco pollen tube growth. Planta 217, 122–130. doi: 10.1007/s00425-002-0965-4
Ren, H., Gao, K., Liu, Y., Sun, D., and Zheng, S. (2017). The role of AtPLC3 and AtPLC9 in thermotolerance in Arabidopsis. Plant Signal. Behav. 12:e1162368. doi: 10.1080/15592324.2016.1162368
Repp, A., Mikami, K., Mittmann, F., and Hartmann, E. (2004). Phosphoinositide-specific phospholipase C is involved in cytokinin and gravity responses in the moss Physcomitrella patens. Plant J. 40, 250–259. doi: 10.1111/j.1365-313X.2004.02205.x
Rietz, S., Dermendjiev, G., Oppermann, E., Tafesse, F. G., Effendi, Y., Holk, A., et al. (2010). Roles of Arabidopsis patatin-related phospholipases a in root development are related to auxin responses and phosphate deficiency. Mol. Plant 3, 524–538. doi: 10.1093/mp/ssp109
Roth, M. G. (2008). Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239. doi: 10.1111/j.1600-0854.2008.00742.x
Roy Choudhury, S., and Pandey, S. (2016). The role of PLDα1 in providing specificity to signal-response coupling by heterotrimeric G-protein components in Arabidopsis. Plant J. 86, 50–61. doi: 10.1111/tpj.13151
Sang, Y., Cui, D., and Wang, X. (2001). Phospholipase D and phosphatidic acid-mediated generation of superoxide in Arabidopsis. Plant Physiol. 126, 1449–1458. doi: 10.1104/pp.126.4.1449
Sassi, M., Ali, O., Boudon, F., Cloarec, G., Abad, U., Cellier, C., et al. (2014). An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr. Biol. 24, 2335–2342. doi: 10.1016/j.cub.2014.08.036
Scandola, S., and Samuel, M. A. (2019). A flower-specific phospholipase D is a stigmatic compatibility factor targeted by the self-incompatibility response in Brassica napus. Curr. Biol. 29, 506–512.e4. doi: 10.1016/j.cub.2018.12.037
Scherer, G. F. E., Labusch, C., and Effendi, Y. (2012). Phospholipases and the network of auxin signal transduction with ABP1 and TIR1 as two receptors: a comprehensive and provocative model. Front. Plant Sci. 3:56. doi: 10.3389/fpls.2012.00056
Scherer, G. F. E., Ryu, S. B., Wang, X., Matos, A. R., and Heitz, T. (2010). Patatin-related phospholipase A: nomenclature, subfamilies and functions in plants. Trends Plant Sci. 15, 693–700. doi: 10.1016/j.tplants.2010.09.005
Selvy, P. E., Lavieri, R. R., Lindsley, C. W., and Brown, H. A. (2011). Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119. doi: 10.1021/cr200296t
Seo, Y. S., Kim, E. Y., and Kim, W. T. (2011). The Arabidopsis sn-1-specific mitochondrial acylhydrolase AtDLAH is positively correlated with seed viability. J. Exp. Bot. 62, 5683–5698. doi: 10.1093/jxb/err250
Seo, Y. S., Kim, E. Y., Mang, H. G., and Kim, W. T. (2008). Heterologous expression, and biochemical and cellular characterization of CaPLA1 encoding a hot pepper phospholipase A1 homolog. Plant J. Cell Mol. Biol. 53, 895–908. doi: 10.1111/j.1365-313X.2007.03380.x
Singh, A., Bhatnagar, N., Pandey, A., and Pandey, G. K. (2015). Plant phospholipase C family: regulation and functional role in lipid signaling. Cell Calcium 58, 139–146. doi: 10.1016/j.ceca.2015.04.003
Su, Y., Li, M., Guo, L., and Wang, X. (2018). Different effects of phospholipase Dζ2 and non-specific phospholipase C4 on lipid remodeling and root hair growth in Arabidopsis response to phosphate deficiency. Plant J. 94, 315–326. doi: 10.1111/tpj.13858
Tabata, R., Ikezaki, M., Fujibe, T., Aida, M., Tian, C. E., Ueno, Y., et al. (2010). Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 51, 64–75. doi: 10.1093/pcp/pcp176
Takáč, T., Šamajová, O., Pechan, T., and Šamaj, J. (2019a). Proteomic analysis of Arabidopsis pldα1 mutants revealed an important role of phospholipase D alpha 1 in chloroplast biogenesis. Front. Plant Sci. 10:89. doi: 10.3389/fpls.2019.00089
Takáč, T., Šamajová, O., Vadovič, P., Pechan, T., and Šamaj, J. (2019b). Shot-gun proteomic analysis on roots of Arabidopsis pldα1 mutants suggesting the involvement of PLDα1 in mitochondrial protein import, vesicular trafficking and glucosinolate biosynthesis. Int. J. Mol. Sci. 20:E82. doi: 10.3390/ijms20010082
Tasma, I. M., Brendel, V., Whitham, S. A., and Bhattacharyya, M. K. (2008). Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiol. Biochem. 46, 627–637. doi: 10.1016/j.plaphy.2008.04.015
Ufer, G., Gertzmann, A., Gasulla, F., Röhrig, H., and Bartels, D. (2017). Identification and characterization of the phosphatidic acid-binding A. thaliana phosphoprotein PLDrp1 that is regulated by PLDα1 in a stress-dependent manner. Plant J. 92, 276–290. doi: 10.1111/tpj.13651
Umezawa, T., Sugiyama, N., Takahashi, F., Anderson, J. C., Ishihama, Y., Peck, S. C., et al. (2013). Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6:rs8. doi: 10.1126/scisignal.2003509
Uraji, M., Katagiri, T., Okuma, E., Ye, W., Hossain, M. A., Masuda, C., et al. (2012). Cooperative function of PLDδ and PLDα1 in abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiol. 159, 450–460. doi: 10.1104/pp.112.195578
Vadovič, P., Šamajová, O., Takáč, T., Novák, D., Zapletalová, V., Colcombet, J., et al. (2019). Biochemical and genetic interactions of phospholipase D alpha 1 and mitogen activated protein kinase 3 affect Arabidopsis stress response. Front. Plant Sci. 10:275. doi: 10.3389/fpls.2019.00275
Wang, G., Ryu, S., and Wang, X. (2012). “Plant phospholipases: an overview” in Lipases and phospholipases. ed. G. Sandoval (Totowa, NJ: Humana Press), 123–137.
Wasternack, C., and Song, S. (2016). Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transciption. J. Exp. Bot. 68, 1303–1321. doi: 10.1093/jxb/erw443
Wimalasekera, R., Pejchar, P., Holk, A., Martinec, J., and Scherer, G. F. E. (2010). Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signaling in Arabidopsis thaliana. Mol. Plant 3, 610–625. doi: 10.1093/mp/ssq005
X. Wang (Ed.) (2014). Phospholipases in plant signaling. (Berlin, Heidelberg: Springer Berlin Heidelberg).
Xia, K., Wang, B., Zhang, J., Li, Y., Yang, H., and Ren, D. (2017). Arabidopsis phosphoinositide-specific phospholipase C 4 negatively regulates seedling salt tolerance: AtPLC4 negatively regulates salt tolerance. Plant Cell Environ. 40, 1317–1331. doi: 10.1111/pce.12918
Yamaguchi, T., Kuroda, M., Yamakawa, H., Ashizawa, T., Hirayae, K., Kurimoto, L., et al. (2009). Suppression of a phospholipase D gene, OsPLDβ1, activates defense responses and increases disease resistance in rice. Plant Physiol. 150, 308–319. doi: 10.1104/pp.108.131979
Yamaoka, S., Shimono, Y., Shirakawa, M., Fukao, Y., Kawase, T., Hatsugai, N., et al. (2013). Identification and dynamics of Arabidopsis adaptor protein-2 complex and its involvement in floral organ development. Plant Cell 25, 2958–2969. doi: 10.1105/tpc.113.114082
Yao, H., Wang, G., Guo, L., and Wang, X. (2013). Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 25, 5030–5042. doi: 10.1105/tpc.113.120162
Yu, L., Nie, J., Cao, C., Jin, Y., Yan, M., Wang, F., et al. (2010). Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytol. 188, 762–773. doi: 10.1111/j.1469-8137.2010.03422.x
Zhang, Q., Lin, F., Mao, T., Nie, J., Yan, M., Yuan, M., et al. (2012). Phosphatidic acid regulates microtubule organization by interacting with MAP65-1 in response to salt stress in Arabidopsis. Plant Cell 24, 4555–4576. doi: 10.1105/tpc.112.104182
Zhang, Q., Qu, Y., Wang, Q., Song, P., Wang, P., Jia, Q., et al. (2017a). Arabidopsis phospholipase D alpha 1-derived phosphatidic acid regulates microtubule organization and cell development under microtubule-interacting drugs treatment. J. Plant Res. 130, 193–202. doi: 10.1007/s10265-016-0870-8
Zhang, Q., Song, P., Qu, Y., Wang, P., Jia, Q., Guo, L., et al. (2017b). Phospholipase Dδ negatively regulates plant thermotolerance by destabilizing cortical microtubules in Arabidopsis: PLD regulation of microtubules and heat shock. Plant Cell Environ. 40, 2220–2235. doi: 10.1111/pce.13023
Zhang, Q., van Wijk, R., Shahbaz, M., Roels, W., van Schooten, B., Vermeer, J. E. M., et al. (2018a). Arabidopsis phospholipase C3 is involved in lateral root initiation and ABA responses in seed germination and stomatal closure. Plant Cell Physiol. 59, 469–486. doi: 10.1093/pcp/pcx194
Zhang, Q., van Wijk, R., Zarza, X., Shahbaz, M., van Hooren, M., Guardia, A., et al. (2018b). Knock-down of Arabidopsis PLC5 reduces primary root growth and secondary root formation while overexpression improves drought tolerance and causes stunted root hair growth. Plant Cell Physiol. 59, 2004–2019. doi: 10.1093/pcp/pcy120
Zhao, J. (2015). Phospholipase D and phosphatidic acid in plant defence response: from protein-protein and lipid-protein interactions to hormone signalling. J. Exp. Bot. 66, 1721–1736. doi: 10.1093/jxb/eru540
Zhao, J., Wang, C., Bedair, M., Welti, R., Sumner, L. W., Baxter, I., et al. (2011). Suppression of phospholipase Dγs confers increased aluminum resistance in Arabidopsis thaliana. PLoS One 6:e28086. doi: 10.1371/journal.pone.0028086
Zheng, S.-Z., Liu, Y.-L., Li, B., Shang, Z.- l., Zhou, R.-G., and Sun, D.-Y. (2012). Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance of Arabidopsis: AtPLC9 plays a role in thermotolerance. Plant J. 69, 689–700. doi: 10.1111/j.1365-313X.2011.04823.x
Keywords: cellular functions, phospholipases, phospholipase A, phospholipase C, phospholipase D, plant development, phosphatidic acid, phytohormones
Citation: Takáč T, Novák D and Šamaj J (2019) Recent Advances in the Cellular and Developmental Biology of Phospholipases in Plants. Front. Plant Sci. 10:362. doi: 10.3389/fpls.2019.00362
Received: 24 October 2018; Accepted: 08 March 2019;
Published: 05 April 2019.
Edited by:
Thorsten Schnurbusch, Leibniz-Institut für Pflanzengenetik und Kulturpflanzenforschung (IPK), GermanyReviewed by:
Swarup Roy Choudhury, Indian Institute of Science Education and Research, IndiaCopyright © 2019 Takáč, Novák and Šamaj. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jozef Šamaj, am96ZWYuc2FtYWpAdXBvbC5jeg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.