
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 13 March 2019
Sec. Plant Breeding
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00288
 Rongsheng Wang1,2
Rongsheng Wang1,2 Guoqiang Jiang2
Guoqiang Jiang2 Xiaomin Feng1,2
Xiaomin Feng1,2 Jianzong Nan1,2
Jianzong Nan1,2 Xiaohui Zhang1,2
Xiaohui Zhang1,2 Qingbo Yuan2
Qingbo Yuan2 Shaoyang Lin1,2*
Shaoyang Lin1,2*As an elite rice variety cultivated in the third accumulative temperature belt in Heilongjiang province, China, Kongyu131 has many excellent traits, such as high quality, high stability, early maturation and cold resistance. However, as with other crop varieties, Kongyu131 has regional restrictions, exhibiting decreased yields when grown at low latitudes. To address these problems, two populations were constructed from cross between japonica and indica varieties. QTL analyses were performed with these two populations to detect regional adaptation related quantitative trait locus. Results in a BC1F6 backcross inbred line population with 168 lines derived from cross between Kongyu131 and GKMP showed a large pleiotropic QTL near 9 Mb on chromosome 7, which significantly delayed the HD of Kongyu131 and increased the plant height (PH), length of main panicle (LMP), number of primary branches (NPB) and grain number of main panicles (GNP). We also found a similar QTL in the population BC3F2 derived from Kongyu131 and GKLPL. Based on the QTL, we developed a gene module named mRA7 with 5 single-nucleotide polymorphism (SNP) markers around the QTL. Through a foreground and background selection based on 197 SNP markers evenly distributed over the 12 chromosomes, we obtained a new plant (a single point substitution line, SPSL) with a new Kongyu131 genome, carrying only a small chromosomal fragment less than 800 kb from GKLPL. The background recovery ratio of the SPSL was 99.8%. Compared with Kongyu131, the SPSL exhibited a significant HD delay of approximately 31 days and increased PH, LMP and GNP values when planted in Heilongjiang province. When cultivated in Guangdong province, HD of SPSL showed only 16 days delay, and less increase in PH, LMP and GNP than in Heilongjiang province. Phenotypic evaluation showed that the SPSL could be moved to south by more than 3 latitude units and cultivated in low-latitude regions. This study exemplifies the feasibility of expanding the regions of cultivation of elite rice varieties via similar methods.
Successful breeding of an elite variety requires a tremendous amount of time and resources. Spreading of a main cultivar to other cultivation regions could have great significance in saving resources. However, as with other crop varieties, rice varieties exhibit specific regional adaptability, which is mainly observed as abnormal heading dates (HDs) and plant architecture when cultivated elsewhere. As a consequence, a specific rice variety can be planted in only a limited cultivation region with specific photoperiod and temperature regime across the growth cycle. Once outside its adaptive environment, a rice variety may grow excessively without heading or prematurely with reduced yield and quality. Regional adaptability has become the main problem restricting the introduction of elite rice varieties to other cultivation regions.
The main determinant of the regional adaptation of rice to different environments is the transition period from vegetative growth to reproductive growth, also known as the heading date (Hayama et al., 2003; Wang et al., 2011). Furthermore, plant height (PH) and length of the main panicle (LMP) are key indicators of adaptation and can be used to decide whether a certain variety can be cultivated elsewhere (Xing and Zhang, 2010). As a short-day plant, rice always exhibits early flowering at low latitudes but delayed flowering at high latitudes. A series of heading date-related quantitative trait loci (QTLs) and genes have been identified, such as Hd1 (Yano, 2000), Hd3a (Kojima et al., 2002; Tamaki et al., 2007), Ehd1 (Doi et al., 2004), Ghd7/Hd4 (Yano et al., 1997; Xue et al., 2008), Hd6 (Takahashi et al., 2001), and Hd16 (Hori et al., 2013). However, there have been only a limited number of reports regarding the improvement of the regional adaptability of rice to environments at high latitudes by manipulation of HD-related QTLs or genes (Yu et al., 2002; Nagata et al., 2015). Although rice varieties in northeast China are famous for their high quality, few of these varieties have been extended to the south. Indeed, all the genetic modifications necessary to improve the adaptation capacity are limited by improvement accuracy. In addition neither traditional breeding nor marker-assisted selection (MAS)-based breeding technology can adjust the HD of a rice variety without changing other excellent traits (Kwon et al., 2002; Zhou et al., 2003). A precise genetic technique is urgently needed to address such problems.
With the rapid development of plant genomics, as whole-genome resequencing and molecular marker technology, we now have an opportunity to address these issues. First, an increasing number of excellent genetic resources regarding natural variations associated with HDs have been identified and compiled for rice breeding (Fan et al., 2006; Zhang et al., 2012; Tang et al., 2013). The dissection of complex HD networks has provided new perspectives for the molecular design of new cultivars with modified photoperiod adaptability that can effectively grow in different cultivation regions (Yano and Sasaki, 1997; Xing and Zhang, 2010; Matsubara et al., 2014). Furthermore, with the development and application of next-generation sequencing and interpretation of the large amounts of data generated (Duitama et al., 2015; Wang et al., 2018), single-nucleotides polymorphisms (SNPs) (Chen et al., 2014; Yu et al., 2014) coupled with high-resolution melting (HRM) analysis for genotyping can improve the ease and effectiveness of the detection of molecular markers. In 2003, “breeding by design” was first proposed as a new breeding strategy (Peleman and van der Voort, 2003). Following with this concept, many new attempts were carried out in recent years (Wang et al., 2011; Zeng et al., 2017). But for the precise and efficient breeding, a gene module, that is a foreign short chromosome segment regulating relevant agronomic trait(s), should be defined together with corresponding SNP markers. With modules developed and the technical foundation mentioned above, the improvement of new variety and aggregation of multi module would be feasible.
Kongyu131 is an elite rice variety of the third accumulated temperature belt in Heilongjiang province, China. This variety has many advantageous traits, such as high quality, high stability, early maturation and cold resistance, which have led to increasing its annual cultivation area since 1990. The cultivation area peaked in 2004, occupying more than half of the total rice cultivated area in Heilongjiang province, and its cumulative extension area has accounted for more than 9.5 million hectares to date (China Rice Data Center, 2018). Kongyu131 exhibits significantly premature development with dramatic height and yield reduction when cultivated in low-latitude regions such as Beijing. In general, increasing adaptation capacity of elite rice varieties by genetic modification of their genome could have a relevant impact for the breeding sector.
In this study, we carried out QTL analysis in two backcross populations and identified a pleiotropic and stable QTL to repair this “bug” in Kongyu131 when grown at low latitudes. After genomic background subtraction, a single point substitution line (SPSL) containing a target chromosome segment was bred. We tested the SPSL in different rice fields and proved that it can be moved to southern regions.
The recurrent parent used in this study was Kongyu131, an elite japonica rice variety that is widely cultivated in the third accumulated temperature belt in Heilongjiang province, China. Kongyu131 is an early maturation variety, with seeding to heading occurring over 98 days, and requires at least 2320°C as an active accumulated temperature. Kongyu131 has an approximate PH of 80 cm, LMP of 16 cm, and GNP of 109. The donor parent GKMP, is an indica cultivar bred from Modan and Koshihikari, which is the main cultivar in Japan. GKMP is not able to head in Heilongjiang province. When cultivated in Beijing, GKMP showed early maturation that required 104 days from seeding to heading, PH of 90 cm, LMP of 20 cm and GNP of 156. The other donor parent GKLPL is also an indica cultivar bred from GKMP but with a different plant type. When cultivated in Heilongjiang province, GKLPL showed late maturation that required 122 days from seeding to heading. This variety had an approximate PH of 96 cm, LMP of 27 cm, and GNP of 286.
All phenotypic experiments were carried out as follow. Presoaking of seeds was conducted in an 8∗12-well plate with a small hole at the bottom. We used 96-well plate with 96 plants as a plot. After sowing, the plates were immersed in water at a constant temperature of 37°C and germinated for approximately 48 h. The seeds were transplanted to nutrient soil to obtain seedlings in bed boxes. Seedlings at the trefoil stage were transplanted to the paddy field in the same arrangement as that in the 96-well plate with one plant per hole, and the plants were spaced with 20 cm within each row and 30 cm between adjacent rows. The area per plot was 5.76 m2 (2.4 m × 2.4 m). The other conditions were the same as those used for local paddy field management.
Kongyu131 was crossed with GKMP, the F1-generation was backcrossed with Kongyu131 to produce the BC1F1-MP population. One seed of each BC1F1-MP line was picked to obtain its inbred seed with a random selection in every generation as according to SSD procedure. Finally, we obtained the backcross inbred line population BC1F6-MP with GKMP as the donor for a total of 168 lines BC1F6-MP. This population was used for QTL analysis.
Kongyu131 was crossed with GKLPL, and then, the F1-generation was backcrossed with Kongyu131 consecutively to produce the BC3F1-LPL population for a total of 162 lines (Figure 1). From this population, we selected a line named BC3F1-LPL55F01. 80 progeny plants generated from the selfing of BC3F1-LPL55F01 were transplanted to the paddy field at the seedling stage and phenotyped in Jiamusi, Heilongjiang province for HD, LMP, and PH and scoring data used for QTL analysis. Simultaneously, one plant from each line of the BC3F1-LPL population was backcrossed with Kongyu131 producing the BC4F1-LPL population of 152 lines. This BC4F1-LPL population was cultivated and genotyped with SNP markers for genotypic selection. Then, based on the genotypic data we selected a plant named BC4F1-LPL222E02, in which a crossover occurred within SNP1 and SNP2 and contained the target QTL of the donor. The nine hundred and sixty progenies obtained by selfing of the BC4F1-LPL222E02 plant were genotyped for markers in the region comprising SNP2 – SNP5. Based on this molecular analysis, plants heterozygous at these markers were selected for genotypic selection in Hannan province. Finally, an SPSL plant named BC4F3-LPL331E09 was obtained. In this plant, a crossover occurred within SNP4 and SNP5, but the homozygous segment between SNP2 to SNP4 were from the donor GKLPL.
The populations used for QTL analysis were planted in Beijing (ZGQ, 39.5°N, 116.5°E) and Jiamusi, Heilongjiang province (LQ, 46.5°N, 130.5°E), separately. Phenotypical validation was carried out in LQ, Changchun, Jilin province (JNQ, 43.5°N, 125.5°E), ZGQ and Guangzhou, Guangdong province (GZQ, 20.5°N, 110.5°E). Different sowing date experiment was carried out in ZGQ. In all field environments, the selfing descendants of the SPSL BC4F3-LPL331E09 and its parents were planted in a randomized block design with three replications. Eight plants in each plot were randomly selected for phenotypic evaluation. The heading date of each plant was recorded when its panicle heading over one third of the panicle length. All agronomic traits were measured three times and followed the standard evaluation system (International Rice Research Institute [IRRI], 2013), including PH, HD, LMP, NPB, GNP, ETP, GN and 1000-grain weight (TGW). Days to heading (DH) was counted from sowing date to heading date. Then, average correlation coefficient and Student’s t-tests were calculated in MS Excel. Arithmetic average with standard deviations were calculated as the phenotype of the SPSL and its parents with 8 plants per replication. Broad-sense heritability (hB2) was calculated with the formula hB2 = VG/VP.
Genomic DNA was extracted following an improved rapid high-throughput DNA isolation method (Stein et al., 2001; Paris and Carter, 2012). One hundred and sixty microliters of precooled Tris–HCl (0.1 M, pH 7.5) and 40 μL of NaOH (0.4 M) were added to a 96-deep-well plate. A piece of fresh leaf was added to the solution buffer with a small steel ball; the plate was covered with a silica gel lid and placed in a vibrator for 3 min; and then, the supernatant was extracted for subsequent polymerase chain reaction (PCR) and high-resolution melting (HRM) analysis after centrifugation.
Polymerase chain reactions were performed in a normal 96-well plate. The reaction system contained 1 μL of 10 × Buffer, 0.4 μL of 2.5 mM dNTPs, 0.1 μL of Taq DNA polymerase, 0.2 μL of 20 × qPCR DNA-binding dye, 10 μM forward primer, 10 μM reverse primer, and 3 μL of genomic DNA template, and ddH2O was added to 10 μL. PCR and HRM analysis were implemented in a qPCR instrument continuously. The PCR procedure consisted of predenaturation at 94°C for 2 min, followed by 40 cycles of 94°C for 10 s and 60°C for 30 s. The HRM procedure consisted of predenaturation at 94°C for 10 s, 60°C for 10 s, and heating to 94°C, with 10 instances of fluorescence detection for every degree of temperature increase.
For the QTL analysis on the population BC1F6-MP, a total of 168 SNP markers evenly distributed on 12 chromosomes were used in 168 individuals. According to the genotype of 195 SNP markers evenly distributed on 12 chromosomes, we screened the BC3F1-LPL population with 162 individuals. An individual named BC3F1-LPL55F01 was used to derive BC3F2-LPL population with its self-bred seeds. Only 37 SNP markers were heterozygous in this population. So, we conducted the QTL analysis with these markers in 80 individuals. Genetic distance were calculated in MapMaker/Exp 3.0b and QTL analyses were conducted by MapMaker/QTL 1.1b (Lander et al., 1987).
Individual selections were performed with SNP1 to SNP5, and SNP3 was detected to make sure about the introduction of the gene module. To excluding segments from the donor, the SNP marker located on the other chromosomes were detected if necessary.
The parents, namely, Kongyu131, GKMP and GKLPL, were resequenced by a HiSeq2000 sequencer. The Ghd7 DNA sequence was downloaded from GenBank1, and the reference sequence used for genomic assemble and gene alignments was downloaded from Oryzabase2 (Nipponbare Build v.4). Sequence search and alignment were conducted using the Localblast system developed by NCBI3.
Based on the sequence alignment between the two parents, some effective SNP positions were selected for SNP marker design. Genome sequences around SNP position were uploaded as supplementary data (SupplementaryData.xlsx). Primers of 18–25 nts were selected around the SNP positions and to give a PCR product with a 40–100 bp length. The SNP markers for the mRA7 module, named SNP1 to SNP5, were located upstream (SNP1 and SNP2), within (SNP3), and downstream (SNP4 and SNP5) of this gene (Supplementary Table S1). The distance from SNP1 to SNP5 was approximately 1 Mb (Figure 2A).
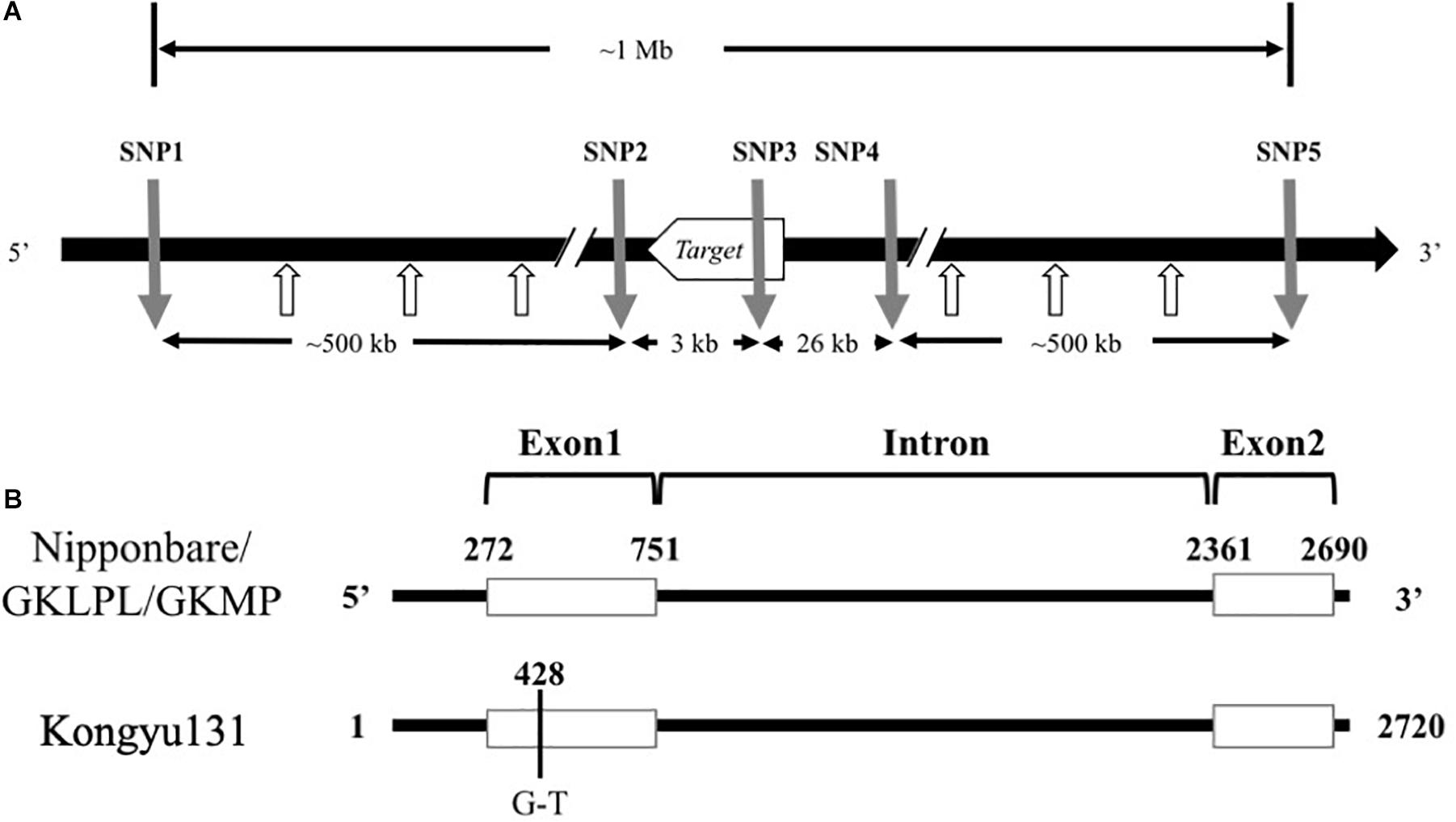
Figure 2. (A) Alignment of Ghd7 sequences from Nipponbare, GKLPL and Kongyu131. The number indicates the position of the Ghd7sequence. The black line indicates the intron region, and the white box indicates the exon region. (B) The relative positions of the SNP markers used for introduction of the mRA7 module. The gray arrows indicate the positions of five SNP markers used for introduction of the module, and the white arrows indicate the SNP marker used for detection of module length.
The genetic maps were performed by MapChart 2.32 (Voorrips, 2002). 168 SNP were used for the BC1F6-MP population. 35 SNP were used for the BC3F2-LPL population. Each of the only one SNP marker on the 3th and the 4th chromosome was omitted.
For QTLs controlling HD useful for improving Kongyu131’s adaptation, we obtained a backcross inbred line (BIL) population BC1F6-MP (168 individuals) using Kongyu131 as the recurrent parent and GKMP as the donor. After planting the population in a paddy field in ZGQ, we surveyed the phenotypes of these individuals by accessing these lines for DH, PH, effective tillers per plant (ETP), LMP, number of primary branches (NPB) and grain number of main panicles (GNP) after maturity. Frequency distributions of different traits showed nearly normal distributions (Supplementary Figure S1). Heritability ranged from 86.28 to 98.77%, which mean the existence of significant effect of genotypic variation on the phenotypic variation (Table 1). A total of 168 SNP markers were used to carry out the QTL analysis. The total length of the linkage map was 4326.8 cM with an average interval of 25.75 cM (Supplementary Figure S2). We found a pleiotropic QTL near 9 Mb on the 7th chromosome. PH, HD, LMP, NPB and GNP were affected by this QTL. The phenotypic explained variance ranged between 40.90 and 59.60% while LOD score ranged from 19.18 to 33.09 indicating that a major effect QTL with a high confidence level was detected (Supplementary Figure S3). In this QTL region, the gene Ghd7 has been identified. The segment derived from the donor GKMP could significantly delay the HD of Kongyu131 and increase the PH, LMP, NPB and GNP (Table 2).
Correlation analysis of traits in this population showed that DH, PH, LMP, NPB, and GNP have a significant relationship between each two of them, all their significant coefficient values were higher than 0.7. ETP showed little relationship with other traits and all their significant coefficient values were lower than 0.25 (Table 3). These suggested the identical conclusion we got from the QTL analysis.
To develop the fine mapping and introgression line, a QTL mapping population BC3F2-LPL was constructed between Kongyu131 and GKLPL. The parent of this population named BC3F1-LPL55F01 was selected for the heterozygous segment on the short arm of the 7th chromosome (Supplementary Figure S4A). 80 descendants of this line were phenotyped in a paddy field in LQ, in the normal season in 2015. According to the phenotypic statistics of the population, the HD and PH exhibited obvious segregation based on statistics and analysis (Figures 3A,B), but the LMP showed a normal distribution (Figure 3C). The heritability of LMP, PH, and DH ranged from 62.32 to 97.58% (Table 1). It is inferred that the introduced segment from the donor GKLPL could lead to the delay of HD and the increase of plant height, similar to the QTL from GKMP. Therefore, we conducted QTL analysis using 37 SNP markers which showed to be heterozygous in the BC3F1-LPL55F01 plant (Figure 3D). Finally, a major QTL controlling DH, PH and LMP was located at the 9 Mb position on the short arm of 7th chromosome, as reported for the BIL population BC1F6-MP (Supplementary Figures S4B–D). This locus explained 86.2% of the phenotypic variance of DH and 71.4% of PH (Table 4). This suggested that the introgression of this locus can lead to delayed HD, and increased PH and LMP in Kongyu131. This QTL located in the same position of a previously reported QTL affecting DH, PH and LMP and cloned as Ghd7 gene (Yano et al., 1997; Xue et al., 2008).
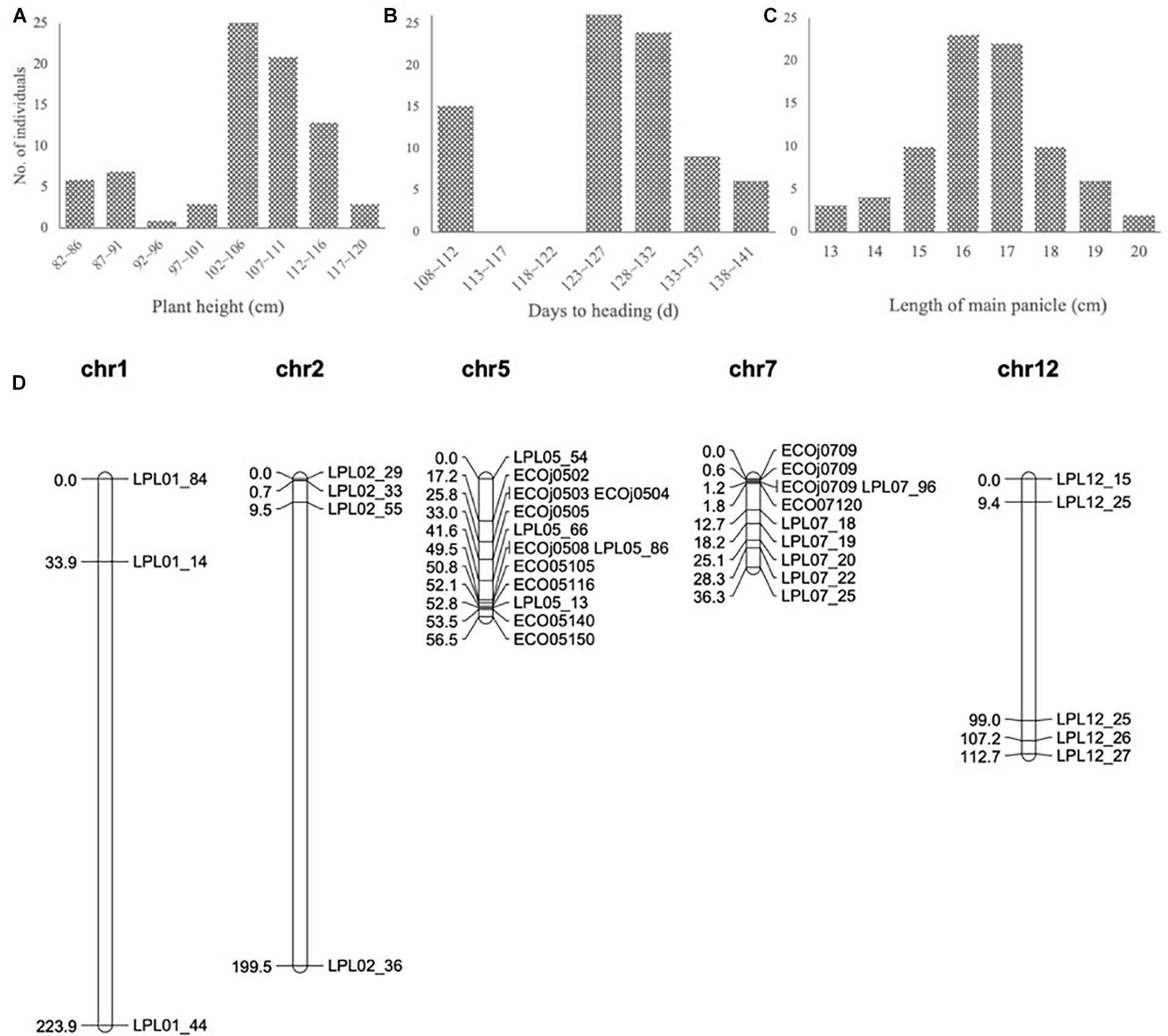
Figure 3. (A–C) Frequency distribution in the QTL population BC3F2-LPL. The vertical axis indicates the number of plants; the horizontal axes indicates the (A) intervals of different plant heights, (B) intervals of different days to heading and (C) intervals of different length of main panicle. (D) Genetic map of QTL population BC3F2-LPL.
Correlation analysis of these three traits used in the QTL analysis showed that LMP has a relationship with PH and DH. PH and DH have a significant relationship with the significant coefficient value 0.8 (Supplementary Table S2).
To reveal whether the phenotypic changes in these QTL populations were caused by the difference of inferred gene sequence, we firstly conducted genome resequencing in the parents Kongyu131, GKLPL and GKMP. BLAST analysis of the Ghd7 gene sequences of these varieties, using the whole-genome sequence of Nipponbare as a reference, showed that this gene was located near 9 Mb on chromosome 7, similar to the location of this gene in Nipponbare, thus being a reliable candidate gene for the QTL. Alignment of the Ghd7 gene sequences of these parents exhibited a single base mutation, from G to T, in the first exon of Kongyu131 Ghd7 at position 428 (Figure 2B). The mutation led to a change from GAG, encoding glutamic acid (Glu), to TAG, which is a stop codon. This mutation may lead to loss of function of this gene. The gene sequences in GKLPL and GKMP were identical.
To ensure precise introduction of the target gene, we designed five SNP markers, namely, SNP1 to SNP5, around this gene according to the sequence alignment information. SNP3 was located in the gene and designed based on the mutant base site. SNP1 and SNP2 were located upstream of the gene, while SNP4 and SNP5 were located downstream (Figure 2A).
To reduce the extension of the introgressed region, upon a further backcross a BC4F1-LPL population of 152 individuals was generated and genotyped with 197 SNP markers evenly distributed on 12 chromosomes. An individual named BC4F1-LPL222E02, with a donor chromosome segment on 7th chromosome and with the least segments on the other chromosomes was selected. In addition to the introduced target segment, it carried also some segments on chromosomes 1, 3, and 7, and the total length of the introduced segments was approximately 28 Mb. Detection of SNPs from SNP1 to SNP5 showed that SNP1 originated from Kongyu131, indicating that a crossover occurred between SNP1 and SNP2. The background recovery ratio was approximately 97.5% (Supplementary Figure S5A).
To further eliminate the linkage drag downstream of the target gene module, nine hundred and sixty progenies of the self-bred BC4F1-LPL222F02 were sowed and screened at the seedling stage with four SNP markers (SNP2 to SNP5) to select an individual in which recombination occurred between SNP4 and SNP5 while ensuring that the SNP3 position originated from the donor. Then, these selected individuals were transplanted to a normal paddy field to obtain self-bred seeds. The same procedure was performed in BC4F2-LPL and BC4F3-LPL population for excluding the segments from the donor, except the target gene module. Finally, we obtained an SPSL named BC4F3-LPL331E09. The segment between SNP2 and SNP4 was homozygous and originated from the donor GKLPL. This gene module was named mRA7. All the other chromosomes were recovered from Kongyu131 with a 99.8% background recovery ratio (Supplementary Figure S5B).
To accurately determinate the length of the introduced sequence, we designed 3 SNP markers between SNP1 and SNP2 as well as between SNP4 and SNP5 separately, with an average marker to marker distance of approximately 120 kb (Figure 2A). We found that all the SNP markers between SNP1 and SNP4-2 exhibited homozygous genotypes from GKLPL, indicating that the segment introduced was shorter than 800 kb.
LQ was not suitable for cultivation of the SPSL due to the prolonged growth period, so SPSL with its parents were compared simultaneously in four environments – LQ, JNQ, ZGQ and GZQ – to analyze phenotypic variance among these lines. In LQ, the HD was delayed by approximately 31 days; the PH increased by 20 cm; the LMP increased by 1 cm; the NPB increased by 6; and the GNP increased by 31 grains. This indicated that introduction of the mRA7 module indeed delayed the HD and increased the PH and LMP (Figures 4A,B) with a positive effect on GNP. Notably, the SPSL exhibited nearly complete maturation when planted in JNQ and was also suitable for the cultivation season in ZGQ (Figures 4A,C,D). However, a decrease in PH was observed in GZQ due to the early HD, which was nearly 27 cm lower than the value of SPSLs in JNQ and LQ. The DH of SPSL was approximately 84 days when planted in ZGQ, 15 days longer than Kongyu131.

Figure 4. Phenotype comparison between SPSL and Kongyu131 in different environments. (A) Kongyu131 (left), SPSL (middle) and GKLPL (right) planted in LQ. The photo was taken on September 3, 2017, and the SPSL exhibited a higher plant height than Kongyu131 and a late heading date. (B) Comparison of the length of the main panicle between the SPSL (left) and Kongyu131 (right). The SPSL exhibited a significantly longer main panicle than Kongyu131. (C) Kongyu131 (left) and the SPSL (right) planted in JNQ. The photo was taken on September 17, 2017, and the SPSL seemed to have undergone maturations. (D) Kongyu131 (left) and the SPSL (right) planted in ZGQ. The photo was taken on September 7, 2017. The scale bar represents 10 cm.
Phenotypic comparison suggested that the DH, PH, LMP, NPB and GNP of the SPSL exhibited significant differences compared with Kongyu131, while for ETP significant differences were found only in GZQ. With decreasing latitudes the HD, PH, NPB, GNP and ETP of the SPSLs all exhibited reduction trend (Table 5).
Differences in DH may be affected by lengths of daytime and temperature. To examine this difference, we evaluated the SPSL with its parents as controls, in a normal paddy field in ZGQ with different sowing dates. The first seeding date was April 10th, with an interval of 14 days. The last seeding date was July 18th, with eight sessions in total. The daytime length in the experimental field peaked in June, with an average of 14.8 h of light per day. This duration gradually decreased to 12.4 h per day in September. Although the different sowing dates, all genotypes exhibited heading between June and September, indicating that all the varieties underwent heading under long-day conditions, as rice plants are affected by daytime length. The results showed that, regardless of seeding date, the DH value of the SPSL with the mRA7 module was greater than that of Kongyu131 (Figure 5). However, there was no linear correlation between daytime length and DH. According to the temperature monitoring data, the daily average temperature first increased but then decreased from May to September. The peak value was observed from July to early August. The DH of the SPSL exhibited no correlation with this temperature trend, but Kongyu131 exhibited an inverse correlation, i.e., the DH decreased at first but then increased. This finding suggests that the SPSL with the mRA7 module was substantially affected by both daytime length and temperature, but it was not temperature sensitive, unlike Kongyu131.
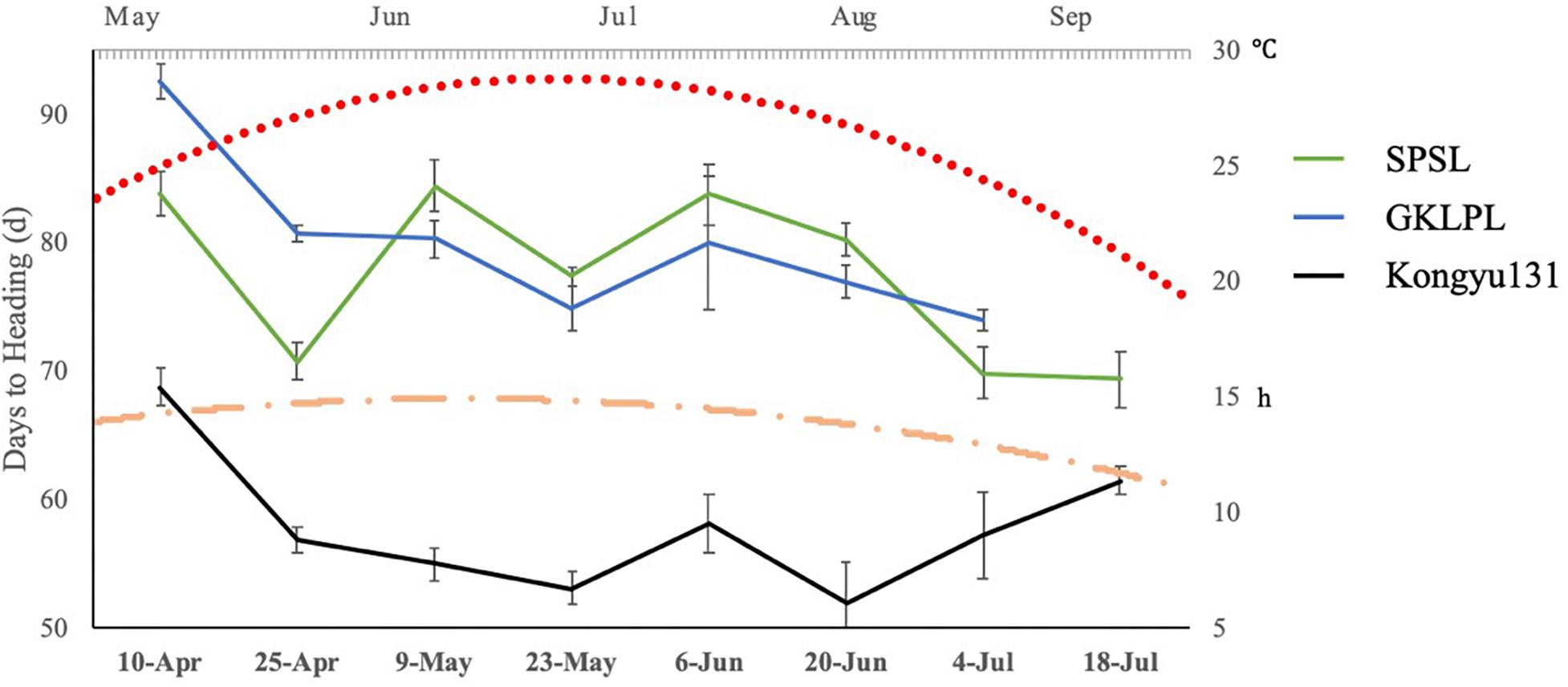
Figure 5. The trend of days to heading values with different sowing dates compared between the SPSL and its parents. The primary horizontal axis indicates the different sowing dates, and the primary vertical axis indicates the days to heading with different sowing dates. The blue line indicates the donor GKLPL (the last time points were missed); the green line indicates the SPSL; and the black line indicates the recurrent parent Kongyu131. Flat-ended arrows indicate standard deviation. The red dot line indicates daily mean temperature from May to September. The orange dot dash line indicates daily mean daytime from May to September.
To evaluate the phenotype of the SPSL in the natural field, the SPSL was planted in JNQ and other low-latitude areas, such as ZGQ and GZQ. The DH value of the SPSL was approximately 125 days in JNQ, which was similar to that of the local variety, exhibiting a significant difference compared to Kongyu131. When planted in ZGQ, the DH value of the SPSL was approximately 84 days, which was 15 days later than that of Kongyu131. Meanwhile, the PH and the LMP increased at both locations. Thus, the SPSL suitable for cultivation between the latitude of JNQ and ZGQ.
The aim of this work was to improve an elite rice variety for its regional adaptation with precise and efficient modularized improvement. In this study, we first conducted a QTL analysis with a BIL population constructed by using the GKMP cultivar as a donor, ensuring the absence of the regional adaptation locus in Kongyu131. Then we used a backcross population BC3F2 constructed by using GKLPL, a cultivar derived from GKMP as a donor to verify the QTL identified above and to attempt the introgression of such a locus in Kongyu131. Based on the position of the locus, we hypothesized that the Ghd7 could be the candidate gene and we inferred that introduction of the Ghd7 from GKLPL or GKMP to Kongyu131 activated its gene expression, thus delaying the HD of Kongyu131, and increasing the PH and yield-related traits, such as LMP and GNP. Then, the foreground and background were both selected from BC4F1 to BC4F2, and we defined mRA7 the introgressed gene module. Finally, the SPSL was obtained with the precisely introduction of the mRA7 module in the BC4F3 generation (Figure 1). Evaluation of the module showed that the HD of SPSL was significantly delayed compared with that of Kongyu131, and the PH and LMP were increased (Table 5). All of these results indicated that introduction of the mRA7 module indeed improved the capacity of adaptation of the original Kongyu131. According to the phenotypic comparison between the SPSL and Kongyu131, we also verified that there were no non-desirable genes that perturbed the expected phenotype.
At the end, our research led to the development of a modular variety by using a single module, extending the cultivation of a main cultivar of Kongyu131 from its original area of cultivation (the third accumulative temperature belt) to low-latitude regions, such as JNQ and ZGQ. This method requires less time and resources than traditional breeding techniques. Notably, this method and technical route can be used as a reference for the transplantation of other main cultivars to non-original areas of cultivation.
Based on genome resequencing and comparison, we could hypothesize that the Ghd7 is the candidate gene of the target QTL/gene module. Previous studies have shown that Hd4/Ghd7 is a pleiotropic gene that controls the HD and yield-related traits such as PH, LMP and GNP. This gene encodes a CCT (CO, CO-LIKE and TIMING OF CAB1) domain protein. With overexpression, this gene can delay the HD under long-day conditions. Simultaneously, the LMP, PH, and GNP can increase significantly (Yano et al., 1997; Xue et al., 2008). Analogously, the SPSL showed a significant increase in PH, LMP, GNP, and NPB in the main panicle compared to Kongyu131. These phenotypic results may further indicate that the two donor parents may carry the functional allele correspond to the Ghd7 gene and thus that its expression could be the responsible of the change in phenotype of the SPSL in respect to the original Kongyu131. However, because the length of the module was approximately 800 kb, some other genes that affect the relevant phenotypes may be included, and further research is required to test this possibility.
The delayed heading date in LQ had the consequences that the SPSL was exposed to low temperature during grain filling stage. That would be direct cause that the TGW of two donor cultivars and the SPSL was lower than Kongyu131, owing to incomplete grain filling under low temperature. In contrast, the SPSL heads at the right time in JNQ and ZGQ and TGW shows no difference between the SPSL and Kongyu131 in ZGQ. As we all known, yield traits could be complex and involve multi-gene actions. By comparing the yield traits of varieties cultivated in these environments, two possible explanations may be deduced. First, the introduction of this module did not affect the grain filling or the accumulation of nutrient substances. Second, the increase of DH led to higher level of photosynthetic product (also known as the source) in ZGQ. The GNP and LMP (also known as the library) increase simultaneous, indicating the presence of an equilibrium between the source and the library.
In both LQ and ZGQ, the ETP exhibited no differences between the SPSL and Kongyu131, indicating the mRA7 module did not affect the number of effective tillers. Therefore, this finding suggests that the extension of the adaptive region of Kongyu131 did not affect its production. The differences of both Kongyu131 and SPSL in these environments could be attributed to various environmental factors.
MAS-based breeding is widely used today based on the selection of linkage markers of target traits, to be combined with phenotypic selections in the field. However, the bred varieties from this technique are sometimes unstable, and the effective genes underlying may not be clear. After a few years’ cultivation, even a preeminent rice variety would be replaced for its defects. Unfortunately, breeders could do nothing to save it, but to develop new varieties with the same procedure. In our research, we successfully improved the genome of Kongyu131 by introducing and accurately controlling the size of the donor chromosome segment. First, we used a number of SNP markers to screen Kongyu131’s chromosomes and eliminate non-target fragments from the donor. Second, we used five SNP markers, namely, SNP1–SNP5, near and within the candidate gene. SNP3 was located in the gene sequence to ensure its introduction. The distance between SNP1 and SNP5 was approximately 1 Mb. With the two-step selection between SNP1 and SNP2 and between SNP4 and SNP5, the introduced segment was well controlled at less than 1 Mb. Traditional MAS-based breeding using a few scattered markers leads to uncertainty in the length of the introduced chromosome segment, often generating a large fragment insertion with the well-known effect of linkage drag.
Except for the precisely control of linkage drags and target genes, the availability of a gene module can allow the rough prediction of its effects upon introgression (Zeng et al., 2017; Kumar et al., 2018). This could be feasible also for high complex trait when main locus/genes responsible of are known or are mapped in the genomes. Therefore, this method can be used for directed and precise improvement of rice varieties.
To date, we have not identified any significant defects during field tests of our SPSL. However, extension of the SPSL to some of non-original cultivation areas requires consideration of the adaptability problems. In addition to the HD, several aspects, such as biotic and abiotic resistance, must be considered. If some defect would emerge after a few years’ cultivation, we could improve the trait by its related genes or QTL using a similar method. Then, two or more modules could be combined to upgrade the SPSL again. Due to the consistency of the genomic background of Kongyu131, we would simply need to hybridize the two SPSLs to obtain a multimodule improved variety. In addition, such kind of materials allow to compare homologous genes, gene interactions and the relationships of the genes with background genomes in a single genomic background. Therefore, our genetic system will be very important in both fundamental research and productive practice.
When attempting to develop a modular variety carrying more than one module, the most important factor to regulate would be the length of the single segment introduced in a variety. Recombination of different SPSLs may generate some unexpected phenotypes, owing to the interactions of genes that were introduced in the linkage drag accompanied with the target gene. The length of the linkage drag would be doubled by the aggregation of two modules. Hence, the control of the length of the introduced segment seems particularly important.
Overall, our research used high-density SNP markers covering the whole genome and introduced the regional adaptation-related mRA7 module from the donor GKLPL into Kongyu131. After elimination of the non-target chromosome fragments and precise control of the target module, an SPSL was obtained. The SPSL delayed the HD and significantly increased the length of the main panicle and the PH when cultivated in LQ, but also in JNQ and ZGQ, in respect to the original parent. We can estimate that this variety could be cultivated till more than 3 latitude units to the south.
SL conceived the experiments. RW carried out most of the experiments, performed the statistical analysis, and wrote the manuscript. XF, JN, and XZ participated in primer design. GJ and QY performed rice hybridization and field management. All authors read and approved the final manuscript.
This work was funded by the “Strategic Priority Research Program” of the Chinese Academy of Sciences (No. XDA08030102–1) and Talent Introduction Subject of State Key Laboratory of Plant Genomics, Institute of Genetics and Developmental Biology, University of Chinese Academy of Sciences (2014C0227–01).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00288/full#supplementary-material
DH, days to heading; ETP, effective tillers per plant; GN, grain number per plant; LQ, plots in Jiamusi, Heilongjiang province; GNP, grain number of main panicles; GZQ, plots in Guangzhou, Guangdong province; HD, heading date; HRM, high-resolution melting; JNQ, plots in Changchun, Jilin province; MAS, marker-assisted selection; NPB, number of primary branches; PH, plant height; QTL, quantitative trait loci; SNP, single-nucleotide polymorphism; SPSL, single point substitute line; ZGQ, plots in Beijing.
Chen, H., Xie, W., He, H., Yu, H., Chen, W., Li, J., et al. (2014). A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 7, 541–553. doi: 10.1093/mp/sst135
China Rice Data Center (2018). Available at: http://www.ricedata.cn/variety/varis/600008.htm
Doi, K., Izawa, T., Fuse, T., Yamanouchi, U., Kubo, T., Shimatani, Z., et al. (2004). Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 18, 926–936. doi: 10.1101/gad.1189604
Duitama, J., Silva, A., Sanabria, Y., Cruz, D. F., Quintero, C., Ballen, C., et al. (2015). Whole genome sequencing of elite rice cultivars as a comprehensive information resource for marker assisted selection. PLoS One 10:e0124617. doi: 10.1371/journal.pone.0124617
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Hayama, R., Yokoi, S., Tamaki, S., Yano, M., and Shimamoto, K. (2003). Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 422, 719–722. doi: 10.1038/nature01549
Hori, K., Ogiso-Tanaka, E., Matsubara, K., Yamanouchi, U., Ebana, K., Yano, M., et al. (2013). Hd16, a gene for casein kinase I, is involved in the control of rice flowering time by modulating the day-length response. Plant J. 76, 36–46. doi: 10.1111/tpj.12268
International Rice Research Institute [IRRI] (2013). Standard Evaluation System for Rice (SES), 5th Edn. Manila: International Rice Research Institute.
Kojima, S., Takahashi, Y., Kobayashi, Y., Monna, L., Sasaki, T., Yano, M., et al. (2002). Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant Cell Physiol. 43, 1096–1105. doi: 10.1093/pcp/pcf156
Kumar, A., Sandhu, N., Dixit, S., Yadav, S., Swamy, B. P. M., Shamsudin, N. A. A., et al. (2018). Marker-assisted selection strategy to pyramid two or more QTLs for quantitative trait-grain yield under drought. Rice 11:35. doi: 10.1186/s12284-018-0227-0
Kwon, Y. S., Kim, K. M., Kim, D. H., Eun, M. Y., and Sohn, J. K. (2002). Marker-assisted introgression of quantitative trait loci associated with plant regeneration ability in anther culture of rice (Oryza sativa L.). Mol. Cells 14, 24–28.
Lander, E. S., Green, P., Abrahamson, J., Barlow, A., Daly, M. J., Lincoln, S. E., et al. (1987). MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. doi: 10.1016/0888-7543(87)90010-3
Matsubara, K., Hori, K., Ogiso-Tanaka, E., and Yano, M. (2014). Cloning of quantitative trait genes from rice reveals conservation and divergence of photoperiod flowering pathways in Arabidopsis and rice. Front. Plant Sci. 5:193. doi: 10.3389/fpls.2014.00193
Nagata, K., Ando, T., Nonoue, Y., Mizubayashi, T., Kitazawa, N., Shomura, A., et al. (2015). Advanced backcross QTL analysis reveals complicated genetic control of rice grain shape in a japonica x indica cross. Breed. Sci. 65, 308–318. doi: 10.1270/jsbbs.65.308
Paris, M., and Carter, M. (2012). Cereal DNA: a rapid high-throughput extraction method for marker assisted selection. Plant Mol. Biol. Rep. 18, 357–360. doi: 10.1007/BF02825063
Peleman, J. D., and van der Voort, J. R. (2003). Breeding by design. Trends Plant Sci. 8, 330–334. doi: 10.1016/S1360-1385(03)00134-1
Stein, N., Herren, G., and Keller, B. (2001). A new DNA extraction method for high-throughput marker analysis in a large-genome species such as Triticum aestivum. Plant Breed. 120, 354–356. doi: 10.1046/j.1439-0523.2001.00615.x
Takahashi, Y., Shomura, A., Sasaki, T., and Yano, M. (2001). Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the alpha subunit of protein kinase CK2. Proc. Natl. Acad. Sci. U.S.A. 98, 7922–7927. doi: 10.1073/pnas.111136798
Tamaki, S., Matsuo, S., Wong, H. L., Yokoi, S., and Shimamoto, K. (2007). Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. doi: 10.1126/science.1141753
Tang, S. Q., Shao, G. N., Wei, X. J., Chen, M. L., Sheng, Z. H., Luo, J., et al. (2013). QTL mapping of grain weight in rice and the validation of the QTL qTGW3.2. Gene 527, 201–206. doi: 10.1016/j.gene.2013.05.063
Voorrips, R. E., (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, J. K., Li, H. H., Zhang, X.-C., Yin, C.-B., Li, Y., Ma, Y.-Z., et al. (2011). Molecular design breeding in crops in China. Acta Agron. Sin. 37, 191–201. doi: 10.3724/SP.J.1006.2011.00191
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Xing, Y., and Zhang, Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yano, M. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi: 10.1105/tpc.12.12.2473
Yano, M., Harushima, Y., Nagamura, Y., Kurata, N., Minobe, Y., Sasaki, T., et al. (1997). Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor. Appl. Genet. 95, 1025–1032. doi: 10.1007/s001220050658
Yano, M., and Sasaki, T. (1997). Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35, 145–153. doi: 10.1023/A:1005764209331
Yu, H., Xie, W., Li, J., Zhou, F., and Zhang, Q. (2014). A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 12, 28–37. doi: 10.1111/pbi.12113
Yu, S. B., Li, J. X., Xu, C. G., Tan, Y. F., Li, X. H., Zhang, Q., et al. (2002). Identification of quantitative trait loci and epistatic interactions for plant height and heading date in rice. Theor. Appl. Genet. 104, 619–625. doi: 10.1007/s00122-001-0772-5
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3:17031. doi: 10.1038/nplants.2017.31
Zhang, Q., Yao, G.-X., Hu, G.-L., Chen, C., Tang, B., Zhang, H.-L., et al. (2012). Fine mapping of qTGW3-1, a QTL for 1000-grain weight on chromosome 3 in rice. J. Integr. Agr. 11, 879–887. doi: 10.1016/S2095-3119(12)60078-1
Keywords: elite rice variety, heading date, SNP marker, QTL, gene module
Citation: Wang R, Jiang G, Feng X, Nan J, Zhang X, Yuan Q and Lin S (2019) Updating the Genome of the Elite Rice Variety Kongyu131 to Expand Its Ecological Adaptation Region. Front. Plant Sci. 10:288. doi: 10.3389/fpls.2019.00288
Received: 30 September 2018; Accepted: 21 February 2019;
Published: 13 March 2019.
Edited by:
Petr Smýkal, Palacký University, Olomouc, CzechiaReviewed by:
Elisabetta Mazzucotelli, Council for Agricultural Research and Economics, ItalyCopyright © 2019 Wang, Jiang, Feng, Nan, Zhang, Yuan and Lin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaoyang Lin, c3lsaW5AZ2VuZXRpY3MuYWMuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.